
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 February 2022
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.797841
This article is part of the Research Topic Evolutionary Genomics of Candidatus Liberibacter spp. and Their Interactions with Plant and Insect-Vector Hosts View all 14 articles
Huanglongbing (HLB), caused by “Candidatus liberibacter asiaticus” (CaLas), is one of the most devastating diseases in citrus but its pathogenesis remains poorly understood. Here, we reported the role of the CaLasSDE115 (CLIBASIA_05115) effector, encoded by CaLas, during pathogen-host interactions. Bioinformatics analyses showed that CaLasSDE115 was 100% conserved in all reported CaLas strains but had sequence differences compared with orthologs from other “Candidatus liberibacter.” Prediction of protein structures suggested that the crystal structure of CaLasSDE115 was very close to that of the invasion-related protein B (IalB), a virulence factor from Bartonella henselae. Alkaline phosphatase (PhoA) assay in E. coli further confirmed that CaLasSDE115 was a Sec-dependent secretory protein while subcellular localization analyses in tobacco showed that the mature protein of SDE115 (mSDE115), without its putative Sec-dependent signal peptide, was distributed in the cytoplasm and the nucleus. Expression levels of CaLasSDE115 in CaLas-infected Asian citrus psyllid (ACP) were much higher (∼45-fold) than those in CaLas-infected Wanjincheng oranges, with the expression in symptomatic leaves being significantly higher than that in asymptomatic ones. Additionally, the overexpression of mSDE115 favored CaLas proliferation during the early stages (2 months) of infection while promoting the development of symptoms. Hormone content and gene expression analysis of transgenic plants also suggested that overexpressing mSDE115 modulated the transcriptional regulation of genes involved in systemic acquired resistance (SAR) response. Overall, our data indicated that CaLasSDE115 effector contributed to the early colonization of citrus by the pathogen and worsened the occurrence of Huanglongbing symptoms, thereby providing a theoretical basis for further exploring the pathogenic mechanisms of Huanglongbing disease in citrus.
Citrus Huanglongbing (HLB) is the most devastating disease which has harmed citrus trees in more than 50 countries and regions around the world, thereby causing serious economic losses to the citrus industry (Gottwald, 2010). Its causative agent is a phloem-limited α-Proteobacterium “Candidatus Liberibacter” (Jagoueix et al., 1994; Bové, 2006) for which three species namely “Candidatus Liberibacter asiaticus (CaLas),” “Candidatus Liberibacter africanus” and “Candidatus Liberibacter americanus” are known to be associated with HLB in citrus (Teixeira et al., 2005; Wulff et al., 2014; Lin et al., 2015). However, to date, HLB pathogens have not been successfully cultured in vitro. Among these, CaLas is the most prevalent one in citrus, with its insect vector being the Asian citrus psyllid (ACP) Diaphorina citri Kuwayama. CaLas resides in sieve elements and spreads through the phloem transport system (Duan et al., 2009). HLB symptoms mainly include flush yellowing, leaf mottling, as well as inverted fruit coloring before eventually resulting in plant death (Bassanezi et al., 2009; Graham et al., 2013). Although there have been many successful cases of HLB management based on the “Three basic measures” (planting HLB-free nursery trees, timely clearing of any HLB-infected trees and controlling psyllid populations as much as possible), neither effective cures against the disease and nor HLB-resistant citrus cultivars have, so far, been identified (Wang et al., 2017).
Host-pathogen interactions are vital to understanding and controlling HLB disease. This interplay between hosts and pathogens involves a myriad of molecular interactions that play important roles both in a pathogen’s ability to suppress or avoid a plant’s immunity as well as in the host’s ability to detect and remove the pathogen. First, the plant perceives pathogen-associated molecular patterns (PAMP) such as fungal chitin and bacterial flagellin through its immune receptors PRRs (pattern recognition receptors) to initiate PAMP-triggered immunity (PTI) that prevents pathogen invasion (Boller and Felix, 2009; Segonzac and Zipfel, 2011). To defeat the PTI responses, pathogens manipulate the host’s cellular functions by secreting diverse effector proteins (Yuan et al., 2021) which are recognized by specialized receptors (usually the plant disease resistance proteins) and this further leads to the activation of the host’s effector-triggered immunity (ETI) to prevent pathogen attack (Jones and Dangl, 2006; Haitao et al., 2015). However, a pathogen can also evolve its effectors to suppress the ETI (Nguyen et al., 2021). In CaLas-citrus interactions, the effectors involved and their functions in pathogen virulence are poorly understood, partly due to the unculturability of the pathogen. Thus, exploring the role of CaLas effectors could be key to understanding the pathogenicity mechanisms of CaLas as well as for identifying citrus resistance genes which could eventually be exploited for developing new control methods and improving HLB resistance in citrus.
Many extracellular plant pathogens suppress plant immunity through the translocation of effector proteins using the type-III secretion system (T3SS) (Jones and Dangl, 2006; Toruno et al., 2016). However, since CaLas does not possess a T3SS (Duan et al., 2009) but instead harbors a complete Sec-dependent secretion system (Duan et al., 2009), it is believed that CaLas secrets Sec-dependent effectors (SDEs) including virulence factors into plant cells through this system (Pitino et al., 2016; Prasad et al., 2016; Pagliaccia et al., 2017). In other phloem-limited pathogens such as phytoplasma, several SDEs have been shown to be critical for pathogenicity (Saskia et al., 2008). Similarly, for CaLas SDEs, 86 putative SDE proteins have been shown to have functional Sec-dependent secretion signal peptides (Prasad et al., 2016). Clark et al. (2018) reported that CaLas SDE1 suppressed citrus immunity by inhibiting the activity of papain-like cysteine proteases (PLCPs). Pang et al. (2020) showed that SDE15 suppressed plant immunity by interacting with the citrus protein CsACD2, a homolog of Arabidopsis ACCELERATED CELL DEATH 2. In fact, overexpression of SDE15 or CsACD2 in Duncan grapefruit was found to suppress plant immunity and promote CaLas growth. Furthermore, the authors indicated that SDE15-CsACD2 interactions repressed citrus PTI by affecting the expression of PTI marker genes such as FRK1, GST1 and WRKY22. Other CaLas SDEs were also reported to regulate plant immunity responses (Liu et al., 2019; Ying et al., 2019; Li et al., 2020). In addition, it has been shown that CaLas also transported non-classically secreted proteins (ncSecPs) into host cells to regulate host immunity. In this context, Du et al. (2021) indicated that 10 ncSecPs had opposing effects on early plant defenses. For instance, SC2_gp095 effectors, encoded by CaLas prophages, could suppress the development of host symptoms through the manipulation of plant H2O2-mediated defense signaling (Jain et al., 2015). Similarly, CaLas SahA could degrade salicylic acid (SA) to suppress plant defenses (Li et al., 2017) while the peroxiredoxin effector LasBCP simultaneously inhibited localized as well as systemic innate immune responses through oxylipin-mediated defense signaling in plants (Jain et al., 2018, 2019).
In this study, we investigated the potential role of a putative virulence factor CaLasSDE115 (CLIBASIA_05115) during CaLas infections. Bioinformatics predictions indicated that CaLasSDE115 was very close to that of a virulence factor IalB from Bartonella henselae, which is not only an invasion-associated locus B but is also involved in pathogen-host invasions (Vayssier-Taussat et al., 2010). The sec-dependent secretory characteristics of CaLasSDE115 was also confirmed by bioinformatics analyses and phoA assays while its involvement in citrus’ responses to CaLas infections were investigated by overexpressing mSDE115 in HLB-susceptible Wanjincheng oranges. Altogether, our data indicated that CaLasSDE115 positively regulated early pathogenic colonization of citrus by modulating the transcriptional regulation of genes involved in SAR responses. The potential mechanisms of CaLasSDE115 were eventually discussed in this study. Overall, it is expected that this study will help to further reveal the pathogenic mechanisms of Huanglongbing disease in the future.
Wanjincheng orange plants (C. sinensis Osbeck) as well as all the transgenic ones used in this study were plantged in a greenhouse, with a 16 h photoperiod of 45 μmol m–2s–1 illumination and a relative humidity of 60%, at the National Citrus Engineering Research Center, Chongqing, China. Citrus trees containing CaLas were also maintained in the greenhouse. Healthy adult ACPs were reared on young leaf flush of CaLas-infected or CaLas-free sweet orange (C. sinensis) seedlings in incubators at a temperature of 25 ± 1°C and after 3-weeks of feeding, the insects were collected, frozen with liquid nitrogen and stored at -80°C. Using the primers Cs16S-F/16s-R (Supplementary Table 1), the presence of the CaLas pathogens in both plants and ACPs was confirmed by PCR.
Using cDNA from CaLas-infected Wanjincheng orange plants as a template, the gene encoding for CaLasSDE115 (CLIBASIA_05115) was amplified with the primer pair T-SDE115-F/R (Supplementary Table 1). The PCR was performed in a final volume of 50 μL, containing 25 μL of Primer STAR Max Premix (TaKaRa, Ojin, Japan), 1.5 μL of each primer (10 mM⋅L–1), 19 μL of H2O and 3 μL of cDNA (0.5 × 10–5 ng⋅L–1). The PCR conditions were as follows: pre-denaturation at 96°C for 5 min, followed by 35 amplification cycles (96°C for 30 s, 56°C for 30 s and 72°C for 30 s), before a final extension at 72°C for 3 min. The PCR product was T-cloned into the pGEM-Teasy vector (Promega, WI, United States). The sequence of CaLasSDE115 was then determined by Sanger sequencing.
The signal peptide (SP) and transmembrane structure of CaLasSDE115 were predicted using the signalP5.0 tool1 and TMHMM Server (v.2.0)2, respectively. The secondary as well as tertiary structures of its mature protein were analyzed using SOPMA3, SWISS-MODEL4 and Phyre5 tools, respectively. Multiple sequence alignment of the CaLasSDE115 protein with homologs from other selected species was also performed using ClustalW2 tool6. A phylogenetic tree was constructed based on the neighbor-joining method using MEGA7.0 software (Kumar et al., 2016).
PhoA assays of CaLasSDE115 were performed, in triplicate, as previously described (Liu et al., 2019). CaLasSDE115, mSDE115 without the SP-coding sequence, as well as SDE115sp containing only the SP-coding sequences were separately inserted into mphoA-containing pET-mphoA (without the SP-coding sequence), to generate different in-frame gene fusions while PET-phoA (containing its SP-coding sequence) and PET-mphoA vectors were used as positive and negative controls, respectively. These different constructs were then transformed into E. coli BL21 cells which were incubated overnight at 37°C on LB medium supplemented with 90 μg/ml of BCIP (5-bromo-4-chloro-3-indolylphosphate) and 75 mM of Na2HPO4 in order to detect phoA activities. The blue transformants indicated that fusion proteins were secreted outside the cells.
The mSDE115 sequence was cloned into pCAMBIA1300-35S-GFP to construct the fusion gene mSDE115:GFP prior to its transfer into A. tumefaciens EHA105. Agroinfiltration of N. benthamiana leaves was performed as described before (Liu et al., 2019) and 3 days post-inoculation, the localization of the fusion protein was determined using an FV3000 confocal microscope equipped with a UV light source (Olympus, Tokyo, Japan). A histone 2B (H2B) fusion with the red fluorescent protein (RFP) was used as a marker for the nucleus (Martin et al., 2009). This test was repeated twice.
For citrus transformation, the mSDE115 gene sequence was first cloned into a pLGN vector (Zou et al., 2017) to construct the p35S:mSDE115 vector in which mSDE115 expression was controlled by a 35S strong constitutive promoter. The vector was then introduced into an EHA105 strain.
Epicotyl segments from Wanjincheng orange seedlings were used for A. tumefaciens-mediated transformation, with the experiment performed as previously described (Peng et al., 2015). Transgenic shoots were then identified by GUS staining as well as PCR analysis before being micrografted onto Troyer citrange [Poncirus trifoliata (L.) Raf. × C. sinensis (L.) Osbeck] seedlings in vitro. The resulting plantlets were further grafted onto Troyer citrange seedlings in a greenhouse, and the expression levels of mSDE115 gene in those transgenic plants were determined by RT-qPCR analysis.
Evaluating the tolerance of transgenic plants to CaLas was performed as previously described by Zou et al. (2017). Three to five plants per independent transgenic line, including wild type (WT) plants, were inoculated by CaLas-infected Wanjincheng orange branches using the shoot top grafting method (Cifuentes-Arenas et al., 2019). All the inoculated plants were then maintained in a greenhouse and regularly checked for any sign of disease incidence.
The CaLas bacterial populations (CaLas cells μg–1 of citrus DNA) were determined as previously described (Zou et al., 2017). Every 2 months, three midrib sections from three different leaves per plant were randomly extracted and pooled into one sample from which DNA was isolated. The amount of CaLas 16S rRNA genes and citrus 18S rRNA genes in the isolated DNA samples were subsequently detected by quantitative PCR (qPCR) using the Cs16S-f/Cs16S-r and Cs18S-f/Cs18S-r primers (Supplementary Table 1), respectively. The qPCR was performed in a final volume of 12 μL, containing 6 μL of the SYBRPRIME qPCR Kit (Bioground Biotech, Chongqing, China), 4.4 μL H2O, 0.3 μL of each primer (10 mM⋅L–1) as well as 1 μL DNA(10 ng⋅μL–1)while the PCR protocol involved a pretreatment (95°C for 2 min) followed by 40 amplification cycles (each at 94°C for 10 s and 60°C for 60 s). Using the citrus 18S gene as the internal reference, the CaLas bacterial populations were calculated as reported by Zou et al. (2017). In this case, the disease intensity in the transgenic lines was evaluated based on the bacterial populations of three to five plants compared with the WT control.
Total RNA isolation from citrus tissues, including leaves, roots and midrib sections were performed using the EASYspin Plant RNA Extraction Kit according to the manufacturer’s instructions (Aidlab, Beijing, China). In the case of ACPs, 30–40 CaLas-infected insects were ground to a powder in liquid nitrogen before immediately extracting total RNA with TRIzol Reagent (Invitrogen, MA, United States). A 1 μg cDNA was then synthesized by reverse transcription (RT) using the PrimeScript ™RT Master Mix (TaKaRa, Ojin, Japan) prior to quantitative reverse transcription-polymerase chain reaction (QRT-PCR using the primers listed in Supplementary Table 1). Gene expression was detected using the SYBRPRIME qPCR Kit (Bioground Biotech, Chongqing, China). The PCR reactions were as follows: pre-denaturation (50°C for 60 s, 95°C for 120 s), followed by 40 cycles of amplification (each at 95°C for 5 s, 60°C for 15 s). The CaLas DNA gyrase subunit A (CaLasgyrA, GenBank No. CP001677.5) gene was employed as an internal reference to investigate the expression of CaLas05115 in CaLas-infected plants and ACPs while the citrus GAPDH gene (Mafra et al., 2012) was used as an internal reference to evaluate gene expression in transgenic plants. All experiments were performed in triplicate. All the relative expression values were calculated by the Ct method (2–Δ Δ Ct) (Livak and Schmittgen, 2001).
Fresh tissues (0.1 g) were collected from three fully mature leaves per line and ground to a fine powder in liquid nitrogen. The powder was then homogenized in 0.9 mL of phosphate saline buffer (PBS) of pH = 7.4, before being centrifuged at 800 rpm for 10 min. The resulting supernatant was transferred to a new, enzyme-free tube and stored in -20°C until assayed.
The SA, methyl salicylate (MeSA) and jasmonic acid (JA) content in the supernatant was determined using plant enzyme-linked immunosorbent assay (ELISA) kits (Jiweibio, Shanghai, China) (Zou et al., 2021). Briefly, the supernatant was first diluted with a 5 × ELISA diluent buffer before adding 50 μL of the diluted supernatant to reaction wells, followed by 100 μl of the detection antibody (horseradish peroxidase-conjugated antibody). After incubating the mixture at 37°C for 1 h, the wells were washed five times with 350 μL of the washing buffer prior to a second incubation with 100 μL of 1 × TMB (3,3′, 5,5′-tetramethylbenzidine) solution at 37°C for 15 min. The reaction was eventually stopped by adding 50 μL of the stop solution. The OD of each well was then measured, at 450 nm, using a Spectra-Max® M2 microplate reader (Molecular Devices Corporation, Menlo Park, CA, United States). The hormone content per gram of fresh weight leaf (μg/g) was calculated using Excel 365. All the assays were repeated three times.
Statistical analyses of all data were conducted in Excel 365, using the Student’s t-test to compare differences between the control and samples at 5% significance level.
Based on the sequence of CaLasSDE115 from CaLas psy62 (Duan et al., 2009), a 558-bp long coding sequence was cloned from CaLas-infected Wanjincheng oranges. This gene encoded 186 amino acid residues and was found to be 100% conserved in all CaLas strains based on the complete genomes which are currently available (Supplementary Figure 1). Multiple alignments were performed by ClustalW2 using CaLasSDE115 homologs from selected, different species (Figures 1A,B). The results showed that CaLasSDE115 had a 41 to 65% similarity in amino acid sequence compared with the selected orthologs from other Candidatus Liberibacters. Phylogenetic analysis also showed that CaLasSDE115 was closely related to the orthologs from “Candidatus Liberibacter africanus” and “Candidatus Liberibacter solanacearum” (Figure 1C). The prediction of tertiary structures further indicated that the crystal structure of CaLasSDE115 was very close to that of the virulence factor IalB from Bartonella henselae (Vayssier-Taussat et al., 2010) despite only 15.6% of sequence identity between them (Supplementary Figures 2, 3). In fact, the amino acid sequence of CaLasSDE115 had a 38% identity to those of the IalB homologs from Rhizobium undicola and Agrobacterium vitis (Figure 1B). These results suggested that CaLasSDE115 was involved in CaLas pathogenicity.
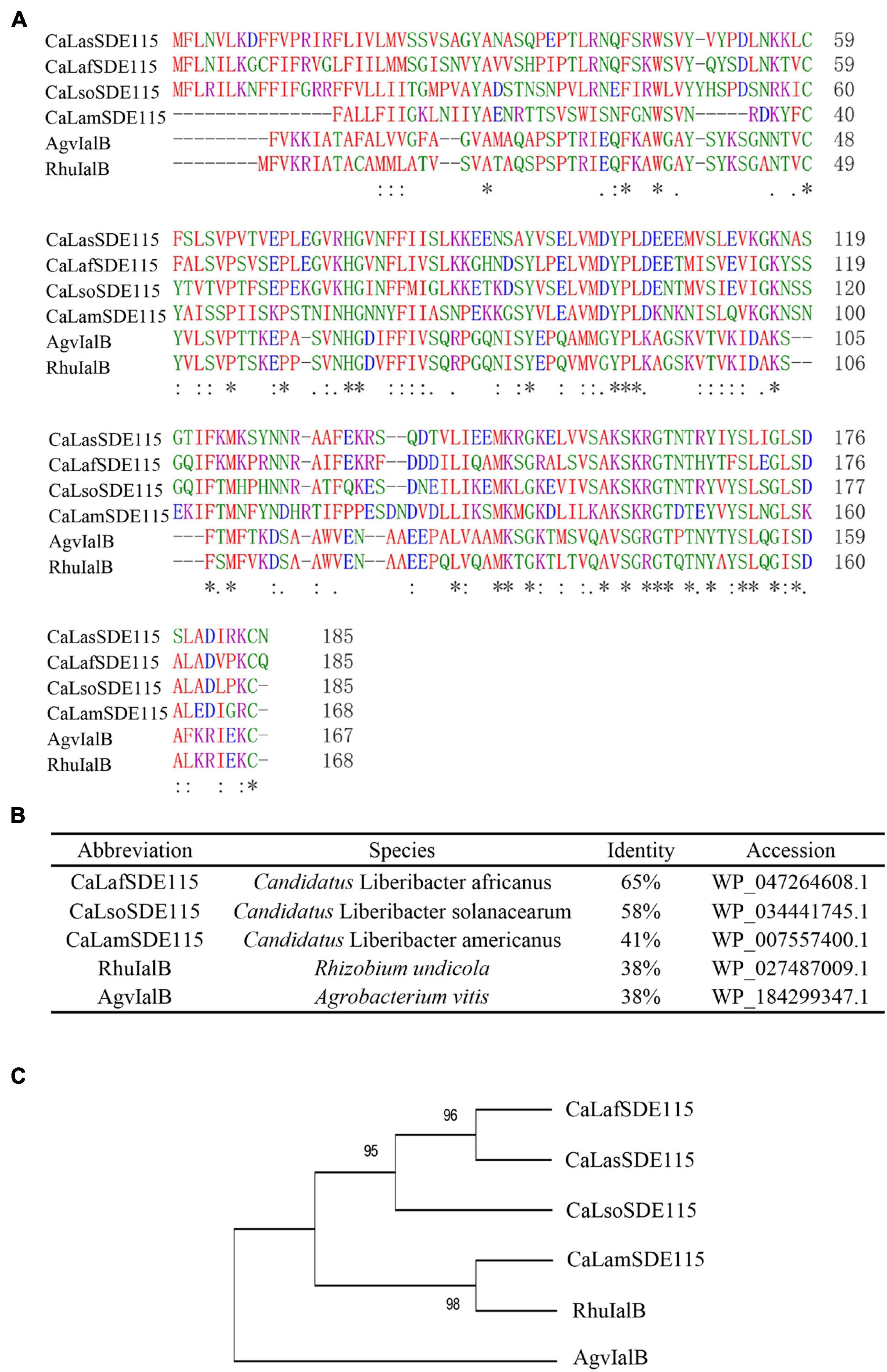
Figure 1. Analysis of CaLasSDE115 protein sequence. (A,B) Multiple sequence alignment of CaLasSDE115 with selected homologs. (C) A phylogenetic tree containing CaLasSDE115 and other selected homologs. The reliability of the tree was estimated using 1000 bootstrap replicates and the bootstrap values (%) are shown above branches. In (A), “*”, “:” and “.” indicate a single completely conservative, highly and low similar among residues, respectively.
SignalP5.0 predicted that CaLasSDE115 was a Sec-dependent presecretory protein containing a putative signal peptide (namely SDE115sp) with a cleavage site between amino acids 32 and 33 (ANA-SQ; probability: 0.885) (Figure 2A). The SDE115sp had a hydrophobic core, flanked by three positively charged residues at the N-terminus and a less positively charged C terminus (Figure 2A), which had the characteristics of a Sec-dependent signal peptide (Tamar and Damon, 2018). Based on the TMHMM Server (v.2.0) analysis, no transmembrane domains were found in the mature CaLasSDE115 protein. To determine the export of this protein via the Sec-dependent secretory system, an E. coli phoA assay was also used (Figures 2B,C). In this case, the data showed that bacteria containing fusions between mphoA (without its native SP) and either the native CaLasSDE115 or only its signal peptide SDE115sp became blue overnight. On the other hand, bacteria containing only mphoA (without its native SP) or its fusion with the mature protein mSDE115 remained white. These results, along with the positive control containing the complete phoA (mphoA + its native SP), confirmed that CaLasSDE115 was a typical Sec-dependent secretory protein and that the signal peptide, as predicted by SignalP5.0, was involved in directing extracellular secretion of proteins in bacteria.
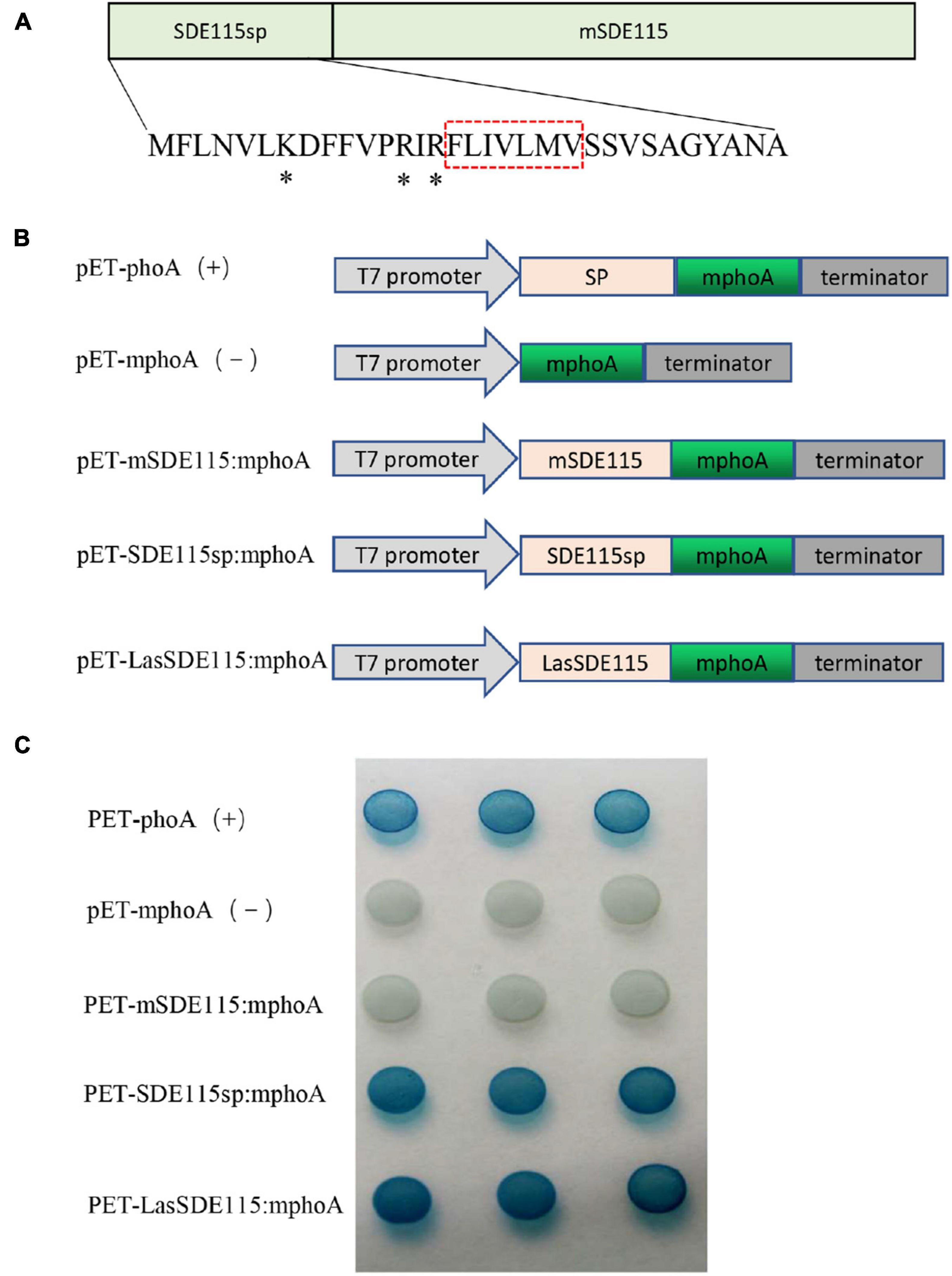
Figure 2. CaLasSDE115 is a Sec-dependent secretory protein. (A) Structure of the CaLasSDE115 protein. Its signal peptide and mature protein were named SDE115sp and mSDE115, respectively. *indicated the positively charged amino acids lysine (K) and arginine (R) while the red box indicated the central hydrophobic core. (B) Schematic diagram of the prokaryotic expression cassettes for phoA assay. (C) PhoA assay for the secretion of CaLasSDE115 in E. coli cells. The data showed that SDE115sp directed the extracellular translocation of mPhoA lacking its native SP and that of CaLasSDE115:mphoA fusion.
Since the above results revealed that the CaLasSDE115 mature protein crossed the bacterial outer membrane into the host cells via a Sec-dependent secretory system, the next step was to observe the subcellular localization of CaLasSDE115 mature proteins in plants. For this purpose, the mSDE115 coding sequence was fused with the GFP reporter gene (Figure 3A). Transient expression tests in N. benthamiana leaves showed that the mSDE115:GFP fusion had similar cellular locations as those of the GFP protein alone, with fluorescence signals detected in the nucleus and cytoplasm (Figure 3B).
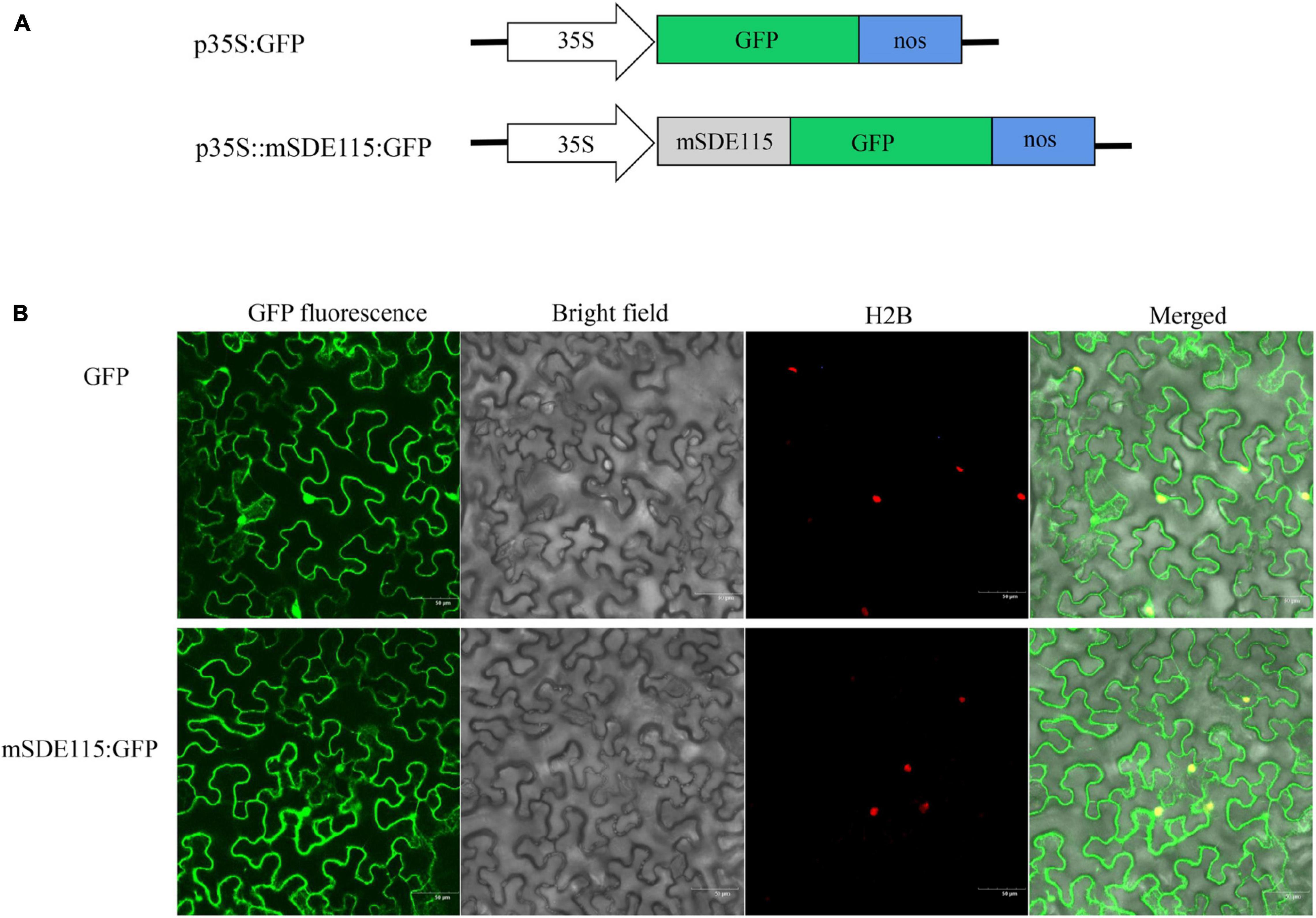
Figure 3. Subcellular location of mSDE115 in tobacco epidermis cell. (A) Schematic diagram of the constructs used for agroinfiltration. A 35S, CaMV 35S promoter; NOS, the nopaline synthase terminator. (B) Subcellular localization of mSDE115:GFP fusion protein as observed by confocal microscopy. A histone 2B (H2B) fusion with the red fluorescent protein (RFP) was used as a marker for the nucleus (Martin et al., 2009).
To compare the expression levels of CaLasSDE115 in citrus and insect hosts, RT-qPCR was performed on the total RNA extracted from CaLas-infected Wanjincheng oranges and ACPs (CT values of Wanjincheng oranges and psyllids were listed in Supplementary Table 2). The results showed that the expression level of CaLasSDE115 in CaLas-infected ACPs was significantly higher (∼45-fold) than that in CaLas-infected Wanjincheng oranges (Figure 4A and Supplementary Table 2), hence indicating that CaLasSDE115 could play a role in CaLas infections by the Asian citrus psyllid. Moreover, the data showed that CaLasSDE115 expression in symptomatic leaves was significantly higher than that in asymptomatic ones, thereby indicating that CaLasSDE115 was involved in the development of symptoms in citrus. However, it was also found that the expression levels between midribs and roots, as well as between mature and young leaves were not significantly different (Figures 4B–D and Supplementary Table 2).
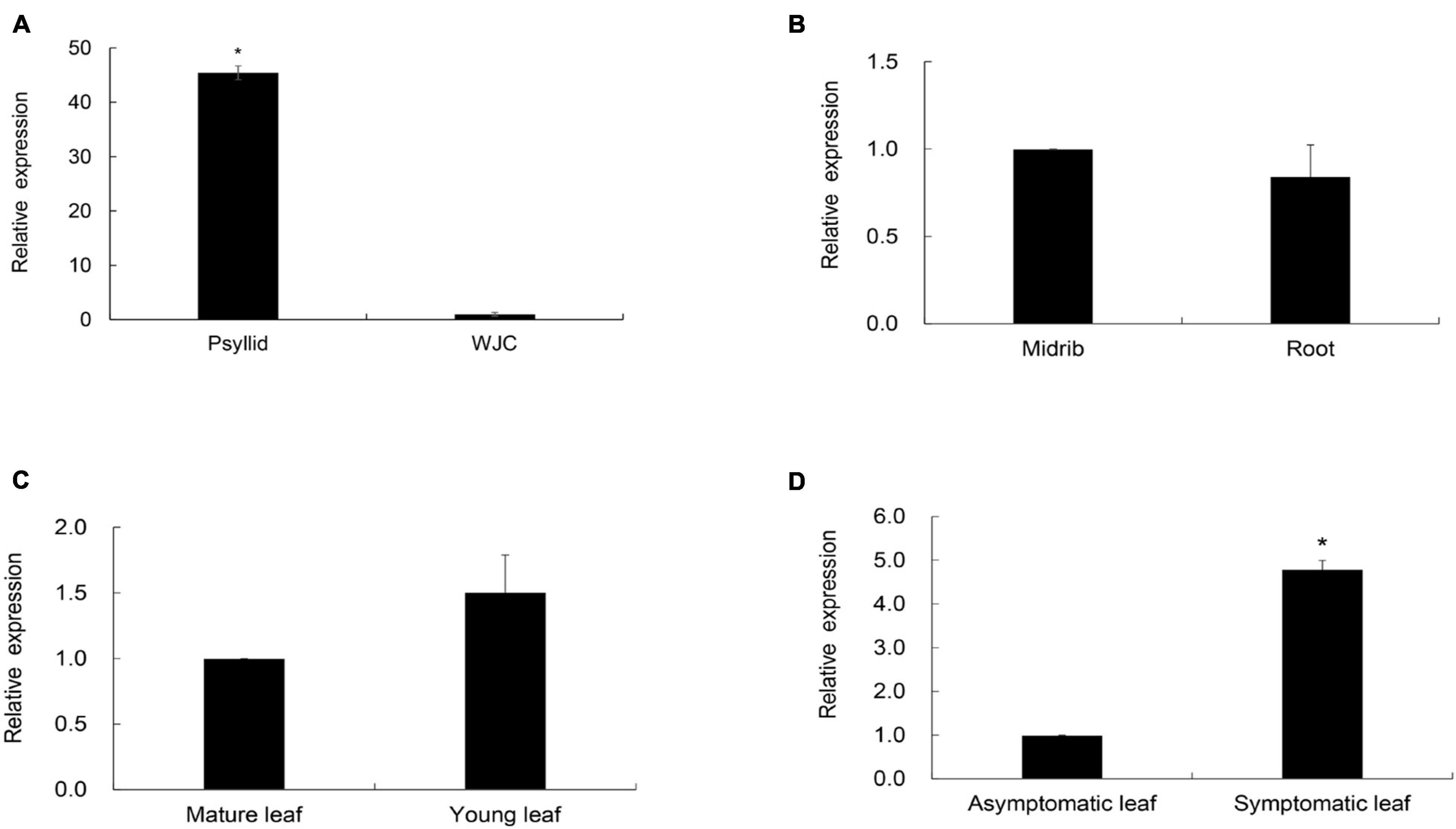
Figure 4. Transcription levels of CaLasSDE115 in Wanjincheng (WJC) orange and psyllids. Psyllid RNA was isolated from psyllid fed by CaLas-infected sweet orange for 3 weeks, and citrus RNA was isolated from Wanjincheng orange tissues infected by CaLas for 1 years. Using CaLasgyrA gene (GenBank No. CP001677.5) gene as internal reference, relative expressions in (A) psyllids, (B) root, (C) young leaf and (D) symptomatic leaf were determined compared to WJC orange, root, midrib, mature leaf and asymptomatic leaf, respectively. The Ct values of CaLasSDE115 and CaLasgyrA are listed in Supplementary Table 2. Values are expressed as means ± standard deviation of three independent tests. *on top of the bars indicates a significant difference (p < 0.05, Student’s t-test).
To understand the functions of CaLasSDE115 in CaLas pathogenicity, the mSDE115 gene, encoding for the mature protein SDE115, was used to construct the plant expression vector p35S:mSDE115 in which mSDE115 expression was controlled by a strong promoter 35S (Figure 5A). Four transgenic plants were identified by PCR (Figure 5B) and qRT-PCR analysis showed that all four had significant overexpression of mSDE115 transcripts, when compared with the wildtype control (Figure 5C). After being planted in a greenhouse, those transgenic plants displayed no obvious differences in phenotypes compared with WT ones.
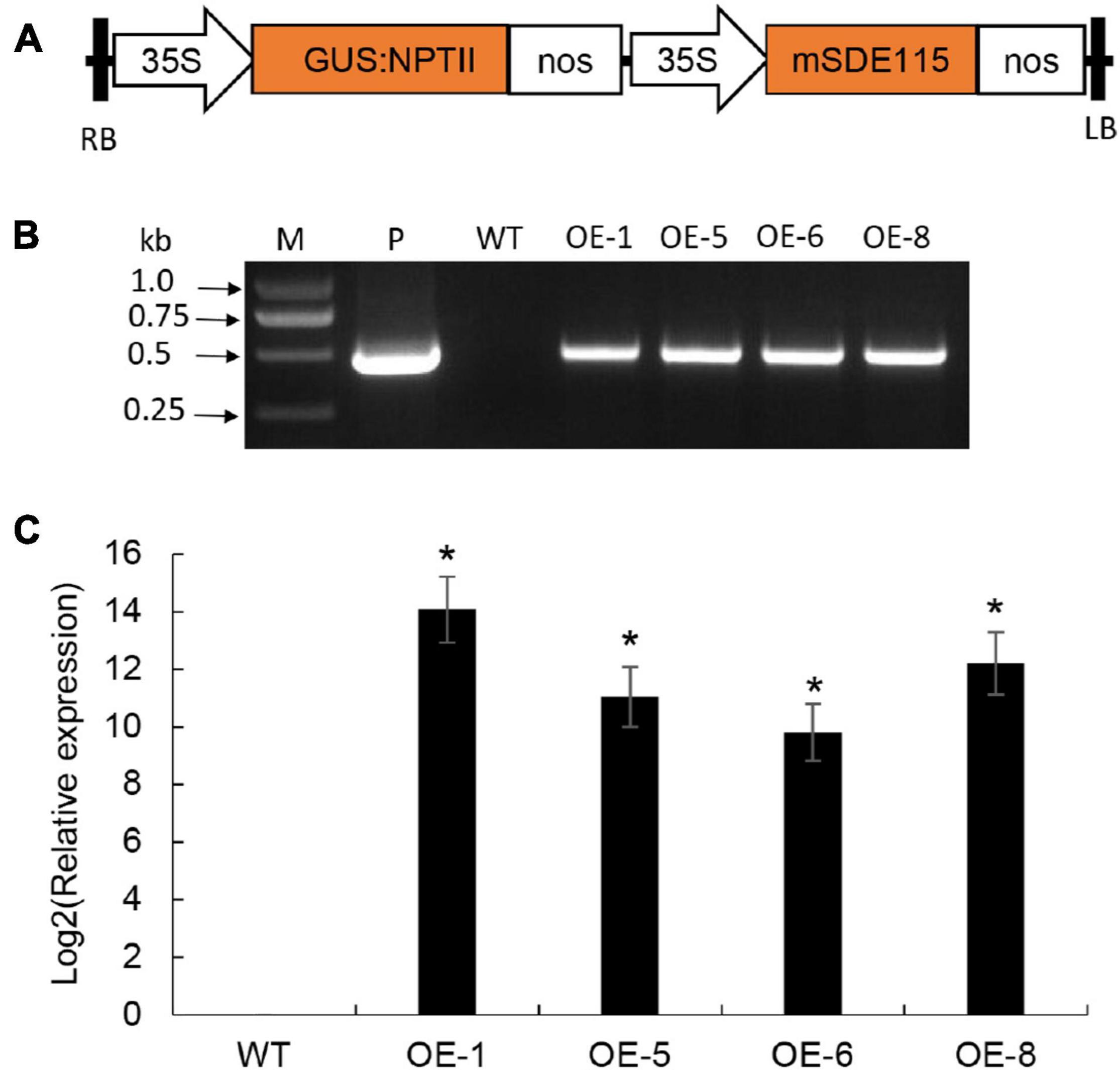
Figure 5. Wanjincheng orange transgenic plants overexpressing mSDE115. (A) T-DNA structure of plant expression vector for the genetic transformation of citrus. A 35S, CaMV 35S promoter; GUS:NPTII, fusion of β-glucuronidase and neomycin phosphotransferase genes (for the screening of citrus transformants); mSDE115, the coding sequence of CaLasSDE115 mature protein; NOS, the nopaline synthase terminator; LB, left border; RB, right border. (B) Identification of transgenic plants by PCR. M, DNA marker, P, p35S:mSDE115 plasmid; WT, wildtype control; OE-#, transgenic plants. (C) Relative expression levels of mSDE115 in transgenic plants. Relative expression of mSDE115 in transgenic plans was normalized against its expression in wildtype control using the citrus GAPDH gene (Mafra et al., 2012) as internal reference. Values are expressed as means ± standard deviation of three independent tests. *on top of the bars indicates significant differences compared to WT control (p < 0.05, Student’s t-test).
To determine the response of transgenic plants to CaLas infections, three to five plants per transgenic line, including the wildtype control, were infected by grafting them with branches containing CaLas. CaLas growth in plant tissues was then detected using qPCR at two, four and 6 months after the infection (Supplementary Table 3), with the data showing that bacterial growth in the transgenic plants was faster than that in WT plants after 2 months of infection (Figure 6A). In this case, the OE-1, OE-6, and OE-8 lines had significantly higher titers of CaLas compared with WT plants (Figure 6A). However, between two and 6 months after infection, CaLas growth accelerated in WT plants compared with transgenic ones, such that the CaLas titers in the latter, were no longer significantly different from those of WT plants at four to 6 months after inoculation (Figure 6A). After 6 months of infection, chlorosis or mottled yellow symptoms were first detected in the new leaves of transgenic plants while no visible symptoms appeared in the WT control (Figure 6B). Our data suggested that overexpression of mSDE115 favored early colonization by CaLas in citrus, which could promote the development of symptoms.
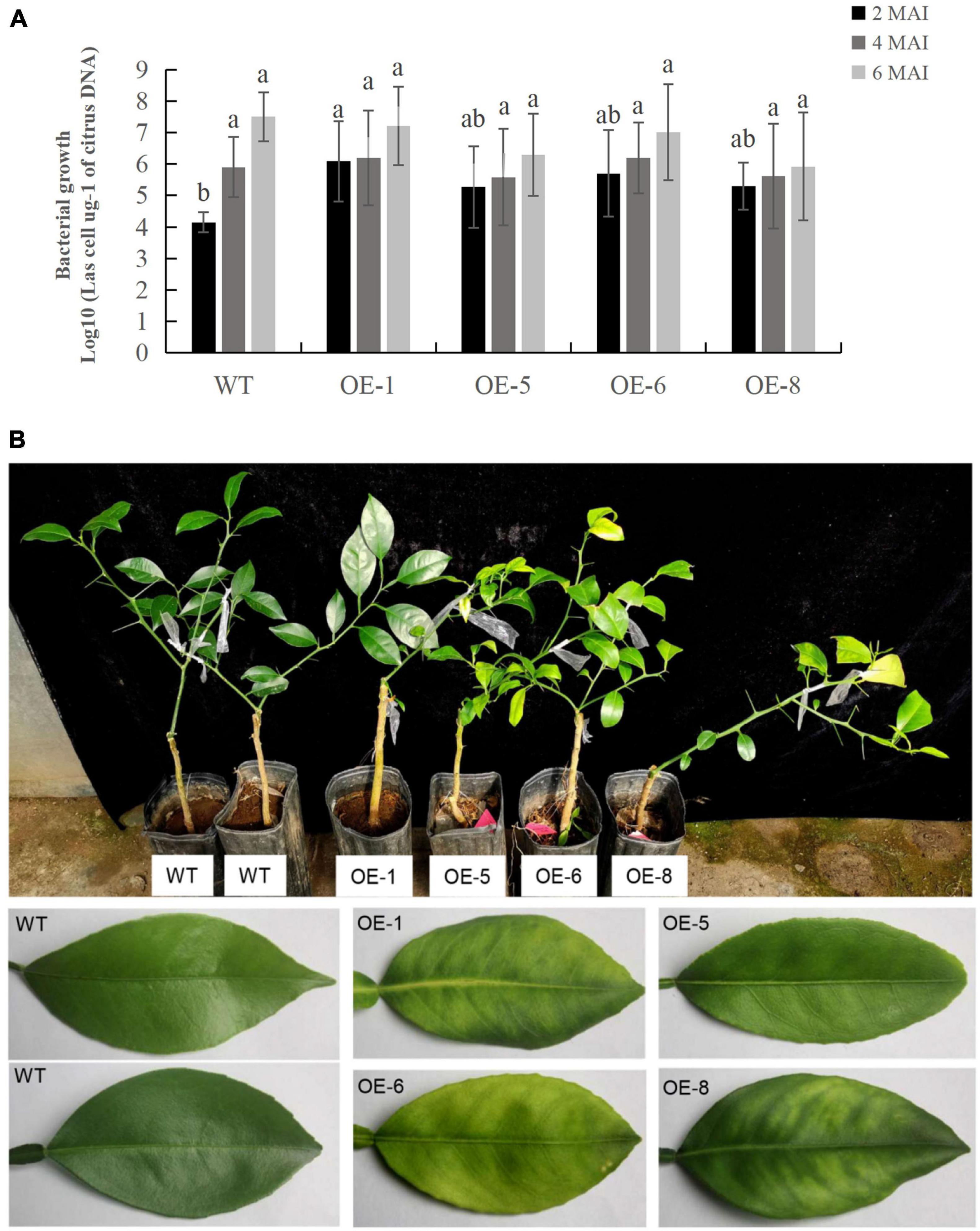
Figure 6. Evaluation of Citrus Huanglongbing (HLB) resistance in transgenic plants grown in a greenhouse. (A) Quantitative analysis of CaLas growth at 2, 4, and 6 months after infection (MAI). The bacterial populations (CaLas cells μg– 1 of citrus DNA) were determined using qPCR. Standard errors were calculated from three to five plants per line. Different letters on top of the bars indicates significant differences from the WT control (p < 0.05, Student’s t test). (B) HLB symptoms in the transgenic plants and wild type (WT) controls after 6 MAI. WT, wild type; OE-#, transgenic plants.
Plant SAR resistance plays important roles in citrus tolerance to HLB (Zou et al., 2021), with SA, MeSA, and JA being essential signals for plant SAR responses (Park et al., 2007; Zhu et al., 2014). To understand the effects of mSDE115 overexpression on SAR response, we first determined changes in the levels of SA, MeSA, and JA for OE1, OE6, and OE8 transgenic plants which were more susceptible to CaLas (Figure 7A). Compared with the WT control, SA and JA levels in all the transgenic lines were downregulated after overexpressing mSDE115 while MeSA content was not affected by the overexpression. Furthermore, the expression of nine SAR-associated genes, including three CsNPR3 genes (CsNPR3-73 for Ciclev10017873m, CsNPR3-15 for Ciclev10031115m, CsNPR3-49 for Ciclev10031749m) and one CsNPR4-08 (Ciclev10033908m) (Wang et al., 2016), and CsPR1, CsPR2, CsPR5, CsWRKY45, and CsWRKY70 (Zou et al., 2021) in transgenic plants were investigated by qRT-PCR. The results showed that the expression levels of all the PR and WRKY genes were significantly lower in OE1, OE6 and OE8 transgenic lines compared with the control. CsNPR3-15 and CsNPR3-49 exhibited enhanced expression while the expression of CsNPR3-73 and CsNPR4-08 was reduced (Figure 7B). The response characteristics of citrus SAR modulated by CaLasSDE115 was illustrated in Figure 7C.
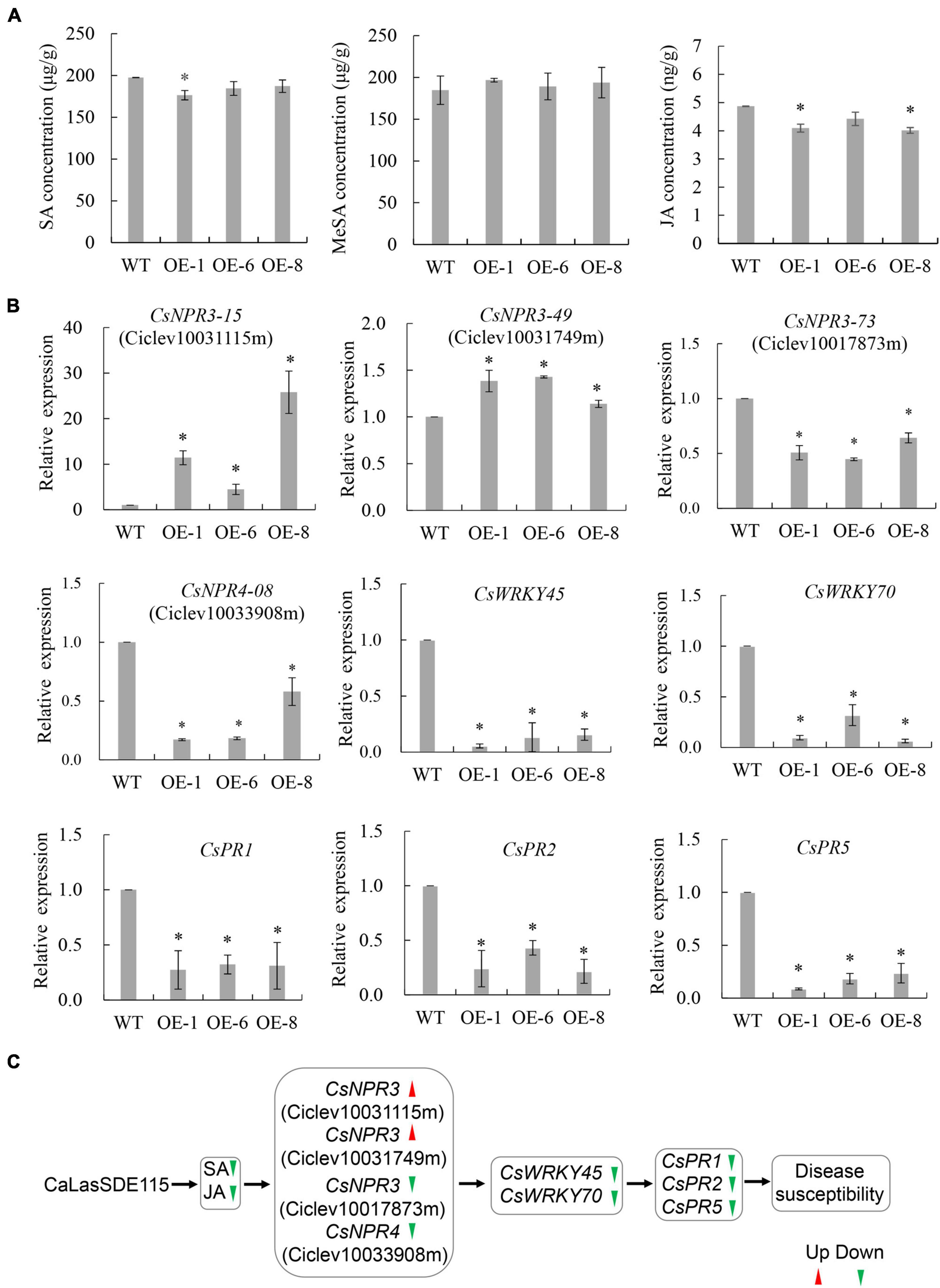
Figure 7. SAR characteristics of transgenic plants overexpressing mSDE115. (A) Characteristics of SA, MeSA, and JA contents in transgenic plants compared to WT control. (B) Expression characteristics of SAR-associated genes in transgenic plants. Compared to WT control, relative expressions were determined using GAPDH gene (Mafra et al., 2012) as internal reference. (C) Illustration of the transcriptional regulation of SAR response modulated by CaLasSDE115 effector in citrus. Red and green arrowheads indicate up-regulated and down-regulated, respectively. Values are expressed as means ± standard deviation of three independent tests. *on top of the bars indicates a significant difference (p < 0.05, T-test). WT, wild type; OE-#, transgenic plants.
Function prediction showed that CaLasSDE115 was very close to that of IalB homologs from Bartonella henselae, Rhizobium undicola, and Agrobacterium vitis. Bartonella IalB (BaIalB) is a 19.9 kDa protein with a putative signal peptide (Deng et al., 2018) and similarly, CaLasSDE115 is a 21.1 kDa protein with a signal peptide. In addition, BaIalB shares high homology with the Ail protein, a virulence marker of Yersinia enterocolitica that plays a major role in cell attachment and invasion (Deng et al., 2016). In this context, it has been shown that BaIalB is also a virulence factor for pathogen-erythrocyte invasion as the transformation of E. coli with BaIalB from Bartonella bacilliformis conferred the ability to invade erythrocytes while the deletion of this gene from both B. birtlesii and B. tribocorum reduced erythrocyte infection (Vayssier-Taussat et al., 2010; Deng et al., 2016). CaLas and Bartonella henselae are also both considered to be intracellular parasitic bacteria. In terms of functions, the expression of CaLasSDE115 was higher in young leaves of CaLas-infected WT plants compared with mature ones (Figure 4C), with young leaves generally considered to be the site where most new infections occur (Martinelli et al., 2013). Our evaluation of resistance confirmed that the overexpression of CaLasSDE115 in transgenic plants triggered CaLas growth mainly during the initial months, hence suggesting that, in the same way as IalB, CaLasSDE115 could contribute to early colonization of citrus by CaLas.
Despite the above similarities between CaLasSDE115 and IalB, it should be noted that the latter’s location in bacteria cell membrane remains unclear. For example, IalB from B. bacilliformis is an inner membrane protein, but that from B. henselae is an outer membrane protein (Coleman and Minnick, 2001; Chenoweth et al., 2004; Vayssier-Taussat et al., 2010; Saenz et al., 2017). Our data showed that CaLasSDE115 was secreted outside bacterial cells through a Sec-dependent secretion system. Prediction analysis of transmembrane domains revealed that the mature CaLasSDE115 protein was not a membrane protein. All the data suggested that the mature CaLasSDE115 protein could cross the bacterial cell membrane into the host cells to regulate host immune response. Subcellular localization analysis in tobacco implied that the CaLasSDE115 mature protein functioned possibly in the nucleus and cytoplasm of the plants. However, it has yet to be determined whether CaLasSDE115 adhered to extracellular surfaces of CaLas cells by binding to some bacterial membrane proteins or whether it was secreted into host cells.
The expression of virulence factors at different stages of the infection process is determined by continuous changes within the host’s environment (Chowdhury et al., 1996). CaLas is a plant pathogen transmitted by the Asian citrus psyllid and during transmission, it differentially expresses genes which are critical for its survival and/or pathogenicity in either host. This was reported by Yan et al. (2013) who compared the expression levels of 381 CaLas genes in plants and psyllids. In this case, the authors noted that 182 genes were up-regulated in plants when compared with psyllids while, at the same time, only 16 genes were up-regulated in the latter, with the majority of these 16 genes being predicted to be involved in cell motility (Yan et al., 2013). In this study, it was shown that the expression levels of CaLasSDE115 were significantly higher in psyllids compared with those in plants. In the same way in which Bartonella species enter mammalian erythrocytes (Deng et al., 2018), CaLas cells enter a psyllid’s body by crossing the gut epithelium before subsequently moving to other internal organs and tissues (Carmo-Sousa et al., 2020). Thus, it is suggested that CaLasSDE115 could also be involved in CaLas’s entry and intracellular movement in the Asian citrus psyllid.
Pathogen effectors have vital roles in maintaining the virulence of bacteria and symptoms of HLB. For instance, transient expression of CaLas5315 in tobacco can induce callose deposition (Pitino et al., 2016), while overexpressing CLIBASIA_03915 and CLIBASIA_04250 SDEs in Nicotiana benthamiana caused phloem necrosis in the senescent leaves (Li et al., 2020). Since the higher expression level of CaLasSDE115 in symptomatic leaves indicated that this effector could be contributing to the symptoms of HLB, the functions of CaLasSDE115 in symptom development was determined via the overexpression of the gene in HLB-susceptible Wanjincheng oranges. In this case, the results showed that CaLasSDE115 overexpression worsened the development of HLB symptoms in transgenic plants after CaLas infection.
Our data indicated that CaLasSDE115 was a virulence effector which participated in the regulation of the citrus SAR defense, with many reports even showing that CaLas triggered citrus susceptibility by suppressing the host’s SAR response (Martinelli et al., 2013; Zou et al., 2019). CaLas resides in the sieve elements and spreads systemically within plants through the phloem transport system (Folimonova and Achor, 2010). At the same time, SA, JA and MeSA also transfer SAR signals through this system to regulate the plant’s defense (Van Bel and Gaupels, 2004; Park et al., 2007). As a result, strong interactions occur between these signals and CaLas, leading to the possibility that CaLas could regulate SA, JA and MeSA-mediated defense response through its effectors in citrus. For example, Li et al. (2017) indicated that CaLas suppressed plant defenses by expressing a SahA to degrade the host’s SA. Similarly, Clark et al. (2018) showed that the CaLas effector SDE1 targeted citrus PLCPs proteases that were involved in SA-induced defense. It was shown that phytopathogen secreted effectors to directly bind NPR1, the master regulator of SA signaling, to inhibit the expression of downstream genes for dampening plant immunity (Chen et al., 2017; Han and Kahmann, 2019; Li et al., 2019). In this study, the overexpression of mSDE115 reduced SA and JA levels but also significantly decreased the expression of CsPR1, CsPR2, CsPR5, CsWRKY45, and CsWRKY70 in transgenic plants. PR2 is a SAR marker and the expression levels of CsPR2, CsPR5, WRKY45, and WRKY70 have been correlated with tolerance to HLB in citrus (Zou et al., 2019). These data indicated that mSDE115 could modulate some regulators of SA signaling to negatively regulate plant defense (Figure 7C). In addition, PR1, PR2, PR5, WRKY45, and WRKY70 have also been reported as being downstream of NPR-mediated signaling (Martinelli et al., 2013; Dutt et al., 2015; Ali et al., 2018). NPR4 is a required part of the basal defense against pathogens and, hence could be involved in the interactions between the pathogens and the SA- or JA-dependent signaling pathways (Liu et al., 2005). Furthermore, at low or basal concentration of SA, NPR3 and NPR4 serve as negative regulators to suppress defense genes, but once the SA concentration is elevated, NPR1-dependent gene expression is activated to defeat pathogen attack (Ding et al., 2018; Ding and Ding, 2020). Meanwhile, the suppression of NPR3 and NPR4 on defense genes is derepressed by increased SA, which further enhanced NPR1-dependent gene expression (Ding et al., 2018; Ding and Ding, 2020). CsNPR3 and CsNPR4 genes displayed different expression levels (two had increased expression and the other two had decreased expression) in the transgenic plants, thereby suggesting that they played different functions in CaLasSDE115-mediated defense responses. Our previous study confirmed that the overexpression of the NPR1-like homology (named as CsNPR3-49 here) from HLB-tolerant “Jackson” grapefruit (Citrus paradisi Macf.) enhanced resistance of Wanjincheng oranges to HLB (Peng et al., 2021). Interestingly, the current data showed that the expression of this gene was also increased by mSDE115 overexpression. Likewise, overexpressing AtNPR1 from Arabidopsis conferred enhanced resistance to HLB in citrus (Dutt et al., 2015). Thus, in future studies, it would be meaningful to acquire a deeper understanding of the functions of these CsNPR3 and CsNPR4 genes in response to CaLas infection.
Our study indicated that the CaLasSDE115 effector was an invasion-associated locus B of “Candidatus liberibacter asiaticus” and it participated in the early bacterial colonization of citrus. It also regulated citrus resistance to HLB through modulating the transcriptional regulation of SAR-related genes (Figure 7C). However, the potential molecular mechanism of this effector during interactions between the pathogen and the host remains to be verified, especially to determine whether the CaLasSDE115 effector targeted specific host proteins or genes and this could form the basis of future studies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
MD and XZ designedthe experiments, analyzed the data, wrote, and revised the manuscript. MD, SW, RQ, LD, LZ, YH, and SC performed the experiments. XZ supervised the research. All authors read and approved the final version of the manuscript.
This work was supported by the National Natural Science Foundation of China (31972393), the Key-Area Research and Development Program of Guangdong Province (2018B020202009), Science and Technology Major Project of Guangxi (Gui Ke AA18118046-6) and the Earmarked Fund for China Agriculture Research System (CARS-27).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Weimin Li (Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China) for providing pET-mphoA and pET-phoA plasmids and Shuo Duan (Gannan Normal University, China) for providing ACPs.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.797841/full#supplementary-material
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018). Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 212-213, 29–37. doi: 10.1016/j.micres.2018.04.008
Bassanezi, R. B., Montesino, L. H., and Stuchi, E. S. (2009). Effects of huanglongbing on fruit quality of sweet orange cultivars in Brazil. Eur. J. Plant Pathol. 125, 565–572. doi: 10.3389/fpls.2021.641457
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant
Bové, J. M. (2006). Huanglongbing: a destructive, newly emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37. doi: 10.1371/journal.pone.0111032
Carmo-Sousa, M., Cortés, M. T. B., and Lopes, J. R. S. (2020). Understanding psyllid transmission of candidatus liberibacter as a basis for managing huanglongbing. Trop. Plant Pathol. 45, 572–585. doi: 10.1007/s00248-016-0733-9
Chen, H., Chen, J., Li, M., Chang, M., Xu, K., Shang, Z., et al. (2017). A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788. doi: 10.1016/j.chom
Chenoweth, M. R., Greene, C. E., Krause, D. C., and Gherardini, F. C. (2004). Predominant outer membrane antigens of bartonella henselae. Infect. Immuni. 72, 3097–3105. doi: 10.1128/IAI.72.6.3097-3105
Chowdhury, R., Sahu, G. K., and Das, J. (1996). Stress response in pathogenic bacteria. J. Biosci. 21, 149–160. doi: 10.1007/BF02703105
Cifuentes-Arenas, J. C., Beattie, G. A. C., and Lopes, S. A. (2019). Murraya paniculata and swinglea glutinosa as short-term transient hosts of ‘ candidatus liberibacter asiaticus’ and implications for the spread of huanglongbing. Phytopathology 109, 2064–2073. doi: 10.1094/PHYTO-06-19-0216-R
Clark, K., Franco, J. Y., Schwizer, S., Pang, Z. Q., Hawara, E., Liebrand, T. W. H., et al. (2018). An effector from the huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 9:1718. doi: 10.1038/s41467-018-04140-9
Coleman, S. A., and Minnick, M. F. (2001). Establishing a direct role for the bartonella bacilliformis invasion-associated locus b (ialb) protein in human erythrocyte parasitism. Infect. Immun. 69, 4373–4381. doi: 10.1128/IAI.69.7.4373-4381.2001
Deng, H., Pang, Q., Xia, H., Le Rhun, D., Naour, E. L., Yang, C., et al. (2016). Identification and functional analysis of invasion associated locus b (ialb) in bartonella species. Microb. Pathog. 98, 171–177. doi: 10.1016/j.micpath.2016.05.007
Deng, H., Pang, Q., Zhao, B., and Vayssier-Taussat, M. (2018). Molecular mechanisms of Bartonella and mammalian erythrocyte interactions: a review. Front. Cell. Infect. Microbiol. 8:431. doi: 10.3389/fcimb.2018.00431
Ding, P., and Ding, Y. (2020). Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 25, 549–565. doi: 10.1016/j.tplants.2020.01.004
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1–14. doi: 10.1016/j.cell.2018.03.044
Du, P., Zhang, C., Zou, X., Zhu, Z., Yan, H., Wuriyanghan, H., et al. (2021). ‘Candidatus Liberibacter asiaticus’ secretes nonclassically cecreted proteins that suppress host hypersensitive cell death and induce expression of plant pathogenesis-related proteins. Appl. Environ. Microbiol. 87:e00019-21. doi: 10.1128/AEM.00019-21
Duan, Y., Zhou, L., Hall, D. G., Li, W., Doddapaneni, H., Lin, H., et al. (2009). Complete genome sequence of citrus huanglongbing bacterium, ‘candidatus liberibacter asiaticus’ obtained through metagenomics. Mol. Plant Microbe Interact. 22, 1011–1020. doi: 10.1094/MPMI-22-8-1011
Dutt, M., Barthe, G., Irey, M., and Grosser, J. (2015). Transgenic citrus expressing an Arabidopsis NPR1 gene exhibit enhanced resistance against huanglongbing (HLB; Citrus Greening). PLoS One 10:e0137134. doi: 10.1371/journal.pone.0137134
Folimonova, S. Y., and Achor, D. S. (2010). Early events of citrus greening (Huanglongbing) disease development at the ultrastructural level. Phytopathology 100, 949–958. doi: 10.1094/PHYTO-100-9-0949
Gottwald, T. R. (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009-114418
Graham, J. H., Johnson, E. G., Gottwald, T. R., and Irey, M. S. (2013). Pre-symptomatic fibrous root decline in citrus trees caused by huanglongbing and potential interaction with phytophthora spp. Plant Dis. 97, 1195–1199. doi: 10.1094/PDIS-01-13-0024-RE
Haitao, C., Kenichi, T., and Jane, E. P. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Han, X., and Kahmann, R. (2019). Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10:822. doi: 10.3389/fpls.2019.00822
Jagoueix, S., Bove, J., and Garnier, M. (1994). The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the proteobacteria. Int. J. Syst. Bacteriol. 44, 379–386. doi: 10.1099/00207713-44-3-379
Jain, M., Fleites, L. A., and Gabriel, D. W. (2015). Prophage-encoded peroxidase in ‘candidatus liberibacter asiaticus’ is a secreted effector that suppresses plant defenses. Mol. Plant Microbe Interact. 28, 1330–1337. doi: 10.1094/MPMI-07-15-0145-R
Jain, M., Munoz-Bodnar, A., and Gabriel, D. W. (2019). ‘Candidatus liberibacter asiaticus’ peroxiredoxin (LasBCP) suppresses oxylipin-mediated defense signaling in citrus. J. Plant Physiol. 236, 61–65. doi: 10.1016/j.jplph.2019.03.001
Jain, M., Munoz-Bodnar, A., Zhang, S., and Gabriel, D. W. (2018). A secreted ‘candidatus liberibacter asiaticus’ peroxiredoxin simultaneously suppresses both localized and systemic innate immune responses in planta. Mol. Plant Microbe Interact. 31, 1312–1322. doi: 10.1094/MPMI-03-18-0068-R
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kumar, S., Stecher, G., and Tamura, K. (2016). Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, H., Ying, X., Shang, L., Redfern, B., Kypraios, N., Xie, X., et al. (2020). Heterologous expression of clibasia_03915/clibasia_04250 by tobacco mosaic virus resulted in phloem necrosis in the senescent leaves of nicotiana benthamiana. Int. J. Mol. Sci. 21:1414. doi: 10.3390/ijms21041414
Li, J., Pang, Z., Trivedi, P., Zhou, X., Ying, X., Jia, H., et al. (2017). ‘Candidatus liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant Microbe Interact. 30, 620–630. doi: 10.1094/MPMI-12-16-0257-R
Li, Q., Chen, Y., Wang, J., Zou, F., Jia, Y. L., Shen, D. Y., et al. (2019). A phytophthora capsici virulence effector associates with NPR1 and suppresses plant immune responses. Phytopathol. Res. 1:6. doi: 10.1186/s42483-019-0013-y
Lin, H., Pietersen, G., Han, C., Read, D. A., Lou, B., Gupta, G., et al. (2015). Complete genome sequence of ‘candidatus liberibacter africanus,’ a bacterium associated with citrus huanglongbing. Genome Announc. 3:e00733-15. doi: 10.1128/genomeA.00733-15
Liu, G., Holub, E. B., Alonso, J. M., Ecker, J. R., and Fobert, P. R. (2005). An Arabidopsis NPR1-like gene. NPR4, is required for disease resistance. Plant J. 41, 304–318. doi: 10.1111/j.1365-313X.2004.02296.x
Liu, X., Fan, Y., Zhang, C., Dai, M., Wang, X., and Li, W. (2019). Nuclear import of a secreted ‘candidatus liberibacter asiaticus’ protein is temperature dependent and contributes to pathogenicity in nicotiana benthamiana. Front. Microbiol. 10:1684. doi: 10.3389/fmicb.2019.01684
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2–Δ Δ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mafra, V., Kubo, K. S., Alves-Ferreira, M., Ribeiro-Alves, M., Stuart, R. M., Boava, L. P., et al. (2012). Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 7:e31263. doi: 10.1371/journal.pone.0031263
Martin, K., Kopperud, K., Chakrabarty, R., Banerjee, R., Brooks, R., and Goodin, M. M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162. doi: 10.1111/j.1365-313X.2009.03850.x
Martinelli, F., Reagan, R. L., Uratsu, S. L., Phu, M. L., Albrecht, U., Zhao, W., et al. (2013). Gene regulatory networks elucidating huanglongbing disease mechanisms. PLoS One 8:e74256. doi: 10.1371/journal.pone.0074256
Nguyen, Q. M., Iswanto, A., Son, G. H., and Sang, H. K. (2021). Recent advances in effector-triggered immunity in plants: new pieces in the puzzle create a different paradigm. Int. J. Mol. Sci. 22:4709. doi: 10.3390/ijms22094709
Pagliaccia, D., Shi, J. X., Pang, Z. Q., Hawara, E., Clark, K., Thapa, S. P., et al. (2017). A pathogen secreted protein as a detection marker for citrus huanglongbing. Front. Microbiol. 8:2041. doi: 10.3389/fmicb.2017.02041
Pang, Z., Zhang, L., Coaker, G. L., Ma, W., and Wang, N. (2020). Citrus CsACD2 is a target of candidatus liberibacter asiaticus in huanglongbing disease. Plant Physiol. 184, 792–805. doi: 10.1104/pp.20.00348
Park, S. W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Peng, A., Xu, L., He, Y., Lei, T., Yao, L., Chen, S., et al. (2015). Efficient production of marker-free transgenic ‘Tarocco’ blood orange (Citrus sinensis Osbeck) with enhanced resistance to citrus canker using a Cre/loxP site-recombination system. Plant Cell Tissue Organ Cult. 123, 1–13. doi: 10.1007/s11240-015-0799-y
Peng, A., Zou, X., He, Y., Chen, S., Liu, X., Zhang, J., et al. (2021). Overexpressing a NPR1-like gene from citrus paradisi enhanced huanglongbing resistance in C. sinensis. Plant Cell Rep. 40, 529–541. doi: 10.1007/s00299-020-02648-3
Pitino, M., Armstrong, C. M., Cano, L. M., and Duan, Y. (2016). Transient expression of candidatus liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci. 7:982. doi: 10.3389/fpls.2016.00982
Prasad, S., Xu, J., Zhang, Y., and Wang, N. (2016). SDE-translocon dependent extracytoplasmic proteins of candidatus liberibacter asiaticus. Front. Microbiol. 7:1989. doi: 10.3389/fmicb.2016.01989
Saenz, H., Engel, P., Stoeckli, M., Lanz, C., Raddatz, G., Vayssier-Taussat, M., et al. (2017). Genomic analysis of bartonella identifies type IV secretion systems as host adaptability factors. Nat. Genet. 39, 1469–1476. doi: 10.1038/ng.2007.38
Saskia, A. H., Kenro, O. S., Ammar, E. D., Shigeyuki, K., Heather, N., and Kingdom, S. N. (2008). Phytoplasmas: bacteria that manipulate plants and insects. Mol. Plant Pathol. 9, 403–423. doi: 10.1111/j.1364-3703.2008.00472.x
Segonzac, C., and Zipfel, C. (2011). Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. doi: 10.1016/j.mib.2010.12.005
Tamar, C. S., and Damon, H. (2018). The way is the goal: how seca transports proteins across the cytoplasmic membrane in bacteria. Fems Microbiol. Lett. 365:fny093. doi: 10.1093/femsle/fny093
Teixeira, D. C., Saillard, C., Eveillard, S., Danet, J. L., Ayres, A. J., and Bové, J. M. (2005). A new liberibacter species, candidatus liberibacter americanus sp. nov. is associated with citrus huanglongbing (greening disease) in São Paulo State, Brazil. Int. J. Syst. Evol. Microbiol. 55, 1857–1862. doi: 10.1099/ijs.0.63677-0
Toruno, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
Van Bel, A. J., and Gaupels, F. (2004). Pathogen-induced resistance and alarm signals in the phloem. Mol. Plant Pathol. 5, 495–504. doi: 10.1111/j.1364-3703.2004.00243.x
Vayssier-Taussat, M., Rhun, D. L., Deng, H. K., Biville, F., Cescau, S., Danchin, A., et al. (2010). The trw type iv secretion system of bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 6:e1000946. doi: 10.1371/journal.ppat.1000946
Wang, N., Pierson, E. A., Setubal, J. C., Xu, J., Levy, J. G., Zhang, Y., et al. (2017). The candidatus liberibacter-host interface: insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 55, 451–482. doi: 10.1146/annurev-phyto-080516-035513
Wang, Y., Zhou, L., Yu, X., Stover, E., Luo, F., and Duan, Y. (2016). Transcriptome profiling of huanglongbing (HLB) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 7:933. doi: 10.3389/fpls.2016.00933
Wulff, N. A., Zhang, S., Setubal, J. C., Almeida, N. F., Martins, E. C., Harakava, R., et al. (2014). The complete genome sequence of ‘candidatus liberibacter americanus’, associated with citrus huanglongbing. Mol. Plant Microbe Interact. 27, 163–176. doi: 10.1094/MPMI-09-13-0292-R
Yan, Q., Sreedharan, A., Wei, S., Wang, J., Pelz-Stelinski, K., Folimonova, S., et al. (2013). Global gene expression changes in candidatus liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol. Plant Pathol. 14, 391–404. doi: 10.1111/mpp.12015
Ying, X., Wan, M., Hu, L., Zhang, J., Li, H., and Lv, D. (2019). Identification of the virulence factors of candidatus liberibacter asiaticus via heterologous expression in nicotiana benthamiana using tobacco mosaic virus. Int. J. Mol. Sci. 20:5575. doi: 10.3390/ijms20225575
Yuan, M. H., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Zhu, F., Xi, D. H., Yuan, S., Xu, F., Zhang, D. W., and Lin, H. H. (2014). Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in nicotiana benthamiana. Mol. Plant Microbe Interact. 27, 567–577. doi: 10.1094/MPMI-11-13-0349-R
Zou, X., Bai, X., Wen, Q., Xie, Z., Wu, L., Peng, A., et al. (2019). Comparative analysis of tolerant and susceptible citrus reveals the role of methyl salicylate signaling in the response to huanglongbing. J. Plant Growth Regul. 38, 1516–1528. doi: 10.1007/s00344-019-09953-6
Zou, X., Jiang, X., Xu, L., Lei, T., Peng, A., He, Y., et al. (2017). Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to huanglongbing. Plant Mol. Biol. 93, 341–353. doi: 10.1007/s11103-016-0565-5
Keywords: Citrus Huanglongbing, Candidatus Liberibacter asiaticus, effector, LasSDE115, overexpression, SAR, early colonization
Citation: Du M, Wang S, Dong L, Qu R, Zheng L, He Y, Chen S and Zou X (2022) Overexpression of a “Candidatus Liberibacter Asiaticus” Effector Gene CaLasSDE115 Contributes to Early Colonization in Citrus sinensis. Front. Microbiol. 12:797841. doi: 10.3389/fmicb.2021.797841
Received: 19 October 2021; Accepted: 20 December 2021;
Published: 21 February 2022.
Edited by:
Kranthi Kiran Mandadi, Texas A&M University, United StatesReviewed by:
Veronica Ancona, Texas A&M University-Kingsville, United StatesCopyright © 2022 Du, Wang, Dong, Qu, Zheng, He, Chen and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuping Zou, em91eGl1cGluZ0BjcmljLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.