
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 December 2022
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1048013
This article is part of the Research Topic Women in Microorganisms in Vertebrate Digestive Systems: 2022 View all 6 articles
The gut microbiota is essential for the development and maintenance of the hosts’ immune system. Disturbances to the gut microbiota in early life stages can result in long-lasting impacts on host health. This study aimed to determine if topical ivermectin treatment for endemic hookworm (Uncinaria sanguinis) infection in endangered Australian sea lion (Neophoca cinerea) pups resulted in gut microbial changes. The gut microbiota was characterised for untreated (control) (n = 23) and treated (n = 23) Australian sea lion pups sampled during the 2019 and 2020/21 breeding seasons at Seal Bay, Kangaroo Island. Samples were collected pre- and post-treatment on up to four occasions over a four-to-five-month period. The gut microbiota of untreated (control) and treated pups in both seasons was dominated by five bacterial phyla, Fusobacteria, Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes. A significant difference in alpha diversity between treatment groups was seen in pups sampled during the 2020/21 breeding season (p = 0.008), with higher richness and diversity in treated pups. Modelling the impact of individual pup identification (ID), capture, pup weight (kg), standard length (cm), age and sex on beta diversity revealed that pup ID accounted for most of the variation (35% in 2019 and 42% in 2020/21), with pup ID, capture, and age being the only significant contributors to microbial variation (p < 0.05). There were no statistically significant differences in the composition of the microbiota between treatment groups in both the 2019 and 2020/21 breeding seasons, indicating that topical ivermectin treatment did not alter the composition of the gut microbiota. To our knowledge, this is the first study to characterise the gut microbiota of free-ranging Australian pinniped pups, compare the composition across multiple time points, and to consider the impact of parasitic treatment on overall diversity and microbial composition of the gut microbiota. Importantly, the lack of compositional changes in the gut microbiota with treatment support the utility of topical ivermectin as a safe and minimally invasive management strategy to enhance pup survival in this endangered species.
The Australian sea lion is Australia’s only endemic pinniped species and has a highly fragmented population, breeding across 80 colonies which extend from the Houtman Abrolhos in Western Australia to The Pages Islands in South Australia (Gales et al., 1994; Kirkwood and Goldsworthy, 2013). The Australian sea lion population has undergone continual decline since commercial harvesting in the 18th and 19th centuries (Ling, 1999). It is currently listed as endangered on both the IUCN Red List (Goldsworthy et al., 2015) and the Environmental Protection and Biodiversity Conservation (EPBC) ACT (Threatened Species Scientific Committee, 2020) with approximately 10,000 free-ranging individuals remaining (Goldsworthy et al., 2021), making it one of the rarest pinniped species in the world. The reasons for this decline are multifactorial, with fisheries interactions (Hamer et al., 2013), entanglement in marine debris (Page et al., 2004; Byard and Machado, 2019), anthropogenic pollution (antibiotic resistant and human-associated bacteria, persistent organic pollutants, and heavy metal contaminants; Fulham et al., 2020, 2022; Taylor et al., 2021, 2022), and disease (Marcus et al., 2014, 2015a) identified as contributing factors.
At two of the largest Australian sea lion breeding colonies, Seal Bay Conservation Park on Kangaroo Island and Dangerous Reef in the Spencer Gulf, high levels of pup mortality have been reported, with rates of up to 41.8 and 44.6%, respectively (Goldsworthy et al., 2007, 2019). Pup mortality within these colonies has been attributed to starvation, conspecific trauma, and stillbirths (McIntosh and Kennedy, 2013). Pups at both colonies are endemically infected with Uncinaria sanguinis, a haematophagous nematode (Marcus et al., 2014) that causes localised intestinal inflammation, anaemia and hypoproteinaemia (Marcus et al., 2015a). Uncinaria sanguinis infection is typically patent for 2–3 months, and after clearance of infection, pups do not become re-infected (Marcus et al., 2014). Given the continual population decline and high rates of pup mortality, recent conservation management for this species has included mitigation of pup mortality through hookworm treatment (Marcus et al., 2015b; Lindsay et al., 2021). Lindsay et al. (2021) determined that topical ivermectin treatment was 96.5% effective at eliminating hookworm with improved haematological parameters and increased bodyweight in treated compared to untreated (control) pups. Hookworm infection is also prevalent in other pinniped species, contributing to differing levels of clinical disease and mortality in northern fur seal (Callorhinus ursinus; DeLong et al., 2009; Lyons et al., 2011), California sea lion (Zalophalus californianus; Spraker et al., 2007), South American fur seal (Arctocephalus australis; Seguel et al., 2013, 2016) and New Zealand sea lion (Phocarctos hookeri; Castinel et al., 2007; Michael et al., 2019) pups. Ivermectin has been administered to New Zealand sea lion, northern fur seal, and South American fur seal pups to reduce hookworm-associated mortality, and in all studies treatment resulted in hookworm clearance and improved pup growth and survival (Castinel et al., 2007; DeLong et al., 2009; Seguel et al., 2016; Michael et al., 2021). However, previous studies have not considered the potential consequences of treatment intervention on the composition of the gut microbiota in pinniped pups.
The gut microbiota of numerous pinniped species has been characterised (Nelson et al., 2013; Smith et al., 2013; Bik et al., 2016; Pacheco-Sandoval et al., 2019; Stoffel et al., 2020; Tian et al., 2020; Toro-Valdivieso et al., 2021), including in adult Australian sea lions (Delport et al., 2016). In most of these pinniped species, the gut microbiota is usually dominated by the Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria phyla, with the relative abundance differing between species and sampling site (Nelson et al., 2013; Bik et al., 2016; Delport et al., 2016; Pacheco-Sandoval et al., 2019). However, many of these studies have been limited to captive or adult pinnipeds, with the gut microbiota of pups only characterised in Juan Fernández fur seals (Arctocephalus philippii; Toro-Valdivieso et al., 2021), spotted seals (Phoca largha; Tian et al., 2020), Australian fur seals (Arctocephalus pusillus doriferus; Smith et al., 2013), and southern and northern elephant seals (Mirounga leonina and Mirounga angustirostris; Nelson et al., 2013; Stoffel et al., 2020).
The mammalian gastrointestinal tract is home to over 100 trillion microorganisms, collectively termed the microbiota (Clemente et al., 2012; Ursell et al., 2012; Thursby and Juge, 2017). Mammalian hosts and their microbiota have co-evolved over millions of years to establish a mutualistic relationship, and the importance of this relationship for host health has been established in recent decades (Hooper et al., 2012; Brestoff and Artis, 2013). Commensal bacteria contribute to digestion and nutrient provision (Hooper et al., 2002), synthesise essential vitamins and minerals (Yatsunenko et al., 2012), protect against pathogen colonisation through colonisation resistance (Bäumler and Sperandio, 2016) and are essential for the development and regulation of the immune system (Cebra, 1999; Brestoff and Artis, 2013).
The impact of intestinal parasites, such as helminths, on the composition of the gut microbiota remains relatively unknown, varying between host and parasite species (Walk et al., 2010; Rausch et al., 2013; Zaiss et al., 2015). Helminth infections in laboratory mice can result in substantial shifts in the gut microbiota composition (Li et al., 2012; Aivelo and Norberg, 2018; Peachey et al., 2018; Lee et al., 2020), which could be a consequence of multiple factors, including helminth secretory products (Zaiss et al., 2015). As intestinal helminths can both directly and indirectly interact with the gut microbiota, the removal of parasites via antiparasitic administration also has the potential to alter the composition of the microbial community (Giacomin et al., 2015; Reynolds et al., 2015). In humans and mice, the removal of intestinal helminths has varying impacts on the gut microbiota; outcomes of parasite treatment range from no associated change (Martin et al., 2018), minimal to moderate change in microbiota composition (Ramanan et al., 2016; Martin et al., 2018), to a shift in the composition of the microbiota to resemble individuals without helminth infection (Jenkins et al., 2018). In wildlife species, treatment with antiparasiticides has been associated with alterations to the faecal gut microbiota and metabolic profile (He et al., 2018; Moustafa et al., 2021). Given the cross-talk between the gut microbiota and the immune system, it is likely that the modulation of the microbial community by intestinal helminths can have both direct and indirect impacts on the hosts’ immune system and immune responses (Giacomin et al., 2015; Zaiss et al., 2015; Martin et al., 2019). Intestinal parasitism is prevalent in many free-ranging wildlife species (Spratt and Beveridge, 2019), for this reason, it is crucial to understand how parasitic infection (or removal thereof) influences gut microbial composition.
Just as microbes in the gut environment have evolved with their hosts, parasites can evolve with their hosts (Ebert and Fields, 2020), and it is likely U. sanguinis has evolved with its host species. Furthermore, interactions between the host, intestinal helminths and the gut microbiota are likely to be complex and multidirectional (Cortés et al., 2019). Whilst ivermectin treatment has been shown to improve Australian sea lion pup health parameters in the short term (Marcus et al., 2015b; Lindsay et al., 2021), the impact of removal of U. sanguinis on the gut microbial community, and potentially on pup health notwithstanding the improvements associated with mitigating hookworm disease, is unknown. The gut microbiota becomes established during early mammalian life stages, with disruptions or alterations potentially impacting the functional capacity of the microbiome, the development of the immune system (Johansson et al., 2012; Brestoff and Artis, 2013), altering the microbial composition, promoting colonisation of pathogenic bacteria, and most importantly, has the potential to influence lifelong host health and disease status (Guinane and Cotter, 2013; Schroeder and Bäckhed, 2016). Given the importance of the gut microbiota and its influence on development during early life stages, investigating whether treatment intervention and parasite elimination alters the gut microbiota of Australian sea lion pups is critical for understanding any potential consequences.
To establish the safety of a topical antiparasitic treatment as a potential management strategy to assist in the recovery of the Australian sea lion, the impact of treatment on the gut microbial community must be understood. To this end, this study aims to characterise and monitor the microbial composition of the gut microbiota of both untreated (control) and treated (and subsequently hookworm-free) Australian sea lion pups.
Faecal samples were collected from neonatal Australian sea lion pups at Seal Bay Conservation Park on Kangaroo Island, South Australia (35.99°S, 137.32°E). Pups were sampled during the 2019 winter (n = 160) and 2020/21 summer (n = 184) breeding seasons as part of an ivermectin treatment trial (Lindsay et al. unpublished).
In brief, pups were captured on up to four occasions, at approximately 4-week intervals. Pups were captured by hand and physically restrained in a ventilated canvas bag designed specifically for pinniped pups. During initial capture (capture 1 – the time point at which treatment intervention occurs but prior to impact of treatment), pups were assigned to an untreated (control) or treated (and subsequently hookworm-free, herein referred to as treated) group based on a randomised number chart, generated using Microsoft Excel. Morphometric data was collected from each pup during each capture event including bodyweight (kg), standard length (cm, measured from tip of the nose to tip of the tail), sex and body condition (poor, fair-thin, good, excellent). The initial capture also included a unique ‘hair cut’ on the dorsolumbar pelage and application of commercial hair dye to the hair cut (Schwarzkopf Nordic Blonde, Henkel Australia, Melbourne, Australia) to facilitate individual pup identification for recapture. Faecal swabs were collected via insertion of a sterile swab (Copan, Brescia, Italy) within a lubricated sheath into the rectum. Swabs were then subsampled into Sterile FecalSwab™ tubes (Copan, Brescia, Italy) and stored at 4°C for up to 2 months, followed by storage at −20°C for up to 2 months, and then at −80°C. Blood samples were collected from the brachial vein as previously described (Fulham et al., 2018, 2022). Up to 1 ml of blood was transferred into a tube containing ethylenediaminetetraacetic acid (EDTA; Sarstedt, Nümbrecht, Germany) for haematological analysis. Blood samples were stored at 4°C and processed within 10 h of collection. Sampling of Australian sea lion pups was approved by the Animal Ethics Committee at the University of Sydney (Protocol Number 2017/1260).
Of the total number of pups sampled in 2019 and 2020/21, a subset of pups (n = 46) that had been captured on four occasions were randomly selected from both the untreated (control) and treated groups by assigning a random number (Microsoft Excel for Mac v16.61.1) to each pup ID for inclusion in the microbial study.
After collection, faecal swabs were kept at 4°C prior to storage at −20°C. Samples were processed within 14 days of collection. The hookworm status of each pup was determined following methods described by Lindsay et al. (2021). In summary, faecal material was transferred onto a clean glass slide and examined via light microscopy for the presence of U. sanguinis eggs. In samples where no eggs were detected, two additional smears were evaluated to confirm negative status. A grading system was used to grade hookworm burden in cases where samples were positive: grade 1 = 1–9 eggs, grade 2 = 10–19 eggs, grade 3 = 20–29 eggs, grade 4 ≥ 30 eggs (Lindsay, pers. comm). Hookworm grade was determined using the mean value for the total number of smears performed for each individual.
Blood samples were processed following methodology described by Lindsay et al. (2021). In brief, the packed cell volume (PCV; L/L) was measured using the microhaematocrit method and total plasma protein (TPP; g/L) was estimated using a hand-held refractometer (Reichert TS Meter, Cambridge Instruments, Buffalo, United States). Blood smears were prepared in duplicate and fixed in 100% methanol (Chem-Supply Pty Ltd., Port Adelaide, South Australia) for 4 min. An aliquot (200 μl) of anticoagulated EDTA blood was transferred to a separate tube for preservation with an equal aliquot (200 μl) of Streck Cell preservative (Streck, Omaha, United States) and stored at 4°C prior to further analysis. Preserved samples were analysed on an automated haematology analyser (Sysmex XT-2000iV, Sysmex, Kobe, Japan) at the Veterinary Pathology Diagnostic Service, Sydney School of Veterinary Science, The University of Sydney, within 2–8 days of sample collection. From automated haematology analysis, the total erythrocyte count (x1012/L), haemoglobin concentration (g/L), mean cell volume (fL), mean cell haemoglobin concentration (g/L), platelet count (x109/L) and total nucleated (leukocyte) cell count (TNCC, ×109/L) were determined. The differential leukocyte count was obtained by differentiating 100 leukocytes for every 10×109/L TNCC to determine the absolute neutrophil, lymphocyte, eosinophil and monocyte counts (×109/L) by multiplying the percentage of each leukocyte by the TNCC. Nucleated red cell (nRBC) count was determined by counting the number of nucleated red blood cells per 100 leukocytes.
As part of an ongoing monitoring program at Seal Bay Conservation Park, the breeding colony is frequently monitored during the breeding season, allowing birth dates to be recorded (Goldsworthy et al., 2014). Date of birth for pups was provided by the Department of Environment and Water (DEW), South Australia, and the South Australian Research and Development Institute (SARDI), as part of ongoing monitoring within each breeding season. For the 2020/21 breeding season, all pups included in the study had known dates of birth and age at each capture was calculated based on known birth date. For the 2019 breeding season, birth dates were only known for a subset of pups (n = 13). For the remaining pups (n = 10) sampled in 2019, a regression analysis was utilised to estimate pup age, following the approach of Bradshaw et al. (2000). The fitted age model for the 2019 data is:
where sex has the value of 1 for males and 0 for females, and BCI (body condition index) is calculated based on a regression of length on weight (Stokes et al., 2020).
From the randomised subset of pups, DNA was extracted from a total of 184 faecal swabs (n = 92 from 2019 and n = 92 from 2020/21). DNA was also extracted from two faecal samples collected from N. cinerea pups in 2019 to be used as control samples to account for variation between sequencing runs.
Faecal DNA was extracted from FecalSwab™ media (200 μl) using the ISOLATE II Fecal DNA Kit (Bioline, Sydney, Australia) following the manufacturer’s protocol. Faecal DNA was tested for PCR competency by a 16S PCR using methods described by Fulham et al. (2020) using forward primer 27F and reverse primer 1492R (Lane, 1991). Amplicons were resolved using gel electrophoresis (2% agarose w/v) with SYBR safe gel stain (Invitrogen, Sydney, Australia) and conducted at 100 V for 30 min with product size approximated using HyperLadder II 50 bp DNA marker (Bioline, Sydney, Australia).
Prior to sequencing, the concentration of DNA (ng/μl) and purity of nucleic acids in all faecal DNA was tested using a NanoDrop® ND-1000 UV–Vis Spectrophotometer (Thermo Fisher Scientific, Massachusetts, United States). DNA was submitted to the Ramaciotti Centre for Genomics (University of New South Wales, Sydney, Australia) for 16SV1-3 amplicon sequencing with an Illumina Miseq v3 2×300 bp sequencing kit using primers 27F and 519R, producing a ~ 530 bp fragment (Lane, 1991).
Demultiplexed paired-end sequences were analysed using Quantitative Insights Into Microbial Ecology 2 (QIIME 2) version 2022.2 software (Bolyen et al., 2019). The DADA2 (Divisive Amplicon Denoising) plugin (Callahan et al., 2016) in QIIME 2 was used to trim and filter sequences for quality including the removal of primers, denoising and chimaera removal. Sequences were clustered into amplicon sequence variants (ASVs), also known as exact sequence variants, following denoising. Operational taxonomic units (OTUs) are also frequently used to define the gut microbiota and clusters sequences either based on the observed sequences or using a reference database (Preheim et al., 2013). Recently, more focus has been on ASVs as they are more sensitive and specific than OTUs, can be reused across studies, and are reproducible for future data sets (Callahan et al., 2017).
Taxonomies were assigned to each ASV using a 16S rRNA V1-V3 classifier trained against the frequently updated SILVA database (release 138; Quast et al., 2013; Yilmaz et al., 2014; Glöckner et al., 2017) using the q2-feature-classifier plugin in QIIME 2 (Rognes et al., 2016). Low abundance ASVs that were not present in at least two samples with a total read count below 20 were filtered out and removed prior to further analysis.
All statistical analyses were conducted in either QIIME 2 or RStudio (v2022.2.3 + 492, Boston Massachusetts). Data was exported from QIIME 2 into R using the QIIME2R package (Hall and Beiko, 2018). Using both QIIME 2 and the vegan package (Oksanen et al., 2022), alpha diversity rarefaction curves were generated to filter samples based on sufficient sampling depth and data was rarefied with the threshold of 5,000 reads used as the cut-off for samples collected in both 2019 and 2020/21. Significance was determined when p > 0.05 for all statistical tests.
Mean pup age in untreated (control) and treated groups were calculated at each capture event for both breeding seasons. A paired sample t-test was used to determine whether there was a significant difference in pup age at each capture between treatment groups and breeding seasons.
For analysis of hookworm burden, the mean hookworm grade from samples collected from untreated (control) and treated pups during the initial capture was determined. A paired sample t-test was utilised to test for significant differences in hookworm grade between treatment groups at the time of first capture.
The alpha (within-sample) diversity was estimated using the phyloseq package (McMurdie and Holmes, 2013). Richness, the total number of bacterial species present, and diversity, the amount of individual bacteria from each species identified, were measured using two metrics: Chao1, which estimates the richness in a sample through the estimation of the total number of species present (Chao, 1984); and the Shannon-Wiener index, which estimates diversity based on richness and abundance (Shannon, 1948). A Shapiro–Wilk test was used to test for data normality. Wilcoxon’s Rank Sum Tests were employed to estimate the statistical differences in alpha diversity metrics due to treatment group (untreated [control] and treated) and breeding seasons (2019 and 2020/21). Statistical differences in alpha diversity pre-treatment (capture 1) and post-treatment (capture 2) for both treatment groups were also tested using Wilcoxon’s Rank Sum Test.
To examine the correlations between host factors and alpha diversity, four linear mixed models (LMMs) were fitted using the lme4 package v1.1-25 (Bates et al., 2015). The first two models were conducted when analysing entire datasets within a season. The first model included treatment group, capture (1-4), pup sex, weight (kg), standard length (cm) and age as predictors and pup ID as a random effect, and the alpha diversity metric (Chao1 or Shannon-Wiener Index) as the response variable. In the second model, treatment group, hookworm status, age (months), TPP, TNCC, leukocyte counts (absolute neutrophil, absolute monocyte, absolute lymphocyte and absolute eosinophil; ×109/L) were predictors, pup ID was a random effect, and the alpha diversity metric was the response variable. Models three and four tested the influence of host factors within treatment groups and were the same as models one and two, respectively, with treatment group excluded.
The variation between samples (beta diversity) was analysed using Bray–Curtis dissimilarity calculations using rarefied data based on the abundance of ASVs and using principal coordinate analysis (PCoA) plots. Using the adonis function in the vegan package (Oksanen et al., 2022), a permutational analysis of variance (PERMANOVA) was employed to test the statistical differences in ASV abundances due to treatment group, hookworm status, weight, standard length, sex, capture, and pup age, and computed with 999 permutations. Four PERMANOVA models were fitted; the first included pup ID, hookworm status and treatment group; the second model included pup ID, treatment group, capture, and age; the third model included treatment group, pup ID, weight, standard length and pup sex; and the final model included pup ID, treatment group, TPP, TNCC and leukocyte counts (neutrophil, lymphocyte, monocyte and eosinophil). The second model was compared post hoc to account for effects of repeated measures. The assumption of multivariate homogeneity was met across all models for PERMANOVA tests (p > 0.05).
Wilcoxon’s Rank Sum Test was used to test for significant differences in the relative abundance of bacterial phyla and families between treatment groups and within and between breeding seasons. Finally, an analysis of composition of microbiomes (ANCOM) was used to test for differences in differentially abundant ASVs between treatment groups (Mandal et al., 2015), identifying any bacterial families that had the highest contribution to any dissimilarities observed in the gut microbial composition of untreated (control) and treated groups.
Randomisation of the sampled pups resulted in the sequencing of samples from 12 untreated (control) and 11 treated pups from the 2019 breeding season, and 11 untreated (control) and 12 treated pups from the 2020/21 breeding season. Treatment with ivermectin resulted in 100% elimination of hookworm infection in the n = 23 Australian sea lion pups in the treated group (Lindsay et al. unpublished).
When comparing mean age of pups between untreated (control) and treated groups, there was no significant difference (p > 0.05) between groups at any of the capture events, in both the 2019 and 2020/21 cohorts (Table 1). There was, however, a significant difference in the age of treated pups at the third capture event between the 2019 and 2020/21 breeding seasons. There was no significant difference in the mean hookworm grade between untreated (control) and treated pups at first capture (p > 0.05) in both breeding seasons.
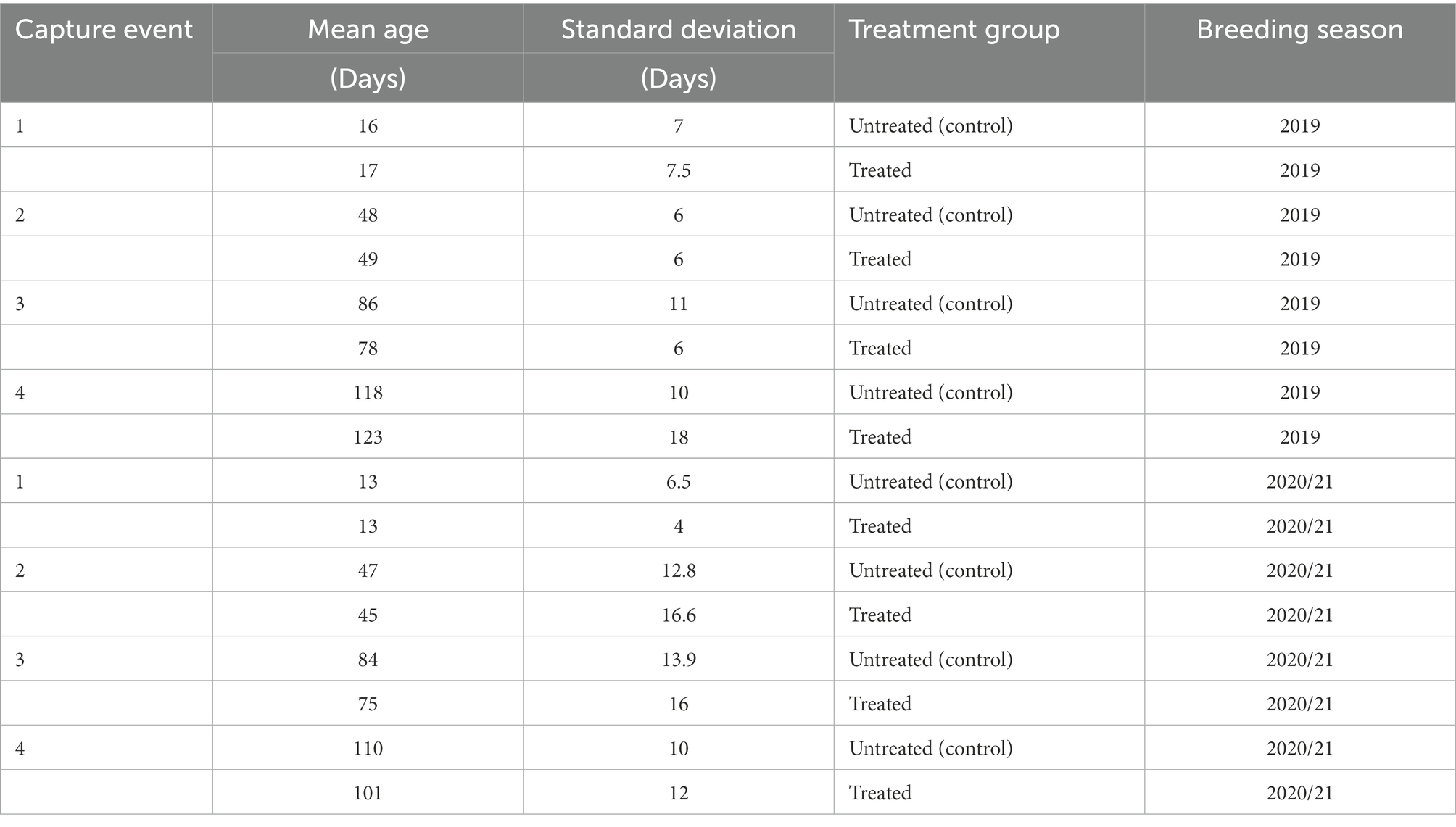
Table 1. Mean age (days) of pups in untreated (control) and treatment groups at each capture event during the 2019 and 2020/21 breeding seasons.
The hypervariable V1 to V3 region of the 16 s rRNA gene was sequenced in a total of 184 samples collected in 2019 and 2020/21, resulting in 12,078,034 and 8,439,797 sequences, respectively. After filtering steps and removal of low quality (n = 32) samples, 75 samples from 2019 (37 untreated [control], 38 treated) and 77 samples (38 untreated [control], 39 treated) from 2020/21 were imported into RStudio for analysis. The filtering of low abundance ASVs resulted in the clustering of 621 and 512 ASVs in the 2019 and 2020/21 datasets, respectively.
Five bacterial phyla, Fusobacteria, Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes, were present in untreated (control) and treated Australian sea lion pups (Figure 1). The majority of ASVs in both untreated (control) and treated pups were assigned to the Fusobacteria phylum (Table 2). In both groups, Fusobacteria had the highest average relative abundance (average relative abundance ± standard deviation = 55.4% ± 27.5 in untreated (control) and 61.3% ± 27.1 in treated), followed by Firmicutes (27.1% ± 12.9 and 20.5% ± 9.2), Proteobacteria (10.3% ± 4.3 and 9.2% ± 5.7), Actinobacteria (6.3% ± 3.3 and 6.5% ± 3.7) and Bacteroidetes (0.1% ± 0.1 in both treatment groups). There was no significant difference in the average relative abundance of each bacterial phyla pre- or post-treatment (capture 1-capture 2; p > 0.05) in both treatment groups and across captures 1 – 4 (p > 0.05). In both untreated (control) and treated pups, the relative abundance of Fusobacteria remained stable across all captures, whilst the abundance of Firmicutes decreased from capture 1 to capture 4 (Figure 1). The abundance of Actinobacteria increased after the second capture and then decreased in captures 3 and 4 in pups from both treatment groups, with the exception of one pup (pupID 19-141) in the untreated (control) group with a much greater abundance of Actinobacteria after capture 4 (Figure 1).
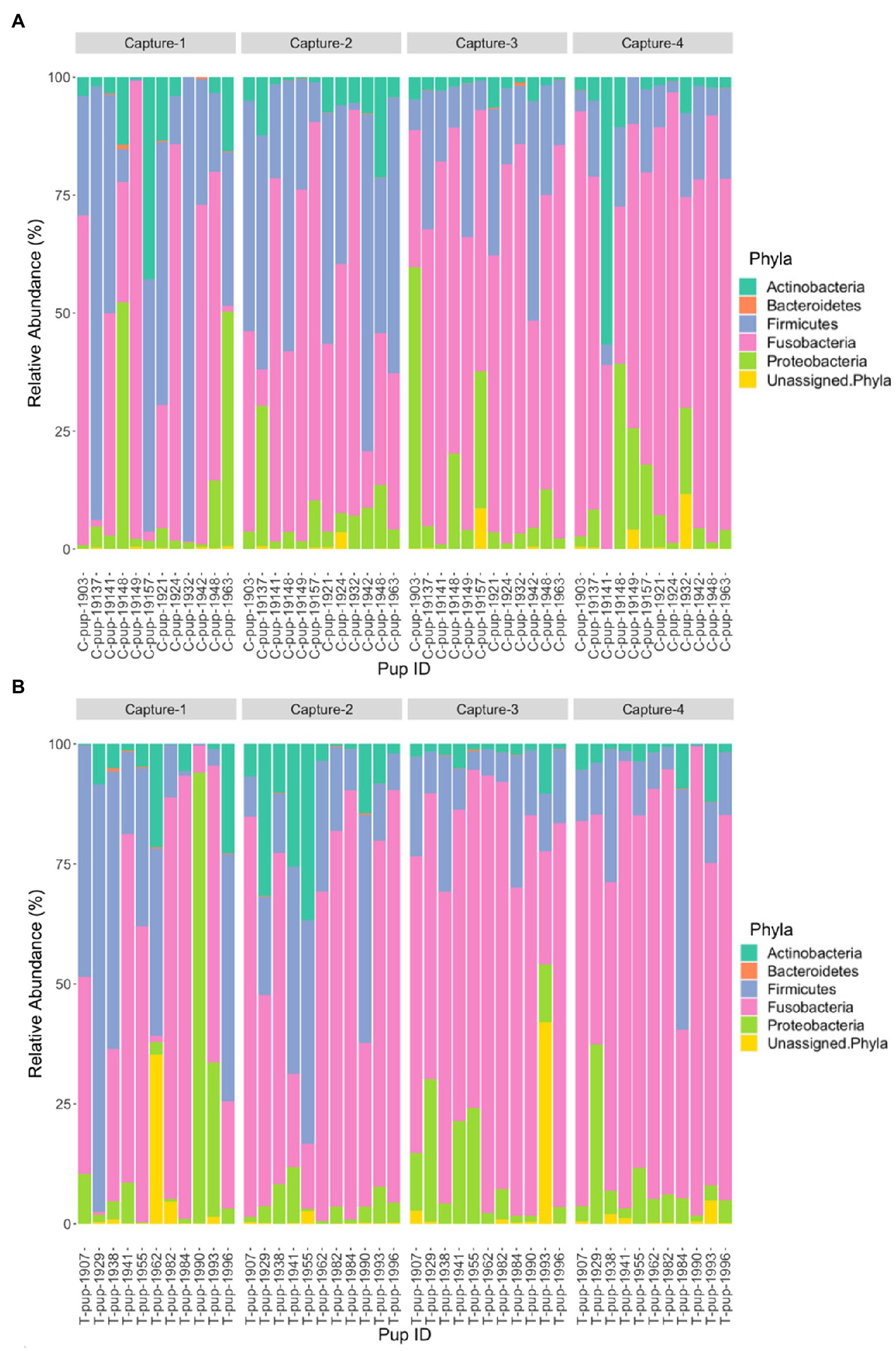
Figure 1. Relative abundance (%) of bacterial phyla in (A) untreated (control) and (B) treated Australian sea lion pups across four captures during the 2019 breeding season.
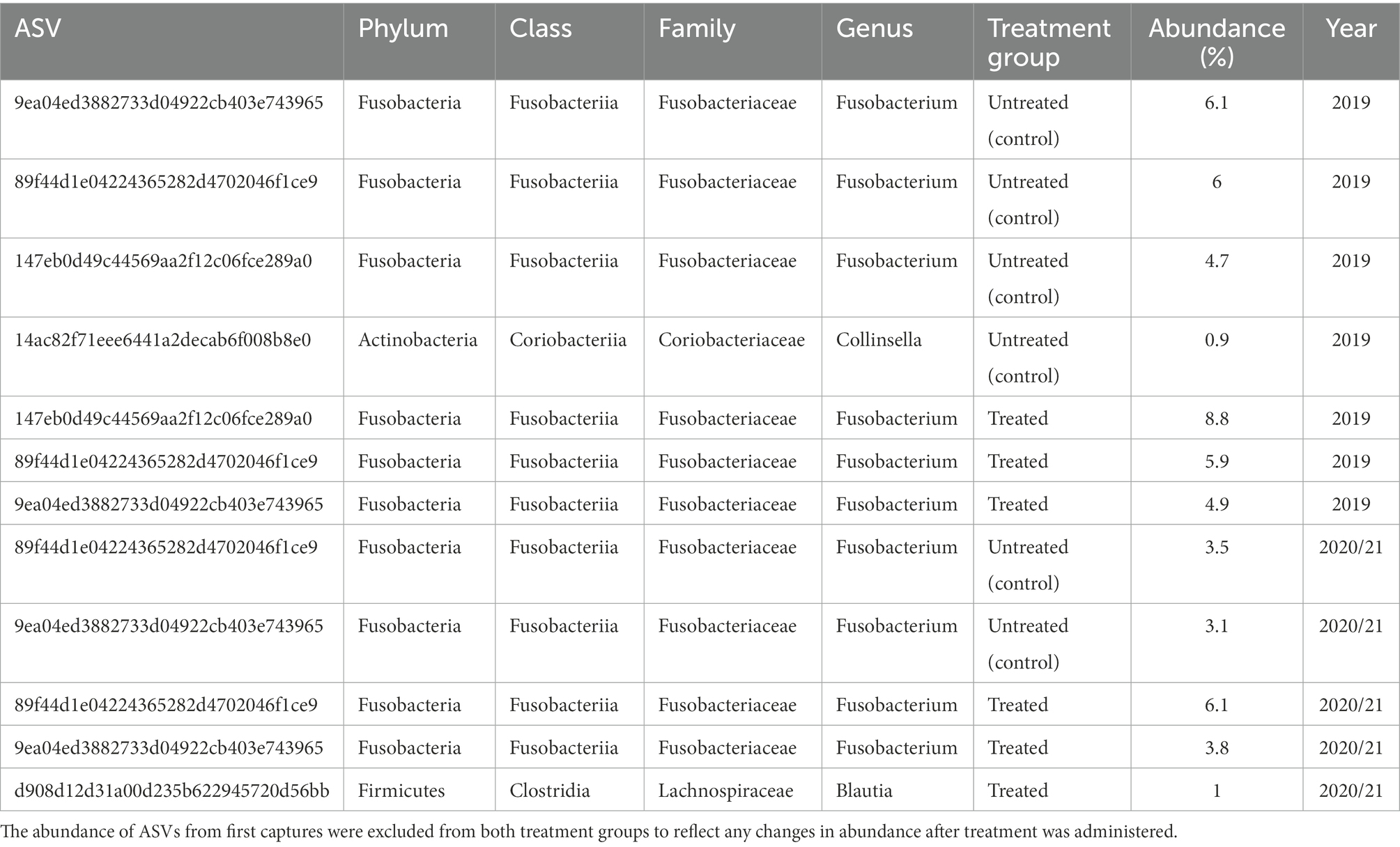
Table 2. Abundance of ASVs in untreated (control) and treated pups in captures 2–4, present in at least 90% of samples in each treatment group.
In both untreated (control) and treated pups, Fusobacteriaceae was the most dominant bacterial family. In untreated (control) pups, ASVs were assigned to five families from four bacterial phyla. The Fusobacteriaceae family occurred at the highest average relative abundance (54.1% ± 27.7), followed by families from the Firmicutes phyla, Clostridiaceae (17.8% ± 7.1) and Ruminococcaceae (7.4% ± 6.8), Coriobacteriaceae (4.4% ± 3.0) from the Actinobacteria phyla, and Succinivibrionaceae (2.7% ± 1.7) from the Proteobacteria phyla. Seven bacterial families from four bacterial phyla were identified in treated pups, with Fusobacteriaceae being similarly dominant (55.2% ± 20.7). The Clostridiaceae family was also the second most abundant in treated pups (13.5% ± 9.4), followed by Coriobacteriaceae (5.7% ± 3.5), Ruminococcaceae (5.7% ± 4.1), Succinivibrionaceae (4.5% ± 2.3), Rhodobacteraceae (2.9% ± 1.1), a member of the Proteobacteria phyla, and Leptotrichiaceae (2.2% ± 0.5) a family belonging to Fusobacteria (Figure 2). There was no significant difference in the average relative abundance of each family between treatment groups (p > 0.05). The Rhodobacteraceae, Leptotrichiaceae, and Succinivibrionaceae families were only identified at an abundance ≥1% in ASVs in treated pups, with Leptotrichiaceae only present in treated pups after capture 4 (Figure 2).
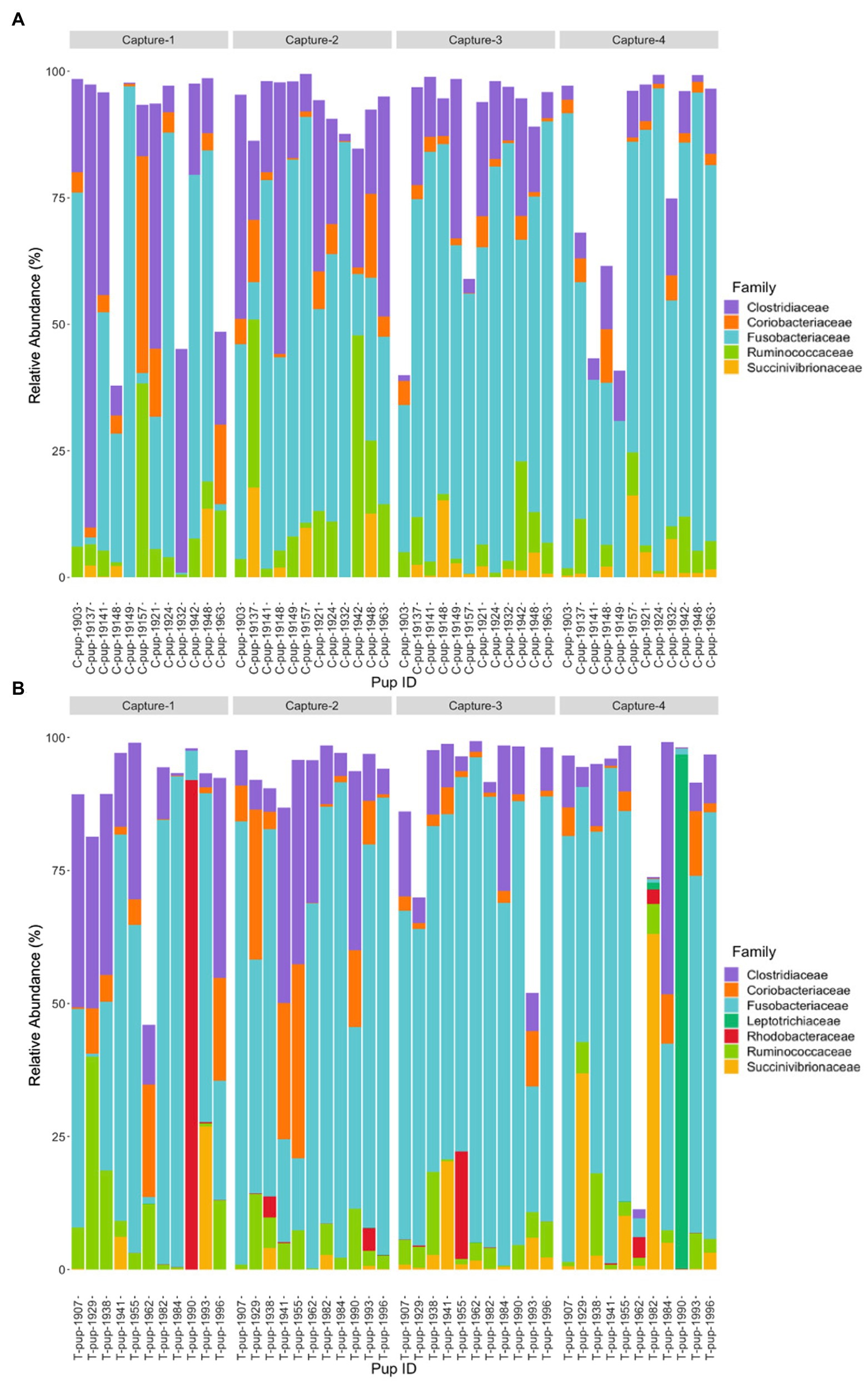
Figure 2. Relative abundance (%) of bacterial families in (A) untreated (control) and (B) treated Australian sea lion pups across all four captures during the 2019 breeding season. Only families that occurred at an abundance ≥1% in each treatment group are shown.
In Australian sea lion pups sampled during the 2020/21 breeding season, five bacterial phyla were identified. The gut microbiota of both untreated (control) and treated pups was dominated by Fusobacteria, which had the highest average relative abundance (38.6% ± 23.2 in untreated [control] and 42.7% ± 20.9 in treated pups), followed by Firmicutes (36.1% ± 20.4 and 36.3% ± 20.3), Proteobacteria (16.7% ± 8.3 and 14.7% ± 6.3), Bacteroidetes (5.0% ± 1.7 and 4.5% ± 1.2) and Actinobacteria (3.4% ± 1.2 and 1.1% ± 0.3). The majority of ASVs in both treatment groups were assigned to the phylum Fusobacteria (Table 2). A small percentage of ASVs could not be assigned to a bacterial phylum in both groups, 0.2% in untreated (control) and 0.7% in treated pups. The average relative abundance of each bacterial phyla did not change when compared pre- and post-treatment in the treated group (p < 0.05). There was no significant difference in the average relative abundance of each bacterial phyla between treatment groups and across captures 1 – 4 (p > 0.05). The relative abundance of Fusobacteria and Firmicutes decreased in both treatment groups across the four captures whilst Proteobacteria and Actinobacteria increased (Figure 3). The abundance of Bacteroidetes was greatest in samples from capture 4 in both treatment groups (Figure 3).
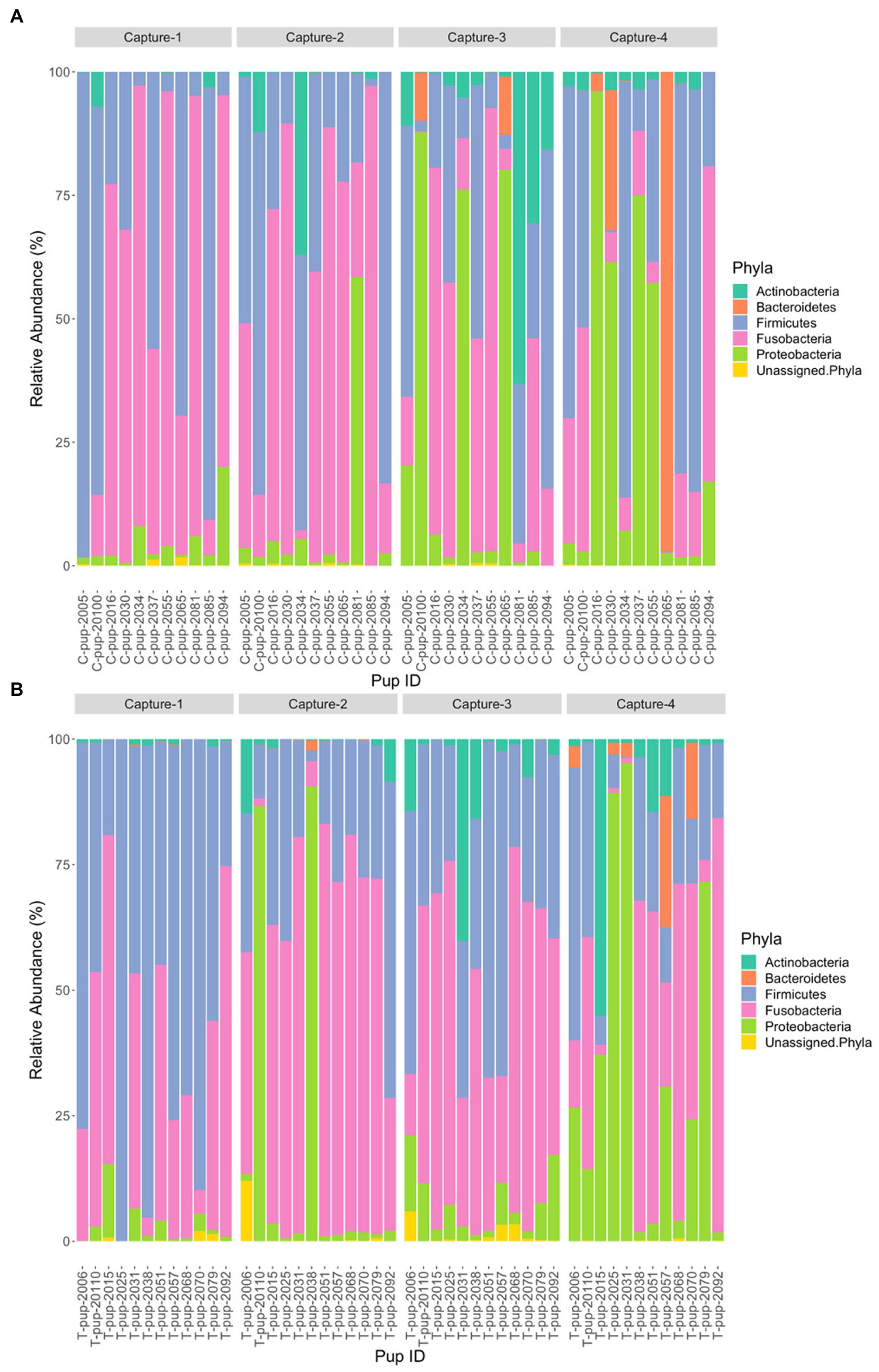
Figure 3. Relative abundance (%) of bacterial phyla in (A) untreated (control) and (B) treated Australian sea lion pups across the four captures during the 2020/21 breeding season.
At the family level, seven bacterial families were present in untreated (control) pups and five families were identified in treated pups that had a total abundance ≥1%. In untreated (control) pups, the average relative abundance was highest for the Fusobacteriaceae family (39.4% ± 23.9), followed by families belonging to the Firmicutes phyla, Lachnospiraceae (19.3% ± 7.6), Clostridiaceae (8.9% ± 3.1) and Ruminococcacae (5.0% ± 2.2), Halomonadaceae (7.5% ± 1.5) and Rhizobioaceae (3.1% ± 0.9) from the Proteobacteria phyla, and Flavobacteriaceae (3.2% ± 1.0) from the Bacteroidetes phyla. The five families assigned to reads from treated pups were similarly abundant, with Fusobacteriaceae being the most dominant (42.4% ± 27.1), followed by Lachnospiraceae (18.9% ± 10.2), Clostridiaceae (10.3% ± 7.7), Halomonadaceae (6.0% ± 5.8) and Ruminococcaceae (6.0% ± 5.0). Whilst the abundance of Fusobacteriaceae decreased from capture 1 to capture 4 in both treatment groups, a more noticeable decrease in Clostridiaceae was apparent in treated pups after capture 1 (Figure 4). The Flavobacteriaceae and Rhizobiaceae families were only present in untreated (control) pups in captures 3 and 4 (Figure 4).
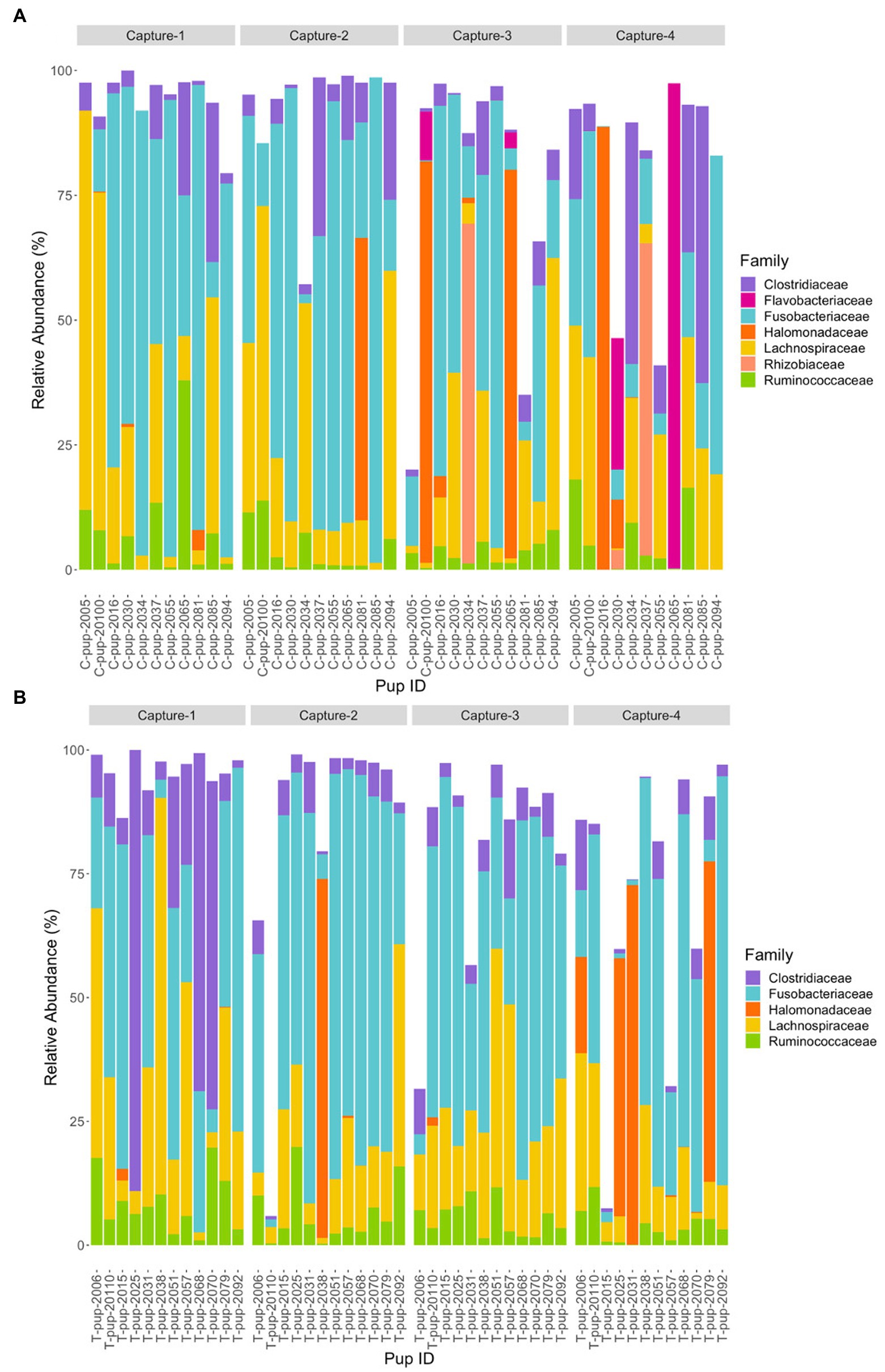
Figure 4. Relative abundance (%) of bacterial families in (A) untreated (control) and (B) treated Australian sea lion pups over four captures, sampled during the 2020/21 breeding season. Only families that occurred at an abundance ≥1% in each treatment group are shown.
Five bacterial phyla were identified in both untreated (control) and treated pups in both breeding seasons. In both 2019 and 2020/21, the majority of ASV counts belonged to three phyla: Fusobacteria (53.5 and 39.0%), Firmicutes (22.1 and 33.8%) and Proteobacteria (9.6 and 14.4%). The Actinobacteria phyla was more dominant in 2019 (5.8 and 2.1%) whilst Bacteroidetes was more dominant in 2020/21 (0.1 and 4.4%). A small percent of ASVs, 1.6 and 0.43%, could not be assigned to a bacterial phylum in 2019 and 2020/21, respectively. There was a significant difference in the relative abundance of Fusobacteria (p < 0.001), Firmicutes (p = 0.0021), Actinobacteria (p < 0.0001) and ASVs that could not be assigned to a phylum (p = 0.001) between breeding seasons. The average relative abundance of Fusobacteria and Actinobacteria were higher in 2019 (58.6 ± 27.2 and 6.30 ± 3.43) compared to 2020/21 (41.4 ± 20.1 and 4.4 ± 2.3), whilst the opposite was observed for Firmicutes (23.8 ± 11.1 in 2019 and 33.8 ± 16.0 in 2020/21). There was no significant difference in the relative abundance of Bacteroidetes (p = 0.11) or Proteobacteria (p = 0.29) across the 2019 and 2020/21 breeding seasons.
When comparing the relative abundance of each phylum in untreated (control) pups across seasons, there was only a significant difference in Fusobacteria (p = 0.021) and Actinobacteria (p = 0.0023), with a greater relative abundance of both phyla in 2019. In treated pups, there was a significant difference between breeding seasons in the relative abundance of Fusobacteria (p < 0.01), Firmicutes (p < 0.01) and Actinobacteria (p < 0.01). The relative abundance of Fusobacteria and Actinobacteria was higher in treated pups in 2019, whilst Firmicutes occurred at a higher relative abundance in 2020/21.
Three families, Fusobacteriaceae, Clostridiaceae and Ruminococcaceae were present in pups in both breeding seasons. There was a significant difference in the relative abundance of Fusobacteriaceae (p = 0.001) and Clostridiaceae (p < 0.001) between the 2019 and 2020/21 breeding seasons. In 2019, the relative abundance of Fusobacteriaceae and Clostridiaceae were significantly higher than in 2020/21 in both untreated (control) pups (p = 0.001 and p < 0.001) and treated pups (p = 0.018 and p = 0.041). For both treatment groups, there was no significant difference in the relative abundance of Ruminococcaceae (p > 0.05).
Two measures of alpha diversity (Chao1 and Shannon-Wiener Index) were used to analyse differences in within-sample diversity between treatment groups and breeding seasons. To determine any immediate differences post-treatment, alpha diversity was compared between capture 1 and capture 2 for each cohort (2019 and 2020/21) as well as within each treatment group.
In the 2019 cohort, the mean diversity, as indicated by the Shannon-Wiener index, was higher in the untreated (control) group compared to the treated group, whilst richness, measured through the Chao1 index, was higher in treated pups (Table 3 and Figure 5). There was no significant difference in alpha diversity between treatment groups based on the Chao1 index (p = 0.157) and the Shannon-Wiener index (p = 0.159). No significant differences in alpha diversity were observed between capture 1 and capture 2 in the untreated (control) and treated groups in the Chao1 index (p = 0.318 and p = 0.558) and the Shannon-Wiener index (p = 1.00 and p = 0.644).
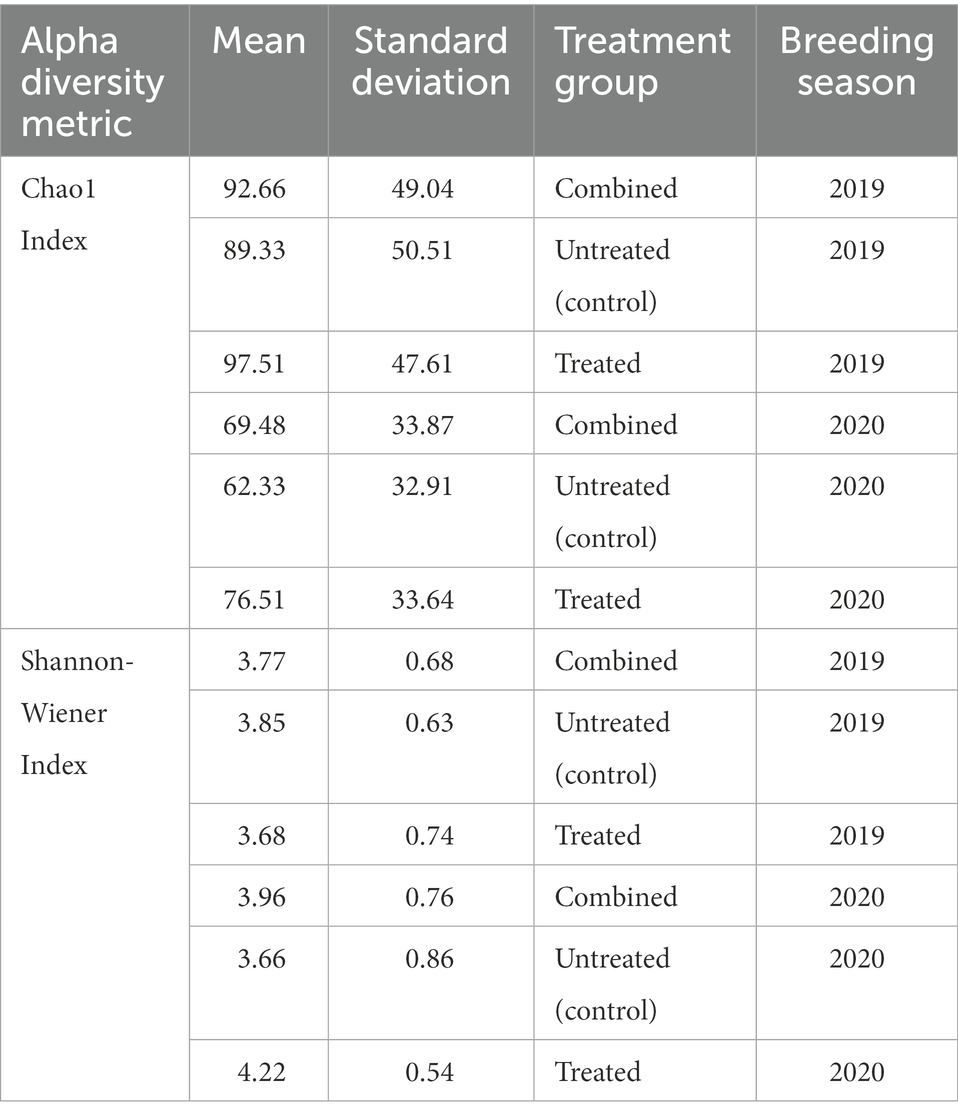
Table 3. Mean Chao1 index and Shannon-Wiener indices for untreated (control) and treated groups, and cohorts as a whole (combined) in both the 2019 and 2020/21 breeding seasons.
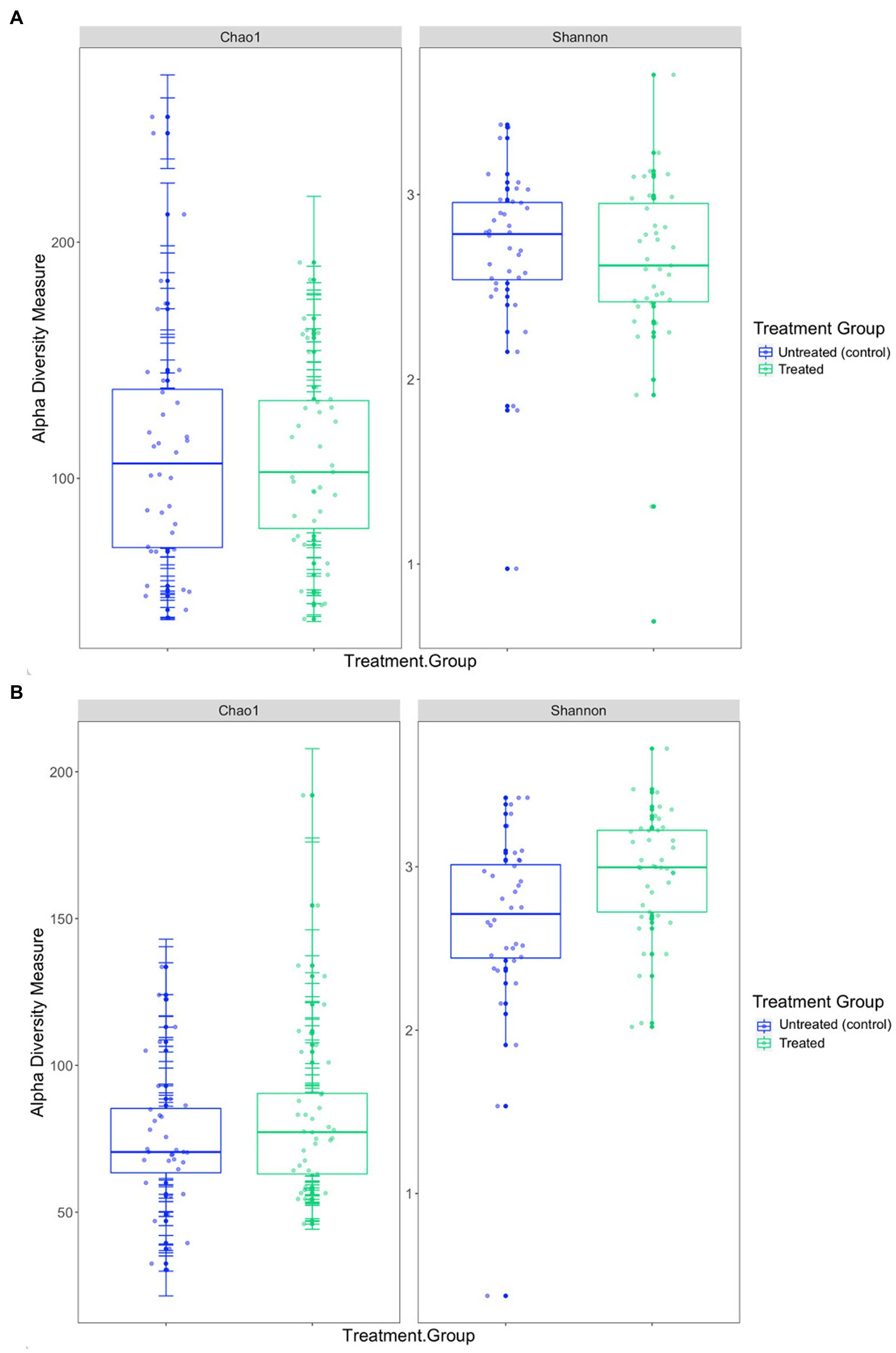
Figure 5. Comparison of the two alpha diversity measures, Chao1 index and the Shannon-Wiener index between treatment groups in the (A) 2019 breeding season and (B) 2020/21 breeding season.
The mean diversity in the 2020/21 cohort was higher in the treated group (Table 3 and Figure 5), which was reflected in the significant difference in the Shannon-Wiener index (p = 0.008). Richness was also, on average, higher in the treated group (Table 3), however, there was no significant difference in the Chao1 index between treatment groups (p = 0.104) or in alpha diversity between capture 1 and capture 2 for both the untreated (control) and treated groups for the Chao1 index (p = 0.250 and p = 0.193) and the Shannon-Wiener index (p = 0.693 and p = 0.977).
There was a significant difference in both the Chao1 index (p = 0.001) and the Shannon-Wiener index (p = 0.017) between the 2019 and 2020/21 breeding seasons. On average, richness was greater in the 2019 cohort whilst diversity was greater in the 2020/21 cohort (Table 3). Diversity and richness were also compared in treatment groups between breeding seasons. In the untreated (control) groups, there was no significant difference in diversity (p = 0.215) between seasons, however, there was a significant difference in richness (p = 0.013), with higher richness observed in the 2019 untreated (control) group (Table 3). There was a significant difference in both the Chao1 index (p = 0.025) and the Shannon-Wiener index (p = 0.001) between the treated groups between breeding seasons, with higher diversity in 2020/21.
When analysing Australian sea lion pups sampled during the 2019 breeding season as a cohort using the first LMM, there was a significant correlation between capture and Chao1 (R2 = 0.134, p = 0.048; Supplementary Table 1). The richness of the gut microbiota fluctuated across captures, with the highest mean richness observed in samples collected during capture 3 (101 ± 5.21). No significant correlations between any of the variables included in the first model and the Shannon-Wiener index were identified (Supplementary Table 1). The correlation between pup morphometrics and alpha diversity was also investigated within each treatment group using the third LMM, which included the same factors as model 1 with treatment group excluded. In untreated (control) pups, there was no significant correlation between the Chao1 index and pup morphometrics, however, there was a significant correlation between capture and the Shannon-Wiener index (R2 = 0.176, p = 0.011; Supplementary Table 2). The mean diversity varied across captures and was highest after capture 3 (4.11 ± 0.38) before decreasing by capture 4 (3.74 ± 0.61). In Australian sea lion pups treated in the 2019 breeding season, there was no significant correlation between pup morphometrics and both the Chao1 index and the Shannon-Wiener index (Supplementary Table 2).
In the 2020/21 cohort, there was a significant correlation between treatment group and both the Chao1 index (R2 = 0.107, p = 0.043) and the Shannon-Wiener index (R2 = 0.198, p = 0.002; Supplementary Table 1). The mean values of richness and diversity were higher in the treated compared to the untreated (control) group (Supplementary Table 1). However, within the untreated (control) and treated groups there was no significant correlation between pup morphometrics and alpha diversity (Supplementary Table 2).
In the 2019 cohort, there was a significant correlation between capture and the Chao1 index (R2 = 0.134, p = 0.013) in the second LMM (Supplementary Table 1); significant correlations between the Chao1 index and any of the other variables, including treatment group, were not identified. As with the results of model 1, there was no significant correlation with any factors and the Shannon-Wiener index. In the fourth model, which determined correlations between haematological parameters and alpha diversity within each treatment group, there were no significant correlations observed for the Chao1 index in either treatment group (Supplementary Table 2). There was a significant correlation between capture and the Shannon-Wiener index in the untreated (control) group (R2 = 0.257, p = 0.019); there were no significant correlations for the Shannon-Wiener index in the treated group. There was no significant correlation or relationship between haematological parameters and alpha diversity in either treatment group in the 2019 breeding season.
Analysis using the second fitted LMM found a significant correlation between treatment and both the Chao1 index (R2 = 0.135, p = 0.015) and the Shannon-Wiener index (R2 = 0.200, p = 0.04) in the 2020/21 breeding season (Supplementary Table 1). Within the untreated (control) and treated groups during the 2020/21 season there was no significant correlation between any factors included in model four and richness or diversity (Supplementary Table 1).
Based on analysis of the differences in ASV abundance through the Bray–Curtis dissimilarity, 13.8% of the variation in the spread of the data could be explained by the first axis, and 8.7% could be explained by the second axis (Figure 6). There was no clear clustering of the data based on treatment group, indicating that dissimilarity between samples was not based on the treatment status of the individual (Figure 6). The results from the PERMANOVA models indicate that in pups sampled during the 2019 breeding season, pup ID was the most significant predictor of microbial similarity and accounted for most of the variation (Table 4). The capture event was also a significant predictor (p = 0.02), along with pup ID (p = 0.001) in the second model. The results from all models, presented in Table 4, determined that treatment group, hookworm status, age, pup weight, standard length and haematological parameters (TPP, TNCC, absolute neutrophil count, absolute lymphocyte count, absolute monocyte count and absolute eosinophil count), were all non-significant predictors of microbial similarity.
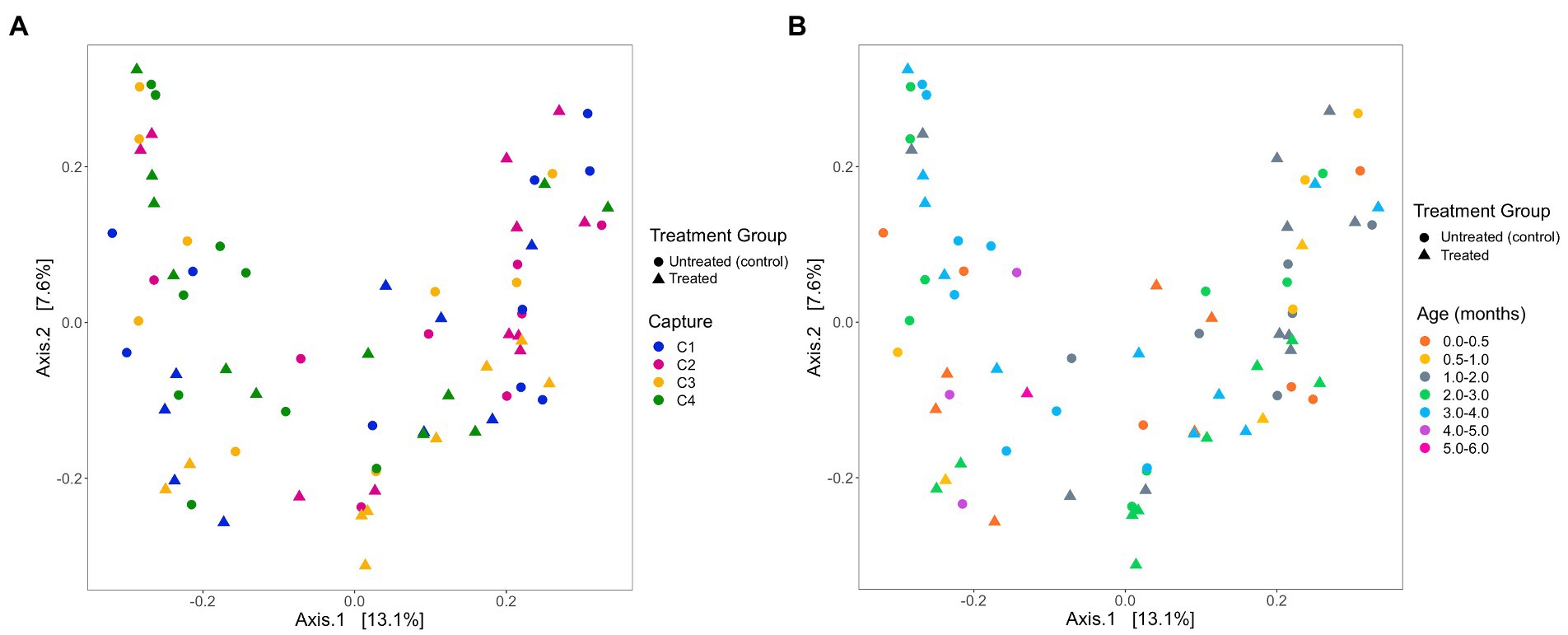
Figure 6. Beta diversity of faecal samples collected from Australian sea lion pups during the 2019 breeding season based on Bray-Curtis dissimilarities across (A) pup age groups (months) and (B) captures. In both plots, circles represent untreated (control) pups and triangles represent treated pups.
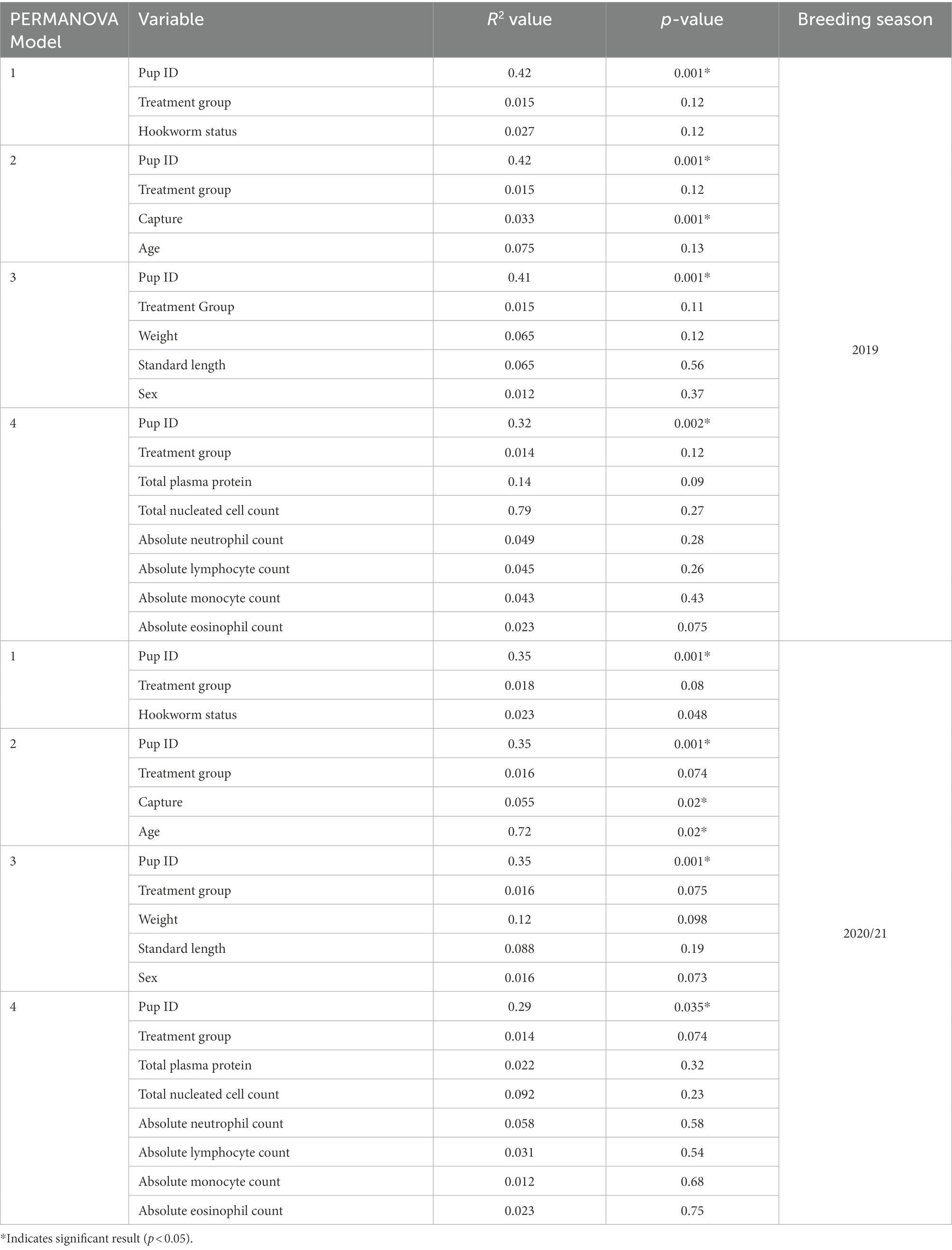
Table 4. Results from PERMANOVA analysis from both 2019 and 2020/21 breeding seasons, with variables included in each model, variation accounted for by each variable (R2) and p-value.
There was a difference in the number of bacterial families identified between treatment groups, with five and seven in untreated (control) and treated groups, respectively. However, ANCOM analysis indicated that these unique features were not present at a high enough abundance to be detected as differentially abundant at the family or genus level between treatment groups.
The analysis of the beta diversity of the gut microbial community of pups sampled during the 2020/21 breeding season revealed that the first and second axis explained 13.1 and 7.6% of the variation, respectively (Figure 7). From the four PERMANOVA models, pup ID was the most significant predictor of similarity and accounted for most of the variation in each model (Table 4). In the second model, both pup age and capture were also significant predictors (p = 0.02). The contribution of capture and age to similarity can be visualised in the Bray–Curtis dissimilarity matrix, with samples collected at similar captures from pups of closer age groups clustering together (Figure 7).
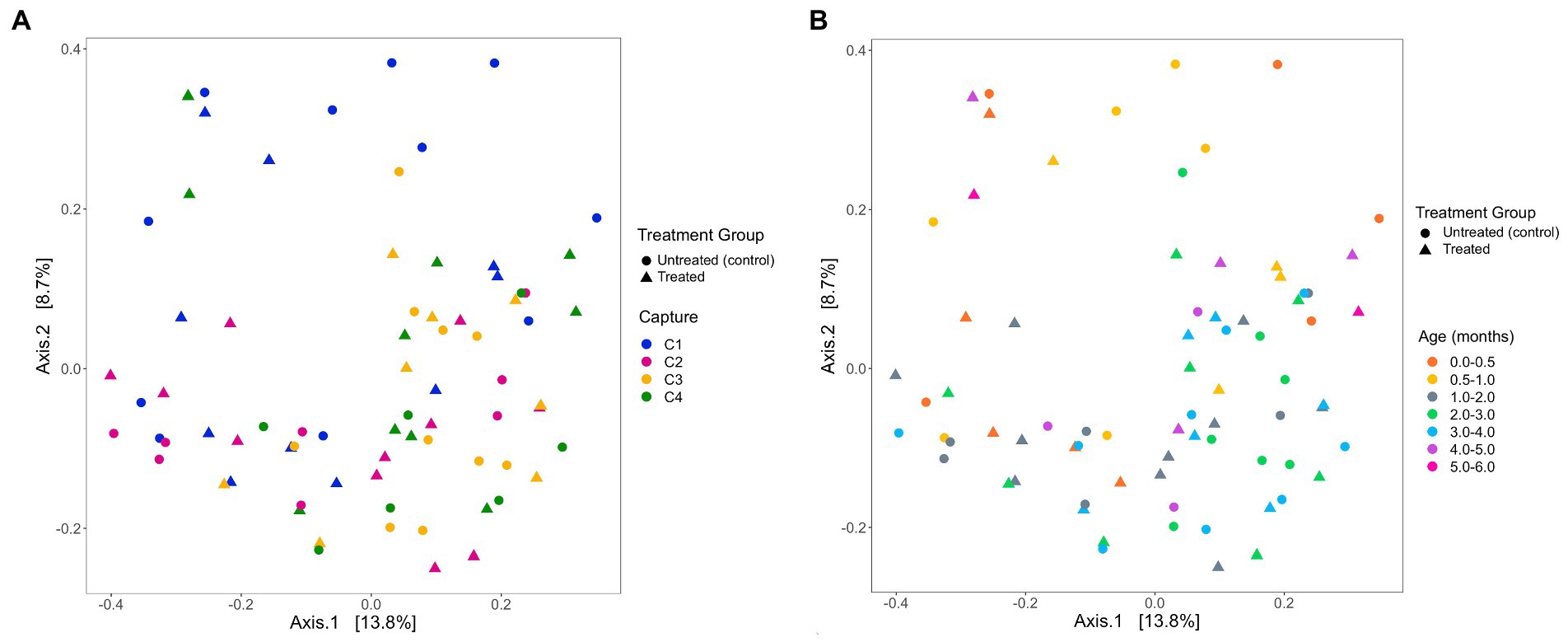
Figure 7. PCoA ordination of Bray-Curtis dissimilarities across (A) captures and (B) pup age groups (months), representing beta diversity of faecal samples collected from Australian sea lion pups during the 2020/21 breeding season. Circles represent untreated (control) pups and triangles represent treated pups.
In the 2020/21 breeding season, seven and five bacterial families were detected in samples collected from untreated (control) and treated pups, respectively. Whilst there was a qualitative difference between the two groups, ANCOM analysis revealed that the features assigned to families only identified in untreated (control) pups were not present at a high enough abundance to be considered differentially abundant between treatment groups.
This study aimed to determine whether topical ivermectin treatment and elimination of U. sanguinis infection altered the gut microbiota in endangered Australian sea lion pups. The findings from this study indicate that topical ivermectin treatment does not result in compositional change in the gut microbiota of Australian sea lion pups, suggesting that treatment does not cause any significant change to the functional capacity of the gut microbiome. This knowledge is crucial when considering the safety and efficacy of antiparasitic treatment in a free-ranging population, given the profound impact that the gut microbiota can have on host development and health through the regulation of the immune system, digestion and by protecting the host against pathogens (Ley et al., 2008a, 2008b; Lozupone et al., 2012; Margaret et al., 2013).
It was hypothesised that treatment with topical ivermectin and subsequent elimination of U. sanguinis would alter the gut microbiota of Australian sea lion pups when compared pre- and post-treatment. However, in both the untreated (control) and treated groups in both breeding seasons, there were no significant differences in the composition of the gut microbiota between pre-treatment (capture 1) and post-treatment (capture 2) time points. Given the endemic nature of U. sanguinis infection in Australian sea lion pups at Seal Bay (Marcus et al., 2014), the composition of a ‘healthy’ gut microbiota or gut microbiota without parasite infestation is unknown. However, as pups are infected with U. sanguinis via the colostrum immediately after birth (Marcus et al., 2014), it is unlikely that the gut microbiota of an Australian sea lion without infection with U. sanguinis would be observed. There were no compositional changes in the gut microbiota seen in association with either the natural elimination of hookworm in untreated (control) pups or via administration of topical ivermectin in treated pups. This absence of a change in microbial composition could suggest that the gut microbiota of Australian sea lion pups has adapted to the presence of this endemic parasite over their long symbiotic association, with the same composition of gut microbiota attained regardless of the speed at which infection is cleared. Whilst understanding the gut microbiota of Australian sea lion pups has adapted to U. sanguinis is an important knowledge gap, it should be noted that the outcomes of U. sanguinis infection for Australian sea lion pups are either to succumb to infection or to naturally clear the parasite at approximately 2 months of age (Marcus et al., 2014). Additionally, no other macroscopic parasites have previously been observed in both live or dead Australian sea lion pups (Marcus et al., 2014).
For both treatment groups in 2019 and 2020/21, the gut microbial communities were characterised by five bacterial phyla: Fusobacteria, Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes, and were dominated by Fusobacteria. These phyla have previously been identified as the main phyla in the gut of numerous pinniped species (Nelson et al., 2013; Smith et al., 2013; Bik et al., 2016; Delport et al., 2016; Medeiros et al., 2016; Pacheco-Sandoval et al., 2019; Stoffel et al., 2020; Tian et al., 2020; Toro-Valdivieso et al., 2021). The relative abundance of each bacterial phylum differs between species and is likely influenced by differences in life histories, geographical ranges, environmental conditions, diet, and sampling techniques.
The outcome of helminth treatment on the gut microbiota of the host has been studied in humans, mice, and numerous wildlife species with varying results (Cooper et al., 2013; Ramanan et al., 2016; He et al., 2018; Jenkins et al., 2018; Peachey et al., 2018; Moustafa et al., 2021). Some human studies observed no effect between treated and untreated individuals pre- and post-treatment (Cooper et al., 2013; Ramanan et al., 2016; Rosa et al., 2018), whilst others found compositional changes and reduced alpha diversity in individuals’ post-treatment (Jenkins et al., 2018). In this study, the only significant difference in alpha diversity correlated with treatment group was observed in Australian sea lion pups treated during the 2020/21 breeding season. Both the Chao1 and Shannon-Wiener indices were higher in treated pups compared to untreated (control) pups. However, in treated pups, there was no significant difference in alpha diversity between pre- and post-treatment time points (capture 1 versus capture 2), suggesting that the removal of hookworm infection after treatment is not the cause of the significant difference observed. Furthermore, hookworm status was not associated with significant changes in alpha diversity in either of the treatment groups.
Despite differences in richness and diversity of microbial community in Australian sea lion pups, there were no significant differences in microbial community composition between treatment groups, which is attributed to the topical method of administration. In Amur tigers (Panthera tigris altaica) treated with oral Fenbendazole and ivermectin, significant changes in both the gut microbiota and faecal metabolic phenotypes were seen, suggesting that treatment both disturbed the microbial community within the gut as well as metabolic homeostasis (He et al., 2018). Significant differences in gut microbial composition and diversity were also observed in Asian elephants (Elephas maximus) administered oral Albendazole treatment (Moustafa et al., 2021) and in humans treated with oral ivermectin for helminth infection (Jenkins et al., 2018).
Parasite presence in the gastrointestinal tracts of humans, livestock and wildlife species have been found to have differing impacts on gut microbial diversity, largely dependent on host and parasite species (Giacomin et al., 2015; Reynolds et al., 2015; Zaiss et al., 2015). In the present study, there was no clear association between the presence of U. sanguinis and taxonomic assignment of ASVs or microbial diversity in both treatment groups regardless of breeding season. Investigations into the relationship between parasite burden and the composition of the gut microbiota have produced mixed results. In western lowland gorillas (Gorilla gorilla gorilla), there was no relationship found between the number of eggs per gram (EPG) in faeces and the overall diversity and composition of the gut microbiota (Vlčková et al., 2018). In contrast, studies in both zebrafish (Danio rerio) and mice documented changes in both alpha diversity and the composition of the gut microbiome corresponding to parasite burden (Gaulke et al., 2019; Guiver et al., 2022). In minimally invasive and non-invasive studies it is difficult to determine the burden of parasite infection as the number of eggs observed in faeces is often not representative of the burden of adult worms in the intestine (Anderson and Schad, 1985; Warnick, 1992). In this study, a semi-quantitative evaluation of hookworm intestinal burden was used. The grading system suggested that the infection intensity of U. sanguinis was not significantly different between untreated (control) and treated pups, prior to the administration of treatment. Despite the lack of microbial composition changes associated with hookworm removal, it is possible that changes to the microbiota are occurring at the site of infection in the small intestine (Marcus et al., 2014) but these changes are not observed in the faecal microbiota. Furthermore, there is the potential for pups with higher parasite burdens to experience greater localised changes at the site of infection, such that the interpretation of these results should take this into consideration.
In pinnipeds, the relative abundance of Fusobacteria appears to be highest in pups and decreases with age (Nelson et al., 2013; Stoffel et al., 2020; Tian et al., 2020). For example, in southern elephant seals, there was a significantly higher abundance of Fusobacteria in pups compared to sub-adults and adults (Nelson et al., 2013), and the relative abundance of Fusobacteria has been found to decrease steadily during the weaning period of Pacific harbour seals (Stoffel et al., 2020). Changes in bacterial phyla abundance was also reported with age in Australian fur seals, with significant shifts observed between pup, juvenile and adult life stages (Smith et al., 2013). In Australian sea lion pups, a shift in the relative abundance of bacterial phyla and families across capture events was observed, although this shift is likely an age-related change, with up to 5 months between first and fourth captures. The results from the Bray–Curtis dissimilarity matrix suggested that the gut microbiota was more similar in pups of the same age, regardless of treatment group. A higher relative abundance of Fusobacteria was also observed in the current study in both treatment groups between the 2019 and 2020/21 breeding seasons, where Fusobacteria was the most abundant bacterial phyla. Bacterial families belonging to the Fusobacteria phylum were also the most dominant in the gut microbiota of Australian sea lion pups between both breeding seasons. The trends observed in the 2020/21 breeding season also suggest that the relative abundance of Fusobacteria decreases with age, with the highest relative abundance occurring in samples collected during the first capture and steadily decreased across subsequent captures, irrespective of treatment group. There was no significant difference in the mean age of untreated (control) and treated pups at each capture event, suggesting that any differences in the composition or diversity of the gut microbiota is not due to differences in age structure between the two groups. The significant difference in the mean age of treated pups at the third capture event between 2019 and 2020/21 is likely due to more targeted sampling of younger pups, in order to maximise treatment effectiveness (Lindsay et al. unpublished).
The unique 18-month breeding cycle of the Australian sea lion (Higgins and Gass, 1993; Gales et al., 1997) means that the 2019 breeding season at Seal Bay occurred during the austral winter, whilst 2020/21 began during summer. As such, it was expected that seasonal and environmental variations could influence the gut microbiota of Australian sea lion pups. There were differences in the relative abundance of the more abundant phyla, Fusobacteria, Firmicutes, and Actinobacteria, and in two bacterial families that were present in both seasons, Fusobacteriaceae and Clostridiaceae. Within the 2019 and 2020/21 breeding seasons, there were no significant changes in the relative abundance of bacterial phyla or families across captures. However, qualitative changes in microbial community composition were observed; in 2019, the relative abundance of Fusobacteria remained relatively stable across all captures in both treatment groups, whilst the abundance of Firmicutes steadily decreased, and Actinobacteria and Proteobacteria increased. The relative abundance of Fusobacteria decreased more markedly in pups sampled during 2020/21 and was accompanied by a decrease in Firmicutes and an increase in Proteobacteria. This was also reflected at the family level, with the relative abundance of Fusobacteriaceae, a family belonging to the Fusobacteria phylum, decreasing from capture 1 to capture 4 in both treatment groups. Despite the significant differences between breeding seasons, the gut microbiota of both untreated (control) and treated Australian sea lion pups was dominated by the same bacteria phyla in 2019 and 2020/21, suggesting that the functional capacity of the gut microbial community is similar across treatment groups and breeding seasons. When comparing richness and diversity of the gut microbiota between seasons, there was again a significant difference; the richness of the gut microbiota was higher in Australian sea lion pups sampled in 2019, whilst diversity was, on average, higher in the 2020/21 cohort. The richness and diversity of the gut microbiota was significantly different in the treated group when compared between 2019 and 2020/21, and only richness differed significantly in the untreated (control) groups between seasons. The significant differences observed between seasons highlights the influence of external environmental conditions on the richness, diversity, and composition of the gut microbiota in Australian sea lion pups. Further investigations are required to determine the specific environmental characteristics contributing to these differences.
Host factors including pup weight, standard length, sex, and haematological parameters were also investigated to elucidate their relationship with treatment and the gut microbiota. In pups from both treatment groups sampled in the 2019 and 2020/21 breeding seasons, weight, standard length, and sex of the pup did not significantly influence the gut microbial composition. In sexually dimorphic pinniped species, differences in relative abundance of bacterial phyla and the relative abundance of ASVs or OTUs have been observed between male and females (Nelson et al., 2013; Stoffel et al., 2020). In adult southern elephant seals, differences between sexes were attributed to differences in diet rather than body mass (Nelson et al., 2013). When investigating the contribution of sex to the gut microbiome in weaned southern elephant seal pups (1–3 months old), Stoffel et al. (2020) found differences between sexes even though the male and female pups were indistinguishable, suggesting that some gut microbes are sex-specific and necessary for future adult feeding strategies. Whilst Australian sea lions are a sexually dimorphic species, Australian sea lion pups sampled as part of this study were a maximum of 4–6 months old and completely dependent upon their mothers for nutrition. Furthermore, Australian sea lion pups are not weaned until 18 months of age (Kirkwood and Goldsworthy, 2013). As a result, sampled pups were likely too young for any sex-associated differences in microbial composition to occur. The haematological parameters of Australian sea lion pups can be used as indicators of overall health and improvement in total leukocyte counts have previously been seen with ivermectin treatment (Marcus et al., 2015a,b; Lindsay et al., 2021). However, there was no significant association between any haematological parameter and alpha diversity, beta diversity, or the relative abundance of any bacterial phyla or family.
Topical ivermectin treatment of Australian sea lion pups has been identified as a potential management strategy to improve the health of free-ranging pups in a declining population (Lindsay et al., 2021), however, unexpected consequences of treatment need to be explored to ensure deleterious outcomes do not ensue. Characterisation of the gut microbiota of free-ranging untreated (control) and treated Australian sea lion pups revealed that topical ivermectin treatment and elimination of U. sanguinis infection did not alter the composition of the gut microbial community. Despite some minor differences, the absence of statistically significant alterations to the composition of the gut microbial community, together with the known benefits of ivermectin treatment for Australian sea lion pup health and growth, suggest that topical ivermectin could be an effective management strategy to mitigate the impact of endemic hookworm disease on population growth in this species.
The data presented in this study are deposited in the Dryad repository, https://doi.org/10.5061/dryad.ngf1vhhxh. Raw sequences are deposited in the European Nucleotide Archive, accession number PRJEB56630.
The animal study was reviewed and approved by the Animal Ethics Committee at the University of Sydney.
MF, RG, and MP contributed to the conceptualization and design of the study. MF and RG collected all samples from pinniped pups. MF completed laboratory analysis of samples and data analysis and drafted the manuscript. All authors participated in revising the manuscript, contributed to the manuscript, and approved the submitted version.
Collection and analysis of samples, including sequencing, was funded by the Ecological Society of Australia through the Holsworth Wildlife Research Endowment. Field work was supported by funds provided by the Hermon Slade Foundation (grant no. HSF 16-03) and Department for Environment and Water, South Australia. The publication costs were funded by the G M Mitchell Bequest, Sydney School of Veterinary Science.
We thank the staff at Seal Bay Conservation Park, Kangaroo Island, Department of Environment and Water (DEW), South Australia, particularly Melanie Stonnill, Melanie Stephens, Alana Binns, and Tanya Rosewarne for field assistance and logistical support; Simon Goldsworthy and the South Australian Research and Development Institute (SARDI) for field assistance. We would also like to thank Scott Lindsay, Shannon Taylor, Matthew Gray, and our many volunteer field assistants for assistance in sample collection and Janet Simpson for logistical support. Differential white cell count and hookworm burden was provided by Scott Lindsay.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1048013/full#supplementary-material
Aivelo, T., and Norberg, A. (2018). Parasite–microbiota interactions potentially affect intestinal communities in wild mammals. J. Anim. Ecol. 87, 438–447. doi: 10.1111/1365-2656.12708
Anderson, R. M., and Schad, G. A. (1985). Hookworm burdens and faecal egg counts: an analysis of the biological basis of variation. Trans. R. Soc. Trop. Med. Hyg. 79, 812–825. doi: 10.1016/0035-9203(85)90128-2
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-model effects using lme4. J. Stat. Softw. 1, 1–48. doi: 10.18637/jss.v067.i01
Bäumler, A. J., and Sperandio, V. (2016). Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93. doi: 10.1038/nature18849
Bik, E. M., Costello, E. K., Switzer, A. D., Callahan, B. J., Holmes, S. P., Wells, R. S., et al. (2016). Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 7:10516. doi: 10.1038/ncomms10516
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bradshaw, C. J. A., Davis, L. S., Lalas, C., and Harcourt, R. G. (2000). Geographic and temporal variation in the condition of pups of the New Zealand fur seal (Arctocephalus forsteri): evidence for density dependence and differences in the marine environment. J. Zool. 252, 41–51. doi: 10.1111/j.1469-7998.2000.tb00818.x
Brestoff, J. R., and Artis, D. (2013). Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 14, 676–684. doi: 10.1038/ni.2640
Byard, R. W., and Machado, A. (2019). Characteristic “neck collar” injuries in Australian sea lions (Neophoca cinerea) caused by marine debris. Forensic Sci. Med. Pathol. 15, 631–634. doi: 10.1007/s12024-018-0060-1
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Castinel, A., Duignan, P. J., Pomroy, W. E., López-Villalobos, N., Gibbs, N. J., Chilvers, B. L., et al. (2007). Neonatal mortality in New Zealand sea lions (Phocarctos hookeri) at Sandy Bay, Enderby Island, Auckland Islands from 1998 to 2005. J. Wildl. Dis. 43, 461–474. doi: 10.7589/0090-3558-43.3.461
Cebra, J. J. (1999). Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69, 1046–1051. doi: 10.1093/ajcn/69.5.1046s
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Clemente, J. C., Ursell, L. K., Parfrey, L. W., and Knight, R. (2012). The impact of the gut microbiota on human health: an integrative view. Cells 148, 1258–1270. doi: 10.1016/j.cell.2012.01.035
Cooper, P., Walker, A. W., Reyes, J., Chico, M., Salter, S. J., Vaca, M., et al. (2013). Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS One 8:e76573. doi: 10.1371/journal.pone.0076573
Cortés, A., Peachey, L. E., Jenkins, T. P., Scotti, R., and Cantacessi, C. (2019). Helminths and microbes within the vertebrate gut – not all studies are created equal. Parasitology 146, 1371–1378. doi: 10.1017/S003118201900088X
DeLong, R. L., Orr, A. J., Jenkinson, R. S., and Lyons, E. T. (2009). Treatment of northern fur seal (Callorhinus ursinus) pups with ivermectin reduces hookworm-induced mortality. Mar. Mamm. Sci. 25, 944–948. doi: 10.1111/j.1748-7692.2008.00274.x
Delport, T. C., Power, M. L., Harcourt, R. G., Webster, K. N., and Tetu, S. G. (2016). Colony location and captivity influence the gut microbial community composition of the Australian sea lion (Neophoca cinerea). Appl. Environ. Microbiol. 82, 3440–3449. doi: 10.1128/AEM.00192-16
Ebert, D., and Fields, P. D. (2020). Host–parasite co-evolution and its genomic signature. Nat. Rev. Genet. 21, 754–768. doi: 10.1038/s41576-020-0269-1
Fulham, M., McDougall, F., Power, M., McIntosh, R. R., and Gray, R. (2022). Carriage of antibiotic resistant bacteria in endangered and declining Australian pinniped pups. PLoS One 17:e0258978. doi: 10.1371/journal.pone.0258978
Fulham, M., Power, M., and Gray, R. (2018). Comparative ecology of Escherichia coli in endangered Australian sea lion (Neophoca cinerea) pups. Infect. Genet. Evol. 62, 262–269. doi: 10.1016/j.meegid.2018.05.002
Fulham, M., Power, M., and Gray, R. (2020). Diversity and distribution of Escherichia coli in three species of free-ranging Australian pinniped pups. Front. Mar. Sci. 7:571171. doi: 10.3389/fmars.2020.571171
Gales, N. J., Shaughnessy, P. D., and Dennis, T. E. (1994). Distribution, abundance and breeding cycle of the Australian sea lion Neophoca cinerea (Mammalia: Pinnipedia). J. Zool. 234, 353–370. doi: 10.1111/j.1469-7998.1994.tb04853.x
Gales, N. J., Williamson, P., Higgins, L. V., Blackberry, M. A., and James, I. (1997). Evidence for a prolonged post implantation period in the Australian sea lion (Neophoca cinerea). J. Reprod. Fertil. 111, 159–163. doi: 10.1530/jrf.0.1110159
Gaulke, C. A., Martins, M. L., Watral, V. G., Humphreys, I. R., Spagnoli, S. T., Kent, M. L., et al. (2019). A longitudinal assessment of host-microbe-parasite interactions resolves the zebrafish gut microbiome’s link to Pseudocapillaria tomentosa infection and pathology. Microbiome 7:10. doi: 10.1186/s40168-019-0622-9
Giacomin, P., Croese, J., Krause, L., Loukas, A., and Cantacessi, C. (2015). Suppression of inflammation by helminths: a role for the gut microbiota? Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 1–6. doi: 10.1098/RSTB.2014.0296
Glöckner, F. O., Yilmaz, P., Quast, C., Gerken, J., Beccati, A., Ciuprina, A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
Goldsworthy, S. D., Gales, N. J., and Chilvers, B. L. (2015). Neophoca cinerea. IUCN Red List Threat. Species 2015 e.T14549A45228341. Gland: IUCN.
Goldsworthy, S. D., Kennedy, C., and Lashmar, K. (2014). Monitoring the status, trends in abundance and key demographic rates of the Australian sea lion population at Seal Bay - Kangaroo Island. SARDI Publication No. F2014/000787-1 SARDI Research Report Series No. 815.
Goldsworthy, S. D., Lowther, A. D., Shaughnessy, P. D., McIntosh, R. R., and Page, B. (2007). Assessment of pup production and the maternal investment strategies of the Australian sea lion Neophoca cinerea at dangerous reef in the 2006-07 breeding season. Adelaide, Australia: SARDI Aquatic Sciences Publication.
Goldsworthy, S. D., Shaughnessy, P. D., Mackay, A. I., Bailleul, F., Holman, D., Lowther, A. D., et al. (2021). Assessment of the status and trends in abundance of a coastal pinniped, the Australian sea lion Neophoca cinerea. Endanger. Species Res. 44, 421–437. doi: 10.3354/ESR01118
Goldsworthy, S. D., Shaughnessy, P. D., Smart, J., Mackay, A. I., Bailleul, F., Reinhold, S.-L., et al. (2019). Monitoring of Seal Bay and other populations on Kangaroo Island: 2017/2018. Report to the Department for Environment and Water. SARDI Publication No. F2014/000322-5 SARDI Research Report Series No. 1018.
Guinane, C. M., and Cotter, P. D. (2013). Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 6, 295–308. doi: 10.1177/1756283X13482996
Guiver, E., Galan, M., Lippens, C., Bellenger, J., Faivre, B., and Sorci, G. (2022). Increasing helminth infection burden depauperates the diversity of the gut microbiota and alters its composition in mice. Curr. Res. Parasitol. Vector Borne Dis. 2:100082. doi: 10.1016/j.crpvbd.2022.100082
Hall, M., and Beiko, R. G. (2018). “16S rRNA gene analysis with QIIME2” Meth. Mol. Biol. 1849, 113–129. doi: 10.1007/978-1-4939-8728-3_8
Hamer, D. J., Goldsworthy, S. D., Costa, D. P., Folwer, S., Page, M., and Sumner, M. D. (2013). The endangered Australian sea lion extensively overlaps with and regularly becomes by-catch in demersal shark gill-nets in South Australian shelf waters. Biol. Conserv. 157, 386–400. doi: 10.1016/j.biocon.2012.07.010
He, F., Zhai, J., Zhang, L., Liu, D., Ma, Y., Rong, K., et al. (2018). Variations in gut microbiota and fecal metabolic phenotype associated with Fenbendazole and Ivermectin tablets by 16S rRNA gene sequencing and LC/MS-based metabolomics in Amur tiger. Biochem. Biophys. Res. Commun. 499, 447–453. doi: 10.1016/j.bbrc.2018.03.158
Higgins, L. V., and Gass, L. (1993). Birth to weaning: parturition, duration of lactation, and attendance cycles of Australian sea lions (Neophoca cinerea). Can. J. Zool. 71, 2047–2055. doi: 10.1139/z93-290
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Hooper, L. V., Midtvedt, T., and Gordon, J. I. (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307. doi: 10.1146/annurev.nutr.22.011602.092259
Jenkins, T. P., Formenti, F., Castro, C., Piubelli, C., Perandin, F., Buonfrate, D., et al. (2018). A comprehensive analysis of the faecal microbiome and metabolome of Strongyloides stercoralis infected volunteers from a non-endemic area. Sci. Rep. 8:15651. doi: 10.1038/s41598-018-33937-3
Johansson, M. A., Saghafian-Hedengren, S., Haileselassie, Y., Roos, S., Troye-Blomberg, M., Nilsson, C., et al. (2012). Early-life gut bacteria associate with IL-4−, IL-10− and IFN-γ production at two years of age. PLoS One 7:e49315. doi: 10.1371/journal.pone.0049315
Kirkwood, Roger., and Goldsworthy, Simon. (2013). Fur Seals and Sea Lions. Collingwood, VIC: CSIRO Publishing.
Lane, D. (1991). “Nucleic acid techniques in bacterial systematics” in Nucleic Acid Techniques in Bacterial Systematics. ed. E. Stackebrandt (Hoboken, NJ: John Wiley and Sons)
Lee, C. Y., Peralta-Sánchez, J. M., Martínez-Bueno, M., Møller, A. P., Rabelo-Ruiz, M., Zamora-Muñoz, C., et al. (2020). The gut microbiota of brood parasite and host nestlings reared within the same environment: disentangling genetic and environmental effects. ISME J. 14, 2691–2702. doi: 10.1038/s41396-020-0719-y
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008a). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., and Gordon, J. I. (2008b). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Li, R. W., Wu, S., Li, W., Navarro, K., Couch, R. D., Hill, D., et al. (2012). Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect. Immun. 80, 2150–2157. doi: 10.1128/IAI.00141-12
Lindsay, S. A., Caraguel, C. G. B., and Gray, R. (2021). Topical ivermectin is a highly effective seal ‘spot-on’: a randomised trial of hookworm and lice treatment in the endangered Australian sea lion (Neophoca cinerea). Int. J. Parasitol. Parasites Wildl. 16, 275–284. doi: 10.1016/j.ijppaw.2021.11.002
Ling, J. K. (1999). Exploitation of fur seals and sea lions from Australian, New Zealand and adjacent subantarctic islands during the eighteenth, nineteenth and twentieth centuries. Aust. Zool. 31, 323–350. doi: 10.7882/AZ.1999.036
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Lyons, E. T., DeLong, R. L., Nadler, S. A., Laake, J. L., Orr, A. J., DeLong, B. L., et al. (2011). Investigations of peritoneal and intestinal infections of adult hookworms (Uncinaria spp.) in northern fur seal (Callorhinus ursinus) and California sea lion (Zalophus californianus) pups on San Miguel Island, California (2003). Parasitol. Res. 109, 581–589. doi: 10.1007/s00436-011-2289-4
Mandal, S., van Treuren, W., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Marcus, A. D., Higgins, D. P., and Gray, R. (2014). Epidemiology of hookworm (Uncinaria sanguinis) infection in free-ranging Australian sea lion (Neophoca cinerea) pups. Parasitol. Res. 113, 3341–3353. doi: 10.1007/s00436-014-3997-3
Marcus, A. D., Higgins, D. P., and Gray, R. (2015a). Health assessment of free-ranging endangered Australian sea lion (Neophoca cinerea) pups: effect of haematophagous parasites on haematological parameters. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 184, 132–143. doi: 10.1016/j.cbpa.2015.02.017
Marcus, A. D., Higgins, D. P., and Gray, R. (2015b). Ivermectin treatment of free-ranging endangered Australian sea lion (Neophoca cinerea) pups: effect on hookworm and lice infection status, haematological parameters, growth, and survival. Parasitol. Res. 114, 2743–2755. doi: 10.1007/s00436-015-4481-4
Margaret, M.-N., Michael, G. H., Thomos, G. B. C., Hannah, V. C., Tomislav, D.-L., Angela, E. D., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Martin, I., Djuardi, Y., Sartono, E., Rosa, B. A., Supali, T., Mitreva, M., et al. (2018). Dynamic changes in human-gut microbiome in relation to a placebo-controlled anthelminthic trial in Indonesia. PLoS Negl. Trop. Dis. 12:e0006620. doi: 10.1371/journal.pntd.0006620
Martin, I., Kaisar, M. M. M., Wiria, A. E., Hamid, F., Djuardi, Y., Sartono, E., et al. (2019). The effect of gut microbiome composition on human immune responses: an exploration of interference by helminth infections. Front. Genet. 10:1028. doi: 10.3389/fgene.2019.01028
McIntosh, R. R., and Kennedy, C. W. (2013). Morphology, sex ratio and cause of death in Australian sea lion (Neophoca cinerea) pups. Aust. Mammal 35, 93–100. doi: 10.1071/AM12037
McMurdie, P. J., and Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Medeiros, A. W., Giongo, A., Valdez, F. P., Blaese de Amorin, D., Tavares, M., d’Azevedo, P. A., et al. (2016). Characterization of the faecal bacterial community of wild young South American (Arctocephalus australis) and Subantarctic fur seals (Arctocephalus tropicalis). FEMS Microbiol. Ecol. 92, 1–8. doi: 10.1093/femsec/fiw029
Michael, S. A., Hayman, D. T. S., Gray, R., and Roe, W. D. (2021). Risk factors for New Zealand sea lion (Phocarctos hookeri) pup mortality: Ivermectin improves survival for conservation management. Front. Mar. Sci. 8:680678. doi: 10.3389/fmars.2021.680678
Michael, S. A., Hayman, D. T. S., Gray, R., Zhang, J., Rogers, L., and Roe, W. D. (2019). Pup mortality in New Zealand sea lions (Phocarctos hookeri) at Enderby Island, Auckland Islands, 2013-18. PLoS One 14:e0225461. doi: 10.1371/journal.pone.0225461
Moustafa, M. A. M., Chel, H. M., Thu, M. J., Bawm, S., Htun, L. L., Win, M. M., et al. (2021). Anthropogenic interferences lead to gut microbiome dysbiosis in Asian elephants and may alter adaptation processes to surrounding environments. Sci. Rep. 11:741. doi: 10.1038/s41598-020-80537-1
Nelson, T. M., Rogers, T. L., Carlini, A. R., and Brown, M. (2013). Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145. doi: 10.1111/1462-2920.12022
Oksanen, J., Simpson, G. L., Guillaume Blanchet, F., Kindt, R., Legendre, P., Minchin, P. R., et al. (2022). vegan: Community Ecology Package. R package version 2.6-2.
Pacheco-Sandoval, A., Schramm, Y., Heckel, G., Brassea-Pérez, E., Martínez-Porchas, M., and Lago-Lestón, A. (2019). The Pacific harbor seal gut microbiota in Mexico: its relationship with diet and functional inferences. PLoS One 14:e0221770. doi: 10.1371/journal.pone.0221770
Page, B., McKenzie, J., McIntosh, R., Baylis, A., Morrissey, A., Calvert, N., et al. (2004). Entanglement of Australian sea lions and New Zealand fur seals in lost fishing gear and other marine debris before and after government and industry attempts to reduce the problem. Mar. Pollut. Bull. 49, 33–42. doi: 10.1016/j.marpolbul.2004.01.006
Peachey, L. E., Molena, R. A., Jenkins, T. P., di Cesare, A., Traversa, D., Hodgkinson, J. E., et al. (2018). The relationships between faecal egg counts and gut microbial composition in UK thoroughbreds infected by cyathostomins. Int. J. Parasitol. 48, 403–412. doi: 10.1016/j.ijpara.2017.11.003
Preheim, S. P., Perrotta, A. R., Martin-Platero, A. M., Gupta, A., and Alm, E. J. (2013). Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl. Environ. Microbiol. 79, 6593–6603. doi: 10.1128/AEM.00342-13
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramanan, D., Bowcutt, R., Lee, S. C., Tang, M. S., Kurtz, Z. D., Ding, Y., et al. (2016). Helminth infection promotes colonization resistance via type 2 immunity. Science 352, 608–612. doi: 10.1126/science.aaf3229
Rausch, S., Held, J., Fischer, A., Heimesaat, M. M., Kühl, A. A., Bereswill, S., et al. (2013). Small intestinal nematode infection of mice is associated with increased enterobacterial loads alongside the intestinal tract. PLoS One 8:e74026. doi: 10.1371/journal.pone.0074026
Reynolds, L. A., Finlay, B. B., and Maizels, R. M. (2015). Cohabitation in the intestine: interactions among helminth parasites, bacterial microbiota, and host immunity. J. Immunol. 195, 4059–4066. doi: 10.4049/jimmunol.1501432
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rosa, B. A., Supali, T., Gankpala, L., Djuardi, Y., Sartono, E., Zhou, Y., et al. (2018). Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 6:33. doi: 10.1186/s40168-018-0416-5
Schroeder, B. O., and Bäckhed, F. (2016). Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089. doi: 10.1038/nm.4185
Seguel, M., Muñoz, F., Navarrete, M. J., Paredes, E., Howerth, E., and Gottdenker, N. (2016). Hookworm infection in South American fur seal (Arctocephalus australis) pups: pathology and factors associated with host tissue damage and mortality. Vet. Pathol. 54, 288–297. doi: 10.1177/0300985816677151
Seguel, M., Pavés, H., Paredes, E., and Schlatter, R. (2013). Causes of mortality in south American fur seal pups (Arctophoca australis gracilis) at Guafo Island, southern Chile (2004–2008). Mar. Mamm. Sci. 29, 36–47. doi: 10.1111/j.1748-7692.2011.00534.x
Shannon, C. E. (1948). A mathematical theory of communication. Bell. Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Smith, S. C., Chalker, A., Dewar, M. L., and Arnould, J. P. Y. (2013). Age-related differences revealed in Australian fur seal Arctocephalus pusillus doriferus gut microbiota. FEMS Microbiol. Ecol. 86, 246–255. doi: 10.1111/1574-6941.12157
Spraker, T. R., DeLong, R. L., Lyons, E. T., and Melin, S. R. (2007). Hookworm enteritis with bacteremia in California sea lion pups on San Miguel Island. J. Wildl. Dis. 43, 179–188. doi: 10.7589/0090-3558-43.2.179
Spratt, D. M., and Beveridge, I. (2019). Wildlife parasitology in Australia: past, present and future. Aust. J. Zool. 66, 286–305. doi: 10.1071/ZO19017
Stoffel, M. A., Acevedo-Whitehouse, K., Morales-Durán, N., Grosser, S., Chakarov, N., Krüger, O., et al. (2020). Early sexual dimorphism in the developing gut microbiome of northern elephant seals. Mol. Ecol. 29, 2109–2122. doi: 10.1111/mec.15385
Stokes, A. L., Terkildsen, M., Lindsay, S. A., Stonnill, M., Fulham, M., Taylor, S., et al. (2020). Growth Curves, Age Prediction and a Body Condition Index in Ivermectin-Treated Neophoca cinerea Pups. Sydney: Doctor of Veterinary Medicine Professionally Focused Project, The University of Sydney.
Taylor, S., Terkildsen, M., McQuilty, R., Lee, D., Wing-Simpson, A., and Gray, R. (2022). Non-essential heavy metals and protective effects of selenium against mercury toxicity in endangered Australian sea lion (Neophoca cinerea) pups with hookworm disease. Environ. Int. 107521, 1–14. doi: 10.1016/j.envint.2022.107521
Taylor, S., Terkildsen, M., Stevenson, G., de Araujo, J., Yu, C., Yates, A., et al. (2021). Per and polyfluoroalkyl substances (PFAS) at high concentrations in neonatal Australian pinnipeds. Sci. Total Environ. 786:147446. doi: 10.1016/j.scitotenv.2021.147446
Threatened Species Scientific Committee (2020). Conservation Advice Neophoca cinerea Australian Sea Lion. Canberra: Department of Agriculture, Water and the Environment.
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/BCJ20160510
Tian, J., Du, J., Han, J., Song, X., and Lu, Z. (2020). Age-related differences in gut microbial community composition of captive spotted seals (Phoca largha). Mar. Mamm. Sci. 36, 1231–1240. doi: 10.1111/mms.12728
Toro-Valdivieso, C., Toro, F., Stubbs, S., Castro-Nallar, E., and Blacklaws, B. (2021). Patterns of the fecal microbiota in the Juan Fernández fur seal (Arctocephalus philippii). Microbiol. Open 10:e1215. doi: 10.1002/mbo3.1215
Ursell, L. K., Metcalf, J. L., Parfrey, L. W., and Knight, R. (2012). Defining the human microbiome. Nutr. Rev. 70, S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x
Vlčková, K., Pafčo, B., Petrželková, K. J., Modrý, D., Todd, A., Yeoman, C. J., et al. (2018). Relationships between gastrointestinal parasite infections and the fecal microbiome in free-ranging western lowland gorillas. Front. Microbiol. 9:1202. doi: 10.3389/FMICB.2018.01202/FULL
Walk, S. T., Blum, A. M., Ewing, S. A.-S., Weinstock, J. V., and Young, V. B. (2010). Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm. Bowel Dis. 16, 1841–1849. doi: 10.1002/ibd.21299
Warnick, L. (1992). Daily variability of equine fecal strongyle egg counts. Cornell Vet. 82, 453–463.
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: microbiome, hookworm, ivermectin, pinniped, endangered
Citation: Fulham M, Power M and Gray R (2022) Gut microbiota of endangered Australian sea lion pups is unchanged by topical ivermectin treatment for endemic hookworm infection. Front. Microbiol. 13:1048013. doi: 10.3389/fmicb.2022.1048013
Received: 19 September 2022; Accepted: 02 December 2022;
Published: 19 December 2022.
Edited by:
Maria Siwek, University of Science and Technology (UTP), PolandReviewed by:
Mauricio Seguel, University of Georgia, United StatesCopyright © 2022 Fulham, Power and Gray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachael Gray, cmFjaGFlbC5ncmF5QHN5ZG5leS5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.