- USCR Bactériologie Moléculaire & Génomique, Institut National des Sciences Appliquées et de Technologie, Université de Carthage, Tunis, Tunisia
The genus Frankia is at present the sole genus in the family Frankiaceae and encompasses filamentous, sporangia-forming actinomycetes principally isolated from root nodules of taxonomically disparate dicotyledonous hosts named actinorhizal plants. Multiple independent phylogenetic analyses agree with the division of the genus Frankia into four well-supported clusters. Within these clusters, Frankia strains are well defined based on host infectivity range, mode of infection, morphology, and their behaviour in culture. In this study, phylogenomics, overall genome related indices (OGRI), together with available data sets for phenotypic and host-plant ranges available for the type strains of Frankia species, were considered. The robustness and the deep radiation observed in Frankia at the subgeneric level, fulfilling the primary principle of phylogenetic systematics, were strengthened by establishing genome criteria for new genus demarcation boundaries. Therefore, the taxonomic elevation of the Frankia clusters to the rank of the genus is proposed. The genus Frankia should be revised to encompass cluster 1 species only and three novel genera, Protofrankia gen. nov., Parafrankia gen. nov., and Pseudofrankia gen. nov., are proposed to accommodate clusters 2, 3, and 4 species, respectively. New combinations for validly named species are also provided.
Introduction
The genus Frankia Brunchorst, 1886 (Approved Lists 1980) (Brunchorst, 1886; Becking, 1970; Skerman et al., 1980; Lechevalier and Lechevalier, 1989) is currently the solitary genus within the family Frankiaceae (Becking, 1970; Hahn et al., 1989; Normand et al., 1996; Stackebrandt et al., 1997) of the order Frankiales (Sen et al., 2014; Normand and Benson, 2015). The genus encompasses soil-inhabiting mesophilic actinomycetes mostly are able to fix dinitrogen and to establish symbiosis with pioneer and economically important plants, collectively named actinorhizal plants (Normand and Fernandez, 2019). Both in culture and the host root nodules, Frankia strains produce branched septate hyphae which, for most analysed strains, can carry multilocular sporangia, while the diazotrophic strains produce vesicles, where nitrogen fixation occurs (Normand et al., 2014; Gtari et al., 2019). Based on the 16S rRNA gene phylogeny (Normand et al., 1996), which was further substantiated by internal transcribed spacer (ITS) 16S-23S rRNA genes (Ghodhbane-Gtari et al., 2010), multilocus sequence analysis (MLSA) (Nouioui et al., 2011; Persson et al., 2011; Sen et al., 2014; Gtari et al., 2015; Pozzi et al., 2018), amplified fragment length polymorphism (AFLP) (Bautista et al., 2011) and whole-genome analyses (Gtari et al., 2019; Nouioui et al., 2019b), four phylogenetic clusters are consistently delineated within the genus, grouping strains with similar cultural behaviour, morphology, host range, and mode of infection (Benson and Silvester, 1993). Cluster 1 members colonise species of Alnus (Betulaceae), Allocasuarina and Casuarina (Casuarinaceae), and Comptonia and Myrica (Myricaceae), while cluster 2 contains strains that infect Coriariaceae, Datiscaceae, Dryadoideae (Rosaceae), and Ceanothus (Rhamnaceae). Cluster 3 contains Frankia associated with Elaeagnaceae, Colletieae (Rhamnaceae), Morella (Myricaceae), and Gynmnostoma (Casuarinaceae). Members of cluster 3 have been also occasionally isolated from Alnus, Allocasuarina, Casuarina, Ceanothus, and Dryadoideae root nodules (Benson and Dawson, 2007). Finally, cluster 4 includes asymbiotic Frankia strains, which are unable to fix nitrogen and/or to re-infect their host plants.
Prior to the genomic era, bacterial taxonomy relied on the polyphasic approach (Colwell, 1970) which integrates morphological, metabolic, and chemotaxonomic makers, 16S rRNA phylogeny, and pairwise dissimilarity. For ambiguous situations, the gold standard DNA–DNA hybridization (DDH) was used for drawing conclusions on species delineation (Wayne et al., 1987). For the genus Frankia, problems of applying traditional bacteriological techniques persisted due primarily to the high proportion of uncultivable strains and, secondly, to the very slow growing rate of most cultivated strains. Metabolic behaviours and wet-lab experimental DNA relatedness were, thus, inconsistent when the polyphasic taxonomic approach was applied in the case of the genus Frankia (Normand and Fernandez, 2008; Gtari et al., 2013).
Taking advantage of incorporating Taxogenomic and omniLog® phenoarray into the polyphasic approach (Gtari et al., 2019), 13 species with validly published names have been described based on accepted thresholds for bacterial species delineation, i.e., 99.0% (with a maximum probability of error of 1.0%) for 16S rRNA similarities (Meier-Kolthoff et al., 2013), 70% for digital DDH (Auch et al., 2010), and 95% for average nucleotide identity (ANI) (Konstantinidis et al., 2006). Ten of these species are facultative symbiotic species from clusters 1, 2, and 3 and include the type species Frankia alni (Nouioui et al., 2016), Frankia canadensis (Normand et al., 2018), Frankia casuarinae (Nouioui et al., 2016), and Frankia torreyi (Nouioui et al., 2019a) of cluster 1; Frankia coriariae (Nouioui et al., 2017b) of cluster 2; and Frankia elaeagni (Nouioui et al., 2016), Frankia discariae (Nouioui et al., 2017d),Frankia irregularis (Nouioui et al., 2018b), Frankia soli (Gtari et al., 2020), and Frankia colletiae (Nouioui et al., 2022) of cluster 3. Cluster 4 includes Frankia asymbiotica (Nouioui et al., 2017c), Frankia inefficax (Nouioui et al., 2017a), and Frankia saprophytica (Nouioui et al., 2018a). Additionally, four candidate species are also defined to accommodate uncultured taxa: Candidatus Frankia datiscae (Persson et al., 2011) and Candidatus Frankia californiensis (Normand et al., 2017) from cluster 2, as well as Candidatus Frankia alpina (Pozzi et al., 2020) and Candidatus Frankia nodulisporulans (Herrera-Belaroussi et al., 2020) from cluster 1.
Bacterial classification at higher taxonomic ranks relies primarily on phylogenetic systematics (Ludwig and Klenk, 2005; Oren and Garrity, 2014) which has been greatly improved through phylogenomics (Dagan, 2011; Oren and Garrity, 2014; Hugenholtz et al., 2021). Genomic criteria for demarcating genus boundaries include conserved indels and proteins signatures (Naushad et al., 2014), comparative pangenome (Caputo et al., 2019), average amino acid identity (AAI) (Konstantinidis and Tiedje, 2007), percentage of conserved proteins (POCP) (Qin et al., 2014), and ANI (Barco et al., 2020). The gold standard overall genome related indices (OGRI) for genus demarcation remains pending inquiry (Sant’Anna et al., 2019).
The commonplace availability of genomic tools and algorithms for phylogenetic purposes and OGRI has motivated our interest to review the taxonomic structure of the genus Frankia in relation to the evolutionary history of its well-known phylogenetic clusters. The results obtained in the present study support the taxonomic elevation of each of the four clusters to the rank of the genus. Hence, three novel genera, Protofrankia gen. nov., Parafrankia gen. nov., and Pseudofrankia gen. nov., to accommodate clusters 2, 3, and 4, respectively, with 9 related new combinations, are proposed.
Materials and methods
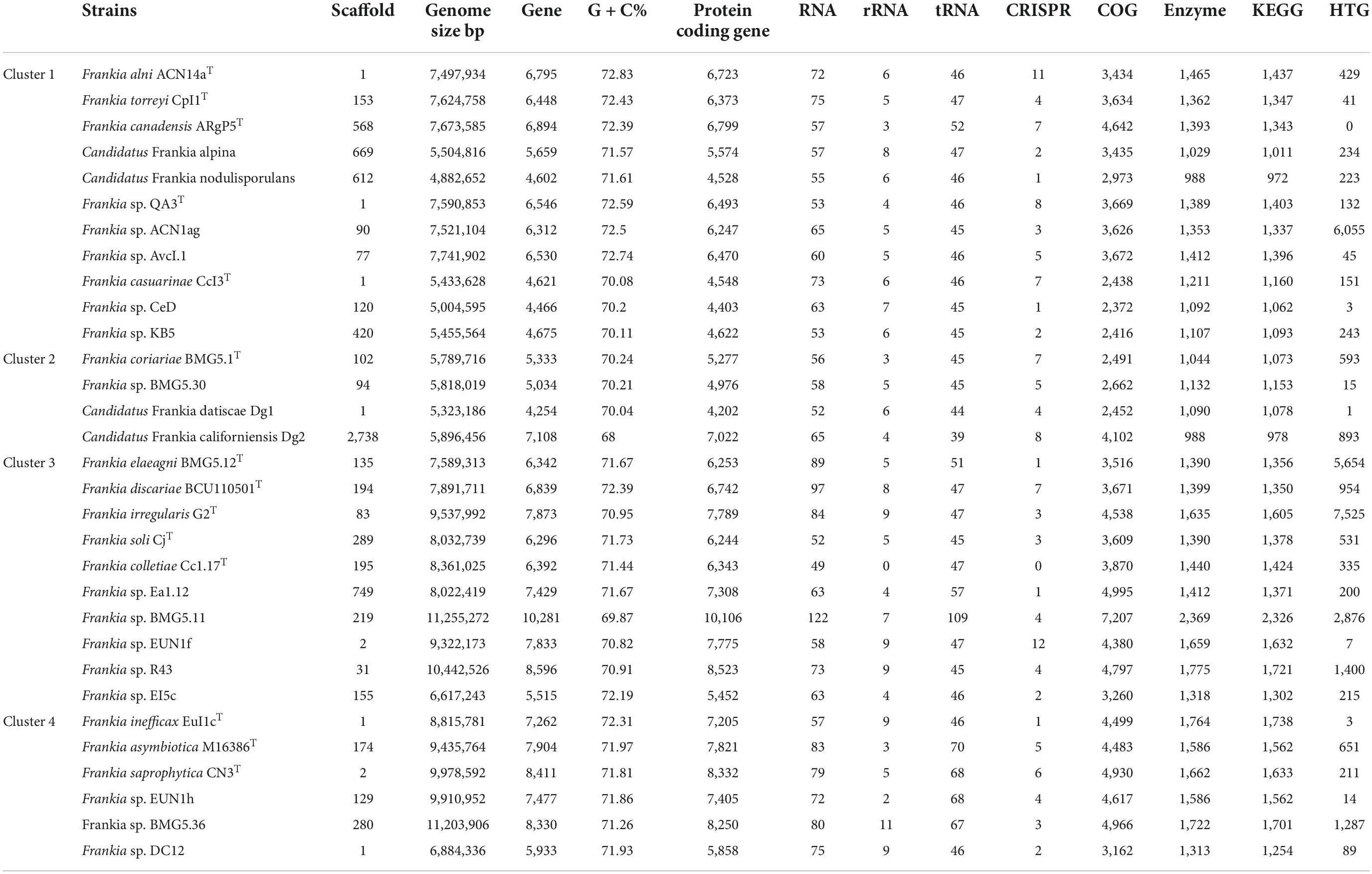
Complete and draft genomes for type strains and candidate species together with other selected published genomes covering current Frankia diversity were used in this study and are listed in Table 1.
Phylogenetic analysis
Phylogenetic analyses based on 16S rRNA gene sequences were carried out using the GGDC web server adapted to single genes (Meier-Kolthoff et al., 2013). Maximum-likelihood (ML) and maximum-parsimony (MP) trees were inferred with RAxML (Stamatakis, 2014) and tree analysis new technology (TNT) (Goloboff et al., 2008), respectively. For ML, rapid bootstrapping was used in conjunction with the autoMRE bootstrapping criterion (Pattengale et al., 2010), followed by a search for the best tree. For MP, 1,000 bootstrapping replicates were used in conjunction with tree-bisection-and-reconnection branch swapping and 10 random addition-sequence replicates. The sequences were checked for compositional bias using the χ2 test as implemented in phylogenetic analysis using parsimony (PAUP)* (Swofford, 2002).
The whole-genome-based taxonomic analysis was performed through the Type Strain Genome Server1 (Meier-Kolthoff and Göker, 2019; Meier-Kolthoff et al., 2022). Pairwise genomic comparisons were calculated and intergenomic distances were inferred under the algorithm “trimming” and distance formula d5 using 100 distance replicates in FastME 2.0 (Lefort et al., 2015).
The pangenome analysis was performed using Roary (Page et al., 2015) implemented in the “Pan” module of the Prokaryotic Genomics and Comparative Genomics Analysis Pipeline (PGCGAP v1.0.21) (Liu et al., 2020). Single-copy analysis of core proteins, alignment of sequences, sequence concatenation, best model, and phylogenetic tree reconstruction based on 303 single copies of core proteins were performed with the “CoreTree” module of PGCGAP v1.0.21.
Overall genome related indices calculations
Overall genome related indices, including pairwise ANI, were calculated through the IMG/M data management and analysis system (Varghese et al., 2015). Pairwise AAI was calculated with the EzAAI tool (v1.1) (Kim et al., 2021) with default settings, which use MMSeqs2 for protein comparisons and consider a minimum query coverage of 50% and a minimum identity of 40% for AAI calculations. Pairwise POCP was calculated according to Qin et al.’s (2014) findings, following the steps described by Adamek et al. (2018) and Margos et al. (2018). Reciprocal BlastP (Altschul, 1997) for each pair of genomes used an E-value of < 1 × 10–5, > 40% sequence identity and > 50% of the query sequence. The pairwise POCP value was then deduced as [(C1 + C2)/(T1 + T2)] × 100, where C1 and C2 represent the conserved proteins numbers between the genome pair, while T1 and T2 are the total numbers of compared proteins in each genome (Qin et al., 2014).
Results and discussion
Phylogenetics and phylogenomics
An update of the 16S rRNA phylogeny was, here made available using the largest possible dataset of sequences (n = 72) from all type strains and candidate species, and those available in public databases or extractable from genome sequences providing a size of ≥ 900 nt (Figure 1). A more robust phylogenetic history was obtained using a single copy of core proteins (Figure 2) and whole-genome sequences (Figure 3), as shown by higher bootstrap and posterior probability values supporting the branching for the four clusters. Otherwise, in congruence with Salam et al. (2020), the most closely related Actinomycetia to Frankiaceae are Acidothermus (Acidothermaceae and Acidothermales) and Jatrophihabitans (Jatrophihabitantaceae and Jatrophihabitantales).
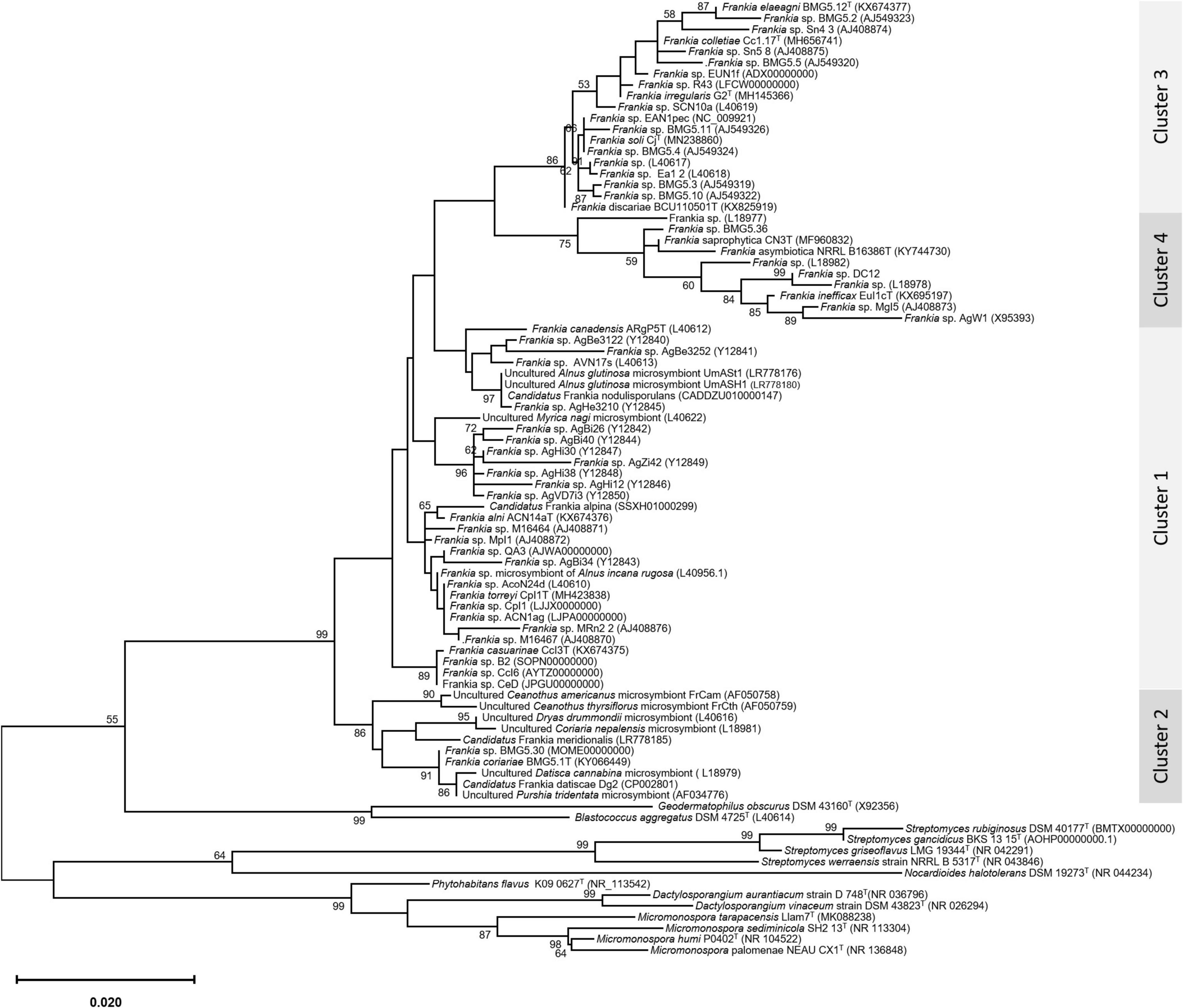
Figure 1. A maximum-likelihood tree based on the 16s rRNA gene sequences of 72 members of the genus Frankia. The maximum-likelihood (ML) tree was inferred using the GTR + GAMMA model and rooted by midpoint rooting, and the branches are scaled in terms of the expected number of substitutions per site. The numbers above the branches support values when larger than 60% bootstrapping.
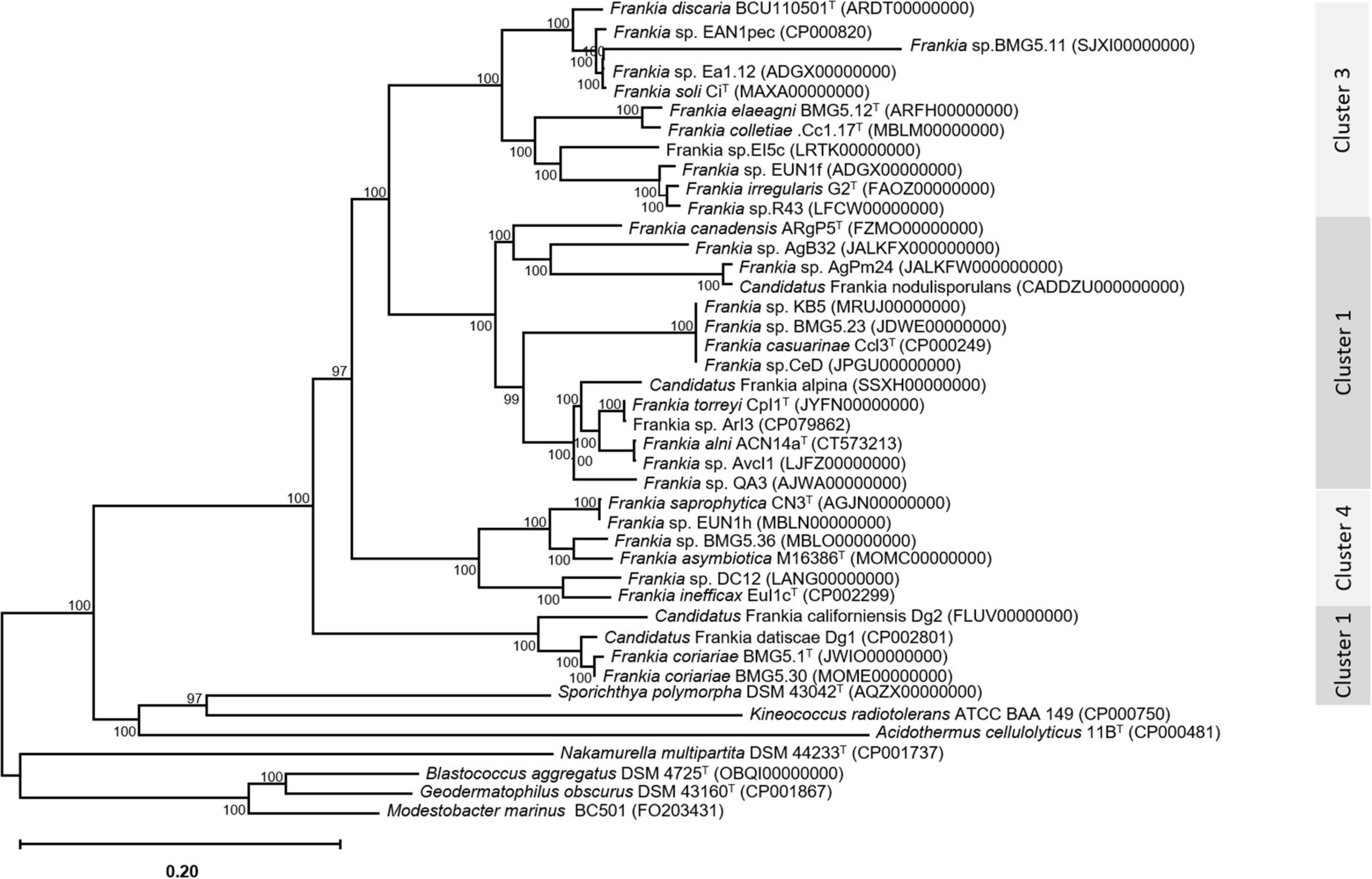
Figure 2. A maximum-likelihood tree inferred by the module “CoreTree” of PGCGAP v1.0.21 (Liu et al., 2020) with the best-fit model JTT + F + R4 and based on the concatenation of single-copy conserved proteins. The midpoint-rooted phylogenetic tree of single-copy core proteins.
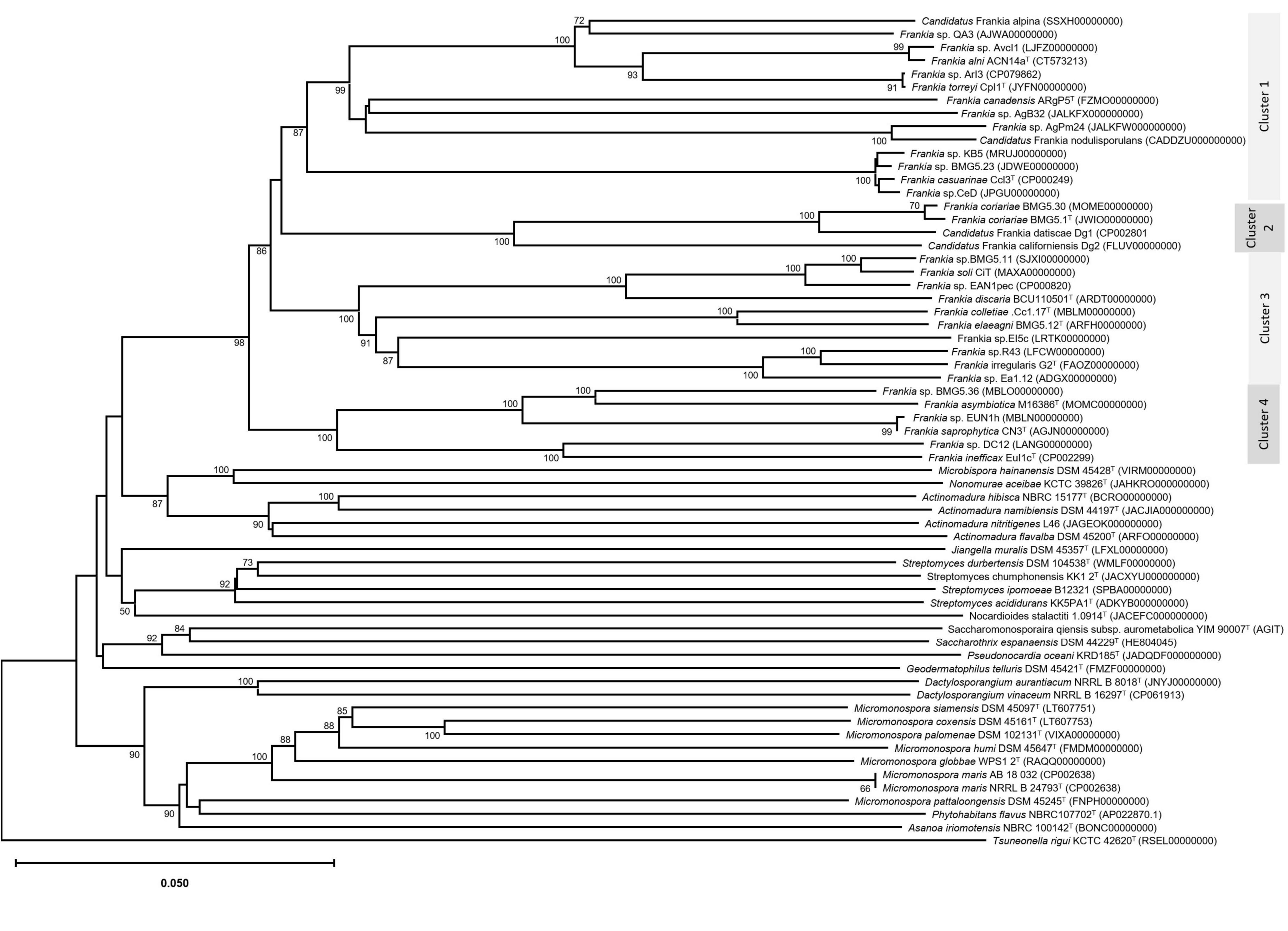
Figure 3. Phylogenomic tree based on genome sequences in the Type Strain Genome Server (TYGS) tree inferred in FastME 2.1.6.1 (Meier-Kolthoff and Göker, 2019) based on genome BLAST distance phylogeny (GBDP). The branch lengths are scaled based GBDP distance formula d5. The tree was rooted at the midpoint (Meier-Kolthoff et al., 2013). GBDP pseudo-bootstrap support values > 60% from 100 replications, with average branch support of 93.7% were shown.
The trees obtained, in this study, showed broadly similar patterns and topology with those previously reported for the 16S rRNA gene (Normand et al., 1996), ITS 16S-23S rRNA (Ghodhbane-Gtari et al., 2010), AFLP (Bautista et al., 2011), or combined data sets; 16S rRNA and glnA (Clawson et al., 2004), GyrB, glnII, and nifH (Nouioui et al., 2011), atpD, dnaA, ftsZ, pgk, and rpoB (Sen et al., 2014; Gtari et al., 2015; Nouioui et al., 2017a,b, 2018a,2018b,2019a; Pozzi et al., 2018), as well as 50 (Sen et al., 2014; Persson et al., 2015) or up to 200 gene sequences (Gtari et al., 2019; Nouioui et al., 2019b). Alongside the trees’ robustness and topology congruencies for phylogenetic splitting, no ambiguous or shifting affiliations between clusters were observed for any of the studied strains regardless of the algorithms or the extent of the genomic region used for inferring Frankia evolutionary history.
The relative positioning of each of the four Frankia clusters, and the timeline assumed for their separation, have been for a long a source of debate. While 16S rRNA (Normand et al., 1996), ITS 16S-23S rRNA (Ghodhbane-Gtari et al., 2010), AFLP (Bautista et al., 2011), and MLSA (Nouioui et al., 2011) based phylogenies placed cluster 4 at the base of the tree, followed by cluster 3 and then clusters 1 and 2 which form sister groups, and other MLSA studies (Pozzi et al., 2018) or concatenation of proteins (Sen et al., 2014; Gtari et al., 2015; Persson et al., 2015) showed cluster 2 as basal, followed by cluster 4 and then the symbiotic cluster 3, and finally, cluster 1 as the most derived. Both situations were here obtained based on the whole genome (Figure 3) and on core proteins (Figure 2), respectively, which could imply a different evolution pressure of the whole genome versus the core genome or a bias in codon usage.
Whole-genome-based criteria
Inter-cluster pairwise AAI (Figure 4 and Supplementary Table 1), POCP (Figure 4 and Supplementary Table 2), and ANI (Supplementary Table 3) values ranged between 66.5 and 72.2, 33.5 and 61.3, and 78 and 81.5%, respectively. A cut-off AAI value at 72.2 ± 0.03%, which is within the 60–80% threshold recommended for the delineation of genera (Konstantinidis and Tiedje, 2005; Rodriguez-R and Konstantinidis, 2014), permitted a congruent regrouping with phylogenetic clusters. The POCP threshold of 50% originally proposed for genus delineation by Qin et al. (2014) has been shown to be inappropriate for multiple taxa (Aliyu et al., 2016; Pannekoek et al., 2016; Li et al., 2017; Lopes-Santos et al., 2017; Orata et al., 2018; Wirth and Whitman, 2018; Park et al., 2022). Sangal et al. (2022) considered this POCP cutoff as overly stringent and proposed its reappraisement to 58–66%. The inter-cluster pairwise POCP values obtained in this study (33.5–61.3%) are within this proposed new range.
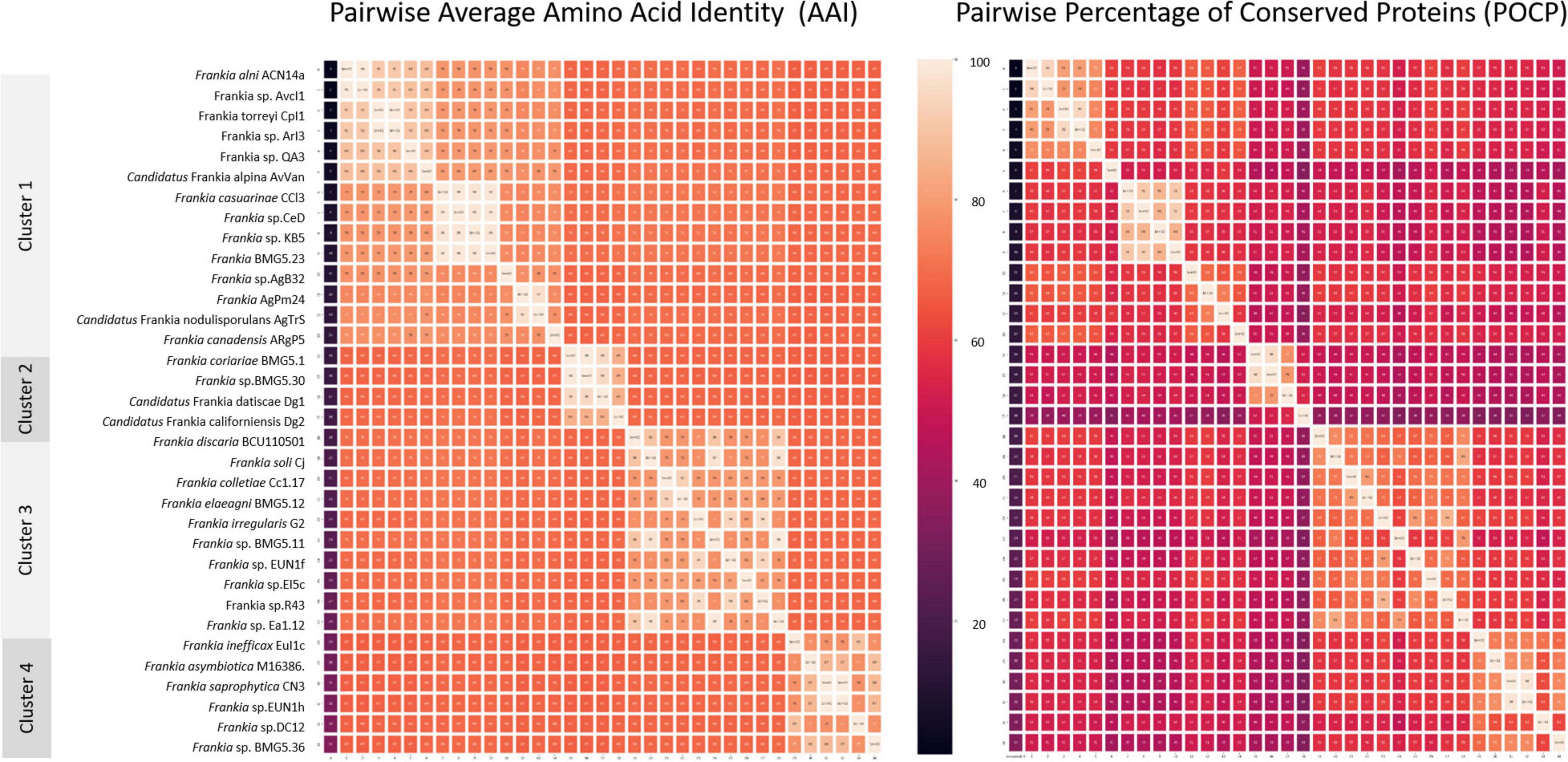
Figure 4. Heat maps based on pairwise average amino acid identity (AAI) and percentage of conserved proteins (POCP) calculated values. Pairwise AAI was calculated with the EzAAI tool (v1.1) using default settings, with a minimum query coverage of 50% and a minimum identity of 40%. The pairwise POCP was calculated considering an E-value of < 1 × 10–5, > 40% sequence identity and > 50% of the query sequence.
When ANI values are compared between more divergent taxa than species, they are prone to saturation and loss of information and, hence, considered inappropriate standards for genus delineation (Konstantinidis and Tiedje, 2007; Kim et al., 2014; Qin et al., 2014; Rodriguez-R and Konstantinidis, 2014; Gosselin et al., 2022; Park et al., 2022). The ANI values in the present study (78–81.5%) were consistently lower than the ANI species thresholds of 95–96.5%. Values closer to the species threshold (89–90%) were, however, seen with the quartic function of 750 genomes analysed (Barco et al., 2020).
Other genomic criteria
Some genes and operon organisation show some distinctiveness for each of the four clusters (Supplementary Figure 1). There are two ribosomal operons for clusters 1 and 2 strains and three for clusters 3 and 4 (Gtari et al., 2007). Nitrogenase complexes encoding (nif) genes are totally absent in the asymbiotic cluster 4 (not retrieved in the draft genome sequence, except for F. asymbiotica strain M16386T). The nif genes are, however, organised in different ways with respect to each cluster (Tisa et al., 2016; Nouioui et al., 2019b). The Ni-Fe hydrogenase or uptake hydrogenase (hup) genes are clustered into two operons for clusters 1 and 3, while only one operon is present for clusters 2 and 4 (Tisa et al., 2016). Another important distinctive gene is murC, related to peptidoglycan biosynthesis. Different copies in the genomes of the four different clusters were found (Berckx et al., 2020). Two copies, murC1 and murC2, are present in clusters 2 and 3, which differ in their orientation with the presence of an open reading frame (ORF) encoding a nitroreductase family deazaflavin-dependent oxidoreductase in cluster 3. Clusters 1 and 4 contain only one copy of murC2 with the ORF encoding a nitroreductase family deazaflavin-dependent oxidoreductase only present in cluster 1.
Overall differentially encoded proteins between genomes of the four clusters were provided in Supplementary Table 4. The presence/absence profiles of the protein clusters for the four phylogenetic clusters were illustrated in Figure 5 and Supplementary Figure 2.
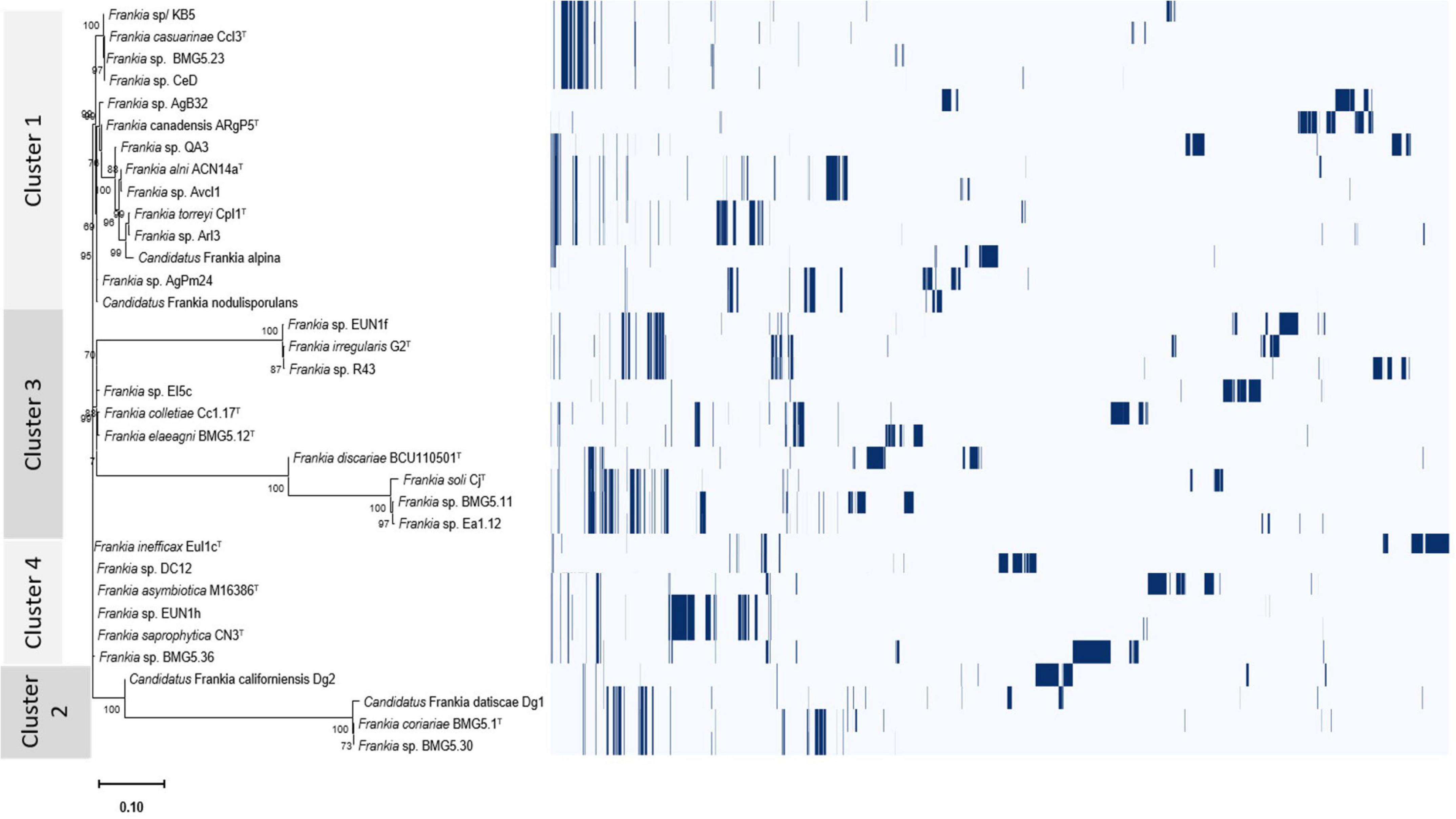
Figure 5. Roary plot showing the tree compared to a matrix with the presence and absence of core and accessory genes identified for the four cluster genomes.
Ecological and phenotypic features
The phylogenetic segregation of Frankia clusters found additional support in the ecological lifestyle interaction with host plants and cultural behaviours among the Frankia strains of each cluster. Frankia strains from cluster 1 (except those nodulating Allocasuarina and Casuarina) and cluster 3 are globally distributed in soils irrespective of the presence or absence of compatible host plants (Smolander and Sundman, 1987; Põlme et al., 2014). The distribution of cluster 1 of Casuarinaceae infective and cluster 2 strains is restricted to the native range of their respective host plants (Simonet et al., 1999). Cluster 4 ineffective strains (nodulating but non-nitrogen fixing) were shown to represent the most prominent Frankia population, exhibiting a higher diversity in prairie soils in the absence of actinorhizal host plants (Ben Tekaya et al., 2017) and wet soils under Alnus glutinosa (Hahn et al., 1988; van Dijk and Sluimer, 1994; Wolters et al., 1997a,b).
Most Frankia strains from clusters 1 and 3 have been cultured in axenic conditions with relative ease. Strains from cluster 4, which have been isolated as a “by-product” in studies aimed at the cultivation of the “true” beneficial microsymbionts in actinorhizal root nodules, show very similar cultural behaviours to other soil actinomycetes (Normand and Chapelon, 1997; Nouioui et al., 2017a,b, 2018a). Cluster 1 contains as-yet-uncultured Frankia microsymbionts, which are characterised by profuse sporulation within nodules tissues (Sp + types; Schwintzer, 1990), and a very higher proportion of cluster 2 strains are as-yet-uncultured. The Candidatus status has been used to accommodate some of these uncultured Frankia which were defined based on genome sequences (Persson et al., 2011, 2015; Nguyen et al., 2016, 2019; Normand et al., 2017; Herrera-Belaroussi et al., 2020; Pozzi et al., 2020; Berckx et al., 2022). Two strains from cluster 2 have been successfully cultivated following a dual approach consisting of comparative genomics and direct physiological assay on nodule tissues (Gtari et al., 2015; Gueddou et al., 2019).
While filamentous hyphae are the primary vegetative state for all growing Frankia strains, the extent of sporangia and vesicle formation varies from cluster to cluster. The sporulation of strains from clusters 1, 3, and 4 may be readily detected in vitro or may depend on the composition of media and cultural conditions (Tisa et al., 1983; Krumholz et al., 2003). The sporulation of the two cultivated strains of cluster 2 seems to be completely suppressed (Gtari et al., 2015; Nouioui et al., 2017b). In general, vesicles containing nitrogenase are formed in response to the limited availability of nitrogen (Fontaine et al., 1984; Murry et al., 1984). Some Frankia strains, belonging to cluster 3, continue to form vesicles even in the presence of a nitrogen source, but the numbers are reduced compared with growth in nitrogen-depleted media (Gauthier, 1983; Meesters et al., 1985). Strains of cluster 4 are unable to fix dinitrogen and thus to form vesicles, with the exception of F. asymbiotica.
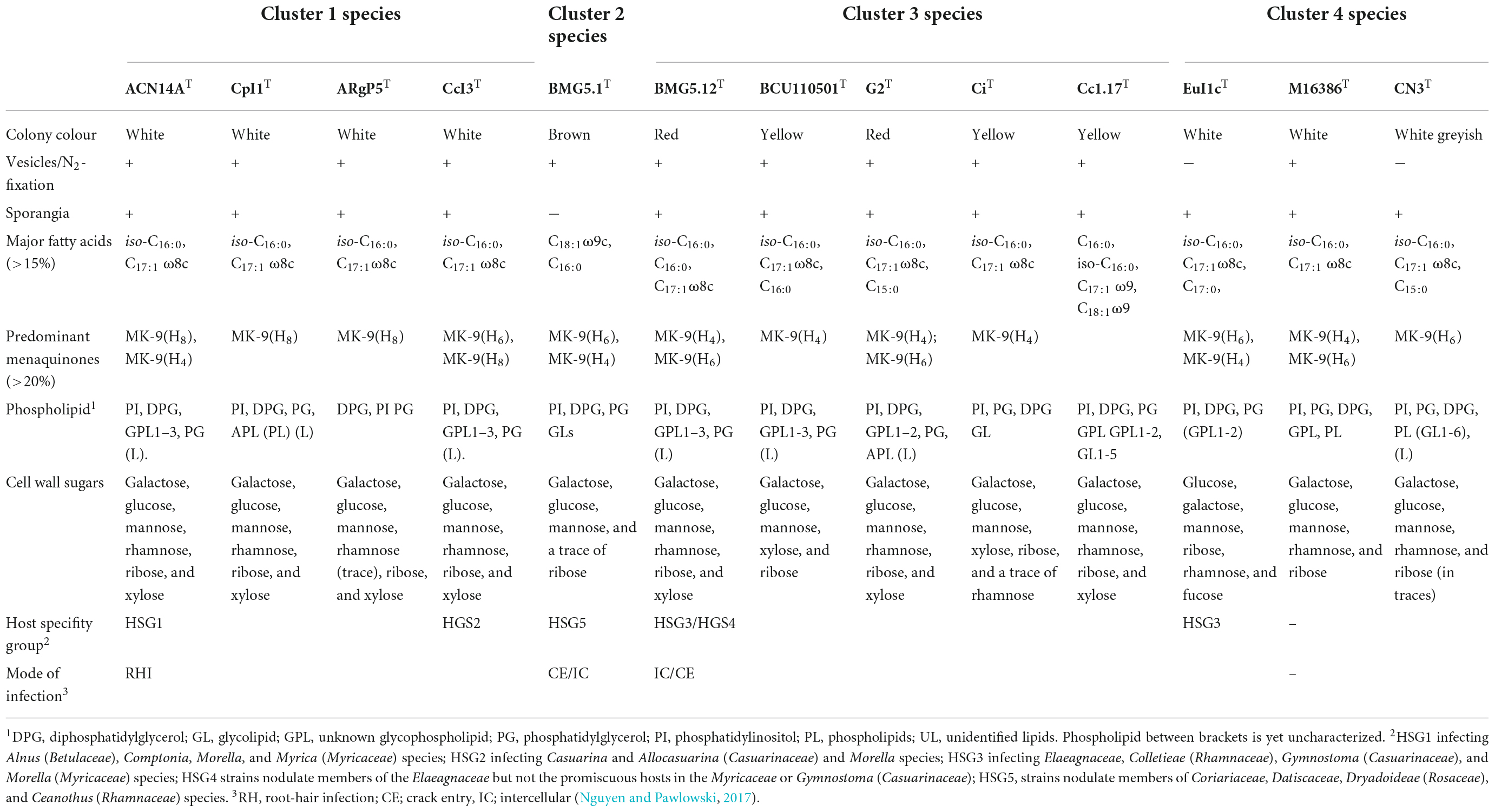
Strains from clusters 1 and 3 grow well in nitrogen-depleted media and metabolise short-chain fatty acids, TCA cycle intermediates, and carbohydrates. Strains in cluster 4 are similar to other saprophytic actinomycetes, more active physiologically, grow more rapidly and utilise a variety of monosaccharides and disaccharides, and produce hydrolytic enzymes, such as pectinases, cellulases, amylases, and proteases (Lechevalier, 1994). The two cultivated strains from cluster 2 are more slowly growing and have an unusual physiological requirement for alkalophilic growth media (Gtari et al., 2015; Nouioui et al., 2017b). Other phenotypic markers, including chemotaxonomy, are provided in Table 2.
Conclusion and description of the new taxa
Evidence of the splitting of Frankia into novel genera is here provided based on phylogenomics and OGRI recommended for bacterial genus boundary demarcation. The taxonomic elevation of phylogenetically distant Frankia clusters is clearly supported through the consistent sequence divergence in phylogenetic trees, OGRI analysis, and other genome-related criteria. The genus Frankia should be revised to accommodate cluster 1 species only, while clusters 2, 3, and 4 are taxonomically elevated to the rank of the genus as Protofrankia gen. nov., Parafrankia gen. nov., and Pseudofrankia gen. nov., respectively, and therefore, new combinations are provided for related species names. In the case of Protofrankia, in addition to the type species, two previously described Candidatus species can be reclassified as members of the genus. Thus, Candidatus Frankia datiscae (Persson et al., 2011) should be renamed Candidatus Protofrankia datiscae and Candidatus Frankia californiensis (Normand et al., 2017) should be renamed Candidatus Protofrankia californiensis.
The taxonomic revision provided, in this study, will help clarify the confusing past classification of the actinorhizal microsymbionts for taxonomic and applied purposes.
Description of Protofrankia gen. nov.
Protofrankia (Pro.to.fran’ki.a. Gr. masc. adj. protos, earlier than, prior to; N.L. fem. n. Frankia a bacterial genus name; N.L. fem. n. Protofrankia, a genus considered here as phylogenetically basal to Frankia).
The genus is defined by the taxonomic elevation of the taxon previously defined as Frankia phylogenetic cluster 2. Host plants include Coriariaceae, Datiscaceae, Dryadoideae (Rosaceae), and Ceanothus (Rhamnaceae) species. Genome sizes in the range of 5.3–5.8 Mb with G + C mol% content of 68.0–70.2%. The type species is Protofrankia coriaria.
Protofrankia coriaria comb. nov.
Protofrankia coriaria (co.ri.a’ri.ae. N.L. gen. fem. n. coriariae, of Coriaria, referring to the origin of isolation of the type strain).
Basonym: Frankia coriaria (Nouioui et al., 2017b,c).
The description of Protofrankia coriaria comb. nov. is identical to that given by Nouioui et al. (2017b) for F. coriaria. The type strain is BMG5.1T (= DSM 100624T = CECT 9032T).
Description of Parafrankia gen. nov.
Parafrankia (Pa.ra.fran’ki.a. Gr. prep. para, beside; N.L. fem. n. Frankia, a bacterial genus name; N.L. fem. n. Parafrankia, beside Frankia).
The genus Parafrankia is defined by the taxonomic elevation of the taxon previously defined as Frankia phylogenetic cluster 3. Host plants include members of Elaeagnaceae, Colletieae, Morella, and Gynmnostoma. Genome sizes range from 6.6 to 11.2 Mb with G + C mol% of 69.7–72.3. The type species is Parafrankia elaeagni. In addition, four species can be reclassified as members of the genus. Frankia discariae (Nouioui et al., 2018b), F. irregularis (Nouioui et al., 2018a), F. soli (Gtari et al., 2020), and F. colletiae (Nouioui et al., 2022) should be named Parafrankia discariae, Parafrankia irregularis, Parafrankia soli, and Parafrankia colletiae, respectively.
Parafrankia elaeagni comb. nov.
Parafrankia elaeagni (e.lae.ag’ni. N.L. gen. masc. n. elaeagni, of Elaeagnus, referring to the source of the isolate).
Basonym: Frankia elaeagni (Nouioui et al., 2016).
The description of Parafrankia elaeagni is the same as that given by Nouioui et al. (2016) for F. elaeagni. The type strain is BMG5.12T (= DSM 46783T = CECT 9031T).
Parafrankia discariae comb. nov.
Parafrankia discariae (dis.ca’ri.ae. N.L. gen. fem. n. discariae, of Discaria, the host plant origin of isolation of the type strain).
Basonym: Frankia discariae (Nouioui et al., 2017d).
The description of Parafrankia discariae is the same as that given by Nouioui et al. (2017d) for F. discariae. The type strain is BCU110501T (= DSM 46785T = CECT 9042T).
Parafrankia irregularis comb. nov.
Parafrankia irregularis (ir.re.gu.la’ris. L. fem. adj. irregularis, of irregular, referring to the inability of the species to infect its original host plant and to infect taxonomically disparate host plants).
Basonym: Frankia irregularis (Nouioui et al., 2018b).
The description of Parafrankia irregularis is the same as that given by Nouioui et al. (2018b) for F. irregularis. The type strain is G2T (= DSM 45899T = CECT 9038T).
Parafrankia soli comb. nov.
Parafrankia soli (so’li. L. gen. neut. n. soli, of soil, referring to the isolation source of the type strain).
Basonym: Frankia soli (Gtari et al., 2020).
The description of Parafrankia soli comb. nov. is the same as that given by Gtari et al. (2020) for F. soli. The type strain is CjT (= DSM 100623T = CECT 9041T = NRRL B-16219T).
Parafrankia colletiae comb. nov.
Parafrankia colletiae (col.le’ti.ae. N.L. gen. n. colletiae of Colletia, referring to the host plant, Colletia, origin of isolation of the strain).
Basonym: Frankia colletiae (Nouioui et al., 2022).
The description of Parafrankia colletiae comb. nov. is the same as that given by Nouioui et al. (2022) for F. colletiae. The type strain is Cc1.17T (= DSM 43829T = CECT 9313T).
Description of Pseudofrankia gen. nov.
Pseudofrankia (Pseu.do.fran’ki.a. Gr. masc. adj. pseudes, false; N.L. fem. n. Frankia, a bacterial genus name; N.L. fem. n. Pseudofrankia, a false Frankia).
Pseudofrankia gen. nov. is defined by the taxonomic elevation of the taxon previously defined as Frankia phylogenetic cluster 4. Members of the genus have been isolated from actinorhizal root nodules and are non-infective and/or non-nitrogen-fixing taxa. The size range of the genomes is 6.6–9.9 Mb with 71.2–72.3 of G + C mol%. The type species is Pseudofrankia inefficax. In addition, two other species can be reclassified as members of the genus. Frankia asymbiotica (Nouioui et al., 2018a) and F. saprophytica (Nouioui et al., 2018) should be named Pseudofrankia asymbiotica and Pseudofrankia saprophytica, respectively.
Pseudofrankia inefficax comb. nov.
Pseudofrankia inefficax (in.ef’fi.cax. L. fem. adj. inefficax, inefficient in reference to the inability of the bacterium to form the effective nitrogen-fixing symbiosis with its plant host).
Basonym: Frankia inefficax (Nouioui et al., 2017a).
The description of Pseudofrankia inefficax comb. nov. is the same as that given by Nouioui et al. (2017a) for F. inefficax. The type strain is EuI1cT (=DSM 45817T = CECT 9037T).
Pseudofrankia asymbiotica comb. nov.
Pseudofrankia asymbiotica (a.sym.bi.o’ti.ca. Gr. pref. a-, not; N.L. fem. adj. symbiotica, living together; N.L. fem. adj. asymbiotica, not symbiotic).
Basonym: Frankia asymbiotica (Nouioui et al., 2017c).
The description of Pseudofrankia asymbiotica comb. nov. is the same as that given by Nouioui et al. (2017c) for F. asymbiotica. The type strain is M16386T (= DSM 100626T = CECT 9040T = NRRL B-16386T).
Pseudofrankia saprophytica comb. nov.
Pseudofrankia saprophytica (sa.pro.phy’ti.ca. Gr. masc. adj. sapros, rotten; Gr. masc. adj. phytikos, belonging to plants; N.L. fem. adj. saprophytica, growing on rotten material, referring to the asymbiotic lifestyle of the type strain).
Basonym: Frankia saprophytica (Nouioui et al., 2018a).
The description of Pseudofrankia saprophytica comb. nov. is the same as that given by Nouioui et al. (2018a) for F. saprophytica. The type strain is CN3T (=DSM 105290T = CECT 9314T).
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MG conceived the study, performed the analyses, and wrote the manuscript.
Acknowledgments
A special thank is addressed to Nawres Mazzazi for her helpful assistance and Professor Faten Ghodhbane-Gtari for her enriching comments. The author thanks Professor Aharon Oren (The Hebrew University of Jerusalem) for expert advice on Latin nomenclature and formatting of protologues.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1041425/full#supplementary-material
Supplementary Figure 1 | Comparative representations of the nif operons in strains representing Frankia clusters 1, 2, and 3 (A), and hup gene cluster (B) and biosynthetic peptidoglycan genes murC (C) in clusters 1, 2, 3, and 4.
Supplementary Figure 2 | Clusters of Orthologous Genes (COG) categories distribution for all studied strains.
Abbreviations
AAI, average amino acid identity; ANI, average nucleotide identity; GGDC, genome-to-genome distance calculator; ML, maximum-likelihood; MP, maximum-parsimony; AFLP, amplified fragment length polymorphism; MLSA, multilocus sequence analysis; POCP, percentage of conserved proteins; TYGS, type strain genome server; LPSN, list of prokaryotic names with standing in nomenclature.
Footnotes
References
Adamek, M., Alanjary, M., Sales-Ortells, H., Goodfellow, M., Bull, A. T., Winkler, A., et al. (2018). Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 19:426. doi: 10.1186/s12864-018-4809-4
Aliyu, H., Lebre, P., Blom, J., Cowan, D., and De Maayer, P. (2016). Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 39, 527–533. doi: 10.1016/j.syapm.2016.09.004
Altschul, S. F. (1997). “Evaluating the statistical significance of multiple distinct local alignments,” in Theoretical and computational methods in genome research, ed. S. Suhai (Boston, MA: Springer), 1–14. doi: 10.1007/978-1-4615-5903-0_1
Auch, A. F., von Jan, M., Klenk, H. P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134.
Barco, R. A., Garrity, G. M., Scott, J. J., Amend, J. P., Nealson, K. H., and Emerson, D. (2020). A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio 11, e02475–19. doi: 10.1128/mBio.02475-19
Bautista, G. H. H., Cruz, H. A., Nesme, X., Valdés, M., Mendoza, H. A., and Fernandez, M. P. (2011). Genomospecies identification and phylogenomic relevance of AFLP analysis of isolated and non-isolated strains of Frankia spp. Syst. Appl. Microbiol. 34, 200–206. doi: 10.1016/j.syapm.2010.10.004
Becking, J. H. (1970). Frankiaceae fam. nov. (Actinomycetales) with one new combination and six new species of the genus Frankia Brunchorst, 1886, 174. Int. J. Syst. Evol. Microbiol. 20, 201–220. doi: 10.1099/00207713-20-2-201
Ben Tekaya, S., Ganesan, A. S., Guerra, T., Dawson, J. O., Forstner, M. R., and Hahn, D. (2017). Sybr green-and TaqMan-based quantitative PCR approaches allow assessment of the abundance and relative distribution of Frankia clusters in soils. Appl. Environ. Microbiol. 83, e02833–16. doi: 10.1128/AEM.02833-16
Benson, D. R., and Dawson, J. O. (2007). Recent advances in the biogeography and genecology of symbiotic Frankia and its host plants. Physiol. Plant. 130, 318–330. doi: 10.1111/j.1399-3054.2007.00934.x
Benson, D. R., and Silvester, W. B. (1993). Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol. Rev. 57, 293–319. doi: 10.1128/mr.57.2.293-319.1993
Berckx, F., Nguyen, T. V., Bandong, C. M., Lin, H. H., Yamanaka, T., Katayama, S., et al. (2022). A tale of two lineages: How the strains of the earliest divergent symbiotic Frankia clade spread over the world. BMC Genomics 23:602. doi: 10.1186/s12864-022-08838-5
Berckx, F., Wibberg, D., Kalinowski, J., and Pawlowski, K. (2020). The peptidoglycan biosynthesis gene murC in Frankia: Actinorhizal vs. plant type. Genes 11:432. doi: 10.3390/genes11040432
Brunchorst, J. (1886). Über einige Wurzelanschwellungen, besonders diejenigen von Alnus und den Elaegnaceen. Bot. Inst. Tubin. 2, 151–177.
Caputo, A., Fournier, P. E., and Raoult, D. (2019). Genome and pan-genome analysis to classify emerging bacteria. Biol. Direct 14, 1–9. doi: 10.1186/s13062-019-0234-0
Clawson, M. L., Bourret, A., and Benson, D. R. (2004). Assessing the phylogeny of Frankia-actinorhizal plant nitrogen-fixing root nodule symbioses with Frankia 16S rRNA and glutamine synthetase gene sequences. Mol. Phylogenet. Evol. 31, 131–138. doi: 10.1016/j.ympev.2003.08.001
Colwell, R. R. (1970). Polyphasic taxonomy of the genus Vibrio: Numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J. Bacteriol. 104, 410–433. doi: 10.1128/jb.104.1.410-433.1970
Dagan, T. (2011). Phylogenomic networks. Trends Microbiol. 19, 483–491. doi: 10.1016/j.tim.2011.07.001
Fontaine, M. S., Lancelle, S. A., and Torrey, J. G. (1984). Initiation and ontogeny of vesicles in cultured Frankia sp. strain HFPArI3. J. Bacteriol. 160, 921–927. doi: 10.1128/jb.160.3.921-927.1984
Gauthier, D. L. (1983). Effect of L-methionine-DL-sulfoximine on acetylene reduction and vesicle formation in derepressed cultures of Frankia strain D11. Can. J. Microbiol. 29, 1003–1006.
Ghodhbane-Gtari, F., Nouioui, I., Chair, M., Boudabous, A., and Gtari, M. (2010). 16S-23S rRNA intergenic spacer region variability in the genus Frankia. Microb. Ecol. 60, 487–495. doi: 10.1007/s00248-010-9641-6
Goloboff, P. A., Farris, J. S., and Nixon, K. C. (2008). TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786. doi: 10.1111/j.1096-0031.2008.00217.x
Gosselin, S., Fullmer, M. S., Feng, Y., and Gogarten, J. P. (2022). Improving phylogenies based on average nucleotide identity, incorporating saturation correction and nonparametric bootstrap support. Syst. Biol. 71, 396–409. doi: 10.1093/sysbio/syab060
Gtari, M., Brusetti, L., Cherif, A., Boudabous, A., and Daffonchio, D. (2007). Heteroduplex structures in 16S-23S rRNA intergenic transcribed spacer PCR products reveal ribosomal interoperonic polymorphisms within single Frankia strains. J. Appl. Microbiol. 103, 1031–1040. doi: 10.1111/j.1365-2672.2007.03329.x
Gtari, M., Ghodhbane-Gtari, F., and Nouioui, I. (2020). Frankia soli sp. nov., an actinobacterium isolated from soil beneath Ceanothus jepsonii. Int. J. Syst. Evol. Microbiol. 70, 1203–1209. doi: 10.1099/ijsem.0.003899
Gtari, M., Ghodhbane-Gtari, F., Nouioui, I., Ktari, A., Hezbri, K., Mimouni, W., et al. (2015). Cultivating the uncultured: Growing the recalcitrant cluster-2 Frankia strains. Sci. Rep. 5:13112. doi: 10.1038/srep13112
Gtari, M., Nouioui, I., Sarkar, I., Ghodhbane-Gtari, F., Tisa, L. S., Sen, A., et al. (2019). An update on the taxonomy of the genus Frankia Brunchorst, 1886, 174AL. Anton. Leeuwenhoek 112, 5–21. doi: 10.1007/s10482-018-1165-y
Gtari, M., Tisa, L. S., and Normand, P. (2013). “Diversity of Frankia strains, actinobacterial symbionts of actinorhizal plants,” in Symbiotic endophytes, ed. R. Aroca (Berlin: Springer), 123–148. doi: 10.1007/978-3-642-39317-4_7
Gueddou, A., Swanson, E., Hezbri, K., Nouioui, I., Ktari, A., Simpson, S., et al. (2019). Draft genome sequence of the symbiotic Frankia sp. strain BMG5.30 isolated from root nodules of Coriaria myrtifolia in Tunisia. Anton. Leeuwenhoek 112, 67–74. doi: 10.1007/s10482-018-1138-1
Hahn, D., Lechevalier, M. P., Fische, A., and Stackebrandt, E. (1989). Evidence for a close phylogenetic relationship between members of the genera Frankia, Geodermatophilus, and “Blastococcus” and emendation of the family Frankiaceae. Syst. Appl. Microbiol. 11, 236–242. doi: 10.1016/S0723-2020(89)80019-0
Hahn, D., Starrenburg, M. J. C., and Akkermans, A. D. L. (1988). Variable compatibility of cloned Alnus glutinosa ecotypes against ineffective Frankia strains. Plant Soil 107, 233–243. doi: 10.1007/BF02370552
Herrera-Belaroussi, A., Normand, P., Pawlowski, K., Fernandez, M. P., Wibberg, D., Kalinowski, J., et al. (2020). Candidatus Frankia nodulisporulans sp. nov., an Alnus glutinosa-infective Frankia species unable to grow in pure culture and able to sporulate in-planta. Syst. Appl. Microbiol. 43:126134.
Hugenholtz, P., Chuvochina, M., Oren, A., Parks, D. H., and Soo, R. M. (2021). Prokaryotic taxonomy and nomenclature in the age of big sequence data. ISME J. 15, 1879–1892. doi: 10.1038/s41396-021-00941-x
Kim, D., Park, S., and Chun, J. (2021). Introducing EzAAI: A pipeline for high throughput calculations of prokaryotic average amino acid identity. J. Microbiol. 59, 476–480. doi: 10.1007/s12275-021-1154-0
Kim, M., Oh, H.-S., Park, S.-C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Konstantinidis, K. T., Ramette, A., and Tiedje, J. M. (2006). The bacterial species definition in the genomic era. Phil. Trans. Soc. B. 361, 1929–1940. doi: 10.1098/rstb.2006.1920
Konstantinidis, K. T., and Tiedje, J. M. (2005). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264. doi: 10.1128/jb.187.18.6258-6264.2005
Konstantinidis, K. T., and Tiedje, J. M. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: Advancements and challenges ahead. Curr. Opin. Microbiol. 10, 504–509. doi: 10.1016/j.mib.2007.08.006
Krumholz, G. D., Chval, M. S., McBride, M. J., and Tisa, L. S. (2003). “Germination and physiological properties of Frankia spores,” in Frankia symbiosis. Developments in plant and soil sciences, Vol. 100, eds P. Normand, J. O. Dawson, and K. Pawlowski (Dordrecht: Springer). doi: 10.1007/978-94-017-1601-7_7
Lechevalier, M. P. (1994). Taxonomy of the genus Frankia (Actinomycetales). Int. J. Syst. Evol. Microbiol. 44, 1–8. doi: 10.1099/00207713-44-1-1
Lechevalier, M. P., and Lechevalier, H. A. (1989). “Genus frankia brunchorst 1886, 174AL,” in Bergey’s manual of systematic bacteriology, Vol. 4, eds S. T. Williams, M. E. Sharpe, and J. G. Holt (Baltimore, MA: The Williams & Wilkins Co), 2410–2417.
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Li, Y., Xue, H., Sang, S.-Q., Lin, C.-L., and Wang, X.-Z. (2017). Phylogenetic analysis of family Neisseriaceae based on genome sequences and description of Populibacter corticis gen. nov., sp. nov., a member of the family Neisseriaceae, isolated from symptomatic bark of Populus× euramericana canker. PLoS One 12:e0174506. doi: 10.1371/journal.pone.0174506
Liu, H., Xin, B., Zheng, J., Zhong, H., Yu, Y., Peng, D., et al. (2020). Build a bioinformatics analysis platform and apply it to routine analysis of microbial genomics and comparative genomics. Protocol. Exchange. doi: 10.21203/rs.2.21224/v2
Lopes-Santos, L., Castro, D. B. A., Ferreira-Tonin, M., Corrêa, D. B. A., Weir, B. S., Park, D., et al. (2017). Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Anton. Leeuwenhoek 110, 727–736. doi: 10.1007/s10482-017-0842-6
Ludwig, W., and Klenk, H. P. (2005). “Overview: A phylogenetic backbone and taxonomic framework for procaryotic systematics,” in Bergey’s manualof systematic bacteriology, eds D. R. Boone and R. W. Castenholz (New York, N.Y: Springer-Verlag), 49–66. doi: 10.1002/9781118960608.bm00020
Margos, G., Gofton, A., Wibberg, D., Dangel, A., Marosevic, D., Loh, S. M., et al. (2018). The genus Borrelia reloaded. PLoS One 13:e0208432. doi: 10.1371/journal.pone.0208432
Meesters, T. M., Van Genesen, S. T., and Akkermans, A. D. L. (1985). Growth, acetylene reduction activity and localization of nitrogenase in relation to vesicle formation in Frankia strains Cc1. 17 and Cp1. 2. Arch. Microbiol. 143, 137–142. doi: 10.1007/BF00411036
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Murry, M. A., Fontaine, M. S., and Tjepkema, J. D. (1984). Oxygen protection of nitrogenase in Frankia sp. HFPArI3. Arch. Microbiol. 139, 162–166. doi: 10.1007/BF00401993
Naushad, H. S., Lee, B., and Gupta, R. S. (2014). Conserved signature indels and signature proteins as novel tools for understanding microbioal phylogeny and systematics: Identification of molecular signatures that are specific for the phytopathogenic genera Dickeya, Pectobacterium and Brenneria. Int. J. Syst. Evol. Microbiol. 64, 366–383. doi: 10.1099/ijs.0.054213-0
Nguyen, T. V., and Pawlowski, K. (2017). “Frankia and actinorhizal plants: Symbiotic nitrogen fixation,” in Rhizotrophs: plant growth promotion to bioremediation, ed. S. Mehnaz (Singapore: Springer), 237–261. doi: 10.1007/978-981-10-4862-3_12
Nguyen, T. V., Wibberg, D., Battenberg, K., Blom, J., Vanden Heuvel, B., Berry, A. M., et al. (2016). An assemblage of Frankia cluster II strains from California contains the canonical nod genes and also the sulfotransferase gene nodH. BMC Genomics 17:796. doi: 10.1186/s12864-016-3140-1
Nguyen, T. V., Wibberg, D., Vigil-Stenman, T., Berckx, F., Battenberg, K., Demchenko, K. N., et al. (2019). Frankia-enriched metagenomes from the earliest diverging symbiotic Frankia cluster: They come in teams. Genome Biol. Evol. 11, 2273–2291. doi: 10.1093/gbe/evz153
Normand, N., Nouioui, I., Pujic, P., Fournier, P., Dubost, A., Schwob, G., et al. (2018). Frankia canadensis sp. nov., isolated from root nodules of Alnus incana subspecies rugosa growing in Canada. Int. J. Evol. Syst. Microbiol. 68, 3001–3011. doi: 10.1099/ijsem.0.002939
Normand, P., and Benson, D. R. (2015). “Order XVI frankiales,” in Bergey’s manual of systematic bacteriology the actinobacteria, 2nd Edn, Vol. 5, eds M. Goodfellow, P. Kämpfer, H. J. Busse, M. E. Trujillo, K. I. Suzuki, W. Ludwig, et al. (New York, NY: Springer), 508–510. doi: 10.1002/9781118960608.obm00010
Normand, P., Benson, D. R., Berry, A. M., and Tisa, L. S. (2014). “Family frankiaceae,” in The prokaryote–actinobacteria, eds E. Rosenberg, E. F. Delong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer-Verlag), 339–356. doi: 10.1007/978-3-642-30138-4_183
Normand, P., and Chapelon, C. (1997). Direct characterization of Frankia and of close phyletic neighbors from an Alnus viridis rhizosphere. Physiol. Plant. 99, 722–731. doi: 10.1111/j.1399-3054.1997.tb05377.x
Normand, P., and Fernandez, M. (2008). “Evolution and diversity of Frankia,” in Prokaryotic symbionts in plants, Vol. 8, ed. K. Pawlowski (Heidelberg: Springer), 103–125. doi: 10.1007/7171_2008_121
Normand, P., and Fernandez, M. P. (2019). “Frankia,” in Bergey’s manual of systematics of archaea and bacteria, eds M. E. Trujillo, S. Dedysh, P. DeVos, B. Hedlund, P. Kämpfer, F. A. Rainey, et al. (Hoboken, NJ: John Wiley & Sons, Inc), doi: 10.1002/9781118960608.gbm00042.pub2
Normand, P., Orso, S., Cournoyer, B., Chapelon, C., Jeannin, P., Dawson, J., et al. (1996). Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int. J. Syst. Evol. Microbiol. 46, 1–9. doi: 10.1099/00207713-46-1-1
Normand, P., Van Nguyen, T., Battenberg, K., Berry, A. M., Heuvel, B. V., Fernandez, M. P., et al. (2017). Proposal of’Candidatus Frankia californiensis’, the uncultured symbiont in nitrogen-fixing root nodules of a phylogenetically broad group of hosts endemic to western North America. Int. J. Syst. Evol. Microbiol. 67, 3706–3715. doi: 10.1099/ijsem.0.002147
Nouioui, I., Cortés-Albayay, C., Carro, L., Castro, J. F., Gtari, M., Ghodhbane-Gtari, F., et al. (2019b). Genomic insights into plant-growth-promoting potentialities of the genus Frankia. Front. Microbiol. 10:1457. doi: 10.3389/fmicb.2019.01457
Nouioui, I., Ghodhbane-Gtari, F., Beauchemin, N. J., Tisa, L. S., and Gtari, M. (2011). Phylogeny of members of the Frankia genus based on gyrB, nifH and glnII sequences. Anton. Leeuwenhoek 100, 579–587. doi: 10.1007/s10482-011-9613-y
Nouioui, I., Ghodhbane-Gtari, F., Jando, M., Klenk, H. P., and Gtari, M. (2022). Frankia colletiae sp. nov., a nitrogen fixing actinobacterium isolated from Colletia Cruciata. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijsem.0.005656
Nouioui, I., Ghodhbane-Gtari, F., Jando, M., Tisa, L. S., Klenk, H.-P., and Gtari, M. (2019a). Frankia torreyi sp. nov., the first actinobacterium of the genus Frankia Brunchorst, 1886, 174AL isolated in axenic culture. Anton. Leeuwenhoek 112, 57–65. doi: 10.1007/s10482-018-1131-8
Nouioui, I., Ghodhbane-Gtari, F., Klenk, H. P., and Gtari, M. (2018a). Frankia saprophytica sp. nov. an atypical non-infective (Nod–) and non-nitrogen fixing (Fix–) actinobacterium isolated from Coriaria nepalensis root nodules. Int. J. Syst. Evol. Microbiol. 68, 1090–1095. doi: 10.1099/ijsem.0.002633
Nouioui, I., Ghodhbane-Gtari, F., Montero-Calasanz, M. D., Göker, M., Meier-Kolthoff, J. P., Schumann, P., et al. (2016). Proposal of a type strain for Frankia alni (Woronin 1866) Von Tubeuf 1895, emended description of Frankia alni, and recognition of Frankia casuarinae sp. nov. and Frankia elaeagni sp. nov. Int. J. Syst. Evol. Microbiol. 66, 5201–5210. doi: 10.1099/ijsem.0.001496
Nouioui, I., Ghodhbane-Gtari, F., Montero-Calasanz, M. D. C., Rohde, M., Tisa, L. S., Gtari, M., et al. (2017a). Frankia inefficax sp. nov., an actinobacterial endophyte inducing ineffective, non nitrogen-fixing, root nodules on its actinorhizal host plants. Anton. Leeuwenhoek 110, 313–320. doi: 10.1007/s10482-016-0801-7
Nouioui, I., Ghodhbane-Gtari, F., Rhode, M., Sangal, V., Klenk, H. P., and Gtari, M. (2018b). Frankia irregularis sp. nov., an actinobacterium unable to nodulate its original host, Casuarina equisetifolia, but effectively nodulates members of the actinorhizal Rhamnales. Int. J. Syst. Evol. Microbiol. 68, 2883–2914. doi: 10.1099/ijsem.0.002914
Nouioui, I., Ghodhbane-Gtari, F., Rohde, M., Klenk, H. P., and Gtari, M. (2017b). Frankia coriariae sp. nov., an infective and effective microsymbiont isolated from Coriaria japonica. Int. J. Syst. Evol. Microbiol. 67, 1266–1270. doi: 10.1099/ijsem.0.001797
Nouioui, I., Gueddou, A., Ghodhbane-Gtari, F., Rhode, M., Gtari, M., and Klenk, H. P. (2017c). Frankia asymbiotica sp. nov., a non infective actinobacterium isolated from Morella californica root nodule. Int. J. Syst. Evol. Microbiol. 67, 4897–4901. doi: 10.1099/ijsem.0.002153
Nouioui, I., Montero-Calasanz, M. D. C., Ghodhbane-Gtari, F., Rohde, M., Tisa, L. S., Klenk, H. P., et al. (2017d). Frankia Discariae sp. nov.: an infective and effective microsymbiont isolated from the root nodule of Discaria trinervis. Arch. Microbiol. 199, 641–647. doi: 10.1007/s00203-017-1337-6
Orata, F. D., Meier-Kolthoff, J. P., Sauvageau, D., and Stein, L. Y. (2018). Phylogenomic analysis of the gammaproteobacterial methanotrophs (Order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 9:3162. doi: 10.3389/fmicb.2018.03162
Oren, A., and Garrity, G. M. (2014). Then and now: A systematic review of the systematics of prokaryotes in the last 80 years. Anton. Leeuwenhoek 106, 43–56. doi: 10.1007/s10482-013-0084-1
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Pannekoek, Y., Qi-Long, Q., Zhang, Y. Z., and van der Ende, A. (2016). Genus delineation of Chlamydiales by analysis of the percentage of conserved proteins justifies the reunifying of the genera Chlamydia and Chlamydophila into one single genus Chlamydia. Pathog. Dis. 74:ftw071. doi: 10.1093/femspd/ftw071
Park, M. J., Kim, Y. J., Park, M., Yu, J., Namirimu, T., Roh, Y. R., et al. (2022). Establishment of genome based criteria for classification of the family Desulfovibrionaceae and proposal of two novel genera, Alkalidesulfovibrio gen. nov. and Salidesulfovibrio gen. nov. Front. Microbiol. 13:738205. doi: 10.3389/fmicb.2022.738205
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R., Moret, B. M., and Stamatakis, A. (2010). How many bootstrap replicates are necessary? J. Comput. Biol. 17, 337–354. doi: 10.1089/cmb.2009.0179
Persson, T., Battenberg, K., Demina, I. V., Vigil-Stenman, T., Vanden Heuvel, B., Pujic, P., et al. (2015). Candidatus Frankia datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS One 10:e0127630. doi: 10.1371/journal.pone.0127630
Persson, T., Benson, D. R., Normand, P., van den Heuvel, B., Pujic, P., Chertkov, O., et al. (2011). Genome sequence of “Candidatus Frankia datiscae” Dg1, the uncultured microsymbiont from nitrogen-fixing root nodules of the dicot Datisca glomerata. J. Bacteriol. 193, 7017–7018. doi: 10.1128/JB.06208-11
Põlme, S., Bahram, M., Kõljalg, U., and Tedersoo, L. (2014). Global biogeography of Alnus-associated Frankia actinobacteria. New Phytol. 204, 979–988. doi: 10.1111/nph.12962
Pozzi, A. C., Bautista-Guerrero, H. H., Abby, S. S., Herrera-Belaroussi, A., Abrouk, D., Normand, P., et al. (2018). Robust Frankia phylogeny, species delineation and intraspecies diversity based on multi-locus sequence analysis (MLSA) and single-locus strain typing (SLST) adapted to a large sample size. Syst. Appl. Microbiol. 41, 311–323. doi: 10.1016/j.syapm.2018.03.002
Pozzi, A. C. M., Herrera-Belaroussi, A., Schwob, G., Bautista-Guerrero, H. H., Bethencourt, L., Fournier, P., et al. (2020). Proposal of’Candidatus Frankia alpina’, the uncultured symbiont of Alnus alnobetula and A. incana that forms spore-containing nitrogen-fixing root nodules. Int. J. Syst. Evol. Microbiol. 70, 5453–5459. doi: 10.1099/ijsem.0.004433
Qin, Q., Xie, B., Zhang, X., Chen, X., Zhou, B., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species: Culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe 9, 111–118. doi: 10.1128/microbe.9.111.1
Salam, N., Jiao, J. Y., Zhang, X. T., and Li, W. J. (2020). Update on the classification of higher ranks in the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 70, 1331–1355. doi: 10.1099/ijsem.0.003920
Sangal, V., Goodfellow, M., Jones, A. L., and Sutcliffe, I. C. (2022). A stable home for an equine pathogen: Valid publication of the binomial Prescottella equi gen. nov., comb. nov., and reclassification of four rhodococcal species into the genus Prescottella. Int. J. Syst. Evol. Microbiol. 72:005551. doi: 10.1099/ijsem.0.005551
Sant’Anna, F., Bach, E., Porto, R. Z., Guella, F., and Hayashi, H. (2019). Genomic metrics made easy: What to do and where to go in the new era of bacterial taxonomy. Crit. Rev. Microbiol. 45, 182–200. doi: 10.1080/1040841X.2019.1569587
Schwintzer, C. R. (1990). “Spore-positive and spore-negative nodules,” in The biology of frankia and actinorhizal plants, eds C. R. Schwintzer and J. D. Tjepkema (New York, NY: Academic Press, Inc), 177–193.
Sen, A., Daubin, V., Abrouk, D., Gifford, I., Berry, A. M., and Normand, P. (2014). The phylogeny of actinobacteria revisited in the light of complete genomes, the orders Frankiales and Micrococcales should be split into coherent entities. Proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov. and Nakamurellales ord. nov. Int. J. Syst. Evol. Microbiol. 64, 3821–3832. doi: 10.1099/ijs.0.063966-0
Simonet, P., Navarro, E., Rouvier, C., Reddell, P., Zimpfer, J., Dommergues, Y., et al. (1999). Co-evolution between Frankia populations and host plants in the family Casuarinaceae and consequent patterns of global dispersal. Environ. Microbiol. 1, 525–533. doi: 10.1046/j.1462-2920.1999.00068.x
Skerman, V. B. D., McGowan, V., and Sneath, P. H. A. (1980). Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30, 225–420. doi: 10.1099/00207713-30-1-225
Smolander, A., and Sundman, V. (1987). Frankia in acid soils of forests devoid of actinorhizal plants. Physiol. Plant. 70, 297–303. doi: 10.1111/j.1399-3054.1987.tb06147.x
Stackebrandt, E., Rainey, F. A., and Ward-Rainey, N. L. (1997). Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Evol. Microbiol. 47, 479–491. doi: 10.1099/00207713-47-2-479
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Swofford, D. L. (2002). PAUP*. phylogenetic analysis using parsimony (*and Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates, doi: 10.1111/j.0014-3820.2002.tb00191.x
Tisa, L., McBride, M., and Ensign, J. C. (1983). Studies of growth and morphology of Frankia strains EAN1pec, EuI1c, CpI1, and ACN1AG. Can. J. Bot. 61, 2768–2773. doi: 10.1139/b83-306
Tisa, L. S., Oshone, R., Sarkar, I., Ktari, A., Sen, A., and Gtari, M. (2016). Genomic approaches toward understanding the actinorhizal symbiosis: An update on the status of the Frankia genomes. Symbiosis 70, 5–16. doi: 10.1007/s13199-016-0390-2
van Dijk, C., and Sluimer, A. (1994). Resistance to an ineffective Frankia strain type in Alnus glutinosa (L.) Gaertn. New Phytol. 128, 497–504. doi: 10.1111/j.1469-8137.1994.tb02996.x
Varghese, N. J., Mukherjee, S., Ivanova, N., Konstantinidis, K. T., Mavrommatis, K., Kyrpides, N. C., et al. (2015). Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43, 6761–6771. doi: 10.1093/nar/gkv657
Wayne, L. G., Brenner, B. D., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464. doi: 10.1099/00207713-37-4-463
Wirth, J. S., and Whitman, W. B. (2018). Phylogenomic analyses of a clade within the Roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 68, 2393–2411. doi: 10.1099/ijsem.0.002833
Wolters, D. J., Akkermans, A. D. L., and Van Dijk, C. (1997a). Ineffective Frankia strains in wet stands of Alnus glutinosa L. Gaertn. in the Netherlands. Soil Biol. Biochem. 29, 1707–1712. doi: 10.1016/S0038-0717(97)00075-8
Keywords: Frankiaceae, bacterial genus boundaries, phylogenomics, pangenome, AAI, POCP, ANI, 16S rRNA gene
Citation: Gtari M (2022) Taxogenomic status of phylogenetically distant Frankia clusters warrants their elevation to the rank of genus: A description of Protofrankia gen. nov., Parafrankia gen. nov., and Pseudofrankia gen. nov. as three novel genera within the family Frankiaceae. Front. Microbiol. 13:1041425. doi: 10.3389/fmicb.2022.1041425
Received: 10 September 2022; Accepted: 12 October 2022;
Published: 08 November 2022.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Michael Goodfellow, Newcastle University, United KingdomHaitham Sghaier, National Center for Nuclear Science and Technology, Tunisia
Copyright © 2022 Gtari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maher Gtari, maher.gtari@insat.rnu.tn
 Maher Gtari
Maher Gtari