- 1State Key Laboratory of Tropical Oceanography, Key Laboratory of Tropical Marine Bio-Resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 3Guangdong Provincial Academy of Environmental Science, Guangzhou, China
- 4School of Biological Sciences, Universiti Sains Malaysia, Penang, Malaysia
- 5Daya Bay Marine Biology Research Station, Chinese Academy of Sciences, Shenzhen, China
In this study, 16S high-throughput and metagenomic sequencing analyses were employed to explore the changes in microbial community and function with the succession of mangroves (Sonneratia alba, Rhizophora apiculata, and Bruguiera parviflora) along the Merbok river estuary in Malaysia. The sediments of the three mangroves harbored their own unique dominant microbial taxa, whereas R. apiculata exhibited the highest microbial diversity. In general, Gammaproteobacteria, Actinobacteria, Alphaproteobacteria, Deltaproteobacteria, and Anaerolineae were the dominant microbial classes, but their abundances varied significantly among the three mangroves. Principal coordinates and redundancy analyses revealed that the specificity of the microbial community was highly correlated with mangrove populations and environmental factors. The results further showed that R. apiculata exhibited the highest carbon-related metabolism, coinciding with the highest organic carbon and microbial diversity. In addition, specific microbial taxa, such as Desulfobacterales and Rhizobiales, contributed the highest functional activities related to carbon metabolism, prokaryote carbon fixation, and methane metabolism. The present results provide a comprehensive understanding of the adaptations and functions of microbes in relation to environmental transition and mangrove succession in intertidal regions. High microbial diversity and carbon metabolism in R. apiculata might in turn facilitate and maintain the formation of climax mangroves in the middle region of the Merbok river estuary.
Introduction
Mangroves represent one of the most productive ecosystems in tropical and subtropical estuaries and shorelines. They possess biological resources and play important roles in carbon fixation, erosion mitigation, and water purification (Giri et al., 2011; Brander et al., 2012). They often occur in marine-terrestrial ecotones with obvious geographical and hydrological heterogeneities, leading to interesting sequential species zonation along continuous gradients. The adaptation and succession of mangroves in intertidal regions are speculated to be closely related to microbes in the sediments. Unfortunately, the potential roles of microbes and their functions in mangrove ecosystems are still poorly understood, although changes in vegetation during mangrove succession and how mangrove plants adapt to intertidal environmental adversities have been well studied (Wang et al., 2019; Cheng et al., 2020).
In recent years, high-throughput sequencing has offered a comprehensive perspective of microbes (Andreote et al., 2012; Alzubaidy et al., 2016; Lin et al., 2019). Benthic microbial community is highly correlated with soil properties (the depth of soil layer, pH, salinity, and nutrient availability) (Abed et al., 2015; Zhou et al., 2017; Tong et al., 2019) and aboveground plants (Bardgett and van der Putten, 2014). Previous findings also showed that microbial diversity and function might vary significantly among different mangrove habitats because of environmental transition and mangrove succession (Bai et al., 2013). Mangrove coverage also regulates the structure and composition of microbial community by altering redox conditions and organic carbon levels in the sediments (Holguin et al., 2001).
Moreover, microbes in sediments also play important biogeochemical roles (e.g., C, N, and S cycles), which can facilitate mangrove survival in intertidal regions (Reis et al., 2017). Owing to the withering and retention of mangrove branches and leaves, mangrove sediments contain a large amount of organic carbon, and most carbon turnover in mangrove ecosystems is carried out by sediment microbes (Alongi, 1988). Benthic microbes may also promote the efficiency of biogeochemical cycles in the sediments, such as C, N, and S cycles (Lin et al., 2019). In addition, anaerobic metabolism can further facilitate the production and consumption of methane and nitrous oxide (Giani et al., 1996; Reis et al., 2017), which can contribute to the emission of greenhouse gases from mangrove wetlands (Rosentreter et al., 2018). However, the responses of microbes to environmental transition and mangrove succession have not been well demonstrated. It is essential to further identify microbial taxa and their metabolic potential in mangrove ecosystems.
Thus, the present study aimed to (i) identify the changes in microbial community and diversity among different mangrove habitats; (ii) explore microbial functions and metabolic potentials in mangrove ecosystems; and (iii) explore the potential correlations among environmental factors, mangrove populations, and benthic microbial communities and functions. The purpose of this study was to evaluate the hypothesis that microbial community and function would respond positively to environmental transitions and might contribute to mangrove survival and succession in intertidal regions. Therefore, surface sediments from three mangrove fields (Sonneratia alba, Rhizophora apiculata, and Bruguiera parviflora) were examined using 16S high-throughput and metagenomic sequencing analyses. The implications of this study should be useful for guiding future research on the roles of the microbial community and their functions in mangrove succession.
Materials and Methods
Study Area and Sample Collection
Sediment samples were collected on November 25, 2019, in a mangrove reserve located in the Merbok river estuary, Malaysia (Figure 1). In the upper estuary (with lower salinity), the habitats were mainly occupied by S. alba and sporadically mixed with Nypa fruticans, whereas the lower estuary had groves of B. parviflora and sporadic Avicennia genera. In the middle region of the Merbok river estuary, the dominant species was R. apiculata. According to tidal transition and mangrove succession, surface sediments were collected from three mangrove populations (S. alba, R. apiculata, and B. parviflora), and five parallel samples were collected from each mangrove population. The samples were placed in liquid nitrogen and frozen for nucleic acid extraction. The chemical parameters of the sediments were determined and described as follows.
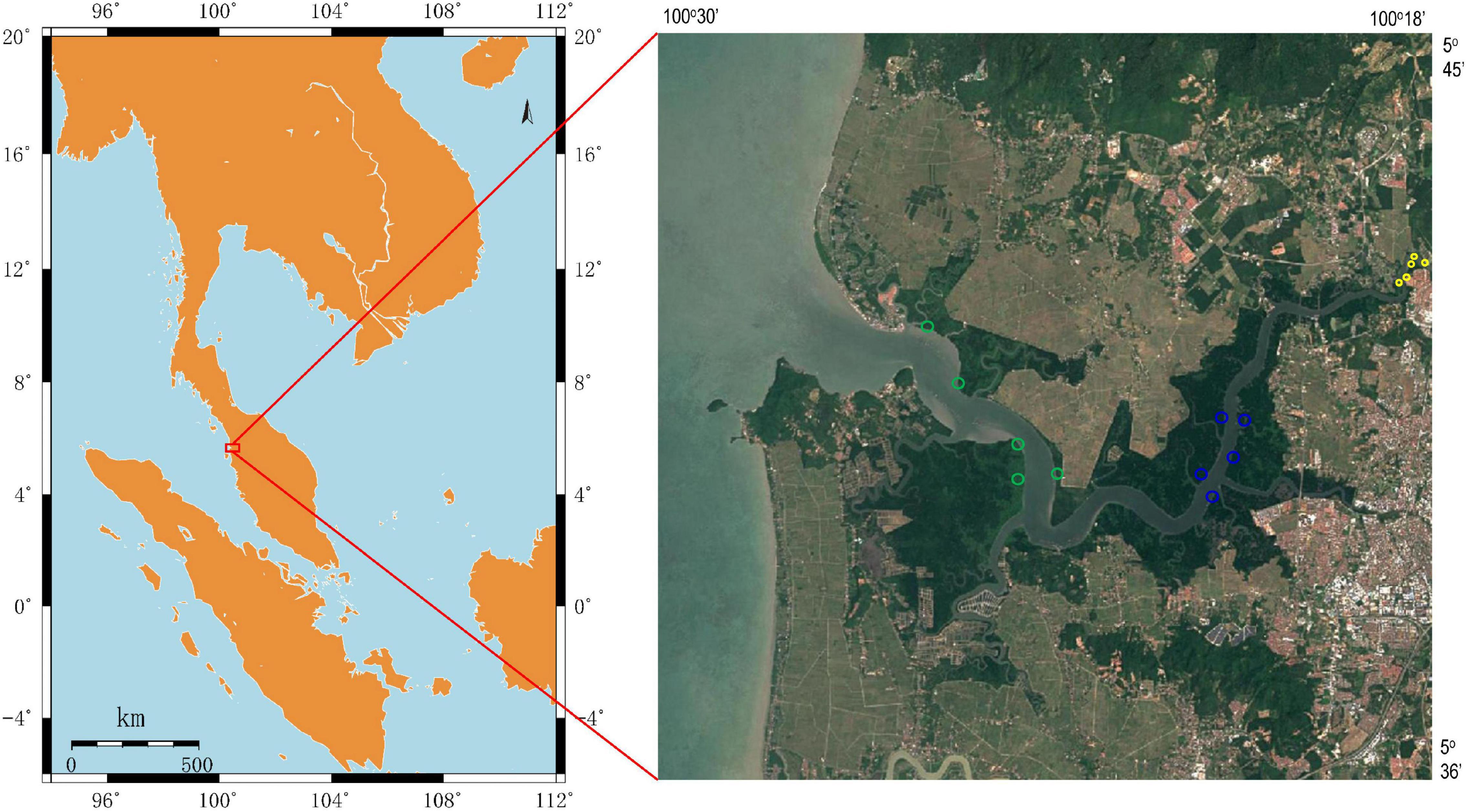
Figure 1. Location of sampling sites in Merbok river estuary, Malaysia. Sediment samples were collected from Sonneratia alba (yellow circle), Rhizophora apiculata (blue circle) and Bruguiera parviflora (green circle).
Determination of the Chemical Parameters of Mangrove Sediments
The sediment properties included salinity, pH, total organic carbon (TOC), total phosphorus (TP), and total nitrogen (TN). Salinity and pH were measured during sampling in the field. The samples were dried naturally and sieved (2 mm). TOC, TP, and TN in the sediments were then analyzed following standard measurements (Huang et al., 2017).
Illumina Sequencing of 16S rRNA Gene and Data Analysis
Sediment samples (1 g) were weighed, and total DNA was extracted using an E.Z.N.A.® Soil DNA Kit (Omega, Norcross, GA, United States). In this study, the V3 and V4 regions of the 16S rRNA gene were sequenced using 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers. PCR was performed in a 25 μl reaction containing the following: 5 × FastPfu buffer (4 μl), 2.5 mM of dNTPs (2 μl), 5 U/μl of FastPfu polymerase (0.5 μl), 5.0 μM of primers (1.0 μl each), and template DNA (10 ng). PCR was performed using the following conditions: pre-denaturation at 95°C for 3 min; 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s; and a final extension at 72°C for 10 min. After amplification, the PCR products were purified and analyzed by paired-end sequencing (2 × 250) using the Illumina MiSeq platform (Illumina, San Diego, CA, United States) according to standard protocols. All sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) Database under accession number PRJNA756333.
For the paired-end reads obtained by MiSeq sequencing, samples were distinguished according to the barcode information and then spliced according to overlap sequences using FLASH (version 1.2.11). After quality control analysis, normalization of clean data was carried out for operational taxonomic unit (OTU) clustering analysis and species taxonomy analysis. CD-HIT tool was used to define tags with sequence similarities >97% as OTU clusters. QIIME software (version 1.9.1) was used to analyze the alpha diversities of the sequences, based on the Shannon index and ACE index. A representative sequence (showing the highest default abundance) in each OTU was selected for species classification using RDP classifier software (version 2.11) with the default threshold 0.8. Two-sided Welch’s t-test was used to analyze the statistical significances of microbial community structure in different samples. Beta diversity was calculated using unweighted UniFrac, and principal coordinates analysis (PCoA) was performed with R software (version 3.3.1). Redundancy analysis (RDA) was performed using the R software (version 3.3.1) vegan package, and statistical significances of RDA were judged by performing the PERMUTEST analysis of variance.
Metagenomic Sequencing Analysis
Qualified DNA samples were diluted in fragmentation buffer and randomly disrupted using a Covaris M220 ultrasonicator (Covaris, Inc., Woburn, MA, United States). The DNA fragments obtained after disruption were used for library construction. The qualified library was sequenced using the Illumina HiSeq 2500 high-throughput sequencing platform with 2 × 150 paired-ends. This platform was also used for data configuration and image analysis with HiSeq Control software. Metagenomic data have been deposited in the NCBI SRA Database (PRJNA766709).
Raw sequence data were trimmed with FASTP1, and low-quality reads, lengths of < 50 base pairs, N-rich reads, and adapter reads were removed. Sequences with different sequencing depths were assembled using Megahit software2, and contigs were obtained using the succinct de Bruijn graph method. MetaGene (Noguchi et al., 2006) was used to predict open reading frames (ORFs) in contigs, and a statistical table of gene predictions was obtained for each sample. The predicted gene sequences of all samples were clustered by CD-HIT3, and the longest gene from each cluster was selected as the representative sequence to construct a non-redundant gene set. With the use of SOAPaligner (Li et al., 2009), the high-quality reads of each sample were compared with the non-redundant gene set (95% identity) to determine the abundance information for genes in corresponding samples. BLASTP (version 2.2.28) was used to compare the non-redundant gene set with the NR database (e-value: 1e–5), and species-annotation results were obtained based on the corresponding taxonomy information of the NR database. The analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) was used for functional annotation according to the BLAST results. Community contributions to functions were determined using the NR database annotation.
Results
Sediment Physicochemical Parameters
The differences in physicochemical properties among the three mangrove populations are shown in Supplementary Figure 1. Overall, the highest TOC and TN levels were detected in the sediments of R. apiculata among the three mangroves studied (p < 0.05). Higher TP and pH, but lower salinity, were observed in the sediments of S. alba than R. apiculata and B. parviflora.
16S rRNA Gene Illumina MiSeq Sequencing
Based on Illumina sequencing, 893,587 sequences were obtained from 15 samples. In total, 4,842 OTUs were observed at a 97% similarity level (Supplementary Table 1). In terms of microbial diversity, R. apiculata exhibited the highest OTUs number and bacterial richness index among the three mangrove populations (p < 0.05; Figure 2B). Higher Shannon index values were also represented for R. apiculata, although the differences were not significant. The Venn diagram shown in Figure 2C revealed a total of 3,532 OTUs in the sediments associated with R. apiculata, whereas 640 specific OTUs were observed. In addition, 1,645 common OTUs were observed among the three mangroves. As shown in Figures 2D–F, the types and relative abundances of dominant OTUs for R. apiculata and B. parviflora increased significantly compared with those for S. alba. Significant differences were also found in dominant OTUs between R. apiculata and B. parviflora.
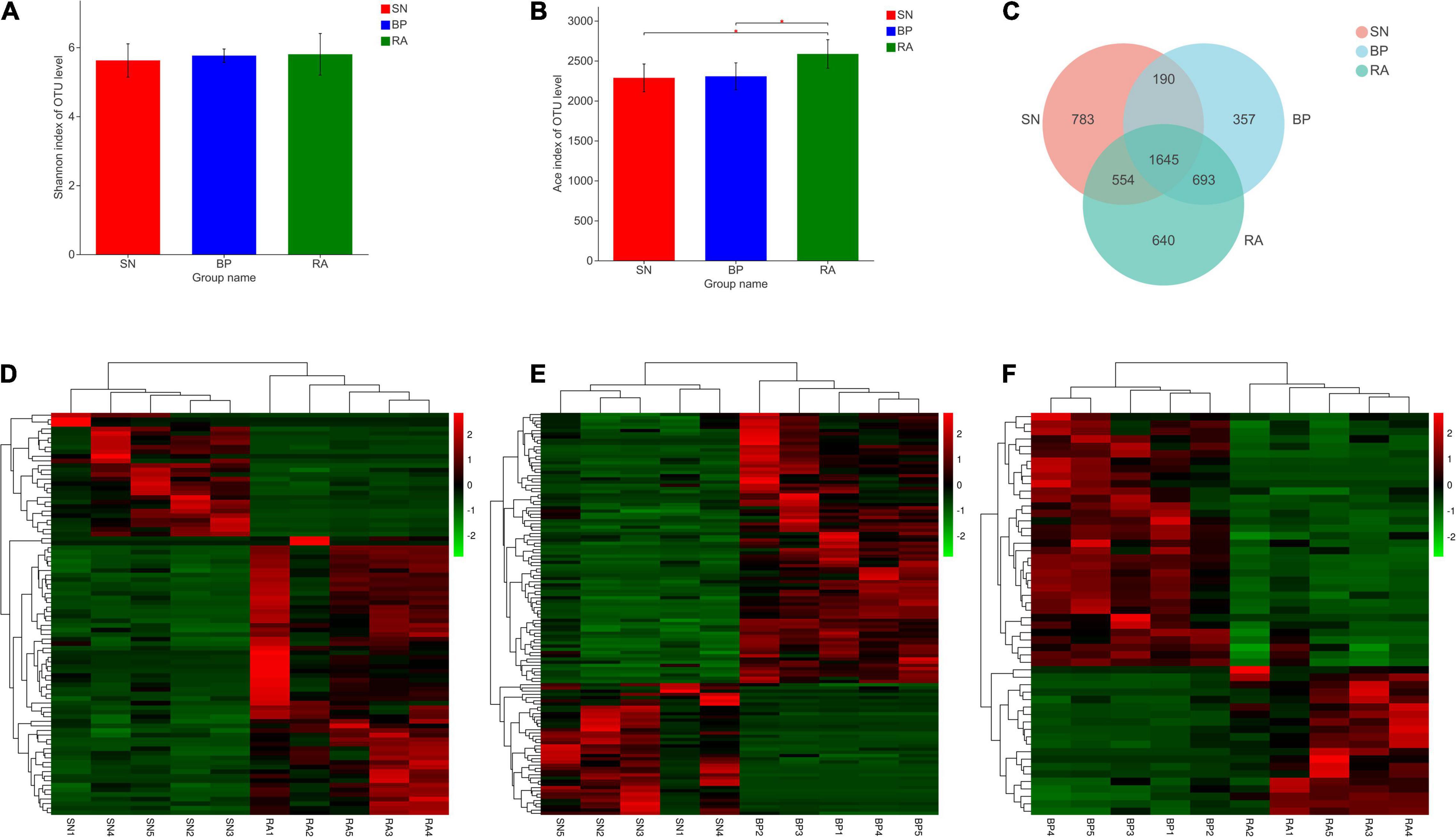
Figure 2. Diversity and significant difference analysis of operational taxonomic units (OTUs). (A) Shannon index of OTU level. (B) ACE index of OTU level. (C) Venn diagram analysis of OTU numbers between samples. (D) Significant difference analysis of OTUs between RA and SN. (E) Significant difference analysis of OTUs between BP and SN. (F) Significant difference analysis of OTUs between RA and BP. SN, Sonneratia alba; RA, Rhizophora apiculata; BP, Bruguiera parviflora.
Bacterial Community Composition in Mangrove Sediments
The abundances of bacterial taxa corresponding to the sediments are shown in Figure 3. Dominant bacteria (>5% at class level, Figure 3A) in mangrove sediments included Actinobacteria, Gammaproteobacteria, Deltaproteobacteria, Alphaproteobacteria, Anaerolineae, Bacilli, and Bacteroidia. However, the abundances of these dominant bacteria varied significantly among the three mangrove populations (Figure 3B). The abundances of Alphaproteobacteria, Deltaproteobacteria, and Bacilli were found to be higher for R. apiculata and B. parviflora, while the highest abundance of Gammaproteobacteria was observed in S. alba among the three mangroves.
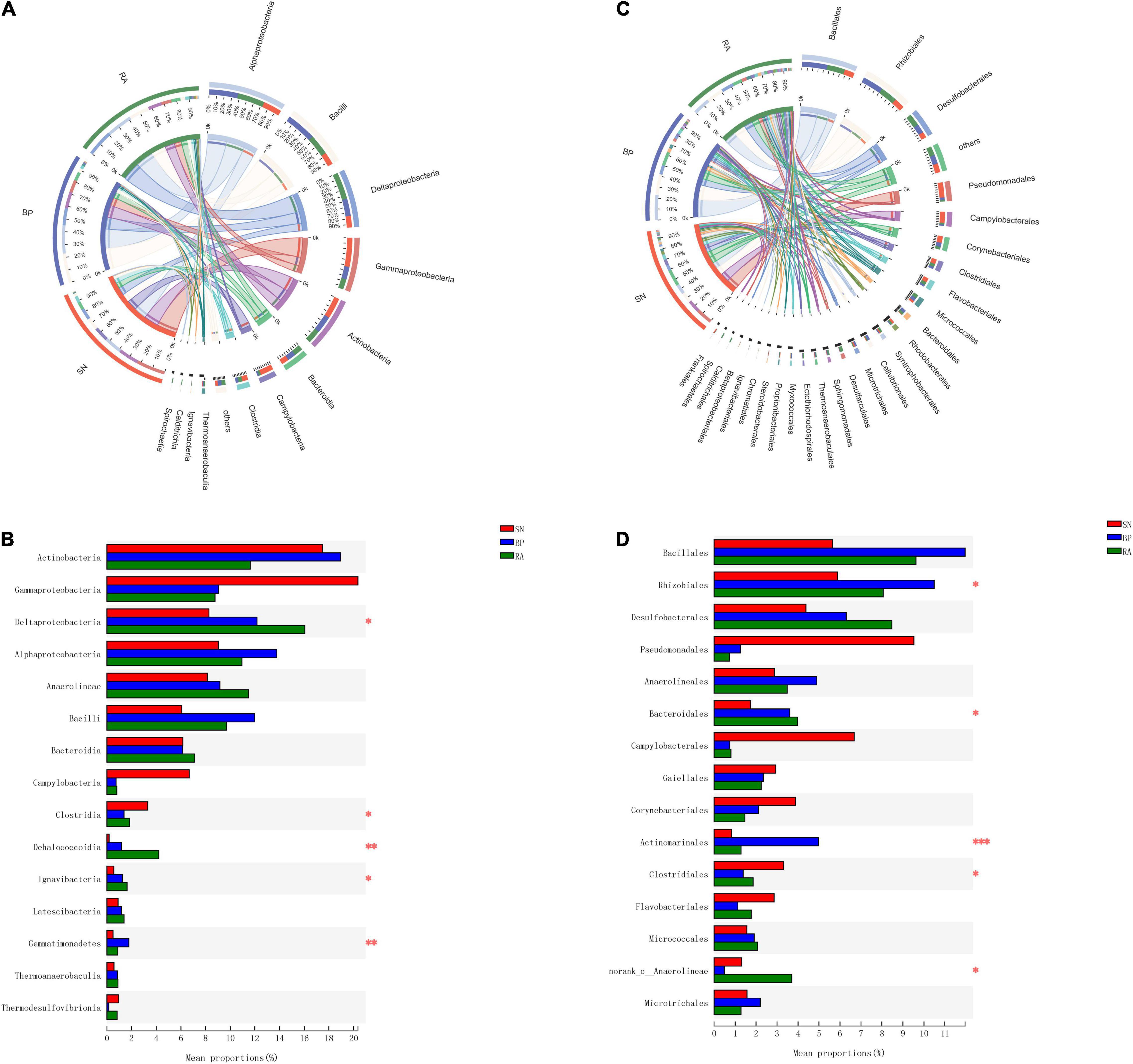
Figure 3. The components of bacterial taxa at class level (A) and order level (B). (C) Significant difference analysis of dominant taxa at class level. (D) Significant difference analysis of dominant taxa at order level. SN, Sonneratia alba; RA, Rhizophora apiculata; BP, Bruguiera parviflora.
The dominant bacterial taxa at the order level (Figure 3C) were Bacillales, Rhizobiales, Desulfobacterales, Pseudomonadales, Anaerolineales, and Bacteroidales. The highest abundances of Desulfobacterales were observed in the sediments associated with R. apiculata, followed by B. parviflora and S. alba (Figure 3D). Relative higher abundances of Bacillales and Rhizobiales were also detected in R. apiculata and B. parviflora than S. alba. In contrast, the highest abundance of Pseudomonadales was observed in S. alba among the three mangroves.
Principal Coordinates Analysis and Redundancy Analysis of Microbial Community
The results from PCoA (Figure 4A) showed that microbial communities from the same mangrove population clustered together well. RDA was performed to explore the relationships between environmental parameters and microbial community (Figure 4B). The microbial communities with R. apiculata and B. parviflora were highly correlated with TOC, TN, and salinity, whereas bacteria with S. alba were highly affected by TP and pH. The results (Figure 4C) showed that the dominant microbial taxa were grouped into two clusters. Cluster I, including Chromatiales, Corynebacteriales, Rhodobacterales, and Anaerolineales, showed positive correlations with TP or pH (p < 0.05). Cluster II, including Desulfobacterales, Rhizobiales, and Micrococcales, exhibited positive trends for correlations with TOC, TN, and salinity. More detailed information of the dominant bacterial taxa at family level is shown in Supplementary Figure 2.
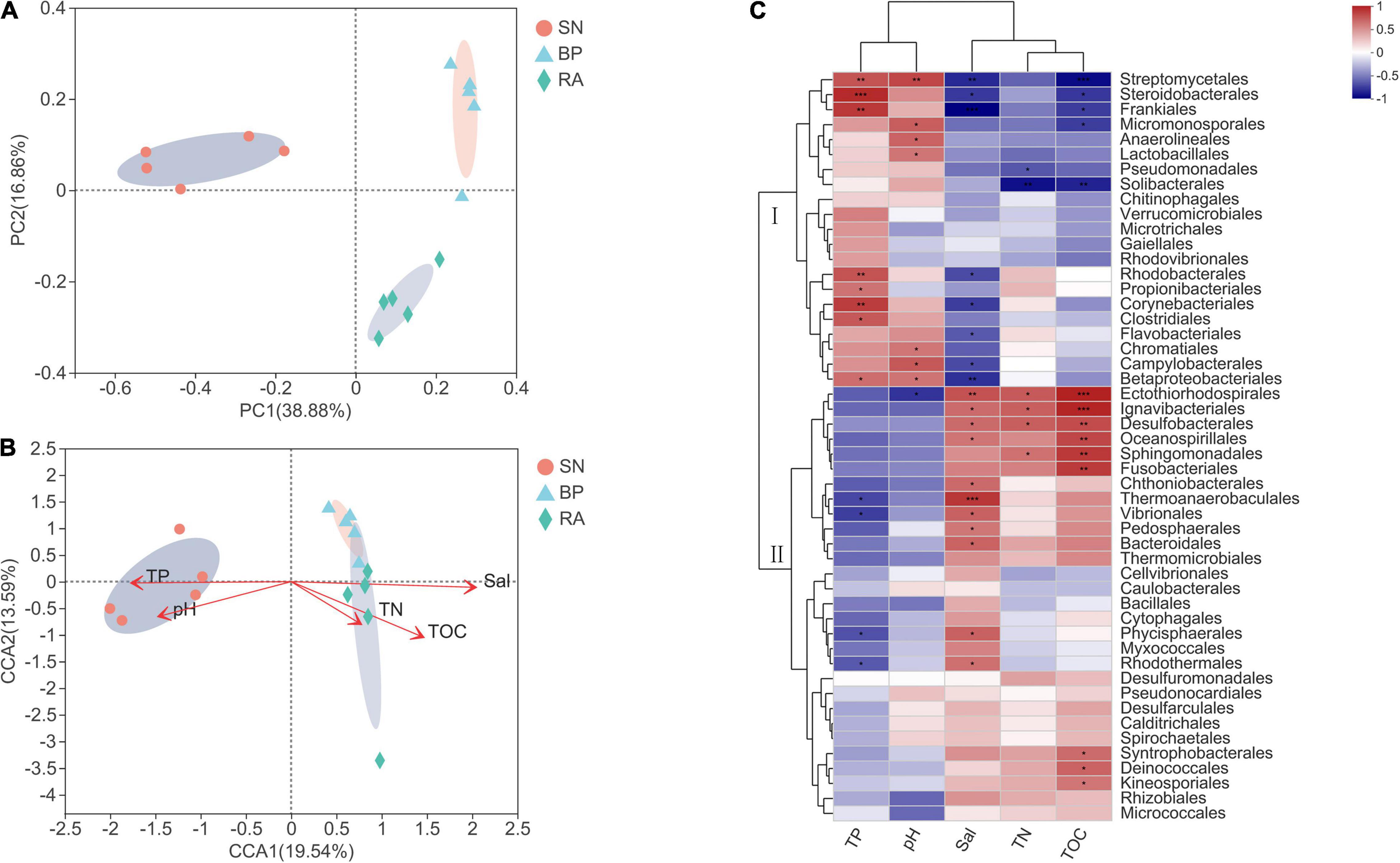
Figure 4. (A) Principal coordinates analysis (PCoA) of bacterial community structure. (B) Canonical correlation analysis of environmental factors and bacterial community. (C) Correlation analysis of dominant bacterial taxa and environmental factors. SN, Sonneratia alba; RA, Rhizophora apiculata; BP, Bruguiera parviflora.
Metagenomic Analysis of Microbial Community Function in Different Mangrove Sediments
Metagenome analysis generated a massive amount of sequence information, ranging from 90,898,652 to 139,622,818 reads among samples. In total, 14,664,335 non-redundant genes were detected in the metagenomes, and 9,005 KEGG Orthogroups (KOs) were identified. The results from Supplementary Figure 3 show a high correlation coefficient between the microbial community and functional diversity, with values of 0.84 and 0.87 for α diversity and β diversity, respectively. The data further illustrated the differences in key metabolic pathways (e.g., carbon-related metabolism) among the three mangroves. The sediments associated with R. apiculata exhibited the highest carbon metabolism, ABC transporters, prokaryote carbon fixation, and methane metabolism among the three mangroves (Figure 5 and Supplementary Figure 4).
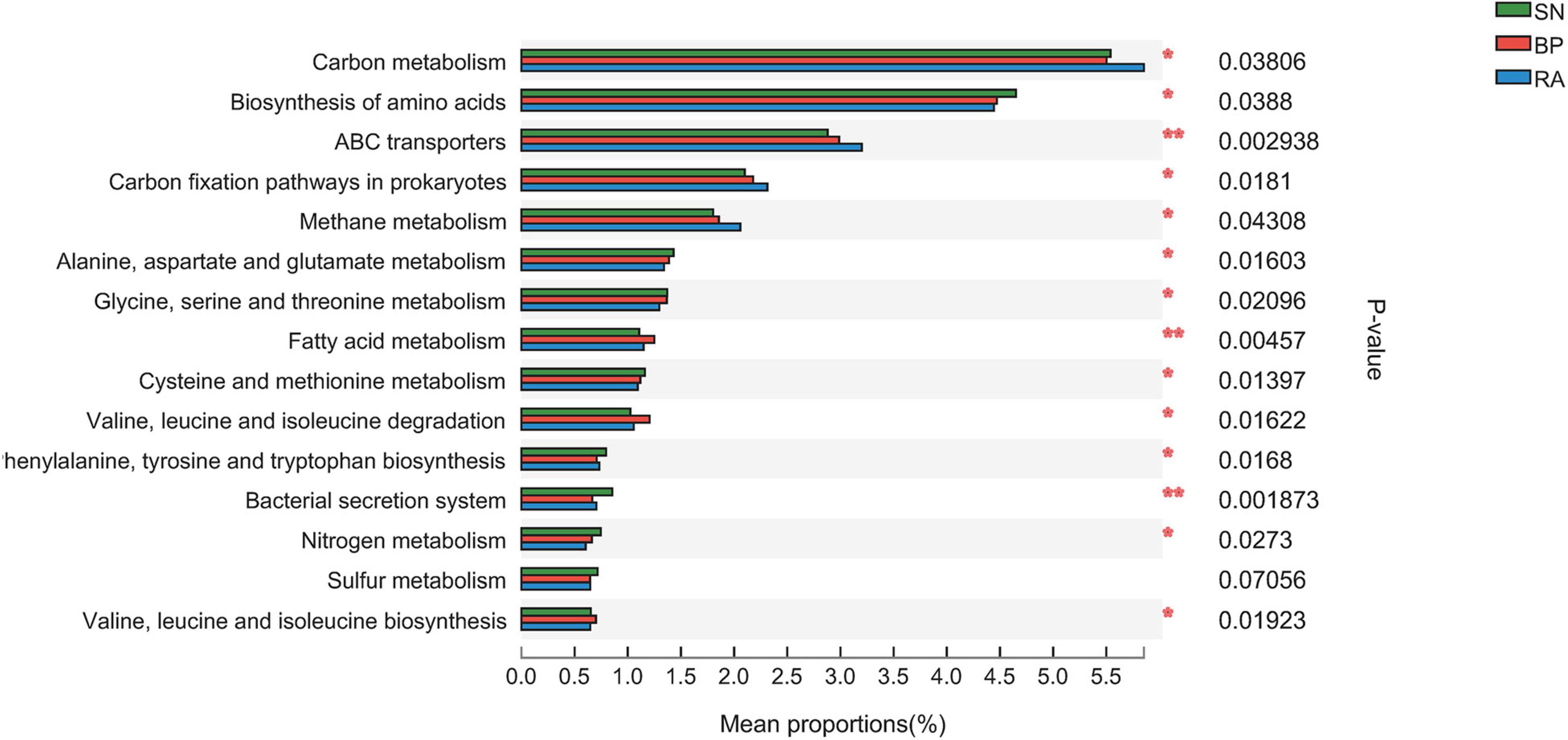
Figure 5. Significant difference analysis of metabolic function based on Kyoto Encyclopedia of Genes and Genomes (KEGG) function annotation. SN, Sonneratia alba; RA, Rhizophora apiculata; BP, Bruguiera parviflora.
Contribution of Microbial Community to Kyoto Encyclopedia of Genes and Genomes Function
As shown in Figure 6 and Supplementary Figure 5, Desulfobacterales (e.g., Desulfobacteraceae) and Rhizobiales (e.g., Xanthobacteraceae) were two major contributors to metabolic functions. The contribution of microbial taxa to the function varied significantly among the different mangrove species. Desulfobacterales consistently had the highest contributions to carbon-related metabolism (e.g., carbon metabolisms, prokaryote carbon fixation, methane metabolism, and ABC transporters) in the sediments associated with R. apiculata. When compared with S. alba, Rhizobiales also exhibited higher contributions to carbon-related metabolism in sediments of R. apiculata and B. parviflora. In addition to Desulfobacterales and Rhizobiales, the highest contributions of Micrococcales and Bacillales to metabolic functions were also observed in sediments associated with R. apiculata. In contrast, the lowest contributions of Corynebacteriales, Burkholderiales, and Planctomycetales to the aforementioned metabolic pathways were observed in the sediments associated with R. apiculata.
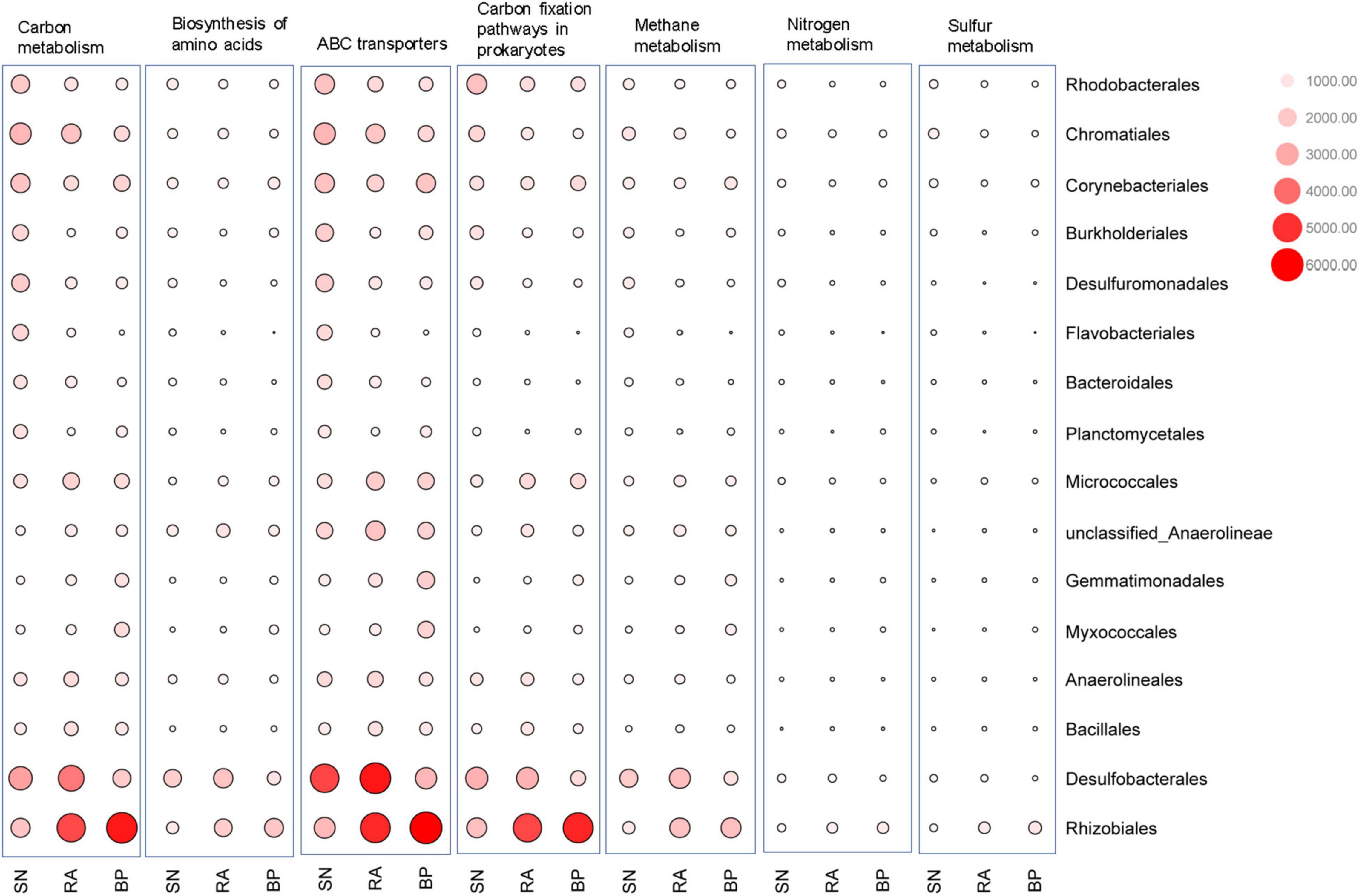
Figure 6. Contribution analysis of dominant microbial taxa to the function at order level. SN, Sonneratia alba; RA, Rhizophora apiculata; BP, Bruguiera parviflora.
Discussion
Microbial Diversity Was Highly Affected by Tidal Transitions and Mangrove Succession
Significant differences were found in the microbial community among the three mangrove populations along the Merbok river estuary, and specific microbial communities were formed in each mangrove population (Figures 2, 3). Our previous data also indicated that microbial diversity was highly influenced by the structure of mangrove populations (Wu et al., 2016). Root exudates and secondary metabolites, which also could serve as carbon sources and antimicrobial substances, vary significantly among mangrove species (Gao et al., 2003; Koh et al., 2013). Changes in root exudates might not only affect benthic microbial density but also strongly affect the structure and function of microbes (Berendsen et al., 2012; Zhuang et al., 2020). Moreover, the present data showed that sediments associated with R. apiculata exhibited the highest microbial diversity. R. apiculata is a dominant mangrove species in South Asia and often develops into a stable and fully successional mangrove community. In this study, R. apiculata occupied the majority of ecological niches in the central Merbok river estuary, whereas the genera of Avicennia, Bruguiera, Sonneratia, and Nypa occupied a smaller habitat in the lower and/or upper estuary. Higher microbial diversity in sediments associated with R. apiculata suggested that mangrove succession could enrich benthic microbial community.
Coinciding with high microbial diversity, the sediments associated with R. apiculata also exhibited higher TOC than those with B. parviflora and S. alba, indicating a higher capacity for plant productivity, carbon fixation, and burial. As an important carbon source for microbes, previous investigators also claimed that the accumulation of organic matter in sediments could promote bacterial diversity (Sjöling et al., 2005; Chen et al., 2016). Moreover, the highest TN was also observed in sediments associated with R. apiculata. The present results were consistent with previous documents (Zhu et al., 2018), in which the shift in bacterial community structure was partly driven by the increased TOC and total organic nitrogen with the succession of mangrove. Nevertheless, other environmental parameters (salinity, pH, and TP) had significant effects on the microbial community (Figure 4B). Salinity and pH have already been reported to have large effects on the microbial communities of mangroves (Ikenaga et al., 2010; Chambers et al., 2016). Moderate salinity was also positively correlated with bacterial abundances and closely linked to community composition and diversity (Morrissey et al., 2014; Crespo-Medina et al., 2016; Tong et al., 2019).
High Carbon-Related Metabolic Potential in Sediments Associated With Rhizophora apiculata
In this study, microbial diversity was significantly higher in sediments associated with R. apiculata, coinciding with the highest TOC. Enhanced microbial diversity would promote the transformation and utilization of organic carbon (Holguin et al., 2001; Sjöling et al., 2005; Berendsen et al., 2012). Thus, it is not surprising that the sediments associated with R. apiculata exhibited a higher carbon metabolic potential (Figure 5). Previous data also showed that bacterial diversity and metabolic potential (especially carbon metabolism) appeared to be enhanced during mangrove succession (Zhu et al., 2018).
The findings of this study further showed that sediments associated with R. apiculata also exhibited the highest abundances of genes involved in carbon-related pathways (Supplementary Figure 4). It is well known that several prokaryotes can assimilate CO2 into organic carbon (Lynn et al., 2017), although the functions of prokaryotes in carbon fixation have not been fully reported in mangrove ecosystems. In this study, Desulfobacterales and Rhizobiales exhibited high contributions in CO2 fixation. The enrichment of these bacteria could result in a higher capacity for prokaryotic carbon fixation, which plays an essential role in carbon storage.
It should be noted that the sediments of R. apiculata also exhibited the strongest potential for methane metabolism (Figure 5). In this study, the high organic carbon contents in R. apiculata sediments might promote the growth of methanogens, contributing to the potential production of CH4. Moreover, CH4 production and emission are aggravated under anaerobic conditions (Fey et al., 2004; Kutzbach et al., 2004). In addition, ABC transporters were also important indicators of microbial functions that reflected the positive activity of carbon and nutrient transformation. In this study, Desulfobacterales and Rhizobiales were the main contributors to ABC transporters. Higher abundances of ABC transporters in sediments associated with R. apiculata indicated that this mangrove had higher carbon and nutrient transportation activities than other mangroves (Wood et al., 2001).
Overall, this study suggested that climax mangroves (e.g., R. apiculata) exhibited a faster turnover rate of organic matter between plants and microbes owing to high carbon utilization and transportation. Microbial carbon fixation contributed to carbon sequestration, whereas the degraded small organic molecules could be conducive to the growth and succession of mangroves. The positive feedback of microbial community and function might in turn contribute to the formation of climax mangrove populations with high productivity (Chen et al., 2016). The inherent correlations among carbon metabolism, environmental transition, and mangrove succession need to be further studied.
Specific Microbial Taxa and Functional Potential in Maintaining Mangrove Survival and Succession
Although mangrove sediments are rich in organic matter, they are generally nutrient-deficient. Nutrient limitations were also widely reported in mangrove forests (Feller et al., 2003; Reef et al., 2010; Wang and Gu, 2013; Cheng et al., 2020). Even worse, anaerobic conditions might further aggravate the enrichment of anaerobic microbes and reductive phytotoxins (e.g., CH4, H2S, and sulfides). It is worth exploring how mangroves maintain survival and succession in such a terrible habitat in intertidal regions. The present results could partly explain this issue from the perspectives of microbial community and function.
Desulfobacterales, a type of sulfate-reducing bacteria, consistently exhibited the highest contributions to metabolic functions in sediments associated with R. apiculata (Figure 6 and Supplementary Figure 5). In this study, Desulfobacterales (e.g., Desulfobacteraceae) was one of the main microbial taxa responsible for the differences among mangroves, whereas R. apiculata also exhibited the highest abundances of these bacteria (Figure 3D and Supplementary Figure 2A). Significantly positive correlations among Desulfobacterales, TOC, and TN were also observed (Figure 4C and Supplementary Figure 2B). Previous investigators also claimed an important role for Desulfobacterales in C, N, and S cycles (Zhu et al., 2018). Increased Desulfobacterales might be beneficial for R. apiculata by alleviating the toxicity of sulfides under anaerobic conditions. The high abundances and strong metabolic potential of Desulfobacterales could also accelerate carbon and nutrient transformation and utilization (Lyimo et al., 2002; Meyer and Kuever, 2007), facilitating mangrove survival and succession in intertidal regions.
The important potential of Rhizobiales in C, N, and S metabolisms was also revealed (Figure 6). Rhizobiales was a well-studied plant symbiont that widely occurred in the rhizosphere of mangrove plants (Gomes et al., 2010). This taxon played a beneficial role for the host by providing various nutrients, phytohormones, and precursors of essential metabolites (Delmotte et al., 2009; Verginer et al., 2010; Garrido-Oter et al., 2018). The findings of this study also revealed that Rhizobiales (e.g., Xanthobacteraceae) was metabolically versatile, especially in terms of carbon-related metabolism (Figure 6 and Supplementary Figure 5). In this study, relative higher abundances of Rhizobiales (e.g., Xanthobacteraceae) were also detected in the sediments of R. apiculata and B. parviflora than those of S. alba (Figures 2A, 3D). Similarly, positive correlation trends among Rhizobiales, TOC, and TN were also observed in this study (Figure 4C).
In addition to Desulfobacterales and Rhizobiales, Micrococcales might also partly contribute to the higher metabolic function in sediments associated with R. apiculata. Although little attention has been paid to this taxon, the present data (Figures 3D, 4C, 6) indicated that Micrococcales might also be important for metabolic functions, which are involved in carbon and nutrient metabolisms. Nevertheless, Bacillus, which was often considered a bionematicide, could promote the growth of plants by protecting roots from pathogens (Mendoza et al., 2008; Mendis et al., 2018). Bacillus was also a dominant bacterial component in this study (Figure 3D); however, relatively low metabolic potentials of this taxon were observed (Figure 6). Multi-omics analyses, such as metagenomics, metatranscriptomics, and metaproteomics, focused on the functions of microbes in mangrove ecosystems should be further conducted.
Conclusion
The present findings provided a broader understanding of the relationships among microbes, environmental transition, and mangrove succession from the perspective of microbial community and function. Benthic microbial community was highly correlated with environmental factors and aboveground mangrove species, whereas the highest microbial diversity and metabolic potential (carbon metabolism, prokaryote carbon fixation, methane metabolism, and ABC transporters) were observed in sediments associated with R. apiculata. Specific microbial taxa (e.g., Desulfobacterales and Rhizobiales) involved in C, N, and S cycles might facilitate mangrove survival and succession in intertidal regions. The present data indicated that mangrove succession could enrich microbial diversity and carbon metabolism. More detailed multi-omics researches focused on the roles of microbes in mangrove succession should be further conducted.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZM carried out manuscript writing and revisions. FS and HC designed the research, writing, review, and editing; YW carried out writing – review. SF contributed to the sample collection and data analysis. LW carried out data analysis. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the International Partnership Program of Chinese Academy of Sciences (133244KYSB20180012), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), China (GML2019ZD0303 and GML2019ZD0305), Science and Technology Basic Resources Investigation Program of China (2017FY100707), Key Research and Development Program of Hainan Province (ZDYF2021XDNY131), National Natural Science Foundation of China (41676086, 41890853, 41906128, and 41406130), and the Project of Guangdong Science and Technology Department (2017A020216008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.764974/full#supplementary-material
Footnotes
- ^ https://github.com/OpenGene/fastp
- ^ https://github.com/voutcn/megahit
- ^ http://www.bioinformatics.org/cd-hit
References
Abed, R. M. M., Al-Kindi, S., and Al-Kharusi, S. (2015). Diversity of bacterial communities along a petroleum contamination gradient in desert soils. Microb. Ecol. 69, 95–105. doi: 10.1007/s00248-014-0475-5
Alongi, D. M. (1988). Bacterial productivity and microbial biomass in tropical mangrove sediments. Microb. Ecol. 15, 59–79. doi: 10.1007/BF02012952
Alzubaidy, H., Essack, M., Malas, T. B., Bokhari, A., Motwalli, O., Kamanu, F. K., et al. (2016). Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene 576(2, Pt 1), 626–636. doi: 10.1016/j.gene.2015.10.032
Andreote, F. D., Jiménez, D. J., Chaves, D., Dias, A. C. F., Luvizotto, D. M., Dini-Andreote, F., et al. (2012). The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One 7:e38600. doi: 10.1371/journal.pone.0038600
Bai, S., Li, J., He, Z., Van Nostrand, J. D., Tian, Y., Lin, G., et al. (2013). GeoChip-based analysis of the functional gene diversity and metabolic potential of soil microbial communities of mangroves. Appl. Microbiol. Biotechnol. 97, 7035–7048. doi: 10.1007/s00253-012-4496-z
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Brander, L. M., Wagtendonk, A. J., Hussain, S. S., McVittie, A., Verburg, P. H., de Groot, R. S., et al. (2012). Ecosystem service values for mangroves in Southeast Asia: a meta-analysis and value transfer application. Ecosyst. Serv. 1, 62–69. doi: 10.1016/j.ecoser.2012.06.003
Chambers, L. G., Guevara, R., Boyer, J. N., Troxler, T. G., and Davis, S. E. (2016). Effects of salinity and inundation on microbial community structure and function in a mangrove peat soil. Wetlands 36, 361–371. doi: 10.1007/s13157-016-0745-8
Chen, Q., Zhao, Q., Li, J., Jian, S., and Ren, H. (2016). Mangrove succession enriches the sediment microbial community in South China. Sci. Rep. 6:27468. doi: 10.1038/srep27468
Cheng, H., Liu, Y., Jiang, Z. Y., and Wang, Y. S. (2020). Radial oxygen loss is correlated with nitrogen nutrition in mangroves. Tree Physiol. 40, 1548–1560. doi: 10.1093/treephys/tpaa089
Crespo-Medina, M., Bowles, M. W., Samarkin, V. A., Hunter, K. S., and Joye, S. B. (2016). Microbial diversity and activity in seafloor brine lake sediments (Alaminos Canyon block 601, Gulf of Mexico). Geobiology 14, 483–498. doi: 10.1111/gbi.12185
Delmotte, N., Knief, C., Chaffron, S., Innerebner, G., Roschitzki, B., Schlapbach, R., et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 16428–16433. doi: 10.1073/pnas.0905240106
Feller, I. C., McKee, K. L., Whigham, D. F., and O’Neill, J. P. (2003). Nitrogen vs. phosphorus limitation across an ecotonal gradient in a mangrove forest. Biogeochemistry 62, 145–175. doi: 10.1023/A:1021166010892
Fey, A., Claus, P., and Conrad, R. (2004). Temporal change of 13C-isotope signatures and methanogenic pathways in rice field soil incubated anoxically at different temperatures. Geochim. Cosmochim. Acta. 68, 293–306. doi: 10.1016/S0016-7037(03)00426-5
Gao, M., Teplitski, M., Robinson, J. B., and Bauer, W. D. (2003). Production of substances by medicago truncatula that affect bacterial quorum sensing. Mol. Plant Microbe Interact. 16, 827–834. doi: 10.1094/mpmi.2003.16.9.827
Garrido-Oter, R., Nakano, R. T., Dombrowski, N., Ma, K. W., McHardy, A. C., and Schulze-Lefert, P. (2018). Modular traits of the rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24, 155–167.e5. doi: 10.1016/j.chom.2018.06.006
Giani, L., Bashan, Y., Holguin, G., and Strangmann, A. (1996). Characteristics and methanogenesis of the Balandra lagoon mangrove soils, Baja California Sur, Mexico. Geoderma 72, 149–160. doi: 10.1016/0016-7061(96)00023-7
Giri, C., Ochieng, E., Tieszen, L. L., Zhu, Z., Singh, A., Loveland, T., et al. (2011). Status and distribution of mangrove forests of the world using earth observation satellite data. Glob. Ecol. Biogeogr. 20, 154–159. doi: 10.1111/j.1466-8238.2010.00584.x
Gomes, N. C. M., Cleary, D. F. R., Pinto, F. N., Egas, C., Almeida, A., Cunha, A., et al. (2010). Taking root: enduring effect of rhizosphere bacterial colonization in mangroves. PLoS One 5:e14065. doi: 10.1371/journal.pone.0014065
Holguin, G., Vazquez, P., and Bashan, Y. (2001). The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol. Fertil. Soils 33, 265–278. doi: 10.1007/s003740000319
Huang, W., Chen, X., Jiang, X., and Zheng, B. (2017). Characterization of sediment bacterial communities in plain lakes with different trophic statuses. Microbiologyopen 6:e00503. doi: 10.1002/mbo3.503
Ikenaga, M., Guevara, R., Dean, A. L., Pisani, C., and Boyer, J. N. (2010). Changes in community structure of sediment bacteria along the Florida coastal everglades marsh-mangrove-seagrass salinity gradient. Microb. Ecol. 59, 284–295. doi: 10.1007/s00248-009-9572-2
Koh, C. L., Sam, C. K., Yin, W. F., Tan, L. Y., Krishnan, T., Chong, Y. M., et al. (2013). Plant-derived natural products as sources of anti-quorum sensing compounds. Sensors 13, 6217–6228.
Kutzbach, L., Wagner, D., and Pfeiffer, E. M. (2004). Effect of microrelief and vegetation on methane emission from wet polygonal tundra, Lena Delta, Northern Siberia. Biogeochemistry 69, 341–362. doi: 10.1023/B:BIOG.0000031053.81520.db
Li, R., Yu, C., Li, Y., Lam, T.-W., Yiu, S.-M., Kristiansen, K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Lin, X., Hetharua, B., Lin, L., Xu, H., Zheng, T., He, Z., et al. (2019). Mangrove sediment microbiome: adaptive microbial assemblages and their routed biogeochemical processes in Yunxiao mangrove national nature reserve, China. Microb. Ecol. 78, 57–69. doi: 10.1007/s00248-018-1261-6
Lyimo, T. J., Pol, A., and den Camp, H. (2002). Sulfate reduction and methanogenesis in sediments of Mtoni mangrove forest, Tanzania. Ambio 31, 614–616.
Lynn, T. M., Ge, T., Yuan, H., Wei, X., Wu, X., Xiao, K., et al. (2017). Soil carbon-fixation rates and associated bacterial diversity and abundance in three natural ecosystems. Microb. Ecol. 73, 645–657. doi: 10.1007/s00248-016-0890-x
Mendis, H. C., Thomas, V. P., Schwientek, P., Salamzade, R., Chien, J. T., Waidyarathne, P., et al. (2018). Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I-1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions. PLoS One 13:e0193119. doi: 10.1371/journal.pone.0193119
Mendoza, A. R., Kiewnick, S., and Sikora, R. A. (2008). In vitro activity of Bacillus firmus against the burrowing nematode Radopholus similis, the root-knot nematode Meloidogyne incognita and the stem nematode Ditylenchus dipsaci. Biocontrol Sci. Technol. 18, 377–389. doi: 10.1080/09583150801952143
Meyer, B., and Kuever, J. (2007). Molecular analysis of the diversity of sulfate-reducing and sulfur-oxidizing prokaryotes in the environment, using apra as functional marker gene. Appl. Environ. Microbiol. 73, 7664–7679. doi: 10.1128/aem.01272-07
Noguchi, H., Park, J., and Takagi, T. (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34, 5623–5630. doi: 10.1093/nar/gkl723
Morrissey, E. M., Gillespie, J. L., Morina, J. C., and Franklin, R. B. (2014). Salinity affects microbial activity and soil organic matter content in tidal wetlands. Glob. Chang. Biol. 20, 1351–1362. doi: 10.1111/gcb.12431
Reef, R., Feller, I. C., and Lovelock, C. E. (2010). Nutrition of mangroves. Tree Physiol. 30, 1148–1160. doi: 10.1093/treephys/tpq048
Reis, C. R. G., Nardoto, G. B., and Oliveira, R. S. (2017). Global overview on nitrogen dynamics in mangroves and consequences of increasing nitrogen availability for these systems. Plant Soil 410, 1–19. doi: 10.1007/s11104-016-3123-7
Rosentreter, J. A., Maher, D. T., Erler, D. V., Murray, R. H., and Eyre, B. D. (2018). Methane emissions partially offset “blue carbon” burial in mangroves. Sci. Adv. 4:eaao4985. doi: 10.1126/sciadv.aao4985
Sjöling, S., Mohammed, S. M., Lyimo, T. J., and Kyaruzi, J. J. (2005). Benthic bacterial diversity and nutrient processes in mangroves: impact of deforestation. Estuar. Coast Shelf Sci. 63, 397–406. doi: 10.1016/j.ecss.2004.12.002
Tong, T., Li, R., Wu, S., and Xie, S. (2019). The distribution of sediment bacterial community in mangroves across China was governed by geographic location and eutrophication. Mar. Pollut. Bull. 140, 198–203. doi: 10.1016/j.marpolbul.2019.01.046
Verginer, M., Siegmund, B., Cardinale, M., Müller, H., Choi, Y., Míguez, C. B., et al. (2010). Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol. Ecol. 74, 136–145. doi: 10.1111/j.1574-6941.2010.00942.x
Wang, W., Li, X., and Wang, M. (2019). Propagule dispersal determines mangrove zonation at intertidal and estuarine scales. Forests 10:245.
Wang, Y. F., and Gu, J. D. (2013). Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl. Microbiol. Biotechnol. 97, 7015–7033. doi: 10.1007/s00253-012-4430-4
Wood, D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., et al. (2001). The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294, 2317–2323. doi: 10.1126/science.1066804
Wu, P., Xiong, X., Xu, Z., Lu, C., Cheng, H., Lyu, X., et al. (2016). Bacterial communities in the rhizospheres of three mangrove tree species from Beilun Estuary, China. PLoS One 11:e0164082. doi: 10.1371/journal.pone.0164082
Zhou, Z., Meng, H., Liu, Y., Gu, J. D., and Li, M. (2017). Stratified bacterial and archaeal community in mangrove and intertidal wetland mudflats revealed by high throughput 16S rRNA gene sequencing. Front. Microbiol. 8:2148. doi: 10.3389/fmicb.2017.02148
Zhu, P., Wang, Y., Shi, T., Zhang, X., Huang, G., and Gong, J. (2018). Intertidal zonation affects diversity and functional potentials of bacteria in surface sediments: a case study of the Golden Bay mangrove, China. Appl. Soil. Ecol. 130, 159–168. doi: 10.1016/j.apsoil.2018.06.003
Keywords: mangrove succession, microbial community and function, carbon metabolism, Rhizophora apiculata, Merbok river estuary
Citation: Mai Z, Ye M, Wang Y, Foong SY, Wang L, Sun F and Cheng H (2021) Characteristics of Microbial Community and Function With the Succession of Mangroves. Front. Microbiol. 12:764974. doi: 10.3389/fmicb.2021.764974
Received: 26 August 2021; Accepted: 05 November 2021;
Published: 07 December 2021.
Edited by:
Diane Purchase, Middlesex University London, United KingdomReviewed by:
Zhichao Zhou, University of Wisconsin–Madison, United StatesAlejandra Prieto-Davó, Universidad Nacional Autónoma de México, Mexico
Copyright © 2021 Mai, Ye, Wang, Foong, Wang, Sun and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fulin Sun, ZmxzdW5Ac2NzaW8uYWMuY24=; Hao Cheng, Y2hlbmdoYW9Ac2NzaW8uYWMuY24=
†These authors have contributed equally to this work
 Zhimao Mai
Zhimao Mai Mai Ye
Mai Ye Youshao Wang
Youshao Wang Swee Yeok Foong
Swee Yeok Foong Lin Wang
Lin Wang Fulin Sun
Fulin Sun Hao Cheng
Hao Cheng