- 1Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, China
- 2Jiangsu Key Laboratory of Zoonosis, Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, China
- 3Joint International Research Laboratory of Agriculture and Agri-Product Safety, Yangzhou University, Yangzhou, China
This study aimed to investigate the prevalence of fosfomycin fosA7 in Salmonella enterica isolates from food animals and retail meat products in China and the impact of fosA7 on bacterial fitness. A total of 360 Salmonella isolates collected from 11 provinces and cities in China were detected for fosA7. All fosA7-positive Salmonella isolates were determined minimum inhibitory concentrations (MICs) and sequenced by Illumina Hiseq. The fosA7 gene of S. Derby isolate HA2-WA5 was knocked out. The full length of fosA7 was cloned into vector pBR322 and then transformed into various hosts. MICs of fosfomycin, growth curves, stability, and fitness of fosA7 were evaluated. The fosA7 gene was identified in S. Derby (ST40, n = 30) and S. Reading (ST1628, n = 5). MICs to fosfomycin of 35 fosA7-positive isolates were 1 to 32 mg/L. All fosA7 were located on chromosomes of Salmonella. The deletion of fosA7 in HA2-WA5 decreased fosfomycin MIC by 16-fold and slightly affected its fitness. The acquisition of plasmid-borne fosA7 enhanced MICs of fosfomycin in Salmonella (1,024-fold) and Escherichia coli (16-fold). The recombinant plasmid pBR322-fosA7 was stable in Salmonella Typhimurium, S. Pullorum, S. Derby, and E. coli, except for Salmonella Enteritidis, and barely affected on the growth of them but significantly increased biological fitness in Salmonella. The spread of specific Salmonella serovars such as S. Derby ST40 will facilitate the dissemination of fosA7. fosA7 can confer high-level fosfomycin resistance and enhance bacterial fitness in Salmonella if transferred on plasmids; thus, it has the potential to be a reservoir of the mobilized fosfomycin resistance gene.
Introduction
In recent decades, multidrug-resistant and extensively drug-resistant Gram-negative pathogens are increasingly reported worldwide, posing a serious threat to clinical medicine (Karaiskos et al., 2019). Available antibiotics are very limited to be used in clinical practice; thus, older antibiotics such as colistin and fosfomycin have been reintroduced to treat infections caused by multidrug-resistant and extensively drug-resistant Gram-negative bacteria (Karaiskos et al., 2019).
Fosfomycin is a bactericidal antibiotic with a broad-spectrum antimicrobial activity against Gram-positive and Gram-negative microorganisms by inhibiting bacterial cell synthesis (Falagas et al., 2016; Sastry and Doi, 2016). In Gram-negative bacteria, resistance to fosfomycin can be mediated by mutations in the chromosomal fosfomycin transporter genes uhpT and glpT, amino acid modification or overexpression of the fosfomycin target MurA, and production of the fosfomycin-modifying enzymes such as FosA and FosC (Falagas et al., 2016; Sastry and Doi, 2016). Among them, glutathione S-transferase FosA encoded by chromosomes or plasmids is the main reason for widespread fosfomycin resistance in Gram-negative bacteria (Sastry and Doi, 2016; Ito et al., 2017).
To date, 10 fosA variants (fosA1∼fosA10) have been identified in Enterobacteriaceae since the first fosA (namely fosA1) described in Serratia marcescens in 1980 (Mendoza et al., 1980; Zurfluh et al., 2020). Fosfomycin resistance has been sporadically reported in Salmonella, one of the leading foodborne pathogens (Lin and Chen, 2015; Qi et al., 2019). So far, fosA3, fosA4, and fosA7 have been detected in Salmonella (Ito et al., 2017; Rehman et al., 2017). FosA7, identified on the chromosomes of Salmonella enterica serovar Heidelberg from broiler chickens in Canada in 2017 (Rehman et al., 2017), has been detected globally in various Salmonella serotypes (e.g., Agona, Brandenburg, and Derby), Escherichia coli, and Citrobacter koseri from various sources (Supplementary Table 1). The fosA7 gene has not yet been detected in Salmonella isolates from China. Thus, in this study, we aimed to investigate the prevalence of fosA7 among Salmonella isolates from food animals, one pig slaughterhouse and retail meat products in China, and to evaluate the impact of fosA7 on bacterial fitness.
Materials and Methods
Bacterial Strains, fosA7 Detection, and Antimicrobial Susceptibility Testing
We obtained 264 S. enterica isolates including serovars London (n = 43), Kentucky (n = 35), Typhimurium (n = 34), Derby (n = 26), Rissen (n = 24), Indiana (n = 19), and others (n = 83) from 1,041 fecal samples from food-producing animals (pigs, chickens, and cattles) and 1,205 retail meat products (chicken meat, pork, and beef) in supermarkets or farmers’ markets in Gansu, Guangdong, Guizhou, Henan, Hubei, Jiangsu, Liaoning, Shandong, and Xinjiang provinces and Shanghai of China from July 2019 to June 2020 (Supplementary Table 2). Eighty-seven Salmonella isolates including serovars Typhimurium (n = 33), Rissen (n = 30), London (n = 14), Derby (n = 4), Reading (n = 4), and Pakistan (n = 2) were obtained from pig carcass swab samples and intestinal content samples in a slaughterhouse in Jiangsu province and nine Salmonella isolates including Wandsworth (n = 2), Lexington (n = 2), Reading (n = 1), Indiana (n = 1), Typhimurium (n = 1), London (n = 1), and Virchow (n = 1) from chicken meat from Chongqing, China, in 2016.
The presence of fosA7 was detected by polymerase chain reaction and sequencing with the primers fosA7-F (5′-ACTTAACGCTTGCTGTC-3′) and fosA7-R (5′-TGCTAAATCTCCCACAT-3′). All fosA7-positive isolates were tested for their susceptibility to ampicillin, cefotaxime, meropenem, gentamicin, amikacin, streptomycin, tetracycline, chloramphenicol, florfenicol, nalidixic acid, ciprofloxacin, fosfomycin (with 25 mg/L glucose-6-phosphate), and sulfamethoxazole/trimethoprim using the agar dilution method and colistin using the microbroth dilution method. The results were interpreted according to CLSI M100, 30th edition (Clinical and Laboratory Standards Institute [CLSI], 2020). Streptomycin (≥32 mg/L) and florfenicol (≥ 32 mg/L) were interpreted according to EUCAST1 epidemiological cutoff values for S. enterica. Fosfomycin (> 32 mg/L) was interpreted according to EUCAST (see text footnote 1) breakpoint tables for Enterobacterales. E. coli American Type Culture Collection (ATCC) 25922 was used as quality control.
Whole-Genome Sequencing and Analysis
The whole genomes of all fosA7-positive Salmonella were sequenced by Illumina Hiseq, and sequence reads were assembled into contigs with SPAdes v.3.13.0. The whole-genome sequences were analyzed for resistance genes, chromosome mutations, and plasmids using Center for Genomic Epidemiology pipelines2. Phylogenetic analysis of 30 fosA7-positive S. Derby isolates and ST71 fosA7-negative S. Derby 14C-S8N4 obtained from pork in Yangzhou, Jiangsu province, China, in 2014 based on 3002 cgMLST locus were performed using cgmlstfinder.py contained in the cgMLSTFinder service (Clausen et al., 2018). iTOL was used to make the phylogenetic tree visual (Letunic and Bork, 2021). Five fosA7-carrying S. Reading isolates in this study and S. Reading CVM N42528 (GenBank accession no. GCA_001481335.1) from pork chop in the United States were analyzed for the core genome regions by Parsnp software. The whole-genome sequencing data have been deposited in the GenBank under accession number PRJNA743999.
In-Frame Deletion of Chromosomal fosA7 in S. Derby
The fosA7 deletion in S. Derby HA2-WA5 that exhibits susceptibility to all tested agents was performed by double exchange of homologous recombination as previously described using primers listed in Supplementary Table 3 (Guo et al., 2020). The deletion mutant HA2-WA5ΔfosA7 was tested for minimum inhibitory concentration (MIC) of fosfomycin (25 mg/L glucose-6-phosphate).
Cloning of fosA7 and Functional Analysis
The fosA7 of HA2-WA5 was amplified by polymerase chain reaction and cloned into the pBR322 vector. The vector pBR322 and the recombinant plasmid pBR322-fosA7 were transformed into E. coli DH5α, Salmonella Typhimurium SL1344, S. Pullorum C79-13, S. Enteritidis P125109, S. Derby 14C-S8N4 (ST71, fosA7-negative), Enterobacter cloacae CMCC45301, and Klebsiella pneumoniae CMCC46117 via electroporation, selected by 32 mg/L ampicillin. The function of fosA7 was determined by testing their MICs of fosfomycin (25 mg/L glucose-6-phosphate).
Growth Curve and Plasmid Stability
The growth curves of all transformants, HA2-WA5 and HA2-WA5ΔfosA7, were determined in LB broth for 14 h at 37°C and 180 rpm. The overnight cultures were adjusted to an OD600 of 1 and then diluted 1:200 (10 μl/20 ml) in fresh LB broth. The OD600 of the cultures was measured every hour using Tecan Sunrise Microplate Reader (TECAN, Switzerland). Experiments were performed in triplicate.
To investigate the stability of pBR322 and pBR322-fosA7 in different hosts, all transformants were maintained for 7 days in daily refreshed (100-fold dilution) LB broth without antibiotics. Cultures were streaked on LB agar plates, and 50 colonies were replica-plated on LB agar plates containing 32 mg/L ampicillin or 256 mg/L fosfomycin with 25 mg/L glucose-6-phosphate on each day.
Growth Competition Experiments in vitro
To investigate the relative fitness (RF) of fosA7, Salmonella isolates SL1344-pBR322-fosA7, C79-13-pBR322-fosA7, 14C-S8N4-pBR322-fosA7, P125109-pBR322-fosA7, and HA2-WA5ΔfosA7 were competed against SL1344-pBR322, C79-13-pBR322, 14C-S8N4-pBR322, P125109-pBR322, and HA2-WA5, respectively, in daily refreshed (1,000-fold dilution) LB broth for 96 h as previously described (Wu et al., 2018). At 48 and 96 h, the total number of bacteria was determined by plating proper diluted cultures on LB agar, and all colonies were replica-plated on LB agar with 16 mg/L fosfomycin and 25 mg/L glucose-6-phosphate. All competition experiments were performed at least three replicates. The RF was calculated as the percentage of transformants or deletion mutant in all isolates. An RF value > 0.5 indicated that the transformants/deletion mutant had a selective advantage over the original strain, whereas a score < 0.5 indicated a fitness cost.
Statistical Analysis
The results were analyzed with GraphPad Prism version 7.0 (GraphPad Software, Inc., La Jolla, CA, United States).
Results
Salmonella Derby ST40 Was the Main Carrier for fosA7
Among 360 Salmonella isolates, fosA7 was detected in 35 strains, with S. Derby (n = 30) being the dominant host. Five S. Reading were also positive for fosA7. The fosA7-carrying Salmonella isolates showed highest resistance to chloramphenicol (n = 31, 88.57%), followed by nalidixic acid (n = 30, 85.71%), tetracycline (n = 29, 82.86%), florfenicol (n = 27, 77.14%), sulfamethoxazole/trimethoprim (n = 27, 77.14%), and ampicillin (n = 27, 77.14%), but none of them were resistant to amikacin, meropenem, and colistin (Table 1). Notably, all fosA7-positive Salmonella isolates were still susceptible to fosfomycin, with MICs ranging between 1 and 32 mg/L (Table 1).
Whole-genome sequencing showed that all S. Derby belonged to ST40, with one being ST40 variant (isolate YZ19MBS1), and five S. Reading strains were classified as ST1628. The fosA7 gene was located on chromosomes of all Salmonella isolates in this study and differed from the firstly identified fosA7 in S. Heidelberg by one (S. Reading) or 12 (S. Derby) nucleotide differences, resulting in one or five amino acid changes. Agreement with the first detected fosA7 in S. Heidelberg ABB07-SB3031 (Rehman et al., 2017), the fosA7 gene was surrounded by putative open reading frames. The genetic environment of fosA7 and its neighboring region (∼24.3 kb) in all S. Derby and S. Reading strains obtained in this study were highly identical (> 98.0%) to the corresponding region of numerous S. Heidelberg strains, including ABB07-SB3031 (GAC_000973785) and S. Stanleyville strain CFSAN059881 (CP075116) (Supplementary Figure 1). In addition to fosA7, 35 Salmonella isolates carried one to 28 resistance genes, conferring resistance or decreasing susceptibility to β-lactams (blaTEM, blaOXA, blaCTX–M, and blaCMY), aminoglycosides [aac(3)-IV, aac(3)-IVa, aph(4)-Ia, aph(3)-Ia, aac(6′)-Iaa, ant(3″)-Ih, aac(6′)-IId, aadA, and strAB], quinolones [qnrS, qnrVC, aac(6′)-Ib-cr, and oqxAB], tetracyclines [tet(A), tet(B), and tet(M)], chloramphenicols (catA1, catB3, and floR), sulfonamides (sul1, sul2, and sul3), trimethoprims (dfrA12 and dfrA14), macrolides (mefB and mphA), and rifampin (arr-3). Mutation within parC (T57S) was found in all fosA7-bearing S. Derby; gyrA (D87N) mutation was also observed in two of them (Figure 1A). No plasmid replicons and mutations in gyrA or parC were detected in S. Reading. Among the 30 fosA7-positive S. Derby, plasmid replicons were identified in 10 of them (Figure 1A).

Figure 1. (A) Phylogenetic tree of fosA7-positive ST40 S. Derby compared with fosA7-negative ST71 S. Derby. Antibiotic resistance genes, mutations, and plasmid replicons are shown. (B) Phylogenetic tree of fosA7-positive S. Reading compared with S. Reading isolate CVM N42528 (GCA_001481335.1). Antibiotic resistance genes are shown. No mutations or plasmid replicons are identified.
Phylogenetic Analysis of fosA7-Positive S. Derby and S. Reading
To further understand their genetic relationship, we performed a phylogenomic analysis on the fosA7-positive S. Derby and S. Reading. The results showed the division of 30 fosA7-positive S. Derby ST40 isolates into four clades (Figure 1A). Clade I only included one isolate, HA2-WA5, obtained from a pig slaughterhouse in 2016 with the least resistance genes. Twelve S. Derby isolates from pork were classified as clade II, and seven of them contained the same resistance genes. Clade III contained two isolates from pork and one isolate from cattle with few resistance genes. Fourteen isolates from various sources, including all S. Derby from cattle and beef, belonged to clade IV; numerous resistance genes were observed in this clade. Five fosA7-positive S. Reading were clustered in two clades (Figure 1B). The strains in the same clade had identical antimicrobial susceptibility profiles and resistance genes. The results indicate that clonal spread has occurred particularly in the same host.
Deletion of fosA7 Reduced Susceptibility of Fosfomycin and Slightly Affected Fitness
Because fosA7 was located on the chromosome and S. Derby was the most common host, we constructed an in-frame fosA7 deletion mutant to assess the impact of chromosomally encoded FosA7 in S. Derby. After deletion of fosA7 in S. Derby HA2-WA5, its MIC of fosfomycin was reduced from 32 to 2 mg/L, further confirming the role of chromosomally located fosA7 in reducing the susceptibility of fosfomycin. The growth curves of HA2-WA5 and HA2-WA5ΔfosA7 were almost identical, suggesting that the fosA7 deletion in the chromosome did not affect the growth (Figure 2A). Additionally, deletion of fosA7 caused slightly decreased fitness at 48 h (P > 0.05) but enhanced fitness at 96 h with a biological advantage (0.01 < P < 0.05) (Figure 3A).
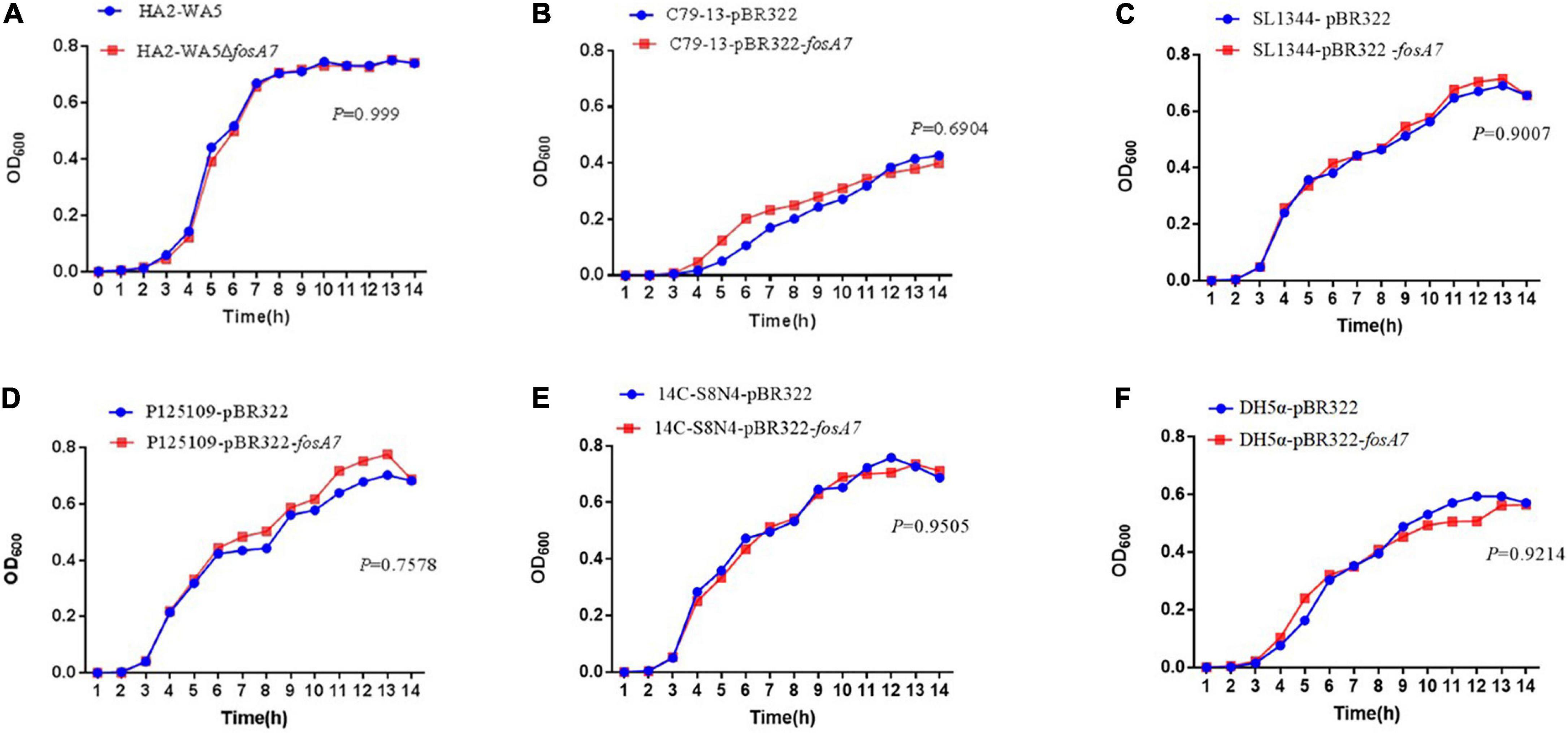
Figure 2. Growth curves of (A) fosA7-positive S. Derby HA2-WA5 and its fosA7 deletion mutant; (B) S. Typhimurium SL1344-pBR322 and SL1344-pBR322-fosA7; (C) S. Pullorum C79-13-pBR322 and C79-13-pBR322-fosA7; (D) S. Enteritidis P125109-pBR322 and P125109-pBR322-fosA7; (E) S. Derby 14C-S8N4-pBR322 and 14C-S8N4-pBR322-fosA7; (F) E. coli DH5α-pBR322 and DH5α-pBR322-fosA7.
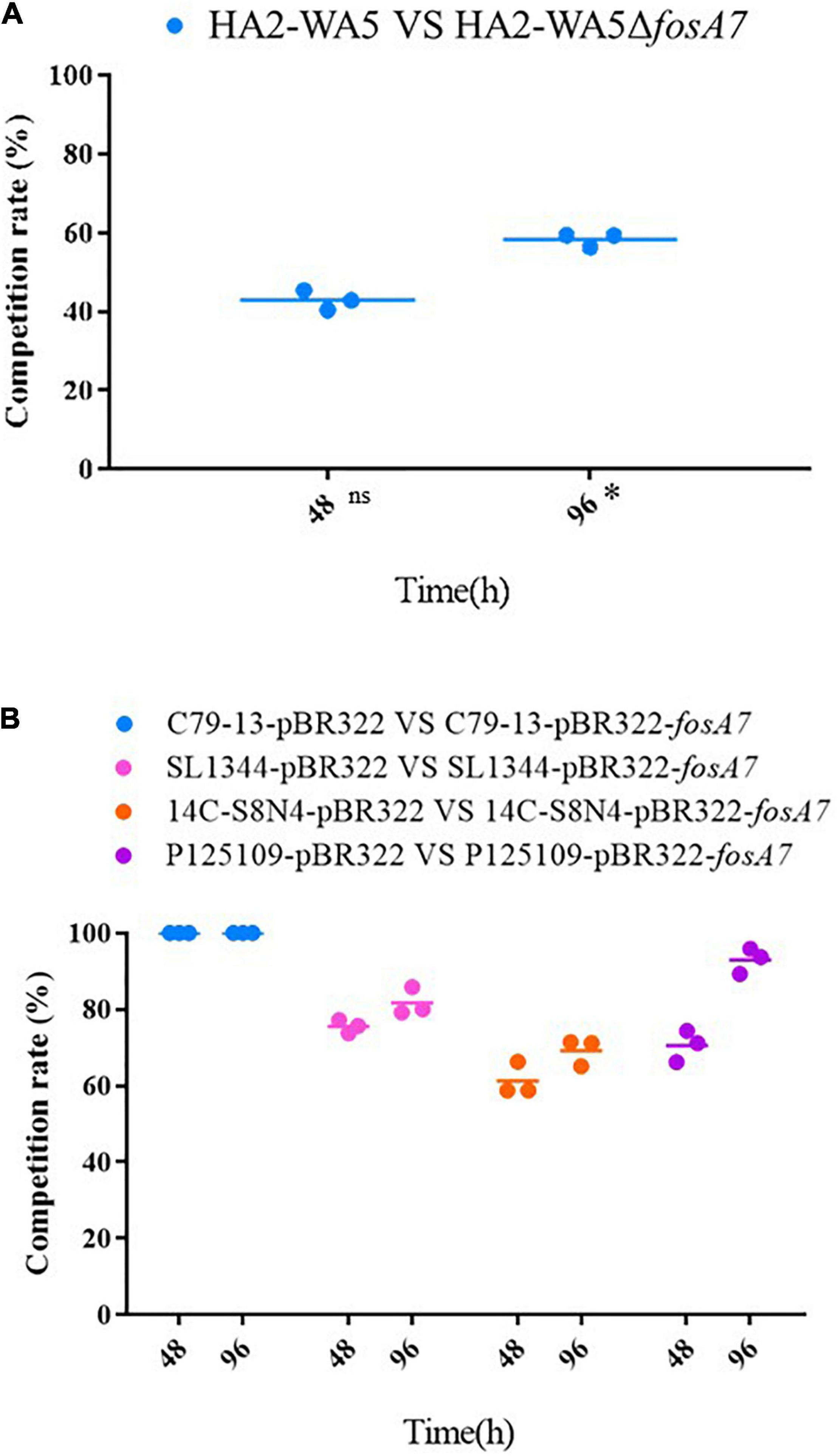
Figure 3. Effects of fosA7 on bacterial relative fitness. (A) fosA7-positive S. Derby HA2-WA5 and its fosA7 deletion mutant, ns indicates difference with no significance, * indicates difference with significance; (B) plasmid-borne fosA7 in S. Pullorum C79-13, S. Typhimurium SL1344, S. Derby 14C-S8N4, and S. Enteritidis P125109. RF was calculated as percentage of transformants or deletion mutant in all isolate.
Plasmid-Borne fosA7 Significantly Increase Minimum Inhibitory Concentrations of Fosfomycin in Salmonella
Chromosomal-encoding FosA7 in S. Derby exhibited low MIC values to fosfomycin. To investigate whether the chromosomal fosA7 in S. Derby could be mobilized to fosA7-negative bacteria and inactivated fosfomycin, we constructed recombinant plasmid pBR322-fosA7 and transformed it to different hosts. The recombinant plasmid pBR322-fosA7 was successfully transformed to E. coli DH5α, S. Typhimurium, S. Pullorum, S. Enteritidis, and S. Derby but failed to K. pneumoniae and E. cloacae after multiple electroporation experiments. Plasmid pBR322-fosA7 was stable for 7 days in S. Typhimurium, S. Pullorum, S. Derby, and E. coli but was lost (∼4%) in S. Enteritidis on the last day of passage (Supplementary Figure 2).
The acquisition of fosA7 significantly increased MICs of fosfomycin in Salmonella (1,024-fold) and conferred fosfomycin resistance, whereas E. coli DH5α was still susceptible after acquiring fosA7 with 16-fold elevated MIC of fosfomycin (Supplementary Table 4).
Plasmid-Borne fosA7 Did Not Affect Bacterial Growth and Increased Fitness of Salmonella
To assess the impact of fosA7 on the bacterial growth, growth curves of E. coli, S. Typhimurium, S. Pullorum, S. Enteritidis, and S. Derby carrying pBR322 or pBR322-fosA7 were compared. The acquisition of fosA7 rarely affected the growth of E. coli and Salmonella (Figures 2B–E).
The RF of fosA7 was determined by growth competition in vitro; E. coli DH5α was not included because FosA7 did not confer fosfomycin resistance in DH5α. The fosA7 gene significantly increased bacterial fitness, particularly in S. Pullorum (Figure 3B). S. Pullorum C79-13-pBR322-fosA7 completely replaced C79-13-pBR322 at 48 h, showing a significant biological benefit. In S. Typhimurium, fosA7 also exhibited fitness advantage (RF = 0.7555 ± 0.0168) at 48 h; no apparent difference was observed between 48 and 96 h (RF = 0.7920 ± 0.0710). Among four Salmonella serotypes, the least enhancement of fitness was shown in S. Derby (RF = 0.6924 ± 0.0435 at 96 h) after acquiring fosA7.
Discussion
Since the identification of fosA7 on the chromosomes of S. Heidelberg from broiler chickens in 2017, it has been detected in multiple Salmonella serotypes, with S. Heidelberg being the most common host (Supplementary Table 1). In this study, fosA7 was detected in S. Derby and S. Reading among 27 serotypes, with S. Derby ST40 being predominant. In Europe, S. Derby has become the sixth most common human serotype and the most common in turkeys, as well as the second most frequently detected in pig carcasses and pigs in 2018 (European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC], 2020). In China, S. Derby has also become one of the leading serovars associated with the pork production chain, with ST40 being predominant (Cai et al., 2016; Jiu et al., 2020). ST40, mainly isolated from the pork sector, is the dominant ST type within S. Derby; however, S. Derby isolates from the poultry mainly belong to ST71, particularly originating from Europe (Sévellec et al., 2019). The fosA7 gene is located on the chromosomes of all tested S. Derby isolates belonging to ST40; the global spread of S. Derby ST40, particularly in pigs and pork, will facilitate the worldwide dissemination of fosA7.
Genes on the chromosomes of various species could act as reservoirs of potential resistance genes; they have the opportunity to be captured by mobile genetic elements (e.g., insertion sequences, transposons, plasmids, and integrative conjugative elements) and to become mobilized resistance genes to horizontally disseminate between different bacteria (Partridge, 2011). For instance, fosA8 may be mobilized from the chromosome of natural fosfomycin-resistant enterobacterial species Leclercia adecarboxylata to IncN plasmid in E. coli, thus conferring high-level fosfomycin resistance in E. coli (Poirel et al., 2019). IncN plasmids carrying fosA8 further spread in E. coli and K. pneumoniae and cointegrated into IncFII or IncR plasmid, respectively (Biggel et al., 2021). Also, the most popular plasmid-mediated fosfomycin resistance gene fosA3 was likely originated from Kluyvera georgiana via IS26-mediated capture and further jumped into various types of plasmids such as IncHI2 and F33:A-: B-, thus rapidly disseminating in Enterobacteriaceae (Ito et al., 2018; Fang et al., 2020; Lv et al., 2020a). It seems that fosA7 is exclusively chromosomal in S. Derby and S. Reading, which accords with previously reported S. Heidelberg (Rehman et al., 2017). Analysis of the genetic structure of fosA7 suggests that it may exist as an intrinsic gene being the progenitor/origin of the fosA7 gene and may have other functions, which need further investigation. Although all fosA7-positive Salmonella isolates in this study were still susceptible to fosfomycin, fosA7 can confer high-level fosfomycin resistance in Salmonella if transferred on plasmids. It is possible due to the expression difference of FosA7 encoding by multicopy plasmid instead of the chromosome. The precise reason will be further investigated. Interestingly, a similar phenomenon was reported in K. georgiana (Ito et al., 2018). As the possible origin of plasmid-mediated fosA3, chromosomally located fosA in K. georgiana strains YDC799 and ATCC 51603 exhibited fosfomycin MICs of 32 and 0.5 mg/L, respectively, but showed high-level resistance to fosfomycin (MIC > 1,024 mg/L) for E. coli after cloning into a plasmid vector (Ito et al., 2018). Thus, although chromosome-located fosA7 only reduces the susceptibility of fosfomycin in Salmonella, it can allow the bacterium to survive under selective pressure, e.g., low level of fosfomycin in the environment. On the other hand, it has the potential to be a reservoir of a mobilized fosfomycin resistance gene. The emergence of fosA7 with a transposase gene downstream on the chromosome of E. coli or fosA7.5 flanked by insertion sequences located on plasmid further supports this hypothesis (Milner et al., 2020; Pan et al., 2021). The fosA7 gene can confer high-level fosfomycin resistance once it is captured by a high copy plasmid or the mobile elements that bring stronger promoters that enhance its expression.
However, E. coli DH5α remained susceptible after the acquisition of pBR322-fosA7, showing a 16-fold elevated MIC of fosfomycin. Similarly, the intrinsic chromosomal fosA of Pseudomonas aeruginosa contributed to a 16-fold increase of fosfomycin MIC in E. coli TOP10 (Ito et al., 2017). A novel FosA7.5 variant was identified and conferred fosfomycin resistance in E. coli. It differed from FosA7 in Salmonella by four amino-acid substitutions, whereas three FosA7.5 variants (one or two amino-acid changes) in E. coli resulted in fosfomycin resistance or susceptibility (Milner et al., 2020). It indicates the critical role of key amino acid differences in FosA7 in fosfomycin resistance within different hosts, which may also explain susceptibility to fosfomycin in fosA7-positive S. Derby and S. Reading in this study.
Generally, antibiotic resistance is associated with a fitness cost that is usually observed as a reduction in bacterial growth rate or competitive ability, such as colistin resistance gene mcr-1 (Andersson and Hughes, 2010; Yang et al., 2017). However, the fitness cost caused by resistance could be alleviated by compensatory mutations (Björkman and Andersson, 2000). Fitness cost and its compensation are two key factors that determine the spread and stability of antibiotic resistance in bacteria (Björkman and Andersson, 2000). For example, the plasmid-encoded efflux pump TMexCD1-TOprJ1 confers resistance to tetracyclines and reduces susceptibility to cephalosporins, aminoglycosides, and quinolones, barely affecting the growth of K. pneumoniae, but significantly reducing the growth of E. coli and S. Typhimurium (Lv et al., 2020b). It may explain why tmexCD1-toprJ1 is mainly described in K. pneumoniae (Lv et al., 2020b; Sun et al., 2020; Hirabayashi et al., 2021). In this study, it appears that the deletion of chromosomally located fosA7 slightly affects the fitness of S. Derby ST40, yet the acquisition of plasmid-borne fosA7 not only confers high-level fosfomycin resistance but also obviously improves the bacterial fitness of Salmonella. It may facilitate the dissemination and persistence of fosA7 in bacterial populations.
To conclude, fosA7 will widely disseminate with the spread of specific Salmonella serovars, such as S. Derby ST40, in our study. Although chromosomally borne fosA7 in S. Derby and S. Reading only reduce susceptibility to fosfomycin, it will help the organisms to survive under low-level of fosfomycin and has the potential to be a reservoir of the mobilized fosfomycin resistance gene. Because it can induce high-level resistance to fosfomycin and enhance bacterial fitness in Salmonella if transferred on plasmids, and plasmid-mediated fosA7.5 has already been detected in E. coli, it needs further surveillance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA743999/.
Author Contributions
JW and XJ conceived the study. YW, HW, JW, C-YM, and P-CS carried out the experiments. JW, YW, and Z-YW analyzed the data. JW wrote the manuscript. Z-MP and XJ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (grant numbers 31902319, 31730094, and 31920103015); the fifth phase of the “333 project” scientific research project in Jiangsu Province (BRA2020002); and the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.781306/full#supplementary-material
Footnotes
References
Andersson, D. I., and Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Biggel, M., Zurfluh, K., Treier, A., Nüesch-Inderbinen, M., and Stephan, R. (2021). Characteristics of fosA-carrying plasmids in E. coli and Klebsiella spp. isolates originating from food and environmental samples. J. Antimicrob. Chemother. 76, 2004–2011. doi: 10.1093/jac/dkab119
Björkman, J., and Andersson, D. I. (2000). The cost of antibiotic resistance from a bacterial perspective. Drug Resist. Updat. 3, 237–245. doi: 10.1054/drup.2000.0147
Cai, Y., Tao, J., Jiao, Y., Fei, X., Zhou, L., Wang, Y., et al. (2016). Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 222, 56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020
Clausen, P. T. L. C., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100, 30th Edn. Wayne, PA: CLSI.
European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC] (2020). The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 18:6007. doi: 10.2903/j.efsa.2020.6007
Falagas, M. E., Vouloumanou, E. K., Samonis, G., and Vardakas, K. Z. (2016). Fosfomycin. Clin. Microbiol. Rev. 29, 321–347. doi: 10.1128/CMR.00068-15
Fang, L. X., Jiang, Q., Deng, G. H., He, B., Sun, R. Y., Zhang, J. F., et al. (2020). Diverse and flexible transmission of fosA3 associated with heterogeneous multidrug resistance regions in Salmonella enterica serovar Typhimurium and Indiana isolates. Antimicrob. Agents Chemother. 64:e02001-19. doi: 10.1128/AAC.02001-19
Guo, Y., Gu, D., Huang, T., Cao, L., Zhu, X., Zhou, Y., et al. (2020). Essential role of Salmonella Enteritidis DNA adenine methylase in modulating inflammasome activation. BMC Microbiol. 20:226. doi: 10.1186/s12866-020-01919-z
Hirabayashi, A., Ha, V. T. T., Nguyen, A. V., Nguygen, S. T., Shibayama, K., and Suzuki, M. (2021). Emergence of a plasmid-borne tigecycline resistance in Klebsiella pneumoniae in Vietnam. J. Med. Microbiol. 70:001320. doi: 10.1099/jmm.0.001320
Ito, R., Mustapha, M. M., Tomich, A. D., Callaghan, J. D., McElheny, C. L., Mettus, R. T., et al. (2017). Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e00749-17. doi: 10.1128/mBio.0074
Ito, R., Pacey, M. P., Mettus, R. T., Sluis-Cremer, N., and Doi, Y. (2018). Origin of the plasmid-mediated fosfomycin resistance gene fosA3. J. Antimicrob. Chemother. 73, 373–376. doi: 10.1093/jac/dkx389
Jiu, Y., Meng, X., Hong, X., Huang, Q., Wang, C., Chen, Z., et al. (2020). Prevalence and characterization of Salmonella in three typical commercial pig abattoirs in Wuhan, China. Foodborne Pathog. Dis. 17, 620–627. doi: 10.1089/fpd.2019.2737
Karaiskos, I., Lagou, S., Pontikis, K., Rapti, V., and Poulakou, G. (2019). The “Old” and the “New” antibiotics for MDR Gram-negative pathogens: for whom, when, and how. Front. Public Health. 7:151. doi: 10.3389/fpubh.2019.00151
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lin, D., and Chen, S. (2015). First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in Salmonella isolates of food origin. Antimicrob. Agents Chemother. 59, 1381–1383. doi: 10.1128/AAC.04750-14
Lv, L., Huang, X., Wang, J., Huang, Y., Gao, X., Liu, Y., et al. (2020a). Multiple plasmid vectors mediate the spread of fosA3 in extended-spectrum-β-lactamase-producing Enterobacterales isolates from retail vegetables in China. mSphere 5:e00507-20. doi: 10.1128/mSphere.00507-20
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020b). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio 11:e02930-19. doi: 10.1128/mBio.02930-19
Mendoza, C., Garcia, J. M., Llaneza, J., Mendez, F. J., Hardisson, C., and Ortiz, J. M. (1980). Plasmid-determined resistance to fosfomycin in Serratia marcescens. Antimicrob. Agents Chemother. 18, 215–219. doi: 10.1128/AAC.18.2.215
Milner, K. A., Bay, D. C., Alexander, D., Walkty, A., Karlowsky, J. A., Mulvey, M. R., et al. (2020). Identification and characterization of a novel FosA7 member from fosfomycin-resistant Escherichia coli clinical isolates from Canadian hospitals. Antimicrob. Agents Chemother. 65:e00865-20. doi: 10.1128/AAC.00865-20
Pan, Y., Hu, B., Bai, X., Yang, X., Cao, L., Liu, Q., et al. (2021). Antimicrobial resistance of non-O157 shiga toxin-producing Escherichia coli isolated from humans and domestic animals. Antibiotics 10:74. doi: 10.3390/antibiotics10010074
Partridge, S. R. (2011). Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 820–855. doi: 10.1111/j.1574-6976.2011.00277.x
Poirel, L., Vuillemin, X., Kieffer, N., Mueller, L., Descombes, M. C., and Nordmann, P. (2019). Identification of FosA8, a plasmid-encoded fosfomycin resistance determinant from Escherichia coli, and its origin in Leclercia adecarboxylata. Antimicrob. Agents Chemother. 63:e01403-19. doi: 10.1128/AAC.01403-19
Qi, X., Li, P., Xu, X., Yuan, Y., Bu, S., and Lin, D. (2019). Epidemiological and molecular investigations on Salmonella responsible for gastrointestinal infections in the southwest of Shanghai from 1998 to 2017. Front. Microbiol. 10:2025. doi: 10.3389/fmicb.2019.02025
Rehman, M. A., Yin, X., Persaud-Lachhman, M. G., and Diarra, M. S. (2017). First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob. Agents Chemother. 61, 410–417. doi: 10.1128/AAC.00410-17
Sastry, S., and Doi, Y. (2016). Fosfomycin: resurgence of an old companion. J. Infect. Chemother. 22, 273–280. doi: 10.1016/j.jiac.2016.01.010
Sévellec, Y., Felten, A., Radomski, N., Granier, S. A., Le Hello, S., Petrovska, L., et al. (2019). Genetic diversity of Salmonella Derby from the poultry sector in Europe. Pathogens 8:46. doi: 10.3390/pathogens8020046
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Wu, R., Yi, L. X., Yu, L. F., Wang, J., Liu, Y., Chen, X., et al. (2018). Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 9:331. doi: 10.3389/fmicb.2018.00331
Yang, Q., Li, M., Spiller, O. B., Andrey, D. O., Hinchliffe, P., Li, H., et al. (2017). Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 8:2054. doi: 10.1038/s41467-017-02149-0
Keywords: fosfomycin resistance, fosA7, Salmonella Derby, chromosome-located, ST40
Citation: Wang J, Wang Y, Wang Z-Y, Wu H, Mei C-Y, Shen P-C, Pan Z-M and Jiao X (2021) Chromosomally Located fosA7 in Salmonella Isolates From China. Front. Microbiol. 12:781306. doi: 10.3389/fmicb.2021.781306
Received: 22 September 2021; Accepted: 25 November 2021;
Published: 28 December 2021.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Ye Feng, Zhejiang University, ChinaRafael Vignoli, Universidad de la República, Uruguay
Copyright © 2021 Wang, Wang, Wang, Wu, Mei, Shen, Pan and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinan Jiao, amlhb0B5enUuZWR1LmNu
†These authors have contributed equally to this work
 Jing Wang
Jing Wang Yan Wang1,2†
Yan Wang1,2† Zhen-Yu Wang
Zhen-Yu Wang Zhi-Ming Pan
Zhi-Ming Pan Xinan Jiao
Xinan Jiao