
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 09 November 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1021837
This article is part of the Research Topic Insights in Microbial Immunology: 2022 View all 9 articles
 Ang Hu1,2†
Ang Hu1,2† Zeming Hu3†
Zeming Hu3† Haohong Zou1,2†
Haohong Zou1,2† Jiankang Zhang1
Jiankang Zhang1 Dongliang Zhang1
Dongliang Zhang1 Hao Wang1
Hao Wang1 Jianing Zhong2*
Jianing Zhong2* Bin Chen1*
Bin Chen1*Microbial infection, caused by fungi, bacteria, viruses, and parasites, significantly contributes to the global death burden and health costs. The innate and adaptive immune systems orchestrate a multifaceted signaling response to invading pathogens as the human antimicrobial system. In this process, caspase recruitment domain-containing protein 9 (CARD9) emerges as a critical intermediary adaptor molecule to participate in regulating a series of antimicrobial immune reactions. Previous publications have confirmed that CARD9 plays a crucial role in fungal, bacterial, viral, and parasitic infections. In this study, we aim to provide an update on the recent clinical and basic studies where the mechanism and function of CARD9 have been further studied and understood. In addition, we summarize the latest treatment and prevention strategies based on CARD9 and discuss the current perspectives and future direction of CARD9.
Caspase recruitment domain-containing protein 9 (CARD9), a novel member of the CARD family, was originally discovered through Millennium Pharmaceuticals' proprietary database searches with previously known CARD family proteins (Bertin et al., 2000). The CARD family is the second largest family of the death domain (DD) superfamily and is characterized by the presence of a caspase-associated recruitment domain (Kao et al., 2015). Of these, the protein composition of CARD9, including the N-terminal CARD domain, coiled-coil domain, and C-terminal tail, is shown in Figure 1 (Ruland and Hartjes, 2019). The N-terminal CARD region (amino acids 7–98) is a protein–protein interaction module that binds to CARD-containing apoptosis proteins to mediate intracellular signaling cascades and regulate cell apoptosis responses to an altered extracellular environment (Bertin et al., 2000). The coiled-coil region (amino acids 140–420) takes charge of the process of protein oligomerization, forming heptad repeats labeled “abcdefg,” with a and d denoting the hydrophobic residues (Hara and Saito, 2009). The C-terminal tail is the designated site where ubiquitin ligase TRIM62 interacts with CARD9. TRIM62 is responsible for the ubiquitination of lysine 125, which is a requisite for the activation of CARD9 (Cao et al., 2015). CARD-containing membrane-associated guanylate kinase (MAGUK/CRAMA) proteins family, including CARD11 (CARMA1), CARD10 (CARMA2), and CARD14 (CARMA3), has been proposed to have a similar function to CARD9 (Bertin et al., 2001; Wang et al., 2001). However, the difference lies in protein structure because CARD9 lacks the C-terminal MAGUK region, which contains PDZ, Src Homology 3, and the Guanylate Kinase domain (Jiang and Lin, 2012). CARD9 is a protein of 536 amino acids with a predicted molecular weight of 62.3 kDa and has been mapped to the 9q34.3 chromosomal region (Bertin et al., 2000). Figure 1 provides the CARD9 protein and mRNA expression of various human tissues, indicating that high expression is mainly observed in immune-related tissues. At the cellular level, CARD9 is abundantly expressed in myeloid cells, such as monocytes, neutrophils, macrophages, and dendritic cells (Hsu et al., 2007).
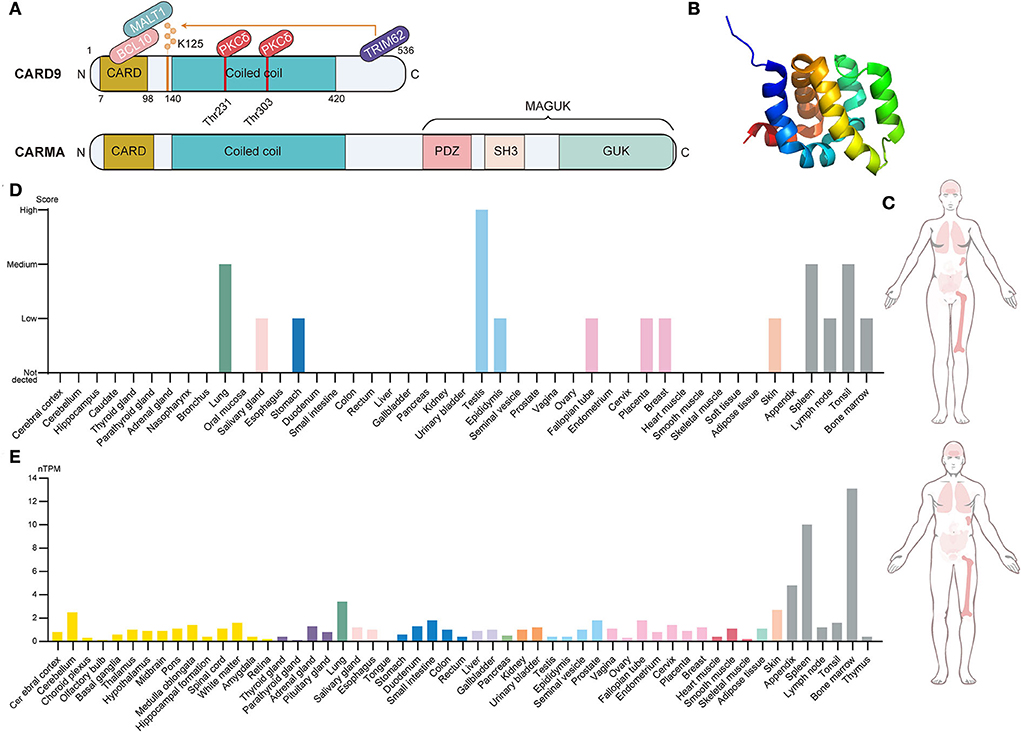
Figure 1. Brief introduction to CARD9. (A) Structural features, protein-interacting domains, and post-translational modification sites of CARD9 and CARMA. CARD9 and CARMA family have a similar structure in which the N-terminal CARD domain, the coiled-coil domain, and the C-terminal tail are shared. Apart from these, CARMA still contains the C-terminal MAGUK region, which includes the PDZ domain, SH3 domain, and GUK domain. PKCδ-mediated phosphorylation (Thr 231 and Thr 303) and TRIM62-mediated ubiquitination (Lys 125) are required for the activation of CARD9. BCL10 interacts with activated CARD9 on the homologous CARD domain, and then they bind to MALT1 to induce the assembly of the trimolecular complex. (B) The NMR solution structure of the CARD9 CARD. Image from the RCSB PDB (RCSB PDB accession code PDB 6E26). (C) Integrated RNA and protein expression of CARD9 in the various normal tissues of a different gender. The color shade of the tissues represents the extent of expression (Data from The Human Protein Atlas). (D) CARD9 protein expression overview categorized by the system. Color coding is based on tissue groups that contain tissues with functional features in common (Data from The Human Protein Atlas). (E) CARD9 RNA expression overview categorized by the system. Color coding is based on tissue groups that contain tissues with functional features in common. nTPM represents the normalized expression of RNA (Data from The Human Protein Atlas). MAGUK, membrane-associated guanylate kinase; SH3, Src Homology 3; GUK, Guanylate Kinase; MALT, mucosa-associated lymphoid tissue lymphoma; BCL10, B cell lymphoma 10; PKCδ, protein kinase Cδ.
Generally, CARD9 acts as a crucial downstream member of pattern recognition receptors (PRRs) and mediates a series of inflammatory cascades against invasive fungi, bacteria, viruses, and parasites. Mutation of the CARD9 gene correlating with lower expression and function loss is an autosomal recessive primary immunodeficiency disorder, and it predisposes individuals to microbial infection. The downstream of PRRs, PRRs/Syk/CARD9, is one of the most classic signaling pathways. When PRRs recognize specific microbial pathogens, PPRs recruit spleen tyrosine kinase (Syk) to activate the protein kinase Cδ (PKCδ), which can coordinate with VAV to phosphorate CARD9 at positions Thr 231 and 303 in the coiled-coil protein domain. Phosphorated CARD9 induces the formation of multiple protein complexes termed CBM in the cytoplasm, which consists of CARD9, B cell lymphoma 10 (BCL10), and mucosa-associated lymphoid tissue lymphoma (MALT1) and acts as a signaling integration center (Hartjes and Ruland, 2019). Under the control of CBM, the nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways are initiated to regulate multiple cellular processes (described below) in downstream cascades (Drummond et al., 2018). A more detailed introduction to the CARD9 signaling network can be seen in the studies by Roth and Ruland (2013) and Wang et al. (2020). At present, the relationship between CARD and host immunity has been extensively studied. Many emerging studies have broadened the scope of the known mechanisms and improved our understanding of the function of CARD9 in microbial infection. Herein, we briefly describe previous findings and review recent studies to further elucidate the inner relationship between CARD9 and infection immunity.
It is well acknowledged that CARD9 and its related signaling network play a pivotal role in the immunology of fungal infection (Drummond et al., 2018). Mechanistically, the C-type lectin receptor (CLR) family, including Dectin-1, Dectin-2, Dectin-3, and Mincle, initiates the signaling cascades against the fungal invasion through ligand-receptor binding. Studies have mainly concentrated on Dectin-1, which can be recognized by fungus components, although the downstream Syk-dependent pathway identifies all these receptors. Indeed, the Syk/CARD9/CBM cascade and the following NF-κB and MAPK signaling pathways are defined as the main effector signaling networks against fungal invasion.
Earlier studies have found that a deficiency of CARD9 makes many fungi susceptible (Wu et al., 2018), such as Cryptococcus neoformans (Yamamoto et al., 2014) and Phialophora verrucose (Wu et al., 2016). However, recent studies have further broadened this range by proving the susceptibility to Aspergillus (Zhang et al., 2021), Pneumocystis (Kottom et al., 2020b), Mucor irregularis (Sun et al., 2021), and Rhizopus arrhizus (Sun et al., 2019). For instance, Guo et al. (2019) reported that individuals with homozygous CARD9 alleles (C/C) were more susceptible to Pallidocercospora crystalline compared to those with heterozygous (G/C) or wild-type (G/G) CARD9 alleles. Another report identified that 17 patients with deep dermatophytosis from Tunisian, Algerian, and Moroccan families displayed autosomal recessive CARD9 deficiency (Lanternier et al., 2013). Furthermore, Rieber et al. (2016) showed that CARD9 deficiency was the first known inherited or acquired a status that was predisposed to extrapulmonary aspergillosis with sparing of the lungs.
Given the association between CARD9 and fungal susceptibility, more and more biologists and medical scientists are struggling to elucidate this intricate mechanism. Recently, Kottom et al. (2020a) performed a comprehensive analysis of the susceptibility of CARD9 KO and immunocompetent hosts to Pneumocystis. They demonstrated that this defect was attributable to a compromised proinflammatory response, in which CARD9−/−macrophages could not perform cell differentiation and express polarization markers, thereby failing to upregulate the expression of CLRs. Intriguingly, the CARD9 KO mice not only exhibited an intact T-helper (Th) cell cytokine but displayed a normal survival rate, the same as that of wild-type (WT) mice. The probable explanation is that the modulation of Th cytokine may be CARD9-independent, and low-level inflammatory response may ameliorate lung injury. Wang et al. (2018) identified a novel CARD9 mutation (p.S23X) in patients susceptible to dematiaceous fungi. They found the impairment of proinflammatory cytokine and chemokine production, neutrophil recruitment, pathway activation (NF-κB and MAPK), and Th-associated (Th 17 and Th 22) responses in patients with CARD9 deficiency. Remarkably, neutrophil phagocytosis and reactive oxygen species (ROS) production remained normal. Similarly, in a study of susceptibility to M. irregularis, the release of neutrophil extracellular traps (NETs) and the expression of IL-6 and TNF-α showed an impaired state in neutrophils, whereas intact phagocytosis and ROS production were observed (Sun et al., 2021). Different from the impaired neutrophil antifungal immunity, the functions of phagocytosis and ROS production are seemingly conflicting. Indeed, these results were in line with the previous studies in which the same situation of Phialophora verrucosa and Candida albicans infection were reported. This may be because both of these functions are free of the control of CARD9 (Gazendam et al., 2014; Liang et al., 2015). Generally, neutrophils constitute the first line of defense against fungal invasion, in which infiltration, phagocytosis, ROS, and NETs participate. Local infection provides the necessary CXC signal to induce neutrophil recruitment indirectly, in which CARD9 has been proven to play a crucial part. Recent studies found that the neutrophil-targeting chemokines CXCL1 and CXCL2 were a low expression in Card9−/− mice, and CARD9 could promote the neutrophils chemotaxis by regulating the production of neutrophil-recruiting IL-1β and CXCL1 in the central nervous system, thereby confirming the importance of CARD9 in neutrophil-induced immune response (Drummond et al., 2019; Sun et al., 2021). In addition to these, the increased susceptibility of Card9−/− mice to R. arrhizus and M. irregularis were reported as well, and they were attributed to impaired cytokine and chemokine production, NF-κB (p65) activation, and Th cell differentiation (Sun et al., 2019, 2021).
B-cell-mediated humoral immunity acts as another crucial line of antifungal defense, in which CARD9 shows an emerging regulatory role. Recently, Doron et al. (2021) identified C. albicans as the antibodies preferentially targeted species and the main trigger of the production of fungicidal IgG. They found that colonization of intestinal fungi depends on CARD9 and CARD+CX3CR1+ macrophages to regulate the generation of antifungal antibodies, proving that the loss of CARD9 functionality induced by mutation was connected with disturbed IgG reaction. However, C. albicans is known as a beneficial commensal in the human gut, although it offers a classic function model of CARD9 in humoral immunity. Further exploration of the humoral immunity of detrimental and invasive fungi is needed to provide more intuitive insights into CARD9 and fungal infection.
Current studies have provided insight into the intricate regulation network and protein interactions of CARD9. For example, Loh et al. (2019) reported that Dok3 coordinated with protein phosphatase 1 (PP1) to dephosphorylate CARD9, ultimately inhibiting the downstream NF-κB and JNK signaling pathway activation and antifungal immune response. Moreover, Tartey et al. (2018) revealed that a previously unknown cross-talk between CARD9 and SHP-1 modulated IL-1α-induced signaling cascade and inflammation response in a mouse model of neutrophilic dermatoses. These studies further enrich the functional mechanisms of CARD9 while indicating its future research direction.
Taken together, CARD9 is an indispensable regulator in antifungal immunity, and CARD deficiency is generally accompanied by fungal susceptibility (new mutations of CARD9 over the past 4 years are listed in Table 1). Potential reasons could be dampened cytokine production, Th-cell differentiation, neutrophil immunity, and pathway activation (Vornholz and Ruland, 2020; Wang et al., 2020; Sheng et al., 2021). Although most of these mechanisms have been documented in previous publications, it is necessary to further explore other fungal species because species-specific mechanisms exist in certain fungi, such as Candida parapsilosis and C. albicans (Zajta et al., 2021). Notably, an emerging view indicates that Syk has a more eminent impact on anti-Candida immunity than CARD9 in the classic Syk/CARD9 antifungal pathway due to the presence of Syk-dependent but CARD9-independent antifungal signaling (Zajta et al., 2021). Therefore, this viewpoint may prompt a new research direction targeting CARD9 and Syk, which are mutually independent mechanisms.
The antibacterial signaling cascades of CARD9 are initiated by toll-like receptors (TLRs), a type of PRR. TLR2 and TLR4 are two main members of the TLR family that recognizes the components of gram-positive and gram-negative bacteria, respectively. For instance, muramyl-dipeptide (MDP) and peptidoglycan for the former (Akira et al., 2006; Kawai and Akira, 2010), and lipopolysaccharide (LPS) for the latter (Kawai and Akira, 2010). Once recognized, TLRs collaborate with myeloid differentiation primary response 88 (MyD88) to recruit interleukin-1 receptor-associated kinase (IRAK1) and receptor-interacting protein 2 (RIP2) before inducing the assembly of the CBM protein complex (Kobayashi et al., 2002; Dong et al., 2006). Nucleotide-binding oligomerization domain 2 (Nod2) interacts with CARD9 to promote the recognition of MDP and the activation of the JNK and p38 signaling pathways and ultimately regulates the production of inflammatory cytokines against bacterial infection. Meanwhile, classic CLR signaling also participates in antibacterial immunity, which is further proved by several recent studies. Previous publications have revealed that Dectin-2 served as a critical regulator in host immunity against Streptococcus pneumoniae infection by affecting phagocytosis of neutrophils but excluding the recruitment of neutrophils (McGreal et al., 2006; Akahori et al., 2016). At present, Ishizuka et al. (2020) found that CARD9 KO mice other than dectin-2 KO mice showed impaired neutrophil recruitment and decreased inflammatory cytokine and chemokine production compared to respective control mice. They indicated that CARD9-mediated signaling was indispensable in anti-pneumococcal immunity through modulating neutrophil function and cytokine production, in which both neutrophil phagocytosis and neutrophil accumulation required the participation of CARD9. However, the regulatory signaling of the former was initiated by Dectin-2, while that of the latter was initiated by another non-Dectin-2 CLR or an unidentified CLR. Notably, Mincle may be a promising candidate for this due to its ability to recognize glucosyl-diacylglycerol, a key component of S. pneumoniae (Behler-Janbeck et al., 2016). Moreover, Prado Acosta et al. (2021) identified Mincle as a receptor for the surface (S)-layer of Lactobacillus brevis. They found that S-layer/Mincle interaction induced a balanced immune response between pro- and anti-inflammatory cytokines through the Mincle/Syk/CARD9 axis. Under this circumstance, the deletion of Mincle, CARD9, and Syk disturbed the balance, resulting in upregulated proinflammatory and downregulated anti-inflammatory cytokines. Meanwhile, an altered CD4+ T cell priming capacity was observed under the Mincle knockdown, implying this signaling pathway was also involved in regulating cellular immunity. In most instances, CARD9 acts as a critical downstream adaptor molecule for signal transduction triggered by PRRs. However, PRRs can still initiate CARD9-independent antibacterial signaling, which interweaves with a CARD9-dependent mechanism to exert a bactericidal effect (Akahori et al., 2016). Therefore, further investigation is still necessary to define the different mechanisms of different receptors.
Although the composition of the virus is merely genetic material and essential enzyme, some PRRs, including DC-SIGN, L-SIGN, Langerin, MMR, DCIR, MDL-1, LSECtin, MGL, TLR2, TLR3, TLR4, TLR7, TLR8, and TLR9, has been confirmed to involve in recognition of viral components (Drummond et al., 2018; Zhou et al., 2021). As the classic downstream molecule of PRR signaling, CARD9 has been acknowledged to play a key role in viral infection. For example, CARD9 interacted with DNA sensor Rad50 and dsDNA to induce the assembly of the dsDNA-Rad50-CARD9 complex. It ultimately caused the activation of NF-κB and the generation of a pro-IL-1β response to the virus (Roth et al., 2014). A similar mechanism can also be observed in RNA virus recognition by RIG-I, which, together with MAVS, CARD9, and Bcl-10, for NF-κB activation (Poeck et al., 2010). Recently, Monteiro et al. (2019) recognized Dectin-1, Dectin-2, and Mincle as several potential receptors to interact with the La Crosse virus (LACV). The evidence is that LACV infection of Mincle−/− and CARD9−/− DCs displayed impaired proinflammatory cytokine production, including IL-6 and TNF-α. However, the deletion of CARD9 and Mincle did not alter the elimination of LACV. Therefore, they concluded that the Mincle/CARD9 axis played an indispensable role in early anti-LACV immunity. In parallel, Pavasutthipaisit et al. (2021) reported that CARD9 had a limited effect on early antiviral reactions and encephalomyelitis virus (TMEV) removal. In their study, the knockout of CARD9 eliminated the production of proinflammatory cytokines and the infiltration of T and B cells. It even triggered a temporary increase of TMEV-infected cells in the brain. Moreover, they still confirmed that CARD9 deficiency did not affect the initiation of CD8+ T cell response. In a Coxsackievirus B3 (CVB3)-induced viral myocarditis mouse model, CARD9 deletion downregulated the mRNA and protein expression of TGF-β, IL-17A, and BCL-10, thereby ameliorating the CARD9-involved potent inflammation response (Sun et al., 2020). Based on these, a conflicting effect of CARD9 in viral infection is unfolding before our eyes. On the one hand, it can modulate the host's innate immune response to viral infection. On the other hand, it can cause severe viral infection and potential immune-pathological damage.
TLR2, TLR4, TLR9, TLR11, Dinctin-2, SIGNR5, and Mincle have been identified as the prominent sensor of parasite pathogens (Ashour, 2015; Kalantari et al., 2018). SIGNR5 has been reported to coordinate with Dectin-2 and Mincle to initiate the FcRγ-Syk-CARD9 signaling and then promote IL-1β and IL-23 production and Th 17-induced immunity against Schistosome Egg (Kalantari et al., 2018). In Toxoplasma gondii, Pandori et al. (2019) found that the Syk-CARD9/MALT-1-NF-κB signaling pathway was activated to regulate NLRP3 inflammasome-mediated IL-1β production in human monocytes against pathogens. Intriguingly, IL-1β is released by monocytes in general upon gasdermin-D-induced pyroptotic cell death (Cookson and Brennan, 2001). In contrast, T. gondii was shown to initiate the release of IL-1β in a pyroptosis-free and gasdermin-D independently. Moreover, Maknitikul et al. (2018) reported that the CARD9 signaling pathway regulates host immune response induced by hemozoin in Plasmodium falciparum-associated acute lung injury, such as the production of IL-1β and the apoptosis of type II alveolar cells. However, a previous publication showed CARD9 did not participate in the Plasmodium berghei-induced pathology in experimental cerebral malaria. Their evidence is that similar pathologic features, including impaired blood-brain barrier, elevated proinflammatory response. Brain-infiltrating CD8+ cells were observed in both WT and Card9−/− mice, although the expression of CARD9 increased during P. berghei infection (Hafalla et al., 2012). Possible reasons for these ambivalent results lie in the tissue specificity between brain and lung and/or fungal species between P. falciparum and P. berghei. Studies on parasites are currently in the minority compared to studies on fungi, bacteria, and viruses. As a crucial regulator of host defense, CARD9 is worthy of more attention and investigation within the scope of antiparasitic immunity.
Considering the central role of CARD9 in the immune reaction against fungi, bacteria, viruses, and parasites (Figure 2), CARD9 has been proposed as a novel therapeutic and vaccine-developing target. Recently, Hung et al. (2018) designed a multivalent Coccidioides posadasii vaccine called “GCP-rCpa1,” which consists of recombinant Coccidioides polypeptide antigen (rCpa1), yeast cell wall particles, and oligonucleotide to strengthen the protective cellular immune response to fungal invasion. In their subsequent study, further investigation was conducted to reveal the fungal vaccine's protective effect and the underlying mechanism. They found that vaccinated mice showed the upregulation of proinflammatory cytokines (TNF-α, IL-6, IL-1β), which was connected with the activation of Dectin-1/CARD9 and Dectin-2/CARD9 signaling. Moreover, they reported that the macrophage production of inflammatory cytokines and the acquisition of Th cells, especially Th 17 cells, were impaired in vaccinated Dectin-1−/−, Dectin-2−/−, and CARD9−/− mice. Therefore, the mechanism by which GCP-rCpa1 activates the CLRs/CARD9 signaling pathway to initiate a potent Th 17 immune response was confirmed (Campuzano et al., 2020). Based on these, a more microbe-specific vaccine may be designed and developed by referring to the pattern of the GCP-rCpa1 vaccine. Regarding treatment, Kottom et al. (2020a) proposed that inhibiting the activity of CARD9 by pharmacological inhibitor BRD5529 might act as an underlying treatment strategy for Pneumocystis jirovecii via macrophage innate immune and anti-inflammatory activity. We have evidence that BRD5529 dramatically inhibited phospho-p38 and phospho-pERK1 signaling and the release of TNF-α upon Pneumocystis β-glucans exposure. In addition to BRD5529, currently known CARD9 inhibitors include BRD4203, BRD8991, and BRD4098 (Leshchiner et al., 2017), and they may also have the potential to become CARD9-targeted therapeutic drugs.
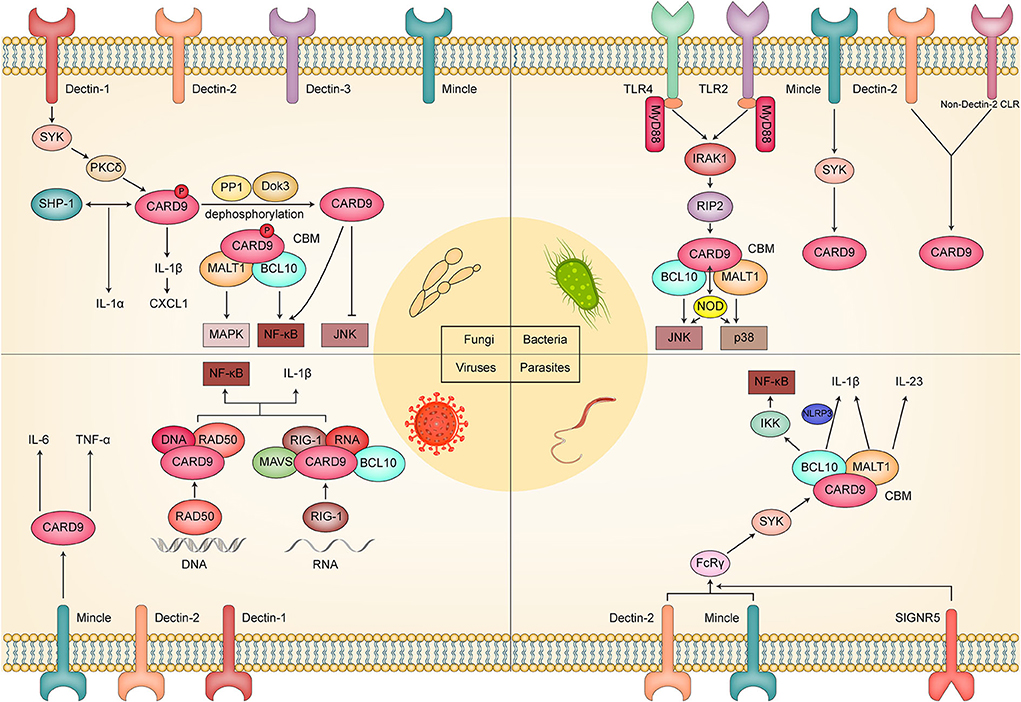
Figure 2. Schematic diagram of the molecular mechanism by which CARD9 plays a critical role in fungal, bacterial, viral, and parasitic infection.
Infections driven by microbial pathogens cause more than 400 million deaths worldwide each year, a higher burden than either cancer or cardiovascular disease (GBD 2016 Causes of Death Collaborators, 2017). Innate and adaptive immunity are necessary for host homeostasis and are two critical ways to remove invasive microbial pathogens. Therefore, the essential role of the immune response induced by PRRs signaling has gained much attention, and a crucial downstream adaptor molecule called “CARD9” is widely involved in this cascade. At present, CARD9 has shown an emerging role in the infection immunity of fungi, bacteria, viruses, and parasites; it is partly based on the results of CARD9 KO mice experiments (Table 2). However, there is still some controversy surrounding its mechanism and effect in this regard. For example, Syk-CARD9 signaling is known as the classic CARD9-associated way to resist fungal invasion. However, Syk-dependent and CARD9-independent, plus Syk-independent, and CARD9-dependent specific antifungal signals, are proven to play an important role in candidiasis infection as well (Zajta et al., 2021). Therefore, future studies must explore the inner relationship between specific and classic pathways more definitively. In addition, is CARD9-induced susceptibility to microbial infection widely applicable to these four species? Indeed, CARD9 deficiency is currently reported to induce susceptibility to bacteria in an animal model but not in a human organism. Standing at a crossroads between immunity and infection, CARD9 is worth studying to elucidate the commonalities and individualities of immune defense in the infectious process of fungi, bacteria, viruses, and parasites. Therefore, CARD9 may offer an ideal opportunity to improve our understanding of the process of microbial infection in the future.
AH, ZH, and HZ wrote the first draft of this article. JZha, DZ, and HW were responsible for searching the scientific literature and drawing the schematic diagram. JZho and BC strictly reviewed and edited the manuscript. All authors approved the final version submitted and agreed on its submission to this journal.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akahori, Y., Miyasaka, T., Toyama, M., Matsumoto, I., Miyahara, A., Zong, T., et al. (2016). Dectin-2-dependent host defense in mice infected with serotype 3 Streptococcus pneumoniae. BMC Immunol. 17, 1. doi: 10.1186/s12865-015-0139-3
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Ashour, D. S. (2015). Toll-like receptor signaling in parasitic infections. Expert Rev. Clin. Immunol. 11, 771–780. doi: 10.1586/1744666X.2015.1037286
Ba, H., Peng, H., Cheng, L., Lin, Y., Li, X., He, X., et al. (2021). Case report: Talaromyces marneffei infection in a chinese child with a complex heterozygous CARD9 Mutation. Front. Immunol. 12, 685546. doi: 10.3389/fimmu.2021.685546
Behler-Janbeck, F., Takano, T., Maus, R., Stolper, J., Jonigk, D., Tort Tarrés, M., et al. (2016). C-type lectin mincle recognizes glucosyl-diacylglycerol of Streptococcus pneumoniae and plays a protective role in pneumococcal pneumonia. PLoS Pathog. 12, e1006038. doi: 10.1371/journal.ppat.1006038
Bertin, J., Guo, Y., Wang, L., Srinivasula, S. M., Jacobson, M. D., Poyet, J. L., et al. (2000). CARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-kappa B. J. Biol. Chem. 275, 41082–41086. doi: 10.1074/jbc.C000726200
Bertin, J., Wang, L., Guo, Y., Jacobson, M. D., Poyet, J. L., Srinivasula, S. M., et al. (2001). CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J. Biol. Chem. 276, 11877–11882. doi: 10.1074/jbc.M010512200
Campuzano, A., Zhang, H., Ostroff, G. R., Dos Santos Dias, L., Wüthrich, M., Klein, B. S., et al. (2020). CARD9-associated dectin-1 and dectin-2 are required for protective immunity of a multivalent vaccine against Coccidioides posadasii infection. J. Immunol. 204, 3296–3306. doi: 10.4049/jimmunol.1900793
Cao, Z., Conway, K. L., Heath, R. J., Rush, J. S., Leshchiner, E. S., Ramirez-Ortiz, Z. G., et al. (2015). Ubiquitin ligase TRIM62 regulates CARD9-mediated anti-fungal immunity and intestinal inflammation. Immunity 43, 715–726. doi: 10.1016/j.immuni.2015.10.005
Cetinkaya, P. G., Ayvaz, D. C., Karaatmaca, B., Gocmen, R., Söylemezoglu, F., Bainter, W., et al. (2018). A young girl with severe cerebral fungal infection due to card 9 deficiency. Clin. Immunol. 191, 21–26. doi: 10.1016/j.clim.2018.01.002
Cookson, B. T., and Brennan, M. A. (2001). Pro-inflammatory programmed cell death. Trends Microbiol. 9, 113–114. doi: 10.1016/S0966-842X(00)01936-3
Dong, W., Liu, Y., Peng, J., Chen, L., Zou, T., Xiao, H., et al. (2006). The IRAK-1-BCL10-MALT1-TRAF6-TAK1 cascade mediates signaling to NF-kappaB from Toll-like receptor 4. J. Biol. Chem. 281, 26029–26040. doi: 10.1074/jbc.M513057200
Doron, I., Leonardi, I., Li, X. V., Fiers, W. D., Semon, A., Bialt-Decelie, M., et al. (2021). Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell 184, 1017–1031.e14. doi: 10.1016/j.cell.2021.01.016
Drummond, R. A., Franco, L. M., and Lionakis, M. S. (2018). Human CARD9: a critical molecule of fungal immune surveillance. Front. Immunol. 9, 1836. doi: 10.3389/fimmu.2018.01836
Drummond, R. A., Swamydas, M., Oikonomou, V., Zhai, B., Dambuza, I. M., Schaefer, B. C., et al. (2019). CARD9(+) microglia promote antifungal immunity via IL-1β- and CXCL1-mediated neutrophil recruitment. Nat. Immunol. 20, 559–570. doi: 10.1038/s41590-019-0377-2
Gazendam, R. P., van Hamme, J. L., Tool, A. T., van Houdt, M., Verkuijlen, P. J., Herbst, M., et al. (2014). Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood 124, 590–597. doi: 10.1182/blood-2014-01-551473
GBD 2016 Causes of Death Collaborators (2017). Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1151–1210. doi: 10.1016/S0140-6736(17)32152-9
Guo, Y., Zhu, Z., Gao, J., Zhang, C., Zhang, X., Dang, E., et al. (2019). The Phytopathogenic fungus pallidocercospora crystallina-caused localized subcutaneous phaeohyphomycosis in a patient with a homozygous missense CARD9 mutation. J. Clin. Immunol. 39, 713–725. doi: 10.1007/s10875-019-00679-4
Hafalla, J. C., Burgold, J., Dorhoi, A., Gross, O., Ruland, J., Kaufmann, S. H., et al. (2012). Experimental cerebral malaria develops independently of caspase recruitment domain-containing protein 9 signaling. Infect. Immun. 80, 1274–1279. doi: 10.1128/IAI.06033-11
Hara, H., and Saito, T. (2009). CARD9 versus CARMA1 in innate and adaptive immunity. Trends Immunol. 30, 234–242. doi: 10.1016/j.it.2009.03.002
Hartjes, L., and Ruland, J. (2019). CARD9 Signaling in intestinal immune homeostasis and oncogenesis. Front. Immunol. 10, 419. doi: 10.3389/fimmu.2019.00419
Hsu, Y. M., Zhang, Y., You, Y., Wang, D., Li, H., Duramad, O., et al. (2007). The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol. 8, 198–205. doi: 10.1038/ni1426
Huang, C., Peng, Y., Zhang, Y., Li, R., Wan, Z., Wang, X., et al. (2019). Deep dermatophytosis caused by Trichophyton rubrum. Lancet Infect. Dis. 19, 1380. doi: 10.1016/S1473-3099(19)30551-1
Hung, C. Y., Zhang, H., Castro-Lopez, N., Ostroff, G. R., Khoshlenar, P., Abraham, A., et al. (2018). Glucan-chitin particles enhance Th17 response and improve protective efficacy of a multivalent antigen (rCpa1) against pulmonary Coccidioides posadasii infection. Infect. Immun. 86:e00070-18. doi: 10.1128/IAI.00070-18
Imanaka, Y., Taniguchi, M., Doi, T., Tsumura, M., Nagaoka, R., Shimomura, M., et al. (2021). Inherited CARD9 deficiency in a child with invasive disease due to exophiala dermatitidis and two older but asymptomatic siblings. J. Clin. Immunol. 41, 975–986. doi: 10.1007/s10875-021-00988-7
Ishizuka, S., Yokoyama, R., Sato, K., Shiroma, R., Nakahira, A., Yamamoto, H., et al. (2020). Effect of CARD9 deficiency on neutrophil-mediated host defense against pulmonary infection with Streptococcus pneumoniae. Infect. Immun. 89, 305–320. doi: 10.1128/IAI.00305-20
Jiang, C., and Lin, X. (2012). Regulation of NF-κB by the CARD proteins. Immunol. Rev. 246, 141–153. doi: 10.1111/j.1600-065X.2012.01110.x
Kalantari, P., Morales, Y., Miller, E. A., Jaramillo, L. D., Ponichtera, H. E., Wuethrich, M. A., et al. (2018). CD209a synergizes with dectin-2 and mincle to drive severe Th17 cell-mediated schistosome egg-induced immunopathology. Cell Rep. 22, 1288–1300. doi: 10.1016/j.celrep.2018.01.001
Kalantri, M., Khopkar, U., Shah, A., Bargir, U. A., Hule, G., Madkaikar, M., et al. (2021). A case of disseminated subcutaneous phaeohyphomycosis caused by Exserohilum rostratum with CARD9 mutation. Indian J. Dermatol. Venereol. Leprol. 88, 59–61. doi: 10.25259/IJDVL_293_19
Kao, W. P., Yang, C. Y., Su, T. W., Wang, Y. T., Lo, Y. C., Lin, S. C., et al. (2015). The versatile roles of CARDs in regulating apoptosis, inflammation, and NF-κB signaling. Apoptosis 20, 174–195. doi: 10.1007/s10495-014-1062-4
Kawai, T., and Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384. doi: 10.1038/ni.1863
Kobayashi, K., Inohara, N., Hernandez, L. D., Galán, J. E., Núñez, G., Janeway, C. A., et al. (2002). RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416, 194–199. doi: 10.1038/416194a
Kottom, T. J., Carmona, E. M., and Limper, A. H. (2020a). Targeting CARD9 with small-molecule therapeutics inhibits innate immune signaling and inflammatory response to Pneumocystis carinii β-glucans. Antimicrob. Agents Chemother. 64, 1210–1220. doi: 10.1128/AAC.01210-20
Kottom, T. J., Nandakumar, V., Hebrink, D. M., Carmona, E. M., and Limper, A. H. (2020b). A critical role for CARD9 in pneumocystis pneumonia host defence. Cell. Microbiol. 22, e13235. doi: 10.1111/cmi.13235
Lai, S. H. Y., Duque, J. S. R., Chung, B. H., Chung, T. W., Leung, D., Ho, R. S., et al. (2021). Invasive cerebral phaeohyphomycosis in a Chinese boy with CARD9 deficiency and showing unique radiological features, managed with surgical excision and antifungal treatment. Int. J. Infect. Dis. 107, 59–61. doi: 10.1016/j.ijid.2021.04.052
Lanternier, F., Pathan, S., Vincent, Q. B., Liu, L., Cypowyj, S., Prando, C., et al. (2013). Deep dermatophytosis and inherited CARD9 deficiency. N. Engl. J. Med. 369, 1704–1714. doi: 10.1056/NEJMoa1208487
Leshchiner, E. S., Rush, J. S., Durney, M. A., Cao, Z., Dančík, V., Chittick, B., et al. (2017). Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 114, 11392–11397. doi: 10.1073/pnas.1705748114
Liang, P., Wang, X., Wang, R., Wan, Z., Han, W., Li, R., et al. (2015). CARD9 deficiencies linked to impaired neutrophil functions against Phialophora verrucosa. Mycopathologia 179, 347–357. doi: 10.1007/s11046-015-9877-2
Loh, J. T., Xu, S., Huo, J. X., Kim, S. S., Wang, Y., Lam, K. P., et al. (2019). Dok3-protein phosphatase 1 interaction attenuates Card9 signaling and neutrophil-dependent antifungal immunity. J. Clin. Invest. 129, 2717–2729. doi: 10.1172/JCI126341
Maknitikul, S., Luplertlop, N., Chaisri, U., Maneerat, Y., and Ampawong, S. (2018). Featured article: immunomodulatory effect of hemozoin on pneumocyte apoptosis via CARD9 pathway, a possibly retarding pulmonary resolution. Exp Biol Med. 243, 395–407. doi: 10.1177/1535370218757458
McGreal, E. P., Rosas, M., Brown, G. D., Zamze, S., Wong, S. Y., et al. (2006). The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology 16, 422–430. doi: 10.1093/glycob/cwj077
Monteiro, J. T., Schön, K., Ebbecke, T., Goethe, R., Ruland, J., Baumgärtner, W., et al. (2019). The CARD9-associated C-type lectin, mincle, recognizes la crosse virus (LACV) but plays a limited role in early antiviral responses against LACV. Viruses 11:303. doi: 10.3390/v11030303
Nazarian, R. M., Lilly, E., Gavino, C., Hamilos, D. L., Felsenstein, D., Vinh, D. C., et al. (2020). Novel CARD9 mutation in a patient with chronic invasive dermatophyte infection (tinea profunda). J. Cutan. Pathol. 47, 166–170. doi: 10.1111/cup.13574
Pandori, W. J., Lima, T. S., Mallya, S., Kao, T. H., Gov, L., Lodoen, M. B., et al. (2019). Toxoplasma gondii activates a Syk-CARD9-NF-κB signaling axis and gasdermin D-independent release of IL-1β during infection of primary human monocytes. PLoS Pathog. 15, e1007923. doi: 10.1371/journal.ppat.1007923
Pavasutthipaisit, S., Stoff, M., Ebbecke, T., Ciurkiewicz, M., Mayer-Lambertz, S., Störk, T., et al. (2021). CARD9 deficiency increases hippocampal injury following acute neurotropic picornavirus infection but does not affect pathogen elimination. Int. J. Mol. Sci. 22, 6982. doi: 10.3390/ijms22136982
Poeck, H., Bscheider, M., Gross, O., Finger, K., Roth, S., Rebsamen, M., et al. (2010). Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat. Immunol. 11, 63–69. doi: 10.1038/ni.1824
Prado Acosta, M., Goyette-Desjardins, G., Scheffel, J., Dudeck, A., Ruland, J., Lepenies, B., et al. (2021). S-layer from Lactobacillus brevis modulates antigen-presenting cell functions via the mincle-Syk-Card9 axis. Front. Immunol. 12, 602067. doi: 10.3389/fimmu.2021.602067
Rieber, N., Gazendam, R. P., Freeman, A. F., Hsu, A. P., Collar, A. L., Sugui, J. A., et al. (2016). Extrapulmonary Aspergillus infection in patients with CARD9 deficiency. JCI Insight 1, e89890. doi: 10.1172/jci.insight.89890
Roth, S., Rottach, A., Lotz-Havla, A. S., Laux, V., Muschaweckh, A., Gersting, S. W., et al. (2014). Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1β production. Nat. Immunol. 15, 538–545. doi: 10.1038/ni.2888
Roth, S., and Ruland, J. (2013). Caspase recruitment domain-containing protein 9 signaling in innate immunity and inflammation. Trends Immunol. 34, 243–250. doi: 10.1016/j.it.2013.02.006
Ruland, J., and Hartjes, L. (2019). CARD-BCL-10-MALT1 signalling in protective and pathological immunity. Nat. Rev. Immunol. 19, 118–134. doi: 10.1038/s41577-018-0087-2
Sari, S., Dalgic, B., Muehlenbachs, A., Deleon-Carnes, M., Goldsmith, C. S., Ekinci, O., et al. (2018). Prototheca zopfii colitis in inherited CARD9 deficiency. J. Infect. Dis. 218, 485–489. doi: 10.1093/infdis/jiy198
Sheng, R., Zhong, X., Yang, Z., and Wang, X. (2021). The role of CARD9 deficiency in neutrophils. Mediators Inflamm. 2021, 6643603. doi: 10.1155/2021/6643603
Sun, C., Zhang, X., Yu, Y., Li, Z., and Xie, Y. (2020). CARD9 mediates T cell inflammatory response in Coxsackievirus B3-induced acute myocarditis. Cardiovasc. Pathol. 49, 107261. doi: 10.1016/j.carpath.2020.107261
Sun, L., Wan, Z., Li, R., and Yu, J. (2019). Impairment of Th cell response in Card9 knockout mice with cutaneous mucormycosis caused by Rhizopus arrhizus. Exp. Dermatol. 28, 1244–1251. doi: 10.1111/exd.14020
Sun, L., Zhang, S., Wan, Z., Li, R., and Yu, J. (2021). In vivo and in vitro impairments in T helper cell and neutrophil responses against Mucor irregularis in Card9 Knockout Mice. Infect. Immun. 89:e00040–21. doi: 10.1128/IAI.00040-21
Tartey, S., Gurung, P., Samir, P., Burton, A., and Kanneganti, T. D. (2018). Cutting edge: dysregulated CARD9 signaling in neutrophils drives inflammation in a mouse model of neutrophilic dermatoses. J. Immunol. 201, 1639–1644. doi: 10.4049/jimmunol.1800760
Vornholz, L., and Ruland, J. (2020). Physiological and pathological functions of CARD9 signaling in the innate immune system. Curr. Top. Microbiol. Immunol. 429, 177–203. doi: 10.1007/82_2020_211
Wang, L., Guo, Y., Huang, W. J., Ke, X., Poyet, J. L., Manji, G. A., et al. (2001). Card10 is a novel caspase recruitment domain/membrane-associated guanylate kinase family member that interacts with BCL10 and activates NF-kappa B. J. Biol. Chem. 276, 21405–21409. doi: 10.1074/jbc.M102488200
Wang, X., Wang, A., Wang, X., Li, R., and Yu, J. (2019). Cutaneous mucormycosis caused by Mucor irregularis in a patient with CARD9 deficiency. Br. J. Dermatol. 180, 213–214. doi: 10.1111/bjd.17144
Wang, X., Zhang, R., Wu, W., Song, Y., Wan, Z., Han, W., et al. (2018). Impaired specific antifungal immunity in CARD9-deficient patients with phaeohyphomycosis. J. Invest. Dermatol. 138, 607–617. doi: 10.1016/j.jid.2017.10.009
Wang, Y., Zhang, D., Hou, Y., Shen, S., and Wang, T. (2020). The adaptor protein CARD9, from fungal immunity to tumorigenesis. Am. J. Cancer Res. 10, 2203–2225.
Wu, W., Zhang, R., Wang, X., Song, Y., and Li, R. (2018). Subcutaneous infection with dematiaceous fungi in Card9 knockout mice reveals association of impair neutrophils and Th cell response. J. Dermatol. Sci. 92, 215–218. doi: 10.1016/j.jdermsci.2018.08.005
Wu, W., Zhang, R., Wang, X., Song, Y., Liu, Z., Han, W., et al. (2016). Impairment of immune response against dematiaceous fungi in Card9 knockout mice. Mycopathologia 181, 631–642. doi: 10.1007/s11046-016-0029-0
Yamamoto, H., Nakamura, Y., Sato, K., Takahashi, Y., Nomura, T., Miyasaka, T., et al. (2014). Defect of CARD9 leads to impaired accumulation of gamma interferon-producing memory phenotype T cells in lungs and increased susceptibility to pulmonary infection with Cryptococcus neoformans. Infect. Immun. 82, 1606–1615. doi: 10.1128/IAI.01089-13
Zajta, E., Csonka, K., Tóth, A., Tiszlavicz, L., Németh, T., Orosz, A., et al. (2021). Signaling through Syk or CARD9 mediates species-specific anti-candida protection in bone marrow chimeric mice. MBio 12, e0160821. doi: 10.1128/mBio.01608-21
Zhang, Y., Huang, C., Song, Y., Ma, Y., Wan, Z., Zhu, X., et al. (2021). Primary cutaneous Aspergillosis in a patient with CARD9 deficiency and Aspergillus susceptibility of Card9 knockout mice. J. Clin. Immunol. 41, 427–440. doi: 10.1007/s10875-020-00909-0
Keywords: CARD9, fungi, bacteria, viruses, parasites
Citation: Hu A, Hu Z, Zou H, Zhang J, Zhang D, Wang H, Zhong J and Chen B (2022) CARD9 in host immunity to fungal, bacterial, viral, and parasitic infections: An update. Front. Microbiol. 13:1021837. doi: 10.3389/fmicb.2022.1021837
Received: 17 August 2022; Accepted: 07 October 2022;
Published: 09 November 2022.
Edited by:
Ian Marriott, University of North Carolina at Charlotte, United StatesReviewed by:
Joshua J. Obar, Dartmouth College, United StatesCopyright © 2022 Hu, Hu, Zou, Zhang, Zhang, Wang, Zhong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Chen, Y2hlbmIxOTcwODI5QDE2My5jb20=; Jianing Zhong, emhvbmduaW5nXzAwM0AxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.