
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 November 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.783195
Our previous study identified a new β-galactosidase in Erwinia sp. E602. To further understand the lactose metabolism in this strain, de novo genome assembly was conducted by using a strategy combining Illumina and PacBio sequencing technology. The whole genome of Erwinia sp. E602 includes a 4.8 Mb chromosome and a 326 kb large plasmid. A total of 4,739 genes, including 4,543 protein-coding genes, 25 rRNAs, 82 tRNAs and 7 other ncRNAs genes were annotated. The plasmid was the largest one characterized in genus Erwinia by far, and it contained a number of genes and pathways responsible for lactose metabolism and regulation. Moreover, a new plasmid-borne lac operon that lacked a typical β-galactoside transacetylase (lacA) gene was identified in the strain. Phylogenetic analysis showed that the genes lacY and lacZ in the operon were under positive selection, indicating the adaptation of lactose metabolism to the environment in Erwinia sp. E602. Our current study demonstrated that the hybrid de novo genome assembly using Illumina and PacBio sequencing technologies, as well as the metabolic pathway analysis, provided a useful strategy for better understanding of the evolution of undiscovered microbial species or strains.
Erwinia is a group of the straight rod-shaped, facultative anaerobic, gram-negative bacterium of the Erwiniaceae family of Enterobacteriaceae. Most of the Erwinia species identified by far are pathogens, saprophytes, or epiphytes of plants. It has been reported that some Erwinia species ferment lactose as a carbon source. Our previous study characterized a β-galactosidase with relatively high activity at low temperature in the Erwinia sp. strain E602 (Xia et al., 2018).
The lac operons typically involve three genes encoding the enzymes that enable bacteria to utilize lactose (Diaz-Hernandez and Santillan, 2010). The gene lacZ encodes the β-galactosidase, an enzyme that degrades lactose into monosaccharides glucose and galactose. Similarly, lacY encodes a membrane-embedded transporter that helps bring lactose into cells. The gene lacA encodes galactoside O-acetyltransferase that catalyzes the transfer of an acetyl group from acetyl-CoA to the 6-hydroxyl of galactopyranosides, with its exact physiological function remaining unclear. In addition, the gene lacI encodes the lac repressor, which is a protein that represses the transcription of lac operon genes by binding to the promoter and preventing the operon from transcription when lactose is not available. In the presence of lactose, the lac repressor is released from the operon to allow RNA transcription (Marbach and Bettenbrock, 2012). Notably, the lac operon confers a competitive advantage for bacterial survival in the environments (Pinto et al., 2021). Therefore, the study of lac operon in the newly identified strain Erwinia sp. E602 may improve our understanding of the role of lactose metabolism in this genus.
High-throughput sequencing and de novo assembly allow obtaining the total genetic information of microbes. It is widely used to dissect the genome sequence, gene composition, and evolutionary features of novel or unknown species. Notably, long-read sequencing technologies, such as PacBio (Rhoads and Au, 2015; Zhang et al., 2021), have dramatically speeded up the understanding of microorganisms. Moreover, the whole genome sequence of microorganisms provides an important basis for post-genomics research of microorganisms, such as subsequent gene mining and functional validation. In order to further investigate the regulation mechanisms of the β-galactosidase expression in the strain Erwinia sp. E602, we sequenced its genome and conducted de novo assembly, using the hybrid Illumina and PacBio sequencing (De Maio et al., 2019). From the assembly of the Erwinia sp. E602 genome, we identified a large plasmid and a new plasmid-borne lac operon containing a lacZ gene encoding the β-galactosidase with low-temperature activity, as well as evidence supporting positive selection pressure of the lac operon.
As summarized in Supplementary Tables 1–3, the Illumina and PacBio reads were used to conduct de novo genome assembly. After hybrid de novo assembly using SPAdes, two circular scaffolds were obtained, including a chromosome sequence of 4,842,717 bp with a GC content of 58.91% (GenBank: CP046582.1) and a large plasmid sequence of 325,969 bp (GenBank: CP046581.1). A total of 4,739 genes, including 4,543 protein-coding, 25 rRNAs, 82 tRNAs, and 7 other ncRNAs genes were annotated as shown in Table 1 and Figures 1A,B. Among these genes, 4,472 genes including 4,296 protein-coding, 25 rRNAs, 82 tRNAs, and 7 other ncRNAs genes were located on the chromosome, while 277 protein-coding genes were located on the large plasmid.
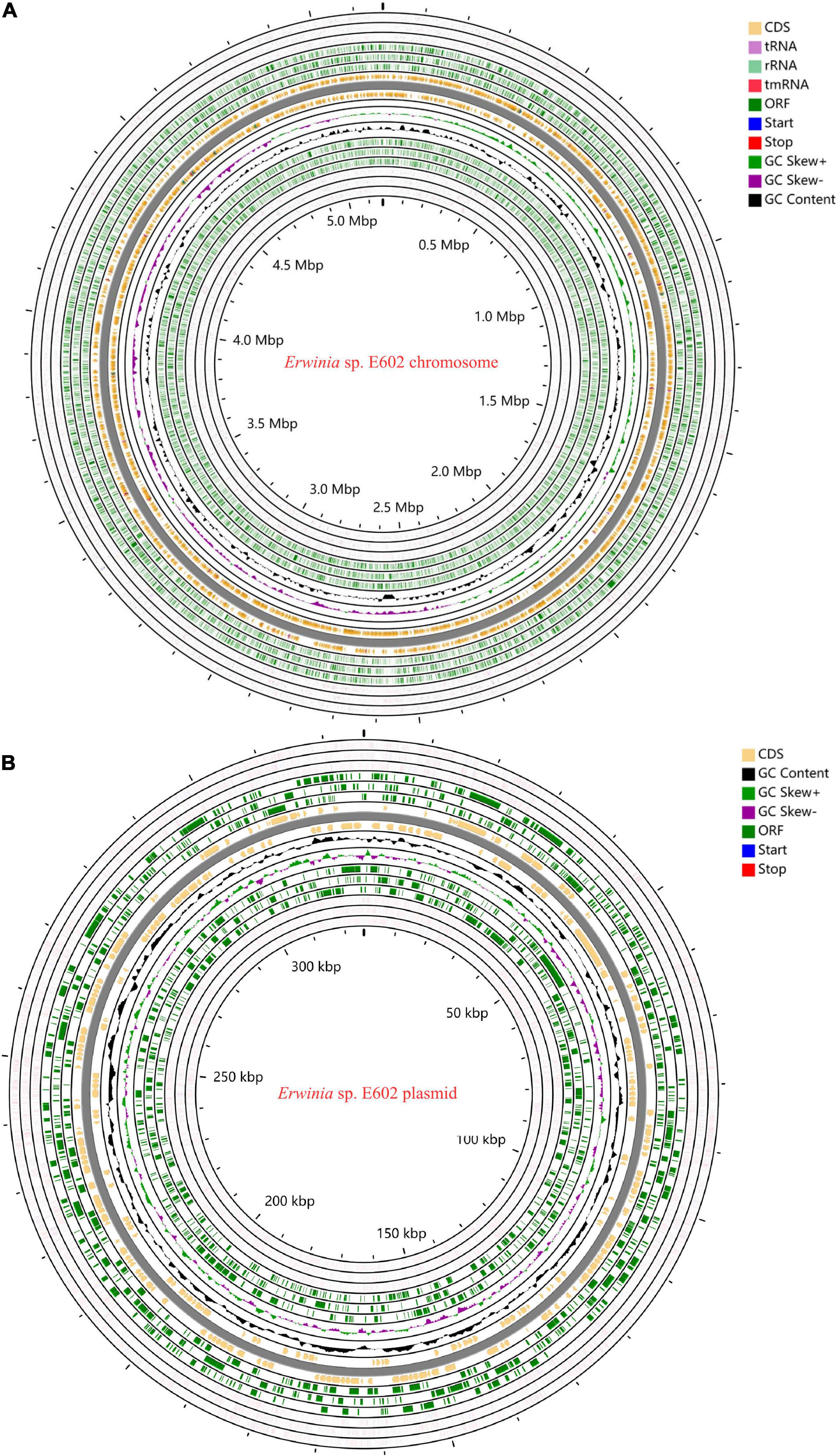
Figure 1. Circos plots of the annotated genome of Erwinia sp. E602. Two circular DNA, including the chromosome (A) and a large plasmid (B) are shown. Different colors represent CDS, rRNA, tRNA, tmRNA, Start, Stop, ORF, GC content, GC Skew+, and GC skew- respectively.
EggNOGv5.0 was then used to predict the functions of these genes. The KEGG pathway enrichment analysis was also performed (Supplementary Material 1). Pathways with a number of genes greater than 5 were shown, and finally, 122 enriched pathways were obtained. The most-enriched pathways (gene counts) included biosynthesis of secondary metabolites (357), microbial metabolism in diverse environments (260), ABC transporters (204), biosynthesis of cofactors (156), biosynthesis of amino acids (132), two-component systems (115), carbon metabolism (98), purine metabolism (71), quorum sensing (63), pyrimidine metabolism (51), ribosome (51), cysteine and methionine metabolism (50), bacterial secretion system (48), and amino sugar and nucleotide sugar metabolism (47). By dbCAN2 database annotation (Zhang et al., 2018), 593 carbohydrases were found on the chromosome and 51 on the plasmid (Supplementary Material 1). Through Resfams database annotation (Gibson et al., 2015), we found 211 resistance genes on the chromosome and 21 resistance genes on the plasmid, respectively (Supplementary Material 1).
From the gene annotation results of de novo assembly, Erwinia sp. E602 was found to contain a number of genes and pathways for lactose metabolism and regulation (Figure 2A). A list of lactose/galactose-related functional genes were shown in Table 2.
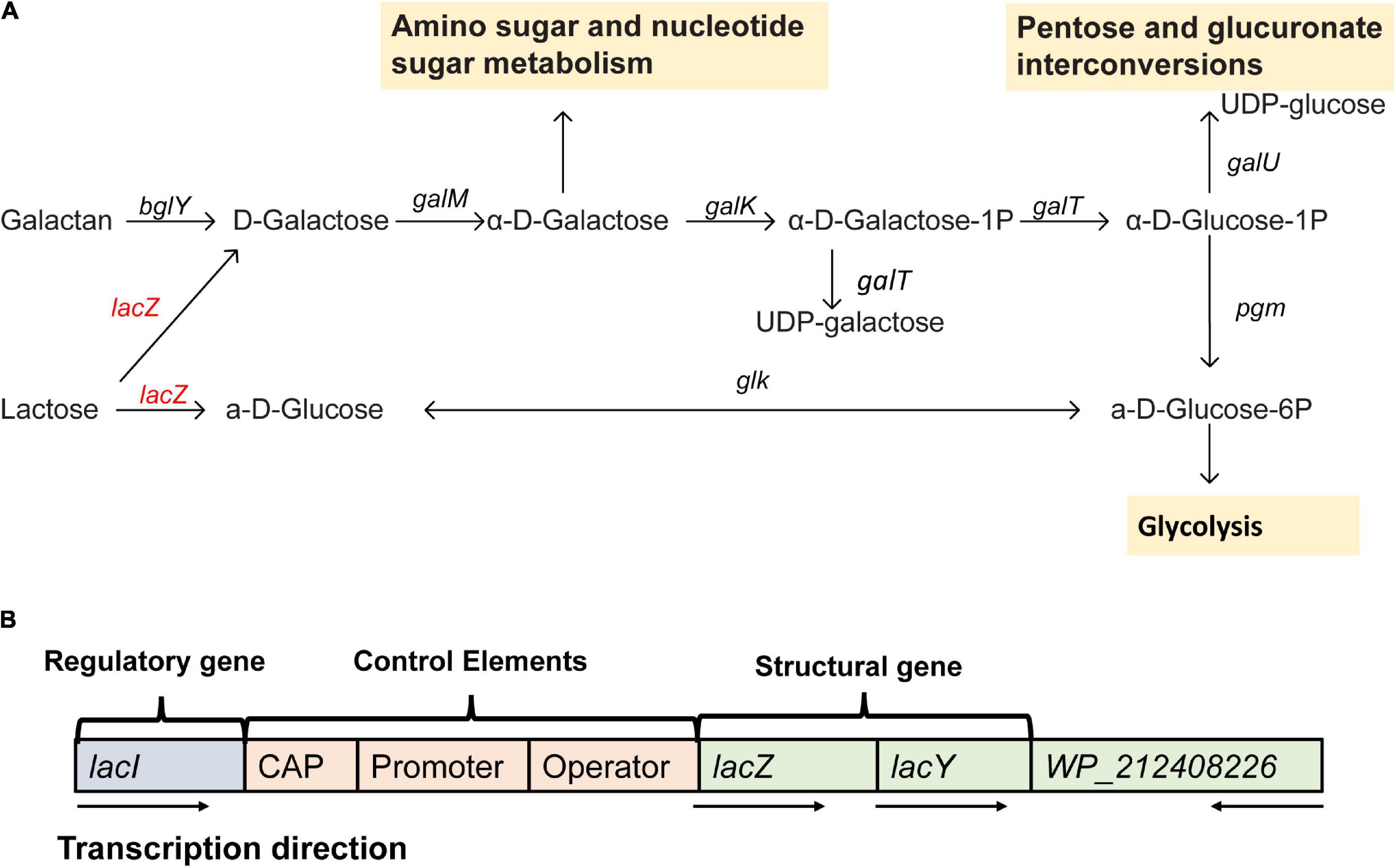
Figure 2. A new plasmid-borne lac operon characterized in Erwinia sp. E602. (A) Lactose-galactose metabolic pathway. Gene names are above the arrow, and the yellow box represents the downstream metabolic pathway. The red color indicates that the gene is located on the plasmid and black means that the gene is located on the chromosome. (B) A structure diagram of the new lac operon and its downstream lacI-like gene encoding the WP_212408226 protein. The arrows indicate the transcription directions of the genes.
Our previous study characterized a new β-galactosidase that exhibited activity at low temperatures, indicating its potential unique biological function (Xia et al., 2018). Intriguingly, the de novo assembly results showed that the exact lacZ gene encoding the new β-galactosidase was not located on the chromosome of Erwinia sp. E602. Instead, further analysis of the sequence context surrounding the lacZ gene found that the gene was a component of a new lac operon on the large plasmid. The lac operons are known to be composed of regulatory genes, manipulated regions, and structural genes (Diaz-Hernandez and Santillan, 2010). The newly identified lac operon in the current study contained lacI, lacZ, lacY, as well as a regulatory element-CAP and a promoter, between the location of lacI and lacZ (Figure 2B). BLAST results of the plasmid-borne lac operon showed that it shares low sequence similarity with other sequences in other Erwinia strains. These results are included in Supplementary Materials. Moreover, this newly discovered lac operon, compared with the classical ones, lacked a lacA gene but had a reversed lacI-like gene encoding protein WP_212408226. Further comparison of the related genes revealed that although other lacZ, lacI and lacY paralogs were also observed in the genome of Erwinia sp. E602, they did not form a canonical lac operon due to the lack of essential structure. Moreover, the lacI and lacY genes of the plasmid-borne lac operon share low similarity with their homologs in the genomes of Erwinia species (Table 3), suggesting that this lac operon was distinct from canonical ones. In addition, the NCBI BLAST search using the nucleic acid database was performed to identify potentially similar operon-related genes, and the results showed that the genes in this operon were more similar to those in genus Citrobacter (Figure 3). Likewise, the lacZ gene of the plasmid-borne lac operon shared a relatively low identity with its paralogs in the other Erwinia species (Table 4). Instead, its lacZ and lacY had high similarities with their homologs in Citrobacter species. The genes of lacI and lacI-like protein (WP_212408226) were compared using the NCBI nucleic acid database, and no sequences with similarities greater than 80% were found. In addition, no lacA was found either on the chromosome of Erwinia sp. E602, or on its large plasmid.
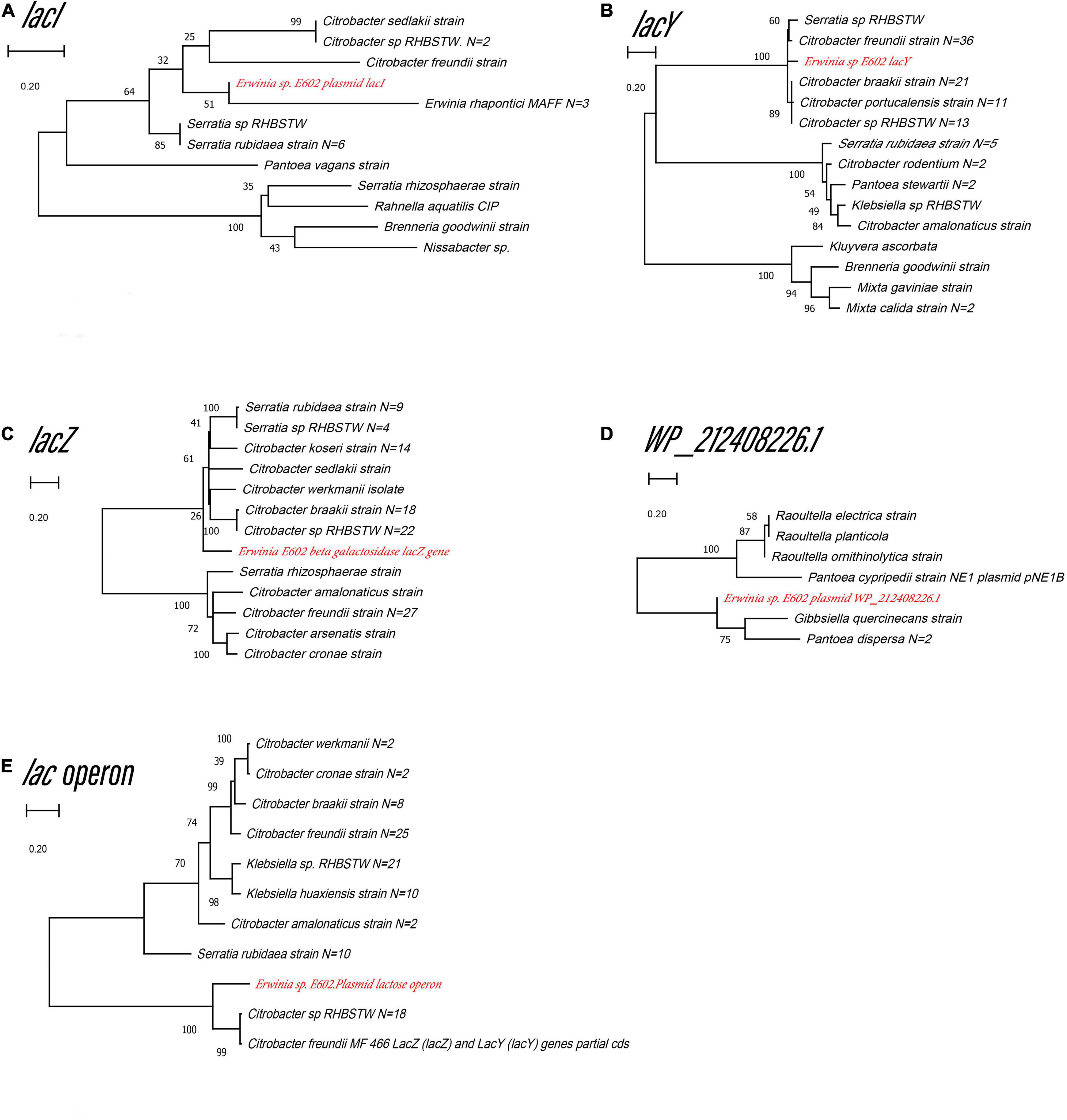
Figure 3. Unrooted maximum likelihood phylogeny of the four plasmid-borne lac operon and lacI-like (WP_212408226) genes. Phylogeny trees of the genes lacI (A), lacY (B), lacZ (C), lacI-like (WP_212408226) (D), lac operon (E) based on gene sequences. These trees were obtained using software MEGA7 by the Neighbor-Joining method. Numbers at branch nodes indicate bootstrap values of 1,000 trials (only bootstrap values above 50% were shown). Bar = 0.20 substitutions per nucleotide site.
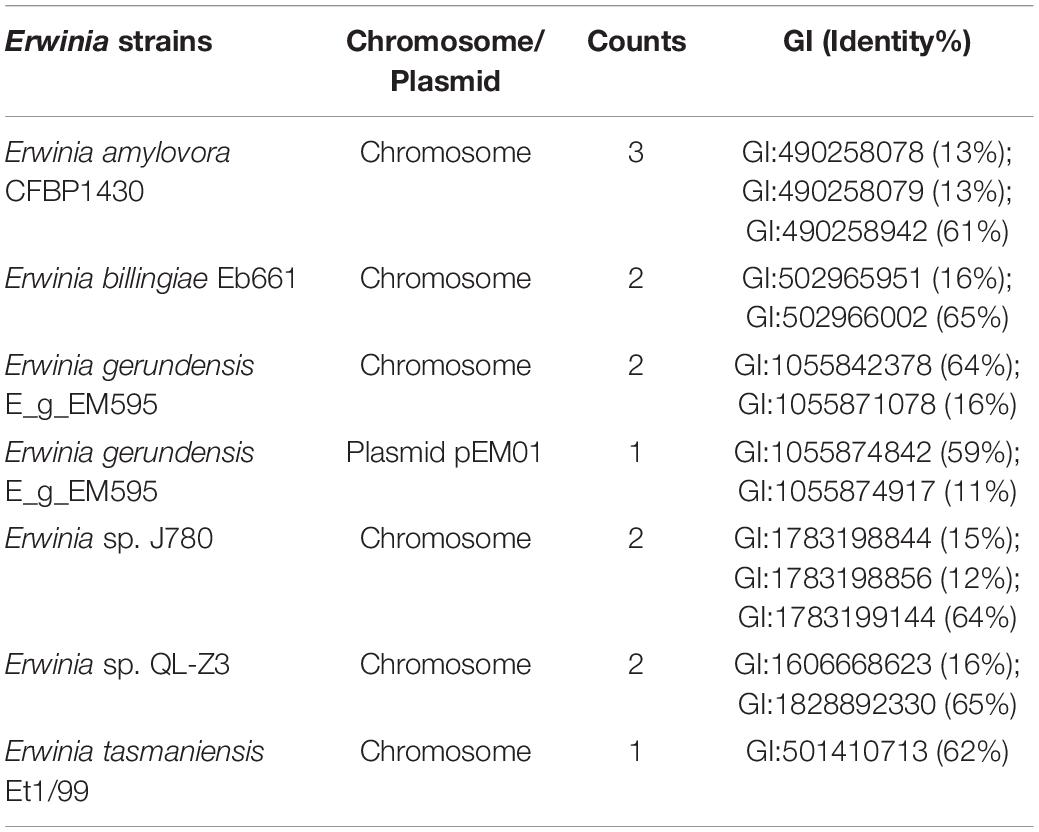
Table 4. Identity of plasmid-borne LacZ in Erwinia sp. E602 with its paralogs in other Erwinia species.
To better understand whether genes in the plasmid-borne lac operon are subject to natural selection during adaptation to the environment, positive selection sites were identified using branch-site models (model A vs. model A null) and site models (M1a vs. M2a; M7 vs. M8) implemented in Phylogenetic Analysis by Maximum Likelihood (PAML) (20). Due to fewer sequences similar to genes lacI and the lacI-like protein (WP_212408226) in other bacteria strains, we chose the top 100 sequences ranked by identity with lacY or lacZ to be included in the two datasets separately for selection pressure analysis. The results of the branch-site model comparison showed that the gene lacY had a site at codon 187 under positive selection (Table 5). Surprisingly, there are multiple sites under strong positive selection in the gene lacZ, indicating the strong positive selection pressure in it (Table 6).
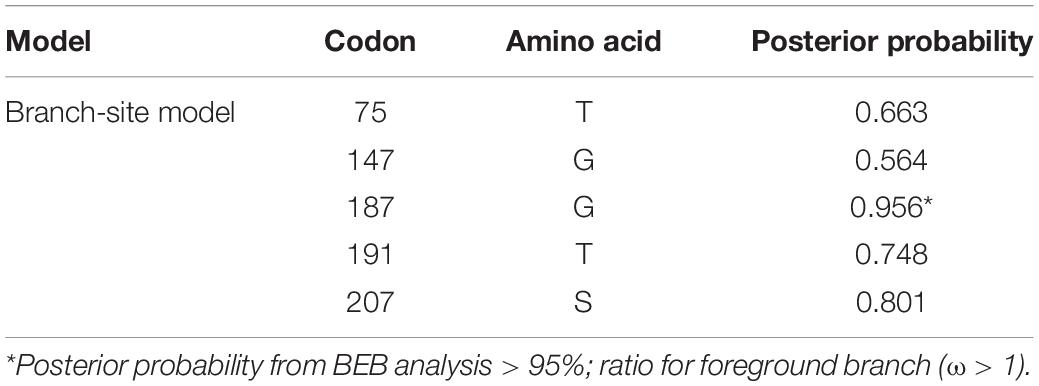
Table 5. Bayes Empirical Bayes analysis of positively selected sites identified in lacY of Erwinia sp. E602 with the branch-site model A.
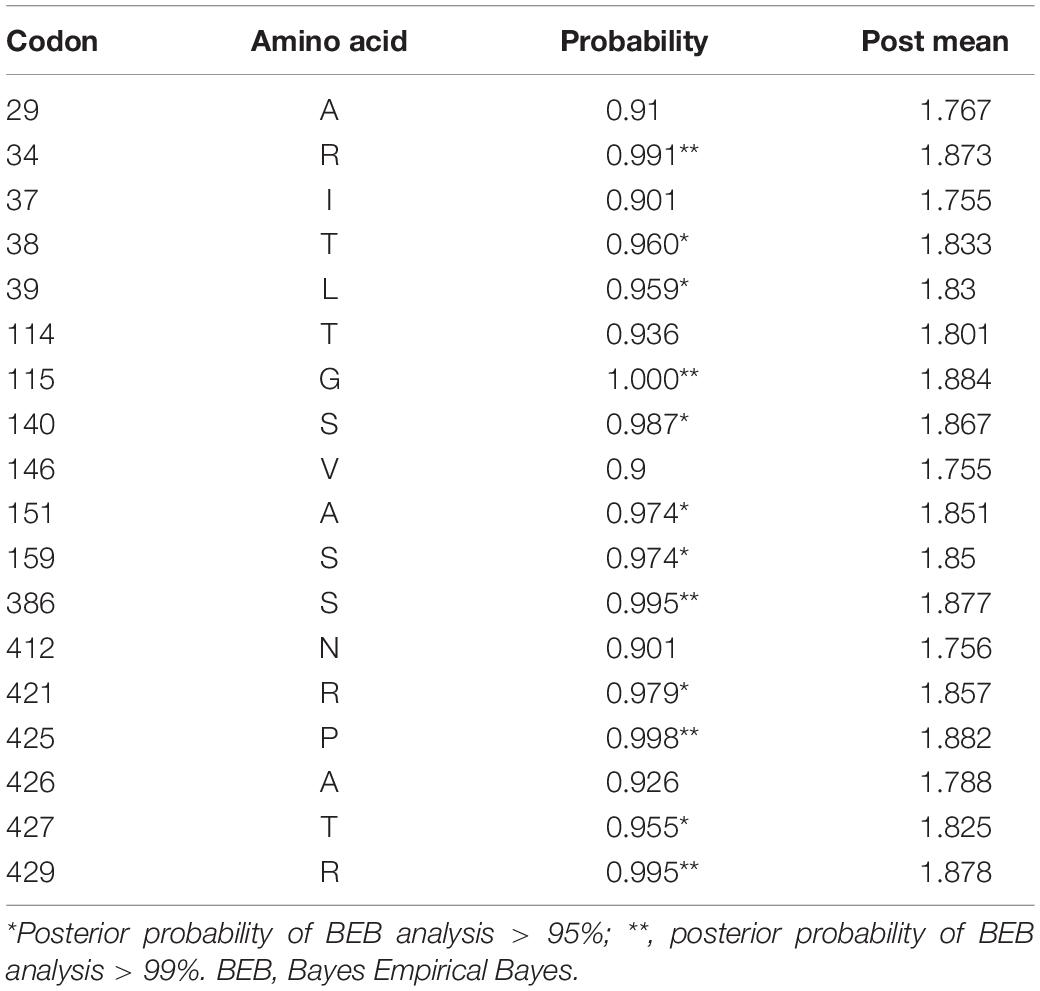
Table 6. Positively selected sites in lacZ of Erwinia sp. E602 identified with site model M2a using Bayes Empirical Bayes analysis.
By using a strategy combining Illumina and PacBio sequencing technology, the de novo assembly of the whole genome of Erwinia sp. E602 was performed. Moreover, our study characterized the largest plasmid in the genus Erwinia by far, and reported a new plasmid-borne lac operon.
From the assembly, a large plasmid with a length of 325,969 bp and a total of 297 genes was characterized. Before our current study, 6 plasmids had been reported in the genus Erwinia, among which the largest plasmid was CP037949 derived from Erwinia sp. QL-Z3. That plasmid was 149,889 bp in length and encoded a total of 124 genes.1 Therefore, the plasmid characterized in our current de novo genome assembly of Erwinia sp. E602 is the largest plasmid found in the genus Erwinia by far. In general, the genes in plasmids might provide microorganisms with potential genetic advantages. Yet the specific role of the large plasmid in Erwinia sp. E602 still remains unclear.
The lac operon is a hallmark gene of the regulatory circuit for bacteria to regulate metabolism according to nutrient conditions in the environment (Leonard et al., 2015; Malakar, 2015; Karkare et al., 2021; Pinto et al., 2021). Through genome function annotation, a large number of genes related to lactose metabolism were found in Erwinia sp. E602, a cold-adapted strain. In particular, a new plasmid-borne lac operon was characterized in this study. The lac operon shared low similarity with homologous genes in Erwinia species. It lacked a typical lacA. Instead, the position of β-galactoside transacetylase was replaced by a lacI-like gene encoding uncharacterized function. The gene lacZ in the newly identified lac operon encodes a β-galactosidase with the characteristics of low-temperature adaptation (Xia et al., 2018), which might help the organism survive in extreme environments. Similarly, identification and isolation of a 127-kb large plasmid in a rat E. coli isolate (EC93), containing the cdiI gene, would show higher toxic potency, thus helping it to exert a competitive advantage (Waneskog et al., 2021). Plasmids are mobile parts of bacterial genomes that carry genes that benefit the survival of the organism and confer selective advantages such as antibiotic resistance, thus helping bacterial communities to quickly adapt to the environment (Bogomazova et al., 2020). Similarly, the large plasmid we found in Erwinia sp. E602 also contains a group of I toxin-antitoxin modules, mainly including vagC, vapC, relE, stbD, yefM, and other related genes.
The plasmid-borne lac operon might enable Erwinia sp. E602 to utilize substrates from environmental resources for lactose metabolism, thereby gaining an adaptive advantage over competitors. Similar functions have been found in E. coli (Harwani, 2014). Nevertheless, our analysis showed that the lac operon in Erwinia sp. E602 had low sequence similarity to that of E. coli and was evolutionarily distant from its E. coli counterparts. Likewise, other studies reported the lac operons in specific strains (Vaughan et al., 1998; Bidart et al., 2018), which also suggested that the lac operon could vary in different habitats. Furthermore, the lacZ and lacY genes on the lac operon in Erwinia sp. E602 were under positive selection, which might benefit the bacterium in terms of survival or competition (Bundalovic-Torma et al., 2020). Meanwhile, the strain Erwinia sp. E602 was isolated from the high latitudes of northeast China and the enzyme encoded by the plasmid-borne lacZ gene exhibited good activity at low temperatures. These findings might be in line with the possible adaptation of this Erwinia strain to the cold environment.
The plasmid-borne lac operon lacked the lacA gene in Erwinia sp. E602. It has been reported that the transacetylase encoded by the lacA gene is not an essential element for lactose catabolism (Lagesen et al., 2007). In contrast, a lacI-like gene was found in the opposite strand downstream of the lac operon in Erwinia sp. E602. It remains to be elucidated whether the lacI-like gene contributes to the regulation of the lac operon.
In the current study, by combining Illumina and PacBio sequencing technologies, a hybrid de novo assembly was performed to obtain a more accurate and complete map of the genome of Erwinia sp. E602, providing a reference basis for subsequent transcriptome studies. Furthermore, a new plasmid-borne lac operon was characterized with evidence supporting evolution and adaptation to the environment of the genus Erwinia. The specific role of the large plasmid on Erwinia sp. E602 needs further studies.
Bacterial culture and DNA extraction of Erwinia sp. E602 were performed as described in our previous report (Xia et al., 2018). The genomic DNA was used for subsequent Illumina and PacBio sequencing.
The library for short read sequencing was prepared from 1 μg Erwinia sp. E602 genomic DNA using NEBNext Ultra DNA Library Prep Kit (NEB, Ipswich, Massachusetts) according to the manufacturer’s instructions, assessed using a Qubit® 2.0 Fluorometer (Invitrogen, Waltham, Massachusetts) and an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, California) for quality control (QC), and sequenced on the Hi-Seq 2500 platform (Illumina, San Diego, California) using a 100 bp paired-end configure. All adapter sequences and low-quality bases contained in the reads were filtered and removed using Trimmomatic (version 0.31), to keep only reads with sequencing read length more than 90 nucleotides (nt), and ensure the proportion of base quality greater than 20 (Q20) was not less than 90%.
The library for single-molecule long read sequencing was also prepared from 5 μg genomic DNA using SMRTbell DNA Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, California) according to the manufacturer’s instructions, assessed using a Qubit® 2.0 Fluorometer and an Agilent 2100 Bioanalyzer for quality control (QC), and sequenced on the PacBio RS System. Raw data was filtered to obtain clean data with a cutoff of minimum polymerase read quality 0.75 and minimum polymerase read length 3,500 bp.
The filtered Illumina reads and PacBio subreads were then used to conduct the hybrid assembly using SPAdes software (v3.5.0) (Bankevich et al., 2012). PacBio subreads were provided to SPAdes using the –PacBio option for gap closure and repeat resolution.
The software Prokka (Seemann, 2014) (v 1.14.6) was used to predict genes in the assembly results, Carbohydrate enzymes were annotated using dbCAN2 (Zhang et al., 2018). Resistance genes were annotated using the Resfams database (Gibson et al., 2015). The prediction of rRNA was performed using rnammer1.2 (Lagesen et al., 2007). The prediction of tRNA was performed by the software tRNAscan-SE-1.23 (Chan and Lowe, 2019). The sRNA annotation was obtained by the software infernal-1.1rc4 (Nawrocki and Eddy, 2013). The CGView server (Stothard and Wishart, 2005) was used as an interactive comparison genomics tool to draw the circos plots.
EggNOG v5.0 (Huerta-Cepas et al., 2019) were used for the functionally annotation of genes. Pathway enrichment analysis was performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and KEGG Orthology (KO) databases.2
The prediction of lac operon was conducted using Operon-mapper (Taboada et al., 2018).
Phylogenetic analysis was performed using software MEGA7 (Kumar et al., 2016) using the Neighbor-Joining method. The 1,000 replicates of bootstrap test were used to evaluate the clustering of taxa at branch nodes.
In order to analyze the selection pressure of the related genes, PAML (v4.1) was used to identify sites under positive selection in the genes with site and branch-site models (Yang, 2007). For the branch-site models, Erwinia. sp. E602 was selected as the foreground branch of the branch-site model, and other bacteria were used as the background branch.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YX and J-HC conceived the project, planned the experiments, and participated in the results discussions. Z-YW performed the bioinformatic analysis and improved the data analysis pipelines. RH participated in the DNA extraction from bacteria. J-HL, Z-XW, and J-DH participated in the data analysis. All authors contributed to the final manuscript.
This work was supported by the Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF) with the No. CX(19)3109, the National First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-14), Taihu Lake Talent Plan, and the Postgraduate Training Innovation Project of Jiangnan University: Project No. JNKY19072.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Special acknowledgments to Prof. Dr. Daming Fan (Jiangnan University) for providing the original soil samples for isolation of the strain Erwinia sp. E602.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.783195/full#supplementary-material
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bidart, G. N., Rodriguez-Diaz, J., Perez-Martinez, G., and Yebra, M. J. (2018). The lactose operon from Lactobacillus casei is involved in the transport and metabolism of the human milk oligosaccharide core-2 N-acetyllactosamine. Sci. Rep. 8:7152. doi: 10.1038/s41598-018-25660-w
Bogomazova, A. N., Gordeeva, V. D., Krylova, E. V., Soltynskaya, I. V., Davydova, E. E., Ivanova, O. E., et al. (2020). Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia. Int. J. Food. Microbiol. 319:108497.
Bundalovic-Torma, C., Whitfield, G. B., Marmont, L. S., Howell, P. L., and Parkinson, J. (2020). A systematic pipeline for classifying bacterial operons reveals the evolutionary landscape of biofilm machineries. PLoS Comput. Biol. 16:e1007721. doi: 10.1371/journal.pcbi.1007721
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14.
De Maio, N., Shaw, L. P., Hubbard, A., George, S., Sanderson, N. D., Swann, J., et al. (2019). Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 5:294. doi: 10.1099/mgen.0.000294
Diaz-Hernandez, O., and Santillan, M. (2010). Bistable behavior of the lac operon in E. coli when induced with a mixture of lactose and TMG. Front. Physiol. 1:22.
Gibson, M. K., Forsberg, K. J., and Dantas, G. (2015). Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216. doi: 10.1038/ismej.2014.106
Harwani, D. (2014). Regulation of gene expression: Cryptic-glucoside (bgl) operon of Escherichia coli as a paradigm. Braz. J. Microbiol. 45, 1139–1144.
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Karkare, K., Lai, H. Y., Azevedo, R. B. R., and Cooper, T. F. (2021). Historical contingency causes divergence in adaptive expression of the lac operon. Mol. Biol. Evol. 38, 2869–2879. doi: 10.1093/molbev/msab077
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Leonard, S. R., Lacher, D. W., and Lampel, K. A. (2015). Acquisition of the lac operon by Salmonella enterica. BMC Microbiol. 15:173. doi: 10.1186/s12866-015-0511-8
Malakar, P. (2015). Pre-induced Lac operon effect on non specific sugars: pre-culture effect is dependent on strength of induction, exponential phase and substrate concentration. Open Microbiol. J. 9, 8–13. doi: 10.2174/1874285801509010008
Marbach, A., and Bettenbrock, K. (2012). lac operon induction in Escherichia coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J. Biotechnol. 157, 82–88.
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. doi: 10.1093/bioinformatics/btt509
Pinto, C., Melo-Miranda, R., Gordo, I., and Sousa, A. (2021). The selective advantage of the lac operon for Escherichia coli is conditional on diet and microbiota composition. Front. Microbiol. 12:709259. doi: 10.3389/fmicb.2021.709259
Rhoads, A., and Au, K. F. (2015). PacBio sequencing and its applications. Genom. Proteom. Bioinf. 13, 278–289. doi: 10.1016/j.gpb.2015.08.002
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Stothard, P., and Wishart, D. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Taboada, B., Estrada, K., Ciria, R., and Merino, E. (2018). Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 34, 4118–4120. doi: 10.1093/bioinformatics/bty496
Vaughan, E. E., Pridmore, R. D., and Mollet, B. (1998). Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 180, 4893–4902.
Waneskog, M., Halvorsen, T., Filek, K., Xu, F., Hammarlof, D. L., Hayes, C. S., et al. (2021). Escherichia coli EC93 deploys two plasmid-encoded class I contact-dependent growth inhibition systems for antagonistic bacterial interactions. Microb. Genom. 7:534. doi: 10.1099/mgen.0.000534
Xia, Y., He, L., Mao, J., Fang, P., Ma, X., and Wang, Z. (2018). Purification, characterization, and gene cloning of a new cold-adapted β-galactosidase from Erwinia sp. E602 isolated in northeast China. J. Dairy Sci. 101, 6946–6954. doi: 10.3168/jds.2018-14605
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Keywords: Erwinia, hybrid sequencing, genome assembly, lac operon, bioinformatic analysis
Citation: Xia Y, Wei Z-Y, He R, Li J-H, Wang Z-X, Huo J-D and Chen J-H (2021) Hybrid de novo Genome Assembly of Erwinia sp. E602 and Bioinformatic Analysis Characterized a New Plasmid-Borne lac Operon Under Positive Selection. Front. Microbiol. 12:783195. doi: 10.3389/fmicb.2021.783195
Received: 25 September 2021; Accepted: 21 October 2021;
Published: 11 November 2021.
Edited by:
Hui Wu, East China University of Science and Technology, ChinaReviewed by:
Rongming Liu, University of Colorado, Boulder, United StatesCopyright © 2021 Xia, Wei, He, Li, Wang, Huo and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Xia, eXV4aWFAamlhbmduYW4uZWR1LmNu; Jian-Huan Chen, Y2poX2Jpb0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.