- 1Institute of Agrophysics, Polish Academy of Sciences, Lublin, Poland
- 2Institute of Environmental Sciences, Leiden University, Leiden, Netherlands
- 3Department of Forest Entomology and Pathology, Faculty of Forestry and Wood Technology, Poznań University of Life Sciences, Poznań, Poland
- 4Department of Microbial Ecology, Groningen Institute for Evolutionary Life Sciences, University of Groningen, Groningen, Netherlands
- 5Institute of Plant Genetics, Polish Academy of Sciences, Poznań, Poland
The soil microbiome contributes to several ecosystem processes. It plays a key role in sustainable agriculture, horticulture and forestry. In contrast to the vast number of studies focusing on soil bacteria, the amount of research concerning soil fungal communities is limited. This is despite the fact that fungi play a crucial role in the cycling of matter and energy on Earth. Fungi constitute a significant part of the pathobiome of plants. Moreover, many of them are indispensable to plant health. This group includes mycorrhizal fungi, superparasites of pathogens, and generalists; they stabilize the soil mycobiome and play a key role in biogeochemical cycles. Several fungal species also contribute to soil bioremediation through their uptake of high amounts of contaminants from the environment. Moreover, fungal mycelia stretch below the ground like blood vessels in the human body, transferring water and nutrients to and from various plants. Recent advances in high-throughput sequencing combined with bioinformatic tools have facilitated detailed studies of the soil mycobiome. This review discusses the beneficial effects of soil mycobiomes and their interactions with other microbes and hosts in both healthy and unhealthy ecosystems. It may be argued that studying the soil mycobiome in such a fashion is an essential step in promoting sustainable and regenerative agriculture.
Introduction
The use of intensive agricultural production worldwide aims to supply sufficient food for humankind. However, the occurrence of sudden ecological disasters and wars show that this apparently stable situation is actually fragile, but overall, we may generally conclude that food security is growing. These benefits have been achieved at the cost of a damaged natural environment, which is causing increased levels of dissatisfaction with regard to modern industrial agriculture (Frouz and Frouzová, 2022). According to the Food and Agriculture Organization, current food systems account for more than one-third of global greenhouse gas emissions (Crippa et al., 2021; FAO, 2021). The other adverse effect is the high degree of deforestation of areas which are significant to the whole planet, such as tropical rainforests. The recent drastic decrease in the area of ‘the lungs of Earth’ has adversely impacted both our climate and biodiversity (Nobre et al., 2016). Intensive farming with excessive ploughing causes land degradation, including its erosion by water and wind. Moreover, this degradation is also significantly changing the soil structure, water availability, and the availability of numerous macro- and microelements taken from the soil by crop plant monocultures (Beylich et al., 2010). The malfunctioning of current food and industrial systems with the resulting high impact on our climate has led to the novel phenomenon of environmental refugees – people who are displaced due to permanent environmental disasters (Myers, 2002).
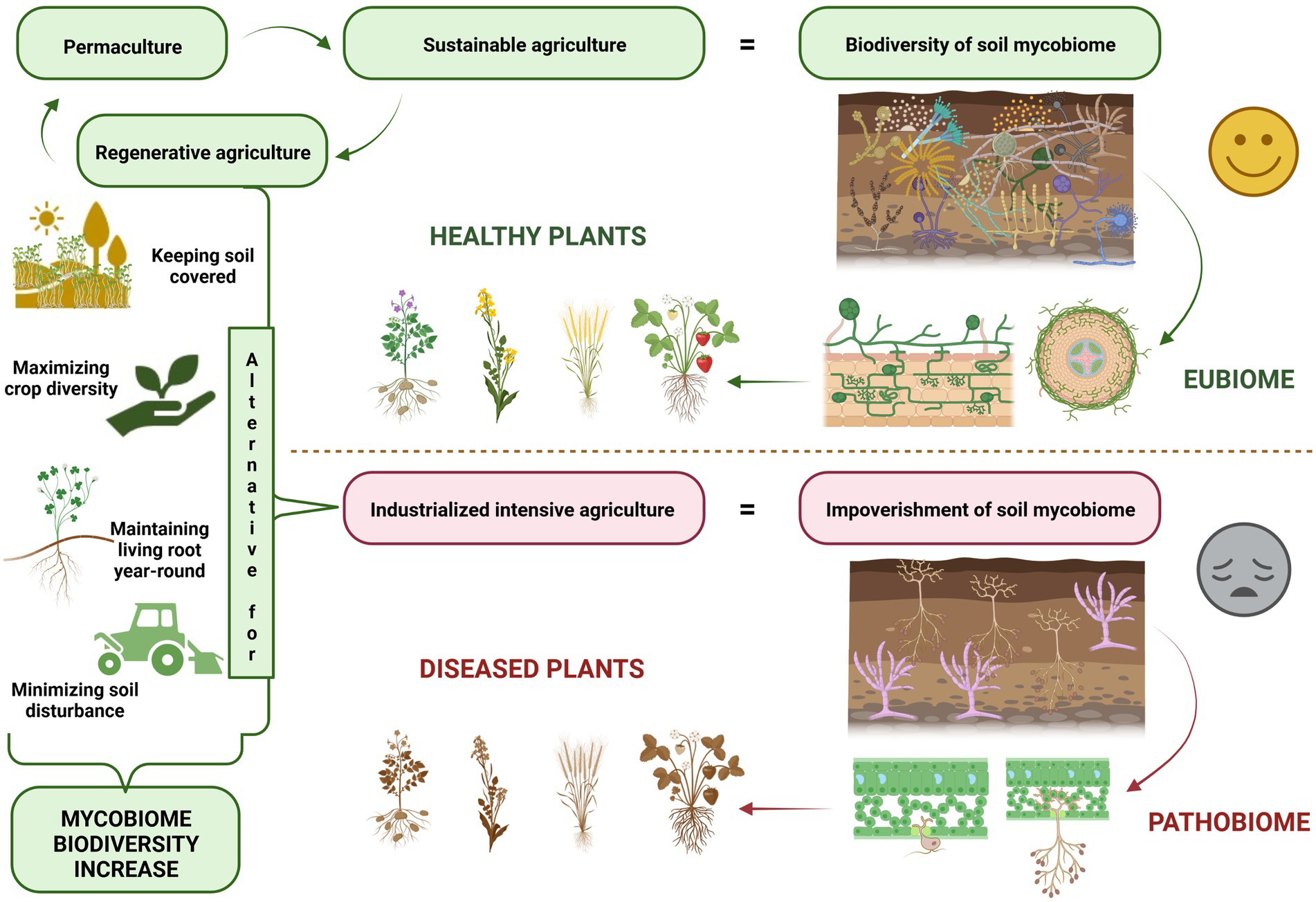
The lack of nutrient cycling in intensive, high-input agriculture, and the use of considerable amounts of pesticides, has an impact on the natural soil microbiome, including its mycobiome (O’Brien, 2013). The soil mycobiome, which is also called the fungal microbiome, is one of the main components of an integrative microbiome which includes different microbial groups which inhabit agroecosystems (Geisen, 2021). At present, the soil mycobiome is still being poorly studied in relative terms as the main focus of recent global research has been connected to the bacterial component of the soil microbiome (Yadav et al., 2021). Despite the limited research undertaken to date, it is well known that fungi support many ecosystem processes and perform functions that are essential for the sustainable development of future agriculture (Fernandes et al., 2022), including plant–soil interactions, organic matter decomposition (Vétrovský et al., 2019), plant health promotion and nutrition (Põlme et al., 2020). A great majority of the agricultural land currently in use requires soil regeneration and the restoration of its biological processes (Gosnell, 2022), which includes restoring all components of the soil mycobiome. The solution to soil restoration in an agricultural setting is environmentally-friendly farming, which is usually referred to as sustainable agriculture. A holistic approach linking agricultural production with an unharmed environment is referred to as regenerative agriculture (Newton et al., 2020).
A clean environment and the maintenance of economic profitability are significant concerns in agriculture, horticulture and forestry (Dubey et al., 2019). Plant pathogenic fungi decrease plant yield and soil quality (Malarczyk et al., 2019), but beneficial fungi have the potential to contribute to soil stability and functioning and thus serve as vital components of regenerative agriculture. Below we provide an overview of mycobiome-based solutions and tools for sustainable and regenerative agriculture. The thrilling perspective is the possibility of neutralising plant diseases by controlling and altering mycobiome shifts in soils. The microbiome-mediated plant protection is an emerging direction of research (Bass et al., 2019), shifting the traditional ‘one pathogen – one disease paradigm’ towards pathobiome healing with microbial inoculants (Berg et al., 2021). Recent studies show that microbial communities in soils are much more diverse than we have previously thought. If so, is the control of myriads of soil microorganisms possible to any extent?
Soil mycobiome – from butterfly effect to Ariadne’s thread
Microbes play essential roles in the ecology and physiology of plants (Dastogeer et al., 2020). Plants and their associated mycobiomes, including soil mycobiota, form complex and dynamic mutual interactions where the plants provide ecological niches and easily utilizable carbon to the mycobiome, which in turn influence plant growth, development and fitness (Vincent et al., 2020). The rhizosphere mycobiomes and plant endophytes suppress disease (Mendes et al., 2011; Frąc et al., 2018) and improve abiotic stress resistance (Lau and Lennon, 2012; Coleman-Derr and Tringe, 2014).
Fungi in the soil and rhizosphere operate in interlocking networks (van der Heijden and Hartmann, 2016), and form mycological hubs in the soil or in association with plants. The application of practices that promote the complexity and connectivity of these networks may serve to enhance the services that the mycobiome provides and this also has the potential to improve agricultural productivity due to positive soil feedback. Therefore, sustainability involves a greater degree of plant reliance on the beneficial functions offered by the soil mycobiome. In sustainable and regenerative agriculture, the dynamic interactions between soil mycobiome, plant mycobiome and plant health should be linked to agricultural practices (EASAC, 2022).
Microbial ecosystem balance, which is referred to as eubiosis, is accompanied by a high degree of suitable microorganisms (Iebba et al., 2016) and it may be regarded as a fundamental concept of ecosystem stability (Figure 1). According to this concept pathogens represent a small but important part of the microorganisms present. Their presence may lead to the loss of biodiversity and the dysbiosis state, which shifts microbe composition and influences pathogen emergence and outbreaks (Chen et al., 2020). Healthy balanced host–microbe interactions of the holobiont are termed eubiosis. At the same time, dysbiosis (pathobiome) refers to a holobiont disease state (Berg et al., 2020). Dysbiosis refers to the reduced capacity of host-microbiome regulation. This loss of function causes a decline in plant health (Arnault et al., 2022).
Dysbiosis can be observed not only when plant disease is advanced but even at the initial stage, before planting, and it can predetermine future plant health (Wei et al., 2019). Detrimental mycobiota identified on time may be neutralised using various tools, leading to microbiome-mediated plant protection. Pathobiome analyses showed changes in community structure caused by pathogens (Dastogeer et al., 2022; Jia et al., 2022), and these small differences trigger great micro- and mycobiome modulations (Berg et al., 2021). Van Dijk et al. (2022) proved that soil microbiomes change aboveground plant-pathogen-insect interactions. In other words, small changes can have a great, non-linear impact on a complex soil microbiome-plant health system, referred to as the ‘butterfly effect’.
Because fungi are important organisms around the world (O'Hanlon, 2017), this concept may be used in the context of mycobiomes, thereby indicating that soil and plant mycobiota determine both the healthy and unhealthy states of agroecosystems. The best example of this is suppressive and conducive soils (Schlatter et al., 2017), in which plants actively promote the selection of beneficial microorganisms which are active against soilborne pathogens (Weller et al., 2002). The application of soil amendments to supply the missing parts of the microbiota, such as Streptomycetes, has received considerable attention (Wiggins and Kinket, 2005a,b; Li et al., 2020). Recent studies have shown that the application of biofertilizers actively induces soil suppressiveness, thereby promoting the control of both bacterial and fungal diseases in tomatoes (Deng et al., 2021, 2022). Modulation of the microbiome using microbial inoculants is a promising tool for smart sustainable agriculture (Berg et al., 2021). Soil mycobiome networks undergo complicated dependencies and a great role of science is to disentangle them and use fungal mycelium as Ariadne’s thread into the desired direction, to obtain a healthy crop.
Beneficial fungi for agricultural crops, fruits, and forests
Among the pathogens of crops, fungi are the most numerous and damaging (Agrios, 2005). A decrease in yield caused by fungi has been documented for all globally important crops, such as wheat, rice, maize, potato, soybean, rapeseed and many others (Savary et al., 2019), this state of affairs has attracted a great deal of attention and caused concern (Jeger et al., 2021). Each crop has at least a few pathogens; while some have several of them, some are important on a worldwide scale and others are damaging at a regional or local scale; this is usually linked to conducive weather conditions. Most of them inhabit the soil and form their mycobiota for at least a part of their life cycle. Moreover, novel pathogens may emerge (Havis et al., 2015) because of adverse climate change, agricultural zones, crop diversity and changes in agronomic practices. Due to the Green Deal policy of the European Union (European Commission, 2020) and the general trend in favour of sustainable agriculture, the current focus of research, and development concerns beneficial microorganisms or their communities, including fungi. At present, the list of beneficial fungi in widespread use in agriculture is short and usually limited to Biological Control Agents developed for horticultural production, typically the generalists which are common in many habitats (Gostinčar et al., 2019) with mycoparasitic properties (Kubicek et al., 2011). The most popular preparations are based on Trichoderma harzianum (Błaszczyk et al., 2014; Dawidziuk et al., 2016), Clonostachys rosea (Peng et al., 2011; Sun et al., 2020), Aureobasidium pullulans (Prasongsuk et al., 2018) and other yeast-like fungi. A good example is Coniothyrium minitans, the super parasite of S. sclerotiorum (Smolinska and Kowalska, 2018).
Soft fruits are affected by various diseases caused by fungal pathogens inducing damage to roots, leaves, crowns and fruit. The most common and damaging diseases of soft fruit include grey mould, anthracnose, crown and root rot, plant wilt and fungi causing postharvest losses (Gubler and Converse, 1993; Garrido et al., 2016; Malarczyk et al., 2020). In recent years, under the pressure of a changing climate, pathogens which in the past, typically occurred mainly in tropical and Mediterranean climate zones, began to appear in the continental climate zone (Debode et al., 2011; Karimi et al., 2016; Torbati et al., 2019; Malarczyk et al., 2020). Key groups of beneficial mycobiota in soft fruit production include arbuscular mycorrhizal fungi (AMF), which for the most part, belong to the orders Glomerales, Archaeosporales, Paraglomerales and Diversisporales (Boyer et al., 2016; Begum et al., 2019). Ericoid mycorrhizal fungi can improve plant resistance to diseases (Jansa and Vosátka, 2000; Bizabani and Dames, 2015; Fehrer et al., 2019; Ważny et al., 2022). Many biopesticides are based on the entomopathogenic fungi (Bamisile et al., 2018; Canassa et al., 2020; Litwin et al., 2020). Fungal endophytes, such as Clonostachys rosea (Cota et al., 2009), Piriformospora indica (Sinclair et al., 2013), Xylaria sp. (Ważny et al., 2022), Penicillium sp. (Hamim et al., 2017), Hannaella coprosmae and Oberwinklerozyma straminea (Nguyen et al., 2021) are used in biocontrol and can have a beneficial impact on fruit yield (Murphy et al., 2019). The most common fungi able to promote plant growth and capable of acting as biocontrol agents belong to Trichoderma (Chen et al., 2019; Pylak et al., 2019; Lombardi et al., 2020; Mącik et al., 2020; Oszust et al., 2021; Rees et al., 2022) and yeasts (Sun et al., 2018; Calderon et al., 2019; De Miccolis Angelini et al., 2019; Freimoser et al., 2019).
Fungi constitute a significant fraction of the forest soil microbiome (Pérez-Izquierdo et al., 2021). Their high degree of ecological and taxonomical diversity significantly impacts the functioning of forest ecosystems (Witzell and Martín, 2018). In forests, saprotrophs, endophytes, biomass degraders, and the decomposers of lignified plant materials perform multiple tasks and form mutualistic relationships with roots (Frąc et al., 2018; Yan et al., 2018; Li et al., 2021). They can positively influence tree health by acting as growth-promoting or protective agents. However, many fungal species are causal agents of forest diseases, although only a few are active pathogens (Witzell and Martín, 2018). Nevertheless, the boundaries between the categories of fungus according to their function are not strictly defined, as some species can shift from one trophic state to another (Delaye et al., 2013).
Fungi are not homogeneously distributed in the forest soil (Lladó et al., 2018). Their activity is driven by the dynamics of ecosystem processes, root exudates, and soil heterogeneity (Baldrian, 2017; Lladó et al., 2018). Communities of litter saprotrophic and root-associated fungi are vertically separated within profiles. Within these layers, the fungal functional groups occupy different spatial niches according to their mode of C assimilation. Also, the competition for space and resources is an essential determinant of these communities (Boddy, 2000; Kennedy, 2010; Bödeker et al., 2016). Changes in climatic conditions, atmospheric CO2 levels, forest management regimes, and nutrient availability may affect the various fungal functional guilds in different ways, leading to shifts in their competitive balance and niches (Bödeker et al., 2016). Decomposers dominate the litter layer, while mycorrhizal fungi dominate the humus and mineral layers (Asplund et al., 2019). Few studies have documented the microbial changes that occur after the emergence of a plant pathogen, especially in forest ecosystems (Stewart et al., 2021). Some species (e.g., Laccaria laccata, Hebeloma crustuliniforme, Paxillus involutus, Trichoderma harzianum, Pisolithus tinctorius, Tricholoma pessundatum) have been shown to play an essential role in decreasing the severity of disease caused by root pathogens (Chakravarty and Hwang, 1991; Suárez et al., 2018; Stewart et al., 2021; Zaki et al., 2021). However, some fungal pathogens can deactivate genes in non-pathogenic species; they are involved in antimicrobial production and further weaken tree symbionts by releasing secondary metabolites (Stewart et al., 2021).
Bioinformatics for mycobiome control in sustainable agriculture
In recent years the development of high-throughput sequencing has enabled scientists around the globe to identify soil fungi (Tedersoo et al., 2014); see Figure 2. Most of the published studies use Illumina MiSeq to characterize the fungi targeting ITS regions, particularly ITS2 (Schoch et al., 2012), which is also deemed to be the best technique in comparative studies (Tedersoo et al., 2015; Frau et al., 2019). There has, however, been a recent call to use longer-read sequencing such as PacBio or Nanopore sequencing and to analyse both ITS2 regions and parts of the 18S and 28S genes (D’Andreano et al., 2021; Runnel et al., 2022). Obtaining longer-read will enable scientists to assign reads to particular species and strains with an increased level of confidence. This will be crucial, especially when linking the identity of a species to its functionality. There are multiple options for bioinformatic analyses and pipelines, and the appropriate choice depends mainly on the sequencing technique used but also on the individual preferences of the scientist. The most commonly used pipelines for the analyses of bacteria and fungi are DADA2 (Callahan et al., 2016) and QIIME2 (Bolyen et al., 2019), which both rely on user-making decisions concerning quality filtering and error learning which is based on the region amplified and organism studied.
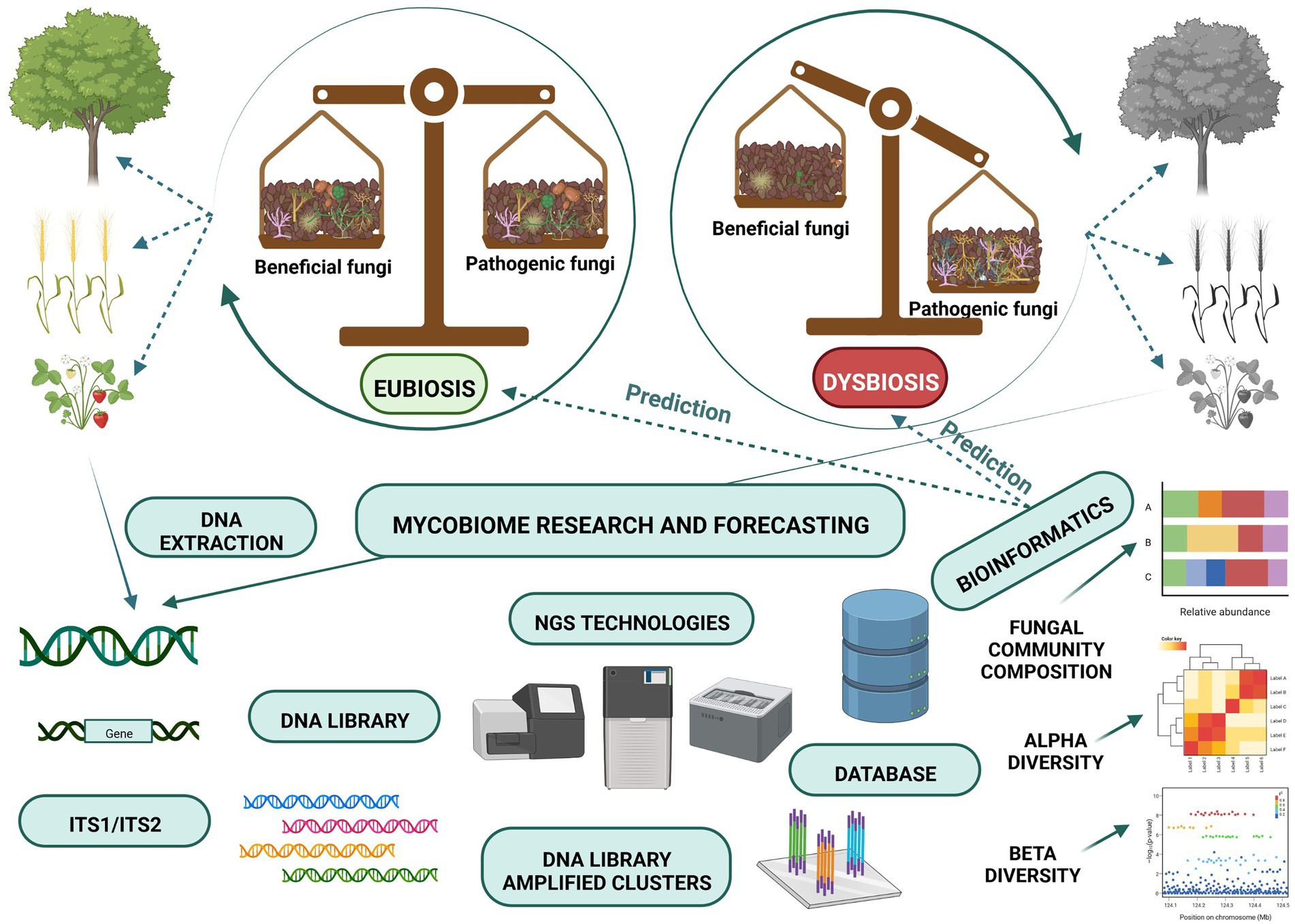
Figure 2. Next Generation Sequencing and bioinformatic programs are modern tools for the monitoring of soil mycobiome composition.
Some pipelines have been designed explicitly for fungi (such as PIPITS; Gweon et al., 2015) in which some decisions and steps are automated. For fungi, it is always advised to extract the ITS region amplified after joining pair-end reads to increase the quality of the data (Bengtsson-Palme et al., 2013). As the ITS region is variable in length (Schoch et al., 2014), the step of filtering reads of a specific size should be omitted. Using a curated database like UNITE (Nilsson et al., 2019) is highly recommended when studying fungi. UNITE relies on the species hypothesis concept, which clusters similar sequences into one hypothetical taxonomic unit. After the sequencing data is paired, filtered, clustered, and assigned the appropriate taxonomy, the hypothesis can be tested using multivariate statistical analyses.
Similarly to bacteria, transformations of data and careful interpretation are required to reach reliable conclusions (Weiss et al., 2017; Knight et al., 2018). Unique to fungi, potential functional guilds (such as pathogens, mutualists and saprotrophs) may be assigned using Funguild (Nguyen et al., 2016) or FungalTraits (Põlme et al., 2020) and the richness and relative abundance of these groups can then be evaluated (Hannula and Träger, 2020). However, for many sequence types or taxonomic units, no information concerning their lifestyle is available, thereby rendering this prediction uncertain. Future studies should focus on the functioning of fungal species as well as the functional sequencing of the soils to shed light on the functional diversity of fungi in the soils.
Concluding remarks
Mycobiomes comprise a large part of the Earth’s biodiversity and play a key role in soils, where they perform numerous functions within the ecosystem. Soil fungi play a crucial role in the environment, and affect plant health as symbiotes, pathogens, or through matter degradation. Agroecosystem mycobiomes are increasingly recognized as beneficial to soil and plant health as they facilitate and even control numerous ecosystem processes. In order to meet the various challenges of maintaining food security and the environment, mycobiome studies connected with plant pathology and protection should implement multidisciplinary approaches, including the use of traditional, molecular and bioinformatic tools. Further research into the identity, abundance, distribution and function of soil mycrobiomes, as well as their different roles in soils, is necessary to understand the dimensions of fungal biodiversity, its impact on plant health and to prevent fungal diseases. It is essential to focus on mycobiome shifts caused by climate change, their interactions with other microbes, and the determining relations between mycobiomes and microbiomes in both healthy and dysfunctional conditions.
The challenge for the future is not only to gain the ability to rapidly recognize the major and trace components of the soil mycobiomes and their functions within networks, as well as their interplay with plant roots and other constituents of the soil but also to gain the ability to redirect them into the desired composition and proportions, thereby healing the ecosystem in question. The coming years will show whether it is just a dream or the first solutions of this kind will start functioning in practice.
Author contributions
MF, EH, JS, MB, and MJ wrote, drafted, read, corrected, improved, revised and accepted the last version of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the following projects: BIOSTRATEG3/344433/16/NCBR/2018 funding by The National Centre for Research and Development within the framework of the BIOSTRATEG Programme, SUSCROP/I/POTATOMETABIOME/01/2019 funding by The National Centre for Research and Development within the framework of the ERA-NET SusCrop Programme and the paper was partially financed within the framework of Ministry of Science and Higher Education programme ‘Regional Initiative of Excellence’ in years 2019-2022, Project No. 005/ RID/2018/19’.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arnault, G., Mony, C., and Vandenkoornhuyse, P. (2022). Plant microbiota dysbiosis and the Anna Karenina principle. Trends Plant Sci. 22, 1360–1385. doi: 10.1016/j.tplants.2022.08.012
Asplund, J., Kauserud, H., Ohlson, M., and Nybakken, L. (2019). Spruce and beech as local determinants of forest fungal community structure in litter, humus and mineral soil. FEMS Microbiol. Ecol. 95:fyi232. doi: 10.1093/femsec/fiy232
Baldrian, P. (2017). Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol. Rev. 41, 109–130. doi: 10.1093/femsre/fuw040
Bamisile, B. S., Dash, C. K., Akutse, K. S., Keppanan, R., and Wang, L. (2018). Fungal endophytes: beyond herbivore management. Front. Microbiol. 9:544. doi: 10.3389/fmicb.2018.00544
Bass, D., Stentiford, G. D., Wang, H. C., Koskella, B., and Tyler, C. R. (2019). The Pathobiome in animal and plant diseases. Trends Ecol. Evol. 34, 996–1008. doi: 10.1016/j.tree.2019.07.012
Begum, N., Qin, C., Ahanger, M. A., Raza, S., Khan, M. I., Ashraf, M., et al. (2019). Role of arbuscular mycorrhizal fungi in plant growth regulation: implications in abiotic stress tolerance. Front. Plant Sci. 10:1068. doi: 10.3389/fpls.2019.01068
Bengtsson-Palme, J., Ryberg, M., Hartmann, M., Branco, S., Wang, Z., Godhe, A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919. doi: 10.1111/2041-210X.12073
Berg, G., Kusstatscher, P., Abdelfattahm, A., Cernava, T., and Smalla, K. (2021). Microbiome modulation—toward a better understanding of plant microbiome response to microbial inoculants. Front. Microbiol. 12:650610. doi: 10.3389/fmicb.2021.650610
Berg, G., Rybakova, D., Fischer, D., Cernaval, T., Vergès, M. C. C., Charles,, et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome. 8:103. doi: 10.1186/s40168-020-00875-0
Beylich, A., Oberholzer, H. R., Schrader, S., Höper, H., and Wilke, B. M. (2010). Evaluation of soil compaction effects on soil biota and soil biological processes in soils. Soil Tillage Res. 109, 133–143. doi: 10.1016/j.still.2010.05.010
Bizabani, C., and Dames, J. (2015). Effects of inoculating Lachnum and Cadophora isolates on the growth of Vaccinium corymbosum. Microbiol. Res. 181, 68–74. doi: 10.1016/j.micres.2015.08.005
Błaszczyk, L., Siwulski, M., Sobieralski, K., Lisiecka, J., and Jędryczka, M. (2014). Trichoderma spp. application and prospects for use in organic farming and industry. J. Plant Prot. Res. 54, 309–317. doi: 10.2478/jppr-2014-0047
Boddy, L. (2000). Interspecific combative interactions between wood-decaying basidiomycetes. FEMS Microbiol. Ecol. 31, 185–194. doi: 10.1111/j.1574-6941.2000.tb00683.x
Bödeker, I. T. M., Lindahl, B. D., Olson, Å., and Clemmensen, K. E. (2016). Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 30, 1967–1978. doi: 10.1111/1365-2435.12677
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Boyer, R. L., Feng, W., Gulbis, N., Hajdu, K., Harrison, R. J., Jeffries, P., et al. (2016). The use of arbuscular mycorrhizal fungi to improve strawberry production in coir substrate. Front. Plant Sci. 7:1237. doi: 10.3389/fpls.2016.01237
Calderon, C. E., Rotem, N., Harris, R., Vela-Corcia, D., and Levy, M. (2019). Pseudozyma aphidis activates reactive oxygen species production, programmed cell death and morphological alterations in the necrotrophic fungus Botrytis cinerea. Mol. Plant Pathol. 20, 562–574. doi: 10.1111/mpp.12775
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Canassa, F., Esteca, F. C. N., Moral, R. A., and Meyling, N. V.,·Klingen, I., and Delalibera, I. (2020). Root inoculation of strawberry with the entomopathogenic fungi Metarhizium robertsii and Beauveria bassiana reduces incidence of the twospotted spider mite and selected insect pests and plant diseases in the field. J. Pest. Sci. 93, 261–274. doi: 10.1007/s10340-019-01147-z
Chakravarty, P., and Hwang, S. F. (1991). Effect of an ectomycorrhizal fungus, Laccaria laccata on fusarium damping-off in Pinus banksiana seedlings. Eur. J. Plant Pathol. 21, 97–106. doi: 10.1111/j.1439-0329.1991.tb00949.x
Chen, L., Boka, B., Kedves, O., Nagy, V. D., Szűcs, A., Champramary, S., et al. (2019). Towards the biological control of devastating Forest pathogens from the genus Armillaria. Forests 10:1013. doi: 10.3390/f10111013
Chen, T., Nomura, K., Wang, X., Sohrabi, R., Xu, J., Yao, L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature. 580, 653–657. doi: 10.1038/s41586-020-2185-O
Coleman-Derr, D., and Tringe, S. G. (2014). Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 5:283. doi: 10.3389/fmicb.2014.00283
Cota, L. V., Maffia, L. A., Mizubuti, E. S. G., and Macedo, P. E. F. (2009). Biological control by Clonostachys rosea as a key component in the integrated management of strawberry gray mold. Biol. Control 50, 222–230. doi: 10.1016/j.biocontrol.2009.04.017
Crippa, M., Solazzo, E., Guizzardi, D., Montforti-Ferrario, F., Tubiello, F. N., and Leip, A. (2021). Food systems are responsible for a third of global anthropogenic GHG emissions. Nature Food 2, 198–209. doi: 10.1038/s43016-021-00225-9. ISSN 2662-1355
D’Andreano, S., Cuscó, A., and Francino, O. (2021). Rapid and real-time identification of fungi up to species level with long amplicon nanopore sequencing from clinical samples. Biol. Methods Protoc. bpaa026, 1–6. doi: 10.1093/biomethods/bpaa026
Dastogeer, K. M. G., Tumpa, F. H., Sultana, A., Akter, M. J., and Chakraborty, A. (2020). Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 23:100161. doi: 10.1016/j.cpb.2020.100161
Dastogeer, K. M. G., Yasuda, M., and Okazaki, S. (2022). Microbiome and pathobiome analyses reveal changes in community structure by foliar pathogen infection in rice. Front. Microbiol. 13:949152. doi: 10.3389/fmicb.2022.949152
Dawidziuk, A., Popiel, D., Kaczmarek, J., Strakowska, J., and Jędryczka, M. (2016). Optimal Trichoderma strains for control of stem canker of brassicas: molecular basis of biocontrol properties and azole resistance. BioControl. 61, 755–761. doi: 10.1007/s10526-016-9743-2
De Miccolis Angelini, R. M., Rotolo, C., Gerin, D., Abate, D., Pollastro, S., and Faretra, F. (2019). Global transcriptome analysis and differentially expressed genes in grapevine after application of the yeast derived defense inducer cerevisane. Pest Manag. Sci. 75, 2020–2033. doi: 10.1002/ps.5317
Debode, J., Van Hemelrijck, W., Heungens, K., Maes, M., and Creemers, P. (2011). First report of Pilidium concavum causing tan-Brown rot on strawberry fruit in Belgium. Plant Dis. 95:1029. doi: 10.1094/PDIS-10-10-0752
Delaye, L., García-Guzmán, G., and Heil, M. (2013). Endophytes versus biotrophic and necrotrophic pathogens—are fungal lifestyles evolutionarily stable traits? Fungal Divers. 60, 125–135. doi: 10.1007/s13225-013-0240-y
Deng, X., Zhang, N., Li, Y., Zhu, C., Qu, B., Liu, H., et al. (2022). Bio-organic soil amendment promotes the suppression of Ralstonia solanacearum by inducing changess in the functionality and composition of rhizosphere bacterial communities. New Phytol. 235, 1558–1574. doi: 10.1111/nph.18221
Deng, X., Zhang, N., Shen, Z., Zhu, C., Liu, H., Xu, Z., et al. (2021). Soil microbiome manipulation triggers direct and possible indirect suppression against Ralstonia solanacearum and fusarium oxysporum. Nat. Biofilms 7:33. doi: 10.1038/s41522-021-00204-9
Dubey, A., Malla, A. M., Khan, F., Chowdhary, K., Yadav, S., Kumar, A., et al. (2019). Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers. Conserv. 28, 2405–2429. doi: 10.1007/s10531-019-01760-5
EASAC (2022). Regenerative agriculture in Europe: a critical analysis of contributions to European Union farm to fork and biodiversity strategies. European Academies Science Advisory Council, policy report 44, 1–70. ISBN: 978-3-8047-4372-4, www.easac.eu
European Commission (2020). A European green Deal. Available at: https://ec.europa.eu/info/strategy/priorities-2019-2024/european-green-deal_en (accessed August 31, 2022).
FAO (2021). News article: food systems account for more than one third of global greehhouse gas emissions. Available at: www.fao.org (accessed April 22, 2021).
Fehrer, J., Réblová, M., Bambasová, V., and Vohník, M. (2019). The root-symbiotic Rhizoscyphus ericae aggregate and Hyaloscypha (Leotiomycetes) are congeneric: phylogenetic and experimental evidence. Stud. Mycol. 92, 195–225. doi: 10.1016/j.simyco.2018.10.004
Fernandes, M. L. P., Bastida, F., Jehmlich, N., Martinović, T., Větrovský, T., Baldrian, P., et al. (2022). Functional soil mycobiome across ecosystems. J. Proteome 252:104428. doi: 10.1016/j.jprot.2021.104428
Frąc, M., Hannula, S. E., Bełka, M., and Jędryczka, M. (2018). Fungal biodiversity and their role in soil health. Front. Microbiol. 9:707. doi: 10.3389/fmicb.2018.00707
Frau, A., Kenny, J. G., Lenzi, L., Campbell, B. J., Ijaz, U. Z., Duckworth, C. A., et al. (2019). DNA extraction and amplicon production strategies deeply influence the outcome of gut mycobiome studies. Sci. Rep. 9:9328. doi: 10.1038/s41598-019-44974-x
Freimoser, F. M., Rueda-Mejia, M. P., Tilocca, B., and Migheli, Q. (2019). Biocontrol yeasts: mechanisms and applications. World J. Microbiol. Biotechnol. 35:154. doi: 10.1007/s11274-019-2728-4
Frouz, J., and Frouzová, J. (2022). How agriculture, forestry and fisheries shape our planet. Cham, Switzerland: Springer Nature.
Garrido, C., González-Rodríguez, V. E., Carbú, M., Husaini, A. M., and Cantoral, J. M. (2016). “Fungal diseases of strawberry and their diagnosis,” in Strawberry: Growth, Development and Diseases. eds. A. M. Husaini and D. Neri. (Wallingford, UK: CABI BooI, CABI International), 157–195.
Geisen, S. (2021). The future of (soil) microbiome studies: current limitations, integration, and perspectives. mSystems 6, e00613–e00621. doi: 10.1128/mSystems.00613-21
Gosnell, H. (2022). Regenerating soil, regenerating soul: an integral approach to understanding agricultural transformation. Sustain. Sci. 17, 603–620. doi: 10.1007/s11625-021-00993-0
Gostinčar, C., Turk, M., Zajc, J., and Gunde-Cimerman, N. (2019). Fifty Aureobasidium pullulans genomes reveal a recombining polyextremotolerant generalist. Environ. Microbiol. 21, 3638–3652. doi: 10.1111/1462-2920.14693
Gubler, D. W., and Converse, R. H. (1993). Disease of Strawberry. American Phytopathological Society, St Paul, Minnesota.
Gweon, H. S., Oliver, A., Taylor, J., Booth, T., Gibbs, M., Read, D. S., et al. (2015). PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 6, 973–980. doi: 10.1111/2041-210X.12399
Hamim, A., Miché, L., Douaik, A., Mrabet, R., Ouhammou, A., Duponnois, R., et al. (2017). Diversity of fungal assemblages in roots of Ericaceae in two Mediterranean contrasting ecosystems. Comptes Rendus – Biol. 340, 226–237. doi: 10.1016/j.crvi.2017.02.003
Hannula, S. E., and Träger, S. (2020). Soil fungal guilds as important drivers of the plant richness–productivity relationship. New Phytol. 226, 947–949. doi: 10.1111/nph.16523
Havis, N. D., Brown, J. K. M., Clemente, G., Frei, P., Jedryczka, M., Kaczmarek, J., et al. (2015). Ramularia collo-cygni - an emerging pathogen of barley crops. Phytopathology 105, 895–904. doi: 10.1094/PHYTO-11-14-0337-FI
Iebba, V., Totino, V., Gagliardi, A., Santangelo, F., Cacciotti, F., Trancassini, M., et al. (2016). Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol. 39, 1–12.
Jansa, J., and Vosátka, M. (2000). In vitro and post vitro inoculation of micropropagated rhododendrons with ericoid mycorrhizal fungi. Appl. Soil Ecol. 15, 125–136. doi: 10.1016/S0929-1393(00)00088-3
Jeger, M., Beresford, R., Bock, C., Brown, N., Fox, A., Newton, A., et al. (2021). Global challenges facing plant pathology: multidisciplinary approaches to meet the food security and environmental challenges in the mid-twenty-first century. CABI Agric Biosci 2:20. doi: 10.1186/s43170-021-00042-x
Jia, M., Sun, X., Chen, M., Liu, S., Zhou, J., and Peng, X. (2022). Deciphering the microbial diversity associated with healthy and wilted Paeonia suffruticosa rhizosphere soil. Front. Microbiol. 13:967601. doi: 10.3389/fmicb.2022.967601
Karimi, K., Arzanlou, M., Babai-Ahari, A., and Pertot, I. (2016). Biological and molecular characterisation of Pilidium lythri, an emerging strawberry pathogen in Iran. Phytopathol. Mediterr. 55, 366–379. doi: 10.14601/Phytopathol_Mediterr-18391
Kennedy, P. (2010). Ectomycorrhizal fungi and interspecific competition: species interactions, community structure, coexistence mechanisms, and future research directions. New Phytol. 187, 895–910. doi: 10.1111/j.1469-8137.2010.03399.x
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., and Druzhinina, J. S. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. doi: 10.1186/gb-2011-12-4-r40
Lau, J. A., and Lennon, J. T. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. PNAS 109, 14058–14062. doi: 10.1073/pnas.1202319109
Li, Y., Guo, Q., He, F., Li, Y., Xue, Q., and Lai, H. (2020). Biocontrol of root diseases and growth promotion of the tuberous plant Aconitum carmichaelii induced by Actinomycetes are related to shifts in the rhizosphere microbiota. Microb. Ecol. 79, 134–147. doi: 10.1007/s00248-019-01388-6
Li, W. Q., Huang, Y. X., Chen, F. S., Liu, Y. Q., Lin, X. F., Zong, Y. Y., et al. (2021). Mixing with broad-leaved trees shapes the rhizosphere soil fungal communities of coniferous tree species in subtropical forests. For. Ecol. Manag. 480:118664. doi: 10.1016/j.foreco.2020.118664
Litwin, A., Nowak, M., and Różalska, S. (2020). Entomopathogenic fungi: unconventional applications. Rev. Environ. Sci. Biotechnol. 19, 23–42. doi: 10.1007/s11157-020-09525-1
Lladó, S., López-Mondéjar, R., and Baldrian, P. (2018). Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 102, 4331–4338. doi: 10.1007/s00253-018-8950-4
Lombardi, N., Caira, S., Troise, A. D., Scaloni, A., Vitaglione, P., Vinale, F., et al. (2020). Trichoderma applications on strawberry plants modulate the physiological processes positively affecting fruit production and quality. Front. Microbiol. 11:1364. doi: 10.3389/fmicb.2020.01364
Mącik, M., Gryta, A., and Frąc, M. (2020). Biofertilizers in agriculture: an overview on concepts, strategies and effects on soil microorganisms. Adv. Agron. 162, 31–87. doi: 10.1016/bs.agron.2020.02.001
Malarczyk, D., Panek, J., and Frąc, M. (2019). Alternative molecular-based diagnostic methods of plant pathogenic fungi affecting berry crops – a review. Molecules 162, 31–87. doi: 10.3390/molecules24071200
Malarczyk, D. G., Panek, J., and Frąc, M. (2020). Triplex real-time pcr approach for the detection of crucial fungal berry pathogens – botrytis spp., colletotrichum spp. and verticillium spp. Int. J. Mol. Sci. 24:1200. doi: 10.3390/ijms21228469
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1202007
Murphy, B. R., Soldi, E., Jadwiszczak, M. J., and Hodkinson, T. R. (2019). Synergy between fungal endophytes improves fruit production in strawberry cultivar. Emer Life Sci Res 5, 29–41. doi: 10.31783/elsr.2019.512941
Myers, N. (2002). Environmental refugees: a growing phenomenon of the 21st century. Philos. Trans. Proc. Royal Soc. 357, 609–613. doi: 10.1098/rstb.2001.0953
Newton, P., Civita, N., Frankel-Goldwater, L., Bartel, K., and Johns, C. (2020). What is regenerative agriculture? A review of scholar and practitioner definitions based on processes and outcomes. Front. Sustain. Food Syst. 4:577723. doi: 10.3389/fsufs.2020.577723
Nguyen, M. P., Lehosmaa, K., Martz, F., Koskimäki, J. J., Pirttilä, A. M., and Häggman, H. (2021). Host species shape the community structure of culturable endophytes in fruits of wild berry species (Vaccinium myrtillus L., Empetrum nigrum L. and Vaccinium vitis-idaea L.). FEMS Microbiol. Ecol. 97:fiab097. doi: 10.1093/femsec/fiab0
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Nobre, C. A., Sampaio, G., Borma, L. S., Castilla-Rubio, J. C., Silva, J. S., and Cardoso, M. (2016). Land-use and climate change risks in the Amazon and the need of a novel sustainable development paradigm. PNAS 113, 10759–10768. doi: 10.1073/pnas.16055161
O’Brien, A. T. (2013). Supporting soil fungi to rebuild soils in agriculture. PAN: philosophy, activism. Nature 10, 24–35. doi: 10.4225/03/5851e79846008
O'Hanlon, R. (2017). Fungi in the Environment, in: Fungi: Biology and Applications, John Wiley & Sons Inc, New York.
Oszust, K., Pylak, M., and Frąc, M. (2021). Trichoderma-based biopreparation with prebiotics supplementation for the naturalization of raspberry plant rhizosphere. Int. J. Mol. Sci. 22:6356. doi: 10.3390/ijms
Peng, G., McGregor, L., Lahlali, R., Gossen, B. D., Hwang, S. F., Adhikari, K. K., et al. (2011). Potential biological control of clubroot on canola and crucifer vegetable crops. Plant Pathol. 60, 566–574. doi: 10.1111/j.1365-3059.2010.02400.x
Pérez-Izquierdo, L., Rincón, A., Lindahl, B. D., and Buée, M. (2021). Fungal community of forest soil: diversity, functions, and services. Forest Microbiol. 1, 231–255. doi: 10.1016/B978-0-12-822542-4.00022-X
Põlme, S., Abarenkov, K., Nilsson, H. R., Lindahl, B. D., Clemmensen, K. E., Kauserud, H., et al. (2020). Fungal traits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 105, 1–16. doi: 10.1007/s13225-020-00466-2
Prasongsuk, S., Lotrakul, P., Ali, I., Bankeeree, W., and Punnapayak, H. (2018). The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 63, 129–140. doi: 10.1007/s12223-017-0561-4
Pylak, M., Oszust, K., and Frąc, M. (2019). Review report on the role of bioproducts, biopreparations, biostimulants and microbial inoculants in organic production of fruit. Rev. Environ. Sci. Biotechnol. 18, 597–616. doi: 10.1007/s11157-019-09500-5
Rees, H. J., Drakulic, J., Cromey, M. G., Bailey, A. M., and Foster, G. D. (2022). Endophytic Trichoderma spp. can protect strawberry and privet plants from infection by the fungus Armillaria mellea. PLoS One 17:e0271622. doi: 10.1371/journal.pone.0271622
Runnel, K., Abarenkov, K., Copoț, O., Mikryukov, V., Kõljalg, U., Saar, I., et al. (2022). DNA barcoding of fungal specimens using PacBio long-read high-throughput sequencing. Mol. Ecol. Resour. 22, 2871–2879. doi: 10.1111/1755-0998.13663
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). Assessing the global burden of pathogens and pests on major food crops. Nature Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schlatter, D., Kinkel, L., Thomashow, L., Weller, D., and Paulitz, T. (2017). Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107, 1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW
Schoch, C. L., Robbertse, B., Robert, V., Vu, D., Cardinali, G., Irinyi, L., et al. (2014). Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database J. Biol. 2014:bau061. doi: 10.1093/database/bau061
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., and Levesque, C. A. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Sinclair, G., Charest, C., Dalpé, Y., and Khanizadeh, S. (2013). Influence of arbuscular mycorrhizal fungi and a root endophyte on the biomass and root morphology of selected strawberry cultivars under salt conditions. Can. J. Plant Sci. 93, 997–999. doi: 10.4141/cjps2012-279
Smolinska, U., and Kowalska, B. (2018). Biological control of the soil-borne fungal pathogen Sclerotinia sclerotiorum - a review. J. Plant Pathol. 100, 1–12. doi: 10.1007/s42161-018-0023-0
Stewart, J. E., Kim, M., Lalande, B., and Klopfenstein, N. B. (2021). Pathobiome and Microbial Communities Associated with Forest Tree Root Diseases, in Forest microbiology, Forest Microbiology, Cambridge Academic Press.
Suárez, J. O., Villada, D., de Rueda, J. A. O., Alves-Santos, F. M., and Diez, J. J. (2018). Effects of Lactarius deliciosus and Rhizopogon roseolus ectomycorrhyzal fungi on seeds and seedlings of scots and stone pines inoculated with fusarium oxysporum and fusarium verticillioides. For. Chron. 94, 126–134. doi: 10.5558/tfc2018-019
Sun, C., Fu, D., Lu, H., Zhang, J., Zheng, X., and Yu, T. (2018). Autoclaved yeast enhances the resistance against Penicillium expansum in postharvest pear fruit and its possible mechanisms of action. Biol. Control. doi: 10.1016/j.biocontrol.2018.01.010
Sun, Z. B., Li, S. D., Ren, Q., Xu, J. L., Lu, X., and Sun, M. H. (2020). Biology and applications of Clonostachys rosea. J. Appl. Microbiol. 129, 486–495. doi: 10.1111/jam.14625
Tedersoo, L., Anslan, S., Bahram, M., Põlme, S., Riit, T., Liiv, I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera,, et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688
Torbati, M., Arzanlou, M., Abed-Ashtiani, F., and Golmohammadi, H. (2019). Occurrence of fruit rot on cornelian cherry caused by Pilidium lythri in Iran. Crop Prot. 125:104884. doi: 10.1016/j.cropro.2019.104884
van der Heijden, M. G. A., and Hartmann, M. (2016). Networking in the plant microbiome. PLoS Biol. 14:e1002378. doi: 10.1371/journal.pbio.1002378
van Dijk, L. J. A., Abdelfattah, A., Ehrlén, J., and Tack, A. J. M. (2022). Soil microbiomes drive aboveground plant–pathogen–insect interactions. Oikos 2022:e09366. doi: 10.1111/oik.09366
Vétrovský, T., Kohout, P., Kopecký, M., Machac, A., Man, M., Bahnmann, B. D., et al. (2019). A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat. Commun. 10:5142. doi: 10.1038/s41467-019-13164-8
Vincent, D., Rafiqi, M., and Job, D. (2020). The multiple facets of plant–fungal interactions revealed through plant and fungal Secretomics. Front. Plant Sci. 10:1626. doi: 10.3389/fpls.2019.01626
Ważny, R., Jędrzejczyk, R. J., Rozpądek, P., Domka, A., and Turnau, K. (2022). Biotization of highbush blueberry with ericoid mycorrhizal and endophytic fungi improves plant growth and vitality. Appl. Microbiol. Biotechnol. 106, 4775–4786. doi: 10.1007/s00253-022-12019-5
Wei, Z., Gu, Y., Friman, V. P., Kowalchuk, G. A., Xu, Y., Shan, Q., et al. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5:eaaw0759. doi: 10.1126/sciadv.aaw0759
Weiss, S., Xu, Z. Z., Peddada, S., Amir, A., Bittinger, K., Gonzalez, A., et al. (2017). Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 5:27. doi: 10.1186/s40168-017-0237-y
Weller, D. M., Raaijsmakers, J. M., McSpadden Gardener, B. B., and Thomashow, L. S. (2002). Microbial populations responsible for specific suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.0300402.110010
Wiggins, B. E., and Kinket, L. L. (2005a). Green manures and crop sequences influence alfalfa root rot and pathogen inhibitory activity among soil-borne streptomycetes. Plant Soil 268, 271–283. doi: 10.1007/s11104-004-0300-x
Wiggins, B. E., and Kinket, L. L. (2005b). Green manures and crop sequences influence potato diseases and pathogen inhibitory activity of indigenous streptomycetes. Phytopathology 95, 178–185. doi: 10.1094/PHYTO-95-0178
Witzell, J., and Martín, J. A. (2018). Endophytes of Forest Trees: Biology and Applications, in: Endophytes and Forest Health BT - Endophytes of Forest Trees: Biology and Applications. New York Springer International Publishing.
Yadav, A. N., Kour, D., Kaur, T., Devi, R., Yadav, A., and Dikilitas, M. (2021). Biodiversity, and biotechnological contribution of beneficial soil microbiomes for nutrient cycling, plant growth improvement and nutrient uptake. Biocat. Agricult. Biotechnol 33:102009. doi: 10.1016/j.bcab.2021.102009
Yan, D. F., Mills, J. G., Gellie, N. J. C., Bissett, A., Lowe, A. J., and Breed, M. F. (2018). High-throughput eDNA monitoring of fungi to track functional recovery in ecological restoration. Biol. Conserv. 217, 113–120. doi: 10.1016/j.biocon.2017.10.035
Keywords: bioinformatics, dysbiosis, eubiosis, fungal microbiome, microbiome-mediated plant protection, phytopathogens, regenerative agriculture
Citation: Frąc M, Hannula ES, Bełka M, Salles JF and Jedryczka M (2022) Soil mycobiome in sustainable agriculture. Front. Microbiol. 13:1033824. doi: 10.3389/fmicb.2022.1033824
Edited by:
Khondoker M. G. Dastogeer, Bangladesh Agricultural University, BangladeshReviewed by:
Daniel Manter, USDA-ARS, United StatesCopyright © 2022 Frąc, Hannula, Bełka, Salles and Jedryczka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malgorzata Jedryczka, bWplZEBpZ3IucG96bmFuLnBs
 Magdalena Frąc
Magdalena Frąc Emilia Silja Hannula
Emilia Silja Hannula Marta Bełka
Marta Bełka Joana Falcao Salles
Joana Falcao Salles Malgorzata Jedryczka
Malgorzata Jedryczka