- 1Tianjin Union Medical Center, TEDA Institute of Biological Sciences and Biotechnology, The Institute of Translational Medicine Research, Nankai University, Tianjin, China
- 2Key Laboratory of Molecular Microbiology and Technology, Nankai University, Ministry of Education, Tianjin, China
Morganella morganii, which is often regarded as a human commensal organism, can be an opportunistic pathogen, causing a variety of clinical infections with serious morbidity and mortality. An efficient and convenient method for subtyping and identifying M. morganii strains in epidemiological surveillance and control is urgently needed. Serotyping based on bacterial surface polysaccharide antigens (O-antigen or K-antigens) is a standard subtyping method for many gram-negative bacteria. Here, through whole genome sequencing and comparative genomics analysis of 27 strains, we developed a molecular serotyping scheme based on the genetic variation of O-antigen gene clusters (O-AGC) in M. morganii, and 11 distinct O-AGC types were identified. A conventional serotyping scheme was also developed by the production of antisera and agglutination experiments, which was shown to be perfectly consistent with the molecular serotyping scheme, confirming that the variation in M. morganii O-AGC correlated with phenotypic O-antigen diversification. Furthermore, a microsphere-based suspension array (MSA) with high specificity was developed based on the specific genes within each O-AGC type. The sensitivity of MSA was determined to be 0.1 ng of genomic DNA and 103 CFU of pure culture. We further analyzed 104 M. morganii genomes available in GenBank, and an additional six novel O-AGC types were identified, indicating that the extension of this molecular serotyping scheme is convenient. Our work provides an important tool for the detection and epidemiological surveillance of M. morganii, and this method has the potential to be widely utilized, especially for bacterial genera/species without an efficient typing approach.
Introduction
Morganella morganii is a Gram-negative bacillus belonging to the Enterobacteriaceae family, and it is a common inhabitant of the environment and intestinal tracts of humans, mammals, and reptiles (Erlanger et al., 2019). Conventionally, M. morganii is recognized as an unusual opportunistic pathogen that is isolated specifically in the urinary tract or wound infections. However, M. morganii has recently been regarded as an increasingly important pathogenic bacterium due to its virulence and increased drug resistance, which causes a variety of clinical infections, such as urinary tract infections, bacteremia and sepsis, and results in a high mortality rate in some infections (Liu et al., 2016; Minnullina et al., 2021).
Gram-negative bacteria produce lipopolysaccharide (LPS), which belongs to a family of structurally related glycolipids located in the outer membranes of bacteria (Megrian et al., 2020). A typical LPS molecule consists of a mix of well-conserved and highly variable structural elements (Supplementary Figure 1), including (i) lipid A, a component in the outer leaflet of the outer membranes that contributes to the barrier properties of bacteria and provides a pathogen-associated molecular pattern (Simpson and Trent, 2019); (ii) the core oligosaccharide (core OS), which is relatively conserved within a species, attaches to lipid A and is involved in the stability of the outer membrane (Whitfield and Trent, 2014); and (iii) the O-antigen or O-polysaccharide (O-PS), which is composed of hypervariable oligosaccharide repeating units (O-units) composed of two to eight different monosaccharide residues (heteroglycans) or identical sugars (homoglycans) in some bacteria (Valvano, 2003). The O-antigen is the most variable portion of LPS and provides the basis for serological specificity.
Serotyping based on the structural diversity of surface polysaccharide antigens, mainly O-antigen and K-antigen, remains the ‘gold standard’ for the detection and identification of strains, including pathogenic strains, in clinical specimens and environmental samples (Jacob et al., 2020; Elder et al., 2021). An O-antigenic scheme of M. morganii was established in the 1970s to 1990s (Vörös and Senior, 1990); however, the relevant antisera and the strains used for those studies are not currently available. The approach of traditional serotyping is limited by its high cost, high time and labor requirements, complicated procedures, cross-reactivity and subjective interpretation (Sumrall et al., 2020; Bian et al., 2021; Chaiden et al., 2021). To address these issues, many DNA-based serotyping methods on variant platforms targeting specific genes involved in the synthesis of surface polysaccharide antigens have been developed (Li et al., 2018, 2020; Zürn et al., 2020; Ouattara et al., 2021). In addition, a molecular serotyping scheme based on sequence variations of surface polysaccharide antigen synthesis genes has been rapidly established for several bacterial species. Moreover, the molecular serotyping scheme is theoretically consistent with the traditional serotyping system, and advances in next-generation sequencing have facilitated the establishment of these schemes (Sun et al., 2011; Guo et al., 2018).
Genes for O-antigen synthesis are normally clustered at the chromosome and named the O-AGC. Genetic variations in O-AGC are the major determinants of differences among the various O-antigens. The O-antigen synthesis genes fall into three major groups. The first group of genes is involved in the synthesis of nucleotide sugar precursors of the O-antigen. The second group, glycosyltransferase genes, is responsible for the sequential transfer of precursor sugars to undecaprenyl phosphate (UndP), thus forming the UndPP-O-unit. The O-antigen processing proteins, which are encoded by genes of the third group (wzx/wzy or wzm/wzt), are involved in translocation across the membrane and polymerization of the O-unit (Samuel and Reeves, 2003). According to the assembly mechanism, the process of O-antigen synthesis can be divided into three major pathways: the Wzx/Wzy-dependent pathway, which is usually used for heteroglycan synthesis (Islam and Lam, 2014), the ABC transporter (encoded by wzm and wzt) pathway, which is common during homoglycan synthesis (Greenfield and Whitfield, 2012), and the synthase pathway that has only been reported in Salmonella enterica O54 (Keenleyside et al., 1994). O-antigen processing genes and glycosyltransferase genes are always specific to individual O-antigens, and they are used in molecular assays to detect strains belonging to different serogroups (Ballmer et al., 2007).
Morganella morganii belongs to the tribe Proteeae along with another two members, Proteus and Providencia. Currently, no DNA-based typing assay, such as multilocus sequence typing (MLST), has been developed in M. morganii. Our group has investigated the O-AGCs of Proteus and Providencia in depth and developed molecular serotyping systems for each (Yu et al., 2017; Du et al., 2018). To our knowledge, the location of O-AGC in the genome of M. morganii remains unclear. Here, by sequencing and comparatively analyzing the genomes of 27 M. morganii strains from Shanghai Disease Control and Prevention, China, we established a molecular serotyping scheme for M. morganii with 11 different putative O-AGC types. The production of antisera against each O-AGC type and agglutination experiments confirmed the accuracy of our molecular serotyping scheme. A MSA based on O-AGC-specific genes was also developed. In summary, our work provides a promising and efficient tool for molecular diagnostics and epidemiological surveillance of M. morganii.
Materials and Methods
Genomic Sequencing and Bioinformatic Analysis
Twenty-seven M. morganii strains (Table 1) were cultured overnight in Luria-Bertani broth at 37°C with shaking. Subsequently, genomic DNA was extracted from 1.5 mL of each of the overnight bacterial cultures using a DNA extraction kit according to the manufacturer’s instructions (Tiangen, Beijing, China). The genomic DNA was sheared, polished, and prepared using the Illumina Sample Preparation Kit. Genomic libraries containing 500-bp paired-end inserts were constructed, and sequencing was then performed with Solexa sequencing technologies (Illumina Inc., San Diego California, United States) to produce approximately 100-fold coverage. The obtained reads were assembled using the de novo genome assembly program Velvet to generate a multicontig draft genome. Next, Artemis (Rutherford et al., 2000) was subjected to annotate genes. BLAST and PSI-BLAST were used to search available databases, including the GenBank1 and Pfam protein motif databases (pfam.sanger.ac.uk), for gene and protein annotation (Mistry et al., 2021), respectively. The TMHMM v2.0 analysis program2 was used to identify potential transmembrane domains within the protein sequences.
Preparation of Antigens and Antisera
All strains used for immunization were cultured in 10 mL of Luria-Bertani agar overnight. The cultures were harvested by centrifugation at 5,000 × g for 20 min, washed in 20 mL of 0.85% NaCl, and suspended in 20 ml of 0.85% NaCl (approximately 109 cells/ml). The cell suspensions were subsequently heated at 121°C for 30 min and then cooled. Only strains that did not exhibit autoagglutination were used as the resultant antigens for immunization. Subsequently, adult New Zealand White rabbits (12 weeks of age) were injected intravenously with the prepared antigens. At 3-day intervals, injections were administered at doses of 0.5 ml, 1 ml, 2 ml, and 4 ml. One week after the final injection, the rabbit was exsanguinated, and the separated antisera were stored at 4°C.
Agglutination Test and Antisera Absorption
Antigens were prepared from the cells cultured overnight in 10 mL of Luria-Bertani agar at 37°C and then mixed with 25 μL of 2-fold diluted antiserum in 0.85% NaCl. The selected titer was the most diluted concentration of antiserum that gave a positive reaction. The cell suspensions used for absorption were washed three times with 0.85% NaCl and resuspended in 3 mL of antiserum. The mixture was incubated at room temperature for 2 h and then centrifuged at 10,000 × g for 15 min, after which the supernatant was collected. The absorbed antiserum was tested against all antigens that reacted with the unabsorbed antiserum. This process was repeated until cross-reactions no longer occurred.
Development of a Microsphere-Based Suspension Array for Molecular Serotyping
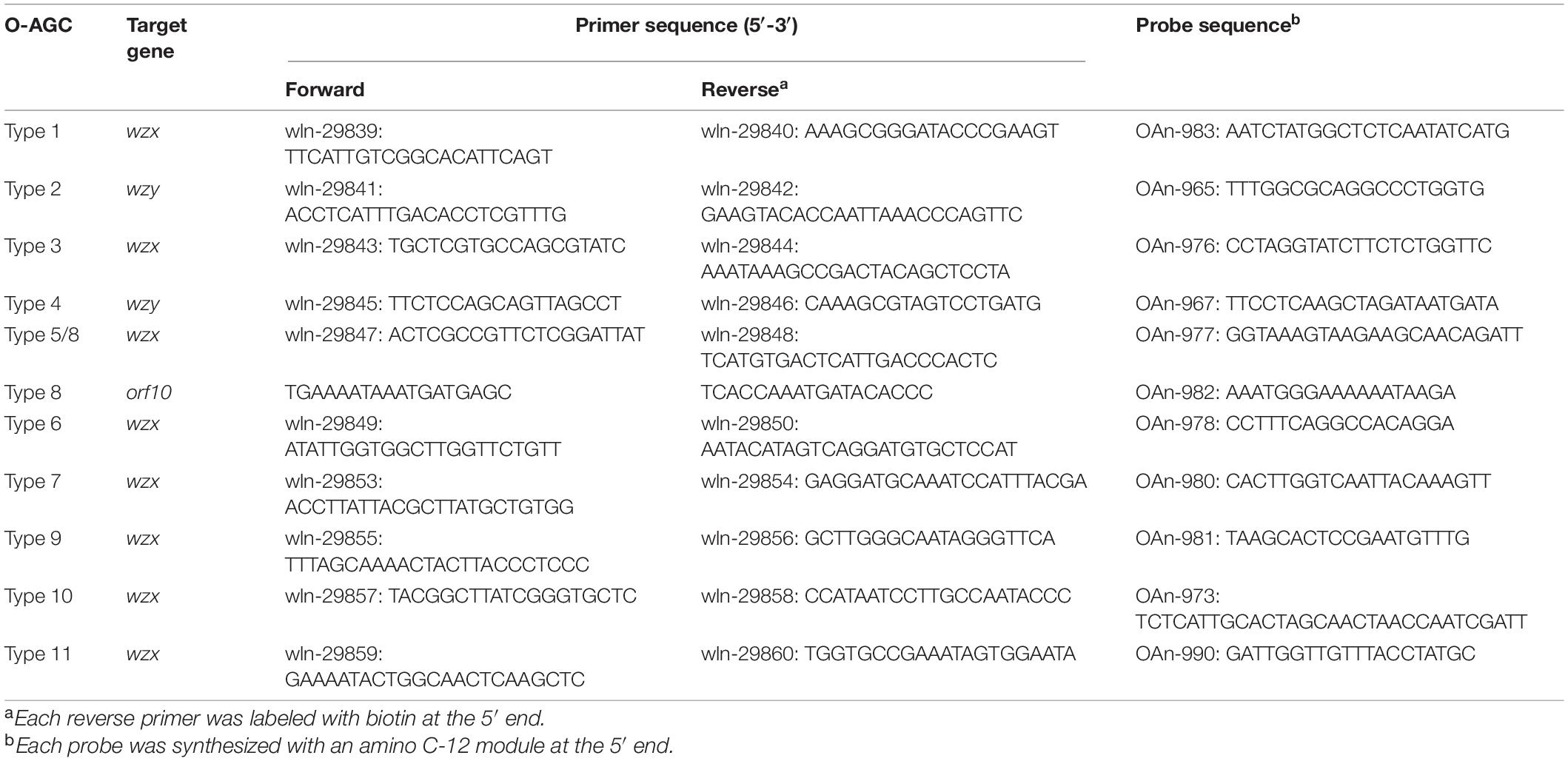
Almost all primers and probes (Table 2) for MSA were designed based on the wzx or wzy gene using Primer Premier v5.0 software (Premier Biosoft International, Palo Alto, CA, United States), except those targeting orf10type 8, which is utilized to differentiate type 8 from type 5. Multiplex PCR amplification was performed in a 50 μL reaction mixture composed of 100 ng genomic DNA, 1 × Goldstar PCR buffer, 0.04 mM deoxynucleoside triphosphates, 0.1 mM each primer, and 1 unit Goldstar DNA polymerase. The PCR parameters were as follows: 95°C for 10 min; 30 cycles at 98°C for 10 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. Probe-microsphere coupling, hybridization and MSA analysis were performed as described previously (Guo et al., 2018). A positive signal was defined as a median fluorescence intensity (MFI) ≥ 200 and a signal/background ratio (S/B ratio = MFI/Blank) > 5.
Results
Location and Genetic Features of O-Antigen Gene Clusters in Morganella morganii
By sequencing and analyzing the genomes of 27 M. morganii isolates, several putative gene clusters associated with polysaccharide synthesis were annotated (data not shown). Among them, a genetic region between two housekeeping genes was analyzed, and the locus of each isolate shares some common features. First, all of the loci are mapped between two flanked genes, cpxA and secB, which encode a two component system sensor kinase and protein-export chaperone, respectively, as in the case of Proteus O-AGC (Yu et al., 2017; Du et al., 2018). Second, each locus contains three classes of genes associated with O-antigen synthesis. Third, each locus shows a lower % GC content (∼35%) than that of the whole genome (∼51%) (Chen et al., 2012). Therefore, we propose that the genetic region between cpxA and secB is a candidate O-AGC for M. morganii, and it may also be recently obtained by lateral transfer from a different species, as in many other bacteria.
In total, 11 types of putative O-AGCs were identified, with their nucleotide sequences ranging in size from 10,850 to 15,182 bp. A conserved gene set trmL-cysE-gpsA is located at the 3′ end of each O-AGC, with the minor exceptions of one gene being inserted between trmL and cysE in type 10 and type 11. Moreover, the wzx gene encoding O-unit flippase and the wzy gene encoding O-antigen polymerase were both found within each O-AGC, indicating that M. morganii probably synthesizes the O-antigen via the Wzx/Wzy-dependent pathway (Figure 1). The characteristics of open reading frames (ORFs) for all putative O-AGCs are summarized in Supplementary Table 1.
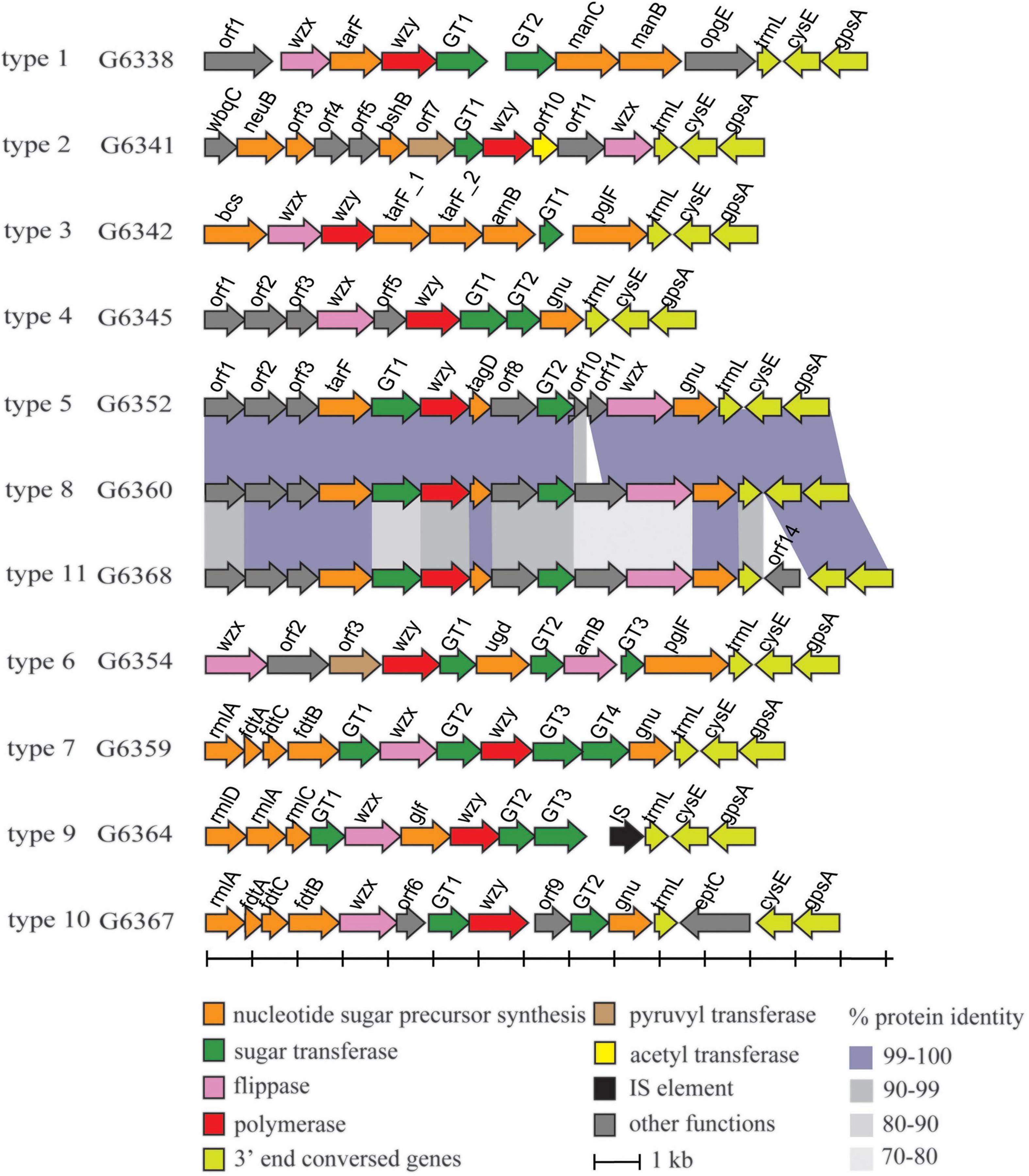
Figure 1. Schematic diagram of the 11 putative O-AGCs characterized from the 27 M. morganii strains. Genes are represented by arrows and colored according to the gene key at the bottom.
Several sugar residues within the O-unit of M. morganii can be predicted based on the presence of corresponding nucleotide sugar precursor synthesis genes in O-AGC. For instance, the manC gene encoding mannose-1-phosphate guanylyltransferase and the manB gene encoding phosphomannomutase were assigned to O-AGC type 1, and their products are involved in the synthesis of GDP-D-mannose along with phosphomannose isomerase ManA (manA gene is always outside the O-AGC) (Samuel and Reeves, 2003). In O-AGC type 9, rmlD/A/C and glf were annotated. RmlA/B/C/D have been identified as responsible for the synthesis of dTDP-L-rhamnose, the nucleotide sugar precursor of L-rhamnose (Allard et al., 2001), and Glf (UDP-galactopyranose mutase) has been identified to catalyze the conversion from UDP-galactose pyranose to UDP-galactose furanose (Nassau et al., 1996). In O-AGC types 7 and 10, a rmlA/ftdA/C/B set is located at the 5’ region. The enzymes encoded by these four genes (glucose-1-phosphate thymidylyltransferase, dTDP-6-deoxy-hex-4-ulose isomerase, dTDP-6-deoxy-D-xylo-hex-3-ulose aminase and dTDP-D-Fuc3N acetylase), together with dTDP-D-glucose 4,6-dehydratase (encoded by rmlB gene), have been identified to be involved in the synthesis of dTDP-3-acetamido-3-deoxy-D-fucose (dTDP-Fuc3NAc) (Pfoestl et al., 2003). It should be noted that rmlB, which is always present along with rmlACD genes, is not found within O-AGC 9, 7, and 10, thus indicating that rmlB may be located elsewhere at the chromosome, and we successively assigned the homolog of rmlB in the genome of strains belonging to O-AGC types 9, 7, and 10.
In addition, several O-AGCs of M. morganii are genetically related. For instance, the sequences of type 5 and type 8 O-AGCs share high-level identity (96 to 100%) except for the genes between GT2 and wzx, respectively. In addition, the O-AGCs of type 8 and type 11 also share high level identity (72 to 100%), with the exception of orf14 inserted between trmL and cysE of type 11 O-AGC. Compared with the nucleotide sugar precursor synthesis genes, the identity of glycosyltransferase genes and processing genes (wzx/wzy) between these two O-AGCs was relatively low (Figure 1), indicating that the products encoded by these genes may determine the specificity of the O-antigen structure of these two types.
Collectively, these data suggest that the region between cpxA and secB in the M. morganii genome exhibits the genetic features of O-AGC and a potential molecular serotyping scheme could be established in M. morganii based on the genetic diversity of O-AGC types.
Conventional Serotyping Scheme Is Correlated Well With the Molecular Serotyping Scheme
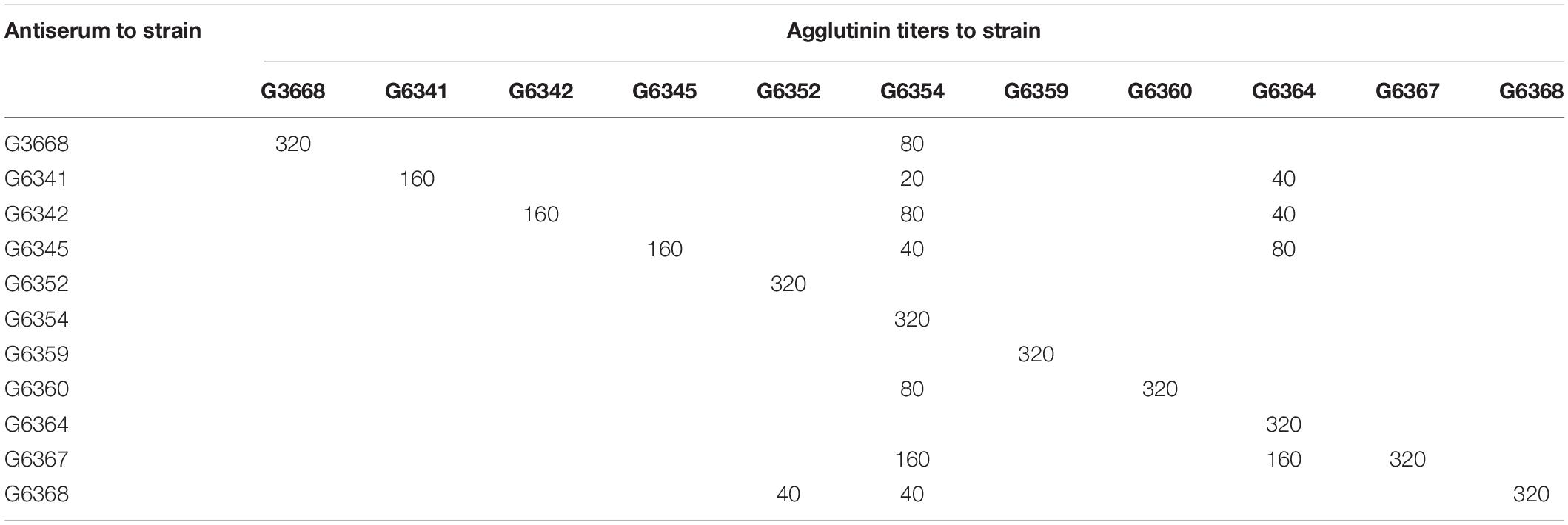
Antiserum against each type strain representing unique O-AGC was prepared, and an agglutination test was performed. The homologous and heterologous titers are summarized in Table 3. Generally, each antiserum reacted to its homologous strain with high titers (160 to 320). Simultaneously, except for the antiserum against G6352, G6354, G6359, and G6364, the remainders agglutinated with heterologous strains and thus needed to be absorbed. After absorption, all the cross reactions disappeared and each absorbed antiserum only generated a positive result with its corresponding homologous strain. Next, we tested 16 other strains with the absorbed antisera. As expected, each of them reacted with only one of the 11 antisera, and the grouping is consistent with the case in the O-AGC allocation. These data suggest that the conventional serotyping scheme is perfectly consistent with the molecular serotyping scheme, thus confirming that the variation in M. morganii O-AGC is correlated with phenotypic O-antigen diversification. We designated G6338, G6341, G6342, G6345, G6352, G6354, G6359, G6360, G6364, G6367, and G6368 as type strains (types 1 to 11) for each O-AGC type.
Development of a Microsphere-Based Suspension Assay
Compared to the nucleotide sugar precursor synthesis genes and glycosyltransferase genes, O-antigen processing genes are always more heterogeneous (Ballmer et al., 2007). Thus, the wzx or wzy gene was used for primer and probe design in our study, and orf10type8 was selected to differentiate type 8 from type 5. The 27 M. morganii strains carrying O-AGC 1 to 11 and other bacteria that are genetically close to M. morganii or frequently isolated from the urinary tract, including Proteus spp. (n = 5), Providencia spp. (n = 4), Escherichia coli (n = 2), Klebsiella pneumoniae (n = 2), Enterococcus faecalis (n = 2), Group B Streptococcus (n = 1), and Staphylococcus aureus (n = 1), were used to test the specificity of the microsphere-based suspension (MAS) assay. Our test showed that each O-AGC-specific probe detected the homologous strains correctly and no hybridization signals were generated against the heterologous strains, indicating the accurate specificity of our assay (Figure 2). Moreover, a sensitivity test was performed using genomic DNA or pure cultures from the type strains containing each O-AGC type. Generally, the genomic DNA of each type strain was serially diluted by 10-fold (10 ng to 0.1 pg) as the template, and the target could be determined using at least 0.1 ng DNA. Our results also showed that the MAS array could generate a positive signal for boiled supernatants from at least 103 CFU of pure bacterial cultures.
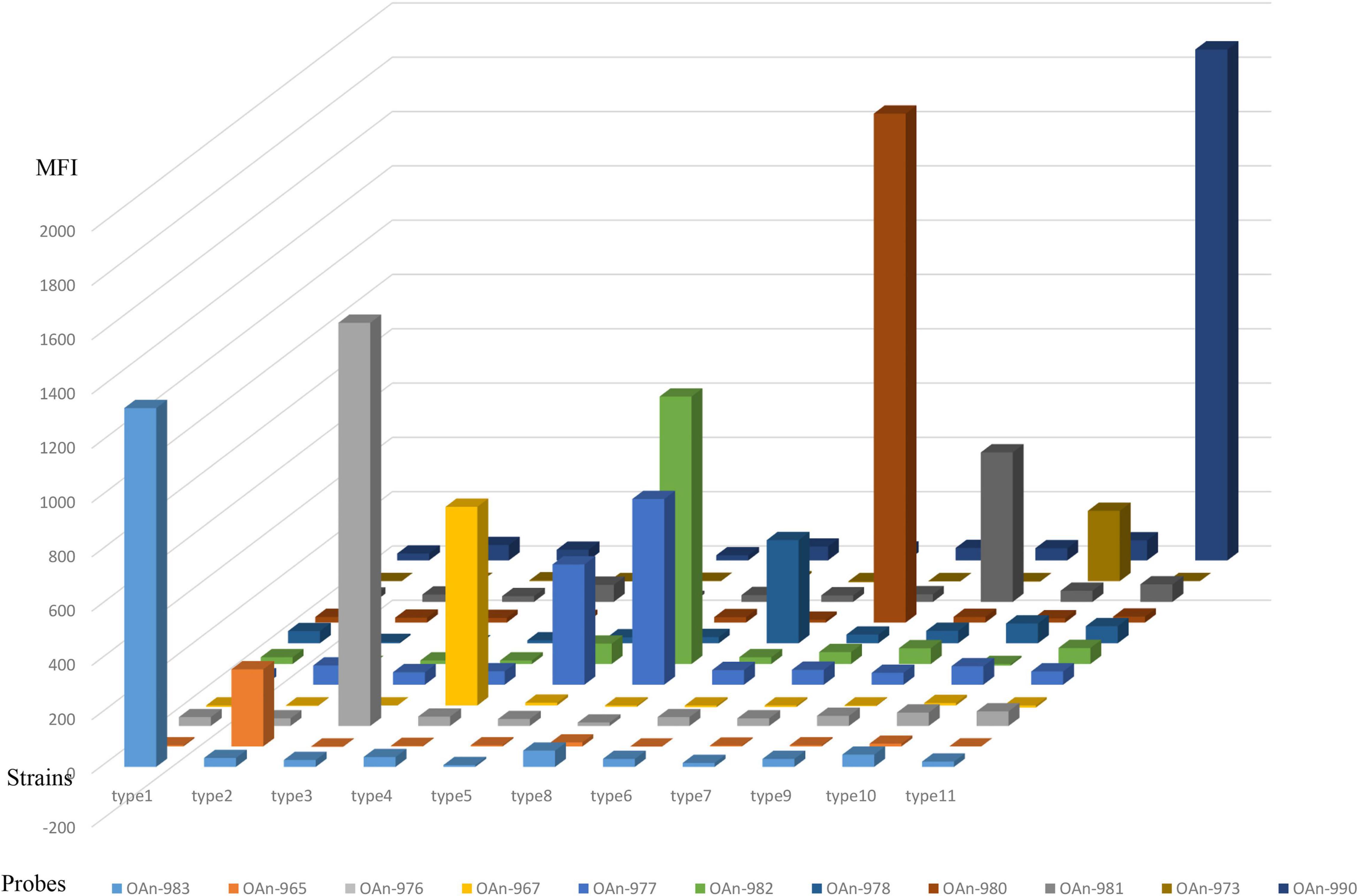
Figure 2. Specific test of the MSA assay. The hybridization signals are presented in terms of median fluorescence intensity (MFI) on the y-axis, and each sample representing the corresponding type strain is indicated on the x-axis. Probe OAn-982 were designed targeting orf10type8 to differentiate type 8 from type 5.
Serotype Allocation Using Genome Data
A program was designed by us to perform the in silico serotyping of M. morganii using the genome data. In brief, a database was first generated based on the wzx or wzy genes and orf10type8, which were all verified by our MSA assay. Next, the 104 M. morganii genome assemblies from GenBank were subjected to a BLASTn search against the database with a coverage cutoff of >90% and an identity cutoff of >99%. Among them, 42 strains could be assigned to one of the eight O-AGC types characterized here, with type 8 (43%) and type 11 (26%) representing the predominant groups, and none of the strains matched to types 2, 4 and 10 (Supplementary Table 2).
Among the remaining 62 genome assemblies, 16 were too fragmented and were thus excluded from the next analysis. The genetic regions between cpxA and secB in 46 strains were extracted and analyzed, and six additional putative O-AGCs (temp1 to 6) were characterized (Figure 3 and Supplementary Table 3).
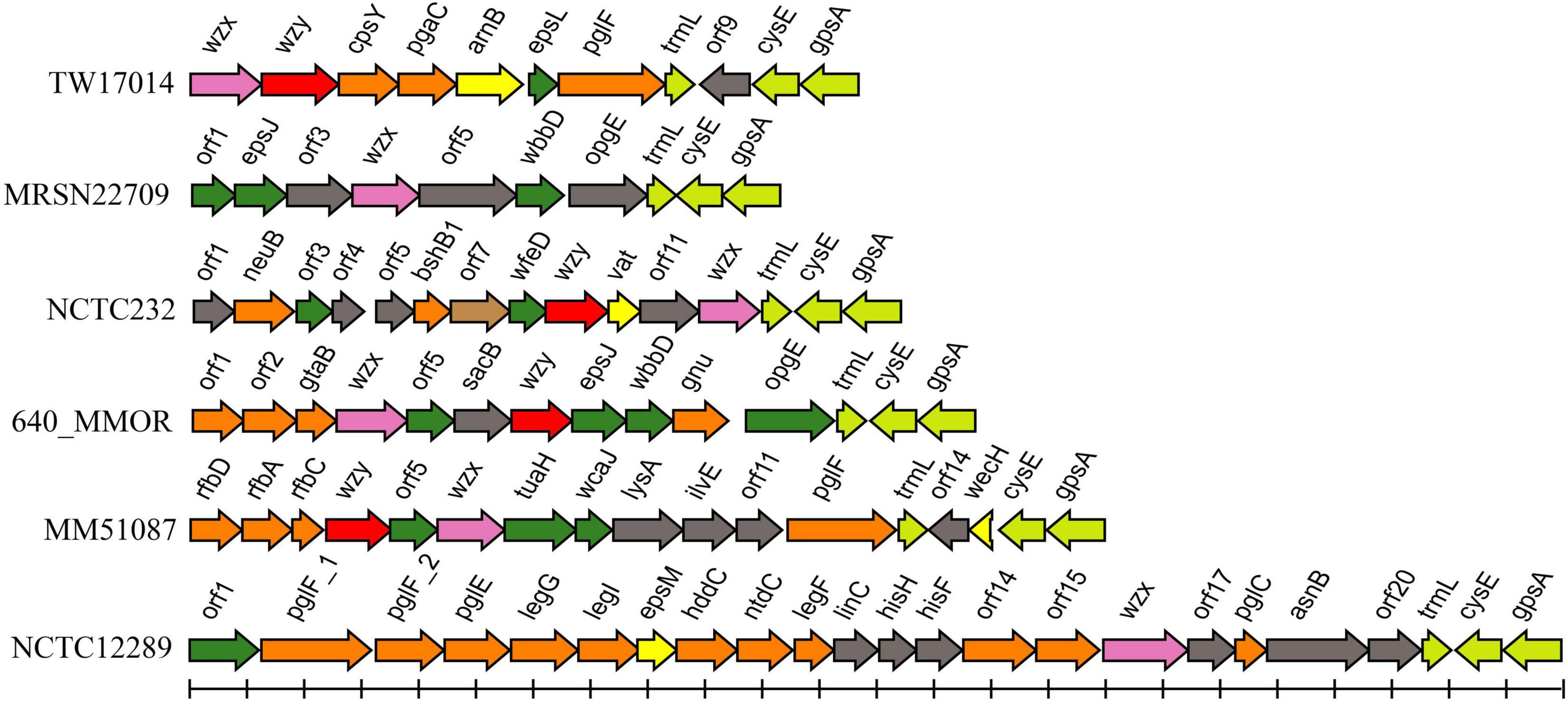
Figure 3. Schematic diagram of the six temporary O-AGCs extracted from M. morganii genomes from GenBank. For the gene key, see Figure 1.
Discussion
According to a recent classification based on genomic phylogeny, Morganella is a type genus of the novel Morganellaceae family, which also includes the genetically adjacent genera Proteus and Providencia (Adeolu et al., 2016). The sharing of the same O-AGC locus by Morganella and Proteus also indicates the close relationship between the two genera. Several methods have been developed for M. morganii subclassification, including biotyping (Rauss and Vörös, 1959), phage typing (Schmidt and Jeffries, 1974), bacteriocin typing (Senior, 1987), and protein profile typing (Senior and Vörös, 1990). Among these techniques, biotyping and phage typing have low distinguishing power. Although much more discriminating, the typing method by bacteriocin testing and protein profiling is time-consuming and laborious. Currently, whole-genome sequencing (WGS)-based approaches, mainly including core genome multilocus sequence typing (cg-MLST) and single-nucleotide polymorphism (SNP)-based approaches, have been presented and applied to pathogenic epidemiological surveillance, thus providing maximal discrimination compared to other methods (Hazen et al., 2016; Njamkepo et al., 2016; Beyrouthy et al., 2018). However, the WGS-based approach has not been applied widely to routine detection, especially in basic infection control agencies, due to its relatively high cost and lengthy turnaround time.
The variation in O-antigen structures provides the basis for serotyping many Gram-negative bacteria, and it has been widely used to classify strains for epidemiological investigation and surveillance and represents the ‘gold standard’ (Liu et al., 2020). Conventional serotyping based on agglutination reactions is also limited by the high prevalence of non-typeable isolates and cross reactions, which is common in clinical isolates. The molecular serotyping system developed by our group correlated well with the traditional antigenic scheme and can offer better resolution (Guo et al., 2018). Accordingly, we believe that fast and reliable PCR-based methodologies targeting serospecific genes, such as MSA technology, are ideal for pathogenic classification and epidemiological purposes.
Among six novel putative O-AGCs, no wzy gene was assigned in strains MRSN22709 and NCTC12289 (Figure 3). It is likely that the functional wzy gene is located outside the O-AGC in these two strains. This atypical feature has been reported in other species. For example, the wzy gene maps far from the O-AGCs in Salmonella serogroups A, B, and D1 (Wang et al., 2002). By screening the genome assemblies, we also observed that the cpxA-secB regions in five genomes possess neither glycosyltransferase genes nor possessing genes. It is likely that in these isolates, the functional O-AGCs reside in other sites of the chromosome, which will be the subject of future studies.
The chemical structure of M. morganii O-antigens should be elucidated in the future to further support our findings. In addition, as O-antigen is an important virulence factor associated with bacterial pathogenesis (March et al., 2013; Sarkar et al., 2014; Caboni et al., 2015), our work also provides the basis for future studies on the role of O-antigen in M. morganii pathogenesis and could lead to a deeper understanding of the virulence mechanisms of this bacterium.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession numbers OK482582–OK482592.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care Committee at Nankai University (Tianjin, China; protocol code 20190325.02).
Author Contributions
LW and BL (correspondence) designed the research. BL (first author), XG, JW, PW, and SL performed the research. LF provided technical support and insights. XG and JW analyzed the data. XG and BL (correspondence) wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (NSFC) Program (31770144, 31820103002, 32070130, 81772148, and 81871624), Young Scholar of Tianjin (20JCJQJC00180), the Natural Science Foundation (Key Project) of Tianjin (20JCZDJC00820), the National Key R&D Program of China (2018YFA0901000), and the National Key Programs for Infectious Diseases of China (2017ZX10303405).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.791165/full#supplementary-material
Footnotes
References
Adeolu, M., Alnajar, S., Naushad, S., and S Gupta, R. (2016). Genomebased phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Allard, S. T., Giraud, M. F., Whitfield, C., Graninger, M., Messner, P., and Naismith, J. H. (2001). The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J. Mol. Biol. 307, 283–295. doi: 10.1006/jmbi.2000.4470
Ballmer, K., Korczak, B. M., Kuhnert, P., Slickers, P., Ehricht, R., and Hächler, H. (2007). Fast DNA serotyping of Escherichia coli by use of an oligonucleotide microarray. J. Clin. Microbiol. 45, 370–379. doi: 10.1128/JCM.01361-06
Beyrouthy, R., Barets, M., Marion, E., Dananché, C., Dauwalder, O., Robin, F., et al. (2018). Novel enterobacter lineage as leading cause of nosocomial outbreak involving carbapenemase-producing strains. Emerg. Infect. Dis. 24, 1505–1515. doi: 10.3201/eid2408.180151
Bian, S., Jia, Y., Zhan, Q., Wong, N. K., Hu, Q., Zhang, W., et al. (2021). VPsero: rapid serotyping of Vibrio parahaemolyticus using serogroup-specific genes based on whole-genome sequencing data. Front. Microbiol. 12:620224. doi: 10.3389/fmicb.2021.620224
Caboni, M., Pédron, T., Rossi, O., Goulding, D., Pickard, D., Citiulo, F., et al. (2015). An O antigen capsule modulates bacterial pathogenesis in Shigella sonnei. PLoS Pathog. 11:e1004749. doi: 10.1371/journal.ppat.1004749
Chaiden, C., Jaresitthikunchai, J., Kerdsin, A., Meekhanon, N., Roytrakul, S., and Nuanualsuwan, S. (2021). Streptococcus suis serotyping by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. PLoS One 16:e0249682. doi: 10.1371/journal.pone.0249682
Chen, Y. T., Peng, H. L., Shia, W. C., Hsu, F. R., Ken, C. F., Tsao, Y. M., et al. (2012). Whole-genome sequencing and identification of Morganella morganii KT pathogenicity-related genes. BMC Genomics 13, Suppl 7(Suppl. 7):S4. doi: 10.1186/1471-2164-13-S7-S4
Du, Y., Li, H., Yin, Z., Rozalski, A., Torzewska, A., Yang, P., et al. (2018). Development of a molecular serotyping scheme and a multiplexed luminex-based array for Providencia. J. Microbiol. Methods 153, 14–23. doi: 10.1016/j.mimet.2018.08.009
Elder, J. R., Fratamico, P. M., Liu, Y., Needleman, D. S., Bagi, L., Tebbs, R., et al. (2021). A targeted sequencing assay for serotyping Escherichia coli using agriseq technology. Front. Microbiol. 11:627997. doi: 10.3389/fmicb.2020.627997
Erlanger, D., Assous, M. V., Wiener-Well, Y., Yinnon, A. M., and Ben-Chetrit, E. (2019). Clinical manifestations, risk factors and prognosis of patients with Morganella morganii sepsis. J. Microbiol. Immunol. Infect. 52, 443–448. doi: 10.1016/j.jmii.2017.08.010
Greenfield, L. K., and Whitfield, C. (2012). Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24. doi: 10.1016/j.carres.2012.02.027
Guo, X., Wang, M., Wang, L., Wang, Y., Chen, T., Wu, P., et al. (2018). Establishment of a Molecular Serotyping Scheme and a Multiplexed Luminex-Based Array for Enterobacter aerogenes. Front. Microbiol. 9:501. doi: 10.3389/fmicb.2018.00501
Hazen, T. H., Donnenberg, M. S., Panchalingam, S., Antonio, M., Hossain, A., Mandomando, I., et al. (2016). Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat. Microbiol. 1:15014. doi: 10.1038/nmicrobiol.2015.14
Islam, S. T., and Lam, J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 60, 697–716. doi: 10.1139/cjm-2014-0595
Jacob, J. J., Rachel, T., Shankar, B. A., Gunasekaran, K., Iyadurai, R., Anandan, S., et al. (2020). MLST based serotype prediction for the accurate identification of non typhoidal Salmonella serovars. Mol. Biol. Rep. 47, 7797–7803. doi: 10.1007/s11033-020-05856-y
Keenleyside, W. J., Perry, M., Maclean, L., Poppe, C., and Whitfield, C. (1994). A plasmid-encoded rfbO:54 gene cluster is required for biosynthesis of the O:54 antigen in Salmonella enterica serovar Borreze. Mol. Microbiol. 11, 437–448. doi: 10.1111/j.1365-2958.1994.tb00325.x
Li, H., Du, Y., Qian, C., Li, L., Jiang, L., Jiang, X., et al. (2018). Establishment of a suspension array for Pseudomonas aeruginosa O-antigen serotyping. J. microbiol. Methods 155, 59–64. doi: 10.1016/j.mimet.2018.11.006
Li, Y., Huang, J., Wang, X., Xu, C., Han, T., and Guo, X. (2020). Genetic characterization of the o-antigen and development of a molecular serotyping scheme for Enterobacter cloacae. Front. Microbiol. 11:727. doi: 10.3389/fmicb.2020.00727
Liu, B., Furevi, A., Perepelov, A. V., Guo, X., Cao, H., Wang, Q., et al. (2020). Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 44, 655–683. doi: 10.1093/femsre/fuz028
Liu, H., Zhu, J., Hu, Q., and Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
March, C., Cano, V., Moranta, D., Llobet, E., Pérez-Gutiérrez, C., Tomás, J. M., et al. (2013). Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS One 8:e56847. doi: 10.1371/journal.pone.0056847
Megrian, D., Taib, N., Witwinowski, J., Beloin, C., and Gribaldo, S. (2020). One or two membranes? Diderm firmicutes challenge the Gram-positive/Gram-negative divide. Mol. Microbiol. 113, 659–671. doi: 10.1111/mmi.14469
Minnullina, L., Kostennikova, Z., Evtugin, V., Akosah, Y., Sharipova, M., and Mardanova, A. (2021). Diversity in the swimming motility and flagellar regulon structure of uropathogenic Morganella morganii strains. Int. Microbiol. doi: 10.1007/s10123-021-00197-7 [Epub ahead of print].
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419.
Nassau, P. M., Martin, S. L., Brown, R. E., Weston, A., Monsey, D., McNeil, M. R., et al. (1996). Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacterial. 178, 1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996
Njamkepo, E., Fawal, N., Tran-Dien, A., Hawkey, J., Strockbine, N., Jenkins, C., et al. (2016). Global phylogeography and evolutionary history of Shigella dysenteriae type 1. Nat. Microbiol. 1:16027. doi: 10.1038/nmicrobiol.2016.27
Ouattara, M., Tamboura, M., Kambiré, D., Lê, K. A., Van Phan, T., Velusamy, S., et al. (2021). Triplex direct quantitative polymerase chain reaction for the identification of Streptococcus pneumoniae serotypes. J. Infect. Dis. 224(Supplement_3) S204–S208. doi: 10.1093/infdis/jiab056
Pfoestl, A., Hofinger, A., Kosma, P., and Messner, P. (2003). Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-galactose in Aneurinibacillus thermoaerophilus L420-91T. J. Boil. Chem. 278, 26410–26417. doi: 10.1074/jbc.M300858200
Rauss, K., and Vörös, S. (1959). The biochemical and serological properties of Proteus morganii. Acta. Microbiol. Acad. Sci. Hung. 6, 233–248.
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualisation and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Samuel, G., and Reeves, P. (2003). Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338, 2503–2519. doi: 10.1016/j.carres.2003.07.009
Sarkar, S., Ulett, G. C., Totsika, M., Phan, M. D., and Schembri, M. A. (2014). Role of capsule and O antigen in the virulence of uropathogenic Escherichia coli. PLoS One 9:e94786. doi: 10.1371/journal.pone.0094786
Schmidt, W. C., and Jeffries, C. D. (1974). Bacteriophage typing of Proteus mirabilis, Proteus vulgaris, and Proteus morganii. Appl. Microbiol. 27, 47–53. doi: 10.1128/am.27.1.47-53.1974
Senior, B. W. (1987). The typing of Morganella morgani by bacteriocin production and sensitivity. J. Med. Microbiol. 23, 33–39. doi: 10.1099/00222615-23-1-33
Senior, B. W., and Vörös, S. (1990). Protein profile typing–a new method of typing Morganella morganii strains. J. Med. Microbiol. 33, 259–264. doi: 10.1099/00222615-33-4-259
Simpson, B. W., and Trent, M. S. (2019). Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416. doi: 10.1038/s41579-019-0201-x
Sumrall, E. T., Röhrig, C., Hupfeld, M., Selvakumar, L., Du, J., Dunne, M., et al. (2020). Glycotyping and specific separation of Listeria monocytogenes with a novel bacteriophage protein tool kit. Appl. Environ. Microbiol. 86, e612–e620. doi: 10.1128/AEM.00612-20
Sun, Y., Wang, M., Liu, H., Wang, J., He, X., Zeng, J., et al. (2011). Development of an O-antigen serotyping scheme for Cronobacter sakazakii. Appl. Environ. Microbiol. 77, 2209–2214. doi: 10.1128/AEM.02229-10
Valvano, M. A. (2003). Export of O-specific lipopolysaccharide. Front. Biosci. 8, s452–s471. doi: 10.2741/1079
Vörös, S., and Senior, B. W. (1990). New O antigens of Morganella morganii and the relationships between haemolysin production. O antigens and morganocin types of strains. Acta. Microbiol. Hung. 37, 341–349.
Wang, L., Andrianopoulos, K., Liu, D., Popoff, M. Y., and Reeves, P. R. (2002). Extensive variation in the O-antigen gene cluster within one Salmonella enterica serogroup reveals an unexpected complex history. J. Bacteriol. 184, 1669–1677. doi: 10.1128/JB.184.6.1669-1677.2002
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi: 10.1146/annurev-biochem-060713-035600
Yu, X., Torzewska, A., Zhang, X., Yin, Z., Drzewiecka, D., Cao, H., et al. (2017). Genetic diversity of the O antigens of Proteus species and the development of a suspension array for molecular serotyping. PLoS One 12:e0183267. doi: 10.1371/journal.pone.0183267
Keywords: Morganella morganii, surface polysaccharide, O-antigen gene cluster, serotyping, microsphere-based suspension array
Citation: Liu B, Guo X, Wang J, Wu P, Li S, Feng L, Liu B and Wang L (2021) Development of a Molecular Serotyping Scheme for Morganella morganii. Front. Microbiol. 12:791165. doi: 10.3389/fmicb.2021.791165
Received: 08 October 2021; Accepted: 02 November 2021;
Published: 23 November 2021.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Olivier Barraud, INSERM U1092 Anti-infectieux Supports Moléculaires des Résistances et Innovations Thérapeutiques, FranceBo Pang, National Institute for Communicable Disease Control and Prevention (CDC), China
Copyright © 2021 Liu, Guo, Wang, Wu, Li, Feng, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Liu, bGl1YmluMTk4MUBuYW5rYWkuZWR1LmNu; Lei Wang, d2FuZ2xlaUBuYW5rYWkuZWR1LmNu
†These authors have contributed equally to this work
 Bin Liu1,2†
Bin Liu1,2† Xi Guo
Xi Guo Bin Liu
Bin Liu Lei Wang
Lei Wang