
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 January 2022
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.783103
This article is part of the Research Topic Combined Toxicity and Single Toxicity of Mycotoxins View all 4 articles
 Md Atiqul Haque1,2†
Md Atiqul Haque1,2† Fei Wang1†
Fei Wang1† Yi Chen1
Yi Chen1 Foysal Hossen3
Foysal Hossen3 Md Aminul Islam3
Md Aminul Islam3 Md Amzad Hossain3
Md Amzad Hossain3 Naila Siddique4
Naila Siddique4 Cheng He1*
Cheng He1* Firoz Ahmed3*
Firoz Ahmed3*The current study provides information on Bacillus spp. contamination along with present status in commercially available poultry and animal feeds as well as animal-derived products in Bangladesh. The research has been conducted to determine if animal feed and its components are a source of Bacillus spp. contamination in feed and food chain. Out of 180 different feeds, milk, egg, and human stool samples, 218 Bacillus spp. were isolated and identified by cultural morphology, microscopic, biochemical, and molecular characteristics where B. cereus, B. subtilis, B. amyloliquefaciens, B. licheniformis, B. thuringiensis, B. megaterium, and B. coagulans accounted for 51, 22, 9.1, 5.9, 5, 3.6, and 2.2%, respectively. Regarding the enumeration of total viable count and total Bacillus count, correspondingly 67 and 39% samples were found to be contaminated with above 10,000 CFU/g, while highest contamination was 85 and 75% in broiler feed, respectively. The total number of bacteria above the regulatory limits in commercially available feeds indicates a poor compliance with regulation and abuse administration in the Bangladeshi market. Moreover, a hospital-based survey showed that food-borne Bacillus spp. contributed to 4.5% human diarrhea cases and 25% food contamination associated with vegetables, rice, RTE food, milk, and egg, accounting for 46, 34, 14, 4, and 2%, respectively. B. cereus was the dominant isolate correspondingly accounting for 56 and 51% egg and milk contamination followed by B. amyloliquefaciens (32%) and B. thuringiensis (12%) in egg and B. subtilis (25%), B. amyloliquefaciens (12%), B. thuringiensis (6.4%), and B. coagulans (3.2%) in milk, respectively. Toxin gene profiling of Bacillus spp. revealed that B. cereus constituted a principal part of virulence, while B. thuringiensis, B. licheniformis, B. megaterium, B. coagulans, and B. subtilis showed genetic diversity and B. amyloliquefaciens had not carried any toxin gene. Detection rate of enterotoxin genes (nheA, nheB, nheC, cytK, hblA, hblC, hblD, and entFM) showed that 55% isolates carried nheABC genes, 80% entFM, and 71% cytK, whereas only 33% of the isolates contained hblACD gene clusters. These virulence genes were posing a threat to human health due to spread across the food and feed chain. Finally, our findings support the hypothesis that B. cereus might contribute to clinical diarrhea, gizzard erosion, and lung infection in duck and poultry, and that it contaminates animal-derived foods resulting in toxicity and antibacterial resistance to humans. Therefore, maximal tolerance limits of Bacillus spp. and their potential risks to the animal industry are urgently needed to clarify. Moreover, Bacillus spp.–induced toxin residual must be altered for human health via food chain transmission.
Bacillus spp. are Gram-positive, rod-shaped, motile (flagellated), aerobic or facultative anaerobic, spore- and biofilm-forming bacteria commonly found in nature, isolated from fermented or unfermented food and feed stuffs, that have caught attention as potential probiotics, which have been patented in the form of a wide range of health supplements. Probiotics are claimed to provide health benefits for the hosts and commonly fed to humans, animals, and plants to increase the production efficiency in livestock, poultry, and aquaculture combating gastrointestinal infections and alternatives to antibiotics (Elshaghabee et al., 2017; Cui et al., 2019; Gupta et al., 2019; Haque et al., 2021). Bacillus spp. have the ability to produce antimicrobial substances as a probiotic and ensure its stability and vitality in different habitats in comparison with conventional commercial probiotics. Bacillus spp., especially B. toyonensis, B. licheniformis, B. subtilis, B. amyloliquefaciens, B. thuringiensis, B. mycoides, B. coagulans, B. clausii, and B. pumilus, have been widely used as probiotics in humans, plants, and animal production (Cui et al., 2019; Gupta et al., 2019; Cao et al., 2020).
While probiotic Bacillus strains have been shown to inhibit the growth of pathogenic bacteria (E. coli, Clostridium, Streptococcus, and Salmonella), certain species, especially those belonging to the B. cereus group, are considered to be unsafe due to the possibility of toxin genes (nhe, hbl, cytK1, ces) and antibiotic resistance genes (tet45, erm34, aadD2, blaBCL–1, catBcl) being transferred from commercial probiotic products (Zhu et al., 2016; Gupta et al., 2019; Zuo et al., 2020; Deng et al., 2021). Although some countries are developing guidelines to ensure the safety of probiotic products, lack of legal inspection or requirement to demonstrate efficacy may pose a threat of getting B. cereus contamination in animal-used probiotics, followed by increasing hazard to food safety. Hence, it is necessary to determine whether Bacillus spp. strain is pathogenic or beneficial, as well as their connection to those with generally recognized as safe (GRAS) status. Correspondingly, various toxins must be investigated to ensure its safety before adding into animal feeds and human medications. Furthermore, B. cereus was a leading cause of food-borne illness in the United States and the third most common food pathogen in China, which causes two distinct gastrointestinal diseases, emesis and diarrhea, as well as many systemic and local complications including hemorrhagic necroses in the central nervous system, fulminant bacteremia, pneumonia, endophthalmitis, and gas gangrene-like cutaneous symptoms in both immune-deficient and immune-competent individuals (Cui et al., 2019; Li et al., 2020; Deng et al., 2021; Haque et al., 2021). In 2018, B. cereus was found in 98 reported outbreaks across EU member States, accounting for 1.9% of all outbreaks, affecting 1,539 people, 111 hospitalizations, and 1 death (Rodrigo et al., 2021). Food-borne B. cereus poisoning includes animal-derived products (poultry, meat and meat products, puddings, sauces, milk and milk products, pasteurized liquid eggs, fish, stew) and plant-based foods (rice, potatoes, cereals, grains, spices, vegetables, salads) (Haque et al., 2021). The survival potential of B. cereus spores in extreme environment poses a significant risk to food safety and also causes economic losses to the food industry. In addition, feed-borne B. cereus contamination worsens extreme diarrhea and malnutrition in poultry by causing gizzard erosion and ulceration syndrome, as well as hemorrhagic inflammation in lungs and immunosuppression co-infection with other pathogens (Zuo et al., 2020; Haque et al., 2021). It can lead to major food safety concerns because the formation of spores, biofilms, and various virulence potentials, like heat-tolerant/labile emetic and/or diarrheal toxins and tissue-destructive enzymes, makes it difficult to fully prevent their presence in food (Haque et al., 2021). A recent study in Bangladesh revealed that residues of antimicrobial used against B. cereus and B. subtilis were found in 68% of liver and 66% of kidney layers, respectively. More interestingly, 33% animal probiotics carried potential pathogens, including human intestinal cya toxin gene in the vicinity of chicken and fish farm in a recent Chinese survey (Li et al., 2020; Hassan et al., 2021). Most Bacillus spp. (B. cereus, B. subtilis, B. licheniformis) are antibiotic resistant and transfer antibiotic resistance genes to humans, implying that antibiotic residues may enter consumer food products and the human food chain (Deng et al., 2021). This can result in the emergence of multidrug-resistant bacterial strains, with the possibility of resistance gene transfer to other pathogenic and non-pathogenic bacteria (Islam et al., 2017; Haque et al., 2021). Several research on the prevalence of Bacillus spp. in various Bangladeshi foods have been conducted, but the isolate’s ability to produce emetic and enterotoxins has yet to be determined. Our hypothesis for this study is to explore the key information on contamination levels and toxigenic profiles of isolated Bacillus spp. in animal feeds, animal-derived products, and human stool that will help us better understand the pathogen’s current status and pathogenic potential in Bangladesh. Our findings will be used to assess the risk and apply food safety measures to ensure food security.
The current study was conducted in Noakhali district (22.70° N, 91.10° E) of Chittagong division, about 151 km from Dhaka, the capital of Bangladesh. A total of 180 samples were collected during September 2019 to June 2020 from various sources, including layer (n = 20), broiler (n = 20), duck (n = 13), fish (n = 15), and cattle (n = 20) feed, table egg (n = 20), milk (n = 20), and human stool (50) in different locations across the district. All these samples were collected from layer farms, broiler farms, duck farms, cattle farms, fish farms, selling outlet stores, and a human hospital (Supplementary Table 1). Aseptic procedures were maintained while collecting these samples, with amounts varying by types: 100 g for feed, 20–30 ml for milk, 1 g for stool, and a whole egg. During sample collection, sterile gloves were used to handle feed, egg, and milk samples from each source. In case of milk samples, spot collection was employed; raw milk was gathered during milking of cow into sterile wide-mouth tubes, corked, and carefully labeled. In case of stool samples, sterile screw cap vials were used to avoid environmental contamination. Each sample was immediately kept in sterile Ziploc plastic bags, transported in an insulated foam box with cold chain (temperature, 4–6°C) to the Microbiology Laboratory, Noakhali Science and Technology University, Noakhali, Bangladesh. Upon arrival, all samples were refrigerated at 4°C until microbiological analysis, which was completed within 24 h after receiving the samples.
A cross-sectional survey was conducted to enroll participants for interviews and 38 farms were randomly chosen, including layer (n = 15), broiler (n = 15), and duck farms (n = 8). Data on the various types of farms, the age and populations of birds, the type of housing, feeds, egg production, feed additives and antibiotics, feed source, and clinical history were collected using a questionnaire survey (Supplementary Appendix I). A structured data collection schedule was also used to obtain information on diarrheal patients and types of food consumed (Supplementary Appendix II). After getting informed cell phone consent, the researcher interviewed patients to collect data. The experimental protocols were approved by an Ethical Reviewing Board on Institutional Animal Care and Use Committee at Noakhali Science and Technology University. Our survey of 38 farms revealed that majority of the farmers were small-scale producers of broiler (53%) and duck farms (85%) with a maximum of 2,000 birds, and medium-scale producers of layer farms (73%) with a maximum of 5,000 birds compared with the multiple ages broiler farms (6.6%); 100% of the surveyed layer and duck farms had an all-in all-out breeding system, while 93% of broiler farms had the same raising system (Supplementary Tables 2–4).
Microbiological qualities of the samples such as total viable count (TVC) and total Bacillus count (TBC) were determined using the method described by USDA/FSIS (Microbiology Laboratory Guidebook [MLG], 2015). All the glassware used in this study were sterilized by autoclaving at 121°C for 15 min and then cooling to 45°C. The system was also used for serial dilution, inoculation and incubation, sub-culture, Gram staining, and identification of isolates. Pure cultures were stored at −80°C in glycerol stocks for further study.
The samples were first cultured into Nutrient Agar (NA) (HiMedia India) for TVC and Mannitol–Egg Yolk–Phenol Red–Polymyxin Agar (MYPA) (HiMedia, India) for TBC. The feed samples were homogenized in peptone water in the beaker as described previously (Gyang et al., 2019) by thoroughly mixing 1 g of each sample in 9 ml of peptone water. In case of table eggs, yolks were treated as described previously (Khan et al., 2016). Briefly, the eggs were soaked in 75% alcohol for 5 min before being sterilized for 5–10 s on a hot flame. The egg yolk was then drained into the sterilized polythene bags and manually homogenized after a small hole was cut on the shell surface. In case of milk, 1 ml of sample was homogenized with 9 ml of sterile buffered peptone water (Oxoid, United Kingdom) using a vortex mixture. In case of stool samples, 1 g of stool is diluted in 9 ml sterile buffered peptone water (Oxoid) and homogenized using a vortex mixture for primary enrichment. Finally, each sample was diluted in phosphate buffered saline (PBS) at a ratio of tenfold (1:10) serial dilution. This was accomplished by adding 1 ml homogenized sample to 9 ml PBS, mixing thoroughly with vortexing and repeating until 10–6 dilution was obtained (Abraha et al., 2017). These acceptable dilutions were cultured on NA (HiMedia) and MYPA (HiMedia) media using the spread plate technique (Microbiology Laboratory Guidebook [MLG], 2015) and incubated overnight at 37°C in the incubator. Following incubation, plates with 25–250 colonies were counted and TVC as well as TBC were expressed as colony-forming units per gram of sample (CFU/g).
From the dilution tubes, 10 μl of each dilution was spot dropped in triplicate onto MYPA (HiMedia) for Bacillus spp. isolation. The plates were incubated at 37°C for 24 h and following incubation colonies on the media were studied morphologically, including size, shape, surface texture, edge, elevation, color, and opacity. To identify and validate the isolated colonies, the Gram staining and following biochemical tests were performed: Triple Sugar Iron (TSI) Test, Motility Indole Urease (MIU) Test, Oxidase Test, Catalase Test, Voges Proskauer (VP) Test, Simon Citrate Agar Test, and Starch Hydrolysis Test as described previously (Zhang et al., 2019). For identification and confirmation, all presumptive colonies of Bacillus spp. were purified and subjected to morphological and biochemical tests as defined in A Color Atlas of Bacillus Species (Parry et al., 1988).
Genomic DNA was extracted by boiling method as described previously (Hu et al., 2021). Briefly, positive colonies grown overnight in nutrient broth at 37°C, then 1 ml was taken in Eppendorf tube followed by centrifuging at 13,000 rpm for 10 min and resuspending in 200 μl sterile Milli-Q water (Sigma Aldrich). Then the resuspended bacterial cell suspension was boiled at 100°C for 10 min, placed on ice immediately for 10 min, and finally centrifuged at 10,000 rpm for 10 min to obtain the genomic DNA of each isolate. The concentration of DNA was determined with a Colibri LB-915 Microvolume Spectrophotometer (Berthold Technologies, Germany) and the sample was diluted to a final concentration of approximately 100 ng/μl. Toxin gene profiling and distribution of emetic and enterotoxin gene of Bacillus spp. was evaluated by PCR. The targeted genes included non-hemolytic (nheA, nheB, nheC) and hemolytic (hblA, hblC, hblD) complexes, CytK, entFM, and Ces. PCR protocols were performed as previously described (Ehling-Schulz et al., 2006; Sergeev et al., 2006; Table 1). Bacillus cereus ATCC 14579 was used as positive control and sterile Milli-Q water (Sigma Aldrich) was used for negative control. The PCR mixture was prepared in a volume of 25 μl, with OneTaqQuick-Load 2 × Master Mix (New England Biolabs Inc., United States), 0.2 μmol L–1 final concentration of each primer, and 2.5 μl of prepared DNA template. PCR was performed on T100 Thermal cycler (Bio-Rad, United States). The PCR products were separated on 1.5% agarose gel (MP Biomedicals LLC, United States) using Mini-Sub Cell GT Horizontal Electrophoresis System (Bio-Rad, United States), stained with ethidium bromide, visualized on a UV transilluminator (Gel Doc EZ), and photographed by gel documentation system.
Data are expressed as mean ± SEM and analyzed using SPSS v.25 software. The one-sample t-test and χ2 test were applied to determine the cumulative difference among the parameters studied. Statistically significant difference was judged as *p < 0.05 and **p < 0.01.
Frequency of Bacillus spp.–positive samples and TVC and TBC of various feeds, eggs, milk, and stool samples are shown in Tables 2, 3, respectively. Positive Bacillus spp. were found in 100% of layer feed, broiler feed, egg, and milk, as compared with 80, 73, 70, and 16% in duck feed, fish feed, cattle feed, and human stools, respectively. In terms of TVC, 67% contaminated samples yielded more than 10,000 CFU/g, with the highest and lowest contamination rates being 85 and 55% in broiler feed (p < 0.01) and egg, respectively. In terms of TBC, 39% contaminated samples were above 10,000 CFU/g, with the highest and lowest contamination rates being 75 and 5% in broiler feed (p < 0.01) and egg, respectively.
Regarding Bacillus spp., B. cereus, B. subtilis, B. amyloliquefaciens, B. licheniformis, B. thuringiensis, B. megaterium, and B. coagulans were isolated from layer, broiler, duck, cattle and fish feed, egg, milk, and human stool. Out of 218 isolates, 112 strains (51%) of B. cereus, 49 strains (22%) of B. subtilis, 20 strains (9.1%) of B. amyloliquefaciens, 13 strains (5.9%) of B. licheniformis, 11 strains (5%) of B. thuringiensis, 8 strains (3.6%) of B. megaterium, and 5 strains (2.2%) of B. coagulans were identified by morphological and biochemical tests (Table 4 and Supplementary Table 5). Obviously, B. cereus, B. subtilis, and B. amyloliquefaciens were dominant distributions in all the tested samples, accounting for 181 isolates (83%) (Table 4).
More interestingly, Bacillus spp. from human diarrhea and food contamination (egg and milk) had 4.5 and 25% positivity, respectively. Regarding human diarrhea, B. cereus was dominant among the isolates (70%), followed by B. subtilis (30%). Likewise, 56% eggs and 51% milk were determined to have B. cereus contamination, followed by B. amyloliquefaciens (32%) and B. thuringiensis (12%) in egg, and B. subtilis (25%), B. amyloliquefaciens (12%), B. thuringiensis (6.4%), and B. coagulans (3.2%), respectively, in milk (Table 4).
Toxin genes of isolated Bacillus spp. by PCR test as indicated in Figures 1A–D, 2A–D showed the specific amplified target band. The toxin gene profiles of Bacillus spp. were classified into 24 different types of isolates suggesting genetic diversity (Table 5). The distribution of virulence genes among 218 Bacillus spp. isolates is shown in Supplementary Tables 6–7 and Figures 3, 4. Positivity of entFM (80%) gene was higher among the enterotoxin genes than other genes of nheB (77%), nheA (71%), cytK (71%), nheC (66%), hblA (57%), hblD (48%), and hblC (39%). Regarding nhe-based gene clusters, 64% of the strains harbored enterotoxigenic gene nheAB compared with 61% nheAC, 59% nheBC, and 55% nheABC, whereas 17% of the isolated did not possess nhe-based genes. For the hbl-based gene clusters, only 44% of the isolates possessed hblAD, 36% hblAC, 33% hblCD, and 33% hblACD, whereas 35% of the isolates did not possess an hbl-based gene. All the Bacillus spp. isolates were negative for emetic toxin gene Ces. Distribution of virulence gene in the B. cereus isolates obtained from animal feed correspondingly possessed 73, 56, 34, 72, 54, 36, 46, and 84% for nheA, nheB, nheC, cytK, hblA, hblC, hblD, and entFM genes. Furthermore, B. cereus carried virulence genes with the rate of 63, 36, 26, 66, 46, 40, 50, and 80% in animal-derived foods and 71, 42, 28, 85, 57, 42, 28, and 71% in human diarrheal case for nheA, nheB, nheC, cytK, hblA, hblC, hblD, and entFM, respectively. In contrast, two other dominant isolates, B. subtilis and B. amyloliquefaciens, had no major toxin gene; only 33% strains of B. subtilis carried hblD in human diarrheal cases and 25, 28, and 33% of nheA were identified from the animal feed, animal-derived food, and human diarrheal cases, respectively (Supplementary Table 8).
Figure 1. Toxin genes of isolated Bacillus spp. by PCR test. (A) (nheA gene, L1: 100 bp marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control); (B) (nheB gene, L1: 100 bp marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control); (C) (nheC gene, L1: 100 bp marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control); (D) (cytK gene, L1: 100 bp marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control).
Figure 2. Toxin genes of isolated Bacillus spp. by PCR test (continued). (A) (hblA gene, L1: 100 bp marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control); (B) (hblC gene, L1: 1 kb marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control); (C) (hblD gene, L1: 1 kb marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control); (D) (entFM gene, L1: 100 bp marker, L2: positive control, L3: sample 1, L4: sample 2, L5: sample 3, L6: sample 4, L7: sample 5, L8: negative control).
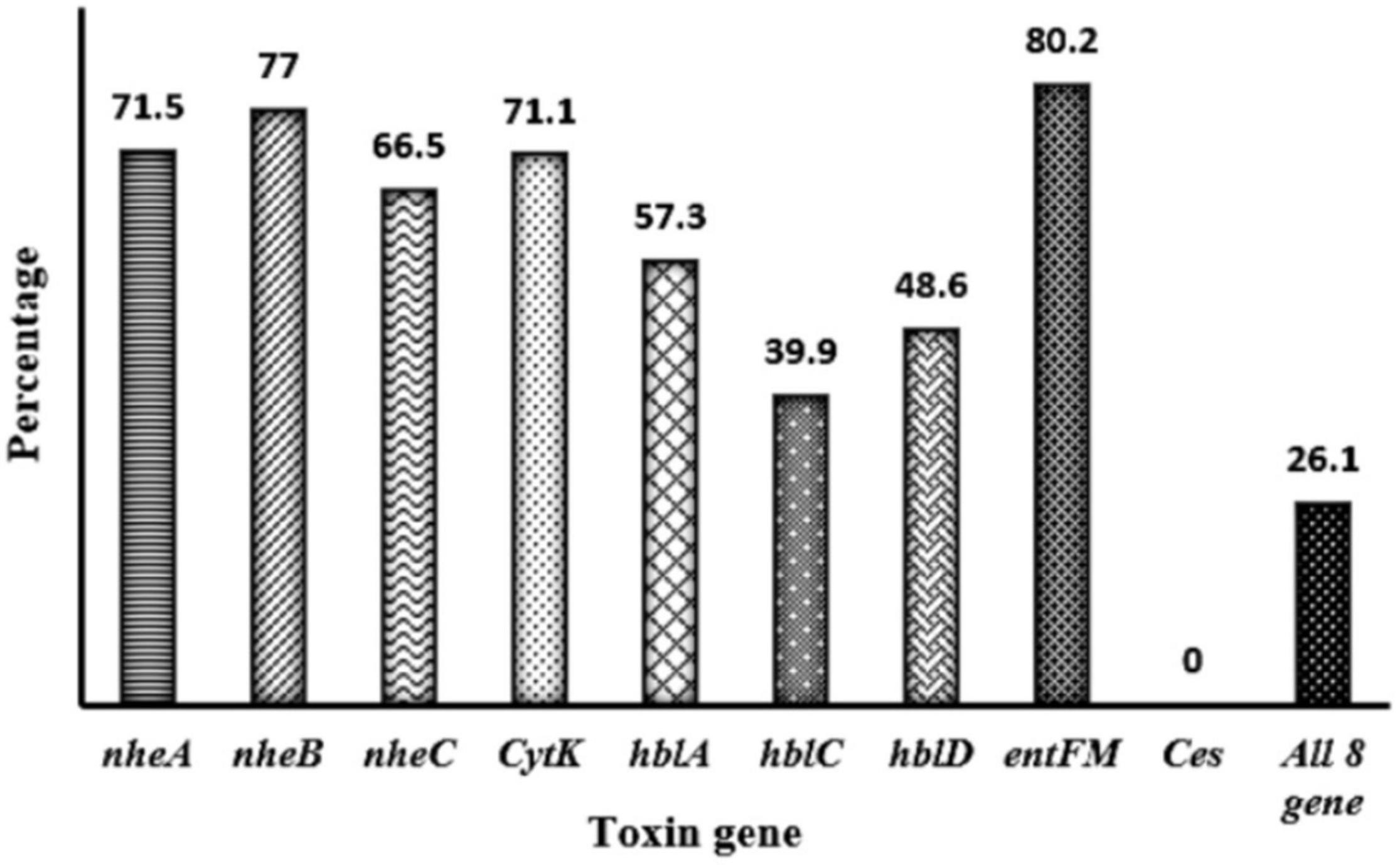
Figure 3. Distribution of toxin gene of Bacillus spp. isolated from animal feed, animal-derived products, and human stool in Bangladesh. The number at the top of the bars represents the positive rate of the corresponding toxin genes. “All eight genes” presents the strains containing all the detected toxin genes.
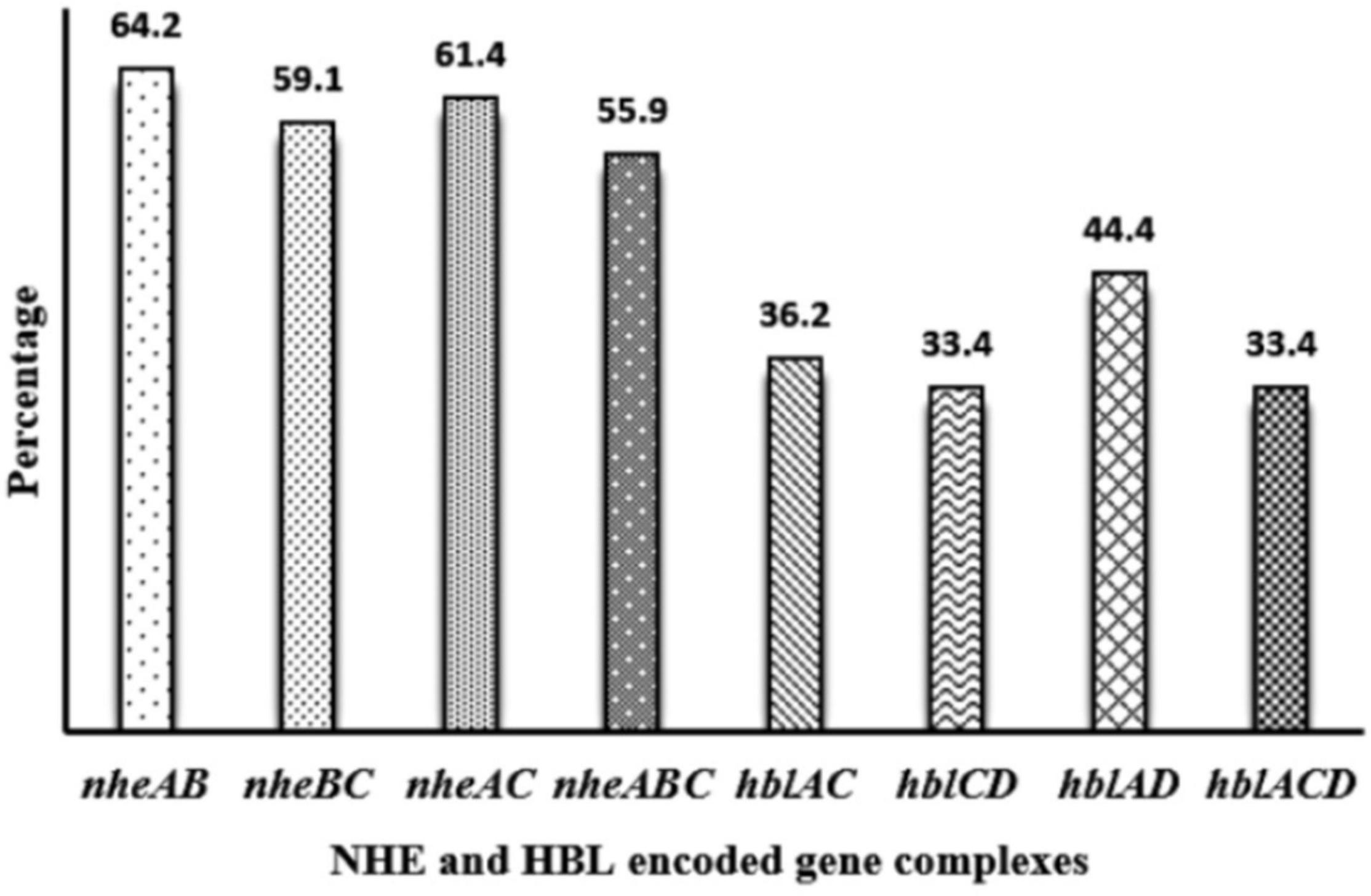
Figure 4. Distribution of nhe-based gene and hbl-based gene cluster of Bacillus spp. isolated from animal feed, animal-derived products, and human stool in Bangladesh. The number at the top of the bars represents the positive rate of the corresponding toxin gene clusters.
Regarding Bacillus spp. contamination in broiler and duck production parameters, at least 20% farms were affected with diarrhea (p < 0.05) with moderate decrease in growth and feed conversion rate. Moreover, mortality (p < 0.05) was recorded to reach 20% in broiler and 12% in duck farm and more than 2% delay of slaughtering time was observed (Table 6). In contrast, Bacillus spp. contamination was associated with layer production parameters; 13% farms were affected with diarrhea (p < 0.01), small decrease in egg production, and low-quality eggs (p < 0.05), as well as a slight increase in culling rate (p < 0.05) and mortality (p < 0.01) in 13% farms of over 2% (Table 7). Regarding hospital-based survey, clinical diarrhea cases due to B. cereus (70%) and B. subtilis (30%) infection were linked to consuming diets while vegetables, rice, RTE food, milk, and eggs were accounted for 46, 34, 14, 4, and 2%, respectively (Tables 4, 8).
Necropsy revealed mortality on two affected ducks during field investigation. Postmortem findings showed ulceration on gizzard and inflammation and hemorrhagic lesions on lungs caused by the bacteria (Supplementary Figures 1, 2). On culture from the gizzard and lungs, B. cereus was found to be responsible for gizzard erosion and lung inflammation (Supplementary Figure 3). Bacillus spp. were isolated from table eggs and human stools (Supplementary Figures 4, 5).
Food-borne diseases are likely one of the most serious public health problems in Bangladesh concerning food safety challenges and their associated economic and social costs (Chowdhury et al., 2021). This is the first study of the Bacillus spp. contamination levels in poultry (layer, broiler, and duck) feed, cattle feed, animal products (milk and eggs), and human stool in Bangladesh. In the present study, 51% of all samples were B. cereus contamination, which was more or less the same as those of previous surveys in other types of foods elsewhere, i.e., 20–48% in Korea, 50% in China, 52% in Spain, and 57% in Mexico (Yu et al., 2019). Moreover, 55% isolates harbored nheABC and 33% isolates carried hblACD gene clusters, respectively, while 80% isolates harbored entFM gene and 71% isolates possessed cytK gene. Our findings confirmed that feed-borne B. cereus might cause clinical diarrhea in duck and poultry by producing diverse toxins, contributing to lower productivity, toxin residual in eggs and milk, and the threat to public health via feed chain. On the other hand, the maximal tolerance limit of B. cereus in most of food is set up by the regulatory authorities, 102– < 103 (Food Standard Australia and New Zealand), <104CFU/g (European Food Safety Authority), <103– ≤ 105 (Health Protection Agency United Kingdom, Center for Food Safety, Hong Kong, China) as mentioned in Haque et al. (2021). The widespread distribution of B. cereus including thermoresistant spores attributable to the ingestion of foods contaminated with this pathogen containing >104 CFU/g is not acceptable and unsafe for human consumption. Bacillus spp. are found in vegetables, rice, animal products (milk and eggs), and RTE food as a result of a variety of factors, including soil or air contact during cultivation, transportation, and distribution; spore or cell transfer in the dairy farm via feed and bedding material; raw milk contamination during the milking procedure, from within or exterior of the udder, and the surface of the milk handling utensils; and subsequent temperature abuse during shipment. The inclusion of cracked and contaminated eggs, cross-contamination, and poor hygienic conditions may result in the occurrence of B. cereus in these foods (Soni et al., 2016; Islam et al., 2017; Yu et al., 2019; Haque et al., 2021). Hence, it is of great concern that food safety criteria for B. cereus were not met in 70% of the positive milk samples and 5% of egg, thus signaling a potentially dangerous risk. More interestingly, 46% human diarrheal cases were attributed to consumption of vegetable diet contaminated with B. cereus, which is similar to findings in several countries (Yu et al., 2019). B. cereus spore adhesion and resistance to external environment can lead to biofilm formation, where vegetative cells may survive on vegetables, when slow cooling and prolonged storage at room temperature have been documented to enable the spores to germinate and develop in cooked rice (Soni et al., 2016; Antequera-Gómez et al., 2021). RTE foods are highly susceptible to contamination during handling, direct contact with food-contact surfaces, and dirty cutting utensils (Valero et al., 2016).
Diarrhea caused by B. cereus is attributed to different enterotoxins produced by the strains in the small intestine including HBL, NHE, and CytK. In this study, eight enterotoxin genes and the presence of entFM gene was highest (80%), which was in agreement with other reports (Yu et al., 2019; Zhao et al., 2020; Hu et al., 2021; Kong et al., 2021; Sornchuer and Tiengtip, 2021). The frequency of nheABC (55%) and hblACD (33%) gene clusters in our study was lower than those reports from different foodstuffs in Thailand, Korea, and China. The frequency of cytK (71%) was higher than that observed on market food in Thailand (68%), China (45%), Italy (41%), and Tunisia (37%) but lower than that of the pasteurized milk (73%), meat and meat products (82%), and vegetables (87%) in China and milk products (75%) in Ghana (Owusu-Kwarteng et al., 2017; Gdoura-Ben Amor et al., 2019; Proroga et al., 2019; Yu et al., 2019; Hu et al., 2021; Kong et al., 2021; Sornchuer and Tiengtip, 2021). Bacillus spp. have been identified on a variety of substrates, including soil, plant-based foods, corn- and soybean-based fermented feedstuffs for livestock and poultry, and fermented functional food for human consumption harboring the blend of Bacillus spp. (Elshaghabee et al., 2017).
The distribution of enterotoxin genes in B. cereus isolated from different feed, animal-derived products, and human stools suggested that food poisoning by the diarrheal toxin-producing strains cannot be neglected and new probiotic candidate of B. cereus group as food and feed additive must be demonstrated with safety evaluation of no toxic potentials. In addition to B. cereus, B. subtilis, B. amyloliquefaciens, B. licheniformis, B. thuringiensis, B. megaterium, and B. coagulans have all been linked to food safety issues (Haque et al., 2021). In Bangladesh, approximately 94% of poultry farmers use antibiotics indiscriminately in their farms as a growth promoter, treatment, and prophylaxis, and they seem to be more cautious about the potential costs of meat and egg production (Hosain et al., 2021). Moreover, the use of antibacterial agents in veterinary practice is not strictly controlled, and farmers abuse antibiotics to control poultry disease. In our survey, at least 20, 60, and 12% of farms were confirmed to abuse antibiotics for medication, prophylaxis, and both purposes without withdrawal time, respectively (Supplementary Tables 2–4). Bacillus spp. contaminated in animal products can act as reservoir for toxic and virulent genes, increasing the likelihood of developing resistant bacteria in humans. Considering the overall situations like B. cereus and Bacillus spp. mediated contaminations and spreading of toxin gens followed by aberrant antibiotic uses and mounting resistance, immediate integrated measures are required for the developing world. Contamination of probiotic-borne Bacillus spp. can be regulated by specific fermentation parameters such as sampling material composition, subsequent culture processes, fermentation properties, post-operation techniques, and utilization of bacterial peptide bacteriocins with a broad range of antibacterial action against B. cereus (Haque et al., 2021). Despite the fact that the Bangladesh Food Safety Authority (BFSA) has recently released food safety regulations and guidelines, there is still much work to be done in terms of raising awareness and putting the animal feeds and food products of animal origin safety standards into practice, securing food safety.
To summarize, this is the first study in Bangladesh to investigate the quantity of Bacillus spp. contamination and toxigenic potentials persistent in animal feed, animal-derived food, and human stool. By detecting and profiling toxin genes as well as completing validated questionnaire surveys, this study assesses the sanitary risk potential of Bacillus spp. strains. The findings revealed that this feed-borne Bacillus spp. group collection has significant toxigenic potentials, contaminating animal feed (poultry, duck, cattle, fish), animal products (egg, milk), and daily food (vegetables, rice, and RTE food). Humans consume vegetative cells and spores of Bacillus spp. on a regular basis via fermented meals and raw vegetables, resulting in diarrhea. Therefore, further research could be allotted to evaluating risk factors, potential pathogenesis, and antibiotic resistance genes of feed-borne Bacillus spp. to ensure sustainable animal production and public health. Finally, it emphasizes the importance of prevention and control strategies such as (1) routine checkup both in animal feedstuffs and human food, and (2) R&D test method that allows for rapid screening and species identification.
All datasets generated for this study were included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Ethical Reviewing Board at Noakhali Science and Technology University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements. The animal study was reviewed and approved by Ethical Reviewing Board at Noakhali Science and Technology University on Institutional Animal Care and Use Committee (ICAUC).
CH, FA, and MHa: conceptualization and methodology. MHa, FH, MI, and MHo: perform the experiments. CH and FA: supervise the project. MHa: data curation and writing – original draft. FW and YC: resources. CH, FA, and NS: writing, review, and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Taishan Scholar Foundation of Shandong Province under Grant No. ts201511084, Yuandou Industry Leading Talent of Shandong Province, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the laboratory support and execution of the research of Microbiology Laboratory, Department of Microbiology, Noakhali Science and Technology University, Noakhali, Bangladesh.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.783103/full#supplementary-material
Abraha, A., Bikila, T., Alemu, S., and Muktar, Y. (2017). Bacillus cereus isolation and load from raw cow milk sold in markets of Haramaya District, eastern Ethiopia. Int. J. Food Contam. 4:15. doi: 10.1186/s40550-017-0060-z
Antequera-Gómez, M. L., Diaz-Martinez, L., Guadix, J. A., Sánchez-Tévar, A. M., Sopeña-Torres, S., Hierrezuelo, J., et al. (2021). Sporulation is dispensable for the vegetable-associated life cycle of the human pathogen Bacillus cereus. Microb. Biotechnol. 14, 1550–1565. doi: 10.1111/1751-7915.13816
Cao, J., Yu, Z., Liu, W., Zhao, J., Zhang, H., Zhai, Q., et al. (2020). Probiotic characteristics of Bacillus coagulans and associated implications for human health and diseases. J. Funct. Foods 64:103643. doi: 10.1016/j.jff.2019.103643
Chowdhury, S., Aleem, M. A., Khan, M. S. I., Hossain, M. E., Ghosh, S., and Rahman, M. Z. (2021). Major zoonotic diseases of public health importance in Bangladesh. Vet. Med. Sci. 7, 1199–1210. doi: 10.1002/vms3.465
Cui, Y., Märtlbauer, E., Dietrich, R., Luo, H., Ding, S., and Zhu, K. (2019). Multifaceted toxin profile, an approach toward a better understanding of probiotic Bacillus cereus. Crit. Rev. Toxicol. 49, 342–356. doi: 10.1080/10408444.2019.1609410
Deng, F., Chen, Y., Sun, T., Wu, Y., Su, Y., Liu, C., et al. (2021). Antimicrobial resistance, virulence characteristics and genotypes of Bacillus spp. from probiotic products of diverse origins. Food Res. Int. 139:109949. doi: 10.1016/j.foodres.2020.109949
Ehling-Schulz, M., Guinebretiere, M. H., Monthán, A., Berge, O., Fricker, M., and Svensson, B. (2006). Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol. Lett. 260, 232–240. doi: 10.1111/j.1574-6968.2006.00320.x
Elshaghabee, F. M. F., Rokana, N., Gulhane, R. D., Sharma, C., and Panwar, H. (2017). Bacillus as potential probiotics: status, concerns, and future perspectives. Front. Microbiol. 8:1490. doi: 10.3389/fmicb.2017.01490
Gdoura-Ben Amor, M., Jan, S., Baron, F., Grosset, N., Culot, A., Gdoura, R., et al. (2019). Toxigenic potential and antimicrobial susceptibility of Bacillus cereus group bacteria from Tunisian foodstuffs. BMC Microbiol. 19:196. doi: 10.1186/s12866-019-1571-y
Gupta, R. C., Srivastava, A., and Lall, R. (2019). Nutraceuticals in Veterinary Medicine. Cham: Springer Nature Switzerland AG, 271–285. doi: 10.1007/978-3-030-04624-8
Gyang, L., Obiekezie, S. O., Owuna, J. E., Adamu, M. O., and Obiekezie, S. O. (2019). Bacterial contamination of poultry feeds, molecular studies and antibacterial resistance profiles of isolates in Keffi metropolis, Nigeria. Int. J. Eng. Sci. 8, 6–14. doi: 10.9790/1813-0811020614
Haque, M. A., Quan, H., Zuo, Z., Khan, A., Siddique, N., and He, C. (2021). Pathogenicity of feed-borne Bacillus cereus and its implication on food safety. Agrobiol. Rec. 3, 1–16. doi: 10.47278/journal.abr/2020.015
Hassan, M. M., El Zowalaty, M. E., Lundkvist, Å, Jarhult, J. D., Nayem, M. R. K., Tanzin, A. Z., et al. (2021). Residual antimicrobial agents in food originating from animals. Trends Food Sci. Technol. 111, 141–150. doi: 10.1016/j.tifs.2021.01.075
Hosain, M. Z., Kabir, S. M. L., and Kamal, M. M. (2021). Antimicrobial uses for livestock production in developing countries. Vet. World 14, 210–221. doi: 10.14202/vetworld.2021.210-221
Hu, Q., Fang, Y., Zhu, J., Xu, W., and Zhu, K. (2021). Characterization of Bacillus species from market foods in Beijing, China. Processes 9:866. doi: 10.3390/pr9050866
Islam, M. S., Khanam, S., and Mohanta, M. K. (2017). Isolation, characterization and identification of bacterial isolates from the poultry environment at Rajshahi metropolis, Bangladesh. J. Entomol. Zool. Stud. 5, 918–926. doi: 10.2139/ssrn.3856330
Khan, A., Rind, R., Shoaib, M., Kamboh, A. A., Mughal, G. A., Lakho, S. A., et al. (2016). Isolation, identification and antibiogram of Escherichia coli from table eggs. J. Anim. Health Prod. 4, 1–5. doi: 10.14737/journal.jahp/2016/4.1.1.5
Kong, L., Yu, S., Yuan, X., Li, C., Yu, P., Wang, J., et al. (2021). An investigation on the occurrence and molecular characterization of Bacillus cereus in meat and meat products in China. Foodborne Pathog. Dis. 18, 306–314. doi: 10.1089/fpd.2020.2885
Li, X., Li, Q., Wang, Y., Han, Z., Qu, G., Shen, Z., et al. (2020). Gastric ulceration and immune suppression in weaned piglets associated with feed-borne Bacillus cereus and Aspergillus fumigatus. Toxins 12:703. doi: 10.3390/toxins12110703
Microbiology Laboratory Guidebook [MLG] (2015). United States Department of Agriculture, Food Safety and Inspection Service, MLG 3.02. Available online at: https://www.fsis.usda.gov/sites/default/files/media_file/2021-03/MLG-3.pdf (accessed October 30, 2021).
Owusu-Kwarteng, J., Wuni, A., Akabanda, F., Tano-Debrah, K., and Jespersen, L. (2017). Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol. 17:65. doi: 10.1186/s12866-017-0975-9
Parry, J. M., Turnbull, P. C. B., and Gibson, J. R. (1988). A Colour Atlas of Bacillus Species (Paperback). Maryland Heights, MO: Mosby.
Proroga, Y. T. R., Capuano, F., Castellano, S., Giordano, A., Mancusi, A., Delibato, E., et al. (2019). Occurrence and toxin gene profile of Bacillus cereus in dairy products. J. Microbiol. Biotech. Food Sci. 9, 58–62. doi: 10.15414/jmbfs.2019.9.1.58-62
Rodrigo, D., Rosell, C. M., and Martinez, A. (2021). Risk of Bacillus cereus in relation to rice and derivatives. Foods 10:302. doi: 10.3390/foods10020302
Sergeev, N., Distler, M., Vargas, M., Chizhikov, V., Herold, K. E., and Rasooly, A. (2006). Microarray analysis of Bacillus cereus group virulence factors. J. Microbiol. Methods 65, 488–502. doi: 10.1016/j.mimet.2005.09.013
Soni, A., Oey, I., Silcock, P., and Bremer, P. (2016). Bacillus spores in the food industry: a review on resistance and response to novel inactivation technologies. Compr. Rev. Food Sci. Food Saf. 15, 1139–1148. doi: 10.1111/1541-4337.12231
Sornchuer, P., and Tiengtip, R. (2021). Prevalence, virulence genes, and antimicrobial resistance of Bacillus cereus isolated from foodstuffs in Pathum Thani Province, Thailand. Pharm. Sci. Asia 48, 194–203. doi: 10.29090/psa.2021.02.19.119
Valero, A., Rodríguez, M.-Y., Posada-Izquierdo, G. D., Pérez-Rodríguez, F., Carrasco, E., and García-Gimeno, R. M. (2016). “Chapter 2: Risk factors influencing microbial contamination in food service centers,” in Significance, Prevention and Control of Food Related Diseases, ed. H. A. Makun (London: Intech), 27–58. doi: 10.5772/63029
Yu, P., Yu, S., Wang, J., Guo, H., Zhang, Y., Liao, X., et al. (2019). Bacillus cereus isolated from vegetables in China: incidence, genetic diversity, virulence genes, and antimicrobial resistance. Front. Microbiol. 10:948. doi: 10.3389/fmicb.2019.00948
Zhang, Q., Zuo, Z., Guo, Y., Zhang, T., Han, Z., Huang, S., et al. (2019). Contaminated feed-borne Bacillus cereus aggravates respiratory distress post avian influenza virus H9N2 infection by inducing pneumonia. Sci. Rep. 9:7231. doi: 10.1038/s41598-019-43660-2
Zhao, S., Chen, J., Fei, P., Feng, H., Wang, Y., Ali, M. A., et al. (2020). Prevalence, molecular characterization, and antibiotic susceptibility of Bacillus cereus isolated from dairy products in China. J. Dairy Sci. 103, 3994–4001. doi: 10.3168/jds.2019-17541
Zhu, K., Hölzel, C. S., Cui, Y., Mayer, R., Wang, Y., Dietrich, R., et al. (2016). Probiotic Bacillus cereus strains, a potential risk for public health in China. Front. Microbiol. 7:718. doi: 10.3389/fmicb.2016.00718
Keywords: food-borne pathogen, Bacillus spp., toxin gene, food chain risk, diarrhea, animal production
Citation: Haque MA, Wang F, Chen Y, Hossen F, Islam MA, Hossain MA, Siddique N, He C and Ahmed F (2022) Bacillus spp. Contamination: A Novel Risk Originated From Animal Feed to Human Food Chains in South-Eastern Bangladesh. Front. Microbiol. 12:783103. doi: 10.3389/fmicb.2021.783103
Received: 25 September 2021; Accepted: 19 November 2021;
Published: 04 January 2022.
Edited by:
Jia-Sheng Wang, University of Georgia, United StatesReviewed by:
Thomas S. Hammack, United States Food and Drug Administration, United StatesCopyright © 2022 Haque, Wang, Chen, Hossen, Islam, Hossain, Siddique, He and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng He, aGVjaGVuZ0BjYXUuZWR1LmNu; Firoz Ahmed, Zmlyb3pAbnN0dS5lZHUuYmQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.