- 1Department of Obstetrics and Gynecology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Xi’an Jiaotong University Health Science Center, Xi’an, China
- 3Institute of Molecular and Translational Medicine, Translational Medicine Institute, Xi’an Jiaotong University Health Science Center, Xi’an, China
- 4The CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 5CAS Center for Excellence in Molecular Cell Science, Shanghai, China
The innate immune system defenses against pathogen infections via patten-recognition receptors (PRRs). PRRs initiate immune responses by recognizing pathogen-associated molecular patterns (PAMPs), including peptidoglycan, lipopolysaccharide, and nucleic acids. Several nucleic acid sensors or families have been identified, such as RIG-I-like receptors (RLRs), Toll-like receptors (TLRs), cyclic GMP-AMP synthase (cGAS), and PYHIN family receptors. In recent years, the PYHIN family cytosolic DNA receptors have increased attention because of their important roles in initiating innate immune responses. The family members in humans include Absent in melanoma 2 (AIM2), IFN-γ inducible protein 16 (IFI16), interferon-inducible protein X (IFIX), and myeloid cell nuclear differentiation antigen (MNDA). The PYHIN family members are also identified in mice, including AIM2, p202, p203, p204, and p205. Herein, we summarize recent advances in understanding the activation and immune regulation mechanisms of the PYHIN family during microbial infection. Furthermore, structural characterizations of AIM2, IFI16, p202, and p204 provide more accurate insights into the signaling mechanisms of PYHIN family receptors. Overall, the molecular details will facilitate the development of reagents to defense against viral infections.
Introduction
The innate immune system can utilize pattern recognition receptors (PRRs) to detect and defense against invading pathogens. Once PRRs recognize pathogen-associated molecular patterns (PAMPs), peptidoglycan, lipopolysaccharide, and nucleic acids induce innate immune responses. A series of PRRs capable of sensing nucleic acids from pathogens have been identified over the last decades. Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), including RIG-I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), are responsible for detecting pathogen-derived RNA in the cytosol (Berke et al., 2013). Endosomal Toll-like receptors (TLRs) include TLR3, TLR7, and TLR8 sense RNA, while TLR9 is a DNA sensor in the endosome (Hemmi et al., 2000; Alexopoulou et al., 2001; Bauer et al., 2001; Ahmad-Nejad et al., 2002). In addition, cyclic GMP-AMP synthase (cGAS) senses cytosolic double-strand DNA (dsDNA) (Sun et al., 2013; Wu et al., 2013). Human PYHIN family receptors Absence in melanoma 2 (AIM2) (Hornung et al., 2009), IFN-γ inducible protein 16 (IFI16), Interferon-inducible protein X (IFIX, or pyrin and HIN domain family member 1, PYHIN1) (Diner et al., 2015), and mouse p204 (Unterholzner et al., 2010; Horan et al., 2013; Zhu et al., 2014) sense DNA in the cytosol. Furthermore, IFI16, p204, and IFIX also sense DNA in the nucleus. Lastly, other cytosolic DNA sensors have been reported, including DNA dependent activator of IFN regulatory factor (DAI, also called Z-DNA-binding protein 1, ZBP1) (Takaoka et al., 2007; Jiao et al., 2020), RNA polymerase III, leucine-rich repeat-containing protein (LneRRF1P1) (Yang et al., 2010), Ku70/Ku80 protein (Zhang X. et al., 2011), DEAH box polypeptide 9 (DHX9) and DEAH box polypeptide 36 (DHX36) (Kim T. et al., 2010), and DDX41 helicase (Zhang Z. et al., 2011).
PYHIN family (also called p200 protein) are DNA sensors activated by pathogens infection and stress conditions such as DNA break. In humans, these family members include AIM2, IFI16, myeloid cell nuclear differentiation antigen (MNDA) (Briggs et al., 1992), IFIX. Sometimes the pyrin domain only protein 3 (POP3) was also considered as a variant of the PYHIN family, which is resulted from a HIN domain deletion (Khare et al., 2014), while members in mouse include AIM2, p202, p203, p204, and p205 (Ludlow et al., 2008). Most PYHIN family members possess an N-terminal pyrin domain (PYD, or PAAD or DAPIN domain) and one or two C-terminal hematopoietic interferon-inducible nuclear antigens with 200 amino acid repeats (HIN or HIN-200) domain (Figure 1). p202 lacks a PYD and only harbors two HIN domains. POP3 only possesses a PYD and lacks a HIN domain. PYD belongs to the death domain (DD) superfamily and forms interactions with other PYD-containing proteins to form higher complexes (Park et al., 2007; Jin and Xiao, 2015). PYD: PYD interactions regulate various cellular processes, ranging from inflammation and immunity to apoptosis and cell cycle (Stehlik, 2007). HIN domains have been classified into three subtypes: A, B, and C. MNDA and IFIX contain a single type A HIN (HINa) domain, whereas IFI16, p202, and p204 have one HINa and one HINb domain. p203 has a single HINb domain, whereas AIM2 has a single type C HIN (HINc) domain. HIN domains are responsible for binding DNA. In addition to DNA sensing and subsequence immune activation, it was reported that PYHIN family members function in cell growth and cell cycle control (Lembo et al., 1998; Dauffy et al., 2006), apoptosis (Aglipay et al., 2003), senescence (Xin et al., 2004), DNA damage response, tumor suppression (Chen et al., 2006), and differentiation and autoimmunity (Dermott et al., 2004).
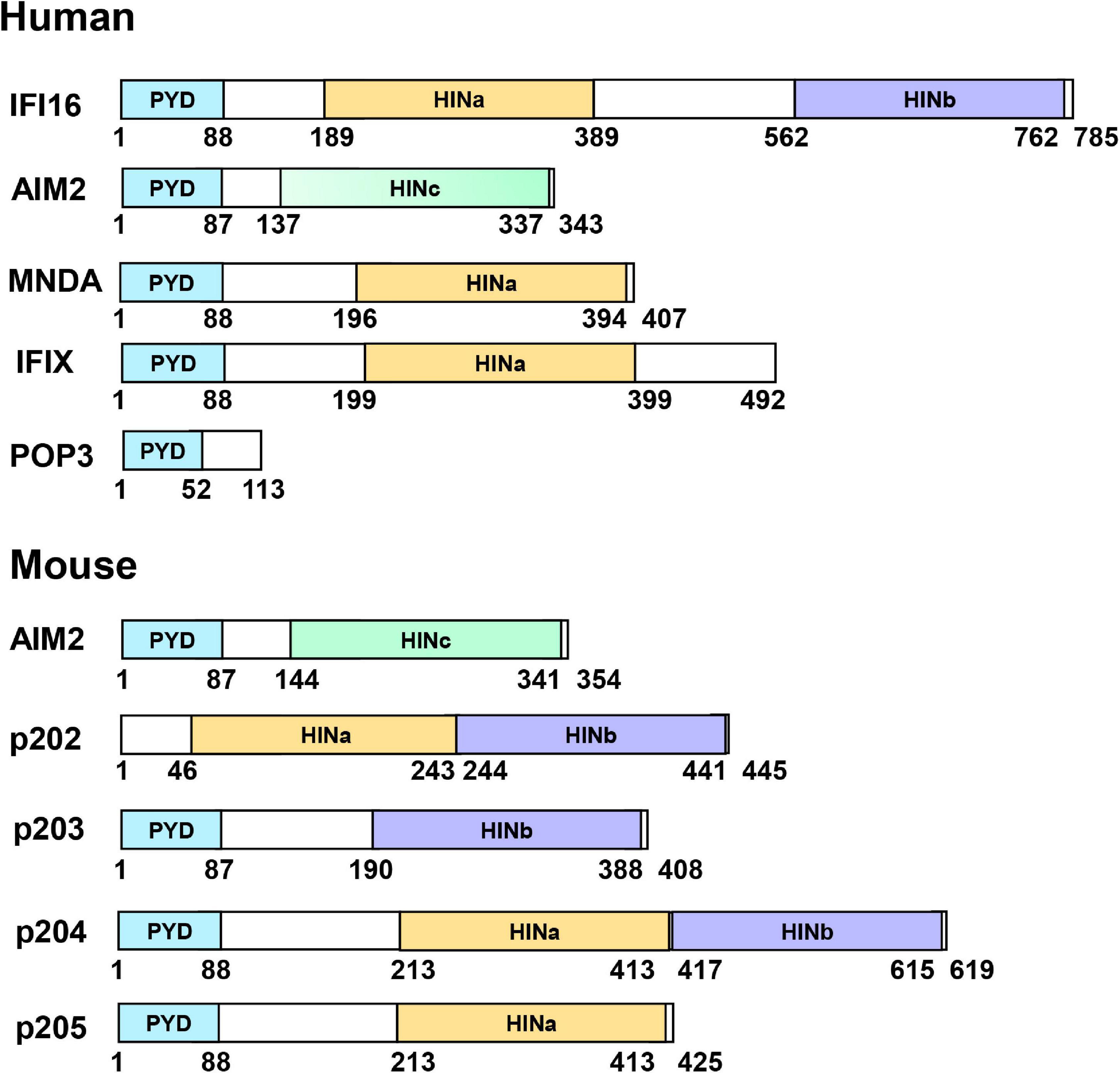
Figure 1. Domain organization of PYHIN family proteins. PYHIN family members possess an N-terminal pyrin domain (PYD) and one or two C-terminal HIN domains, classified as three subtypes; HIN A, HIN B, or HIN C. p202 from mouse lacks PYD and POP3 from human lacks of HIN domains.
In this article, we briefly review the recent advances in understanding the activation and immune regulation mechanisms of the PYHIN family during microbial infection, especially in defending against viral infection. Structural insights into AIM2, IFI16, p202, and p204 provide more accurate DNA recognition mechanisms and signaling transduction pathways. Furthermore, the molecular details will facilitate the development of reagents to defense against pathogen infections.
Absent in Melanoma 2
Absent in melanoma 2 (AIM2) is a 39 kDa protein expressed in the spleen, small intestine, and peripheral leukocytes (DeYoung et al., 1997). It is associated with psoriasis, systemic lupus erythematosus (SLE), chronic kidney disease, diabetes, atherosclerosis, and neuronal diseases (Kimkong et al., 2009; Dombrowski et al., 2011; Yang et al., 2015; Sharma et al., 2019). As a tumor suppressor, the dysregulation of AIM2 is associated with colon and small bowel cancers (Schulmann et al., 2005; Woerner et al., 2007), hepatocellular carcinoma, and prostate cancer (Ponomareva et al., 2013; Ma et al., 2016; Chen et al., 2017), Epstein-Barr virus (EBV)-associated nasopharyngeal carcinoma (Chen et al., 2012), human cutaneous squamous carcinoma (Farshchian et al., 2017), human papillomavirus (HPV)-associated cervical cancer (Milutin Gasperov et al., 2014), non-small cell lung cancer (Kong et al., 2015). Hu et al. (2016) found AIM2 could detect irradiation-induced DNA damage and assemble into inflammasome in the nucleus. AIM2 was later identified as a cytosolic dsDNA sensor that can assemble into inflammasome with ASC (apoptosis-associated speck-like protein containing a CARD) and pro-caspase-1 (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). AIM2 inflammasome can be activated by bacterial pathogens such as Francisella tularensis (Jones et al., 2010), Listeria monocytogenes (Kim S. et al., 2010; Rathinam et al., 2010; Sauer et al., 2010; Warren et al., 2010; Wu et al., 2010; Ge et al., 2012), Streptococcus pneumoniae (Fang et al., 2011), Mycobacterium tuberculosis (Saiga et al., 2012), Staphylococcus aureus (Hanamsagar et al., 2014), and Aspergillus fumigatus (Karki et al., 2015). The activation of AIM2 inflammasome relies on type I interferons during Francisella novicida infection (Belhocine and Monack, 2012; Man et al., 2015). Recently, it was reported that AIM2 could enhance the stability of T regulatory cells (Tregs) during inflammation (Chou et al., 2021). Later, Fidler et al. (2021) found activated AIM2 inflammasome in Jak2V617F macrophages could aggravate atherosclerosis. Lee et al. (2021) reported that AIM2, pyrin, and ZBP1 could form a PANoptosome complex to drive PANoptosis for host defense during HSV-1 and Francisella novicida infections.
AIM2 also senses dsDNA from virus such as HPV 16 (Reinholz et al., 2013), hepatitis B virus (HBV) (Zhen et al., 2014), and EBV (Torii et al., 2017). Human herpesviruses herpes simplex type 1 (HSV-1) and Kaposi’s sarcoma-associated herpesvirus (KSHV) activate the inflammasome in an AIM2-independent manner. Mouse cytomegalovirus (MCMV) and vaccinia virus (VACV) infection induce caspase-1 activation and IL-1β secretion in an AIM2-dependent manner (Hornung et al., 2009; Rathinam et al., 2010). Additionally, several RNA viruses such as Chikungunya virus (CHKV) or West Nile virus (Ekchariyawat et al., 2015) and enterovirus A71 can also activate AIM2 inflammasome, but the mechanism remains unknown (Yogarajah et al., 2017).
Molecular Mechanism of Absent in Melanoma 2 Inflammasome Assembly
Three structures of AIM2 PYD (AIM2PYD) have been reported, including mouse AIM2PYD (mAIM2PYD) (Hou and Niu, 2015), wild-type human AIM2PYD (hAIM2PYD), and its F27G mutant (hAIM2-F27GPYD) (Jin et al., 2013; Lu et al., 2014a). AIM2PYD exhibits a globular structure of a six-helix bundle, sharing the common feature of typical death domains (Figure 2A). Except for the minor differences around α2-α3 helix, the structure of AIM2PYD in the cryo-electron microscopic (Cryo-EM) is similar to its crystal structure, indicating the structural plasticity of AIM2PYD is vital in the PYD: PYD interaction (Lu et al., 2015). Lu et al. (2014b,2015) reported the cryo-EM structure of GFP-hAIM2PYD and human PYD of ASC helical filaments. Electrostatic and hydrophobic interactions are necessary for AIM2PYD polymerization confirmed by EM analysis, pull down, and yeast two-hybrid assay (Lu et al., 2014a; Morrone et al., 2015; Hafner-Bratkovic et al., 2018). It is proposed that the AIM2PYD filament serves as a platform to nucleate ASCPYD filaments (Lu et al., 2014b). Crystal structure of AIM2 HIN domain (AIM2HIN) in complex with dsDNA was determined, and the structure reveals that the HIN domain binds to both the major and minor grooves of dsDNA (Jin et al., 2012). Both oligonucleotide/oligosaccharide (OB) folds and the connecting linker of AIM2HIN participate in dsDNA binding (Figure 2B). It has been reported that a minimum size of ∼ 80 bp dsDNA is required for activating AIM2 and inducing IL-1β production (Jin et al., 2012).
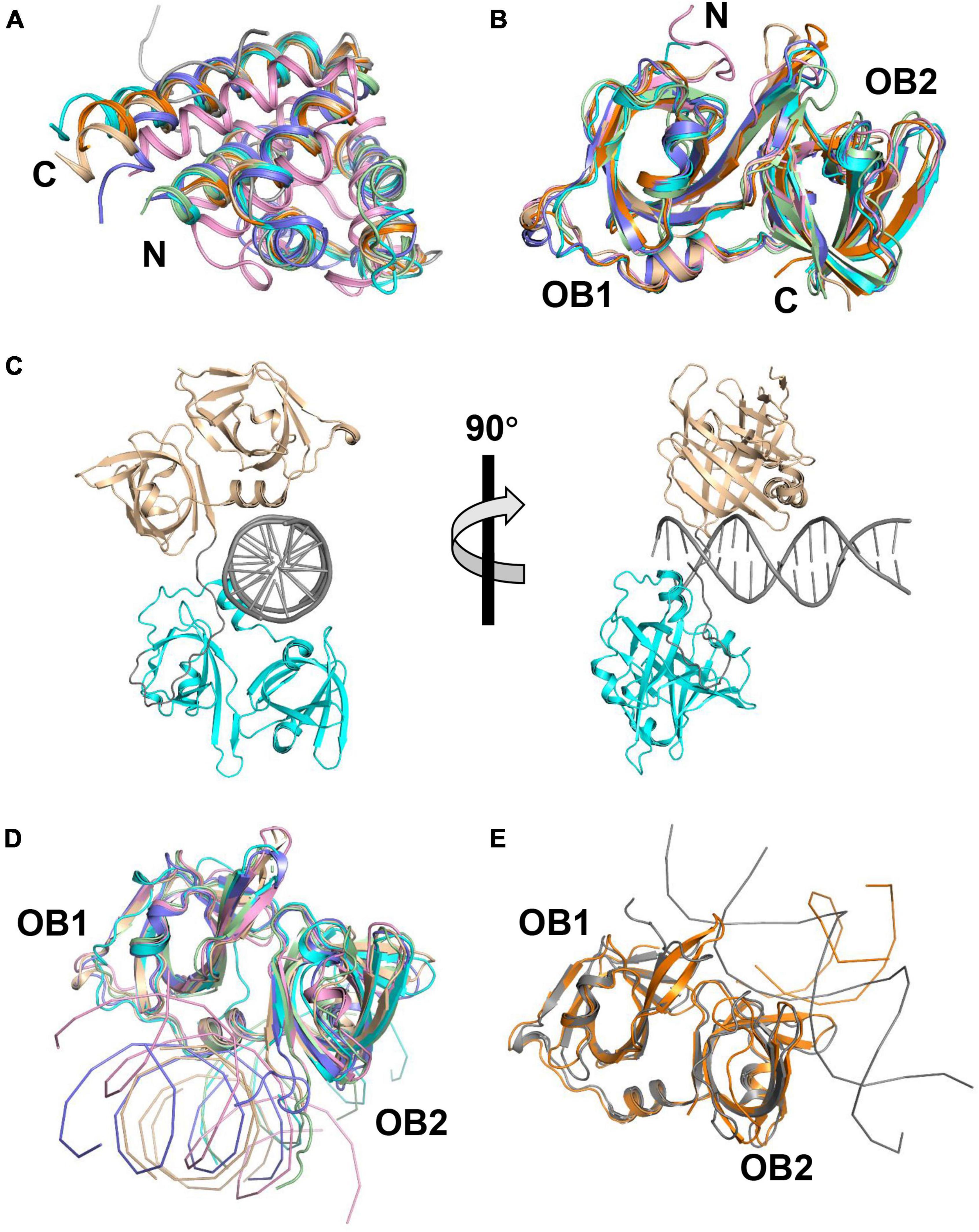
Figure 2. Structures of PYHIN family proteins. (A) Structural superposition of PYD of AIM2, MNDA, and p205. The structure of human AIM2 PYD with MBP (PDB: 3VD8) and its mutant (F27G, PDB: 4O7Q) are shown in cyan and pale green. The structure of mouse AIM2 PYD (PDB: 2N00) is shown in slate. Structures of human MNDA PYD without MBP (PDB: 5WPZ) and with MBP (PDB: 5H7Q, 5WQ6) are shown in pink, orange, and wheat. The solution structure of p205 PYD (PDB: 2YU0) is shown in gray. (B) Structural superposition of HIN domains of IFI16, p202, and p204. Structures of p204 HINa (PDB: 5YZP) and HINb (PDB: 5YZW) are shown in wheat and cyan. Structures of IFI16 HINa (PDB: 2OQ0) and HINb (PDB: 3B6Y) are shown in orange and slate. Structures of p202 HINa (PDB: 4JBJ) and HINb (PDB: 4L5T) are shown in pink and pale green. (C) Crystal structure of p204 HINab: dsDNA complex. Structures of p204 HINa and HINb are shown in wheat and cyan. dsDNA and the linker between HINa and HINb are shown in gray. (D) Similar DNA-binding mode of AIM2, IFI16 HINb, and p204 HINa and HINb. Structures of p204 HINa: DNA (wheat) and HINb: DNA (cyan) are from HINab: dsDNA complex (PDB: 5Z7D). Structures of human AIM2 HIN (PDB: 3RN2), mouse AIM2 HIN (PDB: 4JBM), and IFI16 HINb (PDB: 3RNU) are shown in pink, pale green, and slate, respectively. (E) Similar DNA-binding mode of IFI16 HINa and p202 HINa. Structures of p202 HINa (PDB: 4L5R) and IFI16 HINa (PDB: 4QGU) are shown in gray and orange, respectively.
Absent in melanoma 2 presents in an auto-inhibited state in the absence of DNA, confirmed by in vitro pull-down assay, ITC, and fluorescence polarization inhibition assays (Jin et al., 2012, 2013). Lu et al. (2015) proposed the AIM2HIN: dsDNA filament model, in which AIM2HIN is wrapped around dsDNA filament and the filament diameter is ∼7.5 nm. Each HIN domain interacts with six adjacent HIN domains, contributing to adjacent PYDs and forming short helical filaments. Subsequently, AIM2PYD filaments interact with ASCPYD filaments, and the CARD also organizes into ASCCARD filaments to nucleate caspase-1 filaments. Lastly, caspase-1-activating inflammasome induces the production of mature IL-1β and IL-18.
Regulation of Absent in Melanoma 2
AIM2 inflammasome is activated by the elevated expression of Histone deacetylases 3 (HDAC3) and downregulated by RGFP966 (an HDAC3 inhibitor) (Zhang et al., 2020). Tripartite motif protein 11 (TRIM11) negatively regulates AIM2 by mediating the degradation of AIM2 (Liu et al., 2016; Yang et al., 2017). IFI16 isoform IFI16-β inhibits AIM2 via the competition for dsDNA and interaction with AIM2 (Wang et al., 2018). p202 and POP3 also inhibit AIM2 activation (Roberts et al., 2009; Khare et al., 2014). Additionally, the virus also inhibits AIM2 inflammasome as a defense strategy. Herpes simplex virus 1 (HSV-1) tegument protein VP22 and Human cytomegalovirus (HCMV) protein pUL83 (also named pp65) interact with AIM2 to inhibit the oligomerization and activation of AIM2 (Huang et al., 2017; Maruzuru et al., 2018).
Signal Transduction Pathway of Absent in Melanoma 2
AIM2 is in an auto-inhibited state in the cytoplasm (Figure 3). Once AIM2 senses the invasion of viral dsDNA, its HIN domain interacts with dsDNA. Then AIM2 associates with the adaptor protein ASC through PYD: PYD interaction. ASC further recruits and activates pro-caspase-1 through a homotypic CARD: CARD interaction. Caspase-1, in turn, processes the inactive precursors of IL-1β and IL-18 into mature cytokines. The activation of caspase-1 results in a rapid inflammatory form of cell death. dsDNA also acts as a platform for recruiting multiple AIM2 molecules, promoting the close approximation of AIM2PYD and the formation of AIM2 inflammasome. AIM2 inflammasome, in turn, facilitates the nucleation of ASCPYD, leading to the formation of ASC filaments and activation of caspase-1. However, the signal transduction pathway of AIM2 can be inhibited by p202, IFI16 isoform IFI16-β, and POP3.
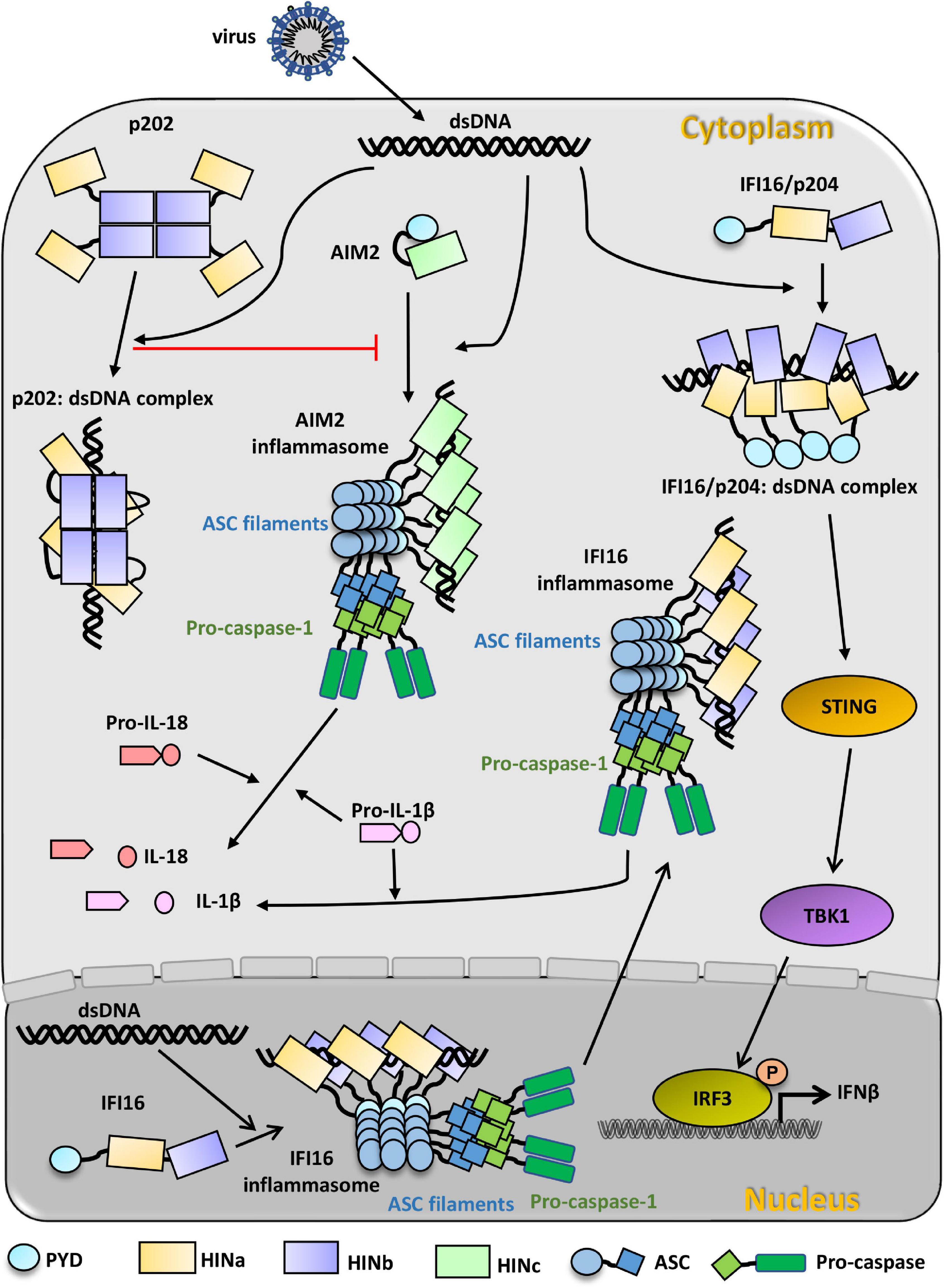
Figure 3. PYHIN family proteins in innate immunity recognition of viral dsDNA. Cytosolic dsDNA from invading virus activates AIM2, which presents an auto-inhibition state before recognizing viral dsDNA. AIM2 HIN domain binds to dsDNA in the cytosol and subsequently binds to the adapter ASC through PYD: PYD interaction. AIM2 inflammasome is formed through AIM2-ASC-procaspase-1 oligomerization. Activated caspase-1 can directly cleave pro-IL-1β and pro-IL-18 to IL-1β and IL-18, which can respond to infection. p202 exists as a tetramer in the cytosol and can also bind to dsDNA from invading viruses. p202 can inhibit the formation of AIM2 inflammasome. IFI16 presents an extended state before recognizing dsDNA. In the nucleus, viral dsDNA binds to IFI16 HIN domains and induces IFI16 oligomerization with the assistant of ASC and pro-caspase-1, and the oligomer migrates to the cytoplasm to cleave pro-IL-1β to IL-1β. In the cytosol, viral dsDNA can also directly initiate the activation of IFI16/p204 independent of the inflammasome. Multiple IFI16/p204 HINab domains synergistically bind to the long dsDNA, resulting in the adjacent PYD aggregate to activate STING and TBK1. Activated TBK1 phosphorylates IRF3 and induces the production of IFNβ to defend against the virus infection.
IFN-γ Inducible Protein 16
IFN-γ inducible protein 16 (IFI16) is an 82 kDa protein and contains two HIN domains (HINa and HINb) (Figure 1). It is engaged in cell cycle control and transcriptional regulation. It can interact with transcription factors Sp1 and p53 (Johnstone et al., 2000; Liao et al., 2011). It can also bind to the promoter region of oncogenes like c-MYC and RAS to repress their transcription (Zhang et al., 2007; Egistelli et al., 2009). By cooperating with p53, IFI16 can inhibit tumorigenesis (Fujiuchi et al., 2004; Choubey and Panchanathan, 2016; Lin et al., 2017). It is also associated with the retinoblastoma tumor-suppressor protein Rb and the transcription factor E2F1 to mediate transcriptional repression (Xin et al., 2003). It can interact with Sp1 to inhibit viral replication (Gariano et al., 2012). The aberrant activity of IFI16 is also associated with several autoimmune disorders, such as SLE and Sjögren syndrome (Seelig et al., 1994; Uchida et al., 2005; Mondini et al., 2007, 2010; Kimkong et al., 2009; Veeranki and Choubey, 2012; Gugliesi et al., 2013; Alunno et al., 2015; Baer et al., 2016). IFI16 was initially identified as a novel DNA sensor by affinity pull-down using cytosolic extracts from human THP-1 monocytes (Unterholzner et al., 2010). It can form inflammasomes sensing DNA viruses replicating in the nucleus (Kerur et al., 2011). It also can response to infection with retroviruses such as human immunodeficiency virus type 1 (HIV-1) in macrophages (Jakobsen et al., 2013) as well as to infection with intracellular bacteria such as Listeria monocytogenes (Hansen et al., 2014) and Francisella novicida (Storek et al., 2015).
IFI16 senses dsDNA from several viruses, including KSHV and VACV (Unterholzner et al., 2010; Kerur et al., 2011; Ansari et al., 2015), HSV-1 (Johnson et al., 2013), EBV (Ansari et al., 2013), and HIV-1 (Jakobsen et al., 2013). It also recognizes ssDNA from HIV-infected CD4+ T cells and nuclear-damaged DNA from etoposide-treated keratinocytes (Monroe et al., 2014; Dunphy et al., 2018). Yang et al. (2020) reported that IFI16 could recognize the covalently closed circular DNA (cccDNA) of HBV and mediate the epigenetic silencing of HBV gene expression (Yang et al., 2020). IFI16 can also reduce viral replication in cells carrying the episomal HPV 18 genome (Lo Cigno et al., 2015) and reduce HCMV mRNAs synthesis (Griffiths et al., 2015). Wichit et al. (2019) found that IFI16 could inhibit the replication of Zika (ZIKV) and CHKV in primary foreskin fibroblasts (Wichit et al., 2019). Additionally, IFI16 was found to participate in the innate immunity via interacting with RIG-I (retinoic acid-inducible gene I) with its PYD, which leads to the activation of RIG-I (Sui et al., 2014; Jiang et al., 2021). Moreover, IFI16-dependent inflammasome was activated by sensing herpes viruses such as KSHV, HSV-1, and EBV (Kerur et al., 2011; Ansari et al., 2013; Johnson et al., 2013). HSV-1 and HCMV infection initialed the IFI16 dependent IFN-β induction via the STING/TBK1 pathway (Unterholzner et al., 2010; Diner et al., 2016).
Structural Basis of IFN-γ Inducible Protein 16 Sensing Viral DNA
It has been reported that HINa and HINb domains of IFI16 bind to both ssDNA and dsDNA in vitro, and the HINb domain possesses a stronger DNA binding affinity (Yan et al., 2008; Unterholzner et al., 2010; Jin et al., 2012). Structural superimposition reveals that the overall topologies of IFI16 HINa (PDB: 2OQ0) and HINb (PDB: 3B6Y) are similar (Figure 2B). Comparing the structures of IFI16 HINa in HINa: DNA complex (PDB: 4QGU) and HINb in HINb: DNA complex (PDB: 3RNU), HINa and HINb bind to DNA at different surfaces (Figures 2D,E), IFI16 HINa binds DNA via the loops from OB folds to tether DNA whereas HINb binds DNA via the linker between OB1 and OB2 folds. Ni et al. (2016) reported that the mutants with impaired DNA-binding ability, IFI16 HINa enhanced, but HINb reduced the production of IFN-β, suggesting IFI16 HINa and HINb might play distinct roles during sensing DNA.
The full-length and HINab domain of IFI16 exists as a monomer in solution confirmed by small-angle X-ray scattering (SAXS) analysis (Liao et al., 2011; Ni et al., 2016). IFI16 HINab forms a large oligomer in the presence of 42 mer dsDNA (Ni et al., 2016). Morrone et al. (2014) found IFI16 binds to dsDNA in a length-dependent manner and assembles into filaments on the long dsDNA. They proposed that HIN domains of IFI16 independently bind to dsDNA like beads on a string, which further induces the conditional proximity of PYD to driven cooperative filament formation of IFI16 upon encountering dsDNA (Morrone et al., 2014).
Regulation of IFN-γ Inducible Protein 16 Pathways
IFI16 possesses an evolutionarily conserved multipartite nuclear localization signal (NLS) for sensing viral DNA in the nucleus (Li et al., 2012). IFI16 can be phosphorylated at unknown sites (Choubey and Lengyel, 1992; Johnstone et al., 1998). Li et al. (2012) identified the phosphorylation sites of IFI16 NLS and indicated that S95, S106, and S153 have little impact on localization. Acetyltransferase p300 and histone deacetylase HDAC can regulate the acetylation status of IFI16 (Li et al., 2012). Li et al. (2012) reported that the overexpression of p300 or the inhibition of HDAC could trigger the cytoplasmic accumulation of IFI16. They also reported that STING negatively regulates IFI16 stability by recruiting the E3 ligase TRIM21 to eliminate the high risk of IFI16 accumulation (Li et al., 2019).
Viruses have evolved several strategies to escape host innate immune responses mediated by IFI16. HBV could negatively regulate the expression of IFI16 in hepatocytes (Yang et al., 2020). HCMV virion protein pUL83 can interact with IFI16 to enhance the transcriptional activity of viral immediate-early promoters (Cristea et al., 2010). Li et al. (2013) found that pUL83 can interact with the PYD of IFI16, resulting in the blocking of antiviral response. pUL38 can also cooperate with pUL97 (HCMV encoded serine/threonine-specific kinase) to phosphorylate IFI16 (Dell’Oste et al., 2014). US28, an HCMV encoded G-protein-coupled receptor, is associated with the degradation of IFI16 in the viral latency (Elder et al., 2019). ICP0, a viral ubiquitin ligase from HSV-1, can induce IFI16 degradation (Orzalli et al., 2012). However, another research reported that ICP0 is neither sufficient nor necessary to degrade IFI16 during HSV-1 infection (Cuchet-Lourenco et al., 2013).
Signal Transduction Pathway of IFN-γ Inducible Protein 16
IFI16 exhibits an extended state in the absence of invasion DNA (Figure 3). In the cytoplasm, IFI16 binds to dsDNA via its C-terminal HINa and HINb domains. Along the long dsDNA, IFI16 forms filaments with the association of PYD. Subsequently, IFI16 filaments recruit STING and TBK1 to phosphorylate STING. The activated STING induces the phosphorylation of IRF3, leading to IFN-I production (Unterholzner et al., 2010; Orzalli et al., 2012; Jakobsen et al., 2013; Liu et al., 2015; Almine et al., 2017). In the nucleus, the binding of exogenous DNA by IFI16 leads to the formation of IFI16-ASC-procaspase-1 inflammasome and cytoplasmic translocation, resulting in the secretion of proinflammatory cytokine IL-1β to defend against viral infection (Kerur et al., 2011; Ansari et al., 2013; Singh et al., 2013).
p202
Mouse p202 was discovered as an interferon-inducible protein in 1989 (Kingsmore et al., 1989). Though there are three homologous p202 genes (p202a, b, and c), only p202a and p202b are expressed (Wang et al., 1999). p202 possesses HINa and HINb domain and lacks PYD (Figure 1). It can negatively regulate transcription factors including MYOD1, myogenin, AP-1, and NF-κB (Min et al., 1996; Datta et al., 1998; Ma et al., 2003). In prostate cancer cells, p202 can suppress apoptosis by deregulated expression of E2F1 (Yan et al., 2003). In sensitizing breast cancer cells, p202 exhibits pro-apoptotic effects by binding to NF-κB (Wen et al., 2000). p202 has been reported to regulate cell cycle progression. It may negatively regulate p53 transcriptional activity by binding 53BP1 (Datta et al., 1996). It can inhibit cell growth by modulating p21 and inhibit cell proliferation by interacting with the retinoblastoma tumor suppressor protein (pRb) and the transcription factor E2F (Choubey et al., 1996; Choubey and Gutterman, 1997; Gutterman and Choubey, 1999). Rozzo et al. (2001) found that p202 is a potential susceptibility candidate gene in autoimmune disease SLE. They also found that p202 can negatively regulate AIM2 inflammasome by binding to DNA alongside AIM2 (Roberts et al., 2009).
Structural Basis of Viral Double-Strand DNA Sensing by p202
Several structures of p202 with and without dsDNA have been reported, revealing the mechanism of p202 sensing viral dsDNA. Crystal structures of p202 HINa (PDB: 4JBJ) and HINb (PDB: 4L5T) are highly similar to the structures of known HIN domains of the PYHIN family (Figure 2B). Four crystal structures of HINa in complex with dsDNA have been reported, including with 20 mer dsDNA (PDB: 4L5R and 4LNQ), 12 mer dsDNA (PDB: 4L5S), and 14 mer dsDNA (PDB: 4JBK) complex structures. Comparing p202 HINa and DNA-bound structures reveals that HINa does not undergo obvious conformational changes upon DNA binding. p202 binds to dsDNA via the loops from both OB1 and OB2 folds, different from AIM2 HIN and IFI16 HINb (Figures 2D,E).
Though p202 HINb alone lacks DNA binding activity, it enhances dsDNA binding for the full-length p202 (Roberts et al., 2009). p202 HINb exists as a tetramer in the solution and presents as dimers in the crystal. p202 HINb structure shows that two molecules form a face-to-face dimer via the same interface analogous to the HINa dsDNA binding site, and two such dimers further oligomerize tail to tail (Li et al., 2014a). The full-length p202 is also a tetramer in vivo and in vitro. It seems that the tetrameric HINb serves as the central platform for HINa to append to and increase the affinity of HINa for targeting dsDNA (Yin et al., 2013).
Additionally, due to p202 HINb interacting with AIM HIN, Yin et al. (2013) proposed that this interaction may result in a spatial separation of the AIM2 PYD and lead to p202 preventing ASC and AIM2 inflammasome activation. Li et al. (2014a) proposed a model of how the full-length p202 protein binds dsDNA from the crystal packing of the p202 HINa: dsDNA complex. Four p202 HINb domains form a tetramer and then tether four p202 HINa domains close, resulting in the simultaneous binding of p202 HINa domains to a dsDNA molecule.
Molecular Mechanism of p202 Inhibiting Absent in Melanoma 2 Activation
To date, two molecular mechanisms of p202 negatively regulating AIM2 inflammasome have been proposed. In the model proposed by Yin et al. (2013), AIM2 HIN domains are spatially separated by the direct interaction between AIM2 HIN and p202 HINb, which prevents AIM2-mediated ASC oligomerization. The other mechanism was proposed by Ru et al. (2013) and Li et al. (2014a), they proposed that p202 inhibits the activation of AIM2 inflammasome by competition for dsDNA. Li et al. (2014a) found that the dsDNA affinity of p202 HINa is approximately fivefold higher than that of AIM2 HIN. The tetrameric p202 also enhances the binding affinity of the HINa domain for dsDNA. When p202 competes for dsDNA bound by AIM2, p202 HINa (higher DNA affinity) can displace AIM2 HIN from dsDNA. Subsequently, the free AIM2 HIN is recruited to and interacts with the closely linked p202 HINb tetramer, which would prevent AIM2 HIN from binding dsDNA and activating the AIM2 inflammasome. Combined with current research, both direct interactions between p202 HINb and AIM2 HIN and the competition of p202 HINa for DNA binding may play a role in p202 inhibiting the activation of AIM2 inflammasome (Li et al., 2014a).
p204
Interferon-inducible protein 204 (p204) with an apparent molecular weight of 72 kDa is a mouse PYHIN protein and is a functional ortholog of human IFI16. p204 possesses PYD, HINa, and HINb domains (Figure 1). It modulates cell proliferation and differentiation (Luan et al., 2008; Zhao et al., 2015). It can directly interact with UBF1 to inhibit ribosomal RNA synthesis and interact with the Rb to coactivate certain transcription factors (Liu et al., 1999; Luan et al., 2007). Yi et al. (2018) found that p204 is necessary for lipopolysaccharide (LPS)-induced Toll-like receptor 4 (TLR4) signaling in pathogen infection. In recent years, p204 has been reported to act as a DNA sensor to defense against pathogen infections. It can sense viral DNA to activate the inflammasome and induce interferon production upon recognizing viral DNA in the cytoplasm and nucleus (Unterholzner et al., 2010; Conrady et al., 2012). It produces IFN-Is in cooperation with cGAS during bacterial infection (Storek et al., 2015). Knockdown of p204 can significantly inhibit IFN-β release in response to infections by Francisella novicida (Storek et al., 2015) and Mycobacterium bovis (Chunfa et al., 2017). Chen et al. (2019) reported that p204 triggers inflammatory responses during Staphylococcus infection. Ryabchenko et al. (2021) found p204 senses viral genomes in the nucleus during the infection of mouse polyomavirus (MPyV).
Structural Basis of p204 Sensing Viral Double-Strand DNA
To date, crystal structures of p204 HINa, HINb, and HINab: dsDNA complex were reported (Tian and Yin, 2019; Fan et al., 2021). The overall structures of p204 HINa (PDB: 5YZP) and HINb (5YZW) are highly similar to those of known HIN domains (Figure 2B). The crystal structure of p204 HINab: dsDNA (PDB: 5Z7D) is the first complex structure of tandem HIN domain bound with dsDNA in the PYHIN family (Figure 2C). There are no obvious conformational changes of p204 HINa and HINb with or without dsDNA. p204 HINa and HINb share a similar dsDNA-binding mode in which the HIN domain binds to DNA mainly via the linker connecting two OB folds. This binding mode is the canonical dsDNA-binding mode of HIN domains in the PYHIN family (Figure 2D). The DNA-binding affinities of p204 HINa and HINb are similar. Furthermore, p204 HINab has a higher DNA-binding affinity than HINa or HINb alone, implying that HINa and HINb synergistically bind to dsDNA (Fan et al., 2021). Size exclusion chromatography and SAXS assay confirmed that p204 HINab alone stays as a monomer in the solution, similar to IFI16. In the p204 HINab: dsDNA complex structure, three HINab molecules form a C-ring-shaped structure for binding dsDNA. The linker between HINa and HINb changes ∼90° angles induced by dsDNA binding, resulting in higher dsDNA affinity (Fan et al., 2021). Interestingly, p204 HINa presents a dimer in the crystal structure, which attributes to dsDNA binding (Fan et al., 2021).
Signal Transduction Pathway of p204
Combining several studies of the ortholog IFI16, the model of p204 recognizes viral dsDNA to activate downstream signaling pathways was proposed (Fan et al., 2021; Figure 3). p204 exhibits an extended conformation in the cytoplasm in the absence of dsDNA. In the presence of viral dsDNA, p204 HINa, and HINb would synergistically bind to dsDNA. Whereafter, more HINab molecules bind to dsDNA and form a C-ring-shaped structure along dsDNA. The dimerization of the HINa domain was stabilized by the binding of dsDNA, which results in the proximity of the adjacent N-terminal PYD domain. As an ortholog of human IFI16, p204 PYD may also aggregate and activate TBK1 and IRF3, inducing the production of IFNβ and proinflammatory cytokines to defend against viruses. The downstream signal pathway of p204 after recognizing viral DNA in the nucleus awaits further studies.
Other Members of the PYHIN Family
Myeloid Cell Nuclear Differentiation Antigen
Myeloid cell nuclear differentiation antigen (MNDA) is a 55 kDa protein and identified in HL-60 cells (Goldberger et al., 1986). It possesses an N-terminal PYD and a C-terminal HINa domain (Figure 1). It is expressed specifically in monocytes, myeloid progenitors, and granulocytes (Goldberger et al., 1984, 1986; Cousar and Briggs, 1990; Briggs et al., 1994). It can dimerize via an imperfect leucine zipper and a basic region (Xie et al., 1997a). Early studies reported that MNDA plays a role in myeloid differentiation and gene transcription. It can bind to the nucleolar proteins nucleolin and nucleophosmin (Xie et al., 1995, 1997b). It can also bind to dsDNA, and the interaction between MNDA and transcription factor YY1 can enhance the affinity of YY1 for its target DNA (Xie et al., 1998; Li et al., 2014b). MNDA plays a role in neutrophil apoptosis (Fotouhi-Ardakani et al., 2010). Latency-associated nuclear antigen (LANA) from KSHV has been shown to associate with MNDA, which may modulate IFN-mediated host defense activities (Fukushi et al., 2003). pUL83 can interact with the PYD of MNDA to block antiviral response (Li et al., 2013). Two crystal structures of MNDA PYD (PDB: 5WPZ and 5H7Q) have been solved, and these structures are highly similar to that of AIM2 PYD (Figure 2A).
Interferon-Inducible Protein X
Interferon-inducible protein X (IFIX, also know as PYHIN1) gene is predicted to encode six protein isoforms (α1, α2, β1, β2, γ1, and γ2) which are localized in the nucleus (Ding et al., 2004). All of them have a PYD and an NLS. Except for IFIXγ, both IFIXα and IFIXβ have HINa domain (Figure 1). Ding et al. (2004) found that IFIXα1 possesses antitumor activity and proposed that it be used as a therapeutic agent. They also found that IFIXα1 can interact with HDM2 to positively regulate p53, which may contribute in part to the antitumor activity (Ding et al., 2006). Li et al. (2013) found that pUL83 can interact with IFIX PYD to block its antiviral response. Diner et al. (2015) found that IFIX can restrict herpesvirus replication via binding viral DNA with the HIN domain and leads to IFN-I response. They also discovered that IFIX could act as an antiviral DNA sensor to detect viral DNA in the nucleus and cytoplasm (Diner et al., 2015). The molecular mechanism of dsDNA recognition and activation for IFIX awaits future studies.
Pyrin Domain Only Protein 3
Pyrin domain only protein 3 (POP3) is sometimes considered a new member of the PYHIN family and was identified in 2014 (Connolly and Bowie, 2014; Khare et al., 2014). It only possesses a PYD and lacks the typical HIN domain of the PYHIN family (Figure 1). It could interact with the PYDs of AIM2 and IFI16, resulting in the inhibition of DNA virus-induced activation of AIM2 and IFI16-inflammasomes (Khare et al., 2014). However, the molecular mechanism of POP3’s interaction with the PYDs of AIM2 and IFI16 remains unknown.
p203
p203 is a nuclear protein in mice and was identified in 1997 (Gribaudo et al., 1997). It possesses PYD and HINb domains (Figure 1). It is expressed in the thymus, bone marrow, spleen, and liver (Gribaudo et al., 1999; Zhang et al., 2008). The level of p203 protein decreased during liver regeneration (Zhang et al., 2008).
p205
p205 harbors a PYD and a C-terminal HINa domain (Figure 1). It is expressed in the thymus, bone marrow, spleen, heart, and skeletal muscle tissue (Asefa et al., 2004; Weiler et al., 1999). It contributes to cell growth and allows progenitor cells to differentiate during myelomonocytic cell differentiation (Dermott et al., 2004). It can directly bind to Rb and p53 and upregulate p21 and Rb (Asefa et al., 2006). It can also inhibit growth in proliferating cells (Asefa et al., 2006). The solution structure of p205 PYD has been solved (PDB: 2YU0) (Figure 2A), which is highly similar to known PYD.
Comparison of HIN Domains in PYHIN Family
To date, the known HIN domain structures include human AIM2 HIN (Jin et al., 2012), human IFI16 HINa and HINb (Liao et al., 2011; Jin et al., 2012; Ni et al., 2016), mouse AIM2 HIN (Ru et al., 2013), mouse p202 HINa and HINb (Ru et al., 2013; Yin et al., 2013), and mouse p204 HINa and HINb (Tian and Yin, 2019; Fan et al., 2021). Although the overall structure of HIN domains is highly conserved, their superposition reveals significant flexibility in the loops in OB folds (Figure 2B). Furthermore, these HIN domains exhibit different surface charges, indicating distinct DNA-binding surfaces (Fan et al., 2021).
Six HIN: dsDNA complex structures have been reported, including human AIM2: dsDNA (PDB: 3RN2) (Jin et al., 2012), mouse AIM2: dsDNA (PDB: 4JBM) (Ru et al., 2013), human IFI16 HINa: dsDNA (PDB: 4QGU) (Ni et al., 2016), human IFI16 HINb: dsDNA (PDB: 3RNU) (Jin et al., 2012), mouse p202 HINa: dsDNA (Ru et al., 2013; Yin et al., 2013), and p204 HINab: dsDNA (PDB: 5Z7D) (Fan et al., 2021). The structural superposition of the HIN: dsDNA complex reveals two distinctly different DNA-binding modes in the PYHIN family (Figures 2D,E). AIM2 HIN, IFI16 HINb, p204 HINa, and HINb employ the linker connecting two OB folds and the surrounding residues to engage dsDNA, whereas IFI16 HINa and p202 HINa interact with dsDNA via an opposite surface formed by the loops of two OB folds.
Therapeutic Approaches to Targeting PYHIN Proteins
Concerning their roles in innate immune responses, PYHIN family members could be utilized as therapeutic targets in autoimmune, inflammatory disorders, and cancers. For example, synthetic oligodeoxynucleotides (ODN) A151 can inhibit immune responses by binding to AIM2, which may treat infectious and autoimmune diseases (Kaminski et al., 2013). LL-37, a human cathelicidin antimicrobial peptide, can interact with DNA to block AIM2 inflammasome activation, which could be used to treat chronic skin disease (Dombrowski et al., 2011). A peptide derived from a virus may aid the design of inhibitors. Such as A46 (known as VIPER), a peptide derived from VACV, could inhibit the TLR signaling pathway (Lysakova-Devine et al., 2010). Thus, analogical viral inhibitors could be designed to target PYHIN proteins. Cai et al. (2020) found that IFI16 can upregulate PD-L1 through the STING-TBK1-NF−κB pathway to promote cervical cancer progression, suggesting IFI16 could be developed as a novel immunotherapy target.
Concluding Remarks and Perspective
Immunological studies, in association with biochemical and structural studies, have revealed the molecular mechanisms of AIM2, IFI16, p202, and p204, sensing viral DNA and resulting in immune responses. However, several questions remain unclear. Firstly, what is the molecular basis for DNA length-dependent response? Morrone et al. (2014) suggested that even in the presence of excess DNA, the formation of IFI16 PYD-driven filament could allow multiple IFI16 to bind adjacent molecules to form signaling foci. Secondly, why IFI16 does not bind to self-DNA? Morrone et al. (2014) suggested that due to the weak binding affinity of HIN domains to self-DNA, HIN domains coupled with filament formation could sufficiently inhibit the interaction with self-DNA. In addition, they also suggested that although the viral genome and host histones package into chromatin after virus invasion, it is more loosely packed than self-DNA. Furthermore, IFI16 filament selectively engages foreign DNA while minimizing its interaction with self-DNA by a competitive mechanism. It is still needed to investigate these fundamental questions further. Thirdly, the mechanism of AIM2 recognizing viral RNA has yet to be investigated. PYHIN proteins participate in response to many pathogens and mediate their clearance. They also contribute to the pathogenesis of autoimmune, auto-inflammatory diseases and cancers. Investigation into the therapeutic approaches targeting PYHIN proteins will greatly aid the development of treating associated diseases.
Author Contributions
XF wrote the manuscript. LJ and TJ reviewed and edited the manuscript. All authors provided funding and contributed to the article, and approved the submitted version.
Funding
XF was supported by the National Natural Science Fund for Young Scholars (31800639). TJ was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29030104), National Natural Science Foundation of China (31870731 and 31971129), and Fundamental Research Funds for the Central Universities. LJ was supported by the National Natural Science Foundation of China (32071213), Shaanxi Provincial Scientific and Technological Activities Funding Project (2020001), and Young Talent Program of Xi’an Jiaotong University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aglipay, J. A., Lee, S. W., Okada, S., Fujiuchi, N., Ohtsuka, T., Kwak, J. C., et al. (2003). A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene 22, 8931–8938. doi: 10.1038/sj.onc.1207057
Ahmad-Nejad, P., Hacker, H., Rutz, M., Bauer, S., Vabulas, R. M., and Wagner, H. (2002). Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur. J. Immunol. 32, 1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958<3.0.CO;2-U
Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738. doi: 10.1038/35099560
Almine, J. F., O’hare, C. A., Dunphy, G., Haga, I. R., Naik, R. J., Atrih, A., et al. (2017). IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 8:14392. doi: 10.1038/ncomms14392
Alunno, A., Caneparo, V., Carubbi, F., Bistoni, O., Caterbi, S., Bartoloni, E., et al. (2015). Interferon gamma-inducible protein 16 in primary Sjogren’s syndrome: a novel player in disease pathogenesis? Arthritis Res. Ther. 17:208. doi: 10.1186/s13075-015-0722-2
Ansari, M. A., Dutta, S., Veettil, M. V., Dutta, D., Iqbal, J., Kumar, B., et al. (2015). Herpesvirus genome recognition induced acetylation of nuclear IFI16 is essential for its cytoplasmic translocation, inflammasome and IFN-beta responses. PLoS Pathog. 11:e1005019. doi: 10.1371/journal.ppat.1005019
Ansari, M. A., Singh, V. V., Dutta, S., Veettil, M. V., Dutta, D., Chikoti, L., et al. (2013). Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J. Virol. 87, 8606–8623. doi: 10.1128/JVI.00805-13
Asefa, B., Dermott, J. M., Kaldis, P., Stefanisko, K., Garfinkel, D. J., and Keller, J. R. (2006). p205, a potential tumor suppressor, inhibits cell proliferation via multiple pathways of cell cycle regulation. FEBS Lett. 580, 1205–1214. doi: 10.1016/j.febslet.2006.01.032
Asefa, B., Klarmann, K. D., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., and Keller, J. R. (2004). The interferon-inducible p200 family of proteins: a perspective on their roles in cell cycle regulation and differentiation. Blood Cells Mol. Dis. 32, 155–167. doi: 10.1016/j.bcmd.2003.10.002
Baer, A. N., Petri, M., Sohn, J., Rosen, A., and Casciola-Rosen, L. (2016). Association of antibodies to interferon-inducible Protein-16 with markers of more severe disease in primary sjogren’s syndrome. Arthritis Care Res. (Hoboken) 68, 254–260. doi: 10.1002/acr.22632
Bauer, S., Kirschning, C. J., Hacker, H., Redecke, V., Hausmann, S., Akira, S., et al. (2001). Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. U.S.A. 98, 9237–9242. doi: 10.1073/pnas.161293498
Belhocine, K., and Monack, D. M. (2012). Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells. Cell Microbiol. 14, 71–80. doi: 10.1111/j.1462-5822.2011.01700.x
Berke, I. C., Li, Y., and Modis, Y. (2013). Structural basis of innate immune recognition of viral RNA. Cell Microbiol. 15, 386–394. doi: 10.1111/cmi.12061
Briggs, J. A., Burrus, G. R., Stickney, B. D., and Briggs, R. C. (1992). Cloning and expression of the human myeloid cell nuclear differentiation antigen: regulation by interferon alpha. J. Cell. Biochem. 49, 82–92. doi: 10.1002/jcb.240490114
Briggs, R., Dworkin, L., Briggs, J., Dessypris, E., Stein, J., Stein, G., et al. (1994). Interferon alpha selectively affects expression of the human myeloid cell nuclear differentiation antigen in late stage cells in the monocytic but not the granulocytic lineage. J. Cell. Biochem. 54, 198–206. doi: 10.1002/jcb.240540208
Bürckstümmer, T., Baumann, C., Blüml, S., Dixit, E., Dürnberger, G., Jahn, H., et al. (2009). An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10, 266–272. doi: 10.1038/ni.1702
Cai, H., Yan, L., Liu, N., Xu, M., and Cai, H. (2020). IFI16 promotes cervical cancer progression by upregulating PD-L1 in immunomicroenvironment through STING-TBK1-NF-kB pathway. Biomed. Pharmacother. 123:109790. doi: 10.1016/j.biopha.2019.109790
Chen, I. F., Ou-Yang, F., Hung, J. Y., Liu, J. C., Wang, H., Wang, S. C., et al. (2006). AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol. Cancer Ther. 5, 1–7. doi: 10.1158/1535-7163.MCT-05-0310
Chen, L. C., Wang, L. J., Tsang, N. M., Ojcius, D. M., Chen, C. C., Ouyang, C. N., et al. (2012). Tumour inflammasome-derived IL-1beta recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol. Med. 4, 1276–1293. doi: 10.1002/emmm.201201569
Chen, S. L., Liu, L. L., Lu, S. X., Luo, R. Z., Wang, C. H., Wang, H., et al. (2017). HBx-mediated decrease of AIM2 contributes to hepatocellular carcinoma metastasis. Mol. Oncol. 11, 1225–1240. doi: 10.1002/1878-0261.12090
Chen, W., Yu, S. X., Zhou, F. H., Zhang, X. J., Gao, W. Y., Li, K. Y., et al. (2019). DNA sensor IFI204 contributes to host defense against Staphylococcus aureus infection in mice. Front. Immunol. 10:474. doi: 10.3389/fimmu.2019.00474
Chou, W. C., Guo, Z., Guo, H., Chen, L., Zhang, G., Liang, K., et al. (2021). AIM2 in regulatory T cells restrains autoimmune diseases. Nature 591, 300–305. doi: 10.1038/s41586-021-03231-w
Choubey, D., and Gutterman, J. U. (1997). Inhibition of E2F-4/DP-1-stimulated transcription by p202. Oncogene 15, 291–301. doi: 10.1038/sj.onc.1201184
Choubey, D., and Lengyel, P. (1992). Interferon action: nucleolar and nucleoplasmic localization of the interferon-inducible 72-kD protein that is encoded by the Ifi 204 gene from the gene 200 cluster. J. Cell Biol. 116, 1333–1341. doi: 10.1083/jcb.116.6.1333
Choubey, D., and Panchanathan, R. (2016). IFI16, an amplifier of DNA-damage response: role in cellular senescence and aging-associated inflammatory diseases. Ageing Res. Rev. 28, 27–36. doi: 10.1016/j.arr.2016.04.002
Choubey, D., Li, S. J., Datta, B., Gutterman, J. U., and Lengyel, P. (1996). Inhibition of E2F-mediated transcription by p202. EMBO J. 15, 5668–5678. doi: 10.1002/j.1460-2075.1996.tb00951.x
Chunfa, L., Xin, S., Qiang, L., Sreevatsan, S., Yang, L., Zhao, D., et al. (2017). The central Role of IFI204 in IFN-beta release and autophagy activation during mycobacterium bovis infection. Front. Cell. Infect. Microbiol. 7:169. doi: 10.3389/fcimb.2017.00169
Connolly, D. J., and Bowie, A. G. (2014). The emerging role of human PYHIN proteins in innate immunity: implications for health and disease. Biochem. Pharmacol. 92, 405–414. doi: 10.1016/j.bcp.2014.08.031
Conrady, C. D., Zheng, M., Fitzgerald, K. A., Liu, C., and Carr, D. J. (2012). Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunol. 5, 173–183. doi: 10.1038/mi.2011.63
Cousar, J. B., and Briggs, R. C. (1990). Expression of human myeloid cell nuclear differentiation antigen (MNDA) in acute leukemias. Leuk. Res. 14, 915–920. doi: 10.1016/0145-2126(90)90182-9
Cristea, I. M., Moorman, N. J., Terhune, S. S., Cuevas, C. D., O’keefe, E. S., Rout, M. P., et al. (2010). Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 84, 7803–7814. doi: 10.1128/JVI.00139-10
Cuchet-Lourenco, D., Anderson, G., Sloan, E., Orr, A., and Everett, R. D. (2013). The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J. Virol. 87, 13422–13432. doi: 10.1128/JVI.02474-13
Datta, B., Li, B., Choubey, D., Nallur, G., and Lengyel, P. (1996). p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J. Biol. Chem. 271, 27544–27555. doi: 10.1074/jbc.271.44.27544
Datta, B., Min, W., Burma, S., and Lengyel, P. (1998). Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Mol. Cell. Biol. 18, 1074–1083. doi: 10.1128/MCB.18.2.1074
Dauffy, J., Mouchiroud, G., and Bourette, R. P. (2006). The interferon-inducible gene, Ifi204, is transcriptionally activated in response to M-CSF, and its expression favors macrophage differentiation in myeloid progenitor cells. J. Leukoc. Biol. 79, 173–183. doi: 10.1189/jlb.0205083
Dell’Oste, V., Gatti, D., Gugliesi, F., De Andrea, M., Bawadekar, M., Lo Cigno, I., et al. (2014). Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J. Virol. 88, 6970–6982. doi: 10.1128/JVI.00384-14
Dermott, J. M., Gooya, J. M., Asefa, B., Weiler, S. R., Smith, M., and Keller, J. R. (2004). Inhibition of growth by p205: a nuclear protein and putative tumor suppressor expressed during myeloid cell differentiation. Stem Cells 22, 832–848. doi: 10.1634/stemcells.22-5-832
DeYoung, K. L., Ray, M. E., Su, Y. A., Anzick, S. L., Johnstone, R. W., Trapani, J. A., et al. (1997). Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene 15, 453–457. doi: 10.1038/sj.onc.1201206
Diner, B. A., Li, T., Greco, T. M., Crow, M. S., Fuesler, J. A., Wang, J., et al. (2015). The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol. Syst. Biol. 11:787. doi: 10.15252/msb.20145808
Diner, B. A., Lum, K. K., Toettcher, J. E., and Cristea, I. M. (2016). Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. mBio 7:e01553-16. doi: 10.1128/mBio.01553-16
Ding, Y., Lee, J. F., Lu, H., Lee, M. H., and Yan, D. H. (2006). Interferon-inducible protein IFIXalpha1 functions as a negative regulator of HDM2. Mol. Cell. Biol. 26, 1979–1996. doi: 10.1128/MCB.26.5.1979-1996.2006
Ding, Y., Wang, L., Su, L. K., Frey, J. A., Shao, R., Hunt, K. K., et al. (2004). Antitumor activity of IFIX, a novel interferon-inducible HIN-200 gene, in breast cancer. Oncogene 23, 4556–4566. doi: 10.1038/sj.onc.1207592
Dombrowski, Y., Peric, M., Koglin, S., Kammerbauer, C., Goss, C., Anz, D., et al. (2011). Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3:82ra38. doi: 10.1126/scitranslmed.3002001
Dunphy, G., Flannery, S. M., Almine, J. F., Connolly, D. J., Paulus, C., Jonsson, K. L., et al. (2018). Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 Mediates NF-kappaB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e5. doi: 10.1016/j.molcel.2018.07.034
Egistelli, L., Chichiarelli, S., Gaucci, E., Eufemi, M., Schinina, M. E., Giorgi, A., et al. (2009). IFI16 and NM23 bind to a common DNA fragment both in the P53 and the cMYC gene promoters. J. Cell. Biochem. 106, 666–672. doi: 10.1002/jcb.22053
Ekchariyawat, P., Hamel, R., Bernard, E., Wichit, S., Surasombatpattana, P., Talignani, L., et al. (2015). Inflammasome signaling pathways exert antiviral effect against Chikungunya virus in human dermal fibroblasts. Infect. Genet. Evol. 32, 401–408. doi: 10.1016/j.meegid.2015.03.025
Elder, E. G., Krishna, B. A., Williamson, J., Lim, E. Y., Poole, E., Sedikides, G. X., et al. (2019). Interferon-responsive genes are targeted during the establishment of human cytomegalovirus latency. mBio 10:e02574-19. doi: 10.1128/mBio.02574-19
Fan, X., Jiang, J., Zhao, D., Chen, F., Ma, H., Smith, P., et al. (2021). Structural mechanism of DNA recognition by the p204 HIN domain. Nucleic Acids Res. 49, 2959–2972. doi: 10.1093/nar/gkab076
Fang, R., Tsuchiya, K., Kawamura, I., Shen, Y., Hara, H., Sakai, S., et al. (2011). Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. J. Immunol. 187, 4890–4899. doi: 10.4049/jimmunol.1100381
Farshchian, M., Nissinen, L., Siljamaki, E., Riihila, P., Piipponen, M., Kivisaari, A., et al. (2017). Tumor cell-specific AIM2 regulates growth and invasion of cutaneous squamous cell carcinoma. Oncotarget 8, 45825–45836. doi: 10.18632/oncotarget.17573
Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J., and Alnemri, E. S. (2009). AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513. doi: 10.1038/nature07710
Fidler, T. P., Xue, C., Yalcinkaya, M., Hardaway, B., Abramowicz, S., Xiao, T., et al. (2021). The AIM2 inflammasome exacerbates atherosclerosis in clonal haematopoiesis. Nature 592, 296–301. doi: 10.1038/s41586-021-03341-5
Fotouhi-Ardakani, N., Kebir, D. E., Pierre-Charles, N., Wang, L., Ahern, S. P., Filep, J. G., et al. (2010). Role for myeloid nuclear differentiation antigen in the regulation of neutrophil apoptosis during sepsis. Am. J. Respir. Crit. Care Med. 182, 341–350. doi: 10.1164/rccm.201001-0075OC
Fujiuchi, N., Aglipay, J. A., Ohtsuka, T., Maehara, N., Sahin, F., Su, G. H., et al. (2004). Requirement of IFI16 for the maximal activation of p53 induced by ionizing radiation. J. Biol. Chem. 279, 20339–20344. doi: 10.1074/jbc.M400344200
Fukushi, M., Higuchi, M., Oie, M., Tetsuka, T., Kasolo, F., Ichiyama, K., et al. (2003). Latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus interacts with human myeloid cell nuclear differentiation antigen induced by interferon alpha. Virus Genes 27, 237–247. doi: 10.1023/a:1026391715071
Gariano, G. R., Dell’Oste, V., Bronzini, M., Gatti, D., Luganini, A., De Andrea, M., et al. (2012). The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 8:e1002498. doi: 10.1371/journal.ppat.1002498
Ge, J., Gong, Y. N., Xu, Y., and Shao, F. (2012). Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc. Natl. Acad. Sci. U.S.A. 109, 6193–6198. doi: 10.1073/pnas.1117490109
Goldberger, A., Brewer, G., Hnilica, L. S., and Briggs, R. C. (1984). Nonhistone protein antigen profiles of five leukemic cell lines reflect the extent of myeloid differentiation. Blood 63, 701–710.
Goldberger, A., Hnilica, L. S., Casey, S. B., and Briggs, R. C. (1986). Properties of a nuclear protein marker of human myeloid cell differentiation. J. Biol. Chem. 261, 4726–4731.
Gribaudo, G., Ravaglia, S., Guandalini, L., Riera, L., Gariglio, M., and Landolfo, S. (1997). Molecular cloning and expression of an interferon-inducible protein encoded by gene 203 from the gene 200 cluster. Eur. J. Biochem. 249, 258–264. doi: 10.1111/j.1432-1033.1997.t01-1-00258.x
Gribaudo, G., Riera, L., Hertel, L., and Landolfo, S. (1999). In vitro and in vivo expression analysis of the interferon-inducible 203 gene. J. Interferon Cytokine Res. 19, 129–136. doi: 10.1089/107999099314270
Griffiths, P., Baraniak, I., and Reeves, M. (2015). The pathogenesis of human cytomegalovirus. J. Pathol. 235, 288–297. doi: 10.1002/path.4437
Gugliesi, F., Bawadekar, M., De Andrea, M., Dell’Oste, V., Caneparo, V., Tincani, A., et al. (2013). Nuclear DNA sensor IFI16 as circulating protein in autoimmune diseases is a signal of damage that impairs endothelial cells through high-affinity membrane binding. PLoS One 8:e63045. doi: 10.1371/journal.pone.0063045
Gutterman, J. U., and Choubey, D. (1999). Retardation of cell proliferation after expression of p202 accompanies an increase in p21(WAF1/CIP1). Cell Growth Differ. 10, 93–100.
Hafner-Bratkovic, I., Susjan, P., Lainscek, D., Tapia-Abellan, A., Cerovic, K., Kadunc, L., et al. (2018). NLRP3 lacking the leucine-rich repeat domain can be fully activated via the canonical inflammasome pathway. Nat. Commun. 9:5182. doi: 10.1038/s41467-018-07573-4
Hanamsagar, R., Aldrich, A., and Kielian, T. (2014). Critical role for the AIM2 inflammasome during acute CNS bacterial infection. J. Neurochem. 129, 704–711. doi: 10.1111/jnc.12669
Hansen, K., Prabakaran, T., Laustsen, A., Jorgensen, S. E., Rahbaek, S. H., Jensen, S. B., et al. (2014). Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 33, 1654–1666. doi: 10.15252/embj.201488029
Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., et al. (2000). A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745. doi: 10.1038/35047123
Horan, K. A., Hansen, K., Jakobsen, M. R., Holm, C. K., Soby, S., Unterholzner, L., et al. (2013). Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J. Immunol. 190, 2311–2319. doi: 10.4049/jimmunol.1202749
Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D. R., et al. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518. doi: 10.1038/nature07725
Hou, X., and Niu, X. (2015). The NMR solution structure of AIM2 PYD domain from Mus musculus reveals a distinct alpha2-alpha3 helix conformation from its human homologues. Biochem. Biophys. Res. Commun. 461, 396–400. doi: 10.1016/j.bbrc.2015.04.046
Hu, B., Jin, C., Li, H. B., Tong, J., Ouyang, X., Cetinbas, N. M., et al. (2016). The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science 354, 765–768. doi: 10.1126/science.aaf7532
Huang, Y., Ma, D., Huang, H., Lu, Y., Liao, Y., Liu, L., et al. (2017). Interaction between HCMV pUL83 and human AIM2 disrupts the activation of the AIM2 inflammasome. Virol. J. 14:34. doi: 10.1186/s12985-016-0673-5
Jakobsen, M. R., Bak, R. O., Andersen, A., Berg, R. K., Jensen, S. B., Tengchuan, J., et al. (2013). IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci. U.S.A. 110, E4571–E4580. doi: 10.1073/pnas.1311669110
Jiang, Z. M., Wei, F. H., Zhang, Y. Y., Wang, T., Gao, W. H., Yu, S. F., et al. (2021). IFI16 directly senses viral RNA and enhances RIG-I transcription and activation to restrict influenza virus infection. Nat. Microbiol. 6, 932–945. doi: 10.1038/s41564-021-00907-x
Jiao, H., Wachsmuth, L., Kumari, S., Schwarzer, R., Lin, J., Eren, R. O., et al. (2020). Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580, 391–395. doi: 10.1038/s41586-020-2129-8
Jin, T., and Xiao, T. S. (2015). Activation and assembly of the inflammasomes through conserved protein domain families. Apoptosis 20, 151–156. doi: 10.1007/s10495-014-1053-5
Jin, T., Perry, A., Jiang, J., Smith, P., Curry, J. A., Unterholzner, L., et al. (2012). Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36, 561–571. doi: 10.1016/j.immuni.2012.02.014
Jin, T., Perry, A., Smith, P., Jiang, J., and Xiao, T. S. (2013). Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J. Biol. Chem. 288, 13225–13235. doi: 10.1074/jbc.M113.468033
Johnson, K. E., Chikoti, L., and Chandran, B. (2013). Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J. Virol. 87, 5005–5018. doi: 10.1128/JVI.00082-13
Johnstone, R. W., Kershaw, M. H., and Trapani, J. A. (1998). Isotypic variants of the interferon-inducible transcriptional repressor IFI 16 arise through differential mRNA splicing. Biochemistry 37, 11924–11931. doi: 10.1021/bi981069a
Johnstone, R. W., Wei, W., Greenway, A., and Trapani, J. A. (2000). Functional interaction between p53 and the interferon-inducible nucleoprotein IFI 16. Oncogene 19, 6033–6042. doi: 10.1038/sj.onc.1204005
Jones, J. W., Kayagaki, N., Broz, P., Henry, T., Newton, K., O’rourke, K., et al. (2010). Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. U.S.A. 107, 9771–9776. doi: 10.1073/pnas.1003738107
Kaminski, J. J., Schattgen, S. A., Tzeng, T. C., Bode, C., Klinman, D. M., and Fitzgerald, K. A. (2013). Synthetic oligodeoxynucleotides containing suppressive TTAGGG motifs inhibit AIM2 inflammasome activation. J. Immunol. 191, 3876–3883. doi: 10.4049/jimmunol.1300530
Karki, R., Man, S. M., Malireddi, R. K. S., Gurung, P., Vogel, P., Lamkanfi, M., et al. (2015). Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe 17, 357–368. doi: 10.1016/j.chom.2015.01.006
Kerur, N., Veettil, M. V., Sharma-Walia, N., Bottero, V., Sadagopan, S., Otageri, P., et al. (2011). IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9, 363–375. doi: 10.1016/j.chom.2011.04.008
Khare, S., Ratsimandresy, R. A., De Almeida, L., Cuda, C. M., Rellick, S. L., Misharin, A. V., et al. (2014). The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat. Immunol. 15, 343–353. doi: 10.1038/ni.2829
Kim, S., Bauernfeind, F., Ablasser, A., Hartmann, G., Fitzgerald, K. A., Latz, E., et al. (2010). Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40, 1545–1551. doi: 10.1002/eji.201040425
Kim, T., Pazhoor, S., Bao, M., Zhang, Z., Hanabuchi, S., Facchinetti, V., et al. (2010). Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 15181–15186. doi: 10.1073/pnas.1006539107
Kimkong, I., Avihingsanon, Y., and Hirankarn, N. (2009). Expression profile of HIN200 in leukocytes and renal biopsy of SLE patients by real-time RT-PCR. Lupus 18, 1066–1072. doi: 10.1177/0961203309106699
Kingsmore, S. F., Snoddy, J., Choubey, D., Lengyel, P., and Seldin, M. F. (1989). Physical mapping of a family of interferon-activated genes, serum amyloid P-component, and alpha-spectrin on mouse chromosome 1. Immunogenetics 30, 169–174. doi: 10.1007/BF02421202
Kong, H., Wang, Y., Zeng, X., Wang, Z., Wang, H., and Xie, W. (2015). Differential expression of inflammasomes in lung cancer cell lines and tissues. Tumour Biol. 36, 7501–7513. doi: 10.1007/s13277-015-3473-4
Lee, S., Karki, R., Wang, Y., Nguyen, L. N., Kalathur, R. C., and Kanneganti, T. D. (2021). AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature 597, 415–419. doi: 10.1038/s41586-021-03875-8
Lembo, M., Sacchi, C., Zappador, C., Bellomo, G., Gaboli, M., Pandolfi, P. P., et al. (1998). Inhibition of cell proliferation by the interferon-inducible 204 gene, a member of the Ifi 200 cluster. Oncogene 16, 1543–1551. doi: 10.1038/sj.onc.1201677
Li, D., Wu, R., Guo, W., Xie, L., Qiao, Z., Chen, S., et al. (2019). STING-mediated IFI16 degradation negatively controls Type I interferon production. Cell Rep. 29, 1249–1260.e4. doi: 10.1016/j.celrep.2019.09.069
Li, H., Wang, J., Wang, J., Cao, L. S., Wang, Z. X., and Wu, J. W. (2014a). Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta Crystallogr. F Struct. Biol. Commun. 70, 21–29. doi: 10.1107/S2053230X1303135X
Li, H., Wang, Z. X., and Wu, J. W. (2014b). Purification, characterization and docking studies of the HIN domain of human myeloid nuclear differentiation antigen (MNDA). Biotechnol. Lett. 36, 899–905. doi: 10.1007/s10529-013-1432-y
Li, T., Chen, J., and Cristea, I. M. (2013). Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14, 591–599. doi: 10.1016/j.chom.2013.10.007
Li, T., Diner, B. A., Chen, J., and Cristea, I. M. (2012). Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci. U.S.A. 109, 10558–10563. doi: 10.1073/pnas.1203447109
Liao, J. C., Lam, R., Brazda, V., Duan, S., Ravichandran, M., Ma, J., et al. (2011). Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure 19, 418–429. doi: 10.1016/j.str.2010.12.015
Lin, W., Zhao, Z., Ni, Z., Zhao, Y., Du, W., and Chen, S. (2017). IFI16 restoration in hepatocellular carcinoma induces tumour inhibition via activation of p53 signals and inflammasome. Cell Prolif. 50:e12392. doi: 10.1111/cpr.12392
Liu, C. J., Wang, H., and Lengyel, P. (1999). The interferon-inducible nucleolar p204 protein binds the ribosomal RNA-specific UBF1 transcription factor and inhibits ribosomal RNA transcription. EMBO J. 18, 2845–2854. doi: 10.1093/emboj/18.10.2845
Liu, S., Cai, X., Wu, J., Cong, Q., Chen, X., Li, T., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630
Liu, T., Tang, Q., Liu, K., Xie, W., Liu, X., Wang, H., et al. (2016). TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-dependent selective autophagy. Cell Rep. 16, 1988–2002. doi: 10.1016/j.celrep.2016.07.019
Lo Cigno, I., De Andrea, M., Borgogna, C., Albertini, S., Landini, M. M., Peretti, A., et al. (2015). The nuclear DNA Sensor IFI16 acts as a restriction factor for human papillomavirus replication through epigenetic modifications of the viral promoters. J. Virol. 89, 7506–7520. doi: 10.1128/JVI.00013-15
Lu, A., Kabaleeswaran, V., Fu, T., Magupalli, V. G., and Wu, H. (2014a). Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J. Mol. Biol. 426, 1420–1427. doi: 10.1016/j.jmb.2013.12.029
Lu, A., Magupalli, V. G., Ruan, J., Yin, Q., Atianand, M. K., Vos, M. R., et al. (2014b). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206. doi: 10.1016/j.cell.2014.02.008
Lu, A., Li, Y., Yin, Q., Ruan, J., Yu, X., Egelman, E., et al. (2015). Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 1:15013. doi: 10.1038/celldisc.2015.13
Luan, Y., Lengyel, P., and Liu, C. J. (2008). p204, a p200 family protein, as a multifunctional regulator of cell proliferation and differentiation. Cytokine Growth Factor Rev. 19, 357–369. doi: 10.1016/j.cytogfr.2008.11.002
Luan, Y., Yu, X. P., Xu, K., Ding, B., Yu, J., Huang, Y., et al. (2007). The retinoblastoma protein is an essential mediator of osteogenesis that links the p204 protein to the Cbfa1 transcription factor thereby increasing its activity. J. Biol. Chem. 282, 16860–16870. doi: 10.1074/jbc.M610943200
Ludlow, L. E., Hii, L. L., Thorpe, J., Newbold, A., Tainton, K. M., Trapani, J. A., et al. (2008). Cloning and characterisation of Ifi206: a new murine HIN-200 family member. J. Cell. Biochem. 103, 1270–1282. doi: 10.1002/jcb.21512
Lysakova-Devine, T., Keogh, B., Harrington, B., Nagpal, K., Halle, A., Golenbock, D. T., et al. (2010). Viral inhibitory peptide of TLR4, a peptide derived from vaccinia protein A46, specifically inhibits TLR4 by directly targeting MyD88 adaptor-like and TRIF-related adaptor molecule. J. Immunol. 185, 4261–4271. doi: 10.4049/jimmunol.1002013
Ma, X. Y., Wang, H., Ding, B., Zhong, H., Ghosh, S., and Lengyel, P. (2003). The interferon-inducible p202a protein modulates NF-kappaB activity by inhibiting the binding to DNA of p50/p65 heterodimers and p65 homodimers while enhancing the binding of p50 homodimers. J. Biol. Chem. 278, 23008–23019. doi: 10.1074/jbc.M302105200
Ma, X., Guo, P., Qiu, Y., Mu, K., Zhu, L., Zhao, W., et al. (2016). Loss of AIM2 expression promotes hepatocarcinoma progression through activation of mTOR-S6K1 pathway. Oncotarget 7, 36185–36197. doi: 10.18632/oncotarget.9154
Man, S. M., Karki, R., Malireddi, R. K., Neale, G., Vogel, P., Yamamoto, M., et al. (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475. doi: 10.1038/ni.3118
Maruzuru, Y., Ichinohe, T., Sato, R., Miyake, K., Okano, T., Suzuki, T., et al. (2018). Herpes simplex Virus 1 VP22 inhibits AIM2-dependent inflammasome activation to enable efficient viral replication. Cell Host Microbe 23, 254–265.e7. doi: 10.1016/j.chom.2017.12.014
Milutin Gasperov, N., Farkas, S. A., Nilsson, T. K., and Grce, M. (2014). Epigenetic activation of immune genes in cervical cancer. Immunol. Lett. 162, 256–257. doi: 10.1016/j.imlet.2014.09.019
Min, W., Ghosh, S., and Lengyel, P. (1996). The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol. Cell. Biol. 16, 359–368. doi: 10.1128/MCB.16.1.359
Mondini, M., Costa, S., Sponza, S., Gugliesi, F., Gariglio, M., and Landolfo, S. (2010). The interferon-inducible HIN-200 gene family in apoptosis and inflammation: implication for autoimmunity. Autoimmunity 43, 226–231. doi: 10.3109/08916930903510922
Mondini, M., Vidali, M., Airo, P., De Andrea, M., Riboldi, P., Meroni, P. L., et al. (2007). Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann. N. Y. Acad. Sci. 1110, 47–56. doi: 10.1196/annals.1423.006
Monroe, K. M., Yang, Z., Johnson, J. R., Geng, X., Doitsh, G., Krogan, N. J., et al. (2014). IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432. doi: 10.1126/science.1243640
Morrone, S. R., Matyszewski, M., Yu, X., Delannoy, M., Egelman, E. H., and Sohn, J. (2015). Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat. Commun. 6:7827. doi: 10.1038/ncomms8827
Morrone, S. R., Wang, T., Constantoulakis, L. M., Hooy, R. M., Delannoy, M. J., and Sohn, J. (2014). Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc. Natl. Acad. Sci. U.S.A. 111, E62–E71. doi: 10.1073/pnas.1313577111
Ni, X., Ru, H., Ma, F., Zhao, L., Shaw, N., Feng, Y., et al. (2016). New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16. J. Mol. Cell Biol. 8, 51–61. doi: 10.1093/jmcb/mjv053
Orzalli, M. H., Deluca, N. A., and Knipe, D. M. (2012). Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U.S.A. 109, E3008–E3017. doi: 10.1073/pnas.1211302109
Park, H. H., Lo, Y. C., Lin, S. C., Wang, L., Yang, J. K., and Wu, H. (2007). The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 25, 561–586. doi: 10.1146/annurev.immunol.25.022106.141656
Ponomareva, L., Liu, H., Duan, X., Dickerson, E., Shen, H., Panchanathan, R., et al. (2013). AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol. Cancer Res. 11, 1193–1202. doi: 10.1158/1541-7786.MCR-13-0145
Rathinam, V. A., Jiang, Z., Waggoner, S. N., Sharma, S., Cole, L. E., Waggoner, L., et al. (2010). The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402. doi: 10.1038/ni.1864
Reinholz, M., Kawakami, Y., Salzer, S., Kreuter, A., Dombrowski, Y., Koglin, S., et al. (2013). HPV16 activates the AIM2 inflammasome in keratinocytes. Arch. Dermatol. Res. 305, 723–732. doi: 10.1007/s00403-013-1375-0
Roberts, T. L., Idris, A., Dunn, J. A., Kelly, G. M., Burnton, C. M., Hodgson, S., et al. (2009). HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323, 1057–1060. doi: 10.1126/science.1169841
Rozzo, S. J., Allard, J. D., Choubey, D., Vyse, T. J., Izui, S., Peltz, G., et al. (2001). Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity 15, 435–443. doi: 10.1016/s1074-7613(01)00196-0
Ru, H., Ni, X., Zhao, L., Crowley, C., Ding, W., Hung, L. W., et al. (2013). Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 23, 855–858. doi: 10.1038/cr.2013.52
Ryabchenko, B., Soldatova, I., Sroller, V., Forstova, J., and Huerfano, S. (2021). Immune sensing of mouse polyomavirus DNA by p204 and cGAS DNA sensors. FEBS J. 288, 5964–5985. doi: 10.1111/febs.15962
Saiga, H., Kitada, S., Shimada, Y., Kamiyama, N., Okuyama, M., Makino, M., et al. (2012). Critical role of AIM2 in Mycobacterium tuberculosis infection. Int. Immunol. 24, 637–644. doi: 10.1093/intimm/dxs062
Sauer, J. D., Witte, C. E., Zemansky, J., Hanson, B., Lauer, P., and Portnoy, D. A. (2010). Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7, 412–419. doi: 10.1016/j.chom.2010.04.004
Schulmann, K., Brasch, F. E., Kunstmann, E., Engel, C., Pagenstecher, C., Vogelsang, H., et al. (2005). HNPCC-associated small bowel cancer: clinical and molecular characteristics. Gastroenterology 128, 590–599. doi: 10.1053/j.gastro.2004.12.051
Seelig, H. P., Ehrfeld, H., and Renz, M. (1994). Interferon-gamma-inducible protein p16. A new target of antinuclear antibodies in patients with systemic lupus erythematosus. Arthritis Rheum. 37, 1672–1683. doi: 10.1002/art.1780371117
Sharma, B. R., Karki, R., and Kanneganti, T. D. (2019). Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur. J. Immunol. 49, 1998–2011. doi: 10.1002/eji.201848070
Singh, V. V., Kerur, N., Bottero, V., Dutta, S., Chakraborty, S., Ansari, M. A., et al. (2013). Kaposi’s sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J. Virol. 87, 4417–4431. doi: 10.1128/JVI.03282-12
Stehlik, C. (2007). The PYRIN domain in signal transduction. Curr. Protein Pept. Sci. 8, 293–310. doi: 10.2174/138920307780831857
Storek, K. M., Gertsvolf, N. A., Ohlson, M. B., and Monack, D. M. (2015). cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J. Immunol. 194, 3236–3245. doi: 10.4049/jimmunol.1402764
Sui, H., Zhou, M., Chen, Q., Lane, H. C., and Imamichi, T. (2014). siRNA enhances DNA-mediated interferon lambda-1 response through crosstalk between RIG-I and IFI16 signalling pathway. Nucleic Acids Res. 42, 583–598. doi: 10.1093/nar/gkt844
Sun, L., Wu, J., Du, F., Chen, X., and Chen, Z. J. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. doi: 10.1126/science.1232458
Takaoka, A., Wang, Z., Choi, M. K., Yanai, H., Negishi, H., Ban, T., et al. (2007). DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505. doi: 10.1038/nature06013
Tian, Y., and Yin, Q. (2019). Structural analysis of the HIN1 domain of interferon-inducible protein 204. Acta Crystallogr. F Struct. Biol. Commun. 75, 455–460. doi: 10.1107/S2053230X19007167
Torii, Y., Kawada, J. I., Murata, T., Yoshiyama, H., Kimura, H., and Ito, Y. (2017). Epstein-Barr virus infection-induced inflammasome activation in human monocytes. PLoS One 12:e0175053. doi: 10.1371/journal.pone.0175053
Uchida, K., Akita, Y., Matsuo, K., Fujiwara, S., Nakagawa, A., Kazaoka, Y., et al. (2005). Identification of specific autoantigens in Sjogren’s syndrome by SEREX. Immunology 116, 53–63. doi: 10.1111/j.1365-2567.2005.02197.x
Unterholzner, L., Keating, S. E., Baran, M., Horan, K. A., Jensen, S. B., Sharma, S., et al. (2010). IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004. doi: 10.1038/ni.1932
Veeranki, S., and Choubey, D. (2012). Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol. Immunol. 49, 567–571. doi: 10.1016/j.molimm.2011.11.004
Wang, H., Chatterjee, G., Meyer, J. J., Liu, C. J., Manjunath, N. A., Bray-Ward, P., et al. (1999). Characteristics of three homologous 202 genes (Ifi202a, Ifi202b, and Ifi202c) from the murine interferon-activatable gene 200 cluster. Genomics 60, 281–294. doi: 10.1006/geno.1999.5923
Wang, P. H., Ye, Z. W., Deng, J. J., Siu, K. L., Gao, W. W., Chaudhary, V., et al. (2018). Inhibition of AIM2 inflammasome activation by a novel transcript isoform of IFI16. EMBO Rep. 19:e45737. doi: 10.15252/embr.201845737
Warren, S. E., Armstrong, A., Hamilton, M. K., Mao, D. P., Leaf, I. A., Miao, E. A., et al. (2010). Cutting edge: cytosolic bacterial DNA activates the inflammasome via Aim2. J. Immunol. 185, 818–821. doi: 10.4049/jimmunol.1000724
Weiler, S. R., Gooya, J. M., Ortiz, M., Tsai, S., Collins, S. J., and Keller, J. R. (1999). D3: a gene induced during myeloid cell differentiation of Linlo c-Kit+ Sca-1(+) progenitor cells. Blood 93, 527–536. doi: 10.1182/blood.v93.2.527
Wen, Y., Yan, D. H., Spohn, B., Deng, J., Lin, S. Y., and Hung, M. C. (2000). Tumor suppression and sensitization to tumor necrosis factor alpha-induced apoptosis by an interferon-inducible protein, p202, in breast cancer cells. Cancer Res. 60, 42–46.
Wichit, S., Hamel, R., Yainoy, S., Gumpangseth, N., Panich, S., Phuadraksa, T., et al. (2019). Interferon-inducible protein (IFI) 16 regulates Chikungunya and Zika virus infection in human skin fibroblasts. EXCLI J. 18, 467–476. doi: 10.17179/excli2019-1271
Woerner, S. M., Kloor, M., Schwitalle, Y., Youmans, H., Doeberitz, M., Gebert, J., et al. (2007). The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes Chromosomes Cancer 46, 1080–1089. doi: 10.1002/gcc.20493
Wu, J., Fernandes-Alnemri, T., and Alnemri, E. S. (2010). Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J. Clin. Immunol. 30, 693–702. doi: 10.1007/s10875-010-9425-2
Wu, J., Sun, L., Chen, X., Du, F., Shi, H., Chen, C., et al. (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. doi: 10.1126/science.1229963
Xie, J., Briggs, J. A., and Briggs, R. C. (1997a). MNDA dimerizes through a complex motif involving an N-terminal basic region. FEBS Lett. 408, 151–155. doi: 10.1016/s0014-5793(97)00404-3
Xie, J., Briggs, J. A., Morris, S. W., Olson, M. O., Kinney, M. C., and Briggs, R. C. (1997b). MNDA binds NPM/B23 and the NPM-MLF1 chimera generated by the t(3;5) associated with myelodysplastic syndrome and acute myeloid leukemia. Exp. Hematol. 25, 1111–1117.
Xie, J., Briggs, J. A., and Briggs, R. C. (1998). Human hematopoietic cell specific nuclear protein MNDA interacts with the multifunctional transcription factor YY1 and stimulates YY1 DNA binding. J. Cell. Biochem. 70, 489–506. doi: 10.1002/(sici)1097-4644(19980915)70:4<489::aid-jcb6>3.0.co;2-f
Xie, J., Briggs, J. A., Olson, M. O., Sipos, K., and Briggs, R. C. (1995). Human myeloid cell nuclear differentiation antigen binds specifically to nucleolin. J. Cell. Biochem. 59, 529–536. doi: 10.1002/jcb.240590412
Xin, H., Curry, J., Johnstone, R. W., Nickoloff, B. J., and Choubey, D. (2003). Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene 22, 4831–4840. doi: 10.1038/sj.onc.1206754
Xin, H., Pereira-Smith, O. M., and Choubey, D. (2004). Role of IFI 16 in cellular senescence of human fibroblasts. Oncogene 23, 6209–6217. doi: 10.1038/sj.onc.1207836
Yan, D. H., Abramian, A., Li, Z., Ding, Y., Wen, Y., Liu, T. J., et al. (2003). P202, an interferon-inducible protein, inhibits E2F1-mediated apoptosis in prostate cancer cells. Biochem. Biophys. Res. Commun. 303, 219–222. doi: 10.1016/s0006-291x(03)00320-6
Yan, H., Dalal, K., Hon, B. K., Youkharibache, P., Lau, D., and Pio, F. (2008). RPA nucleic acid-binding properties of IFI16-HIN200. Biochim. Biophys. Acta 1784, 1087–1097. doi: 10.1016/j.bbapap.2008.04.004
Yang, C. A., Huang, S. T., and Chiang, B. L. (2015). Sex-dependent differential activation of NLRP3 and AIM2 inflammasomes in SLE macrophages. Rheumatology (Oxford) 54, 324–331. doi: 10.1093/rheumatology/keu318
Yang, J., Liu, Z., and Xiao, T. S. (2017). Post-translational regulation of inflammasomes. Cell. Mol. Immunol. 14, 65–79. doi: 10.1038/cmi.2016.29
Yang, P., An, H., Liu, X., Wen, M., Zheng, Y., Rui, Y., et al. (2010). The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 11, 487–494. doi: 10.1038/ni.1876
Yang, Y., Zhao, X., Wang, Z., Shu, W., Li, L., Li, Y., et al. (2020). Nuclear Sensor interferon-inducible Protein 16 inhibits the function of Hepatitis B virus covalently closed circular DNA by Integrating innate immune activation and epigenetic suppression. Hepatology 71, 1154–1169. doi: 10.1002/hep.30897
Yi, Y. S., Jian, J., Gonzalez-Gugel, E., Shi, Y. X., Tian, Q., Fu, W., et al. (2018). p204 is required for canonical lipopolysaccharide-induced TLR4 signaling in mice. EBioMedicine 29, 78–91. doi: 10.1016/j.ebiom.2018.02.012
Yin, Q., Sester, D. P., Tian, Y., Hsiao, Y. S., Lu, A., Cridland, J. A., et al. (2013). Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 4, 327–339. doi: 10.1016/j.celrep.2013.06.024
Yogarajah, T., Ong, K. C., Perera, D., and Wong, K. T. (2017). AIM2 inflammasome-mediated pyroptosis in enterovirus A71-infected neuronal cells restricts viral replication. Sci. Rep. 7:5845. doi: 10.1038/s41598-017-05589-2
Zhang, M. J., Zhao, Q. C., Xia, M. X., Chen, J., Chen, Y. T., Cao, X., et al. (2020). The HDAC3 inhibitor RGFP966 ameliorated ischemic brain damage by downregulating the AIM2 inflammasome. FASEB J. 34, 648–662. doi: 10.1096/fj.201900394RRR
Zhang, X., Brann, T. W., Zhou, M., Yang, J., Oguariri, R. M., Lidie, K. B., et al. (2011). Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 186, 4541–4545. doi: 10.4049/jimmunol.1003389
Zhang, Y., Howell, R. D., Alfonso, D. T., Yu, J., Kong, L., Wittig, J. C., et al. (2007). IFI16 inhibits tumorigenicity and cell proliferation of bone and cartilage tumor cells. Front. Biosci. 12, 4855–4863. doi: 10.2741/2433
Zhang, Y., Tian, Q., Du, Y., Cao, H., Lengyel, P., and Kong, W. (2008). Multiple splicing results in at least two p203 proteins that are expressed in the liver and down-regulated during liver regeneration. Front. Biosci. 13, 2444–2451. doi: 10.2741/2857
Zhang, Z., Yuan, B., Bao, M., Lu, N., Kim, T., and Liu, Y. J. (2011). The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965. doi: 10.1038/ni.2091
Zhao, H., Gonzalezgugel, E., Cheng, L., Richbourgh, B., Nie, L., and Liu, C. (2015). The roles of interferon-inducible p200 family members IFI16 and p204 in innate immune responses, cell differentiation and proliferation. Genes Dis. 2, 46–56. doi: 10.1016/j.gendis.2014.10.003
Zhen, J., Zhang, L., Pan, J., Ma, S., Yu, X., Li, X., et al. (2014). AIM2 mediates inflammation-associated renal damage in hepatitis B virus-associated glomerulonephritis by regulating caspase-1, IL-1beta, and IL-18. Mediators Inflamm. 2014:190860. doi: 10.1155/2014/190860
Keywords: innate immunity, PRR, PAMP, PYHIN family, AIM2, IFI16, p202, p204
Citation: Fan X, Jiao L and Jin T (2022) Activation and Immune Regulation Mechanisms of PYHIN Family During Microbial Infection. Front. Microbiol. 12:809412. doi: 10.3389/fmicb.2021.809412
Received: 05 November 2021; Accepted: 09 December 2021;
Published: 25 January 2022.
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Divaker Choubey, University of Cincinnati, United StatesPu Gao, Institute of Biophysics, Chinese Academy of Sciences (CAS), China
Copyright © 2022 Fan, Jiao and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianying Jiao, amlhb2x5QHhqdHUuZWR1LmNu; Tengchuan Jin, amludEB1c3RjLmVkdS5jbg==
 Xiaojiao Fan
Xiaojiao Fan Lianying Jiao
Lianying Jiao Tengchuan Jin
Tengchuan Jin