- 1Department of Agricultural, Food and Environmental Sciences, Marche Polytechnic University, Ancona, Italy
- 2Laboratory of Plant Protection, National Institute for Agronomic Research of Tunisia, University of Carthage, Ariana, Tunisia
Stagonosporopsis cucurbitacearum is an important seedborne pathogen of squash (Cucurbita maxima). The aim of our work was to develop a rapid and sensitive diagnostic tool for detection and quantification of S. cucurbitacearum in squash seed samples, to be compared with blotter analysis, that is the current official seed test. In blotter analysis, 29 of 31 seed samples were identified as infected, with contamination from 1.5 to 65.4%. A new set of primers (DB1F/R) was validated in silico and in conventional, quantitative real-time PCR (qPCR) and droplet digital (dd) PCR. The limit of detection of S. cucurbitacearum DNA for conventional PCR was ∼1.82 × 10–2 ng, with 17 of 19 seed samples positive. The limit of detection for ddPCR was 3.6 × 10–3 ng, which corresponded to 0.2 copies/μl. Detection carried out with artificial samples revealed no interference in the absolute quantification when the seed samples were diluted to 20 ng. All seed samples that showed S. cucurbitacearum contamination in the blotter analysis were highly correlated with the absolute quantification of S. cucurbitacearum DNA (copies/μl) in ddPCR (R2 = 0.986; p ≤ 0.01). Our ddPCR protocol provided rapid detection and absolute quantification of S. cucurbitacearum, offering a useful support to the standard procedure.
Introduction
Pumpkin, squash, and gourds (Cucurbita spp.) are grown throughout the world with a total production of 22.9 million tonnes, which 20.9% is produced in Europe in 2019 (FAOSTAT, 2021). Italy is ranked 9th in the world with 569.120 tonnes in terms of total production (FAOSTAT, 2021).
Several diseases have economic impacts on the production of the Cucurbita spp. (squash; cucurbit). These include gummy stem blight (GSB) (with foliar symptoms) and black rot (with fruit symptoms), which are caused by three species of the fungi Stagonosporopsis: (i) Stagonosporopsis cucurbitacearum (Fr.) Aveskamp, Gruyter, and Verkley [anamorph Phoma cucurbitacearum (Fr.) Sacc.], synonym Didymella bryoniae (Fuckel) Rehm; (ii) Stagonosporopsis caricae (Syd. and P. Syd.) Aveskamp, Gruyter, and Verkley (synonym Mycosphaerella caricae Syd. and P. Syd.); and (iii) Stagonosporopsis citrulli M. T. Brewer and J. E. Stewart (Stewart et al., 2015).
In the Mediterranean areas as well in Asia, S. cucurbitacearum was described as the main pathogenic fungus of Cucurbita maxima seedling related to GSB, able to reduce both squash yield and quality (Moumni et al., 2019, 2020; Zhao et al., 2020). In warm and humid environments, infections of these fungi can result 15 to 50% in reduction of yields and rapid death of the cucurbit plants (Keinath et al., 1995; Boughalleb et al., 2007; Keinath, 2011; Li et al., 2015; Yao et al., 2016; Moumni et al., 2019; Zitter and Kyle, 1992). These diseases are known to spread in the greenhouse during the growth season through airborne ascospores and by conidia transported in water on the plant surfaces, with further spread by contact between plants, or between plants and man, and onto the host plant (De Neergaard, 1989; Keinath, 1996, 2011). Thus, under favorable environmental conditions, a low level of latently infected seedlings can potentially result in a major disease epidemic (Ling et al., 2010). Moreover, S. cucurbitacearum is both an external and internal seedborne pathogen, which can thus carry inoculum from the seeds to the plants, although it is found mainly on the seed coat (Sudisha et al., 2006).
For these reasons, the production and the movement of seeds represent a particularly efficient vehicle to disperse such seedborne pathogens (Zhang et al., 2018; Moumni et al., 2020; Zhao et al., 2020). Also, as global trade has increased, so has the movement of seeds and other plant materials between countries, thus increasing the risk of the transport and transfer of plant pathogens (Epanchin-Niell and Hastings, 2010; Mancini and Romanazzi, 2014; Mancini et al., 2016). The introduction of exotic pathogens in this way can have catastrophic effects on both natural and agricultural ecosystems, and can result in large economic losses from lost ecosystem services and reduced crop yields (Atallah et al., 2010; Short et al., 2015; Cunniffe et al., 2016; Epanchin-Niell, 2017). Many countries have formulated legislation to limit or prevent the introduction of exotic pathogens into new areas, and these are generally supported by detection techniques (Choudhury et al., 2017). However, the low inoculum levels and the varied distribution of a pathogen within seed lots make the testing of seeds a difficult task.
Moumni et al. (2020) reported that the selection of healthy fruit is not sufficient to reduce the infection by this seedborne pathogen. On the other hand, cultural practices and fungicide application can have important roles in GSB management, although this pathogen has developed resistance to many fungicides that were previously very effective (Finger et al., 2014; Gimode et al., 2020; Newark et al., 2020). Therefore, early diagnosis of this pathogen is a fundamental step in the management of these crop diseases (Ora et al., 2011).
The most common methods currently used for rapid detection of agent related to GSB are based on the use of molecular detection tools. These include conventional polymerase chain reaction (PCR), PCR–enzyme-linked immunosorbent assays (PCR-ELISA) (Somai et al., 2002; Keinath et al., 2003), and quantitative real-time PCR (qPCR) (Somai et al., 2002; Ha et al., 2009; Ling et al., 2010). Recently, loop-mediated isothermal amplification assays have also been designed to detect S. cucurbitacearum in cucurbit seeds (Tian et al., 2016) and for infections in young muskmelon leaves with suspected early symptoms of GSB (Yao et al., 2016).
Droplet digital PCR (ddPCR) is a recent technology that provides both detection and quantification of DNA targets. Unlike other methods, ddPCR does not require standard curves of known concentrations for quantification (Huggett and Whale, 2013). Additionally, ddPCR has been shown to be more sensitive for the detection of low level of inoculum or unevenly distribution of pathogens in infected plants (Dreo et al., 2014; Selvaraj et al., 2018; del Pilar Martínez-Diz et al., 2020). ddPCR also shows high resistance to inhibitors compared to qPCR (Rački et al., 2014). Thus, ddPCR represents an ideal choice for infection testing of nursery propagation materials and seeds (Dingle et al., 2013; Rani et al., 2019).
Droplet digital PCR has been used to identify soil and seedborne pathogens, aiding plant disease diagnosis (Selvaraj et al., 2018; Liu et al., 2020; Lu et al., 2020; Yu et al., 2020).
Considering the emerging importance for squash seeds of S. cucurbitacearum the focus of our research, we set up a rapid and sensitive protocol, based on conventional PCR, validated also in qPCR and ddPCR. Such systems would speed up the diagnosis of GSB infection, which to date has generally been carried out using the blotter test, the official diagnostic method and requires several weeks before the results can be obtained.
Materials and Methods
Dot Blot Analysis and Morphological Identification
Thirty-one seed samples of squash (Cucurbita maxima Duchesne, cv. Bjaoui) were collected from the same number of fruits showing clear symptoms, mild symptoms of GBS, and asymptomatic samples. The fruit samples were collected in nine fields in the northwest of Tunisia in October 2015, 2016, and 2017. Each seed sample was analyzed to verify the presence of S. cucurbitacearum using the standard blotter method of the International Seed Testing Association (Mathur and Kongsdal, 2003). Two hundred seeds per sample were soaked for 5 min in 1% sodium hypochlorite solution, and then triple-rinsed with sterile distilled water. The seeds were dried for 2 min on sterile paper towels under a laminar flow hood. They were then placed in Petri dishes (diameter, 110 mm) on eight overlapping sterile filter paper layers (Whatman N° 4) that were moistened with 5 ml sterile distilled water, and incubated at 25°C under 12 h/12 h day/night artificial light cycles (Master TL-D Super 80 58W/830).
For each sample of seeds, 20 Petri dishes were used, each of which contained 10 seeds. From days 7 to 15 after plating, the Petri dishes were examined daily under a stereomicroscope for the presence of S. cucurbitacearum fungal structures. The pycnidia on the seeds were excised and morphological identification was carried out under a compound microscope. For each sample, the proportion (%) of the seeds infected by S. cucurbitacearum was calculated. Statistical analyses were performed using the software SPSS (version 20; IBM, Armonk, NY, United States). The data were first tested for normality and homogeneity of variance by Levene’s test. Welch’s ANOVA was performed to determine any differences in seed samples, and means were separated using the Games-Howell post hoc test (P < 0.05).
The fungal structures on the seeds that were morphologically identified as S. cucurbitacearum were transferred onto potato dextrose agar (Liofilchem Srl, Roseto degli Abruzzi, Italy) in Petri dishes. After 10 days on the potato dextrose agar at 23 ± 2°C, morphological identification was carried out according to the color and shape of the colonies, combined with the characteristics of the pycnidia and spores.
DNA Extraction and Molecular Characterization of Natural Seed Inoculum
Total DNA was initially extracted from the mycelia of isolate D33 that was previously identified as S. cucurbitacearum by morphological and internal transcribed spacer sequence analysis (Moumni et al., 2019, 2020). In order to have a preliminary molecular characterization of the natural inoculum, the colonies, picked up from blotter test, grown in PDA and identified by morphological characters, were subjected to DNA extraction, and analyzed by a multiplex PCR able to distinguish the three morphologically similar species (S. cucurbitacearum, S. citrulli, and S. caricae) (Brewer et al., 2015).
For the seeds, according to the results from the blotter analysis and molecular characterization of the natural inoculum, the following were selected for further analysis: 17 seed samples as representative of S. cucurbitacearum infection; 2 seed samples (T18 and T101) that appeared not to be infected using the conventional method; and 1 seed sample certified as healthy (IHS) (Seminis, Monsanto Agricoltura, Italy). From each of 19 samples, 100 seeds/sample were pulverized in liquid nitrogen, with DNA extraction carried out starting from 300 mg seed tissue homogenized in 5 ml extraction buffer (3% cetyl trimethylammonium bromide, 100 mM Tris-HCl, 1.4 M NaCl, 20 mM EDTA, 2% polyvinyl pyrrolidone, and 2% sodium metabisulfite) in tissue extraction bags (12 × 15 cm; Bioreba, Switzerland). The lysates were washed with phenol/chloroform (1:1), and then chloroform. The total nucleic acids were precipitated in 1 vol. cold isopropanol, and immediately centrifuged at 13,000 × g for 25 min. The pellets were dried at room temperature, and then resuspended in 60 μl sterile water. The quality and quantity of the extracted DNA were evaluated using a biophotometer (Eppendorf, Hamburg, Germany).
Validation of Stagonosporopsis cucurbitacearum Identification in Conventional Polymerase Chain Reaction and Sequencing
In a previous study based on random amplification of polymorphic DNA markers, Ling et al. (2010) developed primers based on sequence-characterized amplified regions (i.e., the DB17 primer set) with broad-spectrum specificity for S. cucurbitacearum.
Here, the two 559 bp nucleotide sequences from S. cucurbitacearum that are representative of the RGI and RGII molecular types (GenBank accession Nos. GQ872461 and GQ872462) were downloaded in the FASTA format and aligned using ClustalX (version 1.83) (Thompson et al., 1994). Based on the conserved sequence region common to both of these genotypes (i.e., RGI and RGII), we designed a new set of primers using the Primer3 Plus software, defined here as DBF1 (5′-TCGAATGGCTCAGAGAAGGT-3′) and DBR1 (5′-AAGTCCACGTCAGACCCATC-3′), which were then synthesized (Sigma Aldrich Merck, Darmstadt, Germany). The primers were first validated in silico using NCBI Primer-Blast (Ye et al., 2012). The specificity of the primers was then confirmed and tested in PCR with reference strains, previously identified by multisequence analysis of calmodulin,β-tubulin, histone H3, translation elongation factor and internal transcribed spacer regions and available in NCBI database (Moumni et al., 2019, 2020): S. cucurbitacearum (isolates D33, D49, D12, D83, ID1, and ID3), Phoma sp. (isolate Ph39), Alternaria alternata (isolates A38, A15), Fusarium solani (isolate F174), Curvularia specifera (isolate B170), Paramyrothecium roridum (isolate M123), Albifimbria verrucaria (isolate M144), and Stemphylium vesicarium (isolate P164).
Preliminary conventional PCR tests were set up to define the optimal concentrations of MgCl2 (0.8, 1.0, 1.2, 1.5, and 2 mM), primers (0.5 and 1.0 μM), and template. The amplification cycling was: 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 55–60°C for 30 s, and 72°C for 30 s, and with a final extension cycle at 72°C for 7 min. The PCR products were visualized on 1.5% agarose gels in Tris-acetate buffer (40 mM Tris-acetate, 1 mM EDTA, and pH 8.0) after staining with GelRed (Biotium, United States), to confirm the specificity, expected size of the PCR product, and the sensitivity of the diagnostic system.
The specific amplicons were sequenced in both directions by Genewiz (Germany) according to Sanger Method. Nucleotide sequences were carefully checked reading the chromatogram in order to exclude ambiguous peaks, and then formatted in fasta format in order to carry out Megablast, optimized for highly similar sequences, in NCBI database nucleotide collection (nr/nt).
The total DNA extracted from S. cucurbitacearum strain D33 was diluted to different concentrations (285−2.9 × 10–5 ng). In addition to this determination of the right quantity of DNA for the amplification, serial dilutions were set up for seed sample T85 (285, 57, and 20 ng), as a seed sample that was highly contaminated by S. cucurbitacearum (65.4%) according to the blotter analysis.
Molecular Survey of Stagonosporopsis cucurbitacearum on Naturally Infected Seeds by Conventional Polymerase Chain Reaction
The total DNA extracted from 19 of the seed samples with different S. cucurbitacearum infestations were processed through the conventional PCR using the DBF1/R1 primer pair at the optimized mix concentration and thermocycling conditions according to the preliminary tests. DNA of isolate D33 was used as the positive control. After the amplification, five specific amplicons were sequenced and compared with the database.
Validation of Stagonosporopsis cucurbitacearum Identification by qPCR
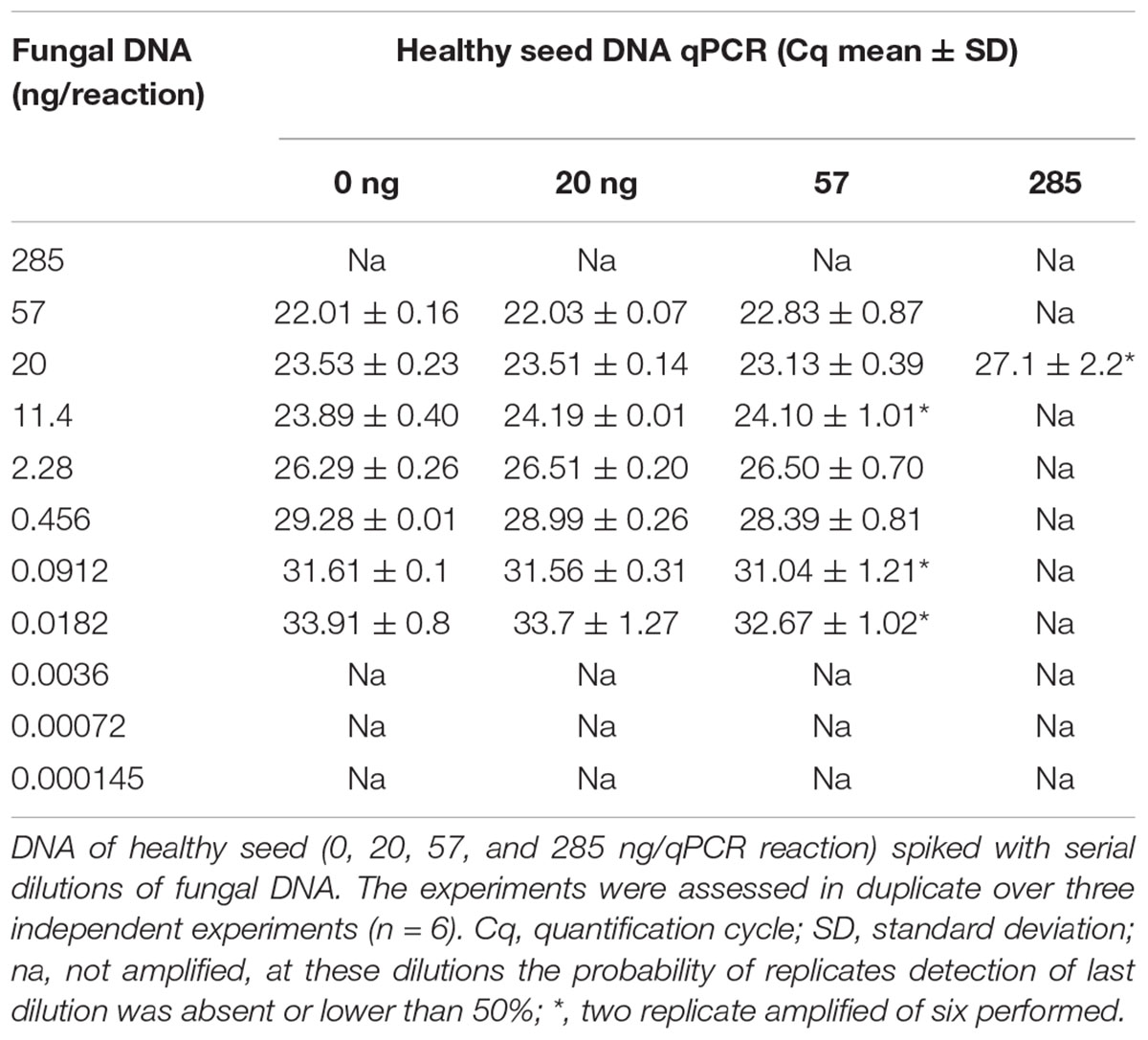
The primer pair DBF1/R1 was tested in qPCR. The analytical sensitivity (limit of detection; LOD, Bustin et al., 2009) was assessed using serial dilutions (285 ng, 57 ng, 20 ng, 11.4 ng, 2.28 ng, 4.56 × 10–1 ng, 9.12 × 10–2 ng, 1.82 × 10–2 ng, 3.6 × 10–3 ng, 7.2 × 10–4 ng, 1.45 × 10–4 ng, and 2.9 × 10–5 ng) of DNA extracted from the mycelia of S. cucurbitacearum isolate D33 spiked with 20, 57, and 285 ng of DNA from healthy seeds in qPCR (Landi et al., 2021). The reactions were carried out in a total volume of 12 μl containing: 2× Master mix iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, United States), and 0.10 μM of the designed primers (DBF1/R1). The reactions were subjected to the following conditions: 3 min at 95°C, followed by 40 cycles of 20 s at 95°C, and 40 s at 60°C. The final step included the melting curve analyses from 65 to 95°C. Two technical replications were performed for three independent experiments (n = 6).
Validation, Molecular Survey, and Absolute Quantification of Stagonosporopsis cucurbitacearum on Naturally Infected Seeds by Digital Droplet Polymerase Chain Reaction
The primer pair DBF1/R1, previously validated in conventional and qPCR, were tested in ddPCR with the same samples previous analyzed for qPCR validation. The ddPCR inhibitors, the optimal concentration of DNA template, the LOD and limit of quantification (LOQ) (Huggett et al., 2013) were determined. Here, 20 μl of the reaction mixture, containing 1 × QX200 ddPCR EvaGreen supermix (Bio-Rad), 150 or 300 nM each primer, and 20 ng of template, was transferred to a DG8 cartridge for droplet generation (QX200 droplet generator; Bio-Rad, Hercules, CA, United States). Then 70 μl droplet generation oil (Bio-Rad) was added to the cartridge, which was placed into the droplet generator. Droplet generation of 40 μl was carefully transferred to the ddPCR 96-well PCR plates (Bio-Rad), and the plates were sealed at 180°C using a PCR plate sealer (PX1; Bio-Rad).
The amplification was performed in a thermal cycler (ICycler; Bio-Rad), with a ramp rate of 2°C/s, with the following protocol: initial denaturation at 95°C for 5 min, then 45 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s (temperature ramp, 2°C/s), and finally, incubation at 98°C for 10 min, and storage at 4°C. After the cycling, the 96-well plates were fixed into a plate holder and positioned in the droplet reader (QX200; Bio-Rad). The droplets of each sample were analyzed sequentially, and the fluorescent signals of each droplet were measured individually by a detector. The droplets were read in the droplet reader, and then the ddPCR data were analyzed using Quantasoft version 1.7, which defined an automatic threshold or with a selected manually defined threshold applied. This incorporated the calculation of the basic parameters of the ddPCR (e.g., concentrations, mean amplitudes of positive, and negative droplets), and the mean copies per partition and total volume of the partitions measured, as defined by the digital MIQE guidelines (Huggett et al., 2013). Two positive droplets were enough to determine a sample as positive, and only the reactions with more than 10,000 accepted droplets were used for analysis.
After a preliminary validation of the technique, the primers DB1F/R1 were used at the concentration of 150 nM and 20 ng of total DNA extracted from 13 of the seed samples, which were representative of the different levels of pathogen contamination (as previously defined with the blotter analysis and conventional PCR) were processed by ddPCR (QX200 system; Bio-Rad, Hercules, CA, United States), according to the manufacturer instructions.
Results
Incidence of Stagonosporopsis cucurbitacearum Using the Blotter Method
The level of squash seed infection detected for S. cucurbitacearum using the blotter test ranged from 0 to 65.4% (Table 1). S. cucurbitacearum was detected for 29 of the 31 seed samples collected (93.6% of samples). More than 20% incidence of seedborne S. cucurbitacearum was detected for 12 of these seed samples (Table 1).
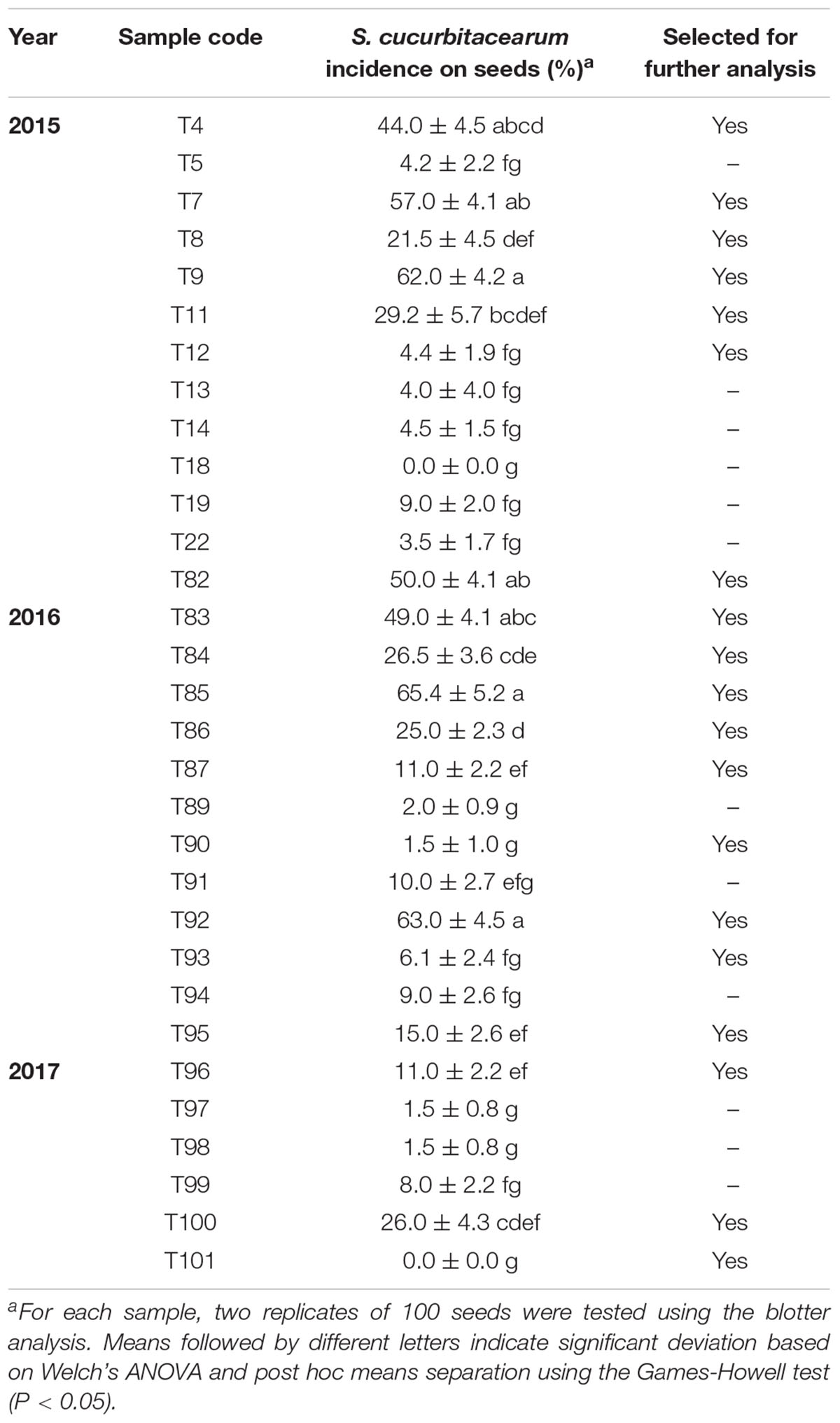
Table 1. Incidence of Stagonosporopsis cucurbitacearum infection using the blotter method for the 31 squash seed samples collected in 2015, 2016, and 2017.
The molecular approach allowed to corroborate the data emerging during the morphological characterization, and speed up the process of identification. By a multiplex PCR able to distinguish the three morphologically similar species, the natural inoculum of seeds in our study was represented only by S. cucurbitacearum (data not shown).
Molecular Detection of Stagonosporopsis cucurbitacearum by Conventional and Quantitative Real-Time Polymerase Chain Reaction
Optimization of Amplification Conditions and Primer Specificity by Conventional Polymerase Chain Reaction
The preliminary tests defined the optimal analysis conditions as 20 ng total DNA, 200 μM dNTPs mixture, 0.5 μM each primer, 1.2 mM MgCl2, 1.25 U Taq polymerase (Promega Corporation, Madison, WI, United States), and 20 ng template, in a total reaction volume of 25 μl. We set up the following cycling conditions: 95°C for 3 min., 35 cycles with 95°C for 30 s, 58°C for 30 s, and 72°C for 30 min, followed by a final extension step at 72°C for 5 min. The specificity tests amplified a 208-bp specific fragment in isolates D33, D12, D49, ID1, and ID3, which had been previously identified as S. cucurbitacearum by multilocus sequence analysis (Moumni et al., 2019, 2020; Supplementary Figure 1). The fragment was sequenced and analyzed by BlastN. All of the fragments shared high nucleotide similarity (>99%) with the sequences available in the database.
No amplification was recorded for the water control and for the other fungal pathogen samples [Phoma sp. (Ph39), Alternaria alternata (A38, A15), F. solani (F174), C. specifera (B170), P. roridum (M123), Albifimbria verrucaria (M144), and S. vesicarium (P164)], which are commonly isolated from squash seeds (Moumni et al., 2020; Supplementary Figure 1).
Limit of Detection for Conventional Polymerase Chain Reaction
The sensitivity of the DBF1/R1 primers was evaluated using serial dilution of DNA extracted from S. cucurbitacearum mycelia. The minimum concentration of target DNA that could be detected with these primers was 9.12 × 10–2 ng (Supplementary Figure 2). An ambiguous amplification was recorded at 1.82 × 10–2 ng. The PCR amplification fragments were strongly visualized when the DNA concentrate of S. cucurbitacearum ranged from 40 to 10 ng, and weakly visualized with 1 ng template. Very low DNA concentration (<1.82 × 10–2 ng) were not amplified by the primers (Supplementary Figure 2).
Once the specificity and sensitivity of the primers for S. cucurbitacearum were established, the 19 seed samples were analyzed. In 17 of these 19, a specific fragment of ∼208 bp was detected using the DBF1/R1 primer pair, as for the reference strain D33 of S. cucurbitacearum (positive control). These primers amplified the DNA from the seeds naturally contaminated by S. cucurbitacearum down to a threshold of 4.5% incidence in 200 seeds, as indicated in the blotter analysis. Faint band was obtained when the seed samples were contaminated at ∼1.5% in the blotter analysis (Figure 1). The PCR primers consistently showed strong bands for the seed samples with incidence from 44.0 to 65.4%, moderate bands between 21.5 and 29.1%, weak bands between 4.2 and 4.5%, and a very faint band with 1.5% infection (Figure 1). No amplification was detected in samples T18 and T101, as also for the water control and the certified healthy seed sample (IHS).
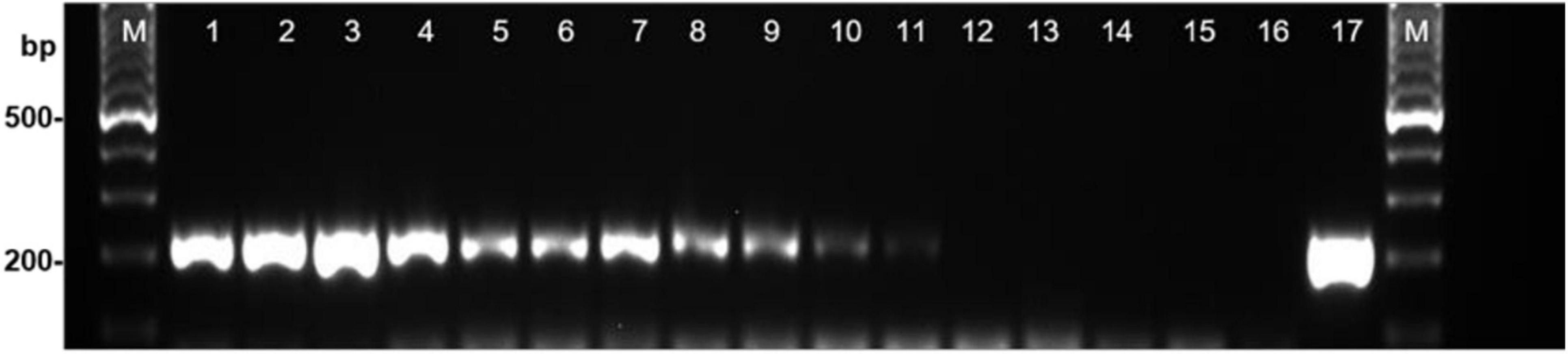
Figure 1. Detection of Stagonosporopsis cucurbitacearum from squash seed samples with different levels of infestation using PCR with primer pair DBF1/DBR1. Lanes 1–12: decreasing incidence of contamination (see Table 1): T85, T92, T9, T4, T86, T95, T96, T87, T93, T12, T90 and T101 (local healthy seed). Lane 13, commercial healthy seed (IHS); lane 14, water control (WC); lane 15, Curvularia spicifera (isolate B170); lane 16, Phoma sp. (isolate Ph39); lane 17, S. cucurbitacearum (isolate D33). M: Ladder, 100-bp.
The amplicons obtained from the DNA extracted from the mycelia (ID3 and ID9) and seed samples (T4and T7) were sequenced, and have been deposited with the NCBI database as MZ218113–MZ218116.
Validation of Stagonosporopsis cucurbitacearum Identification by qPCR
The reliable detection of DNA, extracted from S. cucurbitacearum isolate D33, ranged from 57 ng/reaction (Cq 22.01 ± 0.16) to 0.0182 ng/reaction (Cq 33.91 ± 0.8) (Table 2). The spiked sampled obtained with 20 ng/reaction of healthy seed DNA showed consistent results. For these concentrations, the Cq values was correlated with those obtained during the analysis of the DNA extracted from the mycelia of S. cucurbitacearum (Table 2). A single peak in positive samples suggests a single size product, with the melting temperature (TM) of 85.5°C (Supplementary Figure 3). At these conditions, it is reasonable to indicate as LOD in qPCR at Cq ≤ 33 (Table 2). Not consistent results were observed for S. cucurbitacearum mycelia DNA spiked with 57 and 285 ng of DNA from healthy seed.
Detection of Stagonosporopsis cucurbitacearum by Droplet Digital PCR
The limit of detection of S. cucurbitacearum by ddPCR was 3.6 × 10–3 ng, below which that there was no linear quantification of S. cucurbitacearum (Figures 2A,B). From the spiked samples analysis, a concentration of DNA > 20 ng had a negative influence on the detection of S. cucurbitacearum and on the accuracy of the absolute quantification (Figures 2A,B). The optimum condition was seen for 20 ng DNA from healthy seeds, at which the quantification of S. cucurbitacearum was highly related to the concentration of S. cucurbitacearum DNA added.
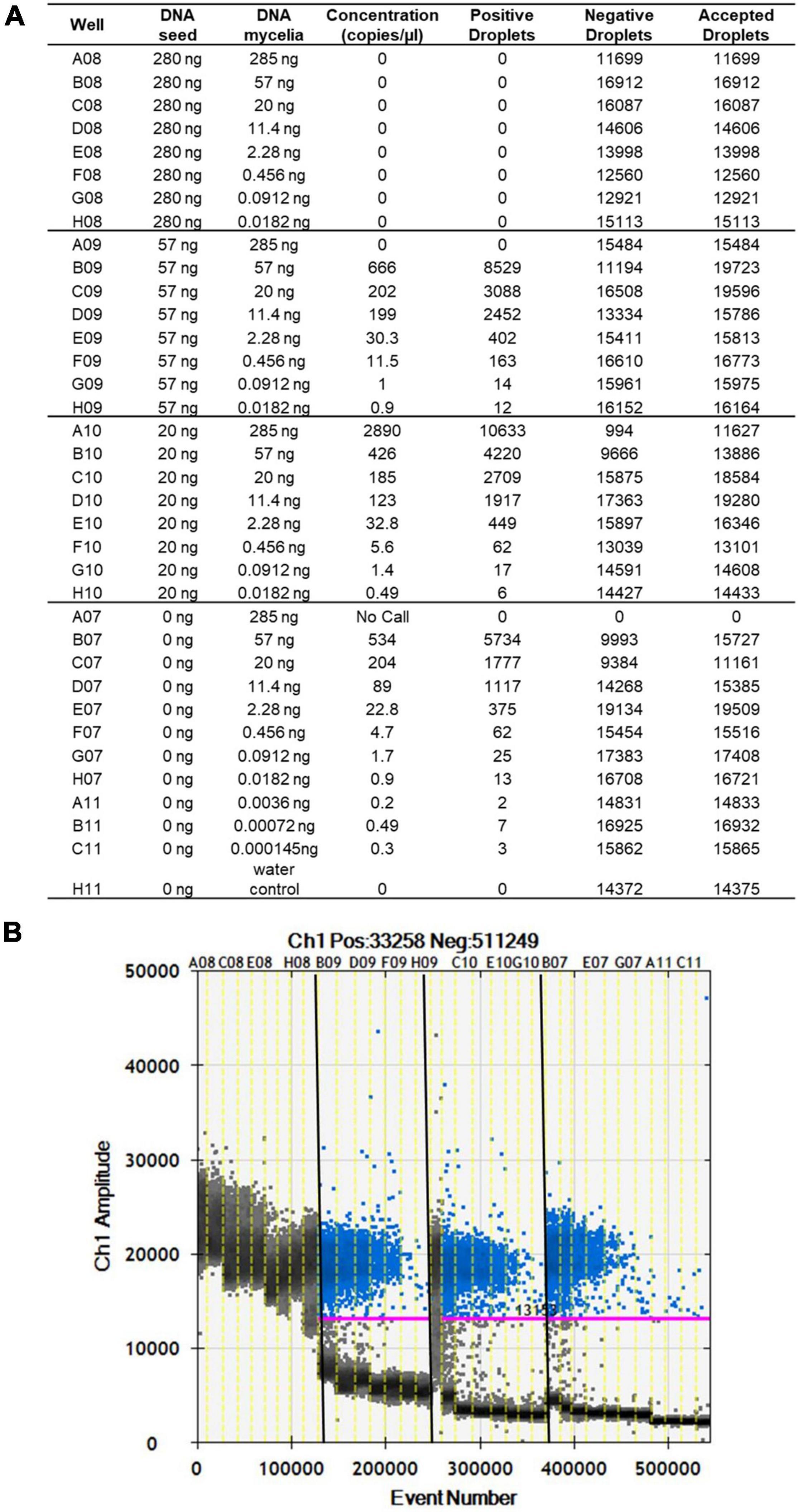
Figure 2. Analysis of spike samples obtained without and with mixing DNA from healthy seeds at different concentrations (as indicated) with DNA mycelia of S. cucurbitacearum at different concentrations (as indicated). (A) Absolute quantification. (B) Fluorescence amplitude. Pink line, threshold above which positive droplets (blue) contain at least one copy of target DNA, and below which negative droplets (gray) contain no copies of target DNA. Different sample sets in (A) are divided by vertical dotted yellow lines in (B). Wells A08 to H08 showed saturation signals.
After optimization of the amplification conditions, the ddPCR was applied to 13 seed samples that were naturally infected with S. cucurbitacearum and had been analyzed previously using the blotter analysis and the conventional PCR. The number of total events corresponding to the amount of droplets generated by the ddPCR ranged from 8,375 (sample T93) to 16,574 (sample T8) (Figures 3A,B). These values were related to the number of events, and ranged from 7.5 copies/μl (sample T85) to 0.2 copies/μl (sample T90). For sample T101, the water control and the healthy seeds sample (IHS), no amplifications were recorded (Figures 3A,B).
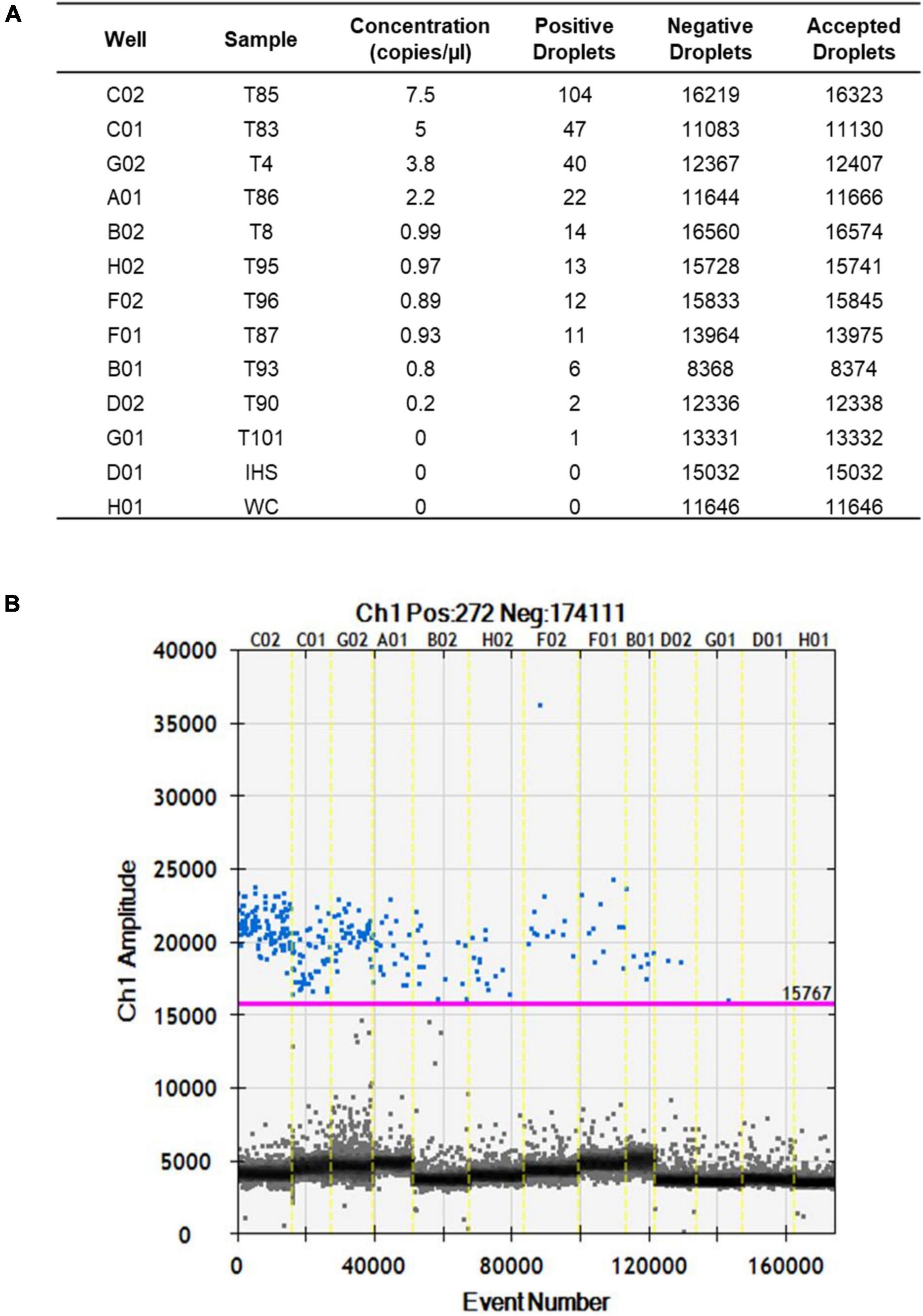
Figure 3. Fluorescence amplitudes of S. cucurbitacearum in the naturally infected seed samples (as indicated) and controls (IHS and WC) using ddPCR. (A) Absolute quantification. (B) Fluorescence amplitude.
Comparisons Between the Blotter Analysis, Conventional Polymerase Chain Reaction, and Droplet Digital PCR
The data from the blotter analysis were analyzed in relation to the data obtained for the ddPCR (Table 2), with high correlation seen (R2 = 0.986, p ≤ 0.01). The concentration of the amplified DNA target expressed as copies/μl was in accordance with seed contamination, in terms of the proportions of seeds infected by S. cucurbitacearum, recorded in blotter analysis. For the water control and the healthy seeds sample (IHS), where no infection was detected in the blotter analysis, no amplicons or positive events were seen for either conventional PCR or ddPCR. In sample T90, which showed seed infection of 1.5% in the blotter analysis, and where conventional PCR showed a light, and ambiguous band, the ddPCR showed an absolute quantification of 0.2 copies/μl. Finally, sample T101 that was negative in the blotter analysis and conventional PCR showed only 1 positive event for ddPCR, so this was also considered as negative in the ddPCR, along with the water control and the healthy seeds sample (Table 3).
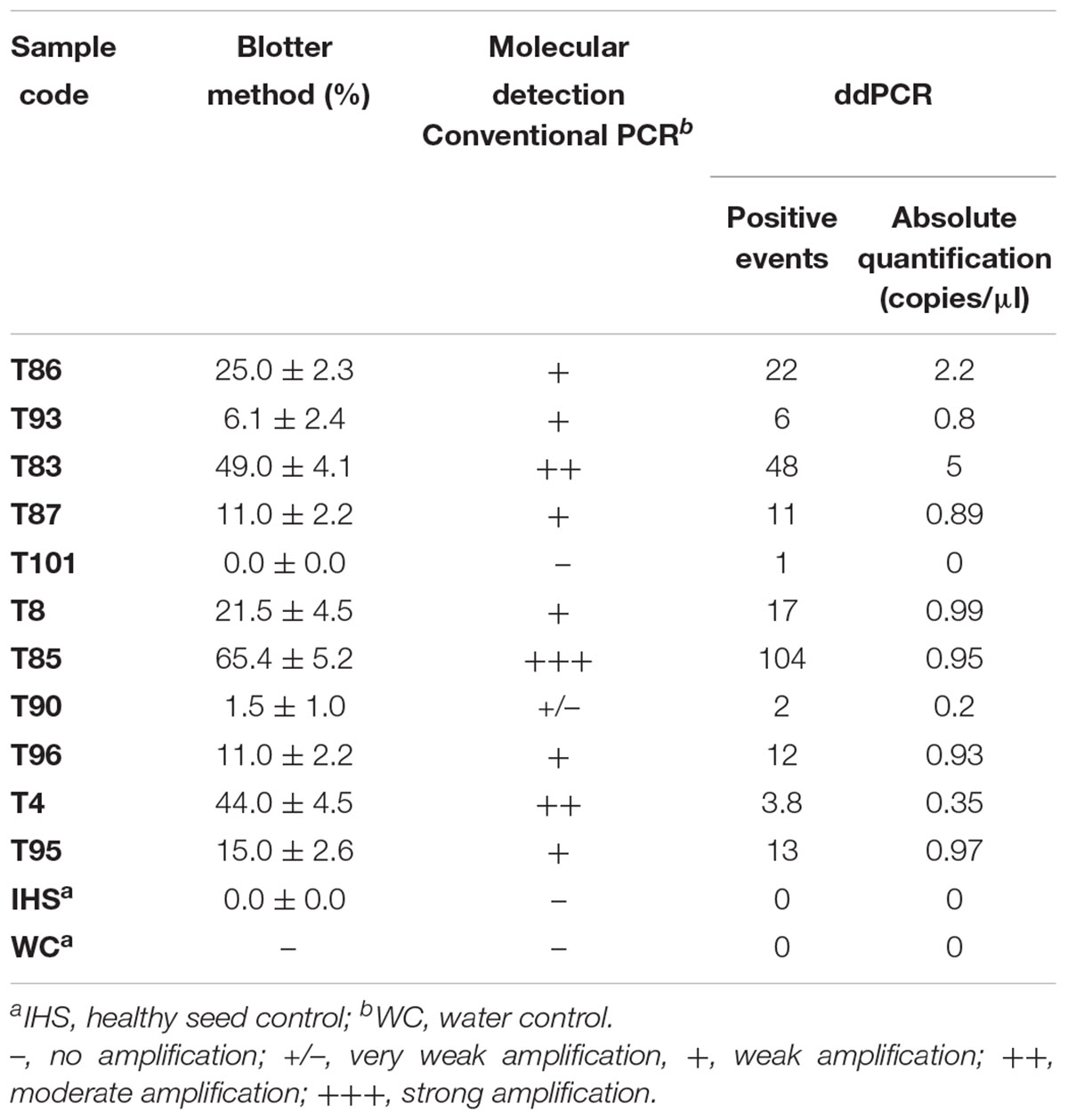
Table 3. Comparative detection of Stagonosporopsis cucurbitacearum in the naturally infected seed samples, according to the blotter analysis and conventional PCR and ddPCR.
Discussion
Healthy seeds are the start of healthy plants, and this is an essential requirement to safeguard the productivity of crops. GSB is a widespread disease and leads to significant losses in yield and quality for cucurbit crops worldwide (Keinath, 2011; Li et al., 2015; Yao et al., 2016; Zhao et al., 2020). In addition, the use of grafted cucurbits further increases the risk of GSB development from seedborne inoculum. Indeed, GSB was observed for grafted watermelon in Tunisia, which caused severe yield losses (Boughalleb et al., 2007).
Stagonosporopsis cucurbitacearum was recovered most frequently from Cucurbita spp. (Rennberger and Keinath, 2018; Zhao et al., 2018), and for squash seed the only fungal pathogen related to GSB in Tunisia and Italy (Moumni et al., 2019, 2020). Many other studies have demonstrated that infected seeds are the primary inoculum for GSB (De Neergaard, 1989; Sudisha et al., 2006; Keinath, 2011). The use of highly sanitized quality seeds decreases the primary inoculum in the field (Ciampi-Guillardi et al., 2020). Therefore, the detection of seedborne fungal pathogens is an important aspect for disease management.
The blotter method was appropriate in this study for the detection of S. cucurbitacearum in these seed samples. However, a drawback of this conventional method is that the morphological identification requires mycological skills and is time consuming; also, fungal contaminants can often mask the development of a pathogen. Indeed, the morphological characterization of S. cucurbitacearum, as well S. citrulli, S. cariacae and Phoma spp. are similar, and so distinguishing between these can be difficult (Keinath et al., 1995; Rennberger and Keinath, 2018). For this reason, in our study after the blotter tests and morphological identification, a preliminary molecular analysis carried out on the S. cucurbitacearum, as proposed by Brewer et al. (2015), allowed to confirm the identity. Rapid and accurate detection of pathogens transmitted by seeds should improve integrated disease management strategies, to control and prevent the spread of diseases caused by these pathogens. Several molecular methods have now been reported to detect pathogens on seeds (Lee et al., 2001; Pryor and Gilbertson, 2001; Samac et al., 1998). To set up efficient substitutes for the more traditional techniques, these methods need to be specific, sensitive, rapid, and adaptable to routine analysis. In the present study, the DBF1/DBR1 primers were designed to detect S. cucurbitacearum in these squash seeds. This set of primers successfully amplified the predicated size of the DNA fragment in infected material. One specific advantage of this PCR detection protocol is that it requires 1 day for completion, compared to the 10 days required for the blotter method. Thus, it can be used to examine both greater numbers and larger sample sizes with high reliability. Indeed, several studies have already reported primer pairs for the identification of S. cucurbitacearum in plant fragments after isolation to purity (Somai et al., 2002; Keinath et al., 2003; Brewer et al., 2015). Our analysis showed that the ddPCR method had a higher sensitivity than qPCR, and it is more reliable for the detection of the pathogen even at lowest titer.
Droplet digital PCR represents an innovative application in the diagnostic field, which is a user-friendly quantification technology, which does not require standard curve for the calibration (Bustin et al., 2009) that can be broadly used in several scientific fields, and its application to plant pathology is growing (Zhao et al., 2016). The present study represents the first approach to assess ddPCR as a reliable tool to detect and quantify pathogenic fungi associated with seeds. Many studies have reported that ddPCR is beneficial in terms of improved sensitivity of pathogen detection, and reduced effects of PCR inhibitors on PCR efficiency (Rački et al., 2014; Dupas et al., 2019; Bae et al., 2020).
The present study was thus designed to assess the diagnostic potential and sensitivity of ddPCR for absolute quantification of S. cucurbitacearum in the seeds of squash, as also compared to conventional PCR. ddPCR and the blotter test showed a high degree of correlation (R2 = 0.986, p ≤ 0.01) here. del Pilar Martínez-Diz et al. (2020) showed that ddPCR was more sensitive than qPCR for detection and quantification of the fungus Ilyonectria liriodendri. Dupas et al. (2019) demonstrated that ddPCR can improve the detection of the bacterium Xylella fastidiosa at low levels of infection and identified positive samples in those defined as negative by real-time PCR. Liu et al. (2020) suggested the use of ddPCR for detection of the fungus Tilletia controversa in soil samples and demonstrated that ddPCR was 100 times more sensitive than conventional PCR. Similarly, the data in the present study show that this ddPCR assay is a reliable alternative for quantification of S. cucurbitacearum on squash seeds. At the moment, for several important seedborne pathogens of vegetable crops, included S. cucurbitacearum, the International Seed Health Initiative and the American Seed Trade Association are attempting to compile pragmatic minimum thresholds based on experimental data, as well as empirical evidence and experience, that can be applied to seed in commerce. Considering that S. cucurbitacearum is transmitted by seeds and even low seed infection can cause medium-high economic losses in the field (Keinath, 2011), in our study we applied on one hand the blot test, an official method suggested by ISTA, on the other hand we explore the application of an absolute quantification method (ddPCR). To the best of our knowledge, this study is the first report of ddPCR for detection of such seedborne fungi. Further studies are required to evaluate and validate this new technology for routine use in the diagnosis of this and other seedborne pathogens. In addition, knowledge improvement of pathogen epidemiology initiated from seeds could lead to improve the management of such disease.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/; MZ218113–MZ218116.
Author Contributions
SM, LL, and GR: conceptualization. SM, VM, and MM: data curation and investigation. SM, MM, LL, and VM: formal analysis. SM and LL: methodology. SM and GR: project administration. SM, GR, and MA: supervision. MM and SM: writing—original draft. SM, MM, LL, MA, and GR: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This project was partially funded by UNIVPM Strategic Project 2016 “Control of plant diseases by natural compounds with quantification of plant pathogens and microbiological biodiversity, for a sustainable production of high fruit quality” and by the project “Strategies for management of diseases of seed-bearing vegetable crops for integrated pest management and organic agriculture (CleanSeed)” funded by Marche Region.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.764447/full#supplementary-material
References
Atallah, Z. K., Maruthachalam, K., du Toit, L., Koike, S. T., Davis, R. M., Klosterman, S. J., et al. (2010). Population analyses of the vascular plant pathogen Verticillium dahliae detect recombination and transcontinental gene flow. Fungal Gen. Biol. 47, 416–422. doi: 10.1016/j.fgb.2010.02.003
Boughalleb, N., El Mahjoub, M., Abad-Campos, P., Pérez-Sierra, A., García-Jiménez, J., and Armengol, J. (2007). First report of gummy stem blight caused by Didymella bryoniae on grafted watermelon in Tunisia. Plant Dis. 91:468. doi: 10.1094/PDIS-91-4-0468B
Brewer, M. T., Rath, M., and Li, H. X. (2015). Genetic diversity and population structure of cucurbit gummy stem blight fungi based on microsatellite markers. Phytopathology 105, 815–824. doi: 10.1094/PHYTO-10-14-0282-R
Bustin, S., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Choudhury, R. A., Garrett, K. A., Klosterman, S. J., Subbarao, K. V., and McRoberts, N. (2017). A framework for optimizing phytosanitary thresholds in seed systems. Phytopathology 107, 1219–1228. doi: 10.1094/PHYTO-04-17-0131-FI
Ciampi-Guillardi, M., Ramiro, J., Moraes, M. H. D. D., Barbieri, M. C. G., and Massola, N. S. Jr. (2020). Multiplex qPCR assay for direct detection and quantification of Colletotrichum truncatum, Corynespora cassiicola, and Sclerotinia sclerotiorum in soybean seeds. Plant Dis. 104, 3002–3009. doi: 10.1094/PDIS-02-20-0231-RE
Cunniffe, N. J., Cobb, R. C., Meentemeyer, R. K., Rizzo, D. M., and Gilligan, C. A. (2016). Modeling when, where, and how to manage a forest epidemic, motivated by sudden oak death in California. Proc. Natl. Acad. Sci. U.S.A. 113, 5640–5645. doi: 10.1073/pnas.1602153113
De Neergaard, E. (1989). Studies of Didytmella bryoniae (Auersw.) Rehm: development in the host. J. Phytopathol. 127, 107–115.
del Pilar Martínez-Diz, M., Andrés-Sodupe, M., Berbegal, M., Bujanda, R., Díaz-Losada, E., and Gramaje, D. (2020). Droplet digital PCR technology for detection of Ilyonectria liriodendri from grapevine environmental samples. Plant Dis. 104, 1144–1150. doi: 10.1094/PDIS-03-19-0529-RE
Dingle, T. C., Sedlak, R. H., Cook, L., and Jerome, K. R. (2013). Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 59, 1670–1672. doi: 10.1373/clinchem.2013.211045
Dreo, T., Pirc, M., Ramšak, Ž., Pavšič, J., Milavec, M., Zel, J., et al. (2014). Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal. Bioanal. Chem. 406, 6513–6528. doi: 10.1007/s00216-014-8084-1
Dupas, E., Legendre, B., Olivier, V., Poliakoff, F., Manceau, C., and Cunty, A. (2019). Comparison of real-time PCR and droplet digital PCR for the detection of Xylella fastidiosa in plants. J. Microbiol. Meth. 162, 86–95. doi: 10.1016/j.mimet.2019.05.010
Epanchin-Niell, R. S., and Hastings, A. (2010). Controlling established invaders: integrating economics and spread dynamics to determine optimal management. Ecol. Lett. 13, 528–541. doi: 10.1111/j.1461-0248.2010.01440.x
Epanchin-Niell, R. S. (2017). Economics of invasive species policy and management. Biol. Invasions 39, 1–22. doi: 10.1016/j.ecolecon.2006.07.024
Finger, M. J., Parkunan, V., Ji, P., and Stevenson, K. L. (2014). Allele-specific PCR for the detection of azoxystrobin resistance in Didymella bryoniae. Plant Dis. 98, 1681–1684. doi: 10.1094/PDIS-02-14-0136-RE
Gimode, W., Bao, K., Fei, Z., and McGregor, C. (2020). QTL associated with gummy stem blight resistance in watermelon. Theor. Appl. Genet. 134, 573–584. doi: 10.1007/s00122-020-03715-9
Ha, Y., Fessehaie, A., Ling, K., Wechter, W., Keinath, A., and Walcott, R. (2009). Simultaneous detection of Acidovorax avenae subsp. citrulli and Didymella bryoniae in cucurbit seedlots using magnetic capture hybridization and real-time polymerase chain reaction. Phytopathology 99, 666–678. doi: 10.1094/PHYTO-99-6-0666
Huggett, J. F., and Whale, A. (2013). Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin. Chem. 59, 1691–1693. doi: 10.1373/clinchem.2013.214742
Huggett, J. F., Foy, C. A., Benes, V., Emslie, K., Garson, J. A., and Haynes, R. (2013). The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 59, 892–902. doi: 10.1373/clinchem.2013.206375
Keinath, A. P., Somai, B. M., and Dean, R. A. (2003). Method of Diagnosing Gummy Stem Blight in Plants Using a Polymerase Chain Reaction Assay. U.S. Patent No: 20010758073. Clemson, SC: Clemson University.
Keinath, A. P. (1996). “Spread of Didymella bryoniae from contaminated watermelon seed and transplants in greenhouse and field environments,” in Recent Research Developments in Plant Pathology, ed. S. G. Pandalai (Trivandrum: Research Signpost), 65–72.
Keinath, A. P. (2011). From native plants in central Europe to cultivated crops worldwide: the emergence of Didymella bryoniae as a cucurbit pathogen. HortScience 46, 532–535.
Keinath, A. P., Farnham, M. W., and Zitter, T. A. (1995). Morphological pathological and genetic differentiation of Didymella bryoniae and Phoma spp. isolated from cucurbits. Phytopathology 85, 364–369. doi: 10.1094/phyto-85-364
Landi, L., Foglia, R., Murolo, S., and Romanazzi, G. (2021). The mycorrizal status in vineyards affected by esca. J. Fungi 7:869. doi: 10.3390/jof7100869
Lee, H. K., Tewari, J. P., and Turkington, T. K. (2001). A PCR-based assay to detect Rhynchosporium secalis in barley seed. Plant Dis. 85, 220–225. doi: 10.1094/PDIS.2001.85.2.220
Li, P. F., Ren, R. S., Yao, X. F., Xu, J. H., Babu, B., Paret, M. L., et al. (2015). Identification and characterization of the causal agent of gummy stem blight from muskmelon and watermelon in east China. J. Phytopathol. 163, 314–319. doi: 10.1111/jph.12277
Ling, K. S., Wechter, W. P., Somai, B. M., Walcott, R. R., and Keinath, A. P. (2010). An improved real-time PCR system for broad-spectrum detection of Didymella bryoniae, the causal agent of gummy stem blight of cucurbits. Seed Sci. Technol. 38, 692–703.
Liu, J., Li, C., Muhae-Ud-Din, G., Liu, T., Chen, W., Zhang, J., et al. (2020). Development of the droplet digital PCR to detect the teliospores of Tilletia controversa Kühn in the soil with greatly enhanced sensitivity. Front. Microbiol. 11:4. doi: 10.3389/fmicb.2020.00004
Lu, Y., Zhang, H.-J., Zhao, Z.-J., Wen, C.-L., Wu, P., Song, S.-H., et al. (2020). Application of droplet digital PCR in detection of seed-transmitted pathogen Acidovorax citrulli. J. Integr. Agric. 19, 561–569.
Mancini, V., and Romanazzi, G. (2014). Seed treatments to control seedborne fungal pathogens of vegetable crops. Pest Manage. Sci. 70, 860–868. doi: 10.1002/ps.3693
Mancini, V., Murolo, S., and Romanazzi, G. (2016). Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol. 65, 691–703.
Mathur, S. B., and Kongsdal, O. (2003). Common Laboratory Seed Health Testing Methods for Detecting Fungi. Bassersdorf: International Seed Testing Association.
Moumni, M., Allagui, M. B., Mancini, V., Murolo, S., Tarchoun, N., and Romanazzi, G. (2020). Morphological and molecular identification of seedborne fungi in squash (Cucurbita maxima, Cucurbita moschata). Plant Dis. 104, 1335–1350. doi: 10.1094/PDIS-04-19-0741-RE
Moumni, M., Mancini, V., Allagui, M. B., Murolo, S., and Romanazzi, G. (2019). Black rot of squash (Cucurbita moschata) caused by Stagonosporopsis cucurbitacearum reported in Italy. Phytopathol. Mediterr. 58, 379–383.
Newark, M. J., Li, P., Yang, X. P., Paret, M. L., and Dufault, N. S. (2020). Comparing Stagonosporopsis spp. fungicide resistance profiles in Florida and east China cucurbit production systems. Plant Dis. 104, 129–136. doi: 10.1094/PDIS-02-19-0370-RE
Ora, N., Faruq, A. N., Islam, M. T., Akhtar, N., and Rahman, M. M. (2011). Detection and identification of seed borne pathogens from some cultivated hybrid rice varieties in Bangladesh. Middle East J. Sci. Res. 10, 482–488.
Pryor, B. M., and Gilbertson, R. L. (2001). Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mtSSU rDNA sequences. Mycol. Res. 11, 1312–1321. doi: 10.1017/s0953756200003002
Rački, N., Dreo, T., Gutierrez-Aguirre, I., Blejec, A., and Ravnikar, M. (2014). Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 10:42.
Rani, A., Donovan, N., and Mantri, N. (2019). The future of plant pathogen diagnostics in a nursery production system. Biosens. Bioelectron. 145:111631. doi: 10.1016/j.bios.2019.111631
Rennberger, G., and Keinath, A. P. (2018). Susceptibility of 14 new cucurbit species to gummy stem blight caused by Stagonosporopsis citrulli under field conditions. Plant Dis. 102, 1365–1375. doi: 10.1094/PDIS-12-17-1953-RE
Samac, D. A., Nix, R. J., and Oleson, A. E. (1998). Transmission frequency of Clavibacter michiganensis subsp. insidiosus to alfalfa seed and identification of the bacterium by PCR. Plant Dis. 82, 1362–1367. doi: 10.1094/PDIS.1998.82.12.1362
Selvaraj, V., Maheshwari, Y., Hajeri, S., Chen, J., McCollum, T. G., and Yokomi, R. (2018). Development of a duplex droplet digital PCR assay for absolute quantitative detection of “Candidatus Liberibacter asiaticus”. PLoS One 13:e0197184. doi: 10.1371/journal.pone.0197184
Short, D. P. G., Gurung, S., Koike, S. T., Klosterman, S. J., and Subbarao, K. V. (2015). Frequency of Verticillium species in commercial spinach fields and transmission of V. dahliae from spinach to subsequent lettuce crops. Phytopathology 105, 80–90. doi: 10.1094/PHYTO-02-14-0046-R
Somai, B. M., Keinath, A. P., and Dean, R. A. (2002). Development of PCR-ELISA for detection and differentiation of Didymella bryoniae from related Phoma species. Plant Dis. 86, 710–716. doi: 10.1094/PDIS.2002.86.7.710
Stewart, J. E., Turner, A. N., and Brewer, M. T. (2015). Evolutionary history and variation in host range of three Stagonosporopsis species causing gummy stem blight of cucurbits. Fungal Biol. 119, 370–382.
Sudisha, J., Niranjana, S. R., Umesha, S., Prakash, H. S., and Shetty, H. S. (2006). Transmission of seed-borne infection of muskmelon by Didymella bryoniae and effect of seed treatments on disease incidence and fruit yield. Biol. Control 37, 196–205.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tian, Y., Liu, D., Zhao, Y., Wu, J., Hu, B., and Walcott, R. R. (2016). Visual detection of Didymella bryoniae in cucurbit seeds using a loop-mediated isothermal amplification assay. Eur. J. Plant Pathol. 147, 255–263. doi: 10.1007/s10658-016-0996-5
Bae, J. M., Wen, X., Kim, T. S., Kwak, Y., Cho, N. Y., Lee, H. S., et al. (2020). Fibroblast growth factor receptor 1 (FGFR1) amplification detected by droplet digital polymerase chain reaction (ddPCR) is a prognostic factor in colorectal cancers. Cancer Res. Treat. 52:74. doi: 10.4143/crt.2019.062
Yao, X., Li, P., Xu, J., Zhang, M., Ren, R., Liu, G., et al. (2016). Rapid and sensitive detection of Didymella bryoniae by visual loop-mediated isothermal amplification assay. Front. Microbiol. 7:1372. doi: 10.3389/fmicb.2016.01372
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134
Yu, L. U., Zhang, H. J., Zhao, Z. J., Wen, C. L., Ping, W. U., Song, L., et al. (2020). Application of droplet digital PCR in detection of seed-transmitted pathogen Acidovorax citrulli. J. Integr. Agric. 19, 561–569. doi: 10.1016/s2095-3119(19)62673-0
Zhang, Y. Y., Zhang, J., Gao, J., Zhang, G., Yu, Y., Zhou, H. Y., et al. (2018). The colonization process of sunflower by a green fluorescent protein-tagged isolate of Verticillium dahliae and its seed transmission. Plant Dis. 102, 1772–1778. doi: 10.1094/PDIS-01-18-0074-RE
Zhao, Q., Wu, J., Zhang, L., Yan, C., Jiang, S., Li, Z., et al. (2020). Genome-scale analyses and characteristics of putative pathogenicity genes of Stagonosporopsis cucurbitacearum, a pumpkin gummy stem blight fungus. Sci. Rep. 10:18065. doi: 10.1038/s41598-020-75235-x
Zhao, Q., Wu, J. Z., and Gong, Z. P. (2018). Identification and characterization of cucurbita gummy stem blight fungi in northeast China. J. Phytopathol. 166, 305–313.
Zhao, Y., Xia, Q., Yin, Y., and Wang, Z. (2016). Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri subsp. citri. PLoS One 11:e0159004. doi: 10.1371/journal.pone.0159004
Keywords: etiology, fungal pathogens, molecular, pathogen detection, techniques
Citation: Murolo S, Moumni M, Mancini V, Allagui MB, Landi L and Romanazzi G (2022) Detection and Quantification of Stagonosporopsis cucurbitacearum in Seeds of Cucurbita maxima Using Droplet Digital Polymerase Chain Reaction. Front. Microbiol. 12:764447. doi: 10.3389/fmicb.2021.764447
Received: 25 August 2021; Accepted: 15 December 2021;
Published: 11 January 2022.
Edited by:
Spiros Paramithiotis, Agricultural University of Athens, GreeceReviewed by:
Vladimiro Guarnaccia, University of Turin, ItalyAnna Cutarelli, Experimental Zooprophylactic Institute of Southern Italy (IZSM), Italy
Copyright © 2022 Murolo, Moumni, Mancini, Allagui, Landi and Romanazzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianfranco Romanazzi, Zy5yb21hbmF6emlAdW5pdnBtLml0
 Sergio Murolo
Sergio Murolo Marwa Moumni
Marwa Moumni Valeria Mancini1
Valeria Mancini1 Mohamed Bechir Allagui
Mohamed Bechir Allagui Lucia Landi
Lucia Landi Gianfranco Romanazzi
Gianfranco Romanazzi