- 1The Engineering Research Center for High-Valued Utilization of Fruit Resources in Western China, Ministry of Education, National Research and Development Center of Apple Processing Technology, College of Food Engineering and Nutritional Science, Shaanxi Normal University, Xian, China
- 2Department of Human Nutrition, Food and Animal Sciences, College of Tropical Agriculture and Human Resources, Honolulu, HI, United States
The commercial active dry yeast strains used for cider production in China are far behind the requirements of the cider industry development in recent decades. In this study, eight yeasts, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, Pichia bruneiensis, and Pichia kudriavzevii, were screened and assessed by growth performance, methanol production, aroma analysis, and their transcriptive characterization. Saccharomyces cerevisiae strains WFC-SC-071 and WFC-SC-072 were identified as promising alternatives for cider production. Strains WFC-SC-071 and WFC-SC-072 showed an excellent growth capacity characterized by 91.6 and 88.8% sugar utilization, respectively. Methanol production by both strains was below 200 mg/L. Key aroma compounds imparting cider appreciably characteristic aroma increased in cider fermented by strains WFC-SC-071 and WFC-SC-072. RT-qPCR analysis suggested that most genes associated with growth capacity, carbohydrate uptake, and aroma production were upregulated in WFC-SC-071 and WFC-SC-072. Overall, two Saccharomyces cerevisiae strains are the optimal starters for cider production to enable the diversification of cider, satisfy the differences in consumer demand, and promote cider industry development.
Introduction
Cider, with an alcohol content of 1.2–8.5%, is the second most-consumed fruit wine globally (Lorenzini et al., 2019; Zhang et al., 2021). As a fruit beverage, apple cider has the characteristics of both apple juice and beer, such as being golden in color, rich in fermented aroma, strong palatability, and mellow mouthfeel. Moreover, cider has nutrients from apples and fermentation products; thus, it has a higher nutritional value (Lee et al., 2019; Zhang et al., 2020; Nie et al., 2021). As a result, cider is becoming a promising product, especially in the eastern Asian markets (Khandpur and Gogate, 2015; Guine et al., 2021). Due to the rapid expansion of the cider market, developing and producing high-quality cider is a priority for the cider industry.
Yeasts play a decisive role in fermentation and eventually determine cider quality. They are the primary contributor to the color, aroma, product features, and styles of ciders (Song et al., 2020). In addition, yeast converts sugars and other substrates to ethanol, carbon dioxide, and other aromatic substances during fermentation (Fejzullahu et al., 2021). Traditional ciders are produced through natural fermentation by autochthonous yeasts. However, the number of wild microbes in mature fruit pericarps and stems far exceeds autochthonous yeasts. These wild microbes often interfere with yeast fermentation, thus affecting the quality of fruit wine (Lorenzini et al., 2019). Moreover, commercial active dry yeast strains are ubiquitously used for winemaking, but little research on unique strains for cider fermentation. Therefore, it is necessary and significant to identify appropriate yeasts for cider fermentation.
Various yeasts have been isolated from different fruits and fermented grains to endow fruit wines with high concentrations of pleasant aromas and distinctive floral fragrances in winemaking (Callejon et al., 2010; Saberi et al., 2012). Saccharomyces cerevisiae, isolated from a vineyard in Piedmont (Italy), was used to produce wine with an alcohol content of 13.29 g/L, as well as a pleasant smell of rose (Costantini et al., 2021). Saccharomyces cerevisiae GP-34 was isolated from the natural fermentation liquid of loquat juice. Compared with commercial Saccharomyces cerevisiae, the contents of alcohols, esters, acids, and phenols in loquat wine fermented by Saccharomyces cerevisiae GP-34 were 31, 86, 25, and 11% higher, respectively (Xu et al., 2019).
Meanwhile, the pleasant flavor of fruit wine is mainly attributed to non-Saccharomyces yeasts, which shape the chemical profile of aromatic compounds and enhance the diversity of aroma (van Wyk et al., 2019; Godoy et al., 2020). Several non-Saccharomyces yeasts have been identified and utilized to confer characteristic flavors to fruit wines. For example, Torulaspora delbrueckii, obtained from Brazilian culture collection, synthesized large amounts of phenethyl acetate during fermentation, thereby conferring a fruity fragrance (Van Mullem et al., 2022). However, data on the promising yeast for cider processing are still scarce, and new strains shall be identified to generate diversified styles of ciders.
Moreover, methanol, a hazard factor of fruit wine fermentation, is derived from the methyl group of pectin hydrated by yeast enzymes (Wu et al., 2017). Methanol is toxic and can cause weakness, vomiting, coma or even death when intaken excessively (Padilla et al., 2016; Ghosh et al., 2021). Consequently, it is critical to comprehensively evaluate the strains and obtain dominant strains capable of improving overall cider quality and diversifying cider styles to meet the needs of a wide range of consumers.
In the present study, 75 yeast strains were isolated from the apples harvested in Changwu, Shaanxi Province, China’s renowned hometown of apples, and fermented grains from Maotai and Xifeng, two well-known Chinese liquors. A simple olfactory test was used to screen the isolated strains, and eight yeasts producing desirable aromas were obtained. Their identities were confirmed by 28S rRNA analysis. These eight yeast strains were then used for cider-making to evaluate their growth capacity, methanol production, and volatile production. Based on these assessments, two yeast strains were identified as promising alternatives for cider fermentation. In summary, this work promotes the diversification of cider, potentially satisfies the differences in consumer demand, and promotes cider industry development.
Materials and methods
Materials
Fresh Red Fuji apples (56.7% fructose, 10.4% glucose, and 32.9% sucrose) were purchased from Changwu, Shaanxi province, China. The apples were kept in cold storage before subsequent studies. The fermented grains were obtained from Maotai (Guizhou) and Xifeng Distilleries (Shaanxi), China. All organic solvents were chromatographic grade and purchased from Sigma-Aldrich (Shanghai, China). Total RNA extractor reagent (B511311) was obtained from Sangon Biotech (Shanghai, China). One-step gDNA Removal kit (AT341-02) and Perfectstart™ Green qPCR SuperMix kit (AQ601-02) were purchased from TransGen Biotech (Beijing, China).
Enrichment and isolation of yeast strains
Apples were soaked in 75% ethanol for 5 min, washed twice with sterile water, then dipped in 2% sodium hypochlorite for 5 min, washed four times with sterile water, and cut into 1 cm3 squares. Five grams of the fermented grains obtained from Maotai and Xifeng Distilleries and 5 g of apple squares were mixed with 100 ml yeast peptone glucose (YPD; 2% glucose, 2% peptone, and 1% yeast extract) at 30°C for 24 h. Then, culture fluids enriched with yeasts were spread separately over YPD agar and incubated at 30°C for 48 h. Next, 75 yeast colonies were selected and streaked on YPD agar plates for purification. This process was repeated in triplicate (Kumari et al., 2020). Finally, each strain was coded and stored at −80°C for subsequent experiments.
Fermentation trial
Apple juice was prepared from peeled apples using a juicer (Hurom, Korea; model HUF8800STS). Vitamin C (1‰) was added to inhibit apple juice oxidation and discoloration. Fresh apple juice was sterilized at 100°C for 30 s before inoculation. All strains obtained in section “Enrichment and isolation of yeast strains” were used to ferment apple juice. Each yeast strain was pre-cultured in 50 ml YPD medium at 30°C for 1 day on a rotary shaker with a speed of 200 rpm, separated by centrifugation (5,000 × g, 4°C, 5 min), washed twice with sterile water, and inoculated in 200 ml sterilized apple juice for cider production at a concentration of approximately 107 CFU/ml. The fermentations were conducted at 30°C for 7 days.
Screening and identification of yeast strains
Twenty students and 10 staff from Shaanxi Normal University were recruited as screeners. They performed a simple olfactory test to describe and grade cider aroma, and strains with a low score and undesirable aroma were excluded (Gutiérrez et al., 2018). Eight strains were selected using this screening test. The specific scoring form and scoring guidelines are shown in Supplementary Table S1.
The eight strains selected by the olfactory test were further identified. These yeasts were inoculated in YPD at 30°C for 1 day. Cell morphology was observed with an optical microscope (Nikon, Japan; model ECLIPSE E100). These strains were identified by 28S rRNA. The genomic DNA of yeast was extracted by the SDS-based method (Xia et al., 2019). The D1/D2 domain of the 28S rRNA was amplified with primers NL1 and NL4 (Mikhailova et al., 2020). The sequences were then compared with those in the GenBank and analyzed using BLAST software. Phylogenetic tree construction of these yeasts was performed using MEGA X software.
Assessment of yeast strains
Sugar utilization
Total sugar content of cider was determined using the phenol−sulfuric acid method. Sample preparation was conducted using a modified existing method (Wu et al., 2022). Briefly, 100 μl of cider was diluted with 1 ml of ultrapure water, and then mixed with 1 ml of 5% phenol and 5 ml of concentrated sulfuric acid. The mixture was incubated in a water bath at 35°C for 30 min. The sample absorbance was measured at 490 nm using a UV–VIS Spectrophotometer (Optima, Japan). A glucose standard solution is prepared as a reference solution. The standard linear equation generated was y = 0.0061x−0.0073, r2 = 0.998.
Methanol production
Methanol content in cider was analyzed using an existing method with modifications (Arslan et al., 2015). The cider was distilled to collect the volatile components and filtered through a 0.25 μm membrane filter (Jinlong, China). Methanol was analyzed using a GCMS-QP2010SE system (Shimadzu, Japan) coupled with a flame ionization detector (FID; Shimadzu, Japan) and a DB-5 capillary column (2 m × 2 mm I.d.; film thickness: 1.5 μm; Chrompack, Netherlands). The oven temperature started at 40°C for 3 min, and increased to 100°C at a rate of 5°C/min, then increased to 200°C at 20°C/min and maintained for 5 min. The single injection analysis time was 25 min. The FID temperature was set at 250°C. Detector flow rates of 20, 400, and 25 ml/min were applied to hydrogen, air, and helium (make-up). A column flow rate of 1 ml/min was applied to helium (as carrier gas). Each sample was measured in triplicate. Methanol was identified according to retention time and quantified via the external standard method.
Aroma analysis
Quantitative descriptive analysis
Quantitative descriptive analysis (QDA) was conducted by 10 evaluators (five males and five females). The evaluation was based on a modified procedure (Zhao et al., 2021). All assessors were trained to be familiar with cider aroma assessment. They were instructed to assess the ciders in eight aroma attributes: acid, sweet, fruity, alcoholic, spicy, smooth, dry, and dense. Five milliliter of cider were transferred in a 20 ml sterile cup, marked with a random number, and scored using a scale of 0–9 (0 = none, 9 = very strong). Sensory evaluation of each sample was carried out in triplicate and at room temperature, and the results for each aroma attribute were averaged and plotted on a radar chart.
Analysis of aroma compounds by HS-SPME-GC-MS
Extraction of aroma compounds in cider was performed using a headspace solid-phase micro-extraction (HS-SPME) according to an existing method with minor modifications (Niu et al., 2019). In brief, 5 ml cider, 2 g sodium chloride, and 10 μl 3-octanol (10 mg/L, internal standard) were placed in a 20 ml micro-reaction vial. The vial was put into a thermostatic water bath at 70°C for 30 min. The 50/30 μm divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS; Sigma-Aldrich, United States; 57348-U) fiber was exposed to the sample headspace (about 1 cm above the liquid surface) at 70°C for 30 min, injected in the GC injector and kept for 3 min, and analyzed using an Agilent 8890 GC system equipped with a 5977B mass spectrometer (MS) according to a modified method (Niu et al., 2019). Chromatographic separations were performed using a DB-wax column (20 m × 0.18 mm, Agilent, Santa Clara, CA, United States). The column flow rate of helium was 1.5 ml/min. Electron energy of 40 Ev and mass range of m/z 30–450 were applied during MS analysis. The initial temperature was maintained at 40°C for 6 min, increased to 100°C at 4°C /min rate, then increased to 230°C at 10°C/min rate, and held for 20 min. Each sample was measured in triplicate.
RT-qPCR
Total cellular RNA was extracted using Trizol reagent (Sangon Biotech, China; B511311-0100) according to the manufacturer’s instructions. Samples were quantified using a Nanophotometer N60 Touch spectrophotometer (Implen GmbH, Germany; T60765). The reverse transcription reaction system was prepared according to the manufacturer’s protocol, containing 1 μg of the RNA sample. Primer sequence, reaction efficiency and threshold were described in Supplementary Table S2. Real-time PCR was performed using a Perfectstart™ Green qPCR SuperMix kit. Amplification parameters include an initial denaturation stage at 95°C for 3 min, followed by 40 cycles of 10 s at 95°C and 63°C for 30 s, and a final extension stage at 72°C for 5 min. A dissociation protocol was carried out to verify the non-specific amplification absence. The mRNA abundance change was calculated using the 2[-Delta Delta C(T)] method (Lorca Mandujano et al., 2022). Beta-actin (ACT1) gene was used as the internal control gene.
Data analysis
Origin 2021 software (Origin Lab Co., Northampton, Massachusetts, United States) was used for graphing. All experiments were conducted in triplicates, and results were expressed as means ± SD. One-way ANOVA, followed by Duncan’s multiple range test, was performed using the SPSS 21.0 software (SPSS Inc., Chicago, IL, United States). The difference between the means was considered significant when p < 0.05.
Results
Eight yeast strains with pleasant aroma were identified from different sources
Yeasts from apples and fermented grains were cultured in an YPD medium, and 75 yeast strains were obtained. We performed a simple olfactory test described in section “Screening and identification of yeast strains” of the Materials and Methods to screen aroma-producing yeasts rapidly. We selected eight isolates based on their high scores during the olfactory test. Furthermore, these eight strains also endowed ciders with predominantly fruity or floral flavors or a combination of fruit and floral flavors. Thus, these eight strains (WFC-014, 045, 047, 048, 051, 054, 071, and 072) were selected for subsequent molecular analysis (Table 1).
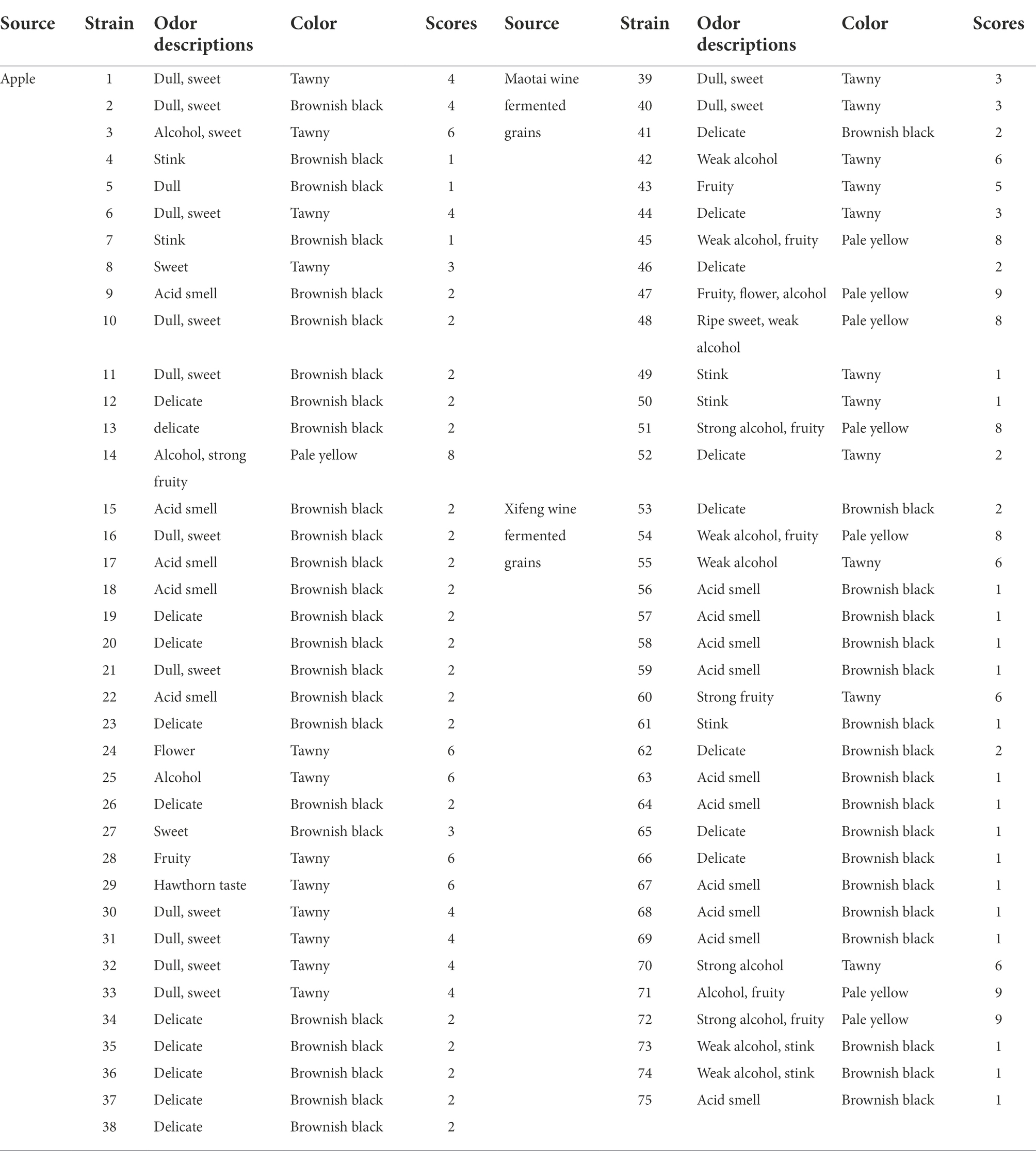
Table 1. Aroma-producing characteristics and sensory analysis of fermented ciders from 75 different yeasts.
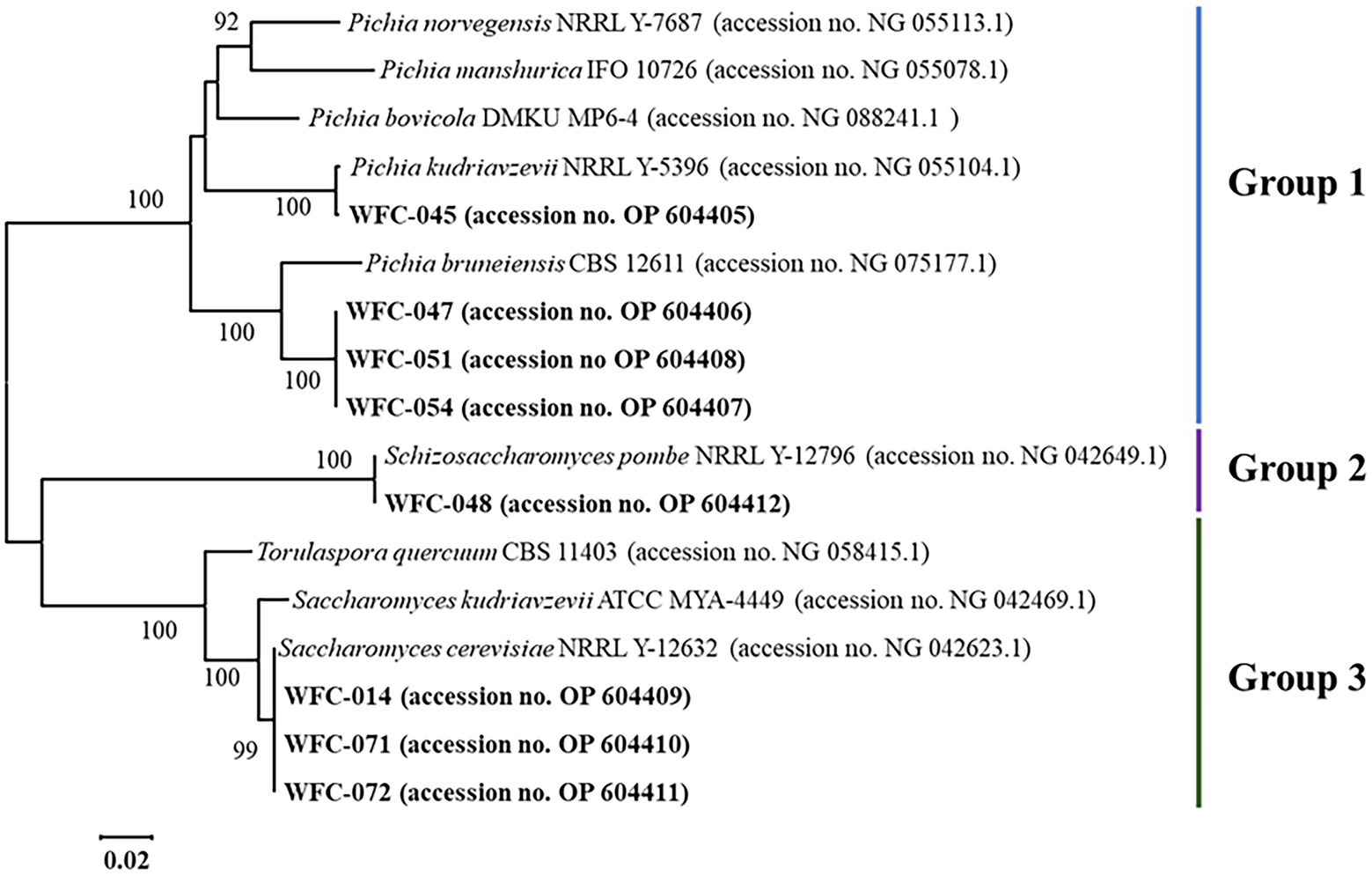
The phylogenetic tree of the yeast species was established by MEGA X software based on 28S rRNA sequences (Figure 1). The type of selected strains fell into three distinct groups. Four strains of group 1 clustered with Pichia type (accession no. NG 075177.1 and NG 055104.1) and shared a clade. Strain WFC-045 (accession no. OP 604405) was more closely related to Pichia kudriavzevii NRRL Y-5396, as the bootstrap value between them was 100. And the three remaining strains were very close to Pichia bruneiensis CBS 12611. A strain of group 2 clustered with the Schizosaccharomyces type (accession no. NG 042649.1) and was very close to Schizosaccharomyces pombe NRRL Y-12796. At the same time, three strains of group 3 clustered with Saccharomyces cerevisiae type (accession no. NG 042623) and were very close to Saccharomyces cerevisiae NRRL Y-12632. The four Pichia yeasts of group 1 were named WFC-PK-045, WFC-PB-047, WFC-PB-051, and WFC-PB-054. The Schizosaccharomyces pombe strain of group 2 was named WFC-SP-048. And the three S. cerevisiae strains of group 3 were named WFC-SC-014, WFC-SC-071, and WFC-SC-072.
Assessment of the fermentation performance of eight yeasts
One of the crucial characteristics of yeast suitable for cider production is its growth ability (Wang et al., 2022). Yeasts use carbon sources (mainly sugar) to ferment and produce high-sensory quality cider (Lorca Mandujano et al., 2022). Therefore, sugar utilization was evaluated to determine the yeast fermentation capacity of these eight yeast strains. The three S. cerevisiae strains showed a similar sugar utilization tendency during the whole fermentation (Figure 2A). Nevertheless, the sugar consumption rate (0–24 h) and total sugar consumption were significantly different (p < 0.05; Figures 2A,B). Strain WFC-SC-072 showed a maximum sugar consumption rate (0–24 h), whereas strain WFC-SC-071 exhibited the highest total sugar consumption. Typically, ethanol accumulation inhibits non-Saccharomyces yeasts’ metabolism, resulting in less sugar utilization (Bao et al., 2021). However, strains WFC-PK-045, WFC-PB-047, and WFC-PB-054 consumed sugar by up to 80% (Figure 2B). The sugar consumption level by strain WFC-PK-045 was close to the S. cerevisiae strains. Strain WFC-SP-048 was the only strain with lower sugar consumption capability. These results indicate that most strains had robust growth capability.
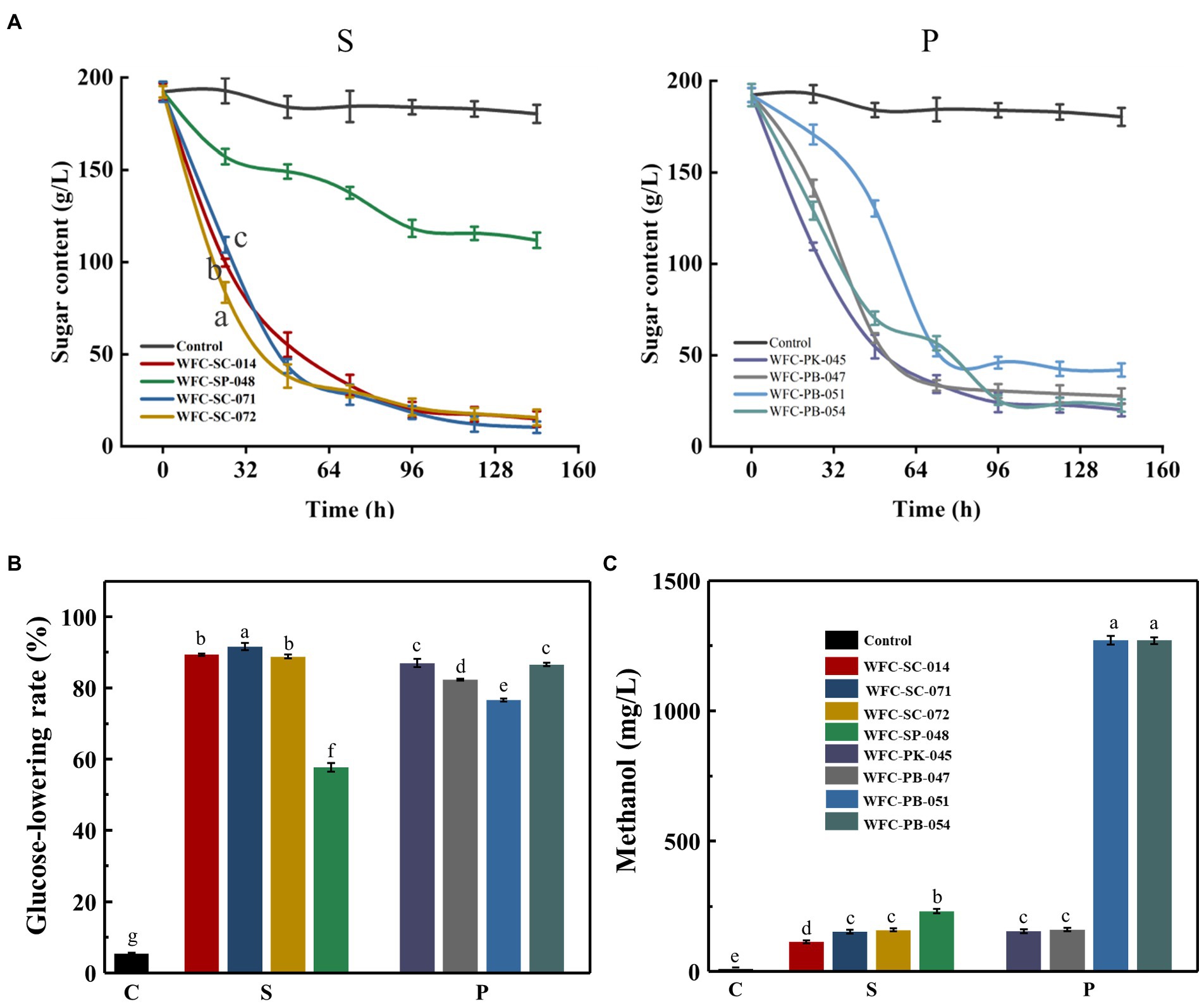
Figure 2. Growth capability of isolated strains on apple juice. (A) Total sugar consumption by Saccharomyces cerevisiae strains and Schizosaccharomyces pombe strain (S), and Pichia yeasts (P). (B) Glucose-lowering rate of apple juice fermented by different strains for 7 days. Means within glucose-lowering rate between different yeasts with different letters (a–g) differ significantly (p < 0.05). Both sugar consumption and glucose-lowering rate experiments were replicated three times. (C) Methanol concentration in ciders fermented by eight yeasts. Means with different letters (a–e) differ significantly (p < 0.05). Most strains had robust growth capability and produced acceptable amounts of methanol.
Methanol content is a critical parameter of cider quality. Its content should be strictly controlled below 400 mg/L (Wei et al., 2020b). Therefore, these yeast strains’ methanol production capacity was measured (Figure 2C). Compared to the Saccharomyces cerevisiae yeasts, Pichia yeast strains produced more methanol. Among all Pichia yeast strains, WFC-PB-051 and WFC-PB-054 had the highest methanol (1,270 mg/L). Most strains produced little methanol, below 300 mg/L. No difference (p > 0.05) in methanol production was observed among Pichia yeasts WFC-PK-045 and WFC-PB-047, Saccharomyces strains WFC-SC-071 and WFC-SC-072. Notably, Saccharomyces strain WFC-SC-014’s methanol production (114.2 mg/L) was the lowest of all strains and lower than commercial Saccharomyces cerevisiae (Chen et al., 2019). In summary, six strains with methanol content below 400 mg/L could be used for cider production.
Sensory evaluation and quantitative descriptive analysis of ciders fermented by eight yeasts
Quantitative descriptive analysis (QDA) was performed to evaluate the aroma traits under different yeast fermentation conditions (Figure 3). Eight aroma attributes, acid, sweet, fruity, alcoholic, spicy, smooth, dry, and dense, were representative of odor attributes. A least significant difference (LSD) analysis shows that “dense,” “acid,” “spicy,” and “fruity” attributes differed among Group S and the “fruity” attribute differed among Group P (p < 0.05). As for the overall aroma profile, fruity and alcohol attributes scores were higher than the other attributes, suggesting that they were the predominant aroma attributes in cider. Regarding the fruity attribute, it was the maximum intensity perceived in cider, which could be mainly interpreted by the synthesis of acetates and fatty acid ethyl esters during fermentation. In addition, the cider fermented by strain WFC-SC-071 exhibited a similar profile compared to that cider fermented by strain WFC-SC-072, with both being similar to commercial ciders. Overall, these results indicate that Saccharomyces cerevisiae strains WFC-SC-071, WFC-SC-072, and Pichia yeast WFC-PB-047 resulted in valuable and desirable characteristics of the cider style.
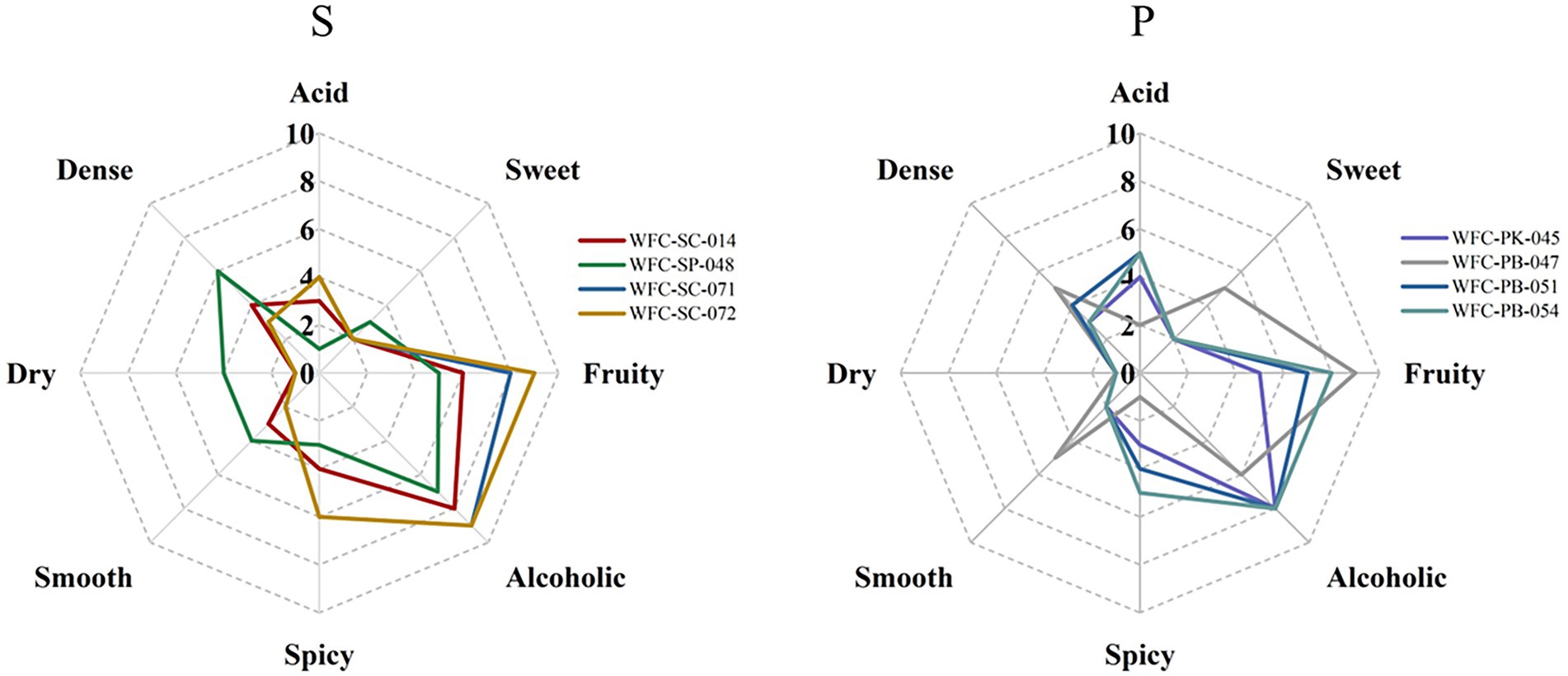
Figure 3. Sensory descriptive analysis of ciders fermented by Saccharomyces cerevisiae strains and Schizosaccharomyces pombe strain (S) or Pichia yeasts (P). Cider fermented by Saccharomyces cerevisiae strains WFC-SC-071, WFC-SC-072, or Pichia yeast WFC-PB-047 has desirable aroma characteristics for the cider style.
SPME-GC–MS analysis of volatile composition profiles of ciders
Aroma and flavor define cider quality, while molecule profiles account for organoleptic properties (Qin et al., 2018). Cider aroma compounds mainly comprise esters, alcohols, fatty acids, acetyls, terpenes, and other trace components (Nespor et al., 2019). The aromatic profiles were drastically improved after fermentation.
Esters are ciders’ predominant group of volatile compounds (Aung et al., 2015). Esters are synthesized via esterification during fermentation; they have a fruity odor. For fruity esters, fermentation improved ester production (Figure 4). Fermentation by these eight yeasts improved esters abundance, especially acetates and fatty acid ethyl esters. As shown in Supplementary Table S3, fermentation by all eight strains elevated acetates and fatty acid ethyl esters species to various degrees. Ester abundance of cider fermented by strain WFC-SC-071 or WFC-PB-047 resulted in four or five times higher than the unfermented group. In addition, esters were significantly increased (p < 0.05) after fermentation by these eight strains. The total ester content of cider fermented by strain WFC-PB-047 was about four times more than the unfermented group. Typically, acetate in WFC-PB-047 was about 10 times more than in the unfermented group.
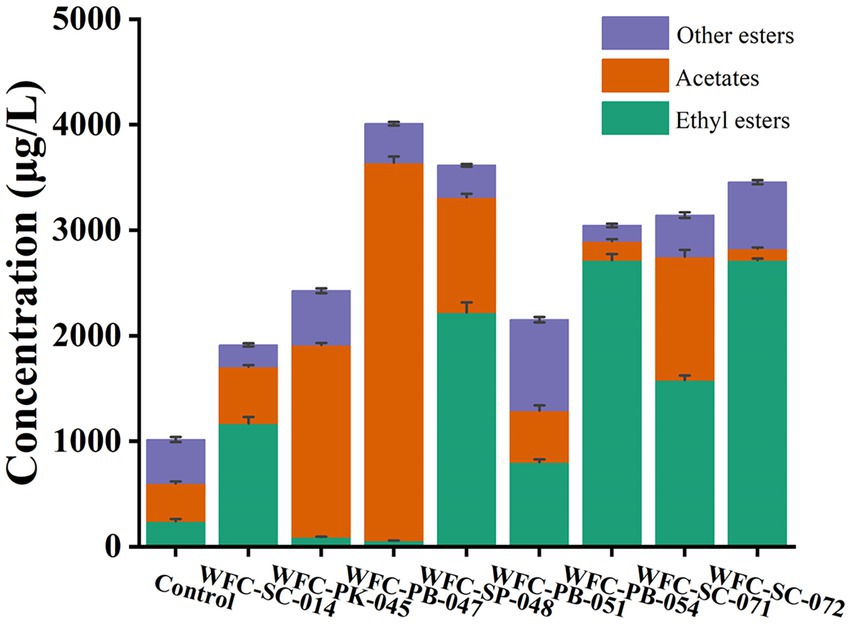
Figure 4. Ester production in ciders fermented by different yeasts. The orange bar indicates average acetate concentration, the green bar represents ethyl esters, and the purple bar shows other esters. All eight strains had outstanding ester production capacity to improve cider sensory quality.
Ciders fermented by different yeast strains showed contrasting ester profiles (Figure 5A). For example, the content of 11 aroma compounds in ciders fermented by strains WFC-SC-071 and WFC-SC-072 were significantly higher (p < 0.05) than in unfermented apple juice. Similarly, seven compounds were accumulated in cider fermented by strain WFC-PB-047. Ethyl esters and acetates, among esters, are the most determinant compounds contributing to cider flavor, such as ethyl acetate, ethyl octanoate, ethyl nonanoate, and isoamyl acetate. Ethyl nonanoate was detected in ciders fermented by strains WFC-SC-014, WFC-SC-071, WFC-SC-072, WFC-SP-048, and WFC-PB-051, imparting ciders’ grape and rose flavor. Ethyl octanoate, with pineapple and sweet flavor, was detected before and after fermentation, although the abundance changed significantly (p < 0.05). However, hexyl hexanoate and hexyl octanoate decreased by 74–100% (p < 0.01) after fermentation (Supplementary Table S3).
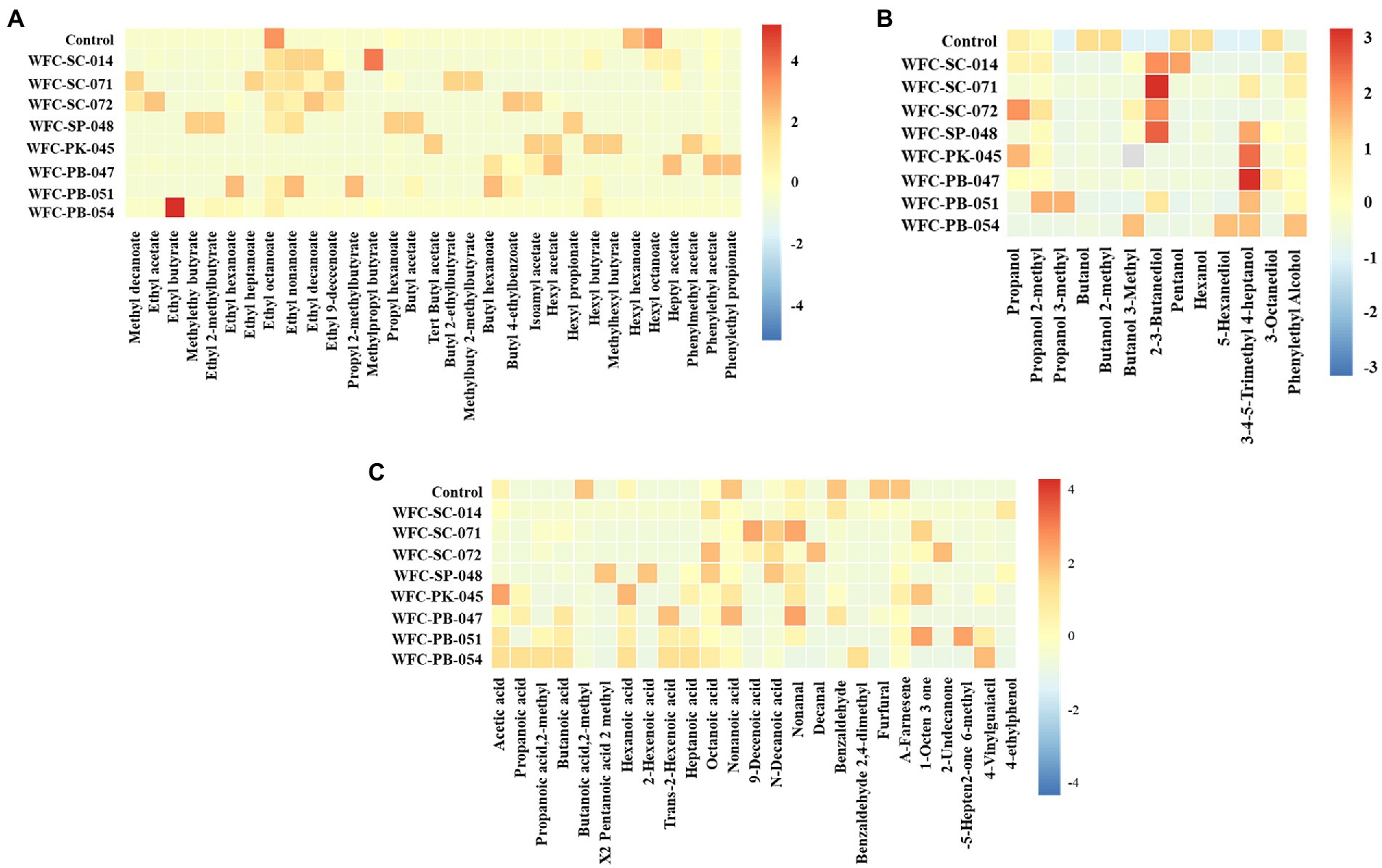
Figure 5. Volatile compounds of the fermented ciders. The heat map describes the aromatic compounds of ciders fermented for 7 days by eight strains. Esters (A), alcohols (B), and other aroma compounds (C) in fermented ciders. Aroma components changed significantly after fermentation. Aroma compounds were prominently expressed in ciders fermented by strains WFC-SC-071 and WFC-SC-072.
Alcohols were the second largest group among the volatile compounds in ciders. Most alcohols are produced from sugar and amino acid catabolism during fermentation (Qin et al., 2018). The alcohol content in apple juice decreased significantly after fermentation (Supplementary Table S3). However, many key alcohols imparting cider appreciably characteristic aroma increased and became an essential component in the resultant ciders (Figure 5B). More than 10 alcohols were highly expressed in several ciders, excluding cider fermented by strain WFC-SC-014. Among alcohols, 2, 3-butanediol, 3-methyl-1-butanol, and phenylethyl alcohol are pivotal substances contributing to cider style. Phenylethyl alcohol, which has honey, spicy, rose, and lilac aromas, was concentrated in all ciders compared to apple juice. 3-Methyl-1-butanol was found most bountiful in ciders fermented by strains WFC-SC-014, WFC-SC-072, WFC-PB-047, and WFC-PB-054, and this compound contributes to a whiskey odor in cider even in low concentration. However, the relative abundance of 1-butanol and 2-methyl-1-butanol was reduced after fermentation.
Acids are mainly produced during fermentation (Wei et al., 2020a). As shown in Figure 5C, ciders contained various volatile acids, while the unfermented apple juice only had a considerable level of 2-methyl butanoic acid. High increments of short-chain fatty acids were observed in Pichia yeast strains, while Saccharomyces cerevisiae yeast strains generated large amounts of medium-chain fatty acids. The aldehydes are generated via alcohol oxidation or acid decarboxylation during fermentation (Xiao et al., 2015). Several aldehydes, including benzaldehyde, nonanal, and decanal, were identified in ciders. Nonanal, a crucial aromatic compound in fruit wines was enriched in seven ciders after fermentation. With almond and burnt sugar odor, benzaldehyde was suggested to foster characteristic sensory properties (Sun et al., 2016). An abundance of benzaldehyde was detected in ciders fermented by strains WFC-SC-014 and WFC-PB-047. Moreover, 1-octen-3-one, a volatile unsaturated ketone with a strong mushroom-like odor, was enriched in four ciders. 6-Methyl-5-hepten-2-one, which has a strong citrus-like odor, was abundant in cider made by strain WFC-PB-051. Extraordinary, phenolic off-flavors like 4-vinylguaiacil and 4-ethylphenol, which have a negative impact on the overall aromatic profile, were detected in cider fermented by strains WFC-PK-045 and WFC-PB-047 (Alvarez Gaona et al., 2021). In conclusion, aromatic profiles were drastically improved after fermentation. The ideal aroma profile was presented in ciders fermented by strains WFC-SC-071 and WFC-SC-072.
Analysis of eight strains’ gene expression during fermentation
Different yeast strains possess different genetic predispositions, thus profoundly influencing fermentation. RNA samples were collected both before and after fermentation to evaluate the transcription level changes of the yeasts during fermentation. Using RT-qPCR, we quantitatively analyzed the gene expression levels related to anaerobic stress resistance, aroma production, carbohydrate uptake, and yeast growth capacity to investigate the characteristic yeast properties. Gene expression levels are presented in Figure 6, and the average values are provided in Supplementary Table S4.
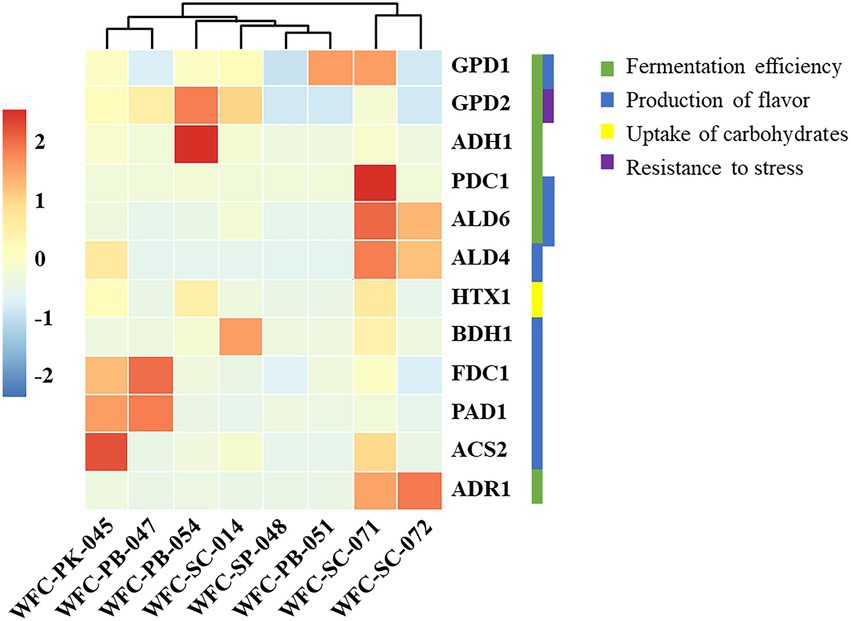
Figure 6. Changes in gene expression. The results are expressed as the median of the difference in gene expression level before and after the fermentation of triplicate samples. Genes in green, blue, yellow, and purple are related to fermentation capacity, flavor production, carbohydrate uptake, and stress resistance, respectively. Strains WFC-SC-071 and WFC-SC-072 demonstrated ideal expression levels of most genes associated with aroma production, carbohydrate uptake, and yeast fermentation capacity.
Gene expression analysis revealed deviated metabolic pathways during fermentation. GPD1 and GPD2, two homologous genes, regulated NAD-dependent glycerol-3-phosphate dehydrogenase (Gpd1p and Gpd2p), act on dihydroxyacetone phosphate to form glycerol-3-phosphate (Vilela, 2021). GPD1 is related to glycerol production during osmotic conditions (Marullo and Dubourdieu, 2022). GPD2 is associated with modulating the balance of NADH-NAD redox under osmotic stress (Sun et al., 2022). GPD1 and GPD2 expression is specifically upregulated in response to anaerobic stress (Baeza et al., 2022). Among the eight strains, both strains WFC-PB-051 and WFC-SC-071 showed higher expression levels of GPD1, while strain WFC-PB-054 exhibited more pronounced upregulation of GPD2 (p < 0.05). The upregulated expression of GPD1 and GPD2 levels in strains WFC-PB-051, WFC-PB-054, and WFC-PB-047 might be interpreted as adapting to the anaerobic environment during fermentation.
Alcohol dehydrogenase I, encoded by ADH1, reduces acetaldehyde to ethanol during fermentation (Raj et al., 2014). ADH1 was upregulated in all eight strains, and comparatively higher expression levels were measured in strain WFC-PB-054. PDC1 gene encoding an isoform of pyruvate decarboxylase was activated among eight strains, and higher expression levels were detected in strain WFC-SC-071. PDC1 is related to the non-oxidative conversion of pyruvate to acetaldehyde and carbon dioxide during fermentation and participates in amino acid catabolism (Dzialo et al., 2017). Yeasts lacking PDC1 activity grow slowly for an inadequate acetyl CoA supply or an imbalanced cytoplasmic redox (Mitsui et al., 2022).
Expression levels of ADR1, ALD4, and ALD6 were prominent in WFC-SC-071 and WFC-SC-072. ADR1, encoding a transcription activator of glucose-repressible alcohol dehydrogenase, was repressed in the absence of glucose, while its activation occurs upon glucose exhaustion (Zhang G. et al., 2022). ALD6, encoding the cytosolic acetaldehyde dehydrogenase, is indispensable for converting acetaldehyde to acetate (Wang et al., 2020). These genes are also intercorrelated as ADR1 activates genes related to glucose fermentation, including ALD4 and ALD6 (Kalender and Çalık, 2020). The expression levels of these genes associated with the respiratory cycle, uptake and metabolism of carbohydrates exhibited an upregulated metabolic profile in strains WFC-SC-071 and WFC-SC-072 during fermentation.
PAD1 and FDC1, participating in the decarboxylation of aromatic compounds and carboxylic acids generated during fermentation, were markedly activated in the WFC-PK-045 and WFC-PB-047 strains. Like ADR1 and ALD4, the expression of PAD1 and FDC1 are also interdependent. Together, the two genes are related to the production of the clove-like off-flavor during fermentation. In addition, PAD1, encoding flavin prenyltransferase, is involved in the decarboxylation of phenylacrylic acids, such as ferulic acid, p-coumaric acid, and cinnamic acid, producing the corresponding vinyl derivatives (Gulmezian et al., 2007; Lin et al., 2015).
Interestingly, the expression levels of ACS2 were prominently upregulated in strains WFC-SC-071 and WFC-PK-045. Among these genes, ACS2 encodes an acetyl-CoA synthetase and may contribute to ethanol tolerance and ethyl-acetate synthesis of yeast cells (Xu et al., 2021a,b). BDH1 gene is overexpressed in strain WFC-SC-014 and is responsible for reducing acetoin to 2,3-butanediol, a neutral-scented compound (Ehsani et al., 2009).
The HXT1 gene encodes carriers for hexose transport and plays as the sugar transporter during yeast growth, specialized for glucose and fructose (Munoz-Miranda et al., 2022). Strains in the absence of HXT1 genes could not grow on glucose or fructose and have no glycolytic flux (Nadai et al., 2021). HXT1 was activated in strains WFC-PK-045, WFC-PB-054, and WFC-SC-071 and exhibited lower expression levels at the end of fermentation.
Strains WFC-SC-071, WFC-SC-072, WFC-PB-047, and WFC-PB-054 exhibited upregulated transcription profiles for most genes associated with aroma production, carbohydrate uptake, and yeast fermentation capacity. The upregulated transcription profiles verified that strains WFC-SC-071 and WFC-SC-072 possess desirable fermentation and aroma properties.
Discussion
Cider is the second most-consumed fruit drink and is becoming a promising drinking product (Cousin et al., 2017). Urgent requirements for diverse and innovative ciders have emerged. Aroma and flavor formed by yeast metabolism inarguably define cider organoleptic quality and consumer acceptance (Bingman et al., 2022). Therefore, it is necessary to identify new yeast strains for cider fermentation to enhance the aroma compound profile. In this study, we identified two Saccharomyces cerevisiae with excellent growth capacity, capable of generating an abundant aroma compound profile. They are selected as promising starters for cider production.
Alcohols
Higher alcohols, with fruity and floral odors, are one of the most integral traits that define the unique aroma profiles of cider (Hinkley et al., 2022). Higher alcohols are synthesized from amino acids via the anabolic pathway or the catabolic (Ehrlich) pathway (Wang et al., 2019). The Ehrlich pathway is a three-step and pivotal biochemical pathway in aroma production. Strains WFC-SC-071 and WFC-SC-072 increased the key higher alcohol content during fermentation, such as phenethyl alcohol. This higher alcohol accumulation in ciders may be attributed to the upregulation of PDC1 and BDH1, as these genes are responsible for reducing aldehydes to alcohols (Varga et al., 2021). At the same time, 3-methyl butanol content in strain WFC-SC-071 was lower than strains WFC-SC-072 and WFC-PB-047 after fermentation, owing to the low expression level of ALD6 in strain WFC-SC-071. The higher expression level of ALD6 will result in increased acids and reduced alcohols, such as isobutanol (Zheng et al., 2020). Moreover, plenty of 2,3-butanediol was measured in ciders fermented by strains WFC-SC-071 and WFC-SC-014, which may transform from acetoin (a buttery-like smell), by the BDH1 overexpression in strains WFC-SC-071 and WFC-SC-014 (Bae et al., 2021). Notably, GPD1 and GPD2 upregulation in strain WFC-SC-071 may tune ethanol and glycerol content to produce cider with lower ethanol content (Vicente et al., 2021).
Methanol is typically detected in ciders and derives from the methyl group of pectin hydrated via yeast enzymes (Pressman et al., 2020). Methanol can also be synthesized via the yeast glycine metabolic pathway. Glycine produces methylamine by the action of yeast glycine decarboxylase, which reacts with nitrite to produce methanol (Liang et al., 2015). In vivo accumulation of methanol will cause blindness, coma, and metabolic disturbances that incur high mortality (Botelho et al., 2020). It has been previously reported that the methanol content of cider fermented by commercial Saccharomyces cerevisiae Fermicru LS2, Zymaflore VL1 and RM1515 was, respectively, 225, 227, and 219 mg/L (Nicolini et al., 2018). Methanol in ciders fermented by the two yeasts selected, 148.9 and 155.8 mg/L, were comparatively lower than commercial yeasts. Methanol content in ciders is closely associated with enzymatic activities during fermentation. Therefore, less methanol in the selected yeasts may be attributed to lower pectin methyl esterase and glycine decarboxylase expression (Blumenthal et al., 2021).
Moreover, Pichia yeasts, categorized as methylotrophic yeasts, rely on methanol or their unique methanol utilization pathways to grow. Therefore, Pichia bruneiensis WFC-PB-047 can induce less methanol accumulation than Saccharomyces cerevisiae. Since they produce less methanol, these two selected yeasts possess the potential for cider production. Nevertheless, lower methanol (25 mg/L) has been observed in cider preprocessed with bentonite and malic in apple juice before fermentation (Bedrinana et al., 2019; Simonato et al., 2021). Therefore, additional treatment could be conducted in subsequent fermentation trials to further reduce methanol production.
Esters
Esters are a vast and cardinal group of compounds in ciders. They confer pleasant fruity and floral aroma nuances to cider, thus improving perceptual quality (Munoz-Gonzalez et al., 2020). The total ester content in cider fermented by commercial Saccharomyces cerevisiae S288C was 2,077 mg/L (Zhang Z. et al., 2022). In this study, the total ester in the two selected yeasts was 3,455 and 3,672 mg/L, respectively, significantly exceeding commercial Saccharomyces cerevisiae. Acetates and fatty acid ethyl esters, the most determinant compounds contributing to the fruity and sweet scent, were dramatically increased in diversity and abundance, with their species increasing from 2 to 9 and 3 to 11, respectively (Simonato et al., 2021). Acetate is derived from pyruvate, the terminal product during glycolysis and plays a central role in carbon metabolism. Pyruvate converts to acetate via two pathways, namely coupling with reactive oxygen species (ROS) or catalyzed by ketoacid dehydrogenases, which have a neomorphic function as pyruvate decarboxylases (Liu et al., 2018). Acetate synthesis relies on catalysis by pyruvate decarboxylase (PDC; Mews et al., 2017; Bradshaw, 2021). Acetaldehyde dehydrogenase (ALD) is also essential in cytosolic acetyl-CoA and lipid production (Koivuranta et al., 2018). High expression of PDC1, ALD4, and ALD6 in the selected yeasts explained the increase in both abundance and species of acetates. Ethyl octanoate, with pineapple and candy flavor, was detected at 0.13 mg/L in Edinburgh Yeast, 0.16 mg/L in Belgian II Yeast, 0.29 mg/L in French Saison Yeast, and 0.2 mg/L in Abbey Yeast (Bingman et al., 2020), whereas, 126.9 mg/L in the WFC-PB-047 evaluated herein. Ethyl nonanoate, absent in apple juice, was highly expressed in the two selected yeasts after fermentation. Ethyl esters, as secondary metabolites in yeasts during fermentation, are synthesized upon ethanol consumption (Knight et al., 2014). Ethanol is generated from acetaldehyde by yeast alcohol dehydrogenase I encoded by the ADH1 gene (Raj et al., 2014). Upregulated expression of PDC1, which is also associated with ethanol production, supplementarily contributes to ciders’ ethanol level (Dickinson et al., 2003). Therefore, the accumulation of fatty acid ethyl esters may be associated with the high expression of ADH1 and PDC1 in the two selected yeasts.
In general, these selected strains positively enhanced apple juice fermentation and improved cider quality in multiple aspects. Additionally, some strains with slight flaws but agreeable overall performance, characterized by inferior cider product quality, could be enhanced and compensated by co-culturing with the other strains of complementary characters. This work promotes the diversification of cider, satisfies the differences in consumer demand, and will promote cider industry development in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LL: conceptualization, data curation, formal analysis, writing–original draft, and visualization. PZ: resources, supervision, writing–reviewing and editing, and methodology. CH: writing–reviewing and editing. DT: data curation. HD: supervision and project administration. YM: resources, project administration, writing–reviewing and editing, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (31972089) and the 2020 Team Innovation Project from the Fundamental Scientific Research Special Capital Fund of the National Universities, China (GK202001008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1042613/full#supplementary-material
SUPPLEMENTARY TABLE 1 | The scoring form and guidelines.
SUPPLEMENTARY TABLE 2 | Sequence of primers used in gene expression analyses.
SUPPLEMENTARY TABLE 3 | Volatile compounds of ciders produced from apple juice fermentation carried out by eight strains. “-” indicates no detection.
SUPPLEMENTARY TABLE 4 | Values of gene expression used in heat map analysis. The values are a media of triplicates.
References
Alvarez Gaona, I. J., Assof, M. V., Jofre, V. P., Combina, M., and Ciklic, I. F. (2021). Mutagenesis, screening and isolation of Brettanomyces bruxellensis mutants with reduced 4-ethylphenol production. World J. Microbiol. Biotechnol. 37:6. doi: 10.1007/s11274-020-02981-5
Arslan, M. M., Zeren, C., Aydin, Z., Akcan, R., Dokuyucu, R., Keten, A., et al. (2015). Analysis of methanol and its derivatives in illegally produced alcoholic beverages. J. Forensic Legal Med. 33, 56–60. doi: 10.1016/j.jflm.2015.04.005
Aung, M. T., Lee, P.-R., Yu, B., and Liu, S.-Q. (2015). Cider fermentation with three Williopsis saturnus yeast strains and volatile changes. Ann. Microbiol. 65, 921–928. doi: 10.1007/s13213-014-0935-7
Bae, S.-J., Kim, S., Park, H. J., Kim, J., Jin, H., Kim, B.-G., et al. (2021). High-yield production of (R)-acetoin in Saccharomyces cerevisiae by deleting genes for NAD(P)H-dependent ketone reductases producing meso-2,3-butanediol and 2,3-dimethylglycerate. Metab. Eng. 66, 68–78. doi: 10.1016/j.ymben.2021.04.001
Baeza, M., Zúñiga, S., Peragallo, V., Gutierrez, F., Crisostomo, S. K. B., Alcaino, J., et al. (2022). Response to cold: a comparative transcriptomic analysis in eight cold-adapted yeasts. Front. Microbiol. 13:828536. doi: 10.3389/fmicb.2022.828536
Bao, Y., Zhang, M., Chen, W., Chen, H., Chen, W., and Zhong, Q. (2021). Screening and evaluation of suitable non-saccharomyces yeast for aroma improvement of fermented mango juice. Food Biosci. 44:101414. doi: 10.1016/j.fbio.2021.101414
Bedrinana, R. P., Lobo, A. P., and Valles, B. S. (2019). Influence of the method of obtaining freeze-enriched juices and year of harvest on the chemical and sensory characteristics of Asturian ice ciders. Food Chem. 274, 376–383. doi: 10.1016/j.foodchem.2018.08.141
Bingman, M. T., Hinkley, J. L., Bradley, C. P. P. II, and Cole, C. A. (2022). Aroma profiles of dry-hopped ciders produced with Citra, galaxy, and mosaic hops. Appl. Sci. 12:310. doi: 10.3390/app12010310
Bingman, M. T., Stellick, C. E., Pelkey, J. P., Scott, J. M., and Cole, C. A. (2020). Monitoring cider aroma development throughout the fermentation process by headspace solid phase microextraction (HS-SPME) gas chromatography–mass spectrometry (GC-MS). Analysis 6:40. doi: 10.3390/beverages6020040
Blumenthal, P., Steger, M. C., Einfalt, D., Rieke-Zapp, J., Quintanilla Bellucci, A., Sommerfeld, K., et al. (2021). Methanol mitigation during manufacturing of fruit spirits with special consideration of novel coffee cherry spirits. Molecules 26:2585.
Botelho, G., Anjos, O., Estevinho, L. M., and Caldeira, I. (2020). Methanol in grape derived, fruit and honey spirits: a critical review on source, quality control, and legal limits. PRO :8. doi: 10.3390/pr8121609
Bradshaw, P. C. (2021). Acetyl-CoA metabolism and histone acetylation in the regulation of aging and lifespan. Antioxidants 10:572. doi: 10.3390/antiox10040572
Callejon, R. M., Clavijo, A., Ortigueira, P., Troncoso, A. M., Paneque, P., and Morales, M. L. (2010). Volatile and sensory profile of organic red wines produced by different selected autochthonous and commercial Saccharomyces cerevisiae strains. Anal. Chim. Acta 660, 68–75. doi: 10.1016/j.aca.2009.09.040
Chen, A.-J., Fu, Y.-Y., Jiang, C., Zhao, J.-L., Liu, X.-P., Liu, L., et al. (2019). Effect of mixed fermentation (Jiuqu and Saccharomyces cerevisiae EC1118) on the quality improvement of kiwi wine. CyTA J. Food 17, 967–975. doi: 10.1080/19476337.2019.1682678
Costantini, A., Cravero, M. C., Panero, L., Bonello, F., Vaudano, E., Pulcini, L., et al. (2021). Wine fermentation performance of indigenous saccharomyces cerevisiae and saccharomyces paradoxus strains isolated in a piedmont vineyard. Beverages 7:30. doi: 10.3390/beverages7020030
Cousin, F. J., Le Guellec, R., Schlusselhuber, M., Dalmasso, M., Laplace, J.-M., and Cretenet, M. (2017). Microorganisms in fermented apple beverages: current knowledge and future directions. Microorganisms 5:39. doi: 10.3390/microorganisms5030039
Dickinson, J. R., Salgado, L. E., and Hewlins, M. J. (2003). The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J. Biol. Chem. 278, 8028–8034. doi: 10.1074/jbc.M211914200
Dzialo, M. C., Park, R., Steensels, J., Lievens, B., and Verstrepen, K. J. (2017). Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41, S95–S128. doi: 10.1093/femsre/fux031
Ehsani, M., Fernández, M. R., Biosca, J. A., Julien, A., and Dequin, S. (2009). Engineering of 2,3-butanediol dehydrogenase to reduce acetoin formation by glycerol-overproducing, low-alcohol Saccharomyces cerevisiae. Appl. Environ. Microbiol. 75, 3196–3205. doi: 10.1128/AEM.02157-08
Fejzullahu, F., Kiss, Z., Kun-Farkas, G., and Kun, S. (2021). Influence of non-saccharomyces strains on chemical characteristics and sensory quality of fruit Spirit. Foods 10:1336. doi: 10.3390/foods10061336
Ghosh, P., Cao, X., Pal, J., Chowdhury, S., Saha, S., Nandi, S., et al. (2021). Mathematical modeling for the prevention of methanol poisoning through ethanol by impulsive way. Different. Equat. Dynam. Syst. 29, 435–452. doi: 10.1007/s12591-018-0420-z
Godoy, L., Acuña-Fontecilla, A., and Catrileo, D. (2020). Formation of Aromatic and Flavor Compounds in Wine: A Perspective of Positive and Negative Contributions of Non-Saccharomyces Yeasts. Chemistry and Biochemistry of Winemaking, Wine Stabilization and Aging IntechOpen.
Guine, R. P. F., Barroca, M. J., Coldea, T. E., Bartkiene, E., and Anjos, O. (2021). Apple fermented products: An overview of technology, Properties and Health Effects. PRO 9:223. doi: 10.3390/pr9020223
Gulmezian, M., Hyman, K. R., Marbois, B. N., Clarke, C. F., and Javor, G. T. (2007). The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467, 144–153. doi: 10.1016/j.abb.2007.08.009
Gutiérrez, A., Boekhout, T., Gojkovic, Z., and Katz, M. J. J. O. T. I. O. B. (2018). Evaluation of non-Saccharomyces yeasts in the fermentation of wine, beer and cider for the development of new beverages. J. Inst. Brew. 124, 389–402. doi: 10.1002/jib.512
Hinkley, J. L., Bingman, M. T., Lee, J. S., Bradley, C. P., and Cole, C. A. (2022). Volatile profile survey of five apple varieties grown in Southwest Colorado from juice to finished, dry-hopped cider. J. Am. Soc. Brew. Chem., 1–10. doi: 10.1080/03610470.2021.2013645
Kalender, Ö., and Çalık, P. J. A. M. (2020). Transcriptional regulatory proteins in central carbon metabolism of Pichia pastoris and Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 104, 7273–7311. doi: 10.1007/s00253-020-10680-2
Khandpur, P., and Gogate, P. R. (2015). Effect of novel ultrasound based processing on the nutrition quality of different fruit and vegetable juices. Ultrason Sonochem 27, 125–136. doi: 10.1016/j.ultsonch.2015.05.008
Knight, M. J., Bull, I. D., and Curnow, P. (2014). The yeast enzyme Eht1 is an octanoyl-CoA:ethanol acyltransferase that also functions as a thioesterase. Yeast 31, 463–474. doi: 10.1002/yea.3046
Koivuranta, K., Castillo, S., Jouhten, P., Ruohonen, L., Penttila, M., and Wiebe, M. G. (2018). Enhanced triacylglycerol production with genetically modified Trichosporon oleaginosus. Front. Microbiol. 9:1337. doi: 10.3389/fmicb.2018.01337
Kumari, A., Kumar, R., Kumar, S., and Siddiqui, S. (2020). Screening the efficiency of various strains of yeast for wine production from grapes. Indian J. Hortic. 77, 559–561. doi: 10.5958/0974-0112.2020.00082.1
Lee, B., Heo, J., Hong, S., Kim, E.-Y., Sohn, Y. J., and Jung, H.-S. (2019). dl-Malic acid as a component of alpha-hydroxy acids: effect on 2,4-dinitrochlorobenzene-induced inflammation in atopic dermatitis-like skin lesions in vitro and in vivo. Immunopharmacol. Immunotoxicol. 41, 614–621. doi: 10.1080/08923973.2019.1680688
Liang, M.-H., Liang, Y.-J., Wu, X.-N., Zhou, S.-S., and Jiang, J.-G. (2015). Mutation breeding of Saccharomyces cerevisiae with lower methanol content and the effects of pectinase, cellulase and glycine in sugar cane spirits. J. Sci. Food Agric. 95, 1949–1955. doi: 10.1002/jsfa.6908
Lin, F., Ferguson, K. L., Boyer, D. R., Lin, X. N., and Marsh, E. N. (2015). Isofunctional enzymes PAD1 and UbiX catalyze formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase. ACS Chem. Biol. 10, 1137–1144. doi: 10.1021/cb5008103
Liu, X., Cooper, D. E., Cluntun, A. A., Warmoes, M. O., Zhao, S., Reid, M. A., et al. (2018). Acetate production from glucose and coupling to mitochondrial metabolism in mammals. Cells 175, 502–513.e13. doi: 10.1016/j.cell.2018.08.040
Lorca Mandujano, G. P., Alves, H. C., Prado, C. D., Martins, J. G. O., Novaes, H. R., De Oliveira, M., et al. (2022). Identification and selection of a new Saccharomyces cerevisiae strain isolated from Brazilian ethanol fermentation process for application in beer production. Food Microbiol. 103:103958. doi: 10.1016/j.fm.2021.103958
Lorenzini, M., Simonato, B., Slaghenaufi, D., Ugliano, M., and Zapparoli, G. (2019). Assessment of yeasts for apple juice fermentation and production of cider volatile compounds. Lwt-Food Sci. Technol. 99, 224–230. doi: 10.1016/j.lwt.2018.09.075
Marullo, P., and Dubourdieu, D. (2022). Yeast Selection for Wine Flavor Modulation Managing Wine Quality Elsevier. Vol 2, 371–426.
Mews, P., Donahue, G., Drake, A. M., Luczak, V., Abel, T., and Berger, S. L. (2017). Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386. doi: 10.1038/nature22405
Mikhailova, E. V., Karpun, N. N., Agumava, A. A., Efremov, A. M., Ravish, C., and Samarina, L. S. (2020). DNA-detection of leaf curl pathogen Taphrina deformans in asymptomatic leaves of peach (Prunus persica (L.) Batsch) in Russia. Res. J. Biotechnol. 15, 126–129.
Mitsui, R., Yamada, R., Matsumoto, T., and Ogino, H. (2022). Bioengineering for the industrial production of 2,3-butanediol by the yeast, Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 38:38. doi: 10.1007/s11274-021-03224-x
Munoz-Gonzalez, C., Perez-Jimenez, M., and Pozo-Bayon, M. A. (2020). Oral persistence of esters is affected by wine matrix composition. Food Res. Int. 135:109286. doi: 10.1016/j.foodres.2020.109286
Munoz-Miranda, L. A., Pereira-Santana, A., Gomez-Angulo, J. H., Christine Gschaedler-Mathis, A., Amaya-Delgado, L., Figueroa-Yanez, L. J., et al. (2022). Identification of genes related to hydrolysis and assimilation of agave fructans in Candida apicola NRRL Y-50540 and Torulaspora delbrueckii NRRL Y-50541 by denovo transcriptome analysis. FEMS Yeast Res. 22:foac005. doi: 10.1093/femsyr/foac005
Nadai, C., Crosato, G., Giacomini, A., and Corich, V. (2021). Different gene expression patterns of hexose transporter genes modulate fermentation performance of four Saccharomyces cerevisiae strains. Fermentation 7:164. doi: 10.3390/fermentation7030164
Nespor, J., Karabin, M., Stulikova, K., and Dostalek, P. (2019). An HS-SPME-GC-MS method for profiling volatile compounds as related to technology used in cider production. Molecules 24:2117. doi: 10.3390/molecules24112117
Nicolini, G., Román, T., Carlin, S., Malacarne, M., Nardin, T., Bertoldi, D., et al. (2018). Characterisation of single-variety still ciders produced with dessert apples in the Italian Alps. J. Inst. Brew. 124, 457–466. doi: 10.1002/jib.510
Nie, Q., Hu, J., Gao, H., Li, M., Sun, Y., Chen, H., et al. (2021). Bioactive dietary fibers selectively promote gut microbiota to exert antidiabetic effects. J. Agric. Food Chem. 69, 7000–7015. doi: 10.1021/acs.jafc.1c01465
Niu, Y., Wang, R., Xiao, Z., Zhu, J., Sun, X., and Wang, P. (2019). Characterization of ester odorants of apple juice by gas chromatography-olfactometry, quantitative measurements, odour threshold, aroma intensity and electronic nose. Food Res. Int. 120, 92–101. doi: 10.1016/j.foodres.2019.01.064
Padilla, B., Gil, J. V., and Manzanares, P. J. F. I. M. (2016). Past and future of non-Saccharomyces yeasts: From spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7:411. doi: 10.3389/fmicb.2016.00411
Pressman, P., Clemens, R., Sahu, S., and Hayes, A. W. (2020). A review of methanol poisoning: a crisis beyond ocular toxicology. Cutan. Ocul. Toxicol. 39, 173–179. doi: 10.1080/15569527.2020.1768402
Qin, Z., Petersen, M. A., and Bredie, W. L. P. (2018). Flavor profiling of apple ciders from the UK and Scandinavian region. Food Res. Int. 105, 713–723. doi: 10.1016/j.foodres.2017.12.003
Raj, S. B., Ramaswamy, S., and Plapp, B. V. (2014). Yeast alcohol dehydrogenase structure and catalysis. Biochemistry 53, 5791–5803. doi: 10.1021/bi5006442
Saberi, S., Cliff, M. A., and Van Vuuren, H. J. J. (2012). Impact of mixed S. cerevisiae strains on the production of volatiles and estimated sensory profiles of chardonnay wines. Food Res. Int. 48, 725–735. doi: 10.1016/j.foodres.2012.06.012
Simonato, B., Lorenzini, M., and Zapparoli, G. (2021). Effects of post-harvest fungal infection of apples on chemical characteristics of cider. Lwt-Food Sci. Technol. 138:110620. doi: 10.1016/j.lwt.2020.110620
Song, Y., Gibney, P., Cheng, L., Liu, S., and Peck, G. (2020). Yeast Assimilable nitrogen concentrations influence yeast gene expression and hydrogen sulfide production during cider fermentation. Front. Microbiol. 11:1264. doi: 10.3389/fmicb.2020.01264
Sun, J., Cui, H., Wu, B., Wang, W., Yang, Q., Zhang, Y., et al. (2022). Genome-wide identification of cotton (Gossypium spp.) Glycerol-3-phosphate dehydrogenase (GPDH) family members and the role of GhGPDH5 in response to drought stress. Plan. Theory 11:592. doi: 10.3390/plants11050592
Sun, S. Y., Gong, H. S., Zhao, Y. P., Liu, W. L., and Jin, C. W. (2016). Sequential culture with Torulaspora delbrueckii and Saccharomyces cerevisiae and management of fermentation temperature to improve cherry wine quality. J. Sci. Food Agric. 96, 1880–1887. doi: 10.1002/jsfa.7293
Van Mullem, J. J., Zhang, J., Dias, D. R., and Schwan, R. F. (2022). Using wild yeasts to modulate the aroma profile of low-alcoholic meads. Braz. J. Microbiol. 53, 1743–1749. doi: 10.1007/s42770-022-00840-z
Van Wyk, N., Grossmann, M., Wendland, J., Von Wallbrunn, C., and Pretorius, I. S. (2019). The whiff of wine yeast innovation: strategies for enhancing aroma production by yeast during wine fermentation. J. Agric. Food Chem. 67, 13496–13505. doi: 10.1021/acs.jafc.9b06191
Varga, V., Štefuca, V., Mihálová, L., Levarski, Z., Struhárňanská, E., Blaško, J., et al. (2021). Recombinant enzymatic redox systems for preparation of aroma compounds by biotransformation. Front. Microbiol. 12:684640. doi: 10.3389/fmicb.2021.684640
Vicente, J., Calderon, F., Santos, A., Marquina, D., and Benito, S. (2021). High potential of Pichia kluyveri and other Pichia species in wine technology. Int. J. Mol. Sci. 22:1196. doi: 10.3390/ijms22031196
Vilela, A. J. F. (2021). An overview of CRISPR-based technologies in wine yeasts to improve wine flavor and safety. Bus. Ferment. 7:5. doi: 10.3390/fermentation7010005
Wang, D., Jiang, Y., Sun-Waterhouse, D.-X., Zhai, H., Guan, H., Rong, X., et al. (2020). MicroRNA-based regulatory mechanisms underlying the synergistic antioxidant action of quercetin and catechin in H2O2-stimulated HepG2 cells: roles of BACH1 in Nrf2-dependent pathways. Free Radic. Biol. Med. 153, 122–131. doi: 10.1016/j.freeradbiomed.2020.04.018
Wang, M., Sun, Z., Wang, Y., Wei, Z., Chen, B., Zhang, H., et al. (2019). The effect of pitching rate on the production of higher alcohols by top-fermenting yeast in wheat beer fermentation. Ann. Microbiol. 69, 713–726. doi: 10.1007/s13213-019-01463-w
Wang, X., Wang, Z., and Feng, T. (2022). Screening of yeast in various vineyard soil and study on its flavor compounds from brewing grape wine. Molecules 27:512.
Wei, X., Francoise, U., Qin, M., Chen, Q., Li, Y., Sun, X., et al. (2020b). Effects of different fermentation and storage conditions on methanol content in Chinese spine grape (Vitis davidii Foex) wine. Cyta J. Food 18, 367–374. doi: 10.1080/19476337.2020.1737238
Wei, J., Zhang, Y., Qiu, Y., Guo, H., Ju, H., Wang, Y., et al. (2020a). Chemical composition, sensorial properties, and aroma-active compounds of ciders fermented with Hanseniaspora osmophila and Torulaspora quercuum in co- and sequential fermentations. Food Chem. 306:125623. doi: 10.1016/j.foodchem.2019.125623
Wu, C.-F., Cheng, H., Chang, W.-C., Wu, M.-C., and Lin, J.-S. (2017). Use of highly Methoxyl-esterified cross-linked alcohol insoluble substance to remove Pectinesterase for the reduction of methanol in sweet Orange wine. J. Pure Appl. Microbiol. 11, 1239–1249. doi: 10.22207/JPAM.11.3.02
Wu, L., Gao, Y., Ren, W.-C., Su, Y., Li, J., Du, Y.-Q., et al. (2022). Rapid determination and origin identification of total polysaccharides contents in Schisandra chinensis by near-infrared spectroscopy. Spectrochim. Acta Part A. Mol. Biomol. Spectroscopy 264:120327. doi: 10.1016/j.saa.2021.120327
Xia, Y., Chen, F., Du, Y., Liu, C., Bu, G., Xin, Y., et al. (2019). A modified SDS-based DNA extraction method from raw soybean. Biosci. Rep. 39:BSR20182271. doi: 10.1042/BSR20182271
Xiao, Z., Zhou, X., Niu, Y., Yu, D., Zhu, J., and Zhu, G. (2015). Optimization and application of headspace-solid-phase micro-extraction coupled with gas chromatography-mass spectrometry for the determination of volatile compounds in cherry wines. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 978-979, 122–130. doi: 10.1016/j.jchromb.2014.12.006
Xu, X., Bao, Y., Wu, B., Lao, F., Hu, X., and Wu, J. (2019). Chemical analysis and flavor properties of blended orange, carrot, apple and Chinese jujube juice fermented by selenium-enriched probiotics. Food Chem. 289, 250–258. doi: 10.1016/j.foodchem.2019.03.068
Xu, W., Jian, Z., and Kong, X. (2021a). RNA-seq-based transcriptomic comparison of Saccharomyces cerevisiae during spontaneous and inoculated fermentations of organic and conventional grapes. Lwt-Food Sci. Technol. 143:111183. doi: 10.1016/j.lwt.2021.111183
Xu, X., Niu, C., Liu, C., Wang, J., Zheng, F., and Li, Q. (2021b). Screening lager yeast with higher ethyl-acetate production by adaptive laboratory evolution in high concentration of acetic acid. World J. Microbiol. Biotechnol. 37:125. doi: 10.1007/s11274-021-03082-7
Zhang, C., Hu, C., Wang, X., Meng, Y., and Guo, Y. (2021). Polyphenols in fermented apple juice: beneficial effects on human health. J. Funct. Foods 76:104294. doi: 10.1016/j.jff.2020.104294
Zhang, X., Jiang, R., Lin, H., and Xu, S. (2020). Feeding tricarboxylic acid cycle intermediates improves lactate consumption and antibody production in Chinese hamster ovary cell cultures. Biotechnol. Prog. 36, e2975. doi: 10.1002/btpr.297
Zhang, Z., Lan, Q., Yu, Y., Zhou, J., and Lu, H. J. F. C. M. S. (2022). Comparative metabolome and transcriptome analyses of the properties of Kluyveromyces marxianus and Saccharomyces yeasts in apple cider fermentation. Food Chem. 4:100095. doi: 10.1016/j.fochms.2022.100095
Zhang, G., Zhang, C., Wang, Z., Wang, Q., Nielsen, J., and Dai, Z. J. M. E. (2022). Dual β-oxidation pathway and transcription factor engineering for methyl ketones production in Saccharomyces cerevisiae. Metab. Eng. 73, 225–234.
Zhao, Y., Wang, P., Zhan, P., Tian, H., Lu, C., and Tian, P. (2021). Aroma characteristics of cloudy kiwifruit juices treated with high hydrostatic pressure and representative thermal processes. Food Res. Int. 139:109841. doi: 10.1016/j.foodres.2020.109841
Keywords: Saccharomyces cerevisiae, Pichia bruneiensis, cider, methanol, alcohols, esters
Citation: Liu L, Zhao PT, Hu CY, Tian D, Deng H and Meng YH (2022) Screening low-methanol and high-aroma produced yeasts for cider fermentation by transcriptive characterization. Front. Microbiol. 13:1042613. doi: 10.3389/fmicb.2022.1042613
Edited by:
Makoto Kimura, Nagoya University, JapanReviewed by:
Tomoyuki Nakagawa, Gifu University, JapanYongsheng Tao, Northwest A&F University, China
Copyright © 2022 Liu, Zhao, Hu, Tian, Deng and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Deng, aG9uZ2RlbkBzbm51LmVkdS5jbg==; Yong Hong Meng, bWVuZ3lvbmdob25nQHNubnUuZWR1LmNu
 Liang Liu1
Liang Liu1 Yong Hong Meng
Yong Hong Meng