- 1College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
- 2Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants, Hangzhou, China
Plant growth-promoting rhizobacteria (PGPR) are well-acknowledged root endophytic bacteria used for plant growth promotion. However, which metabolites produced by PGPR could promote plant growth remains unclear. Additionally, which genes are responsible for plant growth-promoting traits is also not elucidated. Thus, as comprehensive understanding of the mechanism of endophyte in growth promotion is limited, this study aimed to determine the metabolites and genes involved in plant growth-promotion. We isolated an endophytic Rhizobium sp. WYJ-E13 strain from the roots of Curcuma wenyujin Y.H. Chen et C. Ling, a perennial herb and medicinal plant. The tissue culture experiment showed its plant growth-promoting ability. The bacterium colonization in the root was confirmed by scanning electron microscopy and paraffin sectioning. Furthermore, it was noted that the WYJ-E13 strain produced cytokinin, anthranilic acid, and L-phenylalanine by metabolome analysis. Whole-genome analysis of the strain showed that it consists of a circular chromosome of 4,350,227 bp with an overall GC content of 60.34%, of a 2,149,667 bp plasmid1 with 59.86% GC, and of a 406,180 bp plasmid2 with 58.05% GC. Genome annotation identified 4,349 putative protein-coding genes, 51 tRNAs, and 9 rRNAs. The CDSs number allocated to the Kyoto Encyclopedia of Genes and Genomes, Gene Ontology, and Clusters of Orthologous Genes databases were 2027, 3,175 and 3,849, respectively. Comparative genome analysis displayed that Rhizobium sp. WYJ-E13 possesses the collinear region among three species: Rhizobium acidisoli FH23, Rhizobium gallicum R602 and Rhizobium phaseoli R650. We recognized a total set of genes that are possibly related to plant growth promotion, including genes involved in nitrogen metabolism (nifU, gltA, gltB, gltD, glnA, glnD), hormone production (trp ABCDEFS), sulfur metabolism (cysD, cysE, cysK, cysN), phosphate metabolism (pstA, pstC, phoB, phoH, phoU), and root colonization. Collectively, these findings revealed the roles of WYJ-E13 strain in plant growth-promotion. To the best of our knowledge, this was the first study using whole-genome sequencing for Rhizobium sp. WYJ-E13 associated with C. wenyujin. WYJ-E13 strain has a high potential to be used as Curcuma biofertilizer for sustainable agriculture.
Introduction
Endophytes are an endosymbiotic group of microorganisms ubiquitous in nature and well-known to dwell inside plants without causing any apparent harmful effect on the host plant (Kaul et al., 2016; Dubey et al., 2020). Among plant microbiota, endophytes can be found in most plant species, and have been recovered from various tissues, including roots, leaves, stems, flowers, fruits, and seeds (Elmagzob et al., 2019). With the rapid development of science, plant growth-promoting rhizobacteria (PGPR) have attracted more attentions due to their plant growth and development promoting effects.
Certainly, the beneficial relationships conferred by endophytes to host plants are worth revealing, which could be displayed from three different aspects, including growth promotion of their host plants, the secretion of bioactive compounds to increase the stress resistance of the host plants, and the accumulation of important secondary metabolites produced by endophytes (Jia et al., 2016). Particularly, mounting research has paid attentions to the effects of endophytes on plant growth. For example, the application of endophytic fungus Paecilomyces variotii has resulted in improved growth in the development of pepper seedlings (Moreno-Gavíra et al., 2020). Enterobacter roggenkampii ED5 and Pseudomonas aeruginosa B18 have been both able to enhance the growth of sugarcane (Guo et al., 2020; Singh, P., et al., 2021). Bacillus velezensis LDO2 has been able to increase the root length of peanut plants (Chen et al., 2019). Two strains Microbacterium spp. and Streptomyces spp. have significantly promoted ice plant growth compared with the control (Kataoka et al., 2021). Generalist endophyte Phomopsis liquidambaris colonization of Oryza sativa L. has promoted plant growth under nitrogen starvation (Zhou et al., 2022). Notably, the effect of plant growth promotion depends on phosphate solubilization (Liu et al., 2014; Oteino et al., 2015), nitrogen fixation (Guo et al., 2020), hormone indole-3-acetic acid (IAA) synthesis (Hassan, 2017; Harman et al., 2021), and augmenting the resistance of plants to insects and pathogens by producing bioactive metabolites (Etminani and Harighi, 2018). Besides, beneficial endophytes have also been found in many genera, including Stenotrophomonas sp. (Hazarika et al., 2021), Pantoea cypripedii and Kosakonia arachidis (Singh, R. K., et al., 2021), Pennisetum giganteum (Yankey et al., 2022), and Klebsiella aerogenes (Cheng et al., 2022).
Curcuma wenyujin Y.H. Chen et C. Ling is a perennial herb and medicinal plant belonging to the Zingiberaceae family, which is mainly distributed in Wenzhou area of Zhejiang province, China. Its raw rhizomes, steamed rhizomes, and steamed roots constitute three herbal medicines currently listed in the Chinese Pharmacopoeia: pian-jiang-huang, wen-e-zhu and wen-yu-jin, respectively (Li et al., 2021). Accordingly, the research on C. wenyujin has been focused on its various anticancer activities in human renal cell carcinoma (Liu et al., 2019), lung cancer (Wu P. et al., 2021), and hepatocellular carcinoma (Wu et al., 2022). Nevertheless, the interaction between endophytes and Curcuma plant has also been gradually explored. For example, the endophyte P. aeruginosa (BacDOB-E19) have shown multiple plant growth promoting traits, such as increasing plant height and fresh rhizome yield/plant in comparison with untreated control under greenhouse conditions (Vinayarani and Prakash, 2018). Quinalphos-tolerant endophytic Bacillus sp. Fcl1 has been isolated from the rhizome of Curcuma longa, and shown its toxicity-alleviating effect in Vigna unguiculata (Juby et al., 2021). Two species of endophytic Paenibacillus have been identified from the rhizome as indole 3 acetic acid producers, thus have potential growth-regulating effect in rhizomes (Aswathy et al., 2013). However, no plant growth-promoting endophytic rhizobium isolated from C. wenyujin has been reported so far.
The Rhizobiaceae family covers a series of different bacterial genera, including Rhizobium, Sinorhizobium, Bradyrhizobium, Azorhizobium, and Allorhizobium (Manasa et al., 2017). The beneficial symbiotic relationships between the Rhizobium and leguminous crops have been first studied in stem and root nodulating bacteria (Frank, 1889). Generally, most of the rhizobial species are endophytes and colonize intracellularly for root growth promotion by both direct and indirect approaches (Mus et al., 2016). Rhizobia is also employed as PGPR to increase plant growth through the production of hormones, nitrogen fixation, phosphate solubilization, and siderophore development (Pravin et al., 2016). Furthermore, rhizobium is well-known for plant microbiome due to its growth-promoting attributes (Ferreira et al., 2019). Among all rhizobial species, Bradyrhizobium japonicum is suitable for siderophore production, and Rhizobium leguminosarum trifolii is suitable for phosphate solubilization and IAA production (Garcia-Fraile et al., 2012). Hence, the isolation, screening, and exploitation of efficient endophytic bacteria in agricultural practices have great significance.
Which metabolites produced by endophytic rhizobia could promote plant growth remains unknown. Moreover, an in-silico study of various strains has revealed the types of genes and their number that are responsible for the plant growth-promoting traits (Moszer, 1998). Thus, a complete genome study can be used to categorize genes implicated in the positive effects of PGPR, aiding the clarification of plant growth-promoting molecular and functional mechanism (Qin et al., 2017). Therefore, a complete genome sequence and metabolite profiling accessibility of the endophyte will provide an opportunity to understand the mechanism of plant-growth promotion.
This study aimed to (i) isolate and identify the Rhizobium strains from the root of C. wenyujin, (ii) explore their plant growth-promoting ability, (iii) examine the colonization pattern of WYJ-E13 strain by scanning electron microscopy (SEM) and paraffin sectioning, (iv) perform metabolome analysis, and (v) to sequence the WYJ-E13 genome. To the best of our knowledge, this is the first study of the metabolome and whole-genome analysis of endophyte Rhizobium isolated from the C. wenyujin root as a potential biofertilizer to improve plant growth.
Materials and methods
Materials and sample collection
The seedlings of C. wenyujin Y.H. Chen et C. Ling were provided by Zhejiang Wenzhou Tianhe Biotechnology Co., LTD. The seedlings were planted in the climate chamber of Hangzhou Normal University (longitude: 120° 02′ E, latitude: 30° 29′ N), Zhejiang province, China. After growth of 5 months, the tuberous root with a diameter of 2 cm was sampled for further isolation of endophytes.
Furthermore, the plantlets of C. wenyujin Y.H. Chen et C. Ling were also cultivated and obtained via tissue culture in our laboratory.
Isolation and purification of the plant endophytes
The tuberous root sample was rinsed with tap water, soaked in 75% ethanol for 1 min, rinsed with sterilized double-distilled water, disinfected in 0.1% HgCl2 for 15 min, and rinsed three times with sterile water. Subsequently, the sample was cut into small pieces of 5 mm of length, and placed onto the potato dextrose agar (PDA) medium containing 50 mg/l streptomycin with the cutting surface toward the medium. Each petri dish of the medium had four root segments of the tuberous root. Then, the root segments were cultured at 28°C for 5 days. Different colonies were selected and streaked on PDA medium to check the purity. The last rinse water was spread onto the medium and also placed at 28°C in an incubator to check the disinfection method accomplishment.
Morphological and molecular identification of the isolated endophytes
Morphological identification of endophytic bacteria was performed based on colony’s surface morphology, edge shape, and color. Molecular characterization was performed by 16S rDNA sequencing according to our previous study (Song et al., 2017). In detail, bacteria genomic DNA was extracted using the Ezup column bacteria genomic DNA purification Kit (Sangon Biotechnology Co., Ltd., Shanghai, China). The universal primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′) were designed and synthesized. Next, Polymerase chain reaction (PCR) was performed, and the PCR products were recycled, purified, and ligated to the pMD19-T vector. The ligation products were transformed into E. coli DH5α cells. Positive clones were selected, and insertions in plasmid were sequenced. The sequences were analyzed by NCBI BLAST online software to determine the species of isolated endophytic bacteria. Phylogenetic tree was performed using MEGA X software.
Inoculation of Rhizobium sp. strain WYJ-E13 to the plantlets
Prior to inoculation on plants, the bacterium solution of Rhizobium sp. strain WYJ-E13 was prepared. First, WYJ-E13 was added into 100 ml PDA liquid medium and grown at 150 rpm for 12 h at 28°C in constant temperature shaker. After centrifugation at 12000 rpm for 15 min, the obtained sediment was diluted with fresh MS medium to an OD600 value of about 0.02, and the resuspend bacterium solution was obtained. Next, the plantlets of C. wenyujin Y.H. Chen et C. Ling were inoculated on MS solid medium containing 2 mg/l 6-BA and 1 mg/l indoleacetic acid (IAA). Then, 10 μl WYJ-E13 bacterium solution was added onto the base part of the plantlets. Lastly, the plantlets were grown at 22 ± 2°C for 2000 lux with 12 h light condition in a constant temperature incubator. After growth of 15 and 35 days, the growth traits of plantlets were observed.
Root colonization of Rhizobium sp. strain WYJ-E13
The root colonization inside the C. wenyujin Y.H. Chen et C. Ling was confirmed by scanning electron microscopy (SEM) techniques. For SEM analysis, the root tissues were cut into small pieces and fixed in glutaraldehyde solution overnight at 4°C. The samples were washed three times with double-distilled water and dehydrated in 30, 50, 70, 90, 95 and 100% ethanol for 15 min and then isoamyl acetate for 15 min. After drying the samples with a critical point dryer, the colonization of Rhizobium sp. strain WYJ-E13 was observed by SEM.
For paraffin sectioning analysis, the root and bud tissues were fixed in FAA solution for 24 h. Subsequently, the root and bud tissues were washed with 70% ethanol and dehydrated by passing along a graded ethanol series. Xylene was used as a transitional fluid prior to paraffin wax infiltration and embedding. Transverse sections were obtained using LEICA CM 3050 cryostat, and root sections were 8 μm thick. Double staining (1% safranin for 10 min and 1% fast green for 20 s) was used for the preparation of permanent slides. Finally, the images were captured by Olympus CX22LED light microscope.
Extraction of metabolites produced by Rhizobium sp. strain WYJ-E13
The metabolites produced by Rhizobium sp. strain WYJ-E13, named as WYJ-FB, were extracted. First, the sample was freeze-dried and vortexed in 1 ml cold mixture containing 40% acetonitrile, 40% methanol, and 20% water. Subsequently, the sample was sonicated for 1 h in ice bath and incubated at −20°C for 1 h to precipitate protein. After centrifugation at 16000 rpm for 20 min at 4°C, the obtained supernatant was dried in a high-speed vacuum desiccator. The sample was mixed with 100 μl solution of acetonitrile: water (1:1) for 1 min to end derivatization. After centrifugation, 5 μl supernatant was analyzed through liquid chromatography electrospray ionization tandem mass spectrometry. Additionally, the sterilized LB medium of the same batch was set as the control, named as LB.
Chromatographic separation of metabolites
Chromatographic separation was performed on an ultra-high performance liquid chromatography system (Agilent 1,290 Infinity, Waldbronn, Germany) equipped with HILIC column. The column oven was kept at 25°C with a flow rate of 0.3 ml/min. The mobile phase A consisted of water, 25 mM aqueous ammonia, and 25 mM ammonia, while phase B consisted of acetonitrile. Gradient elution conditions were set as follows: 0–0.5 min, 95% B; 0.5–7 min, linear from 95 to 65% B; 7–9 min, linear from 65 to 40% B; 9–10 min, 40% hold B; 10–11.1 min, linear from 40 to 95% B; 11.1–16 min, 95% hold B. The quality control samples were prepared to evaluate system stability.
Mass spectrometric analysis of metabolites and data processing
The metabolites separated by chromatography were then analyzed using a mass spectrometer Triple-TOF 5600 (AB SCIEX, United States) with an electronic spray ionization (ESI) source operated in both positive and negative ion modes. The main parameter was set as following: ion source gas1/2: 60; curtain gas: 30; IonSpray voltage floating: ± 5,500 V; TOF MS m/z range: 60–1,200 Da; TOF scan time: 0.15 s/spectra; product ion scan m/z range: 25–1,200 Da; product ion scan time: 0.03 s/spectra. MS/MS spectra was obtained by information-dependent acquisition with a high sensitivity mode with the following parameters: declustering potential with ±60 V, collision energy of 30 eV, excluding isotopes within 4 Da, and candidate ions to monitor per cycle: 6.
After processing the raw data, the metabolites were identified via a self-built database (BioProfile Technology Co., Ltd., Shanghai, China) according to the accurate mass and MS/MS spectrogram. Furthermore, principal component analysis (PCA), partial least square discriminant analysis (PLS-DA), and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed. Differentially accumulated metabolites (DAMs) were identified according to the following criteria: a variable influence on projection (VIP)> 1 and value of p < 0.05. Analysis on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of DAMs was performed.
DNA extraction, library construction and genome sequencing
Genomic DNA was extracted from the Rhizobium sp. WYJ-E13 using the CTAB method by Shanghai Personalbio Technology Co., Ltd. (Shanghai, China). The concentration of the DNA sample was evaluated using Quant-iT PicoGreen dsDNA Assay Kit, while the integrity was detected by 1% agarose gel. An insertion library of 400 bp pair-end was constructed with Illumina TruSeq DNA Sample Prep Kit according to manufacturer instructions and sequenced using an Illumina MiSeq instrument (Illumina, United States). Larger 20 k bp insertion (S20K) library was prepared by SMRTbell Template Prep Kit 1.0 according to the PacBio protocol, and sequenced using a PacBio Sequel I platform (Pacific Biosciences, United States).
Genome assembly, annotation and gene prediction
The raw reads generated from Illumina and PacBio sequencing were utilized for bioinformatics investigation. For the Illumina short reads, the adaptor sequences were removed by Adapter Removal v2.1.7 (Schubert et al., 2016) and the k-mers for the paired-end libraries were tailed using SOAPec v2.0 (Luo et al., 2012). Furthermore, the sequencing reads were verified using SPAdes v3.9.0 (Bankevich et al., 2012), followed by assembly with A5-miseq v20150522 software (Tritt et al., 2017). Additionally, the PacBio sequencing data was assembled using HGAP4 (Chin et al., 2016) and CANU v1.6 (Koren et al., 2017). Subsequently, the assembled contigs from both Illumina and PacBio reads were merged, and gaps were filled. Finally, the full genome assembly was generated with Pilon v1.22 software (Walker et al., 2017).
GeneMarkS software v4.32 was used to predict protein-coding genes (Besemer et al., 2001). The protein-coding genes were aligned to the NCBI-NR database (Non-Redundant Protein Sequence Database) using diamond software v0.9.10.111 to retrieve annotations with an E-value cutoff of 1e-6 (Buchfink et al., 2015). Cluster of orthologous genes (COGs) was annotated with eggNOG-mapper with eggNOG database v4.5 and an E-value cutoff of 1e-6 (Huerta-Cepas et al., 2017). The KEGG orthology annotation was performed by KASS v2.1 software (Moriya et al., 2007). GO was annotated by InterPro v66.0 software (Finn et al., 2017). The annotation of Swiss-Prot was performed by BLAST with an E-value cutoff of 1e-6. tRNAscan-SE (Lowe and Eddy, 1997), Barrnap software (0.9-dev), and Rfam database (Burge et al., 2013) were used to predict tRNA, rRNA, and other non-coding RNAs, respectively. Clustered regularly interspaced short palindromic repeats (CRISPRs) finder tool was used to predict CRISPR regions (Grissa et al., 2007). The genome circular map of strain WYJ-E13 was drawn and visualized by CGView software (Stothard and Wishart, 2005).
Comparative genome analysis
The average nucleotide identity (ANI) values among 16 genome sequences, including Rhizobium sp. WYJ-E13 and other 15 Rhizobium strains, were calculated using the JSpeciesWS online service (Richter et al., 2016). ANI results were used for hierarchical cluster analysis using TBtools v1.0987663 software.
Furthermore, collinearity of the conserved and highly orthologous genomic regions was analyzed using Mauve 20,150,226 software with default parameters (Darling et al., 2004). The locally collinear blocks (LCBs) showed the conserved and highly similar genomic region. The white areas inside colored regions indicated sequence elements specific to one genome that was not aligned. The height of similarity profile was present inside each block. The colored lines that connect LCBs represented translocations of homologous regions. Blocks above or below the horizontal bar indicated regions that aligned in the forward or reverse orientation, respectively.
Results
Isolation and characterization of endophytes from the root of Curcuma wenyujin
In this study, the tuberous root of medicinal plant C. wenyujin was used as experimental materials (Figure 1A). After 3 days of growth on the PDA medium, different colonies spread from the cut surface of the tuberous root (Figure 1B). Single-colony isolation was repeated at least three times for purification of the endophyte isolates (Figure 1C). Based on the morphology, color, and growth characteristics, a total of 14 strains were isolated (Supplementary Figure S1).
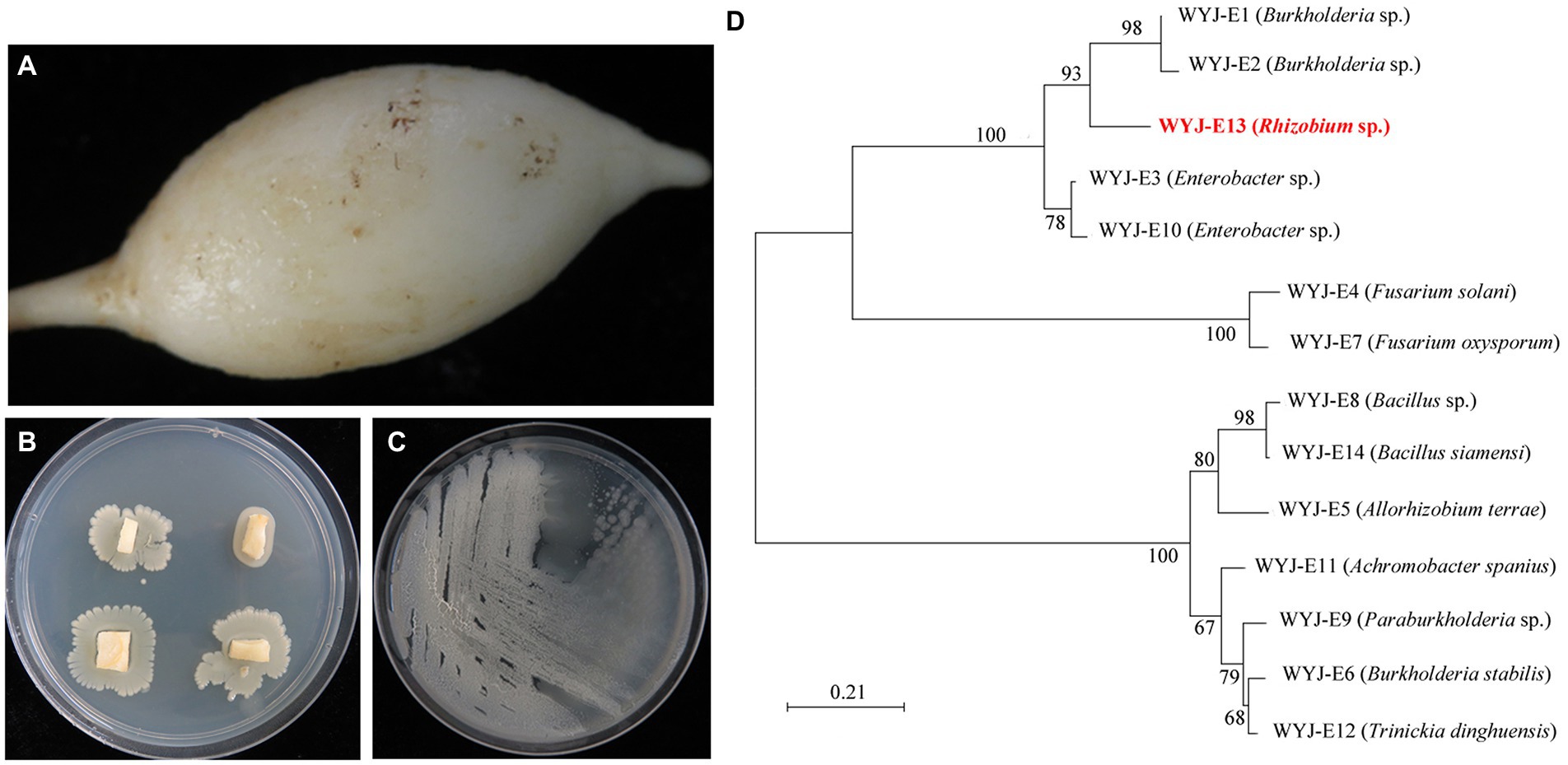
Figure 1. The isolation and characterization of endophytes isolated from the medicinal plant Curcuma wenyujin. (A) The tuberous root of medicinal plant Curcuma wenyujin. (B) The culture of endophytes on PDA medium. (C) The single-colony isolation of endophytes. (D) The phylogenetic tree of isolated 14 endophytes using MEGA X software.
Subsequently, the identification of all 14 strains was performed based on partial 16S rRNA gene sequencing. The obtained strain sequences were compared using the BlastN tool, with NCBI GenBank database nucleotide sequences and similarity values of ≥97% being obtained. The results showed the evolutionary relationships of all 14 strains (Figure 1D).
Considering the possible growth-promoting features, the Rhizobium sp. strain WYJ-E13 was selected for further research. Furthermore, the endophytic Rhizobium sp. strain WYJ-E13 was stored in the China General Microbiological Culture Collection Center at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (strain preservation number CGMCC14808).
Evaluation of plant growth promoting attributes
For evaluating its potential of promoting plant growth, Rhizobium sp. strain WYJ-E13 was inoculated into the plantlets. The results showed a significant increase in growth rate, seedling height, and root length in the inoculated plantlets compared with not-inoculated control plants (p < 0.05), indicating that Rhizobium sp. strain WYJ-E13 could promote the growth of C. wenyujin (Figure 2).
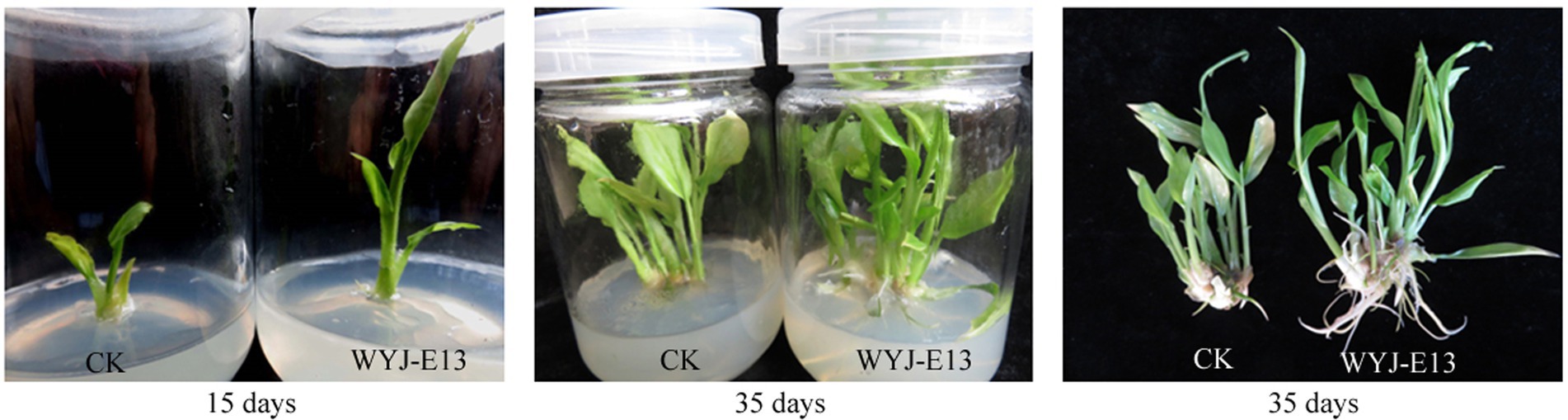
Figure 2. Plant growth promotion assay of Rhizobium sp. WYJ-E13 on the Curcuma wenyujin after 15 days and 35 days of inoculation.
Colonization study of endophytic Rhizobium sp. strain WYJ-E13 on Curcuma wenyujin
The root colonization of Rhizobium sp. strain WYJ-E13 was examined by SEM, as this bacterium confirmed some plant growth promoting traits. These techniques helped study the interaction mechanism of the selected potential strains in C. wenyujin. After 35 days of incubation with the inoculated WYJ-E13 strain, SEM results confirmed the symbiotic colonization of WYJ-E13 in root tissues of C. wenyujin (Figures 3A–D). Furthermore, the morphology of endophytic Rhizobium sp. strain WYJ-E13 was rod-shaped (Figure 3D).
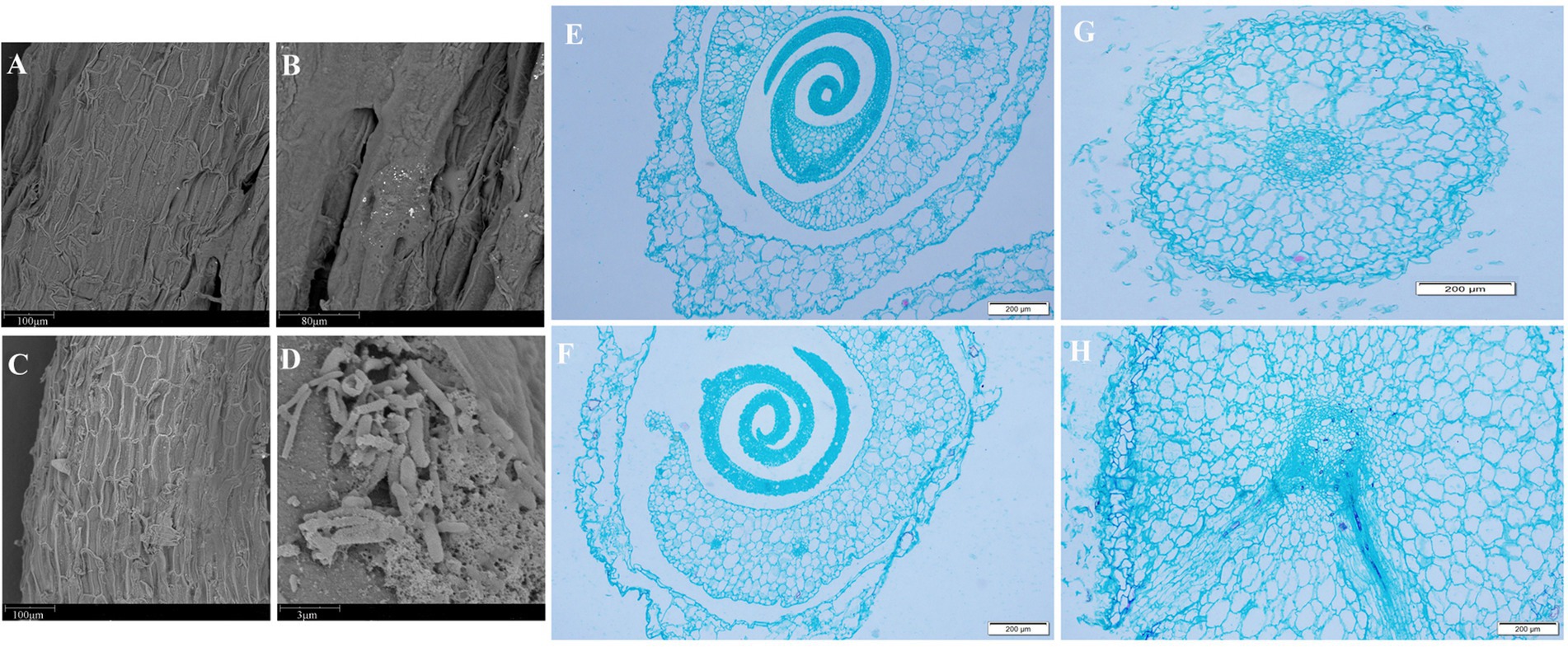
Figure 3. The observation of bacterium colonization by scanning electron microscopy (SEM) and paraffin section techniques. (A,B) The root colonization of Curcuma wenyujin without inoculation of WYJ-E13 by SEM. (C,D) The root colonization of Curcuma wenyujin after inoculation of WYJ-E13 by SEM. (E) The bud colonization without inoculation of WYJ-E13. (F) The bud colonization after inoculation of WYJ-E13. (G) The root colonization without inoculation of WYJ-E13. (H) The root colonization after inoculation of WYJ-E13.
Besides, the paraffin sectioning of root and bud was observed by light microscope. The results showed that bacteria colonization was spotted in root tissues of C. wenyujin while no colonization was observed in the bud (Figures 3E–H), indicating the role of WYJ-E13 in root growth.
Metabolites analysis of endophytic Rhizobium sp. strain WYJ-E13
In this study, the non-targeted metabolomics analysis was performed to explore possible metabolites secreted by the endophytic Rhizobium sp. strain WYJ-E13 that played potential promoting roles in plant growth. During the UPLC–MS/MS analysis process, the reproducibility of the instrument was monitored using the quality control (QC) samples, and the total ion chromatograms (TICs) of the QC samples in the positive (ESI +) and negative (ESI -) ions modes basically overlapped (Supplementary Figure S2). Furthermore, the PCA score plot of QC samples was tightly clustered near the origin (Supplementary Figure S3). These results indicated the stability of the instrument and the reliability of data generated from the LC–MS/MS analysis.
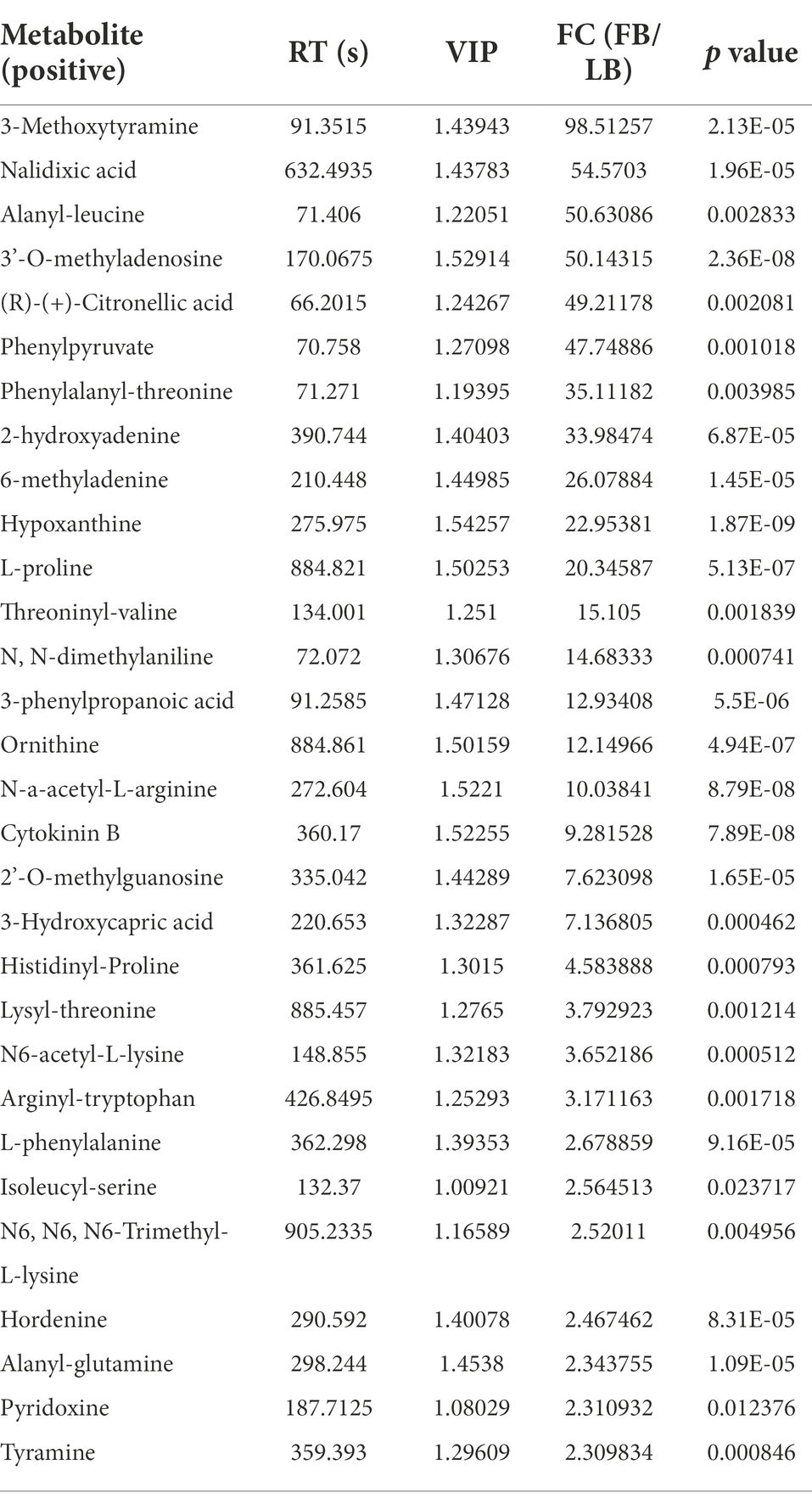
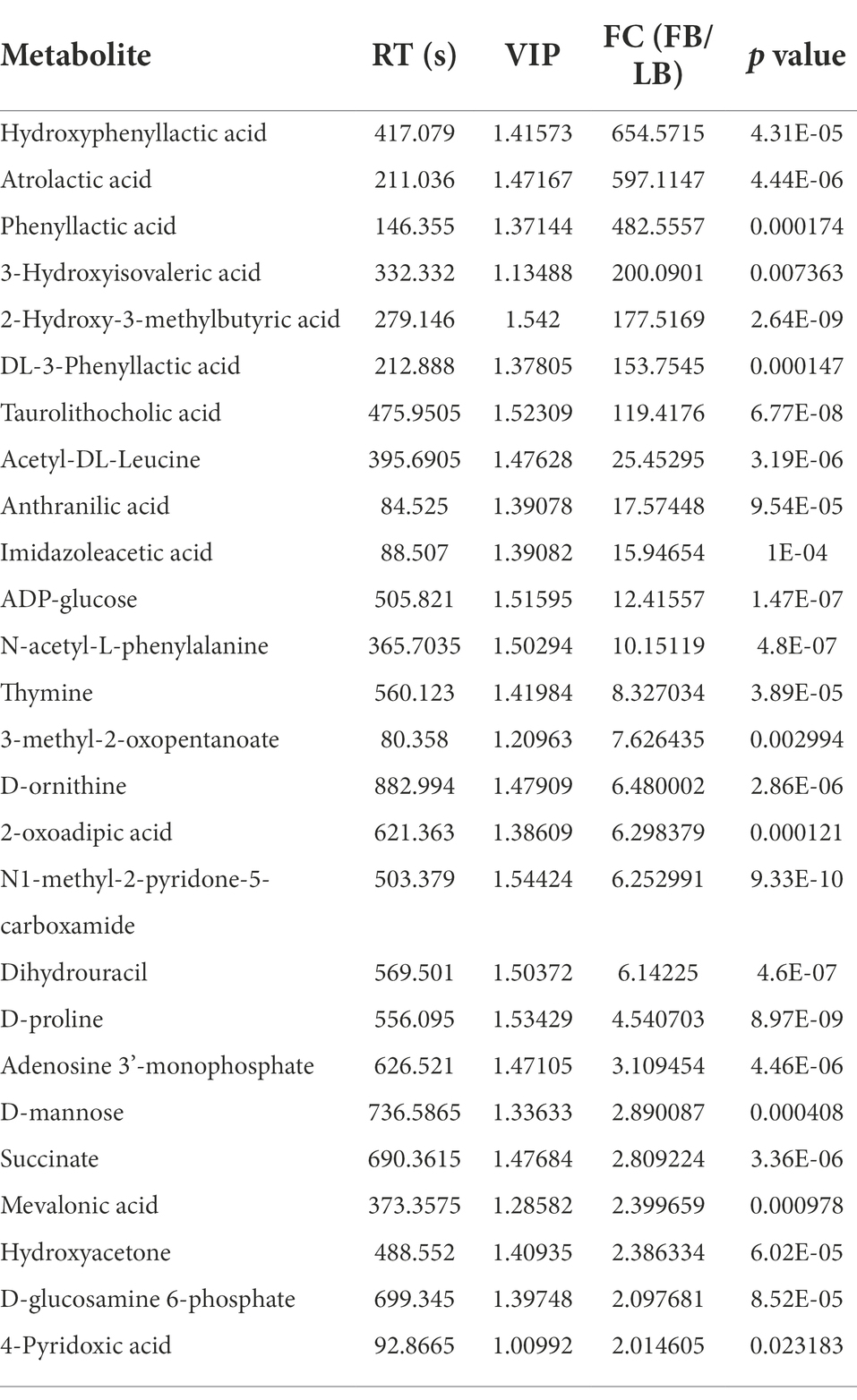
Using UPLC-MS/MS analysis, 13,283 positive ions and 13,303 negative ions were detected. PCA of the corrected and filtered data showed an obvious separation between WYJ-FB and LB medium (Figure 4A). The goodness-of-fit (R2) and predictability (Q2) of the PLS-DA models were higher than 0.95 in both WYJ-FB and LB medium (Figure 4B). In combination with the screening criterion of a VIP > 1.0, value of p < 0.05 and fold change (FC) > 2, a total of 1933 (ESI +) and 2,249 (ESI -) differential ions between WYJ-FB and LB medium were detected. Finally, 30 (ESI +) and 26 (ESI -) differential metabolites were obtained. Expectedly, the content of metabolites in WYJ-FB was higher than that in LB, regarding 3-methoxytyramine, nalidixic acid, phenylpyruvate, 6-methyladenine, hypoxanthine, cytokinin B and pyridoxine in positive ion modes (Table 1). In negative ion modes, the metabolites content of hydroxyphenyllactic acid, atrolactic acid, phenyllactic acid, 3-hydroxyisovaleric acid, 2-hydroxy-3-methylbutyric acid, and taurolithocholic acid were 100 folds higher than that in LB control medium (Table 2). Importantly, KEGG results showed that seven significantly enriched pathways were found, including biosynthesis of amino acids, aminoacyl-tRNA biosynthesis, pantothenate and CoA biosynthesis, ABC transporters, phenylalanine metabolism, purine metabolism and alanine, aspartate and glutamate metabolism (Figure 4C). Furthermore, the heatmap of differential metabolites was displayed (Figure 5). These results indicated that the Rhizobium sp. strain WYJ-E13 could produce various secondary metabolites.
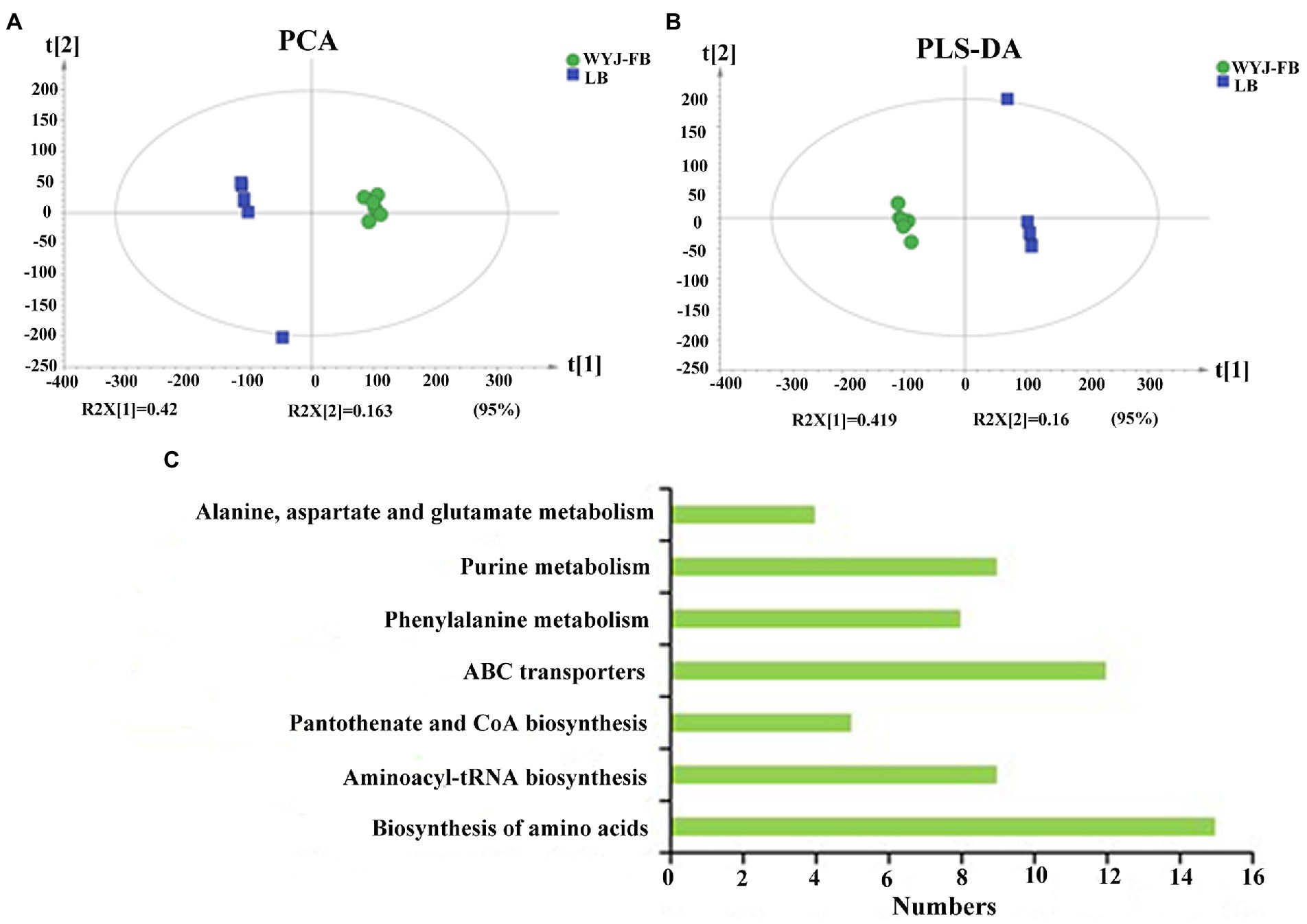
Figure 4. The metabolome analysis between WYJ-FB and LB medium. (A) PCA of the corrected and filtered data showed an obvious separation between WYJ-FB and LB medium. (B) The goodness-of-fit (R2) and predictability (Q2) of the PLS-DA models. (C) The KEGG pathway analysis of the identified differential metabolites produced by Rhizobium sp. WYJ-E13.
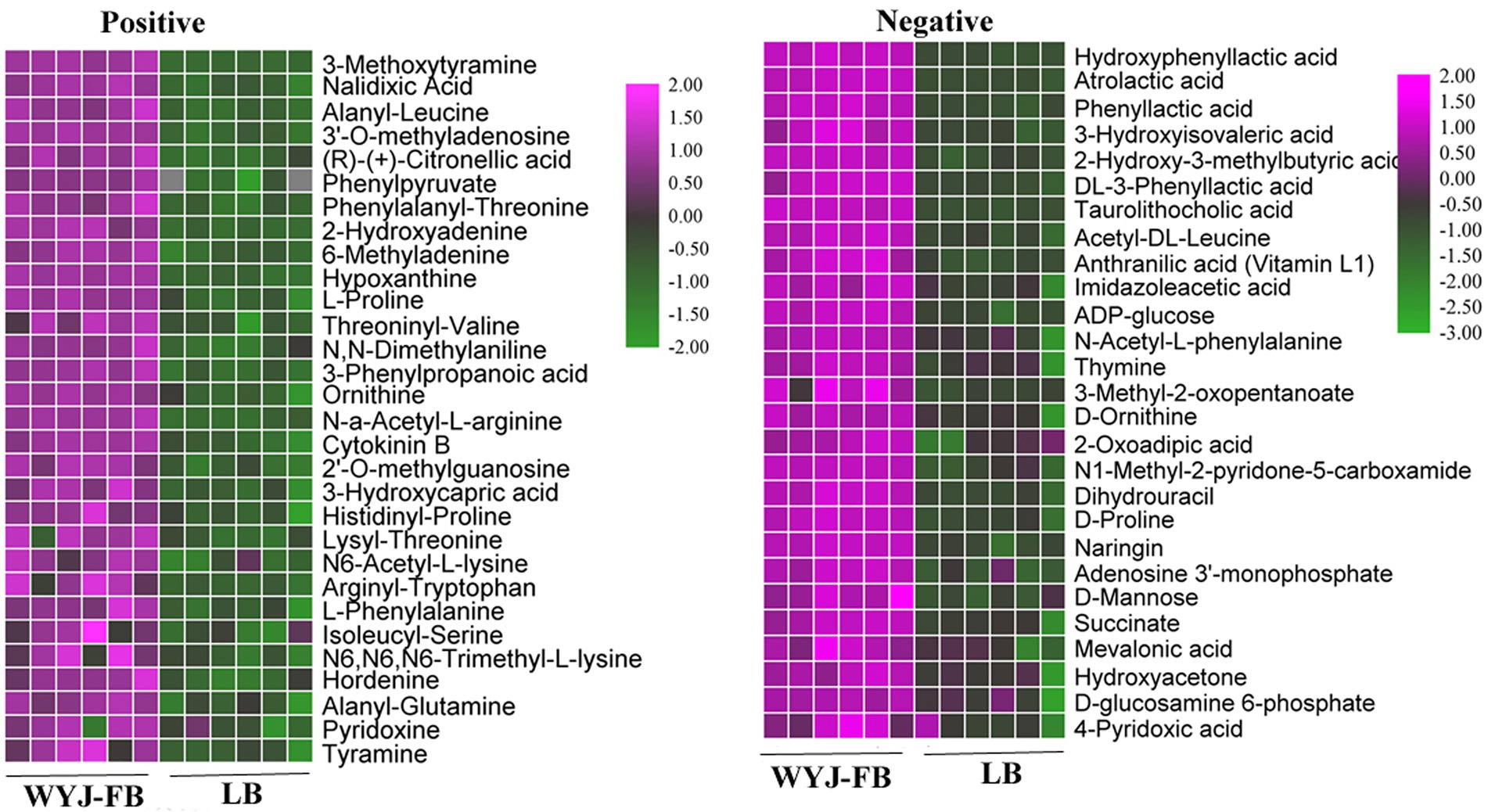
Figure 5. The heatmap of differential metabolites identified in positive ion modes and negative ion modes.
Genome characteristics of Rhizobium sp. strain WYJ-E13
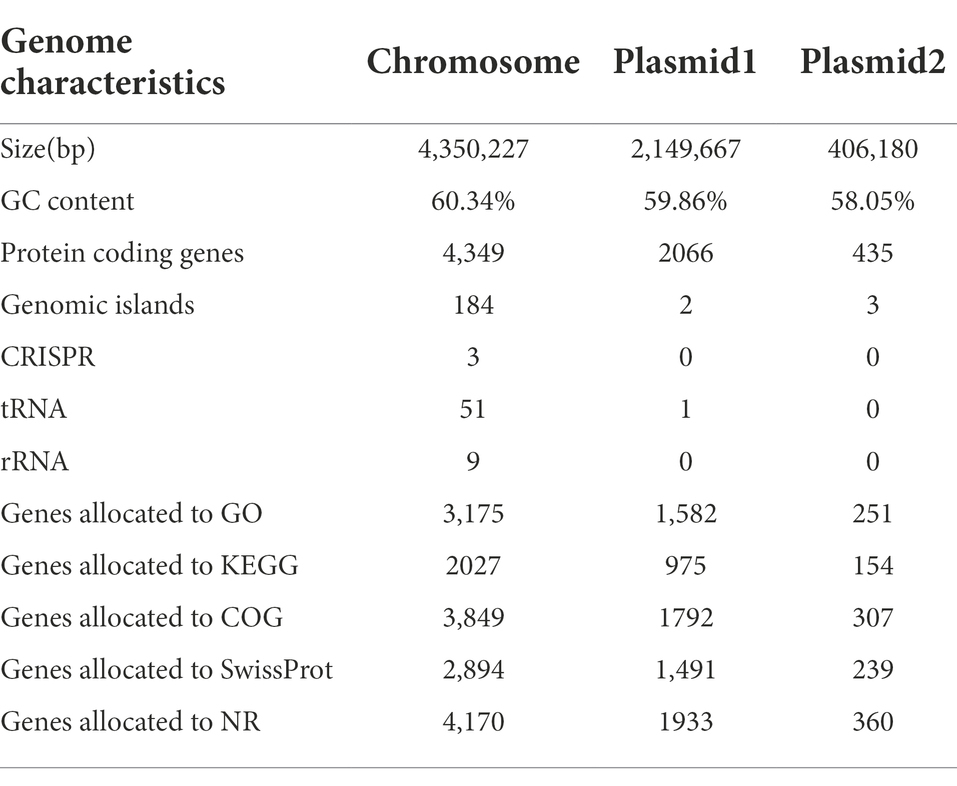
In this study, a total of 3,741,172 raw reads were generated. After trimming, 3,663,822 high-quality reads were obtained on the Illumina MiSeq platform. The percentage of high-quality reads was 97.93%, reflecting the actual nucleotide composition. Furthermore, a total of 133,991 sequences were obtained on the PacBio Sequel I platform. The whole-genome characteristics of Rhizobium sp. strain WYJ-E13 were presented, which comprised of a circular chromosome of 4,350,227 bp with an overall GC content of 60.34%, of a 2,149,667 bp plasmid1 with 59.86% GC, and of a 406,180 bp plasmid2 with 58.05% GC (Table 3). The chromosome of WYJ-E13 displayed a clear GC skew transition, which corresponded to its replication origin and terminus (Figure 6A). There were 4,349 putative protein-coding genes, 51 tRNAs, and 9 rRNAs (Table 3). The characterization of predicted genes against KEGG, GO and COG database were 2027, 3,175 and 3,849 (Supplementary Figures S4–S6; Table 3). Furthermore, the plasmid1 genome annotations estimated protein coding with 2066 genes and plasmid2 with 435 protein coding genes (Figures 6B,C; Table 3).
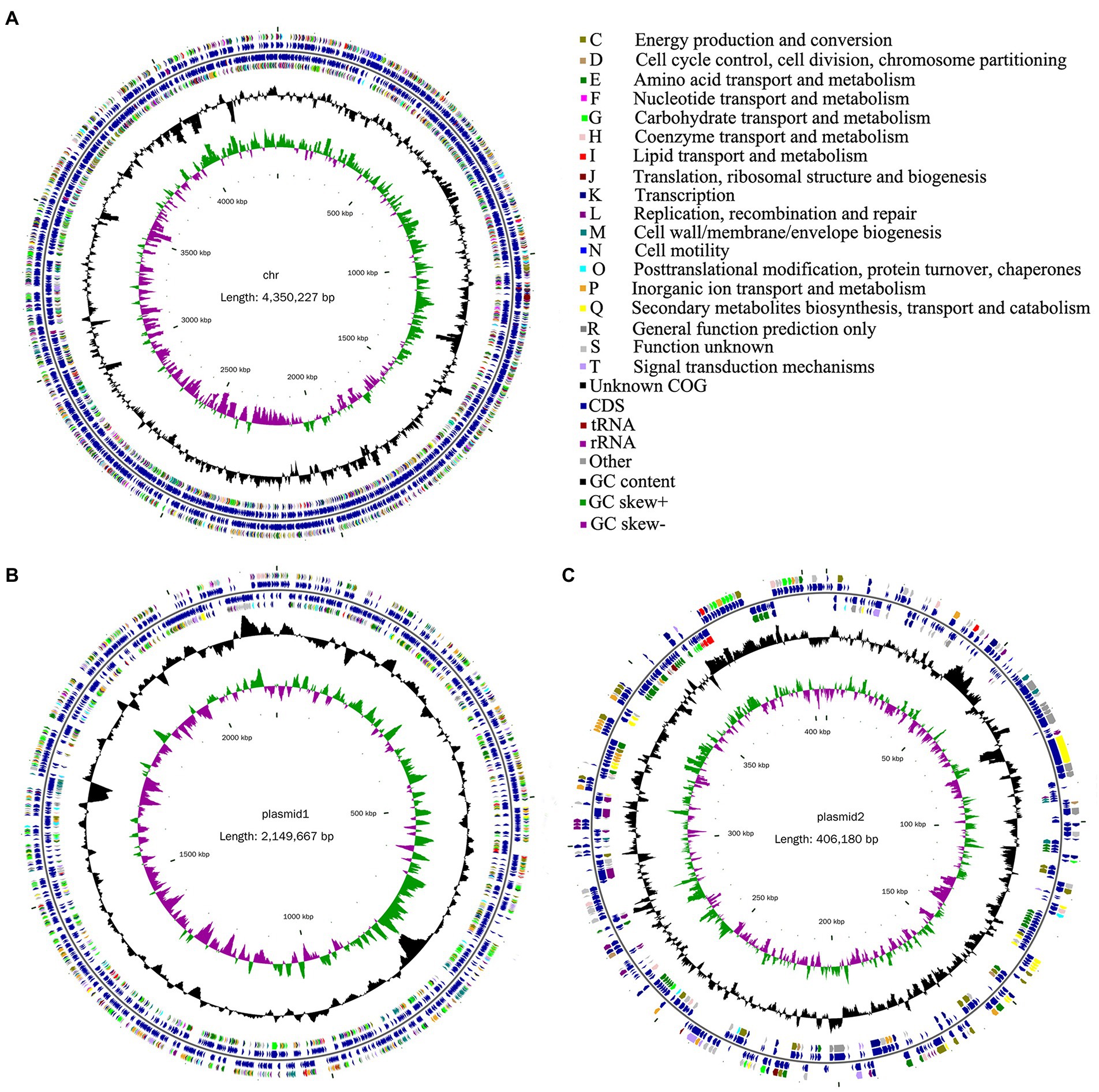
Figure 6. Circular representation of chromosome and plasmid of endophyte Rhizobium sp. WYJ-E13 strain isolated from the root of Curcuma wenyujin. (A–C) chromosome, plasmid1 and plasmid2, respectively; From the outer to the inner concentric circle: circle 1, genomic position in kilobases; circle 2, GC skew; circle 3, GC content; circle 4 and circle 7, the COG belonging to each CDS; circle 5 and circle 6, the position of CDS, tRNA and rRNA in the genome of strain WYJ-E13. Furthermore, C-T show the functional classification of the CDS genes in the chromosome and plasmid with the colors of the COG database.
CRISPRs are parts of prokaryotic DNA containing short base sequence repetitions, and CRISPR-related genes form a CRISPR-Cas system, which is an important defense system for organisms against foreign invaders. A total of three CRISPRs were also predicted from the chromosome sequence of WYJ-E13 with 81 bp shortest and 143 longest direct repeat sequences, no CRISPRs were found in both plasmid1 and plasmid2 (Table 3). Finally, a complete genome sequence of the Rhizobium sp. strain WYJ-E13 was submitted at GenBank with accession number CP076853-CP076855 (BioProject: PRJNA738292, BioSample: SAMN19717677).
Comparative genome analysis
The ANI value of Rhizobium sp. WYJ-E13 and other 15 strains were less than 95%, the highest value was 81.21% for Rhizobium acidisoli FH23 and Rhizobium phaseoli R650. The cluster analysis showed that Rhizobium sp. WYJ-E13 was a novel species (Figure 7A). Besides, three representative strains were selected for comparative genome analysis and the result of collinearity analysis was displayed (Figure 7B). In detail, 61.9% (7,633 genes), 40.8% (5,031 genes) and 60.7% (7,488 genes) of Rhizobium sp. WYJ-E13 have orthologous in the R. acidisoli FH23, Rhizobium gallicum bv. gallicum R602sp strain R602, and R. phaseoli R650, respectively.
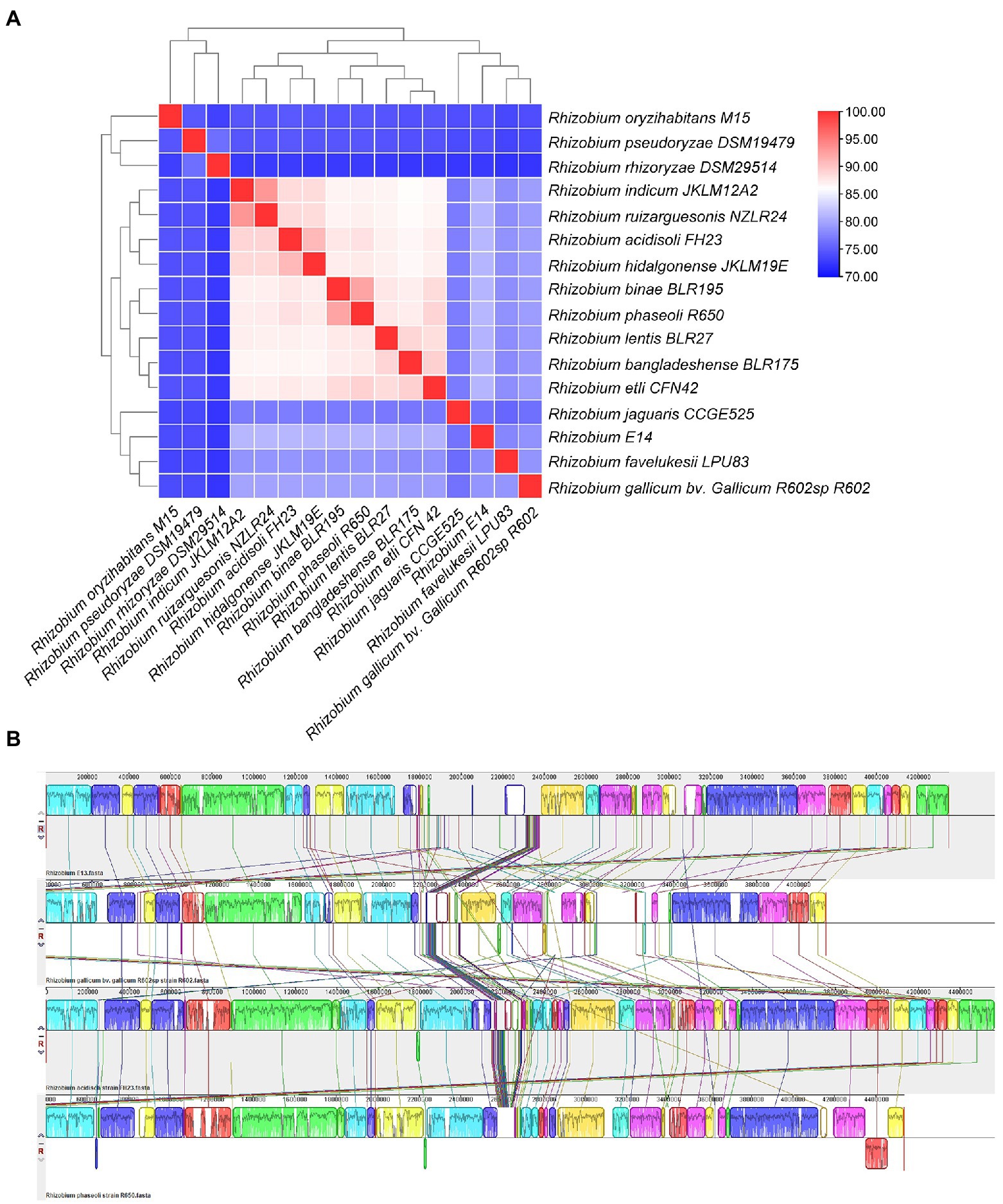
Figure 7. The average nucleotide identity analysis and comparative genome analysis. (A) The heat maps of ANI (average nucleotide identity) between strain WYJ-E13 and other 15 Rhizobium genus. (B) Genome-to-genome alignment of Rhizobium sp. WYJ-E13, Rhizobium acidisoli FH23, Rhizobium gallicum bv. gallicum R602sp strain R602 and Rhizobium phaseoli R650 used a progressive mauve software with a window of 1,000 nucleotides. WYJ-E13 was set as the reference genome. Boxes with the same color indicate the syntenic regions. Boxes below the horizontal line indicate inverted regions. Rearrangements are shown by colored lines.
Genes associated with plant growth promoting in endophytic Rhizobium sp. WYJ-E13 genome
It has been reported that endophytes could promote the fitness and growth of host plants by secreting certain metabolites, such as nutrients and hormones. Thus, investigating the genes coded specific metabolites and involved in related biological processes in the genome of Rhizobium sp. WYJ-E13 is essential.
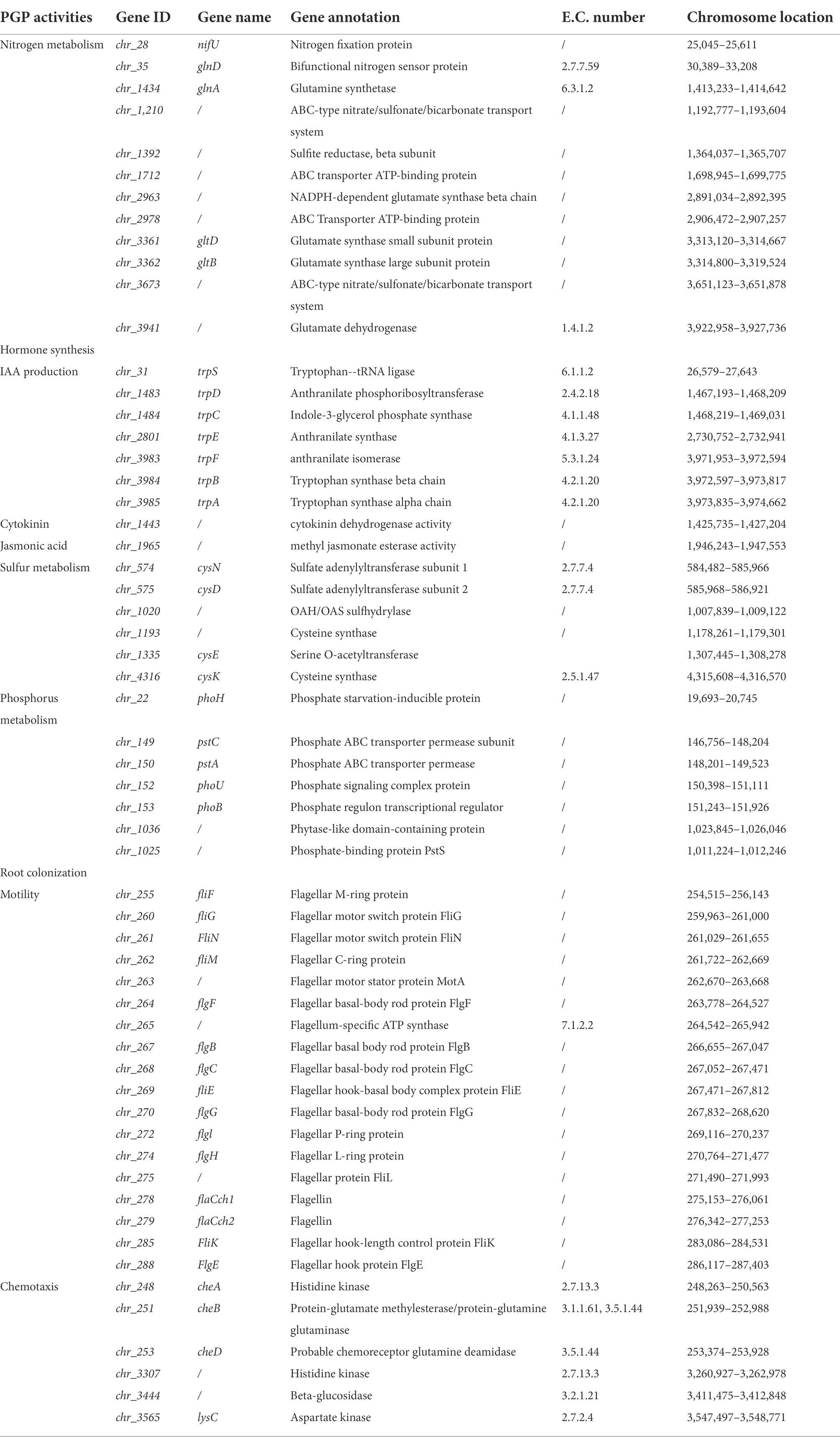
The annotation of the Rhizobium sp. strain WYJ-E13 genome identified 12 genes related to nitrogen metabolism including three nitrogen fixation genes (chr_28, chr_35 and chr_1434), three glutamate synthase large subunit (chr_2963, chr_3361 and chr_3362), four nitrate transporter (chr_1,210, chr_1712, chr_2978 and chr_3673), a ferredoxin-nitrite reductase chr_1392 and a glutamate dehydrogenase chr_3941 (Table 4).
Endophytes could enhance plant growth through the synthesis of hormone. In this study, seven IAA-related genes (chr_31, chr_1483, chr_1484, chr_2801, chr_3983, chr_3984 and chr_3985), a cytokinin gene chr_1443 and a jasmonic acid-related gene chr_1965 were annotated in the genome of WYJ-E13 (Table 4).
Sulfur and phosphorus are vital nutrients for plant development. And a total of six sulfur assimilation-genes (chr_574, chr_575, chr_1020, chr_1193, chr_1335 and chr_4316) and seven phosphorus-genes (chr_22, chr_149, chr_150, chr_152, chr_153, chr_1036 and chr_1025) were annotated in the WYJ-E13 genome, respectively (Table 4).
Furthermore, the plant growth promoting attribute of WYJ-E13 firstly depends on its migration and colonization to the interior of host plants. Accordingly, the 18 motility-related genes and six chemotaxis-related genes were found in the WYJ-E13 genome. And additional information is presented in Table 4.
Discussion
Beneficial relationship between endophytic Rhizobium sp. WYJ-E13 to host plant Curcuma
It is well-known that the symbiotic fungi or bacteria that live asymptomatically within a healthy plant tissue are called endophytes (Trivedi et al., 2020). Nowadays, an increasing body of evidence points to the important roles that endophytes could play in the host plants. Thus, plant-endophyte interactions have gradually become a research hotspot, especially focusing on the benefits to their host.
Certainly, such a beneficial relationship could be displayed from three different aspects, including the promoting growth of their host plants, the secretion of bioactive compounds to increase the stress resistance of host plants, and the accumulation of important secondary metabolites produced by endophytes (Jia et al., 2016). Accordingly, mounting researches have paid attentions to the promoting effects of different endophytes on host plant growth, such as P. variotii (Moreno-Gavíra et al., 2020), E. roggenkampii ED5 (Guo et al., 2020), P. aeruginosa B18 (Singh, P., et al., 2021), B. velezensis LDO2 (Chen et al., 2019), Microbacterium spp. and Streptomyces spp. (Kataoka et al., 2021), Rhizobium pusense MB-17a (Chaudhary et al., 2021), P. liquidambaris (Zhou et al., 2022), Bacillus altitudinis (Zhang et al., 2021; Hasan et al., 2022), and P. giganteum (Yankey et al., 2022).
In this study, the inoculation experiment of Rhizobium sp. WYJ-E13 on the C. wenyujin was firstly performed and found that WYJ-E13 could promote the root growth of its host plant compared with the control (Figure 2). The result is consistent with the common perception that most of the rhizobial species are endophytes and colonize intracellularly for root growth promotion (Mus et al., 2016). Certainly, successful colonization is the prerequisite for endophytes to work (Khare et al., 2018; Harman et al., 2021). After inoculation of WYJ-E13 to its host plant Curcuma, the result of SEM observation confirmed the consensus (Figure 3D). To further explore which metabolites produced by endophytic Rhizobium sp. WYJ-E13 have growth-promoting effects and its encoding genes, the metabolome analysis and genome sequencing were performed. A total of 30 DAMs in positive ion modes were identified (Table 1) and 26 DAMs in negative ion modes (Table 2). Importantly, hormone cytokinin B was detected in metabolome (Table 1). Furthermore, the genome of Rhizobium sp. WYJ-E13 comprised of a chromosome and two plasmids (Figure 6 and Table 3). Moreover, a larger number of genes associated with nitrogen metabolism, hormone production, sulfur/phosphorus metabolism, motility and colonization were annotated (Table 4). These above-mentioned results may be responsible for the plant growth-promoting attributes of Rhizobium sp. WYJ-E13 to its host plant C. wenyujin. In addition, the growth promoting-related genes are discussed below in detail.
Genes associated with nitrogen metabolism may play roles in growth of host plant Curcuma
Notoriously, the most efficient contribution to biologically fixed nitrogen is from symbioses between legumes and rhizobia (Pankievicz et al., 2019; Schulte et al., 2021). Recently, emerging evidence showed that some endophytes carried genes essential for biological nitrogen fixation to convert atmospheric N2 into ammonia and nitrate for secretion to the plant host (Ronson et al., 1981; Dubey et al., 2020; Mohr et al., 2021). For example, the endophytic Stenotrophomonas sp. and Pseudomonas sp. (Hazarika et al., 2021), Pseudomonassp.Y1 (Chen et al., 2022), P. cypripedii AF1 and K. arachidis EF1 (Singh, R. K., et al., 2021) are beneficial endophytes that can enhance the growth of their host plant by nitrogen fixation.
It has been reported that glutamate synthetases are the major enzymes of nitrogen metabolism (Liu et al., 2022). For example, Gene nifU is necessary for nitrogen fixation and takes part in the Fe-S cluster assembly (Smith et al., 2005). Gene glnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum (Zhang et al., 2005). Deletion of type I glutamine synthetase (glnA) could deregulate nitrogen metabolism in Clostridium thermocellum (Rydzak et al., 2017). And the role of glutamate synthase gltBD in nitrogen metabolism of Escherichia coli has been demonstrated (Pahel et al., 1978; Goss et al., 2001). Interestingly, a total of 12 genes were annotated to be related to nitrogen metabolism, such as nifU (nitrogen fixation protein), glnD (bifunctional nitrogen sensor protein), glnA (glutamine synthetase), gltD (glutamate synthase small subunit protein) and gltB (glutamate synthase large subunit protein; Table 4). These genes were annotated in the WYJ-E13 genome, which further proved that the strain is directly connected with nitrogen metabolism.
Genes associated with hormone may play roles in growth of host plant Curcuma
Endophytic bacterial strains promoted plant growth via diverse systems, such as producing IAA, cytokinin and jasmonic acid (Harman et al., 2021; Wu W. et al., 2021). For example, the plant growth-promoting ability of mycorrhizal Fusarium strain KB-3 was enhanced by its IAA producing endohyphal bacterium, K. aerogenes (Cheng et al., 2022). Serratia marcescens PLR enhanced lateral root formation through supplying PLR-derived auxin and enhancing auxin biosynthesis in Arabidopsis (Zhang et al., 2022). Certainly, the occurrence of tryptophan-linked genes was associated with IAA biosynthesis (Gupta et al., 2014; Asaf et al., 2018). For example, the tryptophan synthase β-subunit paralogs TrpB1 and TrpB2 in Thermococcus kodakarensis were both involved in tryptophan biosynthesis and indole salvage (Hiyama et al., 2014). And the genome of P. aeruginosa L10 also harbor tryptophan biosynthesis genes (trp ABCDEFG) that were responsible for IAA synthesis (Wu et al., 2018). Interestingly, seven genes were annotated to be associated with IAA production in the genome of WYJ-E13, such as trpB (tryptophan synthase beta chain), trpC (indole-3-glycerol phosphate synthase), and trpD (anthranilate phosphoribosyltransferase; Table 4).
Besides, cytokinins also play a significant role in plant growth promotion (Gao et al., 2022). For example, cytokinin production by Azospirillum brasilense contributed to increase in growth, yield, antioxidant, and physiological systems of wheat (Zaheer et al., 2022). Expectedly, a gene chr_1443 was annotated to possess cytokinin dehydrogenase activity (Table 4). In addition, Bacillus circulans GN03 altered the microbiota, and promoted cotton seedling growth by increasing the expression of phytohormone jasmonic acid synthesis (Qin et al., 2021). Similarly, a gene chr_1965 was annotated to have methyl jasmonate esterase activity (Table 4). The presence of hormone-associated genes may reveal the possible reason why WYJ-E13 promoted plant growth.
Genes associated with sulfur/phosphorus metabolism may play roles in growth of host plant Curcuma
Sulfur is one of the essential nutrients that is required for the adequate growth and development of plants (Narayan et al., 2022). For example, the phenotype of multiple Arabidopsis sulfur metabolism mutants was partially or completely rescued by the plant growth-promoting bacteria Enterobacter sp. SA187 as much as by the addition of sulfate, implying its regulation of sulfur metabolic pathways (Andrés-Barrao et al., 2021). The combined treatment of JIL321 and NaHS could further improve the growth of rice seedlings, most likely due to the interaction effect between H2S and strain JIL321 (Wang et al., 2022). In addition, the cysP gene was functionally verified with a strain E. coli transformed by expressing B. subtilis cysP gene through a mutated sulfate transport (Mansilla and de Mendoza, 2000). The operon determined by cysP gene in B. subtilis was accountable for sulfur metabolism, for example, the sulfate adenylyltransferase gene (Aguilar-Barajas et al., 2011). Coincidently, two genes cysD and cysN (sulfate adenylyltransferase subunit) were found to be annotated in the genome of WYJ-E13 (Table 4).
Furthermore, phosphorus was another vital and limiting macronutrient for plant growth. Specific bacteria played an important role in supplying accessible inorganic phosphorous in the form of orthophosphate to the plant, owing to phosphate generally existing in the soil in the form of insoluble compounds and plants are only proficient to receive free orthophosphate (Bergkemper et al., 2016). A highly efficient phosphorus solubilizing bacteria strain Pseudomonas sp. JP233 exhibited a significant plant growth-promoting effect on maize development (Yu et al., 2022). And phosphorus-related genes (pstA, pstC, phoB, phoH, phoU) were annotated in the genome of WYJ-E13 (Table 4). Moreover, phytase-producing bacteria were also proved to promote plant growth (Singh et al., 2014). Interestingly, a gene chr_1036 was annotated as phytase-like domain containing protein in the genome of WYJ-E13 (Table 4), which may further imply the possible roles of WYJ-E13 in promoting growth of host plant.
Genes associated with root colonization may play roles in growth of host plant Curcuma
Generally, beneficial microorganism colonized on the tissues of various sections in the plant. The interactions between beneficial microbes of the host plant might play an essential part in the achievement of microbial bioinoculant for promoting the growth of plant. The SEM images confirmed the colonization of Rhizobium sp. WYJ-E13 in the root of C. wenyujin (Figure 3). Furthermore, motility and chemotaxis were important characteristics for endophytes (Taghavi et al., 2010). Recently, the biofilm formation of diazotrophic bacteria Gluconacetobacter diazotrophicus was associated with the stimulation of certain compounds, and promoted its motility and colonization into the host plant, thus further improving biological nitrogen fixation with increased grain yield (Yan et al., 2022). In WYJ-E13 genome, a total of 18 genes were associated with motility (Table 4).
Besides, about half of the sequenced bacterial genomes contained genes encoding chemotactic signaling cascades (Sanchis-López et al., 2021). One of the benefits of chemotaxis accessed to nutrients and promoted its host colonization (Corral-Lugo et al., 2016). For example, the B. velezensis SQR9 was a commercially widely used plant growth-promoting rhizobacterium, its dCache LBD containing chemoreceptor McpA plays a critical role in chemotaxis to root exudates, biofilm formation, and rhizosphere colonization (Shao et al., 2015; Feng et al., 2019). Similarly, six genes were annotated to be associated with chemotaxis in the genome of WYJ-E13 (Table 4), which implied the roles of root colonization in plant growth promotion.
Conclusion
This is the first report of experimental confirmations of the endophyte Rhizobium sp. strain WYJ-E13 as a PGPR isolated from the root of medicinal plant C. wenyujin. The bacterium exhibited plant growth-promoting activities for the enhanced root growth of C. wenyujin. Additionally, hormone cytokinin produced by the strain WYJ-E13 might be one of the reasons in growth-promotion. Most important, the WYJ-E13 genome carried a total set of genes associated with nitrogen metabolism, hormone synthesis, sulfur/phosphorus metabolism and root colonization. So, it can be summarized that the WYJ-E13 strain may be used as a possible alternate for biofertilizer and play a role in growth promotion. However, field trials are required to explain the usability of the Rhizobium sp.WYJ-E13 in the field earlier than it can be established as a plant growth promoter for utilizing in agriculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP076853; https://www.ncbi.nlm.nih.gov/genbank/, CP076854; https://www.ncbi.nlm.nih.gov/genbank/, CP076855.
Author contributions
XH drafted the manuscript. ZZ and ZC provided the bioinformatics analysis. XT isolated the endophytes. JJ and CH provided the assistance in experiments. XH and TX designed the experiment, and provided the overall guidance and revision. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by grants from the National Natural Science Foundation of China (grant no. 31872181) and Hangzhou Science and Technology Development Plan (grant no. 20191203B05 and 20211231Y088).
Acknowledgments
We thank the Shanghai Bioprofile Technology Corporation for metabolome analysis and Shanghai Personalbio Technology Corporation (Shanghai, China) for the whole genome sequencing in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1035167/full#supplementary-material
Supplementary Figure S1
The 14 endophytic strains isolated from the root of medicinal plant Curcuma wenyujin.
Supplementary Figure S2
The total ion chromatograms (TICs) of the QC samples in the positive (ESI +) and negative (ESI -) ions modes.
Supplementary Figure S3
The PCA score plot of QC samples.
Supplementary Figure S4
The characterization of predicted genes against KEGG database.
Supplementary Figure S5
The characterization of predicted genes against GO database.
Supplementary Figure S6
The characterization of predicted genes against COG database.
References
Aguilar-Barajas, E., Díaz-Pérez, C., Ramírez-Díaz, M. I., Riveros-Rosas, H., and Cervantes, C. (2011). Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24, 687–707. doi: 10.1007/s10534-011-9421-x
Andrés-Barrao, C., Alzubaidy, H., Jalal, R., Mariappan, K. G., de Zélicourt, A., Bokhari, A., et al. (2021). Coordinated bacterial and plant sulfur metabolism in Enterobacter sp. SA187 induced plant salt stress tolerance. Proc. Natl. Acad. Sci. U. S. A. 118:e2107417118. doi: 10.1073/pnas.2107417118
Asaf, S., Khan, A. L., Khan, M. A., Al-Harrasi, A., and Lee, I. J. (2018). Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3Biotech 8:389. doi: 10.1007/s13205-018-1403-z
Aswathy, A. J., Jasim, B., Jyothis, M., and Radhakrishnan, E. K. (2013). Identification of two strains of Paenibacillus sp. as indole 3 acetic acid-producing rhizome-associated endophytic bacteria from Curcuma longa. 3 Biotech 3, 219–224. doi: 10.1007/s13205-012-0086-0
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bergkemper, F., Schöler, A., Engel, M., Lang, F., Krüger, J., Schloter, M., et al. (2016). Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ. Microbiol. 18, 1988–2000. doi: 10.1111/1462-2920.13188
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using diamond. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Burge, S. W., Daub, J., Eberhardt, R., Tate, J., Barquist, L., Nawrocki, E. P., et al. (2013). Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232. doi: 10.1093/nar/gks1005
Chaudhary, T., Gera, R., and Shukla, P. (2021). Deciphering the potential of Rhizobium pusense MB-17a, a plant growth-promoting root endophyte, and functional annotation of the genes involved in the metabolic pathway. Front. Bioeng. Biotech. 8:617034. doi: 10.3389/fbioe.2020.617034
Chen, L., Shi, H., Heng, J. Y., Wang, D. X., and Bian, K. (2019). Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol. Res. 218, 41–48. doi: 10.1016/j.micres.2018.10.002
Chen, Z., Zhou, W. N., Sui, X., Xu, N., Zhao, T., Guo, Z. P., et al. (2022). Plant growth-promoting rhizobacteria with ACC deaminase activity enhance maternal lateral root and seedling growth in switchgrass. Front. Plant Sci. 12:800783. doi: 10.3389/fpls.2021.800783
Cheng, S., Jiang, J. W., Tan, L. T., Deng, J. X., Liang, P. Y., Su, H., et al. (2022). Plant growth-promoting ability of mycorrhizal Fusarium strain KB-3 enhanced by its IAA producing endohyphal bacterium Klebsiella aerogenes. Front. Microbiol. 13:855399. doi: 10.3389/fmicb.2022.855399
Chin, C. S., Peluso, P., Sedlazeck, F. J., Nattestad, M., Concepcion, G. T., Clum, A., et al. (2016). Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054. doi: 10.1038/nmeth.4035
Corral-Lugo, A., Torre, J. D., Matilla, M. A., Fernández, M., Morel, B., Espinosa-Urgel, M., et al. (2016). Assessment of the contribution of chemoreceptor-based signalling to biofilm formation. Environ. Microbiol. 18, 3355–3372. doi: 10.1111/1462-2920.13170
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dubey, A., Malla, M. A., Kumar, A., Dayanandan, S., and Khan, M. L. (2020). Plants endophytes: unveiling hidden agenda for bioprospecting toward sustainable agriculture. Crit. Rev. Biotechnol. 40, 1210–1231. doi: 10.1080/07388551.2020.1808584
Elmagzob, A. A. H., Ibrahim, M. M., and Zhang, G. (2019). Seasonal diversity of endophytic bacteria associated with Cinnamomum camphora (L.) Presl. Diversity 11:112. doi: 10.3390/d11070112
Etminani, F., and Harighi, B. (2018). Isolation and identification of endophytic bacteria with plant growth promoting activity and biocontrol potential from wild pistachio trees. Plant Pathol. J. 34, 208–217. doi: 10.5423/PPJ.OA.07.2017.0158
Feng, H. C., Zhang, N., Fu, R. X., Liu, Y. P., Krell, T., Du, W. B., et al. (2019). Recognition of dominant attract ants by key chemoreceptors mediates recruitment of plant growth-promoting rhizobacteria. Environ. Microbiol. 21, 402–415. doi: 10.1111/1462-2920.14472
Ferreira, C. M. H., Soares, H. M. V., and Soares, E. V. (2019). Promising bacterial genera for agricultural practices: an insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 682, 779–799. doi: 10.1016/j.scitotenv.2019.04.225
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Frank, B. (1889). Ueber die pilzsymbiose der leguminosen. Ber. Deutsch. Bot. Ges. 7, 332–346. doi: 10.1111/j.1438-8677.1889.tb05711.x
Gao, Y. U., Zou, H., Wang, B. S., and Yuan, F. (2022). Progress and applications of plant growth-promoting bacteria in salt tolerance of crops. Int. J. Mol. Sci. 23:7036. doi: 10.3390/ijms23137036
Garcia-Fraile, P., Carro, L., Robledo, M., Bahena, M. H. R., Flores-Felix, J. D., Fernandez, M. T., et al. (2012). Rhizobium promotes non-legumes growth and quality in several production steps: towards a biofertilization of edible raw vegetables healthy for humans. PLoS One 7:e38122. doi: 10.1371/journal.pone.0038122
Goss, T. J., Perez-Matos, A., and Bender, R. A. (2001). Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes. J. Bacteriol. 183, 6607–6619. doi: 10.1128/JB.183.22.6607-6619.2001
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Guo, D. J., Singh, R. K., Singh, P., Li, D. P., Sharma, A., Xing, Y. X., et al. (2020). Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 11:580081. doi: 10.3389/fmicb.2020.580081
Gupta, A., Gopal, M., Thomas, G. V., Manikandan, V., Gajewski, J., Thomas, G., et al. (2014). Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS One 9:e104259. doi: 10.1371/journal.pone.0104259
Harman, G., Khadka, R., Doni, F., and Uphoff, N. (2021). Benefits to plant health and productivity from enhancing plant microbial symbiont. Front. Plant Sci. 11:610065. doi: 10.3389/fpls.2020.610065
Hasan, N., Khan, I. U., Farzand, A., Heng, Z., Moosa, A., Saleem, M., et al. (2022). Bacillus altitudinis HNH7 and bacillus velezensis HNH9 promote plant growth through upregulation of growth-promoting genes in upland cotton. J. Appl. Microbiol. 132, 3812–3824. doi: 10.1111/jam.15511
Hassan, S. E. (2017). Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. Adv. Res. 8, 687–695. doi: 10.1016/j.jare.2017.09.001
Hazarika, S. N., Saikia, K., Borah, A., and Thakur, D. (2021). Prospecting endophytic bacteria endowed with plant growth promoting potential isolated from Camellia sinensis. Front. Microbiol. 12:738058. doi: 10.3389/fmicb.2021.738058
Hiyama, T., Sato, T., Imanaka, T., and Atomi, H. (2014). The tryptophan synthase β-subunit paralogs TrpB1 and TrpB2 in Thermococcus kodakarensis are both involved in tryptophan biosynthesis and indole salvage. FEBS J. 281, 3113–3125. doi: 10.1111/febs.12845
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Jia, M., Chen, L., Xin, H. L., Zheng, C. J., Rahman, K., Han, T., et al. (2016). A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front. Microbiol. 7:906. doi: 10.3389/fmicb.2016.00906
Juby, S., Choyikutty, D., Nayana, A. R., Jayachandran, K., and Radhakrishnan, E. K. (2021). Quinalphos tolerant endophytic Bacillus sp. Fcl1 and its toxicity-alleviating effect in Vigna unguiculata. Curr. Microbiol. 78, 904–910. doi: 10.1007/s00284-020-02317-4
Kataoka, R., Akashi, M., Taniguchi, T., Kinose, Y., Yaprak, A. E., and Turgay, O. C. (2021). Metabolomics analyses reveal metabolites affected by plant growth-promoting endophytic bacteria in roots of the halophyte Mesembryanthemum crystallinum. Int. J. Mol. Sci. 22:11813. doi: 10.3390/ijms222111813
Kaul, S., Sharma, T., and Dhar, M. K. (2016). “Omics” tools for better understanding the plant-endophyte interactions. Front. Plant Sci. 7:955. doi: 10.3389/fpls.2016.00955
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. doi: 10.3389/fmicb.2018.02732
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Li, Y. H., Wu, Y. C., Li, Y. M., and Guo, F. J. (2021). Review of the traditional uses, phytochemistry, and pharmacology of Curcuma wenyujin Y. H. Chen et C. Ling. J. Ethnopharmacol. 269:113689. doi: 10.1016/j.jep.2020.113689
Liu, X. J., Hu, B., and Chu, C. C. (2022). Nitrogen assimilation in plants: current status and future prospects. J. Genet. Genomics 49, 394–404. doi: 10.1016/j.jgg.2021.12.006
Liu, F. P., Liu, H. Q., Zhou, H. L., Dong, Z. G., Bai, X. H., Bai, P., et al. (2014). Isolation and characterization of phosphate solubilizing bacteria from betel nut (Areca catechu) and their effects on plant growth and phosphorus mobilization in tropical soils. Biol. Fertil. Soils 50, 927–937. doi: 10.1007/s00374-014-0913-z
Liu, R., Pei, Q., Shou, T., Zhang, W. J., Hu, J., and Li, W. (2019). Apoptotic effect of green synthesized gold nanoparticles from Curcuma wenyujin extract against human renal cell carcinoma A498 cells. Int. J. Nanomed. 14, 4091–4103. doi: 10.2147/IJN.S203222
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Luo, R. B., Liu, B. H., Xie, Y. L., Li, Z. Y., Huang, W. H., Yuan, J. Y., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18
Manasa, K., Reddy, S., and Triveni, S. (2017). Characterization of potential PGPR and antagonistic activities of Rhizobium isolates from different rhizosphere soils. J. Pharmacogn. Phytochem. 6, 51–54. doi: 10.20546/ijcmas.2017.605.316
Mansilla, M. C., and de Mendoza, D. (2000). The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (pit) family. Microbiology 146, 815–821. doi: 10.1099/00221287-146-4-815
Mohr, W., Lehnen, N., Ahmerkamp, S., Marchant, H. K., Graf, J. S., Tschitschko, B., et al. (2021). Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 600, 105–109. doi: 10.1038/s41586-021-04063-4
Moreno-Gavíra, A., Diánez, F., Sánchez-Montesinos, B., and Santos, M. (2020). Paecilomyces variotii as a plant-growth promoter in horticulture. Agronomy 10:597. doi: 10.3390/agronomy10040597
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Moszer, I. (1998). The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 430, 28–36. doi: 10.1016/S0014-5793(98)00620-6
Mus, F., Crook, M. B., Garcia, K., Costas, A. G., Geddes, B. A., Kouri, E. D., et al. (2016). Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 82, 3698–3710. doi: 10.1128/AEM.01055-16
Narayan, O. P., Kumar, P., Yadav, B., Dua, M., and Johri, A. K. (2022). Sulfur nutrition and its role in plant growth and development. Plant Signal. Behav. 7:2030082. doi: 10.1080/15592324.2022.2030082
Oteino, N., Lally, R. D., Kiwanuka, S., Lloyd, A., Ryan, D., Germaine, K. J. N., et al. (2015). Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 6:745. doi: 10.3389/fmicb.2015.00745
Pahel, G., Zelenetz, A. D., and Tyler, B. M. (1978). gltB gene and regulation of nitrogen metabolism by glutamine synthetase in Escherichia coli. J. Bacteriol. 133, 139–148. doi: 10.1128/jb.133.1.139-148.1978
Pankievicz, V. C. S., Irving, T. B., Maia, L. G. S., and Ané, J. M. (2019). Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol. 17:99. doi: 10.1186/s12915-019-0710-0
Pravin, V., Rosazlin, A., Tumirah, K., Ismail, S., and Boyce, A. N. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 21:573. doi: 10.3390/molecules21050573
Qin, S., Feng, W. W., Wang, T. T., Ding, P., Xing, K., and Jiang, J. H. (2017). Plant growth-promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant Soil 416, 117–132. doi: 10.1007/s11104-017-3192-2
Qin, L. J., Tian, P. D., Cui, Q. Y., Hu, S. P., Jian, W., Xie, C. J., et al. (2021). Bacillus circulans GN03 alters the microbiota, promotes cotton seedling growth and disease resistance, and increases the expression of phytohormone synthesis and disease resistance-related genes. Front. Plant Sci. 12:644597. doi: 10.3389/fpls.2021.644597
Richter, M., Rossello-Mora, R., Oliver Glockner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Ronson, C. W., Lyttleton, P., and Robertson, J. G. (1981). C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. U. S. A. 78, 4284–4288. doi: 10.1073/pnas.78.7.4284
Rydzak, T., Garcia, D., Stevenson, D. M., Sladek, M., Klingeman, D. M., Holwerda, E. K., et al. (2017). Deletion of type I glutamine synthetase deregulates nitrogen metabolism and increases ethanol production in Clostridium thermocellum. Metab. Eng. 41, 182–191. doi: 10.1016/j.ymben.2017.04.002
Sanchis-López, C., Cerna-Vargas, J. P., Santamaría-Hernando, S., Ramos, C., Krell, T., Rodríguez-Palenzuela, P., et al. (2021). Prevalence and specificity of chemoreceptor profiles in plant-associated bacteria. mSystems 6:e0095121. doi: 10.1128/mSystems.00951-21
Schubert, M., Lindgreen, S., and Orlando, L. (2016). AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC. Res. Notes 9:88. doi: 10.1186/s13104-016-1900-2
Schulte, C. M. C., Borah, K., Wheatley, R. M., Terpolilli, J. J., Saalbach, G., Crang, N., et al. (2021). Metabolic control of nitrogen fixation in rhizobium-legume symbioses. Sci. Adv. 7:eabh2433. doi: 10.1126/sciadv.abh2433
Shao, J. H., Xu, Z. H., Zhang, N., Shen, Q. R., and Zhang, R. F. (2015). Contribution of indole-3-acetic acid in the plant growth promotion by the rhizospheric strain Bacillus amyloliquefaciens SQR9. Biol. Fert. Soils 51, 321–330. doi: 10.1007/s00374-014-0978-8
Singh, P., Kumar, V., and Agrawal, S. (2014). Evaluation of phytase producing bacteria for their plant growth promoting activities. Int. J. Microbiol. 2014:426483. doi: 10.1155/2014/426483
Singh, R. K., Singh, P., Guo, D. J., Sharma, A., Li, D. P., Li, X. K., et al. (2021). Root-derived endophytic diazotrophic bacteria Pantoea cypripedii AF1 and Kosakonia arachidis EF1 promote nitrogen assimilation and growth in sugarcane. Front. Microbiol. 12:774707. doi: 10.3389/fmicb.2021.774707
Singh, P., Singh, R. K., Guo, D. J., Sharma, A., Singh, R. N., Li, D. P., et al. (2021). Whole genome analysis of sugarcane root-associated endophyte Pseudomonas aeruginosa B18-a plant growth-promoting bacterium with antagonistic potential against Sporisorium scitamineum. Front. Microbiol. 12:628376. doi: 10.3389/fmicb.2021.628376
Smith, A. D., Jameson, G. N. L., Santos, P. C. D., Agar, J. N., Naik, S., Krebs, C., et al. (2005). NifS-mediated assembly of [4Fe-4S] clusters in the N-and C-terminal domains of the NifU scaffold protein. Biochemistry 44, 12955–12969. doi: 10.1021/bi051257i
Song, Y. L., Wu, P., Li, Y. F., Tong, X. X., Zheng, Y. F., Chen, Z. H., et al. (2017). Effect of endophytic fungi on the host plant growth, expression of expansin gene and flavonoid content in Tetrastigma hemsleyanum Diels & Gilg ex Diels. Plant Soil 417, 393–402. doi: 10.1007/s11104-017-3266-1
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Taghavi, S., van der Lelie, D., Hoffman, A., Zhang, Y. B. D., Walla, M., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp.638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Tritt, A., Eisen, J. A., Facciotti, M. T., and Darling, A. E. (2017). An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7:e42304. doi: 10.1371/journal.pone.0042304
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Vinayarani, G., and Prakash, H. S. (2018). Growth promoting rhizospheric and endophytic bacteria from Curcuma longa L. as biocontrol agents against rhizome rot and leaf blight diseases. Plant Pathol. J. 34, 218–235. doi: 10.5423/PPJ.OA.11.2017.0225
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2017). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, G., Li, B. W., Peng, D. L., Zhao, H. Z., Lu, M. Y., Zhang, L. S., et al. (2022). Combined application of H2S and a plant growth promoting strain JIL321 regulates photosynthetic efficacy, soil enzyme activity and growth-promotion in rice under salt stress. Microbiol. Res. 256:126943. doi: 10.1016/j.micres.2021.126943
Wu, W., Chen, W. H., Liu, S. Y., Wu, J. J., Zhu, Y. T., Qin, L. P., et al. (2021). Beneficial relationships between endophytic bacteria and medicinal plants. Front. Plant Sci. 12:646146. doi: 10.3389/fpls.2021.646146
Wu, P., Dong, X. M., Song, G. Q., Wei, M. M., Fang, C., Zheng, F. B., et al. (2021). Bioactivity-guided discovery of quality control markers in rhizomes of Curcuma wenyujin based on spectrum-effect relationship against human lung cancer cells. Phytomedicine 86:153559. doi: 10.1016/j.phymed.2021.153559
Wu, J. J., Tang, X. J., Shi, Y., Ma, C. J., Zhang, H. Y., Zhang, J. H., et al. (2022). Crosstalk of LncRNA HOTAIR and SP1-mediated repression of PDK1 contributes to β-elemene-inhibited proliferation of hepatocellular carcinoma cells. J. Ethnopharmacol. 283:114456. doi: 10.1016/j.jep.2021.114456
Wu, T., Xu, J., Xie, W. J., Yao, Z. G., Yang, H. J., Sun, C. L., et al. (2018). Pseudomonas aeruginosa L10: a hydrocarbon-degrading, biosurfactant-producing, and plant-growth-promoting endophytic bacterium isolated from a reed (Phragmites australis). Front. Microbiol. 9:1087. doi: 10.3389/fmicb.2018.01087
Yan, D. W., Tajima, H., Cline, L. C., Fong, R. Y., Ottaviani, J. I., Shapiro, H. Y., et al. (2022). Genetic modification of flavone biosynthesis in rice enhances biofilm formation of soil diazotrophic bacteria and biological nitrogen fixation. Plant Biotechnol. J. 1–14. doi: 10.1111/pbi.13894
Yankey, R., Omoor, I. N. A., Karanja, J. K., Wang, L., Urga, R. T., Fang, C. H., et al. (2022). Metabolic properties, gene functions, and biosafety analysis reveal the action of three rhizospheric plant growth-promoting bacteria of Jujuncao (Pennisetum giganteum). Environ. Sci. Pollut. Res. Int. 29, 38435–38449. doi: 10.1007/s11356-021-17854-z
Yu, H. Y., Wu, X. Q., Zhang, G. Z., Zhou, F. Y., Harvey, P. R., Wang, L. L., et al. (2022). Identification of the phosphorus-solubilizing bacteria strain JP233 and its effects on soil phosphorus leaching loss and crop growth. Front. Microbiol. 13:892533. doi: 10.3389/fmicb.2022.892533
Zaheer, M. S., Ali, H. H., Iqbal, M. A., Erinle, K. O., Javed, T., Iqbal, J., et al. (2022). Cytokinin production by Azospirillum brasilense contributes to increase in growth, yield, antioxidant, and physiological systems of wheat (Triticum aestivum L.). Front. Microbiol. 13:886041. doi: 10.3389/fmicb.2022.886041
Zhang, Y. P., Pohlmann, E. L., and Roberts, G. P. (2005). GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J. Bacteriol. 187, 1254–1265. doi: 10.1128/JB.187.4.1254-1265.2005
Zhang, D. N., Xu, H. L., Gao, J. Y., Portieles, R., Du, L. H., Gao, X. Y., et al. (2021). Endophytic Bacillus altitudinis strain uses different novelty molecular pathways to enhance plant growth. Front. Microbiol. 12:692313. doi: 10.3389/fmicb.2021.692313
Zhang, C. L., Yu, Z. P., Zhang, M. Y., Li, X. X., Wang, M. J., Li, L. X., et al. (2022). Serratia marcescens PLR enhances lateral root formation through supplying PLR-derived auxin and enhancing auxin biosynthesis in Arabidopsis. J. Exp. Bot. 73, 37611–33725. doi: 10.1093/jxb/erac074
Keywords: Curcuma wenyujin, nitrogen metabolism, Rhizobium sp., genome, plant growth-promoting rhizobacteria
Citation: Huang X, Zeng Z, Chen Z, Tong X, Jiang J, He C and Xiang T (2022) Deciphering the potential of a plant growth promoting endophyte Rhizobium sp. WYJ-E13, and functional annotation of the genes involved in the metabolic pathway. Front. Microbiol. 13:1035167. doi: 10.3389/fmicb.2022.1035167
Edited by:
Rouhallah Sharifi, Razi University, IranReviewed by:
Abozar Ghorbani, Shiraz University, IranAnastasia Venieraki, Agricultural University of Athens, Greece
Copyright © 2022 Huang, Zeng, Chen, Tong, Jiang, He and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taihe Xiang, eHRoY25AMTYzLmNvbQ==
 Xiaoping Huang
Xiaoping Huang Zhanghui Zeng
Zhanghui Zeng Zhehao Chen1,2
Zhehao Chen1,2 Taihe Xiang
Taihe Xiang