
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 October 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.755233
This article is part of the Research Topic Zoonotic Microorganisms and Spread of Acquired Polymyxin Resistance Determinants View all 15 articles
 Yan Zhang1†
Yan Zhang1† Xu Kuang1†
Xu Kuang1† Juan Liu1
Juan Liu1 Ruan-Yang Sun1
Ruan-Yang Sun1 Xing-Ping Li2
Xing-Ping Li2 Jian Sun1,3,4
Jian Sun1,3,4 Xiao-Ping Liao1,3,4
Xiao-Ping Liao1,3,4 Ya-Hong Liu1,3,4
Ya-Hong Liu1,3,4 Yang Yu1,3,4*
Yang Yu1,3,4*We determined the prevalence and transmission characteristics of mcr-1-positive Escherichia coli (MCRPEC) isolates from migratory birds Anser indicus in Guangdong, China. We identified 22 MCRPEC from 303 A. indicus fecal samples (7.3%) in Guangzhou, Zhaoqing, and Futian. The mcr-1 gene coexisted with 24 other types of antibiotic resistance genes (ARG), and 11 ARGs were highly prevalent at levels >50%. The MCRPEC displayed a diversity of sequence types (ST), and 19 distinct STs were identified with ST10, ST1146, and ST1147 as the most prevalent. In addition, these MCRPEC from birds were closely related phylogenetically to those from other sources in China. Whole-genome sequencing analysis demonstrated that mcr-1 was located on IncX4 (n=9, 40.9%), IncI2 (n=5, 22.7%) and IncP (n=1, 4.5%) plasmids and the latter shared an identical plasmid backbone with other sources. These results highlight the significance of migratory birds in the transmission of antibiotic resistance and provide powerful evidence that migratory birds are potential transmitters of antibiotic resistance.
Colistin is a polymyxin antibiotic that has been used in veterinary clinics for many years. It is now applied as a last-resort drug for treatment of carbapenem-resistant Enterobacterial infections (Mendes et al., 2018). The emergence of the plasmid-mediated mobile colistin resistance gene mcr-1 in China and its rapid global spread now poses a serious threat to public health (Liu et al., 2016; Wang et al., 2018). Significantly, these mcr-1 plasmids often harbor other antibiotic resistance genes (ARG) that encode carbapenemase and extended-spectrum β-lactamase (ESBL) resistance (Haenni et al., 2016; Yao et al., 2016). Therefore, the global distribution and spread of mcr-1 gene is concerning.
The World Health Organization (WHO) has developed a global strategy to address antimicrobial resistance based on the “One Health” approach that is based on the close links between human, animal, and environmental health (Dafale et al., 2020). Wildlife can act as ARG reservoirs and transmission vectors between microorganisms and the environment (Allen et al., 2010; Arnold et al., 2016). Therefore, a contaminated environment also plays a key role in the spread of antibiotic resistance since this is where bacteria from different sources are able to exchange ARGs (Aeksiri et al., 2019). In particular, birds play a large part in ARG dissemination due to their environmental exposure through ingested food or polluted water (Jarma et al., 2021). This is especially true for migratory birds that can act as ARG reservoirs and spread antibiotic resistance through migration (Vergara et al., 2017; Elsohaby et al., 2021). For instance, the mcr-1 gene has been isolated from European herring gulls (Ruzauskas and Vaskeviciute, 2016), from Spanish and Portuguese gulls (Ahlstrom et al., 2021) as well as in Thailand and China in migratory birds (Aeksiri et al., 2019; Lake, 2020).
Since colistin is considered the last-resort antibiotic used to treat multidrug-resistant bacteria (Li et al., 2020), the emergence of the mcr-1 gene in migratory birds is especially worrisome. Monitoring the level of antibiotic resistance carried in migratory birds is a necessary step to prevent ARG spread. This study investigated the prevalence and genomic structures of mcr-1 producing Escherichia coli (MCRPEC) isolated from migratory birds Anser indicus in Guangdong, China.
The Institutional Review Board of South China Agricultural University (SCAU-IRB) approved the Samples and bacteria protocols. All A. indicus feces were sampled under authorization from Animal Research Committees of South China Agricultural University (SCAU-IACUC).
A total of 303 feces samples from A. indicus were collected in 2017 in Guangzhou, Zhaoqing, and Futian in Guangdong province. Briefly, all samples were placed to 1.0ml of LB Broth and incubated for 16–18h at 37°C followed by inoculation on MacConkey agar plates containing 2.0mg/L colistin that were incubated for 16h. Two or three different forms red colonies were selected for identification using the MALDI-TOF 80 MS Axima system (Shimadzu-Biotech Corp., Kyoto, Japan) and 16S rRNA sequencing. The colistin-resistant isolates were screened for the presence of the mcr-1 gene by PCR as previously described (Yang et al., 2019a).
The antimicrobial sensitivity of 22 MCRPE for 15 antibiotics was examined by measuring the minimal inhibitory concentration (MIC) using the agar dilution method and interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI M100-S28). Susceptibility to colistin was performed by broth micro-dilution as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST 92 Version 6.0; EUCAST, 2016). Quality control of the procedure was conducted by using the susceptible E. coli ATCC 25922.
Total genomic DNA was extracted from 22 MCRPEC isolates using a Genomic DNA Purification Kit (Tiangen, Beijing, China) as per the manufacturer’s instructions. WGS was performed with the Illumina Hiseq 2500 System (Novogene Guangzhou, China) using the paired-end 2×150-bp sequencing protocol. Raw sequence reads were trimmed using Trim Galore, and the genomes were de novo-assembled into contigs using SPAdes (3.9.0) predefined kmers set. All genome assemblies of 22 E. coli isolates were deposited in GenBank and are registered with BioProject number PRJNA748548.
The CGE platform1 was used for analyses of multilocus sequence typing (MLST-2.0), acquired resistance genes (ResFinder 4.1, all antibiotic resistance databases were selected with a cut-off value of 95% identity and 80% minimum coverage) and plasmid incompatibility groups (PlasmidFinder-2.1 version, using Enterobacteriaceae database with parameters of minimum 95% identity and 85% query coverage). Sequence comparisons between mcr-1 carrying plasmids were performed using Easyfig and Brig (Alikhan et al., 2011). Phylogenetic trees for MCRPEC isolates in this study were constructed using homologous strains from the NCBI database using CSI Phylogeny (v1.4), and E. coli (U4) was used as the reference genome (Kaas et al., 2014). The corresponding characteristics of each isolate visualized using FigTree v1.4.4 and the online tool iTOL.2 Single polynucleotide pairs (SNP) analysis was visualized as a heatmap of the SNP count matrix using BacWGSTdb 2.0 (Feng et al., 2021). The sequences of mcr-1 plasmids were constructed by de novo assembly using Genious (10.0.7). Circular comparisons among mcr-1-related IncX4 and IncI2 plasmids from this study and NCBI database were performed using BLAST Ring Image Generator (BRIG v0.9555).
We recovered 22 (7.3%) MCRPE from 303 A. indicus fecal samples in Guangdong Province, China, that included 10% from Futian (12/120), 5.7% from Zhaoqing (2/35) and 5.4% from Guangzhou (8/148). The MIC values for colistin for these isolates were all ≥4μg/ml. These isolates also possessed high levels of resistance to other common antibiotics including sulfamethoxazole/trimethoprim (95.4%), fosfomycin (81.8%), florfenicol (81.8%), tetracycline (77.2%), ciprofloxacin (34.1%), and olaquindox (34.1%). Smaller numbers of isolates displayed resistance to nalidixic acid (27.3%), cefotaxime (22.7%), streptomycin (18.1%), cefoxitin (13.6%), gentamicin (4.5%), and amikacin (4.5%). Notably, all strains remained sensitive to meropenem and tigecycline (Table 1; Supplementary Figure S2).
We sequenced 22 of MCRPE isolates using the Illumina HiSeq platform, and the average genome size was 4.9Mbp. Interestingly, we identified 19 distinct STs that included ST10 (2/22), ST1146 (2/22), and ST1147 (2/22) and the remaining were single STs that included ST198, ST2351, ST4014, ST58, ST5878, ST155, ST746, ST6725, ST101, ST2280, ST46, ST5229, ST9401, ST162, and ST1258. The strain FT109-1 was a member of a previously unreported new ST (Figure 1).
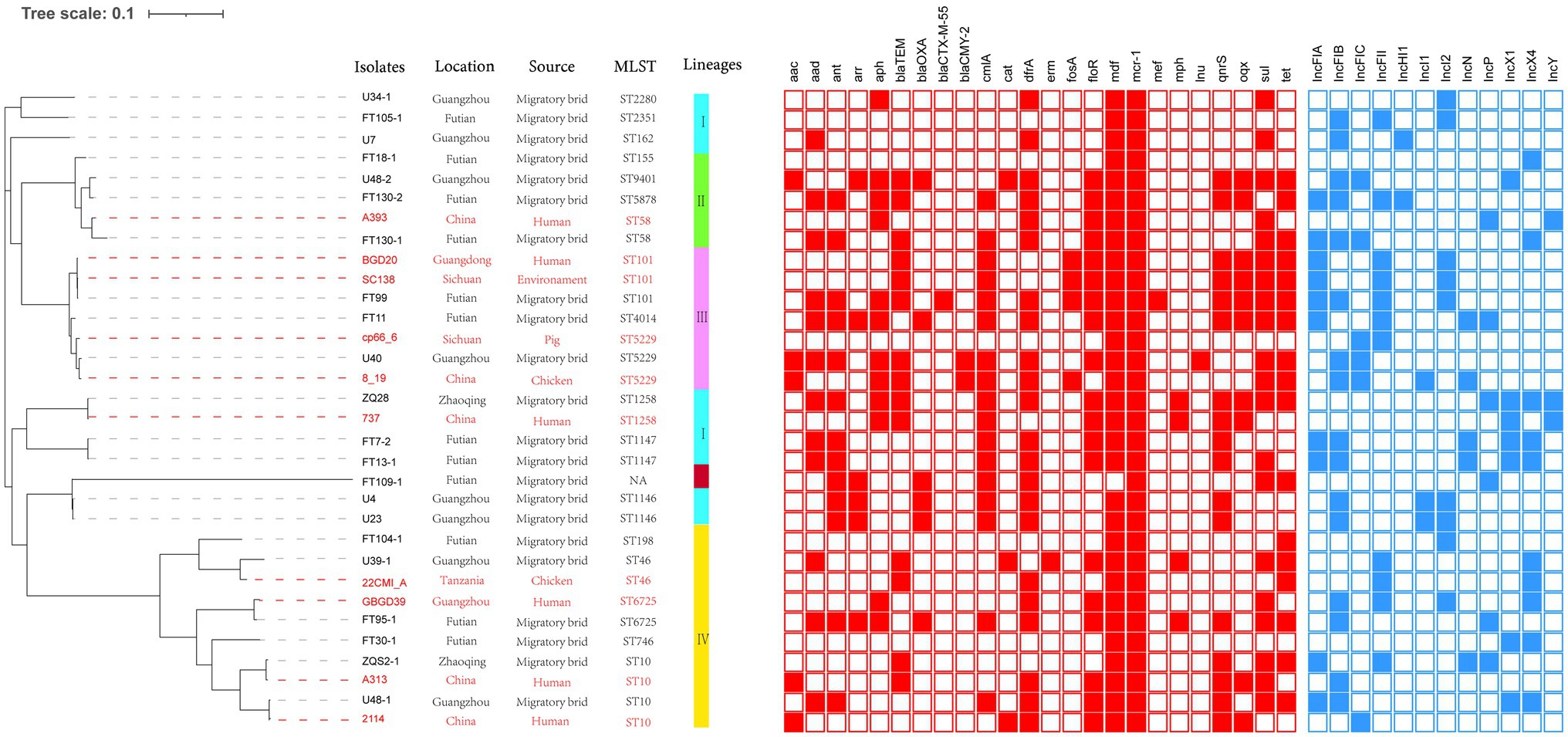
Figure 1. Analysis of MCRPE isolates used in this study. Left Panel: Isolates from migratory birds in Guangdong, China (black font) and from the NCBI database (red font). Right Panel: Genetic relationships among 32 mcr-1 positive Escherichia coli (MCRPEC) isolates using a maximum likelihood tree. Red-filled squares indicate possession of the indicated antibiotic resistance genes (ARG) and blue-filled squares indicated plasmid Inc type.
We reconstructed a phylogenetic tree using the 22 MCRPE isolates along with 10 MCRPE isolates from the NCBI database. All isolates were classified into four clades lineage except FT109-1 (see above). The major lineage IV included 10 (31%) isolates, and ST10 members accounted for 40% of this lineage. In addition, we also found close phylogenetic relationships between our 22 MCRPE from A. indicus and other sources in China that included humans, pigs, chickens, and environmental samples.
To further assess the relationships between these isolates, we performed a SNP analysis that indicated most of our MCRPE isolates had significant SNP differences. Notably, in two cases, there were two isolates possessing a collection of different genomic characteristics that were recovered from the same sample (FT130 and U48; Table 1). For instance, both ST10 (U48-1) and ST9401 (U48-2) were recovered from sample 18FS1 and shared 42,580 SNPs (Supplementary Figure S1).
Additionally, our MCRPE group represented 24 ARG types that conferred resistance to 14 classes of antibiotics including colistin, fosfomycin, streptomycin, nalidixic acid, ciprofloxacin, florfenicol, cefotaxime, cefoxitin, tetracyclines, gentamicin, amikacin, olaquindox, and sulfamethoxazole/trimethoprim. Among these, 11 ARGs were highly prevalent with detection rates >50% and included aad, aph, cmlA, dfrA, floR, mdf, mcr-1, qnrs, oqx, sul, and tet (Table 1; Figure 1).
Whole-genome sequencing analysis indicated that 12 different Inc types were present on plasmids in the 22 MCRPEC isolates (Figure 1). In 15/22 isolates, we could identify the replicon sequence type of mcr-1-harboring plasmids and included IncX4 (n=9, 40.9%), IncI2 (n=5, 22.7%) and IncP (n=1, 4.5%). Alignment of mcr-1-carrying IncX4 and IncI2 plasmids demonstrated that all mcr-1-carrying plasmids shared identical plasmid backbones. More importantly, the backbones of these mcr-1-linked plasmids from A. indicus were highly similar to those recovered from other sources including diverse animals, humans, and poultry. Plasmids used for comparison include the following: human plasmids: pMCR_WCHEC1604 (KY829117), pHNSHP45 (KP347127.1), pGZ49260 (MG210937.1) and animals: pNFGDF93 (MF978388.1), pLWY24J (MN689940.1), pNFGDF49 (MF978387.1), and pHNSHP49 (MF774188.1). Interestingly, the insertion element ISApl1 was not present in our mcr-1-associated IncI2 plasmids and IS26 was ether partially or totally absent from the mcr-1-carrying IncX4 plasmids (Figure 2).
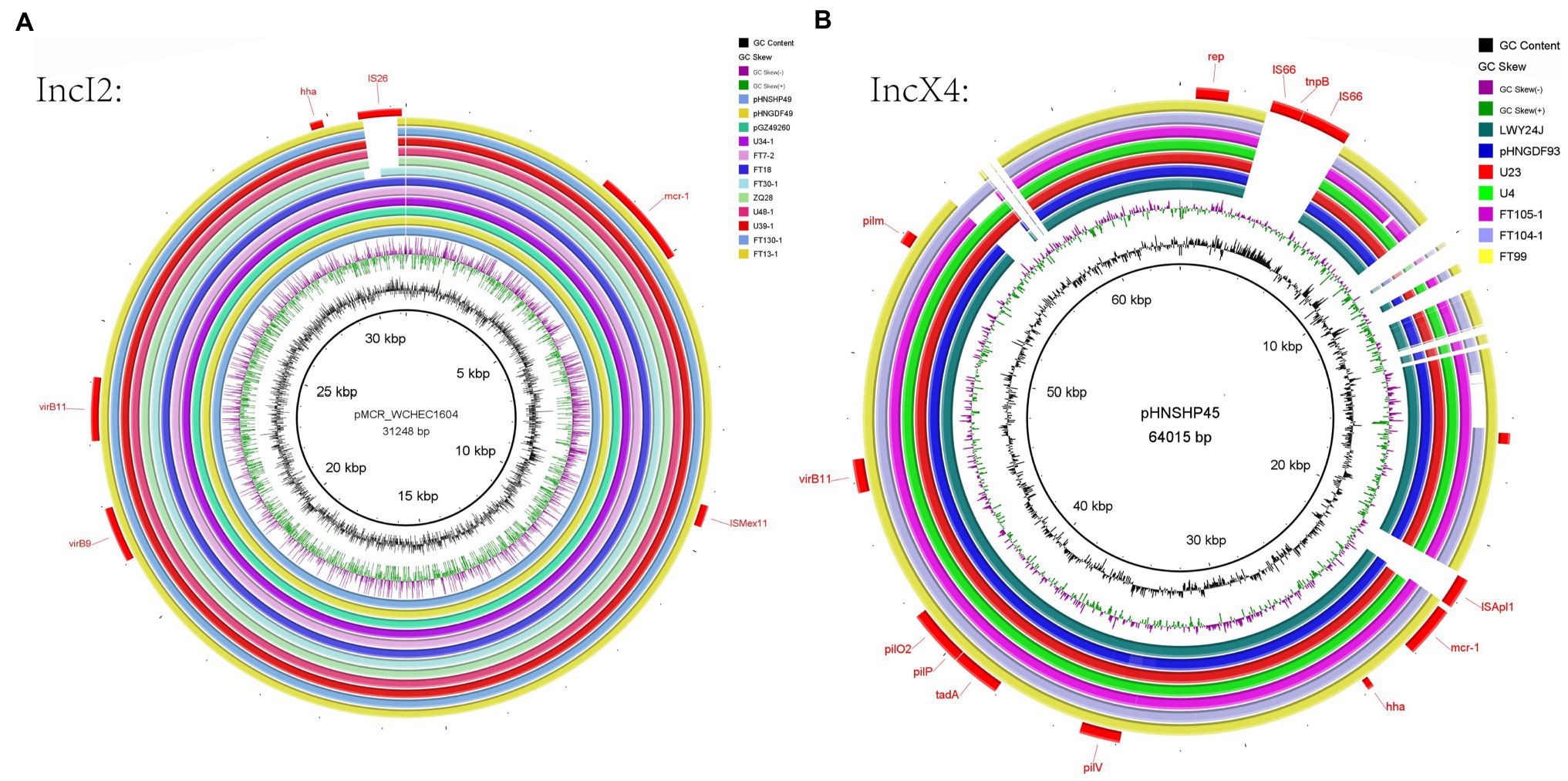
Figure 2. IncI2 plasmid analysis for 21 MCRPEC isolates from migratory birds (A). Alignments of IncI2 plasmids from this study with reference plasmid pHNSHP45. (B) Alignment of IncX4 plasmids with reference plasmid pMCR_WCHEC1604. The sources of these plasmids were migratory birds (FT7-2, FT18, FT13-1, FT30-1, FT99, FT104-1, FT105-1, FT130-1, U4, U23, U34-1, U39-1, U48-1, and ZQ28), human plasmids (pMCR_WCHEC1604, pHNSHP45, and pGZ49260) and animals (pNFGDF93, pLWY24J, pNFGDF49, and pHNSHP49).
Additionally, we also found the presence of broad-host-range IncP plasmids that acted as mcr-1 vectors in this study. The mcr-1-carrying IncP plasmids shared conserved backbones with the plasmids pMCR_1622 and pZR12 taken from E.coli human and pig isolates, respectively (Figure 3). Mcr-1-PAP2 and ISApl1-mcr-1-PAP2 were inserted within the same locus in pMCR_1622 and pZR12 and suggested that this locus is a hot spot for the insertion of ISApl1 (see above).
The plasmid-mediated colistin resistance gene mcr-1 was originally identified in E. coli and has since been found in many countries on all continents (Liu et al., 2016; Matamoros et al., 2017). The speed and extent of the spread of the mcr-1 gene are worrying especially since migratory birds carry foreign ARGs and act as a major source of bacterial antibiotic resistance in local environments (Lake, 2020). Long-distance migratory birds are considered key agents in the spread of antibiotic resistance (Franklin et al., 2020). For antibiotic-resistant bacteria, aquatic systems are major traffic routes between wildlife and humans (Taylor et al., 2011). The resistance genes acquired by birds can be re-introduced in the environment, possibly into different water systems (e.g., by migrating ducks) and might re-infect humans directly via contact with contaminated water or indirectly by the introduction of these genes into the food chain (Marcelino et al., 2019), and long-distance migration of migratory birds may accelerate this spread, which poses a great threat to human safety (Chen et al., 2017; Hadjadj et al., 2017; Monte et al., 2017; Mohsin et al., 2019). Therefore, it is very necessary to monitor the flora of migratory birds. In our study, we isolated 22 MCRPEC isolates in fecal samples from A. indicus in Guangdong, China. This is the first report of the presence of MCRPEC from migratory birds in Guangdong, and our study provides strong evidence that migratory birds are potential transmitters of the colistin resistance gene mcr-1.
Whole-genome sequencing analysis of these mcr-1 positive E. coli strains identified 19 different STs and was consistent with previous reports documenting the ST diversity of MCRPEC (Wang et al., 2018). However, ST10, ST1146, and ST1147 were more prevalent in our isolates and ST10 is frequently found in clinical (Lin et al., 2020), animal (García-Meniño et al., 2018), and environmental samples (Flament-Simon et al., 2020). In addition, we found that the genomes of two MCRPE isolates from the same sample differed by the phylogenetic analysis. This scenario is worrying and indicated that a significant diversity was present in our population of MCRPE isolates from migratory birds. More notably, the emergence and spread of mcr-1 gene in humans, animals, and the environment have been reported globally. In our study, the close phylogenetic relationships for 22 of the MCRPEC isolates from A. indicus and other sources in China indicated the existence of clonal transmission of mcr-1 in migratory bird populations, and the latter are likely responsible for the widespread occurrence of mcr-1. This indicated that migratory birds played an important role in the transmission of the mcr-1 gene.
In addition to mcr-1, we also identified numerous ARGs in MCRPEC isolates such as the plasmid-mediated quinolone resistance gene qnrS1, the tetracycline resistance gene tet, and ESBL-dependent bla CTX-M-55 genes. Many previous studies have reported high levels of ARGs in the intestinal flora of migratory birds (Jarma et al., 2021) and thereby indicated that antibiotic resistance can easily be disseminated via migration (Elsohaby et al., 2021). According to a survey, mcr-1 resistance gene is widespread among different birds, accounting for 50% of the total samples (Cao et al., 2020). Notably, silent transmission of mcr-1 gene was observed, and MCRPE may be difficult to detect whether the mcr-1 gene is only tested for in colistin-resistant isolates (Moura et al., 2016). As the reservoir of resistance genes, the resistance level of the strains carried by migratory birds should continue to be concerned.
Whole-genome sequencing analysis of our isolates further revealed the presence of different mcr-1 harboring plasmids, and the IncX4 and IncI2 types were the most prevalent. The IncX4 and IncI2 type plasmids have a high transferability, and this creates favorable conditions for horizontal transmission of mcr-1 (Yang et al., 2019b). IncX4-type plasmids represent promiscuous plasmids contributing to the intercontinental spread of the mcr-1 gene (Fernandes et al., 2016), which obtained from different bacterial species, belonging to different STs, isolated in different clinical contexts, including wildlife, and found on different continents are highly similar in the plasmid backbone sequences. The ubiquity and transferability of IncX4 plasmids carrying mcr-1 sheds light on the role of this incompatibility group in the global spread of colistin resistance (Sellera et al., 2017). In our study, nine IncX4 plasmids and five IncI2 plasmids carrying mcr-1 in our study shared the same plasmid skeletons as isolates acquired from humans, animals, and the environment as reported elsewhere (Figure 3). This indicated that these plasmid types have been diffusing between humans, animals, and the environment (Matamoros et al., 2017), IncX4 and IncI2 plasmids facilitated the transmission of the mcr-1 gene. The additional presence of an mcr-1-pap2 cassette (Yang et al., 2017) can also function as an independent mobile genetic that could facilitate ARG acquisition and mobilization between bacteria by a mechanism known as “copy-paste and paste” (Partridge et al., 2018). For instance, one or two copies of ISApl1 that flank the mcr-1-pap2 cassette can actively capture and mobilize mcr-1 genes.
Together our data provide evidence for the horizontal transfer of mcr-1 due to migratory bird long-distance migrations. ISApl1 is a common mobile genetic element found in mcr-1 plasmids (Snesrud et al., 2016). Interestingly, ISApl1 and other mobile elements were not present in the mcr-1-related IncX4 and IncI2 plasmid backgrounds in our study isolates. The reason for this was most likely that ISApl1 was lost during subsequent recombination steps following the initial mobilization of mcr-1 (Snesrud et al., 2016). In addition, in this study, mcr-1 was also carried by the broad-host-range IncP type plasmid and shared conserved backbones with E. coli plasmids pMCR_1622 and pZR12 from human and pig samples, respectively. Moreover, IncP plasmids have been reported to mediate the transmission of mcr-1 across various hosts and has the potential to become a dominant mcr-1 carrier (Zhao et al., 2017; Fish, 2018). The emergence of a large group of host plasmids carrying mcr-1 in long-haul migratory birds is of great concern for public health and further suggests that the drug-resistant flora in migratory birds should be monitored.
In conclusion, we identified 22 mcr-1-positive E. coli isolates from migratory birds A. indicus in Guangdong, China. Notably, this is the first study to report the development of diversity in the population of MCRPE isolates from migratory birds in Guangdong. Phylogenetic analysis proved that the development of diversity in the population of MCRPE isolates from migratory birds. WGS analysis further determined that mcr-1 coexisted with other ARGs and also demonstrated diversity in the plasmid population, and the latter provides important epidemiological information for the global dissemination of the mcr-1 gene. These plasmids can serve as vectors for rapid spread of colistin resistance among different hosts over long distances due to bird migration. The antibiotic-resistant flora in migratory birds should therefore be under constant surveillance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA748548.
YZ and X-PLi: sampling. YZ and R-YS: data analysis. YZ: conceptualization, methodology, and writing-first draft. XK and JL: investigation and writing – review and editing. YY, JS, X-PLa, and Y-HL: conceptualization, writing – review and editing, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
This study was jointly supported by the National Natural Science Foundation of China (31730097); the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2019BT02N054); the Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (Grant No. IRT13063); and the innovation Team Project of Guangdong University (2019KCXTD001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.755233/full#supplementary-material
Supplementary Figure S1 | Single polynucleotide pair (SNP)-heatmap analysis of MCRPE isolates used in this study using SNP count matrices.
Supplementary Figure S2 | The antimicrobial sensitivity of 22 MCRPE for 15 antibiotics, yellow indicates that the strain is not sensitive to the drug (CS, colistin; FOS, Fosfomycin; STR, streptomycin; OLA, olaquindox; NAL, nalidixic acid; GEN, gentamicin; AMK, amikacin; FOX, cefoxitin; CTX, cefotaxime; FFC, florfenicol; CIP, ciprofloxacin; TET, tetracycline; MEM, meropenemS/T; TIG, tigecycline; trimethoprim/sulfamethoxazole).
Aeksiri, N., Toanan, W., Sawikan, S., Suwannarit, R., Pungpomin, P., Khieokhajonkhet, A., et al. (2019). First detection and genomic insight into mcr-1 encoding plasmid-mediated colistin-resistance gene in Escherichia coli ST101 isolated from the migratory bird species hirundo rustica in Thailand. Microb. Drug Resist. 25, 1437–1442. doi: 10.1089/mdr.2019.0020
Ahlstrom, C. A., van Toor, M. L., Woksepp, H., Chandler, J. C., Reed, J. A., Reeves, A. B., et al. (2021). Evidence for continental-scale dispersal of antimicrobial resistant bacteria by landfill-foraging gulls. Sci. Total Environ. 764:144551. doi: 10.1016/j.scitotenv.2020.144551
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Arnold, K. E., Williams, N. J., and Bennett, M. (2016). “Disperse abroad in the land”: the role of wildlife in the dissemination of antimicrobial resistance. Biol. Lett. 12:20160137. doi: 10.1098/rsbl.2016.0137
Cao, J., Hu, Y., Liu, F., Wang, Y., Bi, Y., Lv, N., et al. (2020). Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome 8:26. doi: 10.1186/s40168-019-0781-8
Chen, K., Chan, E. W. C., Xie, M., Ye, L., Dong, N., and Chen, S. (2017). Widespread distribution of mcr-1-bearing bacteria in the ecosystem, 2015 to 2016. Eur. Secur. 22:17-00206. doi: 10.2807/1560-7917.ES.2017.22.39.17-00206
Dafale, N. A., Srivastava, S., and Purohit, H. J. (2020). Zoonosis: an emerging link to antibiotic resistance under “one health approach”. Indian J. Microbiol. 60, 139–152. doi: 10.1007/s12088-020-00860-z
Elsohaby, I., Samy, A., Elmoslemany, A., Alorabi, M., Alkafafy, M., Aldoweriej, A., et al. (2021). Migratory wild birds as a potential disseminator of antimicrobial-resistant bacteria around Al-Asfar Lake, eastern Saudi Arabia. Antibiotics 10:260. doi: 10.3390/antibiotics10030260
Feng, Y., Zou, S., Chen, H., Yu, Y., and Ruan, Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49, D644–D650. doi: 10.1093/nar/gkaa821
Fernandes, M. R., McCulloch, J. A., Vianello, M. A., Moura, Q., Pérez-Chaparro, P. J., Esposito, F., et al. (2016). First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob. Agents Chemother. 60, 6415–6417. doi: 10.1128/AAC.01325-16
Flament-Simon, S. C., de Toro, M., Mora, A., García, V., García-Meniño, I., Díaz-Jiménez, D., et al. (2020). Whole genome sequencing and characteristics of mcr-1–harboring plasmids of porcine Escherichia coli isolates belonging to the high-risk clone O25b:H4-ST131 clade B. Front. Microbiol. 11:387. doi: 10.3389/fmicb.2020.00387
Franklin, A. B., Ramey, A. M., Bentler, K. T., Barrett, N. L., McCurdy, L. M., Ahlstrom, C. A., et al. (2020). Gulls as sources of environmental contamination by colistin-resistant bacteria. Sci. Rep. 10:4408. doi: 10.1038/s41598-020-61318-2
García-Meniño, I., García, V., Mora, A., Díaz-Jiménez, D., Flament-Simon, S. C., Alonso, M. P., et al. (2018). Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 9:2659. doi: 10.3389/fmicb.2018.02659
Hadjadj, L., Riziki, T., Zhu, Y., Li, J., Diene, S. M., and Rolain, J. M. (2017). Study of mcr-1 gene-mediated colistin resistance in enterobacteriaceae isolated from humans and animals in different countries. Genes 8:394. doi: 10.3390/genes8120394
Haenni, M., Métayer, V., Gay, E., and Madec, J. Y. (2016). Increasing trends in mcr-1 prevalence among extended-spectrum-β-lactamase-producing Escherichia coli isolates from French calves despite decreasing exposure to colistin. Antimicrob. Agents Chemother. 60, 6433–6434. doi: 10.1128/AAC.01147-16
Jarma, D., Sánchez, M. I., Green, A. J., Peralta-Sánchez, J. M., Hortas, F., Sánchez-Melsió, A., et al. (2021). Faecal microbiota and antibiotic resistance genes in migratory waterbirds with contrasting habitat use. Sci. Total Environ. 783:146872. doi: 10.1016/j.scitotenv.2021.146872
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Lake, Q. (2020). Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID-19. The COVID-19 resource centre is hosted on Elsevier Connect, the company’s public news and information.
Li, B., Yin, F., Zhao, X., Guo, Y., Wang, W., Wang, P., et al. (2020). Colistin resistance gene mcr-1 mediates cell permeability and resistance to hydrophobic antibiotics. Front. Microbiol. 10:3015. doi: 10.3389/fmicb.2019.03015
Lin, Y., Dong, X., Wu, J., Rao, D., Zhang, L., Faraj, Y., et al. (2020). Metadata analysis of mcr-1-bearing plasmids inspired by the sequencing evidence for horizontal transfer of antibiotic resistance genes between polluted river and wild birds. Front. Microbiol. 11:352. doi: 10.3389/fmicb.2020.00352
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Marcelino, V. R., Wille, M., Hurt, A. C., González-Acuña, D., Klaassen, M., Schlub, T. E., et al. (2019). Meta-transcriptomics reveals a diverse antibiotic resistance gene pool in avian microbiomes. BMC Biol. 17:31. doi: 10.1186/s12915-019-0649-1
Matamoros, S., Van Hattem, J. M., Arcilla, M. S., Willemse, N., Melles, D. C., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Mendes, A. C., Novais, Â., Campos, J., Rodrigues, C., Santos, C., Antunes, P., et al. (2018). Mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016–2017. Emerg. Infect. Dis. 24, 762–766. doi: 10.3201/eid2404.171787
Mohsin, M., Azam, M., Ur Rahman, S., Esposito, F., Sellera, F. P., Monte, D. F., et al. (2019). Genomic background of a colistin-resistant and highly virulent MCR-1-positive Escherichia coli ST6395 from a broiler chicken in Pakistan. Pathog. Dis. 77:ftz064. doi: 10.1093/femspd/ftz064
Monte, D. F., Mem, A., Fernandes, M. R., Cerdeira, L., Esposito, F., Galvão, J. A., et al. (2017). Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 61:e02718-16. doi: 10.1128/AAC.02718-16
Moura, Q., Sartori, L., Silva, K. C., Cunha, M. P., Esposito, F., Lopes, R., et al. (2016). Disseminação silenciosa do gene mrc-1 de E. coli resistente a colistina na América do Sul – Surveillance 2016. Euro Surveill. 21, 1–6.
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Ruzauskas, M., and Vaskeviciute, L. (2016). Detection of the mcr-1 gene in Escherichia coli prevalent in the migratory bird species Larus argentatus. J. Antimicrob. Chemother. 71, 2333–2334. doi: 10.1093/jac/dkw245
Sellera, F. P., Fernandes, M. R., Sartori, L., Carvalho, M. P. N., Esposito, F., Nascimento, C. L., et al. (2017). Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J. Antimicrob. Chemother. 72, 1255–1256. doi: 10.1093/jac/dkw543
Snesrud, E., He, S., Chandler, M., Dekker, J. P., Hickman, A. B., McGann, P., et al. (2016). A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60, 6973–6976. doi: 10.1128/AAC.01457-16
Taylor, N. G. H., Verner-Jeffreys, D. W., and Baker-Austin, C. (2011). Aquatic systems: maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol. Evol. 26, 278–284. doi: 10.1016/j.tree.2011.03.004
Vergara, A., Pitart, C., Montalvo, T., Roca, I., Sabaté, S., Hurtado, J. C., et al. (2017). Prevalence of extended-spectrum-β-lactamase-and/or carbapenemase-producing Escherichia coli isolated from yellow-legged gulls from Barcelona, Spain. Antimicrob. Agents Chemother. 61:e02071–16. doi: 10.1128/AAC.02071-16
Wang, R., Van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Yang, Q., Li, M., Spiller, O. B., Andrey, D. O., Hinchliffe, P., Li, H., et al. (2017). Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 8:2054. doi: 10.1038/s41467-017-02149-0
Yang, F., Shen, C., Zheng, X., Liu, Y., Ahmed, M. A. E. G. E. S., Zhao, Z., et al. (2019a). Plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China. Infect. Drug Resist. 12, 385–389. doi: 10.2147/IDR.S194635
Yang, Q. E., Tansawai, U., Andrey, D. O., Wang, S., Wang, Y., Sands, K., et al. (2019b). Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): an increasing public health risk. Environ. Int. 122, 281–290. doi: 10.1016/j.envint.2018.11.021
Yao, X., Doi, Y., Zeng, L., Lv, L., and Liu, J. H. (2016). Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect. Dis. 16, 288–289. doi: 10.1016/S1473-3099(16)00057-8
Keywords: migratory birds, mcr-1, Escherichia coli, transmission, antibiotic resistance
Citation: Zhang Y, Kuang X, Liu J, Sun R-Y, Li X-P, Sun J, Liao X-P, Liu Y-H and Yu Y (2021) Identification of the Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli Isolates From Migratory Birds in Guangdong, China. Front. Microbiol. 12:755233. doi: 10.3389/fmicb.2021.755233
Received: 08 August 2021; Accepted: 23 September 2021;
Published: 21 October 2021.
Edited by:
Azucena Mora Gutiérrez, University of Santiago de Compostela, SpainReviewed by:
Justin Joseph Donato, University of St. Thomas, United StatesCopyright © 2021 Zhang, Kuang, Liu, Sun, Li, Sun, Liao, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Yu, Z3JhY2V5eXVAc2NhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.