Corrigendum: Phylogenetic and genomic analyses of two new species of Clavispora (Metschnikowiaceae, Saccharomycetales) from Central China
- 1College of Life Science and Agricultural Engineering, Nanyang Normal University, Nanyang, China
- 2Research Center of Henan Provincial Agricultural Biomass Resource Engineering and Technology, Nanyang Normal University, Nanyang, China
- 3State Key Laboratory of Motor Vehicle Biofuel Technology, Henan Tianguan Enterprise Group Co., Ltd, Nanyang, China
Species in the genus Clavispora have previously been reported primarily in the northeast and northwest regions of China; the species diversity of Clavispora in central China is not currently clear. In this study, phylogenetic inferences of Clavispora based on sequences of a single-locus (LSU D1/D2) and a two-locus (LSU D1/D2 and ITS) were conducted. Two new species isolated from rotting wood in central China, namely Clavispora xylosa sp. nov. and Clavispora paralusitaniae sp. nov., were delimited and proposed based on morphological and molecular evidence. Cl. xylosa was closely related to C. thailandica CBS 10610T, but with 11.5% divergence in the LSU D1/D2 domains and 11.5% divergence in the ITS regions. Cl. paralusitaniae was a sister to Cl. lusitaniae CBS 6936T from which it differs with 4.7% divergence in the LSU D1/D2 domains and 5.4% divergence in the ITS regions. Description of Cl. xylosa sp. nov. and Cl. paralusitaniae sp. nov. was also supported by morphological comparisons and genomic analyses between the two new species and their closest relatives, C. thailandica CBS 10610T and Cl. lusitaniae CBS 6936T. These results indicate a potentially great diversity of Clavispora spp. inhabiting rotting wood in central China, ripe for future discovery.
Introduction
The genus Clavispora was established by Rodrigues de Miranda and is typified by Clavispora lusitaniae, the teleomorph of Candida lusitaniae (Rodrigues de Miranda, 1979; Lachance and Phaff, 2011). Later, Lodderomyces opuntiae was reclassified as Cl. lusitaniae because its ascospore is clavate in shape (Phaff et al., 1986). Yurkov et al. (2009), identified yeasts isolated from soils and proposed a new member of the genus Clavispora, Clavispora reshetovae, based on its morphology and phylogenetic placement. In addition, due to an inability to form sexual spores, some yeast species that belong to the Clavispora clade as revealed by phylogenetic analysis based on DNA sequences were previously placed in the anamorphic genus Candida (Lu et al., 2004; Jindamorakot et al., 2007; Nguyen et al., 2007; Rosa et al., 2007; James et al., 2009; Fell et al., 2011; Groenewald et al., 2011; Lachance et al., 2011; Nakase et al., 2011; Ribeiro et al., 2011; Limtong and Kaewwichian, 2013; Zhang et al., 2014). With the implementation of the “one fungus, one name” nomenclature, the relationships between Candida and Clavispora species began to be clarified (Daniel et al., 2014; Kurtzman et al., 2018). Daniel et al. (2014) provisionally reassigned 40 Candida species that are not related to the genus Metschnikowia to the genus Clavispora, although their monophyletic origin is doubtful. Recently, Candida fructus has been transferred to Clavispora as a new combination, and Clavispora santaluciae and Candida xylosifermentans have also been included as members of the Clavispora clade (Kurtzman et al., 2018; Kaewwichian et al., 2019; Drumonde-Neves et al., 2020). The genus Clavispora belongs to the family Metschnikowiaceae in the order Saccharomycetales, and is closely related to the genus Metschnikowia based on multigene phylogenetic analyses (Kurtzman and Robnett, 2013; Kurtzman et al., 2018). To date (December 2019), the YeastIP database (Weiss et al., 2013) lists 33 members of this clade, but the actual number of species assigned to the clade has reached 35 (Kaewwichian et al., 2019; Drumonde-Neves et al., 2020): five species of the genus Clavispora and 30 asexual species still assigned to the genus Candida.
The habitats of species belonging to the Clavispora clade are very diverse and they have been found in plants (Lu et al., 2004; Rosa et al., 2007; Groenewald et al., 2011; Lachance et al., 2011; Ribeiro et al., 2011; Limtong and Kaewwichian, 2013; Zhang et al., 2014; Drumonde-Neves et al., 2020), soil (Yurkov et al., 2009; Nakase et al., 2011; Limtong and Kaewwichian, 2013), insects (Nguyen et al., 2007; Lachance et al., 2011), insect frass (Jindamorakot et al., 2007; Lachance et al., 2011; Nakase et al., 2011), fish (Lachance et al., 2011), fresh water (Fell et al., 2011), moss (Lachance et al., 2011; Kaewwichian et al., 2019), and clinical samples (Lachance and Phaff, 2011). Cl. lusitaniae can be pathogenic to humans and is responsible for about 1% of invasive candidiasis cases, particularly in pediatric and hematology–oncology patients (Merz et al., 1992; Favel et al., 2003). Some species in this clade, such as Candida akabanensis, C. xylosifermentans, and Cl opuntiae, were found to ferment D-xylose, which is the second most abundant sugar in plant cell-wall carbohydrates (Phaff et al., 1986; Lachance et al., 2011; Valinhas et al., 2018; Kaewwichian et al., 2019). The ability of yeasts to ferment D-xylose is of interest because such yeasts can be applied to bioethanol production from lignocellulosic biomass (Agbogbo and Wenger, 2007; Urbina et al., 2013). Therefore, Clavispora species are important not only due to their pathogenicity in humans, but also for their potential applications in food and biofuels.
Species in the Clavispora clade have a worldwide distribution, but most were originally identified in Asia (Lu et al., 2004; Jindamorakot et al., 2007; Lachance et al., 2011; Nakase et al., 2011; Limtong and Kaewwichian, 2013; Zhang et al., 2014; Kurtzman et al., 2018; Kaewwichian et al., 2019), Europe (Yurkov et al., 2009; Drumonde-Neves et al., 2020), North America (Nguyen et al., 2007; Fell et al., 2011; Lachance et al., 2011), or South America (Rosa et al., 2007; Groenewald et al., 2011; Ribeiro et al., 2011). Over the last several years, this clade has received a great deal of attention in Asia, with two novel species found in Japan (Lachance et al., 2011; Nakase et al., 2011) and five novel species found in Thailand (Jindamorakot et al., 2007; Nakase et al., 2011; Limtong and Kaewwichian, 2013; Kaewwichian et al., 2019). In contrast, little is known about Clavispora spp. in China. To date, only two species of the Clavispora clade have been reported in China: Candida asparagi and Candida pruni (Lu et al., 2004; Zhang et al., 2014).
The aim of the current study is to explore the species diversity of Clavispora in central China, and more importantly, to construct a more natural taxonomic system of Clavispora, based on phylogenetic analyses. In addition, genomic data that can be analyzed with tools to determine species delineation is explored.
Materials and methods
Sample collection and yeast isolation
Rotting wood samples were collected in two areas of Henan Province in central China, in the Funiu Mountain Nature Reserve (32°45′N, 113°30′E) and in the Baotianman Nature Reserve (33°27′N, 111°48′E). The predominant vegetation was characterized in those areas as a warm-temperate to subtropical forest biome. The climate was warm-temperate, with an annual precipitation volume of 885.6 mm and an average temperature of 15.2°C. Yeasts were isolated from rotting wood samples as previously described (Kaewwichian et al., 2019; Lv et al., 2020). Briefly, each wood sample (1 g) was added to 20 ml sterile D-xylose medium (yeast nitrogen base 0.67%, D-xylose 0.5%, and chloramphenicol 0.02%, pH 5.0 ± 0.2) in a 150 ml Erlenmeyer flask and then cultured at 25°C for 3–10 days on a rotary shaker. Subsequently, 0.1 ml aliquots of the enrichment culture and appropriate decimal dilutions were spread on D-xylose agar plates and then incubated at 25°C for 3–4 days. Representative colonies were selected and the yeasts were isolated through repeated plating on D-xylose agar, and then stored on yeast extract-malt extract (YM; 1% glucose, 0.5% peptone, 0.3% yeast extract, and 0.3% malt extract; pH 5.0 ± 0.2) agar slants at 4°C or in 15% glycerol at −80°C. Strains of the two new species described in this paper are listed in Table 1.
Phenotypic characterization
The morphological, physiological and biochemical characteristics of the two new species were determined using standard methods (Kurtzman et al., 2011). Carbon and nitrogen assimilation tests were performed using liquid media, and growth was observed for up to 4 weeks. Carbon fermentation was tested in a YP base media (1% yeast extract and 2% peptone, pH 5.0 ± 0.2), and Durham tubes were used to visualize carbon dioxide production. Growth was assessed at a range of temperatures (30, 35, 37, and 40°C) by streaking cells onto yeast extract peptone glucose (YPD) agar (2% glucose, 2% peptone, 1% yeast extract, and 2% agar) plates and incubating them for 2 weeks. Formation of true hyphae and pseudohyphae were investigated using the Dalmau plate method on both cornmeal (CM) and 5% malt extract (ME) agar plates. The beginning of the sexual stage was determined by incubating single or mixed cultures of each of the two strains on CM agar, 5% ME agar, potato-dextrose agar (PDA; 20% potato infusion, 2% glucose, 2% agar), and yeast carbon base plus 0.01% ammonium sulfate (YCBAS) agar at 15°C or 25°C for 6 weeks (Yurkov et al., 2009; Lachance and Phaff, 2011; Drumonde-Neves et al., 2020). Photomicrographs were taken using a Leica DM 2500 microscope (Leica Microsystems GmbH, Wetzlar, Germany) with a Leica DFC295 digital microscope color camera using bright field, phase contrast, and differential interference contrast (DIC) optics. Novel taxonomic descriptions and proposed names were deposited in MycoBank (http://www.mycobank.org; 12 September 2022).
DNA extraction, amplification, and sequencing
Genomic DNA was extracted from the yeasts using an Ezup Column Yeast Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China) following the manufacturer’s instructions. The internal transcribed spacer (ITS) regions and the D1/D2 domains of the large subunit (LSU) rRNA gene were amplified using the primer pairs ITS1/ITS4 (White et al., 1990) and NL1/NL4 (Kurtzman and Robnett, 1998), respectively. The PCR program was as follows: initial denaturation at 95°C for 3 min; 35 cycles of 94°C for 40 s, 56°C for 45 s, and 72°C for 1 min; and a final extension of 72°C for 10 min (Shi et al., 2021). PCR products were cleaned and sequenced by Sangon Biotech Inc.
Phylogenetic analysis
Phylogenetic analyses were conducted using the novel sequences generated from the five strains isolated in this study and all reference sequences of related strains available in GenBank (Table 2). Alignments for the LSU D1/D2 and ITS loci were conducted using MAFFT v7.110 with default settings (Katoh and Standley, 2013) and manually corrected where necessary. To establish the identity of the isolates at the species level, phylogenetic analyses were conducted first with each locus individually, then using two loci (ITS and LSU D1/D2) together, that are widely used molecular “barcode” regions for fungi, with LSU D1/D2 particularly suitable for yeasts. Phylogenetic trees were constructed in MEGA11 software (Tamura et al., 2021) for each of the datasets using the maximum likelihood (ML) and neighbor-joining (NJ) methods. Saccharomyces cerevisiae CBS 1171T was used as the outgroup (Kurtzman et al., 2018; Kaewwichian et al., 2019).
Analysis with the ML method was performed using the best-fit substitution model GTR+ I + G (Nei and Kumar, 2000), whereas analysis with the NJ method was performed using the Kimura 2-parameter model (Saitou and Nei, 1987). The confidence levels of the clades were estimated from 1,000 bootstrap replicates (Felsenstein, 1985).
Genome sequencing and assembly
The genomes of the type strains of C. thailandica and Cl. lusitaniae are publicly available, whereas the genomes of the novel strains NYNU 174173T and NYNU 161120T were sequenced for this study. The genomic DNA of each strain was isolated using a phenol: chloroform extraction protocol (Schwartz and Sherlock, 2016). The genomic DNA of the strains were sequenced using long-read PacBio platforms with single-molecule real-time (SMRT) technology. Sequencing libraries were constructed using TruSeq DNA PCR Free (350) kits (Illumina) and run on an Illumina NovaSeq instrument at Beijing BerryGenomics Co., Ltd. (Beijing, China).
To generate whole genome assemblies, paired-end Illumina reads were processed with the meta-assembler pipeline iWGS v1.1 (Zhou et al., 2016). Briefly, this pipeline performed quality-based read trimming followed by k-mer length optimization, then used a range of state-of-the-art assemblers to generate several genome assemblies for each strain. The quality of each assembly was assessed using QUAST v3.1 (Gurevich et al., 2013), and the best assembly for each species was chosen based on N50 statistics and genome size as calculated with Quast v5.0.2 (Gurevich et al., 2013).
Genomic analyses
The DNA G + C content was calculated and protein-coding open reading frames (ORFs) were predicted using Glimmer v3.02 (Delcher et al., 2007). We used RNAmmer v1.2 (Lagesen et al., 2007) and tRNAscan-SE v1.3.1 (Lowe and Eddy, 1997) to predict rRNAs and tRNAs, respectively. The average nucleotide identity (ANI) values were calculated between strains NYNU 174173T and NYNU 161120T genomes and those of the most closely related species using a web-based calculator (Yoon et al., 2017)1 and FastANI (Jain et al., 2018). To calculate the distances between genomes, digital DNA–DNA homology (dDDH) values were estimated using the Genome-to-Genome Distance Calculator 2.1.2 The dDDH values presented here were calculated using Formula 2 (IXY: = sum of identical base pairs over all HSPs), which estimates values based on the identities of high-scoring segment pairs (Meier-Kolthoff et al., 2013).
Results
Phylogenetic analysis
Among the yeasts isolated from rotting wood samples collected in Henan Province, central China, five strains that could not be identified as known yeast species based on the rDNA sequences were selected for further taxonomic characterization. To establish the taxonomic position of the novel strains, phylogenetic analysis was carried out with the LSU D1/D2 sequences of the five novel strains and type strains of members of the Clavispora clade. The resulting phylogenetic trees showed that the five strains represented two new members of the Clavispora clade and could be classified into two taxa (Figures 1, 2). Three strains (NUNU 167235, NUNU 168424, and NYNU 161120) in group NYNU 161120T had identical sequences in both the D1/D2 and ITS regions, which indicated that they are conspecific. Two other strains (NUNU 168193 and NYNU 174173) in group NYNU 174173T had identical sequences in both the D1/D2 and ITS regions, which indicated that they are conspecific. The two new members of the Clavispora clade clearly distinguishable from those of other species, whether undescribed or previously characterized.
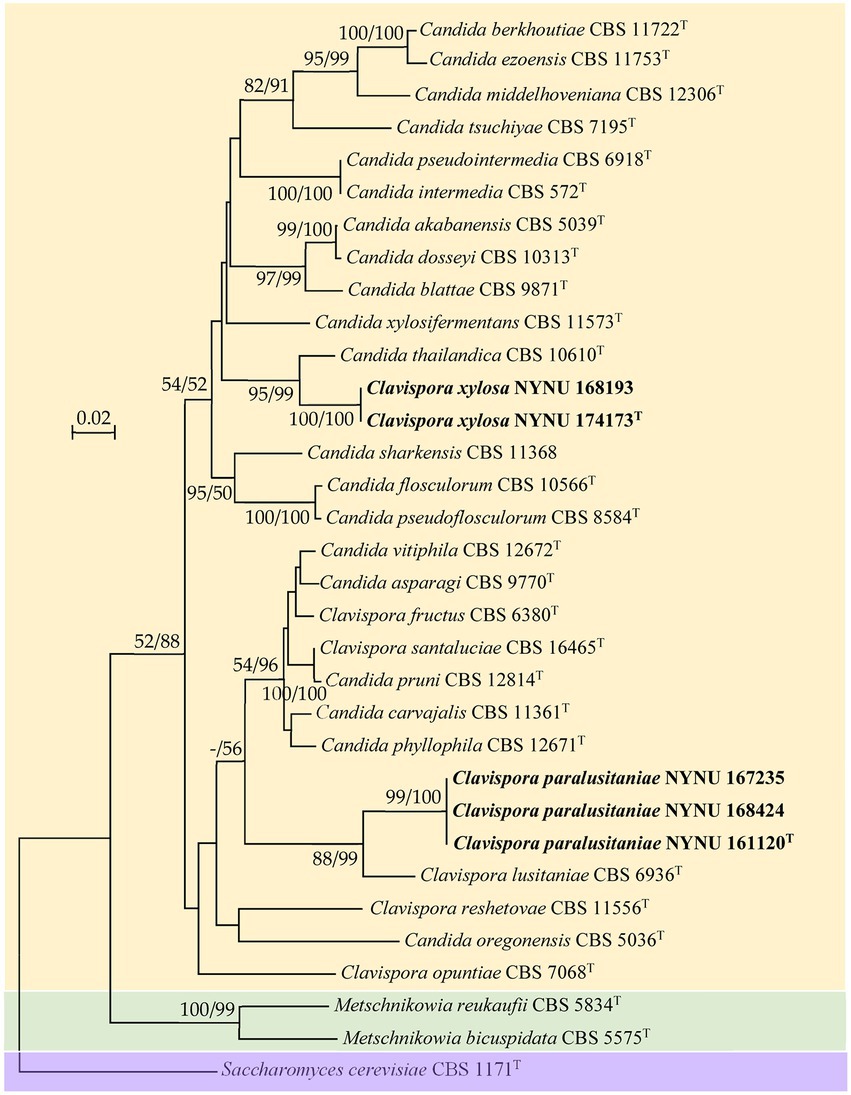
Figure 1. Maximum likelihood (ML) phylogram of Clavispora species based on the D1/D2 domains of the LSU rRNA gene. Saccharomyces cerevisiae CBS 1171T was used as the outgroup. ML and neighbor-joining (NJ) bootstrap support values above 50% are shown at the nodes (ML/NJ). The novel strains described in this study are shown in bold.
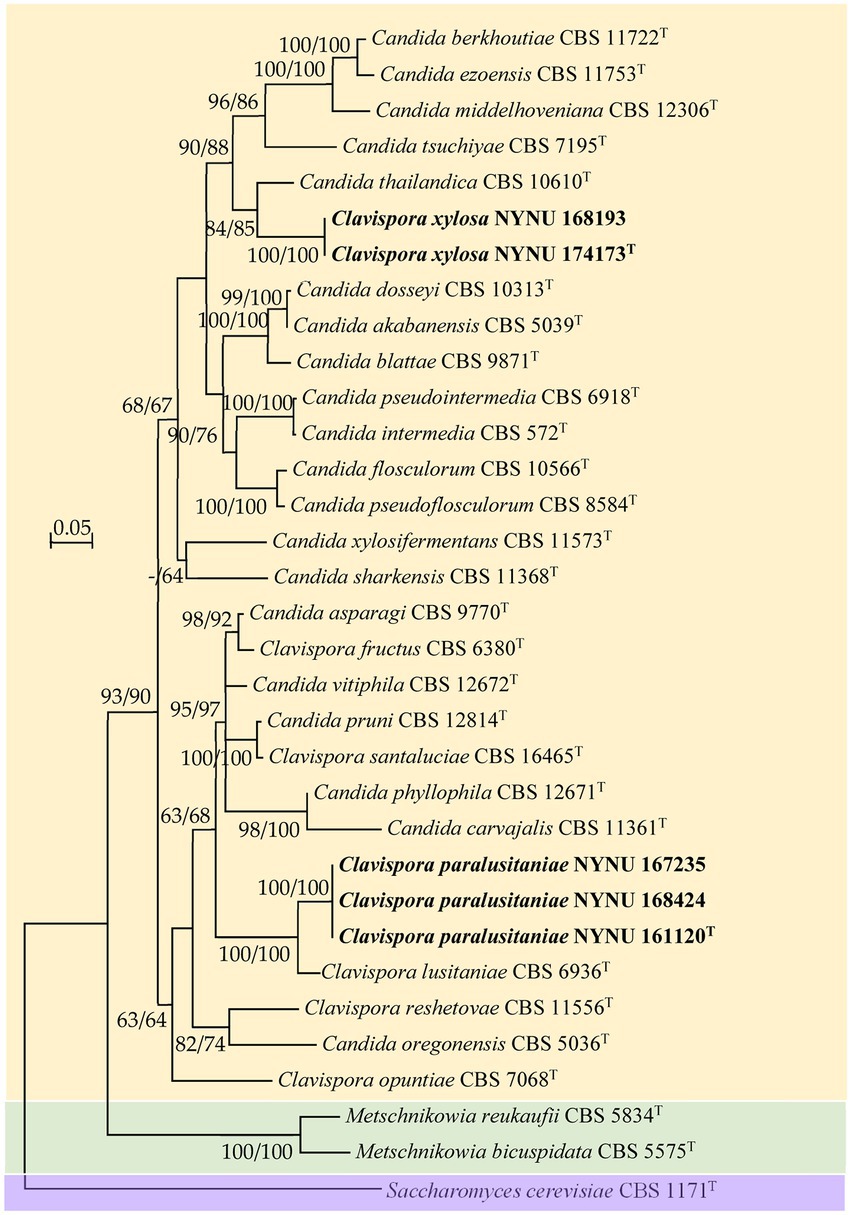
Figure 2. Maximum likelihood (ML) phylogram of Clavispora species based on the combined ITS + LSU D1/D2 sequence data. Saccharomyces cerevisiae CBS 1171T was used as the outgroup. ML and neighbor-joining (NJ) bootstrap support values above 50% are shown at the nodes (ML/NJ). The novel strains described in this study are shown in bold.
Strain NYNU 174173T formed a well-delineated lineage with C. thailandica with a high bootstrap value (95% ML; 99% NJ; Figure 1) in the Candida intermedia subclade. Strain NYNU 174173T differed by 11.5% divergence (24 substitutions and four gaps) from its close relative C. thailandica CBS 10610T in the LSU D1/D2 domains and differed by 11.5% divergence (30 substitutions and 11 gaps) in the ITS regions. In accordance with the guidelines for yeast identification based on nucleotide divergences, yeast strains with 1% or more substitution in the D1/D2 domain or 1–2% nucleotide differences in the ITS region usually represent separate species (Kurtzman and Robnett, 1998; Scorzetti et al., 2002). The differences in both the LSU D1/D2 and ITS sequences were significant enough for this strain to be considered a novel species. The close relationship between strain NYNU 174173T and C. thailandica CBS 10610T was further confirmed by the combined ITS and LSU D1/D2 dataset (84% ML; 85% NJ; Figure 2).
Strain NYNU 161120T clustered with Cl. lusitaniae with strong bootstrap support (88% ML; 99% NJ; Figure 1) in the Cl. lusitaniae subclade. Its nearest phylogenetic neighbor was Cl. lusitaniae CBS 6936T, from which strain NYNU 161120T differed by 4.7% divergence (14 substitutions and two gaps) in the LSU D1/D2 domains and 5.4% divergence (24 substitutions and four gaps) in the ITS regions. According to the criteria mentioned above, the relatively low sequence similarity indicated that strain NYNU 161120T and Cl. lusitaniae CBS 6936T were distinct Clavispora species. The phylogenetic relationships between strain NYNU 161120T and the previously described Clavispora species were further confirmed by the phylogenetic tree built from the combined ITS and LSU D1/D2 dataset (100% ML; 100% NJ; Figure 2).
Phenotypic characterization
Phenotypic characterization was carried out for strains NYNU 174173T and NYNU 161120T using standard methods (Kurtzman et al., 2011). The yeasts shared similar phenotypic characteristics with other species in the Clavispora clade. Colonies were white to cream-colored, buttery, convex, and had an entire margin (Figure 3A, 4A). Cells were ovoid to elongate, proliferated by multilateral budding (Figure 3B, 4B), and formed pseudohyphae but not hyphae (Figure 3C, 4C). They were fermentative and could not assimilate nitrate as a nitrogen source. Their growth in vitamin-free medium was inconsistent with previous descriptions of the Clavispora clade (Jindamorakot et al., 2007; Yurkov et al., 2009; Lachance and Phaff, 2011), but studies of other closely related species demonstrated that this trait must be considered variable in this clade. Neither conjugation nor ascospores were observed in single or mixed cultures on sporulation media, suggesting that these strains represent anamorphs of the genus Clavispora.
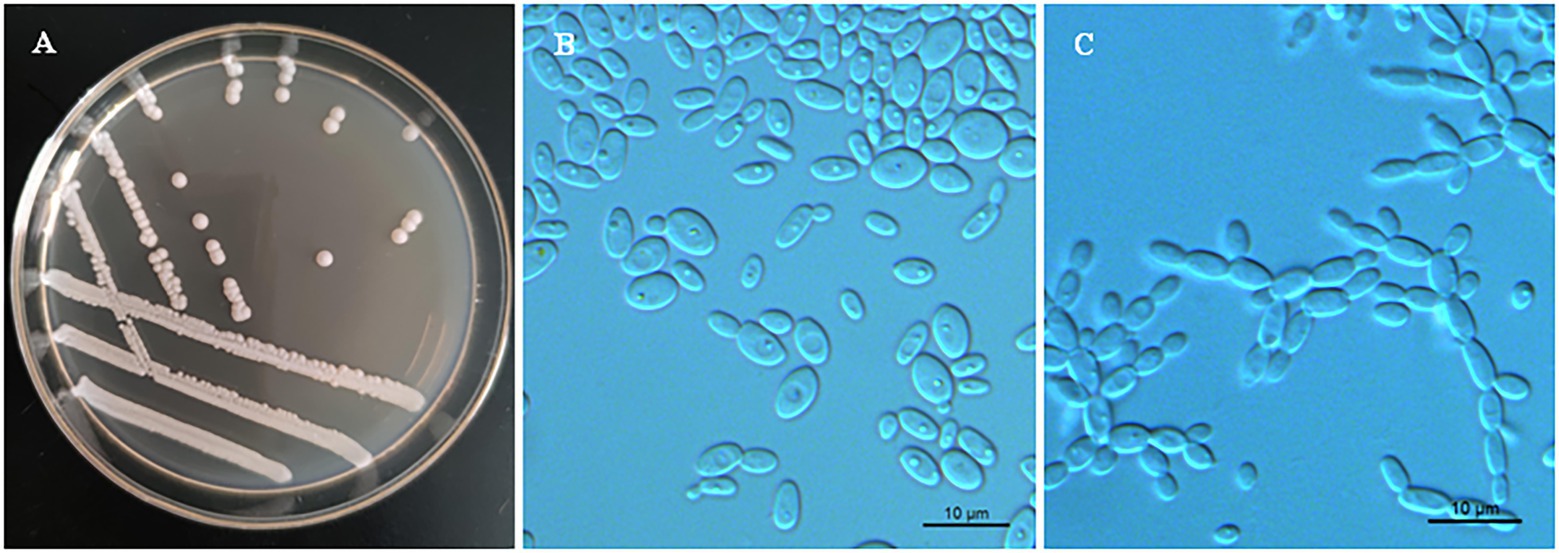
Figure 3. Morphological characteristics of Clavispora xylosa sp. nov. (CICC 33277, holotype). (A) Colony morphology on YM agar after growth for 3 days at 25°C. (B) Budding cells after growth for 3 d in YM broth at 25°C. (C) Pseudohyphae with blastoconidia on CM agar after growth for 7 days at 25°C. Scale bars = 10 μm.
Phenotypic characteristics that differed between strains NYNU 174173T and NYNU 161120T and closely related species in the Clavispora clade are shown in Table 3. Strain NYNU 174173T could be differentiated from the most closely related known species, C. thailandica CBS 10610T (Jindamorakot et al., 2007), based on fermentation of D-xylose; growth at 35°C; ability to assimilate L-arabinose and L-rhamnose; inability to assimilate D-arabinose, 2-keto-D-gluconate, DL-lactate, or cadaverine; and the inability to grow in 0.01% cycloheximide. Strain NYNU 161120T could be morphologically differentiated from its nearest phylogenetic neighbor, Cl. lusitaniae CBS 6936T (Lachance and Phaff, 2011), by the inability to produce one or two clavate ascospores in a liberated ascus. Physiologically, strain NYNU 161120T could be differentiated from Cl. lusitaniae CBS 6936T based on the ability to assimilate inulin; the inability to assimilate ethanol; and the inability to grow in 10% NaCl with 5% glucose and in 0.01% cycloheximide (Table 3).
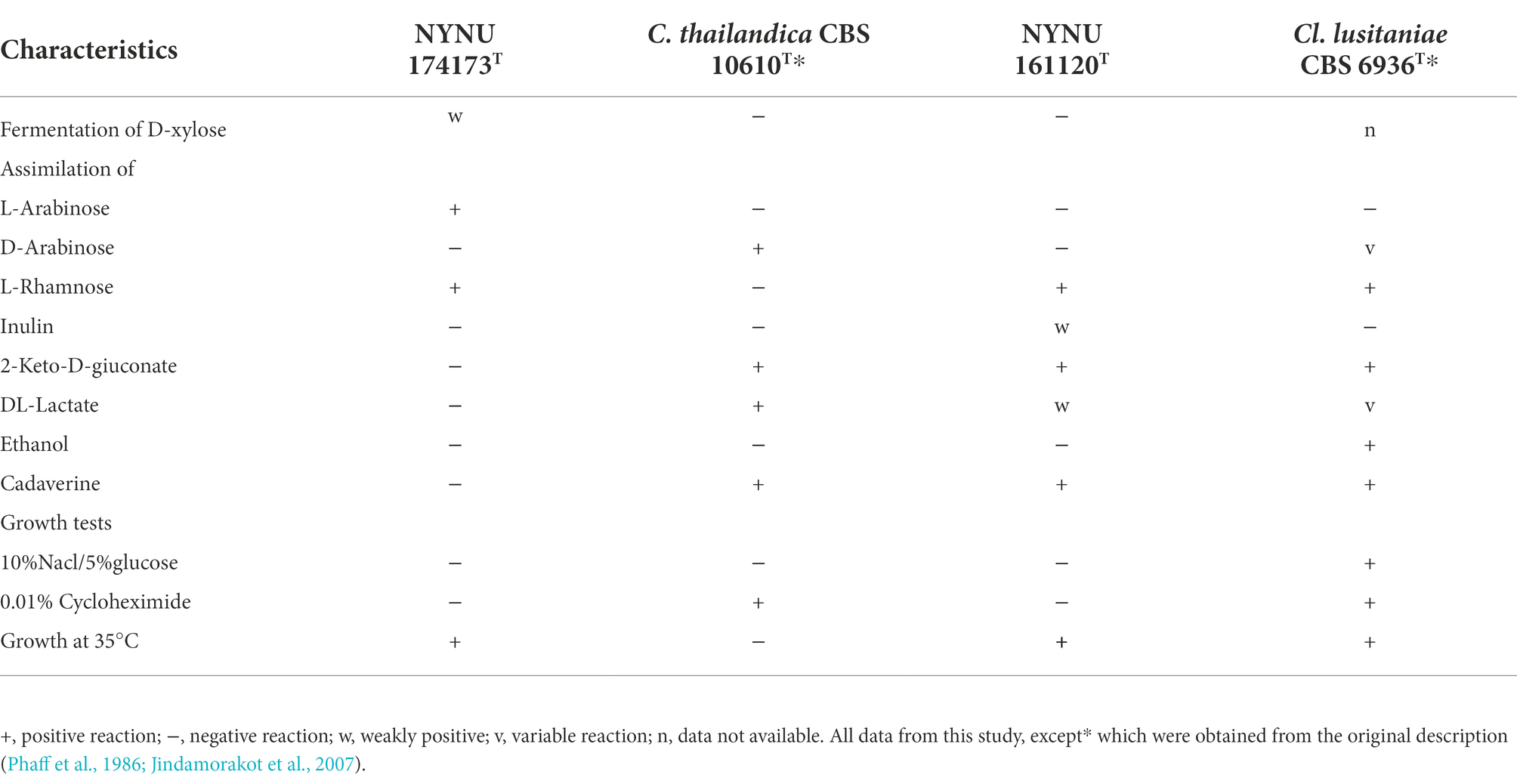
Table 3. Phenotypic characteristics that differed between the novel strains NYNU 174173T and NYNU 161120T and their closely related taxa.
Genomic analyses
The genomes of strains NYNU 174173T and NYNU 161120T were assembled and compared with those of the closest related species, C. thailandica CBS 10610T and Cl. lusitaniae CBS 6936T, respectively. The detailed characteristics of these genomes are shown in Table 4. The genome of strain NYNU 174173T is 15,305,418 bp in size, consisting of 10 scaffolds with an N50 length of 2,408,949 bp, and maximum and minimum scaffold lengths of 3,517,412 and 56,211 bp, respectively. The genome of strain NYNU 161120T is 12,617,314 bp in size, consisting of eight scaffolds with an N50 length of 1,920,963 bp, and maximum and minimum scaffold lengths of 2,542,591 and 780,161 bp, respectively. A total of 4,930 genes, 4,647 ORFs, and 283 RNAs are predicted in strain NYNU 174173T, and a total of 5,085 genes, 4,774 ORFs, and 311 rRNAs are predicted in strain NYNU 161120T. The genomic G + C contents are 50.87 and 44.51% for strains NYNU 174173T and NYNU 161120T, respectively; this is much higher than the G + C content in C. thailandica CBS 10610 T and Cl. lusitaniae CBS 6936T (Table 4).
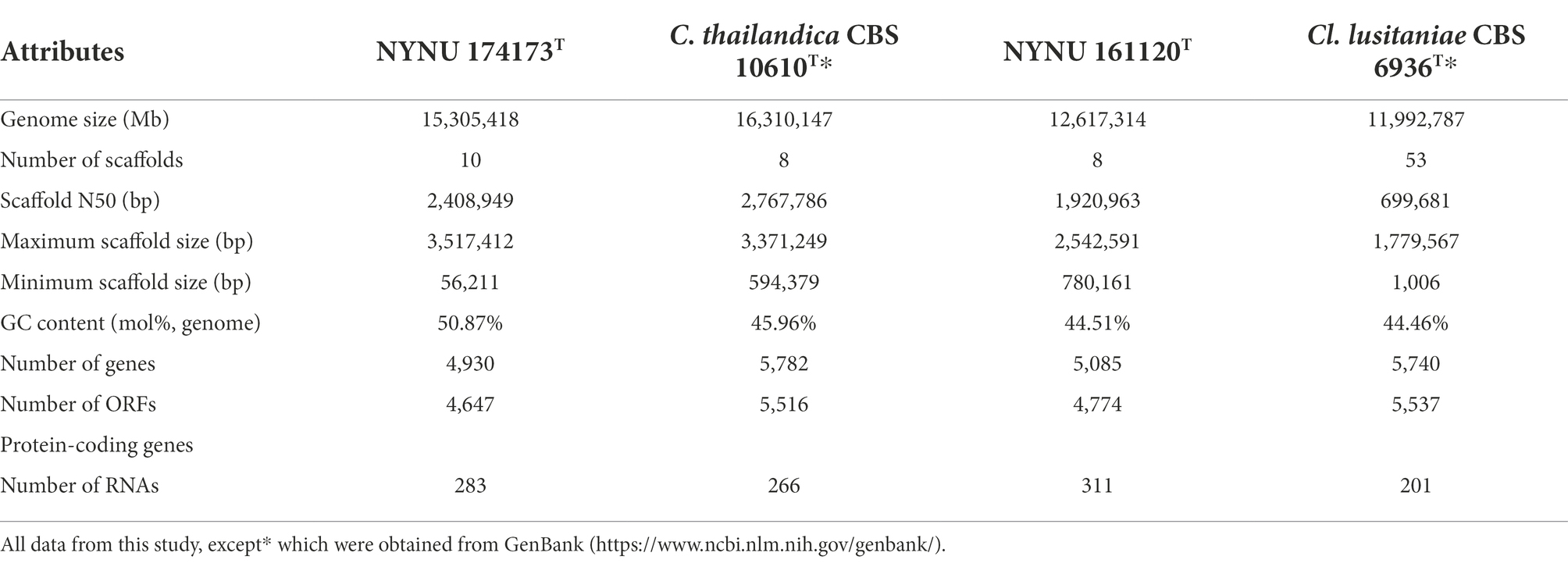
Table 4. Genome properties for strains NYNU 174173T and NYNU 161120T and for the type strains of the most closely related species in the Clavispora clade.
In yeast systematics, genomic data can be analyzed with tools that calculate genome-wide genetic distances between a novel taxon and its closest relatives to determine species delineation (Libkind et al., 2011; Lachance and Lee, 2020; Cadež et al., 2021). For estimation of genetic distances between strains NYNU 174173T and NYNU 161120T and their closest relatives, we used two tools that are available as web interfaces: the ANI calculator and the Genome-to-Genome Distance Calculator. The ANI and DDH values between the strain NYNU 174173T and C. thailandica CBS 10610T were 77.78 and 30.70%, respectively (Table 5); the ANI and DDH values between the strain NYNU 161120T and Cl. lusitaniae CBS 6936T were 94.72 and 53.50%, respectively. All these percentages were lower than 95 and 70%, respectively, which are the defined cut-off limits for species delineation (Lachance and Lee, 2020; Cadež et al., 2021), and further support the inclusion of these two strains as novel taxa in the Clavispora clade.
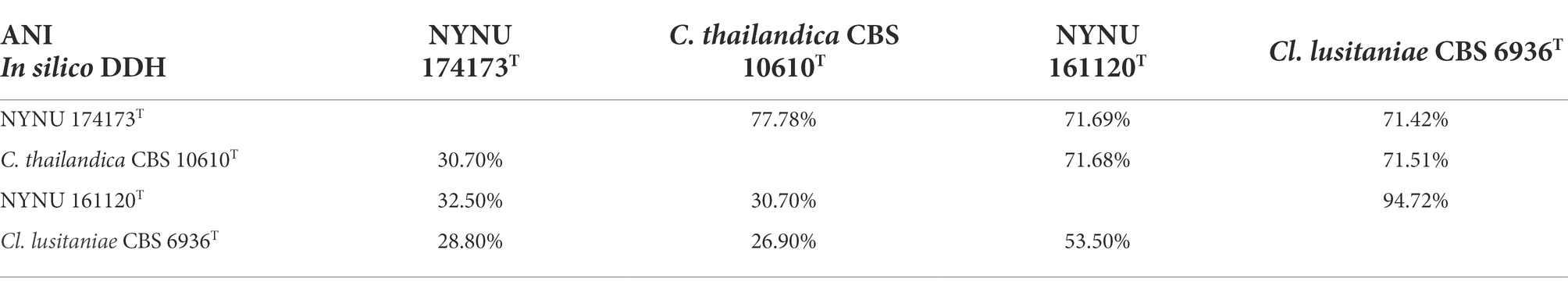
Table 5. Matrix of in silico DNA–DNA hybridization (DDH) and Ortho average nucleotide identity (OrthoANI) percentages between the genomes of strains NYNU 174173T and NYNU 161120T and the type strains of the most closely related species in the Clavispora clade.
Taxonomy
Clavispora xylosa F.L. Hui & C.Y. Chai sp. nov. (Figure 3).
Mycobank No.: 845627.
Type. China, Henan Province, Baotianman Nature Reserve, rotting wood, 3 August, 2016, NYNU 174173 (holotype CICC 33277, culture ex-type CBS 15236).
Etymology. Clavispora xylosa (xy.lo’sa. N.L. masc. adj. xylosa, of or pertaining to xylose, because the type strain can assimilate D-xylose).
Description. In YM broth after 3 days at 25°C, cells are ovoid to elongate (2–5.5 × 3.5–7 μm) and occur singly or in pairs (Figure 3B). Budding is multilateral. In YM broth after a month, a sediment is formed, but a pellicle is not observed. On YM agar after 3 days at 25°C, colonies are white to cream-colored, buttery, convex, and smooth, with entire margins (Figure 3A). After 7 days at 25°C on a Dalmau plate culture with CM agar, hyphae are not produced, but pseudohyphae are present (Figure 3C). Asci or signs of conjugation were not observed on sporulation media.
Glucose, galactose, trehalose and D-xylose (weak) are fermented. Glucose, galactose, L-sorbose, D-glucosamine, D-xylose, L-arabinose, L-rhamnose, sucrose, maltose, trehalose, methyl α-D-glucoside, cellobiose, salicin, arbutin, lactose, melezitose, ribitol, xylitol, D-glucitol, D-mannitol, galactitol, D-glucono-1, 5-lactone, D-gluconate, succinate, and citrate are assimilated. No growth was observed in D-ribose, D-arabinose, melibiose, raffinose, inulin, glycerol, erythritol, myo-inositol, 2-Keto-D-gluconate, D-glucuronate, DL-lactate, methanol or ethanol. In nitrogen-assimilation tests, growth is present on ethylamine, L-lysine, glucosamine, and D-tryptophan, while growth is absent on nitrate, nitrite, cadaverine, creatine, creatinine, and imidazole. Growth is observed at 35°C but not at 37°C. Growth in vitamin-free medium is present, but growth in the presence of 0.01% cycloheximide, 10% NaCl with 5% glucose and 1% acetic acid is absent. Starch-like compounds are not produced. Urease activity and diazonium blue B reactions are negative. The G + C content of the genome of the strain NYNU 174173T is 50.87 mol%, its approximate size is 15.3 Mbp.
Note. Members of Cl. xylosa have intraspecific variability in phenotypic characteristics. Strain NYNU 174173T is able to assimilate maltose, salicin and ribitol, while strain NYNU 168193 do not.
Clavispora paralusitaniae F.L. Hui & C.Y. Chai sp. nov. (Figure 4).
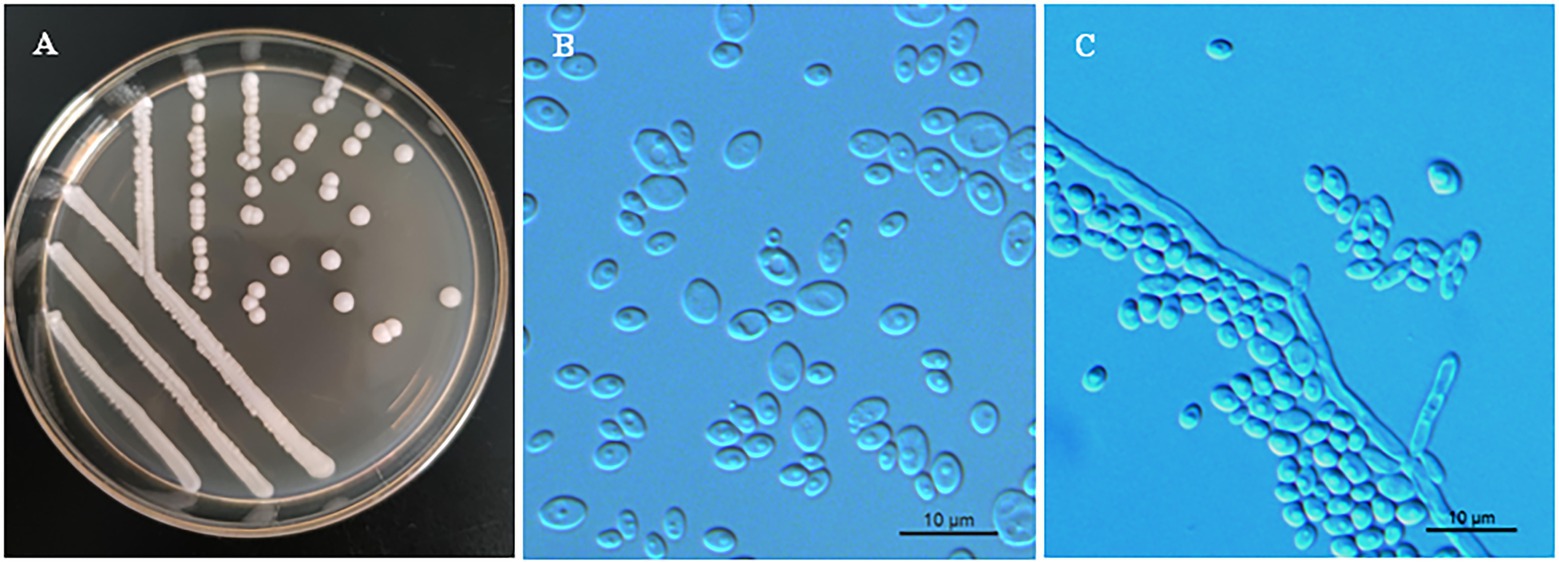
Figure 4. Morphological characteristics of Clavispora paralusitaniae sp. nov. (CICC 33276, holotype). (A) Colony morphology on YM agar after growth for 3 days at 25°C. (B) Budding cells after growth for 3 days in YM broth at 25°C. (C) Pseudohyphae with blastoconidia on CM agar after growth for 7 days at 25°C. Scale bars = 10 μm.
Mycobank No.: 845628.
Type. China, Henan Province, Funiu Mountain Nature Reserve, rotting wood, 2 August, 2016, NYNU 161120 (holotype CICC 33276, culture ex-type CBS 15234).
Etymology. Clavispora paralusitaniae (pa.ra.lu.si.ta.ni’ae. N.L. gen. n. paralusitaniae, similar to lusitaniae, referring to the close relationship to Clavispora lusitaniae).
Description. In YM broth after 3 days at 25°C, cells are ovoid to elliptical (2–7 × 2–8 μm) and occur singly or in pairs (Figure 4B). Budding is multilateral. In YM broth after a month, a sediment is formed, but a pellicle is not observed. On YM agar after 3 days at 25°C, colonies are white to cream-colored, buttery, raised, and smooth, with entire margins (Figure 4A). After 7 days at 25°C on a Dalmau plate culture with CM agar, hyphae are not produced, but pseudohyphae are present (Figure 4C). Asci or signs of conjugation were not observed on sporulation media.
Glucose, galactose, maltose (weak), sucrose, trehalose (weak), and cellobiose are fermented. Glucose, galactose, L-sorbose, D-glucosamine, D-xylose, L-rhamnose, sucrose, maltose, trehalose, methyl α-D-glucoside, cellobiose, salicin, arbutin, melezitose, inulin (weak), glycerol, ribitol, xylitol, D-glucitol, D-mannitol, D-glucono-1, 5-lactone, 2-keto-D-gluconate (weak), D-gluconate, DL-lactate, succinate, and citrate are assimilated. No growth was observed in D-ribose, L-arabinose, D-arabinose, melibiose, lactose, raffinose, erythritol, galactitol, myo-inositol, D-glucuronate, methanol or ethanol. In nitrogen-assimilation tests, growth is present on ethylamine, L-lysine, cadaverine, glucosamine, and D-tryptophan, while growth is absent on nitrate, nitrite, creatine, creatinine, and imidazole. Growth is observed at 37°C but not at 40°C. Growth in vitamin-free medium is present, but growth in the presence of 0.01% cycloheximide, 10% NaCl with 5% glucose and 1% acetic acid is absent. Starch-like compounds are not produced. Urease activity and diazonium blue B reactions are negative. The G + C content of the genome of the strain NYNU 161120T is 44.51 mol%, its approximate size is 12.6 Mbp.
Note. Strains of Cl. paralusitaniae exhibits minor differences in phenotypic characteristics. Strain NYNU 168424 assimilates D-arabinose but strains NYNU 161120T and NYNU 167235 do not. Additionally, fermentation of galactose is positive for two strain NYNU 161120T and NYNU 167235, while that of strain NYNU 168424 is negative.
Discussion
In this study, we isolated five novel yeast strains from rotting wood samples collected in Henan Province, central China. Phylogenetic analyses based on a single-locus (LSU D1/D2) and a two-locus (ITS and LUS D1/D2) approach showed that the five strains formed two robust groups, represented by strains NYNU 174173T and NYNU 161120T, together with their relatives (C. thailandica CBS 10610T and Cl. lusitaniae CBS 6936T), and were obviously separate from other type strains in the Clavispora clade. Pairwise sequence comparison of the LSU D1/D2 domains and the ITS regions of the two novel strains with related species showed that they had lower similarity values than the common threshold for species differentiation in yeast (Kurtzman and Robnett, 1998; Scorzetti et al., 2002). In addition, they shared similar physiological and biochemical characteristics with species in the Clavispora clade, but clearly differed from the closest known species, C. thailandica CBS 10610T and Cl. lusitaniae CBS 6936T (Table 3). Thus, the two isolates represented two novel species in the genus Clavispora. This distinction was supported by genomic analyses. Based on genomic G + C content, strains NYNU 174173T and NYNU 161120T were clearly distinct from C. thailandica CBS 10610T and Cl. lusitaniae CBS 6936T, respectively (Table 4). When strains NYNU 174173T and NYNU 161120T were compared with each other or with their closest relatives, the genomic relatedness with ANI and DDH values was lower than the proposed threshold value for species delineation (Table 5; Lachance and Lee, 2020; Cadež et al., 2021). These results, together with the robust phylogenetic positions and the differences in phenotypic characteristics, indicated that the two isolated strains represented two novel species in the genus Clavispora. We therefore propose two new species, namely Clavispora xylosa sp. nov. and Clavispora paralusitaniae sp. nov., to accommodate these yeasts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
C-YC and YL isolated the strains and performed the taxonomic characterization and genomic analyses. C-YC prepared the draft manuscript, and the tables and figures. Z-LY and F-LH designed the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (Project no. 31570021) and the State Key Laboratory of Motor Vehicle Biofuel Technology, Henan Tianguan Enterprise Group Co., Ltd., China (no. 2018001).
Acknowledgments
The authors are very grateful to their colleagues at the School of Life Science and Agricultural Engineering, Nanyang Normal University, including Lin Zhang, Kai-Fang Liu, and Zhi-Wen Xi for providing specimens; to Ting Lei for help with phylogenetic analysis; and to Ran-Ran Jia and Wen-Ting Hu for help with morphological observations.
Conflict of interest
Z-LY was employed by the company Henan Tianguan Enterprise Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Agbogbo, F. K., and Wenger, K. S. (2007). Production of ethanol from corn Stover hemicellulose hydrolyzate using Pichia stipitis. J. Ind. Microbiol. Biotechnol. 34, 723–727. doi: 10.1007/s10295-007-0247-z
Cadež, N., Bellora, N., Ulloa, R., Tome, M., Petković, H., Groenewald, M., et al. (2021). Hanseniaspora smithiae sp. nov., a novel apiculate yeast species from Patagonian forests that lacks the typical genomic domestication signatures for fermentative environments. Front. Microbiol. 12:679894. doi: 10.3389/fmicb.2021.679894
Daniel, H. M., Lachance, M. A., and Kurtzman, C. P. (2014). On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek 106, 67–84. doi: 10.1007/s10482-014-0170-z
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Drumonde-Neves, J., Čadež, N., Reyes-Domíngue, Y., Gallmetzer, A., Schuller, D., Lima, T., et al. (2020). Clavispora santaluciae f.a., sp. nov., a novel ascomycetous yeast species isolated from grapes. Int. J. Syst. Evol. Microbiol. 70, 6307–6312. doi: 10.1099/ijsem.0.004531
Favel, A., Michel-Nguyen, A., Peyron, F., Martin, C., Thomachot, L., Datry, A., et al. (2003). Colony morphology switching of Candida lusitaniae and acquisition of multidrug resistance during treatment of a renal infection in a newborn: case report and review of the literature. Diagn. Microbiol. Infect. Dis. 47, 331–339. doi: 10.1016/s0732-8893(03)00094-4
Fell, J. W., Statzell-Tallman, A., Scorzetti, G., and Gutiérrez, M. H. (2011). Five new species of yeasts from fresh water and marine habitats in the Florida Everglades. Antonie Van Leeuwenhoek 99, 533–549. doi: 10.1007/s10482-010-9521-6
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Groenewald, M., Robert, V., and Smith, M. T. (2011). Five novel Wickerhamomyces- and Metschnikowia-related yeast species, Wickerhamomyces chaumierensis sp. nov., Candida pseudoflosculorum sp. nov., Candida danieliae sp. nov., Candida robnettiae sp. nov. and Candida eppingiae sp. nov., isolated from plants. Int. J. Syst. Evol. Microbiol. 61, 2015–2022. doi: 10.1099/ijs.0.026062-0
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
James, S. A., Barriga, E. J. C., Bond, C. J., Cross, K., Núñez, N. C., Portero, P. B., et al. (2009). Candida carvajalis sp. nov., an ascomycetous yeast species from the Ecuadorian Amazon jungle. FEMS Yeast Res. 9, 784–788. doi: 10.1111/j.1567-1364.2009.00518.x
Jindamorakot, S., Limtong, S., Yongmanitchai, W., Tuntirungkij, M., Potacharoen, W., Kawasaki, H., et al. (2007). Two new anamorphic yeasts, Candida thailandica sp. nov. and Candida lignicola sp. nov., isolated from insect frass in Thailand. FEMS Yeast Res. 7, 1409–1414. doi: 10.1111/j.1567-1364.2007.00305.x
Kaewwichian, R., Khunnamwong, P., Am-In, S., Jindamorakot, S., Groenewald, M., and Savitree Limtong, S. (2019). Candida xylosifermentans sp. nov., a D-xylose-fermenting yeast species isolated in Thailand. Int. J. Syst. Evol. Microbiol. 69, 2674–2680. doi: 10.1099/ijsem.0.003505
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kurtzman, C. P., Fell, J. W., Boekhout, T., and Robert, V. (2011). “Methods for isolation, phenotypic characterization and maintenance of yeasts” in The Yeasts–A Taxonomic Study. eds. K. Innis, F. CP, and B. JW. 5th ed (Amsterdam, Netherlands: Elsevier), 87–110.
Kurtzman, C. P., and Robnett, C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371. doi: 10.1023/a:1001761008817
Kurtzman, C. P., and Robnett, C. J. (2013). Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res. 13, 23–33. doi: 10.1111/1567-1364.12006
Kurtzman, C. P., Robnett, C. J., Basehoar, E., and Ward, T. J. (2018). Four new species of Metschnikowia and the transfer of seven Candida species to Metschnikowia and Clavispora as new combinations. Antonie Van Leeuwenhoek 111, 2017–2035. doi: 10.1007/s10482-018-1095-8
Lachance, M. A., Boekhout, T., Scorzetti, G., Fell, J. W., and Kurtzman, C. P. (2011). “Candida Berkhout (1923),” in The yeasts – A taxonomic study. eds. K. Innis, F. CP, and B. JW. 5th ed (Amsterdam: Elsevier), 987–1278.
Lachance, M. A., and Lee, D. K. (2020). Tom Hsiang delineating yeast species with genome average nucleotide identity: a calibration of ANI with haplontic, heterothallic Metschnikowia species. Antonie Van Leeuwenhoek 113, 2097–2106. doi: 10.1007/s10482-020-01480-9
Lachance, M. A., and Phaff, H. J. (2011). “Clavispora Rodrigues de Miranda (1979)” in The yeasts – A taxonomic study. eds. K. Innis, F. CP, and B. JW. 5th ed (Amsterdam: Elsevier), 349–353.
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Libkind, D., Tognetti, C., Ruffini, A., Sampaio, J. P., and van Broock, M. R. (2011). Xanthophyllomyces dendrorhous (Phaffia rhodozyma) on stromata of Cyttaria hariotii in Patagonian Nothofagus forests. Rev. Argent. Microbiol. 43, 226–232. doi: 10.1590/S0325-75412011000300011
Limtong, S., and Kaewwichian, R. (2013). Candida phyllophila sp. nov. and Candida vitiphila sp. nov., two novel yeast species from grape phylloplane in Thailand. J. Gen. Appl. Microbiol. 59, 191–197. doi: 10.2323/jgam.59.191
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Lu, H. Z., Jia, J. H., Wang, Q. M., and Bai, F. Y. (2004). Candida asparagi sp. nov., Candida diospyri sp. nov. and Candida qinlingensis sp. nov., novel anamorphic, ascomycetous yeast species. Int. J. Syst. Evol. Microbiol. 54, 1409–1414. doi: 10.1099/ijs.0.03055-0
Lv, S. L., Chai, C. Y., Wang, Y., Yan, Z. L., and Hui, F. L. (2020). Five new additions to the genus Spathaspora (Saccharomycetales, Debaryomycetaceae) from Southwest China. MycoKeys 75, 31–49. doi: 10.3897/mycokeys.75.57192
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14:60. doi: 10.1186/1471-2105-14-60
Merz, W. G., Khazan, U., Jabra-Rizk, M. A., Wu, L. C., Osterhout, G. J., and Lehmann, P. F. (1992). Strain delineation and epidemiology of Candida (Clavispora) lusitaniae. J. Clin. Microbiol. 30, 449–454. doi: 10.1128/jcm.30.2.449-454.1992
Nakase, T., Jindamorakot, S., Am-In, S., Lee, C. F., Imanishi, Y., and Limtong, S. (2011). Three novel species of the anamorphic yeast genus Candida in the Candida intermedia clade found in Japan, Thailand and Taiwan. J. Gen. Appl. Microbiol. 57, 73–81. doi: 10.2323/jgam.57.73
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. Oxford University Press: New York, NY.
Nguyen, N. H., Suh, S. Q., and Blackwell, M. (2007). Five novel Candida species in insect-associated yeast clades isolated from Neuroptera and other insects. Mycologia 99, 842–858. doi: 10.3852/mycologia.99.6.842
Phaff, H. J., Miranda, M., Starmer, W. T., Tredick, J., and Barker, J. S. F. (1986). Clavispora opuntiae, a new heterothallic yeast occurring in necrotic tissue of Opuntia species. Int. J. Syst. Bacteriol. 36, 372–379. doi: 10.1099/00207713-36-3-372
Ribeiro, J. R. A., Carvalho, P. M. B., Cabral, A. S., Macrae, A., Mendonça-Hagler, L. C. S., Berbara, R. L. L., et al. (2011). Candida middelhoveniana sp. nov., a new yeast species found on the rhizoplane of organically cultivated sugarcane. Antonie Van Leeuwenhoek 100, 341–347. doi: 10.1007/s10482-011-9589-7
Rodrigues de Miranda, L. (1979). Clavispora, a new genus of the Saccharomycetales. Antonie Van Leeuwenhoek 45, 479–483. doi: 10.1007/BF00443285
Rosa, C. A., Pagnocca, F. C., Lachance, M. A., Ruivo, C. C. C., Medeiros, A. O., Pimentel, M. R. C., et al. (2007). Candida flosculorum and Candida floris, two novel yeast species associated with tropical flowers. Int. J. Syst. Evol. Microbiol. 57, 2970–2974. doi: 10.1099/ijs.0.65230-0
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schwartz, K., and Sherlock, G. (2016). Preparation of yeast DNA sequencing libraries. Cold Spring Harb. Protoc. 2016:88930. doi: 10.1101/pdb.prot088930
Scorzetti, G., Fell, J. W., Fonseca, A., and Statzell-Tallman, A. (2002). Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2, 495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x
Shi, C. F., Zhang, K. H., Chai, C. Y., Yan, Z. L., and Hui, F. L. (2021). Diversity of the genus Sugiyamaella and description of two new species from rotting wood in China. MycoKeys 77, 27–39. doi: 10.3897/mycokeys.77.60077
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Urbina, H., Frank, R., and Blackwell, M. (2013). Scheffersomyces cryptocercus: a new xylose-fermenting yeast associated with the gut of wood roaches and new combinations in the Sugiyamaella yeast clade. Mycologia 105, 650–660. doi: 10.3852/12-094
Valinhas, R. V., Pantoja, L. A., Maia, A. C. F., Miguel, M. G. C. P., Vanzela, A. P. F. C., Nelson, D. L., et al. (2018). Xylose fermentation to ethanol by new Galactomyces geotrichum and Candida akabanensis strains. Peer J. 6:e4673. doi: 10.7717/peerj.4673
Weiss, S., Samson, F., Navarro, D., and Casaregola, S. (2013). YeastIP: a database for identification and phylogeny of ascomycetous yeasts. FEMS Yeast Res. 13, 117–125. doi: 10.1111/1567-1364.12017
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: A guide to methods and applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322.
Yoon, S. H., Ha, S. M., Lim, J. M., Kwon, S. J., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yurkov, A., Schäfer, A. M., and Begerow, D. (2009). Clavispora reshetovae a. Yurkov, a.M. Schäfer & Begerow, sp. nov. fungal planet 35. Persoonia 23, 182–183. doi: 10.3767/persoonia.2021.46.11
Zhang, D., Lu, C., Zhang, T., Spadaro, D., Liu, D., and Liu, W. (2014). Candida pruni sp. nov. is a new yeast species with antagonistic potential against brown rot of peaches. Arch. Microbiol. 196, 525–530. doi: 10.1007/s00203-014-0999-6
Zhou, X. F., Peris, D., Kominek, J., Kurtzman, C. P., Hittinger, C. T., and Rokas, A. (2016). In silico whole genome sequencer and analyzer (iWGS): a computational pipeline to guide the design and analysis of de novo genome sequencing studies. G3-genes Genom. Genet. 6, 3655–3662. doi: 10.1534/g3.116.034249
Keywords: Clavispora paralusitaniae sp. nov., taxonomy, rotted wood-inhabiting yeast, genomic analyses, phylogeny, Clavispora xylosa sp. nov.
Citation: Chai C-Y, Li Y, Yan Z-L and Hui F-L (2022) Phylogenetic and genomic analyses of two new species of Clavispora (Metschnikowiaceae, Saccharomycetales) from Central China. Front. Microbiol. 13:1019599. doi: 10.3389/fmicb.2022.1019599
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesCopyright © 2022 Chai, Li, Yan and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng-Li Hui, ZmVuZ2xpaHVpQHllYWgubmV0
 Chun-Yue Chai
Chun-Yue Chai Ying Li1
Ying Li1 Feng-Li Hui
Feng-Li Hui