
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 November 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1036451
 Zhenrong Lin1
Zhenrong Lin1 Lina Shi1
Lina Shi1 Xiaoting Wei2
Xiaoting Wei2 Bing Han1
Bing Han1 Cuoji Peng1
Cuoji Peng1 Zeying Yao1,3
Zeying Yao1,3 Qing Xiao1
Qing Xiao1 Xinmin Lu4
Xinmin Lu4 Yanfang Deng5
Yanfang Deng5 Huakun Zhou6
Huakun Zhou6 Kesi Liu1
Kesi Liu1 Xinqing Shao1*
Xinqing Shao1*The alpine meadow on the Qinghai-Tibetan Plateau, which is susceptible to global climate change and human activities, is subject to nutrient addition such as nitrogen (N) and phosphorus (P) to enhance soil available nutrients and ecosystem productivity. Soil bacterial community partly drivers the effects of nutrient additions on ecosystem processes, whereas the factors influencing N and P additions on bacterial community in alpine meadows are not well documented. We conducted a N and P addition experiment in an alpine meadow ecosystem on the Qinghai-Tibetan Plateau with four treatments: untreated control (CK), N addition (N), P addition (P), and NP addition (NP). We employed a high-throughput Illumina Miseq sequencing technology to investigate the response of soil bacterial community to short-term N and P additions. N and P additions decreased soil bacterial richness (OTU numbers and Chao 1 index), and P addition decreased soil bacterial diversity (Shannon and Simpson indices). N addition directly induced the change of soil , and decreased plant diversity. The N and P additions reduced soil bacterial community diversity, whose response was independent with plant diversity. Additionally, nutrient additions altered soil bacterial community composition, which were highly correlated with soil properties (i.e. pH, , and TP) as shown by RDA. Consistently, structural equation modeling results revealed that N addition indirectly acted on soil bacterial community through altering soil available nutrients and pH, while P addition indirectly affected bacterial community by increasing soil P availability. These findings imply that more attention should be paid to soil properties in regulating belowground biodiversity process in alpine meadows under future environmental change scenario.
Nitrogen (N) and phosphorus (P) availability often constrain the growth and metabolism of plant and soil microorganisms in terrestrial ecosystems (Dijkstra et al., 2012; Yuan and Chen, 2015; Jiang et al., 2021). Supplies and inputs of N and P have been greatly affected by pre-industrial revolution and human activities (e.g., fossil combustion and excessive fertilizer applications) (Ma et al., 2016; Yan et al., 2022). Presently, global inputs of N and P have increased by 84.0 and 43.3%, respectively, relative to the input before the 20th century (Peñuelas et al., 2013). Unbalanced N and P inputs are exacerbating P limitation or N and P co-limitation in terrestrial ecosystems, which generates drastic adverse effects on organisms performance (e.g., biodiversity loss) (Hooper et al., 2012) and weakens related ecosystem functions and services (Isbell et al., 2013; Spohn and Schleuss, 2019).
Soil microorganisms are important biotic factors in maintaining plant diversity and determining nutrient cycling (Jiang et al., 2019; Yang et al., 2021). For instance, soil bacteria regulate numerous ecological processes (Delgado-Baquerizo et al., 2017). In the past few decades, an increasing interest has been focused on the influence of N inputs on the soil bacterial community. N enrichment-induced loss of species diversity has been demonstrated to pose threats to both ecosystem stabilization and interactions between above and below ground (Geisseler and Scow, 2014). Previous studies have found that N fertilizer applications reduce soil bacterial diversity and alter the bacterial community composition (Dong et al., 2020; Sui et al., 2022). Ramirez et al. (2012) found in an incubated experiment that consistent N addition altered soil bacterial community composition with the increase in the relative abundance of Actinobacteria and Firmicutes and the reduction in the relative abundance of Acidobacteria and Verrucomicrobia. However, soil bacterial community composition is inconsistently responsive to N additions, possibly due to ecosystem-specific factors or variations in the soil nutrient status, plant community, and climatic factors (Yan et al., 2022).
P addition is paying much attention as a key determinant of the soil bacterial community (Ling et al., 2017; Long et al., 2018). However, there are still inconsistent results about the influence of P addition on soil bacterial community. For instance, in a Tibetan meadow, P input have been found almost no effect on the bacterial community composition (He et al., 2016); in a N-rich forest, P addition alters the diversity of soil bacteria, but do not affect the bacterial richness (Wang et al., 2018a); in a temperate meadow, the positive impact of P addition on soil bacterial richness is observed (Yan et al., 2022). These inconsistent results might be attributed to variation in ecosystem types and local environmental factors. Hence, a comprehensive understanding of the feedbacks in soil bacterial community to N and P additions is essential for evaluating the effects of global change and human activities on terrestrial ecosystem processes.
Nutrient additions can substantially alter soil bacterial community through direct and indirect effects on soil and plant properties (Zeng et al., 2016). N inputs indirectly alter soil bacterial community by affecting pH, resulting in a bacterial niche differentiation in semi-arid steppe (Ling et al., 2017). According to Ramirez et al. (2010), the response of soil microbial respiration to N addition is consistent irrespective of soil type and N form due to direct changes in N availability. The effect of change in soil available nutrient induced by N input on bacterial diversity and richness is also observed (Dimitriu and Grayston, 2009; Yan et al., 2018). Changes in soil P availability following P addition have been shown to be a key environmental factor determining soil bacterial community (Ling et al., 2017). A study conducted by Turner and Joseph Wright (2014) in a tropical forest demonstrated that P addition decreased microbial biomass by reducing soil phosphatase activity. The correlation between plant parameters and microbial community composition has been verified in grassland ecosystems (Sugiyama et al., 2008; Prober et al., 2015). The change of plant community resulted from nutrient inputs also affects soil bacterial community (Raynaud et al., 2021). Wei et al. (2013) demonstrated that N deposition weakened the effect of plant community on microbial community composition in a semiarid grassland. Hence, exploring how soil and plant characteristics affect soil bacterial community can improve our understanding of nutrient-induced ecological processes of ecosystems.
As the major ecosystem on the Qinghai-Tibetan Plateau (QTP), grassland ecosystems account for 68% of the total land area of QTP and about one-fourth of the total natural grassland area of China (Gai et al., 2009). The alpine meadow accounts for 35% of the total grassland area in QTP (Cao et al., 2004). The average rate of N deposition in QTP is up to 10 kg ha−1 yr−1 (Zhang et al., 2018). Furthermore, N and P fertilizers are usually applied to enhance soil available nutrients and the productivity of alpine meadow ecosystems (Yang et al., 2011; He et al., 2016). However, the effects of N and P additions on soil bacterial community composition were not well documented in Tibetan meadows. To evaluate the effects of short-term N, P and NP additions on the bacterial community composition of soil, we carried out a full-factorial N and P additions experiment in an alpine meadow. We hypothesized that: (1) N and P additions reduced soil bacterial diversity and changed community composition in the alpine meadow; (2) the responses of soil bacterial community to short-term N and P additions were regulated by the variation in soil properties and plant diversity.
The experiment was conducted in a nutrient addition platform in an alpine meadow (36°55′N, 100°57′E, 3, 000~3,100 m a.s.l.), which was located in Haiyan County, Qinghai Province, northeastern Qinghai-Tibetan Plateau, China. Highland continental climate is characteristic of this region. The amount of mean precipitation is 400 mm per year and mainly distributes in the growing season between June and August. The average annual temperature is −0.45°C, with extremes of −29°C and 27°C distributed between January and July per year. The soil is clay-loam and classified as Mat-Gryic-Cambisol (Li et al., 2020). The main species of plants in this study area include Leymus secalinus, Stipa purpurea, Poa pratensis, Kobresia humilis, Melissitus ruthenica, Potentilla chinensis, and Artemisia frigida.
The study area was fenced off to exclude large herbivores since 2019. The distribution of plants was fairly homogeneous, with few visible patches of individual species. This experiment was conducted using a completely randomized block experiment consisting of four treatments with four replicates. Each treatment was set up as following: the control plots with no fertilization (referred as CK), N fertilizer with a rate of 100 kg N ha−1 yr−1 (referred as N), P fertilizer with a rate of 80 kg P ha−1 yr−1 (referred as P), and the combination of N and P fertilizer at rates of 100 kg N ha−1 yr−1 and 80 kg P ha−1 yr−1 (referred as NP), respectively (Luo et al., 2019). N and P fertilizers were applied as forms of urea and superphosphate, respectively. After sunset, fertilizers were manually applied to the plot ground surface (for higher moisture levels) on early May, 2019, 2020, and 2021, respectively. Each experimental plot area was 3 m length × 4 m width. A 1 m width buffer strip was designed to avoid disturbance to neighboring plots as much as possible.
We collected soil samples from each plot of four treatments in August 2021. Each plot was sampled by taking five cylindrical soil cores (3.5 cm diameter and 10 cm depth) and mixing them thoroughly to form one composite sample (i.e., 16 composite samples). After collected, samples were put into self-sealing bags and immediately placed in a cool box and transported to the laboratory. In the lab, each soil sample was sieved (2 mm) to remove visible roots and little rocks. And then, they were separated into three sub-samples. And two of the three sub-samples were stored at −80°C until extraction of DNA and 4°C for soil water content and available nutrients analysis, respectively, while the remainder was air-dried for physico-chemical analysis.
Soil pH was measured using a pH probe (Thermo Fisher Scientific Inc., Beverly, MA, USA) in distilled water at a 1:2.5 (wt/vol) ratio. Soil available nitrogen ( and ) were performed with continuous-flow analyzer as previously described (Han et al., 2021). Soil total phosphorus (TP) was determined colorimetrically at 880 nm by digesting samples and reacting them with molybdenum blue.
Plant community composition investigation and sampling were performed at the period of maximum sward biomass of August, 2021. Plant coverage was identified by visual estimation. Briefly, one quadrat (0.5 × 0.5 m) with 100 meanly distributed grid points was placed in each plot. Visual estimates of the relative coverage of each species were estimated in each quadrat. The Shannon-Wiener index (H) was employed to indicate plant diversity, which was calculated by the following Equation (Dong et al., 2020):
where Pi represents the relative biomass of the i species of plant, and S represents the total number of species in each plot.
Aboveground part of plant species was mowed to the ground level and then divided into three functional groups (grasses, legumes, and forbs). After that, the plant belowground biomass was obtained by taking three random soil cores (10 cm diameter, 0–20 cm depth) from each plot and washing them under running water on a 2-mm sieve. The collected above and belowground parts were oven-dried at 65°C for 72 h, and then weighed.
Bacterial DNA extractions from 0.1-g of fresh soil were performed using the Omaga DNA Kit (Omega Bio-Tek, Norcross, GA, USA). Then, the extracted DNA samples was quantified by a spectrophotometer and the 1% agarose gel electrophoresis was used to assess DNA quality. The 16S rRNA gene V3-V4 region was amplified with the primer set 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). The PCR procedures were conducted according to the manufacturer's instructions. The raw sequence data were submitted to the Sequence Read Archive of the National Center for Biotechnology Information, USA (No. PRJNA884813).
We used QIIME2 (v2019.4) software platform to demultiplex the raw 16S RNA sequence data using the q2-demux plugin and then cut the primers using the cutadapt plugin (Martin, 2011). We then call the DADA2 pipeline via q2-dada2 to perform a series of quality control operations including de-priming, quality filtering, denoising, splicing and de-chimerisation (Callahan et al., 2016). Each de-duplicated sequence generated using DADA2 quality control was called ASVs (amplicon sequence variants). After that, the non-single ASVs were compared using MAFFT (Katoh et al., 2002) and further processed using FastTree2 to build a phylogenetic tree (Price et al., 2009). Using the classify-sklearn naive Bayes taxonomy classifier in feature-classifier plugin, taxonomies were assigned to ASVs classified. The QIIME2 diversity plugin was used to assess the diversity (Shannon-Wiener index and Simpson index) of the bacterial community.
The effects on soil properties, plant parameters, and soil bacterial diversity of N addition, P addition, and their interaction were tested using a two-way ANOVA. After that, we employed the least significant difference test (LSD) (at P < 0.05) to examine the difference in soil, plants, and soil microbes among four treatments. Prior to running ANOVA, TP was log-transformed due to its not meeting the normality. The statistical analyses of ANOVA were modeled with SPSS 23.0 software (SPSS, Inc., Armonk, NY, USA). Principal coordinate analysis (PCoA) and permutational MANOVA (“adonis” function in vegan package) were used to assess the effect of each treatment on the soil bacterial community composition.
We conducted a redundancy analysis (RDA) to evaluate the correlation between soil bacterial community composition and soil parameters (pH, TP, and ) and plant community variables (plant coverage, plant Shannon index). Only the vectors of soil parameters and plant community variables with variance inflation factors (VIFs) < 10 were taken into account. We used the “rda” function for model construction, “anova.cca” for significance testing of variables and axes, and “vif.cca” for variance inflation factor analysis. To estimate the importance of each explanatory variable, we used a hierarchical partitioning and a permutation test to derive significance at a level of P < 0.05. RDA analysis was conducted in R, version 4.0.5 (R Core Team, 2019), based on the “vegan” and “rdacca.hp” packages (Lai et al., 2022).
We conducted a structural equation modeling (SEM) to explore direct and indirect relationships among plant diversity, bacterial community, and soil properties under treatments. The bacterial community composition used in the SEM were represented using the first axis of PCoa analysis.The fitness of the model was evaluated using the chi-square test (0< χ2/df≤2 and P > 0.05), the root mean square error of approximation (RMSEA ≤ 0.05), the high goodness-of-fit index (GFI > 0.90) and the comparative fit index (CFI > 0.90) (Zhang et al., 2013). SEM analysis was constructed using AMOS 23.0 (SPSS Inc., Chicago, IL, USA). The critical level of significance for all statistical testing was set at P < 0.05.
Soil pH decreased significantly after the addition of N and NP, while it was unchanged by P addition (Figure 1A). Both N and NP addition increased soil available N, especially concentrations, but were not significant between treatments (Figures 1B,C). P addition alone decreased soil concentration (Figure 1B), while increased soil total P concentration (Figure 1D). Soil total P concentration was significantly increased by N addition but not NP addition (Figure 1D).
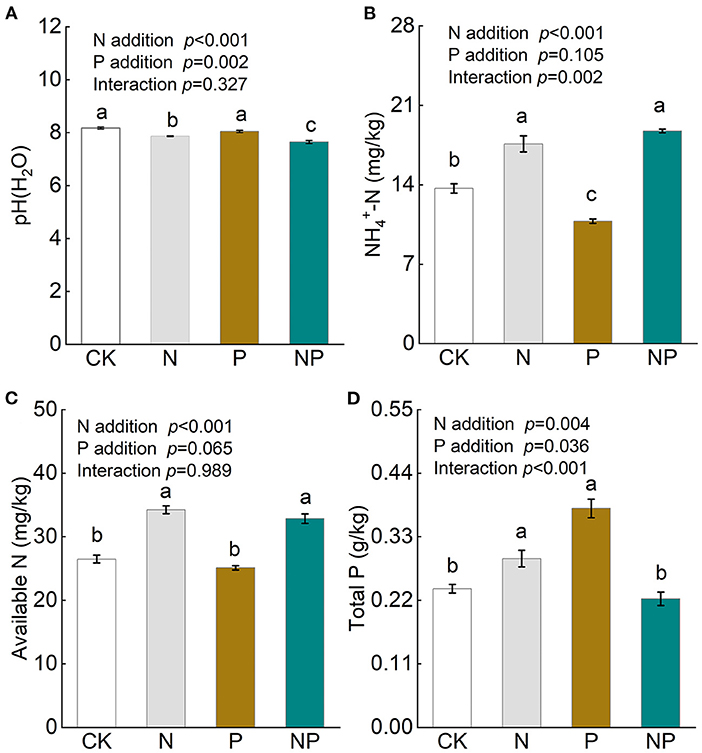
Figure 1. The variation of soil pH (A), soil NH-N (B), soil available N (C), and soil total P (D) under different nutrient additions. Values are means with SE (n = 4). Different lowercase letters represent significant difference among treatments determined by LSD-test (P = 0.05). CK, the control; N, nitrogen addition; P, phosphorous addition; NP, the combined addition of nitrogen and phosphorous.
N and NP addition significantly enhanced plant coverage and reduced plant Shannon index compared to CK and P addition (Figures 2A,B). N and NP addition significantly enhanced the relative aboveground biomass (AGB) of grasses (Supplementary Figure S1). Plant AGB was increased by NP addition compared to CK (Figure 2C). Total belowground biomass of plant was unchanged by N and P additions (Figure 2D).
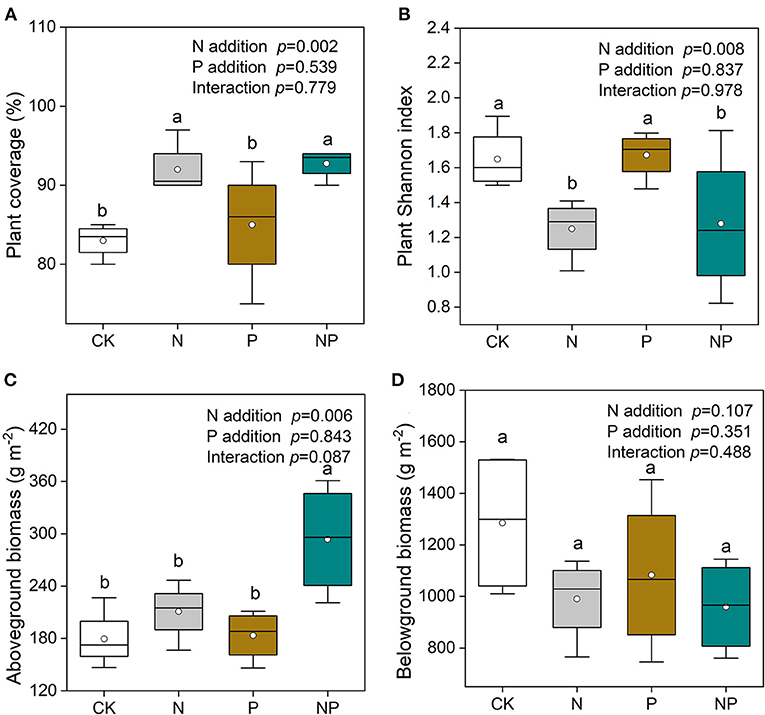
Figure 2. (A) The variation of plant coverage, (B) plant Shannon index, (C) total aboveground biomass, and (D) total belowground biomass under different nutrient additions. Values are means with SE (n = 0.4). Different lowercase letters represent significant difference among treatments determined by LSD-test (P = 0.05). CK, the control; N, nitrogen addition; P, phosphorous addition; NP, the combined addition of nitrogen and phosphorous.
Bacterial Chao1 richness and Shannon diversity rarefaction curves were saturated with enhanced sequence numbers (Supplementary Figure S2), indicating that the sequencing depth was sufficient. N and P additions significantly reduced bacterial richness (OTU numbers and Chao 1 indices). Lower bacterial OTU numbers and Chao1 indices were detected at the N, P, and NP addition treatments in comparison with CK (Figures 3A,B). The Shannon and Simpson indices of bacterial community were decreased significantly by N and P additions (Figures 3C,D).
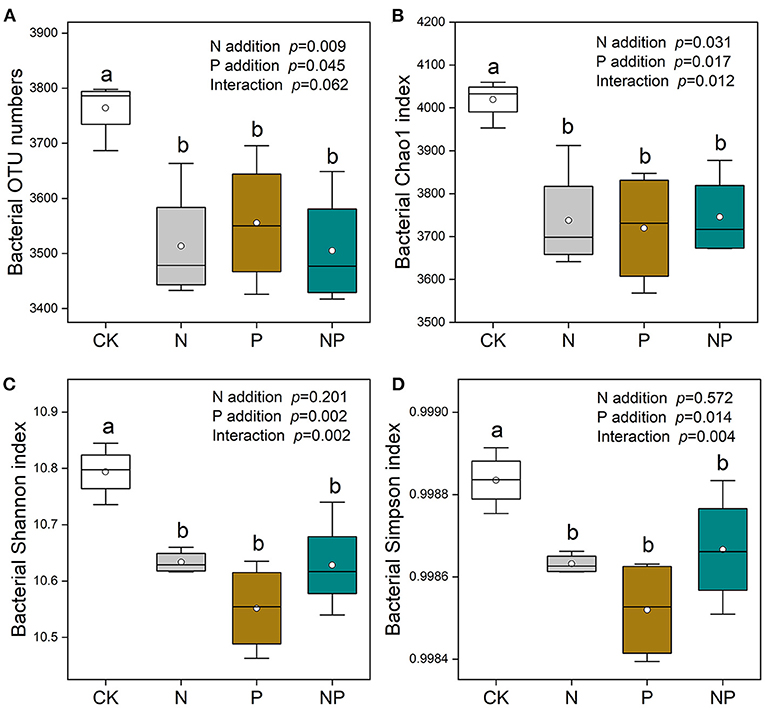
Figure 3. The variation of soil bacterial richness [OUT numbers (A) and Chao1 indices (B)] and diversity [Shannon (C) and Simpson (D) indices] under different nutrient additions. Values are means with SE (n = 4). Different lowercase letters represent significant difference among treatments determined by LSD-test (P = 0.05). CK, the control; N, nitrogen addition; P, phosphorous addition; NP, the combined addition of nitrogen and phosphorous.
Proteobacteria (29.8%), Actinobacteria (28.9%), Acidobacteria (18.9%), and Planctomycetes (7.1%) were the predominant phyla in the soil bacterial community in all treatments (Supplementary Figure S3). The relative abundances of Alphaproteobacterial were significantly increased by P addition (P = 0.038), whereas the addition of N and NP did not act on the relative abundance of the four main bacterial taxa (Supplementary Figure S3). PCoA showed that bacterial community composition was obviously separated by treatments. The first two principal coordinates explained 33.26% of the total variation (PCoA 1 = 22.60%, PCoA 2 = 13.66%) in the bacterial community composition (Supplementary Figure S4). The bacterial community composition was shown to be statistically significant using PERMANOVA (Supplementary Table S1).
Redundancy analysis (RDA) indicated that soil pH, TP, and were the most significant explanatory variables for variation in soil bacterial community composition (Figure 4). There were 26.44% on the first constraint axis and 24.22% on the second constraint axis for all soil variables, which explained 50.66% of the total variation in bacterial community composition (Figure 4).
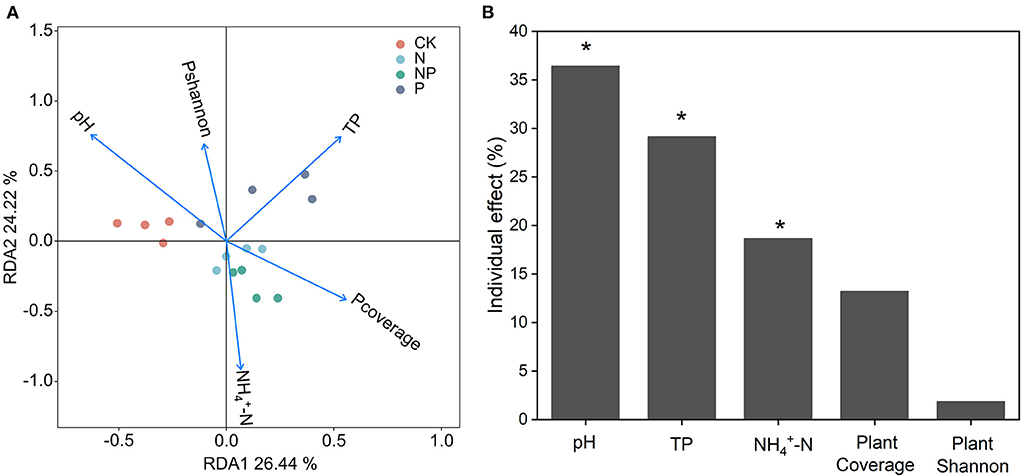
Figure 4. (A) Soil bacterial community composition and its variation partition ordination plot of a redundancy analysis (RDA) under N and P additions treatments. (B) The black bars for the independent explanation of bacterial community composition calculated by hierarchical partitioning. (The percentage of values shows adjusted R2, *indicates P < 0.05). TP, total phosphorus; , ammonium nitrogen; Pcoverage, plant coverage and Pshannon, plant Shannon. Proportions for the bacterial community composition were explained by soil properties and plant parameters.
The structural equation modeling (SEM) revealed the integrated reaction of soil properties, plant parameters, and bacterial community to N and P additions. It provided a good fit between all variables (P = 0.53, χ2/df = 0.95, CFI = 1.000, GFI = 0.800, RMSEA <0.01), which accounted for 74% variation in pH, 84% in TP, 60% in , 32% in plant diversity, 97, 73, and 82% in bacterial composition, richness, and diversity, respectively (Figure 5). Overall, the increase in soil AN and the decrease in soil pH caused N and P additions were primary factors that contributed to the significant influence on the plant and soil bacterial community. N addition indirectly affected plant diversity through direct effect on soil . N and P additions decreased soil bacterial richness through affecting soil pH and TP, respectively. N addition had an indirect effect on bacterial composition through directly affecting , pH, and TP, while P addition indirectly affected soil bacterial composition through increasing soil TP. N addition indirectly affected bacterial diversity via mediating soil properties, and P addition reduced bacterial richness and indirectly regulated bacterial diversity (Figure 5).
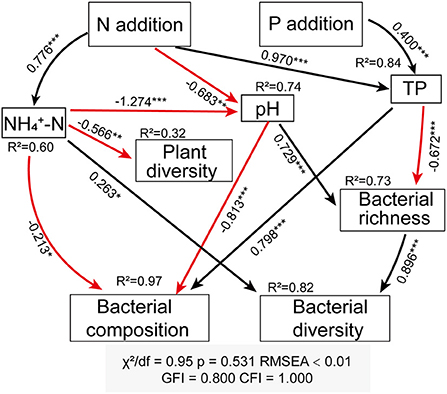
Figure 5. Structural equation modeling (SEM) analysis depict the regulatory pathway of N and P addition on plant diversity, bacterial composition, bacterial richness (Chao 1 index), and bacterial diversity (Shannon index). The solid black arrow lines represent positive effects, solid red arrow lines represent negative effects. Adjacent numbers labeled in the same direction as the arrow represent standardized path coefficients. Values of R2 indicate the proportion of variance explained by each variable in the model. *Indicates P < 0.05, **indicates P < 0.01, and ***indicates P < 0.001.
Nutrient addition could affect soil bacterial community composition by directly or indirectly influencing plant diversity and soil properties (Dong et al., 2020; Chen et al., 2021). The significant response of soil bacterial diversity to N and P additions was observed in this study, suggesting that nutrient addition was an key influential factor (Klironomos et al., 2011; Ling et al., 2017). It is crucial to clarify the relationship among plant diversity, soil properties and bacterial community with regard to assessing the impact of nutrient addition on grassland ecosystem functions. The findings showed that soil bacterial community, including diversity and richness, was indirectly influenced by N and P additions through direct variation in soil available nutrients or acidification status. N and P additions had uncoupling effects on plant diversity and soil bacterial diversity. The results in an alpine meadow highlighted that soil properties, rather than plant diversity, regulated the response of soil bacterial community to short-term nutrient addition.
In the present study, we found that N and P additions induced reductions in bacterial richness and diversity (Figure 3). N addition generated a significantly negative effect on bacterial diversity (Figures 3C,D), which was in accordance with previous studies (Zhou et al., 2015; Wang et al., 2018b). Soil pH can partially explain this phenomenon, and earlier studies found that the diversity of soil bacteria was affected by the decrease in soil pH following N addition (Roem et al., 2002; Wang et al., 2018a). As previously observed in tropical forests and temperate grasslands, bacteria richness decreased as enhancing N input (Zeng et al., 2016; Wang et al., 2018c). This supports our first hypothesis that N and P additions decreases soil bacterial richness. It is noteworthy that both P addition and the combination of N and P reduced soil bacterial diversity (Figures 3C,D), which highlighted that P was a growth-limiting nutrient for soil bacteria in an Tibetan alpine meadows. We observed that P addition significantly reduced the richness of soil bacteria (Figures 3A,B). It can be speculated that expanded P availability might exacerbate competition between soil bacteria and plants for nutrients, resulting in a reduction in soil bacterial richness following P addition (Zhang et al., 2014). In addition, enhanced soil available P resulting from P addition might reduce phosphate-solubilized bacteria, and thereby decreasing bacterial richness (Long et al., 2018). This result was conflicted with a non-significant effect of P addition on bacterial richness from old-growth tropical forests, which concluded that P addition failed to mitigate excessive N-derived soil acidification in an acidic environment (Wang et al., 2018c). These distinct phenomena might be attributed to the difference in ecosystem type, P application amount, and experimental duration.
Proteobacterial, Actinobacteria, Acidobacteria, and Planctomycetes were found to be the main bacterial phyla in alkaline meadow soil, which was in line with recent studies in an temperate meadow (Yan et al., 2022) and Tibetan alpine steppe (Dong et al., 2020). The RDA result demonstrated that the change in soil bacterial community composition was regulated by soil pH, TP, and (Figure 4). This coincides with a previous study that was conducted by Zeng et al. (2016), in which they showed that soil and pH were the important factors acting on soil bacterial community. A strong correlation between soil TP and bacterial community composition was also confirmed in SEM (Figure 5), which suggested that increased soil P is conducive to shape soil bacterial community composition in an alpine meadow (Yan et al., 2022).
As shown in SEM, soil parameters, but not plant diversity, had significant direct and indirect effects on bacterial diversity and richness (Figure 5), which partly supported our second hypothesis. Soil is a powerful ecological filter influencing the composition and diversity of soil bacterial community (Ling et al., 2017). Among the soil parameters mediating bacterial community, pH has been expected as a main factor (Lauber et al., 2009). N addition induced a substantial reduction in soil pH, while a non-significant effect of P addition was observed (Figure 5), which could be considered as a potential reason for the differential responses of bacterial richness and diversity to N and P additions. Soil acidification caused by N addition facilitates acidophilic bacterial growth, which might conversely inhibit the activity of other bacterial taxa (Zhang et al., 2014; Song et al., 2022). N addition reduced soil bacterial richness, which probably resulted from the fact that it narrows the optimal range of pH for bacteriological growth (Rousk et al., 2010; Husson, 2013). SEM also indicated that the declined pH caused a reduction in bacterial richness, and thus reduced bacterial diversity (Figure 5). The high-rate urea input (N input) can also cause ammonia toxicity to soil microbes (Vitousek et al., 1997; Geisseler and Scow, 2014), and then decreases soil bacterial richness. Furthermore, decreased soil pH and increased ammonium ionic strength caused by the high-rate N fertilizer application could be another potential reason for the decrease in bacterial richness (Demoling et al., 2007; Geisseler and Scow, 2014).
In line with recent findings (Zeng et al., 2016; Wang et al., 2018a), we observed that soil bacterial diversity was closely related to available N (Figure 5). Soil N availability, especially , is the preferred nutrient of soil bacteria (Nie et al., 2018). Most bacterial community, especially those in symbiosis with other organisms, maintain high abundances at the ammonium-rich environments (Müller et al., 2006). We also observed that had significantly direct effects on bacterial composition (Figure 5), which was also supported by Zeng et al. (2016) and Wang et al. (2018b). It is well-documented that soil available N is an important factor in shaping the soil bacterial community. Furthermore, the findings demonstrated that the increase in TP due to P addition led to a reduction in soil bacterial richness (Figure 5), potentially because P addition exacerbates the competition between plant and soil bacteria for nutrients. The explanation of soil TP for a variation in bacterial richness was in agreement with a previous study observed in global grasslands, which suggesting that P element might be a limitation factor to bacterial richness (Prober et al., 2015).
Many studies have reported that there is no significant correlation between plant diversity and soil bacteria. For example, a study conducted in tallgrass prairie found that soil microbial diversity cannot be predicted by plant richness (Porazinska et al., 2003). In global temperate grasslands, the association between plant alpha diversity and soil bacterial community is not detected (Prober et al., 2015). Although plant diversity and bacterial diversity were affected by N and P additions, we did not find directly significant correlation (Figure 5). The results were inconsistent with previous studies, which found that soil bacterial diversity increased with enhancing plant diversity (Hiiesalu et al., 2014). The inconsistent results were probably attributed to asynchronous changes in plant and microbial diversity resulting from nutrient additions (Bardgett et al., 2005). The results also supported the view that plant diversity and soil bacterial diversity was largely uncorrelated.
Soil properties directly acted on soil bacterial diversity and community composition, while plant diversity had no effect on bacterial diversity, following the short-term N and P additions in the alpine meadow. N addition directly caused an increase in soil , thus affecting the plant diversity. It indirectly modified the bacterial diversity and composition through altering available nutrients and pH, while P addition indirectly acted on the community by increasing P availability. Moreover, soil bacterial diversity was independent with plant diversity following nutrient additions. These results highlighted that the impacts of short-term nutrient addition on soil bacterial community were mediated by soil properties rather than plant diversity in alpine meadow ecosystems. Future studies should focus on the response of belowground biodiversity in alpine meadows under environmental change. This study provide evidence that soil microbial diversity responds to anthropogenic disturbances, which can help to understand soil belowground ecosystem processes and guide grassland management in future N and P enrichment scenarios.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
XS and KL conceived the study, supervised the writing, and revised the manuscript. ZL led the writing. LS, XW, BH, CP, ZY, QX, XL, YD, and KL contributed sections to the manuscript. HZ and XS provided funding support. All authors read and approved the final submission.
This research was jointly supported by the National Natural Science Foundation of China (Grant Nos. 32171685 and 31971746) and the Second Batch of Forestry and Grassland Ecological Protection and Restoration Funds in 2020: Qilian Mountain National Park Qinghai Area Biodiversity Conservation Project (QHTX-2021-009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1036451/full#supplementary-material
Bardgett, R. D., Bowman, W. D., Kaufmann, R., and Schmidt, S. K. (2005). A temporal approach to linking aboveground and belowground ecology. Trends Ecol. Evol. 20, 634–641. doi: 10.1016/j.tree.2005.08.005
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, G., Tang, Y., Mo, W., Wang, Y., Li, Y., and Zhao, X. (2004). Grazing intensity alters soil respiration in an alpine meadow on the Tibetan plateau. Soil Biol. Biochem. 36, 237–243. doi: 10.1016/j.soilbio.2003.09.010
Chen, L., Xu, H., Wu, S., and Baoyin, T. (2021). Plant and soil properties mediate the response of soil microbial communities to moderate grazing in a semiarid grassland of northern China. J. Environ. Manage. 284, 112005. doi: 10.1016/j.jenvman.2021.112005
Delgado-Baquerizo, M., Trivedi, P., Trivedi, C., Eldridge, D. J., Reich, P. B., Jeffries, T. C., et al. (2017). Microbial richness and composition independently drive soil multifunctionality. Funct. Ecol. 31, 2330–2343. doi: 10.1111/1365-2435.12924
Demoling, F., Figueroa, D., and Bååth, E. (2007). Comparison of factors limiting bacterial growth in different soils. Soil Biol. Biochem. 39, 2485–2495. doi: 10.1016/j.soilbio.2007.05.002
Dijkstra, F. A., Pendall, E., Morgan, J. A., Blumenthal, D. M., Carrillo, Y., LeCain, D. R., et al. (2012). Climate change alters stoichiometry of phosphorus and nitrogen in a semiarid grassland. New Phytol. 196, 807–815. doi: 10.1111/j.1469-8137.2012.04349.x
Dimitriu, P., and Grayston, S. (2009). Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and batural boreal forest soils. Microb. Ecol. 59, 563–573. doi: 10.1007/s00248-009-9590-0
Dong, J., Wang, S., Niu, H., Cui, X., Li, L., Pang, Z., et al. (2020). Responses of soil microbes and their interactions with plant community after nitrogen and phosphorus addition in a Tibetan alpine steppe. J Soil Sediment 20, 2236–2247. doi: 10.1007/s11368-020-02586-3
Gai, J. P., Christie, P., Cai, X. B., Fan, J. Q., Zhang, J. L., Feng, G., et al. (2009). Occurrence and distribution of arbuscular mycorrhizal fungal species in three types of grassland community of the Tibetan Plateau. Ecol. Res. 24, 1345–1350. doi: 10.1007/s11284-009-0618-1
Geisseler, D., and Scow, K. M. (2014). Long-term effects of mineral fertilizers on soil microorganisms – a review. Soil Biol. Biochem. 75, 54–63. doi: 10.1016/j.soilbio.2014.03.023
Han, B., Li, J., Liu, K., Zhang, H., Wei, X., and Shao, X. (2021). Variations in soil properties rather than functional gene abundances dominate soil phosphorus dynamics under short-term nitrogen input. Plant Soil. 469, 227–241. doi: 10.1007/s11104-021-05143-0
He, D., Xiang, X., He, J.-S., Wang, C., Cao, G., Adams, J., et al. (2016). Composition of the soil fungal community is more sensitive to phosphorus than nitrogen addition in the alpine meadow on the Qinghai-Tibetan Plateau. Biol Fert Soils. 52, 1059–1072. doi: 10.1007/s00374-016-1142-4
Hiiesalu, I., Pärtel, M., Davison, J., Gerhold, P., Metsis, M., Moora, M., et al. (2014). Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol. 203, 233–244. doi: 10.1111/nph.12765
Hooper, D. U., Adair, E. C., Cardinale, B. J., Byrnes, J. E. K., Hungate, B. A., Matulich, K. L., et al. (2012). A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature. 486, 105–108. doi: 10.1038/nature11118
Husson, O. (2013). Redox potential (Eh) and pH as drivers of soil/plant/microorganism systems: a transdisciplinary overview pointing to integrative opportunities for agronomy. Plant Soil. 362, 389–417. doi: 10.1007/s11104-012-1429-7
Isbell, F., Reich, P. B., Tilman, D., Hobbie, S. E., Polasky, S., and Binder, S. (2013). Nutrient enrichment, biodiversity loss, and consequent declines in ecosystem productivity. Proc. Natl. Acad. Sci. U.S.A. 110, 11911–11916. doi: 10.1073/pnas.1310880110
Jiang, J., Wang, Y.-P., Liu, F., Du, Y., Zhuang, W., Chang, Z., et al. (2021). Antagonistic and additive interactions dominate the responses of belowground carbon-cycling processes to nitrogen and phosphorus additions. Soil Biol. Biochem. 156, 108216. doi: 10.1016/j.soilbio.2021.108216
Jiang, J., Wang, Y.-P., Yang, Y., Yu, M., Wang, C., and Yan, J. (2019). Interactive effects of nitrogen and phosphorus additions on plant growth vary with ecosystem type. Plant Soil. 440, 523–537. doi: 10.1007/s11104-019-04119-5
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066
Klironomos, J., Zobel, M., Tibbett, M., Stock, W. D., Rillig, M. C., Parrent, J. L., et al. (2011). Forces that structure plant communities: quantifying the importance of the mycorrhizal symbiosis. New Phytol. 189, 366–370. doi: 10.1111/j.1469-8137.2010.03550.x
Lai, J., Zou, Y., Zhang, J., and Peres-Neto, P. R. (2022). Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol. 13, 782–788. doi: 10.1111/2041-210X.13800
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microb 75, 5111–5120. doi: 10.1128/AEM.00335-09
Li, J., Yang, C., Liu, X., Ji, H., and Shao, X. (2020). Soil aggregate size influences the impact of inorganic nitrogen deposition on soil nitrification in an alpine meadow of the Qinghai–Tibet Plateau. PeerJ 8, e8230. doi: 10.7717/peerj.8230
Ling, N., Chen, D., Guo, H., Wei, J., Bai, Y., Shen, Q., et al. (2017). Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 292, 25–33. doi: 10.1016/j.geoderma.2017.01.013
Long, X.-E., Yao, H., Huang, Y., Wei, W., and Zhu, Y.-G. (2018). Phosphate levels influence the utilisation of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Biol. Biochem. 118, 103–114. doi: 10.1016/j.soilbio.2017.12.014
Luo, R., Fan, J., Wang, W., Luo, J., Kuzyakov, Y., He, J.-S., et al. (2019). Nitrogen and phosphorus enrichment accelerates soil organic carbon loss in alpine grassland on the Qinghai-Tibetan Plateau. Sci. Total. Environ. 650, 303–312. doi: 10.1016/j.scitotenv.2018.09.038
Ma, W., Jiang, S., Assemien, F., Qin, M., Ma, B., Xie, Z., et al. (2016). Response of microbial functional groups involved in soil N cycle to N, P and NP fertilization in Tibetan alpine meadows. Soil Biol. Biochem. 101, 195–206. doi: 10.1016/j.soilbio.2016.07.023
Martin, M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17. doi: 10.14806/ej.17.1.200
Müller, T., Walter, B., Wirtz, A., and Burkovski, A. (2006). Ammonium toxicity in bacteria. Curr. Microbiol. 52, 400–406. doi: 10.1007/s00284-005-0370-x
Nie, Y., Wang, M., Zhang, W., Ni, Z., Hashidoko, Y., and Shen, W. (2018). Ammonium nitrogen content is a dominant predictor of bacterial community composition in an acidic forest soil with exogenous nitrogen enrichment. Sci. Total Environ. doi: 10.1016/j.scitotenv.2017.12.142
Peñuelas, J., Poulter, B., Sardans, J., Ciais, P., van der Velde, M., Bopp, L., et al. (2013). Human-induced nitrogen–phosphorus imbalances alter natural and managed ecosystems across the globe. Nat. Commun. 4, 2934. doi: 10.1038/ncomms3934
Porazinska, D., Bardgett, R., Postma-Blaauw, M. B., Hunt, H., Parsons, A., Seastedt, T., et al. (2003). Relationships at the aboveground-belowground interface: Plants, soil biota, and soil processes. Ecol. Monogr. 73. doi: 10.1890/0012-9615(2003)073[0377:RATAIP]2.0.CO
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Prober, S. M., Leff, J. W., Bates, S. T., Borer, E. T., Firn, J., Harpole, W. S., et al. (2015). Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 18, 85–95. doi: 10.1111/ele.12381
R Core Team. (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Ramirez, K. S., Craine, J. M., and Fierer, N. (2010). Nitrogen fertilization inhibits soil microbial respiration regardless of the form of nitrogen applied. Soil Biol. Biochemist. 42, 2336–2338. doi: 10.1016/j.soilbio.2010.08.032
Ramirez, K. S., Craine, J. M., and Fierer, N. (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Global Change Biol 18, 1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x
Raynaud, T., Pivato, B., Siol, M., Spor, A., and Blouin, M. (2021). Soil microbes drive the effect of plant species and genotypic diversity interaction on productivity. Plant Soil 467, 165–180. doi: 10.1007/s11104-021-05071-z
Roem, W. J., Klees, H., and Berendse, F. (2002). Effects of nutrient addition and acidification on plant species diversity and seed germination in heathland. J. Appl. Ecol. 39, 937–948. doi: 10.1046/j.1365-2664.2002.00768.x
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Song, B., Li, Y., Yang, L., Shi, H., Li, L., Bai, W., et al. (2022). Soil acidification under long-term N addition decreases the diversity of soil bacteria and fungi and changes their community composition in a semiarid grassland. Microb. Ecol. doi: 10.1007/s00248-021-01954-x
Spohn, M., and Schleuss, P.-M. (2019). Addition of inorganic phosphorus to soil leads to desorption of organic compounds and thus to increased soil respiration. Soil Biol. Biochem. 130, 220–226. doi: 10.1016/j.soilbio.2018.12.018
Sugiyama, S., Zabed, H. M., and Okubo, A. (2008). Relationships between soil microbial diversity and plant community structure in seminatural grasslands. Grassl. Sci. 54, 117–124. doi: 10.1111/j.1744-697X.2008.00113.x
Sui, P., Tian, P., Wang, Z., Lian, H., Yang, Y., Ma, Z., et al. (2022). Soil properties and microbial communities of spring maize filed in response to tillage with straw incorporation and nitrogen fertilization in northeast China. PeerJ. 10, e13462. doi: 10.7717/peerj.13462
Turner, B. L., and Joseph Wright, S. (2014). The response of microbial biomass and hydrolytic enzymes to a decade of nitrogen, phosphorus, and potassium addition in a lowland tropical rain forest. Biogeochemistry. 117, 115–130. doi: 10.1007/s10533-013-9848-y
Vitousek, P. M., Aber, J. D., Howarth, R. W., Likens, G. E., Matson, P. A., Schindler, D. W., et al. (1997). Human alteration of the global nitrogencycle: sources and consequences. Ecol. Appl. 7, 737–750. doi: 10.1890/1051-0761(1997)007[0737:HAOTGN]2.0.CO
Wang, H., Liu, S., Zhang, X., Mao, Q., Li, X., You, Y., et al. (2018a). Nitrogen addition reduces soil bacterial richness, while phosphorus addition alters community composition in an old-growth N-rich tropical forest in southern China. Soil Biol. Biochem. 127, 22–30. doi: 10.1016/j.soilbio.2018.08.022
Wang, Q., Jiang, X., Guan, D., Wei, D., Zhao, B., Ma, M., et al. (2018b). Long-term fertilization changes bacterial diversity and bacterial communities in the maize rhizosphere of Chinese Mollisols. Agric. Ecosyst. Environ,. Appl. Soil Ecol. 125, 88–96. doi: 10.1016/j.apsoil.2017.12.007
Wang, Q., Wang, C., Yu, W., Turak, A., Chen, D., Huang, Y., et al. (2018c). Effects of nitrogen and phosphorus Inputs on soil bacterial abundance, diversity, and community composition in chinese fir plantations. Front. Microbiol. 9, 1543. doi: 10.3389/fmicb.2018.01543
Wei, C., Yu, Q., Bai, E., Lü, X., Li, Q., Xia, J., et al. (2013). Nitrogen deposition weakens plant–microbe interactions in grassland ecosystems. Global Change Biol. 19, 3688–3697. doi: 10.1111/gcb.12348
Yan, G., Xing, Y., Wang, J., Zhang, Z., Xu, L., Han, S., et al. (2018). Effects of winter snowpack and nitrogen addition on the soil microbial community in a temperate forest in northeastern China. Ecol. Indic. 93, 602–611. doi: 10.1016/j.ecolind.2018.05.048
Yan, Y., Sun, X., Sun, F., Zhao, Y., Sun, W., Guo, J., et al. (2022). Sensitivity of soil fungal and bacterial community compositions to nitrogen and phosphorus additions in a temperate meadow. Plant Soil 471, 477–490. doi: 10.1007/s11104-021-05237-9
Yang, X., Long, Y., Sarkar, B., Li, Y., Lü, G., Ali, A., et al. (2021). Influence of soil microorganisms and physicochemical properties on plant diversity in an arid desert of Western China. J. Forestry Res. 32, 2645–2659. doi: 10.1007/s11676-021-01292-1
Yang, Z., van Ruijven, J., and Du, G. (2011). The effects of long-term fertilization on the temporal stability of alpine meadow communities. Plant Soil. 345, 315–324. doi: 10.1007/s11104-011-0784-0
Yuan, Z. Y., and Chen, H. Y. H. (2015). Decoupling of nitrogen and phosphorus in terrestrial plants associated with global changes. Nat Clim Change. 5, 465–469. doi: 10.1038/nclimate2549
Zeng, J., Liu, X., Song, L., Lin, X., Zhang, H., Shen, C., et al. (2016). Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 92, 41–49. doi: 10.1016/j.soilbio.2015.09.018
Zhang, N. N., Sun, G., Liang, J., Wang, E. T., Shi, C. G., He, J., et al. (2018). Response of ammonium oxidizers to the application of nitrogen fertilizer in an alpine meadow on the Qinghai-Tibetan Plateau. Agric. Ecosyst. Environ,. Appl. Soil Ecol. 124, 266–274. doi: 10.1016/j.apsoil.2017.11.018
Zhang, S., Li, Q., Lü, Y., Zhang, X., and Liang, W. (2013). Contributions of soil biota to C sequestration varied with aggregate fractions under different tillage systems. Soil Biol. Biochem. 62, 147–156. doi: 10.1016/j.soilbio.2013.03.023
Zhang, X., Wei, H., Chen, Q., and Han, X. (2014). The counteractive effects of nitrogen addition and watering on soil bacterial communities in a steppe ecosystem. Soil Biol. Biochem. 72, 26–34. doi: 10.1016/j.soilbio.2014.01.034
Keywords: plant diversity, soil bacterial diversity, soil properties, N and P additions, alpine meadow
Citation: Lin Z, Shi L, Wei X, Han B, Peng C, Yao Z, Xiao Q, Lu X, Deng Y, Zhou H, Liu K and Shao X (2022) Soil properties rather than plant diversity mediate the response of soil bacterial community to N and P additions in an alpine meadow. Front. Microbiol. 13:1036451. doi: 10.3389/fmicb.2022.1036451
Received: 07 September 2022; Accepted: 17 October 2022;
Published: 03 November 2022.
Edited by:
Kai Xue, University of Chinese Academy of Sciences, ChinaReviewed by:
Yu Liu, Northwest A&F University, ChinaCopyright © 2022 Lin, Shi, Wei, Han, Peng, Yao, Xiao, Lu, Deng, Zhou, Liu and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinqing Shao, c2hhb3hpbnFpbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.