
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 12 September 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1003783
This article is part of the Research Topic Rising Stars in Evolutionary and Genomic Microbiology: 2022 View all 9 articles
 Yu-Ling Han1,2
Yu-Ling Han1,2 Xu-Hui Wen1,2
Xu-Hui Wen1,2 Wen Zhao1
Wen Zhao1 Xi-Shan Cao1
Xi-Shan Cao1 Jian-Xun Wen3
Jian-Xun Wen3 Jun-Rui Wang1
Jun-Rui Wang1 Zhi-De Hu1
Zhi-De Hu1 Wen-Qi Zheng1,2*
Wen-Qi Zheng1,2*Carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP), a type of Klebsiella pneumoniae (KP) that exhibits hypervirulence and carbapenem resistance phenotypes, can cause severe infections, both hospital- and community-acquired infections. CR-hvKP has brought great challenges to global public health and is associated with significant morbidity and mortality. There are many mechanisms responsible for the evolution of the hypervirulence and carbapenem resistance phenotypes, such as the horizontal transfer of the plasmid carrying the carbapenem resistance gene to hypervirulent Klebsiella pneumoniae (hvKP) or carbapenemase-producing Klebsiella pneumoniae (CRKP) acquiring a hypervirulence plasmid carrying a virulence-encoding gene. Notably, KP can evolve into CR-hvKP by acquiring a hybrid plasmid carrying both the carbapenem resistance and hypervirulence genes. In this review, we summarize the evolutionary mechanisms of resistance and plasmid-borne virulence as well as the prevalence of CR-hvKP.
Klebsiella pneumoniae (KP) is an opportunistic pathogen that can cause infectious diseases in the urinary tract, respiratory tract, blood and soft tissue (Podschun and Ullmann, 1998). KP can be categorized into classic Klebsiella pneumoniae (cKP) and hypervirulent Klebsiella pneumoniae (hvKP) according to its phenotypic and genotypic characteristics. HvKP has higher virulence than cKP (Shon et al., 2013) and can cause severe infectious diseases, especially pyogenic liver abscess (Wang et al., 1998), endophthalmitis (Liu et al., 1986), and meningitis (Xu et al., 2019). It was first identified in seven patients in 1986 (Liu et al., 1986). These patients suffered from severe liver abscesses and endophthalmitis. Despite antibiotic therapy being initiated in a timely manner, six patients lost vision, and one was visually impaired (Liu et al., 1986). To date, many investigations have reported the prevalence of hvKP (Shon et al., 2013). In recent years, partially due to antibiotic abuse, the prevalence of carbapenem-resistant Klebsiella pneumoniae (CRKP) has increased (Doi and Paterson, 2015; Chung, 2016). HvKP and CRKP can evolve into carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) by acquiring the plasmids carrying the carbapenem resistance gene or virulence-encoding gene, respectively. Notably, cKP can evolve into CR-hvKP by acquiring a hybrid plasmid carrying both the carbapenem resistance and hypervirulence genes (He et al., 2022). Therefore, CR-hvKP exhibits both hypervirulence and carbapenem resistance phenotypes, which may cause severe infections in individuals that are difficult to treat with current antibiotics, should attracted worldwide attention (Gu et al., 2018a). At present, CR-hvKP has spread worldwide (Figure 1) and poses a great threat to human public health. In this study, we summarize the resistance mechanisms and virulence evolution mechanism of CR-hvKP.
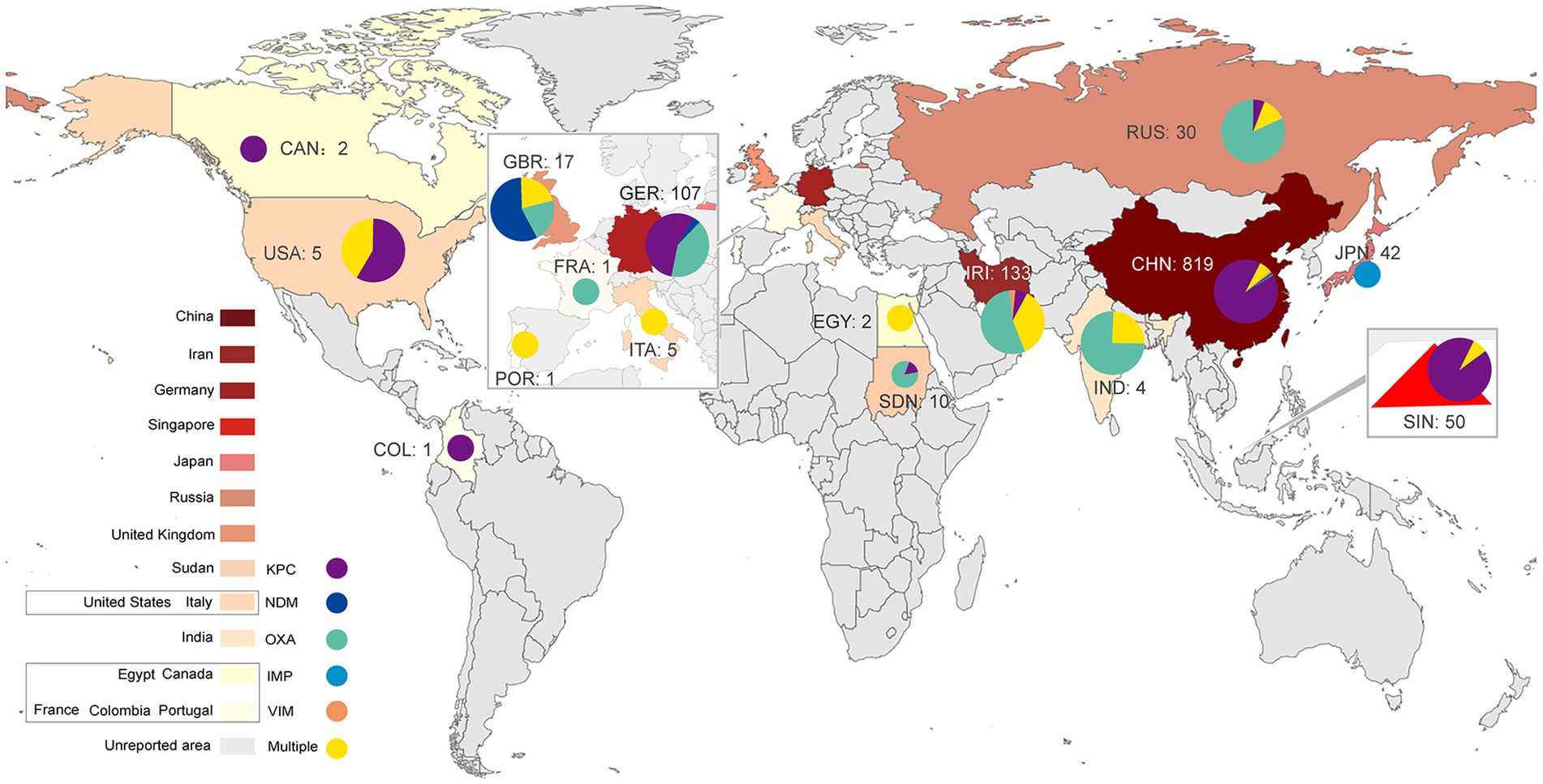
Figure 1. Global distribution of CR-hvKP (2015–2022). KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; VIM, Verona integron-encoded metallo-β-lactamase; IMP, imipenemase; OXA, oxacillinase.
There are three mechanisms responsible for the evolution of CR-hvKP, including carbapenemase production, activation of the efflux pump system and loss of outer membrane proteins (OMPs) expression (Liao et al., 2020). These mechanisms are briefly introduced in this review.
Multidrug resistance genes in plasmids and genomes regulate the resistance characteristics of KP. These genes can encode aminoglycosides, extended-spectrum β-lactamase (ESBL), AmpC β-lactamases or carbapenemases (Tzouvelekis et al., 2012). Carbapenemase production is an essential mechanism of carbapenem resistance in CRKP (Bonomo et al., 2018). According to the Ambler classification, carbapenemases can be categorized into three classes: A, B and D (Queenan and Bush, 2007). Carbapenemase class A can hydrolyze nearly all β-lactam antibiotics (Rasmussen and Bush, 1997). Klebsiella pneumoniae carbapenemases (KPCs) are the primary class A enzymes abundant in CRKP (Brink, 2019). This enzyme was first identified in 1996 in the United States from a KP isolate (Chen et al., 2014). There are many subtypes of KPCs, including KPC-2 to KPC-13 (Tzouvelekis et al., 2012). Notably, the KPC genes, blaKPC, in the genome and plasmid share transmission features, such as horizontal transfer or cloning to other strains mediated by genetic elements (Rasmussen and Bush, 1997; Chen et al., 2014; Chew et al., 2017). When hvKP obtains blaKPC, it can evolve to CR-hvKP with both high virulence and carbapenem-resistance characteristics. Globally, many blaKPC-positive CR-hvKP strains have been reported since 2010 (Lan et al., 2021). A report showed that the most common subtypes of blaKPC in the USA are blaKPC-2 and blaKPC-3. In Asia (especially China) and Europe, the most frequent type is blaKPC-2 (Yang et al., 2021a). The global prevalence of the blaKPC-positive CR-hvKP strain is summarized in Figure 1; Table 1.
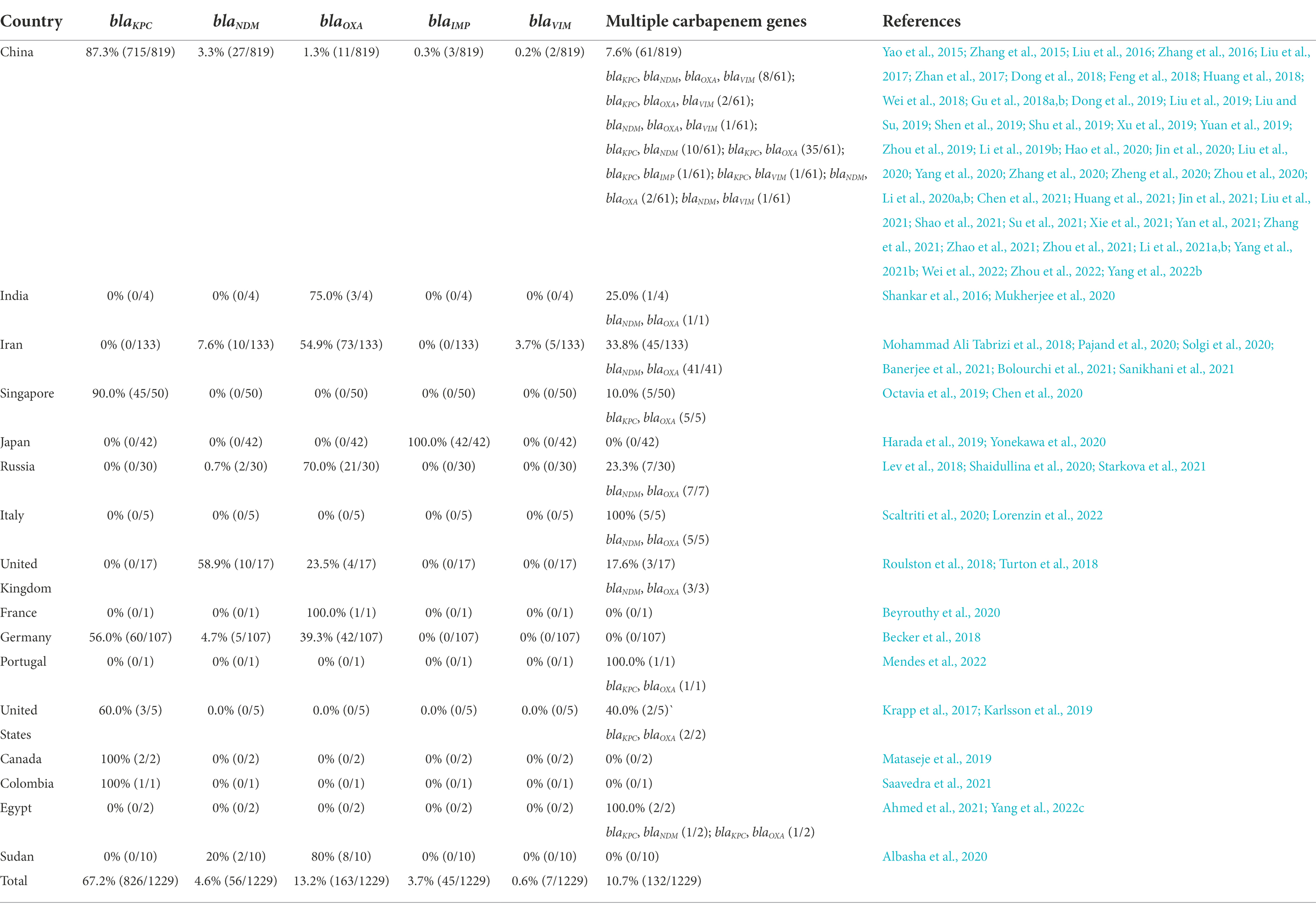
Table 1. Global geographic distribution of isolates obtained carbapenem resistance gene from June 2015 to April 2022.
Class B carbapenemase enzymes, also named metallo-β-lactamases (MBLs), mainly include New Delhi metallo-β-lactamase (NDM-1), Verona integron-encoded metallo-β-lactamase (VIM-1) and imipenemase (IMP). These enzymes are characterized by one or two zinc ions as active centers (Tzouvelekis et al., 2012; Bonomo et al., 2018). MBLs can catalyze the hydrolysis of almost all β-lactam antibiotics except for monobactams (Jeon et al., 2015). Class D enzymes mainly include oxacillinase 48 (OXA-48) and 181 (OXA-181). These enzymes have carbapenemase activity, but their carbapenem hydrolysis activity is weak. Notably, the transmission of class B and class D enzyme genes depends mainly on plasmids (Tzouvelekis et al., 2012; Bonomo et al., 2018), making their distribution more regional. Class B (e.g., NDM-1, VIM-1) and D (e.g., OXA-48, OXA-181) enzymes are primarily distributed in Asia and Europe (Nordmann et al., 2012). In Asian countries such as India and Iran and in European countries such as Russia and Italy, the majority of CR-hvKP isolates were OXA-48-positive strains, followed by NDM-1- and VIM-1-positive strains (Figure 1; Supplementary Table S1). In 2018, an investigation from Iran reported five patients infected with rare strains of K1 and ST23 CR-hvKP, which carry the blaVIM-2 gene (Mohammad Ali Tabrizi et al., 2018). Notably, four of these patients died during hospitalization (Mohammad Ali Tabrizi et al., 2018), indicating the hypervirulence of this type of CR-hvKP. In addition, some Asian countries, such as China and Japan, have occasionally reported IMP-positive CR-hvKP strains (Zhang et al., 2015; Harada et al., 2019).
In conclusion, the distribution of carbapenemases differs according to geographical features. CR-hvKP strains that produce class A enzymes are highly prevalent in Asia and America, and a handful of strains produce class B and D enzymes in Europe (Figure 1; Supplementary Table S1). However, the increase in factors such as global immigration may lead to changes in the linkages between these bacterial resistance mechanisms and regions or cities (Bonomo et al., 2018). Therefore, the areas with low prevalence cannot be ignored when detecting CR-hvKP-related resistance genes.
Furthermore, CR-hvKP strains simultaneously producing two or more carbapenemases can cause serious infectious diseases with high mortality. These types of CR-hvKP have been reported in many countries and regions (Figure 1; Supplementary Table S1). For example, CR-hvKP strains producing NDM and OXA-48 were reported in Italy and Iran in 2020 (Scaltriti et al., 2020; Solgi et al., 2020; Bolourchi et al., 2021). A CR-hvKP strain carrying blaNDM-1, blaNDM-5 and blaOXA-48 carbapenem resistance genes was reported in northern Italy in 2022 (Lorenzin et al., 2022). In particular, a CR-hvKP strain carrying blaKPC-2 and blaOXA-48 genes was reported in Egypt in the same year (Yang et al., 2022c). This CR-hvKP isolate contained two resistance plasmids: pEBSI041-2 (a blaOXA-48 gene carrier) and pEBSI041-3 (a blaKPC-2 gene carrier). pEBSI041-3 can be transferred with the assistance of plasmid pEBSI041-2, and the two plasmids can fuse into a larger plasmid carrying both blaOXA-48 and blaKPC-2 genes during co-transfer. Plasmids carrying different carbapenem resistance genes could be transferred through gene recombination, rearrangement and the formation of fusion plasmids, resulting in more complex resistance mechanisms in CR-hvKP strains (Yang et al., 2022c).
KP can rapidly pump intracellular antibiotics out of the cell through the efflux pump system, thus decreasing the concentration of antibiotics in the bacteria. Activation of the efflux pump system and decreased expression of outer membrane porins (OMPs) are two common mechanisms of carbapenem resistance in CRKP (Figure 2). The tripartite efflux system AcrAB-TolC is the archetypal resistance-nodulation-cell division (RND) efflux pump and contributes to multidrug resistance (Tam et al., 2021). In KP, the AcrAB-TolC efflux pump can be regulated by RamA, an AraC/XylS transcriptional activator (Xu et al., 2021a). Upregulation of ramA enhances the expression of acrAB and tolC, resulting in increased translation of the AcrAB-TolC pump proteins, ultimately leading to multidrug resistance (Nishino and Yamaguchi, 2004). In 2021, 17 CRKP strains that cause urinary tract infections were isolated. The carbapenem resistance mechanism of 11 KPC and/or VIM-positive CRKP strains was mainly related to the overexpression of the ramA gene and upregulation of the acrB and oqxB genes. Only 6 strains exhibited other resistance mechanisms (Lee et al., 2021). Therefore, overexpression of efflux pumps is one of the major resistance mechanisms of CRKP (Du et al., 2018).
Moreover, mutation of the efflux pump is another mechanism for changing the activity of the efflux pump (Figure 2). In 2021, researchers found that the transcription factor ramA could downregulate the efflux pump genes acrB and oqxB after mutation, resulting in a significant decrease in the MIC of tigecycline in carbapenem-susceptible cKP (Xu et al., 2021a). When the efflux pump regulator is mutated, the corresponding antibiotic resistance may also reduce bacterial adaptability and virulence (Du et al., 2018).
OMPs are the channels through which antibiotics enter bacteria. Similar to efflux pumps, changes in OMPs also affect the resistance phenotype of bacteria (Li et al., 2015). OmpC, OmpD, OmpE, OmpF, and PhoE are common outer membrane pore proteins in clinical Enterobacter strains (Vergalli et al., 2020). OmpK36 belonging to the OmpC family and OmpK35 belonging to the OmpF family are common in KP (Pagès et al., 2008). In CRKP, changes in the expression levels of OmpK35 and OmpK36 can have important effects. Low expression of OmpK35 is one of the important mechanisms for the antibiotic resistance of ESBL-producing CRKP (Wang et al., 2009). OmpK36 can reduce the sensitivity of ESBL and AmpC β-lactamase of CRKP and is closely related to the resistance of CRKP to carbapenems (Wang et al., 2009). In a retrospective study with 28 cases of CRKP infections from 9 cities in China, 5 CR-hvKP strains were identified (Zhang et al., 2015). The expression levels of OmpK35 and OmpK36 were decreased in 2 CR-hvKP strains (Zhang et al., 2015), indicating that the low expression levels of OmpK35 and OmpK36 are potential causes of resistance to carbapenems (Zhang et al., 2015).
In addition, in 2009, a study with 28 CRKP strains found that 9 CRKP isolates with high levels of ertapenem resistance consistently lacked both OmpK35 and OmpK36 porins (Doumith et al., 2009). Sequencing of the ompK35 and ompK36 genes revealed diverse types of disruption in most isolates, including point mutations or the presence of insertion sequences (Iss), such as IS1 and IS10 (Doumith et al., 2009), which indicated that the carbapenem resistance characteristics of the CRKP strains are likely to be influenced by these mutations in OMPs (Doumith et al., 2009).
The outermost capsule polysaccharides are key for the hypermucoviscous phenotype of hvKP. The capsular polysaccharide can protect bacteria from harmful environmental factors, such as antimicrobial compounds, and improve their virulence, so hvKP is traditionally thought to have hypermucoviscosity characteristics (Russo and Marr, 2019). Therefore, the string test to detect viscosity is a classic method to identify hvKP. Briefly, the monoclonal colonies on the agar plate are gently picked up with a bacteriological loop, and a length of viscous filament >5 mm is judged as a positive result in the string test (Lan et al., 2021).
Many studies have shown that two capsule regulator genes, mucoid phenotype A (rmpA), and mucoid phenotype A2 (rmpA2), on the virulence plasmid are hvKP-specific virulence factors (Chen and Chen, 2021). Overexpression of the rmpA gene increases the virulence of KP in a mouse infection model (Lin et al., 2020), which indicates that the highly virulent phenotype is associated with high expression of the rmpA gene (Lin et al., 2020). However, some studies have indicated that there is no correlation between hypermucoviscosity and virulence. A retrospective study that included 28 CRKP isolates collected from 9 regions in China showed that although 2 strains were positive in the string test, they did not carry the rmpA gene (Zhang et al., 2015). In addition, 5 CRKP strains carried the rmpA gene but were negative in the string test (Zhang et al., 2015). Therefore, the string test (hypermucoviscosity) is not a sensitive and specific method to identify hvKP. Consequently, the identification of hvKP should include the identification of invasion-related clinical characteristics and other virulence-associated determinants.
Variations in the capsular polysaccharide composition result in different capsules (Choby et al., 2020). The K-antigens encoded by the cps (capsule polysaccharide synthesis) locus belong to the bacterial capsule polysaccharides (CPS) with genetic diversity. It has been reported that there are nine kinds of cps operons (Chung The et al., 2015). Among them, the wzi gene is one of the most important determinants of CPS, and usually, KP serotypes can be identified by detecting the wzi gene (Wyres et al., 2016; Choby et al., 2020).
Recent studies have shown that K-antigens are associated with hvKP infection (Follador et al., 2016). The number of serotypes has been estimated to be 77 for K-antigens (Choby et al., 2020). K1 and K2 are the most common capsule serotypes and the major pathogen capsule serotypes causing invasive infectious diseases (Fung et al., 2002; Chuang et al., 2006; Su et al., 2020). Therefore, capsule serotypes K1 and K2 are typical characteristics of hvKP (Choby et al., 2020). Other capsule serotypes, such as K20 (Zhan et al., 2017), K47 (Huang et al., 2018), K54 (Yuan et al., 2019), K57 (Zhang et al., 2020b) and K64 (Liu et al., 2017; Zhang et al., 2020b), can also cause serious infectious diseases and have strong virulence. Thus, capsule type may be associated with bacterial virulence or can enhance virulence (Russo and Marr, 2019; Choby et al., 2020), but it is not the only virulence-associated determinant of hvKP.
A study with 85 hvKP and 90 cKP strains assessed the diagnostic accuracy rate of K1, K2, K5, K20, K54, and K57 for hvKP (Russo et al., 2018). Although the diagnostic specificity of these capsule serotypes was above 90%, the sensitivity of K1 and K2 of hvKP was only 55 and 20%, respectively. For K5, K20, K54, and K57, the sensitivity was less than 8%. However, it is worth noting that, for the combination of the K1, K2, K5, K20, K54, and K57 capsule types, the sensitivity and specificity were 93 and 88%, respectively, and the accuracy rate was 90% (Russo et al., 2018). Therefore, combining multiple capsule types in clinical treatment can also be used to accurately diagnose hvKP.
HvKP strains can release siderophores (SPs) when they invade the host body. SPs can acquire iron from the binding protein in the host body and then bind to the SP-specific receptor to re-enter KP cells. Eventually, hvKP used the “steal” iron to grow (Russo et al., 2014; Russo and Marr, 2019; Choby et al., 2020). There are four SPs secreted by hvKP, namely enterobactin, salmochelin, yersiniabactin and aerobactin. Site-specific gene disruptions were performed on the four SPs in previous research. Only when the aerobactin gene iucA was destroyed was the ex vivo growth/survival of hvKP in human ascites fluid and serum significantly decreased, and the virulence of hvKP in the mouse infection model was also significantly decreased. In contrast, the other three iron carriers did not show the features mentioned above (Russo et al., 2014, 2015). These results indicate that aerobactin in SPs is the primary determinant of the virulence of hvKP. In addition, a diagnostic accuracy study suggested that the expression levels of peg-344, iroB and iucA can accurately predict the virulence of hvKP (Russo et al., 2018). Peg-344, iroB and iucA have demonstrated >0.95 diagnostic accuracy in identifying hvKP strains. Therefore, SP-related encoding genes, especially iucA, are key markers in the identification of hvKP (Russo et al., 2018).
As mentioned above, cKP can transform into hvKP with a hypervirulence virulence phenotype by acquiring virulence plasmids. These virulence plasmids usually carry many virulence coding genes, such as hypermucoviscous phenotype genes (rmpA and rmpA2), SP-related genes (iucABCD-iutA, iroBCDN, ybtAEPQTUX and entABCDEFS) and genes encoding tellurite and silver resistance (terABCDEWXZ, silCERS). Among them, iroB, iucA, peg-344, p-rmpA and p-rmpA2 are the most accurate and characteristic molecular markers for defining hvKP on virulence plasmids (Hsu et al., 2011; Bulger et al., 2017; Russo et al., 2018). To date, the 224 kb plasmid pNTUH-K2044 on K1-ST23 KP strain NTUH-K2044 and its highly similar plasmid pLVPK on K2-ST86 KP strain CG43 are the most reported virulence plasmids (Chen et al., 2004; Wu et al., 2009). The virulence of KP strains carrying these virulence plasmids (pNTUH-K2044, pLVPK or pLVPK-like virulence plasmid) can be significantly increased (Gu et al., 2018a). In a study, the hvKP strain K2602 carrying the virulence plasmid pK2602 was used as the donor, and the ST11 CRKP strain HS11286 and E. coli J53 were used as the recipient strains for conjugation experiments. It was found that the transconjugants (HS11286-K2606 and J53-K2606) acquired the virulence plasmid pK2606, which can encode aerobactin from K2602, and exhibited increased siderophore production, though the result for HS11286-K2606 was more significant. These two transconjugants could also cause high mortality in Galleria mellonella and mice. Therefore, virulence plasmids are an important cause of the hypervirulence phenotype (Tian et al., 2021). More importantly, many studies have reported fusion plasmids integrating both carbapenem resistance and hypervirulence phenotypes in recent years (Lorenzin et al., 2022; Zhou et al., 2022; Yang et al., 2022b), which signifies that hvKP has evolved into a “superbug.”
Bloodstream infection (BSI) and pneumonia caused by CRKP can have higher mortality than CRKP urinary colonization (Hauck et al., 2016). Some scholars have also carried out relevant studies on CRKP virulence. In 2015, researchers infected mice with KPC-producing ST11 CRKP and ST258 CRKP strains and compared their virulence. The results showed that the virulence of these two strains was low, indicating that KPC production is irrelevant to the virulence of these strains (Chiang et al., 2016). Similarly, in 2017, researchers in the United States isolated 4 CRKP strains from patients with necrotizing skin and soft tissue infections (Krapp et al., 2017). All 4 CRKP strains presented low virulence in the mouse acute pneumonia model. However, in a mouse subcutaneous tissue infection model, one CRKP strain with a positive string test result caused severe skin abscess and finally spread to the liver (Krapp et al., 2017). These results indicate that the virulence level of the CRKP strain with a positive string test result might be tissue-dependent (Krapp et al., 2017). Moreover, a study that included 56 KPC-producing CRKP strains isolated from hospitalized patients in China in 2018 compared the virulence characteristics of different sequence types. Among the CRKP stains, 43 were ST11, 6 were ST147, 4 were ST15, and 3 were ST1456, ST65, and ST23. According to the Galleria mellonella infection model, the virulence of the ST11 CRKP strains was lower than that of other sequence types (Liu et al., 2017). This result indicated that the virulence of CRKPs with different sequence types varies. The virulence characteristic of the ST11 CRKP strain, the most common in Asia, was low before acquisition of the virulence-associated determinants.
The complex and diverse evolutionary mechanisms are the key factors that can improve the virulence and antibiotic resistance of KP stains and increase the relevant morbidity and mortality of the associated infectious diseases. This is the primary reason why the low-virulence cKP and CRKP strains, or the strains with low antibiotic resistance, evolved to “superbug” CR-hvKP with the characteristic of stronger virulence, high carbapenem resistance and global transmission. The following three pathways are the main mechanisms by which cKP, hvKP, and CRKP evolve into CR-hvKP.
Plasmids, transposons, phages and insertion elements carrying movable carbapenem resistance genes in CRKP can be horizontally transferred to hvKP strains to form CR-hvKP. Because of the regional distribution characteristics of carbapenemases (Cui et al., 2019), the carbapenem resistance phenotype acquired by hvKP also showed similar regional characteristics. For example, KPC genes were the most common carbapenemase genes in China and the United States (Cui et al., 2019), and KPC-positive CR-hvKP strains were also reported more frequently in these areas. In 2014, a study in the United States was the first to report a hypermucoviscous ST23 KP strain that acquired the blaKPC-2 gene and evolved into multidrug-resistant CR-hvKP (Cejas et al., 2014). Similarly, five K1 hvKP strains were reported in China, which formed K1 CR-hvKP by acquiring virulence plasmids harboring the blaKPC-2 gene or combining a movable DNA fragment carrying blaKPC-2 (Zhang et al., 2016). Furthermore, similar evolutionary mechanisms were also mentioned in a study in Hong Kong, China (Dong et al., 2019). HvKP strains with ST23 and K1 capsule serotypes acquired blaVIM-1-bearing carbapenem resistance plasmids via horizontal transfer, thus evolving into CR-hvKP (Dong et al., 2019). In addition, NDM and OXA are more common in Russia and India. In 2018, Russian researchers first reported that ST23 and K1 hvKP simultaneously acquired plasmids carrying the extended-spectrum β-lactamase CTX-M-15 gene and carbapenemase OXA-48 gene and evolved into CR-hvKP (Lev et al., 2018). In India, it was also reported that a hvKP strain harboring blaOXA and blaNDM genes encoding carbapenem resistance caused two deaths (Shankar et al., 2016). Notably, a study referenced a rare IMP-positive CR-hvKP isolate, XH210, recovered from human blood in Hangzhou, China (He et al., 2022). This CR-hvKP strain was characterized as having the ST17 KL38/O2 serotype and had the resistance plasmid pXH210-IMP, which carries the blaIMP-4 gene. pXH210-IMP could be transferred from XH210 to E. coli and KP recipients in a conjugation experiment, indicating that the resistance plasmid had transferability. Furthermore, XH210 exhibited hypervirulence in the Galleria mellonella and mouse infection models but lacked the characteristic markers that are frequently associated with hypervirulence. The study demonstrated that hvKP strains without an obvious hypervirulence phenotype could evolve into CR-hvPK by acquiring the blaIMP-4 gene (He et al., 2022). The horizontal transmission of mobile genes is the key factor leading to the continuous evolution of KP into CR-hvKP (Chen et al., 2014). The acquisition of carbapenem resistance genes by these virulent strains indicates that these major hospital pathogens are evolving.
Compared with highly virulent strains, multidrug-resistant strains are more likely to produce a wide range of surface polysaccharide sites by chromosomal recombination and are more likely to acquire virulence plasmids (Wyres et al., 2019). Therefore, the original CRKP easily evolved into CR-hvKP through horizontal transfer of mobile virulence genes. A number of studies have shown that the virulence of CRKP can be significantly enhanced after acquisition of the pLVPK/pLVPK-like virulence plasmid (Gu et al., 2018a; Zhou et al., 2020; Zhang et al., 2020b). For example, an outbreak of ST11 CR-hvKP was reported in a hospital in China in 2018 (Gu et al., 2018a). Five patients died due to severe pneumonia after CR-hvKP infection. The researchers found that all the CR-hvKP isolates were positive in the string test and showed high virulence in a neutrophil killing assay and Galleria mellonella infection model (Gu et al., 2018a). Furthermore, these CR-hvKP isolates all carried a 170 kb pLVPK-like virulence plasmid (Gu et al., 2018a). The virulence plasmid of hvKP can be horizontally transferred to ST11 CRKP alone or can lead to the formation of ST11 CR-hvKP by conjugating with the IncF plasmid (Xu et al., 2021b). Interestingly, Bolourchi et al. (2021) claims that two CR-hvKP strains evolved from acquiring the plasmids carrying carbapenem resistance genes by hvKP strains carrying non-conjugated virulence plasmids. According to the latest study, non-conjugated virulence plasmids can integrate other plasmids carrying conjugative transfer genes to improve their own transmission. When this new conjugation plasmid was transferred to E. coli strain EC600 or other types of KP, its virulence potential in the mouse infection model could be significantly improved (Yang et al., 2022a). After acquiring the virulence plasmid, these CR-hvKP strains presented strong virulence and had the capacity of high virulence transmission (Xu et al., 2021b), which is more likely to cause outbreaks in hospitals or communities.
In recent years, many studies have reported the evolution of KP into CR-hvKP via the acquisition of hybrid plasmids. There are two important mechanisms for the formation of hybrid plasmids. One involves two single-chain fragments changing at special fusion sites to form hybrid plasmids, and the other involves homologous recombination to form hybrid plasmids (Xu et al., 2021b; Figure 2).
Researchers have detected the fusion plasmid p17ZR-91-Vir-KPC in an ST86, K2 CR-HvKP strain in China (Xie et al., 2021). The p17ZR-91-Vir-KPC plasmid was formed by homologous recombination of the pLVPK-like virulence plasmid p17ZR-91-Vir and the circular resistance plasmid p17ZR-91-KPC carrying blaKPC-2 in HR1 and HR2 homologous regions. Notably, the p17ZR-91-Vir-KPC fusion plasmid can be transferred among strains with different sequence types. ST11 hvKP can evolve into ST11 CR-hvKP by acquiring the fusion plasmid, which is an evolutionary mechanism for the most prevalent strain ST11 CR-hvKP (Xie et al., 2021). In addition, one study found a hybrid plasmid, pCRHV-C2244, carrying a series of virulence genes, including iroBCDN, iucABCD-iutA, and rmpA2, and the carbapenem resistance gene blaKPC-2, which was detected in an ST11, K64 CRKP strain in China in 2021. Multiple IS26 sequences on the plasmid can regulate the recombination of the fragments carrying blaKPC-2 and virulence genes, reverse the large sequence fragment on the plasmid, and finally form the hybrid plasmid pCRHV-C2244 (Jin et al., 2021). Thus, recombination between gene fragments with insertion elements is a mechanism for the formation of hybrid plasmids.
In addition, the virulence plasmid can be integrated into chromosomes, allowing bacteria to evolve into a more virulent strain. In 2021, two hvKP strains were isolated from the wound of a severely infected patient (Eger et al., 2021). Whole-genome sequencing revealed that the hypervirulence genes, such as iroBCDN, iucABCD/iutA, rmpA/A2 and peg, were all located in the specific regions inserted into the chromosome. This suggests that the virulence plasmids can be integrated into the chromosome. Further analysis showed that these processes affected the function of chromosomes, and the evolved highly virulent strains could be vertically transmitted (Eger et al., 2021). The virulence plasmids integrated into chromosomes of the above KP strains indicated that hvKP can evolve by transferring its genes horizontally to other strains and can evolve through vertical transmission.
The recent emergence of ST11 is the most frequent clone for CR-hvKP globally, especially in China (Wu et al., 2017; Liao et al., 2020; Wei et al., 2022). ST11 CR-hvKP can cause bacterial liver abscess (Yang et al., 2020; Cai et al., 2022), bacteremia and other infections, and it is a high-risk clinical pathogen that attracts worldwide attention (Wu et al., 2017). Notably, a report from China showed that pneumonia with high mortality was caused by ST11 CR-hvKP (Yang et al., 2022b). Hybrid conjugative virulence plasmids, e.g., a pLVPK-like hybrid plasmid with high virulence and carbapenem resistance genes, are demonstrated to readily convert an ST11 CRKP strain to a CR-hvKP strain via conjugation (Xie et al., 2021; Yang et al., 2022b). Therefore, ST11 CR-hvKP is a challenge for clinicians in various clinical settings and deserves more attention (Gu et al., 2018a).
In recent years, CR-hvKP has been increasingly reported in China (Gu et al., 2018a; Yuan et al., 2019; Su et al., 2020; Yang et al., 2020; Zhang et al., 2020b; Chen and Chen, 2021), the United States (Chiang et al., 2016), India (Shankar et al., 2016), Russia (Shaidullina et al., 2020), Egypt (Ahmed et al., 2021), Italy (Di Pilato et al., 2021), and other countries, represented by the ST11, ST23, and ST258 types. In this study, we summarized the relevant information on the CR-hvKP strains identified in articles published from June 2015 to April 2022. As shown in Table 1, Asia is the main epidemic area of CR-hvKP, represented by China, India and Singapore. China reported the largest number of CR-hvKP strains among these countries, and the most common sequence type was ST11 (Zhang et al., 2017), followed by ST23 (Su et al., 2021), ST25 (Li et al., 2019b) and ST65 (Su et al., 2021). The most common capsular serotypes were K1 (Zhang et al., 2016) and K2 (Su et al., 2020), followed by K64 (Hao et al., 2020). In China, the blaKPC-2 gene in CR-hvKP was widespread, and the blaKPC gene detection rate was as high as 87.3% (715/819), while the detection rates of the blaNDM, blaOXA, blaIMP and blaVIM genes were 3.3% (27/819), 1.3% (11/819), 0.3% (3/819), and 0.2% (2/819), respectively (Table 1). CR-hvKP strains that carry two or more carbapenem resistance genes were also found in China (7.6%, 61/819). In other Asian countries, such as India (Shankar et al., 2016) and Iran (Mohammad Ali Tabrizi et al., 2018; Solgi et al., 2020), blaOXA and blaNDM genes were more common. In Iran, the detection rates of blaOXA and blaNDM were 54.9% (73/133) and 7.6% (10/133), respectively. In Japan (Yonekawa et al., 2020), the main detected enzyme was IMP, and the detection rate was 100% (42/42). The virulence plasmids in CR-hvKP strains were usually reported in China, and the most widespread were the pLVPK (Gu et al., 2018a) and pLVPK-like plasmids (Li et al., 2020b; Chen et al., 2021). In addition, hybrid plasmids pKP70-2 (Dong et al., 2018) and pCRHV-C2244 (Jin et al., 2021) were also reported. Europe and America have the second highest prevalence of CR-hvKP, mainly represented by ST23 and K1/K2 capsular serotypes strains. The primary carbapenem resistance genes in Europe are blaNDM and blaOXA (Becker et al., 2018; Lev et al., 2018; Turton et al., 2018), while blaKPC-2 is more prevalent in America (Karlsson et al., 2019; Mataseje et al., 2019). The pLVPK-like virulence plasmids have also been reported in Europe and the United States (Lev et al., 2018), but there are few reports on the fusion plasmid with both virulence and carbapenem resistance genes. The virulence genes of these CR-hvKP strains are usually located on chromosomes, with few strains carrying virulence plasmids. Moreover, two cases of ST11-type CR-hvKP infection were reported in Egypt in the last 2 years. One strain carried both blaKPC-2 and blaNDM-1 genes (Ahmed et al., 2021), while the other strain carried both blaKPC-2 and blaOXA-48 genes (Yang et al., 2022c). In 2020, 10 CR-hvKP strains carrying both blaNDM and blaOXA-48 genes were reported in Sudan (Albasha et al., 2020).
As a class of bacteria of CRE, most of the current treatment plans for CR-hvKP refer to the treatment guidelines of CRE. Currently, carbapenems are the most widely used first-line antibiotics for CRE (Tzouvelekis et al., 2012). Meropenem, meropenem-vaborbactam and imipenem-relebactam are commonly used in pediatric CRE infection diseases (Chiotos et al., 2020). Overwise, meropenem-vaborbactam can effectively inhibit the growth of CRE strain producing KPC, and has a higher clinical cure rate and lower side effects, especially nephrotoxicity (Pfaller et al., 2018). Pascale et al. noted that high-dose continuous infusion of meropenem is a wise choice to treat CRE in exceptional cases (Pascale et al., 2019). However, the use of carbapenem antibiotics leads to an increasing number of carbapenemase-producing strains and increases the difficulty of clinical treatment (Tzouvelekis et al., 2012). Some new anti-CRE antibacterials are also good options for CRE treatment, including ceftazidime-avibactam (Shields et al., 2018; Chiotos et al., 2020), plazomicin, tigecycline combination therapy (Ni et al., 2016) and colistin (Li et al., 2019a). The use of ceftazidime-avibactam against CRKP strains has shown an excellent therapeutic effect on clinical cure rate and survival rate (Farrell et al., 2014; Yin et al., 2019). However, high ceftazidime hydrolysis activity and OmpK35 porin deficiency can lead to the decreased susceptibility to ceftazidime-avibactam in KPC-producing ST11 CRKP (Shen et al., 2017). Importantly, given the different epidemiological situations in different regions and the characteristics of enzyme production of the strains, clinical medication should be based on specific circumstances (Pogue et al., 2019).
CR-hvKP has become widespread in many countries. Treatment options for CR-hvKP are also relatively limited. The transfer of plasmids carrying removable genes, as well as the fusion and recombination of plasmids between bacteria, are the key factors underlying the high mutation and transmission rates of CR-hvKP. Future studies should also pay more attention to the vertical transmission of the virulence plasmid of CR-hvKP integrated into the chromosome, which may lead to new discoveries in research on the molecular mechanisms of CR-hvKP.
W-QZ designed and supervised the study. Y-LH collected the data from previous studies and drafted the manuscript. X-HW, X-SC, WZ, and J-XW provided administrative support and provided intellectual contributions to the manuscript. J-RW, Z-DH, and W-QZ critically reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the General Program of the Inner Mongolia Medical University Inner Mongolia (YKD2021MS011), the Young Talents of Science and Technology in Universities of Inner Mongolia (NJYT-20-B14), and the Science and Technology innovation team of the Inner Mongolia Medical University Inner Mongolia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1003783/full#supplementary-material
Ahmed, M., Yang, Y., Yang, Y., Yan, B., Chen, G., Hassan, R. M., et al. (2021). Emergence of Hypervirulent Carbapenem-resistant Klebsiella pneumoniae Coharboring a Bla (NDM-1)-carrying virulent plasmid and a Bla (KPC-2)-carrying plasmid in an Egyptian hospital. mSphere 6:21. doi: 10.1128/mSphere.00088-21
Albasha, A. M., Osman, E. H., Abd-Alhalim, S., Alshaib, E. F., Al-Hassan, L., and Altayb, H. N. (2020). Detection of several carbapenems resistant and virulence genes in classical and hyper-virulent strains of Klebsiella pneumoniae isolated from hospitalized neonates and adults in Khartoum. BMC. Res. Notes 13:312. doi: 10.1186/s13104-020-05157-4
Banerjee, T., Wangkheimayum, J., Sharma, S., Kumar, A., and Bhattacharjee, A. (2021). Extensively drug-resistant Hypervirulent Klebsiella pneumoniae from a series of neonatal sepsis in a tertiary care hospital, India. Front Med (Lausanne) 8:645955. doi: 10.3389/fmed.2021.645955
Becker, L., Kaase, M., Pfeifer, Y., Fuchs, S., Reuss, A., von Laer, A., et al. (2018). Genome-based analysis of Carbapenemase-producing Klebsiella pneumoniae isolates from German hospital patients, 2008-2014. Antimicrob. Resist. Infect. Control 7:62. doi: 10.1186/s13756-018-0352-y
Beyrouthy, R., Dalmasso, G., Birer, A., Robin, F., and Bonnet, R. (2020). Carbapenem resistance conferred by OXA-48 in K2-ST86 Hypervirulent Klebsiella pneumoniae, France. Emerg. Infect. Dis. 26, 1529–1533. doi: 10.3201/eid2607.191490
Bolourchi, N., Shahcheraghi, F., Giske, C. G., Nematzadeh, S., Noori Goodarzi, N., Solgi, H., et al. (2021). Comparative genome analysis of colistin-resistant OXA-48-producing Klebsiella pneumoniae clinical strains isolated from two Iranian hospitals. Ann. Clin. Microbiol. Antimicrob. 20:74. doi: 10.1186/s12941-021-00479-y
Bonomo, R. A., Burd, E. M., Conly, J., Limbago, B. M., Poirel, L., Segre, J. A., et al. (2018). Carbapenemase-producing organisms: A global scourge. Clin. Infect. Dis. 66, 1290–1297. doi: 10.1093/cid/cix893
Brink, A. J. (2019). Epidemiology of carbapenem-resistant gram-negative infections globally. Curr. Opin. Infect. Dis. 32, 609–616. doi: 10.1097/QCO.0000000000000608
Bulger, J., MacDonald, U., Olson, R., Beanan, J., and Russo, T. A. (2017). Metabolite transporter PEG344 is required for full virulence of Hypervirulent Klebsiella pneumoniae strain hvKP1 after pulmonary but not subcutaneous challenge. Infect. Immun. 85:17. doi: 10.1128/IAI.00093-17
Cai, Z., Jia, T., Pu, M., Zhang, S., Zhang, J., Geng, R., et al. (2022). Clinical and molecular analysis of ST11-K47 Carbapenem-resistant Hypervirulent Klebsiella pneumoniae: A strain causing liver abscess. Pathogens 11:657. doi: 10.3390/pathogens11060657
Cejas, D., Fernandez Canigia, L., Rincon Cruz, G., Elena, A. X., Maldonado, I., Gutkind, G. O., et al. (2014). First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J. Clin. Microbiol. 52, 3483–3485. doi: 10.1128/JCM.00726-14
Chen, Y. T., Chang, H. Y., Lai, Y. C., Pan, C. C., Tsai, S. F., and Peng, H. L. (2004). Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198. doi: 10.1016/j.gene.2004.05.008
Chen, Y., and Chen, Y. (2021). Clinical challenges with Hypervirulent Klebsiella Pneumoniae (hvKP) in China. J Transl Int Med 9, 71–75. doi: 10.2478/jtim-2021-0004
Chen, R., Liu, Z., Xu, P., Qi, X., Qin, S., Wang, Z., et al. (2021). Deciphering the epidemiological characteristics and molecular features of Bla (KPC-2)- or Bla (NDM-1)-positive Klebsiella pneumoniae isolates in a newly established hospital. Front. Microbiol. 12:741093. doi: 10.3389/fmicb.2021.741093
Chen, Y., Marimuthu, K., Teo, J., Venkatachalam, I., Cherng, B. P. Z., De Wang, L., et al. (2020). Acquisition of Plasmid with Carbapenem-resistance gene Bla(KPC2) in Hypervirulent Klebsiella pneumoniae, Singapore. Emerg. Infect. Dis. 26, 549–559. doi: 10.3201/eid2603.191230
Chen, L., Mathema, B., Chavda, K. D., DeLeo, F. R., Bonomo, R. A., and Kreiswirth, B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
Chew, K. L., Lin, R. T. P., and Teo, J. W. P. (2017). Klebsiella pneumoniae in Singapore: Hypervirulent infections and the Carbapenemase threat. Front. Cell. Infect. Microbiol. 7:515. doi: 10.3389/fcimb.2017.00515
Chiang, T. T., Yang, Y. S., Yeh, K. M., Chiu, S. K., Wang, N. C., Lin, T. Y., et al. (2016). Quantification and comparison of virulence and characteristics of different variants of carbapenemase-producing Klebsiella pneumoniae clinical isolates from Taiwan and the United States. J. Microbiol. Immunol. Infect. 49, 83–90. doi: 10.1016/j.jmii.2015.08.011
Chiotos, K., Hayes, M., Gerber, J. S., and Tamma, P. D. (2020). Treatment of Carbapenem-resistant Enterobacteriaceae infections in children. J Pediatric Infect Dis Soc 9, 56–66. doi: 10.1093/jpids/piz085
Choby, J. E., Howard-Anderson, J., and Weiss, D. S. (2020). Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 287, 283–300. doi: 10.1111/joim.13007
Chuang, Y. P., Fang, C. T., Lai, S. Y., Chang, S. C., and Wang, J. T. (2006). Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193, 645–654. doi: 10.1086/499968
Chung, P. Y. (2016). The emerging problems of Klebsiella pneumoniae infections: carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 363:219. doi: 10.1093/femsle/fnw219
Chung The, H., Karkey, A., Pham Thanh, D., Boinett, C. J., Cain, A. K., Ellington, M., et al. (2015). A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol. Med. 7, 227–239. doi: 10.15252/emmm.201404767
Cui, X., Zhang, H., and Du, H. (2019). Carbapenemases in Enterobacteriaceae: detection and antimicrobial therapy. Front. Microbiol. 10:1823. doi: 10.3389/fmicb.2019.01823
Di Pilato, V., Errico, G., Monaco, M., Giani, T., Del Grosso, M., Antonelli, A., et al. (2021). The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J. Antimicrob. Chemother. 76, 355–361. doi: 10.1093/jac/dkaa431
Doi, Y., and Paterson, D. L. (2015). Carbapenemase-producing Enterobacteriaceae. Semin. Respir. Crit. Care Med. 36, 74–84. doi: 10.1055/s-0035-1544208
Dong, N., Lin, D., Zhang, R., Chan, E. W., and Chen, S. (2018). Carriage of blaKPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J. Antimicrob. Chemother. 73, 3317–3321. doi: 10.1093/jac/dky358
Dong, N., Sun, Q., Huang, Y., Shu, L., Ye, L., Zhang, R., et al. (2019). Evolution of Carbapenem-resistant serotype K1 Hypervirulent Klebsiella pneumoniae by acquisition of Bla (VIM-1)-bearing plasmid. Antimicrob. Agents Chemother. 63:19. doi: 10.1128/AAC.01056-19
Doumith, M., Ellington, M. J., Livermore, D. M., and Woodford, N. (2009). Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63, 659–667. doi: 10.1093/jac/dkp029
Du, D., Wang-Kan, X., Neuberger, A., van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi: 10.1038/s41579-018-0048-6
Eger, E., Heiden, S. E., Becker, K., Rau, A., Geisenhainer, K., Idelevich, E. A., et al. (2021). Hypervirulent Klebsiella pneumoniae sequence type 420 with a chromosomally inserted virulence plasmid. Int. J. Mol. Sci. 22:196. doi: 10.3390/ijms22179196
Farrell, D. J., Sader, H. S., Flamm, R. K., and Jones, R. N. (2014). Ceftolozane/tazobactam activity tested against gram-negative bacterial isolates from hospitalised patients with pneumonia in US and European medical centres (2012). Int. J. Antimicrob. Agents 43, 533–539. doi: 10.1016/j.ijantimicag.2014.01.032
Feng, Y., Lu, Y., Yao, Z., and Zong, Z. (2018). Carbapenem-resistant Hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob. Agents Chemother. 62:17. doi: 10.1128/AAC.02644-17
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom 2:e000073. doi: 10.1099/mgen.0.000073
Fung, C. P., Chang, F. Y., Lee, S. C., Hu, B. S., Kuo, B. I., Liu, C. Y., et al. (2002). A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50, 420–424. doi: 10.1136/gut.50.3.420
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018a). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Gu, D., Lv, H., Sun, Q., Shu, L., and Zhang, R. (2018b). Emergence of tet(A) and Bla(KPC-2) co-carrying plasmid from a ST11 hypervirulent Klebsiella pneumoniae isolate in patient's gut. Int. J. Antimicrob. Agents 52, 307–308. doi: 10.1016/j.ijantimicag.2018.06.003
Hao, M., Shi, X., Lv, J., Niu, S., Cheng, S., Du, H., et al. (2020). In vitro activity of Apramycin against Carbapenem-resistant and Hypervirulent Klebsiella pneumoniae isolates. Front. Microbiol. 11:425. doi: 10.3389/fmicb.2020.00425
Harada, S., Aoki, K., Ishii, Y., Ohno, Y., Nakamura, A., Komatsu, M., et al. (2019). Emergence of IMP-producing hypervirulent Klebsiella pneumoniae carrying a pLVPK-like virulence plasmid. Int. J. Antimicrob. Agents 53, 873–875. doi: 10.1016/j.ijantimicag.2019.05.007
Hauck, C., Cober, E., Richter, S. S., Perez, F., Salata, R. A., Kalayjian, R. C., et al. (2016). Spectrum of excess mortality due to carbapenem-resistant Klebsiella pneumoniae infections. Clin. Microbiol. Infect. 22, 513–519. doi: 10.1016/j.cmi.2016.01.023
He, J., Du, X., Zeng, X., Moran, R. A., van Schaik, W., Zou, Q., et al. (2022). Phenotypic and genotypic characterization of a Hypervirulent Carbapenem-resistant Klebsiella pneumoniae ST17-KL38 clinical isolate harboring the Carbapenemase IMP-4. Microbiol Spectr 10:e0213421. doi: 10.1128/spectrum.02134-21
Hsu, C. R., Lin, T. L., Chen, Y. C., Chou, H. C., and Wang, J. T. (2011). The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology (Reading) 157, 3446–3457. doi: 10.1099/mic.0.050336-0
Huang, Y. H., Chou, S. H., Liang, S. W., Ni, C. E., Lin, Y. T., Huang, Y. W., et al. (2018). Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J. Antimicrob. Chemother. 73, 2039–2046. doi: 10.1093/jac/dky164
Huang, Q. S., Liao, W., Xiong, Z., Li, D., Du, F. L., Xiang, T. X., et al. (2021). Prevalence of the NTEKPC-I on IncF plasmids among Hypervirulent Klebsiella pneumoniae isolates in Jiangxi Province, South China. Front. Microbiol. 12:622280. doi: 10.3389/fmicb.2021.622280
Jeon, J. H., Lee, J. H., Lee, J. J., Park, K. S., Karim, A. M., Lee, C. R., et al. (2015). Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci. 16, 9654–9692. doi: 10.3390/ijms16059654
Jin, L., Liu, Y., Jing, C., Wang, R., Wang, Q., and Wang, H. (2020). Neutrophil extracellular traps (NETs)-mediated killing of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) are impaired in patients with diabetes mellitus. Virulence 11, 1122–1130. doi: 10.1080/21505594.2020.1809325
Jin, L., Wang, R., Gao, H., Wang, Q., and Wang, H. (2021). Identification of a novel hybrid plasmid encoding KPC-2 and virulence factors in Klebsiella pneumoniae sequence type 11. Antimicrob. Agents Chemother. 65:20. doi: 10.1128/AAC.02435-20
Karlsson, M., Stanton, R. A., Ansari, U., McAllister, G., Chan, M. Y., Sula, E., et al. (2019). Identification of a Carbapenemase-producing Hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob. Agents Chemother. 63:19. doi: 10.1128/AAC.00519-19
Krapp, F., Morris, A. R., Ozer, E. A., and Hauser, A. R. (2017). Virulence characteristics of Carbapenem-resistant Klebsiella pneumoniae strains from patients with necrotizing skin and soft tissue infections. Sci. Rep. 7:13533. doi: 10.1038/s41598-017-13524-8
Lan, P., Jiang, Y., Zhou, J., and Yu, Y. (2021). A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist 25, 26–34. doi: 10.1016/j.jgar.2021.02.020
Lee, Y. J., Huang, C. H., Ilsan, N. A., Lee, I. H., and Huang, T. W. (2021). Molecular epidemiology and characterization of Carbapenem-resistant Klebsiella pneumoniae isolated from urine at a teaching Hospital in Taiwan. Microorganisms 9:271. doi: 10.3390/microorganisms9020271
Lev, A. I., Astashkin, E. I., Kislichkina, A. A., Solovieva, E. V., Kombarova, T. I., Korobova, O. V., et al. (2018). Comparative analysis of Klebsiella pneumoniae strains isolated in 2012-2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog Glob Health 112, 142–151. doi: 10.1080/20477724.2018.1460949
Li, Y., Dong, L., Gao, W., Zhen, J., Dong, F., and Yao, K. (2021a). Hypervirulent Klebsiella pneumoniae infections in pediatric populations in Beijing (2017-2019): clinical characteristics, molecular epidemiology and antimicrobial susceptibility. Pediatr. Infect. Dis. J. 40, 1059–1063. doi: 10.1097/INF.0000000000003253
Li, J., Huang, Z. Y., Yu, T., Tao, X. Y., Hu, Y. M., Wang, H. C., et al. (2019b). Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. 19:219. doi: 10.1186/s12866-019-1593-5
Li, Y., Li, D., Xue, J., Ji, X., Shao, X., and Yan, J. (2021b). The epidemiology, virulence and antimicrobial resistance of invasive Klebsiella pneumoniae at a Children’s medical Center in Eastern China. Infect Drug Resist 14, 3737–3752. doi: 10.2147/idr.S323353
Li, C., Li, Y., Zhao, Z., Liu, Q., and Li, B. (2019a). Treatment options and clinical outcomes for carbapenem-resistant Enterobacteriaceae bloodstream infection in a Chinese university hospital. J. Infect. Public Health 12, 26–31. doi: 10.1016/j.jiph.2018.08.002
Li, D., Liao, W., Huang, H. H., Du, F. L., Wei, D. D., Mei, Y. F., et al. (2020b). Emergence of Hypervirulent Ceftazidime/Avibactam-resistant Klebsiella pneumoniae isolates in a Chinese tertiary hospital. Infect Drug Resist 13, 2673–2680. doi: 10.2147/idr.S257477
Li, C., Ma, G., Yang, T., Wen, X., Qin, C., Yue, L., et al. (2020a). A rare carbapenem-resistant hypervirulent K1/ST1265 Klebsiella pneumoniae with an untypeable blaKPC-harboured conjugative plasmid. J Glob Antimicrob Resist 22, 426–433. doi: 10.1016/j.jgar.2020.04.009
Li, X. Z., Plesiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Liao, W., Liu, Y., and Zhang, W. (2020). Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: A review over the last 10 years. J Glob Antimicrob Resist 23, 174–180. doi: 10.1016/j.jgar.2020.09.004
Lin, Z. W., Zheng, J. X., Bai, B., Xu, G. J., Lin, F. J., Chen, Z., et al. (2020). Characteristics of Hypervirulent Klebsiella pneumoniae: does low expression of rmpA contribute to the absence of Hypervirulence? Front. Microbiol. 11:436. doi: 10.3389/fmicb.2020.00436
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Z., Chu, W., Li, X., Tang, W., Ye, J., Zhou, Q., et al. (2021). Genomic features and virulence characteristics of a community-acquired bloodstream infection-causing Hypervirulent Klebsiella pneumoniae ST86 strain harboring KPC-2-encoding IncX6 plasmid. Microb. Drug Resist. 27, 360–368. doi: 10.1089/mdr.2019.0394
Liu, C., Du, P., Zhao, J., Li, B., Wang, C., Sun, L., et al. (2020). Phenotypic and genomic characterization of virulence heterogeneity in multidrug-resistant ST11 Klebsiella pneumoniae during inter-host transmission and evolution. Infect Drug Resist 13, 1713–1721. doi: 10.2147/idr.S243836
Liu, Z., Gu, Y., Li, X., Liu, Y., Ye, Y., Guan, S., et al. (2019). Identification and characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann. Lab. Med. 39, 167–175. doi: 10.3343/alm.2019.39.2.167
Liu, P. P., Liu, Y., Wang, L. H., Wei, D. D., and Wan, L. G. (2016). Draft genome sequence of an NDM-5-producing Klebsiella pneumoniae sequence type 14 strain of serotype K2. Genome Announc. 4:15. doi: 10.1128/genomeA.01610-15
Liu, Y., Liu, P. P., Wang, L. H., Wei, D. D., Wan, L. G., and Zhang, W. (2017). Capsular polysaccharide types and virulence-related traits of epidemic KPC-producing Klebsiella pneumoniae isolates in a Chinese university hospital. Microb. Drug Resist. 23, 901–907. doi: 10.1089/mdr.2016.0222
Liu, B. T., and Su, W. Q. (2019). Whole genome sequencing of NDM-1-producing serotype K1 ST23 hypervirulent Klebsiella pneumoniae in China. J. Med. Microbiol. 68, 866–873. doi: 10.1099/jmm.0.000996
Lorenzin, G., Gona, F., Battaglia, S., Spitaleri, A., Saluzzo, F., Trovato, A., et al. (2022). Detection of NDM-1/5 and OXA-48 co-producing extensively drug-resistant hypervirulent Klebsiella pneumoniae in northern Italy. J Glob Antimicrob Resist 28, 146–150. doi: 10.1016/j.jgar.2022.01.001
Mataseje, L. F., Boyd, D. A., Mulvey, M. R., and Longtin, Y. (2019). Two Hypervirulent Klebsiella pneumoniae isolates producing a Bla (KPC-2) Carbapenemase from a Canadian patient. Antimicrob. Agents Chemother. 63:19. doi: 10.1128/AAC.00517-19
Mendes, G., Ramalho, J. F., Bruschy-Fonseca, A., Lito, L., Duarte, A., Melo-Cristino, J., et al. (2022). First description of Ceftazidime/Avibactam resistance in a ST13 KPC-70-producing Klebsiella pneumoniae strain from Portugal. Antibiotics (Basel) 11:167. doi: 10.3390/antibiotics11020167
Mohammad Ali Tabrizi, A., Badmasti, F., Shahcheraghi, F., and Azizi, O. (2018). Outbreak of hypervirulent Klebsiella pneumoniae harbouring Bla(VIM-2) among mechanically-ventilated drug-poisoning patients with high mortality rate in Iran. J Glob Antimicrob Resist 15, 93–98. doi: 10.1016/j.jgar.2018.06.020
Mukherjee, S., Naha, S., Bhadury, P., Saha, B., Dutta, M., Dutta, S., et al. (2020). Emergence of OXA-232-producing hypervirulent Klebsiella pneumoniae ST23 causing neonatal sepsis. J. Antimicrob. Chemother. 75, 2004–2006. doi: 10.1093/jac/dkaa080
Ni, W., Han, Y., Liu, J., Wei, C., Zhao, J., Cui, J., et al. (2016). Tigecycline treatment for Carbapenem-resistant Enterobacteriaceae infections: A systematic review and meta-analysis. Medicine (Baltimore) 95:e3126. doi: 10.1097/MD.0000000000003126
Nishino, K., and Yamaguchi, A. (2004). Role of histone-like protein H-NS in multidrug resistance of Escherichia coli. J. Bacteriol. 186, 1423–1429. doi: 10.1128/JB.186.5.1423-1429.2004
Nordmann, P., Dortet, L., and Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18, 263–272. doi: 10.1016/j.molmed.2012.03.003
Octavia, S., Kalisvar, M., Venkatachalam, I., Ng, O. T., Xu, W., Sridatta, P. S. R., et al. (2019). Klebsiella pneumoniae and Klebsiella quasipneumoniae define the population structure of blaKPC-2Klebsiella: a 5 year retrospective genomic study in Singapore. J. Antimicrob. Chemother. 74, 3205–3210. doi: 10.1093/jac/dkz332
Pagès, J. M., James, C. E., and Winterhalter, M. (2008). The porin and the permeating antibiotic: a selective diffusion barrier in gram-negative bacteria. Nat. Rev. Microbiol. 6, 893–903. doi: 10.1038/nrmicro1994
Pajand, O., Darabi, N., Arab, M., Ghorbani, R., Bameri, Z., Ebrahimi, A., et al. (2020). The emergence of the hypervirulent Klebsiella pneumoniae (hvKp) strains among circulating clonal complex 147 (CC147) harbouring Bla(NDM/OXA-48) carbapenemases in a tertiary care center of Iran. Ann. Clin. Microbiol. Antimicrob. 19:12. doi: 10.1186/s12941-020-00349-z
Pascale, R., Giannella, M., Bartoletti, M., Viale, P., and Pea, F. (2019). Use of meropenem in treating carbapenem-resistant Enterobacteriaceae infections. Expert Rev. Anti-Infect. Ther. 17, 819–827. doi: 10.1080/14787210.2019.1673731
Pfaller, M. A., Huband, M. D., Mendes, R. E., Flamm, R. K., and Castanheira, M. (2018). In vitro activity of meropenem/vaborbactam and characterisation of carbapenem resistance mechanisms among carbapenem-resistant Enterobacteriaceae from the 2015 meropenem/vaborbactam surveillance programme. Int. J. Antimicrob. Agents 52, 144–150. doi: 10.1016/j.ijantimicag.2018.02.021
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Pogue, J. M., Bonomo, R. A., and Kaye, K. S. (2019). Ceftazidime/Avibactam, Meropenem/Vaborbactam, or both? Clinical and formulary considerations. Clin. Infect. Dis. 68, 519–524. doi: 10.1093/cid/ciy576
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/cmr.00001-07
Rasmussen, B. A., and Bush, K. (1997). Carbapenem-hydrolyzing beta-lactamases. Antimicrob. Agents Chemother. 41, 223–232. doi: 10.1128/AAC.41.2.223
Roulston, K. J., Bharucha, T., Turton, J. F., Hopkins, K. L., and Mack, D. J. F. (2018). A case of NDM-carbapenemase-producing hypervirulent Klebsiella pneumoniae sequence type 23 from the UK. JMM Case Rep 5:e005130. doi: 10.1099/jmmcr.0.005130
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:19. doi: 10.1128/CMR.00001-19
Russo, T. A., Olson, R., Fang, C. T., Stoesser, N., Miller, M., MacDonald, U., et al. (2018). Identification of biomarkers for differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 56:18. doi: 10.1128/JCM.00776-18
Russo, T. A., Olson, R., MacDonald, U., Beanan, J., and Davidson, B. A. (2015). Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 83, 3325–3333. doi: 10.1128/IAI.00430-15
Russo, T. A., Olson, R., Macdonald, U., Metzger, D., Maltese, L. M., Drake, E. J., et al. (2014). Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82, 2356–2367. doi: 10.1128/IAI.01667-13
Saavedra, S. Y., Bernal, J. F., Montilla-Escudero, E., Arevalo, S. A., Prada, D. A., Valencia, M. F., et al. (2021). Complexity of genomic epidemiology of Carbapenem-resistant Klebsiella pneumoniae isolates in Colombia urges the reinforcement of whole genome sequencing-based surveillance programs. Clin. Infect. Dis. 73, S290–S299. doi: 10.1093/cid/ciab777
Sanikhani, R., Moeinirad, M., Solgi, H., Hadadi, A., Shahcheraghi, F., and Badmasti, F. (2021). The face of hypervirulent Klebsiella pneumoniae isolated from clinical samples of two Iranian teaching hospitals. Ann. Clin. Microbiol. Antimicrob. 20:58. doi: 10.1186/s12941-021-00467-2
Scaltriti, E., Piccinelli, G., Corbellini, S., Caruso, A., Latronico, N., and De Francesco, M. A. (2020). Detection of a hypermucoviscous Klebsiella pneumoniae co-producing NDM-5 and OXA-48 carbapenemases with sequence type 383, Brescia, Italy. Int. J. Antimicrob. Agents 56:106130. doi: 10.1016/j.ijantimicag.2020.106130
Shaidullina, E., Shelenkov, A., Yanushevich, Y., Mikhaylova, Y., Shagin, D., Alexandrova, I., et al. (2020). Antimicrobial resistance and genomic characterization of OXA-48- and CTX-M-15-co-producing Hypervirulent Klebsiella pneumoniae ST23 recovered from nosocomial outbreak. Antibiotics (Basel) 9:862. doi: 10.3390/antibiotics9120862
Shankar, C., Nabarro, L. E., Devanga Ragupathi, N. K., Muthuirulandi Sethuvel, D. P., Daniel, J. L., Doss, C. G., et al. (2016). Draft genome sequences of three Hypervirulent Carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announc. 4:16. doi: 10.1128/genomeA.01081-16
Shao, C., Jin, Y., Wang, W., Jiang, M., and Zhao, S. (2021). An outbreak of Carbapenem-resistant Klebsiella pneumoniae of K57 capsular serotype in an emergency intensive care unit of a teaching Hospital in China. Front. Public Health 9:724212. doi: 10.3389/fpubh.2021.724212
Shen, Z., Ding, B., Ye, M., Wang, P., Bi, Y., Wu, S., et al. (2017). High ceftazidime hydrolysis activity and porin OmpK35 deficiency contribute to the decreased susceptibility to ceftazidime/avibactam in KPC-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 72, 1930–1936. doi: 10.1093/jac/dkx066
Shen, Z., Gao, Q., Qin, J., Liu, Y., and Li, M. (2019). Emergence of an NDM-5-producing Hypervirulent Klebsiella pneumoniae sequence type 35 strain with chromosomal integration of an integrative and conjugative element, ICEKp1. Antimicrob. Agents Chemother. 64:19. doi: 10.1128/AAC.01675-19
Shields, R. K., Nguyen, M. H., Chen, L., Press, E. G., Kreiswirth, B. N., and Clancy, C. J. (2018). Pneumonia and renal replacement therapy are risk factors for Ceftazidime-Avibactam treatment failures and resistance among patients with Carbapenem-resistant Enterobacteriaceae infections. Antimicrob. Agents Chemother. 62:17. doi: 10.1128/AAC.02497-17
Shon, A. S., Bajwa, R. P., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Shu, L., Dong, N., Lu, J., Zheng, Z., Hu, J., Zeng, W., et al. (2019). Emergence of OXA-232 Carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob. Agents Chemother. 63:18. doi: 10.1128/AAC.02246-18
Solgi, H., Shahcheraghi, F., Bolourchi, N., and Ahmadi, A. (2020). Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring Bla(NDM-1) and Bla(OXA-48) carbapenemases in Iran. Microb. Pathog. 149:104507. doi: 10.1016/j.micpath.2020.104507
Starkova, P., Lazareva, I., Avdeeva, A., Sulian, O., Likholetova, D., Ageevets, V., et al. (2021). Emergence of hybrid resistance and virulence plasmids harboring New Delhi Metallo-β-lactamase in Klebsiella pneumoniae in Russia. Antibiotics (Basel) 10:691. doi: 10.3390/antibiotics10060691
Su, C., Wu, T., Meng, B., Yue, C., Sun, Y., He, L., et al. (2021). High prevalence of Klebsiella pneumoniae infections in AnHui Province: clinical characteristic and antimicrobial resistance. Infect Drug Resist 14, 5069–5078. doi: 10.2147/idr.S336451
Su, S., Zhang, J., Zhao, Y., Yu, L., Wang, Y., Wang, Y., et al. (2020). Outbreak of KPC-2 Carbapenem-resistant Klebsiella pneumoniae ST76 and Carbapenem-resistant K2 Hypervirulent Klebsiella pneumoniae ST375 strains in Northeast China: molecular and virulent characteristics. BMC Infect. Dis. 20:472. doi: 10.1186/s12879-020-05143-y
Tam, H. K., Foong, W. E., Oswald, C., Herrmann, A., Zeng, H., and Pos, K. M. (2021). Allosteric drug transport mechanism of multidrug transporter AcrB. Nat. Commun. 12:3889. doi: 10.1038/s41467-021-24151-3
Tian, D., Wang, W., Li, M., Chen, W., Zhou, Y., Huang, Y., et al. (2021). Acquisition of the conjugative virulence plasmid from a CG23 Hypervirulent Klebsiella pneumoniae strain enhances bacterial virulence. Front. Cell. Infect. Microbiol. 11:752011. doi: 10.3389/fcimb.2021.752011
Turton, J. F., Payne, Z., Coward, A., Hopkins, K. L., Turton, J. A., Doumith, M., et al. (2018). Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and 'non-hypervirulent' types ST147, ST15 and ST383. J. Med. Microbiol. 67, 118–128. doi: 10.1099/jmm.0.000653
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Vergalli, J., Bodrenko, I. V., Masi, M., Moynié, L., Acosta-Gutiérrez, S., Naismith, J. H., et al. (2020). Porins and small-molecule translocation across the outer membrane of gram-negative bacteria. Nat. Rev. Microbiol. 18, 164–176. doi: 10.1038/s41579-019-0294-2
Wang, X. D., Cai, J. C., Zhou, H. W., Zhang, R., and Chen, G. X. (2009). Reduced susceptibility to carbapenems in Klebsiella pneumoniae clinical isolates associated with plasmid-mediated beta-lactamase production and OmpK36 porin deficiency. J. Med. Microbiol. 58, 1196–1202. doi: 10.1099/jmm.0.008094-0
Wang, J. H., Liu, Y. C., Lee, S. S., Yen, M. Y., Chen, Y. S., Wang, J. H., et al. (1998). Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438. doi: 10.1086/516369
Wei, D. D., Wan, L. G., and Liu, Y. (2018). Draft genome sequence of an NDM-1- and KPC-2-coproducing Hypervirulent Carbapenem-resistant Klebsiella pneumoniae strain isolated from burn wound infections. Genome Announc. 6:18. doi: 10.1128/genomeA.00192-18
Wei, T., Zou, C., Qin, J., Tao, J., Yan, L., Wang, J., et al. (2022). Emergence of Hypervirulent ST11-K64 Klebsiella pneumoniae poses a serious clinical threat in older patients. Front. Public Health 10:765624. doi: 10.3389/fpubh.2022.765624
Wu, K. M., Li, L. H., Yan, J. J., Tsao, N., Liao, T. L., Tsai, H. C., et al. (2009). Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 191, 4492–4501. doi: 10.1128/JB.00315-09
Wu, H., Li, D., Zhou, H., Sun, Y., Guo, L., and Shen, D. (2017). Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb. Pathog. 104, 254–262. doi: 10.1016/j.micpath.2017.01.049
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. doi: 10.1099/mgen.0.000102
Wyres, K. L., Wick, R. R., Judd, L. M., Froumine, R., Tokolyi, A., Gorrie, C. L., et al. (2019). Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 15:e1008114. doi: 10.1371/journal.pgen.1008114
Xie, M., Yang, X., Xu, Q., Ye, L., Chen, K., Zheng, Z., et al. (2021). Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol 4:650. doi: 10.1038/s42003-021-02148-4
Xu, M., Fu, Y., Fang, Y., Xu, H., Kong, H., Liu, Y., et al. (2019). High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in eastern China. Infect Drug Resist 12, 641–653. doi: 10.2147/idr.S191892
Xu, Q., Sheng, Z., Hao, M., Jiang, J., Ye, M., Chen, Y., et al. (2021a). RamA upregulates multidrug resistance efflux pumps AcrAB and OqxAB in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 57:106251. doi: 10.1016/j.ijantimicag.2020.106251
Xu, Y., Zhang, J., Wang, M., Liu, M., Liu, G., Qu, H., et al. (2021b). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13:119. doi: 10.1186/s13073-021-00936-5
Yan, R., Lu, Y., Zhu, Y., Lan, P., Jiang, S., Lu, J., et al. (2021). A sequence type 23 Hypervirulent Klebsiella pneumoniae strain presenting Carbapenem resistance by acquiring an IncP1 Bla (KPC-2) plasmid. Front. Cell. Infect. Microbiol. 11:641830. doi: 10.3389/fcimb.2021.641830
Yang, X., Dong, N., Chan, E. W., Zhang, R., and Chen, S. (2021a). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yang, X., Dong, N., Liu, X., Yang, C., Ye, L., Chan, E. W., et al. (2021b). Co-conjugation of virulence plasmid and KPC plasmid in a clinical Klebsiella pneumoniae strain. Front. Microbiol. 12:739461. doi: 10.3389/fmicb.2021.739461
Yang, Q., Jia, X., Zhou, M., Zhang, H., Yang, W., Kudinha, T., et al. (2020). Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerg Microbes Infect 9, 320–331. doi: 10.1080/22221751.2020.1721334
Yang, X., Liu, X., Xu, Y., Yang, C., Chan, E. W., Shum, H. P., et al. (2022a). Genetic and functional characterization of a conjugative KpVP-2-type virulence plasmid from a clinical Klebsiella pneumoniae strain. Front. Microbiol. 13:914884. doi: 10.3389/fmicb.2022.914884
Yang, X., Sun, Q., Li, J., Jiang, Y., Li, Y., Lin, J., et al. (2022b). Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect 11, 841–849. doi: 10.1080/22221751.2022.2049458
Yang, Y., Yang, Y., Ahmed, M., Qin, M., He, R., Wu, Y., et al. (2022c). Carriage of distinct Bla(KPC-2) and Bla(OXA-48) plasmids in a single ST11 hypervirulent Klebsiella pneumoniae isolate in Egypt. BMC Genomics 23:20. doi: 10.1186/s12864-021-08214-9
Yao, B., Xiao, X., Wang, F., Zhou, L., Zhang, X., and Zhang, J. (2015). Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. doi: 10.1016/j.ijid.2015.06.023
Yin, D., Wu, S., Yang, Y., Shi, Q., Dong, D., Zhu, D., et al. (2019). Results from the China antimicrobial surveillance network (CHINET) in 2017 of the in vitro activities of Ceftazidime-Avibactam and Ceftolozane-Tazobactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63:18. doi: 10.1128/AAC.02431-18
Yonekawa, S., Mizuno, T., Nakano, R., Nakano, A., Suzuki, Y., Asada, T., et al. (2020). Molecular and Epidemiological Characteristics of Carbapenemase-Producing Klebsiella pneumoniae Clinical Isolates in Japan. mSphere 5:20. doi: 10.1128/mSphere.00490-20
Yuan, Y., Li, Y., Wang, G., Li, C., Chang, Y. F., Chen, W., et al. (2019). Bla (NDM-5) carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrob. Resist. Infect. Control 8:140. doi: 10.1186/s13756-019-0596-1
Zhan, L., Wang, S., Guo, Y., Jin, Y., Duan, J., Hao, Z., et al. (2017). Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 isolates with Carbapenem resistance in a tertiary Hospital in China. Front. Cell. Infect. Microbiol. 7:182. doi: 10.3389/fcimb.2017.00182
Zhang, Y., Jin, L., Ouyang, P., Wang, Q., Wang, R., Wang, J., et al. (2020). Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J. Antimicrob. Chemother. 75, 327–336. doi: 10.1093/jac/dkz446
Zhang, R., Lin, D., Chan, E. W., Gu, D., Chen, G. X., and Chen, S. (2016). Emergence of Carbapenem-resistant serotype K1 Hypervirulent Klebsiella pneumoniae strains in China. Antimicrob. Agents Chemother. 60, 709–711. doi: 10.1128/AAC.02173-15
Zhang, R., Liu, L., Zhou, H., Chan, E. W., Li, J., Fang, Y., et al. (2017). Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106. doi: 10.1016/j.ebiom.2017.04.032
Zhang, Y., Wang, X., Wang, S., Sun, S., Li, H., Chen, H., et al. (2021). Emergence of Colistin resistance in Carbapenem-resistant Hypervirulent Klebsiella pneumoniae under the pressure of Tigecycline. Front. Microbiol. 12:756580. doi: 10.3389/fmicb.2021.756580
Zhang, Y., Zeng, J., Liu, W., Zhao, F., Hu, Z., Zhao, C., et al. (2015). Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Inf. Secur. 71, 553–560. doi: 10.1016/j.jinf.2015.07.010
Zhao, J., Zhang, Y., Fan, Y., Han, J., Xiong, Z., Liu, X., et al. (2021). Characterization of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 65 clone from a lung transplant recipient. Emerg Microbes Infect 10, 396–399. doi: 10.1080/22221751.2021.1889932
Zheng, B., Xu, H., Lv, T., Guo, L., Xiao, Y., Huang, C., et al. (2020). Stool samples of acute diarrhea inpatients as a reservoir of ST11 Hypervirulent KPC-2-producing Klebsiella pneumoniae. mSystems 5:20. doi: 10.1128/mSystems.00498-20
Zhou, S., Ren, G., Liu, Y., Liu, X., Zhang, L., Xu, S., et al. (2022). Challenge of evolving Klebsiella pneumoniae infection in patients on hemodialysis: from the classic strain to the carbapenem-resistant hypervirulent one. Int. J. Med. Sci. 19, 416–424. doi: 10.7150/ijms.69577
Zhou, Y., Wang, X., Shen, J., Lu, Z., and Liu, Y. (2019). Endogenous Endophthalmitis caused by Carbapenem-resistant Hypervirulent Klebsiella Pneumoniae: A case report and literature review. Ocul. Immunol. Inflamm. 27, 1099–1104. doi: 10.1080/09273948.2018.1502786
Zhou, C., Wu, Q., He, L., Zhang, H., Xu, M., Yuan, B., et al. (2021). Clinical and molecular characteristics of Carbapenem-resistant Hypervirulent Klebsiella pneumoniae isolates in a tertiary Hospital in Shanghai, China. Infect Drug Resist 14, 2697–2706. doi: 10.2147/IDR.S321704
Keywords: Klebsiella pneumoniae, carbapenem-resistant, hypervirulent, hybrid plasmid, evolution
Citation: Han Y-L, Wen X-H, Zhao W, Cao X-S, Wen J-X, Wang J-R, Hu Z-D and Zheng W-Q (2022) Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 13:1003783. doi: 10.3389/fmicb.2022.1003783
Received: 26 July 2022; Accepted: 24 August 2022;
Published: 12 September 2022.
Edited by:
Ernesto Perez-Rueda, Universidad Nacional Autónoma de México, MexicoReviewed by:
Carlos Henrique Camargo, Adolfo Lutz Institute, BrazilCopyright © 2022 Han, Wen, Zhao, Cao, Wen, Wang, Hu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Qi Zheng, emhlbmd3ZW5xaTIwMTFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.