
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 15 December 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1034674
This article is part of the Research Topic Women in Microbial Physiology and Metabolism: 2022 View all 12 articles
Methanogens are anaerobic archaea which conserve energy by producing methane. Found in nearly every anaerobic environment on earth, methanogens serve important roles in ecology as key organisms of the global carbon cycle, and in industry as a source of renewable biofuels. Environmentally, methanogenic archaea play an essential role in the reintroducing unavailable carbon to the carbon cycle by anaerobically converting low-energy, terminal metabolic degradation products such as one and two-carbon molecules into methane which then returns to the aerobic portion of the carbon cycle. In industry, methanogens are commonly used as an inexpensive source of renewable biofuels as well as serving as a vital component in the treatment of wastewater though this is only the tip of the iceberg with respect to their metabolic potential. In this review we will discuss how the efficient central metabolism of methanoarchaea could be harnessed for future biotechnology applications.
Methanogens are single-celled organisms that conserve energy via the conversion of substrate carbon compounds into methane gas (Ferry, 2012). The majority of methanogens subsist in anaerobic environments by the reduction of one carbon (C1) compounds including carbon dioxide and carbon monoxide, methanol, methylamines, and methyl sulfides as well as the fermentation of acetate (Daniels et al., 1977; Rother and Metcalf, 2004; Buan, 2018). The gaseous methane they produce then bubbles back into the aerobic world where it is consumed by methanotrophic organisms and is returned to the carbon cycle. The methane produced by methanogens is of interest due to methane’s ecological impact resulting from agricultural production by livestock (Johnson and Johnson, 1995) and rice cultivation (Schütz et al., 1989) as well as methane’s benefits as a renewable source of natural gas (Luo and Angelidaki, 2012; Huang et al., 2017) which is a high energy fuel used for heat, electricity generation, and for transportation including as a propellant for rocket engines (Neill et al., 2009; Sheehan, 2021). In nature, methanogenic archaea have been identified in environments spanning the boundaries of life sustaining conditions, from acidic to alkaline (pH 3.0–10.2), thermophilic to psychrophilic (−2°C to 110°C), and including both fresh and saline aquatic environments (Martin and Sousa, 2016). In addition to these environments, methanogens are found symbiotically communing in a wide range of single-and multi-cellular hosts ranging from amoebae (Holmes et al., 2014) and protozoa (Stumm and Zwart, 1986) to termites, (Brune, 1998, 2018) bovines (Whitford et al., 2001), and humans (Fricke et al., 2006; Rajilić-Stojanović et al., 2007).
As more methanogen species are discovered, it is becoming evident that methanogens may be able to use a wider variety of substrates than previously known. Biomethane generation has been observed from subsurface coal beds (Ulrich and Bower, 2008; Mayumi et al., 2016) as well as oceanic oil sinks (Laso-Pérez et al., 2019; Zhou et al., 2022). Methanogens may form syntrophic partnerships with other microorganisms such as hydrocarbon-degrading bacteria, thereby indirectly facilitating the reintroduction of crude oil carbon into a bioavailable state (Zengler et al., 1999; Dolfing et al., 2008; Jones et al., 2008). However, methanogens are suspected of being capable of alkane oxidation independent of any other archaeal or bacterial partner (Borrel et al., 2019; Laso-Pérez et al., 2019). Ecological methane accumulation has been observed in correlation with colonized oil droplets at deep-sea oil seeps. These proposed alkane utilizing methanogens are not limited to short-chain alkanes; Candidatus Methanoliparum has been shown to degrade long-chain hydrocarbons with methanogenesis (Zhou et al., 2022). The mechanism by which these so far uncultured alkanotrophic methanogens are capable of utilizing hydrocarbons is still being investigated, though the phenomenon does not appear to be a rare occurrence. Alkane-degrading methanogens are widely distributed, (Zengler et al., 1999; Laso-Pérez et al., 2019; Zhou et al., 2022) indicating that methanogens are directly or indirectly involved in the bioconversion of crude oil to methane on a large scale and may serve a benefit to bioremediation efforts in anaerobic environments such as deep-sea sediments.
The ability for methanogens to thrive in these diverse environments is testament to their metabolic robustness. Regardless of the environment they inhabit, methanogens share a similar metabolic niche, the bioconversion of low-energy substrates into biomass and high-energy molecules with a high degree of efficiency. All cultured methanogens to date are strict obligate anaerobes and produce methane as an essential byproduct of metabolism (Daniels et al., 1977; Rother and Metcalf, 2004; Ferry, 2012; Buan, 2018). To grow on these energy poor substrates methanogens have adopted a highly efficient pathway for conserving energy called methanogenesis (Figure 1; Thauer, 2012; Gonnerman et al., 2013). In this review we discuss how methanogen metabolism allows these organisms to thrive under strict energetic conditions and how their special metabolic features could be utilized in biotechnology.
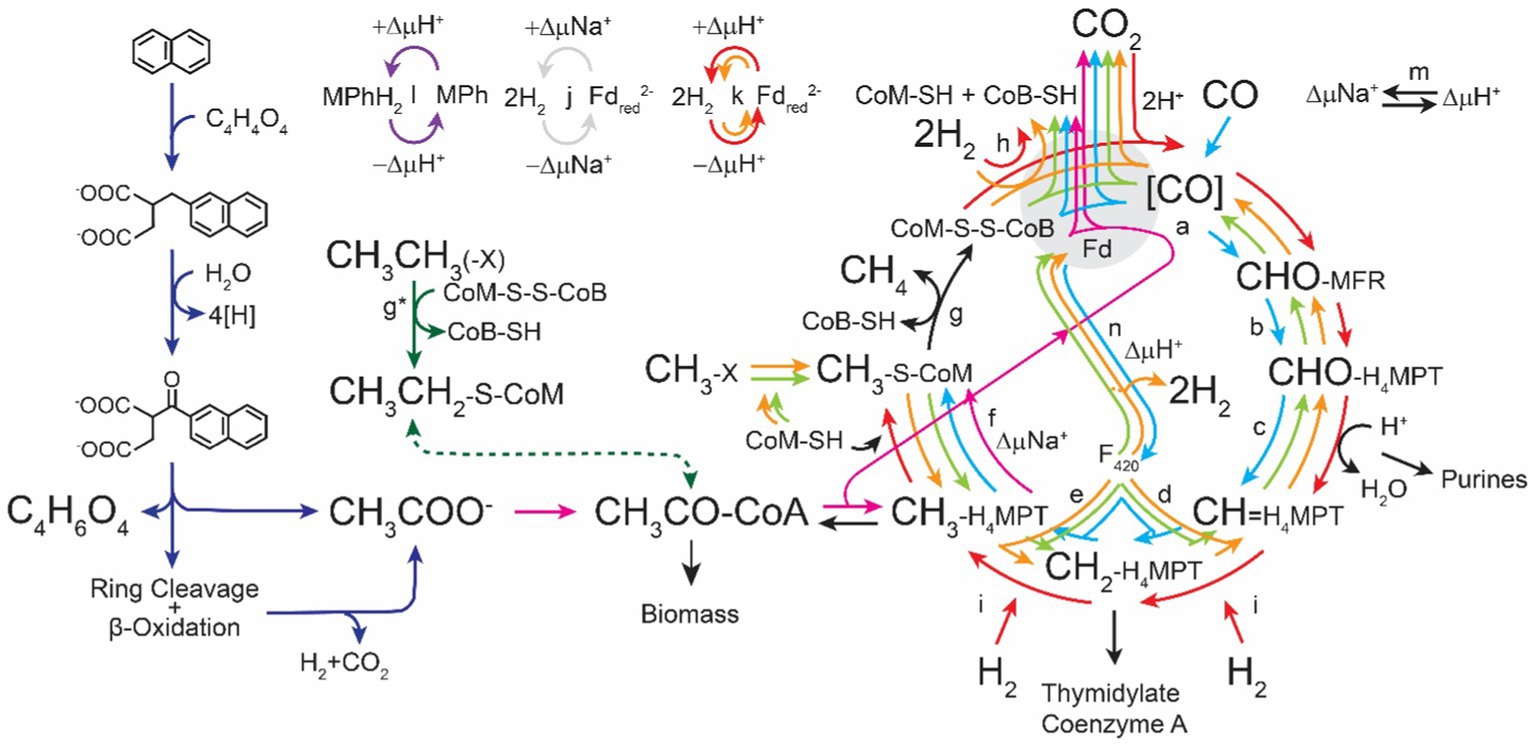
Figure 1. Pathways for methanogenesis (adapted from Buan, 2018). The direction of arrows represents the direction of biochemical reactions. Reactions which are utilized in every methanogenic pathway are represented in black. Hydrogenotrophic methanogenesis (aka. The Wolfe Cycle) (Thauer, 2012) is represented in red. Methyl oxidation is represented in orange. Methylotrophic methanogenesis is represented in green. Acetotrophic methanogenesis is represented in fuchsia. Degradation of polyaromatic hydrocarbons is represented in dark blue (Siegert et al., 2011). Ethylene and long chain alkane reduction is represented in purple (Lemaire and Wagner, 2022). Carboxydotrophic methanogenesis is represented in cyan. CoB-SH, coenzyme B thiol; CoM-SH, coenzyme M thiol; CoM-S-S-CoB, coenzyme M-coenzyme B heterodisulfide; Fd, ferredoxin; Fdred, reduced ferredoxin; H4MPT, tetrahydromethanopterin; MFR, methanofuran; MPh, methanophenazine; MPhH2, reduced methanophenazine. Enzymes involved in methanogenesis: (a) Formyl-methanofuran dehydrogenase (Fmd), (b) Formyl-methanofuran:H4MPT formyl transferase (Ftr), (c) Methenyl-H4MPT cyclohydrolase (Mch), (d) F420-dependent Methylene-H4MPT dehydrogenase (Mtd), (e) F420-dependent Methylene-H4MPT reductase (Mer), (f) Methyl-H4MPT:coenzyme M methyltransferase (Mtr), (g) Methyl-coenzyme M reductase (Mcr), (g*) Atypical methyl-coenzyme M reductase (Mcr),(Wang et al., 2019) (h) Electron-bifurcating hydrogenase:heterodisulfide reductase complex (Mvh:HdrABC), (i) F420-reducing hydrogenase (Frh), (j) Energy-converting sodium pumping ferredoxin hydrogenase, (k) Ferredoxin reducing hydrogenase (Eha/Ech), (l) Proton-translocating methanophenazine:heterodisulfide reductase (HdrED), (m) Sodium–proton antiporter (MrpA), (n) F420 proton-pumping methanophenazine reductase (Fpo).
Despite their ability to live in a wide diversity of habitats, methanogens are united by their unique central metabolism. In the five characterized versions of the methanogenesis pathway, substrates are reduced to methane while formate, primary alcohols/amines/thiols, or H2 are oxidized to CO2 or H2O (Ermler et al., 1997; Buan, 2018). Redox cofactors associated with the methanogenesis pathway are regenerated through formation of a transmembrane ion gradient which is coupled to ATP synthesis via ATP synthase (Costa and Leigh, 2014; Diender et al., 2015). These reactions yield a small amount of energy for the methanogen amounting to between 0.5 and 2 moles of ATP per mole of substrate (Buan, 2018). A result of this low energy yield is a high relative flux through energy conservation pathways, with over 99% of the chemistry within the cell being directly tied to methanogenesis (Feist et al., 2006). The remaining 1–2% of carbon substrate is used to generate biomass for replication. The average macromolecular composition of a methanogen includes 63% protein, 0.1% fatty acid lipids, 5% isoprenoid lipids, 0.5% carbohydrates, 28% nucleic acids, and 4% metabolites and metabolic precursors (Gonnerman et al., 2013). The relatively high abundance of isoprenoid lipids and high protein concentration make them an appealing source of difficult-to-synthesize lipids and molecules from inexpensive C1 compounds or acetate, yields and titers of which could be further enhanced through genetic engineering. The unique properties of methanogenesis and highly efficient energy conservation mechanisms make methanoarchaea ideal organisms for the production of renewable biofuels as the vast majority of feed substrate is converted efficiently to methane.
It should be noted, however, that while methanogenesis is highly conserved and exceedingly efficient, it can also be modified to better serve biotechnological goals without necessarily undermining methanogenic growth. Methanosarcina in particular may be well-suited to metabolic engineering, as they can use multiple methanogenesis pathways and are genetically tractable (Metcalf et al., 1997; Ehlers et al., 2005).
Methanogenesis is inherently limited by substrate availability though this limitation can be overcome by expanding the carbon and energy sources available to methanoarchaea. Methanosarcina acetivorans has been successfully engineered to expand its substrate use and to enhance metabolic efficiency. M. acetivorans is a marine methanogen that can use methylotrophic and acetotrophic methanogenesis pathways, but unusually cannot use H2 for methanogenesis (Sowers et al., 1984; Guss et al., 2009). As a result, M. acetivorans appears to use very efficient intracellular redox balancing mechanisms, thus avoiding loss of H2 reducing equivalents by gas diffusion, which is a possibility for methanogens that use H2 cycling to generate transmembrane proton gradients (Kulkarni et al., 2009). Methylotrophic methanogenesis relies on substrate specific methyltransferases to convert substrates to CH3-CoM for entry into the pathway. It has been demonstrated that heterologous expression of the bacterial broad-specificity esterase from Pseudomonas veronii in M. acetivorans increased esterase activity 80-fold and greatly enhanced growth on methyl acetate and methyl propionate substrates (Lessner et al., 2010). Once substrates have entered the methanogenesis pathway, energy conservation occurs by the regeneration of methanogenic cofactors (Thauer, 2012). Cofactor regeneration is catalyzed by membrane bound, redox-driven enzyme complexes such as Rnf (Schlegel et al., 2012) and HdrED (Duszenko and Buan, 2017), which combine cofactor regeneration with ion transport or by cytoplasmic enzymes such as Fpo (Welte and Deppenmeier, 2011), or the terminal oxidase HdrABC (Catlett et al., 2015; Buckel and Thauer, 2018). By enhancing cofactor regeneration it is possible to stimulate increased methanogenesis. For example, when the cytoplasmic enzyme heterodisulfide reductase (HdrABC) is overexpressed, methane production on methanol is 30% faster without a detectable change in growth rate compared to the parent strain (Catlett et al., 2015). The exogenous addition of methanophenzine (MPh), an electron carrier found in methanogens which fulfils a similar role as quinones in other electron transport chains, was found to significantly increase growth in Methanosarcina spp (Duszenko and Buan, 2017). Additionally, it is possible that the regeneration of methanogenic cofactors could be achieved through pathway engineering (Aldridge et al., 2021). The reduction of the disulfide complex between coenzyme M and coenzyme B is the final step in all methanogenic pathways and is restricted to the heterodisulfide reductases HdrABC and HdrED (Buan and Metcalf, 2010; Yan and Ferry, 2018). Providing an alternative means of cofactor reduction would eliminate this metabolic bottleneck, freeing up cofactors at a greater rate (Aldridge et al., 2021). If a methanogen were engineered to produce a non-native metabolite which allows for the reduction of ferredoxin, F420, coenzyme M, or coenzyme B then production of that metabolite has the potential to increase the rate of methanogenesis while also synthesizing the desired product (Aldridge et al., 2021).
Due to the tight energetic restrictions methanogenesis is proposed to rely heavily on substrate channeling to minimize entropic effects (Costa et al., 2010; Matschiavelli et al., 2012; Catlett et al., 2015; Yan and Ferry, 2018; Watanabe et al., 2021). Substrate channeling allows methanogenesis to function efficiently but presents challenges for metabolic engineers as the metabolite pools for methanogenesis have limited availability outside of the channeled enzyme complexes. To overcome this metabolic obstacle metabolic engineers must choose products which draw from metabolites which are not directly channeled or incorporate the production of their products within methanogenesis. Table 1 lists potential strategies to increase substrate variety, optimize growth rates and culture conditions, or generate new metabolic products by engineering methanogenesis.
Given the efficiency of methanogenesis and the abundance of anaerobic environments around the world, methanogens are distributed across every continent. Yet of the approximately 1 Gt of methane produced by methanogens in the wild each year in anaerobic and microanaerobic environments, roughly half escapes into the aerobic carbon cycle (Conrad, 2009). It is estimated between 43–90% of biogenic methane is oxidized my aerobic methanotrophs at the anaerobic/aerobic interface (Hao et al., 1988; Roslev and King, 1995; Le Mer and Roger, 2001; Conrad, 2009). The remainder of this methane is either trapped within anaerobic environments (as gas or methane gas hydrates) or consumed by methanotrophic archaea and bacteria (Knittel and Boetius, 2009; Thauer, 2011). Given the estimated 70Gt of CO2 fixed by photosynthesis into biomass, methane as a product of methanogenesis accounts for approximately 2% of the annual total carbon utilization (Thauer et al., 2008). Previously it was believed that the anaerobic oxidation of methane (AOM) was possible through the symbiotic exchange of metabolites and electrons between the methanotrophic archaea and sulfate reducers (Alperin and Hoehler, 2009; Summers et al., 2010). Within this process anaerobic methane-oxidizing archaea (ANME) consisting of Methanomicrobiales (ANME-1) and Methanosarcinales (ANME-2 and ANME-3) form granular aggregates with delta-proteobacteria in which electrons are transferred between organisms via multi-heme cytochromes (McGlynn et al., 2015). Metabolic modeling has suggested that iron and sulfate can be co-substrates in AOM (Riedinger et al., 2014) and 16S rRNA gene-sequences for Candidadus Methanoperedens correlated with increased AOM in sulfate-rich anoxic sediments suggesting the occurrence of AOM independent of a bacterial partner (Su et al., 2020). In laboratory conditions it was found that trace amounts of AOM was observed in Methanothermobacter marburgenis (Scheller et al., 2010) and Methanosarcina acetivorans (Moran et al., 2005) though it was not observed that these strains were able to use methane as the major source of carbon and energy for growth. However, by scouring the metagenomes of unculturable ANME-1 samples from aquatic regions with high amounts of AOM, a novel variant of methyl-coenzyme M reductase (Mcr) was discovered which correlated to AOM without the need for a syntrophic sulfate-reducing partner (Meyerdierks et al., 2010; Shima et al., 2012). When the uncultured ANME-1 Mcr was introduced into M. acetivorans it was found that isotope labeled methane was converted into acetate while also facilitating growth, (Soo et al., 2016). Furthermore, methanogen strains containing this ANME-1 Mcr gene can be utilized along with a consortia of microbes including Geobacter sulfurreducens to produce electricity in a microbial fuel cell utilizing only methane as a substrate (McAnulty et al., 2017). As every step of methanogenesis is reversible, reverse methanogenesis is theoretically possible for any methanogen though under most conditions these reactions are non-energy yielding (Thauer, 2012). These observations indicate that the bidirectionality of methanogenesis enables methane to be utilized as growth substrate for methanogens, particularly by Methanosarcina spp. For example: a Methanosarcina culture which has been engineered to produce a high-value terpenoid product is grown using methyl compounds until stationary phase is achieved and biomass accumulation is no longer necessary; this culture could then be induced to produce the terpenoids utilizing potentially any C1 compound or mixtures of compounds including CO, CO2, or CH4 based on substrate availability. This potential extends beyond the production of secreted products, as the biomass of methanogens itself can be utilized as a source of valuable lipids.
Methanogen membranes, like those found in all archaea, are distinct from those found in bacteria and eukarya. In bacterial and eukaryotic organisms lipid membrane structures are composed of fatty acid chains ester liked to glycerol-3-phosphate (G3P) (Koga and Morii, 2007). Archaeal lipids membranes instead utilize isoprenoid alkyl chains ether linked to glycerol-1-phosphate (G1P; Figure 2; Koga and Morii, 2007; Koga, 2012). This fundamental differentiation in membrane composition is the basis of the so called ‘lipid divide’ separating archaea from the other two domains of life (Villanueva et al., 2021). Given the high quantity and the molecular uniformity of lipid membranes, comprising on average 5% of total methanogen dry weight (Gonnerman et al., 2013), and the relatively high metabolic flux through the archaeal mevalonate lipid biosynthesis pathway, high-value isoprenoid lipids are attractive metabolic engineering targets. The isoprenoid lipids used by archaea allow them to tolerate a wide range of environmental stressors. The most abundant archaeal lipid structures are archeol, consisting of a pair of phytanyl chains ether linked to G1P and caldarcheol, a cyclic dimer of archeol. Caldarcheol is of particular biotechnological interest as the cyclized tetraether lipids maintain cellular homeostasis in the presence of extreme pH and thermal stress (Boyd et al., 2013; Siliakus et al., 2017). Archaeal ether linked lipids are more stable than ester linked membranes when exposed to extremes of pH and thermal conditions, and the unique monolayer structure of tetraether linked lipids imparts resistance to degradation to phospholipases (Jacquemet et al., 2009). These stable properties and the intrinsic monolayer formed by caldarcheol represents an enticing alternative to traditional phospholipids in liposome-based commercial applications. One such application is in the delivery of chemotherapeutic compounds via archaeal derived liposomes. It has been found that tetraether linked artificial liposomes reduce leakage of chemotherapeutic compounds by 9-fold compared to conventional eukaryotic derived liposomes, which results in a lower dose required for therapeutic effects (Leriche et al., 2017). The archaeal liposomes themselves also contribute therapeutic effects as archaeal liposomes utilized to transport vaccine components induce robust antigen specific humoral and cellular immune responses exceeding those found from traditional delivery mechanisms (Conlan et al., 2001; Patel and Chen, 2010; Haq et al., 2016; Landi et al., 2017).
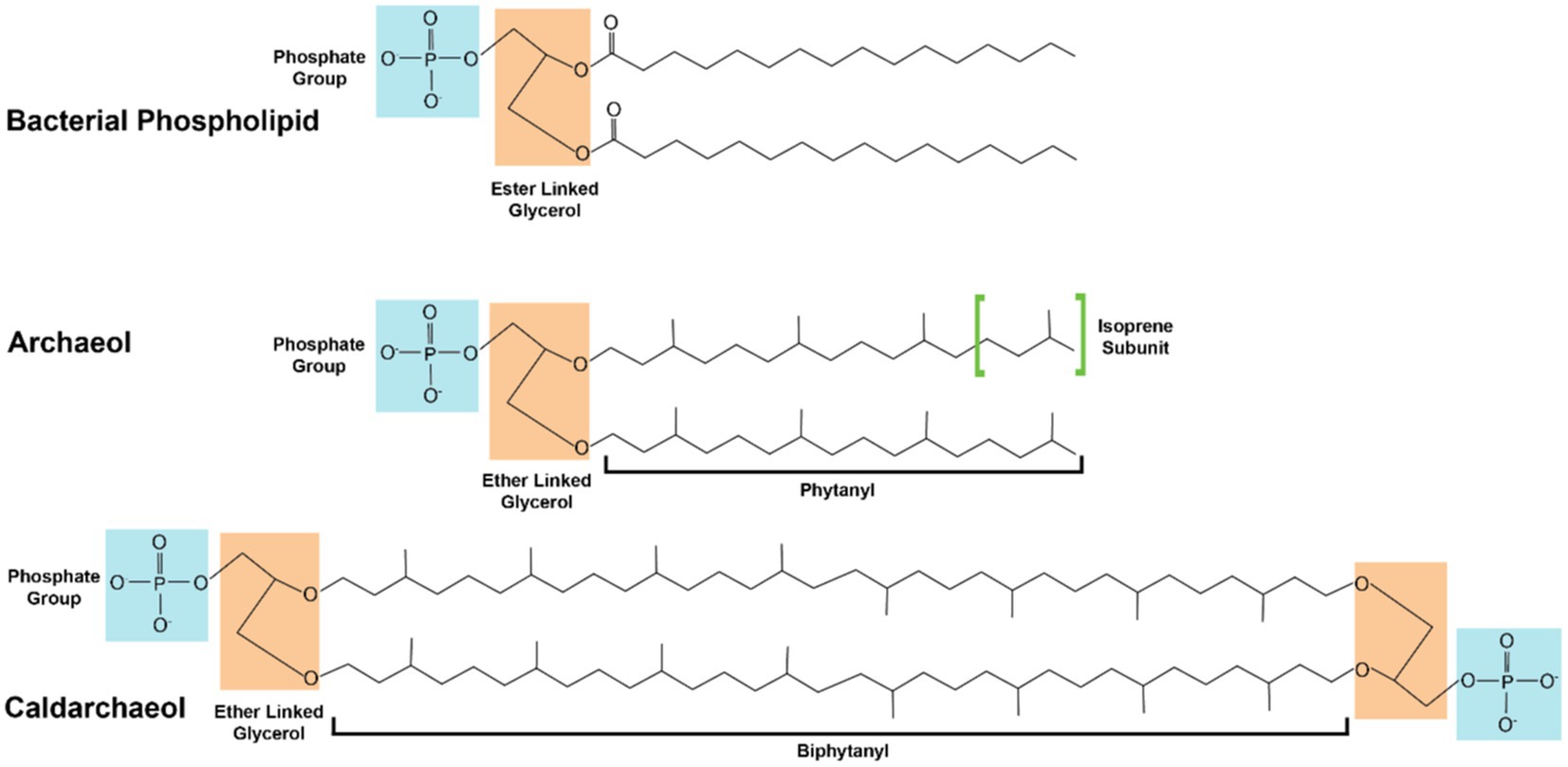
Figure 2. Comparison between structures of bacterial and archaeal lipids. Glycerol molecules are shaded in orange. Phosphate groups are shaded in cyan. The isoprenoid subunits which make up the archaeal lipids are highlighted in green brackets. Fully saturated lipids are shown; organisms may produce versions of unsaturated alkane lipids with multiple double bonds or hydroxyl moieties.
In addition to the direct application of archaeal lipids, the high metabolic flux through the archaeal mevalonate pathway presents an opportunity for low-cost production of terpene compounds. Terpenes are the largest class of natural compounds and have a wide range of commercial applications. Odorant terpenes such as limonene, eucalyptol, and linalool are cornerstones of the $29B flavor and fragrance industry (Markets, 2021). In addition to odorants, terpenes are often the active compound in pharmaceuticals including the anti-cancer drug paclitaxel and the antimalarial artemisinin. Hundreds of natural terpenes have shown promising bioactivity (Gould, 1997; Sgadari et al., 2000; Friedman et al., 2002; Paduch et al., 2007; Ajikumar et al., 2008) yet are limited in application due to their availability. Many of these terpenes are currently harvested from their native plant, fungal, and marine producers which are limited by the endogenous expression levels which are prohibitively low (Long et al., 1998; Sills et al., 1998; Newman and Cragg, 2004) or non-renewably synthesized from petroleum precursors. Organically produced terpenes are primarily produced via compounds derived from one of two isoprenoid synthesis pathways, the mevalonate (MVA) pathway and the deoxyxylose 5-phosphate (DXP) pathway (Lange et al., 2000). These pathways in non-archaeal organisms suffer low carbon flux and depletion of precursors towards non-target compounds (McGarvey and Croteau, 1995; Rodriguez-Concepcion and Boronat, 2002; Vranova et al., 2012). Archaea, however, synthesize the majority of membrane lipids through the mevalonate pathway, accounting for a higher flux as compared to eukaryal or bacterial organisms (Boucher et al., 2004; Jain et al., 2014; Villanueva et al., 2014). As such, there is a naturally higher abundance of metabolic precursors available for the synthesis of isoprenoid and terpene products using methanoarchaea. Concerns over the depletion of these membrane precursors have been alleviated by the synthesis of mono-isoprene from engineered strains of M. acetivorans and Methanosarcina barkeri (Aldridge et al., 2021; Carr et al., 2021). These strains demonstrate that methanogens are able to withstand the metabolic burden of membrane substrate depletion without a significant decrease in growth rate or final carrying capacity, opening the door for further isoprenoid products that could be produced by addition of relatively few genes (Table 2). Inducible promoters such as Ptet could also be used to drive expression of genes for terpenoid biosynthesis in two-stage fermentation processes to increase bioreactor carrying capacity and maximize terpenoid titer and yield (Urlinger et al., 2000; Loew et al., 2010). One challenge is that some terpenes require molecular oxygen for complete biosynthesis and this might be difficult for anaerobic organisms to achieve. However, Methanosarcina acetivorans is remarkably oxygen-tolerant and it is possible to further enhance resistance to oxidative stress through engineering or adaptation (Horne and Lessner, 2013; Jasso-Chávez et al., 2015). Therefore, it is theoretically feasible to use O2 availability as a biosynthetic inducer during terpene fermentation with oxygen-tolerant methanogens.
The use of methanogens in bioproduction is beneficial in a myriad of ways including ease of selection, low cost of media, and flexibility of products (Table 3). Methanogens have been shown to be an excellent source of metabolically active compounds such as coenzyme M (CoM) which acts as a potent chemotherapy adjuvant as the drug mesna (Shaw and Graham, 1987) as well as immune stimulating lipids for vaccine delivery (Patel and Chen, 2010; Haq et al., 2016). Due to their anaerobic metabolism which requires a lack of O2, they are able to produce novel chiral precursors which could later be tailored by chemists through custom oxidation steps and subsequent functionalization.
In large scale industrial fermentations pure aseptic environments are difficult to maintain, and often media and growth conditions are utilized to ensure continuous selection during the fermentation (Mosier and Ladisch, 2011; Doran, 2013). Methanogens circumvent this issue by growing in selective environments free of oxygen using substrates that cannot be used by the majority of common contaminating factors such as lactic acid bacteria and fungi (Skinner and Leathers, 2004; Beckner et al., 2011). Methanogens are prototrophic organisms, able to synthesize all vitamins and cofactors required for growth from inorganic material, allowing for additional selection by limiting available vitamins and nutrients required for contaminating growth by exclusion (Patil and Muskan, 2009; Thauer, 2012; Buan, 2018). While viral predation on methanogens has been observed (Park et al., 2007) there is little evidence that these methanophage/methanovirus particles have a substantial effect on methanogenic digestor performance as viral titers did not correlate with a significant decrease in methane output and methanogen carrying capacity.
Another major challenge in industrial fermentations is the large amounts of fresh water required for E. coli or yeast (Chen, 2012). Methanogens, however, thrive in environments with high salt concentrations, allowing for the utilization of seawater in fermentations. Non-sterile hypersaline environments such ocean water and hydraulic fracking fluids have been demonstrated to select for methylotrophic methanogens such as Methanohalophilus, Methanohalobium, and Methanosarcina spp. while also presenting a high concentration of non-competitive substrates such as methylamines (McGenity, 2010; Guan et al., 2015). Methanogens are utilized worldwide for the production of renewable biogas in non-selective environments with high degrees of contamination such as municipal and agricultural wastewater treatment. In these environments methanogens are exposed to a wide variety of stressors including dramatic shifts in ammonia, osmotic shifts, and exposure to heavy metals (Yan et al., 2020). Many methanogens are natively capable of withstanding these stressors (Macario and Macario, 2003) though as stated above, using genetic tools it is possible to combine or “stack” desirable traits onto a single methanogen strain to gain the maximum benefit from a single organism.
Methanogens are biologically important organisms with a wide-reaching impact both in ecological and biotechnological applications. Their extremely efficient central metabolism makes them an ideal source of renewable biofuels that can be captured through anaerobic digestion or fermentation processes. They are able to grow prototrophically with inexpensive feedstocks and can produce endotoxin-free protein, carbohydrates, and valuable isoprenoid lipids. Their unique membrane composition can be used to expand the biotechnological toolbox for the delivery of chemotherapeutics as well as source for novel terpene compounds previously not available via conventional extraction means. By continuing to investigate the molecular, genetic, and synthetic biology potential of these unique organisms, researchers may unlock a wide range of applications from environmental and ecological management, renewable energy, agriculture, chemical manufacturing, and pharmaceutic industries.
SC conceived and wrote the manuscript. NRB conceived, wrote, and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Science Foundation (IOS-1938948) and the Nebraska Center for Energy Sciences Research (Cycle 15). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the funding agencies.
NRB has disclosed a significant financial interest in RollingCircle Biotech, LLC and Molecular Trait Evolution, LLC.
SC declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ajikumar, P. K., Tyo, K., Carlsen, S., Mucha, O., Phon, T. H., and Stephanopoulos, G. (2008). Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol. Pharm. 5, 167–190. doi: 10.1021/mp700151b
Aldridge, J., Carr, S., Weber, K. A., and Buan, N. R. (2021). Anaerobic production of isoprene by engineered Methanosarcina species archaea. Appl. Environ. Microbiol. 87, e02417–e02420. doi: 10.1128/AEM.02417-20
Alperin, M. J., and Hoehler, T. M. (2009). Anaerobic methane oxidation by archaea/sulfate-reducing bacteria aggregates: 1. Thermodynamic and physical constraints. Am. J. Sci. 309, 869–957. doi: 10.2475/10.2009.02
Anderson, K. L., Apolinario, E. E., and Sowers, K. R. (2012). Desiccation as a long-term survival mechanism for the archaeon Methanosarcina barkeri. Appl. Environ. Microbiol. 78, 1473–1479. doi: 10.1128/AEM.06964-11
Atomi, H., Imanaka, T., and Fukui, T. (2012). Overview of the genetic tools in the archaea. Front. Microbiol. 3:337. doi: 10.3389/fmicb.2012.00337
Balch, W., Fox, G., Magrum, L., Woese, C., and Wolfe, R. (1979). Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260. PMCID:
Bao, J., and Scheller, S. (2021). Efficient CRISPR/Cas12a-based genome editing toolbox for metabolic engineering in Methanococcus maripaludis. bioRxiv. doi: 10.1101/2021.12.29.474413
Beckner, M., Ivey, M. L., and Phister, T. G. (2011). Microbial contamination of fuel ethanol fermentations. Lett. Appl. Microbiol. 53, 387–394. doi: 10.1111/j.1472-765X.2011.03124.x
Blank, C. E., Kessler, P. S., and Leigh, J. A. (1995). Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J. Bacteriol. 177, 5773–5777.
Bohlmann, J., Crock, J., Jetter, R., and Croteau, R. (1998). Terpenoid-based defenses in conifers: cDNA cloning, characterization, and functional expression of wound-inducible (E)-alpha-bisabolene synthase from grand fir (Abies grandis). Proc. Natl. Acad. Sci. U. S. A. 95, 6756–6761. doi: 10.1073/pnas.95.12.6756
Bohlmann, J., Martin, D., Oldham, N. J., and Gershenzon, J. (2000). Terpenoid secondary metabolism in Arabidopsis thaliana: cDNA cloning, characterization, and functional expression of a myrcene/(E)-β-ocimene synthase. Arch. Biochem. Biophys. 375, 261–269. doi: 10.1006/abbi.1999.1669
Bohlmann, J., Steele, C. L., and Croteau, R. (1997). Monoterpene synthases from grand fir (Abies grandis). cDNA isolation, characterization, and functional expression of myrcene synthase, (−)-(4S)-limonene synthase, and (−)-(1S,5S)-pinene synthase. J. Biol. Chem. 272, 21784–21792. doi: 10.1074/jbc.272.35.21784
Borrel, G., Adam, P. S., and Gribaldo, S. (2016). Methanogenesis and the Wood–Ljungdahl pathway: an ancient, versatile, and fragile association. Genome Biol. Evol. 8, 1706–1711. doi: 10.1093/gbe/evw114
Borrel, G., Adam, P. S., McKay, L. J., Chen, L.-X., Sierra-García, I. N., Sieber, C. M., et al. (2019). Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat. Microbiol. 4, 603–613. doi: 10.1038/s41564-019-0363-3
Boucher, Y., Kamekura, M., and Doolittle, W. F. (2004). Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 52, 515–527. doi: 10.1111/j.1365-2958.2004.03992.x
Bouwmeester, H. J., Wallaart, T. E., Janssen, M. H., van Loo, B., Jansen, B. J., Posthumus, M. A., et al. (1999). Amorpha-4,11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry 52, 843–854. doi: 10.1016/s0031-9422(99)00206-x
Boyd, E., Hamilton, T., Wang, J., He, L., and Zhang, C. (2013). The role of tetraether lipid composition in the adaptation of thermophilic archaea to acidity. Front. Microbiol. 4:62. doi: 10.3389/fmicb.2013.00062
Brune, A. (1998). Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16, 16–21. doi: 10.1016/S0167-7799(97)01151-7
Buan, N. R. (2018). Methanogens: pushing the boundaries of biology. Emerg. Topics Life Sci. 2, 629–646. doi: 10.1042/ETLS20180031
Buan, N., Kulkarni, G., and Metcalf, W. (2011). Genetic methods for Methanosarcina species. in Methods in Enzymology (Vol. 494, pp. 23–42). Academic Press.
Buan, N. R., and Metcalf, W. W. (2010). Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75, 843–853. doi: 10.1111/j.1365-2958.2009.06990.x
Buckel, W., and Thauer, R. K. (2018). Flavin-based electron bifurcation, ferredoxin, Flavodoxin, and anaerobic respiration with protons (Ech) or NAD(+) (Rnf) as electron acceptors: a historical review. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00401
Carr, S., Aldridge, J., and Buan, N. R. (2021). Isoprene production from municipal wastewater biosolids by engineered archaeon Methanosarcina acetivorans. Appl. Sci. 11:3342. doi: 10.3390/app11083342
Catlett, J. L., Carr, S., Cashman, M., Smith, M. D., Walter, M., Sakkaff, Z., et al. (2022). R. Metabolic synergy between human symbionts Bacteroides and Methanobrevibacter. Microbiol Spectr. 10:e0106722. doi: 10.1128/spectrum.01067-22 Epub 2022 May 10.;PMC9241691.
Catlett, J. L., Ortiz, A. M., and Buan, N. R. (2015). Rerouting cellular electron flux to increase the rate of biological methane production. Appl. Environ. Microbiol. 81, 6528–6537. doi: 10.1128/AEM.01162-15
Chadwick, G. L., Skennerton, C. T., Laso-Pérez, R., Leu, A. O., Speth, D. R., Yu, H., et al. (2022). Comparative genomics reveals electron transfer and syntrophic mechanisms differentiating methanotrophic and methanogenic archaea. PLoS Biol. 20:e3001508.;PMC9012536. doi: 10.1371/journal.pbio.3001508
Chen, G.-Q. (2012). New challenges and opportunities for industrial biotechnology. Microb. Cell Factories 11, 1–3.;PMC3520750. doi: 10.1186/1475-2859-11-111
Conlan, J. W., Krishnan, L., Willick, G. E., Patel, G. B., and Sprott, G. D. (2001). Immunization of mice with lipopeptide antigens encapsulated in novel liposomes prepared from the polar lipids of various Archaeobacteria elicits rapid and prolonged specific protective immunity against infection with the facultative intracellular pathogen Listeria monocytogenes. Vaccine 19, 3509–3517. doi: 10.1016/s0264-410x(01)00041-x
Conrad, R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292. doi: 10.1111/j.1758-2229.2009.00038.x
Cornforth, J., Cornforth, R. H., Popjak, G., and Yengoyan, L. (1966). Studies on the biosynthesis of cholesterol: XX. Steric course of decarboxylation of 5-pyrophosphomevalonate and of the carbon to carbon bond formation in the biosynthesis of farnesyl pyrophosphate. J. Biol. Chem. 241, 3970–3987. doi: 10.1016/S0021-9258(18)99800-5
Costa, K. C., and Leigh, J. A. (2014). Metabolic versatility in methanogens. Curr. Opin. Biotechnol. 29, 70–75. doi: 10.1016/j.copbio.2014.02.012
Costa, K. C., Wong, P. M., Wang, T., Lie, T. J., Dodsworth, J. A., Swanson, I., et al. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U. S. A. 107, 11050–11055.;PMC2890747. doi: 10.1073/pnas.1003653107
Daniels, L., Fuchs, G., Thauer, R., and Zeikus, J. (1977). Carbon monoxide oxidation by methanogenic bacteria. J. Bacteriol. 132, 118–126.;PMC221834. doi: 10.1128/jb.132.1.118-126.1977
Deguerry, F., Pastore, L., Wu, S., Clark, A., Chappell, J., and Schalk, M. (2006). The diverse sesquiterpene profile of patchouli, Pogostemon cablin, is correlated with a limited number of sesquiterpene synthases. Arch. Biochem. Biophys. 454, 123–136. doi: 10.1016/j.abb.2006.08.006
Diender, M., Stams, A. J., and Sousa, D. Z. (2015). Pathways and bioenergetics of anaerobic carbon monoxide fermentation. Front. Microbiol. 6:1275. doi: 10.3389/fmicb.2015.01275
Dolfing, J., Larter, S. R., and Head, I. M. (2008). Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2, 442–452. doi: 10.1038/ismej.2007.111
Donath, J., and Boland, W. (1995). Biosynthesis of acyclic homoterpenes: enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39, 785–790. doi: 10.1016/0031-9422(95)00082-I
Drancourt, M., Nkamga, V. D., Lakhe, N. A., Régis, J.-M., Dufour, H., Fournier, P.-E., et al. (2017). Evidence of archaeal methanogens in brain abscess. Clin. Infect. Dis. 65, 1–5. doi: 10.1093/cid/cix286
Duszenko, N., and Buan, N. R. (2017). Physiological evidence for isopotential tunneling in the electron transport chain of methane-producing archaea. Appl. Environ. Microbiol. 83, e00950–e00917.;PMC5583484. doi: 10.1128/AEM.00950-17
Ehlers, C., Weidenbach, K., Veit, K., Deppenmeier, U., Metcalf, W. W., and Schmitz, R. A. (2005). Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol. Gen. Genomics. 273, 290–298. doi: 10.1007/s00438-005-1128-7
Ermler, U., Grabarse, W., Shima, S., Goubeaud, M., and Thauer, R. K. (1997). Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278, 1457–1462. doi: 10.1126/science.278.5342.1457
Feist, A. M., Scholten, J. C., Palsson, B. Ø., Brockman, F. J., and Ideker, T. (2006). Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2:2006.0004.;PMC1681478. doi: 10.1038/msb4100046
Ferry, J. G. (2012). Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. Berlin Springer Science & Business Media. ISBN: 9781461523918.
Fink, C., Beblawy, S., Enkerlin, A. M., Mühling, L., Angenent, L. T., and Molitor, B. (2021). A shuttle-vector system allows heterologous gene expression in the thermophilic methanogen Methanothermobacter thermautotrophicus ΔH. MBio 12, e02766–e02721.; PMCID: PMC8609365. doi: 10.1128/mBio.02766-21
Fricke, W. F., Seedorf, H., Henne, A., Krüer, M., Liesegang, H., Hedderich, R., et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 188, 642–658.; PMCID: PMC1347301. doi: 10.1128/JB.188.2.642-658.2006
Friedman, M., Henika, P. R., and Mandrell, R. E. (2002). Bactericidal activities of plant essential oils and some of their isolated constituents against campylobacter jejuni, Escherichia coli, listeria monocytogenes, and salmonella enterica. J. Food Protect. 65, 1545–1560. doi: 10.4315/0362-028x-65.10.1545
Fu, L., Zhou, T., Wang, J., You, L., Lu, Y., Yu, L., et al. (2019). NanoFe3O4 as solid electron shuttles to accelerate acetotrophic methanogenesis by Methanosarcina barkeri. Front. Microbiol. 10:388. doi: 10.3389/fmicb.2019.00388
Gambliel, H., and Croteau, R. (1984). Pinene cyclases I and II. Two enzymes from sage (Salvia officinalis) which catalyze stereospecific cyclizations of geranyl pyrophosphate to monoterpene olefins of opposite configuration. J. Biol. Chem. 259, 740–748.
Gonnerman, M. C., Benedict, M. N., Feist, A. M., Metcalf, W. W., and Price, N. D. (2013). Genomically and biochemically accurate metabolic reconstruction of Methanosarcina barkeri Fusaro, iMG746. Biotechnol. J. 8, 1070–1079. doi: 10.1002/biot.201200266
Gould, M. N. (1997). Cancer chemoprevention and therapy by monoterpenes. Environ. Health Persp. 105, 977–979. doi: 10.2307/3433313
Goyal, N., Zhou, Z., and Karimi, I. A. (2016). Metabolic processes of Methanococcus maripaludis and potential applications. Microb. Cell Factories 15, 1–19. doi: 10.1186/s12934-016-0500-0
Guan, Y., Hikmawan, T., Antunes, A., Ngugi, D., and Stingl, U. (2015). Diversity of methanogens and sulfate-reducing bacteria in the interfaces of five deep-sea anoxic brines of the Red Sea. Res. Microbiol. 166, 688–699. doi: 10.1016/j.resmic.2015.07.002
Guss, A. M., Kulkarni, G., and Metcalf, W. W. (2009). Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J. Bacteriol. 191, 2826–2833. doi: 10.1128/JB.00563-08
Guss, A. M., Rother, M., Zhang, J. K., Kulkkarni, G., and Metcalf, W. W. (2008). New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2, 193–203.;PMC2685592. doi: 10.1155/2008/534081
Hao, W., Scharffe, D., Crutzen, P., and Sanhueza, E. (1988). Production of N2O, CH4, and CO2 from soils in the tropical savanna during the dry season. J. Atmos. Chem. 7, 93–105. doi: 10.1007/BF00048256
Haq, K., Jia, Y., and Krishnan, L. (2016). Archaeal lipid vaccine adjuvants for induction of cell-mediated immunity. Expert Rev. Vaccines 15, 1557–1566. doi: 10.1080/14760584.2016.1195265
Hildenbrand, C., Stock, T., Lange, C., Rother, M., and Soppa, J. (2011). Genome copy numbers and gene conversion in methanogenic archaea. J. Bacteriol. 193, 734–743. doi: 10.1128/JB.01016-10
Holmes, D. E., Giloteaux, L., Orellana, R., Williams, K. H., Robbins, M. J., and Lovley, D. R. (2014). Methane production from protozoan endosymbionts following stimulation of microbial metabolism within subsurface sediments. Front. Microbiol. 5:366. doi: 10.3389/fmicb.2014.00366
Horne, A. J., and Lessner, D. J. (2013). Assessment of the oxidant tolerance of Methanosarcina acetivorans. FEMS Microbiol. Lett. 343, 13–19. doi: 10.1111/1574-6968.12115
Huang, Z., Sednek, C., Urynowicz, M. A., Guo, H., Wang, Q., Fallgren, P., et al. (2017). Low carbon renewable natural gas production from coalbeds and implications for carbon capture and storage. Nat. Commun. 8, 1–11. doi: 10.1038/s41467-017-00611-7
Iijima, Y., Gang, D. R., Fridman, E., Lewinsohn, E., and Pichersky, E. (2004). Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 134, 370–379.;PMC316316. doi: 10.1104/pp.103.032946
Innard, N. (2018). Developing Genetic Tools in Methanococcus maripaludis, Heslington: University of York. Available at: https://etheses.whiterose.ac.uk/20486/
Jacquemet, A., Barbeau, J., Lemiègre, L., and Benvegnu, T. (2009). Archaeal tetraether bipolar lipids: structures, functions and applications. Biochimie 91, 711–717. doi: 10.1016/j.biochi.2009.01.006
Jain, S., Caforio, A., and Driessen, A. J. (2014). Biosynthesis of archaeal membrane ether lipids. Front. Microbiol. 5:641. doi: 10.3389/fmicb.2014.00641
Jasso-Chávez, R., Santiago-Martínez, M. G., Lira-Silva, E., Pineda, E., Zepeda-Rodríguez, A., et al. (2015). Air-adapted Methanosarcina acetivorans shows high methane production and develops resistance against oxygen stress. PLoS One 10:e0117331.; PMCID: PMC4338226. doi: 10.1371/journal.pone.0117331
Jennings, M. E., Schaff, C. W., Horne, A. J., Lessner, F. H., and Lessner, D. J. (2014). Expression of a bacterial catalase in a strictly anaerobic methanogen significantly increases tolerance to hydrogen peroxide but not oxygen. Microbiology 160, 270–278. doi: 10.1099/mic.0.070763-0
Jin, D., Kang, K., Wang, H., Wang, Z., Xue, B., Wang, L., et al. (2017). Effects of dietary supplementation of active dried yeast on fecal methanogenic archaea diversity in dairy cows. Anaerobe 44, 78–86. doi: 10.1016/j.anaerobe.2017.02.007
Johnson, K. A., and Johnson, D. E. (1995). Methane emissions from cattle. J. Anim. Sci. 73, 2483–2492. doi: 10.2527/1995.7382483x
Jones, D., Head, I., Gray, N., Adams, J., Rowan, A., Aitken, C., et al. (2008). Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180. doi: 10.1038/nature06484
Jones, W. J., Nagle, D., and Whitman, W. B. (1987). Methanogens and the diversity of archaebacteria. Microbiol. Rev. 51, 135–177.;PMC373095. doi: 10.1128/mr.51.1.135-177.1987
Jones, W. J., Paynter, M., and Gupta, R. (1983). Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 135, 91–97. doi: 10.1007/BF00425213
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
Koga, Y. (2012). Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea 2012, 1–6. doi: 10.1155/2012/789652
Koga, Y., and Morii, H. (2007). Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 71, 97–120.; PMCID: PMC1847378. doi: 10.1128/MMBR.00033-06
Kohler, P. R., and Metcalf, W. W. (2012). Genetic manipulation of Methanosarcina spp. Front. Microbiol. 3:259.; PMCID: PMC3403347. doi: 10.3389/fmicb.2012.00259
Kulkarni, G., Kridelbaugh, D. M., Guss, A. M., and Metcalf, W. W. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc. Natl. Acad. Sci. U. S. A. 106, 15915–15920. doi: 10.1073/pnas.0905914106
Kurth, J. M., Op den Camp, H. J., and Welte, C. U. (2020). Several ways one goal—methanogenesis from unconventional substrates. Appl. Microbiol. Biotechnol. 104, 6839–6854.; PMCID: PMC7374477. doi: 10.1007/s00253-020-10724-7
Landi, A., Law, J., Hockman, D., Logan, M., Crawford, K., Chen, C., et al. (2017). Superior immunogenicity of HCV envelope glycoproteins when adjuvanted with cyclic-di-AMP, a STING activator or archaeosomes. Vaccine 35, 6949–6956. doi: 10.1016/j.vaccine.2017.10.072
Lange, B. M., Rujan, T., Martin, W., and Croteau, R. (2000). Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. U. S. A. 97, 13172–13177.; PMCID: PMC27197. doi: 10.1073/pnas.240454797
Laso-Pérez, R., Hahn, C., van Vliet, D. M., Tegetmeyer, H. E., Schubotz, F., Smit, N. T., et al. (2019). Anaerobic degradation of non-methane alkanes by “Candidatus Methanoliparia” in hydrocarbon seeps of the Gulf of Mexico. MBio 10, e01814–e01819.; PMCID: PMC6703427. doi: 10.1128/mBio.01814-19
Le Mer, J., and Roger, P. (2001). Production, oxidation, emission and consumption of methane by soils: a review. Eur. J. Soil Biol. 37, 25–50. doi: 10.1016/S1164-5563(01)01067-6
Lemaire, O. N., and Wagner, T. (2022). A structural view of alkyl-coenzyme M reductases, the first step of alkane anaerobic oxidation catalyzed by archaea. Biochemistry 61, 805–821. doi: 10.1021/acs.biochem.2c00135
Leriche, G., Cifelli, J. L., Sibucao, K. C., Patterson, J. P., Koyanagi, T., Gianneschi, N. C., et al. (2017). Characterization of drug encapsulation and retention in archaea-inspired tetraether liposomes. Org. Biomol. Chem. 15, 2157–2162. doi: 10.1039/C6OB02832B
Lessner, D. J., Lhu, L., Wahal, C. S., and Ferry, J. G. (2010). An engineered methanogenic pathway derived from the domains bacteria and archaea. MBio 1, e00243–e00210.; PMCID: PMC2975365. doi: 10.1128/mBio.00243-10
Loew, R., Heinz, N., Hampf, M., Bujard, H., and Gossen, M. (2010). Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 10, 1–13.; PMCID: PMC3002914. doi: 10.1186/1472-6750-10-81
Long, B. H., Carboni, J. M., Wasserman, A. J., Cornell, L. A., Casazza, A. M., Jensen, P. R., et al. (1998). Eleutherobin, a novel cytotoxic agent that induces tubulin polymerization, is similar to paclitaxel (Taxol (R)). Cancer Res. 58, 1111–1115.
Long, F., Wang, L., Lupa, B., and Whitman, W. B. (2017). A flexible system for cultivation of Methanococcus and other formate-utilizing methanogens. Archaea 2017, 1–12. doi: 10.1155/2017/7046026
López Muñoz, M. M., Schönheit, P., and Metcalf, W. W. (2015). Genetic, genomic, and transcriptomic studies of pyruvate metabolism in Methanosarcina barkeri fusaro. J. Bacteriol. 197, 3592–3600. doi: 10.1128/JB.00551-15
Luo, G., and Angelidaki, I. (2012). Integrated biogas upgrading and hydrogen utilization in an anaerobic reactor containing enriched hydrogenotrophic methanogenic culture. Biotechnol. Bioeng. 109, 2729–2736. doi: 10.1002/bit.24557
Macario, E. C. D., and Macario, A. J. (2003). Molecular biology of stress genes in methanogens: potential for bioreactor technology. Biomethanation I, 95–150. doi: 10.1007/3-540-45839-5_4
Markets, M. A. (2021). Flavors & Fragrances Market by ingredients (natural, synthetic), end use (beverage, Savory & Snacks, bakery, dairy products, confectionery, consumer products, fine fragrances), and region (Asia Pacific, North America, Europe) – global forecast to 2026. Available at: https://www.marketsandmarkets.com/Market-Reports/flavors-fragrance-market-175163912.html
Martin, W. F., and Sousa, F. L. (2016). Early microbial evolution: the age of anaerobes. Cold Spring Harb. Perspect. Biol. 8:a018127. doi: 10.1101/cshperspect.a018127
Matschiavelli, N., Oelgeschläger, E., Cocchiararo, B., Finke, J., and Rother, M. (2012). Function and regulation of isoforms of carbon monoxide dehydrogenase/acetyl coenzyme a synthase in Methanosarcina acetivorans. J. Bacteriol. 194, 5377–5387. doi: 10.1128/JB.00881-12
Mayumi, D., Mochimaru, H., Tamaki, H., Yamamoto, K., Yoshioka, H., Suzuki, Y., et al. (2016). Methane production from coal by a single methanogen. Science 354, 222–225. doi: 10.1126/science.aaf8821
McAnulty, M. J., Poosarla, V., Kim, K.-Y., Jasso-Chávez, R., Logan, B. E., and Wood, T. K. (2017). Electricity from methane by reversing methanogenesis. Nat. Commun. 8, 1–8. doi: 10.1038/ncomms15419
McGarvey, D. J., and Croteau, R. (1995). Terpenoid metabolism. Plant Cell 7, 1015–1026. doi: 10.1105/tpc.7.7.1015
McGenity, T. J. (2010). “Methanogens and methanogenesis in hypersaline environments” in Handbook of Hydrocarbon and Lipid Microbiology. ed. K. N. Timmis Berlin, Heidelberg: Springer, 665–680.
McGlynn, S. E., Chadwick, G. L., Kempes, C. P., and Orphan, V. J. (2015). Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535. doi: 10.1038/nature15512
Meigs, T. E., and Simoni, R. D. (1997). Farnesol as a regulator of HMG-CoA reductase degradation: characterization and role of farnesyl pyrophosphatase. Arch. Biochem. Biophys. 345, 1–9. doi: 10.1006/abbi.1997.0200
Metcalf, W. W., Zhang, J. K., Apolinario, E., Sowers, K. R., and Wolfe, R. S. (1997). A genetic system for archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. U. S. A. 94, 2626–2631.; PMCID: PMC20139. doi: 10.1073/pnas.94.6.2626
Meyerdierks, A., Kube, M., Kostadinov, I., Teeling, H., Glöckner, F. O., Reinhardt, R., et al. (2010). Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ. Microbiol. 12, 422–439. doi: 10.1111/j.1462-2920.2009.02083.x
Moran, J. J., House, C. H., Freeman, K. H., and Ferry, J. G. (2005). Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1, 303–309.; PMCID: PMC2685550. doi: 10.1155/2005/650670
Mosier, N. S., and Ladisch, M. R. (2011). Modern Biotechnology: Connecting Innovations in Microbiology and Biochemistry to Engineering Fundamentals. Hoboken: John Wiley & Sons, ISBN: 1118210204.
Nayak, D. D., and Metcalf, W. W. (2017). Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc. Natl. Acad. Sci. 114, 2976–2981. doi: 10.1073/pnas.1618596114
Neill, T., Judd, D., Veith, E., and Rousar, D. (2009). Practical uses of liquid methane in rocket engine applications. Acta Astronaut. 65, 696–705. doi: 10.1016/j.actaastro.2009.01.052
Newman, D. J., and Cragg, G. M. (2004). Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 67, 1216–1238. doi: 10.1021/np040031y
Paduch, R., Kandefer-Szerszen, M., Trytek, M., and Fiedurek, J. (2007). Terpenes: substances useful in human healthcare. Arch. Immunol. Ther. Ex. 55, 315–327. doi: 10.1007/s00005-007-0039-1
Park, M.-O., Ikenaga, H., and Watanabe, K. (2007). Phage diversity in a methanogenic digester. Microb. Ecol. 53, 98–103. doi: 10.1007/s00248-006-9053-9
Patel, G. B., and Chen, W. (2010). Archaeal lipid mucosal vaccine adjuvant and delivery system. Expert Rev. Vaccines 9, 431–440. doi: 10.1586/erv.10.34
Patil, U. K., and Muskan, K. (2009). Essentials of Biotechnology. Bengaluru: IK International Pvt Ltd. ISBN: 9380026528.
Paulo, L. M., Stams, A. J., and Sousa, D. Z. (2015). Methanogens, sulphate and heavy metals: a complex system. Rev. Environ. Sci. Biotechnol. 14, 537–553. doi: 10.1007/s11157-015-9387-1
Pechous, S. W., and Whitaker, B. D. (2004). Cloning and functional expression of an (E, E)-alpha-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219, 84–94. doi: 10.1007/s00425-003-1191-4
Pichersky, E., Raguso, R. A., Lewinsohn, E., and Croteau, R. (1994). Floral Scent Production in Clarkia (Onagraceae) (I. Localization and Developmental Modulation of Monoterpene Emission and Linalool Synthase Activity). Plant Physiol. 106, 1533–1540. doi: 10.1104/pp.106.4.1533
Pritchett, M. A., Zhang, J. K., and Metcalf, W. W. (2004). Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70, 1425–1433. doi: 10.1128/AEM.70.3.1425-1433.2004
Ragab, A., Shaw, D. R., Katuri, K. P., and Saikaly, P. E. (2020). Effects of set cathode potentials on microbial electrosynthesis system performance and biocathode methanogen function at a metatranscriptional level. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-76229-5
Rajilić-Stojanović, M., Smidt, H., and De Vos, W. M. (2007). Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x
Riedinger, N., Formolo, M. J., Lyons, T. W., Henkel, S., Beck, A., and Kasten, S. (2014). An inorganic geochemical argument for coupled anaerobic oxidation of methane and iron reduction in marine sediments. Geobiology 12, 172–181. doi: 10.1111/gbi.12077
Rodriguez-Concepcion, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089. doi: 10.1104/pp.007138
Roslev, P., and King, G. M. (1995). Aerobic and anaerobic starvation metabolism in methanotrophic bacteria. Appl. Environ. Microbiol. 61, 1563–1570.; PMCID: PMC1388422. doi: 10.1128/aem.61.4.1563-1570.1995
Rother, M., and Metcalf, W. W. (2004). Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc. Natl. Acad. Sci. U. S. A. 101, 16929–16934. doi: 10.1073/pnas.0407486101
Rouviere, P. E., and Wolfe, R. (1988). Novel biochemistry of methanogenesis. J. Biol. Chem. 263, 7913–7916.
Sarmiento, F. B., Leigh, J. A., and Whitman, W. B. (2011). Methods in Enzymology Amsterdam: Elsevier. doi: 10.1016/B978-0-12-385112-3.00003-2
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608. doi: 10.1038/nature09015
Schiraldi, C., Giuliano, M., and De Rosa, M. (2002). Perspectives on biotechnological applications of archaea. Archaea 1, 75–86. doi: 10.1155/2002/436561
Schlegel, K., Welte, C., Deppenmeier, U., and Muller, V. (2012). Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279, 4444–4452. doi: 10.1111/febs.12031
Schütz, H., Seiler, W., and Conrad, R. (1989). Processes involved in formation and emission of methane in rice paddies. Biogeochemistry 7, 33–53. doi: 10.1007/BF00000896
Sgadari, C., Toschi, E., Palladino, C., Barillari, G., Carlei, D., Cereseto, A., et al. (2000). Mechanism of paclitaxel activity in Kaposi's sarcoma. J. Immunol. 165, 509–517. doi: 10.4049/jimmunol.165.1.509
Sharon-Asa, L., Shalit, M., Frydman, A., Bar, E., Holland, D., Or, E., et al. (2003). Citrus fruit flavor and aroma biosynthesis: isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J. 36, 664–674. doi: 10.1046/j.1365-313x.2003.01910.x
Shaw, I., and Graham, M. (1987). Mesna—a short review. Cancer Treat. Rev. 14, 67–86. doi: 10.1016/0305-7372(87)90041-7
Sheehan, S. W. (2021). Electrochemical methane production from CO2 for orbital and interplanetary refueling. Iscience 24:102230. doi: 10.1016/j.isci.2021.102230
Shima, S., Krueger, M., Weinert, T., Demmer, U., Kahnt, J., Thauer, R. K., et al. (2012). Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481, 98–101. doi: 10.1038/nature10663
Siegert, M., Cichocka, D., Herrmann, S., Gründger, F., Feisthauer, S., Richnow, H.-H., et al. (2011). Accelerated methanogenesis from aliphatic and aromatic hydrocarbons under iron-and sulfate-reducing conditions. FEMS Microbiol. Lett. 315, 6–16. doi: 10.1111/j.1574-6968.2010.02165.x
Siliakus, M. F., van der Oost, J., and Kengen, S. W. (2017). Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 21, 651–670. doi: 10.1007/s00792-017-0939-x
Sills, A. K., Williams, J. I., Tyler, B. M., Epstein, D. S., Sipos, E. P., Davis, J. D., et al. (1998). Squalamine inhibits angiogenesis and solid tumor growth in vivo and perturbs embryonic vasculature. Cancer Res. 58, 2784–2792.
Silver, G. M., and Fall, R. (1991). Enzymatic synthesis of isoprene from dimethylallyl diphosphate in aspen leaf extracts. Plant Physiol. 97, 1588–1591.; PMCID: PMC1081206. doi: 10.1104/pp.97.4.1588
Sirohi, S., Pandey, N., Singh, B., and Puniya, A. (2010). Rumen methanogens: a review. Indian J. Microbiol. 50, 253–262. doi: 10.1007/s12088-010-0061-6
Skinner, K. A., and Leathers, T. D. (2004). Bacterial contaminants of fuel ethanol production. J. Ind. Microbiol. Biotechnol. 31, 401–408. doi: 10.1007/s10295-004-0159-0
Sogodogo, E., Drancourt, M., and Grine, G. (2019). Methanogens as emerging pathogens in anaerobic abscesses. Eur. J. Clin. Microbiol. Infect. Dis. 38, 811–818. doi: 10.1007/s10096-019-03510-5
Soo, V. W., McAnulty, M. J., Tripathi, A., Zhu, F., Zhang, L., Hatzakis, E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Factories 15, 1–14. doi: 10.1186/s12934-015-0397-z
Sowers, K. R., Baron, S. F., and Ferry, J. G. (1984). Methanosarcina acetivorans sp. nov., an Acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47, 971–978. doi: 10.1128/aem.47.5.971-978.1984
Stumm, C., and Zwart, K. (1986). Symbiosis of protozoa with hydrogen-utilizing methanogens. Microbiol. Sci. 3, 100–105.
Su, G., Zopfi, J., Yao, H., Steinle, L., Niemann, H., and Lehmann, M. F. (2020). Manganese/iron-supported sulfate-dependent anaerobic oxidation of methane by archaea in lake sediments. Limnol. Oceanogr. 65, 863–875. doi: 10.1002/lno.11354
Subedi, B. P., Martin, W. F., Carbone, V., Duin, E. C., Cronin, B., Sauter, J., et al. (2021). Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry. FEMS Microb. 2:xtab012. doi: 10.1093/femsmc/xtab012
Summers, Z. M., Fogarty, H. E., Leang, C., Franks, A. E., Malvankar, N. S., and Lovley, D. R. (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330, 1413–1415. doi: 10.1126/science.1196526
Takai, K., Nakamura, K., Toki, T., Tsunogai, U., Miyazaki, M., Miyazaki, J., et al. (2008). Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. U. S. A. 105, 10949–10954. doi: 10.1073/pnas.0712334105
Tanner, R. S., and Wolfe, R. S. (1988). Nutritional requirements of Methanomicrobium mobile. Appl. Environ. Microbiol. 54, 625–628.; PMCID: PMC202515. doi: 10.1128/aem.54.3.625-628.1988
Thauer, R. K. (2011). Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14, 292–299. doi: 10.1016/j.mib.2011.03.003
Thauer, R. K. (2012). The Wolfe cycle comes full circle. Proc. Natl. Acad. Sci. U. S. A. 109, 15084–15085. doi: 10.1073/pnas.1213193109
Thauer, R. K., Kaster, A.-K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro193
Tumbula, D. L., Makula, R. A., and Whitman, W. B. (1994). Transformation of Methanococcus maripaludis and identification of a Pst I-like restriction system. FEMS Microbiol. Lett. 121, 309–314. doi: 10.1111/j.1574-6968.1994.tb07118.x
Ulrich, G., and Bower, S. (2008). Active methanogenesis and acetate utilization in Powder River Basin coals, United States. Int. J. Coal Geol. 76, 25–33. doi: 10.1016/j.coal.2008.03.006
Urlinger, S., Baron, U., Thellmann, M., Hasan, M. T., Bujard, H., and Hillen, W. (2000). Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. 97, 7963–7968. doi: 10.1073/pnas.130192197
van Der Hoeven, R. S., Monforte, A. J., Breeden, D., Tanksley, S. D., and Steffens, J. C. (2000). Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12, 2283–2294. doi: 10.1105/tpc.12.11.2283
Vandermoten, S., Santini, S., Haubruge, E., Heuze, F., Francis, F., Brasseur, R., et al. (2009). Structural features conferring dual geranyl/farnesyl diphosphate synthase activity to an aphid prenyltransferase. Insect Biochem. Mol. Biol. 39, 707–716. doi: 10.1016/j.ibmb.2009.08.007
Villanueva, L., Damste, J. S., and Schouten, S. (2014). A re-evaluation of the archaeal membrane lipid biosynthetic pathway. Nat. Rev. Microbiol. 12, 438–448. doi: 10.1038/nrmicro3260
Villanueva, L., von Meijenfeldt, F. B., Westbye, A. B., Yadav, S., Hopmans, E. C., Dutilh, B. E., et al. (2021). Bridging the membrane lipid divide: bacteria of the FCB group superphylum have the potential to synthesize archaeal ether lipids. ISME J. 15, 168–182.; PMCID: PMC7852524. doi: 10.1038/s41396-020-00772-2
Vranova, E., Coman, D., and Gruissem, W. (2012). Structure and dynamics of the isoprenoid pathway network. Mol. Plant 5, 318–333. doi: 10.1093/mp/sss015
Wang, C., Wang, C., Liu, J., Xu, Q., Han, Z., Xu, X., et al. (2020). Tolerance of aceticlastic methanogenesis enhanced by magnetite under the condition of ammonia stress. ACS Sustain. Chem. Eng. 8, 1417–1426. doi: 10.1021/acssuschemeng.9b05585
Wang, Y., Wegener, G., Hou, J., Wang, F., and Xiao, X. (2019). Expanding anaerobic alkane metabolism in the domain of archaea. Nat. Microbiol. 4, 595–602. doi: 10.1038/s41564-019-0364-2
Watanabe, T., Pfeil-Gardiner, O., Kahnt, J., Koch, J., Shima, S., and Murphy, B. J. (2021). Three-megadalton complex of methanogenic electron-bifurcating and CO2-fixing enzymes. Science 373, 1151–1156. doi: 10.1126/science.abg5550
Welte, C., and Deppenmeier, U. (2011). Re-evaluation of the function of the F420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J. 278, 1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x
Whitford, M. F., Teather, R. M., and Forster, R. J. (2001). Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1, 1–5.; PMCID: PMC32158. doi: 10.1186/1471-2180-1-5
Wise, M. L., Savage, T. J., Katahira, E., and Croteau, R. (1998). Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1,8-cineole synthase, and (+)-bornyl diphosphate synthase. J. Biol. Chem. 273, 14891–14899. doi: 10.1074/jbc.273.24.14891
Yan, Z., and Ferry, J. G. (2018). Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front. Microbiol. 9:1322. doi: 10.3389/fmicb.2018.01322
Yan, M., Treu, L., Zhu, X., Tian, H., Basile, A., Fotidis, I. A., et al. (2020). Insights into ammonia adaptation and methanogenic precursor oxidation by genome-centric analysis. Environ. Sci. Technol. 54, 12568–12582. doi: 10.1021/acs.est.0c01945
Zengler, K., Richnow, H. H., Rosselló-Mora, R., Michaelis, W., and Widdel, F. (1999). Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401, 266–269. doi: 10.1038/45777
Zhou, Z., Zhang, C.-J., Liu, P.-F., Fu, L., Laso-Pérez, R., Yang, L., et al. (2022). Non-syntrophic methanogenic hydrocarbon degradation by an archaeal species. Nature 601, 257–262. doi: 10.1038/s41586-021-04235-2
Keywords: methanogens, archaea, biotechnology, methane, biofuel, terpenoid chemicals
Citation: Carr S and Buan NR (2022) Insights into the biotechnology potential of Methanosarcina. Front. Microbiol. 13:1034674. doi: 10.3389/fmicb.2022.1034674
Received: 01 September 2022; Accepted: 28 October 2022;
Published: 15 December 2022.
Edited by:
Sabine Kleinsteuber, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
James Ferry, The Pennsylvania State University, United StatesCopyright © 2022 Carr and Buan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole R. Buan, bmJ1YW5AdW5sLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.