
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Microbiol., 04 November 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1039188
This article is part of the Research TopicWomen in Infectious Agents and Disease: 2022View all 10 articles
The host genetics play an important role in the induction of autoimmune response against self-antigens however; several epidemiological and molecular evidences suggest microbial pathogens (viruses and bacteria) are the principal environmental triggers of autoimmunity (Fujinami, 2001; Ascherio and Munger, 2007; Libbey and Fujinami, 2010; Tarakhovsky and Prinjha, 2018; Lester et al., 2019). Reportedly, four main criteria have been associated with molecular mimicry and the underlying human disease, (i) epidemiological evidence associating a pathogen with a disease, (ii) presence of antibodies and/or immune cells against the specific antigens related with the disease, (iii) cross-reactivity of antibodies or immune cells of the host with microbial antigens and, (iv) reproduction by the antigen of the disease process in vivo or in vitro (Ang et al., 2004; Johnson et al., 2017).
Microbes can exhibit four types of molecular mimicry with the host proteins, (i) sequential or structural similarities with the proteins or protein-domains, (ii) similarities with the protein-structures without sequence homology, (iii) architectural similarities with the binding surfaces without sequence homology (interface mimicry) and, (iv) similarities with the short linear motifs (SLiMs) of the proteins (motif mimicry) (Xue et al., 2014; Dolan et al., 2015). SLiMs are short stretches of amino acids (3–10 amino acids) which are functionally diverse and mediate various signaling and protein-protein interactions (PPIs) (Davey et al., 2012; Dinkel et al., 2014; Garg et al., 2022). One another important functional component of a protein is molecular recognition features (MoRFs) 5–25 amino acid long. They are present in the structurally disordered region of protein hence disordered. But once MoRFs binds to the their partner, they attain a well-developed structure (Vacic et al., 2007; Yan et al., 2016). Reportedly, several viruses and bacteria propagate and sustain themselves inside the host by mimicking SLiMs of the host proteins (Sámano-Sánchez and Gibson, 2020).
PPIs and cell signaling play a pivotal role in the functioning of cellular life. Hence, mimicry of the host proteins involved in PPIs and cell signaling greatly helps microbes in modulating/disrupting the host defense mechanisms. Moreover, substitution of microbial proteins in the host PPI networks eventually shifts the paradigm of host-pathogen interactions in the favor of the pathogen. Several eukaryotic host-like SLiMs have been reported in viral proteins which helps in their entry inside the host cell and modulation of host cellular pathways related to transcription regulation, cell cycle, immune response etc. (Davey et al., 2011; Garamszegi et al., 2013; Hagai et al., 2014). Similarly, several eukaryotic host-like SLiMs have also been reported in bacterial proteins which helps them to interfere with signaling pathways involving protein tyrosine kinases (TKs) and mitogen-activated protein kinases (MAPKs), tyrosine phosphorylation etc., (Higashi et al., 2002; Frese et al., 2006; Li et al., 2007; Zhu et al., 2007; Sámano-Sánchez and Gibson, 2020). The emergence of antibiotic resistance in pathogens necessitates discovery of new anti-infective therapies. Recently, protein-based immune-modulatory molecules or drugs which can disrupt the host-pathogen PPIs have been proposed as a novel anti-infective therapy (Sámano-Sánchez and Gibson, 2020). Such therapeutics can interfere with the host-pathogen SLiM-mediated interactions and create a hostile and non-conducive host environment for the survival of the pathogen. Though, mimicry of the eukaryotic host-like SLiMs has been recognized as an important mechanism underlying microbial pathogenicity, presence and characteristics of host-like SLiMs have not been studied in the mimicry peptides (mimitopes) of bacteria and viruses experimentally associated with autoimmune diseases. Thus, the present study was conducted to discern if the experimentally verified microbial mimitopes underlying various autoimmune diseases exhibit motif mimicry with the host and potentially modulate the host PPIs. Additionally, the evolutionary pressure on microbial mimitopes was also determined. This is the first report on potential of autoimmunity-related microbial mimitopes in modulating host protein-protein interactions and their evolutionary characteristics.
In the present study, the information on bacterial, viral and the host mimicry proteins was retrieved from a database of experimentally verified mimicry proteins, miPepBase (updated version assessed on January 2021). The database collates information about only the experimentally verified mimicry proteins/peptides and autoimmune diseases, thereof (Garg et al., 2017).
The presence of eukaryotic host-like SLiMs in the microbial mimicry proteins and mimitopes was predicted using ANCHOR (Dosztányi et al., 2009). A eukaryotic host-like SLiM was considered to be present in the microbial mimitope if at least half of the amino acid of mimitopes overlapped with the SLiM region.
The putative homologous/orthologous sequence of microbial mimicry proteins were identified using BLAST search against NCBI non-redundant (NR) protein database. To obtain a sequence homologous to each microbial protein, the NR database was searched using the microbial protein as query. Search results with query coverage of ≥80 and atleast 80% sequence identity were selected as the criteria for homology and used for further evolutionary analyses. The Orthologous protein clusters of each mimicry protein were aligned using ClustalW version 2 (Larkin et al., 2007). The corresponding gene sequences of these proteins were also aligned on the basis of their codons using pal2nal (Suyama et al., 2006). The ratio between the rate of non-synonymous substitution to the rate of synonymous substitution (ω = dN/dS) was used as a measure of the strength of selection pressure acting on a protein-coding gene. Assuming synonymous mutations are subjected to almost strictly neutral selection, the ω < 1, ω = 1, and ω > 1 represent negative selection, neutral evolution, and positive Darwinian selection, respectively (Yang, 2002). To compute the dN/dS ratio for each amino acid of the microbial mimitope, a site-specific model of the likelihood method was used using the codeml module of the PAML package (Yang, 2007).
Microbial mimicry of the host peptides has been implicated in several autoimmune diseases like multiple sclerosis (Wekerle and Hohlfeld, 2003), type 1 diabetes mellitus (Coppieters et al., 2012), autoimmune uveitis (Wildner and Diedrichs-Möhring, 2003), encephalomyelitis (Phelan et al., 2020), inflammatory bowel disease (Biank et al., 2007; Yusung and Braun, 2014), Crohn's disease (Cunha-Neto and Kalil, 2014; Polymeros et al., 2014), sarcoidosis, etc.
Motif mimicry of the eukaryotic host-like SLiMs has been associated with the pathogenicity of several bacteria and viruses because it helps them to hijack the host processes by rewiring the host PPI networks, signaling pathways, post-translational modifications etc., (Davey et al., 2012; Dinkel et al., 2014). Since, SLiMs are abundantly present in cell signaling and PPI network proteins, drugs targeting SLiM-regulated cellular processes have been proposed as novel therapeutics against bacterial and viral infections (Davey et al., 2011; Via et al., 2015; Sámano-Sánchez and Gibson, 2020). Thus, the present study was conducted to discern if the mimicry peptides (experimentally verified as responsible for autoimmune diseases; collated in miPepBase) contain the functional modules of PPIs, SLiMs and can be explored as novel drug targets.
A total of 150 bacterial mimicry proteins with their corresponding 28 host proteins and, 34 viral mimicry proteins with their corresponding 22 host mimicry proteins were retrieved from the miPepBase. The bacterial and their corresponding host mimicry proteins were named as Bacterial-set proteins. The viral and their corresponding host mimicry proteins were named as Viral-set proteins. The Bacterial-set proteins were involved in 16, while the Viral-set proteins were involved in 12 different types of autoimmune diseases. The detailed information of the host and pathogen, UniProtKB ID and name of the mimicry-protein(s), mimitope sequences, associated autoimmune diseases, and source of information (Pubmed ID) is provided in Supplementary Table 1.
During infection, the pathogens repurpose the host cells in a manner which is conducive to their own survival, proliferation and helpful for evading the host immunity. Thus, if the host cells can be rewired to the uninfected state or the one that is close to it, the disease progression can be halted or slowed down. Thus, drugs/molecules which can inhibit either (i) interactions between microbial host-like SLiMs and host PPI networks or, (ii) the metabolic chokepoints of these PPI networks or, (iii) nodes in the regulatory networks upstream or downstream of the hijacked host-SLiMs can be explored as novel therapeutics. Also termed as host-directed therapy, this approach has proved useful in combating many viruses and bacteria (Sámano-Sánchez and Gibson, 2020). The classical examples of host-directed therapy are the chemical inhibition of the proteasomal pathway which resulted in reduced viral loads in dengue infection and, RNA interference (RNAi) against some human proteins which inhibited replication of HIV and hepatitis C virus (Schwegmann and Brombacher, 2008). Our results revealed that the eukaryotic host-like SLiMs were abundant in the microbial mimicry proteins because 41 bacterial and 20 viral mimicry proteins showed the presence of SLiMs (Supplementary Table 2). On the contrary, only a few microbial mimitopes overlapped with the eukaryotic host-like SLiMs. Of the total 155 bacterial mimitopes, 10 bacterial mimitopes showed SLiMs and of the 43 viral mimitopes only 4 viral mimitopes had SLiMs (Table 1). This suggests that the most of the microbial mimitopes implicated in autoimmune diseases could not potentially rewire the host PPI networks for their own benefit. And, their pathology might have only the autoimmune component due to cross reactivity between the microbial epitopes and host peptides. However, the microbial mimitopes of ten bacteria and three viruses overlapped with the eukaryotic host-like SliMs (Table 1). Seven mimitopes, one each of Agrobacterium tumefaciens, Yersinia enterocolitica, Escherichia coli, Propionibacterium freudenreichii, Streptococcus mutans, Lactobacillus johnsonii, Bifidobacterium longum and two each of Mycobacterium leprae and Group A streptococcus showed the presence of eukaryotic host-like SLiMs in their mimitopes. Among the viruses, one mimitope of Herpes simplex virus and Herpes virus saimiri and two mimitopes of Epstein Barr virus had host-like SLiMs (Table 1). This suggests that these peptide regions might not only be responsible for inducing autoimmune diseases in the host by exhibiting epitope mimicry with the host, motif mimicry of the host-like SLiMs might also aid these bacteria and viruses in interrupting the host cell signaling and PPI networks.
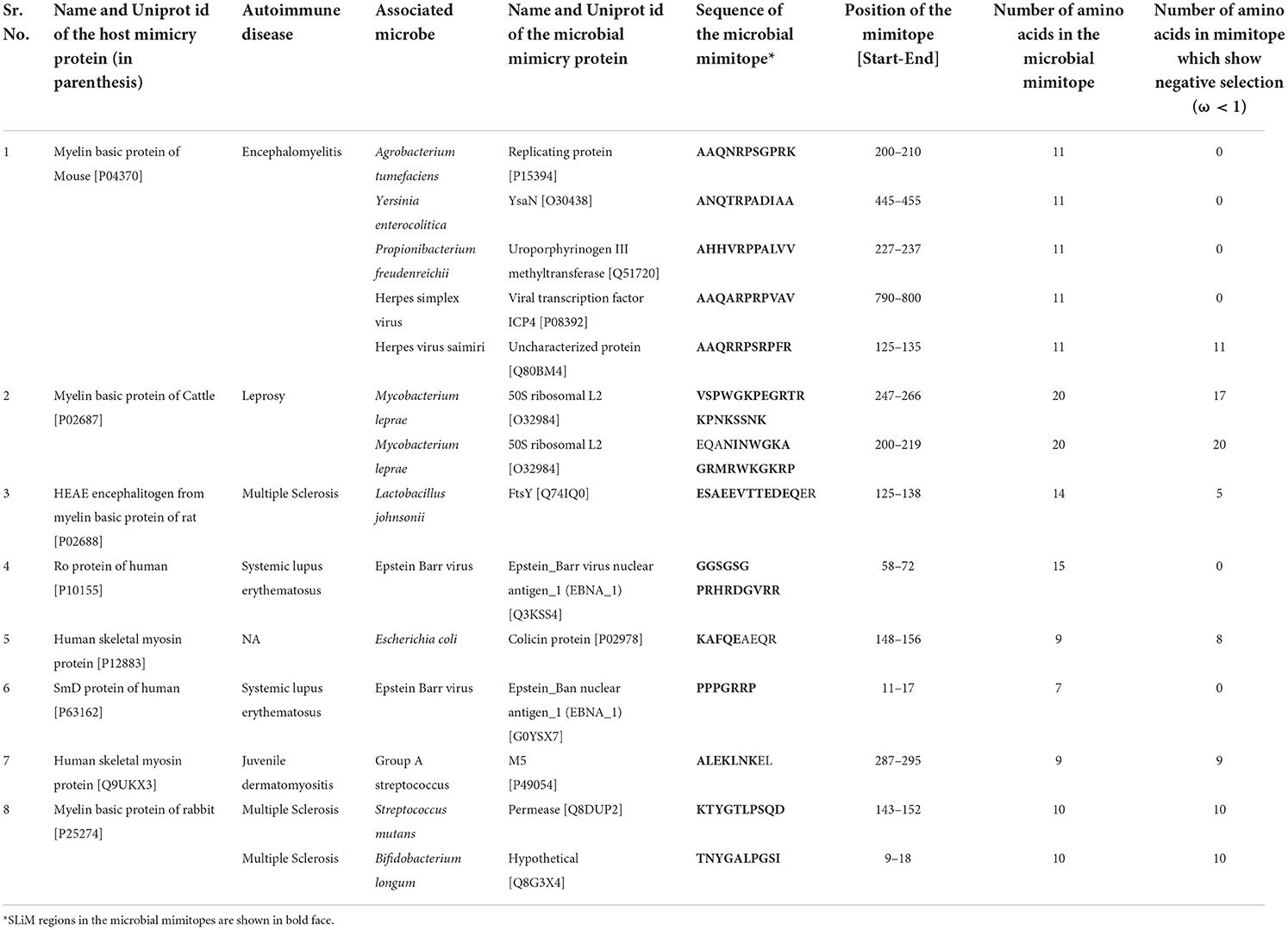
Table 1. Detailed information about the microbial mimitopes which overlapped with the eukaryotic host-like SliMs.
Evaluation of the selection pressure on the microbial mimitopes revealed that 77.85% of the bacterial and 83.54% of the viral amino acids were under negative selection pressure (ω < 1), implying that microbial mimtopes were generally conserved or had a low mutation rate. Comas et al., reported that the T cell epitopes of M. tuberculosis were highly conserved which might be a distinct evolutionary strategy of a highly successful pathogen as M. tuberculosis for immune subversion (Comas et al., 2010). This suggests that other pathogens might also employ a similar strategy for a sustained survival inside the host. However, analysis of the microbial mimitopes which overlapped with the eukaryotic host-like SLiMs revealed that only 46.75% of the bacterial mimitopes and 25% of the viral mimitopes were under negative selection pressure (ω < 1) (Table 1; Supplementary Figure 1). This observation can be explained by the fact that SLiMs are in disordered regions of the proteins hence, the rate of mutation is high and these regions are usually under a positive selection pressure (Uyar et al., 2014). In past we have also shown that low complexity regions of proteins are not always disordered (Kumari et al., 2015).
Conventionally, autoimmune diseases like systemic lupus erythematosus, encephalomyelitis, multiple sclerosis etc., are treated using immunomodulators or immunosuppressants which only try to cure the symptoms but do not eliminate the etiological agent which continues to proliferate inside the host and interfering with vital cellular process(es). On the contrary, protein-based immune-modulatory molecules/drug molecules designed to inhibit the mimitopes (or eukaryotic host-like SLiMs) might also disrupt the host-pathogen PPIs. This implies that inhibitors of microbial mimitopes (or eukaryotic host-like SLiMs) identified in this study would not only help in treating the pathogen-associated autoimmune disease but also eliminate the etiopathological agent associated with the disease.
Finally, the repertoire of mimitopes discovered in this study suggests new paradigms underlying autoimmune diseases and the etiopathology associated with the infection. Such knowledge can provide important clues for the discovery of new drugs/protein-based immune-modulatory molecules against the pathogens.
NS and MK conceived and designed the study. AG performed the work, prepared the illustrations, and wrote initial draft manuscript. All authors contributed to the critical analysis and revision, analyzed the results, and wrote the final versions of manuscript.
The work was carried out using the resources funded by the Indian Council of Medical Research [Grant Nos: VIR(25)/2019/ECD-1 and ISRM/12(33)/2019]. AG was supported by ICMR-JRF scheme [Grant Number: 3/1/3 J.R.F.-2016/LS/HRD-(32262)]. NS was supported by CSIR Senior Research Associateship (Scientists' Pool Scheme) [Grant Number: 13(9089-A)/2019-Pool].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1039188/full#supplementary-material
Ang, C. W., Jacobs, B. C., and Laman, J. D. (2004). The Guillain-Barré syndrome: a true case of molecular mimicry. Trends Immunol. 25, 61–66. doi: 10.1016/j.it.2003.12.004
Ascherio, A., and Munger, K. L. (2007). Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Ann. Neurol. 61, 504–513. doi: 10.1002/ana.21141
Biank, V., Broeckel, U., and Kugathasan, S. (2007). Pediatric inflammatory bowel disease: clinical and molecular genetics. Inflamm. Bowel Dis. 13, 1430–1438. doi: 10.1002/ibd.20213
Comas, I., Chakravartti, J., Small, P. M., Galagan, J., Niemann, S., Kremer, K., et al. (2010). Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42, 498–503. doi: 10.1038/ng.590
Coppieters, K. T., Wiberg, A., and von Herrath, M. G. (2012). Viral infections and molecular mimicry in type 1 diabetes. APMIS 120, 941–949. doi: 10.1111/apm.12011
Cunha-Neto, E., and Kalil, J. (2014). Molecular mimicry and chagas' disease. Mol. Mimicry Microb. Autoimmun. doi: 10.1128/9781555818074.ch18
Davey, N. E., Travé, G., and Gibson, T. J. (2011). How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159–169. doi: 10.1016/j.tibs.2010.10.002
Davey, N. E., Van Roey, K., Weatheritt, R. J., Toedt, G., Uyar, B., Altenberg, B., et al. (2012). Attributes of short linear motifs. Mol. Biosyst. 8, 268–281. doi: 10.1039/C1MB05231D
Dinkel, H., Van Roey, K., Michael, S., Davey, N. E., Weatheritt, R. J., Born, D., et al. (2014). The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 42, D259–D266. doi: 10.1093/nar/gkt1047
Dolan, P. T., Roth, A. P., Xue, B., Sun, R., Dunker, A. K., Uversky, V. N., et al. (2015). Intrinsic disorder mediates hepatitis C virus core-host cell protein interactions: HCV Core MoRFs target cellular proteins. Protein Sci. 24, 221–235. doi: 10.1002/pro.2608
Dosztányi, Z., Mészáros, B., and Simon, I. (2009). ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 25, 2745–2746. doi: 10.1093/bioinformatics/btp518
Frese, S., Schubert, W.-D., Findeis, A. C., Marquardt, T., Roske, Y. S., Stradal, T. E. B., et al. (2006). The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 281, 18236–18245. doi: 10.1074/jbc.M512917200
Fujinami, R. S. (2001). Viruses and autoimmune disease – two sides of the same coin? Trends Microbiol. 9, 377–381. doi: 10.1016/S0966-842X(01)02097-2
Garamszegi, S., Franzosa, E. A., and Xia, Y. (2013). Signatures of pleiotropy, economy and convergent evolution in a domain-resolved map of human–virus protein–protein interaction networks. PLoS Pathog. 9, e1003778. doi: 10.1371/journal.ppat.1003778
Garg, A., Dabburu, G. R., Singhal, N., and Kumar, M. (2022). Investigating the disordered regions (MoRFs, SLiMs and LCRs) and functions of mimicry proteins/peptides in silico. PLoS ONE 17, e0265657. doi: 10.1371/journal.pone.0265657
Garg, A., Kumari, B., Kumar, R., and Kumar, M. (2017). miPepBase: a database of experimentally verified peptides involved in molecular mimicry. Front. Microbiol. 8, 2053. doi: 10.3389/fmicb.2017.02053
Hagai, T., Azia, A., Babu, M. M., and Andino, R. (2014). Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 7, 1729–1739. doi: 10.1016/j.celrep.2014.04.052
Higashi, H., Tsutsumi, R., Fujita, A., Yamazaki, S., Asaka, M., Azuma, T., et al. (2002). Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA. 99, 14428–14433. doi: 10.1073/pnas.222375399
Johnson, T. P., Tyagi, R., Lee, P. R., Lee, M.-H., Johnson, K. R., Kowalak, J., et al. (2017). Nodding syndrome may be an autoimmune reaction to the parasitic worm Onchocerca volvulus. Sci. Transl. Med. 9, eaaf6953. doi: 10.1126/scitranslmed.aaf6953
Kumari, B., Kumar, R., and Kumar, M. (2015). Low complexity and disordered regions of proteins have different structural and amino acid preferences. Mol. Biosyst. 11, 585–594. doi: 10.1039/C4MB00425F
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lester, P. J., Buick, K. H., Baty, J. W., Felden, A., and Haywood, J. (2019). Different bacterial and viral pathogens trigger distinct immune responses in a globally invasive ant. Sci. Rep. 9, 5780. doi: 10.1038/s41598-019-41843-5
Li, H., Xu, H., Zhou, Y., Zhang, J., Long, C., Li, S., et al. (2007). The phosphothreonine lyase activity of a bacterial type III effector family. Science. 315, 1000–1003. doi: 10.1126/science.1138960
Libbey, J. E., and Fujinami, R. S. (2010). Potential triggers of MS. Results Probl. Cell Differ. 51, 21–42. doi: 10.1007/400_2008_12
Phelan, J., Grabowska, A. D., and Sepúlveda, N. (2020). A potential antigenic mimicry between viral and human proteins linking myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) with autoimmunity: the case of HPV immunization. Autoimmun. Rev. 19, 102487. doi: 10.1016/j.autrev.2020.102487
Polymeros, D., Tsiamoulos, Z. P., Koutsoumpas, A. L., Smyk, D. S., Mytilinaiou, M. G., Triantafyllou, K., et al. (2014). Bioinformatic and immunological analysis reveals lack of support for measles virus related mimicry in Crohn's disease. BMC Med. 12, 139. doi: 10.1186/s12916-014-0139-9
Sámano-Sánchez, H., and Gibson, T. J. (2020). Mimicry of short linear motifs by bacterial pathogens: a drugging opportunity. Trends Biochem. Sci. 45, 526–544. doi: 10.1016/j.tibs.2020.03.003
Schwegmann, A., and Brombacher, F. (2008). Host-directed drug targeting of factors hijacked by pathogens. Sci. Signal. 1, re8. doi: 10.1126/scisignal.129re8
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
Tarakhovsky, A., and Prinjha, R. K. (2018). Drawing on disorder: how viruses use histone mimicry to their advantage. J. Exp. Med. 215, 1777–1787. doi: 10.1084/jem.20180099
Uyar, B., Weatheritt, R. J., Dinkel, H., Davey, N. E., and Gibson, T. J. (2014). Proteome-wide analysis of human disease mutations in short linear motifs: neglected players in cancer? Mol. Biosyst. 10, 2626–2642. doi: 10.1039/C4MB00290C
Vacic, V., Oldfield, C. J., Mohan, A., Radivojac, P., Cortese, M. S., Uversky, V. N., et al. (2007). Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 6, 2351–2366. doi: 10.1021/pr0701411
Via, A., Uyar, B., Brun, C., and Zanzoni, A. (2015). How pathogens use linear motifs to perturb host cell networks. Trends Biochem. Sci. 40, 36–48. doi: 10.1016/j.tibs.2014.11.001
Wekerle, H., and Hohlfeld, R. (2003). Molecular mimicry in multiple sclerosis. N. Engl. J. Med. 349, 185–186. doi: 10.1056/NEJMcibr035136
Wildner, G., and Diedrichs-Möhring, M. (2003). Autoimmune uveitis induced by molecular mimicry of peptides from rotavirus, bovine casein and retinal S-antigen. Eur. J. Immunol. 33, 2577–2587. doi: 10.1002/eji.200324058
Xue, B., Blocquel, D., Habchi, J., Uversky, A. V., Kurgan, L., Uversky, V. N., et al. (2014). Structural disorder in viral proteins. Chem. Rev. 114, 6880–6911. doi: 10.1021/cr4005692
Yan, J., Dunker, A. K., Uversky, V. N., and Kurgan, L. (2016). Molecular recognition features (MoRFs) in three domains of life. Mol. Biosyst. 12, 697–710. doi: 10.1039/C5MB00640F
Yang, Z. (2002). Inference of selection from multiple species alignments. Curr. Opin. Genet. Dev. 12, 688–694. doi: 10.1016/S0959-437X(02)00348-9
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yusung, S., and Braun, J. (2014). Molecular mimicry, inflammatory bowel disease, and the vaccine safety debate. BMC Med. 12, 166. doi: 10.1186/s12916-014-0166-6
Keywords: motif mimicry, short linear motifs (SLiMs), autoimmunity, mimitopes, drug targets
Citation: Garg A, Singhal N and Kumar M (2022) Investigating the eukaryotic host-like SLiMs in microbial mimitopes and their potential as novel drug targets for treating autoimmune diseases. Front. Microbiol. 13:1039188. doi: 10.3389/fmicb.2022.1039188
Received: 07 September 2022; Accepted: 17 October 2022;
Published: 04 November 2022.
Edited by:
Svetlana Khaiboullina, University of Nevada, Reno, United StatesReviewed by:
Punit Kaur, All India Institute of Medical Sciences, IndiaCopyright © 2022 Garg, Singhal and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manish Kumar, bWFuaXNoQHNvdXRoLmR1LmFjLmlu; Neelja Singhal, bmVlbGphQHNvdXRoLmR1LmFjLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.