- 1College of Resources and Environment, Academy of Agricultural Sciences, Southwest University, Chongqing, China
- 2Interdisciplinary Research Center for Agriculture Green Development in Yangtze River Basin, Southwest University, Chongqing, China
- 3Sugarcane Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, China
- 4Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, St Lucia, QLD, Australia
- 5College of Resource Sciences, Sichuan Agricultural University, Chengdu, China
Introduction: A better understanding of the regulatory role of microorganisms on soil phosphorous (P) mobilization is critical for developing sustainable fertilization practices and reducing P resource scarcity. The phoD genes regulate soil organic P (Po) mobilization.
Methods: Based on the long-term P application experiments in acid purple soil of maize system in Southwest China (started in 2010), the experiment included five P levels: 0, 16, 33, 49, and 65.5 kg P hm–2 (P0, P16, P33, P49, and P65.5, respectively). The molecular speciation of organic P in soil was determined by 31P-nuclear magnetic resonance (NMR), high-throughput sequencing technology, and real-time qPCR were used to analyze the bacterial community and abundance of phoD-harboring bacterial genes, exploring the bacterial community and abundance characteristics of phoD gene and its relationship with the forms of Po and alkaline phosphatase (ALP) activity in the soil.
Results: The results showed that the orthophosphate monoesters (OM) were the main Po speciation and varied by P fertilization in acid purple soil. ALP activity decreased as P fertilization increased. Co-occurrence network analysis identified the overall network under five P fertilizations. The keystone taxon base on the network showed that Collimonas, Roseateles, Mesorhizobium, and Cellulomonas positively correlated with both OM and Po. The random forest showed that Cellulomonas, Roseateles, and Rhodoplanes were the key predictors for ALP activity. The keystone taxon was a more important predictor than the dominant taxon for ALP, OM, and Po. The structural equation model (SEM) showed that soil organic matter (SOM), available P (AP), and OM were the main factors influencing the ALP by reshaping phoD-harboring bacteria alpha diversity, community composition, and phoD abundance.
Discussion: The phoD-harboring bacterial community composition especially the keystone taxon rather than alpha diversity and abundance dominated the ALP activity, which could promote P utilization over an intensive agroecosystem. These findings improve the understanding of how long-term gradient fertilization influences the community composition and function of P-solubilizing microorganisms in acid purple soil.
Introduction
Excessive mineral P input in agricultural soil beyond the crop’s need to boost productivity is widespread in intensive agroecosystems (MacDonald et al., 2011; Lou et al., 2015; Zhu et al., 2018). P fertilizer is applied in the field soil, the majority of which is fixed in soils rather than absorbed by plants (Kochian, 2012). The residual P caused by the continuous accumulation resulted in the potential eutrophication of surface water (Powers et al., 2016). It is estimated that the global mineral P reserves are evaluated to be completely depleted by 2100 (Penuelas et al., 2013). Therefore, the ecologically friendly management of fertilization for reducing the use of P and improving P use efficiency is urgently needed for both sustainable agricultural production and environmental health.
Organic P is an important part of the soil P pool. The average content of total P in the topsoil of agroecosystems globally is estimated to be 1,762 kg ha–1, of which the Po accounts for 42% (747 kg ha–1) (Stutter et al., 2012; Menezes-Blackburn et al., 2018). The orthophosphate monoesters (OM) content measured by 31P-nuclear magnetic resonance (NMR) showed no consistent results (unchanged, increased, and decreased) with applied long-term Pi fertilization (Gatiboni et al., 2013; Cade-Menun et al., 2017; van der Bom et al., 2019). Further, little is known about the effect of long-term mineral P fertilization on the Po speciation in acid purple soil commonly seen in Southwest China.
Microorganisms play a key role in the biogeochemical degradation of Po, as they can excrete extracellular enzymes such as phosphatase, including alkaline phosphatase (ALP) and acid phosphatase, in soils. Three bacterial homologous genes encoding phosphatase involved in the production of ALP (phoA, phoD, and phoX) have been identified (Kathuria and Martiny, 2011). phoD is the most common ALP gene, which is considered the key ALP gene in soil (Tan et al., 2013), and P supply level is an important factor influencing the community composition of phoD-harboring bacteria. With sufficient soil P, Actinobacteria and Cyanobacteria are the most important groups that mediate Po mineralization in rice soil, while Methylbacillus species mainly facilitate Po mineralization under the condition of sufficient P (Wei et al., 2019). P supply level and carbon source provided by roots synergistically affect the community composition of phoD-harboring bacteria in the rice rhizosphere. Among them, Nostocales dominate under low P conditions, while Rhizobiales and Rhodospirillales dominate under moderate and high P conditions, respectively. These dominant populations mainly use the carbon source provided by plants (Long et al., 2018), which suggests that the soil P supply level and plants co-determine the community composition of phoD-harboring bacteria. It was found that phoD-harboring bacteria promote ALP secretion and thus mineralization of Po in both the traditional intensive agricultural and natural grassland soils (Fraser et al., 2015). Further research found that ALP secreted by microorganisms mainly mineralizes unstable forms of Po (Luo et al., 2017), which showed that the community characteristics of phoD-harboring bacteria are closely related to Po speciation. Therefore, under different P supply levels, the community characteristics of phoD-harboring bacteria (including alpha diversity, beta diversity, community composition, network characteristics, and keystone taxon), Po speciation, ALP activity, and their interactions need more detailed and systematic study.
This study aimed to provide a systematic knowledge of how the microbiota relevant to Po mineralization was affected by gradient P fertilization in acid purple soil. We hypothesized that moderate P fertilizer application would increase the phoD-harboring bacterial α-diversity and promote the complexity and stability of microbial network structure with conditions favorable for Po mineralization in intensive maize crop production. To test this hypothesis, we studied the phoD-harboring bacterial abundance, α-diversity, microbial community composition, structure, network, and keystone taxa in the soil as affected by different rates of P fertilizer applied to maize crops. For this purpose, soil samples were collected from a long-term field experiment that had been continuously fertilized for 10 years, and the phoD gene was used to quantify and identify the soil bacterial abundance, communities, and keystone taxon.
Materials and methods
Field site situation and experiment design
The experimental site is located at the Yaan Experimental Station of the Sichuan Agricultural University, Yaan city, Sichuan Province, China (29°58′59.1″N, 102°58′56.4″E). Yaan city is a typical area of intensive agriculture on acid purple soil in Southwest China, and more than 80% of the agricultural fields in the county follow a wheat-maize rotation. The site is located at an altitude of 600 m, and it has a subtropical monsoon humid climate with annual average precipitation and temperature of 1,732 mm and 13.2°C, respectively. It receives on average 218 days of rainfall annually, with about 50% of precipitation falling in summer and about 30% during the wheat growing season. The soil is a purple moist rudiment soil, and its basic soil properties are shown in Supplementary Table 1.
Experimental design
The field experiment was conducted in October 2010 in a farmland under monoculture maize cropping for 10 years without fertilization just prior to this experiment. During the entire duration of this field experiment, in each rotation year, maize (variety Zhongyu3) was planted at the beginning of April and harvested at the end of August. To build up a range of soil AP levels quickly in the experimental area, 0, 16, 33, 49, and 65.5 kg P ha–1 were applied for maize. Each treatment had three replicated plots, with each plot having an area of 26.6 m2 (2.8 m × 9.5 m).
At the end of each crop season, straw was removed before the next maize was sown, fertilizers were then broadcasted and mixed into the topsoil by plowing: P fertilizer was applied as calcium superphosphate, N as urea (75 kg N ha–1), and K as potassium sulfate (50 kg K ha–1). About 150 kg N ha–1 as urea was top-dressed at the jointing stage for wheat and at the 12-leaf stage for maize. To compensate for the low precipitation in the growing season, maize was flood irrigated with 50 mm of water once at sowing.
Sampling and analysis
Soil samples were taken on the maize silking stage on 2 July 2019. From each plot, soil cores from five different locations were taken from 20 cm depth between plant rows, which were combined as a single replicate sample. The combined soil sample was sieved with 2 mm mesh and then divided into two portions. One portion was stored at –20°C for DNA extraction and subsequent amplicon sequencing, and the other portion was air-dried for the analysis of physiochemical properties. There were a total of 15 soil samples, and each soil sample was analyzed as two parallel samples. Soil available P (AP) was extracted with 0.5 M NaHCO3 at pH 8.5 and then determined by colorimetry (Olsen and Sommers, 1982). The acid digestion method was used to determine the total P in soil (Tiessen and Moir, 1993). Po was determined by the ignition method (Saunders and Williams, 1955). Pi was calculated as the total P minus Po. Soil pH was determined in a 1:2.5 (soil/water ratio) suspension with a pH meter. Soil organic carbon (SOC) was determined by the external-heat potassium dichromate oxidation-colorimetric method (Nelson and Sommers, 1983). Soil total N (TN) was measured by dry combustion with an elemental analyzer (Vario max CN, Elementar, Hanau, Germany). The ALP was determined using the method described by Tabatabai (1994).
Soil P speciation by solution-state 31P-NMR spectroscopy
P speciation in soils was identified by 31P-NMR spectroscopy following the method of Cade-Menun and Preston (1996). The MestReC software (version 4.9.9.9) was used to correct and process the 31P spectra. The free induction decays for solution-state 31P-NMR spectra were transformed by using a four-fold zero filling and a line broadening of 6 Hz. The individual peaks of the signal areas were calculated from a deconvolution process. The MDP was used as the internal standard in the solution for NMR analyses and it also calibrated the frequency axis, standardized data, and performed a quantitative assessment of P speciation. Signals were assigned based on the references (Cade-Menun, 2005; McLaren et al., 2015).
DNA extraction and gene quantification
DNA extract was used as 0.25 g soil per sample with a DNeasy PowerSoil (QIAGEN, Germany) according to the manufacturer’s instructions. A spectrophotometer (Nanodrop ND-1000, Thermo Scientific, Waltham, MA, United States) was used to analyze the quality of extracted DNA. QuantiFluor® dsDNA system (Promega, Madison, WI, United States) was used to determine the concentration of DNA based on fluorometric analysis with a microplate reader (Spectramax M5, Molecular Devices, Sunnyvale, CA, United States). DNA samples were stored at −20°C until they were used for analyses. Real-time quantitative PCR (qPCR) was used to determine the bacterial phoD gene abundance with the primers of ALPS-F730/ALPS-R1101 (CAGTGGGACGACCACGAGGT/GAGGCCGATCGGCATG TCG) (Sakurai et al., 2008). The real-time qPCR system was in a 20-μl reaction system with triplicates. LightCycler 480 (Roche, Basel, Switzerland) was used to quantify the gene copy number. The PCR reaction conditions used for the phoD gene are as follows: 3 min at 94°C, 40 cycles at 95°C for 15 s, 56°C for 30 s, 72°C for 15 s, and finally 1 min at 72°C. The standard curve was prepared in triplicate with six serial 10-fold dilutions of standard plasmids carrying the phoD gene, and the gene copy number was calculated based on the concentration of plasmid DNA and the base pairs therein (vector plus primer). The efficiency of each reaction was 92.41%.
PCR amplification, high-throughput sequencing, and data analysis
The primers ALPS-F730/ALPS-R1101 were used to amplify the bacterial phoD gene (Sakurai et al., 2008), which was the same as in the real-time qPCR experiment. The protocol of the amplification for the bacterial phoD gene was as follows: 3 min at 94°C, followed by 40 cycles at 95°C for 15 s, 30 s at 56°C, 72°C for 15 s, and finally 1 min at 72°C. Universal DNA Purification Kit (Tiangen, Beijing, China) and the QuantiFluor® dsDNA system were used to purify and quantify the PCR products, respectively. The purified amplicons were pooled in equimolar volumes and subjected to paired-end sequencing using the Illumina Hiseq 2500 platform (Novogene, Beijing, China).
All the raw sequencing data were purified by the Quantitative Insights Into Microbial Ecology (QIIME) system. The raw data filtered the reads which contained ambiguous-based nucleotide mismatches within the primer or barcode, and reads shorter than 200 bp would be removed before analysis. UCHIME was used to identify and eliminate chimeric sequences. UCLUST clustering was used to cluster Operational taxonomic units (OTUs) at a cutoff of 75% similarity (Fraser et al., 2015). The RDP classifier was used to retrieve and classify the representative sequences. Singletons were removed during this clustering process. Subsequently, Local Blast 2.2.27+1 was used to taxonomically assign the representative OTUs, and the P-solubilizing bacterial taxon was annotated with critical criteria (E < 1 × 10–10 and sequence identity >99%) (Zheng et al., 2017). In our study, a total of 3,103,247 raw sequences were obtained for high-quality filtered sequences. All sequences were deposited in the NCBI Sequence Read Archive database (accession number PRJNA787763).
Calculations and statistical analysis
One-way analysis of variance was used to examine the significant differences in soil physiochemical properties, abundance, and α-diversity of phoD-harboring bacterial data between the five gradient fertilization treatments and Duncan’s multiple range test at the 0.05 level were used to compare with the mean values with SPSS version 20.0 software package (SPSS, Inc., Chicago, IL, United States). Permutational multivariate analysis of variance was used to investigate the phoD-harboring bacterial community affected by gradient P fertilization (Anderson, 2001), which was performed by a Vegan package in R (version 3.3.1). The effects of gradient fertilization on phoD-harboring bacterial communities were determined by the analysis of similarities. The “analysis of similarities” function statistically tests the significant effect among treatments. The effects of the soil physiochemical on phoD-harboring bacteria were determined by Redundancy analysis (RDA) using Vegan; the RDA model selected the significant environmental variables (P < 0.05) (measured based on 999 permutations). Spearman correlation coefficient analyses were conducted to conclude the networks with phoD-harboring bacterial OTU-OTU interaction as described by Faust et al. (2012) and Pérez-Jaramillo et al. (2019). The OTUs were selected from the top 500 at the taxonomic level to perform the network analysis. The classification of community composition level was genus. Co-occurrence network visualization was achieved with the Gephi platform (version 0.9.2) (Bastian et al., 2009). The importance of Po, OM, and ALP to keystone taxon was estimated by the Random forest model (Trivedi et al., 2016). Mean squared error (MES%) was used to select the significant important variables (Breiman, 2001). A3 package was used to evaluate the R2 values for cross-validation and the significance of the model. The “rfPermute” package was used to assess the significance of variables. The structural equation modeling (SEM) framework was conducted to estimate the relationships among soil physiochemical properties (including P rates, SOM, AP, and OM), phoD gene abundance, alpha diversity, community composition of the phoD-harboring bacteria, and ALP activities using AMOS software (IBM SPSS AMOS 17.0.0). The maximum likelihood estimation and the chi-square test (v2) were used to conduct the best-fit model, the overall goodness of the model fit used the root mean square errors of approximation (RMSEA < 0.05) and comparative fit index (CFI) (>0.90).
Results
Soil properties and P status
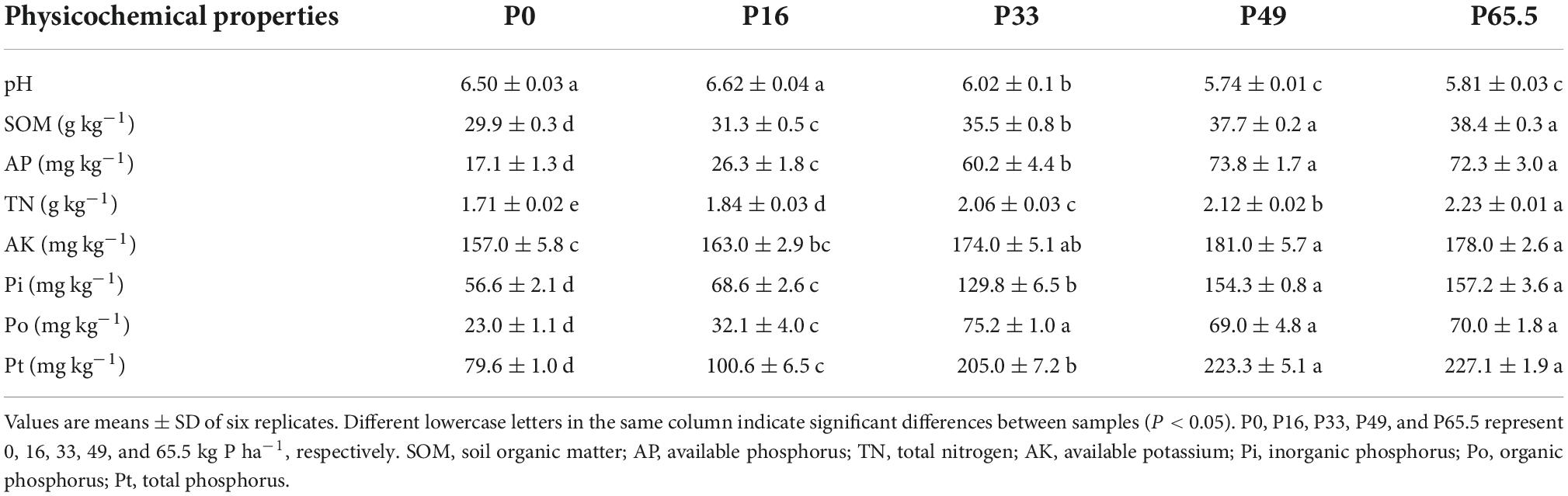
P fertilization had significant effects on soil properties (Table 1). Soil pH decreased significantly as the P fertilization rate increased; SOM, TN, AK, AP, Pi, Po, and Pt were increased as the P fertilization rate increased (Table 1). 31P-NMR spectra for the soil samples are shown in Figure 1A. Po speciation measured by 31P-NMR showed that the OM was the main organic species and it increased with increased P fertilization. A similar pattern was evident for orthophosphate diester (OD) as well (Figure 1B).
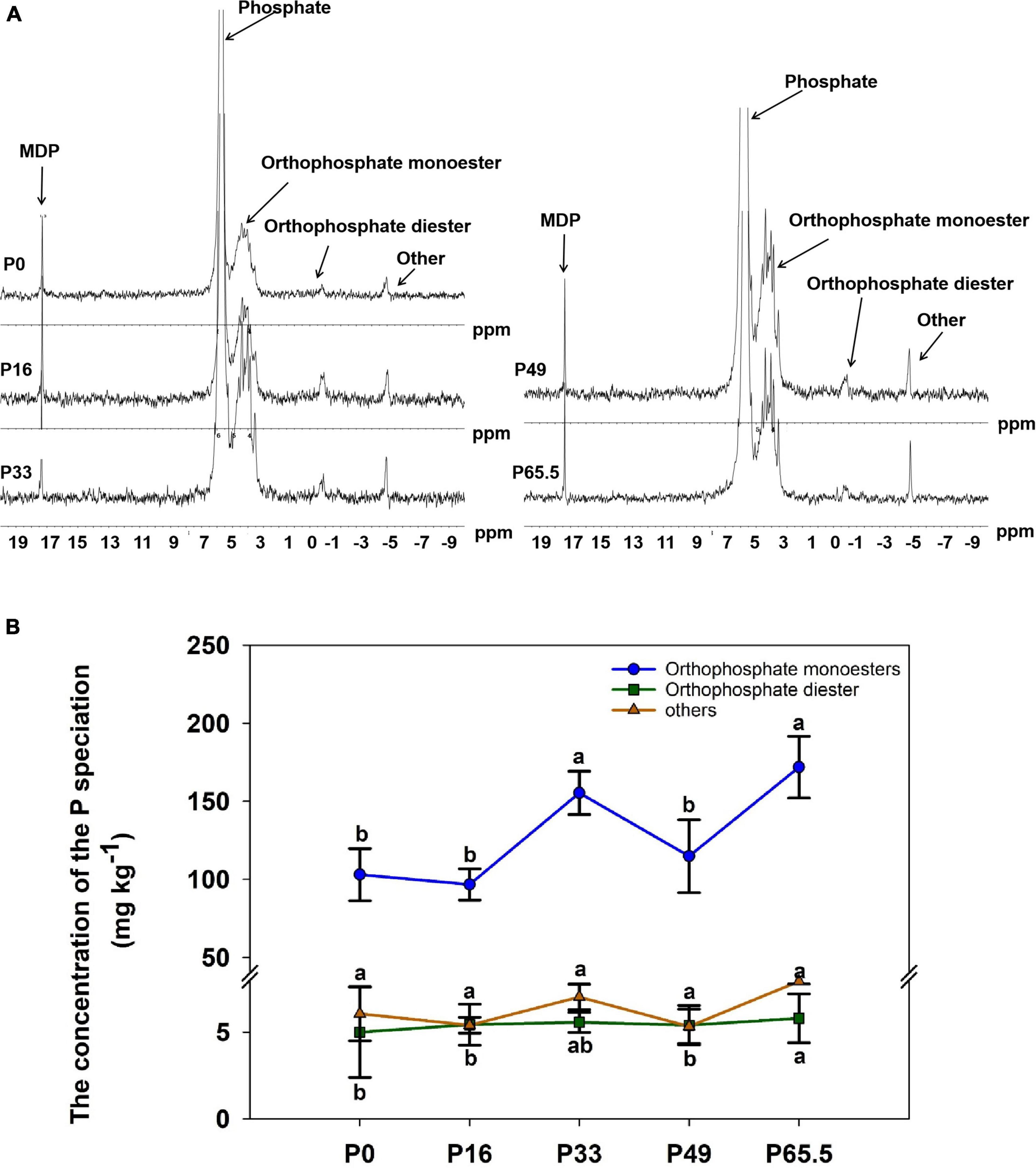
Figure 1. The concentration of the P speciation under gradient P fertilization rates. (A) Solution 31P NMR (nuclear magnetic resonance) spectra on NaOH-EDTA extracts for the five P fertilization rates. Nuclear magnetic resonance can be used to identify the exact molecular forms of P in soils. These include phosphate (δ 5.83 ppm), orthophosphate monoester (OM) (δ 4.75 ppm), orthophosphate diester (OD) (δ –0.44 ppm), and others (δ –4.76 ppm). (B) Line charts based on 31P NMR. The data are based on the concentration of organophosphorus molecular form at five P fertilization rates. P0, P16, P33, P49, and P65.5 represent 0, 16, 33, 49, and 65.5 kg P ha–1 applied, respectively.
Abundance of phoD, community diversity, and composition of phoD-harboring bacterial communities under long-term gradient P fertilization
The phoD gene abundance was not significantly altered by the gradient P fertilization (Figure 2A), whereas the activity of ALP was decreased as P fertilization decreased (Figure 2B). The alpha diversity was estimated by the Sob index and the Shannon–Wiener index. Generally, there were no significant differences in the phoD-harboring Sob index and the Shannon–Wiener index among all five treatments except for the P16 treatment (Figures 3A,B), but the phoD-harboring bacterial community structure was significantly affected by P fertilization (Figure 3C and Supplementary Table 3). Based on the composition of all soil samples, the dominant phoD-harboring bacterial phyla were Proteobacteria and Actinobacteria, reaching up to 84.7 and 14.7%, respectively (Supplementary Figure 1A). The dominant phoD-harboring bacterial class was Alpha-proteobacteria, Beta-proteobacteria, and Actinobacteria, reaching up to 59.5, 19.8, and 14.6%, respectively (Supplementary Figure 1B). A total of 10 genera were aligned, and the most dominant genera belonged to Bradyrhizobium (43.2%), Streptomyces (7.8%), and Burkholderia (5.5%) (Supplementary Figure 2). The relative abundance of Bradyrhizobium was highest in P0 treatment and it decreased with increasing P fertilization (Supplementary Figure 2A). The relative abundance of Streptomyces increased first and then decreased (Supplementary Figure 2B), but there was no significant difference among five P fertilization treatments (Supplementary Figure 2C). The random forest showed that Collimonas, Rhodoplanes, Roseateles, Pseudomonas, and Mesorhizobium were the key predictors for OM and Po (Figure 4). Moreover, Mesorhizobium and Burkholderia were the key predictors of ALP activity (Supplementary Figure 3).
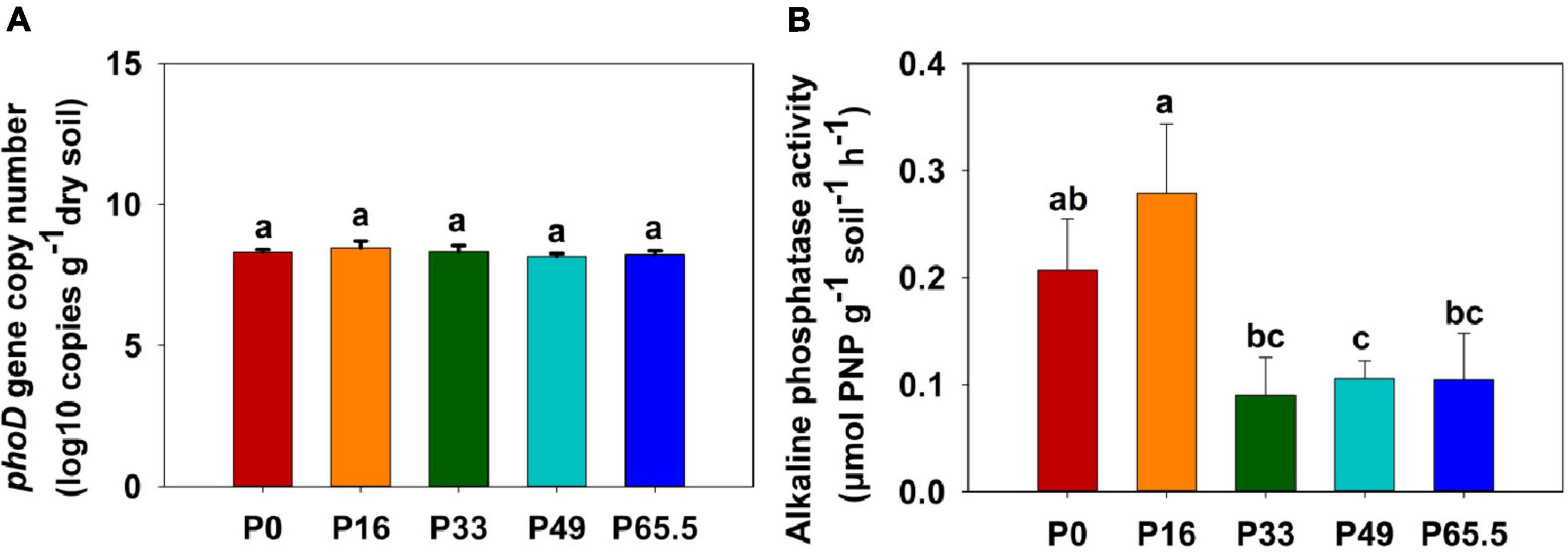
Figure 2. phoD gene abundance (A) and alkaline phosphatase (ALP) (B) in soils treated with five phosphorus fertilization rates. Small letters indicate a significant difference (P < 0.05 after the Duncan test) between P levels. P0, P16, P33, P49, and P65.5 represent 0, 16, 33, 49, and 65.5 kg P ha–1, respectively.
Figure 3. Alpha diversity (A,B) and redundancy analysis (RDA) (C) of phoD-harboring bacteria under gradient P fertilization. In (A,B), small letters indicate significant differences (P < 0.05 after the Duncan test) between P levels. In (C), RDA was used to explore the relationship between the entire phoD-harboring bacterial community and selected soil properties. **Correlation is significant at the 0.01 level. P0, P16, P33, P49, and P65.5 represent 0, 16, 33, 49, and 65.5 kg P ha–1, respectively.
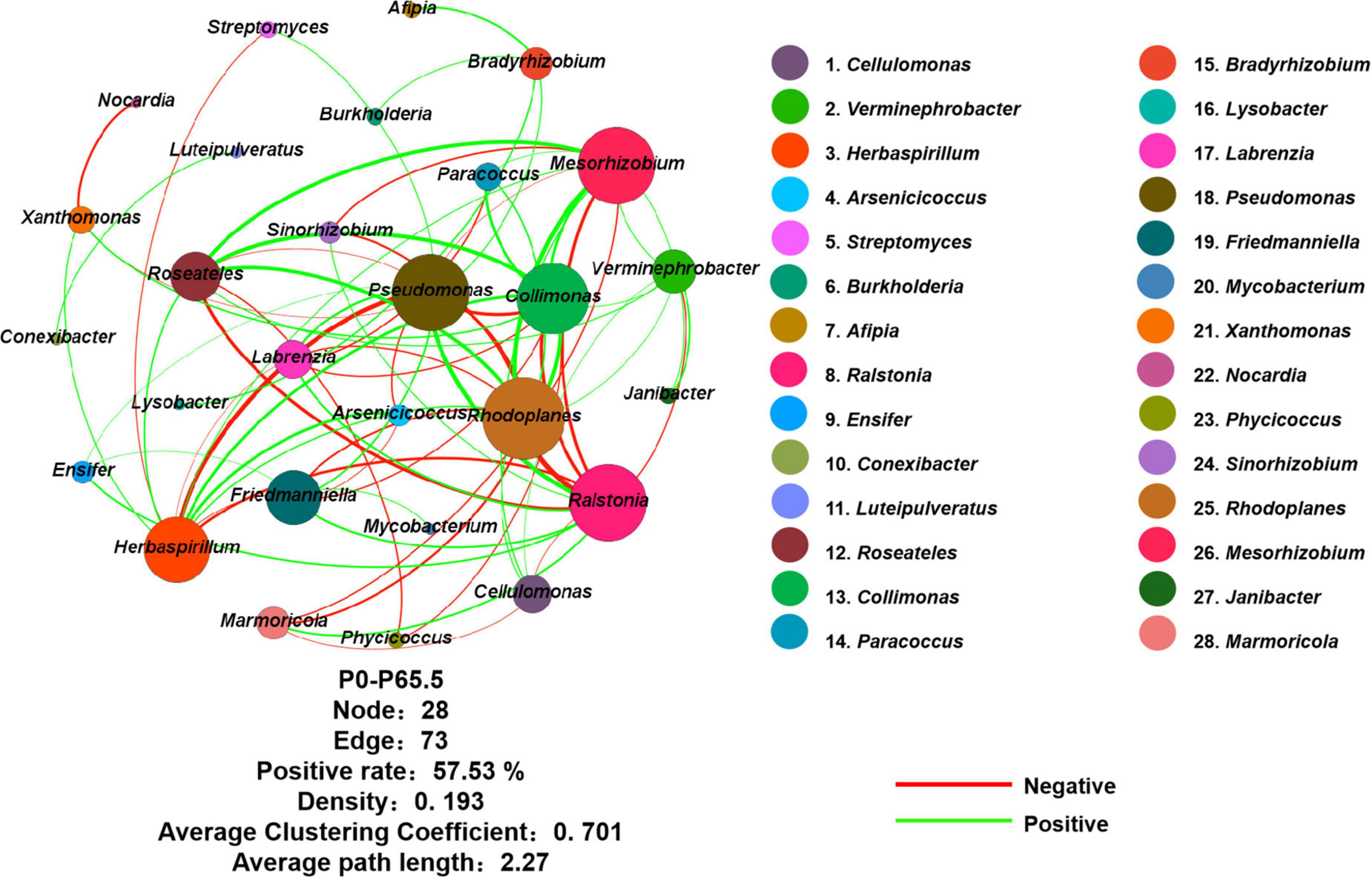
Figure 4. Network of phoD-harboring bacteria based on Spearman correlation analysis from OTU profiles. The network consists of five gradient phosphorus fertilization rates (including P0, P16, P33, P49, and P65.5). The color of the nodes represents the taxa of the genus classification. Red lines represent a positive correlation, and green lines indicate a negative correlation. Node, edge, positive rate, density, average clustering coefficient, and average path length of the topological parameters were shown below the network. P0, P16, P33, P49, and P65.5 represent 0, 16, 33, 49, and 65.5 kg P ha–1, respectively.
Network structure and keystone taxa
Using co-occurrence network analysis, we identified the overall network under five P fertilization (Figure 4). The keystone taxon of the overall network was significantly affected by different rates of P fertilization (Figure 5). Among them, Collimonas, Roseateles, Mesorhizobium, and Cellulomonas were positively correlated with both OM and Po concentrations (Supplementary Figures 4, 5), while Pseudomonas, Ralstonia, and Friedmanniella were positively correlated with ALP (Figure 6). The random forest analysis showed that Rhodoplanes and Roseateles were the key predictors for both OM and Po (Figures 7A,B). Moreover, Cellulomonas, Roseateles, and Rhodoplanes were the key predictors of ALP activity (Figure 7C). Distinct network structures were identified separately for five P fertilization treatments used in this study (Supplementary Figure 6). The number of edges and average degree were increased as P fertilization increased (Supplementary Figure 6). The keystone taxon varied with the P fertilization rate (Supplementary Table 3). The majority of the keystone taxa were positively correlated with P forms including AP, Pi, Po, and Pt for P33, P49, and P65.5 treatments, while they were negatively correlated with SOM (Supplementary Figure 7). Both the Mantel test and Random Forest analysis were conducted to identify the importance of keystone and dominant taxon for Po mineralization (Figure 8). Mantel test showed that the ALP, OM, and Po were significantly correlated with the keystone taxon more than that of the dominant taxon (Figure 8A). Random Forest analysis showed that the keystone taxon was the most important predictor than the dominant taxa for ALP, OM, and Po (Figure 8B).
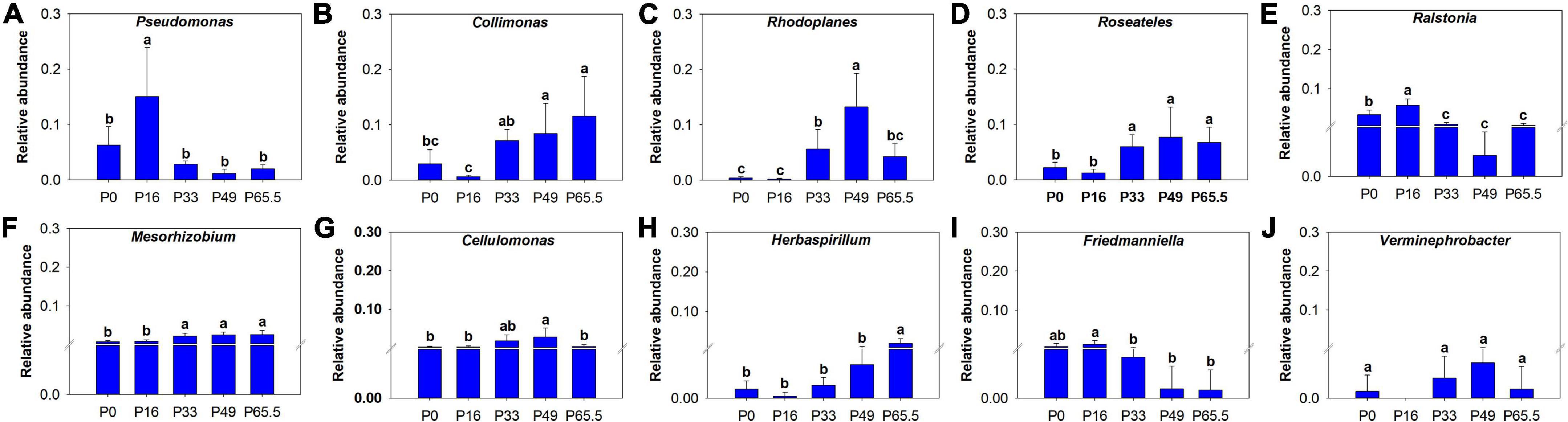
Figure 5. Bar chart of relative abundance of the keystone taxa at genus level under different phosphorus fertilization rates. (A) Pseudomonas; (B) Collimonas; (C) Rhodoplanes; (D) Roseateles; (E) Ralstonia; (F) Mesorhizobium; (G) Cellulomonas; (H) Herbaspirillum; (I) Friedmanniella; (J) Verminephrobacter. The key species were selected by ranking the top 10 “Degree” values based on the network. P0, P16, P33, P49, and P65.5 represent 0, 16, 33, 49, and 65.5 kg P ha–1, respectively.
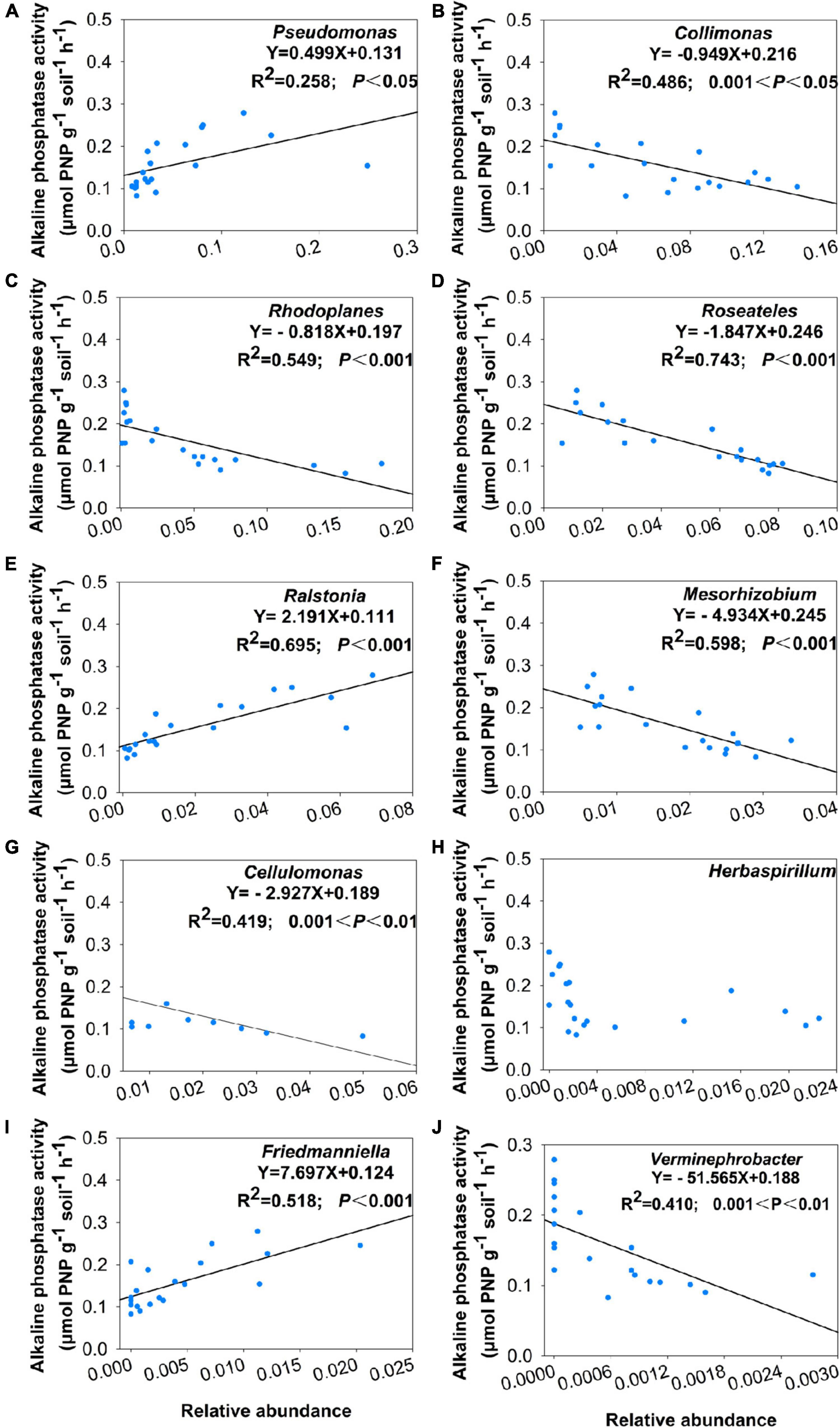
Figure 6. Correlation between alkaline phosphatase activity and relative abundance of keystone taxa which include (A) Pseudomonas, (B) Collimonas, (C) Rhodoplanes, (D) Roseateles, (E) Ralstonia, (F) Mesorhizobium, (G) Cellulomonas, (H) Herbaspirillum, (I) Friedmanniella, and (J) Verminephrobacter.
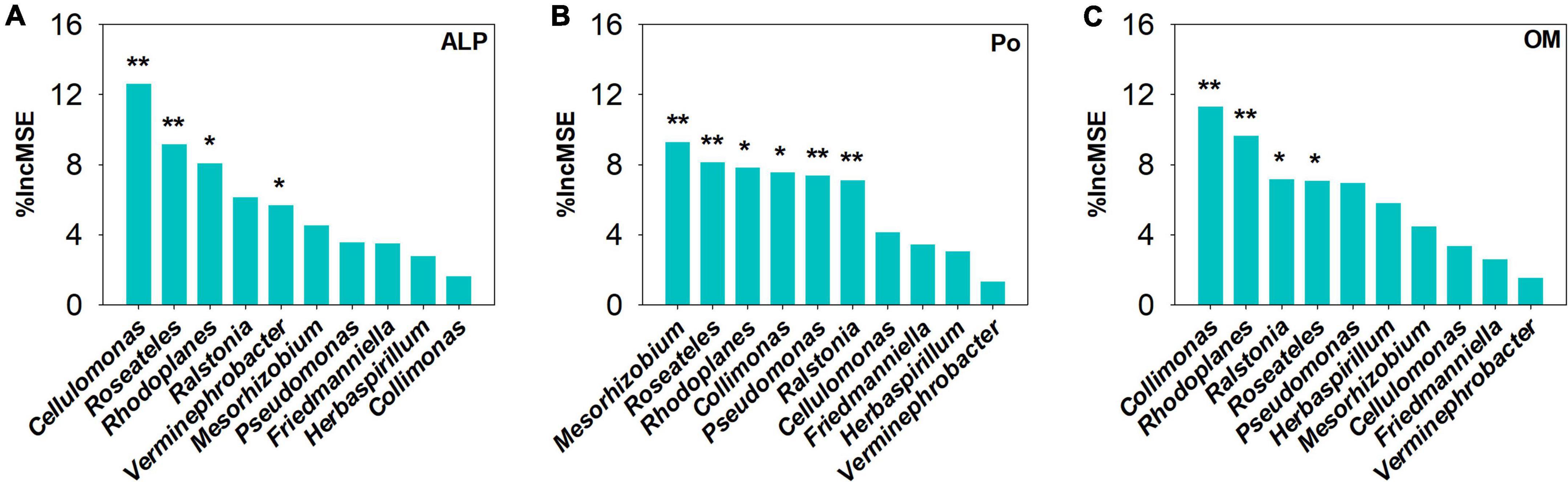
Figure 7. Random Forest analysis to identify the main predictors of keystone taxa on (A) alkaline phosphatases (ALP), (B) organophosphorus (Po), and (C) orthophosphate monoester. The mean decrease in accuracy (%IncMSE) was used to indicate the relative importance of each variable for predicting the soil’s total P concentration. *Correlation is significant at the 0.05 level; **correlation is significant at the 0.01 level.
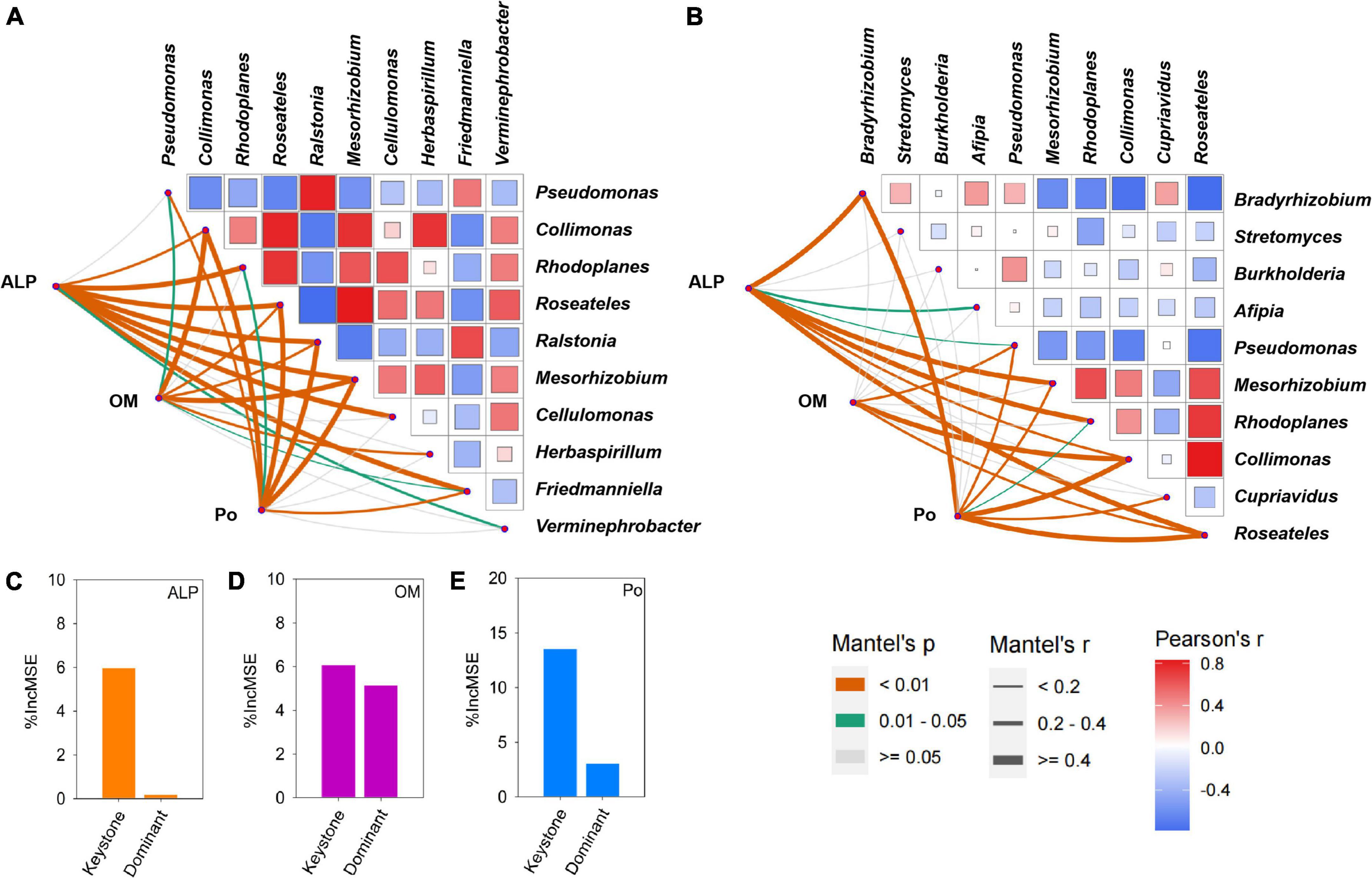
Figure 8. Correlation among alkaline phosphatases (ALP), orthophosphate monoester (OM), and organophosphorus (Po) with the keystone (A) and dominant taxon (B). Pairwise comparisons of the keystone and dominant taxon are separately shown in the heatmap with a color gradient to represent Spearman’s correlation coefficients. Statistically significant correlations among ALP, OM, and Po with the keystone and dominant taxon are shown in the corresponding lower left. Edge width corresponds to Mantel’s coefficients. The orange line means P < 0.01, the green line means 0.01 < P < 0.05, and the gray line means no significant effect. A random forest analysis to identify the importance of keystone taxon and dominant taxon to ALP (C), OM (D), and Po (E).
The main driver of soil properties with phoD abundance and community under long-term gradient P fertilization
A structural equation model (SEM) was constructed to further investigate the relationships among P application rates, phoD-harboring bacteria characteristics (including alpha diversity, community composition, and abundance), soil physicochemical factors (including SOM, AP, and OM), and ALP activity (Figure 9). P fertilization rates were significantly and positively related to SOM (path coefficient = 0.93) and OM (path coefficient = 0.67) and were negatively related to alpha diversity (path coefficient = −1.50). SOM and OM were significantly and positively related to alpha diversity, the path coefficients of 1.37 and 0.70, respectively. However, there was no significant correlation between alpha diversity and ALP activity. ALP activity was significantly negatively affected by phoD abundance (path coefficient = −0.37). The relationship between community composition (path coefficient = −1.07) and ALP activity was more significant than phoD abundance (path coefficient = −0.37) and alpha diversity (path coefficient = 0.18). However, AP was significantly and positively related to the community composition of phoD-harboring bacteria (Figure 9A). ALP activity was negatively affected indirectly by P fertilization rates, SOM, and AP, and positively affected by OM. phoD abundance and community composition had a direct negative effect on ALP activity. Alpha diversity has a direct positive effect on ALP activity (Figure 8B). In addition, the standardized total effect revealed that the P fertilization rate had a strong negative effect on ALP (Figure 9B).
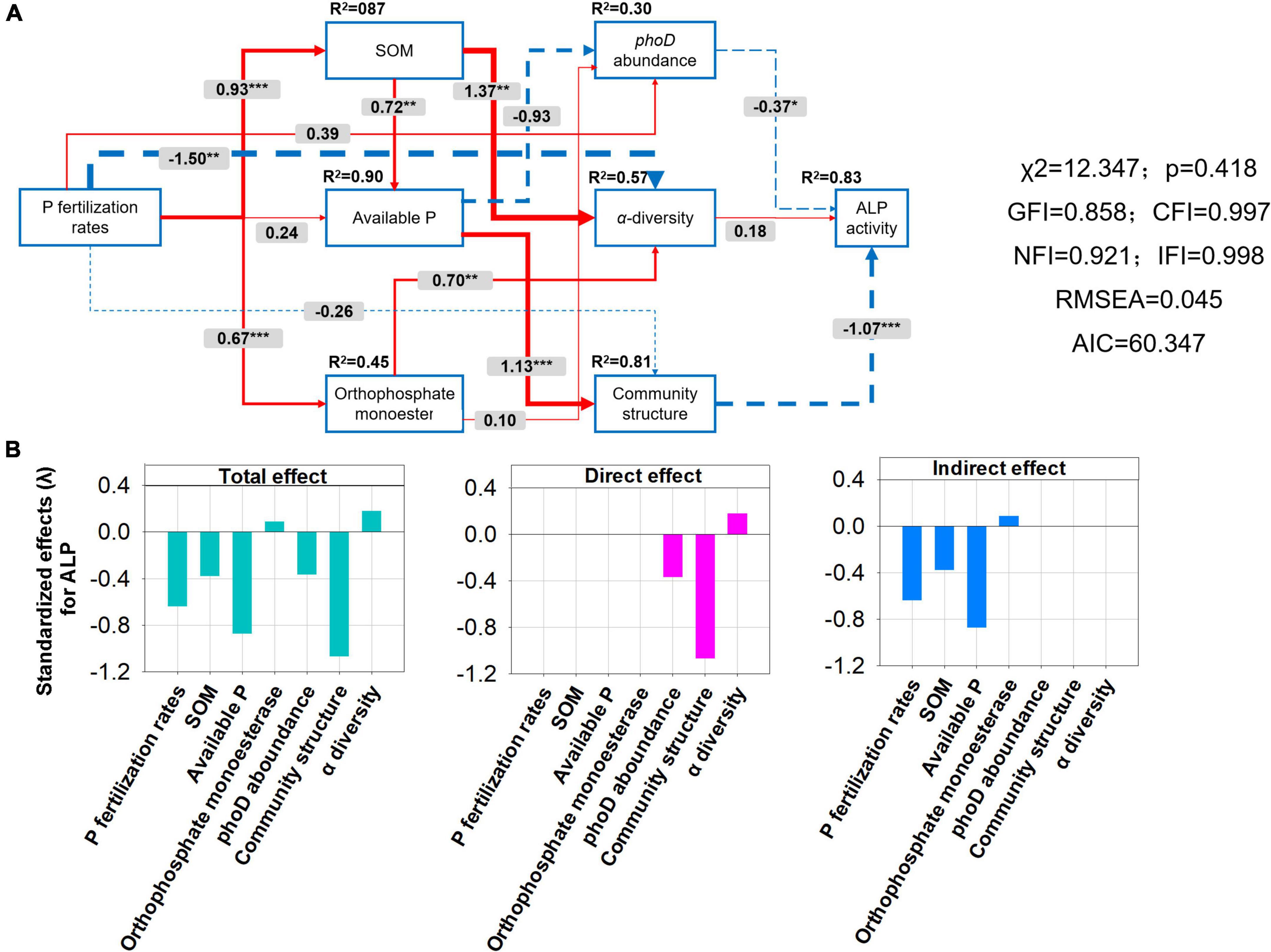
Figure 9. Structural equation model (SEM) (A) and bar plots of effect values (total, direct, indirect) on alkaline phosphatases (ALP) activity (B) showing the influence of various factors on the correlationship among soil properties [including soil organic matter (SOM), available P (AP), and orthophosphate monoesters (OM)], alpha diversity, phoD abundance, community structure, and ALP activity of soil under long-term gradient phosphorus fertilization rates. The width of the arrow indicates the strength of the causal effect. The red and blue arrows indicate the positive and negative relationships between the indicators. The number above the arrow indicates the path coefficient. “*”, “**”, and “*” represent significant path. The percentage above each indicator represents the R2 value, which is the variance-explained ratio of each variable. The final model fits the data well. The model is: (χ2 = 13.343, df = 1.026, CFI = 0.997, IFI = 0.997, RMSEA < 0.043, AIC = 59.343).
Discussion
Long-term gradient P fertilization affecting P speciation and the alkaline phosphatase activity
In the acidic purple soil, the major limiting factor of maize production is low P availability. Hence, the application of the mineral P fertilization is a common agricultural management practice to improve soil P fertility (Hu et al., 2018). In the current study, P fertilization greatly increased the AP and Pt. The soil mineralogy such as the Fe/Al minerals combine with the pH to co-determine the forms and the availability of P in soil (Hou et al., 2018). Additionally, Po and the main component of Po named OM are generally first increased and then remained unchanged with the increase of P supply level in acid purple soil (Table 1 and Figure 1). The evidence has shown that crop input (root residue) and microbial immobilization are the key ways to form Po in soil (Condron et al., 2014). The changes in root residue and Po show a similar pattern during 10 consecutive years of a wheat-maize rotation system in calcareous soil (Deng et al., 2017; Lang et al., 2021), which may be the case in our present study as well.
The indicator of the potential Po mineralization is ALP activity (Chen, 2003; Gao et al., 2022). ALP in soil hydrolyzes Po and then transforms into the AP forms that plants can absorb (Tabatabai, 1994; Turner and Haygarth, 2005). A previous study shows that P fertilization could enhance the activity of ALP (Fraser et al., 2015). However, the activity of ALP is found suppressed after long-term (30 years) fertilization (Zeng et al., 2022), and our results corroborated this finding, shown in Figure 2B. ALP activity in soil generally origin from microbes, and phoD-harboring bacteria secrete ALP, a common enzyme that is widespread in the bacterial kingdom (Ragot et al., 2015). Previous studies indicate that ALP activity is affected by some factors including soil physicochemical properties, the interactions of the soil organism, and the inputs of the nutrient (Bünemann et al., 2011), and the effects of the phoD-harboring bacterial community vary for P fertilizer application among sites in specific environments (Fraser et al., 2015; Ikoyi et al., 2018; Chen et al., 2019).
Long-term gradient P fertilization affecting the abundance and community composition of phoD-harboring bacteria
Our results showed that P fertilization significantly affected the phoD-harboring bacterial community structure and composition, but not the phoD gene abundance. Previous studies demonstrate that the phoD-harboring bacterial community is more important than the gene abundance in regulating the expression of phosphatase (Bi et al., 2020; Wan et al., 2020). Meanwhile, in the agroecosystem, Fraser et al. (2015) show that inorganic (mineral) and organic (manure) P fertilizer inputs solely affect the community of phoD bacteria but not the copy numbers of phoD gene, demonstrating that the community of phoD-harboring bacteria is more sensitive than that of gene abundance. The dominant phoD-harboring bacteria in our study were Proteobacteria and Actinobacteria, which were in agreement with the previous studies (Lagos et al., 2016; Zheng et al., 2017; Hu et al., 2018). The Proteobacteria and Bacilli in the phoD-harboring communities belonging to the copiotrophic bacteria enriched in the abundant P supplies (with P33, P49, and P65.5 treatments). Bradyrhizobium and Methylobacterium belonged to the Rhizobials enriched in the P33–65.5 treatment, which are generally dominant in soil where the AP is relatively high (Ragot et al., 2016; Long et al., 2018). The 18O-DNA-stable isotope probing coupled with high-throughput sequencing was used to characterize the active phoD-harboring bacterial communities, showing that the Bacillus is the dominant taxa in mineral fertilized soil (Liu et al., 2021), which is consistent with our results. Soil acidification is a common phenomenon under the long-term application of mineral fertilizer in an intensive agroecosystem (Guo et al., 2010; Yang et al., 2018), which was consistent with our results (Table 1). The previous study shows that pH is a key factor in determining Po mineralizing-related gene abundance, which consequently affected corresponding microbial diversity (Wan et al., 2020). The excessive input of the mineral fertilizers caused soil acidification, and soil P forms followed the P fertilization rate, which may alter microbial diversity and soil enzyme activity (Hartmann et al., 2015; Liu et al., 2019). Especially, the dominant taxa Bradyrhizobium, nitrogen-fixing symbiotic bacteria which requires a phosphate transport system (Bardin et al., 1996), increases soil AP according to the production of phosphatase or organic acids (Mander et al., 2012; Sharma et al., 2013; Ikoyi et al., 2018). These results reveal that the phoD-harboring bacterial community is more sensitive to P fertilization than the gene abundance.
The effect of the network and keystone taxa on organic P mineralization under long-term gradient P fertilization
The bacterial taxa interaction in niches and the keystone taxon influencing the corresponding bacterial communities can be analyzed by network analysis (Banerjee et al., 2016; Zheng et al., 2017). The keystone taxa obtained from phoD-harboring bacteria varied with P fertilization, and some of them were positively or negatively correlated with Po speciation and ALP (Figures 5, 6). Mesorhizobium, one of the plant growth-promoting rhizobacteria (PGPRs) (Chen et al., 2021) could produce plenty of metabolites that can promote plant growth (Baba et al., 2021). Ding et al. (2021) demonstrated that the concentrations of sugars and organic acids are coupled with the keystone taxon, such as Mesorhizobium and Bradyrhizobium. The genus Collimonas secreted enzymes such as phosphatases, peptidases, chitinases, and nucleases and had a specific role in weathering and mycophagy (Mela et al., 2012). The Collimonas strains can solubilize Pi and produce gluconic acid from glucose, demonstrating that acidification is one of the key mechanisms conducted by these bacteria for mineral weathering (Uroz et al., 2009). These kinds of taxa may have improved Pi dissolution and Po decomposition. The presence of Pseudomonas, Ralstonia, and Friedmanniella positivity correlated with ALP activity, which demonstrates that both genera may play an important role in P deficiency conditions. The previous study showed that ALP activity is revealed to be closely related to phoD-harboring bacteria with less abundance, but not with the diversity and abundance of phoD-harboring bacteria in subalpine forest ecosystems (Li et al., 2021). Collectively, these results suggest that some specific keystone taxon remain the key drivers of soil ALP activity. Furthermore, the importance of the keystone and dominant taxon analysis conducted by the mantel test and the Random Forest analysis (Figure 8) revealed that the keystone taxon contributed more than that of the dominant taxon to Po mineralization.
Long-term gradient P fertilization affecting the relationships between phoD-harboring bacterial communities and soil biogeochemical properties
Orthophosphate monoesters was the main specification present in acid purple soil, which directly affected the alpha diversity and phoD-abundance (Figures 1, 9). The previous study shows that Po availability shapes the diversity of phoD-harboring bacteria in agricultural soil, which highlights the key role that Po plays in the phoD-harboring bacterial community structuring (Wei et al., 2021). Furthermore, previous studies demonstrated that environmental factors such as pH, AP, and SOC determine the soil bacterial community in different ecosystems (Zhao et al., 2014; Zhou et al., 2015; Chen et al., 2019). More specifically, SOM and AP are the main factors determining the soil phoD-harboring bacterial community structures in the wheat fields (Liu et al., 2020). Furthermore, AP and ALP activity shows a significant correlation with the phoD-harboring bacterial community structure, and there is a significant correlation between SOC and the phoD-harboring bacteria community structure in Northeast China of maize field (Chen et al., 2019). Similarly, SOM and AP significantly affected the phoD-harboring bacterial community in our present study (Figure 9), demonstrating their key roles in the phoD-harboring bacterial community in acid purple soil.
Conclusion
In this study, a 10-year field experiment demonstrated that long-term excessive P fertilization significantly decreases soil ALP activity. The keystone taxon contributed more than that of the dominant taxon to Po mineralization. The keystone taxa Pseudomonas, Ralstonia, and Friedmanniella positively correlated with ALP activity. In the acid purple soil, the phoD-harboring bacterial communities were mainly driven by SOM, AP, and Po speciation. P solubilizing bacterial community composition especially the keystone taxon played a key role in regulating phosphatase activity than the alpha diversity and abundance, as well as promoting P utilization in intensive agroecosystems. Thus, our results further proved the potential of soil microorganisms for efficient P fertilization management in an intensive agroecosystem. In summary, our results revealed that applying moderate P fertilization could improve the P solubilizing bacterial community and facilitate P mineralization and utilization, which remarkably contributes toward sustainable agriculture. Future studies will verify these findings in more diverse agroecosystems at large spatial scales.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
ML and XC conceived the study. HL contributed to the data analysis of bioinformatics. ML and HL contributed to the soil sampling and drafting the article. ML, XC, PL, and YC contributed to the critical review and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Natural Science Foundation of China (32002126 and 32272800), the Fundamental Research Funds for the Central Universities (SWU-KR22010), the National Maize Production System in China (CARS-02), and the National Natural Science Foundation of China (U20A2047).
Acknowledgments
We thank Shuikuan Bei for his comments on this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1045919/full#supplementary-material
Footnotes
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral J. Ecol. 26, 32–46.
Baba, Z. A., Hamid, B., Sheikh, T. A., Alotaibi, S. H., El Enshasy, H. A., Ansari, M. J., et al. (2021). Psychrotolerant Mesorhizobium sp. Isolated from temperate and cold desert regions solubilizes potassium and produces multiple plant growth promoting metabolites. Molecules 26:5758. doi: 10.3390/molecules26195758
Banerjee, O., Cicowiez, M., Horridge, J. M., and Vargas, R. (2016). A conceptual framework for integrated economic-environmental modelling. J. Environ. Dev. 25, 276–305. doi: 10.1016/j.wasman.2016.01.017
Bardin, S., Dan, S., Osteras, M., and Finan, T. M. (1996). A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178, 4540–4547. doi: 10.1128/jb.178.15.4540-4547.1996
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3, 361–362. doi: 10.1609/icwsm.v3i1.13937
Bi, Q. F., Li, K. J., Zheng, B. X., Liu, X. P., Li, H. Z., Jin, B. J., et al. (2020). Partial replacement of inorganic phosphorus (P) by organic manure reshapes phosphate mobilizing bacterial community and promotes P bioavailability in a paddy soil. Sci. Total Environ. 703:134977. doi: 10.1016/j.scitotenv.2019.134977
Bünemann, E. K., Prusisz, B., and Ehlers, K. (2011). “Characterization of phosphorus forms in soil microorganisms,” in Soil biology, eds E. Bünemann, A. Oberson, and E. Frossard (Berlin: Springer).
Cade-Menun, B. J. (2005). Characterizing phosphorus in environmental and agricultural samples by 31P nuclear magnetic resonance spectroscopy. Talanta 66, 359–371. doi: 10.1016/j.talanta.2004.12.024
Cade-Menun, B. J., and Preston, C. M. (1996). Comparison of soil extraction procedures for 31P NMR spectroscopy. Soil Sci. 161, 770–785. doi: 10.1097/00010694-199611000-00006
Cade-Menun, B. J., Doody, D. G., Liu, C. W., and Watson, C. J. (2017). Long-term changes in grassland soil phosphorus with fertilizer application and withdrawal. J. Environ. Qual. 46, 537–545. doi: 10.2134/jeq2016.09.0373
Chen, H. (2003). Phosphatase activity and P fractions in soils of an 18-year-old Chinese fir (Cunninghamia lanceolata) plantation. For. Ecol. Manag. 178, 301–310. doi: 10.1016/S0378-1127(02)00478-4
Chen, X., Jiang, N., Condron, L. M., Dunfield, K. E., Chen, Z., Wang, J., et al. (2019). Impact of long-term phosphorus fertilizer inputs on bacterial phoD gene community in a maize field, Northeast China. Sci. Total Environ. 669, 1011–1018. doi: 10.1016/j.scitotenv.2019.03.172
Chen, Z. J., Tian, W., Li, Y. J., Sun, L. N., Chen, Y., Zhang, H., et al. (2021). Responses of rhizosphere bacterial communities, their functions and their network interactions to Cd stress under phytostabilization by Miscanthus spp. Environ. Pollut. 287:117663. doi: 10.1016/j.envpol.2021.117663
Condron, R., Farrokh, C., Jordan, K., McClure, P., Ross, T., and Cerf, O. (2014). Guidelines for experimental design protocol and validation procedure for the measurement of heat resistance of microorganisms in milk. Int. J. Food Microbiol. 192, 20–25. doi: 10.1016/j.ijfoodmicro.2014.09.027
Deng, Y., Feng, G., Chen, X., and Zou, C. (2017). Arbuscular mycorrhizal fungal colonization is considerable at optimal Olsen-P levels for maximized yields in an intensive wheat-maize cropping system. Field Crop. Res. 209, 1–9. doi: 10.1016/j.fcr.2017.04.004
Ding, X., Liu, K., Yan, Q., Liu, X., Chen, N., Wang, G., et al. (2021). Sugar and organic acid availability modulate soil diazotroph community assembly and species co-occurrence patterns on the Tibetan Plateau. Appl. Microbiol. Biotechnol. 105, 8545–8560. doi: 10.1007/s00253-021-11629-9
Faust, K., Sathirapongsasuti, J. F., Izard, J., Segata, N., Gevers, D., Raes, J., et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. doi: 10.1371/journal.pcbi.1002606
Fraser, T. D., Lynch, D. H., Bent, E., Entz, M. H., and Dunfield, K. E. (2015). Soil bacterial phoD gene abundance and expression in response to applied phosphorus and long-term management. Soil Biol. Biochem. 88, 137–147. doi: 10.1016/j.soilbio.2015.04.014
Gao, Y. J., Tariq, A., Zeng, F. J., Sardans, J., Zhang, Z. H., Islam, W., et al. (2022). “Fertile islands” beneath three desert vegetation on soil phosphorus fractions, enzymatic activities, and microbial biomass in the desert-oasis transition zone. Catena 212:106090. doi: 10.1016/j.catena.2022.106090
Gatiboni, L. C., Brunetto, G., Rheinheimer, D. D. S., Kaminski, J., Pandolfo, C. M., Veiga, M., et al. (2013). Spectroscopic quantification of soil phosphorus forms by 31P-NMR after nine years of organic or mineral fertilization. Rev. Bras. Cienc. 37, 640–648. doi: 10.1590/S0100-06832013000300010
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant acidification in major Chinese croplands. Science 327, 1008–1010.
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hou, Y., Wei, S., Ma, W. Q., Roelcke, M., Nieder, R., Shi, S. L., et al. (2018). Changes in nitrogen and phosphorus flows and losses in agricultural systems of three megacities of China, 1990-2014. Resour. Conserv. Recy. 139, 64–75. doi: 10.1016/j.resconrec.2018.07.030
Hu, Y., Xia, Y., Sun, Q., Liu, K., Chen, X., Ge, T., et al. (2018). Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Sci. Total Environ. 628, 53–63. doi: 10.1016/j.scitotenv.2018.01.314
Ikoyi, I., Fowler, A., and Schmalenberger, A. (2018). One-time phosphate fertilizer application to grassland columns modifies the soil microbiota and limits its role in ecosystem services. Sci. Total Environ. 630, 849–858. doi: 10.1016/j.scitotenv.2018.02.263
Kathuria, S., and Martiny, A. C. (2011). Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ. Microbiol. 13, 74–83. doi: 10.1111/j.1462-2920.2010.02310.x
Lagos, L. M., Acuña, J. J., Maruyama, F., Ogram, A., de la Luz Mora, M., and Jorquera, M. A. (2016). Effect of phosphorus addition on total and alkaline phosphomonoesterase-harboring bacterial populations in ryegrass rhizosphere microsites. Biol. Fert. Soils 52, 1007–1019. doi: 10.1007/s00374-016-1137-1
Lang, M., Zou, W. X., Chen, X. X., Zou, C. Q., Zhang, W., Deng, Y., et al. (2021). Soil microbial composition and phoD gene abundance are sensitive to phosphorus level in a long-term wheat-maize crop system. Front. Microbiol. 11:605955. doi: 10.3389/fmicb.2020.605955
Li, J. B., Xie, T., Zhu, H., Zhou, J., Li, C. N., Xiong, W. J., et al. (2021). Alkaline phosphatase activity mediates soil organic phosphorus mineralization in a subalpine forest ecosystem. Geoderma 404:115376. doi: 10.1016/j.geoderma.2021.115376
Liu, J., Ma, Q., Hui, X., Ran, J., Ma, Q., Wang, X., et al. (2020). Long-term high-P fertilizer input decreased the total bacterial diversity but not phoD-harboring bacteria in wheat rhizosphere soil with available-P deficiency. Soil Biol. Biochem. 149:107918. doi: 10.1016/j.soilbio.2020.107918
Liu, W., Ling, N., Luo, G., Guo, J., Zhu, C., Xu, Q., et al. (2021). Active phoD-harboring bacteria are enriched by long-term organic fertilization. Soil Biol. Biochem. 152:108071. doi: 10.1016/j.soilbio.2020.108071
Liu, Y., Hou, H., Ji, J., Lv, Z., Liu, X., Liu, G., et al. (2019). Long-term fertiliser (organic and inorganic) input effects on soil microbiological characteristics in hydromorphic paddy soils in China. Soil Res. 57, 459–466. doi: 10.1071/SR18141
Long, X. E., Yao, H. Y., Huang, Y., Wei, W., and Zhu, Y. G. (2018). Phosphate levels influence the utilisation of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Biol. Biochem. 118, 103–114. doi: 10.1016/j.soilbio.2017.12.014
Lou, H., Yang, S., Zhao, C., Zhou, Q., Bai, J., Hao, F., et al. (2015). Phosphorus risk in an intensive agricultural area in a mid-high latitude region of China. Catena 127, 46–55. doi: 10.1016/j.catena.2014.12.013
Luo, G. W., Ling, N., Nannipieri, P., Chen, H., Raza, W., Wang, M., et al. (2017). Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil fractions. Biol. Fert. Soils 53, 375–388. doi: 10.1007/s00374-017-1183-3
MacDonald, G. K., Bennett, E. M., Potter, P. A., and Ramankutty, N. (2011). Agronomic phosphorus imbalances across the world’s croplands. Proc. Natl. Acad. Sci. U.S.A. 108, 3086–3091. doi: 10.1073/pnas.1010808108
Mander, C., Wakelin, S., Young, S., Condron, L., and O’Callaghan, M. (2012). Incidence and diversity of phosphate-solubilising bacteria are linked to phosphorus status in grassland soils. Soil Biol. Biochem. 44, 93–101. doi: 10.1016/j.soilbio.2011.09.009
McLaren, T. I., Smernik, R. J., Simpson, R. J., McLaughlin, M. J., McBeath, T. M., Guppy, C. N., et al. (2015). Spectral sensitivity of solution 31P NMR spectroscopy is improved by narrowing the soil to solution ratio to 1: 4 for pasture soils of low organic P content. Geoderma 257, 48–57. doi: 10.1016/j.geoderma.2015.02.004
Mela, F., Fritsche, K., de Boer, W., Van den Berg, M., Van Veen, J. A., Maharaj, N. N., et al. (2012). Comparative genomics of bacteria from the genus Collimonas: Linking (dis) similarities in gene content to phenotypic variation and conservation. Environ. Microbiol. Rep. 4, 424–432. doi: 10.1111/j.1758-2229.2012.00336.x
Menezes-Blackburn, D., Giles, C., Darch, T., George, T. S., Blackwell, M., Stutter, M., et al. (2018). Opportunities for mobilizing recalcitrant phosphorus from agricultural soils: A review. Plant Soil 427, 5–16. doi: 10.1007/s11104-017-3362-2
Nelson, D. W., and Sommers, L. E. (1983). “Total carbon, organic carbon, and organic matter,” in Methods of soil analysis: Part 2 chemical and microbiological properties, eds D. L. Sparks, A. L. Page, P. A. Helmke, R. H. Loeppert, P. N. Soltanpour, and M. A. Tabatabai (Wisconsin, WI: American Society of Agronomy), 539–579.
Olsen, S. R., and Sommers, L. E. (1982). “Phosphorus,” in Methods of soil analysis, part 2, chemical and microbiological properties, eds A. L. Page, R. H. Miller, and D. R. Keeney (Wisconsin, WI: American Society of Agronomy), 403–427. doi: 10.2134/agronmonogr9.2.2ed.c24
Penuelas, J., Poulter, B., Sardans, J., Ciais, P., Van Der Velde, M., Bopp, L., et al. (2013). Human-induced nitrogen-phosphorus imbalances alter natural and managed ecosystems across the globe. Nat. Commun. 4:2934. doi: 10.1038/ncomms3934
Pérez-Jaramillo, J. E., de Hollander, M., Ramírez, C. A., Mendes, R., Raaijmakers, J. M., and Carrión, V. J. (2019). Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 7:114. doi: 10.1186/s40168-019-0727-1
Powers, S. M., Bruulsema, T. W., Burt, T. P., Chan, N. I., Elser, J. J., Haygarth, P. M., et al. (2016). Long-term accumulation and transport of anthropogenic phosphorus in three river basins. Nat. Geosci. 9, 353–356.
Ragot, S. A., Kertesz, M. A., and Bünemann, E. K. (2015). phoD alkaline phosphatase gene diversity in soil. Appl. Environ. Microb. 81, 7281–7289. doi: 10.1128/AEM.01823-15
Ragot, S. A., Michael, A., Mészáros, E., Frossard, E., and Bünemann, E. K. (2016). Soil phoD and phoX alkaline phosphatase gene diversity responds to multiple environmental factors. FEMS Microbiol. Ecol. 93:fiw212. doi: 10.1093/femsec/fiw212
Sakurai, M., Wasaki, J., Tomizawa, Y., Shinano, T., and Osaki, M. (2008). Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci. Plant. Nutr. 54, 62–71. doi: 10.1111/j.1747-0765.2007.00210.x
Saunders, W. M. H., and Williams, E. G. (1955). Observations on the determination of total organic phosphorus in soils. J. Soil Sci. 6, 254–267. doi: 10.1111/j.1365-2389.1955.tb00849.x
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587. doi: 10.1186/2193-1801-2-587
Stutter, M. I., Shand, C. A., George, T. S., Blackwell, M. S. A., Bol, R., MacKay, R. L., et al. (2012). Recovering phosphorus from soil: A root solution? Rev. Environ. Sci. Bio. 46, 1977–1978. doi: 10.1021/es2044745
Tabatabai, M. A. (1994). “Methods of soil analysis, part 2-microbiological and biochemical properties,” in Soil enzymes, eds R. W. Weaver, S. Angle, P. Bottomley, D. Bezdicek, S. Smith, A. Tabatabai, et al. (Cambridge, MA: Academic Press), 775–833.
Tan, H., Barret, M., Mooij, M. J., Rice, O., Morrissey, J. P., Dobson, A., et al. (2013). Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phoD phosphorus mineraliser group in pasture soils. Biol. Fert. Soils 49, 661–672. doi: 10.1007/s00374-012-0755-5
Tiessen, H., and Moir, J. O. (1993). “Characterization of available p by sequential extraction,” in Soil sampling and methods of analysis, ed. M. R. Carter (Boca Raton, FL: Lewis Publishers), 75–86.
Trivedi, P., Delgado-Baquerizo, M., Trivedi, C., Hu, H. W., Anderson, I. C., Jeffries, T. C., et al. (2016). Microbial regulation of the soil carbon cycle: Evidence from gene–enzyme relationships. ISME J. 10, 2593–2604. doi: 10.1038/ismej.2016.65
Turner, B. L., and Haygarth, P. M. (2005). Phosphatase activity in temperate pasture soils: Potential regulation of labile organic phosphorus turnover by phosphodiesterase activity. Sci. Total Environ. 344, 27–36. doi: 10.1016/j.scitotenv.2005.02.003
Uroz, S., Calvaruso, C., Turpault, M. P., Sarniguet, A., de Boer, W., Leveau, J. H. J., et al. (2009). Efficient mineral weathering is a distinctive functional trait of the bacterial genus Collimonas. Soil Biol. Biochem. 41, 2178–2186.
van der Bom, F. J. T., Mclaren, T. I., Doolette, A. L., Magid, J., Frossard, E., Oberson, A., et al. (2019). Influence of long-term phosphorus fertilisation history on the availability and chemical nature of soil phosphorus. Geoderma 355:113909.
Wan, W., Li, X., Han, S., Wang, L., Luo, X., Chen, W., et al. (2020). Soil aggregate fractionation and phosphorus fraction driven by long-term fertilization regimes affect the abundance and composition of P-cycling-related bacteria. Soil Till. Res. 196:104475. doi: 10.1016/j.still.2019.104475
Wei, X., Hu, Y., Cai, G., Yao, H., Ye, J., Sun, Q., et al. (2021). Organic phosphorus availability shapes the diversity of phoD-harboring bacteria in agricultural soil. Soil Biol. Biochem. 161:108364. doi: 10.1016/j.soilbio.2021.108364
Wei, X., Hu, Y., Razavi, B. S., Zhou, J., Shen, J., Nannipieri, P., et al. (2019). Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol. Biochem. 131, 62–70. doi: 10.1016/j.soilbio.2018.12.025
Yang, X.-d., Ni, K., Shi, Y.-z., Yi, X.-y., Zhang, Q.-f., Fang, L., et al. (2018). Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agric. Ecosyst. Environ. 252, 74–82. doi: 10.1016/j.agee.2017.10.004
Zeng, Q., Mei, T., Delgado-Baquerizo, M., Wang, M. X., and Tan, W. F. (2022). Suppressed phosphorus-mineralizing bacteria after three decades of fertilization. Agric. Ecosyst. Environ. 323:107679. doi: 10.1016/j.agee.2021.107679
Zhao, J., Ni, T., Li, Y., Xiong, W., Ran, W., Shen, B., et al. (2014). Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS One 9:e85301. doi: 10.1371/journal.pone.0085301
Zheng, B. X., Hao, X. L., Ding, K., Zhou, G. W., Chen, Q. L., Zhang, J. B., et al. (2017). Long-term nitrogen fertilization decreased the abundance of inorganic phosphate solubilizing bacteria in an alkaline soil. Sci. Rep. 7:42284. doi: 10.1038/srep42284
Zhou, Y., Clark, M., Su, J., and Xiao, C. (2015). Litter decomposition and soil microbial community composition in three Korean pine (Pinus koraiensis) forests along an altitudinal gradient. Plant Soil 386, 171–183. doi: 10.1007/s11104-014-2254-y
Keywords: long-term P fertilization, organic P speciation, phoD-harboring bacteria, co-occurrence network, keystone taxa, alkaline phosphatase
Citation: Lang M, Li H, Lakshmanan P, Chen Y and Chen X (2022) phoD-harboring bacterial community composition dominates organic P mineralization under long-term P fertilization in acid purple soil. Front. Microbiol. 13:1045919. doi: 10.3389/fmicb.2022.1045919
Received: 16 September 2022; Accepted: 03 November 2022;
Published: 24 November 2022.
Edited by:
Ruibo Sun, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Shuo Jiao, Northwest A&F University, ChinaWenjie Wan, Wuhan Botanical Garden (CAS), China
Copyright © 2022 Lang, Li, Lakshmanan, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinping Chen, Y2hlbnhwMjAxN0Bzd3UuZWR1LmNu
†These authors have contributed equally to this work
 Ming Lang
Ming Lang Haoming Li
Haoming Li Prakash Lakshmanan
Prakash Lakshmanan Yuanxue Chen
Yuanxue Chen Xinping Chen
Xinping Chen