- 1College of Plant Health and Medicine, Qingdao Agricultural University, Qingdao, China
- 2Engineering Research Center of Fruit and Vegetable Pest Precise Control of Qingdao, Qingdao, China
Gray mold caused by Botrytis cinerea is a devastating disease that leads to huge economic losses worldwide. Autophagy is an evolutionarily conserved process that maintains intracellular homeostasis through self-eating. In this study, we identified and characterized the biological function of the autophagy-related protein Atg6 in B. cinerea. Targeted deletion of the BcATG6 gene showed block of autophagy and several phenotypic defects in aspects of mycelial growth, conidiation, sclerotial formation and virulence. All of the phenotypic defects were restored by targeted gene complementation. Taken together, these results suggest that BcAtg6 plays important roles in the regulation of various cellular processes in B. cinerea.
Introduction
Botrytis cinerea (teleomorph Botryotinia fuckeliana) is a necrotrophic ascomycete fungus that causes serious pre- and postharvest crop losses worldwide to a large scope of plant species such as vegetables, fruits and ornamentals (Williamson et al., 2007; Dean et al., 2012). Due to the lack of resistant varieties, chemical control remains the most effective strategy for gray mold management, but many types of fungicides have failed by the genetic plasticity of B. cinerea (Williamson et al., 2007; Hu et al., 2016). Therefore, understanding the molecular mechanisms underlying development and virulence of B. cinerea will contribute to establish more effective disease control strategies. Moreover, B. cinerea has become a model fungus for molecular study of necrotrophic fungi (Tudzynski and Kokkelink, 2009).
Autophagy is a cellular degradation pathway for coping with nutritional stress and balancing energy sources in the process of cell differentiation (Glick et al., 2010). Autophagy also plays a housekeeping role in removing misfolded or aggregated proteins, clearing damaged organelles (such as endoplasmic reticulum, mitochondria and peroxisomes), and eliminating intracellular pathogens (Mizushima, 2007; Levine and Kroemer, 2008). To date, more than 40 autophagy-related proteins (ATGs) have been identified in yeast as regulating the initiation, nucleation, elongation and fusion of autophagy (Li and Vierstra, 2012; Klionsky et al., 2021). The initiation of autophagy is regulated by two protein complexes: the Unc-51-like autophagy-activating kinase (ULK) complex and the phosphoinositide 3-kinase (PI3K) complex III (Hurley and Young, 2017). The PI3K complex III catalyzes phosphatidylinositol 3-phosphate (PI3P) synthesis and recruits PI3P-binding proteins, especially the ATG18-ATG2 complex, for the initiation of autophagic membranes to autophagosome formation (Kihara et al., 2001; Suzuki et al., 2001). Yeast ATG6/vacuolar protein sorting 30 (VPS30), the ortholog of mammalian Beclin 1, is the key element of the PI3K complex along with VPS34, VPS15, and ATG14 (Kametaka et al., 1998).
The common role of Atg6 in the regulation of autophagy has been verified in yeast, plants and animals, and the yeast Atg6 is also required for the sorting of vacuolar hydrolases (Furuya et al., 2010). Arabidopsis Atg6 regulates normal growth, pollen germination and responses to biotic/abiotic stresses (Fujiki et al., 2007; Qin et al., 2007). Beclin 1 functions as a tumor suppressor in mammals (Qu et al., 2003; Yue et al., 2003). For pathogenic fungi, Atg6 plays an important role in vegetative differentiation and pathogenesis in the rice blast fungus Magnaporthe oryzae and the Fusarium head blight fungus Fusarium graminearum (Kershaw and Talbot, 2009; Lv et al., 2017).
Despite the growing interest in Atg6/Beclin 1, the knowledge about Atg6 in the model fungus B. cinerea remains unknown. In this study, we identified and characterized BcAtg6 in B. cinerea, and determined its role in autophagy, fungal development and pathogenicity.
Materials and Methods
Strains and Culture Conditions
The B. cinerea wild-type strain B05.10 was used as parental strain for genetic modifications. B05.10 and the derivative strains were cultured on potato dextrose agar (PDA), minimal medium (MM), and complete medium (CM), as described previously (Ren et al., 2018a). PDA and sterilized potato fragments were used to determine conidiation. PDA and MM media were used to determine sclerotial formation. MM-N [MM without (NH4)2SO4] was used for induction of autophagy.
Gene Deletion and Complementation
To replace BcATG6 in the wild-type strain B05.10, 1,372-bp upstream and 1,281-bp downstream flanking sequences of BcATG6 were amplifies by PCR from the genomic DNA of B05.10. The resulting amplicons were fused with HPH (hygromycin resistance gene) by using double-joint PCR (Yu et al., 2004). Protoplast preparation and transformation were performed as the previously described method (Gronover et al., 2001). The resulting hygromycin-resistant transformants were preliminarily screened by PCR with primers (Supplementary Table 1), and further confirmed by Southern blotting analysis. The upstream fragment of BcATG6 was used as a probe and labeled with digoxigenin (DIG) using the High Prime DNA Labeling and Detection Starter Kit II, according to the protocol of the manufacturer (Roche Diagnostics, Mannheim, Germany). The genomic DNA was digested with BamH1 endonuclease. For complementation assays, BcAtg6-GFP cassette was generated as described previously (Ren et al., 2018b). Briefly, the entire ORF (open reading frame) of BcATG6 (without stop codon) was amplified and cloned into pNAN-OGG vector containing the GFP allele and the nourseothricin resistance gene. The resulting construct was confirmed by sequencing and transformed into the BcATG6 deletion mutant.
Western Blotting Assay
The total proteins of the GFP-BcAtg8 fusion protein expressing strains under nutrient-rich and nitrogen starvation conditions were extracted as previously described (Gu et al., 2015) and equal amounts of proteins were loaded into each lane of a 10% sodium dodecyl sulfate-polyacrylamide gel. After electrophoresis, proteins were transferred onto Immobilon-P transfer membrane (Millipore, Billerica, MA, United States) with a Bio-Rad electroblotting apparatus. The anti-GFP antibody (Abcam, Cambridge, Cat#Ab32146) and anti-GAPDH antibody (Hangzhou Huaan Biotechnology Co., Ltd., Hangzhou, China, Cat #EM1101) were used at 1: 5,000–1: 10,000 dilution for immunoblot analyses. All experiments were performed at least three times.
Pathogenicity Assay
Infection tests were performed on strawberry fruits and cucumber leaves. Briefly, the tested plant tissues were point-inoculated with 5-mm diameter mycelial plugs of 3-day-old cultures. Before inoculation, the cuticle of hosts was wounded with a sterilized needle tip to facilitate penetration of the fungus into plant tissues. Additionally, water agar plugs without fungal mycelia were used as negative controls (mock). The inoculated samples were placed in a high relative humidity condition (about 95%) at 25°C with 16 h of daylight. These experiments were repeated three times and each time with at least ten samples.
Results
Identification of BcAtg6 in Botrytis cinerea
The BcAtg6 protein coding gene BcATG6 (BCIN_05g05500) was retrieved based on BLASTP search of the genome database of B. cinerea1 with the Saccharomyces cerevisiae Atg6 protein as a query. BcATG6 was predicted to encode a 501 amino-acid protein, which shares 32% identity with S. cerevisiae Atg6 (Figure 1A). Phylogenetic analysis of BcAtg6 with other Atg6 homologs from different fungal species revealed that BcAtg6 is evolved conservatively in fungi kingdom (Figure 1B).
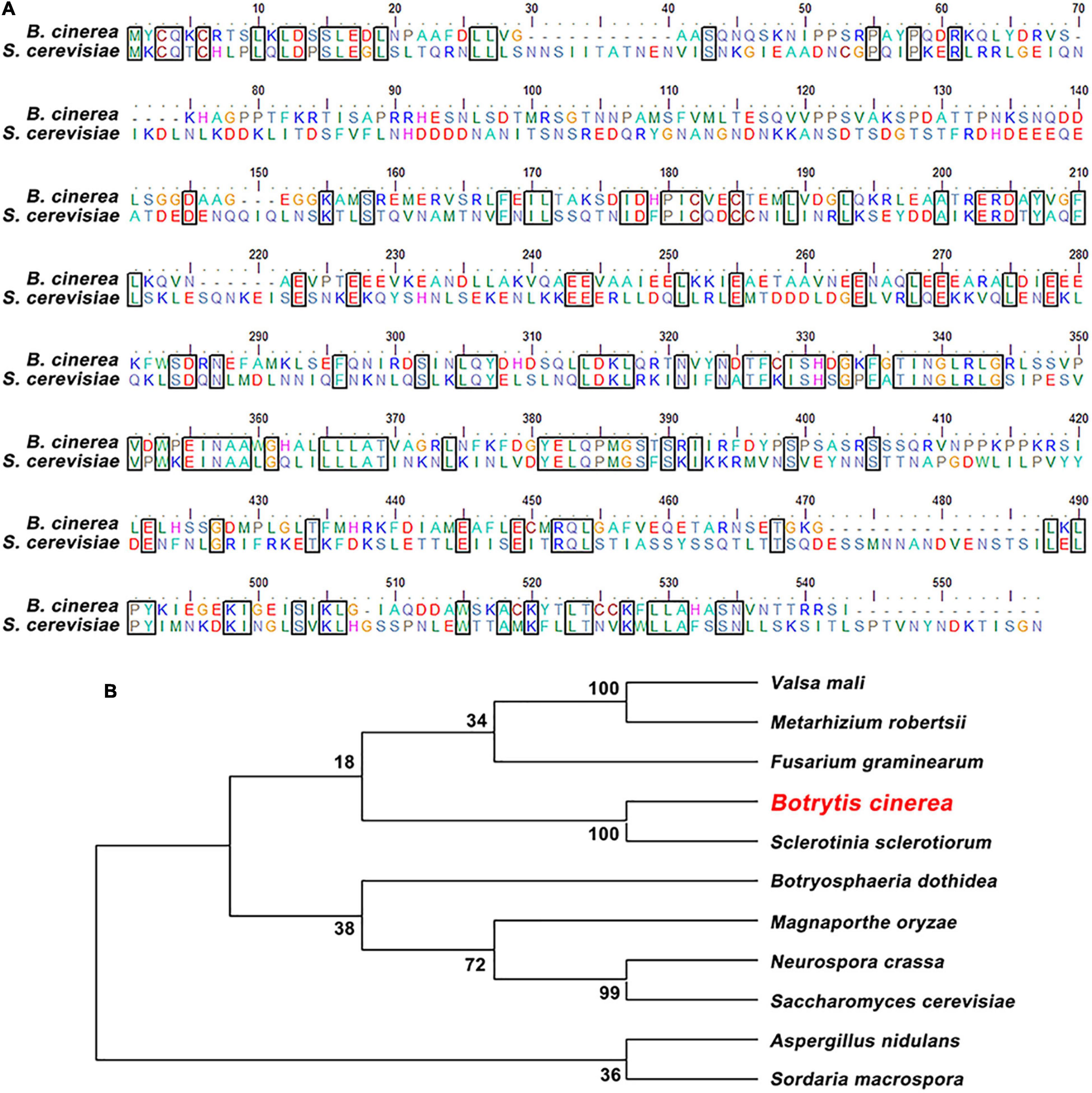
Figure 1. Sequence alignment and phylogenetic analysis of BcAtg6. (A) Protein sequence alignment of Atg6 orthologs from Botrytis cinerea and Saccharomyces cerevisiae. The identical amino acids are in a same box. (B) Phylogenetic analysis of BcAtg6 and its orthologs, including Valsa mali, Metarhizium robertsii, Fusarium graminearum, Sclerotinia sclerotiorum, Botryosphaeria dothidea, Magnaporthe oryzae, Neurospora crassa, Aspergillus nidulans, Sordaria macrospora, and Saccharomyces cerevisiae. The phylogenetic tree was constructed by MEGA7 software using neighbor-joining method.
To investigate the role of BcATG6 in B. cinerea, the targeted gene deletion mutants of BcATG6 were generated using a homologous recombination strategy (Figure 2A). The putative BcATG6 deletion mutants were selected from the hygromycin-resistant transformants by PCR analysis (Figure 2B). Southern blotting further confirmed the right recombination event of ΔBcAtg6 occurred at the BcATG6 locus (Figure 2C).
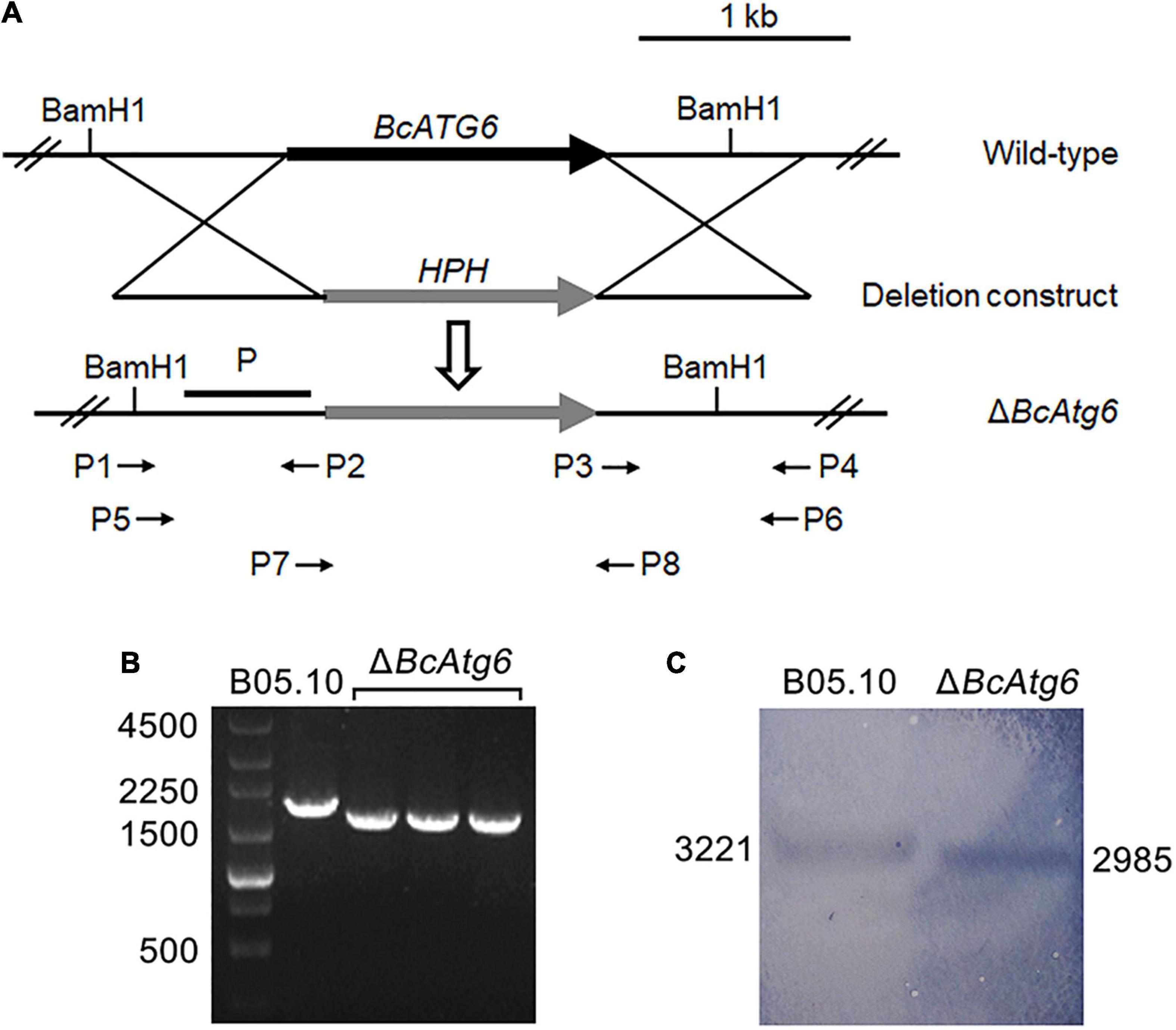
Figure 2. Target gene deletion of BcATG6. (A) Schematic diagram of the BcATG6 homologous replacement strategy. (B) PCR-based screening of the putative BcATG6 deletion mutants using P7/P8 primer. (C) Southern blotting analysis of the BcATG6 deletion mutants using a BcATG6 upstream fragment as probe and the genomic DNA was digested with BamH1 endonuclease.
BcAtg6 Is Required for Autophagy
GFP-BcAtg8 is a useable marker to monitor autophagy in B. cinerea (Ren et al., 2018a). To determine the role of BcAtg6 in autophagy, the proteolysis of GFP-BcAtg8 was analyzed. Under nutrient-rich conditions, the GFP-BcAtg8 fusion protein and free GFP protein were detected by anti-GFP western blotting in the wild-type strain B05.10, and nitrogen starvation promoted the GFP-BcAtg8 proteolysis. However, nitrogen starvation had no effect on the protein contents of GFP-BcAtg8 and GFP in the BcATG6 deletion mutant ΔBcAtg6 (Figure 3). These results indicate that BcAtg6 plays an important role in the regulation of autophagy in B. cinerea.
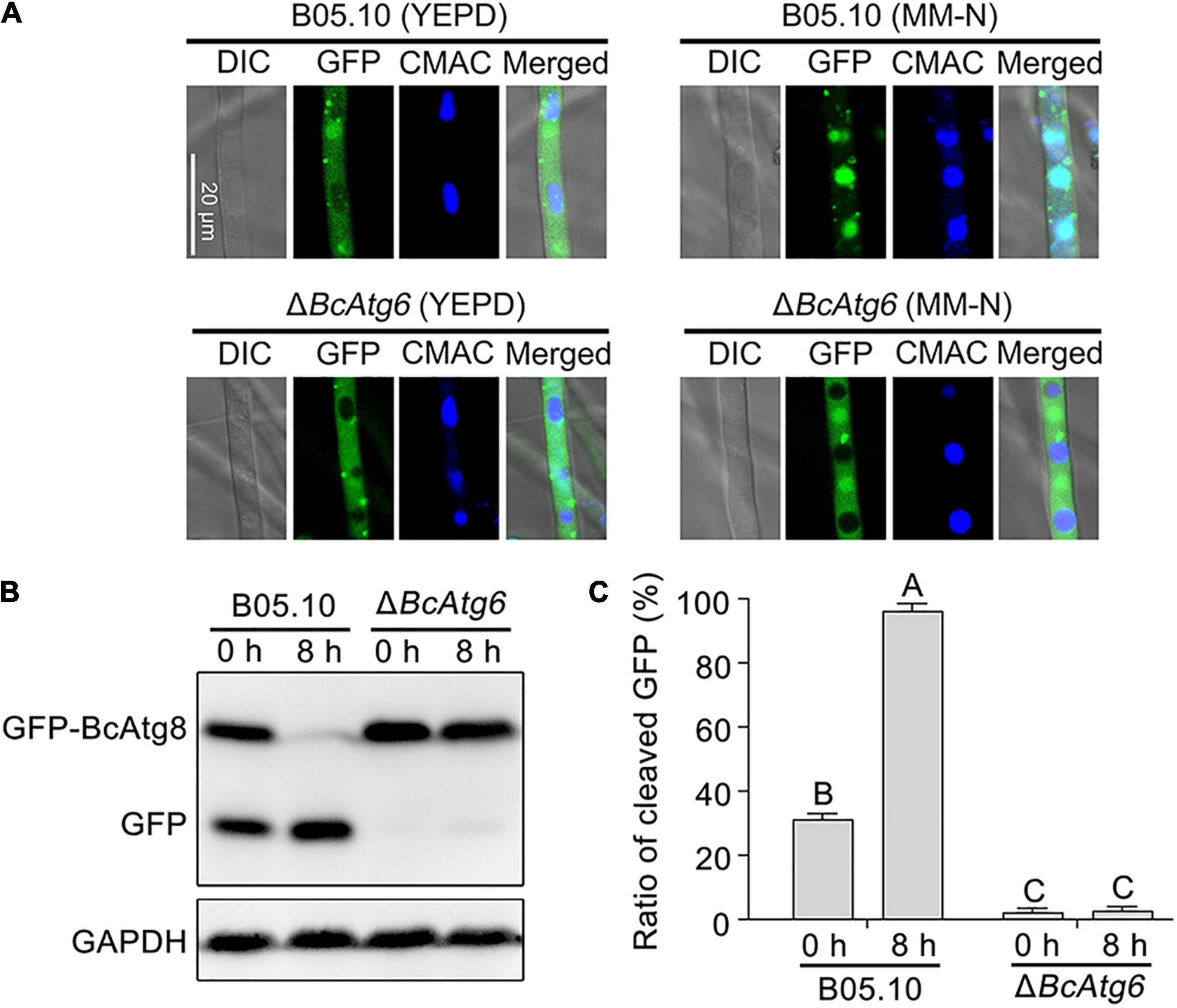
Figure 3. BcAtg6 is essential for autophagy. (A) GFP-BcAtg8 localization in mycelia of the wild-type strain B05.10 and BcATG6 deletion mutant ΔBcAtg6 that treated under nutrient-rich and nitrogen starvation conditions. The vacuoles were stained with CMAC (7-amino-4-chloromethylcoumarin). (B) GFP-BcAtg8 proteolysis of each strain (starvation for 0 and 8 h) was analyzed by Western blotting using an anti-GFP antibody. GAPDH was used as an internal reference. (C) The percentage of GFP on the total of GFP and GFP-BcAtg8. Error bars indicate standard deviation from three independent experiments. Values on the bars followed by the same letter are not significantly different at P = 0.05.
BcAtg6 Is Involved in Vegetative Growth
To determine the role of BcAtg6 in growth, the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C were cultured on PDA, MM and CM media. After incubation at 25°C for 3 days, ΔBcAtg6 exhibited a distinct colony morphology with fewer aerial mycelia (Figure 4A), and a decreased colony radial growth rate compared with B05.10 and ΔBcAtg6-C (Figure 4B). These results indicate that BcAtg6 is important for vegetative growth in B. cinerea.
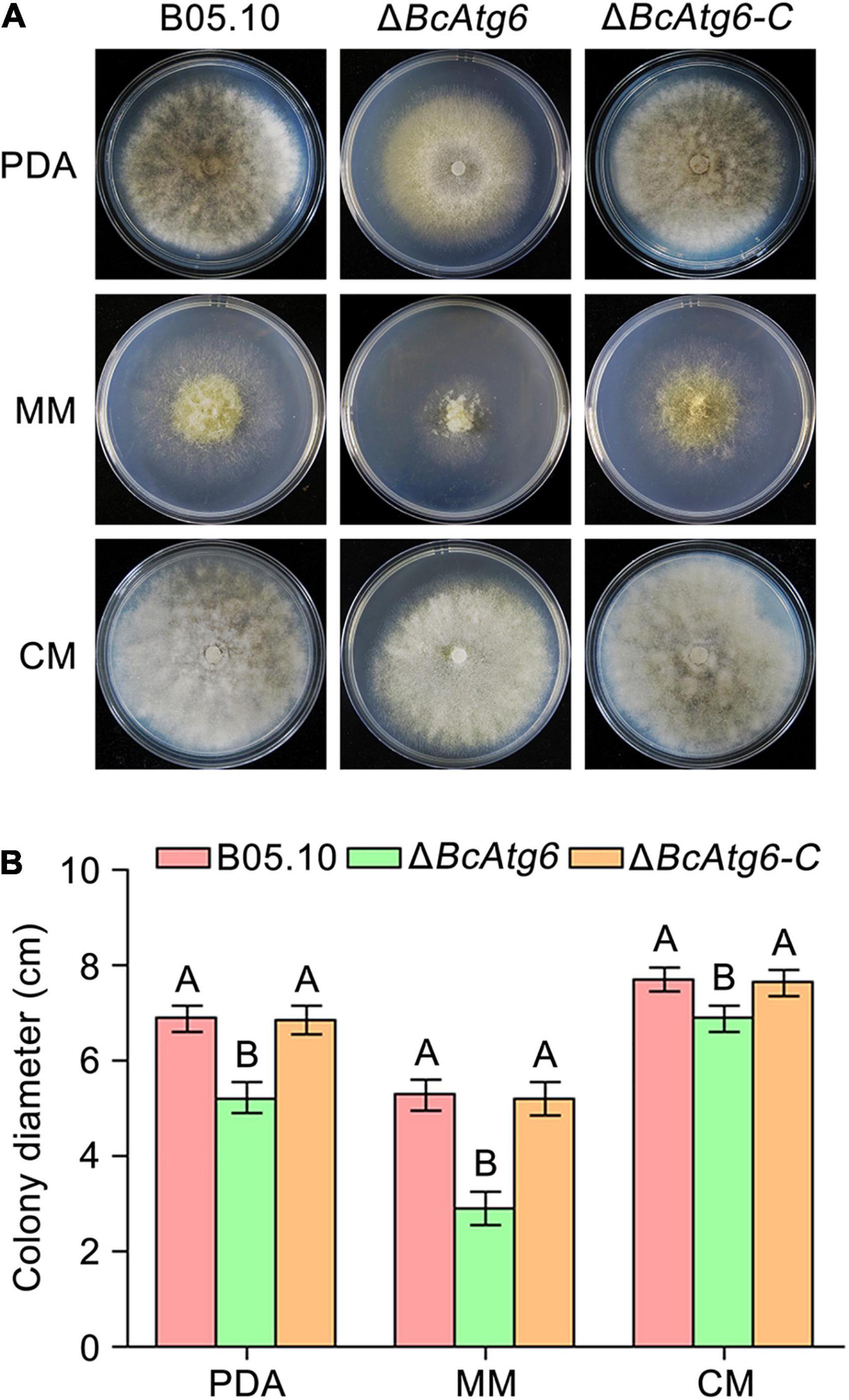
Figure 4. BcAtg6 regulates vegetative growth. (A) Colony morphology of the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C growing on PDA, MM, and CM media at 25°C for 3 days. (B) The mycelial growth rates of each strain on various media. Error bars indicate standard deviation from three independent experiments. Values on the bars followed by the same letter are not significantly different at P = 0.05.
BcAtg6 Is Involved in Conidiation
To determine the role of BcAtg6 in conidiation, the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C were cultured on sterilized potato chips. After incubation at 20°C under white light for 10 days, Both B05.10 and ΔBcAtg6-C formed a dense layer of mycelia covered with lots of conidia, while ΔBcAtg6 failed to form a conidial layer (Figure 5A). The conidiation of ΔBcAtg6 was significantly reduced compared with that of B05.10 and ΔBcAtg6-C (Figure 5B). In addition, ΔBcAtg6 produced some deformed conidia (Figure 5C). These results indicate that BcAtg6 is important for conidiation in B. cinerea.
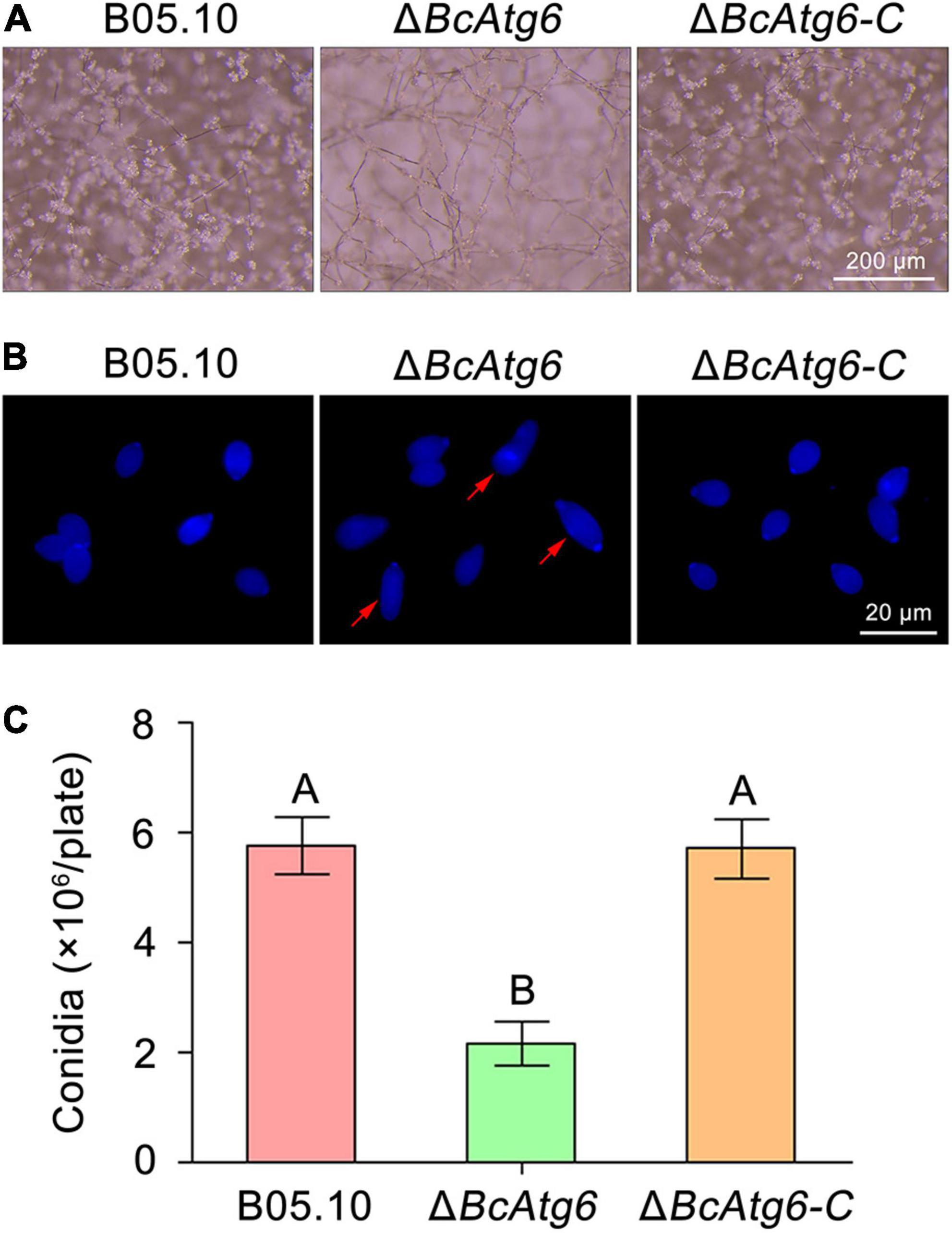
Figure 5. BcAtg6 regulates conidiation. (A) Conidia on conidiophores of the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C were observed after 7 days of incubation on sterilized potato fragments at 25°C with a 12-h photophase. (B) Conidial morphology of each strain. Red arrows indicate abnormal conidia. (C) Quantification of the conidia produced by each strain on sterilized potato fragments. Error bars indicate standard deviation from three independent experiments. Values on the bars followed by the same letter are not significantly different at P = 0.05.
BcAtg6 Is Involved in Sclerotial Formation
Sclerotium is a structure that allows the fungus to survive under unfavorable conditions (such as over the winter) (Williamson et al., 2007). To determine the role of BcAtg6 in sclerotial formation, the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C were culture on PDA and MM media. After incubation at 10°C in the dark for 3 weeks, B05.10 and ΔBcAtg6-C were able to form sclerotia on both PDA and MM media, while ΔBcAtg6 only formed sclerotia on MM medium (Figure 6A), and the amount of sclerotia produced by ΔBcAtg6 on MM medium was significantly reduced compared with that of B05.10 and ΔBcAtg6-C (Figure 6B). These results indicate that BcAtg6 is important for sclerotial formation in B. cinerea.
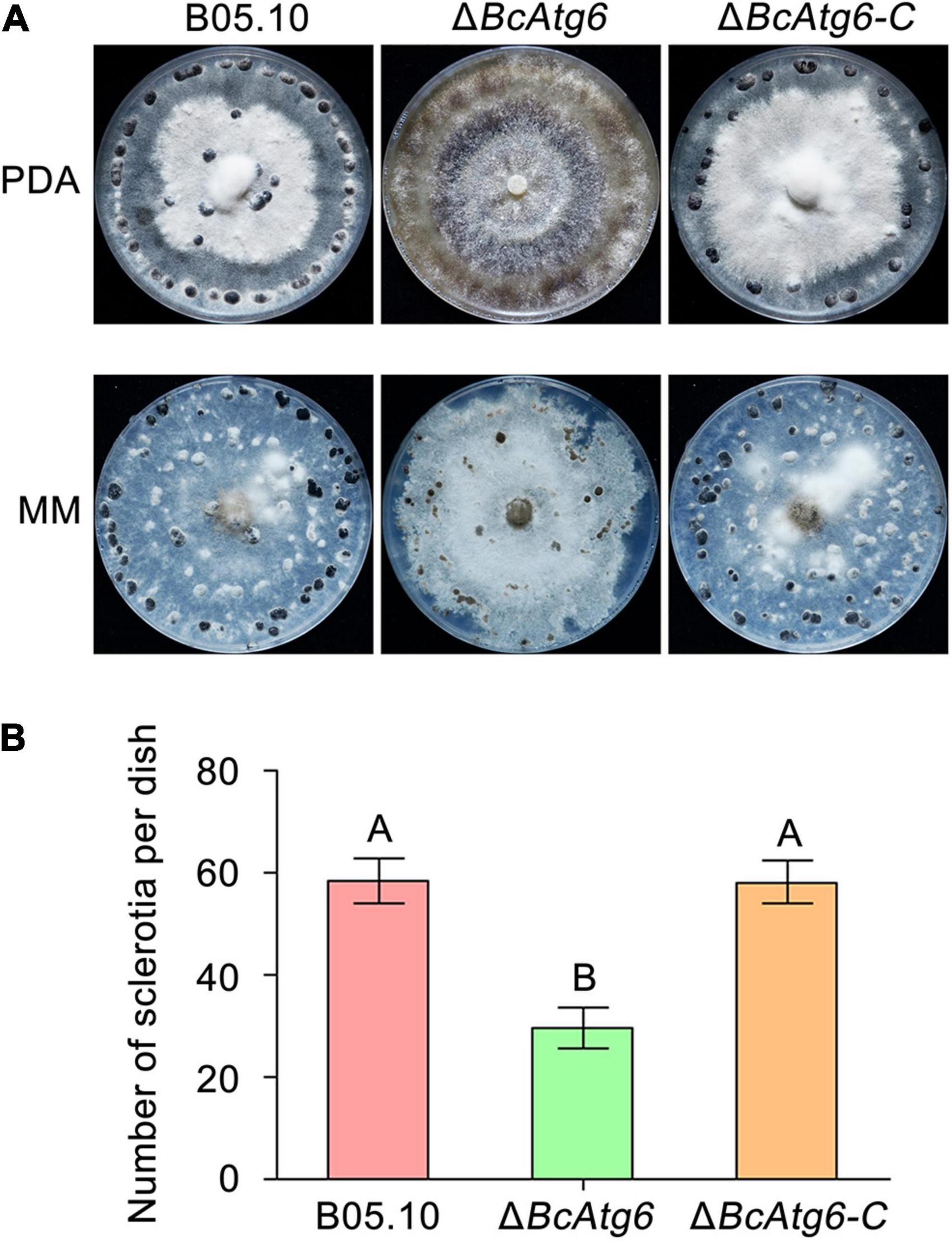
Figure 6. BcAtg6 regulates sclerotial formation. (A) Sclerotial formation of the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C after 3 weeks of incubation on PDA and MM media at 10°C in the dark. (B) The number of sclerotia produced by each strain on MM media. Error bars indicate standard deviation from three independent experiments. Values on the bars followed by the same letter are not significantly different at P = 0.05.
BcAtg6 Is Required for Virulence
Autophagy plays an important role in virulence of pathogenic fungi (Liu et al., 2016). To determine the role of BcAtg6 in virulence of B. cinerea, cucumber leaves and strawberry fruits were inoculated with the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C. After 3 days post-inoculation (dpi), B05.10 and ΔBcAtg6-C caused typical symptoms on both cucumber leaves and strawberry fruits, while ΔBcAtg6 failed to infect those two host tissues (Figures 7A,B). In addition, ΔBcAtg6 lost ability to form infection structures at the initial stage of infection (Figure 7C). These results indicate that BcAtg6 is essential for virulence in B. cinerea.
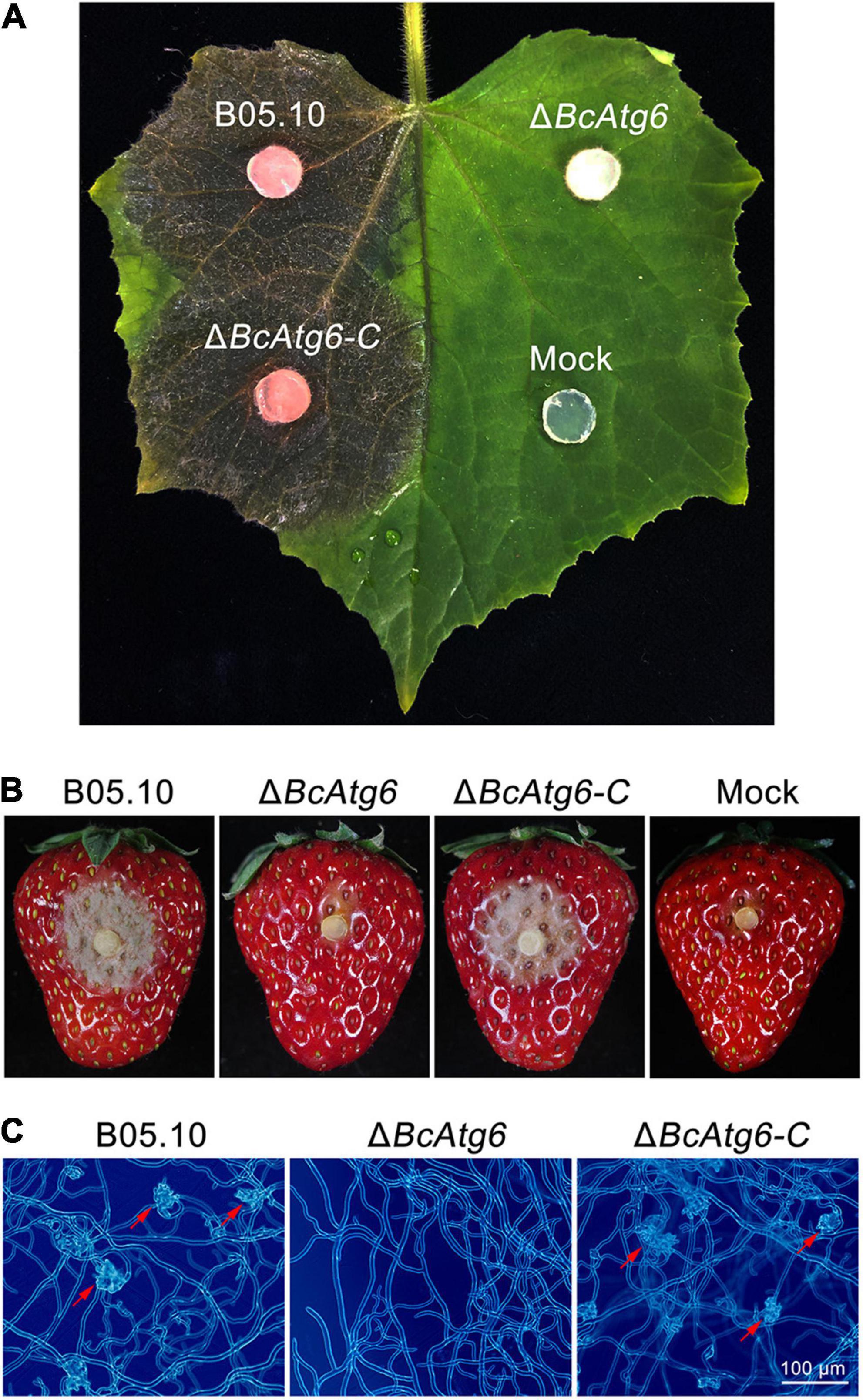
Figure 7. BcAtg6 is required for virulence. (A) Cucumber leaves were inoculated with the wild-type strain B05.10, BcATG6 deletion mutant ΔBcAtg6 and complemented strain ΔBcAtg6-C for 3 days. (B) Strawberry fruits were inoculated with each strain for 3 days. (C) The infection structures formed by each strain in early infection stage. Red arrows indicate infection structures.
Discussion
The most primordial function of autophagy is to recycle proteins and organelles within cells as an adaptation to nutrient deprivation (Klionsky, 2007). With the deepening of research, more and more autophagy functions have been discovered in eukaryotes including plant pathogenic fungi (Yang and Klionsky, 2009). In this study, we focused on BcAtg6, a hitherto uncharacterized protein in the gray mold fungus B. cinerea. Reverse genetic analysis showed that BcAtg6 is essential for autophagy and plays important roles in mycelial growth, conidiation, sclerotial formation and virulence which are consistent with the previous reports of the role of autophagy in B. cinerea (Liu et al., 2019).
As a core component of the class III phosphatidylinositol 3-kinase (PI3K-III) complex, Atg6 is essential for autophagy and the phosphatidylinositol-3-phosphate (PI3P) signaling pathways (Yue et al., 2015). Previously, fungal Atg6 orthologs had been identified and characterized in M. oryzae and F. graminearum, and their necessity in autophagy had been demonstrated (Kershaw and Talbot, 2009; Lv et al., 2017). In this study, loss of BcAtg6 blocked autophagy in B. cinerea suggesting that the function of BcAtg6 in autophagy is evolutionarily conserved.
Endogenous recycling of the cellular constituents by autophagy is crucial for the normal life of fungi and autophagy has been reported to be involved in the regulation of mycelial growth and development in some filamentous fungi (Pollack et al., 2009; Khan et al., 2012). In this study, the ΔBcAtg6 mutant exhibited severe defects in mycelial growth and conidiation, which is consistent with the ΔAtg6 mutants in M. oryzae and F. graminearum (Kershaw and Talbot, 2009; Lv et al., 2017). It is worth noting that the ΔBcAtg6 mutant has the ability to produce sclerotia only on a medium of specific nutrients. These results indicate that autophagy-mediated intracellular recycling plays an important role in supporting the normal vegetative growth and differentiation in B. cinerea.
Autophagy plays an important role in morphogenesis of pathogenic fungi during the initial infection stage (Pollack et al., 2009). Previous studies have shown that the main reason for reduced virulence of the autophagy blocked mutants in M. oryzae and M. robertsii was the appressorium formation defects in the early infection stage (Liu et al., 2007; Duan et al., 2013). In this study, the ΔBcAtg6 mutant cannot form infection structures in the early infection stage and therefore lost pathogenicity. These results indicate that the infection structure morphogenesis mediated by autophagy is required for virulence in B. cinerea.
Conclusion
In conclusion, our results indicate that BcAtg6 is involved in the regulation of fungal development and virulence in B. cinerea.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BL and WR conceived and designed the study. NL and SZ performed the experiments. NL and WR analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32001937), Natural Science Foundation of Shandong Province (ZR2020QC125), and the Scientific Research Fund for High-Level Talents in Qingdao Agricultural University (665/1120060 and 663/1121023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.798363/full#supplementary-material
Footnotes
References
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Duan, Z., Chen, Y., Huang, W., Shang, Y., Chen, P., and Wang, C. (2013). Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy 9, 538–549. doi: 10.4161/auto.23575
Fujiki, Y., Yoshimoto, K., and Ohsumi, Y. (2007). An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143, 1132–1139. doi: 10.1104/pp.106.093864
Furuya, T., Kim, M., Lipinski, M., Li, J., Kim, D., Lu, T., et al. (2010). Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol. Cell 38, 500–511. doi: 10.1016/j.molcel.2010.05.009
Glick, D., Barth, S., and Macleod, K. F. (2010). Autophagy: cellular and molecular mechanisms. J. Pathol. 221, 3–12. doi: 10.1002/path.2697
Gronover, C. S., Kasulke, D., Tudzynski, P., and Tudzynski, B. (2001). The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 14, 1293–1302. doi: 10.1094/MPMI.2001.14.11.1293
Gu, Q., Zhang, C., Liu, X., and Ma, Z. (2015). A transcription factor FgSte12 is required for pathogenicity in Fusarium graminearum. Mol. Plant Pathol. 16, 1–13. doi: 10.1111/mpp.12155
Hu, M. J., Cox, K. D., and Schnabel, G. (2016). Resistance to increasing chemical classes of fungicides by virtue of “selection by association” in Botrytis cinerea. Phytopathology 106, 1513–1520. doi: 10.1094/PHYTO-04-16-0161-R
Hurley, J. H., and Young, L. N. (2017). Mechanisms of autophagy initiation. Annu. Rev. Biochem. 86, 225–244. doi: 10.1146/annurev-biochem-061516-044820
Kametaka, S., Okano, T., Ohsumi, M., and Ohsumi, Y. (1998). Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284–22291. doi: 10.1074/jbc.273.35.22284
Kershaw, M. J., and Talbot, N. J. (2009). Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 106, 15967–15972. doi: 10.1073/pnas.0901477106
Khan, I. A., Lu, J., Liu, X., Rehman, A., and Lin, F. (2012). Multifunction of autophagy-related genes in filamentous fungi. Microbiol. Res. 167, 339–345. doi: 10.1016/j.micres.2012.01.004
Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530. doi: 10.1083/jcb.152.3.519
Klionsky, D. J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nature Rev. Mol. Cell Biol. 8, 931–937. doi: 10.1038/nrm2245
Klionsky, D. J., Abdel-Aziz, A. K., Abdelfatah, S., Abdellatif, M., Abdoli, A., Abel, S., et al. (2021). Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 17, 1–382. doi: 10.1080/15548627.2020.1797280
Levine, B., and Kroemer, G. (2008). Autophagy in the pathogenesis of disease. Cell 132, 27–42. doi: 10.1016/j.cell.2007.12.018
Li, F., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Liu, N., Ren, W., Li, F., Chen, C., and Ma, Z. (2019). Involvement of the cysteine protease BcAtg4 in development and virulence of Botrytis cinerea. Curr. Genet. 65, 293–300. doi: 10.1007/s00294-018-0882-0
Liu, X., Lu, J., Zhang, L., Dong, B., Min, H., and Lin, F. (2007). Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell 6, 997–1005. doi: 10.1128/EC.00011-07
Liu, X., Xu, F., Snyder, J. H., Shi, H., Lu, J., and Lin, F. (2016). Autophagy in plant pathogenic fungi. Semin. Cell Dev. Biol. 57, 128–137.
Lv, W., Wang, C., Yang, N., Que, Y., Talbot, N. J., and Wang, Z. (2017). Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 7:11062. doi: 10.1038/s41598-017-11640-z
Mizushima, N. (2007). Autophagy: process and function. Genes Dev. 21, 2861–2873. doi: 10.1101/gad.1599207
Pollack, J. K., Harris, S. D., and Marten, M. R. (2009). Autophagy in filamentous fungi. Fungal Genet. Biol. 46, 1–8. doi: 10.1016/j.fgb.2008.10.010
Qin, G., Ma, Z., Zhang, L., Xing, S., Hou, X., Deng, J., et al. (2007). Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17, 249–263. doi: 10.1038/cr.2007.7
Qu, X., Yu, J., Bhagat, G., Furuya, N., Hibshoosh, H., Troxel, A., et al. (2003). Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820. doi: 10.1172/JCI20039
Ren, W., Liu, N., Sang, C., Shi, D., Zhou, M., Chen, C., et al. (2018a). The autophagy gene BcATG8 regulates the vegetative differentiation and pathogenicity of Botrytis cinerea. Appl. Environ. Microbiol. 84, e02455–17. doi: 10.1128/AEM.02455-17
Ren, W., Sang, C., Shi, D., Song, X., Zhou, M., and Chen, C. (2018b). Ubiquitin-like activating enzymes BcAtg3 and BcAtg7 participate in development and pathogenesis of Botrytis cinerea. Curr. Genet. 64, 919–930. doi: 10.1007/s00294-018-0810-3
Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. doi: 10.1093/emboj/20.21.5971
Tudzynski, P., and Kokkelink, L. (2009). “Botrytis cinerea: molecular aspects of a necrotrophic life style,” in Plant Relationships, ed. H. B. Deising (Berlin: Springer), 29–50. *editmade. doi: 10.1007/978-3-540-87407-2_2
Williamson, B., Tudzynski, B., Tudzynski, P., and Van Kan, J. A. L. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
Yang, Z., and Klionsky, D. J. (2009). An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335, 1–32. doi: 10.1007/978-3-642-00302-8_1
Yu, J., Hamari, Z., Han, K., Seo, J. A., Reyes-Domínguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Yue, J., Sun, H., Zhang, W., Pei, D., He, Y., and Wang, H. (2015). Wheat homologs of yeast ATG6 function in autophagy and are implicated in powdery mildew immunity. BMC Plant Biol. 15:95. doi: 10.1186/s12870-015-0472-y
Keywords: Botrytis cinerea, autophagy, Atg6, development, pathogenicity
Citation: Liu N, Zhou S, Li B and Ren W (2021) Involvement of the Autophagy Protein Atg6 in Development and Virulence in the Gray Mold Fungus Botrytis cinerea. Front. Microbiol. 12:798363. doi: 10.3389/fmicb.2021.798363
Received: 20 October 2021; Accepted: 25 November 2021;
Published: 14 December 2021.
Edited by:
Fangfang Li, Institute of Plant Protection, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Jiatao Xie, Huazhong Agricultural University, ChinaYanjun Kou, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2021 Liu, Zhou, Li and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weichao Ren, cmVud2VpY2hhb3F3QDE2My5jb20=
 Na Liu
Na Liu Shanyue Zhou1,2
Shanyue Zhou1,2 Weichao Ren
Weichao Ren