- 1State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing, China
- 3Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing, China
- 4College of Life Science, Hebei University, Baoding, China
The genus Weissella is attracting an increasing amount of attention because of its multiple functions and probiotic potential. In particular, the species Weissella confusa is known to have great potential in industrial applications and exhibits numerous biological functions. However, the knowledge on this bacterium in insects is not investigated. Here, we isolated and identified W. confusa as the dominant lactic acid bacteria in the gut of the migratory locust. We named this strain W. confusa LM1, which is the first genome of an insect-derived W. confusa strain with one complete chromosome and one complete plasmid. Among all W. confusa strains, W. confusa LM1 had the largest genome. Its genome was the closest to that of W. confusa 1001271B_151109_G12, a strain from human feces. Our results provided accurate evolutionary relationships of known Weissella species and W. confusa strains. Based on genomic analysis, the pan-genome of W. confusa is in an open state. Most strains of W. confusa had the unique genes, indicating that these strains can adapt to different ecological niches and organisms. However, the variation of strain-specific genes did represent significant correlations with their hosts and ecological niches. These strains were predicted to have low potential to produce secondary metabolites. Furthermore, no antibiotic resistance genes were identified. At the same time, virulence factors associated with toxin production and secretion system were not found, indicating that W. confusa strains were not sufficient to perform virulence. Our study facilitated the discovery of the functions of W. confusa LM1 in locust biology and their potential application to locust management.
Introduction
The bacterial genus Weissella was first established in 1993 and isolated from fermented sausages. It is a member of the well-known group of lactic acid bacteria (LAB) (Collins et al., 1993). Weissella are Gram-positive, catalase-negative, and non-endospore-forming cells with coccoid or rod-shaped morphology, belonging to the family Leuconostocaceae of phylum Firmicutes.
Species and strains of Weissella have been isolated from various ecological niches and have different functions. They have been isolated from various fermented foods, such as yoghurt, sourdough, and kimchi (Wang et al., 2011; Zannini et al., 2018; Mun and Chang, 2020; Valerio et al., 2020), and play important roles in the process of fermentation, thereby influencing the texture and taste of foods. They also exist in the guts of humans and vertebrate animals, such as giant pandas and rainbow trout (Xiong et al., 2019; Mortezaei et al., 2020; Wang et al., 2020), their functions involve reducing depressive-like behavior (Sandes et al., 2020), influencing gut permeability and intestinal epithelial regeneration (Prado et al., 2020), killing harmful bacteria (Dey and Kang, 2020; Lee et al., 2020; Pelyuntha et al., 2020), affecting host metabolism (Elshaghabee et al., 2020), and preventing cancer cell proliferation (Le et al., 2020). Some Weissella species may be beneficial to plants because they inhibit the infection of plant pathogenic fungi, such as Fusarium verticillioides (Quattrini et al., 2020).
Recently, more studies suggested that Weissella species and strains can be used in the food and pharmaceutical industries. Some Weissella species have elicited research interest because of their probiotic, biotechnological, and bacteriocinogenic potential. In particular, some studies have clarified that W. confusa can be used as a direct-fed microbial candidate (Sturino, 2018; Cupi and Elvig-Jørgensen, 2019; Bourdichon et al., 2021) and exist in the breast milk of healthy women (Martin et al., 2007; Albesharat et al., 2011). W. confusa has been added to the International Dairy Federation (IDF) Inventory (Bourdichon et al., 2012), and the Senate Commission on Food Safety (SKLM) also validated its usage in traditional methods of food fermentation (Vogel et al., 2011).
With the rapid development of sequencing techniques, the genomes of diverse bacterial strains have been sequenced. This advancement promoted research on comparative genomics, which helped illustrate the relationships between genotypes and phenotypes (Wang et al., 2018), thereby addressing the effects of isolation source on the genetic variations (Mao et al., 2021), revealing horizontal gene transfer (Singh et al., 2021), and performing taxonomic revision (Lavecchia et al., 2021). As the importance of Weissella species has been recognized, attempts have been made to explore their evolutionary relationship and genomic characteristics. Comparative genomic analyses of some Weissella species, such as Weissella hellenica (Panthee et al., 2019) and Weissella cibaria (Li et al., 2017), have been conducted. By 2021, 38 genome assemblies of W. confusa strains have been deposited into the National Center for Biotechnology Information (NCBI) public database, including 3 complete genomes and 35 draft genomes, and most of them were sequenced in the past 3 years. Comparative genomic analysis of these W. confusa strains will allow us to gain a deeper understanding of W. confusa species and facilitate the potential application of W. confusa strains.
In this study, we identified a strain of W. confusa from the gut of the migratory locust (Locusta migratoria) and completed its whole-genome sequence. We designated the special strain as W. confusa LM1, which stably existed in the gut of the migratory locust, a worldwide locust species (Guo et al., 2020). By performing comparative genomic analysis of Weissella species, we obtained a more accurate evolutionary relationship and the genetic characteristics of different W. confusa strains. Our results provided important insights into the role of this strain in locust biology and offered a potential target for locust control.
Materials and Methods
Bacterial Strain
The bacterial strain used in this study was isolated from the gut of the migratory locust, which was reared as an animal model system at the Institute of Zoology, Chinese Academy of Sciences, Beijing, China. The whole guts of 10 locusts were carefully dissected with sterile devices, homogenized in 10-ml of sterile phosphate-buffered saline (PBS) solution (pH 7.4), and then diluted by sterile PBS solution (1:10,000). Subsequently, 50 ml of the diluted homogenized solution was spread over de Man Rogosa and Sharpe (MRS) medium (Man et al., 1960). A single colony was used to prepare the broth and establish the glycerol stocks. The single colony was also used for 16S rRNA gene amplification by using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′). Amplified products were sequenced and assigned to taxa based on NCBI public databases by using the basic local alignment search tool. Isolated strains were maintained at −80°C as frozen stock cultures in MRS broth.
DNA Extraction and Whole-Genome Sequencing
Weissella confusa LM1 was cultured in MRS broth at 37°C for 12 h. Genomic DNA was extracted by using Qiagen DNeasy Blood & Tissue Kit (Qiagen) according to the instructions of the manufacturer. Harvested DNA quality was checked by agarose gel electrophoresis (0.8%), and DNA quantity was determined by using NanoDrop 1000 (Thermo Fisher Scientific). The whole-genome sequence of W. confusa LM1 was obtained through single-molecule real-time (SMRT) sequencing by using PacBio Sequel platform and through next-generation sequencing (NGS) by using Illumina NovaSeq platform. Libraries for SMRT sequencing were constructed with an insert size of 10 kb by using the SMRTbell Template kit (Pacific Biosciences). Libraries for NGS were constructed with an insert size of 350 bp by using Ultra DNA Library Prep Kit (NEB).
Genome Assembly
Preliminary assembly sequences were obtained on the basis of the raw data from PacBio Sequel platform by using the assembly software PacBio SMRT Link V5.0.1 Low-quality reads of less than 500 bp were filtered to obtain clean data and to ensure the accuracy of the subsequent analyses. The automatic error correction function of SMRT portal was performed. Long reads of over 6,000 bp were selected as the seed sequence, the other reads were aligned to the seed sequence by using Basic Local Alignment with Successive Refinement (BLASR) to improve the accuracy of the seed sequence (Chaisson and Tesler, 2012). After obtaining the initial assembly result, the arrow algorithm was used to correct the variant sites in the initial assembly result by using the variant Caller module of the SMRT Link. NGS data were used to further correct the preliminary assembly sequence by BWA (Li and Durbin, 2009). Cyclization was then confirmed according to overlap (at least 1.5 kb). The initiation site was corrected by BLAST with the DnaA genes and confirmed through GC-Skew.
Gene Prediction and Genome Annotation
The genome of W. confusa LM1 was automatically annotated using the GeneMarkS-2+ based on the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016). Each gene was assigned to a category within the Clusters of Orthologous Groups (COG) database (Galperin et al., 2021). Circular layouts were generated using Circos V0.692 (Krzywinski et al., 2009). Ribosome RNA (rRNA) was identified by using RNAmmer V1.2 (Lagesen et al., 2007), and tRNA genes were predicted by tRNAscan-SE V2.0 (Lowe and Chan, 2016). Carbohydrate-Active enZYmes (CAZy) database (Lombard et al., 2014) was used for carbohydrate metabolism analysis by using DIAMOND with a cutoff e-value of 1e-5 and minimum identity of >40% (Buchfink et al., 2015). The online server of the Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH V6.03) was used for the annotation and analysis of secondary metabolite biosynthesis gene clusters with “relaxed” detection strictness (Blin et al., 2021).
Pathogenicity and Antibiotic Resistance Analysis
The virulence factor database (VFDB) core dataset (setA) was used to curate information about the virulence factors of bacterial pathogens by using DIAMOND with a cutoff e-value of 1e-5 and 70% identity based on the protein data (Chen et al., 2012). Based on the genome data, PathogenFinder V1.14 was used to predict the pathogenicity of the bacteria toward human hosts (Cosentino et al., 2013). Prophage regions of W. confusa strains were detected by using the PHASTER server (Arndt et al., 2016). Phylogenetic tree of intact phages was created by using VICTOR5 (Meier-Kolthoff and Goker, 2017). ResFinder database was used to identify the acquired antibiotic resistance genes based on genome data by using the ResFinder 4.16 with thresholds of 90% identity and 60% gene coverage (Zankari et al., 2012). The Comprehensive Antibiotic Resistance Database (CARD) was also used to identify antibiotic resistance genes by using the Resistance Gene Identifier (RGI) software to predict resistome from protein data based on homology and SNP models with the selection criteria of perfect and strict hits only7 (Alcock et al., 2020).
Comparative Analysis
For the comparative genome analysis, the genomic sequences of each Weissella species and strains were downloaded from NCBI. Average nucleotide identity (ANI) value was calculated by using JSpecies V1.2.1 with BLAST (Richter and Rossello-Mora, 2009) under the default parameters (Goris et al., 2007). The dropoff value for gapped alignment was set to 150, the e-value was set to 1e-15, the identity was set to 30%, and alignment length was set to 1,020 bp. ANI is the average value based on the comparisons of all orthologous protein-encoding genes of pairwise genomes and is a classic method for judging whether strains belong to the same species. Strains with ANI values of over 95% are generally considered to be of the same species (Yoon et al., 2017). The obtained ANI value matrix was clustered and visualized by using R package pheatmap V1.0.12. Multiple genomic sequence alignment visualization was displayed by using Mauve V20150226 (Darling et al., 2004), which is used for multiple genome alignments to identify genome-wide rearrangements and inversions. Minimum locally colinear block (LCB) weight was set as 15 and Muscle V3.6 was used for full alignment (Edgar, 2004).
Pan-Genome Analysis
The pan-genome of W. confusa was constructed by using Bacterial Pan Genome Analysis (BPGA) V1.3.0 with 50% sequence identity cu-off for clustering (Chaudhari et al., 2016). Phylogenetic tree was constructed on the basis of the core genes of different strains through the neighbor-joining method. The effects of isolation ecological niches on the genetic characteristics of W. confusa strains were analyzed via principal component analysis (PCA) according to the gene presence–absence matrix generated from BPGA.
Data Availability
The complete genomic sequences of W. confusa LM1 have been deposited in GenBank under the accession numbers CP080582.1 (chromosome) and CP080583.1 (plasmid). The BioProject accession number for this project was PRJNA752246 (http://www.ncbi.nlm.nih.gov/bioproject/752246).
Results
Weissella confusa LM1 Strain and Its Genome
We used the MRS medium to screen and isolate LAB in the gut of the migratory locust. Approximately 107 colony-forming units (CFUs) were found in the whole gut of locusts. Eighty colonies were randomly selected to identify species composition by sequencing 16S rRNA gene sequence. We found that 74 (92.5%) of the colonies were W. confusa, which stably existed in the locust gut.
We isolated the strain and sequenced the whole genome of W. confusa LM1. The W. confusa LM1 genome consisted of a circular chromosome and a circular plasmid (Figures 1A,B). The size of its genome was 2.53 Mb; the sizes of the circular chromosome and circular plasmid were 2.50 Mb and 25.85 kb, respectively (Table 1). A total of 2,320 protein-coding sequences (CDSs) were predicted by GeneMarkS, and 1,543 genes were annotated in the COG database, which were primarily mapped to the metabolism category, particularly carbohydrate transport and metabolism (Figure 1C). This strain from locust had 114 RNA genes, which are as follows: 84 tRNAs, 10 5S rRNA, 9 16S rRNA, 9 23S rRNA, and 2 sRNA genes.
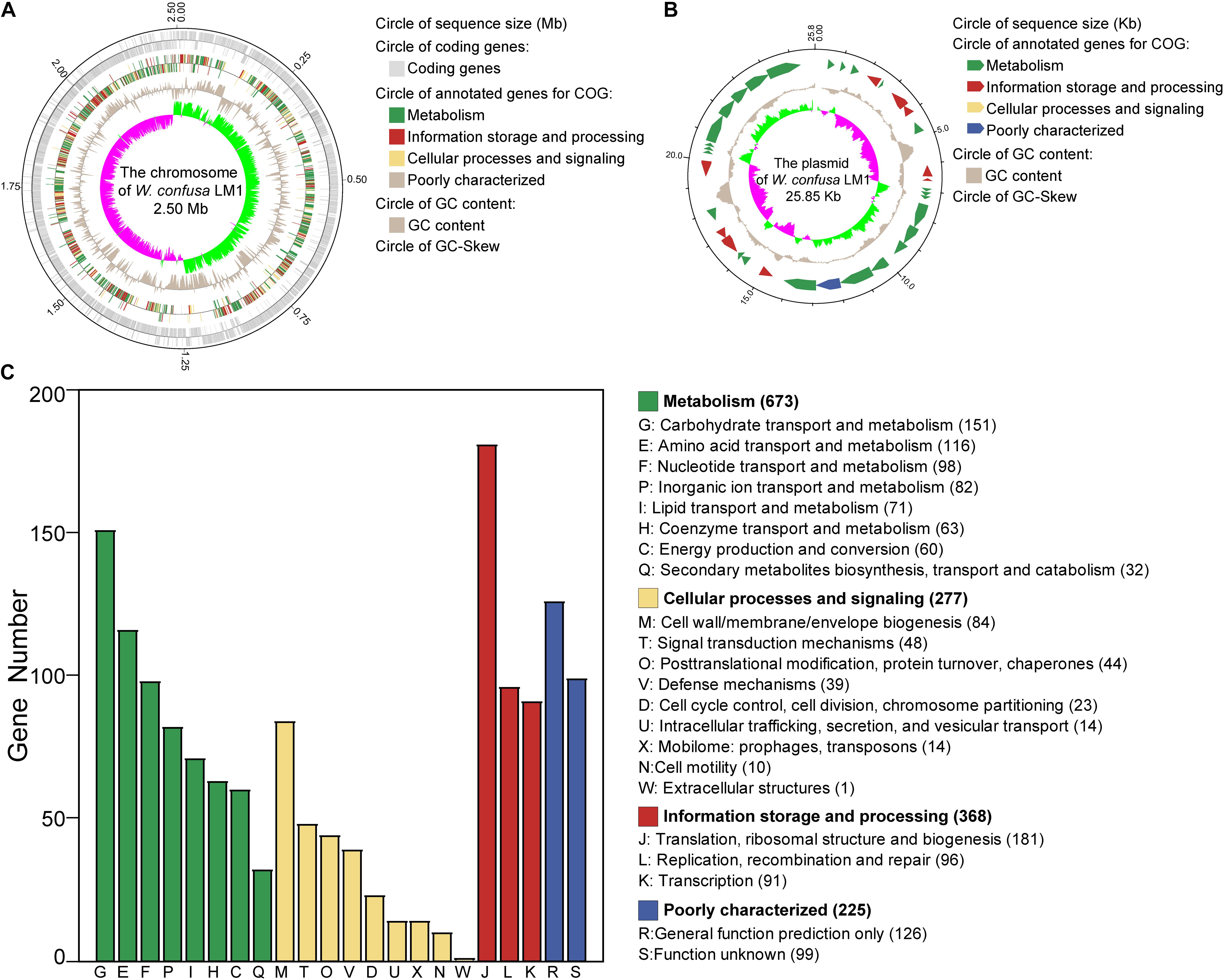
Figure 1. Genome analysis of Weissella confusa LM1. (A) The chromosome features of W. confusa LM1. (B) The plasmid features of W. confusa LM1. (C) Clusters of Orthologous Groups (COG) analysis of W. confusa LM1 genome.
Comparative Genomics of Weissella
There were 28 complete genomes of Weissella genus in the NCBI public databases. Based on the complete genome sequences, we calculated the ANI value between different species and strains (Figure 2). The ANI value reflected the evolutionary relationship between two species, and 95% sequence identity was considered as the species level threshold.
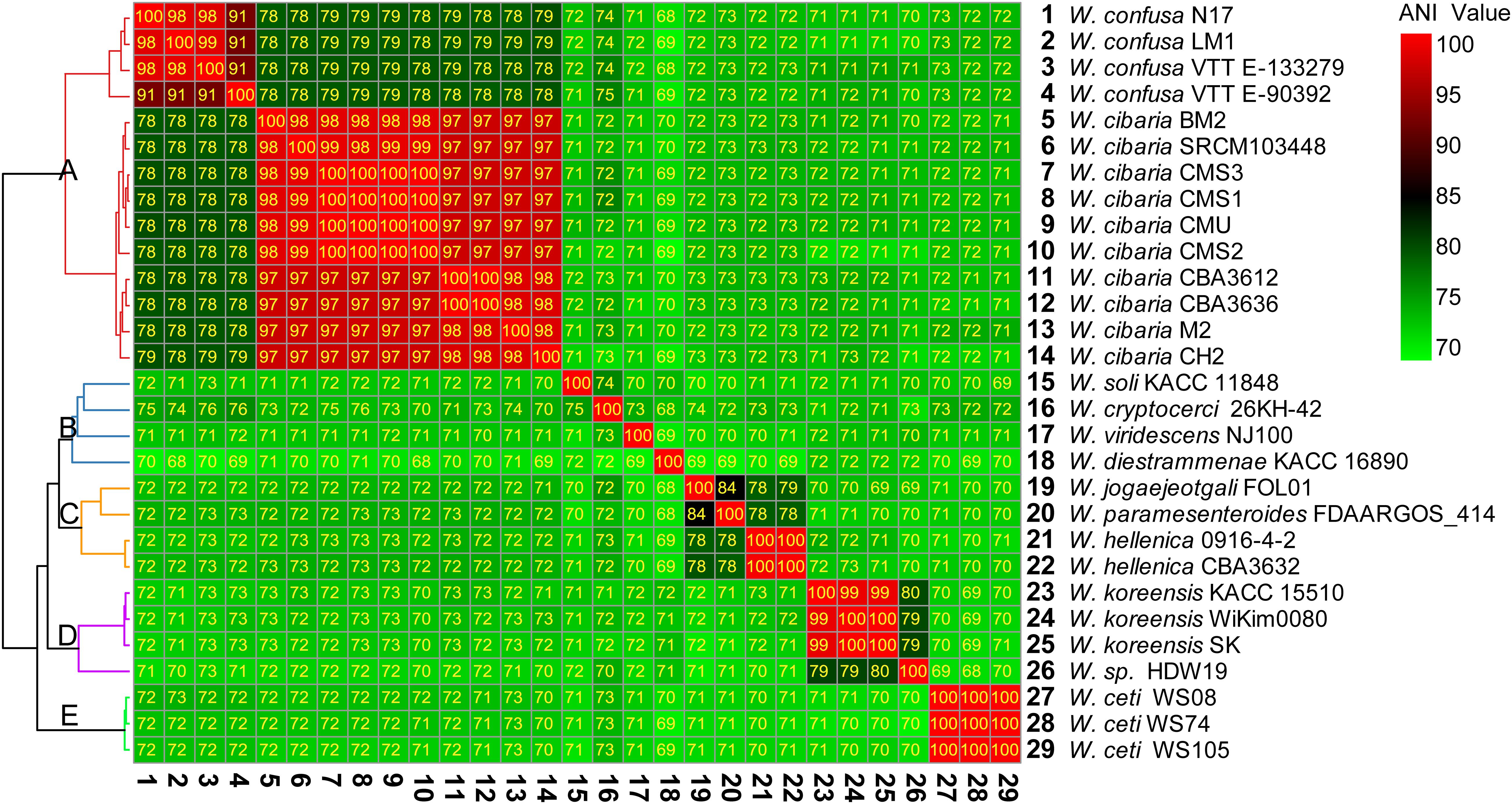
Figure 2. Heatmap of percentage average nucleotide identity (ANI) value between the different species and strains of Weissella. The strain on the vertical axis serves as the reference.
The 29 complete genome assemblies of Weissella were clustered into five clusters, namely, A, B, C, D, and E (Figure 2). Cluster A had 2 species and 14 strains of W. cibaria and W. confusa, whose ANI values were about 78–80% compared with each other. However, the ANI value was less than 75% compared with that of other species. Cluster B had four species and four strains, namely, Weissella soli, Weissella cryptocerci, Weissella viridescens, and Weissella diestrammenae, and all of them had lower ANI values compared with other strains. In particular, the ANI value of W. diestrammenae was only about 70% compared with that of other species, indicating its species-specific status. Cluster C had three species and four strains, namely, Weissella jogaejeotgali, Weissella paramesenteroides, and W. hellenica, and they were found to have a close evolutionary relationship with an ANI value of above 78%. In particular, the ANI value between W. jogaejeotgali and W. paramesenteroides was 84%. Cluster D had two species and four strains, namely, Weissella koreensis and the unidentified species W. sp. HDW19. The species of Weissella sp. HDW19 had a high ANI value of 80% with W. koreensis. Cluster E had 1 species, W. ceti, and three strains. W. ceti had a low ANI value with all other species.
Comparative Genomics of Weissella confusa
By 2021, three complete and 35 draft genomes of W. confusa strains were deposited into the NCBI public database, and 85% of them were sequenced in the past 3 years. According to the information on source and region, W. confusa could be isolated from different niches, such as food, plants, and animals, especially in vertebrate animals (Supplementary Table 1). LM1 was the first insect-derived W. confusa strain to be sequenced. It had the largest genome size (2.53 Mb), which was at least 6% more than those of the other strains (2.18–2.38 Mb). Additionally, LM1 had a lower GC content than the other W. confusa strains.
To analyze the evolutionary relationships among different W. confusa strains, we calculated the ANI value according to the genomic information of 39 strains (Figure 3A) and performed an evolutionary tree analysis based on the CDSs (Figure 3B). Although all strains were named W. confusa, according to ANI values, the ANI values fluctuated between 90 and 100%. According to their ANI values. The 39 strains were classified into four clusters, namely, A, B, C, and D, on the basis of their genome sequences (Figure 3A). Cluster A had 3 strains, which had a relatively low ANI value compared with the 36 other strains (91–92%). The W. confusa VTTE-062653 was remarkably different from all the other strains. However, the ANI value between W. confusa VTTE-90392 and W. confusa 718955 was 99%. Cluster B had 1 strain, W. confusa VTTE-153457, whose ANI value was approximately 95% compared with those of other strains. Cluster C had 29 strains, and they all had high ANI values (98–100%). Cluster D had six strains, whose ANI values were in the range of 96–98%. W. confusa LM1 was the most similar to W. confusa 1001271B_151109_G12, which was isolated from human feces (United States).
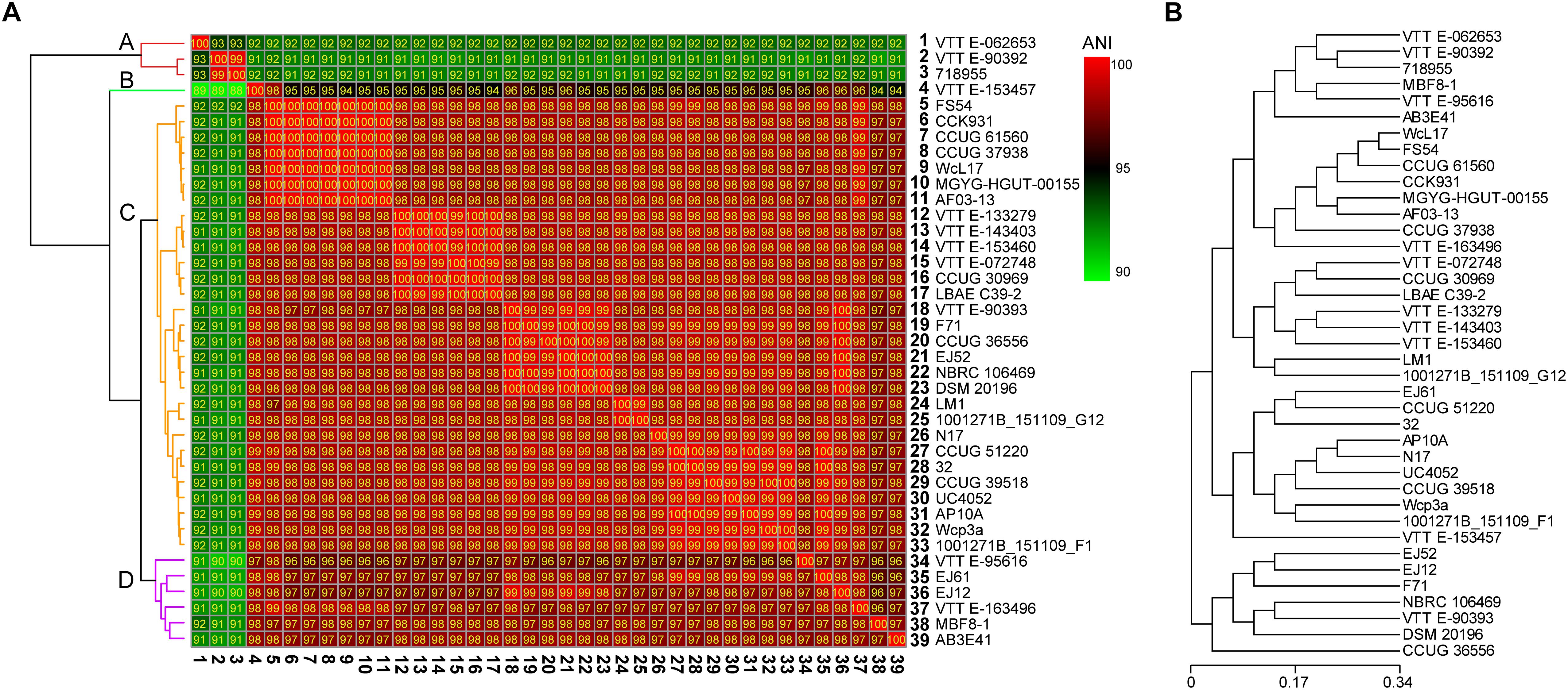
Figure 3. Evolutionary relationships of 39 W. confusa strains. (A) Heatmap of percentage ANI value among different W. confusa strains; the strain on the vertical axis serves as the reference. (B) Evolutionary tree analysis based on the protein-coding genes.
To analyze the genomic structures among different W. confusa strains, we compared all four complete genomes and subjected them to similarity analysis by using the Mauve software (Figure 4). With W. confusa LM1 as the reference, the four genomes exhibited 11 LCBs, and W. confusa LM1 showed more uncolored regions.
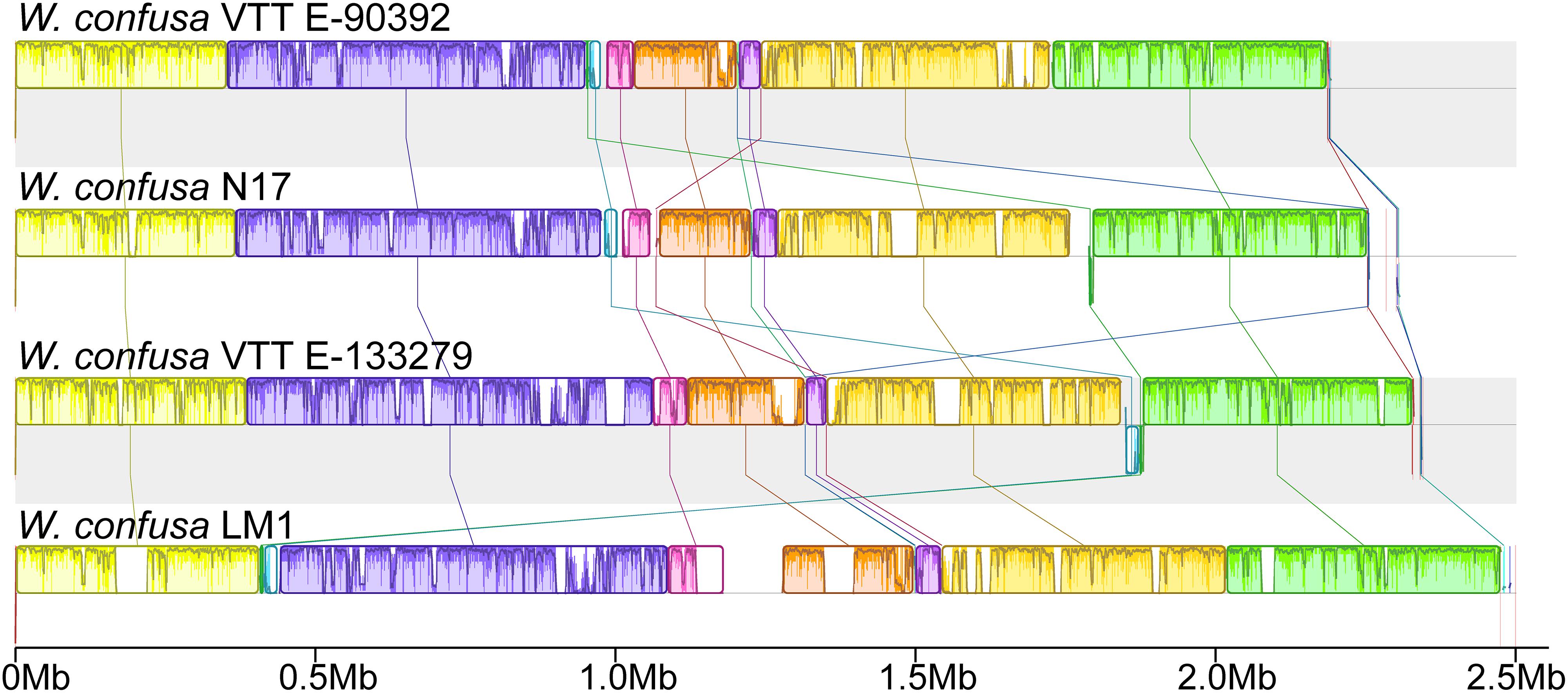
Figure 4. Complete genome alignment of W. confusa strains. MAUVE alignment of the genome sequences of W. confusa VTTE-90392, W. confusa N17, W. confusa VTTE-133279, and W. confusa LM1. Boxes in the same color represent homologous regions [local colinear blocks (LCBs)] between W. confusa genomes. Uncolored regions within the LCBs or in-between LCBs indicate the presence of specific sequences in this strain.
Pan-Genome Analysis of Weissella confusa
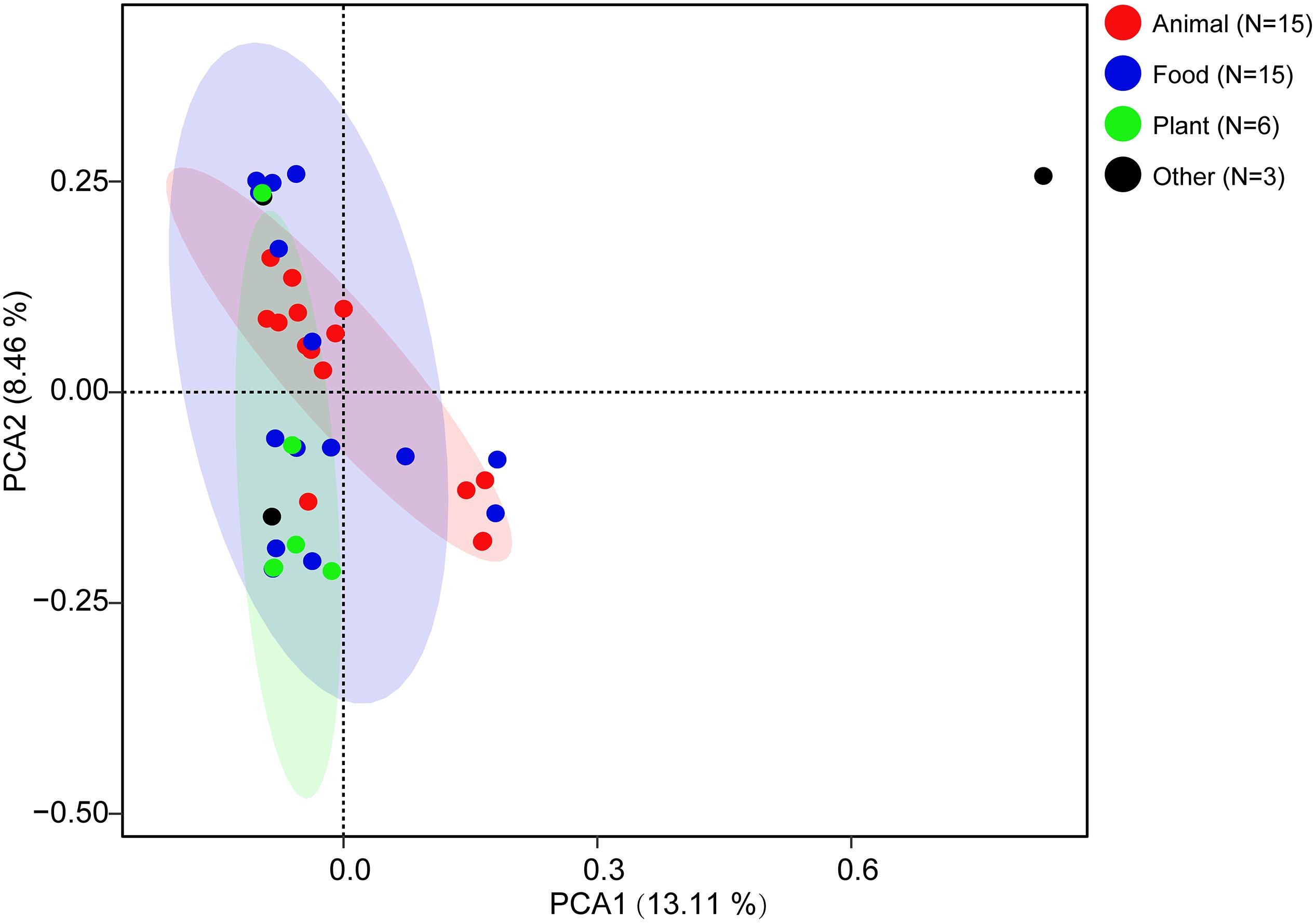
To explore the whole genetic characteristics of W. confusa, we performed pan-genome analysis and obtained the core, accessory, and unique genes of the 39 strains of W. confusa (Figure 5). The 768 genes shared by all W. confusa strains formed the core genome. The 2,246 genes present in more than one strain formed the accessory genome, and 1,193 genes specific to one single strain formed the unique genome. The percentage of unique genes was divergent in different W. confusa strains (Figure 5). Some strains, such as W. confusa FS54 (32%), W. confusa LM1(11%), and W. confusa VTTE-062653 (7%), contained a high percentage of unique genes. By contrast, five strains, including W. confusa MGYG-HGUT-00155, W. confusa AF03-13, W. confusa NBRC 106469, W. confusa VTTE-133279, and W. confusa VTTE-153460, did not have unique genes. These different W. confusa strains were widely distributed in food, animals, and plants. However, little was known about the relationship between their genomic characteristics and their ecological niches. Among the 39 W. confusa strains, 15 strains were isolated from food, 15 strains were isolated from animals, 6 strains were isolated from plants, and 3 strains were isolated from other ecological niches (Supplementary Table 1). On the basis of the gene presence–absence matrix of different W. confusa strains, no close relationship was found between their genetic characteristics and ecological niches (Figure 6). Moreover, we performed the fitting curve f(x) = a × x^b (a = 2,055, b = 0.18) based on the correlation of total gene number and genome number. The curve parameter indicated that the genome of W. confusa was still open (Figure 7). The open pan-genome implied a great potential for discovering novel genes with the increasing number of W. confusa strains sequenced.
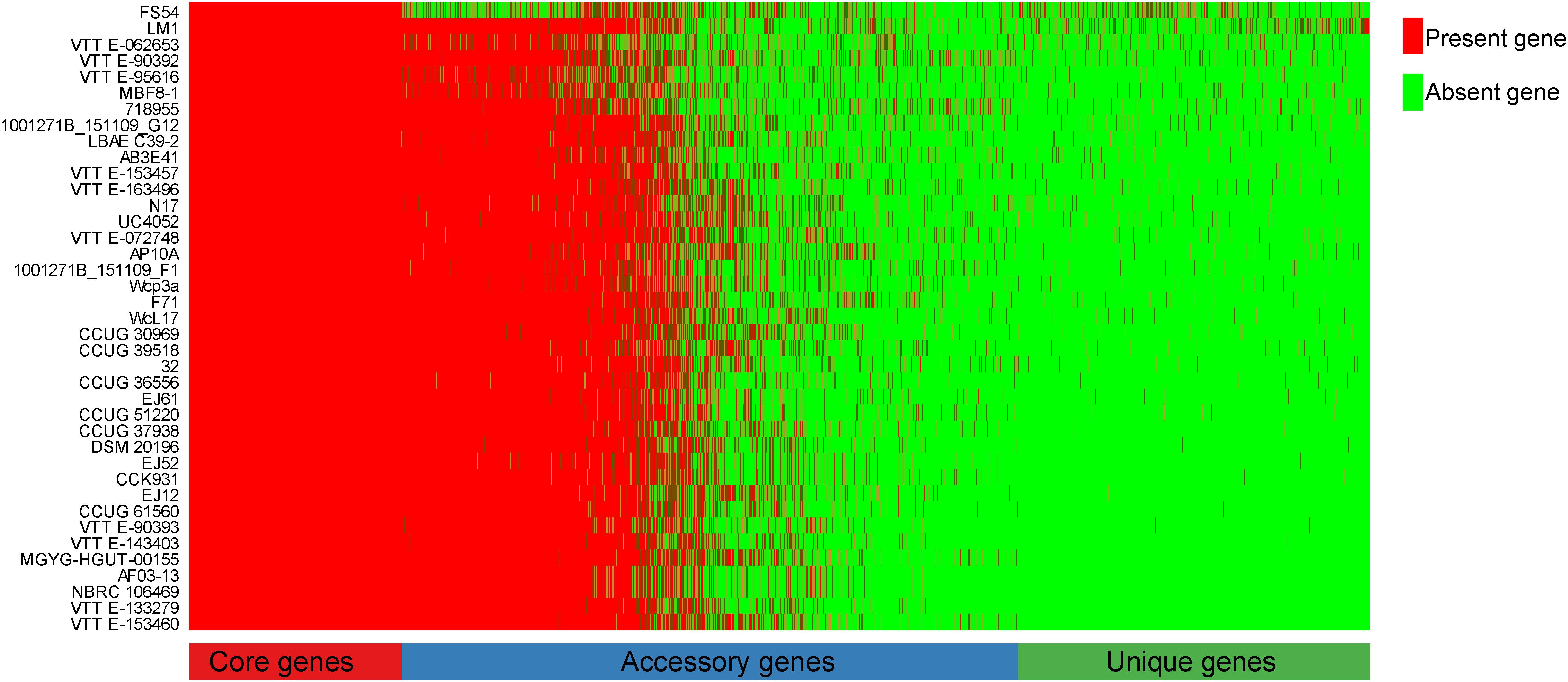
Figure 5. Gene distribution of 39 W. confusa strains based on the gene presence–absence matrix generated from the Bacterial Pan Genome Analysis (BPGA).
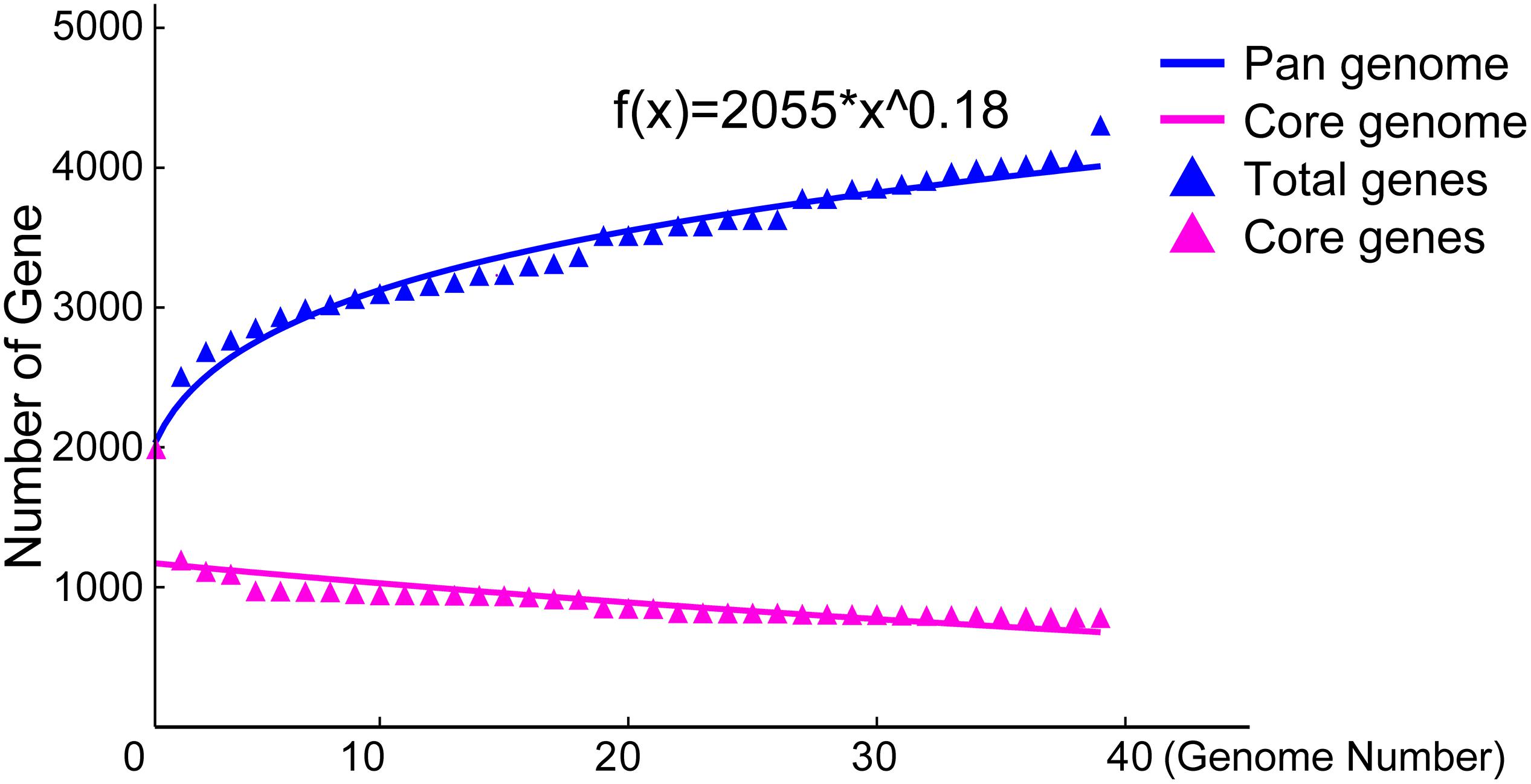
Figure 7. Pan- and core-genome plots of W. confusa based on the results of pan-genome analysis of 39 W. confusa strains.
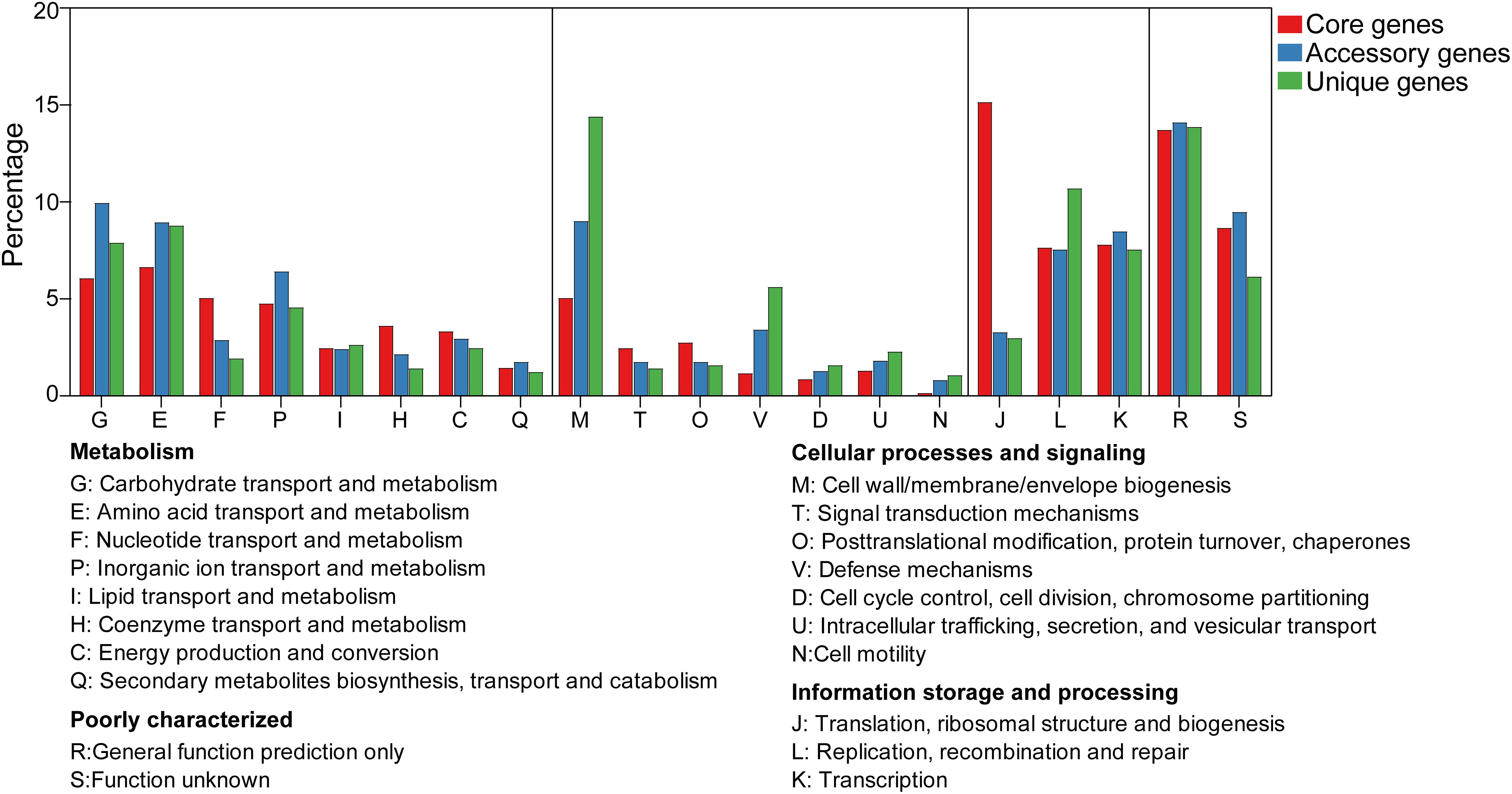
To compare genome functional categories, we used functional assignments from the COG database. In the pan-genome of W. confusa, we found that most core, accessory, and unique genes fell in the metabolism category (Figure 8). Meanwhile, genes with predicted functions and unknown functions were also more abundant (Supplementary Figure 1). Compared with the core genome, the majority of genes in the no-core genome (accessory and unique genomes) were involved in carbohydrate transport and metabolism (G), amino acid transport and metabolism (E), cell wall/membrane/envelope biogenesis (M), defense mechanisms (V), and cell motility (N). By comparison, the vast majority of genes in the core genome were involved in housekeeping functions, such as nucleotide transport and metabolism (F), coenzyme transport and metabolism (H), and translation, ribosomal structure, and biogenesis (J).
To further understand the carbohydrate metabolism in W. confusa, we identified the CAZymes based on the pan-genome. Most genes fell into glycosyltransferases (GTs), glycoside hydrolases (GHs), carbohydrate esterases (CEs), carbohydrate-binding molecules (CBMs), whereas a few genes were annotated in polysaccharide lyases (PLs). All of the genes were not annotated in auxiliary activities (AAs). A few core genome genes were on CAZymes. Compared with the core genome, the accessory genome had more genes on GT, GH, CBM, and CE, the unique genome had more genes on GT, GH, and CE, but fewer genes on CBM (Figure 9A). In addition, different W. confusa strains exhibited various numbers of CAZyme genes (Supplementary Table 2). There were 110 CAZyme genes in W. confusa 718955, but only 85 CAZyme genes in W. confusa MBF8-1. This finding implied that the metabolic abilities of carbohydrates were specialized among W. confusa strains. To further compare the specificity of different strains, we analyzed the CAZyme genes of different strains based on unique genome (Figure 9B). Twenty W. confusa strains contained unique CAZyme genes, and GT (56%) and GH (31%) genes were the most common in the unique CAZyme genes, such as GT2 (28%), GT4 (17%), GH23 (7%), and GH70 (6%).
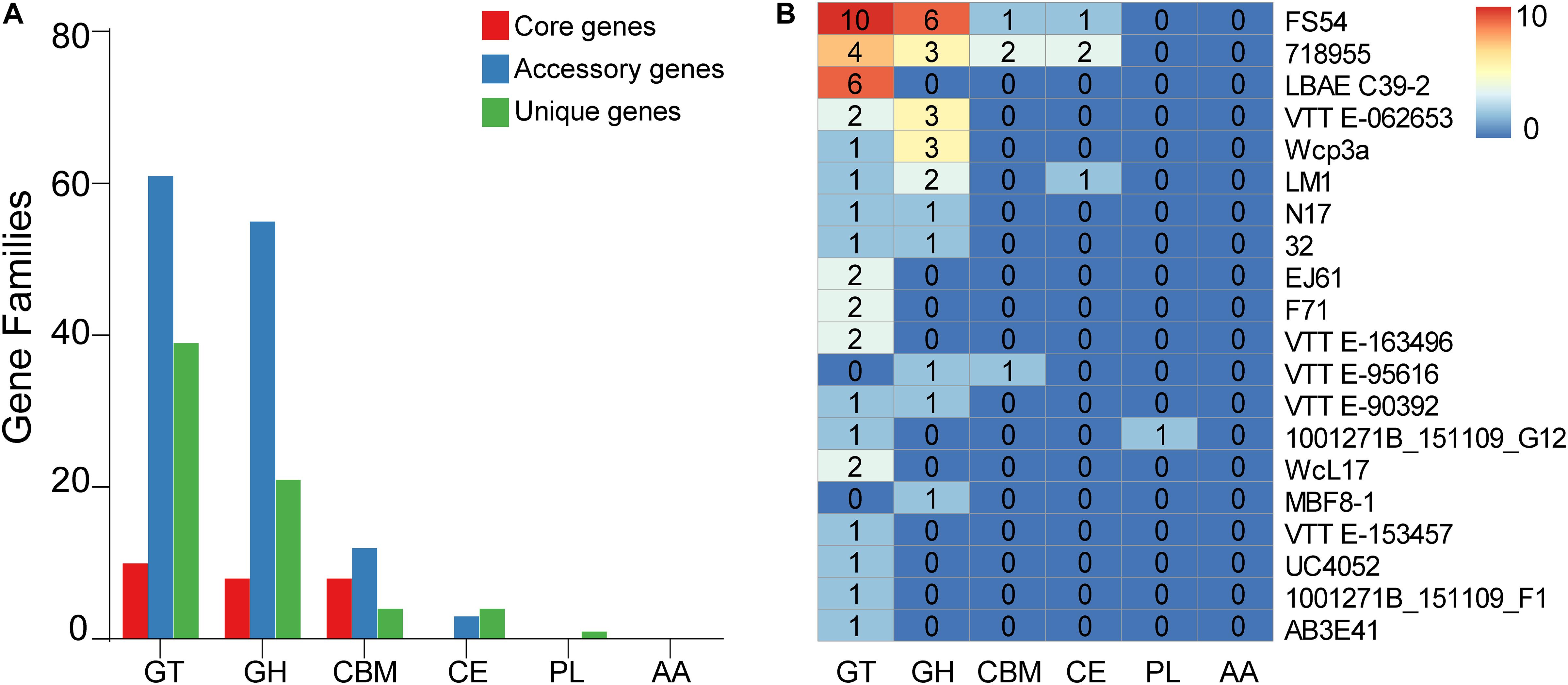
Figure 9. Distribution of pan-genome of W. confusa on CAZymes. (A) The counts of orthologous genes assigned in the core genome, accessory genome, and unique genome on CAZymes. (B) The counts of unique genes of different W. confusa strains on CAZymes.
Secondary Metabolite in Weissella confusa
The genomic repertoire for the secondary metabolite synthesis of the 39 W. confusa strains was assessed by using antiSMASH. W. confusa strains were found to contain a small number of genes that encode key secondary metabolism biosynthesis enzymes. Only 42 gene clusters encoded secondary metabolites in all the 39 W. confusa strains (Supplementary Table 3). Moreover, 42 gene clusters that encoded secondary metabolites were classified into three types, namely, T3PKS (39), arylpolyene (2), and lassopeptide (1). Each strain harbored one T3PKS. The strain W. confusa FS54 and W. confusa LM1 harbored one arylpolyene. The strain W. confusa WcL17 harbored one lassopeptide.
Pathogenicity and Antibiotic Resistance in Weissella confusa
Based on the VFDB core dataset (setA), we scanned the W. confusa genomes for virulence-related factors. In total, we identified 107 putative orthologs involved in the production of virulence factors in 39 W. confusa strains (Supplementary Table 4). These 107 putative orthologs were classified into three virulence-related factors, namely, hyaluronic acid capsule, capsule, and EF-Tu. All W. confusa strains had three virulence-related factors except W. confusa FS54, which only had one capsule. There was no strain-specific virulence factor. According to the function of the three virulence-related factors (Supplementary Table 4), we found that none of the virulence-related factors was associated with toxin production and secretion system, thereby indicating that W. confusa strains were not sufficient to perform virulence. Moreover, we predicted the pathogenicity of W. confusa strains toward human hosts by using PathogenFinder. W. confusa strains were predicted to be non-human pathogens. We estimated the antibiotic resistance of W. confusa strains based on the antibiotic resistance databases, ResFinder and CARD, and no antibiotic resistance genes were recognized in W. confusa strains. We detected the phages in 39 genomes of W. confusa strains. There were 112 phages, including 40 intact phages, 55 incomplete phages, and 17 questionable phages (Supplementary Table 5). There were 30 W. confusa strains with intact phages, which could be divided into two major clusters (Supplementary Figure 2), and we did not find any virulence factors and antibiotic resistance genes in all phages.
Discussion
LAB are generally recognized as safe microorganisms, and they have many beneficial functions in the host gut (Feord, 2002). In this study, we used the MRS medium to culture LAB from the locust gut. W. confusa was found to be a dominant LAB. The W. confusa strains have many functions and are distributed in different niches, such as food, plants, and vertebrate animals. However, a few W. confusa strains are found in insects, except in locust and cricket (Milanovic et al., 2018; Galli et al., 2020). We isolated the locust-derived W. confusa strain, designated it as LM1, and then sequenced its genome. W. confusa LM1 had the largest genome size (Supplementary Table 1) and had a high percentage of unique genes (Figure 5), thereby indicating the specificity of locust-derived W. confusa and providing more genomic information on the pan-genome of W. confusa.
At present, the classification of the bacteria is mostly based on the 16S rRNA gene sequence. However, some species belonging to one genus may have very similar 16S rRNA gene sequences, leading to a poor resolution when distinguishing different species (Case et al., 2007). The ANI analysis with genomic information is helpful in constructing a more precise phylogenetic relationship and performing taxonomic revision. In the present study, W. confusa strains VTTE-062653, VTTE-90392, and 718955 were found to have low ANI value (92%) compared with other W. confusa strains (Figure 3). This 92% ANI value did not meet the standards of species delineation (95%). Thus, we suggested that these three strains should be treated as new species of Weissella or new subspecies of W. confusa. Moreover, ANI analysis were helpful in predicting and analyzing the functions of different species and strains. For instance, W. cibaria and W. confusa were found to have a high ANI value (Figure 2) and possessed many similar characteristics (Fusco et al., 2015; Nel et al., 2019; Rizzello et al., 2019; Quattrini et al., 2020).
The pan-genome status of W. confusa is open (Figure 7), as this species has a high percentage of accessory (53%) and unique (28%) genes. Some accessory and unique genes are from lateral gene transfer, which allows different strains of species to adapt better to specific niches (Caputo et al., 2019). The open pan-genome of W. confusa was consistent with the fact that W. confusa can exist in various niches (Supplementary Table 1). Core genes are important to understand the core factor that maintains the survival and reproduction of this species because they can be used to detect interactions among different species (Yang et al., 2019). The genome of the W. confusa LM1 strain and the pan-genome of W. confusa were helpful in detecting the interaction between W. confusa and the migratory locust.
Comparative genomic analysis revealed a high genomic variation among W. confusa strains (Figure 5). This result implied that different strains are able to perform strain-specific function. The strain-specific function would link with the production of exopolysaccharides (EPSs), which are extracellular macromolecules with wide-ranging biofunctional properties and one of the distinctive phenotypic features of Weissella (Fusco et al., 2015; Teixeira et al., 2021). Among Weissella species, W. confusa is one of the most important EPS producers (Teixeira et al., 2021). However, different W. confusa strains, like W. confusa VP30, XG-3, and KR780676, were found to have various EPSs with different functions (Jin et al., 2019; Kavitake et al., 2020; Zhao et al., 2020). On the other hand, different W. confusa strains were found to have property to produce strain-specific bacteriocin. For instance, W. confusa A3 can produce bacteriocin A3 (Goh and Philip, 2015), and W. confusa MBF8-1 can produce Weissellicin-MBF (Malik et al., 2021). Thus, the strain-specific genes and their function need to further investigate because of EPS and bacteriocin potential applications to the food, medical, and pharmaceutical industries (Valerio et al., 2020).
Some insect pests possessed insecticide resistance for bacteria-mediated degradation (Itoh et al., 2018; Blanton and Peterson, 2020). Exploring the dominant gut bacteria and its potential functions will be helpful in pesticide development, pest management strategies, and pesticide bioremediation. A recent study discovered a new metabolism pathway that the W. confusa Lb.Con strain can degrade chlorpyrifos (Hamoud and Sifour, 2021), which is a commercial pesticide used to control foliar insects, including locusts (Zhang et al., 2019). W. confusa is a dominant gut bacteria of the migratory locust, high abundance of W. confusa may be one of the important reasons for the marginal chlorpyrifos resistance of this pest (Yang et al., 2009). Given that spraying fenitrothion can enhance the abundance of fenitrothion-degrading Burkholderia strains in Riptortus pedestris gut (Kikuchi et al., 2012), the high level of W. confusa in the migratory locust gut is probably the consequence of the increased amount of pesticides or other stresses affecting the migratory locust. These results further indicated that the sustainable management of major insect pests need to consider the role of gut microbiota (Blanton and Peterson, 2020).
As a stable and dominant strain in the migratory locust gut, W. confusa LM1 has the potential to be modified into engineered bacteria with specific functions. With the development of genetic technology, engineered symbiotic microorganisms have been used under different conditions. In mammals, many engineered symbiotics can metabolize harmful substances and serve as a vector to treat certain diseases, and some of them are already in the clinical stage of research (Charbonneau et al., 2020). Furthermore, significant progresses have been achieved in the exploration of the effects of engineered strains on some insects. Some examples include the following: modified Photorhabdus can enhance the control of the agricultural pest in maize root (Machado et al., 2020); modified Snodgrassella alvi can increase the longevity and immunity of beneficial insect (Leonard et al., 2020); and modified Serratia can reduce the transmission of malaria by reducing Plasmodium in mosquitoes (Wang et al., 2017). The migratory locust is an important agricultural pest, and outbreaks of locust plague impose huge losses to agriculture and animal husbandry (Guo et al., 2020). At present, the control of locust plague primarily depends on chemical agents, which act rapidly but may lead to some ecological problems (Roussi, 2020). The stable and safe symbiotic W. confusa LM1 can serve as a potential target for controlling locust plague by inducing this strain to express some metabolites and, thus, decrease the vigorous traits of locust, such as reproduction, flight, and aggregation pheromone. The genomic information of this stable strain in our report lays the foundation for further exploration and application.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.ncbi.nlm.nih.gov/bioproject/, PRJNA752246.
Author Contributions
SY, LK, and FZ conceived and designed the project. SY performed the experiments, analyzed the data, and wrote the manuscript. YW organized and saved the data. LK critically reviewed and curated the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by The State Key Laboratory of Integrated Management of Pest Insects and Rodents (Grant No. ChineseIPM1809) and National Natural Science Foundation of China (Grant No. 32088102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.749218/full#supplementary-material
Supplementary Figure 1 | Distribution of core, accessory, and unique genes on COG category.
Supplementary Figure 2 | Phylogenetic tree of the intact phages identified in W. confusa strains.
Footnotes
- ^ http://www.pacb.com/
- ^ http://circos.ca/
- ^ https://antismash.secondarymetabolites.org/
- ^ https://cge.cbs.dtu.dk/services/PathogenFinder/
- ^ https://ggdc.dsmz.de/victor.php#
- ^ https://cge.cbs.dtu.dk/services/ResFinder/
- ^ https://card.mcmaster.ca/analyze/rgi
References
Albesharat, R., Ehrmann, M. A., Korakli, M., Yazaji, S., and Vogel, R. F. (2011). Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 34, 148–155. doi: 10.1016/j.syapm.2010.12.001
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Blanton, A. G., and Peterson, B. F. (2020). Symbiont-mediated insecticide detoxification as an emerging problem in insect pests. Front. Microbiol. 11:547108. doi: 10.3389/fmicb.2020.547108
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Bourdichon, F., Casaregola, S., Farrokh, C., Frisvad, J. C., Gerds, M. L., Hammes, W. P., et al. (2012). Food fermentations: microorganisms with technological beneficial use. Int. J. Food Microbiol. 154, 87–97. doi: 10.1016/j.ijfoodmicro.2011.12.030
Bourdichon, F., Patrone, V., Fontana, A., Milani, G., and Morelli, L. (2021). Safety demonstration of a microbial species for use in the food chain: Weissella confusa. Int. J. Food Microbiol. 339:109028. doi: 10.1016/j.ijfoodmicro.2020.109028
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nature Methods 12, 59–60. doi: 10.1038/Nmeth.3176
Caputo, A., Fournier, P. E., and Raoult, D. (2019). Genome and pan-genome analysis to classify emerging bacteria. Biol. Direct. 14:5. doi: 10.1186/s13062-019-0234-0
Case, R. J., Boucher, Y., Dahllof, I., Holmstrom, C., Doolittle, W. F., Kjelleberg, S., et al. (2007). Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73, 278–288. doi: 10.1128/AEM.01177-06
Chaisson, M. J., and Tesler, G. (2012). Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. doi: 10.1186/1471-2105-13-238
Charbonneau, M. R., Isabella, V. M., Li, N., and Kurtz, C. B. (2020). Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 11:1738. doi: 10.1038/s41467-020-15508-1
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2016). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, L., Xiong, Z., Sun, L., Yang, J., and Jin, Q. (2012). VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40, D641–D645. doi: 10.1093/nar/gkr989
Collins, M. D., Samelis, J., Metaxopoulos, J., and Wallbanks, S. (1993). Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 75, 595–603. doi: 10.1111/j.1365-2672.1993.tb01600.x
Cosentino, S., Larsen, M. V., Aarestrup, F. M., and Lund, O. (2013). PathogenFinder – distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302
Cupi, D., and Elvig-Jørgensen, S. G. (2019). Safety assessment of Weissella confusa – A direct-fed microbial candidate. Regul. Toxicol. Pharmacol. 107:104414. doi: 10.1016/j.yrtph.2019.104414
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dey, D. K., and Kang, S. C. (2020). Weissella confusa DD_A7 pre-treatment to zebrafish larvae ameliorates the inflammation response against Escherichia coli O157:H7. Microbiol. Res. 237:126489. doi: 10.1016/j.micres.2020.126489
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elshaghabee, F. M. F., Ghadimi, D., Habermann, D., de Vrese, M., Bockelmann, W., Kaatsch, H. J., et al. (2020). Effect of oral administration of Weissella confusa on fecal and plasma ethanol concentrations, lipids and glucose metabolism in wistar rats fed high fructose and fat diet. Hepat. Med. 12, 93–106. doi: 10.2147/HMER.S254195
Feord, J. (2002). Lactic acid bacteria in a changing legislative environment. Antonie Van Leeuwenhoek 82, 353–360.
Fusco, V., Quero, G. M., Cho, G.-S., Kabisch, J., Meske, D., Neve, H., et al. (2015). The genus Weissella: taxonomy, ecology and biotechnological potential. Front. Microbiol. 6:155. doi: 10.3389/fmicb.2015.00155
Galli, V., Venturi, M., Pini, N., and Granchi, L. (2020). Technological feature assessment of lactic acid bacteria isolated from cricket powder’s spontaneous fermentation as potential starters for cricket-wheat bread production. Foods 9:1322. doi: 10.3390/foods9091322
Galperin, M. Y., Wolf, Y. I., Makarova, K. S., Alvarez, R. V., Landsman, D., and Koonin, E. V. (2021). COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49, D274–D281. doi: 10.1093/nar/gkaa1018
Goh, H. F., and Philip, K. (2015). Purification and characterization of bacteriocin produced by Weissella confusa A3 of dairy origin. PLoS One 10:e0140434. doi: 10.1371/journal.pone.0140434
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Guo, X., Yu, Q., Chen, D., Wei, J., Yang, P., Yu, J., et al. (2020). 4-Vinylanisole is an aggregation pheromone in locusts. Nature 584, 584–588. doi: 10.1038/s41586-020-2610-4
Hamoud, N. E., and Sifour, M. (2021). Biodegradation of chlorpyrifos by a Weissella confusa strain and evaluation of some probiotic traits. Arch. Microbiol. 203, 3615–3621. doi: 10.1007/s00203-021-02353-z
Itoh, H., Tago, K., Hayatsu, M., and Kikuchi, Y. (2018). Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 35, 434–454. doi: 10.1039/c7np00051k
Jin, H., Jeong, Y., Yoo, S. H., Johnston, T. V., Ku, S., and Ji, G. E. (2019). Isolation and characterization of high exopolysaccharide-producing Weissella confusa VP30 from young children’s feces. Microb. Cell Fact. 18:110. doi: 10.1186/s12934-019-1158-1
Kavitake, D., Singh, S. P., Kandasamy, S., Devi, P. B., and Shetty, P. H. (2020). Report on aflatoxin-binding activity of galactan exopolysaccharide produced by Weissella confusa KR780676. 3 Biotech 10:181. doi: 10.1007/s13205-020-02173-w
Kikuchi, Y., Hayatsu, M., Hosokawa, T., Nagayama, A., Tago, K., and Fukatsu, T. (2012). Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 8618–8622. doi: 10.1073/pnas.1200231109
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lavecchia, A., Chiara, M., De Virgilio, C., Manzari, C., Pazzani, C., Horner, D., et al. (2021). Comparative genomics suggests a taxonomic revision of the Staphylococcus cohnii species complex. Genome Biol. Evol. 13:evab020. doi: 10.1093/gbe/evab020
Le, B., Ngoc, A. P. T., and Yang, S. H. (2020). Synbiotic fermented soymilk with Weissella cibaria FB069 and xylooligosaccharides prevents proliferation in human colon cancer cells. J. Appl. Microbiol. 128, 1486–1496. doi: 10.1111/jam.14551
Lee, D. S., Lee, S. A., Kim, M., Nam, S. H., and Kang, M. S. (2020). Reduction of halitosis by a tablet containing Weissella cibaria CMU: a randomized, double-blind, placebo-controlled Study. J. Med. Food. 23, 649–657. doi: 10.1089/jmf.2019.4603
Leonard, S. P., Powell, J. E., Perutka, J., Geng, P., Heckmann, L. C., Horak, R. D., et al. (2020). Engineered symbionts activate honey bee immunity and limit pathogens. Science 367, 573–576. doi: 10.1126/science.aax9039
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, S. W., Chen, Y. S., Lee, Y. S., Yang, C. H., Srionnual, S., Wu, H. C., et al. (2017). Comparative genomic analysis of bacteriocin-producing Weissella cibaria 110. Appl. Microbiol. Biotechnol. 101, 1227–1237. doi: 10.1007/s00253-016-8073-8
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Machado, R. A. R., Thönen, L., Arce, C. C. M., Theepan, V., Prada, F., Wüthrich, D., et al. (2020). Engineering bacterial symbionts of nematodes improves their biocontrol potential to counter the western corn rootworm. Nat. Biotechnol. 38, 600–608. doi: 10.1038/s41587-020-0419-1
Malik, A., Yuliantie, E., Suprahman, N. Y., Linardi, T., Widiyanti, A. W., Haldy, J., et al. (2021). Construction and functional analysis of the recombinant bacteriocins Weissellicin-MBF from Weissella confusa MBF8-1. Curr. Pharm. Biotechnol. 22, 115–122. doi: 10.2174/1389201021666200611111040
Man, J., Rogosa, M., and Sharpe, M. E. (1960). A medium for the cultivation of Lactobacilli. J. Appl. Microbiol. 23, 130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x
Mao, B., Yin, R., Li, X., Cui, S., Zhang, H., Zhao, J., et al. (2021). Comparative genomic analysis of Lactiplantibacillus plantarum isolated from different niches. Genes (Basel) 12:241. doi: 10.3390/genes12020241
Martin, R., Heilig, H. G., Zoetendal, E. G., Jimenez, E., Fernandez, L., Smidt, H., et al. (2007). Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol. 158, 31–37. doi: 10.1016/j.resmic.2006.11.004
Meier-Kolthoff, J. P., and Goker, M. (2017). VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33, 3396–3404. doi: 10.1093/bioinformatics/btx440
Milanovic, V., Osimani, A., Roncolini, A., Garofalo, C., Aquilanti, L., Pasquini, M., et al. (2018). Investigation of the dominant microbiota in ready-to-eat grasshoppers and mealworms and quantification of carbapenem resistance genes by qPCR. Front. Microbiol. 9:3036. doi: 10.3389/fmicb.2018.03036
Mortezaei, F., Royan, M., Allaf Noveirian, H., Babakhani, A., Alaie Kordghashlaghi, H., and Balcázar, J. L. (2020). In vitro assessment of potential probiotic characteristics of indigenous Lactococcus lactis and Weissella oryzae isolates from rainbow trout (Oncorhynchus mykiss Walbaum). J. Appl. Microbiol. 129, 1004–1019. doi: 10.1111/jam.14652
Mun, S. Y., and Chang, H. C. (2020). Characterization of Weissella koreensis SK isolated from kimchi fermented at low temperature (around 0 degrees C) based on complete genome sequence and corresponding phenotype. Microorganisms 8:1147. doi: 10.3390/microorganisms8081147
Nel, S., Davis, S. B., Endo, A., and Dicks, L. M. T. (2019). Phylogenetic analyses of pheS, dnaA and atpA genes for identification of Weissella confusa and Weissella cibaria isolated from a South African sugarcane processing factory. Curr. Microbiol. 76, 1138–1146. doi: 10.1007/s00284-019-01740-6
Panthee, S., Paudel, A., Blom, J., Hamamoto, H., and Sekimizu, K. (2019). Complete genome sequence of Weissella hellenica 0916-4-2 and its comparative genomic analysis. Front. Microbiol. 10:1619. doi: 10.3389/fmicb.2019.01619
Pelyuntha, W., Chaiyasut, C., Kantachote, D., and Sirilun, S. (2020). Organic acids and 2,4-Di-tert-butylphenol: major compounds of Weissella confusa WM36 cell-free supernatant against growth, survival and virulence of Salmonella Typhi. PeerJ 8:e8410. doi: 10.7717/peerj.8410
Prado, G. K. S., Torrinha, K. C., Cruz, R. E., Goncalves, A. B. B., Silva, C. A. V., Oliveira, F. M. S., et al. (2020). Weissella paramesenteroides WpK4 ameliorate the experimental amoebic colitis by increasing the expression of MUC-2 and the intestinal epithelial regeneration. J. Appl. Microbiol. 129, 1706–1719. doi: 10.1111/jam.14671
Quattrini, M., Korcari, D., Ricci, G., and Fortina, M. G. (2020). A polyphasic approach to characterize Weissella cibaria and Weissella confusa strains. J Appl Microbiol. 128, 500–512. doi: 10.1111/jam.14483
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rizzello, C. G., Coda, R., Wang, Y., Verni, M., Kajala, I., Katina, K., et al. (2019). Characterization of indigenous Pediococcus pentosaceus, Leuconostoc kimchii, Weissella cibaria and Weissella confusa for faba bean bioprocessing. Int. J. Food Microbiol. 302, 24–34. doi: 10.1016/j.ijfoodmicro.2018.08.014
Roussi, A. (2020). Why gigantic locust swarms are challenging governments and researchers. Nature 579:330. doi: 10.1038/d41586-020-00725-x
Sandes, S., Figueiredo, N., Pedroso, S., Sant’Anna, F., Acurcio, L., Abatemarco Junior, M., et al. (2020). Weissella paramesenteroides WpK4 plays an immunobiotic role in gut-brain axis, reducing gut permeability, anxiety-like and depressive-like behaviors in murine models of colitis and chronic stress. Food Res. Int. 137:109741. doi: 10.1016/j.foodres.2020.109741
Singh, A., Gaur, M., Sharma, V., Khanna, P., Bothra, A., Bhaduri, A., et al. (2021). Comparative genomic analysis of mycobacteriaceae reveals horizontal gene transfer-mediated evolution of the CRISPR-cas system in the mycobacterium tuberculosis complex. mSystems 6:e00934-20. doi: 10.1128/mSystems.00934-20
Sturino, J. M. (2018). Literature-based safety assessment of an agriculture- and animal-associated microorganism: Weissella confusa. Regul. Toxicol. Pharmacol. 95, 142–152. doi: 10.1016/j.yrtph.2018.03.013
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Teixeira, C. G., Fusieger, A., Miliao, G. L., Martins, E., Drider, D., Nero, L. A., et al. (2021). Weissella: an emerging bacterium with promising health benefits. Probiotics Antimicrob. Proteins 13, 915–925. doi: 10.1007/s12602-021-09751-1
Valerio, F., Bavaro, A. R., Di Biase, M., Lonigro, S. L., Logrieco, A. F., Logrieco, A. F., et al. (2020). Effect of Amaranth and Quinoa Flours on exopolysaccharide production and protein profile of liquid sourdough fermented by Weissella cibaria and Lactobacillus plantarum. Front. Microbiol. 11:967. doi: 10.3389/fmicb.2020.00967
Vogel, R. F., Hammes, W. P., Habermeyer, M., Engel, K. H., Knorr, D., Eisenbrand, G., et al. (2011). Microbial food cultures–opinion of the Senate Commission on Food Safety (SKLM) of the German Research Foundation (DFG). Mol. Nutr. Food Res. 55, 654–662. doi: 10.1002/mnfr.201100010
Wang, J. P., Yoo, J. S., Jang, H. D., Lee, J. H., Cho, J. H., and Kim, I. H. (2011). Effect of dietary fermented garlic by Weissella koreensis powder on growth performance, blood characteristics, and immune response of growing pigs challenged with Escherichia coli lipopolysaccharide. J. Anim. Sci. 89, 2123–2131. doi: 10.2527/jas.2010-3186
Wang, S. B., Dos-Santos, A. L. A., Huang, W., Liu, K. C., Oshaghi, M. A., Wei, G., et al. (2017). Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science 357, 1399–1402. doi: 10.1126/science.aan5478
Wang, W., Liu, W., and Chu, W. (2020). Isolation and preliminary screening of potentially probiotic Weissella confusa strains from healthy human feces by culturomics. Microb. Pathog. 147:104356. doi: 10.1016/j.micpath.2020.104356
Wang, Y., Jia, B., Xu, X., Zhang, L., Wei, C., Ou, H., et al. (2018). Comparative genomic analysis and characterization of Two Salmonella enterica serovar enteritidis isolates from poultry with notably different survival abilities in egg whites. Front Microbiol. 9:2111. doi: 10.3389/fmicb.2018.02111
Xiong, L., Ni, X., Niu, L., Zhou, Y., Wang, Q., Khalique, A., et al. (2019). Isolation and preliminary screening of a Weissella confusa strain from giant panda (Ailuropoda melanoleuca). Probiotics Antimicrob. Proteins 11, 535–544. doi: 10.1007/s12602-018-9402-2
Yang, M. L., Zhang, J. Z., Zhu, K. Y., Xuan, T., Liu, X. J., Guo, Y. P., et al. (2009). Mechanisms of organophosphate resistance in a field population of oriental migratory locust, Locusta migratoria manilensis (Meyen). Arch. Insect. Biochem. Physiol. 71, 3–15. doi: 10.1002/arch.20254
Yang, Z. K., Luo, H., Zhang, Y., Wang, B., and Gao, F. (2019). Pan-genomic analysis provides novel insights into the association of E.coli with human host and its minimal genome. Bioinformatics 35, 1987–1991. doi: 10.1093/bioinformatics/bty938
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zannini, E., Jeske, S., Lynch, K. M., and Arendt, E. K. (2018). Development of novel quinoa-based yoghurt fermented with dextran producer Weissella cibaria MG1. Int. J. Food. Microbiol. 268, 19–26. doi: 10.1016/j.ijfoodmicro.2018.01.001
Zhang, X., Jie, D., Liu, J., Zhang, J., Zhang, T., Zhang, J., et al. (2019). Aryl hydrocarbon receptor regulates the expression of LmGSTd7 and is associated with chlorpyrifos susceptibility in Locusta migratoria. Pest.Manag. Sci. 75, 2916–2924. doi: 10.1002/ps.5600
Keywords: Locusta migratoria, gut bacteria, Weissella confusa, comparative genomics, pan-genome
Citation: Yuan S, Wang Y, Zhao F and Kang L (2021) Complete Genome Sequence of Weissella confusa LM1 and Comparative Genomic Analysis. Front. Microbiol. 12:749218. doi: 10.3389/fmicb.2021.749218
Received: 29 July 2021; Accepted: 02 September 2021;
Published: 28 September 2021.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Jingfa Xiao, Beijing Institute of Genomics, Chinese Academy of Sciences, ChinaGabriela Nicoleta Tenea, Universidad Técnica del Norte, Ecuador
Copyright © 2021 Yuan, Wang, Zhao and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Le Kang, bGthbmdAaW96LmFjLmNu
 Shenglei Yuan
Shenglei Yuan Yundan Wang
Yundan Wang Fangqing Zhao
Fangqing Zhao Le Kang
Le Kang