- 1Department of Molecular, Cellular and Biomedical Sciences, University of New Hampshire, Durham, NH, United States
- 2LR Ecologie Pastorale, Institut des Régions Arides, Médenine, Tunisia
- 3USCR Bactériologie Moléculaire and Génomique, Institut National des Sciences Appliquées and de Technologie, Université de Carthage, Tunis Cedex, Tunisia
- 4LR Microbiologie Appliquée, Département de Microbiologie, Faculté des Sciences Naturelles et de la Vie, Université Ferhat Abbas, Sétif, Algeria
- 5Institut Supérieur de Biotechnologie de Sidi Thabet, Université de La Manouba, Biotechnopôle, Sidi Thabet, Sidi Thabet, Tunisia
The actinorhizal plant, Coriaria myrtifolia, is a neurotoxic plant species endemic to the western Mediterranean area, which forms a nitrogen-fixing symbiosis with members of Frankia cluster 2. Contrarily to other Frankia clusters, the occurrence and mode of dispersal for infective cluster 2 units outside of the host plant rhizosphere remains controversial. The present study was designed to investigate the structure of the microbiomes of C. myrtifolia phytosphere, rhizosphere, and soil samples extending outward linearly up to 1 km. Results showed that the epiphyte and endophyte communities were not significantly different from each other for most of the plant tissues. The communities associated with the below-ground tissues (nodule and root) were significantly different from those found on the above-ground tissues (fruit, leaves, and stems) and had a higher community richness. Coriaria myrtifolia phytomicrobiomes were dominated by Cyanobacteria for leaf, stem, and fruit while Actinobacteria and Proteobacteria were dominant in the root and nodule organelles. The nodule, a special niche for nitrogen fixation, was mainly inhabited by Frankia but contained several non-Frankia bacteria. Beside Frankia cluster 2, the presence of clusters 1, 4, and large numbers of cluster 3 strains have been detected in nodules, roots, and rhizospheres of C. myrtifolia. Despite Frankia being found in all plots using plant trapping bioassays with C. myrtifolia seedlings, Frankia cluster 2 was not detected in soil metagenomes showing the limits of detection by this approach. This result also suggests that in the absence of appropriate host plant species, Frankia cluster 2 has a reduced number of infective units present in the soil outward from the rhizosphere.
Introduction
Coriaria myrtifolia is a shrub endemic to the Western Mediterranean found mainly in forests and scrublands (Gtari and Dawson, 2011; Ribeiro-Barros et al., 2019). All plant parts contain a sesquiterpenic lactone named coriamyrtine, which is responsible for poisoning of livestock and human (De Haro et al., 2005; Anadón et al., 2018). Neurotoxicity occurs upon ingestion of stems, branches, leaves, or fruits. Goats have the highest frequency of poisoning with lower rates in cattle and horses. The lag time for symptoms is short, 20 min to 2 h after ingestion of the plant (Faliu et al., 1985). Symptomatically, intoxication by the neurotoxin is characterized by ptyalism, mydriasis, sometimes chills, loss of appetite, and bloating. This event is followed by severe neurological signs including retching, trembling, violent seizures that last 10–15 min, muscular contractions, ataxia, anorexia, tachycardia, intense tachypnea and dyspnea, nystagmus, mydriasis, ptyalism, and bloating (Faliu et al., 1985; Anadón et al., 2018). If not treated, death occurs within 20 min to 2 h after ingestion. The use of C. myrtifolia in tanning and dyeing processes is well documented in medieval texts—mainly from Catalunya (Cardon and Pinto, 2007). The phytomicrobiome or plant microbiome consists of a diverse array of beneficial, neutral, or damaging microorganisms associated with all plant compartments; above (phyllosphere) or below (rhizosphere) ground as epiphytes (on surface), endophytes (within) or nearby plant tissues (Singh and Trivedi, 2017; Sharma et al., 2021). Phytomicrobiome has attracted extensive interest in recent years because of the crucial role it plays in plant growth, health, and ecological function and for current and future biotechnological applications (Turner et al., 2013). Our understanding of the phytomicrobiome has greatly increased using culture-independent approaches with high throughput technologies such as metabarcoding, metagenomics, metatranscriptomics, and metaproteomics. Nevertheless, phytomicrobiomes for native populations of trees and shrubs, especially in forests, have remained less characterized than for other herbaceous and cultivated plant species (Terhonen et al., 2019).
The ability of C. myrtifolia to bear nitrogen-fixing root nodules has been confirmed several times (Bond, 1962; Berg et al., 1999; Nouioui et al., 2014) and its microsymbiont was identified as a member of the genus Frankia (Becking, 1970), which forms a distinct lineage (Nick et al., 1992). Frankia found in other Coriaria species, Datiscaceae, Dryadoideae, and Ceanothus species (Nouioui et al., 2011, 2014) fall within cluster 2 of the four well-defined Frankia clusters (Normand et al., 1996; Ghodhbane-Gtari et al., 2010a). Members of Frankia cluster 2 are difficult to isolate in pure culture (Gtari et al., 2015). Although soil is considered a second ecological niche for Frankia, their free-living existence and dispersal mechanisms for their infective units have not been fully uncovered (Dawson, 2007). Currently, almost all information of the soil habitat was based on indirect approaches using either plant-trapping bioassays (Smolander and Sundman, 1987; Rönkkö et al., 1993; Burleigh and Dawson, 1994) or quantitative-PCR (Samant et al., 2012, 2016). The transient occurrence of the microsymbiont in host rhizosphere via released infective units from decaying nodules and long-distance dispersal outward dispersal from host rhizosphere is likely achieved through wind, water, and biological vectors (Dawson, 2007). Isolation of many Frankia strains in pure culture is strong indirect evidence of the ability of strains to thrive asymbiotically in soils. Infective units from clusters 1, 3 and 4 have been shown to be ubiquitously distributed in soils around the world, (Paschke and Dawson, 1992; Nalin et al., 1997; Gtari et al., 2004) even in newly deposited glacial till, volcanic lava deposits, and young sand dunes prior to colonization by host plants (Young et al., 1992). Moreover, Frankia strains from cluster 1 were found to occur abundantly in non-host rhizosphere (Smolander, 1990) and fresh lava deposit with the volcanophile actinorhizal plant species Myrica and Casuarina (Burleigh and Dawson, 1994; Yamanaka and Okabe, 2006; Kurten et al., 2008). For cluster 2, the situation remains more debatable (Battenberg et al., 2017; Ben Tekaya et al., 2017). It has been previously shown that strains from cluster 2 were able to persist in Alnus glutinosa rhizosphere for a decade in the absence of their compatible host plants (Nouioui et al., 2013). The time course of nodule development in the plant bioassay was relatively very long (18 months) and may be essential to increase Frankia cell units to reach a threshold allowing the initiation of root nodulation. This idea is coherent with claims that the absence of compatible host plant results in a rapid drop of infective units in soil outside of the compatible host rhizosphere (Benson and Silvester, 1993). The recent isolation of members of cluster 2 in axenic conditions provided evidence for saprophytic ability by some members of this group of Frankia strains (Gtari et al., 2015; Gueddou et al., 2019).
The objectives of the present study are to investigate (1) whether the wild status of the toxic actinorhizal species. Coriaria myrtifolia, might be able to shape the phylogenetic structures of its phytomicrobiome and (2) to deepen understanding of the spatial distribution of cluster 2 Frankia strains in the soil nearby (rhizosphere) and more distant from C. myrtifolia trees, as well as spatial patterns of associated soil microbiomes.
Materials and methods
Sampling, Frankia isolation, bioassay, and DNA extraction
The sampling site, “Les Gorges de kharrata Bejaia,” is located in the Mediterranean low-altitude limestone cliffs in Northern Algeria characterized by Rendzina soil under sub-humid bioclimate (average annual rainfall is 881 mm; average annual temperature of 13.9°C, the absolute minimum and maximum temperatures are −9.30 and 41°C, respectively), which considered is appropriates for rupicolous vegetation (Bouchibane et al., 2021). Leaf, stem, fruit, root, and nodule tissues were sampled from three distinct 20 m tall C. myrtifolia plants (Figure 1) and placed in sterile plastic pouches. Latex gloves were worn by personnel and changed frequently during the sampling process. A subset of leaves, stems, fruits, roots, and nodules together with seeds of C. myrtifolia were washed with sterile distilled water and surface sterilized; by shaking in 30% (v/v) H2O2 for 30 min and then aseptically rinsed eight times with sterile water. This first subset of samples represents the endophytic community. A second subset of leaves, stems, fruits, roots, and nodules together with seeds of C. myrtifolia were washed with sterile distilled water several times and were used in the DNA extract process as non-disinfected plant tissues. The second subset of samples represents the epiphytic community. Frankia isolation from root nodules was performed as previously described by Gueddou et al. (2019). Soils (10 g) from the rhizosphere were sampled at a depth of 20 cm starting at the base of the same three C. myrtifolia plants and along 1 km linear series of plots extending from three C. myrtifolia stands. A subset of soils samples was used to inoculate axenic seedlings of C. myrtifolia grown on sterile sand and watered with BD medium (Broughton and Dilworth, 1971) without nitrogen source. One month-old seedlings (n = 10) were inoculated by adding 10 g of each soil sample. Fifteen non inoculated seedlings were used as negative control. Growth was maintained for 10 months, and roots were checked monthly for nodule formation.

Figure 1. Location and geographic information for Algerian phytomicrobiome, soil, rhizosphere samples used in this study. Map insets A–C correspond to the three Coriaria myrtifolia plant replicates.
Extraction of DNA from surface-sterilized and washed nondisinfected plant tissues was performed using Plant DNeasy kits (Qiagen, Hilden, Germany). For soil samples, the DneasyPowerSoil Kit (Qiagen, Valencia, CA, United States) was used. Extracted DNA was treated with 0.5 μl of RNase A (10 mg/ml) for 30 min at 37°C to remove RNA from the sample (Qiagen). For each sample, extractions were performed in triplicate. For the nodule samples, three well developed nodules were used per replicate sample.
PCR amplification and paired-end sequencing of the V4 hypervariable region of the 16S rRNA gene amplificon
Prokaryotic community profiles from plant organs and soils were investigated using the V4 hypervariable region of the 16S subunit rRNA gene. The products were amplified and sequenced following the Earth Microbiome Project 515F/806R protocol (Thompson et al., 2017). Paired-end sequencing of the amplification products was performed using the Illumina HiSeq2500 platform (Illumina, San Diego, CA, United States) as previously described (Louati et al., 2020).
Data processing and analysis of amplicon sequences
The 16S rRNA amplicon sequences of each sample were imported into the Quantitative Insights into Microbial Ecology (QIIME) 2 environment (version qiime2-2017.12) for processing and downstream analysis (Caporaso et al., 2010; Bolyen et al., 2019). Despite repeated sequencing and read pooling, poor quality at the 5’end of reverse reads resulted in decreased pairing and a significant loss of sequencing depth. Therefore, a single-end analysis using only the forward reads was selected. The forward reads were trimmed, and error corrected with DADA2 (Callahan et al., 2016) removing chimeric reads and truncating reads at 240 bp to exclude reads with a Phred quality below 35. Phylogenetic diversity of the resulting features was determined with Multiple Alignment using Fast Fourier Transformation (MAFFT; Katoh and Standley, 2013) and fasttree (Price et al., 2010) using the QIIME2 phylogeny plugin (Bokulich et al., 2018). Taxonomic assignment of the sequence variants in the feature table was accomplished using the QIIME2 feature-classifier plugin (Bokulich et al., 2018). A feature classifier was trained using Naive-Bayes (Pedregosa et al., 2011) and the full-length SILVA 99% identity 16S database (release 138; Quast et al., 2013). The resulting classifier was used with the classify-sklearn method (Pedregosa et al., 2011) to assign taxonomy to the feature table. Genus and phyla level data tables were exported from QIIME2 for diversity analysis in R. For the sake of continuity with previous amplicon-based microbial community studies, we will refer to sequence variants as operational taxonomic units (OTUs; Nguyen et al., 2016).
Alpha diversity in each of the four data sets was determined based on observed operational taxonomic units (OTUs) with taxonomies assigned at both the phylum and genus level. The Shannon Diversity Index (Shannon, 1948) was used to calculate alpha diversity of each sample. Python scripting was used to process and format QIIME2 feature table data which was plotted in R using ggplot2 (Wickham, 2016). Non-metric Multidimensional Scaling (NMDS) analysis (Minchin, 1987) was carried out in R using separate exported QIIME2 feature tables collapsed to genus level taxonomic OTUs. The metaMDS function in the vegan package of R (Dixon, 2003; Oksanen et al., 2018) was used for the NMDS analyses with Bray-Curtis distance (Sørensen, 1948), a trymax of 50 and K of 2. Before NDMS analysis, all feature counts in each sample were relativized by the total number of features in the sample to control for extraction and amplification error. The veganCovEllipse function (Torondel et al., 2016) was used to generate 95% confidence intervals within the ordination space. Ellipses were plotted to represent the confidence interval information and the NMDS data plotted in the sample figure. NDMS figures were generated with ggplot2 (Wickham, 2016) in R. Relative abundance taxonomic bar plots were generated with ggplot2 (Wickham, 2016) using QIIME2 feature tables with taxa collapsed to OTUs at the phylum and genus level.
After taxonomic assignment at the genus level (QIIME2 taxa L6) was completed in QIIME2, reads assigned to the genus Frankia in samples from the roots, rhizosphere, and all three bulk soil types were exported and reassigned to Frankia clade level bins with a custom python script. Specifically, a BLAST database was made using the v4 region of the 45 previously sequenced Frankia strains. The average expect value for alignments between each read and the all the Frankia strain v4 sequences in each clade was calculated. All reads were then assigned to a bin corresponding to the clade with the lowest average expect value for that read. The number of reads in each clade bin was normalized by total reads per sample and used to generate a heat map.
Statistical analysis
For all statistical analyses, the significance of level was set to p < 0.05. R was used for all statistical analysis and visualization.1 Significant differences in alpha diversity were determined using ANOVA. Pairwise Permutational Multivariate ANOVA (PERMANOVA) analyses (Anderson, 2014) were carried out in R using the adonis function in the vegan package (Dixon, 2003; Oksanen et al., 2018) to test the significance of between group differences observed in the NMDS analyses. Bray-Curtis distance (Sørensen, 1948) and Bonferroni correction were used in each PERMANOVA analysis (Anderson, 2014). Simper analysis (Clarke, 1993) was carried out to assess the contribution individual features had on the observed differences between samples. Simper analyses (Clarke, 1993) were performed using the vegan package in R (Dixon, 2003; Oksanen et al., 2018) with the default parameters to gain insight into the taxa driving differences between groups observed in NMDS and PERMANOVA.
Results
Phytomicrobiomes of Coriaria myrtifolia
Table 1 summarizes the sequencing results for C. myrtifolia phytomicrobiome, rhizosphere, and surrounding associated soils. Rarefaction curves indicate that the sequencing depth was adequate to capture most of the observable diversity (Supplementary Figure S1).
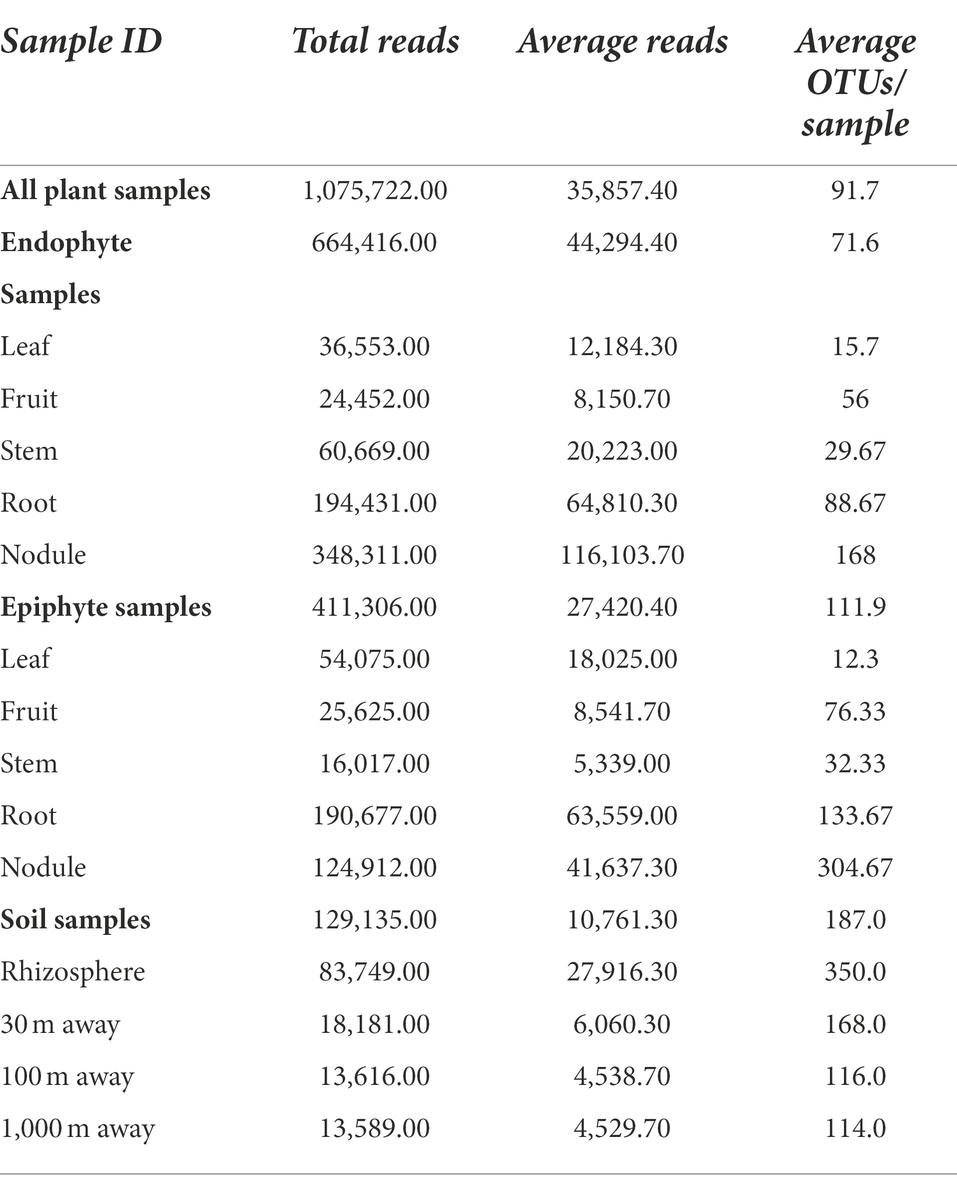
Table 1. Summary of sequencing results for C. myrtifolia phytomicrobiome, rhizosphere and surrounding associated soils.
The measured Shannon diversity metric (Figures 2A,B) of epiphytic and endophytic communities among the various plant organs was not significantly different from each other (Student’s T-test, p > 0.05). Community richness was significantly higher in roots and nodules than for leaves, stems, and fruit (Figures 2A,B). Overall taxonomic compositions of the microbiomes for each plant organ at phylum-level are shown in Figure 3, while Figure 4 shows genus-level distribution. The phyla, Cyanobacteria and Proteobacteria, dominated the leaf and stem microbiome, while fruit contained an abundance of Proteobacteria, Actinobacteria, and Cyanobacteria, which were followed by Firmicutes and Bacteroidetes. Root and nodule communities contained Actinobacteria, Proteobacteria, Firmicutes, Cyanobacteria, Bacteroidetes, and other phyla. Interestingly at family level nodules contained Nocaridiaceae, Phylobacteriaceae, Flammeovirgaceae, Erythrobctaeriaceae, and Rhodospirillaceae, while in root we detected Frankiaceae and Promicromonosporaceae.
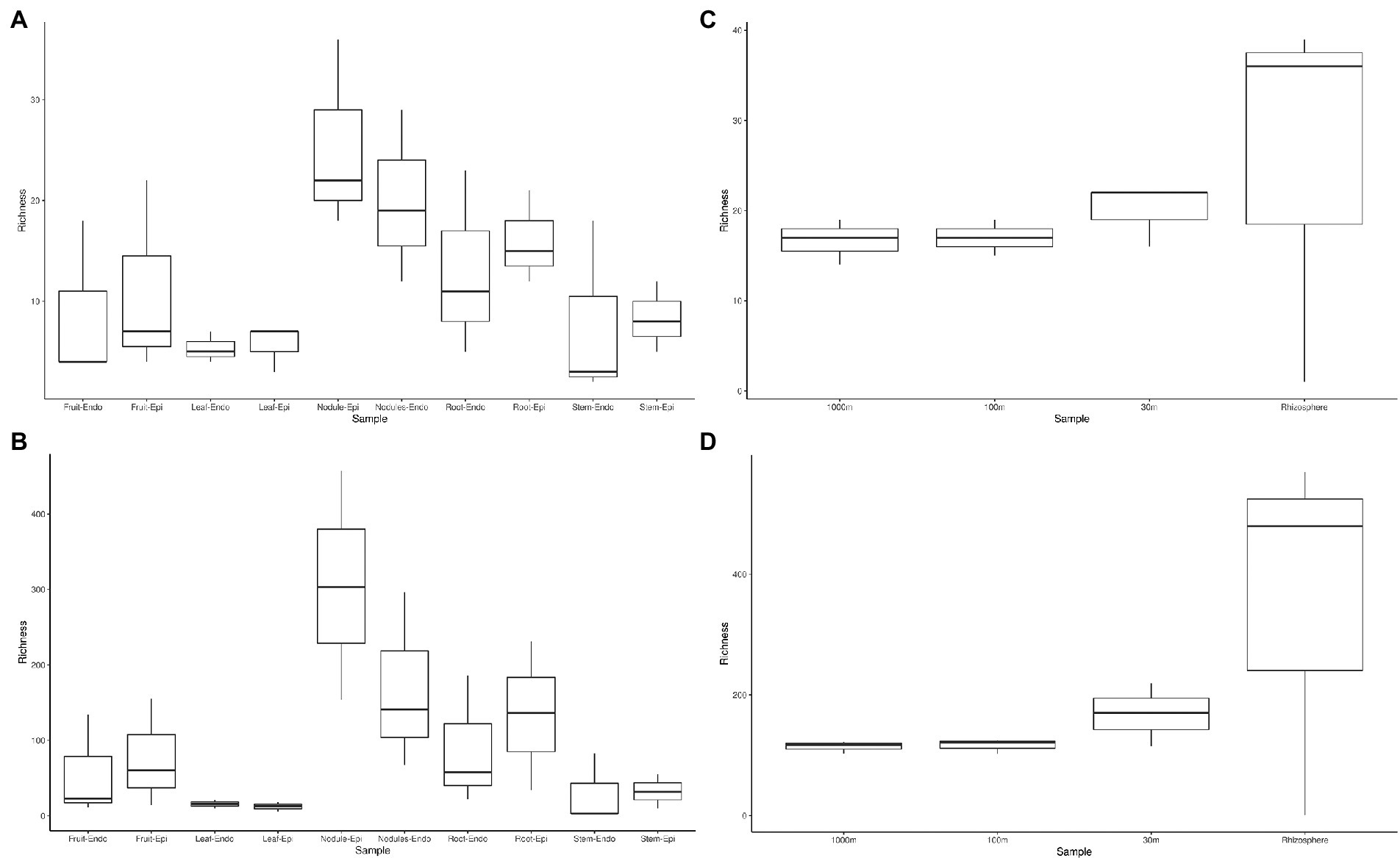
Figure 2. Phylum and genus level alpha diversity of Coriaria myrtifolia phytomicrobiome, soil, and rhizosphere microbiomes. (A,B) represent C. myrtifolia phytomicrobiome, while (C,D) represent C. myrtifolia soil and rhizosphere microbiomes. A and C are at the phylum-level, while panels show the genus-level diversity. Boxes represent 25 to 75th percentile variance around the mean. Whiskers represent 1.5 interval quartile ranges. Data points outside of the whiskers are considered outliers and are plotted separately.

Figure 3. Phylum level taxonomy of Coriaria myrtifolia phytomicrobiome. Average 16S amplicon data show the relative abundance of each phylum after rarefying. Bars represent the average relative abundance of a given taxa across three replicate samples. Both epiphytic (Epi) and endophytic (Endo) communities are presented.
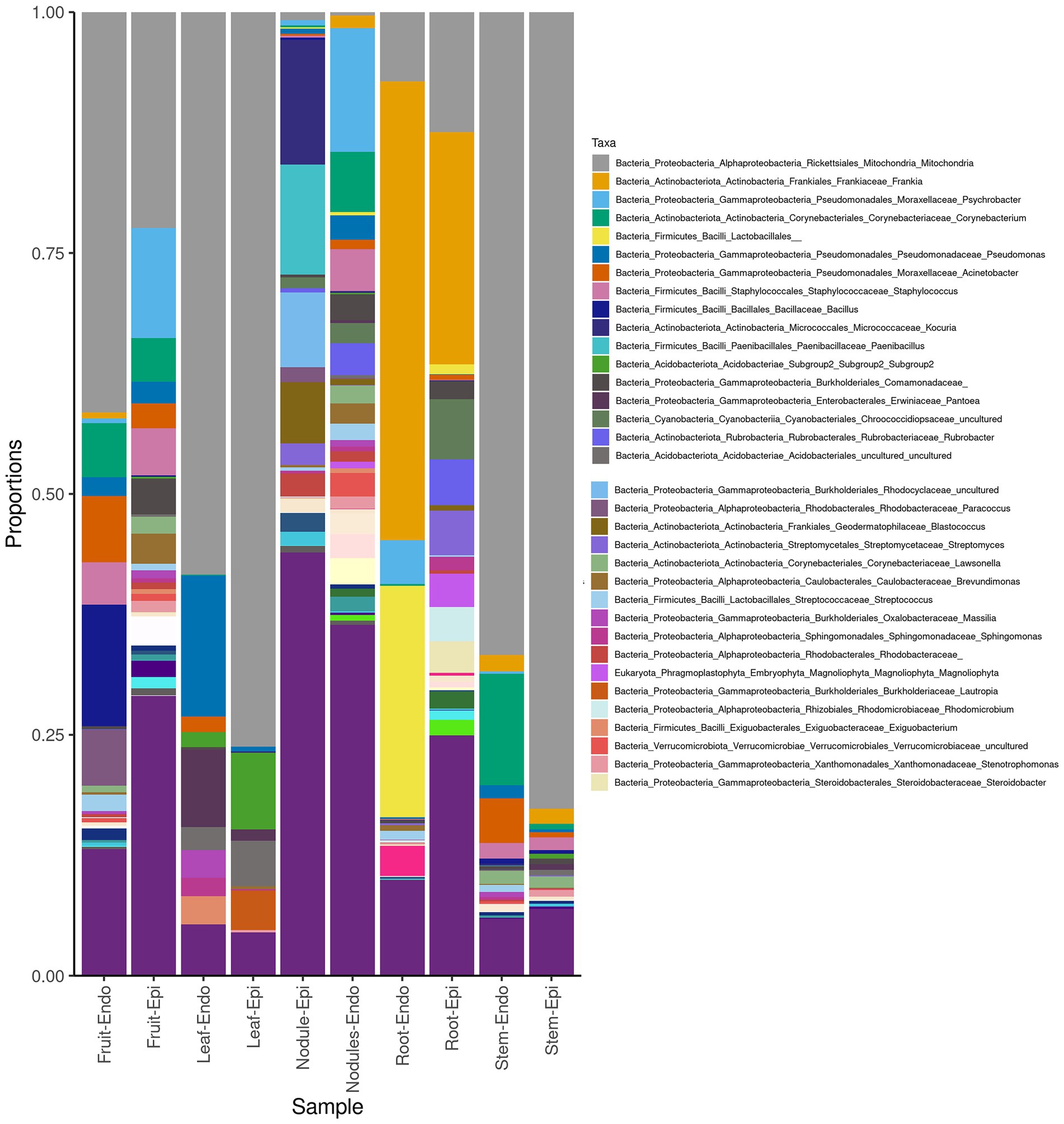
Figure 4. Genus-level taxonomy of Coriaria myrtifolia phyotmicrobiome. Average 16S amplicon data shows he relative abundance of each phylum after rarefying. Bars represent the average relative abundance of a given taxa across three replicate samples. Both epiphytic (Epi) and endophytic (Endo) communities are presented.
Rhizosphere and outward soil plot microbiomes
Sequencing of the C. myrtifolia rhizosphere and nearby soil samples resulted in 129,135 total reads after quality filtering and 4,551 unique features in 12 samples. Features per sample ranged from 203 to 62,808. Table 1 summarizes these results. Rhizosphere soil has the highest number of average OTUs per sample. The remaining soil samples had lower average OTUs per sample but were similar among the different distances. Alpha diversity (Figures 2C,D) measured by the Shannon diversity metric confirmed that the rhizosphere was significantly different from the associated bulk soils (Student’s T- test; p > 0.05) Rarefaction curves indicate that sequencing depth of the soil and rhizosphere samples was adequate to capture most of the diversity present at the sites (Supplementary Figure S2). At phylum level, community structures for soil samples differed only in terms of taxon abundance (Figure 5) and were composed of Proteobacteria and Actinobacteria followed by Acidobacteria, Gemmatimonadetes, Bacteriodetes, Verrucomicrobia, Planctomycetes, and others. Supplementary Figure S3 shows a genus-level distribution.
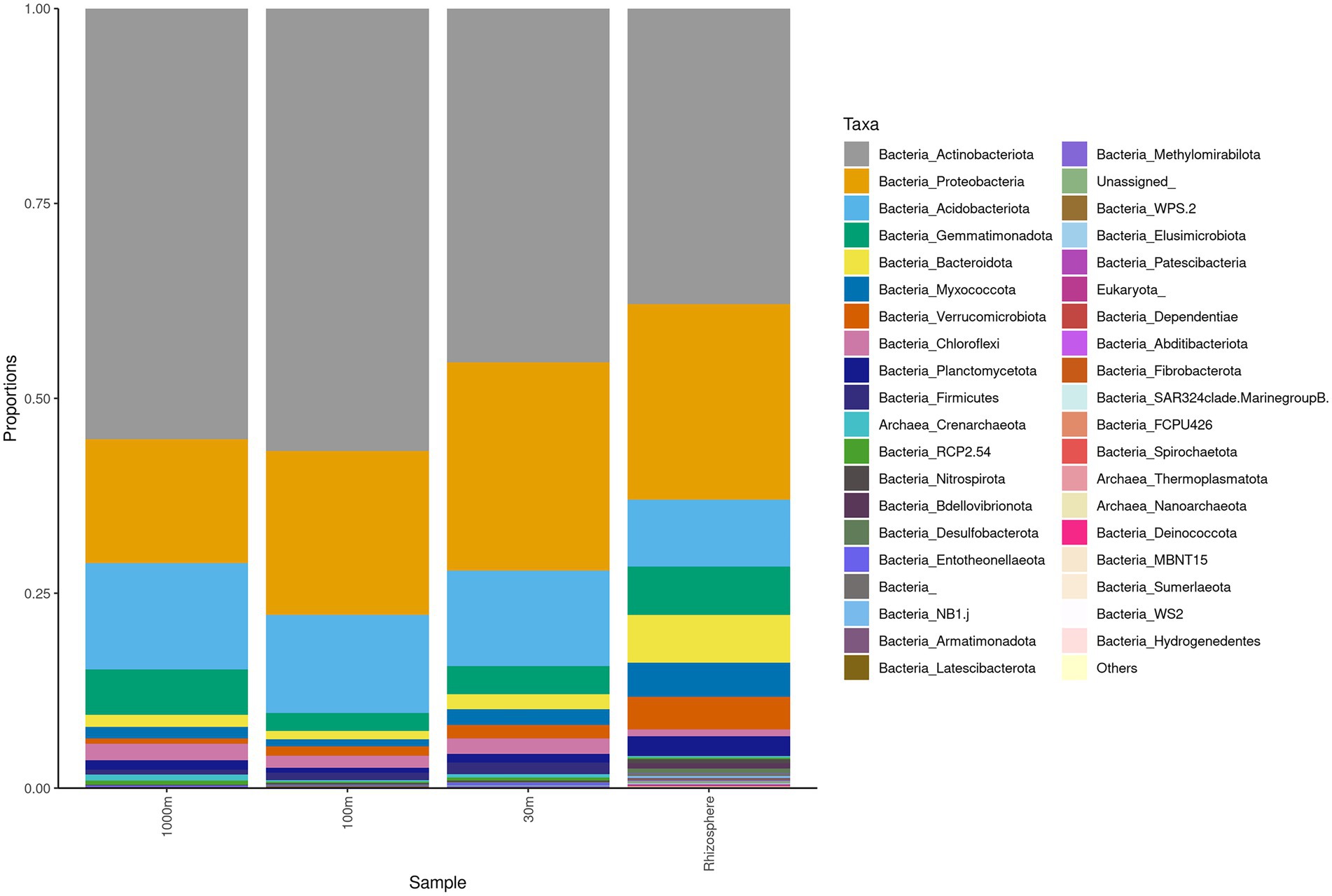
Figure 5. Phylum level taxonomy of Coriaria myrtifolia rhizosphere and nearby soil samples. Average 16S amplicon data shows the relative abundance of each phylum after rarefying. Bars represent the average relative abundance of a given taxa across three replicate samples.
Beta diversity of phytomicrobiome
Beta diversity of phytomicrobiome of the phytobiomes was analyzed to resolve how the samples differed by tissue type, lifestyle (endophyte or epiphyte), or location (above- or below-ground level). The Bray–Curtis distance metric was determined for the samples and ordinated through NMDS analysis. Figure 6 shows the results of this analysis. The tissue clustered together (Figure 6A) and had significant differences between below-ground samples (roots and nodules) and above-ground samples (fruit, leaves, and stems; Figure 6C). However, no difference was identified between endophyte and epihyte samples (Figure 6B). PERMANOVA analysis was performed to determine whether there are significant differences in community structures among the samples (Supplementary Tables S1–S4). Roots were significantly distinct from all other tissue groups individually except nodules. Nodules were significantly distinct from stem and leaf samples, but not from root samples. Fruit and leaf samples were also significantly distinct groups within the ordination. All sample types except stems were individually distinct from the sample types collectively. The NMDS (Figure 7) and PERMANOVA (Supplementary Table S4) analyses of the rhizosphere and associated soil samples from Algeria indicate the only significantly distinct sample type was the rhizosphere.

Figure 6. Non-metric Multidimensional Scaling (NMDS) ordination of C. myrtifolia phytomicrobiome. Ordinations of the phytomicrobiome communities were produced by the Bray-Curts distance metric. (A) shows NMDS for each plant tissue and both epiphytes and endophyte populations. Final ordination stress = 0.139. Procustes rnse <0.001, and max resid = 0.002. Point and ellipse color correspond to tissue type and point fill corresponds to endophyte (unfilled) and epiphyte (filled) samples. (B) shows NMDS for the epiphyte and endophyte communities. Final ordination stress = 0.139, Procrustes rmse <0.001, and max resid = 0.002. Point and ellipse color correspond to epiphyte (blue) and endophyte (red) samples. Point shape and fill correspond to sample replicate. (C) shows NMDS for the above- and below-ground phytomicrobiome communities. Final ordination stress = 0.139, Procrustes rmse <0.001, and max resid = 0.002. Point and ellipse color correspond to above ground samples (AG: fruit, leaf, and stem), and below ground samples (BG: root and nodule). Point shape corresponds to sample types with endophyte and epiphyte samples combined.
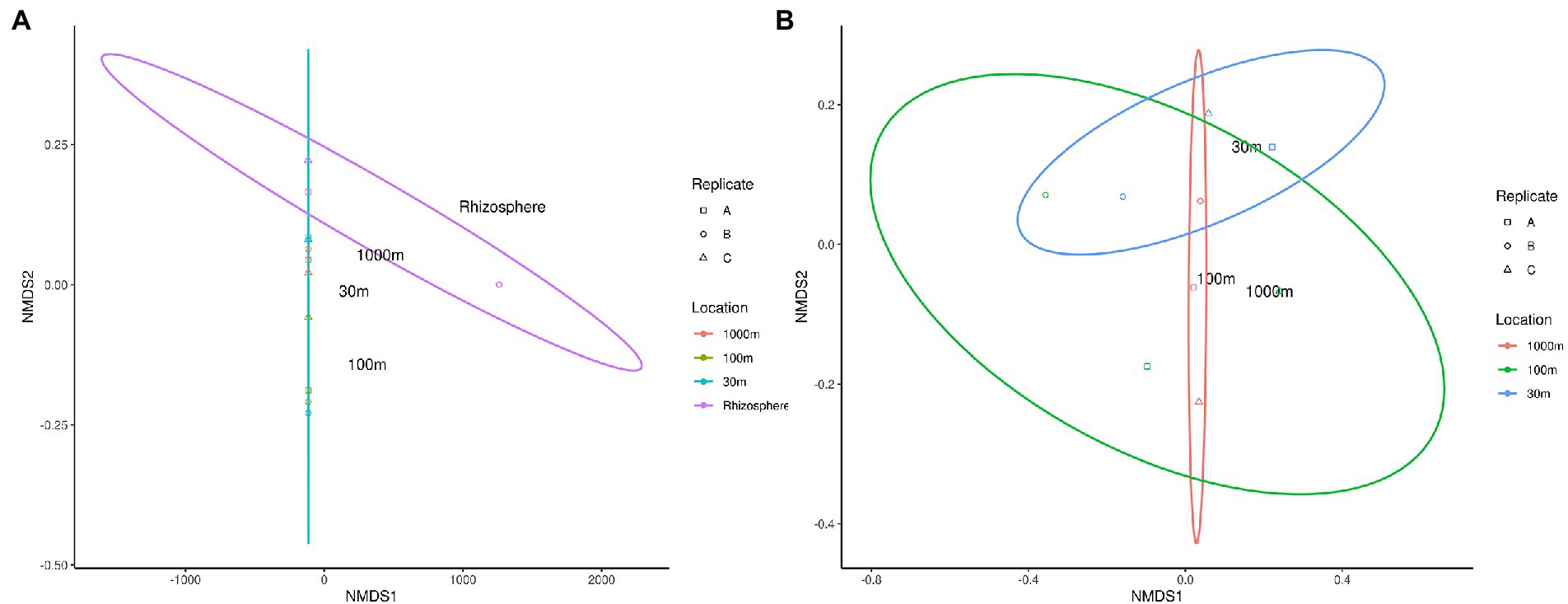
Figure 7. NMDS ordination of C. myrtifolia rhizosphere and nearby soil samples. Ordinations of the microbiome communities were produced by the Bray-Curts distance metric. (A) shows NMDS for the results for C. myrtifolia rhizosphere and nearby soil samples. Final ordination stress—0.107, Procrustes rnse, 0.002, and max resid—0.004. Data point colors correspond to distance from the C. myrtifolia plant. (B) shows the results with only the bulk soil samples and the rhizosphere removed to emphasize differences among those samples. Final stress = 0.130, Procrustes rmse < 0.001, max resid < 0.001. Data point colors correspond to distance from the C. myrtifolia plant. Data point shape corresponds to sample replicates.
Simper analysis showed that Proteobacteria, Firmicutes, Acidobacteria, Actinobacteria, and Bacteroidetes were the most differentiating phyla level taxa between C. myrtifolia fruit and leaf tissue communities. Fruit and root communities, fruit and stem communities, leaf and nodule communities, leaf and stem communities, and nodule and root communities primarily differentiated by the same taxa: Proteobacteria, Firmicutes, Acidobacteria, Actinobacteria, and Bacteroidetes. For between the fruit and nodules communities as well as the nodule and stem communities Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and Verromicrobia were the most differentiating taxa. Leaf and root communities in addition to root and stem communities were differentiated most by Proteobacteria, Firmicutes, Acidobacteria, Actinobacteria, and Phragmoplastophyta. Simpler analysis of the C. myrtifolia phytomicrobiome at the genus level showed that Frankia plays a substantial role in differentiating the phytomicrobiomes of specific plant tissues despite being in a low abundance taxon. The genus Frankia was one of the top two most significant taxa characterizing the differences between fruit and nodule, leaf and root, nodule and root, and root and stem communities.
Actinobacteria, Proteobacteria, Acidobacteria, Gemmatimonadota, and Bacteroidetes were identified by Simper analysis as the most significant phyla contributing to the community differences found between and among all rhizosphere and soil samples from Algeria.
Detection of the different Frankia clusters
Further analysis of the Frankia OTUs by clade level binning (Figure 8) detected Frankia cluster 2 in both endophyte and epiphyte locations in the root and nodule samples, while cluster 1a was detected in root, nodule, rhizosphere, and outward soil samples. Cluster 3 was present in all samples excepting 30 m soil sample. Cluster 1c occurred in root, nodule, and rhizosphere, while cluster 4 was detected in nodule and root. To confirm these results, a plant trapping assay was performed. The mean number of nodules formed on C. myrtifolia seedlings ranged from 5 for rhizospeheric, 2 for 30 m and two nodules for each of 100 m, and 1 km soil samples. Time courses of nodulation were shorter for rhizospheric soil (3 months) than other soil samples (6 months).

Figure 8. Remapped Frankia genus reads to clade level phylogeny. Coriaria myrtifolia rhizosphere, root, nodule, and nearby soil samples are shown. Endophyte and epiphyte communities of the nodule and root samples are shown. Rhizosphere and bulk soil samples and distance from the plant are also presented. Heat-map colors correspond to reads per million for a given set of samples.
Discussion
Coriaria myrtifolia phytomicrobiome
As expected, sequences are mostly assigned to the Bacteria domain while Archaea are not frequently detected (Müller et al., 2016). Overall, the C. myrtifolia phytosphere is composed of Proteobacteria, Actinobacteria, Cyanobacteria, and Firmicutes as the most abundant phyla. Core microbiomes of several plant species including crops, cultivated, wild, and trees include abundant Proteobacteria, Actinobacteria, and Bacteroidetes together with a slighter fraction of Firmicutes (Bulgarelli et al., 2013; Bogas et al., 2015; Müller et al., 2016). The prevalence of bacterial phyla composition varied between above- and below-ground and to lesser extent among plant organs. This result has been attributed to the selective gradient in the soil from the roots to aerial part of plant tissues (Müller et al., 2016). Although epiphyte tissue samples had higher numbers of OTUs than endophytes samples, our results showed that most organ epiphyte and endophyte communities were not significantly different. Several studies reported that epiphytes versus endophytes differ in term of variation in taxon relative abundances (Müller et al., 2016) where epiphytic bacteria are usually more abundant than those of endophytes (James et al., 2002; Wallace et al., 2018). Cyanobacteria were most abundant and diverse at every taxonomic rank in leaf, stem, and fruit. This phylum was also described to predominate in fruit, leaf, flower, and stem of Jingbai Pear trees (Ren et al., 2019), rhizoplane of Spartina alterniflora (Hong et al., 2015), and bark, stem, needle, and root tissues of spruce (Ren et al., 2019), while others have reported that Cyanobacteria are considered absent or infrequently occurring in leaves (Redford et al., 2010; Rigonato et al., 2016). These photosynthetic and for some of them diazotrophic bacteria are wide-spread and aid the health and growth of many plant species (Stanier and Cohen-Bazire, 1977; Ringelberg et al., 2012). Proteobacteria and Actinobacteria were present in leaves, stems, and fruits at lower abundance, which is coherent with the results for several plant species. Previous studies have shown that Alpha- and Gamma-proteobacteria generally predominate other bacterial groups in leaves with variable levels of Beta-proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria presence depending on plant species (Idris et al., 2004; Delmotte et al., 2009; Redford et al., 2010). Similar to Cyanobacteria, Bacteroidetes usually are not detected or detected at low levels in leaves (Lambais et al., 2006; Redford et al., 2010). In this study, Bacteroidetes were not present except in some fruit and nodule samples. Bacteroidetes are ubiquitous colonizers of all types of habitats on Earth (Thomas et al., 2011) producing diverse carbohydrate-active enzymes with large spectrum of substrates from plant, algal, and animal origin that are important in gut-microbiomes (Whitman et al., 1998). The diverse Pedobacter species (Bacteroidetes) are detected in A. glutinosa decaying nodules emphasizing their implication in decomposing process (Mcewan et al., 2017).
Rhizocompartment microbiomes
Root microbiome of C. myrtifolia contained Actinobacteria, Proteobacteria, Firmicutes, Cyanobacteria, Bacteroidetes, and others, which is very similar to composition of Alnus glutinosa root microbiome (Thiem et al., 2018), Casuarina glauca (Ghodhbane-Gtari et al., 2021), several Legume roots (Miranda-Sánchez et al., 2016; Xiao et al., 2017; Zgadzaj et al., 2016), and rice (Edwards et al., 2015; Roman-Reyna et al., 2020).
It is commonly accepted that actinorhizal nodules are a special niche for symbiotic nitrogen fixation, which is primarily colonized by Frankia. However, as it has been shown by the culture-dependent and -independent methods, several non-Frankia coexist with Frankia (Ghodhbane-Gtari et al., 2010b, 2019, 2021; Ghodhbane-Gtari and Tisa, 2014). At phylum level, the C. myrtifolia nodule microbiome differs from root only in term of relative abundance of each phylum and being composed of Bacteroidetes, Proteobacteria, and Actinobacteria. Similar results were shown for A. glutinosa nodule microbiome (Mcewan et al., 2017). At genus rank, Flavobacteria, Nocardia, Rhodococcus, Psychrobacter, and Corynebacteria are the most predominant together with Phyllobacter, Erythrobacter, Erythrobacter, and Flammeovirgacia genera.
Interestingly, the structure of rhizosphere microbiome (Figure 5) at phylum level is similar to root and nodule microbiomes (Figure 3). The root bacterial assemblage is derived mostly from rhizosphere and is established rapidly within a few days after seed germination and subsequently shaped by root exudates and plant genotype (Edwards et al., 2015; Hacquard et al., 2015). This filtration phenomenon is related to the roots affecting the nutrient supply and the physicochemical features of the rhizosphere as well as changes in the environment from soil to endosphere (Gottel et al., 2011; Edwards et al., 2015). Filtration effects were also proposed for legume rhizocompartments through spatial gradients in the following order: nodule > root > rhizosphere > root (Miranda-Sánchez et al., 2016; Xiao et al., 2017).
Frankia cluster 2 dispersal outward from the Coriaria myrtifolia rhizosphere
While the presence of Frankia cluster 2 (the host compatible microsymbionts) and cluster 4 (asymbiotic Frankia that are often isolated from Coriaria nodules; Mirza et al., 1992; Gueddou et al., 2017) was expected, the occurrence of cluster 1 and the super-abundance of cluster 3 in C. myrtifolia nodule, root, and rhizosphere was surprising. As previously stated, clusters 1 and 3 are known to ubiquitously distributed in soils independent to the presence of compatible host-plants (Paschke and Dawson, 1992; Nalin et al., 1997; Gtari et al., 2004). Cluster 4 is also known to be found free-living in soil (Wolters et al., 1997). The presence of cluster 1 was shown to be abundant in Betula pendula rhizosphere (Smolander, 1990). The occurrence of cluster 2 in the A. glutinosa rhizosphere has been demonstrated (Nouioui et al., 2013). Cluster 3 strains have been isolated from the incompatible host Casuarina nodule (Nazaret et al., 1989). Several Frankia cluster 3 strains have been isolated from Ceanothus spp. which like Coriaria spp. is typically infected by Frankia cluster 2 strains (Lechevalier and Ruan, 1984; Lechevalier, 1985; Baker, 1987). Using a glnA sequencing approach, Frankia cluster 3 strains were identified in Ceanothus nodules found in New England (Ktari et al., 2017). Frankia cluster 3 strains were isolated from Coriaria nodules obtained from Algeria but unfortunately, these isolates were lost (M. Gtari unpublished data). Furthermore, Frankia clusters 1 and 3 strains have been shown to dominate or co-dominate in the incompatible host rhizosphere (Samant et al., 2016; Rodriguez et al., 2016). Similar results have been shown for the presence of rhizobia as endophytes in the nodules of legumes other than their hosts (Gage, 2004; Peix et al., 2015; Xiao et al., 2017).
Blastmapping sub-genus level analysis of the metagenome results from 30 and 100 m and 1 km soil samples extending from C. myrtifolia rhizospheres failed to detect cluster 2 contrary to plant trapping bioassay where all seedlings were nodulated. Analysis of host rhizosphere microbiome showed that cluster 2 occupied only a tiny fraction of the microbiome (Battenberg et al., 2017). A quantitative PCR approach indicated that members of cluster 2 might not be present in significant amounts in all soils (Ben Tekaya et al., 2017). Similarly, the microsymbionts of soybean and alfalfa, Ensifer and Bradyrhizobium respectively, were relatively rare in bulk soil (Xiao et al., 2017). This situation was also shown for compatible rhizobial species for Phaseolus vulgaris that is a member of “rare rhizobial biosphere” in the bulk soil but is strongly found enriched on root surfaces and in nodule habitats (Miranda-Sánchez et al., 2016).
Conclusion
Neurotoxicity and wild nature of C. myrtifolia did not greatly shape the phytomicrobiome as it is comparable to several wild or cultivated, herbaceous, or woody plants. The filtration effects of the rhizocompartments (rhizosphere, root, and nodule) was observable in the actinorhizal species C. myrtifolia. These rhizocompartiments are characterized by the occurrence of members of all four Frankia clusters. This fact could be possibly linked to the need of C. myrtifolia to a wider diversity of Frankia microsymbionts with diverse growth-promoting potentialities to cope with the harsher challenges of its wild status. Another possible explanation is that the actinorhizal rhizosphere may also be a primary refuge for the proliferation of all Frankia clusters. Further analysis on other actinorhizal species should clarify this phenomenon. Compared to other Frankia clusters, cluster 2 infective units are present in a less extended amount even in the presence of compatible host species. This amount will further drop outward the rhizosphere of its host plants.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA553505, and PRJNA553534.
Author contributions
MG, FG-G, HC-S, and LT conceived the study. AK, IS, and ES performed the research. ES, FG-G, HC-S, LT, and MG analyzed the data. MG, FG-G, ES, and LT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This material is based upon work supported by the New Hampshire Agricultural Experiment Station, through joint funding of the National Institute of Food and Agriculture, U.S. Department of Agriculture, and the state of New Hampshire. This is Scientific Contribution Number 2947. This project (LST) was supported by the USDA National Institute of Food and Agriculture Hatch 1019869 (LST), the College of Life Science and Agriculture at the University of New Hampshire-Durham, and the Ministère de l’Enseignement Supérieur et de la Recherche Scientifique, Tunisia USCR Bactériologie Moléculaire & Génomique, INSAT-Université de Carthage.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1027317/full#supplementary-material
Footnotes
References
Anadón, A., Martínez-Larrañaga, M. R., Ares, I., and Martínez, M. A. (2018). “Poisonous plants of the Europe,” in Veterinary Toxicology. ed. R. C. Gupta. 3rd ed (Amsterdam: Academic Press), 891–909.
Anderson, M. J. (2014). Permutational multivariate analysis of variance (PERMANOVA). Wiley Statsref 2014, 1–15. doi: 10.1002/9781118445112.stat07841
Baker, D. D. (1987). Relationships among pure cultured strains of Frankia based on host specificity. Physiol. Plant. 70, 245–248. doi: 10.1111/j.1399-3054.1987.tb06139.x
Battenberg, K., Wren, J. A., Hillman, J., Edwards, J., Huang, L., and Berry, A. M. (2017). The influence of the host plant is the major ecological determinant of the presence of nitrogen-fixing root nodule symbiont cluster II Frankia species in soil. Appl. Environ. Microbiol. 83, e02661–e02616. doi: 10.1128/AEM.02661-16
Becking, J. H. (1970). Frankiaceae fam. Nov. (Actinomycetales) with one new combination and six new species of the genus Frankia Brunchorst 1886, 174. Int. J. Syst. Microbiol. 20, 201–220. doi: 10.1099/00207713-20-2-201
Ben Tekaya, S., Ganesan, A. S., Guerra, T., Dawson, J. O., Forstner, M. R., and Hahn, D. (2017). Sybr green-and TaqMan-based quantitative PCR approaches allow assessment of the abundance and relative distribution of Frankia clusters in soils. Appl. Environ. Microbiol. 83, e02833–e02816. doi: 10.1128/AEM.02833-16
Benson, D. R., and Silvester, W. B. (1993). Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol. Rev. 57, 293–319. doi: 10.1128/mr.57.2.293-319.1993
Berg, R. H., Langenstein, B., and Silvester, W. B. (1999). Development in the Datisca-Coriaria nodule type. Can. J. Bot. 77, 1334–1350. doi: 10.1139/b99-076
Bogas, A. C., Ferreira, A. J., Araújo, W. L., Astolfi-Filho, S., Kitajima, E. W., Lacava, P. T., et al. (2015). Endophytic bacterial diversity in the phyllosphere of Amazon Paullinia cupana associated with asymptomatic and symptomatic anthracnose. Springerplus 4:258. doi: 10.1186/s40064-015-1037-0
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2 ' s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bond, G. (1962). Fixation of nitrogen in Coriaria myrtifolia. Nature 193, 1103–1104. doi: 10.1038/1931103a0
Bouchibane, M., Zemouri, M., and Toumi, R. (2021). Contribution à l’étude de la végétation de certains massifs montagneux de la Kabylie des Babors (Nord-Est algérien). Contribution to the study of the vegetation of some mountains to the Kabylia of Babors (northeastern Algeria). Bull. Soc. R. Sci. Liege. 91, 317–360. doi: 10.25518/0037-9565.10696
Broughton, W. J., and Dilworth, M. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125, 1075–1080. doi: 10.1042/bj1251075
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Burleigh, S. H., and Dawson, J. O. (1994). Occurrence of Myrica-nodulating Frankia in Hawaiian volcanic soils. Plant Soil 164, 283–289. doi: 10.1007/BF00010080
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581. doi: 10.1038/Nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cardon, D., and Pinto, A. (2007). Le redoul, herbe des tanneurs et des teinturiers. Collecte, commercialisation et utilisations d’une plante sauvage dans l’espace méridional (XIIIe-XVe siècles). Médiévales 53, 51–64. doi: 10.4000/medievales.3443
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Dawson, J. O. (2007). “Ecology of actinorhizal plants,” in Nitrogen-Fixing Actinorhizal Symbioses. eds. K. L. Pawlowski and W. E. Newton (Dordrecht: Springer), 199–234.
De Haro, L., Pommier, P., Tichadou, L., Hayek-Lanthois, M., and Arditti, J. (2005). Poisoning by Coriaria myrtifolia Linnaeus: a new case report and review of the literature. Toxicon 46, 600–603. doi: 10.1016/j.toxicon.2005.06.026
Delmotte, N., Knief, C., Chaffron, S., Innerebner, G., Roschitzki, B., Schlapbach, R., et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106, 16428–16433. doi: 10.1073/pnas.0905240106
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U. S. A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Faliu, L., Puyt, J. D., and Jean-Blain, C. (1985). Poisoning by Coriaria myrtifolia. Point Veter. 7, 531–532.
Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. doi: 10.1128/MMBR.68.2.280-300.2004
Ghodhbane-Gtari, F., D’Angelo, T., Gueddou, A., Ghazouani, S., Gtari, M., and Tisa, L. S. (2021). Alone yet not alone: Frankia lives under the same roof with other bacteria in actinorhizal nodules. Front. Microbiol. 12:749760. doi: 10.3389/fmicb.2021.749760
Ghodhbane-Gtari, F., Essoussi, I., Chattaoui, M., Chouaia, B., Jaouani, A., Daffonchio, D., et al. (2010a). Isolation and characterization of non-Frankia actinobacteria from root nodules of Alnus glutinosa, Casuarina glauca and Elaeagnus angustifolia. Symbiosis 50, 51–57. doi: 10.1007/s13199-009-0029-7
Ghodhbane-Gtari, F., Nouioui, I., Boudabous, A., and Gtari, M. (2010b). 16S–23S rRNA intergenic spacer region variability in the genus Frankia. Microb. Ecol. 60, 487–495. doi: 10.1007/s00248-010-9641-6
Ghodhbane-Gtari, F., Nouioui, I., Hezbri, K., Lundstedt, E., D’angelo, T., McNutt, Z., et al. (2019). The plant-growth-promoting actinobacteria of the genus Nocardia induces root nodule formation in Casuarina glauca. Antonie Van Leeuwenhoek 112, 75–90. doi: 10.1007/s10482-018-1147-0
Ghodhbane-Gtari, F., and Tisa, L. S. (2014). “Ecology and physiology of non-Frankia actinobacteria from actinorhizal plants,” in Plasticity in Plant-Growth-Promoting and Phytopathogenic Bacteria. ed. E. L. Katsey (New York, NY: Springer), 27–42.
Gottel, N. R., Castro, H. F., Kerley, M., Yang, Z., Pelletier, D. A., Podar, M., et al. (2011). Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 77, 5934–5944. doi: 10.1128/AEM.05255-11
Gtari, M., Brusetti, L., Skander, G., Mora, D., Boudabous, A., and Daffonchio, D. (2004). Isolation of Elaeagnus-compatible Frankia from soils collected in Tunisia. FEMS Microbiol. Letts. 234, 349–355. doi: 10.1111/j.1574-6968.2004.tb09554.x
Gtari, M., and Dawson, J. O. (2011). An overview of actinorhizal plants in Africa. Funct. Plant Biol. 38, 653–661. doi: 10.1071/FP11009
Gtari, M., Ghodhbane-Gtari, F., Nouioui, I., Ktari, A., Hezbri, K., Mimouni, W., et al. (2015). Cultivating the uncultured: growing the recalcitrant cluster-2 Frankia strains. Sci. Rep. 5:13112. doi: 10.1038/srep13112
Gueddou, A., Swanson, E., Hezbri, K., Nouioui, I., Ktari, A., Simpson, S., et al. (2019). Draft genome sequence of the symbiotic Frankia sp. strain BMG5. 30 isolated from root nodules of Coriaria myrtifolia in Tunisia. Antonie Van Leeuwenhoek 112, 67–74. doi: 10.1007/s10482-018-1138-1
Gueddou, A., Swanson, E., Ktari, A., Nouioui, I., Hezbri, K., Ghodhbane-Gtari, F., et al. (2017). Permanent draft genome sequences for three Frankia strains that are atypical, noninfective (nod−) ineffective (fix−) isolates. Genome Announc. 5, e00174–e00117. doi: 10.1128/genomeA.00174-17
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Hong, Y., Liao, D., Hu, A., Wang, H., Chen, J., Khan, S., et al. (2015). Diversity of endophytic and rhizoplane bacterial communities associated with exotic Spartina alterniflora and native mangrove using Illumina amplicon sequencing. Can. J. Microbiol. 61, 723–733. doi: 10.1139/cjm-2015-0079
Idris, R., Trifonova, R., Puschenreiter, M., Wenzel, W. W., and Sessitsch, A. (2004). Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 70, 2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004
James, E. K., Gyaneshwar, P., Mathan, N., Barraquio, W. L., Reddy, P. M., Iannetta, P. P., et al. (2002). Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant-Microbe Interact. 15, 894–906. doi: 10.1094/MPMI.2002.15.9.894
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Ktari, A., Nouioui, I., Furnholm, T., Swanson, E., Ghodhbane-Gtari, F., Tisa, L. S., et al. (2017). Permanent draft genome sequence of Frankia sp. NRRL B-16219 reveals the presence of canonical nod genes, which are highly homologous to those detected in Candidatus Frankia Dg1 genome. Standards in genomic. Science 12:e51. doi: 10.1186/s40793-017-0261-3
Kurten, E. L., Snyder, C. P., Iwata, T., and Vitousek, P. M. (2008). Morella cerifera invasion and nitrogen cycling on a lowland Hawaiian lava flow. Biol. Invasions 10, 19–24. doi: 10.1007/s10530-007-9101-5
Lambais, M. R., Crowley, D. E., Cury, J. C., Bull, R. C., and Rodrigues, R. R. (2006). Bacterial diversity in tree canopies of the Atlantic forest. Science 312:1917. doi: 10.1126/science.1124696
Lechevalier, M. P., and Ruan, J.-S. (1984). Physiological and chemical diversity of Frankia spp. isolated from nodules of Comptonia peregrina (L.) Coult. and Ceanothus americanus L. Plant Soil 78, 15–22. doi: 10.1007/BF02277836
Louati, M., Ennis, N. J., Ghodhbane-Gtari, F., Hezbri, K., Sevigny, J. L., Fahnestock, M. F., et al. (2020). Elucidating the ecological networks in stone-dwelling microbiomes. Environ. Microbiol. 22, 1467–1480. doi: 10.1111/1462-2920.14700
McEwan, N. R., Wilkinson, T., Girdwood, S. E., Snelling, T. J., Collins, T., Dougal, K. L., et al. (2017). Evaluation of the microbiome of decaying alder nodules by next generation sequencing. Endocyt. Cell Res. 28, 14–19. https://zs.thulb.uni-jena.de/receive/jportal_jparticle_00501405
Minchin, P. R. (1987). An evaluation of relative robustness of techniques for ecological ordinations. Vegetatio 69, 89–107. doi: 10.1007/BF00038690
Miranda-Sánchez, F., Rivera, J., and Vinuesa, P. (2016). Diversity patterns of R hizobiaceae communities inhabiting soils, root surfaces and nodules reveal a strong selection of rhizobial partners by legumes. Environ. Microbiol. 18, 2375–2391. doi: 10.1111/1462-2920.13061
Mirza, M. S., Hahn, D., and Akkermans, A. D. L. (1992). Isolation and characterization of Frankia strains from Coriaria nepaknsis. Syst. Appl. Microbiol. 15, 289–295. doi: 10.1016/S0723-2020(11)80103-7
Müller, D. B., Vogel, C., Bai, Y., and Vorholt, J. A. (2016). The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234. doi: 10.1146/annurev-genet-120215-034952
Nalin, R., Normand, P., and Domenach, A. M. (1997). Distribution and N2-fixing activity of Frankia strains in relation to soil depth. Physiol. Plant. 99, 732–738. doi: 10.1111/j.1399-3054.1997.tb05378.x
Nazaret, S., Simonet, P., Normand, P., and Bardin, R. (1989). Genetic diversity among Frankia isolated from Casuarina nodules. Plant Soil 118, 241–247. doi: 10.1007/BF02232812
Nguyen, N. P., Warnow, T., Pop, M., and White, B. (2016). A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microb. 2:16004. doi: 10.1038/Npjbiofilms.2016.4
Nick, G., Paget, E., Simonet, P., Moiroud, A., and Normand, P. (1992). The nodular endophytes of Coriaria spp. form a distinct lineage within the genus Frankia. Mol. Ecol. 1, 175–181. doi: 10.1111/j.1365-294X.1992.tb00173.x
Normand, P., Orso, S., Cournoyer, B., Jeannin, P., Chapelon, C., Dawson, J., et al. (1996). Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int. J. Syst. Microbiol. 46, 1–9.
Nouioui, I., Ghodhbane-Gtari, F., Beauchemin, N. J., Tisa, L. S., and Gtari, M. (2011). Phylogeny of members of the Frankia genus based on gyrB, nifH and glnII sequences. Antonie Van Leeuwenhoek 100, 579–587. doi: 10.1007/s10482-011-9613-y
Nouioui, I., Ghodhbane-Gtari, F., Fernandez, M. P., Boudabous, A., Normand, P., and Gtari, M. (2014). Absence of cospeciation between the uncultured Frankia microsymbionts and the disjunct actinorhizal Coriaria species. Biomed. Res. Int. 2014:924235. doi: 10.1155/2014/924235
Nouioui, I., Sbissi, I., Ghodhbane-Gtari, F., Benbrahim, K. F., Normand, P., and Gtari, M. (2013). First report on the occurrence of the uncultivated cluster 2 Frankia microsymbionts in soil outside the native actinorhizal host range area. J. Biosci. 38, 695–698. doi: 10.1007/s12038-013-9366-z
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2018). Vegan: community ecology package. R package version 2.5-3. Available at: https://CRAN.R-project.org/package=vegan
Paschke, M. W., and Dawson, J. O. (1992). The occurrence of Frankia in tropical forest soils of Costa Rica. Plant Soil 142, 63–67. doi: 10.1007/BF00010175
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011). Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830. doi: 10.5555/1953048.2078195
Peix, A., Ramírez-Bahena, M. H., Velázquez, E., and Bedmar, E. J. (2015). Bacterial associations with legumes. Crit. Rev. Plant Sci. 34, 17–42. doi: 10.1080/07352689.2014.897899
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Redford, A. J., Bowers, R. M., Knight, R., Linhart, Y., and Fierer, N. (2010). The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 12, 2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x
Ren, F., Dong, W., and Yan, D. H. (2019). Endophytic bacterial communities of Jingbai pear trees in North China analysed with Illumina sequencing of 16S rDNA. Arch. Microbiol. 201, 199–208. doi: 10.1007/s00203-018-1597-9
Ribeiro-Barros, A. I., Catarino, S., Moura, I., Ramalho, J. C., Romeiras, M. M., and Ghodhbane-Gtari, F. (2019). Actinorhizal trees and shrubs from Africa: distribution, conservation and uses. Antonie Van Leeuwenhoek 112, 31–46. doi: 10.1007/s10482-018-1174-x
Rigonato, J., Gonçalves, N., Andreote, A. P. D., Lambais, M. R., and Fiore, M. F. (2016). Estimating genetic structure and diversity of cyanobacterial communities in Atlantic forest phyllosphere. Can. J. Microbiol. 62, 953–960. doi: 10.1139/cjm-2016-0229
Ringelberg, D., Foley, K., and Reynolds, C. M. (2012). Bacterial endophyte communities of two wheatgrass varieties following propagation in different growing media. Can. J. Microbiol. 58, 67–80. doi: 10.1139/w11-122
Rodriguez, D., Guerra, T. M., Forstner, M. R., and Hahn, D. (2016). Diversity of Frankia in soil assessed by Illumina sequencing of nifH gene fragments. Systematic and Applied Microbiology 39, 391–397. doi: 10.1016/j.syapm.2016.06.007
Roman-Reyna, V., Pinili, D., Borja, F. N., Quibod, I. L., Groen, S. C., Alexandrov, N., et al. (2020). Characterization of the leaf microbiome from whole-genome sequencing data of the 3000 rice genomes project. Rice 13, 1–8. doi: 10.1186/s12284-020-00432-1
Rönkkö, R., Smolander, A., Nurmiaho-Lassila, E. L., and Haahtela, K. (1993). Frankia in the rhizosphere of nonhost plants: A comparison with root-associated N2-fixing Enterobacter, Klebsiella and Pseudomonas. Plant Soil 153, 85–95. doi: 10.1007/BF00010547
Samant, S., Dawson, J. O., and Hahn, D. (2016). Growth responses of introduced Frankia strains to edaphic factors. Plant Soil 400, 123–132. doi: 10.1007/s11104-015-2720-1
Samant, S., Sha, Q., Iyer, A., Dhabekar, P., and Hahn, D. (2012). Quantification of Frankia in soils using SYBR green based qPCR. Syst. Appl. Microbiol. 35, 191–197. doi: 10.1016/j.syapm.2011.12.004
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x
Sharma, M. M. M., Sharma, P., Kapoor, D., Beniwal, P., and Mehta, S. (2021). “Phytomicrobiome community: an agrarian perspective towards resilient agriculture,” in Plant Performance Under Environmental Stress. ed. A. Husen (Switzerland: Springer), 493–534.
Singh, B. K., and Trivedi, P. (2017). Microbiome and the future for food and nutrient security. Microbial Biotech. 10, 50–53. doi: 10.1111/1751-7915.12592
Smolander, A. (1990). Frankia populations in soils under different tree species—with special emphasis on soils under Betula pendula. Plant Soil 121, 1–10. doi: 10.1007/BF00013091
Smolander, A., and Sundman, V. (1987). Frankia in acid soils of forests devoid of actinorhizal plants. Physiol. Plant. 70, 297–303. doi: 10.1111/j.1399-3054.1987.tb06147.x
Sørensen, T. A. (1948). A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 5, 1–34.
Stanier, R. Y., and Cohen-Bazire, G. (1977). Phototrophic prokaryotes: the cyanobacteria. Annu. Rev. Microbiol. 31, 225–274. doi: 10.1146/annurev.mi.31.100177.001301
Terhonen, E., Blumenstein, K., Kovalchuk, A., and Asiegbu, F. O. (2019). Forest tree microbiomes and associated fungal endophytes: functional roles and impact on forest health. Forests 10:42. doi: 10.3390/f10010042
Thiem, D., Gołębiewski, M., Hulisz, P., Piernik, A., and Hrynkiewicz, K. (2018). How does salinity shape bacterial and fungal microbiomes of Alnus glutinosa roots? Front. Microbiol. 9:651. doi: 10.3389/fmicb.2018.00651
Thomas, F., Hehemann, J. H., Rebuffet, E., Czjzek, M., and Michel, G. (2011). Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2:93. doi: 10.3389/fmicb.2011.00093
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Torondel, B., Ensink, J. H. J., Gundogdu, O., Ijaz, U. Z., Parkhill, J., Abdelahi, F., et al. (2016). Assessment of the influence of intrinsic environmental and geographical factors on the bacterial ecology of pit latrines. Microb. Biotechnol. 9, 209–223. doi: 10.1111/1751-7915.12334
Turner, T. R., Ramakrishnan, K., Walshaw, J., Heavens, D., Alston, M., Swarbreck, D., et al. (2013). Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7, 2248–2258. doi: 10.1038/ismej.2013.119
Wallace, J., Laforest-Lapointe, I., and Kembel, S. W. (2018). Variation in the leaf and root microbiome of sugar maple (Acer saccharum) at an elevational range limit. PeerJ 6:e5293. doi: 10.7717/peerj.5293
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Wolters, D. J., Van Dijk, C., Zoetendal, E. G., and Akkermans, A. D. L. (1997). Phylogenetic characterization of ineffective Frankia in Alnus glutinosa (L.) Gaertn. Nodules from wetland soil inoculants. Mol. Ecol. 6, 971–981. doi: 10.1046/j.1365-294X.1997.00265.x
Xiao, X., Chen, W., Zong, L., Yang, J., Jiao, S., Lin, Y., et al. (2017). Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol. Ecol. 26, 1641–1651. doi: 10.1111/mec.14027
Yamanaka, T., and Okabe, H. (2006). Distribution of Frankia, ectomycorrhizal fungi, and bacteria in soil after the eruption of Miyake-Jima (Izu Islands, Japan) in 2000. J. Forest Res. 11, 21–26. doi: 10.1007/s10310-005-0179-2
Young, D. R., Sande, E., and Peters, G. A. (1992). Spatial relationships of Frankia and Myrica cerifera on a Virginia, USA Barrier Island. Symbiosis 12, 209–220.
Zgadzaj, R., Garrido-Oter, R., Jensen, D. B., Koprivova, A., Schulze-Lefert, P., and Radutoiu, S. (2016). Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 113, E7996–E8005 doi: 10.1073/pnas.1616564113
Keywords: actinorhizal symbiosis, microbiome, endophyte, epiphyte, symbiont, Coriaria myrtifolia, 16S rRNA next-generation-sequencing-metabarcoding, Frankia cluster 2
Citation: Swanson E, Sbissi I, Ktari A, Cherif-Silini H, Ghodhbane-Gtari F, Tisa LS and Gtari M (2022) Decrypting phytomicrobiome of the neurotoxic actinorhizal species, Coriaria myrtifolia, and dispersal boundary of Frankia cluster 2 in soil outward compatible host rhizosphere. Front. Microbiol. 13:1027317. doi: 10.3389/fmicb.2022.1027317
Edited by:
Zhen Wang, Yulin Normal University, ChinaReviewed by:
Debasis Mitra, National Rice Research Institute (ICAR), IndiaKatharina Pawlowski, Stockholm University, Sweden
Copyright © 2022 Swanson, Sbissi, Ktari, Cherif-Silini, Ghodhbane-Gtari, Tisa and Gtari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Louis S. Tisa, bG91aXMudGlzYUB1bmguZWR1; Maher Gtari, bWFoZXIuZ3RhcmlAaW5zYXQucm51LnRu
 Erik Swanson1
Erik Swanson1 Imed Sbissi
Imed Sbissi Hafsa Cherif-Silini
Hafsa Cherif-Silini Faten Ghodhbane-Gtari
Faten Ghodhbane-Gtari Louis S. Tisa
Louis S. Tisa Maher Gtari
Maher Gtari