- Department of Food Science, Canadian Research Institute for Food Safety, University of Guelph, Guelph, ON, Canada
Cronobacter species, in particular C. sakazakii, is an opportunistic bacterial pathogen implicated in the development of potentially debilitating illnesses in infants (<12months old). The combination of a poorly developed immune system and gut microbiota put infants at a higher risk of infection compared to other age groups. Probiotics and prebiotics are incorporated in powdered infant formula and, in addition to strengthening gut physiology and stimulating the growth of commensal gut microbiota, have proven antimicrobial capabilities. Postbiotics in the cell-free supernatant of a microbial culture are derived from probiotics and can also exert health benefits. Synbiotics, a mixture of probiotics and prebiotics, may provide further advantages as probiotics and gut commensals degrade prebiotics into short-chain fatty acids that can provide benefits to the host. Cell-culture and animal models have been widely used to study foodborne pathogens, but sophisticated gut models have been recently developed to better mimic the gut conditions, thus giving a more accurate representation of how various treatments can affect the survival and pathogenicity of foodborne pathogens. This review aims to summarize the current understanding on the connection between Cronobacter infections and infants, as well as highlight the potential efficacy of probiotics, prebiotics, and synbiotics in reducing invasive Cronobacter infections during early infancy.
Introduction
Cronobacter sakazakii has been implicated in the development of neonatal infections including necrotizing enterocolitis (NEC), bacteremia, and meningitis with mortality rates ranging from 40 to 80% for premature (<37weeks gestational age) and/or low-birthweight infants (<2,500g) (Bowen and Braden, 2006; Ray et al., 2007; Friedemann, 2009; Centre for Disease Control, 2020a), and the survivors of C. sakazakii infection may develop chronic sequelae such as neurological impairments (Holý et al., 2019). Powdered infant formula (PIF) is a common source of C. sakazakii infection, but the pathogen can be found in hospitals, homes, and equipment, e.g., enteral feeding tube and baby bottles (Kalyantanda et al., 2015; Centre for Disease Control, 2020b). Cronobacter sakazakii contamination rates of 3–7 and 5% from PIF or PIF processing plants, respectively, have been found (Mardaneh and Dallal, 2016; Fei et al., 2017a; Lu et al., 2019). Recommendations from the World Health Organization (WHO) to minimize contamination of PIF, such as using water >70°C to rehydrate PIF or exclusively breastfeeding infants up to 6months old (World Health Organization, 2007), has not been shown to eliminate the risk of C. sakazakii infection as outbreaks continue to occur, so other preventative strategies are needed.
The infant gut microbiota matures in the first 3years of life, and its composition is affected by many factors including childbirth delivery method, infant diet, gestational age, maternal diet, environment, and genetics (Yatsunenko et al., 2012; Bäckhed et al., 2015; Milani et al., 2017). The infant gut microbiota hosts a few key genera associated with a healthy gut, such as Bifidobacterium and Lactobacillus, which dominate the gut microbiota of 1-year-old breast-fed infants (Bäckhed et al., 2015). PIF poses a greater risk of C. sakazakii infection in infants and cannot establish a healthy gut microbiota as compared to breast-fed infants (Le Doare et al., 2018). Breast milk is estimated to contribute approximately 28% of the infant gut microbiota in an infant’s first 12months of life and contains human milk oligosaccharides (HMOs) that promote a healthy and diverse infant gut microbiota (Mueller et al., 2015; Pannaraj et al., 2017; Le Doare et al., 2018; Robertson et al., 2019). The combination of a diverse gut microbiota and HMOs reduces the risk of illness in infants until the maturation of their gut microbiota occurs following the ingestion of solid foods (Johnson-Henry et al., 2016; Le Doare et al., 2018; Robertson et al., 2019). PIF may be supplemented with probiotics, prebiotics, or a combination of both to partially simulate the complex composition of human breast milk (Ackerberg et al., 2012; Vandenplas et al., 2015), which is a complicated task due to the diverse microbiota and oligosaccharides present. However, current research is being conducted to achieve an ideal cocktail of probiotics and prebiotics that mimics breast milk.
Probiotics have a long history of research and usage, with early recognition and guidance being given by the Food and Agriculture Organization of the United Nations and the World Health organization (FAO/WHO) in 2001/2002 (Food and Agriculture Organization of the United Nations/World Health Organization, 2002), and have demonstrated their ability to inhibit pathogens through direct or indirect antagonism (Mohsin et al., 2015; Tomar et al., 2015; Fijan, 2016). Probiotics produce organic acids, such as lactic acid and acetic acid, and hydrogen peroxide (H2O2), and bacteriocins that can inhibit foodborne pathogens such as Listeria monocytogenes, Escherichia coli, and Salmonella spp. (Makras and De Vuyst, 2006; Kotsou et al., 2008). Furthermore, probiotic strains can release metabolites into the growth medium, collectively referred to as postbiotics, which could play a role in conferring beneficial effects on the host (Tomar et al., 2015). Prebiotics, on the other hand, are predominantly oligosaccharides that contain at least three monomeric units and are neither digested nor absorbed in the small intestine (Venegas et al., 2019). Short-chain fatty acids (SCFAs), for example, butyrate, acetate, and propionate, are produced primarily in the colon by the gut microbiota that metabolizes prebiotics and are considered to be a sub-category of organic acids (Gibson et al., 2017). Acetate and propionate are mainly produced by the Bacteroidetes phyla, while the Firmicute phyla mostly produce butyrate. Bifidobacteria and lactic acid bacteria, for example, can ferment carbohydrates to produce acetate and lactate (Korakli et al., 2002). However, other members of the gut microbiota can act as secondary fermenters to produce other SCFAs (Venegas et al., 2019). SCFAs play an important role in host health by regulating metabolic activities, the immune system, gut epithelial integrity, and by exhibiting antimicrobial properties (Gibson et al., 2017; Robertson et al., 2019). Probiotics and prebiotics have been used to modulate a naïve gut microbiota, but synbiotics have been recommended as an alternative approach to capitalize on their synergism. The concept of synbiotics was introduced by Gibson and Roberfroid (1995) as a mixture of probiotics and prebiotics that improve the survival and colonization of beneficial microbes in the gastrointestinal (GI) tract by selectively stimulating the growth and/or activation of health promoting bacteria and subsequently improving host health. The synergistic effect of prebiotics and probiotics can be found naturally, e.g., breast milk contains prebiotic oligosaccharides and beneficial bacteria that help to stimulate the development of a healthy infant gut (Le doare et al., 2018; Robertson et al., 2019).
Mammalian cell lines are typically used to study the interactions between pathogens, probiotics, and prebiotics, although synbiotics have rarely been studied in this medium (Feeney et al., 2014; Kim et al., 2015; Tomar et al., 2015; Ye et al., 2016; Holý et al., 2019; Jamwal et al., 2019). While useful to visualize the effects of pathogens or probiotics on live cells, cell lines do not closely represent the complex interaction that occurs within the human intestinal environment. Studies on bacterial pathogenesis in vivo using animal models can be challenging to conduct due to ethical, cost, and infrastructure issues. Differences between the animal and human gut have led to the emergence of various in vitro gut models, for example, the Simulator of the Human Intestinal Microbial Ecosystem (SHIME) that provides an opportunity to analyze the pathogen-host and pathogen-gut microbiota interactions in more detail (Van den Abbeele et al., 2010; Van de Wiele et al., 2015).
While the antimicrobial and gut-enhancing properties of probiotics and prebiotics have been frequently studied (Tomar et al., 2015; Fijan, 2016), there is a lack of understanding on the role and advantages of synbiotics. Furthermore, the effects of prebiotics and synbiotics on pathogenic Cronobacter spp. have not been studied and, compared to other enteropathogens, there are few studies into the efficacy of probiotics to reduce Cronobacter (Awaisheh et al., 2013; Weng et al., 2014; Charchoghlyan et al., 2016; Campana et al., 2019; Fan et al., 2019; Jamwal et al., 2019). This review examines (i) the current understanding on the association between pathogenic Cronobacter spp. and infants and (ii) the potential for probiotics, prebiotics, and synbiotics to reduce the morbidity and mortality of invasive Cronobacter infections during early infancy.
Cronobacter Species
Taxonomy and Identification of Cronobacter Species
Cronobacter spp., formerly known as Enterobacter sakazakii, are Gram-negative, rod shaped, motile, opportunistic pathogens initially associated with debilitating infections in infants (Yan et al., 2012). The designated name of Enterobacter sakazakii encompassed all the members of the Cronobacter genus up until 2007 due to limited genomic knowledge (Forsythe, 2018) and E. sakazakii was split into 16 biogroups based on phenotype (Farmer III et al., 1980; Iversen et al., 2006). Eventually, multi-locus sequence typing (MLST) was used to further characterize and differentiate different species of Cronobacter (Baldwin et al., 2009).
Cronobacter Clinical Relevance and Sources
Cronobacter spp. are grouped based on their clinical relevance as shown on Table 1. Among the seven Cronobacter spp., C. sakazakii is the predominant species causing human illness (Sonbol et al., 2013; Fei et al., 2017b). Based on MLST, sequence type profile 4 (ST4) and ST1 strains of C. sakazakii are most often isolated from PIF and hospitalized patients (Joseph and Forsythe, 2011; Joseph et al., 2012). Cronobacter malonaticus was initially classified as a subspecies of C. sakazakii due to their genetic similarities but has since been differentiated from C. sakazakii due to the presence of unique genes required for malonate metabolism (Joseph et al., 2012). More recently, like C. sakazakii, C. malonaticus was found to cause infection in infants by invading and damaging human gut epithelial cells (Alsonosi et al., 2018). However, C. sakazakii has the most open reading frames compared to other Cronobacter spp., potentially indicating a higher probability for C. sakazakii to encode proteins relevant to stress resistance or pathogenicity (Joseph et al., 2012).
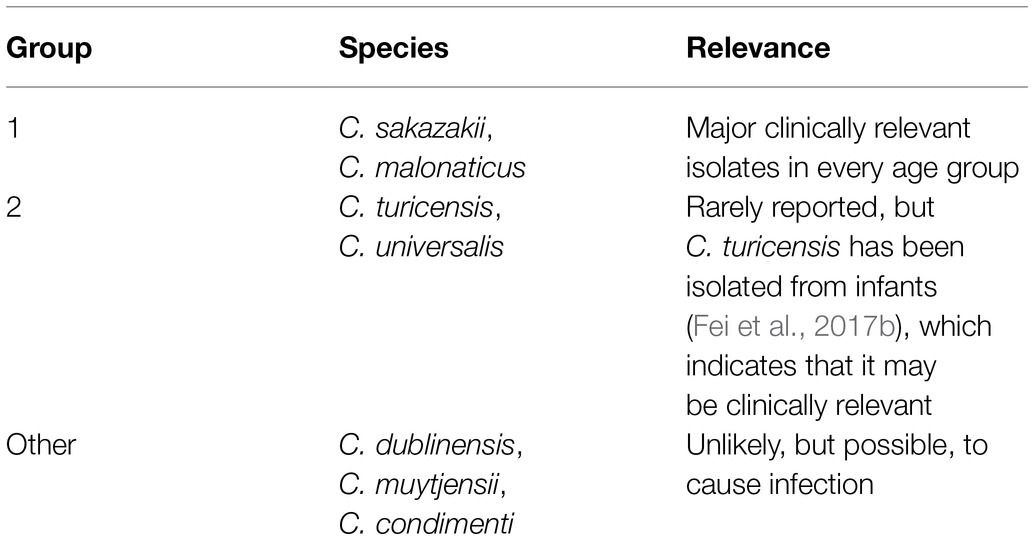
Table 1. Clinical relevance of Cronobacter species (adapted from Forsythe, 2018).
Cronobacter sakazakii is ubiquitous and has been isolated from a wide variety of sources including dried herbs and spices, soil, starches, milk products, and expressed breast milk (Berhilevych and Kasianchuk, 2017; Forsythe, 2018; Jang et al., 2018a). However, C. sakazakii can be found in PIF, factories, hospitals, and homes (Centre for Disease Control, 2020b). PIF may be contaminated with C. sakazakii due to poor hygiene in the PIF production facilities and workflow, such as improper filtration in the spray drying process (Jacobs et al., 2011; Li et al., 2014), but C. sakazakii has been found in hospital air, dust, and within the human body, so careful handling of PIF may not eliminate the risk of infection (Forsythe, 2018). Recently, the genome sequences of C. sakazakii strains have been obtained from various food products, some of which are low-moisture foods of plant origin such as grains, carrots, and mushroom (Jang et al., 2018b). Elkhawaga et al. (2020) found that approximately 9% of the herbs they tested were contaminated with C. sakazakii, which implies a potentially higher risk of C. sakazakii illness occurring in parts of the world, where the use of alternative medicine relies on natural ingredients derived from herbs and other plant products, especially tea leaves, licorice, anise, allspice, and oregano (Jang et al., 2018a; Elkhawaga et al., 2020).
Epidemiology of Cronobacter sakazakii
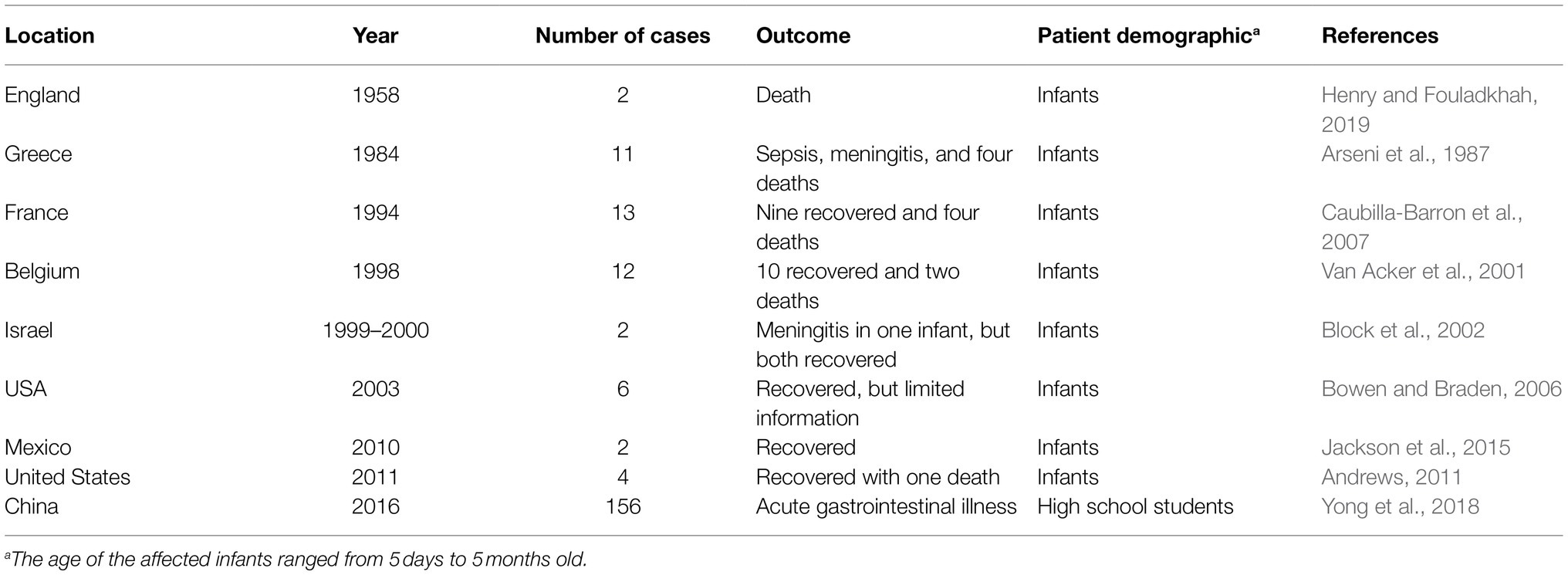
Outbreaks of C. sakazakii are infrequent and most are associated with infants (Table 2). In the United States, the incidence rate of Cronobacter spp. for adults ≥80years old and 70–79years old is 3.93 cases (per 100,000 population) and 2.11 cases, respectively, compared to the 1.81 cases for infants <1year old (Patrick et al., 2014). However, infections in adults are rarer and often occur with mild symptoms unless the individual has underlying health conditions (Tsai et al., 2013; Yong et al., 2018). As C. sakazakii is widespread, the sources of contamination can vary, and reservoirs are unclear. In one outbreak involving high school students, for example, C. sakazakii was isolated from leftover cafeteria food and resulted in acute gastroenteritis in 156 individuals (Yong et al., 2018). Overall, the sources of C. sakazakii infection in adults and the elderly are unclear as the foodborne pathogen can be found in a wide range of environmental sources including plant and animal-derived foods, sewer water, and dust (Tsai et al., 2013; Yong et al., 2018; Ohira et al., 2021).
PIF is a major source of C. sakazakii infection for infants (Centre for Disease Control, 2020b) because heat-sensitive ingredients (e.g., vitamins) are added after milk sterilization and contamination may occur from the manufacturing environment (Kalyantanda et al., 2015). Once contaminated PIF is packaged, C. sakazakii can remain viable for up to 2years (Hu et al., 2018; Parra-Flores et al., 2018a). Furthermore, using water of <70°C to rehydrate PIF might exacerbate the likelihood of contamination (World Health Organization, 2007). Improper handling and/or lack of awareness on how to properly reconstitute PIF can contribute to the survival or contamination of PIF with C. sakazakii. It is recommended that reconstituted PIF be used within 2h of preparation and be discarded if it is not finished within a single feeding (World Health Organization, 2007).
Infants, especially those that are <2months old, of low-birthweight (<1,500–2,500g) and/or born prematurely (<37weeks gestational age), are the main age group associated with C. sakazakii outbreaks (Lai, 2001; Centre for Disease Control, 2020b). Young children up to 3years of age and low-birthweight or premature infants may be afflicted with dysbiosis (Yang et al., 2016), which is described as an imbalanced gut microbiota that has been associated with a range of illnesses including Crohn’s disease, irritable bowel syndrome, and colitis (Biedermann and Rogler, 2015). Infants born via C-section may have diminished transmission from mother-to-infant of key birth canal microbes, such as Bacteroides and Bifidobacterium (Bäckhed et al., 2015), which could increase the infant’s risk of infection caused by pathogens in food and the environment (Durack and Lynch, 2019). Similarly, formula-fed infants can exhibit the delayed establishment of Bifidobacterium and Lactobacillus spp. in the gut, thereby increasing the potential risk of colonization by opportunistic pathogens (Collado et al., 2015).
In the early 2000s, FAO and WHO collaborated on a risk assessment for the microbiological safety of PIF. Salmonella and Cronobacter spp. were classified as Category A pathogens to indicate a clear evidence of causality in PIF contamination (Food and Agriculture Organization of the United Nations/World Health Organization, 2006, 2008). Category B pathogens, including Escherichia, Klebsiella, and Citrobacter genera indicated the plausibility of contamination, while Category C pathogens, such as Clostridium, Staphylococcus, and Listeria genera, were not implicated (Food and Agriculture Organization of the United Nations/World Health Organization, 2006). A risk assessment model for C. sakazakii in PIF was created to quantify the specific risks of C. sakazakii to infants.1 The FAO/WHO model for determining the potential PIF risk in a specific situation is considered as the initial C. sakazakii population, in addition to reconstitution temperature, handling methods, and feeding period.
Pathogenesis of Cronobacter sakazakii
The combination of an underdeveloped immune system and an immature intestinal epithelial barrier has been shown to predispose some infants to colonization and infection of pathogens (Emami et al., 2012). In fact, the infection and subsequent development of necrotizing enterocolitis (NEC), a disease that develops when the tissue in the inner lining of the small or large intestine becomes damaged, inflamed, and necrosized, has been linked to the colonization of an infant’s immature gut by C. sakazakii (Emami et al., 2012; Figure 1). Cronobacter sakazakii can not only suppress the maturation of dendritic cells (DC) but is also able to enter DC to survive and spread, thus avoiding the immune system (Emami et al., 2012). Macrophages and neutrophils have been found to play an important role in eliminating C. sakazakii by regulating the production of DC but provide an ineffective immune response due to the intracellular nature of C. sakazakii infection (Townsend et al., 2007; Emami et al., 2012). NEC can develop due to pro-inflammatory cytokines released during infection, which stimulates the production of nitric oxide synthase and damages intestinal mucosa through nitric oxide (NO) (Emami et al., 2011). Further disruption of the intestinal layers may allow C. sakazakii to reach the brain and/or bloodstream, which can cause meningitis or sepsis, respectively.
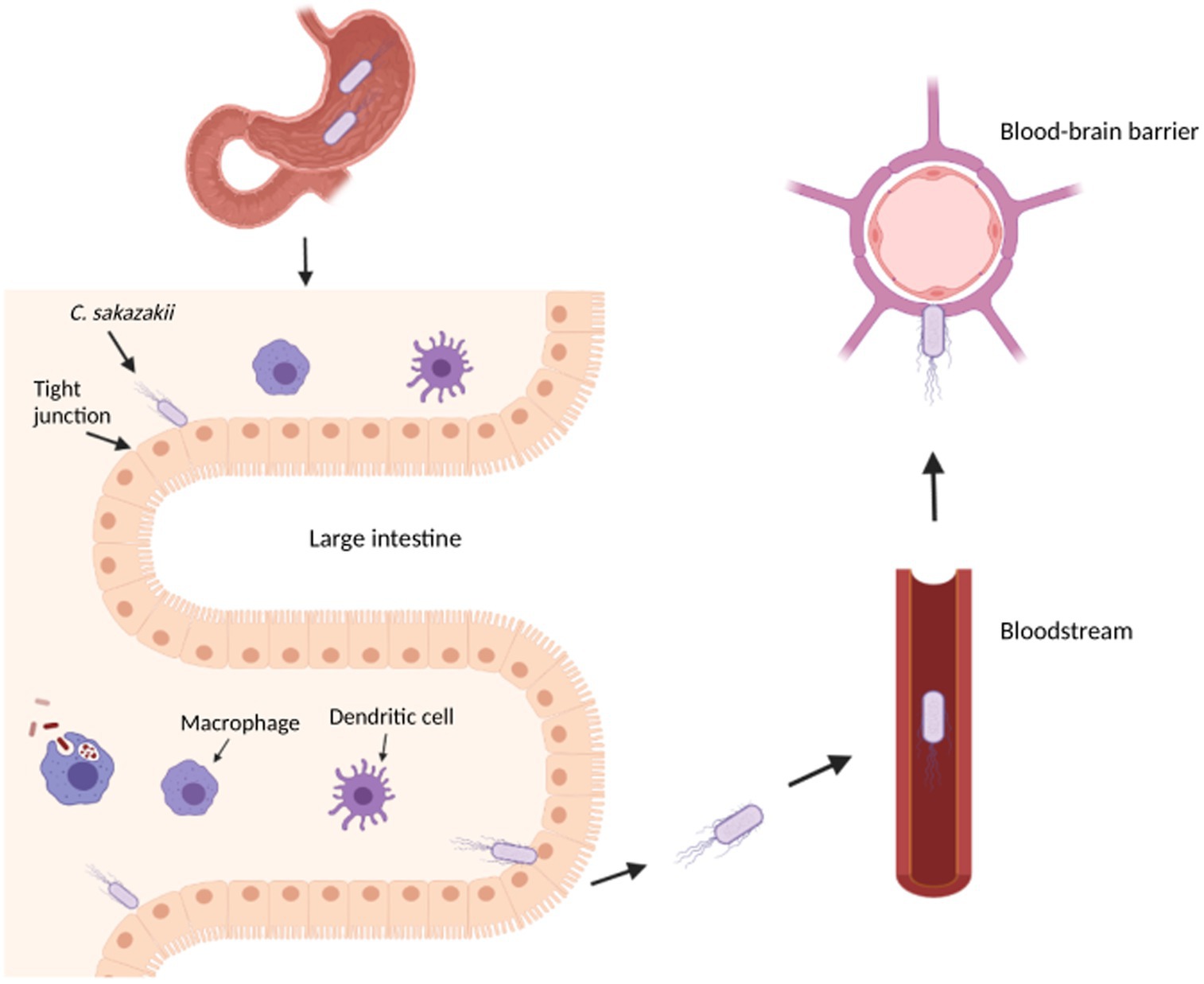
Figure 1. Overview of Cronobacter sakazakii pathogenesis in an infant. This diagram illustrates the transit of C. sakazakii through the stomach and small intestine to reach the large intestine. In the large intestine, C. sakazakii can evade the immune cells to disrupt the epithelial layer, potentially causing necrotizing enterocolitis and entering the bloodstream to cause sepsis. From the bloodstream, C. sakazakii can enter the blood–brain barrier to cause meningitis. The image was created with BioRender.com.
Virulence Factors
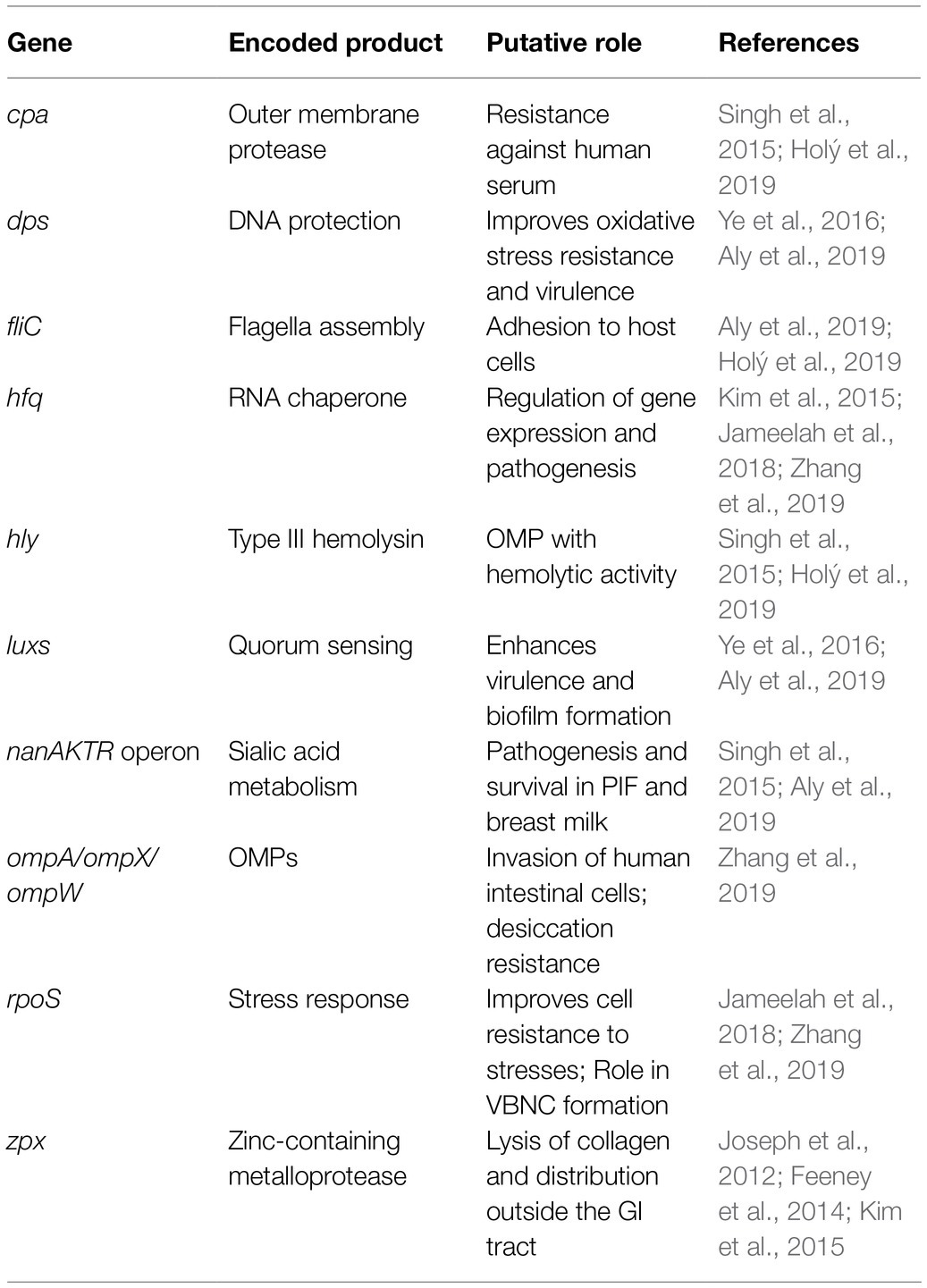
Hfq, also known as Host Factor 1, is a regulatory protein involved in the pathogenesis, communication, and survival of several foodborne pathogens including L. monocytogenes, E. coli, and Salmonella Typhimurium (Kim et al., 2015; Cech et al., 2016). The hfq gene in C. sakazakii was reported to regulate virulence genes, and its absence reduced the infection rate in Caco-2 cells, survival in macrophage-like cells, and oxidative stress resistance (Kim et al., 2015). During desiccation and acid stress conditions, C. sakazakii have an increased expression of hfq by 1 and 3-logs, respectively (Jameelah et al., 2018; Maerani et al., 2020). However, as C. sakazakii entered a viable but not culturable (VBNC) state after desiccation or acid stress conditions, the expression of hfq decreased by approximately 1-log (Jameelah et al., 2018; Maerani et al., 2020).
Outer membrane proteins (OMPs) play a significant role in the virulence of C. sakazakii. The two OMPs, OmpA and OmpX, were found to be essential for C. sakazakii adherence and invasion of intestinal cells (Kim et al., 2015; Singh et al., 2015; Jameelah et al., 2018). The OmpA has also been reported to play a role in facilitating C. sakazakii invasion of the blood–brain barrier (Holý et al., 2019). On the other hand, ompA or ompX C. sakazakii deletion mutants had significantly reduced invasion and adherence rates compared to the wild-type strains (Feeney et al., 2014). The OmpA was also identified as a key protein mediating cell adhesion to fibronectin, a glycoprotein on the extracellular matrix of eukaryotic tissue (Nair et al., 2009).
The enterotoxins and endotoxins from C. sakazakii remain functional during the manufacturing and shelf life of PIF, which may play a role in the pathogenesis of C. sakazakii infection (Jaradat et al., 2014). Cronobacter sakazakii enterotoxins may be similar in function to a lipopolysaccharide by stimulating an inflammatory response in the host, which can lead to NEC (Singh et al., 2015), whereas their endotoxins may be able to help to translocate the pathogen across the intestinal and blood–brain barriers to cause bacteremia and meningitis (Kalyantanda et al., 2015; Shi et al., 2017). Cronobacter sakazakii has many genes related to survival and virulence, but the function of some of them is still under investigation (Table 3).
Sialic acid is found in breast milk, PIF, intestinal mucin, and human brain, as it is a bioactive compound needed for brain development (Joseph et al., 2013; Kalyantanda et al., 2015; Lis-Kuberka and Orczyk-Pawiłowicz, 2019). Whole genome sequencing revealed that within the Cronobacter genus, C. sakazakii is the only species that can use sialic acid as a carbon source, as it possesses the nanAKTR gene cluster (Joseph et al., 2013; Singh et al., 2015). The ability of C. sakazakii to metabolize sialic acid may be a factor in PIF survival and infection of the intestines and brain, but further research is required to validate this hypothesis (Jaradat et al., 2014).
Stress Adaptation
Cronobacter sakazakii is a resilient enteropathogen capable of surviving desiccation, heat, and low pH encountered during food manufacturing processes and in the human GI tract (Feeney et al., 2014; Kim et al., 2015; Jameelah et al., 2018; Parra-flores et al., 2018b; Holý et al., 2019). In addition, its resistance against components in the GI tract, including stomach acid and bile liquids, may be a key factor in the development of illnesses across all age groups, especially in infants due to their lower stomach acidity.
Cronobacter sakazakii was shown to survive desiccation by accumulating electrolytes to increase osmotic pressure inside the cell, which prevents cellular fluids from leaking out (Aly et al., 2019), but there are other biochemical pathways that play a role in desiccation resistance. For example, the upregulation of ompW can contribute to the survival of C. sakazakii in dry conditions while promoting biofilm formation (Aly et al., 2019). Bai et al. (2019) reported that the concentration of trehalose increased inside C. sakazakii cells experiencing desiccation tolerance and found that the accumulation of trehalose also improved the survival of C. sakazakii that were subjected to cold stress. The notion of cross-protection, whereby an acquired tolerance from sublethal stress can cross-protect against other stresses, has been documented in other foodborne pathogens including E. coli (Yurtsev et al., 2016), S. Typhimurium (Leyer and Johnson, 1993), and L. monocytogenes (Begley et al., 2002) but is not well described in Cronobacter spp.
Cronobacter sakazakii biofilms can form in processing environments on a variety of surfaces including silicon, stainless steel, plastic, silicon, and latex during stressful conditions, such as nutrient limitation or desiccation, which contributes to contamination risk in PIF and other products (Kalyantanda et al., 2015; Singh et al., 2015; Aly et al., 2019). Cronobacter spp. have been reported to produce biofilms with the highest cell density among the Enterobacteriaceae (Kalyantanda et al., 2015), further emphasizing the importance of good hygiene and regular cleaning in hospitals and homes. The persistence of mature biofilms in enteral feeding tubes could shed planktonic cells that enter the stomach of infants during feeding, thus contributing to infantile infection (Hurrell et al., 2009; Kalyantanda et al., 2015). Furthermore, C. sakazakii and other pathogens can enter a VBNC state during biofilm formation or PIF production (Forsythe, 2014; Jameelah et al., 2018). Biofilms are more resistant to disinfection methods, and some of the biosynthesis pathways that are required to produce biofilms, such as the colanic acid biosynthesis pathway, have been linked to antibiotic, desiccation, heat, and acid resistance.
Infants have a lower gastric acidity and bile salt concentration as compared to adults (Poquet and Wooster, 2016; Neal-Kluever et al., 2019), which may facilitate the survival of foodborne pathogens in the infant GI tract (Tennant et al., 2008; Sistrunk et al., 2016). Cronobacter sakazakii cells can survive at pH values as low as 4.5, a bile salt concentration of 5%, a salt concentration up to 10%, temperature as high as 60°C, and over 20days of desiccation (Fakruddin et al., 2014; Jameelah et al., 2018). In fact, the sub-lethal GI conditions in an infant may exacerbate C. sakazakii resistance to antimicrobial compounds produced by gut bacteria or probiotics (Hsiao et al., 2010). Furthermore, exposure to sub-lethal acid and thermal conditions may increase the heat resistance of C. sakazakii during rehydration of PIF using water <64°C (Yang et al., 2015), which supports the WHO recommendation of rehydrating PIF using water >70°C.
Biological Models Used To Study Cronobacter Sakazakii Pathogenicity
Cell Culture Models
Pathogens need to adhere to the host cell surfaces to establish infection (Shi et al., 2017). Adherence and invasion of C. sakazakii have been primarily studied using cell lines including Caco-2, HT-29, INT-407, and Hep-2 (Feeney et al., 2014; Kim et al., 2015; Ye et al., 2016; Holý et al., 2019). These models are especially important to simulate the attachment and entry of C. sakazakii to intestinal cells. Human isolates of C. sakazakii have been found to be better at binding to tissue culture cell models as compared to environmental strains (Liu et al., 2012). Following adherence, C. sakazakii increased cell permeability and disrupted tight junctions due to NO production, resulting in apoptosis (Liu et al., 2012). Bacteroides fragilis ZY-312 and citral were shown to reduce C. sakazakii invasion into HT-29 and Caco-2 cell lines, respectively (Shi et al., 2017; Fan et al., 2019). Nair and Venkitanarayanan (2007) showed that OmpA, in addition to the host cytoskeleton, plays an important role in cellular invasion of INT-407. Parra-Flores et al. (2018b) found that C. sakazakii isolates from PIF were able to invade Hep-2 cells, possibly due to the presence of outer membrane proteins. It should be noted, however, that experiments with different tissue culture cell lines need to be interpreted with caution, as C. sakazakii adheres better to certain cell lines (Holý et al., 2019).
Animal Models
Animal models, such as rabbit pups, neonatal rats, mice, and nematodes, are infrequently used to study the pathogenesis of C. sakazakii, because they cannot accurately replicate the in vivo interactions within an infant (Molloy et al., 2010; Hunter and Bean, 2013; Gurien et al., 2018; Fan et al., 2019; Kavita et al., 2020). However, several studies using animal models suggest that probiotic bacteria may be able to reduce the intestinal colonization of C. sakazakii and severity of NEC in vivo (Gurien et al., 2018; Fan et al., 2019; Kavita et al., 2020). Animal studies are useful to indicate whether prophylactic treatments can be further studied in infants. For example, Gurien et al. (2018) showed that as compared to the control group, pre-treatment of rabbit pups with Lactococcus lactis reduced the incidence rate of NEC caused by C. sakazakii.
Gastrointestinal Models
A more advanced approach to study C. sakazakii in the GI tract of humans is by using GI models to assess the pathogen’s survival and subsequent environmental changes in the GI tract. The TNO in vitro GI model (TIM) can mimic intestinal physiology for nutrition or metabolomic studies (Venema, 2015), whereas the SHIME is a multi-compartment dynamic simulator of the human GI tract that can simulate the stomach, small intestine, and the colon (Van den Abbeele et al., 2010). The TIM has been used to study the survival of E. coli or probiotic bacteria (Kheadr et al., 2010; Roussel et al., 2020), but its main advantage is the ability to mimic the luminal conditions in the GI tract through secretions, absorption, and the removal of bioavailable compounds (Minekus, 2015). In contrast, the SHIME system can evaluate interactions between the gut microbiota and/or foodborne pathogens due to its ability to inoculate the colon vessels with fecal samples from different age groups to analyze changes in the gut microbiota or metabolomics based on various experimental parameters (Van de Wiele et al., 2015). However, the SHIME has its own limitations, one of which is the inability to mimic normal peristalsis in the GI tract as it relies on stirrers to mix the GI fluids, so GI interactions and movement dependent on peristalsis may not be accurately portrayed. The TIM and SHIME complement each other to better mimic the complex ecosystem within the GI tract (Roussel et al., 2020), as both models have their unique advantages and disadvantages. Thus far, no studies have been done on the interaction between C. sakazakii and an in vitro gut model. Given the dynamic nature of an infant gut microbiota and its association with the development of infection, the SHIME can be a useful tool to study the changes in gut microbiota in a simulated infant GI tract when exposed to C. sakazakii.
Probiotics, Prebiotics, and Synbiotics
As current measures have proven insufficient to eliminate C. sakazakii contamination and infection, other preventative measures are required. Probiotics, prebiotics, and synbiotics may be promising control measures to inhibit the growth of C. sakazakii in the environment and within the infant GI tract. Furthermore, some probiotic strains can have synergistic effects with prebiotic substrates, i.e., synbiotics, as the prebiotic is degraded into antimicrobial metabolites or used to alter the bacterial surface and improve adhesion to the intestinal epithelial layer (Bocchi et al., 2020). Therefore, synbiotics may be better at mitigating the risk of infection due to foodborne pathogens, such as C. sakazakii, compared to the independent application of probiotics or prebiotics.
Probiotics
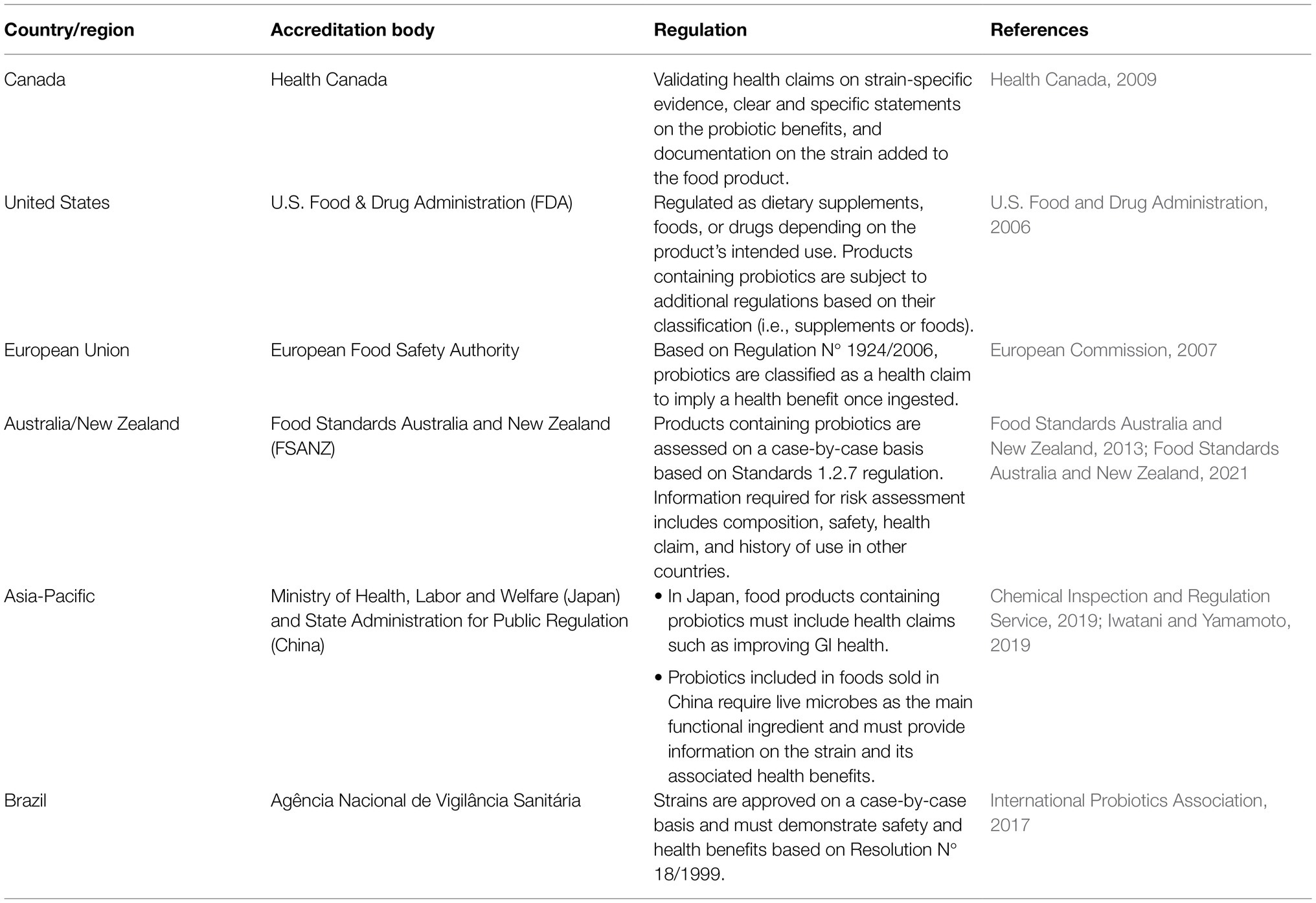
FAO/WHO developed the first definition for probiotics: “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (Food and Agriculture Organization of the United Nations/World Health Organization, 2002). A probiotic strain should have the following characteristics to confer benefits to the host (Food and Agriculture Organization of the United Nations/World Health Organization, 2002; Hill et al., 2014; Nagpal et al., 2018; Binda et al., 2020; Kaur et al., 2021; Rastogi et al., 2021): (1) isolated from a human, (2) lacking in putative virulence genes, (3) sensitive to common antibiotics, (4) tolerant to GI conditions, (5) catalase-negative, (6) able to adhere to the intestinal epithelial membrane, (7) able to compete with native gut microbiota, and (8) able to directly or indirectly inhibit the growth and colonization of potentially pathogenic bacteria. Research into probiotics has quickly expanded, with over 32,000 articles being published between 2002 and 2021 using the term “probiotic,” but proving the health benefits of probiotics, such as improving gut health after the consumption of probiotics, is challenging. Therefore, governments have taken different regulatory approaches on food products containing probiotics (Table 4).
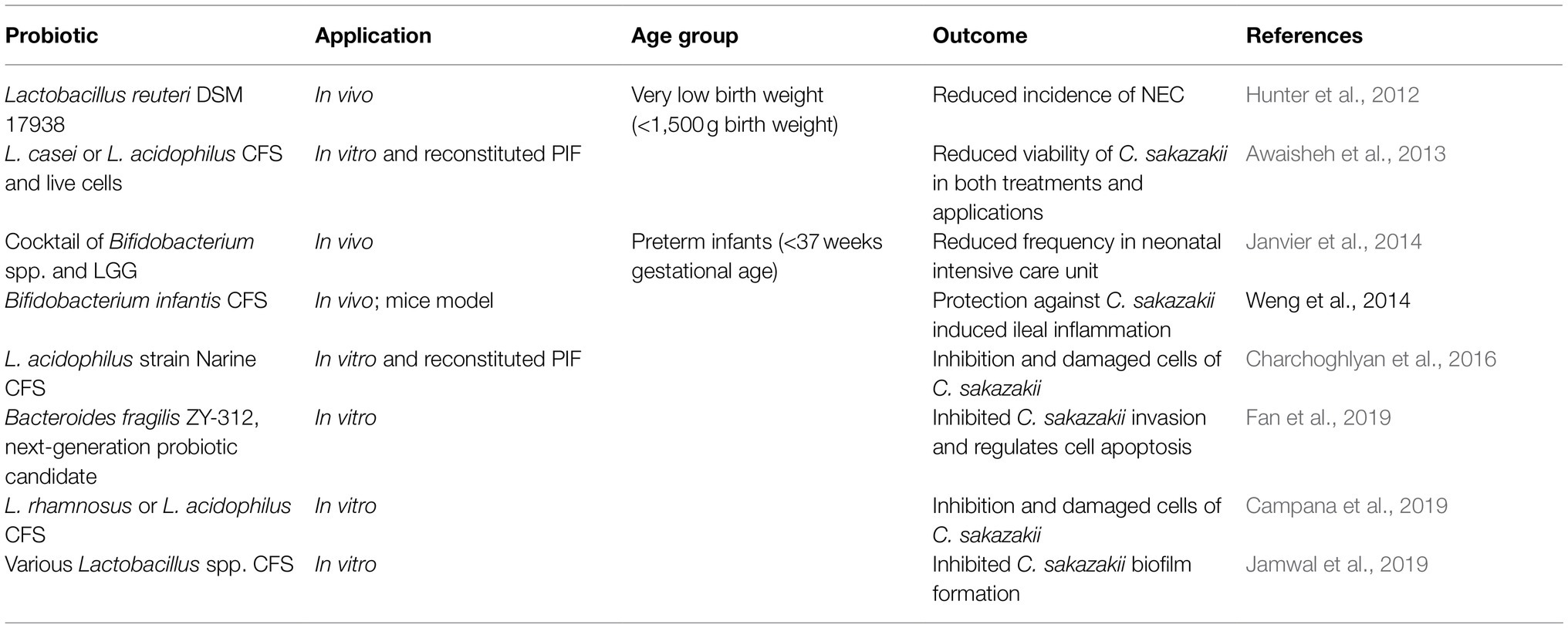
Bifidobacterium and Lactobacillus genera are the earliest examples of probiotics included in food products as they are Generally Recognized as Safe and can impart general health benefits such as supporting a healthy GI tract and immune system (Food and Agriculture Organization of the United Nations/World Health Organization, 2002; Hill et al., 2014; Canadian Food Inspection Agency, 2019). PIF can also be supplemented with probiotics, however, their application and scope of use in Canada is limited. Even among Bifidobacterium and Lactobacillus spp., only Bifidobacterium animalis subsp. lactis (BB-12) and Lactobacillus rhamnosus GG (LGG) are commercially available in PIF sold in Canada (Alberta Health Services, 2017) at a dose of approximately 7–8 log CFU/100ml. The approval of BB-12 and LGG may be due to the vast amounts of research that has been done on these specific strains to validate their safety and potential ability to exert health benefits on the host (Jungersen et al., 2014; Taipale et al., 2016; Flach et al., 2018; Capurso, 2019; Szajewska and Hojsak, 2020). However, the probiotics have not yet been approved by the Health Canada or the CFIA. Canadian probiotic supplements are designed for infants and contain either a single strain or a cocktail of Bifidobacterium and Lactobacillus spp. (AEProbio, 2020). The probiotic supplements have been reported to reduce the occurrence and mortality of NEC in premature and low-birthweight infants by approximately 4–12% (Hunter et al., 2012; Janvier et al., 2014). While the benefits of probiotics to overall health continue to be studied, research has shown several mechanisms by which probiotics can inhibit the growth of foodborne pathogens.
Antimicrobial Capabilities of Probiotics
Probiotics can indirectly mitigate human infections by (i) enhancing epithelial barrier physiology; (ii) inhibiting virulence gene expression; (iii) enhancing the host immune system; (iv) directly attacking pathogens by producing metabolites; (v) competing with the pathogens for nutrients, and (vi) adhering to the surface of GI cells, thus providing competition for attachment (Tomar et al., 2015). In the large intestine, they primarily produce organic acids due to the absence of oxygen (Dittoe et al., 2018). In the stomach and small intestine, the minute concentration of oxygen allows probiotics to produce H2O2 in enough quantities to directly inhibit pathogens; however, low concentrations of H2O2 in the anaerobic environment of the large intestine can only reduce pathogen invasion rather than exerting a bactericidal effect (Knaus et al., 2017). Different probiotic strains are very efficient in producing different compounds to inhibit pathogens. For example, Lactobacillus casei and L. acidophilus were found to produce bacteriocins that could inhibit the growth of C. sakazakii in reconstituted PIF (Awaisheh et al., 2013) and Bifidobacterium spp. produce organic acids, such as acetic acid, lactic acid, and formic acid, that inhibit other pathogens (Makras and De Vuyst, 2006).
The effect of probiotics on the gene expression of C. sakazakii has not been studied thus far, but previous research has shown that probiotics can downregulate virulence gene expression of other enteropathogens such as E. coli O157:H7 and Clostridium difficile (Yun et al., 2014; Mohsin et al., 2015). Escherichia coli Nissle 1917, a known probiotic strain, was able to control the Shiga-toxin gene expression of enterohemorrhagic E. coli O104:H4 and O157:H7 in vitro by downregulating stx2 mRNA transcription (Mohsin et al., 2015). Bifidobacterium longum and other probiotic strains have also been shown to downregulate Shiga toxin gene expression in vitro and in vivo (Carey et al., 2008; Cordonnier et al., 2016). It is possible that the downregulation of Shiga toxin gene expression was due to environmental stresses created by probiotics through the inhibition of the autoinducer-2 or a similar autoinducer system (Park et al., 2017). While the inhibitory effects of probiotics on enteropathogens have been documented, the mechanisms by which they exert their protective effects require further investigation.
Postbiotics
Probiotics are strictly regulated, and concise guidelines for their application in clinical settings (e.g., to reduce NEC) are lacking, thus creating an opportunity to study the application of their by-products to achieve the same beneficial effects (Piqué et al., 2019). Postbiotics have been defined as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host,” thereby containing inactivated microbial cells that may contain metabolites or cellular components to exert said health benefit (Salminen et al., 2021). Postbiotics can originate from a probiotic strain and can be composed of the cell-free supernatant (CFS) consisting of bacteriocins, organic acids, H2O2, and other compounds that can inhibit bacterial cells or biofilm formation (Campana et al., 2019; Jamwal et al., 2019; Piqué et al., 2019; Nataraj et al., 2020). Awaisheh et al. (2013) showed that L. casei and L. acidophilus CFS inhibited the growth of C. sakazakii on agar media and a liquid rehydrated infant formula model, while Lin and Pan (2019) demonstrated that Lactobacillus plantarum NTU 102 CFS contained an active antimicrobial compound, named as LBP102, which was able to inhibit C. sakazakii in vitro. Other Lactobacillus spp. CFS has been shown to weaken the membrane integrity and disrupt the biofilm formation of C. sakazakii in vitro (Charchoghlyan et al., 2016; Campana et al., 2019; Jamwal et al., 2019), indicating the potential for a multi-faceted application of probiotic strains CFS. Several antimicrobial compounds in CFS were heat-resistant, which may be useful as food preservatives for manufacturing foods that are subject to high heat during processing, e.g., spray drying of PIF (Jamwal et al., 2019; Lin and Pan, 2019). Specifically, CFS can contain postbiotic metabolites, such as SCFA, which will be discussed later in this review, and bacteriocins that have antimicrobial properties against pathogenic bacteria and viruses including E. coli, C. difficile, C. sakazakii, Salmonella spp., influenza, and rotavirus (Mantziari et al., 2020; Nataraj et al., 2020). As postbiotics are composed of the bioactive components of an inactivated probiotic, their mechanisms to exert a health benefit on the host are similar to their live counterpart. Postbiotics can modulate the immune system through the production of toll-like receptors, reduce the adhesion of pathogens to the gut epithelium by competitive exclusion, or inhibit the growth of pathogens through bacteriocins (Dunand et al., 2019; Mantziari et al., 2020; Mayorgas et al., 2021). As shown in Table 5, there have been other successful applications of probiotic strains and their CFS in inhibiting C. sakazakii.
Prebiotics
Prebiotics were originally defined as non-digestible carbohydrates that are metabolized by the commensal gut microbiota in the colon (Bezkorovainy, 2001). Recently, the definition of prebiotics has been modified to include substrates used by the GI microbiota that confer health benefits to the host, extending the substrate candidates to include more than just carbohydrates, e.g., phenols and phytochemicals (Gibson et al., 2017). To exert health benefits, prebiotics should resist host digestion, be fermented by gut microbes, and selectively promote the growth of beneficial bacteria, e.g., Lactobacillus and Bifidobacterium spp. (Gibson et al., 2017). In infants, prebiotics can also improve nutrient absorption, decrease the translocation of pathogenic bacteria through gut maturation, and enhance overall GI health (Davis et al., 2017). Common prebiotics included in PIF sold in Canada typically contain GOS, inulin, or polydextrose (PDX) to provide the aforementioned health benefits to infants (Davis et al., 2017). Goat and bovine-milk derived oligosaccharides are currently being investigated for their prebiotic properties and their beneficial effects as part of infant formula (Simeoni et al., 2016; Jakobsen et al., 2019; Leong et al., 2019; Gallier et al., 2020). Goat and cow milk were found to exert bifidogenic effects similar to those from human milk (Simeoni et al., 2016; Jakobsen et al., 2019; Gallier et al., 2020). Additionally, goat milk oligosaccharides reduced the adhesion of E. coli NCTC 10418 and S. Typhimurium to Caco-2 cells (Leong et al., 2019). Bifidobacterium infantis was found to metabolize HMOs, but the enzymes used to break down HMOs are not present in other Bifidobacterium spp. more commonly found in adults such as B. longum and B. lactis (Gibson et al., 2017). Furthermore, prebiotics were shown to be heat-resistant (Huebner et al., 2008; Moore, 2011; Vega and Zuniga-Hansen, 2015), thus providing important nutrients for gut maturation even in heat-treated products, e.g., pasteurized donor breast milk (Mueller et al., 2015).
Prebiotics, including breast milk HMOs, are capable of preventing pathogens and their toxins from adhering to intestinal cells by acting as molecular decoys in the GI tract, indicating why early breast-feeding is a key factor in developing a healthy gut microbiome (Morrow et al., 2005; Le Doare et al., 2018). HMOs are the first prebiotics consumed by infants, comprising approximately 1.1% of breast milk, and are broken down by certain species of bifidobacteria such as Bifidobacterium longum subsp. infantis, B. longum subsp. longum, B. bifidum, and B. breve (Thomson et al., 2018; James et al., 2019; Lawson et al., 2020). However, other species of Bifidobacterium and other gut microbiota can cross-feed on different metabolites, including acetate and fucose, released from HMO degradation (Boehm et al., 2005; Johnson-Henry et al., 2016; Thomson et al., 2018; James et al., 2019; Lawson et al., 2020). Breast feeding is typically done for the first 6months of life (Robertson et al., 2019) and during this time, B. infantis is likely to dominate the infant gut microbiota due to the abundance of HMOs (Vatanen et al., 2019). The abundance of B. infantis and other Bifidobacterium spp. helps to develop a healthy infant gut microbiome in addition to modulating the immune system (Johnson-Henry et al., 2016; Vatanen et al., 2019). Sialic acid on HMOs may play a role in reducing the risk of NEC pathogenesis through complex interactions with the infant immune system, but the mechanisms are unclear (Bode, 2018). One possible method is through the degradation of sialic acid by bifidobacteria due to the production of sialidases located on the nan-nag locus (Egan et al., 2014; Lawson et al., 2020; Wu et al., 2021), which results in competition for nutrients with opportunistic pathogens, such as C. sakazakii and C. difficile, that can also metabolize sialic acid in the colon (Egan et al., 2014).
Prebiotics also can prevent pathogenic colonization and growth, with the latter effect being due to gut microbe metabolism. GOS, for example, contains structures similar to the microvilli present in the colon, and can interfere with pathogen specific receptors and subsequent binding (Tomar et al., 2015). The effects of prebiotics on C. sakazakii and NEC are not well-known, but GOS and PDX have been shown to reduce the adherence of C. sakazakii to the HEp-2 cell line by 42–71% when used separately or combined (Quintero et al., 2011). However, the concentrations of GOS and PDX used in the latter study were higher than those present in PIF (Kent et al., 2015). Srinivasjois et al. (2013) showed that supplementing infant formula with different prebiotic oligosaccharides significantly improved the growth of bifidobacteria but did not decrease the risk of NEC or other illnesses associated with C. sakazakii infection in preterm infants. Additionally, prebiotics can affect C. difficile toxicity differently as demonstrated by analyzing changes in HT-29 cell attachment and growth, thus indicating that carbon source can play a factor in pathogen inhibition (Valdés-Varela et al., 2016). In addition to indirectly decreasing the risk of infection by reducing pathogen adhesion, prebiotics can also provide benefits to the host once metabolized by the gut microbiota.
Role of SCFA in Inhibiting Pathogens and Promoting the Development of a Healthy Gut
Some SCFAs, such as butyrate and propionate, are commonly produced by commensal bacteria in the GI tract and have been extensively studied (Nagpal et al., 2018). Reduced levels of SCFA in the GI tract have been linked to various illnesses including obesity, type II diabetes, and autoimmune diseases in adults, but their association with metabolic and immune conditions in infants is not well studied (Nagpal et al., 2018; Differding et al., 2020). In the human intestinal tract, butyrate and propionate regulate inflammation, epithelial barriers, and intestinal motility and may be able to reduce the risk of various medical conditions such as colitis and intestinal inflammation (Canani et al., 2011; Engels et al., 2016). However, it should be noted that acetate and lactate both have an important role to play as precursors for the production of butyrate and propionate (Venegas et al., 2019). Some gut bacteria, such as Eubacterium hallii, can produce SCFAs from metabolites created by bifidobacterial fermentation, which can play a role in inhibiting pathogens in the gut (Engels et al., 2016; Schwab et al., 2017). The acid conjugates of SCFA, such as acetic acid, can diffuse freely in the cytoplasm of pathogens, and this may ultimately compromise metabolic functions and disturb cell physiology (Sun and O’Riordan, 2013).
In moderate quantities, SCFAs can regulate conditions associated with dysbiosis in infants, e.g., maintain the gut barrier, and reduce inflammation caused by the onset of NEC (Differding et al., 2020; Zheng et al., 2020), but higher concentrations of SCFA may be toxic to infants. The normal concentration of total SCFA in humans range from 20 to 140 mM in the large intestine, but concentrations >300mM may be fatal to infants (Nafday et al., 2005). While the mechanism of SCFA toxicity requires further study, excess quantities of SCFA can cause NEC, as it appears that they can reduce carbohydrate absorption and GI motility, which damages gut mucosal integrity (Lin, 2004) – the latter effect can increase SCFA accumulation and exacerbate intestinal injury (Roy et al., 2018). The potential risk of SCFA toxicity indicates that any underlying health conditions, such as poor digestion in infants, should be considered prior to the application of prebiotics. Conversely, high levels of SCFA in fecal samples may indicate lower microbial diversity in the gut and higher gut permeability, both of which are associated with several health complications including obesity, glycemia, and hypertension (De la Cuesta-Zuluaga et al., 2019). However, there is insufficient evidence on the role of a specific SCFA to maintain a healthy gut, as each metabolite is part of a complex gut ecosystem.
Prebiotics and SCFA can benefit host health, but their success is limited to the gut microbiota present in the host. If an individual lacks the microbes necessary to metabolize the prebiotics consumed, possibly due to dysbiosis, it is unlikely that the prebiotic will confer health benefits to the host (Gibson et al., 2017). In this scenario, the individual may benefit instead from a synbiotic supplementation.
Synbiotics
A synbiotic is defined as “a mixture comprising live microorganisms and substrate(s) selectively utilized by host microorganisms that confers a health benefit on the host” (Swanson et al., 2020). During its conception, synbiotic supplementation was thought to prolong the shelf-life of foods containing live microbes, increase the survival of probiotics, and stimulate metabolism of the gut microbiota (Gibson and Roberfroid, 1995). Other beneficial claims of synbiotics include larger populations of Lactobacillus and Bifidobacterium spp. in the GI tract, improved immune system function, and reduced risk of bacterial infection in higher risk patients (Pandey et al., 2015). Since the gut microbiota and probiotics respond differently to prebiotics, compatibility is an important factor in formulating synbiotics.
When Lactobacillus strains were grown in the presence of different prebiotics, such as inulin, fructooligosaccharide (FOS), and lactulose, L. plantarum demonstrated the most rapid growth with inulin (Nagpal and Kaur, 2011). Differences in oligosaccharide fermentation were shown to exist between L. lactis subsp. lactis strains, as one strain possesses specific enzymes for the transport and hydrolysis of palatinose, yet another strain was unable to ferment palatinose (Pranckute et al., 2014). In another study, Bifidobacterium spp. were demonstrated to be better at fermenting glucooligosaccharides as compared to Lactobacillus spp. possibly due to the presence of numerous genes for carbohydrate metabolism (Grimoud et al., 2010). Similarly, β-galactosidases in bifidobacteria are better at hydrolyzing β(1→6) and β(1→3) linked GOS as compared to lactobacilli (Kittibunchakul et al., 2018). To summarize, different bacterial species and strains within a species produce unique enzymes that make them more compatible with certain prebiotics (Pranckute et al., 2014).
The use of synbiotic supplementation in infants has shown encouraging results (Chua et al., 2017; Vandenplas et al., 2017), but these studies are rarely conducted possibly due to ethical concerns. Ingestion of a synbiotic supplement in preterm infants can decrease the risk of disease by increasing the populations of beneficial gut microbiota and/or reducing pathogen adhesion (Luoto et al., 2014; Kent et al., 2015). Although more studies are needed to validate the findings, synbiotic supplementation in premature infants may have additional benefits, as randomized clinical trials have shown a decrease in NEC cases relative to the control group (Vongbhavit and Underwood, 2016).
Breast milk is known to be the ideal synbiotic as it not only contains beneficial bacteria, such as Lactobacillus, Bifidobacterium, Rothia, Enterococcus, and Veillonella spp., and prebiotic HMOs, but also provides immune factors to stimulate the immunological development of infants (Bertelsen et al., 2016; Le Doare et al., 2018; Robertson et al., 2019; Fehr et al., 2020; Lyons et al., 2020). Breast milk is associated with an increase in the levels of Bifidobacterium and, as the gut matures, Akkermansia muciniphila, Faecalibacterium prausnitzii and Lactobacillus spp. among other obligate anaerobes, also increase in numbers (Robertson et al., 2019). Akkermansia muciniphila and F. prausnitzii have been suggested to be candidates for next-generation probiotics due to their association with a healthy gut and immune system, and potential to reduce NEC (Underwood, 2017; Maftei, 2019).
To the best of our knowledge, there has not been any research on the effect of synbiotics on C. sakazakii, although several studies have shown reduced morbidity or mortality of NEC after synbiotic application (Dilli et al., 2015; Sreenivasa et al., 2015; Nandhini et al., 2016; Pehlevan et al., 2020), and other studies have shown the beneficial effects of synbiotics on other foodborne pathogens such as Salmonella enterica, Shigella sonnei, and enteropathogenic and enterohemorrhagic E. coli (Likotrafiti et al., 2016; Kusmivati and Wahyuningsih, 2018; Shanmugasundaram et al., 2019; Tabashsum et al., 2019; Piatek et al., 2020). The antimicrobial potential of synbiotics is likely dependent on the pathogen-synbiotic combination as E. coli and Campylobacter jejuni were inhibited by synbiotics when cultured in a multi-batch culture system containing a synbiotic with a pathogen, but in a single continuous culture system only C. jejuni was inhibited (Fooks and Gibson, 2003). The excess carbon source in the continuous culture prevented nutrient competition, suggesting that antimicrobial compounds, i.e., SCFA, were produced, and that inhibition efficacy can differ depending on the pathogen-probiotic combination (Fooks and Gibson, 2003). Valdés-Varela et al. (2016) further expanded on the species dependency premise to include substrate dependency. For example, B. longum and B. breve, instead of B. bifidum or B. animalis and in the presence of FOS instead of inulin, inhibited C. difficile (Valdés-Varela et al., 2016). Another study noted that B. longum in the presence of the prebiotic isomaltooligosaccharides inhibited E. coli O157:H7 and E. coli O86 better than Lactobacillus fermentum with FOS (Likotrafiti et al., 2016). The interaction between synbiotics and pathogens is complex and requires sophisticated models to better determine their efficacy as a potential treatment for the control of foodborne pathogens.
Conclusion
Cronobacter can cause severe illness in infants, especially those who are premature or of low-birthweight. Due to their ubiquity and resilience, current preventative measures to control invasive Cronobacter infections during early infancy would benefit from alternative approaches to minimize any potential risks to infants. Probiotics, prebiotics, and synbiotics are well-placed to play a major role in this regard and can potentially act to prevent intestinal colonization of infants by pathogenic Cronobacter spp. or other enteropathogens. However, current research on their efficacy to inactivate or inhibit the growth of pathogenic Cronobacter spp. is limited. While there is evidence that the consumption of probiotics, prebiotics, and synbiotics can reduce the risk of infection through competitive exclusion, the production of antimicrobial compounds, or competition for nutrients, further research is required to validate these hypotheses using more complex in vitro models and in vivo studies. The development of novel models that simulate the human gut provides an exciting opportunity to study in real-time the interactions between Cronobacter spp. and the native infant gut microbiota in vitro.
Author Contributions
AK researched, wrote the topics within the manuscript, and revised the manuscript. VP, JF, and LG revised the manuscript including grammar, syntax, and information gap. All authors contributed to the article and approved the submitted version.
Funding
We would like to acknowledge the financial support from the Canada First Research Excellence Fund (CFREF) through the Food from Thought initiative. Grant #499114.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ackerberg, T. S., Labuschagne, I. L., and Lombard, M. J. (2012). The use of prebiotics and probiotics in infant formula. S. Afr. Fam. Pract. 54, 321–323. doi: 10.1080/20786204.2012.10874243
AEProbio. (2020). Clinical Guide to Probiotic Products Available in Canada. Available at: http://www.probioticchart.ca/PBCPediatricHealth.html?utm_source=pediatric_ind&utm_medium=civ&utm_campaign=CDN (Accessed June 14, 2020).
Alberta Health Services. (2017). Nutrition Guideline Healthy Infants and Young Children Prebiotics and Probiotics. Available at: https://www.albertahealthservices.ca/assets/info/nutrition/if-nfs-ng-healthy-infant-prebiotics-probiotics.pdf (Accessed June 7, 2020).
Alsonosi, A. M., Holy, O., and Forsythe, S. J. (2018). Characterization of the pathogenicity of clinical Cronobacter malonaticus strains based on the tissue culture investigations. Antonie Van Leeuwenhoek 112, 435–450. doi: 10.1007/s10482-018-1178-6
Aly, M. A., Domig, K. J., Kneifel, W., and Reimhult, E. (2019). Whole genome sequencing-based comparison of food isolates of Cronobacter sakazakii. Front. Microbiol. 10:1464. doi: 10.3389/fmicb.2019.01464
Andrews, J. (2011). Cronobacter: FDA, CDC Find No Connection to Infant Formula. Available at: https://www.foodsafetynews.com/2011/12/cronobacter-fda-and-cdc-find-no-connection-to-formula/#.W1jQm9JKiUm (Accessed June 20, 2020).
Arseni, A., Malamou-Ladas, E., Koutsia, C., Xanthou, M., and Trikka, E. (1987). Outbreak of colonization of neonates with Enterobacter sakazakii. J. Hosp. Infect. 9, 143–150. doi: 10.1016/0195-6701(87)90052-1
Awaisheh, S. S., Al-Nabulsi, A. A., Osaili, T. M., Ibrahim, S., and Holley, R. (2013). Inhibition of Cronobacter sakazakii by heat labile bacteriocins produced by probiotic lab isolated from healthy infants. J. Food Sci. 78, 1416–1420. doi: 10.1111/1750-3841.12209
Bäckhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004
Bai, Y., Yu, H., Guo, D., Fei, S., and Shi, C. (2019). Survival and environmental stress resistance of Cronobacter sakazakii exposed to vacuum or air packaging and stored at different temperatures. Front. Microbiol. 10:303. doi: 10.3389/fmicb.2019.00303
Baldwin, A., Loughlin, M., Caubilla-Barron, J., Kucerova, E., Manning, G., Dowson, C., et al. (2009). Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 9:223. doi: 10.1186/1471-2180-9-223
Begley, M., Gahan, C. G., and Hill, C. (2002). Bile stress response in listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68, 6005–6012. doi: 10.1128/AEM.68.12.6005-6012.2002
Berhilevych, O., and Kasianchuk, V. (2017). Identification of Cronobacter spp (Enterobacter sakazakii) from raw milk and environmental samples of dairy farms. East.-Eur. J. Enterp. Technol. 6, 4–10. doi: 10.15587/1729-4061.2017.114637
Bertelsen, R. J., Jensen, E. T., and Ringel-Kulka, T. (2016). Use of probiotics and prebiotics in infant feeding. Best Pract. Res. Clin. Gastroenterol. 30, 39–48. doi: 10.1016/j.bpg.2016.01.001
Bezkorovainy, A. (2001). Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73, 399–405. doi: 10.1093/ajcn/73.2.399s
Biedermann, L., and Rogler, G. (2015). The intestinal microbiota: its role in health and disease. Eur. J. Pediatr. 174, 151–167. doi: 10.1007/s00431-014-2476-2
Binda, S., Hill, C., Johansen, E., Obis, D., Pot, B., Sanders, M. E., et al. (2020). Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front. Microbiol. 11:1662. doi: 10.3389/fmicb.2020.01662
Block, C., Peleg, O., Minster, N., Bar-Oz, B., Simhon, A., Arad, I., et al. (2002). Cluster of neonatal infections in Jerusalem due to unusual biochemical variant of Enterobacter sakazakii. Eur. J. Clin. Microbiol. Infect. Dis. 21, 613–616. doi: 10.1007/s10096-002-0774-5
Bocchi, S., Sagheddu, V., Elli, M., Lim, C. Y., and Morelli, L. (2020). The synergistic interaction between probiotics and food affects their beneficial features. Adv. Nutr. Food Sci. 2020, 1–12. doi: 10.37722/ANAFS.20202
Bode, L. (2018). Human milk oligosaccharides in the prevention of necrotizing enterocolitis: a journey from in vitro and in vivo models to mother-infant cohort studies. Front. Pediatr. 6:385. doi: 10.3389/fped.2018.00385
Boehm, G., Stahl, B., Jelinek, J., Knol, J., Miniello, V., and Moro, G. E. (2005). Prebiotic carbohydrates in human milk and formulas. Acta Paediatr. 94, 18–21. doi: 10.1111/j.1651-2227.2005.tb02149.x
Bowen, A. B., and Braden, C. R. (2006). Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 12, 1185–1189. doi: 10.3201/eid1208.051509
Campana, R., Federici, S., Ciandrini, E., Manti, A., and Baffone, W. (2019). Lactobacillus spp. inhibit the growth of Cronobacter sakazakii ATCC 29544 by altering its membrane integrity. J. Food Sci. Technol. 56, 3962–3967. doi: 10.1007/s13197-019-03928-x
Canadian Food Inspection Agency. (2019). Probiotic claims. Available at: https://www.inspection.gc.ca/food-label-requirements/labelling/industry/health-claims-on-food-labels/eng/1392834838383/1392834887794?chap=10 (Accessed June 13, 2020).
Canani, R. B., Di Costanzo, M., Leone, L., Pedata, M., Meli, R., and Calignano, A. (2011). Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 17, 1519–1528. doi: 10.3748/wjg.v17.i12.1519
Capurso, L. (2019). Thirty years of lactobacillus rhamnosus GG: a review. J. Clin. Gastroenterol. 53, S1–S41. doi: 10.1097/MCG.0000000000001170
Carey, C. M., Kostrzynska, M., Ojha, S., and Thompson, S. (2008). The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J. Microbiol. Methods 73, 125–132. doi: 10.1016/j.mimet.2008.01.014
Caubilla-Barron, J., Hurrell, E., Townsend, S., Cheetham, P., Loc-Carrillo, C., Fayet, O., et al. (2007). Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J. Clin. Microbiol. 45, 3979–3985. doi: 10.1128/JCM.01075-07
Cech, G. M., Szalewska-Pałasz, A., Kubiak, K., Malabirade, A., Grange, W., Arluison, V., et al. (2016). The Escherichia coli Hfq protein: an unattended DNA-transactions regulator. Front. Mol. Biosci. 3, 36. doi: 10.3389/fmolb.2016.00036
Centre for Disease Control. (2020a). Frequently Asked Questions. Available at: https://www.cdc.gov/cronobacter/technical.html (Accessed July 14, 2020).
Centre for Disease Control. (2020b). Cronobacter infection and infants. Available at: https://www.cdc.gov/cronobacter/sources.html (Accessed April 13, 2021).
Charchoghlyan, H., Kwon, H., Hwang, D. J., Lee, J. S., Lee, J., and Kim, M. (2016). Inhibition of Cronobacter sakazakii by lactobacillus acidophilus n.v. Er2 317/402. Korean J. Food Sci. Anim. Resour. 36, 635–640. doi: 10.5851/kosfa.2016.36.5.635
Chemical Inspection and Regulation Service. (2019). SAMR Issues Provisions for Declaration and Review of Probiotic Health Food (Draft) for Public Comments. Available at: http://www.cirs-reach.com/news-and-articles/SAMR-Issues-Provisions-for-Declaration-and-Review-of-Probiotic-Health-Food-Draft-for-Public-Comments.html (Accessed March 25, 2021).
Chua, M. C., Ben-Amor, K., Lay, C., Goh, A. E., Chiang, W. C., Rao, R., et al. (2017). Effect of synbiotic on the gut microbiota of cesarean delivered infants: a randomized, double-blind, multicenter study. J. Pediatr. Gastroenterol. Nutr. 65, 102–106. doi: 10.1097/MPG.0000000000001623
Collado, M. C., Cernada, M., Neu, J., Pérez-Martínez, G., Gormaz, M., and Vento, M. (2015). Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr. Res. 77, 726–731. doi: 10.1038/pr.2015.54
Cordonnier, C., Thévenot, J., Etienne-Mesmin, L., Alric, M., Livrelli, V., and Blanquet-Diot, S. (2016). Probiotic and enterohemorrhagic Escherichia coli: An effective strategy against a deadly enemy? Crit. Rev. Microbiol. 43, 116–132. doi: 10.1080/1040841X.2016.1185602
Davis, E. C., Wang, M., and Donovan, S. M. (2017). The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes 8, 143–171. doi: 10.1080/19490976.2016.1278104
De la Cuesta-Zuluaga, J., Mueller, N. T., Álvarez-Quintero, R., Velásquez-Mejía, E. P., Sierra, J. A., Corrales-Agudelo, V., et al. (2019). Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 11:51. doi: 10.3390/nu11010051
Differding, M. K., Benjamin-Neelon, S. E., Hoyo, C., Østbye, T., and Mueller, N. T. (2020). Timing of complementary feeding is associated with gut microbiota diversity and composition and short chain fatty acid concentrations over the first year of life. BMC Microbiol. 20:56. doi: 10.1186/s12866-020-01723-9
Dilli, D., Aydin, B., Fettah, N. D., Özyazıcı, E., Beken, S., Zenciroğlu, A., et al. (2015). The ProPre-save study: effects of probiotics and prebiotics alone or combined on necrotizing enterocolitis in very low birth weight infants. J. Pediatr. 166, 545–551. doi: 10.1016/j.jpeds.2014.12.004
Dittoe, D. K., Ricke, S. C., and Kiess, A. S. (2018). Organic acids and potential for modifying the avian gastrointestinal tract and reducing pathogens and disease. Front. Vet. Sci. 5:216. doi: 10.3389/fvets.2018.00216
Dunand, E., Burns, P., Binetti, A., Bergamini, C., Peralta, G. H., Forzani, L., et al. (2019). Postbiotics produced at laboratory and industrial level as potential functional food ingredients with the capacity to protect mice against salmonella infection. J. Appl. Microbiol. 127, 219–229. doi: 10.1111/jam.14276
Durack, J., and Lynch, S. V. (2019). The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 216, 20–40. doi: 10.1084/jem.20180448
Egan, M., O'Connell Motherway, M., Ventura, M., and van Sinderen, D. (2014). Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 80, 4414–4426. doi: 10.1128/AEM.01114-14
Elkhawaga, A. A., Hetta, H. F., Osman, N. S., Hosni, A., and El-Mokhtar, M. A. (2020). Emergence of Cronobacter sakazakii in cases of neonatal sepsis in upper Egypt: first report in North Africa. Front. Microbiol. 11:215. doi: 10.3389/fmicb.2020.00215
Emami, C. N., Mittal, R., Wang, L., Ford, H. R., and Prasadarao, N. V. (2011). Recruitment of dendritic cells is responsible for intestinal epithelial damage in the pathogenesis of necrotizing enterocolitis by Cronobacter sakazakii. J. Immunol. 186, 7067–7079. doi: 10.4049/jimmunol.1100108
Emami, C. N., Mittal, R., Wang, L., Ford, H. R., and Prasadarao, N. V. (2012). Role of neutrophils and macrophages in the pathogenesis of necrotizing enterocolitis caused by Cronobacter sakazakii. J. Surg. Res. 172, 18–28. doi: 10.1016/j.jss.2011.04.019
Engels, C., Ruscheweyh, H. J., Beerenwinkel, N., Lacroix, C., and Schwab, C. (2016). The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front. Microbiol. 7:713. doi: 10.3389/fmicb.2016.00713
European Commission. (2007). Guidance on Claims Made on Foods. Available at: https://ec.europa.eu/food/sites/food/files/safety/docs/labelling_nutrition_claim_reg-2006-124_guidance_en.pdf (Accessed March 25, 2021).
Fakruddin, M., Rahaman, M., Ahmed, M. M., and Hoque, M. M. (2014). Stress tolerant virulent strains of Cronobacter sakazakii from food. Biol. Res. 47, 1–12. doi: 10.1186/0717-6287-47-63
Fan, H., Chen, Z., Lin, R., Liu, Y., Wu, X., Puthiyakunnon, S., et al. (2019). Bacteroides fragilis strain ZY-312 defense against Cronobacter sakazakii-induced necrotizing enterocolitis in vitro and in a neonatal rat model. mSystems 4, e00305–e00319. doi: 10.1128/mSystems.00305-19
Farmer, J. J. III, Asbury, M. A., Hickman, F. W., and Brenner, D. J., Enterobacteriaceae Study Group (1980). Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 30, 569–584. doi: 10.1099/00207713-30-3-569
Feeney, A., Kropp, K. A., O’Connor, R., and Sleator, R. D. (2014). Cronobacter sakazakii: stress survival and virulence potential in an opportunistic foodborne pathogen. Gut Microbes 5, 711–718. doi: 10.4161/19490976.2014.983774
Fehr, K., Moossavi, S., Sbihi, H., Boutin, R. C., Bode, L., Robertson, B., et al. (2020). Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers’ milk and the infant gut: the CHILD cohort study. Cell Host Microbe 28, 285–297. doi: 10.1016/j.chom.2020.06.009
Fei, P., Jiang, Y., Feng, J., Forsythe, S. J., Li, R., and Zhou, Y. (2017b). Antibiotic and desiccation resistance of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and processing environments. Front. Microbiol. 8:316. doi: 10.3389/fmicb.2017.00316
Fei, P., Jiang, Y., Jiang, Y., Yuan, X., Yang, T., Chen, J., et al. (2017a). Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter sakazakii isolates from powdered infant formula collected from Chinese retail markets. Front. Microbiol. 8:2026. doi: 10.3389/fmicb.2017.02026.
Fijan, S. (2016). “Antimicrobial effect of probiotics against common pathogens” in Probiotics and Prebiotics in Human Nutrition and Health. eds. V. Rao and L. G. Rao (New York: IntechOpen), 191–221.
Flach, J., van der Waal, M. B., Kardinaal, A. F. M., Schloesser, J., Ruijschop, R. M. A. J., and Claassen, E. (2018). Probiotic research priorities for the healthy adult population: A review on the health benefits of lactobacillus rhamnosus GG and Bifidobacterium animalis subspecies lactis BB-12. Cogent Food Agric. 4:1452839. doi: 10.1080/23311932.2018.1452839
Food and Agriculture Organization of the United Nations/World Health Organization. (2002). Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Available at: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (Accessed June 16, 2020).
Food and Agriculture Organization of the United Nations/World Health Organization. (2006). Enterobacter sakazakii and Salmonella in powdered infant formula: Meeting report. Available at: https://apps.who.int/iris/bitstream/handle/10665/43547/9241563311_eng.pdf?sequence=1 (Accessed June 14, 2020).
Food and Agriculture Organization of the United Nations/World Health Organization. (2008). Enterobacter sakazakii (Cronobacter spp.) in powdered follow-up formulae. Microbiological Risk Assessment Series No. 15. Available at: https://apps.who.int/iris/bitstream/handle/10665/44032/9789241563796_eng.pdf?sequence=1 (Accessed June 14, 2020).
Food Standards Australia and New Zealand. (2013). Risk Analysis in Food Regulation. Available at: https://www.foodstandards.gov.au/publications/riskanalysisfoodregulation/Documents/risk-analysis-food-regulation-intro-pdf1.pdf (Accessed March 25, 2021).
Food Standards Australia and New Zealand. (2021). Notified Food-Health Relationships to Make a Health Claim. Available at: https://www.foodstandards.gov.au/industry/labelling/fhr/Pages/default.aspx?k=probiotic (Accessed March 25, 2021).
Fooks, L. J., and Gibson, G. R. (2003). Mixed culture fermentation studies on the effects of synbiotics on the human intestinal pathogens campylobacter jejuni and Escherichia coli. Anaerobe 9, 231–242. doi: 10.1016/S1075-9964(03)00043-X
Forsythe, S. J. (2014). “Powdered infant formula,” in The Microbiological Safety of Low Water Activity Foods and Spices. ed. J. B. Gurtler (New York, NY: Springer), 177–211.
Forsythe, S. J. (2018). Updates on the Cronobacter genus. Annu. Rev. Food Sci. Technol. 9, 23–44. doi: 10.1146/annurev-food-030117-012246
Friedemann, M. (2009). Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1297–1304. doi: 10.1007/s10096-009-0779-4
Gallier, S., Van den Abbeele, P., and Prosser, C. (2020). Comparison of the bifidogenic effects of goat and cow milk-based infant formulas to human breast milk in an in vitro gut model for 3-month-old infants. Front. Nutr. 7:608495. doi: 10.3389/fnut.2020.608495
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502. doi: 10.1038/nrgastro.2017.75
Gibson, G. R., and Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. doi: 10.1093/jn/125.6.1401
Grimoud, J., Durand, H., Courtin, C., Monsan, P., Ouarné, F., Theodorou, V., et al. (2010). In vitro screening of probiotic lactic acid bacteria and prebiotic glucooligosaccharides to select effective synbiotics. Anaerobe 16, 493–500. doi: 10.1016/j.anaerobe.2010.07.005
Gurien, L. A., Stallings-Archer, K., and Smith, S. D. (2018). Probiotic Lactococcus lactis decreases incidence and severity of necrotizing enterocolitis in a preterm animal model. J. Neonatal-Perinatal Med. 11, 65–69. doi: 10.3233/NPM-181740
Health Canada. (2009). Guidance Document – The Use of Probiotic Microorganisms in Food. Available at: https://www.canada.ca/en/health-canada/services/food-nutrition/legislation-guidelines/guidance-documents/guidance-document-use-probiotic-microorganisms-food-2009.html (Accessed June 13, 2020).
Henry, M., and Fouladkhah, A. (2019). Outbreak history, biofilm formation, and preventive measures for control of Cronobacter sakazakii in infant formula and infant care settings. Microorganisms 7, 1–22. doi: 10.3390/microorganisms7030077
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Holý, O., Cruz-Córdova, A., Xicohtencatl-Cortes, J., Hochel, I., Parra-Flores, J., Petrželová, J., et al. (2019). Occurrence of virulence factors in Cronobacter sakazakii and Cronobacter malonaticus originated from clinical samples. Microb. Pathog. 127, 250–256. doi: 10.1016/j.micpath.2018.12.011
Hsiao, W. L., Ho, W. L., and Chou, C. C. (2010). Sub-lethal heat treatment affects the tolerance of Cronobacter sakazakii BCRC 13988 to various organic acids, simulated gastric juice and bile solution. Int. J. Food Microbiol. 144, 280–284. doi: 10.1016/j.ijfoodmicro.2010.10.006
Hu, S., Yu, Y., and Xiao, X. (2018). Stress resistance, detection and disinfection of Cronobacter spp. in dairy products: a review. Food Control 85, 400–415. doi: 10.1016/j.foodcont.2017.10.014
Huebner, J., Wehling, R. L., Parkhurst, A., and Hutkins, R. W. (2008). Effect of processing conditions on the prebiotic activity of commercial prebiotics. Int. Dairy J. 18, 287–293. doi: 10.1016/j.idairyj.2007.08.013
Hunter, C. J., and Bean, J. F. (2013). Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J. Perinatol. 33, 581–585. doi: 10.1038/jp.2013.26
Hunter, C., Dimaguila, M. A. V. T., Gal, P., Wimmer, J. E., Ransom, J. L., Carlos, R. Q., et al. (2012). Effect of routine probiotic, lactobacillus reuteri DSM 17938, use on rates of necrotizing enterocolitis in neonates with birthweight < 1000 grams: a sequential analysis. BMC Pediatr. 12:142. doi: 10.1186/1471-2431-12-142
Hurrell, E., Kucerova, E., Loughlin, M., Caubilla-Barron, J., and Forsythe, S. J. (2009). Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, salmonella serovars and other Enterobacteriaceae. Int. J. Food Microbiol. 136, 227–231. doi: 10.1016/j.ijfoodmicro.2009.08.007
International Probiotics Association. (2017). IPA guidelines to qualify a microorganism to be termed as ‘probiotic’. Available at: https://internationalprobiotics.org/wp-content/uploads/20170602-IPA-guidelines-to-qualify-a-microorganism-as-probiotic.pdf (Accessed March 25, 2021).
Iversen, C., Waddington, M., Farmer, J. J. III, and Forsythe, S. J. (2006). The biochemical differentiation of Enterobacter sakazakii genotypes. BMC Microbiol. 6:94. doi: 10.1186/1471-2180-6-94
Iwatani, S., and Yamamoto, N. (2019). Functional food products in Japan: a review. Food Sci. Human Wellness 8, 96–101. doi: 10.1016/j.fshw.2019.03.011
Jackson, E. E., Flores, J. P., Fernández-Escartín, E., and Forsythe, S. J. (2015). Reevaluation of a suspected Cronobacter sakazakii outbreak in Mexico. J. Food Prot. 78, 1191–1196. doi: 10.4315/0362-028X.JFP-14-563
Jacobs, C., Braun, P., and Hammer, P. (2011). Reservoir and routes of transmission of Enterobacter sakazakii (Cronobacter spp.) in a milk powder-producing plant. J. Dairy Sci. 94, 3801–3810. doi: 10.3168/jds.2011-4318
Jakobsen, L. M., Sundekilde, U. K., Andersen, H. J., Nielsen, D. S., and Bertram, H. C. (2019). Lactose and bovine milk oligosaccharides synergistically stimulate B. longum subsp. longum growth in a simplified model of the infant gut microbiome. J. Proteome Res. 18, 3086–3098. doi: 10.1021/acs.jproteome.9b00211
Jameelah, M., Dewanti-Hariyadi, R., and Nurjanah, S. (2018). Expression of rpoS, ompA and hfq genes of Cronobacter sakazakii strain Yrt2a during stress and viable but nonculturable state. Food Sci. Biotechnol. 27, 915–920. doi: 10.1007/s10068-018-0313-5
James, K., Bottacini, F., Contreras, J. I. S., Vigoureux, M., Egan, M., Motherway, M. O. C., et al. (2019). Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci. Rep. 9, 1–20. doi: 10.1038/s41598-019-51901-7
Jamwal, A., Sharma, K., Chauhan, R., Bansal, S., and Goel, G. (2019). Evaluation of commercial probiotic lactic cultures against biofilm formation by Cronobacter sakazakii. Intest. Res. 17, 192–201. doi: 10.5217/ir.2018.00106
Jang, H., Addy, N., Ewing, L., Beaubrun, J. J. G., Lee, Y., Woo, J., et al. (2018b). Whole-genome sequences of Cronobacter sakazakii isolates obtained from foods of plant origin and dried-food manufacturing environments. Genome Announc. 6, e00223–e00218. doi: 10.1128/genomeA.00223-18
Jang, H., Woo, J., Lee, Y., Negrete, F., Finkelstein, S., Chase, H. R., et al. (2018a). Draft genomes of Cronobacter sakazakii strains isolated from dried spices bring unique insights into the diversity of plant-associated strains. Stand. Genomic Sci. 13, 1–16. doi: 10.1186/s40793-018-0339-6
Janvier, A., Malo, J., and Barrington, K. J. (2014). Cohort study of probiotics in a north American neonatal intensive care unit. J. Pediatr. 164, 980–985. doi: 10.1016/j.jpeds.2013.11.025
Jaradat, Z. W., Al Mousa, W., Elbetieha, A., Al Nabulsi, A., and Tall, B. D. (2014). Cronobacter spp.–opportunistic food-borne pathogens: a review of their virulence and environmental-adaptive traits. J. Med. Microbiol. 63, 1023–1037. doi: 10.1099/jmm.0.073742-0
Johnson-Henry, K. C., Abrahamsson, T. R., Wu, R. Y., and Sherman, P. M. (2016). Probiotics, prebiotics, and synbiotics for the prevention of necrotizing enterocolitis. Adv. Nutr. 7, 928–937. doi: 10.3945/an.116.012237
Joseph, S., Desai, P., Ji, Y., Cummings, C. A., Shih, R., Degoricija, L., et al. (2012). Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS One 7:e49455. doi: 10.1371/journal.pone.0049455
Joseph, S., and Forsythe, S. J. (2011). Predominance of sequence type 4 in neonatal infections. Emerg. Infect. Dis. 17, 9–11. doi: 10.3201/eid1709.110260
Joseph, S., Hariri, S., Masood, N., and Forsythe, S. (2013). Sialic acid utilization by Cronobacter sakazakii. Microb. Inform. Exp. 3, 1–13. doi: 10.1186/2042-5783-3-3
Jungersen, M., Wind, A., Johansen, E., Christensen, J. E., Stuer-Lauridsen, B., and Eskesen, D. (2014). The science behind the probiotic strain Bifidobacterium animalis subsp. lactis BB-12®. Microorganisms 2, 92–110. doi: 10.3390/microorganisms2020092
Kalyantanda, G., Shumyak, L., and Archibald, L. K. (2015). Cronobacter species contamination of powdered infant formula and the implications for neonatal health. Front. Pediatr. 3:56. doi: 10.3389/fped.2015.00056
Kaur, S., Kaur, R., Rani, N., Sharma, S., and Joshi, M. (2021). “Sources and selection criteria of probiotics,” in Advances in Probiotics for Sustainable Food and Medicine. eds. G. Goel and A. Kumar (Singapore: Springer), 27–43.
Kavita, S., Pooranachithra, M., Singh, N., Prasanth, M. I., Balamurugan, K., and Goel, G. (2020). Lactobacillus gastricus BTM 7 prevents intestinal colonization by biofilm forming Cronobacter sakazakii in Caenorhabditis elegans model host. Antonie Van Leeuwenhoek 113, 1587–1600. doi: 10.1007/s10482-020-01466-7
Kent, R. M., Fitzgerald, G. F., Hill, C., Stanton, C., and Paul Ross, R. (2015). Novel approaches to improve the intrinsic microbiological safety of powdered infant milk formula. Nutrients 7, 1217–1244. doi: 10.3390/nu7021217
Kheadr, E., Zihler, A., Dabour, N., Lacroix, C., Le Blay, G., and Fliss, I. (2010). Study of the physicochemical and biological stability of pediocin PA-1 in the upper gastrointestinal tract conditions using a dynamic in vitro model. J. Appl. Microbiol. 109, 54–64. doi: 10.1111/j.1365-2672.2009.04644.x
Kim, S., Hwang, H., Kim, K. P., Yoon, H., Kang, D. H., and Ryu, S. (2015). Hfq plays important roles in virulence and stress adaptation in Cronobacter sakazakii ATCC 29544. Infect. Immun. 83, 2089–2098. doi: 10.1128/IAI.03161-14
Kittibunchakul, S., Maischberger, T., Domig, K. J., Kneifel, W., Nguyen, H. M., Haltrich, D., et al. (2018). Fermentability of a novel galacto-oligosaccharide mixture by lactobacillus spp. and Bifidobacterium spp. Molecules 23:3352. doi: 10.3390/molecules23123352
Knaus, U. G., Hertzberger, R., Pircalabioru, G. G., Yousefi, S. P. M., and Branco dos Santos, F. (2017). Pathogen control at the intestinal mucosa–H2O2 to the rescue. Gut Microbes 8, 67–74. doi: 10.1080/19490976.2017.1279378
Korakli, M., Gänzle, M. G., and Vogel, R. F. (2002). Metabolism by bifidobacteria and lactic acid bacteria of polysaccharides from wheat and rye, and exopolysaccharides produced by lactobacillus sanfranciscensis. J. Appl. Microbiol. 92, 958–965. doi: 10.1046/j.1365-2672.2002.01607.x
Kotsou, M. G., Mitsou, E. K., Oikonomou, I. G., and Kyriacou, A. A. (2008). In vitro assessment of probiotic properties of lactobacillus strains from infant gut microflora. Food Biotechnol. 22, 1–17. doi: 10.1080/08905430701707844
Kusmivati, N., and Wahyuningsih, T. D. (2018). “Effect of synbiotics Lactobacillus casei AP and inulin extract Dahlia pinnata L. in enteropathogenic Escherichia coli-induced diarrhea,” in 2018 1st International Conference on Bioinformatics, Biotechnology, and Biomedical Engineering-Bioinformatics and Biomedical Engineering, 1, 1–6. IEEE.
Lai, K. K. (2001). Enterobacter sakazakii infections among neonates, infants, children, and adults: case reports and a review of the literature. Medicine 80, 113–122. doi: 10.1097/00005792-200103000-00004
Lawson, M. A., O’Neill, I. J., Kujawska, M., Javvadi, S. G., Wijeyesekera, A., Flegg, Z., et al. (2020). Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 14, 635–648. doi: 10.1038/s41396-019-0553-2
Le Doare, K., Holder, B., Bassett, A., and Pannaraj, P. S. (2018). Mother’s Milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 9:361. doi: 10.3389/fimmu.2018.00361
Leong, A., Liu, Z., Almshawit, H., Zisu, B., Pillidge, C., Rochfort, S., et al. (2019). Oligosaccharides in goats’ milk-based infant formula and their prebiotic and anti-infection properties. Br. J. Nutr. 122, 441–449. doi: 10.1017/S000711451900134X
Leyer, G. J., and Johnson, E. A. (1993). Acid adaptation induces cross-protection against environmental stresses in salmonella typhimurium. Appl. Environ. Microbiol. 59, 1842–1847. doi: 10.1128/aem.59.6.1842-1847.1993
Li, Y., Chen, Q., Zhao, J., Jiang, H., Lu, F., Bie, X., et al. (2014). Isolation, identification, and antimicrobial resistance of Cronobacter spp. isolated from various foods in China. Food Control 37, 109–114. doi: 10.1016/j.foodcont.2013.09.017
Likotrafiti, E., Tuohy, K. M., Gibson, G. R., and Rastall, R. A. (2016). Antimicrobial activity of selected synbiotics targeted for the elderly against pathogenic Escherichia coli strains. Int. J. Food Sci. Nutr. 67, 83–91. doi: 10.3109/09637486.2015.1134444
Lin, J. (2004). Too much short chain fatty acids cause neonatal necrotizing enterocolitis. Med. Hypotheses 62, 291–293. doi: 10.1016/S0306-9877(03)00333-5
Lin, T. H., and Pan, T. M. (2019). Characterization of an antimicrobial substance produced by lactobacillus plantarum NTU 102. J. Microbiol. Immunol. Infect. 52, 409–417. doi: 10.1016/j.jmii.2017.08.003
Lis-Kuberka, J., and Orczyk-Pawiłowicz, M. (2019). Sialylated oligosaccharides and glycoconjugates of human milk. The impact on infant and newborn protection, development and well-being. Nutrients 11:306. doi: 10.3390/nu11020306
Liu, Q., Mittal, R., Emami, C. N., Iversen, C., Ford, H. R., and Prasadarao, N. V. (2012). Human isolates of Cronobacter sakazakii bind efficiently to intestinal epithelial cells in vitro to induce monolayer permeability and apoptosis. J. Surg. Res. 176, 437–447. doi: 10.1016/j.jss.2011.10.030
Lu, Y., Liu, P., Li, C., Sha, M., Fang, J., Gao, X., et al. (2019). Prevalence and genetic diversity of Cronobacter species isolated from four infant formula production factories in China. Front. Microbiol. 10:1938. doi: 10.3389/fmicb.2019.01938
Luoto, R., Ruuskanen, O., Waris, M., Kalliomäki, M., Salminen, S., and Isolauri, E. (2014). Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 133, 405–413. doi: 10.1016/j.jaci.2013.08.020
Lyons, K. E., Ryan, C. A., Dempsey, E. M., Ross, R. P., and Stanton, C. (2020). Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients 12:1039. doi: 10.3390/nu12041039
Maerani, M., Dewanti-Hariyadi, R., and Nurjanah, S. (2020). Expression of stress regulator and virulence genes of Cronobacter sakazakii strain Yrt2a as a response to acid stress. Food Sci. Biotechnol. 29, 1273–1279. doi: 10.1007/s10068-020-00763-1
Maftei, N. M. (2019). “Probiotic, prebiotic and synbiotic products in human health,” in Frontiers and New Trends in the Science of Fermented Food and Beverages. eds. R. L. Solutions-Oviedo and Á. de la C. Pech-Canul (London, UK: IntechOpen), 415–587.
Makras, L., and De Vuyst, L. (2006). The in vitro inhibition of gram-negative pathogenic bacteria by Bifidobacteria is caused by the production of organic acids. Int. Dairy J. 16, 1049–1057. doi: 10.1016/j.idairyj.2005.09.006
Mantziari, A., Salminen, S., Szajewska, H., and Malagón-Rojas, J. N. (2020). Postbiotics against pathogens commonly involved in pediatric infectious diseases. Microorganisms 8:1510. doi: 10.3390/microorganisms8101510
Mardaneh, J., and Dallal, M. M. S. (2016). Study of Cronobacter sakazakii strains isolated from powdered milk infant formula by phenotypic and molecular methods in Iran. Arch. Pediatr. Infect. Dis. 5:e38867. doi: 10.5812/pedinfect.38867
Mayorgas, A., Dotti, I., and Salas, A. (2021). Microbial metabolites, postbiotics, and intestinal epithelial function. Mol. Nutr. Food Res. 65:2000188. doi: 10.1002/mnfr.202000188
Milani, C., Duranti, S., Bottacini, F., Casey, E., Turroni, F., Mahony, J., et al. (2017). The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 81, e00036–e00017. doi: 10.1128/MMBR.00036-17
Minekus, M. (2015). “The TNO gastro-intestinal model (TIM)” in The Impact of Food Bioactives on Health. eds. K. Verhoeckx, P. Cotter, I. López-Expósito, C. Kleiveland, T. Lea, A. Mackie, et al. (Cham: Springer), 37–46.
Mohsin, M., Guenther, S., Schierack, P., Tedin, K., and Wieler, L. H. (2015). Probiotic Escherichia coli Nissle 1917 reduces growth, Shiga toxin expression, release and thus cytotoxicity of enterohemorrhagic Escherichia coli. Int. J. Med. Microbiol. 305, 20–26. doi: 10.1016/j.ijmm.2014.10.003
Molloy, C., Cagney, C., Fanning, S., and Duffy, G. (2010). Survival characteristics of Cronobacter spp. in model bovine gut and in the environment. Foodborne Pathog. Dis. 7, 671–675. doi: 10.1089/fpd.2009.0449
Moore, K. E. (2011). Biological Analysis of prebiotics in various processed food matrices. Dissertations, Theses, & Student Research in Food Science and Technology. Lincoln: University of Nebraska. 17.
Morrow, A. L., Ruiz-Palacios, G. M., Jiang, X., and Newburg, D. S. (2005). Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 135, 1304–1307. doi: 10.1093/jn/135.5.1304
Mueller, N. T., Bakacs, E., Combellick, J., Grigoryan, Z., and Dominguez-Bello, G. M. (2015). The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117. doi: 10.1016/j.molmed.2014.12.002
Nafday, S. M., Chen, W., Peng, L., Babyatsky, M. W., Holzman, I. R., and Lin, J. (2005). Short-chain fatty acids induce colonic mucosal injury in rats with various postnatal ages. Pediatr. Res. 57, 201–204. doi: 10.1203/01.PDR.0000150721.83224.89
Nagpal, R., and Kaur, A. (2011). Synbiotic effect of various prebiotics on in vitro activities of probiotic lactobacilli. Ecol. Food Nutr. 50, 63–68. doi: 10.1080/03670244.2011.539161
Nagpal, R., Wang, S., Ahmadi, S., Hayes, J., Gagliano, J., Subashchandrabose, S., et al. (2018). Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci. Rep. 8:12649. doi: 10.1038/s41598-018-30114-4
Nair, M. K. M., and Venkitanarayanan, K. (2007). Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 62, 664–669. doi: 10.1203/PDR.0b013e3181587864
Nair, M. K. M., Venkitanarayanan, K., Silbart, L. K., and Kim, K. S. (2009). Outer membrane protein A (OmpA) of Cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodborne Pathog. Dis. 6, 495–501. doi: 10.1089/fpd.2008.0228
Nandhini, L. P., Biswal, N., Adhisivam, B., Mandal, J., Bhat, B. V., and Mathai, B. (2016). Synbiotics for decreasing incidence of necrotizing enterocolitis among preterm neonates—a randomized controlled trial. J. Matern. Fetal Neonatal Med. 29, 821–825. doi: 10.3109/14767058.2015.1019854
Nataraj, B. H., Ali, S. A., Behare, P. V., and Yadav, H. (2020). Postbiotics-parabiotics: the new horizons in microbial biotherapy and functional foods. Microb. Cell Factories 19, 1–22. doi: 10.1186/s12934-020-01426-w
Neal-Kluever, A., Fisher, J., Grylack, L., Kakiuchi-Kiyota, S., and Halpern, W. (2019). Physiology of the neonatal gastrointestinal system relevant to the disposition of orally administered medications. Drug Metab. Dispos. 47, 296–313. doi: 10.1124/dmd.118.084418
Ohira, S., Ikeda, E., Kamijo, K., Nagai, T., Tsunemi, K., Uchiyama, N., et al. (2021). Pyosalpinx due to Cronobacter sakazakii in an elderly woman. BMC Womens Health 21:136. doi: 10.1186/s12905-021-01283-8
Pandey, K. R., Naik, S. R., and Vakil, B. V. (2015). Probiotics, prebiotics and synbiotics - a review. J. Food Sci. Technol. 52, 7577–7587. doi: 10.1007/s13197-015-1921-1
Pannaraj, P. S., Li, F., Cerini, C., Bender, J. M., Yang, S., Rollie, A., et al. (2017). Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 171, 647–654. doi: 10.1001/jamapediatrics.2017.0378
Park, H., Lee, K., Yeo, S., Shin, H., and Holzapfel, W. H. (2017). Autoinducer-2 quorum sensing influences viability of Escherichia coli O157: H7 under osmotic and in vitro gastrointestinal stress conditions. Front. Microbiol. 8:1077. doi: 10.3389/fmicb.2017.01077
Parra-Flores, J., Aguirre, J., Juneja, V., Jackson, E. E., Cruz-Córdova, A., Silva-Sanchez, J., et al. (2018b). Virulence and antibiotic resistance profiles of Cronobacter sakazakii and Enterobacter spp. involved in the diarrheic hemorrhagic outbreak in Mexico. Front. Microbiol. 9:2206. doi: 10.3389/fmicb.2018.02206
Parra-Flores, J., Cerda-Leal, F., Contreras, A., Valenzuela-Riffo, N., Rodríguez, A., and Aguirre, J. (2018a). Cronobacter sakazakii and microbiological parameters in dairy formulas associated with a food alert in Chile. Front. Microbiol. 9:1708. doi: 10.3389/fmicb.2018.01708
Patrick, M. E., Mahon, B. E., Greene, S. A., Rounds, J., Cronquist, A., Wymore, K., et al. (2014). Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg. Infect. Dis. 20, 1536–1539. doi: 10.3201/eid2009.140545
Pehlevan, O. S., Benzer, D., Gursoy, T., Karatekin, G., and Ovali, F. (2020). Synbiotics use for preventing sepsis and necrotizing enterocolitis in very low birth weight neonates: a randomized controlled trial. Clin. Exp. Pediatr. 63, 226–231. doi: 10.3345/cep.2019.00381
Piatek, J., Krauss, H., Ciechelska-Rybarczyk, A., Bernatek, M., Wojtyla-Buciora, P., and Sommermeyer, H. (2020). In-vitro growth inhibition of bacterial pathogens by probiotics and a Synbiotic: product composition matters. Int. J. Environ. Res. Public Health 17:3332. doi: 10.3390/ijerph17093332
Piqué, N., Berlanga, M., and Miñana-Galbis, D. (2019). Health benefits of heat-killed (Tyndallized) probiotics: An overview. Int. J. Mol. Sci. 20:2534. doi: 10.3390/ijms20102534
Poquet, L., and Wooster, T. J. (2016). Infant digestion physiology and the relevance of in vitro biochemical models to test infant formula lipid digestion. Mol. Nutr. Food Res. 60, 1876–1895. doi: 10.1002/mnfr.201500883
Pranckute, R., Kaunietis, A., Kuisiene, N., and Čitavičius, D. (2014). Development of synbiotics with inulin, palatinose, α-cyclodextrin and probiotic bacteria. Pol. J. Microbiol. 63, 33–41. doi: 10.33073/pjm-2014-005
Quintero, M., Maldonado, M., Perez-Munoz, M., Jimenez, R., Fangman, T., Rupnow, J., et al. (2011). Adherence inhibition of Cronobacter sakazakii to intestinal epithelial cells by prebiotic oligosaccharides. Curr. Microbiol. 62, 1448–1454. doi: 10.1007/s00284-011-9882-8
Rastogi, S., Mittal, V., and Singh, A. (2021). Selection of potential probiotic bacteria from exclusively breastfed infant Faeces with antagonistic activity Against multidrug-resistant ESKAPE pathogens. Probiotics Antimicrob. Proteins 13, 739–750. doi: 10.1007/s12602-020-09724-w
Ray, P., Das, A., Gautam, V., Jain, N., Narang, A., and Sharma, M. (2007). Enterobacter sakazakii in infants: novel phenomenon in India. Indian J. Med. Microbiol. 25, 408–410. doi: 10.1016/S0255-0857(21)02063-6
Robertson, R. C., Manges, A. R., Finlay, B. B., and Prendergast, A. J. (2019). The human microbiome and Child growth – first 1000 days and Beyond. Trends Microbiol. 27, 131–147. doi: 10.1016/j.tim.2018.09.008
Roussel, C., De Paepe, K., Galia, W., De Bodt, J., Chalancon, S., Leriche, F., et al. (2020). Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol. 18:141. doi: 10.1186/s12915-020-00860-x
Roy, S. K., Meng, Q., Sadowitz, B. D., Kollisch-Singule, M., Yepuri, N., Satalin, J., et al. (2018). Enteral administration of bacteria fermented formula in newborn piglets: A high fidelity model for necrotizing enterocolitis (NEC). PLoS One 13:e0201172. doi: 10.1371/journal.pone.0201172
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M., et al. (2021). The international scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Schwab, C., Ruscheweyh, H. J., Bunesova, V., Pham, V. T., Beerenwinkel, N., and Lacroix, C. (2017). Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front. Microbiol. 8:95. doi: 10.3389/fmicb.2017.00095
Shanmugasundaram, R., Mortada, M., Cosby, D. E., Singh, M., Applegate, T. J., Syed, B., et al. (2019). Synbiotic supplementation to decrease salmonella colonization in the intestine and carcass contamination in broiler birds. PLoS One 14:e0223577. doi: 10.1371/journal.pone.0223577
Shi, C., Sun, Y., Liu, Z., Guo, D., Sun, H., Sun, Z., et al. (2017). Inhibition of Cronobacter sakazakii virulence factors by citral. Sci. Rep. 7:43243. doi: 10.1038/srep43243
Simeoni, U., Berger, B., Junick, J., Blaut, M., Pecquet, S., Rezzonico, E., et al. (2016). Gut microbiota analysis reveals a marked shift to bifidobacteria by a starter infant formula containing a synbiotic of bovine milk-derived oligosaccharides and B ifidobacterium animalis subsp. lactis CNCM I-3446. Environ. Microbiol. 18, 2185–2195. doi: 10.1111/1462-2920.13144
Singh, N., Goel, G., and Raghav, M. (2015). Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 6, 433–440. doi: 10.1080/21505594.2015.1036217
Sistrunk, J. R., Nickerson, K. P., Chanin, R. B., Rasko, D. A., and Faherty, C. S. (2016). Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin. Microbiol. Rev. 29, 819–836. doi: 10.1128/CMR.00031-16
Sonbol, H., Joseph, S., Mcauley, C. M., Craven, H. M., and Forsythe, S. J. (2013). Multilocus sequence typing of Cronobacter spp. from powdered infant formula and milk powder production factories. Int. Dairy J. 30, 1–7. doi: 10.1016/j.idairyj.2012.11.004
Sreenivasa, B., Kumar, P. S., Suresh Babu, M. T., and Ragavendra, K. (2015). Role of synbiotics in the prevention of necrotizing enterocolitis in preterm neonates: a randomized controlled trial. Int. J. Contemp. Pediatr. 2, 127–130. doi: 10.5455/2349-3291.ijcp20150512
Srinivasjois, R., Rao, S., and Patole, S. (2013). Prebiotic supplementation in preterm neonates: updated systematic review and meta-analysis of randomised controlled trials. Clin. Nutr. 32, 958–965. doi: 10.1016/j.clnu.2013.05.009
Sun, Y., and O’Riordan, M. X. (2013). Regulation of bacterial pathogenesis by intestinal short-chain fatty acids. Adv. Appl. Microbiol. 85, 93–118. doi: 10.1016/B978-0-12-407672-3.00003-4
Swanson, K. S., Gibson, G. R., Hutkins, R., Reimer, R. A., Reid, G., Verbeke, K., et al. (2020). The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 17, 687–701. doi: 10.1038/s41575-020-0344-2
Szajewska, H., and Hojsak, I. (2020). Health benefits of lactobacillus rhamnosus GG and Bifidobacterium animalis subspecies lactis BB-12 in children. Postgrad. Med. 132, 441–451. doi: 10.1080/00325481.2020.1731214
Tabashsum, Z., Peng, M., Bernhardt, C., Patel, P., Carrion, M., and Biswas, D. (2019). Synbiotic-like effect of linoleic acid overproducing lactobacillus casei with berry phenolic extracts against pathogenesis of enterohemorrhagic Escherichia coli. Gut Pathog. 11:41. doi: 10.1186/s13099-019-0320-y
Taipale, T. J., Pienihäkkinen, K., Isolauri, E., Jokela, J. T., and Söderling, E. M. (2016). Bifidobacterium animalis subsp. lactis BB-12 in reducing the risk of infections in early childhood. Pediatr. Res. 79, 65–69. doi: 10.1038/pr.2015.174
Tennant, S. M., Hartland, E. L., Phumoonna, T., Lyras, D., Rood, J. I., Robins-Browne, R. M., et al. (2008). Influence of gastric acid on susceptibility to infection with ingested bacterial pathogens. Infect. Immun. 76, 639–645. doi: 10.1128/IAI.01138-07
Thomson, P., Medina, D. A., and Garrido, D. (2018). Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol. 75, 37–46. doi: 10.1016/j.fm.2017.09.001
Tomar, S. K., Anand, S., Sharma, P., Sangwan, V., and Mandal, S. (2015). “Role of probiotic, prebiotics, synbiotics and postbiotics in inhibition of pathogens,” in The Battle against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs. ed. A. Méndez-Vilas (Badajoz: Spain Formatex Research Centre), 717–732.
Townsend, S. M., Hurrell, E., Gonzalez-Gomez, I., Lowe, J., Frye, J. G., Forsythe, S., et al. (2007). Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 153, 3538–3547. doi: 10.1099/mic.0.2007/009316-0
Tsai, H. Y., Liao, C. H., Huang, Y. T., Lee, P. I., and Hsueh, P. R. (2013). Cronobacter infections not from infant formula, Taiwan. Emerg. Infect. Dis. 19, 167–169. doi: 10.3201/eid1901.120774
U.S. Food and Drug Administration. (2006). Guidance for Industry on Complementary and Alternative Medicine Products and Their Regulation by the Food and Drug Administration. Available at: https://www.fda.gov/media/76323/download (Accessed March 25, 2021).
Underwood, M. A. (2017). Impact of probiotics on necrotizing enterocolitis. Semin. Perinatol. 41, 41–51. doi: 10.1053/j.semperi.2016.09.017
Valdés-Varela, L., Hernández-Barranco, A. M., Ruas-Madiedo, P., and Gueimonde, M. (2016). Effect of Bifidobacterium upon Clostridium difficile growth and toxicity when co-cultured in different prebiotic substrates. Front. Microbiol. 7:738. doi: 10.3389/fmicb.2016.00738
Van Acker, J., de Smet, F., Muyldermans, G., Bougatef, A., Naessens, A., and Lauwers, S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39, 293–297. doi: 10.1128/JCM.39.1.293-297.2001
Van de Wiele, T., Van den Abbeele, P., Ossieur, W., Possemiers, S., and Marzorati, M. (2015). “The simulator of the human intestinal microbial ecosystem (SHIME®),” in The Impact of Food Bioactives on Health. eds. K. Verhoeckx, P. Cotter, I. López-Expósito, C. Kleiveland, T. Lea, A. Mackie, et al. (Cham: Springer), 305–317.
Van den Abbeele, P., Grootaert, C., Marzorati, M., Possemiers, S., Verstraete, W., Gérard, P., et al. (2010). Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and clostridium cluster IX. Appl. Environ. Microbiol. 76, 5237–5246. doi: 10.1128/AEM.00759-10
Vandenplas, Y., Analitis, A., Tziouvara, C., Kountzoglou, A., Drakou, A., Tsouvalas, M., et al. (2017). Safety of a new synbiotic starter formula. Pediatr. Gastroenterol. Hepatol. Nutr. 20:167. doi: 10.5223/pghn.2017.20.3.167
Vandenplas, Y., Zakharova, I., and Dmitrieva, Y. (2015). Oligosaccharides in infant formula: more evidence to validate the role of prebiotics. Br. J. Nutr. 113, 1339–1344. doi: 10.1017/S0007114515000823
Vatanen, T., Plichta, D. R., Somani, J., Münch, P. C., Arthur, T. D., Hall, A. B., et al. (2019). Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat. Microbiol. 4, 470–479. doi: 10.1038/s41564-018-0321-5
Vega, R., and Zuniga-Hansen, M. E. (2015). The effect of processing conditions on the stability of fructooligosaccharides in acidic food products. Food Chem. 173, 784–789. doi: 10.1016/j.foodchem.2014.10.119
Venegas, D. P., De La Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs) mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Venema, K. (2015). “The TNO in vitro model of the colon (TIM-2),” in The Impact of Food Bioactives on Health (Cham: Springer), 293–304.
Vongbhavit, K., and Underwood, M. A. (2016). Prevention of necrotizing enterocolitis through manipulation of the intestinal microbiota of the premature infant. Clin. Ther. 38, 716–732. doi: 10.1016/j.clinthera.2016.01.006
Weng, M., Ganguli, K., Zhu, W., Shi, H., and Walker, W. A. (2014). Conditioned medium from Bifidobacteria infantis protects against Cronobacter sakazakii-induced intestinal inflammation in newborn mice. Am. J. Physio. Gastrointest. Liver Physiol. 306, G779–G787. doi: 10.1152/ajpgi.00183.2013
World Health Organization. (2007). Safe preparation, storage and handling of powdered infant formula. Available at: https://www.who.int/foodsafety/publications/micro/pif_guidelines.pdf (Accessed June 13, 2020).
Wu, J., Leilei, Z., Qiang, M., Zhang, H., and Zhan, X. (2021). Metabolic fate of dietary sialic acid and its influence on gut and oral bacteria. Syst. Microbiol. and Biomanuf., 1–9. doi: 10.1007/s43393-021-00047-7
Yan, Q. Q., Condell, O., Power, K., Butler, F., Tall, B. D., and Fanning, S. (2012). Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium. J. Appl. Microbiol. 113, 1–15. doi: 10.1111/j.1365-2672.2012.05281.x
Yang, I., Corwin, E. J., Brennan, P. A., Jordan, S., Murphy, J. R., and Dunlop, A. (2016). The infant microbiome: implications for infant health and neurocognitive development. Nurs. Res. 65, 76–88. doi: 10.1097/NNR.0000000000000133
Yang, H. Y., Kim, S. K., Choi, S. Y., You, D. H., Lee, S. C., Bang, W. S., et al. (2015). Effect of acid, desiccation, and heat stresses on the viability of Cronobacter sakazakii during rehydration of powdered infant formula and in simulated gastric fluid. Food Control 50, 336–341. doi: 10.1016/j.foodcont.2014.09.012
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Ye, Y., Li, H., Ling, N., Han, Y., Wu, Q., Xu, X., et al. (2016). Identification of potential virulence factors of Cronobacter sakazakii isolates by comparative proteomic analysis. Int. J. Food Microbiol. 217, 182–188. doi: 10.1016/j.ijfoodmicro.2015.08.025
Yong, W., Guo, B., Shi, X., Cheng, T., Chen, M., Jiang, X., et al. (2018). An investigation of an acute gastroenteritis outbreak: Cronobacter sakazakii, a potential cause of food-borne illness. Front. Microbiol. 9:2549. doi: 10.3389/fmicb.2018.02549
Yun, B., Oh, S., and Griffiths, M. W. (2014). Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J. Dairy Sci. 97, 4745–4758. doi: 10.3168/jds.2014-7921
Yurtsev, E. A., Conwill, A., and Gore, J. (2016). Oscillatory dynamics in a bacterial cross-protection mutualism. Proc. Natl. Acad. Sci. 113, 6236–6241. doi: 10.1073/pnas.1523317113
Zhang, X., Gao, J., Ling, N., Zeng, H., Tong, L., Zhang, M., et al. (2019). Roles of outer membrane protein W on survival, cellular morphology, and biofilm formation of Cronobacter sakazakii in response to oxidative stress. J. Dairy Sci. 102, 2017–2021. doi: 10.3168/jds.2018-14643
Keywords: Cronobacter, probiotics, prebiotics, synbiotics, short-chain fatty acids, cell-free supernatant, gut model
Citation: Ke A, Parreira VR, Goodridge L and Farber JM (2021) Current and Future Perspectives on the Role of Probiotics, Prebiotics, and Synbiotics in Controlling Pathogenic Cronobacter Spp. in Infants. Front. Microbiol. 12:755083. doi: 10.3389/fmicb.2021.755083
Edited by:
Hang Xiao, University of Massachusetts Amherst, United StatesReviewed by:
Chaoxin Man, Northeast Agricultural University, ChinaVeronique Delcenserie, University of Liège, Belgium
Copyright © 2021 Ke, Parreira, Goodridge and Farber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valeria R. Parreira, dnBhcnJlaXJAdW9ndWVscGguY2E=
 Alfred Ke
Alfred Ke Valeria R. Parreira
Valeria R. Parreira Lawrence Goodridge
Lawrence Goodridge Jeffrey M. Farber
Jeffrey M. Farber