- 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, College of Plant Protection, Yunnan Agricultural University, Kunming, China
- 2College of Agronomy and Biotechnology, Yunnan Agricultural University, Kunming, China
Steroidal glycoalkaloids (SGAs) are secondary metabolites commonly found in members of the family Solanaceae, including potatoes, and are toxic to pests and humans. The predominant SGAs in potato are α-chaconine and α-solanine. We previously reported that Glutamicibacter halophytocola S2, a gut bacterium of the pest Phthorimaea operculella (potato tuber moth), can degrade α-chaconine and α-solanine in potatoes, which can improve the fitness of P. operculella to feed on potatoes with a high content of toxic SGAs. Glutamicibacter halophytocola S2 harbored a gene cluster containing three deglycosylase genes—GE000599, GE000600, and GE000601—that were predicted encode α-rhamnosidase (RhaA), β-glucosidase (GluA), and β-galactosidase (GalA). However, there is limited information is available on the enzyme activities of the three enzymes expressed by this gene cluster and how they degrade the major toxic α-chaconine and α-solanine. In the current study, each enzyme of this gene cluster was produced by a prokaryotic expression approach and the activity of the recombinant enzymes for their target substrate and α-chaconine and α-solanine were evaluated by EPOCH microplate spectrophotometer and liquid chromatography mass spectrometry (LC-MS). The three enzymes had multifunctional activities, with RhaA and GluA could hydrolyze α-rhamnose, β-glucose, and β-galactose, while GalA can hydrolyze β-glucose and β-galactose. The degradation of α-chaconine and α-solanine was consistent with the results of the enzyme activity assays. The final product solanidine could be generated by adding RhaA or GluA alone. In conclusion, this study characterized the multifunctional activity and specific degradation pathway of these three enzymes in G. halophytocola S2. The three multifunctional enzymes have high glycosidic hydrolysis activity and clear gene sequence information, which help facilitates understanding the detoxification mechanism of insect gut microbes. The enzymes have a broad application potential and may be valuable in the removal of toxic SGAs from for potato food consumption.
Introduction
Steroidal glycoalkaloids (SGAs) are natural nitrogen-containing specialized secondary metabolites and commonly exist in the plants of the family Solanaceae (e.g., potato, tomato, eggplant, and so on) (Friedman, 2006; Ryota et al., 2022). α-Solanine and α-chaconine are two major SGAs that account for 95% of the total SGAs content in potato tubers (Ha et al., 2012). These two SGAs consist of a non-polar lipophilic steroid nucleus extended by two fused nitrogen-containing heterocyclic rings at one end and bound to a polar water-soluble carbohydrate component at the other. The side-chain of α-solanine is composed of the trisaccharide solatriose ((α-L-rhap(1-2)[β-D-Glcp(1-3)] β-D-galactopyranosyl) and that of α-chaconine is composed of chacotriose ((α-L-rhap(1-2)[α-L-rhapGlcp1-2)] β-D-glucopyranyl) (Friedman and McDonald, 1997). α-Solanine and α-chaconine are toxic to bacteria, fungi, viruses, insects, animals, and humans, and aids in plant protection (Tingey, 1984; Fewell and Roddick, 1993; Korpan et al., 2004; Yamashoji and Matsuda, 2013; Lin et al., 2018). Much ingestion of SGAs can cause to nausea, fever, diarrhea, headache, and hallucinations (Grunenfelder et al., 2006). The potential human toxicity of SGAs has led to the establishment of guidelines limiting the SGAs content of new cultivars of potatoes before they can be released for commercial use (Liu et al., 2020). Following harvest, the glycoalkaloid content can increases during storage and transportation and under the influence of light, heat, cutting, slicing, sprouting, and exposure to phytopathogens (Deng et al., 2020). At present, the main methods to reduce the content of solanine are applying sprout suppressant (chlorpropham, CIPC), genetic modification, and controlling storage conditions. However, there are obvious deficiencies in both chemical and genetic modification methods (Smith and Bucher, 2012; Sawai et al., 2014). Therefore, it is crucial to identify a safer strategy to reduce the content of solanine in potatoes.
α-Solanine and α-chaconine are reported to be degraded by acid hydrolysis or enzymatic hydrolysis (Friedman et al., 1993; Weltring et al., 1997). Chemical reactions such as acid hydrolysis and two-phase acid hydrolysis require a temperature as high as 85°C and generate a large amount of chemical waste. In addition, because of its low selectivity and yield, the chemical method to control SGAs in potatoes has been abandoned (Li et al., 2019). Some enzymes in potato-derived extracts and extracts of fungus-derived pathogens contain α-solanine and α-chaconine degradation activities (Weltring et al., 1997; Nikolic and Stankovic, 2005; Dahlin et al., 2017), which can remove the carbohydrate by stepwise degradation. And the complete deglycosylation of α-solanine and α-chaconine requires the participation of multiple glycoside hydrolases (GHs). Enzymes are considered natural products, and the discovery of unique functions of enzymes is very beneficial to the development of environmentally friendly and sustainable use of resources (Fernandes, 2010; Cheng et al., 2021). At present, limited information is available on the microbes involved in the degradation of α-solanine and α- chaconine and the use of microbial enzymes. Oda et al. (2002) found that Plectosphaerella cucumerina could only produce α-L-rhamnosidase, which could degrade α-chaconine to β1-chaconine, but could not degrade α-solanine. Jensen et al. (2009) reported that microorganisms in groundwater could stepwise hydrolyze α-solanine into β-solanine, γ-solanine and solanidine, and degrade α-chaconine into β-chaconine, γ-chaconine, and solanidine. The toxicity of these products decreased sequentially, but the microbial species involved in the process were not identified. Hennessy et al. (2018) found that Arthrobacter and Serratia could efficiently degrade the glycosidic alkaloids α-chaconine and α-solanine by three deglycosylation enzymes β-galactosidase (GalA), β-glucosidase (GluA), and α-rhamnosidase (RhaA) belonging to the GH2, GH3, and GH78 protein families, respectively. The enzymatic activity of the gene cluster involved in the complete deglycosylation of both α-chaconine and α-solanine was identified (Hennessy et al., 2020). However, the effects of each enzyme alone or jointly were not evaluated to determine the different steps involved in the degradation of both compounds.
We recently isolated and described Glutamicibacter halophytocola S2, a bacterium capable of complete degradation of SGAs in the Phthorimaea operculella gut, which can improve the fitness of P. operculella to feed on potatoes with a high content of toxic SGAs. Moreover, a gene cluster comprising three genes encoding RhaA, GluA, and GalA was identified in G. halophytocola S2, and these genes had higher expression on α-solanine medium (Wang et al., 2022). The present study aimed to evaluate the role of the enzymes encoded by these three genes, both as each enzyme alone or jointly to determine the different steps involved in the degradation of α-chaconine and α-solanine. The three enzymes described in this paper have potential application value in the removal of toxic glycosidic alkaloids and help us to enhance understanding of the detoxification mechanism of insect gut microorganisms.
Materials and methods
Strains and culture conditions
Bacterial strains were routinely cultured in liquid or solid (1.5% agar) lysogeny broth (LB) medium at 25°C (G. halophytocola S2) or 37°C (Escherichia coli for cloning and expression). When required, the medium was supplemented with 100 μg/mL ampicillin.
Gene cloning and production of recombinant proteins
Genomic DNA of G. halophytocola S2 was extracted using the freeze-thaw method according to Zheng et al. (2017). Primers were designed to target complete gene sequence without any predicted signal peptides (Table 1). Three genes were amplified from G. halophytocola S2 genomic DNA by polymerase chain reaction (PCR) using Q5 High-Fidelity DNA polymerase (NEB, USA) and the respective forward and reverse primers. PCR products were purified using the E.Z.N.A. Cycle-Pure Kit (Omega, USA). Plasmid pET15b (Novagen, USA) harboring the ampicillin selectable marker and an N-terminal HIS tag was modified and subsequently used as a cloning and heterologous expression vector (Hennessy et al., 2020). Plasmid extraction was performed using Plasmid Mini kit I (Omega). Three genes construct was cloned into the NdeI site of a pET15b vector (Novagen). The digestion products were recycled by Gel Extraction Kit (Omega). A One Step Cloning Kit (Vazyme, China) was used for recombination.

Table 1. Primers used for amplification of the target genes from the genomic DNA of Glutamicibacter halophytocola S2.
Vector constructs were transformed into E. coli TOP10 and incubated overnight at 37°C on LB agar with ampicillin (100 μg/mL). Recombinant colonies were picked from plates and cultivated in LB broth with appropriate antibiotics at 37°C overnight with shaking. Recombinant plasmids were purified using the Plasmid Mini kit I (Omega) and verified by DNA sequencing before the production of recombinant enzymes.
For enzyme production, recombinant plasmids (pET15b-000599/000600/000601) were transformed into competent E. coli BL21(DE3)-LysS and incubated overnight at 37°C on LB agar with ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL). Single colonies were picked from the plates and cultivated at 37°C with shaking in 20 mL LB broth with appropriate antibiotics (Mikkel et al., 2018). Isopropyl-β-D-thiogalactopyranoside (IPTG) with a final concentration of 0.5 mM was added to the culture when the optical density at 600 nm (OD600) was 0.3–0.6, and the induction culture was continued at 16°C for 12–16 h. Cells were collected by centrifugation and resuspended in 0.5 mL lysis buffer [50 mM N-(2-hydroxyethyl) piperazine-N’-ethane sulfonic acid (HEPES), pH 7.5, 300 mM NaCl, 2 mM Dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM DNAse]. The cells were broken by sonication, and the supernatant was collected after centrifugation at 18,000 rpm for 30 min at 4°C. Subsequently, 3 mL balanced Ni-beads (Sigma-Aldrich) were added slowly at 4°C to fully bind the protein sample. Then, 15 mL solution A (50 mM HEPES pH 7.5, 300 mM NaCl) was added for the first wash, followed by 20 mL solution B (50 mM HEPES pH 7.5, 300 mM NaCl, 50 mM imidazole) for the second wash. Finally, 10 mL solution C (50 mM HEPES pH 7.5, 300 mM NaCl, 200 mM imidazole) was added for elution and the eluate was collected. Proteins in the eluate were analyzed by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme reactions and activity visualization
Recombinant enzymes were screened for activity against 4-nitrophenyl α-L-rhamnopyranoside (pNP) glycosides, including pNP-α-L-rhamnopyranoside, pNP-β-D-galactopyranoside, and pNP-β-D-glucopyranoside (Yuanye, China) and against the potato SGAs α-chaconine and α-solanine (Extrasynthese, France). For testing the enzyme activity against 1 mM pNP-glycosides, 5 μL of 5 mg/mL recombinant enzyme was added to 95 μL HEPES buffer (pH 7.5) and incubated at 25°C for 1 h, and absorbance at 405 nm was measured using an EPOCH microplate spectrometer (Hennessy et al., 2020). One unit (U) of enzyme activity was defined as the 0.01 change in absorbance value at 405 nm per minute per mg of recombinant enzyme in 100 μL of 50 mM HEPES buffer (pH 7.5) containing 1 mM pNP glycosides. For every sample, the activity was measured three separate times (Li et al., 2019).
For testing enzyme activity against SGAs substrates, the recombinant enzyme sample was diluted to 5 mg/mL with 50 mM HEPES. For α-solanine, 5 μL each recombinant enzyme (RhaA alone, GluA alone, GalA alone, RhaA + GluA, RhaA + GalA, GluA + GalA, and RhaA + GluA + GalA) was added to 95, 90, or 85 μL HEPES buffer (pH 7.5) containing 100 μg/mL α-solanine to give a total reaction volume of 100 μL, then reactions were incubated at 25°C for 1 h. A control reaction was included and comprised 100 μL of 50 mM HEPES buffer containing 100 μg/mL α-solanine. Thus, there were a total of eight treatments, each treatment had three replicates. For α-chaconine, 5 μL recombinant enzyme (RhaA alone, GluA alone, RhaA + GluA) was added to 95 μL or 90 μL HEPES buffer (pH 7.5) containing 100 μg/mL α-chaconine and was incubated at 25°C for 1 h. The control reaction comprised 100 μL of 50 mM HEPES buffer containing 100 μg/mL α-chaconine. Thus, there were a total of four treatments and each treatment had three replicates.
Samples were analyzed on a liquid chromatography mass spectrometry (LCMS)-8040 system (Shimadzu) equipped with a Shim-pack XR-ODS III column. At a flow rate of 0.3 mL/min, 10 μL of each sample was injected onto an ODS column (2.0 mm ID × 75 mm length, 1.6 μm diameter; Shim-pack). The mobile phase was composed of solvent A (0.05% formic acid in water) and solvent B (acetonitrile) used in a gradient mode for the separation. A negative electrospray ionization mode was used for detection (α-solanine m/z 868.05, β1-solanine m/z 706.4, β2-solanine m/z 722.4, γ-solanine m/z 560.4, α-chaconine m/z 852.10, β-chaconine m/z 706.4, γ-chaconine m/z 560.4, solanidine m/z 396.90) (Jensen et al., 2009; Wang et al., 2022). An ion with a specific mass-to-charge ratio generated from the internal standard (the parent ion) was selected and fragmented to obtain its daughter ions. A specific daughter ion was used for generating the chromatogram of the corresponding compound. The peak areas of internal standards and those of the respective target compounds were obtained. The concentration of each compound was quantified by comparing its peak area with the peak area of its respective internal standard (Wang et al., 2022). Levels of α-solanine and α-chaconine were determined by a standard curve.
Statistical analysis
All analyses were performed using Data Processing System (DPS) software version 14.0 (Tang and Feng, 2012). Differences between two groups were compared using Student’s t-test, with P < 0.05 considered a significant difference. Differences between more than two groups were analyzed by one-way analysis of repeated measures with Duncan’s multiple comparisons, with P < 0.05 considered a significant difference. The statistical data were plotted using GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA, USA). Data are presented as the means ± standard deviation (SD) of three independent experiments.
Results
Three glycosidic hydrolase genes were cloned and the recombinant enzyme was purified
To examine the function of the enzyme-encoding genes, three genes—GE000599, GE000600, and GE000601—were cloned into E. coli from the genome of G. halophytocola S2. The lengths of these genes were 2,487, 2,340, and 2,526 bp, respectively (Figure 1A). The purified recombinant proteins RhaA, GluA, and GalA, respectively, were analyzed by 10% SDS-PAGE, and the sizes were approximately 93, 87, and 95 kDa, respectively (Figure 1B).
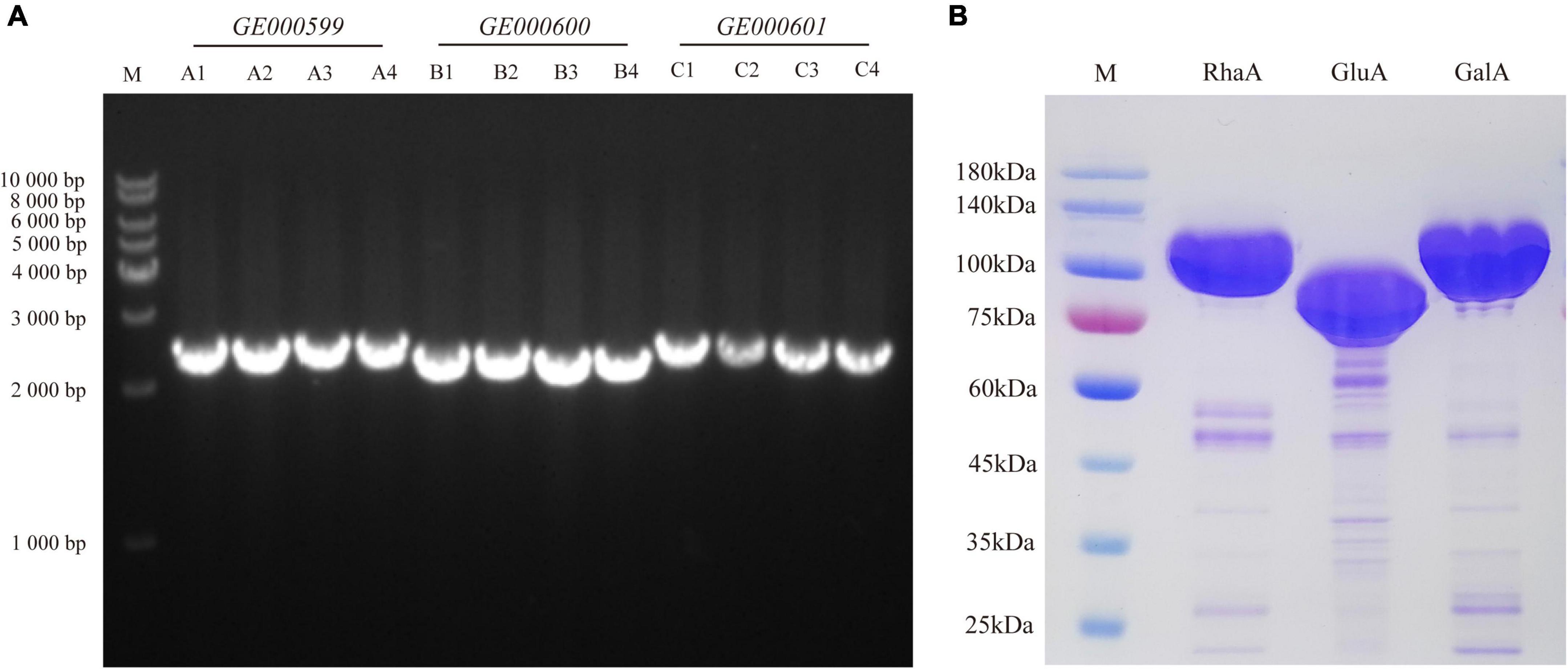
Figure 1. Cloning of glycoside hydrolase genes (GE000599, GE000600, and GE000601) (A) and expression of RhaA, GluA, and GalA in E. coli BL21 (DE3)-LysS (B).
The three enzymes showed a variety of substrate hydrolytic activities
The purified recombinant enzymes expressed from the three GA-degrading genes were tested with various pNP-glycosides, including substrates of α-rhamnosidase, β-glucosidase, and β-galactosidase, to confirm the activities of the three enzymes (Figure 2). Both RhaA and GluA showed hydrolytic activity on pNP-α-L-rhamnopyranoside, pNP-β-D-glucopyranoside, and pNP-β-D-galactopyranoside. RhaA exhibited high hydrolytic activity against pNP-α-L-rhamnopyranoside and pNP-β-D-glucopyranoside, but weak hydrolytic activity against pNP-β-D-galactopyranoside, while GluA exhibited high hydrolytic activity on pNP-β-D-glucopyranoside and pNP-β-D-galactopyranoside, but weak hydrolytic activity on pNP-α-L-rhamnopyranoside. GalA exhibited hydrolytic activity against both pNP-β-D-glucopyranoside and pNP-β-D-galactopyranoside, but the hydrolysis activity on pNP-β-D-galactopyranoside was stronger compared with that on pNP-β-D-glucopyranoside. These results indicated that the purified enzymes had high hydrolytic activity without obvious specificity.
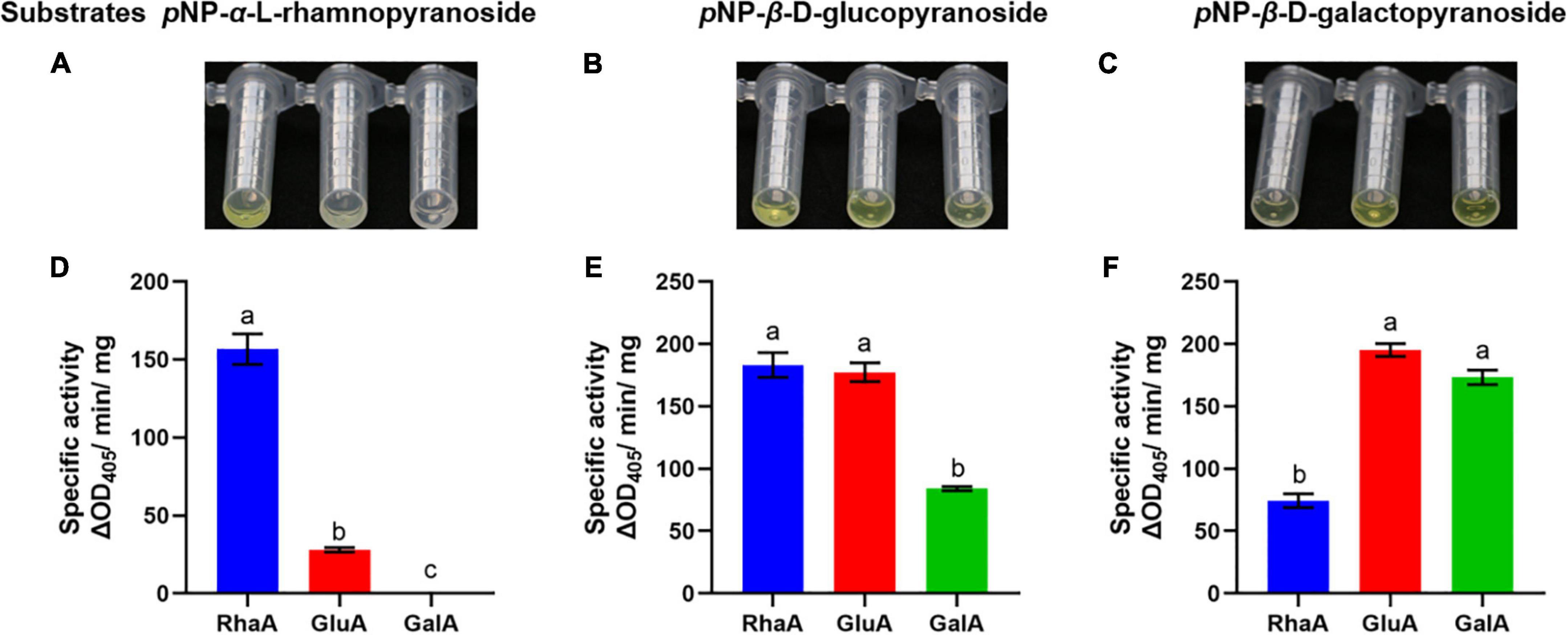
Figure 2. Color reaction and specific activity of recombinant enzymes on the substrates pNP-α-L-rhamnopyranoside (A,D), pNP-β-D-glucopyranoside (B,E) and pNP-β-D-galactopyranoside (C,F). Bars (mean ± SD) labeled with different letters within each treatment are significantly different (P < 0.05, Duncan’s multiple comparison, n = 3).
The three enzymes could degrade α-chaconine and α-solanine alone or jointly
Based on the LC-MS analysis, three monosaccharides from both α-chaconine and α-solanine appear to be removed, producing the intermediate compounds β-chaconine, γ-chaconine, β1-solanine, and γ-solanine, respectively (Figures 3, 4). Compared with the control, the peak areas of α-solanine, β1-solanine, α-chaconine, and β-chaconine were significantly reduced after addition of RhaA (α-solanine: F = 62.40, P < 0.05; β1-solanine: F = 20.47, P < 0.05; α-chaconine: F = 547.09, P < 0.05; β-chaconine: F = 577.05, P < 0.05), while the peak areas of γ-solanine, γ-chaconine, and solanidine were significantly increased (γ-solanine: F = 104.79, P < 0.05; α-solanine—solanidine: F = 128.04, P < 0.05; γ-chaconine: F = 38.70, P < 0.05; α-chaconine—solanidine: F = 143.95, P < 0.05) (Figures 3A, 4A). The reason for this is that RhaA has high hydrolytic activity on α-L-rhamnopyranoside and β-D-glucopyranoside, but low hydrolytic activity on β-D-galactopyranoside, resulting in partial accumulation of γ-solanine. However, the solanidine production results demonstrate that adding RhaA alone can remove three oligosaccharides from α-chaconine and α-solanine.
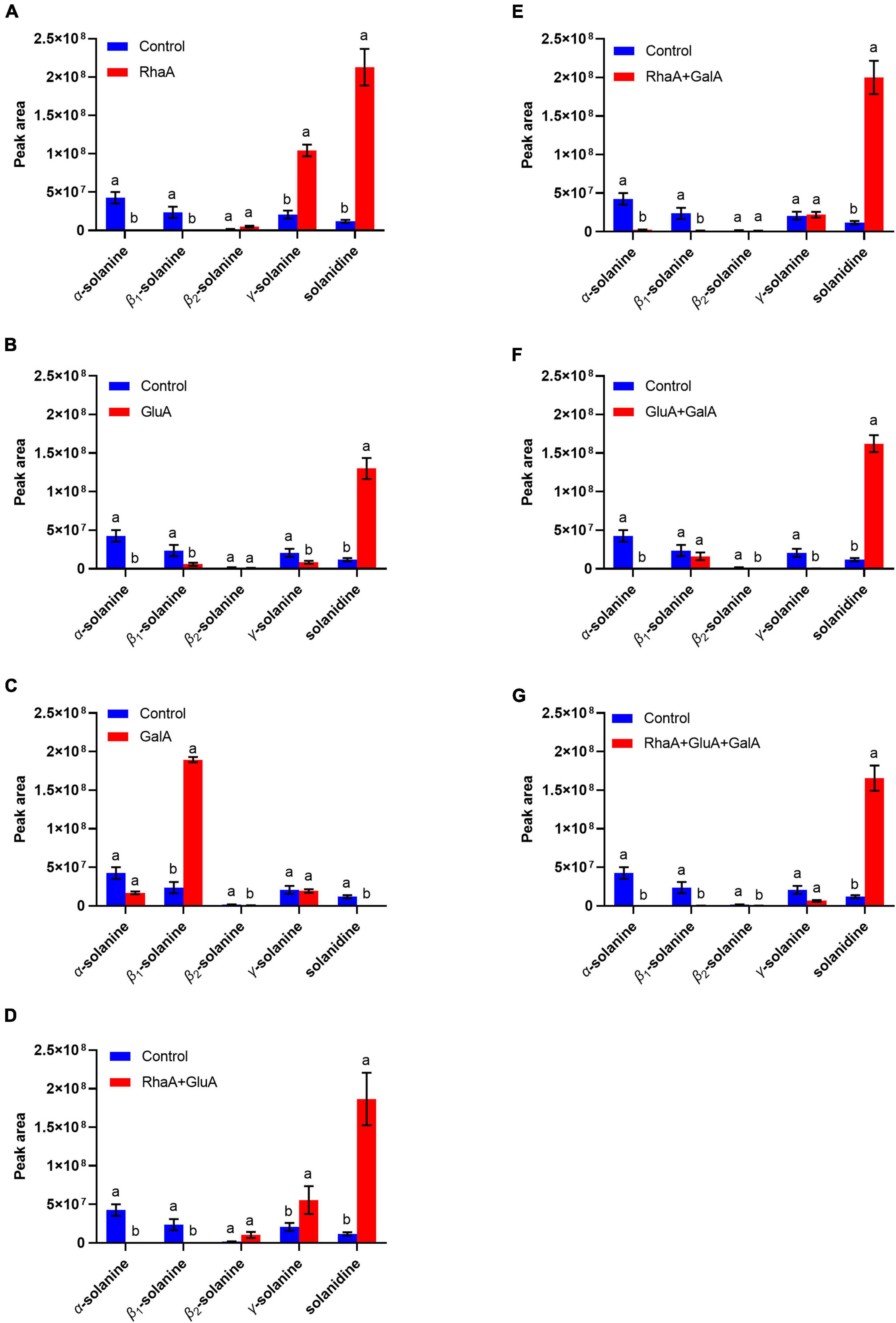
Figure 3. Hydrolysis products released from α-solanine standards by the action of recombinant enzymes of (A) RhaA; (B) GluA; (C) GalA; (D) RhaA + GluA; (E) RhaA + GalA; (F) GluA + GalA; (G) RhaA + GluA + GalA. Bars (mean ± SD) labeled with different letters within each treatment are significantly different (P < 0.05, independent-sample t-tests, n = 3).
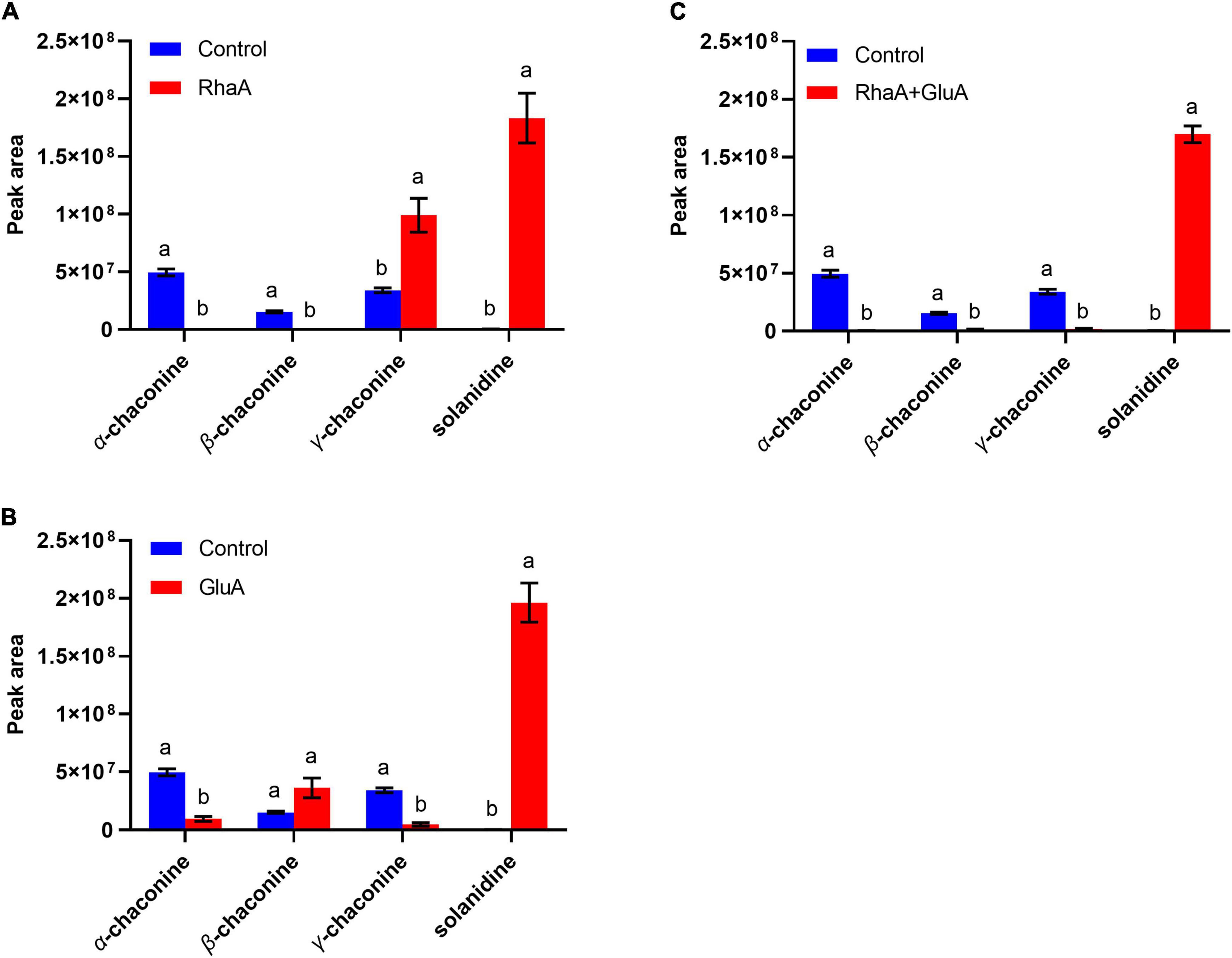
Figure 4. Hydrolysis products released from α-chaconine standards by the action of recombinant enzymes of (A) RhaA; (B) GluA; (C) RhaA + GluA. Bars (mean ± SD) labeled with different letters within each treatment are significantly different (P < 0.05, independent-sample t-tests, n = 3).
Following the addition of GluA, the peak areas of α-solanine, β1-solanine, γ-solanine, α-chaconine, and γ-chaconine were significantly reduced (α-solanine: F = 63.47, P < 0.05; β1-solanine: F = 21.8, P < 0.05; γ-solanine: F = 24.52, P < 0.05; α-chaconine: F = 289.06, P < 0.05; γ-chaconine: F = 167.84, P < 0.05), while that of solanidine was significantly increased (F = 272.46; P < 0.05) (Figures 3B, 4B). These results indicate that the addition of GluA alone can also remove three oligosaccharides from α-chaconine and α-solanine. The addition of GalA alone produced a large amount of the intermediate β1-solanine (F = 1304.30, P < 0.05) (Figure 3C). GalA has high hydrolysis activity on β-D-glucopyranoside and therefore it cannot hydrolyze α-L-rhamnopyranoside, leading to an accumulation of β1-solanine. The peak area of the solanidine was significantly reduced (F = 61.35, P < 0.05), indicating that GalA might degrade solanidine.
Following the addition of RhaA and GluA, the peak areas of α-solanine, β1-solanine, α-chaconine, β-chaconine, and γ-chaconine decreased significantly (α-solanine: F = 63.50, P < 0.05; β1-solanine: F = 19.62, P < 0.05; α-chaconine: F = 553.784, P < 0.05; β-chaconine: F = 1310.155, P < 0.05; γ-chaconine: F = 555.149, P < 0.05), and the peak areas of γ-solanine and solanidine significantly increased (γ-solanine: F = 36.90, P < 0.05; α-solanine—solanidine: F = 56.132, P < 0.05; α-chaconine—solanidine: F = 31.443, P < 0.05) (Figures 3D, 4C). This is because RhaA has high hydrolytic activity on α-L-rhamnopyranoside and β-D-glucopyranoside, but low hydrolytic activity on β-D-galactopyranoside, resulting in partial accumulation of γ-solanine.
With the addition of RhaA and GalA, the peak areas of α-solanine and β1-solanine decreased significantly (α-solanine: F = 55.30, P < 0.05; β1-solanine: F = 20.52, P < 0.05), while that of solanidine was significantly increased (F = 152.01, P < 0.05) (Figure 3E). And after the addition of GluA and GalA, the peak areas of α-solanine and γ-solanine decreased significantly (α-solanine: F = 63.40, P < 0.05; γ-solanine: F = 28.62, P < 0.05), while that of solanidine was significantly increased (F = 287.40, P < 0.05) (Figure 3F). When all three enzymes (RhaA, GluA, and GalA) were added together, the peak areas of α-solanine, β1-solanine, and γ-solanine all decreased (α-solanine: F = 63.664, P < 0.05; β1-solanine: F = 21.121, P < 0.05; γ-solanine: F = 54.952, P < 0.05) and the peak areas of the final product solanidine increased significantly (F = 219.54, P < 0.05) (Figure 3G).
Discussion
Enzyme mining is a promising way to compensate for the insufficiency of present catalysts (Nestl et al., 2011), and exploiting microorganism resources is an important strategy in this approach (Aharoni, 2009; Singh et al., 2019; Cheng et al., 2022). α-L-Rhamnosidase, β-D-galactosidase, and β-D-glucosidase are three common glycosidic hydrolases, which belong to the GH78, GH2, and GH3 glycosidic hydrolase families, respectively (Dong et al., 2021; Cheng et al., 2022). These three enzymes can be obtained from various sources such as microorganisms, plants, and animals. However, according to their source, their properties of the enzymes differ markedly depending on their source (Finocchiaro et al., 1980; Cheng et al., 2021). Compared with other available sources, microorganisms offer numerous advantages including easy handling, higher multiplication rate, and high production yield. Recently, these enzymes have been isolated and purified from a wide range of microorganisms (Panesar et al., 2010; Emanuelle et al., 2022; Kim et al., 2022). In our previous study, we first identified a gene cluster encoding α-L-rhamnosidase, β-D-galactosidase, and β-D-glucosidase in G. halophytocola S2 (Wang et al., 2022). In the current study, three genes (GE000599, GE000600, and GE000601) in the identified gene cluster were cloned and recombinant proteins were expressed on the basis of previous studies, and then the multifunctional activity of these three enzymes was confirmed.
On the basis of the degradation process of α-solanine and α-chaconine summarized by Jensen et al. (2009), the main pathways of degradation of α-solanine by G. halophytocola S2 could be summarized (Figure 5A, right). The first step of degradation of α-solanine by G. halophytocola S2 is to remove the β-glucose on the side chain and generate β1-solanine. In this step, RhaA, GluA, and GalA can all participate, but RhaA and GluA are the key enzymes. In the second step, α-rhamnose was removed to generate γ-solanine. Both RhaA and GluA can participate in this step, but RhaA is the key enzyme. The final step is the removal of β-galactose to produce solanidine. RhaA, GluA, and GalA can all participate in this step, but GluA and GalA are key enzymes. The small amount of β2-solanine generated in some of the treatments in the present study (Figures 3A,D) indicates that there may be a secondary degradation pathway for the breakdown of α-solanine by G. halophytocola S2 (Figure 5A, left). This secondary pathway involves the removal of α-rhamnose from the side chain of α-solanine as the first step, followed by the removal of β-glucose to generate γ-solanine in the second step. In addition, the pathway of degradation of α-chaconine by G. halophytocola S2 can also be summarized and is consistent with many reports (Figure 5B; Friedman et al., 1993; Oda et al., 2002; Koffi et al., 2017). However, the present study found that RhaA and GluA alone could also degrade α-chaconine, that is, the first step was to remove any rhamnose on the side chain of α-chaconine to produce β1-chaconine, and the second step was to remove another rhamnose to produce β2-chaconine. Both RhaA and GluA can participate in these two steps, and RhaA is the key enzyme. Since the m/z of β1-chaconine and β2-chaconine are the same, these compounds were uniformly called β-chaconine in LC-MS analysis. In the final step of the pathway, RhaA and GluA can also participate in the removal of β-glucose, but GluA is the key enzyme.
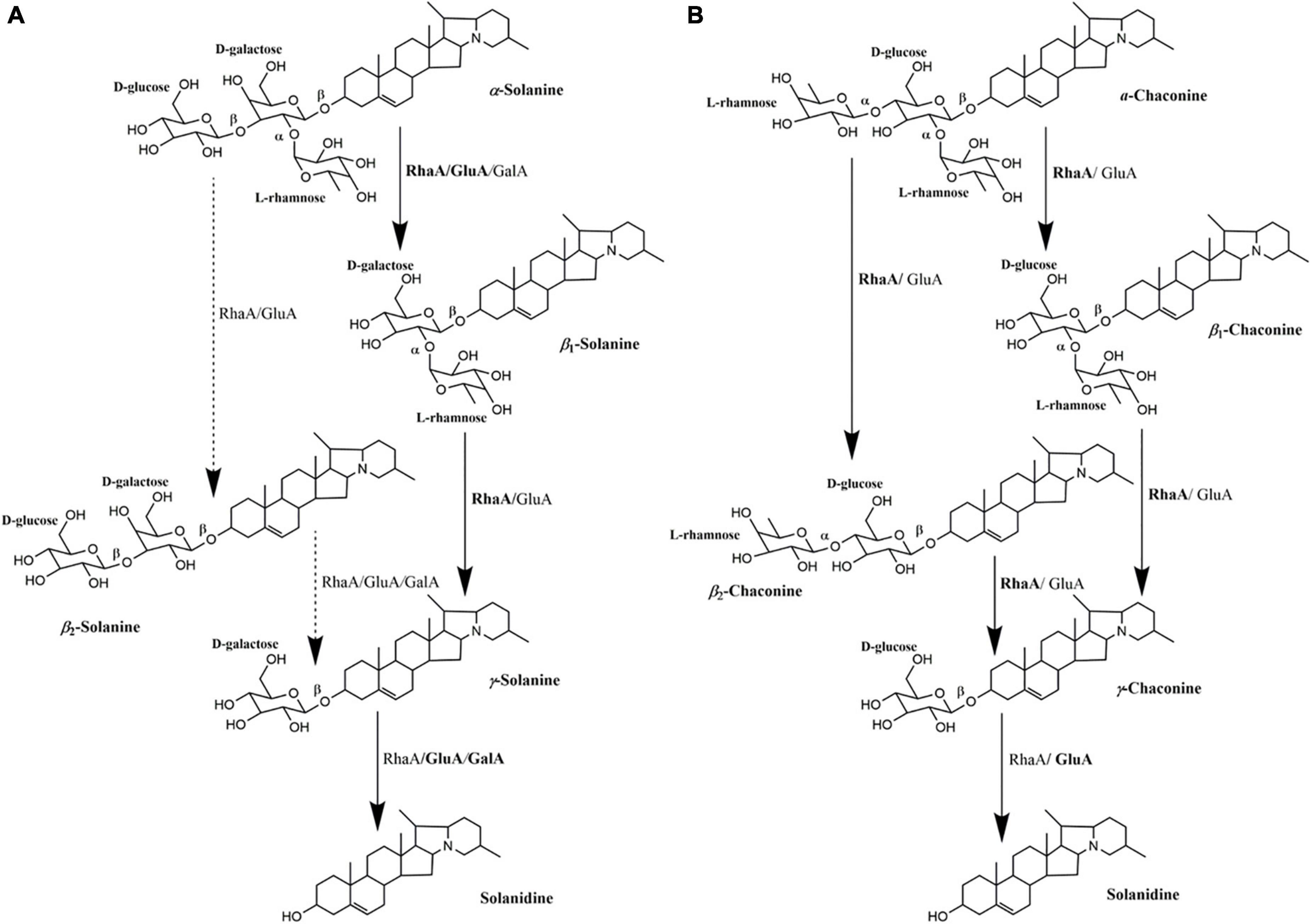
Figure 5. Multifunctional roles of glycoside hydrolases (RhaA, GluA, and GalA) produced by Glutamicibacter halophytocola S2 in the degradation of α-solanine (A) and α-chaconine (B). The degradation process of α-solanine and α-chaconine referred to Jensen et al. (2009). Solid arrows represent the proposed main pathway of G. halophytocola S2. The dashed arrow represents the secondary catabolic pathway. Enzyme names to the right of each arrow is the enzyme that can act on that site.
The crude protein extract produced by potato pathogenic fungus Gibberella pulicaris could degrade α-chaconine and α-solanine, but it was strain specific. For example, strain R-7843 could only degrade α-chaconine, while strain R-6380 could only degrade α-solanine (Weltring et al., 1997). Hennessy et al. (2020) found that the enzyme activities of the three enzymes encoding by the gene cluster in Arthrobacter sp. S41 were involved in the complete deglycosylation of α-chaconine and α-solanine, but the specific functions and degradation pathways of each enzyme were unclear. The present study characterized the multifunctional activities and specific degradation pathways of these three enzymes in G. halophytocola S2. This clarified the molecular mechanism of degradation of α-chaconine and α-solanine by G. halophytocola S2 in the gut of P. operculella. Furthermore, in this study, RhaA, and GluA alone were found to degrade α-solanine and α-chaconine, which had not been reported. Therefore, G. halophytocola has potential to be used to remove α-chaconine and α-solanine in potato foods and is advantageous owing to the high degradation activity, green environmental protection, and a very broad application prospect. The multifunctional enzyme produced by this strain have high glycosidic hydrolase activity and clear sequence information, providing an effective and eco-friendly method for solanidine production. It can reduce the consumption of acid/base, hazardous organic reagents, and raw materials, and has great development potential. Further work will be conducted on product research on the two recombinant enzymes RhaA and GluA. In addition, the next step will be to predict the potential catalytic residues of the enzymes and the binding mechanism of substrates through computational analysis, and aim to understand the catalytic effect of the enzymes on substrates at the molecular level. This will lay a theoretical foundation for the improvement of enzyme properties through protein engineering in the future.
Conclusion
The three enzymes produced by G. halophytocola S2 are multifunctional and result in the efficient degradation of α-solanine and α-chaconine by G. halophytocola S2. This study not only clarified the molecular mechanism of G. halophytocola S2 degradation of α-solanine and α-chaconine but also identified the specific steps in the degradation of α-solanine and α-chaconine by three deglycosylation enzymes. These findings contribute to understanding of the detoxification mechanism of insect gut microbes. Simultaneously, the three multifunctional enzymes have high glycosidic hydrolysis activity and clear gene sequence information, and thus also have application potential in removing toxic SGAs from potato and other edible plants of the family Solanaceae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP102487.
Author contributions
GX and BC conceived the idea, edited the manuscript, and made substantial contribution to the conception of the work. WW and GD performed gene cloning and protein expression and purification, respectively. WW, GY, and KZ performed enzyme activity measurement and SGAs degradation activity measurement. WW conducted statistical analyses and wrote the original manuscript. All authors have read and approved the content of the manuscript and contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (Nos. 32060616 and 31760519) and the Technology Major Project of the China National Tobacco Corporation [No. 110202101049(LS-09)].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aharoni, A. (2009). Mining for new enzymes. Microb. Biotechnol. 2, 128–129. doi: 10.1111/j.1751-7915.2009.00090
Cheng, L. Y., Zhang, H., Cui, H. Y., Cheng, J. M., Wang, W. Y., Wei, B., et al. (2022). A novel α-L-rhamnosidase renders efficient and clean production of icaritin. J. Clean. Prod. 341:130903. doi: 10.1016/j.jclepro.2022.130903
Cheng, L., Zhang, H., Cui, H., Wei, B., and Yuan, Q. (2021). Efficient enzyme-catalyzed production of diosgenin: Inspired by the biotransformation mechanisms of steroid saponins in Talaromyces stollii cly-6. Green Chem. 23, 5896–5910. doi: 10.1039/D0GC04152A
Dahlin, P., Müller, M. C., Ekengren, S., McKee, L. S., and Bulone, V. (2017). The impact of steroidal glycoalkaloids on the physiology of Phytophthora infestans, the causative agent of potato late blight. Mol. Plant Microbe. Interact. 30, 531–542. doi: 10.1094/MPMI-09-16-0186-R
Deng, Y., He, M., Feng, F., Feng, X., Zhang, Y., and Zhang, F. (2020). The distribution and changes of glycoalkaloids in potato tubers under different storage time based on MALDI-TOF mass spectrometry imaging. Talanta 221:121453. doi: 10.1016/j.talanta.2020.121453
Dong, Y., Zhang, S., Lu, C., Xu, J., Pei, J., and Zhao, L. (2021). Immobilization of thermostable β-glucosidase and α-L-rhamnosidase from Dictyoglomus thermophilum DSM3960 and their cooperated biotransformation of total flavonoids extract from epimedium into icaritin. Catal. Lett. 151, 2950–2963. doi: 10.1007/s10562-020-03522-3
Emanuelle, D. S. A. L., Fernandes, P., and De Assis, S. A. (2022). Immobilization of fungal cellulases highlighting β-glucosidase: Techniques, supports, chemical, and physical changes. Protein J. 41, 274–292. doi: 10.1007/s10930-022-10048-7
Fernandes, P. (2010). Enzymes in food processing: A condensed overview on strategies for better biocatalysts. Enzyme Res. 2010:862537. doi: 10.4061/2010/862537
Fewell, A. M., and Roddick, J. G. (1993). Interactive antifungal activity of the glycoalkaloids α-solanine and α-chaconine. Phytochemistry 33, 323–328. doi: 10.1016/0031-9422(93)85511-O
Finocchiaro, T., Olson, N. F., and Richardson, T. (1980). Use of immobilized lactase in milk systems. Adv. Biochem. Eng. 15, 71–88. doi: 10.1007/3540096868_3
Friedman, M. (2006). Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J. Agric. Food Chem. 54, 8655–8681. doi: 10.1021/jf061471t
Friedman, M., and McDonald, G. M. (1997). Potato glycoalkaloids: Chemistry, analysis, safety, and plant physiology. Crit. Rev. Plant Sci. 16, 55–132. doi: 10.1080/07352689709701946
Friedman, M., McDonald, G., and Haddon, W. F. (1993). Kinetics of acid-catalyzed hydrolysis of carbohydrate groups of potato glycoalkaloids α-chaconine and α-solanine. J. Agric. Food Chem. 41, 1397–1406. doi: 10.1021/jf00033a010
Grunenfelder, L. A., Knowles, L. O., Hiller, L. K., and Knowles, N. R. (2006). Glycoalkaloid development during greening of fresh market potatoes (Solanum tuberosum L.). J. Agric. Food Chem. 54, 5847–5854. doi: 10.1021/jf0607359
Ha, M., Kwak, J. H., Kim, Y., and Zee, O. P. (2012). Direct analysis for the distribution of toxic glycoalkaloids in potato tuber tissue using matrix-assisted laser desorption/ionization mass spectrometric imaging. Food Chem. 133, 1155–1162. doi: 10.1016/j.foodchem.2011.11.114
Hennessy, R. C., Jorgensen, N. O. G., Scavenius, C., Enghild, J. J., Greve-Poulsen, M., Sørensen, O. B., et al. (2018). A screening method for the isolation of bacteria capable of degrading toxic steroidal glycoalkaloids present in potato. Front. Microbiol. 9:2648. doi: 10.3389/fmicb.2018.02648
Hennessy, R. C., Nielsen, S. D., Greve-Poulsen, M., Larsen, L. B., and Stougaard, P. (2020). Discovery of a bacterial gene cluster for deglycosylation of the toxic potato steroidal glycoalkaloids α-chaconine and α-solanine. J. Agric. Food Chem. 68, 1390–1396. doi: 10.1021/acs.jafc.9b07632
Jensen, P. H., Jacobsen, O. S., Henriksen, T., Strobel, B. W., and Hansen, H. C. B. (2009). Degradation of the potato glycoalkaloids – α-solanine and α-chaconine in groundwater. Bull. Environ. Contam. Toxicol. 82, 668–672. doi: 10.1007/s00128-009-9698-4
Kim, S. Y., Lee, H. N., Hong, S. J., Kang, H. J., Cho, J. Y., Kim, D., et al. (2022). Enhanced biotransformation of the minor ginsenosides in red ginseng extract by Penicillium decumbens β-glucosidase. Enzyme Microb. Tech. 153:109941. doi: 10.1016/j.enzmictec.2021.109941
Koffi, G. Y., Remaud-Simeon, M., Due, A. E., and Combes, D. (2017). Isolation and chemoenzymatic treatment of glycoalkaloids from green, sprouting and rotting I potatoes for solanidine recovery. Food Chem. 220, 257–265. doi: 10.1016/j.foodchem.2016.10.014
Korpan, Y. I, Nazarenko, E. A., Skryshevskaya, I. V., Martelet, C., Jaffrezic- Renault, N., and El’skaya, A. V. (2004). Potato glycoalkaloids: True safety or false sense of security? Trends Biotechnol. 22, 147–151. doi: 10.1016/j.tibtech.2004.01.009
Li, Q., Wu, T., Zhao, L., Pei, J., Wang, Z., and Xiao, W. (2019). Highly efficient biotransformation of astragaloside IV to cycloastragenol by sugar-stimulated β-glucosidase and β-xylosidase from Dictyoglomus thermophilum. J. Microbiol. Biotechnol. 29, 1882–1893. doi: 10.4014/jmb.1807.07020
Lin, T., Oqani, R. K., Lee, J. E., Kang, J. W., Kim, S. Y., Cho, E. S., et al. (2018). α-Solanine impairs oocyte maturation and quality by inducing autophagy and apoptosis and changing histone modifications in a pig model. Reprod. Toxicol. 75, 96–109. doi: 10.1016/j.reprotox.2017.12.005
Liu, J. M., Wang, S. S., Zheng, X., Jin, N., and Wang, F. Z. (2020). Antimicrobial activity against phytopathogens and inhibitory activity on solanine in potatoes of the endophytic bacteria isolated from potato tubers. Front. Microbiol. 11:570926. doi: 10.3389/fmicb.2020.570926
Mikkel, S. J., Bech, P. K., Hennessy, R. C., Glaring, M. A., Tristan, B., Mirjam, C., et al. (2018). A novel enzyme portfolio for red algal polysaccharide degradation in the marine bacterium Paraglaciecola hydrolytica s66t encoded in a sizeable polysaccharide utilization locus. Front. Microbiol. 9:839. doi: 10.3389/fmicb.2018.00839
Nestl, B. M., Nebel, B. A., and Hauer, B. (2011). Recent progress in industrial biocatalysis. Curr. Opin. Chem. Biol. 15, 187–193. doi: 10.1016/j.cbpa.2010.11.019
Nikolic, N. C., and Stankovic, M. Z. (2005). Hydrolysis of glycoalkaloids from Solanum tuberosum L. haulm by enzymes present in plant material and by enzyme preparation. Potato Res. 48, 25–33. doi: 10.1007/BF02733679
Oda, Y., Saito, K., Ohara-Takada, A., and Mori, M. (2002). Hydrolysis of the potato glycoalkaloid α-chaconine by filamentous fungi. J. Biosci. Bioeng. 94, 321–325. doi: 10.1016/S1389-1723(02)80171-2
Panesar, P. S., Panesar, R., Singh, R. S., Kennedy, J. F., and Kumar, H. (2010). Microbial production, immobilization and applications of β−D-galactosidase. J. Chem. Technol. Biotechnol. 81, 530–543. doi: 10.1002/jctb.1453
Ryota, A., Bunta, W., Junpei, K., Masaru, N., Hyoung, J. L., Naoyuki, U., et al. (2022). Tandem gene duplication of dioxygenases drives the structural diversity of steroidal glycoalkaloids in the tomato clade. Plant Cell Physiol. 63, 981–990. doi: 10.1093/pcp/pcac064
Sawai, S., Ohyama, K., Yasumoto, S., Seki, H., Sakuma, T., Yamamoto, T., et al. (2014). Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26, 3763–3774. doi: 10.1105/tpc.114.130096
Singh, S. K., Mondal, A., Saha, N., and Husain, S. M. (2019). Identification and characterization of an anthrol reductase from Talaromyces islandicus (Penicillium islandicum) WF-38-12. Green Chem. 21, 6594–6599. doi: 10.1039/C9GC03072G
Smith, M. J., and Bucher, G. (2012). Tools to study the degradation and loss of the N-phenyl carbamate chlorpropham–A comprehensive review. Environ. Int. 49, 38–50. doi: 10.1016/j.envint.2012.08.005
Tang, Q. Y., and Feng, M. G. (2012). DPS data processing system for practical statistics. Beijing: China Science Press.
Tingey, W. M. (1984). Glycoalkaloids as pest resistance factors. Am. J. Potato Res. 61, 157–167. doi: 10.1007/BF02854036
Wang, W. Q., Xiao, G. L., Du, G. Z., Chang, L. S., Yang, Y., Ye, J. H., et al. (2022). Glutamicibacter halophytocola- mediated host fitness of potato tuber moth on Solanaceae crops. Pest Manag. Sci. 78, 3920–3930. doi: 10.1002/ps.6955
Weltring, K. M., Wessels, J., and Geyer, R. (1997). Metabolism of the potato saponins α-chaconine and α-solanine by Gibberella pulicaris. Phytochemistry 46, 1005–1009. doi: 10.1016/S0031-9422(97)00388-9
Yamashoji, S., and Matsuda, T. (2013). Synergistic cytotoxicity induced by α-solanine and α-chaconine. Food Chem. 141, 669–674. doi: 10.1016/j.foodchem.2013.03.104
Zheng, Y. Q., Du, G. Z., Li, Y. F., Chen, B., Li, Z. Y., and Xiao, G. L. (2017). Isolation and identification of bacteria from larval gut of the potato tuberworm, Phthorimaea operculella (Zeller) and the degradation for plant-based macromolecular compounds. J. Environ. Entomol. 39, 525–532. doi: 10.3969/j.issn.1674-0858.2017.03.5
Keywords: Glutamicibacter halophytocola, glycoalkaloids degradation, α-chaconine, α-solanine, α-rhamnosidase, β-galactosidase, β-glucosidase
Citation: Wang W, Du G, Yang G, Zhang K, Chen B and Xiao G (2022) A multifunctional enzyme portfolio for α-chaconine and α-solanine degradation in the Phthorimaea operculella gut bacterium Glutamicibacter halophytocola S2 encoded in a trisaccharide utilization locus. Front. Microbiol. 13:1023698. doi: 10.3389/fmicb.2022.1023698
Received: 20 August 2022; Accepted: 20 September 2022;
Published: 12 October 2022.
Edited by:
Jiong Hong, University of Science and Technology of China, ChinaReviewed by:
Yaroslav Korpan, National Academy of Sciences of Ukraine, UkraineRoberta Mariot, Federal University of Rio Grande, Brazil
Copyright © 2022 Wang, Du, Yang, Zhang, Chen and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Chen, Y2hiaW5zQDE2My5jb20=; Guanli Xiao, Z2x4aWFvOUAxNjMuY29t
 Wenqian Wang
Wenqian Wang Guangzu Du1
Guangzu Du1