
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 January 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1045984
This article is part of the Research Topic Antimicrobial Peptides and their Druggability, Bio-safety, Stability, and Resistance View all 23 articles
 Ibeth Guevara-Lora1†
Ibeth Guevara-Lora1† Grazyna Bras2†
Grazyna Bras2† Magdalena Juszczak2
Magdalena Juszczak2 Justyna Karkowska-Kuleta2
Justyna Karkowska-Kuleta2 Andrzej Gorecki3
Andrzej Gorecki3 Marcela Manrique-Moreno4
Marcela Manrique-Moreno4 Jakub Dymek5
Jakub Dymek5 Elzbieta Pyza5
Elzbieta Pyza5 Andrzej Kozik1
Andrzej Kozik1 Maria Rapala-Kozik2*
Maria Rapala-Kozik2*The recent progressive increase in the incidence of invasive fungal infections, especially in immunocompromised patients, makes the search for new therapies crucial in the face of the growing drug resistance of prevalent nosocomial yeast strains. The latest research focuses on the active compounds of natural origin, inhibiting fungal growth, and preventing the formation of fungal biofilms. Antimicrobial peptides are currently the subject of numerous studies concerning effective antifungal therapy. In the present study, the antifungal properties of two synthetic peptides (ΔM3, ΔM4) derived from an insect antimicrobial peptide – cecropin D – were investigated. The fungicidal activity of both compounds was demonstrated against the yeast forms of Candida albicans, Candida tropicalis, and Candida parapsilosis, reaching a MFC99.9 in the micromolar range, while Candida glabrata showed greater resistance to these peptides. The scanning electron microscopy revealed a destabilization of the yeast cell walls upon treatment with both peptides; however, their effectiveness was strongly modified by the presence of salt or plasma in the yeast environment. The transition of C. albicans cells from yeast to filamentous form, as well as the formation of biofilms, was effectively reduced by ΔM4. Mature biofilm viability was inhibited by a higher concentration of this peptide and was accompanied by increased ROS production, activation of the GPX3 and SOD5 genes, and finally, increased membrane permeability. Furthermore, both peptides showed a synergistic effect with caspofungin in inhibiting the metabolic activity of C. albicans cells, and an additive effect was also observed for the mixtures of peptides with amphotericin B. The results indicate the possible potential of the tested peptides in the prevention and treatment of candidiasis.
The development of infections caused by Candida species consistently represents a serious threat, especially in individuals with different immunodeficiencies. Candidiases are generally treated with drugs such as azoles, polyenes, pyrimidine analogs, and echinocandins (Lee et al., 2021). However, when applied to Candida clinical isolates, the effectiveness of these drugs decreases significantly. The constant emergence of fungal strains resistant to classical treatment has prompted the search for new drugs for Candida infections, particularly among products of natural origin, including those from plants, insects, algae, etc. (Aldholmi et al., 2019; Guevara-Lora et al., 2020). These active substances include also antimicrobial peptides (AMPs), many of which have also shown antifungal activity (Bondaryk et al., 2017; Huan et al., 2020). A group of peptides that currently attracts particular attention for their potential role in the fight against candidiasis is cecropins (Brady et al., 2019). These peptides, identified currently in various insects, were originally isolated from the hemolymph of the Hyalophora cecropia moth (Boman et al., 1991) and are actually classified into several subfamilies – cecropins A, B, C, D, E and cecropin-like peptides (Romoli et al., 2017; Peng et al., 2019). These α-helical AMPs generally possess a strongly basic N-terminal region linked to a more hydrophobic C-terminal region through a segment rich in proline and glycine residues and their antimicrobial properties are attributed to their specific chemical structure (Boman et al., 1991; Sato and Feix, 2006). The presence of aromatic residues such as tryptophan and phenylalanine in the amphipathic N-terminal helix was shown to be essential for peptide-pathogen interactions, with subsequent permeabilization of the inner membrane of Gram-negative bacteria (Lee et al., 2015). The hydrophobic C-terminal region synergistically enhances this effect through the interaction with hydrophobic cell membrane. Although cecropins and cecropin-like peptides are more efficient in fighting Gram-negative microorganisms, the antimicrobial effects have also been shown for a broad range of Gram-positive bacteria (for a review, see Brady et al., 2019). Their activity against yeasts and other fungi was also proven, but the effects observed were not satisfactory. Most of the peptides demonstrating anticandidal activity belonged to the cecropin A or B subfamilies and required high concentration doses for effective action. Nevertheless, it was also shown that the use of analogs of these peptides could improve their antifungal properties (De Lucca et al., 2000; Ji et al., 2014), as certain modifications of the peptides, including the substitution of selected amino acid residues with tryptophan or D-amino acids, resulted in improved fungicidal activity due to the increased resistance to microbial proteolysis.
Promising antimicrobial properties were also previously reported for the cecropin D-like peptide (UniProtKB accession number: P85210) from the greater wax moth Galleria mellonella (Cytrynska et al., 2007). Modification of particular residues resulting in an increase of peptide charge and basic residues on the polar face of the molecule compared to a neutral wild-type peptide, enhanced the antibacterial activity of peptides derived from the cecropin D-like natural sequence. One such type of derivative was the synthetic cationic peptide ΔM2 with 39 amino acid residues and a charge of +9, which acts most effectively against Gram-negative bacteria (Oñate-Garzón et al., 2017). The necessity to continue the search for derivatives with enhanced antimicrobial properties, extended also to other bacterial and fungal pathogens, has led to further modifications. In this study, the anticandidal potential of two synthetic peptides specified as ΔM3 (NFFKRIRRAGKRIRKAIISA) and ΔM4 (NFFKRIRRAWKRIWKWIYSA), composed of 20 amino acids and derived from the N-terminal fragments of ΔM2, was analyzed. The ΔM3 peptide has a charge of +8 at physiological pH and was previously demonstrated to possess a high antimicrobial activity against Staphylococcus aureus and low hemolytic activity in human erythrocytes (Manrique-Moreno et al., 2021). The ΔM4 peptide with additional substitution of selected amino acids by three tryptophan and one tyrosine residues and a charge of +7 at physiological pH, presented anticancer properties against human melanoma cells (Santa-González et al., 2020). The helical-wheel projection of ΔM3 and ΔM4 and the predicted α-helical structure of the peptides generated with I-TASSER V5.1 software were presented in previous reports (Santa-González et al., 2020; Manrique-Moreno et al., 2021).
The main aim of this study was first to evaluate the fungicidal activity of ΔM3 and ΔM4 peptides against Candida species determining the minimal fungicidal concentration (MFC). Subsequently, the effects of different concentrations of peptides on Candida albicans hyphae, biofilm formation, and mature biofilm eradication were investigated. The mechanism of action of peptides was verified by examining the permeabilization of fungal cell membranes, pore formation, and occurrence of oxidative stress and apoptosis. Finally, a potential synergism of the tested peptides with conventional antifungal drugs was analyzed.
Peptides ΔM3 (NFFKRIRRAGKRIRKAIISA) and ΔM4 (NFFKRIRRAWKRIWKWIYSA), both synthesized by the solid-phase method, were purchased from GenScript (Piscataway, NJ, United States). Purity of the peptides was verified by analytical high performance liquid chromatography (HPLC) and MALDI-TOF mass spectrometry and was determined as >95% for both peptides.
Peptides at concentration of 1 mg/ml, diluted in 10 mM HEPES pH 7.0 or PBS, without or with 2,2,2-trifluoroethanol (TFE, Sigma-Aldrich, St Louis, MO, United States) at concentrations in the range of 5–25% (commonly used to stabilize secondary structure of polypeptides), and diluted in 10 mM HEPES pH 7.0 with 30 mM SDS (BioShop Canada Inc., Burlington, Canada) (to mimic anionic environment) were prepared. Peptides diluted in 10 mM HEPES pH 7.0 with NaCl at concentrations in the range of 50–200 mM were used to analyze the effect of salt ions on the peptide secondary structure. Circular dichroism (CD) measurements were conducted using a J-715 spectropolarimeter (Jasco, Tokyo, Japan) equipped with an F25 temperature control unit (Julabo, Seelbach, Germany) in quartz cuvettes of 0.01 cm path-length (Hellma, Jena, Germany) at 30°C. The spectra were recorded in the range of 195–250 nm with 1 nm data pitch, 20 nm/min scanning speed, 4 s integrating time, and 2 nm bandwidth and averaged over five acquisitions. All spectra were corrected for the effect from the buffer and all measurements were converted to molar residual ellipticity units. Secondary structure content was determined using JSSE program (Jasco) using CD spectra of α-chymotrypsin A, hemoglobin, lysozyme, myoglobin and ribonuclease A as protein references.
Candida albicans strain 3147 (ATCC® 10231™), Candida tropicalis strain T1 (ATCC® MYA-3404™), Candida parapsilosis strain CDC 317 (ATCC® MYA-4646™) and Candida glabrata strain CBS138 (ATCC® 2001™) were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, United States). Yeast cells (5 × 105/ml at the start) were routinely cultured in YPD medium (1% yeast extract, 2% soybean peptone and 2% glucose) (Sigma-Aldrich) for 18 h at 30°C with shaking (170 rpm) using an orbital rotary shaker MaxQ 6000 (ThermoFisher Scientific, Waltham, MA, United States). Before each experiment, fungal cells were washed three times with the appropriate buffer – phosphate buffered saline pH 7.4 (PBS; Biowest, Nuaillé, France) or 10 mM HEPES pH 7.0 with 5 mM glucose (HEPES/glucose) – and optical density at 600 nm (OD) was measured to estimate the number of cells. To determine the number of colony forming units (CFUs) per ml after treatment with peptide, fungal cells were cultured on solid YPD medium containing 1.5% agar for 24 h at 30°C. Due to the complex experimental setup of this study, a diagram showing the consecutive steps of the experiments is presented (Figure 1).
The analysis of fungicidal activity of ΔM3 and ΔM4 peptides was performed as previously described (Bochenska et al., 2016; van Dijck et al., 2018) with minor modifications. Briefly, 1 × 105 yeast cells of C. albicans were incubated in 100 μl of ΔM3 or ΔM4 solution at concentrations in the range of 0.4–100 μM in 10 mM HEPES pH 7.0, 10 mM HEPES pH 7.0 with 5 mM glucose, PBS or PBS with 50% of human plasma for 2 h at 30°C with shaking (110 rpm) in Eppendorf tubes. Additionally, cells from each strain were incubated in the buffer without peptides under the same conditions. After incubation, 10 μl of the cell suspensions undiluted and diluted (10 and 100-fold) with PBS or sterile water were plated to the YPD agar dishes for determination of CFUs. Similar experiments with yeast cells of C. parapsilosis, C. tropicalis or C. glabrata were performed in 10 mM HEPES pH 7.0 with 5 mM glucose with peptides in the same range of concentrations. The experiments were performed independently at least three times in triplicates. The minimal fungicidal concentrations – MFC90 and MFC99.9 that cause mortality of 90 or 99.9% of fungi, respectively, were determined.
Candida albicans yeast cells (1 × 106/ml) obtained after 16 h of growth in YPD medium were incubated for 2 h at 30°C in a solution of peptides ΔM3 (1.6 μM) and ΔM4 (3.1 μM) in HEPES/glucose. Cells incubated under the same conditions, but without peptides, served as an untreated control. The preparation of the cells for observation with scanning electron microscopy (SEM) was performed similarly to that described by Staniszewska et al. (2013) with minor modifications. Briefly, C. albicans cells were fixed overnight at 4°C in 2.5% glutaraldehyde (Sigma-Aldrich) on round coverslips (⌀13 mm, Bionovo, Legnica, Poland) precoated with 0.001% poly-L-lysine (Sigma-Aldrich), and then washed five times with 0.1 M phosphate buffer, pH 7.2. The further steps included incubation of specimens in 2% osmium tetroxide (Sigma-Aldrich) for 2 h at room temperature and dehydration in a series of ethanol solutions (Linegal Chemicals, Poland), starting from 10 min twice in 50 and 70%, followed by 5 min twice in 95% and 1 min twice in 100% ethanol solution. The samples were then dehydrated with acetone (Merck, Darmstadt, Germany) twice for 30 s, dried by the critical point method in liquid CO2 using an E300 critical point dryer (Quorum Technologies, East Sussex, United Kingdom) and coated with gold in a vacuum evaporator JEOL JFC-1100E (JEOL Ltd., Tokyo, Japan). Visualization was performed at an accelerating voltage of 20 kV with the Hitachi S-4700 scanning electron microscope (Hitachi, Tokyo, Japan) in the Laboratory of Scanning Electron Microscopy and Microanalysis, Institute of Geological Sciences, Jagiellonian University, Krakow, Poland.
Candida albicans yeast cells (1 × 105 per well) were suspended in 100 μl of RPMI 1640 medium (Biowest) containing peptides at concentrations in the range of 3.1–50 μM and 0.8–6.2 μM for ΔM3 and ΔM4, respectively, and placed in triplicates into the wells of 96-well black/clear flat bottom polystyrene high binding microplate (Corning Inc., Corning, NY, United States). Fungal cells were also suspended in medium without peptide as a control of the correct morphological transition. The incubation was carried out at 37°C in an atmosphere of 5% CO2 and 95% humidity for 90 min without shaking. Subsequently, fungal cells were stained with Calcofluor White (Sigma-Aldrich) at a final concentration of 1 μg/ml and Sytox Orange (ThermoFisher Scientific, Waltham, MA, United States) at a final concentration of 1 μM for 10 min in the dark. C. albicans cells were visualized using an Olympus IX73 microscope (Olympus, Tokyo, Japan) equipped with a Hamamatsu Orca Spark camera (Hamamatsu, Hamamatsu City, Japan) and a UPLXAPO60XO lens (Olympus), and the images were analyzed using Olympus CellSens Dimension 3.1 software. To determine the length of the hyphae, three images were taken in each well (40 × magnification) and for each image the length of the hyphae was measured for 20 cells.
The expression of genes characteristic for the filamentous form of C. albicans – encoding agglutinin-like protein 3 (ALS3), hyphally regulated cell wall protein 1 (HYR1) and hyphal wall protein 1 (HWP1) was analyzed after the isolation of total RNA from cells prepared as follows. Yeast cells (2 × 107) were seeded in plates (⌀10 cm) with 10 ml of RPMI medium in the presence of ΔM3 (at concentrations of 12.5 μM and 25 μM), ΔM4 (at concentrations of 1.2 μM and 2.1 μM) and without peptide. Two plates were prepared for each sample. After 90 min of incubation at 37°C in an atmosphere of 5% CO2 and 95% humidity, cells were harvested, washed with PBS and frozen in liquid nitrogen. The expression of genes associated with the generation of oxidative stress (encoding catalase – CAT1, glutathione peroxidase – GPX3, superoxide dismutase 5 – SOD5) was analyzed in mature biofilms. Briefly, yeast cells (5 × 107) were seeded in plates (⌀10 cm) with 10 ml of RPMI medium and incubated for 24 h at 37°C in an atmosphere of 5% CO2 and 95% humidity. Subsequently, the medium was changed to a fresh medium supplemented with 50 μM ΔM3, 6.2 μM ΔM4 or without peptide and the biofilms were incubated for 3 h at 37°C. Two plates were prepared for each sample. The concentration chosen for the analysis of gene expression resulted from the need to perform experiments on living cells in order to obtain undegraded mRNA. Cells were prepared for RNA isolation as described below.
Total RNA from cells was isolated with Tri Reagent (Merck Millipore, Burlington, MA, United States) according to a standard procedure preceded by cell homogenization in the Precellys® Evolution tissue homogenizer (Bertin Technologies SAS, Montigny-le-Bretonneux, France). cDNA was synthesized with the M-MLV Reverse Transcriptase kit (Promega, Madison, WI, United States) according to the manufacturer’s instructions. The expression of the selected genes was quantified with the QuantStudio™ 3 Real-Time PCR System (ThermoFisher Scientific). cDNA amplification was performed at an annealing temperature of 58°C using Kappa SYBR Green Master Mix (Merck Millipore) with specific primers (Table 1). Some primers were projected and verified with Primer3, OligoAnalyzer, and BLAST software. Relative gene expression was determined using the 2-ΔΔCt method normalized to the expression of the ACT1 housekeeping gene.
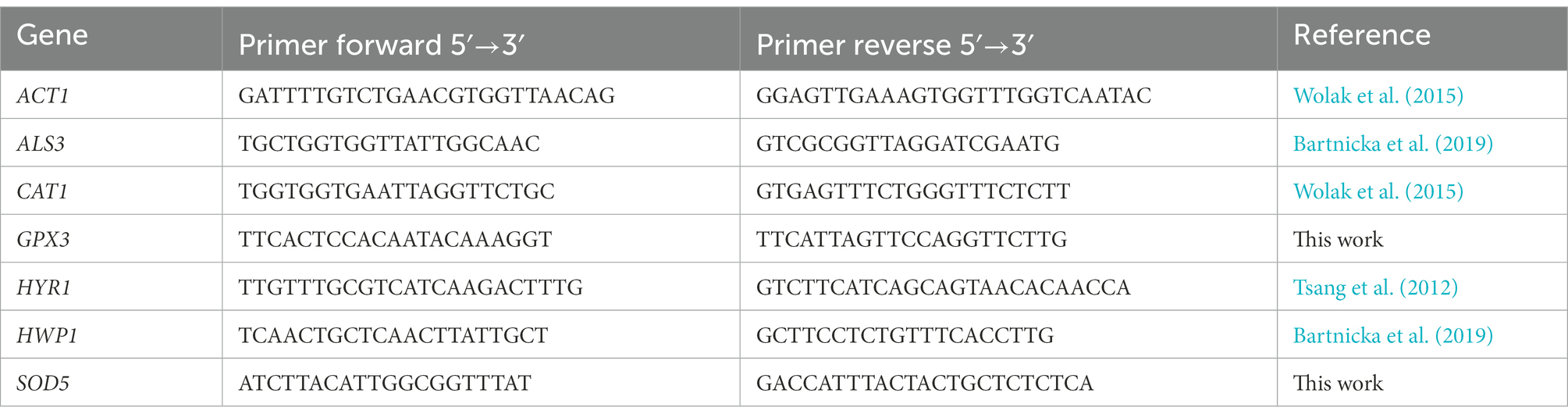
Table 1. The list of primers used for quantification of genes for: ACT1 – actin; ALS3 – agglutinin-like protein 3; CAT1 – catalase; GPX3 – glutathione peroxidase; HYR1 – hyphally regulated cell wall protein 1; HWP1 – hyphal wall protein 1; SOD5 – superoxide dismutase 5.
Candida albicans biofilms were formed generally in 96-well black/clear flat bottom polystyrene high binding microplate (Corning Inc.) after seeding the cells (1 × 105) in 100 μl of RPMI 1640, followed by incubation for 90 min at 37°C in an atmosphere of 5% CO2 and 95% humidity, without shaking to allow cell adhesion. Then non-adherent cells were removed by gentle washing with 200 μl of PBS and 100 μl of fresh RPMI 1640 were added to sessile C. albicans cells for further incubation under the same conditions for 24 h to allow formation of the biofilm.
To determine the impact of peptides on the formation of biofilm, adherent cells, gently washed as described above, were incubated with peptides at a concentration range of 0.4–100 μM in 100 μl of RPMI medium for 24 h at 37°C in an atmosphere of 5% CO2 and 95% humidity. After this time biofilms were washed, and the biofilm metabolic activity was assessed. To examine the effect of peptides on the eradication of the 24-h developed biofilm, a method previously described (Roscetto et al., 2018) was used. The 24-h biofilms (prepared as described above) were washed twice with 200 μl of PBS and then further incubated for 24 h at 37°C in an atmosphere of 5% CO2 and 95% humidity, with 100 μl of RPMI 1640 medium (control) or with ΔM3 or ΔM4 at a concentration ranging from 1.6 μM to 100 μM in the same medium. After incubation, the biofilms were washed twice with 200 μl of PBS and the metabolic activity of C. albicans was measured using the XTT reduction test. All experiments were carried out in triplicate.
To determine the viability of C. albicans, the 2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide (XTT) (ThermoFisher Scientific) reduction assay is used as a routine tool for quantitative measurement of fungal metabolic activity, growth and response to antifungal treatment (van Dijck et al., 2018). The tests were performed as described (Kulig et al., 2022) with some modifications. Briefly, 100 μl of RPMI 1640 medium without phenol red (Biowest) and 50 μl of XTT solution (1 mg/ml; Invitrogen, Waltham, MA, United States), freshly activated with phenazine methosulfate (5 μg/ml; Sigma-Aldrich) in the same medium, were added to C. albicans cells. After 40 min of incubation at 37°C in the dark, 100 μl of supernatants were transferred to the wells of a new 96-well microplate (Sarstedt, Nümbrecht, Germany). The absorbance of the formazan product was measured spectrophotometrically at 450 nm using a Synergy H1 microplate reader (BioTek Instruments, Winooski, VT, United States).
To determine the impact of peptides on the integrity of C. albicans membranes in the mature biofilm, two types of permeabilization assays were performed. In one of them, 24-h biofilms were washed with 200 μl of PBS and then incubated with peptides diluted in RPMI at concentrations in the range of 12.5 μM to 100 μM (ΔM3) and 3.1 μM to 12.5 μM (ΔM4) for 24 h at 37°C in an atmosphere of 5% CO2 and 95% humidity. After incubation, the biofilms were stained with Sytox™ Orange Nucleic Acid Stain (Invitrogen) at a final concentration of 1 μM in the dark for 5 min and visualized using an Olympus IX73 microscope. All images were taken and presented using the same parameters (exposure time, spectrum range), which enabled comparison of the fluorescence of the dye between different preparations. The photographs were taken in the form of the Z-stacs and subjected to the process of 3D deconvolution.
The second type of experiment was performed as described previously by Do Nascimento Dias et al. (2020). The 24-h biofilms were washed three times with 200 μl of PBS and then incubated in 100 μl of PBS for 2 h at 37°C. After this time, 100 μl solution of Rhodamine 6G (Sigma-Aldrich) in PBS (final dye concentration – 10 μM) was added to the wells and the biofilms were incubated for additional 30 min under the same conditions. To remove non-internalized dye, the wells were washed four times with PBS. Then 100 μl of peptide solutions were added to biofilms at concentrations in the range of 1.6 μM to 100 μM in PBS and incubated for 24 h at 37°C in an atmosphere of 5% CO2 and 95% humidity. The untreated biofilms served as controls. After incubation, the supernatants were transferred to the new black microplate and the fluorescence intensity was measured using a Synergy H1 microplate reader with excitation at 550 nm and emission at 580 nm.
Metacaspase activity was determined using the fluorescent method described previously (Shirazi and Kontoyiannis, 2015) with minor modifications. Briefly, the 24-h biofilm of C. albicans was formed in 96 well glass-like microplate (Cellvis, Sunnyvale, CA, United States) and incubated for 24 h at 37°C with ΔM3 or ΔM4 at concentrations in the range of 3.1–12.5 μM or 12.5–100 μM, respectively, diluted in 100 μl of RPMI 1640. After incubation, the biofilms were washed three times with 200 μl of PBS and stained with CaspACE™ FITC-VAD-FMK In Situ Marker (Promega, Madison, WI, United States) at a final concentration of 10 μM for 1 h at room temperature in the dark. The biofilms were washed four times with PBS and the activity of metacaspase was analyzed using the Olympus IX73 microscope.
The generation of intracellular reactive oxygen species (ROS) by C. albicans was determined using a method based on the measurement of the fluorescence of oxidized dihydrorhodamine 123 (DHR 123). The 24-h biofilms of C. albicans, formed as described above, were washed with PBS and then incubated with 100 μl of RPMI 1640 medium (untreated control), or solutions of ΔM3 (50 μM), ΔM4 (6.2 μM), or amphotericin B (2.7 μM; positive control) (Biowest) prepared in RPMI 1640 medium. For the analysis of ROS production, the concentration of peptides corresponding to the apoptotic effect observed with the activity of metacaspases was selected. The incubation was carried out for 3 h at 37°C. Biofilms were then washed with PBS and incubated with non-fluorescent DHR 123 (Invitrogen) at a final concentration of 10 μM for 30 min at 37°C. The fluorescence of oxidized DHR 123 (rhodamine 123) was measured with excitation at 488 nm and emission at 525 nm using a Synergy H1 microplate reader.
The 24-h biofilms of C. albicans were incubated with 100 μl of ΔM3 (50 μM) or ΔM4 (6.2 μM) and amphotericin B (2.7 μM; positive control) solutions in RPMI 1640 medium for 3 h at 37°C. After that time, biofilms were washed with 200 μl of PBS and incubated for 30 min at 37°C with MitoTracker Green TM dye (Invitrogen) at final concentration of 500 nM, used to detect the change in total mitochondrial mass, and MitoTracker Orange (500 nM), used to detect the change in mitochondrial activity. Mitochondria were imaged in the FITC and TRIC channel, using the Olympus IX73 microscope.
Accumulation of cytosolic Ca2+ was measured with the Fura-2 Calcium Flux Assay Kit (Abcam, Cambridge, Great Britain). The 24-h biofilms of C. albicans were incubated for 3 h with 100 μl of ΔM3 (25 μM and 50 μM) or ΔM4 (6.2 μM and 12.5 μM) and H2O2 (10 mM; positive control) solutions prepared in RPMI 1640. The concentration of peptides was chosen close to those for which the effect of apoptosis was obtained in experiments analyzing the activity of metacaspases. After incubation, biofilms were washed with Hanks Balanced Salt Solution (Biowest) with Pluronic® F127 Plus (Abcam). Then 100 μl of Fura-2 AM solution was added to the biofilms and incubated for 1 h at room temperature. The fluorescence was then measured using a Synergy H1 microplate reader with excitation at 340 nm and emission at 510 nm.
The following concentrations of peptides and antifungal drugs were used for the assay: ΔM3 0.4–50 μM, ΔM4 0.4–3.1 μM, amphotericin B 0.2–1.6 μM, fluconazole 0.2–1.6 μM and caspofungin 0.003–0.2 μM (the latter two from Sigma-Aldrich). The combinatory effects of each peptide and each conventional antimycotic drug were tested by the previously described checkerboard method (Guo et al., 2008; Do Nascimento Dias et al., 2020) with minor modification. C. albicans cells (3 × 102) were suspended in 100 μl of RPMI 1640 medium, containing peptide, drug, or their combination, in wells of Falcon® 96-well clear flat bottom TC-treated culture microplate (Corning). Fungal cells only in RPMI 1640 medium without peptides and drugs served as a control. Incubation was carried out for 24 h at 37°C in an atmosphere of 5% CO2 and 95% humidity without shaking. After this time, cell viability was measured using an XTT reduction assay. The analysis was performed in biological triplicate. For each combination, Fractional Inhibitory Concentration Index (FICI) was determined using the following formula:
FICI = FICa + FICb
Where FICa is the ratio of IC90 value for the compound a in combination to the IC90 value of the agent a alone; FICb is the ratio of IC90 value for compound b in combination to the IC90 value of the agent b alone.
The FICI ≤0.5 was interpreted as a synergistic effect, 0.5< FICI ≤1.0 as additive, 1.0< FICI ≤2.0 as indifferent and FICI >2.0 as antagonism.
The toxicity of the peptides was analyzed on primary human dermal fibroblasts (HDF) obtained from Cell Applications Inc. (San Diego, CA, United States). Cells (1 × 105), seeded in a microplate, were treated with peptides with a concentration in the range of 1.6–100 μM for 24 h in culture HDF basal medium (Cell Applications Inc.). After this time, cell viability was assessed with the Alamar blue test (ThermoFisher Scientific). Briefly, after medium removal, 100 μl of Alamar blue reagent were added and after 1 h of incubation the fluorescence was measured with excitation at 560 nm and emission at 590 nm with Synergy H1 microplate reader.
Statistical analysis was performed with the GraphPad Prism 8 software (GraphPad Software, San Diego, CA, United States). To evaluate the significance between groups, the t-test or one-way ANOVA with Dunnett’s multiple comparison post hoc test was performed. The results were considered statistically significant at value of p < 0.05: *p ≤ 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001 or not statistically significant (ns) for p > 0.05.
The interaction between antimicrobial peptides and the pathogen cell wall plays an important role in their fungicidal activity. The achieved effect depends, among others, on environmental factors such as pH, the presence of salt or the presence of substances that can change the hydrophobicity of the peptide (Kandasamy and Larson, 2006). To test the influence of the environment on the fungicidal activity of two cationic peptides, ΔM3 and ΔM4, the viability of C. albicans cells was analyzed in the presence of some commonly used media: HEPES buffer supplemented with 5 mM glucose (HEPES/glucose) and PBS without or with 50% human plasma. The peptides showed different activity against Candida cells depending on the conditions used (Figure 2). The ΔM4 peptide, which is more hydrophobic, was active in the presence of PBS or HEPES/glucose, while the ΔM3 peptide was active only in the HEPES/glucose. Both peptides, ΔM3 and ΔM4 incubated in HEPES/glucose had a strong fungicidal effect against C. albicans after 2 h of incubation at concentrations of 1.6 μM and 3.1 μM, respectively. Peptide ΔM4 in PBS caused a weaker effect compared to that in HEPES/glucose.
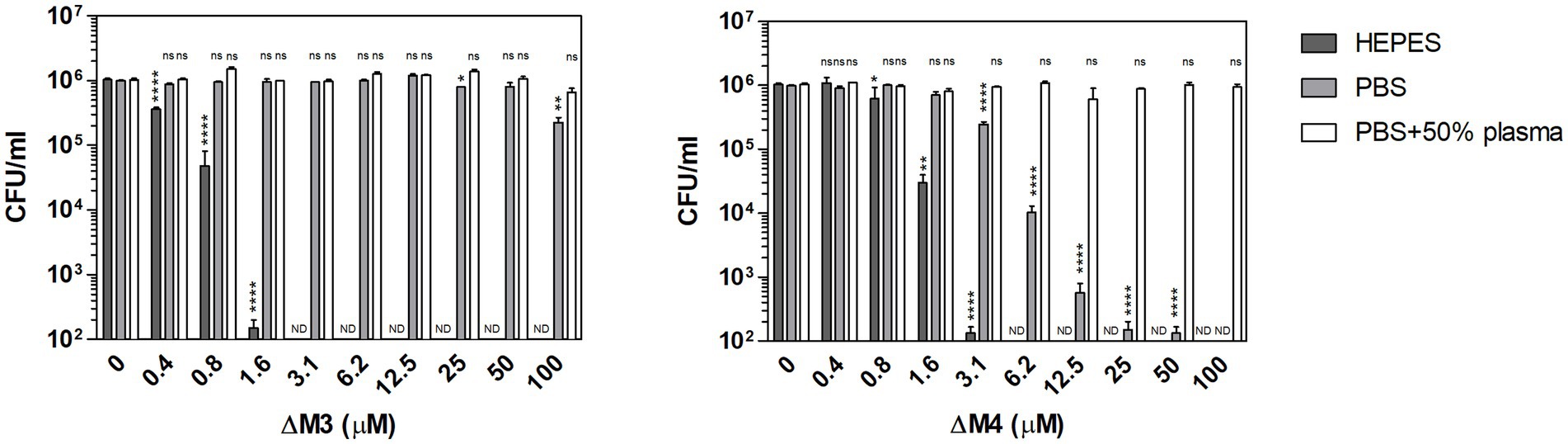
Figure 2. Fungicidal activity of ΔM3 and ΔM4 against C. albicans under various conditions. C. albicans yeast cells (105/100 μl) were incubated with ΔM3 and ΔM4 at concentrations in a range of 0.4–100 μM for 2 h in 10 mM HEPES, PBS, or PBS with 50% human plasma. The cell suspensions were plated on solid YPD medium. After 24 h of cultivation at 30°C, the number of CFUs was determined. The experiment was performed in triplicate. Graphs present mean values ± SD from representative results (n = 4). For statistical analysis one-way ANOVA with Dunnett’s multiple comparisons post hoc test was used (*p < 0.05, **p < 0.005, **** p < 0.0001, ns > 0.05 vs. untreated cells). A value of 102 CFU/ml on the y-axis was the limit of quantification. Values that were below this limit were marked ND.
To clarify the observed effect of both peptides on fungal cell viability, peptide structural studies were performed using circular dichroism measurements (Figure 3; Supplementary Figure S1). In this analysis, we used TFE (2,2,2-trifluoroethanol) as a cosolvent that promotes the stability of the peptide helical structure (Maisetta et al., 2008; Górecki et al., 2015). Nonetheless, upon contact with the fungal cell surface, hydrophobic interactions may also be primary forces driving the formation of the secondary peptide structure. As demonstrated in Figure 3, ΔM3 peptide in buffer solution, containing TFA at increasing concentrations, was less susceptible to adopt a helical structure compared to ΔM4, even in the presence of SDS, popularly used in such tests. The pH changes of the buffers used (pH 7.0 versus pH 7.4) did not exert a significant effect on the peptide helical structure. However, the structural preferences of both peptides differ significantly in the presence of salt, which is particularly relevant for experiments conducted in PBS and in RPMI 1640 medium. The presence of salt in a concentration-dependent manner enhanced the helicity of ΔM4 but had no effect on structural properties of ΔM3. These observations may explain the results obtained for the peptide treatment of fungal cells and their killing effects. Furthermore, supplementation with human plasma resulted in a loss of activity of both peptides, as no significant changes in fungal viability were observed within the range of peptide concentrations tested. Such reduction in the antibacterial activity of the peptides was often observed due to their nonspecific binding to the plasma proteinous components (Sovadinová et al., 2021).
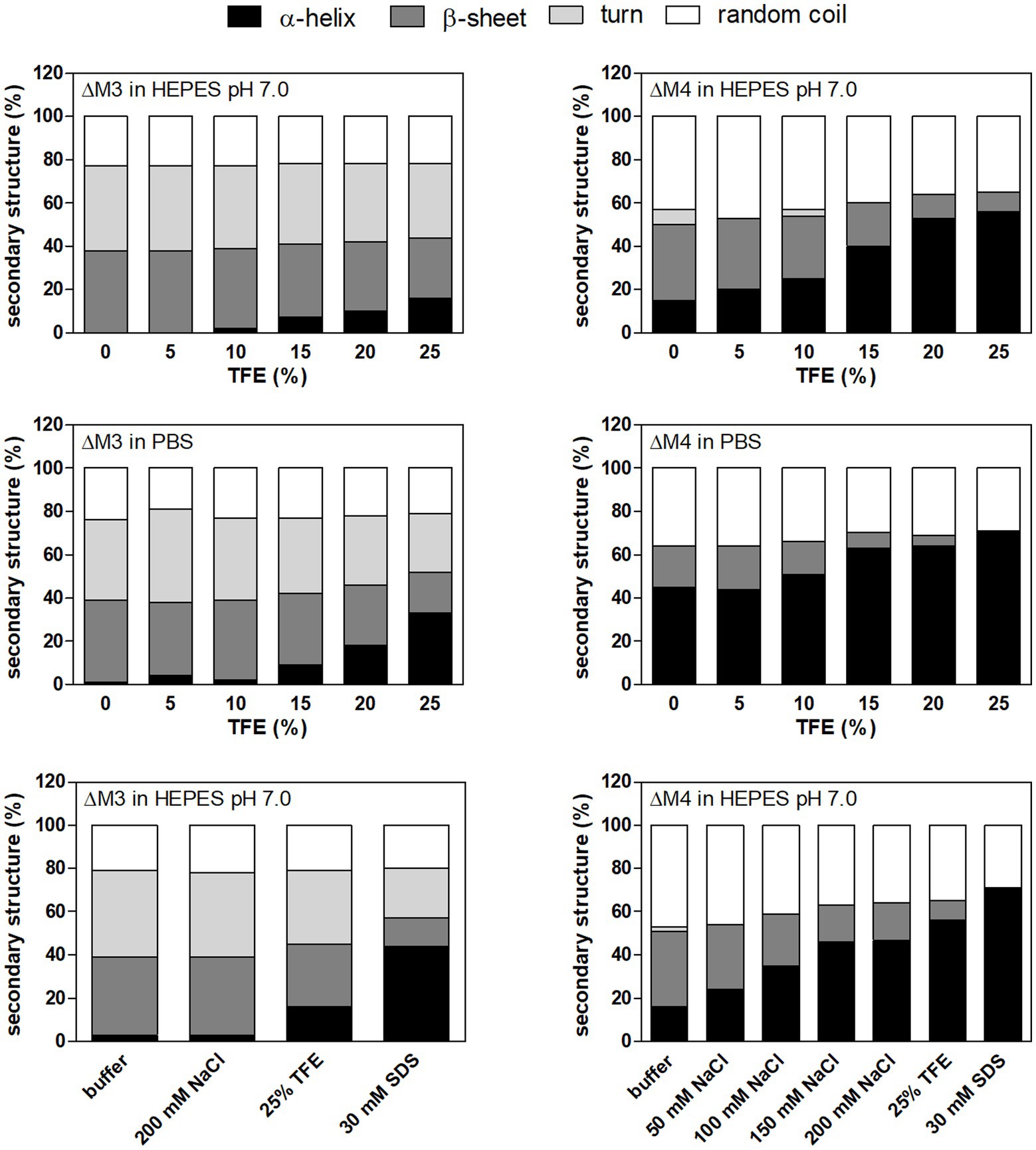
Figure 3. The secondary structure of peptides in different solutions analyzed by circular dichroism (CD) measurements. ΔM3 and ΔM4 peptides (both at a concentration of 1 mg/ml) were prepared in 10 mM HEPES pH 7.0 or PBS. Trifluoroethanol (TFE) at a final concentration in a range of 5–25% was used to enhance secondary structure stability. To check the effect of NaCl on the secondary structure of the peptides, samples were prepared in 10 mM HEPES pH 7.0 with NaCl (50–200 mM). Buffer supplemented with 30 mM SDS was used to mimic microbial membrane. CD spectra of peptides were measured in the range of 195–250 nm and averaged over five acquisitions. Secondary structure content was determined using JSSE program (Jasco) using CD spectra of α-chymotrypsin A, hemoglobin, lysozyme, myoglobin and ribonuclease A proteins as a reference.
Since the investigated peptides showed the best fungicidal properties against C. albicans in HEPES/glucose, the antifungal activity of these compounds against different Candida species was tested under the same conditions. Both peptides showed high efficiency against C. tropicalis and C. parapsilosis, reaching values of minimal fungicidal concentrations (MFCs) in the same range as those determined for C. albicans (Table 2). The values of MFC90 and MFC99.9 were of 0.8 μM and 1.6 μM, respectively, for ΔM3, and of 1.6 μM and 3.1 μM, respectively, for ΔM4. C. glabrata showed higher resistance to these peptides. For the ΔM3 peptide, the value of MFC99.9 was not determined within a range of peptide concentration used, while for ΔM4, which presented greater antifungal activity, MFC99.9 was estimated to be 50 μM. Nevertheless, these observations indicate that both peptides under these conditions may be potential antifungal agents against yeast-like forms of Candida species, especially C. albicans, C. parapsilosis and C. tropicalis.
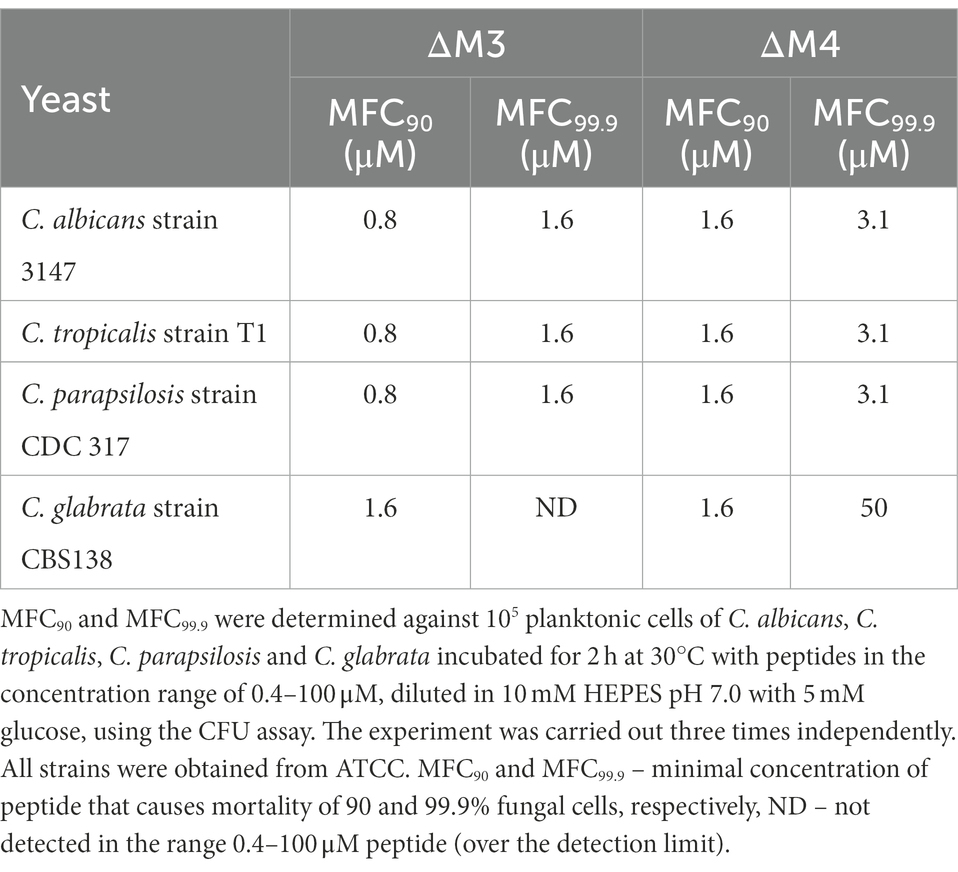
Table 2. Minimal fungicidal concentrations (MFC90 and MFC99.9) of ΔM3 and ΔM4 against yeast cells of Candida species.
The identified effects of ΔM3 and ΔM4 on C. albicans cell viability were also confirmed by SEM analysis of their cell surface structure. The yeast cells showed altered cell surface morphology after incubation with peptides (Figure 4). The untreated cells had a smooth surface and normal shape, whereas treatment with peptides at fungicidal concentration in HEPES/glucose buffer resulted in damage of the cell surface with leakage of intracellular content, marked with arrows on the figures. Similar observations were shown in a recent work, in which C. albicans cells were treated with the cecropin 4-derived peptide C18 (Sun et al., 2022). The cells treated with ΔM4 showed also a rough surface. These quite distinct changes in the cell surface under the influence of both tested peptides may indicate their differential interaction with the cell wall of C. albicans.
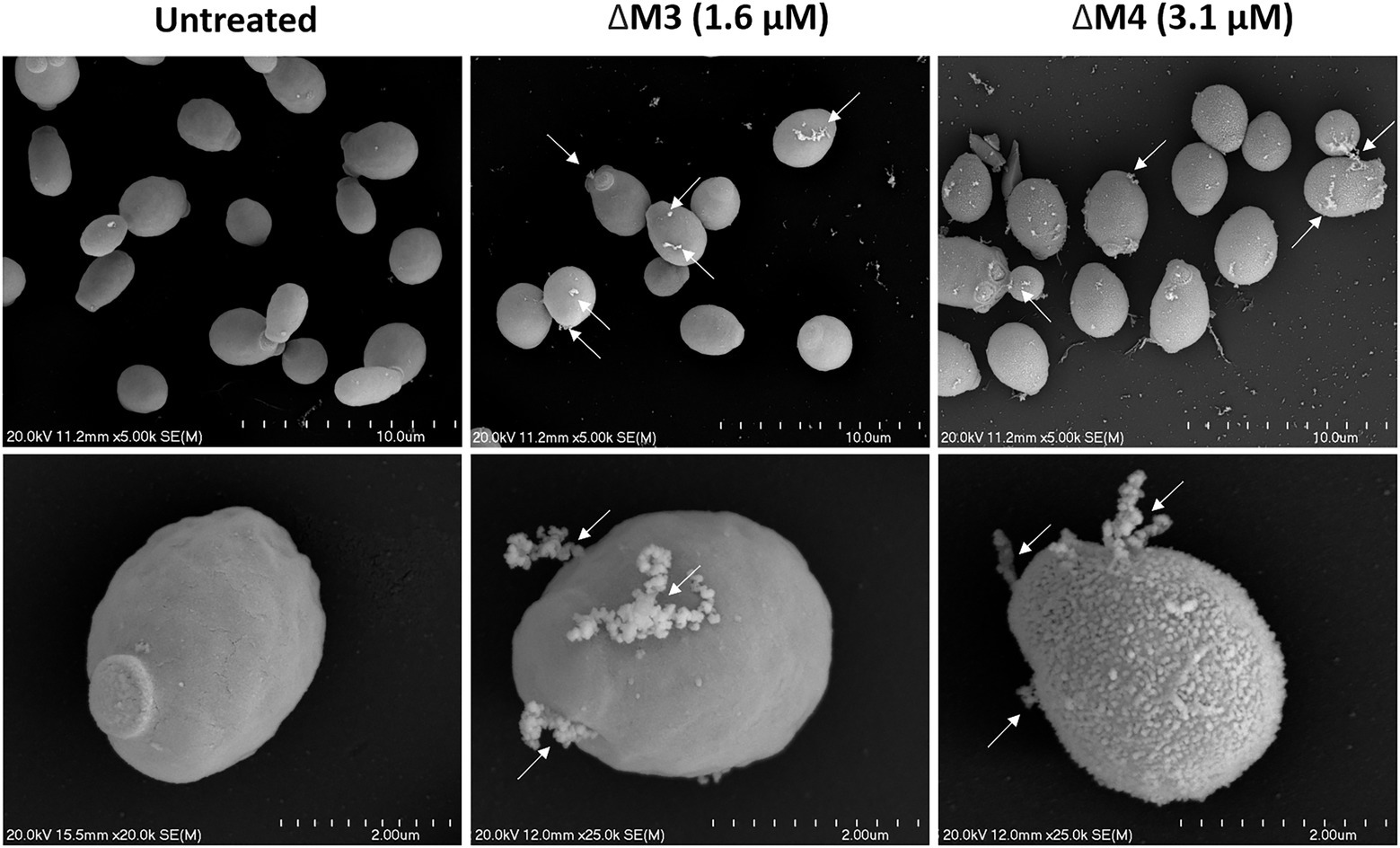
Figure 4. Scanning electron microscopy of the C. albicans cell surface after treatment with ΔM3 and ΔM4. C. albicans yeast cells (105/100 μl) were incubated with ΔM3 (1.6 μM) or ΔM4 (3.1 μM) in 10 mM HEPES pH 7.0 with 5 mM glucose for 2 h at 30°C, then fixed overnight with glutaraldehyde, dehydrated, coated with gold and examined with a scanning electron microscope HITACHI S-4700. Cells incubated in buffer without peptides served as an untreated control. Arrows indicate surface damage that leads to intracellular material leakage.
Candida albicans is characterized by morphological diversity that allows it to adapt to various environmental conditions to survive, and the transition from yeast to hyphae is essential for host cell colonization by C. albicans (Karkowska-Kuleta et al., 2009; Jacobsen et al., 2012). In this study, the effect of both peptides on inhibition of yeast to hyphae transition was investigated using C. albicans cells cultured in RPMI 1640 medium that activates filamentation (Figure 5A). The increasing concentration of ΔM3 peptide caused a progressive decrease in cell filamentation. The average length of cell hyphae at peptide concentrations that do not affect cell permeability – 12.5 μM and 25 μM – was approximately 30 and 50% shorter, respectively, compared to that observed in untreated cells (Figure 5B). Incubation of cells with higher concentrations of this peptide (50 μM) led to an increased influx of Sytox Orange dye, indicating permeabilization of the cells. In turn, ΔM4 peptide already at 2.1 μM concentration inhibited the C. albicans hypha formation, reducing their length by approximately 30%. However, for concentrations of 3.1 μM and 6.2 μM, ΔM4 completely blocked filamentation, probably killing the cells, as demonstrated by cell permeability test performed with Sytox Orange staining (Figure 5A). The observations of the effect of peptides on fungal morphology were also confirmed by the analysis of the expression of genes associated with hypha formation. Cell treatment with ∆M3 (12.5 μM and 25 μM) significantly decreased the expression of genes encoding the main fungal adhesins: ALS3, HYR1 and HWP1 compared to untreated cells (Figure 5C), which corroborated the assumption that ΔM3 contributes to the inhibition of hypha formation. However, for fungal cell treatment with ΔM4, only minor changes in expression of these genes were observed, suggesting that ΔM4 is less effective in inhibiting hypha formation by C. albicans cells.
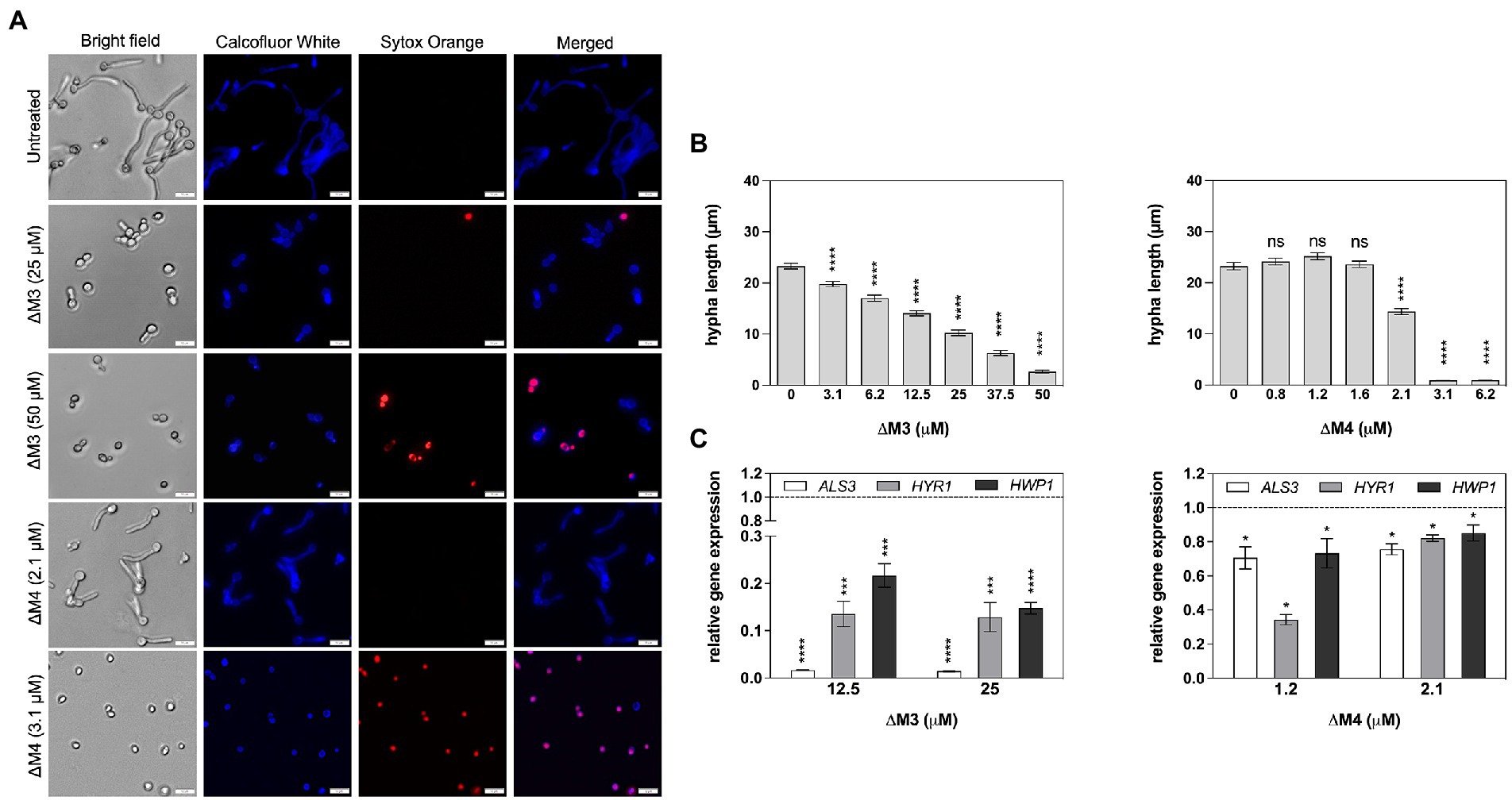
Figure 5. Impact of ΔM3 and ΔM4 on the transition of C. albicans from yeast to filamentous form. (A) C. albicans (105/100 μl) were incubated in RPMI 1640 medium with peptides at concentrations in the range of 3.1–50 μM (ΔM3) or 0.8–6.2 μM (ΔM4) for 90 min at 37°C. The fungi were stained with Calcofluor White (1 μg/ml) or Sytox Orange (1 μM) and visualized using Olympus IX73 microscope. (B) The length of the hyphae (n = 20) was measured using Olympus CellSens programming. For statistical analysis one-way ANOVA with Dunnett’s multiple comparisons post hoc test was used (****p < 0.001). (C) The mRNA expression of ALS3, HYR1 and HWP1 was determined after contact of C. albicans cells (2 × 106/ml) with ΔM3 (12.5 μM and 25 μM) and with ΔM4 (1 μM and 2 μM) for 90 min at 37°C. Bars represent the mean value ± SD from at least three independent experiments. A paired Student’s t test was used for statistical analysis. *p < 0.05, ***p < 0.005 and **** p < 0.0001 vs. untreated cells (dashed line).
The ability to form filaments by Candida spp. is not only required for increased virulence but also plays an important role in biofilm formation (Gulati and Nobile, 2016). The potential of the peptides to inhibit biofilm formation was tested after 24 h of incubation at a wide range of peptide concentrations (Figure 6A). The metabolic activity of cells in the biofilms was determined by the XTT reduction assay and compared to biofilm cells not treated with peptides. As expected, the ΔM4 peptide showed a better ability to completely inhibit biofilm formation already at a concentration of 12.5 μM, while ΔM3 was less effective, achieving a similar effect only at a concentration of 100 μM.
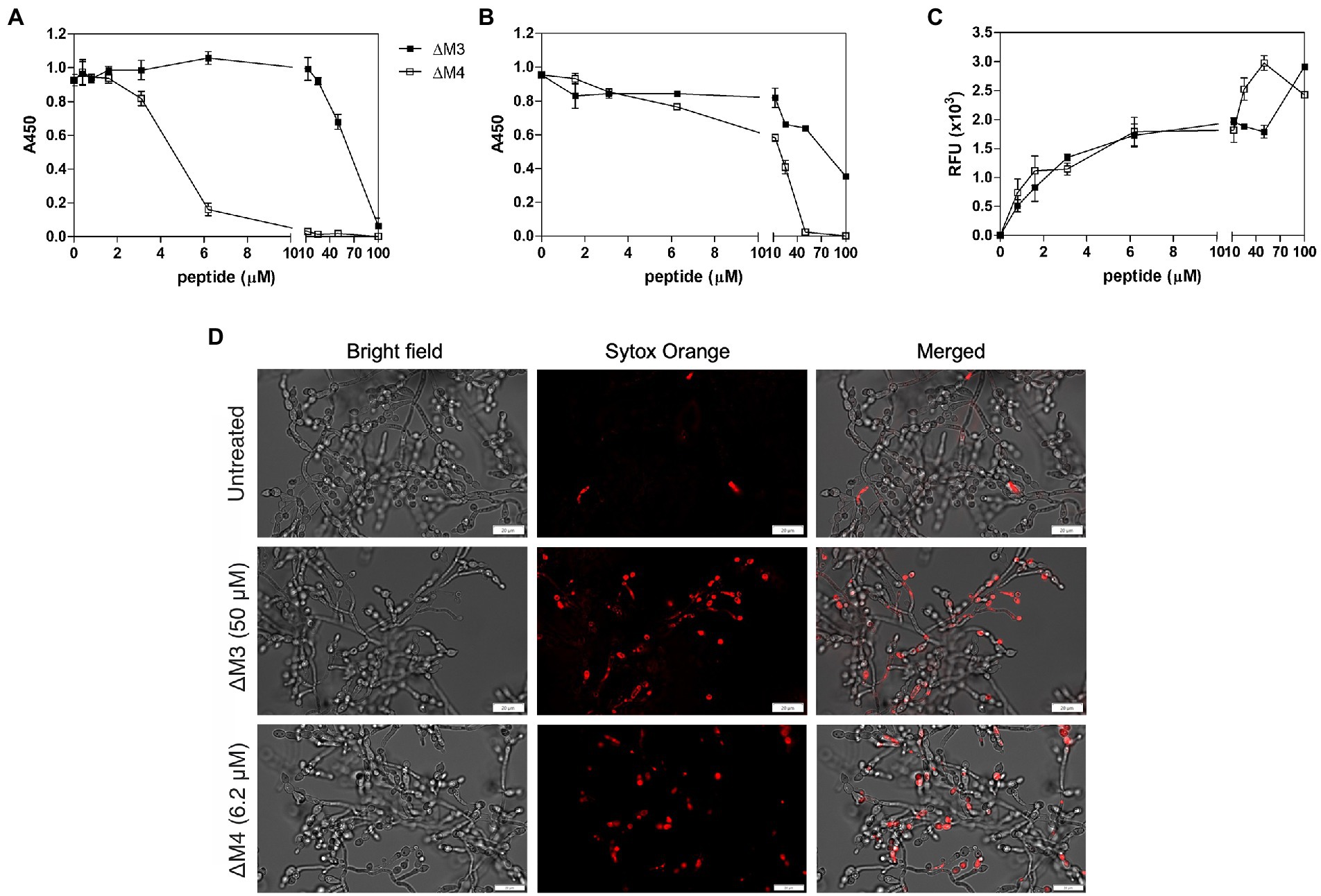
Figure 6. Effect of ΔM3 and ΔM4 on C. albicans biofilm. (A) The effect of peptides on biofilm formation was performed with C. albicans cells (1 × 105/100 μl) incubated in RPMI 1640 medium with peptides at concentrations in a range of 0.4–100 μM for 24 h at 37°C in a microplate wells, after removing unbound cells. The metabolic activity of the biofilm cells was determined using an XTT reduction assay with absorbance measurements at 450 nm (A450). The untreated biofilm served as a control. The mean value ± SD from at least three independent experiments was presented. (B) Changes in metabolic activity of C. albicans biofilm-forming cells were evaluated using an XTT reduction assay after 24-h biofilm treatment with peptides at concentrations in a range of 0.8–100 μM for 24 h at 37°C. (C) Membrane permeability in mature biofilm cells was determined using an adaptation of the Rhodamine G6 efflux assay. The leakage of the fluorescence dye, previously internalized by C. albicans cells from mature biofilm, was measured in the supernatants after 24 h of contact of the biofilm with peptides at concentration range of 0.8–100 μM at 37°C. The results of three independent experiments are presented as means ± SD. RFU – relative fluorescence units. (D) Representative images of the membrane permeabilization study with Sytox Orange dye. The mature C. albicans biofilms, untreated or treated with ΔM3 (50 μM) or ΔM4 (6.2 μM) for 24 h at 37°C, were incubated with 1 μM dye for 5 min in the dark. The biofilms were analyzed using the Olympus IX73 microscope.
Biofilm elimination is an important target in candidiasis, as the formation of this structure is strongly associated with its increased drug resistance (Tsui et al., 2016). The ability of ΔM3 and ΔM4 peptides to eradicate the C. albicans biofilm was revealed with the XTT reduction assay. The mature biofilm was more susceptible to ΔM4 than to ΔM3 (Figure 6B). As estimated after 24 h of biofilm incubation in the presence of ΔM4, the metabolism of cells was completely abolished at a concentration higher than 50 μM, while ΔM3 was unable to achieve this effect at this concentration. At a concentration of 100 μM, ΔM3 reduced cell metabolic activity only by 60%.
To confirm the effect of the peptides on C. albicans biofilm, cell membrane permeabilization was tested with two different assays. In the Rhodamine G6 efflux assay modified according to Do Nascimento Dias et al. (2020), at the beginning cells were loaded with dye, and after washing and incubation with peptides in glucose-free PBS, the efflux of dye was assessed in the supernatant (Figure 6C). The biofilm was affected by both peptides in a dose-dependent manner, although ΔM4 concentrations above 12.5 μM caused a strongly increased membrane permeabilization, while for ΔM3 it was observed at concentrations above 50 μM. The membrane permeability was also studied using Sytox Orange dye, which stain nucleic acids in cells with altered membranes. The dye load was increased with rising peptide concentration, showing a significant change in cell permeability treated with 6.2 μM ΔM4 and 50 μM ΔM3. Representative images from microscopic analysis showed cell dye load for peptides at these concentrations (Figure 6D). These results correlated with that of the Rhodamine G6 test confirming the influence of peptides on the membrane alteration of C. albicans cells forming biofilms; however, ΔM4 was more effective at lower concentrations.
Mechanisms of antifungal activity of peptides are diverse and include not only cell wall disturbances, but also more sophisticated processes associated with the induction of oxidative stress that can lead to cell apoptosis (Vriens et al., 2014; Seyedjavadi et al., 2020). To determine a possible mechanism of C. albicans biofilm destruction, the activity of yeast metacaspases was examined in biofilm treated with ΔΜ3 and ΔM4 at different concentrations of peptides. The metascapase activity was detected with CaspACE™ FITC-VAD-FMK In Situ Marker, an FITC-marked inhibitor of caspases, which irreversible binds to the enzyme, emitting fluorescence in situ. Representative images of microscopic analysis of treated biofilms are shown in Figure 7A. The minimum concentration of peptides, which triggers metacaspase activity in yeast cells and hyphae, was 6.2 μM and 50 μM for ΔM4 and ΔM3, respectively. Pro-apoptotic processes often result in loss of ROS concentration balance in cells and lead to oxidative stress. Here, the effect of ΔΜ3 and ΔM4 peptides on the induction of oxidative stress was investigated in the biofilm of C. albicans. For the analysis of ROS production with dihydrorhodamine 123 (DHR 123), the concentration of the peptide that showed pro-apoptotic activity was selected. Increased amount of intracellular ROS oxidizes non-fluorescent DHR to cationic rhodamine, which exhibits green fluorescence. Both peptides, ΔM3 (50 μM) and ΔM4 (6.2 μM) increased ROS production by approximately 38% compared to the untreated fungal biofilm (Figure 7B). At the same time, the expression of selected genes encoding antioxidant enzymes was tested at the peptide concentration inducing proapoptotic responses. The ΔM3 induced higher gene expression of GPX3 and SOD5 showing a 2-fold and 80-fold increase in mRNA level, respectively. The biofilm stimulation with ΔM4 peptide caused an increase in GPX3 and SOD5 mRNA level by 3.3- and 12.2-fold, respectively (Figure 7C). Additionally, a slight decrease in CAT1 expression was observed when mature biofilms were treated with both peptides.
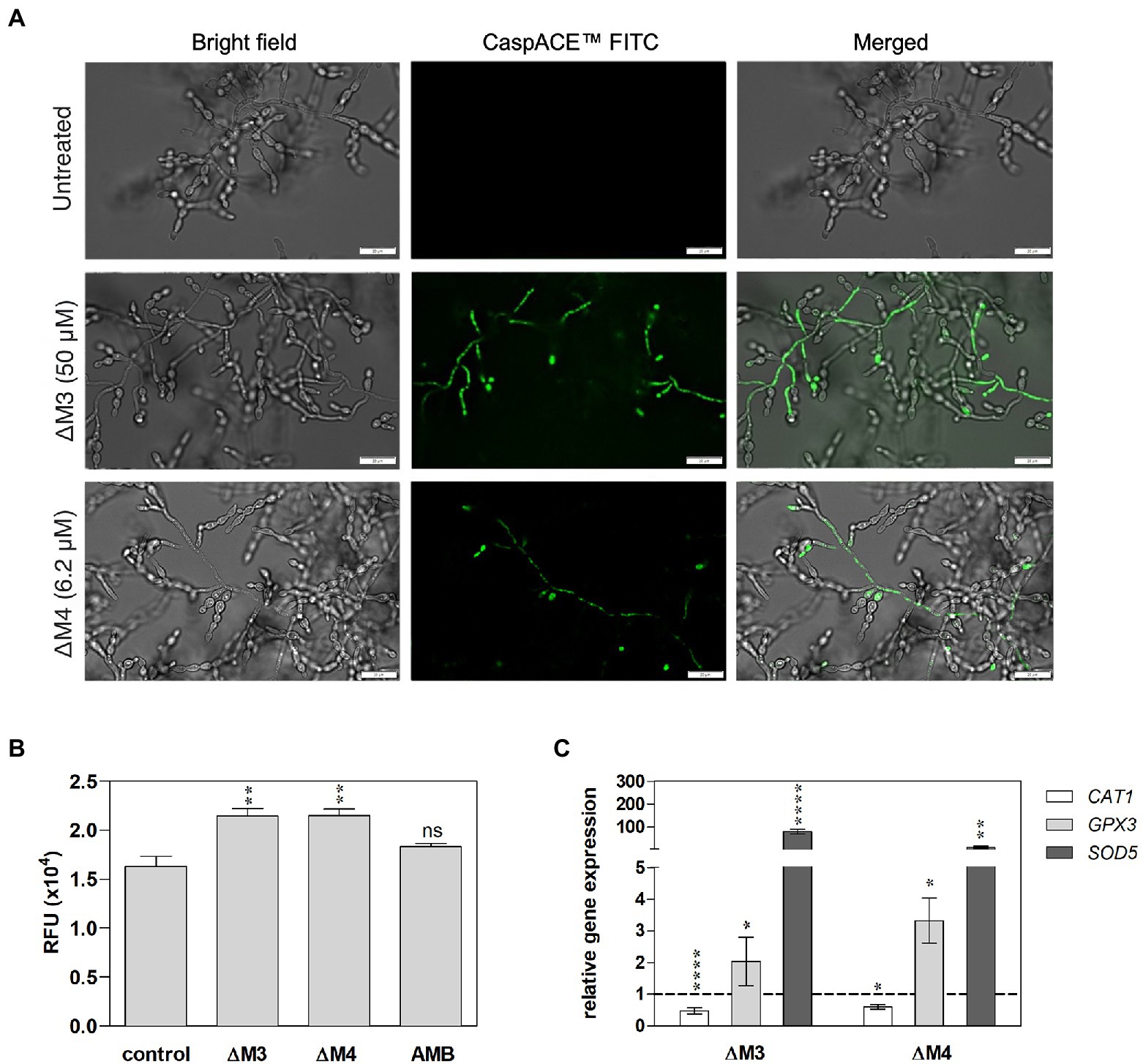
Figure 7. Apoptotic response and oxidative stress induced by ΔM3 and ΔM4 in C. albicans mature biofilm. (A) C. albicans cells (105/ml) were seeded in a 96-well plate and biofilms were grown for 24 h, then peptides were added to the cells in the concentration range of 12.5–100 μM (ΔM3) or 3.1–12.5 μM (ΔM4). The biofilm was grown in the presence of peptides for a further 24 h. The CaspACE ™ FITC-VAD-FMK In Situ Marker kit was used to assess metacaspase activation. The marker (10 μM) was added to the biofilms and incubation was carried out for 1 h at 37°C. Biofilms were imaged with an Olympus IX73 microscope and representative images for ΔM3 (50 μM) and ΔM4 (6.2 μM) are presented. (B) Intracellular ROS production in mature biofilm by peptides was evaluated with DHR test. The 24-h C. albicans biofilms were treated with ΔM3 (50 μM), ΔM4 (6.2 μM) and amphotericin B (2.7 μM) for 3 h at 37°C, washed with PBS and then incubated with DHR 123 (10 μM) for 30 min. The untreated C. albicans biofilm served as a control. The fluorescence of oxidized DHR 123 was measured using the BioTek Synergy H1 microplate reader. The graphs present values as mean ± SD (n = 3), RFU- relative fluorescence units. Statistical analysis was performed with one-way ANOVA with Dunnett’s multiple comparisons post-hoc test. **p < 0.01 (C) Analysis of CAT1, GPX1, and SOD5 gene expression was performed in mature biofilm cells treated with ΔM3 (50 μM) or ΔM4 (6.2 μM) for 3 h at 37°C. The bars represent the mean values ± SD from at least 3 experiments. A paired Student’s t-test was used for statistical analysis. *p < 0.05, **p < 0.01, ****p < 0.0001 vs. untreated cells (dashed line).
To further characterize the action of the peptides on cell mitochondria, their functionality and dynamics were evaluated. Active mitochondria showing normal membrane potential were visualized with MitoTracker Orange, while the entire mitochondrial pool was determined by MitoTracker Green staining (Figure 8A). Representative photographs show a partial loss of mitochondrial activity after incubation with peptides and amphotericin B, as a decrease in MitoTracker Orange fluorescence was noted. In addition, the distribution of active mitochondria was altered in treated cells. Since mitochondrial function depends on intracellular Ca2+ concentration, the cytosolic concentration of these ions in peptide-treated C. albicans biofilm was also investigated. Intracellular Ca2+ mobilization in live cells was determined with a ratiometric calcium assay, in which the fluorescent indicator Fura-2 changes spectral properties when calcium ions are bound. Higher peptide concentrations (50 μM ΔM3 and 12.5 μM ΔM4) resulted in increased cytosolic Ca2+ concentration, while no effect was observed at lower peptide concentrations. The ΔM4 peptide more effectively stimulated cells to Ca2+ production (by approximately 100% compared to untreated cells), while ΔM3 showed an increase in Ca2+ concentration of approximately 70% (Figure 8B). In a positive control, cells were treated with 10 mM H2O2 and demonstrated an increase in Ca2+ concentration of up to 50%. These findings indicate that, at least in part, the antifungal activity of the peptides might be related to their pro-apoptotic properties.
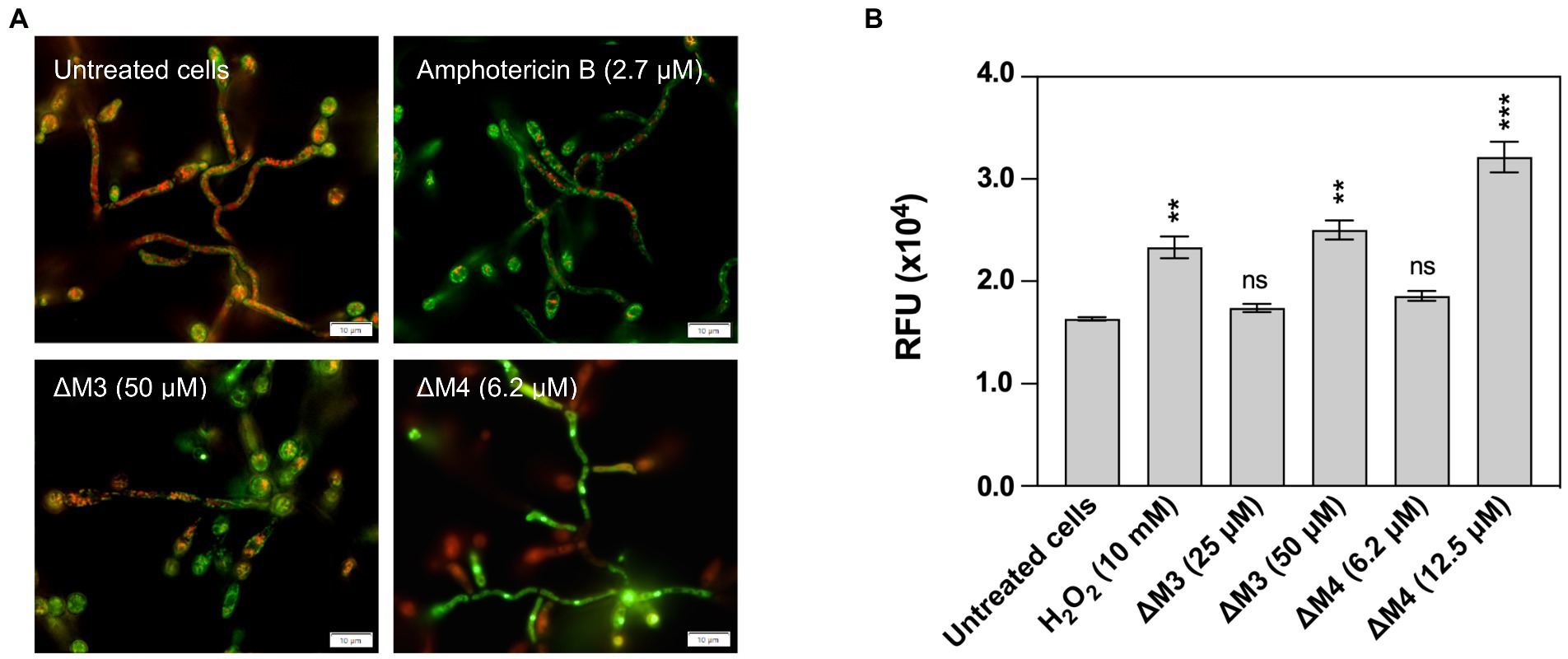
Figure 8. Induction of mitochondria activation and increased intracellular Ca2+ concentration by ΔM3 and ΔM4. (A) The changes in mitochondrial activity after treatment with peptides were detected in 24-h biofilms of C. albicans. Cells were incubated with 50 μM ΔM3, 6.2 μM ΔM4 or 2.7 μM amphotericin B for 3 h. Changes in mitochondrial activity and the entire mitochondrial pool was determined with 0.5 μM Mitotracker Orange and 0.5 μM Mitotracker Green, respectively. Representative images of microscope analysis performed with the Olympus IX73 microscope for each sample are presented. (B) To detect intracellular Ca2+, 24-h biofilms of C. albicans were incubated with ΔM3 (25 μM and 50 μM), ΔM4 (6.2 μM and 12.5 μM) or H2O2 (10 mM, positive control) for 3 h. Next the Fura-2 Calcium Flux Assay Kit was performed according to the manufacturer’s instructions. The bars represent the values of mean ± SD (n = 3), RFU – relative fluorescence unit. Statistical analysis was performed with one-way ANOVA with Dunnett’s multiple comparisons post-hoc test. **p < 0.01, ***p < 0.005.
Increased drug resistance of Candida spp. to conventional antifungal compounds, together with their toxicity, limits the effectiveness of candidiasis treatment (Tsui et al., 2016). In the fight against fungal infections, additional substances are often introduced in combination with conventional drugs to enhance the effect of treatment and to use lower concentrations of these, often toxic, drugs. This study investigated the effect of combined treatment of C. albicans with ΔM3 and ΔM4 peptides and conventional drugs, according to the modified checkerboard microdilution method in which C. albicans cells were seeded into microplate wells in RPMI with or without peptides, drugs or their combination over a wide range of two-fold serial dilutions of both agents, and then incubated for 24 h. To determine the inhibitory concentrations (IC) the XTT metabolic activity assay was used. The capacity for drug/peptide interaction with C. albicans was measured by calculating the fractional inhibitory concentration index (FICI), which was defined in the Materials and Methods section. A FICI value of less than or equal to 0.5 indicates that there is a drug and peptide synergism; a value between 0.5 and 1.0 indicates an additive effect of both substances, while a value between 1.0 and 2.0 indicates that the combination of the two substances is indifferent to cell effects. The FICI values calculated for the combination of ΔM3 and ΔM4 with amphotericin B, caspofungin, or fluconazole are presented in Table 3. Synergy against C. albicans was observed between caspofungin and each peptide. The FICI achieved a value of 0.062 and 0.370 for ΔM3 and ΔM4, respectively. In the case of the caspofungin/ΔM3 combination, the minimal inhibitory concentration that inhibits fungal metabolism by 90% was 0.003 μM and 1.6 μM for caspofungin and peptide, respectively. IC90 values for the caspofungin/ΔM4 combination were 0.012 μM and 0.8 μM for caspofungin and peptide, respectively. Due to the fact that the IC90 for these substances is 0.1 μM for caspofungin and 50 μM or 3.1 μM for ΔM3 and ΔM4, respectively (Table 2), it can be concluded that the treatment of cells with this drug in combination with the peptide strongly lowered the IC90. An additive effect was observed for the combination of peptides with amphotericin B, showing FICI values of 0.508 and 0.625 for ΔM3 and ΔM4, respectively. As with the caspofungin/peptide combination, in this case, the IC90 for the drug combination is reduced compared to the effect achieved by administering these substances alone, reaching values of 0.8 μM for amphotericin B and 0.4 μM for peptides. In turn, the effect of combining peptides with fluconazole turned out to be indifferent. The calculated FICI values were equal to 1.25 for both peptides. In fact, the IC90 values for the peptides were the same as the values obtained when the peptides were used alone. However, IC90 for fluconazole was four-fold lower when administered in combination with each peptide than when administered alone. Inhibition curves for each drug/peptide combination are presented in Supplementary Figure S2. They indicate that both peptides are useful for reducing the dose of conventional drugs such as caspofungin and amphotericin B in inhibiting the growth of C. albicans.
The cytotoxicity of the tested peptides was determined in primary human dermal fibroblasts (HDF) with the Alamar blue assay. Living cells convert the resazurin indicator to highly fluorescent resorufin. The effects of the two peptides on fibroblast viability differed (Figure 9). ΔM3 peptide caused only a slight decrease in cell viability, achieving the major effect at the concentration of 100 μM (by 24%). Differently, the second peptide, ΔM4 was more aggressive toward fibroblasts, causing a 25% decrease in cell viability at the concentration of 12.5 μM. Higher concentrations of this peptide resulted in a strong reduction in cell viability, leading to a complete cell killing at 100 μM.
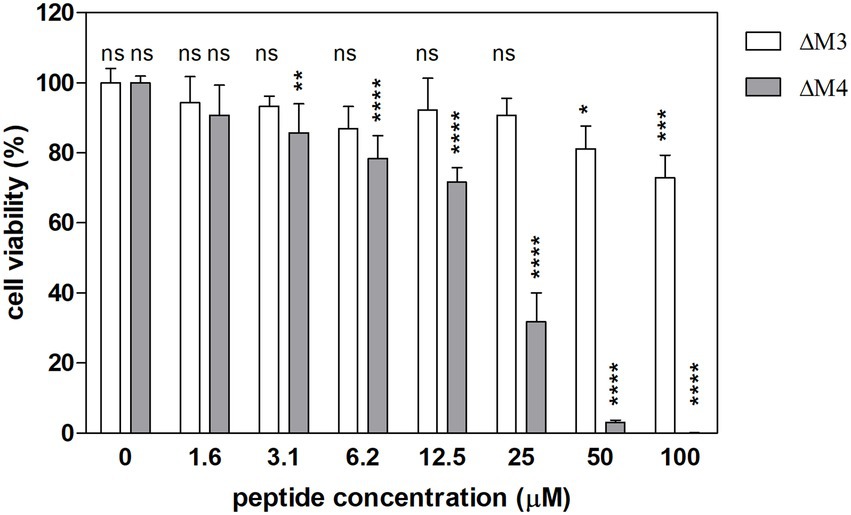
Figure 9. Viability of human dermal fibroblasts (HDF) after treatment with ΔM3 and ΔM4 peptides. The cytotoxicity of the peptides was assessed with Alamar blue test in primary HDF. Cells, seeded on microplate (1 × 105), were treated with peptides at different concentrations for 24 h in culture HDF basal medium. After medium removal, 100 μl of Alamar blue reagent were added. Fluorescence was measured at λex = 560 nm, λem = 590 nm with Synergy H1 microplate reader after 1 h of incubation. The cell viability is presented as the percentage of the untreated cells, the number of which was assumed as 100%. The samples were performed in triplicate. The bars present the mean ± SD (n = 3). Statistical analysis was performed with two-way ANOVA with Tukey’s multiple comparison test. Selected statistical significances were presented vs. untreated cells. *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001.
The incidence of fungal infections and their mortality rates have increased significantly in recent years, particularly for candidiases which are most common (Pfaller et al., 2020). C. albicans still remains the predominant etiological agent of candidiasis; however, non-albicans Candida species, including C. parapsilosis, C. tropicalis, and C. glabrata, are emerging as major opportunistic fungal pathogens, causing increased rate of hospital-acquired infections, with high risk for immunosuppressed patients undergoing surgical procedures or anticancer therapy (Pfaller et al., 2019). These pathogens also often present a reduced susceptibility to commonly used antifungal drugs, due to the increase in frequency of multidrug resistant strains or the formation of biofilms that protect them from conventional therapies (Mayer et al., 2013). Therefore, successful treatment of Candida infections that are often associated with biofilm formation requires novel and more effective antifungal strategies. The selection of efficient antimicrobial peptides possesses some advantages over conventional antifungal agents, the effectivity of which can be reduced by their toxicity toward human host cells. Cecropins have recently become an important source of AMPs, which, thanks to their specific structure and interactions with the lipid membranes of various cells, present antimicrobial and anticancer effects (Hollmann et al., 2018). It has been found that AMPs may use two or more mechanisms to penetrate membranes that depend on the polar and non-polar distribution of amino acids; however, other characteristics such as net charge, amphipathicity, hydrophobicity, percentage of α-helix structure may be important for interaction with the cell wall (Madani et al., 2011). Recent reports have demonstrated the effect of cecropin-derived amphipathic positive peptides on antifungal activity against C. albicans (Peng et al., 2022; Sun et al., 2022). These studies, however, focused on the antifungal activity of peptides that differ in amino acid sequence, charge, and hydrophobicity from the peptides tested in this study.
To test antifungal activity on different forms of Candida cells, we chose two cecropin D-derived cationic peptides, ΔM3 and ΔM4, which are both rich in Lys and Arg residues, while ΔM4 peptide was additionally enriched with three Trp residues and one Tyr residue, thus presenting more hydrophobic properties compared to ΔM3. Differences in amino acid composition resulted in different predisposition of peptides to the formation of α-helical structures in the presence of agents, simulating contact with the microbial cell membrane, as determined by the CD study. Furthermore, the presence of salt in the environment can promote this property with respect to the peptide ΔM4, while such effect was not observed for ΔM3. These structural properties turn directly into the effectiveness of these peptides in contact with yeast cells, especially those that occur in the form of blastospores. In fact, we observed different patterns of the fungal cell surface damage for both investigated peptides by SEM. In accordance with our results, similar observations were reported, showing that incorporation of hydrophobic amino acid residues in AMPs resulted in reduced MIC90 values (Pandit et al., 2020; Ramamourthy et al., 2020). Furthermore, the presence of hydrophobic residues in the transmembrane fragment of antimicrobial peptides was believed to lead to the formation of ion channels within the cell membrane (Buda De Cesare et al., 2020). The amino acid composition also influenced the differences in fungicidal activity of the peptides studied, depending on the presence of ions in the environment. The ΔM3 peptide, which has a charge of +8, appeared to be more susceptible to the salt effect, regardless of the ability to form helical structures, since no activity against C. albicans was observed after treatment of fungal cells with a peptide dissolved in PBS, compared to the strong activity detected when HEPES buffer was used. Such a negative impact of salt on the fungicidal activity of short cationic peptides against C. albicans has previously been demonstrated (Do Nascimento Dias et al., 2020; Ghosh et al., 2021). It may be the result of compensation for the peptide charge or of affecting the local peptide structure by electrostatic interactions of ions with amino acid functional groups (Baldauf et al., 2013). Contrary, ΔM4 was able to kill C. albicans even in PBS but with a lower efficiency compared to the results obtained in HEPES buffer. Such differences can be explained by different mechanisms and/or kinetics of both peptide action, where membrane compromising activity is accompanied by more complex and diverse mechanisms including DNA damage, inhibition of protein synthesis or metabolic pathway, induction of apoptosis, and oxidative stress (Nguyen et al., 2011; Wang et al., 2015; Li et al., 2016). For example, in the case of the application of the tryptophan-rich PuroA peptide, its rapid translocation inside C. albicans cells and its binding to intercellular targets was proposed as a mechanism of yeast inactivation, before subsequent perturbation of the cell membrane (Shagaghi et al., 2017). Another component of the incubation solution that radically inhibited the antifungal activity of the tested peptides was human plasma. Similar observations have also been reported for several AMPs, including LL-37. One study pointed to the possibility of forming complexes between peptides and plasma proteins such as albumin/α1-antitrypsin complex or apolipoprotein AI, resulting in the inactivation of AMPs (Tang et al., 2021).
Preliminary studies on other Candida species have been performed, indicating that the susceptibility of C. albicans, C. tropicalis, and C. parapsilosis cells to both peptides was comparable, while C. glabrata cells were killed at higher concentrations of peptides. This was expected as C. glabrata is known to present a low susceptibility to azoles (Arendrup et al., 2013) and a significant resistance to amphotericin B (Pappas et al., 2016), which was proposed to be associated with the presence of fungal cell wall transporters and their increased activity in the export of antifungal agents (Cavalheiro et al., 2021). Limited susceptibility of C. glabrata to AMPs was also observed in the treatment with histatin 5, magainin 2, cathelicidins, and defensins (Helmerhorst et al., 2005; Del Gaudio et al., 2013).
In the presented study, the observed influence of peptides on fungal cell adhesion and filamentation was also significant, as the regulation of these processes may prevent biofilm formation and fungal cell resistance to treatment. A noticeable inhibition of fungal cell filamentation was observed at a concentration of the peptides lower than fungicidal, and it was manifested by the formation of shorter hyphae and decreased expression of genes encoding proteins characteristic for the filamentous form of C. albicans cells (Hwp1, Hyr1, and Als3). A recent study demonstrated a similar effect on C. albicans by the cecropin-derived peptide C18 (Sun et al., 2022). It should be noted that the arresting of fungal cells in the form of blastospores may facilitate their elimination, even at lower peptide concentration. Such a strategy has been proposed for many antimicrobial peptides (Ogasawara et al., 2007; Akerey et al., 2009; Theberge et al., 2013; Do Nascimento Dias et al., 2020). The development of hyphae by C. albicans cells is a hallmark of the initial steps in biofilm formation (Tsui et al., 2016). Then the consequence of the observed inhibition of hypha production by the peptides was the disruption of biofilm development, which was identified at the same peptide concentration range.
The diagnosis of a yeast infection and the necessity for its treatment often come at the stage of a biofilm formed. Therefore, we also tested the efficacy of both peptides against a mature biofilm, with a developed matrix, which hinders the penetration of various antifungal drugs, forcing an increase in the dose of the drug, which in turn could be toxic to the cells of the treated host (Taff et al., 2013; Cavalheiro and Teixeira, 2018). For ΔM4 peptide, as in the case of many other AMPs (Kodedová and Sychrová, 2017; Do Nascimento Dias et al., 2020), we observed a noticeable decrease in the metabolic activity of mature biofilms at a peptide concentration approximately 15-fold or 8-fold higher compared to its effectiveness against yeast-like or biofilm-initiating cells. However, these concentrations of ΔM4 were close to its cytotoxic properties toward host cells, where HDF viability decreased gradually with increasing ΔM4 concentration, and was eliminated at the ΔM4 concentration of 100 μM. In turn, the ΔM3 peptide showed a lower toxicity in relation to HDF cells; at a concentration of 100 μM fibroblast viability was decreased only by 25%. However, at this concentration, the peptide lost the ability to eradicate the mature biofilm. Regardless of the effect of the peptides on host cells, it can be concluded that at least in the case of ΔM4, all morphological forms of C. albicans can be destroyed. The second peptide, ΔM3, worked better on blastospores and, at high concentrations, can significantly reduce pre-formed and mature biofilms.
Although the effects of AMPs on microorganisms are mainly attributed to their interaction with the cell membrane, other mechanisms have also been identified, including inhibition of cell wall biosynthesis or interaction with DNA (Buda De Cesare et al., 2020). However, certain peptides, such as salivary histatin 5 (Helmerhorst et al., 2001) or the venom wasp protonectin (Wang et al., 2015) involved also ROS production in their antifungal mechanism and induced oxidative stress in yeasts through uncontrolled accumulation of intracellular ROS, which led to cell apoptosis or necrosis (Kim et al., 2020; Seyedjavadi et al., 2020; Zhang et al., 2021). For ΔM3 and ΔM4 peptides, increased cell permeability was also accompanied by increased ROS production within the C. albicans biofilm, suggesting a more complex antifungal mechanism of peptide action. However, C. albicans possesses adaptive responses that allow ROS detoxification, including activation of superoxide dismutase, catalase, and other enzymes involved in the production of ROS scavengers (Jamieson et al., 1996; Dantas et al., 2015). Activation of these reactions was also observed after treatment of the C. albicans biofilm with ΔM4 peptide, where increased expression of the GPX3 and SOD5 genes was detected. However, the expression of CAT1 was reduced, suggesting that ROS detoxification upon ΔM4 treatment may not be completed. The explanation for such a result was indicated by Kaloriti et al. (2014), who showed that simultaneous exposure of C. albicans cells to oxidative and cationic stress may be more effective in yeast killing than oxidative stress alone (Kaloriti et al., 2014). These authors also demonstrated that cationic ions can inhibit catalase, leading to ROS accumulation and inhibition of the Cap1 signaling molecule, which is critical for the response to oxidative stress of C. albicans cells.
Mitochondria are considered the main source of ROS, but uncontrolled ROS production also makes mitochondria a target, influencing mitochondrial membrane functionality. As a result, changes in membrane potential, increased mitochondrial permeability, and finally mitochondrial apoptosis are observed (Choi and Lee, 2015). In this study, mitochondrial functionality was investigated with fluorescence dyes, which identify active mitochondria with normal membrane potential versus the total number of mitochondria in C. albicans cells that form a biofilm, treated with both peptides. In each case, a significant loss of active mitochondria was observed, confirming that peptides contribute to biofilm disruption. These organelles develop different repair mechanisms, including mitochondrial fission-fusion processes, which are important for their proper functioning (Youle and van der Bliek, 2012). A previous study showed that C. albicans can lose its virulence due to the loss of the ability of mitochondria to fuse (Liang et al., 2018). It is possible that biofilm treatment with the peptides can induce such processes, but to confirm this observation, more research is needed.
The fact that the tested peptides increased ROS production and disrupted mitochondrial functionality was confirmed by the detection of increased cytosolic Ca2+ concentration after peptide treatment of C. albicans cells. On the other hand, the accumulation of Ca2+ violates its homeostasis and inhibits mitochondrial respiration, leading to its dysfunction (Rossi et al., 2019). Several studies have demonstrated that high cytosolic Ca2+ concentration causes an increased influx of these ions into mitochondria, resulting in increased permeability and depolarization of the mitochondrial membrane, and enhanced ROS production by C. albicans (Lee and Lee, 2018; Niu et al., 2020; Kwun et al., 2021). Furthermore, it has been shown that deregulated calcium ion homeostasis can induce C. albicans apoptosis (Kwun et al., 2021). Our study demonstrated increased metacaspase activity in the C. albicans biofilm treated with both peptides, which confirms the development of apoptotic processes in the treated biofilm. Previously, the apoptotic effects of various peptides have been demonstrated for Candida yeast cells (Lee and Lee, 2014; Li et al., 2019; Chen and Lan, 2020; Kim et al., 2020). In summary, taking into account increased ROS production and Ca2+ concentration, enhanced mitochondrial dysfunction, and increased metacaspase activity, we can propose a partial pro-apoptotic mechanism in the fungicidal activity of the studied peptides.
Strategies to reduce peptide toxicity include peptide further modification or encapsulation (van Gent et al., 2021). Combining them with conventional antifungal drugs is also a good approach, especially because the use of drug combinations can not only improve the efficiency of both antifungal agents, often with lowering their active concentration range and reducing their cytotoxic effect, but also can decrease the development of the resistance. Therefore, we evaluated the effects of each of the peptides along with amphotericin B, caspofungin and fluconazole that resulted in three different types of effects. In the first type of interaction, synergism was observed only for the combination of peptides with caspofungin, especially strong in the case of ΔM3, for which a 30-fold reduction in IC90 was identified (Table 3). Similar observations have been reported for the combination of caspofungin with many other AMPs (Vriens et al., 2015; Kovács et al., 2019; Shen et al., 2021). Caspofungin inhibits β-1,3-glucan synthase, located in the cell membrane of Candida cells, responsible for an essential component of the fungal cell wall. The reduced content of glucan in the fungal cell wall may facilitate its better penetration by the peptide. The second type of action, additivity, was determined for the combination of the peptides with amphotericin B. For both peptides, a two-fold reduction in IC90 was observed in cooperation with amphotericin B. However, it was found that the concentration of peptides needed to inhibit the formation of C. albicans biofilms to 10% was 125- and 8-fold lower for amphotericin B combination with ΔM3 and ΔM4, respectively, compared to peptides acting alone. Although the interaction between these compounds has not been classified as synergistic, it is relatively strong and after the verification by different types of tests or in vivo studies (Vitale et al., 2005) it might change character, as it was observed previously in the case of caspofungin/amphotericin B combination used against C. parapsilosis (Barchiesi et al., 2007). When peptides were combined with fluconazole, no changes in antifungal properties were observed. Although the determined FICI of 1.25 suggests in both cases an independent effect, where both peptides clearly negatively affect the antifungal activity of fluconazole, as it was detected even at the lowest peptide concentration tested (0.4 μM). However, the negative effect of fluconazole on the antifungal activity of both peptides was not detected (Supplementary Figure S2). Trapping fluconazole by peptides and consequently the drug’s failure to reach its target site without affecting its own antimicrobial activity is one of the possible reasons for the observation of such a phenomenon. It is also possible that the peptides, like some other AMPs (Ma et al., 2020), may increase the expression of Erg11, an essential enzyme in the ergosterol biosynthesis pathway, which is inhibited by fluconazole. Moreover, it is also not excluded that the peptides can increase expression and/or activity of C. albicans efflux pumps and thus increase the fungus resistance to fluconazole but it needs further research.
In conclusion, cecropin D-derived synthetic cationic peptides presented strong antifungal activity against Candida cells, regardless of the morphological form of the fungi. A scheme that demonstrates the most relevant results is presented in Figure 10. As expected, yeast cells of C. albicans were more susceptible to the action of peptides compared to the fungal biofilm. The mode of action of peptides is most likely complex, involving both destabilization of the cell wall and additional intracellular mechanisms such as increased oxidative stress, which contributes to cell death by apoptosis. Moreover, the synergism of tested peptides with caspofungin was demonstrated. That offers an opportunity for a new approach to combating Candida infections by constraining their virulence and dealing with drug resistance.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
IG-L: methodology, investigation, formal analysis, conceptualization, writing-original draft, and writing-review and editing. GB: methodology, investigation, formal analysis, funding acquisition, writing-original draft, and writing-review and editing. MJ and AG: methodology, investigation, and formal analysis. JK-K: investigation and writing-review and editing. MM-M and EP: resources and writing-review and editing. JD: methodology and investigation. AK: writing-review and editing and funding acquisition. MR-K: conceptualization, supervision, funding acquisition, writing-original draft, and writing-review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the National Science Centre of Poland (grant no. 2019/33/B/NZ6/02284 awarded to MR-K) and the Jagiellonian University in Krakow and the Statutory Funds of the Faculty of Biochemistry, Biophysics and Biotechnology (project No. N19/MNS/000035 awarded to GB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1045984/full#supplementary-material
Akerey, B., Le-Lay, C., Fliss, I., Subirade, M., and Rouabhia, M. (2009). In vitro efficacy of nisin Z against Candida albicans adhesion and transition following contact with normal human gingival cells. J. Appl. Microbiol. 107, 1298–1307. doi: 10.1111/j.1365-2672.2009.04312.x
Aldholmi, M., Marchand, P., Ourliac-Garnier, I., Le Pape, P., and Ganesan, A. (2019). A decade of antifungal leads from natural products: 2010-2019. Pharmaceuticals 12:182. doi: 10.3390/ph12040182
Arendrup, M. C., Dzajic, E., Jensen, R. H., Johansen, H. K., Kjaeldgaard, P., Knudsen, J. D., et al. (2013). Epidemiological changes with potential implication for antifungal prescription recommendations for fungaemia: data from a nationwide fungaemia surveillance programme. Clin. Microbiol. Infect. 19, E343–E353. doi: 10.1111/1469-0691.12212
Baldauf, C., Pagel, K., Warnke, S., von Helden, G., Koksch, B., Blum, V., et al. (2013). How cations change peptide structure. Chemistry 19, 11224–11234. doi: 10.1002/chem.201204554
Barchiesi, F., Spreghini, E., Tomassetti, S., Giannini, D., and Scalise, G. (2007). Caspofungin in combination with amphotericin B against Candida parapsilosis. Antimicrob. Agents Chemother. 51, 941–945. doi: 10.1128/AAC.00880-06
Bartnicka, D., Karkowska-Kuleta, J., Zawrotniak, M., Satała, D., Michalik, K., Zielinska,, et al. (2019). Adhesive protein-mediated cross-talk between Candida albicans and Porphyromonas gingivalis in dual species biofilm protects the anaerobic bacterium in unfavorable oxic environment. Sci. Rep. 9:4376. doi: 10.1038/s41598-019-40771-8
Bochenska, O., Rapala-Kozik, M., Wolak, N., Aoki, W., Ueda, M., and Kozik, A. (2016). The action of ten secreted aspartic proteases of pathogenic yeast Candida albicans on major human salivary antimicrobial peptide, histatin 5. Acta Biochim. Pol. 63, 403–410. doi: 10.18388/abp.2016_1318
Boman, H. G., Faye, I., Gudmundsson, G. H., Lee, J. Y., and Lidholm, D. A. (1991). Cell-free immunity in Cecropia. A model system for antibacterial proteins. Eur. J. Biochem. 201, 23–31. doi: 10.1111/j.1432-1033.1991.tb16252.x
Bondaryk, M., Staniszewska, M., Zielińska, P., and Urbańczyk-Lipkowska, Z. (2017). Natural antimicrobial peptides as inspiration for design of a new generation antifungal compounds. J. Fungi Basel 3:46. doi: 10.3390/jof3030046
Brady, D., Grapputo, A., Romoli, O., and Sandrelli, F. (2019). Insect cecropins, antimicrobial peptides with potential therapeutic applications. Int. J. Mol. Sci. 20:5862. doi: 10.3390/ijms20235862
Buda De Cesare, G., Cristy, S. A., Garsin, D. A., and Lorenz, M. C. (2020). Antimicrobial peptides: a new frontier in antifungal therapy. mBio 11, e02123–e02120. doi: 10.1128/mBio.02123-20
Cavalheiro, M., Romão, D., Santos, R., Mil-Homens, D., Pais, P., Costa,, et al. (2021). Role of CgTpo4 in polyamine and antimicrobial peptide resistance: determining virulence in Candida glabrata. Int. J. Mol. Sci. 22:1376. doi: 10.3390/ijms22031376
Cavalheiro, M., and Teixeira, M. C. (2018). Candida biofilms: threats, challenges, and promising strategies. Front. Med. (Lausanne). 5:28. doi: 10.3389/fmed.2018.00028
Chen, R. C., and Lan, C. Y. (2020). Human antimicrobial peptide Hepcidin 25-induced apoptosis in Candida albicans. Microorganisms. 8:585. doi: 10.3390/microorganisms8040585
Choi, H., and Lee, D. G. (2015). Lycopene induces apoptosis in Candida albicans through reactive oxygen species production and mitochondrial dysfunction. Biochimie 115, 108–115. doi: 10.1016/j.biochi.2015.05.009
Cytrynska, M., Mak, P., Zdybicka-Barabas, A., Suder, P., and Jakubowicz, T. (2007). Purification and characterization of eight peptides from galleria mellonella immune hemolymph. Peptides 28, 533–546. doi: 10.1016/j.peptides.2006.11.010
Dantas, A., Day, A., Ikeh, M., Kos, I., Achan, B., and Quinn, J. (2015). Oxidative stress responses in the human fungal pathogen. Candida albicans. Biomolecules. 5, 142–165. doi: 10.3390/biom5010142
De Lucca, A. J., Bland, J. M., Vigo, C. B., Jacks, T. J., Peter, J., and Walsh, T. J. (2000). D-cecropin B: proteolytic resistance, lethality for pathogenic fungi and binding properties. Med. Mycol. 38, 301–308. doi: 10.1080/mmy.38.4.301.308
Del Gaudio, G., Lombardi, L., Maisetta, G., Esin, S., Batoni, G., Sanguinetti, M., et al. (2013). Antifungal activity of the noncytotoxic human peptide Hepcidin 20 against fluconazole-resistant Candida glabrata in human vaginal fluid. Antimicrob. Agents Chemother. 57, 4314–4321. doi: 10.1128/AAC.00904-13
Do Nascimento Dias, J., De Souza Silva, C., De Araújo, A. R., Souza, J., De Holanda Veloso Júnior, P. H., Cabral, W. F., et al. (2020). Mechanisms of action of antimicrobial peptides ToAP2 and NDBP-5.7 against Candida albicans planktonic and biofilm cells. Sci. Rep. 10:10327. doi: 10.1038/s41598-020-67041-2
Ghosh, S., Pandit, G., Debnath, S., Chatterjee, S., and Satpati, P. (2021). Effect of monovalent salt concentration and peptide secondary structure in peptide-micelle binding. RSC Adv. 11, 36836–36849. doi: 10.1039/d1ra06772a
Górecki, A., Bonarek, P., Górka, A. K., Figiel, M., Wilamowski, M., and Dziedzicka-Wasylewska, M. (2015). Intrinsic disorder of human Yin Yang 1 protein. Proteins 83, 1284–1296. doi: 10.1002/prot.24822
Guevara-Lora, I., Bras, G., Karkowska-Kuleta, J., González-González, M., Ceballos, K., Sidlo, W., et al. (2020). Plant-derived substances in the fight against infections caused by Candida species. Int. J. Mol. Sci. 21:6131. doi: 10.3390/ijms21176131
Gulati, M., and Nobile, C. J. (2016). Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 18, 310–321. doi: 10.1016/j.micinf.2016.01.002
Guo, Q., Sun, S., Yu, J., Li, Y., and Cao, L. (2008). Synergistic activity of azoles with amiodarone against clinically resistant Candida albicans tested by chequerboard and time-kill methods. J. Med. Microbiol. 57, 457–462. doi: 10.1099/jmm.0.47651-0
Helmerhorst, E. J., Troxler, R. F., and Oppenheim, F. G. (2001). The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. PNAS 98, 14637–14642. doi: 10.1073/pnas.141366998
Helmerhorst, E. J., Venuleo, C., Beri, A., and Oppenheim, F. G. (2005). Candida glabrata is unusual with respect to its resistance to cationic antifungal proteins. Yeast 22, 705–714. doi: 10.1002/yea.1241
Hollmann, A., Martinez, M., Maturana, P., Semorile, L. C., and Maffia, P. C. (2018). Antimicrobial peptides: interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem. 6:204. doi: 10.3389/fchem.2018.00204
Huan, Y., Kong, Q., Mou, H., and Yi, H. (2020). Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 11:582779. doi: 10.3389/fmicb.2020.582779
Jacobsen, I. D., Wilson, D., Wächtler, B., Brunke, S., Naglik, J. R., and Hube, B. (2012). Candida albicans dimorphism as a therapeutic target. Expert Rev. Anti-Infect. Ther. 10, 85–93. doi: 10.1586/eri.11.152
Jamieson, D. J., Stephen, D. W., and Terrière, E. C. (1996). Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol. Lett. 138, 83–88. doi: 10.1111/j.1574-6968.1996.tb08139.x
Ji, S., Li, W., Zhang, L., Zhang, Y., and Cao, B. (2014). Cecropin A-melittin mutant with improved proteolytic stability and enhanced antimicrobial activity against bacteria and fungi associated with gastroenteritis in vitro. Biochem. Biophys. Res. Commun. 451, 650–655. doi: 10.1016/j.bbrc.2014.08.044
Kaloriti, D., Jacobsen, M., Yin, Z., Patterson, M., Tillmann, A., Smith, D. A., et al. (2014). Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio 5, e01334–e01314. doi: 10.1128/mBio.01334-14
Kandasamy, S. K., and Larson, R. G. (2006). Effect of salt on the interactions of antimicrobial peptides with zwitterionic lipid bilayers. Biochim. Biophys. Acta 1758, 1274–1284. doi: 10.1016/j.bbamem.2006.02.030
Karkowska-Kuleta, J., Rapala-Kozik, M., and Kozik, A. (2009). Fungi pathogenic to humans: molecular bases of virulence of Candida albicans, Cryptococcus neoformans and aspergillus fumigatus. Acta Biochim. Pol. 56, 211–224. doi: 10.18388/abp.2009_2452
Kim, S., Hwang, J. S., and Lee, D. G. (2020). Lactoferricin B like peptide triggers mitochondrial disruption-mediated apoptosis by inhibiting respiration under nitric oxide accumulation in Candida albicans. IUBMB Life 72, 1515–1527. doi: 10.1002/iub.2284
Kodedová, M., and Sychrová, H. (2017). Synthetic antimicrobial peptides of the halictines family disturb the membrane integrity of Candida cells. Biochim. Biophys. Acta Biomembr. 1859, 1851–1858. doi: 10.1016/j.bbamem.2017.06.005
Kovács, R., Nagy, F., Tóth, Z., Bozó, A., Balázs, B., and Majoros, L. (2019). Synergistic effect of nikkomycin Z with caspofungin and micafungin against Candida albicans and Candida parapsilosis biofilms. Lett. Appl. Microbiol. 69, 271–278. doi: 10.1111/lam.13204
Kulig, K., Karnas, E., Woznicka, O., Kuleta, P., Zuba-Surma, E., Pyza, E., et al. (2022). Insight into the properties and Immunoregulatory effect of extracellular vesicles produced by Candida glabrata, Candida parapsilosis, and Candida tropicalis biofilms. Front. Cell. Infect. Microbiol. 12:879237. doi: 10.3389/fcimb.2022.879237
Kwun, M. S., Lee, H. J., and Lee, D. G. (2021). β-Amyrin-induced apoptosis in Candida albicans triggered by calcium. Fungal Biol. 125, 630–636. doi: 10.1016/j.funbio.2021.03.006
Lee, E., Kim, J. K., Jeon, D., Jeong, K. W., Shin, A., and Kim, Y. (2015). Functional roles of aromatic residues and helices of papiliocin in its antimicrobial and anti-inflammatory activities. Sci. Rep. 5:12048. doi: 10.1038/srep12048
Lee, J., and Lee, D. G. (2014). Melittin triggers apoptosis in Candida albicans through the reactive oxygen species-mediated mitochondria/caspase-dependent pathway. FEMS Microbiol. Lett. 355, 36–42. doi: 10.1111/1574-6968.12450
Lee, W., and Lee, D. G. (2018). Reactive oxygen species modulate itraconazole-induced apoptosis via mitochondrial disruption in Candida albicans. Free Rad. Res. 52, 39–50. doi: 10.1080/10715762.2017.1407412
Lee, Y., Puumala, E., Robbins, N., and Cowen, L. E. (2021). Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem. Rev. 121, 3390–3411. doi: 10.1021/acs.chemrev.0c00199
Li, R., Chen, C., Zhang, B., Jing, H., Wang, Z., Wu, C., et al. (2019). The chromogranin A-derived antifungal peptide CGA-N9 induces apoptosis in Candida tropicalis. Biochem. J. 476, 3069–3080. doi: 10.1042/BCJ20190483
Li, L., Sun, J., Xia, S., Tian, X., Cheserek, M. J., and Le, G. (2016). Mechanism of antifungal activity of antimicrobial peptide APP, a cell-penetrating peptide derivative, against Candida albicans: intracellular DNA binding and cell cycle arrest. Appl. Microbiol. Biotechnol. 100, 3245–3253. doi: 10.1007/s00253-015-7265-y
Liang, C., Zhang, B., Cui, L., Li, J., Yu, Q., and Li, M. (2018). Mgm1 is required for maintenance of mitochondrial function and virulence in Candida albicans. Fungal Genet. Biol. 120, 42–52. doi: 10.1016/j.fgb.2018.09.006
Ma, H., Zhao, X., Yang, L., Su, P., Fu, P., Peng, J., et al. (2020). Antimicrobial peptide AMP-17 affects Candida albicans by disrupting its Cell Wall and cell membrane integrity. Infect. Drug Resist. 13, 2509–2520. doi: 10.2147/IDR.S250278
Madani, F., Lindberg, S., Langel, U., Futaki, S., and Gräslund, A. (2011). Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011:414729. doi: 10.1155/2011/414729
Maisetta, G., Di Luca, M., Esin, S., Florio, W., Brancatisano, F. L., Bottai, D., et al. (2008). Evaluation of the inhibitory effects of human serum components on bactericidal activity of human beta defensin 3. Peptides 29, 1–6. doi: 10.1016/j.peptides.2007.10.013
Manrique-Moreno, M., Suwalsky, M., Patiño-González, E., Fandiño-Devia, E., Jemioła-Rzemińska, M., and Strzałka, K. (2021). Interaction of the antimicrobial peptide ΔM3 with the Staphylococcus aureus membrane and molecular models. Biochim. Biophys. Acta Biomembr. 1863:183498. doi: 10.1016/j.bbamem.2020.183498
Mayer, F. L., Wilson, D., and Hube, B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. doi: 10.4161/viru.22913
Nguyen, L. T., Haney, E. F., and Vogel, H. J. (2011). The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29, 464–472. doi: 10.1016/j.tibtech.2011.05.001
Niu, C., Wang, C., Yang, Y., Chen, R., Zhang, J., Chen, H., et al. (2020). Carvacrol induces Candida albicans apoptosis associated with Ca2+/Calcineurin pathway. Front. Cell. Infect. Microbiol. 10:192. doi: 10.3389/fcimb.2020.00192
Ogasawara, A., Komaki, N., Akai, H., Hori, K., Watanabe, H., Watanabe, T., et al. (2007). Hyphal formation of Candida albicans is inhibited by salivary mucin. Biol. Pharm. Bull. 30, 284–286. doi: 10.1248/bpb.30.284
Oñate-Garzón, J., Manrique-Moreno, M., Trier, S., Leidy, C., Torres, R., and Patiño, E. (2017). Antimicrobial activity and interactions of cationic peptides derived from galleria mellonella cecropin D-like peptide with model membranes. J. Antibiot. 70, 238–245. doi: 10.1038/ja.2016.134
Pandit, G., Biswas, K., Ghosh, S., Debnath, S., Bidkar, A. P., Satpati, P., et al. (2020). Rationally designed antimicrobial peptides: insight into the mechanism of eleven residue peptides against microbial infections. Biochim. Biophys. Acta Biomembr. 1862:183177. doi: 10.1016/j.bbamem.2020.183177
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the Management of Candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 62, e1–e50. doi: 10.1093/cid/civ933
Peng, C., Liu, Y., Shui, L., Zhao, Z., Mao, X., and Liu, Z. (2022). Mechanisms of action of the antimicrobial peptide Cecropin in the killing of Candida albicans. Life (Basel) 12:1581. doi: 10.3390/life12101581
Peng, J., Wu, Z., Liu, W., Long, H., Zhu, G., Guo, G., et al. (2019). Antimicrobial functional divergence of the cecropin antibacterial peptide gene family in Musca domestica. Parasit. Vectors 12:537. doi: 10.1186/s13071-019-3793-0
Pfaller, M. A., Carvalhaes, C. G., Smith, C. J., Diekema, D. J., and Castanheira, M. (2020). Bacterial and fungal pathogens isolated from patients with bloodstream infection: frequency of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program (2012-2017). Diagn. Microbiol. Infect. Dis. 97:115016. doi: 10.1016/j.diagmicrobio.2020.115016
Pfaller, M. A., Diekema, D. J., Turnidge, J. D., Castanheira, M., and Jones, R. N. (2019). Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997-2016. Open Forum Infect. Dis. 6, S79–S94. doi: 10.1093/ofid/ofy358
Ramamourthy, G., Park, J., Seo, C., Vogel, H. J., and Park, Y. (2020). Antifungal and Antibiofilm activities and the mechanism of action of repeating lysine-tryptophan peptides against Candida albicans. Microorganisms 8:758. doi: 10.3390/microorganisms8050758
Romoli, O., Saviane, A., Bozzato, A., D'Antona, P., Tettamanti, G., Squartini, A., et al. (2017). Differential sensitivity to infections and antimicrobial peptide-mediated immune response in four silkworm strains with different geographical origin. Sci. Rep. 7:1048. doi: 10.1038/s41598-017-01162-z
Roscetto, E., Contursi, P., Vollaro, A., Fusco, S., Notomista, E., and Catania, M. R. (2018). Antifungal and anti-biofilm activity of the first cryptic antimicrobial peptide from an archaeal protein against Candida spp. clinical isolates. Sci. Rep. 8:17570. doi: 10.1038/s41598-018-35530-0
Rossi, A., Pizzo, P., and Filadi, R. (2019). Calcium, mitochondria and cell metabolism: a functional triangle in bioenergetics. Biochim. Biophys. Acta, Mol. Cell Res. 1866, 1068–1078. doi: 10.1016/j.bbamcr.2018.10.016
Santa-González, G. A., Patiño-González, E., and Manrique-Moreno, M. (2020). Synthetic peptide ΔM4-induced cell death associated with cytoplasmic membrane disruption, mitochondrial dysfunction and cell cycle arrest in human melanoma cells. Molecules 25:5684. doi: 10.3390/molecules25235684
Sato, H., and Feix, J. B. (2006). Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic alpha-helical antimicrobial peptides. Biochim. Biophys. Acta 1758, 1245–1256. doi: 10.1016/j.bbamem.2006.02.021
Seyedjavadi, S. S., Khani, S., Eslamifar, A., Ajdary, S., Goudarzi, M., Halabian, R., et al. (2020). The antifungal peptide MCh-AMP1 derived from Matricaria chamomilla inhibits Candida albicans growth via inducing ROS generation and altering fungal cell membrane permeability. Front. Microbiol. 10:3150. doi: 10.3389/fmicb.2019.03150
Shagaghi, N., Bhave, M., Palombo, E. A., and Clayton, A. H. (2017). Revealing the sequence of interactions of PuroA peptide with Candida albicans cells by live-cell imaging. Sci. Rep. 7:43542. doi: 10.1038/srep43542
Shen, J., Lu, R., Cai, Q., Fan, L., Yan, W., Zhu, Z., et al. (2021). Mangiferin enhances the antifungal activities of caspofungin by destroying polyamine accumulation. Virulence 12, 217–230. doi: 10.1080/21505594.2020.1870079
Shirazi, F., and Kontoyiannis, D. P. (2015). Micafungin triggers caspase-dependent apoptosis in Candida albicans and Candida parapsilosis biofilms, including caspofungin non-susceptible isolates. Virulence 6, 385–394. doi: 10.1080/21505594.2015.1027479
Sovadinová, I., Kuroda, K., and Palermo, E. F. (2021). Unexpected enhancement of antimicrobial polymer activity against Staphylococcus aureus in the presence of fetal bovine serum. Molecules 26:4512. doi: 10.3390/molecules26154512
Staniszewska, M., Bondaryk, M., Swoboda-Kopec, E., Siennicka, K., Sygitowicz, G., and Kurzatkowski, W. (2013). Candida albicans morphologies revealed by scanning electron microscopy analysis. Braz. J. Microbiol. 44, 813–821. doi: 10.1590/S1517-83822013005000056
Sun, C. Q., Peng, J., Yang, L. B., Jiao, Z. L., Zhou, L. X., Tao, R. Y., et al. (2022). A Cecropin-4 derived peptide C18 inhibits Candida albicans by disturbing mitochondrial function. Front. Microbiol. 13:872322. doi: 10.3389/fmicb.2022.872322
Taff, H. T., Mitchell, K. F., Edward, J. A., and Andes, D. R. (2013). Mechanisms of Candida biofilm drug resistance. Future Microbiol. 8, 1325–1337. doi: 10.2217/fmb.13.101
Tang, W. H., Wang, C. F., and Liao, Y. D. (2021). Fetal bovine serum albumin inhibits antimicrobial peptide activity and binds drug only in complex with α1-antitrypsin. Sci. Rep. 11:1267. doi: 10.1038/s41598-020-80540-6
Theberge, S., Semlali, A., Alamri, A., Leung, K. P., and Rouabhia, M. (2013). C. albicans growth, transition, biofilm formation, and gene expression modulation by antimicrobial decapeptide KSL-W. BMC Microbiol. 13:246. doi: 10.1186/1471-2180-13-246
Tsang, P. W., Bandara, H. M., and Fong, W. P. (2012). Purpurin suppresses Candida albicans biofilm formation and hyphal development. PLoS One 7:e50866. doi: 10.1371/journal.pone.0050866
Tsui, C., Kong, E. F., and Jabra-Rizk, M. A. (2016). Pathogenesis of Candida albicans biofilm. Pathog. Dis. 74:ftw018. doi: 10.1093/femspd/ftw018
Van Dijck, P., Sjollema, J., Cammue, B. P., Lagrou, K., Berman, J., d'Enfert, C., et al. (2018). Methodologies for in vitro and in vivo evaluation of efficacy of antifungal and antibiofilm agents and surface coatings against fungal biofilms. Microb. Cell. 5, 300–326. doi: 10.15698/mic2018.07.638
van Gent, M. E., Ali, M., Nibbering, P. H., and Kłodzińska, S. N. (2021). Current advances in lipid and polymeric antimicrobial peptide delivery systems and coatings for the prevention and treatment of bacterial infections. Pharmaceutics 13:1840. doi: 10.3390/pharmaceutics13111840
Vitale, R. G., Afeltra, J., and Dannaoui, E. (2005). Antifungal combinations. Methods Mol. Med. 118, 143–152. doi: 10.1385/1-59259-943-5:143
Vriens, K., Cammue, B. P., and Thevissen, K. (2014). Antifungal plant defensins: mechanisms of action and production. Molecules 19, 12280–12303. doi: 10.3390/molecules190812280
Vriens, K., Cools, T. L., Harvey, P. J., Craik, D. J., Spincemaille, P., Cassiman, D., et al. (2015). Synergistic activity of the plant Defensin HsAFP1 and Caspofungin against Candida albicans biofilms and planktonic cultures. PLoS One 10:e0132701. doi: 10.1371/journal.pone.0132701
Wang, K., Dang, W., Xie, J., Zhu, R., Sun, M., Jia, F., et al. (2015). Antimicrobial peptide protonectin disturbs the membrane integrity and induces ROS production in yeast cells. Biochim. Biophys. Acta 1848, 2365–2373. doi: 10.1016/j.bbamem.2015.07.008
Wolak, N., Tomasi, M., Kozik, A., and Rapala-Kozik, M. (2015). Characterization of thiamine uptake and utilization in Candida spp. subjected to oxidative stress. Acta Biochim. Pol. 62, 445–455. doi: 10.18388/abp.2015_1044
Youle, R. J., and van der Bliek, A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. doi: 10.1126/science.1219855
Keywords: antimicrobial peptides, cecropin, Candida, candidiasis, biofilm
Citation: Guevara-Lora I, Bras G, Juszczak M, Karkowska-Kuleta J, Gorecki A, Manrique-Moreno M, Dymek J, Pyza E, Kozik A and Rapala-Kozik M (2023) Cecropin D-derived synthetic peptides in the fight against Candida albicans cell filamentation and biofilm formation. Front. Microbiol. 13:1045984. doi: 10.3389/fmicb.2022.1045984
Received: 20 September 2022; Accepted: 28 December 2022;
Published: 13 January 2023.
Edited by:
Yue Qu, The Alfred Hospital, AustraliaReviewed by:
Jianhua Wang, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2023 Guevara-Lora, Bras, Juszczak, Karkowska-Kuleta, Gorecki, Manrique-Moreno, Dymek, Pyza, Kozik and Rapala-Kozik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Rapala-Kozik, ✉ bWFyaWEucmFwYWxhLWtvemlrQHVqLmVkdS5wbA==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.