- 1College of Oceanology and Food Science, Quanzhou Normal University, Quanzhou, China
- 2Key Laboratory of Inshore Resources Biotechnology (Quanzhou Normal University), Fujian Province University, Quanzhou, China
- 3Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, China
- 4The Institute of Life Sciences, The Hebrew University of Jerusalem, Edmond J. Safra Campus, Jerusalem, Israel
Members of the family Zoogloeaceae within the order Rhodocyclales are found to play vital roles in terrestrial and aquatic ecosystems by participating in biofloc formation in activated sludge, polycyclic aromatic hydrocarbon degradation, and nitrogen metabolism, such as denitrification and nitrogen fixation. Here, two bacterial strains designated H1-1-2AT and ZN11-R3-1 affiliated to the family Zoogloeaceae were isolated from coastal wetland habitats. The 16S rRNA gene sequences of the two strains were 100% identical and had maximum similarity with Nitrogeniibacter mangrovi M9-3-2T of 98.4% and ≤94.5% with other species. Phylogenetic analysis suggested that the two strains belonged to a single species and formed a novel monophyletic branch affiliated to the genus Nitrogeniibacter. The average nucleotide identity (ANI) value and digital DNA-DNA hybridization (dDDH) estimate between the two strains and N. mangrovi M9-3-2T were 78.5–78.7% and 21.4–21.6%, respectively, indicating that the two strains represent a novel species. The genomes of strain H1-1-2AT (complete genome) and ZN11-R3-1 (draft genome) were 4.7Mbp in length encoding ~4,360 functional genes. The DNA G+C content was 62.7%. Nitrogen fixation genes were found in the two strains, which were responsible for the growth on nitrogen-free medium, whereas denitrification genes found in N. mangrovi M9-3-2T were absent in the two strains. The respiratory quinone was ubiquinone-8. The major polar lipids consisted of phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, and aminophospholipid. The major fatty acids were summed feature 3 (C16:1ω7c and C16:1ω6c), C16:0, C12:0, and C10:0 3-OH. Based on genomic, phenotypic, and chemotaxonomic characterizations, strains H1-1-2AT and ZN11-R3-1 represent a novel species of the genus Nitrogeniibacter, for which the name Nitrogeniibacter aestuarii sp. nov. is proposed. The type strain is H1-1-2AT (=MCCC 1K04284T=KCTC 82672T), and additional strain is ZN11-R3-1 (=MCCC 1A17971=KCTC 82671). Additionally, phylogenomic analysis of the members of the family Zoogloeaceae including type strains and uncultivated bacteria was performed, using the Genome Taxonomic Database toolkit (GTDB-Tk). Combined with the 16S rRNA gene phylogeny, four novel genera, Parazoarcus gen. nov., Pseudazoarcus gen. nov., Pseudothauera gen. nov., and Cognatazoarcus gen. nov., were proposed. This study provided new insights to the taxonomy of the family Zoogloeaceae.
Introduction
The family Zoogloeaceae as a member of the order Rhodocyclales was firstly proposed in 2017 (Boden et al., 2017). Thus far, six genera with validly published names were described,1 including Zoogloea (Shin et al., 1993), Azoarcus (Rabus et al., 2019), Aromatoleum (Rabus et al., 2019), Thauera (Macy et al., 1993), Uliginosibacterium (Weon et al., 2008), and Nitrogeniibacter (Liao et al., 2021). Members of this family were found to play vital roles in terrestrial and aquatic habitats by participating in biofloc formation in activated sludge (such as Zoogloea; Shin et al., 1993), polycyclic aromatic hydrocarbon (PAH) degradation (Thauera; Mechichi et al., 2002), and nitrogen metabolism, such as denitrification (Thauera and Nitrogeniibacter; Liu et al., 2013; Liao et al., 2021) and nitrogen fixation (Azoarcus; Lin et al., 2020).
Previously circumscription of the taxonomy of the family Zoogloeaceae depended largely on phylogeny of 16S rRNA gene sequences, and a small number of species were included (Boden et al., 2017). The family Zoogloeaceae currently includes nearly 50 species with validly published or effectively published names.2 With the advance of next-generation sequencing (NGS) and methods of constructing metagenomic-centric genomes and single-cell genomes used for uncultivated bacteria (Rinke et al., 2013; Parks et al., 2018; Lapidus and Korobeynikov, 2021), a large number of genomes affiliated to the family Zoogloeaceae and the order Rhodocyclales were obtained and released publically in the Genome portal of GenBank. These genomes were obtained from various habitats including wastewater, soil, sediment, and freshwater (Wang et al., 2020). The genomes of uncultivated Zoogloeaceae members expanded our knowledge on their ecological niches and phylogenetic diversity. However, the taxonomic position of several members of the family Zoogloeaceae is still controversial. For instance, the genus Niveibacterium proposed in the family Rhodocyclaceae (Chun et al., 2016) is placed as a member within the Zoogloeaceae in the EzBioCloud Database (Yoon et al., 2017a); Thauera hydrothermalis GD-2T formed a separate branch on the basis of phylogeny of 16S rRNA gene that were distinct from the type species T. selenatis ATCC 55363T (Liao et al., 2021). This may be the result of using a small number of species for phylogenetic analysis based on 16S rRNA gene comparison. Thus, the phylogenetic relationship of the Zoogloeaceae members needs to be reconsidered, especially on the basis of genome sequences. The Genome Taxonomic Database (GTDB) is considered to be a reliable tool to define the bacterial taxonomic ranks using 120 conserved concatenated proteins (Parks et al., 2018) and is used in accurate assignment for not only the described species but also for genomes of uncultivated organisms. Thus, the phylogeny of the family Zoogloeaceae was revisited in this study based on the use of GTDB tools.
Nitrogeniibacter, affiliated to the family Zoogloeaceae, is a recently proposed genus, with a single species, N. mangrovi. The type strain M9-3-2T (=MCCC 1K03313T=JCM 32045T) was isolated from an enrichment culture of mangrove sediment (Liao et al., 2021). The genus is circumscribed on the basis of 16S rRNA gene phylogeny and concatenated core genes (phylogenomic tree) and physiological and chemical characteristics (Liao et al., 2021). The cells are Gram stain-negative and show anaerobic and aerobic growth, rod-shaped, oxidase-positive, and catalase-positive. Ubiquinone-8 (Q-8) is the major respiratory quinone, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phospholipids, and aminophospholipids are major polar lipids, and summed feature 3 (C16:1ω7c and C16:1ω6c), C16:0, C10:0 3-OH, C14:0, and C10:0 are major fatty acids. This genus had the ability of denitrification under both aerobic and anaerobic conditions.
In this study, two isolates designated H1-1-2AT and ZN11-R3-1 were obtained from a sediment sample of a Spartina alterniflora wetland and from styrofoam plastics collected from a mangrove, respectively. The isolates were found to have identical 16S rRNA gene sequences and likely represented a novel species of the genus Nitrogeniibacter within the family Zoogloeaceae. This study aimed to determine the taxonomic status of the two isolates using a polyphasic taxonomic approach. Additionally, the phylogeny of the Zoogloeaceae members was elucidated based on the available genomes to further advance the taxonomy of the family.
Materials and Methods
Bacterial Isolation and Cultivation
Strains H1-1-2AT and ZN11-R3-1 were isolated from a coastal sediment sample and from an enrichment culture inoculated with coastal styrofoam plastics, respectively. The sediment sample was collected from a Spartina alterniflora growing area in a wetland (24°86' N, 118°68' E) in Quanzhou Bay, Quanzhou, PR China, on September 05, 2019. A water-extracted medium (WEM) prepared using the nutrients extracted from the sediment with pure water (w/v=1:1) was used to isolate strain H1-1-2AT (Huang et al., 2020b). The 0.2g sediment sample was subjected to 10-fold serial dilutions and spread on the WEM plates and incubated for 2weeks at 28°C. Strain H1-1-2AT was picked and then streaked onto Marine Broth 2216 (MB, BD) agar plates to obtain a pure culture. For the isolation of strain ZN11-R3-1, styrofoam plastic was collected from a mangrove preservation area (24o27' N, 117o53' E) in Longhai, Zhangzhou, PR China, on November 23, 2019. The plastics were placed into an enrichment medium of 300ml sterile MMC (NaCl 24g/L; MgSO4·7H2O 7.0g/L; NH4NO3 1g/L; KCl 0.7g/L; KH2PO4 2.0g/L; and Na2HPO4·12H2O 3.0g/L, pH=7.4) and maintained at 150rpm shaking at 28°C for 2months. An aliquot (2ml) of enriched culture was then transferred to another 100ml fresh MMC medium containing sterile plastics and cultured for another 2months. Then, the enrichment was repeated as above. The biomass in the third enrichment culture was collected using centrifugation at 6,000rpm for 15min and plated on an MB agar plate and maintained at 30°C. Strains H1-1-2AT and ZN11-R3-1 grew well on MB agar plates and MB medium and were stored at −80°C with 20% glycerol (v/v) in the laboratory.
Phylogeny Analysis Based on 16S rRNA Gene Sequences
The nearly complete 16S rRNA gene sequences of strain H1-1-2AT and strain ZN11-R3-1 were obtained using Sanger sequencing performed as described in a previous study (Huang et al., 2020b). The sequences were also compared with rRNA genes extracted from the genome sequences.
Sequences of the closely related relatives of the two strains were obtained from the EzBioCloud database (Yoon et al., 2017a) and the NCBI nucleotide database.3 Burkholderia cepacia ATCC 25416T was selected as an outgroup. Then, the 16S rRNA gene sequences were aligned and subjected to phylogenetic analysis using two algorithms, neighbor-joining (NJ) and maximum likelihood (ML) methods with 1,000 bootstraps using MEGA 7.0 (Huang et al., 2019). The best model (T92+G+I) with the lowest Bayesian information criterion (BIC) scores was selected.
Genome Sequencing and Gene Annotation
The draft genome sequences of strain H1-1-2AT and strain ZN11-R3-1 were determined using the Illumina HiSeq X-Ten platform (Shanghai Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China). A library of ~400bp fragments was constructed, and paired-end (PE) short reads of ~1 Gb were obtained. The PE reads were firstly trimmed to remove the low base of quality <20 and length <50bp using sickle.4 Then, clean reads were assembled into contigs using SPAdes v 3.8.0 with a serial of k values of 21, 33, 55, 77, 99, 127 and –careful flag (Huang et al., 2019). Then, contigs shorter than 1kb were removed from the assembled contigs. The genome quality was evaluated using QUAST (Gurevich et al., 2013).
The complete genome of type strain H1-1-2AT was obtained using PacBio sequencing with one SMART cell. The 10-kb fragment library was constructed followed the manufacturer’s instructions. The long reads were assembled using the SMRT Link (V6.0.0.47841) of PacBio.
The complete 16S rRNA gene sequence was extracted from the genome sequence using RNAmmer (Lagesen et al., 2007). Genome completeness was evaluated using CheckM v1.0.1 (Parks et al., 2015). Gene annotation was carried out using the RAST server (Aziz et al., 2008) and the KAAS system.5 Functional genes with high similarity to close relatives were searched using the blast+ program with e-value cutoff of 1e-5 (Camacho et al., 2009).
The average nucleotide identity (ANI) values were estimated using OrthoANI computation on the EzBioCloud Database (Yoon et al., 2017b). Digital DNA-DNA hybridization (dDDH) estimates were calculated on the GGDC website.6 Average amino acids identity among genomes was calculated using CompareM v0.1.2.7 The percentage of conserved proteins (POCP), proposed as genus boundary values was also calculated for genomic comparison (Qin et al., 2014).
Phylogenomic Analysis
The genomes affiliated to the order Rhodocyclales were downloaded from the genome portal in NCBI.8 A total of 303 genomes were obtained (until Feb.19, 2021), and the genome quality was checked using CheckM v1.0.1 (Parks et al., 2015). Genomes of <50% completeness and >10% contamination were removed from the following study. In addition, 9 genomes, identified using GTDB-tk v. 0.3.2 (Chaumeil et al., 2019), did not belong to the order Rhodocyclales, and these were removed from the study. Then, the phylogenomic tree of the genomes was inferred using a concatenated alignment of 120 bacterial single-copy genes with GTDB-tk v. 0.3.2 by using FastTree (Parks et al., 2018). The tree was edited using the Interactive Tree of Life (iTOL) online (Letunic and Bork, 2007). In addition, a phylogenomic tree based on the genomes of type strains belonging to the order Rhodocyclales was also constructed using GTDB-Tk.
Phenotypic Properties
Gram staining was carried out using a Gram staining kit (Hangzhou Tianhe Microorganism Reagent, Co., Ltd.). Colony morphology was recorded on a MB agar plate after incubation at 30°C for 3days. Catalase activity was tested by using 3% H2O2 solution. Oxidase activity was tested using the oxidase reagent (1% aqueous solution of N,N,N',N'-tetramethyl-p-phenylenediamine dihydrochloride, bioMérieux, France). Motility was observed by puncturing the cells into 0.5% agar. Growth under the anaerobic condition was tested by inoculating the cells into an anaerobic MB medium for 7days. The growth temperature range, NaCl tolerance range, pH range of the strains, and hydrolysis of substrates were determined as described in our previous study (Huang et al., 2020b). Growth on nitrogen-free medium was tested following the method of Huang et al. with 5g/l NaCl and 5g/l glucose (Huang et al., 2014). N. mangrovi M9-3-2T (=MCCC 1K03313T), obtained from the Marine Culture Collection Center (MCCC), was used as a reference strain.
Physiological and biochemical characterization was carried out using API ZYM, API 20NE, and API 20E kits according to the manufacturer’s instructions (bioMérieux, France). The tested strains and the reference strain were maintained under identical laboratory conditions. Test strips were maintained at 35°C for determining the physiological and biochemical properties.
Chemotaxonomic Characteristics
For the analysis of fatty acids composition, the strains and reference strain were cultured in MB at 35°C for 3days and cells were collected by centrifugation at 8,000rpm for 10min. The cellular fatty acids were saponified, methylated and extracted, and then identified following the standard MIDI protocol (Sherlock Microbial Identification System, version 6B).
For the polar lipids analysis, strain H1-1-2AT was cultured in MB medium at 35°C for 3days, and cells were harvested by using centrifugation as above. Polar lipids were extracted using a chloroform/methanol system and analyzed using one- and two-dimensional TLC using Merck silica gel 60F254 aluminum-backed thin-layer plates. Lipids were detected and identified by spraying the specific reagents (Huang et al., 2020a).
Results and Discussion
Phylogeny of 16S rRNA Gene Sequences
The 16S rRNA gene sequences of strains H1-1-2AT and ZN11-R3-1, obtained by Sanger sequencing or extracted from the genome sequences, had 100% identity, indicating the two strains belonged to same species. The BOX-PCR genotypic fingerprinting profiles of two strains were similar but distinctive (Supplementary Figure 1), which confirmed that they were not clonal. Also, the fingerprinting of the two strains were totally different from N. mangrovi M9-3-2T, indicating they may belong to a novel species different from N. mangrovi.
Sequence similarity search showed that the 16S rRNA gene sequence of strain H1-1-2AT had the maximum similarity (99.6%) with an uncultured bacterium clone IWNB003 (accession number: FR744543), followed by N. mangrovi M9-3-2T (98.4%), and had sequence similarities of ≤94.5% with other species affiliated to the family Zoogloeaceae. The clone IWNB003 was found in nitrate-amended injection seawater from an oil field (Gittel et al., 2012), and N. mangrovi M9-3-2T has the ability of denitrification (Liao et al., 2021), which may indicate that Nitrogeniibacter members play valuable roles in nitrogen cycle in the environment.
Phylogeny of 16S rRNA gene sequence inferred from the ML and NJ methods placed strains H1-1-2AT and ZN11-R3-1 within the genus Nitrogeniibacter as a novel monophyletic line, distinct from N. mangrovi M9-3-2T. This indicated that the two strains could be considered as a novel species of the genus Nitrogeniibacter (Figure 1; Supplementary Figure 2).
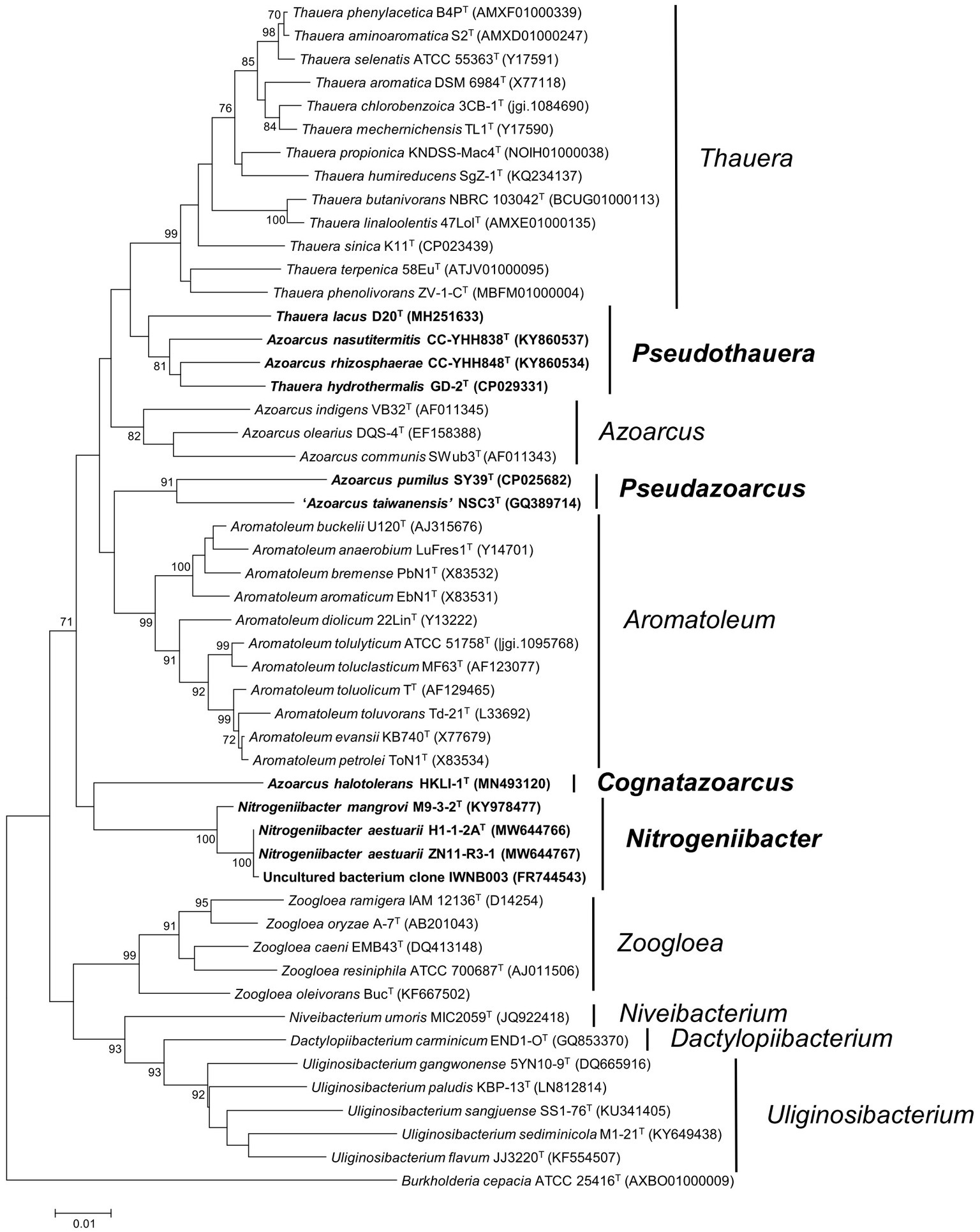
Figure 1. Phylogeny of 16S rRNA gene sequences. The tree was constructed using the neighbor-joining method. Bootstrapping was carried out with 1,000 replicates. Branch node values below 70% are not shown. Burkholderia cepacia ATCC 25416T (AXBO01000009) was selected as the outgroup. Bar, 0.01 represented the nucleotide substitution per position. Members of Nitrogeniibacter, Pseudazoarcus, and Pseudothauera are marked bold.
Phylogeny of 16S rRNA gene sequences indicated that the members of Azoarcus and the members of Thauera were separated into different clades, which were clearly separated from the type species, A. indigens and T. selenatis. Firstly, A. pumilus SY39T and “A. taiwanensis” NSC3T formed a separate cluster, which did not cluster with the type species A. indigens. Here, we named this cluster as a novel genus Pseudazoarcus, which was equal to the group name “Azoarcus_D” of the Genome Taxonomy Database (GTDB; Chaumeil et al., 2019). Thus, A. pumilus should be transferred into the genus Pseudazoarcus. Azoarcus pumilus was designated the type species of this genus and was renamed as Pseudazoarcus pumilus comb. nov. “A. taiwanensis” (a name effectively but not validly published; Lee et al., 2014) was also affiliated to this genus. Secondly, in the phylogenetic clade of the genus Thauera, there were four species, including T. lacus, T. hydrothermalis, A. nasutitermitis, and A. rhizosphaerae that formed a monophyletic cluster. Though this cluster formed a node with other Thauera members, bootstrap support was low (<70% of both ML and NJ; Figure 1; Supplementary Figure 2). The four species may be assigned to a new genus named Pseudothauera, which is equivalent to “Thauera_A” in the GTDB taxonomy. Thus, the four species, T. lacus, T. hydrothermalis, A. nasutitermitis, and A. rhizosphaerae, should be transferred to a novel genus Pseudothauera and renamed as Pseudothauera lacus comb. nov., Pseudothauera hydrothermalis comb. nov., Pseudothauera nasutitermitis comb. nov., and Pseudothauera rhizosphaerae comb. nov., respectively. Thirdly, A. halotolerans HKLI-1T formed an independent line on the phylogenomic tree, which clearly branched with Azoarcus. This species should be reclassified into a novel genus; Cognatazoarcus halotolerans gen. nov., comb. nov. was therefore proposed. Fourthly, A. communis SWub3T did not cluster together with the type species A. indigens and should be transferred into a novel genus. Here, we named this cluster as a novel genus Parazoarcus, which was equal to the genus name “Azoarcus_C” of the Genome Taxonomy Database (GTDB).
Genomic Characteristics
The complete genome of strain H1-1-2AT included one chromosome (4,678,511bp) and one plasmid (66,515bp). The draft genome size of strain ZN11-R3-2 was 4,656,485bp on 42 contigs (>1kb) with N50 value of 343,918bp (Table 1). Three copies of rrn operon (16S-
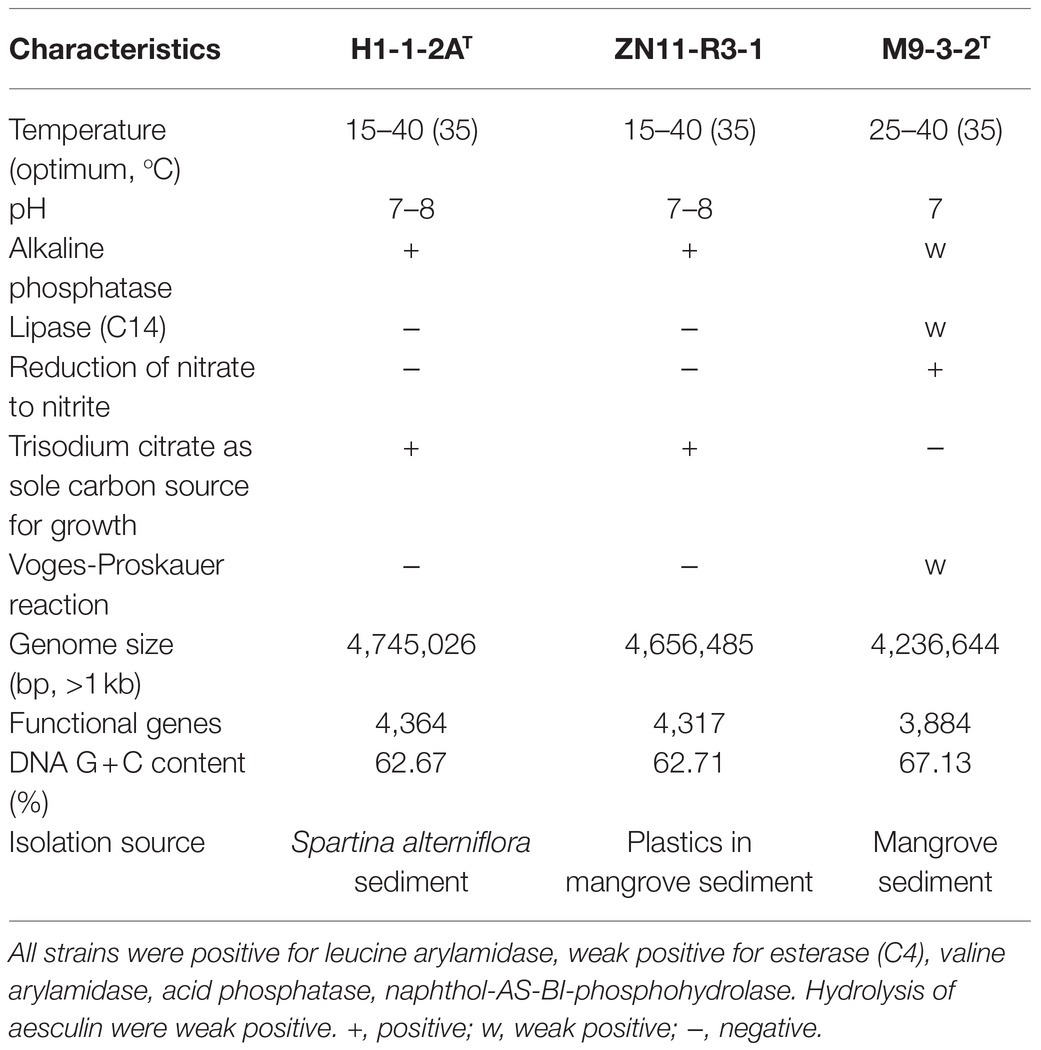
Table 1. Differential characteristics of strain H1-1-2AT and strain ZN11-R3-1 compared to the close relative N. mangrovi M9-3-2T.
23S-5S rRNA genes) were found in the complete genome, and 16S rRNA gene copies were 1,528bp in length and had 100% identity. The genome size of the strains was a little larger than N. mangrovi M9-3-2T (a chromosome of 4,236,644bp; accession number: CP048836). The DNA G+C content of strains H1-1-2AT and ZN11-R3-1 were 62.67 and 62.71%, respectively, which were a little lower than N. mangrovi M9-3-2T (67.13%). Gene prediction showed that there were 4,364 and 3,884 predicted genes in strains H1-1-2AT and M9-3-2T, respectively. dDDH and ANI values between strain H1-1-2AT and ZN11-R3-1 were estimated to be 85.6 and 98.4%, respectively. These values exceeded the threshold of species delineation, which strongly supported that the two strains belonged to the same species (Kim et al., 2014). dDDH and ANI values between strains H1-1-2AT and ZN11-R3-1 and N. mangrovi M9-3-2T were 21.4–21.6% and 78.50–78.72%, indicating that strains H1-1-2AT and ZN11-R3-1 represented a novel species.
Strains H1-1-2AT and ZN11-R3-1 contained nitrogen fixation gene clusters encoding nitrogenase reductase (nifH, KO list: K02588), nitrogenase molybdenum-iron protein (nifD, K02586; nifK, K02591), and related proteins (modABCD; Supplementary Table 1), which were also found in N. mangrovi M9-3-2T (Liao et al., 2021). The nitrogen fixation genes were assumed to enable their growth on nitrogen-free medium. Interestingly, denitrification genes, nirBD (K000362 and K000363), norBC (K004561 and K002305), and narGHIJ (K000370, K000371, K000373, and K000374), were not found in strains H1-1-2AT and ZN11-R3-1, but they were present in N. mangrovi M9-3-2T (Supplementary Table 2), in which denitrification was confirmed in a laboratory experiment (Liao et al., 2021). In addition, the sox system (soxABCDXYZ gene cluster) was present in strain H1-1-2AT (Supplementary Table 3) and in N. mangrovi M9-3-2T (Liao et al., 2021), suggesting that Nitrogeniibacter members may have the ability of sulfur oxidation.
Phylogenomics of the Family Zoogloeaceae
The development of MAG binning and single-cell genomes contributed large numbers of genome sequences of uncultivated bacteria, including members of family Zoogloeaceae and the order Rhodocyclales, which could expand knowledge on the phylogenetic diversity based on core genome analysis. Here, the genomes of the order Rhodocyclales with ≥50% completeness and≤10% contamination were used, of which the genome quality was verified to perform accurate phylogenetic analysis by GTDB-Tk (Bowers et al., 2017). A total of 277 genomes affiliated to the order Rhodocyclales that meet the above standards were used in the phylogenomic analysis. Compared to the 78 and 92 genomes analyzed in phylogenomic studies of the order Rhodocyclales by Wang et al. (2020) and Liao et al. (2021), respectively, our study further expanded the known phylogenetic groups within the order Rhodocyclales. The described species account for a minor part of the phylogenomic tree, indicating that majority of the members of Rhodocyclales are still waiting to be cultivated (Figure 2).
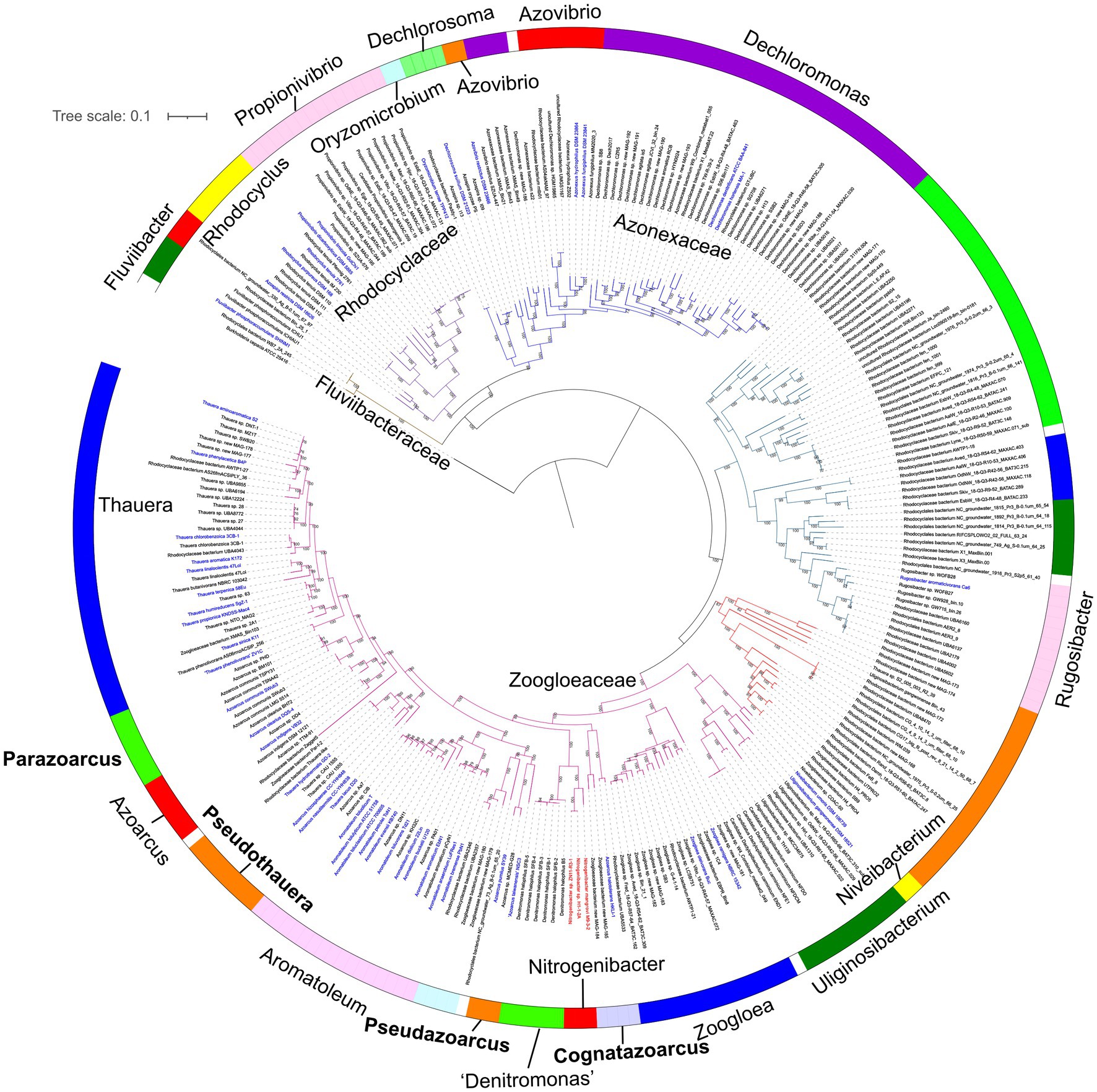
Figure 2. Phylogenomic analysis based on 120 bacterial covered single-copied gene sets of the members affiliated to the order Rhodocyclales using FastTree. The bootstrap values on the node are displayed by >70. Bar, 0.1 represents the nucleotide substitutions per position. The blue names represent validly published species. Red names showed Nitrogeniibacter members. The genus names were shown around the color circle. Four genera proposed in this study are marked bold.
Phylogenomic analysis based on 120 bacterial conserved single-copy genes strongly placed strains H1-1-2AT and ZN11-R3-1 in a sister group of the genus Nitrogeniibacter, which was neighbored by “Denitromonas.” This agreed with the phylogeny based on concatenated core genome sequences (Liao et al., 2021). “Denitromonas” should be transferred into the family Zoogloeaceae and did not belong to the family Rhodocyclaceae.9 In the lineages of the family Zoogloeaceae, the relationship between A. pumilus SY39T and “A. taiwanensis” NSC3T showed congruent topology with 16S rRNA gene phylogeny (Figure 1), which strongly supported the two species should be reclassified into a novel genus, for which we propose the name Pseudazoarcus. Also, in the phylogenomic tree, T. lacus D20T, T. hydrothermalis GD-2T, A. nasutitermitis CC-YHH838T, and A. rhizosphaerae CC-YHH848T formed a monophyletic cluster, which also supported the phylogeny of the 16S rRNA gene. The four species should be assigned to a new genus, for which we propose the name Pseudothauera. In addition, phylogenomic analysis of A. olearius DQS-4T, A. indigens VB32T, and A. communis SWub3T showed topology incongruent with the 16S rRNA gene, possibly due to the small number of sequences used. Thus, it is proposed that A. communis SWub3T be reclassified into a novel genus named Parazoarcus gen. nov. Azoarcus halotolerans HKLI-1T, which is only distantly related to the type species A. indigens, should also be reclassified into a novel genus. Thus, Cognatazoarcus gen. nov. was proposed. Niveibacterium firstly proposed in the family Rhodocyclaceae (Chun et al., 2016) should be transferred to the family Zoogloeaceae based on the phylogenetic analysis. Finally, a family-level lineage including the genus Rugosibacter was clearly separated from the family Zoogloeaceae, indicating that Rugosibacter may represent a novel family.
Figure 3 presents a small phylogenomic tree reconstructed using GTDB-tk, only including the type strains. Two genomes, Thauera selenatis AXT (type species) with high genome contamination and “Zoogloea ramigera” ATCC 19544, possibly incorrectly named, were excluded (Supplementary Table 4). The topology of the small tree was congruent with that of the large tree, which supported the above analysis. AAI values calculated among the 40 members of the family Zoogloeaceae ranged from 60.34 to 94.53% (Figure 4), which exceeded the family boundary of >45% (Konstantinidis et al., 2017). Thus, the members should be considered to belong to the family Zoogloeaceae. Our analysis did not support the proposal of Uliginosibacterium as an independent family (Wang et al., 2020). Compared to POCP values, AAI values demonstrated certain advantages to delineate the genus boundary of the members of the Zoogloeaceae (Figure 4). The calculation of POCP values depends on the similarity of the protein contents of genomes, which had similar genome size (Qin et al., 2014). It is reported that POCP values are also not effective and appropriate for delineating the genera of the families Acetobacteraceae (Rai et al., 2021), Rhodobacteraceae (Suresh et al., 2019), and Methylococcaceae (Orata et al., 2018). For instance, the four species, T. lacus, T. hydrothermalis, A. nasutitermitis, and A. rhizosphaerae, clearly grouped together, ranging from 78.37 to 80.80% of the AAI values for the type strains, which were below the recommended genus cutoff of <80% (Luo et al., 2014). The four species were distinctly separated from Thauera members and other genera (Figure 4). The AAI values of Nitrogeniibacter compared to the genera Thauera, Parazoarcus, Azoarcus, Pseudothauera, Pseudazoarcus, and Cognatazoarcus were 65.5–67.4%, 65.9–66.8%, 65.6–67.0%, 66.2–68.5%, 64.1–66.1%, and 66.4–67.5%, respectively, which were below the genus cutoff of <80% (Luo et al., 2014). Thus, our study expanded the family Zoogloeaceae into 11 genera, including Zoogloea, Azoarcus, Aromatoleum, Thauera, Niveibacterium, Uliginosibacterium, Nitrogeniibacter, Parazoarcus, Cognatazoarcus, Pseudazoarcus, and Pseudothauera.
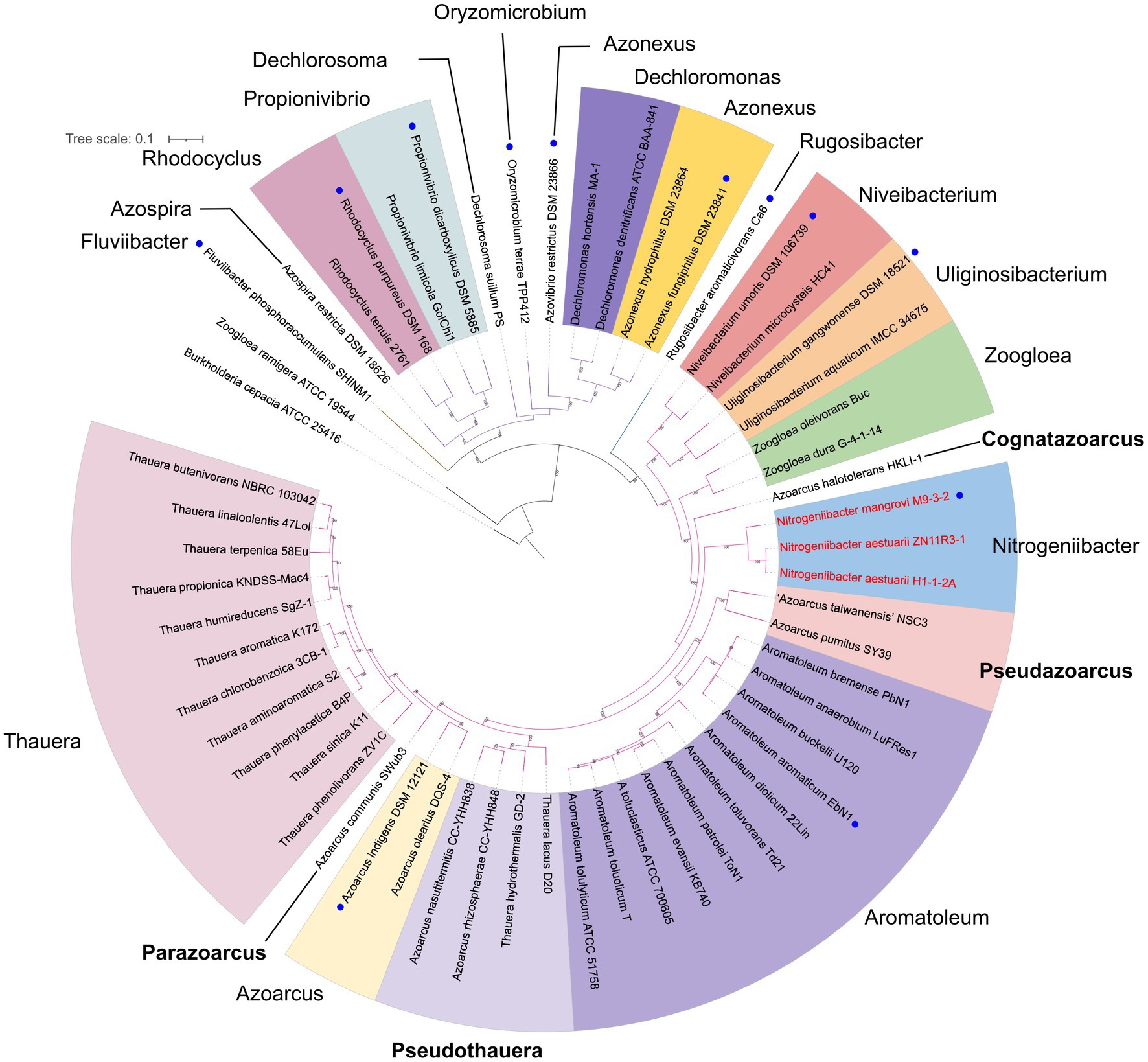
Figure 3. Phylogenomic analysis of the Nitrogeniibacter members and type strains affiliated to the order Rhodocyclales based on 120 bacterial conserved single-copied gene sets of the members. The bootstrap values on the nodes are displayed by >70. Bar, 0.1 represents the nucleotide substitution per position. Blue circles represent type species. Red names show Nitrogeniibacter members. The branch color represents the families, Zoogloeaceae, Azonexaceae, and Rhodocyclaceae of the order Rhodocyclales. Four genera proposed in this study are marked bold.
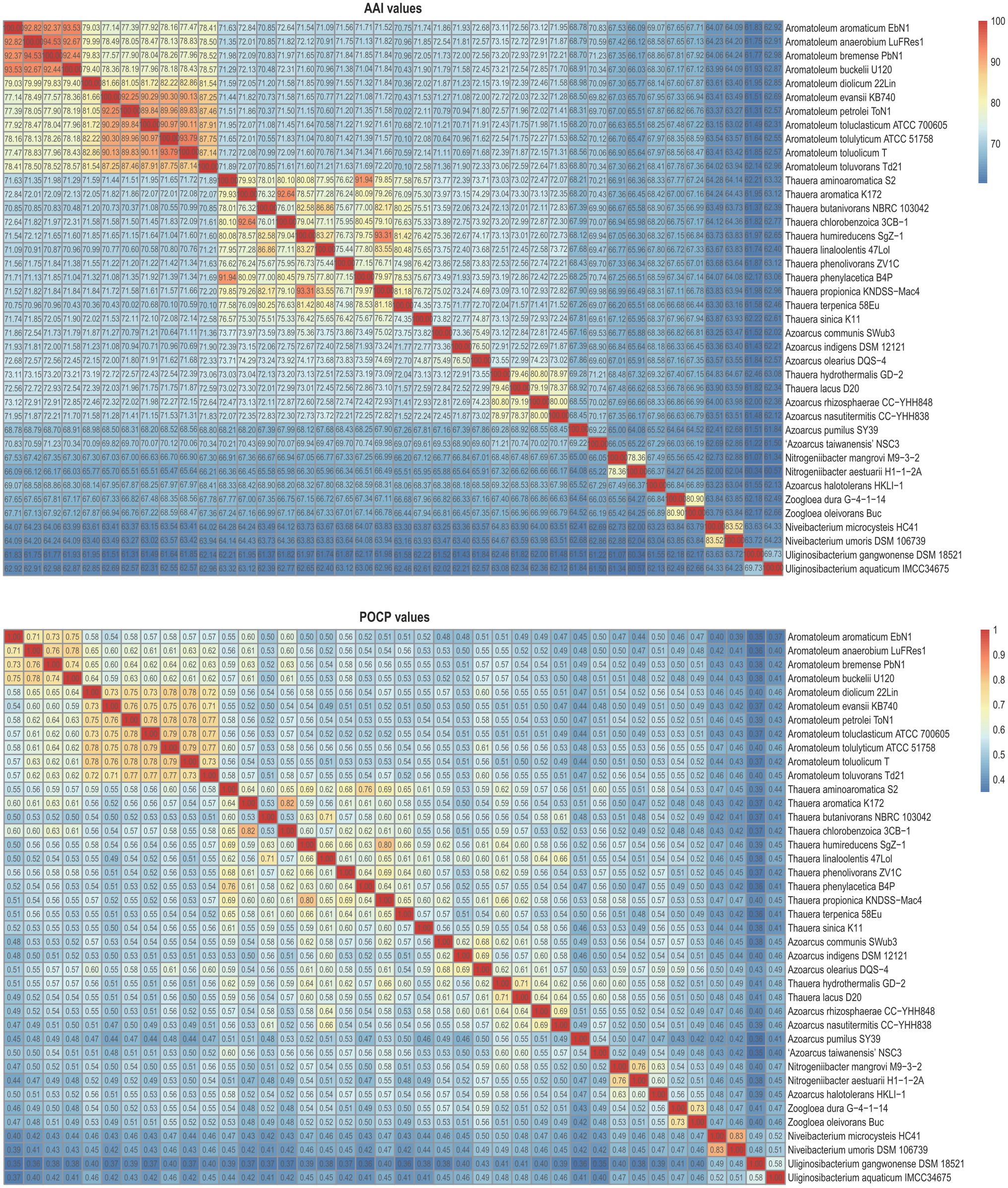
Figure 4. Heatmap showing the AAI values (upper panel) and POCP values (lower panel) among the members of the family Zoogloeaceae.
Phenotypic Properties
Colonies of strains H1-1-2AT and ZN11-R3-1 on MB agar plates at 30°C were round, transparent, convex, and~1mm in diameter. The cells were rod-shaped, motile, and stained Gram-negative. Catalase activity and oxidase activity were found to be positive, similar to N. mangrovi M9-3-2T. The tested strains did not degrade soluble starch, skim milk, carboxymethyl cellulose, and Tweens 40, 60, and 80. Anaerobic growth was not observed for strains H1-1-2AT and ZN11-R3-1. The two strains can grow at 15–40°C with the optimum at 35°C and a pH range of 7.0–8.0 (Table 1). NaCl tolerance was observed at 0–4% (w/v) with the optimum of 0.5%, similar to N. mangrovi M9-3-2T (Table 1). Strains H1-1-2AT and ZN11-R3-1 can grow on nitrogen-free medium, similar to N. mangrovi M9-3-2T. Nitrate cannot be reduced by strains H1-1-2AT and ZN11-R3-1, in contrast to N. mangrovi M9-3-2T which has denitrification ability. Additional biochemical and physiological properties of strains H1-1-2AT and ZN11-R3-1 are listed in the species description.
Chemotaxonomic Properties
The respiratory quinone of strain H1-1-2AT was ubiquinone-8 (Q-8), as in the related N. mangrovi M9-3-2T and other members of family Zoogloeaceae (Liao et al., 2021). The polar lipids consisted of phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG), and phosphatidylglycerol (PG), two unidentified aminophospholipid (APL), one other phospholipid (PL), and one unidentified lipid (L; Supplementary Figure 3). The predominant fatty acids (>5%) of strain H1-1-2AT consisted of summed feature 3 (43.2%), C16:0 (23.0%), C12:0 (9.5%), and C10:0 3-OH (7.6%), similar to strain ZN11-R3-1 (37.9, 26.1, 6.3, and 5.1%, respectively; Table 2). Although the major fatty acids of strain H1-1-2AT and strain ZN11-R3-1 were similar to N. mangrovi M9-3-2T, the presence of minor fatty acids such as C18:0 showed characteristic differences. The major isoprenoid quinone and major fatty acids of Nitrogeniibacter were similar to the closely related genera, Cognatazoarcus, Pseudazoarcus, Pseudothauera, and Parazoarcus, and their polar lipids composition showed somewhat different profiles (Supplementary Table 5).
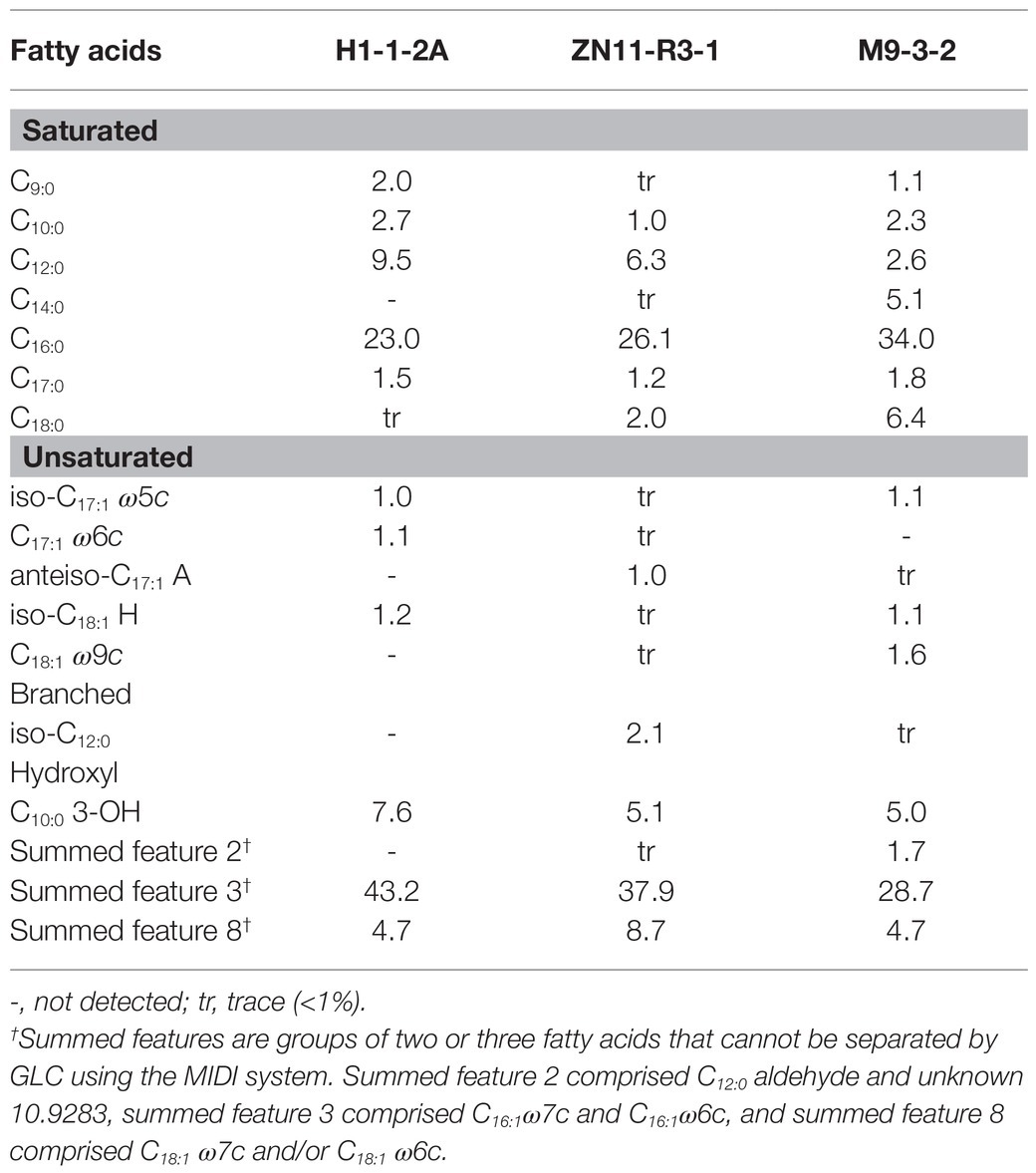
Table 2. Fatty acid profile of strain H1-1-2AT and strain ZN11-R3-1 compared to close relative N. mangrovi M9-3-2T.
Conclusion
Based on the genomic, phylogenetic, phenotypic, and chemotaxonomic characteristics, strains H1-1-2AT and ZN11-R3-1 represent a novel species of the genus Nitrogeniibacter, for which the name Nitrogeniibacter aestuarii sp. nov. is proposed. The type strain is H1-1-2AT (=MCCC 1K04284T=KCTC 82672T); ZN11-R3-1 (=MCCC 1A17971=KCTC 82671) is the second strain of the species. Based on the phylogenetic analysis, four novel genera within the family Zoogloeaceae, Parazoarcus gen. nov., Pseudothauera gen. nov., Pseudazoarcus gen. nov., and Cognatazoarcus gen. nov. were proposed.
Description of Nitrogeniibacter aestuarii sp. nov.
Nitrogeniibacter aestuarii (aes.tu.a’ri.i. L. gen. n. aestuarii, of a coastal wetland, the source of the type strain isolated from wetland cordgrass and mangrove in estuary).
Colonies on MB agar plates cultured for 3days at 30°C are ~1mm, round, transparent, and convex. Cells are Gram stain-negative and rod-shaped. Growth occurs between 15 and 40°C with an optimum at 35°C, at 0–4% NaCl (w/v) with an optimum of 0.5% and a pH range of 7.0–8.0. Catalase-positive and oxidase-positive. Strains can grow on nitrogen-free medium. Nitrate cannot be reduced to nitrite. Positive for alkaline phosphatase, leucine arylamidase; weakly positive for esterase (C4), valine arylamidase, acid phosphatase, and naphtholAS-BI-phosphohydrolase. Hydrolysis of aesculin is weak positive. Malic acid and trisodium citrate can be used as sole carbon sources.
The quinone system is ubiquinone-8. The major fatty acids are summed feature 3 (C16:1ω7c and C16:1ω6c), C16:0, C12:0, and C10:0 3-OH. The major polar lipids include phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG), and phosphatidylglycerol (PG). The genome size is 4.7Mbp with DNA G+C content of 62.7%.
The type strain is H1-1-2AT (=MCCC 1K04284T=KCTC 82672T), isolated from Spartina alterniflora wetland sediment. Another strain is ZN11-R3-1 (=MCCC 1A17971=KCTC 82671), isolated from the enrichment culture inoculated with plastics collected from a wetland mangrove.
The GenBank/EMBL/DDBJ accession numbers of 16S rRNA gene sequence of strains H1-1-2AT and ZN11-R3-1 are MW644766 and MW644767, respectively. The whole-genome sequences of strains H1-1-2AT and ZN11-R3-1 have been deposited at GenBank under the accession numbers CP071321-CP071322 and JAFKAB000000000, respectively.
Emended Description of the Family Zoogloeaceae
In addition to the properties listed in the original description (Boden et al., 2017), the family Zoogloeaceae includes the genera Niveibacterium, Parazoarcus, Pseudothauera, Pseudazoarcus, and Cognatazoarcus. The AAI values among the members range from 60.34 to 94.53%. DNA G+C content is 56.6–68.7%.
Taxonomic Consequences: New Genera
Description of Pseudazoarcus gen. nov.
Pseudazoarcus (Pseud.a.zo.ar΄cus. Gr. masc. adj. pseudes, false; N.L. masc. n. Azoarcus, a bacterial genus name; N.L. masc. n. Pseudazoarcus, false Azoarcus).
The description is as that for Pseudazoarcus pumilus comb. nov., which is the type species. The genus has been separated from Azoarcus based on phylogenetic analyses of 16S rRNA gene and genome sequences. The genomic size is 3.2–4.2Mb. DNA G+C content is 62.8–66.5%.
Description of Pseudothauera gen. nov.
Pseudothauera (Pseu.do.thau΄e.ra. Gr. masc. adj. pseudes, false; N.L. fem. n. Thauera, a bacterial genus name; N.L. fem. n. Pseudothauera, false Thauera).
The description is as that for Pseudothauera hydrothermalis comb. nov., which is the type species. The genus has been separated from Thauera based on phylogenetic analysis of 16S rRNA gene sequence and genome sequences. The genomic size is 3.1Mb-4.7Mb. DNA G+C content is 63.4–68.3%.
Description of Cognatazoarcus gen. nov.
Cognatazoarcus (Cog.nat.a.zo.ar΄cus. L. masc. adj. cognatus, relative, related, kindred; N.L. masc. n. Azoarcus, a bacterial generic name; N.L. masc. n. Cognatazoarcus, related to Azoarcus).
The description is as that for Cognatazoarcus halotolerans comb. nov., which is the type species. The genus has been separated from Azoarcus based on phylogenetic analysis of genome sequences.
Description of Parazoarcus gen. nov.
Parazoarcus (Par.a.zo.ar΄cus. Gr. prep. para beside; N.L. masc. n. Azoarcus, a bacterial genus name; N.L. masc. n. Parazoarcus, beside Azoarcus).
The description is as that for Parazoarcus communis comb. nov., which is the type species. The genus has been separated from Azoarcus based on phylogenetic analysis of genome sequences.
Taxonomic Consequences: New Combinations for Species
Description of Pseudazoarcus pumilus comb. nov.
Pseudazoarcus pumilus (pu΄mi.lus. L. masc. adj. pumilus, small, tiny).
Basonym: Azoarcus pumilus Fu et al. 2019.
The description is as for Azoarcus pumilus (Fu et al., 2019). The type strain is SY39T (=KCTC 62157T=MCCC 1K03430T).
Description of Pseudothauera hydrothermalis comb. nov.
Pseudothauera hydrothermalis (hy.dro.ther.ma΄lis. Gr. neut. n. hydor, water; Gr. masc. adj. thermos, hot; N.L. fem. adj. hydrothermalis, hydrothermal).
Basonym: Thauera hydrothermalis Yang et al. 2018.
The description is as for Thauera hydrothermalis (Yang et al., 2018). The type strain is GD-2T (=NBRC 112472T=CGMCC 1.15527T).
Description of Pseudothauera lacus comb. nov.
Pseudothauera lacus (la’cus. L. gen. n. lacus, of a lake).
Basonym: Thauera lacus Zheng et al. 2019.
The description is as for Thauera lacus (Zheng et al. 2019). The type strain is D20T (=MCCC 1H00305T=KCTC 62586T).
Description of Pseudothauera rhizosphaerae comb. nov.
Pseudothauera rhizosphaerae (rhi.zo.sphae’rae. Gr. fem. n. rhiza, root; Gr. fem. n. sphaira, ball, sphere; N.L. gen. n. rhizosphaerae, from the rhizosphere).
Basonym: Azoarcus rhizosphaerae Lin et al. 2020.
The description is as for Azoarcus rhizosphaerae (Lin et al., 2020). The type strain is CC-YHH848T=BCRC 81060T=JCM 32002T).
Description of Pseudothauera nasutitermitis comb. nov.
Pseudothauera nasutitermitis (na.su.ti.ter’mi.tis. N.L. gen. n. nasutitermitis, of a termite of the genus Nasutitermes).
Basonym: Azoarcus nasutitermitis Lin et al. 2020.
The description is as for Azoarcus nasutitermitis (Lin et al., 2020). The type strain is CC-YHH838T (= BCRC 81059T=JCM 32001T).
Description of Cognatazoarcus halotolerans comb. nov.
Cognatazoarcus halotolerans (ha.lo.to’le.rans. Gr. masc. n. hals, halos, salt; L. pres. part. tolerans, tolerating; N.L. part. adj. halotolerans, salt-tolerating).
Basonym: Azoarcus halotolerans Lin et al. 2020.
The description is as for Azoarcus halotolerans (Li et al., 2020). The type strain is HKLI-1T (= KCTC 72659T=CCTCC AB 2019312T).
Description of Parazoarcus communis comb. nov.
Parazoarcus communis (com.mu’nis. L. masc. adj. communis, usual, common, referring to diverse habitats).
Basonym: Azoarcus communis Reinhold-Hurek et al. 1993.
The description is as for Azoarcus communis (Reinhold-Hurek et al., 1993). The type strain is SWub3T (= ATCC 51397T=DSM 12120T=LMG 9095T).
Significance
A novel species named Nitrogeniibacter aestuarii with two strains affiliated to the family Zoogloeaceae was proposed by using a polyphasic taxonomic approach. The species had the ability of nitrogen fixation, which was assumed to play important roles in the nitrogen cycle of coastal wetlands. Additionally, phylogenetic analysis of the family Zoogloeaceae based on genome sequences of type strains and uncultivated bacteria was performed and four novel genera, Parazoarcus gen. nov., Pseudothauera gen. nov., Pseudazoarcus gen. nov., and Cognatazoarcus gen. nov., were proposed. This study provided new insights into the taxonomy of the family Zoogloeaceae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZH and ZS conceived the study and wrote the manuscript. ZH, RL, FC, and QL conducted the experiments. AO proposed names, wrote and checked etymologies, and edited and corrected the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Marine Microbial Collection Program (2019KJ25) as part of the National Infrastructure of Microbial Resources of China (NIMR 2021-9) and Scientific Research Foundation of Third Institute of Oceanography, Ministry of Natural Resources (2019021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.755908/full#supplementary-material
Supplementary Figure 1 | Electrophoresis pattern of BOX-PCR fingerprinting of the strains. M. DL 2000 DNA marker; 1, strain M9-3-2T; 2, strain H1-1-2AT; 3, strain ZN11-R3-1.
Supplementary Figure 2 | Phylogeny of 16S rRNA gene sequences. The tree was constructed using the maximum likelihood method. Bootstrapping was carried out with 1,000 replicates. Branch node values below 70% are not shown. Burkholderia cepacia ATCC 25416T (AXBO01000009) was selected as the outgroup. Bar, 0.05 represents the nucleotide substitutions per position.
Supplementary Figure 3 | Polar lipid profile of strain H1-1-2AT. PE, phosphatidylethanolamine; DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; APL, aminophospholipid; PL, unidentified phospholipid; L, unidentified lipid.
Supplementary Table 1 | Nitrogen fixation genes predicted from the genome of strain H1-1-2AT.
Supplementary Table 2 | Denitrification genes predicted from the genome of strain M9-3-2T.
Supplementary Table 3 | Sulfur oxidation genes found in the genome of strain H1-1-2AT.
Supplementary Table 4 | Genomic features of the members affiliated to the family Zoogloeaceae.
Footnotes
1. ^https://lpsn.dsmz.de/family/zoogloeaceae
2. ^https://www.ezbiocloud.net/taxonomy?tn=Zoogloeaceae
3. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ^https://github.com/najoshi/sickle
5. ^https://www.genome.jp/tools/kaas/
7. ^https://github.com/dparks1134/CompareM
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Boden, R., Hutt, L. P., and Rae, A. W. (2017). Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the 'Proteobacteria', and four new families within the orders Nitrosomonadales and Rhodocyclales. Int. J. Syst. Evol. Microbiol. 67, 1191–1205. doi: 10.1099/ijsem.0.001927
Bowers, R. M., Kyrpides, N. C., Stepanauskas, R., Harmon-Smith, M., Doud, D., Reddy, T. B. K., et al. (2017). Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731. doi: 10.1038/nbt.3893
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chun, J., Young Kang, J., Cheol Jung, Y., and Yeop Jahng, K. (2016). Niveibacterium umoris gen. nov., sp. nov., isolated from wetland freshwater. Int. J. Syst. Evol. Microbiol. 66, 997–1002. doi: 10.1099/ijsem.0.000826
Fu, G. Y., Yu, X. Y., Yu, X. D., Zhao, Z., Chen, C., Wang, R. J., et al. (2019). Azoarcus pumilus sp. nov., isolated from seawater in Sanya, China. Int. J. Syst. Evol. Microbiol. 69, 1459–1464. doi: 10.1099/ijsem.0.003341
Gittel, A., Kofoed, M. V., Sørensen, K. B., Ingvorsen, K., and Schramm, A. (2012). Succession of Deferribacteres and Epsilonproteobacteria through a nitrate-treated high-temperature oil production facility. Syst. Appl. Microbiol. 35, 165–174. doi: 10.1016/j.syapm.2012.01.003
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Huang, Z., Dong, L., Lai, Q., and Liu, J. (2020a). Spartinivicinus ruber gen. nov., sp. nov., a novel marine gammaproteobacterium producing heptylprodigiosin and cycloheptylprodigiosin as major red pigments. Front. Microbiol. 11:2056. doi: 10.3389/fmicb.2020.02056
Huang, Z., Du, Y., Lai, Q., and Shao, Z. (2019). Nonlabens xiamenensis sp. nov., isolated from coastal seawater. Antonie van Leeuwenhoek 112, 1263–1271. doi: 10.1007/s10482-019-01258-8
Huang, Z., Hu, Y., Lai, Q., and Guo, Y. (2020b). Description of Maribellus sediminis sp. nov., a marine nitrogen-fixing bacterium isolated from sediment of cordgrass and mangrove. Syst. Appl. Microbiol. 43:126099. doi: 10.1016/j.syapm.2020.126099
Huang, X. F., Liu, Y. J., Dong, J. D., Qu, L. Y., Zhang, Y. Y., Wang, F. Z., et al. (2014). Mangrovibacterium diazotrophicum gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from a mangrove sediment, and proposal of Prolixibacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 64, 875–881. doi: 10.1099/ijs.0.052779-0
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Konstantinidis, K. T., Rosselló-Móra, R., and Amann, R. (2017). Uncultivated microbes in need of their own taxonomy. ISME J. 11, 2399–2406. doi: 10.1038/ismej.2017.113
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lapidus, A. L., and Korobeynikov, A. I. (2021). Metagenomic data assembly - the way of decoding unknown microorganisms. Front. Microbiol. 12:613791. doi: 10.3389/fmicb.2021.613791
Lee, D. J., Wong, B. T., and Adav, S. S. (2014). Azoarcus taiwanensis sp. nov., a denitrifying species isolated from a hot spring. Appl. Microbiol. Biotechnol. 98, 1301–1307. doi: 10.1007/s00253-013-4976-9
Letunic, I., and Bork, P. (2007). Interactive tree Of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Li, S., Zhao, L., Han, J., Liu, S., Dai, J., Fu, G., et al. (2020). Azoarcus halotolerans sp. nov., a novel member of Rhodocyclaceae isolated from activated sludge collected in Hong Kong. Int. J. Syst. Evol. Microbiol. 70, 5799–5805. doi: 10.1099/ijsem.0.004476
Liao, H., Qu, M., Hou, X., Lin, X., Li, H., Duan, C. S., et al. (2021). Nitrogeniibacter mangrovi gen. nov., sp. nov., a novel anaerobic and aerobic denitrifying betaproteobacterium and reclassification of Azoarcus pumilus as Aromatoleum pumilum comb. nov. Int. J. Syst. Evol. Microbiol. 71:004946. doi: 10.1099/ijsem.0.004946
Lin, S. Y., Hameed, A., Tsai, C. F., Huang, G. H., Hsu, Y. H., and Young, C. C. (2020). Description of Azoarcus nasutitermitis sp. nov. and Azoarcus rhizosphaerae sp. nov., two nitrogen-fixing species isolated from termite nest and rhizosphere of Ficus religiosa. Antonie van Leeuwenhoek 113, 933–946. doi: 10.1007/s10482-020-01401-w
Liu, B., Mao, Y., Bergaust, L., Bakken, L. R., and Frostegard, A. (2013). Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ. Microbiol. 15, 2816–2828. doi: 10.1111/1462-2920.12142
Luo, C., Rodriguez, R. L., and Konstantinidis, K. T. (2014). MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42:e73. doi: 10.1093/nar/gku169
Macy, J. M., Rech, S., Auling, G., Dorsch, M., Stackebrandt, E., and Sly, L. I. (1993). Thauera selenatis gen. nov., sp. nov., a member of the beta subclass of Proteobacteria with a novel type of anaerobic respiration. Int. J. Syst. Bacteriol. 43, 135–142. doi: 10.1099/00207713-43-1-135
Mechichi, T., Stackebrandt, E., Gad'on, N., and Fuchs, G. (2002). Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 178, 26–35. doi: 10.1007/s00203-002-0422-6
Orata, F. D., Meier-Kolthoff, J. P., Sauvageau, D., and Stein, L. Y. (2018). Phylogenomic analysis of the gammaproteobacterial methanotrophs (order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 9:3162. doi: 10.3389/fmicb.2018.03162
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Rabus, R., Wohlbrand, L., Thies, D., Meyer, M., Reinhold-Hurek, B., and Kämpfer, P. (2019). Aromatoleum gen. nov., a novel genus accommodating the phylogenetic lineage including Azoarcus evansii and related species, and proposal of Aromatoleum aromaticum sp. nov., Aromatoleum petrolei sp. nov., Aromatoleum bremense sp. nov., Aromatoleum toluolicum sp. nov. and Aromatoleum diolicum sp. nov. Int. J. Syst. Evol. Microbiol. 69, 982–997. doi: 10.1099/ijsem.0.003244
Rai, A., Jagadeeshwari, U., Deepshikha, G., Smita, N., Sasikala, C., and Ramana, C. V. (2021). Phylotaxogenomics for the reappraisal of the genus Roseomonas with the creation of six new genera. Front. Microbiol. 12:677842. doi: 10.3389/fmicb.2021.677842
Reinhold-Hurek, B., Hurek, T., Gillis, M., Hoste, B., Vancanneyt, M., Kersters, K., et al. (1993). Azoarcus gen. nov., nitrogen-fixing Proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth), and description of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int. J. Syst. Bacteriol. 43, 574–584. doi: 10.1099/00207713-43-3-574
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437. doi: 10.1038/nature12352
Shin, Y. K., Hiraishi, A., and Sugiyama, J. (1993). Molecular systematics of the genus Zoogloea and emendation of the genus. Int. J. Syst. Bacteriol. 43, 826–831. doi: 10.1099/00207713-43-4-826
Suresh, G., Lodha, T. D., Indu, B., Sasikala, C., and Ramana, C. V. (2019). Taxogenomics resolves conflict in the genus Rhodobacter: a two and half decades pending thought to reclassify the genus Rhodobacter. Front. Microbiol. 10:2480. doi: 10.3389/fmicb.2019.02480
Wang, Z., Li, W., Li, H., Zheng, W., and Guo, F. (2020). Phylogenomics of Rhodocyclales and its distribution in wastewater treatment systems. Sci. Rep. 10:3883. doi: 10.1038/s41598-020-60723-x
Weon, H. Y., Kim, B. Y., Yoo, S. H., Kwon, S. W., Go, S. J., and Stackebrandt, E. (2008). Uliginosibacterium gangwonense gen. nov., sp. nov., isolated from a wetland, Yongneup, in Korea. Int. J. Syst. Evol. Microbiol. 58, 131–135. doi: 10.1099/ijs.0.64567-0
Yang, L., Muhadesi, J. B., Wang, M. M., Wang, B. J., Liu, S. J., and Jiang, C. Y. (2018). Thauera hydrothermalis sp. nov., a thermophilic bacterium isolated from hot spring. Int. J. Syst. Evol. Microbiol. 68, 3163–3168. doi: 10.1099/ijsem.0.002960
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017a). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017b). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Keywords: Nitrogeniibacter, Zoogloeaceae, nitrogen fixation, polyphasic taxonomy, phylogenomic tree
Citation: Huang Z, Liu R, Chen F, Lai Q, Oren A and Shao Z (2021) Nitrogeniibacter aestuarii sp. nov., a Novel Nitrogen-Fixing Bacterium Affiliated to the Family Zoogloeaceae and Phylogeny of the Family Zoogloeaceae Revisited. Front. Microbiol. 12:755908. doi: 10.3389/fmicb.2021.755908
Edited by:
Frank T. Robb, University of Maryland, Baltimore, United StatesReviewed by:
Jeremy Dodsworth, California State University, San Bernardino, United StatesG. Suresh, University of Hyderabad, India
Weiyan Zhang, Ningbo University, China
Copyright © 2021 Huang, Liu, Chen, Lai, Oren and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaobin Huang, emJodWFuZ2VtYWlsQGdtYWlsLmNvbQ==; Zongze Shao, c2hhb3p6QDE2My5jb20=
 Zhaobin Huang
Zhaobin Huang Renju Liu3
Renju Liu3 Qiliang Lai
Qiliang Lai Aharon Oren
Aharon Oren Zongze Shao
Zongze Shao