- 1Research Center of Biotechnology, Russian Academy of Sciences, Moscow, Russia
- 2Institute of Chemistry, University of Tartu, Tartu, Estonia
The sensitivity of anaerobic ammonium-oxidizing (anammox) bacteria to environmental fluctuations is a frequent cause of reactor malfunctions. It was hypothesized that the addition of formate and folate would have a stimulating effect on anammox bacteria, which in turn would lead to the stability of the anammox process under conditions of a sharp increase in ammonium load, i.e., it helps overcome a stress factor. The effect of formate and folate was investigated using a setup consisting of three parallel sequencing batch reactors equipped with a carrier. Two runs of the reactors were performed. The composition of the microbial community was studied by the 16S rRNA gene profiling and metagenomic analysis. Among anammox bacteria, Ca. “Brocadia” spp. dominated during the first run. A stimulatory effect of folate on the daily nitrogen removal rate (dN) was identified. The addition of formate led to progress in dissimilatory nitrate reduction and stimulated the growth of Ca. “Jettenia” spp. The spatial separation of two anammox species was observed in the formate reactor: Ca. “Brocadia” occupied the carrier and Ca. “Jettenia”—the walls of the reactors. Biomass storage at low temperature without feeding led to an interspecies shift in anammox bacteria in favor of Ca. “Jettenia.” During the second run, a domination of Ca. “Jettenia” spp. was recorded along with a stimulating effect of formate, and there was no effect of folate on dN. A comparative genome analysis revealed the patterns suggesting different strategies used by Ca. “Brocadia” and Ca. “Jettenia” spp. to cope with environmental changes.
Introduction
Anaerobic ammonium-oxidizing (anammox) bacteria carry out anoxic oxidation of ammonium with nitrite to dinitrogen gas coupled with the production of small amounts of nitrate (Strous et al., 1998). These bacteria play an important role in the global nitrogen cycle and are used in biotechnology for ammonium removal from wastewater (Kartal et al., 2004; Kuenen, 2020). The sensitivity of anammox bacteria to changes in temperature; pH; and concentrations of dissolved oxygen (DO), ammonium, nitrites, sulfides, and organic compounds is a frequent cause of reactor failures in actual conditions of wastewater treatment facilities (Lackner et al., 2014; Cho et al., 2020). In this respect, it is essential to look for metabolic effectors (regulators) that would allow the anammox community to cope with unfavorable fluctuations in external conditions and help maintain the stability of nitrogen removal from wastewater.
Volatile fatty acids (VFAs) such as formate, acetate, and propionate could serve as potential regulators of the activity of the anammox community. Anammox bacteria can directly assimilate VFAs (Feng et al., 2019; Zhang et al., 2021) and improve their nitrogen metabolism by coupling anaerobic VFA oxidation with the process of dissimilatory nitrate reduction (DNRA). The nitrate reduction by anammox bacteria was shown to be coupled with the oxidation of formate, where nitrate was the electron acceptor and formate—the electron donor. In the DNRA process, anammox bacteria form nitrite and ammonium from nitrate and then use these compounds in a classic chemolithoautotrophic anammox process. VFA oxidation coupled with nitrate reduction also helps anammox bacteria survive under conditions of ammonium and nitrite limitation and reduces their dependence on microorganisms that form these compounds (Kartal et al., 2007). While anammox bacteria grow well with nitrate as the sole nitrogen source, their ability to perform a complete DNRA process was questioned in the present study. Anammox bacteria were shown to express the narG gene for the reduction of nitrate to nitrite in Fe0/Fe2+-dependent partial DNRA, whereas ammonium was produced from nitrite by abiotic reduction, facilitated by Fe0 and Fe2+ (Bi et al., 2020).
When determining the rate of VFA oxidation by various anammox bacteria (Ca. “Brocadia,” Ca. “Kuenenia,” and Ca. “Anammoxoglobus”), the rate of formate oxidation was an order of magnitude higher than that of acetate and propionate in all studied cultures and was maximal for Ca. “Brocadia fulgida” (Kartal et al., 2007, 2008). It was demonstrated that the anammox bacterium Ca. “K. stuttgartiensis” was able to assimilate formate directly via the reductive acetyl-CoA pathway. However, it was not able to use acetate as a carbon or energy source, and the oxidation of acetate was carried out by other bacteria of the bioreactor’s community (Lawson et al., 2020). In contrast to the above-mentioned anammox bacteria, the activity of Ca. “Jettenia” was shown to be inhibited by formate in a concentration of 1 mM (corresponding to 68 mg/l in a form of sodium salt), but not by acetate (Ali et al., 2015). No significant difference in anammox activity was observed with an addition of formate in the concentration range of 25–100 mg/l; it slightly exceeded the activity in the control. Regarding acetate supply, an increase in its concentration to 50 mg/l led to a slight increase in anammox activity, while at a further increase (from 50 to 100 mg/l), the anammox activity was inhibited (Wang L. et al., 2021). The VFA addition at certain concentrations could be beneficial for biotechnology as it promotes higher removal of nitrogen (including nitrate) and organic contaminants and the stabilization of the process (Kartal et al., 2007; Feng et al., 2019; Yin et al., 2019; Li et al., 2020; Zhang et al., 2021). The inhibitory effect of formate on undesirable nitrite-oxidizing bacteria was additionally found (Wang et al., 2020).
Secondary metabolites could be other potential regulators of the activity of the anammox community. Secondary metabolites are not directly required for microbial growth but are important for many biological processes, especially those involved in microbial interactions. The CO2 fixation in anammox bacteria involves a folate-dependent, one-carbon metabolic pathway (the Wood–Ljungdahl route) (Jetten et al., 2009; Kartal et al., 2013). Folate is a necessary subunit for anammox CO2 fixation, including 10-formyltetrahydrofolate, 5,10-methenyltetrahydrofolate, and 5,10-methylenetetrahydrofolate, and it is therefore vital for anammox growth and activity (Wang et al., 2016; Zhao et al., 2018). At the same time, anammox bacteria are not capable of performing their own biosynthesis. Therefore, they have to borrow it from folate-producing prokaryotes as it is the secondary metabolite of many bacterial species. It has been shown that the growth and activity of folate-deficient bacteria increase after co-cultivation with folate-producing bacteria (Leduc et al., 2004). Folate cross-feeding between bacteria of the phyla Armatimonadetes, Proteobacteria, and Planctomycetes (anammox bacteria) was described. It was assumed that such cross-feedings could improve the nitrogen removal rate by increasing anammox growth and activity (Zhao et al., 2018).
The aim of this work was to study the effect of formate and folate on the activity and composition of the anammox community under conditions of sharply increased ammonium loads. Formate serves as an additional substrate for anammox bacteria. Folate being a secondary metabolite of Proteobacteria, can potentially increase the rate of nitrogen removal. Thus, it was hypothesized that the addition of these compounds would have a stimulatory effect on the anammox community enriched with Ca. “Brocadia,” which in turn would lead to the stability of the anammox process under conditions of increased ammonium load, i.e., it would help overcome a stress factor. After the experiment was completed, a mixed biomass sample was stored at +4°C for 3 months, and an interspecies shift between anammox bacteria from Ca. “Brocadia” to Ca. “Jettenia” was further discovered. Interspecies shifts between different anammox bacteria have been frequently observed in laboratory reactors [reviewed by Zhang and Okabe (2020)]. This suggested the existence of genus- or species-specific niche differentiation and competition between anammox bacteria caused by differences in maximum specific growth rates, affinities to limiting substrates (ammonium and nitrite), susceptibility to various compounds, or VFA utilization (Zhang et al., 2021). However, the data explaining the niche differentiation of anammox bacteria are often contradictory, and reasons for one anammox genera or species outcompeting another are still largely unknown (Ali et al., 2018; Zhang and Okabe, 2020). In the present study, the second run of the reactors was performed to find out if the effect of formate and folate on the new Ca. “Jettenia”-enriched anammox community is different from the Ca. “Brocadia”-enriched one and to reveal the probable causes of an interspecies shift by applying a comparative genome analysis.
Materials and Methods
Setup Configuration and Operation
A setup consisted of three sequencing batch reactors (SBRs) operated in parallel, working with complete biomass retention on a fibrous porous carrier (Figure 1A). A detailed design of each reactor was described in Kallistova et al. (2020). Each reactor performed a single-stage partial nitritation/anammox process. The operating mode included a 20-min settling stage, a 30-min stage of feeding by a synthetic medium with simultaneous effluent drainage, and a 310-min stage of aeration and mechanical stirring alternated every 20 min with a 10-min duration of the last stirring phase. The complete cycle duration was 6 h, and the hydraulic retention time (HRT) was 27 h. The operating temperature was 32 ± 2°C, pH was 8.3 ± 0.1, and DO was 0.6 ± 0.2 mg/l. The airflow was gradually increased from 20 ± 0.5 to 30 ± 0.5 l/h during the entire period of the setup operation. The medium composition and preparation are described in Kallistova et al. (2020). An activated sludge containing anammox bacteria (Mardanov et al., 2019a) was used as an inoculum for the reactors’ first run. One liter of the inoculum with a total suspended solid content of 3.8 g/l was added to 3.5 L of the medium to start each reactor. Nitrite was not added to the medium as it was produced in the process of stage I nitrification. The ammonium concentration was increased from 200 to 400 mg N–NH4/l in all reactors after the nitrogen removal efficiency reached >80% (65th day). Formate (in the form of sodium salt) was added to the first reactor at a concentration of 75.5 mg/l, and folate (0.2 mg/l) was added to the second parallel reactor; the third reactor remained the control with no additives.
Figure 1. A setup configuration 3 weeks after a start-up (A) and two types of the activated sludge biofilms: on the carrier (carrier) and the inner part of the walls of the reactor (glass) in the formate reactor during the first run (107th day) (B).
The reactors were stopped after 1.5 months after the additives were supplied (on the 110th day). A sample of biomass mixed from three reactors was stored at +4°C for 3 months. The storage conditions were chosen taking into account the recommendations of Ji and Jin (2014). This biomass was used after the storage to re-inoculate reactors with fresh media (the second run). Formate and folate were added immediately after the inoculation. The ammonium concentration was increased in all reactors from 200 to 500 mg N–NH4/l on the 106th day.
Analytical Methods
Concentrations of ammonium, nitrite, and nitrate were determined in the effluent using the standard methods (Rice and Bridgewater, 2012). The daily amount of removed nitrogen (dN, milligrams per liter) was calculated as the difference between the concentration of ammonium nitrogen (N–NH4) in the inflowing medium and the concentrations of mineral nitrogen species (N–NH4, N–NO2, and N–NO3) in the effluent. The efficiency of nitrogen removal (%) was calculated as the share of removed nitrogen of its concentration in the influent.
Statistical Analysis
Mean values, standard deviations, and Student’s t-test were calculated in MS Excel. The experimental error of nitrogen mineral form concentrations (calculated as the mean deviations of experimental values from the mean value) did not exceed 2.7%.
16S rRNA Sequencing and Analysis
High-throughput sequencing of 16S rRNA gene fragments was used to analyze the community composition in two types of activated sludge biofilms: in biofilms attached to the carrier (further referred to as carrier) and at the inner part of the walls of the reactors (further referred to as glass) in the experimental (in the presence of formate and folate) and in the control reactors (Figure 1B).
Metagenomic DNA was isolated from biofilm samples using the DNeasy Power-Soil Kit (Qiagen, Germany) according to the manufacturer’s recommendations. Two sets of primers were used to amplify the V3–V4 variable region of the 16S rRNA gene: universal primers 341F (CCTAYGGGDBGCWSCAG) and 806R (GGACTACNVGGGTHTCTAAT) (Frey et al., 2016) and specific primers for anammox bacteria, Amx368f (TTCGCAATGCCCGAAAGG) and Amx820r (GGGCACTAA GTAGAGGGGTTTT) (Sonthiphand and Neufeld, 2013). The obtained PCR fragments were used to prepare the libraries with the Nextera XT DNA Library Prep Kit (Illumina, United States) according to the manufacturer’s recommendations. Multiplexing was carried out using the Nextera XT Index Kit v2 (Illumina, United States). PCR fragments were sequenced using Illumina MiSeq. At least 25,000 sequences of the 16S rRNA gene fragments were obtained for each sample. Reads from all samples were pooled together; low-quality reads, singletons, and chimeras were excluded from the analysis. The remaining reads were clustered into operational taxonomic units (OTUs) with a minimal identity of 97%. To determine the shares of OTUs in each sample, original reads (including low-quality and singleton ones) with a minimal identity of 97% were superimposed over the representative OTU sequences. These procedures were carried out using the USEARCH software package (Edgar, 2010). Taxonomic identification based on the 16S rRNA gene sequences was carried out using USEARCH and the SILVA database.
Metagenome Sequencing and Analysis
A combination of Illumina and Oxford Nanopore high-throughput sequencing technologies was used for sequencing the metagenome. At the first stage, the sequencing of a paired-end (2 × 150 bp) NEBNext® Ultra™ II DNA Library using an Illumina HiSeq2500 platform (Illumina, San Diego, CA, United States) resulted in the acquisition of 69,080,752 read pairs. In the obtained reads, the adapter removal and trimming of low-quality sequences (Q < 30) were performed using Cutadapt v.1.8.3 (Martin, 2011) and Sickle v.1.33 (Joshi and Fass, 2011), respectively. Trimmed reads were merged using FLASH v.1.2.11 (Magoč and Salzberg, 2011). The resulting merged and unmerged reads (about 20.7 Gbp in total) were de novo assembled into contigs using metaSPAdes genome assembler v.3.13.0 (Bankevich et al., 2012).
Contigs longer than 1,500 bp were binned into clusters representing metagenome-assembled genome (MAGs) using CONCOCT v.0.4.1 (Alneberg et al., 2014) and MetaBAT v.2.12.1 (Kang et al., 2015). The completeness of the MAGs and their possible contamination (redundancy) were estimated using CheckM v.1.05 (Parks et al., 2015) with lineage-specific marker genes. The assembled MAGs were taxonomically classified using the Genome Taxonomy Database Toolkit (GTDB-Tk) v.0.3.2 and Genome Taxonomy Database (GTDB) (Parks et al., 2018). The contig-to-MAG binning scheme was selected manually by comparing the results from MetaBAT and CONCOCT.
For Nanopore sequencing, the library was prepared using the 1D ligation sequencing kit (SQK-LSK109, Oxford Nanopore, United Kingdom). The sequencing of this library in an R10 flow cell (FLO-MIN110) yielded 788,000 reads with a total length of 3.1 Gbp using the MinION device. Hybrid assembly of Illumina and Nanopore reads was performed using Unicycler v. 0.4.8 (Wick et al., 2017).
Gene search and annotation of MAGs were performed using RAST server 2.0 (Brettin et al., 2015), followed by manual correction of the annotation by comparing the predicted protein sequences with the NCBI databases.
Network Analysis
Co-occurrence networks were inferred based on a Spearman correlation matrix (Langfelder and Horvath, 2012) and constructed using only significant correlation (Barberán et al., 2014). Only OTUs, the relative abundance of which was at least 2% in at least one sample, were included in the analysis. The cutoff for correlation coefficients was determined to be 0.6, and the cutoff for adjusted p-values was 0.001 (Luo and Bhattacharya, 2006). Visualization of co-occurrence network was performed using the Cytoscape v.3.8.2 platform (Shannon et al., 2003; Faust and Raes, 2016).
Results
The First Run of the Reactors
Dynamics of the Concentration of Mineral Nitrogen Species
Figure 2 presents the dynamics of the concentration of mineral nitrogen compounds in the effluent. For the first 2 months, the reactors were operated in the same mode (without the supply of the additives), which can be considered as three independent replicates of one experiment. The concentration of the incoming ammonium was maintained during this period at a constant level of 200 mg N–NH4/l. The ammonium concentration in the effluent varied in the range of 3–86 mg N–NH4/l (Figure 2A). Such fluctuations are associated with the commissioning stage, i.e., the attachment of the biomass on the carrier and adaptation of the community to new conditions. Nitrite was not added to the medium; however, it was present in the effluent, which indicated the occurrence of stage I nitrification in the reactors. For the three reactors observed, the average concentration of nitrite in the effluent varied during the first 2 months of operation in the range of 5–17 mg N–NO2/l (Figure 2B) and that of nitrate in the range of 5–24 mg N–NO3/l (Figure 2C). Elevations in the concentrations of nitrite (37.7 mg N–NO2/l) on the 10th day and nitrate (40.5 mg N–NO3/l) on the 17th day were recorded in the reactor, which later became the control one. These were the maximum values of nitrite and nitrate concentrations for the entire experiment and may indicate an imbalance between stages I and II nitrifiers and anammox bacteria in the community of this reactor. Since the dN did not decrease in this period, and the amount of ammonium in the effluent did not increase, it can be assumed that total nitrogen removal occurred in the future control reactor due to the process of nitri-denitrification, rather than nitritation-anammox. The dN in the reactors varied in the range of 103–166 mg N/l (mean 151.2 ± 20.25 mg N/l) (Figure 2D). The reactors removed 166.3 mg N/l by the 65th day, which corresponded to an ammonium removal efficiency of 83%.
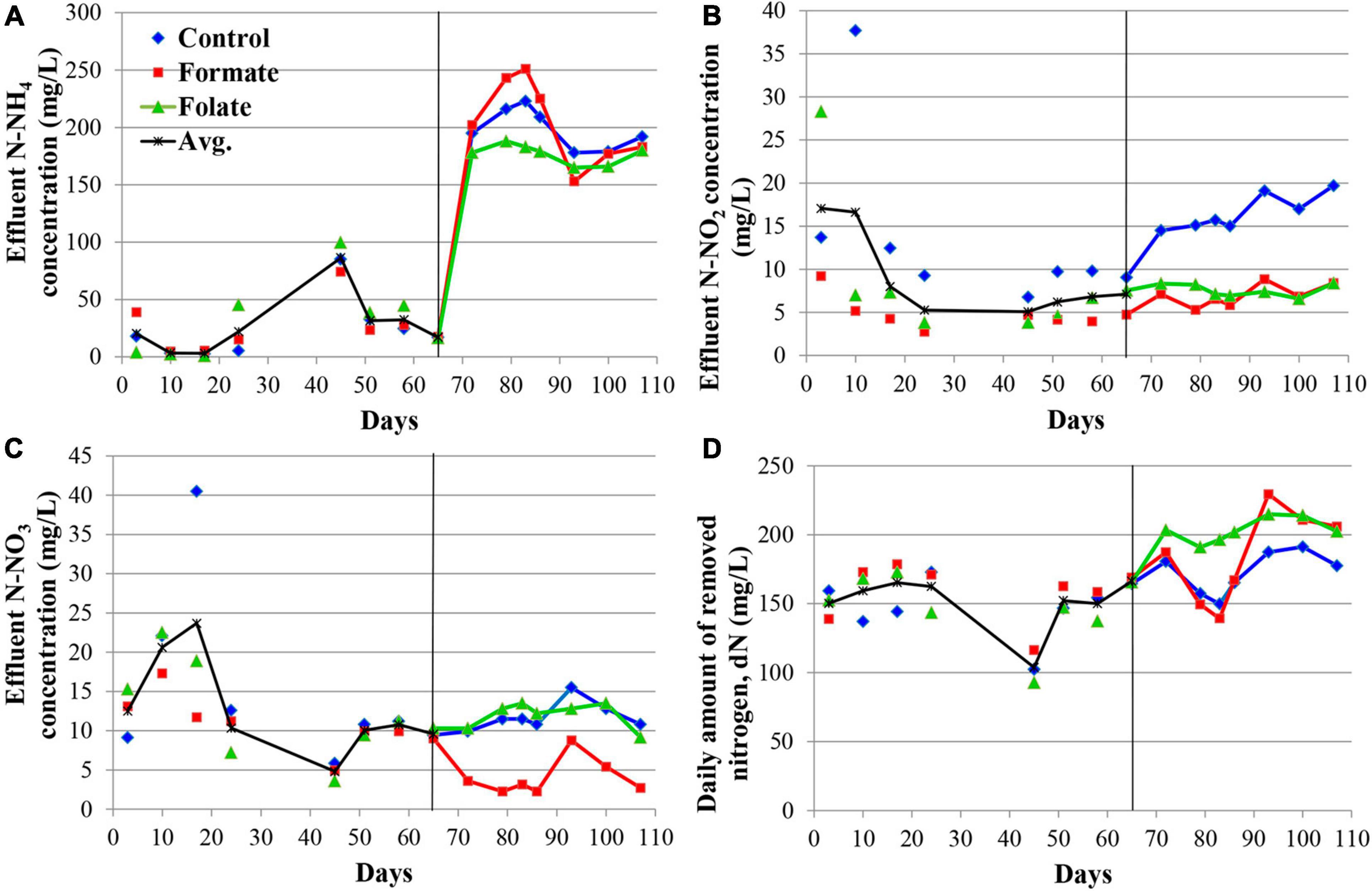
Figure 2. Dynamics of the concentrations of nitrogen compounds in the effluent during the first run of the reactors (milligrams per liter): concentrations of N–NH4 (A), N–NO2 (B), N–NO3 (C), and daily amount of removed nitrogen (dN) (D). A black vertical line separates periods before and after the additives supplied and the ammonium load increased. The data for each reactor are presented as colored dots and lines; a black curve (avg., average) reflects the values averaged for all three reactors.
The nitrogen load was increased from 177.8 to 355.6 mg/l/day in all reactors after the 65th day by increasing the ammonium concentration from 200 to 400 mg N–NH4/l. At the same time, formate was added to the first reactor and folate to the second one; no additives were supplied to the control reactor. The average dN, starting from the 65th day, were 172.7 ± 15.6 mg N/l (in the control), 184.1 ± 33.5 mg N/l (in the formate reactor), and 203.4 ± 8.6 mg N/l (in the folate reactor), which corresponded, however, to a lower overall process efficiency compared to the period before the load increased (40–43 vs 83%). The addition of formate led to a decrease in the nitrate concentration in the effluent from 11 to 4 mg N–NO3/l, which was three times lower than that in the control and in the folate reactors and could be associated with the process of nitrate reduction with formate (DNRA) by anammox bacteria. The nitrite content increased significantly in the control reactor from 9.5 to 19.7 mg N–NO2/l, while in both experimental reactors, this indicator did not change compared to the period before the supply of the additives (staying 7.0 ± 1.3 and 7.6 ± 0.7 mg N–NO2/l in the formate and folate reactor, respectively). In the control reactor, the impact of the stage I nitrifiers and denitrifiers probably increased, and the role of anammox bacteria decreased due to failures at the start-up stage.
In general, the dN was higher for the reactor with the addition of folate than with formate. Folate stimulated the process of nitrogen removal by 18% compared to the control. This stimulation started from day one after its addition, while the effect of formate (dN was 7–11% higher in the formate reactor compared to the control one) manifested itself only after 3 weeks. It has to be noted that a difference of 7–18% between the experimental and control reactors could lie within the limits of the statistical error. Thus, the Student’s t-test was used to analyze the effect of formate and folate. It confirmed that the addition of formate had no effect on the total dN (t-test, p = 0.1706), while the effect of the added folate was statistically significant (t-test, p = 0.0001).
Dynamics of the Composition of Microbial Community
In general, the following representatives of four phyla dominated both in the inoculum and in the biofilms on the carrier and walls of the reactors regardless of additives: Chloroflexi, Bacteroidetes, Planctomycetes, and Proteobacteria. One of the most abundant OTU detected in the inoculum and in the biomass of all reactors (OTU 1) contributed 9–21% to the total number of 16S rRNA gene sequences. It was assigned to the Bacteroidetes/Chlorobi group but was only distantly related to cultured species and had maximal sequence identity (98.69%) with uncultured bacterium partial 16S rRNA gene (GenBank accession no. LR637496.1) obtained from a wastewater treatment system. A minor part of the community was formed by the representatives of the phyla Acidobacteria, Armatimonadetes, Nitrospirae, Spirochaetes, Verrucomicrobia, and Patescibacteria groups. The relative abundance of Archaea in the inoculum was 1.25%, showing the presence of hydrogenotrophic and acetoclastic methanogens of the genera Methanothermobacter and Methanothrix, respectively. It did not exceed 0.06% in biofilms on the carrier during the entire experiment and was not found on the glass (Figure 3). Chloroflexi was the dominant phylum (30–45%) and was represented mainly by the bacteria of the Anaerolineae class (filamentous anaerobic organotrophs), which are often detected in anammox reactors. Among the representatives of the phylum Bacteroidetes, the bulk of the sequences belonged to organotrophic members of the orders Ignavibacteriales and Cytophagales. Representatives of the class Betaproteobacteria belonged to the genera Nitrosomonas (stage I nitrifiers), Comamonas, and Thauera. Denitrifiers were found among the members of the latter two genera.
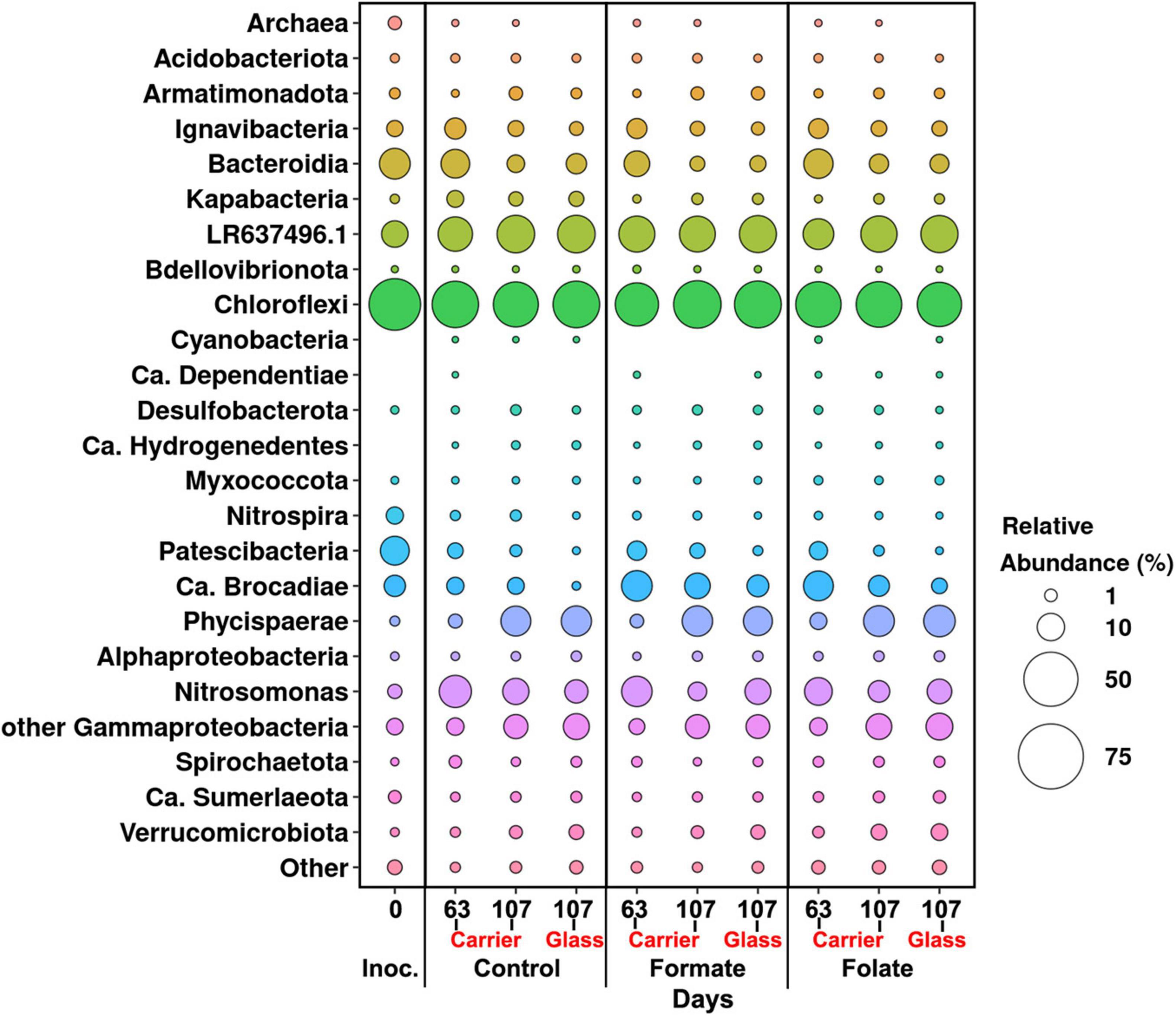
Figure 3. Overall microbial diversity in the inoculum and the biomass on the carrier and on the walls of the reactors (glass) before (the 63rd day) and after (the 107th day) the additives supplied and the ammonium load increased (the first run of the reactors) as revealed by high-throughput sequencing of the 16S rRNA gene fragments.
The relative abundance of sequences affiliated with members of the genus Nitrosomonas increased from 1.6% (in the inoculum) to 10–15% in the biofilms on the carrier in the period before the ammonium concentration was increased and additives were supplied. After the nitrogen loading was increased, their abundance on the carrier was decreased: the minimum abundance of 3.6% was found in the formate reactor and the maximum of 9% in the control one (Figure 4A). At the same time, it was found that Nitrosomonas sequences dominating in the inoculum (OTU 4; 1.24%) were displaced after 2 months of the operation of the reactors by other members of this genus (OTU 6; 11% in the reactors vs. 0.3% in the inoculum). The OTU 4 sequences again became dominant in the reactors after the regime was changed, and the OTU 6 fraction dropped to almost zero, which may be due to the influence of the incoming ammonium concentration.
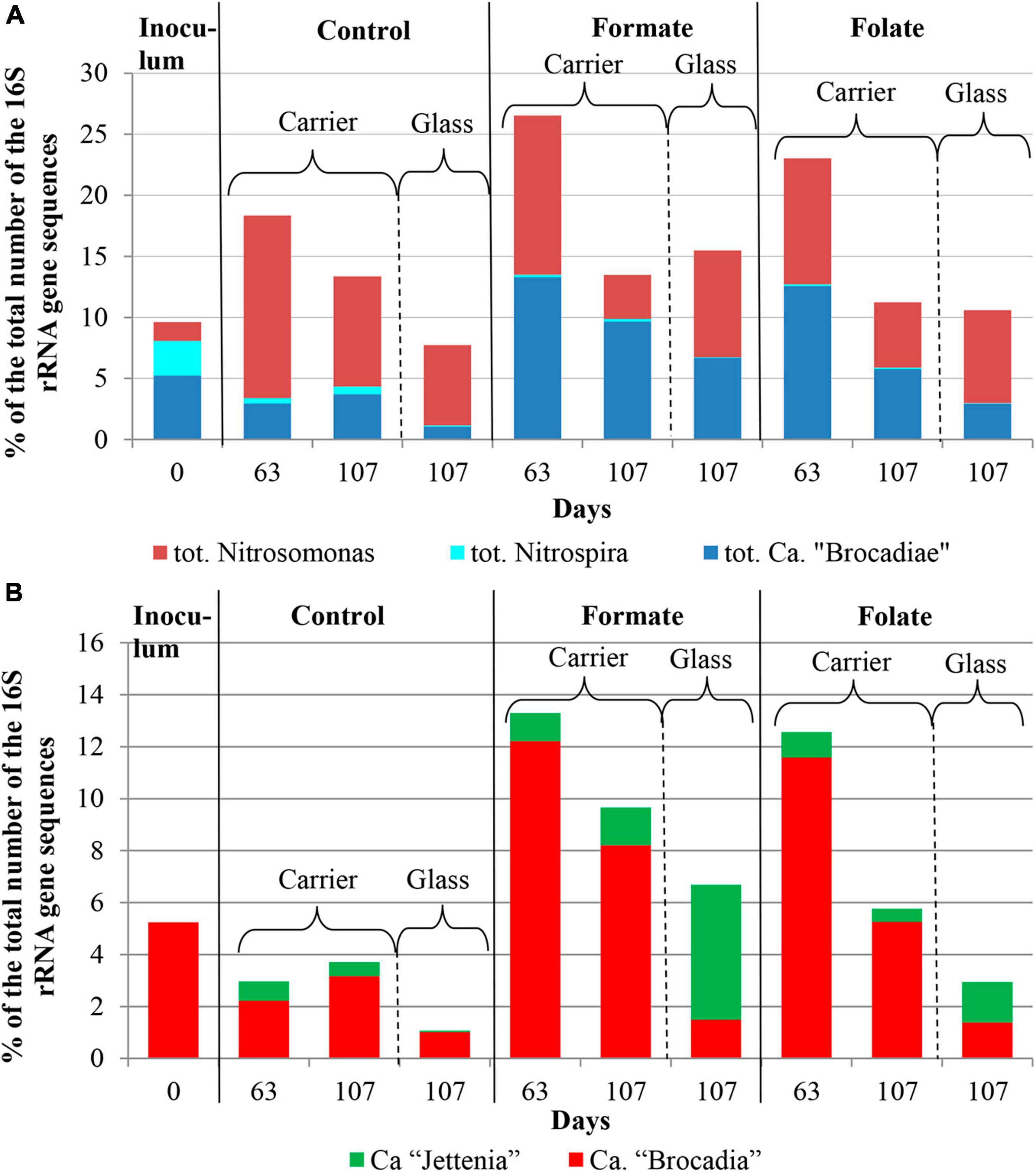
Figure 4. The overall abundance of stage I (genus Nitrosomonas) and stage II (genus Nitrospira) nitrifiers and anammox bacteria (A) and the relative abundance of members of the genera Ca. “Jettenia” and Ca. “Brocadia” (B) in the inoculum and the biomass on the carrier and on the walls of the reactors (glass) before (the 63rd day) and after (the 107th day) the additives supplied and the ammonium load increased (the first run of the reactors).
In addition to the biomass immobilized on the carrier, another type of biofilms developed on the inner surface of the walls of the reactors (referred to as glass). In contrast to the carrier, on which a dense bright orange biomass was formed, biofilms on the glass were slightly pink in color, thin and transparent, easily separated from the glass, and floated to the surface (Figure 1B). These biofilms began to form after the load was increased and were the weakest in the control reactor. The differences between the relative abundance of microbial taxa were more pronounced in the biofilms on the glass than on the carrier (Figure 3). The relative abundance of Nitrosomonas sequences varied on the glass from 6.6 to 8.7% and was maximal in the formate reactor (Figure 4A).
The relative abundance of stage II nitrifiers of the genus Nitrospira reached 2.8% in the inoculum and decreased to <1% on the carrier and <0.1% on the glass. The maximum share of stage II nitrifiers (0.4–0.6%) was observed in the control reactor on the carrier (Figure 4A).
Representatives of the phylum Planctomycetes were among the most abundant members of the community (5.6% in the inoculum and 4.4–21.6% in the reactors). Of these, the relative abundance of sequences affiliated with uncultured representatives of the Phycisphaerae class was 0.4% in the inoculum, 1.3–2.6% in the reactors before the regime was changed, and 10.5–13.4% after the regime was changed. The rest was accounted for anammox bacteria of the Ca. “Brocadiae” family.
In the inoculum, the target family of Ca. “Brocadiae” was represented by sequences (5.2%) affiliated with anammox bacteria of the genus Ca. “Brocadia” (Figure 4B). They dominated in the biofilms on the carrier in all reactors, regardless of the load and the additives. The sequences related to anammox bacteria of the genus Ca “Jettenia” were also found. Their relative abundance was about 1% before the ammonium concentration was increased (Figure 4B). In the reactor, which became the control, the relative abundance of anammox bacteria after 2 months of operation decreased for unknown reasons almost two times compared to the inoculum (from 5.2 to 3%); the share of stage I nitrifiers, on the contrary, increased 10 times (from 1.6 to 15%). These data are consistent with the results of chemical measurements that recorded ejections of nitrite and nitrate in this reactor during the commissioning stage (Figures 2B,C). After the load increased, the abundance of anammox bacteria in the carrier biofilms of the control reactor slightly increased (3 vs. 3.7%) together with a decrease in the share of stage I nitrifiers (15 vs. 9%). In general, a decrease in the relative abundance of anammox bacteria and stage I nitrifiers was recorded in the carrier biofilms of both experimental reactors after an increase in the load and additive supply. The share of stage II nitrifiers remained at a low level. In the formate reactor, the abundance of the sequences related to Ca. “Brocadia” decreased 1.5 times and to Ca. “Jettenia” oppositely increased. In the folate reactor, the most significant reduction (2.2 times) in the abundance of Ca. “Brocadia” was observed, as well as a two times decrease in the share of Ca. “Jettenia” (Figure 4B). Thus, with the introduction of formate against the background of an increase in the ammonium concentration, the Ca. “Jettenia” contribution in carrier biofilms increased maximally (three times compared with the control and folate reactors) with a general decrease in the abundance of Ca. “Brocadia.”
In contrast to the carrier, representatives of the genus Ca. “Jettenia” dominated (5.2%) in biofilms on the glass of the formate reactor; the relative contribution of Ca. “Brocadia” was only 1.5%. The ratio between Ca. “Brocadia” and Ca. “Jettenia” on the glass of the folate reactor was 1:1, and in the control reactor, Ca. “Brocadia” dominated (Figure 4B). Thus, the stimulation of anammox bacteria of the genus Ca. “Jettenia” with formate was revealed, and spatial separation of two species of anammox bacteria inside one reactor was observed.
The Second Run of the Reactors
Dynamics of the Concentration of Mineral Nitrogen Species
A mixed biomass sample from the first run was stored for 3 months at +4°C, followed by its activation for a week in the described mode without the supply of additives under an input ammonium concentration of 200 mg N–NH4/l. After the biomass activation, it was used as an inoculum for the second run of three parallel reactors. Formate was added to the first reactor and folate to the second one; both additives were supplied immediately upon a start-up. The third reactor was a control one (no additives). The ammonium concentration was increased in all reactors from 200 to 500 mg N–NH4/l after 3.5 months of operation. Figure 5 presents data on the dynamics of the concentration of nitrogen compounds in the effluent. The reactors were similar in terms of the concentrations of residual ammonium and dN (Figures 5A,D). The difference between the reactors was revealed when comparing the dynamics of nitrite and nitrate concentrations (Figures 5B,C). With the addition of formate, the average concentrations of nitrite (4.4 ± 1.3 and 11.6 ± 1.6 mg N–NO2/l before and after increasing the concentration of incoming ammonium, respectively) and nitrate (7.1 ± 2.1 and 10.7 ± 0.7 mg N–NO3/l) were expectedly lower than in the folate and control reactors, which could be associated with the DNRA process by anammox bacteria in the presence of formate. The concentration of nitrite steadily increased (from 2.4 to 21.1 mg NO2/l) in the folate reactor during the entire experiment. In the control reactor, the concentration of nitrate began to increase (from 10 to 15 mg N–NO3/l) by the end of the experiment. These variations, however, did not affect dN in both reactors. Before the load was increased, the dN was 115.3 ± 21.4 mg N/l (average value over the entire period) in the control reactor, 123.2 ± 20.0 mg N/l in the formate one, and 121.8 ± 16.6 mg N/l in the folate reactor. The formate reactor slightly pulled ahead in terms of dN after the concentration of incoming ammonium was increased: 201.4 ± 10.2 mg N/l (in the formate reactor) vs. 195.0 ± 12.9 mg N/l (in the folate reactor) and 186.8 ± 3.1 mg N/l (in the control) (Figure 5D). In general, the dN values in the first and second runs differed a little. However, in the second run, folate had no effect on dN (t-test, p = 0.0427 and p = 0.1943 before and after the load increased, respectively), while formate, on the contrary, had a stimulating effect (t-test, p = 0.0026 and p = 0.0082) compared to the control.
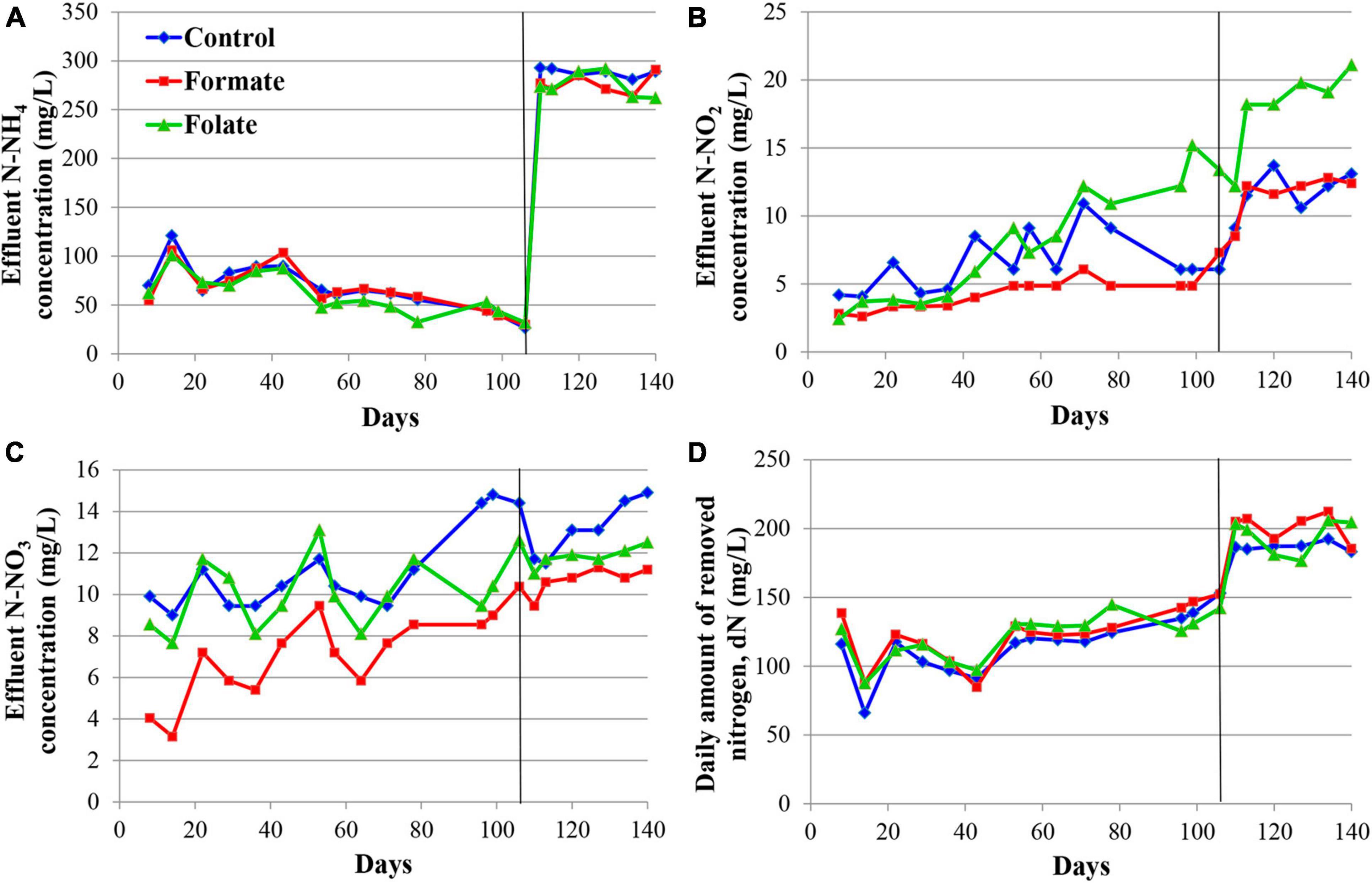
Figure 5. Dynamics of the concentrations of nitrogen compounds in the effluent during the second run of the reactors (milligrams per liter): concentrations of N–NH4 (A), N–NO2 (B), N–NO3 (C), and daily amount of removed nitrogen (dN) (D). A black vertical line separates periods before and after the ammonium load increased.
Dynamics of the Composition of Microbial Community
Figure 6 presents the dynamics of the composition of the microbial community in two types of biofilms (on the carrier and the glass). The overall composition of the community did not differ from that in the first run. There was also no difference in the composition of the community between different reactors (formate, folate, and control). However, the biofilms on the carrier and the glass differed slightly in terms of relative abundances of some taxa. No archaea were found in the biofilms on the glass. The abundance of representatives of the phyla Nitrospirae, Ca. “Hydrogenedentes,” and the Ca. “Brocadiaceae” family decreased, and the relative abundance of representatives of the phylum Spirochaetes, class Bacteroidia, and stage I nitrifiers of the genus Nitrosomonas increased in biofilms on the glass compared to the carrier (Figures 6A,B). The results on the ratio of anammox bacteria of the genera Ca. “Brocadia” and Ca. “Jettenia” in the community were of interest. At the first run, the anammox bacteria in the inoculum were represented almost exclusively by the genus Ca. “Brocadia.” During the experiment, representatives of the genus Ca. “Jettenia” came to light, and their relative abundance increased with the addition of formate (Figure 4B). A shift of Ca. “Brocadia” to Ca. “Jettenia” was documented in the inoculum after the biomass storage, and Ca. “Jettenia” spp. dominated in the reactors during the second run (Figure 7). In the inoculum for the second run, the relative abundance of stage I nitrifiers (Nitrosomonas) was two times lower than that of anammox bacteria (5.4 vs. 10.6%), stage II nitrifiers (Nitrospira) did not exceed 0.5% (Figure 8A), and the ratio of Ca. “Brocadia” to Ca. “Jettenia” was 1:5.6 (Figure 7A). During the operation of the reactors, anammox bacteria predominated among three target microbial groups on the carrier (Figure 8A), with the absolute dominance of sequences affiliated with representatives of the genus Ca. “Jettenia” (Figure 7A). The overall share of anammox bacteria on the carrier was higher in the control than in both experimental reactors (Figure 8A). The abundance of Ca. “Jettenia” decreased on the carrier by two times in all reactors immediately after the increase in nitrogen load; the share of Ca. “Brocadia” oppositely increased in the formate reactor (Figure 7A).
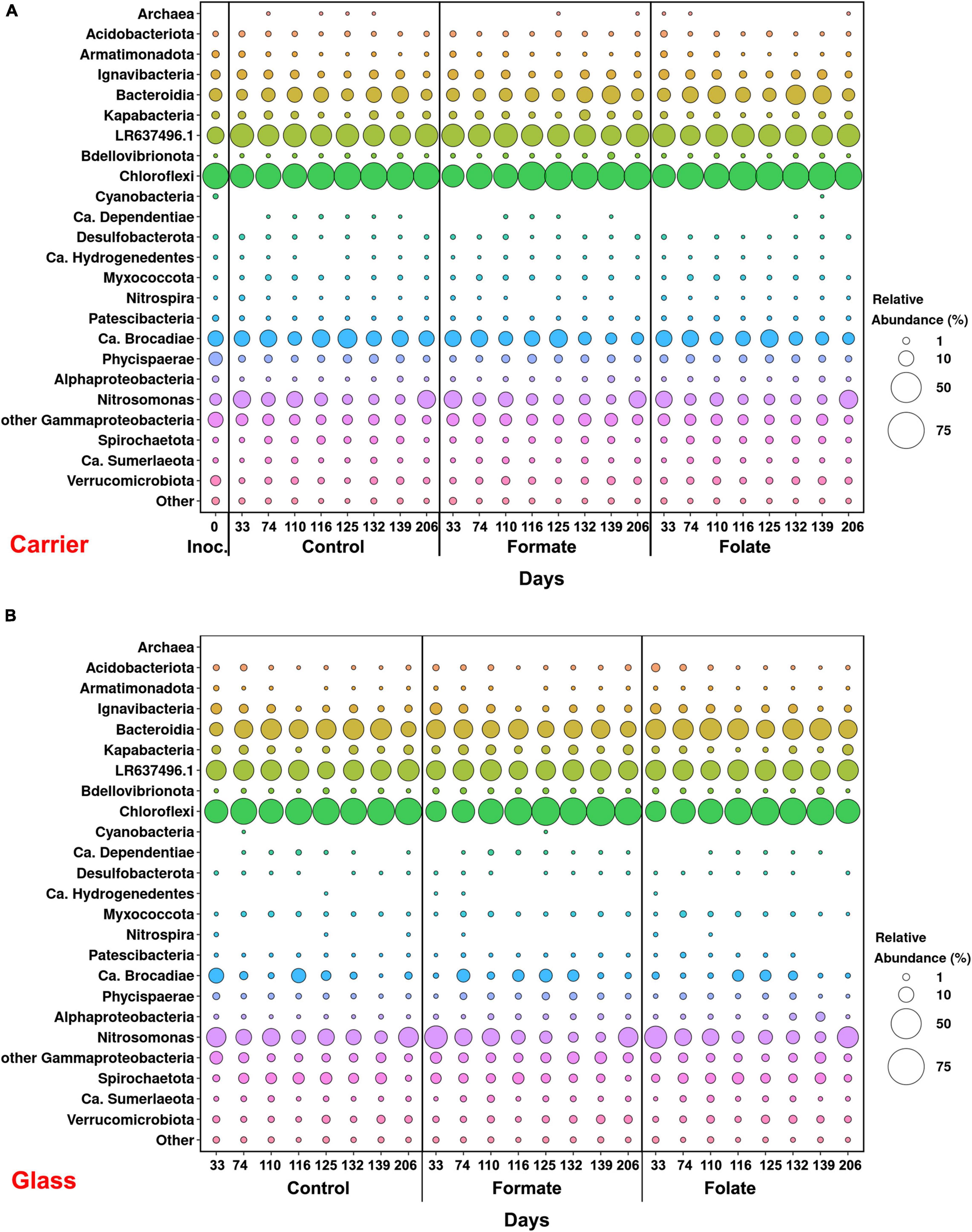
Figure 6. The overall microbial diversity in the inoculum and the biomass on the carrier (A) and on the walls of the reactors (glass) (B) before (0–74 days) and after (110–206 days) the additives supplied and the ammonium load increased (the second run of the reactors) as revealed by high-throughput sequencing of the 16S rRNA gene fragments.
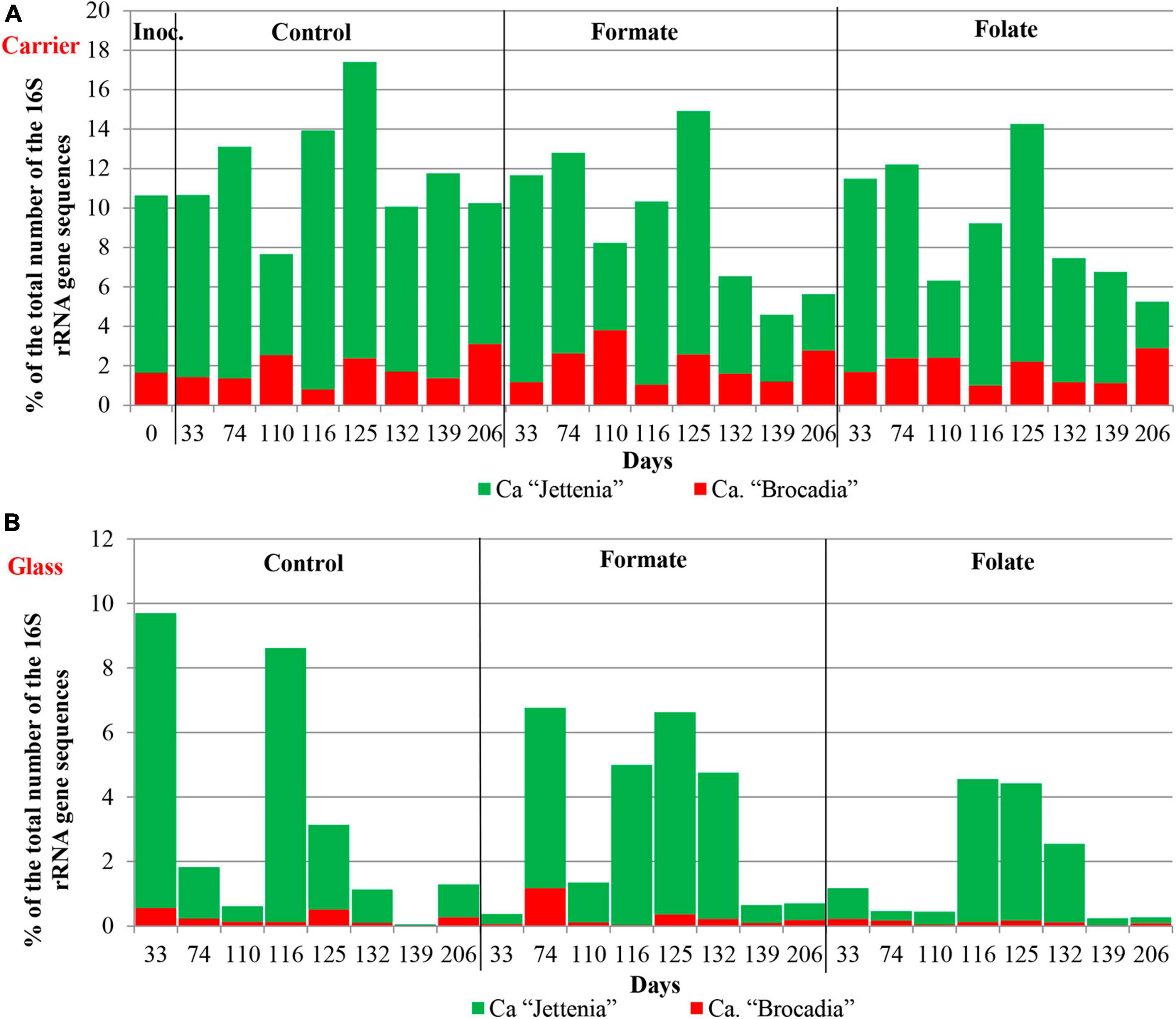
Figure 7. The relative abundance of members of the genera Ca. “Jettenia” and Ca. “Brocadia” in the inoculum and the biomass on the carrier (A) and on the walls of the reactors (glass) (B) before (0–74 days) and after (110–206 days) the additives supplied and the ammonium load increased (the second run of the reactors).
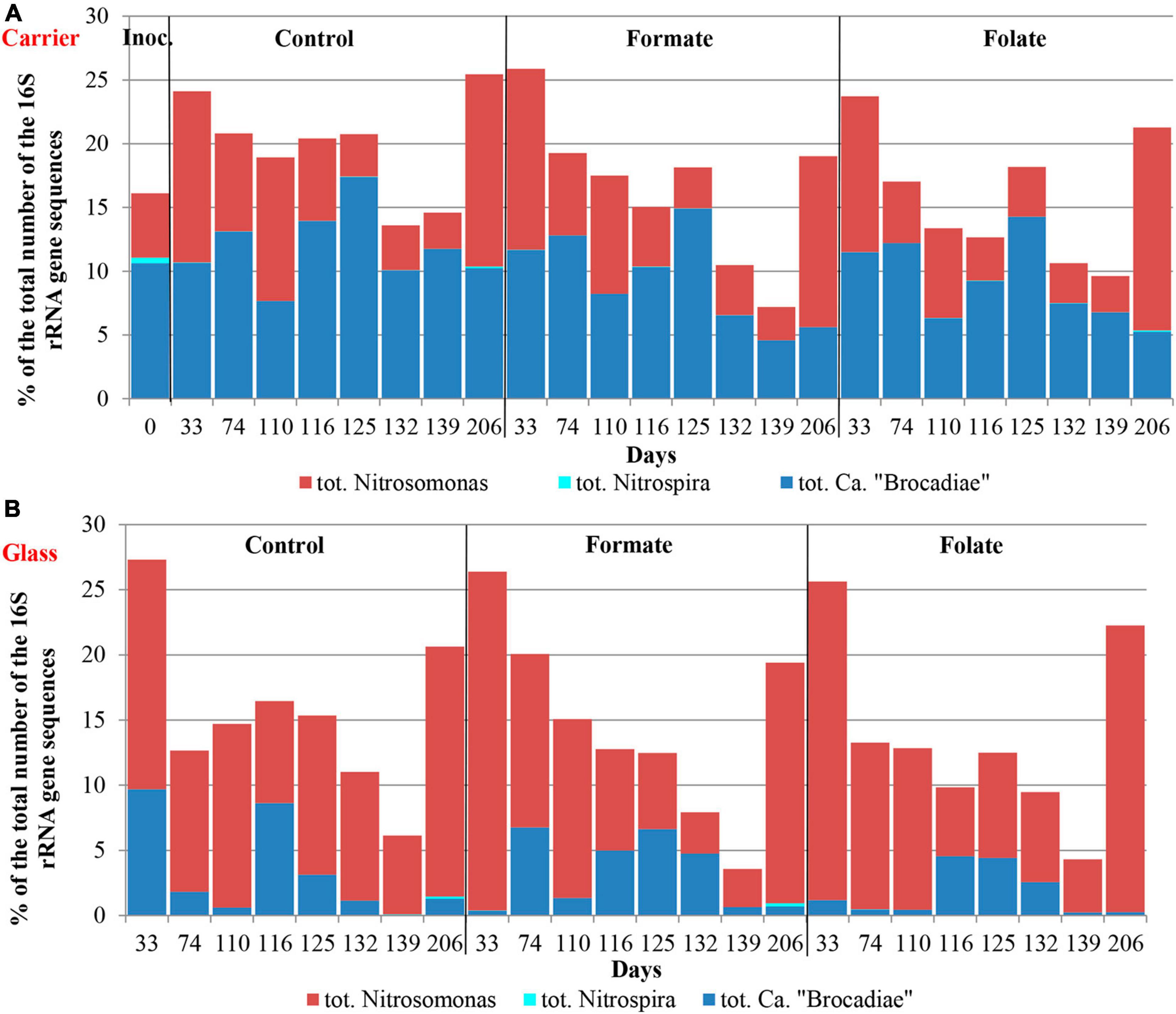
Figure 8. The overall abundance of stage I (genus Nitrosomonas) and stage II (genus Nitrospira) nitrifiers and anammox bacteria (Ca. “Brocadiae” family) in the inoculum and the biomass on the carrier (A) and on the walls of the reactors (glass) (B) before (0–74 days) and after (110–206 days) the additives supplied and the ammonium load increased (the second run of the reactors).
In contrast to the carrier, stage I nitrifiers dominated on the glass (Figure 8B). The abundance of anammox bacteria of the genus Ca. “Brocadia” did not generally exceed 1% in all reactors; the proportion of Ca. “Jettenia” ranged from 0.03 to 9% (Figure 7B). High temporal fluctuations in the relative abundance of anammox bacteria were observed. Their lowest share was recorded in the folate reactor (Figure 8B). Anammox bacteria of the genus Ca. “Brocadia” were almost completely washed out of the glass in all reactors by the end of the experiment; the smallest abundance of Ca. “Brocadia” spp. was observed in the folate reactor (Figure 7B).
Thus, the biomass storage conditions turned out to be unfavorable for the anammox bacteria of the genus Ca. “Brocadia” or, conversely, are favorable for the development of Ca. “Jettenia,” which resulted in an interspecies shift. Potential factors affecting the ratio between the two genera of anammox bacteria in the community could also be, besides the additives (formate and folate), DO, temperature, and nitrogen loading.
Comparative Study of Ca. “Brocadia” and Ca. “Jettenia” Genomes Derived From the Formate Reactor (The Second Run)
A complete metagenome, obtained from the biomass on the carrier of the formate reactor, was sequenced to assemble the composite genomes of the dominant members of the community. A total of 20.7 Gb was sequenced using Illumina technology and 3.1 Gb with Nanopore technology. The obtained sequences were assembled into contigs, which were binned into 39 MAGs with a CheckM assembly completeness of more than 80%. These MAGs accounted for 87% of the whole metagenome (Supplementary Table 1).
Phylogenetic identification of MAGs using genome-wide search in the GTDB database (Parks et al., 2018) revealed that four phyla dominated in the community: Bacteroidetes (36% of the whole metagenome), Chloroflexi (19%), Proteobacteria (13%), and Planctomycetes (11%). A minor part of the MAGs belonged to the phyla Armatimonadetes (2.6%), Verrucomicrobia (2.2%), Ca. “Omnitrophica” (1.3%), and Spirochaetes (0.1%). Among the anammox bacteria, two MAGs affiliated with the members of the genera Ca. “Jettenia” and Ca. “Brocadia” were obtained. The first MAG, designated Bin49, which accounted for 1.7% of the whole metagenome, belonged to the genus Ca. “Jettenia.” It was assembled into a single circular contig of 3,822,769 bp and had a 32.8 × average sequencing coverage. CheckM estimated its completeness in 95.6% with 1.65% possible contamination (redundancy). The second MAG, designated Bin9, which accounted for 3.3% of the whole metagenome, belonged to the genus Ca. “Brocadia.” It was also assembled into a single circular contig of 4,046,273 bp and had a 62.2 × average sequencing coverage. CheckM estimated its completeness in 100% with 2.75% of possible contamination (redundancy). Thus, both MAGs satisfy the proposed criteria for high-quality MAGs (completeness of >90% with contamination of <5% and the presence of all rRNA genes; Bowers et al., 2017). The 16S rRNA gene sequence of Bin49 was 99.28% identical to those of Ca. “Jettenia asiatica” (AB973443; Quan et al., 2008) and that of Bin9 was 100% identical to those of Ca. “Brocadia fulgida” (DQ459989; Kartal et al., 2004). These MAGs are the first complete ring genomes of anammox bacteria of these species.
To study the differences between two genomes of anammox bacteria and to identify the mechanisms of their competition, gene representation profiles were compared using the Clusters of Orthologous Groups of proteins (COG) database (Table 1). The analysis of the gene representation in each COG class showed that there are significantly more genes belonging to the categories of “cell motility,” “defense mechanisms,” and “mobilome: prophages and transposons” in the Bin 9 (Ca. “Brocadia”) genome than in the Bin 49 (Ca. “Jettenia”). Complete pathways for bacterial chemotaxis and flagellar assembly were found in the Bin 9 genome. These genes were absent in the Bin 49 genome; however, Ca. “Jettenia” sp. could form pili and use them to facilitate the attachment of cells to the walls of the reactor. Also, slightly more genes in the Bin 49 genome than in the Bin 9 fall into the categories “energy production and conversion;” “amino acid transport and metabolism;” “coenzyme transport and metabolism;” “posttranslational modification, protein turnover, and chaperones;” “signal transduction mechanisms;” “cell wall/membrane/envelope biogenesis;” and “extracellular structures.” A full set of genes for the synthesis of gas vesicles was discovered in the Bin 49 genome. All genes responsible for the anammox process were found in both genomes (Table 1).
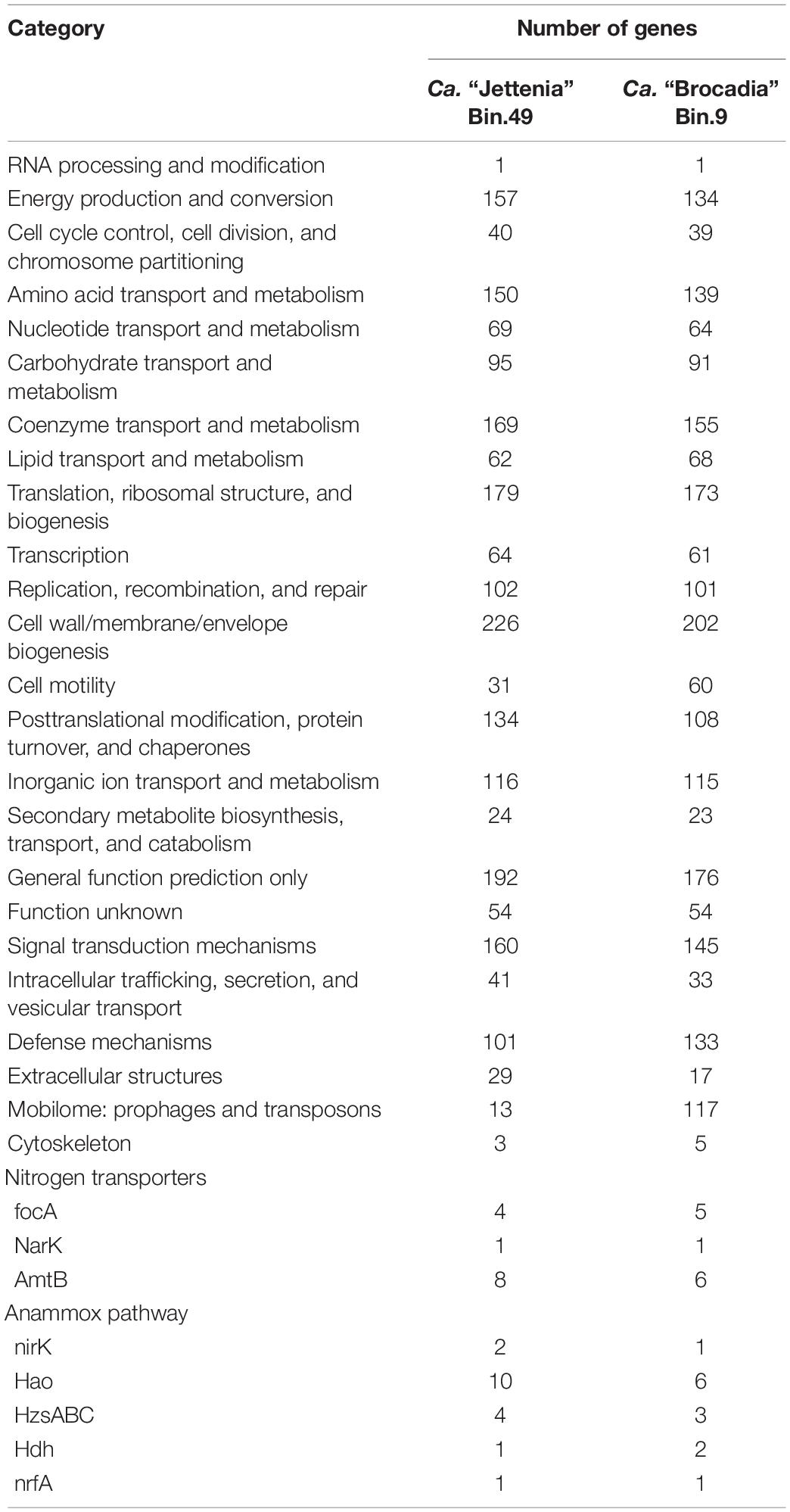
Table 1. Comparative analysis of functional gene numbers on the COG class level between Ca. “Brocadia” and Ca. “Jettenia” genomes.
Co-occurrence and Mutual Exclusion Among Various Microorganisms in the Anammox Community
The structure of microbial community is not accidental and is closely related to the functional differentiation of microorganisms in the studied ecological niche. A network analysis was carried out in order to identify the patterns of interaction between groups of microorganisms in the anammox community. The analysis made it possible to identify co-occurrence and mutual exclusion among various microorganisms in the community (Figure 9).
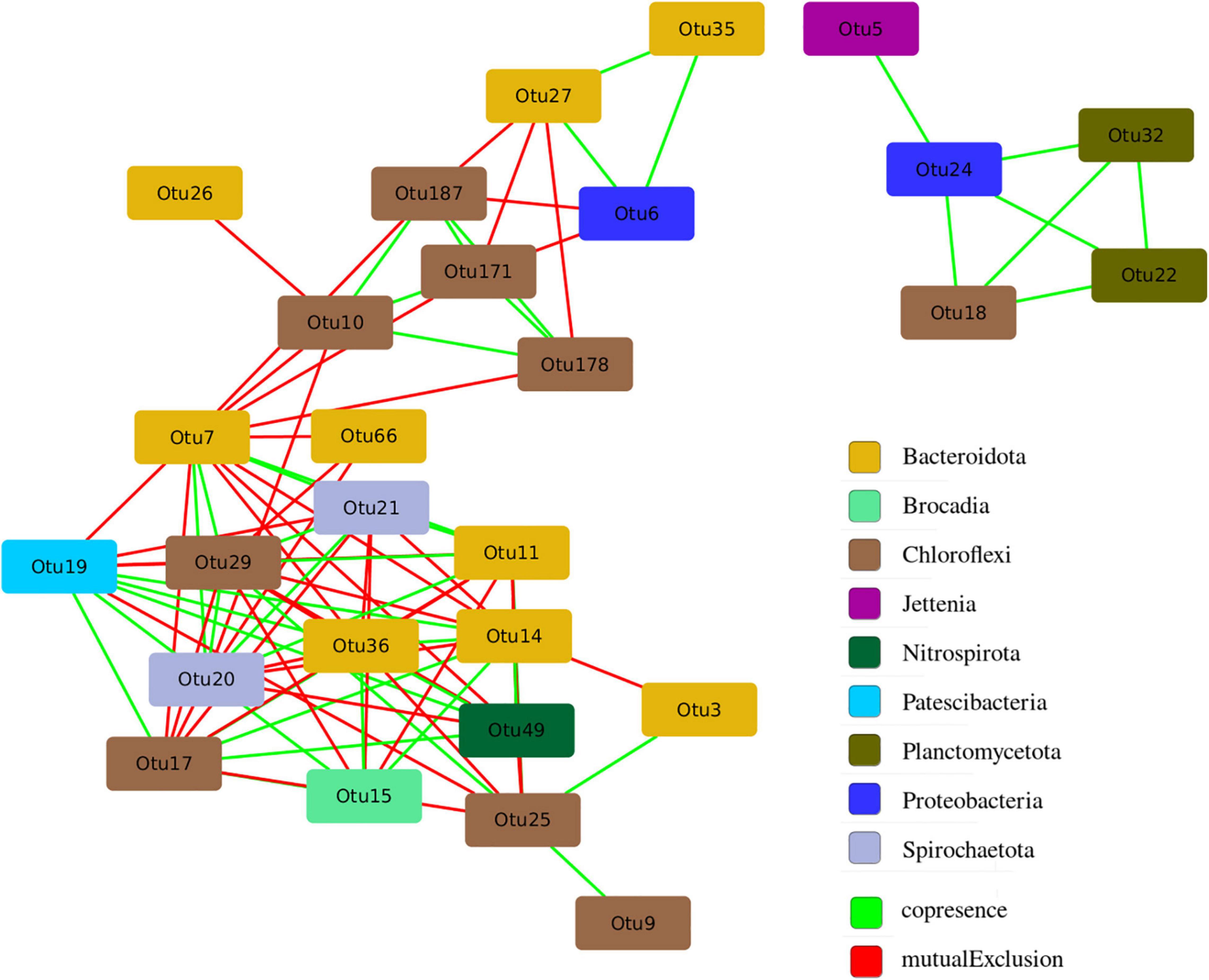
Figure 9. Co-occurrence and mutual exclusion among various microorganisms in the studied anammox community.
All of 28 OTUs with a representation of more than 2% in at least one of the studied samples had statistically significant connections with other OTUs, the maximum number of which was 14 (Supplementary Table 2). It is assumed that OTUs with a large number of connections (“degree”) are key players in the entire community (Berry and Widder, 2014). The largest number of links was found for OTU 7, related to members of the order Sphingobacteriales. The proportion of sequences related to members of the order Sphingobacteriales was significantly higher in the samples taken from glass, which indicates their key role in the formation of biofilm on the walls of the bioreactor. Among the anammox bacteria represented by the genera Ca. “Jettenia” (OTU 5) and Ca. “Brocadia” (OTU 15), the latter OTU 15 had more connections and it is included in the main hub of organisms (Figure 9). At the same time, Ca. “Brocadia” had a positive relationship with heterotrophic bacteria of the phyla Bacteroidetes (OTU 36 and OTU 14) and Chloroflexi (OTU 17) widespread in wastewater treatment plants and with the bacteria of the Patescibacteria group. For the second anammox genus Ca. “Jettenia” (OTU 5), only one positive association with the denitrifying bacterium of the genus Denitratisoma was found.
Discussion
Formate serves as an additional substrate for anammox bacteria (Kartal et al., 2007, 2008; Lawson et al., 2020) and folate being a secondary metabolite of different proteobacteria, can potentially increase the rate of nitrogen removal (Zhao et al., 2018). Thus, it was suggested that the addition of formate and folate would have a stimulatory effect on anammox bacteria, which in turn would lead to the stability of the anammox process under conditions of increased ammonium loads, i.e., it would help overcome a stress factor. A weak stimulatory effect of folate on dN was found during the first run (65–110 days). Formate did not affect the daily nitrogen removal, but its addition led to a decrease in the concentration of nitrate in the effluent compared to the control, which may indicate the progress in DNRA process by anammox bacteria (Kartal et al., 2007). Conversely, a weak stimulatory effect of formate and no effect of folate on dN were found during the second run (0–140 days). The miscellaneous influence of folate and formate on dN during the first and second runs was associated with a different effect of these compounds on the different genera of anammox bacteria that dominated in the community.
Among anammox bacteria, an absolute dominance of representatives of the genus Ca. “Brocadia” was revealed in the inoculum and the biomass on the carrier during the first run. Representatives of the genus Ca. “Brocadia” often gain an advantage over other anammox bacteria in various reactors, due to their significantly higher maximum specific growth rates (Zhang et al., 2017a,b, 2021). The highest μmax among all known anammox bacteria was exhibited by the species Ca. “B. sinica” and Ca. “Brocadia sp.40” (Zhang and Okabe, 2020). The μmax values were determined as 0.33 and 0.18 1/day for immobilized cells of anammox bacteria Ca. “B. sinica” and Ca. “J. caeni,” respectively (Zhang et al., 2017a). The additives (formate and folate) stimulated an increase in the relative abundance of sequences affiliated with representatives of the genus Ca. “Jettenia” and served as a trigger for the subsequent shift of the main anammox genus in the community. Shifts between different anammox bacteria have previously been reported, and the existence of genus- or species-specific niche differentiation and competition caused by differences in maximum specific growth rates, affinities to limiting substrates (ammonium and nitrite), susceptibility to various compounds, or VFA utilization has been suggested (Zhang et al., 2021). In the present study, the addition of formate clearly stimulated the growth of anammox bacteria of the genus Ca. “Jettenia.” However, the niche on the carrier has been already occupied by representatives of the genus Ca. “Brocadia,” and Ca. “Jettenia” failed in outcompeting them. It was suggested that an important role in further proliferation of anammox bacteria of the genus Ca. “Jettenia” in the reactor was played in terms of high adhesion capability and greater DO resistance compared with Ca. “Brocadia.” This allowed Ca. “Jettenia” to anchor and form biofilms on the inner surface of the walls of the reactor (“plastic glass” and polymethyl methacrylate). In the formate reactor, the share of representatives of the genus Ca. “Jettenia” in the biofilms on the glass reached 78% of anammox bacteria, while it did not exceed 15.5% in the biofilms on the carrier. Folate also stimulated the development of Ca. “Jettenia” on the glass, but to a lesser extent than formate. The relative abundance of the Ca. “Jettenia” on the glass was minimal in the control reactor. The results of the present study, however, contradicted the data of Ali et al. (2015) who showed that the anammox activity of Ca. “Jettenia” was inhibited by the addition of formate.
The ability of anammox bacteria of Ca. “Jettenia” to excessively produce extracellular polymeric substances (EPS) could play a key role both in their adhesion to glass and in protection from the toxic effects of DO. Aggregation and adhesion of anammox bacteria occur due to protein-rich EPS (Hou et al., 2015; Chen et al., 2019; Peeters and van Niftrik, 2019). Members of the genus Ca. “Jettenia” produce significantly more EPS proteins than Ca. “Brocadia,” Ca. “Kuenenia,” and Ca. “Scalindua” (Chen et al., 2019). Increased cell aggregation due to increased synthesis of EPS matrix in response to unfavorable environmental conditions was shown for representatives of the species Ca. “Jettenia caeni” (Guo et al., 2017). Anammox bacteria of Ca. “Jettenia,” releasing EPS in excess and enveloping cells with them, could thus protect themselves from the toxic effects of DO or other substances (Kallistova et al., 2020), which gives them an advantage over Ca. “Brocadia” and allows the spatial separation of these two genera in the same reactor. The success of Ca. “Jettenia” in the colonization of the walls of the reactors could also be due to their capability to synthesize type IV pili. It was shown that Ca. “Jettenia” served as initial colonizers, and type IV pili were responsible for the non-specific and irreversible adhesion to the surface (Xiao et al., 2021). Such ability to colonize smooth surfaces due to extra EPS production and formation of the pili could help Ca. “Jettenia” adapt to the competitive environment. In addition, biofilms on the carrier and the glass were influenced by different small-scale hydraulics, which, in turn, impacted the microbial structure and biodiversity (Niederdorfer et al., 2016). Obviously, the supply of ammonium and DO is higher in biofilms on the walls of the reactor than on the carrier; on the latter, substrate gradient and anaerobic zones exist [the stages of biofilm formation and stratification on the carrier are well described in Niederdorfer et al. (2021)]. At the same time, a better DO supply in biofilms on the walls of the reactor creates more favorable conditions for the development of stage I nitrifiers, and, accordingly, anammox bacteria on the glass are provided with nitrite better than in the biofilms on the carrier. This was confirmed by data on the relative abundances of stage I nitrifiers in biofilms on the glass and the carrier.
Anammox metabolism is supported by anoxic conditions or low oxygen concentrations (Strous et al., 1997; Egli et al., 2001; Dalsgaard et al., 2014; Cho et al., 2020). Quite a lot of studies, including transcriptome analyses, have been carried out in recent years to elucidate the effect of various DO concentrations on the activity of anammox bacteria. Most studies revealed a reversible inhibition of anammox bacteria by oxygen, and the inhibitory DO concentration varied over a wide range [reviewed by Seuntjens et al. (2018) and Cho et al. (2020)]. A complete inhibition of anammox activity both at DO < 0.12 mg/l (Strous et al., 1997; Egli et al., 2001; Oshiki et al., 2016; Seuntjens et al., 2018) and at much higher concentrations (1.0 mg/l) (Niederdorfer et al., 2021), as well as retention of activity at DO of 3–3.8 mg/l, was shown for communities enriched with Ca. “Brocadia” (Oshiki et al., 2011; Carvajal-Arroyo et al., 2013; Zekker et al., 2014). This variability could be caused by the inter-genera differences, adaptation to oxygen stress, protection by aerobic microorganisms, the existence of local hypoxic/anoxic zones, and testing procedures (Seuntjens et al., 2018 and the references therein). It has also been shown that anammox bacteria could retain some activity even under aerobic conditions, and the anammox process had a stronger tolerance to higher DO concentrations compared to the denitrification process (Wang L. et al., 2021). The mechanisms and genes involved in oxygen detoxification in anammox bacteria of genera Ca. “Brocadia” and Ca. “Kuenenia” were recently proposed (Ji et al., 2019; Yan et al., 2020; Wang H. et al., 2021). Data on the effect of DO on anammox bacteria of genus Ca. “Jettenia” are currently unavailable.
Once again, the inoculum was enriched by Ca. “Brocadia.” The addition of formate stimulated the growth of Ca. “Jettenia” in the reactor, which, due to their good adhesive capability and possibly to a higher DO resistance, anchored on the walls of the reactors being uncompetitive in the growth on the carrier in relation to Ca. “Brocadia,” so that Ca. “Brocadia” continued to prevail in biofilms on the carrier. However, after mixing the biomass from the three reactors and its storage at +4°C for 3 months, Ca. “Jettenia” became dominant (85% of anammox bacteria in the inoculum for the second run), i.e., a shift between two anammox bacteria occurred precisely at the stage of biomass storage. It can be hypothesized that the selective effect was exerted by the low storage temperature for a rather long time, to which Ca. “Jettenia” was more resistant compared with Ca. “Brocadia.” The literature data, however, do not support this hypothesis. According to previous studies, anammox bacteria belonging to these genera grow well in the temperature range of 20–45°C (Strous et al., 1999; Oshiki et al., 2011; Ali et al., 2015; Nikolaev et al., 2015; Kallistova et al., 2016), and it is representatives of the genus Ca. “Brocadia” that dominate in reactors at low temperatures, down to 5–6°C [reviewed in Tomaszewski et al. (2017) and Cho et al. (2020)]. They also maintain the activity in many freshwater environments at near 0°C, which may indicate their cold-acclimation capacity (Huo et al., 2020 and the references therein). There are little data on the effect of low temperature on anammox bacteria of the genus Ca. “Jettenia;” nevertheless, it has been shown that when the temperature drops from the optimum (35°C) to 25°C, Ca. “Brocadia” has an advantage over Ca. “Jettenia” due to the expression of more cold shock proteins and core enzymes (Huo et al., 2020).
Another selective factor that could lead to an interspecies shift in anammox bacteria is the concentration of ammonium and nitrite, the direct substrates of the anammox process. Ammonium is usually more abundant than nitrite in environments inhabited by anammox bacteria. Therefore, the availability of nitrite becomes more relevant to the niche differentiation among anammox bacteria (Zhang and Okabe, 2020). According to the Michaelis–Menten kinetics, a higher half-saturation constant (Ks), a lower affinity to substrate, and a higher substrate concentration are needed to reach maximum growth rate. Thus, Ks is the key parameter for microbial growth and consequent niche differentiation. Anammox bacteria of the species Ca. “B. caroliniensis” and Ca. “B. fulgida” were shown to have the highest Ks values among anammox bacteria (including Ca. “Jettenia caeni”) for both NH4+ and NO2– (Oshiki et al., 2016; Zhang and Okabe, 2020). This means that these Ca. “Brocadia” species have the lowest affinity to substrate among anammox bacteria and therefore prefer high substrate concentrations. It was found in experiments with three enrichment cultures of anammox bacteria (Ca. “Brocadia,” Ca. “Jettenia,” and Ca. “Kuenenia”) that Ca. “Jettenia” could proliferate only at low nitrogen loading rates (NLRs), i.e., low influent ammonium and nitrite concentrations, whereas Ca. “Brocadia” outcompeted the other two species at higher NLRs. Spatial distribution of Ca. “Jettenia” cells in the inner part of biofilms allowed authors to suggest that Ca. “Jettenia” prefers low-nitrite environments (Zhang et al., 2017b). An intrinsic growth kinetics difference between Ca. “Jettenia” and Ca. “Brocadia” can lead to the proliferation and domination of Ca. “Jettenia” under low NLRs in the presence of acetate (Zhang et al., 2017a,b, 2021). Indeed, a half-saturation constant (Ks) for NH4 in three out of five known Ca. “Brocadia” species is higher than in Ca. “J. caeni” (Oshiki et al., 2016 and the references therein). The observed population shift from Ca. “Brocadia” to Ca. “Kuenenia” was suggested to be caused by the higher affinity for nitrite of representatives of Ca. “Kuenenia” (van der Star et al., 2008) as they have a much lower value of Ks for NO2 than Ca. “Brocadia” (Ding et al., 2013; Oshiki et al., 2016 and the references therein). However, in respect of Ks for NO2, the situation with Ca. “Brocadia” and Ca. “Jettenia” species is not so clear, as three out of five Ca. “Brocadia” species have Ks values for NO2 much lower or comparable to those of Ca. “J. caeni” (Oshiki et al., 2016 and the references therein). However, the value of Ks for NO2 for Ca. “Brocadia sinica” was shown to depend on the type of biomass. It is much lower for biomass in the form of planktonic cells than in the form of homogenized aggregates (Oshiki et al., 2013). It should be noted that there are anammox bacteria (e.g., Ca. “Kuenenia” and Ca. “Scalindua”) that have a higher affinity to the substrate than representatives of the genus Ca. “Jettenia” (Oshiki et al., 2016; Zhang and Okabe, 2020), and therefore, they could outcompete Ca. “Jettenia” under conditions of limited nitrogenous substrates (Xiao et al., 2021).
In the system we studied, the 16S rRNA gene sequence of Ca. “Brocadia” had 100% identity to those of Ca. “Brocadia fulgida” with the highest Ks value reported; thus, it should have an advantage at high substrate concentrations. On the contrary, Ca. “Jettenia” should potentially be more resistant than Ca. “Brocadia” to substrate starvation during long-term biomass storage. Then, there is a probability that most of Ca. “Brocadia” in the biomass died off during the storage with a deficiency of ammonium and nitrite in turn, and after being exposed to favorable conditions, Ca. “Jettenia” took advantage of this, enriched quickly, and further served as an inoculum for the second run. The results of the second run can confirm that Ca. “Brocadia” prefers higher ammonium concentrations than Ca. “Jettenia,” as a quick increase in the proportion of Ca. “Brocadia” and a decrease in the abundance of Ca. “Jettenia” were observed in all reactors after the ammonium load was increased from 177.8 to 444.4 mg N–NH4/l/day (Figure 7A). The opposite results, however, were demonstrated by Zhao et al. (2019), who proposed that Ca. “Brocadia” out-competed Ca. “Jettenia” under low nitrogen concentrations. The authors found that Ca. “Brocadia” genomes encoded the complete pathway for bacterial chemotaxis and flagellar assembly, a more complete two-component signal transduction system, and more redundant marker genes of nitrite reductase compared with Ca. “Jettenia” genomes. Altogether, this led to an advantage of Ca. “Brocadia” over Ca. “Jettenia” at low nitrogen concentrations, as they could improve the access to higher substrate concentrations by movement, they could sense and respond to nitrogen limitation by inducing changes in transcription, and they could adapt quickly and maintain the nitrogen metabolism stability under nitrite limitation. The higher competitiveness for nitrite of Ca. “Brocadia” led to its higher transcript activity and higher nitrogen removal rate (Zhao et al., 2019).
The selection of anammox bacteria species can also be influenced by other microorganisms of the community. Interspecies relationships based on spatial and substrate synergism and competition are established between microorganisms closely coexisting in anammox biofilms. Intraspecific interactions also take place according to the principle of quorum sensing and cross-feeding (Ding et al., 2013; Lawson et al., 2017; Zhao et al., 2018; Tang et al., 2019; Kuenen, 2020). For example, an exchange with secondary metabolites, contributing to EPS production and degradation, and thus to aggregation, and participation in nitrite loop were found between anammox bacteria and bacteria of the phyla Armatimonadetes, Proteobacteria, Chloroflexi, and Chlorobi (Zhao et al., 2018), as well as co-metabolism with Patescibacteria group (Hosokawa et al., 2021). It is assumed that one of the possible ecological roles of the bacteria of the Patescibacteria group in the anammox reactor is to provide lactate and formate to other coexisting bacteria, supporting their growth (Hosokawa et al., 2021). All these bacteria along with other typical members of the anammox communities were found in the present study. A comparison of the obtained sequences with those of culturable microorganisms with known functions in the community as well as with unculturable representatives for which metagenomics and metatranscriptomic data are available revealed bacteria that are directly involved in the nitrogen cycle, e.g., genera Nitrosomonas (stage I nitrifiers), Nitrospira (stage II nitrifiers), Comamonas, Thauera, and Denitratisoma (denitrifying bacteria). However, the vast majority of bacteria found were organotrophs. Their role in the anammox community most likely consists in the decomposition of dead cell biomass of activated sludge and EPS formed by anammox bacteria (Mardanov et al., 2016). Besides, according to metagenomics and metatranscriptomic studies available in the literature, most high-abundance heterotrophic members of the community detected (e.g., Chloroflexi, Ignavibacteria, and Armatimonadetes) have genes encoding different enzymes of nitrogen metabolism (Mardanov et al., 2019b). The network analysis also revealed the largest number of links for bacteria related to members of the order Sphingobacteriales. Members of this order are widespread in wastewater treatment bioreactors (Hu et al., 2012), especially in MBR-type bioreactors (Saunders et al., 2013; Zha et al., 2020). They produce glycosphingolipids, which provide a supportive and protective environment for biofilm dwellers (Al Ashhab et al., 2014). At the same time, the overall composition of the bacterial community remained relatively stable despite the supply of additives, the increase in the ammonium load, and the storage of biomass. Thus, it is not yet possible to link the effect of other microorganisms of the community to the prevalence of a particular anammox species.
The comparative genome analysis performed in this study allows one to speculate on the survival strategies of Ca. “Brocadia” and Ca. “Jettenia” spp., which can help explain the observed interspecies shift. The Ca. “Brocadia” genome encodes a more complete system of cellular chemotaxis and flagellar assembly than the Ca. “Jettenia” genome, which is consistent with the data of Zhao et al. (2019), who showed that all these genes were expressed at the RNA level. Mobility and chemotaxis functions allow members of the genus Ca. “Brocadia” to effectively navigate in a new environment and quickly occupy the most favorable niche. In addition, the large representation of genes for defense mechanisms, as well as phages and transposons, indicates that Ca. “Brocadia” encounters a large number of threats from other organisms of the community and can effectively compete with them. The strategy of Ca. “Brocadia” when environmental conditions change is to, if possible, “escape” from adverse effects, for example, to move into the inner layers of the microbial biofilm. If it is impossible to escape, then Ca. “Brocadia” are likely to die. The Ca. “Jettenia” immobility forces it to better adapt to environmental fluctuations, and on the spot, when it faces an adverse impact, it more or less successfully develops a suitable defense mechanism for the situation. This confirms the greater representation of the genes of categories “cell wall/membrane/envelope biogenesis” and “extracellular structures” in the Ca. “Jettenia” genome than in the Ca. “Brocadia” genome, which is consistent with the assumption that Ca. “Jettenia” can protect itself by the reinforcement of the outer wall, including the extra EPS formation, which not only improves its adhesion to various surfaces but also protects against increased concentrations of adverse substances (e.g., DO, nitrite, and ammonium). In addition, the Ca. “Jettenia” genome revealed a higher abundance of genes for energy transformation and catabolism (categories “energy production and conversion,” “amino acid transport and metabolism,” and “coenzyme transport and metabolism”) and for signal transduction and cell protection (categories “posttranslational modification, protein turnover, and chaperones” and “signal transduction mechanisms”) than the Ca. “Brocadia” genome. This may contribute to the survival of members of the genus Ca. “Jettenia” during the long-term low-temperature storage under starvation. Genes encoding the synthesis of gas vesicles were also discovered in the genome of Ca. “Jettenia.” Although gas vesicles are often regulated by light (Speth et al., 2017), it can be assumed that their formation can stabilize the position of Ca. “Jettenia” cells during biofilm formation on the walls of the reactors and can also protect them from overheating. So, the anammox component of the microbial community evidently exhibits high lability in dependence on the set of environmental conditions (Mardanov et al., 2016).
Conclusion
From the results of the study, the following conclusions can be inferred:
• The addition of formate stimulates the growth of Ca. “Jettenia” spp. in the anammox community dominated by Ca. “Brocadia” spp. Abundant EPS production, which improves cell adhesion to various surfaces and protects the cell against increased concentrations of adverse compounds, could aid Ca. “Jettenia” spp. to gain a foothold on the wall of the reactor and build up biomass. Therefore, a spatial separation of competing anammox species is achieved within the same reactor.
• The higher resistance of Ca. “Jettenia” spp. to the starvation during biomass storage gives it an advantage over Ca. “Brocadia” spp. Altogether, this leads to an interspecies shift from Ca. “Brocadia” to Ca. “Jettenia” in the anammox community along with general sustainability of the anammox process, when the cultivation conditions (additives, load, and storage) change.
• Under the conditions of increased ammonium load, folate has a positive effect on dN in Ca. “Brocadia”-enriched community, and formate has a positive effect on dN in Ca. “Jettenia”-enriched community.
• Genome analysis suggests that Ca. “Brocadia” has the ability to quickly occupy the most favorable niche; it is competitive with other microorganisms of the community and escapes, if possible, from the adverse effects. The immobility of Ca. “Jettenia” forces it to better adapt to environmental fluctuations, and when faced with an adverse impact, it activates suitable defense mechanisms for adaptation and survival.
Data Availability Statement
Raw 16S sequences can be found on the NCBI BioProject accession PRJNA556270.
Author Contributions
AK, YN, and NP designed the study. VG, AD, JB, and AP performed the assembly, reactors maintenance, sampling, and chemical analyses. AB, EG, VK, NR, and AM performed the molecular analyses. AK analyzed the data and wrote the original draft of the manuscript. IZ performed the review and edited the manuscript. All authors have read and approved the submitted version.
Funding
The reactors construction and operation, chemical analyses, and 16S rRNA sequencing were supported by the Russian Foundation for Basic Research (Grant No. 18-29-08008) and the Ministry of Science and Higher Education of the Russian Federation (goszadanie). Metagenomic analysis was supported by the Russian Science Foundation (Grant No. 21-64-00019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.802201/full#supplementary-material
References
Al Ashhab, A., Herzberg, M., and Gillor, O. (2014). Biofouling of reverse-osmosis membranes during tertiary wastewater desalination: microbial community composition. Water Res. 50, 341–349. doi: 10.1016/j.watres.2013.10.044
Ali, M., Oshiki, M., Awata, T., Isobe, K., Kimura, Z., Yoshikawa, H., et al. (2015). Physiological characterization of anaerobic ammonium oxidizing bacterium ‘Candidatus Jettenia caeni’. Environ. Microbiol. 17, 2172–2189. doi: 10.1111/1462-2920.12674
Ali, M., Shaw, D. R., Zhang, L., Haroon, M. F., Narita, Y., Emwas, A. H., et al. (2018). Aggregation ability of three phylogenetically distant anammox bacterial species. Water Res. 143, 10–18. doi: 10.1016/j.watres.2018.06.007
Alneberg, J., Bjarnason, B. S., de Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barberán, A., Ramirez, K. S., Leff, J. W., Bradford, M. A., Wall, D. H., and Fierer, N. (2014). Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 17, 794–802. doi: 10.1111/ele.12282
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Bi, Z., Zhang, W., Ni, M., Song, G., and Huang, Y. (2020). Fe0/Fe2+-Dependent nitrate reduction in anammox consortia questions the enzymatic mechanism of nitrate reduction by anammox bacteria. ACS Sustainable Chem. Eng. 8, 15278–15287. doi: 10.1021/acssuschemeng.0c05152
Bowers, R. M., Kyrpides, N. C., Stepanauskas, R., Harmon-Smith, M., Doud, D., Reddy, T. B. K., et al. (2017). Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731. doi: 10.1038/nbt.3893
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carvajal-Arroyo, J. M., Sun, W., Sierra-Alvarez, R., and Field, J. A. (2013). Inhibition of anaerobic ammonium oxidizing (anammox) enrichment cultures by substrates, metabolites and common wastewater constituents. Chemosphere 91, 22–27. doi: 10.1016/j.chemosphere.2012.11.025
Chen, Z., Meng, Y., Sheng, B., Zhou, Z., Jin, C., and Meng, F. (2019). Linking exoproteome function and structure to anammox biofilm development. Environ. Sci. Technol. 53, 1490-1500. doi: 10.1021/acs.est.8b04397
Cho, S., Kambey, C., and Nguyen, V. K. (2020). Performance of anammox processes for wastewater treatment: a critical review on effects of operational conditions and environmental stresses. Water 12:20. doi: 10.3390/w12010020
Dalsgaard, T., Stewart, F. J., Thamdrup, B., De Brabandere, L., Revsbech, N. P., Ulloa, O., et al. (2014). Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off northern Chile. mBio 5:e01966. doi: 10.1128/mBio.01966-14
Ding, S., Zheng, P., Lu, H., Chen, J., Mahmood, Q., and Abbas, G. (2013). Ecological characteristics of anaerobic ammonia oxidizing bacteria. Appl. Microbiol. Biotechnol. 97, 1841–1849. doi: 10.1007/s00253-013-4697-0
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Egli, K., Fanger, U., Alvarez, P., Siegrist, H., van der Meer, J., and Zehnder, A. (2001). Enrichment and characterization of an anammox bacterium from a rotating biological contactor treating ammonium-rich leachate. Arch. Microbiol. 175, 198–207. doi: 10.1007/s002030100255
Faust, K., and Raes, J. (2016). CoNet app: inference of biological association networks using Cytoscape. F1000Res. 5:1519. doi: 10.12688/f1000research.9050.2
Feng, Y., Zhao, Y., Jiang, B., Zhao, H., Wang, Q., and Liu, S. (2019). Discrepant gene functional potential and crossfeedings of anammox bacteria Ca. Jettenia caeni and Ca. Brocadia sinica in response to acetate. Water Res. 165:114974. doi: 10.1016/j.watres.2019.114974
Frey, B., Rime, T., Phillips, M., Stierli, B., Hajdas, I., Widmer, F., et al. (2016). Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 92:fiw018. doi: 10.1093/femsec/fiw018
Guo, Y., Liu, S., Tang, X., and Yang, F. (2017). Role of c-di-GMP in anammox aggregation and systematic analysis of its turnover protein in Candidatus Jettenia caeni. Water Res. 113, 181-190. doi: 10.1016/j.watres.2017.02.018
Hosokawa, S., Kuroda, K., Narihiro, T., Aoi, Y., Ozaki, N., Ohashi, A., et al. (2021). Cometabolism of the superphylum Patescibacteria with anammox bacteria in a long-term freshwater anammox column reactor. Water 13:208. doi: 10.3390/w13020208
Hou, X., Liu, S., and Zhang, Z. (2015). Role of extracellular polymeric substance in determining the high aggregation ability of anammox sludge. Water Res. 75, 51–62. doi: 10.1016/j.watres.2015.02.031
Hu, M., Wang, X., Wen, X., and Xia, Y. (2012). Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour. Technol. 117, 72–79. doi: 10.1016/j.biortech.2012.04.061
Huo, T., Zhao, Y., Tang, X., Zhao, H., Ni, S., Gao, Q., et al. (2020). Metabolic acclimation of anammox consortia to decreased temperature. Environ. Int. 143:105915. doi: 10.1016/j.envint.2020.105915
Jetten, M. S., van Niftrik, L., Strous, M., Kartal, B., Keltjens, J. T., and Op den Camp, H. J. (2009). Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 44, 65–84. doi: 10.1080/10409230902722783
Ji, X., Wu, Z., Sung, S., and Lee, P. H. (2019). Metagenomics and metatranscriptomics analyses reveal oxygen detoxification and mixotrophic potentials of an enriched anammox culture in a continuous stirred-tank reactor. Water Res. 166:115039. doi: 10.1016/j.watres.2019.115039
Ji, Y.-X., and Jin, R.-C. (2014). Effect of different preservation conditions on the reactivation performance of anammox sludge. Sep. Purif. Technol. 133, 32–39. doi: 10.1016/j.seppur.2014.06.029
Joshi, N. A., and Fass, J. N. (2011). Sickle: A sliding-Window, Adaptive, Quality-Based Trimming Tool For FastQ files (Version 1.33) [Software]. Available online at https://github.com/najoshi/sickle (accessed May 15, 2020).
Kallistova, A. Y., Dorofeev, A. G., Nikolaev, Y. A., Kozlov, M. N., Kevbrina, M. V., and Pimenov, N. V. (2016). Role of anammox bacteria in removal of nitrogen compounds from wastewater. Microbiology 85, 140–156. doi: 10.1134/S0026261716020089
Kallistova, A. Y., Nikolaev, Y. A., Mardanov, A. V., Berestovskaya, Y. Y., Grachev, V. A., Kostrikina, N. A., et al. (2020). Investigation of formation and development of anammox biofilms by light, epifluorescence, and electron microscopy. Microbiology 89, 708–719. doi: 10.1134/S0026261720060077
Kang, D. D., Froula, J., Egan, R., and Wang, Z. (2015). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165
Kartal, B., de Almeida, N. M., Maalcke, W. J., Op den Camp, H. J., Jetten, M. S., and Keltjens, J. T. (2013). How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 37, 428–461. doi: 10.1111/1574-6976.12014
Kartal, B., Kuypers, M. M., Lavik, G., Schalk, J., Op den Camp, H. J. M., Jetten, M. S. M., et al. (2007). Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 9, 635–642. doi: 10.1111/j.1462-2920.2006.01183.x
Kartal, B., van Niftrik, L., Rattray, J., van de Vossenberg, J. L., Schmid, M. C., Sinninghe Damsté, J., et al. (2008). Candidatus “Brocadia fulgida”: an autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol. 63, 46–55. doi: 10.1111/j.1574-6941.2007.00408.x
Kartal, B., van Niftrik, L., Sliekers, O., Schmid, M. C., Schmidt, I., van de Pas-Schoonen, K., et al. (2004). Application, eco-physiology and biodiversity of anaerobic ammonium-oxidizing bacteria. Rev. Environ. Sci. Biotechnol. 3, 255–264. doi: 10.1007/s11157-004-7247-5
Kuenen, J. G. (2020). Anammox and beyond. Environ. Microbiol. 22, 525–536. doi: 10.1111/1462-2920.14904
Lackner, S., Gilbert, E. M., Vlaeminck, S. E., Joss, A., Horn, H., and van Loosdrecht, M. C. M. (2014). Full-scale partial nitritation/anammox experiences - an application survey. Water Res. 55, 292–303. doi: 10.1016/j.watres.2014.02.032
Langfelder, P., and Horvath, S. (2012). Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 46:i11.
Lawson, C. E., Nuijten, G. H. L., de Graaf, R. M., Jacobson, T. B., Pabst, M., Stevenson, D. M., et al. (2020). Autotrophic and mixotrophic metabolism of an anammox bacterium revealed by in vivo 13C and 2H metabolic network mapping. ISME J. 15, 673–687. doi: 10.1038/s41396-020-00805-w
Lawson, C. E., Wu, S., Bhattacharjee, A. S., Hamilton, J. J., McMahon, K. D., Goel, R., et al. (2017). Metabolic network analysis reveals microbial community interactions in anammox granules. Nat. Commun. 8:15416. doi: 10.1038/ncomms15416
Leduc, S., de Troostembergh, J. C., and Lebeault, J. M. (2004). Folate requirements of the 2-keto-L-gulonic acid-producing strain Ketogulonigenium vulgare LMP P-20356 in L-sorbose/CSL medium. Appl. Microbiol. Biotechnol. 65, 163–167. doi: 10.1007/s00253-004-1562-1
Li, J., Li, J., Peng, Y., Wang, S., Zhang, L., Yang, S., et al. (2020). Insight into the impacts of organics on anammox and their potential linking to system performance of sewage partial nitrification-anammox (PN/A): a critical review. Bioresour. Technol. 300:122655. doi: 10.1016/j.biortech.2019.122655
Luo, X., and Bhattacharya, C. B. (2006). Corporate social responsibility, customer satisfaction, and market value. J. Mark. 70, 1–18. doi: 10.1016/j.ridd.2012.05.006
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mardanov, A. V., Beletskii, A. V., Kallistova, A. Y., Kotlyarov, R. Y., Nikolaev, Y. A., Kevbrina, M. V., et al. (2016). Dynamics of the composition of a microbial consortium during start-up of a single-stage constant flow laboratory nitritation/anammox setup. Microbiology 85, 681–692. doi: 10.1134/S002626171606014X
Mardanov, A. V., Kotlyarov, R. Y., Beletsky, A. V., Nikolaev, Y. A., Kallistova, A. Y., Grachev, V. A., et al. (2019a). Metagenomic data of the microbial community of lab-scale nitritation-anammox sequencing-batch bioreactor performing nitrogen removal from synthetic wastewater. Data Brief 27:104722. doi: 10.1016/j.dib.2019.104722
Mardanov, A. V., Beletsky, A. V., Ravin, N. V., Botchkova, E. A., Litti, Y. V., and Nozhevnikova, A. N. (2019b). Genome of a novel bacterium “Candidatus Jettenia ecosi” reconstructed from the metagenome of an anammox bioreactor. Front. Microbiol. 10:2442. doi: 10.3389/fmicb.2019.02442
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Niederdorfer, R., Fragner, L., Yuan, L., Hausherr, D., Wei, J., Magyar, P., et al. (2021). Distinct growth stages controlled by the interplay of deterministic and stochastic processes in functional anammox biofilms. Water Res. 200:117225. doi: 10.1016/j.watres.2021.117225
Niederdorfer, R., Peter, H., and Battin, T. J. (2016). Attached biofilms and suspended aggregates are distinct microbial lifestyles emanating from differing hydraulics. Nat. Microbiol. 1:16178. doi: 10.1038/nmicrobiol.2016.178
Nikolaev, Y. A., Kozlov, M. N., Kevbrina, M. V., Dorofeev, A. G., Pimenov, N. V., Kallistova, A. Y., et al. (2015). Candidatus “Jettenia moscovienalis” sp. nov., a new species of bacteria carrying out anaerobic ammonium oxidation. Microbiology 84, 256–262. doi: 10.1134/S0026261715020101
Oshiki, M., Awata, T., Kindaichi, T., Satoh, H., and Okabe, S. (2013). Cultivation of planktonic anaerobic ammonium oxidation (anammox) bacteria using membrane bioreactor. Microbes Environ. 28, 436–443. doi: 10.1264/jsme2.me13077
Oshiki, M., Satoh, H., and Okabe, S. (2016). Ecology and physiology of anaerobic ammonium oxidizing bacteria. Environ. Microbiol. 18, 2784–2796. doi: 10.1111/1462-2920.13134
Oshiki, M., Shimokawa, M., Fujii, N., Satoh, H., and Okabe, S. (2011). Physiological characteristics of the anaerobic ammonium-oxidizing bacterium Candidatus “Brocadia sinica”. Microbiology 157, 1706–1713. doi: 10.1099/mic.0.048595-0
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Peeters, S. H., and van Niftrik, L. (2019). Trending topics and open questions in anaerobic ammonium oxidation. Curr. Opin. Chem. Biol 49, 45–52. doi: 10.1016/j.cbpa.2018.09.022
Quan, Z. X., Rhee, S. K., Zuo, J. E., Yang, Y., Bae, J. W., Park, J. R., et al. (2008). Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ. Microbiol. 10, 3130–3139. doi: 10.1111/j.1462-2920.2008.01642.x
Rice, E. W., and Bridgewater, L. (eds). (2012). Standard Methods for the Examination of Water and Wastewater, 22nd Edn. Washington, D.C: American Public Health Association.
Saunders, A. M., Larsen, P., and Nielsen, P. H. (2013). Comparison of nutrient-removing microbial communities in activated sludge from full-scale MBRs and conventional plants. Water Sci. Technol. 68, 366–371. doi: 10.2166/wst.2013.183
Seuntjens, D., Carvajal-Arroyo, J. M., Ruopp, M., Bunse, P., De Mulder, C. P., Lochmatter, S., et al. (2018). High-resolution mapping and modeling of anammox recovery from recurrent oxygen exposure. Water Res. 144, 522–531. doi: 10.1016/j.watres.2018.07.024
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sonthiphand, P., and Neufeld, J. D. (2013). Evaluating primers for profiling anaerobic ammonia oxidizing bacteria within freshwater environments. PLoS One 8:e57242. doi: 10.1371/journal.pone.0057242
Speth, D. R., Lagkouvardos, I., Wang, Y., Qian, P. Y., Dutilh, B. E., and Jetten, M. S. (2017). Draft genome of Scalindua rubra, obtained from the interface above the discovery deep brine in the Red Sea, sheds light on potential salt adaptation strategies in anammox bacteria. Microb. Ecol. 74, 1–5. doi: 10.1007/s00248-017-0929-7
Strous, M., Heijnen, J. J., Kuenen, J. G., and Jetten, M. S. M. (1998). The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50, 589–596. doi: 10.1007/s002530051340
Strous, M., Kuenen, J. G., and Jetten, M. S. M. (1999). Key physiology of anaerobic ammonium oxidation. Appl. Microbiol. Biotechnol. 65, 3248–3250. doi: 10.1128/AEM.65.7.3248-3250.1999
Strous, M., Van Gerven, E., Kuenen, J. G., and Jetten, M. (1997). Effects of aerobic and microaerobic conditions on anaerobic ammonium-oxidizing (anammox) sludge. Appl. Environ. Microbiol. 63, 2446–2448. doi: 10.1128/aem.63.6.2446-2448.1997
Tang, X., Guo, Y., Zhu, T., Tao, H., and Liu, S. (2019). Identification of quorum sensing signal AHLs synthases in Candidatus Jettenia caeni and their roles in anammox activity. Chemosphere 225, 608–617. doi: 10.1016/j.chemosphere.2019.02.192
Tomaszewski, M., Cema, G., and Ziembińska-Buczyńska, A. (2017). Influence of temperature and pH on the anammox process: a review and meta-analysis. Chemosphere 182, 203–214. doi: 10.1016/j.chemosphere.2017.05.003
van der Star, W. R. L., Miclea, A. I., van Dongen, U. G. J. M., Muyzer, G., Picioreanu, C., and van Loosdrecht, M. C. M. (2008). The membrane bioreactor: a novel tool to grow anammox bacteria as free cells. Biotechnol. Bioeng. 101, 286–294. doi: 10.1002/bit.21891
Wang, H., Yan, Y., Zhang, L., and Wang, Y. (2021). Response of antioxidant defense to oxidative stress induced by H2O2 and NO in anammox bacteria. Chemosphere 282:131008. doi: 10.1016/j.chemosphere.2021.131008
Wang, J. P., Liu, Y. D., Meng, F. G., and Li, W. (2020). The short and long-term effects of formic acid on rapid nitritation start-up. Environ. Int. 135:105350. doi: 10.1016/j.envint.2019.105350
Wang, L., Hong, Y., Gu, J.-D., Wu, J., Yan, J., and Lin, J.-G. (2021). Influence of critical factors on nitrogen removal contribution by anammox and denitrification in an anammox-inoculated wastewater treatment system. Water Process Eng. 40:101868. doi: 10.1016/j.jwpe.2020.101868
Wang, Y., Hu, X., Jiang, B., Song, Z., and Ma, Y. (2016). Symbiotic relationship analysis of predominant bacteria in a lab-scale anammox UASB bioreactor. Environ. Sci. Pollut. Res. Int. 23, 7615–7626. doi: 10.1007/s11356-015-6016-z
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3:e000132. doi: 10.1099/mgen.0.000132
Xiao, R., Zhu, W., Xu, S., Chai, W., Tong, Y., Zheng, P., et al. (2021). Low strength wastewater anammox start-up by stepwise decrement in influent nitrogen: biofilm formation mechanism and mathematical modelling. Environ Int. 158:106929. doi: 10.1016/j.envint.2021.106929
Yan, Y., Wang, W., Wu, M., Jetten, M. S. M., Guo, J., Ma, J., et al. (2020). Transcriptomics uncovers the response of anammox bacteria to dissolved oxygen inhibition and the subsequent recovery mechanism. Environ. Sci. Technol. 54, 14674–14685. doi: 10.1021/acs.est.0c02842
Yin, X., Zhai, J., Hu, W., Li, Y., Rahaman, M. H., and Mąkinia, J. (2019). A fast start-up of the organotrophic anammox process inoculated with constructed wetland sediment. Ecol. Eng. 138, 454–460. doi: 10.1016/j.ecoleng.2019.07.039
Zekker, I., Rikmann, E., Tenno, T., Seiman, A., Loorits, L., Kroon, K., et al. (2014). Nitritating-anammox biomass tolerant to high dissolved oxygen concentration and C/N ratio in treatment of yeast factory wastewater. Environ Technol. 35, 1565–1576. doi: 10.1080/09593330.2013.874492
Zha, X., Ma, J., and Lu, X. (2020). Use of a low-cost and energy-efficient device for treating low-strength wastewater at low temperatures focusing on nitrogen removal and microbial community. Sci. Total Environ. 722:137916. doi: 10.1016/j.scitotenv.2020.137916
Zhang, L., and Okabe, S. (2020). Ecological niche differentiation among anammox bacteria. Water Res. 171:115468. doi: 10.1016/j.watres.2020.115468
Zhang, L., Narita, Y., Gao, L., Ali, M., Oshiki, M., and Okabe, S. (2017a). Maximum specific growth rate of anammox bacteria revisited. Water Res. 116, 296–303. doi: 10.1016/j.watres.2017.03.027
Zhang, L., Narita, Y., Gao, L., Ali, M., Oshiki, M., Ishii, S., et al. (2017b). Microbial competition among anammox bacteria in nitrite-limited bioreactors. Water Res. 125, 249–258. doi: 10.1016/j.watres.2017.08.052
Zhang, S., Zhang, Z., Xia, S., Ding, N., Liao, X., Yang, R., et al. (2021). The potential contributions to organic carbon utilization in a stable acetate-fed Anammox process under low nitrogen-loading rates. Sci. Total Environ. 784:147150. doi: 10.1016/j.scitotenv.2021.147150
Zhao, Y., Feng, Y., Chen, L., Niu, Z., and Liu, S. (2019). Genome-centered omics insight into the competition and niche differentiation of Ca. Jettenia and Ca. Brocadia affiliated to anammox bacteria. Appl. Microbiol. Biotechnol. 103, 8191–8202. doi: 10.1007/s00253-019-10040-9
Keywords: anammox bacteria, Candidatus “Brocadia”, Candidatus “Jettenia”, formate, folate, interspecies shift
Citation: Kallistova A, Nikolaev Y, Grachev V, Beletsky A, Gruzdev E, Kadnikov V, Dorofeev A, Berestovskaya J, Pelevina A, Zekker I, Ravin N, Pimenov N and Mardanov A (2022) New Insight Into the Interspecies Shift of Anammox Bacteria Ca. “Brocadia” and Ca. “Jettenia” in Reactors Fed With Formate and Folate. Front. Microbiol. 12:802201. doi: 10.3389/fmicb.2021.802201
Received: 26 October 2021; Accepted: 02 December 2021;
Published: 03 February 2022.
Edited by:
Xin Wang, Nankai University, ChinaCopyright © 2022 Kallistova, Nikolaev, Grachev, Beletsky, Gruzdev, Kadnikov, Dorofeev, Berestovskaya, Pelevina, Zekker, Ravin, Pimenov and Mardanov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Kallistova, a2FsbGlzdG9hbm5hQG1haWwucnU=
 Anna Kallistova
Anna Kallistova Yury Nikolaev1
Yury Nikolaev1 Alexey Beletsky
Alexey Beletsky Evgeny Gruzdev
Evgeny Gruzdev Vitaly Kadnikov
Vitaly Kadnikov Alexander Dorofeev
Alexander Dorofeev Julia Berestovskaya
Julia Berestovskaya Ivar Zekker
Ivar Zekker Nikolai Ravin
Nikolai Ravin Nikolai Pimenov
Nikolai Pimenov Andrey Mardanov
Andrey Mardanov