
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 October 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1001865
Agrobacterium tumefaciens, a soil-borne, saprophytic plant pathogen that colonizes plant surfaces and induces tumors in a wide range of dicotyledonous plants by transferring and expressing its T-DNA genes. The limited availabilities and efficacies of current treatments necessitate the exploration of new anti-Agrobacterium agents. We examined the effects of trans-cinnamaldehyde (t-CNMA) and its derivatives on the cell surface hydrophobicity, exopolysaccharide and exo-protease production, swimming motility on agar, and biofilm forming ability of A. tumefaciens. Based on initial biofilm inhibition results and minimum inhibitory concentration (MIC) data, 4-nitro, 4-chloro, and 4-fluoro CNMAs were further tested. 4-Nitro, 4-chloro, and 4-fluoro CNMA at ≥150 μg/ml significantly inhibited biofilm formation by 94–99%. Similarly, biofilm formation on polystyrene or nylon was substantially reduced by 4-nitro and 4-chloro CNMAs as determined by optical microscopy and scanning electron microscopy (SEM) and 3-D spectrum plots. 4-Nitro and 4-chloro CNMAs induced cell shortening and concentration- and time-dependently reduced cell growth. Virulence factors were significantly and dose-dependently suppressed by 4-nitro and 4-chloro CNMAs (P ≤ 0.05). Gene expressional changes were greater after 4-nitro CNMA than t-CNMA treatment, as determined by qRT-PCR. Furthermore, some genes essential for biofilm formation, motility, and virulence genes significantly downregulated by 4-nitro CNMA. Seed germination of Raphanus sativus was not hindered by 4-nitro or 4-fluoro CNMA at concentrations ≤200 μg/ml, but root surface biofilm formation was severely inhibited. This study is the first to report the anti-Agrobacterium biofilm and anti-virulence effects of 4-nitro, 4-chloro, and 4-fluoro CNMAs and t-CNMA and indicates that they should be considered starting points for the development of anti-Agrobacterium agents.
Agrobacterium tumefaciens is a fatal plant pathogenic bacterium responsible for crown-gall disease and contains a Ti-plasmid that is inserted into the plant genome via horizontal gene transfer (Liu and Nester, 2006). This exclusive feature of A. tumefaciens has been well researched and utilized for genetic transformations of plants under laboratory conditions (Nguyen et al., 2021). However, in natural environments, pathogenic agrobacteria may infect a range of important crop plants based on their biovars; (i) A. tumefaciens species complex (biovar I), (ii) Agrobacterium rhizogenes (biovar II), and (iii) Agrobacterium vitis (biovar III) (Slater et al., 2009). Of these, A. tumefaciens is predominantly found living a saprophytic lifestyle in different environments, including the rhizosphere where it thrives, forms biofilms (Heindl et al., 2014), and may infect a broad range of dicotyledonous plant species (>600), induce gall formation, and cause huge crop losses (>5% of economically important crops globally) (Moens, 2009). A. tumefaciens senses chemical signals (rhizospheric signal molecules), such as sugars, organic acids, and amino acids by chemotaxis, and enters host tissues at surface wounds (Liu and Nester, 2006). Furthermore, on receiving signals from plants, Agrobacterium increases the expressions of its virulence genes. The two-component VirA/VirG regulatory system activates virulence genes and assists transfer of Ti-plasmid to host plants (Nabi et al., 2022). Virulent A. tumefaciens transfers and integrates its T-DNA fragment from a Ti-plasmid into the host genome and its subsequent expression increases the production of opines and plant hormones like cytokinin and indole-3-acetic acid (Dessaux and Faure, 2018; Nabi et al., 2022), which enhance the plant growth and induce tumor formation. Opines are utilized by Agrobacterium as nutrients and activate quorum sensing (QS) signaling, which further enhances A. tumefaciens virulence and opine metabolism (Faure and Lang, 2014).
For disease to occur, A. tumefaciens must first physically attach to the host surface, which may occur in a stepwise manner, as follows: (i) initial surface contact by motile flagella, (ii) establishment of transient reversible attachment facilitated by protein adhesins and a range of pili (conjugative and Ctp) (Matthysse, 2014), and (iii) irreversible attachment by bacterial exopolysaccharides (EPS) after biofilm establishment (Thompson et al., 2018). A. tumefaciens can colonize and form biofilms on various abiotic surfaces, plant roots, and wounds, which it reaches by swimming using six flagella located around a single pole (Merritt et al., 2007) and then attaches firmly to cellulose fibrils. Bacterial EPS secretion, pili activity, and biofilm formation vary among soil-borne pathogenic bacteria but are required when pathogens transit to the biofilm mode from planktonic (Muhammad et al., 2020). Generally, antibiotics, copper bactericides, or fosetyl-aluminum are used to control plant pathogenic bacteria, but these measures are less effective at controlling A. tumefaciens infections (Lee et al., 2020), not cost-effective, and not readily available. Agrocin 84 (a biopesticide) produced by genetically modified (GM) Agrobacterium radiobacter (non-pathogenic) strains K84 and K1026, which competitively colonized the roots of several crops, were reported to inhibit the production of leucyl-tRNA synthetase in A. tumefaciens (McCardell and Pootjes, 1976; Kerr and Bullard, 2020). However, its effects on non-targeted useful rhizospheric organisms have not been assessed, and its interactions with agrobacterial species complex (biovar I–III) are unknown. Furthermore, field applications of GM organisms are prohibited in some countries (Kahla et al., 2017). Thus, other novel approaches are needed to prevent A. tumefaciens biofilm formation and virulence and control crown gall disease.
Plant-derived bioactive compounds offer a potential source of anti-Agrobacterium molecules, and cinnamaldehydes derived from the bark of ∼250 species belonging to the genus Cinnamomum are of particular interest (Shreaz et al., 2016). The cinnamaldehyde obtained from essential oils has been categorized as generally regarded as safe (GRAS) by the U.S. Food and Drug Administration (FDA) and has been approved for used in foods (Wei et al., 2011) and given status “A” by the Council of Europe for use in food (Friedman, 2017). Due to its characteristic aroma, color, and taste, trans-cinnamaldehyde (t-CNMA) is used medically and as a flavoring agent (Chun et al., 2013). Furthermore, t-CNMA has been reported to have anti-QS (Zhang et al., 2018), antibiofilm (Yu et al., 2020), and antibacterial effects (Yossa et al., 2014) against several food and clinical pathogens including Erwinia carotovora (Zhang et al., 2018), Pseudomonas fluorescens (Zhang et al., 2018), Campylobacter spp. (Yu et al., 2020), Escherichia coli O157:H7 (Yossa et al., 2014), Salmonella (Yossa et al., 2014), Staphylococcus aureus (Ferro et al., 2016), and the fungus Candida albicans (Chen et al., 2019). Moreover, cinnamaldehydes also have antioxidant, anti-inflammatory, anticancer, and anti-diabetic activities (Rao and Gan, 2014). Nevertheless, the antivirulence and antibiofilm effects of t-CNMA and its derivatives on A. tumefaciens have not been investigated. We hypothesized that t-CNMA and its derivatives might inhibit biofilm formation by A. tumefaciens on abiotic and biotic surfaces, and thus, we investigated the antibacterial and antibiofilm effects of t-CNMA and ten of its derivatives.
Trans-cinnamaldehyde (99%) and 10 of its derivatives: cinnamaldehyde oxime (95%), α-methylcinnamaldehyde (95%), 2-methoxycinnamaldehyde (95%), 2-nitrocinnamaldehyde (98%), 4-bromocinnamaldehyde (95%), 4-methoxycinnamaldehyde (95%), 4-nitrocinnamaldehyde (95%), 4-dimethylaminocinnamaldehyde (98%), 4-fluorocinnamaldehyde (97%), and 4-chlorocinnamaldehyde (95%) were purchased from Sigma-Aldrich (St. Louis, MO, USA) or Combi-Blocks (San Diego, CA, USA). Their molecular weights and chemical structures are given in Table 1. Stocks of 100 mg/ml were prepared in dimethyl sulfoxide (DMSO) and kept at −20°C until required. A total of 0.1% (v/v) DMSO was used as the control for antibacterial and biofilm experiments; at this concentration DMSO did not effect on bacterial growth or biofilm formation.
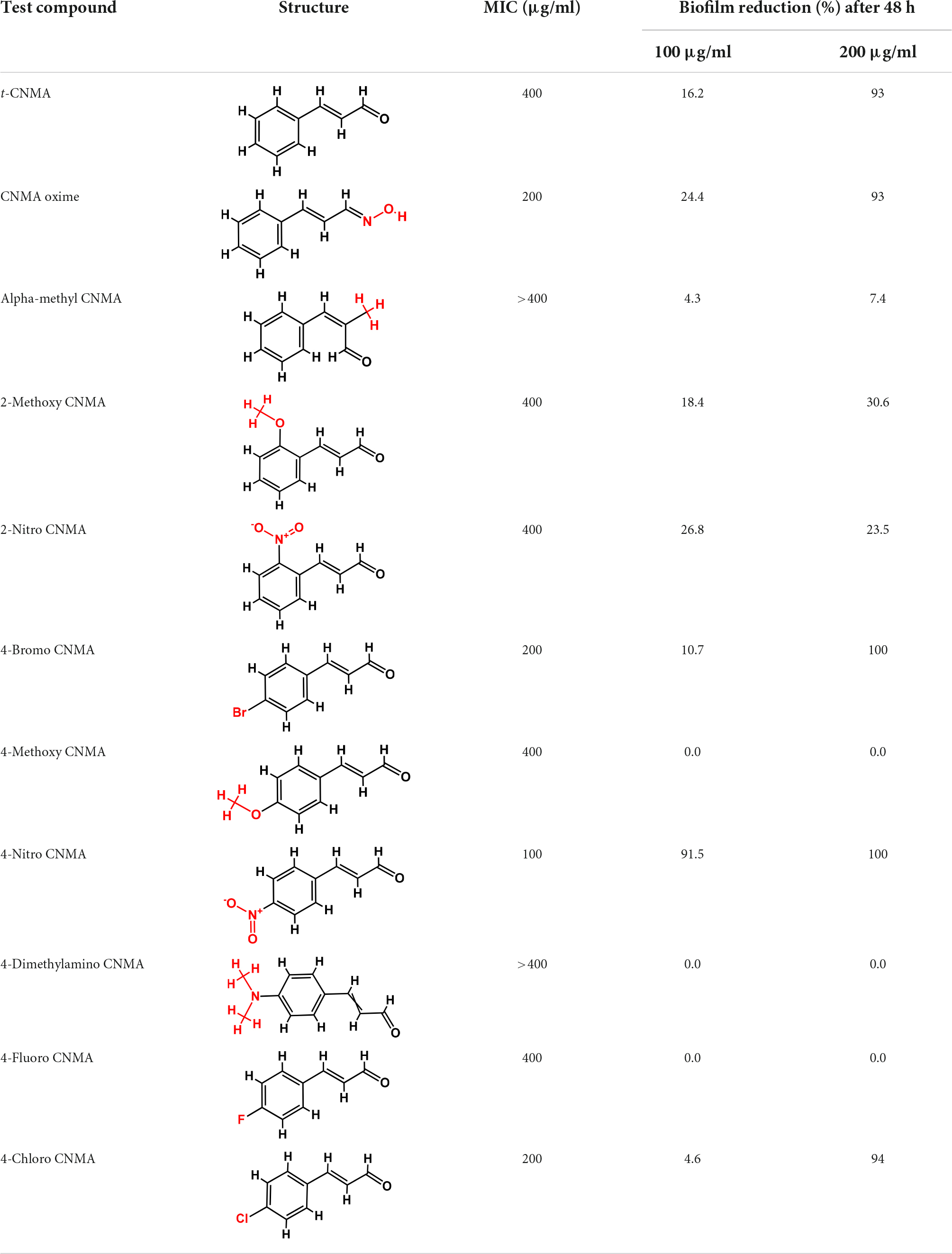
Table 1. Minimum inhibitory concentration and biofilm reduction by t-CNMA derivatives against A. tumefaciens.
Agrobacterium tumefaciens GV2260 was maintained at 30°C on Luria Bertani (LB) agar plates, and for long-term preservation, glycerol (20% v/v) culture stocks in LB were stored at −80°C. For working cultures, two independent colonies from LB agar plates were inoculated in LB broth and incubated at 250 rpm for 24 h at 30°C. At least two independent cultures replicated into three (n = 2 × 3 = 6) were used for experiments.
For antibiofilm screening, colonies of A. tumefaciens grown for 24 h in LB broth were diluted with fresh LB at 1:50, and t-CNMA or its derivatives at 100 or 200 μg/ml in LB were added. Bacterial cells incubated with LB only were considered non-treated controls. Aliquots (300 μl) of these cultures were added to wells of 96-well microtiter plates and incubated for 48 h at 30°C. Then A. tumefaciens biofilm formation was checked using a crystal violet assay (Ramey and Parsek, 2006), with modifications; wells were washed three times with sterile distilled water kept at room temperature, plates were air-dried, and 300 μl of 0.1% crystal violet was added to each well. After incubation for 20 min at room temperature, wells were rinsed three times with sterile distilled water, and 300 μl of 95% ethanol was added. After shaking microtiter plates for 1 min in a plate reader, biofilm absorbance was recorded at 570 nm. To determine minimum inhibitory concentrations (MICs), A. tumefaciens cells were treated with 0–400 μg/ml of t-CNMA or its derivatives in LB diluted at 1:100, and then 300 μl aliquots were incubated in a microtiter plate for 24 h at 30°C. Cell growths were determined by measuring optical density at 620 nm.
Cells of strain GV2260 from 24 h cultures were diluted with LB at 1:50 and 25, 50, 75, 100, 150, 200, or 400 μg/ml of t-CNMA, 4-nitrocinnamaldehyde (4-nitro CNMA), 4-chlorocinnamaldehyde (4-chloro CNMA), or 4-fluorocinnamaldehyde (4-fluoro CNMA) were added. Crystal violet biofilm assays were performed as described in section “Antibiofilm screening and minimum inhibitory concentration determinations” above. A total of six wells and two independent bacterial cultures were used for each test concentration. The experiment was terminated after incubation for 48 h at 30°C under static conditions.
For microscopic observation, biofilm formation by A. tumefaciens was also challenged in 6-well tissue culture plates using t-CNMA, 4-nitro, 4-chloro, or 4-fluoro CNMA. Briefly, 3 ml of bacterial suspensions prepared in LB broth (1:50 culture broth ratio) were mixed with t-CNMA or its derivatives (50–200 μg/ml) and added to wells. After incubation for 48 h at 30°C under static conditions, the media containing planktonic cells was carefully removed, and biofilms were rinsed carefully three times with PBS in the wells. Biofilms were then stained with crystal violet (0.1%) for 20 min at room temperature, rinsed with distilled water, and visualized using the iRiS Digital Cell imaging system (Logos Biosystems, Annandale, VA, USA). Micrographs were captured, and color mesh plots were created using ImageJ software.
Agrobacterium tumefaciens was allowed to form biofilm on nylon membrane surfaces in 96-well plates (Lee et al., 2011). t-CNMA, 4-nitro CNMA, 4-chloro CNMA, or 4-fluoro CNMA (200 μg/ml) were added to bacterial cultures in LB (1:50), and small autoclaved pieces of nylon membrane were added to wells. Plates were incubated for 48 h at 30°C. Membranes were then removed, rinsed with sterile PBS, fixed in a mixture of 2.5% glutaraldehyde and 2% formaldehyde solution in distilled water, left for 30 min at room temperature, and then kept at 4°C overnight (Ahmed et al., 2021). Samples were then dehydrated using an ethanol gradient (30, 50, 70, 90, and 100% for 10 min each), critical point dried, coated with Au or Pt, and visualized by FE-SEM (model S-4200, Hitachi, Tokyo, Japan) at 15 kV. The length of biofilm cells attached to nylon surface were determined by measuring the length of at least 50 cells per test concentration using ImageJ software. A scale was set with the “Analyze” tool of ImageJ and then length of each cell (n = 50) was determined in micrometers (μm).
The swimming motility of A. tumefaciens GV2260 was assessed using peptone-agar (1% peptone, 0.25% agar, and 0.5% NaCl) containing t-CNMA or its derivatives at 25–200 μg/ml. The bacterial inoculum (1 μl) from overnight grown culture was placed at the center of peptone-agar plates and allowed to stand for 72 h at 30°C (Ahmed et al., 2022). Swim diameters were recorded at 48 and 72 h to check cell migration through agar. Data from three replicates were averaged.
Agrobacterium tumefaciens was exposed to t-CNMA or its derivatives at 25, 50, 75, 100, 150, 200, or 400 μg/ml in 1 ml LB at a culture: medium ratio of 1:100 in Eppendorf tubes and incubated at 250 rpm for 24 h at 30°C. Microfuge tubes were then centrifuged at 10,000 rpm for 10 min, and cell pellets were mixed with PBS, washed three times, and resuspended in PBS (1 ml). Optical density (OD) values of suspensions were read at 600 nm and designated A0. The method described earlier for bacterial adhesion to hydrocarbons (BATH) was followed (Rosenberg, 1984) with modifications. Hexadecane was then added to cell suspensions and vigorously vortexed for 1 min and then left for 30 min at room temperature to allow phase separation. Similarly, a blank (1 ml PBS only) was also processed. Aqueous phase OD (600 nm) values were designated Ai. Percent hydrophobicity were calculated using the formula:
Agrobacterium tumefaciens culture mixed with LB at 1:100 was exposed to t-CNMA compounds at 25–400 μg/ml in LB at 250 rpm for 24 h at 30°C. Samples were then spun at 10,000 rpm for 10 min, and supernatants were collected. Supernatants (100 μl) were mixed with an equal volume of azocasein and incubated for 30 min at 37°C when 600 μl of tricarboxylic acid (10%) was added to stop proteolysis. These mixtures were then kept for 30 min at −20°C. After centrifugation at 10,000 rpm for 10 min, 700 μl of the resulting supernatants was added to 700 μl of NaOH, and absorbances were recorded at 440 nm (Sethupathy et al., 2020).
Agrobacterium tumefaciens was grown with or without t-CNMA derivatives at 25, 50, 75, 100, 150, 200, or 400 μg/ml in LB in 1.5 ml microfuge tubes at 250 rpm for 24 h at 30°C. Tubes were centrifuged at 10,000 rpm for 10 min. Supernatants were added with chilled ethanol at a ratio of 1:3 and left undisturbed at 4°C for overnight. EPS precipitates were collected by centrifugation (10,000 rpm for 5 min.) and solubilized in 200 μl of water. A phenol/sulfuric acid mixture (prepared at a ratio of 1:5) was then added to 200 μl of EPS samples, incubated for 30 min. at room temperature, and left at room temperature for 20 min. Absorbances were measured at 490 nm (Ali et al., 2016).
Agrobacterium tumefaciens was grown in LB diluted at 1:100 for 24 h then treated with t-CNMA or its derivatives at different concentrations. These suspensions (300 μl) were then added to the wells of a 96-well plate. Culture growths were monitored every 2 h at 620 nm for 24 h (Ahmed et al., 2019b). The results of two independent cultures in six wells per concentration were averaged and plotted as a function of incubation time and concentration.
Agrobacterium tumefaciens grown for 24 h in LB was diluted at 1:100 with 25–400 μg/ml of t-CNMA, 4-nitro CNMA, 4-chloro CNMA, or 4-fluoro CNMA and incubated at 250 rpm for 24 h at 30°C. Aliquots (100 μl) of appropriate dilutions were then plated on LB agar and incubated for 48 h at 30°C. Colonies were counted, and CFU/ml values were calculated and converted to a log scale:
Survival percentages of A. tumefaciens were also calculated with respect to non-treated controls.
Seeds of Raphanus sativus were germinated on 0.86 g/L MS medium supplemented with 0.7% agar, and after germination, seedlings were grown for 5 days. Seedling roots (n = 5 per test concentration) were placed in 6-well plates in an aseptic environment. Inoculums of A. tumefaciens prepared in 4 ml LB broth at a 1:50 ratio and treated with 200 μg/ml of t-CNMA, nitro CNMA, 4-chloro CNMA, or 4-fluoro CNMA. Biofilm development was initiated at 30°C, and experiments were terminated after 48 h of incubation. Growth media containing planktonic cells and loosely attached biofilms were removed by rinsing the roots with sterile PBS. Root samples were then fixed in 2.5% glutaraldehyde and 2% paraformaldehyde for 30 min at room temperature and then overnight at 4°C. Fixatives were removed, and samples were rinsed with PBS, dehydrated using a graded ethanol series (30, 50, 70, 90, and 100%) for 10 min, and kept in isoamyl acetate. Root samples were dried in a critical point dryer (CPD), sputter-coated with gold or platinum, subjected to SEM (S-4200 Hitachi FE-SEM) at 15 kV, and photographed at different magnifications.
To assess the expressional changes induced by t-CNMA and 4-nitro CNMA, qRT-PCR was used to analyze the expressions of motility (flgE and motA), biofilm (celA, cheA, and phoB), virulence (virE2, chvE, virE0, and virG), stress related response (clpB, dnaK, gsp, marR, soxR, and hspAT2), and efflux pump (emrA, norM, ifeA, and ifeR) genes. A. tumefaciens culture in 25 ml LB with an OD600 nm of 1.0 was incubated with 100 μg/ml of t-CNMA or 4-nitro CNMA at 250 rpm for 8 h at 30°C. An RNase inhibitor (700 μl; RNAlater, Ambion, TX, USA) was added and gently agitated on ice. Centrifugation of untreated and treated cultures was performed at 13,000 rpm for 10 min at 4°C. The RNA was extracted using the Qiagen RNeasy mini kit (Valencia, CA, USA) and concentrations were determined using a nanodrop spectrophotometer (model: Cytiva NanoVue Plus Spectrophotometer, Fisher Scientific, England, UK). Primer sequences of tested genes are provided in Supplementary Table 1. qRT-PCR was conducted using SYBR green master mix and an ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using two independent cultures (Lee et al., 2015).
The impact of t-CNMA and its derivatives on white radish (R. sativus) seed germination was examined (Ahmed et al., 2019a). In brief, R. sativus seeds were soaked in distilled water for 6 h, thoroughly rinsed with distilled water and then ethanol (95%), and surface sterilized using sodium hypochlorite (3% for 10 min). t-CNMA its derivatives at 25–400 μg/ml were added to soft agar (0.7% agar) containing 0.86 g/L Murashige and Skoog (MS) medium. After washing with autoclaved distilled water, 10 seeds/plate/test were placed and incubated at 25°C for 4 days. The seeds showing evidence of germination were counted.
All experiments were performed with two independent bacterial cultures. Data are expressed as means ± standard deviations (SD), and significances of differences were determined using the two-tailed t-test. Statistical significance was accepted for P-values ≤ 0.05 unless otherwise stated. Graphs were prepared using Sigma Plot Ver. 14.0.
The effects of t-CNMA and 10 of its derivatives on A. tumefaciens were investigated. As shown in Table 1, MICs were variable. We selected three derivatives, viz. 4-nitro CNMA, 4-fluoro CNMA, and 4-chloro CNMA based on their MICs, which were 100, 200, and 200 μg/ml, respectively, and their biofilm inhibitory effects at 100 or 200 μg/ml (Table 1). t-CNMA (MIC 400 μg/ml) was used as the control.
Biofilm formation of A. tumefaciens in 96-well plates (Figure 1) was inhibited most by 4-chloro CNMA and 4-nitro CNMA by 94 and 100%, respectively, at 200 μg/ml (Table 1). 4-Fluoro CNMA did not inhibit biofilm at 200 μg/ml but reduced biofilm formation by >90% at 400 μg/ml (Figure 1D) versus the non-treated control. t-CNMA reduced biofilm at ≥150 μg/ml, whereas 4-nitro CNMA reduced it significantly (P ≤ 0.05) at ≥75 μg/ml (Figure 1B). 4-Chloro CNMA inhibited biofilm formation at 200 μg/ml (P ≤ 0.05) (Figure 1C).
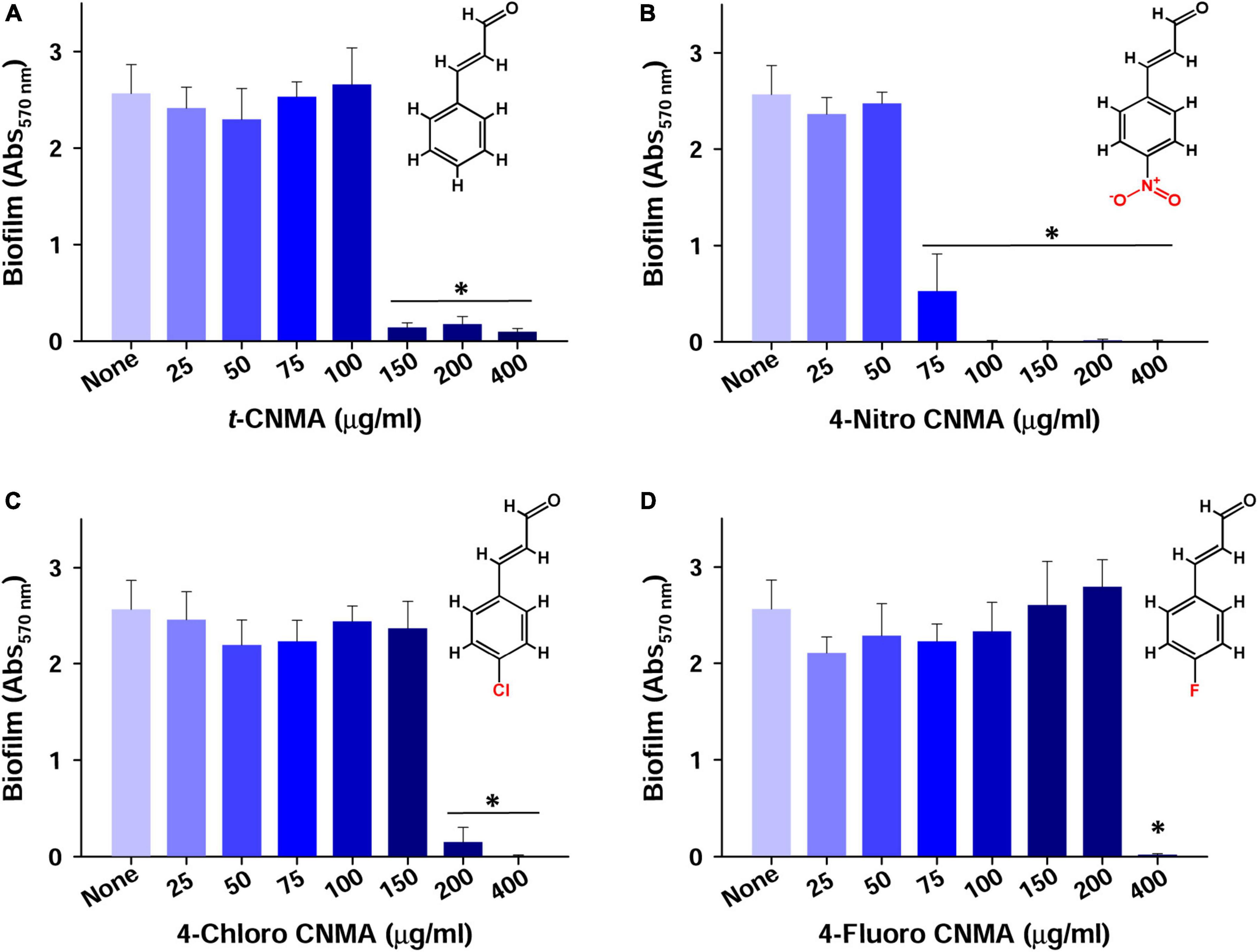
Figure 1. Inhibition of A. tumefaciens biofilm formation by t-CNMA (A), 4-nitro CNMA (B), 4-chloro CNMA (C), and 4-fluoro CNMA (D) after 48 h in polystyrene microtiter plates containing LB. “*” denotes a significant difference by the two-tailed t-test between non-treated and treated cultures.
Biofilm formation by A. tumefaciens was also investigated on flat polystyrene and nylon membranes. Almost all tested compounds dose-dependently reduced biofilm volumes (Figure 2A), and microscopic images showed they reduced biofilm thicknesses (Figure 2B). t-CNMA had the least inhibitory effect at 200 μg/ml, whereas only a few traces of biofilms remained after treatment with 4-nitro CNMA at 150 or 200 μg/ml. 4-Chloro CNMA slightly reduced biofilm formation at 150 μg/ml and completely inhibited it at 200 μg/ml. However, 4-fluoro CNMA did not affect biofilm formation at 200 μg/ml but drastically reduced it at 400 μg/ml. Similarly, 3-D mesh-filled spectrum-LUT plots of biofilms were observed at a scale range of 0–240, after exposing biofilms to different concentrations of t-CNMA, 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA showed dramatic shifts in color to blue at 150–200 μg/ml (Figure 3A). 4-Chloro CNMA (150 μg/ml) and 4-fluoro CNMA (200 μg/ml) had lesser effects on biofilms.
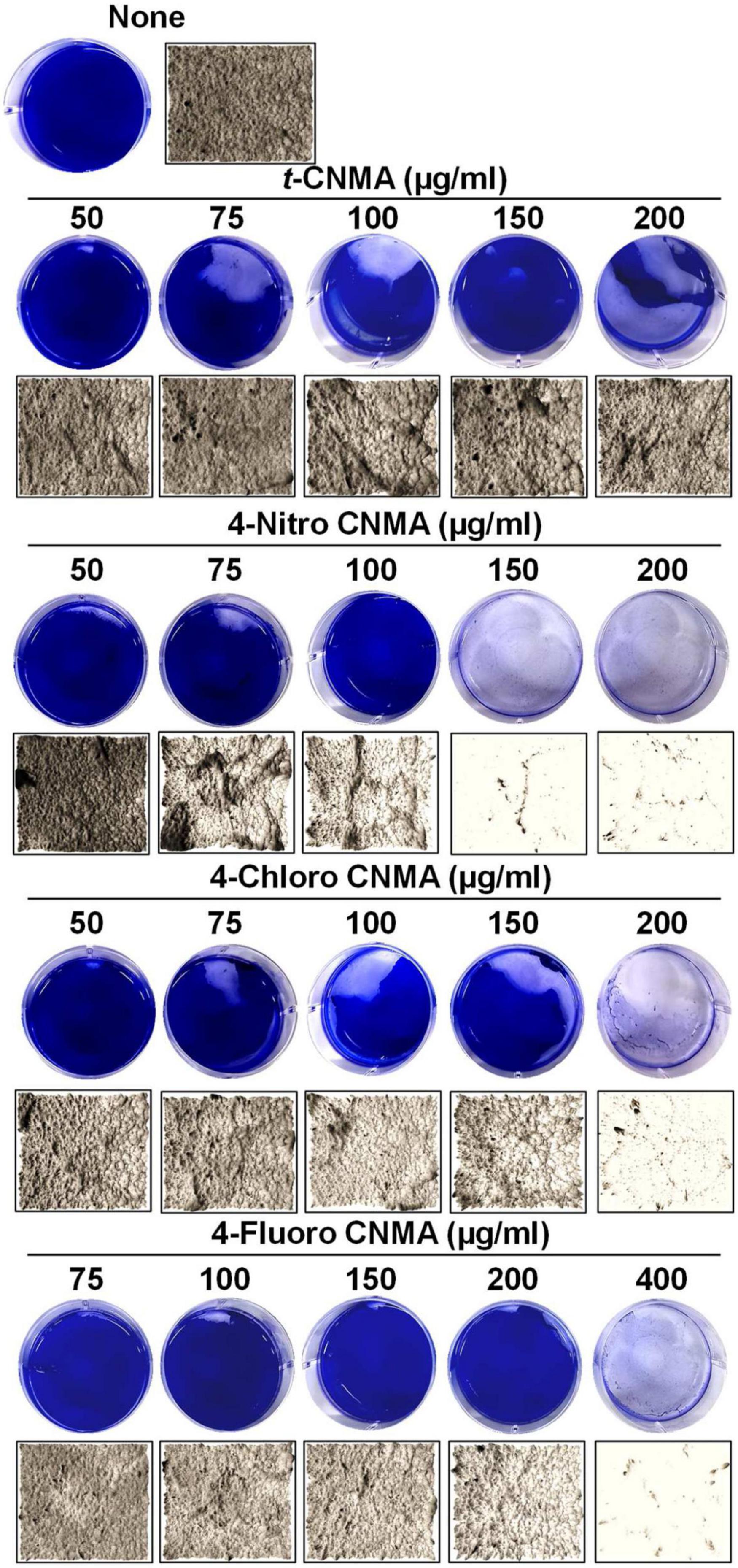
Figure 2. Inhibition of A. tumefaciens biofilm formation on flat polystyrene in 6-well plates as determined by crystal violet staining (A) and optical microscopy at 20× magnification (B) by t-CNMA, 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA in the concentration range (50–400 μg/ml).
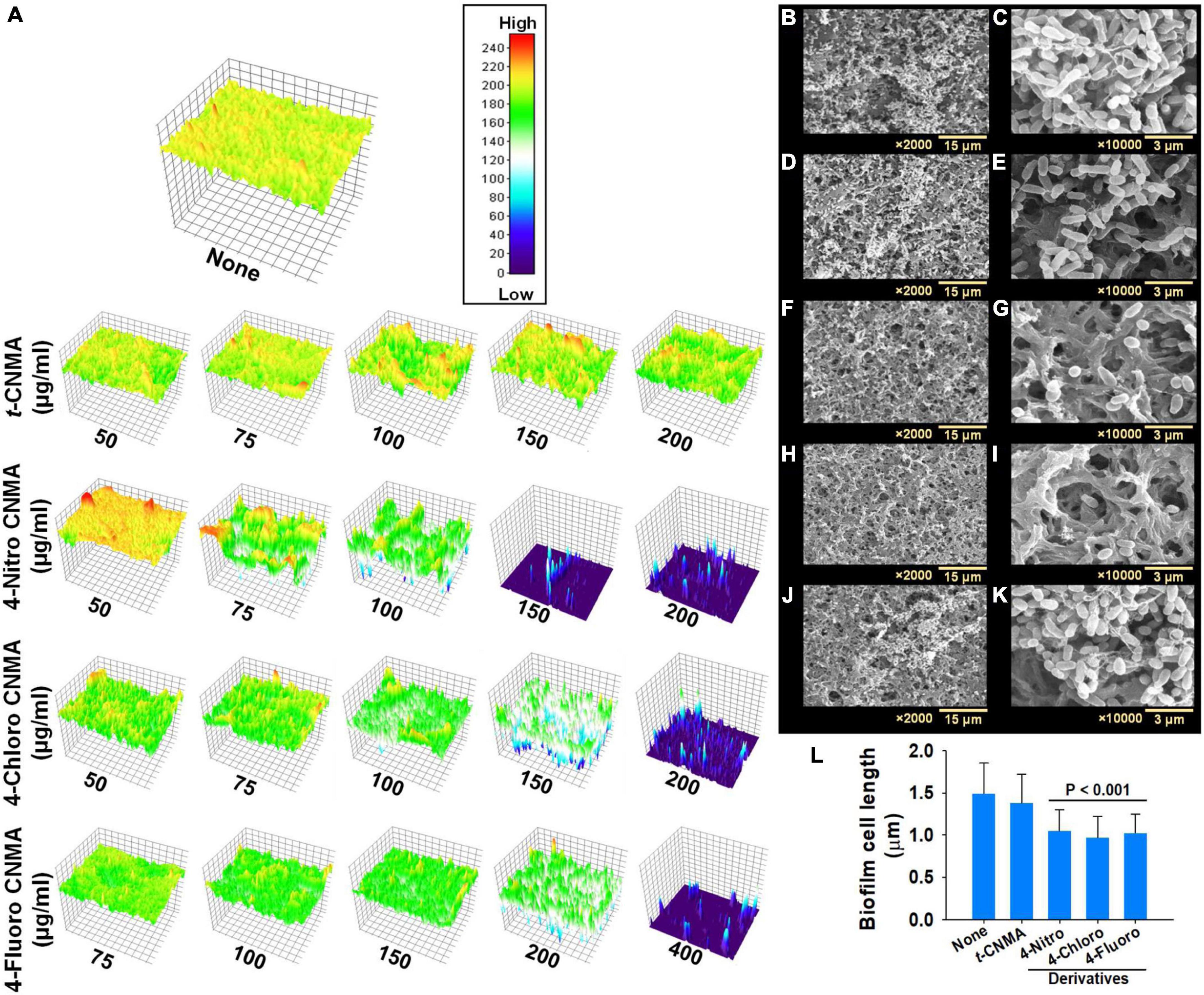
Figure 3. Visual assessment of A. tumefaciens biofilm thicknesses formed on polystyrene after exposure to increasing concentrations of t-CNMA, 4-nitro CNMA, 4-chloro CNMA, or 4-fluoro CNMA. The colored 3-D images show biofilm thicknesses (A) on a low to high (0–200) scale, where red indicates maximum biofilm formation and dark blue no biofilm formation. Biofilms of non-treated (B,C) A. tumefaciens on nylon and its inhibition by t-CNMA (D,E), 4-nitro CNMA (F,G), 4-chloro CNMA (H,I), and 4-fluoro CNMA (J,K) after incubation for 48 h. Micrographs taken at 2,000× and 10,000× revealed complete inhibition by 4-nitro and 4-chloro CNMA and reductions in cell lengths (L), although t-CNMA (E) and 4-fluoro CNMA (K) showed biofilm with the presence of EPS threads.
Scanning electron microscopy (SEM) of biofilms on nylon membranes showed variable reductions in biofilm formation by t-CNMA (Figures 4D,E), 4-nitro CNMA (Figures 4F,G), 4-chloro CNMA (Figures 4H,I), and 4-fluoro CNMA (Figures 4J,K) versus non-treated controls (Figures 4B,C). The specimens treated with 4-nitro CNMA or 4-chloro CNMA were most affected, and fewer cells were attached to nylon surfaces (Figures 4F–I). In addition, shortening of A. tumefaciens cells treated with 4-nitro, 4-chloro, and 4-fluoro derivatives was observed at ×10,000 (Figure 4K). Specifically, the sizes of non-treated A. tumefaciens biofilm cells were 1.5 ± 0.4 μm which decreased slightly by 7.5% (1.4 ± 0.3 μm) after t-CNMA treatment. However, 4-nitro, 4-chloro, and 4-fluoro significantly (P < 0.001) decreased the length by 30% (1 ± 0.25 μm), 35% (0.96 ± 0.25 μm), and 31.3% (1 ± 0.22 μm), respectively (Figure 4L). SEM observations indicated t-CNMA and its derivatives alter the morphology and architecture of A. tumefaciens biofilms.
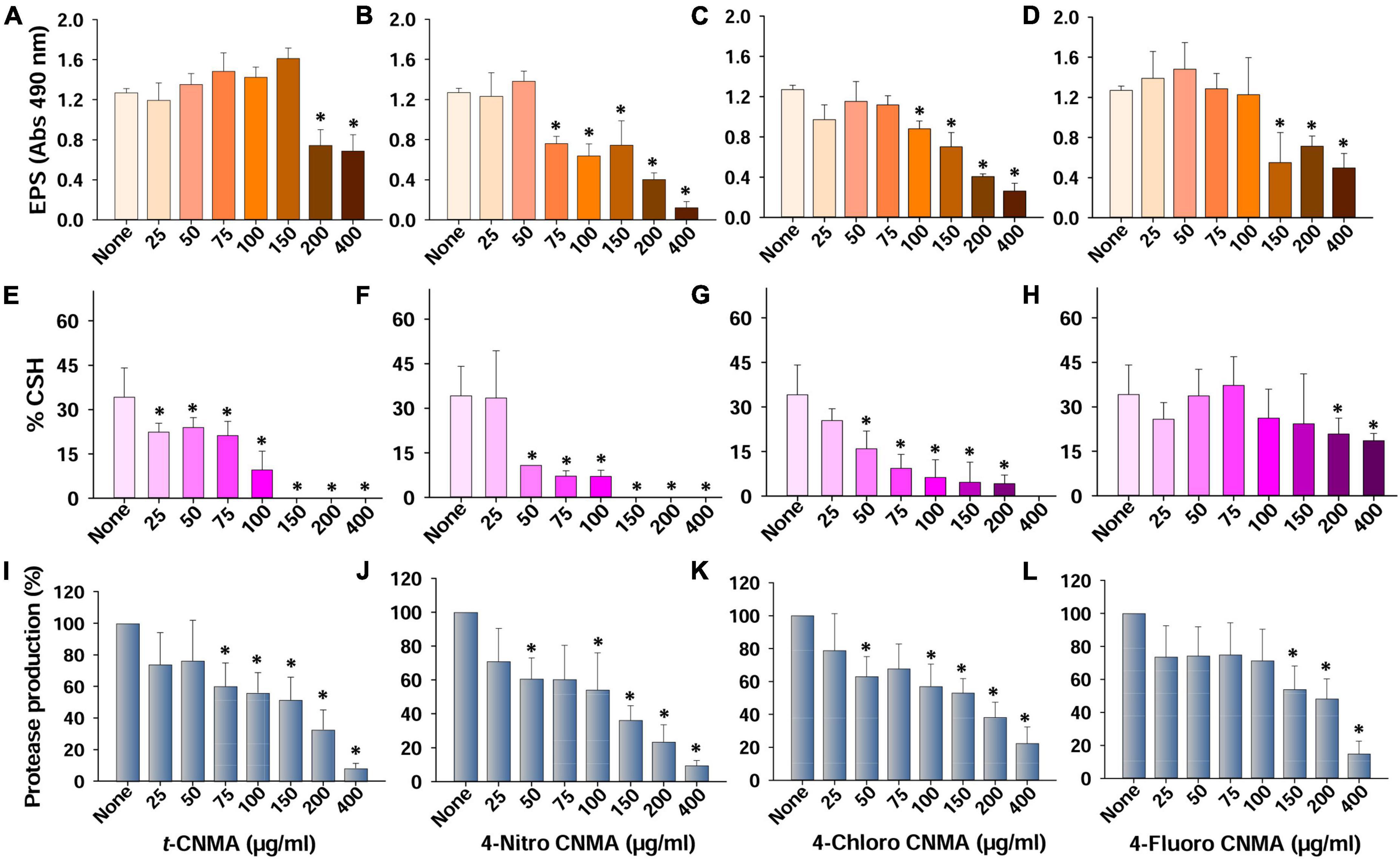
Figure 4. Production of virulence factors and their inhibition by t-CNMA and its 4-nitro, 4-chloro, and 4-fluoro CNMA derivatives. Effects on EPS production (A–D), cell surface hydrophobicity (E–H), and extracellular protease production (I–L). “*” denotes a significant difference by the two-tailed t-test (P ≤ 0.05).
The QS regulates virulence factors considered responsible for A. tumefaciens biofilm formation (Faure and Lang, 2014). Of these, cell surface hydrophobicity and extracellular polymeric substances are critical for bacterial adhesion and successful biofilm formation. Non-treated cells of A. tumefaciens produced significant amounts of EPS; however, treatments with t-CNMA or its derivatives decreased EPS production at relatively high-test concentrations (Figures 4A–D). t-CNMA did not reduce the EPS secretion below 150 μg/ml but decreased it significantly by >40% at concentrations ≥200 μg/ml (Figure 4A). 4-Nitro CNMA at 75 μg/ml caused a similar reduction (40%) and at 200 μg/ml decreased EPS production by 69% (Figure 4B). 4-Chloro CNMA caused a significant reduction at 100 μg/ml followed by a concentration-dependent decrease till 400 μg/ml (Figure 4C). The reduction in EPS production by 4-fluoro CNMA (Figure 4D) was similar to that of t-CNMA. Cell surface hydrophobicity (% CSH) is essential for bacterial attachment to surfaces (Tribedi and Sil, 2014) and was also found to be decreased by t-CNMA and its three derivatives in a concentration-dependent manner (Figures 4E–H). When we compared the inhibitory effects on % CSH at a concentration of 100 μg/ml, 4-nitro CNMA (Figure 4F) and 4-chloro CNMA (Figure 4G) had the greatest effects and reduced CSH by 79% (P ≤ 0.05) and 82% (P ≤ 0.05), respectively, versus non-treated controls. At 150–400 μg/ml, %CSH was zero for t-CNMA (Figure 4E) and 4-nitro CNMA (Figure 4F). As was observed in the EPS assay, 4-fluoro CNMA had least reduction (45% at 200 μg/ml) (Figure 4H).
Production of exo-proteases by A. tumefaciens was dose-dependently reduced by t-CNMA and its derivatives (Figures 4I–L). At 100 μg/ml, t-CNMA, 4-nitro CNMA, and 4-chloro CNMA significantly reduced the protease production. 4-Nitro CNMA most effectively inhibited protease production. In regards of motility, A. tumefaciens exhibited swimming motility on agar plates prepared with 1% peptone, 0.25% agar, and 0.5% NaCl, and this increased with time (48–72 h). The bacterium reached a swimming diameter of 5.7 cm after 72 h incubation in non-treated agar (Figure 5). The addition of t-CNMA or its derivatives to agar at 25–200 μg/ml decreased swimming diameters, and no mobility was observed after treatment with 4-nitro CNMA or 4-chloro CNMA at 150 or 100 μg/ml, respectively (Figure 5). Interestingly, 4-fluoro CNMA inhibited swimming motility at 100 μg/ml (P ≤ 0.05). Furthermore, t-CNMA supplemented agar reduced promoted mobility versus non-treated agar, although even at 200 μg/ml, A. tumefaciens exhibited limited mobility (3.1 cm diameter).
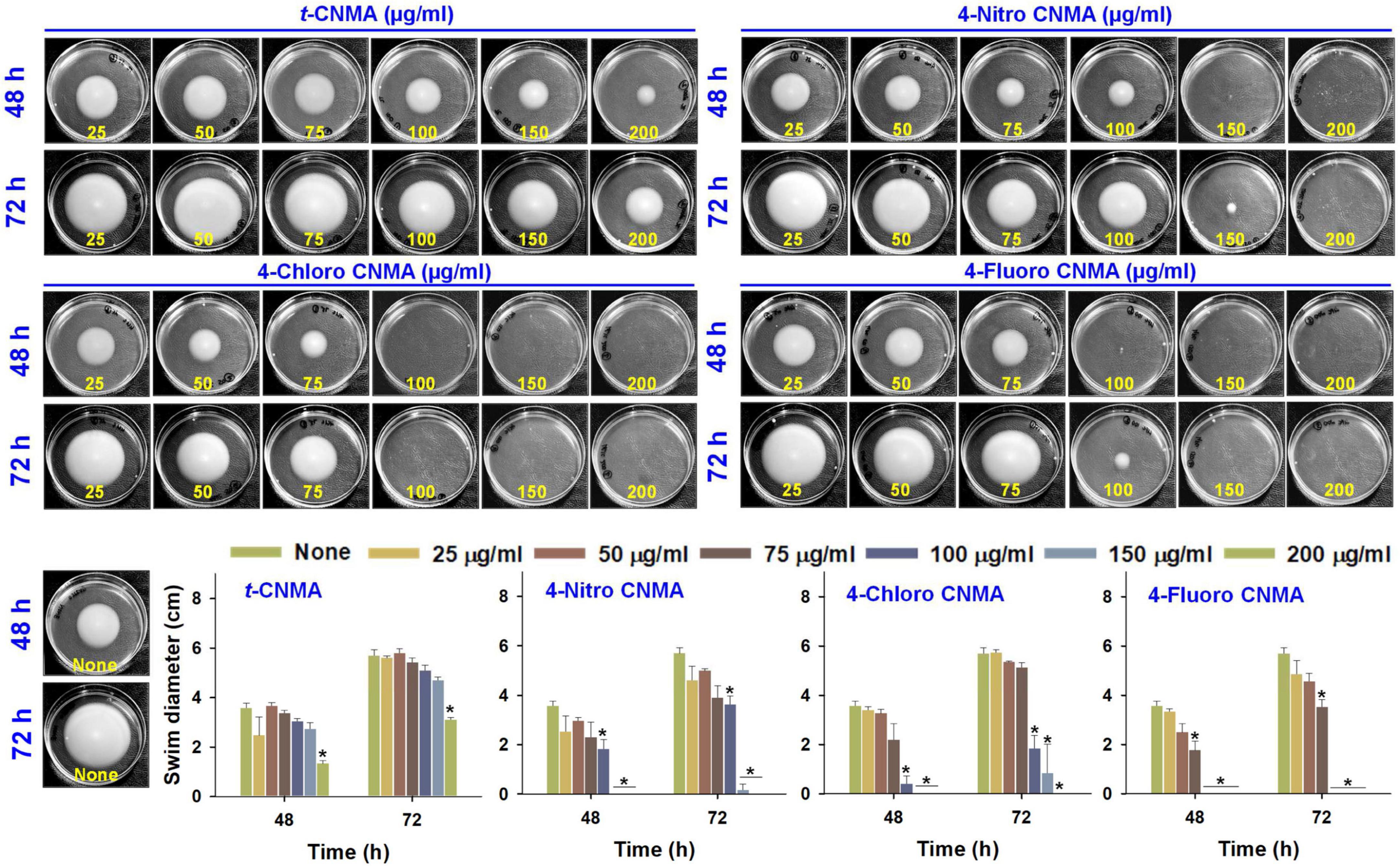
Figure 5. Swimming motility of A. tumefaciens on motility agar (1% peptone, 0.25% agarose, and 0.5% NaCl) and its inhibition by t-CNMA, 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA (25–200 μg/ml) after exposure for 48 and 72 h. “*” denotes a significant difference in swim diameter by the two-tailed t-test (P ≤ 0.05).
The planktonic cell growth of A. tumefaciens was assessed using logarithmic values of CFUs (Figures 6A–D), percent cell survival (Figures 6E–H), and time-dependent growth curves (Figures 6I–L). Total log CFU/ml counts of A. tumefaciens grown in the presence of t-CNMA (Figure 6A), 4-nitro CNMA (Figure 6B), 4-chloro CNMA (Figure 6C), and 4-fluoro CNMA (Figure 6D) were reduced by only 4-nitro CNMA (>6-log reduction) and 4-chloro CNMA (>6.5-log reduction) at 400 μg/ml. 4-Fluoro CNMA also reduced cell survival, but less than the other derivatives; a fraction of cells survived even after treatment with 4-fluoro CNMA at 400 μg/ml. Concentration (25–400 μg/ml) and time (0–24 h) dependent analysis of A. tumefaciens planktonic growth showed t-CNMA (Figure 6I), 4-nitro CNMA (Figure 6J), 4-chloro CNMA (Figure 6K), and 4-fluoro CNMA (Figure 6L) induced concentration-dependent decreases. 4-Nitro CNMA and 4-chloro CNMA inhibited cell growth at 150 and 200 μg/ml, respectively (Figures 6J,K). The possible reason for this cell killing could be the enhanced direct contact of cells with tested compound at 250 rpm shaking.
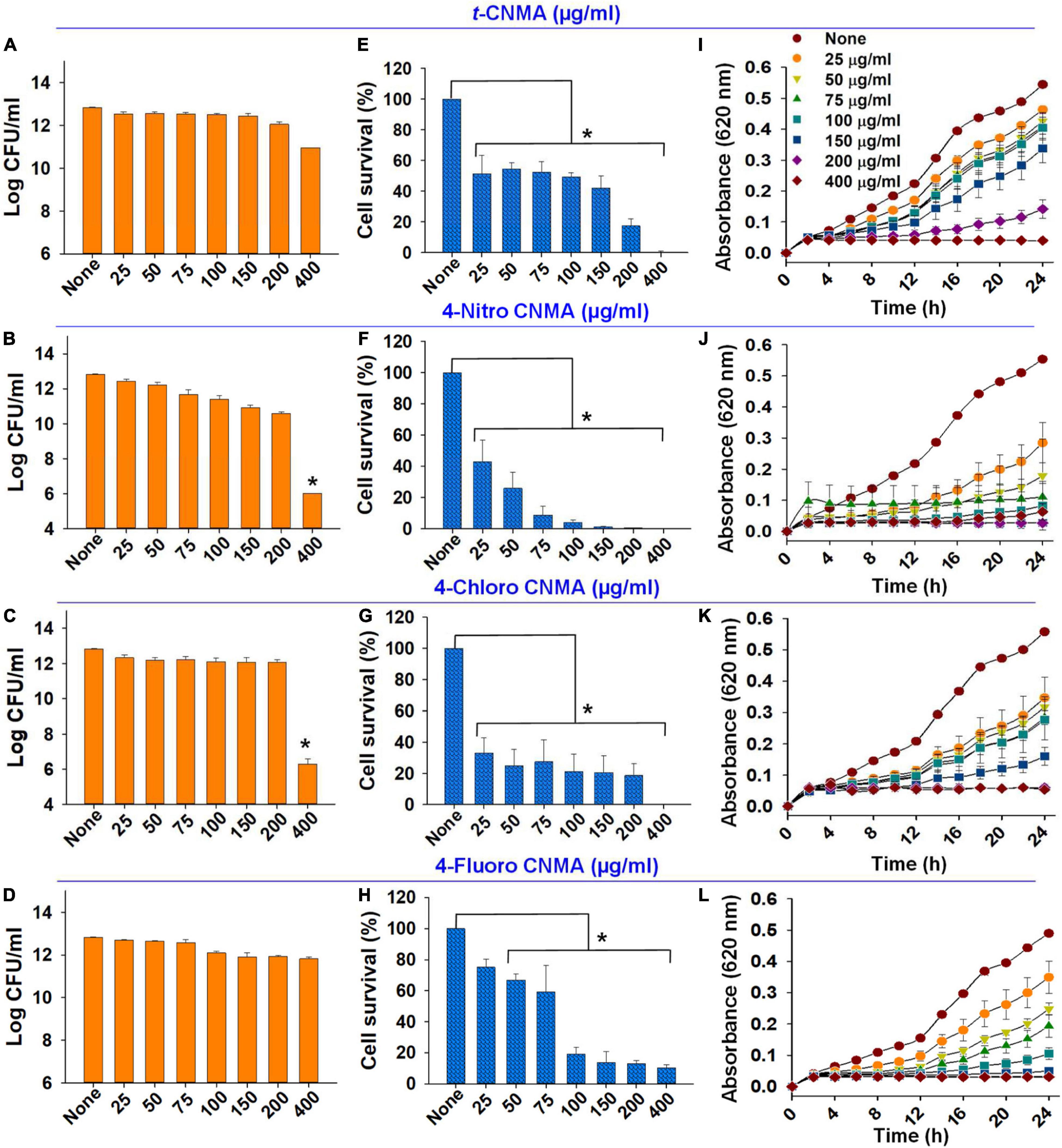
Figure 6. Effects of cinnamaldehydes on A. tumefaciens planktonic growth. Log reduction in CFU counts by t-CNMA (A), 4-nitro CNMA (B), 4-chloro CNMA (C), and 4-fluoro CNMA (D); percent cell survival after treatment with t-CNMA (E), 4-nitro CNMA (F), 4-chloro CNMA (G), or 4-fluoro CNMA (H) for 24 h. Time and concentration-dependent growth inhibition were recorded every 2 h (I–L). “*” denotes a significant difference in planktonic growth by the two-tailed t-test (P ≤ 0.05).
Agrobacterium tumefaciens formed biofilms on R. sativus roots after incubation for 48 h under non-treated and optimized growth conditions (Figures 7A–C). The biofilms produced were dense and showed multiple aggregates of cells embedded in an EPS-like substance (Figure 7C). While observing root surfaces, biofilms were observed in multiple regions after t-CNMA treatment (Figures 7D–F). However, no biofilms were observed on roots after exposure to 4-nitro CNMA (Figures 7G–I), 4-chloro CNMA (Figures 7J–L), or 4-fluoro CNMA (Figures 7M–O), though a few cells were dispersed at some locations on root surfaces (Figures 7I,O). Furthermore, root surfaces in different zones, e.g., root tips, hairs, and meristematic and root elongation zones were undamaged after exposure to 200 μg/ml of the test compounds. SEM findings after 4-fluoro CNMA treatment (Figures 7M–O) differed from nylon membrane results (Figures 3J,K) since 4-fluoro CNMA did not eradicate biofilms on nylon membranes. On the other hand, results for t-CNMA, 4-nitro CNMA, and 4-chloro CNMA treatments of biofilms on nylon (Figure 3) and R. sativus root surfaces (Figure 7) were correlated.
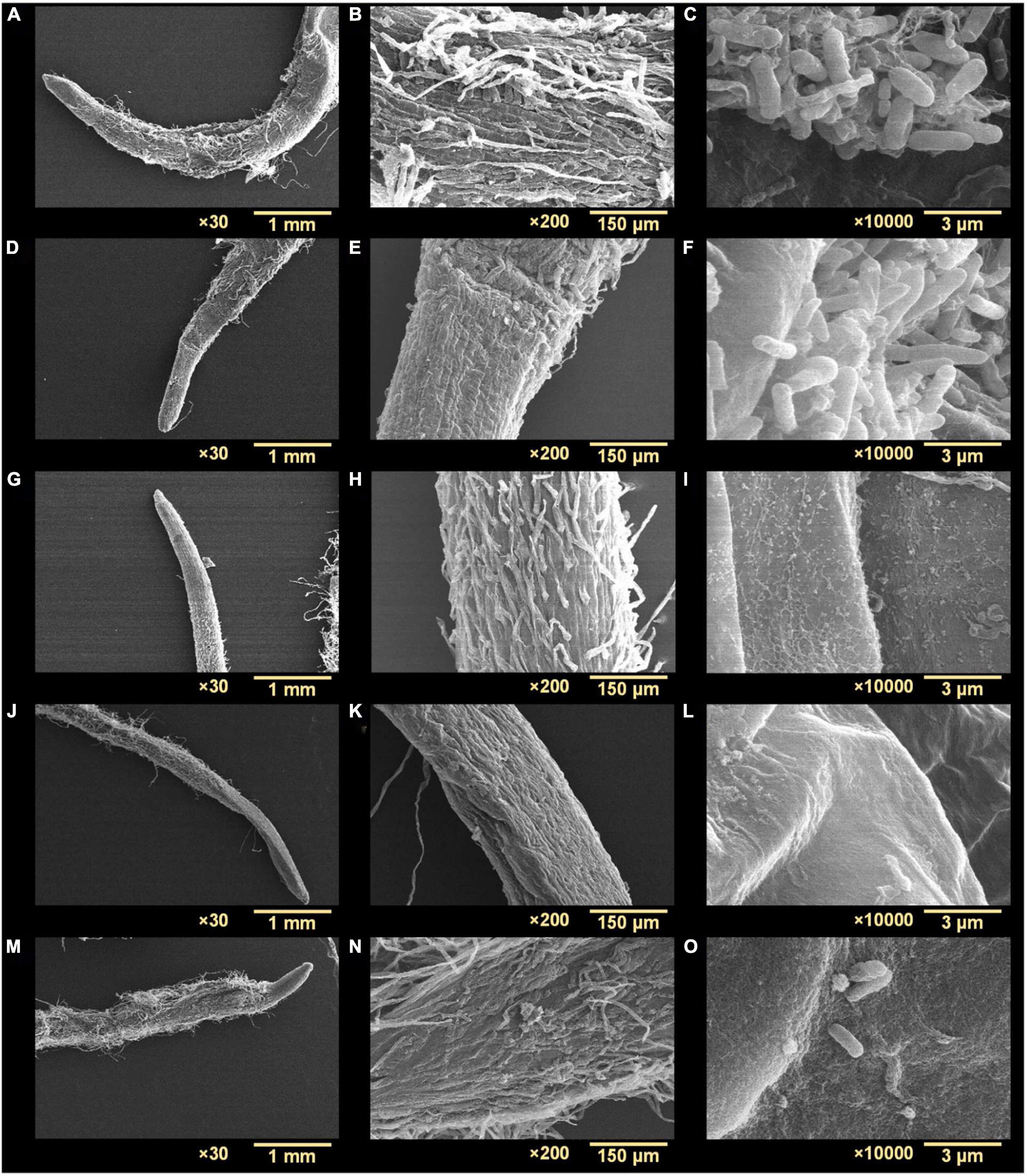
Figure 7. Scanning electron microscopy micrographs of A. tumefaciens biofilms on the surface of R. sativus at three different magnifications (30×, 200×, and 10,000×): non-treated (A–C), treated with t-CNMA (D–F), 4-nitro CNMA (G–I), 4-chloro CNMA (J–L), and 4-fluoro CNMA (M–O).
Biofilm, virulence, stress, motility, and efflux pump regulation genes of A. tumefaciens (OD600 nm = 1.0) assessed after 8 h contact with test compounds in LB at 30°C were variably affected by t-CNMA and 4-nitro CNMA versus non-treated controls while 4-nitro CNMA more significantly affected the gene expression than t-CNMA (Figure 8). Genes subjected to qRT-PCR were selected based on their direct or indirect involvements with these functions. For example, the biofilm formation genes cheA, celA, and phoB encode for the two-component sensor kinase of the Che operon that regulates chemotaxis (Merritt et al., 2007), cellulose synthase required for cellulose production (Matthysse et al., 2005), and production of a regulatory protein for the two-component (PhoR-PhoB) system (Tomlinson et al., 2010), respectively. 4-Nitro CNMA significantly (P ≤ 0.05) downregulated celA, cheA, and phoB by 3.3-, 3.9-, and 3-fold, respectively, versus non-treated controls (Figure 8). Similarly, two flagellar motility genes flgE and motA, which encode for a flagellar hook protein and a constituent of the flagellar motor of A. tumefaciens, were downregulated by 11- and 2.5-fold, respectively, by 4-nitro CNMA (Merritt et al., 2007). Interestingly, t-CNMA only slightly reduced the expression of these genes (Figure 8). Among the virulence genes, virE2 encodes for virulence protein (virE2) that facilitates the import of T-DNA-protein complex in the host nucleus (Li et al., 2020). chvE encodes for a periplasmic-binding protein, which after interacting with the VirA/VirG regulatory system induces the expressions of vir genes (Hu X. et al., 2013). virE0 encodes for a regulator protein that may be directly involved in Agrobacterium-plant interactions (Yuan et al., 2008). virG encodes for a two-component response regulator protein (Yuan et al., 2008) and was significantly downregulated (Figure 8) by t-CNMA only. The expressions of all other virulence genes were either unchanged or non-significantly downregulated. Genes involved in multiple stress responses, namely, clpB, dnaK, gsp, marR, and hspAT2, reported in other studies (Rosen et al., 2001; Tsai et al., 2012; Rittiroongrad et al., 2016) were also tested. Results revealed that dnaK and clpB were downregulated by 2.7- and 28-fold, respectively, and soxR was upregulated (1.72) by 4-nitro CNMA (the expressions of other stress-related genes were slightly changed or unaffected). t-CNMA upregulated the gsp gene, which is associated with general stress, by 2.5-fold, and 4-nitro CNMA reduced the expression of a heat shock protein (hspAT2). Also, some efflux pump genes, namely, emrA, norM, ifeA, and ifeR, were included (Palumbo et al., 1998; Nuonming et al., 2018; Khemthong et al., 2019), but the only effect observed was that 4-nitro CNMA upregulated emrA by 3.7-fold (Figure 8).
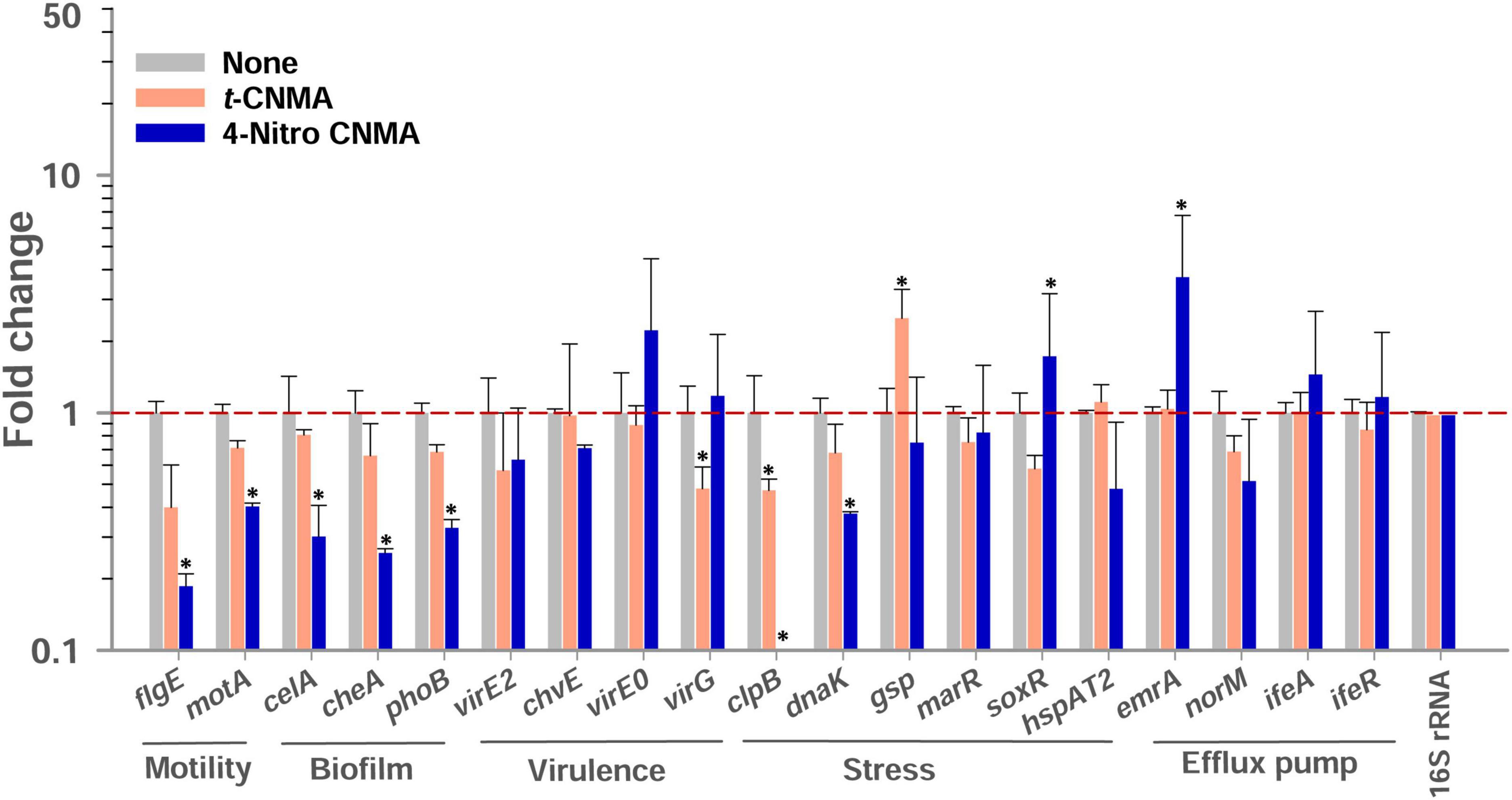
Figure 8. Gene expressional changes in A. tumefaciens induced by 100 μg/ml t-CNMA or 4-nitro CNMA after treatment in LB broth at 250 rpm for 8 h at 30°C. Bars are averages of four reactions performed using two independent cultures. 16S rRNA was used as the housekeeping gene. “*” represents a significant difference (P ≤ 0.05) between non-treated and treated cells. Fold changes in gene expression were calculated using the 2–ΔΔCt method.
The effects of t-CNMA, 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA were also investigated on R. sativus seed germination. The impacts of tested compounds at 25–400 μg/ml on percent seed germination were variable (Supplementary Figure 1). Seed germination was non-significantly reduced by t-CNMA (Supplementary Figure 1A), 4-nitro CNMA (Supplementary Figure 1B), and 4-fluoro CNMA (Supplementary Figure 1D) at concentrations up to 200 μg/ml. However, 4-chloro CNMA caused 70% inhibition at 200 μg/ml (Supplementary Figure 1C). At 400 μg/ml t-CNMA, 4-chloro CNMA, and 4-fluoro CNMA prevented germination, but interestingly, 4-nitro CNMA at this concentration only reduced germination by 41%.
We report the biofilm inhibiting characteristics of t-CNMA and ten derivatives, which were selected because of their dissimilar functional moieties on the aromatic ring or side chain of t-CNMA. A few studies on CNMA derivatives have reported the antityrosinase effects of α-substituted derivatives such as α-methylcinnamaldehyde, α-chlorocinnamaldehyde, and α-bromocinnamaldehyde (Cui et al., 2015). Similarly, fungal growth was inhibited by 2-bromo and 2-chlorocinnamaldehyde (Badawy and Rabea, 2013). Among the ten derivatives investigated in the present study, 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA were subjected to further testing because they exhibited potent antibiofilm effects and low MICs. Badawy and Rabea (2013) reported that chitosan-based derivatives had antifungal activity against seven fungal species. The same authors investigated the effects of chitosan-based derivatives, such as N-(α-methylcinnamyl) chitosan and N-(o-methoxycinnamyl) chitosan, on A. tumefaciens but reported very high MICs of 1,275 and 1,925 μg/ml, respectively. In the present study, the MICs of 4-nitro CNMA, 4-fluoro CNMA, and 4-chloro CNMA were 100, 200, and 200 μg/ml, respectively. Zhu et al. (2009) previously reported t-CNMA thiosemicarbazone had an A. tumefaciens MIC of 100 μg/ml with no report on antibiofilm potential and gene expressional changes.
Biofilms provide microorganisms on abiotic and biotic surfaces with well-structured protective sheaths impermeable to drugs and antibacterial agents (Sharma et al., 2019). Thus, microorganisms in biofilms are more virulent than planktonic cells, and novel solutions are required to address this challenge (Ying et al., 2019). A. tumefaciens adheres to surfaces using its molecular appendages (Thompson et al., 2018) and subsequently forms reversible or irreversible attachments (Heindl et al., 2014) with abiotic (Figures 2, 3) or biotic surfaces (Figure 7). After establishing contact with a surface, A. tumefaciens releases EPS to make this contact reversible and initiates microcolony formation (Heindl et al., 2014). In the present study, 4-nitro CNMA and 4-chloro CNMA significantly inhibited biofilm formation by A. tumefaciens on polystyrene and nylon (Figures 2, 3F–I) and plant root surfaces (Figures 7G–L), as determined by light microscopy and SEM. Similar reductions in bacterial aggregation and microcolony formation by P. fluorescens were observed by light microscopy after cinnamaldehyde exposure. In a previous study, SEM revealed a maximally disrupted biofilm architecture of P. fluorescens at 0.1 μl/ml t-CNMA (Li et al., 2018). We observed gaps and poor volumes of A. tumefaciens biofilms (Figure 3) after treatments with 4-nitro CNMA (150–200 μg/ml), 4-chloro CNMA (200 μg/ml), or 4-fluoro CNMA (400 μg/ml) but not after treatment with t-CNMA. Li et al. (2018) reported that t-CNMA induced fissures in P. fluorescens biofilms, and Kim et al. (2015) observed cinnamon bark oil and t-CNMA at 0.01% v/v reduced enterohemorrhagic E. coli (EHEC) fimbriae formation, which is required for biofilm maturation, and suggested that reduced EHEC fimbriae production by t-CNMA on nylon membranes was largely responsible for biofilm inhibition. Our observations of reductions in A. tumefaciens biofilm formation at ≥150 μg/ml by t-CNMA, 4-nitro CNMA, and 4-chloro CNMA (Figure 1) suggest that –NO2 functional group at the fourth position on the aromatic ring is more detrimental to biofilm formation than –Cl, and that –F is less effective than –NO2 or –Cl. When we compared the gene expressional changes induced by 4-nitro CNMA and t-CNMA, 4-nitro CNMA was found to have substantially more potent effects (Figure 8). Furthermore, our findings regarding the effects of t-CNMA concur with those of Budri et al. (2015) who found that at 106 μg/ml t-CNMA reduced MRSA biofilm formation on stainless steel and polystyrene by 45 and 70%, respectively. Similarly, Albano et al. (2019) found t-CNMA at 300 μg/ml reduced biofilm formation by Staphylococcus epidermidis by 89%.
Biofilm inhibition by t-CNMA might be related to disruption of the QS regulatory system, as has been reported for P. fluorescens (Li et al., 2018) and E. coli (Niu et al., 2006), and this inhibition may be due to the downregulations of curli genes (csgA and csgB) in EHEC (Kim et al., 2015) and biofilm-related adhesion genes (icaA and sarA) in Staphylococcus spp. (Jia et al., 2011). QS signaling controls EPS secretion, motility, protease production, and cell surface hydrophobicity (Li et al., 2014; Tan et al., 2014; Mizan et al., 2016; Pena et al., 2019), and in the present study, these virulence attributes of A. tumefaciens were remarkably and concentration-dependently inhibited by t-CNMA and its derivatives (Figures 4, 5). For t-CNMA, these effects have been suggested to be associated with the aldehyde group (Kot et al., 2015) as also shown for swimming motility of E. coli at 2.17 mM t-CNMA (Niu and Gilbert, 2004). Furthermore, it has been suggested that the presence of halogen (–Cl or –F) or nitro (–NO2) groups in t-CNMA increases its suppressive effects on bacterial virulence (Brackman et al., 2011; Nepali et al., 2018).
We also observed that 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA reduced the lengths of A. tumefaciens cells (Figures 3G,I,K) as compared with non-treated (Figure 3C) and t-CNMA (Figure 3E) treated cells. However, we did not observe the t-CNMA-induced morphological distortions of E. coli and S. aureus cells reported by Shen et al. (2015). Furthermore, exposure to t-CNMA and the three derivatives had markedly impacted cell viability and growth (Figure 6), which could be associated with t-CNMA-induced reductions in intracellular pH (Oussalah et al., 2006), its interactions with membrane proteins (Mousavi et al., 2016), or its effects on cell membrane conductivity (He et al., 2019) or membrane lipid profiles (Wendakoon and Sakaguchi, 1995). In addition, the effects of –NO2, –Cl, and –F containing derivatives may have been influenced by the electronegativities of these groups (Shaikh et al., 2016; Doyle et al., 2019). t-CNMA has also been suggested to act as an ATPase inhibitor and inhibit enzymes involved in cytokine interactions (Shreaz et al., 2016). Furthermore, differences between the cellular uptakes of derivatives and their post-cellular uptake transformations may have modulated their effects.
No biofilm formation was observed on the root surfaces of R. sativus seedlings roots grown in the presence of 4-nitro CNMA, 4-chloro CNMA, or 4-fluoro CNMA (Figures 7G–O). A difference in biofilm volume on root surface and nylon surface was observed in non-treated groups where it was higher on nylon membranes possibly due to more firm attachment of cells on cellulose fibrils (Merritt et al., 2007; Heindl et al., 2014). The inhibition of A. tumefaciens biofilm formation on plant roots by CNMA derivatives has not been previously reported. However, biofilms of Pseudomonas putida KT2440 were reported to be dose-dependently inhibited by t-CNMA (Niu and Gilbert, 2004). A. tumefaciens utilizes adhesive pili, rhicadhesin, and chromosome-encoded factors in addition to universal forces like electrostatic and hydrophobic interactions and Van der Waals forces to attach to plant surfaces (Wheatley and Poole, 2018) and form biofilms. Our qRT-PCR data showed 4-nitro CNMA induced significant changes in the expressions of genes associated with motility (flgE and motA), biofilm formation (celA, phoB, and cheA), stress response (clpB, dnaK, and soxR), and efflux pump (emrA) (Figure 8) compared to t-CNMA. A. tumefaciens downregulated by 4-nitro CNMA induced a series of events that ultimately result in inhibition of bacterial adherence on surfaces, EPS, motility, biofilm as depicted in Figure 9. For example, celA encodes for a regulatory protein of two-component sensor kinase required for chemotaxis, therefore its downregulation reduces the chemotactic response of A. tumefaciens towards rhizospheric chemicals and prevent adherence to plant tissues (Figure 9). Another biofilm gene cheA encodes for cellulose synthase and induces EPS production, therefore its inhibition brings down the EPS matrix of biofilm. The increased transcription of soxR (stress related gene), sensing stress due to 4-nitro CNMA presence induced biofilm formation, however, other genes regulating the biofilm formation directly or indirectly were downregulated. The stress-related clpB and dnaK (or Hsp70) were downregulated and this ATP-dependent protease production and correct folding of proteins were compromised respectively that is suggestive of the possible antibacterial mechanism of 4-nitro CNMA.
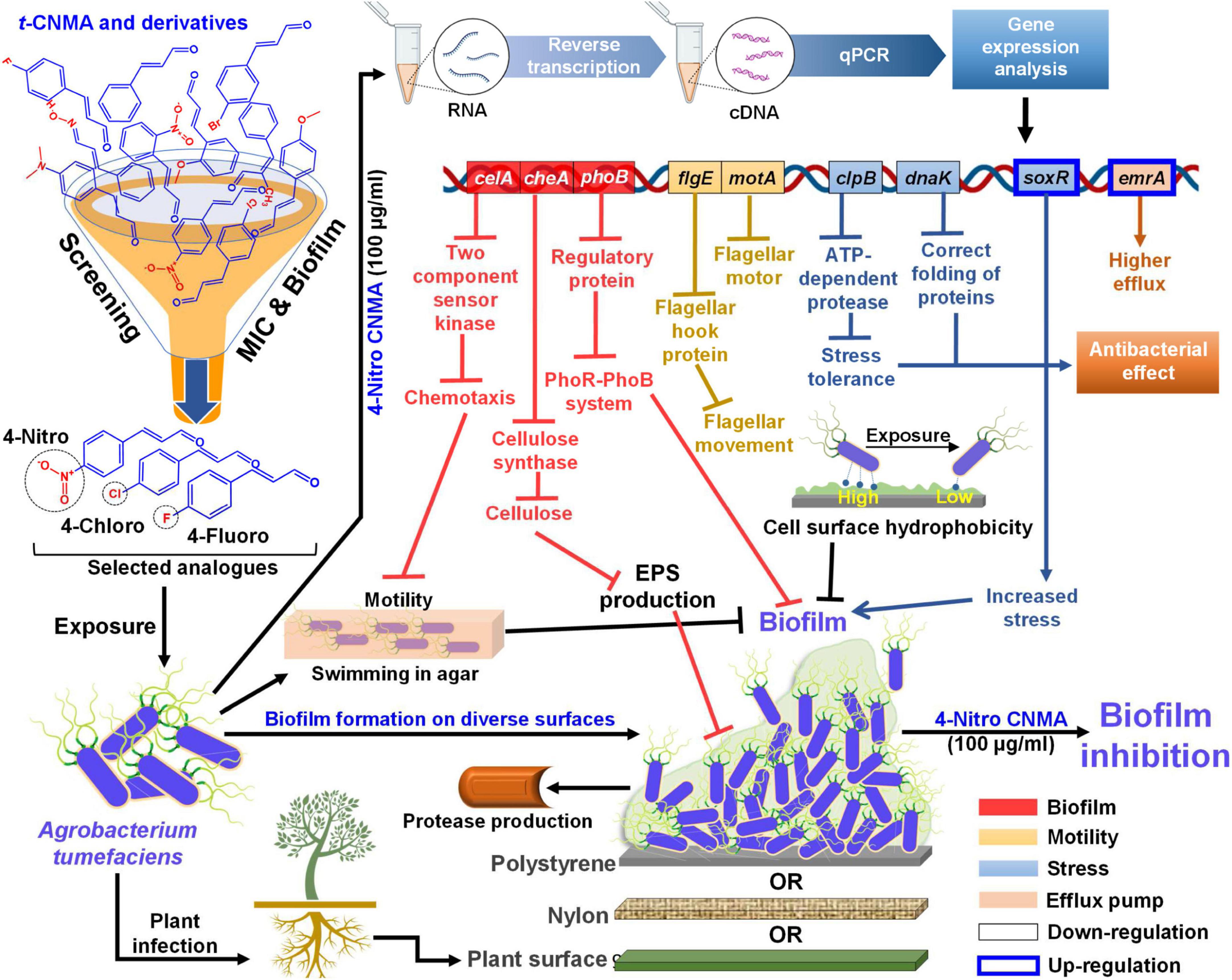
Figure 9. Mechanistic illustration of A. tumefaciens interaction with t-CNMA and its analogs specifically 4-nitro CNMA after screening based on MIC and biofilm inhibition percentages. A. tumefaciens was able to swim in agar by flagellar machinery, showed cell surface hydrophobicity, showed protease and EPS production, EPS and biofilm on three different surfaces nylon, polystyrene, and plant root (displayed beneath the biofilm). Selected derivatives inhibited these virulence factors and biofilm phenotype. Higher alterations in gene expressions were observed by 4-nitro CNMA than t-CNMA. Different color-coded gene boxes represent specific virulence phenotype (i.e. biofilm, motility, stress, and efflux), solid black bordered boxes show downregulation while blue bordered boxes show upregulation of genes. Arrows in different colors represent different group of genes and flat arrow ends stand for inhibition. cDNA, complimentary DNA; qPCR, quantitative polymerase chain reaction; MIC, minimum inhibitory concentration; EPS, exopolysaccharides.
Our findings suggest that the presence of strong electron-withdrawing groups at the fourth position of t-CNMA disrupts the biofilm formation mechanism of A. tumefaciens. The Ca2+ adhesion protein rhicadhesin has been proposed to play a role in A. tumefaciens attachment to plant surfaces (Swart et al., 1994), and it has been suggested that t-CNMA might similarly disrupt Ca2+ homeostasis in Phytophthora capsici (Hu L. et al., 2013). After evaluating the effects of t-CNMA, 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA on A. tumefaciens, we suggest that structure-based activity factors and the presence of a conjugated aldehyde contribute to antibiofilm effects of t-CNMA. Xie et al. (2017) assigned the antifungal activity of t-CNMA and α-methyl CNMA to –CHO and –CH3 group at the ortho position of the aromatic ring. We found the presence of –NO2, –Cl, or –F at the para position had considerable effects on A. tumefaciens biofilms, and that 4-nitro CNMA had a greater suppressive effect on biofilm-associated genetic factors than t-CNMA (Figure 8).
In summary, we evaluated the antibiofilm and antivirulence effects of t-CNMA and 4-nitro CNMA, 4-chloro CNMA, and 4-fluoro CNMA on A. tumefaciens. t-CNMA significantly reduced swimming motility, cell surface hydrophobicity, EPS secretion, and exo-protease production; however, these effects were considerably greater for 4-nitro CNMA and 4-chloro CNMA. We suggest the greater effects of these two derivatives on biofilm formation and growth may have been due to the presence of (i) a conjugated aldehyde group and (ii) an electron-withdrawing group like –NO2 at the para position. Also, qRT-PCR data showed 4-nitro CNMA downregulated the expressions of multiple biofilm formation associated genes, which shows CNMA derivatives target multiple processes and thus are unlikely to induce resistance in A. tumefaciens. Moreover, reductions in A. tumefaciens cell viability, growth, and root surface biofilm formation observed suggest that t-CNMA derivatives with –NO2, –Cl, or –F at position 4 on the aromatic ring provide an excellent starting point for the development of anti-Agrobacterium agents that effectively prevent crown gall disease.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
JL and J-HL: conceptualization, project administration, and funding acquisition. BA and AJ: methodology and software. BA, AJ, and JL: validation and writing the manuscript. JL: resources and supervision. All authors contributed to the article and approved the submitted version.
This work was supported by the Priority Research Center Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A6A1031189), by the Basic Science Research Program of NRF funded by the Ministry of Education (2021R1I1A3A04037486 to J-HL), and by an NRF grant funded by the Korea government (MSIT) (2021R1A2C1008368).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1001865/full#supplementary-material
Ahmed, B., Jailani, A., Lee, J.-H., and Lee, J. (2022). Effect of halogenated indoles on biofilm formation, virulence, and root surface colonization by Agrobacterium tumefaciens. Chemosphere 293:133603. doi: 10.1016/j.chemosphere.2022.133603
Ahmed, B., Solanki, B., Zaidi, A., Khan, M. S., and Musarrat, J. (2019b). Bacterial toxicity of biomimetic green zinc oxide nanoantibiotic: Insights into ZnONP uptake and nanocolloid-bacteria interface. Toxicol. Res. (Camb) 8, 246–261. doi: 10.1039/C8TX00267C
Ahmed, B., Rizvi, A., Zaidi, A., Khan, M. S., and Musarrat, J. (2019a). Understanding the phyto-interaction of heavy metal oxide bulk and nanoparticles: Evaluation of seed germination, growth, bioaccumulation, and metallothionein production. RSC Adv. 9, 4210–4225. doi: 10.1039/c8ra09305a
Ahmed, B., Syed, A., Ali, K., Elgorban, A. M., Khan, A., Lee, J., et al. (2021). Synthesis of gallotannin capped iron oxide nanoparticles and their broad spectrum biological applications. RSC Adv. 11, 9880–9893. doi: 10.1039/d1ra00220a
Albano, M., Crulhas, B. P., Alves, F. C. B., Pereira, A. F. M., Andrade, B. F. M. T., Barbosa, L. N., et al. (2019). Antibacterial and anti-biofilm activities of cinnamaldehyde against S. epidermidis. Microb. Pathog. 126, 231–238. doi: 10.1016/j.micpath.2018.11.009
Ali, K., Dwivedi, S., Azam, A., Saquib, Q., Al-Said, M. S., Alkhedhairy, A. A., et al. (2016). Aloe vera extract functionalized zinc oxide nanoparticles as nanoantibiotics against multi-drug resistant clinical bacterial isolates. J. Colloid Interface Sci. 472, 145–156. doi: 10.1016/j.jcis.2016.03.021
Badawy, M. E. I., and Rabea, E. I. (2013). Synthesis and structure–activity relationship of N-(cinnamyl) chitosan analogs as antimicrobial agents. Int. J. Biol. Macromol. 57, 185–192. doi: 10.1016/j.ijbiomac.2013.03.028
Brackman, G., Celen, S., Hillaert, U., Van Calenbergh, S., Cos, P., Maes, L., et al. (2011). Structure-activity relationship of cinnamaldehyde analogs as inhibitors of AI-2 based quorum sensing and their effect on virulence of Vibrio spp. PLoS One 6:e16084. doi: 10.1371/journal.pone.0016084
Budri, P. E., Silva, N. C. C., Bonsaglia, E. C. R., Júnior, A. F., Júnior, J. P. A., Doyama, J. T., et al. (2015). Effect of essential oils of Syzygium aromaticum and Cinnamomum zeylanicum and their major components on biofilm production in Staphylococcus aureus strains isolated from milk of cows with mastitis. J. Dairy Sci. 98, 5899–5904. doi: 10.3168/jds.2015-9442
Chen, L., Wang, Z., Liu, L., Qu, S., Mao, Y., Peng, X., et al. (2019). Cinnamaldehyde inhibits Candida albicans growth by causing apoptosis and its treatment on vulvovaginal candidiasis and oropharyngeal candidiasis. Appl. Microbiol. Biotechnol. 103, 9037–9055. doi: 10.1007/s00253-019-10119-3
Chun, J.-Y., Kim, K.-B., Shin, J.-B., and Min, S.-G. (2013). Effect of trans-cinnamaldehyde and high pressure treatment on physico-chemical and microbial properties of milk during storage periods. Food Sci. Anim. Resour. 33, 16–23. doi: 10.5851/kosfa.2013.33.1.16
Cui, Y., Liang, G., Hu, Y.-H., Shi, Y., Cai, Y.-X., Gao, H.-J., et al. (2015). Alpha-substituted derivatives of cinnamaldehyde as tyrosinase inhibitors: Inhibitory mechanism and molecular analysis. J. Agric. Food Chem. 63, 716–722. doi: 10.1021/jf505469k
Dessaux, Y., and Faure, D. (2018). Quorum sensing and quorum quenching in Agrobacterium: A “go/no go system”? Genes (Basel) 9, 210. doi: 10.3390/genes9040210
Doyle, A. A., Krämer, T., Kavanagh, K., and Stephens, J. C. (2019). Cinnamaldehydes: Synthesis, antibacterial evaluation, and the effect of molecular structure on antibacterial activity. Results Chem. 1:100013. doi: 10.1016/j.rechem.2019.100013
Faure, D., and Lang, J. (2014). Functions and regulation of quorum-sensing in Agrobacterium tumefaciens. Front. Plant Sci. 5:14. doi: 10.3389/fpls.2014.00014
Ferro, T. A. F., Araújo, J. M. M., dos Santos Pinto, B. L., Dos Santos, J. S., Souza, E. B., Da Silva, B. L. R., et al. (2016). Cinnamaldehyde inhibits Staphylococcus aureus virulence factors and protects against infection in a Galleria mellonella model. Front. Microbiol. 7:2052. doi: 10.3389/fmicb.2016.02052
Friedman, M. (2017). Chemistry, antimicrobial mechanisms, and antibiotic activities of cinnamaldehyde against pathogenic bacteria in animal feeds and human foods. J. Agric. Food Chem. 65, 10406–10423. doi: 10.1021/acs.jafc.7b04344
He, T.-F., Wang, L.-H., Niu, D., Wen, Q., and Zeng, X.-A. (2019). Cinnamaldehyde inhibit Escherichia coli associated with membrane disruption and oxidative damage. Arch. Microbiol. 201, 451–458. doi: 10.1007/s00203-018-1572-5
Heindl, J. E., Wang, Y., Heckel, B. C., Mohari, B., Feirer, N., and Fuqua, C. (2014). Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front. Plant Sci. 5:176. doi: 10.3389/fpls.2014.00176
Hu, L., Wang, D., Liu, L., Chen, J., Xue, Y., and Shi, Z. (2013). Ca2 + efflux is involved in cinnamaldehyde-induced growth inhibition of Phytophthora capsici. PLoS One 8:e76264. doi: 10.1371/journal.pone.0076264
Hu, X., Zhao, J., DeGrado, W. F., and Binns, A. N. (2013). Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc. Natl. Acad. Sci. U.S.A. 110, 678–683. doi: 10.1073/pnas.1215033110
Jia, P., Xue, Y. J., Duan, X. J., and Shao, S. H. (2011). Effect of cinnamaldehyde on biofilm formation and sarA expression by methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 53, 409–416. doi: 10.1111/j.1472-765X.2011.03122.x
Kahla, Y., Zouari-Bouassida, K., Rezgui, F., Trigui, M., and Tounsi, S. (2017). Efficacy of Eucalyptus cinerea as a source of bioactive compounds for curative biocontrol of crown gall caused by Agrobacterium tumefaciens strain B6. Biomed. Res. Int. 2017:9308063. doi: 10.1155/2017/9308063
Kerr, A., and Bullard, G. (2020). Biocontrol of crown gall by Rhizobium rhizogenes: Challenges in biopesticide Commercialisation. Agronomy 10:1126.
Khemthong, S., Nuonming, P., Dokpikul, T., Sukchawalit, R., and Mongkolsuk, S. (2019). Regulation and function of the flavonoid-inducible efflux system, emrR-emrAB, in Agrobacterium tumefaciens C58. Appl. Microbiol. Biotechnol. 103, 5763–5780. doi: 10.1007/s00253-019-09899-5
Kim, Y.-G., Lee, J.-H., Kim, S.-I., Baek, K.-H., and Lee, J. (2015). Cinnamon bark oil and its components inhibit biofilm formation and toxin production. Int. J. Food Microbiol. 195, 30–39. doi: 10.1016/j.ijfoodmicro.2014.11.028
Kot, B., Wicha, J., Piechota, M., Wolska, K., and Gruzewska, A. (2015). Antibiofilm activity of trans-cinnamaldehyde, p-coumaric, and ferulic acids on uropathogenic Escherichia coli. Turkish J. Med. Sci. 45, 919–924. doi: 10.3906/sag-1406-112
Lee, J. E., Jung, M., Lee, S. C., Huh, M. J., Seo, S. M., and Park, I. K. (2020). Antibacterial mode of action of trans-cinnamaldehyde derived from cinnamon bark (Cinnamomum verum) essential oil against Agrobacterium tumefaciens. Pestic. Biochem. Physiol. 165:104546. doi: 10.1016/j.pestbp.2020.02.012
Lee, J. H., Kim, Y. G., Baek, K. H., Cho, M. H., and Lee, J. (2015). The multifaceted roles of the interspecies signalling molecule indole in Agrobacterium tumefaciens. Environ. Microbiol. 17, 1234–1244. doi: 10.1111/1462-2920.12560
Lee, J.-H., Park, J.-H., Kim, J.-A., Neupane, G. P., Cho, M. H., Lee, C.-S., et al. (2011). Low concentrations of honey reduce biofilm formation, quorum sensing, and virulence in Escherichia coli O157: H7. Biofouling 27, 1095–1104. doi: 10.1080/08927014.2011.633704
Li, T., Wang, D., Liu, N., Ma, Y., Ding, T., Mei, Y., et al. (2018). Inhibition of quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas fluorescens by cinnamaldehyde. Int. J. Food Microbiol. 269, 98–106. doi: 10.1016/j.ijfoodmicro.2018.01.023
Li, X., Yang, Q., Peng, L., Tu, H., Lee, L.-Y., Gelvin, S. B., et al. (2020). Agrobacterium-delivered VirE2 interacts with host nucleoporin CG1 to facilitate the nuclear import of VirE2-coated T complex. Proc. Natl. Acad. Sci. U.S.A. 117, 26389–26397. doi: 10.1073/pnas.2009645117
Li, Y., Qu, H.-P., Liu, J.-L., and Wan, H.-Y. (2014). Correlation between group behavior and quorum sensing in Pseudomonas aeruginosa isolated from patients with hospital-acquired pneumonia. J. Thorac. Dis. 6:810. doi: 10.3978/j.issn.2072-1439.2014.03.37
Liu, P., and Nester, E. W. (2006). Indoleacetic acid, a product of transferred DNA, inhibits vir gene expression and growth of Agrobacterium tumefaciens C58. Proc. Natl. Acad. Sci. U.S.A. 103, 4658–4662. doi: 10.1073/pnas.0600366103
Matthysse, A. G. (2014). Attachment of Agrobacterium to plant surfaces. Front. Plant Sci. 5:252. doi: 10.3389/fpls.2014.00252
Matthysse, A. G., Marry, M., Krall, L., Kaye, M., Ramey, B. E., Fuqua, C., et al. (2005). The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 18, 1002–1010. doi: 10.1094/MPMI-18-1002
McCardell, B. A., and Pootjes, C. F. (1976). Chemical nature of agrocin 84 and its effect on a virulent strain of Agrobacterium tumefaciens. Antimicrob. Agents Chemother. 10, 498–502. doi: 10.1128/AAC.10.3.498
Merritt, P. M., Danhorn, T., and Fuqua, C. (2007). Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189, 8005–8014. doi: 10.1128/JB.00566-07
Mizan, M. F. R., Jahid, I. K., Kim, M., Lee, K.-H., Kim, T. J., and Ha, S.-D. (2016). Variability in biofilm formation correlates with hydrophobicity and quorum sensing among Vibrio parahaemolyticus isolates from food contact surfaces and the distribution of the genes involved in biofilm formation. Biofouling 32, 497–509. doi: 10.1080/08927014.2016.1149571
Moens, M. (2009). Meloidogyne Species – a Diverse group of novel and important plant parasites. Root Knot Nematodes 1, 483. doi: 10.1079/9781845934927.0001
Mousavi, F., Bojko, B., Bessonneau, V., and Pawliszyn, J. (2016). Cinnamaldehyde characterization as an antibacterial agent toward E. coli metabolic profile using 96-blade solid-phase microextraction coupled to liquid chromatography–mass spectrometry. J. Proteome Res. 15, 963–975. doi: 10.1021/acs.jproteome.5b00992
Muhammad, M. H., Idris, A. L., Fan, X., Guo, Y., Yu, Y., Jin, X., et al. (2020). Beyond risk: Bacterial biofilms and their regulating approaches. Front. Microbiol. 11:928. doi: 10.3389/fmicb.2020.00928
Nabi, N., Ben Hafsa, A., Gaillard, V., Nesme, X., Chaouachi, M., and Vial, L. (2022). Evolutionary classification of tumor-and root-inducing plasmids based on T-DNAs and virulence regions. Mol. Phylogenet. Evol. 169, 107388. doi: 10.1016/j.ympev.2022.107388
Nepali, K., Lee, H.-Y., and Liou, J.-P. (2018). Nitro-group-containing drugs. J. Med. Chem. 62, 2851–2893. doi: 10.1021/acs.jmedchem.8b00147
Nguyen, T. N. L., Hoang, T. T. H., Nguyen, H. Q., Tu, Q. T., Tran, T. H., Lo, T. M. T., et al. (2021). Agrobacterium tumefaciens–mediated genetic transformation and overexpression of the flavonoid 3’ 5’-hydroxylase gene increases the flavonoid content of the transgenic Aconitum carmichaelii Debx. plant. Vitr. Cell Dev. Biol. 58, 93–102. doi: 10.1007/s11627-021-10190-4
Niu, C., Afre, S., and Gilbert, E. S. (2006). Subinhibitory concentrations of cinnamaldehyde interfere with quorum sensing. Lett. Appl. Microbiol. 43, 489–494. doi: 10.1111/j.1472-765X.2006.02001.x
Niu, C., and Gilbert, E. S. (2004). Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl. Environ. Microbiol. 70, 6951–6956. doi: 10.1128/AEM.70.12.6951-6956.2004
Nuonming, P., Khemthong, S., Dokpikul, T., Sukchawalit, R., and Mongkolsuk, S. (2018). Characterization and regulation of AcrABR, a RND-type multidrug efflux system, in Agrobacterium tumefaciens C58. Microbiol. Res. 214, 146–155. doi: 10.1016/j.micres.2018.06.014
Oussalah, M., Caillet, S., and Lacroix, M. (2006). Mechanism of action of Spanish oregano, Chinese cinnamon, and savory essential oils against cell membranes and walls of Escherichia coli O157: H7 and Listeria monocytogenes. J. Food Prot. 69, 1046–1055. doi: 10.4315/0362-028x-69.5.1046
Palumbo, J. D., Kado, C. I., and Phillips, D. A. (1998). An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J. Bacteriol. 180, 3107–3113. doi: 10.1128/JB.180.12.3107-3113.1998
Pena, R. T., Blasco, L., Ambroa, A., González-Pedrajo, B., Fernández-García, L., López, M., et al. (2019). Relationship between quorum sensing and secretion systems. Front. Microbiol. 10:1100. doi: 10.3389/fmicb.2019.01100
Ramey, B. E., and Parsek, M. R. (2006). Growing and analyzing biofilms in fermenters. Curr. Protoc. Microbiol. 1B–3B. doi: 10.1002/9780471729259.mc01b03s00
Rao, P. V., and Gan, S. H. (2014). Cinnamon: A multifaceted medicinal plant. Evid. Based Complement Altern. Med. 2014, 642942. doi: 10.1155/2014/642942
Rittiroongrad, S., Charoenlap, N., Giengkam, S., Vattanaviboon, P., and Mongkolsuk, S. (2016). Agrobacterium tumefaciens estC, encoding an enzyme containing esterase activity, is regulated by EstR, a Regulator in the MarR Family. PLoS One 11:e0168791. doi: 10.1371/journal.pone.0168791
Rosen, R., Büttner, K., Schmid, R., Hecker, M., and Ron, E. Z. (2001). Stress-induced proteins of Agrobacterium tumefaciens. FEMS Microbiol. Ecol. 35, 277–285.
Rosenberg, M. (1984). Bacterial adherence to hydrocarbons: A useful technique for studying cell surface hydrophobicity. FEMS Microbiol. Lett. 22, 289–295.
Sethupathy, S., Sathiyamoorthi, E., Kim, Y.-G., Lee, J.-H., and Lee, J. (2020). Antibiofilm and antivirulence properties of indoles against Serratia marcescens. Front. Microbiol. 11:584812. doi: 10.3389/fmicb.2020.584812
Shaikh, M. H., Subhedar, D. D., Shingate, B. B., Kalam Khan, F. A., Sangshetti, J. N., Khedkar, V. M., et al. (2016). Synthesis, biological evaluation and molecular docking of novel coumarin incorporated triazoles as antitubercular, antioxidant and antimicrobial agents. Med. Chem. Res. 25, 790–804. doi: 10.1007/s00044-016-1519-9
Sharma, D., Misba, L., and Khan, A. U. (2019). Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8, 1–10. doi: 10.1186/s13756-019-0533-3
Shen, S., Zhang, T., Yuan, Y., Lin, S., Xu, J., and Ye, H. (2015). Effects of cinnamaldehyde on Escherichia coli and Staphylococcus aureus membrane. Food Control 47, 196–202. doi: 10.1016/j.foodcont.2014.07.003
Shreaz, S., Wani, W. A., Behbehani, J. M., Raja, V., Irshad, M., Karched, M., et al. (2016). Cinnamaldehyde and its derivatives, a novel class of antifungal agents. Fitoterapia 112, 116–131. doi: 10.1016/j.fitote.2016.05.016
Slater, S. C., Goldman, B. S., Goodner, B., Setubal, J. C., Farrand, S. K., Nester, E. W., et al. (2009). Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 191, 2501–2511. doi: 10.1128/JB.01779-08
Swart, S., Lugtenberg, B. J., Smit, G., and Kijne, J. W. (1994). Rhicadhesin-mediated attachment and virulence of an Agrobacterium tumefaciens chvB mutant can be restored by growth in a highly osmotic medium. J. Bacteriol. 176, 3816–3819. doi: 10.1128/jb.176.12.3816-3819.1994
Tan, C. H., Koh, K. S., Xie, C., Tay, M., Zhou, Y., Williams, R., et al. (2014). The role of quorum sensing signalling in EPS production and the assembly of a sludge community into aerobic granules. ISME J. 8, 1186–1197. doi: 10.1038/ismej.2013.240
Thompson, M. A., Onyeziri, M. C., and Fuqua, C. (2018). Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. Agrobacterium Biol. 418, 143–184. doi: 10.1007/82_2018_96
Tomlinson, A. D., Ramey-Hartung, B., Day, T. W., Merritt, P. M., and Fuqua, C. (2010). Agrobacterium tumefaciens ExoR represses succinoglycan biosynthesis and is required for biofilm formation and motility. Microbiology 156:2670. doi: 10.1099/mic.0.039032-0
Tribedi, P., and Sil, A. K. (2014). Cell surface hydrophobicity: A key component in the degradation of polyethylene succinate by Pseudomonas sp. AKS 2. J. Appl. Microbiol. 116, 295–303. doi: 10.1111/jam.12375
Tsai, Y.-L., Chiang, Y.-R., Wu, C.-F., Narberhaus, F., and Lai, E.-M. (2012). One out of four: HspL but no other small heat shock protein of Agrobacterium tumefaciens acts as efficient virulence-promoting VirB8 chaperone. PLoS One 7:e49685. doi: 10.1371/journal.pone.0049685
Wei, Q.-Y., Xiong, J.-J., Jiang, H., Zhang, C., and Ye, W. (2011). The antimicrobial activities of the cinnamaldehyde adducts with amino acids. Int. J. Food Microbiol. 150, 164–170. doi: 10.1016/j.ijfoodmicro.2011.07.034
Wendakoon, C. N., and Sakaguchi, M. (1995). Inhibition of amino acid decarboxylase activity of Enterobacter aerogenes by active components in spices. J. Food Prot. 58, 280–283. doi: 10.4315/0362-028X-58.3.280
Wheatley, R. M., and Poole, P. S. (2018). Mechanisms of bacterial attachment to roots. FEMS Microbiol. Rev. 42, 448–461. doi: 10.1093/femsre/fuy014
Xie, Y., Huang, Q., Wang, Z., Cao, H., and Zhang, D. (2017). Structure-activity relationships of cinnamaldehyde and eugenol derivatives against plant pathogenic fungi. Ind. Crops Prod. 97, 388–394. doi: 10.1016/j.indcrop.2016.12.043
Ying, L., Mingzhu, S., Mingju, Y., Ye, X., Yuechen, W., Ying, C., et al. (2019). The inhibition of trans-cinnamaldehyde on the virulence of Candida albicans via enhancing farnesol secretion with low potential for the development of resistance. Biochem. Biophys. Res. Commun. 515, 544–550. doi: 10.1016/j.bbrc.2019.05.165
Yossa, N., Patel, J., Macarisin, D., Millner, P., Murphy, C., Bauchan, G., et al. (2014). Antibacterial Activity of Cinnamaldehyde and Sporan against Escherichia coli O 157: H 7 and Salmonella. J. Food Process. Preserv. 38, 749–757.
Yu, H. H., Song, Y. J., Yu, H., Lee, N., and Paik, H. (2020). Investigating the antimicrobial and antibiofilm effects of cinnamaldehyde against Campylobacter spp. using cell surface characteristics. J. Food Sci. 85, 157–164. doi: 10.1111/1750-3841.14989
Yuan, Z.-C., Liu, P., Saenkham, P., Kerr, K., and Nester, E. W. (2008). Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J. Bacteriol. 190, 494–507. doi: 10.1128/JB.01387-07
Zhang, Y., Kong, J., Xie, Y., Guo, Y., Cheng, Y., Qian, H., et al. (2018). Essential oil components inhibit biofilm formation in Erwinia carotovora and Pseudomonas fluorescens via anti-quorum sensing activity. LWT 92, 133–139.
Keywords: Agrobacterium tumefaciens, biofilm, cinnamaldehyde, plant, virulence
Citation: Ahmed B, Jailani A and Lee J-H and Lee J (2022) Inhibition of growth, biofilm formation, virulence, and surface attachment of Agrobacterium tumefaciens by cinnamaldehyde derivatives. Front. Microbiol. 13:1001865. doi: 10.3389/fmicb.2022.1001865
Received: 24 July 2022; Accepted: 22 September 2022;
Published: 11 October 2022.
Edited by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilReviewed by:
Juan José Valdez Alarcón, Universidad Michoacana de San Nicolás de Hidalgo, MexicoCopyright © 2022 Ahmed, Jailani, Lee and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Hyung Lee, amluaGxlZUB5bnUuYWMua3I=; Jintae Lee, anRsZWVAeW51LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.