- 1Country College of Biological Science and Technology, University of Jinan, Jinan, China
- 2School of Life Sciences, Neijiang Normal University, Neijiang, China
- 3College of Food and Bioengineering, Hezhou University, Hezhou, China
- 4Guangxi Key Laboratory of Health Care Food Science and Technology, Hezhou University, Hezhou, China
Background: Induced by the pathogen Mycobacterium tuberculosis, tuberculosis remains one of the most dangerous infectious diseases in the world. As a special virus, prophage is domesticated by its host and are major contributors to virulence factors for bacterial pathogenicity. The function of prophages and their genes in M. tuberculosis is still unknown.
Methods: Rv2650c is a prophage gene in M. tuberculosis genome. We constructed recombinant Mycobacterium smegmatis (M. smegmatis) to observe bacteria morphology and analyze the resistance to various adverse environments. Recombinant and control strains were used to infect macrophages, respectively. Furthermore, we performed ELISA experiments of infected macrophages.
Results: Rv2650c affected the spread of colonies of M. smegmatis and enhanced the resistance of M. smegmatis to macrophages and various stress agents such as acid, oxidative stress, and surfactant. ELISA experiments revealed that the Rv2650c can inhibit the expression of inflammatory factors TNF-α, IL-10, IL-1β, and IL-6.
Conclusion: This study demonstrates that the prophage gene Rv2650c can inhibit the spread of colonies and the expression of inflammatory factors and promote intracellular survival of M. smegmatis. These results build the foundation for the discovery of virulence factors of M. tuberculosis, and provide novel insights into the function of the prophage in Mycobacterium.
Introduction
According to the 2020 global tuberculosis (TB) report released by WHO (World Health Organization [WHO], 2020), about 2 billion people were latently infected by Mycobacterium tuberculosis and 1.41 million people had fallen ill with TB in 2019. There were about 1.21 million tuberculosis deaths among HIV-negative people and an additional 208 thousand deaths among HIV-positive people. Tuberculosis is still one of the most prevalent and deadly infectious diseases in the world. Therefore, the pathogenesis and virulence factors of M. tuberculosis are still need to be studied. Currently known virulence factors of M. tuberculosis can interfere with phagosome maturation (Jayachandran et al., 2007; Queval et al., 2017); control cell death (Awuh and Flo, 2017); disrupt the production of cytokines (Etna et al., 2014); and can help M. tuberculosis resist various harsh environments (Ehrt et al., 2018) to maintain the survival in macrophages. However, most genes encoding virulence factors of M. tuberculosis are present in the genome of Mycobacterium smegmatis (M. smegmatis), whereby such genes from M. smegmatis can replace their homologous genes in M. tuberculosis and cause disease (Cambier et al., 2014). M. smegmatis is a kind of non-pathogenic environmental saprophytic mycobacteria. Hence, this raises the question of whether there are any unknown specific virulence factors that play a key and irreplaceable role in the pathogenesis and intracellular survival of M. tuberculosis.
As domesticated phage elements that have been integrated into and replicated with the host genome, prophages are involved in the evolution of some bacteria. Prophages can regulate host behavior to improve the host’s fitness to harsh environments. For pathogens, prophages can confer or enhance bacterial virulence and increase the intracellular survival rate and pathogenicity of bacteria (Feiner et al., 2015; Salmond and Fineran, 2015; Argov et al., 2017; Keen and Dantas, 2018). There are two prophages, phiRv1 and phiRv2, in the M. tuberculosis H37Rv genome. They do not exist in the M. smegmatis genome and their functions in the M. tuberculosis genome are still unknown. Our previous study found that some genes of prophage phiRv1 and phiRv2 can be encoded from proteomic data (Deng et al., 2013; Schubert et al., 2013). In this study, we focus on the prophage phiRv2 gene Rv2650c which was predicted to encode the capsid protein (Kapopoulou et al., 2011). Transcriptomic data indicated that Rv2650c is upregulated in the M. tuberculosis-infected macrophage model and persister model (Fan et al., 2016). Those data indicates that Rv2650c is a functional protein and may improve the fitness of Mycobacterium to environmental stresses. Moreover, bacteriophage P4 capsid protein, Psu, was considered to be related to the pathogenicity of various pathogens (Ghosh et al., 2018). In summary, capsid protein, Rv2650c, may play an important role in the response of M. tuberculosis to stress and pathogenicity.
In this study, we construct recombinant M. smegmatis overexpressed Rv2650c and carry out a series of experiments to investigate the role of Rv2650c in the pathogenicity and intracellular survival of Mycobacterium.
Materials and Methods
Bacterial Strains, Plasmid, and Growth Conditions
Escherichia coli (E. coli) DH5α, E. coli BL21, M. smegmatis mc2155, and pNIT-Myc plasmid were preserved by our laboratory. E. coli was grown in Luria-Bertani (LB) medium and was used for plasmid construction and preservation. M. smegmatis mc2155 was grown in either Middlebrook (MB) 7H9 broth or on MB 7H9 agar containing 10 g/L glucose, 2% (v/v) glycerol, and 0.05% (v/v) Tween80. When required, kanamycin (25 μg/mL) and cycloheximide (10 μg/mL) were added. Plasmid pNIT-Myc was constructed from shuttle plasmid pNIT-1 (Pandey et al., 2009; Li et al., 2014). In the plasmid, the inducible nitA promoter (PnitA) controlled the overexpression of expression cassette.
Construction of Recombinant Mycobacterium smegmatis
The Rv2650c gene of phage phiRv2 in the M. tuberculosis H37Rv genome was synthesized in Tsingke Biological Technology Co., Ltd. and ligated into the plasmid pNIT-Myc vector. The multiple cloning sites on both sides were EcoRI and ApaI. Recombinant plasmid pNIT-Myc-Rv2650c was constructed (Supplementary Figure 1), transferred into E. coli and subjected to extraction. The recombinant plasmid and empty pNIT-Myc vector was then separately transferred into M. smegmatis mc2155 using the standard electroporation method of mycobacteria (Pandey et al., 2009). The recombinant M. smegmatis strains were selected on MB 7H9 agar containing kanamycin. The positive recombinant M. smegmatis was selected by colony PCR and sequenced in Tsingke Biological Technology Co., Ltd. The positive clone was named MS_Rv2650c, and the transformed vector strain was named MS_Vec. Western blot was used to detect the positive recombinant strain.
Cellular Aggregation and Colony Morphology
The recombinant M. smegmatis MS_Rv2650c and MS_Vec were inoculated separately to 7H9 medium with 0.05% (v/v) Tween 80 and 28 mM ε-caprolactam and cultured at 37°C until an OD600 of 0.6–0.8 was reached. The cultures were kept at room temperature for 30 min and were observed the accumulation and precipitation.
The MS_Rv2650c and MS_Vec strains were inoculated on MB 7H9 plates with or without 0.05% (v/v) Tween 80 and cultured at 37°C for 5–6 days. The growth of culture colonies was observed.
Scanning Electron Microscope Analysis
The recombinant M. smegmatis MS_Rv2650c and MS_Vec were inoculated separately to 7H9 medium with 28 mM ε-caprolactam and cultured at 37°C until the OD600 reached 0.8–1.0. Cultures were harvested by centrifugation and the harvested precipitation were rinsed by 0.1 M PBS buffer. Then the precipitation was resuspended in 2.5% glutaraldehyde solution. The processed precipitation was dehydrated in a series of ethanol. After naturally drying, the samples were sprayed with gold (MC1000) and were observed by scanning electron microscopy (SEM) (Hitachi regular-8100).
Growth Curve and Stress Analysis of the Recombinant Mycobacterium smegmatis
The recombinant M. smegmatis MS_Rv2650c and MS_Vec were inoculated separately and cultured at 37°C until an OD600 of 0.6–0.8 was reached. 28 mM ε-caprolactam was added to induce the expression of protein. The OD600 was measured every 4 h for a total of 68 h. The growth curve of recombinant M. smegmatis was plotted according to the recorded data.
The growth of recombinant M. smegmatis in response to various stressors was measured according to references (Misra et al., 2011). The log phase of recombinant M. smegmatis was collected by centrifugation, and the bacteria were washed with MB 7H9 at pH 4.5; MB 7H9 at a final concentration of 3% hydrogen peroxide; and MB 7H9 medium at a final concentration of 2% SDS. The bacteria were resuspended in 5 mL of the corresponding conditions of 7H9 broth, and the final OD600 value was controlled at about 0.5. Under the corresponding conditions, the bacteria were cultured and 100 μL of the bacterial liquid was taken at the 0, 3, and 6 h time points to make a ten-fold concentration gradient dilution. The diluted bacteria were inoculated on MB 7H9 plates containing kanamycin and cultured at 37°C for 3–4 days. The number of colonies was counted to determine the average value for drawing. The concentration gradient of each plate was made in three parallels.
Infection of Macrophages With Recombinant Mycobacterium smegmatis
The suspension cultured THP-1 cells were evenly seeded at 1 × 106 cells per well in a 6-well plate, and three replicate wells were taken. The THP-1 cells were differentiated into macrophages in the presence of 100 ng/mL PMA (Sigma, United States) at 37°C under 5% CO2. Macrophages were infected with recombinant M. smegmatis at MOI = 10:1 (bacteria: macrophages). At 13, 25, 48 h after infection, macrophages were washed twice and then lysed with a final concentration of 0.025% (w/v) SDS solution to release M. smegmatis. The cell lysate of each well was diluted by 10 times and then inoculated onto the MB 7H10 plate containing kanamycin. After 3–4 days, the number of colonies on the plate was counted, and the colony forming unit (CFU) was calculated to compare the ability of intracellular survival of recombinant M. smegmatis cells.
Assay for Cytokines
The culture supernatant after infection of THP-1 cells with MS_Rv2650c or MS_Vec was collected at the corresponding time. The concentrations of the tumor necrosis factor-α (TNF-α), 1L-10, IL-1β, and IL-6 in the supernatants were determined using an ELISA kit (eBioscience).
Statistical Analysis
Data of this study were analyzed with paired t-test. Values of p < 0.05 were defined as statistically significant. The error bar in the experiment was expressed as the difference between the three values.
Results
Expression of Prophage Gene Rv2650c in Mycobacterium smegmatis
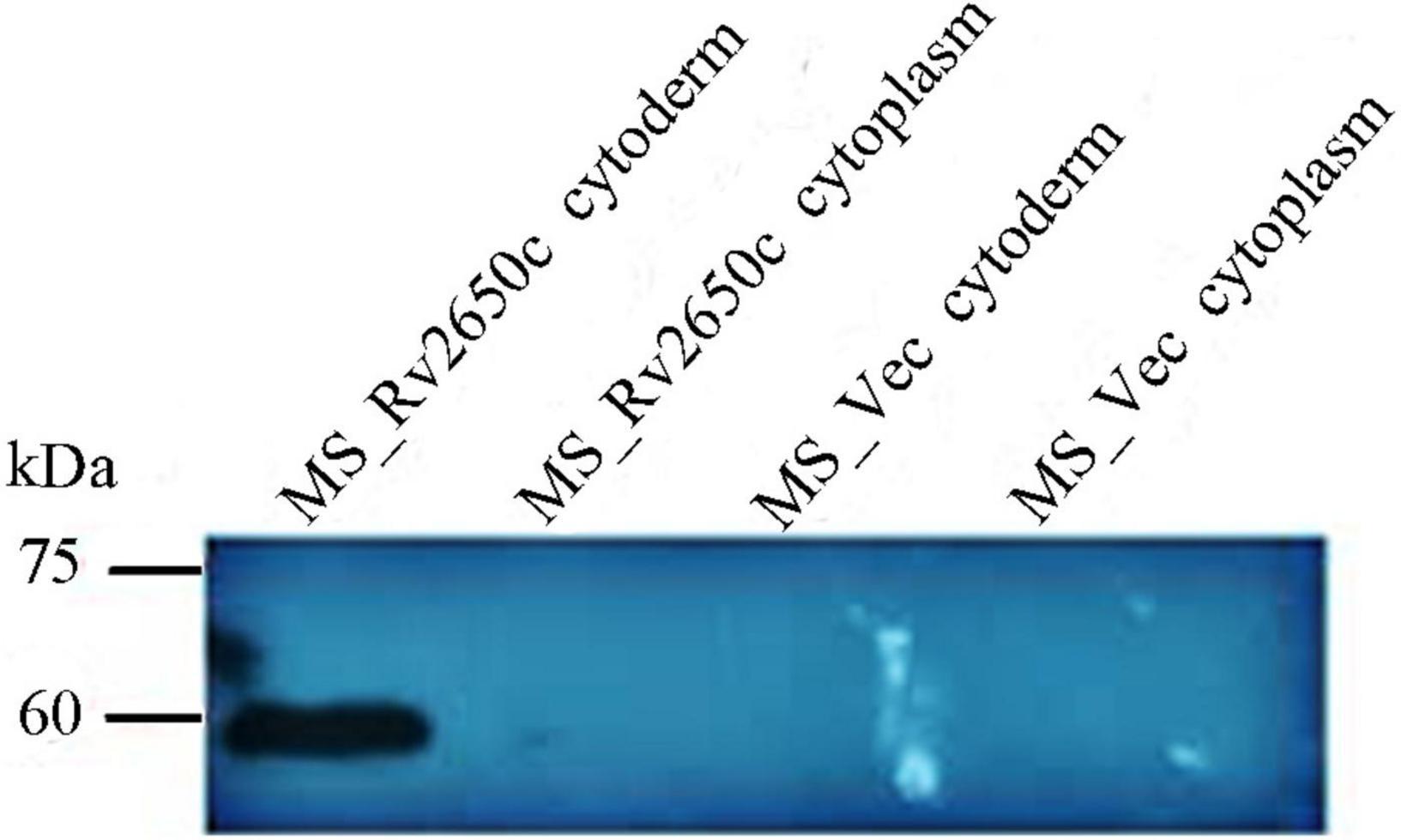
The prophage gene Rv2650c of M. tuberculosis is approximately 1,449 bp in size. We constructed two recombinant M. smegmatis to study the function of Rv2650c. Among them, the recombinant M. smegmatis strain MS_Rv2650c was able to express the M. tuberculosis protein Rv2650c carrying the Myc tag. On the contrary, the recombinant M. smegmatis strain MS_Vec, which carried the empty plasmid pNIT-Myc, served as the negative control. Both MS_Rv2650c and MS_Vec are capable of expressing the aph gene, so both can be grown on kanamycin-containing medium. Western blot analysis confirmed that Rv2650c from M. tuberculosis was successfully expressed in M. smegmatis and localized on the cell wall (Figure 1).
Rv2650c Alters the Diameter Size of Mycobacterium smegmatis Colony
Cell aggregation, single cell morphology, and colony observation experiments were conducted. Comparing the cell aggregation and single cell morphology of MS_Rv2650c and MS_Vec strains, there is no difference between them (Supplementary Figure 2). The results indicated that Rv2650c did not affect the aggregation and morphology of M. smegmatis. The results of colony observation experiments showed that the colony diameter of MS_Rv2650c grown on MB 7H9 plates with Tween80 was significantly lower than that of other strains grown on MB 7H9 plates with or without Tween80 (Figures 2A,B). In addition, the edges of colonies of MS_Rv2650c strain grown on the MB 7H9 plates with Tween80 were more rounded while the edges of colonies of other stains were irregular (Figure 2A). The results suggested that the recombinant protein Rv2650c affected the growth and spread of colonies of M. smegmatis in the presence of Tween80.
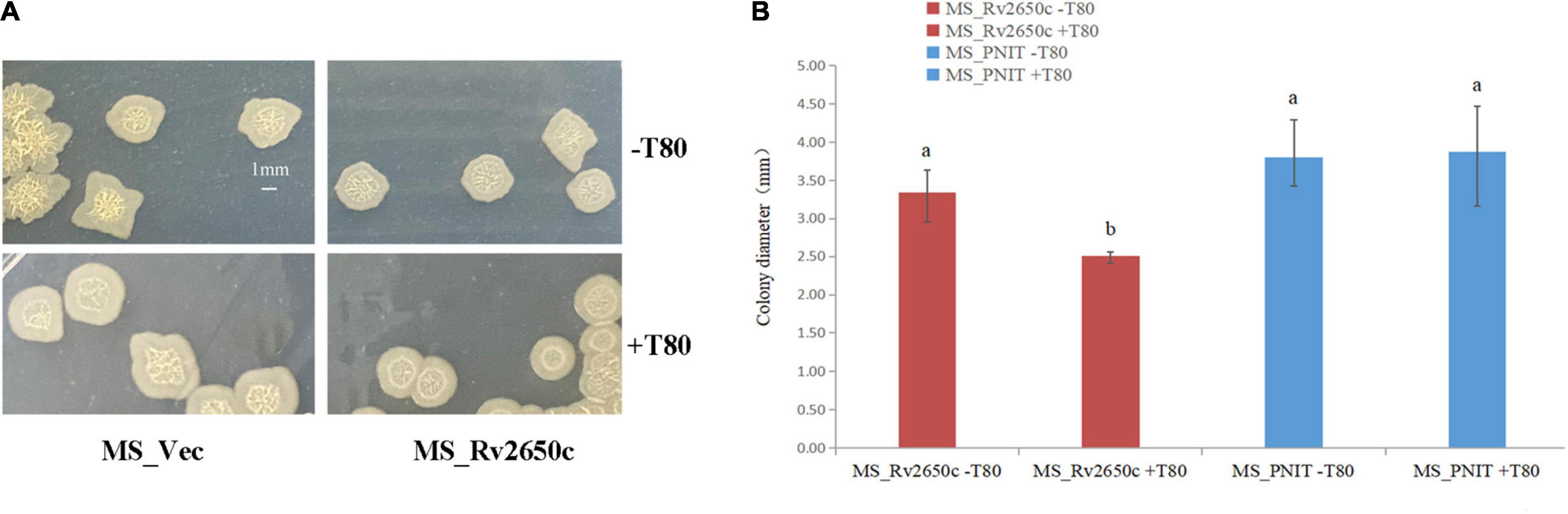
Figure 2. Comparison of the structure of cellular aggregation about the recombinant M. smegmatis MS_Rv2650c and MS_Vec. (A) MS_Rv2650c and MS_Vec strains were inoculated on MB 7H9 plates with or without 0.05% (v/v) Tween 80 and cultured at 37°C for 5–6 days. (B) The diameter of a single colony was observed and counted. Significantly differences (P < 0.05) are represented by different letters.
Rv2650c Enhances the Resistance of Mycobacterium smegmatis to Various Adverse Environments
As an intracellular pathogen, M. tuberculosis mainly parasitizes in macrophages and faces many complex environments such as acidity, oxidative stress, and poor nutrition. To investigate the role of the prophage protein Rv2650c in the response of M. tuberculosis against adverse environmental conditions, we performed growth characteristics analysis of the recombinant M. smegmatis MS_Rv2650c and MS_Vec in stress conditions which mimicked the intracellular environment. As shown in Figure 3A, there is no distinct difference in the in vitro growth kinetic between two strains, indicating that Rv2650c did not change the proliferative activity of Mycobacterium. When the recombinant M. smegmatis was monitored in MB 7H9 at pH 4.5, the survival rates of MS_Rv2650c and MS_Vec were 76.9% and 44.8% (paired t-test, P = 0.012), respectively during a 6 h incubation (Figures 3B,E). When cultured in MB 7H9 with hydrogen peroxide (final concentration 3%) for a 6 h incubation period (Figures 3C,E), the survival rates of MS_Rv2650c and MS_Vec were 232.4% and 57.1% (paired t-test, P = 0.0006). Similarly, MS_Rv2650c had higher rates of survival compared to MS_Vec (11.6% vs. 0.8% after 3 h incubation, paired t-test, P = 0.008; and 9.3% vs. 0.14% after 6 h incubation, paired t-test, P = 0.017) when strains were cultured in MB 7H9 with SDS (final concentration 2%) (Figures 3D,E). Taken together, these results showed that the prophage protein Rv2650c can enhance the resistance of recombinant M. smegmatis against harsh environments. It also indicates that this ability is not dependent upon improving the replication of MS_Rv2650c strain.
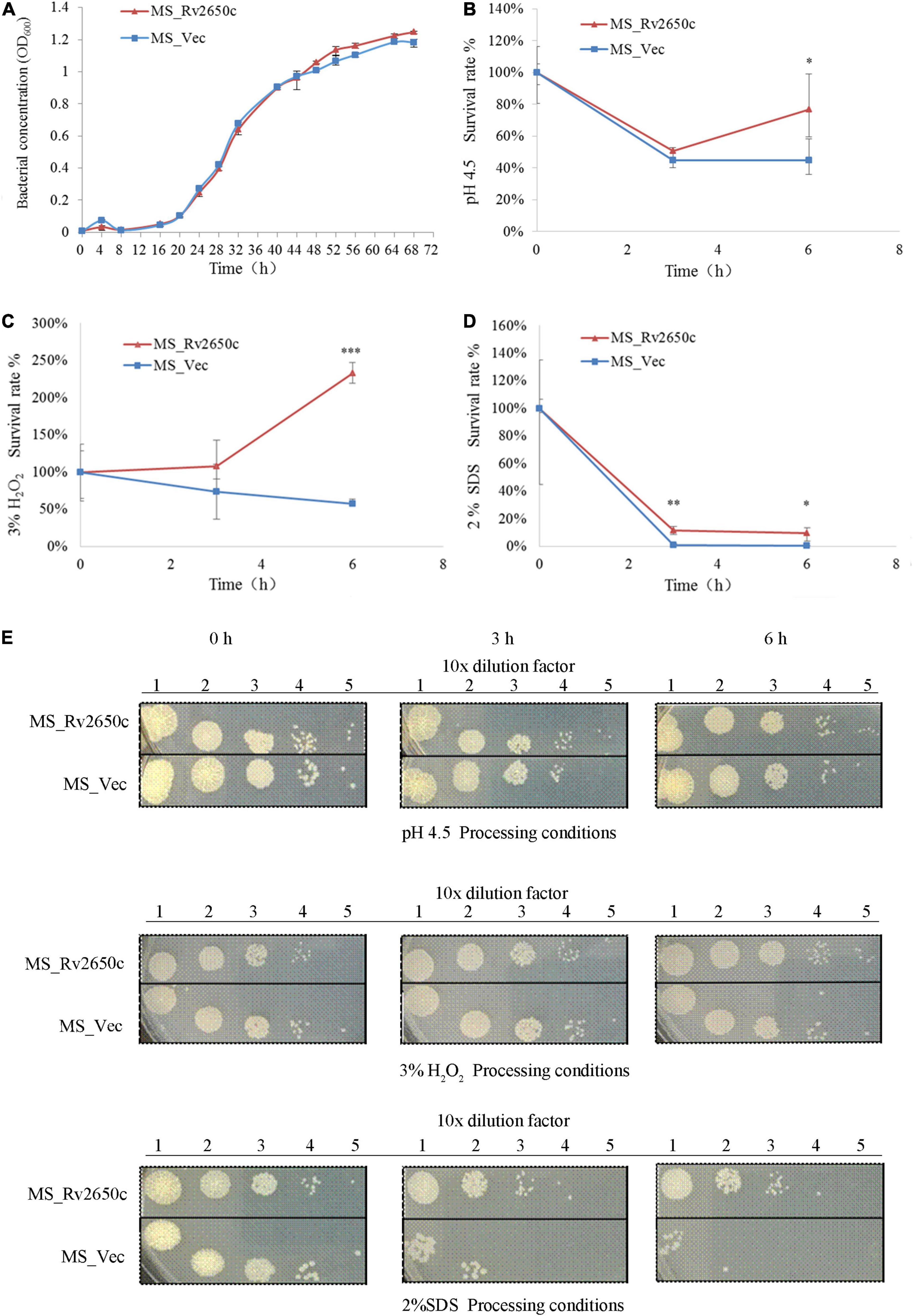
Figure 3. Extracellular growth and stress analysis results of recombinant M. smegmatis. (A) MS_Rv2650c and MS_Vec were inoculated into MB 7H9 broth and cultured. The OD600 value was monitored at 4 h intervals for a total of 68 h. (B) Inoculate MS_Rv2650c and MS_Vec into 7H9 broth at pH 4.5 for 6 h at 37°C. The cultured solution of 0, 3, and 6 h was diluted and coated on MB 7H9 plate, and the number of single colonies was calculated after incubation at 37°C for 3–4 days. (C) MS_Rv2650c and MS_Vec were treated with 3% hydrogen peroxide at 37°C for 6 h in a cultured solution with an OD600 about 0.5, and the cultured solution of 0, 3, and 6 h was diluted and coated on the MB 7H9 plate. The number of single colonies was calculated after incubation for 3–4 days at 37°C. (D) MS_Rv2650c and MS_Vec were treated with 2% SDS for 6 h at 37°C in a cultured solution with an OD600 about 0.5, and the cultured solution of 0, 3, and 6 h was diluted and coated on the MB 7H9 plate. The number of single colonies was calculated after incubation for 3–4 days at 37°C. (E) MS_Rv2650c and MS_Vec were diluted and coated on 7H9 plates under the conditions of pH 4.5, 3% H2O2, 2% SDS, and cultured at 37°C for 3–4 days. Similar results were obtained in 3 independent experiments. (*p < 0.05; **p < 0.01; ***p < 0.001).
Rv2650c Enhances the Intracellular Survival of Mycobacterium in Macrophages
Because the Rv2650c confers the ability of recombinant bacteria to resist harsh environments, we further studied the intracellular survival of MS_Rv2650c and MS_Vec in macrophages. To achieve this, we infected THP-1-derived macrophages with MS_Rv2650c or MS_Vec as previously described (MOI = 10:1). The experimental results showed that the intracellular survival rate of MS_Rv2650c strain was higher than that of the MS_Vec strain after 13, 25, and 48 h post infection (Figure 4). Rv2650c had significant effect on the intracellular survival of M. smegmatis (13.5% of MS_Rv2650c vs. 6.6% of MS_Vec after 13 h post infection, paired t-test, P = 0.043; 6% of MS_Rv2650c vs. 1.1% of MS_Vec after 25 h post infection, paired t-test, P = 0.051; 4% of MS_Rv2650c vs. 0.5% of MS_Vec after 48 h post infection, paired t-test, P = 0.029). This result showed that the expression of Rv2650c can enhance the intracellular survival of M. smegmatis in macrophages.
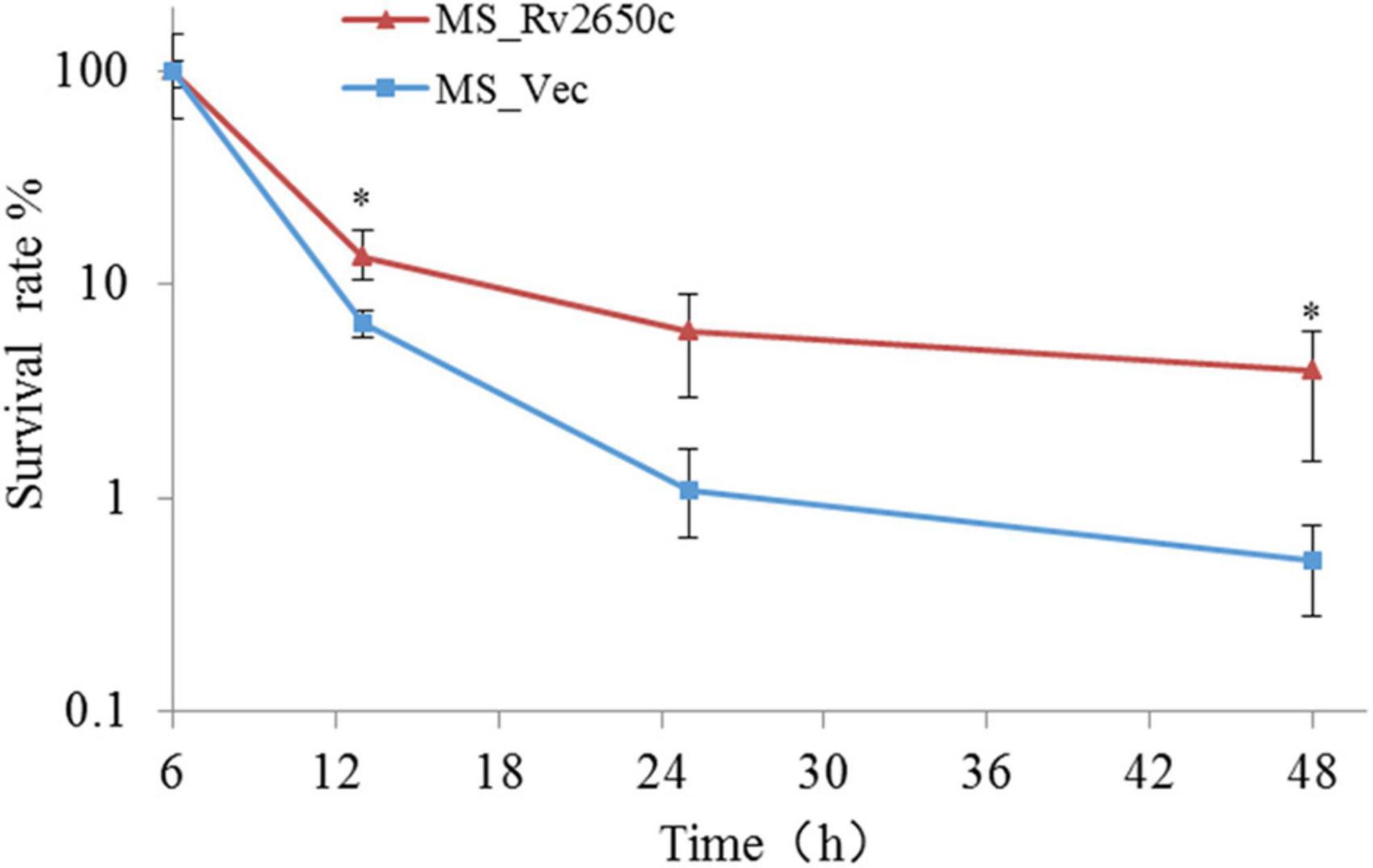
Figure 4. The intracellular survival rate of macrophages infected with recombinant M. smegmatis. THP-1 cells were infected with recombinant M. smegmatis MS_Rv2650c and MS_Vec, respectively, and the cells were lysed with SDS at the corresponding time. The cell lysate was diluted and coated on MB 7H10 plate. The number of single colonies was calculated after incubation for 3–4 days at 37°C. Similar results were obtained in 3 independent experiments. (*p < 0.05).
Rv2650c Suppresses the Secretion of Pro-inflammatory Cytokines in Macrophages
To detect the effect of the prophage protein Rv2650c on pro-inflammatory cytokines in macrophages, we further tested the expression of some cytokines by ELISA. The results showed that the ability of MS_Rv2650c to stimulate the secretion of cytokines TNF-α, IL-1β, IL-6, and IL-10 was lower than that of MS_Vec after 24 h of infection (paired t-test; P = 0.000006, P = 0.004, P = 0.01, P = 0.00002, respectively). The results indicated that the expression of Rv2650c significantly inhibited the secretion of pro-inflammatory factors in macrophages (Figure 5).
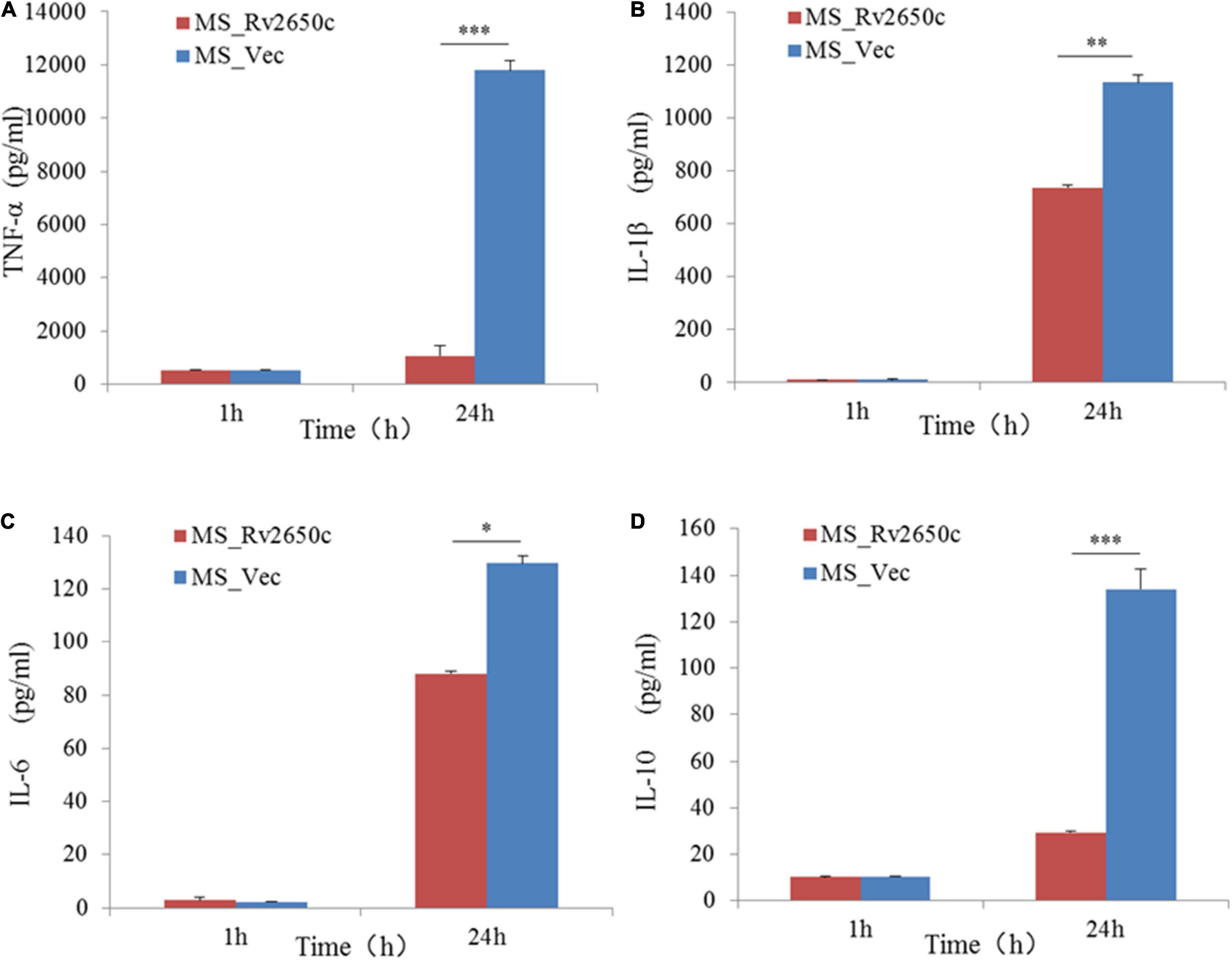
Figure 5. Assay of secretion of cytokines in macrophages. The macrophages were infected with recombinant M. smegmatis MS_Rv2650c and MS_Vec at MOI 10:1, respectively, and the secretion of cytokines TNF-α, IL-10, 1L-1β, and 1L-6 in macrophages was measured by ELSA. The supernatant at a certain time point was collected to detect the secretion of TNF-α (A), 1L-1β (B), 1L-6 (C), and IL-10 (D). Similar results were obtained in 3 independent experiments. (*p < 0.05; **p < 0.01; ***p < 0.001).
Discussion
In this study, we evaluated the function of the prophage gene Rv2650c in M. tuberculosis. The results of morphological experiments suggested Rv2650c affected the spread of colonies of M. smegmatis in the presence of Tween80. MS_Rv2650c showed stronger survivability than MS_Vec did, when they infected macrophages and faced adverse environments such as acidity, oxidative stress, and surfactants. Further, ELISA experiments confirmed that Rv2650c inhibit the expression of inflammatory factors TNF-α, IL-10, 1L-1β, and IL-6. These results demonstrated that the prophage protein Rv2650c promoted the survival of Mycobacterium in macrophages for the first time.
In 1998, it was first revealed that two prophages, phiRv1 and phiRv2, were hidden in the complete genome sequence of M. tuberculosis H37Rv (Cole et al., 1998). As black boxes, the significance and role of these two phages remain unclear. Some studies show that prophage is related to the evolution and adaptation of host bacteria to the harsh environment (Feiner et al., 2015; Argov et al., 2017; Keen and Dantas, 2018). For pathogens, these prophages are prominently related to the pathogenicity of its host (Feiner et al., 2015; Argov et al., 2017). Are prophages phiRv1 and phiRv2 and their phage proteins related to the pathogenicity of M. tuberculosis? Our previous study found that all sequenced strains of M. tuberculosis contained at least one prophage in the genome (Fan et al., 2014, 2016). And prophages do not exist in the M. smegmatis genome. Proteomics studies showed that some prophage genes in M. tuberculosis were translated and had function (Deng et al., 2013; Schubert et al., 2013). The transcriptome or expression profile data of M. tuberculosis under various stresses were explored (Fan et al., 2016), and it was found that some prophage genes were up-regulated under different environmental stresses. They may act as virulence genes or as regulatory genes to help M. tuberculosis resist the harsh environment in macrophages and to improve the pathogenicity of M. tuberculosis.
This study focuses on Rv2650c, a prophage phiRv2 gene. There is not homolog of Rv2650c in the chromosome of M. smegmatis based on blastx. Transcriptome analysis revealed that it was up-regulated in both the M. tuberculosis-infected macrophage model and persister model (Fan et al., 2016). Ka/Ks value (1.33) of Rv2650c gene indicating that the Rv2650c gene was strongly positively selected in M. tuberculosis. It may play an important role in the evolution of M. tuberculosis from environmental saprophytic bacterium to intracellular pathogen, possibly encoding virulence proteins involved in intracellular survival of M. tuberculosis.
Western blot result showed that protein Rv2650c was localized on the cell wall of M. smegmatis. Further microbiological experiments found that its expression enhanced the ability of M. smegmatis to survive in macrophages and the resistance to acidity, oxidative stress, and surfactants. The result of the growth curve showed that the growth kinetics of MS_Rv2650c and MS_Vec were similar, which excluded the possibility that more bacterial cells help M. smegmatis increase resistance to adverse environments. Prophage is a temperate phage genome or its fragment that is inserted into the bacterial chromosome. During the evolution process, bacteria will discard some inactive prophage genes obtained and choose to store prophage genes that can help the host grow or survive in the harsh environment (Obeng et al., 2016). As a prophage gene, Rv2650c encoded a functional protein localized on the cell wall that helped the host bacteria to resist harsh environments. Further, ELISA experiments also confirmed that some pro-inflammatory cytokines such as TNF, IL-10, IL-1β, and IL-6 were inhibited by prophage protein Rv2650c. The expression of inflammatory cytokines in macrophages play an important role in killing and eliminating M. tuberculosis (Lee et al., 2003; Krishnan et al., 2013; Li et al., 2016). We speculated that the M. tuberculosis prophage gene Rv2650c can inhibit the expression of inflammatory factors TNF, IL-1β, and IL-6 in macrophages to reduce the killing of M. smegmatis and promote intracellular survival of bacteria in macrophages.
In short, we have initially discovered that the prophage gene Rv2650c is of great significance for the survival of Mycobacterium. In the future, we will continue to study the function of the prophage gene and explore its mechanism in order to better clarify the mechanism of prophage in M. tuberculosis. This study provides novel insights into the discovery of virulence factors, as well as important new information for the role of prophage in the infection mechanism of M. tuberculosis and also builds the foundation for developing new drugs to control M. tuberculosis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XF and WL participated in the design of the study. XF and ZL analyzed the data and wrote the manuscript. ZL, ZW, HZ, MJ, and ZL helped to modify the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation (31600148 and 81601740), the Natural Science Foundation of Shandong Province (ZR2021MC018), the Shandong Excellent Young Scientist Award Fund (BS2014YY031), the Guangxi Natural Science Fund (2021GXNSFAA220039), and the grant of Neijiang Normal University (17CZ01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.819837/full#supplementary-material
Supplementary Figure 1 | Map of plasmid pNIT-Myc-Rv2650c.
Supplementary Figure 2 | Comparison of the morphology of the recombinant M. smegmatis MS_Rv2650c and MS_Vec. (A) MS_Rv2650c and MS_Vec strains were cultured in 7H9 medium with 0.05% (v/v) Tween 80 and 28 mM ε-caprolactam at 37°C until an OD600 of 0.6–0.8 were reached. The cultures were kept at room temperature for 30 min. (B) The cultures were harvested for scanning electron microscopy (SEM) analysis.
References
Argov, T., Azulay, G., Pasechnek, A., Stadnyuk, O., Ran-Sapir, S., Borovok, I., et al. (2017). Temperate bacteriophages as regulators of host behavior. Curr. Opin. Microbiol. 38, 81–87. doi: 10.1016/j.mib.2017.05.002
Awuh, J. A., and Flo, T. H. (2017). Molecular basis of mycobacterial survival in macrophages. Cell. Mol. Life Sci. 74, 1625–1648.
Cambier, J. C., Stanley, F., and Ramakrishnan, L. (2014). Host evasion and exploitation schemes of mycobacterium tuberculosis. Cell 159, 1497–1509. doi: 10.1016/j.cell.2014.11.024
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Deng, W., Fan, X., Wang, X., He, Y., Li, Z., Song, J., et al. (2013). Mycobacterium tuberculosis biology revealed by proteome profiling and integration of Multi-omics Data—Proteomics Insight into M. tuberculosis systems biology. Curr. Proteomics 10, 261–268.
Ehrt, S., Schnappinger, D., and Rhee, K. Y. (2018). Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 496–507. doi: 10.1038/s41579-018-0013-4
Etna, M. P., Giacomini, E., Severa, M., and Coccia, E. M. (2014). Pro- and anti-inflammatory cytokines in tuberculosis: a two-edged sword in TB pathogenesis. Semin. Immunol. 26, 543–551. doi: 10.1016/j.smim.2014.09.011
Fan, X., Abd Alla, A. A. E., and Xie, J. (2016). Distribution and function of prophage phiRv1 and phiRv2 among Mycobacterium tuberculosis complex. J. Biomol. Structure Dynamics 34, 233–238. doi: 10.1080/07391102.2015.1022602
Fan, X., Xie, L., Li, W., and Xie, J. (2014). Prophage-like elements present in Mycobacterium genomes. BMC Genomics 15:243. doi: 10.1186/1471-2164-15-243
Feiner, R., Argov, T., Rabinovich, L., Sigal, N., Borovok, I., and Herskovits, A. A. (2015). A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 13, 641–650. doi: 10.1038/nrmicro3527
Ghosh, G., Reddy, J., Sambhare, S., and Sen, R. (2018). A bacteriophage capsid protein is an inhibitor of a conserved transcription terminator of various bacterial pathogens. J. Bacteriol. 200, e380–e317. doi: 10.1128/JB.00380-17
Jayachandran, R., Sundaramurthy, V., Combaluzier, B., Mueller, P., Korf, H., Huygen, K., et al. (2007). Survival of mycobacteria in macrophages is mediated by Coronin 1-Dependent activation of calcineurin - sciencedirect. Cell 130, 37–50. doi: 10.1016/j.cell.2007.04.043
Kapopoulou, A., Lew, J. M., and Cole, S. T. (2011). The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis 91, 8–13. doi: 10.1016/j.tube.2010.09.006
Keen, E. C., and Dantas, G. (2018). Close encounters of three kinds: bacteriophages, commensal bacteria, and host immunity. Trends Microbiol. 26, 943–954. doi: 10.1016/j.tim.2018.05.009
Krishnan, N., Robertson, B. D., and Thwaites, G. (2013). Pathways of IL-1β secretion by macrophages infected with clinical Mycobacterium tuberculosis strains. Tuberculosis 93, 538–547. doi: 10.1016/j.tube.2013.05.002
Lee, J. S., Song, C. H., Lim, J. H., Kim, H. J., Park, J. K., Paik, T. H., et al. (2003). The production of tumour necrosis factor-alpha is decreased in peripheral blood mononuclear cells from multidrug-resistant tuberculosis patients following stimulation with the 30-kDa antigen of Mycobacterium tuberculosis. Clin. Exp. Immunol. 132, 443–449. doi: 10.1046/j.1365-2249.2003.02172.x
Li, W., Liu, M., and Xie, J. (2016). Rv3369 Induces cytokine interleukin-1β production and enhances Mycobacterium smegmatis intracellular survival. J. Interferon Cytokine Res. 36, 140–147. doi: 10.1089/jir.2015.0090
Li, W., Zhao, Q., Deng, W., Chen, T., and Liu, M. (2014). Mycobacterium tuberculosis Rv3402c enhances mycobacterial survival within macrophages and modulates the host pro-inflammatory cytokines production via NF-Kappa B/ERK/p38 signaling. PLoS One 9:e94418. doi: 10.1371/journal.pone.0094418
Misra, S. K., Milohanic, E., Aké, F., Mijakovic, I., Deutscher, J., Monnet, V., et al. (2011). Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165. doi: 10.1002/pmic.201100259
Obeng, N., Pratama, A. A., and Elsas, J. D. V. (2016). The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol. 24, 440–449. doi: 10.1016/j.tim.2015.12.009
Pandey, A. K., Raman, S., Proff, R., Joshi, S., Kang, C. M., Rubin, E. J., et al. (2009). Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb) 89, 12–16. doi: 10.1016/j.tube.2008.07.007
Queval, C. J., Song, O. R., Carralot, J. P., Saliou, J. M., Bongiovanni, A., Deloison, G., et al. (2017). Mycobacterium tuberculosis controls phagosomal acidification by Targeting CISH-Mediated signaling. Cell Rep. 20, 3188–3198. doi: 10.1016/j.celrep.2017.08.101
Salmond, G. P. C., and Fineran, P. C. (2015). A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786. doi: 10.1038/nrmicro3564
Schubert, O. T., Mouritsen, J., Ludwig, C., Röst, H. L., Rosenberger, G., Arthur, P. K., et al. (2013). The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13, 602–612. doi: 10.1016/j.chom.2013.04.008
Keywords: Mycobacterium, pathogenic mechanism, prophage, Rv2650c, intracellular survival
Citation: Fan X, Liu Z, Wan Z, Zou H, Ji M, Sun K, Gao R, Li Z and Li W (2022) Prophage Gene Rv2650c Enhances Intracellular Survival of Mycobacterium smegmatis. Front. Microbiol. 12:819837. doi: 10.3389/fmicb.2021.819837
Received: 23 November 2021; Accepted: 29 December 2021;
Published: 17 January 2022.
Edited by:
Heejoon Myung, Hankuk University of Foreign Studies, South KoreaReviewed by:
Quanxin Long, Chongqing Medical University, ChinaPrasit Palittapongarnpim, Mahidol University, Thailand
Copyright © 2022 Fan, Liu, Wan, Zou, Ji, Sun, Gao, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyu Fan, Znh5c25kQDEyNi5jb20=; Zhongfang Li, bGl6aG9uZ2ZhbmcwOEAxMjYuY29t; Wu Li, bGl3dTA1MDNAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Xiangyu Fan
Xiangyu Fan Zichen Liu1†
Zichen Liu1† Zhongfang Li
Zhongfang Li