- 1China Pharmaceutical Culture Collection, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Xinxiang Key Laboratory of Pathogenic Biology, Department of Pathogenic Biology, School of Basic Medical Sciences, Xinxiang Medical University, Xinxiang, China
Particulate matter (PM) has been a threat to the environment and public health in the metropolises of developing industrial countries such as Beijing. The microorganisms associated with PM have an impact on human health if they are exposed to the respiratory tract persistently. There are few reports on the microbial resources collected from PM and their antimicrobial activities. In this study, we greatly expanded the diversity of available commensal organisms by collecting 1,258 bacterial and 456 fungal isolates from 63 PM samples. A total of 77 bacterial genera and 35 fungal genera were included in our pure cultures, with Bacillus as the most prevalent cultured bacterial genus, Aspergillus, and Penicillium as the most prevalent fungal ones. During heavy-haze days, the numbers of colony-forming units (CFUs) and isolates of bacteria and fungi were decreased. Bacillus, Paenibacillus, and Chaetomium were found to be enriched during haze days, while Kocuria, Microbacterium, and Penicillium were found to be enriched during non-haze days. Antimicrobial activity against common pathogens have been found in 40 bacterial representatives and 1 fungal representative. The collection of airborne strains will provide a basis to greatly increase our understanding of the relationship between bacteria and fungi associated with PM and human health.
Introduction
In recent years, frequent haze caused mainly by particulate matter (PM) has been a severe problem threatening public health in north China (Cheng et al., 2013; Cao et al., 2014; Gao et al., 2015; Tan et al., 2016; Yan et al., 2016, 2018; Guo et al., 2020; Yang et al., 2021; Zhao et al., 2021). PM is divided into three categories: total suspended particulates (TSP), PM10 (particulate size smaller than 10 μm), and PM2.5 (particulate size smaller than 2.5 μm), which are a mixture of inorganic, organic, and biological components (Van Dingenen et al., 2004). Various PM fractions show significantly different compositions (Zhang et al., 2006). Previous studies have indicated that PM has an important role in air pollution, visibility reduction, and climate change (Booth et al., 2012; Wang et al., 2012; Randles et al., 2013; Shimadera et al., 2013). More importantly, PM has been shown to increase the morbidity and mortality from stroke (Huang et al., 2019), respiratory disease (Zhang and Cao, 2015), heart disease (Parker et al., 2018; Li et al., 2020a), and lung cancer (Hamra et al., 2014; Li et al., 2020b).
Microorganisms associated with PM are known as bioaerosols in the atmosphere, which contribute up to 25% of aerosolized matter (Jaenicke, 2005). Airborne microorganisms may play an important role in human health, either as human pathogens or allergens (Vermani et al., 2010; Yamamoto et al., 2012; Haas et al., 2013). More microbial pathogens and allergens have been linked to higher levels of PM pollution (Cao et al., 2014). The diversity and community composition of PMs-associated airborne microorganisms have been revealed by high-throughput sequencing in China (Woo et al., 2013; Cao et al., 2014; Wei et al., 2016; Yan et al., 2016, 2018; Zhong et al., 2019; Yu et al., 2020), United States (Bowers et al., 2011a,b, 2013; Be et al., 2015), Italy (Franzetti et al., 2011; Bertolini et al., 2013; Romano et al., 2020), Antarctica (Bottos et al., 2014), and Spain (Barberan et al., 2014). Despite the fact that the culture-independent method can reflect the diversity of airborne microbial community rapidly, the roles and functions of culturable microorganisms which can show the potential ability in human health and atmospheric chemistry are neglected. Using culture-dependent methods, the concentrations and composition of the culturable microbial community in the atmosphere have been investigated (Haas et al., 2013; Alghamdi et al., 2014; Fang, 2014; Li et al., 2015; Niazi et al., 2015; Oh et al., 2015; Dong et al., 2016; Okubo et al., 2017; Yuan et al., 2017). However, the microbial communities in various PM fractions (TSP, PM10, and PM2.5) have rarely been combined for study, and thus are not well understood during haze and non-haze days. We can obtain the microbial strains for further study of the metabolic functions of airborne microorganisms using the culture-dependent method.
There is a continuous challenge for novel antibiotics to overcome the serious problem of evolving pathogens, naturally resistant bacteria and fungi, and multidrug resistance among common microbial pathogens (Alanis, 2005; Sharma et al., 2011; Shah et al., 2017). This crisis has prompted experts to call for the revival of natural product drug discovery (Lewis, 2016). However, the enormous known compounds in the background covered the new ones and presents a serious barrier to discovery (Lewis, 2016). Despite this, recent results like those presented by Maffioli et al. (2017) bring hope to the search for new leads from microbial-extract screening. They reported that a nucleoside-analog inhibitor, pseudouridimycin, inhibited bacterial RNA polymerase and exhibits antibacterial activity against drug-resistant bacterial pathogens (Maffioli et al., 2017). The novel microorganisms with unique metabolic properties may be selected by the extreme conditions with wide-ranging and fluctuating temperatures, high levels of solar irradiation, strong oxidizing, and dryness in the atmosphere (Polymenakou, 2012). Nonetheless, there is a scarce number of studies involving microorganisms collected from the air in antimicrobial activity screening (Sobral et al., 2017).
Microbial resources in the atmosphere are a great treasure, while there are few studies on the isolation of airborne microorganisms, especially microorganisms associated with PM. As we know, no study reported that whether the haze influenced the culturable microorganisms in three types of PM fractions, and anti-microbial activities of microorganisms were screened from the PM. The aim in this study is to address the following questions: (1) what are the concentrations of culturable microbial colonies in various haze levels? (2) Does the culturable microbial community composition differ among various PM samples, or different haze-level samples? (3) Which isolated strains from PM samples show anti-microbial activities against common pathogens?
Materials and Methods
Sample Collection
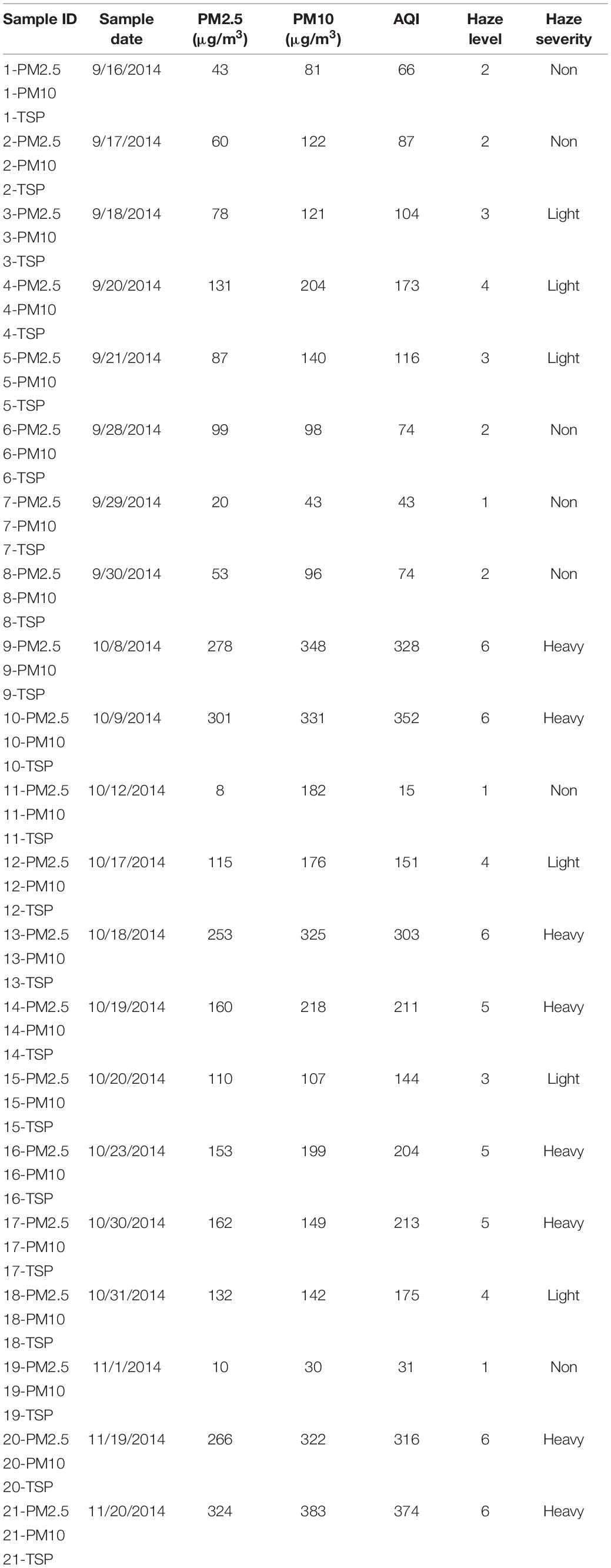
PM2.5 samples (the samples containing particulates smaller than 2.5 μm), PM10 samples (the samples containing particulates smaller than 10 μm), and TSP samples (the samples containing total suspended particulates) were collected from the roof of the Conference Building at the Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences (39°52′43″N, 116°23′21″E, ∼8 m above the ground, ∼400 m from Temple of Heaven Park), an area without major industrial pollution sources nearby. We collected 63 PM samples for 21 days (September 2014 to November 2014) during various haze levels. Sampling was conducted by three portable ambient air samplers (AirMetrics, United States): the impactors were removed from the filter holder of the first for TSP samples, the second was assembled with PM10 impactor for PM10 samples, and the third was assembled with both PM10 and PM2.5 impactors for PM2.5 samples. Ambient air was drawn at an average flow rate of 5 L/min for 24 h per sampling day. PM samples were collected on 47-mm quartz aerosol collection filters (Pall, United States). All the filters were sterilized by autoclaving at 121°C for 20 min before sampling. The filter holders were cleaned with 75% ethanol and all the tools used for changing new filters were autoclaved to avoid contamination (Yan et al., 2016). After sampling, the filters were cut into small pieces and kept in a 2-mL centrifuge tube with 1 mL of saline.
The air quality index (AQI) which is an index for reporting air quality (Yang et al., 2016) was used to indicate the haze level. We defined a day with AQI lower than 100 as a non-haze day, that with AQI in the range of 100–200 as a light-haze day, that with AQI higher than 200 as a heavy-haze day. The environmental parameters such as CO, SO2, and NO2 were recorded from the monitoring data of Temple of Heaven Park site (∼800 m from sampling site) of Beijing Municipal Environmental Monitoring Center1. Temperature (Temp) and relative humidity (RH) were recorded according to the reports of the Chinese National Meteorological Center2. The environmental parameters are listed in Supplementary Table 1.
Isolation and Cultivation of Airborne Bacteria and Fungi
After sampling, the samples were transferred to the laboratory as soon as possible and vortexed for 10 min at first to make the bioaerosol evenly distribute in the saline. Due to the low concentration of airborne microorganisms, 1 mL of PM2.5 samples were diluted with 500 μL of saline, and 500 μL of PM10 samples and 500 μL of TSP samples were diluted with 1 mL of saline, respectively. Then 100 μL of each diluted PM suspension were spread-plated onto four kinds of medium plates, namely; nutrient agar medium (NA) (Haibo, China), tryptic soy agar medium (TSA) (Haibo, China) for the cultivation of bacteria, sabouraud dextrose agar medium (SDA) (OXOID, United Kingdom), and potato dextrose agar medium (PDA) (OXOID, United Kingdom) for cultivation of fungi. Three replicates of each PM sample were spread-plated in each medium. The medium NA and TSA were added with nystatin (0.05 mg/mL) to inhibit the growth of fungi, while SDA and PDA were added with tetracycline (0.05 mg/mL) and streptomycin sulfate (0.05 mg/mL) to inhibit the growth of bacteria. Plates for the cultivation of bacteria were then incubated at 37°C and plates for the cultivation of fungi were incubated at 28°C. After 5 days of incubation, the number of colony-forming units (CFUs) were counted. From each plate, all phenotypically distinct colonies were picked onto fresh media for isolation. Single colonies were picked and re-streaked at least three times to isolate individual strains. Strains were grown in the corresponding medium on the agar slant and temperature conditions and frozen at –80°C in 20% glycerol. A subset of representative isolates (Supplementary Table 2) has been deposited at China Pharmaceutical Culture Collection and the CAMS Collection Center of Pathogenic Microorganisms, Division for Medicinal Microorganisms Related Strains.
Identification of Airborne Bacteria and Fungi
Genomic DNA of the strains on the agar slant was extracted using Chelex-100 method. The bacterial 16S rRNA gene was amplified by PCR (95°C for 5 min, followed by 30 cycles at 95°C for 30 s, 59°C for 60 s, and 72°C for 90 s and a final extension at 72°C for 10 min) using primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′) (Kanagawa, 2003). The fungal internal transcribed spacer (ITS) regions were amplified by PCR (95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 55°C for 45 s, and 72°C for 45 s and a final extension at 72°C for 10 min) using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′- TCCTCCGCTTATTGATATGC-3′) (White et al., 1989). PCR products were purified and Sanger sequenced at Sangon Biotech (Sangon, Shanghai, China). The 60 bp of the beginning 16S rRNA reads were cut off and cut into 660 bp to ensure the accuracy of the sequences. Similarly, the 60 bp of the beginning ITS reads were cut off and cut into 410 bp with Bioedit software. Thus the bacterial sequences were compared with available 16S rRNA gene sequences from GenBank using the BLAST program and a web-based tool at EzTaxon3 as described by Yoon et al. (2017) to determine the approximate phylogenetic affiliation. The top hit from BLAST analysis and GenBank was used to determine fungal species.
Processing and Statistical Analyses of Sequencing Data
Isolates were clustered to operational taxonomic units (OTUs) with 100% similarity cutoff. The OTUs were aligned using the default settings of MAFFT online (Katoh et al., 2017). The evolutionary trees were constructed using MEGA software version 7 on the basis of the neighbor-joining method (Sudhir et al., 2016) and visualized using Interactive Tree Of Life (iTOL4) (Ivica and Peer, 2019). The canonical correspondence analysis (CCA) and permutation test were conducted using R software (version 3.7.0) with the vegan package (R Core Team, 2018). Bubble plots, bar plots, line plots, and violin plots were created using ggplot2 package in the R software. Post hoc tests for ANOVA were performed using Statistical Analysis of Metagenomics Profiles software (STAMP) (Parks et al., 2014) to identify significantly different genera among non-, light, and heavy-haze days.
Screening for Antimicrobial Activity
The antimicrobial activities of the isolates were tested against four type strains of common pathogen, Escherichia coli ATCC 25922, Enterococcus faecalis ATCC 29212, Klebsiella pneumonia ATCC 700603, Candida albicans ATCC 10231, using the Kirby–Bauer method (Patel, 2014). The bacterial representatives (except Actinobacteria) were cultivated in B1 and B2 medium while the representatives of Actinobacteria in A1 and A2 medium at 28°C with shaking at 200 rpm for 72 h. The fungal representatives were cultivated in F1 and F2 medium (Supplementary Table 3) at 28°C with shaking at 200 rpm for 96 h. The fermentation broths were centrifuged at 12,000 rpm for 10 min and the supernatant was transferred to a 1.5 mL sterilized tube carefully without disturbing the bacterial pellets. To prepare test plates of the type strains, E. faecalis ATCC 29212 were cultured onto brain-heart infusion agar (Haibo, China) plates, E. coli ATCC 25922 and K. pneumonia ATCC 700603 were cultured onto Mueller–Hinton agar (AOBOX, China) plates for 18–24 h at 37°C. Candida albicans ATCC 10231 were cultured onto modified Thayer-Martin agar (Haibo, China) plates for 18–24 h at 28°C. Pure colonies from these plates were cultured in corresponding broth for 4–6 h, adjusted to a 0.5 McFarland turbidity standard suspension which was diluted to 1,000 times in the agar medium, and then poured into petri plates. Each sterile disc was impregnated with 40 μL of the fermentation supernatant, placed, and incubated on the corresponding agar at 37°C for 16–24 h (except for Candida albicans, which was cultured at 28°C for 16–24 h) and evaluated for inhibition zones.
Results
The Effects of Haze on the Concentration of Culturable Bacteria and Fungi Associated With Particulate Matter
Quartz aerosol collection filters were arranged from Level 1–6 in Supplementary Figure 1A, which showed more PMs were collected as the increasing of haze level. 63 PM samples (21 samples in PM2.5, PM10, and TSP samples, respectively) were collected during haze and non-haze days, including 9 samples in Level 1-days, 12 samples in Level 2-days, 9 samples in Level 3-days, 9 samples in Level 4-days, 9 samples in Level 5-days, 15 samples in Level 6-days (Table 1 and Supplementary Figure 1B).
The numbers of colonies were calculated in each agar plate and the concentration of the bacteria and fungi in the air were estimated. The most colonies were observed in TSP samples and the fewest in PM2.5 samples (Figure 1A), which indicated that the bigger PMs contained more culturable microbes. Otherwise, the numbers of bacterial colonies are more than the fungal colonies in all types of PM samples (Figure 1A). To explore the effect of haze pollutants on the concentration of culturable bacteria and fungi, we compared the numbers of CFU on various levels of AQI. The results showed that there were different patterns between bacteria and fungi (Figure 1B). The numbers of bacterial colonies descended at first and then increased with the haze-level increasing, which indicated that there were more culturable bacteria during non-haze days and heavy-haze days, and fewest during light-haze days. However, the number of fungal colonies showed a trend of decline, which suggested that fungi were most influenced by heavy-haze (Figure 1B).

Figure 1. Summary of PM samples and isolation numbers of microbial colonies during various haze-level days. (A) Example nutrient agar medium and sabouraud dextrose agar medium showing colony morphologies. (B) Concentrations of microbial colony forming units (CFU) in different PM samples during various haze-level days, Values shown as mean ± standard error (SEM).
Bacterial and Fungal Isolates Differences in Various Media, PM Fractions, and Haze-Levels Samples
Using four different media, we collected 1,258 bacterial and 456 fungal isolates with 637 bacterial isolates from NA medium, 621 bacterial isolates from TSA medium, 261 fungal isolates from PDA medium, and 195 fungal isolates from SDA medium (Figure 2A). Colonies were picked based on phenotypic diversity (representative plates shown in Figure 1A) with 194 bacterial isolates from PM2.5 samples, 443 bacterial isolates from PM10 samples, and 621 bacterial isolates from TSP samples, and with 41 fungal isolates from PM2.5 samples, 185 fungal isolates from PM10 samples, and 230 fungal isolates from TSP samples, which indicated the numbers of isolates increased with the increasing of the PM size and were consistent with the numbers of CFU in PM2.5, PM10, and TSP samples (Figure 2B). Furthermore, we compared the numbers of isolates among non-, light-, and heavy-haze days. With increase of the haze, the microbial isolates were decreased in all fractions of PM samples (Figures 2C,D). Especially, fungal isolates have sharp declines during heavy-haze days (Figure 2D).
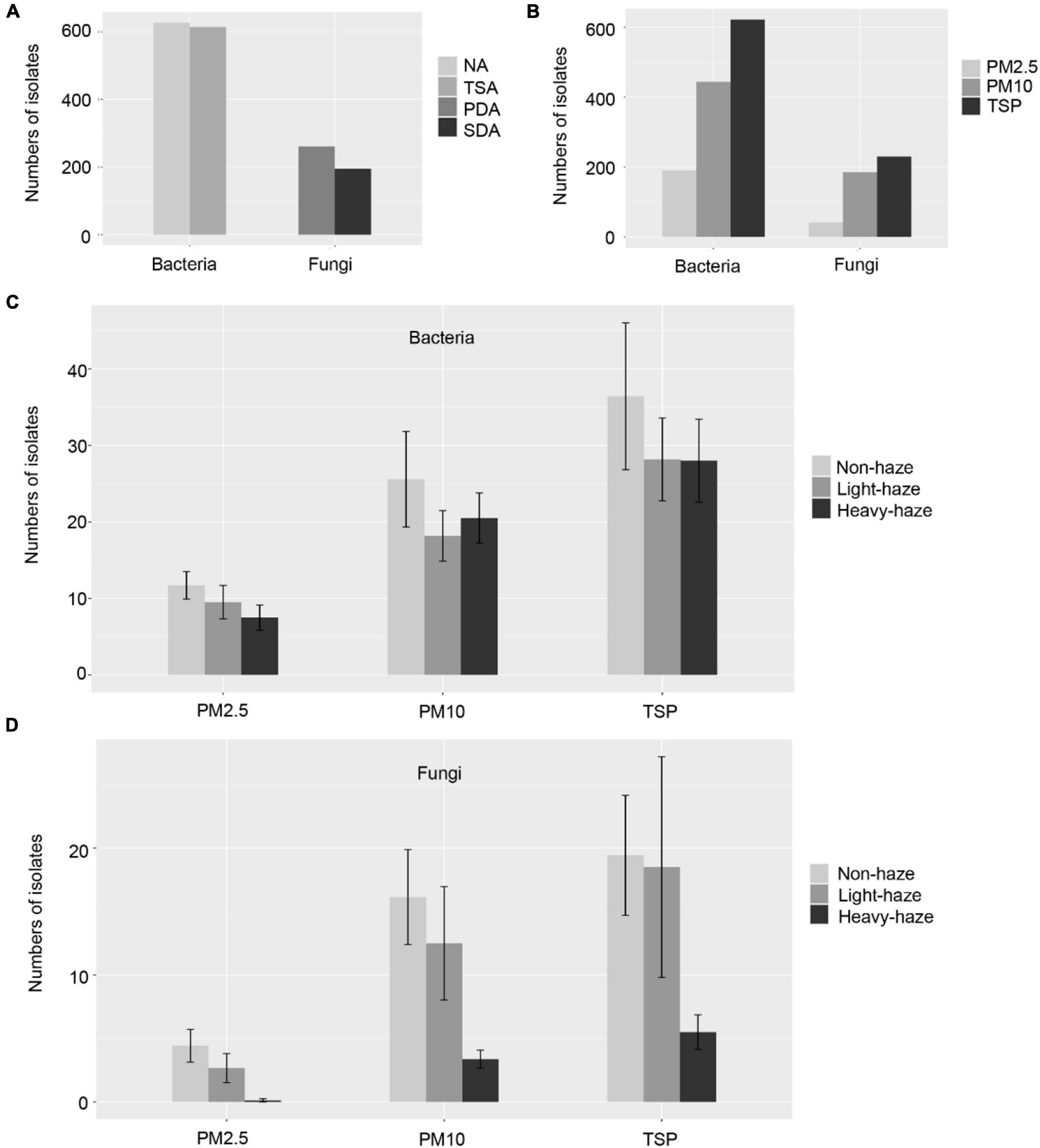
Figure 2. Numbers of microbial isolates in various media (A), PM samples (B), and haze-level samples (C,D). Values are shown as mean ± standard error (SEM) in panels (C,D).
Diverse Bacterial and Fungal Representatives From Microbiome Associated With Particulate Matter
There were 1,258 bacterial isolates clustered into 455 OTUs (100% similarity). The culturable bacterial community associated with PM was dominated by the Firmicutes (930 isolates, 73.9%), followed by the Actinobacteria (283 isolates, 22.5%), Proteobacteria (41 isolates, 3.3%), and Deinococcus-Thermus (4 isolates, 0.32%) (Supplementary Table 4). There were 77 observed genera in the PM samples. Bacillus (750 isolates, 59.6%) was the most abundant genus, followed by Streptomyces (76 isolates, 6.0%), Paenibacillus (57 isolates, 4.5%), Microbacterium (35 isolates, 2.8%), Curtobacterium (34 isolates, 2.7%), Kocuria (34 isolates, 2.7%), and Sporosarcina (24 isolates, 2.0%) (Supplementary Table 5 and Figure 3A).
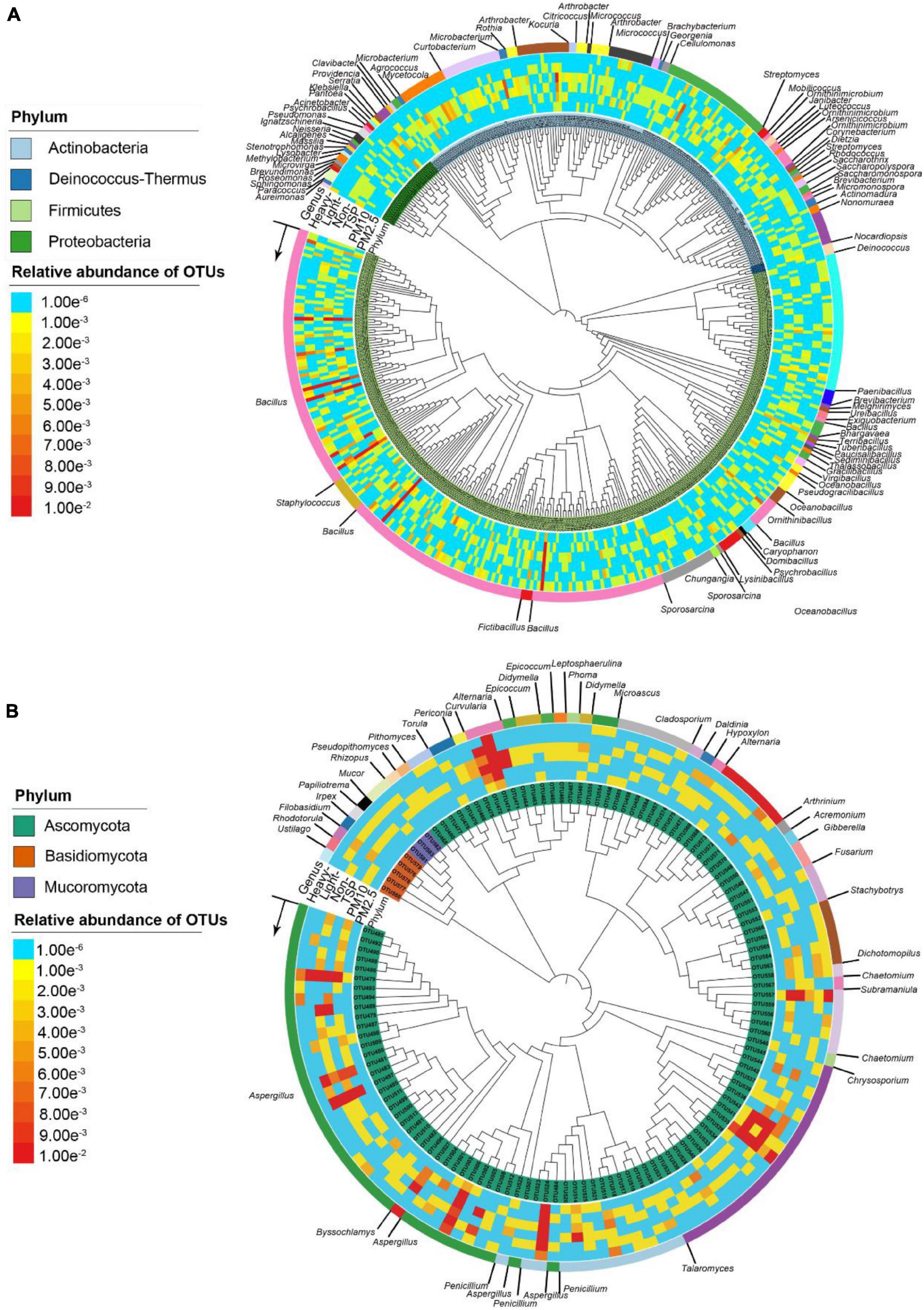
Figure 3. Phylogenetic tree of microbial isolates based on bacterial 16S rRNA sequences (A) and fungal ITS sequences (B) using neighbor-joining methods.
There were 475 fungal isolates clustered into 130 OTUs (100% similarity). The culturable fungal community associated with PM was dominated by the Ascomycota (466 isolates, 98.1%), followed by the Basidiomycota (6 isolates, 1.2%), and Mucoromycota (3 isolates, 0.6%) (Supplementary Table 6). There were 35 observed genera in PM samples. Aspergillus (126 isolates, 26.5%) was the most abundant genus, followed by Penicillium (105 isolates, 22.1%), Talaromyces (95 isolates, 20.0%), Alternaria (59 isolates, 12.4%), Chaetomium (22 isolates, 4.6%), Dichotomopilus (8 isolates, 1.7%), and Arthrinium (8 isolates, 1.7%) (Supplementary Table 7 and Figure 3B).
Culturable Bacterial and Fungal Composition Among Various Particulate Matter Fractions and Among Non-, Light-, and Heavy-Haze Days
Bar plots showed that the most abundant genus Bacillus was enriched during haze days with a haze-level dependent manner (Figure 4A). Surprisingly, Staphylococcus and Klebsiella, which include opportunistic pathogenic strains, were mainly isolated from PM2.5 samples, which increased the risk of infection in the upper respiratory tract (Figure 4A). Furthermore, Staphylococcus was enriched during haze days with a haze-level dependent manner in PM2.5 samples. Conversely, Klebsiella was enriched during non-haze days with a haze-level dependent manner in PM2.5 samples. We further identified the genera with significant differences among non-, light-, and heavy-haze samples using STAMP software (Figure 4B). No genera with significant differences were observed in PM2.5 samples. Compared to non-haze days, Bacillus was significantly enriched during heavy-haze days while Kocuria was significantly decreased during light- and heavy-haze days in PM10 samples (Figure 4B). In TSP samples, Paenibacillus was significantly enriched during heavy-haze days compared to light-haze days; Bacillus and Paenibacillus were significantly enriched during heavy-haze days while Kocuria and Microbacterium were significantly enriched during non-haze days (Figure 4B). CCA (Supplementary Figure 2A) and permutation tests (Supplementary Table 8) were performed to examine the relationships between bacterial community composition and environmental parameters. The concentration of SO2 (r2 = 0.2482, p ≤ 0.001), NO2 (r2 = 0.1447, p ≤ 0.05), CO (r2 = 0.1322, p ≤ 0.05), and the temperature (r2 = 0.1966, p ≤ 0.01) were significantly correlated with the bacterial community composition (Supplementary Figure 2A).
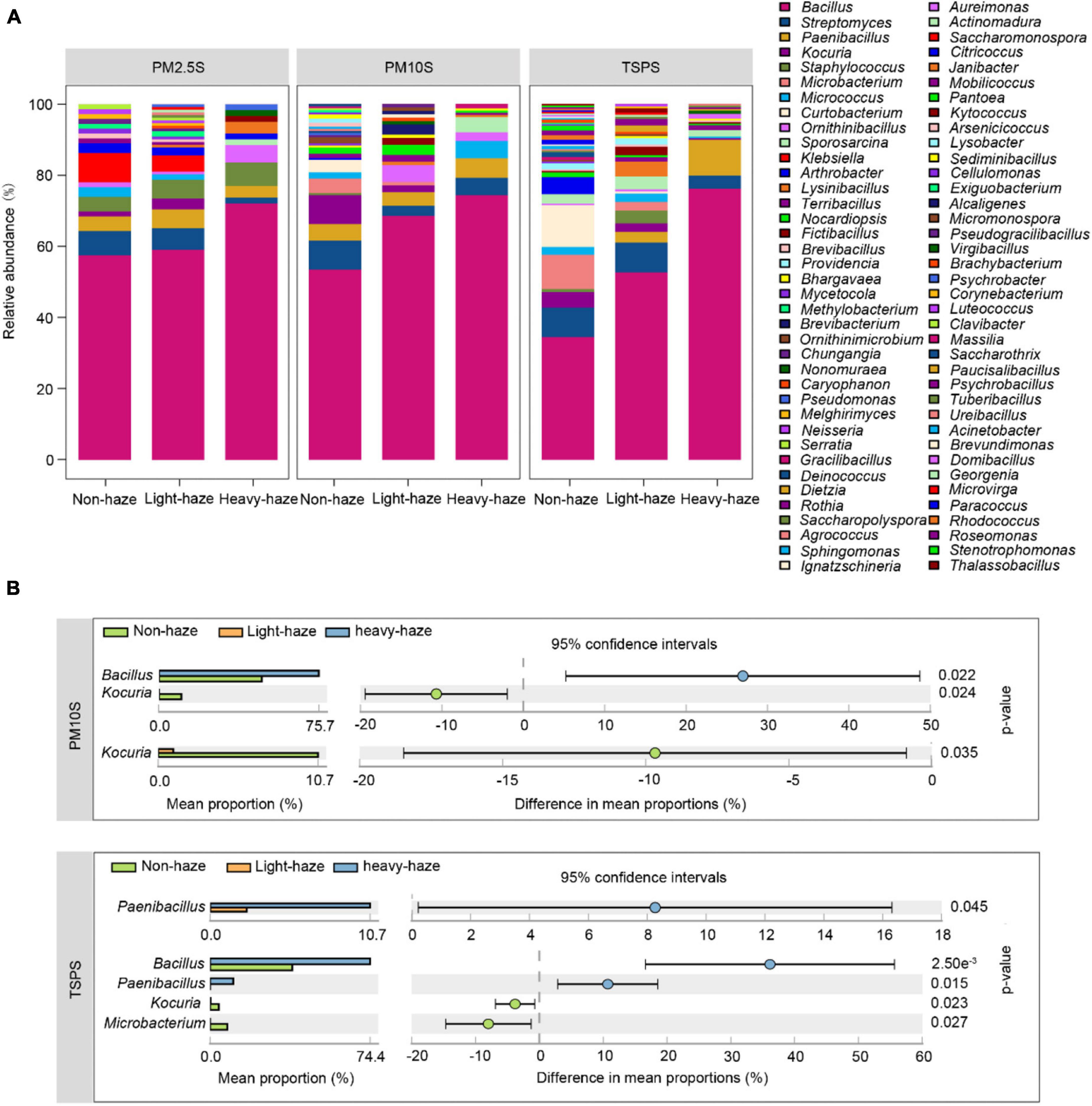
Figure 4. Bacterial diversity and composition among various PMs during non-, light-, and heavy-haze days. (A) Bar plot indicating relative abundances of bacterial genera among PMs. (B) STAMP analysis indicated genera that were significantly different among various haze-level samples.
The most abundant genera, Talaromyces and Aspergillus, have different patterns in various PM samples, which cannot be influenced by haze. For example, we only isolated Talaromyces in PM2.5 samples during heavy-haze days while it was enriched in PM10 samples during haze days (Figure 5A). Penicillium was enriched during non-haze days in a haze-level dependent manner. Especially in PM2.5 samples, Penicillium was only isolated during non-haze days, which indicated that it was easily inhibited by haze pollutants (Figures 5A,B). In addition, Chaetomium was significantly enriched during light- and heavy-haze days compared to non-haze days in TSP samples, which was similar to that in PM10 samples (Figures 5A,B). CCA (Supplementary Figure 2B) and permutation tests (Supplementary Table 9) indicated that the concentration of SO2 (r2 = 0.3973, p ≤ 0.001), CO (r2 = 0.5506, p ≤ 0.001), NO2 (r2 = 0.2679, p ≤ 0.01), and the temperature (r2 = 0.3410, p ≤ 0.01) were significantly correlated with the fungal composition (Supplementary Figure 2B).
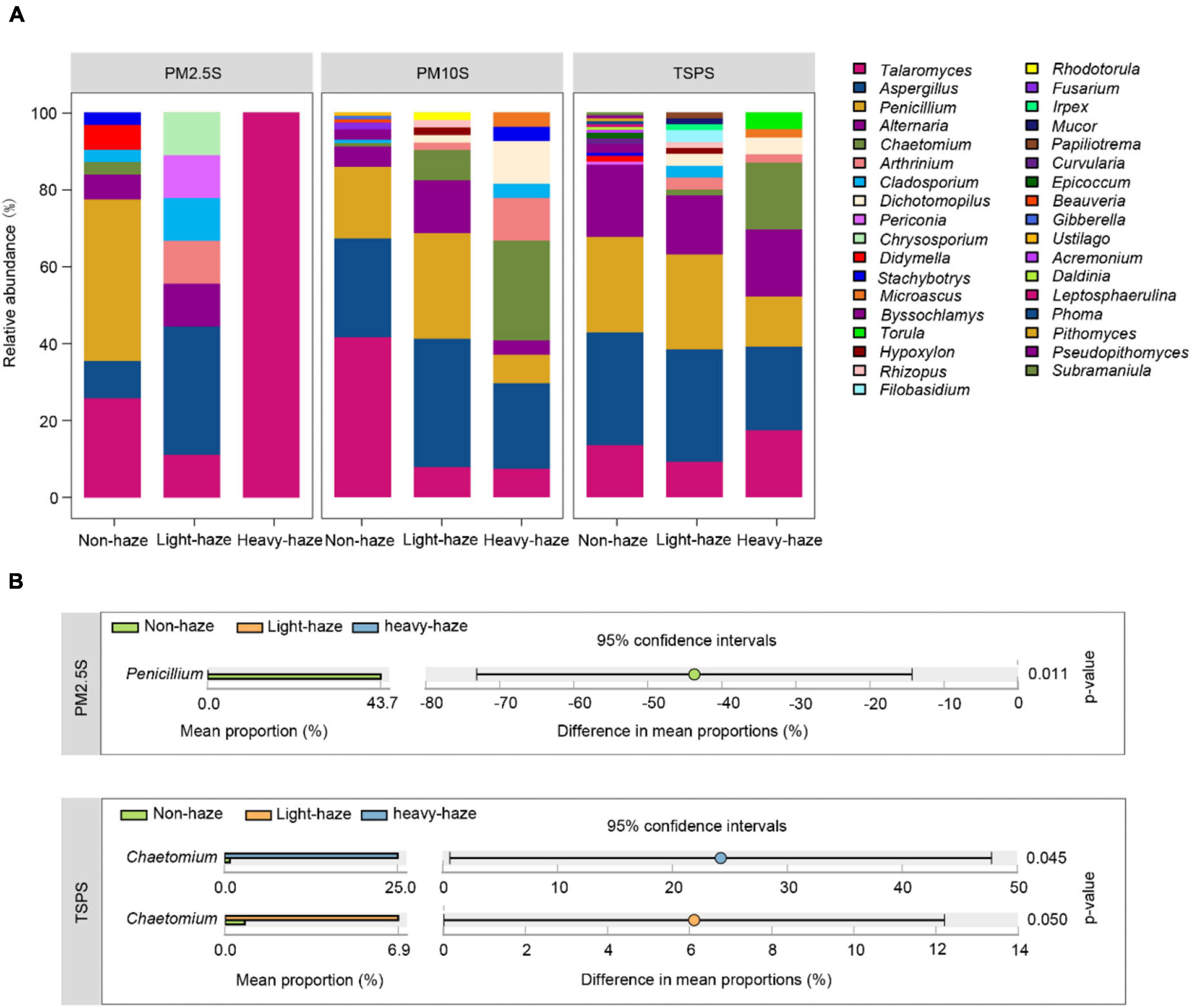
Figure 5. Fungal diversity and composition among various PMs during non-, light-, and heavy-haze days. (A) Bar plot indicating relative abundances of fungal genera among PMs. (B) STAMP analysis indicated genera that were significantly different among various haze-level samples.
Antimicrobial Activities of Bacterial Representatives
To investigate the antimicrobial activity of microbial representatives in PM samples, we screened the microbial representatives of bacterial and fungal isolates. A total of 40 bacterial representatives had antimicrobial activity against E. coli, E. faecalis, K. pneumoniae, and C. albicans, while one fungal representative had antimicrobial activity.
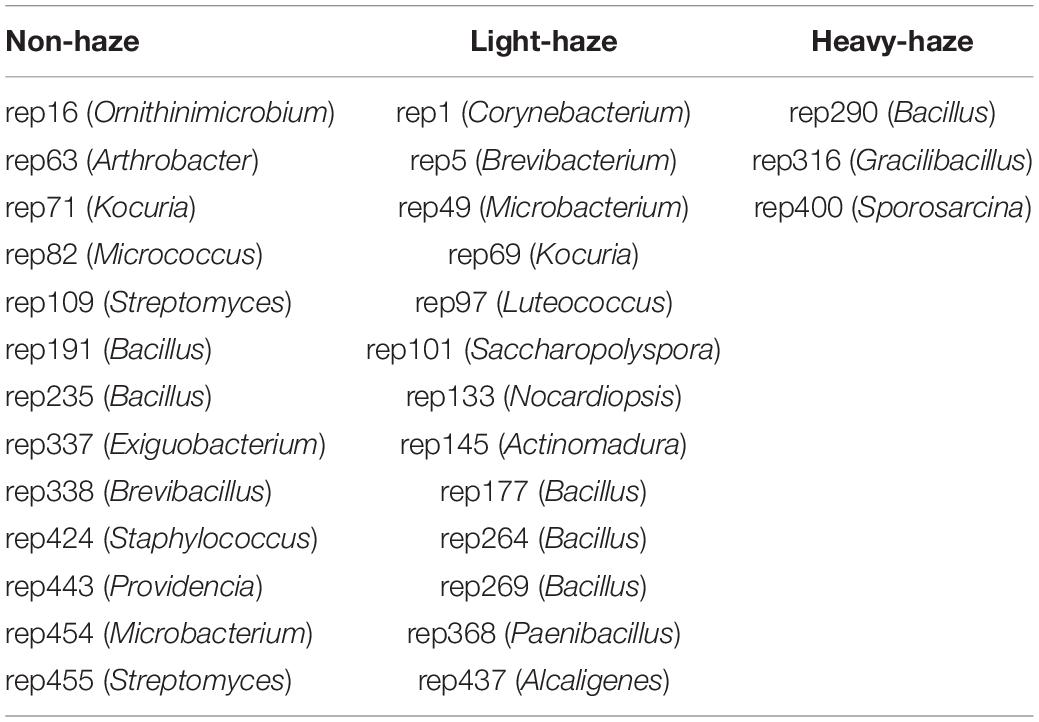
The microbial representatives with antimicrobial activity were dominated by the Firmicutes (21 representatives), followed by the Actinobacteria (17 representatives), and Proteobacteria (2 representatives) (Figure 6A). There were 22 genera with observed antimicrobial activity. Bacillus (15 representatives) was the most abundant genus, followed by Kocuria (3 representatives), Microbacterium (2 representatives), and Streptomyces (2 representatives) (Figure 6A). Anti-E. coli and anti-E. faecalis activities were only detected from fermentation broths of Actinobacteria, such as rep454 (closest to Micrococcus testaceum), rep71 (closest to Kocuria palustris), rep109 (closest to Streptomyces albogriseolus), rep97 (closest to Luteococcus peritonei), and rep101 (closest to Saccharopolyspora gregorii) with anti-E. coli activity, rep101 (closest to Saccharopolyspora gregorii), and rep138 (closest to Streptomyces halstedii) with anti-E. faecalis activity (Figure 6A). Fermentation broths of rep191, rep291, and rep338 (belong to Bacillus) and rep482 (closest to Aspergillus iizukae) have the activity to inhibit K. pneumoniae (Figure 6A). There were 35 representatives belonging to 19 genera that can inhibit the growth of C. albicans. Especially, both fermentation broths of rep82 (closest to Micrococcus antarcticus), rep234 (closest to Bacillus mojavensis), rep264, rep271, rep267, and rep269 (closest to Bacillus siamensis), and rep288 (closest to Bacillus subtilis) showed antimicrobial activity against C. albicans (Figure 6A).
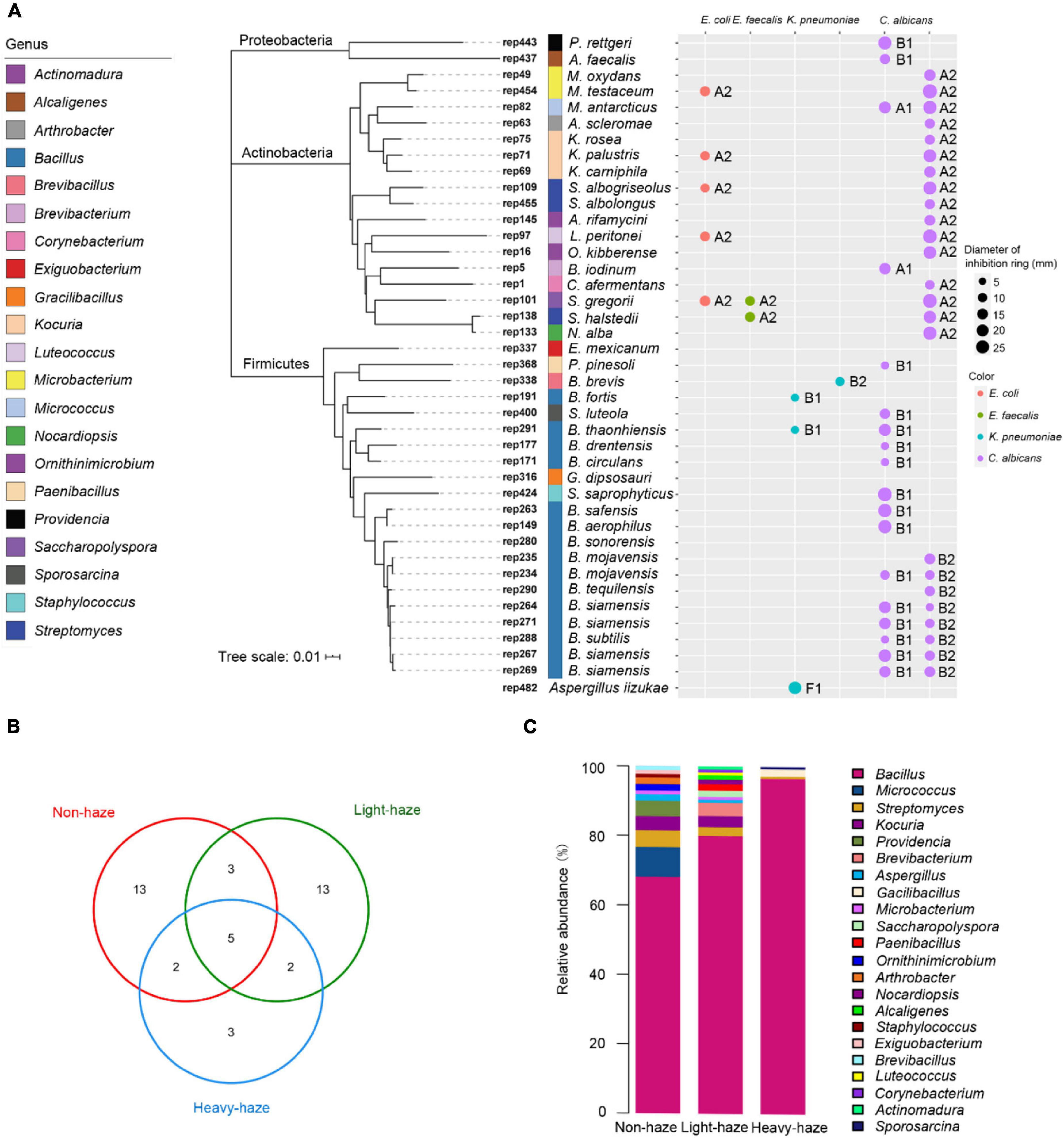
Figure 6. Antimicrobial activity of microbial representatives. (A) Phylogenetic tree of microbial isolates based on bacterial 16S rRNA sequences using neighbor-joining methods and activity patterns of microbial representatives with antimicrobial activity. (B) Venn diagram indicating unique representatives during various haze-level days. (C) Bar plot indicating relative abundances of genera with antimicrobial activity.
The venn diagram indicates the unique representative with antimicrobial activity in various haze-level days. During non-haze days, 13 representatives (2 representatives were classified to Bacillus, 1 representative was classified to Brevibacillus) with antimicrobial activities were observed in PM samples (Figure 6B and Table 2). During light-haze days, 13 representatives (3 representatives were classified to Bacillus, 1 representative was classified to Paenibacillus) with antimicrobial activities were observed in PM samples (Figure 6B and Table 2). The fewest unique representatives with antimicrobial activities were observed during heavy-haze days, only 3 representatives (1 representative were classified to Bacillus, 1 representative was classified to Gracilibacillus) with antimicrobial activities were observed in PM samples, which suggested that the most abundant representatives with antimicrobial activities during heavy-haze days were classified to Bacillus while only a few ones during non- and light-haze days (Figure 6B and Table 2). Additionally, the fewest genera with antimicrobial activities were observed during heavy-haze days, for example, only Bacillus, Streptomyces, Gacilibacillus, and Sporosarcina were observed in PM samples (Figure 6C). To further investigate whether microbial community composition with antimicrobial activities is influenced by environmental parameters, CCA (Supplementary Figure 2C) and permutation test (Supplementary Table 10) were conducted. The results indicated that only the temperature (r2 = 0.2524, p ≤ 0.05) was significantly correlated with the microbial community composition with antimicrobial activities (Supplementary Figure 2C).
Discussion
Haze has been a major issue in north China, posing a substantial threat to public health. Recent studies revealed the diversity and composition of airborne microbial community during haze and non-haze days using culture-independent methods (Woo et al., 2013; Cao et al., 2014; Wei et al., 2016; Yan et al., 2016, 2018; Zhong et al., 2019; Yu et al., 2020). However, the airborne microbial community described with culture-independent methods cannot reflect the true function of microorganisms on human health and the environment. It is critical to investigate the culturable microorganisms in the PM, which enlarge our knowledge on the effects of haze on human health through airborne microorganisms. In this study, we isolated the airborne bacteria and fungi from three types of PM fractions, analyzed the concentration of culturable microorganisms during haze and non-haze days, and further revealed the anti-pathogen activities, which suggested the interactions between culturable microorganisms and common pathogens and help to find potential antimicrobial drugs in the airborne environment.
A lower concentration of fungi than bacteria was observed in the current study, which could be attributable to the samples we collected in autumn and winter. Previous studies reported that the fungal spore concentrations increased during summer and decreased sharply during autumn and winter (Haas et al., 2013; Gao et al., 2015). In our study, the concentrations of culturable bacteria and fungi showed different patterns in various haze levels. The lowest concentrations of airborne bacteria were detected on level 2 or level 3 days, while the highest ones were on level 1 or level 6. Yang et al. (2021) suggested that the mean bioaerosol concentrations were slightly higher during non-haze days than haze days, but there were no significant differences. Using an epifluorescence microscope after staining with the LIVE/DEAD® BacLight™ Bacterial Viability Kit, Gong et al. (2019) reported that the bacterial viability decreased when air pollution occurred and increased again when pollution became severe, which is consistent with our study. According to Gong et al. (2019), during heavy-haze days, the concentrations of total airborne bacteria (including non-culturable and culturable bacteria) were 2–3 times greater than that during light- or non-haze days, explaining why the concentration of culturable bacteria increased. Thus, the growth of bacteria can be inhibited by haze, which accounts for the decrease of bacterial concentration during light-haze days. In addition, the lowest concentration of fungi was observed during heavy-haze days (level 5 and level 6). This is in line with previous findings (Gao et al., 2015), which suggested that haze might influence fungus spore germination.
The bacterial strains we isolated were far more than fungal strains, which confirmed that the fungi were hard to revive and culture in autumn and winter samples. Especially in PM2.5 samples, we only isolated 41 fungal strains compared to 185 and 230 strains in PM10 and TSP samples respectively, which might be due to the fact that most fungal spores have an aerodynamic diameter of > 2 mm (Froehlich-Nowoisky et al., 2009; Raisi et al., 2010; Haas et al., 2013). More microbial strains were isolated from non-haze samples, indicating that haze has an impact on airborne microorganisms. Particularly, we isolated relatively few fungal strains during heavy-haze days, implying that haze decreased more fungal isolates than bacterial ones.
Due to nutrition deficiency, dryness, and high levels of ultraviolet radiation in the air environment, most airborne microorganisms are non-viable in the form of dead cells, cell debris, or DNA fragments, resulting in a bias in the microbial community composition. It is a better strategy to estimate the composition of the airborne microbial community by combining the culture-dependent and culture-independent methods. The culturable bacterial community associated with PM was dominated by the Firmicutes (73.9%), Actinobacteria (22.5%), Proteobacteria (3.3%), and Deinococcus-Thermus (0.32%). With culture-independent methods, our group found that the bacterial community associated with PM was dominated by the Proteobacteria (38.5%), Firmicutes (26.8%), Actinobacteria (17.2%), Bacteroidetes (7.7%), and Deinococcus-Thermus (5.2%) (Yan et al., 2018), which suggested that Firmicutes were enriched by a culture-dependent method. Firmicutes is a phylum that includes all Gram-positive bacteria with a hard cell wall, making them resistant to haze and allowing them to thrive in the air (Garrity et al., 2009). Furthermore, the most abundant genus Bacillus was enriched during haze days in a haze-level dependent manner. Bacillus species have been detected in atmospheric dust (Verdona et al., 2013) and soil (Haas and Défago, 2005) producing oval endospores to adapt to the harsh environmental conditions and remain in a dormant state for a long period. In both PM10 and TSP samples, Kocuria was significantly enriched during non-haze days. In our previous study, Kocuria was identified as a key genus in the airborne PM samples, but no significant differences were observed during haze and non-haze days with culture-independent methods, which indicated that the survival of the genus can be influenced by haze pollutants (Yan et al., 2018). As microbial carriers, PM provided more surfaces for more airborne pathogenic bacteria to adhere to Li et al. (2015, 2017) and Xie et al. (2017). Staphylococcus and Klebsiella, including common pathogenic strains, were mainly isolated from PM2.5 samples, which increased the risk of infection in the upper respiratory tract. Especially, Staphylococcus was increased during haze days, reminding us to pay close attention to the infection by Staphylococcus during haze days. Klebsiella, which are recognized as human pathogens of the respiratory tract (Podschun and Ullmann, 1998), were found to be enriched in PM2.5 samples during non-haze days, implying that infection rates of Klebsiella were increased during non-haze days.
The composition of the culturable fungal community was altered in three types of PM fractions during haze days. Talaromyces was the most abundant genus in the present study, which is also different from the result with culture-independent methods, whereas Cladosporium, Alternaria, Fusarium, and Penicillium dominated the fungal community in our earlier study (Yan et al., 2016). Talaromyces, which is abundant in PM2.5 samples during haze days, contains species that are medically important and raise infection rates in the respiratory tract during hazy days (Yilmaz et al., 2014). For example, T. marneffei can cause a fatal mycosis in especially immunocompromised individuals from East Asian countries such as China, Taiwan, and Vietnam (Deng et al., 1988; Luh, 1998; Hien et al., 2001). Additionally, Penicillium was enriched during non-hazy days, which is similar to the results of a previous study using the culture-dependent method (Li et al., 2015). Haas et al. (2013) discovered that spore concentrations of Penicillium, rise with increased PM during non-haze days (Gao et al., 2015). Our previous study with the culture-independent method has indicated that the relative abundance of Penicillium increased during heavy-haze days (Yan et al., 2016). These findings suggested that haze pollutants might influence the germination or release of fungal spores. Chaetomium was enriched during haze days in the present study. Previous studies also reported Chaetomium stayed in the air (Shelton et al., 2002; David et al., 2013), and it could affect air quality and damage to human health (Nielsen et al., 1998, 1999; Guo et al., 2019), indicating that haze enhanced the disease risk caused by Chaetomium.
Although soils are considered excellent sources for the isolation of microorganisms with diverse potential, the current focus is on exploring previously ignored ecosystems (Lee and Hwang, 2002; Prabavathy et al., 2006; Shah et al., 2017). A scarce number of studies involving microorganisms collected from the air in antimicrobial activity screening (Sobral et al., 2017) and the diverse microbial community associated with PM was observed in the current and previous studies (Yan et al., 2016, 2018; Romano et al., 2020; Yu et al., 2020). The wide-ranging and fluctuating temperatures, high levels of solar irradiation, strong oxidizing, and dryness in the atmosphere may select for novel microorganisms with unique metabolic properties (Polymenakou, 2012). The percentage of representatives with antimicrobial activities decreased during non-haze days and the genera with antimicrobial activities were distributed in various haze-level samples, which indicated that haze pollutants may one of the factors to select for the strains with antimicrobial activities. Though the strains with antimicrobial activities can inhibit the growth of pathogens, the potential drug-resistant bacteria or fungi can be selected in the air, which may promote the dissemination of antibiotic resistance genes (ARGs) and influence public health due to the persistent haze. Zhu et al. (2021) also suggested that the dissemination of ARGs in fresh snow could be exacerbated by air pollution, severely increasing the health risks of both air pollution and ARGs. Bacillus was the most abundant airborne genus with microbial activity in this study. El-Banna (2005) isolated airborne microorganisms and five strains belonging to Bacillus were found to be antagonistic to bacteria or fungi. It is necessary to search for new antibiotics from Bacillus, which is one of the fruitful sources of antibiotics (Emmert et al., 2004; El-Banna, 2005). Actinobacteria, which were the abundant phylum with microbial activity from PM samples in the present study, have been the source of countless drugs and intensively screened as an important source of therapeutically important molecules for over half a century. For example, Streptomyces strains are the richest source of natural products, especially clinically useful antibiotics, antimetabolites, and antitumor agents (Bérdy, 2005; Newman and Cragg, 2007; Olano et al., 2018). Filamentous actinobacteria account for about 45% of all microbial bioactive secondary metabolites with about 80% of these 7,600 compounds being produced by Streptomyces (Bérdy, 2005; Goodfellow et al., 2009). Additionally, only one fungal strain belonging to Aspergillus showed antimicrobial activity, which suggested that the airborne fungi had a weak activity and the genus Aspergillus was the most prolific among fungi (Giordano, 2020).
Conclusion
Our study provided an integrated characterization of the isolation and antimicrobial activities of bacterial and fungal representatives associated with PM during haze and non-haze days using a culture-dependent method. The numbers and composition of culturable bacteria and fungi were influenced by haze pollutants, which further enhances the understanding of the airborne microorganism during haze and non-haze days. We collected 1,258 bacterial isolates and 475 fungal isolates and increased the number of microbial resources for screening active strains. Additionally, 40 bacterial representatives and 1 fungal representative had antimicrobial activities against common pathogens, paving the way for the discovery of novel antibiotics. This study could help researchers better comprehend the airborne microbial resource during haze and non-haze days. Through further genomic characterization and analysis, separation of natural active products, and synthetic biology endeavors, we believe that the collection of airborne strains is a resource that will provide a basis to increase our understanding of the relationship between microbes associated with PM and human health.
Data Availability Statement
The data that support the findings of this study are available in Supplementary Table 2. The raw reads were deposited into the NCBI GeneBank database under accession number SUB10504922 (OK482088–OK482542) and SUB10505231 (OK490148–OK490277).
Author Contributions
L-YY designed the study and revised the manuscript. DY, JS, L-LZ, HW, and X-MF performed sampling. DY performed the laboratory work and wrote the manuscript. TZ and J-LB performed part of the laboratory work and revised the manuscript. DY, Y-QZ, and H-YL analyzed the data. All authors read and approved the final manuscript.
Funding
This research was supported by the National Microbial Resources Center (No. NMRC-2021-3), the National Natural Science Foundation of China (NSFC) (Nos. 81973220 and 32000006), the CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2021-I2M-1-055), the National Science and Technology Project of China (No. 2019ZX09721001-004-006), the Science and Technology Research Project of Henan Province (No. 202102310270), and the Non-profit Central Research Institute Fund of CAMS (2020-PT310-003 and 2019PT350004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.793037/full#supplementary-material
Footnotes
- ^ http://zx.bjmemc.com.cn/
- ^ http://www.nmc.cn/
- ^ https://www.ezbiocloud.net/
- ^ https://itol.embl.de
References
Alanis, A. J. (2005). Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36, 697–705. doi: 10.1016/j.arcmed.2005.06.009
Alghamdi, M. A., Shamy, M., Redal, M. A., Khoder, M., Awad, A. H., and Elserougy, S. (2014). Microorganisms associated particulate matter: a preliminary study. Sci. Total Environ. 479-480, 109–116. doi: 10.1016/j.scitotenv.2014.02.006
Barberan, A., Henley, J., Fierer, N., and Casamayor, E. O. (2014). Structure, inter-annual recurrence, and global-scale connectivity of airborne microbial communities. Sci. Total Environ. 487, 187–195. doi: 10.1016/j.scitotenv.2014.04.030
Be, N. A., Thissen, J. B., Fofanov, V. Y., Allen, J. E., Rojas, M., Golovko, G., et al. (2015). Metagenomic analysis of the airborne environment in urban spaces. Microb. Ecol. 69, 346–355. doi: 10.1007/s00248-014-0517-z
Bertolini, V., Gandolfi, I., Ambrosini, R., Bestetti, G., Innocente, E., Rampazzo, G., et al. (2013). Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl. Microbiol. Biotechnol. 97, 6561–6570. doi: 10.1007/s00253-012-4450-0
Booth, B. B., Dunstone, N. J., Halloran, P. R., Andrews, T., and Bellouin, N. (2012). Aerosols implicated as a prime driver of twentieth-century North Atlantic climate variability. Nature 484, 228–232. doi: 10.1038/nature10946
Bottos, E. M., Woo, A. C., Zawar-Reza, P., Pointing, S. B., and Cary, S. C. (2014). Airborne bacterial populations above desert soils of the mcmurdo dry valleys, Antarctica. Microb. Ecol. 67, 120–128. doi: 10.1007/s00248-013-0296-y
Bowers, R. M., Clements, N., Emerson, J. B., Wiedinmyer, C., Hannigan, M. P., and Fierer, N. (2013). Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ. Sci. Technol. 47, 12097–12106. doi: 10.1021/es402970s
Bowers, R. M., McLetchie, S., Knight, R., and Fierer, N. (2011a). Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5, 601–612. doi: 10.1038/ismej.2010.167
Bowers, R. M., Sullivan, A. P., Costello, E. K., Collett, J. L. Jr., Knight, R., and Fierer, N. (2011b). Sources of bacteria in outdoor air across cities in the midwestern United States. Appl. Environ. Microbiol. 77, 6350–6356. doi: 10.1128/AEM.05498-11
Cao, C., Jiang, W., Wang, B., Fang, J., Lang, J., Tian, G., et al. (2014). Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 48, 1499–1507. doi: 10.1021/es4048472
Cheng, Z., Jiang, J., Fajardo, O., Wang, S., and Hao, J. (2013). Characteristics and health impacts of particulate matter pollution in China (2001–2011). Atmospheric Environ. 65, 186–194. doi: 10.1016/j.atmosenv.2012.10.022
David, R., Mark, W., and David, M. (2013). Chaetoglobosins and azaphilones produced by Canadian strains of Chaetomium globosum isolated from the indoor environment. Mycotoxin Res. 29, 47–54. doi: 10.1007/s12550-012-0144-9
Deng, Z., Ribas, J. L., and Connor, G. D. H. (1988). Infections caused by Penicillium marneffei in China and Southeast Asia : review of eighteen published cases and report of four more Chinese cases. Rev. Infect. Dis. 10, 640–652. doi: 10.1093/clinids/10.3.640
Dong, L., Qi, J., Shao, C., Zhong, X., Gao, D., Cao, W., et al. (2016). Concentration and size distribution of total airborne microbes in hazy and foggy weather. Sci. Total Environ. 541, 1011–1018. doi: 10.1016/j.scitotenv.2015.10.001
El-Banna, N. M. (2005). Effect of carbon source on the antimicrobial activity of the air flora. World J. Microbiol. Biotechnol. 21, 1451–1454. doi: 10.1007/s11274-005-6564-3
Emmert, E. A. B., Klimowicz, A. K., and Thomas, M. G. (2004). Genetics of Zwittermicin a production by Bacillus cereus. Appl. Environ. Microbiol. 70, 104–113. doi: 10.1128/AEM.70.1.104-113.2004
Fang, Z. (2014). Characteristic and concentration distribution of culturable airborne bacteria in residential environments in Beijing, China. Aerosol Air Qual. Res. 14, 943–953. doi: 10.4209/aaqr.2013.04.0109
Franzetti, A., Gandolfi, I., Gaspari, E., Ambrosini, R., and Bestetti, G. (2011). Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl. Microbiol. Biotechnol. 90, 745–753. doi: 10.1007/s00253-010-3048-7
Froehlich-Nowoisky, J., Pickersgill, D. A., Despres, V. R., and Poeschl, U. (2009). High diversity of fungi in air particulate matter. Proc. Natl. Acad. Sci. U.S.A. 106, 12814–12819. doi: 10.1073/pnas.0811003106
Gao, M., Jia, R., Qiu, T., Han, M., Song, Y., and Wang, X. (2015). Seasonal size distribution of airborne culturable bacteria and fungi and preliminary estimation of their deposition in human lungs during non-haze and haze days. Atmospheric Environ. 118, 203–210. doi: 10.1016/j.atmosenv.2015.08.004
Garrity, G., Vos, P. D., Jones, D., Kreig, N., Ludwig, W., Rainey, F. A., et al. (2009). Bergey’s Manual of Systematic Bacteriology. The Firmicutes, Vol. 3. Berlin: Springer.
Giordano, D. (2020). Bioactive molecules from extreme environments. Mar. Drugs 18:640. doi: 10.3390/md18120640
Gong, J., Qi, J., Beibei, E., Yin, Y., and Gao, D. (2019). Concentration, viability and size distribution of bacteria in atmospheric bioaerosols under different types of pollution. Environ. Pollut. 257:113485. doi: 10.1016/j.envpol.2019.113485
Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M. E., Suzuki, K. I., Ludwig, W., et al. (2009). Bergey’s Manual of Systematic Bacteriology. The Actinobacteria, Vol. 5. Berlin: Springer.
Guo, J., Xiong, Y., Shi, C., Liu, C., and Qin, C. (2020). Characteristics of airborne bacterial communities in indoor and outdoor environments during continuous haze events in Beijing: implications for health care. Environ. Int. 139:105721. doi: 10.1016/j.envint.2020.105721
Guo, Q. F., Yin, Z. H., Zhang, J. J., Kang, W. Y., Wang, X. W., Ding, G., et al. (2019). Chaetomadrasins A and B, two new cytotoxic cytochalasans from desert soil-derived fungus chaetomium madrasense 375. Molecules 24:3240. doi: 10.3390/molecules24183240
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Haas, D., Galler, H., Luxner, J., Zarfel, G., Buzina, W., Friedl, H., et al. (2013). The concentrations of culturable microorganisms in relation to particulate matter in urban air. Atmospheric Environ. 65, 215–222. doi: 10.1016/j.atmosenv.2012.10.031
Hamra, G. B., Guha, N., Cohen, A., Laden, F., Raaschou-Nielsen, O., Samet, J. M., et al. (2014). Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ. Health Perspect. 122, 906–911. doi: 10.1289/ehp.1408092
Hien, T. V., Loc, P. P., Hoa, N. T. T., Duong, N. M., Quang, V. M., Mcneil, M. M., et al. (2001). First cases of disseminated penicilliosis marneffei infection among patients with acquired immunodeficiency syndrome in Vietnam. Clin. Infect. Dis. 32, e78–e80. doi: 10.1086/318703
Huang, K., Liang, F., Yang, X., Liu, F., Li, J., Xiao, Q., et al. (2019). Long term exposure to ambient fine particulate matter and incidence of stroke: prospective cohort study from the China-PAR project. BMJ 367:l6720. doi: 10.1136/bmj.l6720
Ivica, L., and Peer, B. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Jaenicke, R. (2005). Abundance of cellular material and proteins in the atmosphere. Science 308, 73–73. doi: 10.1126/science.1106335
Kanagawa, T. (2003). Bias and artifacts in multitemplate polymerase Chain reactions (PCR). J. Biosci. Bioengin. 96, 317–323. doi: 10.1016/S1389-1723(03)90130-7
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Lee, J. Y., and Hwang, B. K. (2002). Diversity of antifungal actinomycetes in various vegetative soils of Korea. Revue Canadienne De Microbiol. 48:407. doi: 10.1139/w02-025
Li, J., Liu, F., Liang, F., Huang, K., Yang, X., Xiao, Q., et al. (2020a). Long-Term effects of high exposure to ambient fine particulate matter on coronary heart disease incidence: a population-based Chinese cohort study. Environ. Sci. Technol. 54, 6812–6821. doi: 10.1021/acs.est.9b06663
Li, J., Lu, X., Liu, F., Liang, F., and Gu, D. (2020b). Chronic effects of high fine particulate matter exposure on lung cancer in China. Am. J. Respiratory Crit. Care Med. 202, 1551–1559. doi: 10.1164/rccm.202001-0002OC
Li, Y., Fu, H., Wang, W., Liu, J., Meng, Q., and Wang, W. (2015). Characteristics of bacterial and fungal aerosols during the autumn haze days in Xi’an, China. Atmospheric Environ. 122, 439–447. doi: 10.1016/j.atmosenv.2015.09.070
Li, Y., Lu, R., Li, W., Xie, Z., and Song, Y. (2017). Concentrations and size distributions of viable bioaerosols under various weather conditions in a typical semi-arid city of Northwest China. J. Aerosol Sci. 106, 83–92.
Luh, K. (1998). Penicillium marneffei fungemia in an AIDS patient: the first case report in Taiwan. Changgeng Yi Xue Za Zhi/Changgeng Ji Nian Yi Yuan = Chang Gung Med. J./Chang Gung Memorial Hospital 21:362.
Maffioli, S. I., Zhang, Y., Degen, D., Carzaniga, T., Del Gatto, G., Serina, S., et al. (2017). Antibacterial nucleoside-analog inhibitor of bacterial RNA polymerase. Cell 169, 1240–1248.
Newman, D. J., and Cragg, G. M. (2007). Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70, 461–477. doi: 10.1021/np068054v
Niazi, S., Hassanvand, M. S., Mahvi, A. H., Nabizadeh, R., Alimohammadi, M., Nabavi, S., et al. (2015). Assessment of bioaerosol contamination (bacteria and fungi) in the largest urban wastewater treatment plant in the Middle East. Environ. Sci. Pollut. Res. Int. 22, 16014–16021. doi: 10.1007/s11356-015-4793-z
Nielsen, K. F., Gravesen, S., Nielsen, P. A., Andersen, B., and Thrane, U. (1999). Production of mycotoxins on artificially and naturally infested building materials. Mycopathologia 145, 43–56. doi: 10.1023/a:1007038211176
Nielsen, K. F., Hansen, M., Larsen, T. O., and Thrane, U. (1998). Production of trichothecene mycotoxins on water damaged gypsum boards in Danish buildings. Int. Biodeterioration Biodegradation 42, 1–7. doi: 10.1289/ehp.99107s3505
Oh, H. J., Jeong, N. N., Chi, W. B., Seo, J. H., Jun, S. M., and Sohn, J. R. (2015). Characterization of particulate matter concentrations and bioaerosol on each floor at a building in Seoul, Korea. Environ. Sci. Pollut. Res. Int. 22, 16040–16050. doi: 10.1007/s11356-015-4810-2
Okubo, T., Osaki, T., Nozaki, E., Uemura, A., Sakai, K., Matushita, M., et al. (2017). Walker occupancy has an impact on changing airborne bacterial communities in an underground pedestrian space, as small-dust particles increased with raising both temperature and humidity. PLoS One 12:e0184980. doi: 10.1371/journal.pone.0184980
Olano, C., Méndez, C., and Salas, J. A. (2018). Antitumor compounds from marine actinomycetes. Mar. Drugs 7, 210–248. doi: 10.3390/md7020210
Parker, J. D., Kravets, N., and Vaidyanathan, A. (2018). Particulate matter air pollution exposure and heart disease mortality risks by race and ethnicity in the United States: 1997 to 2009 national health interview survey with mortality follow-up through 2011. Circulation 137, 1688–1697. doi: 10.1161/CIRCULATIONAHA.117.029376
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123. doi: 10.1093/bioinformatics/btu494
Patel, J. B. (2014). Performance Standards for Antimicrobial Susceptibility Testing : Twenty-Fourth Informational Supplement. Pittsburgh, PA: Clinical and Laboratory Standards Institute.
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Polymenakou, P. N. (2012). Atmosphere: a source of pathogenic or beneficial microbes? Atmosphere 3, 87–102.
Prabavathy, V. R., Mathivanan, N., and Murugesan, K. (2006). Control of blast and sheath blight diseases of rice using antifungal metabolites produced by Streptomyces sp. PM5. Biol. Control 39, 313–319.
R Core Team. (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Raisi, L., Lazaridis, M., and Katsivela, E. (2010). Relationship between airborne microbial and particulate matter concentrations in the ambient air at a Mediterranean site. Global Nest J. 12, 84–91.
Randles, C. A., Colarco, P. R., and da Silva, A. (2013). Direct and semi-direct aerosol effects in the NASA GEOS-5 AGCM: aerosol-climate interactions due to prognostic versus prescribed aerosols. J. Geophys. Res.: Atmospheres 118, 149–169. doi: 10.1029/2012jd018388
Romano, S., Becagli, S., Lucarelli, F., Rispoli, G., and Perrone, M. R. (2020). Airborne Bacteria structure and chemical composition relationships in winter and spring PM10 samples over southeastern Italy. Sci. Total Environ. 730:138899. doi: 10.1016/j.scitotenv.2020.138899
Shah, A. M., Shakeel, U. R., Hussain, A., Mushtaq, S., Rather, M. A., Shah, A., et al. (2017). Antimicrobial investigation of selected soil actinomycetes isolated from unexplored regions of Kashmir Himalayas, India. Microb. Pathog. 110, 93–99. doi: 10.1016/j.micpath.2017.06.017
Sharma, D., Kaur, T., Chadha, B. S., and Manhas, R. K. (2011). Antimicrobial activity of actinomycetes against multidrug resistant Staphylococcus aureus, E. coli and various other pathogens. Tropical J. Pharmaceutical Res. 10, 801–808.
Shelton, B. G., Kirkland, K. H., Flanders, W. D., and Morris, G. K. (2002). Profiles of airborne fungi in buildings and outdoor environments in the United States. Appl. Environ. Microbiol. 68, 1743–1753. doi: 10.1128/AEM.68.4.1743-1753.2002
Shimadera, H., Hayami, H., Morino, Y., Ohara, T., Chatani, S., Hasegawa, S., et al. (2013). Analysis of summertime atmospheric transport of fine particulate matter in Northeast Asia. Asia-Pacific J. Atmospheric Sci. 49, 347–360. doi: 10.1007/s13143-013-0033-y
Sobral, L. V., Melo, K. N., Souza, C. M., Silva, F. S., Silva, G. L. R., Silva, A. L. F., et al. (2017). Antimicrobial and enzymatic activity of anemophilous fungi of a public university in Brazil. Anais da Academia Brasileira de Ciências 89, 2327–2340. doi: 10.1590/0001-3765201720160903
Sudhir, K., Glen, S., and Koichiro, T. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Tan, J., Duan, J., Zhen, N., He, K., and Hao, J. (2016). Chemical characteristics and source of size-fractionated atmospheric particle in haze episode in Beijing. Atmospheric Res. 167, 24–33. doi: 10.1016/j.atmosres.2015.06.015
Van Dingenen, R., Raes, F., Putaud, J.-P., Baltensperger, U., Charron, A., Facchini, M. C., et al. (2004). A European aerosol phenomenology—1: physical characteristics of particulate matter at kerbside, urban, rural and background sites in Europe. Atmospheric Environ. 38, 2561–2577. doi: 10.1016/j.atmosenv.2004.01.040
Verdona, A. M., Garcia-Salamanca, A., Solano, J., de la Rosa, J. D., and Ramos, J. (2013). Chemical and microbiological characterization of atmospheric particulate matter during an intense African dust event in Southern Spain. Environ. Sci. Technol. 47, 3630–3638. doi: 10.1021/es3051235
Vermani, M., Vijayan, V. K., Kausar, M. A., and Agarwal, M. K. (2010). Quantification of airborne aspergillus allergens: redefining the approach. J. Asthma Res. 47, 754–761. doi: 10.3109/02770903.2010.492539
Wang, L., Xu, J., Yang, J., Zhao, X., Wei, W., Cheng, D., et al. (2012). Understanding haze pollution over the southern Hebei area of China using the CMAQ model. Atmospheric Environ. 56, 69–79. doi: 10.1016/j.atmosenv.2012.04.013
Wei, K., Zou, Z., Zheng, Y., Li, J., Shen, F., Wu, C. Y., et al. (2016). Ambient bioaerosol particle dynamics observed during haze and sunny days in Beijing. Sci. Total Environ. 550, 751–759. doi: 10.1016/j.scitotenv.2016.01.137
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1989). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR - Protocols and Applications - A Laboratory Manual, eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T.J. White (Cambridge, MA: Academic Press), 315–322.
Woo, A. C., Brar, M. S., Chan, Y., Lau, M. C. Y., Leung, F. C. C., Scott, J. A., et al. (2013). Temporal variation in airborne microbial populations and microbially-derived allergens in a tropical urban landscape. Atmospheric Environ. 74, 291–300. doi: 10.1016/j.atmosenv.2013.03.047
Xie, Z., Li, Y., Lu, R., Li, W., Fan, C., Liu, P., et al. (2017). Characteristics of total airborne microbes at various air quality levels. J. Aerosol Sci. 116, 57–65.
Yamamoto, N., Bibby, K., Qian, J., Hospodsky, D., Rismani-Yazdi, H., Nazaroff, W. W., et al. (2012). Particle-size distributions and seasonal diversity of allergenic and pathogenic fungi in outdoor air. ISME J. 6, 1801–1811. doi: 10.1038/ismej.2012.30
Yan, D., Zhang, T., Su, J., Zhao, L. L., Wang, H., Fang, X. M., et al. (2016). Diversity and composition of airborne fungal community associated with particulate matters in beijing during haze and non-haze days. Front. Microbiol. 7:487. doi: 10.3389/fmicb.2016.00487
Yan, D., Zhang, T., Su, J., Zhao, L. L., Wang, H., Fang, X. M., et al. (2018). Structural variation of the bacterial community associated with airborne particulate matter in Beijing during haze and non-haze days. Appl. Environ. Microb. 84, AEM.00004–AEM.00018.
Yang, H., Chen, J., Wen, J., Tian, H., and Liu, X. (2016). Composition and sources of PM 2.5 around the heating periods of 2013 and 2014 in Beijing: implications for efficient mitigation measures. Atmospheric Environ. 124, 378–386. doi: 10.1016/j.atmosenv.2015.05.015
Yang, L., Shen, Z., Wang, D., Wei, J., Wang, X., Sun, J., et al. (2021). Diurnal variations of size-resolved bioaerosols during autumn and winter over a semi-arid megacity in Northwest China. Geohealth 5:e2021GH000411. doi: 10.1029/2021GH000411
Yilmaz, N., Visagie, C. M., Houbraken, J., Frisvad, J. C., and Samson, R. A. (2014). Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 78, 175–341. doi: 10.1016/j.simyco.2014.08.001
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., and Chun, J. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yu, R. L., Wang, S. K., Wu, X. L., Shen, L., Liu, Y. D., Li, J. K., et al. (2019). Community structure variation associated with airborne particulate matter at central south of China during hazy and nonhazy days. Atmospheric Pollut. Res. 10, 1536–1542. doi: 10.1016/j.apr.2019.05.002
Yuan, H., Zhang, D., Shi, Y., Li, B., Yang, J., Yu, X., et al. (2017). Cell concentration, viability and culture composition of airborne bacteria during a dust event in Beijing. J. Environ. Sci. (China) 55, 33–40. doi: 10.1016/j.jes.2016.03.033
Zhang, W. J., Sun, Y. L., Zhuang, G. S., and Dong-Qun, X. U. (2006). Characteristics and Seasonal Variations of PM2.5, PM10, and TSP Aerosol in Beijing. Biomed. Environment. Sci. 19:461.
Zhang, Y. L., and Cao, F. (2015). Fine particulate matter (PM2.5) in China at a city level. Sci. Rep. 5:14884.
Zhao, D. L., Tian, P., Zhow, W., Xiao, W., Sheng, J. J., Wang, F., et al. (2021). Evolution and potential source apportionment of atmospheric pollutants of two heavy haze episodes during the COVID-19 lockdown in Beijing, China. Huan Jing Ke Xue 42, 5109–5121. doi: 10.13227/j.hjkx.202104289
Zhong, S., Zhang, L. S., Jiang, X. Y., and Gao, P. (2019). Comparison of chemical composition and airborne bacterial community structure in PM2.5 during haze and non-haze days in the winter in Guilin, China. Sci. Total Environ. 655, 202–210. doi: 10.1016/j.scitotenv.2018.11.268
Keywords: airborne bacteria and fungi, particulate matter, haze, isolate collection, antimicrobial activity
Citation: Yan D, Zhang T, Bai J-L, Su J, Zhao L-L, Wang H, Fang X-M, Zhang Y-Q, Liu H-Y and Yu L-Y (2022) Isolation, Characterization, and Antimicrobial Activity of Bacterial and Fungal Representatives Associated With Particulate Matter During Haze and Non-haze Days. Front. Microbiol. 12:793037. doi: 10.3389/fmicb.2021.793037
Received: 11 October 2021; Accepted: 07 December 2021;
Published: 11 January 2022.
Edited by:
Angelica Bianco, UMR 6016 Laboratoire de Météorologie Physique (LAMP), FranceReviewed by:
Jonathan Vyskocil, Université Clermont Auvergne, FranceAna Monteiro, Escola Superior de Tecnologia da Saúde de Lisboa (ESTeSL), Portugal
Mashura Shammi, Jahangirnagar University, Bangladesh
Yingying Fu, Academy of Military Science of the Chinese People’s Liberation Army, China
Copyright © 2022 Yan, Zhang, Bai, Su, Zhao, Wang, Fang, Zhang, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Yan Yu, eWx5QGNwY2MuYWMuY24=
 Dong Yan
Dong Yan Tao Zhang
Tao Zhang Jing-Lin Bai1
Jing-Lin Bai1 Yu-Qin Zhang
Yu-Qin Zhang Hong-Yu Liu
Hong-Yu Liu Li-Yan Yu
Li-Yan Yu