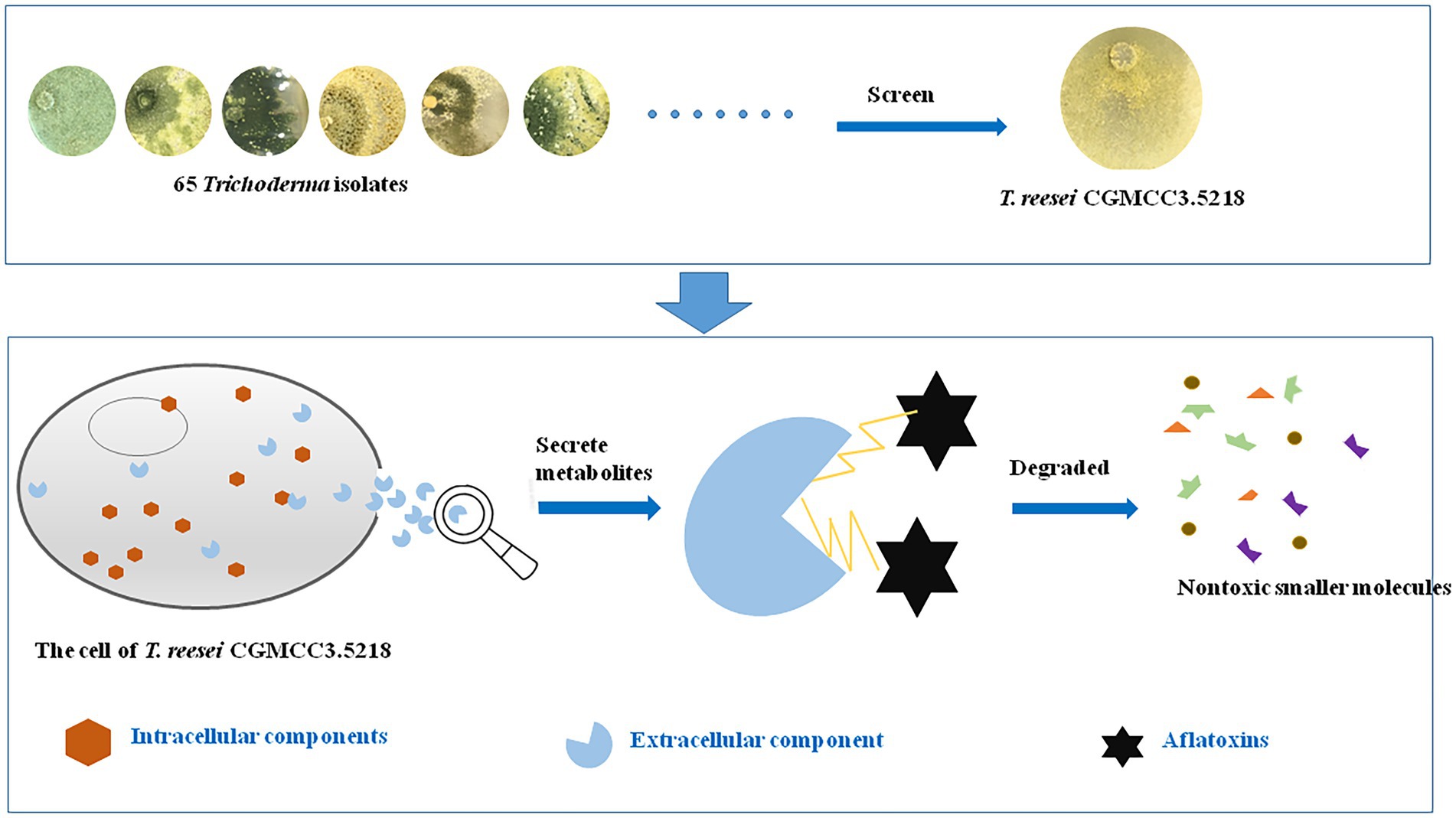
- 1Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
- 2Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, Wuhan, China
- 3Institute of Quality Standard and Testing Technology for Agro-products, Shandong Academy of Agricultural Sciences, Jinan, China
- 4Institutions of Agricultural Product Quality Standard and Testing Research, Tibet Academy of Agricultural and Animal Husbandry Sciences, Lhasa, China
- 5Institute of Sciences of Food Production, National Research Council, Bari, Italy
- 6Hubei Hongshan Lab, Wuhan, China
Aflatoxins, which are produced mainly by Aspergillus flavus and A. parasiticus, are recognized as the most toxic mycotoxins, which are strongly carcinogenic and pose a serious threat to human and animal health. Therefore, strategies to degrade or eliminate aflatoxins in agro-products are urgently needed. We investigated 65 Trichoderma isolates belonging to 23 species for their aflatoxin B1 (AFB1)-degrading capabilities. Trichoderma reesei CGMCC3.5218 had the best performance, and degraded 100% of 50 ng/kg AFB1 within 3 days and 87.6% of 10 μg/kg AFB1 within 5 days in a liquid-medium system. CGMCC3.5218 degraded more than 85.0% of total aflatoxins (aflatoxin B1, B2, G1, and G2) at 108.2–2323.5 ng/kg in artificially and naturally contaminated peanut, maize, and feed within 7 days. Box–Behnken design and response surface methodology showed that the optimal degradation conditions for CGMCC3.5218 were pH 6.7 and 31.3°C for 5.1 days in liquid medium. Possible functional detoxification components were analyzed, indicating that the culture supernatant of CGMCC3.5218 could efficiently degrade AFB1 (500 ng/kg) with a ratio of 91.8%, compared with 19.5 and 8.9% by intracellular components and mycelial adsorption, respectively. The aflatoxin-degrading activity of the fermentation supernatant was sensitive to proteinase K and proteinase K plus sodium dodecyl sulfonate, but was stable at high temperatures, suggesting that thermostable enzymes or proteins in the fermentation supernatant played a major role in AFB1 degradation. Furthermore, toxicological experiments by a micronucleus assay in mouse bone marrow erythrocytes and by intraperitoneal injection and skin irritation tests in mice proved that the degradation products by CGMCC3.5218 were nontoxic. To the best of our knowledge, this is the first comprehensive study on Trichoderma aflatoxin detoxification, and the candidate strain T. reesei CGMCC3.5218 has high efficient and environment-friendly characteristics, and qualifies as a potential biological detoxifier for application in aflatoxin removal from contaminated feeds.
Introduction
Aflatoxins were recognized as a group of small molecules containing difuran rings and lactone rings. The aflatoxins occurring in agricultural products are categorized as B1, B2, G1, and G2 (toxicity grade: AFB1 > AFG1 > AFB2 > AFG2) (Marin et al., 2013; Heshmati et al., 2021). Aflatoxin B1 (AFB1), the most hepatotoxic, mutagenic, and carcinogenic member of the aflatoxin family, has been classified as a Group I carcinogen by the International Agency for Research on Cancer (IARC, 2002). However, agricultural products such as peanuts, maize, wheat, rice and spices are suffering from a high risk of aflatoxin contamination in many countries (Pitt and Hocking, 2006; Keller et al., 2013; Nejad et al., 2014; Eslami et al., 2015). The ingestion of such aflatoxin-contaminated feeds can lead to slow growth, illness, and even death (Bennett and Klich, 2003). Aflatoxins can be biotransformed into metabolites and toxic residues that contaminate meat, eggs, and milk (Nejad et al., 2019; Heshmati et al., 2020). For example, AFB1 and AFB2 can be biotransformed into aflatoxins M1 and M2, respectively, by hydroxylation in animal livers, and subsequently excreted in the urine and milk, posing a serious threat to the safety of dairy foods (Battacone et al., 2005; Kamkar et al., 2014; Nejad et al., 2020).
Many studies have been carried out to explore an effective strategy to degrade aflatoxins to a tolerable level. Although several physical approaches, including γ-radiation treatment and microwave heating (Pankaj et al., 2018), and chemical reactions, e.g., oxidation by ozone (Luo et al., 2013), have achieved some success, these approaches have limitations, such as residual toxicity, losses in nutritional value, and the modification of food or feed properties. The biological degradation of aflatoxins, which mainly includes biodegradation by bacteria, fungi, and yeasts or by their enzymes (Guo et al., 2020; Loi et al., 2020), has been proved to be the most effective, specific, and eco-friendly solution to degrade aflatoxins to nontoxic or less toxic compounds in foods and feeds (Ji et al., 2016).
The fungal genus Trichoderma includes more than 100 species commonly found in soil and root ecosystems, and occasionally in grains and rotten woods. Members of this genus have been well documented to have high value as biocontrol agents, whose mode of action includes the production of secondary metabolites with antibiotic activity (Vinale et al., 2006; Ren et al., 2020). More than 1,000 secondary metabolites have been identified from Trichoderma spp. over the years (Hermosa et al., 2014), some of which have been utilized for biocontrol of plant diseases (Calistru et al., 1997; Braun et al., 2018) or for breaking down complex molecules, such as lignin and cellulose within industrial processes (Juhász et al., 2005; Gusakov, 2011). Although several Trichoderma species, including T. harzianum, T. reesei, T. koningii, and T. asperellum, have been demonstrated to have the ability to degrade hydrocarbon compounds, such as polycyclic aromatic hydrocarbons (PAHs), crude oil, and resins (Zafra and Cortés-Espinosa, 2015), reports on the biodegradation of small molecule contaminants, particularly on degradation of aflatoxins, by Trichoderma spp. are still sparse (Palonen et al., 2004; Pribowo et al., 2012; Hackbart et al., 2014).
Gene sequencing and the analysis of biosynthetic pathways for secondary metabolites have provided insight into the extraordinary capacity of T. reesei to metabolize and degrade small molecular contaminants (Martinez et al., 2008; Zeilinger et al., 2016). The objectives of the present study were to evaluate the AFB1-degradation efficiency of various Trichoderma isolates and to evaluate the T. reesei strains with the strongest AFB1 degradation capability. Furthermore, the mechanism of AFB1 degradation by T. reesei was revealed, which provided a theoretical basis for the degradation of AFB1 in agricultural products by enzymes, and then promote the rapid development of aflatoxin detoxification by the green, efficient and universal enzymes.
Materials and methods
Trichoderma strains, chemicals, and media
A total of 65 Trichoderma isolates from different geographical origins, including China, Italy, the United States, Australia and Borneo, and different sources such as soil, fruit and wood were examined (Supplementary Table S1). In contrast, the sequences of the internal transcribed spacer regions ITS-1 and ITS-2 of the nuclear rDNA are relatively stable, which represents a moderate amount of information and could be used to identify the isolates (Fanelli et al., 2018; Ren et al., 2022). All the isolates were molecularly characterized by analysis of the sequences of the internal transcribed spacer regions ITS-1 and ITS-2 of the nuclear rDNA. The standards of aflatoxins were purchased from Sigma-Aldrich (St. Louis, MO, United States). Water was purified by a Milli-Q purification system (Millipore, Bedford, MA, United States). All other chemicals and reagents were analytically pure or better and purchased from local chemical stores. Yeast extract peptone dextrose (YPD: 5 g yeast extracts, 10 g peptone, 10 g dextrose, and 1 L H2O, pH 7.2) medium was used for the liquid culture of Trichoderma. Potato dextrose agar (PDA) was used for the solid culture of Trichoderma and A. flavus.
Determination the potential Trichoderma isolates capable of detoxifying AFB1
The assay was carried out as described by Hackbart et al. (2014) with some modification. The standard of AFB1 was diluted in YPD medium to a final concentration of 50 ng/kg, and then each well of a 12-well plate (Costar Inc., Cambridge, MA, United States) was filled with 2 ml of the medium and inoculated with one 6-mm-diameter mycelium plug that was excised with a cork-borer from a 5-day-old culture of Trichoderma on PDA. In parallel, controls were made with YPD medium supplemented with the same amount of AFB1 but without inoculation with Trichoderma. After 1, 3, and 7 days of incubation at 28°C under static conditions in the dark, 1 ml of the supernatant was collected from each one of triplicated wells, filtered through a 0.22-μm-filter membrane, and analyzed by HPLC-FLD (high-performance liquid chromatography with fluorescence detector) for determination of AFB1. The AFB1 removal ratio was calculated using the following formula (F1): F1 (%) = (C1 - S1) / C1 × 100, where S1 was the AFB1 concentration in wells supplemented with AFB1 and inoculated with Trichoderma, and C1 was the AFB1 concentration in the control wells.
Effects of various conditions on AFB1 removal ability by Trichoderma reesei CGMCC3.5218
According to the results of the experiment described above, T. reesei CGMCC3.5218 had a great performance in AFB1 removal and was thus used for further study. The effects of incubation time, culture temperature, pH, nutritional sources, and metal ions on AFB1 removal were evaluated as described by Zhang et al. (2014) and Xu et al. (2016). The assay was carried out as follows: tested incubation time with 1, 3, 5, 7, and 10 days; tested pH values with 2.2, 4.5, 5.4, 6.5, 7.2, 8.3, and 9.5, prepared by adjusting the pH of YPD (7.2) with 12 M HCl or 2 M NaOH; tested incubation temperatures with 4, 20, 28, 37, 45, and 50°C; the effects of six growing media were compared: Czapek-Dox broth (CDB, 0.1% NaNO3, 0.1% K2HPO4, 0.05% MgSO4 ▪ 7H2O, 0.05% KCl, 0.001% FeSO4, and 2% sucrose), malt extract broth (MEB, 1.7% malt extract, and 0.3% peptone), nutrient broth (NB, 1% peptone, 0.3% beef extract, and 0.5% NaCl), potato dextrose broth (PDB), liquid Sabouraud broth (LSB, 1% peptone, and 4% dextrose), and YPD; tested metal ions Al3+, Ca2+, Cu2+, Fe2+, K+, Mg2+, Mn2+, Na+, and Zn2+ from AlCl3, CaCl2, CuSO4, FeSO4, KCl, MgCl2, MnSO4, NaCl, and ZnSO4, respectively, all at the concentration of 10 mmol/l (Mwakinyali et al., 2019). The liquid media were prepared as described and then supplemented with AFB1 standard to a final concentration of 500 ng/kg. Two milliliter of each medium were transferred into each well of the 12-well plate, inoculated with one plug of T. reesei CGMCC3.5218. The plates used for testing incubation time were incubated at 28°C for 1, 3, 5, 7, or 10 days. The plates used for testing culture temperatures were incubated at 4, 20, 28, 37, 45, or 50°C for 3 days. The plates used for other tests were incubated at 28°C for 3 days. Controls were prepared in the same way, except that controls were not inoculated with Trichoderma. After different days of incubation, the supernatants were collected, filtered, and analyzed for AFB1 detection. The degradation ratio was calculated with the following formula (F2): F2 (%) = (C2–S2)/C2 × 100, where S2 was the AFB1 concentration in testing wells, and C2 was the AFB1 concentration in control wells.
Box–Behnken design
To investigate cultivation methods that can be used to obtain the optimal efficiency of AFB1 degradation, we used Box–Behnken (BB) design and response surface methodology (RSM) to optimize the values and interaction effects of the factors that affected the AFB1 removal (Mugwagwa and Chimphango, 2019). The incubation time, culture temperature, and pH were identified as three effective factors (X1, X2, and X3, respectively) and were investigated at 3 levels (−1, 0, and 1) (Supplementary Table S2) (Wang et al., 2015b). The experimental design matrix and response values are shown in Supplementary Table S3.
Pattern analysis of AFB1 removal by Trichoderma reesei CGMCC3.5218
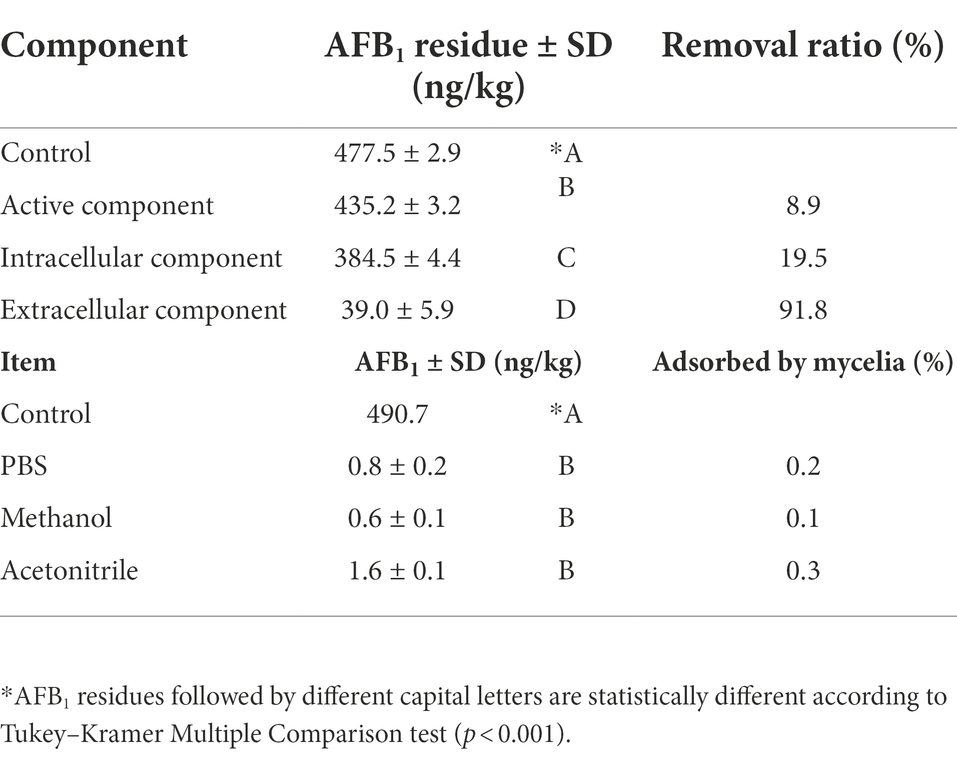
To determine functional detoxification component, we analyzed the effects of the culture supernatant, intracellular components, and mycelium of CGMCC3.5218 on the AFB1 removal ability. The assay was carried out as described in Li et al. (2018) with some modifications. To confirm that the removal of AFB1 did not occur because of the adsorption by the cell wall, a desorption experiment was conducted. AFB1 was supplemented in YPD medium (100 ml) in a flask to a final concentration of 500 ng/kg. Then, five 6-mm-diameter CGMCC3.5218 plugs were added to the flask and incubated for 7 days with constant shaking (125 rpm) at 28°C. The supernatants were collected for AFB1 analysis by HPLC-FLD. The remaining mycelium pellets were also collected, gently rinsed twice with phosphate-buffered saline (PBS, 10 mM sodium phosphate buffer containing 137 mM NaCl and 2.68 mM KCl, pH 7.4) to wash off the medium, and then treated with 25 ml of PBS, methanol, or acetonitrile with constant shaking (200 rpm) for 1 h to elute the AFB1 adsorbed by the pellets. Afterward, the pellets were discarded and the solvents were recovered for AFB1 determination by HPLC-FLD.
Degradation dynamics of AFB1 by Trichoderma reesei CGMCC3.5218 culture supernatant
The time-course of AFB1 degradation by the culture supernatant was studied. The supernatant was supplemented with AFB1 standard to 50, 100, 400, 800, and 1,500 ng/kg and incubated at 37°C under static conditions in the dark. Triplicated samples were collected separately after 0, 12, 24, 48, 72, and 96 h (Mwakinyali et al., 2019). The initial pH of the supernatant was approximately 7 and was then adjusted to different values of pH, viz. 3, 4, 5, 6, 7, 8, and 9. Three milliliter aliquots of the supernatant at different pH values were transferred into new tubes and supplemented with AFB1 standard to the final concentration of 500 ng/kg. Samples were collected after incubated at 37°C in the dark for 24 h. Same aliquots of PBS buffer treated in the same manner of samples were used as a control. The degradation rate of AFB1 was calculated using formula (F3): F3(%) = (C3–S3)/C3 × 100, where C3 refers to the concentration of AFB1 in the control group, and S3 refers to the concentration of AFB1 in the sample group.
Effects of heat, proteinase K, sodium dodecyl sulfonate, and ethylene diamine tetraacetic acid on the activity of culture supernatant to degrade AFB1
To investigate whether AFB1 degradation was caused by enzymatic digestion, the CGMCC3.5218 supernatant was subjected to the following treatments: heating at 121°C for 30 min; separate treatment with 1 mg/ml proteinase K and 1 mg/ml proteinase K plus 1% (w/v) sodium dodecyl sulfonate (SDS) at 55°C for 1 h; treatment with 10 mmol/l EDTA at 37°C for 2 h (Sangare et al., 2014; Wang et al., 2017). PBS buffer was used for the controls and treated in the same way. After the treatments, samples and controls were supplemented with standard AFB1 to the final concentration of 500 ng/kg, kept at 37°C for 24 h under static conditions in the dark, and then analyzed by HPLC-FLD to determine the AFB1 content.
Aflatoxin degradation by Trichoderma reesei CGMCC3.5218 in peanut cake, maize, and completed feed
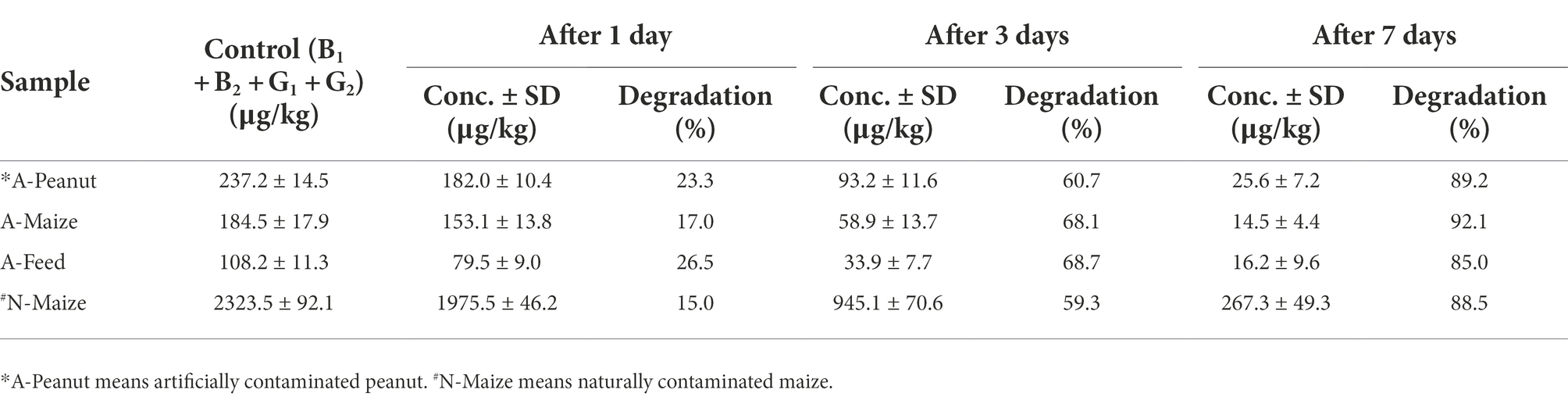
The blank peanut cakes, maize kernels, and animal feed were purchased from the local market. Naturally contaminated maize samples were collected from Shandong Province, China, and stored in our laboratory. The blank samples were inoculated with conidial suspensions of the aflatoxigenic A. flavus strain 3.4408 to produce artificially contaminated samples (Branà et al., 2017). The contaminated samples were autoclaved (121°C for 30 min) and then finely ground into powder. The powder (0.6 g) were transferred into test tubes filled with 20 ml of melted agar (15 g/l) supplemented with 1.0 g/l K2HPO4, 0.5 g/l MgSO4▪7H2O, 0.5 g/l CaCl2, and 0.5 g/l NaCl. After autoclaving (121°C for 15 min), the mixture was thoroughly mixed by a vortex mixer and poured into 9-cm-diameter Petri dishes. Then, every dish was inoculated with three 6-mm-diameter plugs of CGMCC 3.5218. After 1, 3, and 7 days of incubation at 28°C, the samples (approximately 5 g) were excised along the radius of the colony, precisely weighed, transferred to test tubes, and then extracted with 25 ml methanol–water (80:20, v/v) solution. The samples were then vortexed, centrifuged, filtered and analyzed by HPLC-FLD for the determination of aflatoxins B1, B2, G1, and G2.
Toxicity analysis of AFB1 degradation products
The culture supernatant containing AFB1 degradation products were prepared. The toxicity of the supernatant was evaluated by a micronucleus assay in mouse bone marrow erythrocytes (an acute oral toxicity test) and by intraperitoneal injection and skin irritation tests in mice (half male and half female). All of the tests were carried out in accordance with China National Standards GBZ/T 240.11–2011 and China Agriculture Industry Standard NY/T 1109–2017 by the Guangdong Detection Center of Microbiology, which is a Chinese authoritative organization providing toxicity analysis services.
Aflatoxin determination by HPLC-FLD
The aflatoxin determination was carried out according to Ren et al. (2022). Aflatoxins were analyzed by an HPLC-FLD apparatus of the series 1,100 from Agilent Technologies (Santa Clara, CA, United States), with a fluorescence detector and an HPLC C-18 column of 4.6-mm inner diameter, 250-mm length, and 5-μm particle size. The mobile phase was methanol–water (45: 55, v/v) with a 0.8 ml min−1 flow rate and 20 min of total run time. The injection volume was 10 μl, and the excitation and emission wavelengths were 360 and 440 nm, respectively.
Statistical analysis
Data were analyzed by the softwares Origin 9.0 (Origin Lab Corp., Northampton, MA, United States) and GraphPad Instat 3.0 (GraphPad Software, San Diego, CA, United States). One-way analysis of variance (ANOVA) and the Tukey–Kramer test for multiple comparisons were used to analyze the data. Differences were considered extremely significant when p < 0.001, significantly significant when p < 0.01, significant when p < 0. 05 and have no differences when p > 0.05.
Results and discussions
Screening and characterization of the most promising Trichoderma isolates for aflatoxin detoxification
In this study, the HPLC chromatograms showed that the peak areas for AFB1 concentration clearly decreased with the increase of incubation time, indicating the effective removal of AFB1 by Trichoderma (Figure 1). It is noteworthy that we tested the AFB1 removal ratios of the 65 collected Trichoderma isolates to determine the AFB1 removal abilities of different Trichoderma isolates. We found that the 65 Trichoderma isolates possessed different capacity to remove AFB1. After 7 days of incubation, 49 of the 65 isolates could remove more than 70% of AFB1. For species, including T. citrinoviride, T. erinaceum, T. harzianum, T. koningii, T. koningiopsis, and T. viride, the results of one-way ANOVA showed that different strains within the same species had significantly different AFB1 removal capacities, indicating intraspecific variability (Supplementary Table S1).
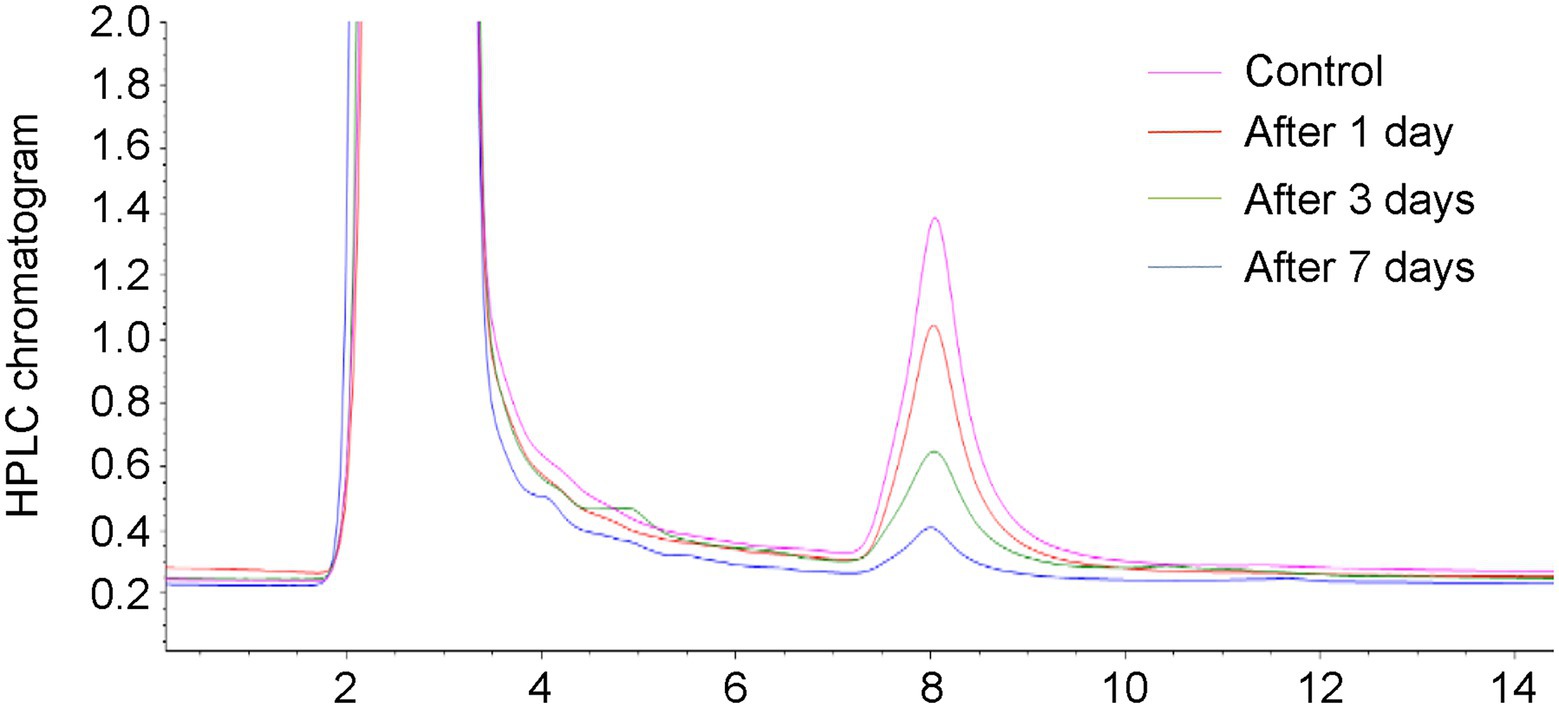
Figure 1. Chromatograms relevant to the aflatoxin B1 (AFB1) (50 ng/kg) in the control and YPD liquid medium after 1, 3, and 7 days of removal by a Trichoderma isolate.
The principles for defining the more efficient Trichoderma isolates were AFB1 removal ratio higher than 80.0% after 3 days of incubation and 100.0% after 7 days of incubation. According to the data in Supplementary Table S1, isolates T. atrobrunneum CGMCC3.1540, T. citrinoviride D20 (xz-5-9), T. harzianum CGMCC3.17876, T. inhamatum D44 (Xz-11-6a), T. longifialidicum D78 (xz-11-5), and T. reesei CGMCC3.5218 met these principles and were considered the top six isolates with high AFB1 removal efficiency. The morphology of colonies of the six strains are shown in Figure 2A. The morphology and visual observations during the experimental process showed that the six strains had similar growth rates, maturity periods, and colony sizes, whereas the colony colors, mycelial structures, and spore morphology were somewhere different (Figure 2A). To analyze the genetic relationships between these species with a higher AFB1 removal efficiency, a phylogenetic tree was first built using MEGA 7.0 software with Penicillium chrysogenum as the root, according to internal transcribed spacer (ITS) sequences of the species. The parameters of the phylogenetic tree are shown in Supplementary Table S4. Phylogenetic analysis indicated that the Trichoderma isolates with similar AFB1 removal abilities were genetically the closest to each other. For example, the top six isolates, which had AFB1 removal ratios higher than 85%, that belong to the species T. atrobrunneum, T. citrinoviride, T. harzianum, T. inhamatum, T. longifialidicum, and T. reesei, respectively, shared approximately 90% ITS sequence identity with each other (Figure 2B).
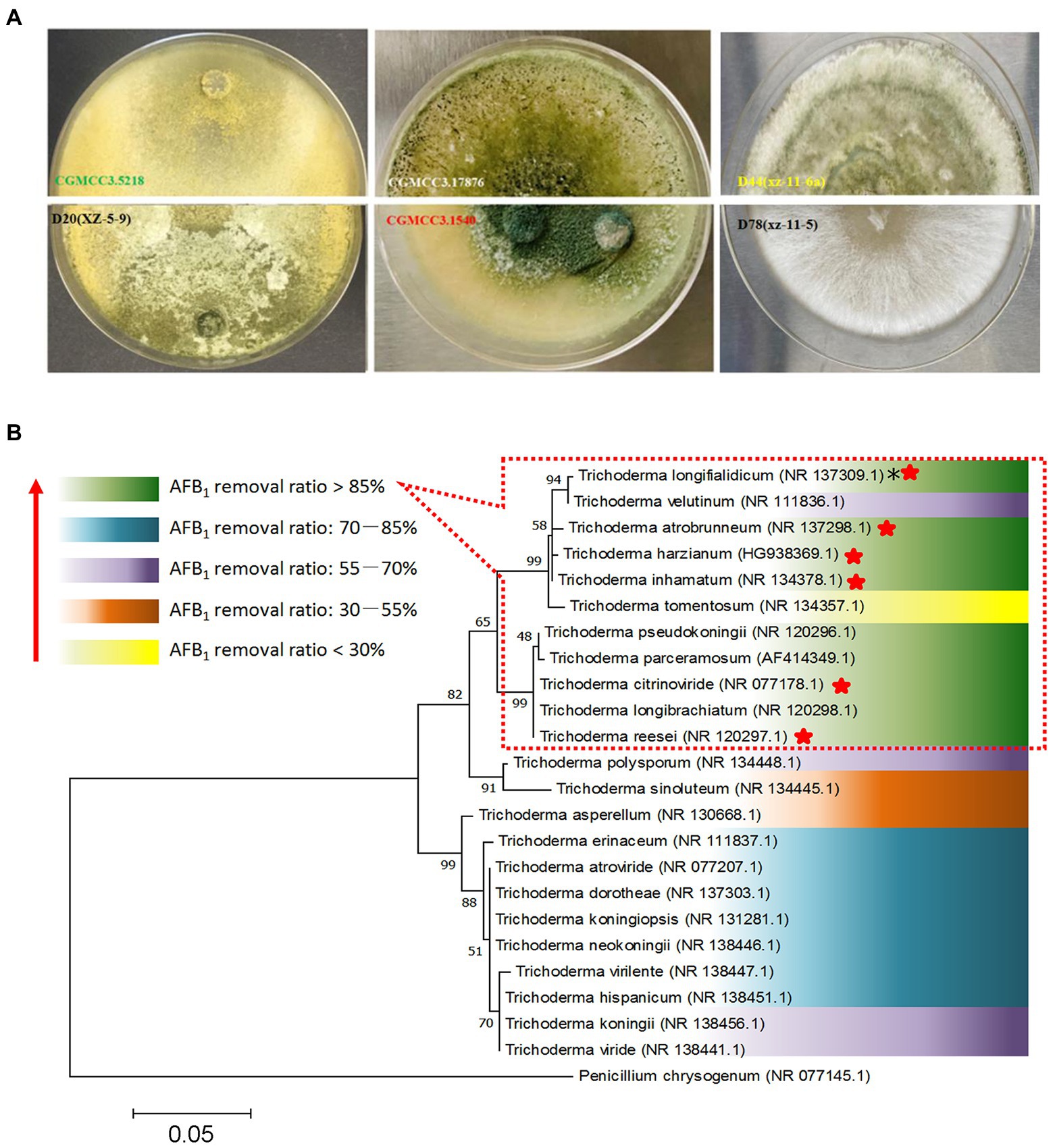
Figure 2. Screening of different Trichoderma isolates for aflatoxin detoxification. (A) The colonial morphology of T. reesei CGMCC3.5218, T. citrinoviride D20 (xz-5-9), T. harzianum CGMCC3.17876, T. atrobrunneum CGMCC3.1540, T. inhamatum D44 (Xz-11-6a), and T. longifialidicum D78 (xz-11-5) cultured in potato dextrose agar (PDA) after 5 days of incubation at 28°C in the dark. (B) The relationships between Trichoderma evolution and aflatoxin B1 (AFB1) removal ability. The abilities of different species to remove AFB1 are divided into five levels, which are represented by different colors according to the mean value of AFB1 removal ratios of different intra-species strains. *NR: The code of the reference internal transcribed spacer (ITS) sequence obtained from the website of the National Center for Biotechnology Information (NCBI). The top six species having a higher AFB1 (50 ng/kg) removal efficiency were marked by red stars.
To further determine which isolate was the most promising potential for aflatoxin detoxification, we tested the AFB1 removal ratios of the top six isolates. The result displayed that the removal ratio by CGMCC3.5218 was 72.1% at 10 μg/kg, which was significantly higher than those of D20 (xz-5-9), CGMCC3.17876, CGMCC3.1540, D44 (Xz-11-6a), and D78 (xz-11-5) (Figure 3A), suggesting that CGMCC3.5218 had the strongest potential to degrade high doses of AFB1. After 5 days of incubation, the removal rates of AFB1 at concentrations of 50 ng/kg, 125 ng/kg, 500 ng/kg, 1 μg/kg, 2.5 μg/kg, 5 μg/kg, 10 μg/kg, and 15 μg/kg were 100.0, 94.9, 95.0, 93.6, 90.4, 90.0, 87.6, and 65.6%, respectively (Figure 3B). Additionally, after 10 days of incubation, the removal rates of 500 ng/kg AFB2, AFG1, and AFG2 were higher than 90% (Figure 3C). On the basis of these results, CGMCC3.5218 proved to be extremely effective in removing AFB1, AFB2, AFG1, and AFG2. Furthermore, the strain could tolerate extreme conditions of high doses of aflatoxins, with no significant inhibitory effects on its removal capacity even at AFB1 concentrations as high as 15 μg/kg. Work by others has demonstrated that various microorganisms and their metabolites have abilities to remove aflatoxins. For example, the cell-free supernatant of microbial consortium TADC7 degraded 95.7% of AFB1 at levels of 5 μg/kg (Wang et al., 2017). The percentages of AFB1 (0.5 μg/kg) bound by Saccharomyces cerevisiae ranged from 81.0 to 99.3% in one study (Gonçalves et al., 2015). Pseudomonas aeruginosa could degrade 100 ng/kg AFB1 by 82.8% (Sangare et al., 2014). The fungus Rhizopus oryzae reduced 100 ng/kg aflatoxin B1 by approximately 100% (Hackbart et al., 2014). In this study, however, T. reesei CGMCC3.5218 was demonstrated to degrade 10 μg/kg AFB1 by approximately 95%. To the best of our knowledge, it was the most capable of removing higher doses of aflatoxins when compared with other strains of bacteria or fungi. The colony of T. reesei CGMCC3.5218 grows fast (2–3 cm/day), and the back of the fresh colony is obviously yellow. Additionally, the strain of T. reesei CGMCC3.5218 can produce yellow-green conidia after growing for at least 3 weeks. After 2 weeks of growth, the hyphae on the colony surface become hard and dense.
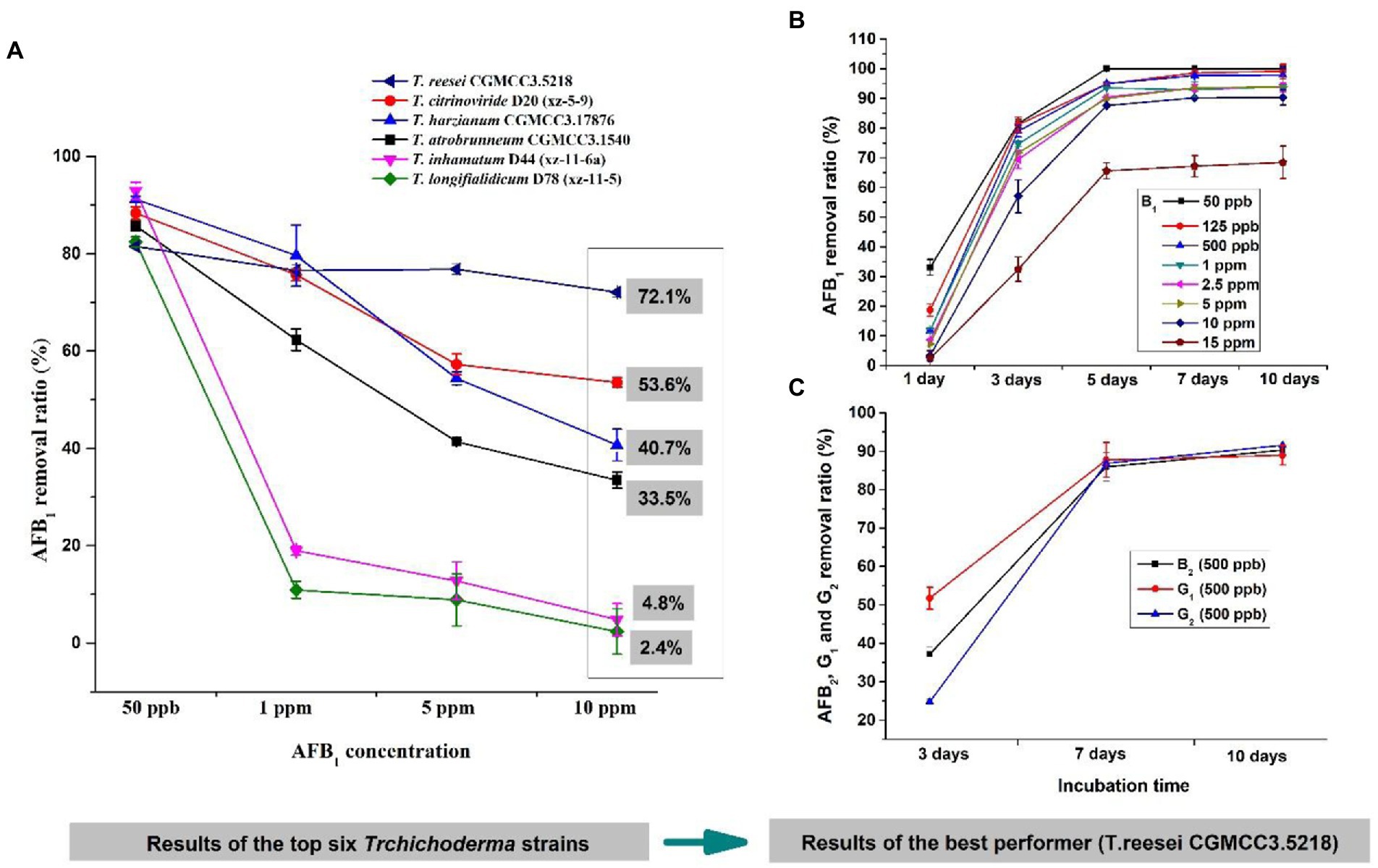
Figure 3. The candidate Trichoderma isolates for aflatoxin detoxification. (A) Removal ratios of aflatoxin B1 (AFB1) by T. reesei CGMCC3.5218, T. citrinoviride D20 (xz-5-9), T. harzianum CGMCC3.17876, T. atrobrunneum CGMCC3.1540, T. inhamatum D44 (xz-11-6a), and T. longifialidicum D78 (xz-11-5) cultured in yeast extract peptone dextrose (YPD) liquid medium, after 3 days of incubation at 28°C in the dark. (B) Removal ratios of AFB1 at concentrations of 50 ng/kg to 15 μg/kg by T. reesei CGMCC3.5218 after 1, 3, 5, 7, and 10 days of incubation. (C) Removal ratios of 500 ng/kg AFB2, AFG1, and AFG2 by T. reesei CGMCC3.5218 cultured in YPD liquid medium, after 3, 7, and 10 days of incubation at 28°C in the dark.
Optimal culture conditions for the Trichoderma reesei CGMCC3.5218 to remove aflatoxins
To determine the effects of incubation time, pH, temperature, medium, and metal ions on the AFB1 removal ability of T. reesei CGMCC3.5218, we tested the AFB1 removal ratios under these culture conditions treatment, respectively. The results showed that with the increase of incubation time, the removal rates of AFB1 gradually increased. The removal rates of AFB1 (500 ng/kg), reached the maximum value at 7 days and then increased almost steadily for up to 10 days (Figure 4A). It has been reported that the biological reduction of AFB1 can be due to binding or degradation (Verheecke et al., 2016; Adebo et al., 2017a,b). However, AFB1 binding is a rapid process and AFB1 can be released back into solutions to some extent with the extension of incubation period (Peltonen et al., 2001; Mwakinyali et al., 2019). Therefore, the removal rates of AFB1 by CGMCC3.5218 in different incubation conditions were shown in Figures 4B–E, indicating temperature and pH had significant effects on AFB1 removal ability. Particularly, NB medium resulted in an increase of AFB1 degradation, whereas the opposite was true with Zn2+.
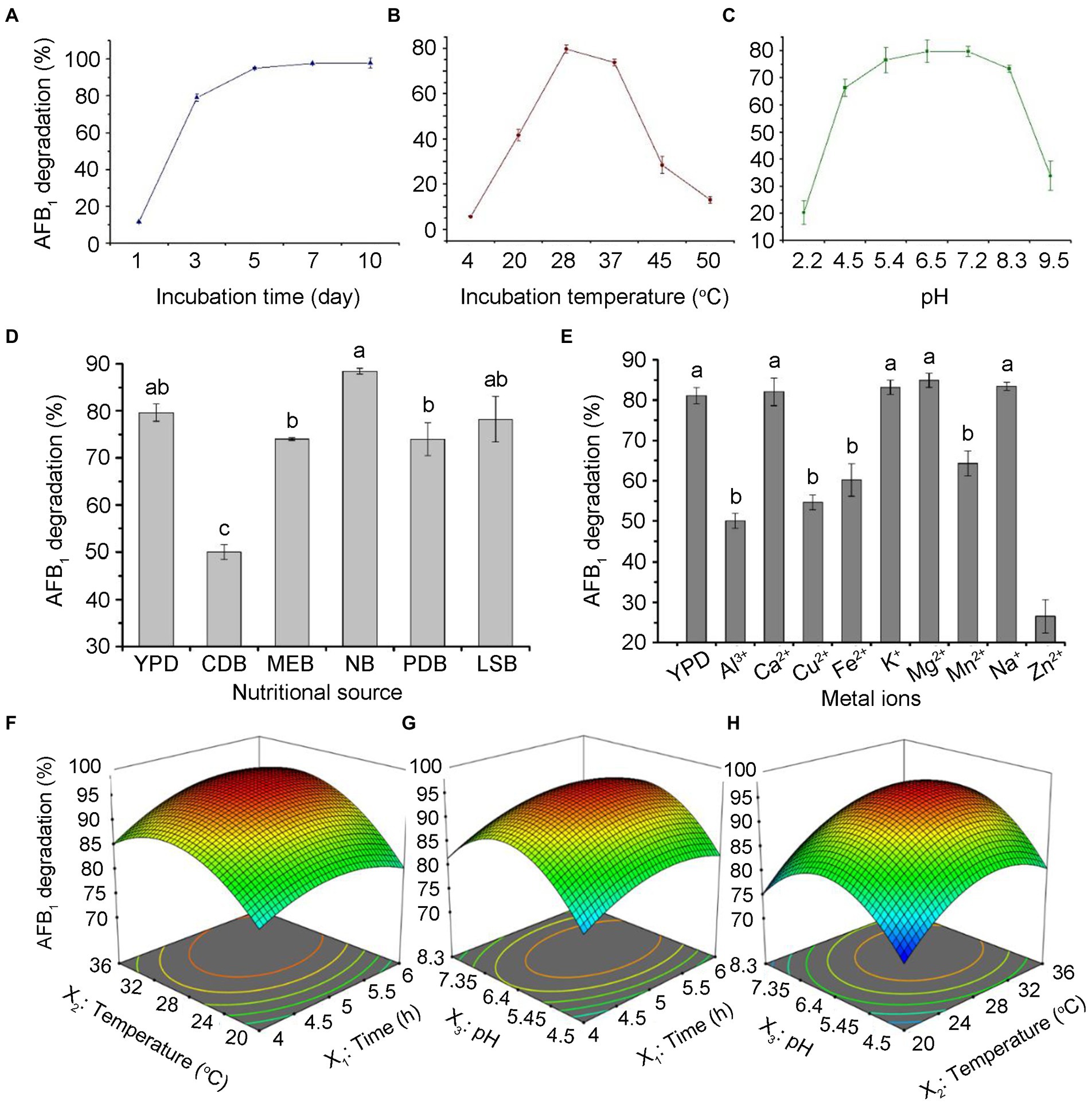
Figure 4. Optimizing culture conditions for CGMCC3.5218 to remove aflatoxins. Effect of (A) incubation time, (B) culture temperature, (C) pH, (D) nutritional sources, and (E) metal ions on the degradation of 500 ng/kg aflatoxin B1 (AFB1) by T. reesei CGMCC3.5218. Response plots between two parameters for the degradation rate of aflatoxin B1: (F) Incubation time (X1) and culture temperature (X2); (G) incubation time (X1) and medium pH (X3); and (H) culturing temperature (X2) and medium pH (X3). The values are the means of three replicates and their standard errors. Bars with different letters are significantly different for p < 0.001 (Tukey–Kramer multiple comparison test).
Meanwhile, to further obtain the optimal efficiency of AFB1 degradation, we used Box–Behnken (BB) design and response surface methodology (RSM) to optimize the values and interaction effects of the factors that affected the AFB1 removal. The Design-Expert 12.0.3 software was used to perform regression analysis on the data in Supplementary Table S3. The detailed analysis of results of the BB test were shown in Supplementary Table S5, and the regression equation was: Y = 95.25 + 2.04×1 + 4.51×2 + 1.74×3 + 1.61X1X2–0.30X1X3 + 0.75X2X3 3.92×12 6.87×22 9.78×32, where Y refers to the predicted value (percentage of AFB1 degradation), X1 refers to the incubation time, X2 refers to the incubation temperature, and X3 refers to the pH of culturing medium. As shown in Supplementary Table S5, the model F-value of 729.80 implied that the model was significant. The lack of fit F-value of 0.15 implied that the lack of fit was not significant relative to the pure error. The R2 value of 0.9989 implied that the model fitted the actual experimental results perfectly. p-values less than 0.05 indicate that model terms are significant. In this case, X1, X2, X3, X1X2, X2X3, X12, X22, and X32 were significant model terms. The response surface and contour map (Figures 4F–H) intuitively reflected the influence of various factors on the response value. The effect of X2 on AFB1 degradation was more significant than X1 and X3, as seen in the curve variation degree and contour density. Taken together, the optimal culture conditions were predicted to be at a pH of 6.7, an incubation temperature of 31.3°C, and an incubation time of 5.1 days, as determined by the software Design-Expert 12.0.3. The AFB1 degradation was 96.2% under the optimal culture conditions.
AFB1 removal patterns by the CGMCC3.5218
To determine the AFB1 removal patterns, we tested the AFB1 removal ability among diverse cell component of T. reesei CGMCC3.5218. As shown in Table 1, the AFB1 removal ratio by extracellular component (91.8%) was significantly higher than that of either active component (8.9%) or intracellular component (19.5%). Additionally, the results also indicated that very small amounts of AFB1 were washed out from the mycelia by PBS (0.8 ± 0.2 ng/kg), methanol (0.6 ± 0.1 ng/kg), and acetonitrile (1.6 ± 0.1 ng/kg) compared with the initial concentration (control) of 490.7 ng/kg (Table 1). However, the AFB1 absorbed by cell walls could be released back into the solution by elution with polar solvents (Peltonen et al., 2001; Haidukowski et al., 2019). These findings indicated that the AFB1 removal was performed by extracellular components rather than intracellular components or adsorption on the cell components. This was in agreement with another report, in which T. reesei strain QM9414 reduced approximately 100% of AFB1 within 96 h through metabolic degradation by extracellular metabolites (Hackbart et al., 2014). Other studies also have reported similar results. The strain Lactobacillus plantarum FJS003 degraded 75.2% AFB1 through the fermentation supernatant, which was higher than that (11.4%) degraded by the cells (Zhu et al., 2021). Aflatoxin B1 degradation was 58.2% after being treated with Aspergillus niger ND-1 culture supernatant for 24 h (Zhang et al., 2014). The cell-free supernatant of isolate Bacillus subtilis JSW-1 degraded aflatoxin B1 effectively, whereas the viable cells and intracellular extracts were far less effective (Xia et al., 2017). It was further confirmed that the degradation mechanism by culture supernatant was enzymatic (Oluwafemi et al., 2017; Wang et al., 2017).
Dynamics of AFB1 degradation by the CGMCC3.5218 culture supernatant
As shown in Figure 5A, most of the degradation by the culture supernatant occurred within 24 h, with calculated AFB1 degradation rates of 85.4–100%. As shown in Figure 5B, the AFB1 degradation by the culture supernatant was temperature-sensitive within the range of 4°C–45°C. The maximum degradation occurred at 45°C (100%). Interestingly, the degradation percentages of AFB1 were approximately the same in the range of 45°C–95°C. This is consistent with the study, in which the degradation activity of the supernatant of microbial consortium TADC7 exhibited thermal stability in range of 45°C–90°C (Wang et al., 2017). These results imply that the enzymes or proteins involved in the AFB1 degradation are thermostable. As shown in Figure 5C, the data indicated that CGMCC3.5218 culture supernatant was able to efficiently degrade AFB1 in a wide range of pH (3.0–9.0), which would increase its practical use in the future. The AFB1 degradation was still high even under strongly acidic and alkali conditions of pH 3.0 and 9.0, respectively. This result imply that pH may contribute to the degradation of AFB1. To investigate this possibility, the effects of different pH values of PBS on the degradation of AFB1 were studied. As shown in Figure 5D, the degradation rates were 17.0, 12.4, 12.1, and 38.4% at pH 3.0, 4.0, 8.0, and 9.0, respectively. This indicated that AFB1 could be destroyed to a certain extent when it was exposed to strongly acidic or strongly alkaline conditions.
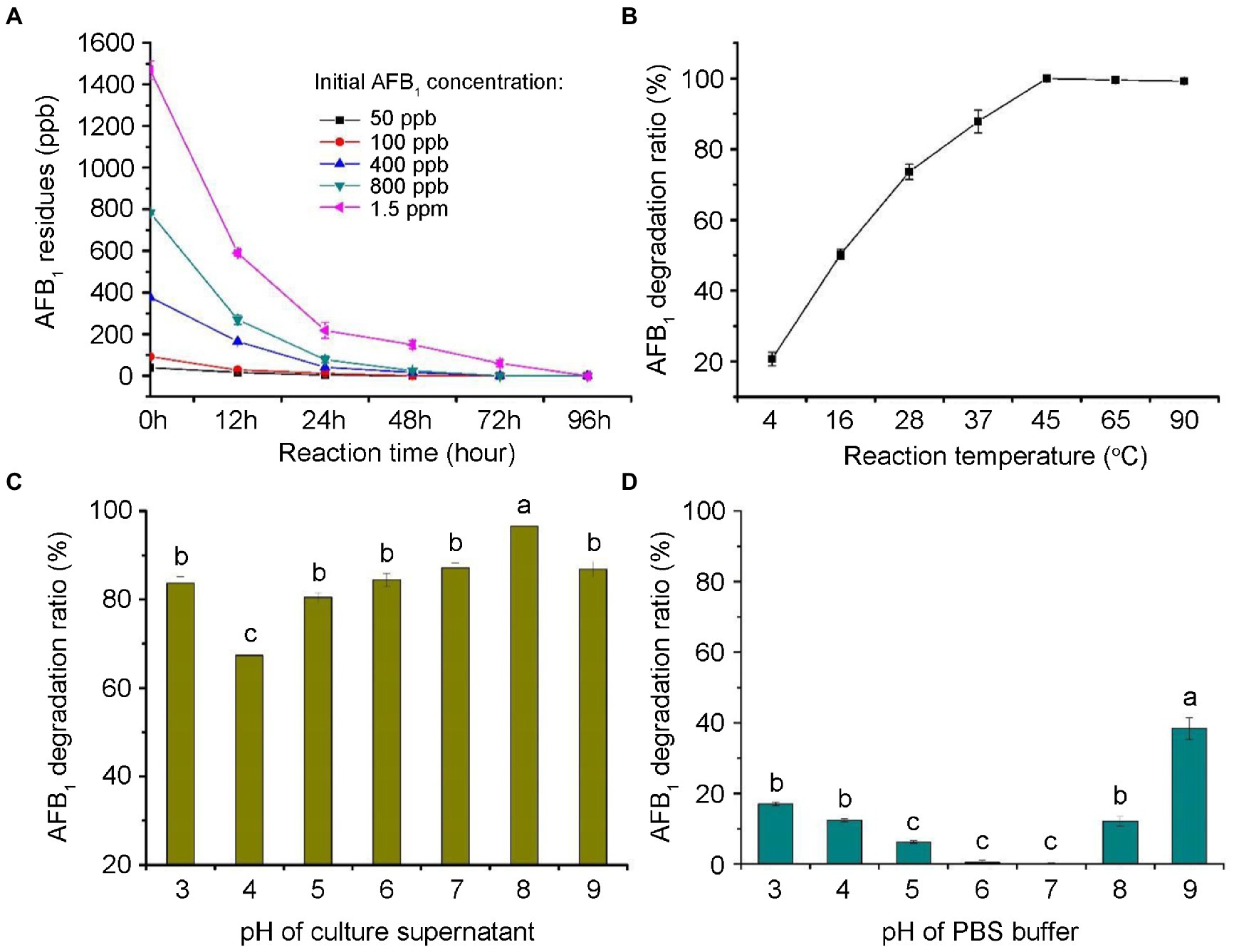
Figure 5. Degradation dynamics of aflatoxin B1 (AFB1) by the culture supernatant of T. reesei CGMCC3.5218. (A) Degradation of 50, 100, 400, 800 ng/kg, and 1.5 μg/kg AFB1 at various reaction time (0, 12, 24, 48, 72, and 96 h). (B) Degradation of 500 ng/kg AFB1 at various temperatures of 4, 16, 28, 37, 45, 65, and 95°C. (C,D) Degradation of AFB1 at a range of pH (3.0–9.0) in culture supernatant and PBS buffer. Bars with different letters are significantly different for p < 0.001 (Tukey–Kramer multiple comparison test). The values are the means of three replicates and their standard errors.
Enzymatic effects of the supernatant on AFB1 degradation
To confirm the actively extracellular components were proteins or enzymes, we investigated the enzymatic effects in CGMCC3.5218 culture supernatant. As shown in Figure 6, about 88.0% of AFB1 were degraded by the active culture supernatant after incubation for 24 h at 37°C. After a treatment with heat at 121°C for 30 min, AFB1 degradation was slightly reduced to 83.3%, suggesting that the enzymes or proteins responsible for AFB1 degradation were extremely thermotolerant. EDTA, as a metal chelating agent, did not appear to destroy the AFB1 degradation activity (87.0%). However, AFB1 degradation declined significantly to 69.6% after treatment with proteinase K, and declined to 29.4% after treatment with proteinase K plus SDS, indicating that enzymes or proteins played a key role in AFB1 degradation. Some studies have reported that T. reesei has a remarkable ability to secrete proteins or enzymes extracellularly (Sirkka and Merja, 1995; Wang et al., 2015a; Liu et al., 2016). To produce thermotolerant enzymes, a thermotolerant laccase gene from Pycnoporus sanguineus and a thermostable β-glucosidase gene from the fungus Periconia sp. were successfully cloned and inserted into the chromosomes of T. reesei (Dashtban and Qin, 2012; Zhao et al., 2018). However, to the best of our knowledge, this is the first report that T. reesei naturally secrete thermotolerant enzymes or proteins involved in contaminant degradation.
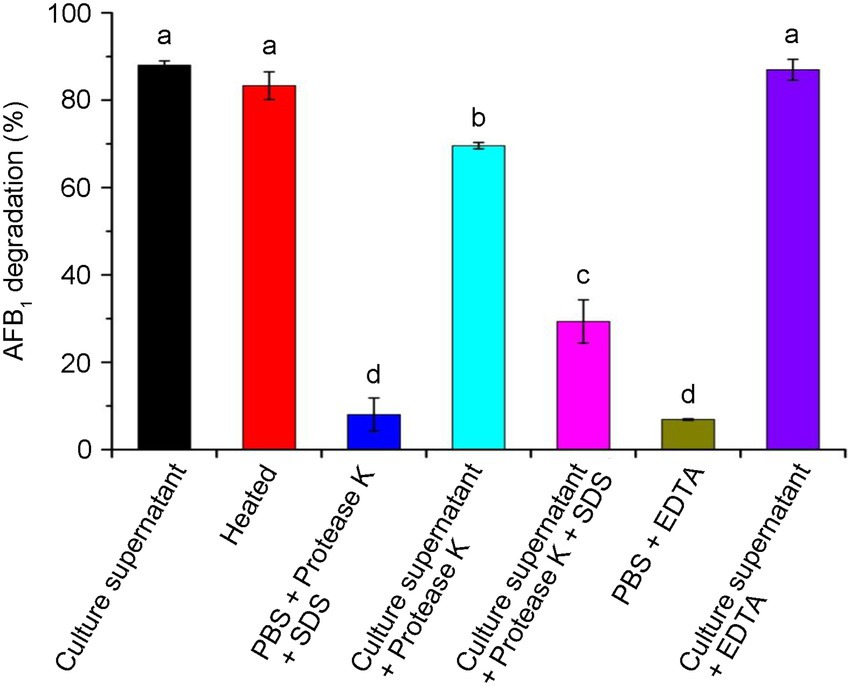
Figure 6. Effects of heat, protease K, protease K plus sodium dodecyl sulfonate (SDS), and Ethylene diamine tetraacetic acid (EDTA) on the aflatoxin B1 (AFB1) degradation activity of the culture supernatant of T. reesei CGMCC3.5218. Bars with different letters are significantly different for p < 0.001 (Tukey–Kramer multiple comparison test). The values are the means of three replicates and their standard errors.
Toxicity analysis of AFB1-degraded products
With regard to aflatoxin degradation, it is conceivable a change in molecular structure may occur, making aflatoxins undetectable, while the toxicity is not necessarily reduced, or is even increased (Detroy and Hesseltine, 1970; Gross-Steinmeyer and Eaton, 2012; Huang et al., 2017). As the aflatoxin degradation potential is not always linked to the reduction of toxicity, it is crucial to determine the toxicity of the degradation products. As shown in Figure 7, the results showed that the micronucleus assay in experimental mouse bone marrow erythrocytes showed no significant differences (p > 0.05) from the negative control (sterile water), although there was a significant difference (p < 0.01) between the assay and the positive control group (cyclophosphamide), for which the rates of micronucleus were 31.4% for males and 27.6% for females. This indicated that the tested substance in the experimental group did not increase the rate of micronucleus in mouse bone marrow erythrocytes. The acute oral toxicity tests showed that no mice died (Supplementary Table S6). Additionally, no obvious organ lesions were found by macroscopic or histopathological examination on the 14th day after intraperitoneal injection (Supplementary Table S7). The results of skin irritation tests showed that no erythema or edema formed in experimental mice (Supplementary Table S8). All of these results indicated that CGMCC3.5218 can transform the AFB1 into nontoxic molecules.
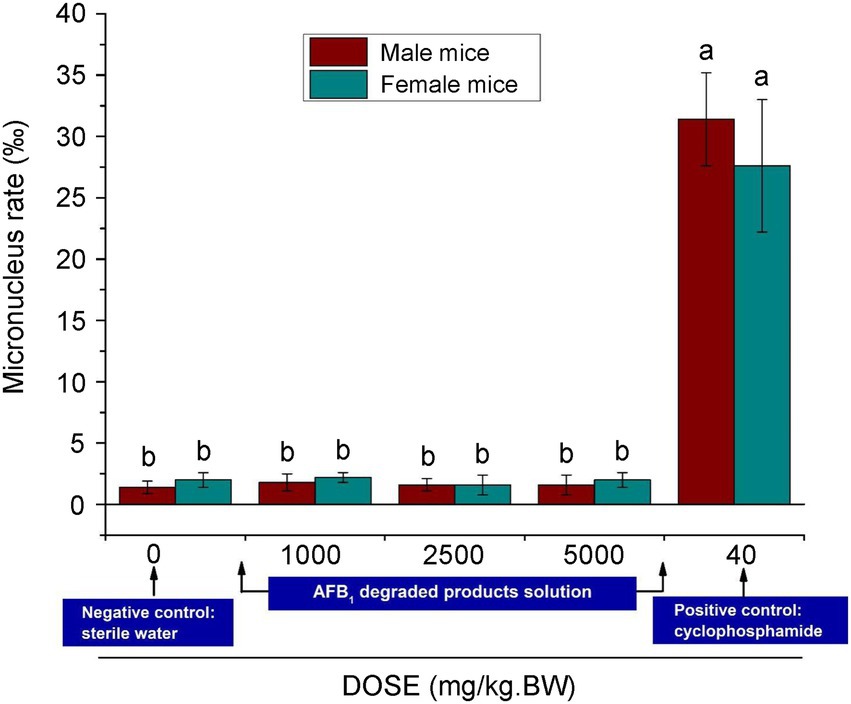
Figure 7. Results of micronucleus test of polychromatic erythrocytes in the bone marrow of male and female mice. Bars with different letters are significantly different for p < 0.001 (Tukey–Kramer multiple comparison test).
Aflatoxin degradation by CGMCC3.5218 in peanut cake, maize, and completed feed
To test the application of T. reesei CGMCC3.5218 and whether there was a loss of viability and detoxification activity in an actual system, we evaluated degradation of AFB1, AFB2, AFG1, and AFG2 in the media containing contaminated peanut cake, maize, and completed feed. As shown in Table 2, when the strain was grown in the media for 7 days, the degradation rates were 85.0–92.1%. Aflatoxin concentrations were significantly reduced compared to the relevant controls. The treatment with CGMCC3.5218 reduced the extremely high aflatoxin content of naturally contaminated maize from 2323.5 μg/kg down to 267.3 μg/kg. These results indicate that CGMCC3.5218 has a noteworthy potential for practical applications for aflatoxin degradation in actual samples.
Conclusion
Trichoderma species are widely found in soils worldwide. Members of this genus have a long history of use as biofertilizer and are generally recognized as safe fungi, whereas very few reports on their use in the biodegradation of small molecular contaminants have been published so far. In this study, AFB1 degrading capabilities of 65 Trichoderma isolates belonging to 23 different species were investigated. T. reesei CGMCC3.5218, which possessed superior efficiency and potential for application in aflatoxin degradation, was selected for further study. The culture conditions for CGMCC3.5218 to degrade AFB1 were optimized; the possible removal mechanisms and the enzymatic effects were explored, which indicated that the key AFB1 degradation activity of the strain were attributed to the thermostable enzymes or proteins in CGMCC3.5218 culture supernatant. Finally, we demonstrated that this strain could be efficiently used in peanut cake, maize and animal feeds without posing any toxicity risk. When considered together, it can be concluded that the strain T. reesei CGMCC3.5218, which has proven to possess strong aflatoxin detoxification ability, has a great potential to be used as a safe and effective biological detoxifier of AFB1 in agro-products, foods and feeds. Further work on the purification and characterization of the thermostable enzymes from T. reesei CGMCC3.5218 supernatant, as well as the identification of the structures of degradation products or degradation pathways and the possible simultaneous degradation of coexisting contaminants, are required to harness the full potential of T. reesei CGMCC3.5218.
Data availability statement
The datasets presented in this article are not readily available because unrestricted. Requests to access the datasets should be directed to XY eGlhb2ZlbmdsMTk4NzAyMDdAMTYzLmNvbQ==.
Author contributions
XY, XR, QZ, and PL conceived and designed the experiments. XY, XR, JF, NW, MH, AL, and QZ performed the experiments. XY, XR, CA, and PL analyzed the data. XY, XR, QZ, and PL wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the key project of National Natural Sciences Foundation of China (Grant No. 32030085), the Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (Grant No. 37000021P11000111320E), the National Natural Science Foundation of China (Grant Nos. 31860473 and 32161143034), the Major project of Hubei Hongshan Laboratory (Grant No. 2021hszd015), and the European Union’s Horizon 2020 Research and innovation program under Grant Agreement No. 678781 (MycoKey). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1003039/full#supplementary-material
References
Adebo, O. A., Njobeh, P. B., Gbashi, S., Nwinyi, O. C., and Mavumengwana, V. (2017a). Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 57, 3208–3217. doi: 10.1080/10408398.2015.1106440
Adebo, O. A., Njobeh, P. B., Sidu, S., Adebiyi, J. A., and Mavumengwana, V. (2017b). Aflatoxin B1 degradation by culture and lysate of a Pontibacter specie. Food Control 80, 99–103. doi: 10.1016/j.foodcont.2017.04.042
Battacone, G., Nudda, A., Palomba, M., Pascale, M., Nicolussi, P., and Pulina, G. (2005). Transfer of aflatoxin B1 from feed to milk and from milk to curd and whey in dairy sheep fed artificially contaminated concentrates. J. Dairy Sci. 88, 3063–3069. doi: 10.3168/jds.S0022-0302(05)72987-8
Bennett, J. W., and Klich, M. (2003). Mycotoxins. Clin. Microbiol. Rev. 16, 497–516. doi: 10.1128/CMR.16.3.497-516.2003
Branà, M. T., Cimmarusti, M. T., Haidukowski, M., Logrieco, A. F., and Altomare, C. (2017). Bioremediation of aflatoxin B1-contaminated maize by king oyster mushroom (Pleurotus eryngii). PLoS One 12:e0182574. doi: 10.1371/journal.pone.0182574
Braun, H., Woitsch, L., Hetzer, B., Geisen, R., Zange, B., and Schmidt-Heydt, M. (2018). Trichoderma harzianum: inhibition of mycotoxin producing fungi and toxin biosynthesis. Int. J. Food Microbiol. 280, 10–16. doi: 10.1016/j.ijfoodmicro.2018.04.021
Calistru, C., McLean, M., and Berjak, P. (1997). In vitro studies on the potential for biological control of Aspergillus flavus and Fusarium moniliforme by Trichoderma species. Mycopathologia 139, 115–121. doi: 10.1023/a:1006867907885
Dashtban, M., and Qin, W. (2012). Overexpression of an exotic thermotolerant β-glucosidase in Trichoderma reesei and its significant increase in cellulolytic activity and saccharification of barley straw. Microb. Cell Factories 11:63. doi: 10.1186/1475-2859-11-63
Detroy, R. W., and Hesseltine, C. W. (1970). Aflatoxicol: structure of a new transformation product of aflatoxin B1. Can. J. Biochem. 48, 830–832. doi: 10.1139/o70-130
Eslami, M., Mashak, Z., Heshmati, A., Shokrzadeh, M., and Nejad, A. S. M. (2015). Determination of aflatoxin B1 levels in Iranian rice by ELISA method. Toxin Rev. 34, 125–128. doi: 10.3109/15569543.2015.1074925
Fanelli, F., Liuzzi, V. C., Logrieco, A. F., and Altomare, C. (2018). Genomic characterization of Trichoderma atrobrunneum (T. harzianum species complex) ITEM 908: insight into the genetic endowment of a multi-target biocontrol strain. BMC Genomics 19:662. doi: 10.1186/s12864-018-5049-3
Gonçalves, B. L., Rosim, R. E., de Oliveira, C. A. F., and Corassin, C. H. (2015). The in vitro ability of different Saccharomyces cerevisiae - based products to bind aflatoxin B1. Food Control 47, 298–300. doi: 10.1016/j.foodcont.2014.07.024
Gross-Steinmeyer, K., and Eaton, D. L. (2012). Dietary modulation of the biotransformation and genotoxicity of aflatoxin B1. Toxicology 299, 69–79. doi: 10.1016/j.tox.2012.05.016
Guo, Y., Qin, X., Tang, Y., Ma, Q., Zhang, J., and Zhao, L. (2020). Cot a laccase, a novel aflatoxin oxidase from bacillus licheniformis, transfroms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 325:126877. doi: 10.1016/j.foodchem.2020.126877
Gusakov, A. V. (2011). Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol. 29, 419–425. doi: 10.1016/j.tibtech.2011.04.004
Hackbart, H. C. S., Machado, A. R., Christ-Ribeiro, A., Prietto, L., and Badiale-Furlong, E. (2014). Reduction of aflatoxins by Rhizopus oryzae and Trichoderma reesei. Mycotoxin Res. 30, 141–149. doi: 10.1007/s12550-014-0202-6
Haidukowski, M., Casamassima, E., Cimmarusti, M. T., Branà, M. T., Longobardi, F., Acquafredda, P., et al. (2019). Aflatoxin B1-adsorbing capability of Pleurotus eryngii mycelium: efficiency and modeling of the process. Front. Microbiol. 10:1386. doi: 10.3389/fmicb.2019.01386
Hermosa, R., Cardoza, R. E., Rubio, M. B., Gutiérrez, S., and Monte, E. (2014). Secondary metabolism and antimicrobial metabolites of Trichoderma. Biotechnol. Biol. Trichoderma, 125–137. doi: 10.1016/B978-0-444-59576-8.00010-2
Heshmati, A., Nejad, A. S. M., and Ghyasvand, T. (2020). The occurrence and risk assessment of aflatoxin M1 in yoghurt samples from Hamadan. Iran. Open Public Health J. 13, 512–517. doi: 10.2174/1874944502013010512
Heshmati, A., Nejad, A. S. M., and Mehri, F. (2021). Occurrence, dietary exposure, and risk assessment of aflatoxins in wheat flour from Iran. Int. J. Environ. Anal. Chem., 1–14. doi: 10.1080/03067319.2021.2011254
Huang, L., Duan, C., Zhao, Y., Gao, L., Niu, C., Xu, J., et al. (2017). Reduction of aflatoxin B1 toxicity by lactobacillus plantarum C88: a potential probiotic strain isolated from Chinese traditional fermented food “tofu”. PLoS One 12:e0170109. doi: 10.1371/journal.pone.0170109
IARC (2002). Aflatoxins. IARC monograph on the evaluation of carcinogenic risks to humans. IARC, World Health Organization, Lyon 82:171.
Ji, C., Fan, Y., and Zhao, L. (2016). Review on biological degradation of mycotoxins. Anim. Nutr. 2, 127–133. doi: 10.1016/j.aninu.2016.07.003
Juhász, T., Szengyel, Z., Réczey, K., Siika-Aho, M., and Viikari, L. (2005). Characterization of cellulases and hemicellulases produced by Trichoderma reesei on various carbon sources. Process Biochem. 40, 3519–3525. doi: 10.1016/j.procbio.2005.03.057
Kamkar, A., Fallah, A. A., and Nejad, A. S. M. (2014). The review of aflatoxin M1 contamination in milk and dairy products produced in Iran. Toxin Rev. 33, 160–168. doi: 10.3109/15569543.2014.922580
Keller, L. A. M., González Pereyra, M. L., Keller, K. M., Alonso, V. A., Oliveira, A. A., Almeida, T. X., et al. (2013). Fungal and mycotoxins contamination in corn silage: monitoring risk before and after fermentation. J. Stored Prod. Res. 52, 42–47. doi: 10.1016/j.jspr.2012.09.001
Li, J., Huang, J., Jin, Y., Wu, C., Shen, D., Zhang, S., et al. (2018). Mechanism and kinetics of degrading aflatoxin B1 by salt tolerant Candida versatilis CGMCC 3790. J. Hazard. Mater. 359, 382–387. doi: 10.1016/j.jhazmat.2018.05.053
Liu, K., Dong, Y., Wang, F., Jiang, B., Wang, M., and Fang, X. (2016). Regulation of cellulase expression, sporulation, and morphogenesis by velvet family proteins in Trichoderma reesei. Appl. Microbiol. Biotechnol. 100, 769–779. doi: 10.1007/s00253-015-7059-2
Loi, M., Renaud, J. B., Rosini, E., Pollegioni, L., Vignali, E., Haidukowski, M., et al. (2020). Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 250:126296. doi: 10.1016/j.chemosphere.2020.126296
Luo, X., Wang, R., Wang, L., Wang, Y., and Chen, Z. (2013). Structure elucidation and toxicity analyses of the degradation products of aflatoxin B1 by aqueous ozone. Food Control 31, 331–336. doi: 10.1016/j.foodcont.2012.10.030
Marin, S., Ramos, A. J., Cano-Sancho, G., and Sanchis, V. (2013). Mycotoxins: occurrence, toxicology, and exposure assessment. Food Chem. Toxicol. 60, 218–237. doi: 10.1016/j.fct.2013.07.047
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26, 553–560. doi: 10.1038/nbt1403
Mugwagwa, L. R., and Chimphango, A. F. A. (2019). Box-Behnken design based multi-objective optimisation of sequential extraction of pectin and anthocyanins from mango peels. Carbohydr. Polym. 219, 29–38. doi: 10.1016/j.carbpol.2019.05.015
Mwakinyali, S. E., Ming, Z., Xie, H., Zhang, Q., and Li, P. (2019). Investigation and characterization of Myroides odoratimimus strain 3J2MO aflatoxin B1 degradation. J. Agric. Food Chem. 67, 4595–4602. doi: 10.1021/acs.jafc.8b06810
Nejad, A. S. M., Ghannad, M. S., and Kamkar, A. (2014). Determination of aflatoxin B1 levels in Iranian and Indian spices by ELISA method. Toxin Rev. 33, 151–154. doi: 10.3109/15569543.2014.942319
Nejad, A. S. M., Heshmati, A., and Ghiasvand, T. (2019). The occurrence and risk assessment of exposure to aflatoxin M1 in ultra-high temperature and pasteurized milk in Hamadan province of Iran. Osong. Public Health Res. Perspect. 10, 228–233. doi: 10.24171/j.phrp.2019.10.4.05
Nejad, A. S. M., Heshmati, A., and Ghiasvand, T. (2020). The occurrence and risk assessment of aflatoxin M1 in cheeses samples from Hamadan. Iran. Iran. J. Pharm. Res. 19, 44–50. doi: 10.22037/ijpr.2020.112399.13754
Palonen, H., Tjerneld, F., Zacchi, G., and Tenkanen, M. (2004). Adsorption of Trichoderma reesei CBH I and EG II and their catalytic domains on steam pretreated softwood and isolated lignin. J. Biotechnol. 107, 65–72. doi: 10.1016/j.jbiotec.2003.09.011
Pankaj, S. K., Shi, H., and Keener, K. M. (2018). A review of novel physical and chemical decontamination technologies for aflatoxin in food. Trends Food Sci. Technol. 71, 73–83. doi: 10.1016/j.tifs.2017.11.007
Peltonen, K., El-Nezami, H., Haskard, C., Ahokas, J., and Salminen, S. (2001). Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria. J. Dairy Sci. 84, 2152–2156. doi: 10.3168/jds.S0022-0302(01)74660-7
Pitt, J. I., and Hocking, A. D. (2006). Mycotoxins in Australia: biocontrol of aflatoxin in peanuts. Mycopathologia 162, 233–243. doi: 10.1007/s11046-006-0059-0
Pribowo, A., Arantes, V., and Saddler, J. N. (2012). The adsorption and enzyme activity profiles of specific Trichoderma reesei cellulase/xylanase components when hydrolyzing steam pretreated corn Stover. Enzym. Microb. Technol. 50, 195–203. doi: 10.1016/j.enzmictec.2011.12.004
Ren, X., Branà, M. T., Haidukowski, M., Gallo, A., Zhang, Q., Logrieco, A. F., et al. (2022). Potential of Trichoderma spp. for biocontrol of aflatoxin-producing Aspergillus flavus. Toxins 14:86. doi: 10.3390/toxins14020086
Ren, X., Zhang, Q., Zhang, W., Mao, J., and Li, P. (2020). Control of aflatoxigenic molds by antagonistic microorganisms: inhibitory behaviors, bioactive compounds, related mechanisms, and influencing factors. Toxins 12:24. doi: 10.3390/toxins12010024
Sangare, L., Zhao, Y., Folly, Y. M. E., Chang, J., Li, J., Selvaraj, J. N., et al. (2014). Aflatoxin B1 degradation by a pseudomonas strain. Toxins 6, 3028–3040. doi: 10.3390/toxins6103028
Sirkka, K., and Merja, P. (1995). Production of recombinant proteins in the filamentous fungus Trichoderma reesei. Curr. Opin. Biotechnol. 6, 534–537. doi: 10.1016/0958-1669(95)80088-3
Verheecke, C., Liboz, T., and Mathieu, F. (2016). Microbial degradation of aflatoxin B1: current status and future advances. Int. J. Food Microbiol. 237, 1–9. doi: 10.1016/j.ijfoodmicro.2016.07.028
Vinale, F., Marra, R., Scala, F., Ghisalberti, E. L., Lorito, M., and Sivasithamparam, K. (2006). Major secondary metabolites produced by two commercial Trichoderma strains active against different phytopathogens. Lett. Appl. Microbiol. 43, 143–148. doi: 10.1111/j.1472-765X.2006.01939.x
Wang, M., Yang, H., Zhang, M., Liu, K., Wang, H., Luo, Y., et al. (2015a). Functional analysis of Trichoderma reesei CKIIα2, a catalytic subunit of casein kinase II. Appl. Microbiol. Biotechnol. 99, 5929–5938. doi: 10.1007/s00253-015-6544-y
Wang, W., Ma, X., Xu, Y., Cao, Y., Jiang, Z., Ding, T., et al. (2015b). Ultrasound-assisted heating extraction of pectin from grapefruit peel: optimization and comparison with the conventional method. Food Chem. 178, 106–114. doi: 10.1016/j.foodchem.2015.01.080
Wang, Y., Zhao, C., Zhang, D., Zhao, M., Zheng, D., Lyu, Y., et al. (2017). Effective degradation of aflatoxin B1 using a novel thermophilic microbial consortium TADC7. Bioresour. Technol. 224, 166–173. doi: 10.1016/j.biortech.2016.11.033
Xia, X., Zhang, Y., Li, M., Garba, B., Zhang, Q., Wang, Y., et al. (2017). Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Control 75, 92–98. doi: 10.1016/j.foodcont.2016.12.036
Xu, J., Wang, H., Zhu, Z., Ji, F., Yin, X., Hong, Q., et al. (2016). Isolation and characterization of bacillus amyloliquefaciens ZDS-1: exploring the degradation of zearalenone by bacillus spp. Food Control 68, 244–250. doi: 10.1016/j.foodcont.2016.03.030
Zafra, G., and Cortés-Espinosa, D. V. (2015). Biodegradation of polycyclic aromatic hydrocarbons by Trichoderma species: a mini review. Environ. Sci. Pollut. Res. Int. 22, 19426–19433. doi: 10.1007/s11356-015-5602-4
Zeilinger, S., Gruber, S., Bansal, R., and Mukherjee, P. K. (2016). Secondary metabolism in Trichoderma-chemistry meets genomics. Fungal Biol. Rev. 30, 74–90. doi: 10.1016/j.fbr.2016.05.001
Zhang, W., Xue, B., Li, M., Mu, Y., Chen, Z., Li, J., et al. (2014). Screening a strain of Aspergillus Niger and optimization of fermentation conditions for degradation of aflatoxin B1. Toxins 6, 3157–3172. doi: 10.3390/toxins6113157
Zhao, J., Zeng, S., Xia, Y., and Xia, L. (2018). Expression of a thermotolerant laccase from Pycnoporus sanguineus in Trichoderma reesei and its application in the degradation of bisphenol A. J. Biosci. Bioeng. 125, 371–376. doi: 10.1016/j.jbiosc.2017.11.010
Keywords: aflatoxins, aflatoxin B1, Aspergillus flavus, Trichoderma reesei CGMCC3.5218, degradation, detoxification
Citation: Yue X, Ren X, Fu J, Wei N, Altomare C, Haidukowski M, Logrieco AF, Zhang Q and Li P (2022) Characterization and mechanism of aflatoxin degradation by a novel strain of Trichoderma reesei CGMCC3.5218. Front. Microbiol. 13:1003039. doi: 10.3389/fmicb.2022.1003039
Edited by:
Qing Kong, Ocean University of China, ChinaReviewed by:
Amir Sasan Mozaffari Nejad, Jiroft University of Medical Sciences, IranCristiane A. Ottoni, São Paulo State University, Brazil
Copyright © 2022 Yue, Ren, Fu, Wei, Altomare, Haidukowski, Logrieco, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudio Altomare, Y2xhdWRpby5hbHRvbWFyZUBpc3BhLmNuci5pdA==; Qi Zhang, emhhbmdxaTAxQGNhYXMuY24=; Peiwu Li, cGVpd3VsaUBvaWxjcm9wcy5jbg==
†These authors have contributed equally to this work
 Xiaofeng Yue
Xiaofeng Yue Xianfeng Ren
Xianfeng Ren Jiayun Fu1,2
Jiayun Fu1,2 Claudio Altomare
Claudio Altomare Miriam Haidukowski
Miriam Haidukowski Antonio F. Logrieco
Antonio F. Logrieco Qi Zhang
Qi Zhang Peiwu Li
Peiwu Li