
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 29 October 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.754140
 Abhilash Kumar Tripathi1,2†
Abhilash Kumar Tripathi1,2† Payal Thakur1,3†
Payal Thakur1,3† Priya Saxena1,3†
Priya Saxena1,3† Shailabh Rauniyar1,2†
Shailabh Rauniyar1,2† Vinoj Gopalakrishnan1,3
Vinoj Gopalakrishnan1,3 Ram Nageena Singh1,2
Ram Nageena Singh1,2 Venkataramana Gadhamshetty2,3,4,5
Venkataramana Gadhamshetty2,3,4,5 Etienne Z. Gnimpieba2,3,6
Etienne Z. Gnimpieba2,3,6 Bharat K. Jasthi2,3,7
Bharat K. Jasthi2,3,7 Rajesh Kumar Sani1,2,3,4,8*
Rajesh Kumar Sani1,2,3,4,8*Sulfate-reducing bacteria (SRB) have a unique ability to respire under anaerobic conditions using sulfate as a terminal electron acceptor, reducing it to hydrogen sulfide. SRB thrives in many natural environments (freshwater sediments and salty marshes), deep subsurface environments (oil wells and hydrothermal vents), and processing facilities in an industrial setting. Owing to their ability to alter the physicochemical properties of underlying metals, SRB can induce fouling, corrosion, and pipeline clogging challenges. Indigenous SRB causes oil souring and associated product loss and, subsequently, the abandonment of impacted oil wells. The sessile cells in biofilms are 1,000 times more resistant to biocides and induce 100-fold greater corrosion than their planktonic counterparts. To effectively combat the challenges posed by SRB, it is essential to understand their molecular mechanisms of biofilm formation and corrosion. Here, we examine the critical genes involved in biofilm formation and microbiologically influenced corrosion and categorize them into various functional categories. The current effort also discusses chemical and biological methods for controlling the SRB biofilms. Finally, we highlight the importance of surface engineering approaches for controlling biofilm formation on underlying metal surfaces.
Sulfur is an abundant element that exists in the forms of pyrite (FeS2), gypsum (CaSO4), and sulfate in the natural environments, including rocks, sediments, and seawater, respectively (Labrado et al., 2019). Some bacteria can use sulfur to generate amino acids (e.g., cysteine and methionine) and enzymes (Muyzer and Stams, 2008) required to generate metabolic energy. Sulfur cycling in the environment is dominated by sulfate-reducing bacteria (SRB) and sulfur-oxidizing bacteria (SOB) (Zhang et al., 2017). SRB gain energy through dissimilatory sulfate reduction, which is the main process of biomineralization of organic matter in marine sediments. SRB respire using sulfate as a terminal electron acceptor, reducing it to hydrogen sulfide. SRB are of primary concern because they accelerate metallic corrosion under mild conditions, including neutral pH, ambient temperature, and absence of oxygen, both in natural and engineered aquatic systems (Lee et al., 1995; Beech and Sunner, 2007). SRB are often responsible for microbiologically influenced corrosion (MIC), accelerating corrosion reactions, or shifting corrosion mechanisms (Venzlaff et al., 2013; Guan et al., 2016).
An essential step used by microorganisms to effectively induce MIC is to grow into a biofilm on underlying metal surfaces. Such biofilms allow them to grow even under hostile conditions and promote redox processes on metal surfaces responsible for accelerating metal corrosion (Procópio, 2019). The formation of biofilm structure and its maintenance are vital factors that influence the kinetics of the MIC process. Biofilms are stratified structures where the deeper layers in contact with the metal surface maintain anoxic conditions. These anoxic niches promote the growth of specific microorganisms, including SRB, that use the metal compounds as resources for their survival (Procópio, 2019). The mechanisms of biofilm formation by SRB involves a plethora of differentially regulated genes (Krumholz et al., 2015; Qi et al., 2016). In general, SRB biofilms are composed of proteins with a minimal exopolysaccharide content (Beech et al., 2005; Clark et al., 2007, 2012). Prior studies on other aerobic or facultative anaerobic microbes have explored differential gene expression and protein profiles in planktonic cells and in biofilms. These studies have revealed that biofilm formation is a complex and yet highly regulated process (Resch et al., 2006; Qi et al., 2016; Sánchez et al., 2019). This review discusses the transcriptional and proteomic variations in SRB biofilm and key genes involved in biofilm formation and subsequent corrosion effects.
While corrosion is a relatively simple process, assessing the relevant in situ and localized mechanism is challenging. Aggravating this problem is the presence of biofilm, which consists of microbial cells adhered to surfaces that influence the electrochemical reactions on the corroding surfaces. These cells release several metabolites and proteins that cause secondary effects on MIC (Beale David et al., 2016). To fully understand these effects, it is essential to analyze changes at the gene and protein level. Until now, most of the review articles have concentrated on electrochemical and thermodynamic aspects of MIC (Li et al., 2018; Lv and Du, 2018). Although the biological and electrochemical mechanisms of MIC in sulfate-rich environments have been extensively reviewed (Enning and Garrelfs, 2014; Gu et al., 2019), an extensive discussion on the molecular and regulatory mechanisms of biofilm formation by SRB is lacking. Biofilm build-up of SRB accelerates metal degradation by creating favorable conditions for microbes to corrode metals (Zhang et al., 2007; Procópio, 2019). The present review describes the process of SRB biofilm formation at the genetic and molecular levels. Noting that the underlying molecular mechanisms of adaptation are influenced by environmental factors (e.g., pH, temperature, and O2 and C levels) and surface characteristics (Toyofuku et al., 2016), we unveil such molecular mechanisms using omics data. We also discuss the underlying physiological adaptation between the biofilm and planktonic growth modes, which is scarce.
A sulfate-reducing bacterium has genes for every activity and function and expresses them in corresponding proteins to form biofilms on different surfaces under various environments. These proteins later induce microbiological corrosion on the metal surfaces. Unlike the planktonic counterparts, the sessile cells in SRB biofilms use a unique combination of genes and proteins to establish a distinct flow of energy and cell associations (Price et al., 2014; De León et al., 2017). Studies based on targeted mutagenesis and gene expression analyses have shown an increased expression of genes coding for exopolysaccharides (EPS) (Clark et al., 2007; Qi et al., 2016). However, it has been reported that SRB biofilm is composed mainly of proteins with minimal EPSs (Beech et al., 2005). There have been very few studies available on genes and proteins on a molecular level in SRB other than Desulfovibrio vulgaris Hildenborough (DVH) and Desulfovibrio alaskensis G20 (DA-G20). DVH and DA-G20 have been used as model organisms for studying biofilm formation and MIC aspects of SRB. A large number of SRB studies have focused on these two species, and thus, information on genes responsible for biofilm formation has also been depicted using them (Clark et al., 2007; Elias et al., 2009; Price et al., 2014; Williamson et al., 2018). It has been found that biofilms of D. vulgaris showed altered levels of transcripts and proteins involved in critical functional categories such as carbon and energy metabolism, amino acid metabolism, stress response, proteases, and ribosomal proteins (Clark et al., 2012; Zhang et al., 2016; De León et al., 2017; Zhu et al., 2019).
Maximum studies on SRB biofilm formation used lactate as an electron donor and sulfate as an electron acceptor. Lactate–sulfate is the standard media used to study SRB biofilm characteristics (Clark et al., 2006; Brileya et al., 2014). However, the type of surfaces for biofilms (metal or non-metal) varies among different studies (Brileya et al., 2014; Dec et al., 2017; Krantz et al., 2019). Lopes et al. (2006) reported that metal (stainless steel) composition plays a significant role in the development of the biofilm of DA G20. They demonstrated that high nickel concentration had a negative effect on cell growth, while low concentration showed a positive impact. In contrast, chromium on the metal surface did not affect cell duplication. Chen et al. (1998) also reported growth retardation of DA G20 in the presence of molybdenum. Previous studies have shown that biofilm cells grown in lactate/sulfate media have different expression profiles than planktonic cells grown under the similar conditions (Clark et al., 2012; Sivakumar et al., 2019).
A transcriptomic study showed that a total of 358 genes downregulated in biofilms as compared to planktonic cells (Clark et al., 2012). Out of these 358 genes, 10% have been predicted to be involved with translation and ribosomal structure/biogenesis. Downexpression was also observed in genes for amino acid transport and metabolism, cell synthesis, and cell division. Besides the down-regulated genes, a higher expression was recorded for the number of gene including hydrogenase and formate dehydrogenases and in genes involved in energy production and conservation, signal transduction, and cell motility in SRB biofilms as compared to planktonic cells. A 26-fold higher rate of formate-dependent methyl viologen reduction was reported in the biofilm, indicating increased gene expression and enzymatic activity. The critical genes in SRB (DA-G20) that have been reported to be essential for biofilm formation are given in Table 1.
Sulfate-reducing bacteria biofilm have been reported to have altered expression of transcripts and proteins at a different level of growth and nutrient availability when compared with planktonic cells (Clark et al., 2012; Krumholz et al., 2015; Qi et al., 2016). It is considered that biofilms have different levels of expression of genes and their corresponding proteins. To sustain the unique state of biofilm growth, the primary requirement is energy generation, and therefore, the expression of genes required for energy conversion and carbon flow is one of the significant activities of the biofilm. It has been found that biofilm cells have differences in the expression of genes of two pyruvate:ferredoxin oxidoreductases [e.g., oor (upregulated) and por (downregulated)] as compared to planktonic cells. Although the expression of pyruvate formate lyase was unchanged, formate dehydrogenase expression (hybA transcript and protein, alpha subunit) was found to increase. Only protein abundance was found to increase for fdnG1 (beta subunit of formate dehydrogenase). At the same time, cytochrome c553 (electron transfer partner for formate dehydrogenase) was found to increase for both mRNA and protein (Sebban-Kreuzer et al., 1998) in DVH. It has also been reported that SRB biofilm has 26-fold higher formate-dependent (dehydrogenase) activity than planktonic cells and assumed that it is because of formate cycling and their role in electron flow (Voordouw, 2002; Heidelberg et al., 2004; Clark et al., 2012). Some other dehydrogenases were also reported in the biofilm, and their roles in proton and electron cycling need to be explored to understand the mechanisms in different states of SRB biofilm.
Carbon uptake and utilization is one of the major requirements to drive energy and biosynthesis of several essential molecules and compounds for bacteria to survive. Previous studies have reported that SRB have several genes, which give flexibility to uptake and utilize different carbon compounds like glycerol and phospholipids and could be processed for amino acid biosynthesis. The observation was supported by the downexpression of glycolytic pathway and gluconeogenesis required for amino acid biosynthesis and amino-sugar biosynthesis, respectively followed by decreased expression of genes responsible for cell wall synthesis and cell division in D. vulgaris (Clark et al., 2012; Egan et al., 2017). It was concluded that D. vulgaris cells use phospholipids released from neighboring dead cells for synthesizing amino acids. The proteins that impact carbohydrate and nitrogen metabolism (Barton and Hamilton, 2007) have been observed and proposed by several researchers. Prior studies have depicted lower carbohydrate content in SRB biofilm than planktonic cells (Clark et al., 2012; Qi et al., 2016). In the study by Qi et al. (2016), the protein content in DVH biofilm cells was almost similar to planktonic cells after 5 days of growth. However, the carbohydrate content in the biofilms decreased significantly at the end of the fifth day, which resulted in a low carbohydrate to protein ratio of 0.11. These results corroborate that SRB biofilms do not produce a carbohydrate-rich biofilm matrix (Clark et al., 2007). In addition to dehydrogenases, fructose-1,6-bisphosphatase was also found downexpressed in DVH biofilm, indicating a decrease in gluconeogenesis rate (Clark et al., 2012) and supporting the hypothesis of slow cell wall synthesis and cell division.
The nitrogen metabolism in SRB allows adaptation in different nutritional habits even under adverse conditions (Lespinat et al., 1987). There is limited information available regarding nitrogen metabolism in SRB biofilm. The studies that focused on transcriptional heterogeneity have found increased expression of nrfA gene (cytochrome c nitrite reductase c552) in the presence of ammonium in the biofilm, but upregulation was also reported when cells were exposed to acetone, ethanol, nitrite, and alkaline pH. A nifU (nitrogenase) gene was found in DVH plasmid, but no change in expression has been recorded (Clark et al., 2012). Therefore, it can be concluded that nitrogen metabolism is still not well understood in SRB biofilms, and the expression of nrfA may be a response to general stress management. More studies are required to unveil the nitrogen metabolism in SRB biofilm.
Biofilm formation involves stress response regulators, and different stress conditions trigger changes in the expression of genes that help microbes survive. The differential expression of the stress response regulators depends upon the environments under which biofilm formation is triggered and the genetic background of the microbe (Jefferson, 2004). Here, we review how different environmental parameters (media type, nutrient deprivation, pH, oxygen radicals, osmotic stress, temperature, and material surface) influence the formation of SRB biofilm. It is possible to infer that the relative expression of various stress-related genes and proteins varies accordingly. For instance, in DVH, the stress response gene DVU2410 coding for superoxide dismutase had increased transcript level (Clark et al., 2012), whereas the same gene was downexpressed in a study reported by Qi et al. (2016). These two studies primarily differed in the type of media and incubation temperature. Clark et al. (2012) used lactate C media (T = 30°C), while the latter used LS4D media and different incubation temperatures (T = 35°C). In addition, a comparison of differential gene expression in other studies also corroborates this hypothesis. For example, mRNA levels for various stress response genes were elevated in the biofilms of certain microorganisms (e.g., Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus). In contrast, for the anaerobic, hyperthermophile Thermotoga maritima, a decrease in the expression of similar genes was documented (Schembri et al., 2003; Pysz et al., 2004; Waite et al., 2006). It is also worth mentioning that the work of Qi et al. (2016) was done on single cells while the transcriptomics analysis of Clark et al. (2012) was the average across biofilm cells in the sample.
For DVH grown in LS4D media at 30°C, various chaperone genes such as uspA (DVU0423) and three hsp20 genes (DVU2442, 2241, and 1471) had been reported with increased expression within the biofilm compared to planktonic cells (Clark et al., 2012). An increase in expression of alkyl hydroperoxide reductase ahpC (DVU2247), msrB (DVU0576), and ahpC/TSA (thiol-specific antioxidant) was also seen in the same study. In addition, relatively slower growth is known for aerobic bacteria due to oxidative stress within the biofilm due to a shift away from oxygen respiration at different biofilm depths (Stewart and Franklin, 2008). However, slower biofilm growth kinetics is attributed to lower intracellular levels of reducing equivalents for SRB biofilms. This condition might make SRB more susceptible to oxidizing agents, and therefore, a defense mechanism is triggered, resulting in overexpression of genes and proteins involved in potential detoxification. For instance, the rubrerythrin gene (rbr) and thioredoxin reductase (trxB) were upregulated in response to oxidative stress, suggesting an important role of these proteins in the oxidative damage resistance response in DVH (Zhang et al., 2006). The upregulation of trxB suggests that thiol-specific redox system might be involved in the oxidative stress response in SRB. Pereira et al. (2008) reported an upregulation of proteins (the [NiFeSe] hydrogenase, formate dehydrogenase(s), and the Hmc membrane complex) associated with periplasmic hydrogenase-dependent mechanism during oxidative stress in DVH. An increase in expression levels for genes and proteins involved in potential detoxification was also noticed during iron limitation due to sequestration and intracellular accumulation of iron (Hindré et al., 2008; Clark et al., 2012). This increase in the expression of detoxification genes and proteins may be due to the ability of some metals to form hydroxyl radical, metallo-oxo, and metallo-peroxo species (Kasprzak, 2002). In addition, genes annotated as proteases such as ion protease (DVU1337), clpX (DVU1336), clpB (DVU1874), htrA peptidase (DVU3278), htpG (DVU2643), and hflC (DVU0683) were downregulated (Clark et al., 2012). One potential reason for this might be a slower turnover of proteins due to the biofilm growth, but more research is needed to confirm this behavior. DVH biofilm cells also showed altered transcript expression profiles for methyl-accepting chemotaxis proteins, histidine kinases, response regulators, and sensory box proteins, as shown in Table 1.
Iron uptake by SRB is necessary for maintaining an optimal concentration of cellular biomass and enzymatic activity. A previous study found that removing iron from the culture medium resulted in a decrease in the concentration of cellular biomass, and adding back iron to the culture medium restored a high level of biomass concentration (Marchal et al., 2001). DVH biofilms are predicted to be iron limited, as shown by the increase in expression of iron transporter subunit feoA (DVU2572) and fepC (DVU0103), a presumptive ABC transporter for iron uptake (Clark et al., 2012). However, similar findings in planktonic cells (Clark et al., 2006) contradict these results. One plausible reason might be that iron acquisition response is a consequence of increased sulfide concentration associated with SRB growth (Clark et al., 2012). In addition, Clark et al. (2012) found that iron storage protein bacterioferritin (DVU1397) and ferritin storage protein (DVU1568) were upregulated within DVH biofilm cells. The upregulation of iron sequestration proteins was also reported in similar biofilm studies (Pysz et al., 2004; Zhang et al., 2007; Hindré et al., 2008). However, under conditions when DVH biofilm was grown on steel, the iron transport systems like feoAB were downregulated (Zhang et al., 2007). Available iron acts differently in the biofilm of different organisms. In some, it induces a thick biofilm formation (Banin et al., 2005), while in others, it plays a detrimental role (Johnson et al., 2005; Hindré et al., 2008).
Among other transporters, Clark et al. (2012) found that four presumptive ABC transporter genes (DVU2387, DVU2384, DVU0484, and DVU2385) were upregulated in DVH biofilm. Other studies on biofilms from different organisms have also reported upregulation of ABC transporter genes (Schembri et al., 2003; Pysz et al., 2004; Shemesh et al., 2007). Subunits of the Sec transport system [secE (DVU2922), secY (DVU1323), and secG (DVU1676), and secD (DVU1819)] responsible for protein secretion across the cytoplasmic membrane were transcriptionally downexpressed at the protein level (Clark et al., 2012; Pena et al., 2019). However, other types of secretion systems have been reported to be amplified, as indicated by the increased abundance of transcripts of a type I system (DVU1013) and increased protein levels of an annotated type III protein secretion system (DVUA0115) (Clark et al., 2012). The role of DVUA0115 has been proven when corresponding gene (present on plasmid pDV1 of DVH) deficient strain (plasmid cured) lost the ability to form biofilm (Clark et al., 2007).
Desulfovibrio biofilms are a complex system with hundreds of genes that are up-/downregulated during the biofilm advancement process. The extracellular proteins present in the biofilm matrix contribute to biofilm structure and stability. Mutational studies have shown that the absence of extracellular proteins in the biofilm matrix results in reduced biofilm formation, stability, and altered biofilm architectures (Kobayashi and Iwano, 2012). The von Willebrand factor domain (vWF) glycoprotein performs various roles in conjunction with different ligands that include cell adhesion, pattern formation, and signal transduction (Colombatti et al., 1993). Clark et al. (2012) found DVU1012 (have a vWF domain) as most abundant and also contain a hemolysin-type calcium-binding domain. The paralog of DVU1012 and DVU1545 was also abundant in extracellular proteins from the biofilm. They also found that two hypothetical/presumptive proteins (DVU0797 and DVU0799) showed increased abundance in both the extracellular and biofilm fraction of proteins but not in mRNA. A study by Zeng et al. (2017) found that both DVU0797 and DVU0799 code for porin proteins. This highlights that the participation of porin proteins in extracellular matrix formation in biofilm structures could play a potential role.
Lipids, including triglycerides and fatty acids, are the main constituents of the cytoplasmic membrane. They are indispensable for bacterial integrity, survival, and growth. Thus, they may play an important role during biofilm formation (Dubois-Brissonnet et al., 2016). However, there are contradicting reports on the fatty acid composition of DVH biofilm. In one study, lpxC (lipid A production), TolB (peptidoglycan-associated lipoprotein), and DVU0869 (undecaprenyl diphosphate synthase) were downregulated (Clark et al., 2012). The downregulation of fatty acid synthesis genes was contradicted in a separate study, where fatty acid biosynthesis in the biofilm cells was upregulated in DVH biofilm cells (Zhang et al., 2016). These contrasting results may be due to the differences in the environment conditions (temperature, pH, and media composition) and the time at which samples were collected. As such, more insights into the fatty acid regulation of DVH biofilm cells are required.
Microbiologically influenced corrosion results from biochemical reactions that release ions or electrons from the metal surface. Microbes stimulate corrosion through the secretion of enzymes, acidic metabolites, and hydrogen consumption that stimulates cathodic reactions (Kip and van Veen, 2015). SRB are anaerobic microorganisms that have been implicated in the corrosion of iron and steel. In the study by Caffrey et al. (2008), the gene expression profile of DVH was analyzed by growing it on iron under cathodic protection conditions (cathodic hydrogen) and comparing it with cells grown with gaseous hydrogen bubbling through the culture. The cells grown under cathodic protection conditions overexpressed two hydrogenases, the hyn-1 genes for [NiFe] hydrogenase and the hyd genes encoding [Fe] hydrogenase. The electron flow from the hydrogenases across the cytoplasmic membrane was also enhanced because the hmc gene encoding high molecular weight cytochromes was overexpressed under the cathodic protection conditions. The corrosion rate of iron electrodes was significantly reduced when hyn-1, hyd, and hmc mutant biofilms were grown, suggesting that these three genes contribute to the corrosion-enhancing metabolism in DVH. Motility is another critical factor influencing biofilm architecture, specifically for overcoming stressful conditions (O’Toole et al., 2000; Houry et al., 2012). Flagellar proteins have been reported to play an essential role in triggering the initial syntrophic interactions (with other microbes) and fitness for biofilm formation (Shimoyama et al., 2009; Williamson et al., 2018). Krumholz et al. (2015) found that DA-G20 mutated for two flagellum genes (flhA and fliF) exhibited no motility, and also the mutant was unable to utilize the source of H2 in media. Furthermore, significantly decreased biofilm formation was observed in tadC (pilus assembly protein) and glycosyltransferase mutants of DA-G20. Mutant of the gene Dde_0681 (Fe–S cluster hydrogenase component) was incapable of syntrophic coupling due to lower growth on H2 or lactate with sulfate as electron acceptor; however, the growth was normal with thiosulfate or sulfite as electron acceptors. Mutant for Fe-only hydrogenase, the quinone reactive complex (qrc), and high molecular weight cytochrome C complex (hmc) was found to be syntrophically deficient and had almost no growth with H2/sulfate. Out of the three periplasmic hydrogenases, only the two Ni–Fe hydrogenases (Dde_2137-38 and Dde_3754-56) were found to be significantly upregulated.
In the study by Wikieł et al. (2014), metabolic activity and biofilm development of DA-G20 was correlated to electrochemical response on carbon steel surfaces. Response regulators/GGDEF domain protein in DA-G20 has an essential role in biofilm formation. A cyclic-di-GMP signaling network regulates various bacterial groups’ biofilm formation and motility (Römling et al., 2013). DVH genome encodes c-di-GMP-modulating response regulators, which are part of a two-component signaling system and confirmed that a transposon mutant (DVU0636: a c-di-GMP-modulating response regulator) strain of DVH lost its ability to form biofilm. The biofilm demonstrated an altered carbohydrate:protein ratio compared to the wild-type strain (Rajeev et al., 2014). In addition, type IV pili also plays a vital role in adherence of cells as an initial step in colonization (Craig et al., 2019), and pilA mutant (lacking a presumptive type IV pilus) in DVH lost its ability to form a biofilm (Brileya et al., 2014). However, the wild-type DVH strain used by Brileya et al. (2014) and Rajeev et al. (2014) is designated as DVH-MO (strain housed at University of Missouri), and the discovery of biofilm deficiency in DVH-MO might change the interpretation of the involvement of these genes (pilA, flhA, fliF, and tadC) in biofilm formation (De León et al., 2017). The DVH-MO strain has a single nucleotide change within the gene DVU1017 [ABC transporter of a type I secretion system (T1SS)], which was discovered to be sufficient in eliminating biofilm formation.
In addition to c-di-GMP-based regulation, sigma factors (σ factors) regulate bacterial transcription and control how efficiently transcription is initiated (Francke et al., 2011; Bush and Dixon, 2012). There are different classes of σ factors present in bacteria, as described previously (Burgess, 2017). In general, σ factors are subunits of all bacterial RNA polymerases (RNAPs) that determine transcriptional initiation specificity through binding to RNAP (Kazakov et al., 2015). One of the sigma factors, called alternative sigma factor σ54, constitutes 41% (37 out of 91) of all response regulators (RR) in DVH when compared to other bacteria where only 10% RR are σ54 dependent (Kazakov et al., 2015; Zhu et al., 2019). The 37 regulons that σ54-dependent regulators control in DVH are involved in nitrogen, carbon, and energy metabolism; transmembrane transport; nitrite stress response; and exopolysaccharide and biofilm synthesis (Kazakov et al., 2015). Zhu et al. (2019) found that in DVH, σ54-dependent regulator (DVU2956) is highly upregulated in planktonic cells and downregulated in biofilm. The σ54-dependent regulator (DVU2956), when expressed in DVH biofilm through the use of promoter that is exclusively expressed in the biofilm mode of growth, reduced biofilm formation by 72 ± 11% in 48 h. On the contrary, knocking out DVU2956 increased biofilm formation by 30.1 ± 0.6%. In addition to suppressing biofilm formation, Zhu et al. (2019) also found that DVU2956, when produced 24 h after biofilm formation, was able to disperse DVH biofilm by 42 ± 4%. Apart from DVH, when the plasmid-expressing DVU2956 was introduced in DA-G20, the biofilm was inhibited by 78 ± 9% after 24 h. This shows that DVU2956 can inhibit biofilm formation in multiple SRB strains. Figure 1 represents a pictorial representation of lactate oxidation and sulfate reduction along with proteins involved in biofilm formation and their cellular location in DA G20 and DVH.
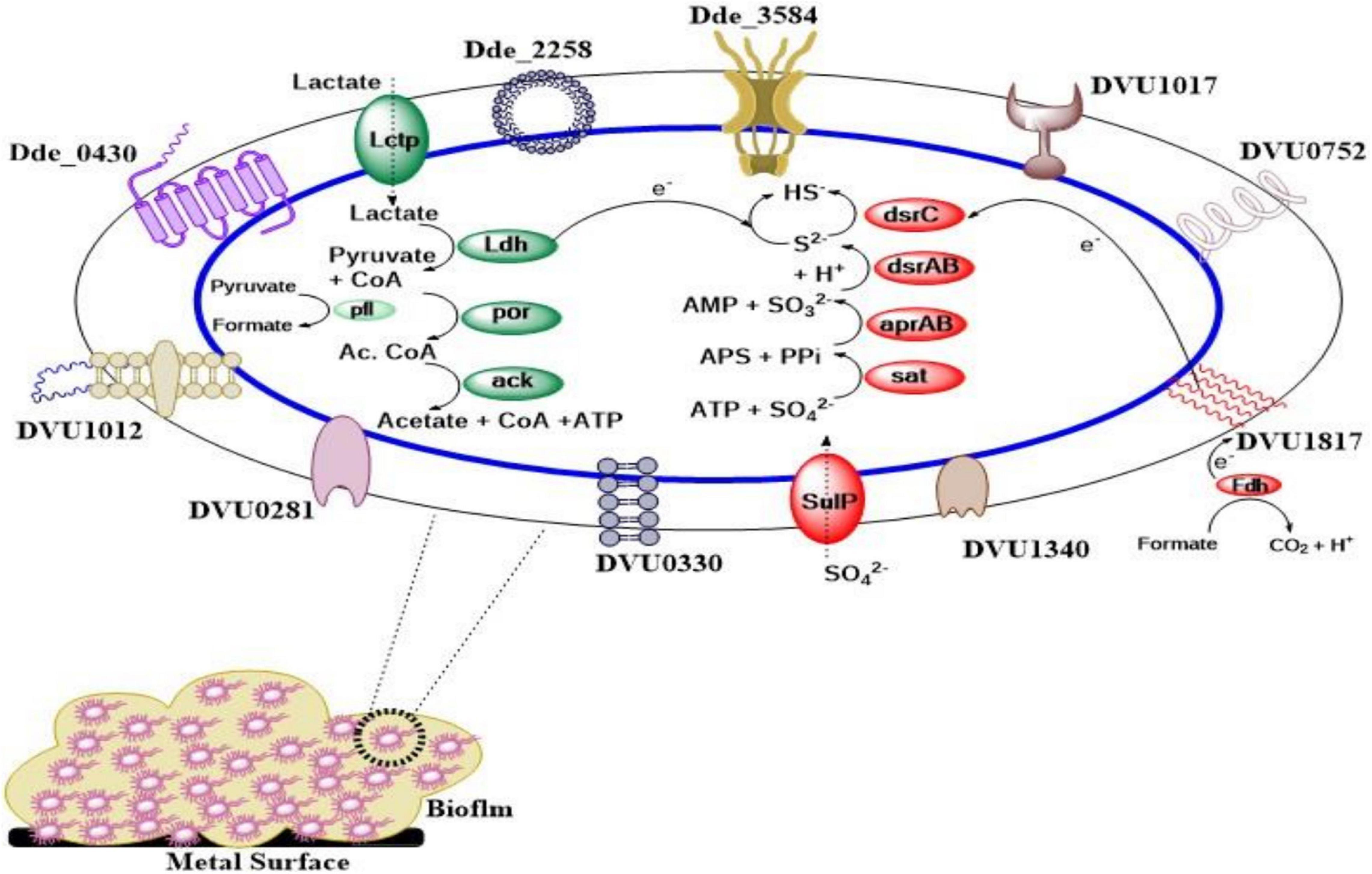
Figure 1. Pictorial representation of biofilm forming and lactate oxidation/sulfate reduction proteins. (Cellular location of all the biofilm formation genes is shown in Supplementary Table 1. Lctp, lactate permease; Ldh, lactate dehydrogenase; por, pyruvate-ferredoxin oxidoreductase; ack, acetate kinase; pfl, pyruvate formate lyase; SulP, sulfate permease; sat, sulfate adenylyltransferase; aprAB, adenosine-5′-phosphosulfate (APS) reductase; dsrAB, dissimilatory sulfate reductase; dsrC, dissimilatory sulfite reductase (Desulfoviridin), gamma subunit; Fdh, formate dehydrogenase).
The previous section discussed the genes in different functional categories that take part in critical cellular processes and stress response systems during biofilm formation by SRB. These genes were either up- or downregulated during the biofilm formation, indicating their role in biofilm development. This section has compiled a list of the reported biofilm-related genes from prior studies and mapped it across 30 common SRB based on sequence similarity (Supplementary Table 1). The organisms were selected based on the availability of quality genomes from the National Center for Biotechnology Information (NCBI). The heatmap was generated based on the presence/absence matrix of the genes, as shown in Figure 2. The genes are organized from top to bottom in the order of presence across the genomes, whereas the genomes are arranged from left to right in the order of the number of shared genes. This means that the organisms on the left-most side of Figure 2 have the maximum number of gene features, and the gene listed on the top is shared by most organisms. The first column of the plot is marked with an up- and downregulated sign. As depicted in the plot, only nine genes are shared across all the SRB, indicating its role in essential functions. The respective functions (gluconeogenesis, protein catalysis, sec transport system, and membrane integrity) are mentioned inside the heatmap.
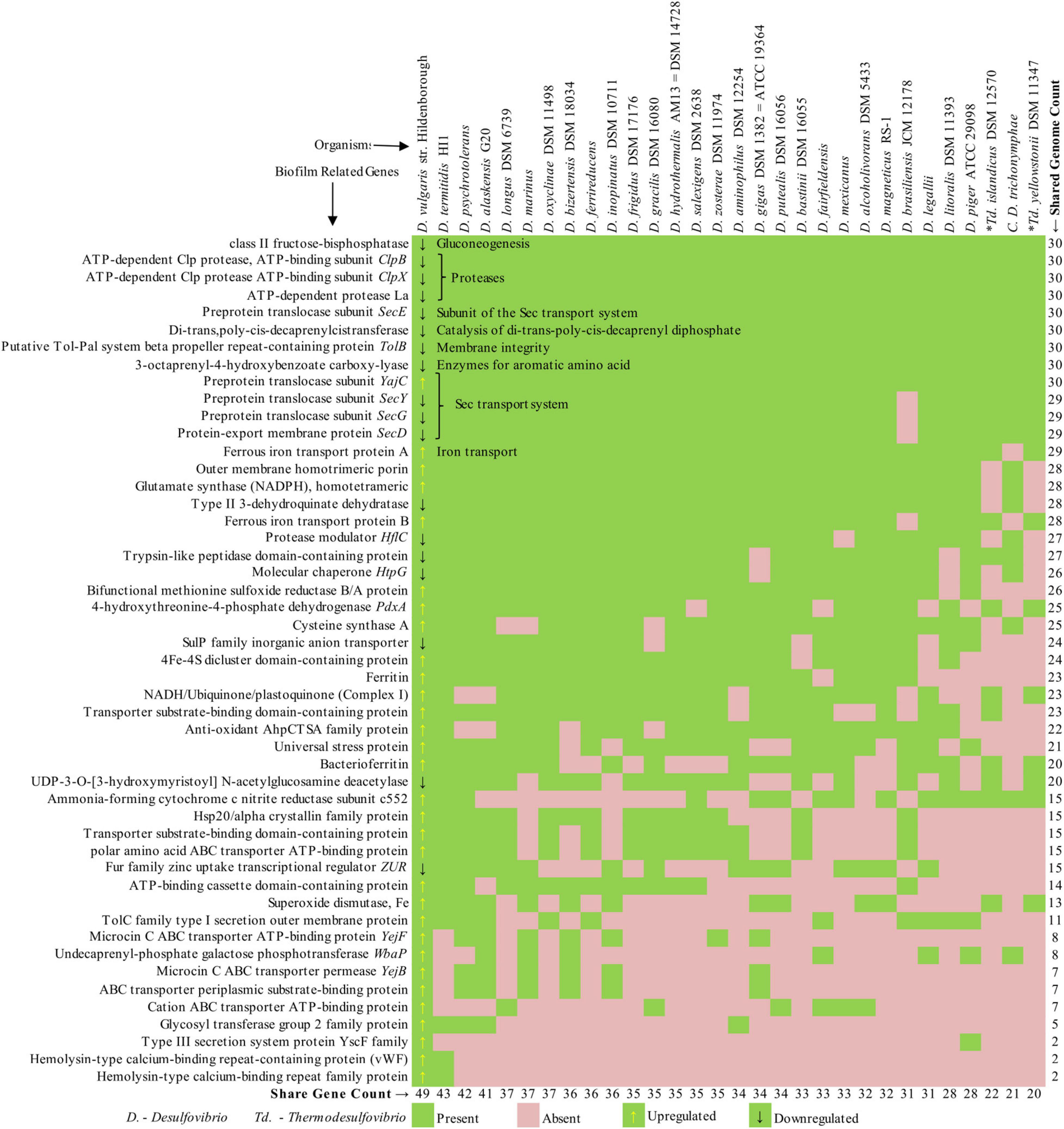
Figure 2. Presence–absence matrix of biofilm genes shared across 30 sulfate-reducing bacteria (SRB) genomes.
Interestingly, eight of the nine genes (except YajC) are reported to be downregulated during the biofilm formation (Clark et al., 2012). As we move down, the list of genes that are shared by a smaller number of organisms is mainly upregulated. Expectedly, the closely related organisms (DVH, DA-G20) shared most of the genes and are positioned on the left half of the plot. On the contrary, the distant organism (Thermodesulfovibrio, Candidatus Desulfovibrio) shared the minimum number of genes and are positioned on the right half. Despite the organisms share many of the listed genes, whether they are up- or down-regulated depends on several unknown factors. This comparative study demonstrates the distribution of active biofilm genes across the genomes of a selected set of sulfate-reducing bacteria, which might be helpful to predict the biofilm nature of other SRB.
The presence of biofilm facilitates direct contact between the metal surface and SRB outer membrane redox proteins or with electroconductive filaments (Beese-Vasbender et al., 2015). Such biofilm matrix establishes anoxic niches in which SRB proliferate, resulting in localized corrosion and pits. The biofilm matrix primarily comprises of rhamnolipids and extracellular polymeric substances (EPS), and the genes responsible for these are controlled by the quorum-sensing (QS) system (Scarascia et al., 2019). QS is a cell-density-dependent communication system in microbes based on the exchange of small molecules called autoinducers. When the density of cells reaches a threshold, such autoinducers bind to receptors to initiate a cascade of reactions that leads to an increase in biofilm formation by overexpressing or suppressing the expression of certain genes (Waters and Bassler, 2005). The molecules used for QS are highly specific, produced and recognized by many different bacterial species. In general, Gram-negative bacteria use small QS molecules known as autoinducers {autoinducer-1 [acyl-homoserine lactone (AHL)] and autoinducer-2}, while Gram-positive bacteria use oligopeptides as QS molecules (Mukherjee and Bassler, 2019). However, studies have detected autoinducer-2 type QS molecules in Gram-positive bacterial species (Surette et al., 1999; Pereira et al., 2013). The presence of autoinducer 2 QS signal molecule in both Gram-positive and Gram-negative bacteria helps mediate interspecies communication between diverse bacterial species (Zhang et al., 2020).
Although the complete mechanism of QS in SRB is still unknown, inferences on the presence of putative QS systems can be made through in vitro cell assay and comparative genomic analysis (Kawaguchi et al., 2008; Scarascia et al., 2016). Some studies using an in vitro cell-free assay have demonstrated the presence of several AHL molecules in active cultures of D. vulgaris and other Desulfovibrio spp. (Kawaguchi et al., 2008; Decho et al., 2010; Scarascia et al., 2016). Diverse AHLs were also detected in a study on the examination of QS molecules in microbial mats with a high abundance of SRB (Decho et al., 2009). Previous studies have suggested that long-chain alkyl AHLs produced by SRB stimulated sulfide oxidation by sulfide-oxidizing bacteria (SOB) (Montgomery et al., 2013). This syntrophic behavior helps SRB generate ATP by gaining access to sulfate through SOB(Scarascia et al., 2016). Genomic approaches have found proteins homologous to LuxR and LuxS in Desulfovibrio magneticus, Desulfovibrio desulfuricans, and DVH (Brady et al., 2004; Williamson et al., 2005; Kimura, 2014). In addition, genome mining of SRB genomes showed that QS proteins homologous to LuxS, LuxP, LuxQ, and LuxO are present in many Desulfovibrio spp. Interestingly, LuxO is reported to be involved in the downstream phosphorylation cascade reactions to upregulate or repress QS-associated genes involved in many phenotypic traits, including biofilm formation (Scarascia et al., 2016). Data mining results from a previous study (Table 2) confirm the presence of QS systems in SRB similar to that present in Vibrio harveyi (Scarascia et al., 2016). In a recent study, AHLs and autoinducer-2 QS molecules were detected in the culture of Desulfovibrio sp. Huiquan 2017 (Li et al., 2021). The presence of autoinducer-2 in other non-desulfovibrio Gram-negative SRB (e.g., Desulfobacter postgatei, Desulfobacterium autotrophicum, and Desulfobulbus propionicus) signifies the use of autoinducer-2 as signal molecule mediating interspecies communication among bacteria. In addition, Gram-positive Desulfotomaculum genomes showed that the majority of them (e.g., Desulfotomaculum kuznetsovii, Desulfotomaculum ruminis, Desulfotomaculum reducens, and Desulfotomaculum acetoxidans) have LuxR-type QS proteins (Scarascia et al., 2016). There are two plausible explanations for the presence of LuxR-type QS proteins in Gram-positive SRB: (1) these proteins may merely be orphan receptors, which have no signal to respond to and therefore are not involved in QS, and (2) these orphan receptors might allow other SRB to sense and respond to QS signal produced by other bacteria present within the microbial community. However, further research is required to prove either of these hypothesis and to establish a direct link between QS in SRB and sulfate reduction and subsequently MIC.
Microbiologically influenced corrosion (a.k.a. biocorrosion) results from a series of coordinated interactions among the bacterial cells and the underlying metal interfaces (Beech and Sunner, 2004) that may generate corrosive metabolites that accelerate corrosion. Due to the generation of corrosive metabolite (hydrogen sulfide), MIC is also known as metabolite corrosion or chemical microbially influenced corrosion (CMIC), whereas corrosion through direct withdrawal of electrons is referred to as electrical microbial influenced corrosion (Enning and Garrelfs, 2014). SRB are non-pathogenic bacteria that use self-secreted enzymes for accelerating the sulfate reduction that indirectly promote oxidation of the underlying metals. Mild steel, stainless steel, carbon steel, copper, and aluminum are examples of industrially relevant metals vulnerable to MIC (Kakooei et al., 2012). Kuehr and Vlugt (1934) first explained the MIC mechanisms using cast iron as model substrates. The proposed mechanism was based on cathodic depolarization (CDP) caused by the hydrogenase-catalyzed reduction of sulfate (SO) by Hads (Figure 3). The increased consumption of cathodic hydrogen increased cathodic reactions leading to high corrosion rates. In CDP theory, water was first partially dissociated into hydrogen and hydroxyl ions (Equation 1), and the exposed cast-iron surfaces were ionized by the electrolytic solution pressure to form ferrous ions (Equation 2). The protons released by dissociation of water (Equation 1) were discharged on the cathode, where they react with electrons (Equation 2) to form hydrogen (Hads) that polarizes the cathode. The hydrogenase enzyme converts H2 to H+ (Equation 4), and the sulfate that acts as depolarizer was reduced by the free electrons and protons (Equations 1 and 2) to generate hydrogen sulfide (Equation 5), which provides energy for SRB metabolism. In the subsequent precipitation reactions (Equations 6 and 7), ferrous ions react with hydrogen sulfide and hydroxyl ions to form FeS and Fe(OH)2.
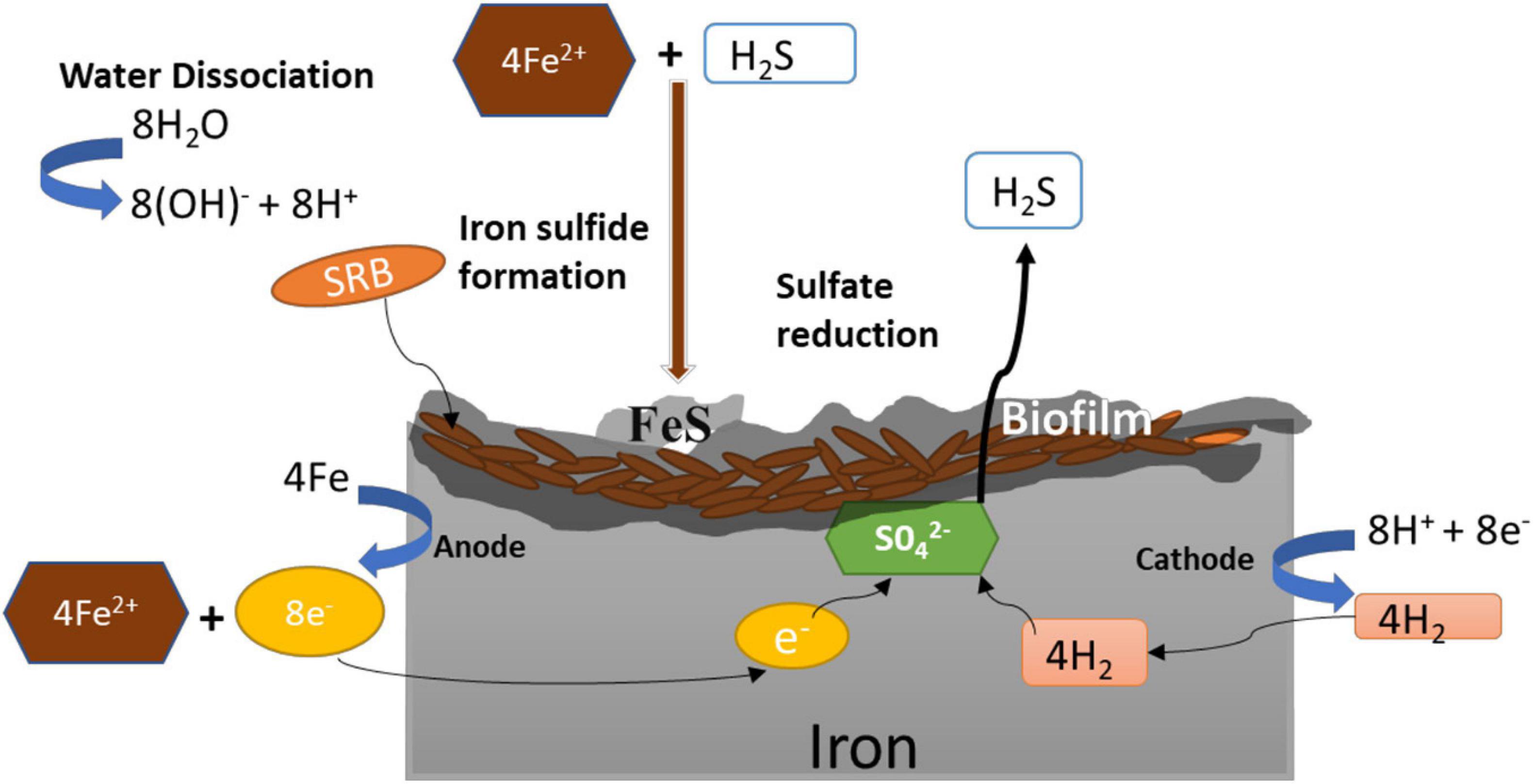
Figure 3. Mechanism of microbiologically influenced corrosion (MIC) of iron by sulfate-reducing bacteria.
| Water dissociation: | 8H2O → 8H+ + 8OH– | (1) |
| Anodic reaction: | 4Fe + 2H → 4Fe2+ + 8e– | 2) |
| Cathodic reaction: | 8H+ + 8 e– → 8Hads | (3) |
| Cathodic depolarization by hydrogenase: | 8Hads → (4H2) → 8H+ + 8 e– | (4) |
| Sulfate reduction by SRB | SO42– + 8e– + 9H+ → HS– + 4H2O | (5) |
| Precipitation: | Fe2+ + H2S → FeS + 2H+ | (6) |
| Precipitation: | 3Fe2+ + 6OH– → 3Fe(OH)2 | (7) |
| Total reaction: | 4Fe + SO42– + 4H2O → FeS + 3Fe(OH)2 + 2OH– | (8) |
Although the CDP theory (Kuehr and Vlugt, 1934) has been questioned, it is still used to describe SRB-induced corrosion mechanism (Procópio, 2019). The primary basis for questioning the CDP mechanism is that the rate-determining step for hydrogen evolution on iron is adsorption rather than desorption (Blackwood, 2020). This means that removing H2 from the surface is not likely to accelerate the cathodic reaction, as it is not the rate-limiting step (Frankenthal and Milner, 1986). In addition, many studies have concluded that both hydrogenase+ and hydrogenase– SRB affect corrosion rate in similar ways (Hardy, 1983; Dinh et al., 2004; Mori et al., 2010). Despite issues with the original CDP theory, cathodic depolarization is consistently observed during corrosion by SRB, alluding to the fact that the kinetics of cathodic reaction rates was indeed increased. This was explained by King and Miller (1971), who suggested that the formation of a conductive film of iron sulfide, which is catalytic to the reduction in protons, results in a positive shift in corrosion potential leading to cathodic depolarization. Furthermore, as most metal sulfide films are cathodic to the base metal, any damage to the film exposes the metal creating localized cathodic and anodic sites (leading to galvanic corrosion) (Syrett, 2013). In addition, microbes in a biofilm arrange themselves in a mutualistic manner that imposes chemical gradients, which creates microenvironments of oxygenated and deoxygenated niches on the metal sulfide layer. The cycling between oxygenated and deoxygenated environments increases corrosion rates, since metal sulfides may act as catalysts to reduce oxygen (Hamilton, 2003; Sherar et al., 2013). The reduction in oxygen by metal sulfides inhibits the formation of the normal protective layer of metal oxides. The erosion of metal oxides due to the reaction between oxygen and metal sulfides exposes anodic sites and initiates pitting corrosion (Hardy and Bown, 1984; Hamilton, 2003).
Another mechanism of biocorrosion reported in 2009 was based on the biocatalytic cathodic sulfate reduction theory (BCSR) (Gu et al., 2009). The BCSR theory analyzed MIC by SRB on carbon steel by combining the concepts of bioenergetics and electrochemical kinetics. Here, MIC is induced by accelerated sulfate reduction at the cathode by the electrons released through iron dissolution at the anode. Under anaerobic conditions, SRB forms biofilm on the surface of the iron. In the initial growth phase under standard conditions, SRB couples oxidation of C substrate such as lactate [e– donor, E°′ = −430 mV vs. SHE (standard hydrogen electrode)] with the reduction in sulfate (e– acceptor, E°′ = −213 mV vs. SHE) (Gu et al., 2009). The standard Gibbs free energy change of the two reactions (ΔG°′ = −168 kJ/mol sulfate) implies that the redox reaction is thermodynamically feasible. SRB couple these two reactions to derive metabolic energy for their growth and metabolism. When the SRB cells evolve to grow into a dense biofilm, the monolayer of cells at the bottom of the biofilm is deprived of C source (e– donor), which results in C starvation. In order to meet their maintenance energy, SRB extracts electrons from elemental iron instead of organic carbon. Thus, the C oxidation is replaced by the elemental iron oxidation with the following reactions (Kakooei et al., 2012).
The increase in corrosion rate due to C starvation is only applicable to metals with iron as the base material (carbon steel and stainless steel). Unlike carbon and stainless steel, C starvation reduced copper corrosion rates by DVH (Dou et al., 2018). The experimental data analysis and thermodynamic study indicated that MIC of Cu by D. vulgaris during C starvation was caused by secreted sulfide as compared to electron harvesting from metal surface in the case of iron. Another study on copper alloy (70Cu–30Ni) found that polarization resistance was reduced for copper alloy in the presence of SRB (Huang et al., 2004). The H2S produced by SRB lowered the reduction potential of anodic reaction, which resulted in an increase in corrosion rate. Moreover, the formation of Cu2S film accelerated localized corrosion due to increased heterogeneity of the copper surface (Dou et al., 2018). In addition to copper and its alloys, aluminum alloys are also widely used in industries and marine engineering facilities (Varshney and Kumar, 2021). Guan et al. (2020) studied the effect of two aluminum alloys (5,052 and Al–Zn–In–Cd aluminum alloy) on SRB metabolic activities and found that both alloys promoted SRB metabolic activity. Compared with 5,052 aluminum, which is known to have good anticorrosion characteristics due to the formation of passive film on its surface, the Al–Zn–In–Cd aluminum alloy is quite active in salt water. In the presence of SRB, the [H] generated at the cathode acts as an electron transfer mediator in the process of EET used by the SRB. This increases cathodic reaction rate due to a large number of H leaving the metal surface. At the same time, the anodic reaction remains stable, which causes electron dissipation from the metal surface (Yu et al., 2013; Guan et al., 2020). The EET mechanism of SRB transports extracellular electrons to the cells cytoplasm for reduction reaction, which indicates that SRB cell acts as biocathode and the bacterial cells directly obtain electrons. This EET mechanism increases corrosion potential of Al–Zn–In–Cd aluminum alloy, which accelerates corrosion rates. The role of EET in corrosion is discussed in detail in the next section.
| Anodic: | 4Fe2+ + 8e– → 4Fe (Iron Dissolution) | E°′ = −447 mV |
| Cathodic: | SO42– + 9H+ + 8e– → HS– + 4H2O (BCSR) | E°′ = −213 mV |
| Total: | SO42– + 9H+ + 4Fe → 4H2O + HS– + 4Fe2+ | Ecell = +234 mV |
Extracellular electron transfer (EET) is a mode of energy conservation by which some microorganisms exchange intracellular electrons with extracellular electron donor/acceptor (Rathinam et al., 2018; Tanaka et al., 2018; Shrestha et al., 2020). Microbial EET potentially endangers corrosion of iron structures. Microbial EET activity augments corrosion via (1) direct uptake of electrons from metallic iron, also known as direct electron transfer MIC (DET-MIC) (Xu et al., 2013; Kato, 2016), and (2) indirect transfer of electrons through the use of electron carriers (mediators), known as mediated electron transfer (MET) (Xu et al., 2013; Zhang et al., 2015). EET microorganisms capable of inducing corrosion are isolated from marine environments and are primarily identified as SRB and methanogenic archaea (Kato et al., 2015). SRB are one of the most prominent microorganisms known to cause anaerobic MIC, mainly via the production of corrosive chemical H2S. However, a number of SRB and other bacterial strains stimulate corrosion via more direct manners. For instance, Jia et al. (2018) reported that lower H2S concentration in the culture medium inoculated with D. vulgaris (ATCC 7757) increased MIC rate on C1018 carbon steel. It was possibly due to the reduction in acidity as the concentration of biogenic H2S in the culture medium decreased. On the contrary, the presence of more biogenic H2S in the culture medium increased acidity, decreased sessile cell count on iron surface, and formed a protective film of Mackinawite on iron surface, thereby inhibiting corrosion. Furthermore, it was also reported that, at pH above 7, direct electron uptake by sessile SRB cells is the dominant mechanism of corrosion.
Some anaerobic microorganisms use DET to utilize zero-valent metallic iron as their electron donor, thereby stimulating iron corrosion. Anaerobic iron corrosion has frequently been reported, and in most cases, it is linked to SRB activity (Dinh et al., 2004). Accelerated cathodic reaction via the consumption of cathodic electrons (cathodic depolarization) is one of the significant causes of MIC. Cathodic depolarization is induced by microbes consuming abiotically generated H2 or by methanogenesis. However, the stimulation of anaerobic corrosion via H2 consumption is controversial. It has been reported that H2 consumption by itself is not sufficient to induce fatal iron corrosion (Blackwood, 2020). A recent study reported that there is no abiotic generation of H2 from stainless steel, further demonstrating that the corrosion mechanism involving H2 as electron carrier is dubious (Tang et al., 2021). Studies have shown that only microorganisms with EET ability (direct or indirect uptake of electrons from metallic iron) can stimulate cathodic reaction and corrode iron aggressively (Venzlaff et al., 2013; Blackwood, 2020; Tang et al., 2021). This manner of MIC, i.e., stimulation of the cathodic reaction by consuming cathodic electrons as the metabolic energy source, is known as electrical MIC (Enning and Garrelfs, 2014). Tang et al. (2021) reported that two Geobacter species (Geobacter sulfurreducens and Geobacter metallireducens) were able to directly extract electrons from stainless steel via DET between the bacteria and iron. When the gene (Gmet 1668) coding for multi-heme c-type cytochrome, which is required for DET, was knocked out, the pitting corrosion drastically reduced. Gmet 1668 could be used in future studies as a molecular target to find microbes with DET capability. Similarly, other SRB strains such as Desulfopila corrodens strain IS4 and Desulfovibrio ferrophilus strain IS5 reduce sulfate with concomitant oxidation of metallic iron much faster than abiotic H2 generation in an organic matter-free medium (Kato, 2016). Compared to DET, Zhang et al. (2015) observed that MET in the presence of electron mediators (10 ppm) such as riboflavin and flavin adenine dinucleotide (FAD) enhanced pitting corrosion of 304 stainless steel by D. vulgaris (ATCC 7757). C-source starvation also plays an important role in electron extraction from iron surface (Xu and Gu, 2014). During C-source starvation, the sessile cells at the bottom might not receive adequate carbon for their survival. As a consequence, the cells at the bottom switch to Fe0 as the terminal electron donor, and the electrons thus released are transported by EET (DET or MET) into the cytoplasm where sulfate reduction takes place. It has been shown that D. vulgaris (ATCC 7757) biofilm grown on iron surface with 90% carbon reduction increased corrosion rate causing a 10-μm maximum pit depth (Xu and Gu, 2014). In addition, the effect of sulfate concentration on biofilm aggressiveness was depicted in a mechanistic modeling study of MIC by SRB (Xu et al., 2016). Sulfate reduction depends on the ability of an SRB biofilm to transfer extracellular electrons from iron surface to SRB cytoplasm. The ability of an SRB biofilm to transfer extracellular electrons is dependent on the type of EET mechanism employed (DET or MET). However, mass and charge transfer becomes increasingly limiting because sulfate has to diffuse through a much larger distance to the bottom of biofilm to get reduced by extracellular electrons. The mechanistic modeling results by Xu et al. (2016) indicated that at high sulfate concentration (7–28 mM), charge transfer becomes limiting, and pitting corrosion depth diminishes. On the other hand, at low sulfate concentration (∼1 mM), mass transfer becomes limiting, and pit growth starts to grow more prominently. So far, EMIC-inducing abilities in SRB have been identified in only a limited number of strains in the deltaproteobacterial families Desulfovibrionaceae and Desulfobulbaceae (Enning and Garrelfs, 2014). Many SRB cannot grow autotrophically and require organic compounds such as acetate as the carbon source and sulfate/H2 as the terminal electron acceptor. Hence, it is postulated that acetate generated by acetogenic bacteria indirectly enhances iron corrosion via stimulation of EMIC-inducing SRB (Mand et al., 2014). It is important to mention that apart from SRB, the biofilm of nitrogen-reducing bacteria (NRB) also caused severe corrosion of C1018 carbon steel (Xu et al., 2013). In case of NRB, extracellular electrons generated from iron oxidation are transported into the cytoplasm for nitrate reduction leading to MIC. In a 1-week lab test with Bacillus licheniformis (NRB), Xu et al. (2013) reported increased corrosion rates with 14.5 μM pit depth and 0.89 mg/cm2 normalized weight loss of C1018 carbon steel. This work substantiates the need to investigate the MIC mechanism of other non-SRB bacteria and study synergistic relation between the two groups of bacteria (NRB and SRB).
Biofilms protect sessile bacteria from biocide attacks because dense biofilms with sessile cells glued together by EPSs increase mass transfer resistances (Chen et al., 1993; Stoodley et al., 1998). In general, an enormous amount of biocide and chemicals are required to treat such dense biofilm. As a matter of concern, there has been an increase in restrictions on environmental regulations and safety concerns; it is therefore highly desirable to make more effective use of biocides at a low concentration (Gaylarde et al., 1999). The use of high biocide concentrations can be circumvented by adding biocide enhancers such as ethylenediamine disuccinate (EDDS). Such enhancers increase the efficacy of biocides used in treatment for biofilm (Fu, 2013; Ashraf et al., 2014).
The biocide, glutaraldehyde, is well known for its unique capability to suppress or lengthen the growth of SRB cells rather than destroy the cells (Wen et al., 2010). In addition, glutaraldehyde can cross-link proteins and, therefore, can also be used as a common fixative (contains 2.5 wt% glutaraldehyde) (Hayat, 1981). However, due to the toxicity of glutaraldehyde to aquatic life, its use is limited (Leung, 2001). When the biodegradable chelator EDDS is supplemented with a low amount of glutaraldehyde, it enhances the penetration of glutaraldehyde to SRB biofilm (Kumari et al., 2020). Wen et al. (2009) observed that sessile SRB was eradicated from the surface of carbon steel with 30 ppm of glutaraldehyde and 2,000 ppm EDDS, while 30 ppm of glutaraldehyde alone was not effective against the sessile cells. Another study found that a combination of methanol (15%), glutaraldehyde (50 ppm), and EDDS (1,000 ppm) completely removed sessile cells from the biofilm. In contrast, without EDDS [only methanol (15%) and glutaraldehyde (50 ppm)], biofilm was still present (Wen et al., 2012). These studies indicate that chelating agents such as EDDS can be used to increase the biocidal efficacy and reduce the environmental impact of biocides such as glutaraldehyde.
Another study (Xu et al., 2012) reported improved biofilm mitigation when D-methionine was used as an enhancer with the biocide tetrakis-hydroxymethyl phosphonium sulfate (THPS). The effectiveness of THPS (50 ppm) in dispersing DVH biofilm grown on C1018 carbon steel increased when it was used in combination with D-methionine (100 ppm). However, even a higher concentration of THPS (1,000 ppm w/w) alone was found ineffective in biofilm mitigation (Xu et al., 2012). Similarly, other biocides such as glyceryl trinitrate (GTN) and caprylic acid (CA) are also capable of mitigating DVH biofilm formation by delaminating the intact DNA resulting in the inhibition of duplication of microbes (Li et al., 2016). Li et al. (2016) observed that probable sessile cell number count reduced to 103 cells/cm2 from 106 cells/cm2 when treated with a cocktail of GTN (25 ppm) and CA (0.1%).
Besides, biocides, mannose, and its analogs such as 2-deoxy-D-glucose are also potential biofilm-mitigating agents. Poosarla et al. (2017) observed up to 50% inhibition in SRB biofilm formation when 30 mM mannose was used. Furthermore, up to 90% of biofilm was inhibited at higher concentrations of mannose (100–500 mM). Since mannose can be a possible alternative carbon source, an analog of mannose, 2-deoxy-D-glucose (500 mM), was used, resulting in 94% biofilm inhibition. In contrast, 60% inhibition was recorded with 500 mM of methyl α-D-mannopyranoside (ADM) and methyl α-D-glucopyranoside (ADG). Interestingly, ADM and ADG only inhibit biofilm formation without compromising cell growth.
Biofilm formation on carbon steel imposes many problems to the petroleum industry like pipe clogging, biofouling, and biocorrosion. Biocorrosion contributes approximately 40% of corrosion of petroleum pipelines. Petroleum industries utilize chemical biocides such as chloride, glutaraldehyde, and quaternary ammonium salts to inhibit SRB biofilm. However, chemical biocides are expensive and toxic to humans and aquatic life due to their persistence in the environment (Summer et al., 2011). The use of antimicrobial substances produced by microorganisms and other plant extracts may be an alternative method to control SRB biofilm growth and MIC.
Many species from the genus Alcaligenes possess antagonistic activity, which inhibits the growth of many bacteria (Austin, 1989; Abd Sharad et al., 2016). Abd Sharad et al. (2016) tested Alcaligenes faecalis for its ability to inhibit SRB biofilm. Crude ethyl acetate extract (0.2–12.8 mg/ml) of A. faecalis showed immediate inhibition against SRB. Similarly, Wood et al. (2018) reported that supernatant of P. aeruginosa could disperse more than 98% of the biofilm. They investigated P. aeruginosa mutants, defective in rhamnolipids production, to determine the biochemical basis of SRB biofilm dispersal and concluded that rhamnolipids play a role in the dispersal of SRB biofilm (Wood et al., 2018). In addition, antimicrobial substances (AMS) produced by Streptomyces lunalinharesii strain 235 have shown to be effective against D. alaskensis NCIMB 13491 biofilm on carbon steel surfaces (Pacheco da Rosa et al., 2013; Rosa et al., 2016). The ability of D. alaskensis to form biofilm was sixfold less in the carbon steel coupons treated with AMS (0.05 g protein/ml) than the non-treated ones (Rosa et al., 2016). However, AMS did not affect biofilm stability when biofilm was already present on the coupon surface, indicating the need to pretreat steel coupons with AMS before inoculation with D. alaskensis. The corrosion rates of AMS (0.05 g protein/ml)-treated steel also decreased to 0.27 mm/year compared to the untreated control (0.38 mm/year). Overall, it was observed that the use of AMS produced by S. lunalinharesii strain 235 to control biofilm formation would be immensely useful in preventing MIC from SRB (Rosa et al., 2016). Another study carried out by Korenblum et al. (2013) elucidated the antimicrobial effect of lemongrass oil (LEO) on SRB. Essential oils are an amalgamation of lipophilic and volatile substances that tend to have the potential for antimicrobial activity. Unlike other natural antimicrobial compounds, essential oils inhibit biofilm growth at the same concentration in planktonic and sessile cells. Therefore, when using essential oil as an antimicrobial agent, the sessile cells cannot protect the organism while forming biofilms. The main component of lemongrass oil is “citral,” which shows antimicrobial activity against several bacteria like Bacillus cereus, S. aureus, and E. coli. LEO treatment causes cell lysis and disruption of the cell wall at different layers of polysaccharides, fatty acids, and phospholipids that ultimately result in cell death. LEO and citral can be used in formulations like synthetic biocides in petroleum industries to prevent SRB-induced metal corrosion and reservoir souring (Korenblum et al., 2013).
The use of other naturally occurring compounds such as plant extracts show potential inhibitory effects against yeast, fungi, and bacteria and can be used to treat MIC (Heisey and Gorham, 1992; Bhola et al., 2014). For example, Neem (Azadirachta indica) extract (4%) was reported to reduce the SRB-induced biocorrosion of pipeline steel (API 5L X80) coupons by approximately 50%. Neem extract significantly reduced the contribution of SRB in the corrosion process by minimizing SRB cells’ growth rate and decreasing the sulfide productions and sessile cell density (i.e., biofilm development) (Bhola et al., 2014). In addition, Neem extracts can form adsorbed intermediates on the metal surface (organo-metallic complexes), thereby providing a protective shield to the metal surface (Okafor et al., 2010). Reduction in SRB-induced corrosion by 50% is a significant achievement, as it will reduce biocide use in petroleum industries.
In addition to AMS and plant extracts, new biocides based on bacteriolytic phages are being investigated (Summer et al., 2011). Phages can lyse bacterial cells in single and multispecies biofilm (Doolittle et al., 1995; Sillankorva et al., 2010). For example, phage T4 can disrupt E. coli biofilm morphology by killing bacterial cells through infection and replication within the bacteria (Lu and Collins, 2007). Summer et al. (2011) isolated SRB-specific phages capable of inhibiting the growth of species from the genera Desulfovibrio. The phage used did not wholly inhibit all SRB, and eventually, the growth was back to normal. However, the usefulness of phage cannot be disregarded, as only a single wild-type phage was used. A cocktail of several phages can be used to ensure complete suppression of bacterial growth (Summer et al., 2011). Another useful strategy for disrupting bacterial biofilm is the enzymatic degradation of EPS. Studies have demonstrated that enzymatic degradation of cell-bound EPS polysaccharides can reduce biofilms of several different species of bacteria (Itoh et al., 2005; Ferriol-González and Domingo-Calap, 2020). In addition, phages can be engineered to express biofilm-degrading enzymes (lysins and depolymerases) to disrupt the biofilm structure. The application of such engineered (enzymatic) phage substantially decreased bacterial biofilm cell counts by ∼4.5 orders of magnitude (∼99.997% removal) (Lu and Collins, 2007). Phage-based methods for biofilm disruption would allow for the design of appropriate and specific biofilm control strategies for different environmental and industrial settings. A brief overview of some chemical and biological compounds used in biofilm inhibition and their mode of action is given in Supplementary Table 2.
Compared to biocides and cathodic protection approaches, the use of protective coatings remains a viable choice for retarding MIC. Such coatings are typically based on metals or polymers. However, most of these coatings were designed to combat abiotic forms of corrosion (Pierre and Roberge, 2012). Such coatings, especially those based on polymers, suffer from disadvantages when exposed to biological environments. Furthermore, the effectiveness of polymer coatings is significantly reduced due to aggressive microbial activity in biofilms on the underlying polymer-coated metal surfaces (Krishnamurthy et al., 2015). Although polymer coatings serve as an effective barrier for atmospheric corrosion prevention, they adhere weakly to metal surfaces and are prone to rapid microbial degradation under immersion conditions (Roy et al., 2002; Zhang et al., 2008; Guo et al., 2018). Microbial activity can induce pin-hole defects in polymer coatings, which can grow in size, attract aggressive ions onto metallic surfaces, and further accelerate the corrosion process (Mayavan et al., 2013; Krishnamurthy et al., 2015). In addition to being prone to degradation and aggravating corrosion in some cases, the thickness of polymeric coatings also might disrupt the functional properties (e.g., electrical and thermal conductivity) and dimensional tolerances of target metals (Geim and Novoselov, 2007; Krishnamurthy et al., 2015; Chilkoor et al., 2019). Here, we discuss an emerging class of protective coatings based on 2D materials, including graphene and hexagonal boron nitride. Atomic layers of 2D materials can grow as conformal layers on metallic surfaces using thin film deposition techniques, including chemical vapor deposition (CVD). These 2D materials are mechanically robust, hydrophobic, chemically inert, thermally, and electrically conductive and can form an impermeable barrier (Balandin et al., 2008; Bunch et al., 2008; Krishnamurthy et al., 2015). Due to these characteristics, these 2D materials can be used as an ultrathin protective coating for preventing both abiotic and biotic forms of corrosion. For instance, Krishnamurthy et al. (2013) found that three to four atomic layers of graphene offer long-term resistance (∼40 days) to bimetallic corrosion of Ni, especially under microbial conditions. Their results indicated that graphene coatings appear to combat MIC by (i) preventing access of microbes to the Ni surface, (ii) forming a protective barrier to minimize charge (Ni2+) transport into the solution, and (iii) protecting the Ni surface from microbial byproducts (e.g., H+) that enhance Ni dissolution. In another study, two commercial polymeric coatings, Parylene-C (PA) and Polyurethane (PU), were compared to ultrathin graphene skin (Gr) coating toward protection against MIC (Krishnamurthy et al., 2015). It was found that MIC on Gr-coated Ni was an order of magnitude lower than PA and PU coated. Gr coating showed ∼10- and ∼100-fold improvement in MIC resistance compared to PA and PU coated. In a more recent study, Chilkoor et al. (2021) found that multilayered Gr coating on Cu and Ni is more suited for MIC prevention when compared to single-layer Gr. Multilayered graphene (MLG) on Cu restricted the MIC by 10- and 1.4-fold compared to single-layer Gr–Cu and bare Cu, respectively. Single-layered graphene on Cu worsened increased MIC by fivefold compared to bare Cu due to the presence of point defects in single-layer graphene, which can accelerate the electrochemical corrosion process (Mayavan et al., 2013). In one another study, atomically thin layers (∼4) of hexagonal boron nitride (hBN) were tested as a protective coating to inhibit MIC of the underlying copper (Cu) surfaces (Chilkoor et al., 2020). The results showed that hBN-coated Cu had six- to sevenfold lower corrosion than bare Cu in abiotic (sulfuric acid and sodium sulfide) and biotic (DAG-G20 bacteria medium) environments. Overall, these studies show that few atomic layers of 2D materials show promise for MIC prevention. However, the use of 2D materials for MIC prevention is a relatively new concept, and more studies are needed to understand nanoscale surface properties, genes activated/deactivated during cell attachment on 2D materials, biofilm growth, and biogenic sulfide attack.
Biofilm formed by SRB induces changes on the metal surface that accelerate the anodic/cathodic process, controlling corrosion rates. The formation of biofilm is tightly regulated by several genes and the prevailing stress conditions. This review summarized the molecular mechanisms of biofilm formation by SRB and its implications in MIC. The critical genes, proteins, and quorum sensing (QS) molecules are highlighted, which were reported to be involved in biofilm formation irrespective of SRB type. Besides, we provided a theoretical understanding of two mechanisms of MIC by SRB: cathodic depolarization theory (CDP) and biocatalytic sulfate reduction theory (BCSR). CDP accelerates corrosion rates by increased consumption of cathodic hydrogen, whereas BCSR accelerates corrosion rates by increased sulfate reduction rate at the cathode. CDP and BCSR use the extracellular electron transfer (EET) pathway of SRB, as the cathodic reaction is stimulated through the EET pathway in SRB. The genes involved in EET are differentially expressed in SRB biofilm, resulting in enhanced MIC of metal surfaces. To understand MIC, it is essential to recognize the genetic level changes in SRB during biofilm formation. At the genetic level, the expression pattern of hydrogenase (Fe and NiFe) and heme-containing cytochromes regulated corrosion rates (Caffrey et al., 2008). The knockout of genes encoding for hydrogenases and heme-containing cytochromes lowered corrosion rates of iron. In addition, the presence of biofilm significantly increased MIC rates; the inhibition of quorum sensing system would prevent biofilm formation and lower down corrosion rates. However, the genetic level correlation between SRB biofilm and MIC is still lacking. In SRB biofilm, significantly upexpressed genes belong to three functional categories: (1) energy production and conversion, (2) signal transduction, and (3) cell motility. Future studies should focus on understanding the role of specific genes and their interacting partners during MIC. This strategy in combination with a greater understanding of QS system would enable us to design more efficient strategies for corrosion mitigation. In addition, we have reviewed various biological and chemical methods that have been used to disperse biofilms formed by SRB. Biological methods are preferred over chemical methods, as chemical biocides are toxic to humans and aquatic life due to their persistence in the environment. In addition to biocides (chemical or biological), studies on the use of various commercial coatings are gaining increased attention. The use of 2D materials such as graphene as the coating has shown exciting results in inhibiting SRB biofilm formation and MIC. Future studies would require understanding the changes in genetic and molecular mechanisms in SRB during biofilm formation on coated surfaces and designing strategies to overcome biofilm formation and MIC.
AT, PT, PS, and SR did literature search and prepared a draft. ViG wrote Section “Role of Quorum Sensing in Sulfate-Reducing Bacteria Biofilm Formation” and reviewed the manuscript. RNS, VeG, EG, and BJ reviewed the manuscript. RKS participated in manuscript design, preparation, and review as well as submissions. All authors contributed to the article and approved the submitted version.
We gratefully acknowledge support from the National Science Foundation (Awards #1736255, #1849206, and #1920954) and the Department of Chemical and Biological Engineering at the South Dakota School of Mines and Technology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to acknowledge Kara De Leon (The University of Oklahoma) for her insightful inputs in improving this work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.754140/full#supplementary-material
Abd Sharad, A., Usup, G., Sahrani, F. K., and Ahmad, A. (2016). “Antimicrobial activity and determination of bioactive components from marine Alcaligenes faecalis extract against a sulfate-reducing bacteria,” in Proceedings of the AIP Conference, (Melville, NY: AIP Publishing LLC).
Ashraf, M. A., Ullah, S., Ahmad, I., Qureshi, A. K., Balkhair, K. S., and Abdur Rehman, M. (2014). Green biocides, a promising technology: current and future applications to industry and industrial processes. J. Sci. Food Agric. 94, 388–403.
Austin, B. (1989). Novel pharmaceutical compounds from marine bacteria. J. Appl. Bacteriol. 67, 461–470. doi: 10.1111/j.1365-2672.1989.tb02517.x
Balandin, A. A., Ghosh, S., Bao, W., Calizo, I., Teweldebrhan, D., Miao, F., et al. (2008). Superior thermal conductivity of single-layer graphene. Nano Lett. 8, 902–907. doi: 10.1021/nl0731872
Banin, E., Vasil, M. L., and Greenberg, E. P. (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102, 11076–11081. doi: 10.1073/pnas.0504266102
Barton, L., and Hamilton, W. (2007). Sulphate-Reducing Bacteria: Environmental and Engineered Systems. Cambridge: Cambridge University Press.
Beale David, J., Karpe Avinash, V., Jadhav, S., Muster Tim, H., and Palombo Enzo, A. (2016). Omics-based approaches and their use in the assessment of microbial-influenced corrosion of metals. Corr. Rev. 34, 1–15.
Beech, I. B., and Sunner, J. (2004). Biocorrosion: towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 15, 181–186. doi: 10.1016/j.copbio.2004.05.001
Beech, I. B., and Sunner, J. A. (2007). “Sulphate-reducing bacteria and their role in corrosion of ferrous materials,” in Sulphate-Reducing Bacteria: Environmental and Engineered Systems, eds L. L. Barton and W. A. Hamilton (Cambridge: Cambridge University Press), 459–482.
Beech, I., Sunner, J., and Hiraoka, K. (2005). Microbe-surface interactions in biofouling and biocorrosion processes. Int. Microbiol. 8, 157–168.
Beese-Vasbender, P. F., Nayak, S., Erbe, A., Stratmann, M., and Mayrhofer, K. J. J. (2015). Electrochemical characterization of direct electron uptake in electrical microbially influenced corrosion of iron by the lithoautotrophic SRB Desulfopila corrodens strain IS4. Electrochim. Acta 167, 321–329. doi: 10.1016/j.electacta.2015.03.184
Bhola, S. M., Alabbas, F. M., Bhola, R., Spear, J. R., Mishra, B., Olson, D. L., et al. (2014). Neem extract as an inhibitor for biocorrosion influenced by sulfate reducing bacteria: a preliminary investigation. Eng. Fail. Anal. 36, 92–103. doi: 10.1016/j.engfailanal.2013.09.015
Blackwood, D. J. (2020). An Electrochemist perspective of microbiologically influenced corrosion. Corr. Mater. Degrad. 1, 59–76. doi: 10.3390/cmd1010005
Brady, S. F., Chao, C. J., and Clardy, J. (2004). Long-chain N-acyltyrosine synthases from environmental DNA. Appl. Environ. Microbiol. 70, 6865–6870. doi: 10.1128/aem.70.11.6865-6870.2004
Brileya, K. A., Camilleri, L. B., Zane, G. M., Wall, J. D., and Fields, M. W. (2014). Biofilm growth mode promotes maximum carrying capacity and community stability during product inhibition syntrophy. Front. Microbiol. 5:693. doi: 10.3389/fmicb.2014.00693
Bunch, J. S., Verbridge, S. S., Alden, J. S., van der Zande, A. M., Parpia, J. M., Craighead, H. G., et al. (2008). Impermeable atomic membranes from graphene Sheets. Nano Lett. 8, 2458–2462. doi: 10.1021/nl801457b
Burgess, R. R. (2017). “Sigma factors✩,” in Reference Module in Life Sciences, eds S. Brenner and J. H. Miller (Amsterdam: Elsevier).
Bush, M., and Dixon, R. (2012). The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbio. Mol. Biol. Rev. 76, 497–529. doi: 10.1128/mmbr.00006-12
Caffrey, S. M., Park, H. S., Been, J., Gordon, P., Sensen, C. W., and Voordouw, G. (2008). Gene expression by the sulfate-reducing bacterium desulfovibrio vulgaris hildenborough grown on an iron electrode under cathodic protection conditions. Appl. Environ. Microbiol. 74:2404. doi: 10.1128/AEM.02469-07
Chen, C. I., Griebe, T., and Characklis, W. G. (1993). Biocide action of monochloramine on biofilm systems of Pseudomonas aeruginosa. Biofouling 7, 1–17. doi: 10.1080/08927019309386240
Chen, G., Ford, T. E., and Clayton, C. R. (1998). Interaction of sulfate-reducing bacteria with molybdenum dissolved from sputter-deposited molybdenum thin films and pure molybdenum powder. J. Colloid Interface Sci. 204, 237–246. doi: 10.1006/jcis.1998.5578
Chilkoor, G., Jawaharraj, K., Vemuri, B., Kutana, A., Tripathi, M., Kota, D., et al. (2020). Hexagonal boron nitride for sulfur corrosion inhibition. ACS Nano 14, 14809–14819. doi: 10.1021/acsnano.0c03625
Chilkoor, G., Shrestha, N., Karanam, S. P., Upadhyayula, V. K. K., and Gadhamshetty, V. (2019). “Graphene coatings for microbial corrosion applications,” in Encyclopedia of Water: Science, Technology, and Society, ed. P. A. Maurice (Hoboken, NJ: John Wiley & Sons, Inc.), 1–25.
Chilkoor, G., Shrestha, N., Kutana, A., Tripathi, M., Robles Hernández, F. C., Yakobson, B. I., et al. (2021). Atomic layers of graphene for microbial corrosion prevention. ACS Nano 15, 447–454. doi: 10.1021/acsnano.0c03987
Clark, M. E., He, Q., He, Z., Huang, K. H., Alm, E. J., Wan, X. F., et al. (2006). Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72:5578. doi: 10.1128/AEM.00284-06
Clark, M. E., He, Z., Redding, A. M., Joachimiak, M. P., Keasling, J. D., Zhou, J. Z., et al. (2012). Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: carbon and energy flow contribute to the distinct biofilm growth state. BMC Genomics 13:138. doi: 10.1186/1471-2164-13-138
Clark, M., Edelmann, R., Duley, M. L., Wall, J., and Fields, M. (2007). Biofilm formation in Desulfovibrio vulgaris Hildenborough is dependent upon protein filaments. Environ. Microbiol. 9, 2844–2854.
Colombatti, A., Bonaldo, P., and Doliana, R. (1993). Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix 13, 297–306. doi: 10.1016/S0934-8832(11)80025-9
Craig, L., Forest, K. T., and Maier, B. (2019). Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440.
De León, K. B., Zane, G. M., Trotter, V. V., Krantz, G. P., Arkin, A. P., Butland, G. P., et al. (2017). Unintended laboratory-driven evolution reveals genetic requirements for biofilm formation by Desulfovibrio vulgaris Hildenborough. mBio 8:e01696-17. doi: 10.1128/mBio.01696-17
Dec, W., Mosiałek, M., Socha, R. P., Jaworska-Kik, M., Simka, W., and Michalska, J. (2017). Characterization of Desulfovibrio desulfuricans biofilm on high-alloyed stainless steel: XPS and electrochemical studies. Mater. Chem. Phys. 195, 28–39. doi: 10.1016/j.matchemphys.2017.04.011
Decho, A. W., Norman, R. S., and Visscher, P. T. (2010). Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 18, 73–80. doi: 10.1016/j.tim.2009.12.008
Decho, A. W., Visscher, P. T., Ferry, J., Kawaguchi, T., He, L., Przekop, K. M., et al. (2009). Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol. 11, 409–420. doi: 10.1111/j.1462-2920.2008.01780.x
Dinh, H. T., Kuever, J., Mußmann, M., Hassel, A. W., Stratmann, M., and Widdel, F. (2004). Iron corrosion by novel anaerobic microorganisms. Nature 427, 829–832. doi: 10.1038/nature02321
Doolittle, M. M., Cooney, J. J., and Caldwell, D. E. (1995). Lytic infection of Escherichia coli biofilms by bacteriophage T4. Can. J. Microbiol. 41, 12–18. doi: 10.1139/m95-002
Dou, W., Jia, R., Jin, P., Liu, J., Chen, S., and Gu, T. (2018). Investigation of the mechanism and characteristics of copper corrosion by sulfate reducing bacteria. Corr. Sci. 144, 237–248. doi: 10.1016/j.corsci.2018.08.055
Dubois-Brissonnet, F., Trotier, E., and Briandet, R. (2016). The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front. Microbiol. 7:1673. doi: 10.3389/fmicb.2016.01673
Egan, A. J. F., Cleverley, R. M., Peters, K., Lewis, R. J., and Vollmer, W. (2017). Regulation of bacterial cell wall growth. FEBS J. 284, 851–867. doi: 10.1111/febs.13959
Elias, D. A., Mukhopadhyay, A., Joachimiak, M. P., Drury, E. C., Redding, A. M., Yen, H.-C. B., et al. (2009). Expression profiling of hypothetical genes in Desulfovibrio vulgaris leads to improved functional annotation. Nucleic Acids Res. 37, 2926–2939. doi: 10.1093/nar/gkp164
Enning, D., and Garrelfs, J. (2014). Corrosion of iron by sulfate-reducing bacteria: new views of an old problem. Appl. Environ. Microbiol. 80:1226. doi: 10.1128/AEM.02848-13
Ferriol-González, C., and Domingo-Calap, P. (2020). Phages for biofilm removal. Antibiotics (Basel, Switzerland) 9, 268. doi: 10.3390/antibiotics9050268
Francke, C., Groot Kormelink, T., Hagemeijer, Y., Overmars, L., Sluijter, V., Moezelaar, R., et al. (2011). Comparative analyses imply that the enigmatic sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385
Frankenthal, R. P., and Milner, P. C. (1986). Technical note: hydrogen evolution kinetics on a high-carbon steel and on tin in seawater. Corrosion 42, 51–53. doi: 10.5006/1.3584879
Fu, W. (2013). Investigation of Type II of Microbiologically Influenced Corrosion (MIC) Mechanism and Mitigation of MIC Using Novel Green Biocide Cocktails. Athens, OH: Ohio University.
Gaylarde, C. C., Bento, F. M., and Kelley, J. (1999). Microbial contamination of stored hydrocarbon fuels and its control. Rev. Microbiol. 30, 1–10.
Geim, A. K., and Novoselov, K. S. (2007). The rise of graphene. Nat. Mater. 6, 183–191. doi: 10.1038/nmat1849
Gu, T., Jia, R., Unsal, T., and Xu, D. (2019). Toward a better understanding of microbiologically influenced corrosion caused by sulfate reducing bacteria. J. Mater. Sci. Technol. 35, 631–636. doi: 10.1016/j.jmst.2018.10.026
Gu, T., Zhao, K., and Nesic, S. (2009). A Practical Mechanistic Model for MIC Based on a Biocatalytic Cathodic Sulfate Reduction Theory. Houston TX: National Association of Corrosion Engineers.
Guan, F., Duan, J., Zhai, X., Wang, N., Zhang, J., Lu, D., et al. (2020). Interaction between sulfate-reducing bacteria and aluminum alloys—Corrosion mechanisms of 5052 and Al-Zn-In-Cd aluminum alloys. J. Mater. Sci. Technol. 36, 55–64. doi: 10.1016/j.jmst.2019.07.009
Guan, F., Zhai, X., Duan, J., Zhang, M., and Hou, B. (2016). Influence of sulfate-reducing bacteria on the corrosion behavior of high strength steel EQ70 under cathodic polarization. PLoS One 11:e0162315. doi: 10.1371/journal.pone.0162315
Guo, J., Yuan, S., Jiang, W., Lv, L., Liang, B., and Pehkonen, S. O. (2018). Polymers for combating biocorrosion. Front. Mater. 5:10. doi: 10.3389/fmats.2018.00010
Hamilton, W. A. (2003). Microbially influenced corrosion as a model system for the study of metal microbe interactions: a unifying electron transfer hypothesis. Biofouling 19, 65–76. doi: 10.1080/0892701021000041078
Hardy, J. A. (1983). Utilisation of cathodic hydrogen by sulphate-reducing bacteria. Br. Corr. J. 18, 190–193. doi: 10.1179/000705983798273642
Hardy, J. A., and Bown, J. L. (1984). The corrosion of mild steel by biogenic sulfide films exposed to air. Corrosion 40, 650–654. doi: 10.5006/1.3593903
Hayat, M. A. (1981). Principles and Techniques of Electron Microscopy. Biological Applications. London: Edward Arnold.
Heidelberg, J. F., Seshadri, R., Haveman, S. A., Hemme, C. L., Paulsen, I. T., Kolonay, J. F., et al. (2004). The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559. doi: 10.1038/nbt959
Heisey, R. M., and Gorham, B. K. (1992). Antimicrobial effects of plant extracts on Streptococcus mutans, Candida albicans, Trichophyton rubrum and other micro-organisms. Lett. Appl. Microbiol. 14, 136–139. doi: 10.1111/j.1472-765X.1992.tb00668.x
Hindré, T., Brüggemann, H., Buchrieser, C., and Héchard, Y. (2008). Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 154, 30–41. doi: 10.1099/mic.0.2007/008698-0
Houry, A., Gohar, M., Deschamps, J., Tischenko, E., Aymerich, S., Gruss, A., et al. (2012). Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 109:13088. doi: 10.1073/pnas.1200791109
Huang, G., Chan, K.-Y., and Fang, H. (2004). Microbiologically induced corrosion of 70Cu30Ni alloy in anaerobic seawater. J. Electrochem. Soc. 151:B434. doi: 10.1149/1.1756153
Itoh, Y., Wang, X., Hinnebusch, B. J., Preston James, F., and Romeo, T. (2005). Depolymerization of β-1,6-N-Acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187, 382–387. doi: 10.1128/JB.187.1.382-387.2005
Jefferson, K. K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x
Jia, R., Tan, J. L., Jin, P., Blackwood, D. J., Xu, D., and Gu, T. (2018). Effects of biogenic H2S on the microbiologically influenced corrosion of C1018 carbon steel by sulfate reducing Desulfovibrio vulgaris biofilm. Corr. Sci. 130, 1–11. doi: 10.1016/j.corsci.2017.10.023
Johnson, M., Cockayne, A., Williams, P. H., and Morrissey, J. A. (2005). Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187, 8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005
Kakooei, S., Ismail, M. C., and Ariwahjoedi, B. (2012). Mechanisms of microbiologically influenced corrosion: a review. World Appl. Sci. J. 17:524.
Kasprzak, K. S. (2002). Oxidative DNA and protein damage in metal-induced toxicity and carcinogenesis1, 3 1this article is part of a series of reviews on “Oxidative DNA Damage and Repair.” the full list of papers may be found on the homepage of the journal. 3guest editor: Miral Dizdaroglu. Free Rad. Biol. Med. 32, 958–967. doi: 10.1016/S0891-5849(02)00809-2
Kato, S. (2016). Microbial extracellular electron transfer and its relevance to iron corrosion. Microb. Biotechnol. 9, 141–148. doi: 10.1111/1751-7915.12340
Kato, S., Yumoto, I., and Kamagata, Y. (2015). Isolation of acetogenic bacteria that induce biocorrosion by utilizing metallic iron as the sole electron donor. Appl. Environ. Microbiol. 81, 67–73. doi: 10.1128/AEM.02767-14
Kawaguchi, T., Chen, Y. P., Norman, R. S., and Decho, A. W. (2008). Rapid screening of quorum-sensing signal N-acyl homoserine lactones by an in vitro cell-free assay. Appl. Environ. Microbiol. 74, 3667–3671. doi: 10.1128/aem.02869-07
Kazakov, A. E., Rajeev, L., Chen, A., Luning, E. G., Dubchak, I., Mukhopadhyay, A., et al. (2015). σ54-dependent regulome in Desulfovibrio vulgaris Hildenborough. BMC Genomics 16:919. doi: 10.1186/s12864-015-2176-y
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Kimura, N. (2014). Metagenomic approaches to understanding phylogenetic diversity in quorum sensing. Virulence 5, 433–442. doi: 10.4161/viru.27850
King, R. A., and Miller, J. D. A. (1971). Corrosion by the sulphate-reducing bacteria. Nature 233, 491–492.
Kip, N., and van Veen, J. A. (2015). The dual role of microbes in corrosion. ISME J. 9, 542–551. doi: 10.1038/ismej.2014.169
Kobayashi, K., and Iwano, M. (2012). BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol. Microbiol. 85, 51–66. doi: 10.1111/j.1365-2958.2012.08094.x
Korenblum, E., Regina de Vasconcelos Goulart, F., de Almeida Rodrigues, I., Abreu, F., Lins, U., Alves, P. B., et al. (2013). Antimicrobial action and anti-corrosion effect against sulfate reducing bacteria by lemongrass (Cymbopogon citratus) essential oil and its major component, the citral. AMB Express 3:44. doi: 10.1186/2191-0855-3-44
Krantz, G. P., Lucas, K., Wunderlich, E. L., Hoang, L. T., Avci, R., Siuzdak, G., et al. (2019). Bulk phase resource ratio alters carbon steel corrosion rates and endogenously produced extracellular electron transfer mediators in a sulfate-reducing biofilm. Biofouling 35, 669–683. doi: 10.1080/08927014.2019.1646731
Krishnamurthy, A., Gadhamshetty, V., Mukherjee, R., Chen, Z., Ren, W., Cheng, H. M., et al. (2013). Passivation of microbial corrosion using a graphene coating. Carbon 56, 45–49. doi: 10.1016/j.carbon.2012.12.060
Krishnamurthy, A., Gadhamshetty, V., Mukherjee, R., Natarajan, B., Eksik, O., Ali Shojaee, S., et al. (2015). Superiority of graphene over polymer coatings for prevention of microbially induced corrosion. Sci. Rep. 5:13858. doi: 10.1038/srep13858
Krumholz, L. R., Bradstock, P., Sheik, C. S., Diao, Y., Gazioglu, O., Gorby, Y., et al. (2015). Syntrophic growth of Desulfovibrio alaskensis requires genes for H2 and formate metabolism as well as those for flagellum and biofilm formation. Appl. Environ. Microbiol. 81, 2339–2348. doi: 10.1128/AEM.03358-14
Kuehr, C. A., and Vlugt, L. S. (1934). The Desulfovibrio vulgaris (Hildenborough) graphitization of cast iron as an electrobiochemical process in anaerobic soil. Water 18, 147–165.
Kumari, N., Rathore, S., Patoli, B. B., and Patoli, A. A. (2020). Bacterial biofilms and ethylenediamine-N, N’-disuccinic acid (EDDS) as potential biofilm inhibitory compound. Proc. Pak. Acad. Sci. B Life Environ. Sci. 57, 85–92.
Labrado, A. L., Brunner, B., Bernasconi, S. M., and Peckmann, J. (2019). Formation of large native sulfur deposits does not require molecular oxygen. Front. Microbiol. 10:24. doi: 10.3389/fmicb.2019.00024
Lee, W., Lewandowski, Z., Nielsen, P. H., and Hamilton, W. A. (1995). Role of sulfate-reducing bacteria in corrosion of mild steel: a review. Biofouling 8, 165–194. doi: 10.1080/08927019509378271
Lespinat, P. A., Berlier, Y. M., Fauque, G. D., Toci, R., Denariaz, G., and LeGall, J. (1987). The relationship between hydrogen metabolism, sulfate reduction and nitrogen fixation in sulfate reducers. J. Ind. Microbiol. 1, 383–388. doi: 10.1007/BF01569336
Leung, H. W. (2001). Ecotoxicology of glutaraldehyde: review of environmental fate and effects studies. Ecotoxicol. Environ. Saf. 49, 26–39. doi: 10.1006/eesa.2000.2031
Li, E., Wu, J., and Zhang, D. (2021). Exogenous autoinducer-2 inhibits biofilm development of Desulfovibrio sp. Huiquan2017. World J. Microbiol. Biotechnol. 37:124. doi: 10.1007/s11274-021-03071-w
Li, Y., Xu, D., Chen, C., Li, X., Jia, R., Zhang, D., et al. (2018). Anaerobic microbiologically influenced corrosion mechanisms interpreted using bioenergetics and bioelectrochemistry: a review. J. Mater. Sci. Technol. 34, 1713–1718. doi: 10.1016/j.jmst.2018.02.023
Li, Y., Zhang, P., Cai, W., Rosenblatt, J., Raad, I. I., Xu, D., et al. (2016). Glyceryl trinitrate and caprylic acid for the mitigation of the Desulfovibrio vulgaris biofilm on C1018 carbon steel. World J. Microbiol. Biotechnol. 32:23.
Lopes, F. A., Morin, P., Oliveira, R., and Melo, L. F. (2006). Interaction of Desulfovibrio desulfuricans biofilms with stainless steel surface and its impact on bacterial metabolism. J. Appl. Microbiol. 101, 1087–1095. doi: 10.1111/j.1365-2672.2006.03001.x
Lu, T. K., and Collins, J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 104:11197. doi: 10.1073/pnas.0704624104
Lv, M., and Du, M. (2018). A review: microbiologically influenced corrosion and the effect of cathodic polarization on typical bacteria. Rev. Environ. Sci. Biotechnol. 17, 431–446. doi: 10.1007/s11157-018-9473-2
Mand, J., Park, H. S., Jack, T. R., and Voordouw, G. (2014). The role of acetogens in microbially influenced corrosion of steel. Front. Microbiol. 5:268. doi: 10.3389/fmicb.2014.00268
Marchal, R., Chaussepied, B., and Warzywoda, M. (2001). Effect of ferrous ion availability on growth of a corroding sulfate-reducing bacterium. Int. Biodeter. Biodegrad. 47, 125–131. doi: 10.1016/S0964-8305(01)00038-5
Mayavan, S., Siva, T., and Sathiyanarayanan, S. (2013). Graphene ink as a corrosion inhibiting blanket for iron in an aggressive chloride environment. RSC Adv. 3, 24868–24871. doi: 10.1039/C3RA43931C
Montgomery, K., Charlesworth, J. C., LeBard, R., Visscher, P. T., and Burns, B. P. (2013). Quorum sensing in extreme environments. Life (Basel, Switzerland) 3, 131–148. doi: 10.3390/life3010131
Mori, K., Tsurumaru, H., and Harayama, S. (2010). Iron corrosion activity of anaerobic hydrogen-consuming microorganisms isolated from oil facilities. J. Biosci. Bioeng. 110, 426–430. doi: 10.1016/j.jbiosc.2010.04.012
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5
Muyzer, G., and Stams, A. J. M. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Okafor, P. C., Ebenso, E. E., and Ekpe, U. J. (2010). Azadirachta indica extracts as corrosion inhibitor for mild steel in acid medium. Int. J. Electrochem. Sci. 5, 978–993.
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Ann. Rev. Microbiol. 54, 49–79.
Pacheco da Rosa, J., Korenblum, E., Franco-Cirigliano, M. N., Abreu, F., Lins, U., Soares, R. M. A., et al. (2013). Streptomyces lunalinharesii strain 235 shows the potential to inhibit bacteria involved in biocorrosion processes. BioMed. Res. Int. 2013:309769. doi: 10.1155/2013/309769
Pena, R. T., Blasco, L., Ambroa, A., González-Pedrajo, B., Fernández-García, L., López, M., et al. (2019). Relationship between quorum sensing and secretion systems. Front. Microbiol. 10:1100. doi: 10.3389/fmicb.2019.01100
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Pereira, P. M., He, Q., Xavier, A. V., Zhou, J., Pereira, I. A. C., and Louro, R. O. (2008). Transcriptional response of Desulfovibrio vulgaris Hildenborough to oxidative stress mimicking environmental conditions. Arch. Microbiol. 189, 451–461. doi: 10.1007/s00203-007-0335-5
Pierre, R., and Roberge, P. D. (2012). Handbook of Corrosion Engineering, 2nd Edn. New York, NY: McGraw-Hill Education.
Poosarla, V. G., Wood, T. L., Zhu, L., Miller, D. S., Yin, B., and Wood, T. K. (2017). Dispersal and inhibitory roles of mannose, 2-deoxy-d-glucose and N-acetylgalactosaminidase on the biofilm of Desulfovibrio vulgaris. Environ. Microbiol. Rep. 9, 779–787.
Price, M. N., Ray, J., Wetmore, K. M., Kuehl, J. V., Bauer, S., Deutschbauer, A. M., et al. (2014). The genetic basis of energy conservation in the sulfate-reducing bacterium Desulfovibrio alaskensis G20. Front. Microbiol. 5:577. doi: 10.3389/fmicb.2014.00577
Procópio, L. (2019). The role of biofilms in the corrosion of steel in marine environments. World J. Microbiol. Biotechnol. 35:73. doi: 10.1007/s11274-019-2647-4
Pysz, M. A., Conners, S. B., Montero, C. I., Shockley, K. R., Johnson, M. R., Ward, D. E., et al. (2004). Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098. doi: 10.1128/AEM.70.10.6098-6112.2004
Qi, Z., Chen, L., and Zhang, W. (2016). Comparison of transcriptional heterogeneity of eight genes between batch Desulfovibrio vulgaris biofilm and planktonic culture at a single-cell level. Front. Microbiol. 7:597. doi: 10.3389/fmicb.2016.00597
Rainey, K., Michalek, S. M., Wen, Z. T., and Wu, H. (2019). Glycosyltransferase-mediated biofilm matrix dynamics and virulence of Streptococcus mutans. Appl. Environ. Microbiol. 85:e02247-18. doi: 10.1128/AEM.02247-18
Rajeev, L., Luning, E. G., Altenburg, S., Zane, G. M., Baidoo, E. E., Catena, M., et al. (2014). Identification of a cyclic-di-GMP-modulating response regulator that impacts biofilm formation in a model sulfate reducing bacterium. Front. Microbiol. 5:382. doi: 10.3389/fmicb.2014.00382
Rathinam, N. K., Tripathi, A. K., Smirnova, A., Beyenal, H., and Sani, R. K. (2018). Engineering rheology of electrolytes using agar for improving the performance of bioelectrochemical systems. Bioresour. Technol. 263, 242–249. doi: 10.1016/j.biortech.2018.04.089
Ren, D., Bedzyk, L. A., Thomas, S. M., Ye, R. W., and Wood, T. K. (2004). Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524. doi: 10.1007/s00253-003-1517-y
Resch, A., Leicht, S., Saric, M., Pásztor, L., Jakob, A., Götz, F., et al. (2006). Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6, 1867–1877. doi: 10.1002/pmic.200500531
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/mmbr.00043-12
Rosa, J. P., Tibúrcio, S. R. G., Marques, J. M., Seldin, L., and Coelho, R. R. R. (2016). Streptomyces lunalinharesii 235 prevents the formation of a sulfate-reducing bacterial biofilm. Braz. J. Microbiol. 47, 603–609. doi: 10.1016/j.bjm.2016.04.013
Roy, D., Simon, G. P., Forsyth, M., and Mardel, J. (2002). Towards a better understanding of the cathodic disbondment performance of polyethylene coatings on steel. Adv. Polym. Technol. 21, 44–58. doi: 10.1002/adv.10010
Sánchez, M. C., Romero-Lastra, P., Ribeiro-Vidal, H., Llama-Palacios, A., Figuero, E., Herrera, D., et al. (2019). Comparative gene expression analysis of planktonic Porphyromonas gingivalis ATCC 33277 in the presence of a growing biofilm versus planktonic cells. BMC Microbiol. 19:58. doi: 10.1186/s12866-019-1423-9
Scarascia, G., Lehmann, R., Machuca, L. L., Morris, C., Cheng, K. Y., Kaksonen, A., et al. (2019). Effect of quorum sensing on the ability of Desulfovibrio vulgaris to form biofilms and to biocorrode carbon steel in saline conditions. Appl. Environ. Microbiol. 86:e01664-19. doi: 10.1128/AEM.01664-19
Scarascia, G., Wang, T., and Hong, P.-Y. (2016). Quorum sensing and the use of quorum quenchers as natural biocides to inhibit sulfate-reducing bacteria. Antibiotics (Basel, Switzerland) 5, 39. doi: 10.3390/antibiotics5040039
Schembri, M. A., Kjærgaard, K., and Klemm, P. (2003). Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48, 253–267. doi: 10.1046/j.1365-2958.2003.03432.x
Sebban-Kreuzer, C., Blackledge, M., Dolla, A., Marion, D., and Guerlesquin, F. (1998). Tyrosine 64 of cytochrome c553 is required for electron exchange with formate dehydrogenase in Desulfovibrio vulgaris Hildenborough. Biochemistry 37, 8331–8340. doi: 10.1021/bi980142u
Shemesh, M., Tam, A., and Steinberg, D. (2007). Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology 153, 1307–1317. doi: 10.1099/mic.0.2006/002030-0
Sherar, B. W. A., Keech, P. G., and Shoesmith, D. W. (2013). The effect of aerobic corrosion on anaerobically-formed sulfide layers on carbon steel in dilute near-neutral pH saline solutions. Corr. Sci. 77, 257–264. doi: 10.1016/j.corsci.2013.08.011
Shimoyama, T., Kato, S., Ishii, S., and Watanabe, K. (2009). Flagellum mediates symbiosis. Science 323:1574. doi: 10.1126/science.1170086
Shrestha, N., Tripathi, A. K., Govil, T., Sani, R. K., Urgun-Demirtas, M., Kasthuri, V., et al. (2020). Electricity from Lignocellulosic substrates by thermophilic Geobacillus species. Sci. Rep. 10:17047. doi: 10.1038/s41598-020-72866-y
Sillankorva, S., Neubauer, P., and Azeredo, J. (2010). Phage control of dual species biofilms of Pseudomonas fluorescens and Staphylococcus lentus. Biofouling 26, 567–575. doi: 10.1080/08927014.2010.494251
Sivakumar, K., Scarascia, G., Zaouri, N., Wang, T., Kaksonen, A. H., and Hong, P.-Y. (2019). Salinity-mediated increment in sulfate reduction, biofilm formation, and quorum sensing: a potential connection between quorum sensing and sulfate reduction? Front. Microbiol. 10:188. doi: 10.3389/fmicb.2019.00188
Stewart, P. S., and Franklin, M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210. doi: 10.1038/nrmicro1838
Stoodley, P., Dodds, I., Boyle, J. D., and Lappin-Scott, H. M. (1998). Influence of hydrodynamics and nutrients on biofilm structure. J. Appl. Microbiol. 85(Suppl. 1), 19s–28s. doi: 10.1111/j.1365-2672.1998.tb05279.x
Summer, E. J., Gill, J. J., Janes, C., and Young, R. F. (2011). “Phage of sulfate reducing bacteria isolated from high saline environment,” in Proceedings of the NACE International Corrosion Conference Series, (Houston, TX: NACE International).
Surette, M. G., Miller, M. B., and Bassler, B. L. (1999). Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U.S.A. 96, 1639–1644. doi: 10.1073/pnas.96.4.1639
Syrett, B. C. (2013). Accelerated corrosion of copper in flowing pure water contaminated with oxygen and sulfide. Corrosion 33, 257–262. doi: 10.5006/0010-9312-33.7.257
Tanaka, K., Yokoe, S., Igarashi, K., Takashino, M., Ishikawa, M., Hori, K., et al. (2018). Extracellular electron transfer via outer membrane cytochromes in a methanotrophic bacterium Methylococcus capsulatus (Bath). Front. Microbiol. 9:2905. doi: 10.3389/fmicb.2018.02905
Tang, H.-Y., Yang, C., Ueki, T., Pittman, C. C., Xu, D., Woodard, T. L., et al. (2021). Stainless steel corrosion via direct iron-to-microbe electron transfer by Geobacter species. ISME J. 15, 3084–3093. doi: 10.1038/s41396-021-00990-2
Toyofuku, M., Inaba, T., Kiyokawa, T., Obana, N., Yawata, Y., and Nomura, N. (2016). Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12. doi: 10.1080/09168451.2015.1058701
Varshney, D., and Kumar, K. (2021). Application and use of different aluminium alloys with respect to workability, strength and welding parameter optimization. Ain Shams Eng. J. 12, 1143–1152. doi: 10.1016/j.asej.2020.05.013
Venzlaff, H., Enning, D., Srinivasan, J., Mayrhofer, K. J. J., Hassel, A. W., Widdel, F., et al. (2013). Accelerated cathodic reaction in microbial corrosion of iron due to direct electron uptake by sulfate-reducing bacteria. Corr. Sci. 66, 88–96. doi: 10.1016/j.corsci.2012.09.006
Voordouw, G. (2002). Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184, 5903–5911. doi: 10.1128/jb.184.21.5903-5911.2002
Waite, R. D., Paccanaro, A., Papakonstantinopoulou, A., Hurst, J. M., Saqi, M., Littler, E., et al. (2006). Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. doi: 10.1186/1471-2164-7-162
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Wen, J., Xu, D., Gu, T., and Raad, I. (2012). A green triple biocide cocktail consisting of a biocide, EDDS and methanol for the mitigation of planktonic and sessile sulfate-reducing bacteria. World J. Microbiol. Biotechnol. 28, 431–435. doi: 10.1007/s11274-011-0832-1
Wen, J., Zhao, K., Gu, T., and Raad, I. (2010). Chelators enhanced biocide inhibition of planktonic sulfate-reducing bacterial growth. World J. Microbiol. Biotechnol. 26, 1053–1057.
Wen, J., Zhao, K., Gu, T., and Raad, I. I. (2009). A green biocide enhancer for the treatment of sulfate-reducing bacteria (SRB) biofilms on carbon steel surfaces using glutaraldehyde. Int. Biodeterior. Biodegradation 63, 1102–1106.
Wikieł, A. J., Datsenko, I., Vera, M., and Sand, W. (2014). Impact of Desulfovibrio alaskensis biofilms on corrosion behaviour of carbon steel in marine environment. Bioelectrochemistry 97, 52–60. doi: 10.1016/j.bioelechem.2013.09.008
Williamson, A. J., Carlson, H. K., Kuehl, J. V., Huang, L. L., Iavarone, A. T., Deutschbauer, A., et al. (2018). Dissimilatory sulfate reduction under high pressure by Desulfovibrio alaskensis G20. Front. Microbiol. 9:1465. doi: 10.3389/fmicb.2018.01465
Williamson, L. L., Borlee, B. R., Schloss, P. D., Guan, C., Allen, H. K., and Handelsman, J. (2005). Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 71, 6335–6344. doi: 10.1128/aem.71.10.6335-6344.2005
Wood, T. L., Gong, T., Zhu, L., Miller, J., Miller, D. S., Yin, B., et al. (2018). Rhamnolipids from Pseudomonas aeruginosa disperse the biofilms of sulfate-reducing bacteria. NPJ Biofilms Microb. 4, 1–8.
Xu, D., and Gu, T. (2014). Carbon source starvation triggered more aggressive corrosion against carbon steel by the Desulfovibrio vulgaris biofilm. Int. Biodeterior. Biodegradation 91, 74–81. doi: 10.1016/j.ibiod.2014.03.014
Xu, D., Li, Y., and Gu, T. (2012). A synergistic D-tyrosine and tetrakis hydroxymethyl phosphonium sulfate biocide combination for the mitigation of an SRB biofilm. World J. Microbiol. Biotechnol. 28, 3067–3074.
Xu, D., Li, Y., and Gu, T. (2016). Mechanistic modeling of biocorrosion caused by biofilms of sulfate reducing bacteria and acid producing bacteria. Bioelectrochemistry 110, 52–58. doi: 10.1016/j.bioelechem.2016.03.003
Xu, D., Li, Y., Song, F., and Gu, T. (2013). Laboratory investigation of microbiologically influenced corrosion of C1018 carbon steel by nitrate reducing bacterium Bacillus licheniformis. Corr. Sci. 77, 385–390. doi: 10.1016/j.corsci.2013.07.044
Yu, L., Duan, J., Du, X., Huang, Y., and Hou, B. (2013). Accelerated anaerobic corrosion of electroactive sulfate-reducing bacteria by electrochemical impedance spectroscopy and chronoamperometry. Electrochem. Commun. 26, 101–104. doi: 10.1016/j.elecom.2012.10.022
Zeng, L., Wooton, E., Stahl, D. A., and Walian, P. J. (2017). Identification and characterization of the major porin of Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 199:e00286-17. doi: 10.1128/JB.00286-17
Zhang, L., Li, S., Liu, X., Wang, Z., Jiang, M., Wang, R., et al. (2020). Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11:5371. doi: 10.1038/s41467-020-19243-5
Zhang, P., Xu, D., Li, Y., Yang, K., and Gu, T. (2015). Electron mediators accelerate the microbiologically influenced corrosion of 304 stainless steel by the Desulfovibrio vulgaris biofilm. Bioelectrochemistry 101, 14–21. doi: 10.1016/j.bioelechem.2014.06.010
Zhang, W., Culley, D. E., Hogan, M., Vitiritti, L., and Brockman, F. J. (2006). Oxidative stress and heat-shock responses in Desulfovibrio vulgaris by genome-wide transcriptomic analysis. Antonie Van Leeuwenhoek 90, 41–55. doi: 10.1007/s10482-006-9059-9
Zhang, W., Culley, D. E., Nie, L., and Scholten, J. C. (2007). Comparative transcriptome analysis of Desulfovibrio vulgaris grown in planktonic culture and mature biofilm on a steel surface. Appl. Microbiol. Biotechnol. 76, 447–457. doi: 10.1007/s00253-007-1014-9
Zhang, W., Picu, R. C., and Koratkar, N. (2008). The effect of carbon nanotube dimensions and dispersion on the fatigue behavior of epoxy nanocomposites. Nanotechnology 19:285709. doi: 10.1088/0957-4484/19/28/285709
Zhang, Y., Pei, G., Chen, L., and Zhang, W. (2016). Metabolic dynamics of Desulfovibrio vulgaris biofilm grown on a steel surface. Biofouling 32, 725–736.
Zhang, Y., Wang, X., Zhen, Y., Mi, T., He, H., and Yu, Z. (2017). Microbial diversity and community structure of sulfate-reducing and sulfur-oxidizing bacteria in sediment cores from the East China Sea. Front. Microbiol. 8:2133.
Keywords: sulfate reducing bacteria, biofilm, microbiologically influenced corrosion, comparative genomics, biocides, 2D-materials, quorum sensing, biocorrosion mechanism
Citation: Tripathi AK, Thakur P, Saxena P, Rauniyar S, Gopalakrishnan V, Singh RN, Gadhamshetty V, Gnimpieba EZ, Jasthi BK and Sani RK (2021) Gene Sets and Mechanisms of Sulfate-Reducing Bacteria Biofilm Formation and Quorum Sensing With Impact on Corrosion. Front. Microbiol. 12:754140. doi: 10.3389/fmicb.2021.754140
Received: 06 August 2021; Accepted: 24 September 2021;
Published: 29 October 2021.
Edited by:
Ales Lapanje, Institut “Jožef Stefan” (IJS), SloveniaReviewed by:
Sarangam Majumdar, University of L’Aquila, ItalyCopyright © 2021 Tripathi, Thakur, Saxena, Rauniyar, Gopalakrishnan, Singh, Gadhamshetty, Gnimpieba, Jasthi and Sani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajesh Kumar Sani, UmFqZXNoLnNhbmlAc2RzbXQuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.