
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 December 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.804333
This article is part of the Research Topic Innovative Biocontrol Strategies to Manage Crop and Pest Diseases View all 17 articles
 Men Thi Ngo1,2
Men Thi Ngo1,2 Minh Van Nguyen1,2
Minh Van Nguyen1,2 Jae Woo Han1
Jae Woo Han1 Bomin Kim1,2
Bomin Kim1,2 Yun Kyung Kim1,2
Yun Kyung Kim1,2 Myung Soo Park3
Myung Soo Park3 Hun Kim1,2*
Hun Kim1,2* Gyung Ja Choi1,2*
Gyung Ja Choi1,2*Microbial metabolites have been recognized as an important source for the discovery of new antifungal agents because of their diverse chemical structures with novel modes of action. In the course of our screening for new antifungal agents from microbes, we found that culture filtrates of two fungal species Aspergillus candidus SFC20200425-M11 and Aspergillus montenegroi SFC20200425-M27 have the potentials to reduce the development of fungal plant diseases such as tomato late blight and wheat leaf rust. From these two Aspergillus spp., we isolated a total of seven active compounds, including two new compounds (4 and 6), and identified their chemical structures based on the NMR spectral analyses: sphaeropsidin A (1), (R)-formosusin A (2), (R)-variotin (3), candidusin (4), asperlin (5), montenegrol (6), and protulactone A (7). Based on the results of the in vitro bioassays of 11 plant pathogenic fungi and bacteria, sphaeropsidin A (1), (R)-formosusin A (2), (R)-variotin (3), and asperlin (5) exhibited a wide range of antimicrobial activity. Furthermore, when plants were treated with sphaeropsidin A (1) and (R)-formosusin A (2) at a concentration of 500 μg/ml, sphaeropsidin A (1) exhibited an efficacy disease control value of 96 and 90% compared to non-treated control against tomato late blight and wheat leaf rust, and (R)-formosusin A (2) strongly reduced the development of tomato gray mold by 82%. Asperlin (5) at a concentration of 500 μg/ml effectively controlled the development of tomato late blight and wheat leaf rust with a disease control value of 95%. Given that culture filtrates and active compounds derived from two Aspergillus spp. exhibited disease control efficacies, our results suggest that the Aspergillus-produced antifungal compounds could be useful for the development of new natural fungicides.
The world population is expected to reach up to 9.7 billion people by 2050, postulating that the required agricultural food production increases up to at least 50–110% (Godfray et al., 2010; Raymaekers et al., 2020). Considering that the additional agricultural area is limited, the reduction of crop yield losses caused by plant pathogens has gained the most attention contributing to food security (Mauser et al., 2015; Raymaekers et al., 2020). Although the use of synthetic pesticides has been recognized as one of the most effective methods to control plant diseases, the overuse of chemical pesticides has led to severe problems, such as resistance, toxicity in humans and animals, and environmental pollution (Hüter, 2011). To compensate for the shortcomings of chemical pesticides, the development and use of biological pesticides based on natural resources exhibiting a promising antimicrobial activity have been crucial in agriculture (Dayan et al., 2009; Choi et al., 2010).
To date, there are over 23,000 known microbial secondary metabolites that are significant sources for life-saving drugs, and 42% of which are produced by fungi (Demain, 2014; Chandra et al., 2020). The genus Aspergillus has been recognized as an enormous source of lead compounds with promising diverse structures and biological activities (Sadorn et al., 2016). Notably, from 2015 to 2019, 362 secondary metabolites were isolated from different Aspergillus species, including alkaloids, butenolides, and cytochalasins, which showed diverse biological activities such as antimicrobial, anti-inflammatory, and anticancer activities (El-hawary et al., 2020). Regarding the antimicrobial activity, it has been reported that aspetritones and candidusin derivatives isolated from the culture of Aspergillus tritici SP2-8-1 exhibited antibacterial activity against the methicillin-resistant strain Staphylococcus aureus (Wang et al., 2017). Two new compounds, aspergillethers A and B, from the endophytic fungus Aspergillus versicolor exhibited significant in vitro antibacterial and antifungal activities toward S. aureus, Bacillus cereus, and Candida albicans (Mohamed et al., 2020). Despite efforts to find secondary metabolites showing antimicrobial activity, to date, relatively few Aspergillus species have been considered as biological agents for plant protection (Zhao et al., 2018; El-Sayed and Ali, 2020).
With the production of antimicrobial metabolites, some fungal species have been reported as a potent material for controlling plant diseases. For example, the basidiomycete fungus Crinipellis rhizomaticola culture filtrate and its active compounds crinipellins suppressed the development of rice blast and pepper anthracnose caused by Magnaporthe oryzae and Collectotrichum coccodes, respectively (Han et al., 2018). Jang et al. (2016) also showed that a culture filtrate of Aspergillus niger F22 was highly active against a root-knot nematode Meloidogyne incognita by which a nematicidal component oxalic acid affected the mortality of second-stage juveniles and the inhibition of egg hatching. Later, the strain A. niger F22 was registered as a natural nematicidal agent in the Korean market (Lee et al., 2017).
The marine environment has been investigated for new natural resources containing bioactive compounds with benefits for the health of humans, animals, and plants (Sohn and Oh, 2010). For this endeavor, marine-derived resources have gained much attention in the past decades (Buttachon et al., 2018). In particular, marine-derived fungi have been considered as a rich source of secondary metabolites with promising antimicrobial effects (Neuhaus et al., 2019; Wang et al., 2021), representing unprecedented scaffolds for further drug design for specific modes of action (Xu et al., 2015; Willems et al., 2020). In the current study, our main goals were (1) to find Aspergillus species derived from a marine environment that have in vitro and in vivo antimicrobial properties against plant pathogens and (2) to identify the active metabolites from the selected Aspergillus species. Based on the in vitro antimicrobial activity and plant disease control efficacies of the fungal cultures containing the identified active compounds, our results could provide valuable information to develop new biological control agents for crops.
The eight strains of Aspergillus species used in this study were kindly provided by Dr. Myung Soo Park (Marine Fungal Resource Bank, Seoul National University; Supplementary Table S1). Of these strains, SFC20200425-M11 and SFC20200425-M27 showing a promising plant disease control efficacy were deposited as a patent microorganism to the Korean Agricultural Culture Collection (KACC, Wanju, South Korea).
For the in vitro antifungal activity assay, six plant pathogenic fungi provided by the KACC were used: Alternaria brassicicola (KACC 40036), Botrytis cinerea (KACC 48736), C. coccodes (KACC 48737), Fusarium oxysporum (KACC 40043), M. oryzae (KACC 46552), and Phytophthora infestans (KACC 48738). Additionally, we used two obligate parasitic fungi Puccinia triticina and Blumeria graminis f. sp. hordei, which were maintained on their host plants, for the disease control efficacy assay (Choi et al., 2010; Shin et al., 2017; Han et al., 2018). For the antibacterial activity assay, the five following bacterial species were used: Agrobacterium tumefaciens SL2434, Clavibacter michiganensis SL4135, Pseudomonas syringae SL308, Ralstonia solanacearum SL1944, and Erwinia amylovora TS3128. All bacteria were provided by the National Academy of Agricultural Sciences (Wanju, Korea), except for R. solanacearum provided by Dr. SW Lee of Dong-A University (Vu et al., 2017). The fungi and bacteria were maintained on potato dextrose agar (PDA; BD Difco, Sparks, MD, United States) medium and tryptic soy agar (TSA; BD Difco) medium, respectively, and kept at 4°C before use.
For the isolation of genomic DNA (gDNA), each fungal species was grown in 50 ml of potato dextrose broth (PDB; BD Difco) medium at 25°C for 4 days on a rotary shaker (150 rpm). The gDNA was extracted using the cetyltrimethylammonium bromide (CTAB) procedure as previously described (Han et al., 2018). For phylogenetic analysis, the calmodulin (CaM) gene was amplified by the primer set CF1D (5'–CAGGTCTCCGAGTACAAG–3') and CF4 (5'–CAGGTCTCCGAGTACAAGTTTYTGCATCATRAGYTGGAC–3'; Lee et al., 2016). The resulting amplicon was purified using the GeneAll ExpinTM PCR purification kit (GeneAll, Seoul, South Korea) and then analyzed using corresponding PCR primers by Macrogen (Seoul, Korea). The resulting sequences were analyzed with the BLASTn program of the NCBI.1 The sequences were aligned using ClustalW implemented in MEGA version X, and distances were estimated based on the Tamura-Nei model (Tamura and Nei, 1993). A phylogenetic tree was generated using the neighbor-joining method with 1,000 bootstrap analyses (Saitou and Nei, 1987).
Twenty mycelial disks (8 mm in diameter) of each fungal strain, SFC20200425-M11 and SFC20200425-M27, were inoculated into 400 ml PDB medium in a 2 L-baffled Erlenmeyer flask and incubated on a rotary shaker at 150 rpm and 25°C for 10 days. The culture broths of SFC20200425-M11 (3.6 L) and SFC20200425-M27 (1.6 L) were centrifuged at 10,000 × g for 30 min and then filtered through two layers of Whatman No. 1 filter paper (Maidstone, United Kingdom). The culture filtrates were partitioned with an equal volume of ethyl acetate (EtOAc) and n-butanol (BuOH), sequentially. Each layer was concentrated to dryness by a rotary evaporator (Rotavapor R-300; Büchi, Flawil, Switzerland).
From the culture filtrate of SFC20200425-M11, the EtOAc (370 mg), BuOH (640 mg), and water (6.8 g) extracts were obtained. The EtOAc extract was applied onto a silica gel column (40–63 μm; Merck, Darmstadt, Germany), using an isocratic elution of n-hexane/EtOAc (3:1, v/v) to give five fractions (E1–E5). The E3 and E4 fractions were pure compounds 1 (51 mg) and 2 (35 mg), respectively. Fraction E5 (152 mg) was further separated by preparative thin-layer chromatography (TLC) using Kieselgel 60 F254 glass plates (Merck). The TLC plates were developed with n-hexane/EtOAc (60:40, v/v) to give five fractions (E51–E55). Fraction E55 was a pure compound 4 (13 mg), and fraction E52 was further purified by high-pressure liquid chromatography (HPLC) using the Shimadzu LC-6 AD system (Kyoto, Japan). The Capcell Pak C18 column (20 × 250 mm, 5 μm; Shiseido, Tokyo, Japan) was used for preparative HPLC and eluted with 68% aqueous methanol at a flow rate of 5 ml/min to give compound 3 (2 mg).
Among the obtained EtOAc (338 mg), BuOH (340 mg), and water (3.1 g) extracts from the culture broth of SFC20200425-M27, the EtOAc extract was purified by preparative HPLC. The column was eluted with 30% aqueous methanol to give compound 5 (91.2 mg). Next, the BuOH extract was separated by an Isolera One mid-pressure liquid chromatography (MPLC) system (Biotage, Uppsala, Sweden) equipped with the Biotage SNAP Ultra C18 cartridge (60 g). The column was eluted with a linear gradient of aqueous methanol (2–100%, v/v) to give two fractions (B1 and B2). Fraction B2 was pure compound 7 (21 mg). The fraction B1 (70 mg) was further purified by preparative HPLC. The column was eluted with 8% aqueous methanol to give compound 6 (22 mg). The isolation schemes for all compounds 1–7 are presented in Supplementary Figures S1 and S2.
Chemical structures of the purified compounds were determined by spectroscopic analyses and comparison with previous literature data. High-resolution electrospray ionization mass spectrometry (HRESIMS) data were obtained by the Synapt G2 system (Waters, Milford, MA, United States). The 1D and 2D nuclear magnetic resonance (NMR) spectra were recorded by a Bruker Advance 500 MHz spectrometer (Rheinstetten, Germany) in chloroform-d, methanol-d4, or pyridine-d5 (Cambridge Isotope Laboratories, Andover, MA, United States). Chemical shifts were referenced to the solvent peaks (δH 7.26 and δC 77.2 for chloroform-d; δH 4.87 and δC 49.0 for methanol-d4; δH 8.71 and δC 149.9 for pyridine-d5).
To determine the absolute configuration of the secondary alcohol compound 4, Mosher’s method using α-methoxy-α-trifluoromethylphenylacetic acid (MTPA) esters was performed as previously described by Nguyen et al. (2018). Briefly, the (R/S)-MTPA esters of 4 (4a and 4b) were prepared using Mosher’s esterification method. Compounds 4 (0.5 mg) and 4-(dimethylamino)-pyridine (0.2 mg) were mixed into a 5 ml vial, and then, the mixture was dried in vacuo. Pyridine-d5 (0.5 ml) and (R)-MPTA or (S)-MPTA (Sigma-Aldrich, St Louis, MO, United States; 6.0 μl) were immediately put into the vial, and then the vial was sealed and shaken to mix evenly. The reaction was carried out at room temperature for 12 h. The reactant was transferred into an NMR tube to measure the 1H-NMR and 1H–1H COSY spectra.
The values for the minimum inhibitory concentration (MIC) of the purified compounds 1–7 were determined against plant pathogenic fungi and bacteria by the broth microdilution method using 96-well microtiter plates modified according to previous methods for the testing of potential antimicrobial natural products (Espinel-Ingroff et al., 2005; Vu et al., 2017; Ngo et al., 2019). Briefly, fungal spore suspensions (5 × 104 spores/ml) or bacterial cell suspensions (2 × 105 CFU/ml) were added to each well of a 96-well microtiter plate containing PDB or tryptic soy broth (TSB; BD Difco) medium, respectively. The purified compounds dissolved in methanol were added and then serially two-fold diluted to reach the final concentrations ranging from 0.06 to 250 μg/ml. The microliter plates were incubated for 1–2 days, and the MIC values were determined by visual inspection of complete growth inhibition (Espinel-Ingroff et al., 2005). Blasticidin-S and oxytetracycline were used as positive controls for the antifungal and antibacterial assays, respectively. The medium containing 1% methanol was used as a negative control.
Six plant diseases caused by fungi were used for the plant disease control efficacy assay: rice blast (RCB, caused by M. oryzae), tomato gray mold (TGM, caused by B. cinerea), tomato late blight (TLB, caused by P. infestans), wheat leaf rust (WLR, caused by P. triticina), barley powdery mildew (BPM, caused by B. graminis f. sp. hordei), and pepper anthracnose (PAN, caused by C. coccodes). We performed the plant disease control efficacy assay as previously described (Lee et al., 2007; Ngo et al., 2021). Briefly, rice (Oryza sativa L, cv. Chucheong), tomato (Solanum lycopersicum cv. Seokwang), wheat (Triticum aestivum cv. Geumgang), barley (Hordeum sativum cv. Hanyoung), and pepper (Capsicum annuum cv. Hyangchon) were used as host plants, which were grown in a greenhouse at 25 ± 5°C for 3–4 weeks. One day before pathogen inoculation, the culture filtrates were directly applied onto the plant by spraying. The plants were also treated with the solvent extracts (1,000 μg/ml) and pure compounds (125, 250, and 500 μg/ml), which were dissolved in 5% aqueous methanol (v/v), using the same method for the culture filtrates. When the culture filtrates, solvent extracts, and pure compounds were applied to the plants, the samples contained 0.025% Tween 20 (w/v) as a wetting agent. Chemical fungicides (blasticidin S, validamycin, fludioxonil, dimethomorph, flusilazole, and dithianon) and 5% aqueous methanol were used as positive and negative controls, respectively. The treated plants were inoculated with spore suspensions (5 × 104 spores/ml) of each fungal pathogen and incubated as previously described (Ngo et al., 2021). The experiment was conducted twice with three replicates for each treatment. The disease control efficacy was calculated with the following equation: control efficacy (%) = 100 × [1 − B/A], where A is the mean lesion area (%) on the leaves of the control plants, and B is the mean lesion area (%) on the leaves of the treated plants (Lee et al., 2007).
Data were subjected to one-way ANOVA, and the means of the treatments were separated by Duncan’s multiple range test (p < 0.05) using the R-software (Version 4.0.5). All values are expressed as the mean ± standard deviation. Significant differences (p < 0.05) were indicated with different small letters in each bar.
Marine-derived fungi have been considered as a rich source of bioactive compounds with promising antimicrobial effects and plant disease control efficacy (Xu et al., 2015; Wang et al., 2021). In this study, a total of eight marine-derived Aspergillus spp. was isolated from different sites of the Korean coast (Supplementary Table S1). After cultivation on PDA medium for 7 days, eight strains of marine-derived Aspergillus showed different colony morphologies (Figure 1A). The CaM gene-based phylogenetic tree was constructed for the identification of the Aspergillus species (Figure 1B). In a previous study, two Aspergillus strains SFC20160112-M06 and SFC20160407-M10 were identified as Aspergillus caesiellus and A. venenatus, respectively (Lee et al., 2016). Here, six Aspergillus strains SFC20160610-M03, SFC20200425-M27, SFC20160907-M26, SFC20160317-M19, SFC20200425-M11, and SFC20160317-M26 were identified as A. jensenii, A. montenegroi, A. luchuensis, A. welwitschiae, A. candidus, and A. montevidensis, respectively (Figure 1B).
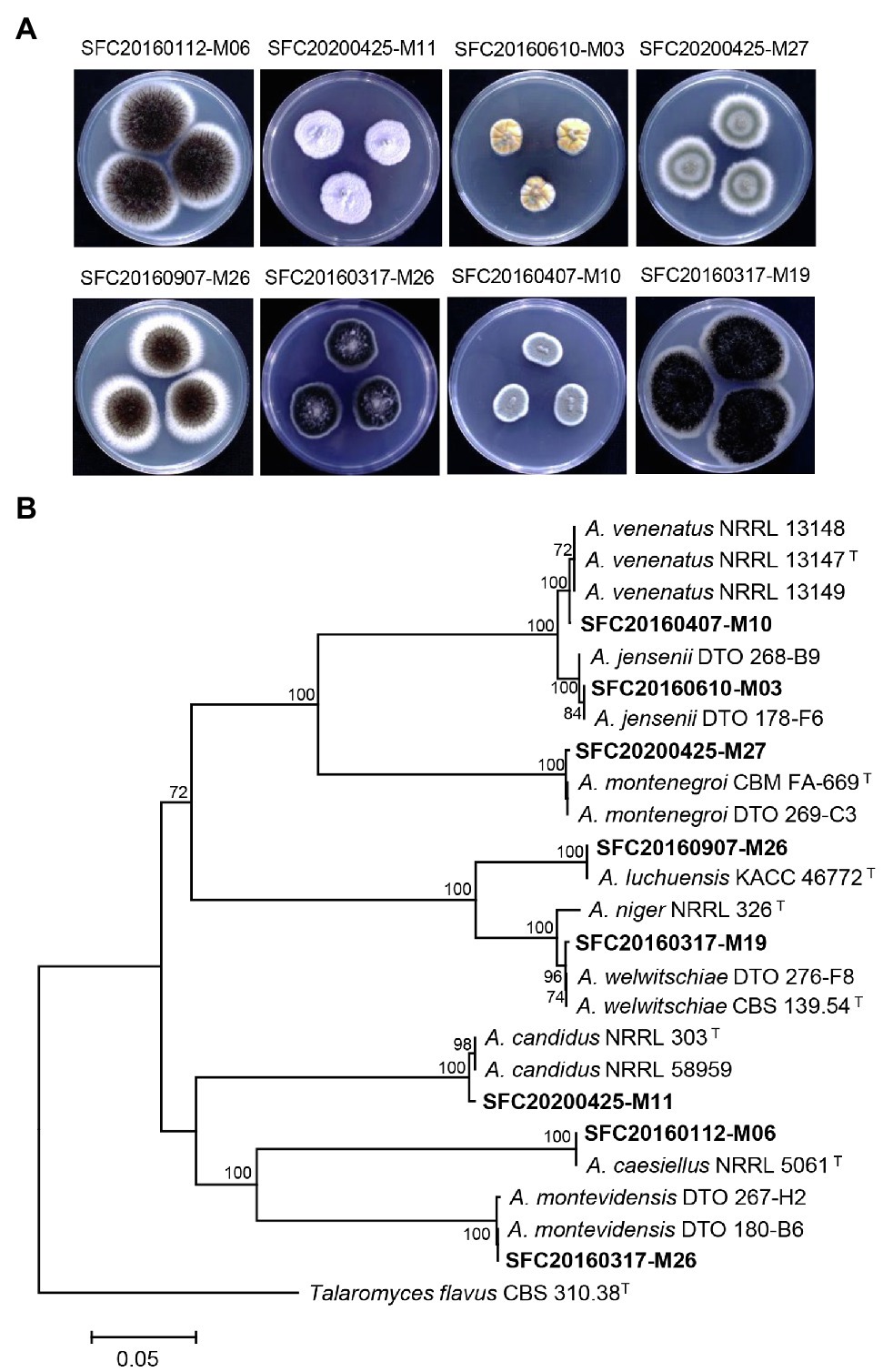
Figure 1. The colony morphology (A) and phylogenetic tree (B) of eight Aspergillus strains isolated from different sites of the Korean coast. Aspergillus isolates were cultivated on potato dextrose agar medium at 25°C for 7 days. The calmodulin gene (CaM)-based phylogenetic analysis was performed using a neighbor-joining method with 1,000 bootstrap samplings. Bootstrap scores (>70%) are presented at the nodes. The scale bar indicates the number of nucleotide substitutions per site. T indicates type strains.
To explore the potential of these Aspergillus strains in plant disease control, the disease control efficacy of the culture filtrates was investigated against six plant diseases RCB, TGM, TLB, WLR, BPM, and PAN (Table 1). When the plants were treated with each culture filtrate, the culture filtrate of A. candidus SFC20200425-M11 significantly reduced the disease development of TGM and PAN with control values of 82 and 91%, respectively, compared to the non-treatment control (Table 1). In contrast, there were weak or no effects on RCB, TLB, WLR, and BPM. The culture filtrate of A. montenegroi SFC20200425-M27 exhibited disease control values of 98 and 100% for TLB and WLR, respectively, whereas the development of RCB, TGM, BPM, and PAN were weak or not inhibited by this culture filtrate (Table 1). The culture filtrates of A. caesiellus SFC20160112-M06, A. jensenii SFC20160610-M03, and A. montevidensis SFC20160317-M26 showed weak or no disease control efficacies (Table 1). Moreover, the culture filtrates of A. luchuensis SFC20160907-M26, A. venenatus SFC20160407-M10, and A. welwitschiae SFC20160317-M19 exhibited phytotoxic symptoms on the foliage parts of the plants (Table 1). Thus, among the eight Aspergillus strains, A. candidus SFC20200425-M11 and A. montenegroi SFC20200425-M27 were selected as potential biocontrol agents in which they may produce antifungal compounds for plant diseases control.
To identify active compounds in the culture filtrates, we explored the disease control efficacy of the solvent extracts derived from the culture filtrates. When each extract was sprayed at a concentration of 1,000 μg/ml onto the plants 24 h before the inoculation of the fungal pathogens, the EtOAc extract of A. candidus SFC20200425-M11 exhibited a broad spectrum of disease control efficacy against TGM, TLB, WLR, and PAN with control values of 100, 94, 73, and 91%, respectively, compared to the non-treatment controls (Table 1). However, the BuOH and water extracts had no significant effects on all tested plant diseases (Table 1). These results suggest that the EtOAc extract of A. candidus SFC20200425-M11 might contain antifungal substances.
In contrast to the solvent extracts of the A. candidus culture, both the EtOAc and BuOH extracts of A. montenegroi SFC20200425-M27 effectively controlled plant diseases. The EtOAc showed control values of 79, 84, 100, and 85% against RCB, TLB, WLR, and PAN, respectively (Table 1). The BuOH extract also exhibited disease control efficacies against RCB, TLB, WLR, and PAN with control values of 85, 82, 100, and 85%, respectively (Table 1). The water extract had no effect on all tested plant diseases (Table 1). These results suggest that A. candidus SFC20200425-M11 and A. montenegroi SFC20200425-M27 produce antifungal substances that have lipophilic properties.
Based on the results of the disease control efficacy assays, the EtOAc extract of A. candidus SFC20200425-M11 and the EtOAc and BuOH extracts of A. montenegroi SFC20200425-M27 were further separated by various chromatographic procedures with the guidance of in vitro antifungal assays against B. cinerea or P. infestans. Compounds 1–4 were isolated from the EtOAc extract of A. candidus SFC20200425-M11, and compounds 5–7 were isolated from the EtOAc and BuOH extracts of A. montenegroi SFC20200425-M27. By comparing our spectroscopic data (Supplementary Table S2) of the isolated compounds with those reported in the literature (Yonehara et al., 1959; Ellestad et al., 1972; Mizuba et al., 1975; Sohn and Oh, 2010; Mizushina et al., 2014), five of them were identified as known compounds: sphaeropsidin A (1), (R)-formosusin A (2), (R)-variotin (3), asperlin (5), and protulactone A (7). For the structural determination of the new compounds 4 and 6, MS and NMR spectroscopic analyses were performed (Table 2; Supplementary Figures S3–S17). All the chemical structures of compounds 1–7 are shown in Figure 2.
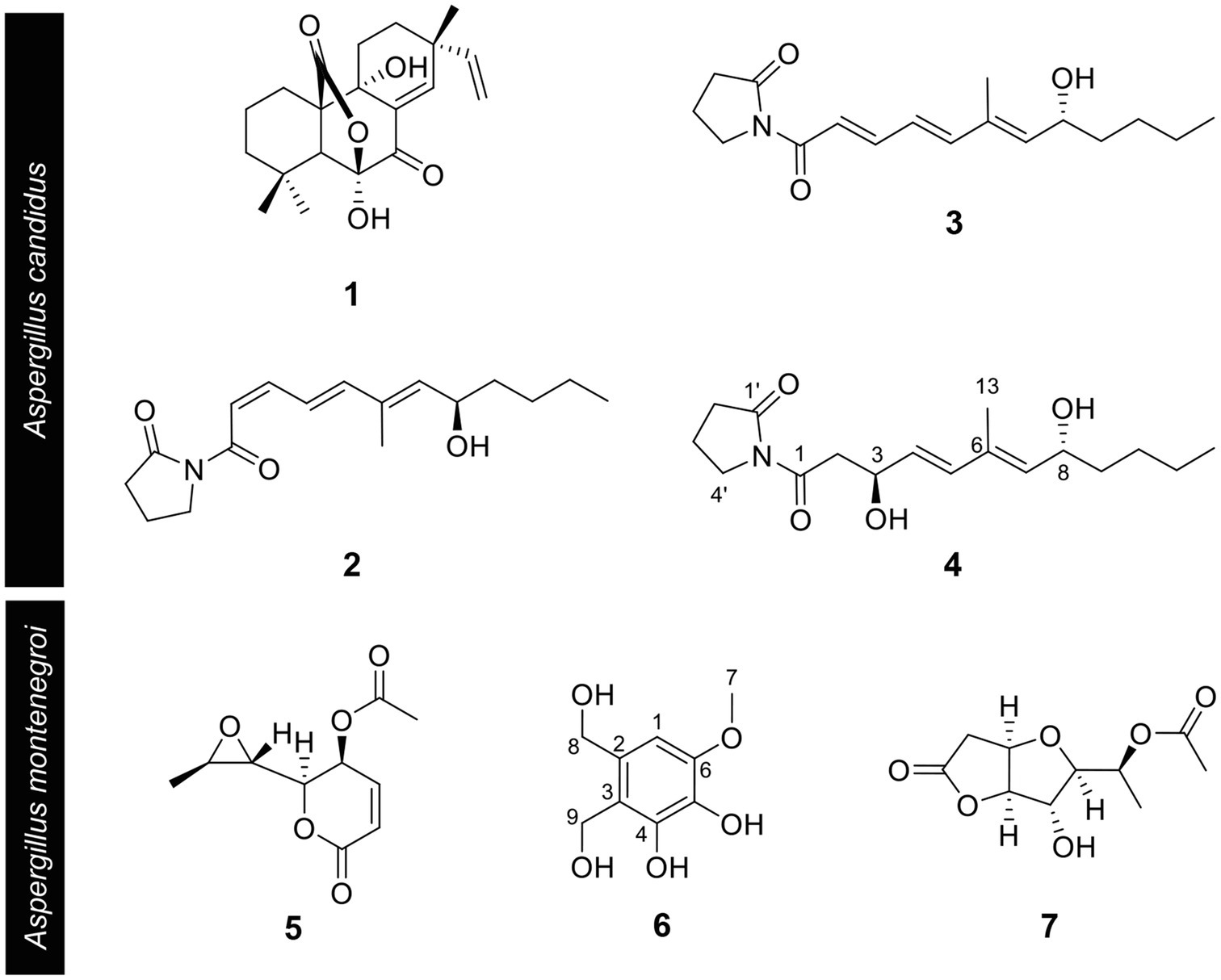
Figure 2. Chemical structures of compounds 1–7 isolated from Aspergillus candidus SFC20200425-M11 and Aspergillus montenegroi SFC20200425-M27. Sphaeropsidin A (1), (R)-formosusin A (2), (R)-variotin (3), and candidusin (4) were isolated from Aspergillus candidus. Asperlin (5), montenegrol (6), and protulactone A (7) were isolated from A. montenegroi.
The HRESIMS spectrum of compound 4 showed a quasi-molecular ion at m/z 332.1833 [M + Na]+, which was consistent with the molecular formula C17H27NO4 (calculated m/z 332.1838 for C17H27NO4Na; Supplementary Figure S3). The 13C-NMR and HSQC spectra revealed two methyls (δC 13.0 and 14.1), seven sp3 methylenes (δC 17.2, 22.7, 27.5, 33.6, 37.3, 44.1, and 45.4), three olefinic methines (δC 129.5, 135.0, and 135.4), two oxygenated methines (δC 68.5, 68.6), one olefinic quaternary carbon (δC 134.3), and two ketone carbons (δC 173.1 and 175.8; Table 2; Supplementary Figures S4–S6). The N-substituted γ-lactam moiety of compound 4 was confirmed by 1H–1H COSY correlations among H-2′, H-3′, and H-4′ along with the HMBC correlations from H-2′, H-3′, and H-4′ to C-1′ (Supplementary Figures S7 and S8). It was also confirmed by the HMBC correlations from H-13 to C-5, C-6, and C-7, together with 1H–1H COSY correlations among H-2, H-3, H-4, and H-5 and among H-7, H-8, H-9, H-10, H-11, and H-12. Two parts of the structure were connected through ketone carbon C-1, which was confirmed by the correlations in the HMBC experiment from H-2, H-3, and H-4′ to C-1. The coupling constants between H-4 and H-5 (J = 15.7 Hz) indicated the trans (E) geometry of the double bond (Table 2). Another double bond was determined to be E based on NOESY correlations from H-4 to H-13 and from H-5 to H-7 (Figure 3A). The planar structure of compound 4 was similar to that of R-variotin (3), except for the presence of one aliphatic methylene at C-2 and a hydroxyl group at C-3 instead of the double bond at C-2. The absolute configuration at C-3 and C-8 was established by the NOESY experiment and Mosher’s method (Supplementary Figures S9–S13). In the NOESY experiment, correlations were observed from H-5 to H-3, from H-5 to H-7, from H-13 to H-4, and from H-13 to H-8 (Figure 3A). The secondary alcohol groups were reacted with R-(−)- and S-(+)-α-methoxy-α-(trifluoromethyl)phenylacetyl chloride (MTPA) to give S- and R-MTPA esters, 4a and 4b, respectively (Figure 3B). The 1H NMR chemical shift differences (Δδ S - R) between 4a and 4b are shown in Figure 3B. Positive Δδ S - R values for H-4 and H-5 together with negative Δδ S - R values for H-2′, H-3′, H-4′, and H-2 indicated the S configuration of C-3 (Figure 3B). The absolute configuration of C-8 was determined as R based on positive Δδ S - R values for H-9, H-10, H-11, and H-12 and a negative Δδ S - R value for H-7 (Figure 3B). Thus, the structure of 4 was elucidated as 1-((3S,4E,6E,8R)-3,8-dihydroxy-6-methyldodeca-4,6-dienoyl)pyrrolidin-2-one and designated as candidusin.
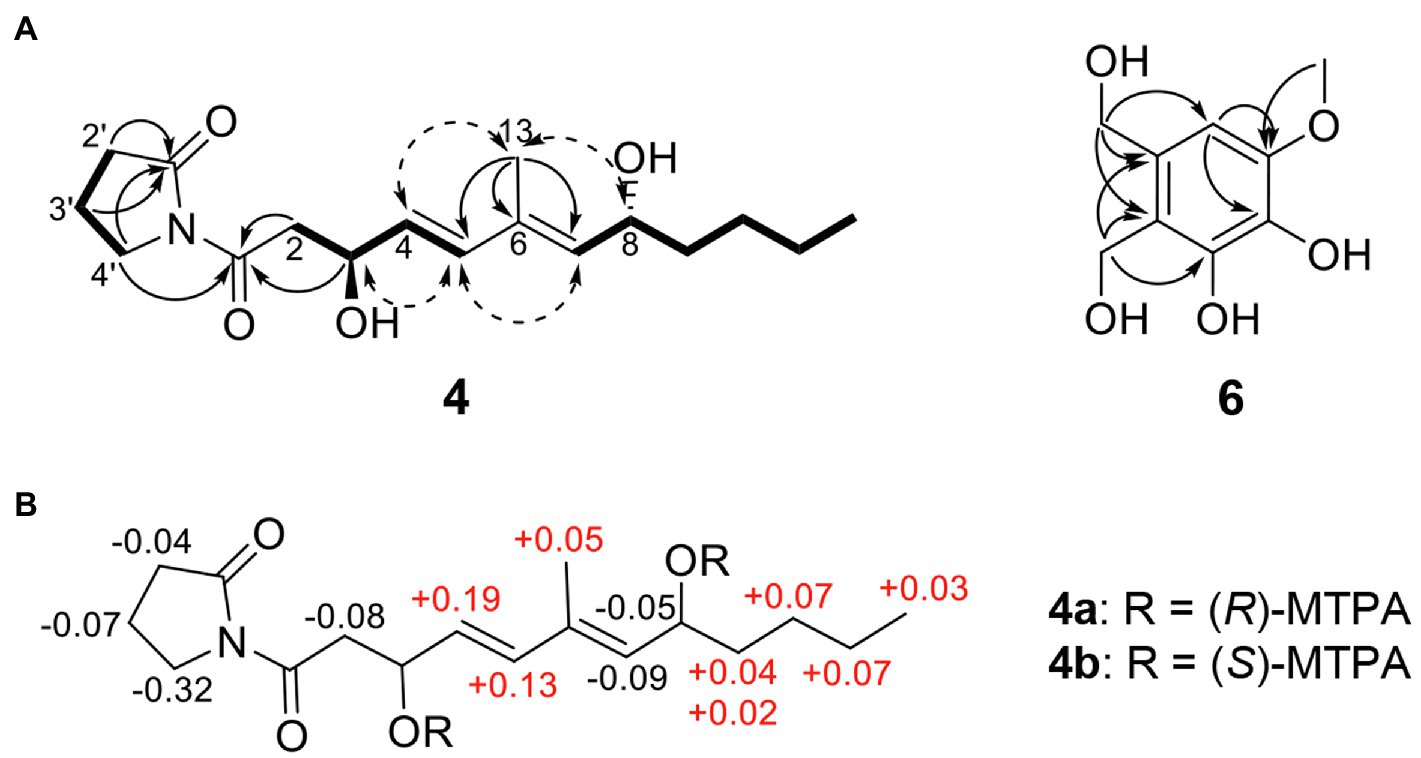
Figure 3. Structural identification of new compounds. (A) Key HMBC (arrow), COSY (bold line), and NOESY (dotted arrow) correlations of compounds 4 and 6. (B) The Δ δS – δR values for (R)- and (S)-MTPA esters of compound 4.
The molecular formula of compound 6 was determined to be C9H12O5 based on the HRESIMS peaks of m/z 223.0581 [M + Na]+ and 205.0476 [M + Na − H2O]+ (calculated 223.0582 and 205.0477 for C9H12O5Na and C9H10O4Na; Supplementary Figure S14). The 1H NMR spectrum of compound 6 exhibited signals for one aromatic proton at δH 6.53, two oxygenated methylene groups at δH 4.71 and 4.58, and one methoxy group at δH 3.81 (Table 2; Supplementary Figure S15). The 13C NMR spectrum of compound 6 contained nine signals, including one methoxy (δC 56.4), two oxygenated methylenes (δC 56.6 and 63.6), one aromatic methine (δC 104.9), and five quaternary aromatic carbons (δC 119.9, 132.2, 134.5, 145.8, and 148.6; Table 2; Supplementary Figure S16). These data suggest that compound 6 has a penta-substituted benzene ring. The positions of five substituents were determined by the HMBC correlations as follows: from an oxygenated methylene proton H-8 (δH 4.58) to C-1, C-2, and C-3; from an oxygenated methylene proton H-9 (δH 4.73) to C-2, C-3, and C-4; from an aromatic methine proton H-1 (δH 6.53) to C-5 and C-6; and a methoxy proton H-7 (δH 3.81) to C-6 (Supplementary Figure S17). Those data suggested that the substituted two oxygenated methylenes, two hydroxyls, and one methoxy group were placed on C-2, C-3, C-4, C-5, and C-6, respectively. Therefore, the structure of compound 6 was elucidated as 2,3-dihydroxymethyl-4,5-dihydroxy-6-methoxybenzene and named montenegrol.
Candidusin (4): colorless oil; = −10 (c = 0.1, methanol); IR (ATR) νmax/cm: 3,371, 2,972, 2,926, 1730, 1,645, 1,549, 1,439, 1,369, 1,242, and 1,173; 1H- and 13C-NMR (500 and 125 MHz, chloroform-d) data: see Table 2; Supplementary Figures S4 and S5; HRESIMS: m/z 332.1833 [M + Na]+ (calculated m/z 332.1838 for C17H27NO4Na; Supplementary Figure S3).
(S)-MTPA ester of 4 (4a): 1H NMR (Pyridine-d5, 500 MHz) δH 6.6244 (H-5), 6.0912 (H-4), 5.9078 (H-8), 5.4669 (H-7), 3.3667 (H-2), 3.1911 (H-4′), 2.4020 (H-2′), 1.9070 (H-13), 1.9069 (H-3′), 1.7134 (H-9a), 1.5270 (H-9b), 1.1980 (H-10, H-11), and 0.7707 (H-12).
(R)-MTPA ester of 4 (4b): 1H NMR (Pyridine-d5, 500 MHz): δH 6.4908 (H-5), 5.9625 (H-8), 5.9035 (H-4), 5.5533 (H-7), 3.5116 (H-4′), 3.4503 (H-2), 2.4464 (H-2′), 1.9755 (H-3′), 1.8536 (H-13), 1.6724 (H-9a), 1.5073 (H-9b), 1.1267 (H-10, H-11), and 0.7374 (H-12).
Motenegrol (6): yellow oil; 1H- and 13C-NMR (500 and 125 MHz, methanol-d4) data: see Table 2; Supplementary Figures S15 and S16; HRESIMS: m/z 223.0687 [M + Na]+ (calculated m/z 223.0582 for C9H12O5Na; Supplementary Figure S14).
The isolated compounds 1–7 were evaluated for their in vitro antifungal and antibacterial property against six plant pathogenic fungi (A. brassicicola, B. cinerea, C. coccodes, F. oxysporum, M. oryzae, and P. infestans) and five plant pathogenic bacteria (A. tumefaciens, C. michiganensis, P. syringae, R. solanacearum, and E. amylovora). All MIC values of the isolated compounds 1–7 are presented in Table 3. Of compounds 1–4 identified from the culture filtrate of A. candidus SFC20200425-M11, sphaeropsidin A (1), (R)-formosusin A (2), and (R)-variotin (3) exhibited an antifungal activity against all tested fungal pathogens, but not for the bacterial pathogens. In particular, sphaeropsidin A (1) exhibited promising antifungal activities against P. infestans, C. coccodes, A. brassicicola, and B. cinerea with MIC values of 0.3, 8, 16, and 63 μg/ml, respectively. (R)-formosusin A (2) showed the most potential antifungal activity against C. coccodes with a MIC value of 1 μg/ml, followed by B. cinerea, M. oryzae, F. oxysporum, and A. brassicicola with MIC values of 4, 4, 16, and 16 μg/ml, respectively. (R)-variotin (3) exhibited promising antifungal activities against C. coccodes, A. brassicicola, and B. cinerea with MIC values of 8, 16, and 16 μg/ml, respectively. Among the fungal pathogens tested, C. coccodes was the most sensitive to sphaeropsidin A (1), (R)-formosusin A (2), and (R)-variotin (3) with MIC values ranging from 1 to 8 μg/ml. Despite the structural similarity with formosusin A (2) and (R)-variotin (3), the new compound candidusin (4) exclusively showed a moderate antifungal activity against A. brassicicola with a MIC value of 250 μg/ml.
Of three isolated compounds 5–7 from the culture filtrate of A. montenegroi SFC20200425-M27, asperlin (5) was the most effective in suppressing the growth of P. infestans, M. oryzae, and C. coccodes (MICs = 1, 31, and 250 μg/ml, respectively). Montenegrol (6) and protulactone A (7) showed a moderate activity to inhibit the growth of P. infestants with MIC values of 250 and 125 μg/ml, respectively. Intriguingly, among all the tested compounds in this study, only asperlin (5) showed an antibacterial activity against C. michiganensis and E. amylovora with MIC values of 125 and 250 μg/ml, respectively.
Considering the in vitro antimicrobial activity and yield of the isolated compounds, we examined the disease control efficacy of sphaeropsidin A (1), (R)-formosusin A (2), and asperlin (5). When plants were treated with each compound, sphaeropsidin A (1) strongly reduced the disease development of TLB by at least 94% at all the treated concentrations compared to the non-treatment control. Sphaeropsidin A (1) also exhibited disease control values of 53 and 90% against WLR at a concentration of 250 and 500 μg/ml, respectively (Figure 4). Despite these vigorous disease control activities against TLB and WLR, sphaeropsidin A (1) showed weak or no disease control effects against RCB, TGM, BPM, and PAN at a high concentration of 500 μg/ml (Supplementary Table S3). In the case of (R)-formosusin A (2), it reduced in a concentration dependent manner the development of TGM with control values of 21, 57, and 82% at concentrations of 125, 250, and 500 μg/ml, respectively (Figure 5). Similar to sphaeropsidin A (1), asperlin (5) also reduced the development of TLB with control values of more than 90% at concentrations of 125–500 μg/ml and exhibited disease control values of 83 and 95% against WLR at the concentration of 250 and 500 μg/ml, respectively (Figure 6). In addition to the plant disease control efficacies, there were no phytotoxic symptoms observed on the treated plants (data not shown).
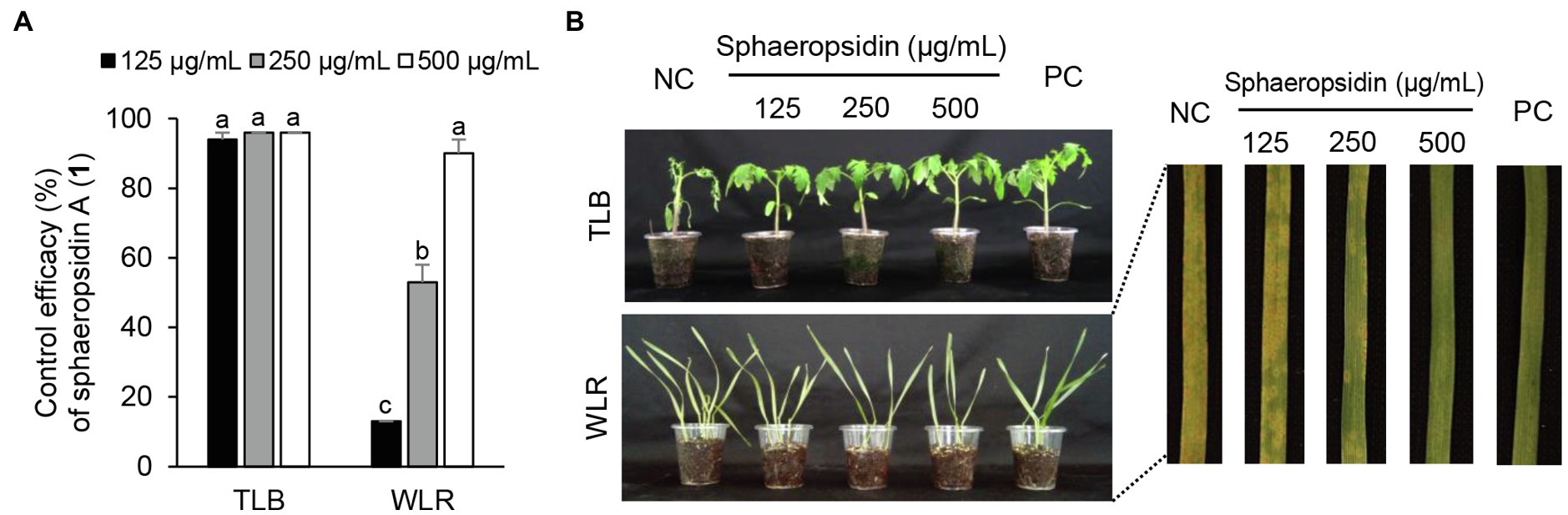
Figure 4. Effects of sphaeropsidin A (1) isolated from Aspergillus candidus SFC20200425-M11 on the development of tomato late blight (TLB) and wheat leaf rust (WLR) caused by Phytophthora infestans and Puccinia triticina. (A) Control efficacy of sphaeropsidin A (1) against TLB and WLR. The bars represent the mean ± standard deviation of two runs with three replicates. Different small letters in each bar indicate a significant difference at p < 0.05 (Duncan’s multiple range test). (B) Representatives of plants treated with sphaeropsidin (1) at a concentration of 125, 250, and 500 μg/ml. Plants were inoculated with sporangia or spores of P. infestans or P. triticina 1 day after treatment with sphaeropsidin (1). Treatment with Tween 20 solution containing 5% methanol and chemical fungicides (dimethomorph for TLB and flusilazole for WLR) were prepared as negative and positive controls (NC and PC), respectively.
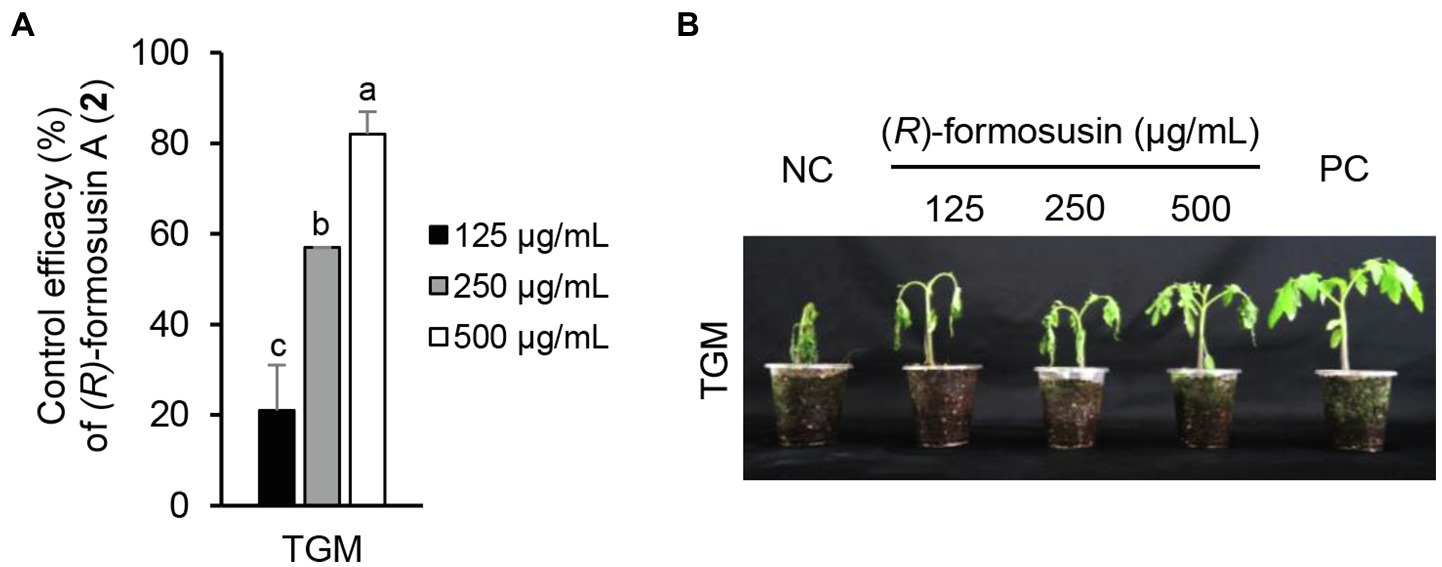
Figure 5. Effect of (R)-formosusin (2) isolated from Aspergillus candidus SFC20200425-M11 on the development of tomato gray mold (TGM) caused by Botrytis cinerea. (A) Control efficacy of (R)-formosusin (2) against TGM. The bars represent the mean ± standard deviation of two runs with three replicates. Different small letters in each bar indicate a significant difference at p < 0.05 (Duncan’s multiple range test). (B) Representatives of plants treated with (R)-formosusin (2) at a concentration of 125, 250, and 500 μg/ml. Plants were inoculated with spores of B. cinerea 1 day after treatment with (R)-formosusin (2). Treatment with the Tween 20 solution containing 5% methanol and a chemical fungicide (fenhexamide) were prepared as negative and positive controls (NC and PC), respectively.
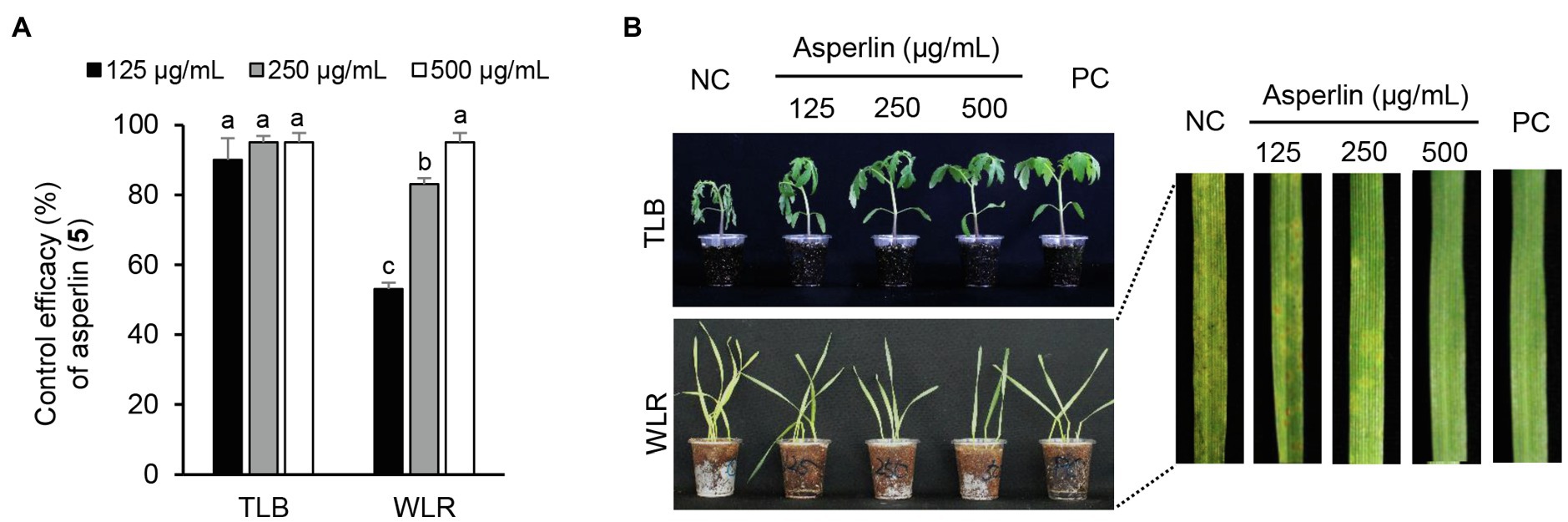
Figure 6. Effects of asperlin (5) isolated from Aspergillus montenegroi SFC20200425-M27 on the development of tomato late blight (TLB) and wheat leaf rust (WLR) caused by Phytophthora infestans and Puccinia triticina. (A) Control efficacy of asperlin (5) against TLB and WLR. The bars represent the mean ± standard deviation of two runs with three replicates. Different small letters in each bar indicate a significant difference at p < 0.05 (Duncan’s multiple range test). (B) Representatives of plants treated with asperlin (5) at a concentration of 125, 250, and 500 μg/ml. Plants were inoculated with sporangia or spores of P. infestans or P. triticina 1 day after treatment with asperlin (5). Treatment with Tween 20 solution containing 5% methanol and chemical fungicides (dimethomorph for TLB and flusilazole for WLR) were prepared as negative and positive controls (NC and PC), respectively.
In the current study, sphaeropsidin A (1) was isolated as one of the major antifungal compounds produced by A. candidus SFC20200425-M11. Sphaeropsidin A (1), an unrearranged pimarane diterpene, was first isolated from the fermentation of the fungus Aspergillus chevalieri (Ellestad et al., 1972). Since then, sphaeropsidin A (1) and its derivatives have been identified from various fungal species such as Aspergilllus porosus, Sphaeropsis sapinea f. sp. cupressi, and Diplodia spp. (Evidente et al., 1997; Neuhaus et al., 2019). In terms of antimicrobial activity, there were several studies presenting the potent antifungal activity of sphaeropsidin A (1) against plant and human pathogenic fungi (Evidente et al., 1996, 1997; Sparapano et al., 2004; Weber et al., 2007). Based on the structure–activity relationship studies, Sparapano et al. (2004) showed that the tricyclic pimarane system (particularly C-ring) and the vinyl group at C-13 are essential for the activity of sphaeropsidin A. In this study, we also showed that sphaeropsidin A (1) exhibits in vitro and in vivo antifungal activity against plant pathogenic fungi (Table 3; Figure 4). To the best of our knowledge, this is the first study to present the in vivo disease control efficacy of sphaeropsidin A (1) against TLB by P. infestans and WLR by P. triticina (Figure 4). Beyond the antifungal activity of sphaeropsidin A (1), it has also been reported to be effective against plant and human pathogenic bacteria such as Staphylococcus haemolyticus, Pseudomonas aeruginosa, and Xanthomonas oryzae pv. oryzae (Evidente et al., 2011; Roscetto et al., 2020). However, in this study, no antibacterial activity of sphaeropsidin A (1) was observed against plant pathogenic bacteria at a concentration of 250 μg/ml.
The other active compounds of A. candidus SFC20200425-M11 were (R)-formosusin A (2) and (R)-variotin (3). (R)-formosusin A (2) is a cis-olefin analog of (R)-variotin (3; Yonehara et al., 1959; Omolo et al., 2000; Mizushina et al., 2014; Yajima et al., 2016). (R)-formosusin A (2) was first isolated from Paecilomyces formosus (Mizushina et al., 2014), and its absolute configuration of (R)-formosusin A (2) was established by Yajima et al. (2016). (R)-formosusin A (2) has been known to inhibit a mammalian DNA polymerase involved in the DNA repair pathway (Beard and Wilson, 2006; Mizushina et al., 2014), but its antifungal activity has not been investigated. In contrast, (R)-variotin (3) has been reported as a broad-spectrum antifungal compound (Yonehara et al., 1959). Considering the structural similarity of (R)-formosusin A (2) and (R)-variotin (3), it may be evident that (R)-formosusin A (2) exhibits an antifungal activity against plant pathogenic fungi. As expected, we found that (R)-formosusin A (2) has an antifungal activity against plant pathogenic fungi, and it was more active than (R)-variotin (3) against B. cinerea, C. coccodes, and M. oryzae (Table 3). However, although the new derivative candidusin (4) in this study was structurally similar to (R)-formosusin A (2) and (R)-variotin (3) containing an enone moiety, we did not observe an antifungal activity at a concentration of 250 μg/ml, except for A. brassicicola (Table 3). This observation could be supported by the antimicrobial results of the diarylheptanoids from the black and green alder, which showed more significant activity of compounds containing the enone moiety in the aliphatic chain (Novaković et al., 2015).
Of the isolated compounds 5–7 from the culture broth of A. montenegroi SFC20200425-M27, asperlin (5) showed the most potent activity against the plant pathogenic fungi P. infestans and M. oryzae, and against the plant pathogenic bacteria C. michiganensis and E. amylovora (Table 3). Asperlin (5) is a polyketide antibiotic that was first isolated from Aspergillu nidulans, and its various biological activities such as antifungal, anti-inflammatory, and anti-atherosclerotic have been extensively studied (Argoudelis and Zieserl, 1966; Komai, 2003; Lee et al., 2011; Zhou et al., 2017; Wu et al., 2019). Here, we report for the first time that asperlin (5) exhibits an in vivo antifungal activity against TLB and WLR.
Evaluation of antagonistic microbes is vital for a better understanding of the ecological significance of the biocontrol of plant diseases. In this study, our results showed that A. candidus SFC20200425-M11 and A. montenegroi SFC20200425-M27 isolated from a marine environment exhibit a biocontrol potential for the first time. Although a variety of secondary metabolites and their biological activities have been reported from A. candidus, there is limited information on A. candidus and its secondary metabolites in plant disease control efficacy. Given that A. candidus and A. montenegroi culture filtrates effectively control various plant diseases, we isolated and identified two new compounds (4 and 6), along with five known compounds (1–3, 5, and 7). The in vitro results revealed the broad antifungal spectrum of sphaeropsidin A (1), (R)-formosusin A (2), (R)-variotin (3), and asperlin (5), and these natural compounds exhibited plant disease control efficacies. In addition to the antifungal activity, asperlin (5) exclusively showed an antibacterial activity against C. michiganensis and E. amylovora. Taken together, our results suggest that Aspergillus spp. and their antimicrobial compounds have great potential to be developed as new biocontrol agents or used as active ingredients for natural pesticides in agricultural fields.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
HK and GC provided the study idea and supervision. MN, BK, YK, and MP contributed to the experimental performance. MN, JH, and HK contributed to the manuscript preparation. MN, JH, HK, and GC contributed to the structure elucidation, revising, and proofreading of the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Cooperative Research Program for Agricultural Science and Technology Development (project PJ016028), Rural Development Administration, Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank to Dr. Jung Hoon Choi (Department of Biochemical Analysis, Korea Basic Science Institute) for recording HRESIMS data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.804333/full#supplementary-material
Argoudelis, A. D., and Zieserl, J. F. (1966). The structure of U-13,933, a new antibiotic. Tetrahedron Lett. 7, 1969–1973. doi: 10.1016/S0040-4039(00)76280-0
Beard, W. A., and Wilson, S. H. (2006). Structure and mechanism of DNA polymerase β. Chem. Rev. 106, 361–382. doi: 10.1021/cr0404904
Buttachon, S., Ramos, A. A., Inácio, Â., Dethoup, T., Gales, L., Lee, M., et al. (2018). Bis-indolyl benzenoids, hydroxypyrrolidine derivatives and other constituents from cultures of the marine sponge-associated fungus Aspergillus candidus KUFA0062. Mar. Drugs 16:119. doi: 10.3390/md16040119
Chandra, P., Sharma, R. K., and Arora, D. S. (2020). Antioxidant compounds from microbial sources: A review. Food Res. Int. 129:108849. doi: 10.1016/j.foodres.2019.108849
Choi, G. J., Jang, K. S., Choi, Y. H., Yu, J. H., and Kim, J.-C. (2010). Antifungal activity of lower alkyl fatty acid esters against powdery mildews. Plant Pathol. J. 26, 360–366. doi: 10.5423/PPJ.2010.26.4.360
Dayan, F. E., Cantrell, C. L., and Duke, S. O. (2009). Natural products in crop protection. Biorg. Med. Chem. 17, 4022–4034. doi: 10.1016/j.bmc.2009.01.046
Demain, A. L. (2014). Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 41, 185–201. doi: 10.1007/s10295-013-1325-z
El-hawary, S. S., Moawad, A. S., Bahr, H. S., Abdelmohsen, U. R., and Mohammed, R. (2020). Natural product diversity from the endophytic fungi of the genus Aspergillus. RSC Adv. 10, 22058–22079. doi: 10.1039/D0RA04290K
Ellestad, G., Kunstmann, M., Mirando, P., and Morton, G. (1972). Structures of fungal diterpene antibiotics LL-S491β and -γ. J. Am. Chem. Soc. 94, 6206–6208. doi: 10.1021/ja00772a054
El-Sayed, A. S., and Ali, G. S. (2020). Aspergillus flavipes is a novel efficient biocontrol agent of Phytophthora parasitica. Biol. Control 140:104072. doi: 10.1016/j.biocontrol.2019.104072
Espinel-Ingroff, A., Fothergill, A., Ghannoum, M., Manavathu, E., Ostrosky-Zeichner, L., Pfaller, M., et al. (2005). Quality control and reference guidelines for CLSI broth microdilution susceptibility method (M 38-A document) for amphotericin B, itraconazole, posaconazole, and voriconazole. J. Clin. Microbiol. 43, 5243–5246. doi: 10.1128/JCM.43.10.5243-5246.2005
Evidente, A., Sparapano, L., Fierro, O., Bruno, G., Giordano, F., and Motta, A. (1997). Sphaeropsidins B and C, phytotoxic pimarane diterpenes from Sphaeropsis sapinea f. sp. cupressi and Diplodia mutila. Phytochemistry 45, 705–713. doi: 10.1016/S0031-9422(97)00006-X
Evidente, A., Sparapano, L., Motta, A., Giordano, F., Fierro, O., and Frisullo, S. (1996). A phytotoxic pimarane diterpene of Sphaeropsis sapinea f. sp. cupressi, the pathogen of a canker disease of cypress. Phytochemistry 42, 1541–1546. doi: 10.1016/0031-9422(96)00206-3
Evidente, A., Venturi, V., Masi, M., Degrassi, G., Cimmino, A., Maddau, L., et al. (2011). In vitro antibacterial activity of sphaeropsidins and chemical derivatives toward Xanthomonas oryzae pv. oryzae, the causal agent of rice bacterial blight. J. Nat. Prod. 74, 2520–2525. doi: 10.1021/np200625m
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: The challenge of feeding 9 billion people. Science 327, 812–818. doi: 10.1126/science.1185383
Han, J. W., Oh, M., Lee, Y. J., Choi, J., Choi, G. J., and Kim, H. (2018). Crinipellins A and I, two diterpenoids from the basidiomycete fungus Crinipellis rhizomaticola, as potential natural fungicides. Molecules 23:2377. doi: 10.3390/molecules23092377
Hüter, O. F. (2011). Use of natural products in the crop protection industry. Phytochem. Rev. 10, 185–194. doi: 10.1007/s11101-010-9168-y
Jang, J. Y., Choi, Y. H., Shin, T. S., Kim, T. H., Shin, K. S., Park, H. W., et al. (2016). Biological control of Meloidogyne incognita by Aspergillus Niger F22 producing oxalic acid. PLoS One 11:e0156230. doi: 10.1371/journal.pone.0156230
Komai, S. (2003). Antifungal activity of pyranone and furanone derivatives, isolated from Aspergillus sp. IFM51759, against Aspergillus fumigatus. Mycotoxins 53, 11–18. doi: 10.2520/myco.53.11
Lee, D. S., Jeong, G. S., Li, B., Lee, S. U., Oh, H. C., and Kim, Y. C. (2011). Asperlin from the marine-derived fungus Aspergillus sp. SF-5044 exerts anti-inflammatory effects through heme oxygenase-1 expression in murine macrophages. J. Pharmacol. Sci. 116, 283–295. doi: 10.1254/jphs.10219FP
Lee, H. R., Jung, J., Riu, M., and Ryu, C. M. (2017). A new frontier for biological control against plant pathogenic nematodes and insect pests I: by microbes. Res. Plant Dis. 23, 114–149. doi: 10.5423/RPD.2017.23.2.114
Lee, S., Park, M. S., and Lim, Y. W. (2016). Diversity of marine-derived Aspergillus from tidal mudflats and sea sand in Korea. Mycobiology 44, 237–247. doi: 10.5941/MYCO.2016.44.4.237
Lee, T. H., Yeh, M. H., Chang, C. I., Lee, C. K., Shao, Y. Y., and Kuo, Y. H. (2007). New lignans from the heartwood of Cunninghamia lanceolata. Biosci. Biotechnol. Biochem. 71, 2075–2078. doi: 10.1271/bbb.70162
Mauser, W., Klepper, G., Zabel, F., Delzeit, R., Hank, T., Putzenlechner, B., et al. (2015). Global biomass production potentials exceed expected future demand without the need for cropland expansion. Nat. Commun. 6:8946. doi: 10.1038/ncomms9946
Mizuba, S., Lee, K., and Jiu, J. (1975). Three antimicrobial metabolites from Aspergillus caespitosus. Can. J. Microbiol. 21, 1781–1787. doi: 10.1139/m75-259
Mizushina, Y., Suzuki-Fukudome, H., Takeuchi, T., Takemoto, K., Kuriyama, I., Yoshida, H., et al. (2014). Formosusin A, a novel specific inhibitor of mammalian DNA polymerase β from the fungus Paecilomyces formosus. Biorg. Med. Chem. 22, 1070–1076. doi: 10.1016/j.bmc.2013.12.038
Mohamed, G. A., Ibrahim, S. R., and Asfour, H. Z. (2020). Antimicrobial metabolites from the endophytic fungus Aspergillus versicolor. Phytochem. Lett. 35, 152–155. doi: 10.1016/j.phytol.2019.12.003
Neuhaus, G. F., Adpressa, D. A., Bruhn, T., and Loesgen, S. (2019). Polyketides from marine-derived Aspergillus porosus: challenges and opportunities for determining absolute configuration. J. Nat. Prod. 82, 2780–2789. doi: 10.1021/acs.jnatprod.9b00416
Ngo, M. T., Han, J. W., Nguyen, M. V., Dang, Q. L., Kim, H., and Choi, G. J. (2021). Antifungal properties of natural products from Pterocarya tonkinensis against phytopathogenic fungi. Pest Manag. Sci. 77, 1864–1872. doi: 10.1002/ps.6211
Ngo, M. T., Han, J. W., Yoon, S., Bae, S., Kim, S. Y., Kim, H., et al. (2019). Discovery of new triterpenoid saponins isolated from Maesa japonica with antifungal activity against rice blast fungus Magnaporthe oryzae. J. Agric. Food Chem. 67, 7706–7715. doi: 10.1021/acs.jafc.9b02236
Nguyen, V. M., Han, B. S., Choi, H. Y., Byun, J., Park, J. S., and Kim, W. G. (2018). Genkwalathins A and B, new lathyrane-type diterpenes from Daphne genkwa. Nat. Prod. Res. 32, 1782–1790. doi: 10.1080/14786419.2017.1402322
Novaković, M., Novaković, I., Cvetković, M., Sladić, D., and Tešević, V. (2015). Antimicrobial activity of the diarylheptanoids from the black and green alder. Braz. J. Bot. 38, 441–446. doi: 10.1007/s40415-015-0151-0
Omolo, J. O., Anke, H., Chhabra, S., and Sterner, O. (2000). New variotin analogues from Aspergillus viridi-nutans. J. Nat. Prod. 63, 975–977. doi: 10.1021/np990509b
Raymaekers, K., Ponet, L., Holtappels, D., Berckmans, B., and Cammue, B. P. A. (2020). Screening for novel biocontrol agents applicable in plant disease management – A review. Biol. Control 144:104240. doi: 10.1016/j.biocontrol.2020.104240
Roscetto, E., Masi, M., Esposito, M., Di Lecce, R., Delicato, A., Maddau, L., et al. (2020). Anti-biofilm activity of the fungal phytotoxin sphaeropsidin A against clinical isolates of antibiotic-resistant bacteria. Toxins 12:444. doi: 10.3390/toxins12070444
Sadorn, K., Saepua, S., Boonyuen, N., Laksanacharoen, P., Rachtawee, P., Prabpai, S., et al. (2016). Allahabadolactones A and B from the endophytic fungus, Aspergillus allahabadii BCC45335. Tetrahedron 72, 489–495. doi: 10.1016/j.tet.2015.11.056
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shin, T. S., Yu, N. H., Lee, J., Choi, G. J., Kim, J. C., and Shin, C. S. (2017). Development of a biofungicide using a mycoparasitic fungus Simplicillium lamellicola BCP and its control efficacy against gray mold diseases of tomato and ginseng. Plant Pathol. J. 33, 337–344. doi: 10.5423/PPJ.FT.04.2017.0087
Sohn, J. H., and Oh, H. C. (2010). Protulactones A and B: two new polyketides from the marine-derived fungus Aspergillus sp. SF-5044. Bull. Kor. Chem. Soc. 31, 1695–1698. doi: 10.5012/bkcs.2010.31.6.1695
Sparapano, L., Bruno, G., Fierro, O., and Evidente, A. (2004). Studies on structure–activity relationship of sphaeropsidins A–F, phytotoxins produced by Sphaeropsis sapinea f. sp. cupressi. Phytochemistry 65, 189–198. doi: 10.1016/j.phytochem.2003.11.006
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Vu, T. T., Kim, H., Tran, V. K., Vu, H. D., Hoang, T. X., Han, J. W., et al. (2017). Antibacterial activity of tannins isolated from Sapium baccatum extract and use for control of tomato bacterial wilt. PLoS One 12:e0181499. doi: 10.1371/journal.pone.0181499
Wang, W., Liao, Y., Tang, C., Huang, X., Luo, Z., Chen, J., et al. (2017). Cytotoxic and antibacterial compounds from the coral-derived fungus Aspergillus tritici SP2-8-1. Mar. Drugs 15, 348. doi: 10.3390/md15110348
Wang, C., Tang, S., and Cao, S. (2021). Antimicrobial compounds from marine fungi. Phytochem. Rev. 20, 85–117. doi: 10.1007/s11101-020-09705-5
Weber, R. W. S., Kappe, R., Paululat, T., Mösker, E., and Anke, H. (2007). Anti-Candida metabolites from endophytic fungi. Phytochemistry 68, 886–892. doi: 10.1016/j.phytochem.2006.12.017
Willems, T., De Mol, M. L., De Bruycker, A., De Maeseneire, S. L., and Soetaert, W. K. (2020). Alkaloids from marine fungi: promising antimicrobials. Antibiotics 9:340. doi: 10.3390/antibiotics9060340
Wu, C., Zhou, Y., Qi, G., Liu, D., Cao, X., Yu, J., et al. (2019). Asperlin stimulates energy expenditure and modulates gut microbiota in HFD-fed mice. Mar. Drugs 17:38. doi: 10.3390/md17010038
Xu, L., Meng, W., Cao, C., Wang, J., Shan, W., and Wang, Q. (2015). Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 13, 3479–3513. doi: 10.3390/md13063479
Yajima, A., Iizuka, Y., Katsuta, R., and Nukada, T. (2016). Synthesis and absolute configuration of formosusin A, a specific inhibitor of mammalian DNA polymerase β. Tetrahedron Lett. 57, 2012–2015. doi: 10.1016/j.tetlet.2016.03.094
Yonehara, H., Takeuchi, S., Umezawa, H., and Sumiki, Y. (1959). Variotin, a new antifungal antibiotic, produced by Paecilomyces varioti Bainier var. antibioticus. J. Antibiot. 12, 109–110.
Zhao, J., Liu, W., Liu, D., Lu, C., Zhang, D., Wu, H., et al. (2018). Identification and evaluation of Aspergillus tubingensis as a potential biocontrol agent against grey mould on tomato. J. Gen. Plant Pathol. 84, 148–159. doi: 10.1007/s10327-018-0764-9
Keywords: Aspergillus candidus, Aspergillus montenegroi, plant disease, biocontrol, antimicrobial compound
Citation: Ngo MT, Nguyen MV, Han JW, Kim B, Kim YK, Park MS, Kim H and Choi GJ (2021) Biocontrol Potential of Aspergillus Species Producing Antimicrobial Metabolites. Front. Microbiol. 12:804333. doi: 10.3389/fmicb.2021.804333
Received: 29 October 2021; Accepted: 30 November 2021;
Published: 23 December 2021.
Edited by:
Jochen Fischer, Institut für Biotechnologie und Wirkstoff-Forschung (IBWF), GermanyReviewed by:
Laith Khalil Tawfeeq Al-Ani, Universiti Sains Malaysia, MalaysiaCopyright © 2021 Ngo, Nguyen, Han, Kim, Kim, Park, Kim and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hun Kim, aHVua2ltQGtyaWN0LnJlLmty; Gyung Ja Choi, a2pjaG9pQGtyaWN0LnJlLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.