- 1MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI, United States
- 2Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI, United States
- 3Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, United States
The orange carotenoid protein (OCP) family of proteins are light-activated proteins that function in dissipating excess energy absorbed by accessory light-harvesting complexes, i.e., phycobilisomes (PBSs), in cyanobacteria. Some cyanobacteria contain multiple homologs of the OCP-encoding gene (ocp). Fremyella diplosiphon, a cyanobacterium studied for light-dependent regulation of PBSs during complementary chromatic acclimation (CCA), contains several OCP homologs – two full-length OCPs, three Helical Carotenoid Proteins (HCPs) with homology to the N-terminus of OCP, and one C-terminal domain-like carotenoid protein (CCP) with homology to the C-terminus of OCP. We examined whether these homologs are distinctly regulated in response to different environmental factors, which could indicate distinct functions. We observed distinct patterns of expression for some OCP, HCP, and CCP encoding genes, and have evidence that light-dependent aspects of ocp homolog expression are regulated by photoreceptor RcaE which controls CCA. RcaE-dependent transcriptional regulator RcaC is also involved in the photoregulation of some hcp genes. Apart from light, additional environmental factors associated with cellular redox regulation impact the mRNA levels of ocp homologs, including salt, cold, and disruption of electron transport. Analyses of conserved sequences in the promoters of ocp homologs were conducted to gain additional insight into regulation of these genes. Several conserved regulatory elements were found across multiple ocp homolog promoters that potentially control differential transcriptional regulation in response to a range of environmental cues. The impact of distinct environmental cues on differential accumulation of ocp homolog transcripts indicates potential functional diversification of this gene family in cyanobacteria. These genes likely enable dynamic cellular protection in response to diverse environmental stress conditions in F. diplosiphon.
Introduction
Oxygenic photosynthesis, a process comprised of a series of light-driven redox reactions that results in the production of ATP and NADPH and ultimately carbohydrates, is a characteristic feature of the metabolism of cyanobacteria. Under some conditions, such as excessive light exposure or during nutrient deprivation, cyanobacteria can become unable to effectively utilize the light energy absorbed for photosynthesis. To modulate the potential negative impacts of overexcitation, while maintaining reasonable levels of photosynthesis, many photosynthetic organisms exhibit cellular responses that allow them to tune the pigment content and size of their light-harvesting complexes in response to the external photoenvironment. Fremyella diplosiphon (also known as Tolypothrix sp. PCC 7601; Yerrapragada et al., 2015) is a cyanobacterium that exhibits a form of light-dependent regulation, or photomorphogenesis, known as chromatic acclimation (Montgomery, 2016, 2017; Sanfilippo et al., 2019). Fremyella diplosiphon exhibits a particular form of chromatic acclimation known as complementary chromatic acclimation (CCA) that involves tuning cellular pigmentation and morphology to changes in available green versus red light (Bennett and Bogorad, 1973; Bordowitz and Montgomery, 2008). Complementary chromatic acclimation is regulated by the red/green-responsive photoreceptor RcaE (Kehoe and Grossman, 1996; Bordowitz and Montgomery, 2008).
Complementary chromatic acclimation is critical to maintain light-harvesting complexes tuned to available light quality and quantity. Despite this, cells in dynamic environments can encounter an imbalance between absorbed and utilized energy that can lead to an overly reduced photosynthetic electron transport pathway and the production of harmful reactive oxygen species (ROS; Busch and Montgomery, 2015a). The intracellular accumulation of ROS can result in cellular damage, including lipid oxidation or protein damage. This damage can ultimately result in reduced cellular fitness. To combat such potentially harmful outcomes, photosynthetic organisms possess mechanisms to protect cells from overexcitation by light. These photoprotective mechanisms include non-photochemical quenching (NPQ), or dissipation of excess absorbed light energy as heat (Montgomery, 2014).
Cyanobacteria, including F. diplosiphon, specifically use NPQ of the phycobilisome (PBS), the light-harvesting cyanobacterial antenna, under high blue-green light to protect against excess light absorption, which would cause cellular damage from the generation of ROS. NPQ is mediated in cyanobacteria by the orange carotenoid protein (OCP), which is found encoded in the genomes of numerous cyanobacteria (Melnicki et al., 2016; Bao et al., 2017a; Kerfeld et al., 2017). The OCP protein serves to mitigate absorption of excess light and to avoid the generation of ROS and associated cellular damage, through both NPQ and direct singlet oxygen (1O2) scavenging (Wilson et al., 2006, 2008; Kirilovsky and Kerfeld, 2013; Sedoud et al., 2014). Orange carotenoid protein, the isolated carotenoid-binding α-helical domain of OCP known as the red carotenoid protein (RCP; Kerfeld et al., 2003) and some Helical Carotenoid Proteins (HCPs) with homology to the N-terminal domain of OCP can function as 1O2 quenchers (Lopez-Igual et al., 2016). Recent analysis of a collection of cyanobacterial genomes has revealed that many cyanobacteria possess multiple homologs of ocp, including full-length variants or proteins homologous to either the N-terminal domain or C-terminal domain of OCP, and that very few cyanobacteria possess the same set of OCP homologs (Melnicki et al., 2016). Characterization of four ocp N-terminal domain homologs (also called helical carotenoid proteins or HCPs) from the cyanobacterium Anabaena sp. PCC 7120 revealed functional diversity between these proteins. One HCP protein was able to quench phycobilisome fluorescence in a manner similar to the full length OCP, whereas two were good scavengers of singlet oxygen and the fourth showed neither activity (Lopez-Igual et al., 2016). Six ocp homologs are found in F. diplosiphon, which include two full-length ocp homologs, three hcp homologs and one C-terminal domain-like carotenoid protein-encoding (ccp) gene (Bao et al., 2017b; Kerfeld et al., 2017). The physiological roles of the six F. diplosiphon ocp homologs are still under investigation. However, biochemical analyses have demonstrated distinct temperature-dependent properties and quenching of PBS fluorescence for full length OCP1 and OCP2 from F. diplosiphon (Bao et al., 2017b), despite comparable spectroscopic properties and photoactivation mechanisms (Kuznetsova et al., 2020). Additionally, initial biochemical analyses of carotenoproteins HCP2, HCP3, and CCP2 described distinct carotenoid-binding properties and excited-state dynamics (Dominguez-Martin et al., 2019, 2020; Khan et al., 2020).
Under environmental stress conditions such as cold or salt stress, photosynthetic cells often exhibit increased propensity for damage from excess light exposure. Under such conditions, cells are expected to have an increased need to protect themselves from overexcitation of PBSs. One response to such stress is for organisms to reduce cellular photosynthetic pigment content. OCP homologs are hypothesized to play critical roles in protecting cells under salt stress, cold, or other stresses.
We hypothesized that the six distinct carotenoproteins encoded by ocp homologs in F. diplosiphon have unique functions. Prior analyses reported transcriptional regulation of some ocp homolog genes, as well as distinct modes of regulation based on gene expression data showing that OCP2 is regulated at the level of mRNA accumulation, whereas OCP1 primarily appears to be regulated post-transcriptionally or alternatively by post-translational control by fluorescence recovery protein (FRP) or protein oligomerization (Bao et al., 2017a). The expression of ocp1, which is differentially regulated by sinusoidal versus fluctuating light, is also associated with differential organismal fitness under dynamic conditions (Agostoni et al., 2016). For this study, we used quantitative real-time PCR (qPCR) to measure the expression levels of the six ocp homologs and the frp gene in F. diplosiphon cultures grown under various conditions and in various mutant strains to investigate their environmental and genetic regulation. The results of this study show that patterns of expression for the ocp homologs and the frp gene fall into three classes. The first class being ocp1 and frp, the second class being ocp2 and the third class being hcp1, hcp2, hcp3, and ccp2. The distinct expression patterns suggest specialized roles for these homologs in response to different environmental conditions associated with photoacclimation of cells to dynamic light conditions to promote photoprotection, thereby supporting increased photosynthetic efficiency and organismal productivity.
Materials and Methods
Strains and Growth
A short-filament strain of F. diplosiphon UTEX 481, known as SF33, was used as wild-type (WT; Cobley et al., 1993); additionally F. diplosiphon strains deficient in rcaE (Kehoe and Grossman, 1996) and rcaC (Bordowitz and Montgomery, 2008) were used in this study. Cultures were maintained in BG-11 medium (Allen, 1968) with 20 mM HEPES at pH 8.0 (hereafter, BG-11/HEPES) under white fluorescent light (WL; F20T12-P/AQ) at 15 μmol m–2 s–1 at 28°C while being shaken at 175 rpm. For experiments involving red or green light, cultures were grown under red light (RL; λmax at 660 nm, LED wholesalers, model 2506RD, Hayward, CA) or green light (GL; λmax at 530 nm; Geneva Scientific LLC, Williams Bay, WI) LEDs at 10, 20, 50, or 100 μmol m–2 s–1 as indicated. Light intensities were measured with a LI-250A light meter (LI-COR, Lincoln, NE) equipped with a quantum sensor (model LI-190SA).
Cultures grown for experiments were diluted to an initial optical density at 750 nm (OD750) of ∼0.1 in a volume of 50 ml of BG-11/HEPES or other variants of BG-11/HEPES medium as indicated and then grown for the indicated time period (e.g., 24 h up to ∼1 week) under the indicated light conditions. Generally 18–24 h before harvesting cells for RNA extraction, the OD750 of each culture was checked and adjusted to ∼0.6. To measure the rapid response to stress treatments and avoid long-term growth impacts due to stress, stress treatments were applied for up to 24 h. For experiments involving cold stress, cultures were transferred to a growth chamber with a temperature of 15 or 8°C for the final 18–24 h of growth. Other samples were treated with either 10 μM of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) (v/v) or 0.03 μM of methyl viologen (MV) (v/v) for the final 24 h before harvesting cells. For salt stress or nitrogen-limitation, samples growing in BG-11/HEPES medium were harvested once the concentration reached 0.6 OD and resuspended in media with 0.2 M salt (NaCl) or lacking nitrogen then incubated for an additional 24 h. The control samples were resuspended in the original media and grown for the additional 24 h. Cell pellets were harvested by chilling samples to ∼5°C by swirling each flask in liquid nitrogen and then pelleted by centrifugation at 5,000 rpm at 4°C for 10 mins. The resulting pellets were snap frozen with liquid nitrogen.
RNA Extraction and Quantitative Real-Time PCR
RNA was extracted from harvested samples of F. diplosiphon as previously described (Seib and Kehoe, 2002; Stowe-Evans et al., 2004) and treated with a TURBO DNA-free kit (Ambion, Austin, TX). Concentrations of isolated RNA were determined with a NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA). For each sample, 0.5 μg of RNA in 16 μl of ddH2O was treated with 4 μl of QuantaBio Reverse Transcriptase Master Mix (Quantabio, Beverly, MA) according to the manufacturer’s instruction. An additional aliquot of each sample was treated identically without the addition of the RT master mix for a No-RT control. After reverse transcription, each sample was diluted eight-fold. Four μl of cDNA was used for each qPCR reaction with 6 μl of Fast SYBR Green Master Mix (Applied Biosystems, Waltham, MA). qPCR was performed using a QuantStudio 7 Flex Real Time PCR System (Thermo Fisher Scientific, Waltham, MA). orf10B, a gene that has been shown to be consistently expressed under GL and RL (Stowe-Evans et al., 2004), was used as an internal reference gene. For a positive control in salt-stress conditions, we used FdTSPO3 which is a full-length homolog of TSPO with a known role in the organism’s physiological response to salt stress (Busch and Montgomery, 2017). For the samples grown under nitrogen limitation and salt stress, the reference gene used was rnpA. Three technical replicates and at least four biological replicates were used for each condition. Primer sets are listed in Table 1. All qPCR procedures and analyses were performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009).
Promoter Analysis
Using the gene search tool of the Integrated Microbial Genomes and Microbiomes (IMG/M) system available from the Department of Energy’s (DOE’s) Joint Genome Institute (JGI), the upstream intergenic DNA sequence was isolated for each ocp homolog to determine the region containing the gene promoter. The upstream region of ocp2 was an exception; the intergenic region consisted of too few base pairs, requiring the sequence used to determine the promoter region to overlap with the 3′ end of the upstream gene. The SoftBerry program to predict bacterial promoters, i.e., BPROM (Solovyev and Salamov, 2011), was used to predict the operon sequence once the target sequence upstream of the ORF start was determined. The algorithm of the BPROM program predicts transcription start positions for bacterial genes regulated by sigma70 promoters. The output includes the position of predicted promoters as well as transcription factor binding sites for each predicted promoter.
Results
Relative Expression of Orange Carotenoid Protein Homologs in Fremyella diplosiphon Under Varied Light Conditions
To investigate whether ocp homologs are controlled at the transcriptional level in response to changes in light availability, cultures of WT F. diplosiphon were grown under various intensities of green light (GL) and red light (RL) and RNA was harvested so that the mRNA levels of ocp homologs could be measured using qPCR. An observed tight correlation between the expression of ocp1 and frp was not surprising given their adjacent positions in the F. diplosiphon genome and the fact that the FRP protein is essential for regulating the function of OCP1 (Bao et al., 2017b). Relative expression of ocp1 and frp trended higher in low RL relative to low GL, and levels somewhat decreased with increased light intensity, although these differences were not significant (Figure 1A).
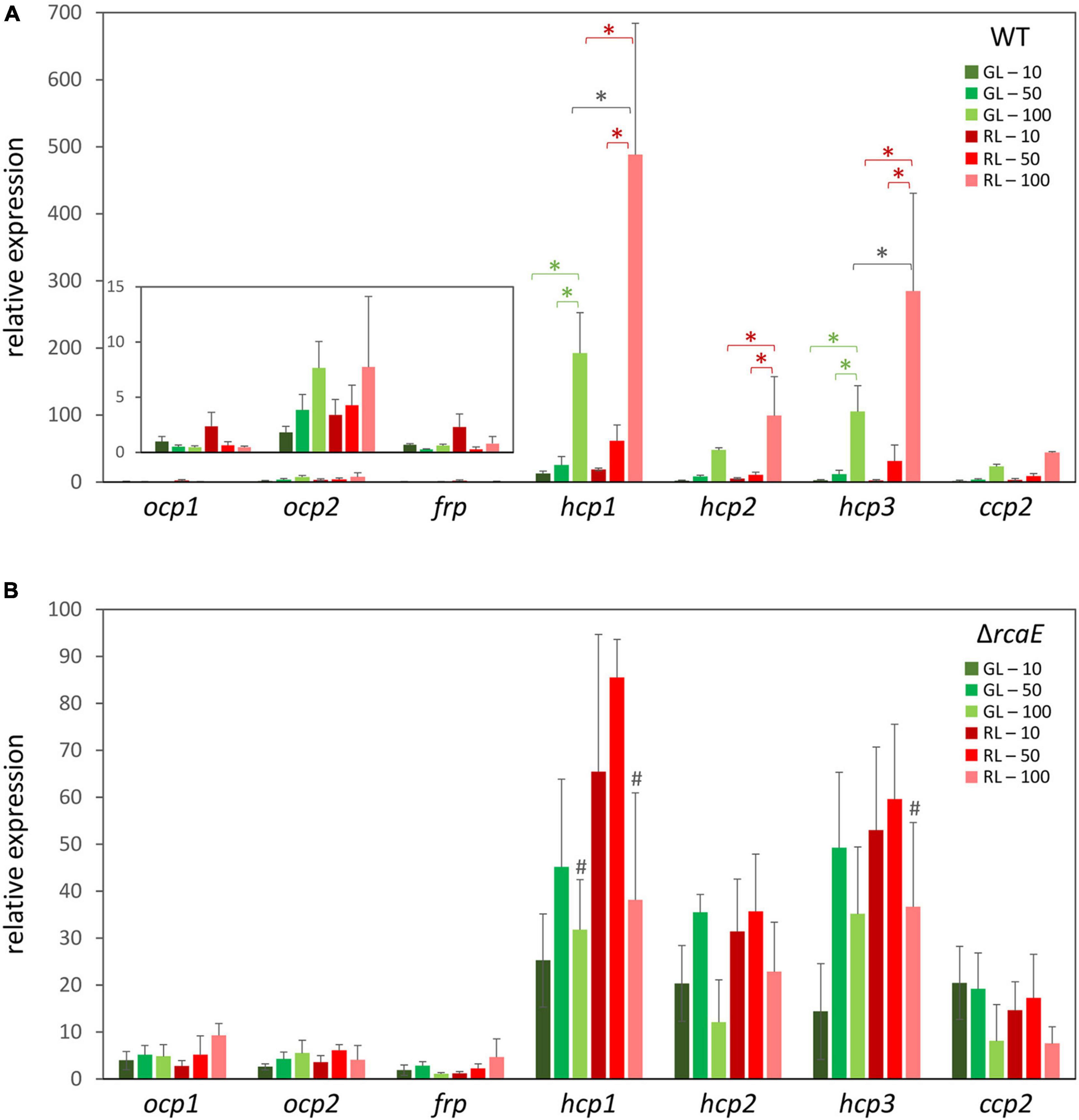
Figure 1. Expression of orange carotenoid protein (ocp) homologs under varying light conditions. Cultures of wild type (A) and ΔrcaE (B) Fremyella diplosiphon were grown under green light (GL) and red light (RL) at light intensities of 10 (dark green and dark red), 50 (green and red), and 100 (light green and light red) μmol m–2 s–1. Relative expression of ocp1, ocp2, frp, hcp1, hcp2, hcp3, and ccp2 were measured using quantitative real-time PCR (qPCR). Expression under each condition is shown relative to expression under 10 μmol m–2 s–1 of green light. Error bars represent the standard deviation of each measurement. *p < 0.05, with dark gray stars indicating a difference for a strain in GL compared to RL at a specific light intensity, and red or green stars indicating a difference between different intensities of RL or GL, respectively; #p < 0.05 representing a difference between WT and ΔrcaE in identical light conditions as determined by a ANOVA with Tukey-Kramer HSD post hoc test.
Distinctly, expression of ocp2 trended upward under increased intensities of either GL or RL (Figure 1A). These differences were not significant, perhaps due to larger standard error at higher light intensities that represent conditions under which cells experience stress. Each of the remaining ocp homologs, hcp1, hcp2, hcp3, and ccp2, shared a common pattern of changes in expression that was consistent across several experimental conditions. For each of these genes growth under RL trended higher than GL, with a significant increase in expression for RL relative to samples grown GL at the highest light intensity specifically for hcp1 and hcp3 (Figure 1A). Likewise, an increase in light intensity caused an increase in mRNA levels for hcp1 and hcp3 in GL and all three hcp genes in RL. Although mRNA levels trended higher for ccp2 at high intensity GL and RL, these differences were not significant.
Relative Expression of Orange Carotenoid Protein Homologs in Complementary Chromatic Acclimation-Deficient Strains of Fremyella diplosiphon
Given the apparent distinctions of the impact of red versus green light on the expression levels of some ocp homologs, we investigated whether known CCA regulator RcaE plays a role in the photoregulation of these light-dependent genes. Prior RNA-sequencing-based analysis indicated light- and RcaE-dependent differences in mRNA counts for ocp homologs, at least for those homologs for which mRNA counts were detected (Table 2). Here, cultures of the CCA photoreceptor-deficient ΔrcaE strain of F. diplosiphon were grown under the same light conditions as the previous experiments with WT (Figure 1A) and RNA was extracted for qPCR analyses. Expression of ocp1, ocp2, and frp showed limited variation under various intensities of green or red light in the ΔrcaE strain (Figure 1B). Of note, the RL vs. GL difference in hcp1 and hcp3 and increased expression in response to increased light intensity observed for all three hcp genes in WT (Figure 1A) were no longer observed in the ΔrcaE mutant (Figure 1B). Specifically, the ΔrcaE strain exhibit lower levels of hcp1 mRNA in both GL and RL compared to WT, and lower levels of hcp3 mRNA in RL compared to WT for strains grown at 100 μmol m–2 s–1 (Figure 1B). Of note, samples of ΔrcaE grown at 100 μmol m–2 s–1 showed poor rates of growth and cultures were unable to reach the OD750 of 0.6 at which the other samples were harvested.
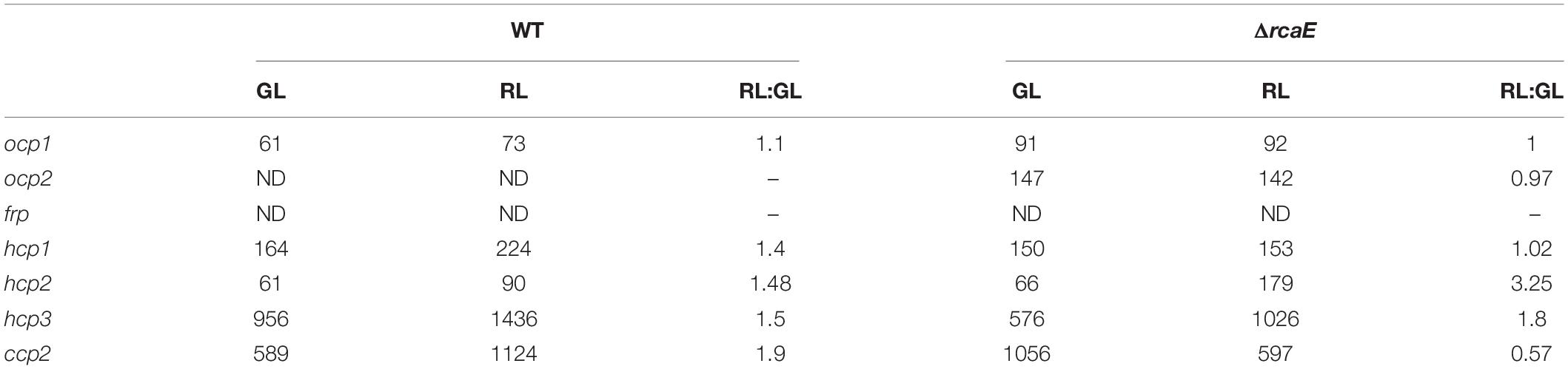
Table 2. RNA-sequencing-based analysis of expression of orange carotenoid protein (OCP), fluorescence recovery protein (FRP), helical carotenoid proteins (HCP), and C-terminal domain-like carotenoid protein (CCP) homologs in wild-type (WT) and RcaE photoreceptor-deficient strains of chromatically-acclimating Fremyella diplosiphon grown under green light (GL), red light (RL), or for RL vs. GL (RL:GL) counts for each strain.
Two effectors, RcaF (Kehoe and Grossman, 1997), which is a small response regulator that lacks an output domain, and RcaC (Kehoe and Grossman, 1996; Li and Kehoe, 2005; Alvey et al., 2007; Li et al., 2008), which is a DNA-binding transcription factor, function downstream of RcaE in the photoregulation of pigmentation. Given the loss of light-dependent regulation of select hcp genes in the ΔrcaE strain and the central role of DNA-binding factor RcaC in direct transcriptional regulation, we also assessed transcript accumulation for ocp homologs in a ΔrcaC strain of F. diplosiphon. Whereas the levels of mRNA accumulation for ocp1, ocp2, and frp were not significantly impacted compared to WT grown in identical conditions, levels of hcp1 and hcp3 were significantly different in the ΔrcaC strain compared to WT in GL conditions (Figure 2).
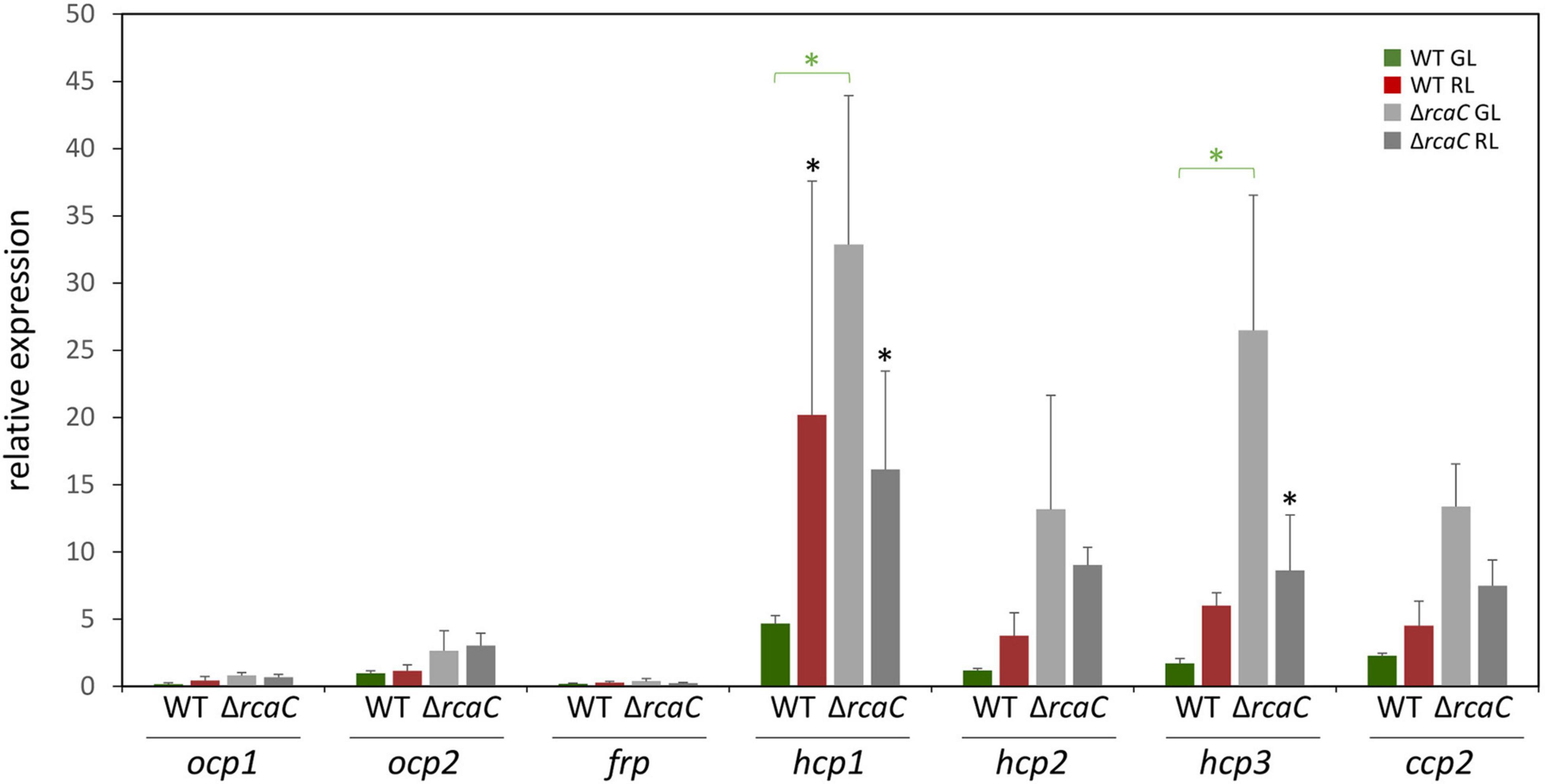
Figure 2. Expression of ocp homologs in the ΔrcaC strain of F. diplosiphon. Wild type and ΔrcaC strains of F. diplosiphon cultures were grown under green and red light at light intensities of 20 μmol m–2 s–1. Relative expression of ocp1, ocp2, frp, hcp1, hcp2, hcp3, and ccp2 were measured using qPCR, comparing samples from the ΔrcaC mutant to samples from SF33. (±SD, n = 4). *p < 0.05, with dark gray stars indicating a difference for a strain in GL compared to RL, and green stars indicating a difference between WT and the ΔrcaC mutant grown in GL as determined by a ANOVA with Tukey-Kramer HSD post hoc test.
Relative Expression of Orange Carotenoid Protein Homologs in Low Temperature Conditions
Previous research demonstrated that the ability of OCP1 and OCP2 to reach and maintain their active (OCPR) state is temperature dependent (Bao et al., 2017b). Nearly 100% of OCP1 protein suspended in solution reaches the activated OCPR state at both 15 and 8°C; however, the process is significantly slower for OCP1 at the lower temperature. OCP2 exhibits a distinct response to cold, with a limited portion of OCP2 (34%) reaching the activated state at 15°C, while 92% of OCP2 reaches the activated OCPR state at 8°C (Bao et al., 2017b). To determine if the temperature-dependent characteristics of OCP1 and OCP2 protein are reflected in the expression of ocp homologs, qRT-PCR was performed with samples of F. diplosiphon grown at 28°C then transitioned to 8°C or 15°C for 24 h. At 8°C, ocp1 and frp were significantly upregulated ∼160 fold compared to the control samples that were left at 28°C, while ocp2 expression was significantly increased ∼260 fold (Figure 3A). This cold-dependent upregulation was the largest change in expression observed for any of the ocp family genes in this study. The three hcp homologs and ccp2 were significantly downregulated ∼2 fold, or more for hcp2, at 8°C compared to the control samples (Figure 3A). At 15°C, ocp1 was upregulated 7-fold and frp was upregulated greater than 20-fold, whereas ocp2 was not impacted significantly (Figure 3B). None of the three hcp genes exhibited a significant change in mRNA levels, whereas ccp2 was downregulated 2-fold in response to 15°C.
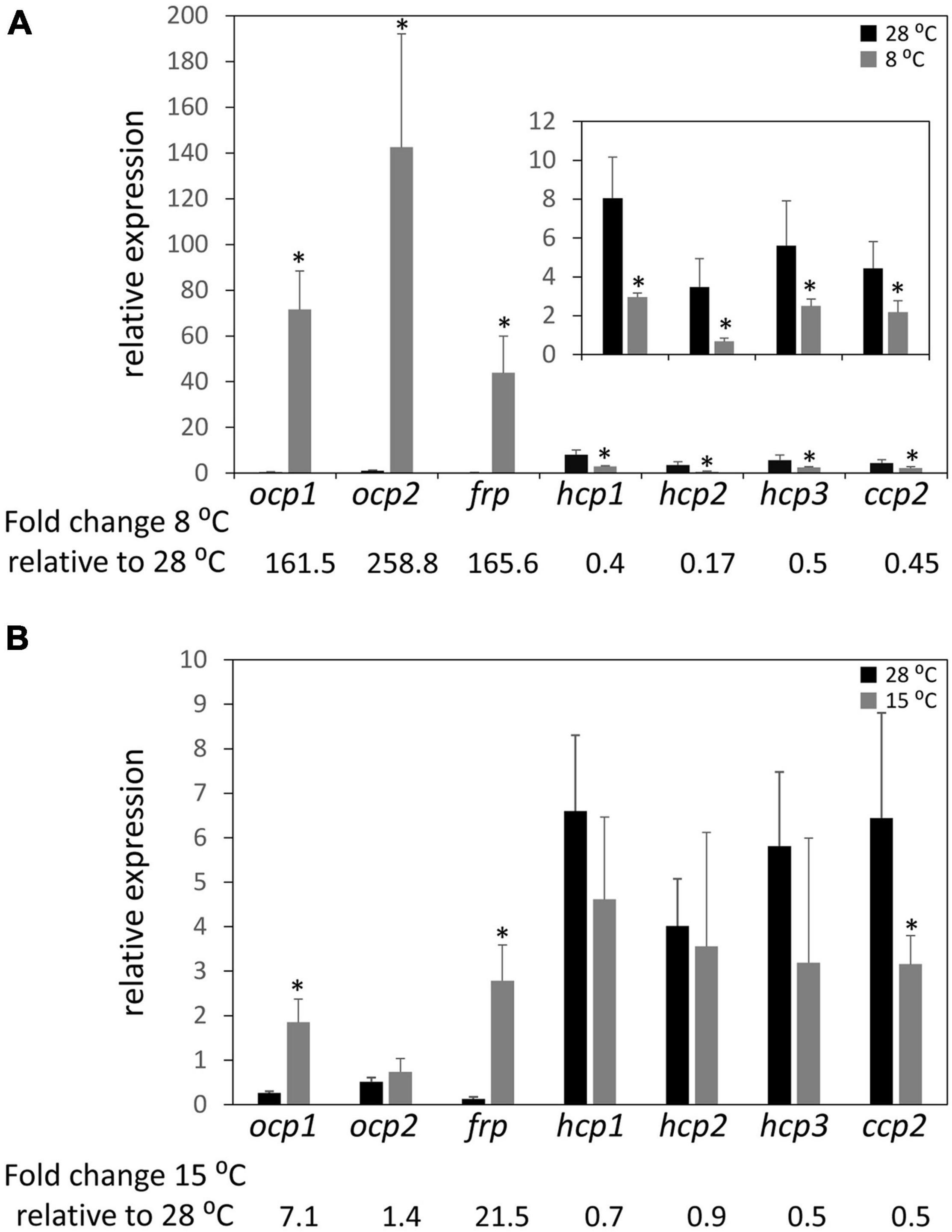
Figure 3. Cold-dependent expression of ocp homologs. Relative expression of ocp1, ocp2, frp, hcp1, hcp2, hcp3, and ccp2 were measured using qPCR, comparing samples incubated for 24 h at (A) 8°C or (B) 15°C to control samples maintained at 28°C (±SD, n = 6). *p < 0.05 as determined by a two tailed Student’s t-test for the comparison of the treated and untreated samples.
Relative Expression of Orange Carotenoid Protein Homologs Under Salt Stress
Given the association of salt stress with an upregulation of ocp expression in Synechocystis sp. PCC 6803 in prior research (Fulda et al., 2006), we measured levels of mRNA for ocp homologs in salt-stressed F. diplosiphon cultures. With the exception of ocp1 and ocp2 that were not impacted and frp that exhibited lower mRNA accumulation in the presence of salt, expression of each hcp gene and ccp2 was significantly higher in WT cultures under salt stress relative to control conditions, similar to the F. diplosiphon salt-stress marker gene tspO3 (Figure 4). The three homologs of the N-terminal domain were the most highly over-expressed—more than 30-fold upregulation was observed for these genes.
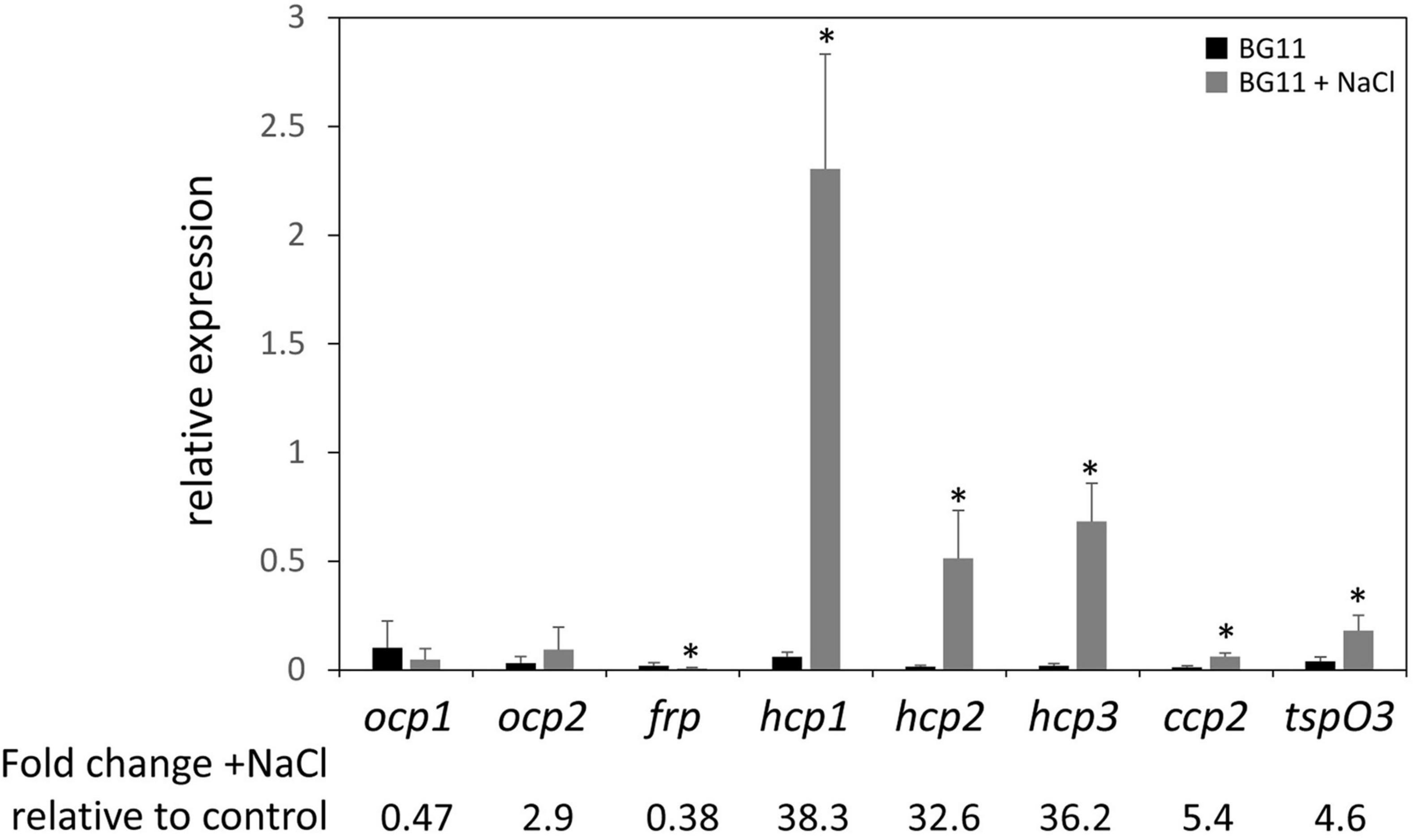
Figure 4. Expression of ocp homologs in response to salt stress. The expression levels of ocp1, ocp2, frp, hcp1, hcp2, hcp3, and ccp2 were measured using qPCR, comparing samples incubated for 24 h in BG-11/HEPES + 0.2 M NaCl to control samples grown in BG-11/HEPES (±SD, n = 6). *p < 0.05 as determined by a two tailed Student’s t-test for the comparison of the treated and untreated samples.
Relative Expression of Orange Carotenoid Protein Homologs Under Nitrogen-Limitation Stress
Nitrogen (N) limitation has been previously associated with altered OCP protein accumulation in a proteomic study with Microcystis aeruginosa (Yue et al., 2015). Additionally, PBSs are degraded in response to N limitation (Collier and Grossman, 1994), suggesting that one of the roles for OCP and homologs in interacting with PBS to dissipate excess absorption of light energy may not be needed to the same degree under these conditions. Together these prior findings related to OCP and PBSs in N-limited conditions suggested to us that ocp homolog expression may be altered in response to N limitation. Based on qPCR analyses, expression of each ocp homolog trended down in WT cells exposed to N limitation compared to growth in N-replete medium. However, the difference was significant only for ocp1, frp, and hcp3 (Figure 5).
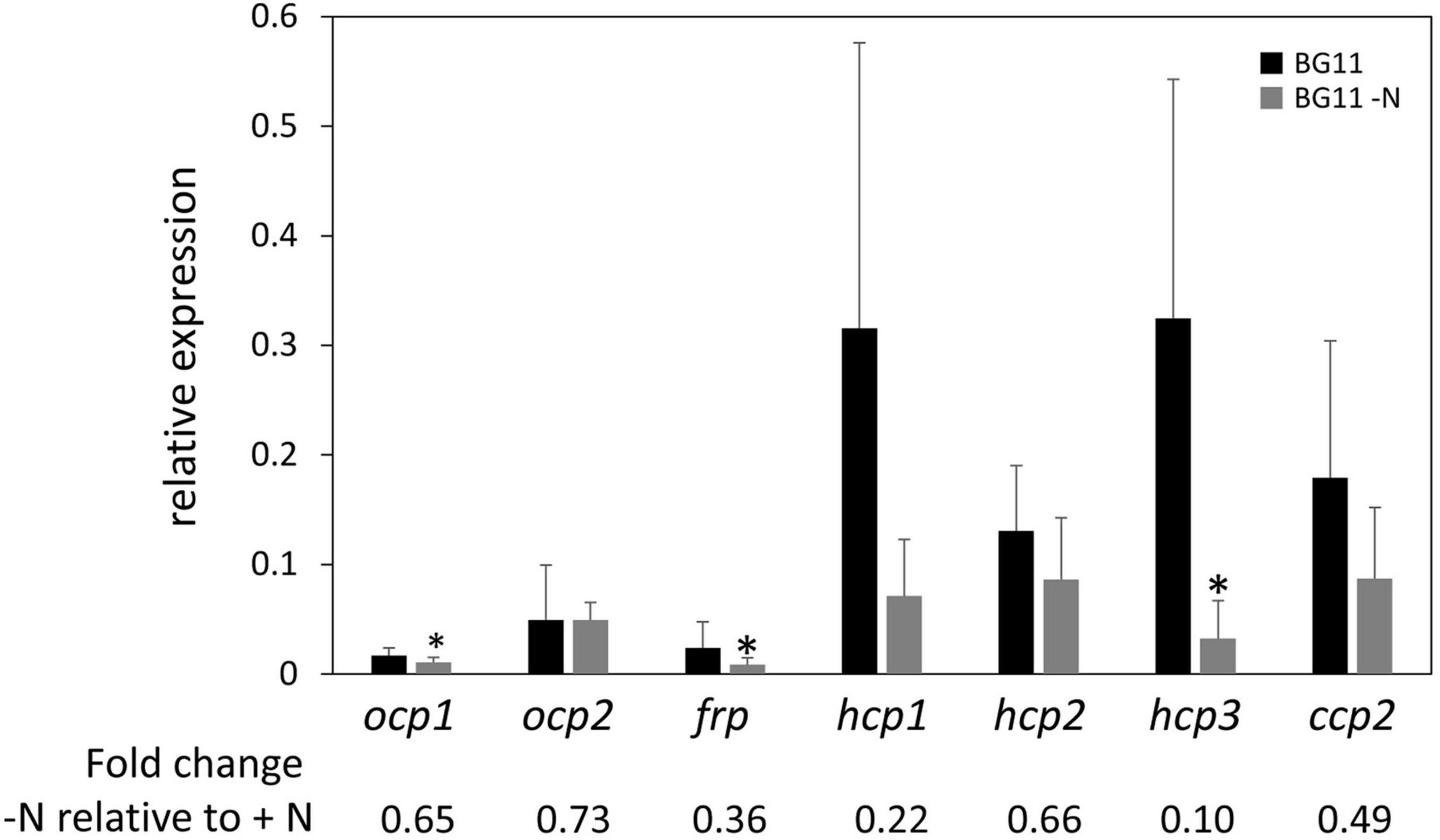
Figure 5. Expression of ocp homologs in response to nitrogen limitation stress. The expression levels of ocp1, ocp2, frp, hcp1, hcp2, hcp3, and ccp2 were measured using qPCR, comparing samples incubated for 24 h in BG-11/HEPES without nitrogen to control samples maintained in BG-11/HEPES with nitrogen (±SD, n = 6). *p < 0.05 as determined by a two tailed Student’s t-test for the comparison of the treated and untreated samples.
Relative Expression of Orange Carotenoid Protein Homologs in Presence of Methyl Viologen and 3-(3,4-Dichlorophenyl)-1,1-Dimethylurea
The upregulation of some ocp homologs in the ΔrcaE strain, which exhibits high ROS accumulation relative to WT (Singh and Montgomery, 2012), as well as the general trend of increased expression of hcp genes in conditions that generally correspond to higher levels of cellular ROS levels led us to further examine the expression of ocp homologs in response to induced redox stress. To probe the impact of oxidative stress on ocp homologs, we treated cells with methyl viologen (MV; Fujii et al., 1990). MV acts as an artificial electron acceptor from photosystems, and thereby results in a disruption in electron transport activity that is associated with ROS generation and subsequently oxidative stress in plants (Kim and Lee, 2002) and cyanobacteria (Busch and Montgomery, 2015b; Oh and Montgomery, 2019). We conducted qPCR experiments with WT samples treated with 0.03 μM of MV and incubated for 24 h to induce oxidative stress. Expression of ocp homologs in the presence of MV was generally increased relative to their expression in untreated samples, although the difference observed was only significant for hcp1 and hcp3 (Figure 6A). The greatest increase in expression was a ∼2- to 3-fold increase observed for hcp1 and hcp3.
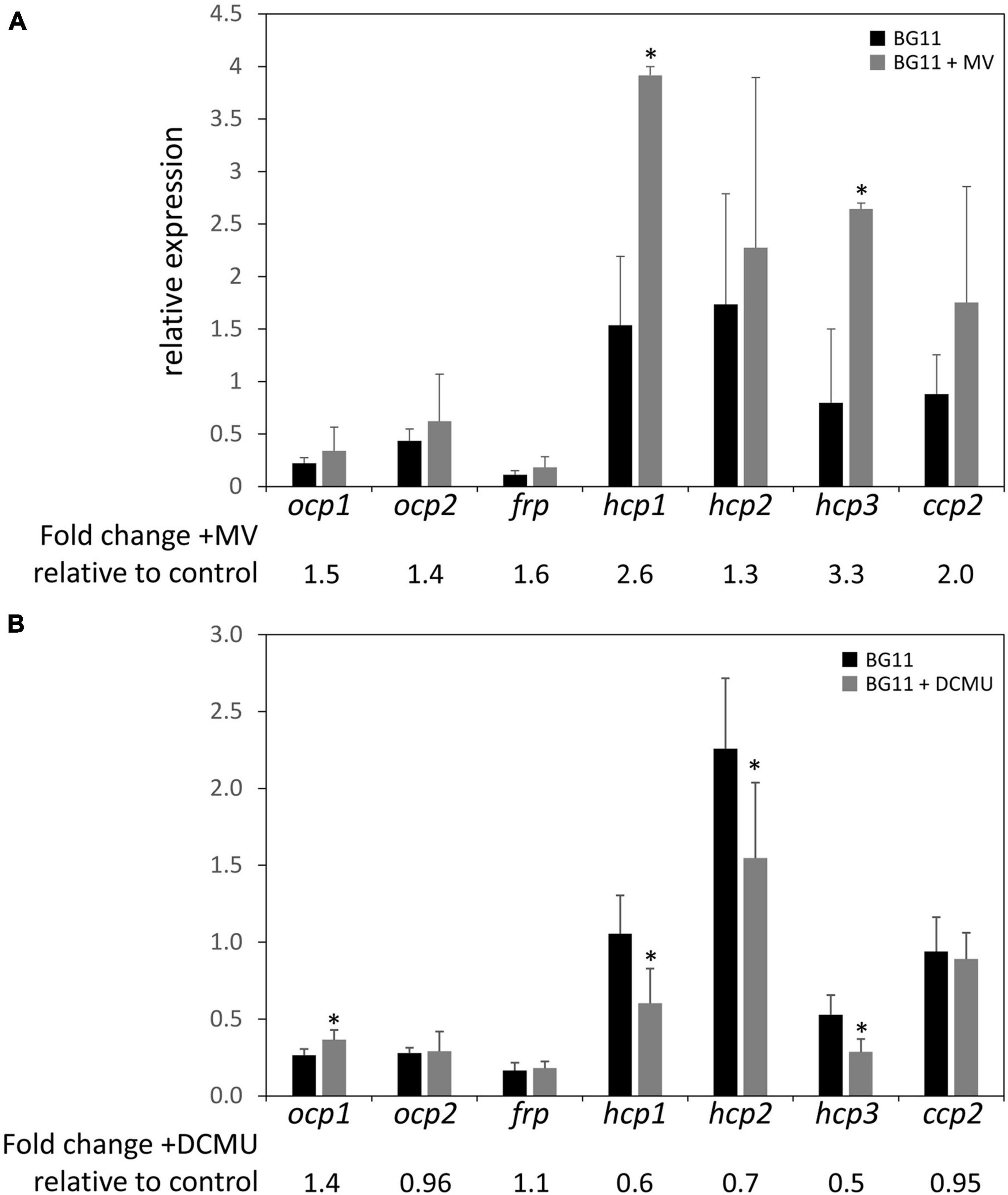
Figure 6. Expression of ocp homologs in the presence of methyl viologen or DCMU. The expression levels of ocp1, ocp2, frp, hcp1, hcp2, hcp3, and ccp2 were measured using qPCR, comparing samples incubated in the presence of 0.03 μM methyl viologen (A) or 10 μM DCMU (B) for 24 h to untreated samples (±SD, n = 4–6). *p < 0.05 as determined by a two tailed Student’s t-test for the comparison of the treated and untreated samples.
We also simulated redox stress by treating WT cultures with 10 μM of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and incubating for 24 h. DCMU is an herbicide that blocks the transfer of electrons from Photosystem II to the plastoquinone pool, leading to a more oxidized photosynthetic electron transport chain. The expression of ocp1 increased significantly in the presence of DCMU compared to untreated WT samples and there was a noted decrease in the expression of hcp1, hcp2, and hcp3 to between 50 and 70% of their expression levels in untreated samples (Figure 6B). The expression of ocp2, frp, or ccp2 was not significantly affected by DCMU treatment (Figure 6B). A previous study described a decrease in ocp expression in the presence of 10 μM DCMU in Synechocystis sp. PCC 6803 (Maksimov et al., 2017). This shows a point of difference in the regulation of Synechocystis ocp and F. diplosiphon ocp1, but similarity between hcp homologs in F. diplosiphon with Synechocystis ocp in response to DCMU treatment.
Promoter Analyses
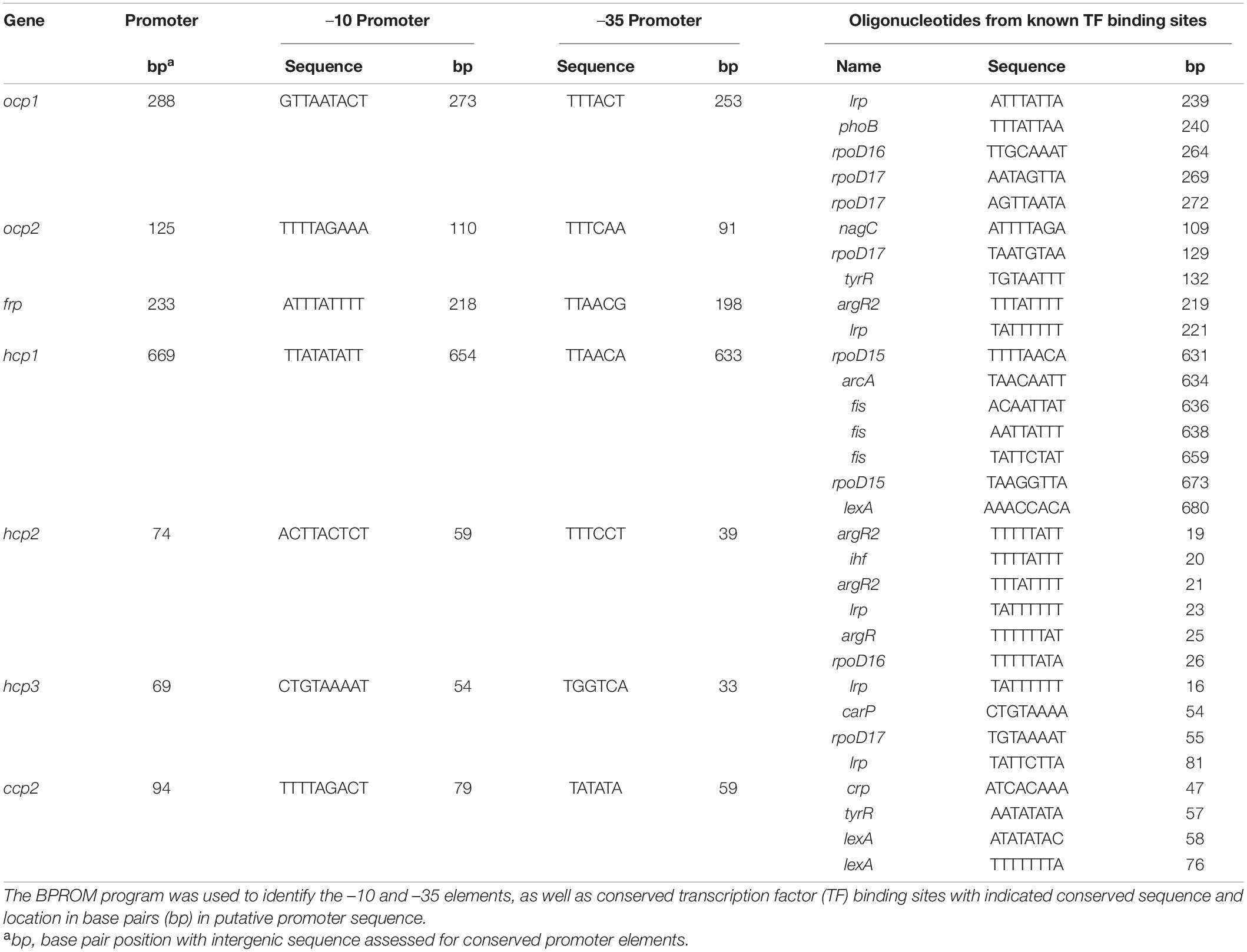
To determine if some of the distinct impacts of different growth conditions on expression of ocp homologs were correlated with distinct transcriptional regulatory elements associated with the promoter of each gene, we conducted sequence based promoter analyses to identify conserved transcriptional elements in the gene promoters (Figure 7 and Table 3). The most common transcription factor binding sites were for rpoD15/16/17, with a binding site for one of these three transcription factors found in the promoter region of every gene with the exception of frp and ccp2 (Figure 7). RpoD is an RNA polymerase sigma factor that functions to support initiation of transcription (Tanaka et al., 1992). Also found across multiple genes, including ocp1, frp, hcp2, and hcp3, was a lrp site that is frequently found to engage a leucine repressor protein that impacts cellular metabolism (Brinkman et al., 2003). Several other transcription factor binding sites were unique to the promoter region of only one or two of the ocp genes analyzed. An argR2 element was found in both hcp2 and frp, which is notable as this factor has been previously implicated in an unknown carbon dioxide-dependent pathway as ΔargR2 mutants were unable to grow with CO2 enrichment (Nicoloff et al., 2004). Also, a lexA site was found in both hcp1 and ccp2. The lexA gene encodes a transcriptional repressor involved in bacterial responses to DNA damage (Butala et al., 2009). The promoter region of frp had the lowest number of promoter elements.
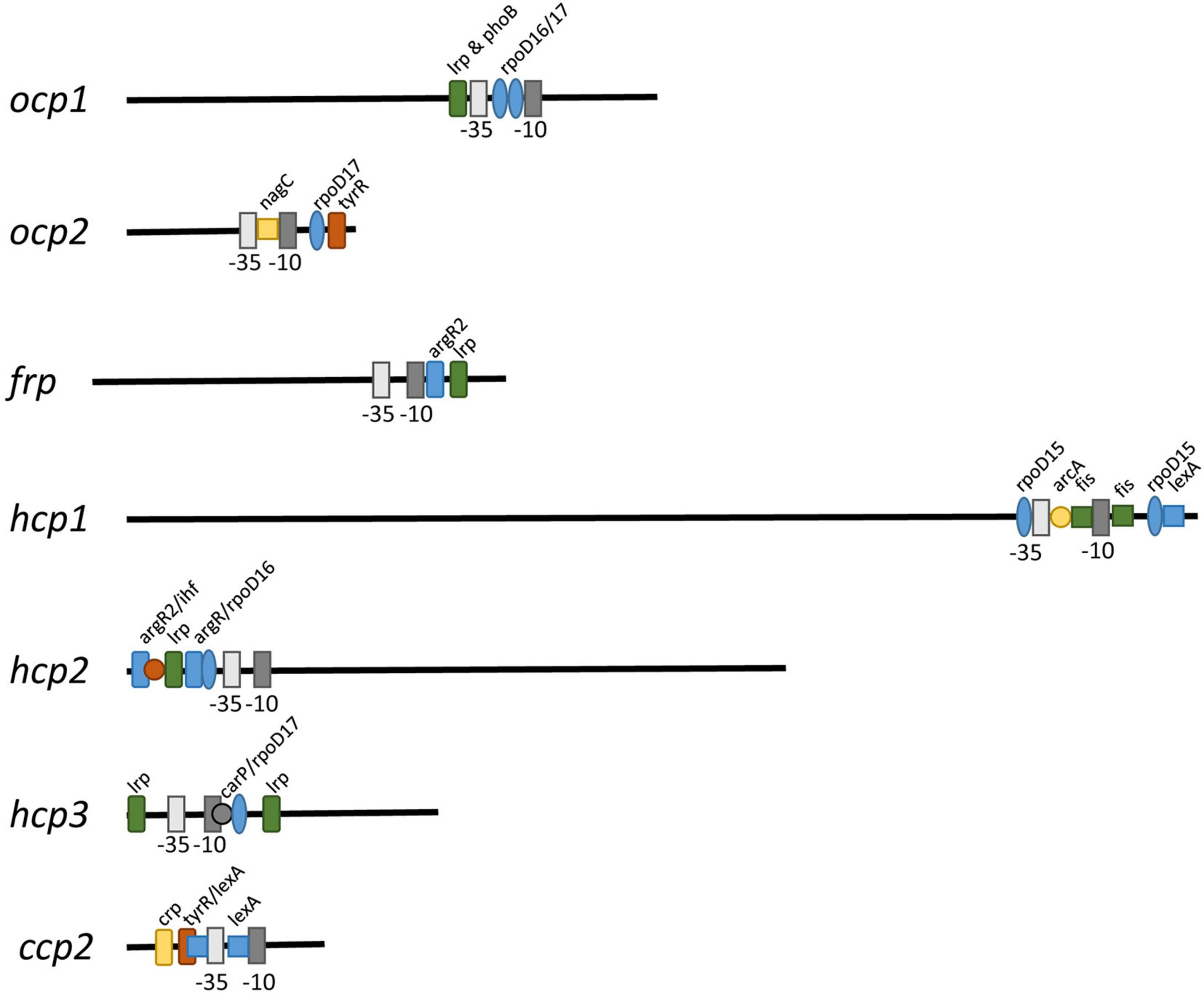
Figure 7. Promoter elements found in ocp homologs. The intergenic region upstream of each ocp homolog was analyzed using BPROM (Solovyev and Salamov, 2011) to predict transcription start positions (–10 and –35 sites) and transcription factor binding sites for each predicted promoter. Distinct binding sites are denoted with identical shapes and colors across distinct promoters.
Discussion
Some cyanobacteria have diversified the family of orange carotenoid protein (ocp) gene homologs. Many of the model species used for cyanobacterial species, including Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942, contain either one or no ocp genes. Thus, our understanding of the roles of multiple ocp homologs in a single species is limited. Initial insights into the potential biochemical activity of OCP homologs have begun to emerge. These findings include evidence that some OCP-related homologs bind PBSs to dissipate excess light energy absorption and others may function to directly quench ROS in cells (Sedoud et al., 2014; Lopez-Igual et al., 2016; Dominguez-Martin et al., 2019, 2020; Khan et al., 2020). Yet, we know very little about whether the expression of these genes are differentially controlled by distinct environmental cues that may offer insights into distinct or overlapping functions of these homologs in vivo.
The ocp gene has previously been shown to have increased expression under several stress conditions in Synechocystis sp. PCC 6803, including higher light intensity in studies conducted with samples grown under 20 and 300 μmol m–2 s–1 (Hihara et al., 2001). We also note differences in expression of ocp homologs in F. diplosiphon in response to distinct wavelengths and intensities of lights. While there are some differences relative to those reported for Synechocystis, the difference seen in the differential expression of ocp1 in F. diplosiphon could be due to the lower light intensity used in our studies or associated with differences in the regulatory mechanisms controlling ocp1 expression due to the presence of multiple ocp homologs in F. diplosiphon. This observation is especially true given an increase in ocp2, as well as hcp1/2/3 mRNA levels in response to increased light intensity in WT.
It is notable that each of the conditions that results in increased accumulation of mRNA for the ocp genes (cold stress and DCMU) and the hcp genes (red vs. green, high light vs. low light, salt stress, and MV treatment) are also associated with higher cellular ROS levels (Singh and Montgomery, 2012). In other experiments, expression of ocp has been shown to increase under conditions associated with high ROS, such as high light (Hihara et al., 2001), iron starvation (Wilson et al., 2007), and the presence of another photosynthesis inhibitor DBMIB (Maksimov et al., 2017). Upregulation of ocp homologs under higher levels of ROS could be beneficial given that both OCP and HCP proteins have been shown to quench singlet oxygen and PBS fluorescence (Sedoud et al., 2014; Lopez-Igual et al., 2016).
The importance of regulating ROS levels in cells are critical. For example, PBS are a target of ROS in cyanobacteria (Liu et al., 2005). Thus, given the likelihood that overexcitation of PBSs or photosystems by light may result in ROS generation due to an inability to maintain photosynthetic efficiency that could subsequently result in PBS damage and impaired photosynthetic efficiency, a role for ocp homologs in both quenching excess light energy and ameliorating ROS accumulation is not surprising. The diversification of ocp homologs in some cyanobacteria is likely related to providing cellular protection related to PBS excitation and oxidative stress under dynamic conditions. OCPs and HCPs thus likely function to provide broad protection against oxidative stress and associated cellular damage across a range of stressful conditions that cells may encounter.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
DP and PD designed and conducted the research, analyzed and interpreted the data, and contributed to writing the manuscript. BM designed the research, analyzed and interpreted the data, and wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science Foundation (IOS-1557324) and the Office of Science of the US Department of Energy (DE-FG02-91ER20021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to Cheryl Kerfeld and members of her laboratory for critical discussion of the biochemical functions of OCP homologs and discussion of some of the gene expression data included in the text.
References
Agostoni, M., Lucker, B. F., Smith, M. Y., Kanazawa, A., Blanchard, G. J., Kramer, D. M., et al. (2016). Competition-based phenotyping reveals a fitness cost for maintaining phycobilisomes under fluctuating light in the cyanobacterium Fremyella diplosiphon. Algal Res 15, 110–119. doi: 10.1016/j.algal.2016.02.005
Allen, M. M. (1968). Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4. doi: 10.1111/j.1529-8817.1968.tb04667.x
Alvey, R. M., Bezy, R. P., Frankenberg-Dinkel, N., and Kehoe, D. M. (2007). A light regulated OmpR-class promoter element co-ordinates light-harvesting protein and chromophore biosynthetic enzyme gene expression. Mol. Microbiol. 64, 319–332. doi: 10.1111/j.1365-2958.2007.05656.x
Bao, H., Melnicki, M. R., and Kerfeld, C. A. (2017a). Structure and functions of Orange Carotenoid Protein homologs in cyanobacteria. Curr. Opin. Plant Biol. 37, 1–9. doi: 10.1016/j.pbi.2017.03.010
Bao, H., Melnicki, M. R., Pawlowski, E. G., Sutter, M., Agostoni, M., Lechno-Yossef, S., et al. (2017b). Additional families of orange carotenoid proteins in the photoprotective system of cyanobacteria. Nat. Plants 3:17089. doi: 10.1038/nplants.2017.89
Bennett, A., and Bogorad, L. (1973). Complementary chromatic adaptation in a filamentous blue-green alga. J. Cell Biol. 58, 419–435. doi: 10.1083/jcb.58.2.419
Bordowitz, J. R., and Montgomery, B. L. (2008). Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J. Bacteriol. 190, 4069–4074. doi: 10.1128/jb.00018-08
Brinkman, A. B., Ettema, T. J., De Vos, W. M., and Van Der Oost, J. (2003). The Lrp family of transcriptional regulators. Mol. Microbiol. 48, 287–294. doi: 10.1046/j.1365-2958.2003.03442.x
Busch, A. W. U., and Montgomery, B. L. (2015a). Interdependence of tetrapyrrole metabolism, the generation of oxidative stress and the mitigative oxidative stress response. Redox Biol. 4, 260–271. doi: 10.1016/j.redox.2015.01.010
Busch, A. W. U., and Montgomery, B. L. (2015b). The tryptophan-rich sensory protein (TSPO) is involved in stress-related and light-dependent processes in the cyanobacterium Fremyella diplosiphon. Front. Microbiol. 6:1393. doi: 10.3389/fmicb.2015.01393
Busch, A. W. U., and Montgomery, B. L. (2017). Distinct light-, stress-, and nutrient-dependent regulation of multiple tryptophan-rich sensory protein (TSPO) genes in the cyanobacterium Fremyella diplosiphon. Plant Signal Behav. 12:e1293221. doi: 10.1080/15592324.2017.1293221
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Butala, M., Zgur-Bertok, D., and Busby, S. J. (2009). The bacterial LexA transcriptional repressor. Cell Mol. Life Sci. 66, 82–93. doi: 10.1007/s00018-008-8378-6
Cobley, J. G., Zerweck, E., Reyes, R., Mody, A., Seludo-Unson, J. R., Jaeger, H., et al. (1993). Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium. Fremyella Diplosiphon. Plasmid 30, 90–105. doi: 10.1006/plas.1993.1037
Collier, J. L., and Grossman, A. R. (1994). A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13, 1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x
Dominguez-Martin, M. A., Hammel, M., Gupta, S., Lechno-Yossef, S., Sutter, M., Rosenberg, D. J., et al. (2020). Structural analysis of a new carotenoid-binding protein: the C-terminal domain homolog of the OCP. Sci. Rep. 10:15564. doi: 10.1038/s41598-020-72383-y
Dominguez-Martin, M. A., Polívka, T., Sutter, M., Ferlez, B., Lechno-Yossef, S., Montgomery, B. L., et al. (2019). Structural and spectroscopic characterization of HCP2. Biochim. Biophys. Acta Bioenerg. 1860, 414–424. doi: 10.1016/j.bbabio.2019.03.004
Fujii, T., Yokoyama, E.-I., Inoue, K., and Sakurai, H. (1990). The sites of electron donation of Photosystem I to methyl viologen. Biochim. Biophys. Acta 1015, 41–48. doi: 10.1016/0005-2728(90)90213-N
Fulda, S., Mikkat, S., Huang, F., Huckauf, J., Marin, K., Norling, B., et al. (2006). Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6, 2733–2745. doi: 10.1002/pmic.200500538
Hihara, Y., Kamei, A., Kanehisa, M., Kaplan, A., and Ikeuchi, M. (2001). DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13, 793–806. doi: 10.1105/tpc.13.4.793
Kehoe, D. M., and Grossman, A. R. (1996). Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273, 1409–1412. doi: 10.1126/science.273.5280.1409
Kehoe, D. M., and Grossman, A. R. (1997). New classes of mutants in complementary chromatic adaptation provide evidence for a novel four-step phosphorelay system. J. Bacteriol. 179, 3914–3921. doi: 10.1128/jb.179.12.3914-3921.1997
Kerfeld, C. A., Melnicki, M. R., Sutter, M., and Dominguez-Martin, M. A. (2017). Structure, function and evolution of the cyanobacterial orange carotenoid protein and its homologs. New Phytol. 215, 937–951. doi: 10.1111/nph.14670
Kerfeld, C. A., Sawaya, M. R., Brahmandam, V., Cascio, D., Ho, K. K., Trevithick-Sutton, C. C., et al. (2003). The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure 11, 55–65. doi: 10.1016/s0969-2126(02)00936-x
Khan, T., Dominguez-Martin, M. A., Símova, I., Fuciman, M., Kerfeld, C. A., and Polívka, T. (2020). Excited-state properties of canthaxanthin in cyanobacterial carotenoid-binding proteins HCP2 and HCP3. J. Phys. Chem. B 124, 4896–4905. doi: 10.1021/acs.jpcb.0c03137
Kim, J.-H., and Lee, C.-H. (2002). Decrease of photochemical efficiency induced by methyl viologen in rice (Oryza sativa L.) leaves is partly due to the down-regulation of PSII. J. Photosci. 9, 65–70.
Kirilovsky, D., and Kerfeld, C. A. (2013). The orange carotenoid protein: a blue-green light photoactive protein. Photochem. Photobiol. Sci. 12, 1135–1143. doi: 10.1039/c3pp25406b
Kuznetsova, V., Dominguez-Martin, M. A., Bao, H., Gupta, S., Sutter, M., Kloz, M., et al. (2020). Comparative ultrafast spectroscopy and structural analysis of OCP1 and OCP2 from Tolypothrix. Biochim. Biophys. Acta Bioenerg. 1861:148120. doi: 10.1016/j.bbabio.2019.148120
Li, L., Alvey, R. M., Bezy, R. P., and Kehoe, D. M. (2008). Inverse transcriptional activities during complementary chromatic adaptation are controlled by the response regulator RcaC binding to red and green light-responsive promoters. Mol. Microbiol. 68, 286–297. doi: 10.1111/j.1365-2958.2008.06151.x
Li, L., and Kehoe, D. M. (2005). In vivo analysis of the roles of conserved aspartate and histidine residues within a complex response regulator. Mol. Microbiol. 55, 1538–1552. doi: 10.1111/j.1365-2958.2005.04491.x
Liu, X. G., Zhao, J. J., and Wu, Q. Y. (2005). Oxidative stress and metal ions effects on the cores of phycobilisomes in Synechocystis sp. PCC 6803. FEBS Lett. 579, 4571–4576. doi: 10.1016/j.febslet.2005.07.020
Lopez-Igual, R., Wilson, A., Leverenz, R. L., Melnicki, M. R., Bourcier De Carbon, C., Sutter, M., et al. (2016). Different functions of the paralogs to the N-Terminal domain of the orange carotenoid protein in the cyanobacterium anabaena sp. PCC 7120. Plant Physiol. 171, 1852–1866. doi: 10.1104/pp.16.00502
Maksimov, E. G., Mironov, K. S., Trofimova, M. S., Nechaeva, N. L., Todorenko, D. A., Klementiev, K. E., et al. (2017). Membrane fluidity controls redox-regulated cold stress responses in cyanobacteria. Photosyn. Res. 133, 215–223. doi: 10.1007/s11120-017-0337-3
Melnicki, M. R., Leverenz, R. L., Sutter, M., Lopez-Igual, R., Wilson, A., Pawlowski, E. G., et al. (2016). Structure, diversity, and evolution of a new family of soluble carotenoid-binding proteins in cyanobacteria. Mol. Plant 9, 1379–1394. doi: 10.1016/j.molp.2016.06.009
Montgomery, B. L. (2014). The regulation of light sensing and light harvesting impacts the use of cyanobacteria as biotechnology platforms. Front. Bioeng. Biotechnol. 2:22. doi: 10.3389/fbioe.2014.00022
Montgomery, B. L. (2016). Mechanisms and fitness implications of photomorphogenesis during chromatic acclimation in cyanobacteria. J. Exp. Bot. 67, 4079–4090. doi: 10.1093/jxb/erw206
Montgomery, B. L. (2017). Seeing new light: recent insights into the occurrence and regulation of chromatic acclimation in cyanobacteria. Curr. Opin. Plant Biol. 37, 18–23. doi: 10.1016/j.pbi.2017.03.009
Nicoloff, H., Arsène-Ploetze, F., Malandain, C., Kleerebezem, M., and Bringel, F. (2004). Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 186, 6059–6069. doi: 10.1128/jb.186.18.6059-6069.2004
Oh, S., and Montgomery, B. L. (2019). Roles of CpcF and CpcG1 in peroxiredoxin-mediated oxidative stress responses and cellular fitness in the cyanobacterium Synechocystis sp. PCC 6803. Frontiers in Microbiology 10:1059. doi: 10.3389/fmicb.2019.01059
Sanfilippo, J. E., Garczarek, L., Partensky, F., and Kehoe, D. M. (2019). Chromatic acclimation in cyanobacteria: a diverse and widespread process for optimizing photosynthesis. Ann. Rev. Microbiol. 73, 407–433. doi: 10.1146/annurev-micro-020518-115738
Sedoud, A., Lopez-Igual, R., Ur Rehman, A., Wilson, A., Perreau, F., Boulay, C., et al. (2014). The cyanobacterial photoactive orange carotenoid protein is an excellent singlet oxygen quencher. Plant Cell 26, 1781–1791. doi: 10.1105/tpc.114.123802
Seib, L. O., and Kehoe, D. M. (2002). A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J. Bacteriol. 184, 962–970. doi: 10.1128/jb.184.4.962-970.2002
Singh, S. P., and Montgomery, B. L. (2012). Reactive oxygen species are involved in the morphology-determining mechanism of Fremyella diplosiphon cells during complementary chromatic adaptation. Microbiology 158, 2235–2245. doi: 10.1099/mic.0.060475-0
Solovyev, V., and Salamov, A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed. R. W. Li (Hauppauge, NY: Nova Science Publishers), 61–78.
Stowe-Evans, E. L., Ford, J., and Kehoe, D. M. (2004). Genomic DNA microarray analysis: identification of new genes regulated by light color in the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 186, 4338–4349. doi: 10.1128/JB.186.13.4338-4349.2004
Tanaka, K., Masuda, S., and Takahashi, H. (1992). Multiple rpoD-related genes of cyanobacteria. Biosci. Biotechnol. Biochem. 56, 1113–1117. doi: 10.1271/bbb.56.1113
Wilson, A., Ajlani, G., Verbavatz, J. M., Vass, I., Kerfeld, C. A., and Kirilovsky, D. (2006). A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell 18, 992–1007. doi: 10.1105/tpc.105.040121
Wilson, A., Boulay, C., Wilde, A., Kerfeld, C. A., and Kirilovsky, D. (2007). Light-induced energy dissipation in iron-starved cyanobacteria: roles of OCP and IsiA proteins. Plant Cell 19, 656–672. doi: 10.1105/tpc.106.045351
Wilson, A., Punginelli, C., Gall, A., Bonetti, C., Alexandre, M., Routaboul, J. M., et al. (2008). A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl. Acad. Sci. USA 105, 12075–12080. doi: 10.1073/pnas.0804636105
Yerrapragada, S., Shukla, A., Hallsworth-Pepin, K., Choi, K., Wollam, A., Clifton, S., et al. (2015). Extreme sensory complexity encoded in the 10-Megabase draft genome sequence of the chromatically acclimating cyanobacterium Tolypothrix sp. PCC 7601. Genome Announc. 3, e315–e355. doi: 10.1128/genomeA.00355-15
Keywords: cyanobacteria, non-photochemical quenching (NPQ), orange carotenoid protein (OCP), photosynthesis, stress
Citation: Petrescu DI, Dilbeck PL and Montgomery BL (2021) Environmental Tuning of Homologs of the Orange Carotenoid Protein-Encoding Gene in the Cyanobacterium Fremyella diplosiphon. Front. Microbiol. 12:819604. doi: 10.3389/fmicb.2021.819604
Received: 21 November 2021; Accepted: 07 December 2021;
Published: 24 December 2021.
Edited by:
Kathleen Scott, University of South Florida, United StatesReviewed by:
Shailendra Pratap Singh, Banaras Hindu University, IndiaHaijun Liu, Washington University in St. Louis, United States
Copyright © 2021 Petrescu, Dilbeck and Montgomery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beronda L. Montgomery, bW9udGcxMzNAbXN1LmVkdQ==
 D. Isabel Petrescu1,2
D. Isabel Petrescu1,2 Beronda L. Montgomery
Beronda L. Montgomery