- 1School of Public Health, Zhengzhou University, Zhengzhou, China
- 2Chinese PLA Center for Disease Control and Prevention, Beijing, China
- 3College of Veterinary Medicine, Shanxi Agricultural University, Taigu, China
Salmonella contamination of eggs and egg shells has been identified as a public health problem worldwide. Here, we reported an outbreak of severe gastrointestinal symptoms caused by Salmonella enterica serovar Enteritidis (S. enteritidis) in China. We evaluated the outbreak by using epidemiological surveys, routine laboratory testing methods, and whole genome sequencing (WGS). This outbreak occurred in a canteen in Beijing, during March 9–11, 2021, 225 of the 324 diners who have eaten at the canteen showed gastrointestinal symptoms. The outbreak had characteristical epidemiological and clinical features. It caused a very high attack rate (69.4%) in a short incubation time. All patients developed diarrhea and high fever, accompanied by abdominal pain (62.3%), nausea (50.4%), and vomiting (62.7%). The average frequency of diarrhea was 12.4 times/day, and the highest frequency of diarrhea was as high as 50 times/day. The average fever temperature was 39.4°C, and the highest fever temperature was 42°C. Twenty strains of S. enteritidis were recovered, including 19 from the patients samples, and one from remained egg fried rice. Antibiotic susceptibility test showed that the 20 outbreak strains all had the same resistance pattern. PFGE results demonstrated that all 20 strains bore completely identical bands. Phylogenetic analysis based on WGS revealed that all 20 outbreak strains were tightly clustered together. So the pathogenic source of this food poisoning incident may was contaminated egg fried rice. Resistance gene analysis showed that the outbreak strains are all multi-drug resistant strains. Virulence gene analysis indicated that these outbreak strains carried a large number of virulence genes, including 2 types of Salmonella pathogenicity islands (SPI-1 and SPI-2). Other important virulence genes were also carried by the outbreak strains, such as pefABCD, rck and shdA. And the shdA gene was not in other strains located in the same evolutionary branch as the outbreak strain. We speculated that this is a significant reason for the serious symptoms of gastroenteritis in this outbreak. This outbreak caused by S. enteritidis suggested government should strengthen monitoring of the prevalence of outbreak clone strains, and take measures to mitigate the public health threat posed by contaminated eggs.
Introduction
World Health Organization (WHO) estimated the global burden of foodborne diseases, the results showed that almost 1 in 10 people fall ill every year from eating contaminated food and 420,000 die as a result (Dewey-Mattia et al., 2018). Salmonellosis is one of the most frequently reported foodborne diseases worldwide. In particular, disease caused by non-typhoid Salmonella is a global public health problem, whether in a high-income country or a low-income country (Feasey et al., 2016). Each year, approximately 40,000 Salmonella infections are reported to the United States Centers for Disease Control and Prevention (CDCs) (Vaughn et al., 2020). Salmonella enterica serovar Enteritidis (S. enteritidis) is the predominant Salmonella serotype accounting for between 40 and 60% of laboratory-confirmed illnesses of salmonellosis in recent years (Quick et al., 2015). Salmonella enteritidis typically cause a self-limiting gastroenteritis with the symptoms of diarrhea, fever, abdominal cramps, and dehydration (Jiang et al., 2020). Salmonellosis is mainly caused by eating eggs and egg products contaminated with S. enteritidis (90%) and has become a serious health problem. It has been attributed to this serovar’s unusual ability to colonize ovarian tissue of hens and to be able to present within the contents of intact shell eggs (Chousalkar et al., 2018).
Here we reported a severe gastroenteritis outbreak of S. enteritidis linked to contaminated egg fried rice. There were 225 cases of diarrhea and fever in a short period of time in a canteen in Beijing within 3 days. Epidemiological investigations and laboratory tests confirmed that the outbreak was caused by S. enteritidis and was related to the undercooked egg fried rice. At present, such a large-scale outbreak with severe clinical symptoms of S. enteritidis caused by undercooked eggs is rarely reported (Li et al., 2020). Therefore, we reported the outbreak and examined its molecular characteristics using whole genome sequencing (WGS).
Materials and Methods
Outbreak Investigation and Sample Collection
After dinner on March 9, 2021 (18:00), many diarrhea cases occurred in a canteen in Beijing, China. We launched an epidemiological field investigation and concomitant laboratory research in the first time. A suspected salmonellosis case for this outbreak was defined as the onset of diarrhea (>3 times per day), vomiting, or abdominal pain occurring in the staff members who have eaten at the canteen in Beijing during March 9–11, 2021. A confirmed case was that the RT-PCR result is positive for Salmonella or our laboratory isolates Salmonella from stool samples or rectal swabs (Gidudu et al., 2011).
We collected details of food exposure histories, clinical manifestations, and demographic data. To identify the possible factors that were associated with the outbreak, we conducted a food hygiene survey, focusing on the food preparation process in the base canteen, and collected samples from patients, food and environment. A total of 67 stool specimens and 9 anal swabs from the base staff and canteen service staff were collected, 69 remained food samples, 13 raw eggs, a sample of tap water, and five copies of environmental spreads such as knives were collected. All samples were placed in sterile plastic sample bags, and kept on ice until transported to the laboratory for being tested.
Pathogen Identification
The collected samples were sent to our laboratory for pathogen isolation and identification. The nucleic acid of samples was extracted with the KingFisher Flex Automatic nucleic acid extractor (Thermo Fisher Scientific, West Sussex, United Kingdom) and LabServ Prefilled Viral NA kit-Flex (Thermo Fisher Scientific, West Sussex, United Kingdom). Twenty-four diarrhea pathogens were screened by using real-time polymerase chain reaction (RT-PCR) with the twenty-four diarrhea pathogens nucleic acid detection kit (BioGerm, Shanghai, China), including Norovirus Type I, Norovirus Type II, Campylobacter coli, Rotavirus Type A, Rotavirus Type B, Rotavirus Type C, Enteric adenovirus, Human Astrovirus, sapovirus, Vibrio parahaemolyticus, Listeria monocytogenes, Aeromonas hydrophila, Vibrio cholerae, Bacillus cereus, Yersinia pseudotuberculosis, Salmonella, Campylobacter jejuni, Vibrio fluvialis, Diarrhea-causing Escherichia coli, Staphylococcus aureus, Vibrio mimicus, Yersinia enterocolitica, Shigella, Plesiomonas shigelloides. Configuration detection system: upstream primer (10 mol/L) 0.625 μL, downstream primer (10 mol/L) 0.625 μL, probe (10 mol/L) 0.5 μL, Enzyme mix 1 μL, RT-QPCR master mix 12.5 μL, water 6.75 μL, DNA template 3 μL. The reaction conditions are as follows: 50°C 30 min, 1 cycle; 95°C 3 min 1 cycle; 95°C 15 s, 55°C 40 s, 40 cycles.
According to Chinese National Standard method (GB 4789.4-2016), the patient’s stool samples were enriched in Selenite Brilliant Green broth (SBG, CHROMagar, Paris, France) at 37°C for 16–22 h. For each food sample, 25 g were transferred to a sterile plastic bag containing 225 mL of Buffered Peptone Water (BPW, Haibo, Qingdao, China) and incubated at 37°C for 18 h, then each pre-enriched homogenate (1 mL) was aseptically added to 10 mL of lactose broth and incubated at 37°C for 24 h. Enriched samples were cultured in Salmonella-Shigella agar medium (SS, Haibo, Qingdao, China) and MacConkey agar medium (MAC, LandBrigde, Beijing, China) at 37°C for 16–18 h. The typical colony morphology of Salmonella on selective SS medium is a colorless and transparent colony with a black center. Based on the above criteria, suspected bacteria were selected and inoculated to Luria-Bertani solid agar for second-generation culture, which were incubated at 37°C for 16–18 h, then they were identified using the commercial biochemical test kit (API 20E system; bioMérieux Vitek, Marcy-L’Etoile, France) according to the manufacturer’s instructions. Serogroup identification was performed using the slide agglutination method according to the Kauffmann-White protocol, as described in the manufacturer’s instructions. The isolated Salmonella strains were serotyped on a glass slide using a microtiter agglutination test for the O and H antigens (SSI, Copenhagen, Denmark). The 16S rDNA sequence was obtained by PCR amplification and sequencing, and then the obtained sequence was compared with the NCBI database to identify strains.
Pulsed-Field Gel Electrophoresis
The pulsed field gel electrophoresis (PFGE) of Salmonella is carried out in accordance with PulseNet’s 1-day standardized PFGE protocol, with slight modifications (Camarda et al., 2015). XbaI (Takara, Dalian, China) was used for restriction endonuclease digestion. The electrophoresis was run on the CHEFER MAPPER variable angle system (Bio-Rad, CA, United States), the parameters were set to 2.16–63.8 s, and the duration was 19 h. The Gel Doc 2000 system (BioRad) was used to capture the images and convert them into TIF format files for further analysis. The collected images were imported into the BioNumerics software (V6.0) database for processing and analysis, and compared with the international standard strain H9812 to calibrate the band position. Cluster analysis uses the arithmetic mean unweighted paired group method (UPGMA).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was determined based on the minimum inhibitory concentration (MIC) of each of the 14 common antimicrobial agents, using the Sensititre broth microdilution system and CMV2AGNF plates (Thermo Fisher Scientific, Inc., West Sussex, United Kingdom). The drug sensitivity test plates contained 14 different antibiotics: ceftriaxone (CRO), tetracycline (TET), ceftiofur sodium (XNL), cefoxitin (FOX), gentamicin (GEN), ampicillin (AMP), chloramphenicol (CHL), ciprofloxacin (CIP), meperidine/sulfamethoxazole (SXT), sulfamethoxazole (FIS). Escherichia coli ATCC 25922 was used for quality control. Susceptibility tests were interpreted using Clinical and Laboratory Standards Institute (CLSI) guidelines (Melvin et al., 2021).
Genome Sequencing and Bioinformatics Analysis
The outbreak strains were submitted for whole-genome sequencing. Libraries were prepared using the TruePrepTM DNA Library Prep Kit V2 for Illumina (Vazyme). Using a single “transposase” enzymatic reaction, sample DNA is simultaneously fragmented and tagged with adapters, an optimized, limited-cycle PCR protocol amplifies tagged DNA and adds sequencing indexes. Individual libraries were assessed on the QIAxcel Advanced Automatic nucleic acid analyzer, and then were quantitated using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, United States) spectrophotometer, and verified by agarose gel electrophoresis. At last, the library was sequenced on an Illumina HiSeq Novaseq 6000 platform (Illumina Inc., San Diego, CA, United States) and 150 bp paired-end reads were generated.
Multi-locus sequence typing (MLST) of the outbreak strains was performed according to PUBMLST1 (Jolley et al., 2018). We downloaded the complete genome sequences of 513 S. enteritidis strains from the NCBI database in April 2021. Core genome single nucleotide polymorphism (SNP) was analyzed using Parsnp (Treangen et al., 2014). The maximum likelihood (ML) method was used to construct phylogenetic tree. And the lineages of the phylogenetic tree were further defined using Bayesian analysis of the population structure (BAPS; version 6.0; Bayesian Statistics Group) (Cheng et al., 2013). In addition, the sequences of outbreak strains and the 513 public strains were analyzed for antimicrobial resistance (AMR) genes and virulence genes. We refer to the relevant literature previously published by our group and set the threshold to 90% (Li et al., 2019). The presence of AMR genes was predicted using the Resistance Gene Identifier (RGI) application of comprehensive antibiotic resistance database (CARD) (Jia et al., 2017), and virulence gene was determined by BLAST against Virulence Factor Database (VFDB) (Liu et al., 2019). Heatmap was drawn using ITOL (v4) (Letunic and Bork, 2019).
Results
Epidemiologic Investigation
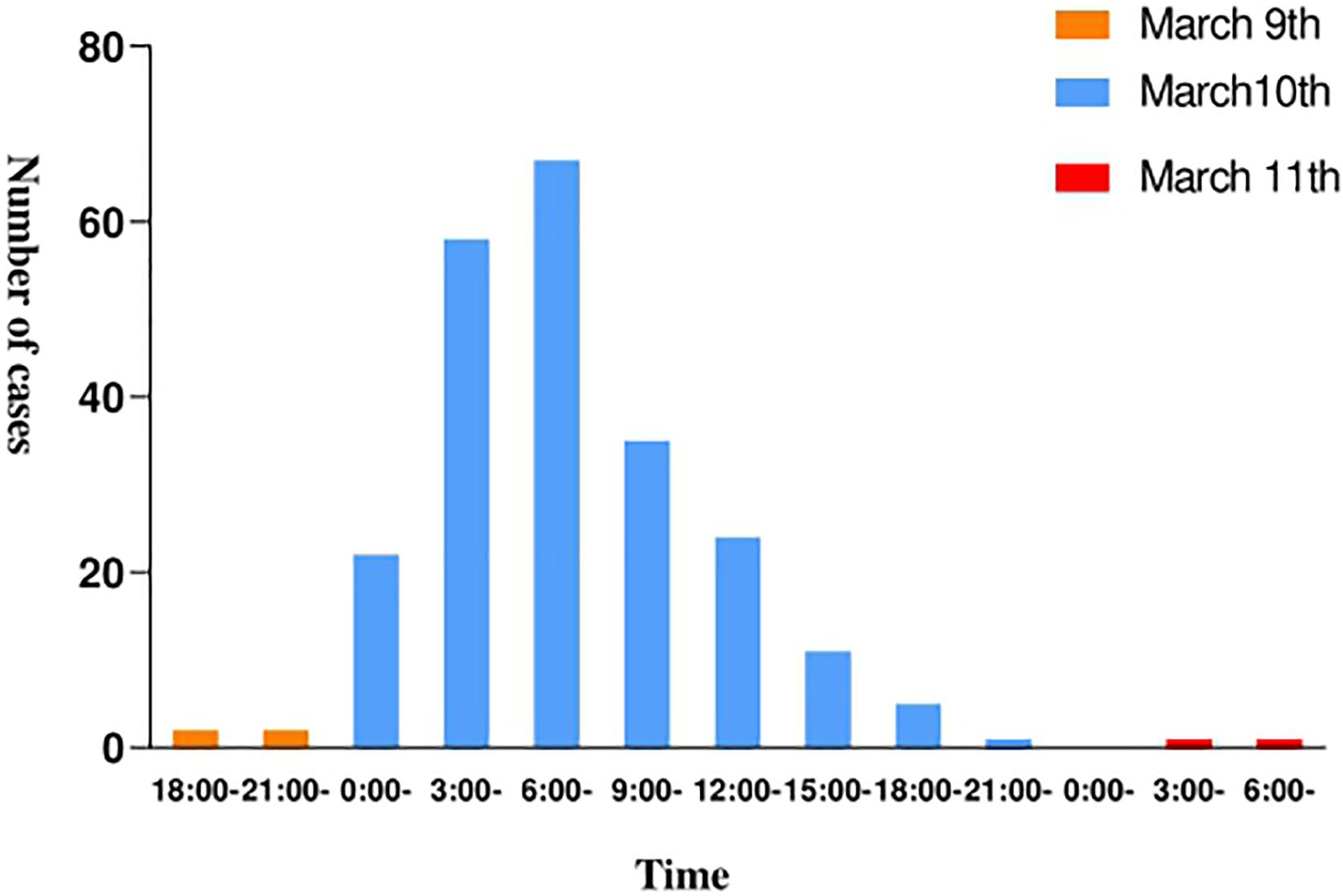
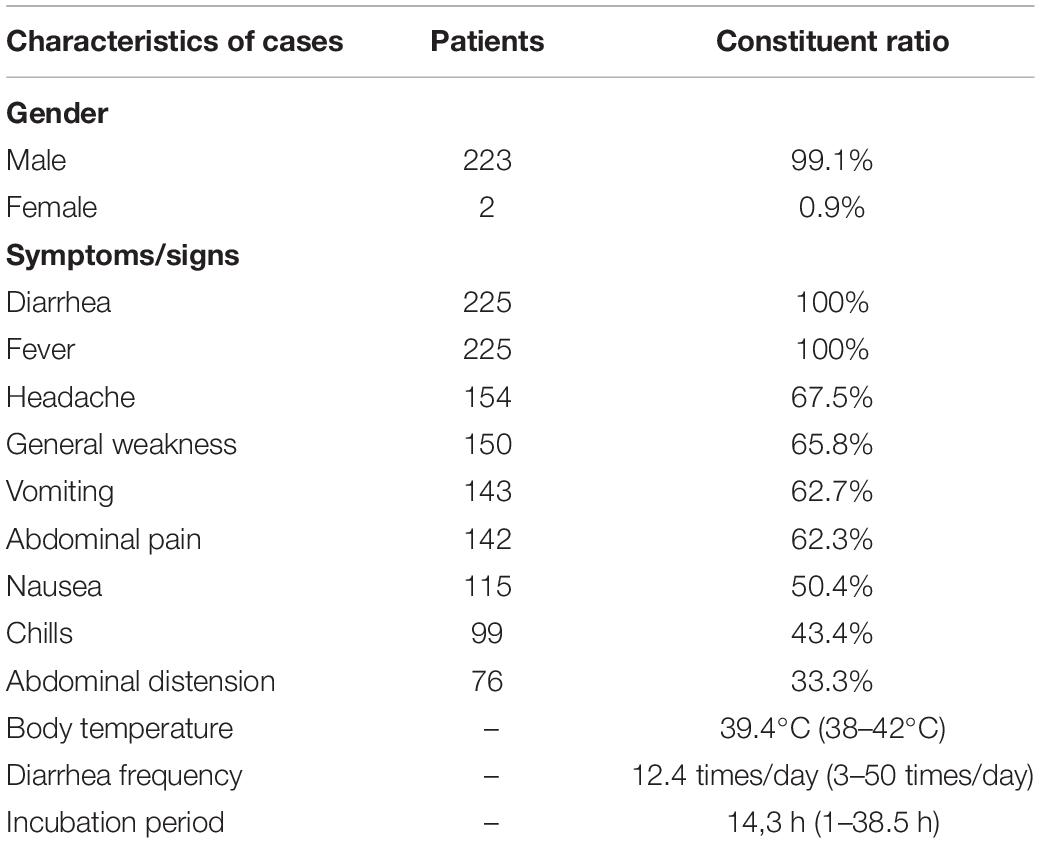
The epidemic occurred in a canteen in Beijing. All personnel of the base had dinner in the canteen, 225 of the 324 diners had diarrhea, and the overall attack rate is 69.4%. The patients included 223 males and 2 females, and the average age is 24.1 years. The shortest incubation period was 1 h, the longest incubation period was 38.5 h and the average incubation period was 14.3 h (Figure 1). The first case occurred diarrhea symptoms at 19:00 on March 9th, the last case was at 8 o’clock on March 11th. A large number of diarrhea cases occurred after 4 a.m. on the March 10th, accompanied by abdominal pain (62.3%), nausea (50.4%), and vomiting (62.7%). Most patients had diarrhea more than 10 times a day, with the highest diarrhea frequency of 50 times/day. After the symptoms of diarrhea, all cases developed high fever, 81.3% of diarrhea cases developed high fever within 4.5 h, and some cases reached 42°C (Table 1). Food hygiene survey showed that it only took 1 min and 50 s for the egg fried rice to go from frying the raw materials to the finished product. The finished product was placed in a basin, which contained residual raw egg liquid.
Laboratory Investigation
Real-time polymerase chain reaction results showed that 42 of the 67 stool samples were positive for Salmonella and 8 were weakly positive for Salmonella. Five of the nine anal swabs were positive for Salmonella. Four of Sixty-nine food samples were positive for Salmonella, included two from egg fried rice, one from spicy chicken, and one from preserved egg lean meat porridge. Seven of thirteen eggs were weakly positive for Salmonella. Whereas five environmental samples were negative for Salmonella, and other intestinal pathogens tests were also negative (Supplementary Table 1). A total of 20 strains were isolated, 16 strains were isolated from base staff, 3 strains were isolated from canteen service staff, and 1 strain was from egg fried rice. Biochemical identification of the isolated strains showed that they were all Salmonella. The results of serotyping and 16s rRNA sequencing both showed that 20 outbreak strains were S. enteritidis.
Pulsed-Field Gel Electrophoresis
We performed PFGE cluster analysis of 20 outbreak strains and 10 strains of S. enteritidis preserved in our laboratory, and the 20 outbreak S. enteritidis strains all had identical PFGE bands. The result demonstrated that it is an outbreak and the egg fried rice is the cause of the outbreak. The PFGE bands of thirty strains formed three branches, and 20 outbreak strains were similar to the strains from Shenzhen exhibiting a similarity of over 90%, indicating that S. enteritidis was closely related to each other in different regions of my country (Figure 2).
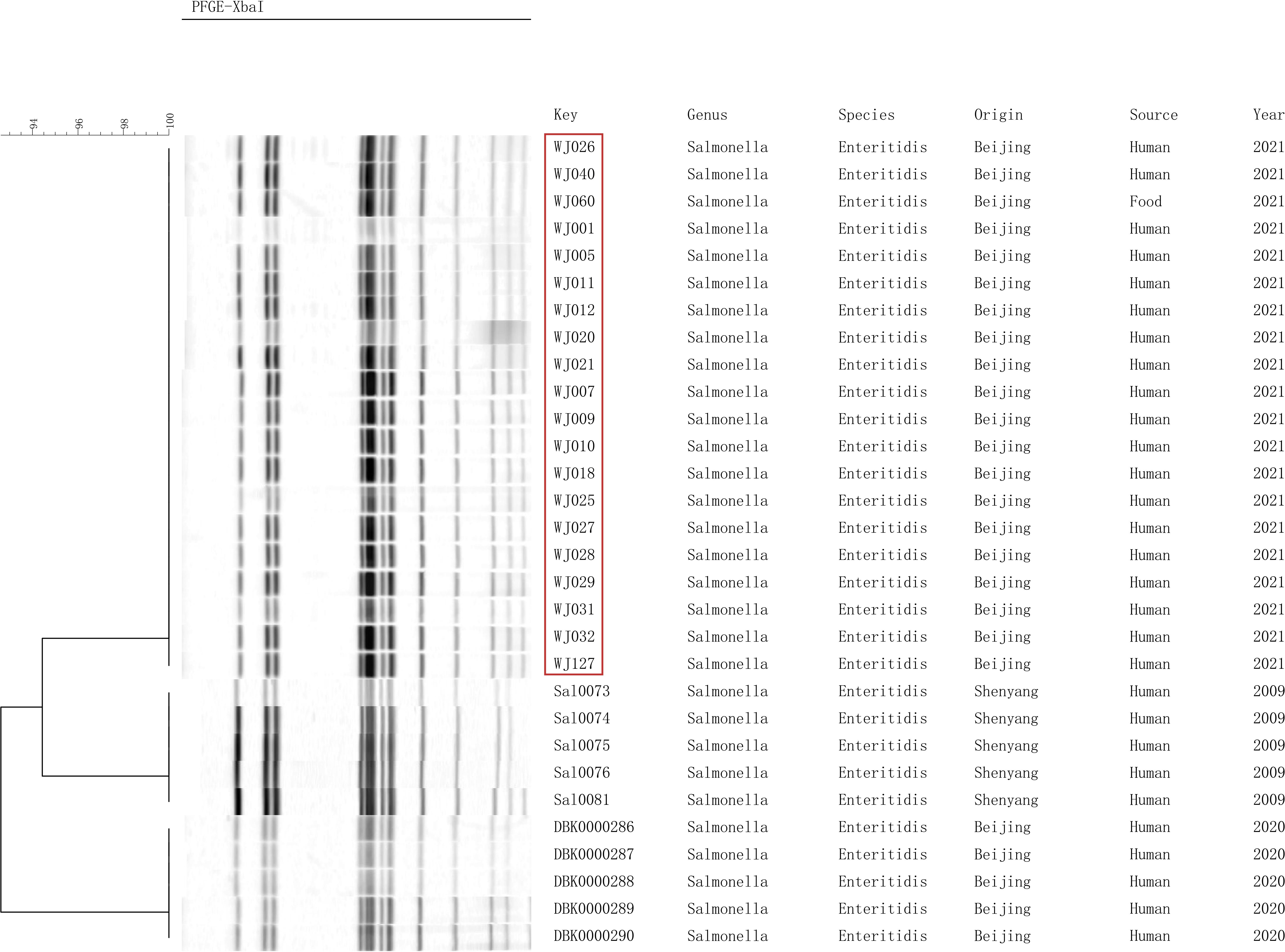
Figure 2. Pulsed-field gel electrophoresis (PFGE) patterns created by digestion with the enzyme XbaI.
Antimicrobial Susceptibility Testing
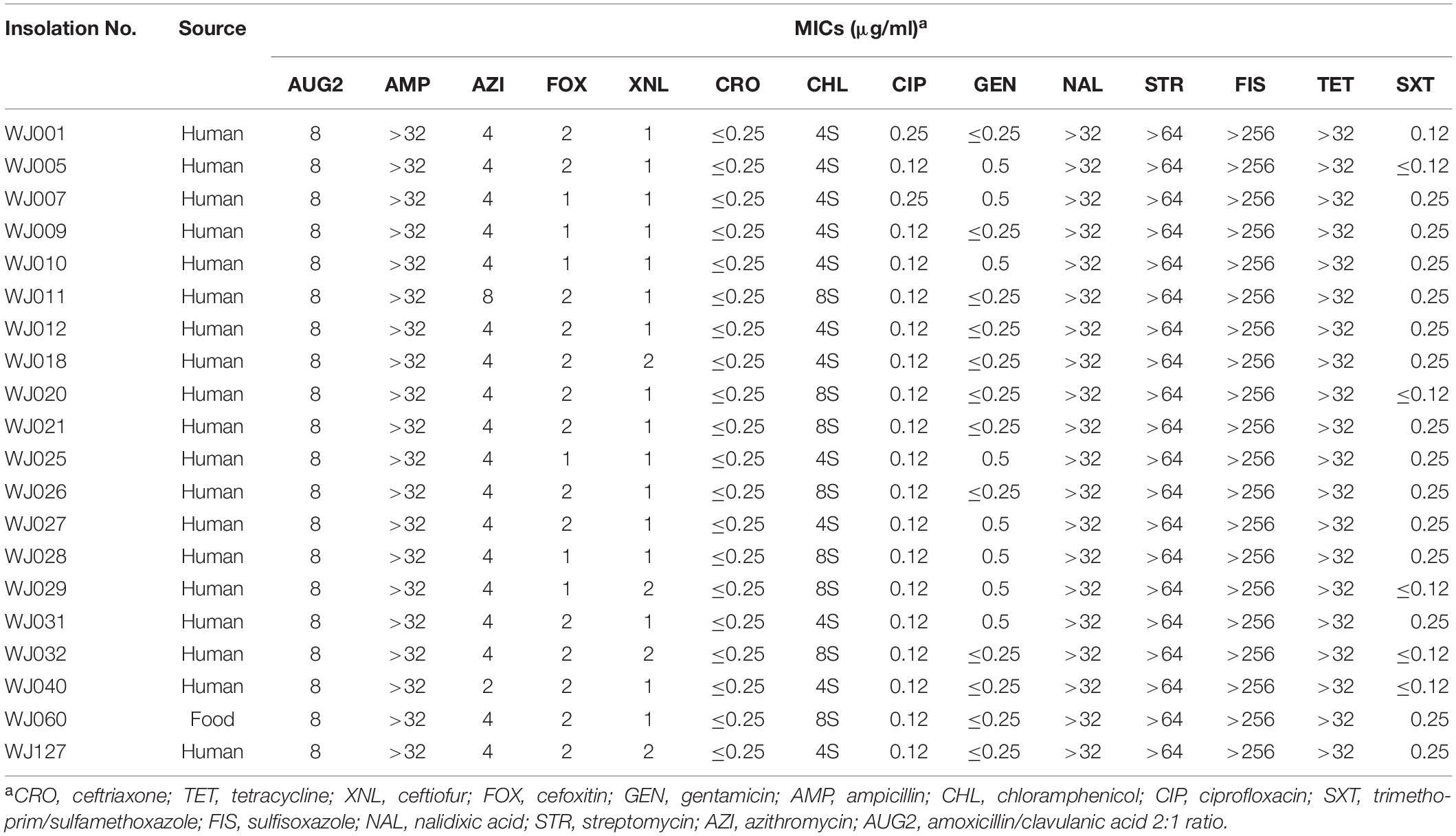
All 20 outbreak isolates displayed the same multidrug resistance (MDR) profiles (Table 2). The outbreak strains were all resistant to ampicillin (minimum inhibitory concentration, MIC; 32 μg/mL), sulfisoxazole (FIS; 512 μg/mL), nalidixic acid (NAL; 32 μg/mL), tetracycline (TET; 16 μg/mL), and streptomycin (STR; 64 μg/mL), whereas sensitive to ciprofloxacin (CIP; 1 μg/mL), gentamicin (GEN; 4 μg/mL) and ceftriaxone (CRO; 1 μg/mL), azithromycin (AZI; 8 μg/mL), cefoxitin (FOX; 8 μg/mL), ceftiofur (XNL; 2 μg/mL), chloramphenicol (CHL; 8 μg/mL), and sulfamethoxazole (SXT; 2 μg/mL) antibiotics.
Phylogenetic Analyses
Phylogenetic analyses showed that the SNP locus difference within the 533 strains was more than 20,000 loci. But the SNP locus difference within the 20 outbreak strains was less than 50 loci, band closely clustered together, which supported that it was indeed an outbreak of S. enteritidis infection. The outbreak strains formed a novel small sub-branch separately, which was located inside the main branch of the phylogenetic tree, suggesting that these strains may originated from a single clone (Figure 3). MLST analysis indicated that all 20 strains of the outbreak belonged to sequence type 11 (ST11; Supplementary Table 2).
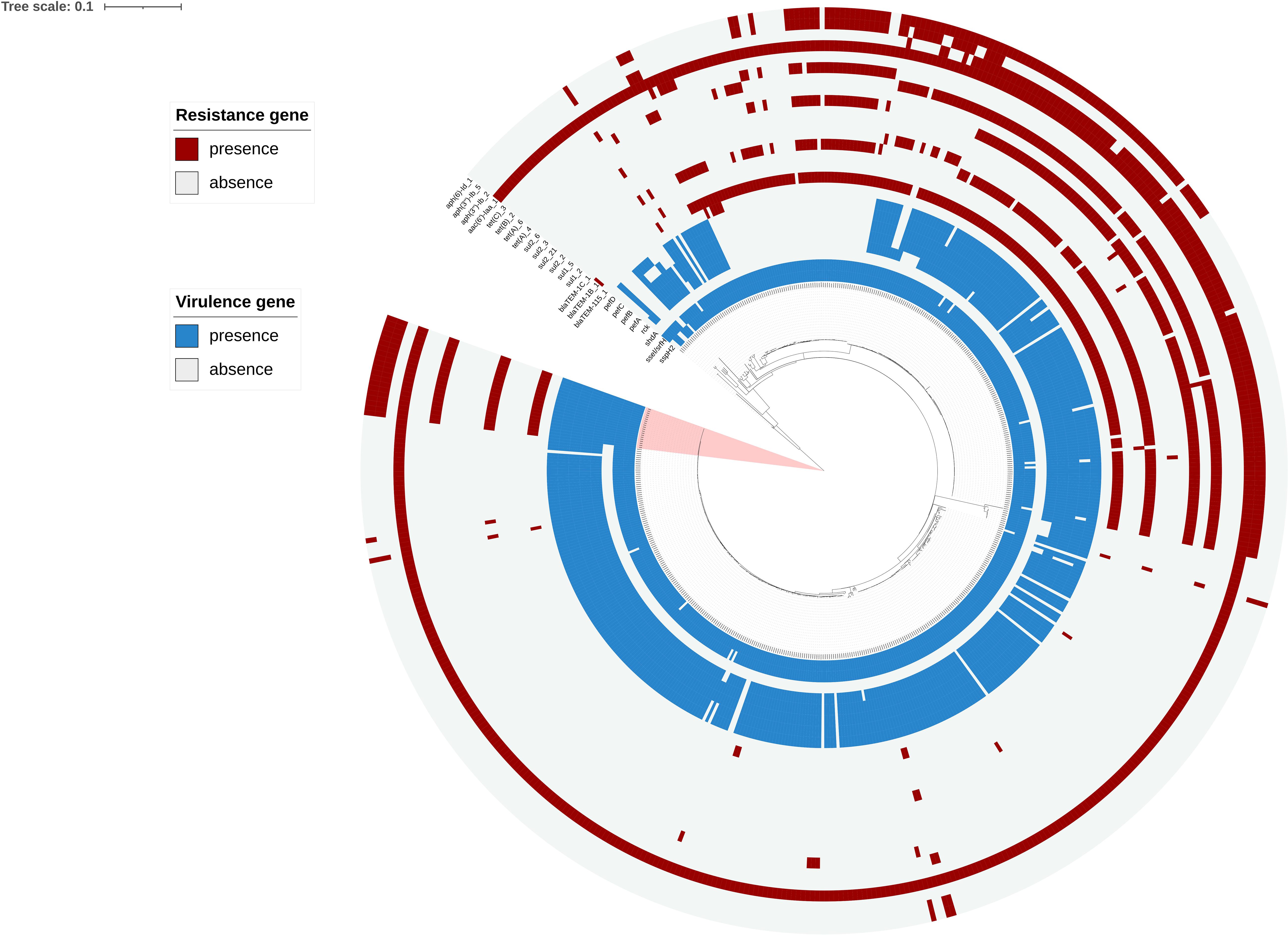
Figure 3. Phylogenetic analysis based on 533 genomes including 20 genomes from the outbreak strains and 513 public genomes. The outbreak strains are light pink areas. Presence and absence of resistance genes were represented by dark red and light gray colors, respectively. Presence and absence of virulence genes is represented by dark blue and light gray colors, respectively.
Analysis of Drug Resistance and Virulence Genes
We performed AMR genes analysis on 533 S. enteritidis strains in Figure 2, which were extracted from WGS analysis. The AMR genes carried by the 20 outbreak strains are exactly the same, including three groups of aminoglycoside resistance genes, namely aac(6′)-Iaa_1, aph(3″)-Ib_5, aph(6)-Id_1, one sulfonamide antibiotic gene, namely sul2, and one group of β-lactamase gene, namely blaTEM–1B_1 (Supplementary Table 2).
Analysis of the virulence genes of the 533 strains is shown in Supplementary Table 2, where we found that these virulence genes were identical in all outbreaks. All outbreak strains had two types of Salmonella pathogenicity islands (SPI-1 and SPI-2), and four types of regulatory function-related genes (fur, phoP, phoQ, and rpoS). The identified virulence genes include many genes related to bacterial adhesion and intestinal colonization, such as csgA-G, lpfA-E, misL, ratB, shdA, and sinH, as well as the T3SS gene sseI/srfH and bacteriophage encoded ssspH2. Interestingly, the plasmidic fimbrial operon pefABCD genes, the plasmid mediated gene rck and adherence gene shdA were detected from outbreak strains, but these genes were missed in the other strains which also located in the main branch (Figure 3).
Discussion
Here, we reported a food poisoning incident with severe gastroenteritis symptoms. Epidemiological surveys showed that 225 people exhibiting gastrointestinal symptoms all had eaten in the same canteen and presented with symptoms within 3 days. All isolated outbreak strains showed the same antibiotic resistance patterns. PFGE is considered as the “gold standard” for bacterial typing and outbreak detection (Neoh et al., 2019). PFGE results demonstrated that all 20 strains bore completely identical bands, thus confirming this was an outbreak. Previous studies have confirmed that WGS analysis provides better resolution than PFGE in determining the outbreak (Turabelidze et al., 2013). In the phylogenetic analysis, all 20 strains were tightly clustered together, with less than 50 SNP loci found to be inconsistent. These results confirmed that this incident was indeed an outbreak of S. enteritidis infection. Moreover, the 20 strains located in a main branch and formed a novel small sub-branch, it may be a newly evolved clone, which deserves our attention and in-depth study.
The epidemiological investigation found that when the egg fried rice was processed for dinner, the shell of the egg was not cleaned before used, and the cooking time of the egg fried rice was too short to guarantee food safety (1 min 50 s). Besides, the processed egg fried rice was placed in a basin, which contained the residual raw materials without cleaning. We speculated that these improper operation practices was the main cause of this outbreak. Twenty strains of S. enteritidis were recovered from the patients samples and remained egg fried rice. There are two kinds of ingredients used in egg fried rice, including boiled rice and raw eggs. The RT-PCR results showed that eggs were positive for Salmonella. The leftover rice was negative for Salmonella. So the contaminated egg fried rice was related to raw eggs carrying S. enteritidis. Therefore, this food poisoning incident was successfully traced back to the source, which was contaminated egg fried rice. Although Salmonella was not isolated from the eggs, the RT-PCR results showed that the surface of the eggshell was positive for Salmonella. So the contaminated egg fried rice may be related to raw eggs carrying S. enteritidis. The outbreak had characteristical epidemiological and clinical features. It caused a very high attack rate (69.4%) in a short incubation time, most of the patients are healthy young adults, the average age of who was only 24.1. Moreover, this outbreak caused severe clinical symptoms, and all patients with diarrhea symptoms developed high fever. The average frequency of diarrhea is 12.4 times/day, and the highest frequency of diarrhea was as high as 50 times/day. The average body temperature was 39.4°C, and the highest body temperature was as high as 42°C (Table 1). These epidemiological characteristics are rarely reported in other outbreaks caused by S. enteritidis (Inns et al., 2015; Pearce et al., 2018). China is the world’s largest egg producer and a major egg consumer (NBS. China, 2020). Thus, it is important to ensure the quality and safety of eggs. Moreover, the outbreak was related to unsanitary and irregular operations, which attract the government’s attention to strengthen health education.
It is well known that the abuse of antibiotics may lead to the development of multi-drug resistance in Salmonella, leading to increased medical costs and failure of clinical treatment (Li et al., 2020). All 20 isolated strains showed an ASSuT tetra-resistant pattern consistent with the results of resistance genes analysis. All of them encoded the chromosomal aminoglycoside acetyl-transferase gene [aac(6′)-Iaa gene] associated with resistance to STR. The blaTEM gene carried by the outbreak strains conferred resistance to AMP. The outbreak isolates co-harbored the sul2 gene, implicated in the resistance to FIS, and the tet(A) gene encoding for Tet resistance. In addition, outbreak strains also showed resistance to NAL. These five antibiotics were the most common antibiotics for which S. enterica exhibited resistance, especially in China (Liang et al., 2015; Li et al., 2020). The results of virulence gene analysis showed the isolates presented simultaneously a minimum of 114 virulence-related genes, which are related to the pathogenic potential of Salmonella isolates. The majority of important gene-encoded virulence factors are mainly located on pathogenicity islands (SPIs), which are highly conserved in Salmonella (Michael et al., 2006). Several genes are important in Salmonella virulence, included invA, spvC spaN, sipB, and sopB, are associated with the ability to invade the intestinal epithelial cells (Klein et al., 2000; Liebl et al., 2017; Zuo et al., 2020; Krishna and Dhanashree, 2021). The outbreak strains also carried a series of genes associated with adhesion (lefA-E), colonization (RatB and SinH) and biofilm formation (csgA-G), which promote invasion and survival of strains in unsuitable environments (Desai et al., 2019; Elkenany et al., 2019).
The outbreak strains carried a lot of virulence genes including several special virulence genes. We speculate that it is an important cause of severe gastroenteritis symptoms caused by this outbreak. The 6 virulence genes are pefABCD, rck, and shdA. Pef fimbriae biogenesis depends pef operon, located on the virulence plasmid of Salmonella (Ledeboer et al., 2006; Hurtado-Escobar et al., 2019). Already reported in the literature, pef mutants of S. Typhimurium were shown to be attenuated in their ability to form mature biofilms on a chicken intestinal tissue (Ledeboer et al., 2006). And pefA was a gene encoding major fimbrial subunit, which was the most-segregative virulence factor (Alshalchi et al., 2017). Rck protein belongs to the Ail/Lom protein family that consists of several bacterial or phage outer membrane proteins (OMP), these proteins are involved in the expression of pathogen virulence and rck play a key role in invasion of different host cells (Rosselin et al., 2010; Mambu et al., 2017). ShdA gene is unique in the 20 outbreak strains, while deleted in the other 513 strains. The role of shdA in S. typhimurium is mainly participated in adherence/invasion of fibronectin-producing cells (Dos et al., 2021), and shdA-mediated binding of the extracellular matrix contributes to persistent intestinal carriage (Kingsley et al., 2000; Urrutia et al., 2014). Interestingly, the study found that shdA gene were only carried in outbreak strains, but not in other strains located in the same evolutionary branch as the outbreak strain. However, the mechanism effect of shdA in S. enterica has not been reported before, and this will be the focus of our next exploration.
Conclusion
We described an outbreak in China caused by S. enteritidis, which resulted in more than 200 people to develop severe gastroenteritis symptoms in a short period of time. The outbreak was mainly caused by contaminated egg fried rice, which is a kind of delicacy with Chinese characteristics. WGS analysis results showed that 20 isolated strains carried consistent resistance genes and multiple virulence factors. Moreover, several significant virulence genes were carried in outbreak strains, included pefABCD, rck and shdA. We speculate that this is an important reason for the serious symptoms of gastroenteritis in this epidemic. The prevalence of the outbreak clone strains should be monitored. The results of this study justified that it is necessary to strengthen the education of food processors and consumers about the risk of cross-contamination of raw eggs and food, and to improve hygiene measures to prevent diseases caused by S. enteritidis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA764371).
Author Contributions
YZ wrote the main manuscript and participated in all experiments. SQ, HY, and YX designed the study and reviewed the manuscript. HQ, DD, XLiu, and YW conducted epidemiological investigation on this outbreak. HL (9th author), CY, HL (11th author), XD, QW, HW, MY participated in data collection. ST, ML, XLi, and YH participated in experiments. ZZ, KL, KZ, LW, and HS contributed to the bioinformatics data analysis. SQ, HY, and YX gave final approval of the version to be submitted. All authors made substantial contributions to preparation and submission of manuscript.
Funding
This work was supported by the Mega-Projects of Science and Technology Research (grant numbers 2018ZX10101003-001-002 and 2018ZX10714002-003-011), the National Key R&D Program of China (grant numbers 2017YFC1600100 and 2018YFC1603801), and the National Nature Science Foundation of China (grant number 81872678).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank administrators and participants for their support during the study. The authors would like to thank the Chinese PLA Centre for Disease Control and Prevention for providing experimental strains for this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.779749/full#supplementary-material
Supplementary Figure 1 | The vertical version of phylogenetic analysis based on 533 genomes including 20 genomes from the outbreak strains and 513 public genomes. The outbreak strains are light pink areas. Presence and absence of resistance genes were represented by dark red and light gray colors, respectively. Presence and absence of virulence genes were represented by dark blue and light gray colors, respectively.
Footnotes
References
Alshalchi, S., Hayer, S. S., An, R., Munoz-Aguayo, J., Flores-Figueroa, C., Nguyen, R., et al. (2017). The possible influence of non-synonymous point mutations within the FimA adhesin of non-typhoidal Salmonella (NTS) isolates in the process of host adaptation. Front. Microbiol. 8:2030. doi: 10.3389/fmicb.2017.02030
Camarda, A., Circella, E., Pupillo, A., Legretto, M., Marino, M., Pugliese, N., et al. (2015). Pulsed-field gel electrophoresis of Salmonella enterica. Methods Mol. Biol. 1301, 191–210. doi: 10.1007/978-1-4939-2599-5_16
Cheng, L., Connor, T. R., Siren, J., Aanensen, D. M., and Corander, J. (2013). Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 30, 1224–1228. doi: 10.1093/molbev/mst028
Chousalkar, K., Gast, R., Martelli, F., and Pande, V. (2018). Review of egg-related salmonellosis and reduction strategies in United States, Australia, United Kingdom and New Zealand. Crit. Rev. Microbiol. 44, 290–303. doi: 10.1080/1040841X.2017.1368998
Desai, S. K., Padmanabhan, A., Harshe, S., Zaidel-Bar, R., and Kenney, L. J. (2019). Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 116, 12462–12467. doi: 10.1073/pnas.1822018116
Dewey-Mattia, D., Manikonda, K., Hall, A. J., Wise, M. E., and Crowe, S. J. (2018). Surveillance for Foodborne Disease Outbreaks – United States, 2009–2015. MMWR Surveill. Summ. 67, 1–11. doi: 10.15585/mmwr.ss6710a1
Dos, S. A., Ferrari, R. G., Panzenhagen, P., Rodrigues, G. L., and Conte-Junior, C. A. (2021). Virulence genes identification and characterization revealed the presence of the Yersinia High Pathogenicity Island (HPI) in Salmonella from Brazil. Gene 787:145646. doi: 10.1016/j.gene.2021.145646
Elkenany, R., Elsayed, M. M., Zakaria, A. I., El-Sayed, S. A., and Rizk, M. A. (2019). Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Vet. Res. 15:124. doi: 10.1186/s12917-019-1867-z
Feasey, N. A., Hadfield, J., Keddy, K. H., Dallman, T. J., Jacobs, J., Deng, X., et al. (2016). Distinct Salmonella enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat. Genet. 48, 1211–1217. doi: 10.1038/ng.3644
Gidudu, J., Sack, D. A., Pina, M., Hudson, M. J., Kohl, K. S., Bishop, P., et al. (2011). Diarrhea: case definition and guidelines for collection, analysis, and presentation of immunization safety data. Vaccine 29, 1053–1071. doi: 10.1016/j.vaccine.2010.11.065
Hurtado-Escobar, G. A., Grepinet, O., Raymond, P., Abed, N., Velge, P., Virlogeux-Payant, I., et al. (2019). H-NS is the major repressor of Salmonella typhimurium pef fimbriae expression. Virulence 10, 849–867. doi: 10.1080/21505594.2019.1682752
Inns, T., Lane, C., Peters, T., Dallman, T., Chatt, C., Crook, P., et al. (2015). A multi-country Salmonella enteritidis phage type 14b outbreak associated with eggs from a German producer: ‘Near real-time’ application of whole genome sequencing and food chain investigations, United Kingdom, May to September 2014. Euro Surveill. 20:21098. doi: 10.2807/1560-7917.es2015.20.16.21098
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, M., Zhu, F., Yang, C., Deng, Y., Kwan, P. S. L., Lin, Y., et al. (2020). Whole-genome analysis of Salmonella enterica serovar enteritidis isolates in outbreak linked to online food delivery, Shenzhen, China, 2018. Emerg. Infect. Dis. 26, 789–792. doi: 10.3201/eid2604.191446
Jolley, K. A., Bray, J. E., and Maiden, M. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Kingsley, R. A., Amsterdam, K. V., Kramer, N., and Bäumler, A. J. (2000). The shdA gene is restricted to serotypes of Salmonella enterica subspecies i and contributes to efficient and prolonged fecal shedding. Infect. Immun. 68:2720. doi: 10.1128/IAI.68.5.2720-2727.2000
Klein, J. R., Fahlen, T. F., and Jones, B. D. (2000). Transcriptional organization and function of invasion genes within Salmonella enterica serovar typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68, 3368–3376. doi: 10.1128/IAI.68.6.3368-3376.2000
Krishna, D., and Dhanashree, B. (2021). Antibiogram, virulence genes, and biofilm-forming ability of clinical Salmonella enterica serovars: an in vitro study. Microb. Drug Resist. 27, 871–878. doi: 10.1089/mdr.2020.0419
Ledeboer, N. A., Frye, J. G., McClelland, M., and Jones, B. D. (2006). Salmonella enterica serovar typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74, 3156–3169. doi: 10.1128/IAI.01428-05
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, J., Bi, Z., Ma, S., Chen, B., Cai, C., He, J., et al. (2019). Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ. Health Perspect. 127:107009. doi: 10.1289/EHP5251
Li, Y., Yang, X., Zhang, H., Jia, H., and Liu, X. (2020). Prevalence and antimicrobial susceptibility of Salmonella in the commercial eggs in China. Int. J. Food Microbiol. 325:108623. doi: 10.1016/j.ijfoodmicro.2020.108623
Liang, Z., Ke, B., Deng, X., Liang, J., and Ran, L. (2015). Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009-2012. BMC Infect. Dis. 15:53. doi: 10.1186/s12879-015-0784-4
Liebl, D., Qi, X., Zhe, Y., Barnett, T. C., and Teasdale, R. D. (2017). SopB-mediated recruitment of SNX18 facilitates Salmonella typhimurium internalization by the host cell. Front. Cell Infect. Microbiol. 7:257. doi: 10.3389/fcimb.2017.00257
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Mambu, J., Virlogeux-Payant, I., Holbert, S., Grepinet, O., Velge, P., Wiedemann, A., et al. (2017). An updated view on the rck invasin of Salmonella: still much to discover. Front. Cell Infect. Microbiol. 7:500. doi: 10.3389/fcimb.2017.00500
Melvin, P., Weinstein, M., James, S., and Lewis, P. F. II (2021). Performance Standards for Antimicrobial Susceptibility. Wayne, PA: Clinical and Laboratory Standards Institute.
Michael, G. B., Butaye, P., Cloeckaert, A., and Schwarz, S. (2006). Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes Infect. 8, 1898–1914. doi: 10.1016/j.micinf.2005.12.019
Neoh, H., Tan, X., Sapri, H. F., and Tan, T. L. (2019). Pulsed-field gel electrophoresis (PFGE): a review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 74:103935. doi: 10.1016/j.meegid.2019.103935
Pearce, M. E., Alikhan, N., Dallman, T. J., Zhou, Z., Grant, K., and Maiden, M. C. J. (2018). Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar enteritidis outbreak. Int. J. Food Microbiol. 274, 1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023
Quick, J., Ashton, P., Calus, S., Chatt, C., Gossain, S., Hawker, J., et al. (2015). Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol. 16:114. doi: 10.1186/s13059-015-0677-2
Rosselin, M., Virlogeux-Payant, I., Roy, C., Bottreau, E., Sizaret, P. Y., Mijouin, L., et al. (2010). Rck of Salmonella enterica, subspecies enterica serovar enteritidis, mediates zipper-like internalization. Cell Res. 20, 647–664. doi: 10.1038/cr.2010.45
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Turabelidze, G., Lawrence, S. J., Gao, H., Sodergren, E., Weinstock, G. M., Abubucker, S., et al. (2013). Precise dissection of an Escherichia coli o157:h7 outbreak by single nucleotide polymorphism analysis. J. Clin. Microbiol. 51, 3950–3954. doi: 10.1128/JCM.01930-13
Urrutia, I. M., Fuentes, J. A., Valenzuela, L. M., Ortega, A. P., Hidalgo, A. A., and Mora, G. C. (2014). Salmonella typhi shdA: pseudogene or allelic variant? Infect. Genet. Evol. 26, 146–152. doi: 10.1016/j.meegid.2014.05.013
Vaughn, E. L., Vo, Q. T., Vostok, J., Stiles, T., Lang, A., Brown, C. M., et al. (2020). Linking epidemiology and whole-genome sequencing to investigate Salmonella outbreak, Massachusetts, USA, 2018. Emerg. Infect. Dis. 26, 1538–1541. doi: 10.3201/eid2607.200048
Keywords: Salmonella enterica serovar enteritidis, outbreak, severe gastroenteritis, whole genome sequencing, virulence gene, phylogenetic analysis
Citation: Zhang Y, Liu K, Zhang Z, Tian S, Liu X, Qi H, Dong D, Wang Y, Liu M, Li X, Han Y, Zhu K, Liu H, Yang C, Liu H, Du X, Wang Q, Wang H, Yang M, Wang L, Song H, Yang H, Xiang Y and Qiu S (2021) A Severe Gastroenteritis Outbreak of Salmonella enterica Serovar Enteritidis Linked to Contaminated Egg Fried Rice, China, 2021. Front. Microbiol. 12:779749. doi: 10.3389/fmicb.2021.779749
Received: 19 September 2021; Accepted: 22 October 2021;
Published: 22 November 2021.
Edited by:
Xiaoyuan Wang, Jiangnan University, ChinaReviewed by:
Chunlei Shi, Shanghai Jiao Tong University, ChinaJianmin Zhang, South China Agricultural University, China
Copyright © 2021 Zhang, Liu, Zhang, Tian, Liu, Qi, Dong, Wang, Liu, Li, Han, Zhu, Liu, Yang, Liu, Du, Wang, Wang, Yang, Wang, Song, Yang, Xiang and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Yang, eWh5QHp6dS5lZHUuY24=; Ying Xiang, aWNpY2xlOTI4QDEyNi5jb20=; Shaofu Qiu, cWl1c2hmMDYxM0Bob3RtYWlsLmNvbQ==
 Yaowen Zhang1,2
Yaowen Zhang1,2 Hongbo Liu
Hongbo Liu Chaojie Yang
Chaojie Yang Mingjuan Yang
Mingjuan Yang Ligui Wang
Ligui Wang Hongbin Song
Hongbin Song Haiyan Yang
Haiyan Yang Shaofu Qiu
Shaofu Qiu