
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 September 2021
Sec. Infectious Agents and Disease
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.750940
This article is part of the Research Topic Regulation of Gene Expression in Enteropathogenic Bacteria, Volume III View all 14 articles
Salmonella alters cellular processes as a strategy to improve its intracellular fitness during host infection. Alternative σ factors are known to rewire cellular transcriptional regulation in response to environmental stressors. σs factor encoded by the rpoS gene is a key regulator required for eliciting the general stress response in many proteobacteria. In this study, Salmonella Typhimurium deprived of an outer membrane protein YcfR was attenuated in intracellular survival and exhibited downregulation in Salmonella pathogenicity island-2 (SPI-2) genes. This decreased SPI-2 expression caused by the outer membrane perturbation was abolished in the absence of rpoS. Interestingly, regardless of the defects in the outer membrane integrity, RpoS overproduction decreased transcription from the common promoter of ssrA and ssrB, which encode a two-component regulatory system for SPI-2. RpoS was found to compete with RpoD for binding to the PssrA region, and its binding activity with RNA polymerase (RNAP) to form Eσs holoenzyme was stimulated by the small regulatory protein Crl. This study demonstrates that Salmonella undergoing RpoS-associated stress responses due to impaired envelope integrity may reciprocally downregulate the expression of SPI-2 genes to reduce its virulence.
The bacterial RNA polymerase (RNAP) holoenzyme is a provisional complex between a multi-subunit RNAP core enzyme (E, α2ββ'ω) and an σ factor. The σ factor forming the Eσ complex directs promoter-specific transcription initiation and then dissociates from the core enzyme E after transcription initiation. In many proteobacteria, σD (σ70), encoded by rpoD, functions as a major σ subunit responsible for the transcription of constitutive promoters. Under unfavorable conditions, bacteria exploit alternative σ factors to redistribute RNAP core enzyme specificity toward discrete subsets of genes whose products help survive and adapt to environmental stressors (Bang et al., 2005). Salmonella enterica serovar Typhimurium possesses five alternative σ factors, including extra-cytoplasmic stress-specific σE (σ24, RpoE), flagella-chemotaxis-specific σF (σ28, FliA), heat-shock response-specific σH (σ32, RpoH), stationary-phase nutrient-starvation-specific σS (σ38, RpoS), and nitrogen-starvation-specific σN (σ54, RpoN). The abundance of alternative σ factors available for Eσ complex formation is regulated not only by environmental signals (Shimada et al., 2017) but also by the interaction with two types of inhibitory proteins, anti-σ factors, and adaptor proteins (Trevino-Quintanilla et al., 2013). Different σ factors operate discrete regulatory circuits containing cognate genes and operons in response to specific environmental cues, but some transcriptional regulons are coordinated by multiple σ factors that function in a regulatory cascade or by competitive interactions. In response to the diverse stimuli encountered by bacterial pathogens upon host infection, multiple alternative σ factors interact with each other to promote bacterial adaptation in hostile conditions. σE can activate one of the rpoH promoters (Hiratsu et al., 1995; Vanaporn et al., 2008) and σH, in turn, stimulates the transcription of Hfq (Muffler et al., 1996), which is required for efficient translation of rpoS mRNA (Bang et al., 2005), indicating sequential activation of multiple regulons by a regulatory cascade of σE, σH, and σS under certain circumstances. Besides, there is a trade-off between self-preservation and nutritional competence and genes required for membrane integrity maintenance and genes associated with metabolism are reciprocally controlled by competitive action between σ factors (Ferenci, 2005; Levi-Meyrueis et al., 2015). In the context of competitive action between σ factors, σ70 and σS recognize almost identical −35 and −10 promoter elements, especially the −10 region (Hengge-Aronis, 2002b). Therefore, competitive binding of EσS to the overlapping promoter regions may occlude transcription initiation by Eσ70, inducing the transcription of a repertoire of genes by σS under stressful environments (Levi-Meyrueis et al., 2015).
Many genes whose promoters bind to both σ70 and σS show stronger transcription activities with σ70 binding than with σS binding, implying a negative role of σS in gene expression (Levi-Meyrueis et al., 2015; Grove et al., 2017; Yin et al., 2018). Interestingly, nullifying the negative effects of σS is beneficial to bacterial growth in the absence of environmental stressors (Zambrano et al., 1993; Notley-McRobb et al., 2002). The attenuated expression associated with EσS may confer fitness advantages to bacteria during unfavorable conditions. σS is induced under nutrient-depleted stationary phase or in response to various stressors, and its activity in Salmonella is known to alter transcription or protein production of more than 20% of its genome (Levi-Meyrueis et al., 2014; Lago et al., 2017). σS upregulates or downregulates the expression of a myriad of genes involved in carbohydrate and amino acid metabolism, stress resistance, and membrane integrity directly or indirectly. In contrast to the essential roles of σS in bacterial stress resistance, the requirement of σS for bacterial virulence varies between bacterial species (Dong and Schellhorn, 2010). Salmonella Typhimurium lacking rpoS gene showed reduced virulence, and σS factor was found to activate the transcription of spvR and spvABCD virulence plasmid genes (Fang et al., 1992; Kowarz et al., 1994).
In this study, we induced outer membrane perturbation on S. Typhimurium by deleting ycfR to stimulate σS-mediated adaptation responses. YcfR is a putative outer membrane protein that is expressed under stressful conditions in enteric pathogens and is known as a multiple stress resistance protein (Zhang et al., 2007; Salazar et al., 2013). Our previous study demonstrated that the deletion of ycfR caused structural alterations in lipopolysaccharide and destabilized Salmonella envelope integrity (Kim and Yoon, 2019). Salmonella devoid of YcfR tremendously increased rpoS transcription and showed an increase in curli fibers, cellulose, and c-di-GMP production and a decrease in motility, implicating comprehensive transcriptional alterations by σS in response to stress on the cellular envelope (Kim and Yoon, 2019). Besides the known repertoires of σS regulatory circuits, such as biofilm formation, this study revealed that virulence genes of Salmonella pathogenicity island-2 (SPI-2) were downregulated by σS. SPI-2 is a locus responsible for the type III secretion system (T3SS) injectisome-mediated delivery of virulence factors from Salmonella to host cells and is critical for bacterial survival and replication inside host cells (Jennings et al., 2017). The negative role of σS in SPI-2 regulation was influenced by a small regulatory protein Crl. Crl is known to be required for σS-dependent transcriptional initiation at the promoters of adrA and csgD genes, whose products activate curli and cellulose production (Robbe-Saule et al., 2006). The role of σS in Salmonella virulence regulation was elucidated by examining the interaction between σS and the ssrAB promoter, which encodes the two-component regulatory system SsrAB for SPI-2.
Salmonella enterica serovar Typhimurium ATCC 14028 was used as the parent strain. Salmonella mutants of ΔycfR and ΔrpoS were constructed using the phage lambda (λ) Red recombination system as described in previous studies (Yoon et al., 2009; Kim and Yoon, 2019) and a mutant lacking both ycfR and rpoS was constructed using P22 HT105/1 int-201-mediated transduction (Kwoh and Kemper, 1978). The phage λ Red recombination system was also used for the construction of Salmonella strains producing HA-tagged SPI-2 proteins (SseC and SsaN) as described in the previous study (Kim et al., 2018). In brief, the kanamycin resistance (kan) cassette of pKD13-2HA was amplified by PCR using primers designed to provide 40-nucleotide sequences homologous to target genes at both termini of the resultant PCR products. The PCR products were introduced into Salmonella cells harboring pKD46 to insert the HA-coding sequences with a kan cassette prior to the stop codon sequences. The kan marker was subsequently removed using pCP20 providing a flip recombinase. Primers used for the construction of HA-tagged SPI-2 genes are listed in Supplementary Table 1. Escherichia coli DH5α strain was used for plasmid cloning and protein purification.
To express rpoS in trans, the rpoS gene was cloned into pACYC184 (Chang and Cohen, 1978) and pBbA2sk-RFP vectors (Lee et al., 2011). For the construction of pRpoS, the rpoS CDS and its promoter region were amplified by PCR using primers pRpoS-CF and pRpoS-CR and inserted into pACYC184 through BamHI and SalI restriction enzyme sites. In cloning pRpoS2, the rpoS gene was amplified using PCR with pRpoS-CF2 and pRpoS-CR2 primers, digested with EcoRI and BglII, and ligated with EcoRI/BglII digested pBbA2sk-RFP plasmid. Primer sequences are listed in Supplementary Table 1.
To construct transcriptional lacZ fusion to the PssrA and PssrB regions, the promoter regions of ssrA (from −253 to +209) and ssrB (from −90 to +303) were amplified by PCR using primers (Supplementary Table 1) of pssrA-lacZ-CF and pssrA-lacZ-CR for ssrA and primers (Supplementary Table 1) pssrB-lacZ-CF and pssrB-lacZ-CR for ssrB, as described by Feng et al. (2003). The amplified promoter regions were cloned into the pRS415 plasmid (Simons et al., 1987) using EcoRI and SalI restriction enzyme sites.
RpoS, RpoD, and Crl proteins were tagged with His6 at their N-termini by cloning three genes into the pUHE21-lacIq plasmid (Soncini et al., 1995) via EcoRI and HindIII and inducing their expression using IPTG. The primers used for the construction of His6 tagged proteins are listed in Supplementary Table 1. All restriction enzymes and ligases were purchased from Takara Bio, Inc. (CA, United States).
Salmonella cells were cultivated in Luria-Bertani (LB) medium broth or acidic minimal medium (AMM) broth at 220rpm at 37°C, as described in previous studies (Yoon et al., 2009, 2011). For AMM cultivation, bacterial cells at the stationary growth phase in LB medium broth were washed twice with PBS, diluted in pH 7.0 minimal medium broth at a 1:100 ratio, and cultivated overnight. Pre-cultured Salmonella cells in minimal medium broth (pH 7.0) were diluted in minimal medium broth (pH 5.0) at a 1:20 ratio and cultivated for 3h to mimic intracellular conditions (Yoon et al., 2009). Antibiotics were purchased from Sigma-Aldrich (MO, United States) and used when required: ampicillin (Amp, 50μg/ml), chloramphenicol (Cm, 35μg/ml), kanamycin (Kan, 50μg/ml), and anhydrotetracycline (aTc, 0.2 or 0.5ng/ml).
To assess bacterial invasiveness, HeLa human epithelial cell line (ATCC CCL-2) was infected as described in the previous study (Kim et al., 2018). HeLa cells were seeded in 24-well plate at 2×105 cells/well and incubated in Dulbecco’s modified Eagle’s medium (DMEM; Corning cellgro, Thermo Scientific Inc., IL, United States) supplemented with 4.5g/L glucose (Thermo Scientific Inc.) and 10% fetal bovine serum (FBS; Gibco, Thermo Scientific Inc.) at 37°C with 5% CO2. After overnight incubation, HeLa cells were treated with Salmonella cells grown for 2.5h in LB medium broth at a multiplicity of infection (MOI) of 100 and centrifuged at 500×g for 5min. At 30min post-infection, the infected cells were washed twice with PBS and replenished with fresh DMEM containing 100μg/ml gentamicin for 1.5h to remove extracellular Salmonella cells. The infected HeLa cells were washed three times with PBS and lysed with 1% Triton X-100. The cell lysates were diluted and plated on LB agar to count intracellular Salmonella cells.
Salmonella survival inside macrophages was examined as described elsewhere (Yoon et al., 2009; Kim and Yoon, 2019). Murine macrophage RAW264.7 (ATCC TIB-71) cells were seeded in 24-well plate at 5×105 cells/well and incubated in DMEM containing 4.5g/L glucose and 10% FBS at 37°C with 5% CO2 overnight. Monolayered-macrophage cells were infected with Salmonella cells grown overnight in LB medium broth at MOI 100, as described for HeLa cell infection. After 30min of infection, the extracellular bacteria were removed by replacing the medium with DMEM containing 100μg/ml gentamicin for 1.5h. The infected macrophages were washed with PBS three times and incubated in fresh DMEM containing 20μg/ml gentamicin for additional 8h. To enumerate intracellular bacteria, RAW264.7 cells were lysed, and the lysates were spread on LB agar as described above.
Bacterial total RNA was isolated from Salmonella cultivated in LB medium and AMM broth or RAW264.7 cells infected with Salmonella. Bacterial cells cultivated in vitro were treated with RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany) and subjected to total RNA extraction using RNeasy mini kit (Qiagen). For RNA extraction from intracellular bacteria, infected macrophage cells were treated with RNAlater™ Stabilization Solution (Invitrogen, Thermo Scientific Inc.) and processed with RNeasy mini kit according to the manufacturer’s recommendations. Isolated total RNA was treated with RNase-free DNase (Ambion, TX, United States) at 37°C for 30min and used to synthesize cDNA using RNA to cDNA EcoDryTM Premix (Takara Bio United States, Inc.). cDNA corresponding to 10ng of input RNA was used as a template in each qRT-PCR, and the primer sequences are listed in Supplementary Table 2. qRT-PCR was conducted using the StepOnePlus Real-time PCR system (Applied Biosystems, MA, United States) with Power SYBR Green PCR Master Mix (Applied Biosystems), and the levels of amplified PCR products were normalized to those of gyrB (Yoon et al., 2009).
The β-galactosidase assay was conducted using the Miller method (Smale, 2010). Bacterial cells were cultivated in LB medium broth, and β-galactosidase activity normalized to the number of input bacteria was represented by Miller units. Miller units were computed as follows: Miller unit=[1,000×(OD420–1.75×OD550)]/(t×V×OD600), where t is time (min) and V is volume (ml).
Bacterial cells were pelleted and resuspended in 1× Laemmli sample buffer (Bio-Rad Laboratories, Inc., CA, United States). The aliquots were loaded on 10% SDS-PAGE gels, and the separated proteins were transferred to PVDF membranes (Bio-Rad Laboratories, Inc.). The membrane was blocked with 5% skim milk solution and treated with anti-RpoS antibody (anti-E. coli RNA Sigma S antibody, BioLegend, CA, United States) at a 1:2,000 dilution ratio or anti-DnaK antibody (Enzo Life Science, NY, United States) at a 1:10,000 dilution ratio in combination with horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad Laboratories, Inc.) at a 1:3,000 dilution ratio. SPI-2 proteins tagged with HA were identified using anti-HA antibody (1:10,000 dilution; Sigma, United States) as a primary antibody. Immunoblotting was conducted using ECL™ Western Blotting Detection Reagents kit (GE Healthcare, Thermo Scientific Inc.), and the blot images were visualized using the ChemiDoc™ MP System (Bio-Rad Laboratories, Inc.). The intensity of the blot images was analyzed using ImageJ software.1
The Chromatin immunoprecipitation (ChIP) assay was performed as previously described (Gu et al., 2016; Yin et al., 2018) with minor modifications. Briefly, Salmonella cells cultivated in the stationary growth phase in LB medium broth were fixed with 1% formaldehyde solution for 10min and subsequently treated with 100mM glycine for 5min. Cells were washed with cold PBS and resuspended in SDS lysis buffer (50mM Tris–HCl, 10mM EDTA, 1% SDS, and pH 8.0) containing 1× protease inhibitor. After 10min of incubation, the cell extract was sonicated to fragment genomic DNA into 200bp to 1kb and centrifuged at 12,000×g for 10min. The supernatant solution was used as input DNA, and the aliquots were further processed for pre-clearing and immunoprecipitation (IP) samples. The lysate solution containing DNA-protein complexes was pre-incubated with Protein A/G Plus-Agarose (Santa Cruz Biotechnology, Inc. TX, United States) at 4°C for 2h to remove DNA or proteins non-specifically bound to Protein A/G Plus-Agarose and centrifuged at 800×g for 3min. The resultant pellet fraction was used as a pre-clearing sample, and the supernatant solution was further incubated with the anti-RpoS antibody at 4°C overnight, followed by Protein A/G Plus-Agarose at 4°C for 2h, and centrifuged at 800×g for 3min to immunoprecipitate DNA-RpoS complexes bound to the agarose. The pellet fraction was used as an IP sample. The pre-clearing and IP samples were washed with LiCl wash buffer (100mM Tris–HCl, pH 8.0, 2% Triton X-100, and 250mM LiCl), twice with high-salt buffer (100mM Tris–HCl, pH 8.0, 600mM NaCl, and 2% Triton X-100), twice with low-salt buffer (100mM Tris–HCl, pH 8.0, 300mM NaCl, and 2% Triton X-100), and with TE wash buffer (10mM Tris–HCl, pH 8.0, and 1mM EDTA). The precipitated DNA-protein complexes were eluted with ChIP elution buffer (50mM Tris–HCl, pH 8.0, 10mM EDTA, and 1% SDS) and incubated with 0.2M NaCl at 65°C overnight to resolve DNA-protein cross-links. All samples were treated with RNase A (10mg/ml) at 37°C for 30min and further incubated with a protease solution (1M Tris–HCl, pH 8.0, 500mM EDTA, proteinase K, and 5M NaCl) at 65°C for 4h. DNA from pre-clearing and IP samples was extracted using phenol: chloroform: isoamyl alcohol (25: 24: 1) solution, precipitated with EtOH and NaOAc (pH 5.2), and resuspended in distilled water.
DNA cross-linked to RpoS was analyzed using quantitative PCR (qPCR), as previously described (Hermans et al., 2016). Relative enrichment (RE) of the promoter of interest was computed using differences in Ct values (ΔCt) with gyrB gene as an endogenous control as follows: RE=2−(∆CtIP−∆Ct Pre-clearing), where ∆CtIP is Ctpromoter test −CtgyrB for the IP samples and ∆CtPre-clearing is Ctpromoter test −CtgyrB for the pre-clearing samples. Aliquots of DNA purified from IP and pre-clearing samples and serial dilutions of input DNA were used as templates in qPCR, and the qPCR primers are listed in Supplementary Table 3. Amplified PCR products were analyzed using the StepOnePlus Real-time PCR system with Power SYBR Green reagent.
Escherichia coli strains producing His6-tagged RpoS, RpoD, and Crl proteins were cultivated in LB medium broth, and the proteins were induced by adding 0.05mM (RpoS and RpoD) or 1mM (Crl) isopropyl β-D-1-thiogalactopyranoside for 7 or 3h at 30°C. Bacterial cells were centrifuged at 10,000×g for 10min and resuspended in lysis buffer (50mM NaH2PO4, 300mM NaCl, 20mM imidazole, and pH 8.0) containing 1mg/ml lysozyme. After 30min incubation on ice, the cells were sonicated and centrifuged at 10,000×g and 4°C for 20min. The resultant soluble lysate fraction was treated with Ni2+-nitrilotriacetic acid (Ni2+-NTA) agarose beads (Qiagen) at 4°C for 1h with rotation and loaded onto a Ni2+-NTA agarose affinity column (Qiagen). The column was washed with washing buffer (50mM NaH2PO4, 300mM NaCl, 40mM imidazole, and pH 8.0) three times, and the proteins were eluted with elution buffer (50mM NaH2PO4, 300mM NaCl, 300mM imidazole, and pH 8.0). The eluted protein fraction was packed into SnakeSkin™ Dialysis tubing with 10K MWCO (Thermo Scientific Inc.) and subjected to dialysis at 4°C in dialysis buffer (20mM Tris–HCl, pH 8.0, 150mM NaCl, 0.1mM EDTA, 5mM DTT, and 20% glycerol). Purified proteins were quantified using Bradford assay.
The binding between σ factors and the PssrA region was investigated using His6-RpoS or His6-RpoD in combination with His6-Crl. The PssrA region was PCR-amplified using primers ssrA-electrophoretic mobility shift assay (EMSA)-F and ssrA-EMSA-R. The csgBA promoter region amplified using primers cgsBA-EMSA-F and cgsBA-EMSA-R was employed as a positive control, whereas the STM14_1978 (putative ABC transporter permease component) CDS region amplified using primers STM14_1978 EMSA-F and STM14_1978 EMSA-R was used as a negative control. The primer sequences used in the EMSA are listed in Supplementary Table 3. EMSA was performed as described previously (Bougdour et al., 2004; Storvik and Foster, 2010) with the following modifications. To reconstitute the RNAP holoenzyme, 20nM RNAP core enzyme (E. coli RNAP Core Enzyme; NEB, MA, United States) was incubated with 300nM His6-RpoS or His6-RpoD in a binding buffer (200mM Tris–HCl, pH 8.0, 30mM KCl, 10mM MgCl2, 50mM NaCl, 1mM DTT, 1mM EDTA, and BSA 20μg/ml) at 30°C for 45min. DNA of 20ng was incubated with the reconstituted RNAP holoenzyme in a binding buffer (50mM Tris–HCl, pH 8.0, 200mM KCl, 3mM MgCl2, 1mM DTT, 0.1mM EDTA, BSA μg/ml, and Poly (di-dc) 12ng/μl) at 25°C for 30min. The reactant was analyzed by electrophoresis using 5% native polyacrylamide gel, and DNA fragments were stained with EtBr solution and detected using the ChemiDoc MP System.
In the competitive binding assay between His6-RpoS and His6-RpoD, one σ factor was used at a constant concentration of 75nM and the other competitor σ factor was used at incremental concentrations from 12.5 to 150nM. After RNAP holoenzyme reconstitution with different concentrations of σ factors, DNA corresponding to the PssrA region was added to the binding reaction and analyzed as described above. To localize His6-RpoS after electrophoresis on a native polyacrylamide gel, proteins on the gel were transferred to a PVDF membrane and processed as described in the immunoblot assay above. Anti-E. coli RNA sigma S antibody was used as a primary antibody at a 1:2,000 dilution ratio, and HRP-conjugated goat anti-mouse IgG was used as a secondary antibody at a 1:3,000 dilution ratio.
In the competition assay using His6-RpoS in combination with His6-Crl, His6-RpoS (25, 50, and 280nM) was pre-incubated with 280nM His6-Crl at 25°C for 15min and then used to compete with 50nM His6-RpoD in the RNAP holoenzyme reconstitution reaction. After the addition of PssrA region, the locations of PssrA DNA and His6-RpoS were identified using EtBr staining and immunoblotting methods, respectively, as described above.
All assays were repeated at least three times, and the average values were presented with their SDs. To determine the significant differences, Student’s t-test was applied, and the value of p was calculated.
YcfR, which is expressed in response to multiple stress conditions, is a putative outer membrane protein important for stress resistance in enteric pathogens such as Salmonella spp. and E. coli (Zhang et al., 2007; Salazar et al., 2013). In this study, we observed that Salmonella lacking YcfR was significantly attenuated in virulence during host cell infection. The lack of YcfR did not influence bacterial growth in vitro, but significantly reduced the ability of bacteria to invade host epithelial cells and survive inside phagocytic cells (Figure 1). The transcription of SPI-1 genes, which produce a distinct T3SS (T3SS1) and promote Salmonella invasion into host cells (Raffatellu et al., 2005), decreased remarkably in the ΔycfR strain (Figure 2A). Besides the attenuated SPI-1 expression, the physiological changes caused by the lack of YcfR, including cellular aggregation and reduced motility (Kim and Yoon, 2019), might impair bacterial invasion ability. Interestingly, the lack of YcfR also decreased the transcription of SPI-2 genes not only inside macrophage cells (Supplementary Figure 1A) but also in LB and AMM cells in vitro (Figure 2B; Supplementary Figure 1B, respectively), which partially reproduce the intestinal lumen and intracellular milieux, respectively (Beuzon et al., 1999; Yoon et al., 2011).
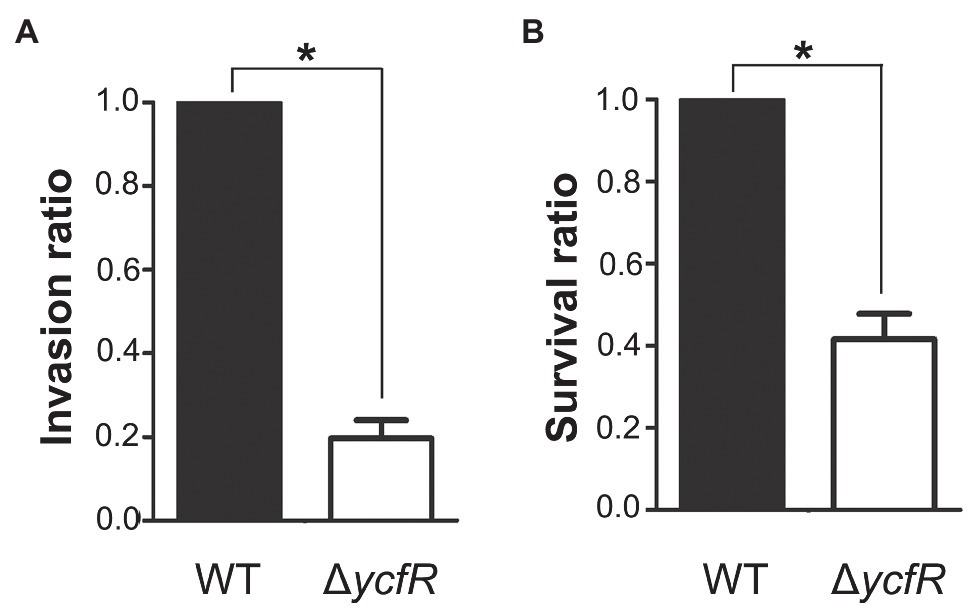
Figure 1. Invasion and survival of a ΔycfR mutant in the host cell. (A) Invasive ability of wild-type and ΔycfR mutant strains was assessed by infecting HeLa cells and counting intracellular bacteria 2h post-infection. The bar indicates relative invasion ability compared to wild-type Salmonella. (B) Survival inside RAW264.7 macrophages was compared between wild-type and ΔycfR mutant strains 10h post-infection. The numbers phagocytosed by macrophages were found comparable between two strains (data not shown) and relative survival ratios are depicted. A significant difference (value of p<0.05) is denoted with an asterisk.
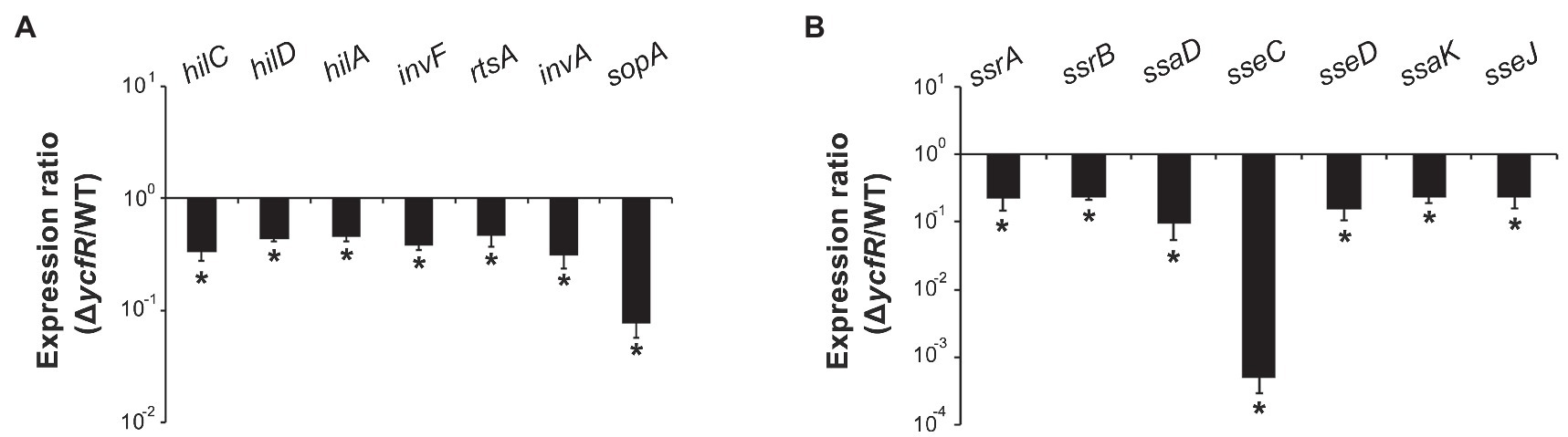
Figure 2. Expression of SPI-1 and Salmonella pathogenicity island-2 (SPI-2) genes in ΔycfR mutant. (A) Transcription levels of SPI-1 genes were examined using RNA isolated from Salmonella strains grown in LB medium broth for 2h. The Ct values of qRT-PCR were normalized using those of gyrB and the fold-change between wild-type and ΔycfR mutant strains was plotted. (B) To analyze the expression of SPI-2 genes, Salmonella strains were cultivated in LB medium broth for 10h and subjected to RNA extraction. Ct values of each gene were subtracted from those of gyrB for normalization, and the fold-change (ΔycfR/wild-type) was calculated. An asterisk indicates a difference of a value of p<0.05.
In order to figure out a transcriptional regulator that coordinates bacterial virulence in response to outer membrane perturbation, we assessed the expression of 21 regulators associated with SPI-1 or SPI-2 regulation in the ΔycfR strain (Supplementary Figure 2) and found that rpoS showed a dramatic increase in its transcription. The levels of RpoS were compared between wild-type and ΔycfR strains in LB and AMM conditions. RpoS increased in the ΔycfR strain grown in both media (1.5-fold in LB; 2.7-fold in AMM; Figure 3). To examine whether an increase in RpoS could downregulate virulence genes associated with SPI-2 T3SS (T3SS2), the transcription of ssrAB encoding the two-component regulatory system for T3SS2 and its cognate effectors was compared. Salmonella deprived of RpoS slightly increased the expression of ssrAB, but the introduction of pRpoS expressing rpoS under its own promoter significantly decreased the transcription of ssrAB, implicating overall downregulation of their cognate T3SS2-associated genes by RpoS (Figure 4A). In addition, the decreased transcription of ssrAB in the absence of YcfR was nullified by the additional rpoS deletion, suggesting the possibility of σS-mediated SPI-2 downregulation in the ΔycfR strain (Figure 4A). The ssrA and ssrB genes, located adjacent to each other, encode a sensor kinase and its response regulator, respectively, and are regarded to be transcribed in a polycistronic mRNA under the same promoter (Bustamante et al., 2008; Fass and Groisman, 2009). However, the identification of a distinct promoter upstream of ssrB revealed the possibility that the expression of ssrA and ssrB could be uncoupled depending on the growth conditions (Feng et al., 2003, 2004). Therefore, the negative role of RpoS was reexamined using lacZ transcriptional fusion constructs, where the promoters of ssrA and ssrB were separately analyzed (Figure 4B). The promoter strength of ssrA was much stronger than that of ssrB in wild-type Salmonella harboring intact rpoS and ycfR genes, and deletion of rpoS alone did not alter ssrA or ssrB transcription. However, ycfR deletion, which led to an increase in RpoS, abolished ssrA transcription but not ssrB, and the additional rpoS deletion in ΔycfR mutant derepressed ssrA only, indicating differential regulation of ssrA and ssrB by σS (Figure 4B). Again, overexpression of RpoS by the introduction of pRpoS2 decreased PssrA::lacZ expression in proportion to aTc concentration. These results suggest that σS at high concentrations dampen transcription activity at the promoter region upstream of ssrAB.
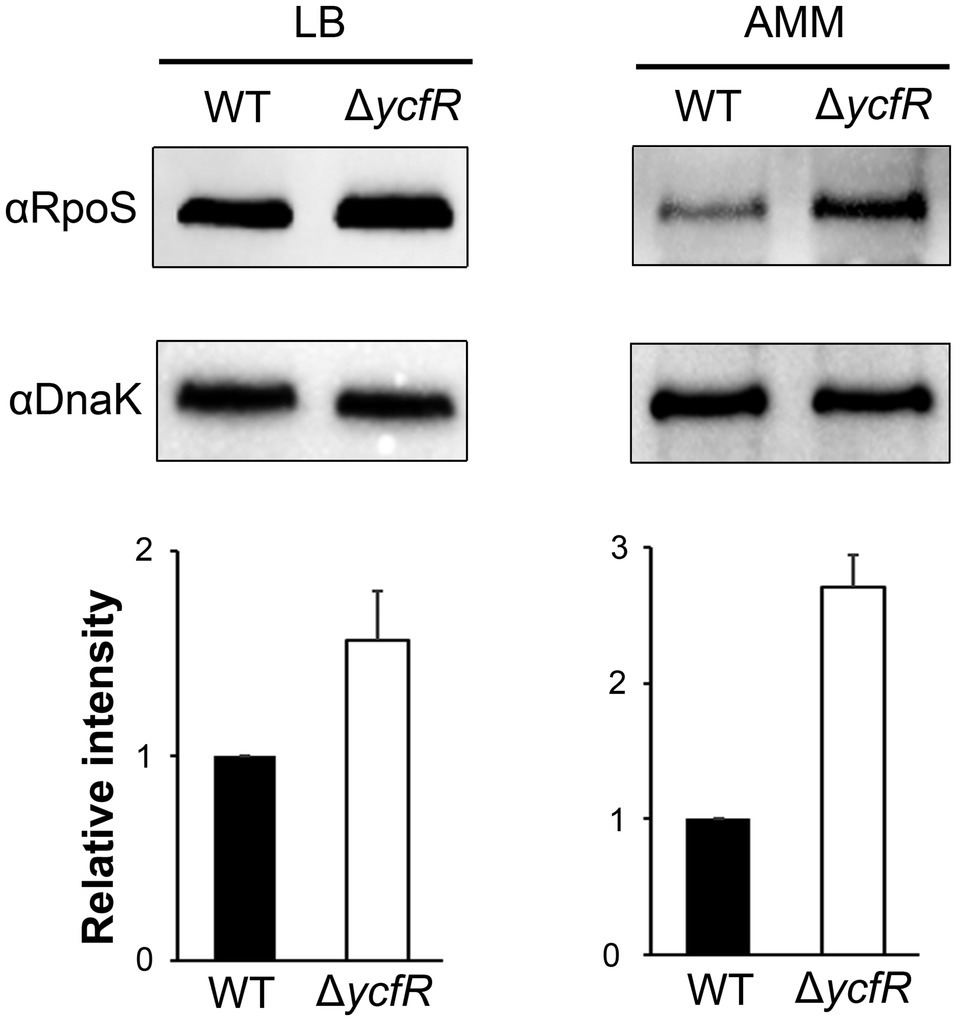
Figure 3. Expression of RpoS in ΔycfR mutant. Salmonella wild-type and ΔycfR mutant strains were cultivated in LB medium broth for 10h or acidic minimal medium (AMM) broth for 3h, and the expression of RpoS was compared using immunoblot assay with anti-RpoS antibody. The cytosolic protein DnaK was used as a control to standardize the protein amounts between the lanes. The abundance of RpoS was normalized to that of DnaK using ImageJ, and the ratios from three independent assays are depicted below the representative blot images.
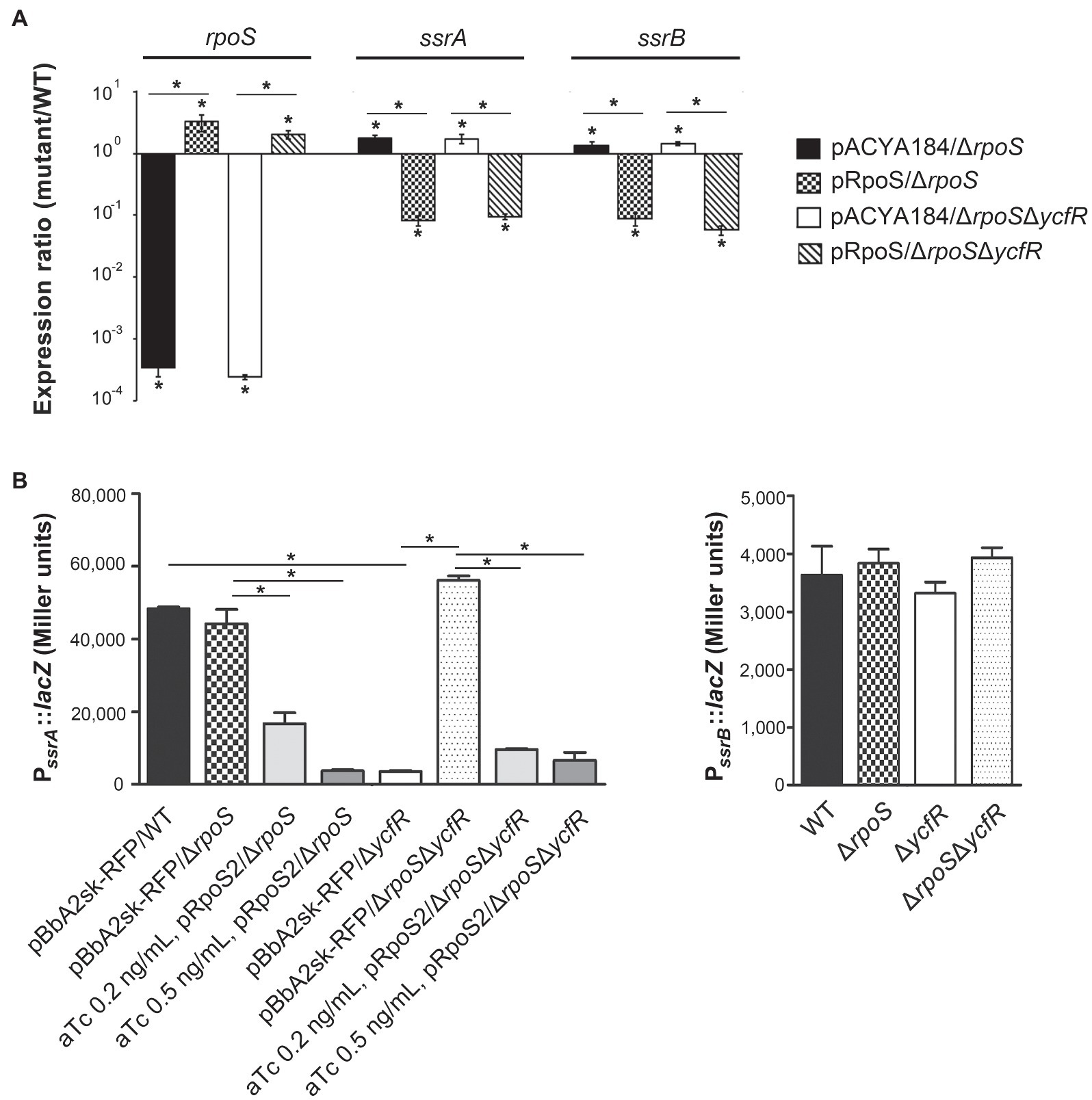
Figure 4. Negative regulation of ssrA by RpoS. (A) Transcription levels of rpoS, ssrA, and ssrB were measured using qRT-PCR. Salmonella strains, including wild-type, ΔrpoS, and ΔrpoSΔycfR strains, were transformed with pRpoS or pACYC184 and cultivated to the stationary growth phase in LB medium broth. Ct values of rpoS, ssrA, and ssrB were normalized using those of gyrB gene. Expression levels of rpoS, ssrA, and ssrB from each strain were compared with those from wild-type strain harboring pACYC184 and the fold-change (mutant/wild-type) was plotted. Value of p with p<0.05 is denoted with an asterisk. (B) Transcription from PssrA and PssrB was measured using lacZ transcriptional fusions. Plasmids pSsrA::lacZ (left) and pSsrB::lacZ (right) were introduced into wild-type, ΔrpoS, ΔycfR, and ΔrpoSΔycfR strains. To overexpress RpoS, pRpoS2 and its empty plasmid pBbA2sk-RFP were introduced into wild-type and mutant strains, and aTc of 0.2 or 0.5ng/ml was added in the LB medium broth cultures. β-galactosidase assay was conducted with bacterial cells at the stationary growth phase. An asterisk indicates a value of p<0.05.
To examine whether σS directly controls the transcription of ssrA, a ChIP assay was performed on Salmonella cells in the stationary growth phase using σS as a bait. DNA fragments bound to σS were co-precipitated using anti-RpoS antibody and used as templates in PCR using primers targeting the promoter regions of ssrA and ssrB (Figure 5A; Supplementary Figure 2). DNA fragments containing the PssrA region were bound to σS and amplified by PCR, but the PssrB region did not co-precipitate with σS (Figure 5B). When five different primer sets from R1-F/R to R5-F/R were used to dissect the ssrA promoter region, only two primer sets, R3-F/R and R4-F/R, resulted in significant PCR amplification (Figure 5C), inferring that σS binds to DNA sequences covering −61 to +136bp at least from the transcription start site of ssrA (Feng et al., 2003). It is believed that the ssrA promoter requires RNAP holoenzyme harnessing σ70, and the consensus −10 and −35 regions for σ70 binding were also predicted (Ramachandran et al., 2012; Banda et al., 2019). Our results raised the possibility that the ssrA promoter could recruit σS as well as σ70. The possibility of σS binding to the PssrA region was also proposed in silico in a previous study (Ramachandran et al., 2012). We further investigated transcription initiation at PssrA, which is controlled by mechanical interaction with σ factors.
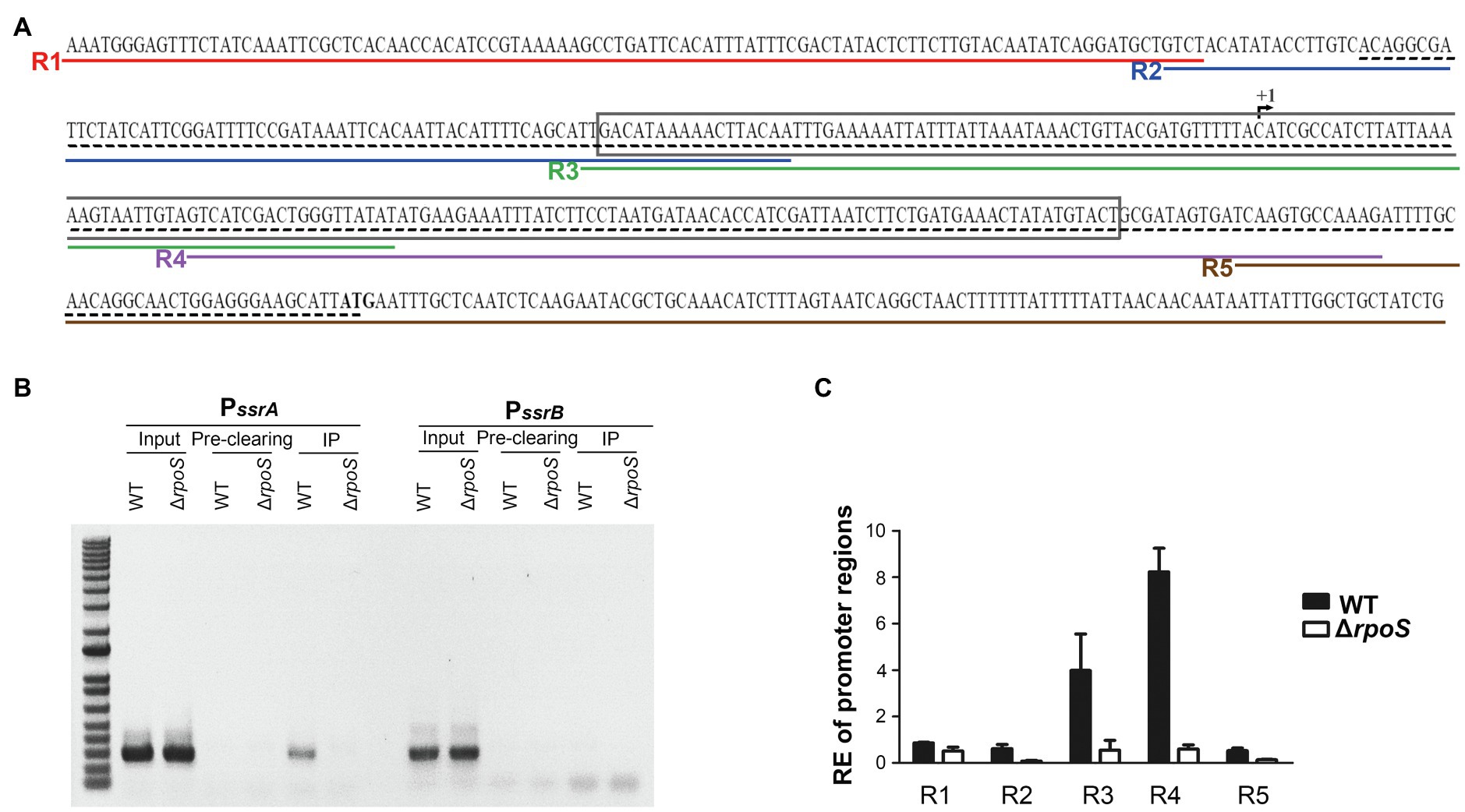
Figure 5. Binding of σS to the ssrA promoter. (A) Scheme for Chromatin immunoprecipitation (ChIP)-PCR and ChIP-quantitative PCR (qPCR). The +1 site of ssrA transcription is indicated with a broken arrow and its start codon is in bold. The region amplified by ChIP-PCR is underlined with a black dashed line and that amplified and used in electrophoretic mobility shift assay (EMSA) is boxed. Five regions amplified in ChIP-qPCR are underlined in different colors: R1 in red by primers R1-F and R1-R, R2 in blue by primers R2-F and R2-R, R3 in green by primers R3-F and R3-R, R4 in purple by primers R4-F and R4-R, and R5 in brown by primers R5-F and R5-R. (B) DNA-protein cross-links were fixed in Salmonella wild-type and ΔrpoS mutant strains using formaldehyde. DNA fragments bound to σS were immunoprecipitated using anti-RpoS antibody and subjected to PCR using primers specific to the PssrA (PssrA-ChIP-F/R) and PssrB (PssrB-ChIP-F/R) regions. Input total DNA and pre-clearing (DNA non-specifically bound to Protein A/G agarose) were analyzed in parallel. The ChIP-PCR products were separated by 1% agarose gel electrophoresis. (C) DNA fragments precipitated in IP and pre-clearing were used as templates in ChIP-qPCR using five different primer sets (R1-F/R, R2-F/R, R3-F/R, R4-F/R, and R5-F/R). Enrichment of the PssrA region with σS was relatively computed using the following formula: relative enrichment (RE)=2−(∆CtIP−∆Ct Pre-clearing), where ∆CtIP is CtPssrA −Ct gyrB for the IP samples and ∆CtPre-clearing is CtPssrA −CtgyrB for the pre-clearing samples. gyrB was used as an endogenous control.
The promoter recognition sequences for σS and σ70 are nearly identical, and a strong functional similarity between σS and σ70 has been suggested. Many σS-regulated genes, such as the csgBA operon, can be transcribed by either σS or σ70 in vitro (Arnqvist et al., 1994; Typas et al., 2007b). The possibility of biphasic ssrA transcription initiation by σS and σ70 was examined in vitro. A DNA fragment of 172bp encompassing the PssrA region targeted by ssrA regulators was incubated with each σ factor (His6-RpoS or His6-RpoD) in the presence or absence of RNAP core enzyme E. The PcsgBA region recognized by either σS or σ70 was used as a positive control, while a DNA fragment of the STM14_1978 gene devoid of the canonical sequences recognized by σS and σ70 was used as a negative control. The core enzyme alone could form a complex with DNA fragments of the PssrA or PcsgBA regions in a non-specific manner, as predicted elsewhere, and the addition of either σ factor (σS or σ70) retarded the mobility of the DNA-protein complex. This indicated that σ factor was engaged in the complex formation between RNAP holoenzyme (Eσ; α2ββ′σω) and the DNA fragments (Figure 6). Interestingly, the addition of σS produced two bands, presumably a lower one between the core enzyme E and the PssrA and an upper one between the EσS and the PssrA, whereas σ70 incorporation produced a single shifted band, representing robust complex formation between Eσ70 and PssrA (compare Figure 6A, lane 5 and Figure 6B, lane 5). These results stimulated us to compare the binding affinities between EσS and Eσ70 at the PssrA region.
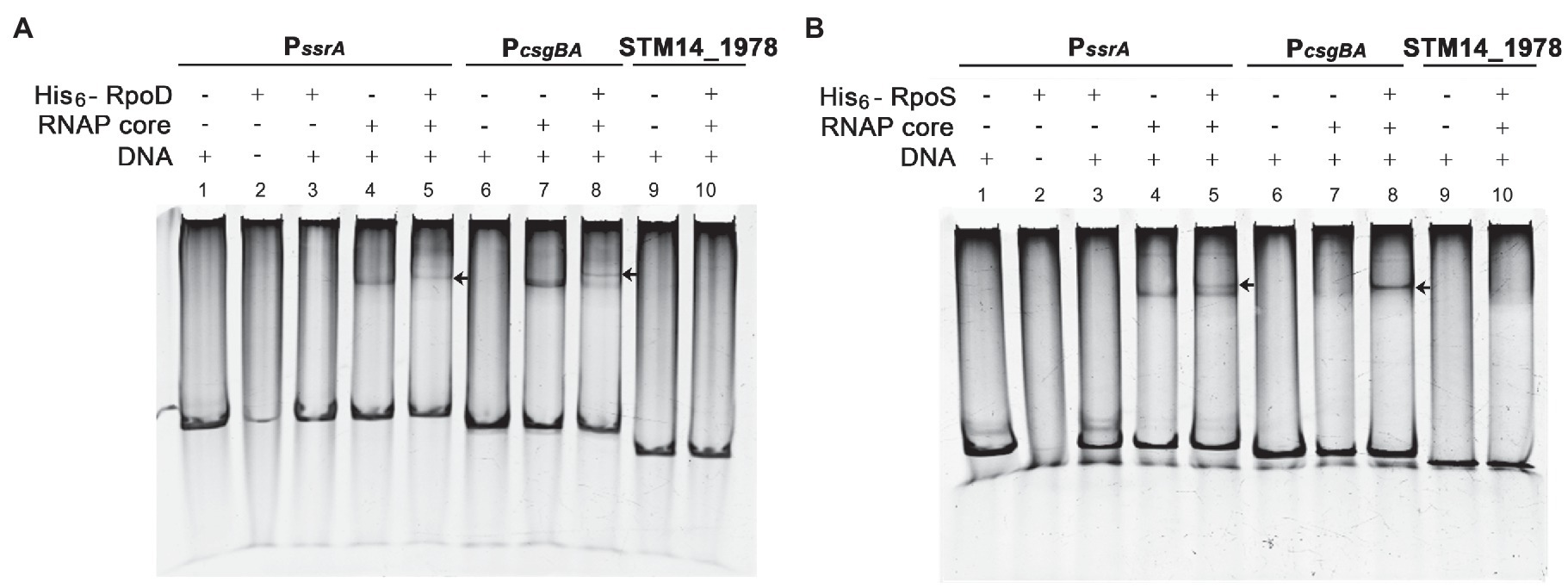
Figure 6. Interaction of PssrA with either σS or σ70 in vitro. Binding of His6-RpoD (A) and His6-RpoS (B) to the ssrA promoter region was tested in vitro using EMSA. DNA fragments of the PssrA region (178bp), PcsgBA region (158bp; a positive control), and STM14_1978 CDS region (148bp; a negative control) were incubated with RNA polymerase (RNAP) core enzyme in combination with His6-RpoD or His6-RpoS. The DNA-protein complexes were loaded onto a 5% native polyacrylamide gel and stained with EtBr. An arrow indicates a shifted band comprising PssrA or PcsgBA DNA cross-linked with the RNAP holoenzyme.
The PssrA DNA fragment was incubated with the core enzyme E and different concentrations of σ factors (His6-RpoS and His6-RpoD), and the levels of EσS associated with the PssrA region were determined using an anti-RpoS antibody. When His6-RpoS was used at a constant concentration of 75nM, but His6-RpoD was increased from 0 to 50nM, the band representing the complex between EσS and PssrA gradually diminished and disappeared at 50nM His6-RpoD (Figure 7A). On the other hand, when His6-RpoD was maintained at 75nM but His6-RpoS was increased from 0 to 150nM, EσS failed to bind to the PssrA region even at a 2-fold higher concentration of His6-RpoS than His6-RpoD (Figure 7B). This result suggests that the PssrA region preferentially recruits Eσ70 when EσS and Eσ70 are present at equivalent concentrations in vitro.
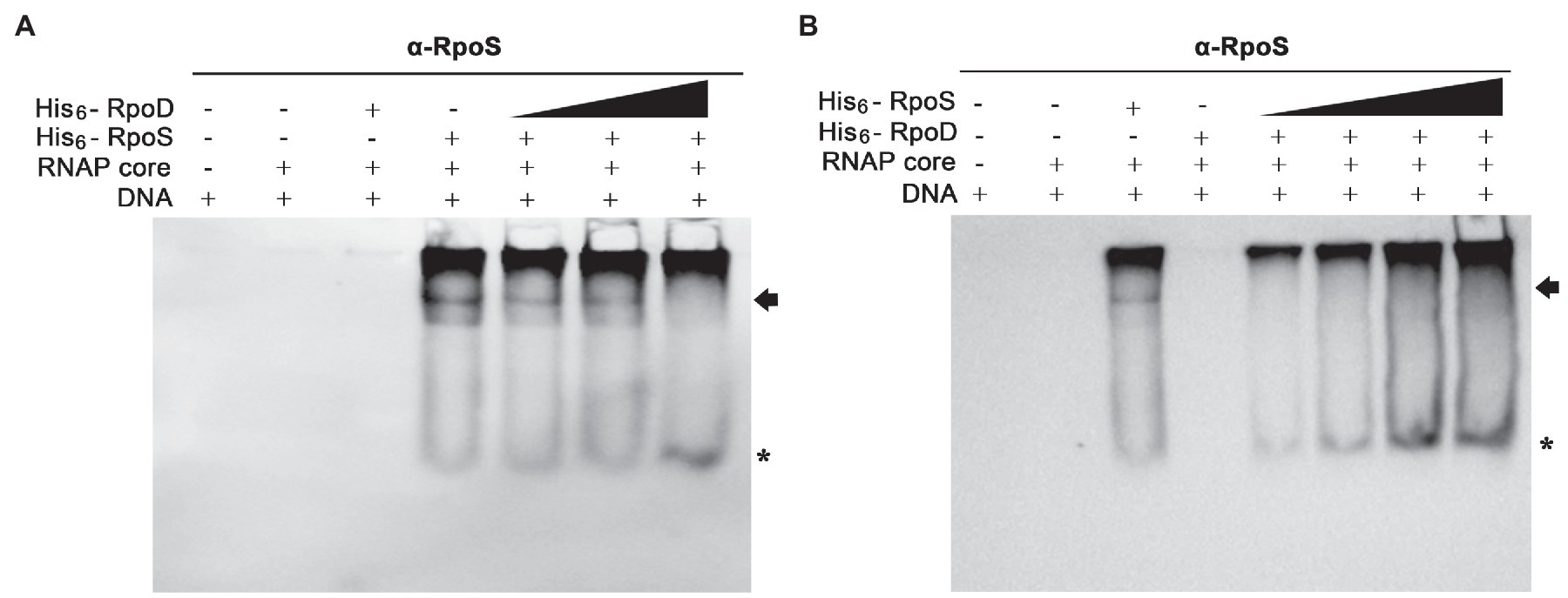
Figure 7. Competitive binding to the PssrA region between σS and σ70 in vitro. (A) Competitive EMSA was conducted with His6-RpoS at a constant concentration of 75nM and with increasing concentrations (0, 12.5, 25, and 50nM) of His6-RpoD as a competitor. RNAP core enzyme was pre-incubated with different combinations of two σ factors and incubated with DNA containing the PssrA region. DNA bands were stained using EtBr and shown in Supplementary Figure 3A. His6-RpoS was localized using subsequent immunoblotting with an anti-RpoS antibody. His6-RpoS comprising EσS cross-linked with the PssrA region is indicated with an arrow and free His6-RpoS is localized with an asterisk. (B) Competition EMSA was applied using His6-RpoD at a constant concentration of 75nM and with increasing concentrations (0, 25, 50, 100, and 150nM) of His6-RpoS as a competitor. After reconstitution of RNAP holoenzyme with different concentrations of σ factors, the interaction between RNAP holoenzyme and PssrA region was analyzed using native gel electrophoresis followed by DNA staining (Supplementary Figure 3B) and immunoblotting with an anti-RpoS antibody. The location of His6-RpoS comprising EσS-PssrA complex is indicated with an arrow and free His6-RpoS not associated with DNA is indicated with an asterisk.
For investigating the possibility that σS replaces σ70 and lowers the ssrA transcription, we searched for a co-regulator that could promote σS activity under stressful conditions and Crl was chosen as a candidate co-regulator of σS-mediated ssrA transcriptional regulation. Crl is a small protein known to interact directly with σS in vitro (Bougdour et al., 2004). The PssrA fragment was incubated with His6-tagged σ factors (σS and σ70) and Crl individually or in combination, and the σS bound to PssrA was localized using an anti-RpoS antibody. In the absence of competition with σ70, Crl addition enabled σS (50nM) to form a complex between EσS and the PssrA region, whereas σS alone at 50nM were insufficient to form the EσS-PssrA complex (Figure 8: compare lanes 4 and 5). In the absence of Crl, His6-RpoS even at 280nM was defeated in the competition with 50nM His6-RpoD and failed to form the protein-DNA complex (Figure 8, lane 8). However, pre-incubation of His6-RpoS with Crl rendered His6-RpoS competitive in forming the complex between Eσ and PssrA, showing a shifted band (Figure 8, lane 10). Crl binding to σS might facilitate the formation of the RNAP holoenzyme incorporating σS instead of σ70, as suggested previously (Gaal et al., 2006; Typas et al., 2007a).
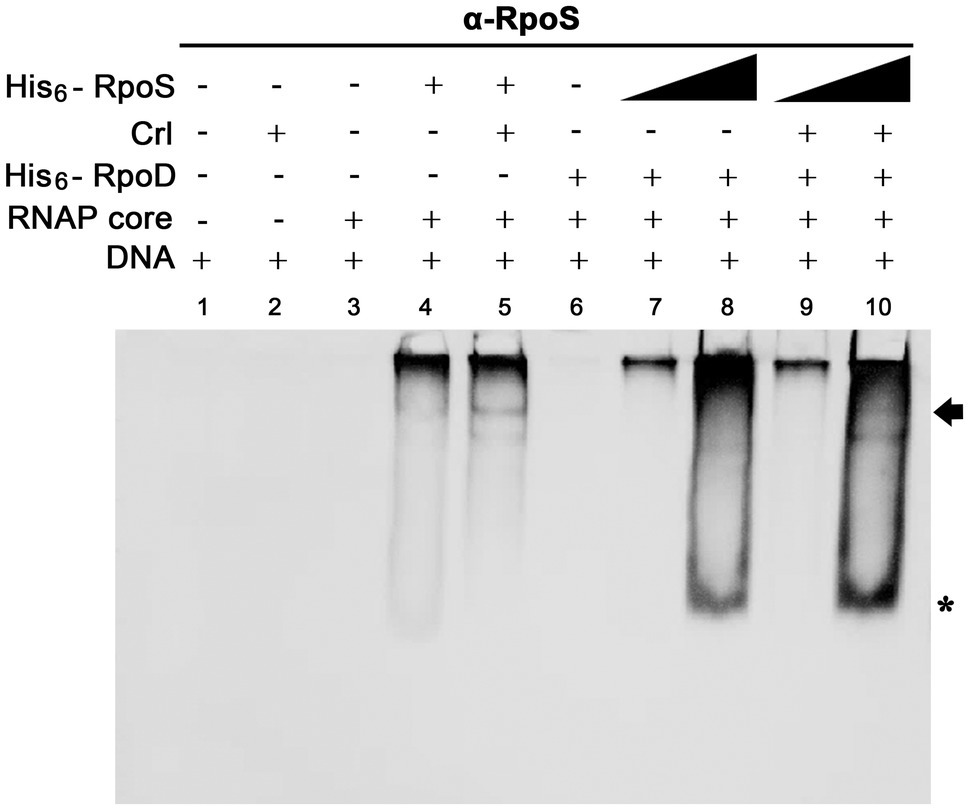
Figure 8. Effect of Crl on the binding affinity of σS to the PssrA region in vitro. Competitive binding between His6-RpoS and His6-RpoD to the PssrA region was re-examined in the presence of His6-Crl. In the absence of His6-RpoD (lanes 1–5), the RNAP core enzyme was incubated with 50nM His6-RpoS, which was pre-incubated with 280nM His6-Crl. In the competition between His6-RpoS and His6-RpoD (lanes 6–10), His6-RpoD was maintained at 50nM, while His6-RpoS was used at 25 and 280nM in combination with 280nM His6-Crl. RNAP holoenzyme was reconstituted using different combinations of His6-Crl, His6-RpoS, and His6-RpoD and incubated with PssrA region DNA. After gel electrophoresis, DNA was analyzed by EtBr staining (Supplementary Figure 4), and His6-RpoS was localized using immunoblotting with an anti-RpoS antibody. His6-RpoS comprising EσS cross-linked with the PssrA region is indicated with an arrow, and free His6-RpoS is indicated by an asterisk.
Our results comparing the ability of σS and σ70 to form the Eσ-PssrA complex in vitro demonstrated that Eσ70 bound to the PssrA region preferentially than EσS. Given the limited cellular resources of the RNAP core enzyme E, ssrA transcription may be dampened when σS stimulated by drastic stressors diverts the RNAP core enzyme E to its cognate regulatory circuit, which is critical for surviving the challenging stressors. SPI-2 genes controlled by SsrAB regulators are known to be activated under hostile conditions such as a nutrition-deprived environment and intracellular milieu (Beuzon et al., 1999; Deiwick et al., 1999), which are prone to stimulate σS-mediated adaptation responses. We investigated the transcriptional response of SPI-2 genes when σS levels surged in response to stress and EσS-mediated transcriptional initiation overwhelmed the transcriptional activity of other Eσ complexes. Salmonella wild-type and ΔrpoS strains were added to macrophage cells, and the transcription levels of rpoS, rpoD, and SPI-2 genes were compared at 2, 4, and 10h after phagocytosis. σS was overexpressed by introducing pRpoS into the ΔrpoS mutant. The absence of rpoS increased rpoD transcriptional levels at 4h post-infection (Figure 9). Comparing mRNA levels of SPI-2 genes between wild-type and ΔrpoS strains showed that most SPI-2 genes increased their transcription in the absence of σS and addition of pRpoS nullified these alterations (Figure 9), indicating a negative role of σS in SPI-2 transcription. In accordance with the transcriptional regulation by σS, the levels of T3SS2-associated proteins were decreased by the overexpression of σS (Supplementary Figure 6). However, the transcriptional response to σS abundance was different among the SPI-2 genes. Many genes, including sscB, sseFG, ssaG, sseJ, and sspH2, showed negative transcriptional regulation by σS abundance throughout the assay, whereas sseCD genes encoding the translocon components of T3SS2 (Chakravortty et al., 2005) showed minimal transcriptional alteration by σS abundance (Figure 9). Differential requirements among T3SS effectors depending on time and site during infection have been proposed in previous studies (Brawn et al., 2007; Nunez-Hernandez et al., 2014). The differential influence of σS between T3SS2-associated genes suggests that SsrA-mediated regulation is not the only mechanism by which σS participates in controlling SPI-2 T3SS-associated genes.
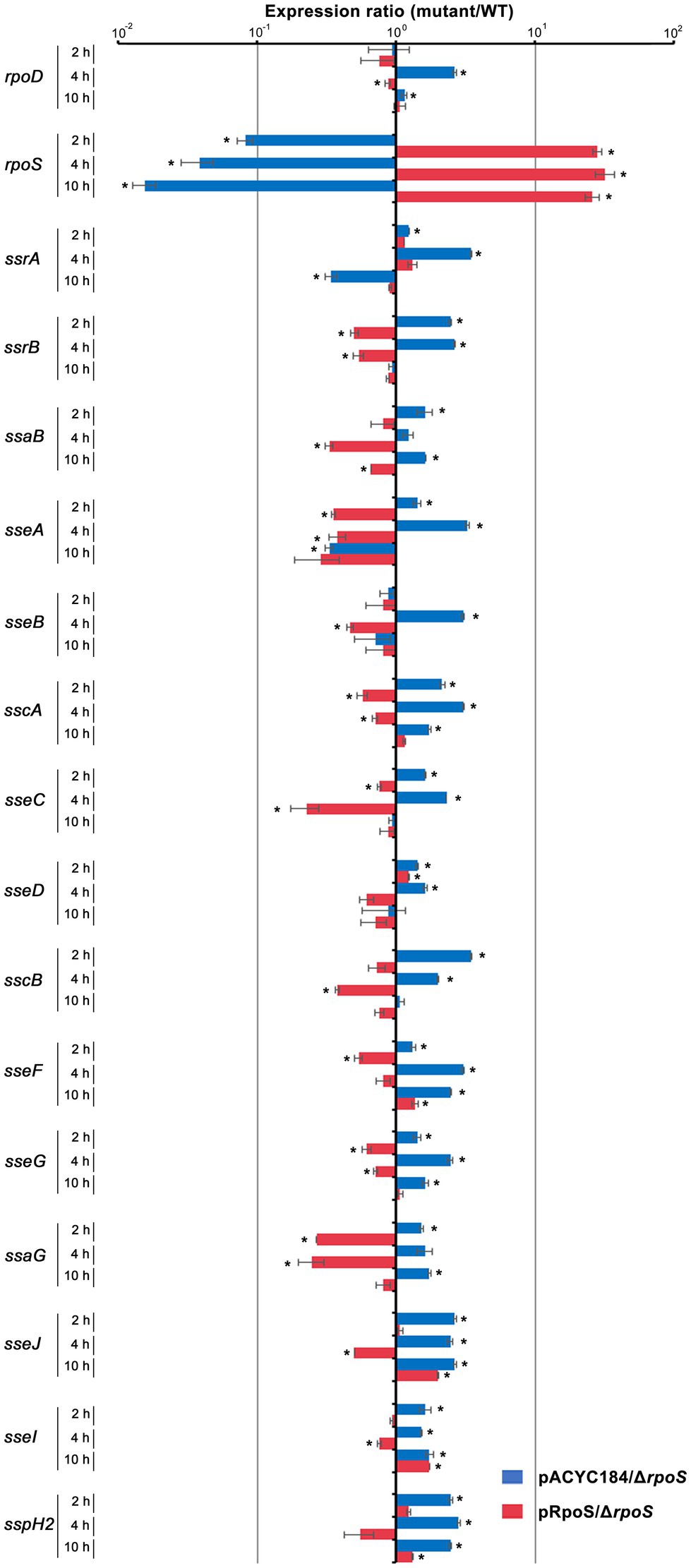
Figure 9. Transcriptional regulation of SPI-2 genes by σS. Salmonella wild-type and ΔrpoS mutant strains containing pRpoS or pACYC184 were added to RAW264.7, and total RNA was isolated at 2, 4, and 10h post-infection and used to measure the transcription levels of genes, including rpoD, rpoS, and SPI-2 T3SS-associated genes. The Ct values for each gene were normalized to those of gyrB. Expression levels of each gene from ΔrpoS mutant strains containing pRpoS or pACYC184 were compared with those from wild-type strain containing pACYC184 and the fold-change (mutant/wild-type) was plotted. An asterisk indicates a value of p<0.05 in comparison with wild-type strain harboring pACYC184.
During host infection, Salmonella undergoes various stress conditions, such as gastric acidity, bile salts, oxidative stress, and nutrient starvation. Alternative σ factors are prominent regulatory proteins that enable bacteria to cope with diverse stresses by redirecting RNAP core enzymes to the transcription of genes required for survival and adaptation in these conditions. RpoS or σS, a σ factor comprising the RNAP holoenzyme, is known to activate the transcription of genes associated with general stress resistance (Hengge-Aronis, 2002a). However, the regulatory roles of σS are not only restricted to stress-resistance genes. In Salmonella, σS was found to directly or indirectly control the expression of genes that make up more than 20% of the genome (Levi-Meyrueis et al., 2014; Lago et al., 2017), implying its multifaceted roles ranging from physiological remodeling against cellular damage to metabolic regulation of sugars, amino acids, and fatty acids (Ibanez-Ruiz et al., 2000; Lago et al., 2017). In addition, σS is involved in Salmonella virulence regulation. Rice et al. (2015) observed genes comprising SPI-1 and SPI-2, which are essential for Salmonella invasion into host cells and intracellular survival, were upregulated in the absence of σS, indicating a negative role of σS in SPI-1 and SPI-2 expression. We found that σS could bind to the PssrA region directly (Figure 5), and a surplus of σS repressed its transcription. This led to an overall downregulation of SPI-2 T3SS-associated genes (Figure 9). Direct negative regulation by σS was recently reported in the transcription of esrB in Edwardsiella piscicida (Yin et al., 2018). Edwardsiella piscicida, which is phylogenetically close to Salmonella enterica, exploits T3SS and T6SS to translocate virulence factors into host cells, and the expression of these virulence machineries is activated by the two-component regulatory system EsrAB, homologs for SsrAB in S. enterica (Wang et al., 2009; Yin et al., 2018). Edwardsiella piscicida σS was proposed to mediate a trade-off between stress adaptation and virulence by inhibiting esrB expression through a direct interaction between σS and the PesrB region (Yin et al., 2018). Binding of the Eσ complex to gene promoter regions is typically presumed to activate transcription initiation. Therefore, the negative regulation by σS may be attributed to the competition between σ factors for binding to the RNAP core enzyme (Farewell et al., 1998; Hsu, 2002). A surge in σS caused by bacterial adaptation to general stresses can exclusively occupy the pool of core enzyme E and impede transcriptional events mediated by other σ factors. However, alternatively to this passive regulation via competitive binding between σ factors, adhesion of the EσS complex to promoter regions may sterically hinder binding of the Eσ70 complex and directly attenuate transcription, as demonstrated in E. piscicida esrB gene (Yin et al., 2018) and S. enterica serovar Typhimurium sdh gene (Levi-Meyrueis et al., 2015).
The promoter region of ssrA is occupied by multiple regulators, including HilD, SlyA, OmpR, and H-NS, and its transcription is controlled by the competitive binding of these regulators to the overlapping DNA (Banda et al., 2019). The consensus promoter sequences recognized by σ70 are also accessible to H-NS, whose binding blocks the access of Eσ70 and transcriptional activators such as OmpR. Anti-repressors such as HilD and SlyA relieve H-NS-mediated silencing by competitive binding to the PssrA region (Banda et al., 2019). Considering the similar recognition motifs at −10 and −35 elements between σS and σ70 and the functional inter-compatibility between two σ factors in some genes, the downregulation of ssrAB by binding of σS to the PssrA region can be achievable through several mechanisms. Firstly, binding of EσS to the PssrA region may not exert transcriptional initiation, but instead sterically hinder Eσ70-mediated transcription as demonstrated in the E. piscicida esrB gene (Yin et al., 2018). A σ factor associated with RNAP core enzyme directs transcription initiation at a specific promoter region but is assumed to dissociate upon transition from transcription initiation to transcription elongation because of a steric clash between the growing RNA product and the σ factor (Hsu, 2002). σS that is not released on time may impede promoter escape of the core enzyme E and hinder transcription elongation. Alternatively, the binding of EσS to PssrA region may produce incorrect transcripts, as shown in the transcriptional regulation of crl gene (Zafar et al., 2014). The crl gene with overlapping promoters sensed by two different σ factors of σ70 and σN may shut down its expression by association with EσN. σN increases in response to nitrogen limitation, forming a DNA-EσN complex at the crl promoter region, but its binding results in a long noncoding RNA transcript lacking a ribosome binding site, thereby preventing Eσ70 from binding to the overlapped promoter and producing translatable crl mRNA (Zafar et al., 2014). We observed that DNA fragments bound to EσS in vivo covered a long region from the known +1 site of ssrA transcript to the start codon for SsrA (Figure 5). To differentiate between these two possibilities, it is important to examine whether EσS bound to the PssrA region can lead to ssrA transcription and whether the resultant transcript can successfully be translated. Another possibility is the competitive EσS binding among promoters with different binding affinities due to recognition motif preference and topological characteristics. Considering that the cellular σS concentration is low even in the stationary phase of growth (Jishage et al., 1996) and its affinity for RNAP core enzyme is the lowest among σ factors in vitro (Maeda et al., 2000), the PssrA region occluded by multiple regulators may be less competent in recruiting EσS and other promoter sites, which are preferentially responsive to σS, may outcompete the ssrA promoter.
In order to cope with limited resources, bacteria allocate cellular resources between reproduction and maintenance in response to environmental cues. In the absence of nutrient depletion and hostile stressors, bacteria proliferate and deploy resources for reproduction. Under these favorable conditions, σ70 is exclusively used for the transcription initiation of housekeeping genes. On the other hand, bacteria challenged by stressful stimuli divert cellular resources to maintenance and resistance, replacing σ70 with alternative σ factors for comprehensive transcription alteration. σS orchestrates the expression of a large number of genes under conditions of starvation and general stress caused by pH, temperature, and osmolarity. SPI-2 T3SS and its cognate effectors critical for Salmonella intracellular survival and replication are thought to be induced by unfavorable stimuli encountered inside host cells (Lober et al., 2006; Fass and Groisman, 2009), which would also likely promote σS-mediated stress adaptation processes. However, we observed that excessive σS production rather decreased the transcription of SPI-2 and its associated genes. Virulence effectors translocated via SPI-2 T3SS help intracellular Salmonella to compromise the host defense systems and facilitate intracellular proliferation and cell-to-cell spread (Grant et al., 2012; Jennings et al., 2017). However, overgrowth of Salmonella, which is less competent to manage hostile stresses, poses a disadvantage to long-term persistence inside hosts because the intense immune responses provoked by the proliferation may eliminate defective bacteria rapidly after all (Nunez-Hernandez et al., 2014). Salmonella executing σS-mediated stress adaptation may attenuate aggressive virulence ascribed to SPI-2 to achieve a trade-off between stress adaptation and virulence. Notably, the growth rates of intracellular Salmonella vary depending on the infected cell types; Salmonella proliferated exclusively in CD18-expressing phagocytes in vivo (Richter-Dahlfors et al., 1997), while restraining its growth in non-professional phagocytes such as subepithelial fibroblasts (Cano et al., 2001). Therefore, the importance of SPI-2 T3SS for Salmonella survival varies depending on infection foci or cell types. For example, SifA, an effector translocated via SPI-2 T3SS, is essential for bacterial growth inside macrophages but is dispensable for survival inside fibroblast cells (Nunez-Hernandez et al., 2014). Interestingly, Grant et al. showed that Salmonella lacking SPI-2 T3SS remained inside phagocytes at a high replication rate but failed to leave the infected cells, suggesting a new role for SPI-2 T3SS in bacterial dissemination to other sites (Grant et al., 2012). Premature escape from infected host cells may impose unaffordable expenses on Salmonella to resist severe host defense systems and constrain its successful host colonization. In this context, it is an energy-effective strategy for Salmonella to employ σS as a dual-purpose regulator that aids in adaptation and resistance against unfavorable conditions and lowers unnecessary virulence attributable to SPI-2 at the same time.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SK designed and conducted the experiment and interpreted the data. EK performed and analyzed the experiment. HY conceived and coordinated the study. SK and HY wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by a grant (2019R1A6A1A11051471) from the Priority Research Centers Program funded by the National Research Foundation of Korea (NRF) and a grant (2021R1F1A1058498) of the Basic Science Research Program through the NRF funded by the Korean government (MSIT).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any produ that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.750940/full#supplementary-material
Arnqvist, A., Olsen, A., and Normark, S. (1994). Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13, 1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x
Banda, M. M., Zavala-Alvarado, C., Perez-Morales, D., and Bustamante, V. H. (2019). SlyA and HilD counteract H-NS-mediated repression on the ssrAB virulence operon of Salmonella enterica serovar Typhimurium and thus promote its activation by OmpR. J. Bacteriol. 201, e00530–e00618. doi: 10.1128/JB.00530-18
Bang, I. S., Frye, J. G., Mcclelland, M., Velayudhan, J., and Fang, F. C. (2005). Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol. Microbiol. 56, 811–823. doi: 10.1111/j.1365-2958.2005.04580.x
Beuzon, C. R., Banks, G., Deiwick, J., Hensel, M., and Holden, D. W. (1999). pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33, 806–816. doi: 10.1046/j.1365-2958.1999.01527.x
Bougdour, A., Lelong, C., and Geiselmann, J. (2004). Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279, 19540–19550. doi: 10.1074/jbc.M314145200
Brawn, L. C., Hayward, R. D., and Koronakis, V. (2007). Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe 1, 63–75. doi: 10.1016/j.chom.2007.02.001
Bustamante, V. H., Martinez, L. C., Santana, F. J., Knodler, L. A., Steele-Mortimer, O., and Puente, J. L. (2008). HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105, 14591–14596. doi: 10.1073/pnas.0801205105
Cano, D. A., Martinez-Moya, M., Pucciarelli, M. G., Groisman, E. A., Casadesus, J., and Garcia-Del Portillo, F. (2001). Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69, 6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001
Chakravortty, D., Rohde, M., Jager, L., Deiwick, J., and Hensel, M. (2005). Formation of a novel surface structure encoded by Salmonella pathogenicity island 2. EMBO J. 24, 2043–2052. doi: 10.1038/sj.emboj.7600676
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978
Deiwick, J., Nikolaus, T., Erdogan, S., and Hensel, M. (1999). Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31, 1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x
Dong, T., and Schellhorn, H. E. (2010). Role of RpoS in virulence of pathogens. Infect. Immun. 78, 887–897. doi: 10.1128/IAI.00882-09
Fang, F. C., Libby, S. J., Buchmeier, N. A., Loewen, P. C., Switala, J., Harwood, J., et al. (1992). The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89, 11978–11982. doi: 10.1073/pnas.89.24.11978
Farewell, A., Kvint, K., and Nystrom, T. (1998). Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29, 1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x
Fass, E., and Groisman, E. A. (2009). Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12, 199–204. doi: 10.1016/j.mib.2009.01.004
Feng, X., Oropeza, R., and Kenney, L. J. (2003). Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 48, 1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x
Feng, X., Walthers, D., Oropeza, R., and Kenney, L. J. (2004). The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol. Microbiol. 54, 823–835. doi: 10.1111/j.1365-2958.2004.04317.x
Ferenci, T. (2005). Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57, 1–8. doi: 10.1111/j.1365-2958.2005.04649.x
Gaal, T., Mandel, M. J., Silhavy, T. J., and Gourse, R. L. (2006). Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 188, 7966–7970. doi: 10.1128/JB.01266-06
Grant, A. J., Morgan, F. J., Mckinley, T. J., Foster, G. L., Maskell, D. J., and Mastroeni, P. (2012). Attenuated Salmonella Typhimurium lacking the pathogenicity island-2 type 3 secretion system grow to high bacterial numbers inside phagocytes in mice. PLoS Pathog. 8:e1003070. doi: 10.1371/journal.ppat.1003070
Grove, A. P., Liveris, D., Iyer, R., Petzke, M., Rudman, J., Caimano, M. J., et al. (2017). Two distinct mechanisms govern RpoS-mediated repression of tick-phase genes during mammalian host adaptation by Borrelia burgdorferi, the Lyme disease spirochete. mBio 8, e01204–e01217. doi: 10.1128/mBio.01204-17
Gu, D., Liu, H., Yang, Z., Zhang, Y., and Wang, Q. (2016). Chromatin immunoprecipitation sequencing technology reveals global regulatory roles of low-cell-density quorum-sensing regulator AphA in the pathogen vibrio alginolyticus. J. Bacteriol. 198, 2985–2999. doi: 10.1128/JB.00520-16
Hengge-Aronis, R. (2002a). Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395. doi: 10.1128/MMBR.66.3.373-395.2002
Hengge-Aronis, R. (2002b). Stationary phase gene regulation: what makes an Escherichia coli promoter sigmaS-selective? Curr. Opin. Microbiol. 5, 591–595. doi: 10.1016/s1369-5274(02)00372-7
Hermans, K., Roberfroid, S., Thijs, I. M., Kint, G., De Coster, D., Marchal, K., et al. (2016). FabR regulates Salmonella biofilm formation via its direct target FabB. BMC Genomics 17:253. doi: 10.1186/s12864-016-2387-x
Hiratsu, K., Amemura, M., Nashimoto, H., Shinagawa, H., and Makino, K. (1995). The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J. Bacteriol. 177, 2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995
Hsu, L. M. (2002). Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta 1577, 191–207. doi: 10.1016/s0167-4781(02)00452-9
Ibanez-Ruiz, M., Robbe-Saule, V., Hermant, D., Labrude, S., and Norel, F. (2000). Identification of RpoS (sigma(S))-regulated genes in Salmonella enterica serovar typhimurium. J. Bacteriol. 182, 5749–5756. doi: 10.1128/JB.182.20.5749-5756.2000
Jennings, E., Thurston, T. L. M., and Holden, D. W. (2017). Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22, 217–231. doi: 10.1016/j.chom.2017.07.009
Jishage, M., Iwata, A., Ueda, S., and Ishihama, A. (1996). Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178, 5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996
Kim, S. I., Kim, S., Kim, E., Hwang, S. Y., and Yoon, H. (2018). Secretion of Salmonella pathogenicity island 1-encoded type III secretion system effectors by outer membrane vesicles in Salmonella enterica serovar typhimurium. Front. Microbiol. 9:2810. doi: 10.3389/fmicb.2018.02810
Kim, S. I., and Yoon, H. (2019). Roles of YcfR in biofilm formation in Salmonella Typhimurium ATCC 14028. Mol. Plant-Microbe Interact. 32, 708–716. doi: 10.1094/MPMI-06-18-0166-R
Kowarz, L., Coynault, C., Robbe-Saule, V., and Norel, F. (1994). The Salmonella Typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J. Bacteriol. 176, 6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994
Kwoh, D. Y., and Kemper, J. (1978). Bacteriophage P22-mediated specialized transduction in Salmonella typhimurium: identification of different types of specialized transducing particles. J. Virol. 27, 535–550. doi: 10.1128/jvi.27.3.535-550.1978
Lago, M., Monteil, V., Douche, T., Guglielmini, J., Criscuolo, A., Maufrais, C., et al. (2017). Proteome remodelling by the stress sigma factor RpoS/sigma(S) in Salmonella: identification of small proteins and evidence for post-transcriptional regulation. Sci. Rep. 7:2127. doi: 10.1038/s41598-017-02362-3
Lee, T. S., Krupa, R. A., Zhang, F., Hajimorad, M., Holtz, W. J., Prasad, N., et al. (2011). BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 5:12. doi: 10.1186/1754-1611-5-12
Levi-Meyrueis, C., Monteil, V., Sismeiro, O., Dillies, M. A., Kolb, A., Monot, M., et al. (2015). Repressor activity of the RpoS/sigmaS-dependent RNA polymerase requires DNA binding. Nucleic Acids Res. 43, 1456–1468. doi: 10.1093/nar/gku1379
Levi-Meyrueis, C., Monteil, V., Sismeiro, O., Dillies, M. A., Monot, M., Jagla, B., et al. (2014). Expanding the RpoS/sigmaS-network by RNA sequencing and identification of sigmaS-controlled small RNAs in Salmonella. PLoS One 9:e96918. doi: 10.1371/journal.pone.0096918
Lober, S., Jackel, D., Kaiser, N., and Hensel, M. (2006). Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296, 435–447. doi: 10.1016/j.ijmm.2006.05.001
Maeda, H., Fujita, N., and Ishihama, A. (2000). Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28, 3497–3503. doi: 10.1093/nar/28.18.3497
Muffler, A., Fischer, D., and Hengge-Aronis, R. (1996). The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10, 1143–1151. doi: 10.1101/gad.10.9.1143
Notley-McRobb, L., King, T., and Ferenci, T. (2002). rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184, 806–811. doi: 10.1128/JB.184.3.806-811.2002
Nunez-Hernandez, C., Alonso, A., Pucciarelli, M. G., Casadesus, J., and Garcia-Del Portillo, F. (2014). Dormant intracellular Salmonella enterica serovar Typhimurium discriminates among Salmonella pathogenicity island 2 effectors to persist inside fibroblasts. Infect. Immun. 82, 221–232. doi: 10.1128/IAI.01304-13
Raffatellu, M., Wilson, R. P., Chessa, D., Andrews-Polymenis, H., Tran, Q. T., Lawhon, S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73, 146–154. doi: 10.1128/IAI.73.1.146-154.2005
Ramachandran, V. K., Shearer, N., Jacob, J. J., Sharma, C. M., and Thompson, A. (2012). The architecture and ppGpp-dependent expression of the primary transcriptome of Salmonella Typhimurium during invasion gene expression. BMC Genomics 13:25. doi: 10.1186/1471-2164-13-25
Rice, C. J., Ramachandran, V. K., Shearer, N., and Thompson, A. (2015). Transcriptional and post-transcriptional modulation of SPI1 and SPI2 expression by ppGpp, RpoS and DksA in Salmonella enterica sv typhimurium. PLoS One 10:e0127523. doi: 10.1371/journal.pone.0127523
Richter-Dahlfors, A., Buchan, A. M., and Finlay, B. B. (1997). Murine salmonellosis studied by confocal microscopy: Salmonella Typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186, 569–580. doi: 10.1084/jem.186.4.569
Robbe-Saule, V., Jaumouille, V., Prevost, M. C., Guadagnini, S., Talhouarne, C., Mathout, H., et al. (2006). Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar typhimurium. J. Bacteriol. 188, 3983–3994. doi: 10.1128/JB.00033-06
Salazar, J. K., Deng, K., Tortorello, M. L., Brandl, M. T., Wang, H., and Zhang, W. (2013). Genes ycfR, sirA and yigG contribute to the surface attachment of Salmonella enterica Typhimurium and saintpaul to fresh produce. PLoS One 8:e57272. doi: 10.1371/journal.pone.0057272
Shimada, T., Tanaka, K., and Ishihama, A. (2017). The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase. PLoS One 12:e0179181. doi: 10.1371/journal.pone.0179181
Simons, R. W., Houman, F., and Kleckner, N. (1987). Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96. doi: 10.1016/0378-1119(87)90095-3
Smale, S. T. (2010). Beta-galactosidase assay. Cold Spring Harb Protoc 2010:pdb.prot5423. doi: 10.1101/pdb.prot5423
Soncini, F. C., Vescovi, E. G., and Groisman, E. A. (1995). Transcriptional autoregulation of the Salmonella Typhimurium phoPQ operon. J. Bacteriol. 177, 4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995
Storvik, K. A., and Foster, P. L. (2010). RpoS, the stress response sigma factor, plays a dual role in the regulation of Escherichia coli's error-prone DNA polymerase IV. J. Bacteriol. 192, 3639–3644. doi: 10.1128/JB.00358-10
Trevino-Quintanilla, L. G., Freyre-Gonzalez, J. A., and Martinez-Flores, I. (2013). Anti-sigma factors in E. coli: common regulatory mechanisms controlling sigma factors availability. Curr. Genom. 14, 378–387. doi: 10.2174/1389202911314060007
Typas, A., Barembruch, C., Possling, A., and Hengge, R. (2007a). Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of sigmas activity and levels. EMBO J. 26, 1569–1578. doi: 10.1038/sj.emboj.7601629
Typas, A., Becker, G., and Hengge, R. (2007b). The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 63, 1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x
Vanaporn, M., Vattanaviboon, P., Thongboonkerd, V., and Korbsrisate, S. (2008). The rpoE operon regulates heat stress response in Burkholderia pseudomallei. FEMS Microbiol. Lett. 284, 191–196. doi: 10.1111/j.1574-6968.2008.01216.x
Wang, Q., Yang, M., Xiao, J., Wu, H., Wang, X., Lv, Y., et al. (2009). Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLoS One 4:e7646. doi: 10.1371/journal.pone.0007646
Yin, K., Guan, Y., Ma, R., Wei, L., Liu, B., Liu, X., et al. (2018). Critical role for a promoter discriminator in RpoS control of virulence in Edwardsiella piscicida. PLoS Pathog. 14:e1007272. doi: 10.1371/journal.ppat.1007272
Yoon, H., Ansong, C., Mcdermott, J. E., Gritsenko, M., Smith, R. D., Heffron, F., et al. (2011). Systems analysis of multiple regulator perturbations allows discovery of virulence factors in Salmonella. BMC Syst. Biol. 5:100. doi: 10.1186/1752-0509-5-100
Yoon, H., Mcdermott, J. E., Porwollik, S., Mcclelland, M., and Heffron, F. (2009). Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar typhimurium. PLoS Pathog. 5:e1000306. doi: 10.1371/journal.ppat.1000306
Zafar, M. A., Carabetta, V. J., Mandel, M. J., and Silhavy, T. J. (2014). Transcriptional occlusion caused by overlapping promoters. Proc. Natl. Acad. Sci. U. S. A. 111, 1557–1561. doi: 10.1073/pnas.1323413111
Zambrano, M. M., Siegele, D. A., Almiron, M., Tormo, A., and Kolter, R. (1993). Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259, 1757–1760. doi: 10.1126/science.7681219
Keywords: Salmonella Typhimurium, RpoS (σS), ssrA, Salmonella pathogenicity island-2, virulence
Citation: Kim S, Kim E and Yoon H (2021) σS-Mediated Stress Response Induced by Outer Membrane Perturbation Dampens Virulence in Salmonella enterica serovar Typhimurium. Front. Microbiol. 12:750940. doi: 10.3389/fmicb.2021.750940
Received: 31 July 2021; Accepted: 30 August 2021;
Published: 30 September 2021.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Sangyong Lim, Korea Atomic Energy Research Institute (KAERI), South KoreaCopyright © 2021 Kim, Kim and Yoon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyunjin Yoon, eW9vbmhAYWpvdS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.