- 1Division of Healthcare Quality Promotion, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 2Goldbelt C6, LLC, Chesapeake, VA, United States
- 3Division of Tuberculosis Elimination, National Center for HIV/AIDS, Viral Hepatitis, STD and TB Prevention, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 4Office of the Director, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 5Biotechnology Core Facility Branch, Division of Scientific Resources, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 6ASRT Incorporated, Atlanta, GA, United States
- 7Department of Pathology, University of Maryland School of Medicine, Baltimore, MD, United States
Enterococcus faecalis and faecium with resistance to daptomycin and/or linezolid are emerging globally. We present the genomic characterization of daptomycin- and linezolid-resistant E. faecalis and E. faecium surveillance isolates from the United States, 2013–2016. Daptomycin resistance was low among E. faecalis (2/364, 0.5%) and E. faecium (17/344, 5%). The majority (71%, 12/17) of daptomycin-resistant E. faecium isolates belonged to the emerging ST736 clone and contained mutations in liaFSR and cls previously associated with resistance. However, 1/2 E. faecalis and 3/17 E. faecium did not contain these mutations previously associated with daptomycin resistance. Linezolid resistance was rare among E. faecalis (1/364, 0.3%) and E. faecium (2/344, 0.6%). These two E. faecium isolates, one of which was also resistant to daptomycin and vancomycin, contained the 23S rRNA nucleotide mutation (G2576T) associated with linezolid resistance. Long-read sequencing revealed the linezolid-resistant E. faecalis isolate contained chromosomal- and plasmid-encoded copies of optrA. The chromosomal optrA was located on the recently described Tn6674 multiresistance transposon. The second copy of optrA was encoded on an ∼65 kb mosaic plasmid, with component regions sharing high sequence identity to optrA-encoding multiresistance plasmids of animal origin. The optrA-encoding plasmid contained open reading frames predicted to encode proteins associated with a pheromone-responsive plasmid transfer system, and filter mating experiments confirmed the plasmid was conjugative. Continued surveillance of enterococci is necessary to assess the prevalence and trends of daptomycin and linezolid resistance in the United States, characterize resistance mechanisms and how they transfer, and monitor for emerging sequence types associated with resistance.
Introduction
Enterococci are an important cause of healthcare-associated infections (HAIs) in the United States, including bloodstream, surgical site, and urinary tract infections. Approximately 30% of enterococcal HAIs are caused by vancomycin-resistant enterococci (VRE), which are categorized as a serious public health threat, requiring prompt and sustained action (CDC, 2019). Enterococci resistant to penicillin, ampicillin, and vancomycin (VAN) require other treatments such as daptomycin (DAP) and linezolid (LZD) (Garcia-Solache and Rice, 2019). However, resistance to these other agents is also being reported (Arias et al., 2011; Pfaller et al., 2017).
Daptomycin is a lipopeptide antibiotic that binds to the bacterial cell membrane (CM), disrupting essential envelope functions and resulting in cell death (Prater et al., 2019). In Enterococcus faecalis, daptomycin resistance (DAP-R) is a result of CM changes that redistribute phospholipids away from the septum, diverting DAP from the cell (Khan et al., 2019). In Enterococcus faecium, the environment can influence how DAP-R evolves, and both repulsion and DAP diversion mechanisms can occur (Prater et al., 2019). In both E. faecalis and E. faecium, these CM responses are most commonly associated with initial mutations in genes encoding the 3-component LiaFSR (lipid-II–interacting antibiotics) stress response system that regulate cell envelope integrity, followed by subsequent mutations in genes involved in phospholipid metabolism (Khan et al., 2019), including cls, encoding a cardiolipin (CL) synthase; and gdpD, encoding a putative glycerophosphodiesterase (Prater et al., 2019). The primary modulator of the LiaFSR system in E. faecalis, LiaX, was recently characterized and found to sense DAP and trigger protective cell membrane remodeling (Khan et al., 2019). A recent report identified the first clinical case of an E. faecalis isolate with a LiaX loss-of-function mutation resulting in DAP-R (Ota et al., 2021). While the mutations in the LiaFSR system are the most common mechanism associated with DAP-R, resistance is driven by complex changes in the membrane stress response network and additional pathways exist. For example, the YycFG and YxdJK 2-component systems that regulate cell wall homeostasis have also been implicated in enterococcal DAP-R (Miller et al., 2019, 2020). Limited data are available on the clonal distribution of DAP-R enterococcal clinical isolates in the United States, but a clone, ST736, associated with DAP-R has recently been reported in New York City (Wang et al., 2014, 2018).
Linezolid is an oxazolidinone antibiotic effective against Gram-positive bacteria, including enterococci and Staphylococcus aureus. In enterococci, resistance to LZD is often caused by mutations in the V domain of the 23S rRNA gene (Hasman et al., 2019), with the G2576T nucleotide mutation (Beukers et al., 2018) being the most common (Bi et al., 2018). Recently, the resistance genes cfr, cfr(B), cfr(D), optrA, and poxtA, encoding transferable resistance to oxazolidinones, have been described in enterococci (Liu et al., 2012, 2014; Deshpande et al., 2015; Wang et al., 2015; Bender et al., 2016; Antonelli et al., 2018; Guerin et al., 2020). The optrA gene encodes an ATP-binding cassette (ABC)-F protein that mediates resistance through the ribosomal protection mechanism (Sharkey et al., 2016). First characterized in E. faecalis and E. faecium isolated in China from humans and food-producing animals in 2009 (Wang et al., 2015), optrA has since been identified on the chromosome or on various-sized plasmids, sometimes in combination with cfr, cfr(B), or poxtA within the same strain or plasmid (Li et al., 2019). When encoded on the chromosome, optrA is typically located on a recently characterized Tn554-family transposon, Tn6674 (Li et al., 2019), adjacent to the resistance gene fexA, which confers resistance to phenicols (Freitas et al., 2020). Tn6674 insertion occurs at a conserved location in the radC gene, which encodes a putative DNA repair protein, and this ΔradC integration site is considered a hotspot for chromosomal optrA integration (Freitas et al., 2020). When encoded on a plasmid, the optrA gene is typically flanked (upstream and/or downstream) by insertion sequence elements of the IS1216 (IS6 family) or ISL3 family type (He et al., 2016; Freitas et al., 2020). The optrA gene has rapidly emerged as a major contributor to the spread of LZD resistance worldwide (Deshpande et al., 2018), but has been only recently detected in human clinical (Pfaller et al., 2017; Deshpande et al., 2018) and animal isolates (Tyson et al., 2018) in the United States. Limited data are available regarding prevalence of optrA among clinical or animal linezolid- resistant (LZD-R) enterococcal isolates in the United States; however, the available studies indicate enterococci harboring optrA remain low (Pfaller et al., 2017; Deshpande et al., 2018; Tyson et al., 2018; Wardenburg et al., 2019). For example, a recent study of United States (U.S.) clinical E. faecium LZD-R isolates revealed they were more likely to harbor mutations in the V domain of the 23S rRNA gene than to carry optrA (Wardenburg et al., 2019).
The U.S. Centers for Disease Control and Prevention’s (CDC), Division of Healthcare Quality Promotion (DHQP) Sentinel Surveillance system performs surveillance of isolates collected from geographically diverse regions of the United States to study their antimicrobial resistance profiles and perform phenotype-genotype correlation studies. The goal of this surveillance is to improve the knowledge of susceptibility patterns and mechanisms of resistance among bacteria causing HAIs. Isolates are collected from patients hospitalized in acute care or long-term care facilities. Here, we present the characterization of E. faecalis and E. faecium isolates collected during the DHQP Sentinel Surveillance system’s 2013–2016 collection period determined to be DAP-R or LZD-R by reference antimicrobial susceptibility testing (AST). Whole genome sequencing (WGS) was used to identify if there were predominant multi-locus sequence typing (MLST) clones associated with DAP-R or LZD-R, and if mechanisms or mutations previously associated with DAP-R or LZD-R could be identified.
Materials and Methods
Bacterial Isolates, Growth Conditions, and Identification
All isolates included in this study were selected from a Sentinel Surveillance collection of 708 enterococci collected during 2013–2016 from eight states (CA, IA, MD, NC, NM, NY, PA, and WA) and resistance to DAP and/or LZD was determined by antimicrobial susceptibility testing (AST) (described below). Isolates were cultured on BD BBL Trypticase soy agar II with 5% sheep blood (SBA) (Thermo Fisher Scientific, Waltham, MA, United States) at 37°C in ambient air from glycerol stocks stored at −70°C. Identification was confirmed with matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry using a MALDI Biotyper, version 3.1 (Bruker Daltronics, Bremen, Germany).
Antimicrobial Susceptibility Testing
Broth microdilution (BMD) AST was performed on all isolates following the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2018, 2021). BMD panels were prepared in-house, consisting of 14 antibiotics at varying twofold serial dilutions in sterile 96-well microtiter plates (Caplugs, Rancho Dominguez, CA, United States) and stored at −70°C until use. Each well contained 100 μl Cation-adjusted Mueller-Hinton Broth (CAMHB, BD Difco, Sparks, MD, United States) and for DAP there was a supplement of 50 μg/ml calcium chloride. The following antimicrobial concentrations were included in the BMD panels: DAP (0.25–16 μg/ml), LZD (0.5–16 μg/ml), and VAN (0.25–64 μg/ml), see Supplementary Table 1 for the full list of antibiotics and concentrations tested in the BMD panel. Enterococcus faecalis ATCC 29212 and Staphylococcus aureus ATCC 29213 were used as quality control strains. Samples and controls were streaked to purity on SBA and colonies were suspended in 5 ml REMEL 0.85% sterile saline (Thermo Fisher Scientific, Waltham, MA, United States) to a turbidity equal to a 0.5 McFarland standard. Panels were inoculated with a 95-pin sterile inoculator (10 μl pickup; Caplugs, Buffalo, NY, United States), incubated at 35°C and read and interpreted according to CLSI guidelines (CLSI, 2021).
Whole Genome Sequencing
All DAP-R or LZD-R isolates underwent WGS. Genomic DNA was extracted from colonies cultured overnight on SBA using the Promega Maxwell® 16 Cell Low Elution Volume DNA Purification Kit and Maxwell® 16 MDx Instrument (Madison, WI, United States) and sheared using the Covaris ME220 Focused-ultrasonicator™ (Woburn, MA, United States). Indexed libraries were prepared using the Tecan Ovation® Ultralow System V2 Assay Kit (San Carlos, CA, United States) and the PerkinElmer Zephyr® G3 NGS Workstation (Waltham, MA, United States). Libraries were analyzed using the Standard Sensitivity NGS Fragment Analysis Kit and Advanced Analytical Fragment Analyzer System (Ankeny, IA, United States). Sequencing was performed using the MiSeq® Reagent Kit v2 (500 cycle) and Illumina MiSeq® System (San Diego, CA, United States) generating 2 × 250 paired-end reads. The Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, United States) was used to extract high molecular weight DNA from the optrA-positive E. faecalis AR-0780 following manufacturer’s instructions. DNA sheared to 20 kb utilizing needle shearing was used to generate large SMRTbell libraries using the standard 20-kb library protocols of the Pacific Biosciences SMRTbell template prep kit 1.0 (PacBio, Menlo Park, CA, United States). The libraries were further size selected utilizing BluePippin (Sage Scientific, Beverly, MA, United States) with a cutoff size of 10 kb. The finished library was bound to proprietary P6v2 polymerase and sequenced on a PacBio RS II sequencer using C4v2 chemistry for 360-min movies. PacBio reads were assembled de novo with HGAP (v3). Assemblies were polished with PacBio reads using Quiver, then polished with Illumina reads using Pilon (v 1.22) (Chin et al., 2013; Walker et al., 2014; Hunt et al., 2015). Sequence data were analyzed using the QuAISAR-H pipeline; a description of custom scripts and publicly available tools and versions utilized by QuAISAR-H is available at https://github.com/DHQP/QuAISAR_singularity. Sequence types (STs) were assigned using the pubMLST E. faecalis or E. faecium multi-locus sequence typing schemes (Homan et al., 2002; Ruiz-Garbajosa et al., 2006; Jolley et al., 2018) and clonal complexes (CCs) were assigned using the pubMLST database or as previously described in the literature, see below. Antimicrobial resistance genes were identified using SRST2 (v0.2.0) (Inouye et al., 2014) and GAMMA (Stanton et al., 2021) against a non-redundant combined database of the AR databases ResFinder (last updated 10/1/2019) (Zankari et al., 2012), ARG-ANNOT (v6, July 2019) (Gupta et al., 2014), and NCBI’s AMRFinder (last updated 11/5/2019) (Feldgarden et al., 2019) (all accessed 12/27/2019); minimum of 98% sequence identity and 90% sequence coverage was used. Assembled genomes were submitted to the NCBI Prokaryotic Genome Annotation Pipeline for annotation. An E. faecium whole genome multi-locus sequence typing (wgMLST) schema consisting of 5,489 accessory loci was used to analyze the ST739 isolates (BioNumerics, v7.6).1 The wgMLST phylogeny was constructed using Interactive Tree Of Life (iTOL, v5) (Letunic and Bork, 2021). To compare plasmids, assemblies were annotated with Prokka (v1.12) (Seemann, 2014) for consistency, aligned with NCBI Blast (v2.2.22) (Camacho et al., 2009), and visualized with EasyFig (v2.2.2) (Sullivan et al., 2011). CGE PlasmidFinder 2.0 was used to type pAR-0780 (Carattoli et al., 2014).
Analysis of Whole Genome Sequencing Data
Analysis of nucleotide mutations associated with LZD resistance within the 23S rRNA gene were performed using the recommendations provided by Beukers et al. (2018). Briefly, the 23S rRNA gene from E. faecium strain Aus0004 (GenBank accession number: NR_103056) was used as the reference sequence and each sample’s reads were mapped to the reference gene using Bowtie 2 (Langmead and Salzberg, 2012), then processed using SAMTools (Li et al., 2009) and then SNP-called using VarScan (v2.4.1) (Koboldt et al., 2012) using mpileup2cns with the following settings: –min-coverage 8 –min-reads2 2 –min-var-freq 0.01. WGS data from the LZD-R isolates were also analyzed for the detection of 23S rRNA mutations, and the optrA, cfr, cfr(B) and poxtA genes using LRE-finder 1.02 (Hasman et al., 2019). Analysis of assemblies for the presence of mutations associated with DAP resistance and analysis of the optrA regions on AR-0780’s chromosome and plasmid were performed using CLC Genomics Workbench 11.3 The liaF, liaS, liaR, gdpD, and yycG gene sequences from the E. faecium strain DO (accession CP003583), and the cls gene sequence from the E. faecium strain UW7606 × 64/3 TC1 (accession CP013009) were used to identify mutations in study strains as described by Wang et al. (2018). E. faecalis, strain S316 (accession ADDP01000082) was used to identify mutations in liaF, liaS, liaR, cls, and gdpD coding regions. The liaX coding region from E. faecium strain R494 (accession JH807905) and E. faecalis strain V583 (accession AE016830) were used to identify liaX mutations. The E. faecium HOU503 (accession CP040706) strain’s pgsA coding region was used as a reference to identify mutations in E. faecium study strains.
Conjugation Experiments
Filter mating experiments were performed to assess the transferability of the E. faecalis AR-0780 plasmid, pAR-0780, that encodes optrA. Isolated colonies of the donor, E. faecalis AR-0780 (optrA +, LZD-resistant, fusidic acid-susceptible) and the recipient E. faecalis JH2-2 (fusidic acid-resistant, LZD-susceptible), were inoculated into 5 ml of BD BBL Myoflask Trypticase Soy Broth (TSB, BD, Franklin Lakes, NJ, United States) containing 5 μl LZD (25 μg/ml) or 5 μl fusidic acid (3 μg/ml), respectively, and incubated overnight at 37°C. One hundred microliters of each overnight culture were transferred to 5 ml TSB, vortexed briefly, and incubated at 37°C for ∼5 h to mid-log phase. The turbidity of each culture was adjusted to the same optical density in sterile 0.85% saline (REMEL, Thermo Fisher Scientific, Waltham, MA, United States) using a Dade Behring Microscan turbidity meter (Siemens, Munich, Germany). Twenty microliters of the AR-0780 suspension was mixed with 180 μl of JH2-2 for a 1:10 ratio of donor to recipient. The conjugation mixture was applied to the center of a Corning 0.22-μm pore bottle top filter (Thermo Fisher Scientific, Waltham, MA, United States) and collected by vacuum filtration. The filter was removed from the filter unit, placed in the middle of a SBA plate, without antimicrobials, and incubated at 37°C for 24 h. The filter was then transferred to a sterile 50 ml conical tube containing 5 ml 0.85% saline and vortexed vigorously to remove growth. Serial dilutions of the conjugation mixture were prepared in sterile 0.85% saline and 100 μl of each dilution were plated in triplicate onto trypticase soy agar (TSA, BD) plates containing the following antimicrobial agents for selection: TSA with 3 μg/ml LZD (donor selection); TSA with 25 μg/ml fusidic acid (recipient selection); and TSA with 3 μg/ml LZD and 25 μg/ml fusidic acid (transconjugant selection). Following incubation at 37°C for 36–48 h, donor, recipient, and transconjugant colonies were counted to calculate the conjugation frequency (transconjugants per donor). Ten transconjugant colonies were randomly selected and propagated on TSA plates containing 3 μg/ml LZD and 25 μg/ml fusidic acid. PCR was performed to verify the presence of optrA in the transconjugants as described above.
optrA PCR
A cell lysate was prepared (Conrad et al., 1996) and used for PCR to verify the presence of optrA as previously described (Wang et al., 2015), using primers A-F (5′-AGGTGGTCAGCGAACTAA-3′) and A-R (5′-ATCAACTGTTCCCATTCA-3′) that amplify a 1,395 bp internal segment of optrA. PCR reaction mixtures consisted of 25 μl HotStarTaq Master Mix (QIAGEN, Germantown, MD), 1 μl of each primer (20 μM), 18 μl RNAse-free water (Promega, Madison, WI, United States), and 5 μl of each cell lysate. PCR reaction conditions were 95°C 15 min; 35 cycles of 94°C 1 min, 55°C 1 min, 72°C 1 min; and a final extension of 72°C for 7 min.
Confirmation of Plasmid Transfer by S1 Nuclease-Pulsed-Field Gel Electrophoresis
Two transconjugants that were PCR-positive for optrA and donor and recipient controls were analyzed by S1 nuclease-pulsed-field gel electrophoresis (PFGE) as previously described,4 with the following modifications for S1 nuclease digestion. Agarose plug slices were cut approximately 1.5 mm wide and digested in a reaction mix consisting of 134 μl molecular grade water, 15 μl l0X restriction buffer (Takara Bio, Mountain View, CA, United States), and 20U S1 nuclease (Takara Bio, Mountain View, CA, United States). Samples were incubated for 30 min at 37°C, then subjected to contour-clamped homogenous electric field (CHEF) PFGE using the CHEF XA system (Bio-Rad Laboratories, Hercules, CA). The MidRange PFG Marker (New England Biolabs, Inc., Ipswich, MA, United States) was included as a size marker. Electrophoresis was carried out at 6 V/cm, 14°C for 20 h, 120° included angle, with switch times of 1 and 25 s. The gel was stained in reverse osmosis water containing GelRed® (Biotium, Fremont, CA, United States) at 1:10,000 and photographed using a UV photoillumination system (Analytik Jena, Jena, Germany).
Results
Antimicrobial Susceptibility of Daptomycin- or Linezolid-Resistant Isolates
During the 2013–2016 isolate collection period, 708 enterococci, identified as E. faecalis (n = 364) or E. faecium (n = 344) by MALDI-TOF, were tested using BMD AST. Nineteen isolates, (2/364, 0.5% E. faecalis and 17/344, 4.9%, E. faecium) were DAP-R, with MICs ranging from 8 to > 16 μg/ml (Table 1). Three isolates, 1/364 (0.3%) E. faecalis and 2/344 (0.6%) E. faecium, were LZD-R, with MICs ranging from 8 to 16 μg/ml, including one E. faecium isolate also resistant to DAP and VAN (Table 2). All three E. faecalis isolates included in this study were VAN-S, while 10/18 E. faecium isolates were VAN-R and eight were VAN-S (Tables 1, 2 and Supplementary Table 1). The full antimicrobial susceptibility profiles for all isolates and antimicrobials tested are summarized in Supplementary Table 1.
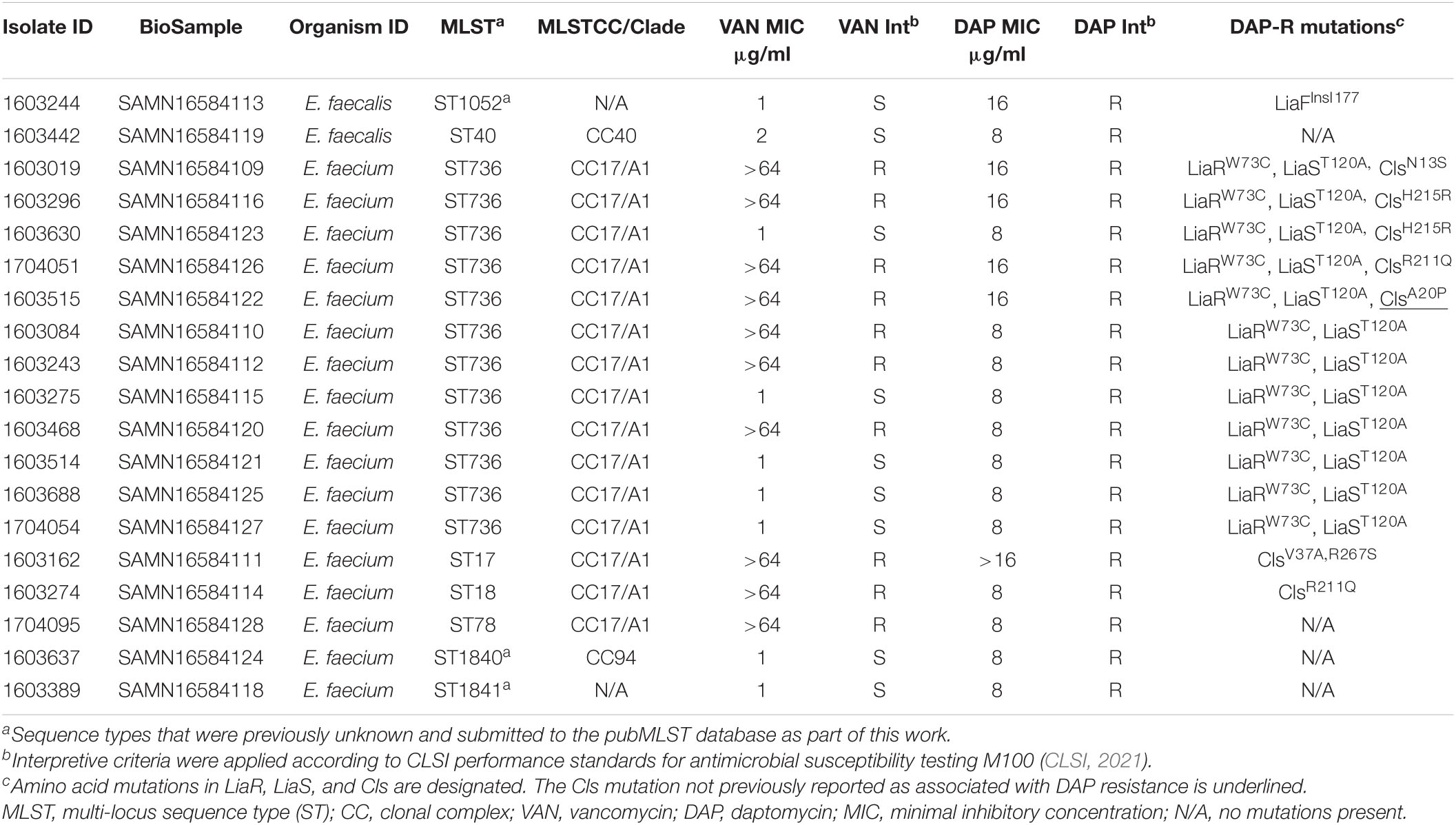
Table 1. Characteristics of E. faecalis and E. faecium CDC Sentinel Surveillance isolates with daptomycin resistance.
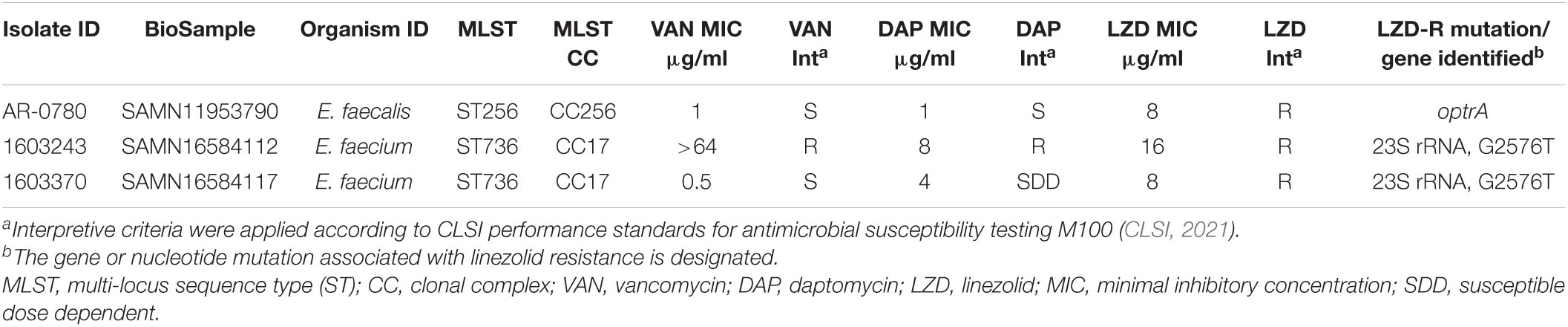
Table 2. Characteristics of E. faecalis and E. faecium CDC Sentinel Surveillance isolates with linezolid resistance.
Whole Genome Sequence Analysis of Daptomycin-Resistant Enterococci
Whole genome sequencing was performed on all isolates and the resulting assemblies were used to determine MLSTs. Of the two DAP-R E. faecalis isolates, one had an unknown ST. The novel profile was submitted to the pubMLST database5 and assigned ST1052 but did not have an associated CC (Table 1). This isolate contained an amino acid insertion, LiaFIns I177, previously associated with DAP-R (Miller et al., 2013). The other DAP-R E. faecalis isolate, 1603442, was ST40, CC40 (Mikalsen et al., 2015), and did not contain mutations previously associated with DAP-R in liaF, liaS liaR, liaX, cls, or gdpD.
Of the 17 DAP-R E. faecium isolates, two had unknown STs and the novel profiles were submitted to the pubMLST database6 and assigned ST1840, which belonged to CC94, and ST1841, which did not have an associated CC (Table 1). Twelve of 17 (71%) E. faecium isolates were ST736 (CC17/Clade A1) (Wang et al., 2018; Top et al., 2020). The remaining three E. faecium isolates were ST17, ST18, and ST78, which also belong to CC17/Clade A1.
All 12 ST736 E. faecium isolates contained the LiaRW73C and LiaST120A amino acid mutations (Table 1) previously associated with DAP-R ST736 isolates (Wang et al., 2018). Five ST736 isolates also contained mutations in Cls, including those previously associated with DAP-R, ClsN13S (1603019), ClsH215R (1603296 and 1603630), and ClsR211Q (1704051) (Wang et al., 2018; Prater et al., 2021). ST736 isolate 1603515 contained an alanine to proline substitution in Cls, ClsA20P, not previously associated with DAP-R. Seven of the 12 (58%) DAP-R ST736 isolates were also VAN-resistant, and all contained vanA. Whole genome MLST was performed to assess the diversity of ST736 E. faecium isolates with respect to state collected, AST and DAP-R mutation profile, and the resulting phylogeny is provided in Figure 1. DAP-R ST736 isolates were collected from WA (n = 5), MD (n = 3), NY (n = 2), IA (n = 1), and PA (n = 1).
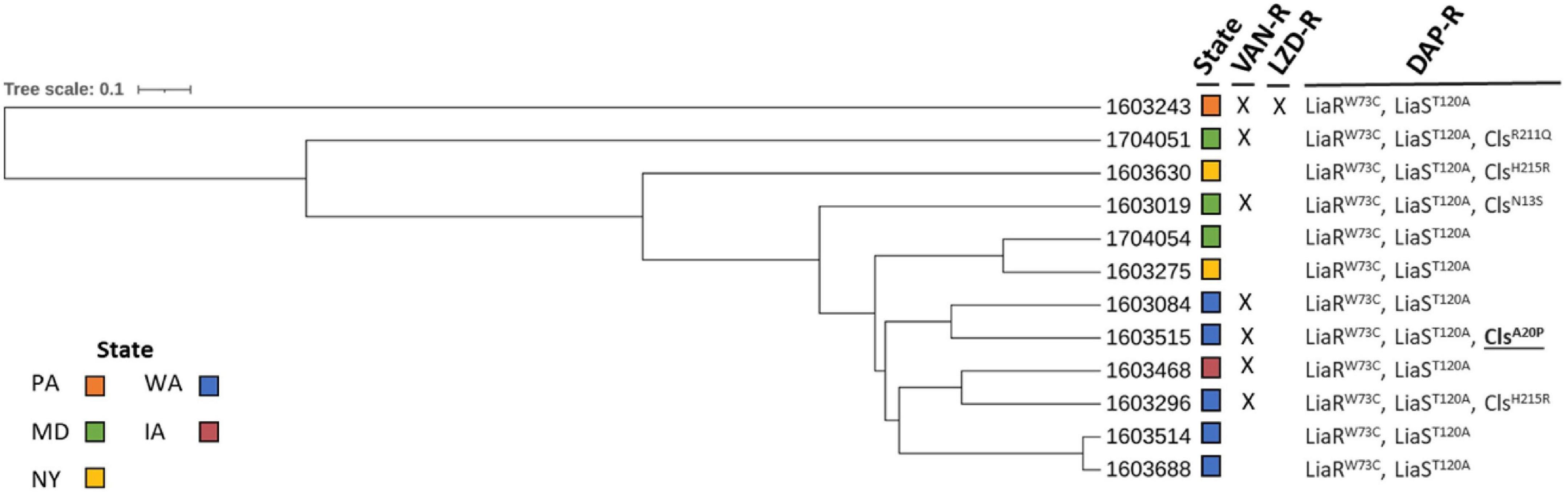
Figure 1. Whole genome multi-locus sequence typing of daptomycin-resistant ST736 E. faecium isolates. Vancomycin-resistant (VAN-R), linezolid-resistant (LZD-R), and daptomycin-resistant (DAP-R) profiles are indicated (X: resistant). Amino acid mutations in LiaR, LiaS, and Cls are designated (Cls mutation not previously reported as associated with DAP resistance is underlined and in bold). States from which the isolates were collected are indicated (colored boxes).
The five non-ST736 E. faecium DAP-R isolates did not contain the LiaRW73C and LiaST120A mutations seen in all DAP-R ST736 isolates. Isolate 1603274 (ST18, CC17), contained the ClsR211Q seen in 1704051 (ST736) that was previously associated with DAP-R (Prater et al., 2021). Isolate 1603162 (ST17) contained two mutations in Cls (ClsV37A, R267S) previously associated with DAP-R (Palmer et al., 2011; Prater et al., 2021), but did not contain mutations in LiaF, LiaS, or LiaR. The remaining three E. faecium DAP-R isolates (1603637/ST1840, 1603389/ST1841, and 1704095/ST78) did not contain mutations previously associated with DAP-R in the genes analyzed.
Whole Genome Sequence Analysis of Linezolid-Resistant Enterococci
Whole genome sequencing data were assessed to identify the presence of genes and mutations known to be associated with LZD resistance, including screening for the presence of cfr and cfr variants, optrA, poxtA, and mutations in the 23S rRNA gene. The LZD-R ST736 E. faecium isolates, 1603243 (DAP-R and VAN-R) and 1603370 (DAP-susceptible dose dependent, and VAN-S) contained the 23S rRNA G2576T nucleotide mutation previously associated with resistance (Table 2; Prystowsky et al., 2001; Beukers et al., 2018). The only LZD-R E. faecalis isolate identified, AR-0780, was ST256 (CC256) (Wang et al., 2018) and contained optrA (Table 2). To analyze the genetic environment of optrA, AR-0780 was sequenced using both short-read (MiSeq) and long-read (PacBio) technologies to create an annotated hybrid assembly. The complete genome consisted of a 2.7 Mb circular chromosome and a single 65,096 bp plasmid, designated pAR-0780. There were two distinct copies of optrA identified, one on the chromosome and one on the plasmid. The optrA encoded on the chromosome contained two nucleotide variants, G1779A (a synonymous mutation at position K595K) and C1833T (a synonymous mutation at position I611I), that correspond to the optrA variant designated as optrA_5 in the Center for Genomic Epidemiology’s database (see text footnote 2, accessed January 2021) (Hasman et al., 2019). An NCBI BLASTN search using the chromosomal optrA region of isolate AR-0780 as a query sequence identified a 12,933 bp region with 100% query coverage and ≥ 99.8% nucleotide sequence identity with other chromosomal optrA gene clusters in GenBank, including E. faecalis strains A101 (MH018572.1), TZ2 (MH225421.1), C25 (MK251150.1), and E1731 (MK737778), which were recently characterized and designated Tn6674, a novel optrA-carrying multiresistance transposon of the Tn554 family/Group I (Li et al., 2019; Freitas et al., 2020). This transposon carries the optrA gene, as well as the spectinomycin resistance gene spc, the chloramphenicol and florfenicol resistance gene fexA, and the macrolide-lincosamide-streptogramin B resistance gene erm(A) (Figure 2). The Tn6674 transposon was inserted into the radC gene, which is a common integration site for Tn554-family transposons (Li et al., 2019; Freitas et al., 2020). The boundaries of AR-0780’s Tn6674 were defined by hexanucleotide sequences at the left (5′-AATCCG-3′) and right (5′-GATATA-3′) junction, which were similar to junction sequences of other Tn554-family transposons (Li et al., 2019).
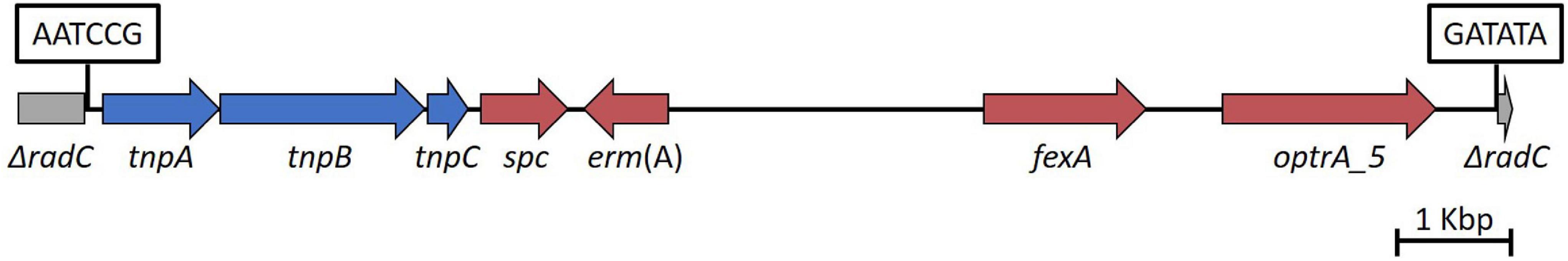
Figure 2. Diagram of AR-0780’s chromosomal transposon, Tn6674 (12,933 bp). optrA along with the resistance genes spc, erm(A), and fexA, are shown in red. Transposase genes, tnpA, tnpB, and tnpC, are shown in blue. The left and right hexanucleotide sequences marking the left and right junction of the Tn6674 radC integration site are shown in boxes. The disrupted radC (ΔradC) is shown in gray.
The second copy of optrA, encoded on pAR-0780 was unique from the chromosomally encoded optrA_5, and contained one mutation at nucleotide position A1441C (non-synonymous mutation, T481P). The position of this mutation has been described before (Freitas et al., 2020) in optrA alleles, but only in isolates containing additional optrA mutations; therefore, no variant designation was available in the Center for Genomic Epidemiology’s database (Hasman et al., 2019). BLASTN analysis of the regions flanking the plasmid-encoded optrA revealed pAR-0780 contained a near identical (100% query coverage, 99.95% nucleotide sequence identity) 14,349 bp optrA gene cluster flanked on the left- and right-hand sides by IS1216 elements in the same orientation previously described on a fragment of an optrA-encoding plasmid p10-2-2 (KT862775.1) from an E. faecalis isolate (ST59) recovered from a pig in China (He et al., 2016; Figure 3). The optrA gene encoded on p10-2-2 contains two nucleotide variants: the A1441C (non-synonymous mutation, T481P) variant seen in pAR-0780 and the T526G (non-synonymous mutation, D176Y), which correspond to optrA_7 in the Center for Genomic Epidemiology’s database. The pAR-0780 plasmid contained the impB-fexA-optrA backbone that is common in Group II optrA positive isolates as described by Freitas et al. (2020), with impB predicted to encode a type VI secretion protein. The pAR-0780 optrA region also contained the erm(A) resistance gene. The remaining pAR-0780 sequence was nearly identical (100% query coverage, 99.88% nucleotide sequence identity) to a 51,543 bp region of the 84,468 bp plasmid, pE508, a pheromone-responsive conjugative multiresistance plasmid recently characterized from E. faecalis strain E508 (ST256) (Figure 3). The pE508 plasmid was isolated from a swine fecal sample in China in 2015 and, in addition to optrA and fexA, is predicted to encode the aminoglycoside resistance gene aac(A)-aph(D) locus, and the tetracycline resistance genes tet(L) and tet(O/W/32/O) (Shang et al., 2019). Plasmid typing of pAR-0780 revealed a copy of repA2 within the pAR-0780-pE508 common region that shared 99.8% nucleotide sequence identity with repA2 from E. faecalis pTEF2 (accession AE016831) and belonged to the rep9 plasmid family (Jensen et al., 2010; Zou et al., 2020). The pAR-0780-pE508 common region also contained the sex pheromone gene cluster, including prgY/traB, prgZ/traC, prgR, prgA/sea1, prgB/asc10, prgU, prgC, and genes associated with conjugal transfer, e.g., a conjugal transfer protein and type IV secretion system proteins (Shang et al., 2019; Zou et al., 2020). The complete pAR-0780 plasmid hybrid assembly (PacBio and Illumina) was deposited in GenBank under accession CP063981.
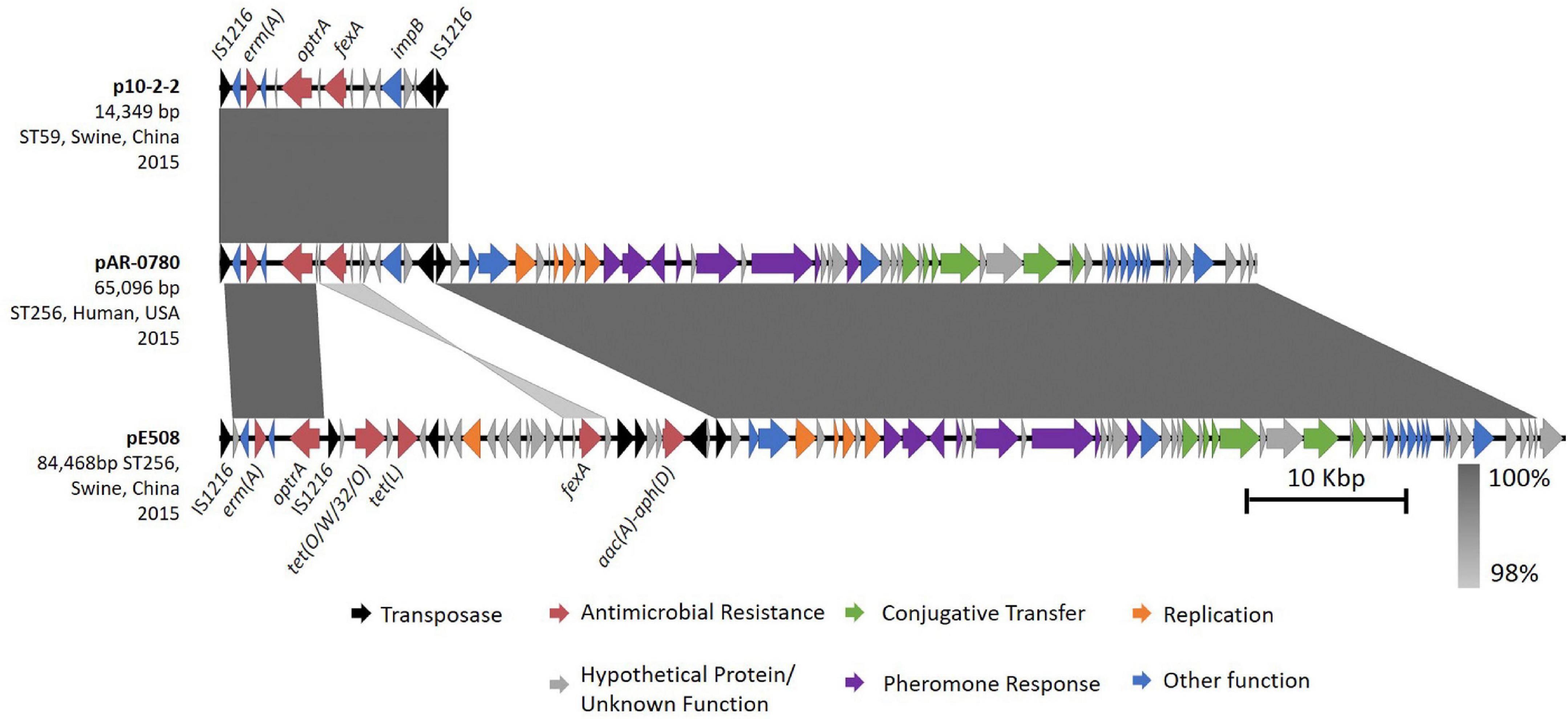
Figure 3. Comparative analysis of pAR-0780 with similar optrA-encoding plasmids. pAR-0780 contained a near identical copy (100% query coverage, 99.95% identity) of a 14,349 bp optrA gene cluster flanked on the left- and right-hand sides by IS1216R elements (black) previously described on plasmid p10-2-2 from an E. faecalis isolate (ST59) recovered from a pig in China in 2015. The remaining 51,543 bp of pAR-0780 was nearly identical (100% coverage, 99.88% identity) to a 51,543 region of the 84,468 bp plasmid, pE508, a pheromone-responsive conjugative multiresistance plasmid from E. faecalis strain E508 (ST256), isolated from a swine fecal sample in 2015.
Transferability of the optrA-Encoding Plasmid, pAR-0780
To assess the transferability of pAR-0780, filter mating experiments were performed using E. faecalis AR-0780 (LZD-R, MIC 8 μg/ml) as the donor and E. faecalis JH2-2 (fusidic acid-resistant and LZD-S, MIC 2 μg/ml) as the recipient. Selection of transconjugants that were LZD- and fusidic acid-resistant revealed pAR-0780 had a conjugation frequency of 1.1 × 10–1 (transconjugants per donor). Two transconjugants (LZD-R, 8 μg/ml and fusidic acid-resistant) were selected for confirmation of optrA transfer. Transconjugant 1 and transconjugant 2 were PCR-positive for the ∼1.4 kb optrA gene product (Supplementary Figure 1A). S1 nuclease-PFGE was used to linearize and size the plasmid content of the donor, recipient, and transconjugants (Supplementary Figure 1B). The recipient, JH2-2 contained no plasmids, but an ∼65 kb band corresponding to the pAR-0780 plasmid was present in both donor and recipient samples, indicating plasmid transfer.
Discussion
Here we characterize DAP-R and LZD-R isolates from CDC’s Sentinel Surveillance collection of 708 enterococci collected during 2013–2016. While the number of DAP-R or LZD-R isolates was low, the presence of the emerging E. faecium ST736 associated with DAP-R, and the acquired LZD mechanism of resistance, optrA, with the propensity to efficiently spread via horizontal gene transfer raises concerns.
The rate of DAP-R E. faecalis was 0.5% and the rate of E. faecium was 4.9%. Resistance to DAP can occur through multiple pathways (Diaz et al., 2014; Wang et al., 2018; Prater et al., 2021), and all mechanisms have not been fully elucidated. While primary mutations in the LiaFSR system and subsequent mutations in the phospholipid metabolism enzyme, Cls, were identified for the majority of DAP-R isolates (15/19, 79%) in this collection, mutations previously associated with DAP-R were not identified in four isolates (one E. faecalis, ST40 and three E. faecium, ST78, ST1840, and ST1841). One novel mutation was identified in isolate 1603515, which contained an alanine to proline substitution in Cls (ClsA20P). While the contribution of this mutation to DAP-R is unknown, an alanine to aspartic acid substitution in Cls at that same position (ClsA20D) was previously characterized in DAP-R isolates (Wang et al., 2018).
Limited data are available on the clonal distribution of DAP-R enterococcal clinical isolates in the United States (Wang et al., 2014). Of the 19 DAP-R isolates (two E. faecalis and 17 E. faecium), WGS revealed the majority (71%, 12/17) of DAP-R E. faecium isolates belonged to the emerging clone, ST736. Daptomycin-resistant and VAN-resistant ST736 E. faecium were first identified in a hospital in New York (Wang et al., 2014), expanded throughout New York, and have been reported in Washington, Texas, and Maryland, as well as Canada, the Caribbean, Germany, and South America (Wang et al., 2018).
In the present collection of ST736 isolates, 7/12 (58%) were also VAN-R, with all isolates harboring vanA. These ST736 strains of E. faecium contain mutations in the LiaFSR system and sometimes subsequent mutations in the related phospholipid metabolism enzymes that predispose them to DAP resistance. E. faecium ST736 isolates are believed to share a unique genetic background that predisposes them to DAP-R and dissemination in the healthcare setting (Wang et al., 2018). Their growing geographic distribution in the United States and internationally has raised concerns about the global dissemination of this DAP-R clone (Wang et al., 2018). The DAP-R ST736 isolates characterized in this study were isolated from hospitalized patients from Iowa, Maryland, New York, Pennsylvania, and Washington. This is consistent with reports that ST736 is expanding in the United States.
All DAP-R ST736 isolates contained mutations in the LiaFSR system, LiaRW73C and LiaST120A, previously described in ST736 isolates, while a subset (5/12, 42%) contained subsequent mutations in Cls. Prior exposure to DAP while patients were hospitalized has been associated with acquisition of these subsequent mutations in Cls, along with higher DAP MICs than isolates with mutations in LiaFSR alone (Wang et al., 2018). A limitation of this work is the lack of exposure or treatment data to determine if this association exists for these surveillance isolates. The majority of ST736 DAP-R isolates with Cls mutations, 4/5 (80%), showed higher MICs (16 μg/ml) than those with LiaFSR mutations alone, which all displayed MICs of 8 μg/ml. All remaining DAP-R non-ST736 isolates had MICs of 8 μg/ml. One ST736 isolate, 1603370, that was selected for inclusion in this study because it was LZD-R, was found to contain the same LiaFSR mutations (LiaRW73C, LiaST120A) seen in all ST736 isolates and was DAP-susceptible dose dependent (MIC of 4 μg/ml) according to CLSI interpretive guidelines (CLSI, 2021). Previous studies have also reported DAP-susceptible isolates with either LiaFSR or Cls mutations that alone are not sufficient to confer resistance to DAP, while the combination of mutations in both LiaFSR and Cls have been sufficient to confer DAP-R (Wang et al., 2018). A limitation of this study is that repeat or alternate antimicrobial susceptibility testing methods, e.g., gradient diffusion strips, were not performed and isolates near the MIC breakpoint for resistance, but without known DAP-R genotypes, could potentially be susceptible upon repeat testing or testing with an alternate method. In addition, only isolates determined to be resistant to DAP or LZD were sequenced; therefore, the number of DAP-susceptible E. faecium isolates from this collection that contain mutations in either LiaFSR or Cls that may predispose them to becoming resistant upon exposure to DAP is unknown.
Linezolid resistance was rare among the 2013–2016 Sentinel isolate collection, with only a single LZD-R E. faecalis isolate and two LZD-R E. faecium isolates identified. A recent survey of clinical E. faecium from the United States determined 30 LZD-R isolates harbored the G2576T 23S rRNA nucleotide mutation, instead of other acquired genes associated with LZD-R, such as optrA, cfr, or poxtA (Wardenburg et al., 2019). Both E. faecium isolates contained the common mutation associated with LZD resistance, the G2576T 23S rRNA mutation (Hasman et al., 2019). In addition, these E. faecium isolates belonged to the emerging healthcare-associated ST736 clone described above and contained the LiaRW73C and LiaST120A mutations associated with DAP-R, including isolate 1603370 that was DAP-susceptible dose dependent, and 1603243, which is the only isolate in this study resistant to DAP, LZD, and VAN.
Long-read sequencing revealed the LZD-R E. faecalis isolate contained two copies of the ABC-F family ribosomal protection protein, optrA, one on the chromosome and one on an approximately 65 kb plasmid. Worldwide, there has been rapid emergence of LZD-R clinical and agricultural isolates carrying optrA since it was first report in 2015; however, the prevalence of optrA in the United States has remained low (Pfaller et al., 2017; Deshpande et al., 2018), which was consistent with this collection, where only a single optrA-harboring E. faecalis isolate was identified. To the best of our knowledge, all reports of optrA isolates collected in the United States have been plasmid-encoded (Pfaller et al., 2017; Deshpande et al., 2018; Tyson et al., 2018). Isolates containing two copies of optrA appear to be rare but have been described. For example, E. faecalis strain C25 isolated from a pig in China between 2016 and 2017, harbored chromosomal- and plasmid-encoded copies of optrA. Here we characterize the first U.S. clinical isolate, with both a chromosomal- and plasmid-encoded optrA. This VAN-S isolate was submitted to the CDC and FDA Antibiotic Resistance Isolate Bank, designated AR-0780, and is publicly available as part of the ‘‘Isolates with New or Novel Antibiotic Resistance’’ panel7.
The chromosomal copy of optrA is encoded on a recently characterized non-conjugative multiresistance Tn554-family transposon designated Tn6674 and first characterized in the E. faecalis strain E1731, which was isolated in 2018 from a swine fecal sample in China. Subsequent analysis of publicly available sequence data by Freitas et al. (2020) revealed Tn6674 has likely driven the spread of optrA among hospitalized patients (with examples from France, China, and Greece), healthy humans (with examples from China and Malaysia), and food-producing animals (with examples from China, Malaysia, and Tunisia) throughout 2012–2018. To the best of our knowledge this represents the first description of a clinical isolate carrying the chromosomal Tn6674 transposon in the United States (Li et al., 2019; Freitas et al., 2020). While Tn6674 was found to be non-conjugative, Li et al. (2019) detected circular intermediate forms of the transposon in E. faecalis strain E1731, indicating the transposon was active in E1731 and providing a potential mechanism for the dissemination of chromosomally-encoded optrA in enterococci. Tn6674 not only encodes optrA, conferring resistance to oxazolidinones like LZD that are used to treat human enterococcal infections, but also encodes fexA, which confers resistance to phenicols that are used in food-producing animals (Li et al., 2019), presenting the concerning possibility of co-selection, maintenance, and spread of both resistance factors.
The plasmid encoding optrA, pAR-0780, also contained the antimicrobial resistance genes erm(A) and the impB-fexA-optrA segment, which was originally described in the first identified optrA plasmid, pE349, and various other plasmids identified in human and pig strains in China (He et al., 2016; Freitas et al., 2020). The pAR-0780 optrA region was > 99.9% identical to the optrA gene cluster previously described on plasmid p10-2-2 from an E. faecalis isolated from a pig in China (He et al., 2016). Additional plasmid-encoded optrA regions with similar gene arrangements and sequence identities have also been identified on plasmids from clinical isolates, including 739884 (China) and 912300 (United States), although the complete plasmid sequences for these isolates were not available (Deshpande et al., 2018). Similar to AR-0780’s chromosomal optrA region, the co-localization of optrA on pAR-0780 with other resistance genes, supports the transfer, co-selection, and persistence of optrA on both the chromosome and plasmid in this isolate when under selective pressure of phenicols and oxazolidinones. This again highlights the problem of antibiotic use selecting for resistance against multiple classes of antibiotics. Use of chloramphenicol may select for resistance not only to chloramphenicol but also to LZD.
The backbone of pAR-0780 was nearly identical to pE508, a conjugative multiresistance plasmid recently characterized from E. faecalis strain E508 (Shang et al., 2019). This shared region contained genes required for a highly efficient conjugation process referred to as the pheromone-responsive plasmid (PRP) transfer system. These PRPs are unique to enterococci and have transfer rates that reach or exceed 10–1 transconjugants per donor (or one transconjugant per 10 donor cells) in vivo and under laboratory conditions (Hirt et al., 2005; Sterling et al., 2020). pAR-0780 belonged to the rep9-type plasmid family, which have been characterized as sex-pheromone or PRPs (Jensen et al., 2010; Zou et al., 2020), and these plasmids have been recently associated with dissemination of optrA in clinical E. faecalis isolates in China (Shang et al., 2019; Zou et al., 2020). pAR-0780 had a high conjugation frequency of 1.1 × 10–1 (transconjugants per donor), which was 10.5% higher than the transfer frequency of 3.7 × 10–2 reported for pE508 (Shang et al., 2019).
pAR-0780 is a conjugative pheromone-responsive mosaic plasmid with high sequence similarity to plasmids belonging to animal isolates from diverse locations, suggesting pAR-0780 is the result of a horizontal gene transfer and/or a recombination event, potentially involving an animal reservoir. The use of long-read sequencing was critical in resolving the genetic landscape of AR-0780’s multiple copies of optrA and deciphering that the two copies were different optrA alleles. The chromosomal optrA on the Tn6674 transposon and the copy of optrA encoded on the conjugative PRP were likely acquired during independent genetic events.
AR-0780 was isolated from a patient in the United States in 2015, the same year optrA was first reported (Wang et al., 2015). There are limited data available on the prevalence of LZD-R optrA- positive isolates in the United States, but work presented here, as well as other surveillance systems targeting clinical and animal isolates (Pfaller et al., 2017; Deshpande et al., 2018; Tyson et al., 2018) suggest optrA in the United States is rare. However, reports of rapid global dissemination of optrA-carrying enterococci, beyond the Asia-Pacific region (Bender et al., 2018; Deshpande et al., 2018; Sassi et al., 2019; Egan et al., 2020), as well as their location on active and highly efficient mobile genetic elements, presents the possibility that the prevalence of optrA in the United States is underestimated and national surveillance efforts to better understand optrA prevalence are needed.
Linezolid and daptomycin are important antibiotics for the treatment of infections caused by VRE. Resistance to these drugs was rare among the CDC’s 2013–2016 Sentinel Surveillance collection. This work found that a single DAP-R E. faecium clone, ST736, may be driving DAP-R resistance. We also identified a LZD-R isolate with optrA encoded not only on a chromosomal transposon, but also on a highly transmissible plasmid. This work highlights the importance of continued surveillance to monitor the emergence and spread of DAP-R and LZD-R clones and antimicrobial resistance mechanisms in the United States.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: WGS short read data for all isolates in this study were submitted to the NCBI Sequence Read Archive (SRA) with the BioSample numbers SAMN16584109–SAMN16584128. The complete chromosome hybrid assembly (PacBio and Illumina) for AR-0780 was deposited in GenBank under BioProject accession number PRJNA523425, and short read data were submitted to the NCBI SRA under BioSample number SAMN11953790.
Author Contributions
ASG, MK, and JDL conceived the project. ASG, JDL, LMS, WZ, and DC contributed to the design of experiments. LMS, WZ, DC, GM, TOE, VA, VAS, MS, and JP collected the data and performed the experiments. ASG, AGK, DC, and DB performed sequence analysis. ASG, AGK, LMS, and DC created the tables and figures. ASG led development of the manuscript. All authors contributed to drafting, revising, and approving the final submission.
Funding
This work was made possible, in part, through the CDC’s Safety and Healthcare Epidemiology Prevention Research Development (SHEPheRD) Program funding awarded to JKJ (contract 200-2011-42064).
Conflict of Interest
LMS, TOE, and AGK were employed by the company Goldbelt C6, LLC. JP was employed by the company ASRT Incorporated.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention/The Agency for Toxic Substances and Disease Registry. Use of trade names is for identification only and does not imply endorsement by the U.S. Centers for Disease Control and Prevention, the Agency for Toxic Substances and Disease Registry, the Public Health Service, or the U.S. Department of Health and Human Services. We wish to thank Brandi M. Limbago, Bonnie Hatcher, and Christine Lascols for their contributions and support of CDC’s Sentinel Surveillance Program. We thank Paige Gable for valuable discussions, and Richard Stanton and Nick Vlachos for bioinformatics support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.807398/full#supplementary-material
Footnotes
- ^ https://www.applied-maths.com/
- ^ https://cge.cbs.dtu.dk/services/LRE-finder/
- ^ https://digitalinsights.qiagen.com
- ^ https://www.cdc.gov/hai/pdfs/labsettings/unified_pfge_protocol.pdf
- ^ https://pubmlst.org/organisms/enterococcus-faecalis
- ^ https://pubmlst.org/bigsdb?db=pubmlst_efaecium_isolates
- ^ https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=10
References
Antonelli, A., D’Andrea, M. M., Brenciani, A., Galeotti, C. L., Morroni, G., Pollini, S., et al. (2018). Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Arias, C. A., Panesso, D., Mcgrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138
Bender, J. K., Fleige, C., Klare, I., Fiedler, S., Mischnik, A., Mutters, N. T., et al. (2016). Detection of a cfr(B) variant in German Enterococcus faecium clinical isolates and the impact on linezolid resistance in Enterococcus spp. PLoS One 11:e0167042. doi: 10.1371/journal.pone.0167042
Bender, J. K., Fleige, C., Lange, D., Klare, I., and Werner, G. (2018). Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 52, 819–827. doi: 10.1016/j.ijantimicag.2018.09.009
Beukers, A. G., Hasman, H., Hegstad, K., and Van Hal, S. J. (2018). Recommendations to address the difficulties encountered when determining linezolid resistance from whole-genome sequencing data. Antimicrob. Agents Chemother. 62:e00613-18. doi: 10.1128/AAC.00613-18
Bi, R., Qin, T., Fan, W., Ma, P., and Gu, B. (2018). The emerging problem of linezolid-resistant enterococci. J. Glob. Antimicrob. Resist. 13, 11–19. doi: 10.1016/j.jgar.2017.10.018
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
CLSI (2018). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. CLSI Standard M07, 11th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2021). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100, 31st Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Conrad, S., Oethinger, M., Kaifel, K., Klotz, G., Marre, R., and Kern, W. V. (1996). gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 38, 443–455. doi: 10.1093/jac/38.3.443
Deshpande, L. M., Ashcraft, D. S., Kahn, H. P., Pankey, G., Jones, R. N., Farrell, D. J., et al. (2015). Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as part of the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 59, 6256–6261. doi: 10.1128/aac.01473-15
Deshpande, L. M., Castanheira, M., Flamm, R. K., and Mendes, R. E. (2018). Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 73, 2314–2322. doi: 10.1093/jac/dky188
Diaz, L., Tran, T. T., Munita, J. M., Miller, W. R., Rincon, S., Carvajal, L. P., et al. (2014). Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob. Agents Chemother. 58, 4527–4534. doi: 10.1128/AAC.02686-14
Egan, S. A., Shore, A. C., O’Connell, B., Brennan, G. I., and Coleman, D. C. (2020). Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 75, 1704–1711. doi: 10.1093/jac/dkaa075
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Using the NCBI AMRFinder tool to determine antimicrobial resistance genotype-phenotype correlations within a collection of NARMS isolates. bioRxiv [Preprint]. doi: 10.1101/550707
Freitas, A. R., Tedim, A. P., Novais, C., Lanza, V. F., and Peixe, L. (2020). Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 6:e000350. doi: 10.1099/mgen.0.000350
Garcia-Solache, M., and Rice, L. B. (2019). The Enterococcus: a model of adaptability to its environment. Clin. Microbiol. Rev. 32:e000 58-18.
Guerin, F., Sassi, M., Dejoies, L., Zouari, A., Schutz, S., Potrel, S., et al. (2020). Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J. Antimicrob. Chemother. 75, 1699–1703. doi: 10.1093/jac/dkaa125
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Lopez-Rojas, R., Kempf, M., Landraud, L., et al. (2014). ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. doi: 10.1128/AAC.01310-13
Hasman, H., Clausen, P., Kaya, H., Hansen, F., Knudsen, J. D., Wang, M., et al. (2019). LRE-Finder, a web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in enterococci from whole-genome sequences. J. Antimicrob. Chemother. 74, 1473–1476. doi: 10.1093/jac/dkz092
He, T., Shen, Y., Schwarz, S., Cai, J., Lv, Y., Li, J., et al. (2016). Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 71, 1466–1473. doi: 10.1093/jac/dkw016
Hirt, H., Manias, D. A., Bryan, E. M., Klein, J. R., Marklund, J. K., Staddon, J. H., et al. (2005). Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J. Bacteriol. 187, 1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005
Homan, W. L., Tribe, D., Poznanski, S., Li, M., Hogg, G., Spalburg, E., et al. (2002). Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40, 1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002
Hunt, M., Silva, N. D., Otto, T. D., Parkhill, J., Keane, J. A., and Harris, S. R. (2015). Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. doi: 10.1186/s13059-015-0849-0
Inouye, M., Dashnow, H., Raven, L. A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6:90. doi: 10.1186/s13073-014-0090-6
Jensen, L. B., Garcia-Migura, L., Valenzuela, A. J., Lohr, M., Hasman, H., and Aarestrup, F. M. (2010). A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 80, 25–43. doi: 10.1016/j.mimet.2009.10.012
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Khan, A., Davlieva, M., Panesso, D., Rincon, S., Miller, W. R., Diaz, L., et al. (2019). Antimicrobial sensing coupled with cell membrane remodeling mediates antibiotic resistance and virulence in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 116, 26925–26932. doi: 10.1073/pnas.1916037116
Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., Mclellan, M. D., Lin, L., et al. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576. doi: 10.1101/gr.129684.111
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, D., Li, X. Y., Schwarz, S., Yang, M., Zhang, S. M., Hao, W., et al. (2019). Tn6674 is a novel enterococcal optrA-carrying multiresistance transposon of the Tn554 family. Antimicrob. Agents Chemother. 63:e00809-19.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, Y., Wang, Y., Schwarz, S., Wang, S., Chen, L., Wu, C., et al. (2014). Investigation of a multiresistance gene cfr that fails to mediate resistance to phenicols and oxazolidinones in Enterococcus faecalis. J. Antimicrob. Chemother. 69, 892–898. doi: 10.1093/jac/dkt459
Liu, Y., Wang, Y., Wu, C., Shen, Z., Schwarz, S., Du, X. D., et al. (2012). First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 56, 1650–1654. doi: 10.1128/AAC.06091-11
Mikalsen, T., Pedersen, T., Willems, R., Coque, T. M., Werner, G., Sadowy, E., et al. (2015). Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genomics 16:282. doi: 10.1186/s12864-015-1407-6
Miller, C., Kong, J., Tran, T. T., Arias, C. A., Saxer, G., and Shamoo, Y. (2013). Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob. Agents Chemother. 57, 5373–5383. doi: 10.1128/AAC.01473-13
Miller, W. R., Murray, B. E., Rice, L. B., and Arias, C. A. (2020). Resistance in vancomycin-resistant enterococci. Infect. Dis. Clin. North Am. 3, 751–771.
Miller, W. R., Tran, T. T., Diaz, L., Rios, R., Khan, A., Reyes, J., et al. (2019). LiaR-independent pathways to daptomycin resistance in Enterococcus faecalis reveal a multilayer defense against cell envelope antibiotics. Mol. Microbiol. 111, 811–824. doi: 10.1111/mmi.14193
Ota, Y., Furuhashi, K., Hayashi, W., Hirai, N., Ishikawa, J., Nagura, O., et al. (2021). Daptomycin resistant Enterococcus faecalis has a mutation in liaX, which encodes a surface protein that inhibits the systems and cell membrane remodeling. J. Infect. Chemother. 27, 90–93. doi: 10.1016/j.jiac.2020.09.004
Palmer, K. L., Daniel, A., Hardy, C., Silverman, J., and Gilmore, M. S. (2011). Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55, 3345–3356. doi: 10.1128/aac.00207-11
Pfaller, M. A., Mendes, R. E., Streit, J. M., Hogan, P. A., and Flamm, R. K. (2017). Five-year summary of in vitro activity and resistance mechanisms of linezolid against clinically important Gram-positive cocci in the United States from the LEADER Surveillance Program (2011 to 2015). Antimicrob. Agents Chemother. 61:e00609-17. doi: 10.1128/AAC.00609-17
Prater, A. G., Mehta, H. H., Beabout, K., Supandy, A., Miller, W. R., Tran, T. T., et al. (2021). Daptomycin resistance in Enterococcus faecium can be delayed by disruption of the LiaFSR stress response pathway. Antimicrob. Agents Chemother. 65:e01317-20. doi: 10.1128/AAC.01317-20
Prater, A. G., Mehta, H. H., Kosgei, A. J., Miller, W. R., Tran, T. T., Arias, C. A., et al. (2019). Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob. Agents Chemother. 63:e00790-19. doi: 10.1128/AAC.00790-19
Prystowsky, J., Siddiqui, F., Chosay, J., Shinabarger, D. L., Millichap, J., Peterson, L. R., et al. (2001). Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45, 2154–2156. doi: 10.1128/AAC.45.7.2154-2156.2001
Ruiz-Garbajosa, P., Bonten, M. J., Robinson, D. A., Top, J., Nallapareddy, S. R., Torres, C., et al. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44, 2220–2228. doi: 10.1128/JCM.02596-05
Sassi, M., Guerin, F., Zouari, A., Beyrouthy, R., Auzou, M., Fines-Guyon, M., et al. (2019). Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006-16. J. Antimicrob. Chemother. 74, 1469–1472. doi: 10.1093/jac/dkz097
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shang, Y., Li, D., Shan, X., Schwarz, S., Zhang, S. M., Chen, Y. X., et al. (2019). Analysis of two pheromone-responsive conjugative multiresistance plasmids carrying the novel mobile optrA locus from Enterococcus faecalis. Infect. Drug Resist. 12, 2355–2362. doi: 10.2147/IDR.S206295
Sharkey, L. K. R., Edwards, T. A., and O’Neill, A. J. (2016). ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 7:e01975. doi: 10.1128/mBio.01975-15
Stanton, R. A., Vlachos, N., and Halpin, A. L. (2021). GAMMA: a tool for the rapid identification, classification and annotation of translated gene matches from sequencing data. Bioinformatics btab607. doi: 10.1093/bioinformatics/btab607
Sterling, A. J., Snelling, W. J., Naughton, P. J., Ternan, N. G., and Dooley, J. S. G. (2020). Competent but complex communication: the phenomena of pheromone-responsive plasmids. PLoS Pathog. 16:e1008310. doi: 10.1371/journal.ppat.1008310
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Top, J., Arredondo-Alonso, S., Schürch, A. C., Puranen, S., Pesonen, M., Pensar, J., et al. (2020). Genomic rearrangements uncovered by genome-wide co-evolution analysis of a major nosocomial pathogen, Enterococcus faecium. Microb. Genom. 6:mgen000488. doi: 10.1099/mgen.0.000488
Tyson, G. H., Sabo, J. L., Hoffmann, M., Hsu, C. H., Mukherjee, S., Hernandez, J., et al. (2018). Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J. Antimicrob. Chemother. 73, 3254–3258. doi: 10.1093/jac/dky369
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, G., Kamalakaran, S., Dhand, A., Huang, W., Ojaimi, C., Zhuge, J., et al. (2014). Identification of a novel clone, ST736, among Enterococcus faecium clinical isolates and its association with daptomycin nonsusceptibility. Antimicrob. Agents Chemother. 58, 4848–4854. doi: 10.1128/AAC.02683-14
Wang, G., Yu, F., Lin, H., Murugesan, K., Huang, W., Hoss, A. G., et al. (2018). Evolution and mutations predisposing to daptomycin resistance in vancomycin-resistant Enterococcus faecium ST736 strains. PLoS One 13:e0209785. doi: 10.1371/journal.pone.0209785
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wardenburg, K. E., Potter, R. F., D’souza, A. W., Hussain, T., Wallace, M. A., Andleeb, S., et al. (2019). Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J. Antimicrob. Chemother. 74, 3445–3452. doi: 10.1093/jac/dkz367
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: Enterococcus faecalis, Enterococcus faecium, daptomycin, linezolid, conjugation, optrA, transmission, pheromone responsive plasmid
Citation: Gargis AS, Spicer LM, Kent AG, Zhu W, Campbell D, McAllister G, Ewing TO, Albrecht V, Stevens VA, Sheth M, Padilla J, Batra D, Johnson JK, Halpin AL, Rasheed JK, Elkins CA, Karlsson M and Lutgring JD (2022) Sentinel Surveillance Reveals Emerging Daptomycin-Resistant ST736 Enterococcus faecium and Multiple Mechanisms of Linezolid Resistance in Enterococci in the United States. Front. Microbiol. 12:807398. doi: 10.3389/fmicb.2021.807398
Received: 02 November 2021; Accepted: 09 December 2021;
Published: 01 February 2022.
Edited by:
David Christopher Coleman, Dublin Dental University Hospital, IrelandReviewed by:
Guido Werner, Robert Koch Institute (RKI), GermanyDeirdre Fitzgerald-Hughes, Royal College of Surgeons in Ireland, Ireland
Copyright © 2022 Gargis, Spicer, Kent, Zhu, Campbell, McAllister, Ewing, Albrecht, Stevens, Sheth, Padilla, Batra, Johnson, Halpin, Rasheed, Elkins, Karlsson and Lutgring. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amy S. Gargis, QUdhcmdpc0BjZGMuZ292
 Amy S. Gargis
Amy S. Gargis Lori M. Spicer1,2
Lori M. Spicer1,2 Alyssa G. Kent
Alyssa G. Kent Davina Campbell
Davina Campbell Jasmine Padilla
Jasmine Padilla Joseph D. Lutgring
Joseph D. Lutgring