
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 December 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1035602
This article is part of the Research Topic Beneficial Microbe-Plant Interactions Under Biotic/Abiotic Stress Conditions View all 23 articles
 Asharani Patel1
Asharani Patel1 Kuleshwar Prasad Sahu1
Kuleshwar Prasad Sahu1 Sahil Mehta1
Sahil Mehta1 Alexander Balamurugan1
Alexander Balamurugan1 Mukesh Kumar1
Mukesh Kumar1 Neelam Sheoran1
Neelam Sheoran1 Shanu Kumar1
Shanu Kumar1 Charishma Krishnappa1
Charishma Krishnappa1 Mushineni Ashajyothi1
Mushineni Ashajyothi1 Aditi Kundu1
Aditi Kundu1 Tushar Goyal1
Tushar Goyal1 Prabhakaran Narayanasamy2
Prabhakaran Narayanasamy2 Aundy Kumar1*†
Aundy Kumar1*†Genetic and functional characteristics of rice leaf endophytic actinobacterial member, Microbacterium are described. Morphotyping, multilocus sequence analysis and transmission electron microscopy indicated the species identity of the endophytic bacterium, OsEnb-ALM-D18, as Microbacterium testaceum. The endophytic Microbacterium showed probiotic solubilization of plant nutrients/minerals, produced hydrolytic enzyme/phytohormones, and showed endophytism in rice seedlings. Further, the endophytic colonization by M. testaceum OsEnb-ALM-D18 was confirmed using reporter gene coding for green fluorescence protein. Microbacterium OsEnb-ALM-D18 showed volatilome-mediated antibiosis (95.5% mycelial inhibition) on Magnaporthe oryzae. Chemical profiling of M. testaceum OsEnb-ALM-D18 volatilome revealed the abundance of 9-Octadecenoic acid, Hexadecanoic acid, 4-Methyl-2-pentanol, and 2,5-Dihydro-thiophene. Upon endobacterization of rice seedlings, M. testaceum altered shoot and root phenotype suggestive of activated defense. Over 80.0% blast disease severity reduction was observed on the susceptible rice cultivar Pusa Basmati-1 upon foliar spray with M. testaceum. qPCR-based gene expression analysis showed induction of OsCERK1, OsPAD4, OsNPR1.3, and OsFMO1 suggestive of endophytic immunocompetence against blast disease. Moreover, M. testaceum OsEnb-ALM-D18 conferred immunocompetence, and antifungal antibiosis can be the future integrated blast management strategy.
Rice (Oryza sativa L.; Family Poaceae) is the cereal staple that serves as the primary source of sustenance for more than half of the world’s population (Shah et al., 2021). Rice cultivation is constrained by several biotic and abiotic stresses causing yield and economic losses worldwide (Fahad et al., 2019; Savary et al., 2019). Among the biotic stresses, diseases incited by phytopathogenic fungi and bacteria have resulted in a significant production loss impacting the global food supply chain (Miah et al., 2013; Godfray et al., 2016; Jiehua et al., 2019; Neupane and Bhusal, 2021). Notably, rice blast disease incited by the hemibiotrophic ascomycete Magnaporthe is responsible for pre-harvest losses of about 10–30% (Rossman et al., 1990; Howard and Valent, 1996; Shan et al., 2013; Kirtphaiboon et al., 2021). The disease symptoms are elliptical-necrotic lesions on the leaf with diamond-shaped gray centers and a reddish-brown margin which is noticed on the vegetative leaf and reproductive panicle (Asibi et al., 2019). Traditionally, the blast mitigation strategies depend on the deployment of host resistance and the application of fungicide (Mehta et al., 2021). However, both approaches are inadequate to sustain rice production. The use of fungicides led to the persistence of harmful residues on grain, which prompted several nations to reject shipments of rice and resulted in financial losses for both traders and farmers (Zhang, 2007). The host resistance is not durable and often ineffective in many regions owing to the high genetic and pathogenic diversity of Magnaporthe (Hubballi et al., 2022). As a result, developing effective and sustainable strategies to combat blast disease assumes significance. In recent years, there is a demand for eco-friendly biocontrol methods for rice blast disease management (Skamnioti and Gurr, 2009; Chaiharn et al., 2020; Sahu et al., 2021a).
Biological control system exploiting naturally occurring endophytic microbes is one of the choices due to their innate ability to interact with the susceptible host niches and the suppression of pathogenic agents by direct antibiosis and indirect defense activation (Liu et al., 2007; Mano and Morisaki, 2008). In addition to probiotic effects on host plants, several endophytic bacteria have been reported to induce systemic response against pathogens in many dicots and monocot crops including rice (Sheoran et al., 2015, 2016; Agisha et al., 2017; Hirakue and Sugiyama, 2018; Li et al., 2019; Sarkar et al., 2019; Ashajyothi et al., 2020; Matsumoto et al., 2021; Patel et al., 2021). Recently, we published the antagonistic and host defense elicitation potential of rice-endophytic flavobacterial isolate Chryseobacterium endophyticum against rice blast (Kumar et al., 2021a). It is known that plant ecological functions including its defense against the pathogen are carried out by the synergistic effects of the plant microbiome, there are ample chances that other rice endophytic bacteria can have blasticidal potential. Considering this point, we focused on isolating other endophytes from rice foliage and evaluated their antifungal potential against Magnaporthe oryzae.
Among the rice endophytic bacterial communities, the genus Microbacterium belonging to the class, Actinobacteria is characterized by small, irregular, short, slender, rod-shaped Gram-positive bacterium (Komagata et al., 1964; Yamada and Komagata, 1972; Collins et al., 1983; Takeuchi and Hatano, 1998). In the past, a series of research publications have highlighted the Microbacterium as a promising microbial inoculant in agriculture that displays biostimulant activity on crops and antibiosis against pathogens. Microbacterium is also prolific for the production of volatile organic compounds that contributed to growth promotion and development (Barnett et al., 2006; Cordovez et al., 2018). Moreover, Microbacterium sp. isolated from rice as well as other plants has been used as an effective biocontrol agent against various plant pathogens by the production of a pectinolytic enzyme, antimicrobial substances, insoluble and soluble exopolysaccharides or degradation of toxic substances (Ueno et al., 1983; Matsuyama et al., 1999; Dikin et al., 2007; Cottyn et al., 2009; Silva et al., 2012; Walitang et al., 2017). The interaction of three different taxonomic groups of bacteria such as Pantoea, Exiguobacterium, and Microbacterium could reduce the disease severity caused by Rhizoctonia solani AG-8 (Barnett et al., 2006). Along with Bacillus amyloliquefaciens, Microbacterium reduced Fusarium verticillioides and the mycotoxin fumonisin B1 and B2 in maize grains (Sartori et al., 2012). Before the present study, leaf epiphytic Microbacterium testaceum was reported to elicit defense responses leading to blast disease suppression upon phyllo bacterization of rice cultivar Pusa Basmati-1 (Sahu et al., 2021b). However, published literature on the biocontrol potential of M. testaceum against blast disease is scanty. Therefore, an in-depth genetic and functional characterization of rice endophytic M. testaceum was carried out to develop a biostimulant for sustainable rice production. We further attempted to elucidate the mode of action of M. testaceum so that it can be incorporated into eco-friendly rice blast disease management options in the future.
The bacterial strain OsEnb-ALM-D18 used in the present study was isolated from the leaf endosphere of rice (O. sativa L.) cultivar HPR2143 grown at Almora (Uttarakhand) as a natural endophyte (Kumar, 2021). 16S rRNA gene sequence was used for identification as M. testaceum and the sequence was submitted to NCBI GenBank with accession number MN889362.1 For routine lab work, culture was preserved at −80°C as glycerol stock was retrieved by streaking on plates of nutrient agar medium [NA, gL–1 Beef extract 3.0; Peptone 5.0; NaCl 5.0; Agar 15.0; pH 7.0 ± 0.2] and incubated at 28°C for 48 h.
Using the negative-staining method (Tranum-Jensen, 1988), the bacterium M. testaceum OsEnb-ALM-D18 (hereinafter M. testaceum D18) was visualized and imaged in the transmission electron microscope (TEM) at magnifications of more than 60K X (Joel, Tokyo, Japan).
Briefly, M. testaceum D18 was grown in nutrient broth with constant agitation (100 rpm). Bacterial culture at mid-log phase (1.5 mL; 1.0 ODA600 nm) was centrifuged at 12,000 rpm for 3.0 min. Pelleted bacterial cells were washed with sterile saline and used for extracting genomic DNA. CTAB-method with slight modification was used to isolate bacterial genomic DNA (Sheoran et al., 2015; Munjal et al., 2016; Eke et al., 2019; Sahu et al., 2021b). Before PCR amplification, the integrity, purity, and yield of genomic DNA were tested spectrophotometrically (Biophotometer, Eppendorf, Hamburg, Germany) and electrophoretically. Finally, a 100 ng μL–1 concentration of the genomic DNA was prepared and used as an amplification template.
A total of eight Microbacterium-specific housekeeping genes were taken for this study. The primers were designed using Primer3plus, and the same were synthesized and used.2 Supplementary Table 1 lists the oligo primers that were used in the PCR amplification. PCR reaction mixture contains-Promega PCR buffer 1X; MgCl2 1.5 mM; dNTPs 200 mM; Taq polymerase 1.0 U; forward and reverse primers 10-pmol each; genomic DNA 100 ng; and mQ water to bring the total volume to 50 μL. The following temperature conditions parameters were used for PCR amplification in a thermal cycler (MasterCycler ProS, Eppendorf, Hamburg, Germany): initial denaturation at 95°C for 5 min, followed by 35 cycles of 94°C for 1.0 min; annealing temperature as per Supplementary Table 1 for 1.0 min; extension at 72°C for 1.0 min, followed by one-cycle of 72°C for 5.0 min; and final cooling at 4.0°C.
Then, the respective PCR products were resolved in 1.0% agarose gel followed by excision and elution using a gel elution kit according to the manufacturer’s instructions (Promega Corporation, Madison, WI, USA). The PCR products were subjected to bidirectional sequencing using Sanger’s dideoxy chain termination technique to obtain the maximum gene sequence coverage. Contigs were assembled with DNA-Baser v5,3 followed by curation with Finch TV,4 and BLAST analysis was used to compare them to GenBank sequences.5 All the gene sequences were assigned accession numbers and deposited in the NCBI GenBank database.
Using Mega X with 1000 bootstrap replications the Maximum Likelihood algorithm and Hasegawa-Kishino-Yano model were used to perform multi-gene sequence-based phylogenetic analysis (Felsenstein, 1985; Hasegawa et al., 1985; Kumar et al., 2018). The gene sequences of Microbacterium sp. isolated from the diverse natural environments were retrieved from NCBI databases (see text footnote 1). CLC Sequence Viewer was used to normalize the nucleotide sequences to a constant length of 767 to 959 base positions (CLC Sequence Viewer 8).
A stable green fluorescent protein (GFP) gene was inserted into the genome of the M. testaceum D18 strain by the tri-parental mating method. Before the transformation, spontaneous rifamycin-resistant M. testaceum D18 was selected by plating on to nutrient agar medium amended with rifamycin (50 μg mL–1) and incubated for 48–72 h at 28°C. The rifamycin-resistant colony was subsequently used in the transformation experiment essentially to counter-select against Escherichia coli XL1 Blue in the conjugation experiment. We used a Tn7-based GFP construct “pBKminiTn7gfp2Gm10” maintained in E. coli XL1 Blue (Gentamycin 20 g mL–1) and a helper plasmid “pUXBF13Amp100” maintained in E. coli XL1 Blue (Ampicillin 100 g mL–1) to tag the bacteria. The tri-parental-mating was conducted to insert the GFP gene into the bacterial genome (Sheoran et al., 2015). The transformed colonies were selected on NA plates amended with Gentamycin (20 μg mL–1) and Rifamycin (50 μg mL–1) incubated at 28°C for 48–72 h.
Putatively transformed colonies that appeared on the antibiotic-amended selection plates were sub-cultured on fresh NA plates amended with the same antibiotics. The colonies that appeared were then subjected to PCR confirmation for the insertion using GFP-specific forward and reverse primers (Ashajyothi et al., 2020). Briefly, primer pairs gfp-Rt-F-5′GGCCGATGCAAAGTGCCGATAAA3′ and gfp-Rt-R-5′AGGGCGAAGAATCTCGTGCTTTCA3′ were used in PCR reaction using GoTaq PCR kit of Promega (GoTaq Buffer–1X, DMSO–6%, dNTPs–200 mM, MgCl2–1.5 mM, forward/reverse primers–10 picomoles each, Taq polymerase–1 U) at an initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 53°C for 30 s and extension at 72°C for 30 s followed by a final extension at 72°C for 5 min. In a 2% agarose gel, a 142-bp PCR amplicon was observed and documented (Quantity One Image Analysis system, Bio-Rad, CA, USA). The colonies that yielded expected amplicons in PCR-reaction were image analyzed under a confocal microscope [confocal laser scanning microscopy (CLSM) DM6000, Leica Microsystems, Wetzlar, Germany] for checking the stable expression of the green fluorescent protein. Further, the transformed bacterial strain with stable expression of gfp was preserved as glycerol stock for downstream activities and designated as M. testaceum D18::gfp.
Endophytic colonization was studied using genetically tagged M. testaceum D18::gfp in the rice cv. Pusa Basmati-1. M. testaceum D18::gfp was cultured on double antibiotic-amended NA plates as described earlier. Surface-sterilized seeds of rice were soaked in a bacterial suspension of 108 CFU mL–1 (1.0 OD at A600) for 24 h and allowed to germinate and grow in Petri dishes incubated under greenhouse conditions (RH > 90% and temperature 28–30°C) for 7-days. Seeds soaked in sterile double distilled water served as mock. The saplings were, then, transplanted into small pots with sterilized farm soil and grown under greenhouse conditions.
Microbacterium testaceum D18::gfp inoculated 1-month-old rice plantlets were taken, and thin slices of various plant parts were fixed in p-formaldehyde (4.0%) for 12 h at 4°C. Leaf, stem, and root cross-sections were scanned, and imaged at multiple sites using CLSM (DM6000, Leica Microsystems, Wetzlar, Germany). The images were processed and examined to determine bacterial localization inside the plant tissue.
PCR-based test of the endophytic bacteria M. testaceum D18::gfp was performed using gfp-specific primers (Rt F-5′GGCCGATGCAAAGTGCCGATAAA3′ and Rt R-5′ AGGGCGAAGAATCTCGTGCTTTCA3′). For the tissue colonization study, the root, shoot, and leaves of the treated plants were crushed separately and 1 mL water extract of each tissue was centrifuged to get the pellet. The resulting pellet was used for the isolation of total genomic DNA using the protocol of bacterial DNA isolation as described earlier. The composition of the reaction mixture, as well as reaction conditions, was the same as that used for the selection of gfp transformants. The PCR amplicon was separated on agarose gel amended with ethidium bromide and visualized with a UV trans-illuminator (BioRad Laboratories, CA, USA).
Microbacterium testaceum D18 was investigated for a range of plant probiotic characteristics. All experiments were done in triplicates with 10 μL bacterial suspension in sterile water as treatment and with appropriate control (10 μL sterile water without bacteria). Phosphate solubilization activity was accessed using an agar medium containing tricalcium phosphate (CaP; 3 gL–1), as the method described by Chen et al. (2006). The method described by Aleksandrov et al. (1967) was used to determine the potassium (K) solubilizing activity by using an agar medium containing inorganic potassium source Potassium Aluminosilicate (AlKO6Si2). Similarly, the solubilization of mineral zinc was detected in a modified Pikovskaya agar medium according to Pikovskaya (1948) containing insoluble zinc sourced from ZnO. The ability to produce siderophore was assayed by adopting the method described by Louden et al. (2011) using CAS agar medium as substrate.
The technique described by Patten and Glick (1996) was used to test indole-3-acetic acid (IAA) production using L-tryptophan in DF salt minimal medium and Salkowski et al.’s (1996) reagent. The ability to produce ammonia was assayed in Nutrient Broth after adding Nessler’s reagent as described by Zhou and Boyd (2016).
Microbacterium testaceum D18 was also analyzed for its ability to produce a range of defense-related enzymes under in vitro conditions. By using the agar well diffusion method, the bacterium M. testaceum D18 was inoculated on casein agar, carboxymethyl cellulose (CMC) agar, starch agar, pectin agar, and xylan agar medium to evaluate the production of protease, cellulase, amylase, pectinase, and xylanase, respectively (Ruiz et al., 2009).
Briefly, in the nutrient agar medium, 1.0% of each substrate was used to make substrate media plates. On the solidified media, a well with a diameter of 5.0-mm was made with a cork borer, and the bacterial cell suspension (10 μl of 0.5 ODA600) was added. The plates were then incubated for 48 h at 28°C. A zone of clearing around the wells was measured, and a minimum diameter of 15 mm was considered a positive result. For analysis of enzyme activity such as pectinase and amylase, plates were flooded with iodine–potassium iodide solution for 15 min and then treated for 10 min with sodium chloride (1.0 mol L–1) for de-staining.
By flooding the plates with Congo Red solution (1.0%) for 15 min and then de-staining with sodium chloride (1 mol L–1) for 10 min, cellulase and xylanase enzyme activity were identified. In casein agar, the formation of clear zones around the wells was utilized to measure the protease production. To assess the production of chitinase enzyme nutrient agar media amended with 1% (v/v) colloidal chitin was used and inoculated with bacterial suspension (5 μL) and incubated for 1 week. Later, the plates were flooded with Congo red (1%). The formation of an orange zone surrounding the colony was marked as positive.
To assess the antagonistic activity of secretory metabolites released by M. testaceum D18 against the mycelial growth of Magnaporthe oryzae (hereafter M. oryzae), a dual culture confrontation assay was conducted in small Petri plates (40.0 mm). A modified TSA-PDA medium [Tryptic soy agar (SRL, Mumbai, India) Potato dextrose agar (HiMedia Laboratories, Mumbai, India)] was poured onto Petri plates and freshly grown mycelial disk (0.5 cm diameter) of the fungus was inoculated at one corner of the plate. The bacterial inoculum on the agar surface was streaked parallel to the mycelial disk and incubated at 28°C for 5–7 days. The mycelial growth was measured and compared against the mock plate to calculate the % inhibition zone. The experiment was performed twice with three replications each. For calculating the mycelial inhibition, the following formula was used.
where, C = Colony diameter in control, and T = Colony diameter in treatment.
In vitro antagonistic activity of bacterial volatile organic compounds released by M. testaceum D18 isolate was investigated on M. oryzae-1637 using the Sheoran et al. (2015) methodology. In the center of a PDA plate, a mycelial disk from a freshly grown fungal culture was placed. Similarly, another Petri plate poured with a Tryptic soy agar plate (HiMedia Laoratories, Mumbai, India) was inoculated with 20 μL of 108 CFU mL–1 mid-log phase bacterial culture. To allow the fungus to be directly exposed to bacterial volatile, the lids of both plates were separated and the inoculated plates were placed face-to-face and fastened with parafilm, Petri seal tape, and klin film so that no volatile should escape from inside during the experiment. Three replications per treatment with proper control were incubated at 28°C for 5–7 days. Further, colony diameter was measured and % mycelial inhibition was calculated using the same formula.
Under in vitro conditions, the volatiles generated by M. testaceum D18 were able to block fungal growth. This observation led to the investigation of the bacterial isolate’s volatile profile. Bacterial volatile organic compounds were extracted in HPLC grade hexane using the solvent extraction method (Joana Gil-Chávez et al., 2013) from a 72-h-old bacterial culture for this study. Hexane, which was used as a solvent and dissolved with bacterial organic compounds, was also analyzed using gas chromatography-mass spectroscopic (GC-MS).
An HP-5MS column (60 m 0.25 mm; /0.25 m, Thermo Co., USA) coupled to a triple-axis mass spectrometer (ThermoFisher Scientific, MA, USA) was used for GC-MS analysis. The injection volume was kept at 1.0 L with flow mode in split control. The flow rate of helium carrier gas was set at 1.0 mL min–1, with a head pressure of 10 psi. GC-MS was performed with the following condition; the oven temperature was maintained at 40°C for the first minute. The temperature was then raised to 120°C at an increasing rate of 3°C min–1 and then maintained for 2 min.
Finally, the temperature rose by 4°C min–1, reaching its maximum at 280°C. The movie lasted a total of 65 min. Electron ionization 70 eV, Ion source 180°C, full scan mode (50–550 mass units), E.M voltage 1376, transfer line temperature 280°C, and solvent delay 3.0 min, were the MS acquisition parameters. At a scanning time of 1.0 s and in the mass range of 50–550 AMU, the ionization energy was 70 eV. By comparing mass spectra, compounds were identified. The Mass Spectra Library of the National Institute of Standards and Technologies (NIST) was utilized as a reference for determining the fundamental components.
Tobacco (Nicotiana tabacum) plants were grown in aclimate-controlled greenhouse. Two to 3-month-old plants with fully expanded 7–8 leaves were used in the experiment. Pre-inoculation conditioning was given to the plants under greenhouse conditions at a temperature of 20°C and 50–60% relative humidity and a 12 h light/dark period for 2.0-days. Fully expanded leaves that were attached to the plants were used in all experiments.
Most of the phytopathogenic bacteria are known to cause hypersensitive reactions (HR) on tobacco leaves which are visible as water-soaked necrotic lesions upon leaf infiltration (Klement, 1963). Similarly, most non-pathogenic bacteria will not cause any hypersensitive reaction. M. testaceum D18 on Nutrient agar and suspension of 1 × 108 CFU/mL (1.0 OD at A600 nm) were prepared in sterile distilled water. It was infiltrated on potted N. tabacum leaves using a sterile hypodermal syringe. The plants were kept for incubation at 25–30°C under glasshouse conditions (12 h of dark/light photoperiods). Likewise, for negative control sterile double distilled water was infiltrated on leaves. Leaf infiltrated with plant pathogenic bacterium, Ralstonia solanacearum, served as a positive control. Induced responses of tobacco leaves were recorded after 24 h post-inoculation (hpi).
Firstly, the seed priming assay was conducted using M. testaceum D18 to test the effect on germination %, root/shoot length, fresh weight, and dry weight in the rice, cv. Pusa Basmati-1 and BPT5404. The bacterium M. testaceum D18 was cultured on NA mixed with the two antibiotics described earlier. Rice seeds were surface sterilized and soaked in sterile distilled water for 24 h. Bacterial suspension of cell concentrations at 108 and 107 cells mL–1 were prepared using mid-log phase culture and seeds were allowed to grow on it for 7 days under green-house conditions (RH > 90% and temperature 28–30°C). The observations were taken for germination %, shoot length, root length, number of roots, fresh weight, and dry weight at 7 days post-inoculation. For mock, seeds were incubated only on sterile deionized water. The experiment was performed in three replications with 20 seeds each and repeated twice.
Blast susceptible varieties, Pusa Basmati-1 and BPT5204 were seed bacterized with two cell densities 107 and 108 CFU mL–1 (0.1 OD and 1.0 OD at A600, respectively) of bacterial suspension and allowed to germinate for 5 days. After germination, the seedlings were transplanted into small two-inch pots with sterile farm soil and grown for 18–21 days till they attained 3-leaf stage under greenhouse conditions (28 ± 2°C with 85 ± 10% relative humidity and 14/12 h of light/dark cycle). M. oryzae was cultured on rice straw extract sucrose agar medium at 25°C for 7–8 days and was used for foliar inoculation in the experiment. 48 h before pathogen inoculation, a prophylactic spray of bacterial cell suspension was given above the foliage using a glass atomizer.
The conidial suspension (∼2 × 105 conidia mL–1) amended with Tween 20 (0.05%) was prepared and sprayed over the foliage of 3-week-old seedlings (Rajashekara et al., 2017). Seedlings kept for positive control were not sprayed with bacteria while plants sprayed with tricyclazole (0.1%) served as a positive control. Further, plants were incubated for conditioning in the dark for 24 h at 22°C ± 2°C with 90% RH in a climate-controlled greenhouse. After that plants were kept for 7–10 days under greenhouse conditions with a 12 h dark/light cycle, temperature 22°C ± 2°C, 90% RH with leaf wetness maintained by spraying water three times a day.
The severity of the blast was measured 7–10 days post-inoculation using a 0–5 disease rating scale as described by Mackill and Bonman (1992). The disease scores were used for the calculation of disease severity using the following formula.
Likewise, the % reduction in disease severity against control was calculated using the following formula.
where C = Disease Severity in control, and T = Disease Severity in treatment.
Following confirmation of the blast suppression capacity, qPCR tests were carried out to determine the influence of bacterial supplementation on the expression of genes implicated in rice defense pathways.
Briefly, germinated seeds of Pusa Basmati-1 as well as BPT5204, each bacterized with 107 and 108 CFU mL–1 bacterial suspension separately were sampled after 7 days post bacterization at the seedling stage. Further, the seedlings were planted in small pots with sterile soil. At the three-leaf stage, a booster dose of respective bacterial suspension (same concentration as seed treatment) was given and sampling was done after 24 h (before challenge inoculation by a pathogen). Samples were quickly snap-frozen in liquid nitrogen to cease all cellular metabolic activity and then kept at −80°C until needed. The SV Tool RNA isolation system was used to isolate total RNA according to the manufacturer’s instructions (Promega Corporation, Madison, WI, USA). NanoDrop 2000 (Thermo Scientific, MA, USA) and agarose gel electrophoresis were used to determine the quantity and quality of RNA.
Rice genes associated with defense-related pathways such as OsCERK1 (Kouzai et al., 2014), OsFMO1 (Koch et al., 2006; Mishina and Zeier, 2006), OsCEBiP (Akamatsu et al., 2013), OsNPR1 (Sugano et al., 2010), OsPAD4 (Ke et al., 2014) and OsEDS1 (Ke et al., 2019), playing a key role in rice innate immunity were selected (Supplementary Table 2). qPCR primers for the above defense genes are given in Supplementary Table 3. qPCR was performed using GoTaq® 1-Step RT-qPCR System (Promega Corporation, Madison, WI, USA) in Real-Time Thermal Cycler (Light Cycler 96, Roche Life Science, Penzberg, Germany).
The following were qPCR reaction conditions; one step of reverse transcription at 37°C for 15 min followed by reverse transcriptase inactivation step of 95°C for 10 min followed by 30 cycles of 95°C for 10 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s followed by three-step melting of 95°C for 10 s, 63°C for 60 s, and 97°C for 1.0 s and then final cooling at 37°C for 30 s. Later, cyclic threshold data points were analyzed for the determination of gene expression relative to reference housekeeping OsActin gene using the software LightCycler®96 Roche. The mean Ct values were considered for the calculation of 2–ΔΔCT to estimate the fold changes in gene expression.
Using the statistical tool GraphPad Prism 9,6 all of the experimental data with good biological and technical replications were pooled and analyzed for the one-way analysis of variance (ANOVA) and two-way ANOVA at p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***) levels of significance with Post-hoc analysis. Furthermore, the statistical analyses for qRT-PCR data were performed using WASP-Web Agri Stat Package 2.0 to group the significant results into different clusters. The data sets with the same alphabet were significantly similar to each other while significantly non-similar data sets were designated with different alphabets. The ns denotes non-significant.
Rice endophytic bacterial isolate OsEnb-ALM-D18 which was identified as M. testaceum D18 by 16S rRNA gene sequencing was used in the present study. Firstly, the identity of the isolate was reconfirmed by using multiple approaches to support the 16S rRNA gene sequencing data. On nutrient agar media M. testaceum D18 appeared as typical small round, convex, on-fluid yellow colonies while in NA added with 2,3,5-triphenyl tetrazolium chloride (TTC) it appeared with the same shape and size of colonies but are bright red (Supplementary Figure 1A). The identification of the endophytic bacterium was again confirmed through TEM imaging (Supplementary Figure 1B) which indicated the different structures like oval, rod, and V shape structures typical of Microbacterium.
A total of eight genes of Microbacterium were used for phylogenetic analysis as well as identity confirmation of the rice endophyte OsEnb-ALM-D18. nBLAST results of the different genes are presented in Supplementary Figure 1C; Table 1 where all of the gene sequences identify the isolate as M. testaceum. Further, all the gene sequences were submitted to NCBI GenBank. Further, the accession numbers were assigned (Accession numbers ON157417-ON157424). However, phylogenetic analysis was performed for eight gene sequences with the sequences of closely related bacterial species obtained from NCBI GenBank databases further confirming the identity of rice endophyte OsEnb-ALM-D18 as M. testaceum with more than 97% bootstrap value (Figure 1; Table 1; Supplementary Table 4).
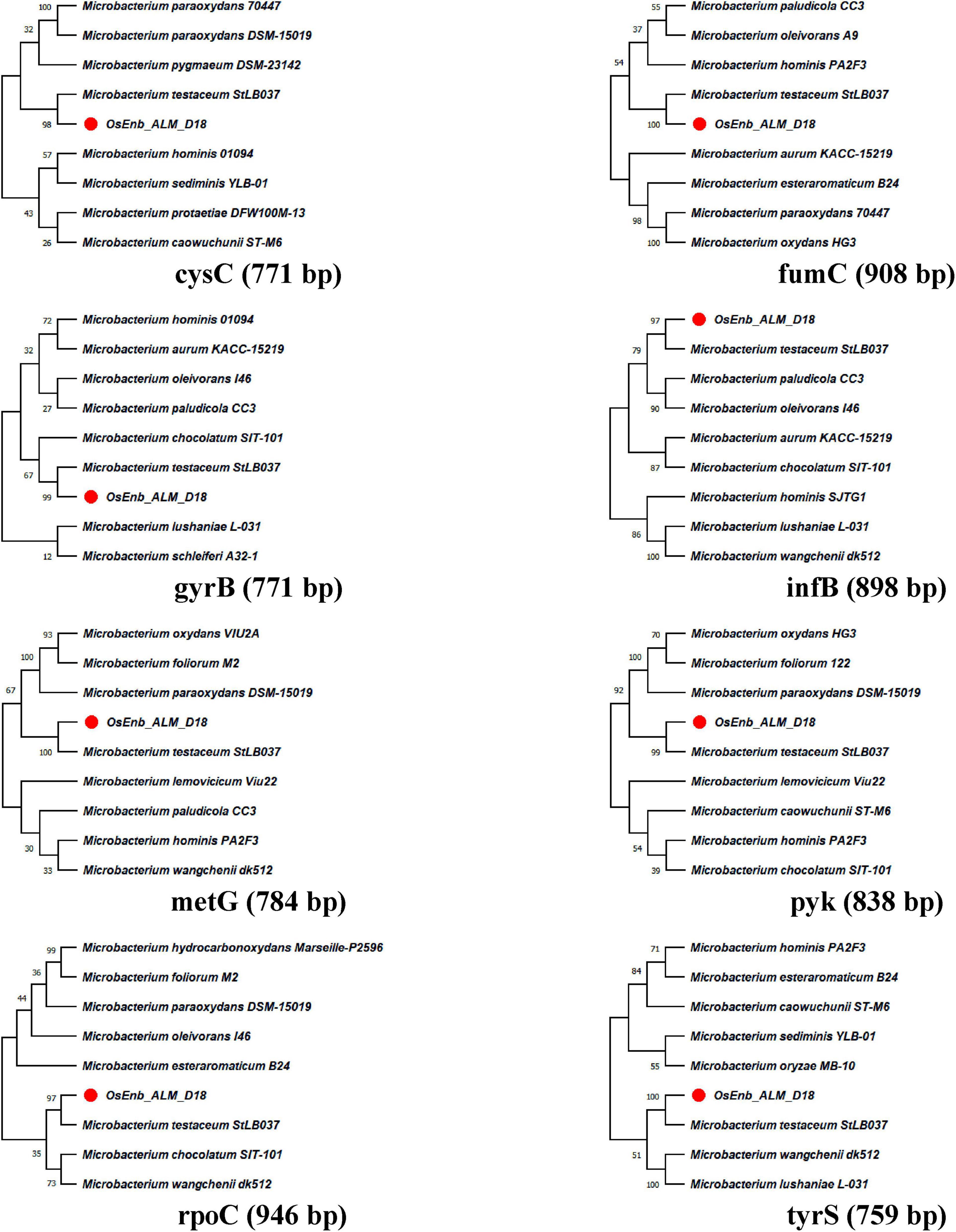
Figure 1. Molecular phylogenetic analysis of Microbacterium species by maximum likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method and the Hasegawa-Kishino-Yano model (Hasegawa et al., 1985). The % of trees in which the associated taxa clustered together is shown above the branches. This analysis involved the nucleotide sequence of nine isolates. Evolutionary analyses were conducted in MEGA11 (Kumar et al., 2018).
To assay the endophytism of M. testaceum D18, firstly, intrinsic rifamycin antibiotic resistance of the bacterium was exploited to select the colonies resistant to rifamycin. These resistant colonies were then used for gfp transformation using the triparental mating method and final transformed cells were selected on a double antibiotic (gentamycin + rifamycin) amended plates (Supplementary Figures 2A,B). The clonal nature of resistant colonies was further confirmed with the help of BOX-PCR fingerprinting and an identical amplicon profile was obtained (Supplementary Figure 2C).
Over 50 putative transformed colonies have found on the double antibiotic selection plates which were again confirmed with PCR as well as CLSM (Supplementary Figure 3). The colonies which showed a high level of gfp expression and were positive for gfp amplification were preserved as glycerol stock and used for downstream experiments. Transformed M. testaceum D18::gfp expressed gfp and showed resistance against gentamycin as well as rifamycin. Antibiotic resistance of the M. testaceum D18::gfp was used in the estimation of the endophytic population in the rice endogenous tissue.
Plantlets of rice cultivar Pusa Basmati-1 emerged from seeds inoculated with M. testaceum D18::gfp (1.0 OD at A600) and were washed thoroughly in sterile distilled water and thin sections were fixed in 4% paraformaldehyde at 4°C for 12 h. Finally, the tissue scanning and imaging were performed in the confocal laser scanning microscope (CLSM; DM6000, Leica Microsystems) at different sites on the sections. The endogenous colonization of M. testaceum D18::gfp was confirmed in rice roots, stems, and the leaf using CLSM imaging of rice seedlings.
Further, PCR amplification using gfp gene-specific primers resulted in 142 bp amplicon in 30 days old rice seedlings that emerged from bacterized seed that confirmed the endophytic nature of M. testaceum D18::gfp (Figure 2). Amplification was not observed in untreated rice seedlings.

Figure 2. Endophytism of Microbacterium testaceum D18::gfp in rice plants as revealed by colony count method and confocal laser scanning microscopy. (A) Genetic transformation of M. testaceum D18 using Tn7-based neutral integration of gfp gene. (B) Plant count method exploiting gentamycin and rifamycin resistance conferred by Tn7-based vector (pBKminiTn7gfp2Gm10). (C) CLSM imaging of endophytically colonized M. testaceum D18::gfp.
Plant probiotic nutrient solubilization and production of hydrolytic enzymes, siderophore, and phytohormone were tested. M. testaceum D18 tested positive for solubilization of phosphorus, potassium, and zinc, and production of siderophore, ammonia, and IAA. In the case of enzymatic activity, the bacterial isolate tested prolific for cellulase and chitinase production and negative for pectinase, xylanase, amylase, and protease (Figure 3; Supplementary Table 5).

Figure 3. Plant probiotic features of M. testaceum D18. (A) Mineral solubilization assays. (B) Production of IAA and Ammonia. (C) Production of hydrolytic enzymes.
Endophytic bacterium M. testaceum D18 was found to inhibit the mycelial growth of M. oryzae primarily by its volatilome. Here, the volatile compounds showed 95.5% mycelial inhibition as against 29.9% inhibition by secreted metabolome. Further, the microscopic examination of M. oryzae conidia after bacterial exposure revealed abnormal structure compared to the unexposed conidia in mock suggestive of antibiotic effect on conidial development. However, the volatilome was found to be fungistatic rather than fungicidal as the mycelial growth re-emerged after the removal of volatile stress (Figure 4).
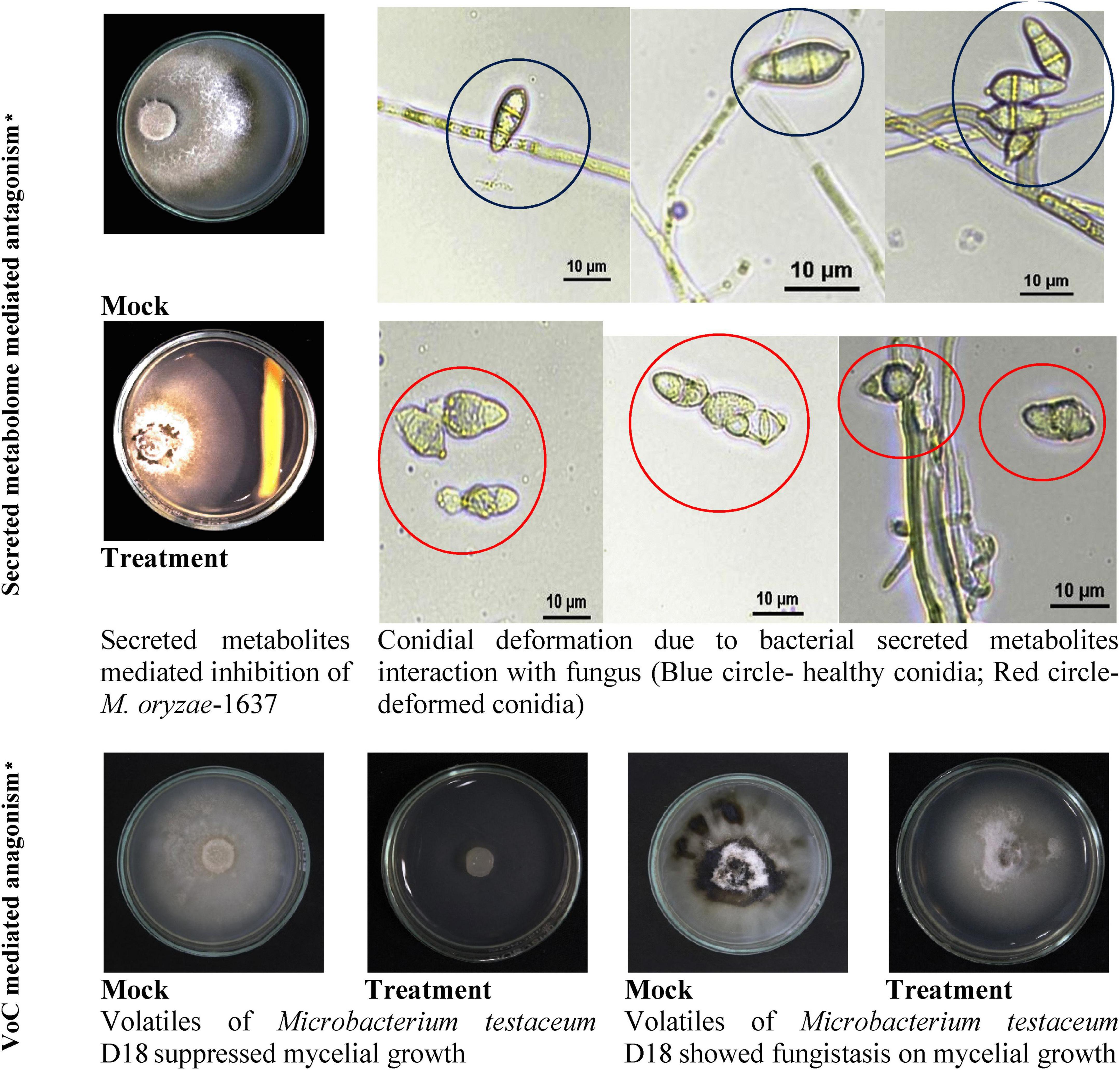
Figure 4. Secreted and volatile metabolite mediated antagonism of M. testaceum D18 on M. oryzae-1637. *% reduction in mycelial growth of Magnaporthe oryzae was observed over mock in both secreted metabolite as well as volatile mediated dual culture assays. In addition to this deformation in conidial morphology was observed in the secreted metabolite-mediated assay.
After observing the inhibitory activity of the volatilome of M. testaceum D18 on rice blast fungus, the volatiles was extracted and analyzed using the solvent extraction method followed by fractionation in GC-MS. The gas chromatography showed the abundance of a wide variety of compounds in the volatilome of M. testaceum D18. A total of 11 chemical compounds were detected in the hexane extract of volatilome. Two compounds namely, 9-octadecenoic acid (32.53%) and hexadecanoic acid (30.06%) contributed over 60% of the chemicals of the volatilome. Other compounds were in the range of 0.26–5.7% in the volatile (Supplementary Figure 4; Supplementary Table 6).
To test this tobacco hypersensitivity assay was conducted on intact tobacco leaves. M. testaceum D18 did not induce necrotic responses in tobacco leaves even after 48 h of leaf infiltration. However, the leaf infiltrated by the pathogen R. solanacearum showed prominent signs of rapid necrosis owing to the hypersensitive reaction, suggesting that M. testaceum D18 is potentially non-pathogenic in plants (Supplementary Figure 5).
The germinated seedlings on bacterial suspension were taken for observation. In the case of shoot length, 108 CFU mL–1 on PB1 was found to increase significantly while in other treatments there was no difference observed. However, maximum root length was observed at 107 CFU mL–1 on BPT5204 whereas the average number of roots was similar in all treatments. The fresh weight, dry weight as well as net weight in all treatments was found to increase slightly in all treatments (Figure 5; Table 2).
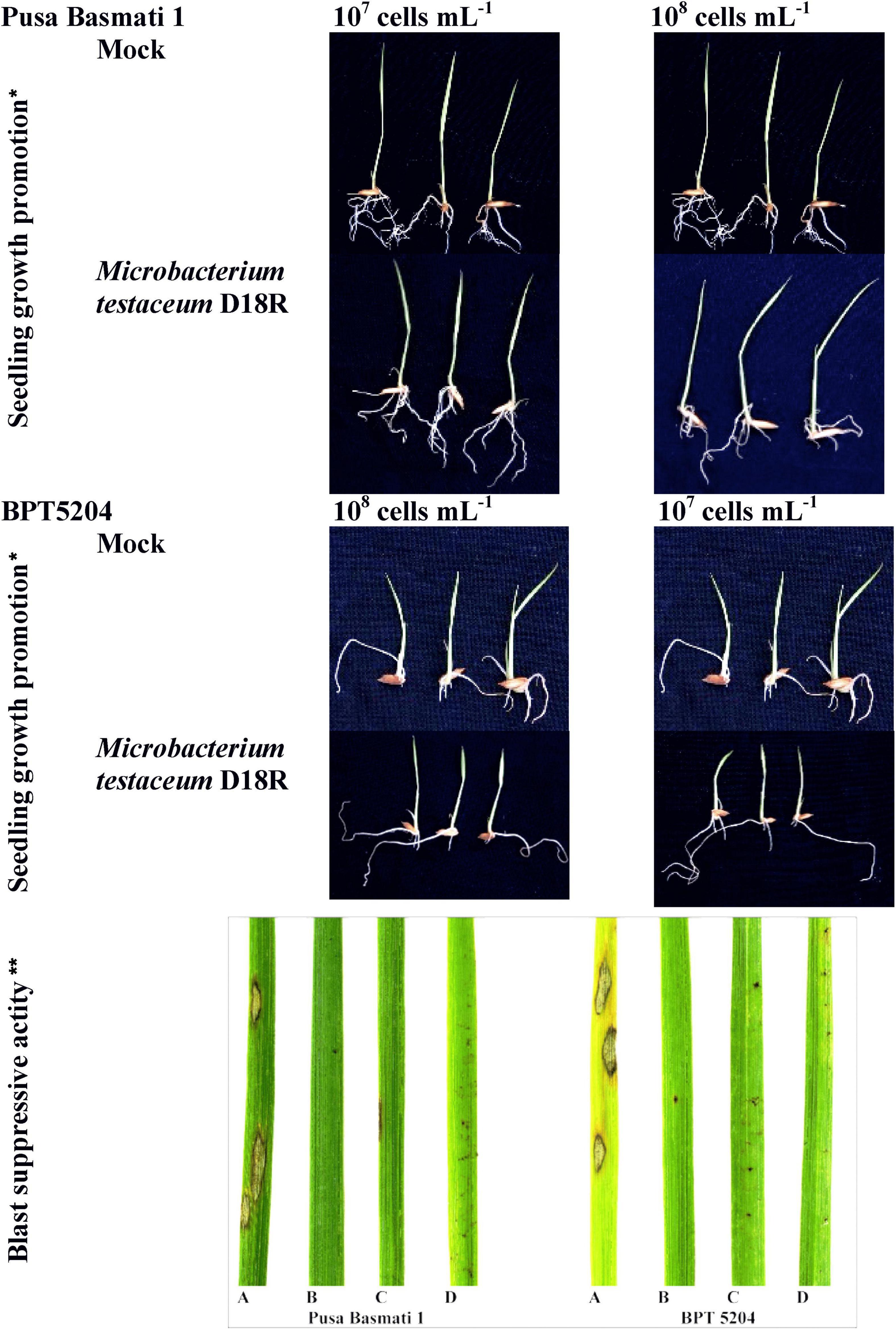
Figure 5. Microbacterium testaceum D18 induced growth promotion in rice seedlings, and blast disease suppressive activity. *M. testaceum stimulated the growth of rice seedlings upon bacterization; **M. testaceum D18 triggered a hypersensitivity type of reaction instead of large lesions observed in mock; the hypersensitive reaction indicates over-expression of defense genes. The number of lesions and size of lesions were found reduced in bacterized plantlets; plant responses are scored as per Mackill and Bonman. (A) Mock. (B) M. testaceum D18 (108 CFU mL–1). (C) M. testaceum D18 (107 CFU mL–1). (D) D. Tricyclazole.
Rice cultivars Pusa Basmati-1, and BPT-5204 were used as susceptible checks for elucidating the blast disease suppressive effects of M. testaceum D18 at 107 and 108 CFU mL–1 applied as seedling treatment as well as a prophylactic foliar spray. The blast disease severity and incidence were scored according to the blast score chart recommended by Mackill and Bonman (1992) (Supplementary Figure 6). The bacterial isolate M. testaceum D18 suppressed blast disease in rice cultivars at tested doses. A significantly less blast severity score was observed at 108 CFU mL–1 on both the varieties PB1 (8.48% PDI) and BPT5204 (6.61% PDI) as compared to untreated plants (45.71–48.03% PDI on PB1 and BPT5204). An over 82% reduction in blast severity over the mock was found in all the treatments (Figure 5; Table 2).
Bacterized rice seedlings were subjected to a gene expression study by qPCR. Several defense-related genes such as OsCERK1, OsCEBiP, OsNPR1, OsPAD4, OsEDS1, and OsFMO1 (Supplementary Table 2) showed a minimal to a high level of induction in bacterized rice seedlings compared with the mock where the seedlings were not treated with bacteria and OsActin was used as reference genes in this study. Post-hoc test confirmed that the bacterization led to a significant increase in the expression pattern of defense-related genes (at P ≤ 0.05 or P ≤ 0.01 or P ≤ 0.001) in both rice genotypes over mock plants. The expression of defense genes was even more pronounced after the booster dose application of bacteria on the phyllosphere. Maximum fold upregulation was observed for the OsFMO1 gene after booster dose application at 108 CFU mL–1 on BPT5204. Similarly, OsCERK1 was maximally expressed in PB1 with 108 CFU mL–1 foliar applications. Moreover, out of all assessed genes, OsNPR1 was not significantly impacted by bacterial treatments, as analyzed by ANOVA. Besides, at the seedling stage, there was not much difference in the expression pattern of most of the genes while foliar application of 108 CFU mL–1 was found effective in eliciting the expression of defense-related genes (Figure 6; Supplementary Table 7).
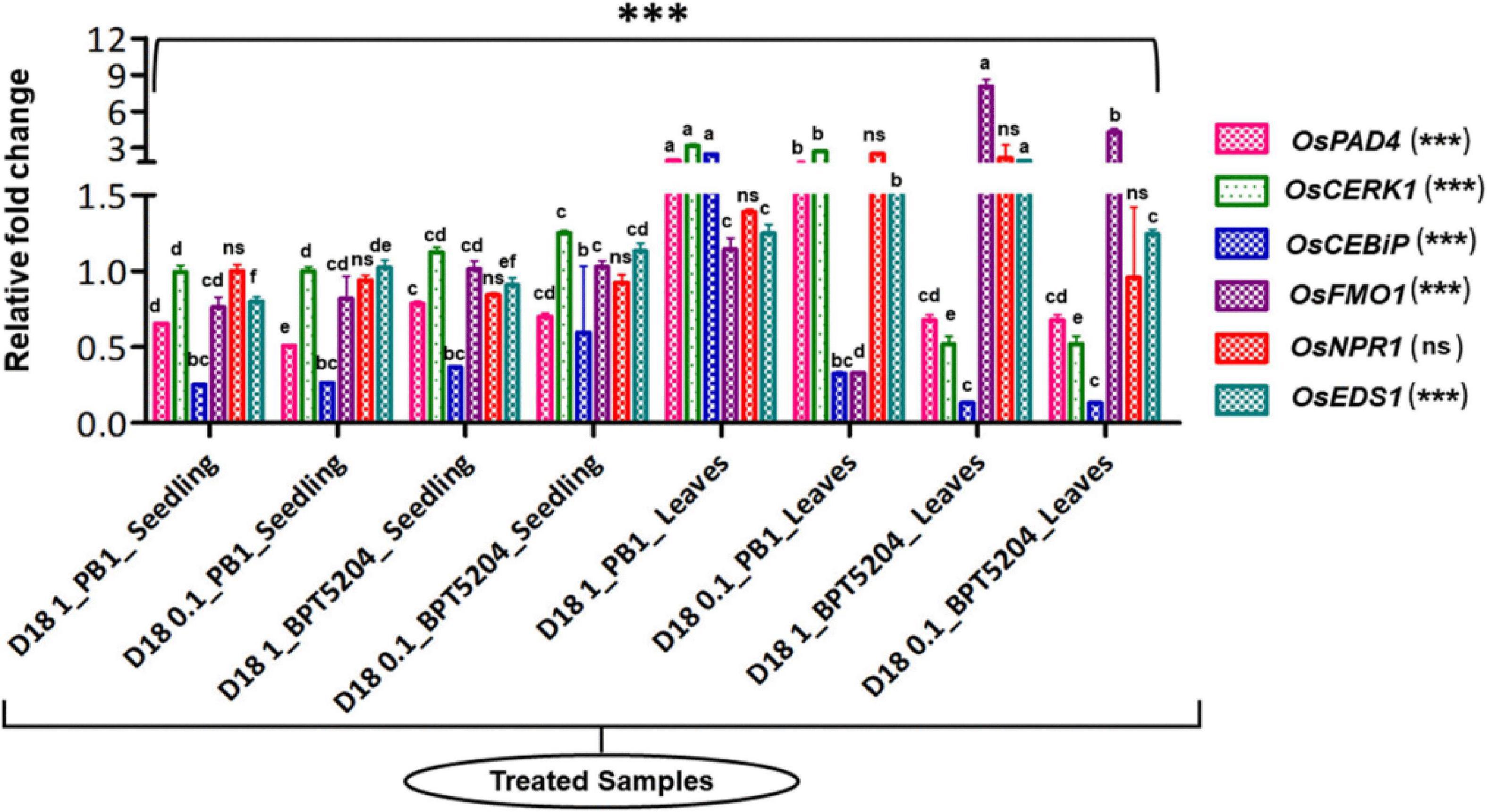
Figure 6. Transcriptional response of defense genes in rice upon bacterization by M. testaceum. qPCR-based transcriptional analysis of defense gene expression in rice seedlings upon endobacterization. The fold change values obtained for the defense genes were imported into the GraphPad Prism program (https://www.graphpad.com/scientific-software/prism), and two-way ANOVA was conducted using Bonferroni post-hoc test for determining the statistical significance at *p > 0.05, **p ≤ 0.001, and ***p ≤ 0.0001. Refer to Supplementary Table 7 for data pertaining to fold changes in gene expression. Different letters represent the different significant groups, based on completely randomized group test done using p-values.
Fungal phytopathogen inciting crop diseases is a threat to agricultural production and food security in the world. Over the decades, two major strategies, i.e., resistance gene introgression and the use of fungicides, either alone or in combination have emerged as effective options for the management of fungal diseases. However, both strategies are inadequate for effective implementation owing to the emergence of new variants with renewed virulence (New Pathotypes) and resistance to fungicides [anti-microbial resistant (AMR)-Type]. The recent ban on fungicides for agricultural applications has created a void in commercial disease management in crop production systems. For instance, the ban on tricyclazole for rice blast management renewed the interest in other options for disease management. As a result, new strategies for combating fungal diseases in crop plants, particularly through the deployment of microbial biological control agents have gained acceptance (Sellem et al., 2017).
Here, we explored and evaluated endophytic bacteria isolated from rice leaves as antagonistic organisms of M. oryzae for biological control of blast disease. Phenotypically the Microbacterium colonies were tiny with pale yellow on nutrient agar medium. Microscopically the cells were small, short slender rods that occurred either in a single culture or in groups during the young stage. However, upon maturation, the cells become shorter and coccoid, and the ends of the cells are attached and produce a V-shaped appearance. The young cells are typically Gram-positive that upon maturation display variable Gram reactions. They are non-endospore producers, and often aflagellate or at times produce lateral flagella.
The species identity of the bacterium was confirmed as M. testaceum through16S rRNA gene sequence as well as multilocus sequence analysis (MLSA) (Fidalgo et al., 2016). For MLSA, we have used Microbacterium-specific eight genetic loci such as rpoC, pyk, gyrB, tyrS, infB, cysS, metG, and fumC. The endophytic M. testaceum isolated from rice leaf was found clustered with a potato leaf endophytic M. testaceum StLB037 (Morohoshi et al., 2009). Before the present study, M. testaceum was reported in diverse habitats like water (Epova et al., 2022), soil (Yang et al., 2014), tannery effluents (Elahi et al., 2019), phyllosphere (Sahu et al., 2021a,b), plant endosphere (Kumar et al., 2021b), and sometimes as an opportunistic pathogen in clinical samples (Leal et al., 2016). We further studied the endophytism of the bacterium in rice plants using gfp-tagged M. testaceum D18::gfp that could be visualized in the root, stem as well as leaf tissues.
Having established the species identity and confirmed the endophytism of M. testaceum, we have functionally characterized the bacterium for several traits such as plant probiosis, pathogen antibiosis; and blast suppressive activity in planta. The endophytic M. testaceum D18 promoted the growth of rice seedlings possibly through multipronged mechanisms such as phytohormone IAA production, solubilization of N, P, K, Fe, Zn, and secretion of hydrolytic enzymes as well as siderophores. Among them, IAA is an auxin that promotes cell proliferation, and elongation, and induces seedling growth (Verma J. P. et al., 2014). The data suggest that M. testaceum D18 promoted the availability of the major plant nutrients N, P, and K which contributed to growth promotion.
The production of ammonia is an important feature of biostimulants as ammonia production plays a key role in plant growth and development by contributing to N nutrition (Yadav et al., 2010; Mukherjee et al., 2020). P nutrition is needed for root development. In soil maximum proportion of phosphorus is unavailable owing to an insoluble state (Miller et al., 2010). Endophytic M. testaceum D18 showed P nutrition solubilization, -one of the key nutrients for plant growth. Previously, Misra et al. (2012) showed the ability of endophytes to solubilize the insoluble phosphate for enhancing leaf area and the consequently improved rate of photosynthesis (Reich et al., 2009). M. testaceum D18 was also found to solubilize K, the third most essential mineral nutrient for plant growth (Bahadur et al., 2016; Mukherjee et al., 2019).
In addition, M. testaceum D18 was also found to synthesize siderophores for iron-chelating Fe to make it unavailable to pathogenic organisms (Seong and Shin, 1996; Saha et al., 2016). Recent studies have suggested that siderophores can induce the defense mechanism of the plant via activating SA- and JA-signaling pathways (Aznar and Dellagi, 2015). In addition to siderophore, the Zn solubilization potential of the endophytic Microbacterium has been reported as a plant growth-promoting trait (Saravanan et al., 2004; Ali et al., 2018; Verma et al., 2022). Priming with M. testaceum D18 enhanced dry and fresh biomass, root and shoot length growth as well as their branching.
Microbacterium testaceum D18 secreted hydrolytic enzymes such as cellulase that are required during endophytic colonization and the endogenous spread (James et al., 2002; Lodewyckx et al., 2002; Walitang et al., 2017). Interestingly, M. testaceum D18 tested positive for other hydrolytic enzymes such as amylase, proteinase, and chitinase. Among them, the chitinolytic activity is particularly relevant as it potentially confers antifungal biocontrol traits against fungal pathogens like M. oryzae that contain chitin in the cell wall (Quecine et al., 2008). Our findings on probiotic features of M. testaceum D18 contribute toward increased shoot and root length, as well as enhanced biomass previously observed with other plant growth-promoting microorganisms (Verma P. et al., 2014; Mukherjee et al., 2020).
The rice endophytic M. testaceum D18 displayed excellent antifungal activity mediated majorly by volatile metabolome against rice blast fungus M. oryzae 1637. The effect of antifungal activities could be observed on mycelial growth and conidial structure. While the mycelia growth was inhibited due to the toxic effect of metabolites, the conidial shape appeared to be deformed. Having observed volatile mediated antifungal activity, we further characterized the volatilome of M. testaceum D18 by GC-MS revealing the high abundance of hexadecanoic acid and 9-Octadecenoic acid contributing to over 60% of the volatile chemicals. Among them, hexadecanoic acid emitted by Trichoderma viride has been shown to have antifungal activity against Macrophomina phaseolina (Khan et al., 2021). Apart from microbial origin, the hexadecanoic acid secreted by the plant Sonchus oleracous also showed antimicrobial properties (Banaras et al., 2020). 9-Octadecenoic acid (Oleic acid) has been the most abundant anti-bacterial unsaturated fatty acid naturally found in staphylococcal abscesses as well as on the skin surface (Speert et al., 1979; Dye and Kapral, 1981). The antimicrobial nature of other chemicals observed in M. testaceum D18 has been reported in the recent past; Tetradecanoic acid (Gajbhiye and Kapadnis, 2021; Nakkeeran et al., 2020), Propenoic acid butyl ester (Garnier et al., 2020), 2-Hexanone (Rocha et al., 2004), Trimethyl-heptadien-4-one (Kassam et al., 2021), 4-Dimethyl-1-heptene (Mannaa and Kim, 2018), 4-Methyl-2-pentanol (Petrović et al., 2013), and 3-Hexanone (Wang et al., 2020).
Blast suppressive activity of M. testaceum D18 was tested on seedlings that emerged from Microbacterium primed seeds of blast susceptible varieties PB1 and BPT5204 that showed significantly less disease severity as compared to untreated control plants. Previously, the biocontrol potential of endophytic Microbacterium StLB037 strains isolated from potato leaf against the soft rot pathogen Pectobacterium carotovorum subsp. carotovorum is reported (Wang et al., 2010). Recently Freitas et al. (2019) reported biological control of Fusarium root rot in cassava using Microbacterium imperial MAIIF2a. More recently Sahu et al. (2021a) showed a reduction in blast disease upon spraying of phyllosphere-adapted Microbacterium oleivorans. Other than the Microbacterium genus, antifungal bacterial genera such as Achromobacter, Streptomyces, and Pseudomonas have been reported against M. oryzae which reduced mycelial growth as well as disease development (Jha et al., 2011; Joe et al., 2012; Xu et al., 2017). Moreover, the perusal of records reveals that the present report on blast suppressiveness of endophytic M. testaceum appears to be novel.
The direct antagonism of the pathogen by endophytic M. testaceum D18, as well as the induction of defense-related pathways in the rice plant, could be responsible for disease suppression. Gene expression analysis using qRT-PCR revealed that M. testaceum D18 significantly upregulated the defense-related genes such as OsFMO, OsCEBiP, OsNPR1, OsCERK1, OsEDS1, and OsPAD4. Cultivar level difference was also observed between the two varieties. Notably, the highest level of induction was observed in the OsFMO1 gene after bacterial suspension spray on BPT5204 plants. Similarly, PB1 sprayed with 108 bacterial cells showed upregulation of all genes with the highest for OsCERK1 (>3 fold). Among the genes, OsCEBiP and OsCERK1 are receptor-like protein (RLP) and receptor-like kinase (RLK), respectively. Chitin and peptidoglycan perception has been reported for the activation of MAMP-triggered immune (MTI) responses in plants (Akamatsu et al., 2013; Kouzai et al., 2014). OsPAD4 and OsEDS1 genes are markers for jasmonic acid-mediated ISR in plants. OsPAD4 is associated with the production of phytoalexin Momilactone-Ain rice plants (Hasegawa et al., 1985; Ke et al., 2014, 2019). Defense genes such as OsNPR1 and OsFMO were marginally upregulated only after booster application, which is a key regulator of systemic acquired resistance mediated by salicylic acid (Sugano et al., 2010).
Having confirmed the plant probiotic, and pathogen antibiotic features as well as endophytism of M. testaceum D18, we performed a tobacco infiltration assay to determine the phytopathogenic nature, if any, and the consequent biological safety of the endophytic bacterium (Klement, 1963). M. testaceum D18 did not show any pathogenic reactions on tobacco suggestive of the non-pathogenic nature of the strain. Biosafety of plant-associated bacterium targeted for agricultural application as bioinoculants is of paramount importance (Kucheryava et al., 1999; Medina-Salazar et al., 2020; Sadeghi and Khodakaramian, 2020).
Taken together, it is concluded that rice endophytic M. testaceum D18 showed unique plant probiotic features as well as pathogen antibiotic activities. The endophytic strain suppressed rice blast disease similar to that of the fungicide, tricyclazole. Coupled with the ability to elicit host defense, the Microbacterium mediated biocontrol can be a supplementary strategy for blast disease management in rice farming. A perusal of published records revealed that this is the first report demonstrating the biocontrol and biostimulation activity of M. testaceum against blast disease.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
AP: conceptualization, methodology, data curation, formal analysis, investigation, writing—original draft, and fund acquisition. KPS: methodology, validation, formal analysis, and writing—original draft. SM: validation and writing—review and editing. AB, CK, TG, SK, and MA: validation. MK: validation and resources. NS and PN: methodology and supervision. AdK: methodology, resources, and supervision. AuK: conceptualization, methodology, formal analysis, resources, data curation, writing—review and editing, visualization, supervision, project administration, and funding acquisition. All authors listed have made a substantial and intellectual contribution to the manuscript writing, reviewing, and editing, have read and approved the publication of the manuscript in its current form.
This study received funding from T-Stanes & Company Ltd., Coimbatore. The funder was not involved in the study design, collection, analysis, and interpretation of data, the writing of this article or the decision to submit it for publication.
We thank the Director, IARI, and Dean, PG School, Indian Council of Agricultural Research–Indian Agricultural Research Institute, New Delhi, for the logistic support and encouragement. We gratefully acknowledge the Biotechnology Industry Research Assistance Council (BIRAC) and the Department of Biotechnology (DBT) for providing research grants in the form of SITARE (Students Innovations for Translation & Advancement of Research Explorations) GYTI (Gandhian Young Technological Innovation) Award, 2021 (project ID: BT/BIRAC/SITARE-GYTI363) for the research work. AuK is thankful to SERB, DST, Government of India, New Delhi, and T-Stanes & Company Ltd., Coimbatore for funding support (SERB File Number: IRR/2018/000012).
Author PN was employed by T-Stanes & Company Ltd., Coimbatore, India.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1035602/full#supplementary-material
Agisha, V., Eapen, S., Monica, V., Sheoran, N., Munjal, V., Suseelabhai, R., et al. (2017). Plant endophytic Pseudomonas putida BP25 induces expression of defense genes in black pepper roots: Deciphering through suppression subtractive hybridization analysis. Physiol. Mol. Plant Pathol. 100, 106–116. doi: 10.1016/j.pmpp.2017.07.006
Akamatsu, A., Wong, H. L., Fujiwara, M., Okuda, J., Nishide, K., Uno, K., et al. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476. doi: 10.1016/j.chom.2013.03.007
Aleksandrov, V., Blagodyr, R., and Ilev, I. (1967). Liberation of phosphoric acid from apatite by silicate bacteria. Mikrobiol. Z. 29, 111–114. doi: 10.1016/j.ijbiomac.2021.08.220
Ali, S., Isaacson, J., Kroner, Y., Saldias, S., Kandasamy, S., and Lazarovits, G. (2018). Corn sap bacterial endophytes and their potential in plant growth-promotion. Environ. Sustain. 1, 341–355. doi: 10.1007/s42398-018-00030-4
Ashajyothi, M., Kumar, A., Sheoran, N., Ganesan, P., Gogoi, R., Subbaiyan, G. K., et al. (2020). Black pepper (Piper nigrum L.). associated endophytic Pseudomonas putida BP25 alters root phenotype and induces defense in rice (Oryza sativa L.) against blast disease incited by Magnaporthe oryzae. Biol. Control 143:104181. doi: 10.1016/j.biocontrol.2019.104181
Asibi, A. E., Chai, Q., and Coulter, J. A. (2019). Rice blast: A disease with implications for global food security. Agronomy 9:451. doi: 10.3390/agronomy9080451
Aznar, A., and Dellagi, A. (2015). New insights into the role of siderophores as triggers of plant immunity: What can we learn from animals? J. Exp. Bot. 66, 3001–3010. doi: 10.1093/jxb/erv155
Bahadur, I., Maurya, B. R., Kumar, A., Meena, V. S., and Raghuwanshi, R. (2016). “Towards the soil sustainability and potassium-solubilizing microorganisms,” in Potassium solubilizing microorganisms for sustainable agriculture, eds V. S. Meena, B. R. Maurya, J. P. Verma, and R. S. Meena (Pune: Springer), 255–266. doi: 10.1007/978-81-322-2776-2_18
Banaras, S., Javaid, A., and Khan, I. H. (2020). Potential antifungal constituents of Sonchus oleraceous against Macrophomina phaseolina. Int. J. Agric. Biol. 24, 1376–1382. doi: 10.17957/IJAB/15.1573
Barnett, S. J., Roget, D. K., and Ryder, M. H. (2006). Suppression of Rhizoctonia solani AG-8 induced disease on wheat by the interaction between Pantoea, Exiguobacterium, and Microbacteria. Aust. J. Soil Res. 44, 331–342. doi: 10.1071/SR05113
Chaiharn, M., Theantana, T., and Pathom-Aree, W. (2020). Evaluation of biocontrol activities of Streptomyces spp. against rice blast disease fungi. Pathogens 9:126. doi: 10.3390/pathogens9020126
Chen, Y., Rekha, P., Arun, A., Shen, F., Lai, W. A., and Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 34, 33–41. doi: 10.1016/j.apsoil.2005.12.002
Collins, M. D., Jones, D., Keddie, R. M., Kroppenstedt, R. M., and Schleifer, K. H. (1983). Classification of some coryneform bacteria in a new genus Aureobacterium. Syst. Appl. Microbiol. 4, 236–252. doi: 10.1016/S0723-2020(83)80053-8
Cordovez, V., Schop, S., Hordijk, K., Dupré de Boulois, H., Coppens, F., Hanssen, I., et al. (2018). Priming of plant growth promotion by volatiles of root-associated Microbacterium spp. Appl. Environ. Microbiol. 84:e01865-18. doi: 10.1128/AEM.01865-18
Cottyn, B., Debode, J., Regalado, E., Mew, T., and Swings, J. (2009). Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J. Appl. Microbiol. 107, 885–897. doi: 10.1111/j.1365-2672.2009.04268.x
Dikin, A., Sijam, K., Kadir, J., and Seman, I. A. (2007). Mode of action of antimicrobial substances from Burkholderia multivorans and Microbacterium testaceum against Schizophyllum commune Fr. Int. J. Agric. Biol. 9, 311–314.
Dye, E., and Kapral, F. (1981). Survival of Staphylococcus aureus in intraperitoneal abscesses. J. Med. Microbiol. 14, 185–194. doi: 10.1099/00222615-14-2-185
Eke, P., Kumar, A., Sahu, K. P., Wakam, L. N., Sheoran, N., Ashajyothi, M., et al. (2019). Endophytic bacteria of desert triangular spurge (Euphorbia antiquorum L.) confer drought tolerance and induce growth promotion in tomato (Solanum lycopersicum L.). Microbiol. Res. 228:126302. doi: 10.1016/j.micres.2019.126302
Elahi, A., Ajaz, M., Rehman, A., Vuilleumier, S., Khan, Z., and Hussain, S. Z. (2019). Isolation, characterization, and multiple heavy metal-resistant and hexavalent chromium-reducing Microbacterium testaceum B-HS2 from tannery effluent. J. King Saud Univ. Sci. 31, 1437–1444. doi: 10.1016/j.jksus.2019.02.007
Epova, E. Y., Shevelev, A. B., Akbayev, R. M., Biryukova, Y. K., Zylkova, M. V., Bogdanova, E. S., et al. (2022). Heterotrophic microbiota from the oligotrophic waters of lake Vostok, Antarctica. Int. J. Environ. Res. Public Health 19:4025. doi: 10.3390/ijerph19074025
Fahad, S., Adnan, M., Noor, M., Arif, M., Alam, M., Khan, I. A., et al. (2019). “Major constraints for global rice production,” in Advances in rice research for abiotic stress tolerance, eds M. Hasanuzzaman, M. Fujita, K. Nahar, and J. K. Biswas (London: Elsevier), 1–22. doi: 10.1016/B978-0-12-814332-2.00001-0
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fidalgo, C., Riesco, R., Henriques, I., Trujillo, M. E., and Alves, A. (2016). Microbacterium diaminobutyricum sp. nov., isolated from Halimione portulacoides, which contains diaminobutyric acid in its cell wall, and emended description of the genus Microbacterium. Int. J. Syst. Evol. Microbiol. 66, 4492–4500. doi: 10.1099/ijsem.0.001379
Freitas, M. A., Medeiros, F. H., Melo, I. S., Pereira, P. F., Peñaflor, M. F. G., Bento, J., et al. (2019). Stem inoculation with bacterial strains Bacillus amyloliquefaciens (GB03) and Microbacterium imperiale (MAIIF2a) mitigates Fusarium root rot in cassava. Phytoparasitica 47, 135–142. doi: 10.1007/s12600-018-0706-2
Gajbhiye, M., and Kapadnis, B. (2021). Lactococcus lactis subsp. cremoris of plant origin produces antifungal Cyclo-(Leu-Pro) and tetradecanoic acid. Indian J. Microbiol. 61, 74–80. doi: 10.1007/s12088-020-00917-z
Garnier, L., Penland, M., Thierry, A., Maillard, M. B., Jardin, J., Coton, M., et al. (2020). Antifungal activity of fermented dairy ingredients: Identification of antifungal compounds. Int. J. Food Microbiol. 322:108574. doi: 10.1016/j.ijfoodmicro.2020.108574
Godfray, H. C. J., Mason-D’Croz, D., and Robinson, S. (2016). Food system consequences of a fungal disease epidemic in a major crop. Philos. Trans. R. Soc. B Biol. Sci. 371:20150467. doi: 10.1098/rstb.2015.0467
Hasegawa, M., Kishino, H., and Yano, T. A. (1985). Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. doi: 10.1007/BF02101694
Hirakue, A., and Sugiyama, S. (2018). Relationship between foliar endophytes and apple cultivar disease resistance in an organic orchard. Biol. Control 127, 139–144. doi: 10.1016/j.biocontrol.2018.09.007
Howard, R. J., and Valent, B. (1996). Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491–512. doi: 10.1146/annurev.micro.50.1.491
Hubballi, M., Pandey, A. K., and Giriyappa, R. (2022). Genetic variation of infection degrees to blast disease among rice genotypes. J. Phytopathol. 170, 107–123. doi: 10.1111/jph.13060
James, E. K., Gyaneshwar, P., Mathan, N., Barraquio, W. L., Reddy, P. M., Iannetta, P. P., et al. (2002). Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant Microbe Interact. 15, 894–906. doi: 10.1094/MPMI.2002.15.9.894
Jha, Y., Subramanian, R., and Patel, S. (2011). Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiol. Plant. 33, 797–802. doi: 10.1007/s11738-010-0604-9
Jiehua, Q., Shuai, M., Yizhen, D., Shiwen, H., and Yanjun, K. (2019). Ustilaginoidea virens: A fungus infects rice flower and threats world rice production. Rice Sci. 26, 199–206. doi: 10.1016/j.rsci.2018.10.007
Joana Gil-Chávez, G., Villa, J. A., Fernando Ayala-Zavala, J., Basilio Heredia, J., Sepulveda, D., Yahia, E. M., et al. (2013). Technologies for extraction and production of bioactive compounds to be used as nutraceuticals and food ingredients: An overview. Compr. Rev. Food Sci. Food Saf. 12, 5–23. doi: 10.1111/1541-4337.12005
Joe, M. M., Islam, M. R., Karthikeyan, B., Bradeepa, K., Sivakumaar, P. K., and Sa, T. (2012). Resistance responses of rice to rice blast fungus after seed treatment with the endophytic Achromobacter xylosoxidans AUM54 strains. Crop Prot. 42, 141–148. doi: 10.1016/j.cropro.2012.07.006
Kassam, R., Yadav, J., Chawla, G., Kundu, A., Hada, A., Jaiswal, N., et al. (2021). Identification, characterization, and evaluation of nematophagous fungal species of Arthrobotrys and Tolypocladium for the management of Meloidogyne incognita. Front. Microbiol. 12:790223. doi: 10.3389/fmicb.2021.790223
Ke, Y., Kang, Y., Wu, M., Liu, H., Hui, S., Zhang, Q., et al. (2019). Jasmonic acid-involved OsEDS1 signaling in Rice-bacteria interactions. Rice 12:25. doi: 10.1186/s12284-019-0283-0
Ke, Y., Liu, H., Li, X., Xiao, J., and Wang, S. (2014). Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host-pathogen interactions. Plant J. 78, 619–631. doi: 10.1111/tpj.12500
Khan, I. H., Javaid, A., and Ahmed, D. (2021). Trichoderma viride controls Macrophomina phaseolina through its DNA disintegration and production of antifungal compounds. Int. J. Agric. Biol. 25, 888–894. doi: 10.17957/IJAB/15.1743
Kirtphaiboon, S., Humphries, U., Khan, A., and Yusuf, A. (2021). Model of rice blast disease under tropical climate conditions. Chaos Soliton Fract 143:110530. doi: 10.1016/j.chaos.2020.110530
Klement, Z. (1963). Rapid detection of the pathogenicity of phytopathogenic pseudomonads. Nature 199, 299–300. doi: 10.1038/199299b0
Koch, M., Vorwerk, S., Masur, C., Sharifi-Sirchi, G., Olivieri, N., and Schlaich, N. L. (2006). A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 47, 629–639. doi: 10.1111/j.1365-313X.2006.02813.x
Komagata, K., Iizuka, H., and Uchino, C. (1964). Taxonomical comparison of four new species of Brevibacterium isolated from rice and the other related bacteria. J. Agric. Chem. Soc. Jpn. 38, 503–509.
Kouzai, Y., Mochizuki, S., Nakajima, K., Desaki, Y., Hayafune, M., Miyazaki, H., et al. (2014). Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant Microbe Interact. 27, 975–982. doi: 10.1094/MPMI-03-14-0068-R
Kucheryava, N., Fiss, M., Auling, G., and Kroppenstedt, R. M. (1999). Isolation and characterization of epiphytic bacteria from the phyllosphere of apple, antagonistic in vitro to Venturia inaequalis, the causal agent of apple scab. Syst. Appl. Microbiol. 22, 472–478. doi: 10.1016/S0723-2020(99)80057-5
Kumar, M. (2021). Structural and functional analysis of rice endophytic microbiome for antimicrobial compounds against rice blast caused by Magnaporthe oryzae. Doctoral dissertation. New Delhi: Indian Agricultural Research Institute, 1–312.
Kumar, M., Charishma, K., Sahu, K. P., Sheoran, N., Patel, A., Kundu, A., et al. (2021a). Rice leaf associated Chryseobacterium species: An untapped antagonistic flavobacterium displays volatile mediated suppression of rice blast disease. Biol. Control 161:104703. doi: 10.1016/j.biocontrol.2021.104703
Kumar, M., Kumar, A., Sahu, K. P., Patel, A., Reddy, B., Sheoran, N., et al. (2021b). Deciphering core-microbiome of rice leaf endosphere: Revelation by metagenomic and microbiological analysis of aromatic and non-aromatic genotypes grown in three geographical zones. Microbiol. Res. 246:126704. doi: 10.1016/j.micres.2021.126704
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Leal, S. M. Jr., Jones, M., and Gilligan, P. H. (2016). Clinical significance of commensal gram-positive rods routinely isolated from patient samples. J. Clin. Microbiol. 54, 2928–2936. doi: 10.1128/JCM.01393-16
Li, Q., Guo, R., Li, Y., Hartman, W. H., Li, S., Zhang, Z., et al. (2019). Insight into the bacterial endophytic communities of peach cultivars related to crown gall disease resistance. Appl. Environ. Microbiol. 85:e02931-18. doi: 10.1128/AEM.02931-18
Liu, C., Chen, X., Liu, T., Lian, B., Gu, Y., Caer, V., et al. (2007). Study of the antifungal activity of Acinetobacter baumannii LCH001 in vitro and identification of its antifungal components. Appl. Microbiol. Biotechnol. 76, 459–466. doi: 10.1007/s00253-007-1010-0
Lodewyckx, C., Vangronsveld, J., Porteous, F., Moore, E. R., Taghavi, S., Mezgeay, M., et al. (2002). Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 21, 583–606. doi: 10.1080/0735-260291044377
Louden, B. C., Haarmann, D., and Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Educ. 12, 51–53. doi: 10.1128/jmbe.v12i1.249
Mackill, D., and Bonman, J. (1992). Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82, 746–749.
Mannaa, M., and Kim, K. D. (2018). Biocontrol activity of volatile-producing Bacillus megaterium and Pseudomonas protegens against Aspergillus and Penicillium spp. predominant in stored rice grains: Study II. Mycobiology 46, 52–63. doi: 10.1080/12298093.2018.1454015
Mano, H., and Morisaki, H. (2008). Endophytic bacteria in the rice plant. Microbes Environ. 24, 109–117. doi: 10.1264/jsme2.23.109
Matsumoto, H., Fan, X., Wang, Y., Kusstatscher, P., Duan, J., Wu, S., et al. (2021). Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 7, 60–72. doi: 10.1038/s41477-020-00826-5
Matsuyama, H., Kawasaki, K., Yumoto, I., and Shida, O. (1999). Microbacterium kitamiense sp. nov., a new polysaccharide-producing bacterium isolated from the wastewater of a sugar-beet factory. Int. J. Syst. Evol. Microbiol. 49, 1353–1357. doi: 10.1099/00207713-49-4-1353
Medina-Salazar, S. A., Rodríguez-Aguilar, M., Vallejo-Pérez, M. R., Flores-Ramírez, R., Marín-Sánchez, J., Aguilar-Benítez, G., et al. (2020). Biodiversity of epiphytic Pseudomonas strains isolated from leaves of pepper and lettuce. Biologia 75, 773–784. doi: 10.2478/s11756-019-00392-y
Mehta, S., Kumar, A., Achary, V. M. M., Ganesan, P., Rathi, N., Singh, A., et al. (2021). Antifungal activity of glyphosate against fungal blast disease on glyphosate-tolerant OsmEPSPS transgenic rice. Plant Sci. 311:111009. doi: 10.1016/j.plantsci.2021.111009
Miah, G., Rafii, M., Ismail, M., Puteh, A., Rahim, H., Asfaliza, R., et al. (2013). Blast resistance in rice: A review of conventional breeding to molecular approaches. Mol. Biol. Rep. 40, 2369–2388. doi: 10.1007/s11033-012-2318-0
Miller, S. H., Browne, P., Prigent-Combaret, C., Combes-Meynet, E., Morrissey, J. P., and O’Gara, F. (2010). Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ. Microbiol. Rep. 2, 403–411. doi: 10.1111/j.1758-2229.2009.00105.x
Mishina, T. E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666–1675. doi: 10.1104/pp.106.081257
Misra, N., Gupta, G., and Jha, P. N. (2012). Assessment of mineral phosphate-solubilizing properties and molecular characterization of zinc-tolerant bacteria. J. Basic Microbiol. 52, 549–558. doi: 10.1002/jobm.201100257
Morohoshi, T., Someya, N., and Ikeda, T. (2009). Novel N-acylhomoserine lactone-degrading bacteria isolated from the leaf surface of Solanum tuberosum and their quorum-quenching properties. Biosci. Biotechnol. Biochem. 73, 2124–2127. doi: 10.1271/bbb.90283
Mukherjee, A., Gaurav, A. K., Singh, S., Chouhan, G. K., Kumar, A., and Das, S. (2019). Role of potassium (K) solubilising microbes (KSM) in growth and induction of resistance against biotic and abiotic stress in plant: A book review. Clim. Chang. Environ. Sustain. 7, 212–214.
Mukherjee, A., Singh, B. K., and Verma, J. P. (2020). Harnessing chickpea (Cicer arietinum L.) seed endophytes for enhancing plant growth attributes and bio-controlling against Fusarium sp. Microbiol. Res. 237:126469. doi: 10.1016/j.micres.2020.126469
Munjal, V., Nadakkakath, A. V., Sheoran, N., Kundu, A., Venugopal, V., Subaharan, K., et al. (2016). Genotyping and identification of broad spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium BP17. Biol. Control 92, 66–76. doi: 10.1016/j.biocontrol.2015.09.005
Nakkeeran, S., Surya, T., and Vinodkumar, S. (2020). Antifungal potential of plant growth promoting Bacillus species against blossom blight of rose. J. Plant Growth Regul. 39, 99–111. doi: 10.1007/s00344-019-09966-1
Neupane, N., and Bhusal, K. (2021). A review of blast disease of rice in Nepal. J. Plant Pathol. Microbiol. 11:528. doi: 10.35248/2157-7471.20.12.528
Patel, A., Kumar, A., Sheoran, N., Kumar, M., Sahu, K. P., Ganeshan, P., et al. (2021). Antifungal and defense elicitor activities of pyrazines identified in endophytic Pseudomonas putida BP25 against fungal blast incited by Magnaporthe oryzae in rice. J. Plant Dis. Protect. 128, 261–272. doi: 10.1007/s41348-020-00373-3
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. doi: 10.1139/m96-032
Petrović, J., Glamočlija, J., Stojković, D. S., Ćirić, A., Nikolić, M., Bukvički, D., et al. (2013). Laetiporus sulphureus, edible mushroom from Serbia: Investigation on volatile compounds, in vitro antimicrobial activity and in situ control of Aspergillus flavus in tomato paste. Food Chem. Toxicol. 59, 297–302. doi: 10.1016/j.fct.2013.06.021
Pikovskaya, R. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Prikl. Biokhim. Mikrobiol. 17, 362–370.
Quecine, M., Araujo, W., Marcon, J., Gai, C., Azevedo, J. D., and Pizzirani-Kleiner, A. (2008). Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett. Appl. Microbiol. 47, 486–491. doi: 10.1111/j.1472-765X.2008.02428.x
Rajashekara, H., Prakash, G., Pandian, R., Sarkel, S., Dubey, A., Sharma, P., et al. (2017). An efficient technique for isolation and mass multiplication of Magnaporthe oryzae from blast infected samples. Indian Phytopathol. 69, 260–265.
Reich, P. B., Oleksyn, J., and Wright, I. J. (2009). Leaf phosphorus influences the photosynthesis–nitrogen relation: A cross-biome analysis of 314 species. Oecologia 160, 207–212. doi: 10.1007/s00442-009-1291-3
Rocha, A. D., de Oliveira, A. B., de Souza Filho, J. D., Lombardi, J. A., and Braga, F. C. (2004). Antifungal constituents of Clytostoma ramentaceum and Mansoa hirsuta. Phytother. Res. 18, 463–467. doi: 10.1002/ptr.1452
Rossman, A. Y., Howard, R. J., and Valent, B. (1990). Pyricularia grisea the correct name for the rice blast disease fungus. Mycologia 82, 509–512. doi: 10.1080/00275514.1990.12025916
Ruiz, M. V., Silva, P., and Laciar, A. (2009). Comparison of microplate, agar drop and well diffusion plate methods for evaluating hemolytic activity of Listeria monocytogenes. Afr. J. Microbiol. Res. 3, 319–324.
Sadeghi, K., and Khodakaramian, G. (2020). Characteristics and ice nucleation activity of sugarcane epiphytic and endophytic bacteria and their role in host frostbite. Sugar Tech. 22, 291–302. doi: 10.1007/s12355-019-00759-0
Saha, M., Sarkar, S., Sarkar, B., Sharma, B. K., Bhattacharjee, S., and Tribedi, P. (2016). Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. 23, 3984–3999. doi: 10.1007/s11356-015-4294-0
Sahu, K. P., Kumar, A., Patel, A., Kumar, M., Gopalakrishnan, S., Prakash, G., et al. (2021a). Rice blast lesions: An unexplored phyllosphere microhabitat for novel antagonistic bacterial species against Magnaporthe oryzae. Microb. Ecol. 81, 731–745. doi: 10.1007/s00248-020-01617-3
Sahu, K. P., Patel, A., Kumar, M., Sheoran, N., Mehta, S., Reddy, B., et al. (2021b). Integrated metabarcoding and culturomic-based microbiome profiling of rice phyllosphere reveal diverse and functional bacterial communities for blast disease suppression. Front. Microbiol. 12:780458. doi: 10.3389/fmicb.2021.780458
Salkowski, C. A., Barber, S. A., Detore, G. R., and Vogel, S. N. (1996). Differential dysregulation of nitric oxide production in macrophages with targeted disruptions in IFN regulatory factor-1 and-2 genes. J. Immunol. 156, 3107–3110.
Saravanan, V. S., Subramoniam, S. R., and Raj, S. A. (2004). Assessing in vitro solubilization potential of different zinc solubilizing bacterial (ZSB) isolates. Braz. J. Microbiol. 35, 121–125. doi: 10.1590/S1517-83822004000100020
Sarkar, D., Rovenich, H., Jeena, G., Nizam, S., Tissier, A., Balcke, G. U., et al. (2019). The inconspicuous gatekeeper: Endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytol. 224, 886–901. doi: 10.1111/nph.15904
Sartori, M., Nesci, A., and Etcheverry, M. (2012). Production of Fusarium verticillioides biocontrol agents, Bacillus amyloliquefaciens and Microbacterium oleovorans, using different growth media: Evaluation of biomass and viability after freeze-drying. Food Addit. Contam. Part A 29, 287–292. doi: 10.1080/19440049.2011.563369
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Sellem, I., Triki, M. A., Elleuch, L., Cheffi, M., Chakchouk, A., Smaoui, S., et al. (2017). The use of newly isolated Streptomyces strain TN258 as potential biocontrol agent of potato tubers leak caused by Pythium ultimum. J. Basic Microbiol. 57, 393–401. doi: 10.1002/jobm.201600604
Seong, K. Y., and Shin, P. G. (1996). Effect of siderophore on biological control of plant pathogens and promotion of plant growth by Pseudomonas fluorescens ps88. Appl. Biol. Chem. 39, 20–24.
Shah, T. M., Tasawwar, S., Bhat, M. A., and Otterpohl, R. (2021). Intercropping in rice farming under the system of rice intensification—an agroecological strategy for weed control, better yield, increased returns, and social–ecological sustainability. Agronomy 11:1010. doi: 10.3390/agronomy11051010
Shan, H., Zhao, M., Chen, D., Cheng, J., Li, J., Feng, Z., et al. (2013). Biocontrol of rice blast by the phenaminomethylacetic acid producer of Bacillus methylotrophicus strain BC79. Crop Prot. 44, 29–37. doi: 10.1016/j.cropro.2012.10.012
Sheoran, N., Kumar, A., Munjal, V., Nadakkakath, A. V., and Eapen, S. J. (2016). Pseudomonas putida BP25 alters root phenotype and triggers salicylic acid signaling as a feedback loop in regulating endophytic colonization in Arabidopsis thaliana. Physiol. Mol. Plant Pathol. 93, 99–111. doi: 10.1016/j.pmpp.2016.01.008
Sheoran, N., Nadakkakath, A. V., Munjal, V., Kundu, A., Subaharan, K., Venugopal, V., et al. (2015). Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds. Microbiol. Res. 173, 66–78. doi: 10.1016/j.micres.2015.02.001
Silva, H. S., Tozzi, J. P., Terrasan, C. R., and Bettiol, W. (2012). Endophytic microorganisms from coffee tissues as plant growth promoters and biocontrol agents of coffee leaf rust. Biol. Control 63, 62–67. doi: 10.1016/j.biocontrol.2012.06.005
Skamnioti, P., and Gurr, S. J. (2009). Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150. doi: 10.1016/j.tibtech.2008.12.002
Speert, D. P., Wannamaker, L. W., Gray, E. D., and Clawson, C. C. (1979). Bactericidal effect of oleic acid on group A streptococci: Mechanism of action. Infect. Immun. 26, 1202–1210. doi: 10.1128/iai.26.3.1202-1210.1979
Sugano, S., Jiang, C. J., Miyazawa, S. I., Masumoto, C., Yazawa, K., Hayashi, N., et al. (2010). Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 74, 549–562. doi: 10.1007/s11103-010-9695-3
Takeuchi, M., and Hatano, K. (1998). Proposal of six new species in the genus Microbacterium and transfer of Flavobacterium marinotypicum ZoBell and Upham to the genus Microbacterium as Microbacterium maritypicum comb. nov. Int. J. Syst. Evol. Microbiol. 48, 973–982. doi: 10.1099/00207713-48-3-973
Tranum-Jensen, J. (1988). ““[50] Electron microscopy: Assays involving negative staining,” in Methods in enzymology, ed. S. Harshman (London: Elsevier), 357–374. doi: 10.1016/s0076-6879(88)65053-1
Ueno, Y., Nakayama, K., Ishii, K., Tashiro, F., Minoda, Y., Omori, T., et al. (1983). Metabolism of T-2 toxin in Curtobacterium sp. strain 114-2. Appl. Environ. Microbiol. 46, 120–127.
Verma, J. P., Yadav, J., Tiwari, K. N., and Jaiswal, D. K. (2014). Evaluation of plant growth promoting activities of microbial strains and their effect on growth and yield of chickpea (Cicer arietinum L.) in India. Soil Biol. Biochem. 70, 33–37. doi: 10.1016/j.soilbio.2013.12.001
Verma, P., Yadav, A. N., Kazy, S. K., Saxena, A. K., and Suman, A. (2014). Evaluating the diversity and phylogeny of plant growth promoting bacteria associated with wheat (Triticum aestivum) growing in central zone of India. Int. J. Curr. Microbiol. Appl. Sci. 3, 432–447.
Verma, S., Kumar, M., Kumar, A., Das, S., Chakdar, H., Varma, A., et al. (2022). Diversity of bacterial endophytes of maize (Zea mays) and their functional potential for micronutrient biofortification. Curr. Microbiol. 79:6. doi: 10.1007/s00284-021-02702-7
Walitang, D. I., Kim, K., Madhaiyan, M., Kim, Y. K., Kang, Y., and Sa, T. (2017). Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of Rice. BMC Microbiol. 17:209. doi: 10.1186/s12866-017-1117-0
Wang, E., Fan, F., Li, Y., Liu, X., Lu, Y., and Song, A. (2020). Noncontact inhibitory of volatile organic compounds from rice root bacteria on Rhizopus microsporus. Health Environ. Res. Online 53, 1986–1996. doi: 10.3864/j.issn.0578-1752.2020.10.006
Wang, W. Z., Morohoshi, T., Ikenoya, M., Someya, N., and Ikeda, T. (2010). AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 76, 2524–2530. doi: 10.1128/AEM.02738-09
Xu, T., Li, Y., Zeng, X., Yang, X., Yang, Y., Yuan, S., et al. (2017). Isolation and evaluation of endophytic Streptomyces endus OsiSh-2 with potential application for biocontrol of rice blast disease. J. Sci. Food Agric. 97, 1149–1157. doi: 10.1002/jsfa.7841
Yadav, J., Verma, J. P., and Tiwari, K. N. (2010). Effect of plant growth promoting rhizobacteria on seed germination and plant growth chickpea (Cicer arietinum L.) under in vitro conditions. Biol. Forum 2, 15–18.
Yamada, K., and Komagata, K. (1972). Taxonomic studies on coryneform bacteria V. Classification of coryneform bacteria. J. Gen. Appl. Microbiol. 18, 417–431. doi: 10.2323/jgam.18.399
Yang, J. K., Zhang, J. J., Yu, H. Y., Cheng, J. W., and Miao, L. H. (2014). Community composition and cellulase activity of cellulolytic bacteria from forest soils planted with broad-leaved deciduous and evergreen trees. Appl. Microbiol. Biotechnol. 98, 1449–1458. doi: 10.1007/s00253-013-5130-4
Zhang, Q. (2007). Strategies for developing green super rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Keywords: rice, endophytic Microbacterium, defense genes, volatiles, blast
Citation: Patel A, Sahu KP, Mehta S, Balamurugan A, Kumar M, Sheoran N, Kumar S, Krishnappa C, Ashajyothi M, Kundu A, Goyal T, Narayanasamy P and Kumar A (2022) Rice leaf endophytic Microbacterium testaceum: Antifungal actinobacterium confers immunocompetence against rice blast disease. Front. Microbiol. 13:1035602. doi: 10.3389/fmicb.2022.1035602
Received: 03 September 2022; Accepted: 07 November 2022;
Published: 21 December 2022.
Edited by:
Yang Song, Utrecht University, NetherlandsReviewed by:
Shaikhul Islam, Bangladesh Agricultural Research Council, BangladeshCopyright © 2022 Patel, Sahu, Mehta, Balamurugan, Kumar, Sheoran, Kumar, Krishnappa, Ashajyothi, Kundu, Goyal, Narayanasamy and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aundy Kumar, a3VtYXJAaWFyaS5yZXMuaW4=; a2F1bmR5QHlhaG9vLmNvbQ==; orcid.org/0000-0002-7401-9885
†Present Address: Aundy Kumar, Department of Botany, Hansraj College, University of Delhi, Delhi, India
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.