- 1MOE Joint International Research Laboratory of Animal Health and Food Safety, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, Jiangsu, China
- 2Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
Bovine herpesvirus type 1 (BHV-1) causes bovine respiratory disease that poses a significant threat to the cattle industry. The prevalence of BHV-1 has recently increased in China. However, the lack of information about the prevalent isolates limits the control of the disease. In this study, a novel strain of BHV-1 was isolated from nasal swabs of Holstein cows in 2020 in China, designated as BHV SHJS. The genome of BHV strain SHJS is 135, 102 bp in length and highly similar to strain SP1777 (KM258883.1) with an identity of 99.64%. Mutations, insertions, or deletions mainly occur in UL27, UL44, and US8, etc., relative to the different genomic coordinates. Phylogenetic tree of UL44 (gC) showed that BHV strain SHJS belongs to BHV-1.2b cluster. The result showed that the strain had a different evolutionary origin from those prevalent in China. This study will enrich our knowledge regarding BHV outbreak strains in China and contribute to the prevention and pathogenic studies of BHV-1.2.
Introduction
Bovine herpesvirus type 1 (BHV-1) was a key pathogen causing a contagious bovine respiratory disease or abortion that poses a threat and significant economic losses in cattle industry (Brake and Studdert, 1985; Hodgson et al., 2005; Jones and Chowdhury, 2007). BHV-1 belongs to the family Herpesviridae, subfamily Alphaherpesvirinae (Nandi et al., 2009). It was divided into BHV-1.1 and 1.2 types, which was further classified into two gene subtypes: BHV 1.2a and 1.2b (Metzler et al., 1985; Zhou et al., 2020).
Bovine herpesvirus type 1 exhibits a double-stranded DNA of approximately 135 kb in size (Engels et al., 1987). The BHV-1 genome contains a unique long sequence region (UL), a unique short sequences region (US), an internal repeat sequences region (IR) between UL and US, and an inverted terminal repeat sequences region (TR) (Leung-Tack, et al., 1994; d'Offay et al., 2016). It encodes about 73 open reading frames (ORFs) (Engels et al., 1986). Glycoprotein B (gB), glycoprotein C (gC), and glycoprotein D (gD) encoded by UL27, UL44, and US6, respectively, were the major attachment proteins, which could induce a high level of neutralizing antibodies against BHV-1 (Babiuk et al., 1987; Van Drunen Littel-van den Hurk et al., 1990; Liang et al., 1991). Inactivated or attenuated vaccines are used against BHV-1 conventionally (Deshpande et al., 2002; O'Toole et al., 2012; Marawan et al., 2021). However, they only partially protect from wild-type BHV-1 infection (O'Toole et al., 2014; Fulton et al., 2015, 2016).
In China, the prevalence of BHV-1 presented fluctuations of 5 to 68.7% from 2012 to 2017 in cattle (Zhong et al., 2012; Pi et al., 2014; Mo and Liu, 2017). A meta-analysis showed the prevalence of BHV infection even increased up to 40% in China (Chen et al., 2018). To date, information about the prevalent isolates, and biological characterization of BHV-1 strains were scarce in China. Therefore, isolation of epidemic strains, analysis of genome of isolates on evolutionary status, and exploration of biological characterization were very meaningful to enrich our knowledge regarding BHV outbreak strains.
In 2020, a local farm of Holstein cows developed severe respiratory inflammation and abortion symptoms in Shanghai, China. We suspected that those cows might be infected with BHV-1. In this study, a novel strain of BHV-1.2b was isolated from the nasal swabs. The genome of the isolate was sequenced and the biological characteristics of the virus were systemically investigated. This study will contribute to investigating the evolution of BHV-1.2b and aid in its future prevention or management.
Materials and methods
Cells and virus isolation
Madin-Darby bovine kidney cells (MDBK) were purchased from the American Type Culture Collection and stored in the laboratory. Cells were incubated using Dulbecco’s Modified Eagle’s Medium (DMEM) (BI, China) with 8% fetal bovine serum (FBS) (PAN Biotech, Germany) at 37°C in a 5% CO2 incubator. Thirty-three nasal swabs were collected from Holstein cows from a bovine farm in Jinshan District, Shanghai, China, in 2020. BHV-1 US7 positive samples were homogenized with DMEM and centrifuged using 6, 000 × g for 5 min. The supernatant was filtered by a filter unit and then inoculated onto MDBK cell monolayers grown for virus isolation by monitoring cytopathic effects (CPEs).
Polymerase chain reaction
DNA was extracted from nasal swabs and the supernatant of the infected cells using the Ezup column viral DNA kit (Sangon Biotech, China). Specific primers were designed to amplify BHV-1 US7, US8, UL48, and UL49 genes (Supplementary Table S1). PCR reactions were amplified using DNA polymerase (Vazyme Biotech, China) under the conditions: 95°C for 3 min, 35 cycles at 95°C for 15 s, 68°C for 1 min, and 72°C for 2 min (Waters and Shapter, 2014).
Plaque formation assay
MDBK cells were infected with the virus of a serial 10-fold dilution. Following 2-h incubation, the cells were washed thrice using PBS buffer. Then, the infected cells were overlaid using 1% agarose (Invitrogen, United States) that was low-melting point and supplemented with 2% FBS. Then, the cells overlaid with agarose were placed 4°C for 5 min. Next, the cells were incubated at 37°C and observed by plaque formation assay. Fifty plaques were randomly selected for virus passage and purification (Moe et al., 1981). The diameter was measured using the software of Image J (National Institutes of Health).
Growth curve and virus titration
The growth kinetics of the virus was determined by infecting MDBK cell monolayers with the virus at a multiplicity of infection (MOI = 1) for 2 h post-infection (hpi). Then, cells were washed thrice with PBS and replaced with DMEM with 2% FBS. The cells and supernatant of infection were collected at different time points (6, 12, 24, 36, and 48 hpi). Following three freeze–thaw cycles at −80°C, the pellets and supernatants were prepared at centrifugation of 12, 000 × g for 3 min. The TCID50 of these samples were determined using 96-well plates coated with MDBK cell monolayers. Cells were infected with the virus of a serial 10-fold dilutions. The CPEs of the cells were observed daily. The TCID50 were determined according to the Reed and Muench method (Reed and Muench, 1938).
Genomic sequencing and phylogenetic analysis
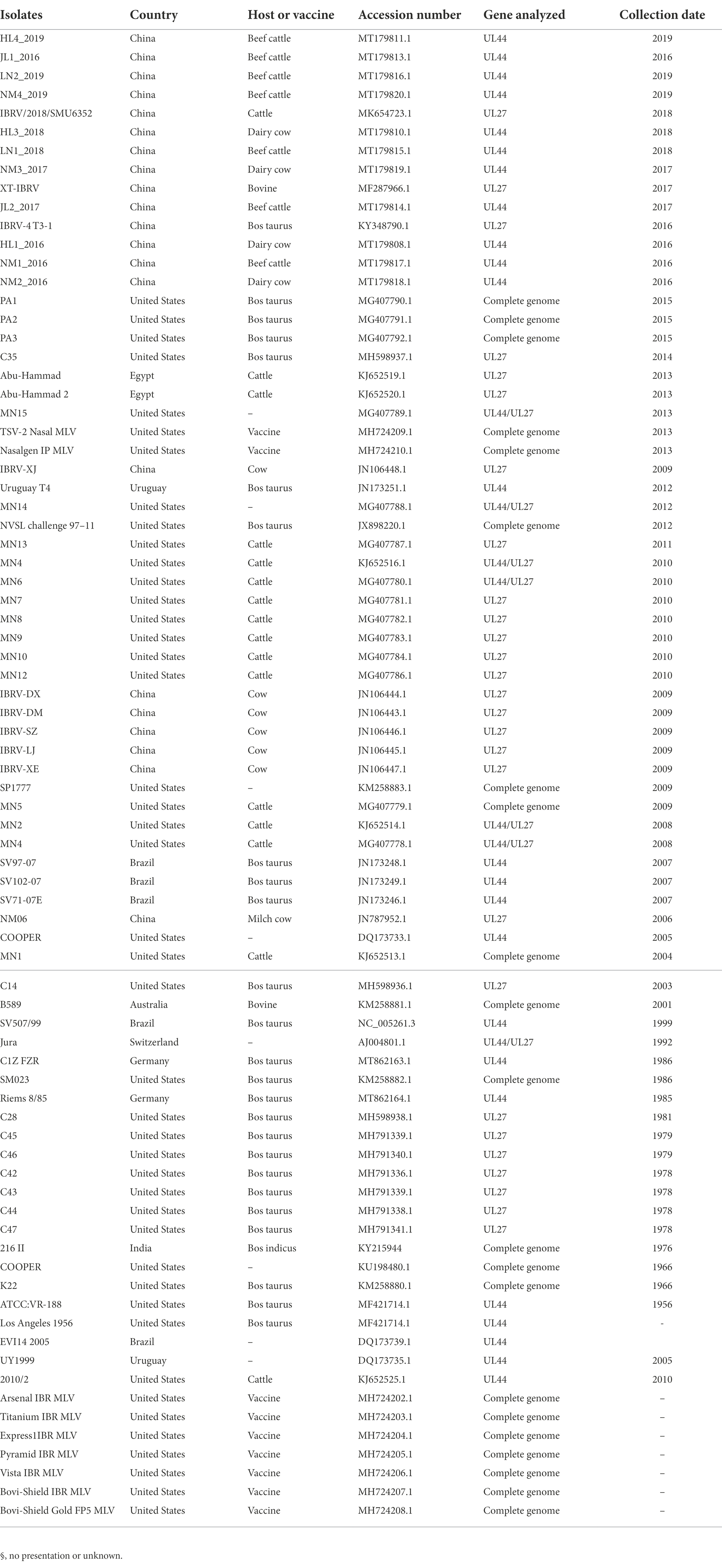
Viral DNA was extracted using Viral DNA kit (Omega, United States) from the supernatant of infected cells referring to the instructions. The full-genome sequence was obtained using the second-generation sequencing platform of the GenScript company. The full-genome sequence was compared with the published BHV-1 complete genomes in the NCBI database. The full genome of the isolated BHV-1 strain was analyzed using the viral genome with the highest homology as a reference. Phylogenetic analyses based on BHV-1 UL44 (gC) and UL27 (gB) sequences were performed using MEGA software (version 7.0.26). The reference sequences of UL44 (gC) and UL27 (gB) genes retrieved from GenBank for phylogenetic analysis are listed in Table 1.
Alignment and prediction of structure
The amino acid sequences were analyzed by alignment online.1 Protein-encoded UL42 and UL46 secondary structures were predicted by PredictProtein.2 Protein domains were predicted by SMART.3
Western-blotting analysis
The cells extracts were processed using Sodium Dodecyl Sulfate – Polyacrylamide Gel Electrophoresis (SDS-PAGE). At room temperature, proteins were electrophoresed to polyvinylidene fluoride (PVDF) membranes (Pall Corp, China) and blocked with 5% non-fat milk in PBS for 1 h. The membranes were incubated overnight at 4°C with BHV-VP22 polysera (prepared in our lab) and β-actin antibody (CST, United States). Then, membranes were washed thrice in PBST and incubated with secondary antibody (Sigma, United States). Membrane signals were detected using a Pierce™ SuperSignal West Pico Chemiluminescent Substrate using a Chemiluminescence Immunoassay image analysis system (Tanon 5,200, China).
Immunofluorescence assay
Viral replication was investigated on coverslips with MDBK cell monolayer at 6, 12, 18, 24, 36, and 48 hpi. Monolayers were incubated and then fixed with 4% formaldehyde at 37°C for 35 min. Then, the cells were washed with PBS buffer and blocked with PBS with 5% milk of non-fat at 37°C for 30 min. Cells were washed with PBS buffer and incubated with BHV-VP22 polysera (1,500 dilution) for 45 min, followed by washing with PBS and staining with 4′, 6-diamidino-2-phenylindole (DAPI). Images were captured under an ECHO fluorescence microscope (RVL-100-g, United States).
Statistical analysis
Data were collated and presented as mean ± standard deviation (SD). Differences of the groups were determined by t-test with GraphPad Prism software (version 7.0). A value of p smaller than 0.05 was considered statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001).
Results
Isolation and biological characteristics of isolated virus
Four samples were identified as positive-BHV from 33 nasal swabs (Supplementary Figure S1). Positive samples were inoculated on MDBK cells to obtain the virus. Only one sample showed visible CPEs with round and enlarged cells shrunk or detached at 36 hpi (Figure 1A). After three purification cycles, 1 to 2 mm diameter plaques were formed in infected cells compared with the mock group (Figure 1B). Three fragments were amplified from the infected cells by PCR (Figure 1C). Further sequence analysis revealed that the three amplified fragments showed high similarity to BHV-1 US7/US8 (3,156 bp), UL48 (1,524 bp), and UL49 (776 bp) (Supplementary Figure S2), respectively. The data suggested that the isolated virus was BHV-1.2b, referred to as BHV SHJS. To investigate the biological characteristics of isolated strain, the growth curve was determined on cells infected with the purified BHV SHJS (MOI = 1) at 6, 12, 18, 24, 30, 36, 42, and 48 hpi. The results showed that the virus titer was 8.02 × 107 TCID50/mL at 48 hpi (Figure 1D). Western blotting and IFA analysis showed that virus infection could be detected at 12 hpi and 6 hpi, respectively (Figures 1E,F).
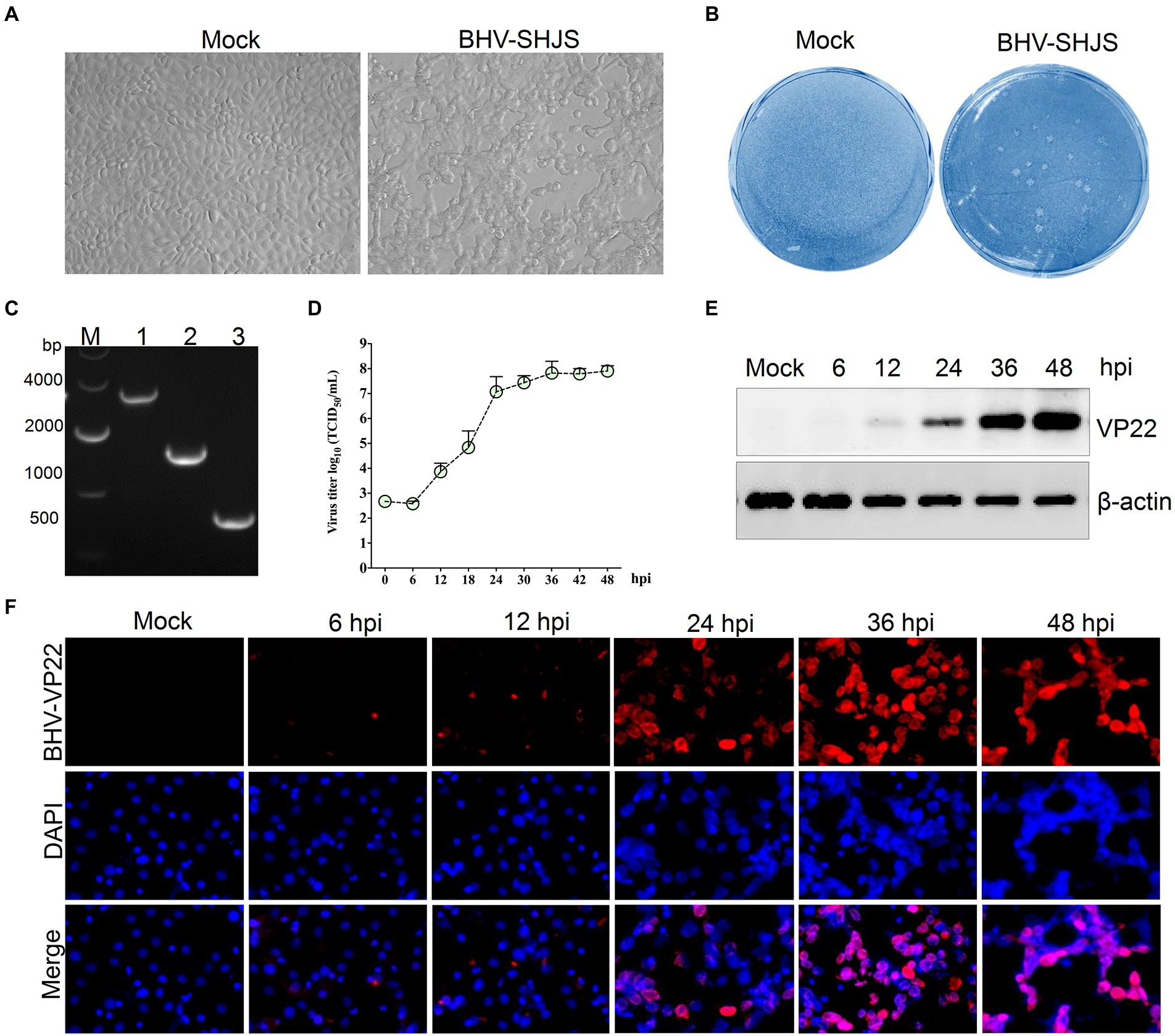
Figure 1. Isolation, identification, and biological characterization of the isolated BHV SHJS strain. (A) Isolation of virus on MDBK cells. Virus was isolated from the nasal swab. CPE was observed on BHV SHJS-infected and mock cells. (B) Plaque assay. MDBK cells were infected with the isolated virus. (C) Amplification of the conserved gene in infected cells. The fragments were amplified from extracted DNA from the supernatant of infected cells using specific primers for BHV-1 US7/US8 (lane 1), UL48 (lane 2), UL49 (lane 3). (D) Growth curves of BHV SHJS in MDBK cells. Cells were infected with BHV SHJS (MOI = 1). The TCID50 values were determined by the Reed and Muench method. Dynamic changes of viral infection were verified by western blotting (E) and IFA (F). The cells infected with the virus (MOI = 1) were incubated and recognized using VP22 polysera.
Characterization of BHV SHJS genome
The full genome of BHV strain SHJS was sequenced by second-generation sequencing. After assembly, the genome of a 135, 102 bp size of BHV strain SHJS was obtained. The average coverage depth of the assembled sequences was 3332.6×. The genome of strain SHJS has been submitted to NCBI (GenBank accession number: OP035381). Although BHV-1.2b is prevalent in China, no full-genome map of the BHV-1.2b isolates has been reported. We found that the BHV SHJS genome was collinear with BHV-1 K22 (KM258880.1) genome (d'Offay et al., 2016). The full genome of the BHV strain SHJS virus included a unique long sequence region (UL), a unique short sequences region (US), an inverted repeat sequences region (IR), and an inverted terminal repeat sequences region (TR). Sequence analysis showed that the BHV SHJS genome encodes 72 ORFs. Among them, 32 ORFs encode viral structure proteins, five ORFs encode related enzymes, and six ORFs encode regulatory function-related proteins. These are marked in different colors in the map of BHV SHJS genome (Figure 2). The position information of the genes revealed in the Supplementary Table S2.
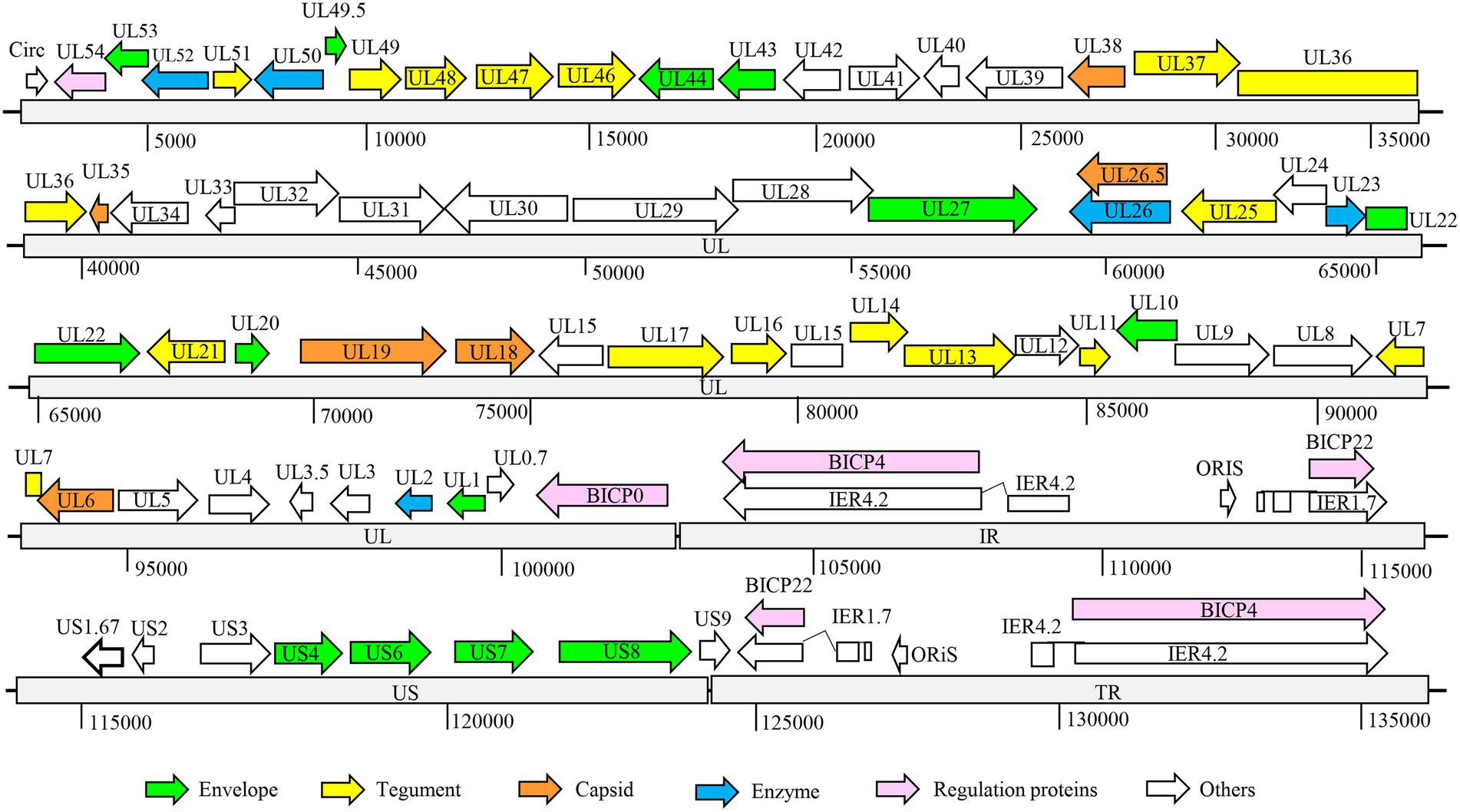
Figure 2. Organization of BHV SHJS full-genome map. Genome of BHV SHJS contains unique long sequence region (UL) and unique short (US) sequences region. An internal repeat sequence region exists between the UL and US region, called the IR region. Inverted repeat sequences, called the TR region, are present at the end of the genome US region.
Analysis of BHV SHJS genome
BHV strain SHJS genome shared 98.57 to 99.64% nucleotide identity with 25 BHV-1 strains (including American, Australian, and Indian strains) available in GenBank, and the highest identity was shown with strain SP1777 (KM258883.1) with a homolog of 99.64%. Genome sequence alignment showed that mutations, insertions, or deletions were observed in BHV SHJS UL12, UL19, UL27, UL37, UL42, UL44, UL46, UL47, US6, US7, and US8 genes compared to its different genomic coordinates (Figure 3A).
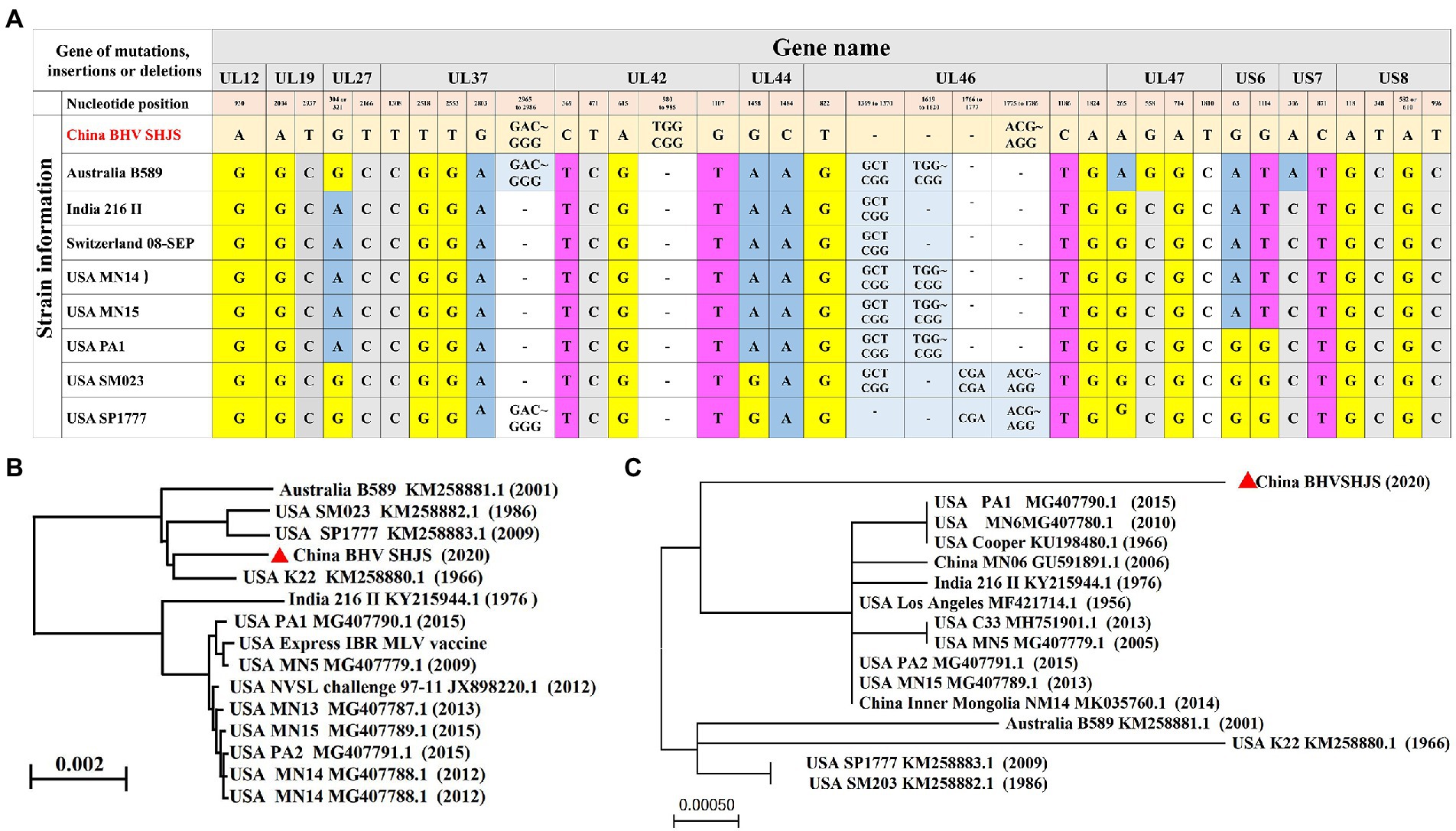
Figure 3. Phylogenetic analysis of the complete genome and US8 (gE) sequence. (A) Analysis of the complete genome sequence for BHV SHJS. (B) Phylogenetic analysis of full-genome sequence of BHV SHJS determined using the Neighbor Joining method in molecular evolutionary genetics on NCBI. (C) Phylogenetic analysis of US8 (gE) sequence using Maximum likehood method by MEGA software.
Amino acid alignment of these sequences showed that 41 Val (V) and 204 Gly (G) were mutated to Ile (I) and Ser (S) in BHV strain SHJS US8 (gE), respectively (Supplementary Figure S3A). Two insertions, including 333 Ser (S) and 334 Ala (A), and one mutation at 369 H (His) that mutated to Q (Gln) were observed in BHV SHJS UL42 (Supplementary Figure S3B). Two deletion sites at 456 to 457, 552 to 571, and 648 to 653, two insertion regions located at 611 to 614 and 649 to 656, and one mutation site at 514 (Phe) that mutated to C (Cys) were found in the sequence of BHV SHJS UL46 (Supplementary Figure S3C). The data suggested that the genomic differences of BHV SHJS compared with that of the previously reported BHV strains occur at US8, UL42, and UL46 genes.
Phylogenetic analysis of the genome and US8 gene of strain SHJS
The phylogenetic tree was constructed and analyzed to explore the genetic and evolutionary status of BHV strain SHJS. Phylogenetic analysis of the BHV SHJS genome showed that BHV SHJS was clustered with the K22 strain and far away from the other BHV-1 strains (Figure 3B). Due to the lack of a full genome for BHV-1.2b strains in China, we used the published BHV-1 US8 (gE) gene based on the nucleotide sequences to construct the phylogenetic tree of gE. Analysis of gE showed that BHV SHJS was displayed on a separate clade far away from the previously reported MN06 and NM14 strains isolated in China and the strains reported in the United States, India, and Australia. These results suggested that China’s newly identified BHV SHJS might be a new outbreak strain (Figure 3C).
Phylogenetic analysis based on UL44 (gC) gene
Bovine herpesvirus type 1 could be divided into BHV-1.1 and 1.2 types according to carboxy-terminal region (451 nucleotides) conserved moderately of BHV UL44 (gC) gene (Rijsewijk et al., 1999; Spilki et al., 2005; Traesel et al., 2015). BHV-1 UL44 (gC) also was suitable for phylogenetic analysis to explore evolutionary status (Chowdhury, 1995, 1997; Zhou et al., 2020). To classify the gene subtypes of BHV SHJS, the phylogenetic tree of UL44 (gC) was constructed based on 38 reference nucleotides sequence (451 nucleotides). The tree showed that isolated strains were clustered in three major clades, including BHV-1, BHV-2, and BHV-5 strains (Figure 4). BHV clade 1 was further divided into two separate lineages: lineage 1 clustering isolates of 1.2b subtypes were mainly composed of BHV SHJS (2020), Liaoning strains (2018, 2019), Jilin strains (2016, 2017, 2019), Heilongjiang strains (2018, 2019), and Inner Mongolia strains (2019) isolated in China and strains isolated in United States (1986, 2009); lineage 2 consisted of strains isolated in America, India (1976), and Switzerland (1992). BHV clade 2 consisted of strains isolated in Germany. BHV clade 5 contained strains isolated in Brazil (1999, 2007) and Uruguay (2012). In this tree, we found that BHV strain SHJS was clustered with Liaoning LN2 (2019) on one branch in the lineage close to strains isolated in China. Based on these analyses, we confirmed that BHV strain SHJS belongs to BHV-1.2 strain.
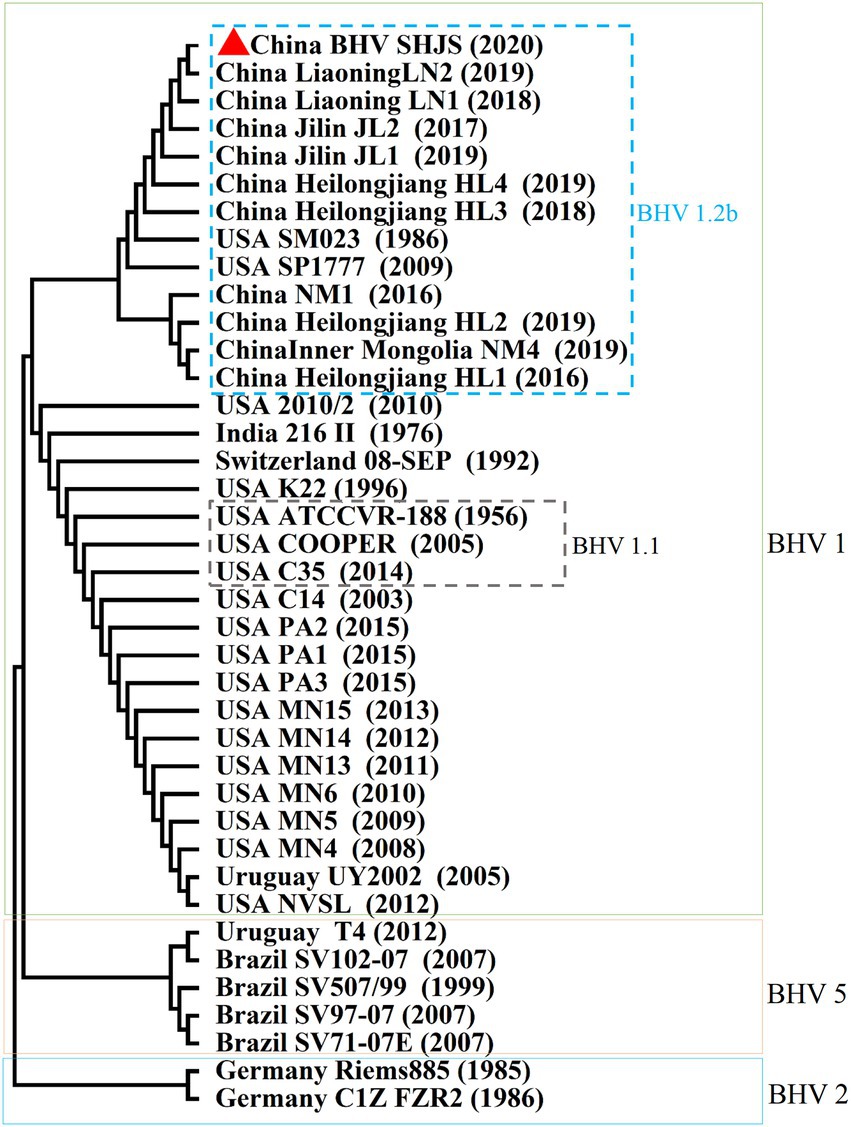
Figure 4. Phylogenetic tree of 451 nucleotides of partial gC gene. Thirty-eight partial nucleotide sequences (451 nucleotides) of the BHV-1 strains and partial gC gene of BHV SHJS sequences are available in GenBank and were compared using MEGA7 software. The sequences were aligned using the Clustal W method and the phylogenetic tree was generated by Maximum-likelihood analysis.
Phylogenetic analysis of major immunogenic genes
BHV-1 UL27(gB), UL44 (gC), and US6 (gD) encode the key immunogenic proteins that could induce neutralizing antibodies (Babiuk et al., 1987; Van Drunen Littel-van den Hurk et al., 1990; Liang et al., 1991; Yoon and Eo, 2007). To analyse whether the available BHV-1 vaccines could protect against BHV SHJS outbreaks by conducting phylogenetic tree based on the nucleotide and amino acid sequences of these genes of the available BHV strains and commercial vaccine strains. Phylogenetic analysis of gB nucleotide sequence showed that the strains were clustered in three clades (1, 2, and 3) (Figure 5). For the phylogenetic tree based on gB, BHV SHJS was closely clustered with China IBRV (2009, 2018), indicating that IBRV (2009, 2018) might be the closest ancestor of the BHV SHJS. Clade 1 consisted of BHV SHJS and other strains isolated in China, the United States, and Egypt, far from the available vaccine strains displayed in clade 2 (Figure 5). Furthermore, phylogenetic analysis of amino acid sequence of gB showed that BHV strain SHJS and American isolates were irregularly clustered in different branches with vaccine-like strain (Supplementary Figure S5). These analyses suggested that the available vaccines might not provide complete protection against BHV SHJS and might provide partial protection against other BHV-1 strains in China. This can also partially explain why BHV-1 vaccine strains only provide partial protection from the new outbreaks of BHV 1 in China or the United States.
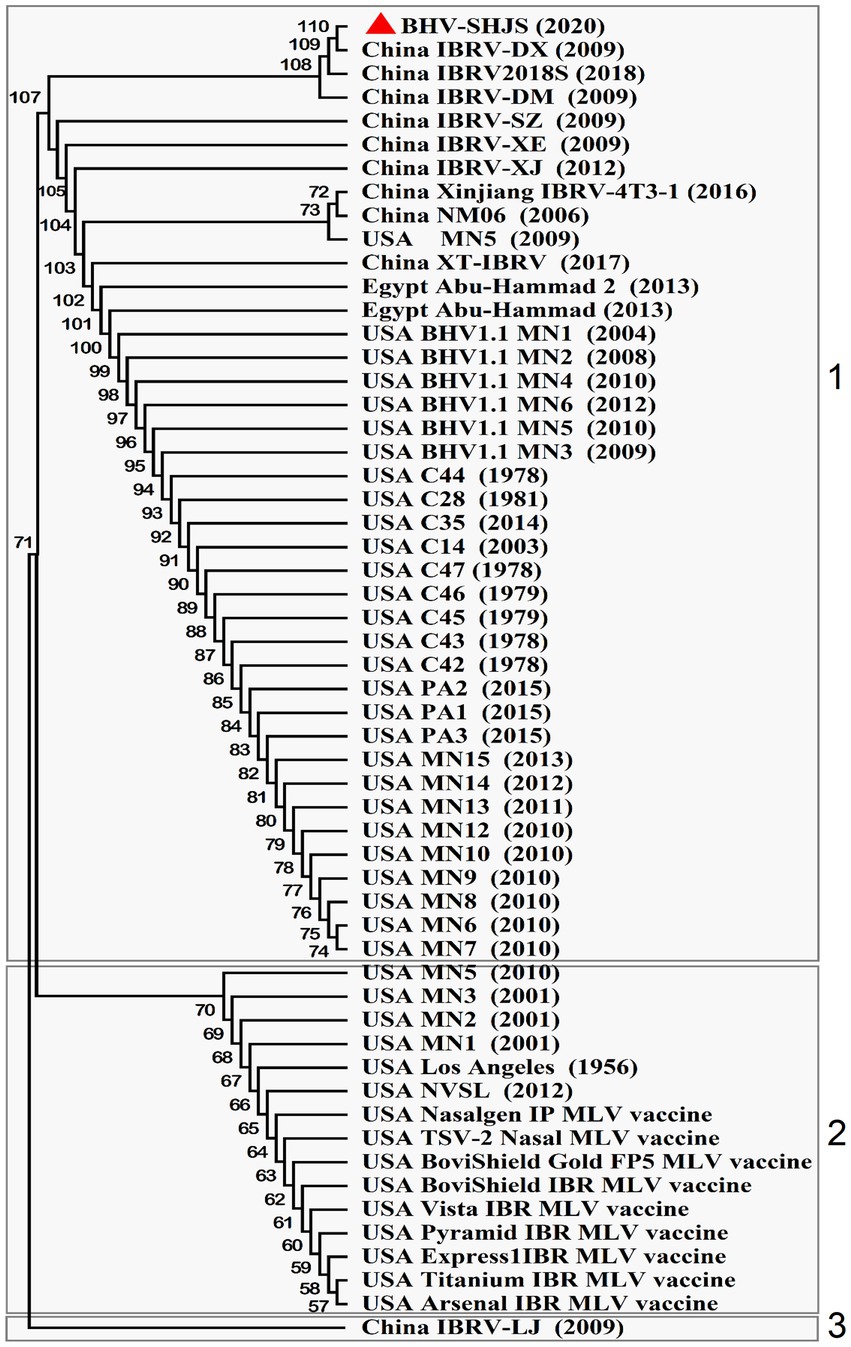
Figure 5. Phylogenetic tree analysis of BHV SHJS UL27 (gB) gene-based sequence. Fifty-six nucleotide sequences of BHV-1 gB are available in GenBank and were compared using MEGA7 software. The sequences of gB were aligned using the Clustal W method and the phylogenetic tree was generated by Maximum-likelihood analysis.
Discussion
Recently, infection and transmission of BHV-1 is common in cattle in China (Mo and Liu, 2017; Zhou et al., 2020). In 2018, the prevalence was 40% in cattle occurred in China (Chen et al., 2018). However, information about the prevalent of Chinese isolates is scarce. In 2020, a BHV 1.2 strain was isolated, referred to as BHV SHJS, from the nasal swabs of Holstein cows in Shanghai, China. Complete genome analyses showed that BHV SHJS has the highest nucleotide identity (99.64%) with strain SP1777 (KM258883.1) (Figure 2). The phylogenetic analysis of gC demonstrated that BHV SHJS belongs to BHV-1.2 strain. Genomic analysis presented that BHV SHJS was a new outbreak field strain in China.
We also found that mutation, deletion, or insertion occurs in some amino acids of BHV SHJS UL42 and UL46 proteins (Supplementary Figures S3B,C). BHV-1 UL46 was reported to impact viral replication (Hou et al., 2019). HSV UL46 had an important role in antiviral innate immune response (You et al., 2019). There are few in-depth studies on BHV-1 UL46 and UL42 proteins. We focused on speculating the function of UL46 and UL42-encoded proteins. We predicted the structure of proteins and showed that UL42-encoded protein consisted of 17.65% alpha helix (H), 26.72% extended β strand (E), and 55.64% loop (L) (Supplementary Figure S4A), and UL46-encoded protein consisted of 44.74% alpha helix (H), 1.8% extended β strand (E), and 53.4% loop (L) (Supplementary Figure S5A) on secondary structure. UL42-encoded protein contained Malic-M, PKS-KR domains (Supplementary Figure S4B), and UL46-encoded VP16 contains HhH1, PKS-TE, ICA69, and GDNF domains (Supplementary Figure S4C). Insertions of BHV SHJS UL42 occurred in the predicted PKS-KR domain, which was associated with bacterial polyketide synthases and catalyzes. Deletions and mutations of BHV SHJS UL46 occurred in an Islet cell autoantigen ICA69 domain and GDNF domain which is a potent survival factor for sympathetic, sensory, and central nervous system neurons (each functional domain description is derived from the database on prediction). Based on these predictions, we speculated that the functions of these two proteins might be similar to these functional domains. However, mutation, deletion, or insertion of these domains might influence protein functions.
Currently, commercial inactivated or attenuated live vaccines and recombinant subunit vaccines are used to immunize cattle to gain protection against BHV-1 (Cowley et al., 2011; Biswas et al., 2013; Fulton et al., 2013; Gonçalves et al., 2021). However, these vaccines only partially protect against BHV-1, as observed in China or the United States (O'Toole et al., 2012). Phylogenetic analyses of immunogenic genes (gB) revealed that BHV SHJS and some other Chinese outbreaks and some American vaccine strains were displayed on different branches (Figure 5). Based on these analyses, we speculate that the available vaccines in China might not protect BHV SHJS and might only provide partial protection against other BHV-1 strains in China. This also partially explains why BHV-1 vaccine strains only provide partial protection from the new outbreaks of BHV 1.2 in China or the United States.
It has been reported that homologous recombination often occurs in the herpesvirus species: Pseudorabies virus (PRV), Varicella-zoster virus, and herpes simplex virus type 1 (HSV-1) (Kintner et al., 1995; Schynts et al., 2003; Thiry et al., 2005; Norberg et al., 2015; Bo et al., 2021). Thus, there was a huge responsibility placed on virus infection prevention. Recombination events between BHV-1 and BHV-5 are also reported (Meurens et al., 2004). Recombination of live-attenuated BHV-1 was first documented in Australia in 2008 (Lee et al., 2012). Whether BHV SHJS isolated also occurred recombination after vaccination, we attempted and performed a recombination prediction analysis with nine commercial vaccine strains. In prediction, we found that BHV SHJS has a potential recombinant region in the genome of BHV SHJS (Supplementary Figure S6). We speculated that BHV SHJS might occur recombination with vaccine in the future prevention, which needs more evidence to prove that a recombination event occurs in the future.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number OP035381, which can be found at: https://www.ncbi.nlm.nih.gov/nuccore/OP035381.
Author contributions
WG, Y-SJ, and HC conceived and designed the experiments. WG and JX performed the experiments. JL and WG analyzed the data. HC and Y-SJ helped with reagents, materials and analytical tool. WG, HC, and JL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This experiment was supported by the National Nature Science Foundation of China (grant numbers 32170161 and U19A2039).
Acknowledgments
This research work was carried out under the National Nature Science Foundation and working in Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The authors would like to thank these.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1033008/full#supplementary-material
Footnotes
References
Babiuk, L. A., L'Italien, J., Van Drunen Littel-van den Hurk, S., Zamb, T., Lawman, J. P., Hughes, G., et al. (1987). Protection of cattle from bovine herpesvirus type I (BHV-1) infection by immunization with individual viral glycoproteins. Virology 159, 57–66. doi: 10.1016/0042-6822(87)90347-3
Biswas, S., Bandyopadhyay, S., Dimri, U., and Patra, P. H. (2013). Bovine herpesvirus-1 (BHV-1) – a re-emerging concern in livestock: a revisit to its biology, epidemiology, diagnosis, and prophylaxis. Vet. Q. 33, 68–81. doi: 10.1080/01652176.2013.799301
Bo, Z., Miao, Y., Xi, R., Gao, X., Miao, D., Chen, H., et al. (2021). Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound. Emerg. Dis. 68, 1454–1464. doi: 10.1111/tbed.13813
Brake, F., and Studdert, M. J. (1985). Molecular epidemiology and pathogenesis of ruminant herpesviruses including bovine, buffalo and caprine herpesviruses l and bovine encephalitis herpesvirus. Aust. Vet. J. 62, 331–334. doi: 10.1111/j.1751-0813.1985.tb07652.x
Chen, X., Wang, X., Qi, Y., Wen, X., Li, C., Liu, X., et al. (2018). Meta-analysis of prevalence of bovine herpes virus 1 in cattle in mainland China. Acta Trop. 187, 37–43. doi: 10.1016/j.actatropica.2018.07.024
Chowdhury, S. I. (1995). Molecular basis of antigenic variation between the glycoproteins C of respiratory bovine herpesvirus 1 (BHV-1) and neurovirulent BHV-5. Virology 213, 558–568. doi: 10.1016/s0042-6822(95)80137-5
Chowdhury, S. I. (1997). Fine mapping of bovine herpesvirus 1 (BHV-1) glycoprotein C neutralizing epitopes by type-specific monoclonal antibodies and synthetic peptides. Vet. Microbiol. 58, 309–314. doi: 10.1016/s0378-1135(97)00146-6
Cowley, D. J., Clegg, T. A., Doherty, M. L., and More, S. J. (2011). Aspects of bovine herpesvirus-1 infection in dairy and beef herds in the Republic of Ireland. Acta Vet. Scand. 53:40. doi: 10.1186/1751-0147-53-40
Deshpande, M. S., Ambagala, T. C., Hegde, N. R., Hariharan, M. J., Navaratnam, M., and Srikumaran, S. (2002). Induction of cytotoxic T-lymphocytes specific for bovine herpesvirus-1 by DNA immunization. Vaccine 20, 3744–3751. doi: 10.1016/s0264-410x(02)00375-4
d'Offay, J. M., Eberle, R., Fulton, R. W., and Kirkland, P. D. (2016). Complete genomic sequence and comparative analysis of four genital and respiratory isolates of bovine herpesvirus subtype 1.2b (BoHV-1.2b), including the prototype virus strain K22. Arch. Virol. 161, 3269–3274. doi: 10.1007/s00705-016-3026-1
Engels, M., Loepfe, E., Wild, P., Schraner, E., and Wyler, R. (1987). The genome of caprine herpesvirus 1: genome structure and relatedness to bovine herpesvirus 1. J. Gen. Virol. 68, 2019–2023. doi: 10.1099/0022-1317-68-7-2019
Engels, M., Giuliani, C., Wild, P., Beck, T. M., Loepfe, E., and Wyler, R. (1986). The genome of bovine herpesvirus 1 strains exhibiting a neuropatho- genic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res 6, 57–73. doi: 10.1016/0168-1702(86)90057-2
Fulton, R. W., d'Offay, J. M., Dubovi, E. J., and Eberle, R. (2016). Bovine herpesvirus-1: genetic diversity of field strains from cattle with respiratory disease, genital, fetal disease and systemic neonatal disease and their relationship to vaccine strains. Virus Res. 223, 115–121. doi: 10.1016/j.virusres.2016.06.017
Fulton, R. W., d'Offay, J. M., and Eberle, R. (2013). Bovine herpesvirus-1: comparison and differentiation of vaccine and field strains based on genomic sequence variation. Vaccine 31, 1471–1479. doi: 10.1016/j.vaccine.2013.01.013
Fulton, R. W., d'Offay, J. M., Eberle, R., Moeller, R. B., Campen, H. V., O'Toole, D., et al. (2015). Bovine herpesvirus-1: evaluation of genetic diversity of subtypes derived from field strains of varied clinical syndromes and their relationship to vaccine strains. Vaccine 33, 549–558. doi: 10.1016/j.vaccine.2014.11.033
Gonçalves, V. S., Santos, F. D. S., Dos Santos Junior, A. G., Piraine, R. E. A., Rodrigues, P. R. C., Brasil, C. L., et al. (2021). Recombinant bovine IL17A acts as an adjuvant for bovine herpesvirus vaccine. Res. Vet. Sci. 136, 185–191. doi: 10.1016/j.rvsc.2021.02.014
Hodgson, P. D., Aich, P., Manuja, A., Hokamp, K., Roche, F. M., Brinkman, F. S., et al. (2005). Effect of stress on viral-bacterial synergy in bovine respiratory disease: novel mechanisms to regulate inflammation. Comp. Funct. Genomics 6, 244–250. doi: 10.1002/cfg.474
Hou, P., Zhao, M., He, W., He, H., and Wang, H. (2019). Cellular microRNA bta-miR-2361 inhibits bovine herpesvirus 1 replication by directly targeting EGR1 gene. Vet. Microbiol. 233, 174–183. doi: 10.1016/j.vetmic.2019.05.004
Jones, C., and Chowdhury, S. (2007). A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex and development of improved vaccines. Anim. Health Res. Rev. 8, 187–205. doi: 10.1017/s146625230700134x
Kintner, R. L., Allan, R. W., and Brandt, C. R. (1995). Recombinants are isolated at high frequency following in vivo mixed ocular infection with two avirulent herpes simplex virus type 1 strains. Arch. Virol. 140, 231–244. doi: 10.1007/bf01309859
Lee, S. W., Markham, P. F., Coppo, M. J., Legione, A. R., Markham, J. F., Noormohammadi, A. H., et al. (2012). Attenuated vaccines can recombine to form virulent field viruses. Science 337:188. doi: 10.1126/science.1217134
Liang, X. P., Babiuk, L. A., Van Drunen Littel-van den Hurk, S., Fitzpatrick, D. R., and Zamb, T. J. (1991). Bovine herpesvirus 1 attachment to permissive cells is mediated by its major glycoproteins gI, gIII, and gIV. J. Virol. 65, 1124–1132. doi: 10.1128/jvi.65.3.1124-1132.1991
Leung-Tack, P., Audonnet, J. C., and Riviere, M. (1994). The complete DNA sequence and the genetic organization of the short unique region (US) of the bovine herpes- virus type 1. Virology, 199, 409–421. doi: 10.1006/viro.1994.1139
Marawan, M. A., Deng, M., Wang, C., Chen, Y., Hu, C., Chen, J., et al. (2021). Characterization of BoHV-1 gG−/tk−/gE-mutant in differential protein expression, virulence, and immunity. Vet. Sci. 8:253. doi: 10.3390/vetsci8110253
Metzler, A. E., Matile, H., Gassmann, U., Engels, M., and Wyler, R. (1985). European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch. Virol. 85, 57–69. doi: 10.1007/bf01317006
Meurens, F., Keil, G. M., Muylkens, B., Gogev, S., Schynts, F., Negro, S., et al. (2004). Interspecific recombination between two ruminant alphaherpesviruses, bovine herpesviruses 1 and 5. J. Virol. 78, 9828–9836. doi: 10.1128/jvi.78.18.9828-9836.2004
Mo, J., and Liu, Q. (2017). Comparative analysis on the large-scale development of dairy industry in China and Japan. Jiangsu Agric. Sci. 45, 303–307. doi: 10.16172/j.cnki.114768.2017.11.023
Moe, J. B., Lambert, R. D., and Lupton, H. W. (1981). Plaque assay for Ebola virus. J. Clin. Microbiol. 13, 791–793. doi: 10.1128/jcm.13.4.791-793.1981
Nandi, S., Kumar, M., Manohar, M., and Chauhan, R. S. (2009). Bovine herpes virus infections in cattle. Anim. Health Res. Rev. 10, 85–98. doi: 10.1017/s1466252309990028
Norberg, P., Depledge, D. P., Kundu, S., Atkinson, C., Brown, J., Haque, T., et al. (2015). Recombination of globally circulating varicella-zoster virus. J. Virol. 89, 7133–7146. doi: 10.1128/jvi.00437-15
O'Toole, D., Chase, C. C., Miller, M. M., and Campen, H. V. (2014). Kennedy, the early sixties, and visitation by the angel of death. Vet. Pathol. 51, 1051–1062. doi: 10.1177/0300985814548515
O'Toole, D., Miller, M. M., Cavender, J. L., and Cornish, T. E. (2012). Pathology in practice: abortion in the heifers of this report was a result of BoHV-1 infection. J. Am. Vet. Med. Assoc. 241, 189–191. doi: 10.2460/javma.241.2.189
Pi, Q., Chen, S. Z., and Hong, N. N. (2014). Serological investigation of infectious rhinotracheitis, sub-tuberculosis and new sporidia in Guizhou province. Prog. Vet. Met. 10, 129–132. doi: 10.16437/j.cnki.1007-5038.2014.10.023
Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty per cent endpoints 12. Am. J. Epidemiol. 27, 493–497. doi: 10.1093/oxfordjournals.aje.a118408
Rijsewijk, F. A., Kaashoek, M. J., Langeveld, J. P., Meloen, R., Judek, J., Bie Ñkowska-Szewczyk, K., et al. (1999). Epitopes on glycoprotein C of bovine herpesvirus-1 (BHV-1) that allow differentiation between BHV-1.1 and BHV-1.2 strains. J. Gen. Virol. 80, 1477–1483. doi: 10.1099/0022-1317-80-6-1477
Schynts, F., Meurens, F., Detry, B., Vanderplasschen, A., and Thiry, E. (2003). Rise and survival of bovine herpesvirus 1 recombinants after primary infection and reactivation from latency. J. Virol. 77, 12535–12542. doi: 10.1128/jvi.77.23.12535-12542.2003
Spilki, F. R., Esteves, P. A., da Silva, A. D., Franco, A. C., Rijsewijk, F. A., and Roehe, P. M. (2005). A monoclonal antibody-based ELISA allows discrimination between responses induced by bovine herpesvirus subtypes 1 (BoHV-1.1) and 2 (BoHV-1.2). J. Virol. Methods 129, 191–193. doi: 10.1016/j.jviromet.2005.06.008
Thiry, E., Meurens, F., Muylkens, B., McVoy, M., Gogev, S., Thiry, J., et al. (2005). Recombination in alphaherpesviruses. Rev. Med. Virol. 15, 89–103. doi: 10.1002/rmv.451
Traesel, C. K., Bernardes, L. M., Spilki, F. R., Weiblen, R., and Flores, E. F. (2015). Sequence analysis of the 5′ third of glycoprotein C gene of south American bovine herpesviruses 1 and 5. Braz. J. Med. Biol. Res. 48, 470–478. doi: 10.1590/1414-431x20144266
Van Drunen Littel-van den Hurk, S., Gifford, G. A., and Babiuk, L. A. (1990). Epitope specificity of the protective immune response induced by individual bovine herpesvirus-1 glycoproteins. Vaccine 8, 358–368. doi: 10.1016/0264-410x(90)90095-4
Waters, D. L., and Shapter, F. M. (2014). The polymerase chain reaction (PCR): general methods. Methods Mol. Biol. 1099, 65–75. doi: 10.1007/978-1-62703-715-0_7
Yoon, H. A., and Eo, S. K. (2007). Differential polarization of immune responses by genetic cotransfer of chemokines changes the protective immunity of DNA vaccine against pseudorabies virus. Immunology 120, 182–191. doi: 10.1111/j.1365-2567.2006.02490.x
You, H., Zheng, S., Huang, Z., Lin, Y., Shen, Q., and Zheng, C. (2019). Herpes simplex virus 1 tegument protein UL46 inhibits TANK-binding kinase 1-mediated signaling. MBio 10:e00919-19. doi: 10.1128/mBio.00919-19
Zhong, C., Pan, L., and Liu, Y. (2012). Serological investigation of IBRV and BVDV in a dairy farm in Bengbu city. China Dairy Cattle 21, 36–38.
Keywords: bovine herpesvirus type 1.2, isolation, full genome, phylogenetic analysis, BHV-1.2
Citation: Guo W, Xie J, Liu J, Chen H and Jung YS (2022) The full-genome characterization and phylogenetic analysis of bovine herpesvirus type 1.2 isolated in China. Front. Microbiol. 13:1033008. doi: 10.3389/fmicb.2022.1033008
Edited by:
Chengming Wang, Auburn University, United StatesReviewed by:
Xin Yin, Chinese Academy of Agricultural Sciences (CAAS), ChinaHeng Wang, South China Agricultural University, China
Copyright © 2022 Guo, Xie, Liu, Chen and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Sam Jung, eXNqdW5nQG5qYXUuZWR1LmNu
†ORCID: Weiqiang Guo https://orcid.org/0000-0001-8273-0338
 Weiqiang Guo1†
Weiqiang Guo1† Jingyi Liu
Jingyi Liu Hongjun Chen
Hongjun Chen Yong-Sam Jung
Yong-Sam Jung