
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 October 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1022006
This article is part of the Research Topic Coronavirus: Broad-Spectrum Anti-viral Targets and Treatments View all 10 articles
LCB1 is a 56-mer miniprotein computationally designed to target the spike (S) receptor-binding motif of SARS-CoV-2 with potent in vitro and in vivo inhibitory activities (Cao et al., 2020; Case et al., 2021). However, the rapid emergence and epidemic of viral variants have greatly impacted the effectiveness of S protein-targeting vaccines and antivirals. In this study, we chemically synthesized a peptide-based LCB1 inhibitor and characterized the resistance profile and underlying mechanism of SARS-CoV-2 variants. Among five variants of concern (VOCs), we found that pseudoviruses of Beta, Gamma, and Omicron were highly resistant to the LCB1 inhibition, whereas the pseudoviruses of Alpha and Delta as well as the variant of interest (VOI) Lambda only caused mild resistance. By generating a group of mutant viruses carrying single or combination mutations, we verified that K417N and N501Y substitutions in RBD critically determined the high resistance phenotype of VOCs. Furthermore, a large panel of 85 pseudoviruses with naturally occurring RBD point-mutations were generated and applied to LCB1, which identified that E406Q, K417N, and L455F conferred high-levels of resistance, when Y505W caused a ∼6-fold resistance fold-change. We also showed that the resistance mutations could greatly weaken the binding affinity of LCB1 to RBD and thus attenuated its blocking capacity on the interaction between RBD and the cell receptor ACE2. In conclusion, our data have provided crucial information for understanding the mechanism of SARS-CoV-2 resistance to LCB1 and will guide the design strategy of novel LCB1-based antivirals against divergent VOCs and evolutionary mutants.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) caused the global pandemic of coronavirus disease 2019 (COVID-19), which has recently resulted in more than 551 million confirmed cases with about 6.4 million deaths.1 During the spread of the virus, many variants of concern (VOCs) emerged with significantly changed infectivity and pathogenicity, leading to new waves of infection that have posed daunting challenges (Callaway, 2021; Garcia-Beltran et al., 2021; He et al., 2021; Hoffmann et al., 2021; Kuzmina et al., 2021). The previous four VOCs are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2); the fifth one, Omicron (B.1.1.529), was first reported from Southern Africa in late November 2021. Genome-sequencing data indicate that Omicron variant evolves with the largest number of mutations, including 32 mutations located within spike (S) protein that cover the key mutations in the receptor binding domain (RBD) or motif (RBM), such as K417N, E484A, and N501Y (Callaway, 2021; He et al., 2021; Tao et al., 2021; Yamasoba et al., 2022). Significantly, Omicron is highly transmissible and can spread several times faster than any previous variants; thus, it has quickly outcompeted Delta variant to dominate the epidemic, caused a large number of breakthrough infection or re-infection, and seriously impaired the clinical efficacies of preventive vaccines and therapeutic antibodies (He et al., 2021; Guo et al., 2022; Markov et al., 2022).
Since the outbreak of COVID-19, many efforts have been devoted to the development of antivirals that block different steps of SARS-CoV-2 life-cycle, including viral entry, replication, assembly, budding, and releasing (Yang and Rao, 2021; Yan et al., 2022). Notably, virus entry inhibitors that can inhibit the interaction between S protein and the human cellular receptor angiotensin-converting enzyme 2 (ACE2) are considered a promising strategy (Plavec et al., 2021; Sabbah et al., 2021; Xiang et al., 2021). By applying computational de novo design approaches, Cao and coworkers developed a group of miniproteins, which bound the spike RBD with affinities ranging from 100 pM to 10 nM and inhibited SARS-CoV-2 infection with 50% inhibitory concentration (IC50) values between 24 pM and 35 nM (Cao et al., 2020). LCB1, a lead miniprotein designed with 56 amino acids, showed the potent in vitro antiviral activity, and its modified versions efficiently blocked SARS-CoV-2 infection in human ACE2 (hACE2)-expressing transgenic mice when administrated as both pre-exposure prophylaxis (PrEP) and post-exposure therapy, providing an ideal candidate for drug development (Case et al., 2021). Considering the COVID-19 epidemic caused by divergent VOCs and still ongoing evolutionary mutations, it is fundamentally important to characterize LCB1 for its drug resistance and underlying mechanism. In this report, we describe our data to specifically address this question, which can guide the design strategy of novel LCB1-based antivirals against divergent VOCs and evolutionary mutants.
A 56-mer LCB1 peptide was synthesized on rink amide 4-methylbenzhydrylamine (MBHA) resin using a standard solid-phase 9-flurorenylmethoxycarbonyl (FMOC) protocol as described previously (Yu et al., 2021a). Plasmids encoding the mutant S proteins of SARS-CoV-2 (Alpha, Beta, Gamma, Delta, Lambda, and Omicron) were a kind gift from Linqi Zhang at the Tsinghua University (Beijing, China). HEK293T and Huh-7 cells were purchased from the American type culture collection (ATCC) (Rockville, MD, USA); 293T/ACE2 cells stably expressing human ACE2 were generated and preserved in our laboratory. Cells were cultured in complete growth medium consisting of Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml of penicillin-streptomycin, 2 mM L-glutamine, and 1mM sodium pyruvate under 37oC and 5% CO2.
Circular dichroism (CD) spectroscopy was applied to determine the secondary structure and thermostability of LCB1 peptide as described previously (Zhu et al., 2020). Briefly, LCB1 was dissolved in phosphate-buffered saline (PBS; pH 7.2) with a final concentration of 20 μM and incubated at 37oC for 30 min. CD spectra were obtained on Jasco spectropolarimeter (model J-815) a using a 1 nm bandwidth with a 1 nm step resolution from 195 to 270 nm at room temperature. The spectra were corrected by subtracting a solvent blank, and the α-helical content was calculated from the CD signal by dividing the mean residue ellipticity [θ] at 222 nm by with a value of −33,000 deg cm2 dmol–1, corresponding to a 100% helix. Thermal denaturation was done by monitoring the ellipticity change at 222 nm from 20 to 98oC at a rate of 2oC/min, and the melting temperature (Tm) was defined as the midpoint of the thermal unfolding transition.
Spike mutants were generated by site-directed mutagenesis as described previously (Yu et al., 2021a). In brief, the forward and reverse primers with 16∼28 nucleotides were designed with specific mutations and occupied the same starting and ending positions on the opposite strands of a codon-optimized S gene cloned in a pcDNA3.1 vector. DNA synthesis was conducted by PCR in a 50-μl reaction volume using 100 ng of denatured plasmid template, 50 pM upper and lower primers, and 5 U of the high-fidelity polymerase PrimeStar (TaKaRa, Dalian, China). PCR amplification was done for one cycle of denaturation at 98°C for 5 min, followed by 25 cycles of 98°C for 10 s and 68°C for 9 min, with a final extension at 72°C for 10 min. The amplicons were treated with restriction enzyme DpnI for 3 h at 37°C, and DpnI-resistant molecules were recovered by transforming E. coli stable3 competent cells with antibiotic resistance. The required mutations were confirmed by DNA sequencing of a single clone.
Infectivity of various SARS-CoV-2 pseudoviruses on 293T/ACE2 or Huh-7 cells was measured by a single-cycle infection assay as described previously (Yu et al., 2021a). Briefly, pseudoviruses were packaged by cotransfecting HEK293T cells with a wild-type (WT) or mutant S protein-expressing plasmid and pNL4-3.luc.RE plasmid that encodes an Env-defective HIV-1NL4–3 genome with luciferase as a reporter. Cell supernatants containing virions were collected after transfection 48 h and stored at −80oC. To determine the inhibitory activity of LCB1 inhibitor, a serially three-fold diluted peptide was incubated with an equal volume of pseudoviruses at 37oC for 1 h, and the peptide-virus mixture was then added to 293T/ACE2 or Huh-7 target cells at a density of 104 cells/100 μl per well in a 96-well culture plate. After incubation at 37oC for 48 h, cells were harvested, lysed in reporter lysis buffer, and measured for luciferase activity using luciferase assay reagents and a luminescence counter (Promega, Madison, WI, USA).
Biolayer interferometry (BLI) was used to measure the binding and blocking activities of LCB1 peptide. In brief, a recombinant His-tagged RBD protein (Sino Biological, Beijing, China) was loaded on a NTA biosensor (ForteBio, San Francisco, CA, USA) at a concentration of 10 μg/ml for 120 s in phosphate buffer saline (PBS), and LCB1 peptide was gradient-diluted in PBST buffer (PBS plus 0.2% Tween 20). The binding kinetics was guided by associating in analyte substrates for 120s and disassociating in PBST alone for 300s.
The blocking activity of LCB1 peptide on the interaction of ACE2 and RBD proteins was also determined by flow cytometry. Briefly, an RBD protein at 2 μg/ml was mixed with serially diluted LCB1 and incubated at 4°C for 1 h. The mixture was then added to 5 × 105 of 293T/ACE2 cells and incubated at 4°C for 1 h. After being washed twice with PBS, the cells were incubated with 1:500 diluted Alexa Fluor 488-labeled rabbit anti-His tag antibody (Cell Signaling Technology, Danvers, MA, USA) at 4°C for 1 h. After two washes with PBS, cells were resuspended by FACS buffer and analyzed with a FACS CantoII instrument (Becton-Dickinson, Mountain View, CA, USA).
The percent inhibition of virus infection and 50% inhibitory concentration (IC50) of LCB1 inhibitor were calculated using GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA, USA). Statistical comparison of divergent pseudovirus infections were conducted by one-way ANOVA with Dunnett’s multiple comparisons test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; ns, not significant).
The previously reported LCB1 miniprotein was recombinantly expressed and purified from E. coli. Herein, we chemically synthesized a 56-mer LCB1 peptide using a standard solid-phase FMOC protocol. The peptide was acetylated at the N terminus and amidated at the C terminus, purified to a 95.15% homogeneity by reverse-phase high-performance liquid chromatography (HPLC) and characterized for amino acids with mass spectrometry (Supplementary Figure 1). The concentration of LCB1 peptide was determined by UV absorbance and a theoretically calculated molar extinction coefficient based on the tryptophan and tyrosine residues. We first characterized the structural properties of LCB1 by CD spectroscopy. As shown in Figures 1A,B, CD spectra of LCB1 displayed typical double minima at 208 and 222 nm, which indicated an α-helical content of ∼82%; however, its thermal unfolding transition could not be precisely determined due to a greater than 95oC melting temperature (Tm). Next, the antiviral activity of LCB1 was measured by a pseudovirus-based single-cycle infection assay. To this end, pseudoviruses carrying wild-type (WT) S protein of the original SARS-CoV-2 Wuhan-Hu-1 strain or a single D614G mutation (B1 strain) were packaged and characterized. As shown in Figures 1C,D, LCB1 potently inhibited infections of the WT and D614G pseudoviruses with mean IC50 values of 0.191 and of 0.062 nM, respectively, on 293T/ACE2 cells and of 0.22 and 0.073 nM, respectively, on Huh-7 cells. In comparison, LCB1 was ∼3-fold more active in inhibiting the D614G mutant relative to its inhibition on WT strain. Taken together, these results validated the structural integrity and functionality of LCB1 as a peptide-based inhibitor.
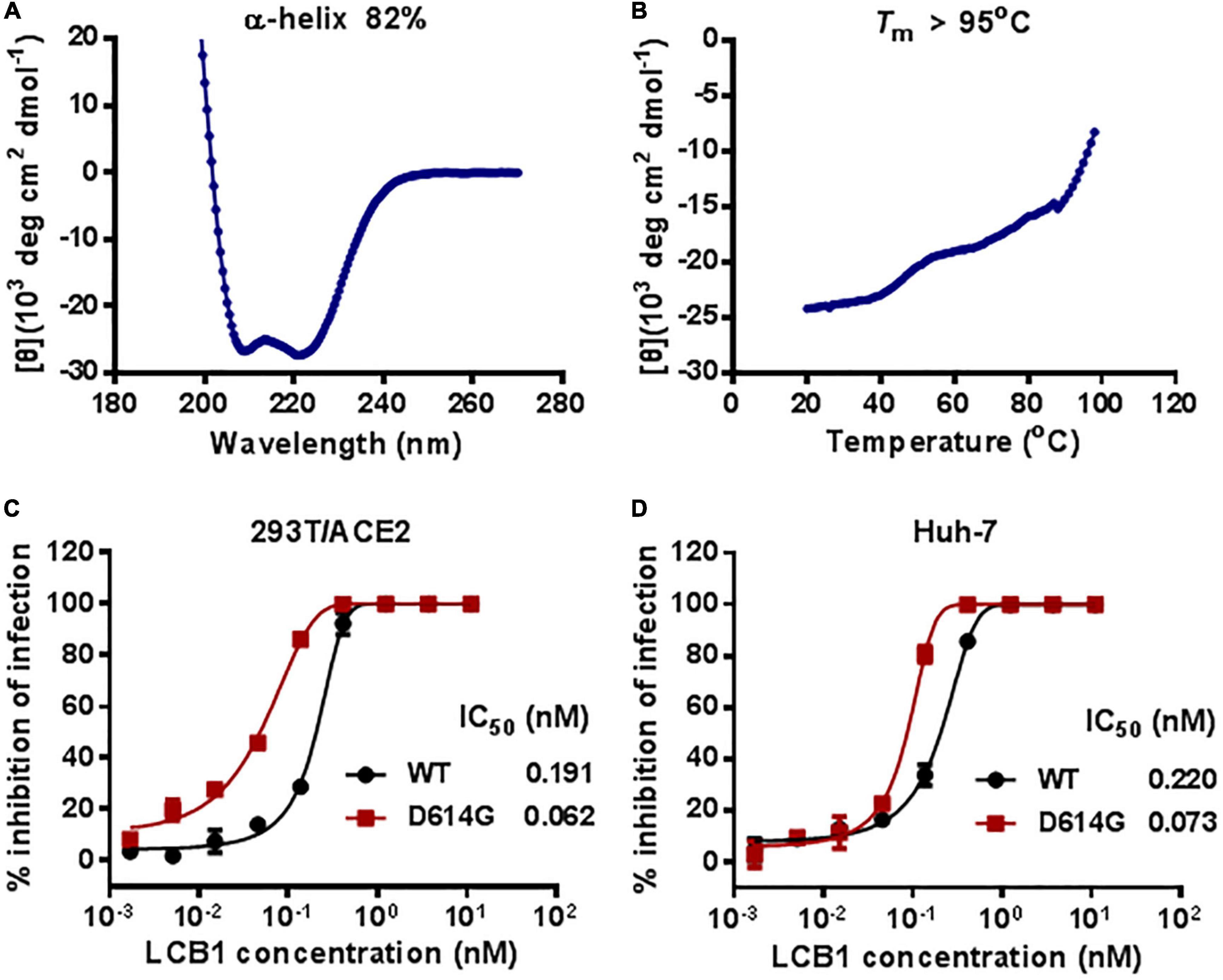
Figure 1. Structural and functional characterization of LCB1 peptide. The α-helicity (A) and thermostability (B) of LCB1 peptide at a concentration of 20 μM were determined by Circular dichroism (CD) spectroscopy. The inhibitory activities of LCB1 against infections of wild-type (WT) SARS-CoV-2 and its D614G mutant on 293T/ACE2 cells (C) and Huh-7cells (D) were measured by a single-cycle infection assay. Samples were tested in triplicate and repeated, and data are presented as means.
We recently reported the functionalities of S proteins derived from divergent SARS-CoV-2 variants to mediated cell-cell fusion and infectivity, as well as their susceptibility to the inhibition of fusion-inhibitory lipopeptides. Herein, we sought to characterize LCB1 peptide for its inhibitory activity on five VOCs and one VOI (Lambda, C.37). The corresponding pseudoviruses were therefore generated and used in the single-cycle infection assay. As shown in Figures 2A,B and Table 1, LCB1 infected Alpha, Beta, Gamma, Delta, Lambda, and Omicron variants with mean IC50 values of 0.899, 901.8, 204.367, 0.569, 0.301, and 956.5 nM, respectively, on 293T/ACE2 cells and of 0.69, 447.083, 70.405, 0.958, 0.47, and 761.217 nM, respectively, on Huh-7 cells. Because all the VOCs were evolved with a D614G background, here we used D614G virus as a reference strain for calculating the fold-changes of resistance. As shown, Alpha, Beta, Gamma, Delta, Lambda, and Omicron displayed about 15-, 14, 545-, 3, 296-, 9-, 5-, and 15,427-fold resistance on 293T/ACE2 cells and about 9-, 6, 124-, 964-, 13-, 6-, and 10,428-fold resistance on Huh-7 cells, respectively. Therefore, the Beta, Gamma, and Omicron variants had very high levels of resistance to the LCB1 inhibition, whereas the Alpha, Delta, and Lambda variants exhibited mild resistance.
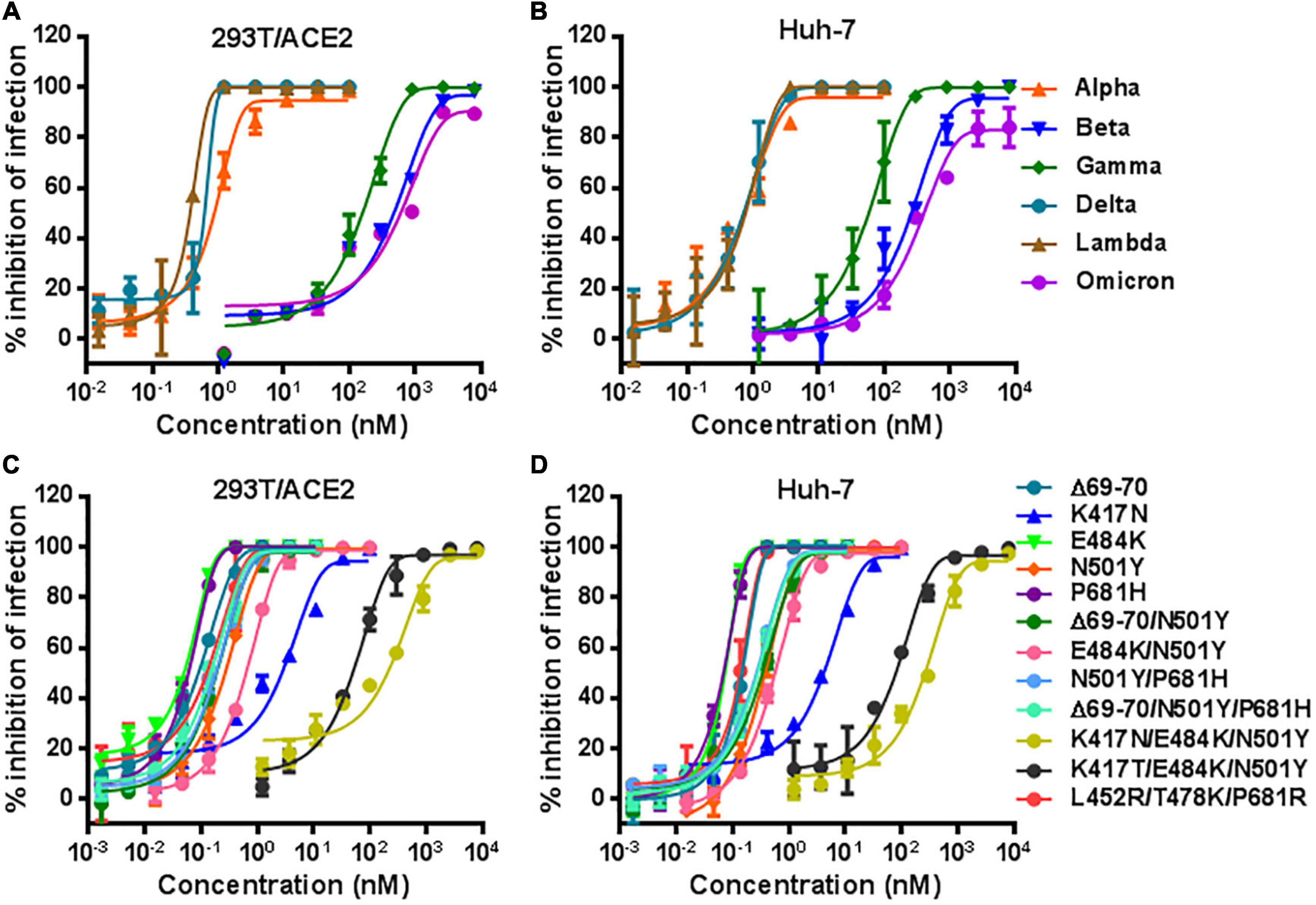
Figure 2. Inhibitory activity of LCB1 against divergent SARS-CoV-2 variants. The activities of LCB1 peptide in inhibiting VOCs and VOI (A,B) as well as the panel of mutant pseudoviruses carrying single or combined mutations (C,D) were measured by the single-cycle infection assay. Samples were tested in triplicate, the experiments were repeated three times, and data are expressed as the means with standard deviations (SD) and presented in Table 1.
In order to identify the mutations responsible for the VOC resistance, we further generated a panel of S protein mutants carrying the key mutations by VOCs. The corresponding pseudoviruses were generated and their infectivity was characterized on both 293T/ACE2 and Huh-7 cells by a single-cycle infection assay (Supplementary Figure 2). In the inhibition of pseudoviruses with single mutations, we found that LCB1 inhibited the infection of K417N mutant with IC50 of 2.774 nM on 293T/ACE2 and 4.898 nM on Huh-7 cells, which indicated 45-fold or 67-fold resistance changes over D614G reference (Figures 2C,D). While N501Y mutant showed mild resistance (∼4-fold), the single mutation of E484K or P681H and the mutant with Δ69-70 deletion did not confer resistance, which were also supported by the IC50 fold-changes of three combination mutations (Δ69-70/N501Y, N501Y/P681H, and Δ69-70/N501Y/P681H). Significantly, the E484K/N501Y mutant was more resistant than the N501Y mutant and its combination with K417N or K417T sharply enhanced the resistance phenotype, as evidenced by the fold-changes of two triple mutations (K417N/E484K/N501Y and K417T/E484K/N501Y). In contrast, the triple mutant L452R/T478K/P681R had no obvious resistance to the LCB1 inhibition. These results demonstrated that K417N or K417T mutation in RBD plays a key role in the resistance profiles of SARS-CoV-2 VOCs to LCB1 inhibitor. In line with this conclusion, the Beta, Gamma, and Omicron variants contain K417N or K417T, whereas the Alpha, Delta, and Lambda variants lack this mutation.
Considering the ongoing mutations of SARS-CoV-2 variants, we are interested in characterizing the effects of naturally occurring mutations on the inhibitory activity of LCB1. Thus, a total of 85 substitutions, which naturally occurred in the spike RBD sequence with relatively higher frequencies, were selected and the corresponding S protein mutants were generated. First, we analyzed the functionality of the S protein mutants to mediate pseudovirus infections on both 293T/ACE2 and Huh-7 cells. As shown in Supplementary Figure 3, many of the mutants displayed significantly decreased infectivity, whereas several mutations resulted in enhanced infections (L452M, I468T, Y505W, and Y508H). Herein, we determined the susceptibility of 81 pseudoviruses to the LCB1 inhibition on Huh-7 cells by the single-cycle infection assay. As indicated by the IC50 values in Table 2, there were three mutants (E406Q, K417T, and L455F) displaying high-levels of resistance with the fold-changes of IC50 at ∼31, ∼25, and ∼121, respectively, whereas the Y505W mutant conferred a mild resistance with a ∼6-fold increased IC50 value. In this experiment, we also verified that L452R and T478K mutations, which appeared in the L452R/T478K/P681R mutant above, had no resistance to LCB1, whereas some mutants (P463S, V483F, A520V, and P521R) behaved with certain degrees of increased susceptibility.
To explore the mechanism underlying the LCB1 resistance, we applied biolayer interferometry (BLI) to analyze the binding affinity of LCB1 to the RBD proteins with or without resistance mutations (Figure 3). Comparing to the equilibrium dissociation constant (Kd) of wild-type RBD (WT-RBD) at 3.397 nM, only the RBD proteins carrying a single K417N or double L452R/T478K mutations exhibited slightly reduced binding affinities with Kd values of 6.805 and 6.77 nM, respectively, none of other single mutations (E406Q, K417T, L455F, and N501Y) significantly impaired the LCB1 binding. Notably, the RBD proteins with the combination mutations of K417N/E484K/N501Y or K417T/E484K/N501 had dramatically decreased binding capacities as indicated by their Kd values at 49.11 and 15.83 nM, respectively.
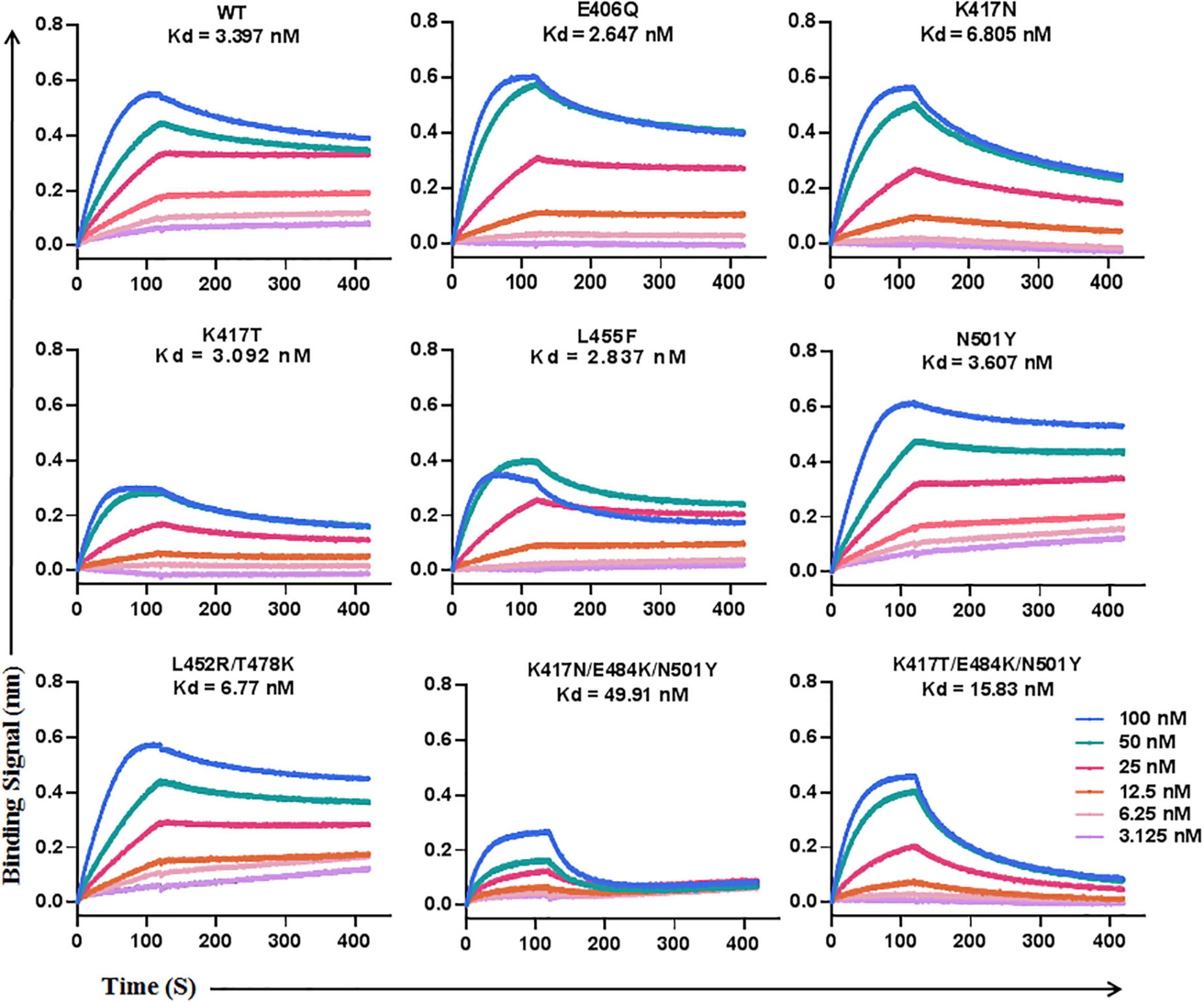
Figure 3. The binding affinity of LCB1 with RBD proteins determined by biolayer interferometry. A recombinant His-tagged RBD protein with wild-type sequence or mutations was loaded onto an NTA biosensors and equilibrated before the baseline was set to zero at t = 0. The binding kinetics was guided by associating in different concentrations of LCB1 for 120 s and disassociating for 300 s. The equilibrium dissociation constant (Kd) was calculated.
We next determined the blocking activity of LCB1 on the binding of RBD protein with ACE2 expressed on 293T/ACE2 cells by flow cytometry. Consistent with its binding affinity above, LCB1 could effectively blocked the binding of RBD proteins with single or double mutations, but it exhibited a dramatically decreased blocking activity on the RBD with K417N/E484K/N501Y or K417T/E484K/N501Y (Figure 4).
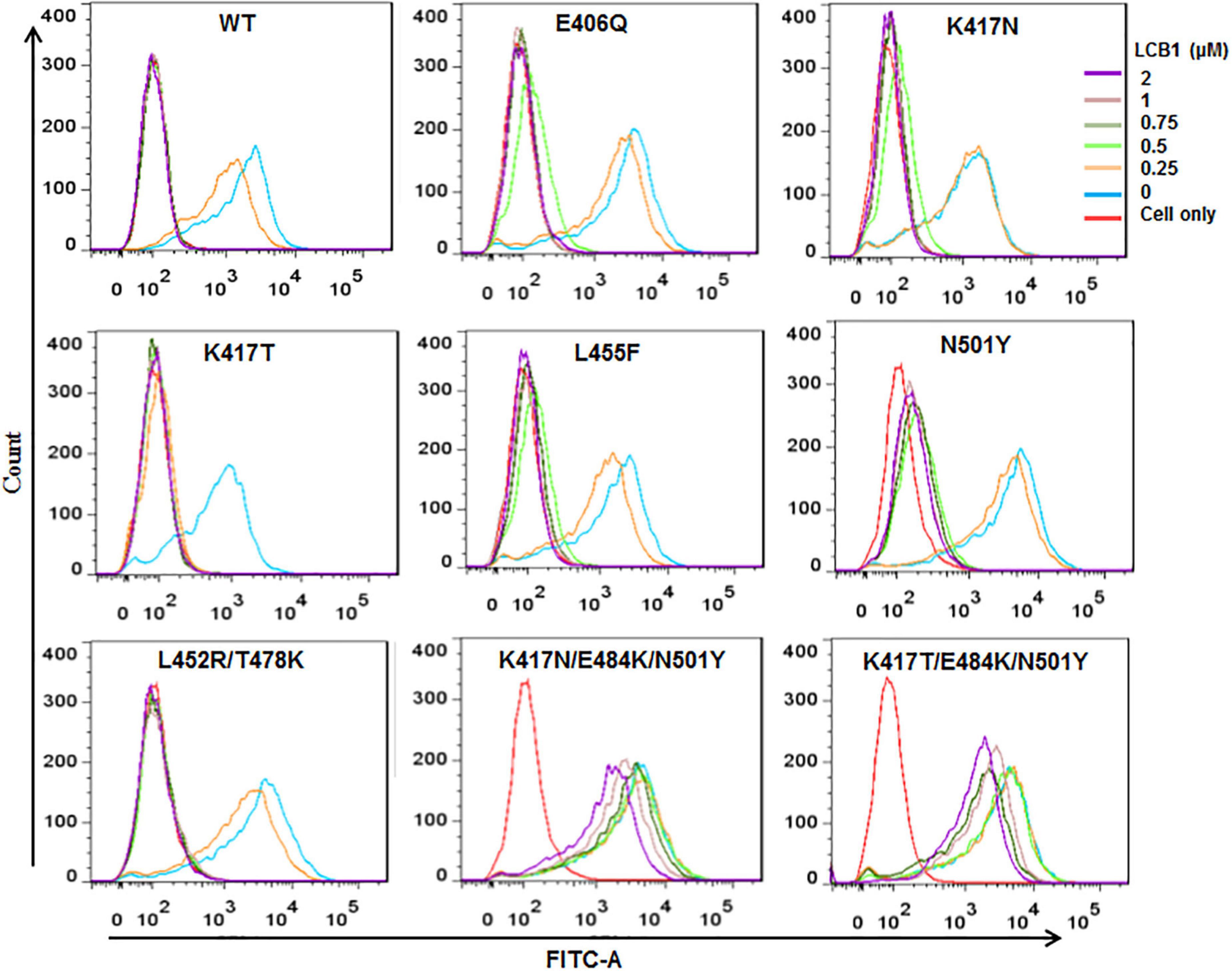
Figure 4. The blocking activity of LCB1 on the binding of RBD protein with the cell receptor ACE2. An His-tagged RBD protein was incubated with serially diluted LCB1 and then added to 293T/ACE2 cells for incubation. The bound RBD was detected by an Alexa Fluor 488-labeled rabbit anti-His tag antibody with FACS analysis.
In this study, we focused on characterizing LCB1 inhibitor for its antiviral activities against divergent SARS-CoV-2 variants and achieved significant findings. First, we successfully synthesized and purified a 56-mer LCB1 peptide, which exhibited high α-helicity, thermostability and antiviral activity. Second, we found that while the Alpha, Delta, and Lambda variants only caused mild resistance to LCB1, the Beta, Gamma, and Omicron variants were highly resistant to the LCB1 inhibition, giving the resistance fold-changes greater than 15,000 or 3,000. Third, with a panel of SARS-CoV-2 mutants carrying the single, double or triple mutations contained in VOCs, K417N, and N501Y in the spike RBD were verified to be critical determinants for the observed resistance phenotype. Fourth, we further generated a large panel of 85 naturally occurring RBD point-mutations, which identified E406Q, K417T and L455F conferring high-levels of resistance, whereas Y505W was responsible for a mild resistance. Moreover, the results by BLI and flow cytometry demonstrated that the resistance mutations could greatly impair the RBD-binding affinity of LCB1 and thus attenuated its blocking function on the RBD-ACE2 interaction. In conclusion, our studies highlight the resistance profiles of divergent SARS-CoV-2 VOCs and natural RBD mutants as well as the underlying mechanisms, which would facilitate the development of LCB1-based antiviral drugs.
Since its first report, LCB1 inhibitor has specially attracted our attention due to its computer-aided design strategy as a miniprotein and high inhibitory potency on SARS-CoV-2 infection by targeting the spike RBD to block virus entrance (Cao et al., 2020). We are also encouraged by the preventive and therapeutic efficacies of modified LCB1 proteins in animal infection models (Case et al., 2021). Unfortunately, SARS-CoV-2 spread with persistent mutations, resulting in the emergence of divergent VOCs and VOIs that caused new waves of worldwide epidemic. Significantly, many of evolved mutations in RBD can shape the binding conformation and interacting affinity of S protein with the cell receptor ACE2 and thus affect virus’s biological properties, e.g., transmission ability and disease severity. The mutations have also affected the performances of vaccines, therapeutic drugs, and diagnostic tools significantly (Garcia-Beltran et al., 2021; Hoffmann et al., 2021; Kuzmina et al., 2021; Fan et al., 2022; Guo et al., 2022; Sun et al., 2022; Zhu et al., 2022). Omicron has emerged as the fifth VOC after Alpha, Beta, Gamma, and Delta, and it has evolved into distinct lineages: BA.1 (B.1.1.529) was responsible for the initial surge but almost replaced by BA.2 in April 2022; while BA.3 remains at low frequency, BA.4 and BA.5 have currently replaced BA.2 and are becoming prevalent globally (Qu et al., 2022; Tegally et al., 2022). In protein level, BA.4 and BA.5 bear identical S proteins, being most comparable to BA.2 but have additional mutations (Δ67-70, L452R, F486V) and wild type amino acid at position Q493, which critically determine their increased fitness and immune evasion than the earlier BA.1 and BA.2 lineages (Cao et al., 2022). In this study, we identified that single K417N or K417T mutation is a key determinant to the resistance, which occurred in Beta, Gamma and Omicron (VOCs highly resistant to LCB1) but not in Alpha, Delta, and Lambda variants (VOCs or VOI mildly resistant to LCB1). Except for Delta, other four VOCs have an N501Y mutation, which is responsible for a low resistance level when being presented in the Alpha variant alone but can markedly boost the resistance levels of three highly resistant VOCs as a combined mutation with K417N/T. The Delta variant evolved with two RBD mutations (T478K and L452R); while T478K was not associated with the LCB1 resistance, L452R did rendered the virus slightly resistant, as evidenced by ∼2-fold IC50 changes (Table 2). L452R also appeared in Delta, Omicron, and many other variants, such as Kappa (B.1.617.1), Epsilon (B.1.429), and B.1.617.3. Our studies with the large panel of naturally occurring RBD mutants also found E406Q, L455F, and Y505W being resistant to LCB1, which are not present in the current VOCs. Given that all the characterized mutations in LCB1-resistant VOCs, including K417N, K417T, T478K, L452R, and N501Y, naturally emerged during the evolution process, whether E406Q, L455F, and Y505W mutations will appear in future VOCs to cause LCB1 resistance is an intriguing question.
In order to overcome the resistance problem by LCB1, the same group of authors continued their efforts to design multivalent proteins by using a cell-free expression workflow, which combines an in vitro DNA assembly step followed by polymerase chain reaction (PCR) to generate linear expression templates that are used to drive cell-free protein synthesis and enable rapid prototyping of new minibinder designs (Hunt et al., 2022). Promisingly, a number of constructs containing LCB1 or other miniproteins displayed dramatically increased binding and inhibitory activities against divergent VOCs and related mutant viruses, and of them a homo-trimeric version of 75-residue ACE2 mimic AHB2 (TRI2-2), which was designed to geometrically match the trimeric spike architecture, conferred prophylactic and therapeutic protection against SARS-CoV-2 challenge when administered intranasally in mice, providing evidence to support LCB1-based drug development with high and broad-spectrum antiviral activities (Hunt et al., 2022). In addition to developing viral entry inhibitors that target membrane fusion step (Zhu et al., 2019, 2020, 2021, 2022; Yu et al., 2021a,b; Xue et al., 2022), our research team is also working to rationally construct LCB1-based inhibitors that have resistance to viral escape and antigenic drift, and several bispecific fusion proteins have been validated with highly potent activity against diverse SARS-CoV-2 VOCs and mutants (unpublished data). Very recently, it was also reported that the length of LCB1 could be reduced to 35 amino acids without interfering its inhibitory capacity, providing a flexible template for LCB1-based design strategies (Weissenborn et al., 2022).
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
TW, YZ, NL, YH, and HC performed the experiments. TW, YZ, and YXH analyzed the data. YH supervised the study and wrote the manuscript with TW and YZ. All authors read and approved the submitted version.
This work was supported by grants from the CAMS Innovation Fund for Medical Sciences (2021-I2M-1037) and the National Natural Science Foundation of China (82002150 and 81630061).
We thank Linqi Zhang at the Tsinghua University (Beijing, China) for providing the plasmids encoding the mutant S proteins of SARS-CoV-2 VOCs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1022006/full#supplementary-material
Supplementary Figure 1 | Synthesis and characterization of LCB1 peptide. (A) LCB1 was chemically synthesized on rink amide 4-methylbenzhydrylamine (MBHA) resin using a standard solid-phase 9-flurorenylmethoxycarbonyl (FMOC) and its purity was determined by reverse-phase HPLC. (B) The peptide was characterized by mass spectrometry, indicating a molecular weight of 6,852.5Da.
Supplementary Figure 2 | Infectivity of divergent SARS-CoV-2 variants. The infectivity of SARS-CoV-2 pseudoviruses with single or multiple mutations on 293T/ACE (A) or Huh-7 (B) cells was determined by a single-cycle infection assay. D614G mutant was treated as a reference, thus its luciferase activity (RLU) was standard as 100% and the relative infectivity of various mutants was calculated accordingly. The experiments were repeated at least three times and columns are expressed as the means ± SD. Statistical analysis was conducted to compare the differences between the D614G reference and diverse mutants.
Supplementary Figure 3 | Infectivity of naturally occurring SARS-CoV-2 mutants. The infectivity of 85 SARS-CoV-2 pseudoviruses carrying natural RBD point-mutations was determined on 293T/ACE (A) or Huh-7 (B) cells by a single-cycle infection assay. Similarly, D614G mutant was treated as a reference, thus its luciferase activity (RLU) was standard as 100% and the relative infectivity of various mutants was calculated accordingly. The experiments were repeated at least three times and columns are expressed as the means ± SD. Statistical analysis was conducted to compare the differences between the D614G reference and diverse mutants.
Callaway, E. (2021). Heavily mutated Omicron variant puts scientists on alert. Nature 600:21. doi: 10.1038/d41586-021-03552-w
Cao, L., Goreshnik, I., Coventry, B., Case, J. B., Miller, L., Kozodoy, L., et al. (2020). De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431. doi: 10.1126/science.abd9909
Cao, Y., Yisimayi, A., Jian, F., Song, W., Xiao, T., Wang, L., et al. (2022). BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608, 593–602. doi: 10.1038/s41586-022-04980-y
Case, J. B., Chen, R. E., Cao, L., Ying, B., Winkler, E. S., Johnson, M., et al. (2021). Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease. Cell Host Microbe 29, 1151–1161e1155. doi: 10.1016/j.chom.2021.06.008
Fan, Y., Li, X., Zhang, L., Wan, S., Zhang, L., and Zhou, F. (2022). SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 7:141. doi: 10.1038/s41392-022-00997-x
Garcia-Beltran, W. F., Lam, E. C., St Denis, K., Nitido, A. D., Garcia, Z. H., Hauser, B. M., et al. (2021). Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 184, 2372–2383e2379. doi: 10.1016/j.cell.2021.03.013
Guo, Y., Han, J., Zhang, Y., He, J., Yu, W., Zhang, X., et al. (2022). SARS-CoV-2 Omicron Variant: Epidemiological Features, Biological Characteristics, and Clinical Significance. Front. Immunol. 13:877101. doi: 10.3389/fimmu.2022.877101
He, X., Hong, W., Pan, X., Lu, G., and Wei, X. (2021). SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm 2, 838–845. doi: 10.1002/mco2.110
Hoffmann, M., Arora, P., Gross, R., Seidel, A., Hornich, B. F., Hahn, A. S., et al. (2021). SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184, 2384–2393e2312. doi: 10.1016/j.cell.2021.03.036
Hunt, A. C., Case, J. B., Park, Y. J., Cao, L., Wu, K., Walls, A. C., et al. (2022). Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Sci. Transl. Med. 14:eabn1252. doi: 10.1126/scitranslmed.abn1252
Kuzmina, A., Khalaila, Y., Voloshin, O., Keren-Naus, A., Boehm-Cohen, L., Raviv, Y., et al. (2021). SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe 29, 522–528e522. doi: 10.1016/j.chom.2021.03.008
Markov, P. V., Katzourakis, A., and Stilianakis, N. I. (2022). Antigenic evolution will lead to new SARS-CoV-2 variants with unpredictable severity. Nat. Rev. Microbiol. 20, 251–252. doi: 10.1038/s41579-022-00722-z
Plavec, Z., Pohner, I., Poso, A., and Butcher, S. J. (2021). Virus structure and structure-based antivirals. Curr. Opin. Virol. 51, 16–24.
Qu, P., Faraone, J., Evans, J. P., Zou, X., Zheng, Y. M., Carlin, C., et al. (2022). Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. N. Engl. J. Med. 386, 2526–2528. doi: 10.1056/NEJMc2206725
Sabbah, D. A., Hajjo, R., Bardaweel, S. K., and Zhong, H. A. (2021). An Updated Review on Betacoronavirus viral entry inhibitors: Learning from past discoveries to Advance COVID-19 drug discovery. Curr. Top. Med. Chem. 21, 571–596. doi: 10.2174/1568026621666210119111409
Sun, C., Xie, C., Bu, G. L., Zhong, L. Y., and Zeng, M. S. (2022). Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants. Signal Transduct. Target. Ther. 7, 202. doi: 10.1038/s41392-022-01039-2
Tao, K., Tzou, P. L., Nouhin, J., Gupta, R. K., de Oliveira, T., Kosakovsky Pond, S. L., et al. (2021). The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 22, 757–773.
Tegally, H., Moir, M., Everatt, J., Giovanetti, M., Scheepers, C., Wilkinson, E., et al. (2022). Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. [Online ahead of print] doi: 10.1038/s41591-022-01911-2
Weissenborn, L., Richel, E., Huseman, H., Welzer, J., Beck, S., Schafer, S., et al. (2022). Smaller, Stronger, More Stable: Peptide Variants of a SARS-CoV-2 neutralizing miniprotein. Int. J. Mol. Sci. 23:6309. doi: 10.3390/ijms23116309
Xiang, Y., Wang, M., Chen, H., and Chen, L. (2021). Potential therapeutic approaches for the early entry of SARS-CoV-2 by interrupting the interaction between the spike protein on SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2). Biochem. Pharmacol. 192:114724. doi: 10.1016/j.bcp.2021.114724
Xue, J., Chong, H., Zhu, Y., Zhang, J., Tong, L., Lu, J., et al. (2022). Efficient treatment and pre-exposure prophylaxis in rhesus macaques by an HIV fusion-inhibitory lipopeptide. Cell 185, 131–144e118. doi: 10.1016/j.cell.2021.11.032
Yamasoba, D., Kimura, I., Nasser, H., Morioka, Y., Nao, N., Ito, J., et al. (2022). Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 185, 2103–2115e2119. doi: 10.1016/j.cell.2022.04.035
Yan, W., Zheng, Y., Zeng, X., He, B., and Cheng, W. (2022). Structural biology of SARS-CoV-2: open the door for novel therapies. Signal Transduct. Target Ther. 7:26. doi: 10.1038/s41392-022-00884-5
Yang, H., and Rao, Z. (2021). Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 19, 685–700. doi: 10.1038/s41579-021-00630-8
Yu, D., Zhu, Y., Jiao, T., Wu, T., Xiao, X., Qin, B., et al. (2021a). Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants. Emerg. Microbes Infect. 10, 1227–1240. doi: 10.1080/22221751.2021.1937329
Yu, D., Zhu, Y., Yan, H., Wu, T., Chong, H., and He, Y. (2021b). Pan-coronavirus fusion inhibitors possess potent inhibitory activity against HIV-1, HIV-2, and simian immunodeficiency virus. Emerg. Microbes Infect. 10, 810–821. doi: 10.1080/22221751.2021.1917309
Zhu, Y., Chong, H., Yu, D., Guo, Y., Zhou, Y., and He, Y. (2019). Design and Characterization of Cholesterylated Peptide HIV-1/2 Fusion Inhibitors with Extremely Potent and Long-Lasting Antiviral Activity. J. Virol. 93, e02312–e02318. doi: 10.1128/JVI.02312-18
Zhu, Y., Dong, X., Liu, N., Wu, T., Chong, H., Lei, X., et al. (2022). SARS-CoV-2 fusion-inhibitory lipopeptides maintain high potency against divergent variants of concern (VOCs) including Omicron. Emerg. Microbes Infect. 11, 1819–1827. doi: 10.1080/22221751.2022.2098060
Zhu, Y., Yu, D., Hu, Y., Wu, T., Chong, H., and He, Y. (2021). SARS-CoV-2-derived fusion inhibitor lipopeptides exhibit highly potent and broad-spectrum activity against divergent human coronaviruses. Signal Transduct. Target. Ther. 6:294. doi: 10.1038/s41392-021-00698-x
Keywords: SARS-CoV-2, variants of concern, entry inhibitor, LCB1, drug resistance
Citation: Wu T, Zhu Y, Liu N, Hu Y, Chong H and He Y (2022) Resistance profile and mechanism of severe acute respiratory syndrome coronavirus-2 variants to LCB1 inhibitor targeting the spike receptor-binding motif. Front. Microbiol. 13:1022006. doi: 10.3389/fmicb.2022.1022006
Received: 18 August 2022; Accepted: 30 August 2022;
Published: 11 October 2022.
Edited by:
Wanbo Tai, Shenzhen Bay Laboratory, ChinaReviewed by:
Fei Yu, Hebei Agricultural University, ChinaCopyright © 2022 Wu, Zhu, Liu, Hu, Chong and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxian He, eWhlQGlwYi5wdW1jLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.