- 1Section of Food Inspection, Faculty of Veterinary Medicine, School of Specialization in Inspection of Foods of Animal Origin, “G. Tiecco” University of Teramo, Teramo, Italy
- 2Section of Food Microbiology, Faculty of Bioscience and Technology for Food, Agriculture and Environment, University of Teramo, Teramo, Italy
Antimicrobial resistance (AMR) is a global concern, and new approaches are needed to circumvent animal and food-borne resistant pathogens. Among the new strategies, the combination of antibiotics with natural compounds such as essential oils (EOs) could be an alternative to challenge bacterial resistance. The present study evaluates the phenotypic and genotypic antibiotic resistance of 36 Salmonella enterica (16 S. Typhimurium, 3 monophasic variant S. Typhimurium, 8 S. Enteritidis, 6 S. Rissen, 1 S. Typhi, and 2 S. Derby) strains, isolated from the swine production chain. The isolates displayed phenotypic resistance to gentamicin, amikacin, tobramycin, and tetracycline, while the resistance genes most commonly detected were parC, catA, nfsB, nfsA, blaTEM, tetA, and tetB. Then 31/36 Salmonella isolates were chosen to evaluate resistance to tetracycline and Thymus vulgaris, Eugenia caryophyllata, and Corydothymus capitatus EOs by determining minimum inhibitory concentrations (MICs). Finally, the synergistic effect between tetracycline and each EOs was evaluated by the checkerboard method, calculating the fractional inhibitory concentration (FIC) index. Among the EOs, C. capitatus displayed the best bioactivity in terms of MICs, with the lowest values (0.31 and 0.625 μl/ml). On the contrary, the strains showed the ability to grow in the presence of the maximum concentration of tetracycline employed (256 μg/ml). While not displaying a real synergism according to the FIC index, the combination of tetracycline compounds and the three EOs resulted in a significant reduction in the MIC values to tetracycline (4 μg/ml), suggesting a restoration of the susceptibility to the antibiotic in Salmonella spp.
Introduction
Salmonella spp. is widespread in the environment, but the main reservoir is the intestinal tract of livestock animals and particularly pig, poultry, and cattle. The pathogen can be transmitted to humans through the food chain (Prasertsee et al., 2016). In Europe and in the United States, contaminated pork and pork products are important sources for salmonellosis in humans (Broadway et al., 2021; EFSA, 2021). Salmonella infections in humans can be divided in two main forms, including invasive typhoidal salmonellosis and non-typhoidal salmonellosis. The former, caused by S. enterica (serotype Typhi and Paratyphi A, B), causes enteric fever, gastroenteritis, and bacteremia. The latter can be caused by several Salmonella serovars. Among these, non-typhoid serotypes, such as S. enterica Typhimurium, has a broad vertebrate host range and causes various symptoms that usually include diarrheal disease (Andrews and Ryan, 2015).
On the other hand, salmonellosis in swine is caused by ubiquitous Salmonella serovars that can occur as a symptomatic disease in a wide range of hosts and, more frequently, as a self-limiting gastroenteritis. Typical symptoms in pigs are enteric, but infected animals are frequently asymptomatic (Bonardi et al., 2016). The presence of infected pigs that acquire a healthy carrier status may pose a threat to public health (Bonardi et al., 2016) and can lead to cross-contamination of carcasses. For this reason, during pig production and particularly in lactation and post-weaning, an extensive administration of oral antibiotics, such as penicillin and tetracycline, occurs (Lekagul et al., 2019). Nevertheless, the use of antimicrobials either to treat or to prevent infections, as well as growth promoters in farm animals, is a major contributing factor for the development of antimicrobial resistance (AMR), potentially leading to the widespread transmission of antimicrobial-resistant bacteria through the food chain (Andrews and Ryan, 2015). Therefore, Salmonella spp. displays the capability of spreading antibiotic resistance by transfer-associated genes, thus, causing the increase in incidence and severity of the disease. In fact, treatment of salmonellosis in humans and animals has become more difficult due to the emergence of multidrug-resistant Salmonella spp. strains (Lekagul et al., 2019).
Antimicrobial resistance is defined as a biological phenomenon of adaptation of some microorganisms that acquire the ability to survive or grow in the presence of an antimicrobial agent (Palma et al., 2020). The ability to resist is due to genetic mutations or acquisition via lateral gene transfer of resistance genes. Despite subsequent restrictions and bans on the use of different antimicrobials in agriculture, human and veterinary medicine, the resistance acquired by the microorganism is retained and is potentially transmissible (World Health Organization [WHO], 2020). Nowadays, alternative treatments to counteract AMR have been evaluated, for example, the use of natural compounds such as essential oils (EOs) (Trifan et al., 2020; Maggio et al., 2021). EOs are oily systems containing a mixture of different bioactive molecules derived by aromatic plants (Rossi et al., 2020). The phytocomplex contained in EOs interacts with multiple bacterial cellular targets, instead of adopting a particular single mode of action, thus, preventing pathogens from acquiring resistance (Yang et al., 2018). EO antimicrobial efficacy is associated with the main compounds; however, the EOs are a consortium of different compounds, each with its own effectiveness. For example, bioactive monoterpenes, such as thymol and carvacrol, which are found mainly in Origanum and Thymus spp. EOs, possess the ability to destabilize the outer membrane of Gram-negative bacteria, causing an increase in membrane permeability that is a mechanism of antimicrobial action (Lambert et al., 2001). Moreover, phenylpropanoids, such as eugenol, frequently found in clove EO (Moemenbellah-Fard et al., 2020), can modify the fatty acid profile of the cell membrane (Marchese et al., 2017). The destabilization of the cell membrane increases the susceptibility of the bacteria toward other antimicrobial compounds. Therefore, the combination of an antibiotic treatment with EOs could allow the natural compounds to permeate the cell membrane under the action of antibiotics, reducing the concentration of the bioactive compounds employed (Yang et al., 2018) and restoring bacterial susceptibility to treatments.
Considering the above reasons, this study first aimed at evaluating the resistance of 36 Salmonella spp. strains from the swine production chain to different antibiotics, generally employed in livestock. Afterward, the potential effect of EOs in restoring the susceptibility of the strains to the antibiotics was investigated. In particular, a selection of tetracycline-resistant isolates was subjected to a combination of different antibiotics and Corydothymus capitatus, Eugenia caryophyllata, and Thymus vulgaris EOs.
Materials and Methods
Bacterial Strains
A total of 36 Salmonella spp. (16 S. Typhimurium, 3 monophasic variant S. Typhimurium, 8 S. Enteritidis, 6 S. Rissen, 1 S. Typhi, and 2 S. Derby) strains, belonging to the biobank of the Department of Food Inspection of the University of Teramo (Italy), previously isolated from the swine production chain and identified, were included in this study (Di Ciccio et al., 2016; Lauteri et al., 2021).
Antimicrobial Susceptibility Testing
The card VITEK 2 AST GN65 was used according to the instructions of the manufacturer (bioMérieux, 2013a) to evaluate antibiograms and determine minimum inhibitory concentrations (MICs). Fifteen antimicrobial agents were tested, in detail: ampicillin, amoxicillin and clavulanic acid, imipenem, cefpodoxime, ceftiofur, tobramycin, piperacillin, gentamicin, amikacin, enrofloxacin, marbofloxacin, chloramphenicol, tetracycline, nitrofurantoin, and trimethoprim-sulfamethoxazole.
The turbidity of the bacterial suspensions was adjusted with a densitometer (DENSICHEK, bioMerieux, Marcy-l’Etoile, France) to match a 0.5–0.63 McFarland standard; then 145 μl of suspension was added to 3 ml of VITEK 0.45% saline solution (bioMérieux, 2013b). The time range between suspension preparation and card filling was less than 30 min to avoid changes in turbidity.
Afterward, the VITEK 2 AST GN65 antimicrobial susceptibility cards and bacterial suspension in tubes, both contained in a cassette, were manually loaded into the VITEK 2 system. Each test card was automatically filled with a bacterial suspension, sealed, incubated, and read by kinetic fluorescence measurement. The reporting time for the direct testing of susceptibility against the 15 antibiotics for culture isolates by the VITEK 2 system ranged from 8.5 to 10.5 h.
Detection of Antibiotic Resistance Genes
The presence of AMR genes was investigated using conventional polymerase chain reaction (PCR) (Kikuvi et al., 2010). The uniplex PCR amplification conditions consisted of initial denaturation at 94°C for 5 min, with 30 cycles of denaturation at 94°C for 30 s, annealing at different temperatures according to the different primers for 30 s, extension at 72°C for 1 min, and a final cycle of amplification at 72°C for 10 min. The specific primers used in the PCR amplifications, the annealing temperatures, and the amplicon sizes are reported in Table 1. PCR products were resolved by agarose gel electrophoresis.

Table 1. Target antibiotic, genes, PCR primers, forward and reverse sequence, annealing temperature of the primers, amplicon size, and reference used to evaluate the presence of antibiotic-resistant genes.
Inocula and Growth Media
For the subsequent analyses, 31 Salmonella strains (15 S. Typhimurium, 3 monophasic variant S. Typhimurium, 6 S. Enteritidis, 6 S. Rissen, and 1 S. Typhi) were selected on account of their resistance to tetracycline, as described before. The inocula were prepared in Mueller–Hinton broth (MH, Oxoid Thermo Fisher Scientific, Rodano, Milan, Italy) and incubated at 37°C for 18 h until early stationary phase. Cells were then harvested by centrifugation and washed three times with phosphate buffer saline (PBS) 50 mM, pH 7.4. The inocula were standardized to OD620 nm 0.1–0.2 (5 × 107 cells/ml) and then diluted to 5 × 106 cells/ml.
Antimicrobial Solutions
Commercial and food-grade T. vulgaris and E. caryophyllata EOs were kindly provided by Flora S.r.l. (Pisa, Italy), while C. capitatus EO was supplied by Exentiae S.r.l. Soc. Agricola (Catania, Italy). According to the results from the analyses carried out by the producers, the EO chemotypes were thymol (46.65%), eugenol (76.2%), and carvacrol (70%), for T. vulgaris, E. caryophyllata, and C. capitatus, respectively. EO emulsions were diluted to 80 μl/ml in PBS and 1% Tween 80 (Sigma-Aldrich, Milan, Italy). Lyophilized tetracycline (≥98%) was provided by Sigma-Aldrich (Milan, Italy).
Minimum Inhibitory Concentration/Minimum Bactericidal Concentration Assays
The MICs were determined for the EO emulsions and the antibiotic solutions following the CLSI guidelines/CLSI protocol (CLSI, 2016) in a 96-well microtiter plate (Corning Incorporated, Kennebunk, ME, United States). The antibacterial activity was examined after incubation at 37°C for 72 h in static conditions. MICs were determined after 24, 48, and 72 h, evidenced by the absence of red discoloration by 2,3,5-triphenyltetrazolium chloride (Sigma-Aldrich, Milan, Italy), added in the growth media in a ratio of 0.1%. Subsequently, the minimum bactericidal concentration (MBC) was determined after 24, 48, and 72 h at 37°C by plating out onto MH agar plates.
Checkerboard Test
The synergy between tetracycline and each EO was tested by the checkerboard method, a two-dimensional matrix of serial concentrations of the compounds under examination (Magi et al., 2015). By the checkerboard test, it was possible to calculate a fractional inhibitory concentration (FIC) index, according to the formulas (Magi et al., 2015):
Moreover, the FIC index values were interpreted in agreement with Fratini et al. (2017): synergistic effect (FIC index ≤ 1.0), commutative effect (FIC index = 1), no interaction (FIC index > 1.0–2.0), and antagonistic effect (FIC index > 2.0).
Statistical Analysis
The data of MIC and MBC assays were expressed as the means of three different repetitions. The datasets of the MICs of tetracycline (μg/ml) from the in vitro analysis in combination with the three EOs (μl/ml) and alone were correlated through the principal component analysis (PCA), using the XLSTAT 2014 software (Redmond, WA, United States).
Results
Antimicrobial Susceptibility of Salmonella Isolates
All Salmonella isolates selected for this study displayed resistance to gentamicin, amikacin, and tobramycin.
A total of 86.1% (31/36) of the Salmonella isolates were resistant to tetracycline, while 55.5% (20/36) were resistant to ampicillin and piperacillin. Twenty-five percent (9/36) of the strains showed resistance to trimethoprim, 5.5% (2/36) to chloramphenicol, and only 2.8% (1/36) to amoxicillin–clavulanic acid and nitrofurantoin.
Moreover, 16 Salmonella isolates (44.4%) showed intermediate resistance to nitrofurantoin and 36.1% (13/36) to chloramphenicol. A total of 8.3% (3/36) of the isolates exhibited intermediate resistance against amoxicillin–clavulanic acid, while 5.5% (2/36) had intermediate resistance to enrofloxacin and 2.8% (1/36) to ceftiofur.
Several strains showed multiple AMR, namely, 27.8% (10/36) of the strains had resistance to three antimicrobial classes, 25% (9/36) were resistant to four antimicrobial classes, and 2.8% (1/36) displayed resistance to even six antimicrobial classes.
The results of the antimicrobial susceptibility testing are reported in Table 2.
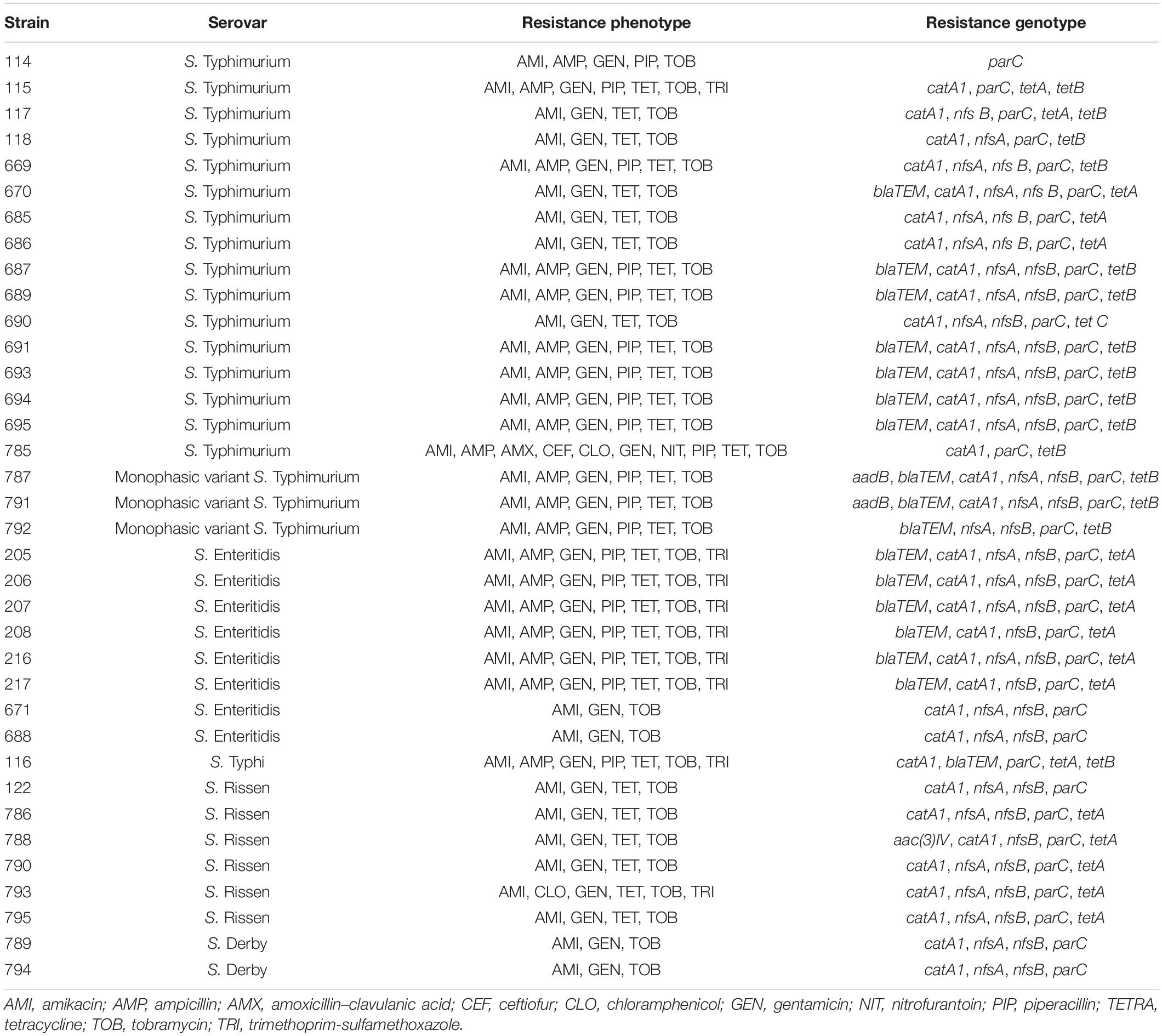
Table 2. Overview of the antibiotic resistance shown by the 36 investigated isolates of Salmonella spp.
Detection of Antibiotic Resistance Genes
All Salmonella isolates recovered from the swine production chain were investigated for the presence of AMR genes by PCR. The most frequently detected resistance genes were parC (36/36, 100%), catA1 (34/36, 94.4%), nfsB (31/36, 86.1%), nfsA (28/36, 77.7%), blaTEM (17/36, 47.2%), tetA (17/36, 47.2%), and tetB (15/36, 41.6%). The genes tetC, aac(3)IV, and aadB were detected only in one isolate, corresponding to 2.8% of the total, while aadA2 and blaPSE were not detected. The association of phenotypic resistance and the presence of AMR genes was variable among the different Salmonella serovars. For example, while phenotypic resistance to one or more antimicrobials was observed for both S. Derby (n = 2) and S. Rissen (n = 5), the corresponding resistance genes were not detected by PCR. On the contrary, S. Rissen isolate 122, which tested positive for the presence of catA1, nfsA, nfsB, and parC, did not show any AMR (Table 2).
Minimum Inhibitory Concentration and Minimum Bactericidal Concentration Determination of Tetracycline and Essential Oils
Minimum inhibitory concentrations and MBCs for tetracycline and EOs were determined after 48 h of incubation at 37°C by broth microdilution assay (Table 3). The MIC of tetracycline was 256 μg/ml for each Salmonella strain. According to the Clinical and Laboratory Standards Institute guidelines (CLSI guidelines, supplement M100S), the strains were classified as resistant to tetracyclines [as per the MIC breakpoints for tetracycline, strain result susceptible (≤4 μg/ml), intermediate (8 μg/ml), and resistant (≥16 μg/ml)]. Regarding the MICs of the EOs, C. capitatus displayed the best bioactivity, with a range of values between <0.31 and 10 μl/ml. In particular, the lowest MIC values were those observed more frequently in the strains analyzed and, in detail, 0.31 μl/ml for 5 isolates (16.1%) and 0.625 μl/ml for 23 isolates (74.2%) out of 31. Nevertheless, T. vulgaris EO exhibited the lowest MIC values ranging from <0.31 to 5 μl/ml, although the MIC values observed more frequently were 0.625 and 1.25 μl/ml, with 7 (22.6%) and 18 (58%) isolates out of 31, respectively. E. caryophyllata EO showed the lowest effectiveness, with MIC values between 0.31 and 20 μl/ml. Moreover, the highest MIC values were common in the set of strains, with 10 and 20 μl/ml for 6 (19.3%) and 16 isolates out of 31 (51.6%), respectively.
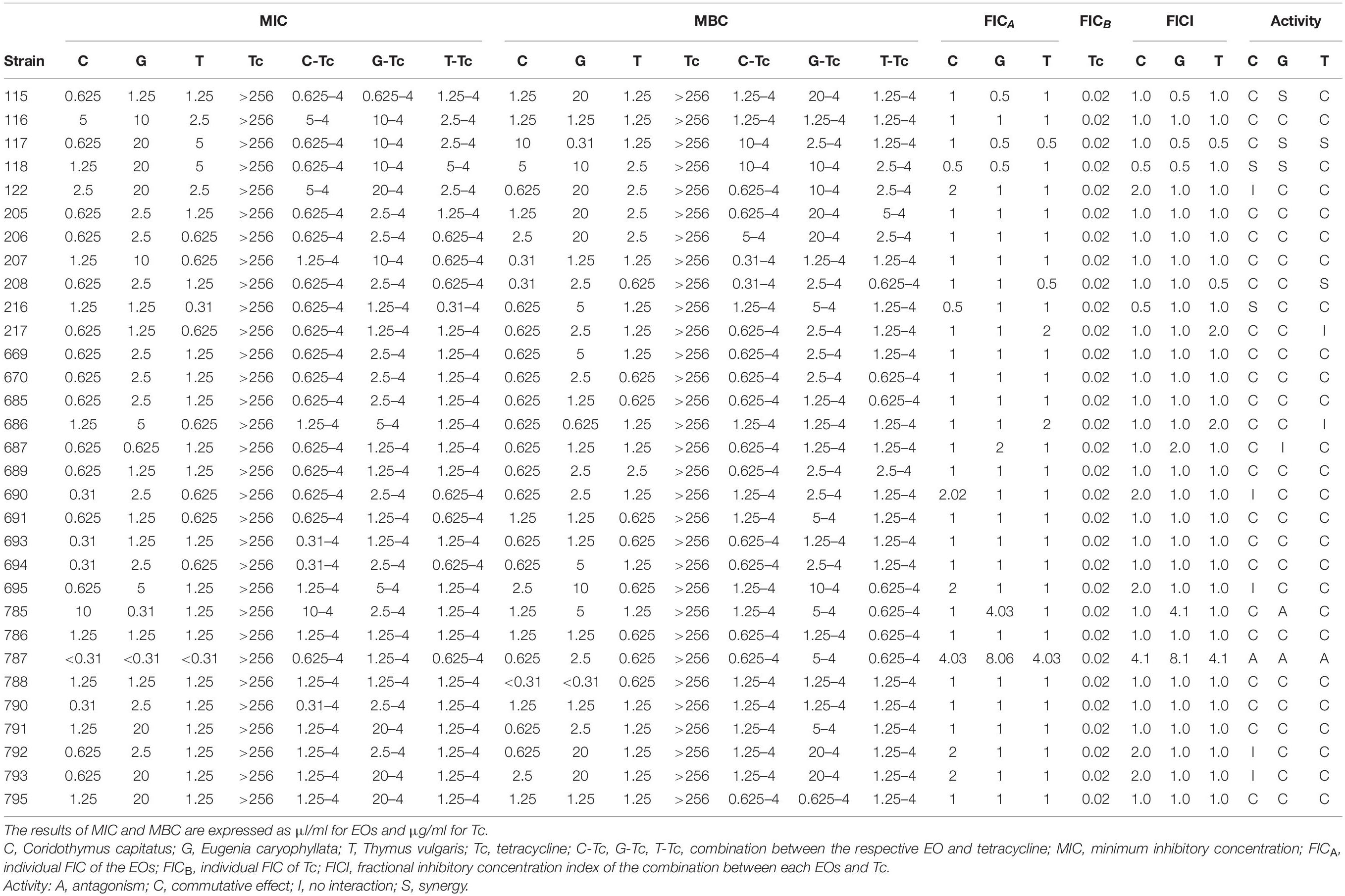
Table 3. Minimum inhibitory concentration (MIC), MBC values, and FIC index of Coridothymus capitatus, Eugenia caryophyllata and Thymus vulgaris EOs alone and in combination with tetracycline against Salmonella strains, determined by broth dilution technique and checkerboard method after 48 h of incubation at 37°C.
Combined Antimicrobial Effect of Tetracycline and Essential Oils
In the checkerboard assay, the combination of tetracycline with the three EOs, C. capitatus, E. caryophyllata, and T. vulgaris was tested (Table 3). The combinations resulted in the clear reduction of the MIC value for the antibiotic (from 256 to 4 μg/ml) for each Salmonella strain. Regarding the EOs, the MIC values, both in combination and alone, were the same in most of the strains. Only 19.3% (6/31) and 6.4% (2/31) of the strains exhibited different values of MICs for C. capitatus, which were, respectively, higher and lower than the EO alone. For E. caryophyllata EO in combination, 9.7% (3/31) and 9.7% (3/31) of the strains displayed the increase and the decrease, respectively, of the MIC value. Finally, for T. vulgaris EO in combination with the antibiotic, 6.4% (2/31) and 9.7% (3/31) of the strains had MIC values higher and lower, respectively, compared with the same EO alone.
Regarding the MBC assays, the behavior of the Salmonella strains in the presence of tetracycline was the same as that observed for the MICs. The MBC for each strain was 256 μg/ml when the antibiotic was applied alone, whereas the MBC reached 4 μg/ml in the presence of the combination of tetracycline and EOs (Table 3). Also, in this case, the combination of the antibiotic with the EOs displayed restoration of susceptibility to tetracycline in Salmonella isolates. MBCs of C. capitatus, E. caryophyllata, and T. vulgaris EOs showed a similar trend with respect to MICs, with the best bioactivity of C. capitatus compared with the other EOs.
In the checkerboard assay, the FIC index values were calculated by considering all the combinations of tetracycline with each EO in which there was no visible growth.
A synergistic effect (FICI ≤ 1.0) was detected in 6.5% (2/31), 9.6% (3/31), and 6.5% (2/31) of the strains, in the antibiotic with C. capitatus, E. caryophyllata, and T. vulgaris EOs, respectively. However, in most cases, the effect was found to be commutative (FICI = 1). In fact, the FICI values detected between tetracycline and C. capitatus, E. caryophyllata, and T. vulgaris EOs ranged between 1.02 and 2.0 for 90.3% (28/31), 80.6% (25/31), and 90.3% (28/31) of the strains, respectively. The indifferent effect (FICI > 1.0–2.0) was observed only in 16.1% (5/31) and 6.5% (2/31) of the strains in the presence of tetracycline with C. capitatus and T. vulgaris EOs, respectively. Finally, the antagonistic effect between the EOs and tetracycline was detected only in one Salmonella strain.
Principal Component Analysis
To assess the relationships between the type of treatment and bioactivity on the Salmonella strains, the dataset obtained from the MIC analysis was subjected to PCA. The PCA biplot (Figure 1) showed that the two principal components explained 81.10% of the total variance with the first axis (PC1) that contributed with 50.49% of the total variance and the second axis (PC2) with 30.61% of the total variance. The loading plot displayed the discrimination of the type of antimicrobial treatments along the PC2, showing the separation of C. capitatus from the other EOs, alone and in combination with tetracycline. The high value reached from each variable of the loading plot indicated the best bioactivity at the highest concentrations of the compounds. The score plot showed the distribution of the strains along the PC1, where they were mainly grouped in one cluster. As observed in the biplot, most of the Salmonella strains clustered with resistance to low concentrations of the various antimicrobial treatments (red circle). However, a smaller part of the strains (7/31) was distributed differently, showing a greater resistance to the EO treatment, closest to which it was positioned.
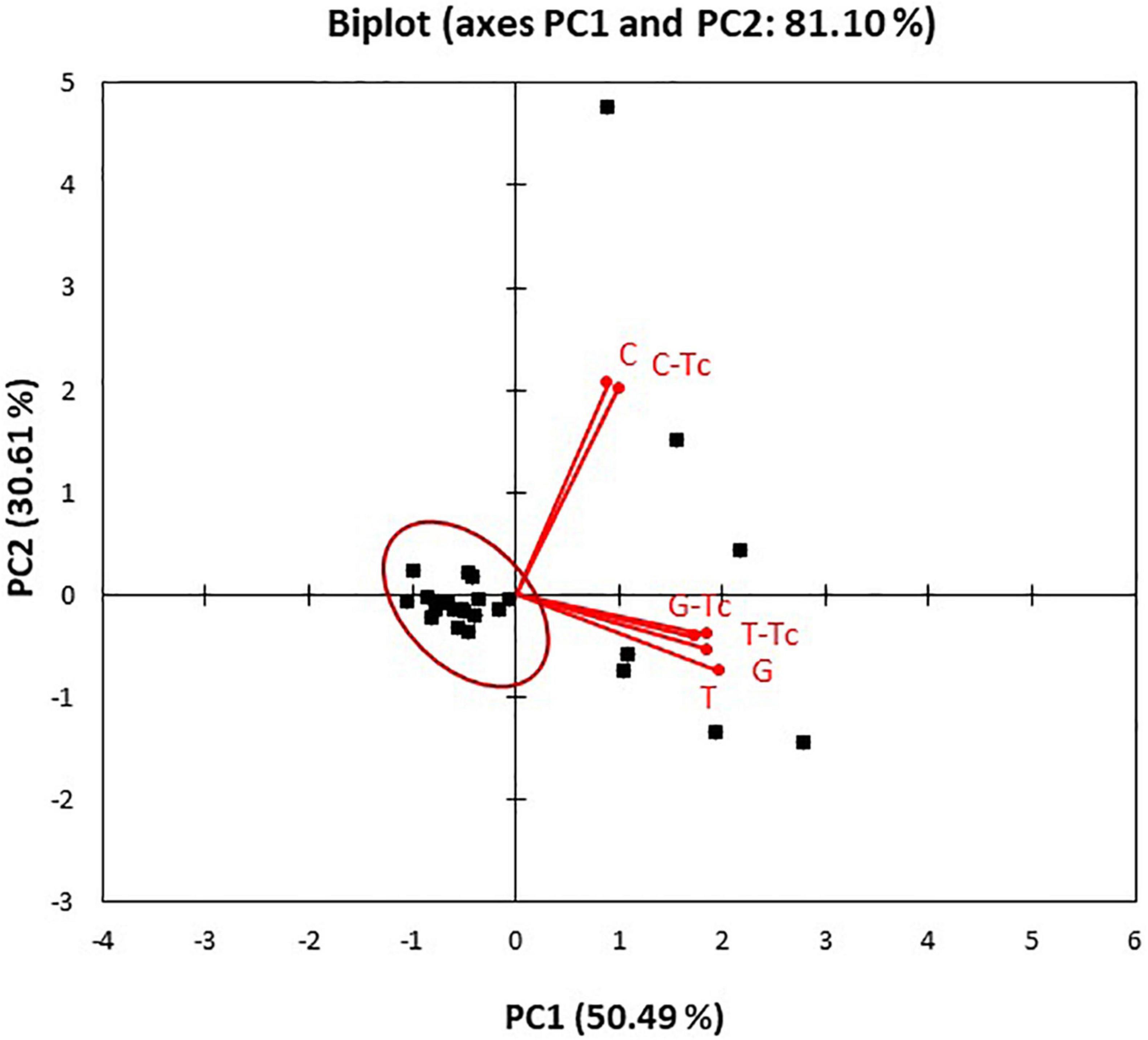
Figure 1. Principal component analysis (PCA) biplot (scores and loadings) based on antibacterial activity of Coridothymus capitatus, Eugenia caryophyllata, and Thymus vulgaris EOs alone or in combination with tetracyclines on the Salmonella strains. C. capitatus (C), E. caryophyllata (G), T. vulgaris (T), and tetracycline (Tc).
The PCA results confirmed the different effect of the EO treatments on the Salmonella strains, in particular, for C. capitatus, as already observed in MIC determinations (Table 3).
Discussion
Antimicrobial compounds have been used to treat bacterial infections in humans and animals since the middle of the 20th century. The selective pressure has been causing the emergence of resistance, which is genetically encoded and subsequently inherited by the progeny of resistant pathogens (Munk et al., 2018). Due to the genetic nature of resistance and the ability to select resistant organisms through the use of antimicrobials in animals, their presence in animal products is considered a potential source of AMR in humans (Andrews and Ryan, 2015). Antibiotic-resistant bacteria have been found in food products (ready-to-eat, cooked meat), in cattle, poultry, swine, and goats in different stages of production (EFSA, 2021). In fact, in our study, all Salmonella spp. isolates showed phenotypic and/or genotypic resistance to at least one class of antibiotic examined. In detail, our results showed that Salmonella isolates were resistant to ampicillin, piperacillin, tetracycline, gentamicin, amikacin, and tobramycin (Table 2). The European Union Summary Report on AMR in zoonotic and indicator bacteria from humans, animals, and food in 2018/2019 showed resistance levels to ampicillin, sulfonamide, and tetracycline greater than 20%, and particularly, in Italy, 68–72% of the isolates were resistant to the abovementioned antibiotics. Ampicillin and tetracycline are commonly used in swine livestock as first-choice therapeutic antibiotics (Lekagul et al., 2019). They are also used as growth promoters, although in Europe, antibiotics have been banned as feed additives since 2006 (Lekagul et al., 2019). In particular, tetracyclines have been mainly used in animal health and in swine livestock, against Gram-positive and Gram-negative infections (Munk et al., 2018; Lekagul et al., 2019). Probably as a consequence, in our research, 31 Salmonella strains (15 S. Typhimurium, 3 Monophasic variant S. Typhimurium, 6 S. Enteritidis, 6 S. Rissen, and 1 S. Typhi) showed phenotypic resistance to tetracycline. According to our results, the AMR phenotype that is more present in the swine production chain is ampicillin, streptomycin, tetracycline, and chloramphenicol (Munk et al., 2018). Furthermore, all strains were sensitive to third-generation cephalosporins and fluoroquinolones, identified as “critically important antimicrobials” (CIA). In particular, all strains were sensitive to cefpodoxime, marbofloxacin, and enrofloxacin, while only 2.8% (1/36) was resistant to ceftiofur. In Europe, the resistance to third-generation cephalosporins is reported to be greater than 10%, and in Italy, more than 5% of the resistant strains were detected in animal samples (EFSA, 2021).
Regarding the antibiotic resistance genes, our results showed that most commonly detected were parC (100%), catA (94.4%), nfsB (86.1%), nfsA (77.7%), blaTEM (47.2%), tetA (47.2%), and tetB (41.6%). Our results are in agreement with other authors, who indicate that Italian pigs show the highest AMR levels in Europe (Munk et al., 2018). Other resistance genes, detected in a small proportion of samples (2.8%), were tetC, aac(3)IV, and aadB, as shown in Table 2.
The association of phenotypic resistance and the presence of AMR genes has been demonstrated to be variable among Salmonella serovars (Deekshit et al., 2012; McDermott et al., 2016). Similarly, divergent phenotypic and genotypic antimicrobial findings were obtained in our study. In fact, whereas phenotypic resistance to one or more antimicrobials was observed for both S. Derby (n = 2) and S. Rissen (n = 5), the corresponding resistance genes were not detected by PCR. On the contrary, for other strains, the opposite situation was observed. Deekshit et al. (2012), demonstrated that ubiquitous strains of non-typhoidal Salmonella can have silent AMR genes and that the correlation between phenotypic and genotypic resistance is not always possible (Deekshit et al., 2012). Some phenotypic and genotypic discrepancies observed may have been possible because not all the resistance genes were tested. Nevertheless, according to literature, some resistance mechanisms still remain unidentified (McDermott et al., 2016). For these reasons, when a resistance mechanism is detected in the genome, while the isolate is phenotypically susceptible, the interpretation criteria of the antibiogram may also be questioned (Lepuschitz et al., 2019).
Antibiotic resistance represents a current global concern, widespread in different fields of pharmaceutical sciences. As mentioned above, the possibility of combining antibiotics with EOs can be an alternative to overcome AMR in bacteria (Solarte et al., 2017). In veterinary clinical practice, data concerning EO treatments in vitro and in vivo do not draw a complete picture as in human medicine (Ebani and Mancianti, 2020). Nevertheless, the positive outcomes of EO treatments have been correlated with both their direct antimicrobial effects and their aspecific antioxidant and anti-inflammatory effects (Miguel et al., 2020), along with the immunomodulatory activity (Valdivieso-Ugarte et al., 2019).
Regarding our results (Table 3), all of the studied Salmonella isolates displayed MIC ≥ 256 μg/ml to tetracycline. Conversely, the three EOs and, in particular, C. capitatus and T. vulgaris EOs inhibited bacterial growth (Table 3). The Salmonella spp. strains were not able to grow even in the presence of the lowest concentrations of the two EOs (0.31 and 0.625 μl/ml), showing low levels of resistance to these compounds. Moreover, even in the presence of E. caryophyllata EO, the strains showed a reduction in growth capacity, as evidenced by the MIC values, although with values higher than the other two EOs (52% of strains showed MIC values of 20 μl/ml).
The antimicrobial activity of C. capitatus, T. vulgaris, and E. caryophyllata EOs can be attributed to the whole phytocomplex; nevertheless, the principal components carvacrol, thymol, and eugenol are known to exert antimicrobial effects. In detail, the mode of action of these natural compounds mainly involves the microbial membrane. The inhibitory activity of the natural compounds has been related to their hydrophobicity, which influences their partition in the cytoplasmic membrane (Lanciotti et al., 2004). The increased toxicity on the cytoplasmic membrane is directly correlated to the higher hydrophobicity level of the natural compound. Arfa et al. (2006) affirmed that carvacrol, due to its high hydrophobicity, exerts the highest antimicrobial activity. Contrarily, eugenol shows a lower efficacy that could be attributed to its lower hydrophobicity. Carvacrol is the major compound of C. capitatus EOs, the botanical species also called Thymbra capitata (Verdeguer et al., 2020). The best effectiveness of this EO, observed in terms of MIC values (Table 3) and confirmed by the different discrimination in PCA biplot (Figure 1), is probably due mainly to carvacrol. Carvacrol and thymol are characterized by the presence of a hydroxyl group and a system of delocalized electrons that are important for the antimicrobial activity of these phenolic compounds. This chemical structure allows carvacrol and thymol to act as proton exchangers, able to reduce the transmembrane gradient. The consequence is the collapse of the proton motive force and the depletion of the ATP pool, which can lead to cell death (Arfa et al., 2006).
The interactions between natural compounds, such as carvacrol and cell membranes, are described to affect both the lipid ordering and the bilayer stability, resulting in membrane integrity decrease (Arfa et al., 2006) and modification of the efflux pump activities (Bolla et al., 2011). Efflux is a mechanism that protects bacterial cells by expelling toxic compounds, such as antibiotics, before they can reach the intracellular targets (Davies, 1994). In Gram-negative bacteria, a tripartite efflux system is necessary to expel the drug to the outer medium: a protein localized in the cytoplasmic membrane, another in the periplasmic space, and a third in the outer membrane (de Sousa Oliveira et al., 2016). The efflux pumps are responsible for drug resistance in pathogenic bacteria, representing one of the main targets to overcome microbial resistance. Five families of membrane-spanning efflux proteins are recognized: major facilitators (MFs), small multidrug resistance (SMR), resistance nodulation cell division (RND), ATP-binding cassette (ABC), and multidrug and toxic compound extrusion (MATE) (Trifan et al., 2020). Active efflux systems have been commonly observed in the Salmonella genus and include tetA and tetB, the genes associated with tetracycline resistance (Frye and Jackson, 2013).
Miladi et al. (2017) observed that carvacrol, thymol, and eugenol act as efflux pump inhibitors in S. Typhimurium, causing the accumulation of the antibiotic, thereby acting synergistically. Synergy occurs when the combined effect of two or more substances is greater than the sum of the individual agents, in terms of enhanced therapeutic actions on the same target (Zhou et al., 2016). As demonstrated by our results (Table 3), synergistic effects between EOs and tetracycline were observed in a few cases and only when the FIC index was ≤1.0. Pirintsos et al. (2020) argued that the main reason for employing combinations of active substances, with synergistic interactions, is to reduce the administered amount of each compound and to increase the biological activity of a preparation/mixture against a specific target. Although the FICI classification did not highlight the synergy between the two types of antimicrobials, the effectiveness of the different compounds was evident. In fact, Table 3 displays the general decrease in tetracycline concentration (from 256 to 4 μg/ml) to which the Salmonella spp. strains were able to resist, only in combination with the EOs. This evidence suggests a restoration of susceptibility in Salmonella spp. to the antibiotic, as a consequence of the presence of EOs. As mentioned before, the inhibition of the efflux pumps by the EOs could cause antibiotic accumulation. In addition, the presence of EOs with a destabilizing effect on the bacterial membrane could facilitate antibiotic penetration into the cytoplasm and the easier reaching of target sites, such as the ribosome. In fact, tetracycline affects the 30S subunit of the bacterial ribosome, thus, inhibiting protein synthesis (de Sousa Oliveira et al., 2016).
The combinations of EOs and antimicrobial compounds could be an important instrument to reduce or reverse AMR (Trifan et al., 2020). The synergistic effect between EOs and antibiotics against multidrug-resistant bacteria has been described by other authors (Fadli et al., 2016). However, there is a lack of studies about the combination of EOs and tetracycline against multidrug-resistant Salmonella isolates (Trifan et al., 2020), in spite of the abundance of multiresistant strains and of the key role of tetracycline in animal husbandry.
Conclusion
This study investigated a new approach to overcome multidrug resistance in Salmonella spp. isolated from the swine production chain. With this is mind, the combination of antibiotics and EOs was evaluated, by using T. vulgaris, E. caryophyllata, and C. capitatus EOs. Our results confirmed the evidence of the widespread AMR. In fact, Salmonella spp. exhibited a complex pattern of AMR, which underlines the need of new weapons to overcome multidrug resistance in the swine production chain. The combination of natural compounds with antibiotics represents a possible strategy. In fact, the most relevant result of the study was that the use of the EOs in combination with tetracycline showed the ability to restore the antibiotic effect of tetracycline in Salmonella strains. In spite of the importance of the topic, the studies related to the combinations of EOs and tetracyclines against Salmonella multidrug-resistant strains are lacking. In this still unexplored scenario, our results can represent a starting point. In this perspective, our research can be a piece of the puzzle in which the current dataset is not complete but is a starting point for further investigations. The potential future studies could evaluate the regulation of resistance genes following treatment with EOs and the investigation of the modulation of cell membrane proteins by proteomics approaches.
Data Availability Statement
The data presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
AS, AP, and AV designed the study. CL, FM, and AF performed the experiments and wrote the manuscript. AS was responsible of the data validation. AP and AV gave important intellectual advice. All the authors checked, read, and approve the final version of manuscripts.
Funding
This research project was funded entirely by the University of Teramo, School of Specialization in Inspection of Foods of Animal Origin “G. Tiecco”.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andrews, J. R., and Ryan, E. T. (2015). Diagnostics for invasive Salmonella infections: current challenges and future directions. Vaccine 33, 8–15. doi: 10.1016/j.vaccine.2015.02.030
Arfa, B., Combes, A., Preziosi-Belloy, S., Gontard, L., and Chalier, P. N. (2006). Antimicrobial activity of carvacrol related to its chemical structure. Lett. Appl. Microbiol. 43, 149–154. doi: 10.1111/j.1472-765X.2006.01938.x
Bolla, J. M., Alibert-Franco, S., Handzlik, J., Chevalier, J., Mahamoud, A., Boyer, G., et al. (2011). Strategies for bypassing the membrane barrier in multidrug resistant Gram-negative bacteria. FEBS Lett. 585, 1682–1690. doi: 10.1016/j.febslet.2011.04.054
Bonardi, S., Alpigiani, I., Bruini, I., Barilli, E., Brindani, F., Morganti, M., et al. (2016). Detection of Salmonella enterica in pigs at slaughter and comparison with human isolates in Italy. Inter. J. Food Microbiol. 218, 44–50. doi: 10.1016/j.ijfoodmicro.2015.11.005
Broadway, P. R., Brooks, J. C., Mollenkopf, D. F., Alexandra Calle, M., Loneragan, G. H., Miller, M. F., et al. (2021). Prevalence and antimicrobial susceptibility of Salmonella serovars isolated from U.S. Retail Ground Pork. Foodborne Pathog. Dis. 18, 219–227. doi: 10.1089/fpd.2020.2853
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing, 26th Edn, Wayne, PA: CLSI.
Davies, J. (1994). Inactivation of antibiotics and the dissemination of resistance genes. Science 264, 375–382. doi: 10.1126/science.8153624
de Sousa Oliveira, K., de Lima, L. A., Cobacho, N. B., Dias, S. C., and Franco, O. L. (2016). “Mechanisms of antibacterial resistance: shedding some light on these obscure processes,” in Antibiotic Resistance. Mechanisms and New Antimicrobial Approaches, eds K. Kon and M. Rai (London: Academic Press), 19–37.
Deekshit, V. K., Kumar, B. K., Rai, P., Srikumar, S., and Karunasagar, I. (2012). Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood-associated nontyphoidal isolates of Salmonella in south-west coast of India. J. App. Microbiol. 112, 1113–1122. doi: 10.1111/j.1365-2672.2012.05290.x
Di Ciccio, P., Ossiprandi, M. C., Zanardi, E., Ghidini, S., Belluzzi, G., Vergara, A., et al. (2016). Microbiological contamination in three large-scale pig slaughterhouses in Northern Italy. Ital. J. Food Saf. 5, 219–223. doi: 10.4081/ijfs.2016.6151
Ebani, V. V., and Mancianti, F. (2020). Use of essential oils in veterinary medicine to combat bacterial and fungal infections. Vet. Sci. 7:193. doi: 10.3390/vetsci7040193
EFSA (2021). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 19:6490. doi: 10.2903/j.efsa.2021.6490
El-Tayeb, M., Ibrahim, A. S. S., Al-Salamah, A. A., Almaary, K., and Elbadawi, Y. B. (2017). Prevalence, serotyping and antimicrobials resistance mechanism of Salmonella enterica isolated from clinical and environmental samples in Saudi Arabia. Braz. J. Microbiol. 48, 499–508. doi: 10.1016/j.bjm.2016.09.021
Fadli, M., Pagès, J. M., Mezrioui, N. E., Abbad, A., and Hassani, L. (2016). Artemisia herba-alba Asso and Cymbopogon citratus (DC.) Stapf essential oils and their capability to restore antibiotics efficacy. Ind. Crops Prod. 89, 399–404. doi: 10.1016/j.indcrop.2016.05.039
Fratini, F., Mancini, S., Turchi, B., Friscia, E., Pistelli, L., Giusti, G., et al. (2017). A novel interpretation of the fractional inhibitory concentration index: the case Origanum vulgare L. and Leptospermum scoparium J. R. et G. Forst essential oils against Staphylococcus aureus strains. Microbiol. Res. 195, 11–17. doi: 10.1016/j.micres.2016.11.005
Frye, J. G., and Jackson, C. R. (2013). Generic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enterococcus spp. isolated from U.S food animals. Front. Microbiol. 4:135. doi: 10.3389/fmicb.2013.00135
Garcia, V., Montero, I., Bances, M., Rodicio, R., and Rodicio, R. (2016). Indigence and genetic bases of nitrofurantoin resistance in clinical isolates of two successful multidrug-resistant clone if Salmonella enterica Serovar Typhimurium: pandemic “DT 104” and pUO-StVR 2. Microb. Drug Resist. 2, 405–412. doi: 10.1089/mdr.2016.0227
Kikuvi, G. M., Ombui, J. N., and Mitema, E. S. (2010). Serotypes and antimicrobial residence profiles of Salmonella isolates from pigs at slaughter in Kenya. J. Infect. Dev. Ctri. 4, 243–248. doi: 10.3855/jidc.446
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009). Antimicrobial Resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75, 559–566. doi: 10.1128/AEM.01821-08
Lambert, R. J., Skandamis, P. N., Coote, P. J., and Nychas, G. J. (2001). A study of the minimum inhibitory concentration and mode of action of oregano essential oil, thymol and carvacrol. J. Appl. Microbiol. 91, 453–462. doi: 10.1046/j.1365-2672.2001.01428.x
Lanciotti, R., Gianotti, A., Patrignani, F., Belletti, N., Guerzoni, E., and Gardini, F. (2004). Use of natural aroma compounds to improve shelf-life and safety of minimally processed fruits. Trends Food Sci. Technol. 15, 201–208. doi: 10.1016/j.tifs.2003.10.004
Lauteri, C., Festino, A. R., Conter, M., and Vergara, A. (2021). “Prevalence and antimicrobial resistance profile in Salmonella spp. isolates from swine food chain,” in Proceedings of the XXX AIVI Congress, September 2021.
Lekagul, A., Angcharoensathien, V., and Yeung, S. (2019). Patterns of antibiotic use in global pig production: a systematic review. Vet. Animal Sci. 7, 2–33. doi: 10.1016/j.vas.2019.100058
Lepuschitz, S., Schill, S., Stoeger, A., Pekard-Amenitsch, S., Huhulescu, S., Inreiter, N., et al. (2019). Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci Total Environ. 662, 227–235. doi: 10.1016/j.scitotenv.2019.01.179
Maggio, F., Rossi, C., Lòpez, C., Valbonetti, L., Desideri, G., Paparella, A., et al. (2021). A single exposure to a sublethal concentration of Origanum vulgare essential oil initiates response against food stressors and restoration of antibiotic susceptibility in Listeria monocytogenes. Food Control 132:108562. doi: 10.1016/j.foodcont.2021.108562
Magi, G., Marini, E., and Facinelli, B. (2015). Antimicrobial activity of essential oils and carvacrol, and synergy of carvacrol and erythromycin, against clinical, erythromycin-resistant Group a Streptococci. Front. Microbiol. 6:165. doi: 10.3389/fmicb.2015.00165
Marchese, A., Barbieri, R., Coppo, E., Orhan, I. E., Daglia, M., Nabavi, S. F., et al. (2017). Antimicrobial activity of eugenol and essential oils containing eugenol: a mechanistic viewpoint. Crit. Rev. Microbiol. 43, 668–689. doi: 10.1080/1040841X.2017.1295225
McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y., Li, C., Folster, J. P., et al. (2016). Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60, 5515–5520. doi: 10.1128/AAC.01030-16
Miguel, M. G., Lourenço, J. P., and Faleiro, M. L. (2020). Superparamagnetic iron oxide nanoparticles and essential oils: a new tool for biological applications. Int. J. Mol. Sci. 21, 1–24. doi: 10.3390/ijms21186633
Miladi, H., Zmantar, T., Kouidhi, B., Chaabouni, Y., Mahdouani, K., Bakhrouf, A., et al. (2017). Use of carvacrol, thymol, and eugenol for biofilm eradication and resistance modifying susceptibility of Salmonella enterica serovar Typhimurium strains to nalidixic acid. Microb. Pathog. 104, 56–63. doi: 10.1016/j.micpath.2017.01.012
Moemenbellah-Fard, M. D., Abdollahi, A., Ghanbariasad, A., and Osanloo, M. (2020). Antibacterial and leishmanicidal activities of Syzygium aromaticum essential oil versus its major ingredient, eugenol. Flav. Fragr. J. 35, 534–540. doi: 10.1002/ffj.3595
Munk, P., Knudsen, B. E., Lukjacenko, O., Ribeiro Duarte, A. S., Van Gompel, L., Luiken, R. E. C., et al. (2018). Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 3, 898–908. doi: 10.1038/s41564-018-0192-9
Palma, E., Tilocca, B., and Roncada, P. (2020). Antimicrobial resistance in veterinary medicine: an overview. Int. J. Mol. Sci. 21:1914. doi: 10.3390/ijms21061914
Pirintsos, S. A., Bariotakis, M., Kampa, M., Sourvinos, G., Lionis, C., and Castanas, E. (2020). The therapeutic potential of the essential oil of Thymbra capitata (L.) Cav., Origanum dictamnus L. and Salvia fruticosa Mill. And a case of plant-based pharmaceutical development. Front. Pharmacol. 11:522213. doi: 10.3389/fphar.2020.522213
Prasertsee, T., Khantaprab, N., Yamsakul, P., Santiyanont, P., Chokesajjawatee, N., and Patchanee, P. (2016). Repetitive sequence-based PCR fingerprinting and the relationship of antimicrobial-resistance characteristics and corresponding genes among Salmonella strains from pig production. Asia Pac. J. Tropical. Dis. 6, 390–395. doi: 10.1016/S2222-1808(15)61054-4
Rossi, C., Chaves-López, C., Serio, A., Casaccia, M., Maggio, F., and Paparella, A. (2020). Effectiveness and mechanisms of essential oils for biofilm control on food-contact surfaces: an updated review. Crit. Rev. Food Sci. Nutr. 30, 1–20. doi: 10.1080/10408398.2020.1851169
Solarte, A. L., Astorga, R. J., Aguiar, F., Galán-Relaño, Á, Maldonado, A., and Huerta, B. (2017). Combination of antimicrobials and essential oils as an alternative for the control of Salmonella enterica multiresistant strains related to foodborne disease. Foodborne Pathog. Dis. 14, 558–563. doi: 10.1089/fpd.2017.2295
Trifan, A., Luca, S. V., Greige-Gerges, H., Miron, A., Gille, E., and Aprotosoaie, A. C. (2020). Recent advance in tackling microbial multidrug resistance with essential oil: combinatorial and nano-based strategies. Crit. Rev. Microbiol. 46, 338–357. doi: 10.1080/1040841X.2020.1782339
Valdivieso-Ugarte, M., Gomez-Llorente, C., Plaza-Díaz, J., and Gil, Á (2019). Antimicrobial, antioxidant, and immunomodulatory properties of essential oils: a systematic review. Nutrients 11:2786. doi: 10.3390/nu11112786
Verdeguer, M., Castañeda, L. G., Torres-Pagan, N., Llorens-Molina, J. A., and Carrubba, A. (2020). Control of Erigeron bonariensis with Thymbra capitata, Mentha piperita, Eucalyptus camaldulensis, and Santolina chamaecyparissus Essential Oils. Molecules 25:562. doi: 10.3390/molecules25030562
World Health Organization [WHO] (2020). Antimicrobial Resistance Report by the Director-General. EXECUTIVE BOARD EB148/11. 148th session. Geneva: WHO.
Yang, S. K., Yusoff, K., Mai, C. W., Lim, W. M., Lim, S. H. E., Asmahani, A., et al. (2018). Mode of action of cinnamon bark (Cinnamomum verum) essential oil and the combinatory bactericidal activity with meropenem against KPC-producing Klebsiella pneumoniae. Asian J. Med. Biomed. 9, 1–7.
Keywords: antimicrobial resistance, Salmonella spp., essential oil, Thymus vulgaris, Eugenia caryophyllata, Corydothymus capitatus, tetracycline, swine production chain
Citation: Lauteri C, Maggio F, Serio A, Festino AR, Paparella A and Vergara A (2022) Overcoming Multidrug Resistance in Salmonella spp. Isolates Obtained From the Swine Food Chain by Using Essential Oils: An in vitro Study. Front. Microbiol. 12:808286. doi: 10.3389/fmicb.2021.808286
Received: 03 November 2021; Accepted: 13 December 2021;
Published: 09 February 2022.
Edited by:
Evandro L. de Souza, Federal University of Paraíba, BrazilReviewed by:
Roberto Guedes, Federal University of Minas Gerais, BrazilAbdennaceur Hassen, Centre de Recherches et des Technologies des Eaux, Tunisia
Nicolás Francisco Cordeiro, Universidad de la República, Uruguay
Copyright © 2022 Lauteri, Maggio, Serio, Festino, Paparella and Vergara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Maggio, Zm1hZ2dpb0B1bml0ZS5pdA==
 Carlotta Lauteri
Carlotta Lauteri Francesca Maggio
Francesca Maggio Annalisa Serio
Annalisa Serio Anna Rita Festino
Anna Rita Festino Antonello Paparella
Antonello Paparella Alberto Vergara
Alberto Vergara