
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 November 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.748890
This article is part of the Research Topic Approaches to address Resistance, Drug Discovery, and Vaccine development in Mycobacterium Tuberculosis: Challenges and opportunities View all 9 articles
 Arun Sharma1†
Arun Sharma1† Kalpana Sagar2,3†
Kalpana Sagar2,3† Neeraj Kumar Chauhan1
Neeraj Kumar Chauhan1 Balaji Venkataraman2
Balaji Venkataraman2 Nidhi Gupta2
Nidhi Gupta2 Tannu Priya Gosain1
Tannu Priya Gosain1 Nikhil Bhalla2
Nikhil Bhalla2 Ramandeep Singh1*
Ramandeep Singh1* Amita Gupta2,3*
Amita Gupta2,3*The extraordinary expansion of Toxin Antitoxin (TA) modules in the genome of Mycobacterium tuberculosis has received significant attention over the last few decades. The cumulative evidence suggests that TA systems are activated in response to stress conditions and are essential for M. tuberculosis pathogenesis. In M. tuberculosis, Rv1955-Rv1956-Rv1957 constitutes the only tripartite TAC (Toxin Antitoxin Chaperone) module. In this locus, Rv1955 (HigB1) encodes for the toxin and Rv1956 (HigA1) encodes for antitoxin. Rv1957 encodes for a SecB-like chaperone that regulates HigBA1 toxin antitoxin system by preventing HigA1 degradation. Here, we have investigated the physiological role of HigB1 toxin in stress adaptation and pathogenesis of Mycobacterium tuberculosis. qPCR studies revealed that higBA1 is upregulated in nutrient limiting conditions and upon exposure to levofloxacin. We also show that the promoter activity of higBA1 locus in M. tuberculosis is (p)ppGpp dependent. We observed that HigB1 locus is non-essential for M. tuberculosis growth under different stress conditions in vitro. However, guinea pigs infected with higB1 deletion strain exhibited significantly reduced bacterial loads and pathological damage in comparison to the animals infected with the parental strain. Transcriptome analysis suggested that deletion of higB1 reduced the expression of genes involved in virulence, detoxification and adaptation. The present study describes the role of higB1 toxin in M. tuberculosis physiology and highlights the importance of higBA1 locus during infection in host tissues.
Tuberculosis (TB), caused by Mycobacterium tuberculosis (M. tuberculosis) is a major health concern and infects nearly one-third of the world population. The failure of BCG vaccine to impart protection in adult population and HIV co-infection has negative impact over the control of global TB cases. There is a significant increase in the number of patients infected with the M. tuberculosis strain resistant to front-line TB drugs such as isoniazid and rifampicin. Studies have shown that the proportion of TB patients infected with multi-drug resistant strains lies in the range of 4.6–25% (Lange et al., 2019). Control of the spread of drug-resistant TB and eradication of TB is hampered by the limited efficacy of therapeutic approaches against drug resistant M. tuberculosis strains and our poor understanding of the strategies used by the pathogen for survival inside the human host. M. tuberculosis has emerged as a highly successful intracellular pathogen due to its ability to sense extracellular stimuli and reprogram metabolic pathways that enables it to survive in host tissues under varied stress conditions.
Toxin antitoxin (TA) systems are mostly two component modules that are widely present in the genome of prokaryotes and have been implicated in bacterial stress adaptation, persister formation and virulence (Page and Peti, 2016; Harms et al., 2018; Slayden et al., 2018). TA module encodes a toxin that generally inhibits bacterial growth in a bacteriostatic manner by inhibiting an essential cellular function (Schuster and Bertram, 2013; Kedzierska and Hayes, 2016; Page and Peti, 2016; Harms et al., 2018; Fraikin et al., 2020; Kamruzzaman et al., 2021). The antitoxin component of the TA operon neutralizes the activity of this toxin. Based on the mechanisms for neutralization of toxin by the cognate antitoxin, the TA systems have been classified into eight types (Lobato-Marquez et al., 2016; Choi et al., 2018; Harms et al., 2018; Song and Wood, 2020; Wang et al., 2021). The M. tuberculosis genome encodes for majorly Type II TA systems, where the antitoxin forms a tight complex with toxin and abrogates the activity associated with toxin (Pandey and Gerdes, 2005; Ramage et al., 2009; Sala et al., 2014; Tandon et al., 2019). VapBC family that encodes for the VapC toxin and VapB antitoxin is the most abundant subfamily of TA systems in M. tuberculosis (Ahidjo et al., 2011). VapC toxins are characterized by the presence of PIN domain and have been shown to inhibit M. tuberculosis growth by targeting either tRNA or rRNA or mRNA (Sharp et al., 2012; Cruz et al., 2015; Winther et al., 2016). The inducible expression of VapC toxins inhibits the growth of M. tuberculosis or M. smegmatis or E. coli in a bacteriostatic manner (Ramage et al., 2009; Winther et al., 2016; Agarwal et al., 2018). Several studies have shown that the expression of a “subset” of TA systems is increased in stress conditions such as nutrient deprivation, low oxygen and in macrophages (Ramage et al., 2009; Gupta et al., 2017; Agarwal et al., 2018). Previously, we have shown that deletion of vapBC3, vapBC4, vapBC11, and vapC22 in the genome of M. tuberculosis impairs its growth in guinea pigs (Agarwal et al., 2018, 2020; Deep et al., 2018). However, parental, ΔvapC28 mutant and ΔvapC21 strain displayed comparable growth kinetics in guinea pigs and mice, respectively (Agarwal et al., 2018; Sharma et al., 2020). MazF toxins belonging to MazEF TA systems are sequence specific endonucleases that are cumulatively required for M. tuberculosis to establish infection in host tissues (Tiwari et al., 2015). RelE toxins belonging to the RelBE TA system have been shown to be individually non-essential for M. tuberculosis virulence in mice tissues but contribute to antibiotic tolerance in a drug-specific manner (Singh et al., 2010).
HigBA TA system was originally identified on Proteus vulgaris plasmid, Rts1 with unique gene arrangement as HigA antitoxin is present downstream of HigB toxin (Tian et al., 1996). HigB belongs to RelE subfamily of toxins and cleaves mRNA in a ribosome dependent manner in V. cholerae, P. vulgaris, and E. coli (Christensen-Dalsgaard and Gerdes, 2006; Hurley and Woychik, 2009; Christensen-Dalsgaard et al., 2010). HigBA TA module from A. baumannii is expressed during stationary phase and under iron deficient conditions (Armalyte et al., 2018). In P. aeruginosa, activation of HigB toxin influences the levels of intracellular c-di-GMP and virulence factors like pyocyanin and pyochelin (Wood and Wood, 2016). Furthermore, HigB toxin also promotes the killing of immune cells by increasing the expression of type III secretion system in ciprofloxacin induced persisters in P. aeruginosa (Li et al., 2016; Wood and Wood, 2016; Zhang et al., 2018). Besides other bicistronic TA operons, M. tuberculosis encodes for two HigBA TA loci, HigBA2 (Rv2022c-Rv2021c) and HigBA3 (Rv3182-Rv3183). The genome of M. tuberculosis also encodes for a tripartite Toxin Antitoxin Chaperone (TAC) system. TAC system comprises of HigB1 toxin (Rv1955), HigA1 antitoxin (Rv1956) and SecB like chaperone (Rv1957). SecB like chaperone prevents HigA1 aggregation and degradation by interacting with Chad like sequences present within HigA1 (Fivian-Hughes and Davis, 2010; Bordes et al., 2016; Guillet et al., 2019). Studies in E. coli and M. smegmatis have shown that overexpression of HigB1 toxin inhibits bacterial growth which is restored upon co-expression of cognate antitoxin, HigA1 (Gupta, 2009; Ramage et al., 2009). It has been shown that HigB1 and HigA1 are co-transcribed with upstream genes Rv1954A, Rv1954c and downstream gene, Rv1957. The locus comprises of two promoters, the P2 promoter controls the expression of Rv1954A-Rv1957 locus, whereas, the P1 promoter is inducible in DNA damaging conditions and controls the expression of Rv1955-Rv1957 only. Previously, it has also been reported that HigA1 possesses helix-turn-helix motif at the amino-terminus, binds to the motif ATATAGG(N)6CCTATAT and represses the expression of Rv1954A-Rv1957 locus (Fivian-Hughes and Davis, 2010). Schuessler et al. (2013) have shown that inducible expression of higB1 decreased IdeR and Zur transcript levels and also cleaves tmRNA. Recently, Texier et al. (2021) has shown that ClpXP1P2 protease complex is involved in HigA1 degradation and proposed a model for HigB1 toxin activation.
In the present study, we have performed experiments to investigate the physiological role of HigB1 in M. tuberculosis. Here, we report that HigB1 toxin is upregulated in M. tuberculosis under nutrient limiting conditions and upon exposure to levofloxacin. Further, we also demonstrate that in comparison to the parental strain, the growth of ΔhigB1 mutant strain was impaired in guinea pigs. We also observed that reduced tissue damage in lung sections of ΔhigB1 mutant strain infected guinea pigs in comparison to the sections from guinea pigs infected with the parental strain. The expression of genes involved in virulence, detoxification and adaptation were reduced in the ΔhigB1 mutant strain in comparison to the wild type strain. Taken together, in this study, we have investigated the role of HigB1 toxin in physiology and pathogenesis of M. tuberculosis.
The E. coli and mycobacterial strains were cultured at 200 rpm, 37°C in Luria Bertani medium and Middlebrook 7H9 medium supplemented with 0.2% glycerol, 0.05% Tween-80 and 1x ADS, respectively. For CFU enumeration, an aliquot was removed at designated time points, diluted 10-folds and plated on Middlebrook 7H11 agar supplemented with 1x OADS plates at 37°C for 3–4 weeks. Unless mentioned, all reagents and chemicals used in the study were purchased from Sigma Aldrich, Merck. M. tuberculosis higB1 gene was deleted from the genome of M. tuberculosis using temperature sensitive mycobacteriophages as described previously (Bardarov et al., 2002). Briefly, pYUB854ΔhigB1 construct was prepared via cloning 800bp upstream (F-gaggccttacgtcctggacaccaacgtggtg, R-gtctagaacccatggcggctggatcaggggg) and downstream (F- gaagcttagagccttcggcgacaccccaccga, R-gactagtactcgaaatcagcggtg gctacgtc) regions of higB1 gene in cosmid pYUB854 flanking the hygromycin resistance cassette. The recombinant cosmid was digested with PacI and packaged in phagemid, phAE87 using Gigapack III Gold Packaging Extract. The recombinant cosmid was electroporated in M. smegmatis to generate high titer temperature sensitive mycobacteriophages. The high titer phages were used to transfect M. tuberculosis H37Rv strain to generate ΔhigB1 mutant strain. The deletion of higB1 gene was confirmed by performing whole genome sequencing using the Nextera XT kit and associated protocols on MiSeq (Illumina). The complemented strain was constructed by cloning higB1 gene with its upstream region in the integrative mycobacterium expression vector pMV306K. The recombinant pMV306K-higB1 was electroporated in ΔhigB1 mutant strain and transformants were selected on 7H11 agar plates containing hygromycin and kanamycin.
In order to determine higB1 and higA1 expression levels in disease relevant stress conditions, total RNA was isolated from M. tuberculosis H37Rv strain exposed to various stress conditions. These conditions were (i) oxidative stress (5 mM H2O2), nitrosative (5 mM NaNO2, 7H9 medium, pH-5.2), nutritional stress (1x Tris buffer saline with 0.05% Tween 80), isoniazid treatment (10 μg/ml) and levofloxacin treatment (10 μg/ml). In order to measure intracellular expression levels, total RNA was isolated from J774.1 macrophage infected with M. tuberculosis. The isolated RNA from different conditions was DNase I treated, cDNA synthesized and qPCR was performed as previously described (Agarwal et al., 2018).
For promoter activity assays, upstream region of higB1 was Polymerase Chain Reaction (PCR) amplified and cloned in an EGFP-based promoter reporter vector, pSCK301T3. The recombinant plasmid was electroporated into wild type, ΔhigB1, Δppk1, and ΔrelA strains and transformants were selected on Middlebrook 7H11 medium supplemented with kanamycin and hygromycin. For measurement of promoter activity, strains were cultured in MB7H9 medium till different stages of growth and fluorescence measurements were determined using a Spectramax M5 plate reader (Molecular devices, Inc., United States) with excitation at 490 nm and emission at 520 nm.
In vitro growth characteristics of parental, ΔhigB1 mutant and complemented strains (ΔhigB1-CT) was determined in MB7H9 medium by measuring OD600 nm at regular intervals. For in vitro stress experiments, early-log phase cultures of various strains were exposed to either 5 mM H2O2, 5 mM NaNO2, 0.25% SDS, or 2.5 mg/ml lysozyme for 24 or 72 h. For nutritional starvation, early-log phase cultures of various strains were harvested, washed and resuspended in 1x tris buffered saline containing 0.05% Tween 80 (1x TBST-80) for either 7 or 14 days. The biofilm formation and colony morphology experiments for various strains were performed as previously described (Agarwal et al., 2018; Arora et al., 2018). For in vitro drug-susceptibility assays, mid-log phase cultures of various strains were exposed to drugs that possess a different mechanism of action. The drugs used in the study were isoniazid (cell wall inhibitor), rifampicin (transcription inhibitor), and levofloxacin (replication inhibitor).
In vivo guinea pig experiments were performed as per the guidelines provided by Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA, Govt of India). The experiments were conducted with prior permission of the institutional animal ethics committee of University of Delhi, South Campus. Single cell suspension was prepared from mid-log phase cultures of various strains and aerosol infection was performed using Madison aerosol exposure chamber. The aerosol infection resulted in implantation of 50–100 bacilli in lung tissues at day 1 post-infection. The bacterial loads and histopathology analysis were performed at day 28 and day 56 post-infection. For CFU enumeration, both lungs and spleens were homogenized in 2 ml saline and 100 μl of 10.0-fold serial dilutions was plated on MB7H11 plates in duplicates. The upper left lobe of infected animals was fixed with 10% formalin and stained with hematoxylin and eosin for histopathology analysis as described previously (Singh et al., 2013, 2016).
For gene expression profiling, total RNA was isolated from wild type H37Rv, ΔhigB1 mutant and complemented strains as previously described (Singh et al., 2013). The isolated RNA was treated with DNase I (Thermo Fischer, United States) and quantified using Nanodrop 2000c spectrophotometer (Thermo Scientific, United States). The purity and integrity of RNA samples were checked on Agilent 2100 Bio analyzer (Agilent Technologies Inc., United States). Further, 25 ng of RNA was amplified and labeled using Low input Quick Amp WT Labeling kit (Agilent Technologies, United States) as described previously (Venkataraman et al., 2014). The labeled cRNA was purified using RNeasy columns (Qiagen, United States) and total yields were quantified on Nanodrop 2000c spectrophotometer. The hybridization was performed using Gene expression hybridization kit as per manufacturer’s recommendation (Agilent Technologies, United States). Hybridizations were performed in triplicates. The slides were washed after hybridization as per manufacturer’s instructions and scanned using the Agilent Microarray Scanner at a resolution of 5 μM. The settings used for scanner were: Agilent HD_GX_1 color (61 × 21.6 mm), TIFF 20-bit, Photomultiplier tube (PMT) gain 100%. The scanned image was analyzed using Agilent Feature Extraction software (v10.5). The raw data obtained from microarray experiment was normalized and analyzed using GeneSpring GX v.11.5 software. Normalization of the raw data was performed by taking the 50th percentile for each sample. Baseline correction was applied to the median of all samples. The normalized data has been submitted to NCBI’s Gene Expression Omnibus database (GEO) and can be queried via accession number GSE179403. The differential expression analysis of samples was performed. Genes that showed a twofold or higher change with a P value of < 0.05 (unpaired Student t-test) were considered to be differentially expressed. Gene Ontology and Pathway analysis of differentially expressed genes was done using DAVID tool1 and Panther Classification system2. Functional and Protein Interaction Network was performed using StringDB3. Clustering of the biologically enriched genes was done using Heatmapper online tool4. Gene regulatory network of enriched pathways and genes was performed using Pathreg algorithm (Theomics International Pvt Ltd, Bangalore, India) and visualized using Cytoscape V2.8.3.
Statistical analysis and generation of graphs was done using Prism 8 software (Version 8.4.3; GraphPad software Inc, CA, United States). Differences between groups were compared using two-tailed t-test and were considered significant at P-value of < 0.05. The David analysis was also as per the statistical criteria.
Mycobacterium tuberculosis higBA1 locus is unusual as the toxin (HigB1, Rv1955) and antitoxin (HigA1, Rv1956) are co-transcribed along with the upstream gene, Rv1954A and the downstream gene Rv1957 (Cole et al., 1998; Figure 1A). Previously, it has been shown that SecB regulates the activity of HigBA1 locus as it prevents aggregation and degradation of HigA1 antitoxin (Sala et al., 2013; Bordes et al., 2016). It has also been shown that HigB1 overexpression results in growth arrest in M. tuberculosis and E. coli (Gupta, 2009; Schuessler et al., 2013). In order to determine the role of HigB1 protein in M. tuberculosis physiology, ΔhigB1 mutant strain was constructed using temperature sensitive mycobacteriophages as described in Materials and Methods (Figure 1A, Bardarov et al., 2002). The generation of ΔhigB1 mutant strain was validated by PCR (data not shown) and whole genome sequencing. As shown in Figure 1B, no sequence reads aligning to the Rv1955 region were obtained in the mutant genome compared to the wild type H37Rv strain genome, confirming that the Rv1955 (higB1) sequence was absent in the mutant strain. Further, we did not identify any other secondary mutations in the genomic DNA sequence of the mutant strain. For construction of complemented strain, pMV306K-higB1 was electroporated into ΔhigB1 mutant strain. Next, the growth patterns of various strains were measured in vitro in liquid medium. We did not observe any significant differences in the growth patterns of various strains by measuring either absorbance or bacterial numbers at regular intervals (Figures 2A,B). The bacterial counts of H37Rv, ΔhigB1 mutant and complemented strains after 10 days were ∼ 9.2 × 108, 1.56 × 109, and 1.81 × 109, respectively (Figure 2B). In Pseudomonas aeruginosa, excess of HigB toxin has been shown to reduce the production of various virulence factors and biofilm formation (Wood and Wood, 2016). TA systems have also been implicated in biofilm formation and quorum sensing (Ren et al., 2004; Wang et al., 2011; Wang and Wood, 2011; Sun et al., 2017; Fu et al., 2018). Next, the ability of wild type, mutant and complemented strains to form biofilms was compared (Figure 2C). We observed that wild type, ΔhigB1 mutant and ΔhigB1 complemented strain were comparable in their ability to form biofilms in vitro (Figure 2C). Also, the colony morphology of the ΔhigB1 mutant strain was similar to that observed for the parental strain on Middlebrook 7H11 medium (Figure 2D).
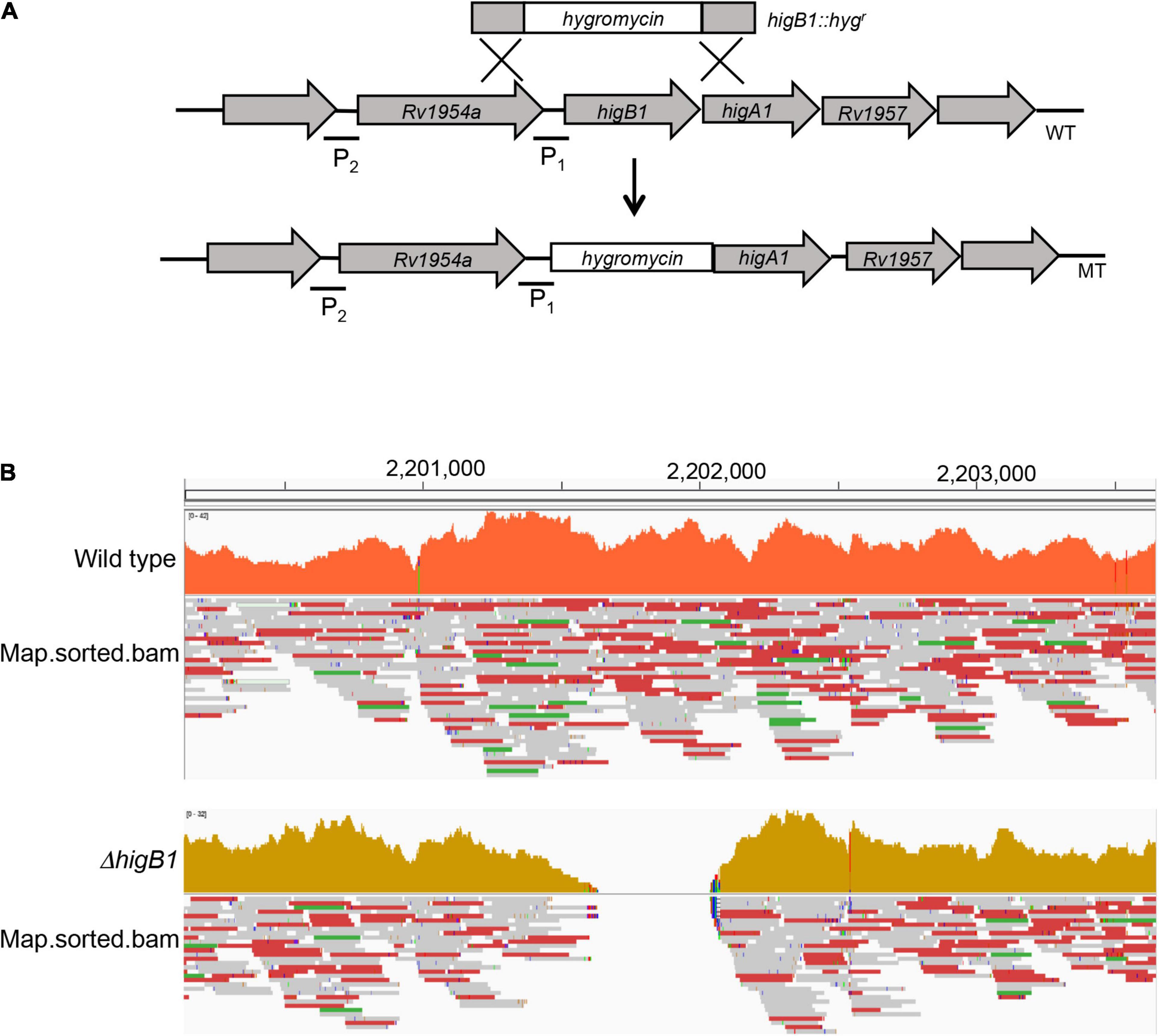
Figure 1. Illustrative representation of higBA1 operon in the wild type and mutant strain of M. tuberculosis. (A) The open reading frame of the higB1 gene in the genome of M. tuberculosis was replaced with the hygromycin resistance gene using temperature sensitive mycobacteriophages. P1 and P2 stands for promoter region 1 and promoter region 2, respectively. (B) The replacement of higB1 open reading frame by hygromycin cassette in the ΔhigB1 mutant strain was confirmed by the whole genome sequencing.
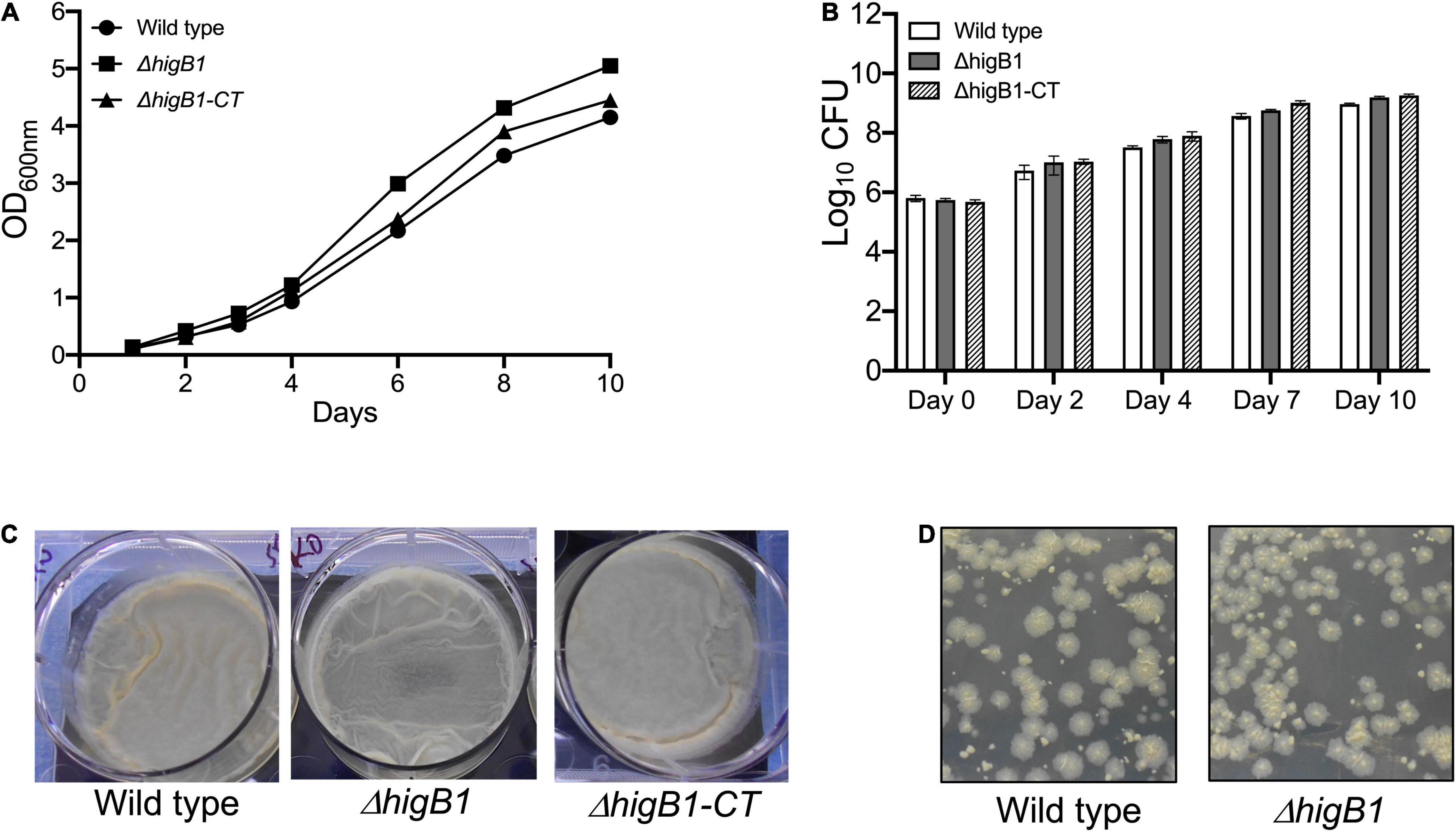
Figure 2. Deletion of HigB1 does not affect in vitro growth of M. tuberculosis. (A,B) The growth patterns of various strains were compared by measuring absorbance at 600 nm (A) and CFU enumeration (B). The biofilm formation (C) and colony morphology (D) was performed as described in section “Materials and Methods.” The data presented shown in (A,C,D) panels is representative of two independent experiments. The data shown in panel (B) is mean ± SE of Log10 CFU obtained from two independent experiments performed in duplicates.
Previous studies have shown that the TAC operon in M. tuberculosis is upregulated upon exposure to DNA damaging agents, heat shock, nutritional stress and low oxygen conditions (Betts et al., 2002; Stewart et al., 2002; Rand et al., 2003; Rustad et al., 2008). We also determined the relative levels of higB1 toxin and higA1 antitoxin by qPCR using gene specific primers after exposure to different in vitro stress conditions as described in Materials and methods. In concordance with previous studies, we observed that the expression of higB1 was increased by ∼3.8-fold in M. tuberculosis upon exposure to nutritional stress. In contrast, the transcript levels of higA1 were increased by 1.5-fold in nutritionally starved growth conditions (Figure 3A). No differences in the transcript levels of higB1 and higA1 was observed after exposure to either oxidative or nitrosative stress (Figure 3A). As shown in Figure 3A, the transcript levels of higB1 were also increased by ∼2.8-fold upon exposure to levofloxacin. The expression of higA1 remained unchanged in levofloxacin treated M. tuberculosis cultures. Further, we also measured the transcript levels of higB1 and higA1 in macrophages at 24 h post-infection. As shown in Figure 3A, we did not observe any significant changes in the transcript levels of higB1 and higA1 in M. tuberculosis infected macrophages.
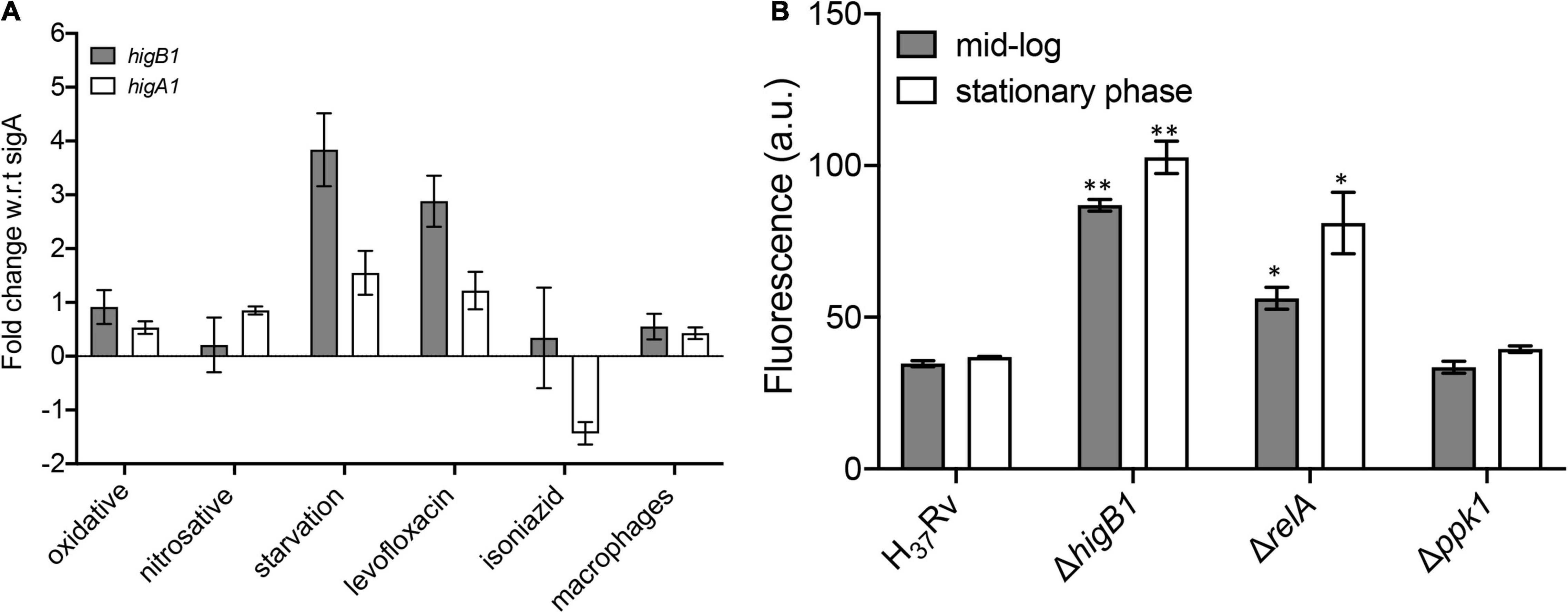
Figure 3. qPCR studies to determine the relative levels of higB1 and higA1 in different stress conditions. (A) The transcript levels of higB1 and higA1 were determined using gene specific primers as described in section “Materials and Methods.” The data obtained was normalized to the levels of sigA, housekeeping gene and shown as mean ± SE obtained from three independent experiments. (B) Promoter activity assays. The promoter activity was measured in various strains at different stages of growth as described in section “Materials and Methods.” The data shown in mean ± SE of promoter activity obtained in various strains. Statistically significant differences were observed for the indicated groups, ∗P < 0.05, ∗∗P < 0.01.
Studies have shown that bacteria adapt to nutrient limiting conditions by changing the transcriptome profile to support its prolonged survival (Rohde et al., 2012). The change in transcription profiles in bacterial pathogens is associated with the synthesis of two intracellular alarmones guanosine 5′,3′ bispyrophosphate (ppGpp) and guanosine pentaphosphate (p)ppGpp. In bacteria, the (p)ppGpp cellular levels are regulated by the enzymatic activities of RelA (alarmone synthetase) and SpoT (alarmone synthetase and hydrolase) (Ronneau and Hallez, 2019). The genome of M. tuberculosis encodes for a single RelA which is responsible to maintain the cellular pools of (p)ppGpp alarmone (Primm et al., 2000). Studies have shown that RelA protein from M. tuberculosis is essential for its long-term survival under starvation and to establish infection in mice tissues (Primm et al., 2000; Weiss and Stallings, 2013). (p)ppGpp levels also regulates the intracellular levels of inorganic polyphosphate (PolyP). The levels of PolyP in bacteria pathogens are regulated by the polyphosphate kinase 1 (PPK-1), Exopolyphosphatases and Polyphosphate kinase 2 (PPK-2). Dysregulation in PolyP levels is associated with attenuation of various intracellular pathogens in animal models (Kornberg et al., 1999; Singh et al., 2013). The (p)ppGpp alarmone and PolyP levels are known to accumulate during stress conditions and these molecules regulate bacterial stress response specifically under nutrient starvation. As higBA1 locus was simultaneously induced when H37Rv M. tuberculosis was exposed to nutritional limiting conditions, we further analyzed the promoter activity of higBA1 locus in ΔrelA and Δppk1 mutant strains. For promoter activity assay, pSCK301T3, harboring eGFP downstream of the higBA1 promoter region was electroporated in either wild type or ΔhigB1 or ΔrelA or Δppk1 strains and fluorescence was determined at mid-log and stationary stages of growth for various strains. We noticed, that in comparison to the wild type strain, the promoter activity was increased by ∼2.0-fold in stationary phase cultures of ΔrelA strain of M. tuberculosis (Figure 3B, ∗P < 0.05). Further, 1.6-fold increase in promoter activity was also seen in mid-log phase cultures of ΔrelA strain as compared to the wild type strain (Figure 3B, ∗P < 0.05). As shown in Figure 3B, we noticed that in comparison to the parental strain, the promoter activity was increased by 2.5-fold and 2.7-fold in ΔhigB1 strain during mid-log and stationary phase of growth, respectively (∗∗P < 0.01). These observations suggested that HigB1 toxin acts as a negative regulator of TAC operon expression. The P2 promoter but not the P1 promoter is reported to be regulated by the HigA1 antitoxin (Fivian-Hughes and Davis, 2010). Our data suggests that the HigA1-HigB1 complex negatively regulates the operon. As shown in Figure 3B, no differences were observed in the activity of the promoter of higBA1 TAC operon during different stages of growth between parental and Δppk1 mutant strain. The observed increase in the promoter activity of higBA1 TAC operon in ΔhigB1 and ΔrelA mutant strains depicts that both higBA1 and relA gene products regulate the expression of higBA1 promoter.
In order to survive, the pathogen should be able to sense, adapt and respond to exogenous stress conditions (Fang et al., 2016; Flint et al., 2016). M. tuberculosis possess the unique ability to adapt to different environmental conditions inside host tissues during infection. TA systems are generally considered as stress responsive elements as they are differentially regulated under various stress conditions. Under specific stress condition, antitoxin protein is degraded by cellular proteases and free toxin can inhibit the bacterial growth by targeting the essential cellular processes which further facilitate the bacterial survival under these conditions. Previous studies have shown that TAC operon of M. tuberculosis is significantly induced in response to heat shock, nutritional starvation, hypoxia and persistence (Betts et al., 2002; Stewart et al., 2002; Rand et al., 2003; Rustad et al., 2008). In concordance, we also observed that the transcripts of higB1 were increased upon exposure to nutritional stress and levofloxacin, therefore, we next investigated the role of HigB1 toxin in the M. tuberculosis stress adaptation and drug tolerance. We compared the survival of wild type, ΔhigB1 mutant and complemented M. tuberculosis strains upon exposure to various in vitro stress conditions. Despite being upregulated in nutrient limiting growth conditions, we observed that the survival of mutant strain was comparable to the parental strain in these conditions (Figure 4A). Also, in other stress conditions tested, the survival of ΔhigB1 strain was comparable to that observed for the parental strain (Figures 4B–E). Taken together, we conclude that HigB1 toxin does not influence stress adaptation of M. tuberculosis in vitro.
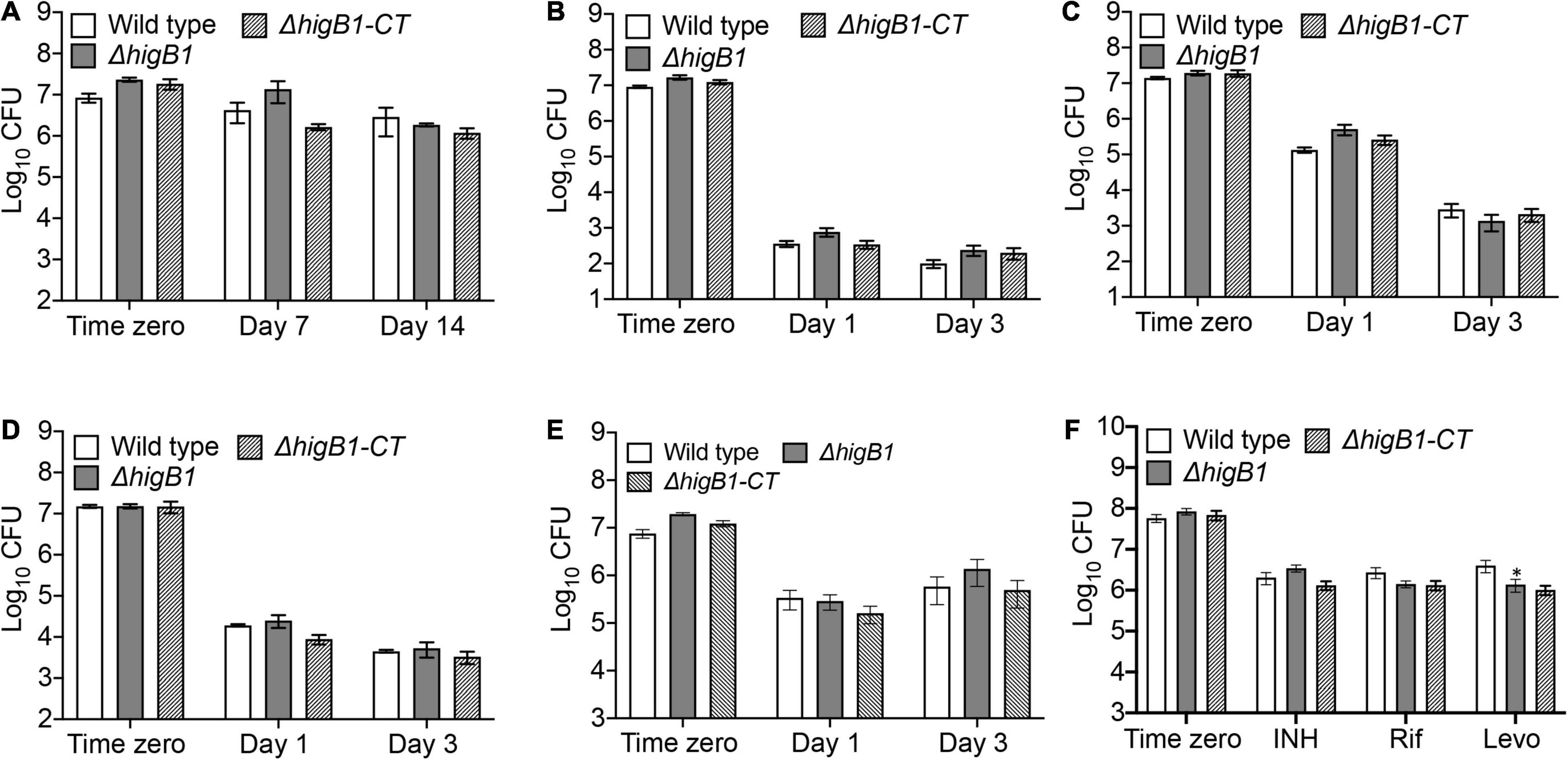
Figure 4. HigB1 is dispensable for M. tuberculosis growth in different stress conditions or drugs in vitro. The survival of various strains was compared in different stress conditions such as nutritional (A) or oxidative (B) or nitrosative (C) or cell wall disrupting agents, SDS (D), lysozyme (E) or after exposure to drugs (F). The data shown in these panels is mean ± SE of Log10CFU obtained from two independent experiments performed in either duplicates or triplicates. Statistically significant differences were observed for the indicated groups, ∗P < 0.05.
Antibiotic persistence is the ability of the bacterial subpopulation to survive the antibiotic treatment. Bacterial persister population makes the TB treatment more difficult and long. Several genes have been implicated in persister formation including TA systems but the role of TA systems in antibiotics mediated persistence is highly questionable. There have been studies which report that overexpression of toxins results in metabolic shutdown that helps the bacterial subpopulation to persist in the presence of antibiotics (Keren et al., 2004a, b, 2011; Tripathi et al., 2014). In M. tuberculosis, it has been shown that MazF toxins (MazF3, MazF6, and MazF9) contribute cumulatively to drug persistence in the presence of levofloxacin and rifampicin (Tiwari et al., 2015). However, deletion of either VapBC3 or VapBC4 or VapBC11 or VapC21 or VapC28 or VapC22 in the genome of M. tuberculosis did not contribute to drug persistence in vitro (Agarwal et al., 2018, 2020; Deep et al., 2018; Sharma et al., 2020). HigBA1 and HigBA2 TA module were also shown to be overexpressed in M. tuberculosis persisters (Keren et al., 2011). In P. aeruginosa, overexpression of HigB toxin was shown to increase the bacterial survival by 1000-fold after exposure to ciprofloxacin (Li et al., 2016). In order to determine the role of HigB1 in drug persistence, we compared the survival of various strains upon exposure to drugs with different mechanism of action. In accordance with qPCR results, we observed that ΔhigB1 mutant strain was susceptible to levofloxacin by 3.0-fold after 7 days of exposure in comparison to the parental strain in vitro (Figure 4F, ∗P < 0.05). As shown in Figure 4F, the deletion of HigB1 did not affect the survival of M. tuberculosis upon exposure to either isoniazid or rifampicin. We also observed that both parental and ΔhigB1 strain displayed comparable MIC99 values against isoniazid, rifampicin, levofloxacin and ethambutol (Supplementary Table 1). Taken together, these studies suggest that the mutant strain was more susceptible to killing upon exposure to levofloxacin. However, the phenotype was not completely restored in the complemented strain.
Based on the in vivo growth phenotype, M. tuberculosis strains have been classified as severe growth in vivo (sgiv) or growth in vivo (giv) or persistence (per) or altered pathology mutants (Hingley-Wilson et al., 2003). TA systems have been implicated in bacterial pathogenesis. We have previously reported that MazF toxins (MazF3, MazF6, and MazF9) contribute to M. tuberculosis pathogenesis (Tiwari et al., 2015). Also, in comparison to parental strain, deletion of either vapBC3 or vapBC4 or vapBC11 or vapC22 attenuates the growth of M. tuberculosis in guinea pigs (Agarwal et al., 2018, 2020; Deep et al., 2018). Recently, it has been reported that deletion of higB toxin reduces the virulence of E. piscicida in fish tissues (Xie et al., 2021). We next investigated the role of HigB1 in M. tuberculosis virulence using guinea pig model of infection. The animals were infected with either parental or ΔhigB1 mutant or complemented M. tuberculosis strains via aerosol route and disease progression was determined during acute (28 days) and chronic stage (56 days) of infection. In concordance with earlier reports, we observed discrete lesions in lung tissues of wild type strain infected guinea pigs (Figures 5A,B). In comparison, significant fewer number of lesions were seen in ΔhigB1 mutant strain infected guinea pigs. The bacterial counts in the lungs of wild type strain infected animals was log10 5.81 and log10 5.27 at 28 days and 56 days post-infection, respectively (Figures 5C,D). We observed that in comparison to wild type strain, the growth ΔhigB1 mutant strain was impaired in lung tissues by ∼ 42.0-fold and 31.0-fold, respectively, during acute and chronic stage of infection (Figures 5C,D, ∗∗P < 0.01 and ∗∗∗P < 0.001). The in vivo growth defect for ΔhigB1 mutant strain was more prominent at chronic stage specifically in spleens of infected animals. In concordance with the lung data, the bacterial numbers in spleens of parental strain and ΔhigB1 mutant strain infected guinea pigs was log10 4.6 and log10 3.22, respectively at 4 weeks post-infection (Figure 5C, ∗P < 0.05). The reduction in splenic bacillary loads of ΔhigB1 infected animals increased to ∼242.0 folds after 56 days post-infection (Figure 5D, ∗P < 0.01). The complementation of ΔhigB1 mutant strain only partially restored the growth defect in spleens of guinea pigs at both time points (Figures 5C,D). Taken together, these observations suggest that TAC locus is required to establish chronic stage of infection in guinea pigs.
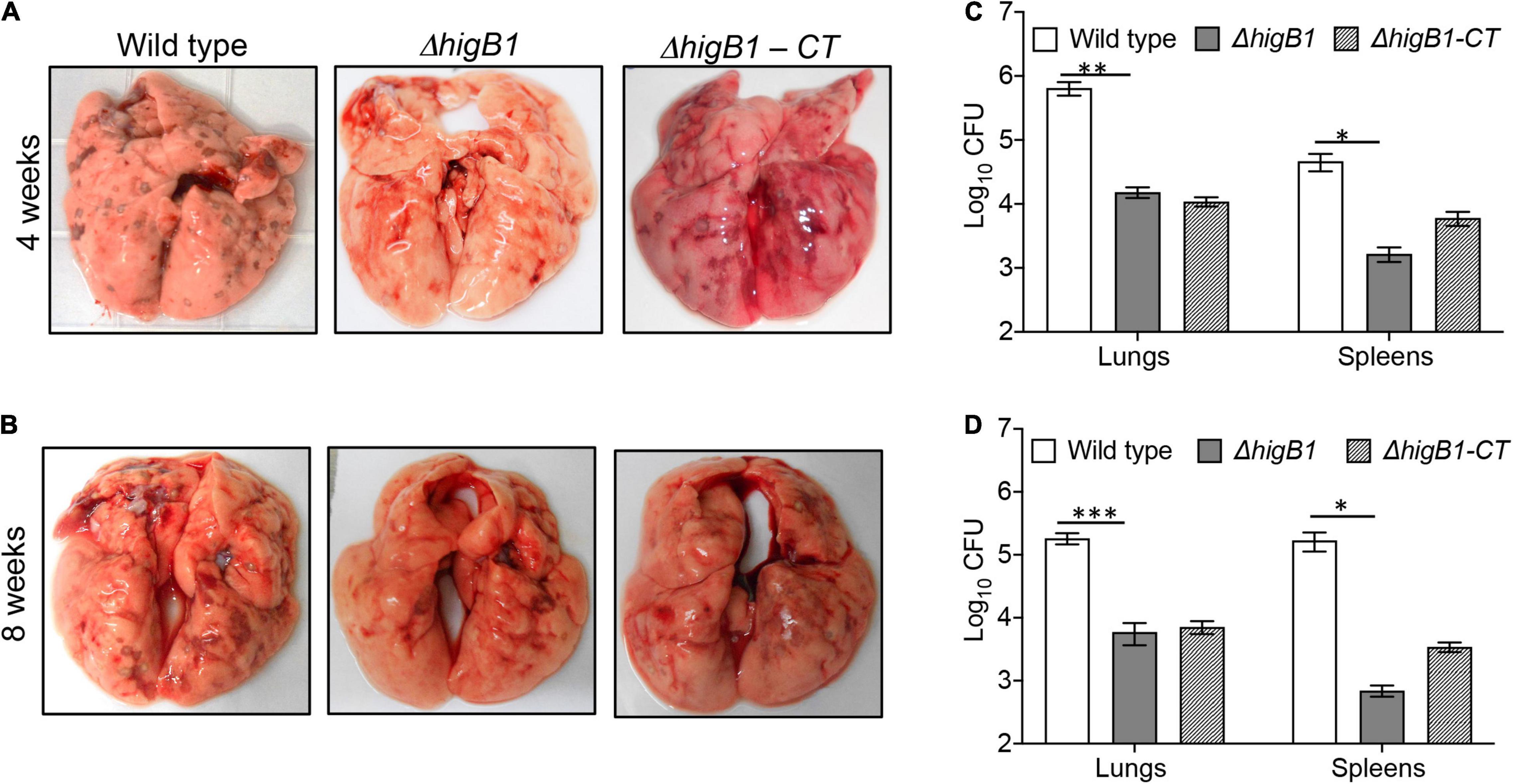
Figure 5. HigB1 locus is important for M. tuberculosis pathogenesis in guinea pigs. (A,B) This panel depicts representative lung images from guinea pigs infected with either wild type or ΔhigB1 mutant or higB1 complemented strains at 4 weeks (A) and 8 weeks (B) post-infection. (C,D) This panel depicts the lungs and splenic bacillary loads in guinea pigs infected with various strains at 4 weeks (C) and 8 weeks (D) post-infection. The data shown in this panel is mean ± SE of Log10CFU obtained from 6 or 7 animals per group per time point. Statistically significant differences were observed for the indicated groups, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Further, we performed histopathology analysis of tissue sections obtained from lungs of guinea pigs infected with various strains of M. tuberculosis at both 4- and 8-weeks post-infection. In concordance with CFU enumeration data, tissue damage was significantly decreased in the tissue sections from ΔhigB1 mutant infected guinea pigs. In comparison, the tissue sections from guinea pigs infected with the parental M. tuberculosis strain showed heavy tissue damage in both acute and chronic phase of infection (Figure 6). As shown in Figure 6, cellular infiltration of lymphocytes and macrophages was seen in the sections from animals infected with the wild type strain. In comparison, lung sections from ΔhigB1 strain infected guinea pigs displayed more alveolar space and less damage of lung parenchyma (Figure 6). At 8 weeks post-infection, necrotic areas were present within granulomas signifying extensive tissue damage in sections from parental strain infected guinea pigs (Figure 6). In concordance with CFU data, no necrosis was seen in sections from animals infected with ΔhigB1 mutant strain at 56 days post-infection. We observed normal lung parenchymal space in tissue sections from guinea pigs infected with the ΔhigB1 mutant strain (Figure 6). In ΔhigB1 complemented strain infected guinea pigs, intermediate levels of tissue damage were observed. Overall, in vivo CFU enumeration and histopathological analysis demonstrates the importance of HigB1 in establishment of successful M. tuberculosis infection in the guinea pigs.
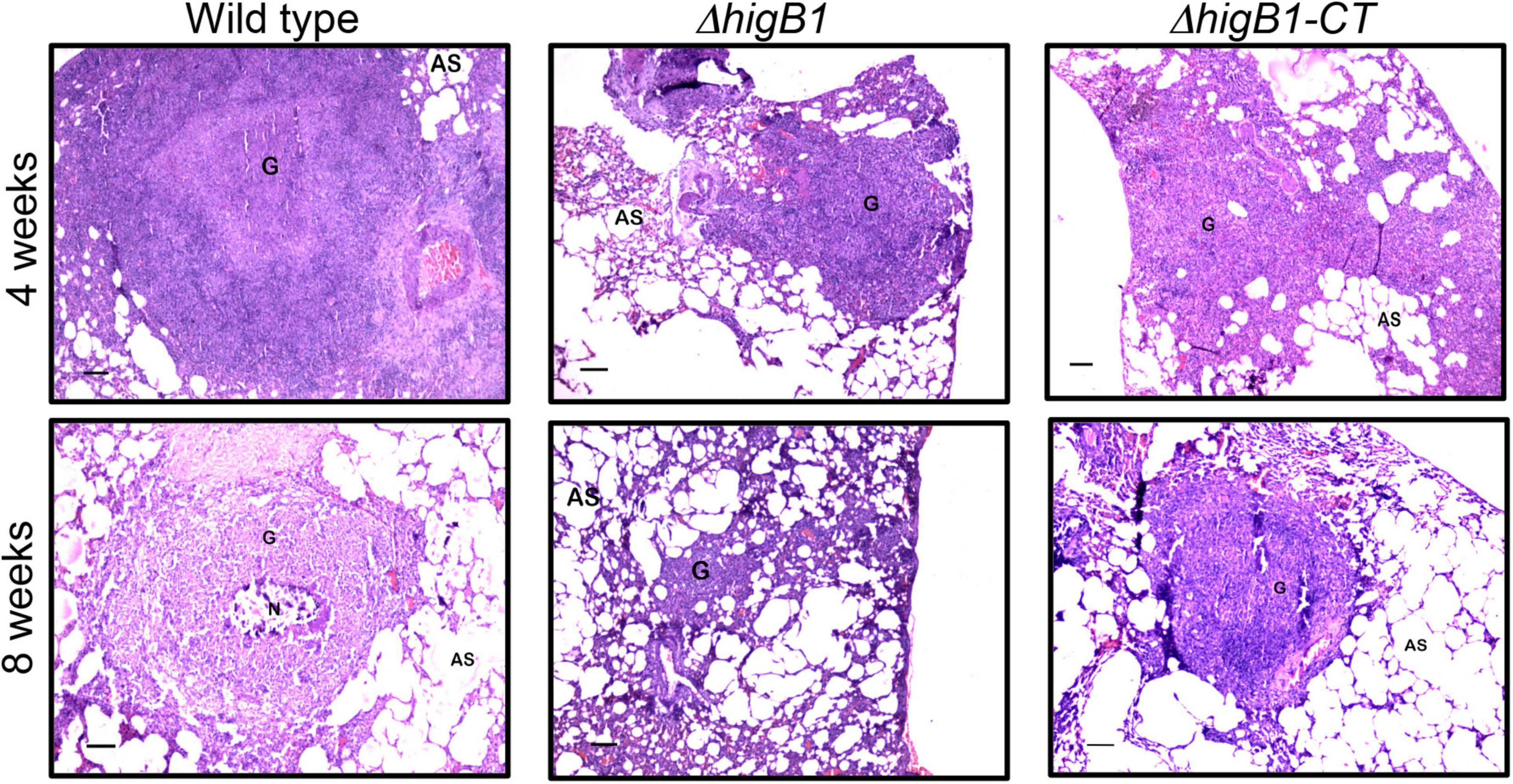
Figure 6. Histopathological analysis of the lung sections obtained from the guinea pigs infected with various strains. The representative photomicrographs of the H&E stained sections (40x magnification) obtained from guinea pigs infected with various strains at 28- and 56-days post-infection. Scale bar – 100 microns.
In order to gain mechanistic insights for the attenuated phenotype of ΔhigB1 mutant strain of M. tuberculosis, we performed microarray experiments to compare the global transcriptome profile of H37Rv, ΔhigB1 mutant and complemented strain. For microarray experiments, total RNA was isolated from mid-log phase cultures of the strains in either duplicates or triplicates. Using a cut-off value of twofold and P-value < 0.05, the fold change was calculated for the genes differentially expressed in the ΔhigB1 mutant vis-à-vis the wild type H37Rv (Table 1). As expected, higB1 transcript levels were reduced by 9.67-fold in the mutant strain (Table 1). Also, the transcript levels of higA1 were reduced by 2.18-fold in the mutant strain (Table 1). The transcript levels of Rv1957 did not show any significant change in the mutant strain. The transcript levels of higB1 were increased by ∼5.32-fold in complemented strain in comparison to the wild type strain (Supplementary Table 2). Also, there was only a marginal increase in the transcript levels of higA1 and Rv1957 in the complemented strain in comparison to the wild type strain. However, the differential expression of genes observed in the mutant strain was not fully restored in the complemented strain. Unsupervised hierarchical clustering of the samples also showed that the profiles obtained in complemented strain clustered separately from the profiles obtained in the wild type strain (Figures 7A,C). This finding is in concordance with the guinea pig data wherein we did not observe complete restoration of the attenuated phenotype in the complemented strain. It is important to mention here that the complemented strain harbors the integrative pMV306K:higB1 while the H37Rv wild type and higB1 knockout strain lack this site-specific integrative plasmid. To the best of our knowledge, there is no evidence of interference of this plasmid in the gene expression profiles of M. tuberculosis.
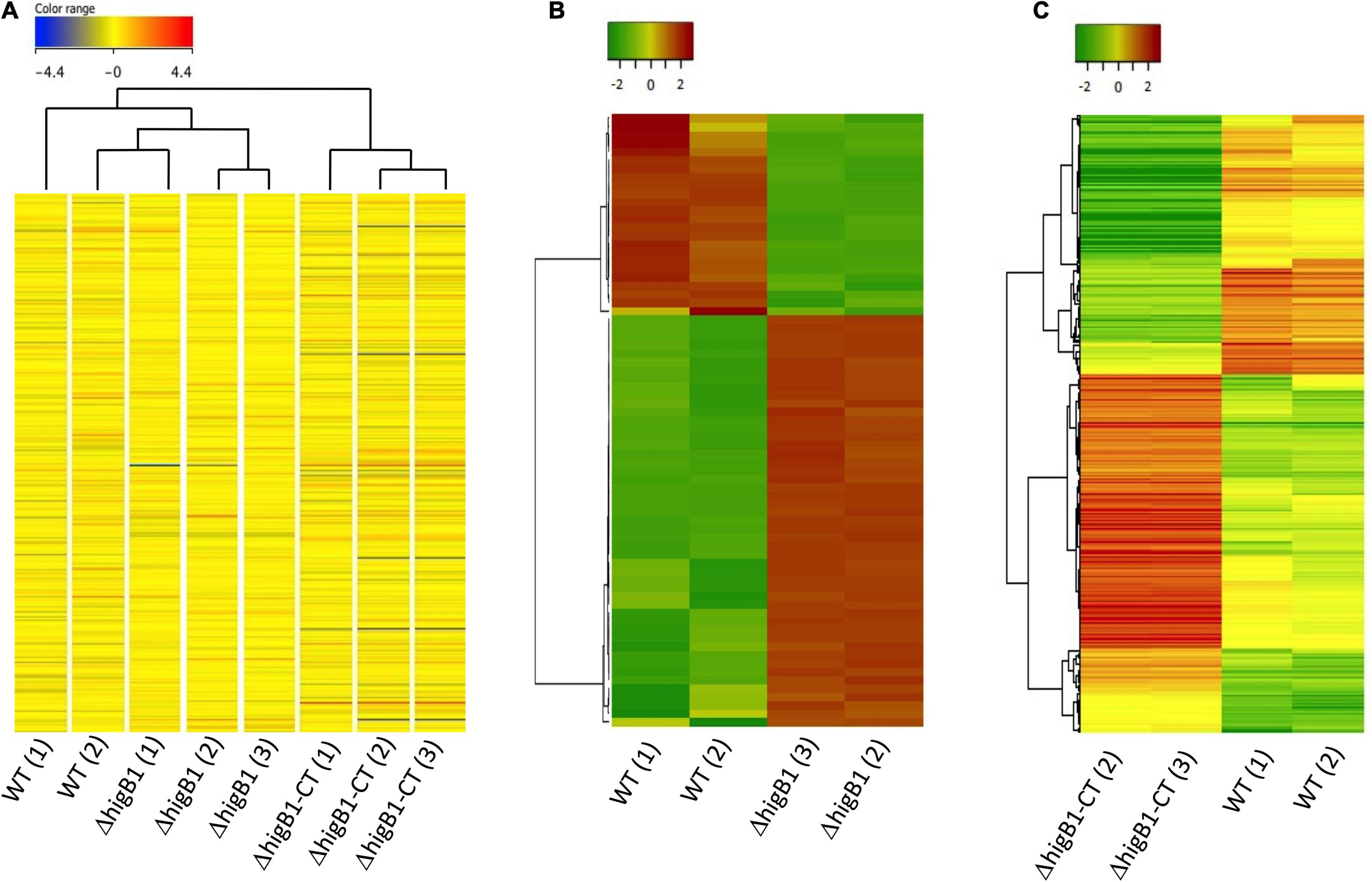
Figure 7. Global transcriptome profile of H37Rv, ΔhigB1 mutant and complemented strains through microarray analysis. (A) The hierarchical cluster analysis, representing the gene expression profile of all genes in sample replicates. Expression values are color coded based upon the extent of expression where red color indicates high level of expression and blue color indicates low level of expression. (B,C) The Unsupervised hierarchical clustering of differentially expressed genes in ΔhigB1 mutant vs. wild type strain (B) or complemented strain vs. wild type (C). Expression values are color coded based upon the extent of expression where red color indicates high level of expression and green color indicates low level of expression. The clustering was done using Pearson distance algorithm with average linkage rule.
Since one of the triplicates for the mutant strain (ΔhigB1-1) and the complemented strain (ΔhigB1-CT-1) appeared as an outlier in the hierarchical clustering therefore they were not included in the unsupervised hierarchical clustering analysis of the data (Figures 7B,C). We noticed that the relative transcript levels of 73 genes were differentially regulated in ΔhigB1 mutant strain compared to the wild type strain (Table 1). Among these, the transcripts of Rv2987c, Rv2988c, Rv2989, and Rv1361c were significantly increased by ∼5.0-fold while the transcript levels of Rv0053, Rv2624c, Rv3135, Rv0250c, and Rv2631 were increased by ∼3.0-fold in ΔhigB1 mutant strain in comparison to the parental strain (Table 1). Further, we also observed that the transcript levels of ribosomal proteins such as Rv0055, Rv0056, Rv0651, Rv0652, Rv0700, Rv0714, Rv0715, Rv0717, and Rv3924c were increased in the ΔhigB1 mutant strain as compared to the wild type strain (Table 1). Transcriptional profiling of ribosomal proteins and its associated proteins in ΔhigB1 mutant strain bore close similarities to that induced by relE3 overexpression and exposure to protein translation inhibitors (Boshoff et al., 2004; Singh et al., 2010). This corroborates with the fact that HigB1 is a translation inhibitor. In addition, the transcript levels Rv0315 were upregulated by twofold in ΔhigB1 mutant strain (Table 1). Rv0315 encodes for an immunostimulatory M. tuberculosis antigen which activates the dendritic cells and drives the Th1 cell response upon M. tuberculosis infection (Byun et al., 2012). The transcripts encoding for Rv3027c (GCN5-related N-acetyltransferase), Rv3628 (inorganic pyrophosphatase) and Rv3340 (cystathionine [beta]-lyase) were upregulated by 2. 9-, 2. 0-, and 2.3-fold, respectively, in the ΔhigB1 mutant strain. We also noticed that transcripts of genes such as Rv2007c, Rv2624c, Rv2625c, Rv2631, and Rv3128c belonging to the DosR regulon were also increased in the mutant strain (Park et al., 2003; Table 1). Among these, Rv2624c has been previously reported to be highly immunogenic antigen as and it has been shown to induce higher levels of IFN-γ and TNFα (Bertholet et al., 2011; Chegou et al., 2012).
Further, the transcript levels of 24 genes were down regulated in the mutant strain in comparison to the wild type strain (Figure 7B and Table 1). Among these, 30 and 21% of the proteins belong to the functional category of intermediary metabolism and respiration and virulence, detoxification and adaptation, respectively. The transcript levels of Rv0341 (iniB, isoniazid inducible protein), Rv0914c, Rv3139 (keto acyl-CoA thiolase) were reduced by 8.6-fold, 3.3-fold, and 3.0-fold respectively in the ΔhigB1 mutant strain (Table 1). We also noticed that deletion of higB1 in the genome of M. tuberculosis reduced the expression of β-propeller gene, Rv1057 by 2.88-fold (Table 1). Previous studies have shown that Rv1057 regulates ESAT-6 secretion and intracellular growth of M. tuberculosis (Fu et al., 2018). Also, the transcript levels for Rv3084, Rv3085, Rv3086, and Rv3087 that belong to the acid responsive mymA operon (Rv3083–Rv3089) were significantly reduced by ∼2.0-fold in the mutant strain in comparison to the parental strain (Table 1; Singh A. et al., 2003; Singh et al., 2005). The expression of Rv0311, a protein shown to be essential for M. tuberculosis to establish extrapulmonary TB infection was also down regulated by ∼2.0 fold in the mutant strain (Be et al., 2008). Further, GO-enrichment analysis was performed using DAVID tool and the most enriched pathways associated with these differentially expressed genes were identified as shown in Figure 8A. These gene sets were then used to create their regulatory network shown in Figure 8B. As evident, the key pathways affected by the higB1 deletion were translation, transcription and oxidoreductase family. While a large number of translation-associated proteins and transcription factors were upregulated in the mutant strain, reduced expression of the genes belonging to the oxidoreductase family was observed in the ΔhigB1 mutant strain. We also observed that in comparison to the parental strain the expression of enzymes involved in translation and transcription pathways was not affected in the complemented strain. These observations indicates that HigB1 expression in the complemented strain restored the expression of proteins belonging to these pathways. However, the expression of enzymes belonging to oxidative phosphorylation pathway was compromised in the complemented strain as compared to the parental strain (Figure 8C).
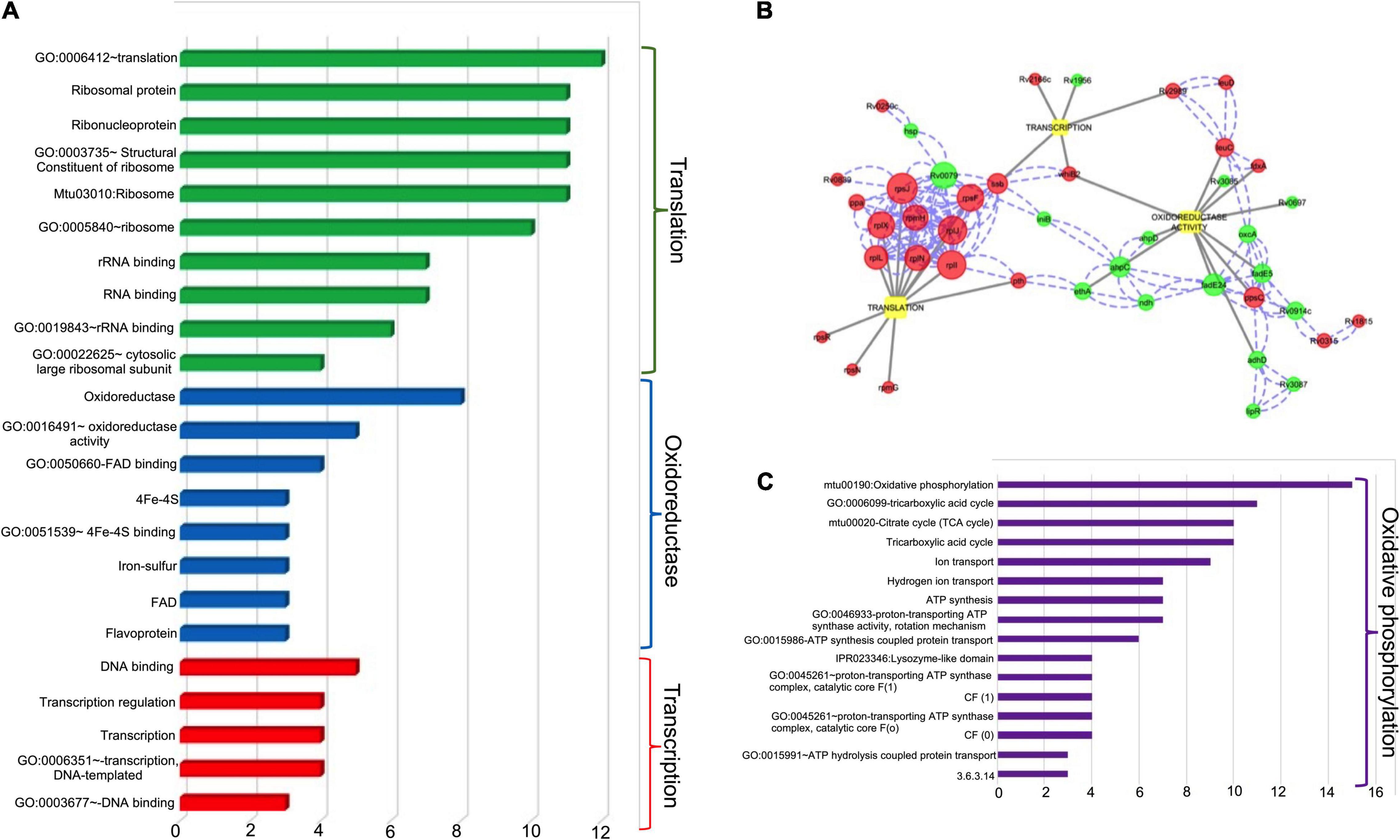
Figure 8. (A) Various Gene Ontology (GO) and Pathways enriched for the three major biological functions altered in the ΔhigB1 mutant in comparison to the wild type strain. X axis indicates the number of DEGs under each GO/Pathway. (B) The biological regulatory network for the differentially expressed genes. The red color and green circles represent upregulated and down regulated genes in the ΔhigB1 mutant strain in comparison to the parental strain. The blue dotted lines indicate physical protein-protein interaction and gray solid lines indicate regulation of specific biological functions. The size of the nodes indicates the connectivity score (bigger the size – higher the score). (C) Various Gene Ontology (GO) and Pathways enriched for the major biological function altered in the complemented strain in comparison to the wild type strain. X axis indicates the number of DEGs under each GO/Pathway.
Chromosomal encoded TA systems are induced in different stress conditions. These have been implicated to help bacteria adapt to different stress conditions by downregulating metabolism and potentiating transition into dormant like stage (Unterholzner et al., 2013; Chan et al., 2016). In addition to slowing down of bacterial metabolism, TA systems are also shown to be essential for persistence and bacterial pathogenesis (Wen et al., 2014; Yang and Walsh, 2017; Alonso, 2021). The large number of TA systems in the genome of M. tuberculosis makes it difficult to perceive the involvement of individual TA systems in the pathogen biology. Previous studies have demonstrated that MazF toxins contribute cumulatively and VapBC3, VapBC4, VapBC11, and VapC22 are essential for the M. tuberculosis pathogenesis (Tiwari et al., 2015; Agarwal et al., 2018, 2020; Deep et al., 2018). Here, in this study we have investigated the role of HigB1 toxin in M. tuberculosis physiology and pathogenesis.
In M. tuberculosis, HigB1 cleaves tmRNA, inhibits the growth of bacteria in a bacteriostatic manner and this is abrogated by high levels of HigA1 (Schuessler et al., 2013). The transcript levels of higB1 are also increased in M. tuberculosis after exposure to nutrient limiting growth conditions and drugs (Betts et al., 2002; Keren et al., 2011). However, its role in bacterial adaptation to these conditions is still not understood. It is still not clear how upregulation of HigB1 to adapt to starvation or exposure to drugs is beneficial to the bacteria or if there is loss of survival/competency in the absence or repression of the toxin. In concordance with previous studies, the transcript levels of higB1 were increased in nutrient limiting growth conditions. However, no upregulation of higB1 expression was observed in other stress conditions evaluated in the study. The transcript levels of higB1 were also upregulated in levofloxacin-treated samples but not after exposure to isoniazid. We observed differential induction of the higB1 and higA1 belonging to higBA TA system upon exposure to levofloxacin and nutritional stress. This might be attributed to differential stability of toxin and antitoxin transcripts in these stress conditions. Similar post-transcriptional regulation of TA system has also been reported in E. coli and M. tuberculosis in different growth conditions (Korch et al., 2009; Singh et al., 2010; Kasari et al., 2013; Ramirez et al., 2013; Tiwari et al., 2015). Under nutrient limiting conditions, M. tuberculosis activates the highly conserved stringent response through guanosine pentaphosphate (p)ppGpp (Primm et al., 2000; Weiss and Stallings, 2013). Previous studies have shown that (p)ppGpp-mediated stringent response in bacteria, in combination with TA activity, can act as a regulated switch to a persistent phenotype (Tian et al., 2017). In M. tuberculosis, RelA and PPK-1 are the main enzymes involved in (p)ppGpp and inorganic polyphosphate biosynthesis (Primm et al., 2000; Singh et al., 2013). Since, we observed the increased expression of higBA1 locus under nutrient limiting conditions, we determined the promoter activity of TAC locus in parental, ΔhigB1, ΔrelA and Δppk1 mutant strain. The increased promoter activity in ΔrelA strain suggests that higBA1 locus promoter is negatively regulated by (p)ppGpp/relA gene product levels in the cells. Also, we speculate that higBA1 locus promoter is negatively regulated by HigB1 either alone or in complex with HigA1 antitoxin as reported previously for other bacterial TA systems (Dienemann et al., 2011; Kang et al., 2017; Nikolic, 2019). This data supports autoregulation and cross regulation between stringent response and TAC system in M. tuberculosis.
To further elucidate the role of HigB1 toxin toward in vitro and in vivo fitness of M. tuberculosis, we constructed ΔhigB1 mutant strain using temperature sensitive mycobacteriophages. The construction of ΔhigB1 strain was confirmed by Next generation sequencing. The colony morphology and ability to form biofilms was comparable between the parental and ΔhigB1 mutant strain. Despite being upregulated in nutrient limiting growth conditions, we observed that the survival of both wild type and ΔhigB1 mutant strain was comparable in nutrient limiting and other stress conditions. In concordance with qPCR results, in comparison to the parental strain, we observed that ΔhigB1 strain was compromised for growth upon exposure to levofloxacin. Further, we observed that despite being non-essential in vitro, higB1 is required for the pathogenesis of M. tuberculosis in guinea pigs. Histopathological analysis revealed necrotic granulomatous tissue in lung sections from animals infected with the parental strain. In comparison, normal parenchyma space was seen in sections from animals infected with the ΔhigB1 mutant strain. The observed attenuation phenotype associated with the mutant strain was more prominent in spleen specifically at chronic stage of infection. The histopathology sections obtain from ΔhigB1 mutant strain infected guinea pigs appeared more similar to sections from uninfected animals as reported earlier (Singh R. et al., 2003; Patel et al., 2011; Cai et al., 2019). The phenotype associated with ΔhigB1 mutant strain was similar to that observed for M. tuberculosis strains deficient in either CarD or PerM or MymA or Icl1 or PcaA. These strains were also attenuated for growth in spleens and the phenotype was more drastic in chronic stage of infection (Glickman et al., 2000; McKinney et al., 2000; Singh et al., 2005; Weiss et al., 2012; Goodsmith et al., 2015). The success of M. tuberculosis as an intracellular pathogen lies in its ability to persist in later stages of infection despite the induction of host adaptive immune response. These observations suggest that HigB1 is important for disease progression and dissemination in guinea pig model of infection.
To gain further mechanistic insights into the attenuation of ΔhigB1 mutant strain in guinea pigs, we compared the global transcriptome profile of H37Rv, ΔhigB1 mutant and complemented strains. We observed that the transcripts of ribosomal proteins of both smaller and larger subunits of ribosome were upregulated in the ΔhigB1 mutant strain. Studies have shown that these ribosomal proteins are able to elicit a strong CD4+ immune response that might be associated with the faster clearance of the mutant strain in host tissues (Johnson et al., 2017; Kennedy et al., 2018). In addition to this, expression level of higA1 (Rv1956), an adjacent gene of higB1 toxin was also reduced in the ΔhigB1 deletion strain. However, transcripts levels of SecB- chaperone protein Rv1957 was not significantly changed in the mutant strain. Microarray studies revealed that the expression of higB1 was restored in the complemented strain by fivefold in comparison to the wild type strain, whereas only marginal change was observed in the levels of higA1 and Rv1957. Despite the restoration of higB1 levels in complemented strain, expression of other genes such as Rv0311, Rv0315, Rv0341, ribosomal proteins (Rv0651, Rv0652, Rv0700, Rv0714), Rv0914c, Rv1057, and mymA operon (Rv3083-Rv3089) was not restored in the complemented strain. In both cases (ΔhigB1 knock out and complemented strains) higB1 levels were either significantly depleted (9.67-fold) or significantly increased (5.18-fold) in comparison to the parental strain. Previous studies have shown that toxin and antitoxin stoichiometry is important for their autoregulation and activation (Vandervelde et al., 2017; Fraikin et al., 2020). We speculate that changes in intracellular toxin antitoxin ratios in both mutant and complemented strain might be responsible for the observed attenuated phenotype and partial restoration of disease pathology in guinea pigs infected with the higB1 complemented strain.
Taken together, we have performed experiments to elucidate the role of HigB1 toxin in M. tuberculosis physiology and pathogenesis. We show that HigB1 of M. tuberculosis is important to establish infection in guinea pigs. Microarray analysis revealed that deletion of higB1 leads to increase in the transcripts of ribosomal proteins and reduction in expression of genes involved in virulence, detoxification and adaptation. This might be responsible for the observed attenuated phenotype of ΔhigB1 mutant strain. Lack of complementation of the mutant strain could be attributed to altered intracellular ratios of toxin, antitoxin and observed differences in the transcription profiles of wild type and complemented strains. In conclusion, HigB1 is vital for M. tuberculosis pathogenesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by University of Delhi South Campus.
RS and AG conceived the study and designed the work plan. AS, KS, BV, NG, NC, and TG performed the cloning and microbiology assays. AS, NC, and TG performed the animal experiments. BV isolated the genomic DNA. AG performed the NGS. KS, AG, and AS performed the microarray studies and analysis. NB carried out the analysis of the NGS data. RS, AS, KS, and AG analyzed the data, interpreted them, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors acknowledge the funding received from Department of Biotechnology, India (Grant ID; BT/PR5510/MED/29/513/2012). RS acknowledge the funding received from DBT-Wellcome India Alliance as a Senior Fellow (IA/S/19/2/504646). The authors acknowledge the funding received from Translational Health Science and Technology Institute under Translational Research Program and from UGC-SAP scheme to Department of Biochemistry, UDSC. pSC301GFP vector, that has been modified and used in this study, was kindly provided to AG by Dr. Yossef Av-Gay (University of British Columbia, Vancouver, BC, Canada). AS acknowledges research fellowship received from Indian Council of Medical Research. NG was recipient of research fellowship from Council of Scientific and Industrial Research. RS is a recipient of Ramalingaswami fellowship and National Bioscience Award from Department of Biotechnology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
TG is thankful to Department of Biotechnology for her fellowship. NC is thankful to Department of Science and Technology for funding under NPDF-scheme. The authors are thankful to staff members of Infection disease research facility and University of Delhi South Campus BSL-3 facility for technical help. The authors are also thankful to DBT-Supported Genomics Facility at South campus for Next Generation Sequencing and Gene Expression Microarray studies. The authors acknowledge help Mr. Madavan Vasudevan in analysis of Microarray data and lab attendants Mr. Rajesh and Mr. Sher Singh for technical help.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.748890/full#supplementary-material
Agarwal, S., Sharma, A., Bouzeyen, R., Deep, A., Sharma, H., Mangalaparthi, K. K., et al. (2020). VapBC22 toxin-antitoxin system from Mycobacterium tuberculosis is required for pathogenesis and modulation of host immune response. Sci. Adv. 6:eaba6944. doi: 10.1126/sciadv.aba6944
Agarwal, S., Tiwari, P., Deep, A., Kidwai, S., Gupta, S., Thakur, K. G., et al. (2018). System-Wide analysis unravels the differential regulation and in vivo essentiality of virulence-associated proteins b and c toxin-antitoxin systems of Mycobacterium tuberculosis. J. Infect. Dis. 217, 1809–1820. doi: 10.1093/infdis/jiy109
Ahidjo, B. A., Kuhnert, D., McKenzie, J. L., Machowski, E. E., Gordhan, B. G., Arcus, V., et al. (2011). VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS One 6:e21738. doi: 10.1371/journal.pone.0021738
Alonso, J. C. (2021). Toxin-Antitoxin systems in pathogenic bacteria. Toxins (Basel) 13:74. doi: 10.3390/toxins13020074
Armalyte, J., Jurenas, D., Krasauskas, R., Cepauskas, A., and Suziedeliene, E. (2018). The higBA toxin-antitoxin module from the opportunistic pathogen Acinetobacter baumannii - regulation, activity, and evolution. Front. Microbiol. 9:732. doi: 10.3389/fmicb.2018.00732
Arora, G., Chaudhary, D., Kidwai, S., Sharma, D., and Singh, R. (2018). CitE enzymes are essential for Mycobacterium tuberculosis to establish infection in macrophages and guinea pigs. Front. Cell. Infect. Microbiol. 8:385. doi: 10.3389/fcimb.2018.00385
Bardarov, S., Bardarov, S., Pavelka, M. S., Sambandamurthy, V., Larsen, M., Tufariello, J., et al. (2002). Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology (Reading) 148, 3007–3017. doi: 10.1099/00221287-148-10-3007
Be, N. A., Lamichhane, G., Grosset, J., Tyagi, S., Cheng, Q. J., Kim, K. S., et al. (2008). Murine model to study the invasion and survival of Mycobacterium tuberculosis in the central nervous system. J. Infect. Dis. 198, 1520–1528. doi: 10.1086/592447
Bertholet, S., Horne, D. J., Laughlin, E. M., Savlov, M., Tucakovic, I., Coler, R. N., et al. (2011). Effect of chemotherapy on whole-blood cytokine responses to Mycobacterium tuberculosis antigens in a small cohort of patients with pulmonary tuberculosis. Clin. Vaccine Immunol. 18, 1378–1386. doi: 10.1128/CVI.05037-11
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Bordes, P., Sala, A. J., Ayala, S., Texier, P., Slama, N., Cirinesi, A. M., et al. (2016). Chaperone addiction of toxin-antitoxin systems. Nat. Commun. 7:13339.
Boshoff, H. I., Myers, T. G., Copp, B. R., McNeil, M. R., Wilson, M. A., and Barry, C. E. III (2004). The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279, 40174–40184. doi: 10.1074/jbc.M406796200
Byun, E. H., Kim, W. S., Shin, A. R., Kim, J. S., Whang, J., Won, C. J., et al. (2012). Rv0315, a novel immunostimulatory antigen of Mycobacterium tuberculosis, activates dendritic cells and drives Th1 immune responses. J. Mol. Med. (Berl.) 90, 285–298. doi: 10.1007/s00109-011-0819-2
Cai, X., Yu, N., Ma, J., Li, W. Y., Xu, M., Li, E., et al. (2019). Altered pulmonary capillary permeability in immunosuppressed guinea pigs infected with Legionella pneumophila serogroup 1. Exp. Ther. Med. 18, 4368–4378. doi: 10.3892/etm.2019.8102
Chan, W. T., Espinosa, M., and Yeo, C. C. (2016). Keeping the wolves at bay: antitoxins of prokaryotic Type II toxin-antitoxin systems. Front. Mol. Biosci. 3:9. doi: 10.3389/fmolb.2016.00009
Chegou, N. N., Black, G. F., Loxton, A. G., Stanley, K., Essone, P. N., Klein, M. R., et al. (2012). Potential of novel Mycobacterium tuberculosis infection phase-dependent antigens in the diagnosis of TB disease in a high burden setting. BMC Infect. Dis. 12:10. doi: 10.1186/1471-2334-12-10
Choi, J. S., Kim, W., Suk, S., Park, H., Bak, G., Yoon, J., et al. (2018). The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 15, 1319–1335. doi: 10.1080/15476286.2018.1532252
Christensen-Dalsgaard, M., and Gerdes, K. (2006). Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62, 397–411. doi: 10.1111/j.1365-2958.2006.05385.x
Christensen-Dalsgaard, M., Jorgensen, M. G., and Gerdes, K. (2010). Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75, 333–348. doi: 10.1111/j.1365-2958.2009.06969.x
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544.
Cruz, J. W., Sharp, J. D., Hoffer, E. D., Maehigashi, T., Vvedenskaya, I. O., Konkimalla, A., et al. (2015). Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 6:7480. doi: 10.1038/ncomms8480
Deep, A., Tiwari, P., Agarwal, S., Kaundal, S., Kidwai, S., Singh, R., et al. (2018). Structural, functional and biological insights into the role of Mycobacterium tuberculosis VapBC11 toxin-antitoxin system: targeting a tRNase to tackle mycobacterial adaptation. Nucleic Acids Res. 46, 11639–11655. doi: 10.1093/nar/gky924
Dienemann, C., Boggild, A., Winther, K. S., Gerdes, K., and Brodersen, D. E. (2011). Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J. Mol. Biol. 414, 713–722. doi: 10.1016/j.jmb.2011.10.024
Fang, F. C., Frawley, E. R., Tapscott, T., and Vazquez-Torres, A. (2016). Bacterial stress responses during host infection. Cell Host Microbe 20, 133–143. doi: 10.1016/j.chom.2016.07.009
Fivian-Hughes, A. S., and Davis, E. O. (2010). Analyzing the regulatory role of the HigA antitoxin within Mycobacterium tuberculosis. J. Bacteriol. 192, 4348–4356. doi: 10.1128/JB.00454-10
Flint, A., Butcher, J., and Stintzi, A. (2016). Stress responses, adaptation, and virulence of bacterial pathogens during host gastrointestinal colonization. Microbiol. Spectr. 4, 1–18.
Fraikin, N., Goormaghtigh, F., and Van Melderen, L. (2020). Type II toxin-antitoxin systems: evolution and revolutions. J. Bacteriol. 202:e00763-19.
Fu, J., Zong, G., Zhang, P., Gu, Y., and Cao, G. (2018). Deletion of the beta-Propeller protein gene Rv1057 Reduces ESAT-6 secretion and intracellular growth of Mycobacterium tuberculosis. Curr. Microbiol. 75, 401–409. doi: 10.1007/s00284-017-1394-8
Glickman, M. S., Cox, J. S., and Jacobs, W. R. Jr. (2000). A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5, 717–727. doi: 10.1016/s1097-2765(00)80250-6
Goodsmith, N., Guo, X. V., Vandal, O. H., Vaubourgeix, J., Wang, R., Botella, H., et al. (2015). Disruption of an M. tuberculosis membrane protein causes a magnesium-dependent cell division defect and failure to persist in mice. PLoS Pathog. 11:e1004645. doi: 10.1371/journal.ppat.1004645
Guillet, V., Bordes, P., Bon, C., Marcoux, J., Gervais, V., Sala, A. J., et al. (2019). Structural insights into chaperone addiction of toxin-antitoxin systems. Nat. Commun. 10:782.
Gupta, A. (2009). Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 290, 45–53. doi: 10.1111/j.1574-6968.2008.01400.x
Gupta, A., Venkataraman, B., Vasudevan, M., and Gopinath Bankar, K. (2017). Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 7:5868.
Harms, A., Brodersen, D. E., Mitarai, N., and Gerdes, K. (2018). Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell 70, 768–784. doi: 10.1016/j.molcel.2018.01.003
Hingley-Wilson, S. M., Sambandamurthy, V. K., and Jacobs, W. R. Jr. (2003). Survival perspectives from the world’s most successful pathogen, Mycobacterium tuberculosis. Nat. Immunol. 4, 949–955. doi: 10.1038/ni981
Hurley, J. M., and Woychik, N. A. (2009). Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613. doi: 10.1074/jbc.M109.008763
Johnson, A. J., Kennedy, S. C., Lindestam Arlehamn, C. S., Goldberg, M. F., Saini, N. K., Xu, J., et al. (2017). Identification of mycobacterial RplJ/L10 and RpsA/S1 proteins as novel targets for CD4(+) T cells. Infect. Immun. 85:e01023-16. doi: 10.1128/IAI.01023-16
Kamruzzaman, M., Wu, A. Y., and Iredell, J. R. (2021). Biological functions of Type II toxin-antitoxin systems in bacteria. Microorganisms 9:1276. doi: 10.3390/microorganisms9061276
Kang, S. M., Kim, D. H., Lee, K. Y., Park, S. J., Yoon, H. J., Lee, S. J., et al. (2017). Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides. Nucleic Acids Res. 45, 8564–8580. doi: 10.1093/nar/gkx489
Kasari, V., Mets, T., Tenson, T., and Kaldalu, N. (2013). Transcriptional cross-activation between toxin-antitoxin systems of Escherichia coli. BMC Microbiol. 13:45. doi: 10.1186/1471-2180-13-45
Kedzierska, B., and Hayes, F. (2016). Emerging roles of toxin-antitoxin modules in bacterial pathogenesis. Molecules 21:790. doi: 10.3390/molecules21060790
Kennedy, S. C., Johnson, A. J., Bharrhan, S., Lindestam Arlehamn, C. S., Xu, J., Garforth, S. J., et al. (2018). Identification of mycobacterial ribosomal proteins as targets for CD4(+) T cells that enhance protective immunity in tuberculosis. Infect. Immun. 86:e00009-18. doi: 10.1128/IAI.00009-18
Keren, I., Kaldalu, N., Spoering, A., Wang, Y., and Lewis, K. (2004a). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18. doi: 10.1016/s0378-1097(03)00856-5
Keren, I., Minami, S., Rubin, E., and Lewis, K. (2011). Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2:e00100-11.
Keren, I., Shah, D., Spoering, A., Kaldalu, N., and Lewis, K. (2004b). Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004
Korch, S. B., Contreras, H., and Clark-Curtiss, J. E. (2009). Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 191, 1618–1630. doi: 10.1128/JB.01318-08
Kornberg, A., Rao, N. N., and Ault-Riche, D. (1999). Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68, 89–125. doi: 10.1146/annurev.biochem.68.1.89
Lange, C., Dheda, K., Chesov, D., Mandalakas, A. M., Udwadia, Z., and Horsburgh, C. R. Jr. (2019). Management of drug-resistant tuberculosis. Lancet 394, 953–966.
Li, M., Long, Y., Liu, Y., Liu, Y., Chen, R., Shi, J., et al. (2016). HigB of Pseudomonas aeruginosa enhances killing of phagocytes by up-regulating the type III secretion system in ciprofloxacin induced persister cells. Front. Cell. Infect. Microbiol. 6:125. doi: 10.3389/fcimb.2016.00125
Lobato-Marquez, D., Diaz-Orejas, R., and Garcia-Del Portillo, F. (2016). Toxin-antitoxins and bacterial virulence. FEMS Microbiol. Rev. 40, 592–609. doi: 10.1093/femsre/fuw022
McKinney, J. D., Honer zu Bentrup, K., Munoz-Elias, E. J., Miczak, A., Chen, B., Chan, W. T., et al. (2000). Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738. doi: 10.1038/35021074
Nikolic, N. (2019). Autoregulation of bacterial gene expression: lessons from the MazEF toxin-antitoxin system. Curr. Genet. 65, 133–138. doi: 10.1007/s00294-018-0879-8
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Pandey, D. P., and Gerdes, K. (2005). Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976. doi: 10.1093/nar/gki201
Park, H. D., Guinn, K. M., Harrell, M. I., Liao, R., Voskuil, M. I., Tompa, M., et al. (2003). Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843. doi: 10.1046/j.1365-2958.2003.03474.x
Patel, P. B., Patel, T. K., Baxi, S. N., Acharya, H. R., and Tripathi, C. (2011). Antitubercular effect of 8-[(4-Chloro phenyl) sulfonyl]-7-Hydroxy-4-Methyl-2H-chromen-2-One in guinea pigs. J. Pharmacol. Pharmacother. 2, 253–260. doi: 10.4103/0976-500X.85951
Primm, T. P., Andersen, S. J., Mizrahi, V., Avarbock, D., Rubin, H., and Barry, C. E. III (2000). The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182, 4889–4898. doi: 10.1128/jb.182.17.4889-4898.2000
Ramage, H. R., Connolly, L. E., and Cox, J. S. (2009). Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767
Ramirez, M. V., Dawson, C. C., Crew, R., England, K., and Slayden, R. A. (2013). MazF6 toxin of Mycobacterium tuberculosis demonstrates antitoxin specificity and is coupled to regulation of cell growth by a Soj-like protein. BMC Microbiol. 13:240. doi: 10.1186/1471-2180-13-240
Rand, L., Hinds, J., Springer, B., Sander, P., Buxton, R. S., and Davis, E. O. (2003). The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 50, 1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x
Ren, D., Bedzyk, L. A., Thomas, S. M., Ye, R. W., and Wood, T. K. (2004). Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64, 515–524.
Rohde, K. H., Veiga, D. F., Caldwell, S., Balazsi, G., and Russell, D. G. (2012). Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 8:e1002769. doi: 10.1371/journal.ppat.1002769
Ronneau, S., and Hallez, R. (2019). Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol. Rev. 43, 389–400. doi: 10.1093/femsre/fuz009
Rustad, T. R., Harrell, M. I., Liao, R., and Sherman, D. R. (2008). The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502. doi: 10.1371/journal.pone.0001502
Sala, A., Bordes, P., and Genevaux, P. (2014). Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6, 1002–1020. doi: 10.3390/toxins6031002
Sala, A., Calderon, V., Bordes, P., and Genevaux, P. (2013). TAC from Mycobacterium tuberculosis: a paradigm for stress-responsive toxin-antitoxin systems controlled by SecB-like chaperones. Cell Stress Chaperones 18, 129–135. doi: 10.1007/s12192-012-0396-5
Schuessler, D. L., Cortes, T., Fivian-Hughes, A. S., Lougheed, K. E., Harvey, E., Buxton, R. S., et al. (2013). Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 90, 195–207. doi: 10.1111/mmi.12358
Schuster, C. F., and Bertram, R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. doi: 10.1111/1574-6968.12074
Sharma, A., Chattopadhyay, G., Chopra, P., Bhasin, M., Thakur, C., Agarwal, S., et al. (2020). VapC21 toxin contributes to drug-tolerance and interacts with non-cognate VapB32 antitoxin in Mycobacterium tuberculosis. Front. Microbiol. 11:2037. doi: 10.3389/fmicb.2020.02037
Sharp, J. D., Cruz, J. W., Raman, S., Inouye, M., Husson, R. N., and Woychik, N. A. (2012). Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 287, 12835–12847. doi: 10.1074/jbc.M112.340109
Singh, A., Gupta, R., Vishwakarma, R. A., Narayanan, P. R., Paramasivan, C. N., Ramanathan, V. D., et al. (2005). Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. J. Bacteriol. 187, 4173–4186. doi: 10.1128/jb.187.12.4173-4186.2005
Singh, A., Jain, S., Gupta, S., Das, T., and Tyagi, A. K. (2003). mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol. Lett. 227, 53–63. doi: 10.1016/S0378-1097(03)00648-7
Singh, M., Tiwari, P., Arora, G., Agarwal, S., Kidwai, S., and Singh, R. (2016). Establishing virulence associated polyphosphate kinase 2 as a drug target for Mycobacterium tuberculosis. Sci. Rep. 6:26900.
Singh, R., Barry, C. E. III, and Boshoff, H. I. (2010). The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 192, 1279–1291. doi: 10.1128/JB.01285-09
Singh, R., Rao, V., Shakila, H., Gupta, R., Khera, A., Dhar, N., et al. (2003). Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 50, 751–762. doi: 10.1046/j.1365-2958.2003.03712.x
Singh, R., Singh, M., Arora, G., Kumar, S., Tiwari, P., and Kidwai, S. (2013). Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J. Bacteriol. 195, 2839–2851. doi: 10.1128/JB.00038-13
Slayden, R. A., Dawson, C. C., and Cummings, J. E. (2018). Toxin-antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog. Dis. 76:fty039.
Song, S., and Wood, T. K. (2020). A primary physiological role of toxin/antitoxin systems is phage inhibition. Front. Microbiol. 11:1895. doi: 10.3389/fmicb.2020.01895
Stewart, G. R., Wernisch, L., Stabler, R., Mangan, J. A., Hinds, J., Laing, K. G., et al. (2002). Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology (Reading) 148, 3129–3138. doi: 10.1099/00221287-148-10-3129
Sun, C., Guo, Y., Tang, K., Wen, Z., Li, B., Zeng, Z., et al. (2017). MqsR/MqsA toxin/antitoxin system regulates persistence and biofilm formation in Pseudomonas putida KT2440. Front. Microbiol. 8:840. doi: 10.3389/fmicb.2017.00840
Tandon, H., Sharma, A., Sandhya, S., Srinivasan, N., and Singh, R. (2019). Mycobacterium tuberculosis Rv0366c-Rv0367c encodes a non-canonical PezAT-like toxin-antitoxin pair. Sci. Rep. 9:1163. doi: 10.1038/s41598-018-37473-y
Texier, P., Bordes, P., Nagpal, J., Sala, A. J., Mansour, M., Cirinesi, A. M., et al. (2021). ClpXP-mediated degradation of the TAC antitoxin is neutralized by the SecB-like chaperone in Mycobacterium tuberculosis. J. Mol. Biol. 433:166815. doi: 10.1016/j.jmb.2021.166815
Tian, C., Semsey, S., and Mitarai, N. (2017). Synchronized switching of multiple toxin-antitoxin modules by (p)ppGpp fluctuation. Nucleic Acids Res. 45, 8180–8189. doi: 10.1093/nar/gkx552
Tian, Q. B., Ohnishi, M., Tabuchi, A., and Terawaki, Y. (1996). A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220, 280–284. doi: 10.1006/bbrc.1996.0396
Tiwari, P., Arora, G., Singh, M., Kidwai, S., Narayan, O. P., and Singh, R. (2015). MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6:6059.
Tripathi, A., Dewan, P. C., Siddique, S. A., and Varadarajan, R. (2014). MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 289, 4191–4205. doi: 10.1074/jbc.M113.510511
Unterholzner, S. J., Poppenberger, B., and Rozhon, W. (2013). Toxin-antitoxin systems: biology, identification, and application. Mob. Genet. Elements 3:e26219. doi: 10.4161/mge.26219
Vandervelde, A., Drobnak, I., Hadzi, S., Sterckx, Y. G., Welte, T., De Greve, H., et al. (2017). Molecular mechanism governing ratio-dependent transcription regulation in the ccdAB operon. Nucleic Acids Res. 45, 2937–2950. doi: 10.1093/nar/gkx108
Venkataraman, B., Vasudevan, M., and Gupta, A. (2014). A new microarray platform for whole-genome expression profiling of Mycobacterium tuberculosis. J. Microbiol. Methods 97, 34–43. doi: 10.1016/j.mimet.2013.12.009
Wang, X., and Wood, T. K. (2011). Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77, 5577–5583. doi: 10.1128/AEM.05068-11
Wang, X., Kim, Y., Hong, S. H., Ma, Q., Brown, B. L., Pu, M., et al. (2011). Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366. doi: 10.1038/nchembio.560
Wang, X., Yao, J., Sun, Y. C., and Wood, T. K. (2021). Type VII toxin/antitoxin classification system for antitoxins that enzymatically neutralize toxins. Trends Microbiol. 29, 388–393. doi: 10.1016/j.tim.2020.12.001
Weiss, L. A., and Stallings, C. L. (2013). Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J. Bacteriol. 195, 5629–5638. doi: 10.1128/JB.00759-13
Weiss, L. A., Harrison, P. G., Nickels, B. E., Glickman, M. S., Campbell, E. A., Darst, S. A., et al. (2012). Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J. Bacteriol. 194, 5621–5631. doi: 10.1128/JB.00879-12
Wen, Y., Behiels, E., and Devreese, B. (2014). Toxin-Antitoxin systems: their role in persistence, biofilm formation, and pathogenicity. Pathog. Dis. 70, 240–249. doi: 10.1111/2049-632x.12145
Winther, K., Tree, J. J., Tollervey, D., and Gerdes, K. (2016). VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res. 44, 9860–9871. doi: 10.1093/nar/gkw781
Wood, T. L., and Wood, T. K. (2016). The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen 5, 499–511. doi: 10.1002/mbo3.346
Xie, J., Zhao, Q., Huang, H., Fang, Z., and Hu, Y. (2021). Edwardsiella piscicida HigB: a type II toxin that is essential to oxidative resistance, biofilm formation, serum survival, intracellular propagation, and host infection. Aquaculture. 535:736382. doi: 10.1016/j.aquaculture.2021.736382
Yang, Q. E., and Walsh, T. R. (2017). Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 41, 343–353. doi: 10.1093/femsre/fux006
Zhang, Y., Xia, B., Li, M., Shi, J., Long, Y., Jin, Y., et al. (2018). HigB reciprocally controls biofilm formation and the expression of Type III secretion system genes through influencing the intracellular c-di-GMP level in Pseudomonas aeruginosa. Toxins (Basel) 10:424. doi: 10.3390/toxins10110424
Keywords: Mycobacterium tuberculosis, HigBA1, toxin antitoxin loci, virulence, stringent response
Citation: Sharma A, Sagar K, Chauhan NK, Venkataraman B, Gupta N, Gosain TP, Bhalla N, Singh R and Gupta A (2021) HigB1 Toxin in Mycobacterium tuberculosis Is Upregulated During Stress and Required to Establish Infection in Guinea Pigs. Front. Microbiol. 12:748890. doi: 10.3389/fmicb.2021.748890
Received: 28 July 2021; Accepted: 29 October 2021;
Published: 30 November 2021.
Edited by:
Divakar Sharma, University of Delhi, IndiaReviewed by:
Pierre Genevaux, FR3743 Centre de Biologie Intégrative (CBI), FranceCopyright © 2021 Sharma, Sagar, Chauhan, Venkataraman, Gupta, Gosain, Bhalla, Singh and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramandeep Singh, cmFtYW5kZWVwQHRoc3RpLnJlcy5pbg==; Amita Gupta, YW1pdGFndXB0YUBzb3V0aC5kdS5hYy5pbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.