- 1Department of Biotechnology, Quaid-i-Azam University, Islamabad, Pakistan
- 2Department of Biosciences, COMSATS University, Islamabad, Pakistan
- 3Center for Biotechnology and Microbiology, University of Swat, Swat, Pakistan
Virus-like particles (VLPs) are nanostructures that possess diverse applications in therapeutics, immunization, and diagnostics. With the recent advancements in biomedical engineering technologies, commercially available VLP-based vaccines are being extensively used to combat infectious diseases, whereas many more are in different stages of development in clinical studies. Because of their desired characteristics in terms of efficacy, safety, and diversity, VLP-based approaches might become more recurrent in the years to come. However, some production and fabrication challenges must be addressed before VLP-based approaches can be widely used in therapeutics. This review offers insight into the recent VLP-based vaccines development, with an emphasis on their characteristics, expression systems, and potential applicability as ideal candidates to combat emerging virulent pathogens. Finally, the potential of VLP-based vaccine as viable and efficient immunizing agents to induce immunity against virulent infectious agents, including, SARS-CoV-2 and protein nanoparticle-based vaccines has been elaborated. Thus, VLP vaccines may serve as an effective alternative to conventional vaccine strategies in combating emerging infectious diseases.
Introduction
The majority of currently available vaccines are predominantly based on either inactivated (killed) or live attenuated approaches. Although these traditional vaccines have been used effectively against various infectious diseases, some of these have several limitations, which include their lower potential to induce a stronger immune response and poor efficacy (Yan et al., 2020). Recent outbreaks of infectious diseases have manifested the need for the development of robust vaccines to overcome these limitations. The main challenge is to develop new technological approaches that enhance immunity without jeopardizing safety, efficacy, and tolerability. Recent advancements in DNA, mRNA, and recombinant viral-vector based vaccines present effective vaccine development methods for difficult-to-target pathogens and control of infectious disease outbreaks (Francis, 2018; Aida et al., 2021).
Virus-like particle (VLP) technology presents an alternative platform for developing effective vaccines to combat infectious diseases of serious concern, and it is moving in parallel with mRNA and viral-vector based vaccines (Lampinen et al., 2021). VLPs are also far more immunogenic as compared to other subunit vaccines as they present repetitive antigenic epitopes on their surface in a more authentic confirmation that the immune system can readily detect. Subunit vaccines, on the other hand, have poor immunogenicity due to misfolding of targeted antigen or insufficient presentation to the immunological system (Noad and Roy, 2003). Moreover, they require adjuvants and repeated doses of vaccination to evoke a sufficient immune response (Brisse et al., 2020). Fraenkel-Conrat and Williams (1955) first described the term virus-like particles by reassembling tobacco mosaic virus (TMV) particles from their purified RNA and protein components. The potential of these nanostructures to induce a potent immune response was then further studied later. The VLP platform can overcome various problems that are usually associated with traditional vaccines; specifically, the infectious nature related to live attenuated vaccines, reversion to a virulent form, risk of mutation, reduced immunogenicity of inactivated vaccine, unstable toxicity, low yield, and lengthy formulation time (Mohsen et al., 2018; Figure 1).
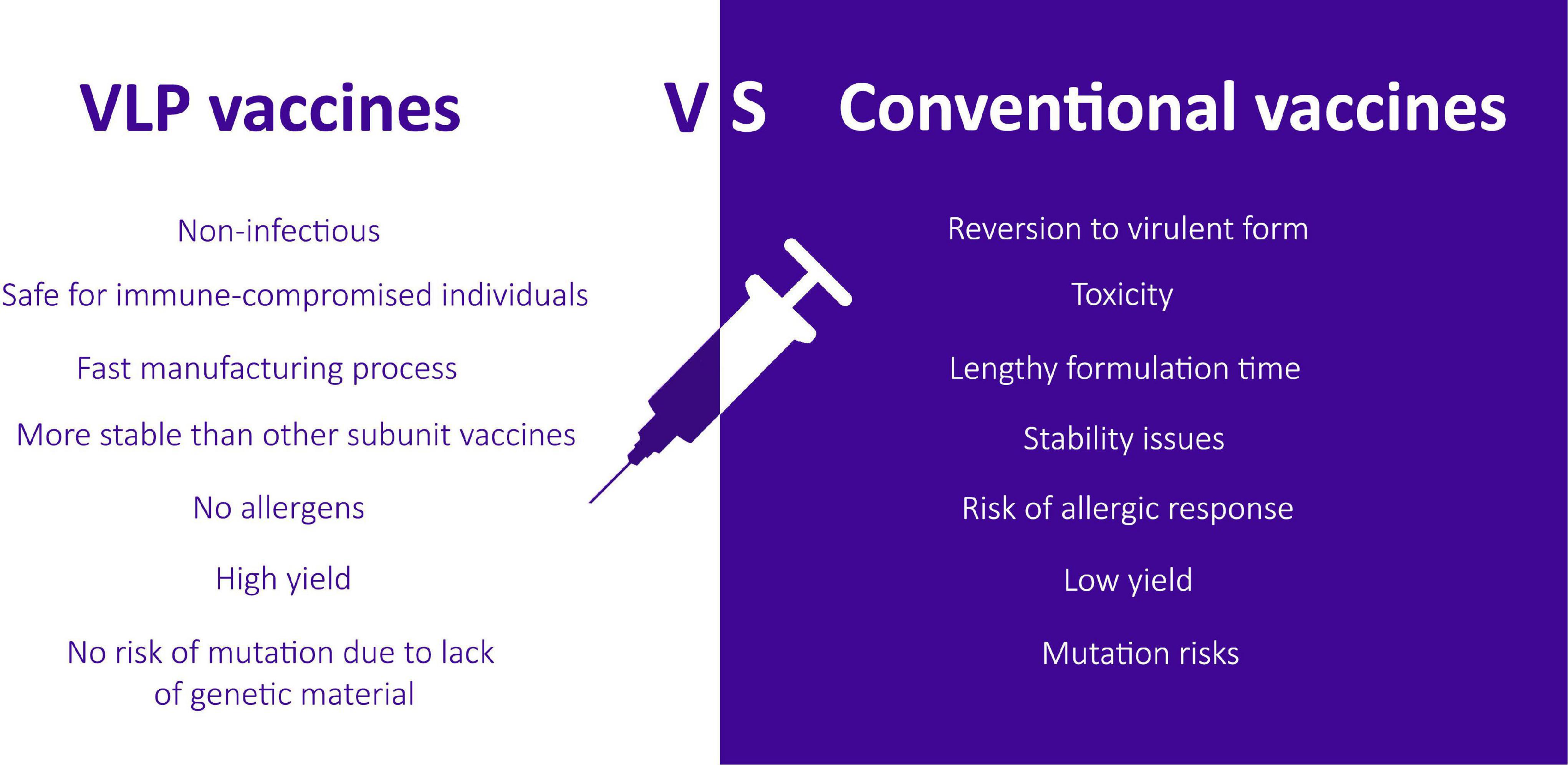
Figure 1. A comparison between VLP-based vaccines and the risks associated with conventional vaccines.
These bioinspired nanostructures have repetitive and highly dense antigens from different virulent agents that aids in triggering a strong immune response. Moreover, these highly immunogenic molecules have the self-assembling property of viral proteins (Urakami et al., 2017; Pechsrichuang et al., 2021). They are biocompatible and have the potential of structural flexibility during their synthesis (Chung et al., 2020). They can be modified either chemically or genetically and have higher stability, uniformity, and functionality, which are considered an effective tool in various biomedical applications (Qian et al., 2020; Figure 2). They are categorized as enveloped or non-enveloped VLPs based on the presence or absence of a lipid membrane (Jeong and Seong, 2017).
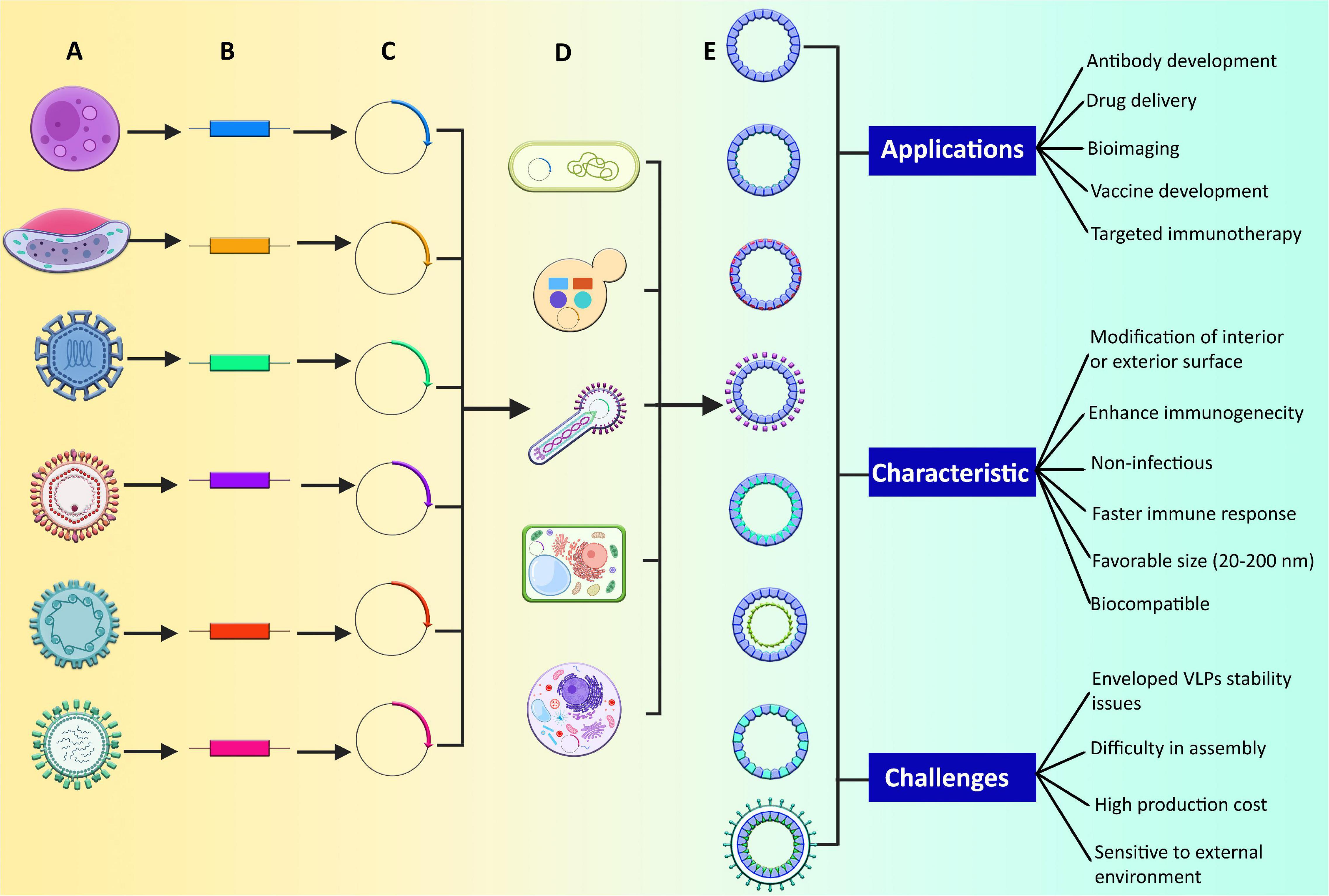
Figure 2. Production of different types of VLPs and their applications, characteristics, and challenges. (A) Different human pathogenic viruses and parasites, (B) identification of genes that form the structural features of pathogens and can result in the formation of VLPs, (C) incorporation of identified genes in expression vectors such as plasmids, (D) vectors are allowed to express in various expression systems, (E) formation of different VLP types, such as enveloped, non-enveloped, and chimeric VLPs. The non-enveloped VLPs can be of two types: single protein or multiprotein. In multiprotein VLPs, there may be a single layer, multiple layers, and some are mosaic as well. The chimeric VLPs can be modified internally, externally, or can be modified by chemical conjugation.
However, some of the key challenges associated with VLPs are lower stability, difficult downstream processing, high production costs, and sensitivity to environmental conditions (Bryant et al., 2007; Vicente et al., 2011). Many diverse VLPs have been synthesized in various expression systems (ESs), such as bacteria, yeasts, mammalian cells, insect cells, and plants (Nooraei et al., 2021). The VLP-based vaccines can potentially be used for the treatment of various infectious diseases, including HIV, influenza, hepatitis B (Spice et al., 2020), hepatitis E, malaria (Qian et al., 2020), Ebola virus (Tripathy et al., 2021), SARS-CoV-2 (Yang et al., 2021), Zika virus, Dengue, and foot and mouth disease, among others (Balke and Zeltins, 2020). Several vaccines based on VLPs have already been licensed and are commercially available in markets including Engerix-B® (GlaxoSmithKline) and Recombivax HB® (Merck & Co) against HBV, Gardasil® (Merck & Co) and Cervarix® (GlaxoSmithKline) against HPV, Hecolin® (Xiamen Innovax Biotech Co.) against HEV (Dai et al., 2018), and Mosquirix™ (GlaxoSmithKline Inc.) against malaria (Mohsen et al., 2017). This review focuses on the basic and advanced technical aspects of the VLP vaccine development. Furthermore, the use of different ESs for VLP production and the development of potential vaccines against various infectious diseases, most notably SARS-CoV-2, have also been discussed. Finally, protein nanoparticles as scaffolds for bearing antigens in the development of vaccines have been elaborated.
Characteristics of Virus-Like Particles
VLPs, in general, are potential candidates as efficient vaccines because of their distinct characteristics (Figure 2). They are potent immune-stimulatory molecules displaying a highly dense viral surface proteins in an appropriate conformation and a highly repeated way, eliciting strong T and B cell acquired immune responses. Mostly VLPs are derived from viral coat or envelope proteins, although core proteins can also be used (Sarkar et al., 2019). VLPs are naturally biocompatible and not contagious because they lack viral genetic material and hence cannot replicate. They are also thought to be safer (cannot revert to wild type) than conventional live attenuated vaccines (Guo et al., 2019). Moreover, they are highly versatile molecules that varies in their size, with most ranging from 20 to 200 nm. The size range is optimal to drain them freely into lymphatic nodes and for easier uptake by antigen-presenting cells (APCs), particularly dendritic cells (DCs), followed by antigen processing and presentation by major histocompatibility complex (MHC) class II molecules (Syomin and Ilyin, 2019). They are highly organized and can be self-assembled into different geometric symmetry, generally in the form of icosahedral, helical symmetry, rod shape structure, or globular in shape, depending on the virus’s source (Comas-Garcia et al., 2020).
VLPs have been synthesized in a wide range of ESs, including prokaryotic (bacteria) and eukaryotic (insect cells, mammalian cell lines, plant cells, or yeast). The functionality of VLPs can be increased through modifying their exterior or interior surface by displaying the heterologous epitopes of interest using different methods like peptide conjugation, genetic fusion, and chemical crosslinking (Mohsen et al., 2017). The VLP technology offers a significant advantage since it is a faster method of synthesizing vaccines. A new VLP vaccine against a specific strain can be prepared within 12–14 weeks after the strain is sequenced, whereas conventional vaccines usually require 24–32 weeks for the manufacturing processes. These vaccines are free of egg protein, which will give huge relief to individuals who are prone to allergies, as well as stronger protection against diseases than conventional vaccines (López-Macías, 2012).
Types of Virus-Like Particles Based on Structure
Based on their structural complexity VLPs can be classified into two groups: enveloped and non-enveloped VLPs. Both groups display foreign antigens (Figure 2).
Non-enveloped Virus-Like Particles
These VLPs are often made up of single or many self-assembled components of the targeted pathogen or viral protein structures. There is no host cell membrane (lipid envelope) in these newly formed VLPs. The expression of the major viral nucleocapsid proteins is primarily responsible for the formation of these VLPs. Non-enveloped VLPs are still being investigated as preferable candidates for developing subunit vaccines against several pathogenic diseases as they are easier to produce and purify (Naskalska and Pyrć, 2015). Furthermore, these VLPs are smaller in size that allows them to easily cross the tissue barrier and drain to lymph nodes (Keikha et al., 2021). The major structural component of the virion is formed by the single, virally encoded protein and is derived from pathogens, such as caliciviruses, papillomaviruses, and parvoviruses (Mohsen et al., 2017), whereas multi-protein non-enveloped VLPs are much more complex containing multiple interacting capsid proteins. They show several striking structural characteristics, such as many complicated concentric layers of different capsid proteins. They are more difficult to make than those that are made up of only one main capsid protein (CP) and derived from infectious bursal disease virus, poliovirus, and retroviruses (Lua et al., 2014).
Enveloped Virus-Like Particles
In comparison to non-enveloped VLPs, enveloped VLPs (eVLPs) have a far more complicated composition. These nano-structures consist of a cell membrane acquired from the host cell called an envelope, with viral proteins present on the outer surface (Dai et al., 2018). One or more than one glycoprotein spikes are embedded in their lipid bilayers and act as a target antigen for producing neutralizing antibodies. These eVLPs show a higher degree of flexibility as they target antigenic epitopes from the same or heterologous viruses. For example, the eVLPs have been developed that contain Gag protein from SIV and Env protein from HIV (Kushnir et al., 2012). Although it may affect downstream applications because of the presence of the host protein. The eVLPs have been used to develop vaccines against viral diseases, such as the hantaan virus, hepatitis C virus (HCV), influenza A, and retroviruses (Wetzel et al., 2019a). These large eVLPs have a size greater than 100 nm, so there is a chance that they might aggregate at the site of injection and will not reach the lymph nodes that limit their application (Keikha et al., 2021). In comparison, lipid nanoparticles carrying mRNA vaccines have a size range of 100 nm or less and show high efficacy in delivering vaccines to the targeted site (Editorial., 2021; Żak and Zangi, 2021).
Chimeric Virus-Like Particles
Chimeric virus-like particles (cVLPs) are considered an effective tool for developing vaccines that provide broader, more powerful, and comprehensive protection against emerging infectious diseases (Wetzel et al., 2019a). The complex, multi-protein macrostructures contain epitopes of different viruses (Latham and Galarza, 2001). cVLPs can be created by constructing recombinant DNA molecules that encode both the relevant viral protein and a foreign peptide or protein. These genetically engineered cVLPs display a high number of repetitive sequences on their surface that can be loaded with exogenous antigens from other viruses via chemical conjugation or genetic fusion (Caldeira et al., 2020). Different chimeric vaccines have undergone clinical trials, including the VLP-based vaccine (M2–HBcAg) against hepatitis, an antimalarial vaccine (MalariVax) (Nooraei et al., 2021), anti-influenza A, anti-HIV (Rutgers et al., 1988), and the nicotine-Qb VLP vaccine to reduced nicotine level in the blood of smokers (Maurer et al., 2005). These chimeric particles are advantageous as they substantially increase immune response and antibody titer in response to foreign antigens. Following administration of cVLPs, they induce strong cytolytic T lymphocyte immune responses (Qian et al., 2020). These vaccines are also targeted against non-infectious diseases, such as hypertension, Alzheimer’s, nicotine addiction, allergies, and diabetes (Huang et al., 2017). The upstream and downstream processing yield of cVLPs is usually low, and their in vivo stability is also quite uncertain (Buonaguro and Buonaguro, 2014).
Virus-Like Particles as Immunogens
It has already been demonstrated that VLPs potentially confer high immunogenicity and antigenicity than subunit vaccines. The potency of these particles has the potential to significantly induce cellular and humoral immunity (Mohsen et al., 2017; Figure 3). In response to the VLPs, various maturation markers like CD40, CD80, and CD86, are expressed on the surface of the DCs, which are responsible for the activation of DCs (Quan et al., 2016). In the first step, DCs are activated by binding VLPs to the specific pattern present on the DCs surface called pattern recognition receptors (PRRs) i.e., Toll-like receptors (TLR2) (Sartorius et al., 2021). Following this, internalization of VLPs takes place in the cytosol of DCs and are presented to cytotoxic T cells and helper T cells by MHC class I and class II molecules, respectively (Zepeda-Cervantes et al., 2020; Keikha et al., 2021). VLPs can stimulate not only B cells to mediate antibody response, but they can also stimulate CD4+ and CD8+ cells proliferation (Roy and Noad, 2008). Some studies show that exogenous antigen can also reach to MHC class 1 pathway through a process called cross penetration (Storni and Bachmann, 2004).
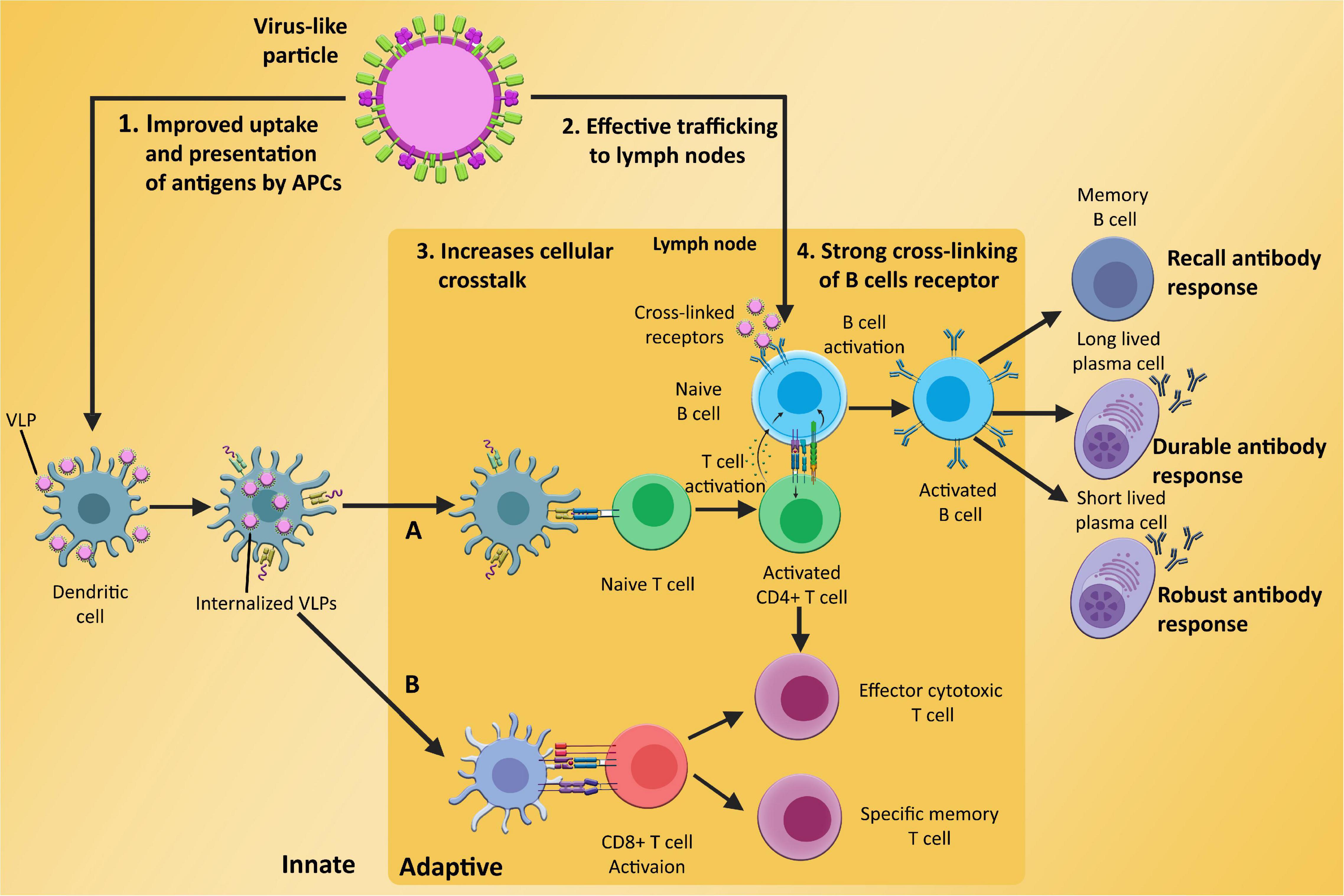
Figure 3. Induction of innate and adaptive immunological responses (A) humoral immunity; (B) cell-mediated immunity) by VLPs, (1) enhanced absorption and presentation of antigens based on VLP by APCs such as dendritic cells, which inform T cells about potential risks, (2) efficient VLP trafficking to lymph nodes, a crucial site for adaptive immunological responses, (3) improved cellular communication between B cells, T cells, and APCs, and (4) the ability of VLP-based antigen to effectively cross-link and activate B cells receptors, which develop into memory cells and long and short lived plasma cells after antigen exposure.
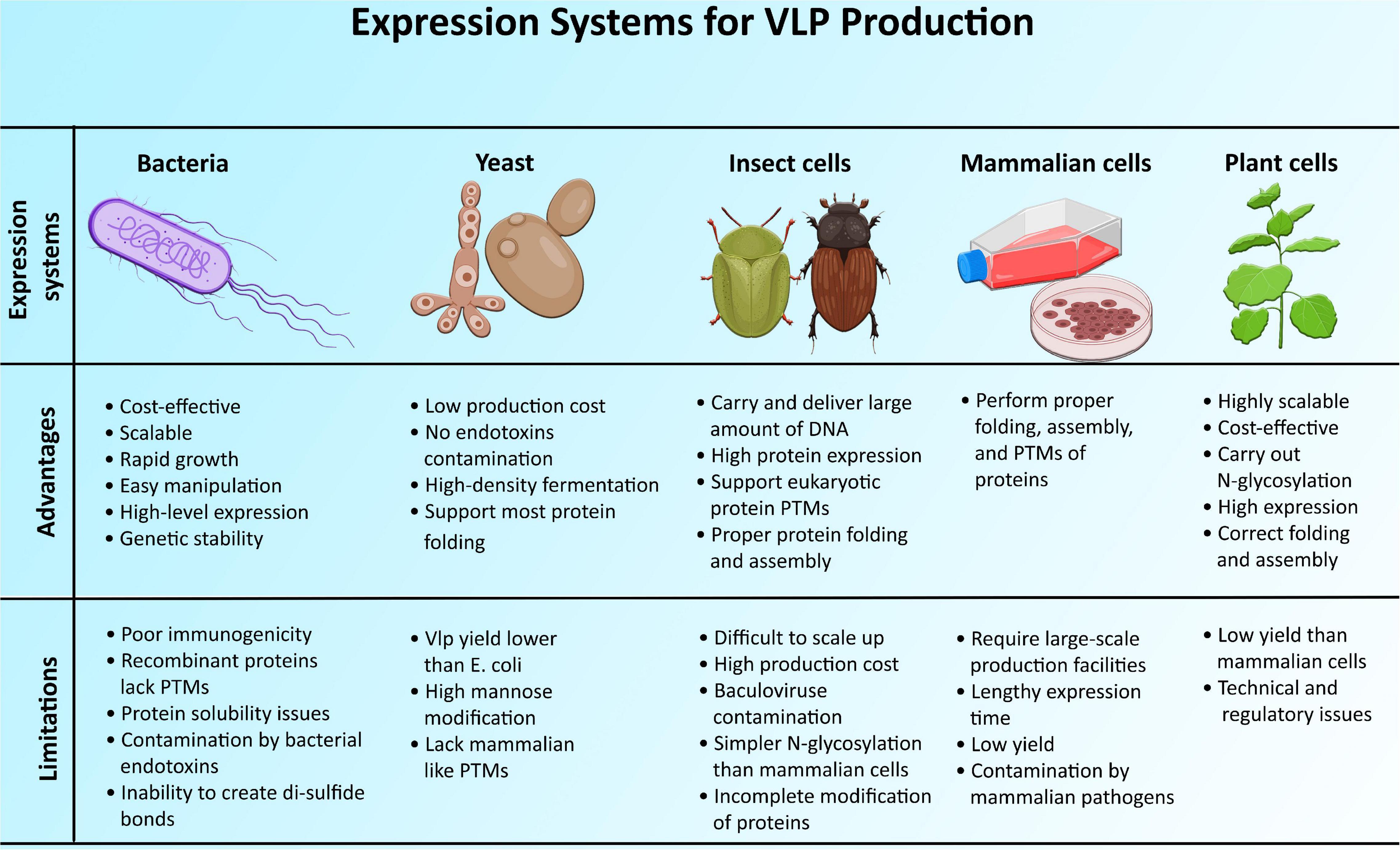
Figure 4. Advantages and limitations of different expression systems for the development of virus-like particles.
Moreover, activation of B cells can induce Th cell expansion and differentiation via toll-like receptor (TLR) signals or homologous interactions, which control the production of cytokines. In order to enhance the efficacy of VLPs, different molecules like Toll-like receptor ligands, biologically active mediators, or other cell receptors can also be attached to VLPs (Keikha et al., 2021).
Commercially Approved Vaccines Based on Virus-Like Particles
The first recombinant VLPs were synthesized from viral coat protein, two genes from the hepatitis B virus (HBV) (HBsAg and HBcAg), and the tobacco mosaic virus (TMV) (Zeltins, 2013). The first commercial VLP-based vaccine produced by recombinant DNA method was approved by the US Food and Drug Administration (FDA) in 1986s. These are the yeast produced HBsAg vaccines that were named as Recombivax HB® (Lagoutte et al., 2016). Later, in 2006, the second VLP based vaccine Gardasil® against human papillomavirus (HPV) vaccine was licensed by the FDA (Koutsky, 2009). Following this, several VLP vaccines against HPV and HBV have been approved, with some demonstrating efficacy in clinical and preclinical trials. The analysis of studies showed that at least 110 VLPs have been produced from viruses of 35 distinct families (Qian et al., 2020). Several VLP based vaccines targeting different viruses including Norwalk Virus, HIV, Ebola Virus, SARS-CoV-2 Virus, Respiratory Syncytial Virus (RSV), Influenza Virus are still under different clinical trial stages (Nooraei et al., 2021). However, just a few VLP vaccines have made it to the market, showing their commercial feasibility, and majority of them are effective against non-enveloped viruses (Guo et al., 2019).
Challenges Specific to Virus-Like Particle-Based Vaccine Platforms
At Present, VLPs are as effective as conventional vaccines, with the additional benefit of being safer. Nevertheless, several obstacles in the successful development of VLP-based vaccines need to be addressed. The main challenge is to identify issues related to downstream processing in the production of clinically viable VLPs (Charlton Hume et al., 2019) for their timely management and economic feasibility.
Stability of Enveloped Virus-Like Particles
VLPs are generally considered more stable than subunit vaccines (Dai et al., 2018). However, when the environmental conditions change, especially during downstream processing, VLPs can become very unstable as they lack the genetic material of the virus. Despite the fact that multiple VLP vaccines are already available in the marketplace, some of the candidates’ vaccines have stability issues (Mohsen et al., 2018). Generally, eVLPs are often more susceptible to external environmental conditions as compared to non-enveloped VLPs (Dai et al., 2018). Variations in conditions such as a change in temperature, shear stress, dissolved oxygen, fluid dynamics, agitation rate, and chemical treatment may all have an impact on the particle’s integrity and stability (Roldao et al., 2011). Moreover, this structural breakdown significantly reduces the immunogenicity of eVLPs. It also interferes with cell growth and the production of metabolic proteins, which has an impact the VLP production. It has been one of the key obstacles to using them as an alternative for a live virus in vaccine manufacture. However, several modifications have been made to increase their thermostability. One typical procedure is the insertion of stabilizing mutations (Dai et al., 2018). A study conducted by Marsian et al. (2017) showed that when stabilizing mutation was induced within the coat proteins of poliovirus type 3 VLP, it become more stable than wild type VLPs. It modified capsid precursor and the viral protease without disturbing its antigenic epitopes and structural confirmation. In another study stability of chimeric (HBcAg-VLP) was increased by the addition of C-terminal linker-hexahistidine-peptide (Schumacher et al., 2018).
High Production Cost
Some VLP-based vaccines are much more complex and, as a result, have a higher cost of production. The numerous impurities associated with eVLPs pose a daunting challenge. During downstream processing (DSP) various impurities like host cell debris (HCD), host cell protein (HCP), and host cell DNA (HCD) are co-purified as in the case of enveloped baculovirus particles. These contaminations can cause undesirable side effects in vaccines if not removed properly during DSP (van Oers, 2011). Large scale production and purification of VLPs require different processes like density gradients or even chromatography to make the final formulated product. These complex processes are very costly and time-consuming (Wetzel et al., 2019a). This also leads to difficulty in industrial scale production and requires several quality control efforts as various downstream processing steps may worsen the VLPs quality (Diamos et al., 2019). A more robust and better analytical methods are required to ensure the quality and quantity of the product that can facilitate their clinical and pharmaceutical utilization (Moleirinho et al., 2020). Different strategies such as clone screening, high-throughput screening, bioreactor engineering, material/matrix screening, filtration, flow-through or size-exclusion chromatography, and polishing have been implemented during upstream and downstream processing for scalable and cost-effective industrial manufacturing of VLPs (Vicente et al., 2011; Lagoutte et al., 2016).
Difficulty in Assembly
The genetic fusion of the sequences of epitope into VLPs can sometimes be challenging, as VLPs may lose their self-assembly property or cause particle misfolding (Guo et al., 2019). Genetically fusion of antigen to capsid protein of virus often hinders either antigen assembly or VLP, making the technique laborious. Therefore, time-intensive planning and optimization are required for individually testing every single antigen (Lampinen et al., 2021).
Expression Host Systems for Virus-Like Particle Production
Different expression systems (ESs) were utilized to make VLP vaccines, including plant, mammalian, insect, yeast, and bacteria (Figure 4).
Bacteria
Most of the bacterial systems are focused on well-studied industrial strains and expression vectors of Escherichia coli (Zeltins, 2013). Bacterial cell cultures were investigated as a platform for VLP development, with benefits in terms of cost and scalability (Kim et al., 2014). The other beneficial features of using bacterial systems for VLP production include (a) easy manipulation, (b) high-level expression, (c) fast growth rate, (d) genetic stability, and (e) simplicity of expression (Masavuli et al., 2017).
Huo et al. (2018) used pCold expression vector to express and purify norovirus (NoV) VLPs in E. coli strain (BL21) and demonstrated the similar binding pattern for VLPs assembled in E. coli as for Sf9 cells assembled VLPs. Similarly, in a recent study, the full-length CP of type 2 porcine circovirus and VP2 protein of porcine parvovirus were expressed in E. coli, which were self-assembled into VLPs. The study suggested that the expression of the CP and VP2 in E. coli is possible for the mass development of VLP vaccines (Liu et al., 2020). Yazdani et al. (2019) investigated the possibility of utilizing VLPs of grapevine fanleaf virus (GFLV) as a potential vector for presenting the L2 epitope of HPV. The antigenic determinant sequence was incorporated genetically into the GFLV capsid protein’s “αB-αB” domain C, which was then overexpressed in E. coli and Pichia pastoris. In E. coli, the highest expression yield was observed. For Hepatitis E virus (HEV), the ORF2 protein region 368–606 aa was purified in vitro from the insoluble E. coli fraction that is assembled into VLPs. This HEV VLP promises 100% effectiveness in clinical trials against symptomatic HEV and is approved as a vaccine for commercial use in China (Gupta et al., 2020). In another study, infectious hypodermal and hematopoietic necrosis virus (IHHNV) VLPs from CP of recombinant IHHNV were reconstructed in E. coli and showed excellent physical stability (Kiatmetha et al., 2018).
Bacterial systems are not always the optimal VLP development strategy because of several factors, including (a) poor immunogenicity, (b) inability to develop recombinant proteins with mammalian−like post-translational modifications (PTMs), (c) issues of protein solubility, (d) inability to create the correct disulfide bonds, and (e) presence of bacterial endotoxins/or lipopolysaccharides in preparation of recombinant proteins (Shirbaghaee and Bolhassani, 2016; Masavuli et al., 2017).
Brito and Singh (2011) have highlighted the acceptable level of endotoxins for various types of vaccines. There are various methods by which endotoxins are removed during the purification step, such as immobilized sepharose, surfactants, activated carbon, ultrafiltration, and anion exchange chromatography. However, the use of these techniques often leads to a considerable reduction in yield, an increase in cost, or a lack in the biological activity of the target protein. Therefore, ClearColi™, a genetically engineered E. coli strain with a genetically modified LPS which does not elicit an endotoxic reaction in humans, has been developed (Mamat et al., 2013).
Yeast
Eukaryotic ESs are a compelling alternative to prokaryotic ones, particularly when it comes to addressing the issue of bacterial endotoxins in vaccine production (Shirbaghaee and Bolhassani, 2016). VLPs can be produced more cost-effectively by recombinant expression of proteins in yeast cells. Manipulation of genes in yeasts is comparatively straightforward, and the transformed cells can grow to extremely high densities until the expression of recombinant proteins is induced. This allows commercial-scale fermenters to produce VLPs in large volumes (Stephen et al., 2018). Some mammalian viruses’ structural genes expressed in yeast can form the VLPs (Shirbaghaee and Bolhassani, 2016).
The FDA has approved some yeast-derived VLP vaccines, including Gardasil® and Gardasil9® against HPV and Mosquirix™ against P. falciparum (Nooraei et al., 2021). Some studies have investigated VLP production using yeast as an ES. Wetzel et al. (2018) established a robust and cost-effective yeast model for the production of cVLPs. The duck HBV membrane integral small surface protein (dS) was selected as the scaffold for VLP, and the safe and industrially applied Hansenula polymorpha yeast as the heterologous expression host. 8 distinct antigens of high molecular weight were derived from four viruses that infect animals and are genetically linked to a protein dS, and then the recombinant isolates were identified and purified. The fusion proteins were highly expressed in all cases, and it was possible to generate chimeric VLP comprising both proteins after co-production with protein dS. The production system based on yeast allows for a low-cost product that is not restricted to small-scale basic research. In another study, Alireza et al. (2018) produced chimeric protein L1/L2 VLPs in the P. Pastoris system by first inserting a cross-neutralizing epitope from the gene HPV-16 L2 into the gene L1 HPV-16. Following that, the chimeric L1/L2 HPV-16 had been introduced into the plasmid (pPICZA) and expressed in P. pastoris. The ELISA results for L1-HPV-16 Ab as well as L2-HPV-16 Ab detection indicated a positive reaction with sensitivity comparable to the commercial testing kit. Similarly, in the P. pastoris ES, Gupta et al. (2020) produced a recombinant VLP against HEV that included 112–608 aa region of the protein ORF2. The results showed that for the development of 112–608 aa VLP, the P. pastoris ES seems to be a superior and safer alternative to the baculovirus (Bv) ES.
During viral infection, enteroviruses like poliovirus, generate empty capsids that are antigenically indistinguishable from that of mature virions. The recombinant synthesis of such capsids with the help of heterologous systems like yeast has enormous potential as candidates for the VLP vaccine. Sherry et al. (2020) showed VLP production in P. pastoris through co-expression of the viral protease 3CD and the structural precursor protein P1.
Construction of yeast ESs is much more challenging than bacterial ESs, especially the Pichia and Hansenula strains. Furthermore, the VLP yield is lower than that of E. coli. Another disadvantage of the yeast ES is its lack of resemblance to mammalian ESs in protein PTMs, particularly glycosylation (Shirbaghaee and Bolhassani, 2016). Their glycoforms are mostly of the high type of mannose, which is undesirable for most pharmaceutical glycoproteins (GPs) (Kim et al., 2014).
Insect Cells
Baculovirus-based protein expression in insect cell lines has appeared as an effective tool for developing complicated protein-based biologics for a variety of purposes, extending from multiprotein complexes to the development of proteins for therapeutic use, such as VLPs (Sari-Ak et al., 2019). Bv is unusual among widely used viral vectors in its ability to tolerate heterologous DNA in large amounts and deliver it faithfully to the desired host cell (Gupta et al., 2019). Gopal and Schneemann (2018) described detailed procedures for producing recombinant baculoviruses (rBVs), screening for VLP expression in insect cell lines, and purifying VLPs.
The Bv-insect cell system is a two-step procedure in which insect cells are originally grown to the required cell concentration before being infected with rBVs for protein expression (Roldão et al., 2010). Recombinant protein and VLP production using Bv expression vector systems (EVSs) is fast, versatile, and provide substantial yields (Strobl et al., 2020). This system can produce exceptionally high protein production levels, combined with complex eukaryotic protein PTMs, which may be crucial for the proper self-assembly and release of some VLPs (Chaves et al., 2018).
MultiBac is an innovative Bv EVS that comprises of a viral genome that has been engineered to suit specific purposes. Recently, a team of scientists described the development of a MultiBac-based VLP-factory, dependent on the CP (M1) of influenza virus and its utilization in generating an arsenal of influenza-derived VLPs with functional modifications in influenza virus hemagglutinin (HA), which are expected to regulate the VLP-derived immunological response (Sari-Ak et al., 2019).
The insect cell-Bv EVSs have proven to be as effective as conventional egg- and cell-based approaches in influenza virus vaccine production with additional features like high production yields, and short production times. Moreover, rBVs construction has become faster, easier, and more flexible, allowing for the fusion of genes from different types and/or subtypes of influenza viruses inside the same expression vector. Sequeira et al. (2018) successfully created a robust High Five cell-based insect platform that combines stable expression with Bv-mediated expression to generate multivalent influenza VLPs. The capability of this modular approach has been proven by infecting the High-Five cell line with 2 distinct HA proteins of subtype H3 (called HA2 and HA1) with a Bv expressing M1 and 3 additional HA proteins of subtype H3 (called HA5, HA4, and HA3), to generate pentavalent VLPs (H3).
The Zaire Ebola virus serotype (ZEBOV) is the most virulent and has the highest mortality rates among other serotypes. ZEBOV-VLPs development have been achieved in insect and mammalian cell lines via co-expression of 3 viral structural proteins, the nucleocapsid protein (NP), the matrix structural protein (VP40), and the glycoprotein (GP). A technique for generating ZEBOV-VLPs in insect cell line was reported by Pastor et al. (2019), which basically consists of employing a high multiplicity of infection (MOI) of bac-GP and bac-VP40, and limiting the NP expression, either via preventing infection or by lowering the bac-NP MOI, was the most suitable for developing VLP.
The main limitations of the insect cell ES are (a) protein contamination by enveloped baculoviruses, (b) difficult to scale-up, and (c) simpler N-glycosylation than mammalian cells (Shirbaghaee and Bolhassani, 2016; Masavuli et al., 2017).
Mammalian Cells
For more than two decades, various mammalian cell lines such as murine myeloma (Sp2/0, NS0), Chinese hamster ovary (CHO), murine C127, baby hamster kidney (BHK21), HT-1080, and HEK293 were established as a possible source of commercialized protein therapeutics for medical purposes, due to their capability to properly fold, assemble, and post-translational modification to proteins (Dumont et al., 2016; Shirbaghaee and Bolhassani, 2016). Systems based on mammalian cell culture offer many benefits, including consistency and flexibility during the development process. It also helps glycosylated proteins to be recovered with compositions of lipid membrane similar to the virus’s host. It has been reported that the stable transfection of viral genes into mammalian cell lines, including 293 or Vero cells, results in VLP production (Buffin et al., 2019).
Several studies reported the efficient production of VLPs from mammalian cell cultures. Hsin et al. (2018) developed a MERS VLP system using Huh7 cells as an expression system for understanding virus infection and morphogenesis. In another study, influenza VLPs encoding neuraminidase (NA), hemagglutinin (HA), and matrix M1 proteins had been expressed in Vero, 293 T, or CHO-K1 cell lines, using transient transfection. Preclinical studies in BALB/c mice revealed that influenza VLPs, when given intramuscularly, were significantly immunogenic at low dosages, inducing functional Abs against NA and HA (Buffin et al., 2019). Wu et al. (2010) designed a scalable method for the effective production of various subtypes of influenza VLPs expressed in mammalian cell line. The study demonstrated that these mammalian influenza VLPs were very similar to the original viruses in particle size, structure, host factor composition, and viral antigen glycosylation. Similarly, in another work, an inducible cell line of human embryonic kidney HEK-293 expressing NA and HA was developed and utilized to generate VLPs following transient transfection with a plasmid encoding HIV-1 Gag (Venereo-Sanchez et al., 2017). A protocol developed for the synthesis of HIV-1 Gag VLP in mammalian cell suspension cultures via transient gene expression showed that the large proportion of Gag-GFPs present in the supernatants of cell culture was fully assembled into VLPs of the predicted morphology and size consistent with immature particles of HIV-1 (Cervera et al., 2013).
Expression systems based on mammalian cells require large-scale production facilities, like fermentation bioreactors, which are prohibitively expensive to construct. As a result, the high cost of production is a challenging part of the cell-based mammalian ES (Kim et al., 2014). The other limitations include lengthy-expression time, low yield, and vulnerability to infections with mammalian pathogens (Masavuli et al., 2017).
Plants
“Molecular farming” is a term used to describe the utilization of plants or plant cells to produce recombinant proteins or other biologic drugs for use as cosmeceuticals, biopharmaceuticals, therapeutics, vaccines, and other reagents (Rybicki, 2020). Plants provide an enticing alternative system for the manufacture of VLP vaccines due to their potential to generate significant amounts of recombinant proteins at a minimal cost, their eukaryotic production machinery for the PTM and correct protein folding and assembly, and their low risk to introduce adventitious human pathogens. Plants do not demand the installation of costly fermentation facilities for the production of biomass, nor do they necessitate the establishment of duplicate facilities for scale-up production. Unlike microbial fermentation, plants have the ability to carry out N-glycosylation as a glycoprotein PTM (Chen and Lai, 2013). Engineered VLP-forming human or animal virus capsid proteins expressed in plant cells include human norovirus CP, HPV L1 protein, hepatitis B core and surface antigens, HIV Gag polyprotein, and HA protein of influenza virus (Rybicki, 2020).
Several studies have extensively discussed the concepts for constructing VLPs in various plant ESs, efficient growth of VLPs in plant hosts, and the self-assembly of multiple structural proteins of viruses in plants. Santi et al. (2008) produced properly assembled recombinant Norwalk virus VLPs in leaves of Nicotiana benthamiana utilizing a TMV-derived transient ES. Young et al. (2015) generated trackable hemagglutinin based VLPs that allowed them to examine particle assembly in plants and the interaction of VLPs inside the mammalian immunological system. van Zyl et al. (2016) investigated the production of bluetongue virus VLPs in N. benthamiana through Agrobacterium-mediated transient expression, which is an inexpensive system. Similarly, Veerapen et al. (2018) transiently expressed VLPs of the foot-and-mouth disease virus in N. benthamiana. Diamos and Mason (2018) produced norovirus VLPs in a plant-based system using modified geminiviral vectors. Recently, Rosales-Mendoza et al. (2020) presented a perspective in developing VLP vaccines based on plants against SARS-CoV-2, which is responsible for the COVID-19 pandemic.
Plants for the production of the VLP platform are not entirely acceptable due to comparatively lower production levels of VLP than mammalian ESs and plant-specific N-glycosylation of glycoproteins (Kim et al., 2014). Nonetheless, the recent creation of novel plant ESs, as well as advancements in plant glycoengineering, have both resolved these challenges (Chen and Lai, 2013). Recent advancements in plant glycoengineering permit human-like modification of glycol and optimization of desirable glycan structures to improve the functionality and safety of recombinant pharmaceutical glycoproteins (Kim et al., 2014).
Development of Virus-Like Particle Vaccines Against Emerging Infectious Diseases
Several VLP vaccines have been produced and are being used against different viral and parasitic infections in recent years (Table 1). The development of VLP vaccines against zika virus, chikungunya virus, influenza virus, and human papillomavirus have been discussed below.
Zika Virus
Zika virus (ZIKV) is a small-enveloped mosquito-borne neurotropic positive-strand RNA virus of the family Flaviviridae (Lin et al., 2018; Shanmugam et al., 2019). ZIKV infections were associated with acute prenatal abnormalities like microcephaly in neonates born to infected mothers, and also Guillain-Barré syndrome (GBS) in adult people (Alvim et al., 2019). Currently, no cure or vaccine is commercially available for effective therapy or treatment, so vaccine development against ZIKV is of great importance (Boigard et al., 2017).
ZIKV has an RNA genome with only one open reading frame, and a solitary polyprotein is formed. This protein-strand is cut into 7 non-structural proteins (NS5, NS4B, NS4A, NS3, NS2B, NS2A, and NS1) and 3 structural proteins (C, prM, and E) by cellular and viral proteases. E-protein is engaged in the binding of viral particles as well as its fusion (Boigard et al., 2017). E-protein has three domains structure, like enveloped-domain-I also known as ED-I, then ED-II, and ED-III. Humoral response mainly targets the fusion-loop belonging to ED-II. Antibodies against fusion-loop epitopes enhance the uptake of DENV by Fcγ-receptors (Shanmugam et al., 2019). So, this protein may serve as a possible target for vaccine development (Shanmugam et al., 2019).
Many VLPs for ZIKV have been developed (Boigard et al., 2017). Garg et al. (2019) compared different VLPs-vaccine for ZIKV in mice and showed that CprME-VLPs (Capsid-preMembrane-Envelope) gave better results than prME-VLPs (preMembrane-Envelope). Cell lines could not be generated using CprME- because co-expression of protease NS2B-3 is needed. In order to get rid of this barrier, a bicistronic vector was generated that uses IRES-sequence to produce both NS2B-3 and CprME-VLPs. Alvim et al. (2019) demonstrated the continuous expression of ZIKV-VLPs by HEK293-cells. In short, the cell lines constitutively producing Zika-VLPs are ideal for developing a vaccine.
Chikungunya Virus
It’s a severe and periodic infectious disease caused by CHIKV (chikungunya-virus) transmitted by a carrier mosquito. Symptoms are high fever, skin rash, etc. No vaccine is available at present, but many VLP-vaccine candidates are under development in different stages (Zhang et al., 2019). Protection from multiple strains of this virus can be conferred by self-assembled VLPs produced as a result of selective expression of CHIKV envelope and capsid proteins (Kramer et al., 2013).
Arévalo et al. (2019) reported 100% protection in adult mice against CHIKV infection when unadjuvanted CHIKV-VLPs were used. Similarly, Metz et al. (2013) compared three different vaccines in mice that were produced in insects by recombinant baculoviruses produced sE1, sE2 CHIKV, and CHIKV (VLPs). One-half of E1 and E2 immunized mice survived to show incomplete protection when compared with VLP-immunized mice.
Akahata and Nabel (2012) uncovered that variable CHIKV structural proteins expression results in VLPs, which mimic replication-competent alphaviruses. This vaccine caused the induction of neutralizing antibodies in large amounts against multiple strains of this virus and conferring complete protection. VLPs were easily produced from CHIKV 37997-strains compared to CHIKV-OPY-1 strain- VLPs despite high amino-acid-sequence similarity. Knowledge of mechanisms involved in CHIKV VLP production will help in other vaccine development also enhancing their range of application for other pathogens. Furthermore, VLP-production was affected by amino acid 234 of E2 in an acid-sensitive region. Mutations in the acid-sensitive region and pH changes also enhance the yield of VLPs.
Influenza Virus
Influenza virus (Iv) infections are the leading cause of chronic human respiratory symptoms, leading to severe public health outcomes about endemic and seasonal infections and even result in unpredictable pandemic outbreaks (Lai et al., 2019). Iv is an enveloped, segmented, negative-sense RNA virus, which belongs to the family Orthomyxoviridae. To date, four types of Ivs have been identified, classified according to the presence of their core proteins: A, B, C, and D. Three Iv types, A, B, and C are pathogenic to human cells and cause severe infections, with Iv type A and B being the most prevalent circulating types. The main surface glycoproteins of influenza viruses are neuraminidase (NA) and hemagglutinin (HA) (To and Torres, 2019).
HA, the key antigen involved in infection, attaches to the residues of sialic acid on the cell membrane surface, facilitating the Iv to enter the host cell. NA appears to be less frequent on the viral surface as compared to HA, with a generally observed NA:HA ratio of 1:4. Its enzymatic function is critical in the cleavage of sialic acid, thereby facilitating viral release from the infected host cell surface. NA activation also enables the successful penetration by influenza to mucus through a mechanism involving cleavage of sialic (Buffin et al., 2019). Kim et al. (2019) investigated the cross-protective efficiency and immunogenicity of VLP containing NA (N1 VLP) derived from the 2009 H1N1 influenza viral pandemic and compared it to inactivated split influenza vaccine. Mouse immunized with the N1 VLPs was able to induce virus-specific Ab responses as well as cross-reactive NA inhibition activity, while strain-specific hemagglutination inhibition test was induced by inactivated split vaccination. Mice vaccinated with N1 VLPs led to the development of cross-protective immunity to antigenically various Ivs, as measured by changes in bodyweight, pulmonary viral titers, infiltration of innate leukocytes, cytokines and Ab secretory cells, and germinal center B-cells. Furthermore, in naïve mice, the immune sera of N1 VLPs conferred cross-protection. Immunity induced by N1 VLPs was neither impaired nor diminished in mice lacking the Fc receptor γ-chain. These findings indicate that NA representing VLPs, along with the current vaccination of influenza, may be further improved and exploited as an important candidate for cross-protective vaccines.
Recently, Kirsteina et al. (2020) studied and examined the efficacy and immunogenicity of an array of widely protective prototypes of Iv vaccine focused on both influenza triple matrix protein 2 ion channel (3M2e) and tri-stalk antigens incorporated into phage AP205 VLPs. VLPs that contained the 3M2e antigen alone stimulated protection in mice toward both standard homologous as well as heterologous virus challenge. The combination of both conserved antigens of the influenza virus into an individual VLP resulted in complete protection against a high dose of homologous influenza H1N1 infection in mice.
Human Papillomaviruses
Human papillomaviruses (HPVs) are members of the Papillomaviridae family of tiny, circular, double-stranded DNA viruses that cause cervical cancer, the world’s second most fatal disease in women after breast cancer (Uddin et al., 2019; Burley et al., 2020). Currently, two VLP-based vaccines are commercially available for inhibiting warts and treating cervical cancers caused by HPV. These include Cervarix by GlaxoSmithKline (GSK) Pharmaceuticals and Gardasil by Merck Pharmaceuticals. Both vaccines consist of the immunogenic major CP (L1) VLPs of HPV 16 and 18, with Gardasil also having 6 and 11 VLPs (Uddin et al., 2019).
The drawback of using L1 protein as an antigen for the VLP vaccine is that it is not conserved among various HPV serotypes. Conversely, the minor CP (L2) is highly conserved across all HPV serotypes and has long been considered a major potential target antigen for developing an HPV vaccination with broad protection. In contrast to CP (L1), the CP (L2) cannot be used for VLP production and is, therefore, less immunogenic (Yadav et al., 2020). The reasons for the low immunogenicity of L2 proteins are due to their slight representation relative to L1, as well as the fact that L2 is mainly buried beneath the surface of the capsid, where it contacts the surface of the cell in vitro or the basement membrane in vivo and induces a conformational shift (Huber et al., 2017).
Several methods have been applied to improve and enhance the L2 peptides low immunogenicities, such as concatemeric fusion peptides consisting of L2 epitopes of various types of HPV, combined with immune activating TLR agonists or using repeated surface arrays of epitopes of L2 on particulate immunostimulatory platforms, such as VLP of TMV, bacteriophages MS2 or PP7, Adeno-associated virus, Lactobacillus casei, or bacterial thioredoxin (Huber et al., 2017).
Another promising strategy to improve L2 low immunogenicity is the presentation of L2 epitopes on the surface via VLPs assembled from HPV L1-L2 chimeric proteins. It was achieved through the genetic insertion of highly conserved B cell epitope RG1 of HPV16 L2 into the surface loop (DE) of the protein HPV16 L1 leading to its multivalent (360x) immunogenic display on the surface of VLP, whereas the conformational neutralization antigenic determinants of the HPV16 L1 scaffold remained mostly preserved. Immunizations with this chimeric RG1-VLP caused high type-specific HPV16 titers and extensive cross-neutralization of heterologous mucosal and distantly associated cutaneous HPVs (Schellenbacher et al., 2009, 2013).
Pouyanfard et al. (2018) described the thermostable thioredoxin vaccine development based on a single-peptide capable of carrying L2 polytopes from up to 11 various types of HPV. The antigens of the L2 polytope exhibit exceptional capabilities regarding the robustness and protection of the elicited immunological responses. To further boost and enhance immunogenicity, the polytope antigen L2 of thioredoxin was fused with a heptamerization domain. Protective responses to all 14 oncogenic types of HPV, as well as the lower risk HPV types (6 and 11), and a range of cutaneous HPVs were achieved in the final design of the vaccine.
Virus-Like Particles Against Severe Acute Respiratory Syndrome-Coronavirus-2
With worldwide causalities crossing 2 million people and more than 20 million individuals affected in more than 200 countries this past year, the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) has emerged as a nuisance across the globe. The COVID-19 global pandemic resulting from SARS-CoV-2 has impacted billions of individuals and emerged as the first major global catastrophe after the 2009 H1N1-pandemic (Alvi et al., 2020). Currently, some commercially available antiviral drugs including Remdesivir® (Gilead Sciences), Paxlovid® (Pfizer), Molnupiravir® (Merck and Co.) have been approved by the FDA and WHO for treatment of patients infected with SARS-CoV-2. Due to the limitations including development, cost, and distribution of targeted drug therapies against the virus, researchers are aiming their focus and efforts on developing vaccines for the long-term fight against COVID-19.
VLPs against SARS-CoV-2 can help combat the widespread pandemic. VLPs can act as therapeutic agents and viable vaccines against this viral disease, with the addition of serving as a feasible diagnostic tool (Fuenmayor et al., 2017). SARS-CoV-2 has a slow mechanism of action as after entering host it takes between 5 and 15 days for a person to display symptoms because the virus enters host cells via endosomal pathway and is not efficient in evading the immune system whereas, vaccines against other coronaviruses (CoVs) strains that cause SARS and Middle East Respiratory Syndrome (MERS) were shown to be effective and active in animal models against SARS-CoV-2 (Cohen, 2020). Like other CoVs, SARS-CoV-2 consists of four proteins that are structurally conserved among different viral serotypes: Spike (S), Nucleoprotein (N), Envelope (E), and Membrane (M) proteins. The precise contribution of the above proteins and their relevant interaction patterns are crucial for the production and assembly of VLPs. M, N, and E are vital for the production VLPs for SARS-CoV-2 (Boson et al., 2020; Figure 5). The S-protein is one such protein that is common among different coronavirus strains and may serve as a potential vaccine development target. VLPs for MERS-CoV have been produced by displaying the S-protein in the insect host cells ES (Kato et al., 2019; Samrat et al., 2020).
Figure 5. Proposed production system and mechanism of action of SARS-CoV2 virus-like particle vaccine. Plasmids encoding the structural proteins (S, N, M, and E) of the SARS-CoV2 can be transfected into an appropriate mammalian cell line. The assembled VLPs are then collected, purified, and administered to humans. The administration of VLPs stimulates both innate and adaptive immunological responses. If the original SARS-CoV2 enters the human body in the future, memory B cells activate and release antibodies against it. Similarly, the activated CD8+ T cells recognize and kill virus-infected cells.
Before developing SARS-CoV-2 VLP-based vaccines, previous work on VLPs against the preceding and closely related strains must first be considered. VLPs have previously been demonstrated to be effective in eliciting specific humoral and cellular immune defenses against SARS-CoV in mice by production in rBV ES (Lu et al., 2010). Ho et al. (2004) reported the synthesis of VLPs for SARS-CoV. These VLPS were assembled in the insect ES by the co-infection with rBV. VLPs prepared via the use of protein corona formation have shown effective vaccination and have led to the development of significant immune responses in the avian model of CoV infection (Chen et al., 2016). Similarly, plasmids containing 4 structural viral proteins (S, N, E, and M) of SARS-CoV were transferred into Vero E6 cells to produce VLPs. These VLPs may serve as an effective tool for studying the pathogenesis of SARS-CoV with the host cell (Hsieh et al., 2005). Recently, a COVID-19 mRNA vaccine expressing SARS-CoV-2 VLPs has shown the induction of a robust antiviral immunological response in the mouse model (Lu et al., 2020).
Currently, the NVX-CoV2372 is the only commercially available VLPs based vaccine against SARS-CoV-2. This vaccine is manufactured by the display of SARS-CoV-2 S-protein in rBV, which are then used for mass production by infecting moth cell expression system. This vaccine has shown various levels of effectiveness in different trials across separate countries (Heath et al., 2021; Shinde et al., 2021). Currently, five teams are also working separately on VLP based vaccines against COVID-19 (Bogani et al., 2020; Callaway, 2020), and clinical trials are underway with efforts being made in developing viable VLPs based vaccines. Biometric VLPs have been developed which can aid in accurate diagnosis of SARS-CoV-2 by acting as positive controls during RT-PCR procedure (Chan et al., 2020). Ghorbani et al. (2020) evaluated the immunogenic properties of various SARS-CoV-2 spike derived epitopes which had already been reported to induce a specific immunogenic response, by using immunoinformatic analysis. Finally, a set of screened epitopes were suggested for which a VLPs-based vaccines against SARS-CoV-2 could be synthesized in different plant species by using molecular farming approaches.
Different mammalian ESs were utilized for the synthesis of SARS-CoV-2 VLPs. Results indicate that SARS-CoV-2 VLPs produced from Vero E6 cell line are more durable and integrated than those derived from HEK-293T cells (Xu et al., 2020). Recently, a VLP vaccine based on plant ES against SARS-CoV-2 has been reported. Findings of clinical phase I trials of VLP vaccine (CoVLP) against SARS-CoV-2 have indicated significant development of IL-4 and IFN-γ immune responses in individuals. The CoVLP vaccine is manufactured by transient expression of S protein of SARS-CoV-2 in the plant ES (N. benthamiana). The trimeric S GPs are exhibited on the surface of self-assembling VLPs that imitate the size and shape of the SARS-CoV-2 (Ward et al., 2020, 2021). Similarly, a combination minispike VLP vaccine for SARS-CoV-2 has also demonstrated high level of immunization in mice after a single dosage. The vaccine elicited the development of neutralizing Abs and protected the K18-hACE2 mice from COVID-19 similar to the patients suffering from COVID-19 (Hennrich et al., 2021).
A capsid VLP centered SARS-CoV-2 vaccine (ABNCoV2) has shown efficient neutralization of SARS-CoV-2 in mice models. The ABNCoV2 vaccine is made by displaying the receptor-binding domain of the SARS-CoV-2 S-protein in insect (Drosophila) cells (Fougeoux et al., 2020). Researchers are also aiming toward developing a DNA-based vaccine that can be delivered through the nasal cavity to the targeted tissue, which will cause the production of SARS-CoV-2 VLPs resulting in a strong immune response in individuals (Samrat et al., 2020).
Protein Nanoparticles for Vaccine Development
Protein nanoparticles (NPs) can help in the development of vaccines for immuno-evasive pathogens such as HIV, influenza, malaria, and can aid in the fight against emerging virulent strains by either acting alone or acting as carriers for targeted drug delivery (Hong et al., 2020; Roth et al., 2021). The engineering of protein molecules for antigen representation with the aid of NPs to be used in vaccine development for generating immune response is an increasingly popular and rapid field in therapeutic and drug development (Jain et al., 2018). Protein-based VLPs act as natural NPs that can be extensively used in vaccine development due to ease in design, self-assembly, and high stability (Nguyen and Tolia, 2021). Three different methods are used to attach antigens for presentation on NPs, (1) tag coupling (van Oosten et al., 2021), (2) chemical conjugation (Lu et al., 2021), and (3) genetic fusion (Antanasijevic et al., 2020). These techniques enable the platform to be decorated with a variety of antigens, leading to increased size and presentation (Nguyen and Tolia, 2021). Most antigens often do not self-assemble into NPs, like those utilized in influenza subunit vaccinations. Self-assembly can be achieved in such circumstances by attaching these antigens to an oligomeric protein platform (Nguyen and Tolia, 2021). There are many naturally occurring oligomeric proteins such as lumazine synthase (LS) (Ladenstein and Morgunova, 2020), ferritin (Rodrigues et al., 2021), dihydrolipoyl acetyltransferase (E2p) (He et al., 2016), non-structural protein 10 (nsp10) (Carter et al., 2020), encapsulin (Lagoutte et al., 2018), and heat shock proteins (HSPs) (Richert et al., 2012) that have been developed for platform design for designing targeted therapeutics. The display of viral glycoproteins using NPs is effective in developing antigen-specific antibodies (Ueda et al., 2020). Antigen-displaying protein NPs are highly effective for generating an immune response to specific antigens and can be used separately or in combination with vaccines (López-Sagaseta et al., 2016).
Among the wide array of platforms available for the display of antigens, ferritin has emerged as a major protein complex that can be combined with NPs for vaccine development (Diaz-Arévalo and Zeng, 2020). Due to its high pH and thermal stability, it allows for easy annealability with surface molecules (Rodrigues et al., 2021). In recent years, ferritin has been studied in preclinical studies as a viable vaccine platform for numerous viral infectious diseases, including HIV, H1N1, HCV, HBV, HFMD, Epstein–Barr, rotavirus, and respiratory diseases caused by coronaviruses (Butkovich et al., 2021). Kelly et al. (2020) compared the immunogenicity between ferritin NPs displaying influenza HA and soluble protein (HA) in vivo. HA-ferritin NPs showed higher immunogenicity and greater protection against a viral challenge as compared to soluble protein (HA).
Recently, a structure-based design of self-assembling protein NP immunogen that produces protective and potent antibody responses against SARS-CoV-2 has demonstrated the development of innate immunity in in vivo studies in mice (Walls et al., 2020). Similarly, a ferritin NP-based SARS-CoV-2 vaccine induced an effective antibody response in mice that reportedly lasted for at least 7 months post-immunization contributing toward sufficient development of immunity (Wang et al., 2021).
Though protein-NPs can serve as viable candidates for vaccine development, in vivo applications of NPs are often restricted by several challenges due to their organic nature (Poon and Patel, 2020), including cellular toxicity, inflammatory responses, and insufficient delivery to the target site (Kianfar, 2021). The term “protein corona” formation has been coined to summarize this unfavorable interaction between protein and NPs in in vivo conditions. The surface protein attached to NPs affects their biological behavior and changes their functionality, which occasionally results in gain or loss-of-function (Karch et al., 2018). The protein corona formation is a complex process that involves complex dynamics and kinetics between two interacting entities (Kapadia et al., 2019). Some cases have been reported where structural changes have occurred after interaction with NP surface evidently altering the NPs native function. Thus, to overcome such challenges it is critical to understand protein conformational changes and the unfolding process to accelerate the biomedical applications of NPs (Park, 2020).
Conclusion
Over the past two decades, resistance in some pathogens against commercially available antimicrobial drugs has increased drastically, mainly due to unregulated over-the-counter sales, misuse and overuse of drugs, and genetic adaptations. The process of drug development is a rigorous task and requires ample time, funding, and repeated trials to commercialize a viable product that could aid in the fight against numerous emerging pathogens. Due to the time-consuming limitation, especially access of patients to appropriate drugs is hindered, whereas the resistance in pathogens is consistently increasing. Therefore, preventive measures are of significant importance to address the emergence of infectious diseases. In the efforts to combat such diseases, VLPs have come to light in the healthcare industry, with multiple applications ranging from vaccine development, drug delivery systems, and molecular diagnostics. VLP based approaches provide an alternative to the available conventional methods for vaccine developments. Currently, some VLP-based vaccines are commercially available against perilous pathogens like ZIKV, HCV, HBV, and HPV. Meanwhile, efforts are underway in the production and designing of efficient VLP vaccines against various emerging virulent pathogens, including SARS-CoV-2. However, there are still some major hurdles that need to be addressed before VLP-based therapeutics can compete with conventional drug therapies in terms of cost and effectiveness. Nonetheless, VLP-based medicinal strategies may be used extensively in the future along with pharmaceutics to aid in the fight against numerous infectious diseases.
Author Contributions
HT and SB conceptualized and drafted the manuscript. HT, SB, and SA wrote the manuscript. HT was responsible for designing and illustrating the figures in the manuscript. MA reviewed and edited the manuscript. BA supervised and proofread the manuscript. All authors have read and agreed to the submitted version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aida, V., Pliasas, V. C., Neasham, P. J., North, J. F., McWhorter, K. L., Glover, S. R., et al. (2021). Novel vaccine technologies in veterinary medicine: a herald to human medicine vaccines. Front. Vet. Sci. 8:340. doi: 10.3389/fvets.2021.654289
Akahata, W., and Nabel, G. J. (2012). A specific domain of the Chikungunya virus E2 protein regulates particle formation in human cells: implications for alphavirus vaccine design. J. Virol. 86, 8879–8883. doi: 10.1128/JVI.00370-12
Alireza, S., Khosrow, A., Bahman, A. K., Alireza, S. Z., Ataollah, G., and Manoocher, M. (2018). VLP production from recombinant L1/L2 HPV-16 protein expressed in pichia pastoris. Protein Peptide Lett. 25, 783–790. doi: 10.2174/0929866525666180809124633
Alvi, M. M., Sivasankaran, S., and Singh, M. (2020). Pharmacological and non-pharmacological efforts at prevention, mitigation, and treatment for COVID-19. J. Drug Target. 28, 742–754. doi: 10.1080/1061186X.2020.1793990
Alvim, R. G., Itabaiana, I. Jr., and Castilho, L. R. (2019). Zika virus-like particles (VLPs): stable cell lines and continuous perfusion processes as a new potential vaccine manufacturing platform. Vaccine 37, 6970–6977.
Antanasijevic, A., Ueda, G., Brouwer, P. J., Copps, J., Huang, D., Allen, J. D., et al. (2020). Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens. PLoS Pathog. 16:e1008665. doi: 10.1371/journal.ppat.1008665
Arévalo, M. T., Huang, Y., Jones, C. A., and Ross, T. M. (2019). Vaccination with a chikungunya virus-like particle vaccine exacerbates disease in aged mice. PLoS Negl. Trop. Dis. 13:e0007316. doi: 10.1371/journal.pntd.0007316
Balke, I., and Zeltins, A. (2020). Recent advances in the use of plant virus-like particles as vaccines. Viruses 12:270. doi: 10.3390/v12030270
Bogani, G., Raspagliesi, F., Ditto, A., and de la Fuente, J. (2020). The adoption of viral capsid-derived virus-like particles (VLPs) for disease prevention and treatments. Vaccines 8:432. doi: 10.3390/vaccines8030432
Boigard, H., Alimova, A., Martin, G. R., Katz, A., Gottlieb, P., and Galarza, J. M. (2017). Zika virus-like particle (VLP) based vaccine. PLoS Negl. Trop. Dis. 11:e0005608. doi: 10.1371/journal.pntd.0005608
Boson, B., Legros, V., Zhou, B., Siret, E., Mathieu, C., Cosset, F. L., et al. (2020). The SARS-CoV-2 Envelope and Membrane proteins modulate maturation and retention of the Spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 296:100111. doi: 10.1074/jbc.RA120.016175
Brisse, M., Vrba, S. M., Kirk, N., Liang, Y., and Ly, H. (2020). Emerging concepts and technologies in vaccine development. Front. Immunol. 11:2578. doi: 10.3389/fimmu.2020.583077
Brito, L. A., and Singh, M. (2011). Commentary: acceptable levels of endotoxin in vaccine formulations during preclinical research. J. Pharm.l Sci. 100, 34–37. doi: 10.1002/jps.22267
Bryant, J. E., Calvert, A. E., Mesesan, K., Crabtree, M. B., Volpe, K. E., Silengo, S., et al. (2007). Glycosylation of the dengue 2 virus E protein at N67 is critical for virus growth in vitro but not for growth in intrathoracically inoculated Aedes aegypti mosquitoes. Virology 366, 415–423. doi: 10.1016/j.virol.2007.05.007
Buffin, S., Peubez, I., Barrière, F., Nicolaï, M. C., Tapia, T., Dhir, V., et al. (2019). Influenza A and B virus-like particles produced in mammalian cells are highly immunogenic and induce functional antibodies. Vaccine 37, 6857–6867. doi: 10.1016/j.vaccine.2019.09.057
Buonaguro, F. M., and Buonaguro, L. (2014). Virus-like Particles in Vaccine Development. London: Future Medicine Limited, 3–4.
Burley, M., Roberts, S., and Parish, J. L. (2020). Epigenetic regulation of human papillomavirus transcription in the productive virus life cycle. Semin. Immunopathol. 42, 159–171. doi: 10.1007/s00281-019-00773-0
Butkovich, N., Li, E., Ramirez, A., Burkhardt, A. M., and Wang, S. W. (2021). Advancements in protein nanoparticle vaccine platforms to combat infectious disease. Wiley Interdiscip. Rev. 13:e1681. doi: 10.1002/wnan.1681
Caldeira, J. C., Perrine, M., Pericle, F., and Cavallo, F. (2020). Virus-like particles as an immunogenic platform for cancer vaccines. Viruses 12:488. doi: 10.3390/v12050488
Callaway, E. (2020). The race for coronavirus vaccines: a graphical guide. Nature 580:576. doi: 10.1038/d41586-020-01221-y
Carter, D. C., Wright, B., Jerome, W. G., Rose, J. P., and Wilson, E. (2020). A unique protein self-assembling nanoparticle with significant advantages in vaccine development and production. J. Nanomater. 2020:4297937.
Cervera, L., Gutiérrez-Granados, S., Martínez, M., Blanco, J., Gòdia, F., and Segura, M. M. (2013). Generation of HIV-1 Gag VLPs by transient transfection of HEK 293 suspension cell cultures using an optimized animal-derived component free medium. J. Biotechnol. 166, 152–165. doi: 10.1016/j.jbiotec.2013.05.001
Chan, S. K., Du, P., Ignacio, C., Mehta, S., Newton, I. G., and Steinmetz, N. F. (2020). Biomimetic virus-like particles as severe acute respiratory syndrome coronavirus 2 diagnostic tools. ACS Nano 15, 1259–1272. doi: 10.1021/acsnano.0c08430
Chang, Y. H., Chiao, D. J., Hsu, Y. L., Lin, C. C., Wu, H. L., Shu, P. Y., et al. (2020). Mosquito cell-derived Japanese encephalitis virus-like particles induce specific humoral and cellular immune responses in mice. Viruses 12:336. doi: 10.3390/v12030336
Charlton Hume, H. K., Vidigal, J., Carrondo, M. J., Middelberg, A. P., Roldão, A., and Lua, L. H. (2019). Synthetic biology for bioengineering virus-like particle vaccines. Biotechnol. Bioeng. 116, 919–935. doi: 10.1002/bit.26890
Chaves, L. C., Ribeiro, B. M., and Blissard, G. W. (2018). Production of GP64-free virus-like particles from baculovirus-infected insect cells. J. Gen. Virol. 99, 265–274. doi: 10.1099/jgv.0.001002
Chen, H. W., Huang, C. Y., Lin, S. Y., Fang, Z. S., Hsu, C. H., Lin, J. C., et al. (2016). Synthetic virus-like particles prepared via protein corona formation enable effective vaccination in an avian model of coronavirus infection. Biomaterials 106, 111–118. doi: 10.1016/j.biomaterials.2016.08.018
Chen, Q., and Lai, H. (2013). Plant-derived virus-like particles as vaccines. Hum. Vaccin. immunotherap. 9, 26–49. doi: 10.4161/hv.22218
Chen, W., Kang, T., Yuan, R., Shao, C., and Jing, S. (2020). Immunogenicity and protective potency of Norovirus GII. 17 virus-like particle-based vaccine. Biotechnol. Lett. 42, 1211–1218. doi: 10.1007/s10529-020-02837-w
Choi, W. H., and Park, J. S. (2020). Immunogenicity and protective effect of a virus-like particle containing the SAG1 antigen of toxoplasma gondii as a potential vaccine candidate for toxoplasmosis. Biomedicines 8:91. doi: 10.3390/biomedicines8040091
Christiansen, D., Earnest-Silveira, L., Grubor-Bauk, B., Wijesundara, D. K., Boo, I., Ramsland, P. A., et al. (2019). Pre-clinical evaluation of a quadrivalent HCV VLP vaccine in pigs following microneedle delivery. Sci. Rep. 9:9251. doi: 10.1038/s41598-019-45461-z
Chung, Y. H., Cai, H., and Steinmetz, N. F. (2020). Viral nanoparticles for drug delivery, imaging, immunotherapy, and theranostic applications. Adv. Drug Deliv. Rev. 156, 214–235. doi: 10.1016/j.addr.2020.06.024
Cohen, J. (2020). Vaccine designers take first shots at COVID-19. Science 368, 14–16. doi: 10.1126/science.368.6486.14
Comas-Garcia, M., Colunga-Saucedo, M., and Rosales-Mendoza, S. (2020). The role of virus-like particles in medical biotechnology. Mol. Pharm. 17, 4407–4420. doi: 10.1021/acs.molpharmaceut.0c00828
Dai, S., Wang, H., and Deng, F. (2018). Advances and challenges in enveloped virus-like particle (VLP)-based vaccines. J. Immunol. Sci. 2, 36–41.
Diamos, A. G., Larios, D., Brown, L., Kilbourne, J., Kim, H. S., Saxena, D., et al. (2019). Vaccine synergy with virus-like particle and immune complex platforms for delivery of human papillomavirus L2 antigen. Vaccine 37, 137–144. doi: 10.1016/j.vaccine.2018.11.021
Diamos, A. G., and Mason, H. S. (2018). High-level expression and enrichment of norovirus virus-like particles in plants using modified geminiviral vectors. Protein Expr. Purif. 151, 86–92. doi: 10.1016/j.pep.2018.06.011
Diaz-Arévalo, D., and Zeng, M. (2020). Nanoparticle-based vaccines: opportunities and limitations. Nanopharmaceuticals 2020, 135–150. doi: 10.1016/b978-0-12-817778-5.00007-5
Dumont, J., Euwart, D., Mei, B., Estes, S., and Kshirsagar, R. (2016). Human cell lines for biopharmaceutical manufacturing: history, status, and future perspectives. Crit. Rev. Biotechnol. 36, 1110–1122. doi: 10.3109/07388551.2015.1084266
Earnest-Silveira, L., Chua, B., Chin, R., Christiansen, D., Johnson, D., Herrmann, S., et al. (2016). Characterization of a hepatitis C virus-like particle vaccine produced in a human hepatocyte-derived cell line. J. Gen. Virol. 97, 1865–1876. doi: 10.1099/jgv.0.000493
Fougeoux, C., Goksøyr, L., Idorn, M., Soroka, V., Myeni, S. K., Dagil, R., et al. (2020). Capsid-like particles decorated with the SARS2-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 12:324. doi: 10.1038/s41467-020-20251-8
Fraenkel-Conrat, H., and Williams, R. C. (1955). Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. U.S.A. 41:690. doi: 10.1073/pnas.41.10.690
Francis, M. J. (2018). Recent advances in vaccine technologies, The Veterinary Clinics of North America. Small Anim. Pract. 48:231. doi: 10.1016/j.cvsm.2017.10.002
Fuenmayor, J., Gòdia, F., and Cervera, L. (2017). Production of virus-like particles for vaccines. New biotechnology 39, 174–180.
Garg, H., Mehmetoglu-Gurbuz, T., and Joshi, A. (2020). Virus Like Particles (VLP) as multivalent vaccine candidate against Chikungunya, Japanese encephalitis, yellow fever and Zika virus. Sci. Rep. 10:4017. doi: 10.1038/s41598-020-61103-1
Garg, H., Mehmetoglu-Gurbuz, T., Ruddy, G. M., and Joshi, A. (2019). Capsid containing virus like particle vaccine against Zika virus made from a stable cell line. Vaccine 37, 7123–7131. doi: 10.1016/j.vaccine.2019.09.093
Ghorbani, A., Zare, F., Sazegari, S., Afsharifar, A., Eskandari, M. H., and Pormohammad, A. (2020). Development of a novel platform of virus-like particle (VLP) based vaccine against coronavirus 2019 (SARS-CoV-2) by exposing of epitopes: an immunoinformatics approach. New Microb. New Infect. 38:100786. doi: 10.1016/j.nmni.2020.100786
Gopal, R., and Schneemann, A. (2018). “Production and application of insect virus-based VLPs,” in Virus–Derived Nanoparticles for Advanced Technologies, eds C. Wege and G. P. Lomonossoff (New York, NY: Humana Press), 125–141. doi: 10.1007/978-1-4939-7808-3_8
Govasli, M. L., Diaz, Y., and Puntervoll, P. (2019). Virus-like particle-display of the enterotoxigenic Escherichia coli heat-stable toxoid STh-A14T elicits neutralizing antibodies in mice. Vaccine 37, 6405–6414. doi: 10.1016/j.vaccine.2019.09.004
Guo, J., Zhou, A., Sun, X., Sha, W., Ai, K., Pan, G., et al. (2019). Immunogenicity of a virus-like-particle vaccine containing multiple antigenic epitopes of toxoplasma gondii against acute and chronic toxoplasmosis in mice. Front. Immunol. 10:592. doi: 10.3389/fimmu.2019.00592
Gupta, J., Kaul, S., Srivastava, A., Kaushik, N., Ghosh, S., Sharma, C., et al. (2020). Expression, purification and characterization of the hepatitis e virus like-particles in the Pichia pastoris. Front. Microbiol. 11:141. doi: 10.1016/j.pep.2013.09.016
Gupta, K., Tölzer, C., Sari-Ak, D., Fitzgerald, D. J., Schaffitzel, C., and Berger, I. (2019). Multibac: baculovirus-mediated multigene DNA cargo delivery in insect and mammalian cells. Viruses 11:198. doi: 10.3390/v11030198
Hankaniemi, M. M., Stone, V. M., Andrejeff, T., Heinimäki, S., Sioofy-Khojine, A. B., Marjomäki, V., et al. (2019). Formalin treatment increases the stability and immunogenicity of coxsackievirus B1 VLP vaccine. Antiviral Res. 171:104595. doi: 10.1016/j.antiviral.2019.104595
Hassine, I. H., Gharbi, J., Hamrita, B., Almalki, M. A., Rodríguez, J. F., and Ben M’hadheb, M. (2020). Characterization of Coxsackievirus B4 virus-like particles VLP produced by the recombinant baculovirus-insect cell system expressing the major capsid protein. Mol. Biol. Rep. 47, 2835–2843. doi: 10.1007/s11033-020-05333-6
He, L., De Val, N., Morris, C. D., Vora, N., Thinnes, T. C., Kong, L., et al. (2016). Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat. Commun. 7, 1–15. doi: 10.1038/ncomms12041
Heath, P. T., Galiza, E. P., Baxter, D. N., Boffito, M., Browne, D., Burns, F., et al. (2021). Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. New Engl. J. Med. 385, 1172–1183.
Hennrich, A. A., Sawatsky, B., Santos-Mandujano, R., Banda, D. H., Oberhuber, M., Schopf, A., et al. (2021). Safe and effective two-in-one replicon-and-VLP minispike vaccine for COVID-19: protection of mice after a single immunization. PLoS Pathog. 17:e1009064. doi: 10.1371/journal.ppat.1009064
Ho, Y., Lin, P. H., Liu, C. Y., Lee, S. P., and Chao, Y. C. (2004). Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 318, 833–838. doi: 10.1016/j.bbrc.2004.04.111
Hong, S., Choi, D. W., Kim, H. N., Park, C. G., Lee, W., and Park, H. H. (2020). Protein-based nanoparticles as drug delivery systems. Pharmaceutics 12:604.
Hsieh, P. K., Chang, S. C., Huang, C. C., Lee, T. T., Hsiao, C. W., Kou, Y. H., et al. (2005). Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 79, 13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005
Hsin, W. C., Chang, C. H., Chang, C. Y., Peng, W. H., Chien, C. L., Chang, M. F., et al. (2018). Nucleocapsid protein-dependent assembly of the RNA packaging signal of Middle East respiratory syndrome coronavirus. J. Biomed. Sci. 25, 1–12. doi: 10.1186/s12929-018-0449-x
Huang, X., Wang, X., Zhang, J., Xia, N., and Zhao, Q. (2017). Escherichia coli-derived virus-like particles in vaccine development. NPJ Vaccines 2, 1–9. doi: 10.1038/s41541-017-0006-8
Huber, B., Schellenbacher, C., Shafti-Keramat, S., Jindra, C., Christensen, N., and Kirnbauer, R. (2017). Chimeric L2-based virus-like particle (VLP) vaccines targeting cutaneous human papillomaviruses (HPV). PLoS One 12:e0169533. doi: 10.1371/journal.pone.0169533
Huo, Y., Wan, X., Ling, T., Wu, J., Wang, W., and Shen, S. (2018). Expression and purification of norovirus virus like particles in Escherichia coli and their immunogenicity in mice. Mol. Immunol. 93, 278–284. doi: 10.1016/j.molimm.2017.07.014
Jain, A., Singh, S. K., Arya, S. K., Kundu, S. C., and Kapoor, S. (2018). Protein nanoparticles: promising platforms for drug delivery applications. ACS Biomater. Sci. Eng. 4, 3939–3961. doi: 10.1021/acsbiomaterials.8b01098
Janitzek, C. M., Peabody, J., Thrane, S., Carlsen, P. H., Theander, T. G., Salanti, A., et al. (2019). A proof-of-concept study for the design of a VLP-based combinatorial HPV and placental malaria vaccine. Sci. Rep. 9:5260. doi: 10.1038/s41598-019-41522-5
Jeong, H., and Seong, B. L. (2017). Exploiting virus-like particles as innovative vaccines against emerging viral infections. J. Microbiol. 55, 220–230. doi: 10.1007/s12275-017-7058-3
Kapadia, C. H., Tian, S., Perry, J. L., Luft, J. C., and DeSimone, J. M. (2019). Role of linker length and antigen density in nanoparticle peptide vaccine. ACS Omega 4, 5547–5555. doi: 10.1021/acsomega.8b03391
Karch, C. P., Matyas, G. R., Burkhard, P., and Beck, Z. (2018). Self-assembling protein nanoparticles: implications for HIV-1 vaccine development. Nanomedicine 13, 2121–2125. doi: 10.2217/nnm-2018-0222
Kato, T., Takami, Y., Deo, V. K., and Park, E. Y. (2019). Preparation of virus-like particle mimetic nanovesicles displaying the S protein of Middle East respiratory syndrome coronavirus using insect cells. J. Biotechnol. 306, 177–184. doi: 10.1016/j.jbiotec.2019.10.007
Keikha, R., Daliri, K., and Jebali, A. (2021). The Use of Nanobiotechnology in Immunology and Vaccination. Vaccines 9:74. doi: 10.3390/vaccines9020074
Kelly, H. G., Tan, H. X., Juno, J. A., Esterbauer, R., Ju, Y., Jiang, W., et al. (2020). Self-assembling influenza nanoparticle vaccines drive extended germinal center activity and memory B cell maturation. JCI Insight 5:e136653. doi: 10.1172/jci.insight.136653
Kianfar, E. (2021). Protein nanoparticles in drug delivery: animal protein, plant proteins and protein cages, albumin nanoparticles. J. Nanobiotechnol. 19, 1–32. doi: 10.1186/s12951-021-00896-3
Kiatmetha, P., Chotwiwatthanakun, C., Jariyapong, P., Santimanawong, W., Ounjai, P., and Weerachatyanukul, W. (2018). Nanocontainer designed from an infectious hypodermal and hematopoietic necrosis virus (IHHNV) has excellent physical stability and ability to deliver shrimp tissues. PeerJ 6:e6079. doi: 10.7717/peerj.6079
Kim, H. S., Jeon, J. H., Lee, K. J., and Ko, K. (2014). N-glycosylation modification of plant-derived virus-like particles: an application in vaccines. BioMed Res. Int. 2014:249519. doi: 10.1155/2014/249519
Kim, K. H., Lee, Y. T., Park, S., Jung, Y. J., Lee, Y., Ko, E. J., et al. (2019). Neuraminidase expressing virus-like particle vaccine provides effective cross protection against influenza virus. Virology 535, 179–188. doi: 10.1016/j.virol.2019.07.008
Kingston, N. J., Kurtovic, L., Walsh, R., Joe, C., Lovrecz, G., Locarnini, S., et al. (2019). Hepatitis B virus-like particles expressing Plasmodium falciparum epitopes induce complement-fixing antibodies against the circumsporozoite protein. Vaccine 37, 1674–1684. doi: 10.1016/j.vaccine.2019.01.056
Kirsteina, A., Akopjana, I., Bogans, J., Lieknina, I., Jansons, J., Skrastina, D., et al. (2020). Construction and immunogenicity of a novel multivalent vaccine prototype based on conserved influenza virus antigens. Vaccines 8:197. doi: 10.3390/vaccines8020197
Koutsky, L. (2009). The epidemiology behind the HPV vaccine discovery. Ann. Epidemiol. 19, 239–244. doi: 10.1016/j.annepidem.2009.01.023
Kramer, R. M., Zeng, Y., Sahni, N., Kueltzo, L. A., Schwartz, R. M., Srivastava, I. K., et al. (2013). Development of a stable virus-like particle vaccine formulation against Chikungunya virus and investigation of the effects of polyanions. J. Pharm. Sci. 102, 4305–4314. doi: 10.1002/jps.23749
Kushnir, N., Streatfield, S. J., and Yusibov, V. (2012). Virus-like particles as a highly efficient vaccine platform: diversity of targets and production systems and advances in clinical development. Vaccine 31, 58–83. doi: 10.1016/j.vaccine.2012.10.083
Ladenstein, R., and Morgunova, E. (2020). Second career of a biosynthetic enzyme: lumazine synthase as a virus-like nanoparticle in vaccine development. Biotechnol. Rep. 27:e00494. doi: 10.1016/j.btre.2020.e00494
Lagoutte, P., Mignon, C., Donnat, S., Stadthagen, G., Mast, J., Sodoyer, R., et al. (2016). Scalable chromatography-based purification of virus-like particle carrier for epitope based influenza A vaccine produced in Escherichia coli. J. Virol. Methods 232, 8–11. doi: 10.1016/j.jviromet.2016.02.011
Lagoutte, P., Mignon, C., Stadthagen, G., Potisopon, S., Donnat, S., Mast, J., et al. (2018). Simultaneous surface display and cargo loading of encapsulin nanocompartments and their use for rational vaccine design. Vaccine 36, 3622–3628. doi: 10.1016/j.vaccine.2018.05.034
Lai, C. C., Cheng, Y. C., Chen, P. W., Lin, T. H., Tzeng, T. T., Lu, C. C., et al. (2019). Process development for pandemic influenza VLP vaccine production using a baculovirus expression system. J. Biol. Eng. 13:78. doi: 10.1186/s13036-019-0206-z
Lampinen, V., Heinimäki, S., Laitinen, O. H., Pesu, M., Hankaniemi, M. M., Blazevic, V., et al. (2021). Modular vaccine platform based on the norovirus-like particle. J. Nanobiotechnol. 19, 1–14. doi: 10.1186/s12951-021-00772-0
Latham, T., and Galarza, J. M. (2001). Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75, 6154–6165. doi: 10.1128/JVI.75.13.6154-6165.2001
Lee, D. H., Chu, K. B., Kang, H. J., Lee, S. H., Chopra, M., Choi, H. J., et al. (2019). Protection induced by malaria virus-like particles containing codon-optimized AMA-1 of Plasmodium berghei. Malaria J. 18, 1–12. doi: 10.1186/s12936-019-3017-2
Li, Y., Han, L., Zhao, Y., Zheng, X., Wang, H., Gai, W., et al. (2020). Immunogenicity assessment of rift valley fever virus virus-like particles in BALB/c mice. Front. Vet. Sci. 7:62. doi: 10.3389/fvets.2020.00062
Lin, H. H., Yip, B. S., Huang, L. M., and Wu, S. C. (2018). Zika virus structural biology and progress in vaccine development. Biotechnol. Adv. 36, 47–53. doi: 10.1016/j.biotechadv.2017.09.004
Liu, G., Qiao, X., Chang, C., Hua, T., Wang, J., Tang, B., et al. (2020). Reduction of postweaning multisystemic wasting syndrome-associated clinical symptoms by virus-like particle vaccine against porcine parvovirus and porcine circovirus type 2. Viral Immunol. 33, 444–456. doi: 10.1089/vim.2019.0201
López-Macías, C. (2012). Virus-like particle (VLP)-based vaccines for pandemic influenza: performance of a VLP vaccine during the 2009 influenza pandemic. Hum. Vaccin. Immunotherap. 8, 411–414. doi: 10.4161/hv.18757
López-Sagaseta, J., Malito, E., Rappuoli, R., and Bottomley, M. J. (2016). Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 14, 58–68. doi: 10.1016/j.csbj.2015.11.001
Lu, B., Huang, Y., Huang, L., Li, B., Zheng, Z., Chen, Z., et al. (2010). Effect of mucosal and systemic immunization with virus-like particles of severe acute respiratory syndrome coronavirus in mice. Immunology 13, 254–261. doi: 10.1111/j.1365-2567.2010.03231.x
Lu, J., Lu, G., Tan, S., Xia, J., Xiong, H., Yu, X., et al. (2020). A COVID-19 mRNA vaccine encoding SARS-CoV-2 virus-like particles induces a strong antiviral-like immune response in mice. Cell Res. 30, 936–939. doi: 10.1038/s41422-020-00392-7
Lu, L., Duong, V. T., Shalash, A. O., Skwarczynski, M., and Toth, I. (2021). Chemical conjugation strategies for the development of protein-based subunit nanovaccines. Vaccines 9:563. doi: 10.3390/vaccines9060563
Lua, L. H., Connors, N. K., Sainsbury, F., Chuan, Y. P., Wibowo, N., and Middelberg, A. P. (2014). Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 111, 425–440. doi: 10.1002/bit.25159
Mamat, U., Woodard, R. W., Wilke, K., Souvignier, C., Mead, D., Steinmetz, E., et al. (2013). Endotoxin-free protein production—ClearColi™ technology. Nat. Methods 10, 916–916. doi: 10.1038/nmeth.f.367
Marsian, J., Fox, H., Bahar, M. W., Kotecha, A., Fry, E. E., Stuart, D. I et al (2017). Plant-made polio type 3 stabilized VLPs—a candidate synthetic polio vaccine. Nat. Commun. 8, 1–9. doi: 10.1038/s41467-017-00090-w
Masavuli, M. G., Wijesundara, D. K., Torresi, J., Gowans, E. J., and Grubor-Bauk, B. (2017). Preclinical development and production of virus-like particles as vaccine candidates for hepatitis C. Front. Microbiol. 8:2413. doi: 10.3389/fmicb.2017.02413
Maurer, P., Jennings, G. T., Willers, J., Rohner, F., Lindman, Y., Roubicek, K., et al. (2005). A therapeutic vaccine for nicotine dependence: preclinical efficacy, and Phase I safety and immunogenicity. Eur. J. Immunol. 35, 2031–2040. doi: 10.1002/eji.200526285
Metz, S. W., Martina, B. E., van den Doel, P., Geertsema, C., Osterhaus, A. D., Vlak, J. M., et al. (2013). Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine 31, 6092–6096. doi: 10.1016/j.vaccine.2013.09.045
Mohsen, M. O., Gomes, A. C., Vogel, M., and Bachmann, M. F. (2018). Interaction of viral capsid-derived virus-like particles (VLPs) with the innate immune system. Vaccines 6:37. doi: 10.3390/vaccines6030037
Mohsen, M. O., Zha, L., Cabral-Miranda, G., and Bachmann, M. F. (2017). Major findings and recent advances in virus–like particle (VLP)-based vaccines. Semin. Immunol. 34, 123–132. doi: 10.1016/j.smim.2017.08.014
Mokoena, N. B., Moetlhoa, B., Rutkowska, D. A., Mamputha, S., Dibakwane, V. S., Tsekoa, T. L., et al. (2019). Plant-produced Bluetongue chimaeric VLP vaccine candidates elicit serotype-specific immunity in sheep. Vaccine 37, 6068–6075.
Moleirinho, M. G., Silva, R. J., Alves, P. M., Carrondo, M. J., and Peixoto, C. (2020). Current challenges in biotherapeutic particles manufacturing. Expert Opin. Biol. Ther. 20, 451–465. doi: 10.1080/14712598.2020.1693541
Naskalska, A., and Pyrć, K. (2015). Virus like particles as immunogens and universal nanocarriers. Polish J. Microbiol. 64, 3–13.
Nguyen, B., and Tolia, N. H. (2021). Protein-based antigen presentation platforms for nanoparticle vaccines. NPJ Vaccin. 6, 1–11. doi: 10.1038/s41541-021-00330-7
Noad, R., and Roy, P. (2003). Virus-like particles as immunogens. Trends Microbiol. 11, 438–444. doi: 10.1016/s0966-842x(03)00208-7
Nooraei, S., Bahrulolum, H., Hoseini, Z. S., Katalani, C., Hajizade, A., Easton, A. J., et al. (2021). Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 19, 1–27. doi: 10.1186/s12951-021-00806-7
Park, S. J. (2020). Protein–nanoparticle interaction: corona formation and conformational changes in proteins on nanoparticles. Int. J. Nanomed. 15:5783. doi: 10.2147/IJN.S254808
Pastor, A. R., González-Domínguez, G., Díaz-Salinas, M. A., Ramírez, O. T., and Palomares, L. A. (2019). Defining the multiplicity and time of infection for the production of Zaire Ebola virus-like particles in the insect cell-baculovirus expression system. Vaccine 37, 6962–6969. doi: 10.1016/j.vaccine.2019.06.029
Pechsrichuang, P., Namwongnao, S., and Jacquet, A. (2021). Bioengineering of Virus-like Particles for the Prevention or Treatment of Allergic Diseases. Allergy Asthma Immunol. Res. 13:23. doi: 10.4168/aair.2021.13.1.23
Poon, C., and Patel, A. A. (2020). Organic and inorganic nanoparticle vaccines for prevention of infectious diseases. Nano Express 1:012001.
Pouyanfard, S., Spagnoli, G., Bulli, L., Balz, K., Yang, F., Odenwald, C., et al. (2018). Minor capsid protein L2 polytope induces broad protection against oncogenic and mucosal human papillomaviruses. J. Virol. 92: e01930-17. doi: 10.1128/JVI.01930-17
Qian, C., Liu, X., Xu, Q., Wang, Z., Chen, J., Li, T., et al. (2020). Recent progress on the versatility of virus-like particles. Vaccines 8:139. doi: 10.3390/vaccines8010139
Quan, F. S., Lee, Y. T., Kim, K. H., Kim, M. C., and Kang, S. M. (2016). Progress in developing virus-like particle influenza vaccines. Expert rev. Vaccin. 15, 1281–1293. doi: 10.1080/14760584.2016.1175942
Richert, L. E., Servid, A. E., Harmsen, A. L., Rynda-Apple, A., Han, S., Wiley, J. A., et al. (2012). A virus-like particle vaccine platform elicits heightened and hastened local lung mucosal antibody production after a single dose. Vaccine 30, 3653–3665. doi: 10.1016/j.vaccine.2012.03.035
Rodrigues, M. Q., Alves, P. M., and Roldão, A. (2021). Functionalizing ferritin nanoparticles for vaccine development. Pharmaceutics 13:1621. doi: 10.3390/pharmaceutics13101621
Roldão, A., Mellado, M. C. M., Castilho, L. R., Carrondo, M. J., and Alves, P. M. (2010). Virus-like particles in vaccine development. Expert Rev. Vaccin. 9, 1149–1176.
Roldao, A., Silva, A. C., Mellado, M. C. M., Alves, P. M., and Carrondo, M. J. T. (2011). Viruses and virus-like particles in biotechnology: fundamentals and applications. Compr. Biotechnol. 1, 633–656.
Rosales-Mendoza, S., Márquez-Escobar, V. A., González-Ortega, O., Nieto-Gómez, R., and Arévalo-Villalobos, J. I. (2020). What does plant-based vaccine technology offer to the fight against COVID-19? Vaccines 8:183. doi: 10.3390/vaccines8020183
Roth, G. A., Picece, V. C., Ou, B. S., Luo, W., Pulendran, B., and Appel, E. A. (2021). Designing spatial and temporal control of vaccine responses. Nature Reviews Materials 1–22. doi: 10.1038/s41578-021-00372-2
Roy, P., and Noad, R. (2008). Virus like particles as a vaccine delivery system: myths and facts. Hum. Vaccin. 4, 5–12. doi: 10.4161/hv.4.1.5559
Rutgers, T., Gordon, D., Gathoye, A. M., Hollingdale, M., Hockmeyer, W., Rosenberg, M., et al. (1988). Hepatitis B surface antigen as carrier matrix for the repetitive epitope of the circumsporozoite protein of Plasmodium falciparum. Biol. Technol. 6, 1065–1070.
Rutkowska, D. A., Mokoena, N. B., Tsekoa, T. L., Dibakwane, V. S., and O’Kennedy, M. M. (2019). Plant-produced chimeric virus-like particles-a new generation vaccine against African horse sickness. BMC Vet. Res. 15:432. doi: 10.1186/s12917-019-2184-2
Rybicki, E. P. (2020). Plant molecular farming of virus-like nanoparticles as vaccines and reagents. Wiley Interdiscip. Rev. 12:e1587. doi: 10.1002/wnan.1587
Samrat, S. K., Tharappel, A. M., Li, Z., and Li, H. (2020). Prospect of SARS-CoV-2 spike protein: potential role in vaccine and therapeutic development. Virus res. 288:198141. doi: 10.1016/j.virusres.2020.198141
Santi, L., Batchelor, L., Huang, Z., Hjelm, B., Kilbourne, J., Arntzen, C. J., et al. (2008). An efficient plant viral expression system generating orally immunogenic Norwalk virus-like particles. Vaccine 26, 1846–1854. doi: 10.1016/j.vaccine.2008.01.053
Sari-Ak, D., Bahrami, S., Laska, M. J., Drncova, P., Fitzgerald, D. J., Schaffitzel, C., et al. (2019). “High-throughput production of influenza virus-like particle (VLP) array by using VLP-factory™, a multibac baculoviral genome customized for enveloped VLP expression,” in High-Throughput Protein Production and Purification, ed. R. Vincentelli (New York, NY: Humana), 213–226. doi: 10.1007/978-1-4939-9624-7_10
Sarkar, B., Islam, S. S., Zohora, U. S., and Ullah, M. A. (2019). Virus like particles-A recent advancement in vaccine development. Microbiol. Soc. Korea 55, 327–343.
Sartorius, R., Trovato, M., Manco, R., D’Apice, L., and De Berardinis, P. (2021). Exploiting viral sensing mediated by Toll-like receptors to design innovative vaccines. NPJ Vaccin. 6, 1–15. doi: 10.1038/s41541-021-00391-8
Schädler, J., Sigrist, B., Meier, S. M., Albini, S., and Wolfrum, N. (2019). Virus-like particles in a new vaccination approach against infectious laryngotracheitis. J. Gen. Virol. 100, 1013–1026. doi: 10.1099/jgv.0.001272
Schellenbacher, C., Kwak, K., Fink, D., Shafti-Keramat, S., Huber, B., Jindra, C., et al. (2013). Efficacy of RG1-VLP vaccination against infections with genital and cutaneous human papillomaviruses. J. Invest. Dermatol. 133, 2706–2713. doi: 10.1038/jid.2013.253
Schellenbacher, C., Roden, R., and Kirnbauer, R. (2009). Chimeric L1-L2 virus-like particles as potential broad-spectrum human papillomavirus vaccines. J. Virol. 83, 10085–10095. doi: 10.1128/JVI.01088-09
Schumacher, J., Bacic, T., Staritzbichler, R., Daneschdar, M., Klamp, T., Arnold, P., et al. (2018). Enhanced stability of a chimeric hepatitis B core antigen virus-like-particle (HBcAg-VLP) by a C-terminal linker-hexahistidine-peptide. J. Nanobiotechnol. 16, 1–21. doi: 10.1186/s12951-018-0363-0
Sequeira, D. P., Correia, R., Carrondo, M. J., Roldão, A., Teixeira, A. P., and Alves, P. M. (2018). Combining stable insect cell lines with baculovirus-mediated expression for multi-HA influenza VLP production. Vaccine 36, 3112–3123. doi: 10.1016/j.vaccine.2017.02.043
Shanmugam, R. K., Ramasamy, V., Shukla, R., Arora, U., Swaminathan, S., and Khanna, N. (2019). Pichia pastoris-expressed Zika virus envelope domain III on a virus-like particle platform: design, production and immunological evaluation. Pathog. Dis. 77:ftz026. doi: 10.1093/femspd/ftz026
Sherry, L., Grehan, K., Snowden, J. S., Knight, M. L., Adeyemi, O. O., Rowlands, D. J., et al. (2020). Comparative molecular biology approaches for the production of poliovirus virus-like particles using Pichia pastoris. Msphere 5, 1–12. doi: 10.1128/mSphere.00838-19
Shinde, V., Bhikha, S., Hoosain, Z., Archary, M., Bhorat, Q., Fairlie, L., et al. (2021). Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B. 1.351 Variant. New Engl. J. Med. 384, 1899–1909. doi: 10.1056/NEJMoa2103055
Shirbaghaee, Z., and Bolhassani, A. (2016). Different applications of virus-like particles in biology and medicine: vaccination and delivery systems. Biopolymers 105, 113–132. doi: 10.1002/bip.22759
Spice, A. J., Aw, R., Bracewell, D. G., and Polizzi, K. M. (2020). Synthesis and assembly of hepatitis B virus-like particles in a Pichia pastoris cell-free system. Front. Bioeng. Biotechnol. 8:72. doi: 10.3389/fbioe.2020.00072
Stephen, S. L., Beales, L., Peyret, H., Roe, A., Stonehouse, N. J., and Rowlands, D. J. (2018). “Recombinant expression of tandem–HBc virus-like particles (VLPs),” in Virus–Derived Nanoparticles for Advanced Technologies, eds C. Wege and G. Lomonossoff (New York, NY: Humana Press), 97–123. doi: 10.1007/978-1-4939-7808-3_7
Storni, T., and Bachmann, M. F. (2004). Loading of MHC class I and II presentation pathways by exogenous antigens: a quantitative in vivo comparison. J. Immunol. 172, 6129–6135. doi: 10.4049/jimmunol.172.10.6129
Strobl, F., Ghorbanpour, S. M., Palmberger, D., and Striedner, G. (2020). Evaluation of screening platforms for virus-like particle production with the baculovirus expression vector system in insect cells. Sci. Rep. 10:1065. doi: 10.1038/s41598-020-57761-w
Syomin, B. V., and Ilyin, Y. V. (2019). Virus-like particles as an instrument of vaccine production. Mol. Biol. 53, 323–334.
Tripathy, S., Dassarma, B., Bhattacharya, M., and Matsabisa, M. G. (2021). Plant-based vaccine research development against viral diseases with emphasis on Ebola virus disease: a review study. Curr. Opin. Pharmacol. 60, 261–267. doi: 10.1016/j.coph.2021.08.001
Uddin, M. N., Henry, B., Carter, K. D., Roni, M. A., and Kouzi, S. S. (2019). A novel formulation strategy to deliver combined DNA and VLP based HPV vaccine. J. Pharm. Pharm. Sci. 22, 536–547. doi: 10.18433/jpps30768
Ueda, G., Antanasijevic, A., Fallas, J. A., Sheffler, W., Copps, J., Ellis, D., et al. (2020). Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife 9:e57659. doi: 10.7554/eLife.57659
Urakami, A., Sakurai, A., Ishikawa, M., Yap, M. L., Flores-Garcia, Y., Haseda, Y., et al. (2017). Development of a novel virus-like particle vaccine platform that mimics the immature form of alphavirus. Clin. Vaccin. Immunol. 24: e00090-17. doi: 10.1128/CVI.00090-17
van Oers, M. M. (2011). Opportunities and challenges for the baculovirus expression system. J. Invertebr. Pathol. 107, S3–S15. doi: 10.1016/j.jip.2011.05.001
van Oosten, L., Altenburg, J. J., Fougeroux, C., Geertsema, C., van den End, F., Evers, W. A., et al. (2021). Two-component nanoparticle vaccine displaying glycosylated spike S1 domain induces neutralizing antibody response against SARS-CoV-2 variants. mbio 12: e01813-21. doi: 10.1128/mBio.01813-21
van Zyl, A. R., Meyers, A. E., and Rybicki, E. P. (2016). Transient Bluetongue virus serotype 8 capsid protein expression in Nicotiana benthamiana. Biotechnol. Rep. 9, 15–24. doi: 10.1016/j.btre.2015.12.001
Veerapen, V. P., van Zyl, A. R., Rybicki, E. P., and Meyers, A. E. (2018). Transient expression of heat-and acid-resistant foot-and-mouth disease virus P1-2A mutants in Nicotiana benthamiana. Virus Res. 256, 45–49. doi: 10.1016/j.virusres.2018.08.004
Venereo-Sanchez, A., Simoneau, M., Lanthier, S., Chahal, P., Bourget, L., Ansorge, S., et al. (2017). Process intensification for high yield production of influenza H1N1 Gag virus-like particles using an inducible HEK-293 stable cell line. Vaccine 35, 4220–4228. doi: 10.1016/j.vaccine.2017.06.024
Vicente, T., Roldão, A., Peixoto, C., Carrondo, M. J., and Alves, P. M. (2011). Large-scale production and purification of VLP-based vaccines. J. Invertebr. Pathol. 107, S42–S48. doi: 10.1016/j.jip.2011.05.004
Walls, A. C., Fiala, B., Schäfer, A., Wrenn, S., Pham, M. N., Murphy, M., et al. (2020). Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.
Wang, W., Huang, B., Zhu, Y., Tan, W., and Zhu, M. (2021). Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell. Mol. Immunol. 18, 749–751. doi: 10.1038/s41423-021-00643-6
Ward, B. J., Gobeil, P., Seguin, A., Atkins, J., Boulay, I., Charbonneau, P. Y., et al. (2020). Phase 1 trial of a candidate recombinant virus-like particle vaccine for Covid-19 disease produced in plants. medRxiv[Preprint] doi: 10.1101/2020.11.04.20226282
Ward, B. J., Gobeil, P., Séguin, A., Atkins, J., Boulay, I., Charbonneau, P. Y., et al. (2021). Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 27, 1071–1078. doi: 10.1038/s41591-021-01370-1
Wetzel, D., Barbian, A., Jenzelewski, V., Schembecker, G., Merz, J., and Piontek, M. (2019a). Bioprocess optimization for purification of chimeric VLP displaying BVDV E2 antigens produced in yeast Hansenula polymorpha. J. Biotechnol. 306, 203–212. doi: 10.1016/j.jbiotec.2019.10.008
Wetzel, D., Chan, J. A., Suckow, M., Barbian, A., Weniger, M., Jenzelewski, V., et al. (2019b). Display of malaria transmission-blocking antigens on chimeric duck hepatitis B virus-derived virus-like particles produced in Hansenula polymorpha. PLoS One 14:e0221394. doi: 10.1371/journal.pone.0221394
Wetzel, D., Rolf, T., Suckow, M., Kranz, A., Barbian, A., Chan, J. A., et al. (2018). Establishment of a yeast-based VLP platform for antigen presentation. Microb. Cell Fact. 17, 1–17. doi: 10.1186/s12934-018-0868-0
Wu, C. Y., Yeh, Y. C., Yang, Y. C., Chou, C., Liu, M. T., Wu, H. S., et al. (2010). Mammalian expression of virus-like particles for advanced mimicry of authentic influenza virus. PLoS One 5:e9784. doi: 10.1371/journal.pone.0009784
Xu, R., Shi, M., Li, J., Song, P., and Li, N. (2020). Construction of SARS-CoV-2 virus-like particles by mammalian expression system. Front. Bioeng. Biotechnol. 8:862.
Yadav, R., Zhai, L., and Tumban, E. (2020). Virus-like particle-based L2 vaccines against HPVs: where are we today? Viruses 12:18. doi: 10.3390/v12010018
Yan, F., Banadyga, L., Zhao, Y., Zhao, Z., Schiffman, Z., Huang, P., et al. (2019). Peste des petits ruminants virus-like particles induce a potent humoral and cellular immune response in goats. Viruses 11:918. doi: 10.3390/v11100918
Yan, X., Zhou, M., Yu, S., Jin, Z., and Zhao, K. (2020). An overview of biodegradable nanomaterials and applications in vaccines. Vaccine 38, 1096–1104. doi: 10.1016/j.vaccine.2019.11.031
Yang, Y., Shi, W., Abiona, O. M., Nazzari, A., Olia, A. S., Ou, L., et al. (2021). Newcastle disease virus-like particles displaying prefusion-stabilized SARS-CoV-2 spikes elicit potent neutralizing responses. Vaccines 9:73. doi: 10.3390/vaccines9020073
Yang, Z., Gao, F., Wang, X., Shi, L., Zhou, Z., Jiang, Y., et al. (2019). Development and characterization of an enterovirus 71 (EV71) virus-like particles (VLPs) vaccine produced in Pichia pastoris. Hum. Vaccin. Immunotherap. 16, 1602–1610. doi: 10.1080/21645515.2019.1649554
Yazdani, R., Shams-Bakhsh, M., Hassani-Mehraban, A., Arab, S. S., Thelen, N., Thiry, M., et al. (2019). Production and characterization of virus-like particles of grapevine fanleaf virus presenting L2 epitope of human papillomavirus minor capsid protein. BMC Biotechnol. 19:81. doi: 10.1186/s12896-019-0566-y
Young, K. R., Arthus-Cartier, G., Yam, K. K., Lavoie, P. O., Landry, N., D’Aoust, M. A., et al. (2015). Generation and characterization of a trackable plant-made influenza H5 virus-like particle (VLP) containing enhanced green fluorescent protein (eGFP). FASEB J. 29, 3817–3827. doi: 10.1096/fj.15-270421
Żak, M. M., and Zangi, L. (2021). Lipid nanoparticles for organ-specific mRNA therapeutic delivery. Pharmaceutics 13:1675. doi: 10.3390/pharmaceutics13101675
Zeltins, A. (2013). Construction and characterization of virus-like particles: a review. Mol. Biotechnol. 53, 92–107. doi: 10.1007/s12033-012-9598-4
Zepeda-Cervantes, J., Ramírez-Jarquín, J. O., and Vaca, L. (2020). Interaction between virus-like particles (VLPs) and pattern recognition receptors (PRRs) from dendritic cells (DCs): toward better engineering of VLPs. Front. Immunol. 11:1100. doi: 10.3389/fimmu.2020.01100
Keywords: virus-like particles, emerging infectious diseases, vaccine, vaccine development, expression system, virus, SARS-CoV2
Citation: Tariq H, Batool S, Asif S, Ali M and Abbasi BH (2022) Virus-Like Particles: Revolutionary Platforms for Developing Vaccines Against Emerging Infectious Diseases. Front. Microbiol. 12:790121. doi: 10.3389/fmicb.2021.790121
Received: 06 October 2021; Accepted: 10 December 2021;
Published: 03 January 2022.
Edited by:
Jianwei Wang, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Kelly Lee, University of Washington, United StatesLee Sherry, University of Leeds, United Kingdom
Copyright © 2022 Tariq, Batool, Asif, Ali and Abbasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bilal Haider Abbasi, YmhhYmJhc2lAcWF1LmVkdS5waw==
 Hasnat Tariq
Hasnat Tariq Sannia Batool
Sannia Batool Saaim Asif
Saaim Asif Mohammad Ali3
Mohammad Ali3 Bilal Haider Abbasi
Bilal Haider Abbasi