
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 January 2023
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1032900
This article is part of the Research Topic Interactions Between Filamentous Plant Pathogens with their Host Plants and Biocontrol Agents View all 16 articles
 Shuai Yang1†
Shuai Yang1† Jie Li2†
Jie Li2† Jie Lu3
Jie Lu3 Ling Wang4
Ling Wang4 Fanxiang Min1
Fanxiang Min1 Mei Guo1
Mei Guo1 Qi Wei1
Qi Wei1 Wenzhong Wang1
Wenzhong Wang1 Xuezhi Dong1
Xuezhi Dong1 Yanzhi Mao1
Yanzhi Mao1 Linshuang Hu1
Linshuang Hu1 Xiaodan Wang2*
Xiaodan Wang2*Introduction: Calcium sensor calcineurin B-like proteins (CBLs) and their interacting partners, CBL-interacting protein kinases (CIPKs), have emerged as a complex network in response to abiotic and biotic stress perception. However, little is known about how CBL-CIPK complexes function in potatoes.
Methods: In this study, we identified the components of one potato signaling complex, StCBL4–StCIPK2, and characterized its function in defense against Rhizoctonia solani causing stem canker in potato.
Results: Expressions of both StCBL4 and StCIPK2 from potato were coordinately induced upon R. solani infection and following exposure to the defense genes. Furthermore, transient overexpression of StCBL4 and StCIPK2 individually and synergistically increased the tolerance of potato plants to R. solani in Nicotiana benthamiana. Additionally, the transgenic potato has also been shown to enhance resistance significantly. In contrast, susceptibility to R. solani was exhibited in N. benthamiana following virus-induced gene silencing of NbCBL and NbCIPK2. Evidence revealed that StCBL4 could interact in yeast and in planta with StCIPK2. StCBL4 and StCIPK2 transcription was induced upon R. solani infection and this expression in response to the pathogen was enhanced in StCBL4- and StCIPK2-transgenic potato. Moreover, accumulated expression of pathogenesis-related (PR) genes and reactive oxygen species (ROS) was significantly upregulated and enhanced in both StCBL4- and StCIPK2- transgenic potato.
Discussion: Accordingly, StCBL4 and StCIPK2 were involved in regulating the immune response to defend the potato plant against R. solani. Together, our data demonstrate that StCBL4 functions in concert with StCIPK2, as positive regulators of immunity, contributing to combating stem canker disease in potato.
Plants have evolved tightly regulated signaling pathways that allow them to respond to threats they encounter during exposure to complex and varying environments (Verma et al., 2021). To cope with numerous environmental stimuli, plants rely on a series of complex signal transduction mechanisms to perceive, transduce, and respond to different stresses (Yin et al., 2017). As a secondary messenger in plants, calcium (Ca2+) links various signal pathways together and participates in many biochemical reactions, with altered Ca2+ levels observed when plants are exposed to biotic and abiotic stressors (Xu et al., 2022). Importantly, transient changes in intracellular Ca2+ concentration are captured by Ca2+ sensors that subsequently regulate signaling pathways involved in plant growth and development. Ca2+ sensors include two types of Ca2+ sensor proteins with distinct structural features and functions: sensor relay proteins and sensor responder proteins. Sensor relay proteins include calmodulin-like proteins (CMLs) and calcineurin B-like proteins (CBLs), while sensor responder proteins include calmodulins (CAMs) and calcium-dependent protein kinases (CDPKs; Tang et al., 2020). Sensor relays (e.g., CMLs and CBLs) lack kinase activity but can specifically target downstream proteins to transmit perceived calcium signals generated in response to various environmental stimuli and developmental processes. In contrast, sensor responder proteins (e.g., CaMs and CDPKs) possess kinase activity and all of the abovementioned sensor relay activities.
As a family of Ca2+ sensor proteins, CBLs are generally found in plants, but not in animals or fungi. CBLs contain a crucial structural component consisting of common elongation factor hand domains (EF hands). EF hands function as calcium-binding sites that capture Ca2+ while engaging in specific binding interactions with NAF/FISL domains present within C-terminal regions of CBL-interacting protein kinases (CIPKs). CIPKs comprise a group of plant-specific serine/threonine (Ser/Thr) kinases that belong to the SnRK3 protein family. These kinases play crucial roles in signal transduction via CBL–CIPK modules (Ma et al., 2020) to thereby control affinities and activities of numerous ion transporters. In contrast, the N-terminal MGCXXS/T motif in some CBL proteins mediates lipid modification via myristoylation and palmitoylation that direct CBL-CIPK complex migration to precise cellular membrane target regions where they become anchored to the membrane (Weinl and Kudla, 2009). Arabidopsis CBLs include CBL1, 4, 5, and 9, which localize to the plasma membrane (Batistic et al., 2010).
Calcineurin B-like protein–interacting protein kinase complexes of many plant species, including model plants, crop plants, and several horticultural plants, have been extensively studiedconcerning their regulation of diverse responses to abiotic stresses (Dong et al., 2021). One such complex, which was first identified in Arabidopsis thaliana, was initially found to participate in the Salt Overly Sensitive (SOS) signaling pathway, whereby CBL4 (SOS3) was shown to interact and form a complex with CIPK24 (SOS2). Once formed, the CBL4-CIPK24 complex was observed to migrate to the plasma membrane, where it activated the Na+/H+ antiporter (SOS1) located on the plasma membrane and the vacuolar H+-ATPase, resulting in enhanced salt tolerance (Ishitani et al., 2000). Subsequently, other complexes were discovered in Arabidopsis (CBL1/CBL9-CIPK23) that were observed to localize to the plasma membrane and regulate K+ in roots and stomatal guard cells by modulating K+ channel Arabidopsis K+ Transporter 1 (AKT1) activity (Cheong et al., 2007). Furthermore, overexpression of OsCBL8 and OsCIPK15 in rice was found to enhance salt tolerance (Xiang et al., 2007). Conversely, expression of OsCIPK31 in tobacco plants was found to participate in the generation of diverse signals in response to cold, salt, light, cytokinins, and sugars (Kim et al., 2003). More recently, TaCIPK23 has been shown to participate in wheat abscisic acid (ABA) and drought stress responses by mediating crosstalk between ABA-induced signaling and other pathways associated with adaptation to drought (Cui et al., 2018). In potato, StCIPK10 has been shown to enhance both cellular scavenging of ROS and cellular modulation of corresponding osmoregulatory substances to strengthen plant drought and osmotic stress tolerance (Ma et al., 2021). In addition, differential expression of maize ZmCBL4, pea PsCBL, Chinese cabbage BrCBL, and grapevine VvCBL have been reported to occur after exposure of plants to various abiotic stresses (Wang et al., 2007; Jung et al., 2017; Xi et al., 2017).
Compared with studies of abiotic stress responses, studies of biotic stress responses have rarely been reported, although a few studies can be found in the literature. One such study using cultured cells of rice plants demonstrated that an interaction between OsCIPK14/15 and OsCBL4 played a crucial role in the microbe-associated molecular pattern-induced defense signaling pathway (Kurusu et al., 2010). CBL10 and CIPK6 were found to promote ROS generation as part of the tomato immune defense response triggered by an interaction between Pseudomonas syringae pv tomato DC3000 and Nicotiana benthamiana (De la Torre et al., 2013). OsCIPK30 was shown to play a critical role in enhancing rice tolerance to rice stripe virus (Liu et al., 2017), while the TaCBL4–TaCIPK5 complex was observed to positively modulate wheat resistance to stripe rust fungus through a ROS-dependent mechanism (Liu et al., 2018). Moreover, the knockdown of CaCIPK1 expression increased the susceptibility of pepper to Phytophthora capsici, reduced root activity, and altered the expression of defense-related genes (Ma et al., 2019). Taken together, the abovementioned results obtained from the few research studies reported to date suggest that CBL-CIPK complexes participate in biotic stress responses.
Rhizoctonia solani (teleomorph: Thanatephorus cucumeris) is one of the most dominant soil-borne necrotrophic fungal pathogens, due to its broad host range and the lack of crop resistance to this pathogen (Foley et al., 2013). Rhizoctonia solani is complex and divided into 14 anastomosis groups (AG1–AG13 and AGBI; Yang et al., 2017). AG3, the most destructive group, causes Rhizoctonia disease on potato (Solanum tuberosum L.; Woodhall et al., 2013), the fourth most cultivated food crop globally. In recent years, black scurf and stem canker have emerged to become the most severe soil-borne diseases, as evidenced by increasingly reduced crop yields and quality due to these diseases each year. However, efforts to combat these diseases have been hindered by a lack of knowledge about the necrotrophic pathogen infection process and the mechanisms by which plants prevent disease-induced cell death (Yang et al., 2017). In our previous study, transcriptome analysis was performed via RNA-Seq to investigate the potato response to R. solani infection, leading to the identification of two key genes, which encoded CBL4 and CIPK2 that exhibited up-regulated expression during infection (Yang et al., 2018). In this study, complete DNA sequences of the genes encoding these two proteins were cloned then their expression patterns were characterized in potato. After that, these genes were overexpressed in potato and the heterologous host N. benthamiana to determine whether increased expression of these genes could increase plant resistance to Rhizoctonia disease. StCBL4 and StCIPK2 were involved in regulating the immune response to defend the potato plant against R. solani. Therefore, the functional research of StCBL4 in concert with StCIPK2, as both are positive regulators of immunity, will contribute to combating stem canker disease in potato.
The virulent R. solani strain (anastomosis group: AG-3PT) used in this study was obtained from Heilongjiang Academy of Agricultural Sciences (HAAS) and cultured in potato dextrose agar (potato 200 g, Dextrose 20 g, agar 17 g, and water 1 L). Cultures were maintained on PDA medium for 1 week at 25°C, and then 5-mm-diameter mycelium plugs were obtained for inoculation of plants. In addition, Agrobacterium tumefaciens GV3101 was cultured in LB medium (10 g tryptone, 10 g NaCl, and 5 g yeast extract per liter).
Various potato germplasm resources studied here were grown in a climate-controlled chamber under 16 h light/8 h dark conditions at 23°C. The list of plant varieties is presented in Supplementary Table 1. In addition, N. benthamiana plants were grown in a climate-controlled chamber under 14 h light/10 h dark conditions and kept at 25°C and 20°C during light and dark cycles, respectively. All plants were grown on separate trays for subsequent RNA extraction, inoculation, and RT-PCR expression analysis. All experiments involving both control and treated plants were carried out using three independent biological replicate samples.
Total RNA was extracted from potato leaves using TRIzol™ Reagent (Invitrogen Co., United Kingdom) according to the manufacturer’s instructions. RNA concentrations were measured using a Thermo NanoDrop 1000 spectrophotometer system (Thermo Fisher Scientific Co., United States). cDNA was synthesized using the GoScript™ Reverse Transcription System (Promega Co., United States), and then was used as a template for subsequent PCR amplifications. Based on transcript sequences, cDNA sequences encoding ORFs of StCBL4 and StCIPK2 were amplified with primers designed using CE Design V1.04 (Supplementary Table 2) according to amino acid sequence alignments conducted using SMART.1 Next, multiple sequence alignments were performed using DNAMAN 8.0 software (Lynnon Biosoft Co., United States). Next, a phylogenetic tree was constructed based on alignments of amino acid sequences via the neighbor-joining (NJ) method using MEGA 5.0 software. Finally, bootstrap analysis with 1,000 replicates was performed to assess the statistical reliability of each branch of the tree.
For transient expression, complete coding sequences (CDSs) of StCBL4 and StCIPK2 were cloned into the pCAMBIA1300-35S-HA-RBS vector to generate constructs designated 35S:HA-StCBL4 and 35S:HA-StCIPK2, respectively. The recombinant constructs were next introduced into A. tumefaciens strain GV3101, which was then used to transform N. benthamiana plants via the Agrobacterium-mediated transformation method, with the transformation of empty vector (EV) 35S:HA serving as a control. To generate constructs for the luciferase complementation assay (LCA), the StCBL4-coding sequence was cloned into the pCAMBIA1300-35S-Cluc-RBS vector, and the StCIPK2-coding sequence was cloned into the pCAMBIA1300-35S-HA-Nluc-RBS vector. To construct the bait vector for the yeast two-hybrid assay, CDSs of StCBL4 and StCIPK2 were cloned into pGBKT7 (DNA-binding domain, BD) and pGADT7 (activation domain, AD) vectors, respectively. Meanwhile, pBWA(V)KS-StCBL4 and pBWA(V)KS-StCIPK2 constructs were generated and transformed into A. tumefaciens strain EHA105 using Agrobacterium-mediated transformation protocols for potato obtained from Wuhan Biorun Bio-Tech Co. Ltd. (Wuhan, China). Regenerated transgenic plants were transplanted in a standard MS medium (Murashige and Skoog, 1962) containing 50 mg/L kanamycin (Sangon Biotech Co., China). The primers used in this study are listed in Supporting Information Supplementary Table 2.
The images of subcellular localization of GFP- and RFP-tagged proteins were visualized through a Leica TCS SP8 confocal microscope (Leica Co., Germany) in N. benthamiana leaves expressing transiently p35S-CBL4-GFP and p35S-CIPK2-RFP constructs. Fluorescence signals were detected at 48 h after injection. GFP and RFP fluorescence was excited using argon lasers at 488 and 545 nm, and the emissions were collected within 495–545 and 580–620 nm, respectively. Relevant fluorescence data analysis and subsequent image processing of the indicated profile were conducted using Leica Application Suite X (LAS-X) software.
For interaction analyses of StCBL4–StCIPK2, MatchMaker yeast two-hybrid assays were performed (Coolaber Co., China). After vector construction, paired pGADT7 (activation domain, AD) and pGBKT7 (DNA-binding domain, BD) plasmids were co-transformed into yeast strain AH109 via the lithium acetate method according to the Yeast Protocols Handbook. The positive control was the diploid hybrid yeast containing pGBKT7-53 and pGADT7-T. Yeast colonies were grown at 30°C for 2–4 days on selection medium before being photographed. After yeast cell numbers/ml were initially quantified using a hemocytometer, they were collected and diluted to 102 and 101 cells/ml in sterile ultrapure water for serial dilutions.
The luciferase complementation assay was performed as described previously (Zhou et al., 2018) after Cluc and Nluc constructs were introduced into N. benthamiana leaves via the Agrobacterium-mediated transient expression-based method followed by co-expression of cloned genes in leaf cells for 2 days. Next, leaf disks in wells of a 96-well plate were incubated with 1 mM luciferin (Biovision Co., United States) then luminescence was measured using a microplate luminometer (Perkin Elmer Co., United States).
Two days after Agrobacterium-mediated transient injection of p35S-StCIPK2-HA constructs into N. benthamiana, leaves were collected for Co-IP experiments. Total proteins were extracted from agroinfiltrated leaves using protein immunoprecipitation (IP) buffer, which contains 50 mM HEPES (pH 7.5), 150 mM KCl, 5 mM EDTA, 0.5% Triton X-100, 1 mM DTT, and a proteinase inhibitor cocktail. Samples were centrifuged and the supernatant was transferred to another 1.5 ml centrifuge tube, and about 45 μl was taken for input analysis. The remaining extracts were then incubated with 10 μl agarose-conjugated anti-GFP antibody (Lablead Co., China) for 3 h at 4°C with shaking, followed by washing beads four times with IP buffer. Finally, about 50 μl elution buffer was added for output analysis. Immunoprecipitated proteins were separated via SDS-PAGE then proteins of interest were detected on immunoblots via probing membranes with anti-HA and anti-GFP antibodies.
The R. solani AG-3 strain was used for plant infection and was cultured on a PDA medium at 25°C for 3 days. 5-mm-diameter mycelium plugs were incubated onto the abaxial side of detached leaves of intact N. benthamiana plants stored on moist tissue in sealed boxes for 24–48 h. Then inoculated leaves were photographed under UV light, and lesion areas were measured using ImageJ2 software. For S. tuberosum experiments, mycelium inoculations were performed using in vitro cultured plantlets, as described previously (Yang et al., 2017).
To understand orthologous gene functions in N. benthamiana, phylogenetic relationships among CBLs and CIPKs from S. tuberosum, N. benthamiana, and A. thaliana were analyzed. Virus-induced gene silencing (VIGS) constructs were generated by cloning 300-bp PCR fragments of orthologous genes from N. benthamiana cDNA into the pBinary tobacco rattle virus (TRV) vector (Liu et al., 2002) between the EcoRI and KpnI sites in the antisense orientation. A TRV construct expressing GFP, as described previously (McLellan et al., 2013), served as the negative control and a TRV construct expressing a cloned fragment of N. benthamiana phytoene desaturase (NbPDS) served as the positive control. Primer sequences are shown in Supplementary Table 2. Plants were either used in assays or to check gene silencing levels via quantitative RT-PCR 2–3 weeks later.
Potato plants overexpressing the abovementioned genes were grown in a climate-controlled chamber under 16 h light/8 h dark conditions at 23°C. After 3–4 weeks of growth, detached leaves were assayed for H2O2 production. H2O2 was detected via staining of inoculated and uninoculated leaves with 1 mg/ml 3,3′-diaminobenzidine (DAB) for 8 h in the dark, and then the leaves were destained with ethanol prior to observation (Lu et al., 2021).
Total RNA was isolated from N. benthamiana as described above. For S. tuberosum samples, RNA isolation was conducted after plants were inoculated with R. solani at 0, 6, 12, 24, 48, and 96 hpi (cv. Youjin-885) or 3 dpi (for different potato germplasm resources). Total RNA (5 μg) was reverse-transcribed using the GoScript™ Reverse Transcription System to generate a cDNA template. Next, cDNA was diluted tenfold for subsequent qRT-PCR analysis, using an ABI7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA) and TransStart Top Green qPCR SuperMix (TransGen, Beijing, China). Relative gene expression levels were calculated by normalization against internal controls NbActin (AY179605) and StActin (DQ252512) for N. benthamiana and S. tuberosum, respectively. All qRT-PCR experiments were performed at least three times. Relative quantification of expression levels was achieved using the 2-ΔΔCt method.
Data were obtained from three replicates and analyzed by unpaired two-tailed Student’s t-test with GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, United States). Values with p < 0.05 were considered statistically significant.
In previous transcriptome analysis, preliminary screening of StCBL4 and StCIPK2 expression was conducted to determine whether infection of potato with R. solani could induce expression of these two genes. Then, relative levels of StCBL4 and StCIPK2 gene transcripts in cv. Youjin-885 plants were measured at 0, 6, 12, 24, 36, 48, and 96 hpi. The results revealed that the expression of StCBL4 and StCIPK2 was induced and upregulated significantly during the infection of R. solani. More specifically, the expression level of StCBL4 was up-regulated and peaked at 24 hpi, 11.18-fold higher than the control level (Figure 1A). StCIPK2 reached a peak level of expression 14.2-fold higher than the control level as early as 6 hpi (Figure 1B). Meanwhile, the lesion symptoms and relative levels of StCBL4 and StCIPK2 gene transcripts in various potato germplasm resources (with different levels of resistance to R. solani) at 7 dpi were measured. As shown in Figure 1C, 5 potato germplasm resources displayed the disease except the varieties of 133, 153, and 812. Compared with the mild symptom of cv. 827, the other four potato germplasm resources exhibited no less than a 5-fold disease level on stem of potato plantlet. Moreover, real-time PCR results revealed that expression levels of both StCBL4 and StCIPK2 were significantly up-regulated in asymptomatic potato plantlets compared with symptomatic plantlets. The latter exhibited no significant change in the expression of these genes except for StCBL4 in potato cv. Z5 and StCIPK2 in potato cv.106 (Figures 1D,E). Based on these observations, we speculated that StCBL4 and StCIPK2 might be involved in the host response to R. solani infection.
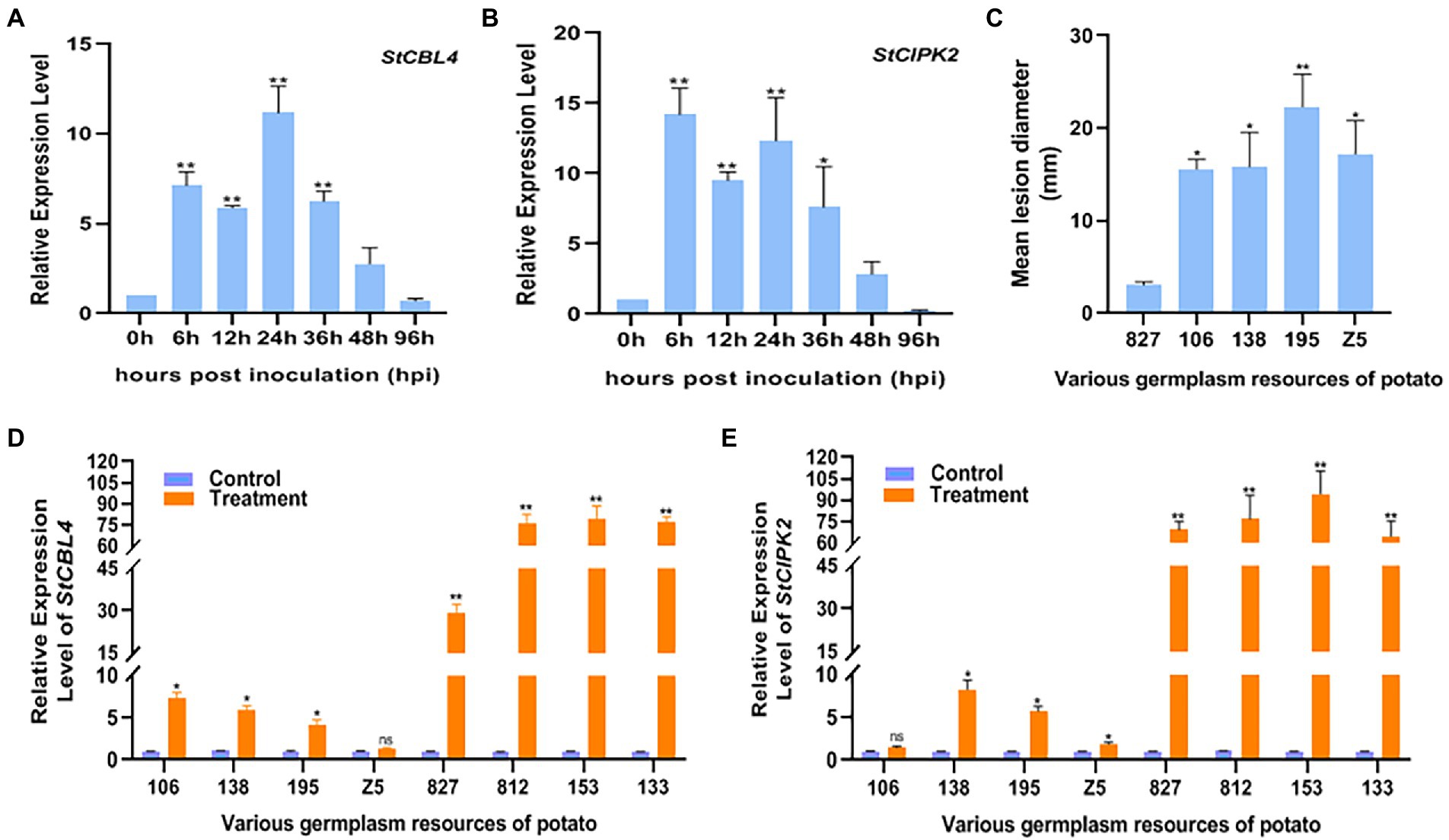
Figure 1. Expression profile of StCBL4 and StCIPK2 genes infected by Rhizoctonia solani in plantlet. (A) The relative expression level of the StCBL4 gene at different hours post-inoculation. (B) The relative expression level of the StCIPK2 gene at different hours post-inoculation. (C) Mean lesion diameter measured on various germplasm resources of potato after R. solani inoculation at 7 days post-inoculation. (D) The relative expression level of the StCBL4 gene in treatment plantlets compared with control plantlets at 7 days post-inoculation. (E) The relative expression level of the StCIPK2 gene in treatment plantlets compared with control plantlets at 7 days post-inoculation. The assay was performed by real-time RT-PCR. The relative mRNA levels were calculated with respect to the expression level of the WT plantlets without pathogen infection. All the experiments were repeated three times, and similar results were obtained. The data represent the means ± SD of triplicate measurements. Asterisks above the columns represent significance based on an unpaired, two-tailed Student’s t-test relative to the control. *p < 0.05; **p < 0.01.
Based on transcript sequences identified via RNA-seq, ORF sequences of StCBL4 and StCIPK2 were cloned and deposited into the GenBank database under accession numbers ON383960 and ON324205, respectively. The ORF of the StCBL4 gene is 663 bp in length and contains 4 EF-hand conserved domains and an FPSF motif that interacts with CIPKs (Supplementary Figure 1A). The predicted StCBL4 ORF contains 220 aa, a polypeptide molecular mass of 25.32 kDa, and an isoelectric point (pI) of 4.77. Results of the phylogenetic analysis indicated that StCBL4 is highly homologous to CBL4 from Capsicum annuum (Supplementary Figure 1B). StCIPK2 is 1,413 bp in length and contains a NAF/FISL motif and a PPI motif (Supplementary Figure 2A). The predicted StCIPK2 ORF contains 470 aa, a polypeptide molecular mass of 53.01 kDa, and an isoelectric point (pI) of 8.66. Results obtained through homology analysis indicated that StCIPK2 has high homology to CIPK of Hordeum vulgare (Supplementary Figure 2B).
To investigate whether StCBL4 and StCIPK2 enhanced host resistance against R. solani infection, we transiently expressed StCBL4-HA and StCIPK2-HA in N. benthamiana plants using Agrobacterium-mediated transient expression, with pCambia1300-35S-HA empty vector (EV) as control. The results showed that StCBL4-HA and StCIPK2-HA exhibited remarkably smaller lesion areas than the EV control (Figures 2A,C), and was about 51 and 50% lower than corresponding levels in control, respectively (Figures 2B,D). These findings indicated that StCBL4 and StCIPK2 could positively enhance N. benthamiana resistance to R. solani infection. Furthermore, western blotting elucidated the stability of StCBL4-HA and StCIPK2-HA expressed in N. benthamiana (Figures 2B,D).
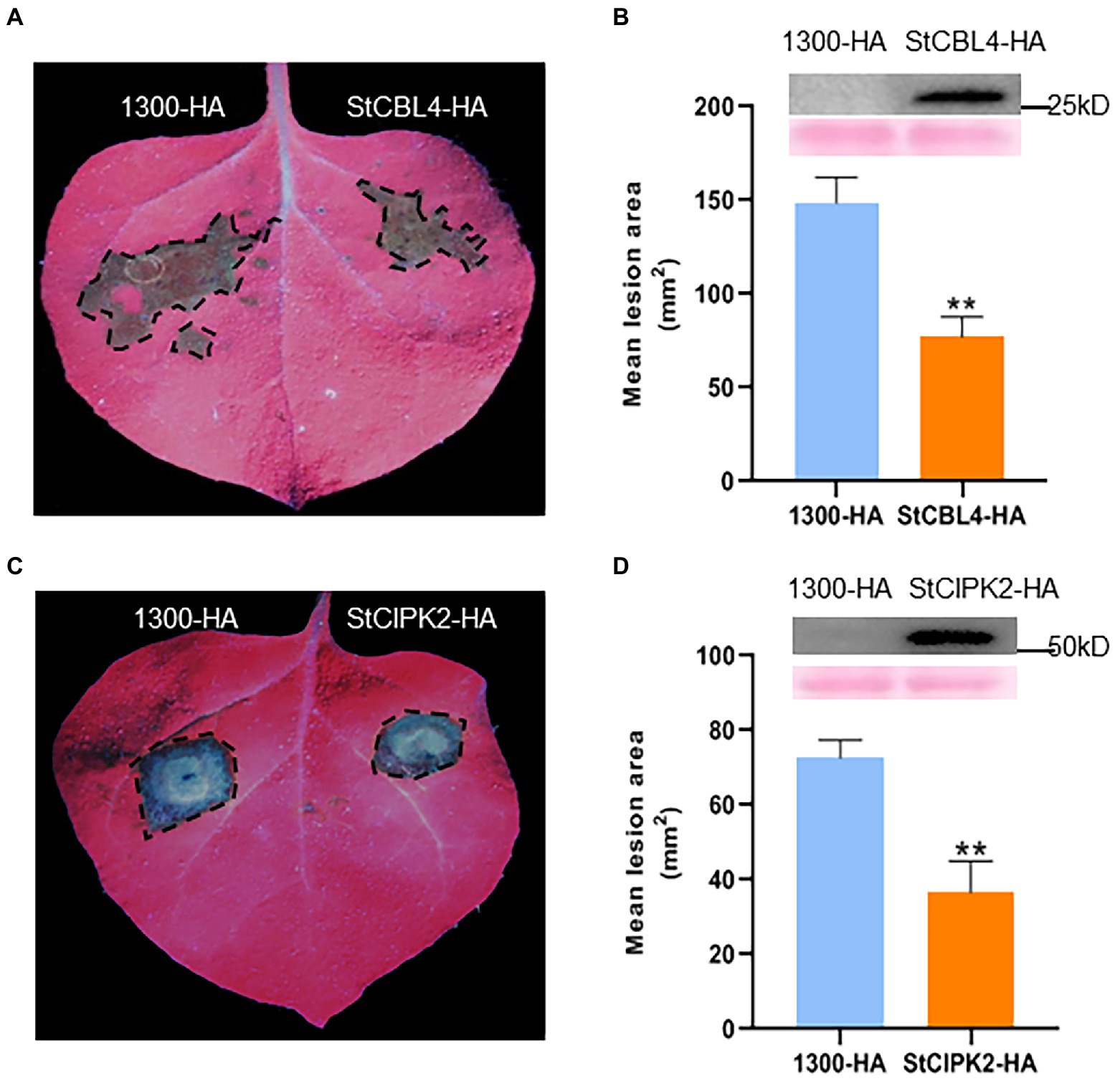
Figure 2. Transient expression of StCBL4 and StCIPK2 enhances the resistance to Rhizoctonia solani in Nicotiana benthamiana. StCBL4-HA, StCIPK2-HA, and 1,300-HA were expressed in N. benthamiana by Agrobacterium-mediated transient expression for 1 day. The leaves were inoculated by R. solani AG-3 and photographed under UV light 36 h later. (A,C) Representative leaf images (under UV light) by StCBL4 and StCIPK2 overexpression showing the extent of R. solani colonization on N. benthamiana leaves expressing each construct as indicated. (B,D) Immunoblotting showed the expression of StCBL4-HA and StCIPK2-HA. The lesion areas were measured and their sizes were calculated. The data represent the means ± SD of measurements (n > 8). The asterisks above the columns represent significance based on an unpaired, two-tailed Student’s t-test relative to the EV. *p < 0.05, **p < 0.01.
In order to analyze the functions of StCBL4 and StCIPK2, VIGS was conducted to silence the expression of orthologous genes NbCBL and NbCIPK2 which was designed using the SGN VIGS Tool3 in N. benthamiana (Supplementary Figures 3A,B, 4A,B). NbPDS VIGS plants began to show photobleaching after 10 days. Efficiencies of NbCBL and NbCIPK2 silencing were confirmed by qRT-PCR of mRNA obtained from leaves that emerged 3 weeks after infiltration of plants with Agrobacterium transformed with TRV2:NbCBL and TRV2:NbCIPK2 constructs. Results of qRT-PCR analysis confirmed that NbCBL and NbCIPK2 were well silenced in VIGS plant leaves, with average expression levels in silenced plants found to be about 22 and 25%, respectively, lower than corresponding levels in negative control plants (TRV2:GFP; Figure 3B). Leaves of NbCBL and NbCIPK2 VIGS plants exhibited visible disease symptoms at R. solani inoculation sites for 2 days. Lesion areas were measured at 60 hpi (Figure 3A). The results showed that NbCBL and NbCIPK2 VIGS plants significantly increased disease lesions compared with GFP VIGS control plants (Figures 3C,D). Taken together, these results indicate that VIGS of NbCBL and NbCIPK2 significantly enhance plant susceptibility to R. solani.
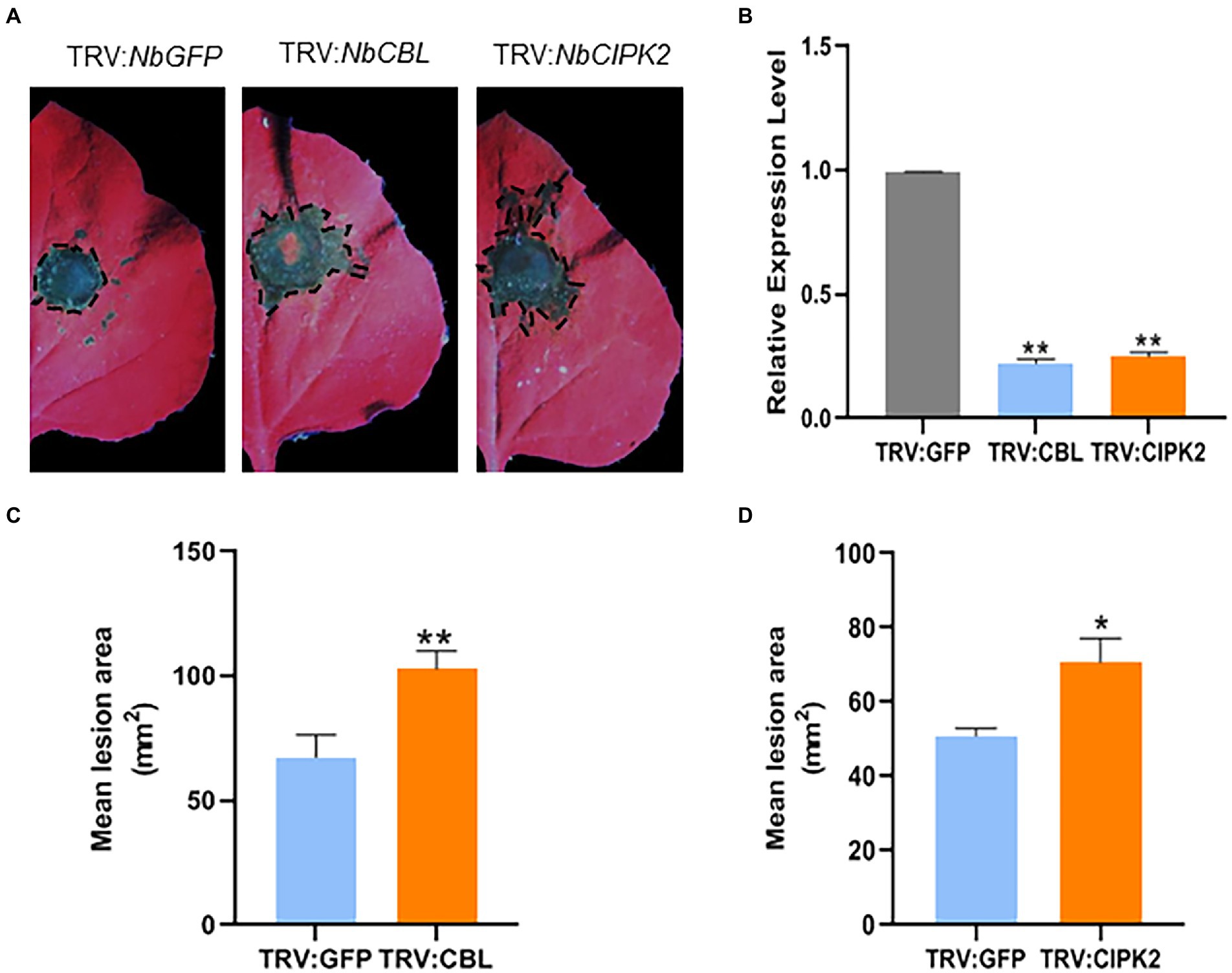
Figure 3. Virus-induced gene silencing (VIGS) of NbCBL and NbCIPK2 significantly promotes R. solani colonization in N. benthamiana. (A) Representative leaf images (under UV light) showing the extent of R. solani colonization on N. benthamiana leaves expressing each construct as indicated. (B) TRV:NbCBL and TRV:NbCIPK2 constructs showed high silencing efficiency in N. benthamiana comparing with the TRV:GFP control vector. (C,D) Mean lesion area measured 60 h after R. solani inoculation. Graph showing that silencing of TRV:NbCBL and TRV:NbCIPK2 constructs significantly increases R. solani lesion diameter compared with TRV:GFP control vector. The data represent the means ± SD of measurements (n > 20). The asterisks above the columns represent significance based on an unpaired, two-tailed Student’s t-test relative to the V-GFP. *p < 0.05, **p < 0.01.
Yeast two-hybrid assays were performed to investigate the proposed interaction between identified proteins. Yeast growth on a minimal selective medium lacking histidine, leucine, tryptophan, and adenine (-His/−Leu/−Trp/−Ade) indicated a physical interaction between StCBL4 and StCIPK2 (Figure 4A). In addition, Co-IP assay and luciferase complementation assay in N. benthamiana by Arobacterium-mediated transient expression was conducted to validate the interaction further. Co-IP results confirmed the Y2H results using GFP beads with anti-HA antibody, StCIPK2-HA protein was successfuliy detected in output samples while GFP was used as a negative control (Figure 4B). In addition, the split luciferase complementation assay showed that StCBL4 interacted strongly with StCIPK2 and the immunoblot analysis showed that all proteins were normally expressed (Figure 4C). All things considered, these results demonstrated that StCBL4 and StCIPK2 interact in vivo.
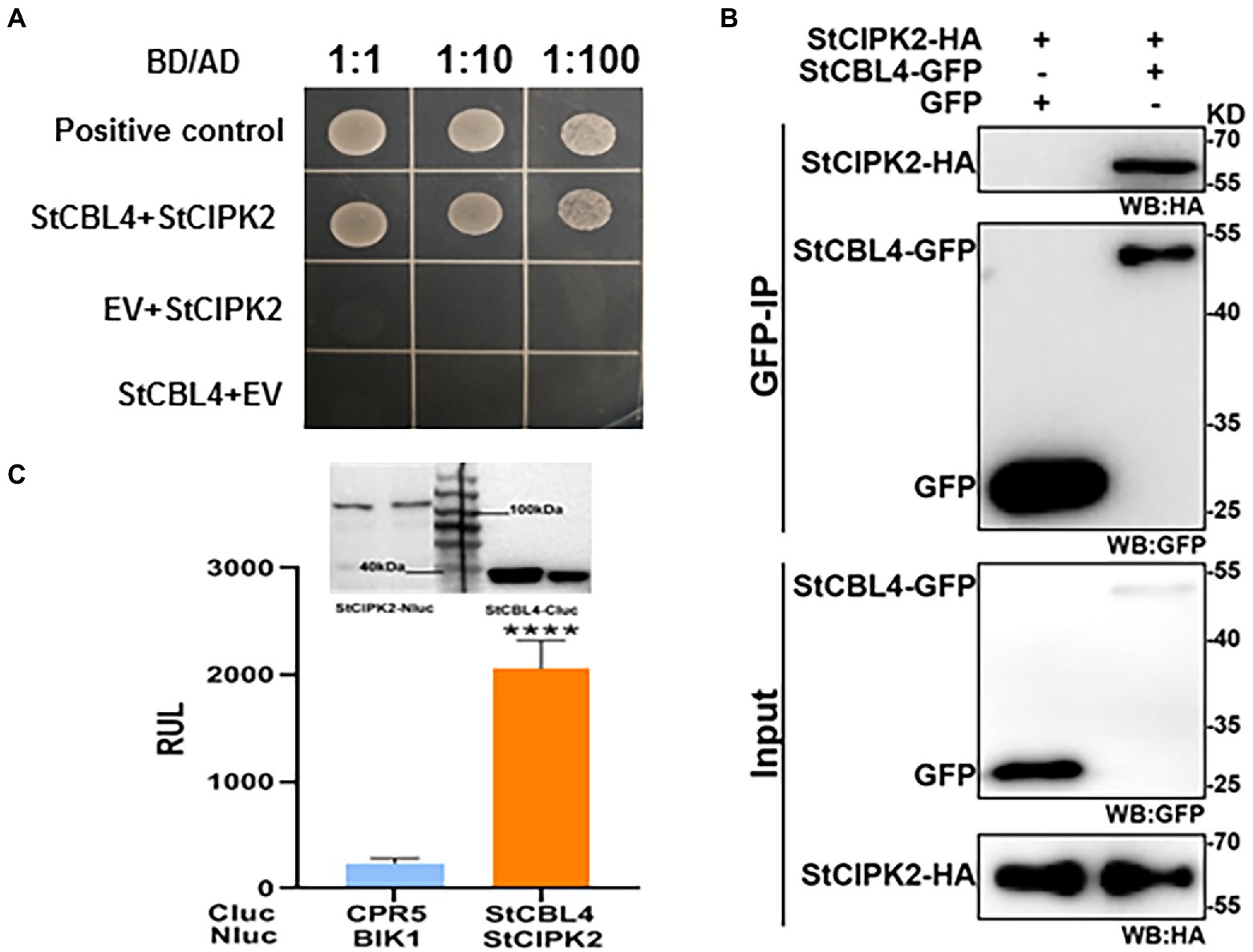
Figure 4. Validation of the protein interaction between StCBL4 and StCIPK2. (A) Yeast two-hybrid analysis of interactions between StCBL4 and StCIPK2. Yeast transformants co-expressing different bait and prey constructs were assayed on SD-Leu-Trp-His-Ade selective medium. BD-53 and AD-T were also co-transformed to serve as a positive control. (B) Co-IP assays were performed using an agarose-conjugated anti-GFP antibody. Input and bound proteins were detected via immunoblot using α-HA and α-GFP antibodies, respectively. (C) StCBL4 interacts with StCIPK2 by luciferase complementation assay and western blot. StCBL4-Cluc and StCIPK2-HA-Nluc constructs were transiently expressed in N. benthamiana plants, and the relative luminescence unit (RLU) was measured 2 days later. Cluc-CPR5 and BAK1-HA-Nluc were used as negative control (Liang et al., 2016). Immunoblotting showed the expression of StCBL4-Cluc and StCIPK2-HA-Nluc. The data represent the means ± SD of measurements (n > 20). The asterisks above the columns represent significance based on an unpaired, two-tailed Student’s t-test relative to the V-GFP. ****p < 0.0001.
To investigate the relationship between StCBL4 and StCIPK2 to R. solani resistance, StCBL4-GFP were co-expressed transiently with StCIPK2-HA in N. benthamiana for 24 h, then inoculated R. solani for 36 h. pCAMBIA1300-HA and pCAMBIA1300-GFP co-expression was performed as a control, and the lesion areas of leaves were photographed at 36 hpi by UV light. As shown results in Figure 5A, leaf lesion areas of co-expressing StCBL4-GFP and StCIPK2-HA exhibited significantly smaller and lower (about 20%) than corresponding levels of the control (Figure 5B). These results revealed a higher difference compared to the single-expression of two genes, and indicated that StCBL4 plays a positive role in the function of StCIPK2 on R. solani resistance.
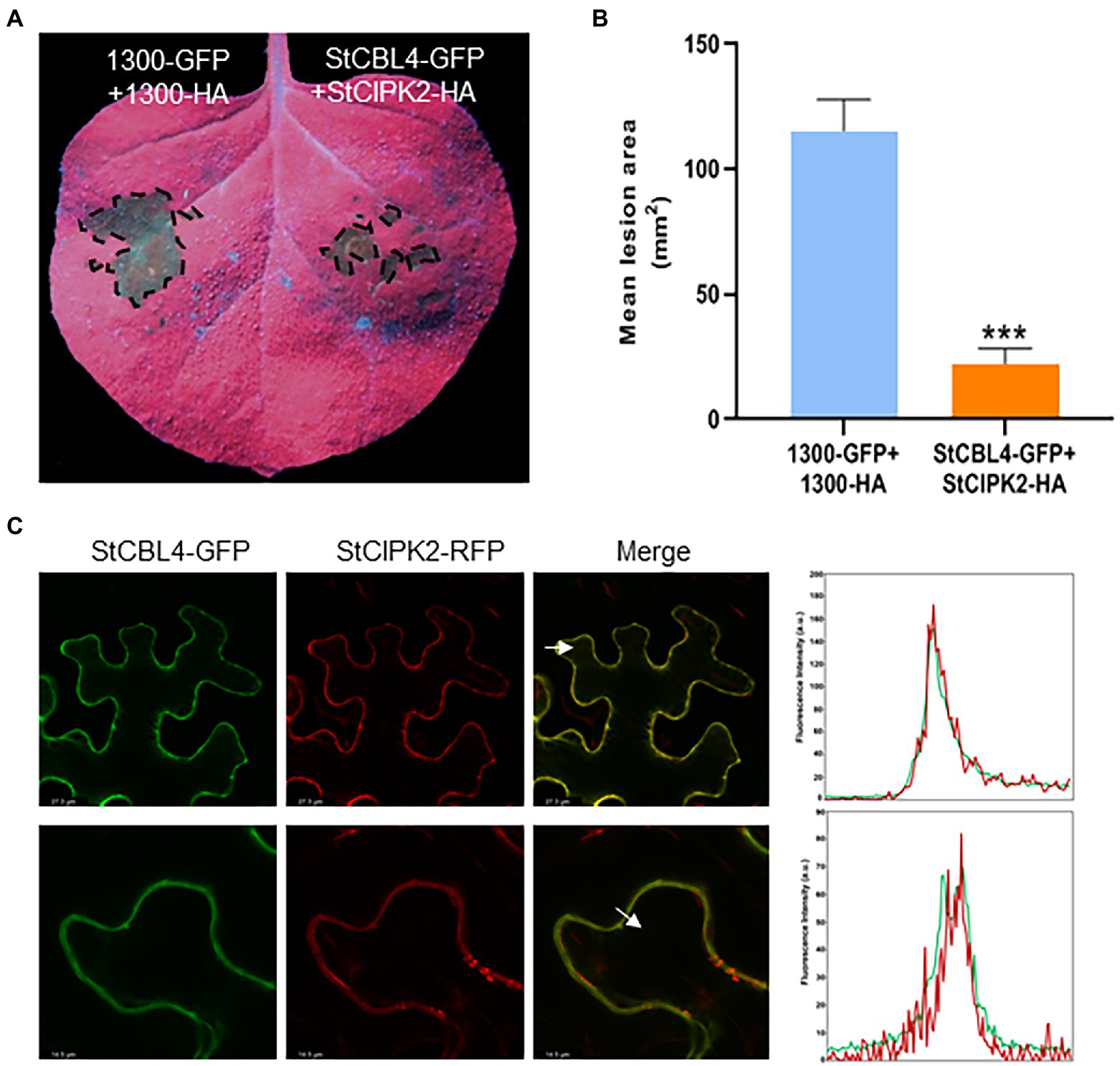
Figure 5. Analysis of transient co-expression and colocalization of StCBL4 and StCIPK2. (A) Transient co-expression of StCBL4 and StCIPK2 enhances the resistance to R. solani in N. benthamiana. Representative leaf images (under UV light) by StCBL4 and StCIPK2 co-overexpression showing the extent of R. solani colonization on N. benthamiana leaves expressing each construct as indicated. (B) The lesion areas were measured and their sizes were calculated by ImageJ software. The data represent the means±SD of measurements (n > 8). The asterisks above the columns represent significance based on an unpaired, two-tailed Student’s t-test relative to the control. *p < 0.05. (C) Confocal laser scanning microscopy showing subcellular co-localization of StCBL4-GFP and StCIPK2-RFP fusion proteins expressed transiently in N. benthamiana leaves. The bottom row is the magnification of the top row. White arrowheads indicate the location of a profile across the membranes and the direction of the fluorescence intensity plot of the profile (right). y axis: Fluorescence intensity. x axis: Distance. Green line, GFP fluorescence. Red line, RFP fluorescence. ***p < 0.001.
Meanwhile, to test the subcellular localization of StCBL4 and StCIPK2, we fused a GFP tag to the C-terminus of StCBL4 and an RFP tag to the C-terminus of StCIPK2, respectively, and viewed the Agrobacterium-mediated expression in N. benthamiana using confocal microscopy. Confocal images showed that the co-expression of StCBL4-GFP and StCIPK2-RFP displayed colocalization in the plasma membrane. Furthermore, the related intensity plots presented coincidence peaks between GFP and RFP fluorescence, indicating the co-localization of StCBL4 and StCIPK2 (Figure 5C).
Given that resistance to R. solani was enhanced in N. benthamiana that overexpressed StCBL4 and StCIPK2, we next generated transgenic potato cultivar Désirée lines expressing StCBL4 and StCIPK2 to investigate the functions of these genes further. As shown process in Figure 6A, transgenic plants were obtained and confirmed via RT-PCR detection of hygromycin transcripts. Empty pBWA(V)KS vector-transformed plants served as wild-type (WT) controls.
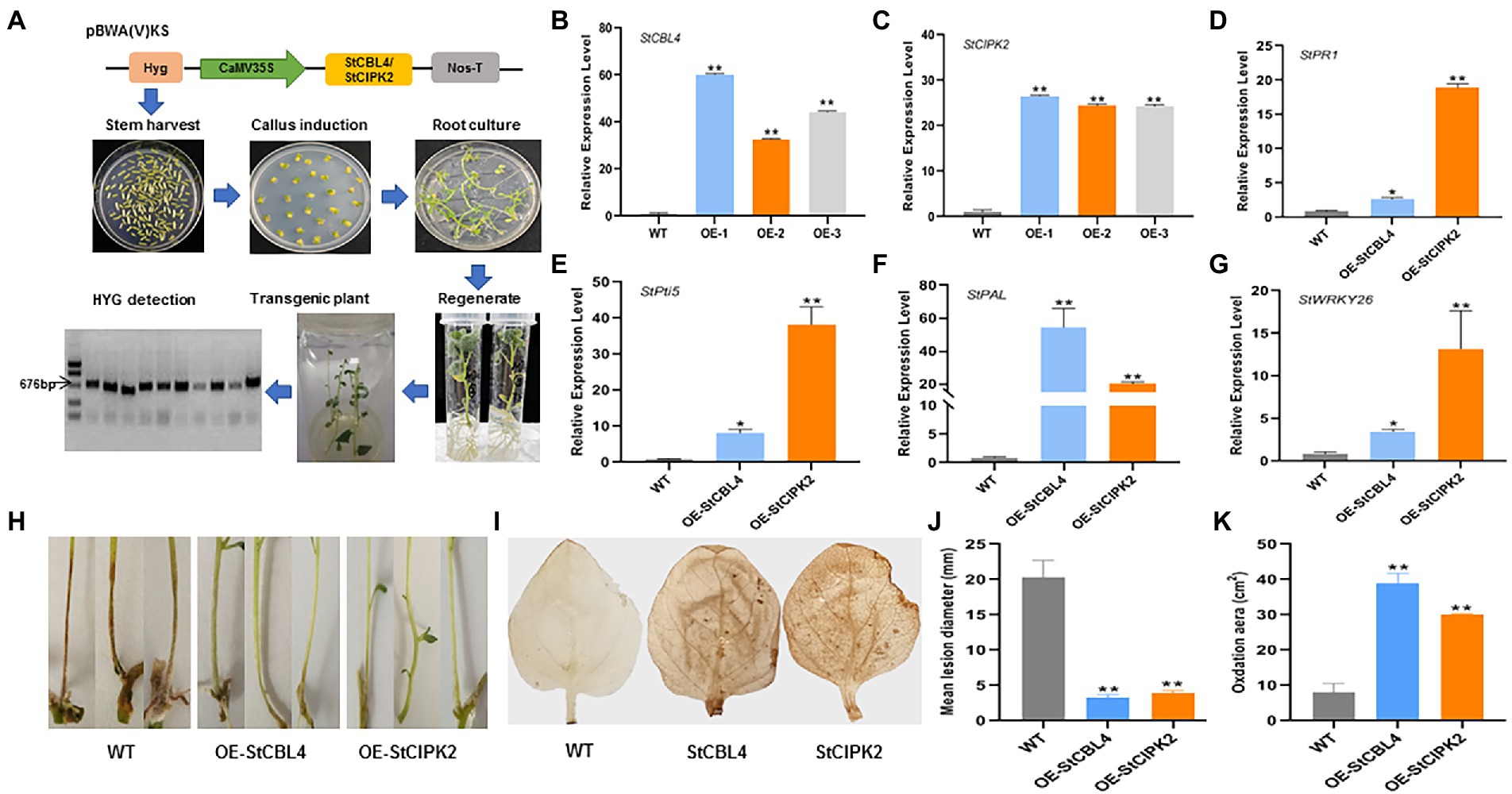
Figure 6. Generation and function analysis of Transgenic Plants on resistance to R. solani. (A) Generation of Transgenic Plants. Skeleton of pBWA(V)KS vectors was used for overexpressing StCBL4 or StCIPK2 gene. The schematic diagram shows the process of inducing plant. Target HYG was detected for the overexpressed positive plant. (B,C) Overexpression profile of StCBL4 and StCIPK2 gene in transgenic plant (OE-1, OE-2, and OE-3) by real-time RT-PCR. The relative mRNA levels were calculated with respect to the expression level of the corresponding gene in the WT plant. (D) The expression levels of StPR1 were higher in StCBL4- and StCIPK2-overexpressing plants than WT plants after R. solani infection. (E) The expression levels of StPti5 were higher in StCBL4- and StCIPK2-overexpressing plants than WT plants after R. solani infection. (F) The expression levels of StPAL were higher in StCBL4- and StCIPK2-overexpressing plants than WT plants after R. solani infection. (G) The expression levels of StWRKY26 were higher in StCBL4- and StCIPK2-overexpressing plants than WT plants after R. solani infection. (H) Symptoms were observed on stems inoculated with R. solani. (I) DAB staining of H2O2 accumulation in leaves of a transgenic plant. (J) Mean lesion diameter measured after R. solani inoculation at 10 days post-inoculation. (K) The amount of H2O2 produced was measured by calculating the DAB-stained area, in which the dark yellow flecks indicated the ROS burst sites. All the experiments were repeated three times, and similar results were obtained. The data represent the means ± SD of triplicate measurements (n > 10). Asterisks above the columns represent significance based on an unpaired, two-tailed Student’s t-test relative to the control. *p < 0.05; **p < 0.01.
The results of the qRT-PCR analysis revealed >30 and > 20-fold upregulation of StCBL4 and StCIPK2 expression in overexpressing plants, respectively (Figures 6B,C). Subsequently, WT and overexpressing transgenic plantlets were incubated with 5-mm-diameter mycelium plugs. Then the plants were grown under identical conditions, and were observed for symptom development after 3 dpi. Compared with the visible symptom of WT plantlets, fewer transgenic plantlets showed canker symptoms at 10 dpi (Figure 6H). As shown in Figure 6J, lesion diameters were measured and showed to be about 16 and 20% lower than corresponding levels in WT. These findings indicated that overexpression of these genes in transgenic plantlets could potentially delay canker development to alter the course of the R. solani infection.
Accumulation of ROS, a significant plant regulatory factor, is an important indicator of plant resistance, especially during the early stage of infection. ROS plays important roles in early signaling events initiated by cellular metabolic perturbation and environmental stimuli (Lehmann et al., 2015). To verify whether StCBL4 and StCIPK2 are involved in H2O2 signaling, ROS bursts were induced in both StCBL4- and StCIPK2-overexpressing plants then H2O2 production was analyzed by analyzing images of DAB-stained leaves. As shown in Figure 6I, a ROS burst was observed by the dark yellow flecks in each overexpressing plants. The amount of H2O2 produced was measured to be about 4.84- and 3.74-fold higher than the corresponding levels in WT by calculating the DAB-stained area, respectively. The results revealed that both StCBL4 and StCIPK2 participate in H2O2 signaling.
To understand molecular regulation underlying increased resistance to the R. solani pathogen, expression levels of plant defense genes were measured in infected StCBL4- and StCIPK2-overexpressing plants at 3 dpi. Defense genes that were studied included pathogenesis-related protein 1 (StPR1), Pto interaction protein 5 (StPti5), phenylalanine ammonia lyase (StPAL), pathogenesis-related protein 10 (StPR10), WRKY transcription factor 26 (StWRKY26), and their expression patterns were examined by qRT-PCR. The results revealed that expression levels of StPR1 were upregulated by 2.67- and 18.9-fold in StCBL4- and StCIPK2-overexpressing plants compared with levels in WT plants. Meanwhile, expression levels of StPti5, StWRKY26, and StPAL were found to be 8.14- and 38.17-fold, 3.49- and 13.18-fold, and 54.46- and 20.14-fold higher in StCBL4 and StCIPK2-overexpressing transgenic plants, respectively. Furthermore, compared with StCBL4-overexpressing plants, higher PR gene expression levels (except for that of StPAL) were observed in StCIPK2-overexpressing plants (Figures 6D–G).
Rhizoctonia solani causes stem canker and black scurf, one of the most severe soil-borne fungal diseases of potato that dramatically reduce yield and quality. Breeding for disease-resistant varieties is viewed as the most effective and eco-friendly method for disease control, with potato disease resistance shown in a previously reported study to rely on R. solani infection-induced triggering of plant pathways that induce production of hormones jasmonic acid (JA) and salicylic acid (SA; Zrenner et al., 2021). However, as no endogenous resistance gene against Rhizoctonia disease has yet been identified in potato, the molecular response of potato to R. solani attack remains largely unknown. Our present study found that StCBL4 and StCIPK2 positively enhance potato anti-fungal responses for combating Rhizoctonia disease.
Although the role of the CBL-CIPK complex in regulating the response against abiotic stress is well documented, it remains unclear whether this complex also regulates responses against biotic stress. In a previous study, several CBL–CIPK interaction partners were hypothesized (based on limited evidence) to play roles in biotic and/or oxidative stress responses and developmental pathways (Pandey et al., 2014). For example, the interaction between OsCIPK14/15 and OsCBL4 plays a crucial role in enhancing rice resistance to Trichoderma viride ethylene-inducing xylanase (Kurusu et al., 2010). The TaCBL4-TaCIPK5 complex acts to promote ROS signaling to enhance the resistance of wheat to Puccinia triiformis f. sp. tritici (Liu et al., 2018). Since CBL4 has been reported to participate in biotic stress resistance functionally, we investigated whether StCBL4 may interact with StCIPK2 to perform similar functions in potato. In this study, transient overexpression of StCBL4 and StCIPK2 individually and synergistically increased the tolerance of potato plants to R. solani in N. benthamiana, meanwhile transgenic potato significantly enhanced resistance as well (Figures 2 and 5B,C,H,J); thus implying that these two genes play crucial roles in defensive responses and innate immunity in potato.
To further confirm that StCBL4 and StCIPK2 genes mediate the interaction between plant and R. solani, TRV-VIGS was used to successfully knock down the expression of StCBL4 and StCIPK2 orthologs NbCBL and NbCIPK2, respectively, that are found in N. benthamiana (Supplementary Figure 3). Notably, silencing of these genes in detached leaves followed by inoculation of leaves with R. solani led to larger lesions in plants with knocked down NbCBL and NbCIPK2 expression compared to unsilenced controls. This result indicates that the knockdown of NbCBL and NbCIPK2 expression increased sensitivity to R. solani infection (Figures 3C,D). Importantly, similar results were observed in wheat, where knockdown of TaCBL4 and TaCIPK5 expression led to a reduced wheat defense response against stripe rust fungus (Liu et al., 2018).
The abovementioned results notwithstanding, the mechanism responsible for the effects of these two proteins on plant resistance remains unknown. However, StCIPK2, an interaction partner of StCBL4, was identified based on results obtained via Y2H assays that were subsequently confirmed via LCA and Co-IP assays (Figure 4). Furthermore, previously reported results have shown that combinations of CBLs and CIPKs functions act as signaling nodes in response to environmental stimuli (Thoday-Kennedy et al., 2015). However, the CIPK protein sequence lacks localization motifs, subcellular localization of CBL-CIPK modules depends on myristoylation and acylation modifications of CBL sites (Mao et al., 2022). In this study, StCBL4 was found to interact with StCIPK2 to specifically recruit StCIPK2 to the plasma membrane in N. benthamiana cells, thus indicating that StCIPK2 interacted with a potentially myristoylated StCBL4 site as a necessary and sufficient condition for StCIPK2 anchoring to the plasma membrane to occur.
Production and accumulation of defense-related proteins in plants in response to biotic stresses are well-known crucial plant defense mechanisms (Wang et al., 2022). Salicylic acid (SA), a key signaling plant resistance molecule involved in resistance to biotrophs, induces the expression of pathogenesis-related (PR) proteins that induce systemic acquired resistance (SAR). As compared with their expression in WT plants, expression levels of PR protein-encoding genes (e.g., PR1, Pti5, and WRKY26) were up-regulated significantly in both StCBL4- and StCIPK2-transgenic plants, thus indicating that StCBL4 and StCIPK2 participated in potato immune response to defend plants against the pathogen (Figures 6D,E,G). PAL is a critical gene in the phenylalanine metabolic pathway that effectively regulates the biosynthesis of corresponding secondary metabolites (Wang et al., 2021). In this study, high-level expression of StPAL revealed the role of an important downstream signaling node as a potential molecular mechanism underlying plant resistance to R. solani disease (Figure 6F).
The accumulation of reactive oxygen species (ROS) is a widespread defense mechanism in higher plants against pathogen attack and sometimes is the cause of cell death that facilitates attack by necrotrophic pathogens (Tian et al., 2021). As well known, necrotrophic fungi with a wide host range can kill host plant cells and use the contents to support their growth. However, despite their substantial role in crop losses, there is much less known about these pathogens and their ability to infect. As a necrotrophic fungus, R. solani could cause physical changes including stunted root growth of the host, transcriptional changes and ROS production (Foley et al., 2016). ROS has multiple functions in plant defense serving as a signaling molecule. It has been proved that H2O2, one of the important ROS productions, plays a role in plant defense as a messenger in signal transduction pathways linked to oxidative stress signaling (Huang et al., 2021). In our study, StCBL4 and StCIPK2 showed the highest induction in transgenic potato plants coinciding with the induction of H2O2 accumulation (Figures 6B,C,I,K). This observation agrees with previous studies that demonstrated that the function of many CBL-CIPK complexes in abiotic and biotic stress signaling is associated with the burst of ROS. Therefore, research focusing on the accumulation of ROS of fungal necrotrophs will also help to understand their pathogenic mechanisms on plants. Indeed, lots of previous research focusing on the RBOH family has confirmed the interaction with CIPKs (Sanyal et al., 2020). Thus, we also speculate that the ROS generation mediated by the StCBL4-StCIPK2 complex during potato resistance to R. solani potentially depends on RBOH proteins. Moreover, identifying which remember of RBOH family plays a role with StCIPK2 should be determined in further studies.
In conclusion, this studyhas illustrated that StCBL4 and StCIPK2 are involved in regulating the potato defensive immune response against R. solani. Furthermore, our results demonstrate that StCBL4 functions in concert with StCIPK2, as positive regulators of immunity and contribute to combating stem canker disease in potato.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
SY and XW conceived and designed the experiments. SY, JLi, JLu, QW, WW, XD, and YM performed the experiments. SY, FM, MG, LH, and LW analyzed the data. SY, JLi, and XW wrote and revised the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Heilongjiang Province (YQ2019C022) and Key-Area Research and Development Program of Guangdong Province (2020B020219002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1032900/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Amino acid sequence alignment and phylogenetic tree analysis of potato StCBL4 with other crops CBL4. (A) Amino acid sequence alignment. (B) Phylogenetic tree analysis. StCBL4(ON383960): Solanum tuberosum CBL4; AtCBL4(AF192886): Arabidopsis thaliana CBL4; CaCBL4(XM_016694246): Capsicum annuum CBL4; CsCBL4(XM_004146172): Cucumis sativus CBL4; OsCBL4(DQ201198): Oryza sativa CBL4; TaCBL4(KU736850): Triticum aestivum CBL4; ZmCBL4(EU973091): Zea mays CBL4; VvCBL4(KT266265): Vitis vinifera CBL4; HvCBL4(HM175879): Hordeum vulgare CBL4.
SUPPLEMENTARY FIGURE 2 | Amino acid sequence alignment and phylogenetic tree analysis of potato StCIPK2 with other crops CIPK2. (A) Amino acid sequence alignment. (B) Phylogenetic tree analysis. StCIPK2 (ON324205): Solanum tuberosum CIPK2; AtCIPK2 (AF192886): Arabidopsis thaliana CIPK2; OsCIPK2 (EU703793): Oryza sativa CIPK2; TaCIPK2 (KU736853): Triticum aestivum CIPK2; HvCIPK2 (KP638475): Hordeum vulgare CIPK2; NtCIPK2 (XM_016647791): Nicotiana tabacum CIPK2.
SUPPLEMENTARY FIGURE 3 | Phylogenetic relationship of CBLs and CIPKs from potato, Arabidopsis and tobacco. The Neighbor-joining (NJ) tree was constructed using MEGA 5.0 software. The black triangle and circle represent StCBL4 and StCIPK2 from potato, respectively. (A) Phylogenetic tree analysis of CBLs from potato, Arabidopsis and tobacco. (B) Phylogenetic tree analysis of CIPKs from potato, Arabidopsis and tobacco.
SUPPLEMENTARY FIGURE 4 | Sequence alignment of StCBL4 with NbCBL and StCIPK2 with NbCIPK2. Sequence alignment was performed by DNAMAN 6.0 software. The red line represents the VIGS site searched by SGN VIGS Tool.
SUPPLEMENTARY TABLE 1 | The list of various potato germplasm resources.
SUPPLEMENTARY TABLE 2 | Primers used in this study.
Batistic, O., Waadt, R., Steinhorst, L., Held, K., and Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. doi: 10.1111/j.1365-313X.2009.04045.x
Cheong, Y. H., Pandey, G. K., Grant, J. J., Batistic, O., Li, L., Kim, B. G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52, 223–239. doi: 10.1111/j.1365-313x.2007.03236.x
Cui, X. Y., Du, Y. T., Fu, J. D., Yu, T. F., Wang, C. T., Ming, C., et al. (2018). Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 18, 93–13. doi: 10.1186/s12870-018-1306-5
De la Torre, F., Gutierrez-Beltran, E., Pareja-Jaime, Y., Chakravarthy, S., Martin, G. B., and del Pozo, O. (2013). The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25, 2748–2764. doi: 10.1105/tpc.113.113530
Dong, Q. Y., Bai, B. W., Almutairi, B., and Kudla, J. (2021). Emerging roles of the CBL-CIPK calcium signaling network as key regulatory hub in plant nutrition. J. Plant Physiol. 257:153335. doi: 10.1016/j.jplph.2020.153335
Foley, R. C., Gleason, C. A., Anderson, J. P., Hamann, T., and Singh, K. B. (2013). Genetic and genomic analysis of Rhizoctonia solani interactions with Arabidopsis; evidence of resistance mediated through NADPH oxidases. PLoS One 8:e56814. doi: 10.1371/journal.pone.0056814
Foley, R. C., Kidd, B. N., Hane, J. K., Anderson, J. P., and Singh, K. B. (2016). Reactive oxygen species play a role in the infection of the necrotrophic fungi, Rhizoctonia solani in Wheat. PLoS One 11:e0152548. doi: 10.1371/journal.pone.0152548
Huang, Z. Q., Lu, J. J., Liu, R. W., Wang, P., Hu, Y. W., and Fang, A. F. (2021). SsCat2 encodes a catalase that is critical for the antioxidant response, QoI fungicide sensitivity, and pathogenicity of Sclerotinia sclerotiorum. Fungal Genet. Biol. 149:103530. doi: 10.1016/j.fgb.2021.103530
Ishitani, M., Liu, J., Halfter, U., Kim, C. S., Shi, W., and Zhu, J. K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12, 1667–1677. doi: 10.2307/3871181
Jung, H. J., Kayum, M. A., Thamilarasan, S. K., Nath, U. K., Park, J. I., Chung, M. Y., et al. (2017). Molecular characterisation and expression profiling of calcineurin B-like (CBL) genes in Chinese cabbage under abiotic stresses. Funct. Plant Biol. 44, 739–750. doi: 10.1071/FP16437
Kim, K. N., Lee, J. S., Han, H., Choi, S. A., Go, S. J., and Yoon, I. S. (2003). Isolation and characterization of a novel rice Ca2C-regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. Plant Mol. Biol. 52, 1191–1202. doi: 10.1023/B:PLAN.0000004330.62660.a2
Kurusu, T., Hamada, J., Nokajima, H., Kitagawa, Y., Kiyoduka, M., Takahashi, A., et al. (2010). Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 153, 678–692. doi: 10.1104/pp.109.151852
Lehmann, S., Serrano, M., L’Haridon, F., Tjamos, S. E., and Metraux, J. P. (2015). Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112, 54–62. doi: 10.1016/j.phytochem.2014.08.027
Liang, X., Ding, P., Lian, K., Wang, J., Ma, M., Li, L., et al. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. elife 5:e13568. doi: 10.7554/eLife.13568
Liu, P., Duan, Y., Liu, C., Xue, Q., Guo, J., Qi, T., et al. (2018). The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J. Exp. Bot. 69, 4443–4457. doi: 10.1093/jxb/ery227
Liu, Z., Li, X., Sun, F., Zhou, T., and Zhou, Y. (2017). Overexpression of OsCIPK30 enhances plant tolerance to Rice stripe virus. Front. Microbiol. 8:2322. doi: 10.3389/fmicb.2017.02322
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313x.2002.01297.x
Lu, J., Liu, T., Zhang, X., Li, J., Wang, X., Liang, X., et al. (2021). Comparison of the distinct, host-specific response of three solanaceae hosts induced by Phytophthora infestans. Int. J. Mol. Sci. 22:11000. doi: 10.3390/ijms222011000
Ma, X., Gai, W. X., Qiao, Y. M., Muhammad, A., Wei, A. M., Luo, D. X., et al. (2019). Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genomics 20:775. doi: 10.1186/s12864-019-6125-z
Ma, X., Li, Q. H., Yu, Y. N., Qiao, Y. M., and Gong, Z. H. (2020). The CBL–CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 21:5668. doi: 10.3390/ijms21165668
Ma, R., Liu, W., Li, S., Zhu, X., Yang, J., Zhang, N., et al. (2021). Genome-wide identification, characterization and expression analysis of the CIPK gene family in potato (Solanum tuberosum L.) and the role of StCIPK10 in response to drought and osmotic stress. Int. J. Mol. Sci. 22:13535. doi: 10.3390/ijms222413535
Mao, J. J., Mo, Z. J., Yuan, G., Xiang, H. Y., Visser, R. G. F., Bai, Y. L., et al. (2022). The CBL-CIPK network is involved in the physiological crosstalk between plant growth and stress adaptation. Plant Cell Environ. 1-11. doi: 10.1111/pce.14396
McLellan, H., Boevink, P. C., Armstrong, M. R., Pritchard, L., Gomez, S., Morales, J., et al. (2013). An RxLR effector from Phytophthora infestans prevents relocalisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 9:e1003670. doi: 10.1371/journal.ppat.1003670
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Pandey, G. K., Kanwar, P., and Pandey, A. (2014). Global Comparative Analysis of CBL-CIPK Gene Families in Plants. Heidelberg: Springer
Sanyal, S. K., Mahiwal, S., Nambiar, D. M., and Pandey, G. K. (2020). CBL-CIPK module-mediated phosphoregulation: facts and hypothesis. Biochem. J. 477, 853–871. doi: 10.1042/BCJ20190339
Tang, R. J., Wang, C., Li, K., and Luan, S. (2020). The CBL–CIPK calcium signaling network: unified paradigm from 20 years of discoveries. Trends Plant Sci. 25, 604–617. doi: 10.1016/j.tplants.2020.01.009
Thoday-Kennedy, E. L., Jacobs, A. K., and Roy, S. J. (2015). The role of the CBL-CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 76, 3–12. doi: 10.1007/s10725-015-0034-1
Tian, L., Li, J., Huang, C., Zhang, D., Xu, Y., Yang, X., et al. (2021). Cu/Zn superoxide dismutase (VdSOD1) mediates reactive oxygen species detoxification and modulates virulence in Verticillium dahliae. Mol. Plant Pathol. 22, 1092–1108. doi: 10.1111/mpp.13099
Verma, P., Sanyal, S. K., and Pandey, G. K. (2021). Ca2+–CBL–CIPK: a modulator system for efficient nutrient acquisition. Plant Cell Rep. 40, 2111–2122. doi: 10.1007/s00299-021-02772-8
Wang, M., Gu, D., Liu, T., Wang, Z., Guo, X., Hou, W., et al. (2007). Overexpression of a putative maize calcineurin B-like protein in Arabidopsis confers salt tolerance. Plant Mol. Biol. 65, 733–746. doi: 10.1007/s11103-007-9238-8
Wang, L., Lu, H., Zhan, J., Shang, Q., Wang, L., Yin, W., et al. (2022). Pathogenesis-related protein-4 (PR-4) gene family in Qingke (Hordeum vulgare L. var. nudum): genome-wide identification, structural analysis and expression profile under stresses. Mol. Biol. Rep. 49, 9397–9408. doi: 10.1007/s11033-022-07794-3
Wang, X. D., Zheng, K. K., Cheng, W. Y., Li, J., Liang, X. X., Shen, J., et al. (2021). Field application of star polymer-delivered chitosan to amplify plant defense against potato late blight. Chem. Eng. J. 417:129327. doi: 10.1016/j.cej.2021.129327
Weinl, S., and Kudla, J. (2009). The CBL–CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi: 10.1111/j.1469-8137.2009.02938.x
Woodhall, J. W., Adams, I. P., Peters, J. C., Harper, G., and Boonham, N. (2013). A new quantitative real-time PCR assay for Rhizoctonia solani AG3-PT and the detection of AGs of Rhizoctonia solani associated with potato in soil and tuber samples in Great Britain. Eur. J. Plant Pathol. 136, 273–280. doi: 10.1007/s10658-012-0161-8
Xi, Y., Liu, J., Dong, C., and Cheng, Z. M. (2017). The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 8:978. doi: 10.3389/fpls.2017.00978
Xiang, Y., Huang, Y., and Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428. doi: 10.1104/pp.107.101295
Xu, G. Y., Moeder, W., Yoshioka, K., and Shan, L. B. (2022). A tale of many families: calcium channels in plant. Plant Cell 34, 1551–1567. doi: 10.1093/plcell/koac033
Yang, S., Min, F. X., Wang, W. Z., Wei, Q., Dong, X. Z., Wang, X. D., et al. (2018). Transcriptome analysis of Rhizoctonia solani anastomosis group 5 early invasion in potato. Int. J. Agric. Biol. 20, 1751–1758. doi: 10.17957/IJAB/15.0684
Yang, S., Min, F. X., Wang, W. Z., Wei, Q., Guo, M., Gao, Y. F., et al. (2017). Anastomosis group and pathogenicity of Rhizoctonia solani associated with stem canker and black scurf of potato in Heilongjiang province of China. Am. J. Potato Res. 94, 95–104. doi: 10.1007/s12230-016-9535-3
Yin, X., Wang, Q. L., Chen, Q., Xiang, N., Yang, Y. Q., and Yang, Y. P. (2017). Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip (Brassica rapa var. rapa). Front. Plant Sci. 8:1191. doi: 10.3389/fpls.2017.01191
Zhou, Z., Bi, G., and Zhou, J. M. (2018). Luciferase complementation assay for protein-protein interactions in plants. Curr. Protoc. Plant Biol. 3, 42–50. doi: 10.1002/cppb.20066
Keywords: Calcineurin B-like protein, CBL-interacting protein kinases, disease resistance, expression profile, regulation, Rhizoctonia solani
Citation: Yang S, Li J, Lu J, Wang L, Min F, Guo M, Wei Q, Wang W, Dong X, Mao Y, Hu L and Wang X (2023) Potato calcineurin B-like protein CBL4, interacting with calcineurin B-like protein-interacting protein kinase CIPK2, positively regulates plant resistance to stem canker caused by Rhizoctonia solani. Front. Microbiol. 13:1032900. doi: 10.3389/fmicb.2022.1032900
Received: 31 August 2022; Accepted: 17 November 2022;
Published: 04 January 2023.
Edited by:
Xiuling Yang, Institute of Plant Protection (CAAS), ChinaReviewed by:
Zhaohui Chu, Wuhan University, ChinaCopyright © 2023 Yang, Li, Lu, Wang, Min, Guo, Wei, Wang, Dong, Mao, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodan Wang, eGR3YW5nQGNhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.