- Department of Biology, Lakehead University, Thunder Bay, ON, Canada
The present study identified the pectinase-producing bacterium isolated from the contaminated broth as Bacillus sp. on 16S rDNA sequence analysis. The bacterium illustrated water-like droplets on the colony grown on the Sabouraud dextrose agar plate. It also exhibited multi-enzymes activities, such as pectinase, polygalacturonase, xylanase, and cellulase by using various agro-wastes as low-cost substrates. The orange peel was observed to be the best substrate among the agro-wastes used for maximum multi-enzymes (pectinase, polygalacturonase, xylanase, and cellulase). However, the bacterium demonstrated its capability to produce different enzymes according to the different substrates/agro-wastes used. The Plackett–Burman design was used to determine the essential influencing factors, while the Box Behnken design response surface methodology was for optimizing cultural conditions. At their optimal conditions (40°C incubation temperature, 24 h of incubation period, 1% w/v orange peel, and 2% v/v inoculum volume), the bacterium exhibited the maximum pectinase (9.49 ± 1.25 U/ml) and xylanase (16.27 ± 0.52 U/ml) activities. Furthermore, the study explored the ability of the bacterium to produce bacterial lipids and observed about 25% bacterial lipid content on a dry weight basis. Therefore, the bacterium is a good candidate for producing important multi-enzymes and subsequent agro-waste degradation controlling the environment, and facilitating waste management. Also, the bacterium can be a potential feedstock in producing renewable biofuel.
Introduction
Agro-wastes are low-cost, renewable, and sustainable resources for industrially important enzyme production (Bharathiraja et al., 2017; Ravindran et al., 2018; Shrestha et al., 2020). Agricultural waste is usually produced during processing, pre-harvesting, post-harvesting, marketing procedures, and household activities (Kodagoda and Marapana, 2017; Shrestha et al., 2021a). The loss of agricultural products during these processes may decrease the products and increase production costs. Additionally, the agro-wastes pollute the environment and create disposal problems (Sadh et al., 2018; Shrestha et al., 2021a). Population growth and waste generation are directly proportional and it is expected the waste generation per year will increase by 83% by 2050.1 Dumping or landfill is the common method of waste disposal. However, it is not safe. The air near or surrounding the area of the dumping site may have air pollution due to the gasses produced and the suspended particles in the air (Siddiqua et al., 2022). The leachate from landfill may contain toxic byproducts and may contaminate water bodies or soil which finally may affect living things including human and aquatic life (Parvin and Tareq, 2021; Siddiqua et al., 2022). At the same time, there is a continuous rise in demand for enzymes, and the pure carbon and nitrogen source used in enzyme production is expensive (Ravindran et al., 2018; Shrestha et al., 2022). Therefore, these agro-wastes can be used as low-cost substrates to produce polysaccharides hydrolyzing enzymes as they contain different polysaccharides such as cellulose, xylose, and pectin in different compositions (Sadh et al., 2018; Mellinas et al., 2020; Shrestha et al., 2020, 2022).
Besides, the multi-enzymes biocatalyst technology and production of enzyme cocktails from a single organism are gaining interest (Amadi et al., 2022). The most common and commercially important enzymes are cellulase, pectinase, and xylanase, used for broad biotechnological applications. Pectinases and xylanases have been used in wastewater treatment, brewing technology, animal feed preparation, textile, pulp and paper industries, and food processing industries (Thite et al., 2020; Nawawi et al., 2022). Cellulolytic enzymes and other enzymes apply to cell wall disruption, juice extraction, and lipid extraction (Guo et al., 2017). In addition, the multi-enzymes help protects the environment by degrading various plant and agro-wastes (Ravindran et al., 2018; Thite et al., 2020; Amadi et al., 2022; Shrestha et al., 2022). Furthermore, enzymes production depends on the microorganisms exploited, and there are only a few studies for multi-enzymes productions from a single bacterium (Laothanachareon et al., 2022; Ozzeybek and Cekmecelioglu, 2022; Shrestha et al., 2022). Producing multi-enzymes by a single bacterium using different agro-wastes is cost-effective and time-efficient because using a single pure carbohydrate as substrate can only induce a single specific enzyme which is expensive and time-consuming (Wang et al., 2019; Thite et al., 2020; Nawawi et al., 2022; Shrestha et al., 2022).
Furthermore, the uninterrupted increase in the world’s population is diminishing fossil fuel reserves, so there is a need to explore alternative energy sources (Yusuf, 2019). In this aspect, some oleaginous microorganisms are natural oil producers. They accumulate about 20% w/v of lipid on a dry weight basis and are the most promising feedstock for lipid and oleochemical production (Patel et al., 2020). Therefore, this study focuses on the isolation and identification of the bacterium that increased the pectinase activity significantly. Also, the current study exploits the bacterium for multi-enzymes production by utilizing different agro-wastes and optimization of the fermentation conditions. Furthermore, the bacterium was exploited to know its capability of producing lipids due to the unique colony of the bacterium with water-like droplets on Sabouraud dextrose agar plate.
Materials and methods
Isolation and identification of bacteria
The bacterium was isolated from the contaminated broth. A loopful of the contaminated broth was streaked on Sabouraud dextrose agar (SDA) and nutrient agar (NA) plates and incubated at room temperature and 35°C, respectively, for 2–3 days. After the growth of colonies, a single colony was sub-cultured many times on SDA and NA plates to get the pure isolated colonies. Once the pure isolated colonies were observed on the agar plate, genomic DNA was extracted following the SDS-CTAB/NaCl method.
The 16S rDNA genes of the isolate were amplified by Taq DNA polymerase with universal primer sets: 16S rDNA forward primer 5′-AGAGTTTGATCCTGGCTCAG-3′ and reverse primer 5′-GGTTACCTTGTTACGACTT-3. The amplification systems for the 16S PCR reaction mixture contained 2 × Taq PCR master mix of 12 μl (10 × Taq DNA polymerase buffer, 10 mM dNTPs, 25 mM of MgCl2, 1 U of Taq DNA polymerase), 3 μl of 10 μM forward and reverse primers, 2 μl of the genomic DNA template, and 4 μl of distilled water, making a total volume of 24 μl. Furthermore, the PCR reaction conditions used were denaturation at 94°C for 5 min and the cycle starting at 94°C for 30 s, followed by annealing at 54°C for 30 s, extending at 72°C for 1.5 min for 33 cycles, and finally extending at 72°C for 10 min. Then the PCR products were determined by 1% (w/v) agarose gel electrophoresis. The 16S rDNA target fragments from the gel were cut, DNA was extracted using a gel extraction minipreps kit (Bio Basic), and the extracted DNA was sent for DNA sequencing.
Gene sequencing and phylogenetic analysis
The 16S rDNA sequences of the isolate provided by the sequencing company were compared with the known sequences found in the National Center for Biotechnology Information (NCBI) database using the basic local alignment search tool (BLASTn). The isolate was identified based on the percentage similarity with the known species sequences in the database. All the sequences were collected and parallelized using the Clustalw module in BioEdit v. 7.0.9.0 (Hall, 1999) with default settings. Phylogenetic analysis was performed using a Neighbor-Joining (NJ) tree with 1,000 bootstraps using MEGA 7 (Kumar et al., 2016).
Screening tests
Different screening tests for pectinase, cellulase, xylanase, and amylase were performed by culturing the bacterium on agar plates containing pectin, cellulose, xylan, and starch, respectively. After the growth of the bacterium, potassium iodide solution for pectinase, Congo red for cellulase and xylanase, and iodine solution for amylase screening test were flooded. The clear halo zone around the colonies indicated the presence of respective enzymes (Meddeb-Mouelhi et al., 2014; Takcı and Turkmen, 2016; Al Mousa et al., 2022).
Pectinase activity
The speck of pure isolated colony, which showed the clear pectinolytic zone on screening pectin agar plate, was subjected to prepare seed culture. Then, 1% v/v seed culture was inoculated into a flask with 50 ml of yeast extract pectin media (YEP) composed of 0.3% yeast extract, 1% pectin, 0.2% KH2PO4, and 0.2% K2HPO4 in distilled water. The inoculated flasks were incubated at 35°C for 4 days at 200 rpm. Samples from inoculated flasks were collected at regular intervals of 24 h, and enzyme activities were assayed and compared. Pectinase activity was calculated by measuring the reducing sugar content released from the substrate following the 3,5-dinitrosalicylic acid (DNS) method (Miller, 1959), as mentioned in a previous study (Shrestha et al., 2021b). In brief, 10 μl of crude enzyme extract was added to 20 μl of 1% citrus pectin solution as a substrate solution in the wells of a microplate, incubated in a 50°C water bath for 10 min, cooled, and 60 μl of DNS reagent was added. Then the microplate was covered and heated in boiling water for 5 min, followed by cooling down to room temperature. To the mixture, 200 μl of distilled water was added, and the absorbance was recorded at 540 nm to calculate the amount of reducing sugar released. Each enzymatic activity was expressed as the amount of enzyme that releases 1 μmol of galacturonic acid in 1 min under the mentioned conditions.
Effect of incubation period, temperature, pH, inoculum volume, and pectin concentration in pectinase production
The bacterium was cultured in YEP, pectinase production media, for different incubation periods (24–120 h). At different incubation periods, the cultured broth was aseptically taken in a sterile Eppendorf tube and centrifuged. The cell-free supernatant was used for enzyme activity assay. The fermentation condition for the bacteria regarding incubation temperatures and pH was studied by culturing at different incubation temperatures (30°C, 35°C, 40°C, and 45°C) and pH (5, 6, 7, 8, 9, and 10). Similarly, the different inoculum volume (0.5, 1, 2, 3, and 4% v/v), and different pectin concentrations (0.5, 1, 1.5, and 2% w/v) were added to the media, to study the enzyme activity for 96 h.
Agro-waste preparation and multi-enzyme production
Different agro-wastes which are easily and locally available were selected. Orange peel, banana peel, pomegranate peel, and pumpkin pulp+seeds were from the waste of those fruits and vegetables bought from the market. Barley straw and maple leaf were commercially available. Canola straw was accumulated from a local farm, and brewer’s spent grains from a local brewing company (Sleeping Giant Brewing Co., Thunder Bay, Ontario, Canada). All those agro-wastes were dried, ground in a coffee grinder, and washed with hot water several times to remove contaminant and simple sugar. The presence of reducing sugar was determined by DNS method. Once the samples were free of reducing sugar, they were dried on a hot air oven at 50°C for 48 h (till constant weight). The dried agro-waste powders were kept in airtight containers for further use.
For multi-enzymes production using agro-wastes, the bacterium was cultured on the media containing 1% w/v agro-wastes as the carbon source instead of 1% w/v pectin in the YEP pectinase production media. Polysaccharides such as pectinase, polygalacturonase (PGase), xylanase, and cellulase activities were determined every 24 h. The citrus pectin, polygalacturonic acid, beechwood xylan, and carboxymethyl cellulose (CMC) were used as the respective substrates.
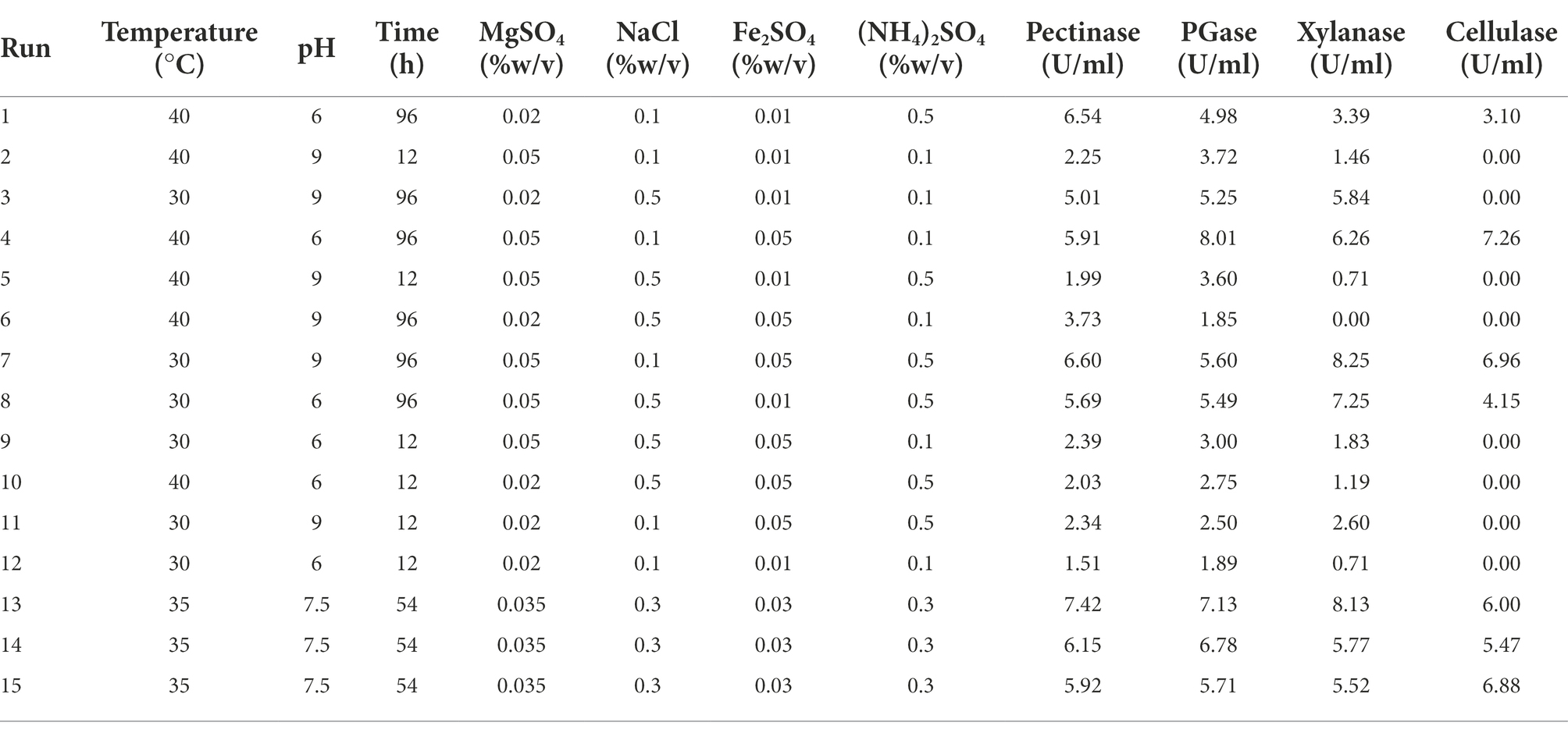
Optimization of cultural conditions for the maximum enzyme production
Orange peel was illustrated as the best substrate among the agro-wastes used in this study, so optimization of enzyme production was performed by using orange peel. Plackett–Burman design included seven different factors at two levels; incubation temperature, pH, incubation period, MgSO4, NaCl, FeSO4, and (NH4)2SO4 to screen the main influencing factor, including 15 experimental runs with three central points. The high level (+1) indicates the maximum concentration, and the low level (−1) indicates the minimum concentration of the variables (Table 1). The Box–Behnken design (BBD) response surface methodology was used to optimize the cultural conditions for maximum enzyme production using the most significant factors from the Plackett–Burman design. Here, enzyme activity was considered response variable, whereas incubation period (h), orange peel concentration (% w/v), and inoculum volume (% v/v) were three independent variables. The BBD used all the factors at three levels assigned as −1, 0, and +1 for the lowest, central, and highest value (Table 2). Both Plackett–Burman and BBD experimental designs were generated by using the Minitab 16 software.
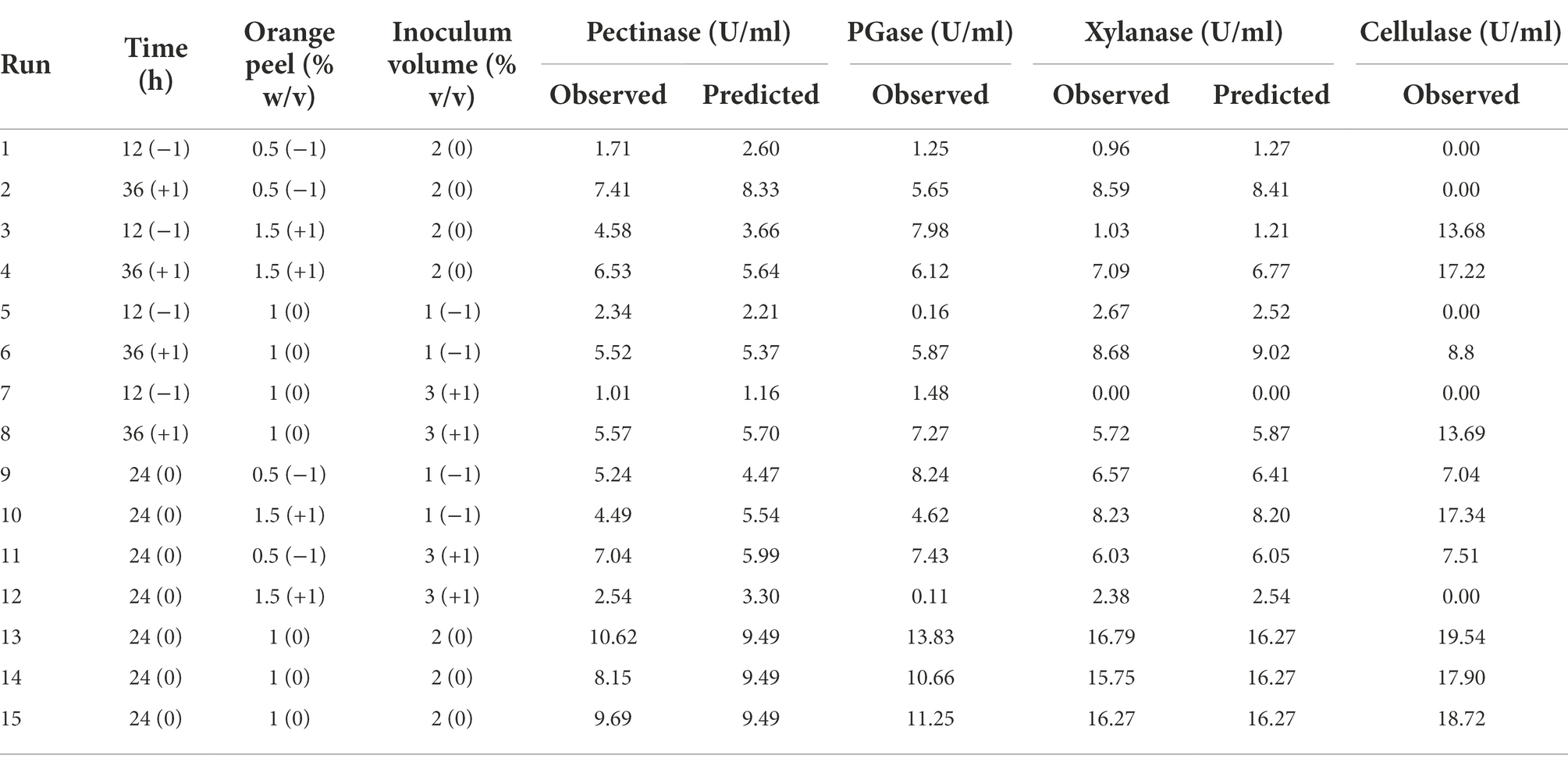
Table 2. Box–Behnken Design for enzymes production by using orange peel as the substrate [−1, 0, and +1 are the codes for the variables].
Determination of lipid content
The bacterium was cultured in yeast extract peptone (YEP’) culture media (consisting of 0.25, 0.25, 0.15, 2% w/v of yeast extract, peptone, MgSO4, dextrose, respectively) and Mineral salt medium (MSM) culture media (consisting of 0.9, 0.15, 0.02, 0.01, 0.00012, 0.002, and 0.005% w/v of Na2HPO4, KH2PO4, MgSO4, (NH4)2SO4, ferric citrate, CaCl2, and NaHCO3, respectively). Every 24 h, lipid content was determined by Bligh and Dyer (1959) with little modification. In brief, the biomass was first harvested by centrifugation at 10,000 g for 10 min, and 1 gm of the pellet was sonicated and then homogenized with a 20 ml mixture containing chloroform, methanol, and water (1:2:0.8 ratio), followed by 20 min of shaking in an orbital shaker at ambient room temperature. The homogenate was centrifuged at low speed (2,000 g) to separate two phases. The upper phase was siphoned off, and the lower chloroform phase containing lipids was evaporated. Finally, the lipids extracted were quantified by weighing, and lipid content (%) was calculated as,
Furthermore, lipid content was determined at two different media (YEP’ and MSM), different incubation periods (24 to 96 h), temperature (30°C–45°C) and pH values (6–9) to optimize the lipid content from the bacterium.
Statistical analysis
All the tests were performed in triplicates, and the data were expressed as mean with standard deviation (mean ± SD). One-way analysis of variance (ANOVA) was used to assess the statistical significance, followed by the Tukey–Kramer comparison. The data is regarded as statistically significant when the p-value of the experimental data is less than 0.05.
Results
Isolation and identification of the bacterium
After subculturing many times, pure and isolated colony was observed. The colony morphology of an isolated colony on the Sabouraud dextrose agar (SDA), Casein starch agar (CSA), and Nutrient agar (NA) plates were observed (Supplementary Figure 1). The colony of the bacterium was pale in color, irregular in shape, has many colorless waterlike droplets on SDA agar. The peripheral of the colony was soft to pick, whereas the inner part was hard. The unique colony characteristics with the occurrence of waterlike droplets suspected the bacterium of producing lipids.
Then the bacterium was screened for pectinase, cellulase, xylanase, lipase, and amylase enzymes. The clear zone around the colony was observed, indicating the bacterium was strongly positive for pectinase and amylase screening tests (Figures 1A,B), whereas weakly positive for cellulase, xylanase and lipase.
The bacterium was identified as Bacillus sp. (accession number OP673585) after isolation of the bacterium, DNA extraction, gel electrophoresis, 16S rDNA sequencing, and phylogenetic analysis. The 16S rDNA amplification and detection were performed as mentioned in the method, and the result of the 16S rDNA after gel electrophoresis depicted the fragments at around 1,500 bp (Supplementary Figure 2). The sequence similarity of the isolate was compared with the known species and similar sequences in the NCBI database. The phylogenetic tree of 16S rDNA sequences based on the Neighbor-Joining algorithm was constructed, as shown in Figure 2. The constructed phylogenetic tree depicted the studied bacterium (Bacillus sp. C19) is closely related to the other Bacillus sp. (Bacillus megaterium, Bacillus siamensis, Bacillus amyloliquefaciens, and Bacillus velezensis; Figure 2).
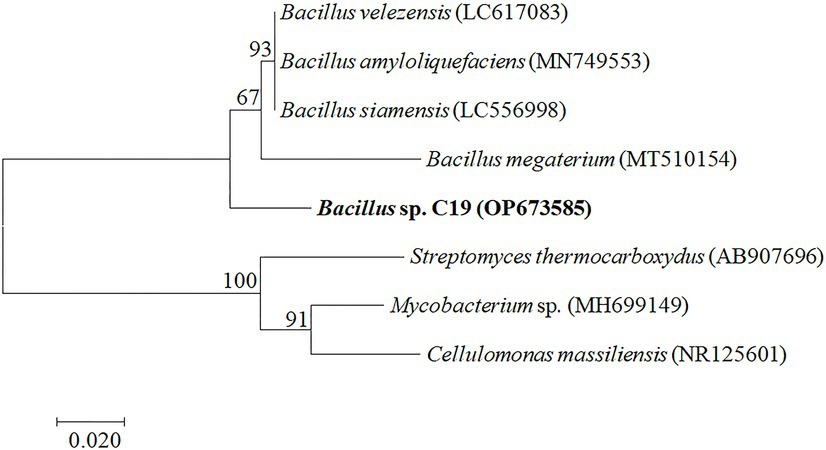
Figure 2. Phylogenetic relationships of the bacterium constructed by Neighbor-Joining (NJ) tree based on 16S rDNA gene sequences. Numbers present on branches of the NJ tree are bootstrap support values.
The uncorrected genetic divergence between the bacterium and topotypic Bacillus siamensis, Bacillus velezensis, and Bacillus amyloliquefaciens was 3.5%, whereas Mycobacterium sp., Cellulomonas massiliensis, and Streptomyces thermocboxydus were 20, 21, and 21%, respectively, (shown in Supplementary Table 1).
Effect of incubation period, temperature, inoculum volume, pH, and pectin concentration in pectinase activity
The maximum pectinase activity was observed in incubation period of 24 h, 40°C, 1% v/v inoculum volume, alkaline pH (7 and 9), and pectin 2% w/v (Figure 3). The values observed were statistically significant (p ≤ 0.05) when analyzed with one-way analysis of variance (ANOVA) followed by Tukey’s comparisons.
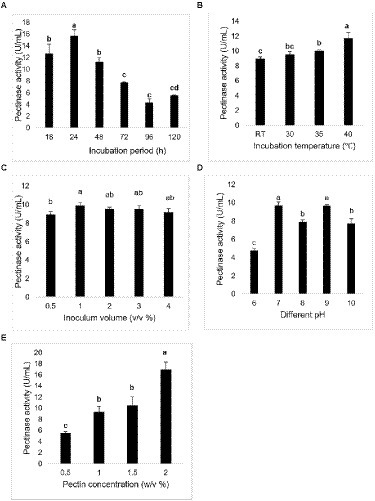
Figure 3. Pectinase activity at different (A) incubation period (h), (B) temperature (°C), (C) inoculum volume (% v/v), (D) pH, and (E) pectin concentration (% w/v). The bar represented the mean pectinase activity with the standard deviation error bar at different conditions. Lowercase alphabets indicate that the values are statistically significant at p ≤ 0.05.
Potential of Bacillus sp. to produce multi-enzymes using agro-wastes
Figure 4 depicts the pectinase activity by Bacillus sp. using eight agro-wastes. The bacterium could not show xylanase activity from barley straw and pumpkin pulp + seeds, while cellulase activity was only observed from orange peel.
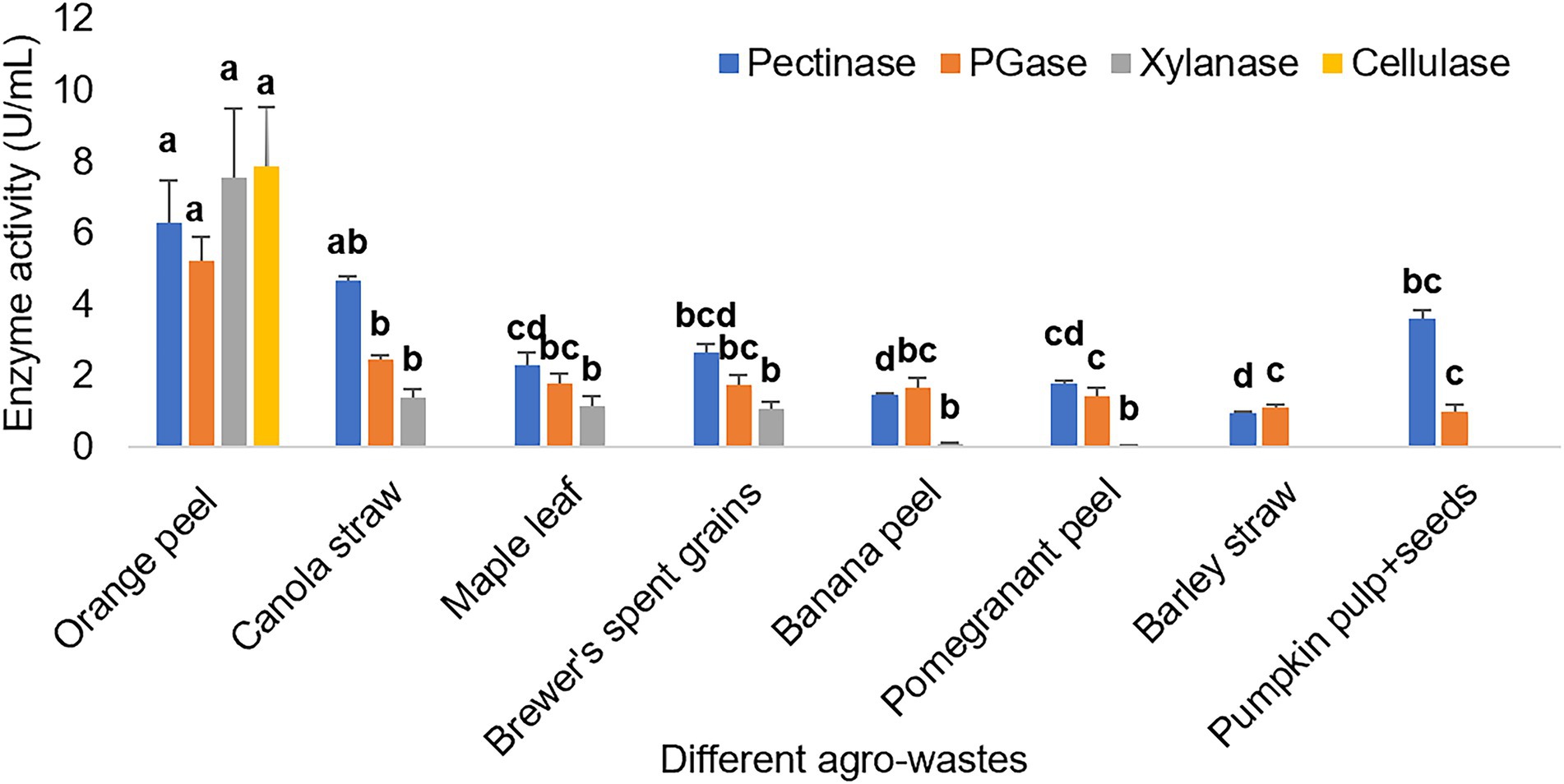
Figure 4. Different enzyme activities exhibited by Bacillus sp. using different agro-wastes as carbon sources. The bar represented the mean enzyme activity with the standard deviation error bar. Lowercase alphabets indicate that the values are statistically significant at p ≤ 0.05.
Optimization of the cultural conditions for maximum enzyme activities using orange peel
After the observations of different agro-wastes utilization to produce the enzymes, the orange peel was found to be the best substrate for multi-enzymes production. Therefore, the orange peel was selected to optimize the cultural conditions.
Plackett–Burman design
Plackett–Burman design was used for the initial statistical screening of seven cultural components (variables) and the enzyme activities (response), and the results are depicted in Table 1. The detailed analysis of the individual factors revealed that the incubation period as an influencing factor for pectinase, PGase, xylanase and cellulase activity with the lowest p-value. At the same time, all the six factors, like incubation period, pH, NaCl, Fe2SO4, MgSO4, and (NH4)2SO4 played important roles in cellulase activity. However, they were not significantly influential for other enzyme activities (Supplementary Table 2; Supplementary Figure 4).
Box–Behnken design
The optimization of cultural conditions was again designed using Box–Behnken design (BBD) and analyzed in Minitab 16 software. Enzyme activities exhibited by the bacterium using BBD illustrated in Table 2.
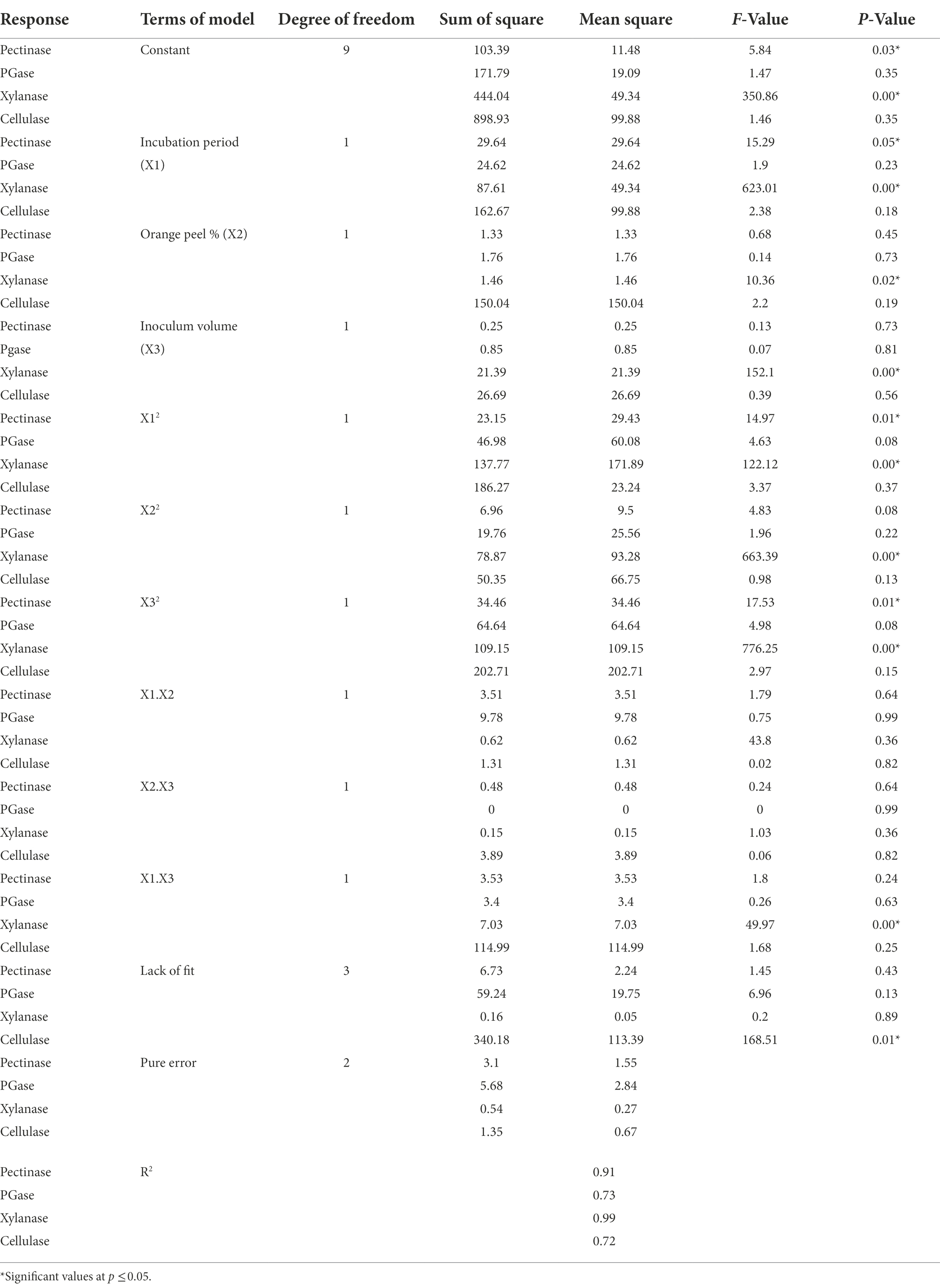
The accuracy and statistical significance of each term in the model were evaluated using Analysis of variance (ANOVA). The BBD experimental results of the current study were fitted with a second-order polynomial equation using a multiple regression technique and expressed as:
Pectinase activity (U/mL) = 9.49 + (1.92 • X1) + (−0.41 • X2) + (−0.18 • X3) + (−0.94 • X1 • X2) + (0.35 • X1 • X3) + (−0.94 • X2 • X3) + (−2.82 • X12) + (−1.60 • X22) + (−3.05 • X32).
PGase activity (U/mL) = 11.91 + (1.75 • X1) + (−0.47 • X2) + (−0.32 • X3) + (−1.56 • X1 • X2) + (0.02 • X1 • X3) + (−0.92 • X2 • X3) + (−4.03 • X12) + (−2.63• X22) + (−4.18 • X32).
Xylanase activity (U/mL) = 16.27 + (3.31 • X1) + (−0.43 • X2) + (−1.64 • X3) + (−0.39 • X1 • X2) + (0.19 • X1 • X3) + (−1.33 • X2 • X3) + (−6.82 • X12) + (−5.03• X22) + (−5.44 • X32).
Cellulase activity (U/mL) = 18.72 + (4.51 • X1) + (4.33 • X2) + (−1.83 • X3) + (0.57• X1 • X2) + (0.98 • X1 • X3) + (−5.36 • X2 • X3) + (−7.89 • X12) + (−4.25• X22) + (−7.41 • X32).
X1 represents time, X2 orange peel concentration, and X3 inoculum volume.
The p-values for pectinase, PGase, xylanase and cellulase activity were 0.03, 0.35, 0.000, 0.35 and lack of fit 0.434, 0.128, 0.892, and 0.01, respectively. The R2 values for pectinase, PGase, xylanase and cellulase were observed as 0.91, 0.73, 0.99, and 0.72, respectively. The regression p-value and lack of fit observed from the analysis of variance revealed that the quadratic equations were relatively reliable for evaluating bacterium’s pectinase and xylanase activities (Table 3).
The Contour plot Figures 5A,D showed the interaction of orange peel concentration and incubation period, (Figures 5B,E) the interaction of inoculum volume and orange peel concentration, and (Figures 5C,F) showed the interaction of inoculum volume and incubation period for pectinase and xylanase activities, respectively.
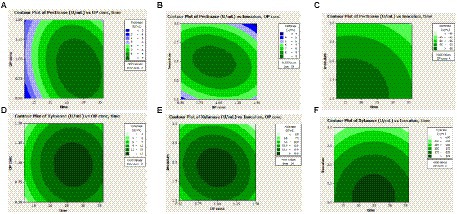
Figure 5. Contour plot showing the interaction effects of orange peel concentration (OP conc), Incubation period (hour), and inoculum volume (Inoculum) on pectinase (A–C) and xylanase (D–F) activities.
Figure 5 depicted that both pectinase and xylanase activities increased with an increase in orange peel concentration and incubation period to certain values and decreased in activities after the optimal values. Figures 5C,F illustrated a low inoculum volume and moderate incubation period gave the maximum pectinase and xylanase activities.
From the response optimizer, the optimal conditions observed for pectinase activity were incubation period of 29 h, 1% w/v orange peel concentration, and 2% v/v inoculum volume. In contrast, the optimal conditions for xylanase activity were 27 h, 1% w/v orange peel concentration, and 2% v/v inoculum volume (Supplementary Figure 5).
The model correctness was determined by performing the experiments under optimal conditions (from response optimizer), and the pectinase and xylanase activities observed were 8.62 ± 1.56 U/ml and 15.02 ± 1.23 U/ml, respectively. However, from BBD, 9.49 U/ml and 16.27 U/ml were revealed as the maximum pectinase and xylanase activities, respectively. At the same time, 9.90 U/ml and 16.76 U/ml were suggested for pectinase and xylanase by BBD optimizer. The enzyme activities thus observed were almost similar; thus, the models for pectinase and xylanase activities were valid and precise.
Lipid production by the bacterium at different conditions
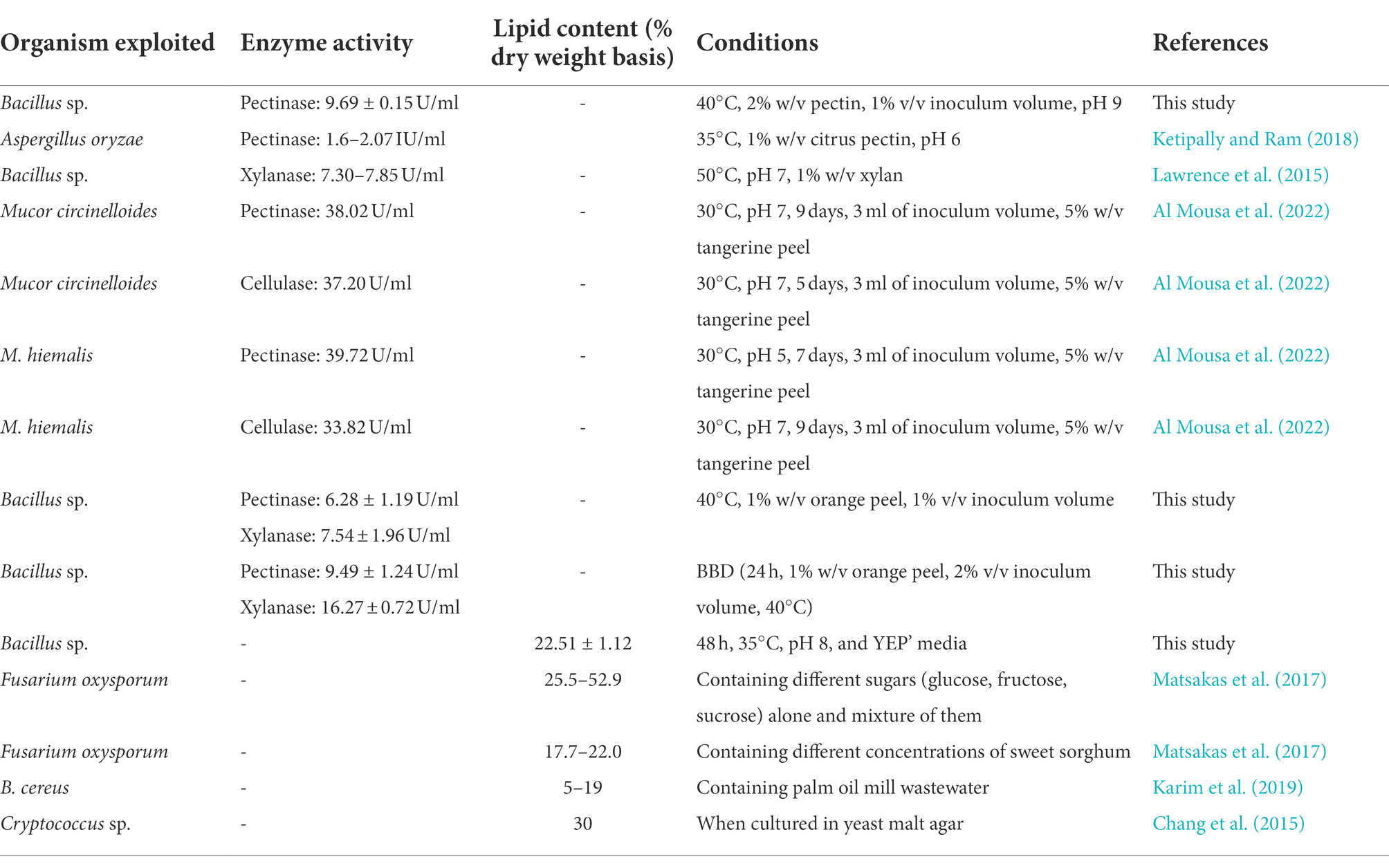
The lipid content of the bacterium was around 25% when grown in yeast extract peptone (YEP’) media. Further, the lipid content from the bacterium were studied using parameters like different incubation periods and pH values. The highest lipid contents by the bacterium were observed at incubation period of 48 h, temperature 35°C and pH 8, as illustrated in following figures (Figure 6).
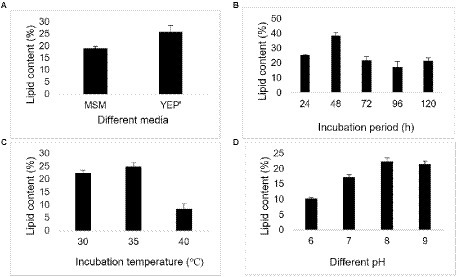
Figure 6. Lipid content exhibited by Bacillus sp. at different conditions (A) different media, (B) different incubation period (h), (C) temperature (°C), and (D) pH.
Discussion
The production of industrially important, novel, and robust enzymes for biotechnological applications is increasing. Therefore, there is a need to isolate and identify the bacteria from natural diversities and optimize the cultural conditions for maximum enzyme production. In our current study, we isolated the bacterium and screened for enzymes. The presence of a relative enzyme hydrolyzes the substrate around the colony, and a clear hydrolysis zone around the colony is observed. However, the substrate not hydrolyzed by relative enzymes forms a complex and does not show a clear zone (Soares et al., 1999; Meddeb-Mouelhi et al., 2014; Shrestha et al., 2021b; Al Mousa et al., 2022). On this basis, the bacterium was pectinase, amylase, cellulase, and xylanase producer.
The contaminant of the broth was identified as bacterium. Only 16S rDNA got amplified and showed the band at 1,500 bp region. Further, the bacterium was identified as Bacillus sp. because the phylogenetic tree illustrated the bacterium closely related to the Bacillus sp. (Bacillus megaterium, Bacillus siamensis, Bacillus amyloliquefaciens, and Bacillus velezensis). Furthermore, the uncorrected genetic divergence helped to confirm the bacterium as Bacillus sp.
Bacillus sp. are the dominant bacteria (Schallmey et al., 2004) and they have irregular colony (De Vos et al., 2015). Our study too revealed the irregular colony of Bacillus sp. Additionally, we observed clear water-like droplets on the colony which is unique character. Each bacterium has specific optimal conditions for the maximum enzyme activity. For instance, the study illustrated the optimal conditions by Bacillus sp. for the maximum xylanase activity were 50°C, 72 h, pH 7.0 ± 0.2, and 1% xylan concentration (Lawrence et al., 2015). Another study recorded the maximum pectinase activity by Aspergillus sp. at 35°C, pH 6.5, 1% citrus pectin and after 192 h of incubation period (Ketipally and Ram, 2018). Similarly, Streptomyces thermocoprophilus demonstrated maximum cellulase and xylanase activity at 40°C, pH 6.5, 120 h, and 1% alkaline peroxide pretreated empty fruit bunch (Sinjaroonsak et al., 2020). And our study observed the highest pectinase activity by Bacillus sp. in 24 h, 40°C, 1% v/v inoculum volume, alkaline pH (7 and 9), and pectin 2% w/v. The variation in the optimal conditions for enzyme activities may be due to the differences in the microorganisms used. The optimal conditions affect the growth and metabolic rate of the organisms exploited, and the maximum activity is illustrated at the most favorable conditions depending on the organisms.
The cheapest and readily available agro-industrial wastes can be used for industrially important enzyme production (Bharathiraja et al., 2017). The various agro-wastes, when used as the carbon source for enzyme production, the bacterium demonstrated the highest enzyme activities from orange peel. This may be because the bacterium must have found all the favorable conditions from orange peel only to demonstrate all the studied enzyme activities. Also, the orange peel may have a lower lignin concentration, making it inaccessible to pectin, hemicellulose and cellulose for the bacterium (Al Mousa et al., 2022). The bacterium exhibited xylanase and pectinase activity from canola straw, maple leaf, brewer’s spent grains, banana peel, and pomegranate peel. Further, the bacterium showed only pectinase activity from barley straw and pumpkin pulp+seeds. Thus, this study depicts the bacterium’s ability to use orange peel more effectively and efficiently than other agro-wastes and demonstrates orange peel as the best substrate for Bacillus sp. Similarly, another study demonstrated that maximum pectinase and cellulase activity by Mucor circinelloides and M. hiemalis using tangerine peel due to the low concentration of lignin, tissue structure flexibility, and easy access to pectin, cellulose and hemicellulose (Al Mousa et al., 2022). Another study reported that banana peel with high pectin and starch content induced most of the target enzymes by Aspergillus niger. In contrast, cellulose-rich sugarcane bagasse induced beta-glucanase and xylanase. At the same time, starch-rich cassava pulp induced amylase and other enzymes but was comparatively lower than banana peel (Laothanachareon et al., 2022). Such variation might be due to the various chemical compositions and carbon sources of agro-wastes which induce the target enzyme production by a microorganism differently.
The different concentrations (0.5, 1, 1.5, and 2% w/v) of agro-wastes were studied for enzyme activities. The enzyme activities were increased with an increase in the concentration of almost all agro-wastes used (orange peel, barley straw, pumpkin pulp + seeds, banana peel, barley spent grains, canola straw, and maple leaf). In contrast, a high concentration of pomegranate peel decreased the pectinase and PGase activities (Supplementary Figures 3A–C). This result relates that the bacterium’s ability to illustrate enzyme activity is directly proportional to agro-waste concentrations except for pomegranate peel. The higher the concentration of agro-wastes, the higher the chance of exposure to relative carbon source (pectin for pectinase, and hemicellulose for xylanase) present in that agro-waste.
Plackett–Burman design is a powerful and unique design to screen, identify and evaluate important variables that affect the response of the experimental tests. Thus, the present study used the Plackett–Burman design as an initial statistical screening of seven cultural components (variables) for enzyme activities (response). Our study revealed incubation period as an influencing factor for pectinase, PGase, xylanase and cellulase activity from this design, with the p-value < 0.05. However, the cellulase activity was only affected by all six factors and was not significantly influential for other enzyme activities. Likewise, a study used the Plackett–Burman design and reported the pectinase activity by Bacillus sonoresis was strongly affected by the pectin mass fraction, pH, and MgSO4 among eight different parameters studied (Mohandas et al., 2018).
Therefore, BBD was again used for optimizing the cultural condition considering incubation period, orange peel concentration and inoculum volume as the independent variables. Generally, the p-value lesser than 0.05 ensures the terms are statistically significant for each coefficient. The relationship between parameters was found significant for pectinase and xylanase but not for PGase and cellulase activities. Further, the statistical significance was checked by F-test to evaluate the coefficient of determination (R2). The lesser R2 value is not good, so the model was not too good for PGase and cellulase activity. However, the model was perfect for pectinase and xylanase activities.
Due to the depletion of fossil fuel reserves, different alternate sources are being explored (Yusuf, 2019). The microorganisms have potential as an alternative for lipid production as they can accumulate the oil/lipid in them (Patel et al., 2020). The present study explored the capacity of bacterium to produce lipids because it depicted the water like droplets on the SDA media. The study illustrated that the lipid content was higher in YEP’ media, possibly due to the higher cell growth, and the media favored the cell growth. In contrast, MSM inhibited the cell growth resulting in a decrease in lipid content. Our study illustrated that the lipid content continuously increased until 48 h of incubation period and decreased onward, which may be related to cell growth. Similarly, the incubation temperature of 35°C and pH 8 favored the cell growth and resulted in higher lipid content at the respective temperature and pH. However, the bacterial lipid accumulation, lipid composition and even the cell membrane compositions of the same species vary with the environmental conditions of exposure (Sohlenkamp and Geiger, 2015) (Table 4). A study demonstrated that the lipid production was in the range of 25.5%–52.9% of cell dry weight by Fusarium oxysporum using the synthetic media containing different sugars (glucose, fructose, and sucrose) alone and mixture of them (Matsakas et al., 2017). Whereas, Bacillus cereus accumulated lipid 5%–19% on a dry weight basis using palm oil mill wastewater (Karim et al., 2019). Another study reported that Lipomyces starkey could potentially be used as a lipid source and accumulate lipid (12%–29.5%) when cultured in oil mill wastewater-containing media (Yousuf et al., 2010). In addition, a study illustrated the microbial lipid from the yeast Cryptococcus sp. using corncob hydrolysate as a raw material (Chang et al., 2015). Therefore, microorganisms can be a potential source of lipid production by bioremediating the wastes. Further, studies relating to the isolation and identification of high lipid-containing microorganisms, the optimization of the culturing condition, compositional analysis, and more are recommended for maximum microbial lipid production.
Conclusion
The pectinase-producing bacterium isolated from the contaminated broth was identified as Bacillus sp. from 16S rDNA sequence analysis. The bacterium produced different polysaccharides degrading enzymes, such as pectinase, polygalacturonase, xylanase, and cellulase. However, the different enzyme activities vary with agro-wastes used as low-cost substrates. The response surface methodology illustrated Bacillus sp. to exhibit maximum pectinase at 40°C, 29 h of incubation period, 1% w/v orange peel concentration, and 2% v/v inoculum volume. In contrast, the optimal conditions for xylanase activity were 40°C, 27 h, 1% w/v orange peel concentration, and 2% v/v inoculum volume. Furthermore, the bacterium has the potential to produce bacterial lipids. Therefore, the bacterium is a good candidate for producing biotechnologically important multi-enzymes and agro-waste degradation. Since the bacterium illustrated lipid content, it can be a potential feedstock in producing renewable biofuels and environmental resilience.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SS designed the experiments, performed majority of experiments, analyzed the data, and drafted the manuscript. JK, CC, and AK helped and coordinated in the entire study. WQ provided the resources, supervised, and did major review of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the Natural Science and Engineering Research Council of Canada (NSERC; RGPIN-2017-05366) to WQ and Ontario Graduate Scholarship (OGS) to SS.
Acknowledgments
The authors express their sincere gratitude to all the lab members and everyone who helped directly or indirectly to complete this study and to the National Science and Engineering Research Council of Canada (NSERC) for financial assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1049692/full#supplementary-material
Footnotes
References
Al Mousa, A. A., Hassane, A. M. A., Gomaa, A. E. R. F., Aljuriss, J. A., Dahmash, N. D., and Abo-Dahab, N. F. (2022). Response-surface statistical optimization of submerged fermentation for pectinase and Cellulase production by Mucor circinelloides and M. hiemalis. Fermentation 8:5. doi: 10.3390/fermentation8050205
Amadi, O. C., Awodiran, I. P., Moneke, A. N., Nwagu, T. N., Egong, J. E., and Chukwu, G. C. (2022). Concurrent production of cellulase, xylanase, pectinase and immobilization by combined cross-linked enzyme aggregate strategy-advancing tri-enzyme biocatalysis. Bioresour. Technol. Rep. 18:101019. doi: 10.1016/j.biteb.2022.101019
Bharathiraja, S., Suriya, J., Krishnan, M., Manivasagan, P., and Kim, S. K. (2017). Production of enzymes from agricultural wastes and their potential industrial applications. 1st Edn. eds. S.-K. Kim and F. Toldra. Amsterdam: Elsevier Inc.
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total extration and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/y59-099
Chang, Y. H., Chang, K. S., Lee, C. F., Hsu, C. L., Huang, C. W., and Der Jang, H. (2015). Microbial lipid production by oleaginous yeast Cryptococcus sp. in the batch cultures using corncob hydrolysate as carbon source. Biomass Bioenergy 72, 95–103. doi: 10.1016/j.biombioe.2014.11.012
Guo, H., Chen, H., Fan, L., Linklater, A., Zheng, B., Jiang, D., et al. (2017). Enzymes produced by biomass-degrading bacteria can efficiently hydrolyze algal cell walls and facilitate lipid extraction. Renew. Energy 109, 195–201. doi: 10.1016/j.renene.2017.03.025
Hall, T. A. (1999). BIoEdit: a user-friendly bilogical sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Karim, A., Islam, M. A., Yousuf, A., Khan, M. M. R., and Faizal, C. K. M. (2019). Microbial lipid accumulation through bioremediation of palm oil mill wastewater by Bacillus cereus. ACS Sustain. Chem. Eng. 7, 14500–14508. doi: 10.1021/acssuschemeng.9b01960
Ketipally, R., and Ram, M. R. (2018). Optimization of pectinase production by Aspergillus Oryzae RR 103. Curr. Agric. Res. J. 6, 37–44. doi: 10.12944/carj.6.1.05
Kodagoda, K., and Marapana, R. (2017). Utilization of fruit processing by-products for industrial applications: A review. Int. J. Food Sci. Nutr. 2, 24–30.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laothanachareon, T., Bunterngsook, B., and Champreda, V. (2022). Profiling multi-enzyme activities of Aspergillus Niger strains growing on various agro-industrial residues. 3 Biotech. 12, 17–16. doi: 10.1007/s13205-021-03086-y
Lawrence, R., Kumar, Y., Kumar, S. A., and Satendra, S. (2015). Production and optimization of Xylanase by Thermophilic bacillus sp. Isolated from soil. J. Pure Appl. Microbiol. 9, 1117–1128.
Logan, N. A., and De Vos, P. (2015). “Cohn 1872, 174,” in Bergey’s manual of systematics of Archae and bacteria eds. M. E. Trujillo, S. Dedysh, P. De Vos, B. Hedlund, P. Kampfer, and F. A. Rainey, et al. (John Wiley & Sons Inc.), 164.
Matsakas, L., Giannakou, M., and Vörös, D. (2017). Effect of synthetic and natural media on lipid production from Fusarium oxysporum. Electron. J. Biotechnol. 30, 95–102. doi: 10.1016/j.ejbt.2017.10.003
Meddeb-Mouelhi, F., Moisan, J. K., and Beauregard, M. (2014). A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzyme Microb. Technol. 66, 16–19. doi: 10.1016/j.enzmictec.2014.07.004
Mellinas, C., Ramos, M., Jiménez, A., and Garrigós, M. C. (2020). Recent trends in the use of pectin from agro-waste residues as a natural-based biopolymer for food packaging applications. Materials 13. doi: 10.3390/ma13030673
Miller, G. L. (1959). Use of Dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Mohandas, A., Raveendran, S., Parameswaran, B., Abraham, A., Athira, R. S. R., Mathew, A. K., et al. (2018). Production of pectinase from bacillus sonorensis MPTD1. Food Technol. Biotechnol. 56, 110–116. doi: 10.17113/ftb.56.01.18.5477
Nawawi, M. H., Ismail, K. I., Sa, N., Mohamad, R., Tahir, P., Zuriyati, A., et al. (2022). Optimisation of xylanase – pectinase cocktail production with bacillus amyloliquefaciens ADI2 using a low-cost substrate via statistical strategy. Fermentation 8:119. doi: 10.3390/fermentation8030119
Ozzeybek, M., and Cekmecelioglu, D. (2022). Formulation of apple pomace, orange peel, and hazelnut shell mix for co-production of bacterial pectinase and cellulase enzymes by mixture design method. Biomass Convers. Biorefinery, 1–9. doi: 10.1007/s13399-022-02409-0
Parvin, F., and Tareq, S. M. (2021). Impact of landfill leachate contamination on surface and groundwater of Bangladesh: a systematic review and possible public health risks assessment. Appl. Water Sci. 11, 100–117. doi: 10.1007/s13201-021-01431-3
Patel, A., Karageorgou, D., Rova, E., Katapodis, P., Rova, U., Christakopoulos, P., et al. (2020). An overview of potential oleaginous microorganisms and their role in biodiesel and omega-3 fatty acid-based industries. Microorganisms 8. doi: 10.3390/microorganisms8030434
Ravindran, R., Hassan, S. S., Williams, G. A., and Jaiswal, A. K. (2018). A review on bioconversion of agro-industrial wastes to industrially important enzymes. Bioengineering 5, 1–20. doi: 10.3390/bioengineering5040093
Sadh, P. K., Duhan, S., and Duhan, J. S. (2018). Agro-industrial wastes and their utilization using solid state fermentation: a review. Bioresour. Bioprocess. 5, 1–15. doi: 10.1186/s40643-017-0187-z
Schallmey, M., Singh, A., and Ward, O. P. (2004). Developments in the use of bacillus species for industrial production. Can. J. Microbiol. 50, 1–17. doi: 10.1139/w03-076
Shrestha, S., Chio, C., Khatiwada, J. R., Laurel, A., and Kognou, M. (2022). Formulation of the agro-waste mixture for multi-enzyme (pectinase, xylanase, and cellulase) production by mixture design method exploiting Streptomyces sp. Bioresour. Technol. Rep. 19:101142. doi: 10.1016/j.biteb.2022.101142
Shrestha, S., Khatiwada, J. R., Sharma, H. K., and Qin, W. (2021a). “Bioconversion of Fruits and Vegetables Wastes into Value-Added Products,” in Advances in Science, Technology and Innovation, eds. Inamuddin and A. Khan. (Switzerland: Springer Nature), 145–163.
Shrestha, S., Khatiwada, J. R., Zhang, X., Chio, C., Kognou, A. L. M., Chen, F., et al. (2021b). Screening and molecular identification of novel Pectinolytic Bacteria from Forest soil. Fermentation 7:40. doi: 10.3390/fermentation7010040
Shrestha, S., Mokale, A. L., Zhang, J., and Qin, W. (2020). Different facets of lignocellulosic biomass including pectin and its perspectives. Waste Biomass Valoriz. 12, 4805–4823. doi: 10.1007/s12649-020-01305-w
Siddiqua, A., Hahladakis, J. N., and Al-Attiya, W. A. K. A. (2022). An overview of the environmental pollution and health effects associated with waste landfilling and open dumping. Environ. Sci. Pollut. Res. 29, 58514–58536. doi: 10.1007/s11356-022-21578-z
Sinjaroonsak, S., Chaiyaso, T., and Kittikun, A. (2020). Optimization of Cellulase and Xylanase productions by Streptomyces thermocoprophilus TC13W using low cost pretreated oil palm empty fruit bunch. Waste Biomass Valoriz. 11, 3925–3936. doi: 10.1007/s12649-019-00720-y
Soares, M. M. C. N., Da Silva, R., and Gomes, E. (1999). Screening of bacterial strains for pectinolytic activity: characterization of the polygalacturonase produced by bacillus sp. Rev. Microbiol. 30, 299–303. doi: 10.1590/S0001-37141999000400002
Sohlenkamp, C., and Geiger, O. (2015). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159. doi: 10.1093/femsre/fuv008
Takcı, H. A. M., and Turkmen, F. U. (2016). Extracellular pectinase production and purification from a newly isolated Bacillus subtilis strain. Int. J. Food Prop. 19, 2443–2450. doi: 10.1080/10942912.2015.1123270
Thite, V. S., Nerurkar, A. S., and Baxi, N. N. (2020). Optimization of concurrent production of xylanolytic and pectinolytic enzymes by bacillus safensis M35 and Bacillus altitudinis J208 using agro-industrial biomass through response surface methodology. Sci. Rep. 10, 3824–3812. doi: 10.1038/s41598-020-60760-6
Wang, J., Chio, C., Chen, X., Su, E., Cao, F., Jin, Y., et al. (2019). Efficient saccharification of agave biomass using Aspergillus Niger produced low-cost enzyme cocktail with hyperactive pectinase activity. Bioresour. Technol. 272, 26–33. doi: 10.1016/j.biortech.2018.09.069
Yousuf, A., Sannino, F., Addorisio, V., and Pirozzi, D. (2010). Microbial conversion of olive oil mill wastewaters into lipids suitable for biodiesel production. J. Agric. Food Chem. 58, 8630–8635. doi: 10.1021/jf101282t
Keywords: Bacillus sp., multi-enzymes, agro-wastes, optimization, lipid content
Citation: Shrestha S, Chio C, Khatiwada JR, Kognou ALM and Qin W (2022) Optimization of multiple enzymes production by fermentation using lipid-producing Bacillus sp. Front. Microbiol. 13:1049692. doi: 10.3389/fmicb.2022.1049692
Edited by:
Sikandar I. Mulla, REVA University, IndiaReviewed by:
Sivakumar Nallusamy, Sultan Qaboos University, OmanAbdallah M. A. Hassane, Al-Azhar University, Egypt
Jasneet Grewal, University of Warsaw, Poland
Copyright © 2022 Shrestha, Chio, Khatiwada, Kognou and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wensheng Qin, d3FpbkBsYWtlaGVhZHUuY2E=
 Sarita Shrestha
Sarita Shrestha Wensheng Qin
Wensheng Qin