
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 16 February 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.773664
This article is part of the Research TopicAntimicrobial Resistance in Foodborne Pathogens: ecology, epidemiology, and mechanismsView all 12 articles
 Yue Zhao1,2‡
Yue Zhao1,2‡ Zhengzheng Cao1,2‡
Zhengzheng Cao1,2‡ Luqing Cui1,2
Luqing Cui1,2 Tianyu Hu1,2
Tianyu Hu1,2 Kaixuan Guo1,2
Kaixuan Guo1,2 Fan Zhang1,2
Fan Zhang1,2 Xiangru Wang1
Xiangru Wang1 Zhong Peng1
Zhong Peng1 Quan Liu1,2
Quan Liu1,2 Menghong Dai1,2*†
Menghong Dai1,2*†This study aimed to determine the effect of enrofloxacin (ENR) on the transfer of the plasmid-mediated quinolone resistance (PMQR) gene qnrS from opportunistic pathogen Escherichia coli (E2) to Salmonella Enteritidis (SE211) and to analyze the resistance characteristics of SE211-qnrS isolates. The plasmid carrying qnrS gene of E2 was sequenced by Oxford Nanopore technology. The plasmid carrying qnrS gene belonged to incompatibility group IncY. In vitro, the transfer experiment of IncY plasmid was performed by the liquid medium conjugation method. The conjugation transfer frequency of the IncY plasmid was 0.008 ± 0.0006 in the absence of ENR, 0.012 ± 0.003 in 1/32 MICENR, 0.01 ± 0.008 in 1/8 MICENR, and 0.03 ± 0.015 (Mean±SD) in 1/2 MICENR, respectively. After inoculation of E. coli E2 and SE211, chickens were treated with different doses of ENR (3.03, 10, and 50 mg/kg b.w.) for 7 days consecutively. To screen the SE211-qnrS strains from intestinal tract of chickens, the resistance genes and susceptibility of isolates were identified. The amount of E. coli E2 and the copy number of qnrS gene in the chicken intestinal tract were determined by colony counting and qPCR, respectively. In vivo, more SE211-qnrS strains were isolated from the treated group compared with the untreated group. SE211-qnrS strains not only obtained IncY plasmid, but also showed similar resistance phenotype as E2. In conclusion, ENR treatment can promote the spread of a IncY-resistance plasmid carrying the qnrS fluoroquinolone-resistance gene in Escherichia coli and the development of drug-resistant bacteria.
Escherichia coli (abbreviated as E. coli) is a common opportunistic pathogen in the gastrointestinal tract. An estimated 44 million ETEC-related diarrheal diseases occur annually, resulting in 113,000 deaths in 2015 (Roussel et al., 2020), and the mortality is 3–5% in E. coli infection.1 Non-typhoidal Salmonellae, one of the leading causes of bacterial diarrhea worldwide, are estimated to cause approximately 153 million cases of gastroenteritis and 57,000 deaths globally each year.2 Fluoroquinolones (FQs) are commonly used in the treatment of colibacillosis and salmonellosis and can induce resistance of intestinal bacteria (Li J. et al., 2019). QnrS gene, a plasmid-mediated FQ-resistance gene, was the most prevalent quinolone gene in E. coli strains isolated from poultry feces (Gosling et al., 2012). Antibiotic resistance genes (ARGs) can be the agent of an outbreak by transferring resistance to multiple unrelated pathogens (Lerminiaux and Cameron, 2019). In the case of antibiotic abuse, whether opportunistic pathogens transfer ARGs to pathogens has attracted widespread attention. E. coli was used as a vector to transmit the qnrS gene, which could make pathogenic bacteria obtain drug-resistance genes and reach the level of clinical drug resistance (Gosling et al., 2012). Furthermore, the plasmid carrying qnrS gene in E. coli also contained other ARGs (Veldman et al., 2014; Slettemeas et al., 2019; Koyama et al., 2020), which could cotransfer with the qnrS gene. Salmonella Enteritidis 211 (abbreviated as SE211) is highly pathogenic (Cui et al., 2020, 2021), and it is sensitive to enrofloxacin. E. coli E2 carrying qnrS gene is a multidrug resistant (MDR) strain (Li et al., 2017). Previous research results show that the qnrS gene promotes the transfer efficiency of ARGs from E. coli E2 to E. coli strain EC600 in vitro, suggesting the intraspecies transfer capacity of resistance genes (opportunistic pathogen E. coli E2 to model recipient E. coli EC600). However, the interspecies transfer (opportunistic pathogen E. coli to pathogenic Salmonella) capacity of the qnrS-carrying E. coli under the antibiotic selection pressure in broiler chicken was unexplored. ENR can favor the broiler gastrointestinal (GI) tract acting as a niche for selection of MDR commensal coliforms (Li et al., 2017). Furthermore, the intestinal tract of chicken provides a habitat for different bacteria. The multifactorial and complex relationships of the microbiota ecosystem of the gut contribute to the spread of these bacteria between animals and humans (Tewari et al., 2019). Therefore, we analyzed the effect of enrofloxacin on the transfer of qnrS gene from E. coli E2 to SE211 both in vitro and in vivo and the resistance characteristic of SE211 carrying qnrS gene (SE211-qnrS). The transfer of qnrS gene from E. coli E2 to SE211 in the absence of ENR and in the presence of subminimal inhibitory concentration (sub-MIC) of ENR was performed by the liquid medium conjugation method. The chickens were gavaged with a bacteria suspension of E. coli E2 and SE211 before ENR administration. SE211-qnrS isolates were screened on plates supplemented with ENR. The colonization levels of E. coli E2 and SE211-qnrS, copy number of qnrS, were determined by traditional culture methods and molecular biology identification techniques.
The study in vivo was carried out in accordance with the guidelines established by the China Regulations for the Administration of Affairs Concerning Experimental Animals (1988) and Regulations for the Administration of Affairs Concerning Experimental Animals in Hubei province (2005) (Project No.2017YFC1600100 and Animal Welfare Assurance No. HZAUCH-2020-0005). All work was made to treat the experimental animals ethically and to minimize suffering in this study.
E. coli E2 containing the qnrS gene was used as a donor strain (MICENR = 128 mg/L). Salmonella SE211 was used as a recipient strain (MICENR = 0.25 mg/L). They were preserved by the Cooperative Innovation Center for Sustainable Pig Production (HZAU), Huazhong Agricultural University.
The standards of enrofloxacin, chloramphenicol, ampicillin, trimethoprim, and sulfamethoxazole were purchased from Dr. Ehrenstorfer (Germany). Ciprofloxacin standard was obtained from MedChemExpress (New Jersey, United States). Tetracycline standard was from China Institute of Veterinary Drug Control (Beijing, China), and 2 × EasyTaq PCR SuperMix and Phanta super-fidelity DNA Polymerase were purchased from Vazyme (Nanjing, China). The pUCm-T vector, IPTG, and X-Gal were from Beyotime Biotechnology (Nantong, China). The primers of this study were all synthesized by Genscript (Nanjing, China).
E. coli E2 was grown in LB broth, and the culture was centrifuged and quickly frozen in liquid nitrogen. Relying on Wuhan Bena Technology Service Limited Company, the genomic DNA was extracted by sodium dodecyl sulfate (SDS) and purified with a 13323 kit. The plasmid sequencing was performed by the Oxford Nanopore Technologies DNA sequencing platform. Annotation of plasmid type and resistance genes were carried out on the website.3 According to the plasmid sequencing result, NCBI was used to design the primers for plasmid replicon and resistance genes. Comparison of IncY plasmids was created by BRIG tools.4 The comparison of IncY plasmids was performed in the following order (inner to outer circles): pTET-GZEC065 (GenBank accession no. CP048027), pTetA_020022 (CP032890), pE2 (CP086663).
The conjugation experiment was conducted using the liquid mating procedure according to previous report (Lambrecht et al., 2018). E. coli E2 and SE211 with 4:1 volume ratio were cocultivated for 4 h in LB broth supplemented with 0, 1/2 MICENR (0.0125 mg/L), 1/8 MICENR (0.003 mg/L), and 1/32 MICENR (0.0007 mg/L). The coculture was diluted 104 times by LB broth and spread on the chromogenic Salmonella agar (second generation) (Hopebio, Qingdao, China). The plates were supplemented with 0.25 mg/L of ENR to screen the SE211-qnrS strains. The conjugation transfer efficiency = .
The animal trial was performed in the animal room at Ke Qian of HuaZhong Agricultural University. Twenty specific-pathogen-free (SPF) male chickens (1-day-old) were purchased from Bejing Boehringer Ingelheim Vital Biotechnology Limited Company and kept in four individual PQ3 type stainless steel poultry isolators (Suzhou Suhang Technology Equipment Co., Ltd., China), which can prevent pollution from external environmental factors. They were fed with sterile water and SPF feed (Beijing Ke Ao Xie Li Feed Limited Company, China).
On day 7, all chickens were inoculated with 0.5 mL of the donor strain (∼109 CFU/mL). On day 10, all chickens were inoculated with 0.5 mL of the recipient strain (∼109 CFU/mL). The chickens were gavaged with bacterial suspension. After successful colonization of the intestine with the donor and recipient bacteria on day 12, the 20 chickens were equally divided into four groups. Then, they were treated with different dosages of ENR (10% enrofloxacin oral solution, Bayer, Germany). The first group (group 3.03) was given a prophylactic dose (3.03 mg/kg b.w.) (Li et al., 2017). The second group (group 10) was given the ENR (10 mg/kg b.w.) as a clinical recommended dose. The third group (group 50) was given a high dose (50 mg/kg b.w.), which could effectively inhibit pathogenic bacteria. The fourth group did not receive treatment and served as a non-treated control group (group NTC). The different dosages of ENR were given for 7 days consecutively (Figure 1).
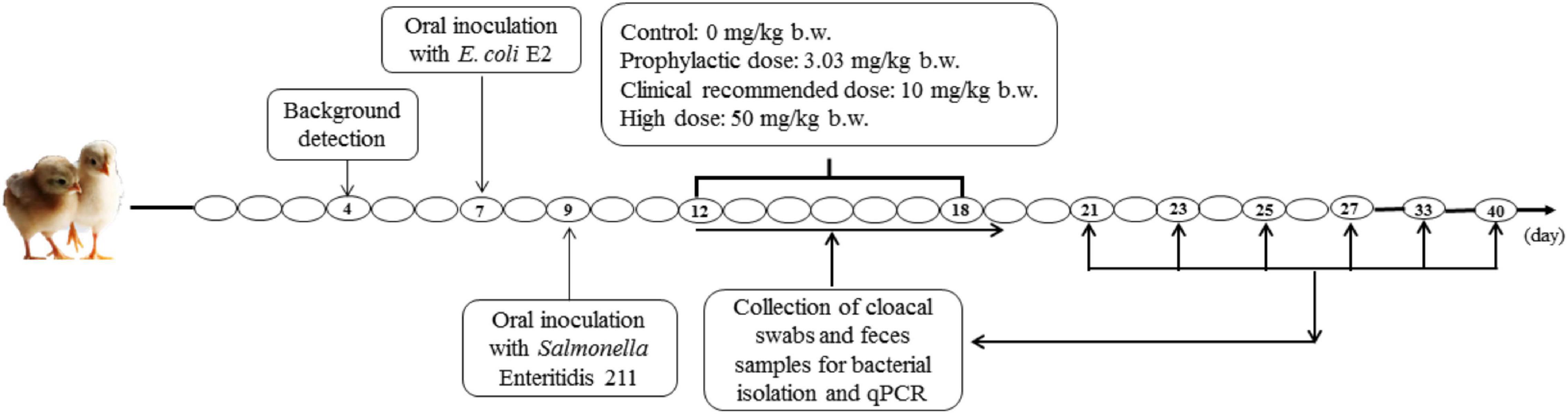
Figure 1. Experimental design. The 12–18 days represent ENR treatment for 7 days. The 21, 23, 25, 27, 33, 40 days represent 2, 4, 6, 8, 14, 21 days after termination of ENR treatment, respectively.
Prior to the inoculation with E. coli E2 and SE211, cloacal swabs were taken from each chicken, then cultivated on chromogenic Salmonella agar (second generation) plates and in LB broth (Hopebio, Qingdao, China). QnrS and repA genes were identified by PCR. None of the chickens was found to be positive for Salmonella, resistance gene qnrS, and IncY plasmid. No contamination of Salmonella, IncY plasmid, and qnrS gene were observed in the drinking water and SPF feed. Cloacal swabs and fecal samples were collected on day 12 (1 day before ENR treatment); days 13–19 (2, 4, 6, 8 days of the treatment of ENR); and days 21, 23, 25, 27, 33, and 40 (1 and 2 weeks after stopping ENR treatment), respectively (Figure 1).
Two swabs were taken from each chicken. One swab was emulsified in 2 mL of selenite cystine broth (SC) (Hopebio, Qingdao, China) and then further inoculated on chromogenic Salmonella agar (second generation) plates supplemented with 0.25 mg/L of ENR. Salmonella represented a typical purple single colony on the chromogenic Salmonella agar (second generation). The colonies were randomly screened for further amplification of qnrS and repA genes to identify SE211-qnrS strains. In the meantime, all the Salmonella and putative SE211-qnrS isolates were stored at –20°C. Another swab was weighed, which was emulsified in 1 mL of sterile 0.9% NaCl and then further diluted 105 times. An aliquot (50 μL) of the appropriate dilution was spread onto eosin-methylene blue agar (Hopebio, Qingdao, China) plates supplemented with ENR (32 mg/L). These plates were incubated at 37°C overnight to detect the colonization level of E. coli E2.
Snapgene software (Version 3.1.1) was used to design primers (5′–3′) for amplification of the full length (657 bp) of qnrS gene (F-5′-ATGGAAACCTACAATCATACATATCGG-3′ and R-5′-TTAGTCAGGATAAACAACAATACCC-3′). PCR product of qnrS gene was obtained by recycling from 1% agarose gels. Then, the recombinant DNA of pUCm-T-qnrS was obtained by ligation and transformation (DH5α). The plasmid pUCm-T-qnrS was extracted with E.Z.N.A.® Plasmid Mini Kit II (Wuhan Tianyuan Huida Biotechnology Limited Company, China). The copy number of qnrS gene was calculated by the equation: Copy number (copies/μL) = 6.02 × 1023 × cDNA (g/mL)/MW, molecular weight (MW) = (plasmid vector length + insertion fragment length) × 660 dalton/bp. Then, pUCm-T-qnrS of 1.17 × 108, 1.17 × 107, 1.17 × 106, 1.17 × 105, and 1.17 × 104 copies were used as the standards to determine the corresponding CT values. CT = –KlgC0+b was used as the standard curve formula, and C0 represented the copy number of qnrS gene.
Total DNA was extracted from 210 mg of fecal samples collected from chickens using by commercial extraction kits (Rapid extraction kit for fecal genomic DNA, Aidlab, Beijing, China) according to the manufacturer’s instruction and our modification. We increased the number of samples through the AC adsorption column and the proteinase K.
Quantitative PCR (qPCR) was performed by SYBR Green (Vazyme, Nanjing, China) detection in triplicate using a QuantStudio 3 real-time PCR detection system (Thermo Fisher Scientific, Germany) by the following procedure: 1 cycle at 95°C for 30 s, 40 cycles at 95°C for 10 s, 57°C for 10 s, 72°C for 30 s. The specificity of the PCR products was confirmed by melting curve. The copy number was calculated by equation of standard curve.
All of the isolates were confirmed to be SE211 strains and then tested the IncY plasmid by the PCR method using the primer pairs (5′–3′) for qnrS (F-5′-CATACATATCGGCACCACAAC-3′ and R-5′-CAGGATAAACAACAATACCCAGT-3′), repA (F-5′-AATTCAAACAACACTGTGCAGCCTG-3′ and R-5′-GCGAGAATGGACGATTACAAAACTTT-3′). All of the SE211-qnrS strains were tested for the resistance genes located in the IncY plasmid by PCR method using the primers (5′–3′) for blaTEM–135 (F-5′-TTGATCGTTGGGAACCGGAG-3′ R-5′-AAT AAACCAGCCAGCCGGAA-3′), tet(A) (F-5′-CATTTCGCTT GCCGCATTTG-3′ and R-5′-TCATTCCGAGCATGAGTGCC-3′), floR (F-5′-CGGATTCAGCTTTGCCTTCG-3′ R-5′-GCCAA TGTCCCGACGATACT-3′), and dfrA-14 (F-5′-CAACGATG TTACGCAGCAGG-3′ R-5′-CAATCGCGGAAAAGGCG TAG-3′).
MICs of four antibiotics (enrofloxacin, ampicillin, chloramphenicol, tetracycline) for all SE211-qnrS strains were determined by the broth microdilution method, and their antibiotic resistance level was interpreted by the Clinical and Laboratory Standards Institute guidelines (CLSI) (CLSI, 2020). In particular, the resistant breakpoint of ampicillin, chloramphenicol, and tetracycline for Salmonella strains was interpreted by the CLSI criteria (ampicillin ≥ 32 mg/L, chloramphenicol ≥ 32 mg/L, tetracycline ≥ 16 mg/L), whereas no resistant breakpoint of ENR was interpreted. Ciprofloxacin was interpreted based on the CLSI breakpoint (R ≥ 1 mg/L). E. coli ATCC 25,922 served as a quality control strain.
In this study, the plasmid stability experiment was different from the other report (Wein et al., 2019), which was evaluated by serial passages. In this study, we aimed to obtain colonies of SE211-qnrS from the glycerol bacteria. Method 1: The SE211-qnrS glycerol bacteria were cultured in LB broth supplemented with ENR (0.25 mg/L) at 37°C for 12–16 h, and then 20 μL culture was spread on chromogenic Salmonella agar (second generation) plates supplemented with ENR (1 mg/L) and incubated at 37°C for 18–24 h. Then, colonies were randomly selected on the resistant plates as a template for PCR identification. Method 2: The glycerol bacteria of SE211-qnrS was supplemented with ENR (0.25 mg/L) and stored in –20°C for 48 h. Cells of SE211-qnrS from the glycerol stock were supplemented with ENR (0.25 mg/L) and stored at –20°C for 48 h. The cells were cultured in LB broth supplemented with ENR (0.25 mg/L) at 37°C for 12 h, and 50 μL of the culture was spread on LB agar plates supplemented with (0.25 mg/L) and incubated again at 37°C for 12 h. Cells were harvested from these plates and resuspended in LB broth supplemented with (0.25 mg/L) and used as a template for PCR. QnrS and repA genes were amplified from resuscitative and eluotropic bacteria solutions to identify the SE211-qnrS strains.
Using Graphpad Prism 7.0 statistical software, the value was expressed by mean ± SD, and the differences among different time points were analyzed by Student’s t-test. *P ≤ 0.05 was set as the significance level, and **P ≤ 0.01 was set as the extremely significance difference.
The plasmid (pE2) harbored in E. coli E2 had a size of 94, 190 bp with an average G + C content of 49.59%. The replicon gene repA showed 100% identity with the replicon of IncY plasmids in the PlasmidFinder database. The plasmid distribution system parA and virB genes were associated with the self-transmitting of IncY plasmids. An oriT sequence was found between the positions 89,095 and 89,379 by oriTfinder. Multiple mobile elements were distributed on pE2, such as insertion sequences (IS) IS421, IS1, ISKpn19, IS26, IS5, IS91; integron functional element IntI1; and transposon Tn3. Additional resistance genes were found on the plasmid, such as tet(A), dfrA-14, blaTEM–135, and floR. The sequence of plasmid pE2 was highly similar to that of pTET-GZEC065 and pTetA_020022 (85% query coverage and 100% sequence identity). These plasmids all contained the floR and tet(A) resistance genes, including insertion elements (Figure 2).
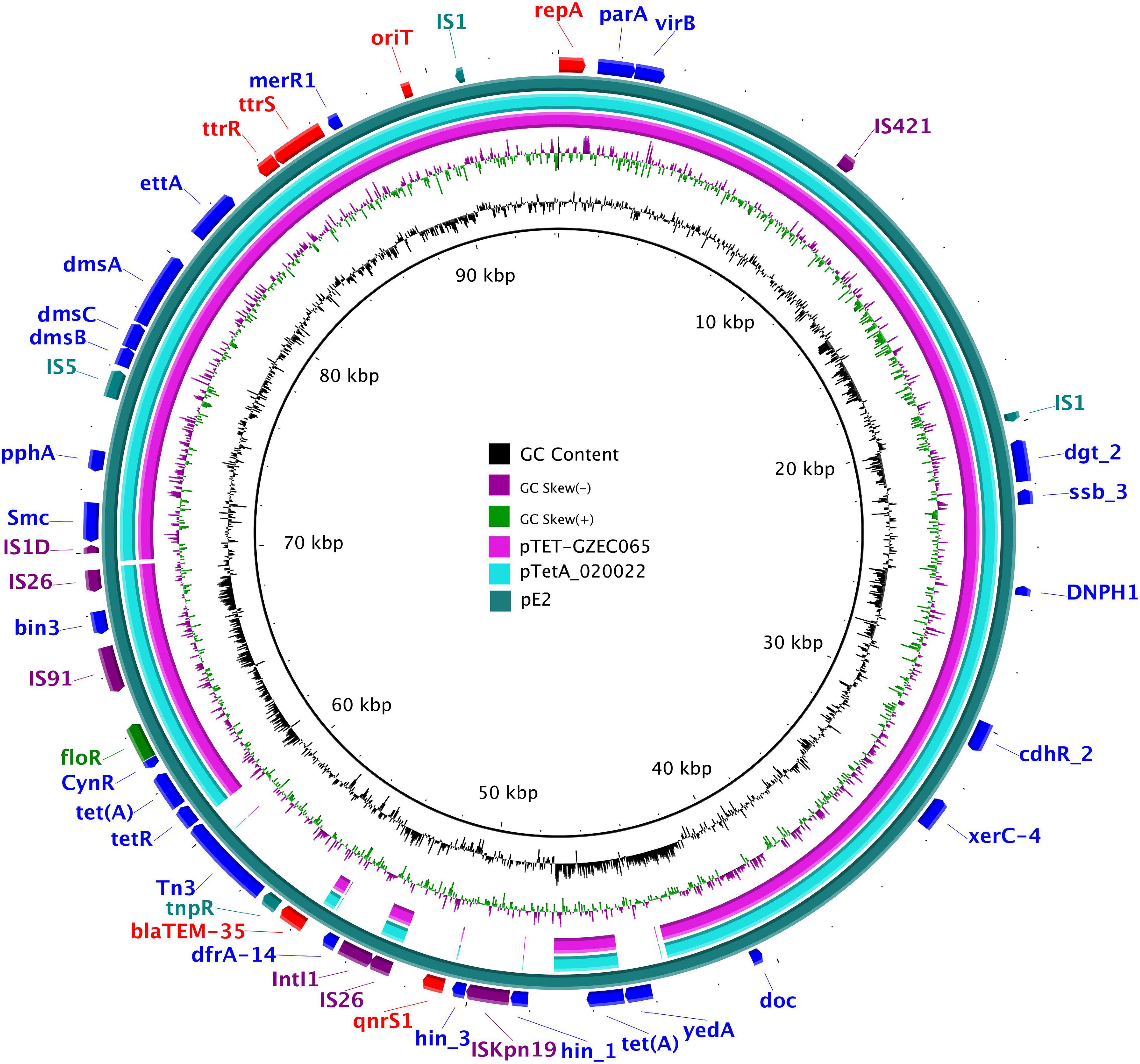
Figure 2. The complete sequence of pE2 (the outer circle) was used as a reference plasmid. The circular maps were generated using the BRIG software, and plasmids were included in the following order (inner to outer circles): pTET-GZEC065 (CP048027), pTetA_020022 (CP032890.1), pE2 (CP086663).
No significant changes in the transfer frequency of qnrS gene from resistant E. coli E2 to susceptible SE211 was observed under 0, 1/2 MICENR (0.125 mg/L), 1/8 MICENR (0.03 mg/L), and 1/32 MICENR (0.007 mg/L) conditions. However, the transfer frequency increased with increasing concentration of ENR (Table 1).
Prior to inoculation, cloacal swabs of the chickens showed no E. coli carrying qnrS gene and Salmonella as judged by the EMB agar plates supplemented with ENR (32 mg/L) and the chromogenic Salmonella agar (second generation) plates. Before ENR treatment, E. coli E2 and SE211 strains had colonized the GI tracts of chickens. E. coli E2 reached levels of 105∼107 CFU per g of feces (Figure 3). SE211 reached levels of 103∼105 CFU per mL of feces. The chickens infected with the SE211 strain showed a somnolent state and cold sensitivity and also passed out green and white loose feces. The E. coli E2 strain was still detected in the ENR treatment period and within 3 weeks after termination of ENR treatment. In the NTC group, however, E. coli E2 was not detected on day 21 after termination of ENR treatment (Figure 3). This result suggests that the resistant E. coli E2 strain could persist in the intestinal tract of chickens under the selection pressure of ENR. In this study, although E. coli E2 was isolated from the GI tracts of chickens (Li J. et al., 2019), it was easily excreted from the chicken intestine without the selective pressure of antibiotics. Then, the copy number of the qnrS gene was determined to evaluate the prevalence of the resistance gene in the chicken intestinal tract. The standard curve is CT = –3.237 lgC0 + 37.172, R2 = 0.998, Efficiency = 103.693%. This standard curve is credible. In the NTC group, the copy number of qnrS gene was higher than the initial detection level on days 1 (1 day after ENR treatment), 3, 7, S.4 (4 days after termination of ENR treatment), S.6, S.14, and lower than the initial detection level on days 5, S.2, S.8, S.21, indicating that the qnrS gene could persist in feces (Figure 4). In the prophylactic dose group, the copy number of the qnrS gene in the intestinal tracts of chickens showed a downward trend within 3 days of ENR treatment. From days 3 to S.4, the copy number of qnrS gene tended to a stable state. From days S.4 to S.21, the copy number of qnrS gene increased slightly, but it was still lower than the level before ENR treatment. In the clinically recommended dose group, the copy number of qnrS gene was lower than that time point before ENR treatment except on days 7 and S.2. In the high-dose group, the copy number of qnrS gene was higher than that time point before ENR treatment from the duration of ENR treatment to day S.4. Within 5 days of ENR treatment, the prophylactic and therapeutic doses of ENR reduced the copy number of qnrS resistance gene in chicken intestinal microflora. The high dose of ENR increased the copy number of the qnrS gene. Similarly, in the clinically recommended dose group, the copy number of the qnrS gene had an increasing trend during 5–7 days of ENR treatment. Therefore, high-dose (50 mg/kg b.w.) and long-term clinically recommended dose (>5 days) ENR treatment increases the risk of qnrS gene transmission. All of the Salmonella and putative SE211-qnrS strains were screened from selective agar plates supplemented with ENR (0.25 mg/L). In ENR treatment duration, no SE211strain was isolated from the high-dosage group. However, the SE211 and putative SE211-qnrS strains were isolated from the other groups. The number of putative SE211-qnrS strains were changeful. Prior to ENR treatment, no SE211-qnrS was detected on ENR-supplemented plates. After 1 day of ENR treatment, one SE211-qnrS strain was acquired in the prophylactic dose group. After 2 days of ENR treatment, three SE211-qnrS strains were obtained in the clinically recommended dose group. One SE211-qnrS strain was isolated from the non-treated control group. After termination of ENR treatment, four SE211-qnrS strains were obtained from the clinically recommended dose group (Table 2).
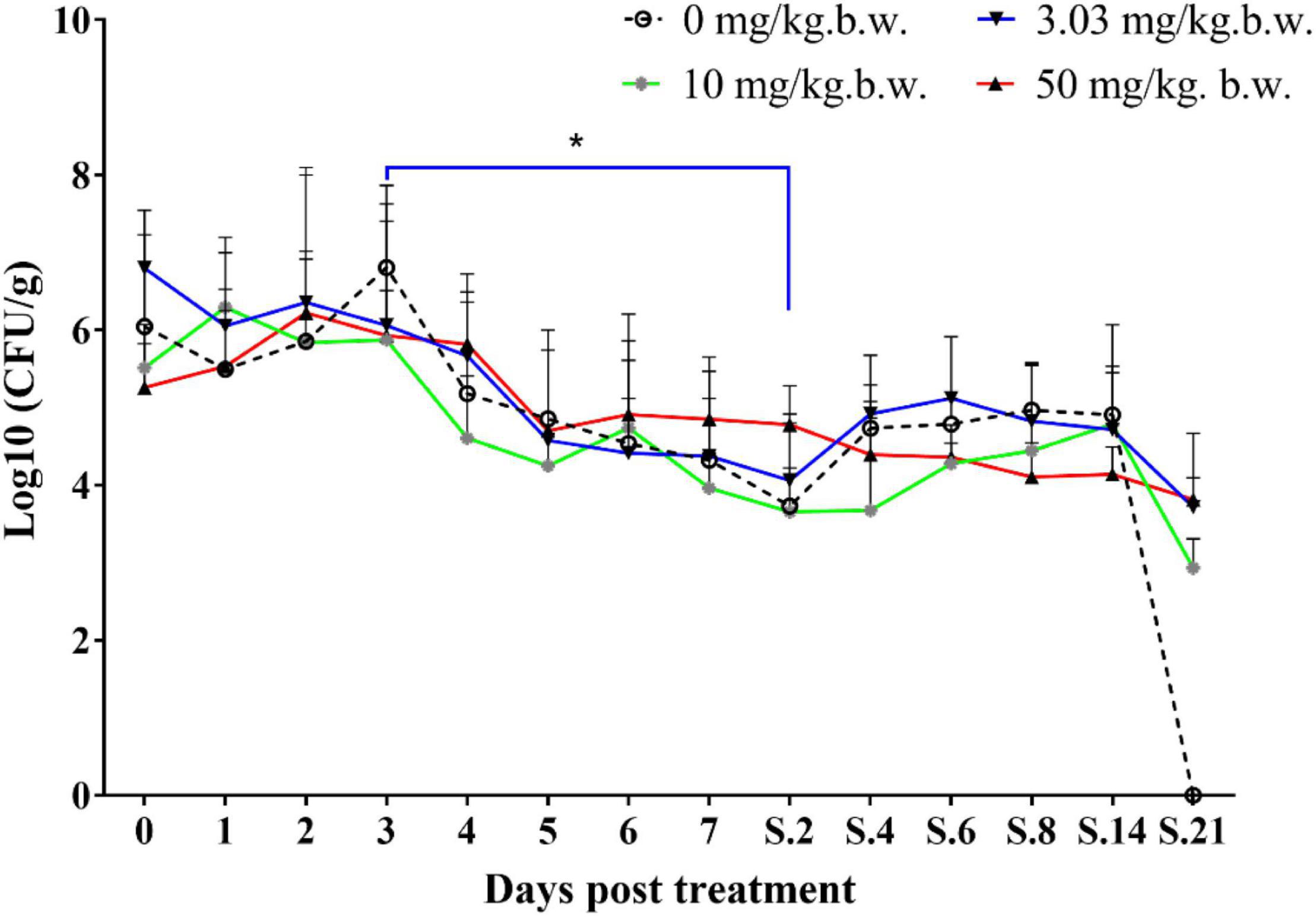
Figure 3. Colonization level of drug-resistant E. coli E2 in chicken intestine. The 0–7 represent days for ENR treatment. S represents days after termination of ENR treatment. S.2 represents 2 days after termination of ENR treatment. The *p ≤ 0.05 that was set as significance level.
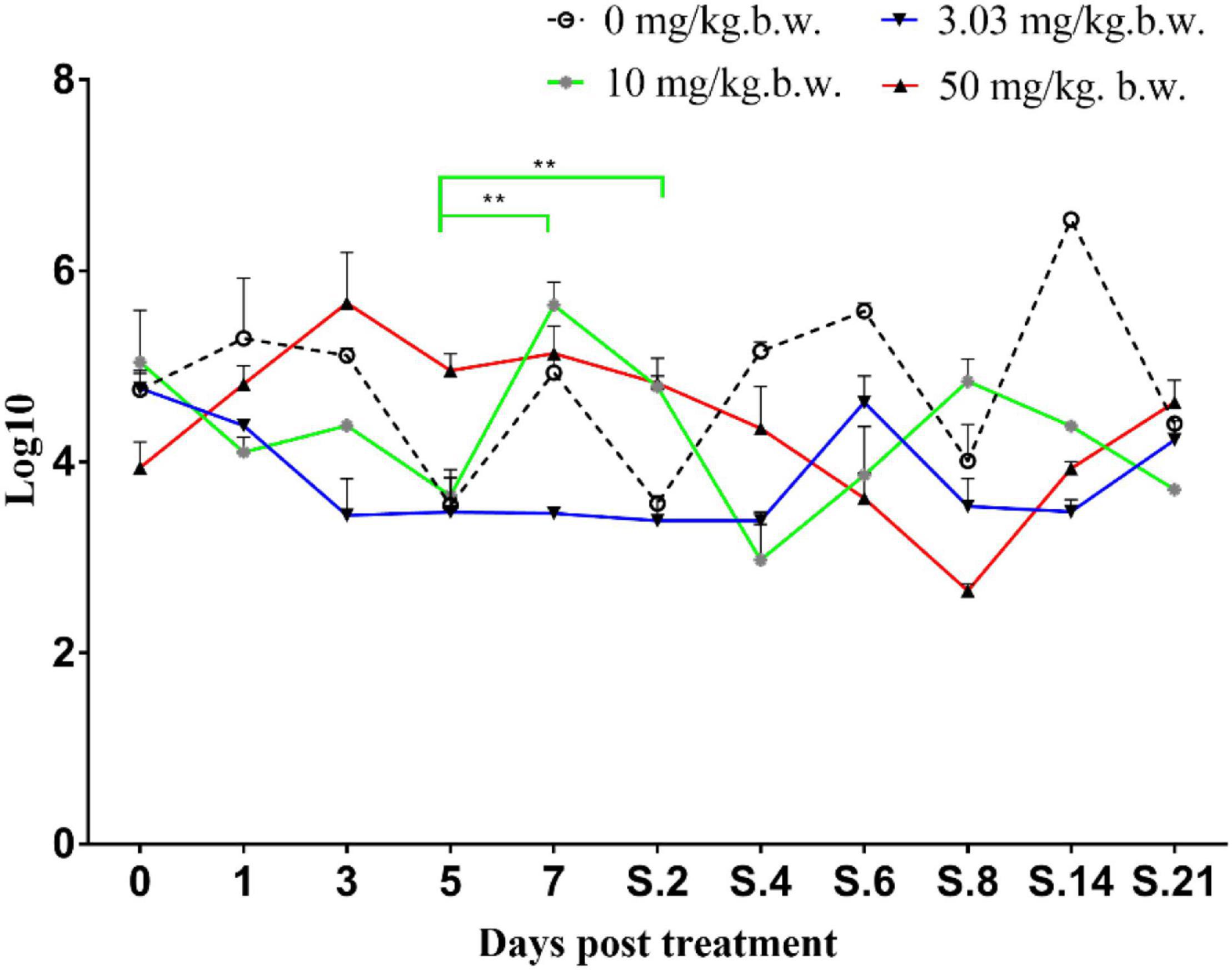
Figure 4. Copy number of qnrS gene in fecal genome. The 0–7 represent days for ENR treatment. S represents days after termination of ENR treatment. S.2 represents 2 days after termination of ENR treatment. The **p ≤ 0.01 that was set as extremely significance difference.
The resistance phenotypes of SE211-qnrS strains were similar to that of the donor strain E. coli E2, which exhibited the MDR phenotype as E. coli E2 (Table 3). 9 SE211-qnrS strains also obtained the blaTEM–35, dfrA-14, floR, and tet(A) genes located on the IncY plasmid.
The SE211-qnrS single colony was not recovered from the glycerol bacteria of 26 SE211-qnrS (17 in vitro and 9 in vivo). Although we identified two SE211-qnrS strains with positive repA and dfrA-14 genes in the screening process, they were still sensitive to trimethoprim/sulfamethoxazole when they were tested for the sensitivity to trimethoprim/sulfamethoxazole by glycerol bacteria resuscitation again. It suggested that the two strains also lost the IncY plasmid.
PMQR genes can be transmitted among bacteria through horizontal gene transfer (HGT). In this study, transfer of qnrS gene from E. coli E2 to SE211 was observed both in vitro and in vivo. Under the laboratory condition, the transfer frequency slightly increased with the increasing concentration of ENR, which might be correlated with the upregulation of conjugation-associated gene expression (Shun-Mei et al., 2018). In this experimental model of intestinal colonization in chickens with MDR E. coli E2 and sensitive SE211strains, more SE211-qnrS strains were isolated from the ENR-treated group than that from the non-treated control group. This suggests that the resistance gene qnrS can be easily transferred to Salmonella strains in vivo under selective pressure of antibiotics. However, one SE211-qnrS strain was also obtained from the NTC group. This indicates that transfer of qnrS gene in the intestinal tract of chicken might occur among different bacteria under natural conditions (Gosling et al., 2012), and this kind of horizontal transfer behavior was independent with antibiotic pressure (Le Devendec et al., 2011).
The abuse of antibiotics in poultry and the residues of antibiotics in animals and the environment play a considerable role in the development of resistance among zoonotic food-borne microorganisms (Racewicz et al., 2020). The half-life of ENR is 59.1 days/115.0 days, 88.9 days, and 190.8 days in the dark/light, aerobic, and anaerobic conditions, respectively (Slana and Sollner-Dolenc, 2016). Thus, ENR can exist in chicken feces for a long time. The use of FQs can lead to the increase of resistant strains (Marshall and Levy, 2011). The horizontal transfer of resistance plasmids was the main reason for increasing the resistance level. The treatment of ENR promotes the transfer of the qnrS gene to the SE211strain in the intestinal environment of chickens. The isolates of SE211-qnrS from the ENR treatment groups were more than that from the NTC group, which confirms the hypothesis that ENR promotes qnrS gene transfer. In addition, antibiotics can promote the transfer of resistance genes in chickens (Chen et al., 2016). Similarly, horizontal transfer of plasmid-encoded resistance determinants is reported in the animal intestinal tract under both the presence and absence of antibiotic selective pressure, and the usage of florfenicol and ENR can facilitate the transmission of resistance gene oqxAB (Chen et al., 2016). Trimethoprim significantly increased both the HGT and vertical gene transfer frequencies (Li B. et al., 2019). Furthermore, the persistence of antibiotics may lead to the maintenance of resistant bacteria and resistance genes. The colonization results of E. coli E2 indicates that ENR can maintain the persistence of E. coli E2 carrying qnrS gene. It also indicates that treatment with ENR can increase the number of resistant E. coli in chicken gut (Roth et al., 2017). Therefore, it is necessary to continuously strengthen the monitoring of resistance genes in clinical strains and take effective measures to eliminate resistance genes and plasmids so as to prevent the spread of resistance genes from aggravating clinical drug resistance.
The other resistance genes coexisting with the qnrS gene on the IncY plasmid were generally β-lactam-resistance genes. In the reported IncY plasmid carrying qnrS gene, it usually came from E. coli in the food chain (Roschanski et al., 2017) and healthy people (Mshana et al., 2016). Similar to this study, the IncY plasmid carried MDR genes to mediate MDR. IncY plasmid as a repository of MDR genes, E. coli can pose a threat to human health through the food chain. Here, we found that the qnrS gene was cotransferred with the other resistance genes (blaTEM–35, tetA, floR, and dfrA-14) on the IncY plasmid. In E. coli, cotransfer of the qnrS gene with β-lactam-resistance genes is reported (Wu et al., 2008; Jiang et al., 2012, 2014). pTET-GZEC065, pTetA_020022, pE2 plasmids had high similarities, carrying different resistance genes, which might be Tn3-family members that confer resistance to antibiotics (Nicolas et al., 2015). Recently, there are many identified Tn3 family members with different combinations of antibiotic-resistance determinants (Stokes and Gillings, 2011; Nordmann et al., 2012). Antimicrobial susceptibility tests show that the resistance spectrum of the SE211-qnrS strain was similar to that of donor strain E. coli E2 (Table 3). The resistance phenotypes were consistent with the resistance genotypes. In addition, we also obtained a drug-resistant Salmonella strain (MICENR = 4 mg/L) from the intestinal tract of chicken without the IncY plasmid and qnrS gene. We speculate that the resistance mechanism may be attributed to mutations in the parE gene. The emergence and increase of MDR bacteria pose a great threat to public health.
The level of conjugated plasmid-mediated drug resistance in Salmonella was found to be slightly higher than that in E. coli (Chen et al., 2018). Our research also confirms this finding. In this study, MICENR of SE211-qnrS strains were 64 or 128 mg/L, whereas MICENR of EC600-qnrS strain was 8 mg/L (Li J. et al., 2019). The plasmids generally impose a fitness cost on their hosts (Carroll and Wong, 2018). The replication of plasmids in the host bacteria caused metabolic load, and the expression of plasmid-encoded genes was one of the important reasons resulting in the metabolic load of plasmids in the host bacteria (Silva et al., 2012). The expression of plasmid-encoded resistance genes led to the stress response of the host bacteria, resulting in the loss of plasmids (Rozkov et al., 2004). In this study, IncY plasmid carried qnrS1, tet(A), floR, dfrA-14, and blaTEM–135 resistance genes and mediated the resistance of ENR, TET, CHL, TMP, and AMP. Compared with the reported IncY plasmid carrying qnrS, the pE2 resistance spectrum was larger. The expression of MDR genes in IncY plasmid undoubtedly increased the metabolic burden of the host bacteria. In addition, the plasmid also had several insertion elements IS421, IS1, ISkpn19, IS26, IS5, and IS91; integron functional elements IntI1; and transposon Tn3, the expression of IS elements and transposons caused the instability of plasmid DNA structure (Haddadin and Harcum, 2005). The copy number of plasmids was also one of the factors determining the metabolic load of plasmids, and the selective pressure of antibiotics was an important condition for maintaining the stability of resistance plasmids. Plasmids with high copy numbers, especially those containing the β-lactam-resistance gene, were unstable and lost quickly in Salmonella without antibiotic selection pressure both in vivo and in vitro (Zhang et al., 2005). In this study, we did not obtain SE211-qnrS colonies on plates supplemented with ENR. Although the repA and dfrA-14 genes of two SE211-qnrS strains were positive in the screening process, they were still sensitive to trimethoprim/sulfamethoxazole following glycerol bacteria resuscitation (data not shown). We speculate that antibiotic-free glycerol preserved SE211-qnrS strains might easily lose the IncY plasmid. In this study, loss of the IncY plasmid in SE211 might be caused by the absence of antibiotic selective pressure. Thus, antibiotics should be appropriately added to the glycerol bacteria of Salmonella transconjugants.
The QnrS gene located on IncY plasmid can transfer from E. coli E2 to Salmonella SE211 both under laboratory conditions and in the chicken intestinal environment. Sub-MIC of ENR and the clinically recommended dose of ENR can promote conjugation transfer of the qnrS gene. The selective pressure exerted by ENR on the intestinal environment of chickens contributed to the persistence of resistance E. coli and qnrS genes, thus increasing the risk of resistance gene transmission as well as the resistance gene reservoir.
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a manuscript that does not adhere to our open data policies.
The animal study was reviewed and approved by the study in vivo was carried out in accordance with the guidelines established by the China Regulations for the Administration of Affairs Concerning Experimental Animals (1988) and Regulations for the Administration of Affairs Concerning Experimental Animals in Hubei province (2005) (Project No. 2017YFC1600100 and Animal Welfare Assurance No. HZAUCH- 2020-0005). Written informed consent was obtained from the owners for the participation of their animals in this study.
MD conceived and designed the study, wrote, reviewed, and edited the manuscript. YZ, ZC, LC, TH, KG, and FZ performed the experiments. YZ and ZC wrote the draft manuscript. All authors participated in the interpretation of the results and read and approved the manuscript.
This work was supported by the National Key R&D Program of China (grant no. 2017YFC1600100), Walmart Foundation (Project no. 61626817) of Walmart Food Safety Collaboration Center and the National Natural Science Foundation of China (NSFC) (grant no. 31772736).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Carroll, A. C., and Wong, A. (2018). Plasmid persistence: costs, benefits, and the plasmid paradox. Can. J. Microbiol. 64, 293–304. doi: 10.1139/cjm-2017-0609
Chen, K., Dong, N., Zhao, S., Liu, L., Li, R., Xie, M., et al. (2018). Identification and characterization of conjugative plasmids that encode ciprofloxacin resistance in Salmonella. Antimicrob Agents Chemotherapy 8, e518–e575. doi: 10.1128/AAC.00575-18
Chen, Y., Sun, J., Liao, X., Shao, Y., Li, L., Fang, L., et al. (2016). Impact of enrofloxacin and florfenicol therapy on the spread of OqxAB gene and intestinal microbiota in chickens. Vet. Microbiol. 192, 1–9. doi: 10.1093/jac/dkn033
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30th Edn. Wayne, PA: CLSI. CLSI supplement M100.
Cui, L., Wang, X., Huang, D., Zhao, Y., Feng, J., Lu, Q., et al. (2020). CRISPR-cas3 of Salmonella upregulates bacterial biofilm formation and virulence to host cells by targeting quorum-sensing systems. Pathogens 9:53. doi: 10.3390/pathogens9010053
Cui, L., Wang, X., Zhao, Y., Peng, Z., Gao, P., Cao, Z., et al. (2021). Virulence comparison of Salmonella enterica Subsp. enterica isolates from chicken and whole genome analysis of the high.virulent strain S. enteritidis 211. Microorganisms 9:2239.
Gosling, R. J., Clouting, C. S., Randall, L. P., Horton, R. A., and Davies, R. H. (2012). Ciprofloxacin resistance in E. coli isolated from turkeys in Great Britain. Avian Pathol. 41, 83–89. doi: 10.1080/03079457.2011.640659
Haddadin, F. T., and Harcum, S. W. (2005). Transcriptome profiles for high-cell-density recombinant and wild-type Escherichia coli. Biotechnol. Bioeng. 90, 127–153. doi: 10.1002/bit.20340
Jiang, H. X., Tang, D., Liu, Y. H., Zhang, X. H., Zeng, Z. L., Xu, L., et al. (2012). Prevalence and characteristics of beta-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67, 2350–2353. doi: 10.1093/jac/dks250
Jiang, X., Li, J., Zhang, Y., Yan, H., Wang, Y., Shi, L., et al. (2014). Detection of plasmid-mediated quinolone resistance determinants and qnrS expression in Enterobacteriaceae clinical isolates. J. Infect. Dev. Countr. 8, 1625–1629. doi: 10.3855/jidc.4617
Koyama, S., Murase, T., and Ozaki, H. (2020). Research note: longitudinal monitoring of chicken houses in a commercial layer farm for antimicrobial resistance in Escherichia coli with special reference to plasmid-mediated quinolone resistance. Poultry Sci. 99, 1150–1155. doi: 10.1016/j.psj.2019.09.005
Lambrecht, E., Van Meervenne, E., Boon, N., Van de Wiele, T., Wattiau, P., Herman, L., et al. (2018). Characterization of cefotaxime- and ciprofloxacin-resistant commensal Escherichia coli originating from Belgian farm animals indicates high antibiotic resistance transfer rates. Microb. Drug Resist. 24, 707–717. doi: 10.1089/mdr.2017.0226
Le Devendec, L., Bouder, A., Dheilly, A., Hellard, G., and Kempf, I. (2011). Persistence and spread of qnr, extended-spectrum beta-lactamase, and ampC resistance genes in the digestive tract of chickens. Microb. Drug Resist. 17, 129–134. doi: 10.1089/mdr.2010.0096
Lerminiaux, N. A., and Cameron, A. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi: 10.1139/cjm-2018-0275
Li, B., Qiu, Y., Song, Y., Lin, H., and Yin, H. (2019). Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 131L105007. doi: 10.1016/j.envint.2019.105007
Li, J., Hao, H., Cheng, G., Wang, X., Ahmed, S., Shabbir, M. A. B., et al. (2017). The effects of different enrofloxacin dosages on clinical efficacy and resistance development in chickens experimentally infected with Salmonella Typhimurium. Sci. Rep. 7:11676. doi: 10.1038/s41598-017-12294-7
Li, J., Hao, H., Dai, M., Zhang, H., Ning, J., Cheng, G., et al. (2019). Resistance and virulence mechanisms of Escherichia coli selected by enrofloxacin in chicken. Antimicrob. Agents Chemother. 63:e182418. doi: 10.1128/AAC.01824-18
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Mshana, S. E., Falgenhauer, L., Mirambo, M. M., Mushi, M. F., Moremi, N., Julius, R., et al. (2016). Predictors of blaCTX–M–15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect Dis. 16:187. doi: 10.1186/s12879-016-1527-x
Nicolas, E., Lambin, M., Dandoy, D., Galloy, C., Nguyen, N., Oger, C. A., et al. (2015). The Tn3-family of replicative transposons. Microbiol. Spectr. 3, 693–726. doi: 10.1128/microbiolspec.MDNA3-0060-2014
Nordmann, P., Dortet, L., and Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18, 263–272. doi: 10.1016/j.molmed.2012.03.003
Racewicz, P., Majewski, M., Madeja, Z. E., Lukomska, A., and Kubiak, M. (2020). Role of integrons in the proliferation of multiple drug resistance in selected bacteria occurring in poultry production. Br. Poult. Sci. 61, 122–131. doi: 10.1080/00071668.2019.1697426
Roschanski, N., Guenther, S., Vu, T., Fischer, J., Semmler, T., Huehn, S., et al. (2017). VIM-1 carbapenemase-producing Escherichia coli isolated from retail seafood, Germany 2016. Euro. Surveill. 22:17-00032. doi: 10.2807/1560-7917.ES.2017.22.43.17-00032
Roth, N., Mayrhofer, S., Gierus, M., Weingut, C., Schwarz, C., Doupovec, B., et al. (2017). Effect of an organic acids based feed additive and enrofloxacin on the prevalence of antibiotic-resistant E. coli in cecum of broilers. Poult. Sci. 96, 4053–4060. doi: 10.3382/ps/pex232
Roussel, C., De Paepe, K., Galia, W., De Bodt, J., Chalancon, S., Leriche, F., et al. (2020). Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 18:141. doi: 10.1186/s12915-020-00860-x
Rozkov, A., Avignone-Rossa, C. A., Ertl, P. F., Jones, P., O’Kennedy, R. D., Smith, J. J., et al. (2004). Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol. Bioeng. 88, 909–915. doi: 10.1002/bit.20327
Shun-Mei, E., Zeng, J., Yuan, H., Lu, Y., Cai, R., and Chen, C. (2018). Sub-inhibitory concentrations of fluoroquinolones increase conjugation frequency. Microb. Pathogenesis 114, 57–62. doi: 10.1016/j.micpath.2017.11.036
Silva, F., Queiroz, J. A., and Domingues, F. C. (2012). Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 30, 691–708. doi: 10.1016/j.biotechadv.2011.12.005
Slana, M., and Sollner-Dolenc, M. (2016). Enrofloxacin degradation in broiler chicken manure under various laboratory conditions. Environ. Sci. Pollut. Res. Int. 23, 4422–4429. doi: 10.1007/s11356-015-5624-y
Slettemeas, J. S., Sunde, M., Ulstad, C. R., Norstrom, M., Wester, A. L., and Urdahl, A. M. (2019). Occurrence and characterization of quinolone resistant Escherichia coli from Norwegian turkey meat and complete sequence of an IncX1 plasmid encoding qnrS1. PLoS One 14:e212936. doi: 10.1371/journal.pone.0212936
Stokes, H. W., and Gillings, M. R. (2011). Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819. doi: 10.1111/j.1574-6976.2011.00273.x
Tewari, R., Mitra, S., Ganaie, F., Das, S., Chakraborty, A., Venugopal, N., et al. (2019). Dissemination and characterisation of Escherichia coli producing extended-spectrum beta-lactamases, AmpC beta-lactamases and metallo-beta-lactamases from livestock and poultry in Northeast India: a molecular surveillance approach. J. Glob. Antimicrob. Resist. 17, 209–215. doi: 10.1016/j.jgar.2018.12.025
Veldman, K., Kant, A., Dierikx, C., van Essen-Zandbergen, A., Wit, B., and Mevius, D. (2014). Enterobacteriaceae resistant to third-generation cephalosporins and quinolones in fresh culinary herbs imported from Southeast Asia. Int. J. Food Microbiol. 177, 72–77. doi: 10.1016/j.ijfoodmicro.2014.02.014
Wein, T., Hulter, N. F., Mizrahi, I., and Dagan, T. (2019). Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 10:2595. doi: 10.1038/s41467-019-10600-7
Wu, J. J., Wen-Chien, K., Wu, H. M., and Yan, J. J. (2008). Prevalence of Qnr determinants among bloodstream isolates of Escherichia coli and Klebsiella pneumoniae in a Taiwanese Hospital, 1999–2005. J. Antimicrob. Chemoth. 6, 1234–1239. doi: 10.1093/jac/dkn111
Keywords: qnrS, Escherichia coli E2, Salmonella enterica serovar enteritidis SE211, enrofloxacin, resistance transfer
Citation: Zhao Y, Cao Z, Cui L, Hu T, Guo K, Zhang F, Wang X, Peng Z, Liu Q and Dai M (2022) Enrofloxacin Promotes Plasmid-Mediated Conjugation Transfer of Fluoroquinolone-Resistance Gene qnrS. Front. Microbiol. 12:773664. doi: 10.3389/fmicb.2021.773664
Received: 10 September 2021; Accepted: 17 December 2021;
Published: 16 February 2022.
Edited by:
Bao-Tao Liu, Qingdao Agricultural University, ChinaReviewed by:
Valentine Usongo, Health Canada, CanadaCopyright © 2022 Zhao, Cao, Cui, Hu, Guo, Zhang, Wang, Peng, Liu and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Menghong Dai, ZGFpbWVuZ2hvbmdAbWFpbC5oemF1LmVkdS5jbg==
†Present address: Menghong Dai, The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan, China
‡These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.