
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 January 2022
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.811343
This article is part of the Research TopicEvolutionary Mechanisms of Infectious Diseases, Volume IIView all 12 articles
 Jin Li1
Jin Li1 Wei-wei Hu2
Wei-wei Hu2 Guo-xin Qu3
Guo-xin Qu3 Xiao-rong Li1
Xiao-rong Li1 Yi Xiang1
Yi Xiang1 Peng Jiang1
Peng Jiang1 Jiang-qiao Luo1
Jiang-qiao Luo1 Wen-huan He1
Wen-huan He1 Yu-jia Jin1
Yu-jia Jin1 Qiong Shi1*
Qiong Shi1*
Burkholderia thailandensis is a clinically underestimated conditional pathogen in the genus Burkholderia, the pathogenicity of the infection caused by B. thailandensis remains poorly understood. According to previous studies, Type-VI secretion system (T6SS) is a protein secreting device widely existing in Gram-negative bacilli. Valine-glycine repeat protein G (VgrG) is not only an important component of T6SS, but also a virulence factor of many Gram-negative bacilli. In one of our previous studies, a unique T6SS vgrG gene (vgrG2 gene) was present in a virulent B. thailandensis strain BPM (BPM), but not in the relatively avirulent B. thailandensis strain E264 (E264). Meanwhile, transcriptome analysis of BPM and E264 showed that the vgrG2 gene was strongly expressed in BPM, but not in E264. Therefore, we identified the function of the vgrG2 gene by constructing the mutant and complemented strains in this study. In vitro, the vgrG2 gene was observed to be involved in the interactions with host cells. The animal model experiment showed that the deletion of vgrG2 gene significantly led to the decrease in the lethality of BPM and impaired its ability to trigger host immune response. In conclusion, our study provides a new perspective for studying the pathogenicity of B. thailandensis and lays the foundation for discovering the potential T6SS effectors.
Burkholderia thailandensis is a clinically underestimated conditional pathogen in the genus Burkholderia. It is very similar to Burkholderia pseudomallei in terms of colony morphology, immunogenicity and antimicrobial susceptibility (Brett et al., 1998; Ngamdee et al., 2015; Garcia, 2017). To date, only a few studies have described the isolation of B. thailandensis from invasive human infections (Lertpatanasuwan et al., 1999; Glass et al., 2006; Ginther et al., 2015; Gee et al., 2018), and the pathogenic mechanism of the infections caused by B. thailandensis remains poorly understood. Therefore, it is necessary to study the pathogenesis of B. thailandensis to more effectively prevent infections caused by B. thailandensis. In one of our previous studies, the virulent B. thailandensis strain BPM was isolated from a blood and sputum specimen of a 76-year-old man with sepsis who died in China (Chang et al., 2017). The biological and biochemical characteristics of BPM are very similar to those of B. pseudomallei, and the clinical symptoms and imaging findings of patients infected with this strain are consistent with acute melioidosis, manifesting as acute high fever, productive cough with white sputum and breathing difficulty (Wiersinga et al., 2012; Tang et al., 2018; Gassiep et al., 2020). However, biochemical identification test results showed that BPM was positive for arabinose assimilation, which is consistent with the biochemical characteristics of B. thailandensis. To confirm the biochemical identification results, 16S rRNA gene sequence and whole-genome sequencing analyses were performed. The results showed that the sequence of the 16S rRNA gene was 100% consistent with that of the B. thailandensis E264 (E264) 16S rRNA gene (GenBank No. CP008785.1, CP008786.1) and that the sequence obtained by whole-genome sequencing was more than 96% homologous to the genome sequence of E264 (GenBank No. CP000085.1, CP000086.1). Finally, the BPM strain was identified as B. thailandensis based on the NT nucleic acid sequence database, and the genome sequences of BPM have been deposited in the GenBank database under accession numbers CP050020-CP050021. In one of our previous studies, we compared the virulence of BPM and E264 in BALB/c mice, and the results indicated that the virulence of BPM was significantly higher than that of E264, which confirmed that BPM is a virulent pathogen (Chang et al., 2017). Additionally, an integrated type VI secretion system (T6SS) gene cluster was found in BPM by bioinformatic analysis. However, the pathogenicity of the T6SS involved in BPM infection is poorly understood.
The T6SS is an important virulence factor that plays a key role in microbial competition and bacterial infection (Fridman et al., 2020). It can deliver toxic effectors to bacterial and eukaryotic neighbors and plays an important role in both bacterial competition and virulence (Hsieh et al., 2019). Valine-glycine repeat protein G (VgrG) has been reported to be an important component of the functional T6SS of B. pseudomallei and is involved in the occurrence of acute melioidosis (Schwarz et al., 2014). However, the functions of the T6SS vgrG gene in the development of B. thailandensis infections remain unknown. In one of our previous studies, a unique T6SS vgrG gene (vgrG2 gene) was present in a virulent BPM, but not in the relatively avirulent E264. Meanwhile, transcriptome analysis of BPM and E264 showed that the vgrG2 gene was strongly expressed in BPM, but not in E264. However, the function of vgrG2 gene remains unknown. Therefore, this study investigated the function of the putative T6SS vgrG2 gene in BPM by knocking out the vgrG2 gene and producing a vgrG2 gene complementation construct. The mutant and complemented strains will be used to determine the function of the T6SS vgrG2 gene by investigating the changes in the virulence of BPM. Altogether, this study aimed to lay a foundation for discovering potential T6SS effectors of B. thailandensis and provide a new perspective on the study of host cell signal transduction and immune defense mechanisms.
All bacterial strains used in this study are listed in Table 1. All strains were cultured on sheep blood agar plates (Thermo, United States) at 37°C for 16–20 h, and isolated colonies were inoculated into 10 mL Luria-Bertani (LB) broth (Solarbio, China), which was then stirred at 37°C for 12 h. The working cultures were prepared by transferring 100 μL of a 12 h culture to 10 mL LB broth (1:100 dilution), which was then allowed to stand at 37°C for 8 h. The stationary-phase bacteria were diluted to 106∼108 colony-forming units (CFU)/mL in LB broth, and phenotypic characteristics were evaluated (Jiang et al., 2016). These final suspensions were plated onto LB agar to accurately determine the number of CFUs per milliliter.
The total RNA of BPM and E264 was extracted with TRIzol reagent (Invitrogen, United States). The quantity and purity of the extracted RNA were assessed using a NanoDrop ND-1000 spectrophotometer (Thermo, United States). RNA-seq libraries were created using the Illumina TruSeq Stranded mRNA Library Prep Kit (Illumina, Inc., United States) according to the manufacturer’s protocol. Sequencing was performed at Shenzhen Hai-yi Biotechnology Co., Ltd. using an Illumina MiSeq System benchtop sequencing instrument (read length: 75 bp, read type: paired end) (Zhu et al., 2016). The raw sequence data were filtered by removing reads containing adapters, reads containing poly N sequences, and low-quality reads. The clean reads were aligned to the genomes of BPM and E264 by using Bowtie2-2.2.3 (Kovacs-Simon et al., 2019). DESeq was used to identify differentially expressed genes (Zhu et al., 2016).
To test the role of the T6SS vgrG2 gene in the pathogenesis of B. thailandensis and its contribution to the development of B. thailandensis infection, knockout mutants of a key component (vgrG2) of the T6SS were constructed by double crossover recombination through allelic replacement of the suicide plasmid pLP12cm as described previously (Luo et al., 2015). The knockout mutant was designated ΔvgrG2. The vgrG2 gene was amplified from the B. thailandensis BPM genome and then ligated into plasmid pTac-tetM to construct the complementation expression plasmid pTac-tetM-vgrG2. Finally, the complementation plasmids were transferred into mutants to generate complemented strains (ΔvgrG2/pvgrG2). All mutant and complemented strains were verified using PCR (Supplementary Figure 1) and DNA sequencing (data not shown).
The strain was cultured on sheep blood agar at 37°C for 18–20 h and then transferred to LB broth for shaking culture at 180 rpm at 37°C. The growth characteristics of the BPM, mutant, and complemented strains were determined via optical density measurements (Eppendorf BioPhotometer, Germany) performed at 600 nm (OD600), and colony formation units (CFUs) were counted over a 24-h period as described previously (Zhu et al., 2020). Then, the antimicrobial susceptibilities of the BPM, mutant, and complemented strains were initially tested with a Vitek-2 Compact automatic microbiological assay system (BioMérieux, French). The experimental methods were performed according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) for P. aeruginosa (Bobenchik et al., 2017). Fresh bacterial colonies extracted directly from sheep blood agar were incubated at 37°C for 18–24 h and then resuspended in sterile saline to obtain a suspension of 0.5 McFarland turbidity. E. coli ATCC 25922 and P. aeruginosa ATCC 2785 were used as quality controls. The antimicrobial susceptibility testing results were explained in accordance with the CLSI M45 guidelines for B. pseudomallei. Each assay was performed three times.
All animal experiments were approved by the research board of the Ethics Committee of the Third Military Medical University under permit number AMUMEC-20201085. To determine the 50% lethal dose (LD50), five-week-old, pathogen-free, female BALB/c mice were obtained from Daping Hospital Animal Center. Ten BALB/c mice were used as a sample population for the survival rate of BALB/c mice infected with BPM, mutant, and complemented strains. Phosphate-buffered saline (PBS) was used as negative control. Ten BALB/c mice were selected for each bacterial concentration to determine the LD50. Two-fold serial dilution of the bacteria was performed from a starting concentration of 8 × 107CFU/mL to 5 × 106CFU/mL, and BALB/c mice were infected intravenously with 0.1 mL of each concentration. Symptoms and mortality rates were observed for seven days. The exact inoculation dose was confirmed on LB agar, and the LD50 was calculated as described by Barnes (Barnes and Ketheesan, 2005).
To examine the differences in the pathological changes caused by the tested strains, livers and lungs were collected from BALB/C mice infected with the BPM, mutant and complemented strains at designated times (4, 8, 12, and 16 h post infection). Tissue samples were fixed in 10% buffered formalin. Paraffin-embedded tissue sections were stained with hematoxylin and eosin according to the standard protocol and examined by light microscopy (Zhao et al., 2011).
To assess the function of the vgrG2 gene in inflammation, serum samples (infected with BPM, mutant, and complemented strains) were collected, and the levels of IL-1β, IL-6 and TNF-α were measured using Mouse Precoated ELISA kits (Dakewei Biotech Co., Ltd). Each assay was performed three times.
Human whole-blood samples used in the experiment were taken from 10 healthy individuals. The whole blood bactericidal assay was performed as previously described with minor modifications (Zong et al., 2019). Briefly, a bacterial inoculum of 100 μL (adjusted to 106 CFU/mL) prepared from the mid-log phase was diluted with PBS and added to 900 μL of fresh whole blood contained in 24-well plates (Corning, United States), and the mixtures were incubated at 37°C. After incubation for 3 h, the bacteria were plated onto LB agar and counted. The survival rates of the BPM, mutant, and complemented strains were expressed by using the following formula: (CFU/mL)t=3h)/(CFU/mL)t=0h) × 100%. Each assay was performed three times.
The cell invasion assay was similar to that previously performed (Pijuan et al., 2019). RAW264.7 cells were incubated at 37°C with 5% CO2 in 24-well plates at a concentration of 5 × 105 cells per well. RAW264.7 cells were grown on DMEM (Gibco GlutaMAX™, United States) containing glucose, glutamine, and 10% fetal bovine serum. B. thailandensis suspensions were added to the cells at an MOI of 10 or 100, followed by centrifugation at 500 g for 5 min and incubation at 37°C with 5% CO2 for 1 h to determine invasion. One-hour post infection (hpi), the monolayers were washed twice with PBS and lysed with 0.1% Triton X-100 (Sigma, United States) in PBS, and serial dilutions were plated and incubated at 37°C for 48 h. The invasion percentages of the BPM, mutant, and complemented strains were calculated as follows: (invasion CFU/total inoculum CFU) × 100 (Lewis et al., 2017). To determine intracellular survival after initial invasion, after 1 h, the monolayers were washed twice with PBS and replenished with complete medium containing 250 μg/ml chloromycetin. At 3 h post inoculation, the monolayers were washed twice with PBS and then lysed with 0.1% Triton X-100 in PBS, and serial dilutions were plated and incubated at 37°C for 48 h. The percent survival of the BPM, mutant, and complemented strains was calculated as (survival CFU/invasion CFU) × 100 (Lewis et al., 2017). Each assay was performed three times.
The cytotoxicity of the bacteria to RAW264.7 cells was tested by CCK-8 assays. Bacteria in the stationary phase resuspended in fresh medium were added to 96-well plates (Corning, United States) (MOI = 10). RAW264.7 cells were washed with PBS, resuspended in DMEM and plated in 96-well plates at a concentration of 5000 cells/well. Next, CCK-8 assay kit (MCE, China) reagents were added to the wells according to the manufacturer’s instructions. The optical density at 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Varioskan LUX, China) to assess cell viability. Cytotoxicity was expressed according to the following formula: cytotoxicity (%) = (test sample – low control)/(high control – low control) × 100 (Tang et al., 2021). Each assay was performed three times.
Statistical analyses were performed using GraphPad Prism 7 (San Diego, United States). One-way ANOVA with the log-rank test was used to compare BPM to the mutant and complemented strains. We also used Tukey’s multiple comparison test to compare each strain to all other strains. Significant differences between groups are indicated: * (P < 0.05), ** (P < 0.01) and *** (P < 0.001).
We sequenced and analyzed the transcriptomes of BPM and E264 and submitted the transcriptome data to the NCBI database to obtain the sequence and annotation information of the transcriptome sequencing assembly (number: GSE147369). Relative to E264, there was no difference in the expression of six BPM homologs of vgrG genes (BPM03563, BPM03564, BPM03921, BPM04575, BPM05231, and BPM05382), while the expression of BPM02934 (vgrG2) was upregulated, and the expression of BPM01336 and BPM05892 was downregulated (Table 2). The heatmap of vgrG gene expression is shown in Figure 1, which indicated that the vgrG2 gene is a unique virulence factor. The VgrG protein is a needle-like structure of the T6SS and is homologous to the T4 bacteriophage cell-puncturing device, which contributes to the development of acute melioidosis (Schwarz et al., 2014). Therefore, the vgrG2 gene was selected and subjected to further experiments in our study.
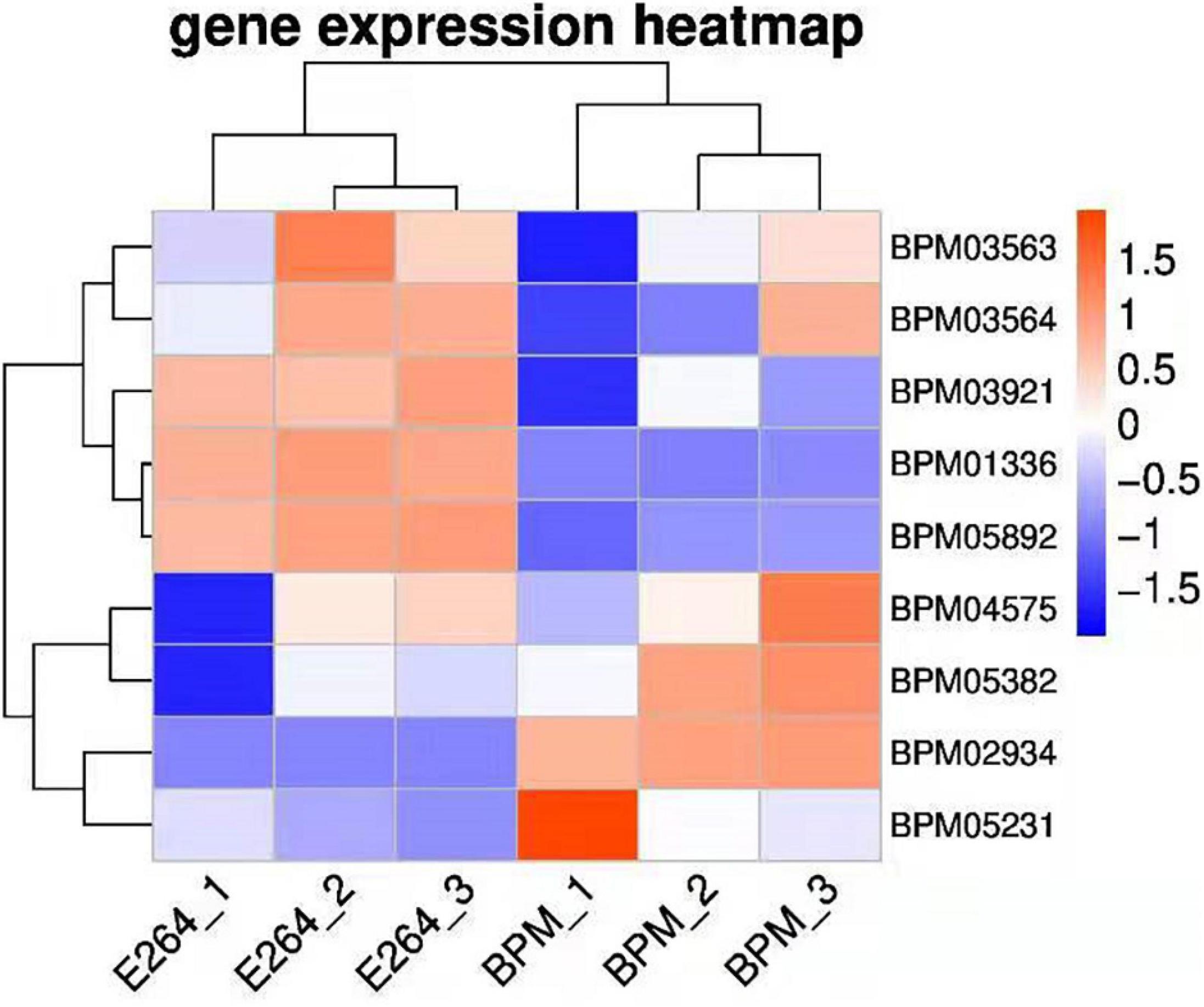
Figure 1. Heatmap of vgrG gene expression in BPM and E264. Colors in the heatmap represent gene expression levels among samples. The BPM and E264 results came from three repeated samples.
Growth rates were plotted according to the measured OD600 values and CFUs as described previously (Zong et al., 2019). No growth rate difference was found when the mutant and complemented strains were compared with BPM (Supplementary Figure 2). The antimicrobial susceptibility results of the mutant and complemented strains related to six antibiotics were consistent with those of BPM, as shown in Supplementary Table 1. All of these strains were sensitive to amoxicillin/clavulanate, ceftazidime, imipenem, tetracycline, doxycycline, and trimethoprim/sulfamethoxazole.
To determine whether the deletion of the vgrG2 gene impairs the virulence of BPM, the survival rate and LD50 of BALB/c mice infected with the BPM, mutant and complemented strains were compared. After infection for 7 days, the survival rate of BALB/c mice infected with BPM and ΔvgrG2/pvgrG2 was significantly lower than that of BALB/c mice infected with ΔvgrG2 (Figure 2, *P < 0.05). The LD50 results showed that the LD50 of ΔvgrG2 was 1.61 × 107 CFU (Table 3), which indicates low virulence. In contrast, BPM and ΔvgrG2/pvgrG2 showed relatively high virulence, with LD50 values of 8.35 × 106 CFU and 8.87 × 106 CFU, respectively (Table 3). Their phenotypic characteristics indicated that the vgrG2 gene was involved in the virulence of BPM in BALB/c mice.
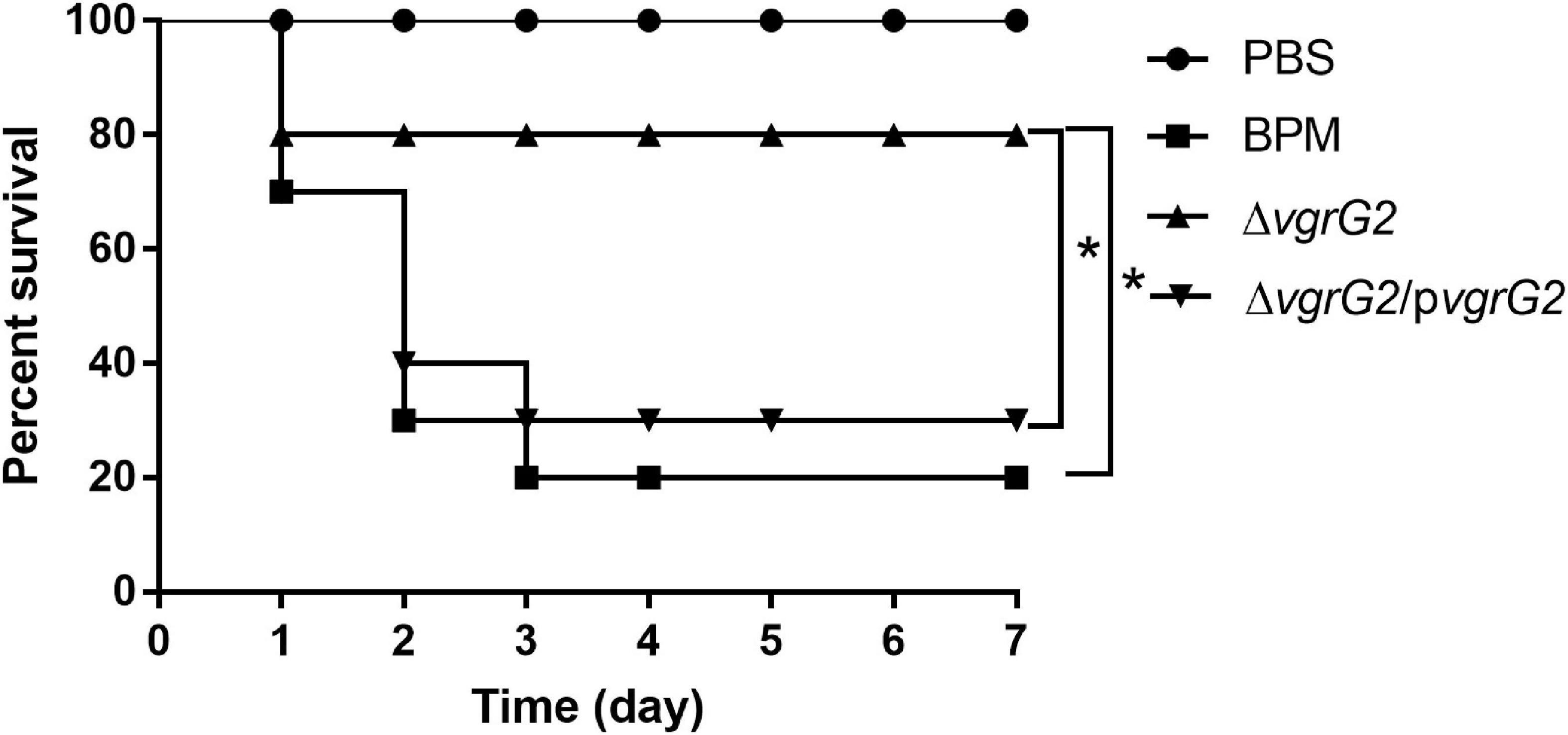
Figure 2. Survival rate of BALB/c mice infected with BPM, mutant and complemented strains. The mortality of BALB/c mice after the intraperitoneal injection of all strains was observed over 7 days. Data points represent the percentage of BALB/c mouse survival in each group (n = 10 mice per strain and 1 × 107CFU per mouse). After infection for 7 days, the survival rate of BALB/c mice infected with PBS and ΔvgrG was significantly lower than that of BALB/c mice infected with BPM *(P < 0.05).
During the first 8 h after infection, BALB/C mice infected with PBS and the three indicator strains showed no significant histopathological changes in the liver or lungs (data not shown). At 24 h after infection, different histopathological changes were observed in the lungs and livers of BALB/c mice infected with PBS and the three indicated strains. As shown in Figure 3, after BALB/C mice were infected with the three indicated strains, a small number of inflammatory cells infiltrated the lung tissue, central vein and convergence area, and liver tissue necrosis and partial destruction of the liver cell structure were observed.
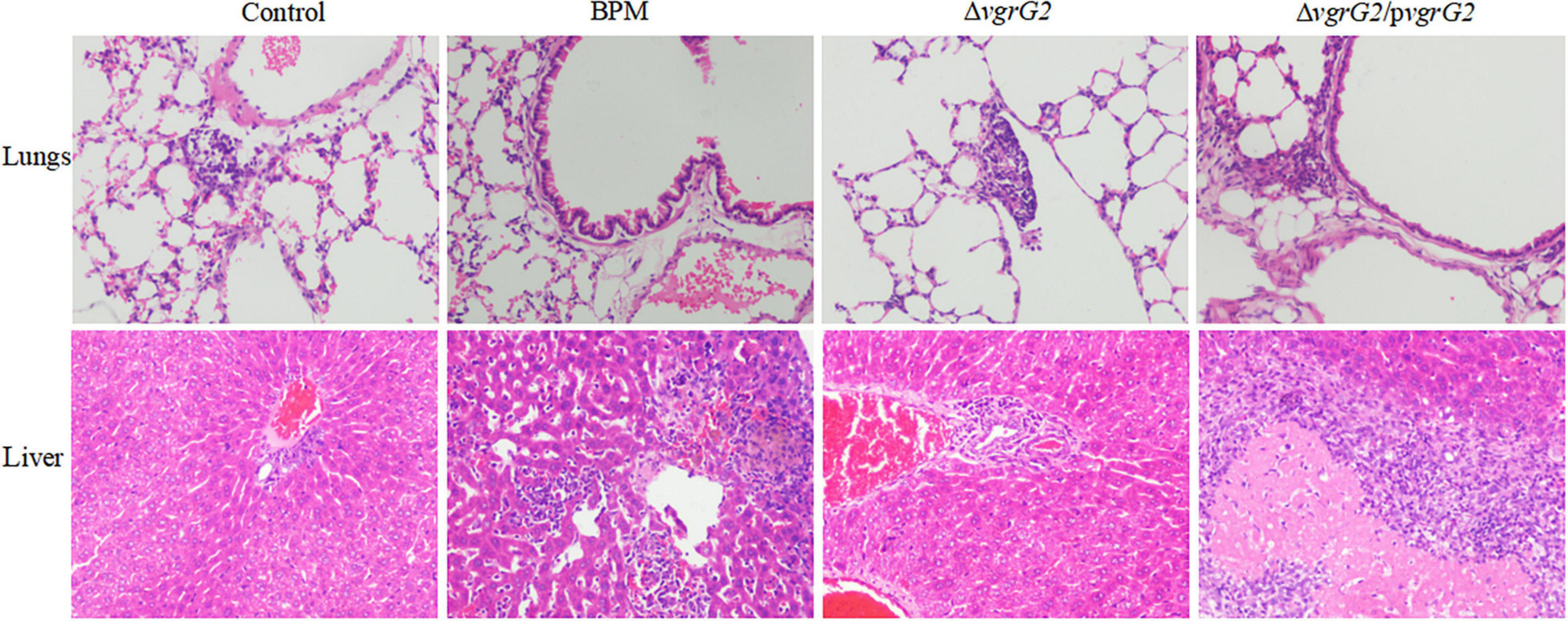
Figure 3. Pathological characterization of lungs and liver tissues of BALB/c mice infected with BPM, mutant and complemented strains. Lungs and liver tissues of BALB/c mice infected with BPM, mutant, complemented strains and PBS (control) were prepared for light microscopy analysis and examined for differences in pathological changes (hematoxylin and eosin staining; original magnification × 200).
To determine whether the vgrG2 gene is involved in the expression of proinflammatory cytokines, serum samples were collected from intravenously infected BALB/c mice for analysis of proinflammatory cytokines. Proinflammatory cytokines were detected in BALB/c mice 8 h after infection with the BPM, mutant and complemented strains. As shown in Figure 4, the production of TNF-α triggered by BPM and ΔvgrG2/pvgrG2 was significantly higher than that triggered by ΔvgrG2 (*** P < 0.001). The levels of IL-1β and IL-6 induced by BPM and ΔvgrG2/pvgrG2 were relatively higher than those induced by ΔvgrG2 (* P < 0.05).
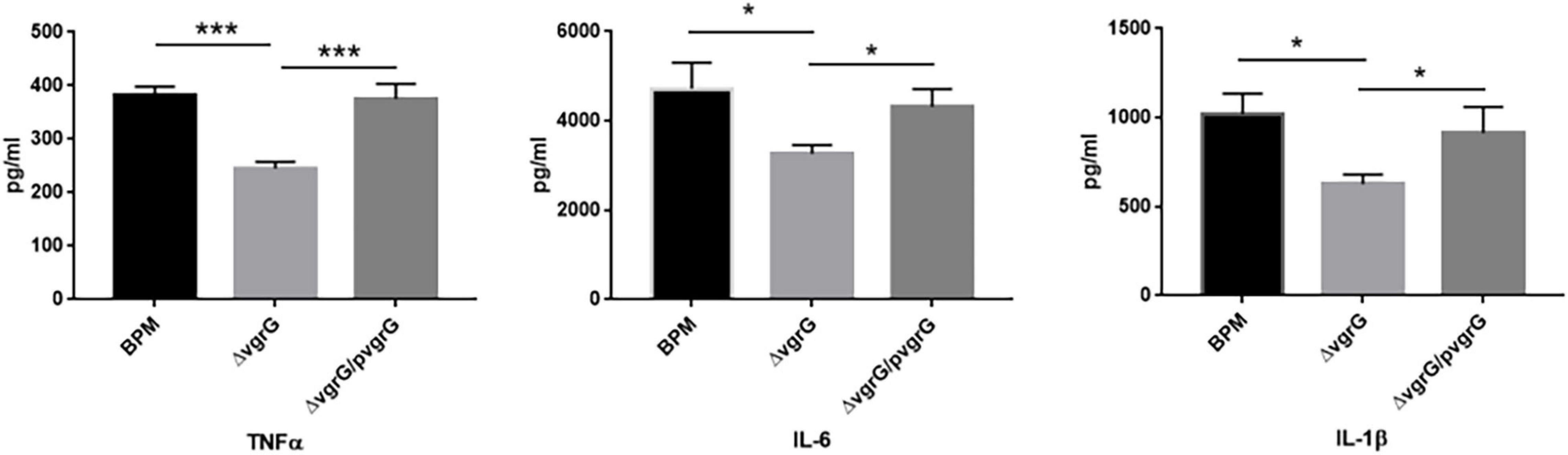
Figure 4. Serum levels of cytokines in BLAB/c mice 16 h after infection with BPM, mutant and complemented strains. Serum IL-1β, IL-6, and TNF-α levels in BALB/c mice 16 h after infection with the BPM, mutant and complement strains. All data are from three independent experiments. Significant differences between groups are indicated: *(P < 0.05) and ***(P < 0.001).
To evaluate the function of the vgrG2 gene in the evasion of innate immune responses, we measured the survival rates of the BPM, mutant and complemented strains in whole blood collected from healthy individuals. The experimental results showed that the survival rate of ΔvgrG2 was significantly lower than those of BPM and ΔvgrG2/pvgrG2 (Figure 5, ** P < 0.01).
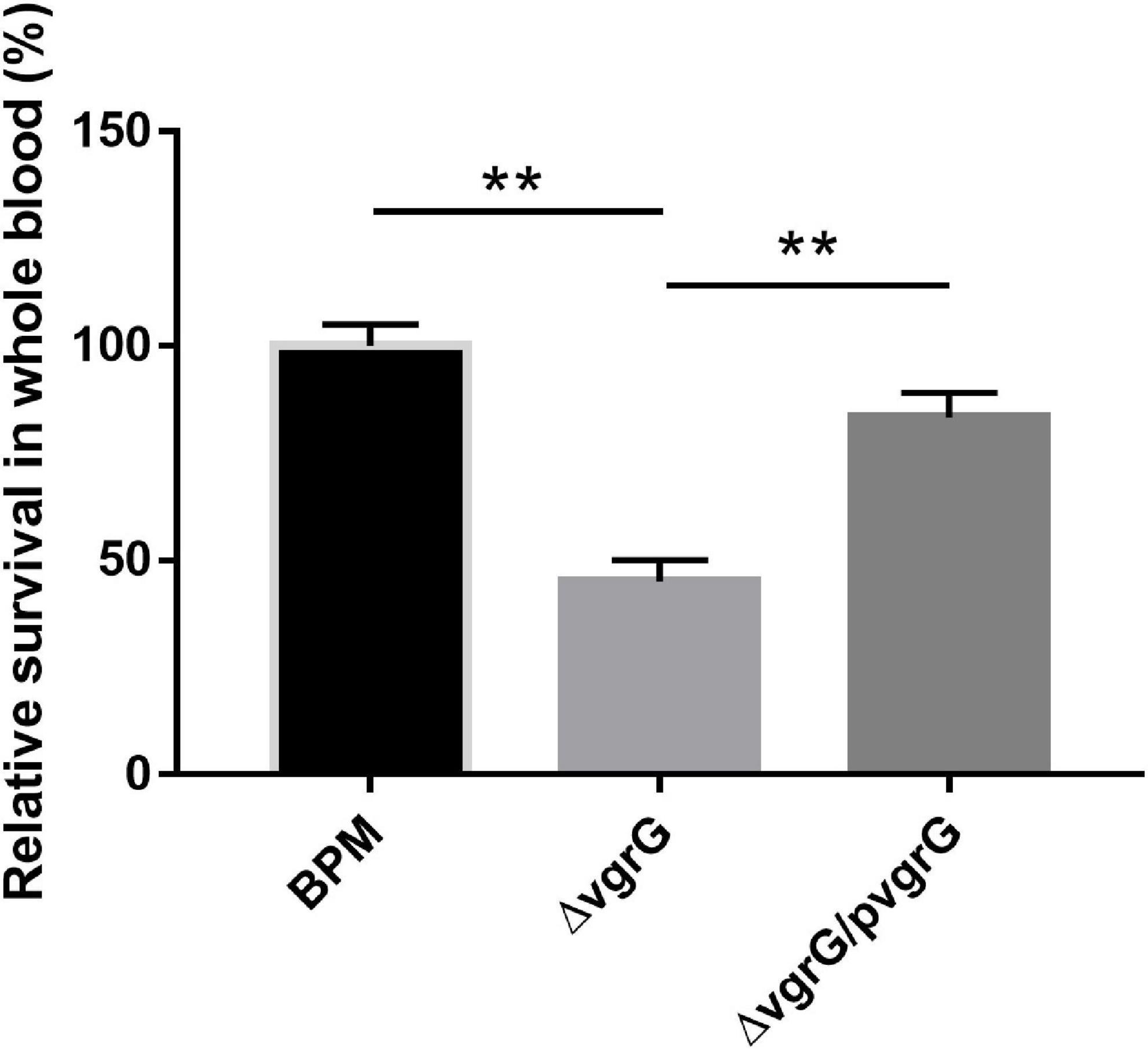
Figure 5. Whole-blood bactericidal experiments of BPM, mutant and complemented strains. Survival rates of the BPM, mutant and complemented strains in human whole blood. The survival rates are expressed relative to those of BPM (100%). Means and SDs of three independent experiments performed in triplicate were calculated. Significant differences between groups are indicated: **(P < 0.01).
To further investigate whether the vgrG2 gene completely or partially impairs T6SS activity, we compared the cell invasion, intracellular survival and cytotoxicity of the mutant and complemented strains with those of BPM. The results showed that the cell invasion, intracellular survival and cytotoxicity of ΔvgrG2 were significantly lower than those of the BPM and ΔvgrG2/pvgrG2 (Figure 6), and no significant difference in cell invasion, intracellular survival or cytotoxicity was found between the BPM and ΔvgrG2/pvgrG2 (Figure 6), suggesting that the deletion of vgrG2 affected the cell invasion, intracellular survival and cytotoxicity of BPM. Since vgrG gene has been reported as a virulence factor of functional T6SS in B. pseudomallei, it can be concluded that the deletion of vgrG2 may impair the overall activity of T6SS by affecting the assembly of T6SS in BPM.
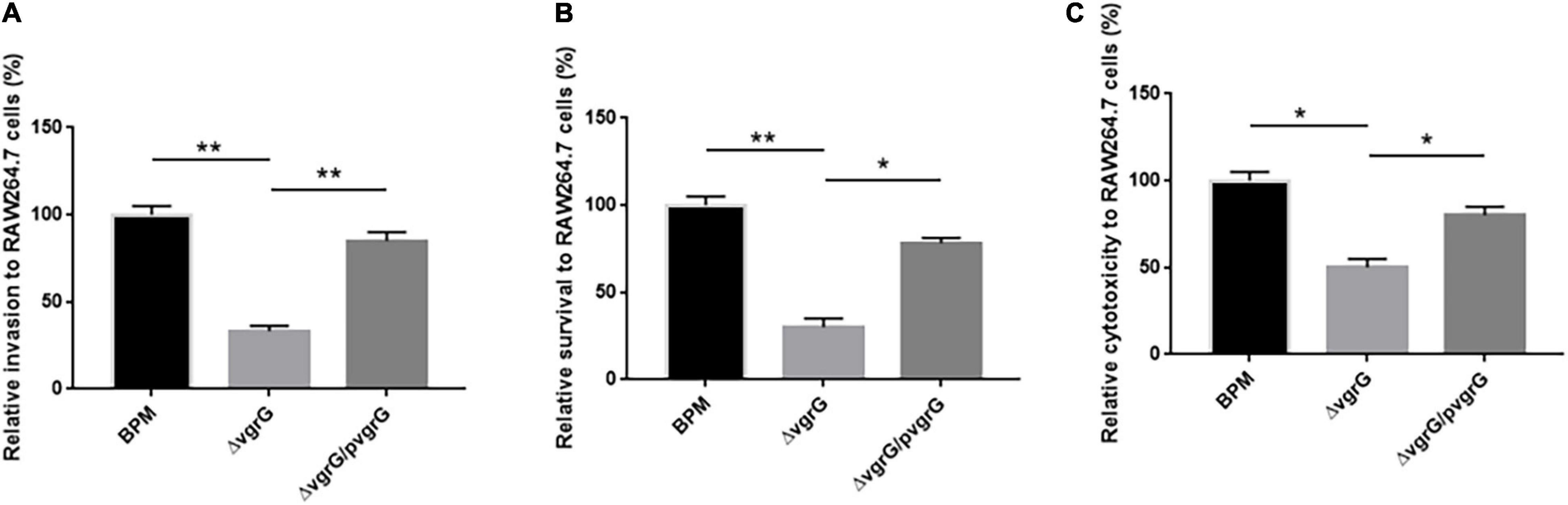
Figure 6. Interaction between bacteria and RAW264.7 cells. (A–C) indicate the invasion ability, intracellular survival ability, and the cytotoxicity, respectively, of the BPM, mutant and complemented strains in RAW264.7 cells. Rates of invasion, survival and cytotoxicity are expressed relative to those of BPM (100%). The means and SDs of three independent experiments in conducted triplicate were calculated. Significant differences between groups are indicated: *(P < 0.05) and **(P < 0.01).
T6SS widely occurs in approximately 25% of all sequenced Gram-negative bacteria, including members of the genera Vibrio, Pseudomonas, Burkholderia, Serratia, Edwardsiella, and Enterobacter (Chieng et al., 2015; Gerc et al., 2015; Wood et al., 2019; Crisan and Hammer, 2020). T6SS plays an important role in pathogenicity, competition, proliferation, and cooperation (Chen et al., 2015). The T6SS is structurally, functionally, and evolutionarily related to contractile injection systems (CISs), a broad family of machines with a spring-like mechanism for delivering macromolecules into target cells (Douzi et al., 2018; Navarro-Garcia et al., 2019). A series of Burkholderia virulence factors, including secreted toxins, adhesins, iron acquisition systems, T6SS, and BLF1, have been reported (Bernhards et al., 2017; Lennings et al., 2018; Rust et al., 2018). Recent studies have indicated that T6SS plays an important role in the competition and pathogenicity of Burkholderia (Chieng et al., 2015). Our previous studies have shown that the clinical symptoms and imaging findings of patients with BPM infection are consistent with those of acute melioidosis (Chang et al., 2017). Therefore, we believe that hypervirulent B. thailandensis may pose a significant threat to human public health, and it is important to study the potential virulence-associated genes involved in BPM. In one of our previous studies, nine vgrG genes were found in the BPM genome. Further sequence analysis showed that only the vgrG2 gene was specific to BPM. Additionally, transcriptome analysis of BPM and E264 showed that the vgrG2 gene was strongly expressed in BPM but not in E264. Therefore, we hypothesized that the vgrG2 gene is involved in the function of the T6SS. To test our hypothesis and describe the role of the vgrG2 gene in BPM, a series of experiments were carried out.
To study the effect of the vgrG2 gene on the pathogenicity of BPM, knockout mutants and complemented strains of the vgrG2 gene were developed from BPM, and there were no significant differences in growth characteristics and antimicrobial sensitivity between them. In addition, the survival rate and LD50 of BALB/c mice infected with these strains were compared. In animal model experiments, we found that the survival rate and LD50 of BALB/c mice infected with ΔvgrG2 were higher than those of BALB/c mice infected with BPM, which demonstrated that the deletion of the vgrG2 gene significantly weakened the virulence of BPM. It has been reported that the T6SS can activate the inflammasome and cause inflammation (Aubert et al., 2016; Ratner et al., 2017; Loeven et al., 2021). To further study the function of the vgrG2 gene in BPM pathogenesis, the serum levels of TNF-a, IL-1β and IL-6 in BALB/c mice were detected. We found that the level of IL-1β was significantly reduced after the deletion of the vgrG2 gene. These findings indicated that the vgrG2 gene of the T6SS in BPM plays an important role in the pathogenicity of BPM, which was consistent with previous reports (Aubert et al., 2016). Unexpectedly, no significant differences in inflammatory cell infiltration were observed in the lungs and livers of the mice after stimulation with these different strains, particularly at 8 h after infection. The results of animal model experiments showed that the deletion of the vgrG2 gene led to the elimination of BPM lethality and a decrease in serum cytokine levels in BALB/c mouse serum.
The evasion of innate immune responses was reported to be very important for the survival and pathogenicity of P. aeruginosa and B. pseudomallei (Gong et al., 2011; Alonso et al., 2020). In this study, our goal was to assess whether the BPM and complemented strains differ from ΔvgrG2 in terms of their virulence and lethality. We accomplished this by measuring the survival rates of the indicated strains in whole blood. The experimental results indicated that the vgrG2 gene may participate in the immune evasion of BPM and play a key role in the evasion of innate immune responses in whole blood. Bacterial adherence to and interaction with RAW264.7 cells are prerequisites for the induction of bacterial infection (Bruballa et al., 2020). We observed the invasion and survival abilities of the BPM, mutant and complemented strains and compared them to the abilities of RAW264.7 cells. We observed some differences when we used an MOI of 10 in our studies. Similarly, only the vgrG2 gene affected the adherence and invasion abilities of BPM, which was consistent with the above results. In addition, previous studies have reported that the vgrG gene in E. coli and B. pseudomallei induced cell toxicity (Hopf et al., 2014; Cianfanelli et al., 2016). Therefore, the cytotoxicity of the BPM, mutant and complemented strains was compared, and the results were consistent with the results obtained from the examination of whole blood killing, adherence and invasion. It could be concluded that the vgrG2 gene located within the T6SS plays a role in BPM pathogenicity, which is consistent with the hypothesis that the vgrG2 gene is functional.
In the current study, we were unable to establish a correlation, and the virulence phenotypes of the BPM, mutant and complemented strains were similar in vitro, although differences in mortality were observed in the in vivo intravenous model of infection. Due to the wide variability of Burkholderia virulence properties, we strongly recommend that the selection of the tissue culture cells used in in vitro studies should be directly related to the cells found in the organ to which the dose will be delivered in vivo (Rao et al., 2020). Therefore, our study attempted to standardize the cell types used for in vitro and in vivo studies to provide a more meaningful comparison.
In conclusion, our study showed that only the vgrG2 gene was involved in the whole blood killing of BPM, which promoted the adhesion and invasion of BPM to host cells and enhanced its pathogenicity in the host. Further studies could focus on exploring potential T6SS effectors to facilitate the development of effective antimicrobial agents for the treatment of B. thailandensis infection.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the research board of the Ethics Committee of the Third Military Medical University under permit number AMUMEC-20201085.
JL, W-WH, and G-XQ performed the laboratory measurements. PJ, J-QL, W-HH, and Y-JJ made substantial contributions to the conception and design. X-RL, YX, JL, and QS participated in the experimental design and data analysis. JL drafted the manuscript. All authors read and approved the final manuscript.
This study was supported by the Youth Project of the National Natural Science Foundation of China (82002113), Chongqing Medical Scientific Research Project (Joint Project of Chongqing Health Commission and Science and Technology Bureau) (No. 2022QNXM034), and Chongqing Natural Science Foundation (cstc2021jcyj-msxmX0269).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.811343/full#supplementary-material
Supplementary Figure 1 | Detection of vgrG2 gene in BPM, mutant and complemented strains.
Supplementary Figure 2 | Growth characteristics of BPM, mutant and complemented strains.
Supplementary Table 1 | The antibiotic susceptibility testing results of BPM, mutant and complemented strains.
BPM, Burkholderia thailandensis BPM; B. pseudomallei, Burkholderia pseudomallei; E. coli, Escherichia coli; P. aeruginosa, Pseudomonas aeruginosa.
Alonso, B., Fernández-Barat, L., Di Domenico, E. G., Marín, M., Cercenado, E., Merino, I., et al. (2020). Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect. Dis. 20:909. doi: 10.1186/s12879-020-05534-1
Aubert, D. F., Xu, H., Yang, J., Shi, X., Gao, W., Li, L., et al. (2016). A burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19, 664–674. doi: 10.1016/j.chom.2016.04.004
Barnes, J. L., and Ketheesan, N. (2005). Route of infection in melioidosis. Emerg. Infect. Dis. 11, 638–639. doi: 10.3201/eid1104.041051
Bernhards, R. C., Cote, C. K., Amemiya, K., Waag, D. M., Klimko, C. P., Worsham, P. L., et al. (2017). Characterization of in vitro phenotypes of Burkholderia pseudomallei and Burkholderia mallei strains potentially associated with persistent infection in mice. Arch. Microbiol. 199, 277–301. doi: 10.1007/s00203-016-1303-8
Bobenchik, A. M., Deak, E., Hindler, J. A., Charlton, C. L., and Humphries, R. M. (2017). Performance of Vitek 2 for antimicrobial susceptibility testing of Acinetobacter baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia with Vitek 2 (2009 FDA) and CLSI M100S 26th Edition Breakpoints. J. Clin. Microbiol. 55, 450–456. doi: 10.1128/JCM.01859-16
Brett, P. J., DeShazer, D., and Woods, D. E. (1998). Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48, 317–320. doi: 10.1099/00207713-48-1-317
Bruballa, A. C., Shiromizu, C. M., Bernal, A. M., Pineda, G. E., Sabbione, F., Trevani, A. S., et al. (2020). Role of shiga toxins in cytotoxicity and immunomodulatory effects of Escherichia coli O157:H7 during host-bacterial interactions in vitro. Toxins 12:48. doi: 10.3390/toxins12010048
Chang, K., Luo, J., Xu, H., Li, M., Zhang, F., Li, J., et al. (2017). Human Infection with Burkholderia thailandensis, China, 2013. Emerg. Infect. Dis. 23, 1416–1418. doi: 10.3201/eid2308.170048
Chen, L., Zou, Y., She, P., and Wu, Y. (2015). Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 172, 19–25. doi: 10.1016/j.micres.2015.01.004
Chieng, S., Mohamed, R., and Nathan, S. (2015). Transcriptome analysis of Burkholderia pseudomallei T6SS identifies Hcp1 as a potential serodiagnostic marker. Microb. Pathog. 79, 47–56. doi: 10.1016/j.micpath.2015.01.006
Cianfanelli, F. R., Alcoforado Diniz, J., Guo, M., De Cesare, V., Trost, M., and Coulthurst, S. J. (2016). VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12:e1005735. doi: 10.1371/journal.ppat.1005735
Crisan, C. V., and Hammer, B. K. (2020). The Vibrio cholerae type VI secretion system: toxins, regulators and consequences. Environ. Microbiol. 22, 4112–4122. doi: 10.1111/1462-2920.14976
Douzi, B., Logger, L., Spinelli, S., Blangy, S., Cambillau, C., and Cascales, E. (2018). Structure-function analysis of the C-terminal domain of the type VI secretion TssB tail sheath subunit. J. Mol. Biol. 430, 297–309. doi: 10.1016/j.jmb.2017.11.015
Fridman, C. M., Keppel, K., Gerlic, M., Bosis, E., and Salomon, D. (2020). A comparative genomics methodology reveals a widespread family of membrane-disrupting T6SS effectors. Nat. Commun. 11:1085. doi: 10.1038/s41467-020-14951-4
Garcia, E. C. (2017). Burkholderia thailandensis: genetic Manipulation. Curr. Protoc. Microbiol. 45, 4C.2.1–4C.2.15. doi: 10.1002/cpmc.27
Gassiep, I., Armstrong, M., and Norton, R. (2020). Human melioidosis. Clin. Microbiol. Rev. 33, e00006–19. doi: 10.1128/CMR.00006-19
Gee, J. E., Elrod, M. G., Gulvik, C. A., Haselow, D. T., Waters, C., Liu, L., et al. (2018). Burkholderia thailandensis isolated from infected wound, Arkansas, USA. Emerg. Infect. Dis. 24, 2091–2094. doi: 10.3201/eid2411.180821
Gerc, A. J., Diepold, A., Trunk, K., Porter, M., Rickman, C., Armitage, J. P., et al. (2015). Visualization of the serratia type VI secretion system reveals unprovoked attacks and dynamic assembly. Cell Rep. 12, 2131–2142. doi: 10.1016/j.celrep.2015.08.053
Ginther, J. L., Mayo, M., Warrington, S. D., Kaestli, M., Mullins, T., Wagner, D. M., et al. (2015). Identification of burkholderia pseudomallei near-neighbor species in the northern territory of Australia. PLoS Negl. Trop. Dis. 9:e0003892. doi: 10.1371/journal.pntd.0003892
Glass, M. B., Gee, J. E., Steigerwalt, A. G., Cavuoti, D., Barton, T., Hardy, R. D., et al. (2006). Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J. Clin. Microbiol. 44, 4601–4604. doi: 10.1128/JCM.01585-06
Gong, L., Cullinane, M., Treerat, P., Ramm, G., Prescott, M., Adler, B., et al. (2011). The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One 6:e17852. doi: 10.1371/journal.pone.0017852
Hopf, V., Göhler, A., Eske-Pogodda, K., Bast, A., Steinmetz, I., and Breitbach, K. (2014). BPSS1504, a cluster 1 type VI secretion gene, is involved in intracellular survival and virulence of Burkholderia pseudomallei. Infect. Immun. 82, 2006–2015. doi: 10.1128/IAI.01544-14
Hsieh, P. F., Lu, Y. R., Lin, T. L., Lai, L. Y., and Wang, J. T. (2019). Klebsiella pneumoniae Type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647. doi: 10.1093/infdis/jiy534
Jiang, X., Yang, Y., Zhou, J., Zhu, L., Gu, Y., Zhang, X., et al. (2016). Roles of the putative type IV-like secretion system key component VirD4 and PrsA in pathogenesis of streptococcus suis type 2. Front. Cell. Infect. Microbiol. 6:172. doi: 10.3389/fcimb.2016.00172
Kovacs-Simon, A., Hemsley, C. M., Scott, A. E., Prior, J. L., and Titball, R. W. (2019). Burkholderia thailandensis strain E555 is a surrogate for the investigation of Burkholderia pseudomallei replication and survival in macrophages. BMC Microbiol. 19:97. doi: 10.1186/s12866-019-1469-8
Lennings, J., West, T. E., and Schwarz, S. (2018). The Burkholderia type VI secretion system 5: composition, regulation and role in virulence. Front. Microbiol. 9:3339. doi: 10.3389/fmicb.2018.03339
Lertpatanasuwan, N., Sermsri, K., Petkaseam, A., Trakulsomboon, S., Thamlikitkul, V., and Suputtamongkol, Y. (1999). Arabinose-positive Burkholderia pseudomallei infection in humans: case report. Clin. Infect. Dis. 28, 927–928. doi: 10.1086/517253
Lewis, E., Kilgore, P. B., Mott, T. M., Pradenas, G. A., and Torres, A. G. (2017). Comparing in vitro and in vivo virulence phenotypes of Burkholderia pseudomallei type G strains. PLoS One 12:e0175983. doi: 10.1371/journal.pone.0175983
Loeven, N. A., Perault, A. I., Cotter, P. A., Hodges, C. A., Schwartzman, J. D., Hampton, T. H., et al. (2021). The Burkholderia cenocepacia type VI secretion system effector TecA is a virulence factor in mouse models of lung infection. mBio 12:e0209821. doi: 10.1128/mBio.02098-21
Luo, P., He, X., Liu, Q., and Hu, C. (2015). Developing universal genetic tools for rapid and efficient deletion mutation in vibrio species based on suicide T-vectors carrying a novel counterselectable marker, vmi480. PLoS One 10:e0144465. doi: 10.1371/journal.pone.0144465
Navarro-Garcia, F., Ruiz-Perez, F., and Cataldi, Á, and Larzábal, M. (2019). Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front. Microbiol. 10:1965. doi: 10.3389/fmicb.2019.01965
Ngamdee, W., Tandhavanant, S., Wikraiphat, C., Reamtong, O., Wuthiekanun, V., Salje, J., et al. (2015). Competition between Burkholderia pseudomallei and B. thailandensis. BMC Microbiol. 15:56. doi: 10.1186/s12866-015-0395-7
Pijuan, J., Barceló, C., Moreno, D. F., Maiques, O., Sisó, P., Marti, R. M., et al. (2019). In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol. 7:107. doi: 10.3389/fcell.2019.00107
Rao, C., Mao, C., Xia, Y., Zhang, M., Hu, Z., Yuan, S., et al. (2020). Transcriptome analysis reveals unfolded protein response was induced during the early stage of burkholderia pseudomallei infection in A549 cells. Front. Genet. 11:585203. doi: 10.3389/fgene.2020.585203
Ratner, D., Orning, M. P., and Lien, E. (2017). Bacterial secretion systems and regulation of inflammasome activation. J. Leukoc. Biol. 101, 165–181. doi: 10.1189/jlb.4MR0716-330R
Rust, A., Shah, S., Hautbergue, G. M., and Davletov, B. (2018). Burkholderia lethal factor 1, a novel anti-cancer toxin, demonstrates selective cytotoxicity in MYCN-amplified neuroblastoma cells. Toxins 10:261. doi: 10.3390/toxins10070261
Schwarz, S., Singh, P., Robertson, J. D., LeRoux, M., Skerrett, S. J., Goodlett, D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. doi: 10.1128/IAI.01368-13
Tang, X., Li, G., Shi, L., Su, F., Qian, M., Liu, Z., et al. (2021). Combined intermittent fasting and ERK inhibition enhance the anti-tumor effects of chemotherapy via the GSK3β-SIRT7 axis. Nat. Commun. 12:5058. doi: 10.1038/s41467-021-25274-3
Tang, Y., Deng, J., Zhang, J., Zhong, X., Qiu, Y., Zhang, H., et al. (2018). Epidemiological and clinical features of melioidosis: a report of seven cases from Southern Inland China. Am. J. Trop. Med. Hyg. 98, 1296–1299. doi: 10.4269/ajtmh.17-0128
Wiersinga, W. J., Currie, B. J., and Peacock, S. J. (2012). Melioidosis. N. Engl. J. Med. 367, 1035–1044. doi: 10.1056/NEJMra1204699
Wood, T. E., Howard, S. A., Förster, A., Nolan, L. M., Manoli, E., Bullen, N. P., et al. (2019). The Pseudomonas aeruginosa T6SS delivers a periplasmic toxin that disrupts bacterial cell morphology. Cell Rep. 29, 187–201.e7. doi: 10.1016/j.celrep.2019.08.094
Zhao, Y., Liu, G., Li, S., Wang, M., Song, J., Wang, J., et al. (2011). Role of a type IV-like secretion system of Streptococcus suis 2 in the development of streptococcal toxic shock syndrome. J. Infect. Dis. 204, 274–281. doi: 10.1093/infdis/jir261
Zhu, Q., Chen, X., Liu, Y., Wang, R., Chen, J., and Chen, Y. (2020). Virulence, antimicrobial susceptibility, molecular and epidemiological characteristics of a new serotype of vibrio parahaemolyticus from diarrhea patients. Front. Microbiol. 11:2025. doi: 10.3389/fmicb.2020.02025
Zhu, Y., Chen, P., Bao, Y., Men, Y., Zeng, Y., Yang, J., et al. (2016). Complete genome sequence and transcriptomic analysis of a novel marine strain Bacillus weihaiensis reveals the mechanism of brown algae degradation. Sci. Rep. 6:38248. doi: 10.1038/srep38248
Zong, B., Zhang, Y., Wang, X., Liu, M., Zhang, T., Zhu, Y., et al. (2019). Characterization of multiple type-VI secretion system (T6SS) VgrG proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence 10, 118–132. doi: 10.1080/21505594.2019.1573491
Keywords: BPM, pathogenicity, virulence factor, T6SS, vgrG2 gene
Citation: Li J, Hu W-w, Qu G-x, Li X-r, Xiang Y, Jiang P, Luo J-q, He W-h, Jin Y-j and Shi Q (2022) Characterization of a Type VI Secretion System vgrG2 Gene in the Pathogenicity of Burkholderia thailandensis BPM. Front. Microbiol. 12:811343. doi: 10.3389/fmicb.2021.811343
Received: 08 November 2021; Accepted: 13 December 2021;
Published: 05 January 2022.
Edited by:
Yufeng Wang, University of Texas at San Antonio, United StatesReviewed by:
Qingpi Yan, Jimei University, ChinaCopyright © 2022 Li, Hu, Qu, Li, Xiang, Jiang, Luo, He, Jin and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Shi, c2hpcWlvbmdAY3FtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.