- 1Jiangsu Key Laboratory of Zoonosis/Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, China
- 3Joint International Research Laboratory of Agriculture and Agri-product Safety of the Ministry of Education, Yangzhou University, Yangzhou, China
Vibrio parahaemolyticus is a foodborne pathogen that can colonize the small intestine of the host and cause diarrhea. The alternative sigma factor RpoN plays a vital role in regulating motility, carbon utilization and affects host colonization in V. parahaemolyticus RIMD2210633. In this study, transcriptome and phenotypic analysis further expanded our understanding of the RpoN regulon in V. parahaemolyticus. A deletion mutant of rpoN (ΔrpoN) was subjected to RNA-seq for systemic identification of the RpoN-controlled genes. Compared with the wild-type (WT), 399 genes were differentially expressed in the ΔrpoN strain. Moreover, 264 genes were down-regulated in the ΔrpoN strain, including those associated with nitrogen utilization (VP0118), glutamine synthetase (VP0121), formate dehydrogenase (VP1511 and VP1513-VP1515), quorum sensing (opaR and luxZ), polar flagellar systems, and type VI secretion system 2 (T6SS2). Quantitative real-time reverse transcription PCR (qRT-PCR) and electrophoretic mobility shift assay (EMSA) further confirmed that RpoN could directly bind to the promoters of these genes associated with polar flagellar systems (flgB and fliE), lateral flagellar systems (flgB2 and lafA), T6SS2 (hcp2 and VPA1044) and glutamine synthetase (VP0121), and then positively regulate the expression of these systems. A RpoN-binding motif was identified in V. parahaemolyticus using the MEME suite and verified by the EMSA. Besides, the deletion of rpoN caused a significant decrease in hemolytic activity, adhesion, and cytotoxicity. Our results provide new cues to better understand the regulatory networks of RpoN protein to motility, T6SS2, and metabolism in V. parahaemolyticus.
Introduction
Vibrio parahaemolyticus is a halophilic Gram-negative bacterium that can cause seafood-borne gastroenteritis due to the consumption of raw or uncooked seafood in Japan, the United States, Brazil, and China (Nair et al., 2007; Raszl et al., 2016; Chen et al., 2017). Gastrointestinal pathogens can overcome the host immune defense system and environmental stresses, which is closely linked to their virulence. The main virulence of V. parahaemolyticus are thermostable direct hemolysin (TDH), TDH-related hemolysin (TRH), motility, biofilm, type III secretion system (T3SS1 and T3SS2) and type VI secretion system (T6SS1 and T6SS2; Love, 2017; Li et al., 2019), which are tightly regulated by the sigma factors and transcriptional regulators (Liu et al., 2021a; Zhang et al., 2021a). Besides, the environmental and intestinal factors can act as cues to induce the expression of genetic components involved in bacterial survival and virulence (Merrell and Camilli, 2002; Cai et al., 2021; Yin et al., 2021). Sigma factors are the most commonly identified transcriptional regulators that could sensor the environmental signals and interact with specific double-stranded DNA promoters of the genes to regulate the expression of these genes responsible for overcoming environmental stresses and conferring virulence (Davis et al., 2017; Wang et al., 2021; Wu et al., 2021).
RpoN is an alternative sigma factor that belongs to the sigma 54 families. This protein contains three domains: a DNA-enhancer-binding domain, an effector ATPase domain, and a receiver domain that could be phosphorylated to respond to environmental signals (Danson et al., 2019; Soules et al., 2020). RpoN protein acts as an alternative sigma factor in transcribing genes with diverse physiological roles in different bacteria. RpoN can recognize the −24 (GG) and −12(GC) elements in promoter regions and regulate the expression of genes responsible for metabolism and virulence, which is conserved in E. coli, Vibrio cholerae, and Pseudomonas aeruginosa (Zhao et al., 2010; Dong and Mekalanos, 2012; Shao et al., 2018; Liu et al., 2021b). In V. cholerae, RpoN can positively regulate the expression of 144 genes in 82 operons, including motility, T6SS, nitrogen utilization, formate dehydrogenase synthesis and phage shock protein synthesis (Dong and Mekalanos, 2012). Notably, 37 RpoN-controlled operons contain the conserved −24 and −12 elements in V. cholerae (Dong and Mekalanos, 2012). In P. aeruginosa, RpoN positively regulated 133 genes involved in translation, motility, protein folding, secondary metabolite biosynthetic process, T6SS, and QS system (Shao et al., 2018). Moreover, RpoN can directly bind to promoters in QS (lasI, rhlI, and pqsR) and T6SS (hcpA and hcpB), thereby affecting the functions of QS and T6SS in P. aeruginosa (Shao et al., 2018; Lloyd et al., 2019). In V. parahaemolyticus, the phenotypical analysis showed that RpoN plays an essential role in motility, utilization of carbon source (mucus) and affects host colonization (Whitaker et al., 2014). However, the regulon of RpoN has not been fully defined in V. parahaemolyticus.
Loss of rpoN enhances the colonization of V. parahaemolyticus in part due to the decrease of motility in the ΔrpoN strain (Whitaker et al., 2014). Furthermore, bacterial motility plays an essential role in colonization, adhesion, and biofilm formation (Brescia et al., 2020; Khan et al., 2020; Buchner et al., 2021; Mea et al., 2021). V. parahaemolyticus contains dual flagellar systems to adapt to different circumstances: the polar flagellar system was used when growing in liquids and the lateral flagellar system was used when growing on surfaces or in dense environments (McCarter, 2004). The expression of flagellar genes is highly regulated by QS, c-di-GMP, and transcriptional regulators in V. parahaemolyticus (Khan et al., 2020). Loss of RpoN leads to reduced motility in V. parahaemolyticus and other pathogens, including V. cholerae, P. aeruginosa, Campylobacter jejuni, Yersinia pseudotuberculosis, and Shewanella baltica (Syed et al., 2009; Whitaker et al., 2014; Lloyd et al., 2017; Mahmud et al., 2020; Sher et al., 2020; Fang et al., 2021).
Type VI secretion system is widespread in many Gram-negative bacteria, which could directly secrete the toxins to the other bacterial or eukaryotic cells (Mougous et al., 2006; Pukatzki et al., 2006; Bao et al., 2020). The V. parahaemolyticus contains two type VI secretion systems, T6SS1 and T6SS2 (Izutsu et al., 2008). T6SS1 is encoded by a gene cluster predominantly found in clinical isolates, which is active in warm marine-like conditions and deliver the effectors into the neighbour bacterial cell to mediate the antibacterial activity (Yu et al., 2012; Ceccarelli et al., 2013; Salomon et al., 2013). T6SS2 is found in all V. parahaemolyticus isolates, which could be active at cold temperatures and low salt conditions and contribute to the adhesion of V. parahaemolyticus to the HeLa cells (Wettstadt, 2020). The TfoY and the other two regulators, VP1391 and VP1407 could positively regulate T6SS1, whereas the H-NS negatively regulate T6SS1 (Ben-Yaakov and Salomon, 2019). T6SS2 is negatively regulated by H-NS and positively regulated by CalR (Salomon et al., 2014; Zhang et al., 2017a). In addition, the QS regulators AphA and OpaR also could directly regulate the expression of T6SS1 and T6SS2 (Zhang et al., 2017b).
This study focused on revealing the pathways regulated by RpoN in V. parahaemolyticus. RNA-seq was used to identify the regulon of RpoN followed by qRT-PCR validation. EMSA was then used to confirm the direct binding between RpoN and the promoter regions of polar flagellar gene clusters, lateral flagellar gene clusters, T6SS2, and metabolism-associated genes. Our results also confirmed the RpoN-controlled promoters, including flgB, fliE, flgB2, lafK, hcp2, VPA1044, and VP0121. Furthermore, V. parahaemolyticus phenotypic analysis indicated that RpoN could regulate the hemolytic activity, adhesion, and cytotoxicity. Our findings provided new insights into the detailed regulatory networks of the RpoN protein to metabolic and virulence-associated pathways in V. parahaemolyticus.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The strains and plasmids used in the present study are listed in Supplementary Table S1. V. parahaemolyticus RIMD2210633 was used in the experiments. E. coli DH5α λpir and E. coli SM10 λpir were used for cloning and conjugation, respectively. All the V. parahaemolyticus and E. coli strains were cultured at 37°C in Luria-Bertani (LB) broth supplemented with 1% NaCl. The following antibiotics were added when required: carbenicillin (Carb, 100 μg/ml) and chloramphenicol (Cm, 40 μg/ml). In addition, Isopropyl β-D-1-thiogalactopyranoside (IPTG, 1 mmol/ml) was used to induce the expression of RpoN in rpoN+ and BL21/pET32a::rpoN strain.
Construction of the rpoN in-frame deletion mutant and complemented strains
The rpoN (VP2670) in-frame deletion mutant strain was constructed according to a previous method using the suicide vector pDM4 (Zhou et al., 2010), the primers used were listed in Supplementary Table S2. In brief, the upstream and downstream fragments of rpoN were amplified from the genome of V. parahaemolyticus with primers rpoN up-F/R and rpoN down-F/R and cloned into pDM4 with Sac I/Sal I sites by a ClonExpress Multis One Step Cloning Kit (Vazyme, Nanjing, China). Then, the positive recombinant plasmid pDM4::ΔrpoN was transformed into the WT strain by conjugation and cultured on an LB agar plate containing Carb and Cm. Following this, the second cross-over recombination was detected in the LB agar plate with 15% sucrose. Finally, the rpoN deletion mutant strain was verified by PCR with the primers rpoN out-F/R and rpoN in-F/R and sequencing analysis.
The ORF of rpoN was amplified with primers rpoN com-F/R and cloned into the pMMB207 plasmid with Xba I/Hind III sites by a ClonExpress Multis One Step Cloning Kit (Vazyme, Nanjing, China). Then, the positive recombinant plasmid pMMB207::rpoN was transformed into the ΔrpoN strain and selected on an LB agar plate containing Carb and Cm. The complemented strain was confirmed by PCR with primers pMMB207-F/R and named rpoN+.
RNA-seq analysis
The V. parahaemolyticus WT and ∆rpoN strains were cultured on LB agar plates for 15 h. One clone was picked and inoculated into LB broth growing at 37°C for 12 h. The cultured bacteria were diluted to 1:100 in new LB broth for 5–6 h at the late logarithmic growth phase (Supplementary Figure S4). Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). First strand cDNA was synthesized from the rRNA-depleted RNA samples and purified using an RNA Clean. After second strand cDNA synthesis, we performed end repair, 3′ end adenylation and adapter ligation, and the library was amplified by PCR. The three parallel RNA samples were sequenced using Illumina HiSeq (GENEWIZ, Suzhou, China). Statistical analysis was performed as described previously (Tang et al., 2021).
qRT-PCR
The qRT-PCR was performed as previously described (Gu et al., 2020). The WT, ΔrpoN, ΔrpoNΔopaR, ΔrpoNΔqrr2, and ΔrpoNopaR+ strains were cultured at 37°C in LB medium overnight and diluted 1:100 in new LB medium for 5–6 h or in the BHI agar for 48 h. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). Genomic DNA was removed using RNase-free DNase I. Equal amounts of RNA (1 μg) were used to generate the first-strand cDNA using the PrimeScript RT Reagent Kit with a gDNA eraser (Takara, Tokyo, Japan). The specific primers used for qRT-PCR are listed in Supplementary Table S2. The reactions were performed on the ABI PRISM 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) with a FastStart Universal SYBR Green Master (Roche, Mannheim, Germany). The transcript levels of each sample were normalized to those of gyrB using the 2−ΔΔCt method. Three independent experiments were performed, and each experiment was run in triplicate.
Motility assay
Overnight cultures of WT, ΔrpoN, rpoN+, ΔrpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2, and ΔrpoNopaR+ strains were adjusted to an optical density at 600 nm (OD600) of 1.0, following which 5 μl volumes of the diluted cultures were spotted on different plates. LB medium with 0.3% agar was used for swimming motility assays, and BHI medium with 1.5% agar was used for swarming motility assays. The LB plates were cultured at the 37°C for 12 h, and the BHI plates were cultured at 30°C for 24 h. IPTG was used to induce the expression of RpoN protein in rpoN+ and ΔrpoN/pMMB207 strains. Each experiment was performed three times.
Overexpression and purification of the RpoN protein
The ORF region of rpoN was amplified with primers RpoN-F/R and cloned into the pET32a plasmid with sites of XhoI and BamHI. The positive recombinant plasmid pET32a::rpoN was transformed into E. coli BL21 (DE3), and verified by PCR with primers pET32a-F/R. The specific primers are listed in Supplementary Table S2. BL21/pET32a::rpoN was cultured in LB broth, and IPTG was added to induce the expression of RpoN until the OD600 value was between 0.4 and 0.6. Then, the bacteria were cultured at 120 rpm and 16°C for 16 h. The cell cultures were collected, washed, lysed, and subjected to the purification of the RpoN-His protein using the His Bind Purification Kit (Novagen, Darmstadt, Germany).
EMSA
EMSA was performed as described previously (Gu et al., 2016). The primers used for EMSA are listed in Supplementary Table S2. The DNA probes were amplified by the primers with the FAM fluorescent label and purified using the Agarose Gel DNA Extraction Kit (Tiangen, Beijing, China). Each EMSA reaction mixture (20 μl) consisted of 10 ng of DNA probes, 4 μl of EMSA buffer (10 mM NaCl, 0.1 mM DTT, 0.1 mM EDTA, 10 mM Tris, pH 7.4), 1 μl of poly-dIdC, different concentrations of the RpoN-His protein, and ddH2O. The mixture was incubated at 25°C for 0.5 h and separated on a 6% native PAGE gel. Finally, the gel was scanned using Typhoon FLA 9500 (GE Healthcare, Uppsala, Sweden).
Adhesion assay
The adhesion assay was performed as described previously (Qiu et al., 2020). The cultures of WT, ΔrpoN, and rpoN+ strains were pelleted by centrifugation, washed, and resuspended in dulbecco phosphate-buffered saline (DPBS). HeLa cell monolayers were infected at a multiplicity of infection (MOI) of 1:100. After incubation at 37°C under 5% CO2 for 2 h, the HeLa cells were washed twice with DPBS and lysed with 0.01% Triton-X 100. The lysates and bacteria were serially diluted and counted on LB agar plates. The adherence rate was calculated as the number of bacterial cells adhered/the number of bacterial cells input.
Hemolytic activity assay
Hemolytic activity was assessed as described previously (Rattanama et al., 2012; Kongrueng et al., 2018). Overnight cultures of WT, ΔrpoN, rpoN+, ΔrpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2, and ΔrpoNopaR+ strains were centrifuged, washed thrice with PBS, and resuspended in DPBS at a final concentration of 0.5 × 109 CFU/ml. Following this, 5 μl of the bacterial suspension was added to the wells in the blood agar plates and cultured at 37°C for 12–24 h. The diameter of clear zone around the well was indicated as the hemolytic activity. The experiments were repeated at least three times.
Cytotoxicity assay
The infection of HeLa cells and the release of lactate dehydrogenase (LDH) were assessed as described previously (Erwin et al., 2012). Overnight cultures of WT, ΔrpoN, rpoN+, ΔrpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2, and ΔrpoNopaR+ strains were diluted 1:100 in fresh LB medium and shaken at 37°C for 4 h. Following this, the cultures were centrifuged, and pellets were resuspended in dulbecco’s modified eagle medium (DMEM) containing 10% fatal bovine serum (FBS). The bacteria were diluted to a concentration of 108 CFU, and the bacterial suspensions were inoculated into each well of a 12-well plate containing 106 HeLa cells/well to achieve an MOI of 100 CFU/cell. After infection for 3 h, the LDH activity of the supernatants was measured using the LDH Cytotoxicity Assay Kit (Beyotime, Haimen, China), according to the manufacturer’s protocol.
Results
Identification the regulon of RpoN in V. parahaemolyticus RIMD2210633
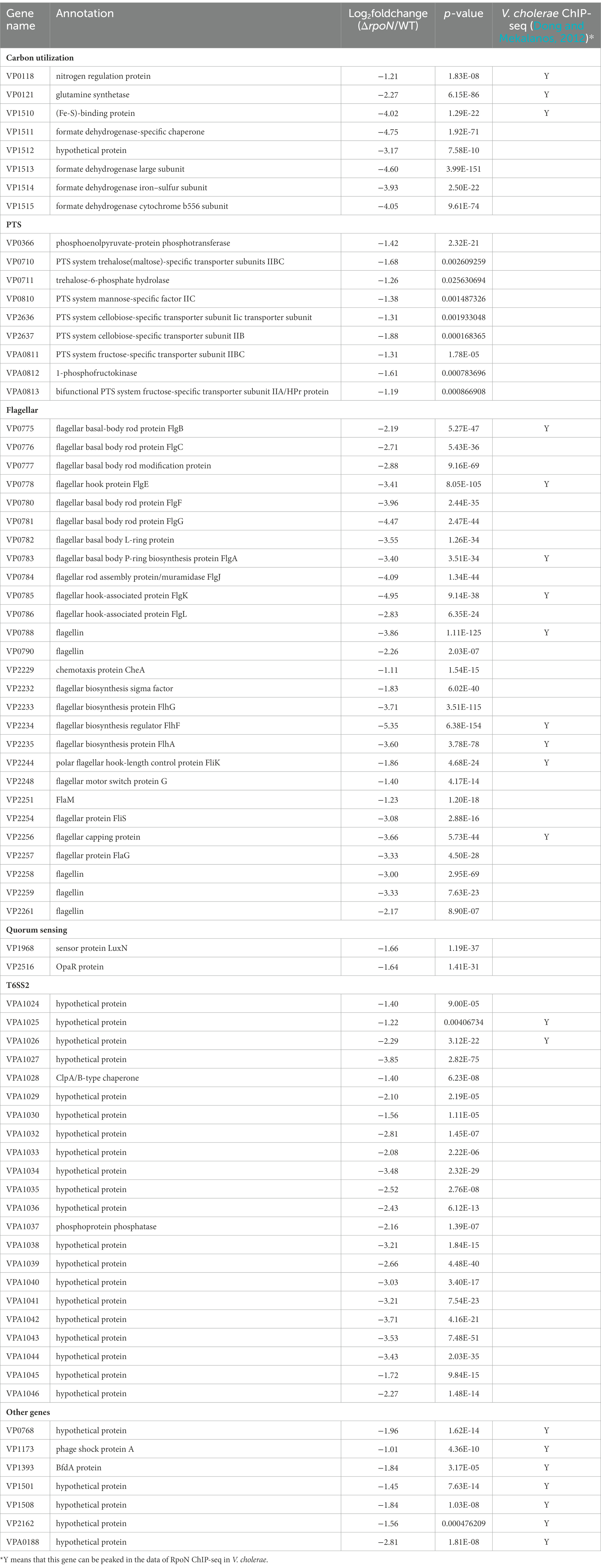
We assessed the regulon of RpoN in V. parahaemolyticus by the RNA-seq analysis. Comparison of the transcriptomes for the WT and ΔrpoN strains grown in LB medium revealed that the expression of 399 genes significantly differed between the ΔrpoN and WT strains (Log2Fold Change ≥1 or ≤−1, p-value < 0.05). As shown in Figure 1A, 135 and 264 genes were up-regulated and down-regulated in the ΔrpoN strain, respectively. As expected, rpoN was not detected in ΔrpoN and highly expressed in WT, suggesting that the RNA-seq data is reliable (Figure 1B). Figure 1B described the expression patterns of genes that are potentially associated with metabolic and virulence, including metabolism, phosphotransferase system (PTS), polar flagellar system, quorum sensing, T3SS1 and T6SS2.
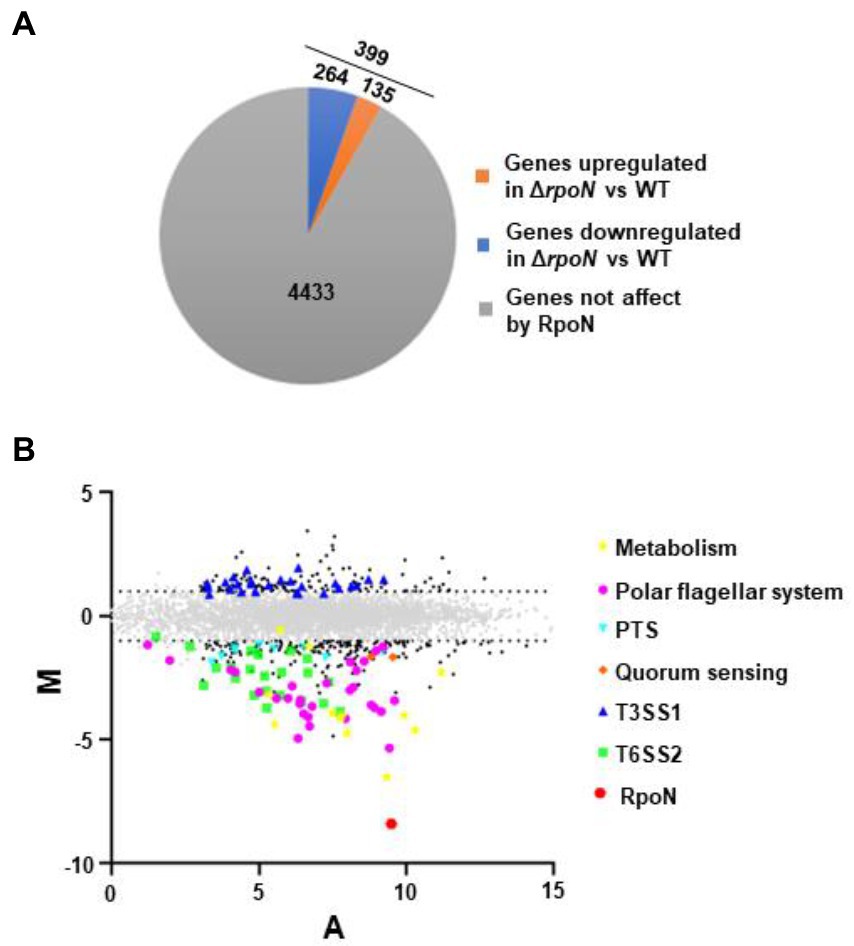
Figure 1. The regulon of RpoN in V. parahaemolyticus. (A) Pie charts show the genes whose expression significantly differed between ΔrpoN and WT strains in LB liquid medium. (B) MA plots showing changes that were observed in gene expression at ΔrpoN compared to WT. The genes associated with metabolism and virulence are highlighted. The log2 values for the ratios of abundances for each transcript are shown between ΔrpoN and WT (M, y axis) and plotted against the average log2 for abundance for that transcript under both conditions (A, x axis). Solid black dots, Padj < 0.05.
Compared with the WT strain, the QS master regulator protein OpaR and the sensor protein LuxN were significantly down-regulated in the ΔrpoN strain. Furthermore, many carbon source-associated genes were significantly regulated by RpoN, such as the nitrogen regulatory protein (VP0118), glutamine synthetase (VP0121), formate dehydrogenase (VP1511 and VP1513-VP1515), and PTS system (Table 1). In addition, virulence-associated genes were also regulated by RpoN. Compared with the WT, the genes related to T6SS2, flagellar systems, and biofilm formation were down-regulated, while those related to the type III secretion system 1 (T3SS1) and siderophore-dependent iron uptake systems were up-regulated in the ΔrpoN strain. The above results indicate that RpoN is an important sigma factor regulating metabolic and virulence-associated pathways in V. parahaemolyticus.
RpoN could regulate the expression of polar flagellar genes to mediate swimming motility in V. parahaemolyticus. RNA-seq results revealed that the polar flagellar genes were down-regulated in the ΔrpoN strain compared with the WT. Assessment of the polar flagellar gene clusters in V. parahaemolyticus revealed that 43 genes were divided into two clusters: polar flagellar gene cluster I (14 genes) and polar flagellar gene cluster II (29 genes; Figure 2A). The transcript levels of the two polar flagellar gene clusters identified by RNA-seq are shown in Table 1. All of the genes located in the polar flagellar cluster I were significantly down-regulated in the ΔrpoN strain. Some of the genes located in polar flagellar cluster II were also significantly down-regulated in the ΔrpoN strain, suggesting that the RpoN protein could positively regulate the expression of polar flagellar genes.
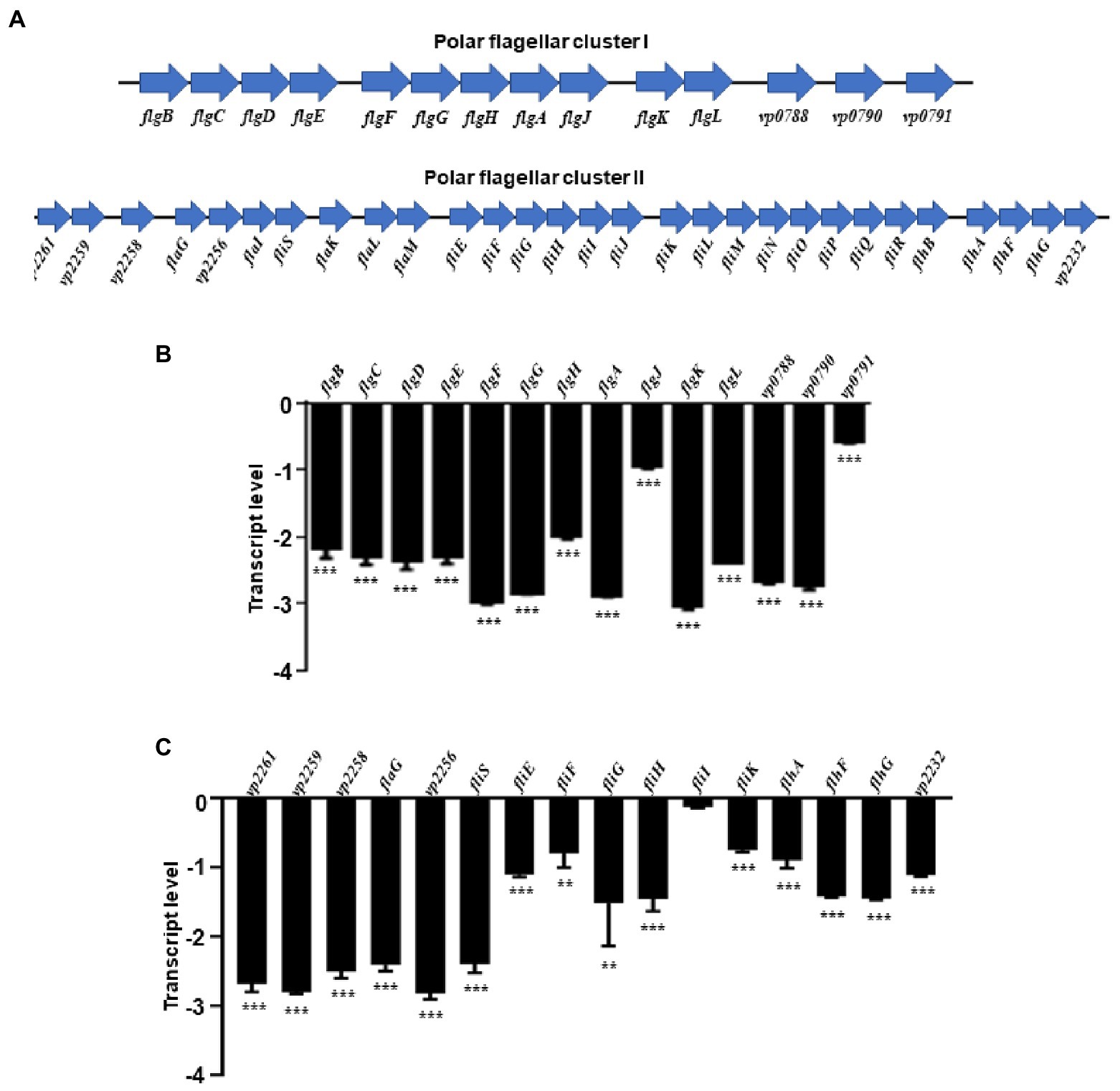
Figure 2. The RpoN positively regulates the expression of flagellar clusters. (A) Genetic map of polar flagellar cluster I and cluster II genes in V. parahaemolyticus. (B,C) qRT-PCR analysis of the transcription levels of polar flagellar cluster I genes (B) and polar flagellar cluster II genes (C). The data are presented as the mean ± SD (n = 3). **p < 0.01, ***p < 0.001, Student’s t test analyzes ΔrpoN compared to WT.
All of the polar flagellar cluster I genes and some of the cluster II genes were subjected to qRT-PCR analysis to verify the results of RNA-seq. All polar flagellar cluster I genes in the ΔrpoN strain were significantly down-regulated compared with the WT strain (Figure 2B). In particular, VP2261, VP2259, VP2258, flaG, VP2256, fliS, fliEFGHI, fliK, flhAFG, and VP2232 located in polar flagellar cluster II were significantly down-regulated by RpoN (Figure 2C). Furthermore, motility analysis revealed that the swimming ability was lost in the ΔrpoN strain, while the swimming ability in rpoN complementary (rpoN+) strain was restored to the similar level observed in the WT (Supplementary Figure S1A), which is consistent with the previous study (Whitaker et al., 2014). Collectively, these results indicate that RpoN is a sigma factor that could regulate the expression of polar flagellar genes to mediate swimming motility in V. parahaemolyticus.
RpoN directly binds to the promoters of the polar flagellar genes flgB and fliE
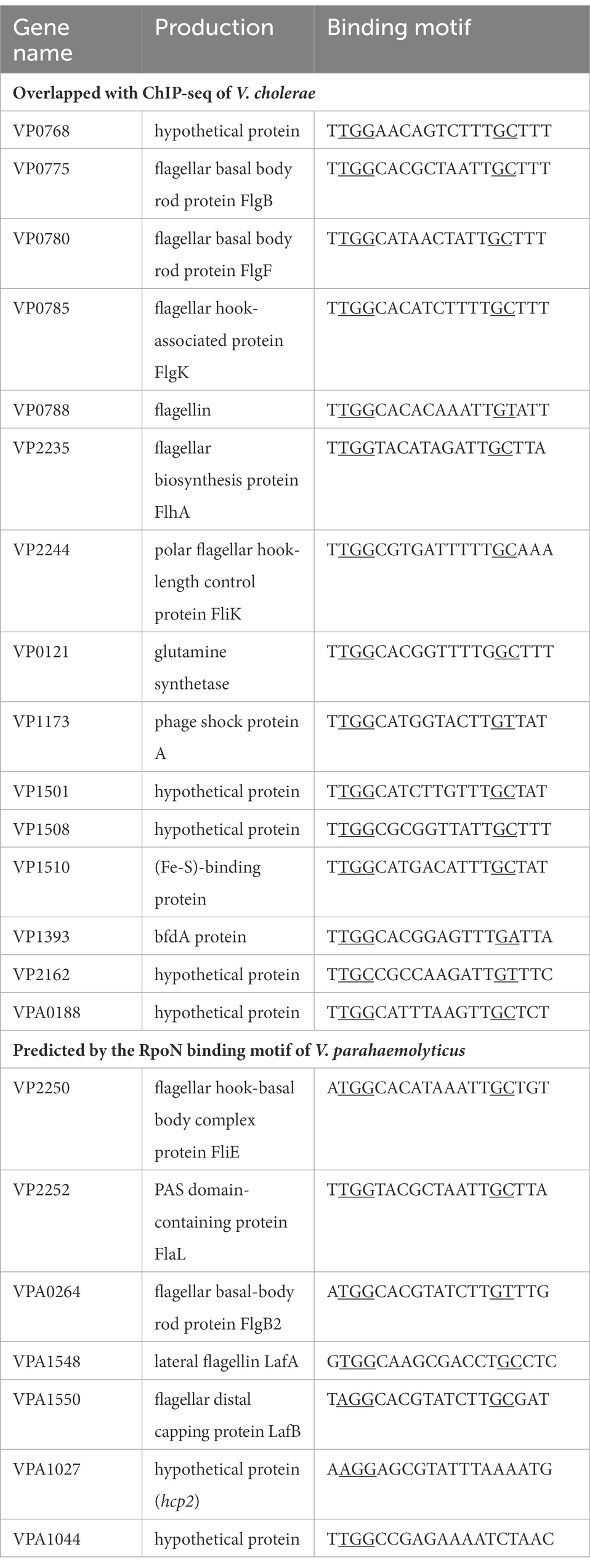
In V. cholerae, 68 RpoN-binding peaks were identified by ChIP-seq (Dong and Mekalanos, 2012). Therefore, we used the 68 RpoN-binidng peaks blast against the genome of V. parahaemolyticus and 21 genes were found significantly down-regulated in ΔrpoN by RNA-seq (Table 1). Then, the promoter regions of these 21 genes were used to generate the RpoN-binding motif by MEME-Suit tool.1 As shown in Figure 3A, the −24 box (GG) and −12 box (GC) were identified as two conserved motifs, and 15 of 21 genes contained the conserved RpoN binding sites (Table 2). The fliE (VP2250) and flaL (VP2252) genes were no difference expressed between ΔrpoN and WT in RNA-seq, whereas peaked in ChIP-seq of V. cholerae and contained the conserved RpoN-binding sites in V. parahaemolyticus (Table 2). The promoters of flgB (VP0775), flgF (VP0780), flgK (VP0785), flhA (VP2235), flaL (VP2252), and fliE (VP2250) involved in the polar flagellar gene clusters were selected to confirm by EMSA. The results showed that RpoN could directly bind to the promoters of flgB (Figure 3B) and fliE (Figure 3C) in a concentration-dependent manner; however, it could not directly bind to the other promoters (Supplementary Figures S2A–D). RpoN did not bind to the promoter of gyrB at the highest concentration (Supplementary Figure S2E), which was used as a negative control.
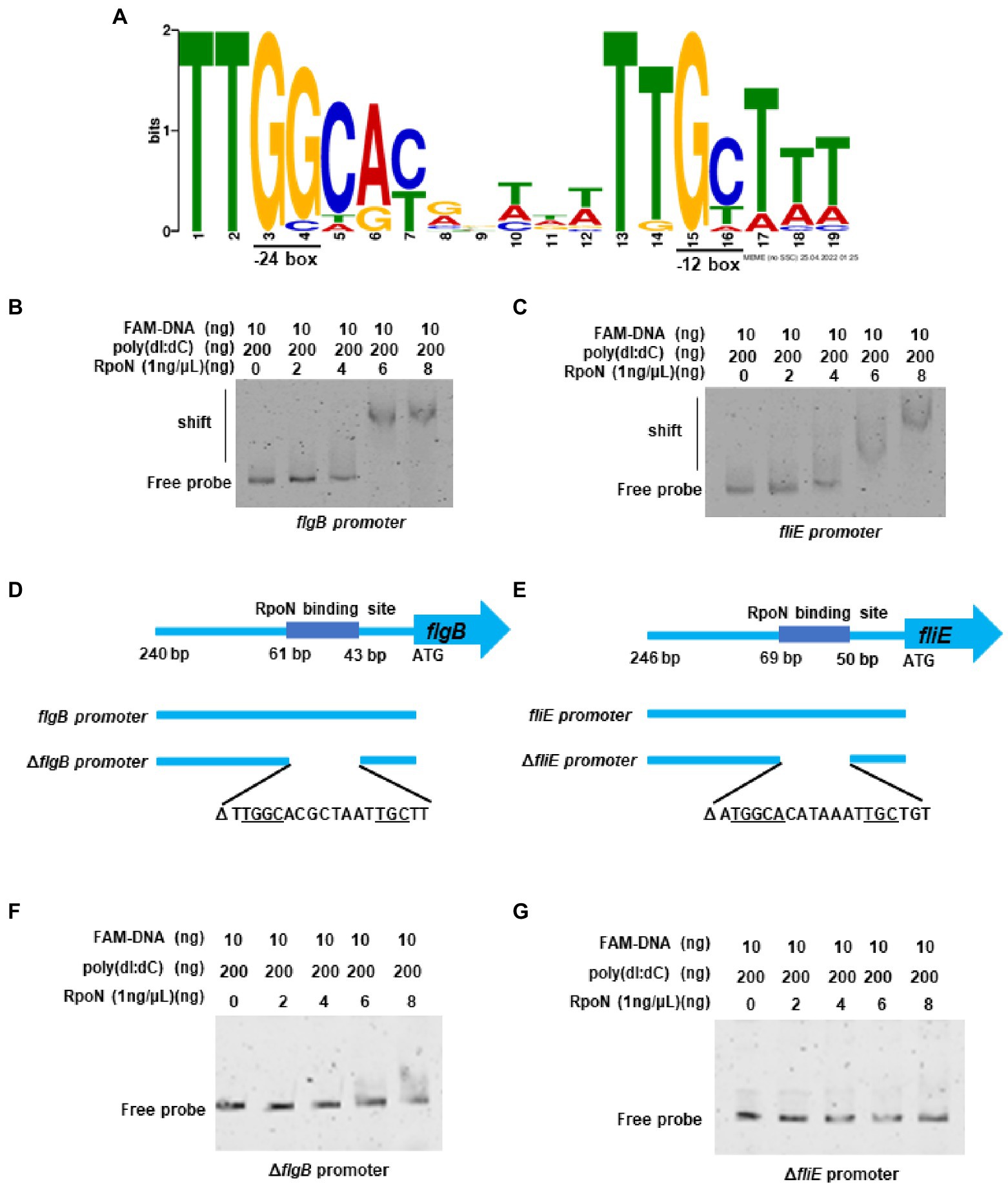
Figure 3. The RpoN directly binds to the promoters of flgB and fliE. (A) The binding motif of RpoN was generated by MEME-Suit tool based on the 21 promoter regions of the gene differentially expressed in our RNA-seq and overlapped with the ChIP-seq of V. cholerae. The height of each base indicates the frequency of occurrence at each location. (B,C) EMSA revealed that RpoN directly binds to the promoter regions of flgB (b) and fliE (C). (D) The promoter region of the flgB. The RpoN binding sites and the probes of flgB promoter (240 bp) and the ∆flgB promoter with deletion of RpoN binding site ranging from bp 43–61 relative to start site ATG. (E) The promoter region of the fliE. The RpoN binding sites and the probes of fliE promoter (246 bp) and the ∆fliE promoter with deletion of RpoN binding site ranging from bp 50–69 relative to start site ATG. (F,G) EMSA was performed to assess the binding of RpoN to specific RpoN binding site-deleted promoters ∆flgB (F) and ∆fliE (G).
The RpoN-binding site in the promoter of flgB and fliE is shown in Figures 3D,E. Next, a mutant DNA probe was constructed by deleting the conserved binding site, and the EMSA results revealed that RpoN could not directly bind to the mutant DNA probe of the ΔflgB promoter (Figure 3F) and ΔfliE promoter (Figure 3G). Our results indicate that the RpoN protein can bind to the conserved −24 box and − 12 box in the promoters of flgB and fliE to regulate the expression of the polar flagellar gene clusters.
RpoN directly binds to the promoters of flgB2 and lafA to regulate the swarming motility of V. parahaemolyticus
Previous studies have shown that the V. parahaemolyticus contains two flagellar systems (McCarter, 2004). However, RNA-seq of the bacteria cultured in LB medium could not identify the transcription level of genes responsible for the lateral flagellar system, which mediates bacterial swarming motility (McCarter, 2004). The swarming ability of the ΔrpoN strain was significantly lower than that of the WT and rpoN+ strain (Supplementary Figure S1B), which is consistent with the previously study (Whitaker et al., 2014). Our previous study also shown that the lateral flagellar systems contained two clusters, lateral flagellar cluster I and lateral flagellar cluster II (Gu et al., 2019). Then the qRT-CPR was used to verify the regulation of RpoN to lateral flagellar gene clusters, and the results showed that the expression of flgM, flgB2C2D2, lafK, motY, fliMN, lafA, and lafBCD genes were significantly down-regulated in the ΔrpoN strain compared to WT (Figures 4A,B). In addition, the RpoN-binding motif of V. parahaemolyticus were used to blast against the lateral flagellar gene clusters, and found that promoters of flgB2, lafA, and lafB contain the potential RpoN binding sites. Then, the above three promoters and the flgM promoter were selected to confirm by EMSA. The results showed that the RpoN protein could directly bound to the promoters of flgB2 (Figure 4C) and lafA (Figure 4D), whereas it could not directly bound to the promoters of lafB (Figure 4E) and flgM (Figure 4F). These results indicated that the RpoN protein could directly bind to the promoters of flgB2 and lafA to regulate the expression of lateral flagellar gene cluster and mediate the swarming motility of V. parahaemolyticus.
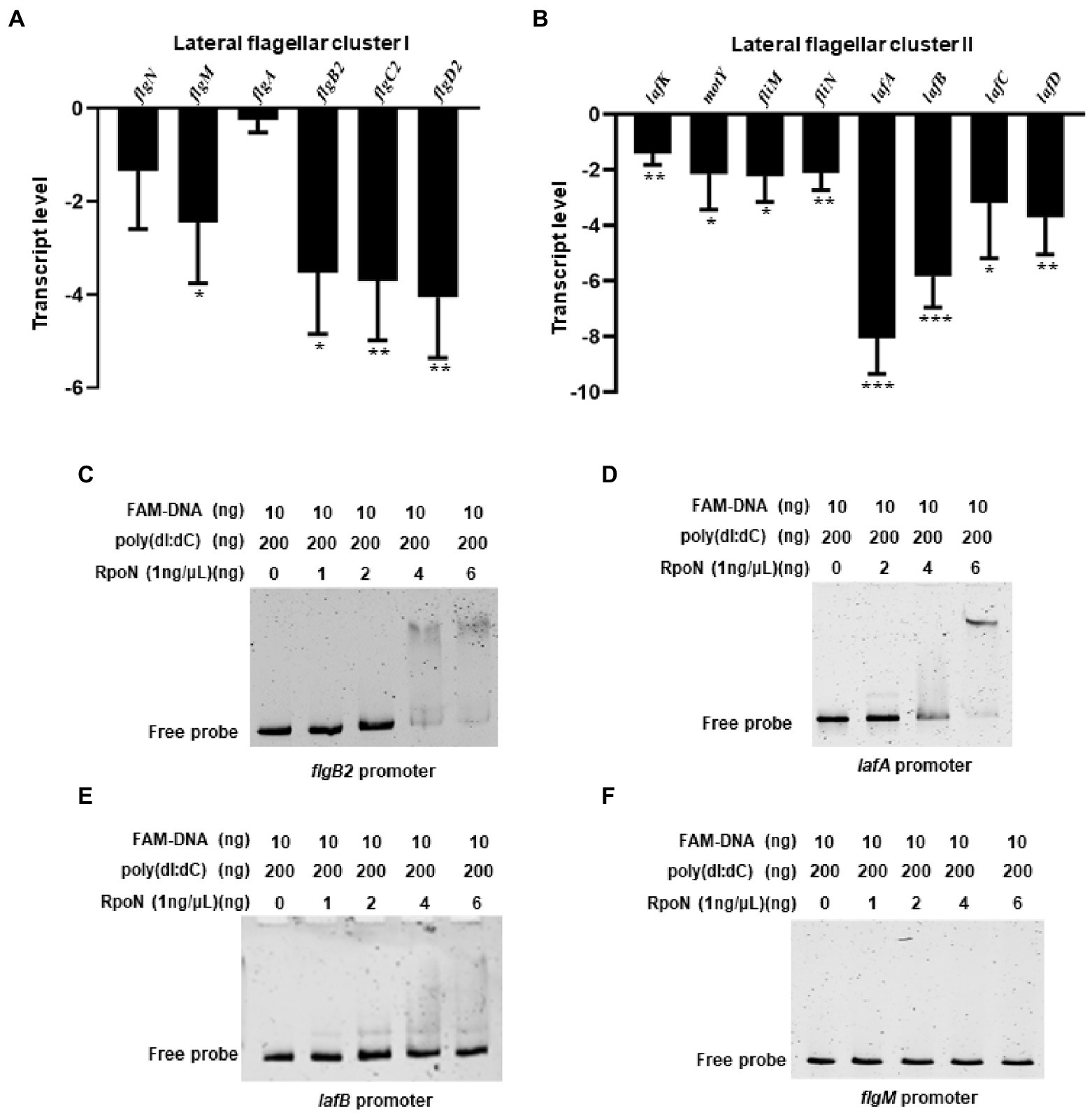
Figure 4. RpoN directly binds to the promoters of flgB2 and lafA to regulate the expression of lateral flagellar genes in V. parahaemolyticus. (A,B) qRT-PCR analysis of the transcription levels of lateral flagellar cluster I (A) and lateral flagellar cluster II (B) genes in ΔrpoN compared to WT. The data are presented as the mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test analyzes ΔrpoN compared to WT. (C–F) EMSA analysis the specifically binds of RpoN to the promoters of flgB2 (C), lafA (D), lafB (E), and flgM (F).
RpoN directly regulates the expression of T6SS2 to mediate the adhesion of V. parahaemolyticus
RNA-seq results also showed that the T6SS2 gene cluster was significantly down-regulated in the ΔrpoN strain compared to WT (Table 1). qRT-PCR results confirmed that RpoN could regulate the expression of T6SS2 genes (Figure 5A). EMSA results found that the RpoN protein could directly bind to the promoters of hcp2 and VPA1044 to regulate the expression of T6SS2 (Figures 5B,C). The previous study has revealed that the T6SS2 predominately contributes to the adhesion of V. parahaemolyticus to host cells (Yu et al., 2012). Therefore, the adhesion rates of WT and ΔrpoN strains to HeLa cells were also determined in this study. The results showed that the adhesion rate of the ΔrpoN strain was significantly lower than that of the WT strain, while the adhesion rate of the rpoN+ strain was restored to the same level as WT (Figure 5D). These results indicated that the RpoN protein could directly regulate the expression of T6SS2 to mediate the adhesion of V. parahaemolyticus to HeLa cells.
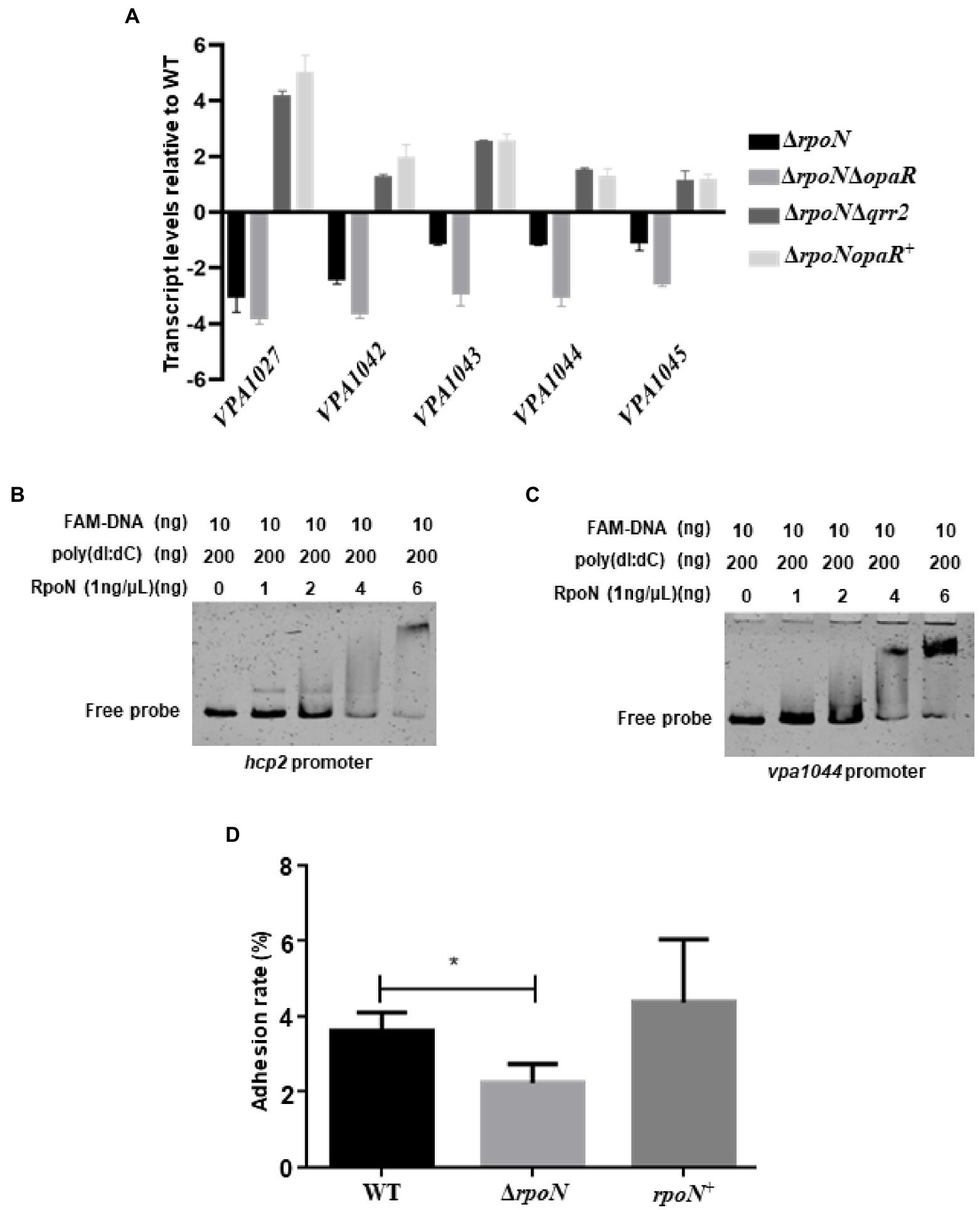
Figure 5. The RpoN directly regulates the expression of T6SS2 to mediate the adhesion of V. parahaemolyticus to HeLa cells. (A) qRT-PCR analysis of the expression levels of genes located in T6SS2 in WT, ΔrpoN, ΔrpoNΔopaR, ΔrpoNΔqrr2 and ΔrpoNopaR+ strains. The data are presented as the mean ± SD (n = 3). Student’s t test analyzes the different mutant strains compared to WT, and all the p-value lower than 0.001. (B,C) EMSA analysis of the specific binding of RpoN protein to the promoters of hcp2 (B) and VPA1044 (C). (D) Adhesion analysis of WT, ΔrpoN, and rpoN+ strains to HeLa cell monolayers. The data are presented as the mean ± SD (n = 3), *p < 0.05, Student’s t test analyzes ΔrpoN compared to WT.
RpoN regulates the expression of metabolic genes in V. parahaemolyticus
RNA-seq analysis showed that the RpoN protein could regulate the expression of nitrogen regulation protein (VP0118), glutamine synthetase (VP0121), and the formate dehydrogenase gene cluster (VP1510-VP1515; Table 1). Then, the qRT-PCR also confirmed that the expression of these genes was down-regulated in the ΔrpoN strain compared to WT (Figures 6A,B). Furthermore, the VP0121 and VP1510 promoters contained the predicted RpoN binding sites (Table 2), whereas the VP0118 promoter did not contain the RpoN binding sites. The EMSA results showed that the RpoN protein could not bind to the promoters of VP1510 (Figure 6C) and VP0118 (Figure 6D), but it could directly bind to the promoter of VP0121 (Figure 6E). These results indicated that the RpoN directly bound to the promoter of VP0121 to regulate the expression of the genes associated with glutamine synthetase, and indirectly regulate the expression of genes associated with nitrogen regulation and the formate dehydrogenase in V. parahaemolyticus.
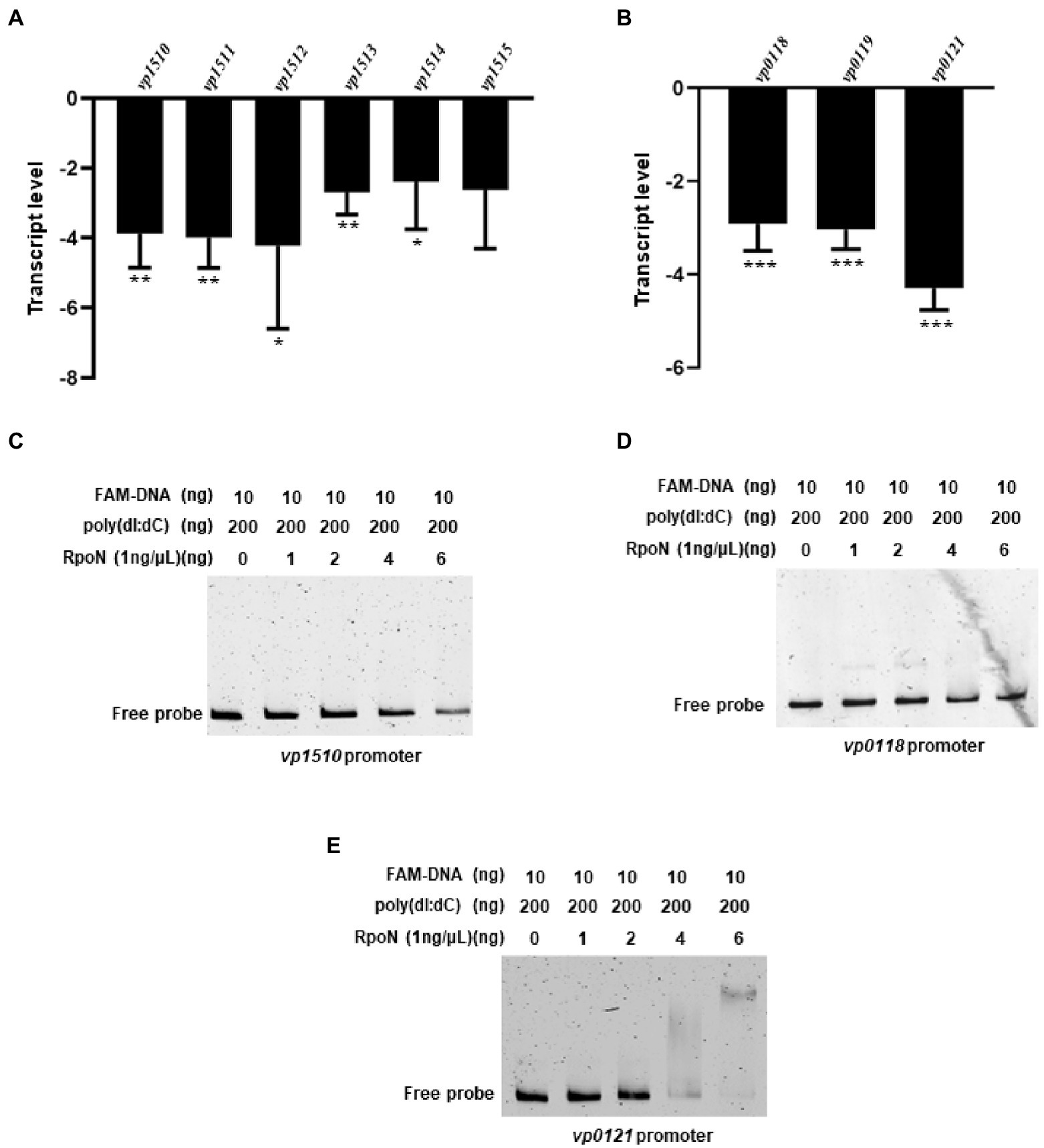
Figure 6. The RpoN regulates the expression of metabolism-associated genes. (A) qRT-PCR analysis of the expression levels of formate dehydrogenase genes (VP1510-VP1515) in ΔrpoN compared to WT. (B) qRT-PCR analysis of the expression levels of nitrogen regulatory protein (VP0118-VP0119) and glutamine synthetase (VP0121) associated genes in ΔrpoN compared to WT. The data are presented as the mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t test analyzes ΔrpoN compared to WT. (C–E) EMSA analysis of the specific binding of RpoN protein to the promoters of VP1510 (C) and VP0118 (D) and VP0121 (E).
RpoN played essential roles in hemolytic activity and cytotoxicity.
In addition, our results also showed that the hemolytic activity of the ΔrpoN strain was significantly lower than that of the WT and the complemented strain (Figure 7A). To further assess the function of RpoN in the host cell, the cytotoxicity of the WT and ΔrpoN mutant strains toward HeLa cells was evaluated. The cytotoxicity of the ΔrpoN strain was significantly lower than that of the WT and the rpoN+ strain (Figure 7B). These results suggest that RpoN plays an essential role in regulating hemolytic activity, and cytotoxicity toward HeLa cells.
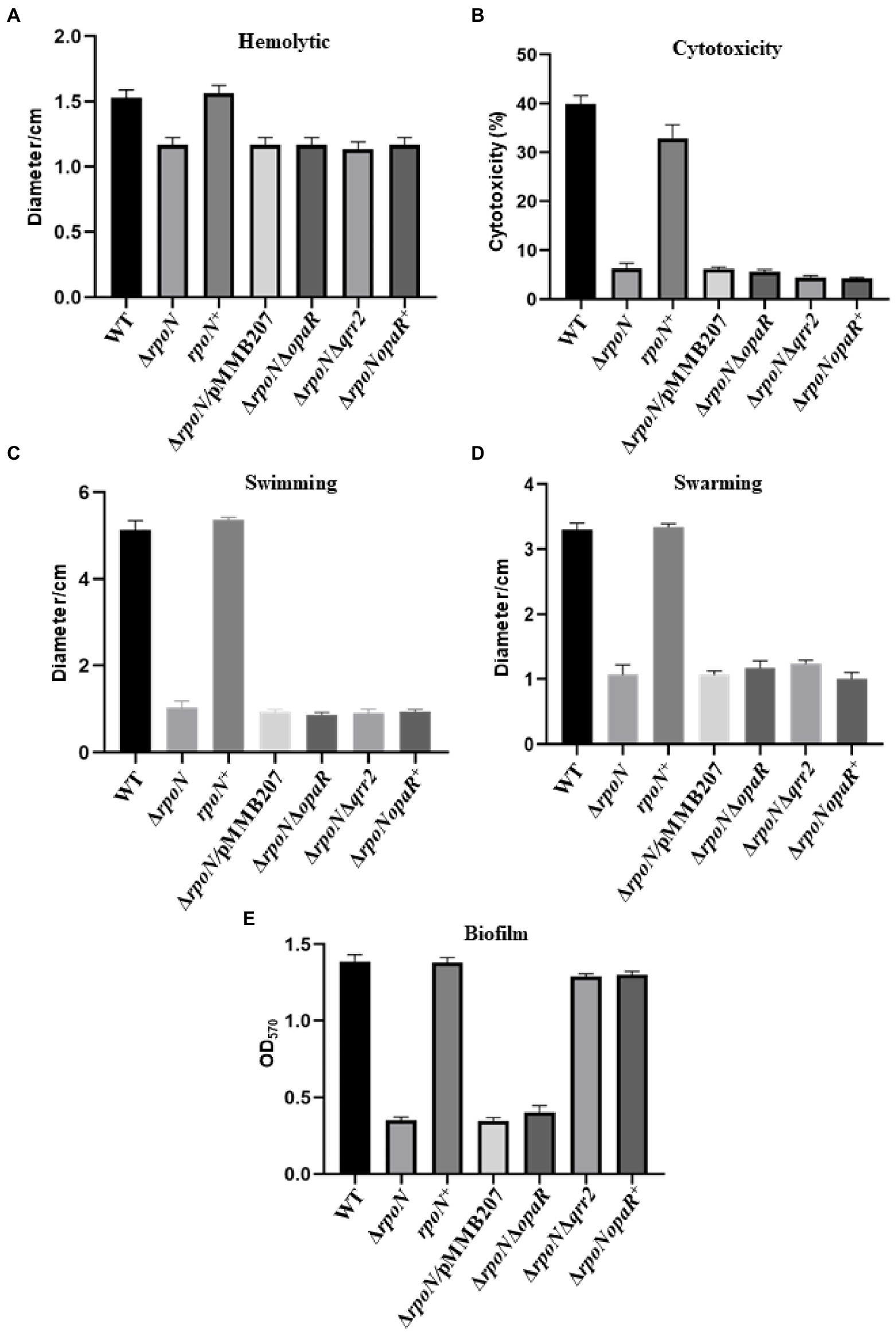
Figure 7. The RpoN regulates the phenotypes in an OpaR-dependent or OpaR-independent way. (A) Hemolytic activity assay of WT, ∆rpoN, rpoN+, ∆rpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2 and ΔrpoNopaR+ strains grown on sheep blood agar plates at 37°C. (B) Assessment of cytotoxicity of WT, ∆rpoN, rpoN+, ∆rpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2 and ΔrpoNopaR+ strains towards HeLa cell monolayers. (C,D) Analyze of swimming (C) and swarming (D) of WT, ∆rpoN, rpoN+, ∆rpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2 and ΔrpoNopaR+ strains in LB medium or BHI medium, respectively. (E) Biofilm formation of WT, ∆rpoN, rpoN+, ∆rpoN/pMMB207, ΔrpoNΔopaR, ΔrpoNΔqrr2 and ΔrpoNopaR+ strains. The data are presented as the mean ± SD (n = 3), Student’s t test analyzes different mutant strains compared to WT, and all the p-value lower than 0.001.
Role of OpaR in the regulation of RpoN to virulence-associated phenotypes
Our RNA-seq results showed that the RpoN protein could regulate the expression of opaR and was confirmed by qRT-PCR (Supplementary Figure S4A), and EMSA results showed that RpoN protein could not directly bound to the promoter of opaR (Supplementary Figure S4B). A previous study showed that the regulation of RpoN to opaR was dependent on the qrr2 (Tague et al., 2022). Thus, we constructed the ΔrpoNΔopaR, ΔrpoNΔqrr2, and ΔrpoNopaR+ strains to investigate whether the regulation of RpoN to hemolytic, cytotoxicity, and motility was through OpaR. The expression levels of opaR in these strains were shown in Supplementary Figure S4A. The hemolytic activity of ΔrpoNΔopaR, ΔrpoNΔqrr2, and ΔrpoNopaR+ strains were significantly lower than that in WT, and similar to the ΔrpoN (Figure 7A), indicating that regulation of RpoN to hemolytic was not associated with OpaR. The similar results were found in cytotoxicity and motility (Figures 7B–D). The above results indicated that RpoN regulates hemolytic activity, cytotoxicity, and motility of Vibrio parahaemolyticus without a relationship to OpaR.
Next, we also investigate whether the regulation of RpoN to T6SS2 and biofilm was through OpaR. As shown in the Figure 5A, the expression of T6SS2 genes (VPA1027, VPA1042, VPA1043, VPA1044, and VPA1045) were significantly increased in the ΔrpoNΔqrr2 and ΔrpoNopaR+ strains compare to WT, indicating that the regulation of RpoN to T6SS2 was related to OpaR. Furthermore, the biofilm formation was decreased in the ΔrpoN and ΔrpoNΔopaR strains, while it was restored to the level of WT in ΔrpoNΔqrr2 and ΔrpoNopaR+ strains (Figure 7E). These results indicated that the regulation of RpoN to T6SS2 and biofilm formation of Vibrio parahaemolyticus with a close relationship to OpaR.
Discussion
In V. parahaemolyticus, RpoN has been reported to regulate carbon utilization and affect host colonization (Whitaker et al., 2014). In other bacteria, RpoN protein also has been reported to regulate the expression of virulence-associated genes, such as those associated with flagellar systems, biofilm formation, QS, and T6SS (Zhao et al., 2010; Dong and Mekalanos, 2012; Shao et al., 2018; Liu et al., 2021b). The present study used RNA-seq to investigate the genes regulated by the alternative sigma factor RpoN in V. parahaemolyticus. RNA-seq results revealed that the expression of 399 genes significantly differed between the ΔrpoN and WT strains. Moreover, 264 genes were positively regulated by RpoN, including those associated with carbon utilization, QS, flagellar systems, and T6SS (Figure 1). Similarly, 70 genes were positively regulated by RpoN in Escherichia coli K-12, including genes associated with bacterial motility and nitrogen metabolism (Zhao et al., 2010). In V. cholerae, 144 genes have been reported to be positively regulated by RpoN, including those associated with motility, T6SS, nitrogen utilization, and phage shock protein synthesis (Dong and Mekalanos, 2012). In P. aeruginosa, the genes taking up approximately 20% of the genome is regulated by RpoN (Damron et al., 2012). The current observations suggest that the sigma factor RpoN plays an essential role in various biological processes in different bacteria and diverse hosts. Similar studies have been conducted in the V. cholerae and E. coli (Zhao et al., 2010; Dong and Mekalanos, 2012). In addition to defining the RpoN regulon in V. parahaemolyticus, we also identified two new RpoN-binding promoters (flgB2 and lafA) and, for the first time, showed that RpoN plays an essential role in hemolytic activity, adhesion, and cytotoxicity. These results provide additional insight into the potential role of RpoN in pathogenesis.
V. parahaemolyticus contains two distinct flagellar systems for expression under different circumstances and facilitates the infection process (Merrell et al., 1984; McCarter, 2004). Compared with the WT strain, swimming motility and swarming motility were defective in the ΔrpoN strain in the previous study (Whitaker et al., 2014). Furthermore, our qRT-PCR data confirmed that RpoN could regulate the expression of the polar flagellar clusters and lateral flagellar clusters to mediate motility in V. parahaemolyticus (Figures 2, 4). The previous study has demonstrated that RpoN directly regulates the expression of flagellar systems in V. cholerae (Dong and Mekalanos, 2012). Based on the conserved binding motif of RpoN in V. cholerae, we searched for the potential RpoN-binding sites in the promoters of flagellar gene clusters in V. parahaemolyticus. Our results revealed that the promoters of flgB, flgF, flgK, flhA, fliE, and flaL in the polar gene clusters contained RpoN-binding sites, and the promoters of lateral flagellar genes flgB2, lafA and lafB contained the conserved RpoN-binding sites (Table 2). Nevertheless, the EMSA results revealed that the RpoN protein directly bind to the promoters of flgB, fliE, flgB2, and lafA (Figures 3B,C, 4C,D). Notably, a previous study reported that flaK is a σ54-dependent regulator of polar flagellar Class II genes in V. parahaemolyticus (McCarter, 2004). In addition, flgB, flgF, flgA, flgK, fliD, fliE, fliK, flhA, motY, and flgT have been reported to be RpoN-binding promoters in V. cholerae (Dong and Mekalanos, 2012). Therefore, we confirmed that RpoN could bind to the promoters of flgB and fliE in polar flagellar gene clusters by EMSA, and identified two new RpoN-binding promoters of flgB2 and lafA in lateral flagellar gene clusters.
Two T6SS loci (T6SS1 and T6SS2) have been identified in V. parahaemolyticus. T6SSs are tightly regulated by transcription regulators and sigma factors (Wettstadt, 2020; Yin et al., 2020; Shao et al., 2021). H-NS, QS, ToxR, TfoY, QsvR, and CalR have been identified as regulators to control the expression or secretion of T6SS in V. parahaemolyticus (Zhang et al., 2017a,b; Qiu et al., 2020). RpoN was also detected to directly control the expression of hcp but not that of the major cluster of T6SS in V. cholerae (Dong and Mekalanos, 2012). However, our RNA-seq data and qRT-PCR analysis revealed that the T6SS2 genes were down-regulated in the ΔrpoN strain compared to the WT strain (Table 1; Figure 5A). EMSA results also indicated that the RpoN protein could directly bind to the promoters of hcp2 and VPA1044 (Figures 5B,C). However, RpoN-binding sites in the promoters of hcp2 and VPA1044 were incomplete, containing only the conserved −24 box (GG) but without the conserved −12 box (Table 2). Furthermore, it was reported that RpoN could directly bind to the promoters of hcpA and hcpB to positively control T6SS expression in P. aeruginosa, which also only contain the conserved −24 box (GC; Shao et al., 2018). The −24 element is an attachment determinant for RpoN, whereas the −12 element is variable (Wang and Gralla, 1988; Barrios et al., 1999), which may explain the RpoN bound to the less conserved binding sites in the promoters of hcp2 and VPA1044. Therefore, our results indicated that RpoN protein not only directly controls the expression of hcp2, but also directly binds to the promoter of VPA1044 to regulate the transcription of T6SS2 gene cluster in V. parahaemolyticus.
In V. cholerae, RpoN was identified directly regulate the metabolism-associated genes responsible for nitrogen utilization and formate dehydrogenase by ChIP-seq (Dong and Mekalanos, 2012). Our RNA-seq results revealed that the genes associated with nitrogen regulatory protein (VP0118), glutamine synthetase (VP0121), and formate dehydrogenase (VP1510-VP1515) were significantly down-regulated in the ΔrpoN strain in comparison with the WT strain (Table 1). BLAST analysis revealed the presence of RpoN-binding sites in the promoters of VP0118, VP0121, and VP1510 (Table 2). Thus, we speculated that RpoN could directly regulate the expression of these genes in V. parahaemolyticus. The EMSA results indicated that the RpoN protein could bind to the promoter of VP0121 for glutamine synthetase (Figure 6E), and V. parahaemolyticus RpoN mutant could not grow in M9G medium containing ammonium as a sole nitrogen source (Whitaker et al., 2014). Our results confirmed that the RpoN protein could directly bind to the promoter of VP0121 and regulate its expression for nitrogen utilization in V. parahaemolyticus. Besides, no binding shift was found in the promoters of VP0118 and VP1510 with the highest concentration of RpoN protein (Figures 6C,D). The previous studies demonstrated that co-factors could be necessary for the protein to bind the promoters in vitro (Bitoun et al., 2012). A previous study used the σ54-RNA polymerase complex to analyze the binding of RpoN with promoters (Berbard et al., 2011). These may explain why RpoN protein could not bind the promoters containing the conserved binding sites. In addition, there may be an indirect regulation event. A previous study has shown that the deletion of rpoN results in changes in the expression of genes that are directly controlled by other sigma factors (Dong et al., 2011). For example, the deletion of rpoN increases the expression of genes controlled by another sigma factor RpoS in E. coli (Dong et al., 2011). In our RNA-seq data, another sigma factor VP2210 was significantly up-regulated in the ΔrpoN strain compared to WT. VP2210 is known to directly regulate the expression of exsC and positively regulate the expression of T3SS1 (Gu et al., 2020). These data would suggest that RopN indirectly controls T3SS1 gene expression via down-regulating VP2210.
Moreover, the QS high cell density master regulator OpaR and sensor protein LuxN were also positively regulated by RpoN in V. parahaemolyticus (Table 1; Supplementary Figure S3). The RpoN protein could directly bind to the lasI promoter and positively regulate the las-QS system in P. aeruginosa (Shao et al., 2018). On the contrary, RpoN together with LuxO could activate the small regulatory RNA to negatively control the transcription of hapR at low cell density (LCD) in V. cholerae (Cheng et al., 2018). In V. alginolyticus, RpoN protein could positively regulate the transcription of luxR at LCD, whereas it could negatively regulate the transcription of luxR at high cell density (HCD; Zhang et al., 2021b). In V. parahaemolyticus, the expression of opaR was off at LCD and on at HCD, and the sRNA qrr2 could inhibit the expression of opaR (Gode-Potratz and McCarter, 2011). In this study, the RNA-seq and qRT-PCR results showed that RpoN could positively regulate the expression of opaR dependent on the qrr2 in V. parahaemolyticus. The above studies indicate that the regulation of QS by RpoN differs in different pathogens. In addition, RpoN can regulate virulence-associated genes responsible for motility, biofilm formation, T6SS, and host colonization in V. cholerae, P. aeruginosa, and C. jejuni (Dong and Mekalanos, 2012; Shao et al., 2018; Sher et al., 2020). Our RNA-seq data and qRT-PCR results also revealed that RpoN could positively regulate the expression of flagellar systems, T6SS2 and metabolic-associated genes (Table 1; Figures 2–6). Above all, our results further supporting that RpoN plays a global regulatory role in metabolic and virulence-associated pathways in V. parahaemolyticus.
Our results shown that RpoN can regulate the expression of virulence-associated genes responsible for hemolytic activity, cytotoxicity, motility, biofilm formation, and T6SS2, and most of these phenotypes are found to be controlled by OpaR (Lu et al., 2019, 2021; Tague et al., 2022; Wu et al., 2022). OpaR is a master regulator of quorum sensing that is known to directly regulate the expression of polar and lateral flagellar genes to inhibit swimming and swarming motility (Lu et al., 2019; Lu et al., 2021). Our results also showed that the RpoN could directly bound to the promoters of flgB, fliE, flgB2, and lafA to induce the expression of flagellar genes (Figures 3, 4). The expression of OpaR was decreased in the ΔrpoNΔopaR compared to ΔrpoN, but the swimming and swarming motility was no different in these strains, indicating that both RpoN and OpaR could regulate the motility and RpoN plays a more important role in motility. Furthermore, both OpaR and RpoN could directly bind to hcp2 and VPA1044 promoters to positively regulate the expression of T6SS2 (Wu et al., 2022). Our results further found that the expression of T6SS2 genes in ΔrpoNΔopaR was lower than that in ΔrpoN, whereas the expression of these genes was increased in the ΔrpoNΔqrr2 and ΔrpoNopaR+ strains (Figure 5A), indicating that both RpoN and OpaR could directly regulate the expression of T6SS2, and RpoN regulate T6SS2 with a close relationship to OpaR.
Conclusion
In this study, Figure 8 shown the RpoN-controlled virulence pathways in the flagellar systems, biofilm formation, T6SS2, hemolytic, cytotoxicity, and QS systems in V. parahaemolyticus. Besides, RpoN could regulate the metabolism pathway, including nitrogen regulatory protein, glutamine synthetase, and formate dehydrogenase. Furthermore, RpoN displayed direct binding to the promoters and controlled the expression of flgB, fliE, flgB2, lafA, hcp2, VPA1044, and VP0121, and mediated the motility, T6SS2, and glutamine utilization in V. parahaemolyticus. In addition, RpoN also contributed to the hemolytic activity, adhesion, and cytotoxicity of V. parahaemolyticus. Thus, our study suggests that RpoN is a global regulator that controls a large group of metabolic and virulence-associated pathways in V. parahaemolyticus, further supporting the conserved function of RpoN in many bacteria.
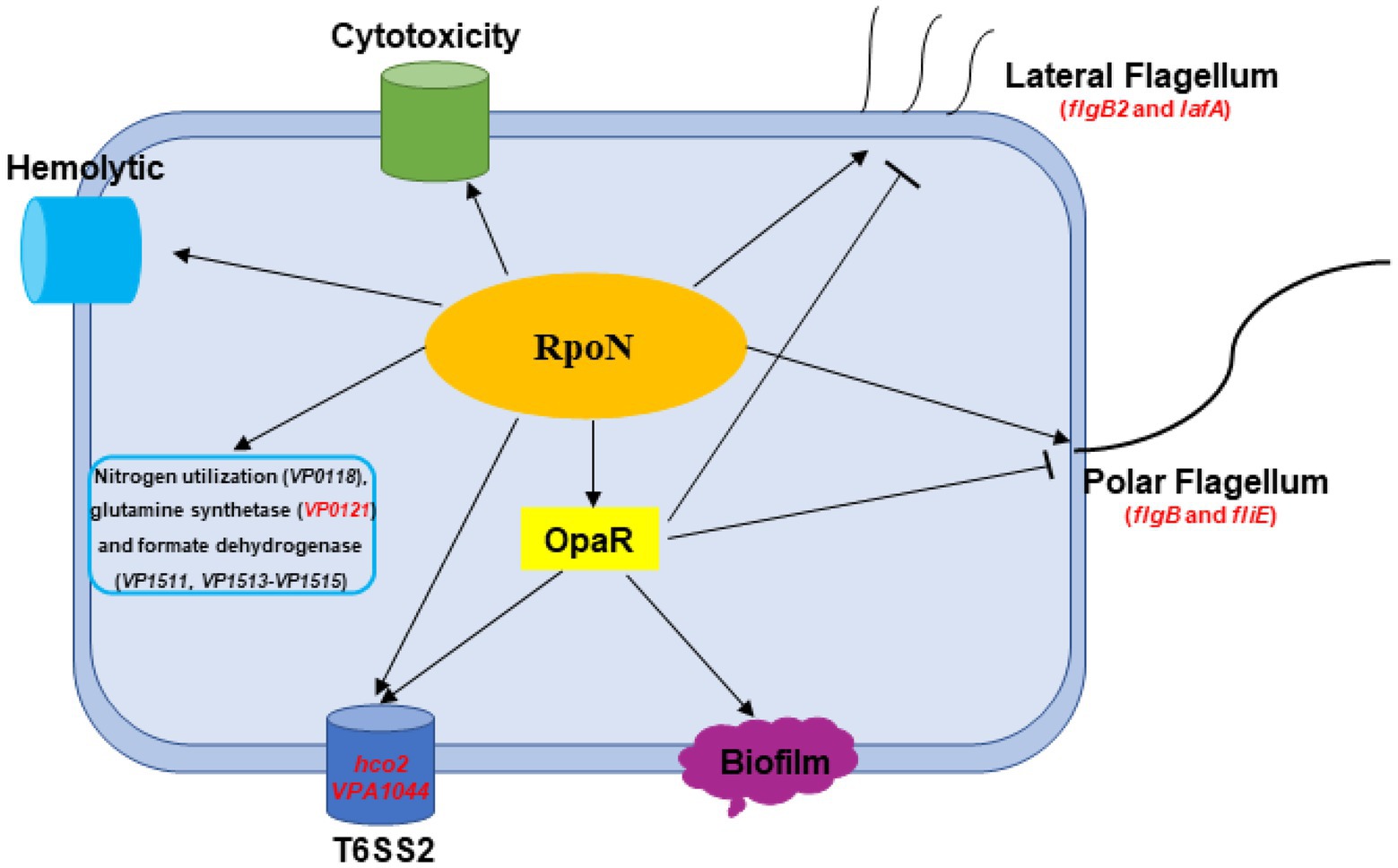
Figure 8. A schematic of the global regulatory networks of RpoN in V. parahaemolyticus. The RpoN directly binds to promoter regions of flgB, fliE, flgB2, and lafA, and positively regulates swimming and swarming motility in V. parahaemolyticus. RpoN protein also directly bind to promoters of hcp2 and VPA1044 to regulate the expression of T6SS2. Furthermore, RpoN protein could directly regulate the expression of VP0121 to be responsible for nitrogen utilization. In addition, RpoN regulates the hemolytic activity, cytotoxicity, and motility in an OpaR-independent way, whereas RpoN regulates the T6SS2 and biofilm in an OpaR-dependent way in Vibrio parahaemolyticus. The red genes indicated the promoters directly bound by RpoN protein. The arrows indicate activation, while the bar-ended lines indicate repression.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
DG, YZ, and KW contributed to the conception, design and performed the experiments. DG, YZ, KW, and ML were responsible for the acquisition of the data analyzed in this study. DG, KW, and XJ were involved in the analysis and interpretation associated with this work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32070127); the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1025960/full#supplementary-material
Footnotes
References
Bao, H., Wang, S., Zhao, J. H., and Liu, S. L. (2020). Salmonella secretion systems: differential roles in pathogen-host interactions. Microbiol. Res. 241:126591. doi: 10.1016/j.micres.2020.126591
Barrios, H., Valderrama, B., and Morett, E. (1999). Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27, 4305–4313. doi: 10.1093/nar/27.22.4305
Ben-Yaakov, R., and Salomon, D. (2019). The regulatory network of Vibrio parahaemolyticus type VI secretion system 1. Environ. Microbiol. 21, 2248–2260. doi: 10.1111/1462-2920.14594
Berbard, C., Beunet, Y. R., Gacioli, M., Lloubès, R., and Cascales, E. (2011). Regulation of type VI secretion gene clusters by sigma 54 and cognate enhancer binding proteins. J. Bacteriol. 193, 2158–2167. doi: 10.1128/JB.00029-11
Bitoun, J. P., Liao, S., Yao, X., Xie, G. G., and Wen, Z. T. (2012). The redox-sensing regulator rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. PLoS One 7:e44766. doi: 10.1371/journal.pone.0044766
Brescia, F., Marchetti-Deschmann, M., Musetti, R., Perazzolli, M., Pertot, I., and Puopolo, G. (2020). The rhizosphere signature on the cell motility, biofilm formation and secondary metabolite production of a plant-associated Lysobacter strain. Microbiol. Res. 234:126424. doi: 10.1016/j.micres.2020.126424
Buchner, A. J., Muller, K., Mehmood, J., and Tam, D. (2021). Hopping trajectories due to long-range interactions determine surface accumulation of microalgae. Proc. Natl. Acad. Sci. U. S. A. 118:e2102095118. doi: 10.1073/pnas.2102095118
Cai, R., Gao, F., Pan, J., Hao, X., Yu, Z., Qu, Y., et al. (2021). The transcriptional regulator Zur regulates the expression of ZnuABC and T6SS4 in response to stresses in Yersinia pseudotuberculosis. Microbiol. Res. 249:126787. doi: 10.1016/j.micres.2021.126787
Ceccarelli, D., Hasan, N. A., Huq, A., and Colwell, R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97. doi: 10.3389/fcimb.2013.00097
Chen, J., Zhang, R., Qi, X., Zhou, B., Wang, J., Chen, Y., et al. (2017). Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus during 2010-2014 in Zhejiang province, China. Food Control 77, 110–115. doi: 10.1016/j.foodcont.2017.02.004
Cheng, A. T., Zamorano-Sánchez, D., Teschler, J. K., Wu, D., and Yildiz, F. H. (2018). NtrC adds a new node to the complex regulatory network of biofilm formation and vps expression in Vibrio cholerae. J. Bacteriol. 200, e00025–e00018. doi: 10.1128/JB.00025-18
Damron, F. H., Owings, J. P., Okkotsu, Y., Varga, J. J., Schurr, J. R., Goldberg, J. B., et al. (2012). Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J. Bacteriol. 194, 1317–1330. doi: 10.1128/JB.06105-11
Danson, A. E., Jovanovic, M., Buck, M., and Zhang, X. (2019). Mechanisms of σ54-dependent transcription initiation and regulation. J. Mol. Biol. 431, 3960–3974. doi: 10.1016/j.jmb.2019.04.022
Davis, M. C., Kesthely, C. A., Franklin, E. A., and MacLellan, S. R. (2017). The essential activities of the bacterial sigma factor. Can. J. Microbiol. 63, 89–99. doi: 10.1139/cjm-2016-0576
Dong, T. G., and Mekalanos, J. J. (2012). Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40, 7766–7775. doi: 10.1093/nar/gks567
Dong, T., Yu, R., and Schellhorn, H. (2011). Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79, 375–386. doi: 10.1111/j.1365-2958.2010.07449.x
Erwin, D. P., Nydam, S. D., and Call, D. R. (2012). Vibrio parahaemolyticus ExsE is requisite for initial adhesion and subsequent type III secretion system 1-dependent autophagy in HeLa cells. Microbiology 158, 2303–2314. doi: 10.1099/mic.0.059931-0
Fang, L., Bi, W., Chen, S., Zhu, J., and Liu, X. (2021). Regulatory function of sigma factors RpoS/RpoN in adaptation and spoilage potential of Shewanella baltica. Food Microbiol. 97:103755. doi: 10.1016/j.fm.2021.103755
Gode-Potratz, C. J., and McCarter, L. L. (2011). Quorum sensing and silencing in Vibrio parahaemolyticus. J. Bacteriol. 193, 4224–4237. doi: 10.1128/JB.00432-11
Gu, D., Guo, M., Yang, M., Zhang, Y., Zhou, X., and Wang, Q. (2016). A σE-mediated temperature gauge controls a switch from LuxR-mediated virulence gene expression to thermal stress adaptation in vibrio alginolyticus. PLoS Pathog. 12:e1005645. doi: 10.1371/journal.ppat.1005645
Gu, D., Meng, H., Li, Y., Ge, H., and Jiao, X. (2019). A GntR family transcription factor (VPA1701) for swarming motility and colonization of Vibrio parahaemolyticus. Pathogens 8:235. doi: 10.3390/pathogens8040235
Gu, D., Zhang, Y., Wang, Q., and Zhou, X. (2020). S-nitrosylation-mediated activation of a histidine kinase represses the type 3 secretion system and promotes virulence of an enteric pathogen. Nat. Commun. 11:5777. doi: 10.1038/s41467-020-19506-1
Izutsu, K., Kurokawa, K., Tashiro, K., Kuhara, S., Hayashi, T., Honda, T., et al. (2008). Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect. Immun. 76, 1016–1023. doi: 10.1128/IAI.01535-07
Khan, F., Tabassum, N., Anand, R., and Kim, Y. (2020). Motility of vibrio spp.: regulation and controlling strategies. Appl. Microbiol. Biotechnol. 104, 8187–8208. doi: 10.1007/s00253-020-10794-7
Kongrueng, J., Srinitiwarawong, K., Nishibuchi, M., Mittraparp-Arthorn, P., and Vuddhakul, V. (2018). Characterization and CRISPR-based genotyping of clinical trh-positive Vibrio parahaemolyticus. Gut Pathog. 10:48. doi: 10.1186/s13099-018-0275-4
Li, L., Meng, H., Gu, D., Li, Y., and Jia, M. (2019). Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 222, 43–51. doi: 10.1016/j.micres.2019.03.003
Liu, X., Ye, Y., Zhu, Y., Wang, L., Yuan, L., Zhu, J., et al. (2021b). Involvement of RpoN in Regulating Motility, Biofilm, Resistance, and Spoilage Potential of Pseudomonas fluorescens. Front. Microbiol. 12:641844. doi: 10.3389/fmicb.2021.641844
Liu, M., Zhu, X., Zhang, C., and Zhao, Z. (2021a). LuxQ-LuxU-LuxO pathway regulates biofilm formation by V. parahaemolyticus. Microbiol. Res. 250:126791. doi: 10.1016/j.micres.2021.126791
Lloyd, M. G., Lundgren, B. R., Hall, C. W., Gagnon, L. B., Mah, T., Moffat, J. F., et al. (2017). Targeting the alternative sigma factor RpoN to combat virulence in Pseudomonas aeruginosa. Sci. Rep. 7:12615. doi: 10.1038/s41598-017-12667-y
Lloyd, M. G., Vossler, J. L., Nomura, C. T., and Moffat, J. F. (2019). Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain. Sci. Rep. 9:6677. doi: 10.1038/s41598-019-43060-6
Love, C. R. (2017). Ecological fitness and virulence features of Vibrio parahaemolyticus in estuarine environments. Appl. Microbiol. Biotechnol. 101, 1781–1794. doi: 10.1007/s00253-017-8096-9
Lu, R., Sun, J., Qiu, Y., Zhang, M., Xue, X., Li, X., et al. (2021). The quorum sensing regulator OpaR is a repressor of polar flagellum genes in Vibrio parahaemolyticus. J. Microbiol. 59, 651–657. doi: 10.1007/s12275-021-0629-3
Lu, R., Tang, H., Qiu, Y., Yang, W., Yang, H., Zhou, D., et al. (2019). Quorum sensing regulates the transcription of lateral flagellar genes in Vibrio parahaemolyticus. Future Microbiol. 14, 1043–1053. doi: 10.2217/fmb-2019-0048
Mahmud, A. K. M. F., Nilsson, K., Fahlgren, A., Navais, R., Choudhury, R., Avican, K., et al. (2020). Genome-scale mapping reveals complex regulatory activities of RpoN in Yersinia pseudotuberculosis. mSystems 5, e01006–e01020. doi: 10.1128/mSystems.01006-20
McCarter, L. L. (2004). Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7, 18–29. doi: 10.1159/000077866
Mea, H. J., Yong, P. V. C., and Wong, E. H. (2021). An overview of Acinetobacter baumannii pathogenesis: motility, adherence and biofilm formation. Microbiol. Res. 247:126722. doi: 10.1016/j.micres.2021.126722
Merrell, D. S., and Camilli, A. (2002). Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 5, 51–55. doi: 10.1016/s1369-5274(02)00285-0
Merrell, B. R., Walker, R. I., and Joseph, S. W. (1984). In vitro and in vivo pathologic effects of Vibrio parahaemolyticus on human epithelial cells. Can. J. Microbiol. 30, 381–388. doi: 10.1139/m84-056
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Dutta, B., Takeda, Y., and Sack, D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Qiu, Y., Hu, L., Yang, W., Yin, Z., Zhou, D., Yang, H., et al. (2020). The type VI secretion system 2 of Vibrio parahaemolyticus is regulated by QsvR. Microb. Pathog. 149:104579. doi: 10.1016/j.micpath.2020.104579
Raszl, S. M., Froelich, B. A., Vieira, C. R., Blackwood, A. D., and Noble, R. T. (2016). Vibrio parahaemolyticus and Vibrio vulnificus in South America: water, seafood and human infections. J. Appl. Microbiol. 121, 1201–1222. doi: 10.1111/jam.13246
Rattanama, P., Thompson, J. R., Kongkerd, N., Srinitiwarawong, K., Vuddhakul, V., and Mekalanos, J. J. (2012). Sigma E regulators control hemolytic activity and virulence in a shrimp pathogenic Vibrio harveyi. PLoS One 7:e32523. doi: 10.1371/journal.pone.0032523
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 8:e61086. doi: 10.1371/journal.pone.0061086
Salomon, D., Klimko, J. A., and Orth, K. (2014). H-NS regulates the Vibrio parahaemolyticus type VI secretion system 1. Microbiology 160, 1867–1873. doi: 10.1099/mic.0.080028-0
Shao, S., Li, C., Zhao, L., Zhang, Y., Yin, K., and Wang, Q. (2021). Interplay between ferric uptake regulator fur and horizontally acquired virulence regulator EsrB coordinates virulence gene expression in Edwardsiella piscicida. Microbiol. Res. 253:126892. doi: 10.1016/j.micres.2021.126892
Shao, X., Zhang, X., Zhang, Y., Zhu, M., Yang, P., Yuan, J., et al. (2018). RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 200, e00205–e00218. doi: 10.1128/JB.00205-18
Sher, A. A., Jerome, J. P., Bell, J. A., Yu, J., Kim, H. Y., Barrick, J. E., et al. (2020). Experimental evolution of Campylobacter jejuni leads to loss of motility, rpoN (σ54) deletion and genome reduction. Front. Microbiol. 11:579989. doi: 10.3389/fmicb.2020.579989
Soules, K. R., Labrie, S. D., May, B. H., and Hefty, P. S. (2020). Sigma 54-regulated transcription is associated with membrane reorganization and type III secretion effectors during conversion to infectious forms of chlamydia trachomatis. MBio 11, e01725–e01720. doi: 10.1128/mBio.01725-20
Syed, K. A., Beyhan, S., Correa, N., Queen, J., Liu, J., Peng, F., et al. (2009). The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191, 6555–6570. doi: 10.1128/JB.00949-09
Tague, J. G., Hong, J., Kalburge, S. S., and Boyd, F. E. (2022). Regulatory small RNA qrr2 is expressed independently of sigma factor-54 and can function as the sole qrr small RNA to control quorum sensing in Vibrio parahaemolyticus. J. Bacteriol. 204:e0035021. doi: 10.1128/JB.00350-21
Tang, Y., Li, F., Gu, D., Wang, W., Huang, J., and Jiao, X. (2021). Antimicrobial effect and the mechanism of diallyl trisulfide against campylobacter jejuni. Antibiotics 10:246. doi: 10.3390/antibiotics10030246
Wang, L., and Gralla, J. D. (1988). Multiple in vivo roles for the −12-region elements of sigma 54 promoters. J. Bacteriol. 180, 5626–5631. doi: 10.1128/JB.180.21.5626-5631.1998
Wang, H., Yang, Z., Swingle, B., and Kvitko, B. H. (2021). AlgU, a conserved sigma factor regulating abiotic stress tolerance and promoting virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 34, 326–336. doi: 10.1094/MPMI-09-20-0254-CR
Wettstadt, S. (2020). Should I kill or should I go: T6SS regulation networks in Vibrio. Environ. Microbiol. 22, 1–4. doi: 10.1111/1462-2920.14830
Whitaker, W. B., Richards, G. P., and Boyd, E. F. (2014). Loss of sigma factor RpoN increases intestinal colonization of Vibrio parahaemolyticus in an adult mouse model. Infect. Immun. 82, 544–556. doi: 10.1128/IAI.01210-13
Wu, K., Long, Y., Liu, Q., Wang, W., Fan, G., Long, H., et al. (2022). CqsA-introduced quorum sensing inhibits type VI secretion system 2 through an OpaR-dependent pathway in Vibrio parahaemolyticus. Microb. Pathog. 162:105334. doi: 10.1016/j.micpath.2021.105334
Wu, P., Wang, Z., Zhu, Q., Xie, Z., Mei, Y., Liang, Y., et al. (2021). Stress preadaptation and overexpression of rpoS and hfq genes increase stress resistance of Pseudomonas fluorescens ATCC12525. Microbiol. Res. 250:126804. doi: 10.1016/j.micres.2021.126804
Yin, K., Zhang, J., Ma, J., Jin, P., Ma, Y., Zhang, Y., et al. (2020). MviN mediates the regulation of environmental osmotic pressure on esrB to control the virulence in the marine fish pathogen Edwardsiella piscicida. Microbiol. Res. 239:126528. doi: 10.1016/j.micres.2020.126528
Yin, W., Zhang, N., Xu, H., Gong, X., Long, H., Ren, W., et al. (2021). Stress adaptation and virulence in vibrio alginolyticus is mediated by two (p)ppGpp synthetase genes, relA and spot. Microbiol. Res. 253:126883. doi: 10.1016/j.micres.2021.126883
Yu, Y., Yang, H., Li, J., Zhang, P., Wu, B., Zhu, B., et al. (2012). Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 194, 827–835. doi: 10.1007/s00203-012-0816-z
Zhang, Y., Gao, H., Osei-Adjei, G., Zhang, Y., Yang, W., Yang, H., et al. (2017b). Transcriptional regulation of the type VI secretion system 1 genes by quorum sensing and ToxR in Vibrio parahaemolyticus. Front. Microbiol. 8:2005. doi: 10.3389/fmicb.2017.02005
Zhang, Y., Liu, H., Dan, G., Lu, X., Zhou, X., and Xia, X. (2021a). Transcriptomic analysis of PhoR reveals its role in regulation of swarming motility and T3SS expression in Vibrio parahaemolyticus. Microbiol. Res. 235:126448. doi: 10.1016/j.micres.2020.126448
Zhang, L., Osei-Adjei, G., Zhang, Y., Gao, H., Yang, W., Zhou, D., et al. (2017a). CalR is required for the expression of T6SS2 and the adhesion of Vibrio parahaemolyticus to HeLa cells. Arch. Microbiol. 199, 931–938. doi: 10.1007/s00203-017-1361-6
Zhang, N., Zhang, S., Ren, W., Gong, X., Long, H., Zhang, X., et al. (2021b). Roles of rpoN in biofilm formation of vibrio alginolyticus HN08155 at different cell densities. Microbiol. Res. 247:126728. doi: 10.1016/j.micres.2021.126728
Zhao, K., Liu, M., and Burgess, R. R. (2010). Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 38, 1273–1283. doi: 10.1093/nar/gkp1123
Zhou, X., Konkel, M. E., and Call, D. R. (2010). Vp1659 is a Vibrio parahaemolyticus type III secretion system 1 protein that contributes to translocation of effector proteins needed to induce cytolysis, autophagy, and disruption of actin structure in HeLa cells. J. Bacteriol. 192, 3491–3502. doi: 10.1128/JB.01493-09
Keywords: RpoN, motility, T6SS2, metabolism, Vibrio parahaemolyticus
Citation: Gu D, Zhang Y, Wang K, Li M and Jiao X (2022) Characterization of the RpoN regulon reveals the regulation of motility, T6SS2 and metabolism in Vibrio parahaemolyticus. Front. Microbiol. 13:1025960. doi: 10.3389/fmicb.2022.1025960
Edited by:
Mitsuo Ogura, Tokai University, JapanReviewed by:
Menghua Yang, Zhejiang Agriculture and Forestry University, ChinaTom Defoirdt, Ghent University, Belgium
Copyright © 2022 Gu, Zhang, Wang, Li and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinan Jiao,  amlhb3hhQHl6dS5lZHUuY24=
amlhb3hhQHl6dS5lZHUuY24=
 Dan Gu
Dan Gu Youkun Zhang
Youkun Zhang Kangru Wang1,2,3
Kangru Wang1,2,3